| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5TVN | A | Amine | 5-Hydroxytryptamine | 5-HT2B | Homo Sapiens | LSD | - | - | 2.9 | 2017-02-01 | doi.org/10.1016/j.cell.2016.12.033 |
|

A 2D representation of the interactions of 7LD in 5TVN
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:W131 | 3.8 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:D135 | 22.64 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:V136 | 13.67 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:S139 | 6.26 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:L209 | 3.46 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:F217 | 9.14 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:S222 | 3.76 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:F340 | 24.38 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:F341 | 8.13 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:N344 | 5.73 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:V366 | 6.21 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:D135 | R:R:V107 | 5.84 | 1 | Yes | No | 8 | 7 | 1 | 2 | | R:R:V107 | R:R:Y370 | 3.79 | 1 | No | Yes | 7 | 8 | 2 | 2 | | R:R:I110 | R:R:W131 | 12.92 | 0 | No | Yes | 5 | 6 | 2 | 1 | | R:R:T114 | R:R:W131 | 3.64 | 0 | No | Yes | 5 | 6 | 2 | 1 | | R:R:W121 | R:R:W131 | 6.56 | 1 | Yes | Yes | 7 | 6 | 2 | 1 | | R:R:C207 | R:R:W121 | 9.14 | 1 | No | Yes | 9 | 7 | 2 | 2 | | R:R:L132 | R:R:W131 | 3.42 | 0 | No | Yes | 5 | 6 | 2 | 1 | | R:R:C207 | R:R:W131 | 3.92 | 1 | No | Yes | 9 | 6 | 2 | 1 | | R:R:D135 | R:R:S139 | 4.42 | 1 | Yes | No | 8 | 7 | 1 | 1 | | R:R:D135 | R:R:Y370 | 5.75 | 1 | Yes | Yes | 8 | 8 | 1 | 2 | | R:R:A187 | R:R:V136 | 3.39 | 0 | No | No | 8 | 7 | 2 | 1 | | R:R:F217 | R:R:V136 | 3.93 | 1 | Yes | No | 7 | 7 | 1 | 1 | | R:R:F217 | R:R:P189 | 10.11 | 1 | Yes | No | 7 | 8 | 1 | 2 | | R:R:F217 | R:R:V190 | 5.24 | 1 | Yes | No | 7 | 6 | 1 | 2 | | R:R:F214 | R:R:L209 | 9.74 | 1 | Yes | No | 5 | 5 | 2 | 1 | | R:R:F214 | R:R:F217 | 4.29 | 1 | Yes | Yes | 5 | 7 | 2 | 1 | | R:R:M218 | R:R:N344 | 8.41 | 0 | No | Yes | 6 | 6 | 2 | 1 | | R:R:F341 | R:R:S222 | 6.61 | 1 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:N344 | R:R:S222 | 4.47 | 1 | Yes | Yes | 6 | 7 | 1 | 1 | | R:R:I345 | R:R:S222 | 4.64 | 0 | No | Yes | 6 | 7 | 2 | 1 | | R:R:F226 | R:R:F341 | 19.29 | 0 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:F340 | R:R:W337 | 7.02 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:F341 | R:R:W337 | 6.01 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:F340 | R:R:F341 | 13.93 | 1 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:F340 | R:R:N344 | 4.83 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | R:R:L362 | R:R:N344 | 5.49 | 0 | No | Yes | 5 | 6 | 2 | 1 | | R:R:V366 | R:R:W367 | 3.68 | 1 | No | Yes | 6 | 7 | 1 | 2 | | R:R:V366 | R:R:Y370 | 10.09 | 1 | No | Yes | 6 | 8 | 1 | 2 | | R:R:W367 | R:R:Y370 | 7.72 | 1 | Yes | Yes | 7 | 8 | 2 | 2 | | L:L:?1 | R:R:G221 | 1.43 | 1 | Yes | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 12.00 | | Average Number Of Links With An Hub | 7.00 | | Average Interaction Strength | 9.05 | | Average Nodes In Shell | 30.00 | | Average Hubs In Shell | 14.00 | | Average Links In Shell | 41.00 | | Average Links Mediated by Hubs In Shell | 40.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6WGT | A | Amine | 5-Hydroxytryptamine | 5-HT2A | Homo Sapiens | LSD | - | - | 3.4 | 2020-09-23 | doi.org/10.1016/j.cell.2020.08.024 |
|

A 2D representation of the interactions of 7LD in 6WGT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:W151 | 4.75 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:D155 | 24.91 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:V156 | 18.64 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:S159 | 10.02 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:L229 | 5.77 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:G238 | 4.28 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:S239 | 8.77 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:S242 | 5.01 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:F339 | 14.22 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:F340 | 10.16 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:N343 | 5.73 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:V366 | 3.73 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:V127 | R:R:W151 | 3.68 | 1 | Yes | Yes | 8 | 5 | 2 | 1 | | R:R:D155 | R:R:V127 | 5.84 | 1 | Yes | Yes | 8 | 8 | 1 | 2 | | R:R:V127 | R:R:Y370 | 3.79 | 1 | Yes | Yes | 8 | 7 | 2 | 2 | | R:R:V130 | R:R:W151 | 14.71 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:W141 | R:R:W151 | 10.31 | 2 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:I152 | R:R:I210 | 4.42 | 0 | No | Yes | 5 | 6 | 2 | 2 | | R:R:I152 | R:R:L229 | 7.14 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:D155 | R:R:V156 | 4.38 | 1 | Yes | Yes | 8 | 6 | 1 | 1 | | R:R:D155 | R:R:S159 | 4.42 | 1 | Yes | No | 8 | 7 | 1 | 1 | | R:R:D155 | R:R:Y370 | 4.6 | 1 | Yes | Yes | 8 | 7 | 1 | 2 | | R:R:S207 | R:R:V156 | 3.23 | 0 | No | Yes | 7 | 6 | 2 | 1 | | R:R:I210 | R:R:V156 | 4.61 | 1 | Yes | Yes | 6 | 6 | 2 | 1 | | R:R:S242 | R:R:T160 | 7.99 | 0 | No | No | 6 | 7 | 1 | 2 | | R:R:F234 | R:R:I210 | 7.54 | 0 | Yes | Yes | 7 | 6 | 2 | 2 | | R:R:K223 | R:R:L228 | 5.64 | 0 | No | No | 3 | 4 | 2 | 1 | | R:R:L228 | R:R:S226 | 4.5 | 0 | No | No | 4 | 4 | 1 | 2 | | R:R:F234 | R:R:L229 | 6.09 | 0 | Yes | No | 7 | 4 | 2 | 1 | | R:R:F234 | R:R:G238 | 4.52 | 0 | Yes | No | 7 | 6 | 2 | 1 | | R:R:N343 | R:R:V235 | 5.91 | 1 | Yes | No | 7 | 6 | 1 | 2 | | R:R:G238 | R:R:I237 | 3.53 | 0 | No | No | 6 | 4 | 1 | 2 | | R:R:F340 | R:R:S239 | 7.93 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:I344 | R:R:S239 | 3.1 | 0 | No | No | 5 | 6 | 2 | 1 | | R:R:F243 | R:R:F340 | 21.43 | 0 | No | Yes | 8 | 7 | 2 | 1 | | R:R:F339 | R:R:W336 | 7.02 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:F340 | R:R:W336 | 9.02 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:W336 | R:R:Y370 | 3.86 | 1 | Yes | Yes | 8 | 7 | 2 | 2 | | R:R:F339 | R:R:F340 | 12.86 | 1 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:F339 | R:R:N343 | 13.29 | 1 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:F339 | R:R:L362 | 3.65 | 1 | Yes | No | 7 | 3 | 1 | 2 | | R:R:F339 | R:R:V366 | 6.55 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | R:R:L362 | R:R:N343 | 4.12 | 1 | No | Yes | 3 | 7 | 2 | 1 | | R:R:V366 | R:R:W367 | 3.68 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:V366 | R:R:Y370 | 8.83 | 1 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:W367 | R:R:Y370 | 12.54 | 1 | Yes | Yes | 6 | 7 | 2 | 2 | | L:L:?1 | R:R:L228 | 2.31 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:S242 | R:R:V241 | 1.62 | 0 | No | No | 6 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 13.00 | | Average Number Of Links With An Hub | 7.00 | | Average Interaction Strength | 9.10 | | Average Nodes In Shell | 33.00 | | Average Hubs In Shell | 15.00 | | Average Links In Shell | 48.00 | | Average Links Mediated by Hubs In Shell | 41.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7SRQ | A | Amine | 5-Hydroxytryptamine | 5-HT2B | Homo Sapiens | LSD | - | - | 2.7 | 2022-09-21 | doi.org/10.1016/j.neuron.2022.08.006 |
|

A 2D representation of the interactions of 7LD in 7SRQ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:D135 | 15.85 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:V136 | 6.21 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:T140 | 3.69 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:A225 | 2.63 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:F340 | 21.33 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:N344 | 3.44 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:V366 | 11.18 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V107 | R:R:W131 | 4.9 | 1 | No | Yes | 7 | 6 | 2 | 2 | | R:R:D135 | R:R:V107 | 5.84 | 1 | Yes | No | 8 | 7 | 1 | 2 | | R:R:V107 | R:R:Y370 | 3.79 | 1 | No | Yes | 7 | 8 | 2 | 2 | | R:R:D135 | R:R:W131 | 3.35 | 1 | Yes | Yes | 8 | 6 | 1 | 2 | | R:R:D135 | R:R:Y370 | 6.9 | 1 | Yes | Yes | 8 | 8 | 1 | 2 | | R:R:A187 | R:R:V136 | 3.39 | 6 | No | No | 8 | 7 | 2 | 1 | | R:R:A225 | R:R:T140 | 3.36 | 1 | No | No | 7 | 7 | 1 | 1 | | R:R:M218 | R:R:N344 | 7.01 | 0 | No | No | 6 | 6 | 2 | 1 | | R:R:F341 | R:R:S222 | 3.96 | 0 | Yes | No | 7 | 7 | 2 | 2 | | R:R:N344 | R:R:S222 | 4.47 | 0 | No | No | 6 | 7 | 1 | 2 | | R:R:F340 | R:R:F341 | 9.65 | 1 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:F340 | R:R:L362 | 3.65 | 1 | Yes | No | 7 | 5 | 1 | 2 | | R:R:F340 | R:R:V366 | 3.93 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:V366 | R:R:Y370 | 5.05 | 1 | No | Yes | 6 | 8 | 1 | 2 | | L:L:?1 | R:R:S139 | 2.5 | 1 | Yes | No | 0 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 8.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 8.35 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 22.00 | | Average Links Mediated by Hubs In Shell | 18.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7SRR | A | Amine | 5-Hydroxytryptamine | 5-HT2B | Homo Sapiens | LSD | - | chim(NtGi2L-Gs-CtGq)/Beta1/Gamma2 | 2.9 | 2022-09-21 | doi.org/10.1016/j.neuron.2022.08.006 |
|
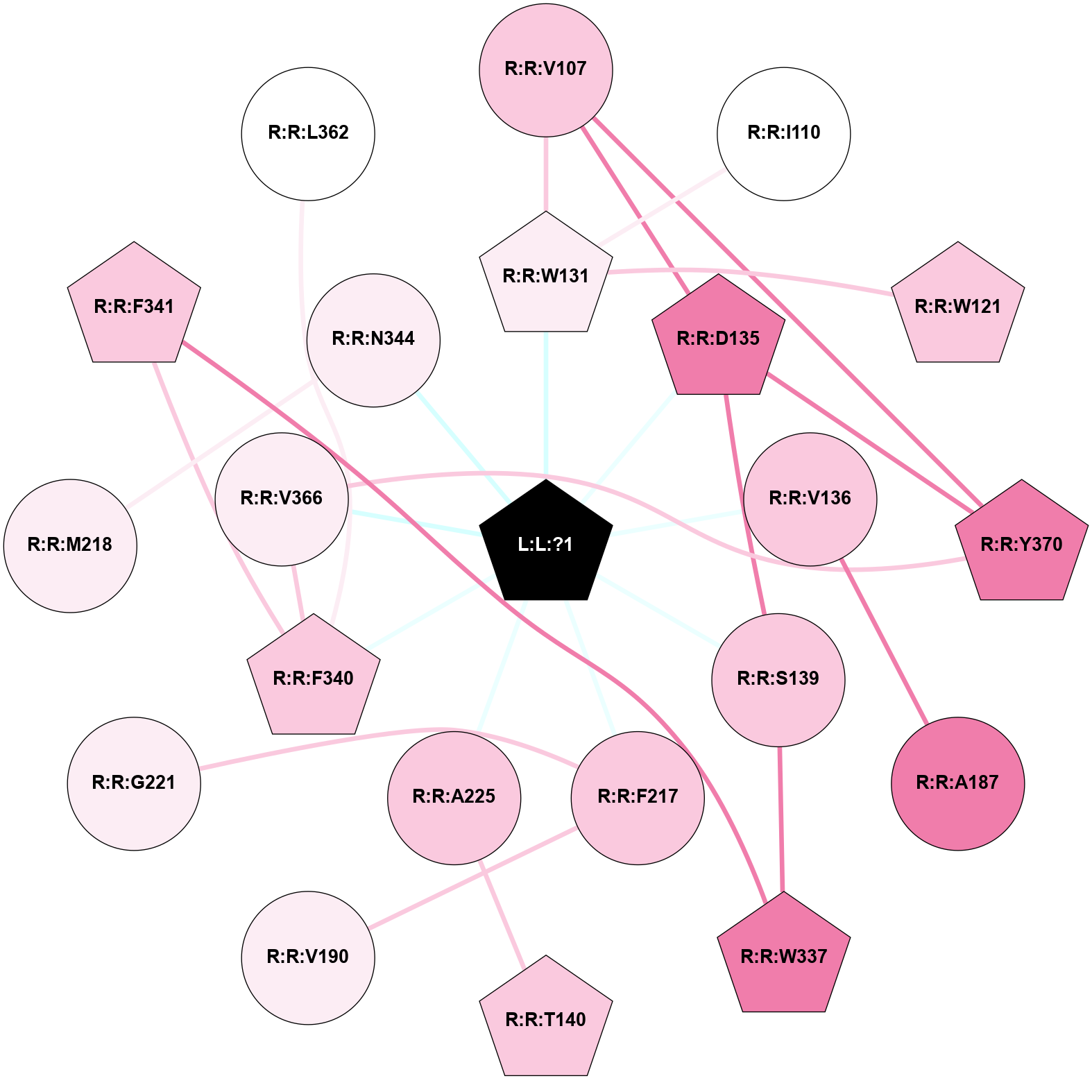
A 2D representation of the interactions of 7LD in 7SRR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:V107 | R:R:W131 | 6.13 | 5 | No | Yes | 7 | 6 | 2 | 1 | | R:R:D135 | R:R:V107 | 2.92 | 5 | Yes | No | 8 | 7 | 1 | 2 | | R:R:V107 | R:R:Y370 | 3.79 | 5 | No | Yes | 7 | 8 | 2 | 2 | | R:R:I110 | R:R:W131 | 7.05 | 0 | No | Yes | 5 | 6 | 2 | 1 | | R:R:W121 | R:R:W131 | 4.69 | 13 | Yes | Yes | 7 | 6 | 2 | 1 | | L:L:?1 | R:R:W131 | 2.85 | 5 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:D135 | R:R:S139 | 4.42 | 5 | Yes | No | 8 | 7 | 1 | 1 | | R:R:D135 | R:R:Y370 | 10.34 | 5 | Yes | Yes | 8 | 8 | 1 | 2 | | L:L:?1 | R:R:D135 | 19.24 | 5 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:A187 | R:R:V136 | 3.39 | 0 | No | No | 8 | 7 | 2 | 1 | | L:L:?1 | R:R:V136 | 16.15 | 5 | Yes | No | 0 | 7 | 0 | 1 | | R:R:S139 | R:R:W337 | 2.47 | 5 | No | Yes | 7 | 8 | 1 | 2 | | L:L:?1 | R:R:S139 | 6.26 | 5 | Yes | No | 0 | 7 | 0 | 1 | | R:R:A225 | R:R:T140 | 3.36 | 0 | No | Yes | 7 | 7 | 1 | 2 | | R:R:F217 | R:R:V190 | 3.93 | 0 | No | No | 7 | 6 | 1 | 2 | | R:R:F217 | R:R:G221 | 3.01 | 0 | No | No | 7 | 6 | 1 | 2 | | L:L:?1 | R:R:F217 | 3.05 | 5 | Yes | No | 0 | 7 | 0 | 1 | | R:R:M218 | R:R:N344 | 7.01 | 0 | No | No | 6 | 6 | 2 | 1 | | L:L:?1 | R:R:A225 | 2.63 | 5 | Yes | No | 0 | 7 | 0 | 1 | | R:R:F341 | R:R:W337 | 8.02 | 5 | Yes | Yes | 7 | 8 | 2 | 2 | | R:R:F340 | R:R:F341 | 5.36 | 5 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:F340 | R:R:L362 | 4.87 | 5 | Yes | No | 7 | 5 | 1 | 2 | | R:R:F340 | R:R:V366 | 3.93 | 5 | Yes | No | 7 | 6 | 1 | 1 | | L:L:?1 | R:R:F340 | 16.26 | 5 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:V366 | R:R:Y370 | 6.31 | 5 | No | Yes | 6 | 8 | 1 | 2 | | L:L:?1 | R:R:V366 | 4.97 | 5 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N344 | 2.29 | 5 | Yes | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 9.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 8.19 | | Average Nodes In Shell | 22.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 27.00 | | Average Links Mediated by Hubs In Shell | 23.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7SRS | A | Amine | 5-Hydroxytryptamine | 5-HT2B | Homo Sapiens | LSD | - | Arrestin2 | 3.3 | 2022-09-21 | doi.org/10.1016/j.neuron.2022.08.006 |
|
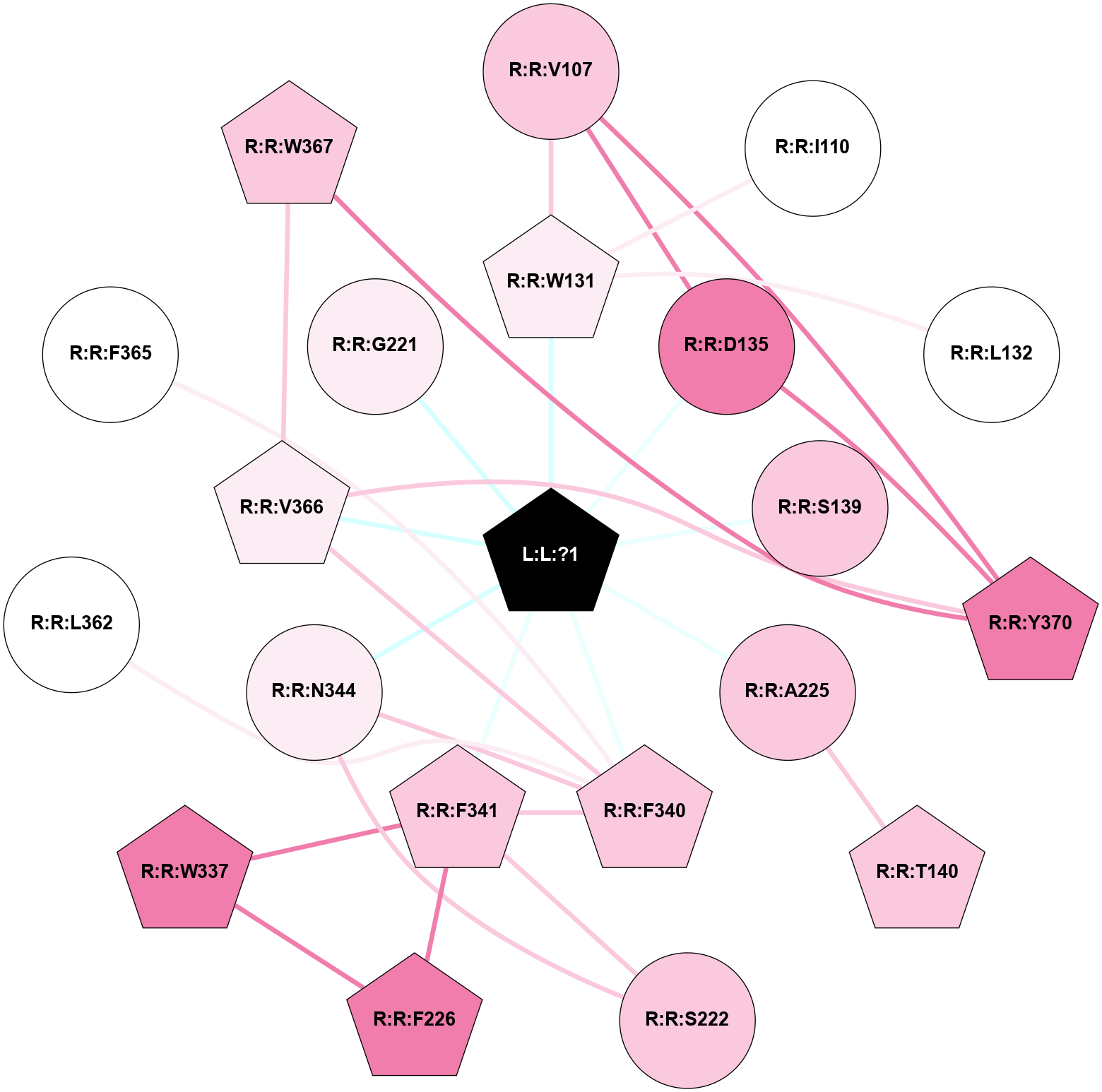
A 2D representation of the interactions of 7LD in 7SRS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:W131 | 1.9 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:D135 | 12.45 | 2 | Yes | No | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:S139 | 3.76 | 2 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:A225 | 2.63 | 2 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:F340 | 11.18 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:F341 | 7.11 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:N344 | 2.29 | 2 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:V366 | 2.49 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:V107 | R:R:W131 | 3.68 | 2 | No | Yes | 7 | 6 | 2 | 1 | | R:R:D135 | R:R:V107 | 2.92 | 2 | No | No | 8 | 7 | 1 | 2 | | R:R:V107 | R:R:Y370 | 5.05 | 2 | No | Yes | 7 | 8 | 2 | 2 | | R:R:I110 | R:R:W131 | 12.92 | 0 | No | Yes | 5 | 6 | 2 | 1 | | R:R:L132 | R:R:W131 | 2.28 | 0 | No | Yes | 5 | 6 | 2 | 1 | | R:R:D135 | R:R:Y370 | 6.9 | 2 | No | Yes | 8 | 8 | 1 | 2 | | R:R:A225 | R:R:T140 | 3.36 | 0 | No | Yes | 7 | 7 | 1 | 2 | | R:R:F341 | R:R:S222 | 3.96 | 2 | Yes | No | 7 | 7 | 1 | 2 | | R:R:N344 | R:R:S222 | 4.47 | 2 | No | No | 6 | 7 | 1 | 2 | | R:R:F226 | R:R:W337 | 4.01 | 2 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:F226 | R:R:F341 | 17.15 | 2 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:F341 | R:R:W337 | 3.01 | 2 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:F340 | R:R:F341 | 7.5 | 2 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:F340 | R:R:N344 | 2.42 | 2 | Yes | No | 7 | 6 | 1 | 1 | | R:R:F340 | R:R:L362 | 8.53 | 2 | Yes | No | 7 | 5 | 1 | 2 | | R:R:F340 | R:R:F365 | 2.14 | 2 | Yes | No | 7 | 5 | 1 | 2 | | R:R:F340 | R:R:V366 | 2.62 | 2 | Yes | Yes | 7 | 6 | 1 | 1 | | R:R:V366 | R:R:W367 | 3.68 | 2 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:V366 | R:R:Y370 | 5.05 | 2 | Yes | Yes | 6 | 8 | 1 | 2 | | R:R:W367 | R:R:Y370 | 7.72 | 2 | Yes | Yes | 7 | 8 | 2 | 2 | | L:L:?1 | R:R:G221 | 1.43 | 2 | Yes | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 9.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 5.03 | | Average Nodes In Shell | 21.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 29.00 | | Average Links Mediated by Hubs In Shell | 27.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7WC6 | A | Amine | 5-Hydroxytryptamine | 5-HT2A | Homo Sapiens | LSD | - | - | 2.6 | 2022-01-26 | doi.org/10.1126/science.abl8615 |
|
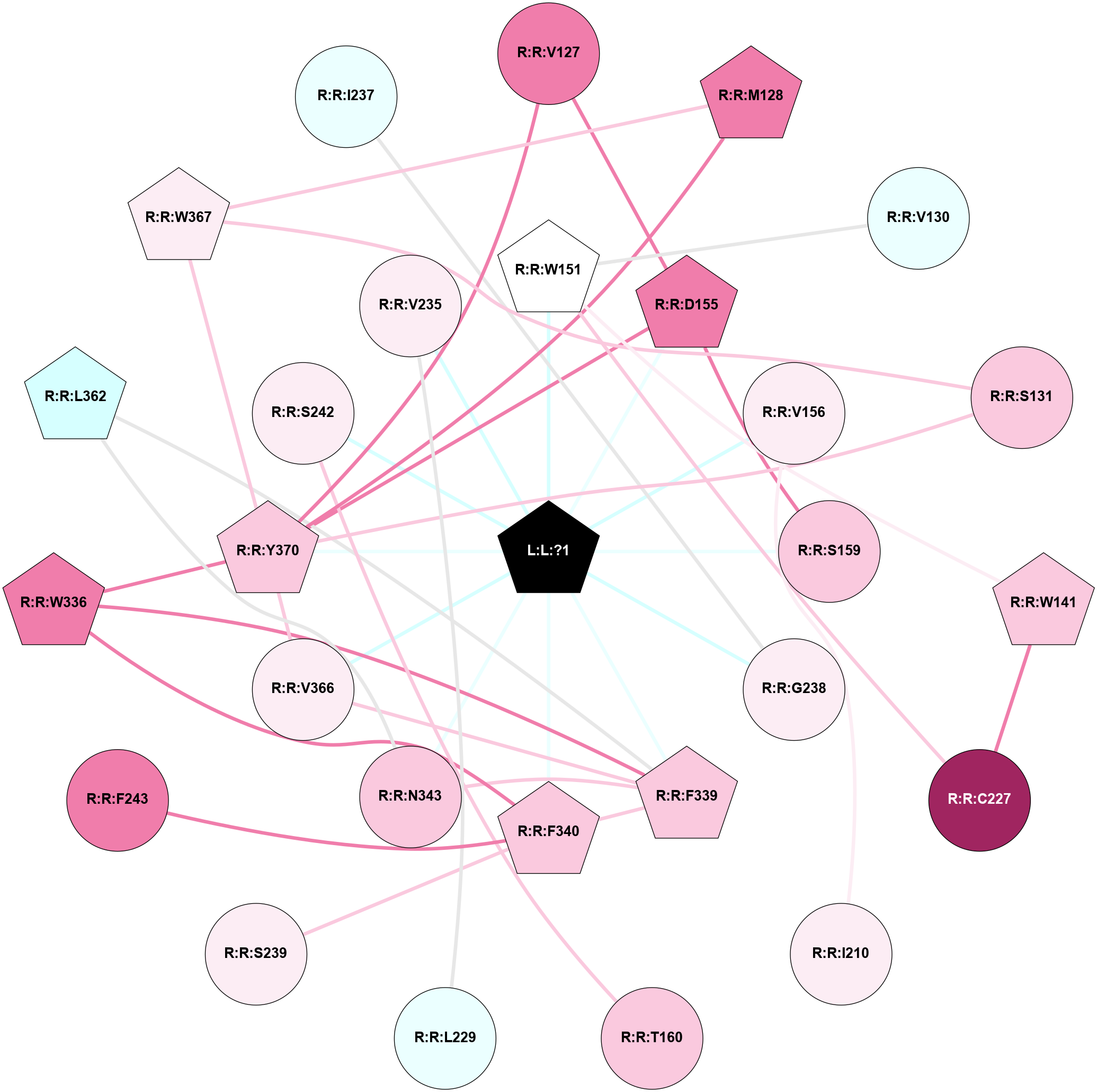
A 2D representation of the interactions of 7LD in 7WC6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:W151 | 15.2 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:D155 | 28.3 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:V156 | 14.91 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:S159 | 6.26 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:G238 | 4.28 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:F339 | 17.27 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:F340 | 11.18 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:N343 | 4.58 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:V366 | 9.94 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:Y370 | 3.91 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:D155 | R:R:V127 | 7.3 | 1 | Yes | No | 8 | 8 | 1 | 2 | | R:R:V127 | R:R:Y370 | 3.79 | 1 | No | Yes | 8 | 7 | 2 | 1 | | R:R:M128 | R:R:W367 | 5.82 | 1 | Yes | Yes | 8 | 6 | 2 | 2 | | R:R:M128 | R:R:Y370 | 10.78 | 1 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:V130 | R:R:W151 | 12.26 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:S131 | R:R:W367 | 7.41 | 1 | No | Yes | 7 | 6 | 2 | 2 | | R:R:S131 | R:R:Y370 | 5.09 | 1 | No | Yes | 7 | 7 | 2 | 1 | | R:R:W141 | R:R:W151 | 9.37 | 1 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:C227 | R:R:W141 | 13.06 | 1 | No | Yes | 9 | 7 | 2 | 2 | | R:R:C227 | R:R:W151 | 5.22 | 1 | No | Yes | 9 | 5 | 2 | 1 | | R:R:D155 | R:R:S159 | 4.42 | 1 | Yes | No | 8 | 7 | 1 | 1 | | R:R:D155 | R:R:Y370 | 4.6 | 1 | Yes | Yes | 8 | 7 | 1 | 1 | | R:R:I210 | R:R:V156 | 4.61 | 0 | No | No | 6 | 6 | 2 | 1 | | R:R:S242 | R:R:T160 | 9.59 | 0 | No | No | 6 | 7 | 1 | 2 | | R:R:L229 | R:R:V235 | 4.47 | 0 | No | No | 4 | 6 | 2 | 1 | | R:R:F340 | R:R:S239 | 3.96 | 1 | Yes | No | 7 | 6 | 1 | 2 | | R:R:F243 | R:R:F340 | 26.79 | 0 | No | Yes | 8 | 7 | 2 | 1 | | R:R:F339 | R:R:W336 | 6.01 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:F340 | R:R:W336 | 6.01 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:W336 | R:R:Y370 | 3.86 | 1 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:F339 | R:R:F340 | 9.65 | 1 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:F339 | R:R:N343 | 9.67 | 1 | Yes | No | 7 | 7 | 1 | 1 | | R:R:F339 | R:R:L362 | 4.87 | 1 | Yes | Yes | 7 | 3 | 1 | 2 | | R:R:F339 | R:R:V366 | 5.24 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:L362 | R:R:N343 | 5.49 | 1 | Yes | No | 3 | 7 | 2 | 1 | | R:R:V366 | R:R:Y370 | 8.83 | 1 | No | Yes | 6 | 7 | 1 | 1 | | R:R:W367 | R:R:Y370 | 18.33 | 1 | Yes | Yes | 6 | 7 | 2 | 1 | | L:L:?1 | R:R:S242 | 3.76 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:V235 | 3.73 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:G238 | R:R:I237 | 1.76 | 0 | No | No | 6 | 4 | 1 | 2 | | R:R:V235 | R:R:V347 | 1.6 | 0 | No | No | 6 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 12.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 10.28 | | Average Nodes In Shell | 29.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 41.00 | | Average Links Mediated by Hubs In Shell | 36.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8FYT | A | Amine | 5-Hydroxytryptamine | 5-HT1A | Homo Sapiens | LSD | - | Gi1/Beta1/Gamma1 | 2.64 | 2024-05-15 | doi.org/10.1038/s41586-024-07403-2 |
|
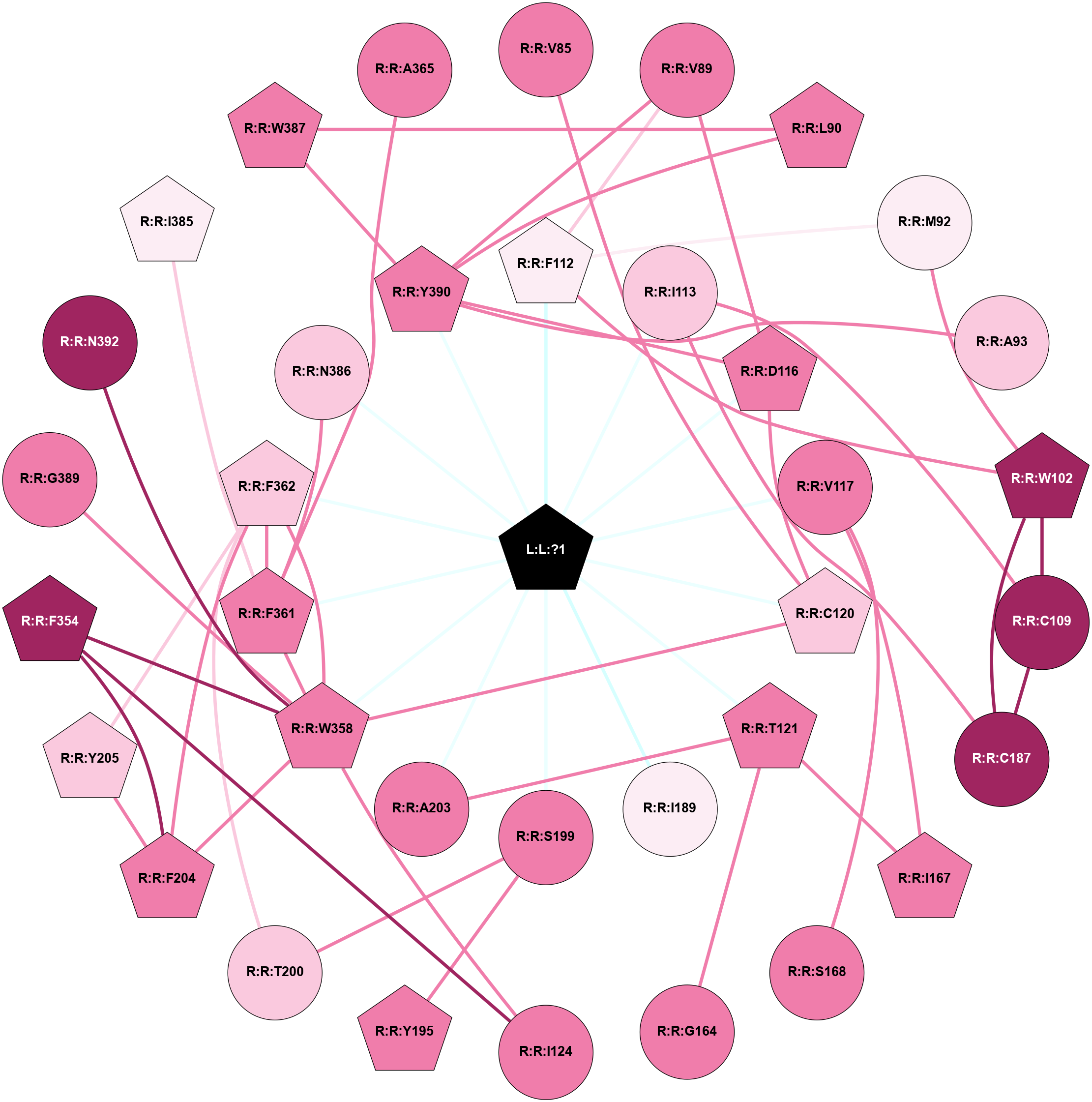
A 2D representation of the interactions of 7LD in 8FYT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:F112 | 3.81 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:I113 | 3.35 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:D116 | 30.77 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:V117 | 17.47 | 1 | Yes | No | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:C120 | 4.96 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:T121 | 4.61 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:I189 | 8.93 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:S199 | 4.7 | 1 | Yes | No | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:A203 | 4.93 | 1 | Yes | No | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:W358 | 4.45 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:F361 | 20.95 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:F362 | 10.47 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:N386 | 8.59 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:Y390 | 10.08 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:C120 | R:R:V85 | 3.42 | 1 | Yes | No | 7 | 8 | 1 | 2 | | R:R:F112 | R:R:V89 | 5.24 | 1 | Yes | No | 6 | 8 | 1 | 2 | | R:R:D116 | R:R:V89 | 7.3 | 1 | Yes | No | 8 | 8 | 1 | 2 | | R:R:V89 | R:R:Y390 | 5.05 | 1 | No | Yes | 8 | 8 | 2 | 1 | | R:R:L90 | R:R:W387 | 3.42 | 1 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:L90 | R:R:Y390 | 4.69 | 1 | Yes | Yes | 8 | 8 | 2 | 1 | | R:R:M92 | R:R:W102 | 3.49 | 1 | No | Yes | 6 | 9 | 2 | 2 | | R:R:F112 | R:R:M92 | 11.2 | 1 | Yes | No | 6 | 6 | 1 | 2 | | R:R:A93 | R:R:Y390 | 4 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:C109 | R:R:W102 | 6.53 | 1 | No | Yes | 9 | 9 | 2 | 2 | | R:R:F112 | R:R:W102 | 10.02 | 1 | Yes | Yes | 6 | 9 | 1 | 2 | | R:R:C187 | R:R:W102 | 10.45 | 1 | No | Yes | 9 | 9 | 2 | 2 | | R:R:C109 | R:R:I113 | 3.27 | 1 | No | No | 9 | 7 | 2 | 1 | | R:R:C109 | R:R:C187 | 7.28 | 1 | No | No | 9 | 9 | 2 | 2 | | R:R:C187 | R:R:I113 | 3.27 | 1 | No | No | 9 | 7 | 2 | 1 | | R:R:C120 | R:R:D116 | 3.11 | 1 | Yes | Yes | 7 | 8 | 1 | 1 | | R:R:D116 | R:R:Y390 | 11.49 | 1 | Yes | Yes | 8 | 8 | 1 | 1 | | R:R:I167 | R:R:V117 | 3.07 | 0 | Yes | No | 8 | 8 | 2 | 1 | | R:R:S168 | R:R:V117 | 3.23 | 0 | No | No | 8 | 8 | 2 | 1 | | R:R:C120 | R:R:W358 | 5.22 | 1 | Yes | Yes | 7 | 8 | 1 | 1 | | R:R:G164 | R:R:T121 | 3.64 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:I167 | R:R:T121 | 6.08 | 0 | Yes | Yes | 8 | 8 | 2 | 1 | | R:R:A203 | R:R:T121 | 5.03 | 1 | No | Yes | 8 | 8 | 1 | 1 | | R:R:F354 | R:R:I124 | 5.02 | 1 | Yes | No | 9 | 8 | 2 | 2 | | R:R:I124 | R:R:W358 | 14.09 | 1 | No | Yes | 8 | 8 | 2 | 1 | | R:R:S199 | R:R:Y195 | 12.72 | 0 | No | Yes | 8 | 8 | 1 | 2 | | R:R:S199 | R:R:T200 | 3.2 | 0 | No | No | 8 | 7 | 1 | 2 | | R:R:F362 | R:R:T200 | 5.19 | 1 | Yes | No | 7 | 7 | 1 | 2 | | R:R:F204 | R:R:Y205 | 9.28 | 1 | Yes | Yes | 8 | 7 | 2 | 2 | | R:R:F204 | R:R:F354 | 8.57 | 1 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:F204 | R:R:W358 | 3.01 | 1 | Yes | Yes | 8 | 8 | 2 | 1 | | R:R:F204 | R:R:F362 | 20.36 | 1 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:F362 | R:R:Y205 | 3.09 | 1 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:F354 | R:R:W358 | 4.01 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F361 | R:R:W358 | 3.01 | 1 | Yes | Yes | 8 | 8 | 1 | 1 | | R:R:F362 | R:R:W358 | 8.02 | 1 | Yes | Yes | 7 | 8 | 1 | 1 | | R:R:G389 | R:R:W358 | 8.44 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:N392 | R:R:W358 | 12.43 | 0 | No | Yes | 9 | 8 | 2 | 1 | | R:R:F361 | R:R:F362 | 12.86 | 1 | Yes | Yes | 8 | 7 | 1 | 1 | | R:R:F361 | R:R:I385 | 3.77 | 1 | Yes | Yes | 8 | 6 | 1 | 2 | | R:R:F361 | R:R:N386 | 7.25 | 1 | Yes | No | 8 | 7 | 1 | 1 | | R:R:W387 | R:R:Y390 | 19.29 | 1 | Yes | Yes | 8 | 8 | 2 | 1 | | R:R:A365 | R:R:F361 | 2.77 | 0 | No | Yes | 8 | 8 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 14.00 | | Average Number Of Links With An Hub | 8.00 | | Average Interaction Strength | 9.86 | | Average Nodes In Shell | 37.00 | | Average Hubs In Shell | 18.00 | | Average Links In Shell | 57.00 | | Average Links Mediated by Hubs In Shell | 52.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8JXR | A | Amine | Dopamine | D1 | Homo sapiens | LSD | - | - | 3.57 | 2024-09-04 | doi.org/10.1016/j.neuron.2024.07.003 |
|
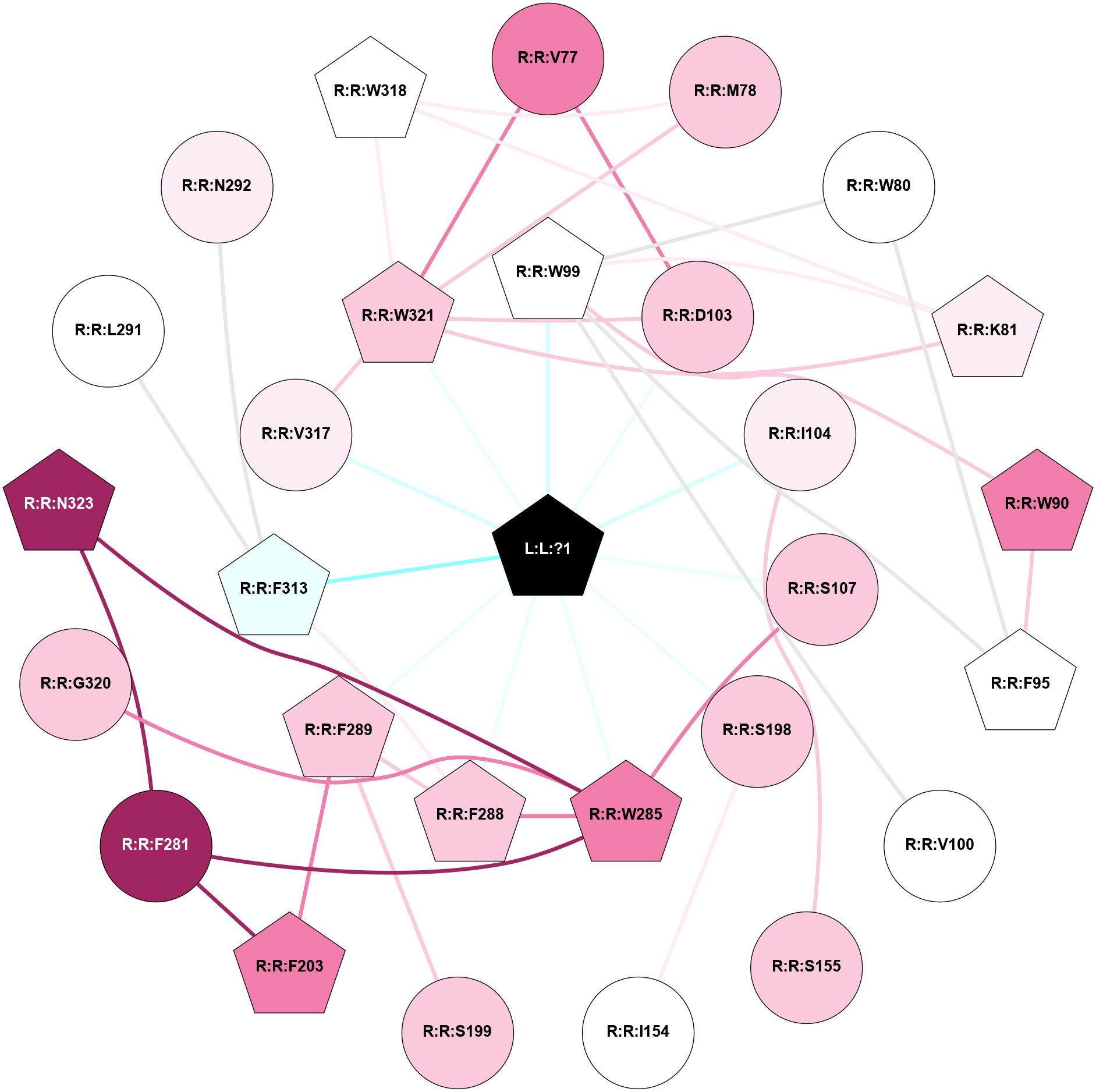
A 2D representation of the interactions of 7LD in 8JXR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:W99 | 4.45 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:D103 | 15.91 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:I104 | 12.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:S107 | 9.39 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:S198 | 4.7 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:W285 | 6.23 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:F288 | 37.13 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:F289 | 8.57 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:F313 | 11.43 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:V317 | 13.98 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:W321 | 3.56 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:D103 | R:R:V77 | 5.84 | 1 | No | No | 7 | 8 | 1 | 2 | | R:R:V77 | R:R:W321 | 4.9 | 1 | No | Yes | 8 | 7 | 2 | 1 | | R:R:M78 | R:R:W318 | 5.82 | 1 | No | Yes | 7 | 5 | 2 | 2 | | R:R:M78 | R:R:W321 | 9.31 | 1 | No | Yes | 7 | 7 | 2 | 1 | | R:R:F95 | R:R:W80 | 5.01 | 1 | Yes | No | 5 | 5 | 2 | 2 | | R:R:W80 | R:R:W99 | 5.62 | 1 | No | Yes | 5 | 5 | 2 | 1 | | R:R:K81 | R:R:W99 | 5.8 | 1 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:K81 | R:R:W318 | 3.48 | 1 | Yes | Yes | 6 | 5 | 2 | 2 | | R:R:K81 | R:R:W321 | 4.64 | 1 | Yes | Yes | 6 | 7 | 2 | 1 | | R:R:F95 | R:R:W90 | 5.01 | 1 | Yes | Yes | 5 | 8 | 2 | 2 | | R:R:W90 | R:R:W99 | 7.5 | 1 | Yes | Yes | 8 | 5 | 2 | 1 | | R:R:F95 | R:R:W99 | 4.01 | 1 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:V100 | R:R:W99 | 3.68 | 0 | No | Yes | 5 | 5 | 2 | 1 | | R:R:D103 | R:R:W321 | 6.7 | 1 | No | Yes | 7 | 7 | 1 | 1 | | R:R:I104 | R:R:S155 | 6.19 | 0 | No | No | 6 | 7 | 1 | 2 | | R:R:S107 | R:R:W285 | 6.18 | 1 | No | Yes | 7 | 8 | 1 | 1 | | R:R:I154 | R:R:S198 | 4.64 | 0 | No | No | 5 | 7 | 2 | 1 | | R:R:F289 | R:R:S199 | 6.61 | 1 | Yes | No | 7 | 7 | 1 | 2 | | R:R:F203 | R:R:F281 | 4.29 | 0 | Yes | No | 8 | 9 | 2 | 2 | | R:R:F203 | R:R:F289 | 24.65 | 0 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:F281 | R:R:W285 | 10.02 | 1 | No | Yes | 9 | 8 | 2 | 1 | | R:R:F281 | R:R:N323 | 7.25 | 1 | No | Yes | 9 | 9 | 2 | 2 | | R:R:F288 | R:R:W285 | 6.01 | 1 | Yes | Yes | 7 | 8 | 1 | 1 | | R:R:G320 | R:R:W285 | 9.85 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N323 | R:R:W285 | 9.04 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F288 | R:R:F289 | 11.79 | 1 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:F288 | R:R:F313 | 6.43 | 1 | Yes | Yes | 7 | 4 | 1 | 1 | | R:R:F313 | R:R:L291 | 4.87 | 1 | Yes | No | 4 | 5 | 1 | 2 | | R:R:F313 | R:R:N292 | 4.83 | 1 | Yes | No | 4 | 6 | 1 | 2 | | R:R:V317 | R:R:W321 | 9.81 | 1 | No | Yes | 6 | 7 | 1 | 1 | | R:R:W318 | R:R:W321 | 14.06 | 1 | Yes | Yes | 5 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 11.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 11.60 | | Average Nodes In Shell | 29.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 42.00 | | Average Links Mediated by Hubs In Shell | 39.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8ZSP | A | Amine | Trace Amine Receptors | TA1 | Homo Sapiens | LSD | - | chim(NtGi1L-Gs-CtGq)/Beta1/Gamma2 | 3.14 | 2024-07-24 | doi.org/10.1016/j.celrep.2024.114505 |
|

A 2D representation of the interactions of 7LD in 8ZSP
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L72 | R:R:M77 | 4.24 | 0 | No | Yes | 7 | 7 | 2 | 2 | | R:R:L72 | R:R:S107 | 4.5 | 0 | No | Yes | 7 | 6 | 2 | 1 | | R:R:H99 | R:R:V76 | 4.15 | 1 | Yes | No | 5 | 7 | 2 | 2 | | R:R:D103 | R:R:V76 | 11.69 | 1 | Yes | No | 6 | 7 | 1 | 2 | | R:R:V76 | R:R:Y294 | 3.79 | 1 | No | Yes | 7 | 7 | 2 | 1 | | R:R:M77 | R:R:Y294 | 15.57 | 0 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:R83 | R:R:S80 | 7.91 | 1 | No | No | 4 | 6 | 1 | 2 | | R:R:H99 | R:R:S80 | 6.97 | 1 | Yes | No | 5 | 6 | 2 | 2 | | R:R:S80 | R:R:Y294 | 7.63 | 1 | No | Yes | 6 | 7 | 2 | 1 | | R:R:H99 | R:R:R83 | 4.51 | 1 | Yes | No | 5 | 4 | 2 | 1 | | L:L:?1 | R:R:R83 | 7.09 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:T100 | 3.69 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:D103 | R:R:S107 | 4.42 | 1 | Yes | Yes | 6 | 6 | 1 | 1 | | R:R:D103 | R:R:Y294 | 10.34 | 1 | Yes | Yes | 6 | 7 | 1 | 1 | | L:L:?1 | R:R:D103 | 15.85 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:F154 | R:R:I104 | 3.77 | 1 | Yes | Yes | 4 | 4 | 2 | 1 | | R:R:I104 | R:R:M158 | 4.37 | 1 | Yes | Yes | 4 | 5 | 1 | 2 | | L:L:?1 | R:R:I104 | 17.86 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:S107 | R:R:W264 | 4.94 | 1 | Yes | Yes | 6 | 8 | 1 | 1 | | L:L:?1 | R:R:S107 | 11.27 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:P151 | R:R:S108 | 3.56 | 0 | Yes | No | 8 | 5 | 2 | 1 | | R:R:S108 | R:R:S198 | 6.52 | 1 | No | No | 5 | 6 | 1 | 1 | | L:L:?1 | R:R:S108 | 5.01 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:I111 | R:R:W264 | 10.57 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:F154 | R:R:M158 | 7.46 | 1 | Yes | Yes | 4 | 5 | 2 | 2 | | R:R:F154 | R:R:T194 | 3.89 | 1 | Yes | No | 4 | 5 | 2 | 1 | | R:R:F186 | R:R:M158 | 3.73 | 1 | Yes | Yes | 1 | 5 | 1 | 2 | | R:R:R179 | R:R:S183 | 7.91 | 0 | No | No | 2 | 4 | 2 | 1 | | L:L:?1 | R:R:S183 | 3.76 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:F186 | R:R:V184 | 11.8 | 1 | Yes | No | 1 | 4 | 1 | 1 | | L:L:?1 | R:R:V184 | 8.7 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:F186 | R:R:S190 | 6.61 | 1 | Yes | No | 1 | 6 | 1 | 2 | | R:R:F186 | R:R:G191 | 4.52 | 1 | Yes | No | 1 | 4 | 1 | 2 | | R:R:F186 | R:R:T194 | 3.89 | 1 | Yes | No | 1 | 5 | 1 | 1 | | L:L:?1 | R:R:F186 | 6.1 | 1 | Yes | Yes | 0 | 1 | 0 | 1 | | L:L:?1 | R:R:T194 | 7.38 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:F195 | R:R:F199 | 6.43 | 1 | Yes | Yes | 5 | 8 | 2 | 2 | | R:R:F195 | R:R:F268 | 4.29 | 1 | Yes | Yes | 5 | 7 | 2 | 1 | | R:R:F268 | R:R:S198 | 3.96 | 1 | Yes | No | 7 | 6 | 1 | 1 | | L:L:?1 | R:R:S198 | 10.02 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:F199 | R:R:F268 | 22.51 | 1 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:F267 | R:R:W264 | 6.01 | 1 | Yes | Yes | 5 | 8 | 1 | 1 | | R:R:F268 | R:R:W264 | 12.03 | 1 | Yes | Yes | 7 | 8 | 1 | 1 | | R:R:G293 | R:R:W264 | 8.44 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N296 | R:R:W264 | 10.17 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | L:L:?1 | R:R:W264 | 9.5 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:F267 | R:R:F268 | 9.65 | 1 | Yes | Yes | 5 | 7 | 1 | 1 | | R:R:F267 | R:R:T271 | 9.08 | 1 | Yes | No | 5 | 5 | 1 | 2 | | R:R:F267 | R:R:L289 | 4.87 | 1 | Yes | No | 5 | 5 | 1 | 2 | | R:R:F267 | R:R:I290 | 3.77 | 1 | Yes | No | 5 | 5 | 1 | 1 | | L:L:?1 | R:R:F267 | 14.22 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:F268 | 13.21 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:I290 | 11.91 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:W291 | R:R:Y294 | 13.5 | 0 | Yes | Yes | 4 | 7 | 2 | 1 | | L:L:?1 | R:R:Y294 | 3.91 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:N286 | 3.44 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:A155 | R:R:I104 | 3.25 | 0 | No | Yes | 6 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 17.00 | | Average Number Of Links With An Hub | 8.00 | | Average Interaction Strength | 9.00 | | Average Nodes In Shell | 38.00 | | Average Hubs In Shell | 18.00 | | Average Links In Shell | 57.00 | | Average Links Mediated by Hubs In Shell | 54.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9AS3 | A | Amine | 5-Hydroxytryptamine | 5-HT2A | Homo Sapiens | LSD | - | - | 3.18 | 2025-04-02 | doi.org/10.1038/s41467-025-57956-7 |
|
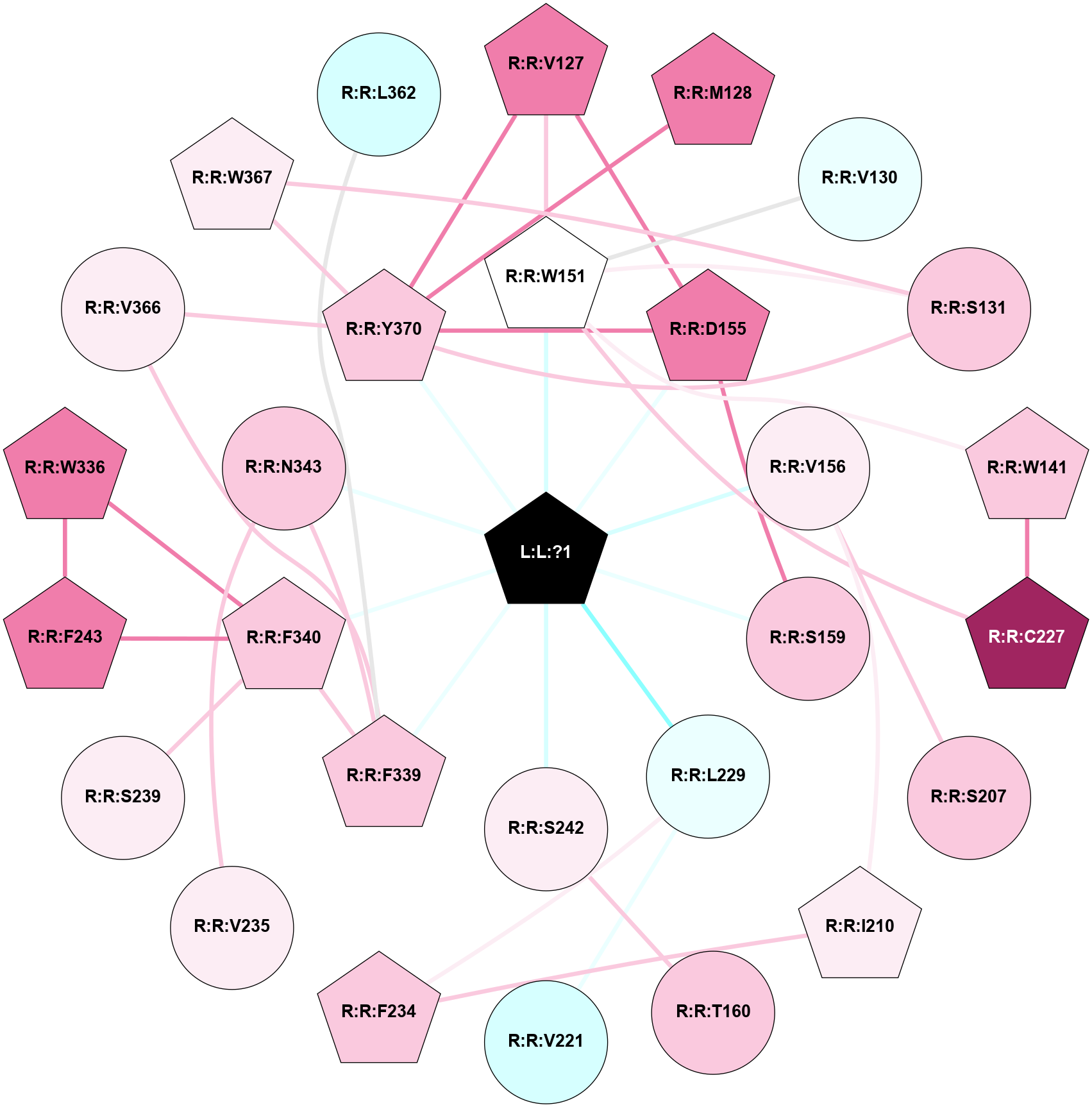
A 2D representation of the interactions of 7LD in 9AS3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:W151 | 10.68 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:D155 | 25.46 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:V156 | 12.81 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:S159 | 9.39 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:L229 | 3.25 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:S242 | 4.7 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:F339 | 11.43 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:F340 | 7.62 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:N343 | 3.22 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:Y370 | 5.5 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:V127 | R:R:W151 | 7.36 | 1 | Yes | Yes | 8 | 5 | 2 | 1 | | R:R:D155 | R:R:V127 | 7.3 | 1 | Yes | Yes | 8 | 8 | 1 | 2 | | R:R:V127 | R:R:Y370 | 3.79 | 1 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:M128 | R:R:Y370 | 11.97 | 0 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:V130 | R:R:W151 | 12.26 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:S131 | R:R:W151 | 6.18 | 1 | No | Yes | 7 | 5 | 2 | 1 | | R:R:S131 | R:R:W367 | 3.71 | 1 | No | Yes | 7 | 6 | 2 | 2 | | R:R:S131 | R:R:Y370 | 5.09 | 1 | No | Yes | 7 | 7 | 2 | 1 | | R:R:W141 | R:R:W151 | 12.18 | 1 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:C227 | R:R:W141 | 10.45 | 1 | Yes | Yes | 9 | 7 | 2 | 2 | | R:R:C227 | R:R:W151 | 3.92 | 1 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:D155 | R:R:S159 | 2.94 | 1 | Yes | No | 8 | 7 | 1 | 1 | | R:R:D155 | R:R:Y370 | 8.05 | 1 | Yes | Yes | 8 | 7 | 1 | 1 | | R:R:S207 | R:R:V156 | 3.23 | 4 | No | No | 7 | 6 | 2 | 1 | | R:R:I210 | R:R:V156 | 4.61 | 0 | Yes | No | 6 | 6 | 2 | 1 | | R:R:S242 | R:R:T160 | 7.99 | 0 | No | No | 6 | 7 | 1 | 2 | | R:R:F234 | R:R:I210 | 5.02 | 0 | Yes | Yes | 7 | 6 | 2 | 2 | | R:R:L229 | R:R:V221 | 5.96 | 0 | No | No | 4 | 3 | 1 | 2 | | R:R:F234 | R:R:L229 | 6.09 | 0 | Yes | No | 7 | 4 | 2 | 1 | | R:R:N343 | R:R:V235 | 2.96 | 1 | No | No | 7 | 6 | 1 | 2 | | R:R:F340 | R:R:S239 | 3.96 | 1 | Yes | No | 7 | 6 | 1 | 2 | | R:R:F243 | R:R:W336 | 3.01 | 1 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:F243 | R:R:F340 | 24.65 | 1 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:F340 | R:R:W336 | 4.01 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:F339 | R:R:F340 | 7.5 | 1 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:F339 | R:R:N343 | 9.67 | 1 | Yes | No | 7 | 7 | 1 | 1 | | R:R:F339 | R:R:V366 | 3.93 | 1 | Yes | No | 7 | 6 | 1 | 2 | | R:R:V366 | R:R:Y370 | 6.31 | 0 | No | Yes | 6 | 7 | 2 | 1 | | R:R:W367 | R:R:Y370 | 16.4 | 1 | Yes | Yes | 6 | 7 | 2 | 1 | | R:R:F339 | R:R:L362 | 2.44 | 1 | Yes | No | 7 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 9.41 | | Average Nodes In Shell | 29.00 | | Average Hubs In Shell | 15.00 | | Average Links In Shell | 40.00 | | Average Links Mediated by Hubs In Shell | 36.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9AS4 | A | Amine | 5-Hydroxytryptamine | 5-HT2A | Homo Sapiens | LSD | - | chim(NtGi1-Gs-CtGq)/Beta1/Gamma2 | 3.06 | 2025-04-02 | doi.org/10.1038/s41467-025-57956-7 |
|
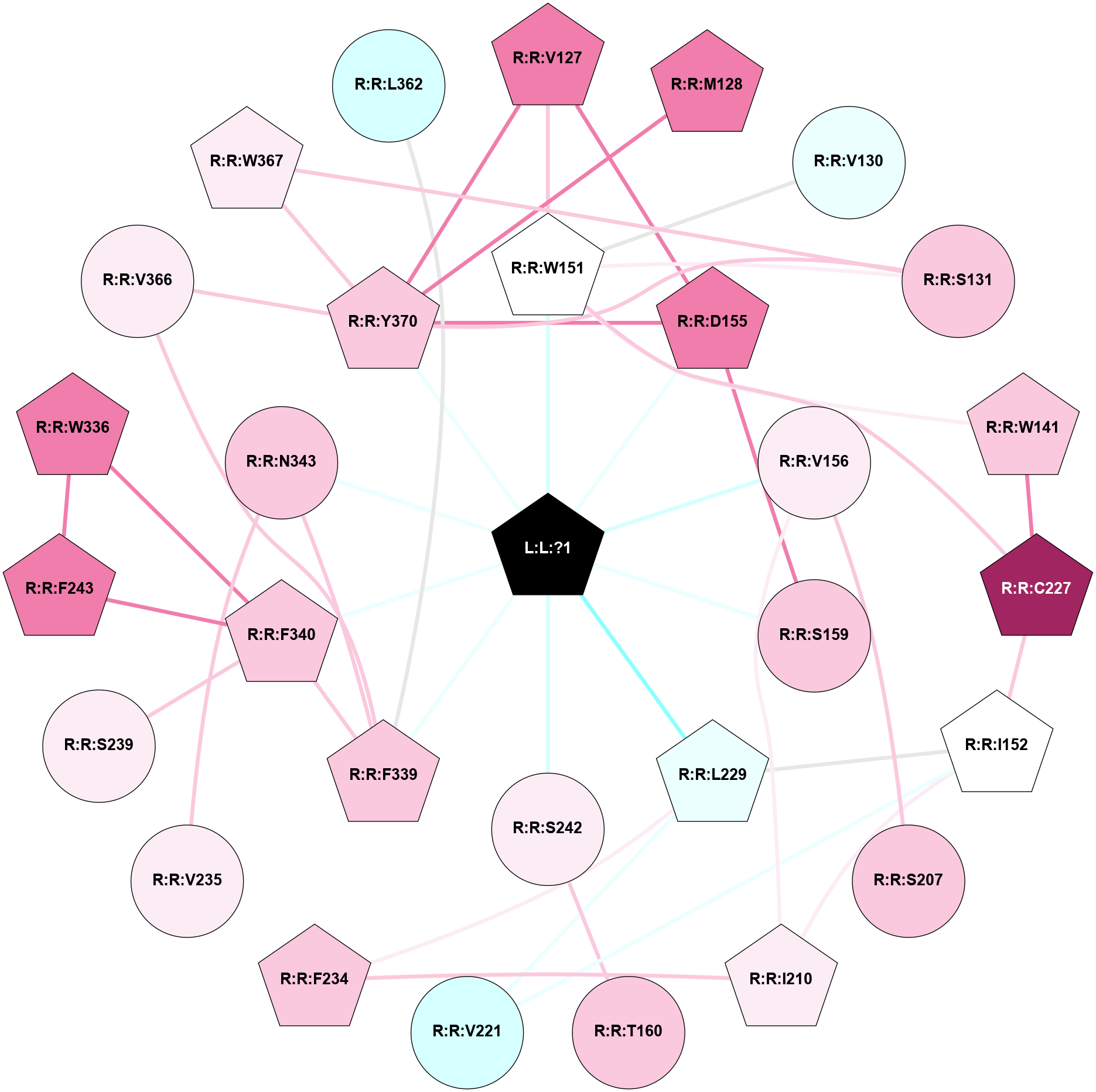
A 2D representation of the interactions of 7LD in 9AS4
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:W151 | 10.68 | 5 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:D155 | 25.46 | 5 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:V156 | 12.81 | 5 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:S159 | 9.39 | 5 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:L229 | 3.25 | 5 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:S242 | 4.7 | 5 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:F339 | 11.43 | 5 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:F340 | 7.62 | 5 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:N343 | 3.22 | 5 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?1 | R:R:Y370 | 5.5 | 5 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:V127 | R:R:W151 | 7.36 | 5 | Yes | Yes | 8 | 5 | 2 | 1 | | R:R:D155 | R:R:V127 | 7.3 | 5 | Yes | Yes | 8 | 8 | 1 | 2 | | R:R:V127 | R:R:Y370 | 3.79 | 5 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:M128 | R:R:Y370 | 11.97 | 0 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:V130 | R:R:W151 | 12.26 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:S131 | R:R:W151 | 6.18 | 5 | No | Yes | 7 | 5 | 2 | 1 | | R:R:S131 | R:R:W367 | 3.71 | 5 | No | Yes | 7 | 6 | 2 | 2 | | R:R:S131 | R:R:Y370 | 5.09 | 5 | No | Yes | 7 | 7 | 2 | 1 | | R:R:W141 | R:R:W151 | 12.18 | 5 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:C227 | R:R:W141 | 10.45 | 5 | Yes | Yes | 9 | 7 | 2 | 2 | | R:R:C227 | R:R:W151 | 3.92 | 5 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:I152 | R:R:I210 | 2.94 | 5 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:I152 | R:R:V221 | 3.07 | 5 | Yes | No | 5 | 3 | 2 | 2 | | R:R:C227 | R:R:I152 | 3.27 | 5 | Yes | Yes | 9 | 5 | 2 | 2 | | R:R:I152 | R:R:L229 | 2.85 | 5 | Yes | Yes | 5 | 4 | 2 | 1 | | R:R:D155 | R:R:S159 | 2.94 | 5 | Yes | No | 8 | 7 | 1 | 1 | | R:R:D155 | R:R:Y370 | 8.05 | 5 | Yes | Yes | 8 | 7 | 1 | 1 | | R:R:S207 | R:R:V156 | 3.23 | 21 | No | No | 7 | 6 | 2 | 1 | | R:R:I210 | R:R:V156 | 4.61 | 0 | Yes | No | 6 | 6 | 2 | 1 | | R:R:S242 | R:R:T160 | 7.99 | 0 | No | No | 6 | 7 | 1 | 2 | | R:R:F234 | R:R:I210 | 5.02 | 0 | Yes | Yes | 7 | 6 | 2 | 2 | | R:R:L229 | R:R:V221 | 5.96 | 5 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F234 | R:R:L229 | 6.09 | 0 | Yes | Yes | 7 | 4 | 2 | 1 | | R:R:N343 | R:R:V235 | 2.96 | 5 | No | No | 7 | 6 | 1 | 2 | | R:R:F340 | R:R:S239 | 3.96 | 5 | Yes | No | 7 | 6 | 1 | 2 | | R:R:F243 | R:R:W336 | 3.01 | 5 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:F243 | R:R:F340 | 24.65 | 5 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:F340 | R:R:W336 | 4.01 | 5 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:F339 | R:R:F340 | 7.5 | 5 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:F339 | R:R:N343 | 9.67 | 5 | Yes | No | 7 | 7 | 1 | 1 | | R:R:F339 | R:R:V366 | 3.93 | 5 | Yes | No | 7 | 6 | 1 | 2 | | R:R:V366 | R:R:Y370 | 6.31 | 0 | No | Yes | 6 | 7 | 2 | 1 | | R:R:W367 | R:R:Y370 | 16.4 | 5 | Yes | Yes | 6 | 7 | 2 | 1 | | R:R:F339 | R:R:L362 | 2.44 | 5 | Yes | No | 7 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 9.41 | | Average Nodes In Shell | 30.00 | | Average Hubs In Shell | 17.00 | | Average Links In Shell | 44.00 | | Average Links Mediated by Hubs In Shell | 41.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|










