| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6W2X | C | Aminoacid | GABAB | GABAB1; GABAB2 | Homo sapiens | PubChem 5311042 | PubChem 23727970 | - | 3.6 | 2020-07-01 | 10.1038/s41586-020-2469-4 |
|

A 2D representation of the interactions of L9Q in 6W2X
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:A640 | R:R:L670 | 1.58 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:A643 | R:R:L670 | 1.58 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:P646 | R:R:V662 | 3.53 | 3 | Yes | No | 6 | 5 | 2 | 2 | | R:R:P646 | R:R:R666 | 2.88 | 3 | Yes | No | 6 | 8 | 2 | 1 | | R:R:R666 | R:R:V662 | 9.15 | 3 | No | No | 8 | 5 | 1 | 2 | | R:R:R666 | W:W:?905 | 7.58 | 3 | No | Yes | 8 | 0 | 1 | 0 | | R:R:L667 | R:R:L725 | 4.15 | 0 | Yes | No | 5 | 8 | 1 | 2 | | R:R:L667 | R:R:W728 | 3.42 | 0 | Yes | Yes | 5 | 9 | 1 | 2 | | R:R:L667 | R:R:Q729 | 3.99 | 0 | Yes | No | 5 | 6 | 1 | 2 | | R:R:L667 | W:W:?905 | 1.73 | 0 | Yes | Yes | 5 | 0 | 1 | 0 | | R:R:G671 | R:R:L670 | 1.71 | 3 | No | Yes | 6 | 8 | 1 | 1 | | R:R:L670 | W:W:?905 | 7.77 | 3 | Yes | Yes | 8 | 0 | 1 | 0 | | R:R:G671 | R:R:L725 | 1.71 | 3 | No | No | 6 | 8 | 1 | 2 | | R:R:G671 | W:W:?905 | 2.13 | 3 | No | Yes | 6 | 0 | 1 | 0 | | R:R:F674 | R:R:K777 | 4.96 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:F674 | W:W:?905 | 9.12 | 0 | No | Yes | 9 | 0 | 1 | 0 | | R:R:E739 | R:R:L758 | 5.3 | 3 | No | Yes | 4 | 5 | 2 | 2 | | R:R:E739 | R:R:H760 | 7.39 | 3 | No | Yes | 4 | 4 | 2 | 1 | | R:R:H760 | R:R:L758 | 12.86 | 3 | Yes | Yes | 4 | 5 | 1 | 2 | | R:R:H760 | R:R:S762 | 4.18 | 3 | Yes | No | 4 | 5 | 1 | 2 | | R:R:H760 | W:W:?905 | 2.41 | 3 | Yes | Yes | 4 | 0 | 1 | 0 | | R:R:F773 | R:R:K777 | 4.96 | 0 | No | Yes | 6 | 9 | 1 | 2 | | R:R:F773 | W:W:?905 | 6.84 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:A820 | R:R:Y774 | 4 | 0 | No | No | 7 | 6 | 2 | 1 | | R:R:Y774 | W:W:?905 | 2.93 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:C816 | R:R:L781 | 3.17 | 0 | No | Yes | 8 | 8 | 1 | 2 | | R:R:C816 | R:R:V812 | 1.71 | 0 | No | No | 8 | 8 | 1 | 2 | | R:R:C816 | W:W:?905 | 4.95 | 0 | No | Yes | 8 | 0 | 1 | 0 | | R:R:A840 | R:R:T819 | 3.36 | 0 | No | No | 6 | 8 | 2 | 1 | | R:R:T819 | W:W:?905 | 2.76 | 0 | No | Yes | 8 | 0 | 1 | 0 | | R:R:A837 | R:R:T823 | 1.68 | 0 | No | No | 5 | 7 | 2 | 1 | | R:R:T823 | W:W:?905 | 1.84 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:S844 | R:R:S845 | 1.63 | 0 | No | Yes | 7 | 8 | 1 | 2 | | R:R:S844 | W:W:?905 | 2.81 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:L815 | R:R:S844 | 1.5 | 0 | No | No | 7 | 7 | 2 | 1 | | R:R:G775 | R:R:Y774 | 1.45 | 0 | No | No | 4 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 12.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 4.41 | | Average Nodes In Shell | 32.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 36.00 | | Average Links Mediated by Hubs In Shell | 28.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
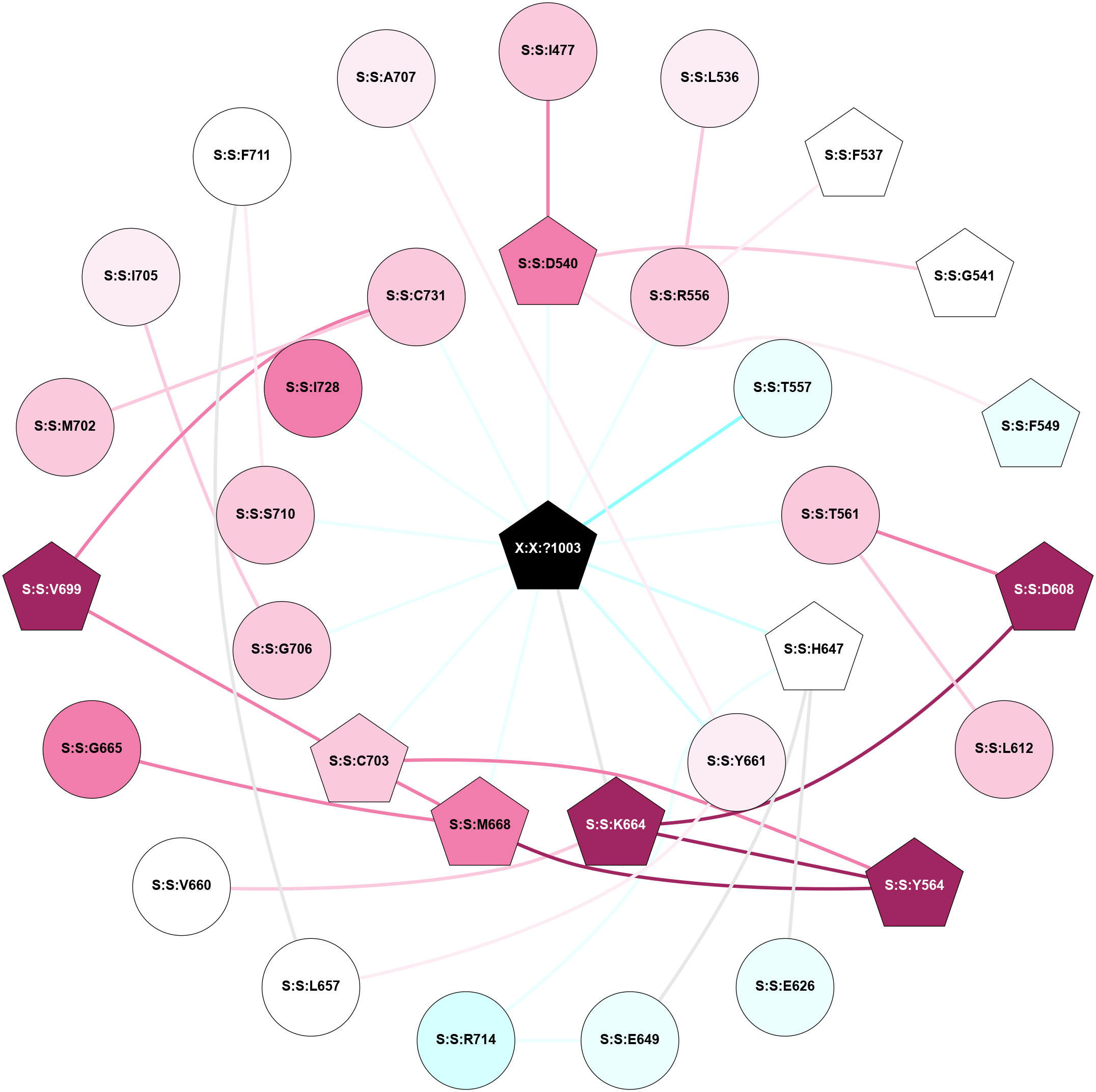
A 2D representation of the interactions of L9Q in 6W2X
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:D540 | S:S:I477 | 5.6 | 0 | Yes | No | 8 | 7 | 1 | 2 | | S:S:L536 | S:S:R556 | 7.29 | 0 | No | No | 6 | 7 | 2 | 1 | | S:S:F537 | S:S:R556 | 7.48 | 0 | Yes | No | 5 | 7 | 2 | 1 | | S:S:D540 | S:S:G541 | 3.35 | 0 | Yes | Yes | 8 | 5 | 1 | 2 | | S:S:D540 | S:S:F549 | 3.58 | 0 | Yes | Yes | 8 | 4 | 1 | 2 | | S:S:D540 | X:X:?1003 | 1.69 | 0 | Yes | Yes | 8 | 0 | 1 | 0 | | S:S:R556 | X:X:?1003 | 4.55 | 0 | No | Yes | 7 | 0 | 1 | 0 | | S:S:T557 | X:X:?1003 | 3.68 | 0 | No | Yes | 4 | 0 | 1 | 0 | | S:S:D608 | S:S:T561 | 4.34 | 59 | Yes | No | 9 | 7 | 2 | 1 | | S:S:L612 | S:S:T561 | 7.37 | 0 | No | No | 7 | 7 | 2 | 1 | | S:S:T561 | X:X:?1003 | 1.84 | 0 | No | Yes | 7 | 0 | 1 | 0 | | S:S:K664 | S:S:Y564 | 9.55 | 0 | Yes | Yes | 9 | 9 | 1 | 2 | | S:S:M668 | S:S:Y564 | 9.58 | 9 | Yes | Yes | 8 | 9 | 1 | 2 | | S:S:C703 | S:S:Y564 | 2.69 | 9 | Yes | Yes | 7 | 9 | 1 | 2 | | S:S:D608 | S:S:K664 | 4.15 | 59 | Yes | Yes | 9 | 9 | 2 | 1 | | S:S:E626 | S:S:H647 | 17.23 | 0 | No | Yes | 4 | 5 | 2 | 1 | | S:S:E649 | S:S:H647 | 9.85 | 9 | No | Yes | 4 | 5 | 2 | 1 | | S:S:H647 | S:S:R714 | 11.28 | 9 | Yes | No | 5 | 3 | 1 | 2 | | S:S:H647 | X:X:?1003 | 4.81 | 9 | Yes | Yes | 5 | 0 | 1 | 0 | | S:S:E649 | S:S:R714 | 3.49 | 9 | No | No | 4 | 3 | 2 | 2 | | S:S:L657 | S:S:Y661 | 10.55 | 0 | No | No | 5 | 6 | 2 | 1 | | S:S:F711 | S:S:L657 | 4.87 | 0 | No | No | 5 | 5 | 2 | 2 | | S:S:K664 | S:S:V660 | 3.04 | 0 | Yes | No | 9 | 5 | 1 | 2 | | S:S:Y661 | X:X:?1003 | 2.19 | 0 | No | Yes | 6 | 0 | 1 | 0 | | S:S:K664 | X:X:?1003 | 6.16 | 0 | Yes | Yes | 9 | 0 | 1 | 0 | | S:S:G665 | S:S:M668 | 3.49 | 0 | No | Yes | 8 | 8 | 2 | 1 | | S:S:C703 | S:S:M668 | 4.86 | 9 | Yes | Yes | 7 | 8 | 1 | 1 | | S:S:M668 | X:X:?1003 | 3.53 | 9 | Yes | Yes | 8 | 0 | 1 | 0 | | S:S:C703 | S:S:V699 | 1.71 | 9 | Yes | Yes | 7 | 9 | 1 | 2 | | S:S:C731 | S:S:V699 | 5.12 | 0 | No | Yes | 7 | 9 | 1 | 2 | | S:S:C731 | S:S:M702 | 8.1 | 0 | No | No | 7 | 7 | 1 | 2 | | S:S:C703 | X:X:?1003 | 5.94 | 9 | Yes | Yes | 7 | 0 | 1 | 0 | | S:S:G706 | S:S:I705 | 1.76 | 0 | No | No | 7 | 6 | 1 | 2 | | S:S:G706 | X:X:?1003 | 2.13 | 0 | No | Yes | 7 | 0 | 1 | 0 | | S:S:F711 | S:S:S710 | 3.96 | 0 | No | No | 5 | 7 | 2 | 1 | | S:S:S710 | X:X:?1003 | 2.81 | 0 | No | Yes | 7 | 0 | 1 | 0 | | S:S:I728 | X:X:?1003 | 2.67 | 0 | No | Yes | 8 | 0 | 1 | 0 | | S:S:C731 | X:X:?1003 | 1.98 | 0 | No | Yes | 7 | 0 | 1 | 0 | | S:S:A707 | S:S:Y661 | 1.33 | 0 | No | No | 6 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 13.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 3.38 | | Average Nodes In Shell | 33.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 39.00 | | Average Links Mediated by Hubs In Shell | 30.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6W2Y | C | Aminoacid | GABAB | GABAB1; GABAB1 | Homo sapiens | PubChem 5311042 | PubChem 23727970 | - | 3.2 | 2020-07-01 | 10.1038/s41586-020-2469-4 |
|
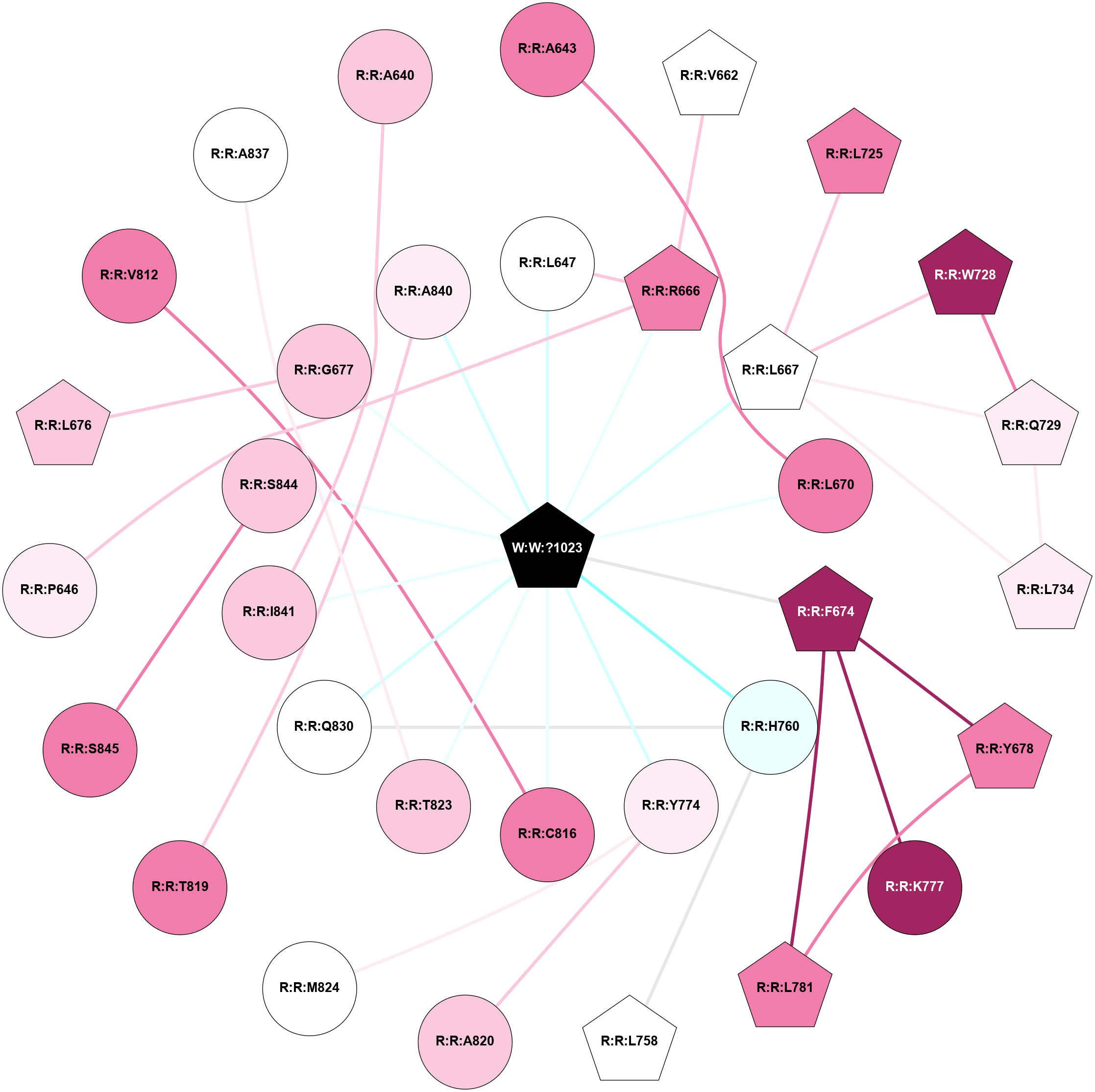
A 2D representation of the interactions of L9Q in 6W2Y
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:A643 | R:R:L670 | 3.15 | 0 | No | No | 8 | 8 | 2 | 1 | | R:R:L647 | R:R:R666 | 4.86 | 3 | No | Yes | 5 | 8 | 1 | 1 | | R:R:L647 | W:W:?1023 | 4.32 | 3 | No | Yes | 5 | 0 | 1 | 0 | | R:R:R666 | R:R:V662 | 6.54 | 3 | Yes | Yes | 8 | 5 | 1 | 2 | | R:R:R666 | W:W:?1023 | 6.06 | 3 | Yes | Yes | 8 | 0 | 1 | 0 | | R:R:L667 | R:R:L725 | 5.54 | 3 | Yes | Yes | 5 | 8 | 1 | 2 | | R:R:L667 | R:R:W728 | 10.25 | 3 | Yes | Yes | 5 | 9 | 1 | 2 | | R:R:L667 | R:R:Q729 | 3.99 | 3 | Yes | Yes | 5 | 6 | 1 | 2 | | R:R:L667 | R:R:L734 | 4.15 | 3 | Yes | Yes | 5 | 6 | 1 | 2 | | R:R:L667 | W:W:?1023 | 3.45 | 3 | Yes | Yes | 5 | 0 | 1 | 0 | | R:R:L670 | W:W:?1023 | 11.23 | 0 | No | Yes | 8 | 0 | 1 | 0 | | R:R:F674 | R:R:Y678 | 6.19 | 3 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F674 | R:R:K777 | 4.96 | 3 | Yes | No | 9 | 9 | 1 | 2 | | R:R:F674 | R:R:L781 | 3.65 | 3 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F674 | W:W:?1023 | 11.4 | 3 | Yes | Yes | 9 | 0 | 1 | 0 | | R:R:L781 | R:R:Y678 | 15.24 | 3 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:Q729 | R:R:W728 | 7.67 | 3 | Yes | Yes | 6 | 9 | 2 | 2 | | R:R:L734 | R:R:Q729 | 3.99 | 3 | Yes | Yes | 6 | 6 | 2 | 2 | | R:R:H760 | R:R:L758 | 10.29 | 3 | No | Yes | 4 | 5 | 1 | 2 | | R:R:H760 | R:R:Q830 | 8.65 | 3 | No | No | 4 | 5 | 1 | 1 | | R:R:H760 | W:W:?1023 | 6.42 | 3 | No | Yes | 4 | 0 | 1 | 0 | | R:R:A820 | R:R:Y774 | 9.34 | 0 | No | No | 7 | 6 | 2 | 1 | | R:R:M824 | R:R:Y774 | 3.59 | 0 | No | No | 5 | 6 | 2 | 1 | | R:R:Y774 | W:W:?1023 | 8.78 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:C816 | W:W:?1023 | 9.9 | 0 | No | Yes | 8 | 0 | 1 | 0 | | R:R:A840 | R:R:T819 | 5.03 | 0 | No | No | 6 | 8 | 1 | 2 | | R:R:T823 | W:W:?1023 | 9.2 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:Q830 | W:W:?1023 | 5.81 | 3 | No | Yes | 5 | 0 | 1 | 0 | | R:R:I841 | W:W:?1023 | 3.56 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:S844 | R:R:S845 | 3.26 | 0 | No | No | 7 | 8 | 1 | 2 | | R:R:S844 | W:W:?1023 | 8.43 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:P646 | R:R:R666 | 2.88 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:G677 | W:W:?1023 | 2.13 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:A840 | W:W:?1023 | 1.97 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:G677 | R:R:L676 | 1.71 | 0 | No | Yes | 7 | 7 | 1 | 2 | | R:R:C816 | R:R:V812 | 1.71 | 0 | No | No | 8 | 8 | 1 | 2 | | R:R:A837 | R:R:T823 | 1.68 | 0 | No | No | 5 | 7 | 2 | 1 | | R:R:A640 | R:R:I841 | 1.62 | 0 | No | No | 7 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 14.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 6.62 | | Average Nodes In Shell | 34.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 38.00 | | Average Links Mediated by Hubs In Shell | 29.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
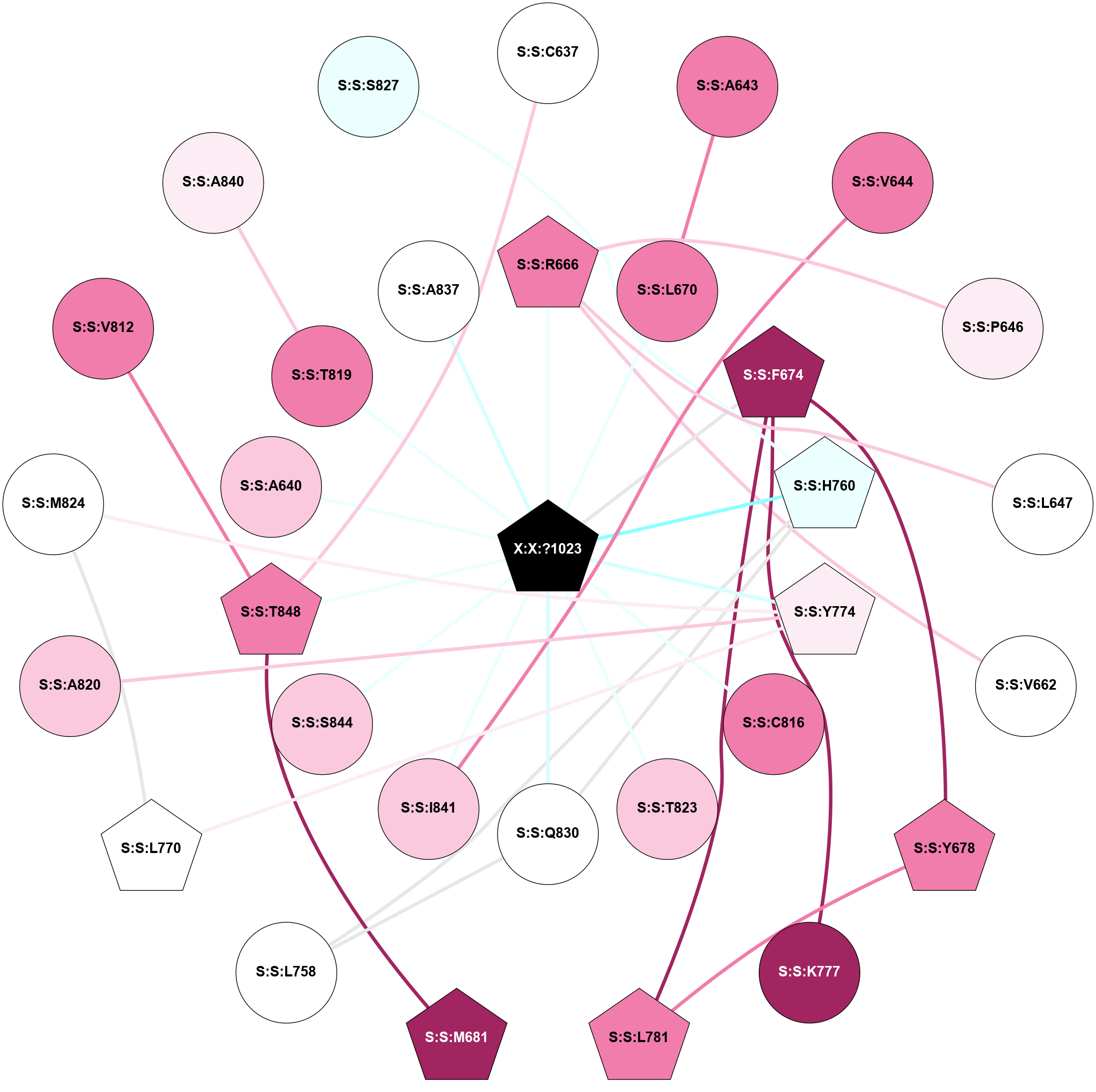
A 2D representation of the interactions of L9Q in 6W2Y
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:C637 | S:S:T848 | 3.38 | 0 | No | Yes | 5 | 8 | 2 | 1 | | S:S:A643 | S:S:L670 | 3.15 | 0 | No | No | 8 | 8 | 2 | 1 | | S:S:I841 | S:S:V644 | 3.07 | 0 | No | No | 7 | 8 | 1 | 2 | | S:S:P646 | S:S:R666 | 4.32 | 0 | No | Yes | 6 | 8 | 2 | 1 | | S:S:L647 | S:S:R666 | 3.64 | 0 | No | Yes | 5 | 8 | 2 | 1 | | S:S:R666 | S:S:V662 | 6.54 | 0 | Yes | No | 8 | 5 | 1 | 2 | | S:S:R666 | X:X:?1023 | 12.88 | 0 | Yes | Yes | 8 | 0 | 1 | 0 | | S:S:L670 | X:X:?1023 | 7.77 | 0 | No | Yes | 8 | 0 | 1 | 0 | | S:S:F674 | S:S:Y678 | 4.13 | 4 | Yes | Yes | 9 | 8 | 1 | 2 | | S:S:F674 | S:S:K777 | 4.96 | 4 | Yes | No | 9 | 9 | 1 | 2 | | S:S:F674 | S:S:L781 | 3.65 | 4 | Yes | Yes | 9 | 8 | 1 | 2 | | S:S:F674 | X:X:?1023 | 8.36 | 4 | Yes | Yes | 9 | 0 | 1 | 0 | | S:S:L781 | S:S:Y678 | 12.89 | 4 | Yes | Yes | 8 | 8 | 2 | 2 | | S:S:M681 | S:S:T848 | 3.01 | 4 | Yes | Yes | 9 | 8 | 2 | 1 | | S:S:H760 | S:S:L758 | 20.57 | 4 | Yes | No | 4 | 5 | 1 | 2 | | S:S:L758 | S:S:Q830 | 6.65 | 4 | No | No | 5 | 5 | 2 | 1 | | S:S:H760 | S:S:Q830 | 3.71 | 4 | Yes | No | 4 | 5 | 1 | 1 | | S:S:H760 | X:X:?1023 | 6.42 | 4 | Yes | Yes | 4 | 0 | 1 | 0 | | S:S:L770 | S:S:Y774 | 5.86 | 4 | Yes | Yes | 5 | 6 | 2 | 1 | | S:S:L770 | S:S:M824 | 5.65 | 4 | Yes | No | 5 | 5 | 2 | 2 | | S:S:A820 | S:S:Y774 | 9.34 | 0 | No | Yes | 7 | 6 | 2 | 1 | | S:S:M824 | S:S:Y774 | 3.59 | 4 | No | Yes | 5 | 6 | 2 | 1 | | S:S:Y774 | X:X:?1023 | 16.09 | 4 | Yes | Yes | 6 | 0 | 1 | 0 | | S:S:T848 | S:S:V812 | 3.17 | 0 | Yes | No | 8 | 8 | 1 | 2 | | S:S:C816 | X:X:?1023 | 12.87 | 0 | No | Yes | 8 | 0 | 1 | 0 | | S:S:A840 | S:S:T819 | 5.03 | 0 | No | No | 6 | 8 | 2 | 1 | | S:S:T823 | X:X:?1023 | 4.6 | 0 | No | Yes | 7 | 0 | 1 | 0 | | S:S:Q830 | X:X:?1023 | 5.81 | 4 | No | Yes | 5 | 0 | 1 | 0 | | S:S:I841 | X:X:?1023 | 4.45 | 0 | No | Yes | 7 | 0 | 1 | 0 | | S:S:S844 | X:X:?1023 | 8.43 | 0 | No | Yes | 7 | 0 | 1 | 0 | | S:S:T848 | X:X:?1023 | 3.68 | 0 | Yes | Yes | 8 | 0 | 1 | 0 | | S:S:A640 | X:X:?1023 | 2.95 | 0 | No | Yes | 7 | 0 | 1 | 0 | | S:S:T819 | X:X:?1023 | 2.76 | 0 | No | Yes | 8 | 0 | 1 | 0 | | S:S:A837 | X:X:?1023 | 1.97 | 0 | No | Yes | 5 | 0 | 1 | 0 | | S:S:H760 | S:S:S827 | 1.39 | 4 | Yes | No | 4 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 14.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 7.07 | | Average Nodes In Shell | 32.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 35.00 | | Average Links Mediated by Hubs In Shell | 31.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|



