| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6IBL | A | Amine | Adrenergic | Beta1 | Meleagris Gallopavo | Arformoterol | - | - | 2.7 | 2019-01-09 | doi.org/10.1038/s41586-020-2419-1 |
|
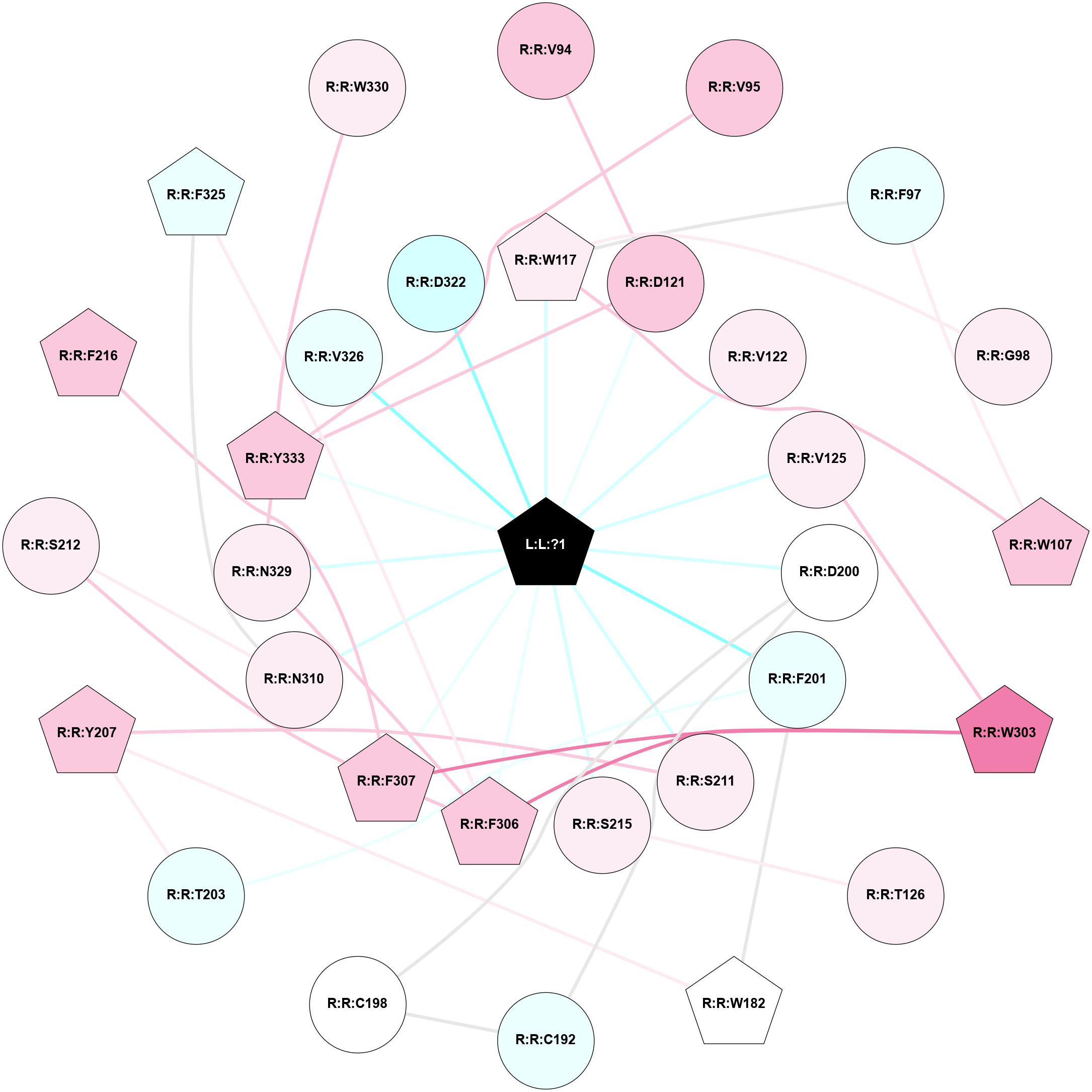
A 2D representation of the interactions of H98 in 6IBL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D121 | R:R:V94 | 7.3 | 1 | No | No | 7 | 7 | 1 | 2 | | R:R:V95 | R:R:Y333 | 5.05 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:F97 | R:R:W107 | 5.01 | 1 | No | Yes | 4 | 7 | 2 | 2 | | R:R:F97 | R:R:W117 | 8.02 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:G98 | R:R:W117 | 4.22 | 0 | No | Yes | 6 | 6 | 2 | 1 | | R:R:W107 | R:R:W117 | 9.37 | 1 | Yes | Yes | 7 | 6 | 2 | 1 | | L:L:?1 | R:R:W117 | 4.49 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:D121 | R:R:Y333 | 10.34 | 1 | No | Yes | 7 | 7 | 1 | 1 | | L:L:?1 | R:R:D121 | 19.62 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:V122 | 12.73 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V125 | R:R:W303 | 9.81 | 0 | No | Yes | 6 | 8 | 1 | 2 | | L:L:?1 | R:R:V125 | 6.85 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:S215 | R:R:T126 | 9.59 | 0 | No | No | 6 | 6 | 1 | 2 | | R:R:F201 | R:R:W182 | 4.01 | 0 | No | Yes | 4 | 5 | 1 | 2 | | R:R:W182 | R:R:Y207 | 5.79 | 3 | Yes | Yes | 5 | 7 | 2 | 2 | | R:R:C192 | R:R:C198 | 7.28 | 1 | No | No | 4 | 5 | 2 | 2 | | R:R:C192 | R:R:D200 | 4.67 | 1 | No | No | 4 | 5 | 2 | 1 | | R:R:C198 | R:R:D200 | 6.22 | 1 | No | No | 5 | 5 | 2 | 1 | | L:L:?1 | R:R:D200 | 4.46 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:F201 | R:R:T203 | 5.19 | 0 | No | No | 4 | 4 | 1 | 2 | | L:L:?1 | R:R:F201 | 20.01 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:T203 | R:R:Y207 | 4.99 | 0 | No | Yes | 4 | 7 | 2 | 2 | | R:R:S211 | R:R:Y207 | 11.45 | 0 | No | Yes | 6 | 7 | 1 | 2 | | L:L:?1 | R:R:S211 | 10.86 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:F307 | R:R:S212 | 5.28 | 1 | Yes | No | 7 | 6 | 1 | 2 | | R:R:N310 | R:R:S212 | 5.96 | 0 | No | No | 6 | 6 | 1 | 2 | | L:L:?1 | R:R:S215 | 5.92 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:F216 | R:R:F307 | 20.36 | 1 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:F306 | R:R:W303 | 7.02 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:F307 | R:R:W303 | 10.02 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:F306 | R:R:F307 | 10.72 | 1 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:F306 | R:R:F325 | 6.43 | 1 | Yes | Yes | 7 | 4 | 1 | 2 | | R:R:F306 | R:R:N329 | 15.71 | 1 | Yes | No | 7 | 6 | 1 | 1 | | L:L:?1 | R:R:F306 | 5.6 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:F307 | 8.81 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:F325 | R:R:N310 | 4.83 | 0 | Yes | No | 4 | 6 | 2 | 1 | | L:L:?1 | R:R:N310 | 6.32 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:N329 | R:R:Y333 | 15.12 | 1 | No | Yes | 6 | 7 | 1 | 1 | | L:L:?1 | R:R:N329 | 16.24 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:W330 | R:R:Y333 | 6.75 | 0 | No | Yes | 6 | 7 | 2 | 1 | | L:L:?1 | R:R:Y333 | 4.62 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:V326 | 2.94 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:D322 | 1.78 | 1 | Yes | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 15.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 8.75 | | Average Nodes In Shell | 32.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 43.00 | | Average Links Mediated by Hubs In Shell | 36.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6TKO | A | Amine | Adrenergic | Beta1 | Meleagris Gallopavo | Arformoterol | - | Arrestin2 | 3.3 | 2020-06-17 | doi.org/10.1038/s41586-020-2419-1 |
|
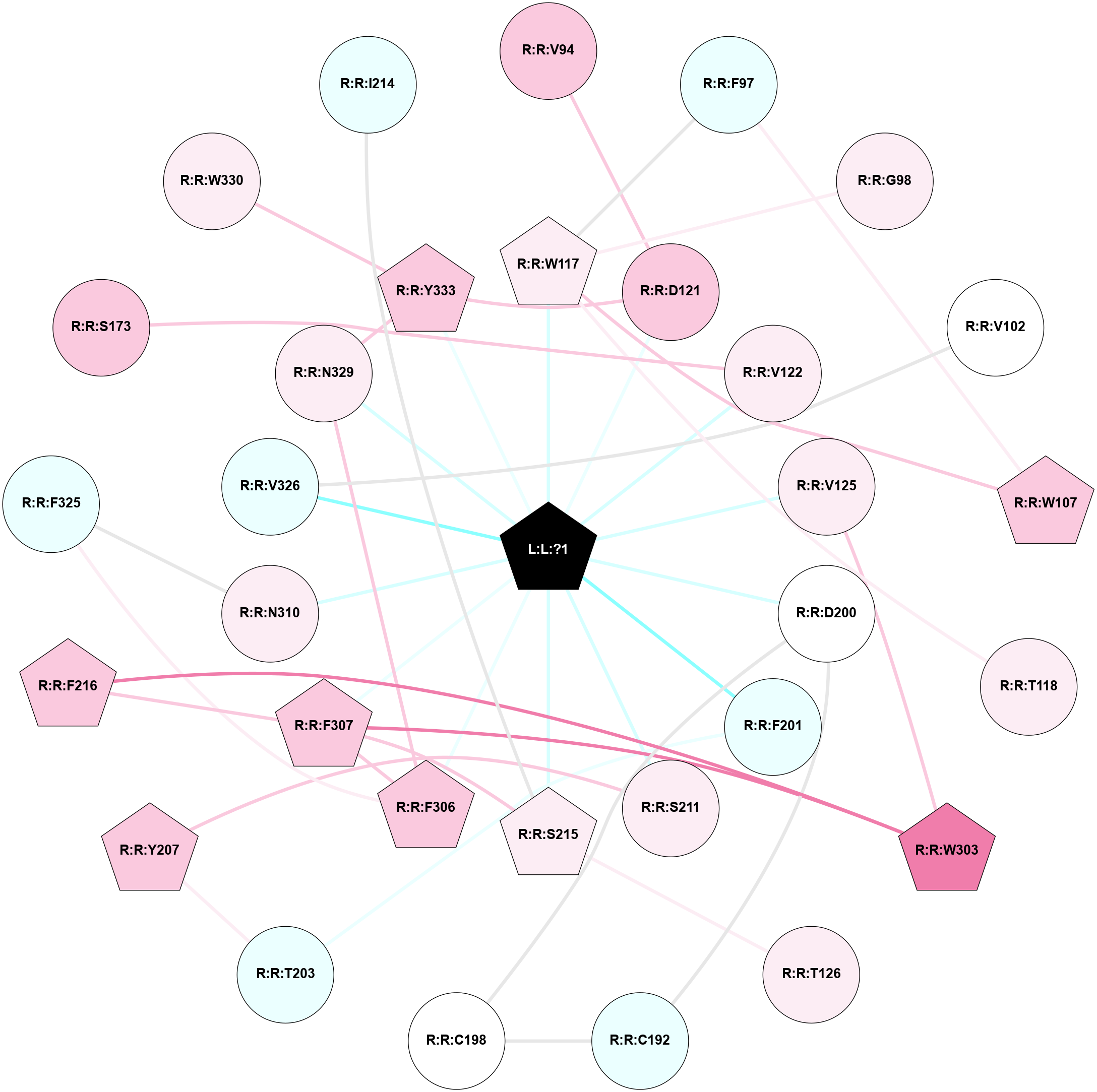
A 2D representation of the interactions of H98 in 6TKO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:W117 | 5.24 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:D121 | 14.27 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:V122 | 6.85 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:V125 | 6.85 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:D200 | 8.03 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:F201 | 23.21 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:S211 | 7.89 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:S215 | 3.95 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:F306 | 11.21 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:F307 | 5.6 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:N310 | 5.41 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:V326 | 4.9 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:N329 | 15.34 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:Y333 | 6.93 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:D121 | R:R:V94 | 5.84 | 1 | No | No | 7 | 7 | 1 | 2 | | R:R:F97 | R:R:W107 | 4.01 | 1 | No | Yes | 4 | 7 | 2 | 2 | | R:R:F97 | R:R:W117 | 8.02 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:G98 | R:R:W117 | 4.22 | 0 | No | Yes | 6 | 6 | 2 | 1 | | R:R:V102 | R:R:V326 | 6.41 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:W107 | R:R:W117 | 19.68 | 1 | Yes | Yes | 7 | 6 | 2 | 1 | | R:R:T118 | R:R:W117 | 4.85 | 0 | No | Yes | 6 | 6 | 2 | 1 | | R:R:D121 | R:R:Y333 | 10.34 | 1 | No | Yes | 7 | 7 | 1 | 1 | | R:R:V125 | R:R:W303 | 8.58 | 0 | No | Yes | 6 | 8 | 1 | 2 | | R:R:S215 | R:R:T126 | 4.8 | 1 | Yes | No | 6 | 6 | 1 | 2 | | R:R:C192 | R:R:C198 | 7.28 | 1 | No | No | 4 | 5 | 2 | 2 | | R:R:C192 | R:R:D200 | 4.67 | 1 | No | No | 4 | 5 | 2 | 1 | | R:R:C198 | R:R:D200 | 6.22 | 1 | No | No | 5 | 5 | 2 | 1 | | R:R:F201 | R:R:T203 | 6.49 | 0 | No | No | 4 | 4 | 1 | 2 | | R:R:T203 | R:R:Y207 | 4.99 | 0 | No | Yes | 4 | 7 | 2 | 2 | | R:R:S211 | R:R:Y207 | 5.09 | 0 | No | Yes | 6 | 7 | 1 | 2 | | R:R:F307 | R:R:S215 | 3.96 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | R:R:F216 | R:R:W303 | 4.01 | 1 | Yes | Yes | 7 | 8 | 2 | 2 | | R:R:F216 | R:R:F307 | 27.86 | 1 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:F307 | R:R:W303 | 5.01 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:F306 | R:R:F307 | 13.93 | 1 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:F306 | R:R:F325 | 8.57 | 1 | Yes | No | 7 | 4 | 1 | 2 | | R:R:F306 | R:R:N329 | 14.5 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:F325 | R:R:N310 | 7.25 | 0 | No | No | 4 | 6 | 2 | 1 | | R:R:N329 | R:R:Y333 | 13.96 | 1 | No | Yes | 6 | 7 | 1 | 1 | | R:R:S173 | R:R:V122 | 3.23 | 0 | No | No | 7 | 6 | 2 | 1 | | R:R:N310 | R:R:S212 | 2.98 | 0 | No | No | 6 | 6 | 1 | 2 | | R:R:W330 | R:R:Y333 | 2.89 | 0 | No | Yes | 6 | 7 | 2 | 1 | | R:R:I214 | R:R:S215 | 1.55 | 0 | No | Yes | 4 | 6 | 2 | 1 | | L:L:?1 | R:R:K322 | 0.93 | 1 | Yes | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 15.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 8.44 | | Average Nodes In Shell | 34.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 44.00 | | Average Links Mediated by Hubs In Shell | 35.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7BZ2 | A | Amine | Adrenergic | Beta2 | Homo Sapiens | Arformoterol | - | Gs/Beta1/Gamma2 | 3.82 | 2020-08-05 | doi.org/10.1038/s41421-020-0176-9 |
|
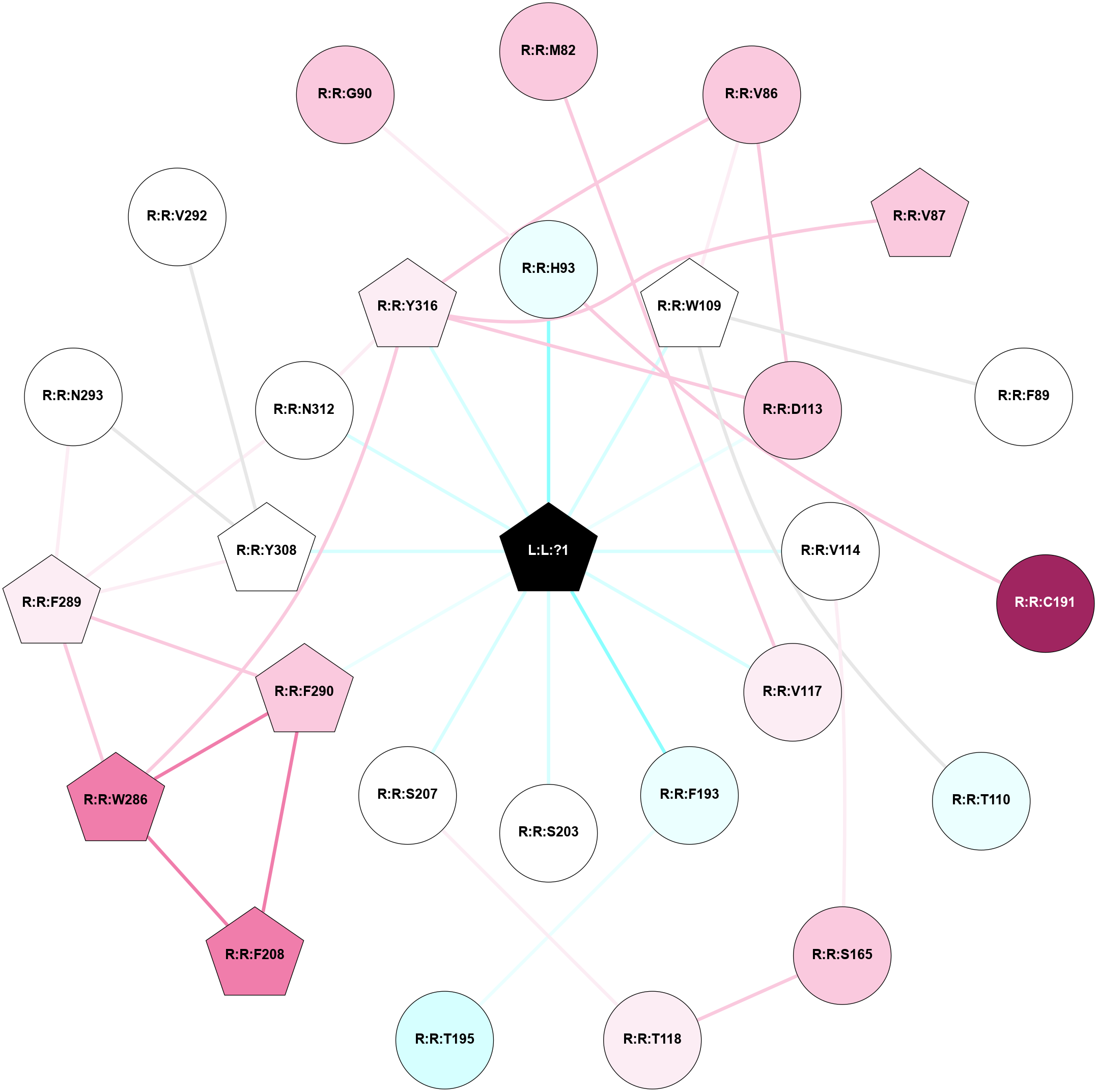
A 2D representation of the interactions of H98 in 7BZ2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:H93 | 5.07 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:W109 | 5.24 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:D113 | 4.46 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:V114 | 7.83 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:V117 | 6.85 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:D192 | 4.46 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:F193 | 10.41 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:T195 | 2.91 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:S203 | 7.89 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:S207 | 4.93 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:F290 | 6.4 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:Y308 | 3.08 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:N312 | 10.83 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:Y316 | 6.16 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:M82 | R:R:V117 | 4.56 | 0 | No | No | 7 | 6 | 2 | 1 | | R:R:V86 | R:R:W109 | 6.13 | 1 | No | Yes | 7 | 5 | 2 | 1 | | R:R:D113 | R:R:V86 | 16.07 | 1 | No | No | 7 | 7 | 1 | 2 | | R:R:V86 | R:R:Y316 | 3.79 | 1 | No | Yes | 7 | 6 | 2 | 1 | | R:R:V87 | R:R:Y316 | 3.79 | 0 | Yes | Yes | 7 | 6 | 2 | 1 | | R:R:F89 | R:R:W109 | 7.02 | 0 | No | Yes | 5 | 5 | 2 | 1 | | R:R:H93 | R:R:W99 | 7.41 | 1 | Yes | Yes | 4 | 7 | 1 | 2 | | R:R:C191 | R:R:H93 | 5.9 | 1 | No | Yes | 9 | 4 | 2 | 1 | | R:R:W109 | R:R:W99 | 6.56 | 0 | Yes | Yes | 5 | 7 | 1 | 2 | | R:R:C191 | R:R:W99 | 14.37 | 1 | No | Yes | 9 | 7 | 2 | 2 | | R:R:T110 | R:R:W109 | 3.64 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:D113 | R:R:Y316 | 12.64 | 1 | No | Yes | 7 | 6 | 1 | 1 | | R:R:S165 | R:R:V114 | 9.7 | 0 | No | No | 7 | 5 | 2 | 1 | | R:R:S165 | R:R:T118 | 3.2 | 0 | No | No | 7 | 6 | 2 | 2 | | R:R:S207 | R:R:T118 | 6.4 | 0 | No | No | 5 | 6 | 1 | 2 | | R:R:D192 | R:R:F194 | 14.33 | 0 | No | No | 3 | 1 | 1 | 2 | | R:R:F193 | R:R:T195 | 6.49 | 1 | No | Yes | 4 | 3 | 1 | 1 | | R:R:T195 | R:R:Y199 | 7.49 | 1 | Yes | Yes | 3 | 6 | 1 | 2 | | R:R:A200 | R:R:T195 | 5.03 | 0 | No | Yes | 5 | 3 | 2 | 1 | | R:R:F208 | R:R:W286 | 4.01 | 1 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:F208 | R:R:F290 | 12.86 | 1 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:F289 | R:R:W286 | 3.01 | 1 | Yes | Yes | 6 | 8 | 2 | 2 | | R:R:F290 | R:R:W286 | 5.01 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:W286 | R:R:Y316 | 2.89 | 1 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:F289 | R:R:F290 | 6.43 | 1 | Yes | Yes | 6 | 7 | 2 | 1 | | R:R:F289 | R:R:N293 | 8.46 | 1 | Yes | No | 6 | 5 | 2 | 2 | | R:R:F289 | R:R:Y308 | 5.16 | 1 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:F289 | R:R:N312 | 13.29 | 1 | Yes | No | 6 | 5 | 2 | 1 | | R:R:N293 | R:R:Y308 | 8.14 | 1 | No | Yes | 5 | 5 | 2 | 1 | | R:R:N312 | R:R:Y316 | 15.12 | 1 | No | Yes | 5 | 6 | 1 | 1 | | R:R:V292 | R:R:Y308 | 2.52 | 0 | No | Yes | 5 | 5 | 2 | 1 | | R:R:G90 | R:R:H93 | 1.59 | 0 | No | Yes | 7 | 4 | 2 | 1 | | R:R:C190 | R:R:D192 | 1.56 | 0 | No | No | 3 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 14.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 6.18 | | Average Nodes In Shell | 34.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 47.00 | | Average Links Mediated by Hubs In Shell | 40.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8JJ8 | A | Amine | Adrenergic | Beta2 | Homo Sapiens | Formoterol | - | - | 3.2 | 2023-09-06 | doi.org/10.1038/s41589-023-01389-0 |
|
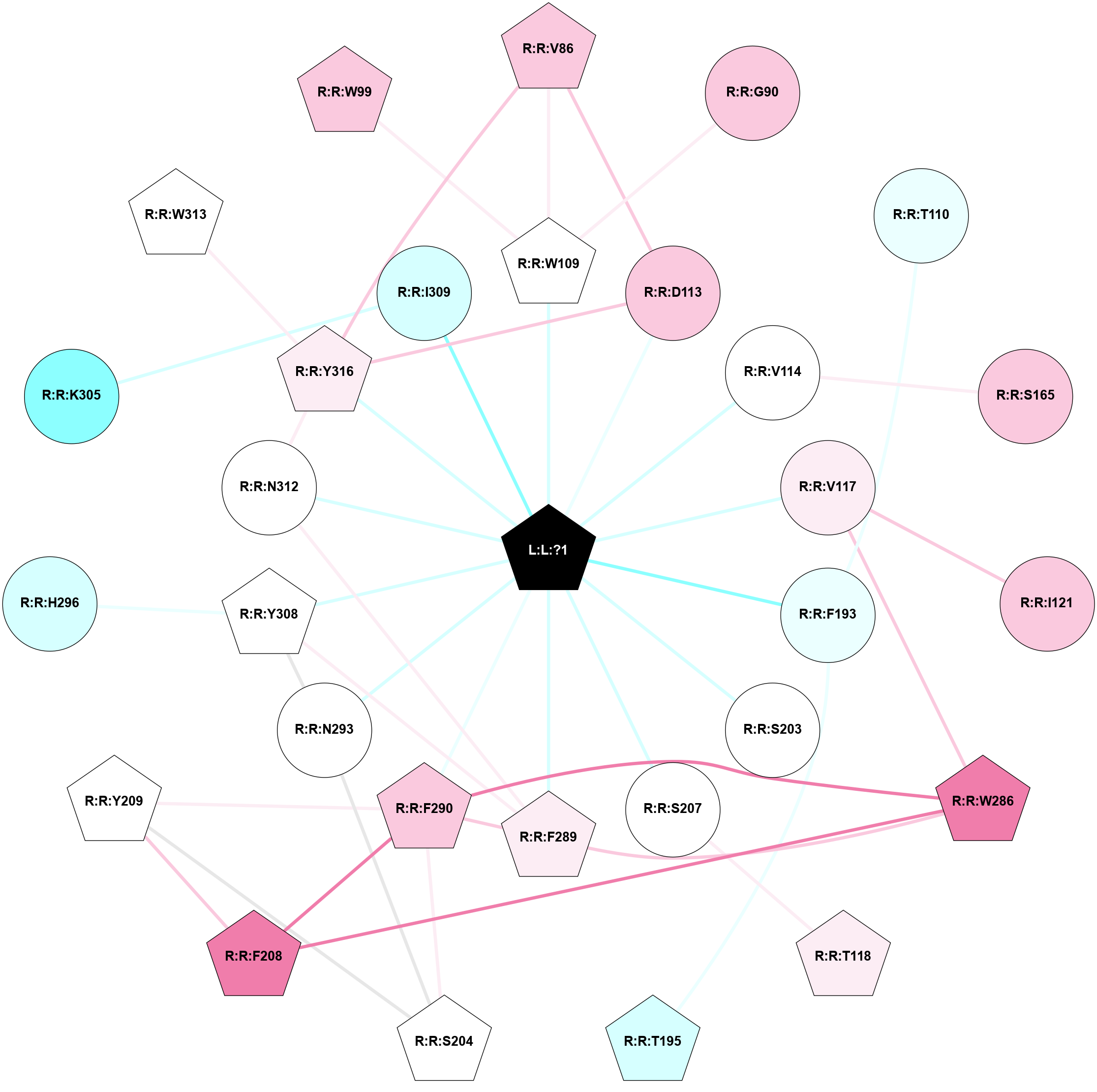
A 2D representation of the interactions of H98 in 8JJ8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:V86 | R:R:W109 | 4.9 | 2 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:D113 | R:R:V86 | 11.69 | 2 | No | Yes | 7 | 7 | 1 | 2 | | R:R:V86 | R:R:Y316 | 5.05 | 2 | Yes | Yes | 7 | 6 | 2 | 1 | | R:R:G90 | R:R:W109 | 4.22 | 0 | No | Yes | 7 | 5 | 2 | 1 | | L:L:?1 | R:R:W109 | 5.24 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:F193 | R:R:T110 | 5.19 | 0 | No | No | 4 | 4 | 1 | 2 | | R:R:D113 | R:R:Y316 | 12.64 | 2 | No | Yes | 7 | 6 | 1 | 1 | | L:L:?1 | R:R:D113 | 16.95 | 2 | Yes | No | 0 | 7 | 0 | 1 | | R:R:S165 | R:R:V114 | 3.23 | 1 | No | No | 7 | 5 | 2 | 1 | | L:L:?1 | R:R:V114 | 6.85 | 2 | Yes | No | 0 | 5 | 0 | 1 | | R:R:I121 | R:R:V117 | 3.07 | 0 | No | No | 7 | 6 | 2 | 1 | | R:R:V117 | R:R:W286 | 9.81 | 0 | No | Yes | 6 | 8 | 1 | 2 | | L:L:?1 | R:R:V117 | 6.85 | 2 | Yes | No | 0 | 6 | 0 | 1 | | R:R:S207 | R:R:T118 | 9.59 | 0 | No | Yes | 5 | 6 | 1 | 2 | | R:R:F193 | R:R:T195 | 5.19 | 0 | No | Yes | 4 | 3 | 1 | 2 | | L:L:?1 | R:R:F193 | 22.41 | 2 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:S203 | 10.86 | 2 | Yes | No | 0 | 5 | 0 | 1 | | R:R:S204 | R:R:Y209 | 3.82 | 2 | Yes | Yes | 5 | 5 | 2 | 2 | | R:R:F290 | R:R:S204 | 5.28 | 2 | Yes | Yes | 7 | 5 | 1 | 2 | | R:R:N293 | R:R:S204 | 4.47 | 2 | No | Yes | 5 | 5 | 1 | 2 | | L:L:?1 | R:R:S207 | 4.93 | 2 | Yes | No | 0 | 5 | 0 | 1 | | R:R:F208 | R:R:Y209 | 7.22 | 2 | Yes | Yes | 8 | 5 | 2 | 2 | | R:R:F208 | R:R:W286 | 3.01 | 2 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:F208 | R:R:F290 | 21.43 | 2 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:F290 | R:R:Y209 | 3.09 | 2 | Yes | Yes | 7 | 5 | 1 | 2 | | R:R:F289 | R:R:W286 | 7.02 | 2 | Yes | Yes | 6 | 8 | 1 | 2 | | R:R:F290 | R:R:W286 | 5.01 | 2 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:F289 | R:R:F290 | 7.5 | 2 | Yes | Yes | 6 | 7 | 1 | 1 | | R:R:F289 | R:R:Y308 | 9.28 | 2 | Yes | Yes | 6 | 5 | 1 | 1 | | R:R:F289 | R:R:N312 | 9.67 | 2 | Yes | No | 6 | 5 | 1 | 1 | | L:L:?1 | R:R:F289 | 6.4 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:F290 | 4.8 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:N293 | R:R:Y308 | 9.3 | 2 | No | Yes | 5 | 5 | 1 | 1 | | L:L:?1 | R:R:N293 | 5.41 | 2 | Yes | No | 0 | 5 | 0 | 1 | | R:R:H296 | R:R:Y308 | 8.71 | 0 | No | Yes | 3 | 5 | 2 | 1 | | R:R:I309 | R:R:K305 | 5.82 | 0 | No | No | 3 | 2 | 1 | 2 | | L:L:?1 | R:R:Y308 | 3.08 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:N312 | R:R:Y316 | 12.79 | 2 | No | Yes | 5 | 6 | 1 | 1 | | L:L:?1 | R:R:N312 | 11.73 | 2 | Yes | No | 0 | 5 | 0 | 1 | | R:R:W313 | R:R:Y316 | 11.58 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | L:L:?1 | R:R:Y316 | 4.62 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:W109 | R:R:W99 | 2.81 | 0 | Yes | Yes | 5 | 7 | 1 | 2 | | L:L:?1 | R:R:I309 | 2.81 | 2 | Yes | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 14.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 8.07 | | Average Nodes In Shell | 30.00 | | Average Hubs In Shell | 15.00 | | Average Links In Shell | 43.00 | | Average Links Mediated by Hubs In Shell | 39.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|



