| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5XJM | A | Peptide | Angiotensin | AT2 | Homo Sapiens | (Sar1,Ile8)-Angiotensin II | - | - | 3.2 | 2018-07-11 | doi.org/10.1038/s41594-018-0079-8 |
|
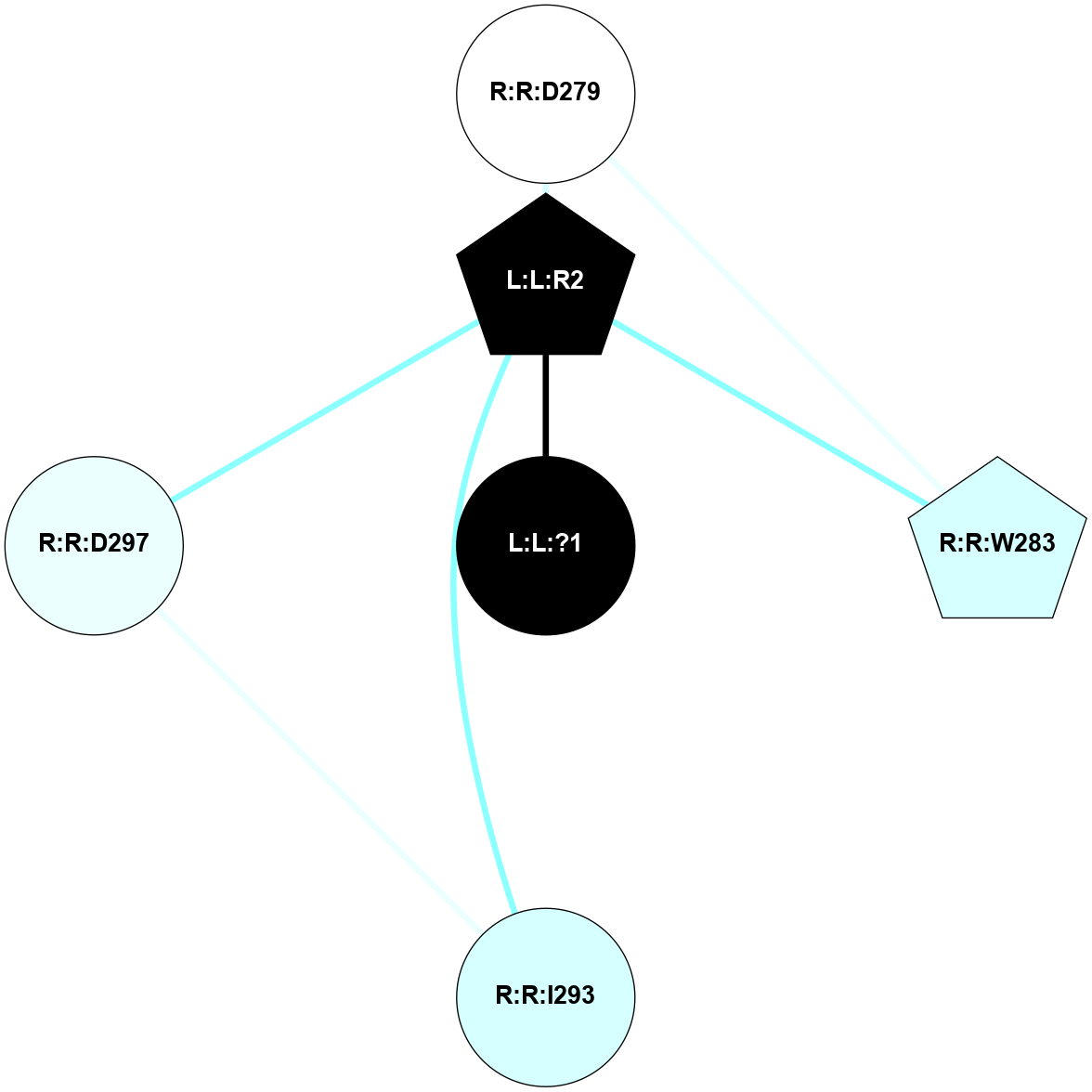
A 2D representation of the interactions of SAR in 5XJM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D279 | R:R:W283 | 7.82 | 5 | No | Yes | 5 | 3 | 2 | 2 | | L:L:R2 | R:R:D279 | 5.96 | 5 | Yes | No | 0 | 5 | 1 | 2 | | L:L:R2 | R:R:W283 | 15.99 | 5 | Yes | Yes | 0 | 3 | 1 | 2 | | R:R:D297 | R:R:I293 | 4.2 | 5 | No | No | 4 | 3 | 2 | 2 | | L:L:R2 | R:R:I293 | 6.26 | 5 | Yes | No | 0 | 3 | 1 | 2 | | L:L:R2 | R:R:D297 | 5.96 | 5 | Yes | No | 0 | 4 | 1 | 2 | | L:L:?1 | L:L:R2 | 5.02 | 0 | No | Yes | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.02 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6DO1 | A | Peptide | Angiotensin | AT1 | Homo Sapiens | Angiotensin-like peptide S1I8 | - | - | 2.9 | 2019-01-30 | doi.org/10.1016/j.cell.2018.12.006 |
|
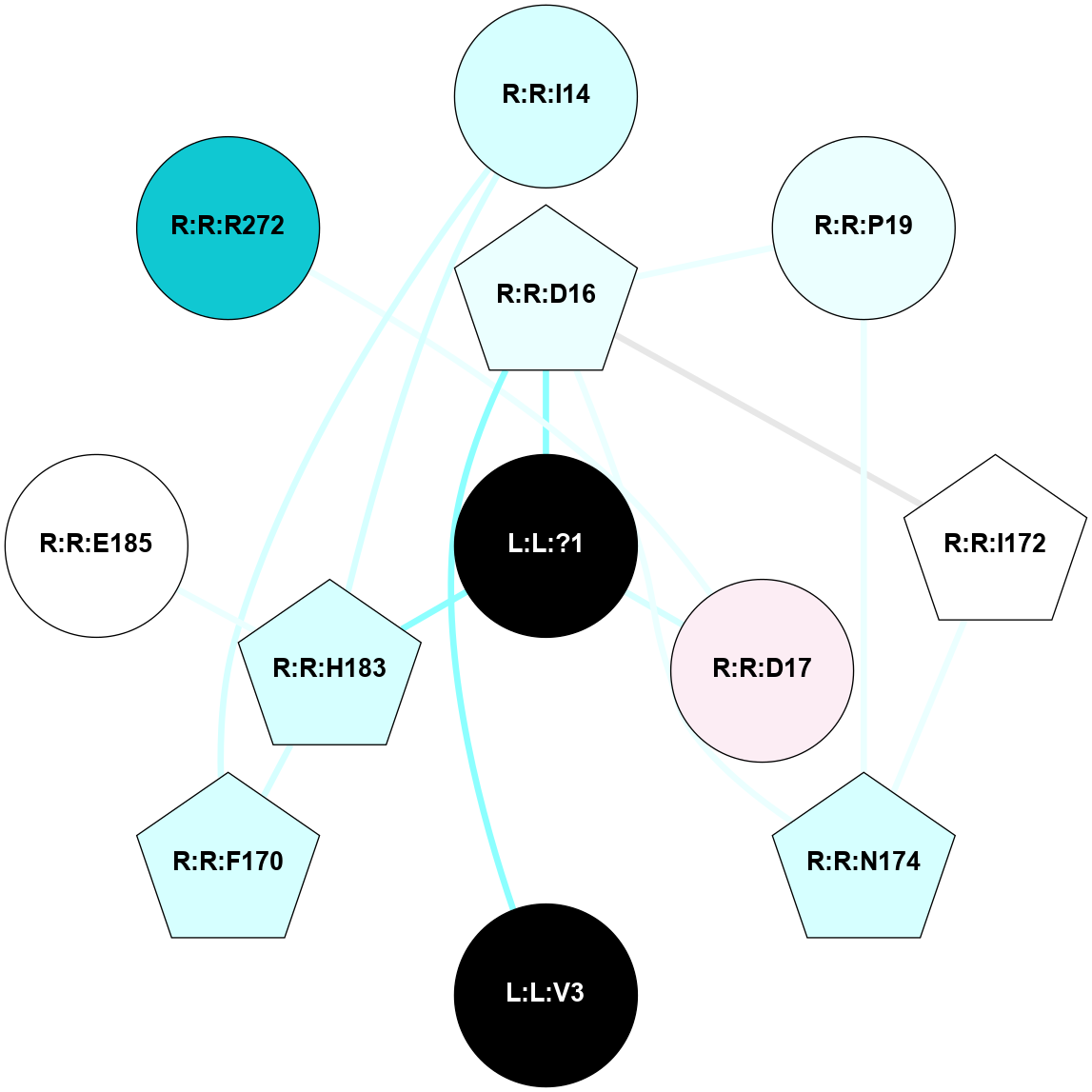
A 2D representation of the interactions of SAR in 6DO1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:D16 | 5.61 | 0 | No | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:D17 | 23.85 | 0 | No | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:H183 | 6.64 | 0 | No | No | 0 | 3 | 0 | 1 | | L:L:V3 | R:R:D16 | 11.69 | 0 | No | Yes | 0 | 4 | 2 | 1 | | R:R:F170 | R:R:I14 | 8.79 | 5 | Yes | No | 3 | 3 | 2 | 2 | | R:R:H183 | R:R:I14 | 18.56 | 5 | No | No | 3 | 3 | 1 | 2 | | R:R:D16 | R:R:P19 | 8.05 | 3 | Yes | No | 4 | 4 | 1 | 2 | | R:R:D16 | R:R:I172 | 4.2 | 3 | Yes | Yes | 4 | 5 | 1 | 2 | | R:R:D16 | R:R:N174 | 4.04 | 3 | Yes | Yes | 4 | 3 | 1 | 2 | | R:R:N174 | R:R:P19 | 3.26 | 3 | Yes | No | 3 | 4 | 2 | 2 | | R:R:F170 | R:R:H183 | 3.39 | 5 | Yes | No | 3 | 3 | 2 | 1 | | R:R:I172 | R:R:N174 | 4.25 | 3 | Yes | Yes | 5 | 3 | 2 | 2 | | R:R:D17 | R:R:R272 | 2.38 | 0 | No | No | 6 | 1 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 12.03 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6OS1 | A | Peptide | Angiotensin | AT1 | Homo Sapiens | TRV026 | - | - | 2.79 | 2020-02-19 | doi.org/10.1126/science.aay9813 |
|
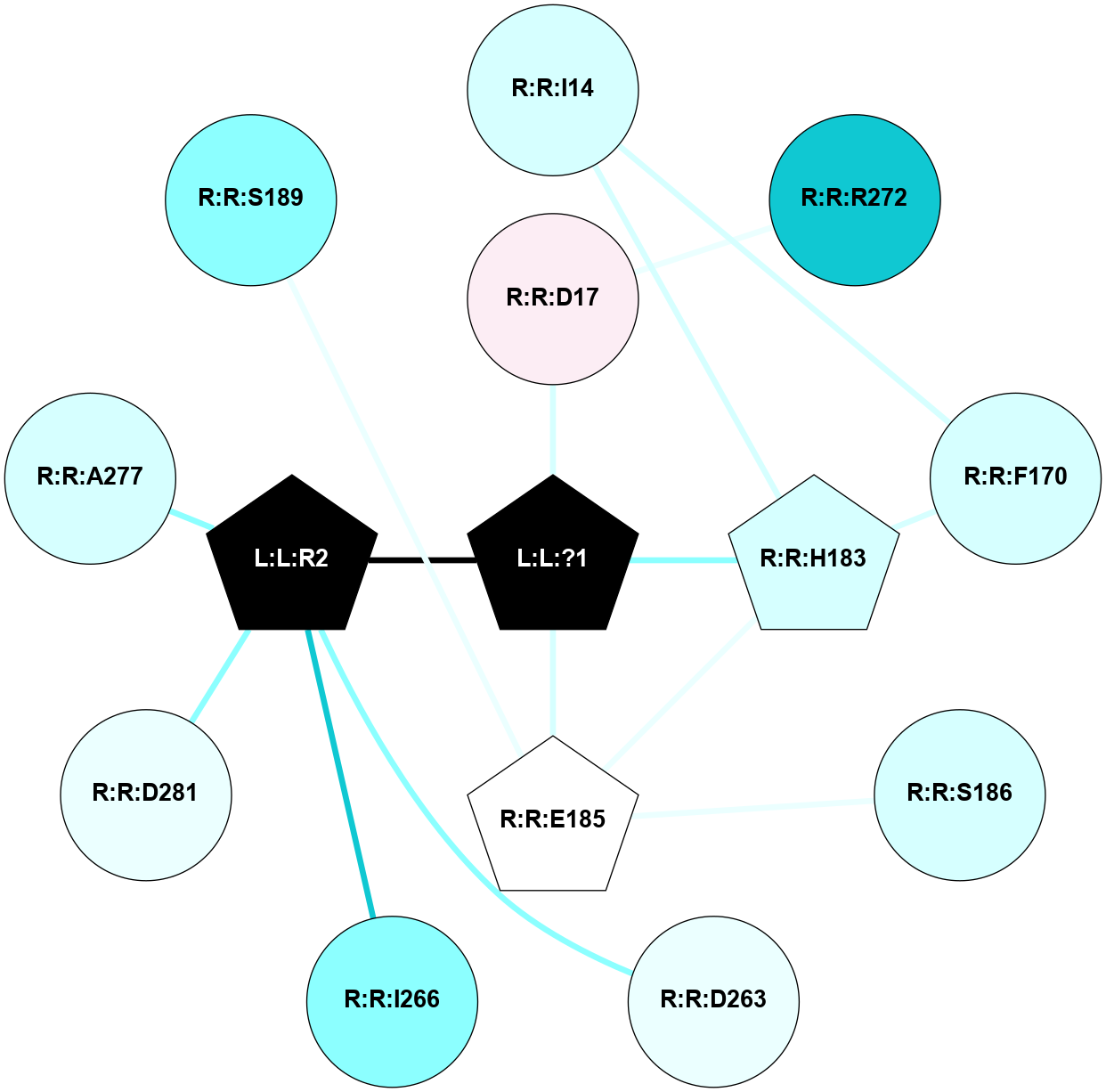
A 2D representation of the interactions of SAR in 6OS1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:D17 | 23.85 | 0 | No | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:H183 | 13.29 | 0 | No | Yes | 0 | 3 | 0 | 1 | | R:R:F170 | R:R:I14 | 13.82 | 5 | No | No | 3 | 3 | 2 | 2 | | R:R:H183 | R:R:I14 | 15.91 | 5 | Yes | No | 3 | 3 | 1 | 2 | | R:R:F170 | R:R:H183 | 4.53 | 5 | No | Yes | 3 | 3 | 2 | 1 | | R:R:E185 | R:R:H183 | 11.08 | 0 | No | Yes | 5 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 18.57 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6OS2 | A | Peptide | Angiotensin | AT1 | Homo Sapiens | TRV026 | - | - | 2.7 | 2020-02-19 | doi.org/10.1126/science.aay9813 |
|
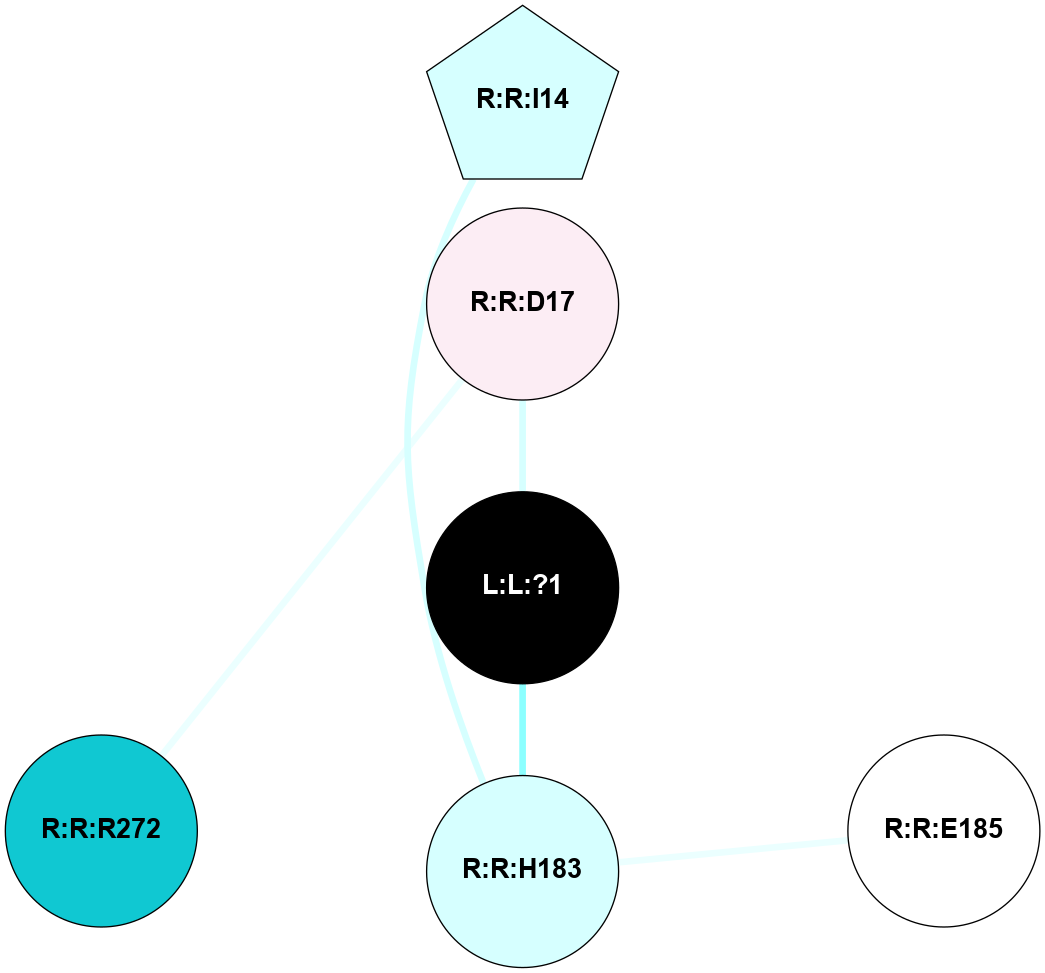
A 2D representation of the interactions of SAR in 6OS2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:D17 | 22.44 | 0 | No | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:H183 | 14.62 | 0 | No | No | 0 | 3 | 0 | 1 | | R:R:H183 | R:R:I14 | 18.56 | 0 | No | Yes | 3 | 3 | 1 | 2 | | R:R:E185 | R:R:H183 | 4.92 | 0 | No | No | 5 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.53 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6PT2 | A | Peptide | Opioid | DOP | Homo Sapiens | KGCHM07 | - | - | 2.8 | 2019-12-11 | doi.org/10.1126/sciadv.aax9115 |
|
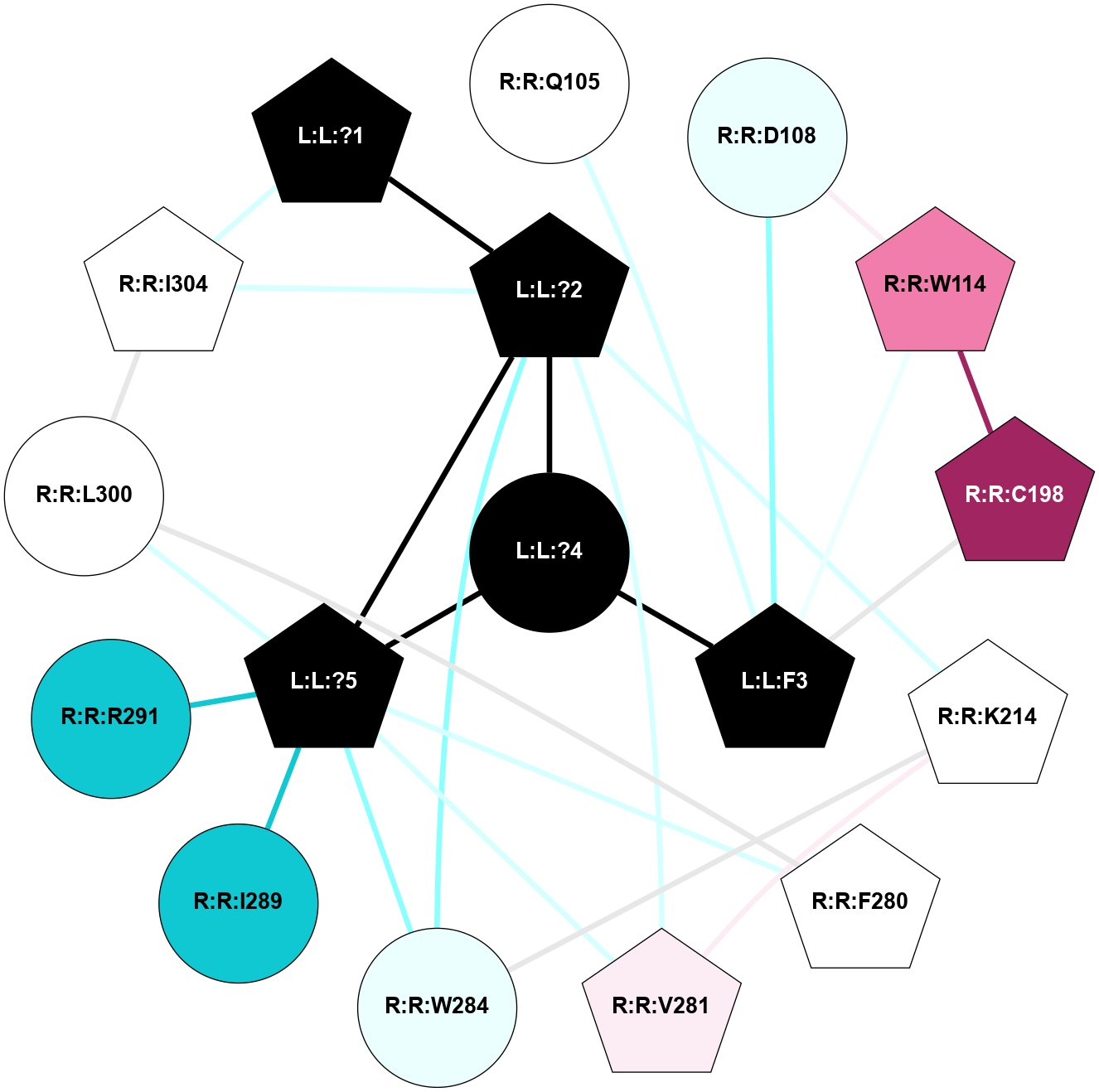
A 2D representation of the interactions of SAR in 6PT2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | L:L:?2 | 23 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?1 | R:R:I304 | 3.3 | 1 | Yes | Yes | 0 | 5 | 2 | 2 | | L:L:?2 | L:L:?4 | 17.11 | 1 | Yes | No | 0 | 0 | 1 | 0 | | L:L:?2 | L:L:?5 | 12.45 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?2 | R:R:K214 | 3.37 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:?2 | R:R:V281 | 3.56 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?2 | R:R:W284 | 3.63 | 1 | Yes | No | 0 | 4 | 1 | 2 | | L:L:?2 | R:R:I304 | 3.41 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:?4 | L:L:F3 | 10.07 | 1 | No | Yes | 0 | 0 | 0 | 1 | | L:L:F3 | R:R:Q105 | 15.23 | 1 | Yes | No | 0 | 5 | 1 | 2 | | L:L:F3 | R:R:D108 | 9.55 | 1 | Yes | No | 0 | 4 | 1 | 2 | | L:L:F3 | R:R:W114 | 4.01 | 1 | Yes | Yes | 0 | 8 | 1 | 2 | | L:L:F3 | R:R:C198 | 4.19 | 1 | Yes | Yes | 0 | 9 | 1 | 2 | | L:L:?4 | L:L:?5 | 15.19 | 1 | No | Yes | 0 | 0 | 0 | 1 | | L:L:?5 | R:R:F280 | 8.89 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:?5 | R:R:V281 | 6.92 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?5 | R:R:W284 | 49.14 | 1 | Yes | No | 0 | 4 | 1 | 2 | | L:L:?5 | R:R:I289 | 5.68 | 1 | Yes | No | 0 | 1 | 1 | 2 | | L:L:?5 | R:R:R291 | 10.48 | 1 | Yes | No | 0 | 1 | 1 | 2 | | L:L:?5 | R:R:L300 | 16.54 | 1 | Yes | No | 0 | 5 | 1 | 2 | | R:R:D108 | R:R:W114 | 8.93 | 1 | No | Yes | 4 | 8 | 2 | 2 | | R:R:C198 | R:R:W114 | 11.75 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:K214 | R:R:V281 | 4.55 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:K214 | R:R:W284 | 3.48 | 1 | Yes | No | 5 | 4 | 2 | 2 | | R:R:F280 | R:R:L300 | 7.31 | 1 | Yes | No | 5 | 5 | 2 | 2 | | R:R:I304 | R:R:L300 | 5.71 | 1 | Yes | No | 5 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 14.12 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 26.00 | | Average Links Mediated by Hubs In Shell | 26.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7C6A | A | Peptide | Angiotensin | AT2 | Homo Sapiens | (Sar1,Ile8)-Angiotensin II | - | - | 3.4 | 2020-07-29 | doi.org/10.1038/s41598-020-68355-x |
|
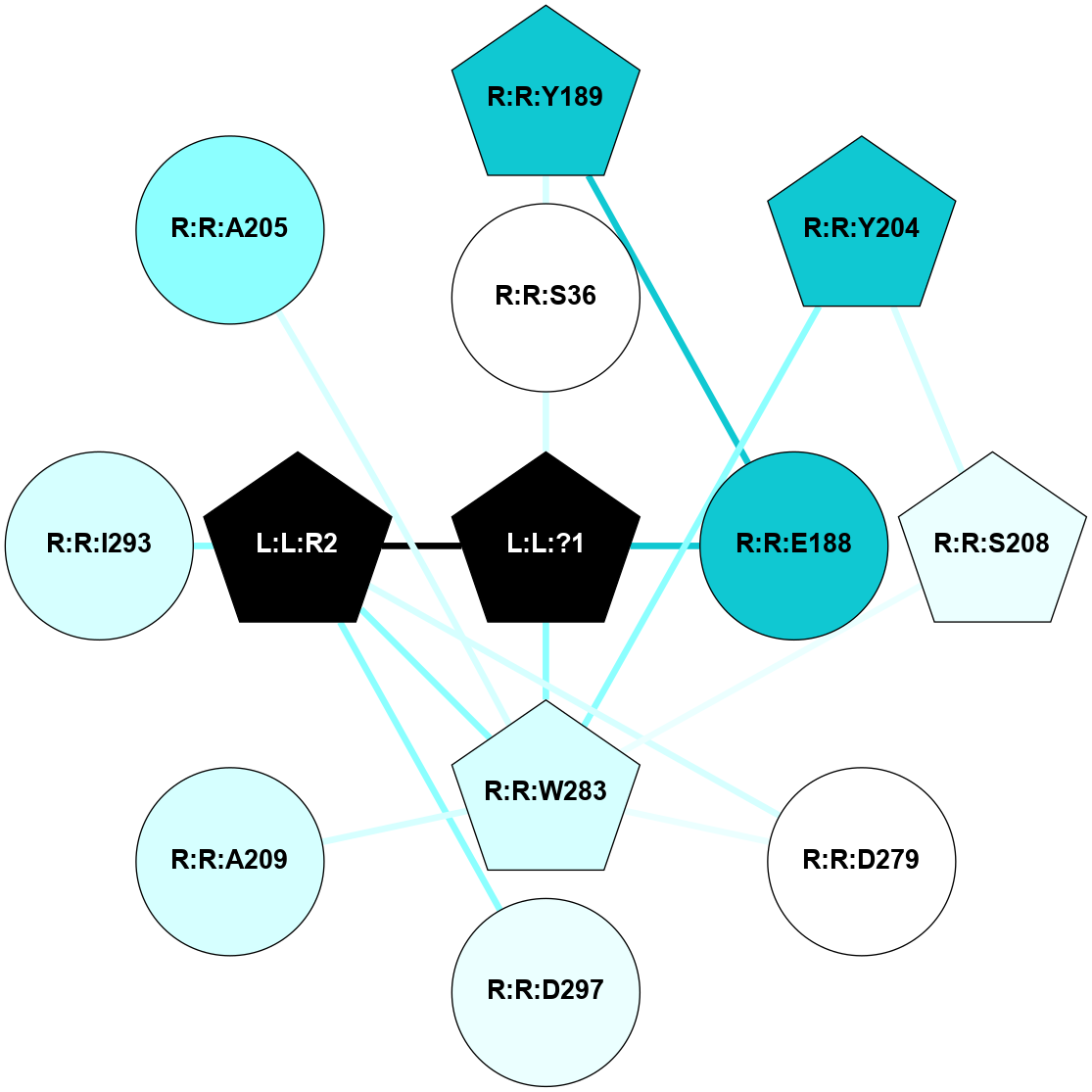
A 2D representation of the interactions of SAR in 7C6A
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:S36 | R:R:Y189 | 7.63 | 0 | No | Yes | 5 | 1 | 1 | 2 | | L:L:?1 | R:R:S36 | 4.66 | 2 | Yes | No | 0 | 5 | 0 | 1 | | R:R:E188 | R:R:Y189 | 5.61 | 0 | No | Yes | 1 | 1 | 1 | 2 | | L:L:?1 | R:R:E188 | 5.48 | 2 | Yes | No | 0 | 1 | 0 | 1 | | R:R:S208 | R:R:Y204 | 5.09 | 2 | Yes | Yes | 4 | 1 | 2 | 2 | | R:R:W283 | R:R:Y204 | 5.79 | 2 | Yes | Yes | 3 | 1 | 1 | 2 | | R:R:S208 | R:R:W283 | 14.83 | 2 | Yes | Yes | 4 | 3 | 2 | 1 | | R:R:D279 | R:R:W283 | 7.82 | 2 | No | Yes | 5 | 3 | 2 | 1 | | L:L:R2 | R:R:D279 | 4.76 | 2 | Yes | No | 0 | 5 | 1 | 2 | | L:L:?1 | R:R:W283 | 3.53 | 2 | Yes | Yes | 0 | 3 | 0 | 1 | | L:L:R2 | R:R:W283 | 8 | 2 | Yes | Yes | 0 | 3 | 1 | 1 | | L:L:R2 | R:R:D297 | 11.91 | 2 | Yes | No | 0 | 4 | 1 | 2 | | L:L:?1 | L:L:R2 | 3.77 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | R:R:A209 | R:R:W283 | 2.59 | 0 | No | Yes | 3 | 3 | 2 | 1 | | L:L:R2 | R:R:I293 | 2.51 | 2 | Yes | No | 0 | 3 | 1 | 2 | | R:R:A205 | R:R:W283 | 1.3 | 0 | No | Yes | 2 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 4.36 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7F6G | A | Peptide | Angiotensin | AT1 | Homo Sapiens | SAR1-AngII | - | Gq/Beta1/Gamma2 | 2.9 | 2023-03-29 | doi.org/10.15252/embj.2022112940 |
|
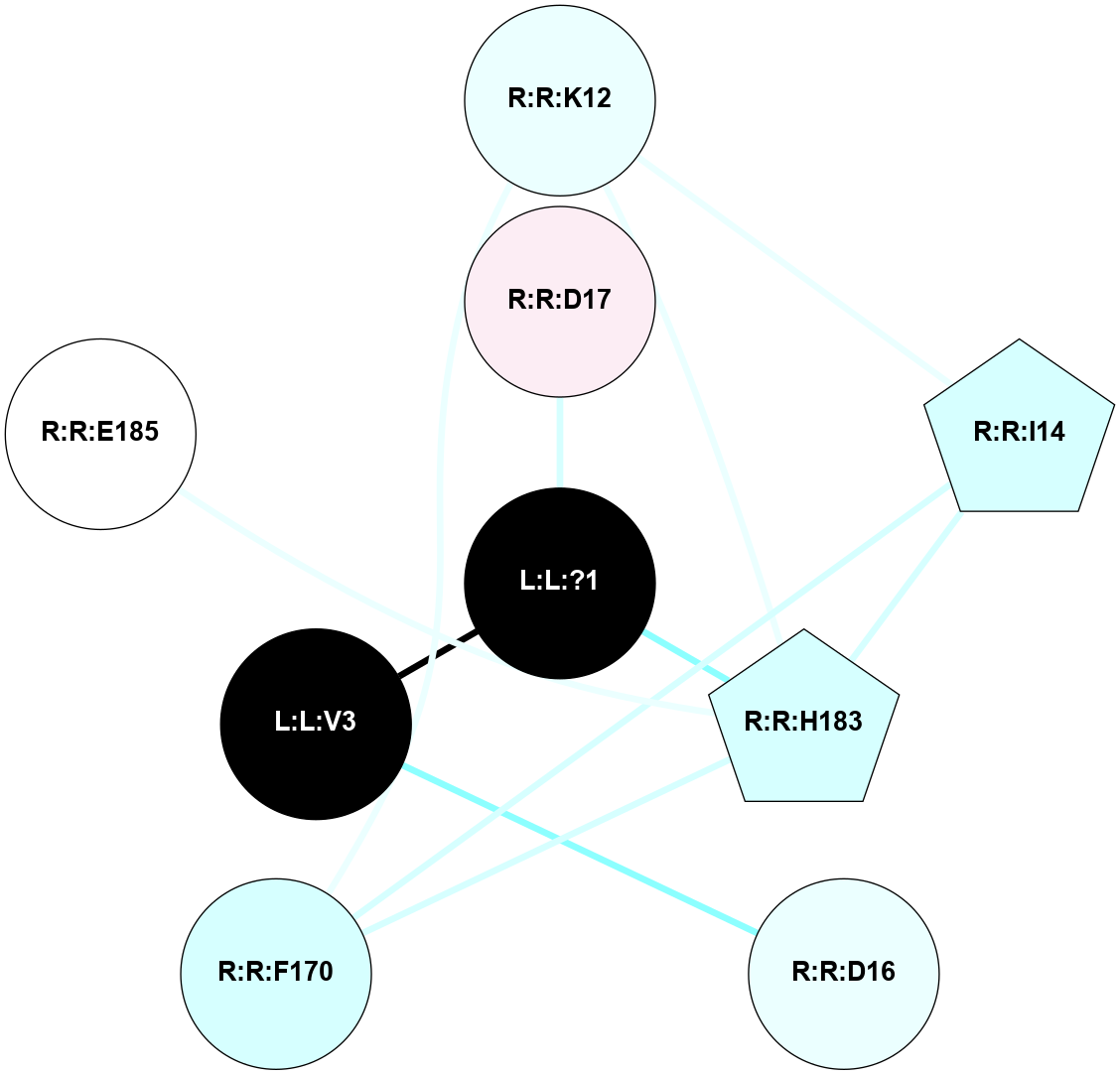
A 2D representation of the interactions of SAR in 7F6G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I14 | R:R:K12 | 5.82 | 19 | Yes | No | 3 | 4 | 2 | 2 | | R:R:F170 | R:R:K12 | 6.2 | 19 | No | No | 3 | 4 | 2 | 2 | | R:R:H183 | R:R:K12 | 2.62 | 19 | Yes | No | 3 | 4 | 1 | 2 | | R:R:F170 | R:R:I14 | 10.05 | 19 | No | Yes | 3 | 3 | 2 | 2 | | R:R:H183 | R:R:I14 | 13.26 | 19 | Yes | Yes | 3 | 3 | 1 | 2 | | L:L:V3 | R:R:D16 | 5.84 | 0 | No | No | 0 | 4 | 1 | 2 | | L:L:?1 | R:R:D17 | 4.21 | 0 | No | No | 0 | 6 | 0 | 1 | | R:R:F170 | R:R:H183 | 3.39 | 19 | No | Yes | 3 | 3 | 2 | 1 | | R:R:E185 | R:R:H183 | 4.92 | 0 | No | Yes | 5 | 3 | 2 | 1 | | L:L:?1 | R:R:H183 | 14.62 | 0 | No | Yes | 0 | 3 | 0 | 1 | | L:L:?1 | L:L:V3 | 4.62 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 7.82 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|






