| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4MQS | A | Amine | Acetylcholine (muscarinic) | M2 | Homo Sapiens | Iperoxo | - | - | 3.5 | 2013-11-27 | doi.org/10.1038/nature12735 |
|

A 2D representation of the interactions of IXO in 4MQS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:D103 | 8.71 | 1 | Yes | No | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y104 | 15.89 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:S107 | 5.36 | 1 | Yes | No | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:N108 | 3.92 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:W155 | 8.94 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:A194 | 5.62 | 1 | Yes | No | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:W400 | 18.69 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y403 | 11.71 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:N404 | 8.81 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:Y426 | 10.87 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:C429 | 4.53 | 1 | Yes | No | 0 | 8 | 0 | 1 | | R:R:I72 | R:R:S76 | 6.19 | 0 | Yes | No | 8 | 8 | 2 | 2 | | R:R:I72 | R:R:S107 | 4.64 | 0 | Yes | No | 8 | 8 | 2 | 1 | | R:R:D103 | R:R:S76 | 11.78 | 1 | No | No | 9 | 8 | 1 | 2 | | R:R:S76 | R:R:Y430 | 8.9 | 1 | No | Yes | 8 | 9 | 2 | 2 | | R:R:Y426 | R:R:Y80 | 25.81 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:Y430 | R:R:Y80 | 5.96 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:L100 | R:R:Y104 | 4.69 | 1 | No | Yes | 7 | 8 | 2 | 1 | | R:R:I178 | R:R:L100 | 7.14 | 1 | Yes | No | 7 | 7 | 2 | 2 | | R:R:D103 | R:R:Y430 | 11.49 | 1 | No | Yes | 9 | 9 | 1 | 2 | | R:R:W155 | R:R:Y104 | 11.58 | 1 | Yes | Yes | 8 | 8 | 1 | 1 | | R:R:I159 | R:R:Y104 | 4.84 | 1 | No | Yes | 8 | 8 | 2 | 1 | | R:R:I178 | R:R:Y104 | 13.3 | 1 | Yes | Yes | 7 | 8 | 2 | 1 | | R:R:F181 | R:R:Y104 | 4.13 | 1 | Yes | Yes | 5 | 8 | 2 | 1 | | R:R:Y104 | R:R:Y403 | 6.95 | 1 | Yes | Yes | 8 | 8 | 1 | 1 | | R:R:Y104 | R:R:Y426 | 6.95 | 1 | Yes | Yes | 8 | 7 | 1 | 1 | | R:R:S107 | R:R:W400 | 7.41 | 1 | No | Yes | 8 | 9 | 1 | 1 | | R:R:M112 | R:R:N108 | 7.01 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:N108 | R:R:S151 | 8.94 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:N108 | R:R:W155 | 14.69 | 1 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:F396 | R:R:V111 | 6.55 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:V111 | R:R:W400 | 7.36 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:T190 | R:R:W155 | 7.28 | 0 | No | Yes | 9 | 8 | 2 | 1 | | R:R:A194 | R:R:W155 | 3.89 | 1 | No | Yes | 8 | 8 | 1 | 1 | | R:R:I159 | R:R:I178 | 5.89 | 1 | No | Yes | 8 | 7 | 2 | 2 | | R:R:F181 | R:R:I178 | 11.3 | 1 | Yes | Yes | 5 | 7 | 2 | 2 | | R:R:F181 | R:R:V407 | 3.93 | 1 | Yes | No | 5 | 8 | 2 | 2 | | R:R:F195 | R:R:Y196 | 12.38 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:F195 | R:R:F396 | 5.36 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:F195 | R:R:N404 | 18.12 | 1 | Yes | Yes | 9 | 7 | 2 | 1 | | R:R:N404 | R:R:Y196 | 4.65 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:F396 | R:R:W400 | 8.02 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:T399 | R:R:W400 | 4.85 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:N432 | R:R:T399 | 7.31 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:C429 | R:R:W400 | 7.84 | 1 | No | Yes | 8 | 9 | 1 | 1 | | R:R:N432 | R:R:W400 | 14.69 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:N404 | R:R:Y403 | 8.14 | 1 | Yes | Yes | 7 | 8 | 1 | 1 | | R:R:V407 | R:R:Y403 | 8.83 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:W422 | R:R:Y403 | 3.86 | 1 | No | Yes | 6 | 8 | 2 | 1 | | R:R:Y403 | R:R:Y426 | 3.97 | 1 | Yes | Yes | 8 | 7 | 1 | 1 | | R:R:C429 | R:R:Y403 | 4.03 | 1 | No | Yes | 8 | 8 | 1 | 1 | | R:R:W422 | R:R:Y426 | 5.79 | 1 | No | Yes | 6 | 7 | 2 | 1 | | R:R:Y426 | R:R:Y430 | 4.96 | 1 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:L154 | R:R:W155 | 2.28 | 0 | No | Yes | 8 | 8 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 11.00 | | Average Number Of Links With An Hub | 7.00 | | Average Interaction Strength | 9.37 | | Average Nodes In Shell | 32.00 | | Average Hubs In Shell | 17.00 | | Average Links In Shell | 54.00 | | Average Links Mediated by Hubs In Shell | 53.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4MQT | A | Amine | Acetylcholine (muscarinic) | M2 | Homo Sapiens | Iperoxo | LY2119620 | - | 3.7 | 2013-11-27 | doi.org/10.1038/nature12735 |
|
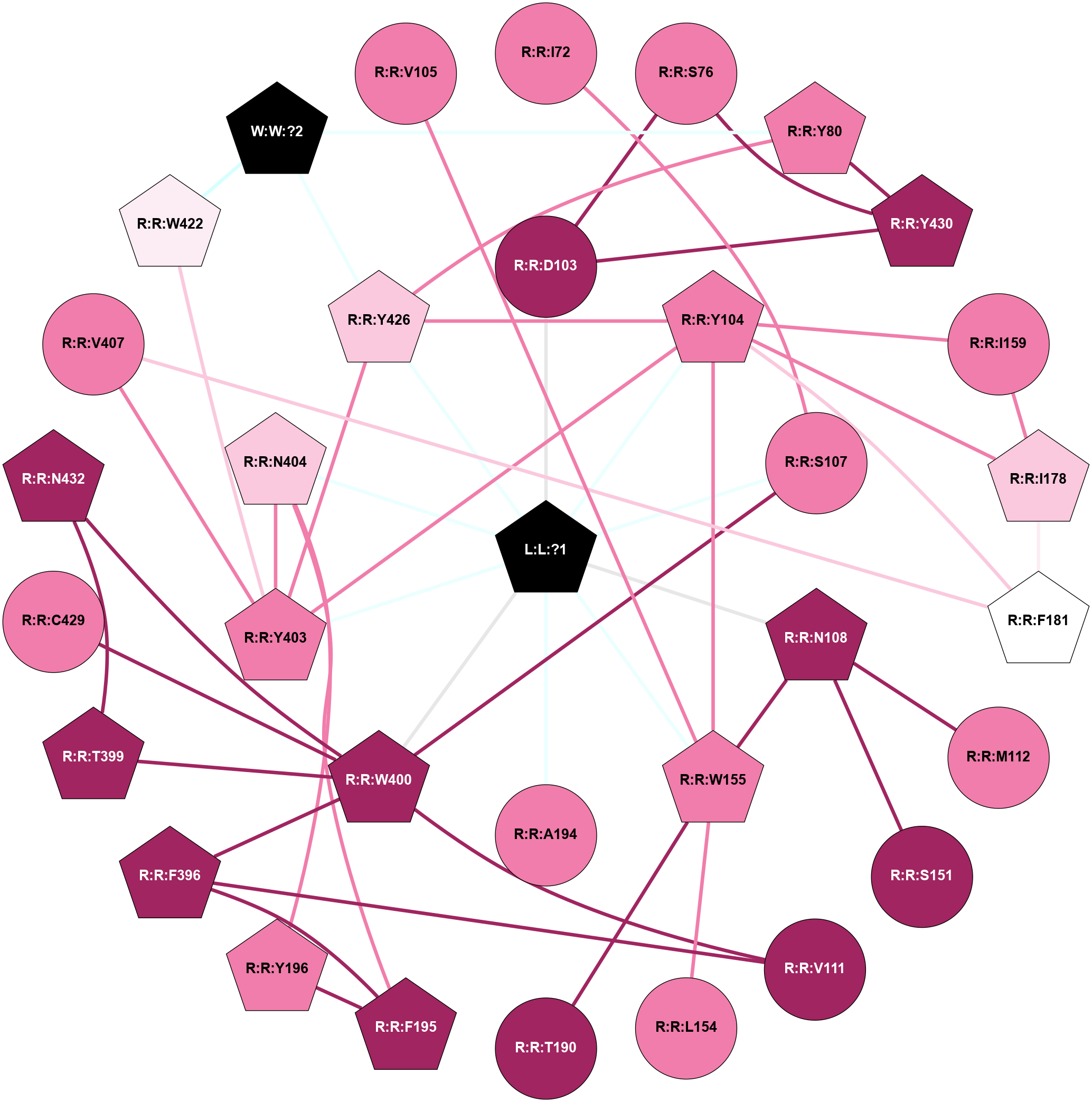
A 2D representation of the interactions of IXO in 4MQT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:D103 | 9.68 | 1 | Yes | No | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y104 | 18.4 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:S107 | 8.57 | 1 | Yes | No | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:N108 | 6.86 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:W155 | 8.94 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:A194 | 5.62 | 1 | Yes | No | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:W400 | 13 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y403 | 14.22 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:N404 | 8.81 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:Y426 | 9.2 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:I72 | R:R:S107 | 4.64 | 0 | No | No | 8 | 8 | 2 | 1 | | R:R:D103 | R:R:S76 | 10.31 | 1 | No | No | 9 | 8 | 1 | 2 | | R:R:S76 | R:R:Y430 | 10.17 | 1 | No | Yes | 8 | 9 | 2 | 2 | | R:R:Y426 | R:R:Y80 | 19.86 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:Y430 | R:R:Y80 | 5.96 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:Y80 | W:W:?2 | 10.34 | 1 | Yes | Yes | 8 | 0 | 2 | 2 | | R:R:D103 | R:R:Y430 | 10.34 | 1 | No | Yes | 9 | 9 | 1 | 2 | | R:R:W155 | R:R:Y104 | 11.58 | 1 | Yes | Yes | 8 | 8 | 1 | 1 | | R:R:I159 | R:R:Y104 | 4.84 | 1 | No | Yes | 8 | 8 | 2 | 1 | | R:R:I178 | R:R:Y104 | 4.84 | 1 | Yes | Yes | 7 | 8 | 2 | 1 | | R:R:Y104 | R:R:Y403 | 7.94 | 1 | Yes | Yes | 8 | 8 | 1 | 1 | | R:R:Y104 | R:R:Y426 | 4.96 | 1 | Yes | Yes | 8 | 7 | 1 | 1 | | R:R:S107 | R:R:W400 | 6.18 | 1 | No | Yes | 8 | 9 | 1 | 1 | | R:R:M112 | R:R:N108 | 8.41 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:N108 | R:R:S151 | 8.94 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:N108 | R:R:W155 | 12.43 | 1 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:F396 | R:R:V111 | 7.87 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:V111 | R:R:W400 | 8.58 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:L154 | R:R:W155 | 10.25 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:T190 | R:R:W155 | 7.28 | 0 | No | Yes | 9 | 8 | 2 | 1 | | R:R:I159 | R:R:I178 | 7.36 | 1 | No | Yes | 8 | 7 | 2 | 2 | | R:R:F195 | R:R:Y196 | 14.44 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:F195 | R:R:F396 | 5.36 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:F195 | R:R:N404 | 19.33 | 1 | Yes | Yes | 9 | 7 | 2 | 1 | | R:R:N404 | R:R:Y196 | 4.65 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:F396 | R:R:W400 | 8.02 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:T399 | R:R:W400 | 6.06 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:N432 | R:R:T399 | 11.7 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:C429 | R:R:W400 | 6.53 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:N432 | R:R:W400 | 14.69 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:N404 | R:R:Y403 | 8.14 | 1 | Yes | Yes | 7 | 8 | 1 | 1 | | R:R:V407 | R:R:Y403 | 6.31 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:W422 | R:R:Y403 | 4.82 | 1 | Yes | Yes | 6 | 8 | 2 | 1 | | R:R:Y403 | R:R:Y426 | 4.96 | 1 | Yes | Yes | 8 | 7 | 1 | 1 | | R:R:W422 | W:W:?2 | 31.82 | 1 | Yes | Yes | 6 | 0 | 2 | 2 | | R:R:Y426 | W:W:?2 | 4.31 | 1 | Yes | Yes | 7 | 0 | 1 | 2 | | R:R:V105 | R:R:W155 | 1.23 | 0 | No | Yes | 8 | 8 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 7.00 | | Average Interaction Strength | 10.33 | | Average Nodes In Shell | 32.00 | | Average Hubs In Shell | 18.00 | | Average Links In Shell | 47.00 | | Average Links Mediated by Hubs In Shell | 45.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6OIJ | A | Amine | Acetylcholine (muscarinic) | M1 | Homo Sapiens | Iperoxo | - | chim(NtGi1L-G11)/Beta1/Gamma2 | 3.3 | 2019-05-08 | doi.org/10.1126/science.aaw5188 |
|
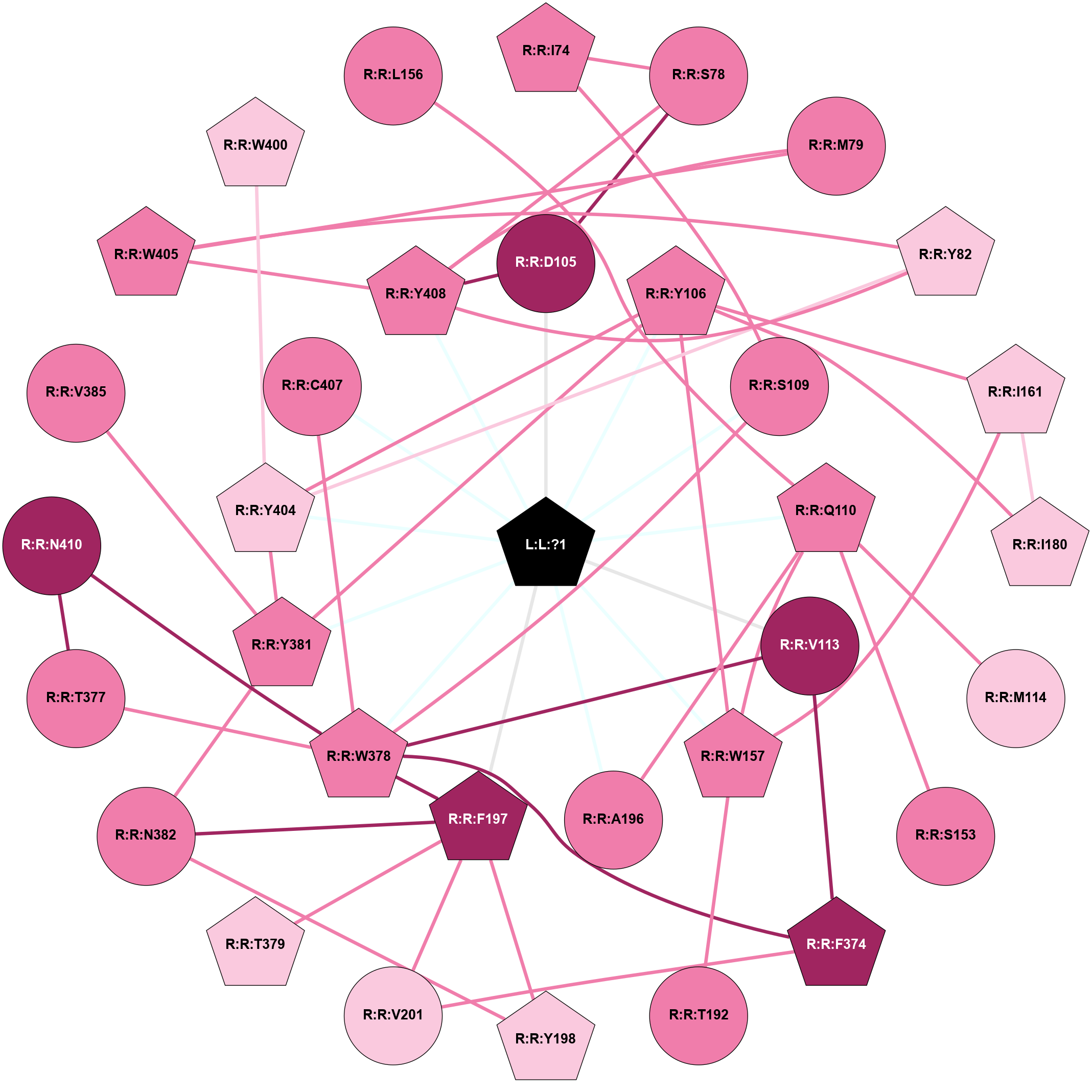
A 2D representation of the interactions of IXO in 6OIJ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I74 | R:R:S78 | 3.1 | 0 | Yes | No | 8 | 8 | 2 | 2 | | R:R:I74 | R:R:S109 | 4.64 | 0 | Yes | No | 8 | 8 | 2 | 1 | | R:R:D105 | R:R:S78 | 10.31 | 1 | No | No | 9 | 8 | 1 | 2 | | R:R:S78 | R:R:Y408 | 3.82 | 1 | No | Yes | 8 | 8 | 2 | 1 | | R:R:M79 | R:R:W405 | 8.14 | 1 | No | Yes | 8 | 8 | 2 | 2 | | R:R:M79 | R:R:Y408 | 11.97 | 1 | No | Yes | 8 | 8 | 2 | 1 | | R:R:Y404 | R:R:Y82 | 11.91 | 1 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:W405 | R:R:Y82 | 4.82 | 1 | Yes | Yes | 8 | 7 | 2 | 2 | | R:R:Y408 | R:R:Y82 | 6.95 | 1 | Yes | Yes | 8 | 7 | 1 | 2 | | R:R:D105 | R:R:Y408 | 9.2 | 1 | No | Yes | 9 | 8 | 1 | 1 | | L:L:?1 | R:R:D105 | 9.68 | 1 | Yes | No | 0 | 9 | 0 | 1 | | R:R:W157 | R:R:Y106 | 16.4 | 1 | Yes | Yes | 8 | 8 | 1 | 1 | | R:R:I161 | R:R:Y106 | 3.63 | 1 | Yes | Yes | 7 | 8 | 2 | 1 | | R:R:I180 | R:R:Y106 | 7.25 | 1 | Yes | Yes | 7 | 8 | 2 | 1 | | R:R:Y106 | R:R:Y381 | 7.94 | 1 | Yes | Yes | 8 | 8 | 1 | 1 | | R:R:Y106 | R:R:Y404 | 6.95 | 1 | Yes | Yes | 8 | 7 | 1 | 1 | | L:L:?1 | R:R:Y106 | 13.38 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:S109 | R:R:W378 | 3.71 | 1 | No | Yes | 8 | 8 | 1 | 1 | | L:L:?1 | R:R:S109 | 11.78 | 1 | Yes | No | 0 | 8 | 0 | 1 | | R:R:M114 | R:R:Q110 | 5.44 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:Q110 | R:R:S153 | 5.78 | 1 | Yes | No | 8 | 8 | 1 | 2 | | R:R:Q110 | R:R:W157 | 8.76 | 1 | Yes | Yes | 8 | 8 | 1 | 1 | | R:R:A196 | R:R:Q110 | 4.55 | 1 | No | Yes | 8 | 8 | 1 | 1 | | L:L:?1 | R:R:Q110 | 10.44 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:F374 | R:R:V113 | 3.93 | 1 | Yes | No | 9 | 9 | 2 | 1 | | R:R:V113 | R:R:W378 | 4.9 | 1 | No | Yes | 9 | 8 | 1 | 1 | | L:L:?1 | R:R:V113 | 5.31 | 1 | Yes | No | 0 | 9 | 0 | 1 | | R:R:I161 | R:R:W157 | 3.52 | 1 | Yes | Yes | 7 | 8 | 2 | 1 | | R:R:T192 | R:R:W157 | 4.85 | 0 | No | Yes | 8 | 8 | 2 | 1 | | L:L:?1 | R:R:W157 | 7.31 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:I161 | R:R:I180 | 5.89 | 1 | Yes | Yes | 7 | 7 | 2 | 2 | | L:L:?1 | R:R:A196 | 3.37 | 1 | Yes | No | 0 | 8 | 0 | 1 | | R:R:F197 | R:R:Y198 | 11.35 | 1 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F197 | R:R:V201 | 7.87 | 1 | Yes | No | 9 | 7 | 1 | 2 | | R:R:F197 | R:R:W378 | 6.01 | 1 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:F197 | R:R:T379 | 3.89 | 1 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F197 | R:R:N382 | 25.37 | 1 | Yes | No | 9 | 8 | 1 | 2 | | L:L:?1 | R:R:F197 | 5.21 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:N382 | R:R:Y198 | 3.49 | 1 | No | Yes | 8 | 7 | 2 | 2 | | R:R:F374 | R:R:V201 | 3.93 | 1 | Yes | No | 9 | 7 | 2 | 2 | | R:R:F374 | R:R:W378 | 8.02 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:T377 | R:R:W378 | 3.64 | 1 | No | Yes | 8 | 8 | 2 | 1 | | R:R:N410 | R:R:T377 | 7.31 | 1 | No | No | 9 | 8 | 2 | 2 | | R:R:C407 | R:R:W378 | 16.98 | 1 | No | Yes | 8 | 8 | 1 | 1 | | R:R:N410 | R:R:W378 | 10.17 | 1 | No | Yes | 9 | 8 | 2 | 1 | | L:L:?1 | R:R:W378 | 16.25 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:N382 | R:R:Y381 | 8.14 | 1 | No | Yes | 8 | 8 | 2 | 1 | | R:R:V385 | R:R:Y381 | 6.31 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:Y381 | R:R:Y404 | 3.97 | 1 | Yes | Yes | 8 | 7 | 1 | 1 | | L:L:?1 | R:R:Y381 | 8.36 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:Y404 | 10.03 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:W405 | R:R:Y408 | 18.33 | 1 | Yes | Yes | 8 | 8 | 2 | 1 | | L:L:?1 | R:R:C407 | 4.53 | 1 | Yes | No | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:Y408 | 5.85 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:W400 | R:R:Y404 | 2.89 | 0 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:L156 | R:R:Q110 | 2.66 | 0 | No | Yes | 8 | 8 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 13.00 | | Average Number Of Links With An Hub | 8.00 | | Average Interaction Strength | 8.58 | | Average Nodes In Shell | 34.00 | | Average Hubs In Shell | 18.00 | | Average Links In Shell | 56.00 | | Average Links Mediated by Hubs In Shell | 54.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6OIK | A | Amine | Acetylcholine (muscarinic) | M2 | Homo Sapiens | Iperoxo | LY2119620 | chim(NtGi1-Go)/Beta1/Gamma2 | 3.6 | 2019-05-08 | doi.org/10.1126/science.aaw5188 |
|

A 2D representation of the interactions of IXO in 6OIK
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I72 | R:R:S76 | 7.74 | 2 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:D103 | R:R:I72 | 2.8 | 2 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:I72 | R:R:S107 | 4.64 | 2 | Yes | No | 8 | 8 | 2 | 1 | | R:R:I72 | R:R:Y430 | 2.42 | 2 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:D103 | R:R:S76 | 10.31 | 2 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:S76 | R:R:Y430 | 3.82 | 2 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:M77 | R:R:Y430 | 4.79 | 0 | No | Yes | 9 | 9 | 2 | 1 | | R:R:Y426 | R:R:Y80 | 15.89 | 2 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:Y430 | R:R:Y80 | 2.98 | 2 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:L100 | R:R:Y104 | 7.03 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:D103 | R:R:Y430 | 12.64 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:W155 | R:R:Y104 | 5.79 | 2 | Yes | Yes | 8 | 8 | 1 | 1 | | R:R:Y104 | R:R:Y403 | 9.93 | 2 | Yes | Yes | 8 | 8 | 1 | 1 | | R:R:Y104 | R:R:Y426 | 9.93 | 2 | Yes | Yes | 8 | 7 | 1 | 1 | | L:L:?1 | R:R:Y104 | 12.54 | 2 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:S107 | R:R:W400 | 4.94 | 2 | No | Yes | 8 | 9 | 1 | 1 | | L:L:?1 | R:R:S107 | 11.78 | 2 | Yes | No | 0 | 8 | 0 | 1 | | R:R:N108 | R:R:V111 | 2.96 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:N108 | R:R:S151 | 8.94 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:L154 | R:R:N108 | 4.12 | 2 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:N108 | R:R:W155 | 7.91 | 2 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:A194 | R:R:N108 | 3.13 | 2 | No | Yes | 8 | 9 | 1 | 1 | | L:L:?1 | R:R:N108 | 6.86 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:F396 | R:R:V111 | 2.62 | 2 | Yes | No | 9 | 9 | 2 | 2 | | R:R:L154 | R:R:W155 | 5.69 | 2 | Yes | Yes | 8 | 8 | 2 | 1 | | R:R:L154 | R:R:T190 | 4.42 | 2 | Yes | No | 8 | 9 | 2 | 2 | | R:R:T190 | R:R:W155 | 10.92 | 2 | No | Yes | 9 | 8 | 2 | 1 | | R:R:A194 | R:R:W155 | 3.89 | 2 | No | Yes | 8 | 8 | 1 | 1 | | L:L:?1 | R:R:W155 | 7.31 | 2 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:A194 | 5.62 | 2 | Yes | No | 0 | 8 | 0 | 1 | | R:R:F195 | R:R:Y196 | 5.16 | 2 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F195 | R:R:V199 | 3.93 | 2 | Yes | No | 9 | 7 | 1 | 2 | | R:R:F195 | R:R:F396 | 5.36 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:F195 | R:R:W400 | 3.01 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:A401 | R:R:F195 | 2.77 | 0 | No | Yes | 7 | 9 | 2 | 1 | | R:R:F195 | R:R:N404 | 15.71 | 2 | Yes | No | 9 | 7 | 1 | 1 | | L:L:?1 | R:R:F195 | 3.48 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:F396 | R:R:V199 | 3.93 | 2 | Yes | No | 9 | 7 | 2 | 2 | | R:R:F396 | R:R:W400 | 7.02 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:T399 | R:R:W400 | 4.85 | 2 | No | Yes | 9 | 9 | 2 | 1 | | R:R:N432 | R:R:T399 | 11.7 | 2 | Yes | No | 9 | 9 | 2 | 2 | | R:R:W400 | R:R:Y403 | 2.89 | 2 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:N432 | R:R:W400 | 10.17 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | | L:L:?1 | R:R:W400 | 30.87 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:N404 | R:R:Y403 | 9.3 | 2 | No | Yes | 7 | 8 | 1 | 1 | | R:R:Y403 | R:R:Y426 | 2.98 | 2 | Yes | Yes | 8 | 7 | 1 | 1 | | L:L:?1 | R:R:Y403 | 15.89 | 2 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:N404 | 3.92 | 2 | Yes | No | 0 | 7 | 0 | 1 | | R:R:Y426 | R:R:Y430 | 10.92 | 2 | Yes | Yes | 7 | 9 | 1 | 1 | | L:L:?1 | R:R:Y426 | 13.38 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:C429 | 5.66 | 2 | Yes | No | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:Y430 | 6.69 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 12.00 | | Average Number Of Links With An Hub | 8.00 | | Average Interaction Strength | 10.33 | | Average Nodes In Shell | 29.00 | | Average Hubs In Shell | 17.00 | | Average Links In Shell | 52.00 | | Average Links Mediated by Hubs In Shell | 52.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TRK | A | Amine | Acetylcholine (muscarinic) | M4 | Homo Sapiens | Iperoxo | - | Gi1/Beta1/Gamma2 | 2.8 | 2023-05-17 | doi.org/10.2139/ssrn.4034884 |
|

A 2D representation of the interactions of IXO in 7TRK
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I81 | R:R:S85 | 6.19 | 0 | Yes | No | 9 | 9 | 2 | 2 | | R:R:I81 | R:R:S116 | 4.64 | 0 | Yes | No | 9 | 8 | 2 | 1 | | R:R:D112 | R:R:S85 | 10.31 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:S85 | R:R:Y443 | 6.36 | 2 | No | Yes | 9 | 9 | 2 | 2 | | R:R:W108 | R:R:Y89 | 5.79 | 2 | Yes | Yes | 7 | 8 | 2 | 2 | | R:R:Y439 | R:R:Y89 | 22.84 | 2 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:Y443 | R:R:Y89 | 4.96 | 2 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:D112 | R:R:W108 | 5.58 | 2 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:D112 | R:R:Y443 | 10.34 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | L:L:?1 | R:R:D112 | 3.87 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:W164 | R:R:Y113 | 14.47 | 2 | Yes | Yes | 8 | 9 | 1 | 1 | | R:R:I168 | R:R:Y113 | 3.63 | 2 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:I187 | R:R:Y113 | 4.84 | 2 | Yes | Yes | 7 | 9 | 2 | 1 | | R:R:Y113 | R:R:Y416 | 7.94 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:Y113 | R:R:Y439 | 6.95 | 2 | Yes | Yes | 9 | 7 | 1 | 1 | | L:L:?1 | R:R:Y113 | 10.87 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:S116 | 9.64 | 2 | Yes | No | 0 | 8 | 0 | 1 | | R:R:M121 | R:R:N117 | 4.21 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:N117 | R:R:S160 | 4.47 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:N117 | R:R:W164 | 9.04 | 2 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:A203 | R:R:N117 | 3.13 | 2 | No | Yes | 9 | 9 | 1 | 1 | | L:L:?1 | R:R:N117 | 10.77 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:F409 | R:R:V120 | 7.87 | 2 | Yes | No | 9 | 9 | 2 | 1 | | R:R:V120 | R:R:W413 | 6.13 | 2 | No | Yes | 9 | 9 | 1 | 1 | | L:L:?1 | R:R:V120 | 3.19 | 2 | Yes | No | 0 | 9 | 0 | 1 | | R:R:L163 | R:R:W164 | 5.69 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:I168 | R:R:W164 | 3.52 | 2 | Yes | Yes | 8 | 8 | 2 | 1 | | R:R:T199 | R:R:W164 | 8.49 | 0 | No | Yes | 9 | 8 | 2 | 1 | | R:R:A203 | R:R:W164 | 3.89 | 2 | No | Yes | 9 | 8 | 1 | 1 | | L:L:?1 | R:R:W164 | 10.56 | 2 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:I168 | R:R:I187 | 4.42 | 2 | Yes | Yes | 8 | 7 | 2 | 2 | | L:L:?1 | R:R:A203 | 5.62 | 2 | Yes | No | 0 | 9 | 0 | 1 | | R:R:F204 | R:R:Y205 | 11.35 | 2 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:F204 | R:R:F409 | 5.36 | 2 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:F204 | R:R:N417 | 18.12 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:N417 | R:R:Y205 | 3.49 | 2 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F409 | R:R:W413 | 13.03 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:T412 | R:R:W413 | 3.64 | 2 | No | Yes | 9 | 9 | 2 | 1 | | R:R:N445 | R:R:T412 | 13.16 | 2 | Yes | No | 9 | 9 | 2 | 2 | | R:R:C442 | R:R:W413 | 7.84 | 2 | No | Yes | 9 | 9 | 1 | 1 | | R:R:N445 | R:R:W413 | 13.56 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | | L:L:?1 | R:R:W413 | 16.25 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:N417 | R:R:Y416 | 6.98 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:V420 | R:R:Y416 | 7.57 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:C442 | R:R:Y416 | 4.03 | 2 | No | Yes | 9 | 9 | 1 | 1 | | L:L:?1 | R:R:Y416 | 7.53 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:N417 | 3.92 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:W435 | R:R:Y439 | 5.79 | 0 | No | Yes | 6 | 7 | 2 | 1 | | R:R:Y439 | R:R:Y443 | 7.94 | 2 | Yes | Yes | 7 | 9 | 1 | 2 | | L:L:?1 | R:R:Y439 | 7.53 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:C442 | 6.79 | 2 | Yes | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 12.00 | | Average Number Of Links With An Hub | 8.00 | | Average Interaction Strength | 8.04 | | Average Nodes In Shell | 31.00 | | Average Hubs In Shell | 19.00 | | Average Links In Shell | 51.00 | | Average Links Mediated by Hubs In Shell | 51.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TRP | A | Amine | Acetylcholine (muscarinic) | M4 | Homo Sapiens | Iperoxo | LY2033298 | Gi1/Beta1/Gamma2 | 2.4 | 2023-05-17 | doi.org/10.2139/ssrn.4034884 |
|

A 2D representation of the interactions of IXO in 7TRP
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:D112 | 8.71 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y113 | 15.05 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:S116 | 11.78 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:N117 | 8.81 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:W164 | 8.12 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:A203 | 4.5 | 1 | Yes | No | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:W413 | 12.19 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y416 | 7.53 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:N417 | 5.88 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y439 | 3.34 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:C442 | 9.06 | 1 | Yes | No | 0 | 9 | 0 | 1 | | R:R:I81 | R:R:S85 | 6.19 | 0 | Yes | No | 9 | 9 | 2 | 2 | | R:R:I81 | R:R:S116 | 3.1 | 0 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:D112 | R:R:S85 | 11.78 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:S85 | R:R:Y443 | 3.82 | 1 | No | Yes | 9 | 9 | 2 | 2 | | R:R:W108 | R:R:Y89 | 3.86 | 1 | Yes | Yes | 7 | 8 | 2 | 2 | | R:R:Y439 | R:R:Y89 | 22.84 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:Y443 | R:R:Y89 | 5.96 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:Y89 | W:W:?1 | 7.84 | 1 | Yes | Yes | 8 | 0 | 2 | 2 | | R:R:D112 | R:R:W108 | 4.47 | 1 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:D112 | R:R:S116 | 4.42 | 1 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:D112 | R:R:Y443 | 8.05 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:W164 | R:R:Y113 | 14.47 | 1 | Yes | Yes | 8 | 9 | 1 | 1 | | R:R:I187 | R:R:Y113 | 8.46 | 1 | Yes | Yes | 7 | 9 | 2 | 1 | | R:R:Y113 | R:R:Y416 | 10.92 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:Y113 | R:R:Y439 | 3.97 | 1 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:S116 | R:R:W413 | 3.71 | 1 | Yes | Yes | 8 | 9 | 1 | 1 | | R:R:M121 | R:R:N117 | 5.61 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:N117 | R:R:S160 | 4.47 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:L163 | R:R:N117 | 4.12 | 1 | No | Yes | 8 | 9 | 2 | 1 | | R:R:N117 | R:R:W164 | 9.04 | 1 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:A203 | R:R:N117 | 3.13 | 1 | No | Yes | 9 | 9 | 1 | 1 | | R:R:F409 | R:R:V120 | 5.24 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:V120 | R:R:W413 | 3.68 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:L163 | R:R:W164 | 10.25 | 1 | No | Yes | 8 | 8 | 2 | 1 | | R:R:F189 | R:R:W164 | 3.01 | 1 | Yes | Yes | 8 | 8 | 2 | 1 | | R:R:T199 | R:R:W164 | 7.28 | 0 | No | Yes | 9 | 8 | 2 | 1 | | R:R:A203 | R:R:W164 | 3.89 | 1 | No | Yes | 9 | 8 | 1 | 1 | | R:R:F189 | R:R:I187 | 7.54 | 1 | Yes | Yes | 8 | 7 | 2 | 2 | | R:R:F204 | R:R:Y205 | 10.32 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:F204 | R:R:F409 | 5.36 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:F204 | R:R:W413 | 3.01 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:F204 | R:R:N417 | 16.92 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:N417 | R:R:Y205 | 4.65 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F409 | R:R:W413 | 8.02 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:T412 | R:R:W413 | 4.85 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:N445 | R:R:T412 | 14.62 | 1 | No | No | 9 | 9 | 2 | 2 | | R:R:C442 | R:R:W413 | 6.53 | 1 | No | Yes | 9 | 9 | 1 | 1 | | R:R:N445 | R:R:W413 | 14.69 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:N417 | R:R:Y416 | 3.49 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:V420 | R:R:Y416 | 8.83 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:W435 | R:R:Y416 | 4.82 | 1 | Yes | Yes | 6 | 9 | 2 | 1 | | R:R:C442 | R:R:Y416 | 4.03 | 1 | No | Yes | 9 | 9 | 1 | 1 | | R:R:W435 | W:W:?1 | 33.83 | 1 | Yes | Yes | 6 | 0 | 2 | 2 | | R:R:Y439 | R:R:Y443 | 4.96 | 1 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:Y439 | W:W:?1 | 4.35 | 1 | Yes | Yes | 7 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 11.00 | | Average Number Of Links With An Hub | 9.00 | | Average Interaction Strength | 8.63 | | Average Nodes In Shell | 32.00 | | Average Hubs In Shell | 21.00 | | Average Links In Shell | 56.00 | | Average Links Mediated by Hubs In Shell | 55.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TRQ | A | Amine | Acetylcholine (muscarinic) | M4 | Homo Sapiens | Iperoxo | VU0467154 | Gi1/Beta1/Gamma2 | 2.5 | 2023-05-17 | doi.org/10.2139/ssrn.4034884 |
|
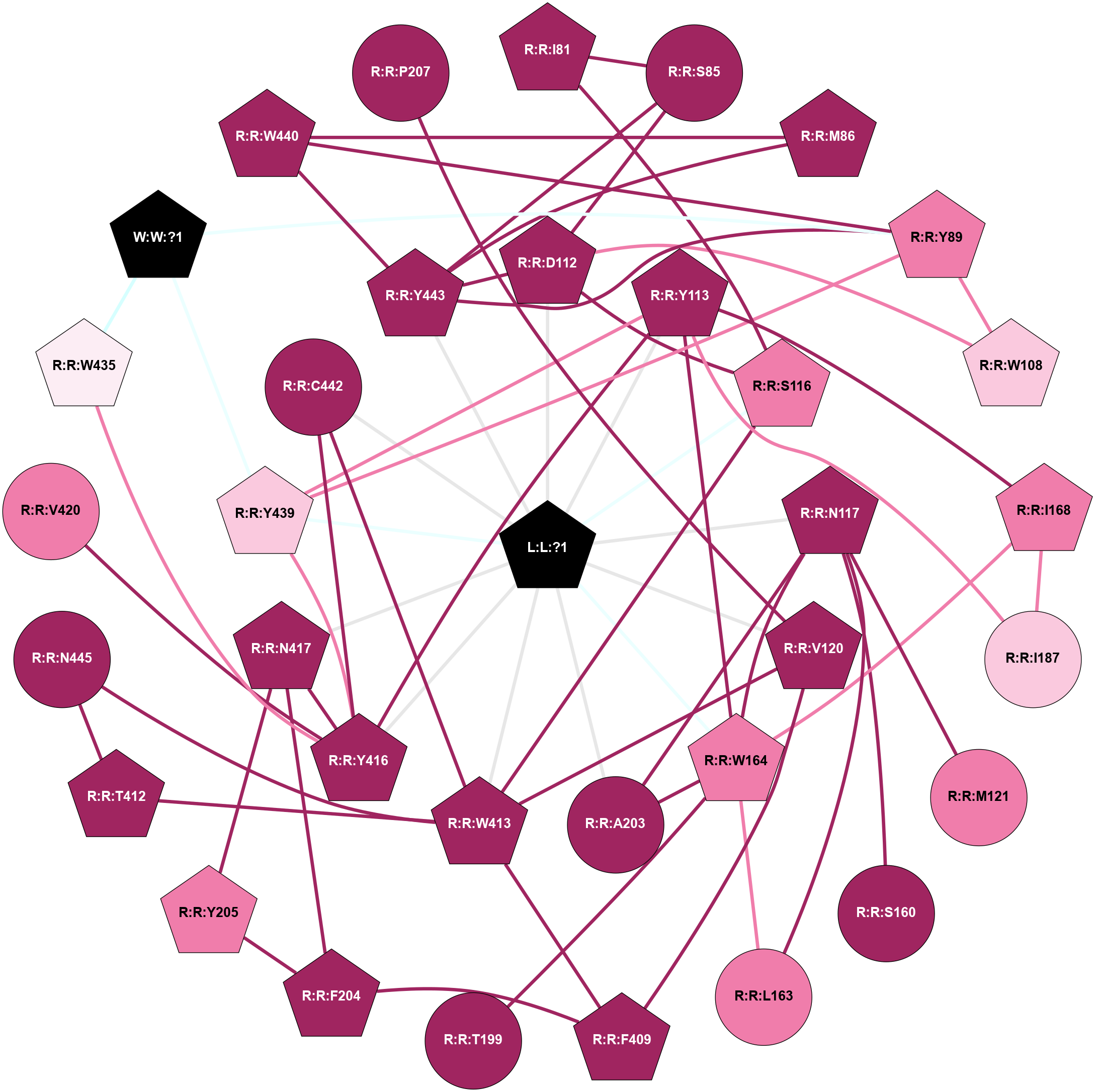
A 2D representation of the interactions of IXO in 7TRQ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:D112 | 9.68 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y113 | 10.87 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:S116 | 8.57 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:N117 | 6.86 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:V120 | 5.31 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:W164 | 4.87 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:A203 | 6.74 | 1 | Yes | No | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:W413 | 19.5 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y416 | 7.53 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:N417 | 6.86 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y439 | 10.87 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:C442 | 5.66 | 1 | Yes | No | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y443 | 4.18 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:I81 | R:R:S85 | 4.64 | 0 | Yes | No | 9 | 9 | 2 | 2 | | R:R:I81 | R:R:S116 | 3.1 | 0 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:D112 | R:R:S85 | 11.78 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:S85 | R:R:Y443 | 3.82 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:M86 | R:R:W440 | 4.65 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:M86 | R:R:Y443 | 7.18 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:W108 | R:R:Y89 | 3.86 | 1 | Yes | Yes | 7 | 8 | 2 | 2 | | R:R:Y439 | R:R:Y89 | 11.91 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:W440 | R:R:Y89 | 8.68 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:Y443 | R:R:Y89 | 4.96 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:Y89 | W:W:?1 | 4.73 | 1 | Yes | Yes | 8 | 0 | 2 | 2 | | R:R:D112 | R:R:W108 | 4.47 | 1 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:D112 | R:R:S116 | 4.42 | 1 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:D112 | R:R:Y443 | 10.34 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:W164 | R:R:Y113 | 14.47 | 1 | Yes | Yes | 8 | 9 | 1 | 1 | | R:R:I168 | R:R:Y113 | 4.84 | 1 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:I187 | R:R:Y113 | 12.09 | 1 | Yes | Yes | 7 | 9 | 2 | 1 | | R:R:Y113 | R:R:Y416 | 7.94 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:Y113 | R:R:Y439 | 5.96 | 1 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:S116 | R:R:W413 | 4.94 | 1 | Yes | Yes | 8 | 9 | 1 | 1 | | R:R:M121 | R:R:N117 | 5.61 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:N117 | R:R:S160 | 4.47 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:L163 | R:R:N117 | 4.12 | 1 | No | Yes | 8 | 9 | 2 | 1 | | R:R:N117 | R:R:W164 | 9.04 | 1 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:A203 | R:R:N117 | 3.13 | 1 | No | Yes | 9 | 9 | 1 | 1 | | R:R:F409 | R:R:V120 | 6.55 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:V120 | R:R:W413 | 6.13 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:L163 | R:R:W164 | 5.69 | 1 | No | Yes | 8 | 8 | 2 | 1 | | R:R:I168 | R:R:W164 | 3.52 | 1 | Yes | Yes | 8 | 8 | 2 | 1 | | R:R:F189 | R:R:W164 | 3.01 | 1 | Yes | Yes | 8 | 8 | 2 | 1 | | R:R:T199 | R:R:W164 | 8.49 | 0 | No | Yes | 9 | 8 | 2 | 1 | | R:R:A203 | R:R:W164 | 3.89 | 1 | No | Yes | 9 | 8 | 1 | 1 | | R:R:I168 | R:R:I187 | 5.89 | 1 | Yes | Yes | 8 | 7 | 2 | 2 | | R:R:F189 | R:R:I168 | 3.77 | 1 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:F189 | R:R:I187 | 7.54 | 1 | Yes | Yes | 8 | 7 | 2 | 2 | | R:R:F204 | R:R:Y205 | 11.35 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:F204 | R:R:F409 | 5.36 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:F204 | R:R:W413 | 3.01 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:F204 | R:R:N417 | 18.12 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:N417 | R:R:Y205 | 4.65 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F409 | R:R:W413 | 8.02 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:T412 | R:R:W413 | 4.85 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:N445 | R:R:T412 | 13.16 | 1 | No | Yes | 9 | 9 | 2 | 2 | | R:R:C442 | R:R:W413 | 9.14 | 1 | No | Yes | 9 | 9 | 1 | 1 | | R:R:N445 | R:R:W413 | 10.17 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:N417 | R:R:Y416 | 4.65 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:V420 | R:R:Y416 | 6.31 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:W435 | R:R:Y416 | 3.86 | 1 | Yes | Yes | 6 | 9 | 2 | 1 | | R:R:Y416 | R:R:Y439 | 6.95 | 1 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:C442 | R:R:Y416 | 4.03 | 1 | No | Yes | 9 | 9 | 1 | 1 | | R:R:W435 | R:R:Y439 | 2.89 | 1 | Yes | Yes | 6 | 7 | 2 | 1 | | R:R:W435 | W:W:?1 | 29.85 | 1 | Yes | Yes | 6 | 0 | 2 | 2 | | R:R:Y439 | W:W:?1 | 6.3 | 1 | Yes | Yes | 7 | 0 | 1 | 2 | | R:R:W440 | R:R:Y443 | 6.75 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:P207 | R:R:V120 | 1.77 | 0 | No | Yes | 9 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 13.00 | | Average Number Of Links With An Hub | 11.00 | | Average Interaction Strength | 8.27 | | Average Nodes In Shell | 36.00 | | Average Hubs In Shell | 26.00 | | Average Links In Shell | 68.00 | | Average Links Mediated by Hubs In Shell | 68.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7V68 | A | Amine | Acetylcholine (muscarinic) | M4 | Homo Sapiens | Iperoxo | LY2119620 | Gi1/Beta1/Gamma2 | 3.4 | 2022-05-11 | doi.org/10.1038/s41467-022-30595-y |
|
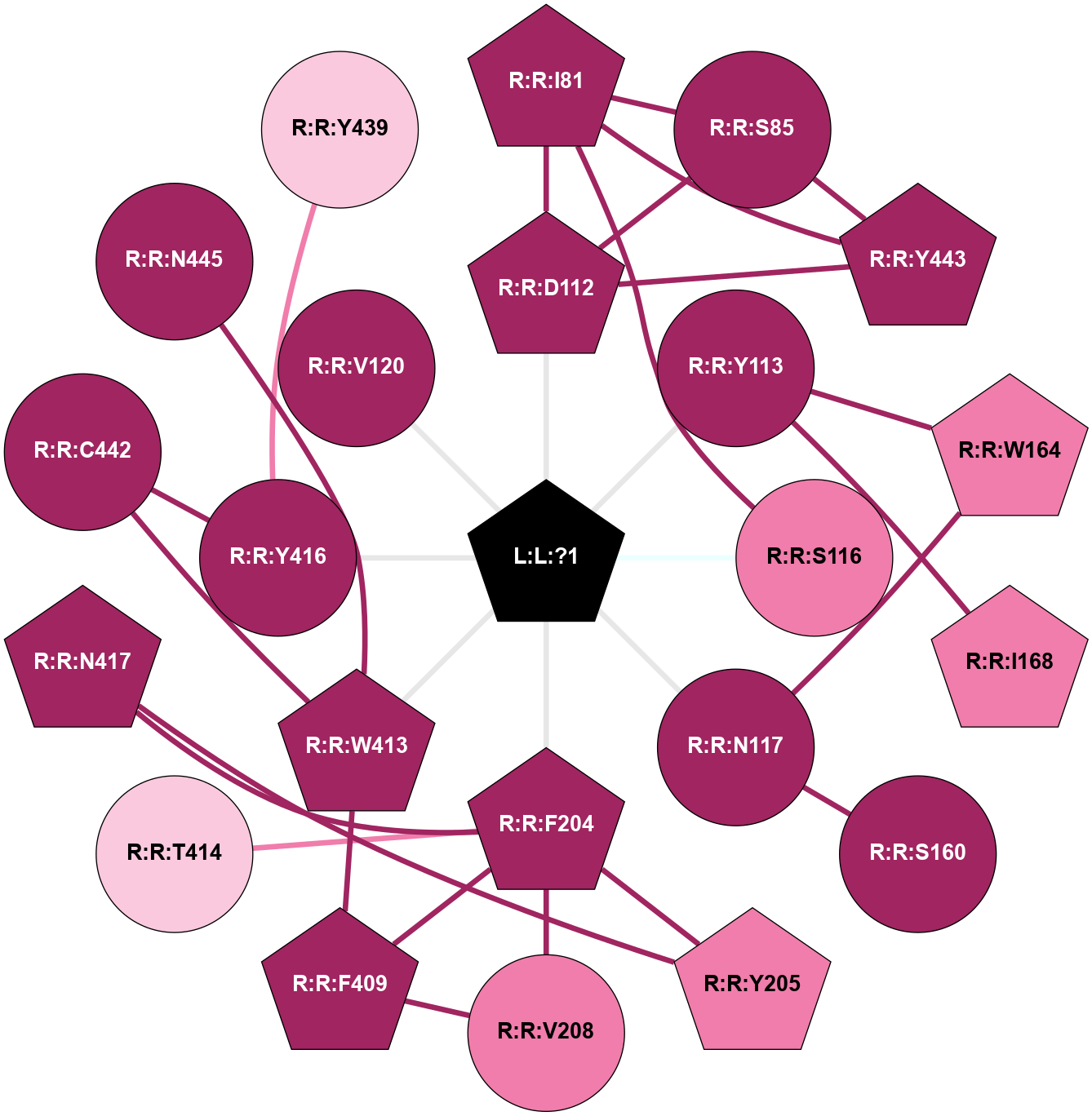
A 2D representation of the interactions of IXO in 7V68
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I81 | R:R:S85 | 4.64 | 21 | Yes | No | 9 | 9 | 2 | 2 | | R:R:D112 | R:R:I81 | 5.6 | 21 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:I81 | R:R:S116 | 4.64 | 21 | Yes | No | 9 | 8 | 2 | 1 | | R:R:I81 | R:R:Y443 | 4.84 | 21 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:D112 | R:R:S85 | 10.31 | 21 | Yes | No | 9 | 9 | 1 | 2 | | R:R:S85 | R:R:Y443 | 10.17 | 21 | No | Yes | 9 | 9 | 2 | 2 | | R:R:D112 | R:R:Y443 | 11.49 | 21 | Yes | Yes | 9 | 9 | 1 | 2 | | L:L:?1 | R:R:D112 | 3.87 | 0 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:W164 | R:R:Y113 | 29.9 | 0 | Yes | No | 8 | 9 | 2 | 1 | | R:R:I168 | R:R:Y113 | 6.04 | 4 | Yes | No | 8 | 9 | 2 | 1 | | L:L:?1 | R:R:Y113 | 15.89 | 0 | Yes | No | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:S116 | 6.43 | 0 | Yes | No | 0 | 8 | 0 | 1 | | R:R:N117 | R:R:S160 | 5.96 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:N117 | R:R:W164 | 9.04 | 0 | No | Yes | 9 | 8 | 1 | 2 | | L:L:?1 | R:R:N117 | 5.88 | 0 | Yes | No | 0 | 9 | 0 | 1 | | R:R:F204 | R:R:Y205 | 9.28 | 17 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F204 | R:R:V208 | 7.87 | 17 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F204 | R:R:F409 | 4.29 | 17 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:F204 | R:R:T414 | 5.19 | 17 | Yes | No | 9 | 7 | 1 | 2 | | R:R:F204 | R:R:N417 | 13.29 | 17 | Yes | Yes | 9 | 9 | 1 | 2 | | L:L:?1 | R:R:F204 | 6.08 | 0 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:N417 | R:R:Y205 | 6.98 | 17 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:F409 | R:R:V208 | 3.93 | 17 | Yes | No | 9 | 8 | 2 | 2 | | R:R:F409 | R:R:W413 | 9.02 | 17 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:C442 | R:R:W413 | 10.45 | 0 | No | Yes | 9 | 9 | 2 | 1 | | R:R:N445 | R:R:W413 | 18.08 | 0 | No | Yes | 9 | 9 | 2 | 1 | | L:L:?1 | R:R:W413 | 7.31 | 0 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:Y416 | R:R:Y439 | 6.95 | 0 | No | No | 9 | 7 | 1 | 2 | | R:R:C442 | R:R:Y416 | 4.03 | 0 | No | No | 9 | 9 | 2 | 1 | | L:L:?1 | R:R:Y416 | 10.87 | 0 | Yes | No | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:V120 | 3.19 | 0 | Yes | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 8.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 7.44 | | Average Nodes In Shell | 23.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 31.00 | | Average Links Mediated by Hubs In Shell | 28.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8E9Z | A | Amine | Acetylcholine (muscarinic) | M3 | Homo Sapiens | Iperoxo | - | chim(NtGi2L-Gs-CtGq)/Beta1/Gamma2 | 2.69 | 2022-11-30 | doi.org/10.1038/s41586-022-05489-0 |
|
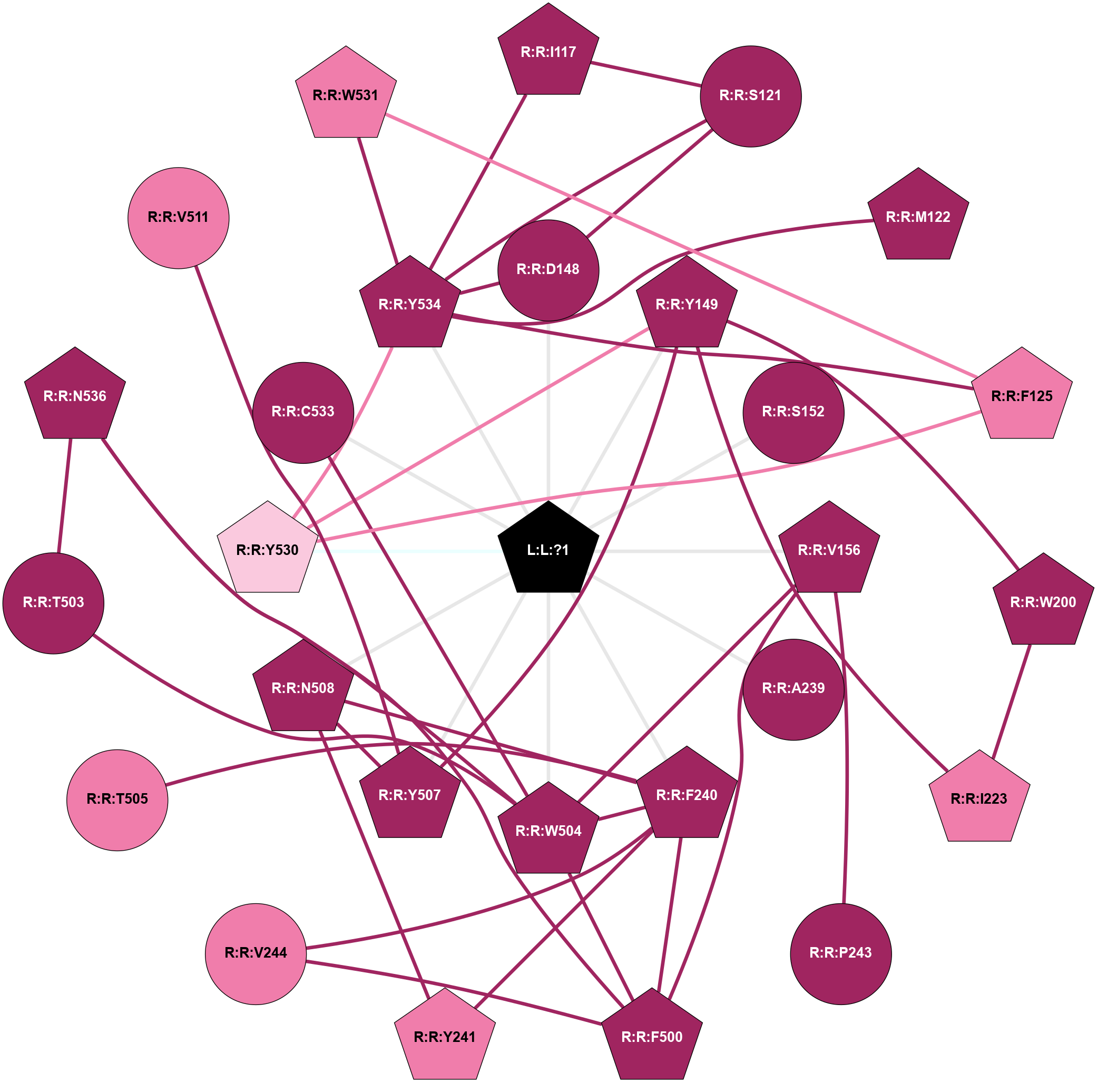
A 2D representation of the interactions of IXO in 8E9Z
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I117 | R:R:S121 | 7.74 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:I117 | R:R:Y534 | 4.84 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:D148 | R:R:S121 | 10.31 | 1 | No | No | 9 | 9 | 1 | 2 | | R:R:S121 | R:R:Y534 | 5.09 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:M122 | R:R:Y534 | 8.38 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:F125 | R:R:Y530 | 11.35 | 1 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:F125 | R:R:W531 | 4.01 | 1 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:F125 | R:R:Y534 | 4.13 | 1 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:D148 | R:R:Y534 | 10.34 | 1 | No | Yes | 9 | 9 | 1 | 1 | | L:L:?1 | R:R:D148 | 6.78 | 1 | Yes | No | 0 | 9 | 0 | 1 | | R:R:W200 | R:R:Y149 | 20.26 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:I223 | R:R:Y149 | 8.46 | 1 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:Y149 | R:R:Y507 | 3.97 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:Y149 | R:R:Y530 | 3.97 | 1 | Yes | Yes | 9 | 7 | 1 | 1 | | L:L:?1 | R:R:Y149 | 6.69 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:S152 | 7.5 | 1 | Yes | No | 0 | 9 | 0 | 1 | | R:R:P243 | R:R:V156 | 3.53 | 0 | No | Yes | 9 | 9 | 2 | 1 | | R:R:F500 | R:R:V156 | 7.87 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:V156 | R:R:W504 | 4.9 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | L:L:?1 | R:R:V156 | 4.25 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:I223 | R:R:W200 | 5.87 | 1 | Yes | Yes | 8 | 9 | 2 | 2 | | L:L:?1 | R:R:A239 | 5.62 | 1 | Yes | No | 0 | 9 | 0 | 1 | | R:R:F240 | R:R:Y241 | 6.19 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F240 | R:R:V244 | 3.93 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F240 | R:R:F500 | 4.29 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:F240 | R:R:W504 | 5.01 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:F240 | R:R:T505 | 3.89 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F240 | R:R:N508 | 16.92 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | L:L:?1 | R:R:F240 | 5.21 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:N508 | R:R:Y241 | 6.98 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F500 | R:R:V244 | 3.93 | 1 | Yes | No | 9 | 8 | 2 | 2 | | R:R:F500 | R:R:W504 | 5.01 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:F500 | R:R:N536 | 3.62 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:T503 | R:R:W504 | 4.85 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:N536 | R:R:T503 | 4.39 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:C533 | R:R:W504 | 10.45 | 1 | No | Yes | 9 | 9 | 1 | 1 | | R:R:N536 | R:R:W504 | 9.04 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | L:L:?1 | R:R:W504 | 18.69 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:N508 | R:R:Y507 | 4.65 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:V511 | R:R:Y507 | 3.79 | 0 | No | Yes | 8 | 9 | 2 | 1 | | L:L:?1 | R:R:Y507 | 7.53 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:N508 | 4.9 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:Y530 | R:R:Y534 | 6.95 | 1 | Yes | Yes | 7 | 9 | 1 | 1 | | L:L:?1 | R:R:Y530 | 14.22 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:W531 | R:R:Y534 | 4.82 | 1 | Yes | Yes | 8 | 9 | 2 | 1 | | L:L:?1 | R:R:C533 | 10.19 | 1 | Yes | No | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y534 | 5.02 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 12.00 | | Average Number Of Links With An Hub | 8.00 | | Average Interaction Strength | 8.05 | | Average Nodes In Shell | 28.00 | | Average Hubs In Shell | 18.00 | | Average Links In Shell | 47.00 | | Average Links Mediated by Hubs In Shell | 46.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8EA0 | A | Amine | Acetylcholine (muscarinic) | M3 | Homo Sapiens | Iperoxo | - | - | 2.56 | 2022-11-30 | doi.org/10.1038/s41586-022-05489-0 |
|
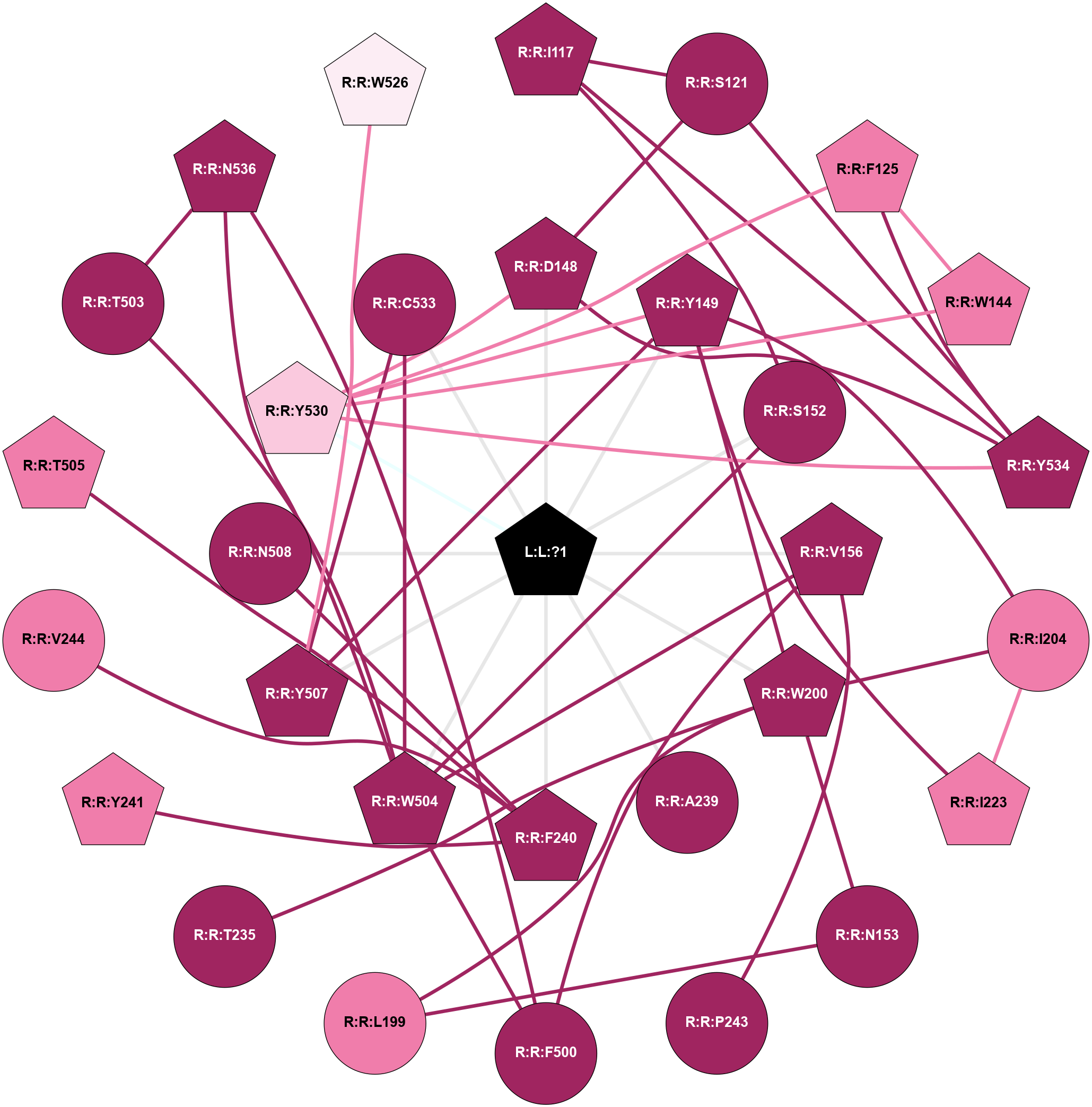
A 2D representation of the interactions of IXO in 8EA0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:D148 | 4.84 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y149 | 13.38 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:S152 | 10.71 | 1 | Yes | No | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:V156 | 4.25 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:W200 | 6.5 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:A239 | 6.74 | 1 | Yes | No | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:F240 | 3.48 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:W504 | 21.12 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y507 | 8.36 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:N508 | 6.86 | 1 | Yes | No | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y530 | 11.71 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:C533 | 6.79 | 1 | Yes | No | 0 | 9 | 0 | 1 | | L:L:?1 | R:R:Y534 | 3.34 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:I117 | R:R:S121 | 7.74 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:I117 | R:R:S152 | 4.64 | 1 | Yes | No | 9 | 9 | 2 | 1 | | R:R:I117 | R:R:Y534 | 4.84 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:D148 | R:R:S121 | 11.78 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:S121 | R:R:Y534 | 5.09 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:M122 | R:R:W531 | 8.14 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:M122 | R:R:Y534 | 7.18 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:F125 | R:R:W144 | 5.01 | 1 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:F125 | R:R:Y530 | 11.35 | 1 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:F125 | R:R:W531 | 6.01 | 1 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:F125 | R:R:Y534 | 5.16 | 1 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:W144 | R:R:Y530 | 3.86 | 1 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:D148 | R:R:Y530 | 3.45 | 1 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:D148 | R:R:Y534 | 11.49 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:W200 | R:R:Y149 | 15.43 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:I204 | R:R:Y149 | 4.84 | 1 | No | Yes | 8 | 9 | 2 | 1 | | R:R:I223 | R:R:Y149 | 13.3 | 1 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:Y149 | R:R:Y507 | 5.96 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:Y149 | R:R:Y530 | 5.96 | 1 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:S152 | R:R:W504 | 3.71 | 1 | No | Yes | 9 | 9 | 1 | 1 | | R:R:L199 | R:R:N153 | 4.12 | 1 | No | No | 8 | 9 | 2 | 2 | | R:R:N153 | R:R:W200 | 9.04 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:P243 | R:R:V156 | 3.53 | 0 | No | Yes | 9 | 9 | 2 | 1 | | R:R:F500 | R:R:V156 | 7.87 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:V156 | R:R:W504 | 4.9 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:L199 | R:R:W200 | 7.97 | 1 | No | Yes | 8 | 9 | 2 | 1 | | R:R:I204 | R:R:W200 | 4.7 | 1 | No | Yes | 8 | 9 | 2 | 1 | | R:R:T235 | R:R:W200 | 8.49 | 0 | No | Yes | 9 | 9 | 2 | 1 | | R:R:I204 | R:R:I223 | 4.42 | 1 | No | Yes | 8 | 8 | 2 | 2 | | R:R:F240 | R:R:Y241 | 9.28 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F240 | R:R:V244 | 7.87 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F240 | R:R:T505 | 3.89 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F240 | R:R:N508 | 21.75 | 1 | Yes | No | 9 | 9 | 1 | 1 | | R:R:F500 | R:R:W504 | 11.02 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:F500 | R:R:N536 | 4.83 | 1 | No | Yes | 9 | 9 | 2 | 2 | | R:R:T503 | R:R:W504 | 3.64 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:N536 | R:R:T503 | 5.85 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:C533 | R:R:W504 | 11.75 | 1 | No | Yes | 9 | 9 | 1 | 1 | | R:R:N536 | R:R:W504 | 9.04 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:W526 | R:R:Y507 | 3.86 | 0 | Yes | Yes | 6 | 9 | 2 | 1 | | R:R:C533 | R:R:Y507 | 4.03 | 1 | No | Yes | 9 | 9 | 1 | 1 | | R:R:Y530 | R:R:Y534 | 7.94 | 1 | Yes | Yes | 7 | 9 | 1 | 1 | | R:R:W531 | R:R:Y534 | 5.79 | 1 | Yes | Yes | 8 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 13.00 | | Average Number Of Links With An Hub | 9.00 | | Average Interaction Strength | 8.31 | | Average Nodes In Shell | 33.00 | | Average Hubs In Shell | 20.00 | | Average Links In Shell | 56.00 | | Average Links Mediated by Hubs In Shell | 55.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|









