| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7FIG | A | Protein | Glycoprotein Hormone | LH | Homo Sapiens | hCG | - | chim(NtGi1L-Gs)/Beta1/Gamma2 | 3.9 | 2021-09-29 | doi.org/10.1038/s41586-021-03924-2 |
|
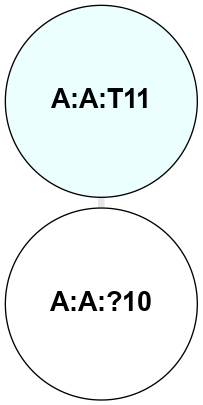
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:?10 | A:A:T11 | 9.47 | 0 | No | No | 5 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 9.47 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:I207 | B:B:W99 | 4.7 | 0 | No | Yes | 9 | 9 | 2 | 1 | | A:A:F222 | B:B:W99 | 23.05 | 0 | No | Yes | 6 | 9 | 2 | 1 | | A:A:?237 | B:B:?59 | 7.21 | 1 | Yes | Yes | 8 | 8 | 0 | 1 | | A:A:?237 | B:B:Q75 | 17.1 | 1 | Yes | Yes | 8 | 9 | 0 | 1 | | A:A:?237 | B:B:W99 | 7.32 | 1 | Yes | Yes | 8 | 9 | 0 | 1 | | A:A:?237 | B:B:M101 | 4.54 | 1 | Yes | Yes | 8 | 9 | 0 | 1 | | A:A:?237 | B:B:L117 | 7.41 | 1 | Yes | Yes | 8 | 9 | 0 | 1 | | A:A:F238 | B:B:W99 | 16.04 | 1 | No | Yes | 9 | 9 | 2 | 1 | | A:A:D240 | B:B:?57 | 4.86 | 0 | No | No | 7 | 9 | 2 | 2 | | A:A:D240 | B:B:W99 | 4.47 | 0 | No | Yes | 7 | 9 | 2 | 1 | | B:B:?57 | B:B:Q75 | 7.15 | 0 | No | Yes | 9 | 9 | 2 | 1 | | B:B:?59 | B:B:I58 | 5.55 | 1 | Yes | Yes | 8 | 9 | 1 | 2 | | B:B:?59 | B:B:Q75 | 19.42 | 1 | Yes | Yes | 8 | 9 | 1 | 1 | | B:B:?59 | B:B:M101 | 5.5 | 1 | Yes | Yes | 8 | 9 | 1 | 1 | | B:B:?59 | B:B:S316 | 20.45 | 1 | Yes | No | 8 | 9 | 1 | 2 | | B:B:?317 | B:B:?59 | 7.21 | 1 | Yes | Yes | 7 | 8 | 2 | 1 | | B:B:?59 | B:B:W332 | 7.75 | 1 | Yes | Yes | 8 | 9 | 1 | 2 | | B:B:Q75 | B:B:W99 | 9.86 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | B:B:L117 | B:B:M101 | 5.65 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | B:B:?145 | B:B:M101 | 5.5 | 1 | Yes | Yes | 8 | 9 | 2 | 1 | | B:B:?116 | B:B:L117 | 4.83 | 1 | No | Yes | 9 | 9 | 2 | 1 | | B:B:?116 | B:B:?145 | 5.87 | 1 | No | Yes | 9 | 8 | 2 | 2 | | B:B:?145 | B:B:L117 | 5.38 | 1 | Yes | Yes | 8 | 9 | 2 | 1 | | B:B:?317 | B:B:S316 | 4.82 | 1 | Yes | No | 7 | 9 | 2 | 2 | | B:B:S316 | B:B:W332 | 6.18 | 1 | No | Yes | 9 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 8.72 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 25.00 | | Average Links Mediated by Hubs In Shell | 24.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:N278 | A:A:W277 | 4.52 | 0 | No | Yes | 8 | 6 | 2 | 1 | | A:A:V287 | A:A:W277 | 4.9 | 0 | No | Yes | 5 | 6 | 2 | 1 | | A:A:S349 | A:A:W277 | 4.94 | 3 | No | Yes | 8 | 6 | 1 | 1 | | A:A:H357 | A:A:W277 | 29.62 | 3 | No | Yes | 7 | 6 | 2 | 1 | | A:A:?358 | A:A:W277 | 6.65 | 3 | Yes | Yes | 4 | 6 | 1 | 1 | | A:A:?359 | A:A:W277 | 7.32 | 3 | Yes | Yes | 6 | 6 | 0 | 1 | | A:A:?358 | A:A:S286 | 11.68 | 3 | Yes | No | 4 | 8 | 1 | 2 | | A:A:?360 | A:A:S286 | 5.84 | 3 | Yes | No | 8 | 8 | 1 | 2 | | A:A:I382 | A:A:S286 | 9.29 | 3 | Yes | No | 7 | 8 | 2 | 2 | | A:A:?360 | A:A:L289 | 5.38 | 3 | Yes | No | 8 | 9 | 1 | 2 | | A:A:?359 | A:A:S349 | 4.82 | 3 | Yes | No | 6 | 8 | 0 | 1 | | A:A:?358 | A:A:D354 | 7.92 | 3 | Yes | No | 4 | 3 | 1 | 2 | | A:A:?358 | A:A:H357 | 5 | 3 | Yes | No | 4 | 7 | 1 | 2 | | A:A:?358 | A:A:?359 | 21.62 | 3 | Yes | Yes | 4 | 6 | 1 | 0 | | A:A:?358 | A:A:?360 | 17.02 | 3 | Yes | Yes | 4 | 8 | 1 | 1 | | A:A:?358 | A:A:R385 | 11.82 | 3 | Yes | No | 4 | 6 | 1 | 2 | | A:A:?359 | A:A:?360 | 40.37 | 3 | Yes | Yes | 6 | 8 | 0 | 1 | | A:A:?360 | A:A:P361 | 14.37 | 3 | Yes | No | 8 | 4 | 1 | 2 | | A:A:?360 | A:A:D378 | 15.84 | 3 | Yes | No | 8 | 7 | 1 | 2 | | A:A:?360 | A:A:D381 | 11.88 | 3 | Yes | No | 8 | 8 | 1 | 2 | | A:A:?360 | A:A:I382 | 8.33 | 3 | Yes | Yes | 8 | 7 | 1 | 2 | | A:A:D381 | A:A:R385 | 13.1 | 3 | No | No | 8 | 6 | 2 | 2 | | A:A:S349 | A:A:T350 | 3.2 | 3 | No | No | 8 | 4 | 1 | 2 | | A:A:?358 | R:R:A560 | 3.06 | 3 | Yes | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 18.53 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 24.00 | | Average Links Mediated by Hubs In Shell | 22.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
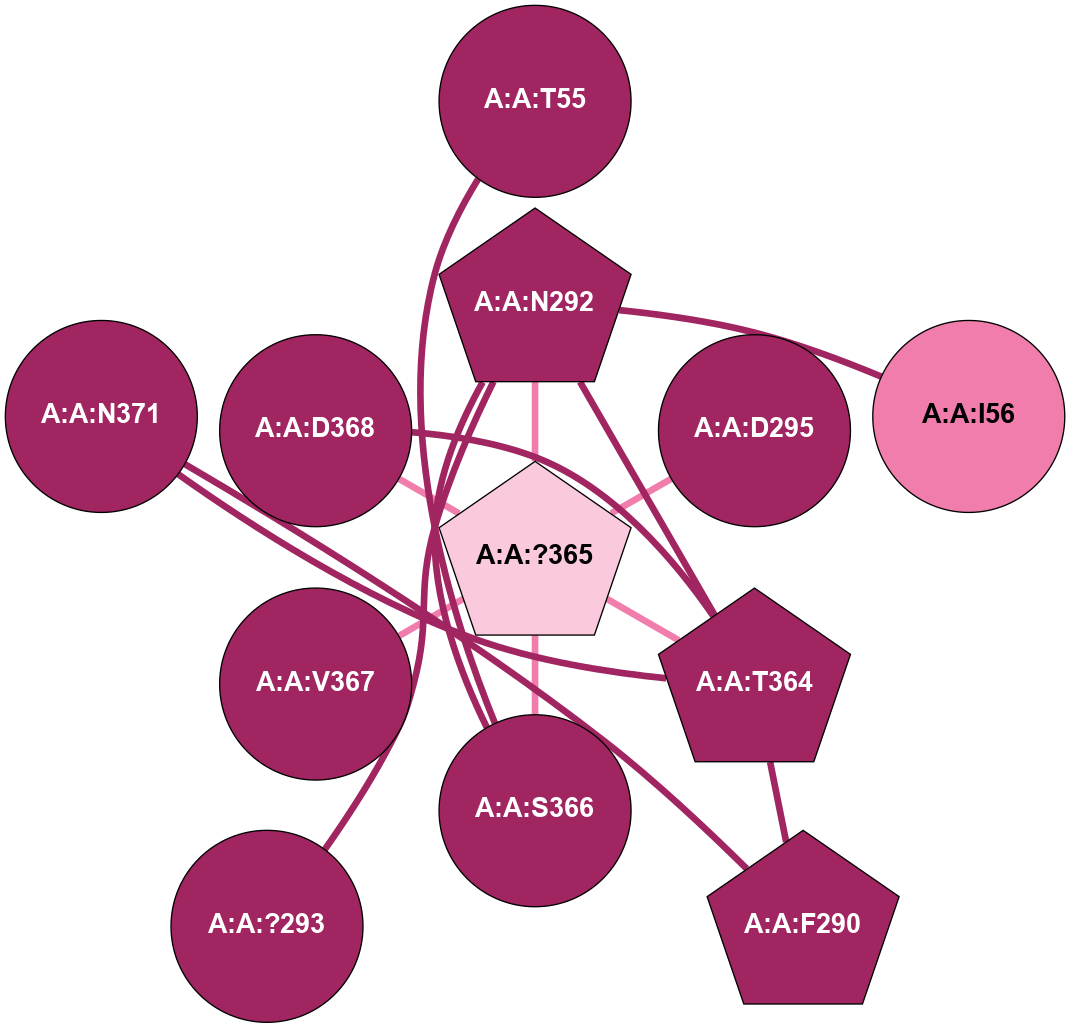
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:S366 | A:A:T55 | 9.59 | 8 | No | No | 9 | 9 | 1 | 2 | | A:A:I56 | A:A:N292 | 5.66 | 0 | No | Yes | 8 | 9 | 2 | 1 | | A:A:F290 | A:A:T364 | 7.78 | 8 | Yes | Yes | 9 | 9 | 2 | 1 | | A:A:F290 | A:A:N371 | 10.87 | 8 | Yes | No | 9 | 9 | 2 | 2 | | A:A:?293 | A:A:N292 | 8.19 | 0 | No | Yes | 9 | 9 | 2 | 1 | | A:A:N292 | A:A:T364 | 4.39 | 8 | Yes | Yes | 9 | 9 | 1 | 1 | | A:A:?365 | A:A:N292 | 4.41 | 8 | Yes | Yes | 7 | 9 | 0 | 1 | | A:A:N292 | A:A:S366 | 4.47 | 8 | Yes | No | 9 | 9 | 1 | 1 | | A:A:?365 | A:A:D295 | 7.27 | 8 | Yes | No | 7 | 9 | 0 | 1 | | A:A:?365 | A:A:T364 | 14.21 | 8 | Yes | Yes | 7 | 9 | 0 | 1 | | A:A:D368 | A:A:T364 | 7.23 | 8 | No | Yes | 9 | 9 | 1 | 1 | | A:A:N371 | A:A:T364 | 11.7 | 8 | No | Yes | 9 | 9 | 2 | 1 | | A:A:?365 | A:A:S366 | 4.82 | 8 | Yes | No | 7 | 9 | 0 | 1 | | A:A:?365 | A:A:V367 | 4.79 | 8 | Yes | No | 7 | 9 | 0 | 1 | | A:A:?365 | A:A:D368 | 20.35 | 8 | Yes | No | 7 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.31 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
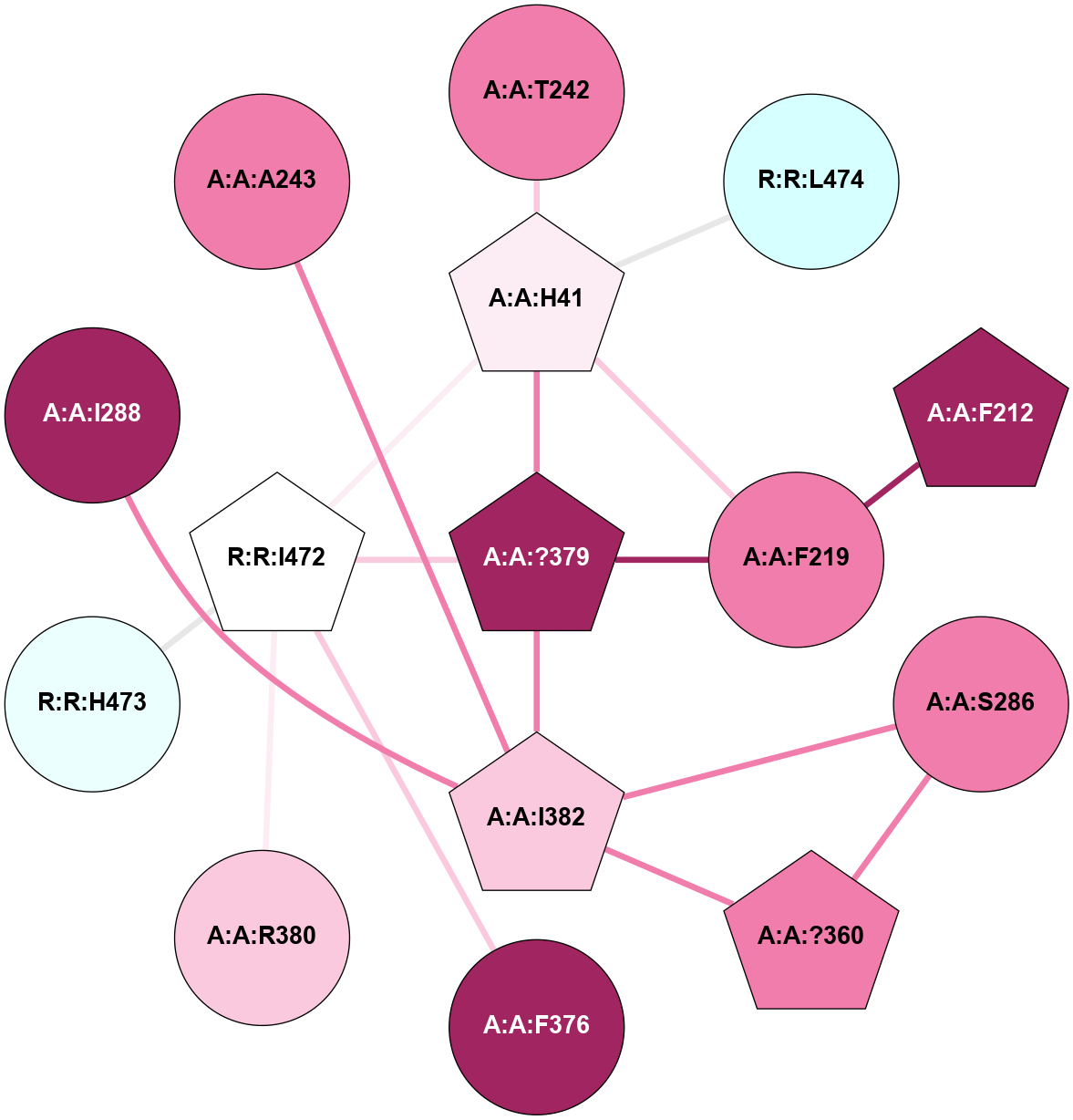
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:F219 | A:A:H41 | 5.66 | 3 | No | Yes | 8 | 6 | 1 | 1 | | A:A:H41 | A:A:T242 | 5.48 | 3 | Yes | No | 6 | 8 | 1 | 2 | | A:A:?379 | A:A:H41 | 4.13 | 3 | Yes | Yes | 9 | 6 | 0 | 1 | | A:A:H41 | R:R:I472 | 7.95 | 3 | Yes | Yes | 6 | 5 | 1 | 1 | | A:A:H41 | R:R:L474 | 5.14 | 3 | Yes | No | 6 | 3 | 1 | 2 | | A:A:F212 | A:A:F219 | 17.15 | 0 | Yes | No | 9 | 8 | 2 | 1 | | A:A:?379 | A:A:F219 | 5.22 | 3 | Yes | No | 9 | 8 | 0 | 1 | | A:A:?360 | A:A:S286 | 5.84 | 3 | Yes | No | 8 | 8 | 2 | 2 | | A:A:I382 | A:A:S286 | 9.29 | 3 | Yes | No | 7 | 8 | 1 | 2 | | A:A:?360 | A:A:I382 | 8.33 | 3 | Yes | Yes | 8 | 7 | 2 | 1 | | A:A:F376 | R:R:I472 | 7.54 | 3 | No | Yes | 9 | 5 | 2 | 1 | | A:A:?379 | A:A:I382 | 6.11 | 3 | Yes | Yes | 9 | 7 | 0 | 1 | | A:A:?379 | R:R:I472 | 6.11 | 3 | Yes | Yes | 9 | 5 | 0 | 1 | | A:A:R380 | R:R:I472 | 7.52 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:H473 | R:R:I472 | 5.3 | 0 | No | Yes | 4 | 5 | 2 | 1 | | A:A:I288 | A:A:I382 | 2.94 | 0 | No | Yes | 9 | 7 | 2 | 1 | | A:A:A243 | A:A:I382 | 1.62 | 0 | No | Yes | 8 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 5.39 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
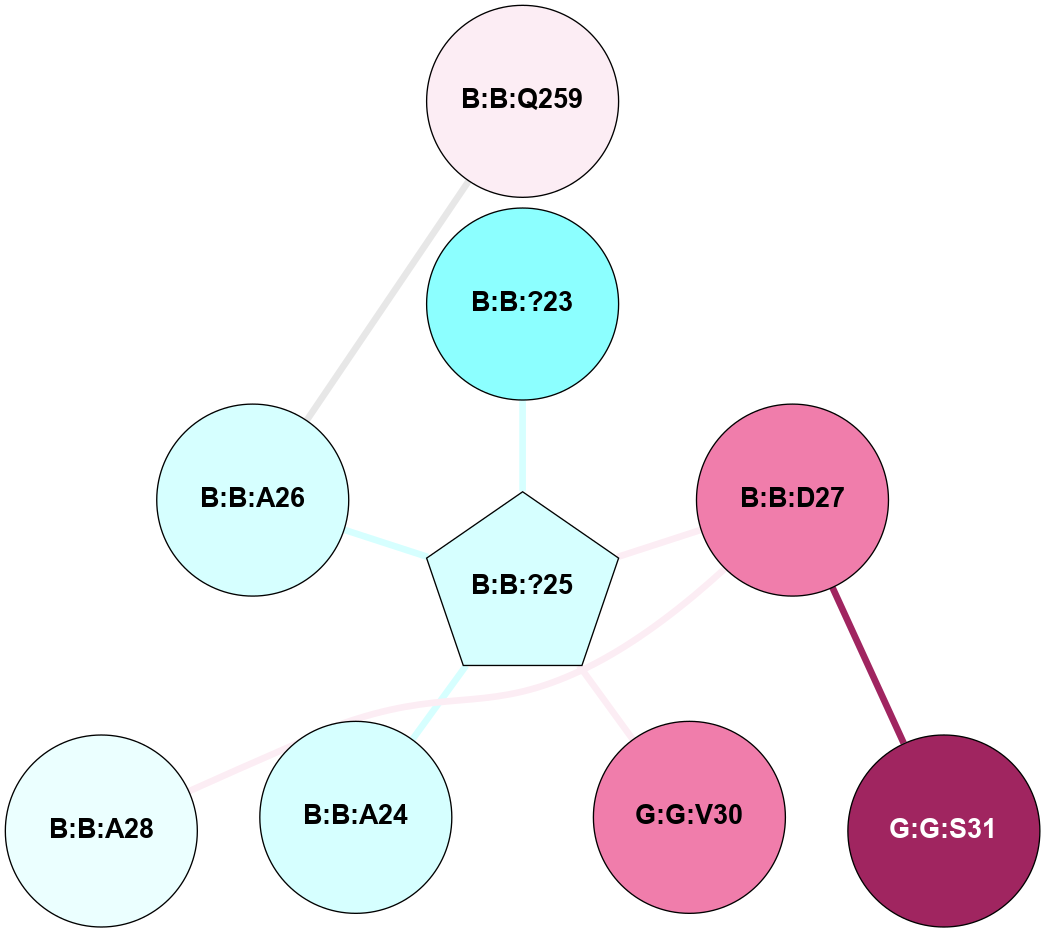
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:?23 | B:B:?25 | 4.42 | 0 | No | Yes | 2 | 3 | 1 | 0 | | B:B:?25 | B:B:D27 | 5.81 | 0 | Yes | No | 3 | 8 | 0 | 1 | | B:B:?25 | G:G:V30 | 4.79 | 0 | Yes | No | 3 | 8 | 0 | 1 | | B:B:A26 | B:B:Q259 | 4.55 | 0 | No | No | 3 | 6 | 1 | 2 | | B:B:D27 | G:G:S31 | 8.83 | 0 | No | No | 8 | 9 | 1 | 2 | | B:B:?25 | B:B:A24 | 3.37 | 0 | Yes | No | 3 | 3 | 0 | 1 | | B:B:?25 | B:B:A26 | 3.37 | 0 | Yes | No | 3 | 3 | 0 | 1 | | B:B:A28 | B:B:D27 | 3.09 | 0 | No | No | 4 | 8 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 4.35 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
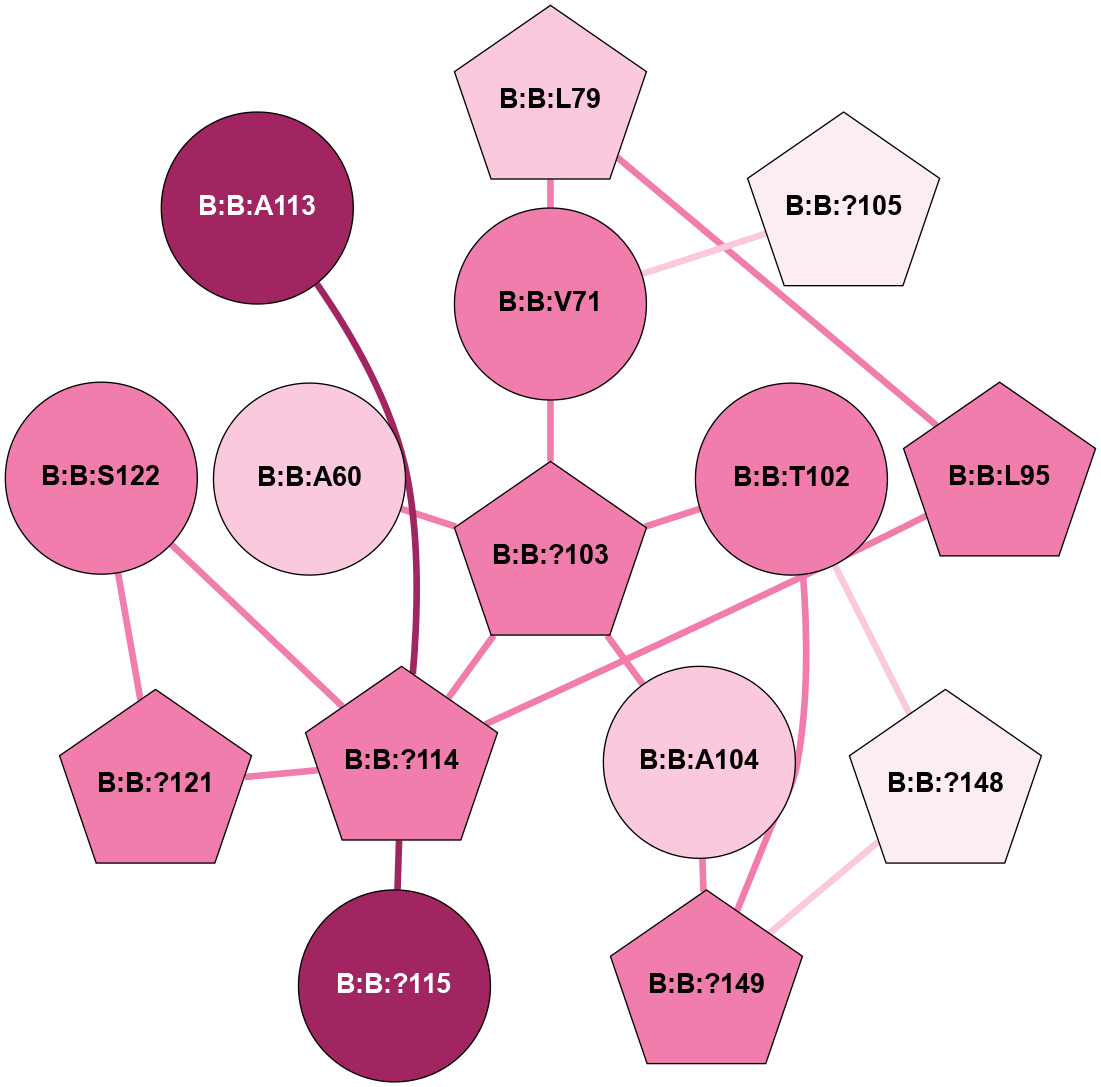
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:L79 | B:B:V71 | 5.96 | 0 | Yes | No | 7 | 8 | 2 | 1 | | B:B:?103 | B:B:V71 | 4.79 | 0 | Yes | No | 8 | 8 | 0 | 1 | | B:B:?105 | B:B:V71 | 10.14 | 5 | Yes | No | 6 | 8 | 2 | 1 | | B:B:L79 | B:B:L95 | 4.15 | 0 | Yes | Yes | 7 | 8 | 2 | 2 | | B:B:?114 | B:B:L95 | 4.45 | 5 | Yes | Yes | 8 | 8 | 1 | 2 | | B:B:?103 | B:B:T102 | 11.05 | 0 | Yes | No | 8 | 8 | 0 | 1 | | B:B:?148 | B:B:T102 | 18.94 | 5 | Yes | No | 6 | 8 | 2 | 1 | | B:B:?149 | B:B:T102 | 11.05 | 5 | Yes | No | 8 | 8 | 2 | 1 | | B:B:?103 | B:B:A104 | 5.06 | 0 | Yes | No | 8 | 7 | 0 | 1 | | B:B:?103 | B:B:?114 | 7.94 | 0 | Yes | Yes | 8 | 8 | 0 | 1 | | B:B:?149 | B:B:A104 | 5.06 | 5 | Yes | No | 8 | 7 | 2 | 1 | | B:B:?114 | B:B:?115 | 16.8 | 5 | Yes | No | 8 | 9 | 1 | 2 | | B:B:?114 | B:B:?121 | 6.35 | 5 | Yes | Yes | 8 | 8 | 1 | 2 | | B:B:?114 | B:B:S122 | 8.04 | 5 | Yes | No | 8 | 8 | 1 | 2 | | B:B:?121 | B:B:S122 | 6.43 | 5 | Yes | No | 8 | 8 | 2 | 2 | | B:B:?148 | B:B:?149 | 31.75 | 5 | Yes | Yes | 6 | 8 | 2 | 2 | | B:B:?103 | B:B:A60 | 3.37 | 0 | Yes | No | 8 | 7 | 0 | 1 | | B:B:?114 | B:B:A113 | 3.37 | 5 | Yes | No | 8 | 9 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.44 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 18.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
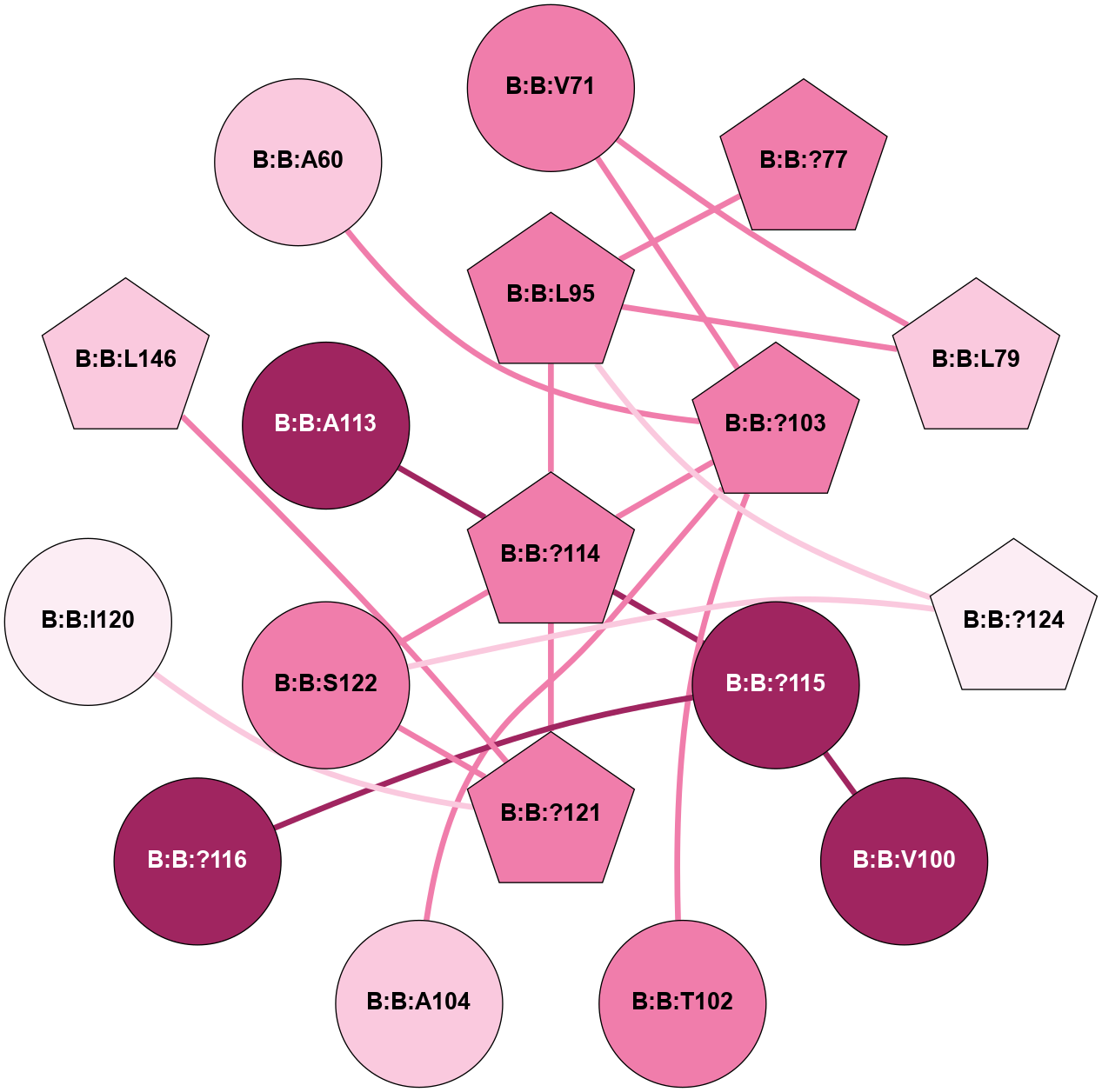
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:L79 | B:B:V71 | 5.96 | 0 | Yes | No | 7 | 8 | 2 | 2 | | B:B:?103 | B:B:V71 | 4.79 | 0 | Yes | No | 8 | 8 | 1 | 2 | | B:B:?77 | B:B:L95 | 9.66 | 1 | Yes | Yes | 8 | 8 | 2 | 1 | | B:B:L79 | B:B:L95 | 4.15 | 0 | Yes | Yes | 7 | 8 | 2 | 1 | | B:B:?114 | B:B:L95 | 4.45 | 5 | Yes | Yes | 8 | 8 | 0 | 1 | | B:B:?124 | B:B:L95 | 6.73 | 5 | Yes | Yes | 6 | 8 | 2 | 1 | | B:B:?115 | B:B:V100 | 5.2 | 0 | No | No | 9 | 9 | 1 | 2 | | B:B:?103 | B:B:T102 | 11.05 | 0 | Yes | No | 8 | 8 | 1 | 2 | | B:B:?103 | B:B:A104 | 5.06 | 0 | Yes | No | 8 | 7 | 1 | 2 | | B:B:?103 | B:B:?114 | 7.94 | 0 | Yes | Yes | 8 | 8 | 1 | 0 | | B:B:?114 | B:B:?115 | 16.8 | 5 | Yes | No | 8 | 9 | 0 | 1 | | B:B:?114 | B:B:?121 | 6.35 | 5 | Yes | Yes | 8 | 8 | 0 | 1 | | B:B:?114 | B:B:S122 | 8.04 | 5 | Yes | No | 8 | 8 | 0 | 1 | | B:B:?115 | B:B:?116 | 10.53 | 0 | No | No | 9 | 9 | 1 | 2 | | B:B:?121 | B:B:I120 | 9.17 | 5 | Yes | No | 8 | 6 | 1 | 2 | | B:B:?121 | B:B:S122 | 6.43 | 5 | Yes | No | 8 | 8 | 1 | 1 | | B:B:?121 | B:B:L146 | 5.93 | 5 | Yes | Yes | 8 | 7 | 1 | 2 | | B:B:?124 | B:B:S122 | 10.22 | 5 | Yes | No | 6 | 8 | 2 | 1 | | B:B:?103 | B:B:A60 | 3.37 | 0 | Yes | No | 8 | 7 | 1 | 2 | | B:B:?114 | B:B:A113 | 3.37 | 5 | Yes | No | 8 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 7.82 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 20.00 | | Average Links Mediated by Hubs In Shell | 18.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
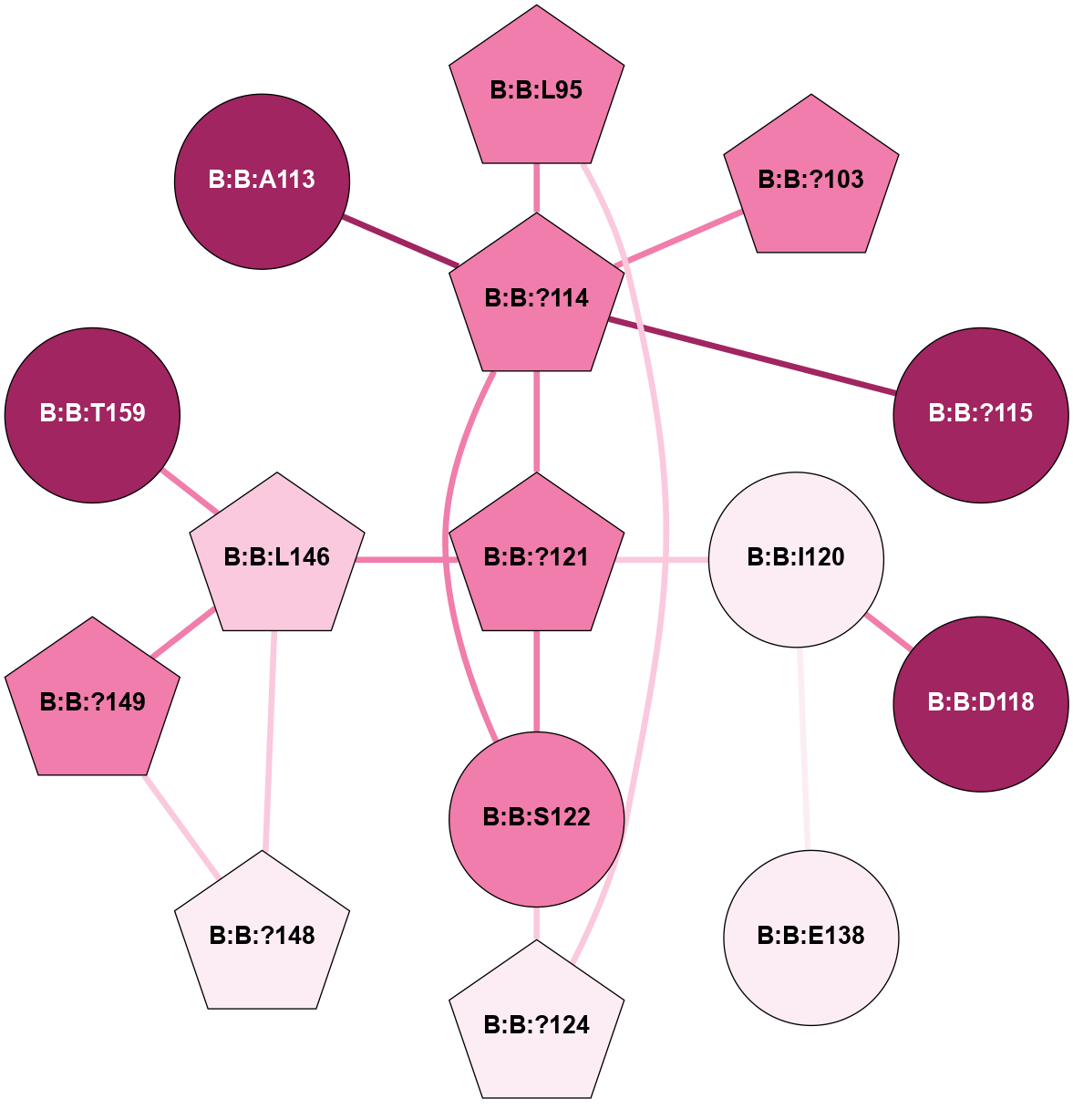
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:?114 | B:B:L95 | 4.45 | 5 | Yes | Yes | 8 | 8 | 1 | 2 | | B:B:?124 | B:B:L95 | 6.73 | 5 | Yes | Yes | 6 | 8 | 2 | 2 | | B:B:?103 | B:B:?114 | 7.94 | 0 | Yes | Yes | 8 | 8 | 2 | 1 | | B:B:?114 | B:B:?115 | 16.8 | 5 | Yes | No | 8 | 9 | 1 | 2 | | B:B:?114 | B:B:?121 | 6.35 | 5 | Yes | Yes | 8 | 8 | 1 | 0 | | B:B:?114 | B:B:S122 | 8.04 | 5 | Yes | No | 8 | 8 | 1 | 1 | | B:B:D118 | B:B:I120 | 9.8 | 0 | No | No | 9 | 6 | 2 | 1 | | B:B:?121 | B:B:I120 | 9.17 | 5 | Yes | No | 8 | 6 | 0 | 1 | | B:B:E138 | B:B:I120 | 13.66 | 0 | No | No | 6 | 6 | 2 | 1 | | B:B:?121 | B:B:S122 | 6.43 | 5 | Yes | No | 8 | 8 | 0 | 1 | | B:B:?121 | B:B:L146 | 5.93 | 5 | Yes | Yes | 8 | 7 | 0 | 1 | | B:B:?124 | B:B:S122 | 10.22 | 5 | Yes | No | 6 | 8 | 2 | 1 | | B:B:?148 | B:B:L146 | 5.93 | 5 | Yes | Yes | 6 | 7 | 2 | 1 | | B:B:?149 | B:B:L146 | 5.93 | 5 | Yes | Yes | 8 | 7 | 2 | 1 | | B:B:L146 | B:B:T159 | 8.84 | 5 | Yes | No | 7 | 9 | 1 | 2 | | B:B:?148 | B:B:?149 | 31.75 | 5 | Yes | Yes | 6 | 8 | 2 | 2 | | B:B:?114 | B:B:A113 | 3.37 | 5 | Yes | No | 8 | 9 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 6.97 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
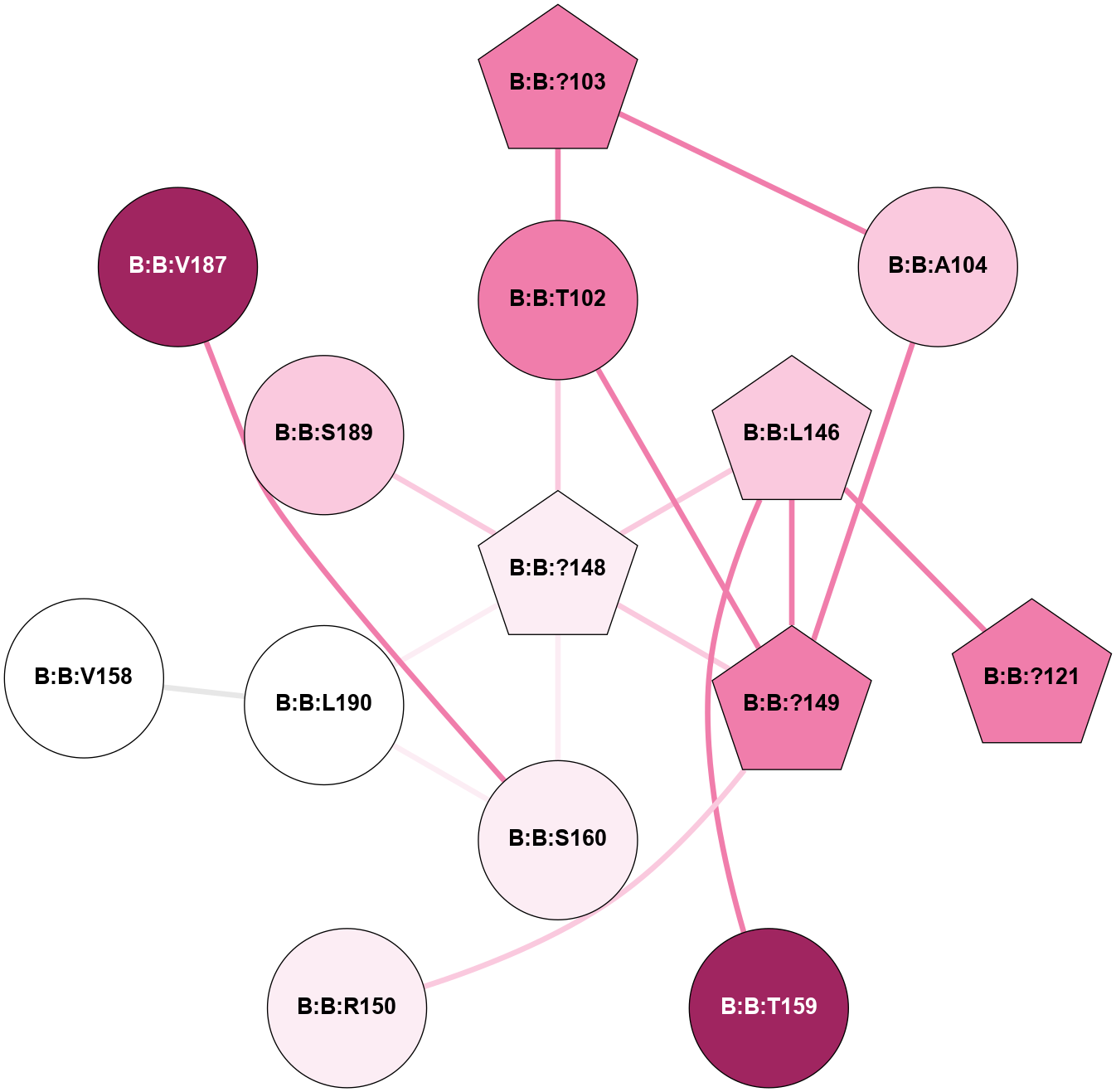
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:?103 | B:B:T102 | 11.05 | 0 | Yes | No | 8 | 8 | 2 | 1 | | B:B:?148 | B:B:T102 | 18.94 | 5 | Yes | No | 6 | 8 | 0 | 1 | | B:B:?149 | B:B:T102 | 11.05 | 5 | Yes | No | 8 | 8 | 1 | 1 | | B:B:?103 | B:B:A104 | 5.06 | 0 | Yes | No | 8 | 7 | 2 | 2 | | B:B:?149 | B:B:A104 | 5.06 | 5 | Yes | No | 8 | 7 | 1 | 2 | | B:B:?121 | B:B:L146 | 5.93 | 5 | Yes | Yes | 8 | 7 | 2 | 1 | | B:B:?148 | B:B:L146 | 5.93 | 5 | Yes | Yes | 6 | 7 | 0 | 1 | | B:B:?149 | B:B:L146 | 5.93 | 5 | Yes | Yes | 8 | 7 | 1 | 1 | | B:B:L146 | B:B:T159 | 8.84 | 5 | Yes | No | 7 | 9 | 1 | 2 | | B:B:?148 | B:B:?149 | 31.75 | 5 | Yes | Yes | 6 | 8 | 0 | 1 | | B:B:?148 | B:B:S160 | 8.04 | 5 | Yes | No | 6 | 6 | 0 | 1 | | B:B:?148 | B:B:L190 | 5.93 | 5 | Yes | No | 6 | 5 | 0 | 1 | | B:B:?149 | B:B:R150 | 14.31 | 5 | Yes | No | 8 | 6 | 1 | 2 | | B:B:L190 | B:B:V158 | 4.47 | 5 | No | No | 5 | 5 | 1 | 2 | | B:B:S160 | B:B:V187 | 8.08 | 5 | No | No | 6 | 9 | 1 | 2 | | B:B:L190 | B:B:S160 | 6.01 | 5 | No | No | 5 | 6 | 1 | 1 | | B:B:?148 | B:B:S189 | 3.22 | 5 | Yes | No | 6 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 12.30 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:H62 | B:B:R150 | 6.77 | 0 | No | No | 6 | 6 | 2 | 1 | | B:B:?103 | B:B:T102 | 11.05 | 0 | Yes | No | 8 | 8 | 2 | 1 | | B:B:?148 | B:B:T102 | 18.94 | 5 | Yes | No | 6 | 8 | 1 | 1 | | B:B:?149 | B:B:T102 | 11.05 | 5 | Yes | No | 8 | 8 | 0 | 1 | | B:B:?103 | B:B:A104 | 5.06 | 0 | Yes | No | 8 | 7 | 2 | 1 | | B:B:?149 | B:B:A104 | 5.06 | 5 | Yes | No | 8 | 7 | 0 | 1 | | B:B:?121 | B:B:L146 | 5.93 | 5 | Yes | Yes | 8 | 7 | 2 | 1 | | B:B:?148 | B:B:L146 | 5.93 | 5 | Yes | Yes | 6 | 7 | 1 | 1 | | B:B:?149 | B:B:L146 | 5.93 | 5 | Yes | Yes | 8 | 7 | 0 | 1 | | B:B:L146 | B:B:T159 | 8.84 | 5 | Yes | No | 7 | 9 | 1 | 2 | | B:B:?148 | B:B:?149 | 31.75 | 5 | Yes | Yes | 6 | 8 | 1 | 0 | | B:B:?148 | B:B:S160 | 8.04 | 5 | Yes | No | 6 | 6 | 1 | 2 | | B:B:?148 | B:B:L190 | 5.93 | 5 | Yes | No | 6 | 5 | 1 | 2 | | B:B:?149 | B:B:R150 | 14.31 | 5 | Yes | No | 8 | 6 | 0 | 1 | | B:B:L192 | B:B:R150 | 4.86 | 0 | No | No | 5 | 6 | 2 | 1 | | B:B:L190 | B:B:S160 | 6.01 | 5 | No | No | 5 | 6 | 2 | 2 | | B:B:?148 | B:B:S189 | 3.22 | 5 | Yes | No | 6 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 13.62 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
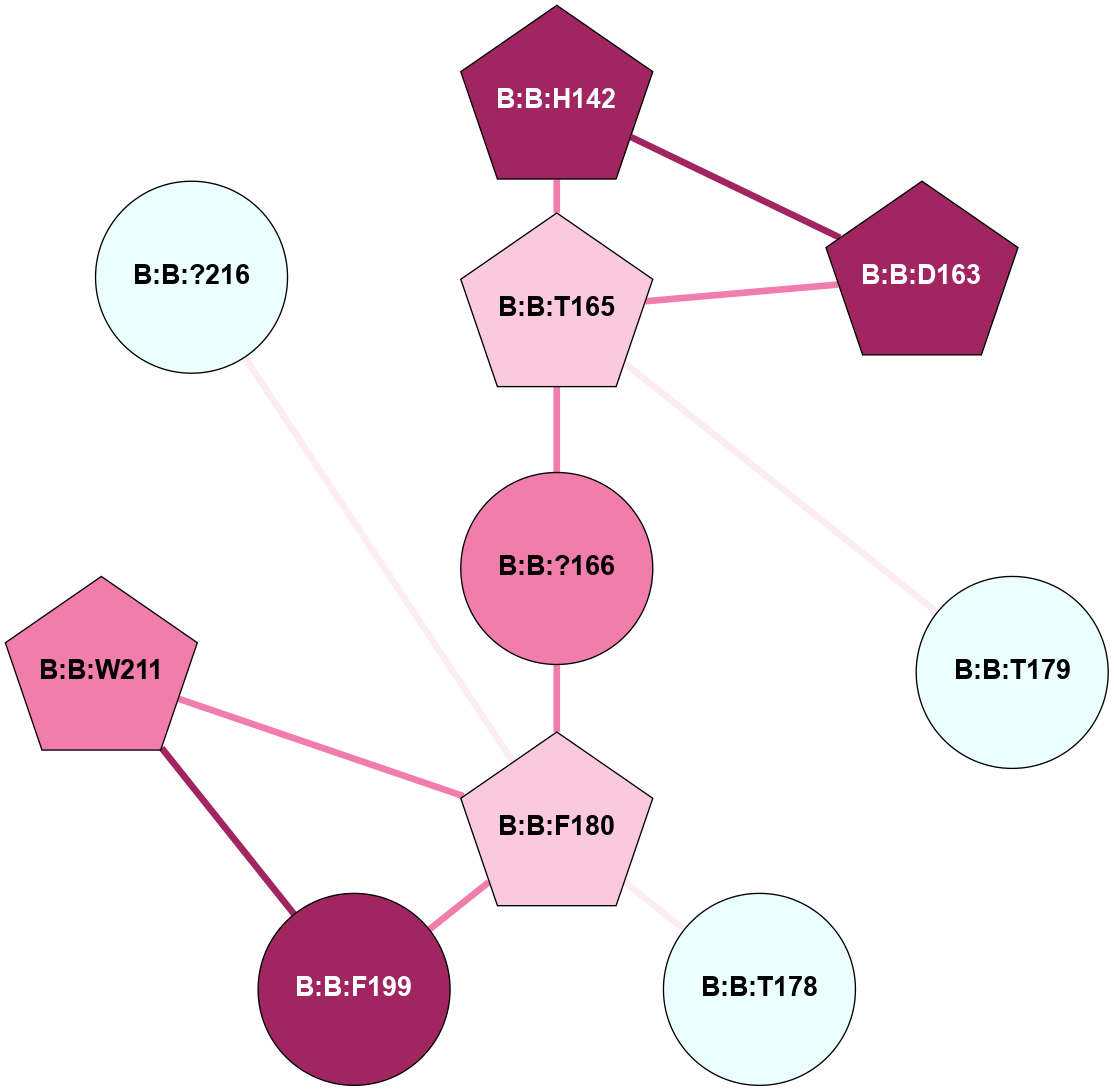
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:D163 | B:B:H142 | 11.35 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | B:B:H142 | B:B:T165 | 5.48 | 1 | Yes | Yes | 9 | 7 | 2 | 1 | | B:B:D163 | B:B:T165 | 10.12 | 1 | Yes | Yes | 9 | 7 | 2 | 1 | | B:B:?166 | B:B:T165 | 11.05 | 0 | No | Yes | 8 | 7 | 0 | 1 | | B:B:T165 | B:B:T179 | 4.71 | 1 | Yes | No | 7 | 4 | 1 | 2 | | B:B:?166 | B:B:F180 | 11.74 | 0 | No | Yes | 8 | 7 | 0 | 1 | | B:B:F180 | B:B:T178 | 7.78 | 1 | Yes | No | 7 | 4 | 1 | 2 | | B:B:F180 | B:B:F199 | 12.86 | 1 | Yes | No | 7 | 9 | 1 | 2 | | B:B:F180 | B:B:W211 | 15.03 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | B:B:?216 | B:B:F180 | 6.37 | 0 | No | Yes | 4 | 7 | 2 | 1 | | B:B:F199 | B:B:W211 | 14.03 | 1 | No | Yes | 9 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 11.39 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:E230 | A:A:R228 | 11.63 | 1 | No | Yes | 9 | 9 | 2 | 2 | | A:A:R228 | B:B:?162 | 7.42 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | A:A:R228 | B:B:?185 | 8.48 | 1 | Yes | Yes | 9 | 5 | 2 | 2 | | A:A:R228 | B:B:D186 | 4.76 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | A:A:E230 | A:A:R232 | 4.65 | 1 | No | Yes | 9 | 8 | 2 | 1 | | A:A:E230 | B:B:?185 | 5.78 | 1 | No | Yes | 9 | 5 | 2 | 2 | | A:A:?233 | A:A:R232 | 4.35 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | A:A:R232 | B:B:?204 | 10.41 | 1 | Yes | Yes | 8 | 7 | 1 | 0 | | A:A:R232 | B:B:D228 | 14.29 | 1 | Yes | No | 8 | 9 | 1 | 1 | | A:A:?233 | B:B:M188 | 6.75 | 1 | Yes | No | 9 | 8 | 2 | 1 | | A:A:?233 | B:B:D228 | 5.67 | 1 | Yes | No | 9 | 9 | 2 | 1 | | B:B:?145 | B:B:S147 | 8.76 | 1 | Yes | No | 8 | 9 | 2 | 2 | | B:B:?145 | B:B:?162 | 16.44 | 1 | Yes | Yes | 8 | 9 | 2 | 2 | | B:B:?145 | B:B:D186 | 13.2 | 1 | Yes | Yes | 8 | 9 | 2 | 1 | | B:B:M188 | B:B:S147 | 4.6 | 0 | No | No | 8 | 9 | 1 | 2 | | B:B:?162 | B:B:D186 | 11.84 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | B:B:A203 | B:B:H183 | 7.32 | 1 | No | Yes | 8 | 9 | 1 | 2 | | B:B:D205 | B:B:T184 | 5.78 | 1 | Yes | No | 9 | 3 | 1 | 2 | | B:B:?185 | B:B:D186 | 5.92 | 1 | Yes | Yes | 5 | 9 | 2 | 1 | | B:B:?204 | B:B:D186 | 5.81 | 1 | Yes | Yes | 7 | 9 | 0 | 1 | | B:B:?204 | B:B:M188 | 6.06 | 1 | Yes | No | 7 | 8 | 0 | 1 | | B:B:?204 | B:B:A203 | 5.06 | 1 | Yes | No | 7 | 8 | 0 | 1 | | B:B:A203 | B:B:D205 | 6.18 | 1 | No | Yes | 8 | 9 | 1 | 1 | | B:B:?204 | B:B:D205 | 4.36 | 1 | Yes | Yes | 7 | 9 | 0 | 1 | | B:B:?204 | B:B:D228 | 23.25 | 1 | Yes | No | 7 | 9 | 0 | 1 | | B:B:D205 | B:B:S207 | 8.83 | 1 | Yes | No | 9 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 9.16 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 26.00 | | Average Links Mediated by Hubs In Shell | 25.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
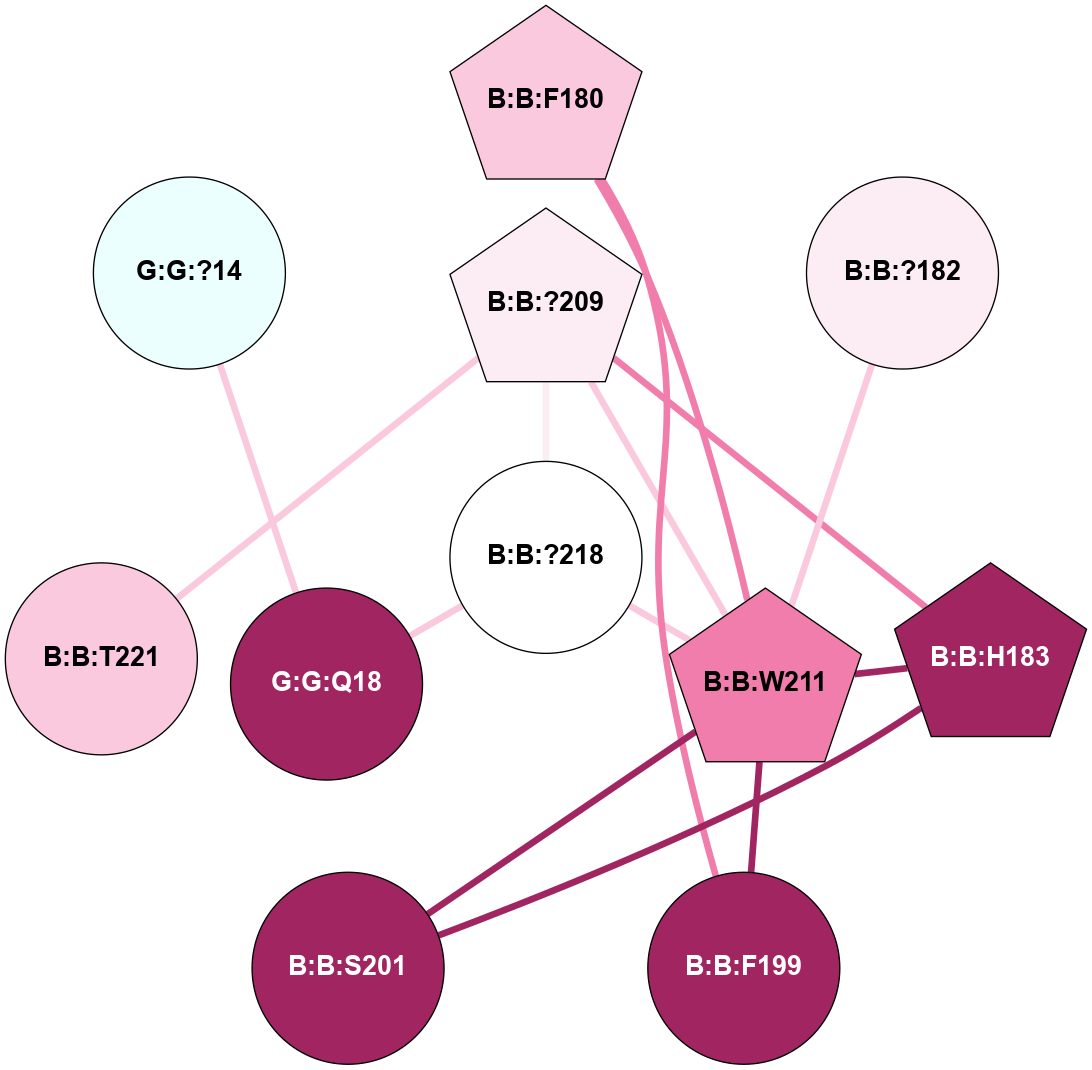
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:F180 | B:B:F199 | 12.86 | 1 | Yes | No | 7 | 9 | 2 | 2 | | B:B:F180 | B:B:W211 | 15.03 | 1 | Yes | Yes | 7 | 8 | 2 | 1 | | B:B:?182 | B:B:W211 | 4.97 | 0 | No | Yes | 6 | 8 | 2 | 1 | | B:B:H183 | B:B:S201 | 5.58 | 1 | Yes | No | 9 | 9 | 2 | 2 | | B:B:?209 | B:B:H183 | 13.04 | 1 | Yes | Yes | 6 | 9 | 1 | 2 | | B:B:H183 | B:B:W211 | 4.23 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | B:B:F199 | B:B:W211 | 14.03 | 1 | No | Yes | 9 | 8 | 2 | 1 | | B:B:S201 | B:B:W211 | 7.41 | 1 | No | Yes | 9 | 8 | 2 | 1 | | B:B:?209 | B:B:W211 | 8.83 | 1 | Yes | Yes | 6 | 8 | 1 | 1 | | B:B:?209 | B:B:?218 | 6.19 | 1 | Yes | No | 6 | 5 | 1 | 0 | | B:B:?209 | B:B:T221 | 7.91 | 1 | Yes | No | 6 | 7 | 1 | 2 | | B:B:?218 | B:B:W211 | 15.86 | 1 | No | Yes | 5 | 8 | 0 | 1 | | B:B:?218 | G:G:Q18 | 4.28 | 1 | No | No | 5 | 9 | 0 | 1 | | G:G:?14 | G:G:Q18 | 6.35 | 0 | No | No | 4 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 8.78 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:?233 | B:B:S191 | 12.86 | 0 | Yes | No | 3 | 8 | 0 | 1 | | B:B:F234 | B:B:S191 | 7.93 | 0 | Yes | No | 5 | 8 | 2 | 1 | | B:B:I232 | B:B:V200 | 9.22 | 1 | Yes | Yes | 8 | 6 | 1 | 2 | | B:B:F234 | B:B:V200 | 11.8 | 0 | Yes | Yes | 5 | 6 | 2 | 2 | | B:B:F241 | B:B:V200 | 9.18 | 1 | Yes | Yes | 6 | 6 | 2 | 2 | | B:B:?233 | B:B:I232 | 6.11 | 0 | Yes | Yes | 3 | 8 | 0 | 1 | | B:B:F241 | B:B:I232 | 8.79 | 1 | Yes | Yes | 6 | 8 | 2 | 1 | | B:B:I232 | B:B:T243 | 4.56 | 1 | Yes | Yes | 8 | 8 | 1 | 2 | | B:B:?233 | B:B:S277 | 4.82 | 0 | Yes | No | 3 | 5 | 0 | 1 | | B:B:?233 | B:B:F278 | 7.83 | 0 | Yes | Yes | 3 | 7 | 0 | 1 | | B:B:F235 | B:B:P236 | 10.11 | 7 | Yes | Yes | 6 | 7 | 2 | 2 | | B:B:F235 | B:B:F278 | 6.43 | 7 | Yes | Yes | 6 | 7 | 2 | 1 | | B:B:?282 | B:B:F235 | 6.37 | 7 | Yes | Yes | 9 | 6 | 2 | 2 | | B:B:F278 | B:B:P236 | 4.33 | 7 | Yes | Yes | 7 | 7 | 1 | 2 | | B:B:?282 | B:B:P236 | 5.73 | 7 | Yes | Yes | 9 | 7 | 2 | 2 | | B:B:A242 | B:B:F278 | 4.16 | 7 | No | Yes | 4 | 7 | 2 | 1 | | B:B:A242 | B:B:L285 | 4.73 | 7 | No | No | 4 | 6 | 2 | 2 | | B:B:S277 | B:B:V320 | 4.85 | 0 | No | No | 5 | 7 | 1 | 2 | | B:B:?282 | B:B:F278 | 7.43 | 7 | Yes | Yes | 9 | 7 | 2 | 1 | | B:B:F278 | B:B:L285 | 13.4 | 7 | Yes | No | 7 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 7.90 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 20.00 | | Average Links Mediated by Hubs In Shell | 18.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
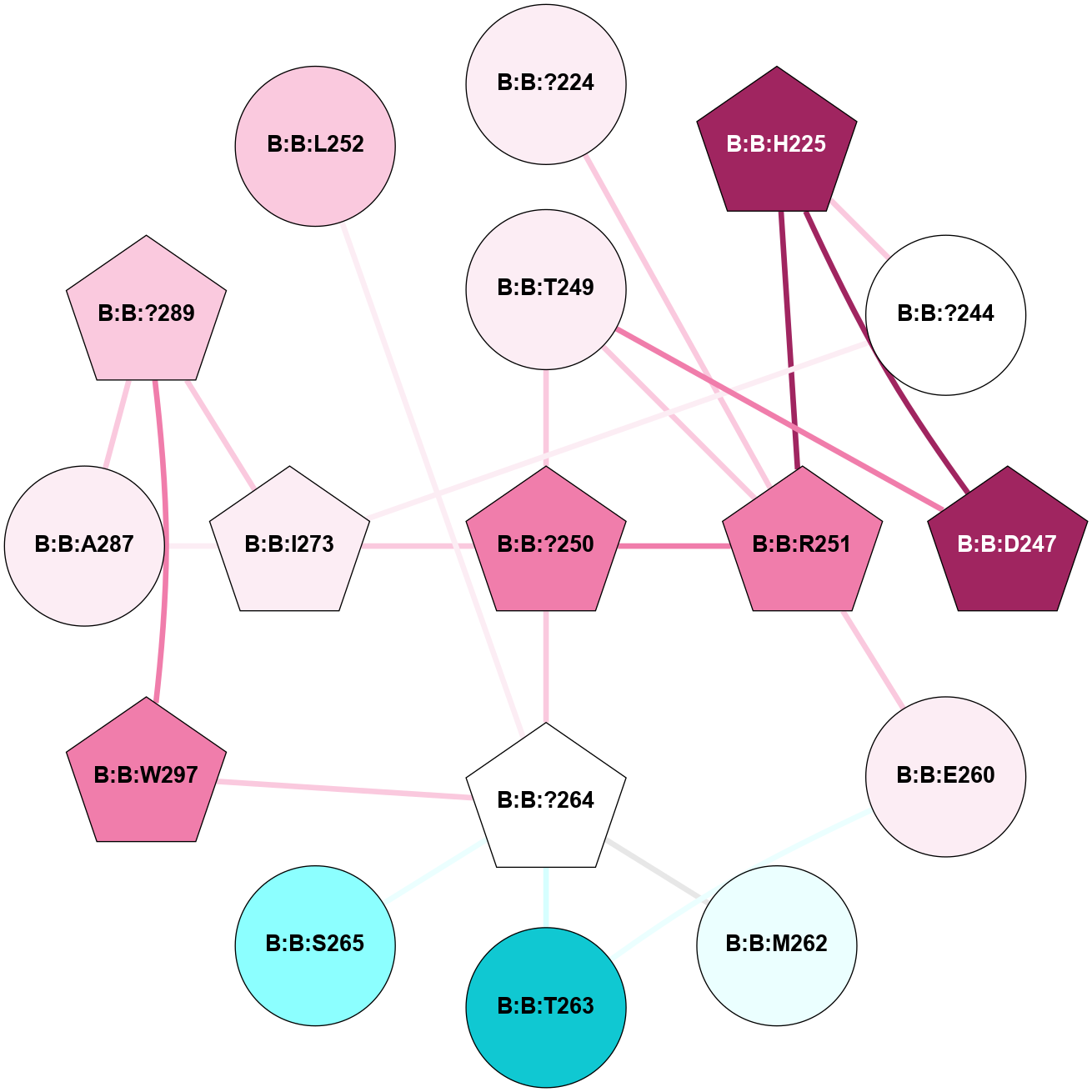
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:?224 | B:B:R251 | 12.71 | 0 | No | Yes | 6 | 8 | 2 | 1 | | B:B:?244 | B:B:H225 | 5.61 | 0 | No | Yes | 5 | 9 | 2 | 2 | | B:B:D247 | B:B:H225 | 10.08 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | B:B:H225 | B:B:R251 | 23.7 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | B:B:?244 | B:B:I273 | 7.47 | 0 | No | Yes | 5 | 6 | 2 | 1 | | B:B:D247 | B:B:T249 | 11.56 | 1 | Yes | No | 9 | 6 | 2 | 1 | | B:B:?250 | B:B:T249 | 12.63 | 1 | Yes | No | 8 | 6 | 0 | 1 | | B:B:R251 | B:B:T249 | 9.06 | 1 | Yes | No | 8 | 6 | 1 | 1 | | B:B:?250 | B:B:R251 | 6.5 | 1 | Yes | Yes | 8 | 8 | 0 | 1 | | B:B:?250 | B:B:?264 | 28.83 | 1 | Yes | Yes | 8 | 5 | 0 | 1 | | B:B:?250 | B:B:I273 | 6.11 | 1 | Yes | Yes | 8 | 6 | 0 | 1 | | B:B:E260 | B:B:R251 | 20.93 | 0 | No | Yes | 6 | 8 | 2 | 1 | | B:B:E260 | B:B:T263 | 9.88 | 0 | No | No | 6 | 1 | 2 | 2 | | B:B:?264 | B:B:M262 | 19.25 | 0 | Yes | No | 5 | 4 | 1 | 2 | | B:B:?264 | B:B:T263 | 8.6 | 0 | Yes | No | 5 | 1 | 1 | 2 | | B:B:?264 | B:B:S265 | 7.3 | 0 | Yes | No | 5 | 2 | 1 | 2 | | B:B:?264 | B:B:W297 | 22.15 | 0 | Yes | Yes | 5 | 8 | 1 | 2 | | B:B:A287 | B:B:I273 | 4.87 | 1 | No | Yes | 6 | 6 | 2 | 1 | | B:B:?289 | B:B:I273 | 12.49 | 1 | Yes | Yes | 7 | 6 | 2 | 1 | | B:B:?289 | B:B:A287 | 6.13 | 1 | Yes | No | 7 | 6 | 2 | 2 | | B:B:?289 | B:B:W297 | 5.54 | 1 | Yes | Yes | 7 | 8 | 2 | 2 | | B:B:?264 | B:B:L252 | 4.04 | 0 | Yes | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 13.52 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 22.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
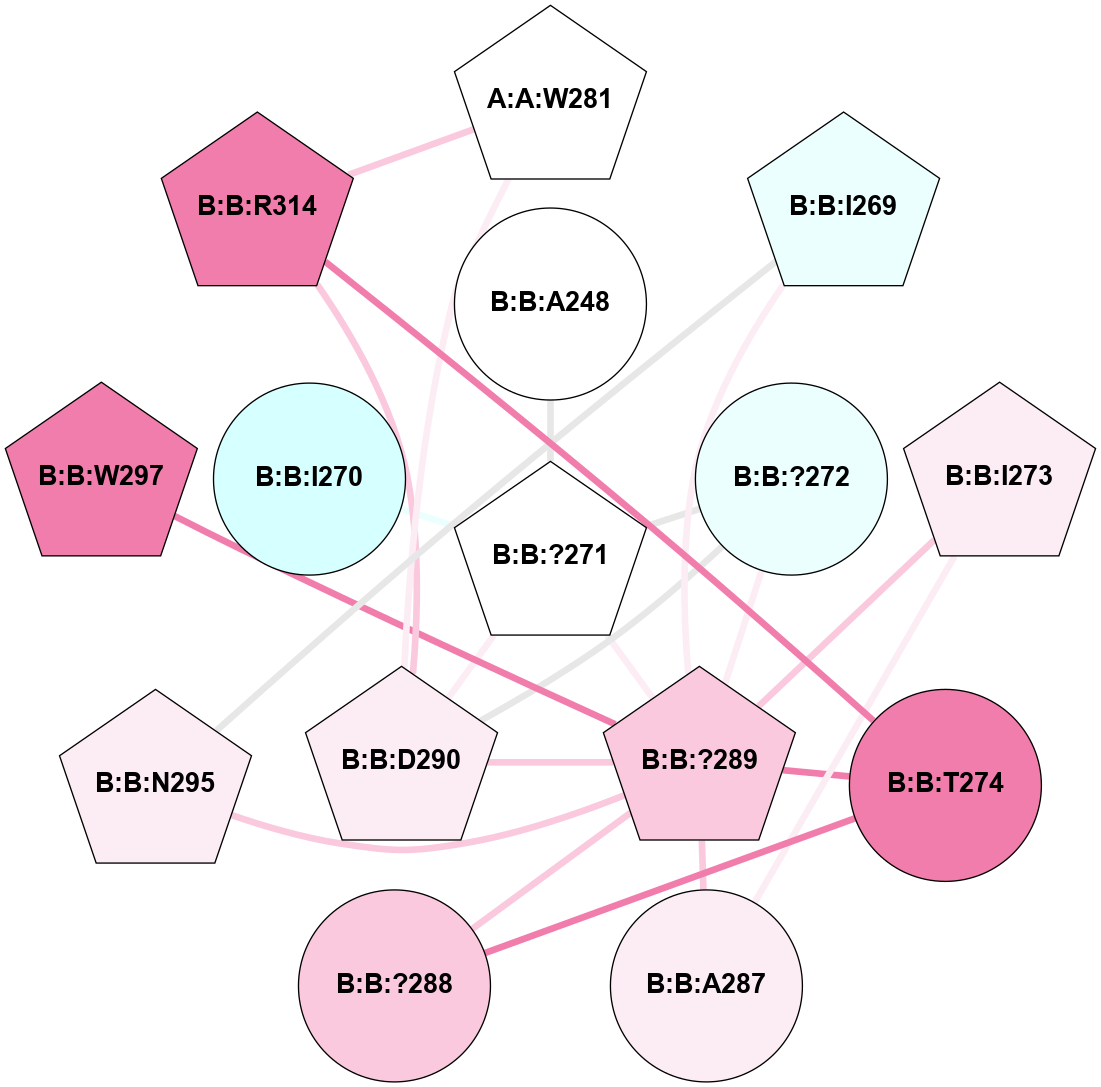
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:W281 | B:B:D290 | 8.93 | 1 | Yes | Yes | 5 | 6 | 2 | 1 | | A:A:W281 | B:B:R314 | 9 | 1 | Yes | Yes | 5 | 8 | 2 | 2 | | B:B:?271 | B:B:A248 | 5.06 | 1 | Yes | No | 5 | 5 | 0 | 1 | | B:B:?289 | B:B:I269 | 4.16 | 1 | Yes | Yes | 7 | 4 | 1 | 2 | | B:B:I269 | B:B:N295 | 4.25 | 1 | Yes | Yes | 4 | 6 | 2 | 2 | | B:B:?271 | B:B:?272 | 20.68 | 1 | Yes | No | 5 | 4 | 0 | 1 | | B:B:?271 | B:B:?289 | 11.53 | 1 | Yes | Yes | 5 | 7 | 0 | 1 | | B:B:?271 | B:B:D290 | 8.72 | 1 | Yes | Yes | 5 | 6 | 0 | 1 | | B:B:?272 | B:B:?289 | 7.04 | 1 | No | Yes | 4 | 7 | 1 | 1 | | B:B:?272 | B:B:D290 | 10.65 | 1 | No | Yes | 4 | 6 | 1 | 1 | | B:B:A287 | B:B:I273 | 4.87 | 1 | No | Yes | 6 | 6 | 2 | 2 | | B:B:?289 | B:B:I273 | 12.49 | 1 | Yes | Yes | 7 | 6 | 1 | 2 | | B:B:?288 | B:B:T274 | 6.43 | 1 | No | No | 7 | 8 | 2 | 2 | | B:B:?289 | B:B:T274 | 8.6 | 1 | Yes | No | 7 | 8 | 1 | 2 | | B:B:R314 | B:B:T274 | 11.64 | 1 | Yes | No | 8 | 8 | 2 | 2 | | B:B:?289 | B:B:A287 | 6.13 | 1 | Yes | No | 7 | 6 | 1 | 2 | | B:B:?288 | B:B:?289 | 27 | 1 | No | Yes | 7 | 7 | 2 | 1 | | B:B:?289 | B:B:D290 | 6.6 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | B:B:?289 | B:B:N295 | 18.7 | 1 | Yes | Yes | 7 | 6 | 1 | 2 | | B:B:?289 | B:B:W297 | 5.54 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | B:B:D290 | B:B:R314 | 8.34 | 1 | Yes | Yes | 6 | 8 | 1 | 2 | | B:B:?271 | B:B:I270 | 3.06 | 1 | Yes | No | 5 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.81 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 22.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
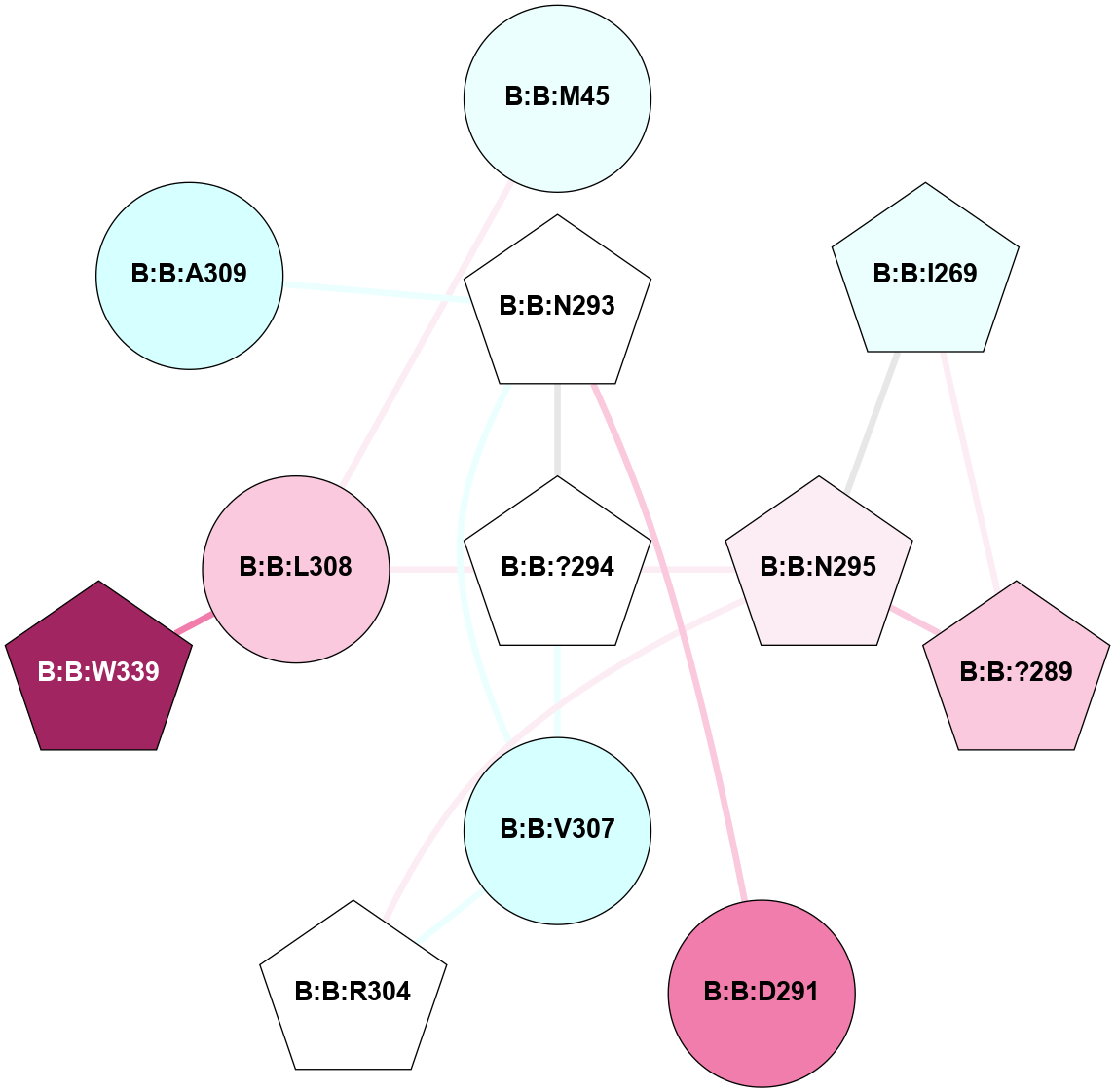
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:L308 | B:B:M45 | 4.24 | 0 | No | No | 7 | 4 | 1 | 2 | | B:B:?289 | B:B:I269 | 4.16 | 1 | Yes | Yes | 7 | 4 | 2 | 2 | | B:B:I269 | B:B:N295 | 4.25 | 1 | Yes | Yes | 4 | 6 | 2 | 1 | | B:B:?289 | B:B:N295 | 18.7 | 1 | Yes | Yes | 7 | 6 | 2 | 1 | | B:B:D291 | B:B:N293 | 12.12 | 0 | No | Yes | 8 | 5 | 2 | 1 | | B:B:?294 | B:B:N293 | 5.88 | 1 | Yes | Yes | 5 | 5 | 0 | 1 | | B:B:N293 | B:B:V307 | 4.43 | 1 | Yes | No | 5 | 3 | 1 | 1 | | B:B:?294 | B:B:N295 | 11.76 | 1 | Yes | Yes | 5 | 6 | 0 | 1 | | B:B:?294 | B:B:V307 | 4.79 | 1 | Yes | No | 5 | 3 | 0 | 1 | | B:B:?294 | B:B:L308 | 7.41 | 1 | Yes | No | 5 | 7 | 0 | 1 | | B:B:N295 | B:B:R304 | 12.05 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | B:B:R304 | B:B:V307 | 5.23 | 1 | Yes | No | 5 | 3 | 2 | 1 | | B:B:L308 | B:B:W339 | 19.36 | 0 | No | Yes | 7 | 9 | 1 | 2 | | B:B:A309 | B:B:N293 | 3.13 | 0 | No | Yes | 3 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 7.46 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
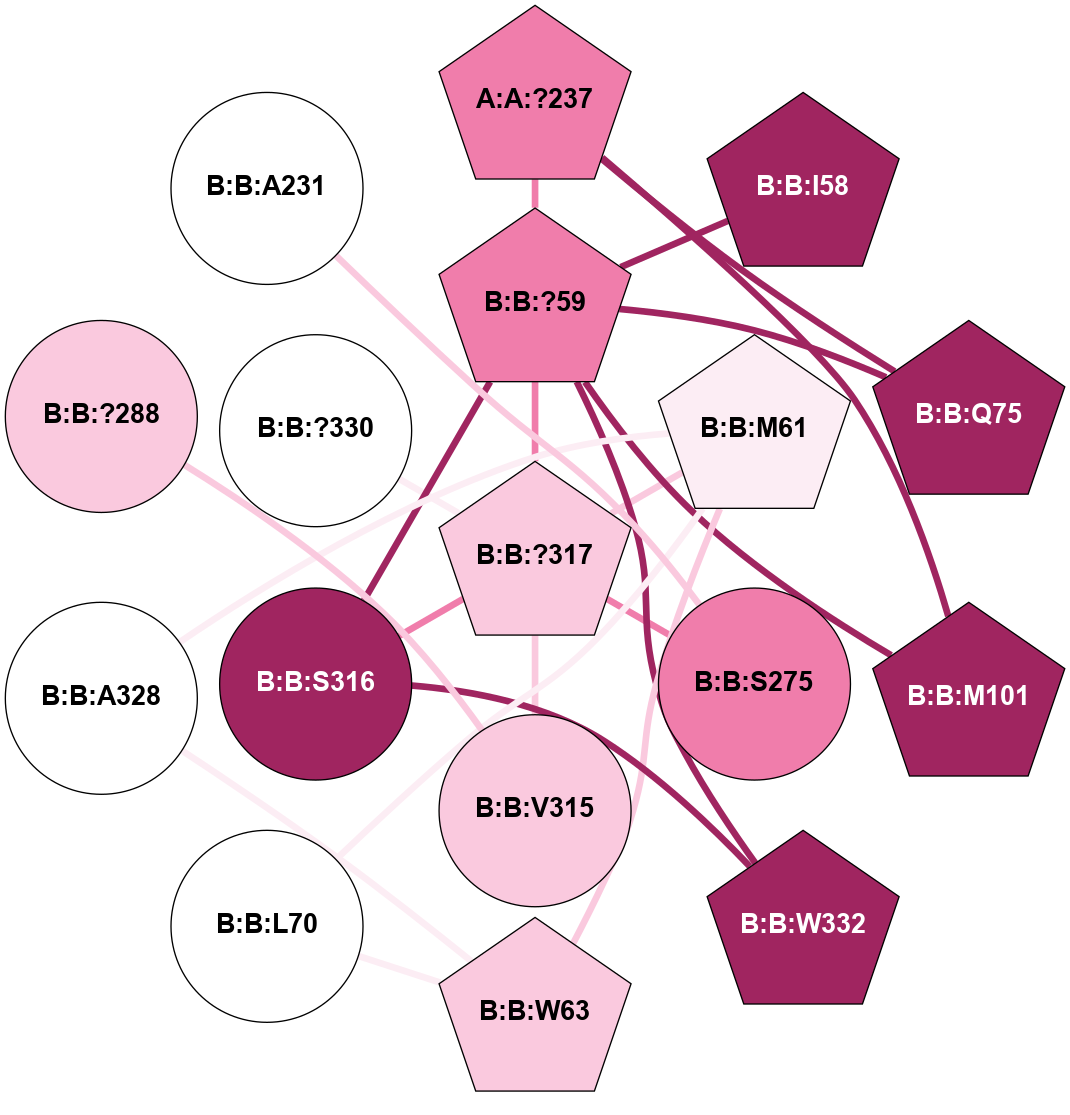
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:?237 | B:B:?59 | 7.21 | 1 | Yes | Yes | 8 | 8 | 2 | 1 | | A:A:?237 | B:B:Q75 | 17.1 | 1 | Yes | Yes | 8 | 9 | 2 | 2 | | A:A:?237 | B:B:M101 | 4.54 | 1 | Yes | Yes | 8 | 9 | 2 | 2 | | B:B:?59 | B:B:I58 | 5.55 | 1 | Yes | Yes | 8 | 9 | 1 | 2 | | B:B:?59 | B:B:Q75 | 19.42 | 1 | Yes | Yes | 8 | 9 | 1 | 2 | | B:B:?59 | B:B:M101 | 5.5 | 1 | Yes | Yes | 8 | 9 | 1 | 2 | | B:B:?59 | B:B:S316 | 20.45 | 1 | Yes | No | 8 | 9 | 1 | 1 | | B:B:?317 | B:B:?59 | 7.21 | 1 | Yes | Yes | 7 | 8 | 0 | 1 | | B:B:?59 | B:B:W332 | 7.75 | 1 | Yes | Yes | 8 | 9 | 1 | 2 | | B:B:M61 | B:B:W63 | 8.14 | 1 | Yes | Yes | 6 | 7 | 1 | 2 | | B:B:L70 | B:B:M61 | 5.65 | 1 | No | Yes | 5 | 6 | 2 | 1 | | B:B:?317 | B:B:M61 | 4.54 | 1 | Yes | Yes | 7 | 6 | 0 | 1 | | B:B:A328 | B:B:M61 | 4.83 | 1 | No | Yes | 5 | 6 | 2 | 1 | | B:B:L70 | B:B:W63 | 10.25 | 1 | No | Yes | 5 | 7 | 2 | 2 | | B:B:A328 | B:B:W63 | 5.19 | 1 | No | Yes | 5 | 7 | 2 | 2 | | B:B:?317 | B:B:S275 | 6.43 | 1 | Yes | No | 7 | 8 | 0 | 1 | | B:B:?288 | B:B:V315 | 5.2 | 1 | No | No | 7 | 7 | 2 | 1 | | B:B:?317 | B:B:V315 | 4.79 | 1 | Yes | No | 7 | 7 | 0 | 1 | | B:B:?317 | B:B:S316 | 4.82 | 1 | Yes | No | 7 | 9 | 0 | 1 | | B:B:S316 | B:B:W332 | 6.18 | 1 | No | Yes | 9 | 9 | 1 | 2 | | B:B:?317 | B:B:?330 | 18.1 | 1 | Yes | No | 7 | 5 | 0 | 1 | | B:B:A231 | B:B:S275 | 3.42 | 0 | No | No | 5 | 8 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 7.65 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 22.00 | | Average Links Mediated by Hubs In Shell | 20.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:I37 | B:B:R283 | 5.01 | 0 | No | Yes | 2 | 9 | 2 | 1 | | B:B:R283 | B:B:V40 | 7.85 | 7 | Yes | Yes | 9 | 3 | 1 | 2 | | B:B:D298 | B:B:V40 | 8.76 | 7 | No | Yes | 9 | 3 | 2 | 2 | | B:B:F235 | B:B:P236 | 10.11 | 7 | Yes | Yes | 6 | 7 | 2 | 2 | | B:B:F235 | B:B:N237 | 13.29 | 7 | Yes | Yes | 6 | 5 | 2 | 2 | | B:B:F235 | G:G:?40 | 10.66 | 7 | Yes | Yes | 6 | 6 | 2 | 1 | | B:B:P236 | G:G:?40 | 7.99 | 7 | Yes | Yes | 7 | 6 | 2 | 1 | | B:B:N237 | G:G:?40 | 14.69 | 7 | Yes | Yes | 5 | 6 | 2 | 1 | | B:B:D298 | B:B:R283 | 14.29 | 7 | No | Yes | 9 | 9 | 2 | 1 | | B:B:L300 | B:B:R283 | 12.15 | 7 | No | Yes | 6 | 9 | 1 | 1 | | B:B:R283 | G:G:?41 | 5.2 | 7 | Yes | Yes | 9 | 7 | 1 | 0 | | B:B:L300 | G:G:?41 | 5.93 | 7 | No | Yes | 6 | 7 | 1 | 0 | | G:G:?40 | G:G:?41 | 28.83 | 7 | Yes | Yes | 6 | 7 | 1 | 0 | | G:G:?40 | G:G:A43 | 4.6 | 7 | Yes | No | 6 | 4 | 1 | 2 | | G:G:?40 | G:G:H44 | 25.01 | 7 | Yes | No | 6 | 6 | 1 | 2 | | G:G:?41 | G:G:E42 | 4.26 | 7 | Yes | No | 7 | 1 | 0 | 1 | | G:G:A43 | G:G:H44 | 4.39 | 7 | No | No | 4 | 6 | 2 | 2 | | G:G:?40 | G:G:A39 | 3.06 | 7 | Yes | No | 6 | 3 | 1 | 2 | | G:G:?41 | G:G:A45 | 1.69 | 7 | Yes | No | 7 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.18 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 19.00 | | Average Links Mediated by Hubs In Shell | 18.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
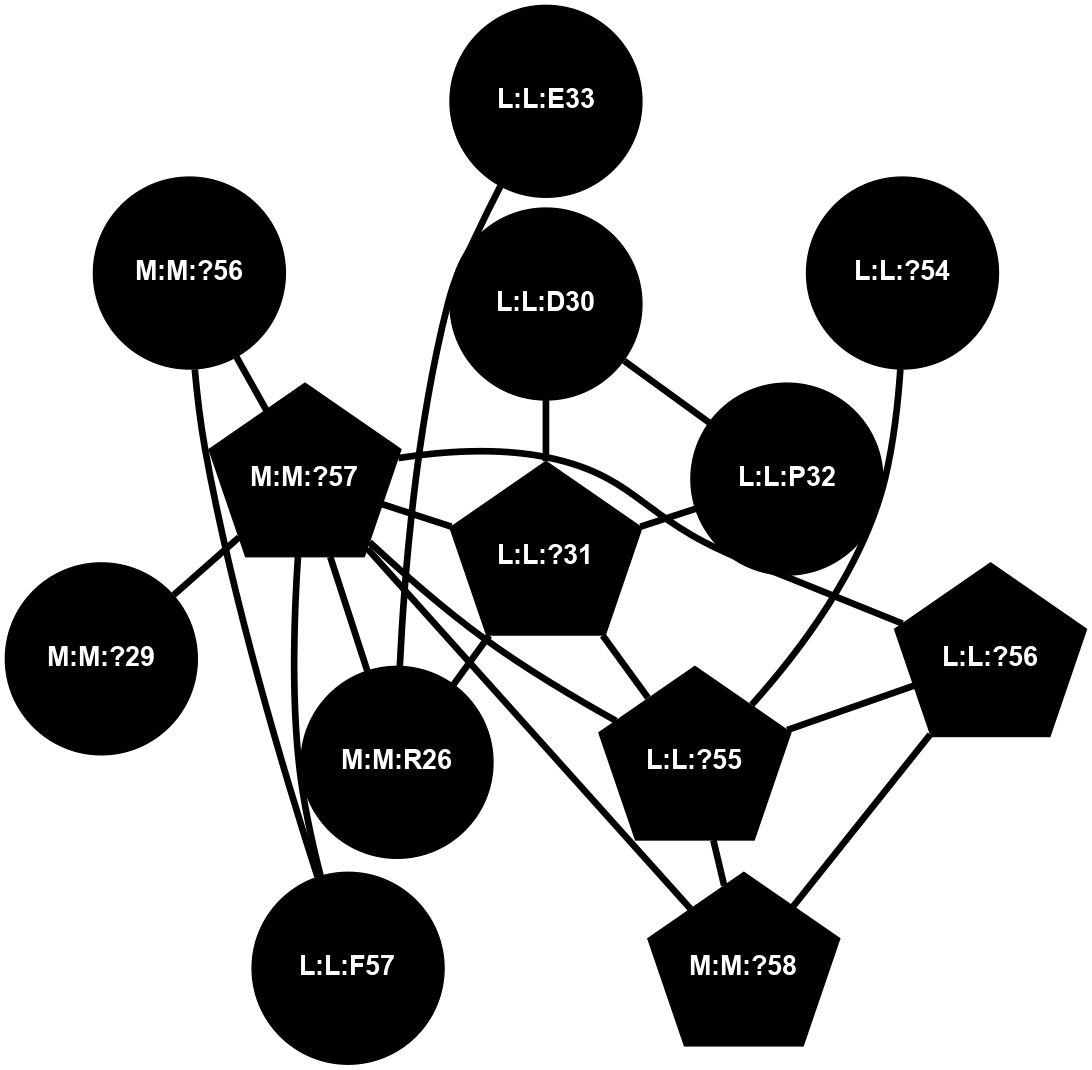
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?31 | L:L:D30 | 10.17 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:D30 | L:L:P32 | 4.83 | 2 | No | No | 0 | 0 | 1 | 1 | | L:L:?31 | L:L:P32 | 15.83 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?31 | L:L:?55 | 19.05 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?31 | N:N:R26 | 20.81 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?31 | N:N:?57 | 7.21 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:E33 | N:N:R26 | 9.3 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:?54 | L:L:?55 | 16.8 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?55 | L:L:?56 | 26.98 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?55 | N:N:?57 | 11.53 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?55 | N:N:?58 | 14.29 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?56 | N:N:?57 | 27.39 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?56 | N:N:?58 | 15.87 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:F57 | N:N:?56 | 11.68 | 2 | No | No | 0 | 0 | 2 | 2 | | L:L:F57 | N:N:?57 | 18.95 | 2 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:R26 | 16.54 | 2 | Yes | No | 0 | 0 | 1 | 1 | | N:N:?29 | N:N:?57 | 12.97 | 0 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?56 | N:N:?57 | 21.13 | 2 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:?58 | 21.62 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 14.61 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 19.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
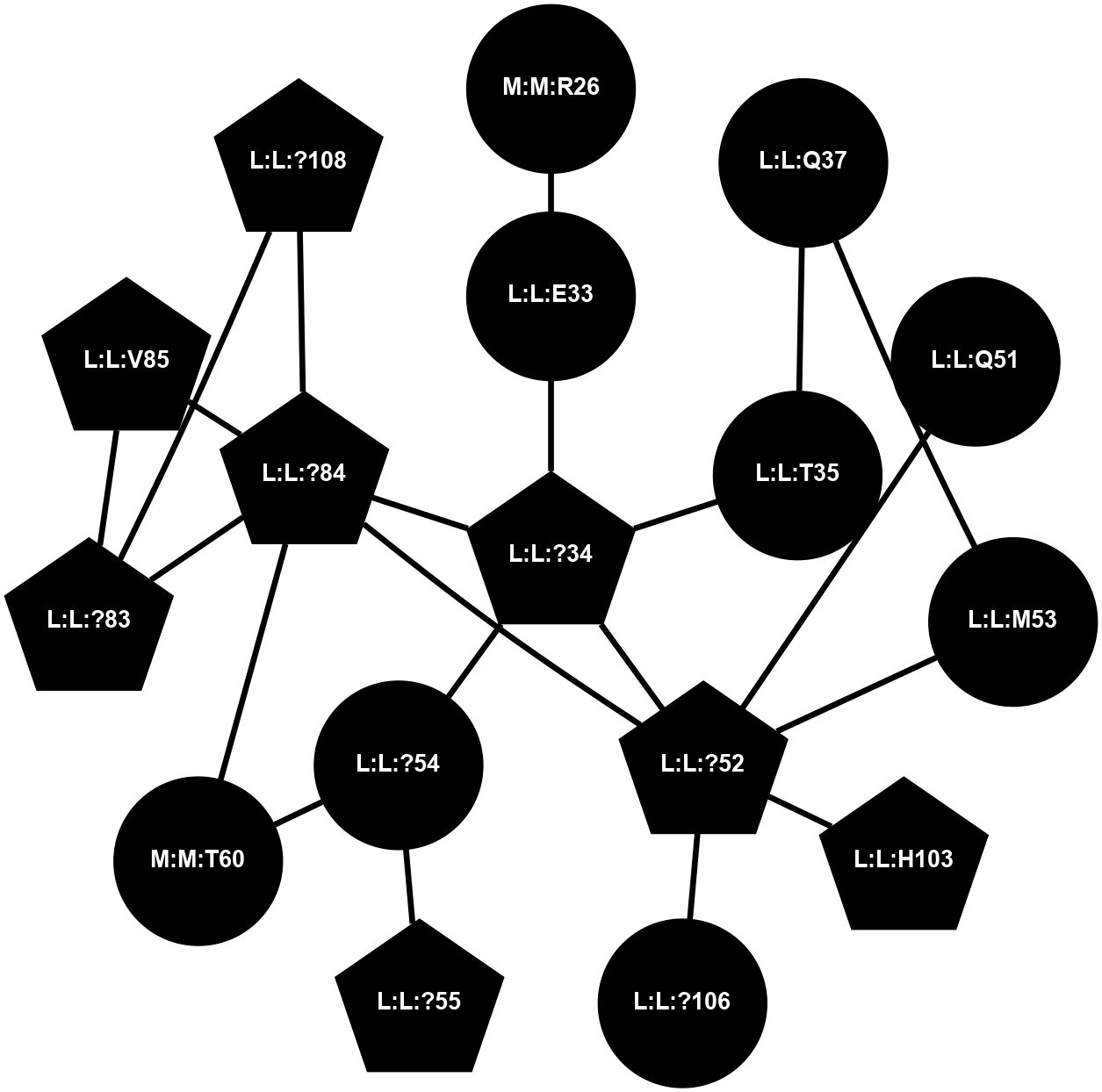
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?34 | L:L:E33 | 4.26 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:E33 | N:N:R26 | 9.3 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:?34 | L:L:T35 | 11.05 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?34 | L:L:?52 | 6.35 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?34 | L:L:?54 | 14.22 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?34 | L:L:?84 | 9.52 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:Q37 | L:L:T35 | 4.25 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:M53 | L:L:Q37 | 12.23 | 0 | No | No | 0 | 0 | 2 | 2 | | L:L:?52 | L:L:Q51 | 7.13 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?52 | L:L:M53 | 6.06 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?52 | L:L:?84 | 6.35 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?52 | L:L:H103 | 12.39 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?106 | L:L:?52 | 12.7 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?54 | L:L:?55 | 16.8 | 0 | No | Yes | 0 | 0 | 1 | 2 | | L:L:?54 | N:N:T60 | 5.14 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:?83 | L:L:?84 | 23.81 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?83 | L:L:V85 | 4.79 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?108 | L:L:?83 | 20.63 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?84 | L:L:V85 | 4.79 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:?84 | 14.29 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?84 | N:N:T60 | 6.31 | 2 | Yes | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.08 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 21.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
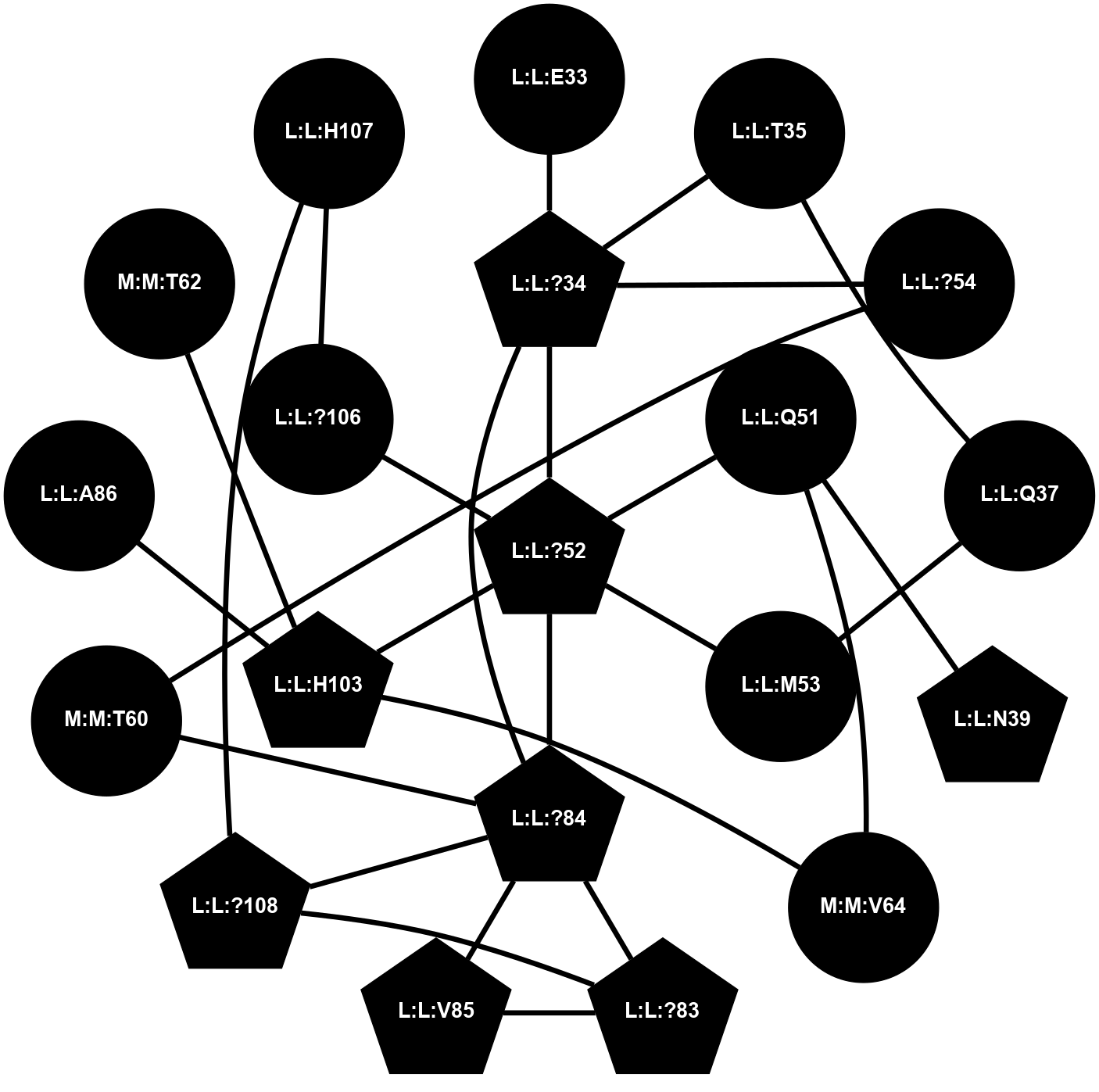
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?34 | L:L:E33 | 4.26 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?34 | L:L:T35 | 11.05 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?34 | L:L:?52 | 6.35 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?34 | L:L:?54 | 14.22 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?34 | L:L:?84 | 9.52 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:Q37 | L:L:T35 | 4.25 | 0 | No | No | 0 | 0 | 2 | 2 | | L:L:M53 | L:L:Q37 | 12.23 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:N39 | L:L:Q51 | 5.28 | 26 | Yes | No | 0 | 0 | 2 | 1 | | L:L:?52 | L:L:Q51 | 7.13 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:Q51 | N:N:V64 | 7.16 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:?52 | L:L:M53 | 6.06 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?52 | L:L:?84 | 6.35 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?52 | L:L:H103 | 12.39 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?106 | L:L:?52 | 12.7 | 0 | No | Yes | 0 | 0 | 1 | 0 | | L:L:?54 | N:N:T60 | 5.14 | 0 | No | No | 0 | 0 | 2 | 2 | | L:L:?83 | L:L:?84 | 23.81 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?83 | L:L:V85 | 4.79 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?108 | L:L:?83 | 20.63 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?84 | L:L:V85 | 4.79 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:?84 | 14.29 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?84 | N:N:T60 | 6.31 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:A86 | L:L:H103 | 8.78 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:H103 | N:N:T62 | 4.11 | 0 | Yes | No | 0 | 0 | 1 | 2 | | L:L:H103 | N:N:V64 | 9.69 | 0 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?106 | L:L:H107 | 5.51 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:H107 | 11.01 | 2 | Yes | No | 0 | 0 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 8.50 | | Average Nodes In Shell | 20.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 26.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
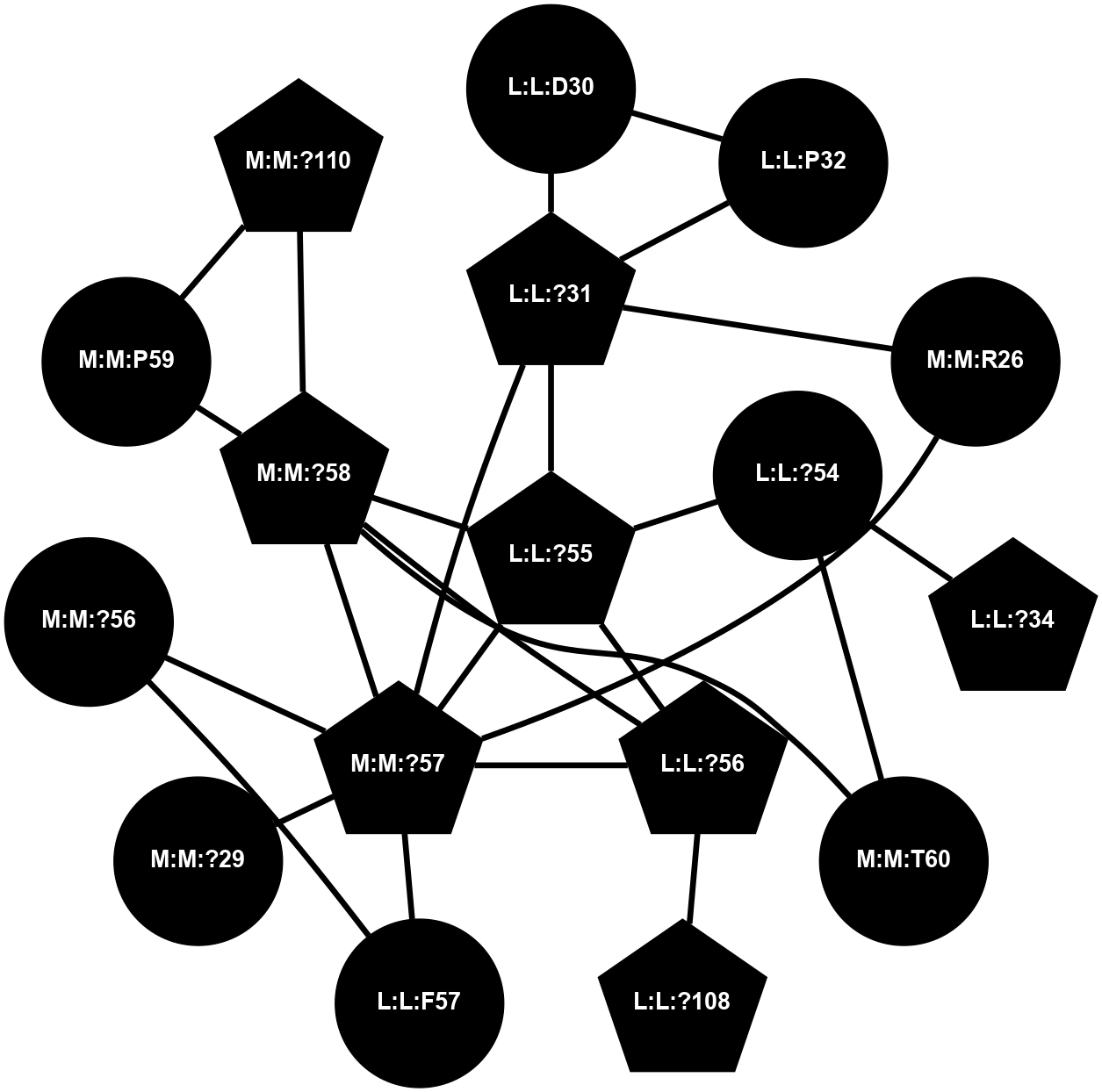
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?31 | L:L:D30 | 10.17 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:D30 | L:L:P32 | 4.83 | 2 | No | No | 0 | 0 | 2 | 2 | | L:L:?31 | L:L:P32 | 15.83 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?31 | L:L:?55 | 19.05 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?31 | N:N:R26 | 20.81 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?31 | N:N:?57 | 7.21 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?34 | L:L:?54 | 14.22 | 2 | Yes | No | 0 | 0 | 2 | 1 | | L:L:?54 | L:L:?55 | 16.8 | 0 | No | Yes | 0 | 0 | 1 | 0 | | L:L:?54 | N:N:T60 | 5.14 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:?55 | L:L:?56 | 26.98 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?55 | N:N:?57 | 11.53 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?55 | N:N:?58 | 14.29 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?108 | L:L:?56 | 9.52 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?56 | N:N:?57 | 27.39 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?56 | N:N:?58 | 15.87 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:F57 | N:N:?56 | 11.68 | 2 | No | No | 0 | 0 | 2 | 2 | | L:L:F57 | N:N:?57 | 18.95 | 2 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:R26 | 16.54 | 2 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?29 | N:N:?57 | 12.97 | 0 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?56 | N:N:?57 | 21.13 | 2 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:?58 | 21.62 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | N:N:?58 | N:N:P59 | 19.34 | 2 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?58 | N:N:T60 | 4.74 | 2 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?110 | N:N:?58 | 12.7 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?110 | N:N:P59 | 5.28 | 2 | Yes | No | 0 | 0 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 17.73 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 25.00 | | Average Links Mediated by Hubs In Shell | 22.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
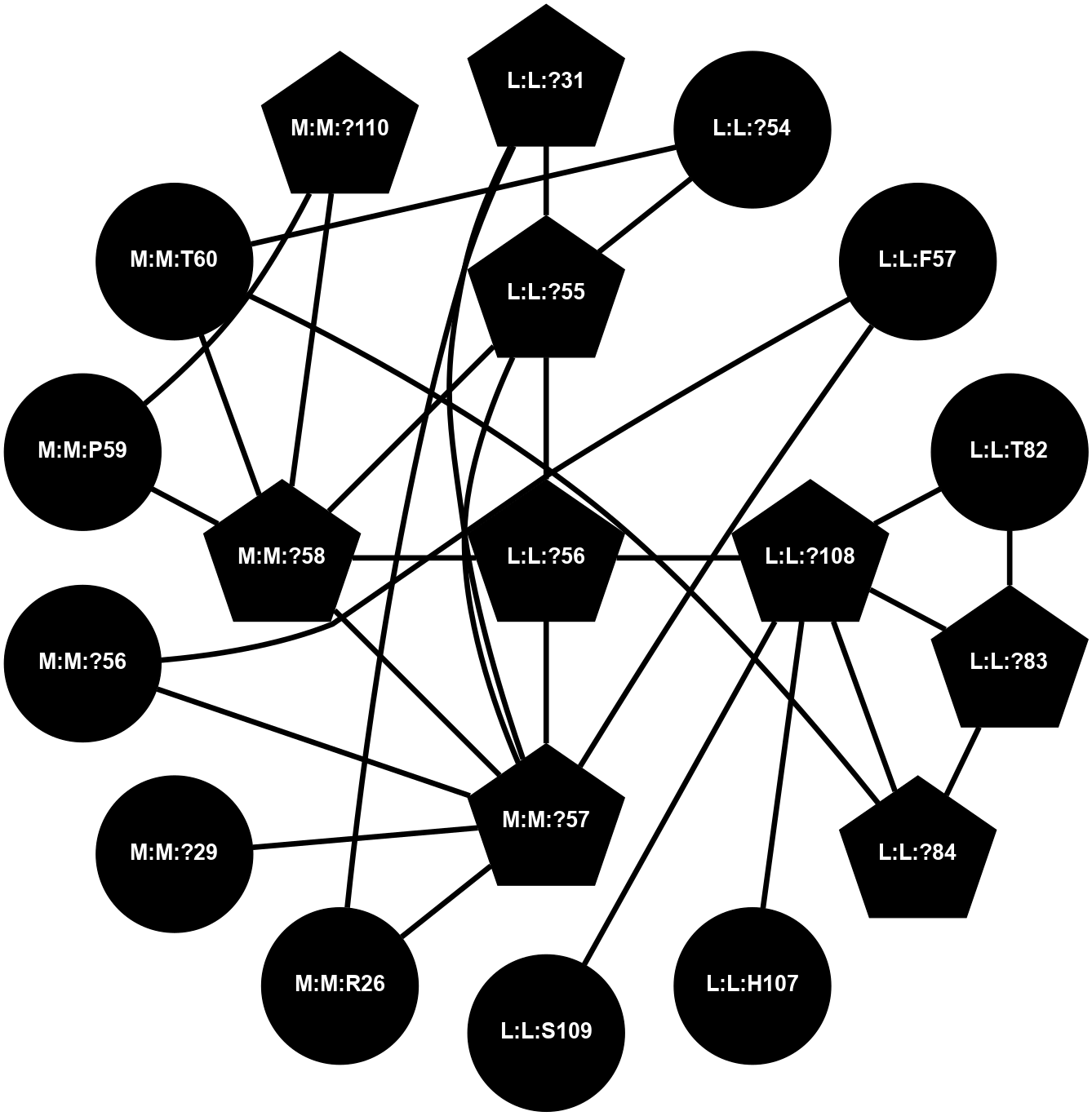
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?31 | L:L:?55 | 19.05 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?31 | N:N:R26 | 20.81 | 2 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?31 | N:N:?57 | 7.21 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?54 | L:L:?55 | 16.8 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?54 | N:N:T60 | 5.14 | 0 | No | No | 0 | 0 | 2 | 2 | | L:L:?55 | L:L:?56 | 26.98 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?55 | N:N:?57 | 11.53 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?55 | N:N:?58 | 14.29 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?108 | L:L:?56 | 9.52 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?56 | N:N:?57 | 27.39 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?56 | N:N:?58 | 15.87 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:F57 | N:N:?56 | 11.68 | 2 | No | No | 0 | 0 | 2 | 2 | | L:L:F57 | N:N:?57 | 18.95 | 2 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?83 | L:L:T82 | 4.74 | 2 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?108 | L:L:T82 | 7.89 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?83 | L:L:?84 | 23.81 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?108 | L:L:?83 | 20.63 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:?84 | 14.29 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?84 | N:N:T60 | 6.31 | 2 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?108 | L:L:H107 | 11.01 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:S109 | 6.43 | 2 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?57 | N:N:R26 | 16.54 | 2 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?29 | N:N:?57 | 12.97 | 0 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?56 | N:N:?57 | 21.13 | 2 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:?58 | 21.62 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | N:N:?58 | N:N:P59 | 19.34 | 2 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?58 | N:N:T60 | 4.74 | 2 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?110 | N:N:?58 | 12.7 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?110 | N:N:P59 | 5.28 | 2 | Yes | No | 0 | 0 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 19.94 | | Average Nodes In Shell | 19.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 29.00 | | Average Links Mediated by Hubs In Shell | 27.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?34 | L:L:?52 | 6.35 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?34 | L:L:?84 | 9.52 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?52 | L:L:?84 | 6.35 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?108 | L:L:?56 | 9.52 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?83 | L:L:T82 | 4.74 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?108 | L:L:T82 | 7.89 | 2 | Yes | No | 0 | 0 | 1 | 1 | | L:L:?83 | L:L:?84 | 23.81 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?83 | L:L:V85 | 4.79 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?108 | L:L:?83 | 20.63 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?111 | L:L:?83 | 17.46 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?83 | N:N:Q74 | 11.4 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?84 | L:L:V85 | 4.79 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?108 | L:L:?84 | 14.29 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?84 | N:N:T60 | 6.31 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:S109 | L:L:V85 | 4.85 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?112 | L:L:V85 | 4.35 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?108 | L:L:H107 | 11.01 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:S109 | 6.43 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?111 | L:L:T110 | 9.47 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?112 | L:L:T110 | 4.3 | 2 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?111 | L:L:?112 | 28.83 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?111 | L:L:H114 | 22.03 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?111 | N:N:Q74 | 5.7 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?111 | R:R:?127 | 4.32 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?112 | L:L:?113 | 39.28 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?112 | L:L:H114 | 7.5 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?112 | N:N:Q74 | 6.47 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?112 | R:R:?127 | 18.33 | 2 | Yes | Yes | 0 | 6 | 2 | 2 | | L:L:?113 | L:L:H114 | 7.5 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?113 | N:N:Q74 | 12.95 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 13.81 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 30.00 | | Average Links Mediated by Hubs In Shell | 30.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
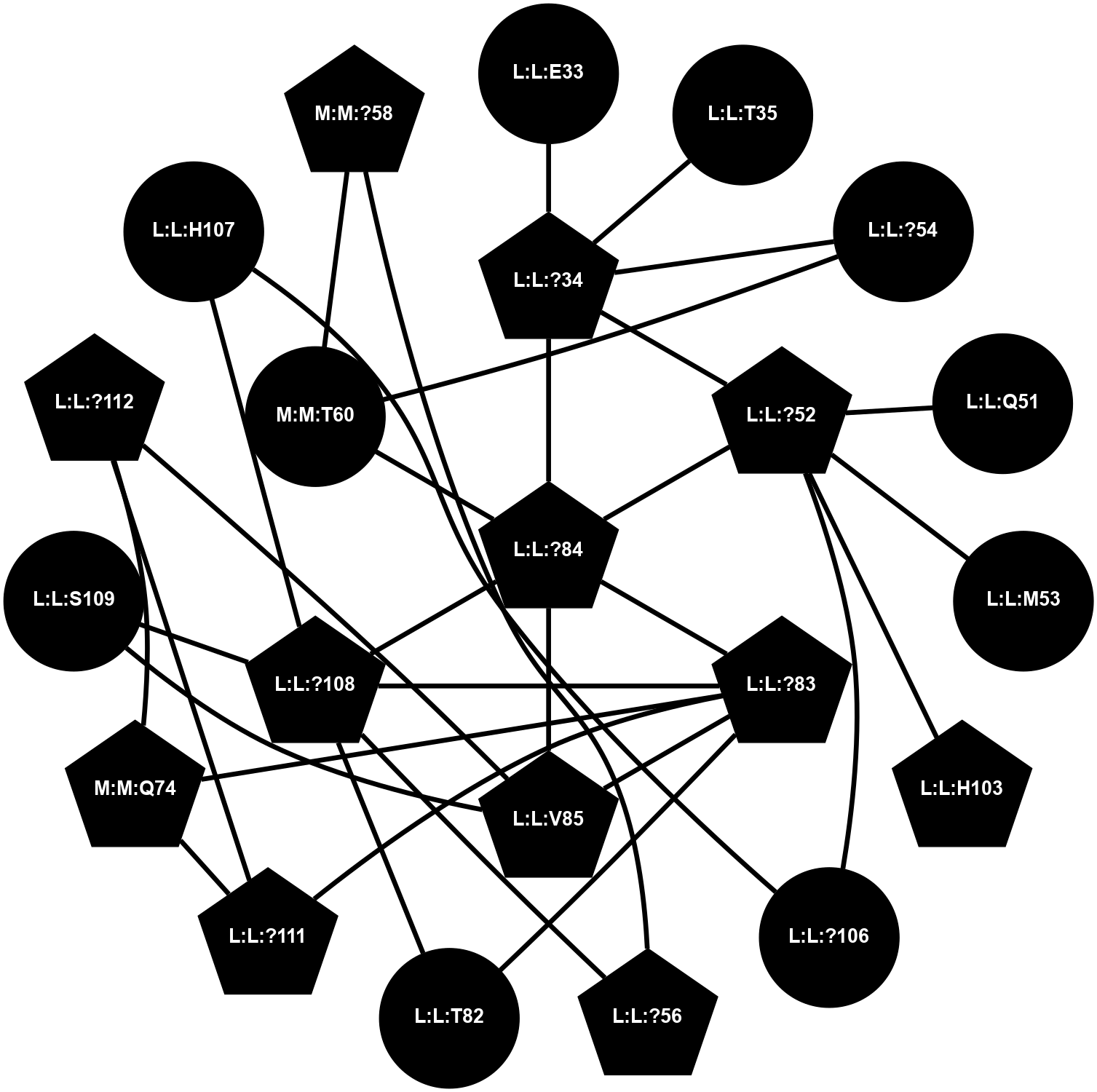
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?34 | L:L:E33 | 4.26 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?34 | L:L:T35 | 11.05 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?34 | L:L:?52 | 6.35 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?34 | L:L:?54 | 14.22 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?34 | L:L:?84 | 9.52 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?52 | L:L:Q51 | 7.13 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?52 | L:L:M53 | 6.06 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?52 | L:L:?84 | 6.35 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?52 | L:L:H103 | 12.39 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?106 | L:L:?52 | 12.7 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?54 | N:N:T60 | 5.14 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:?108 | L:L:?56 | 9.52 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?56 | N:N:?58 | 15.87 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?83 | L:L:T82 | 4.74 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:T82 | 7.89 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?83 | L:L:?84 | 23.81 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?83 | L:L:V85 | 4.79 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?108 | L:L:?83 | 20.63 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?111 | L:L:?83 | 17.46 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?83 | N:N:Q74 | 11.4 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?84 | L:L:V85 | 4.79 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?108 | L:L:?84 | 14.29 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?84 | N:N:T60 | 6.31 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:S109 | L:L:V85 | 4.85 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?112 | L:L:V85 | 4.35 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?106 | L:L:H107 | 5.51 | 0 | No | No | 0 | 0 | 2 | 2 | | L:L:?108 | L:L:H107 | 11.01 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:S109 | 6.43 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?111 | L:L:?112 | 28.83 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?111 | N:N:Q74 | 5.7 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?112 | N:N:Q74 | 6.47 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?58 | N:N:T60 | 4.74 | 2 | Yes | No | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 10.84 | | Average Nodes In Shell | 22.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 32.00 | | Average Links Mediated by Hubs In Shell | 30.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
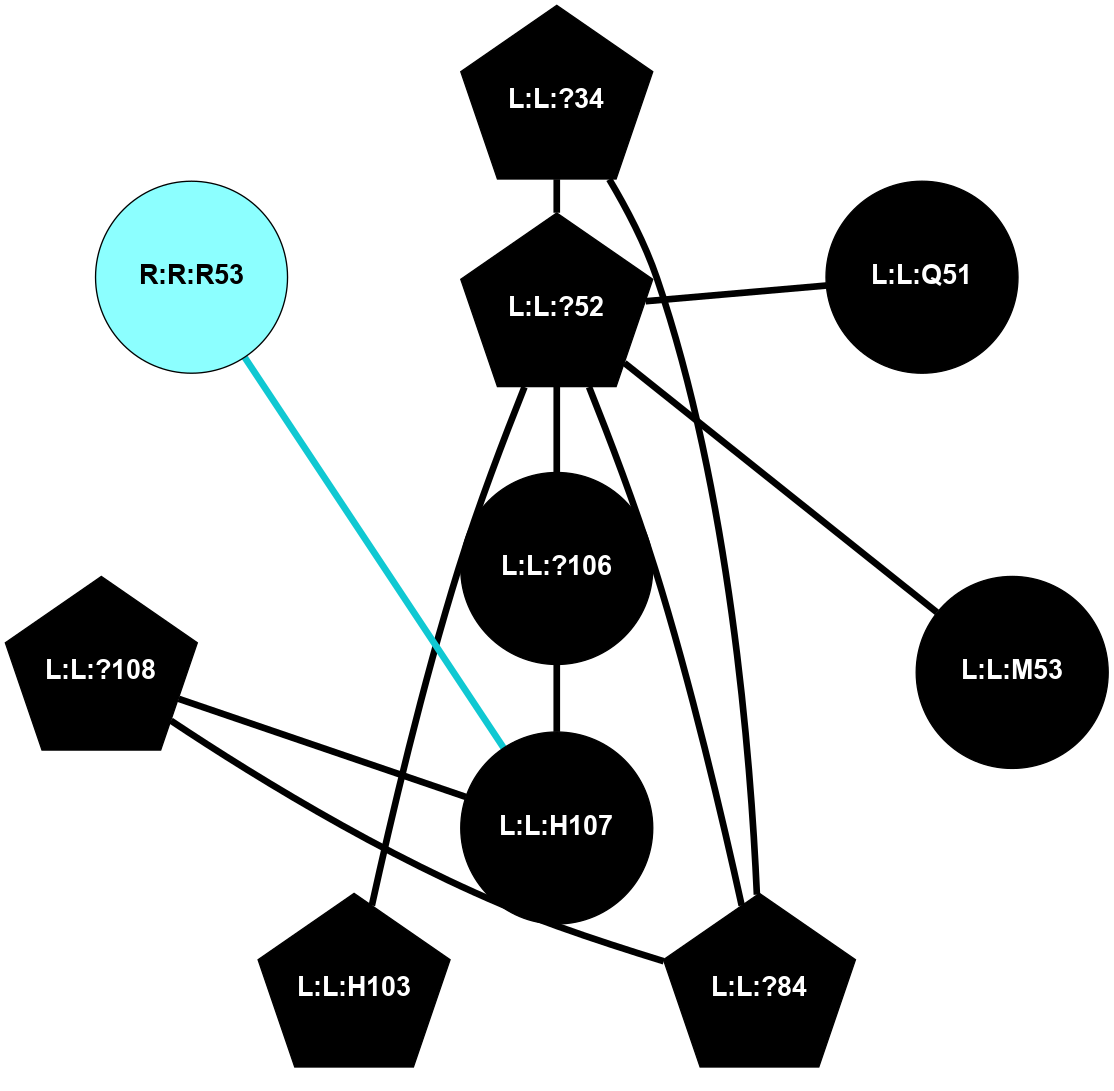
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?34 | L:L:?52 | 6.35 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?34 | L:L:?84 | 9.52 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?52 | L:L:Q51 | 7.13 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?52 | L:L:M53 | 6.06 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?52 | L:L:?84 | 6.35 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?52 | L:L:H103 | 12.39 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?106 | L:L:?52 | 12.7 | 0 | No | Yes | 0 | 0 | 0 | 1 | | L:L:?108 | L:L:?84 | 14.29 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?106 | L:L:H107 | 5.51 | 0 | No | No | 0 | 0 | 0 | 1 | | L:L:?108 | L:L:H107 | 11.01 | 2 | Yes | No | 0 | 0 | 2 | 1 | | L:L:H107 | R:R:R53 | 2.26 | 0 | No | No | 0 | 2 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 9.11 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
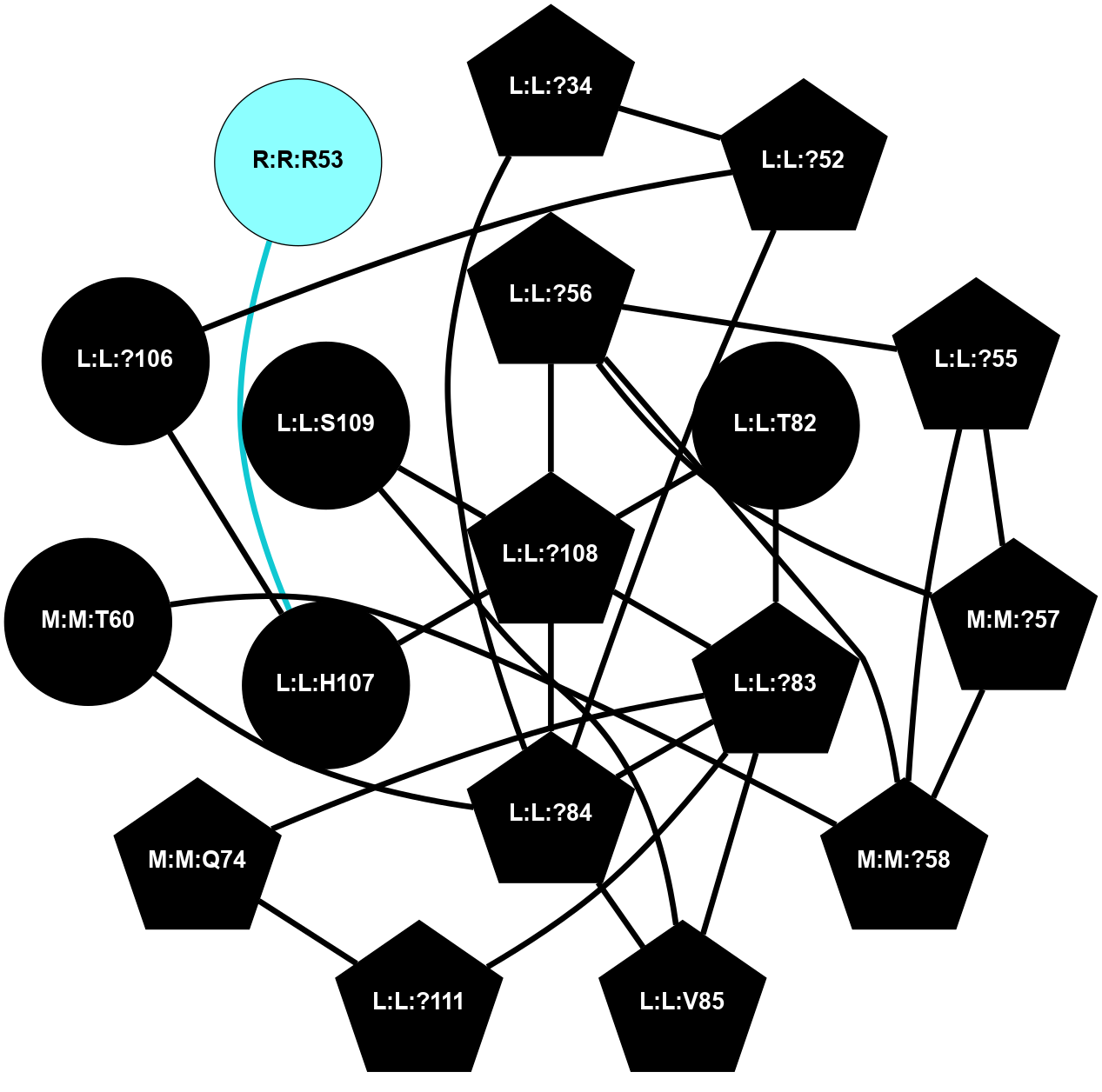
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?34 | L:L:?52 | 6.35 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?34 | L:L:?84 | 9.52 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?52 | L:L:?84 | 6.35 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?106 | L:L:?52 | 12.7 | 0 | No | Yes | 0 | 0 | 2 | 2 | | L:L:?55 | L:L:?56 | 26.98 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?55 | N:N:?57 | 11.53 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?55 | N:N:?58 | 14.29 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?108 | L:L:?56 | 9.52 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?56 | N:N:?57 | 27.39 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?56 | N:N:?58 | 15.87 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?83 | L:L:T82 | 4.74 | 2 | Yes | No | 0 | 0 | 1 | 1 | | L:L:?108 | L:L:T82 | 7.89 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?83 | L:L:?84 | 23.81 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?83 | L:L:V85 | 4.79 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:?83 | 20.63 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?111 | L:L:?83 | 17.46 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?83 | N:N:Q74 | 11.4 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?84 | L:L:V85 | 4.79 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:?84 | 14.29 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?84 | N:N:T60 | 6.31 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:S109 | L:L:V85 | 4.85 | 0 | No | Yes | 0 | 0 | 1 | 2 | | L:L:?106 | L:L:H107 | 5.51 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:?108 | L:L:H107 | 11.01 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?108 | L:L:S109 | 6.43 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?111 | N:N:Q74 | 5.7 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?57 | N:N:?58 | 21.62 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?58 | N:N:T60 | 4.74 | 2 | Yes | No | 0 | 0 | 2 | 2 | | L:L:H107 | R:R:R53 | 2.26 | 0 | No | No | 0 | 2 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 11.63 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 28.00 | | Average Links Mediated by Hubs In Shell | 26.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
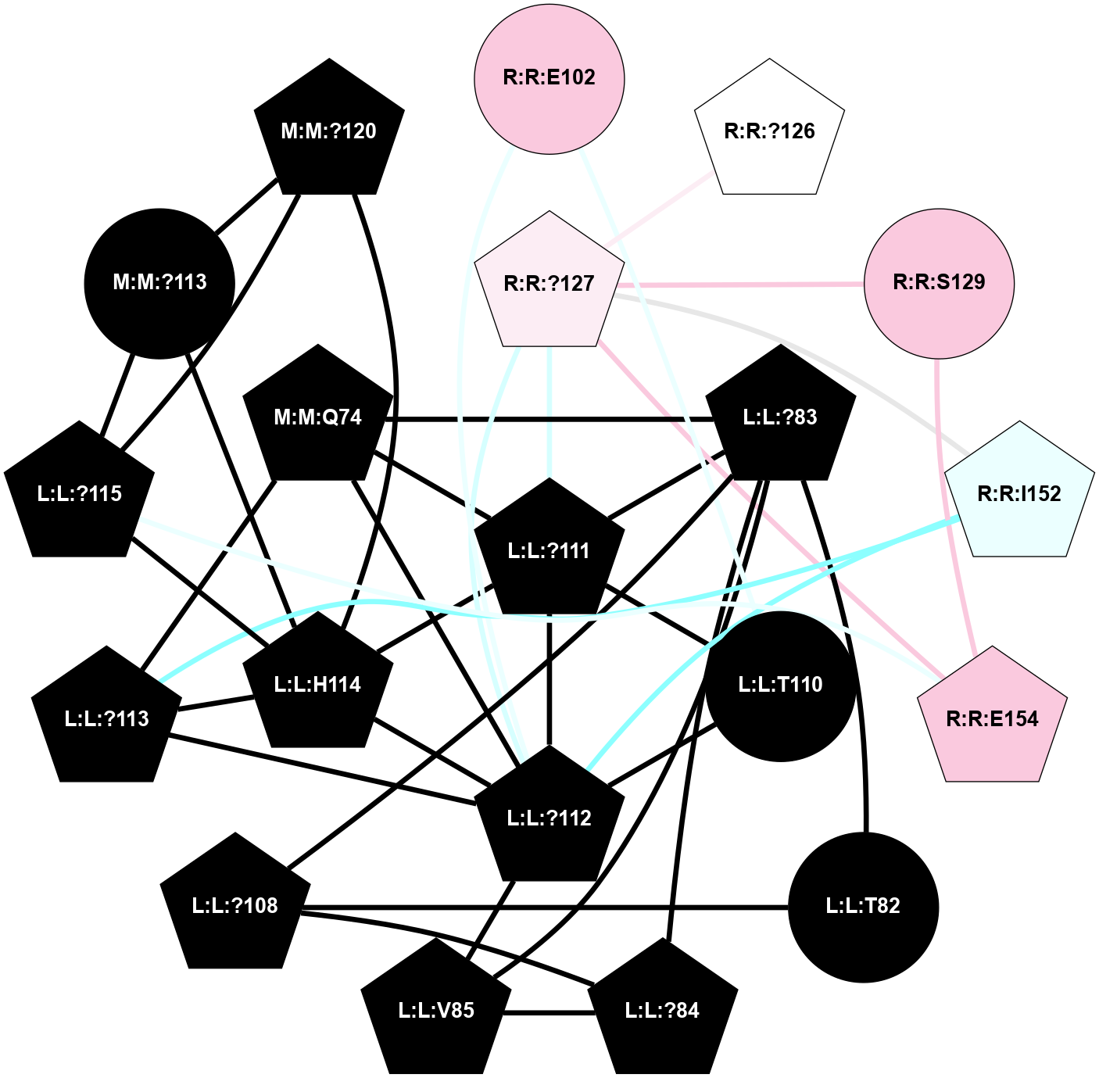
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?83 | L:L:T82 | 4.74 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:T82 | 7.89 | 2 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?83 | L:L:?84 | 23.81 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?83 | L:L:V85 | 4.79 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:?83 | 20.63 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?111 | L:L:?83 | 17.46 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?83 | N:N:Q74 | 11.4 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?84 | L:L:V85 | 4.79 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?108 | L:L:?84 | 14.29 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?112 | L:L:V85 | 4.35 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?111 | L:L:T110 | 9.47 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?112 | L:L:T110 | 4.3 | 2 | Yes | No | 0 | 0 | 1 | 1 | | L:L:T110 | R:R:E102 | 8.47 | 2 | No | No | 0 | 7 | 1 | 2 | | L:L:?111 | L:L:?112 | 28.83 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?111 | L:L:H114 | 22.03 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?111 | N:N:Q74 | 5.7 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?111 | R:R:?127 | 4.32 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?112 | L:L:?113 | 39.28 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?112 | L:L:H114 | 7.5 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?112 | N:N:Q74 | 6.47 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?112 | R:R:E102 | 10.31 | 2 | Yes | No | 0 | 7 | 1 | 2 | | L:L:?112 | R:R:?127 | 18.33 | 2 | Yes | Yes | 0 | 6 | 1 | 1 | | L:L:?112 | R:R:I152 | 4.16 | 2 | Yes | Yes | 0 | 4 | 1 | 2 | | L:L:?113 | L:L:H114 | 7.5 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?113 | N:N:Q74 | 12.95 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?113 | R:R:I152 | 6.94 | 2 | Yes | Yes | 0 | 4 | 2 | 2 | | L:L:?115 | L:L:H114 | 8.44 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:H114 | N:N:?113 | 5.51 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:H114 | N:N:?120 | 15.14 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?115 | N:N:?113 | 6.19 | 2 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?115 | N:N:?120 | 6.19 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?115 | R:R:E154 | 7.11 | 2 | Yes | Yes | 0 | 7 | 2 | 2 | | N:N:?113 | N:N:?120 | 7.94 | 2 | No | Yes | 0 | 0 | 2 | 2 | | R:R:?126 | R:R:?127 | 12.85 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:?127 | R:R:S129 | 10.22 | 2 | Yes | No | 6 | 7 | 1 | 2 | | R:R:?127 | R:R:I152 | 5.55 | 2 | Yes | Yes | 6 | 4 | 1 | 2 | | R:R:?127 | R:R:E154 | 14.18 | 2 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:E154 | R:R:S129 | 10.06 | 2 | Yes | No | 7 | 7 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 14.63 | | Average Nodes In Shell | 20.00 | | Average Hubs In Shell | 15.00 | | Average Links In Shell | 38.00 | | Average Links Mediated by Hubs In Shell | 37.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
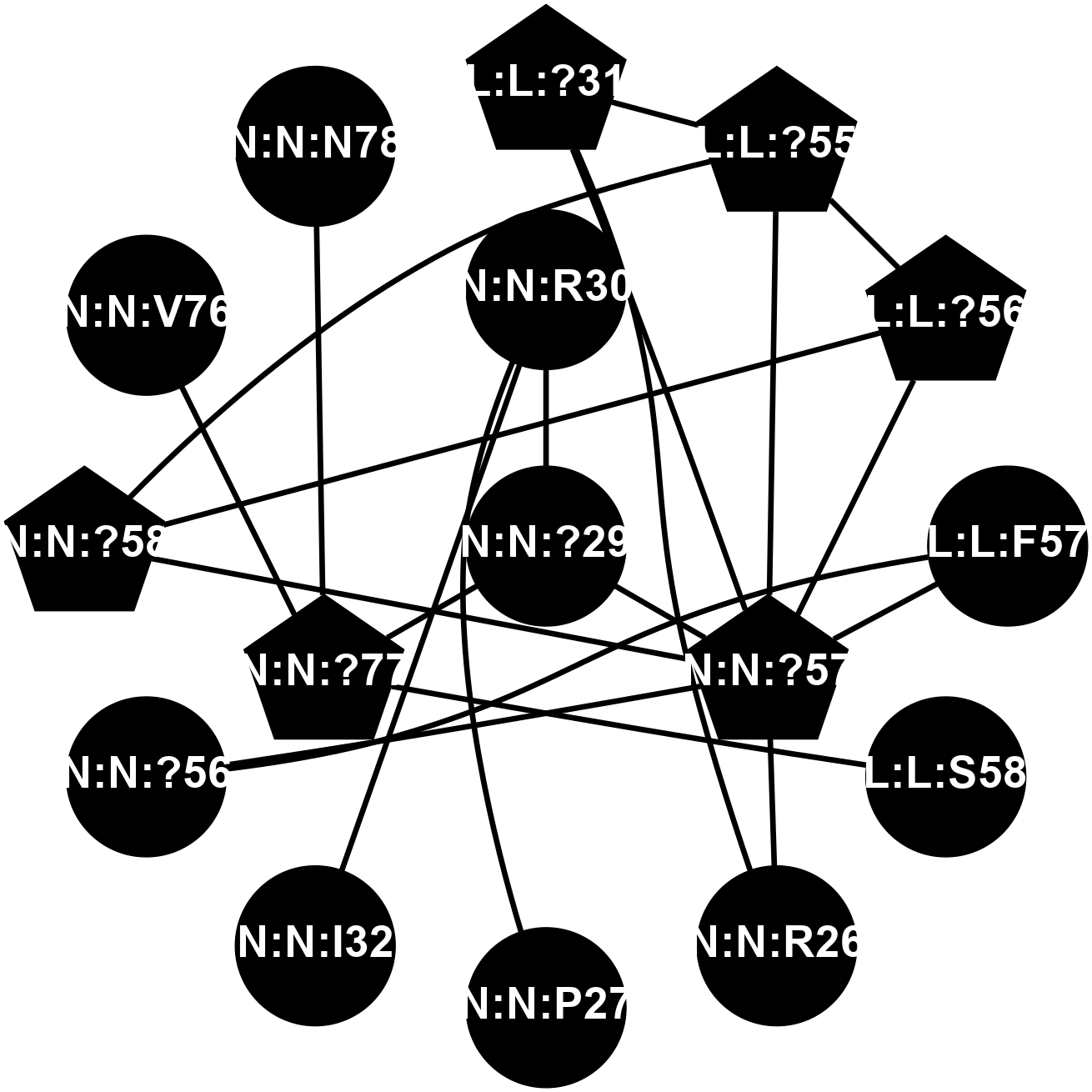
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?31 | L:L:?55 | 19.05 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?31 | N:N:R26 | 20.81 | 2 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?31 | N:N:?57 | 7.21 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?55 | L:L:?56 | 26.98 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?55 | N:N:?57 | 11.53 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?55 | N:N:?58 | 14.29 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?56 | N:N:?57 | 27.39 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?56 | N:N:?58 | 15.87 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:F57 | N:N:?56 | 11.68 | 2 | No | No | 0 | 0 | 2 | 2 | | L:L:F57 | N:N:?57 | 18.95 | 2 | No | Yes | 0 | 0 | 2 | 1 | | L:L:S58 | N:N:?77 | 6.43 | 0 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:R26 | 16.54 | 2 | Yes | No | 0 | 0 | 1 | 2 | | N:N:P27 | N:N:R30 | 10.09 | 0 | No | No | 0 | 0 | 2 | 1 | | N:N:?29 | N:N:R30 | 5.2 | 0 | No | No | 0 | 0 | 0 | 1 | | N:N:?29 | N:N:?57 | 12.97 | 0 | No | Yes | 0 | 0 | 0 | 1 | | N:N:?29 | N:N:?77 | 14.29 | 0 | No | Yes | 0 | 0 | 0 | 1 | | N:N:I32 | N:N:R30 | 7.52 | 0 | No | No | 0 | 0 | 2 | 1 | | N:N:?56 | N:N:?57 | 21.13 | 2 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:?58 | 21.62 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?77 | N:N:V76 | 7.98 | 0 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?77 | N:N:N78 | 4.41 | 0 | Yes | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 10.82 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 21.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
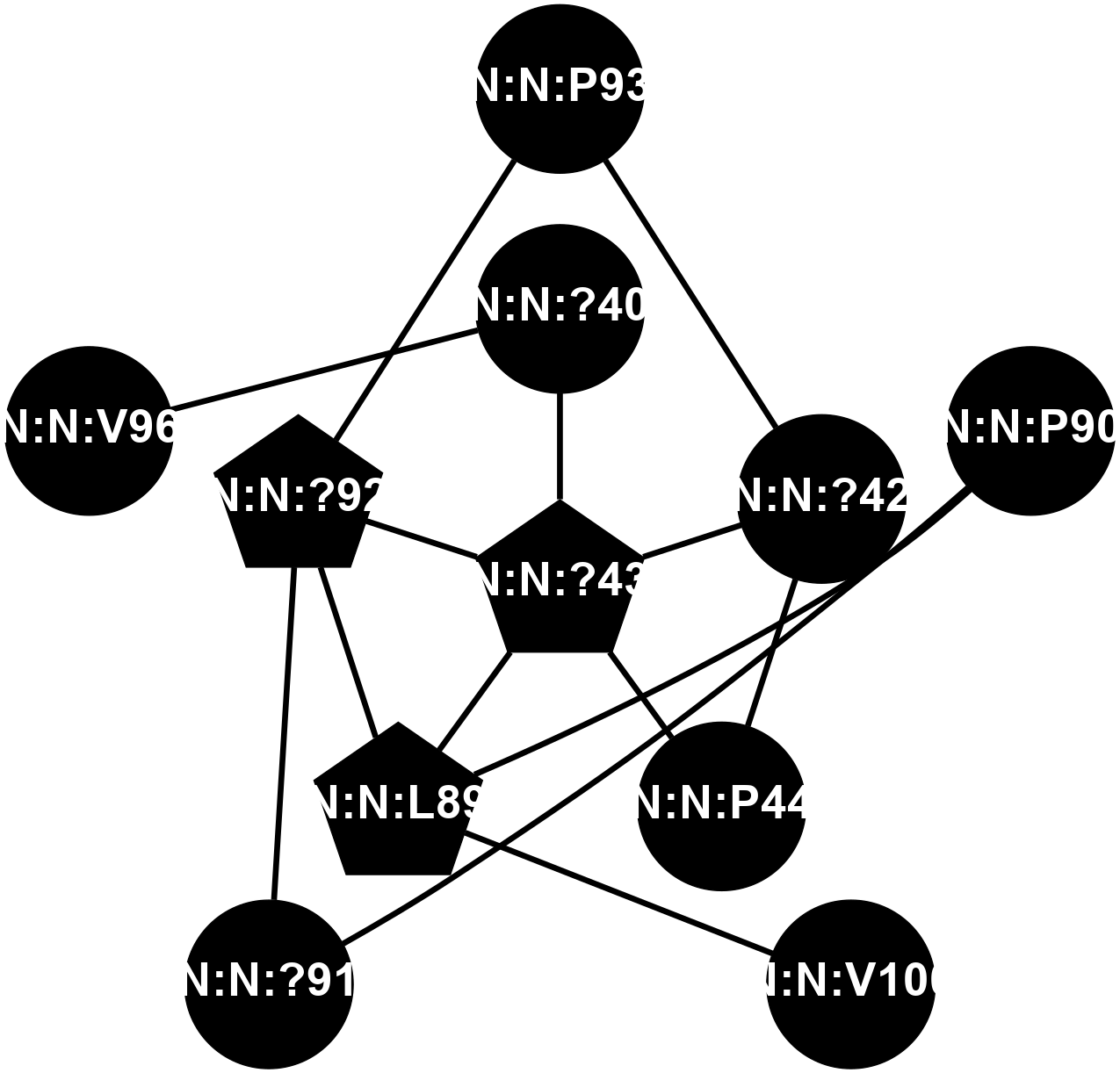
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | N:N:?40 | N:N:?43 | 10.61 | 0 | No | Yes | 0 | 0 | 1 | 0 | | N:N:?42 | N:N:?43 | 19.39 | 16 | No | Yes | 0 | 0 | 1 | 0 | | N:N:?42 | N:N:P44 | 4.3 | 16 | No | No | 0 | 0 | 1 | 1 | | N:N:?42 | N:N:P93 | 8.59 | 16 | No | No | 0 | 0 | 1 | 2 | | N:N:?43 | N:N:P44 | 17.58 | 16 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?43 | N:N:L89 | 4.45 | 16 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?43 | N:N:?92 | 15.87 | 16 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:L89 | N:N:P90 | 4.93 | 16 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?92 | N:N:L89 | 11.86 | 16 | Yes | Yes | 0 | 0 | 1 | 1 | | N:N:L89 | N:N:V100 | 5.96 | 16 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?91 | N:N:P90 | 4.3 | 0 | No | No | 0 | 0 | 2 | 2 | | N:N:?91 | N:N:?92 | 21.97 | 0 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?92 | N:N:P93 | 15.83 | 16 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?40 | N:N:V96 | 1.78 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 13.58 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:R66 | N:N:E39 | 6.98 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:R66 | N:N:D131 | 7.15 | 0 | No | No | 0 | 0 | 2 | 2 | | N:N:?130 | N:N:A37 | 5.06 | 0 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?46 | N:N:E39 | 11.35 | 0 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?46 | N:N:V45 | 9.57 | 0 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?46 | N:N:I47 | 9.17 | 0 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?130 | N:N:?46 | 14.29 | 0 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:I47 | N:N:P90 | 6.77 | 0 | No | No | 0 | 0 | 1 | 2 | | N:N:?130 | N:N:T129 | 12.63 | 0 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?130 | N:N:D131 | 7.27 | 0 | Yes | No | 0 | 0 | 1 | 2 | | L:L:L65 | N:N:E39 | 3.98 | 0 | No | No | 0 | 0 | 2 | 1 | | N:N:I47 | N:N:V38 | 3.07 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 11.10 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?61 | L:L:P62 | 15.97 | 9 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?61 | L:L:V77 | 4.35 | 9 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?61 | N:N:I53 | 6.94 | 9 | Yes | No | 0 | 0 | 2 | 1 | | L:L:?61 | N:N:?79 | 10.47 | 9 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:P62 | N:N:?79 | 4.79 | 9 | No | Yes | 0 | 0 | 2 | 1 | | L:L:V77 | N:N:?79 | 4.35 | 9 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?108 | N:N:P31 | 8.79 | 2 | Yes | No | 0 | 0 | 1 | 2 | | N:N:A34 | N:N:I53 | 6.5 | 0 | No | No | 0 | 0 | 2 | 1 | | N:N:?54 | N:N:I53 | 9.17 | 0 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?54 | N:N:?79 | 23.07 | 0 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?108 | N:N:?54 | 14.29 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:?79 | N:N:N78 | 18.7 | 9 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?79 | N:N:V82 | 5.8 | 9 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?79 | N:N:A105 | 4.6 | 9 | Yes | No | 0 | 0 | 1 | 2 | | N:N:R80 | N:N:S107 | 11.86 | 2 | Yes | No | 0 | 0 | 2 | 2 | | N:N:?108 | N:N:R80 | 5.2 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:Q109 | N:N:R80 | 12.85 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?108 | N:N:S107 | 4.82 | 2 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?108 | N:N:Q109 | 5.7 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:A60 | L:L:S79 | 3.42 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:A60 | N:N:?54 | 3.37 | 0 | No | Yes | 0 | 0 | 1 | 0 | | N:N:?54 | N:N:A55 | 3.37 | 0 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 10.65 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 22.00 | | Average Links Mediated by Hubs In Shell | 20.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
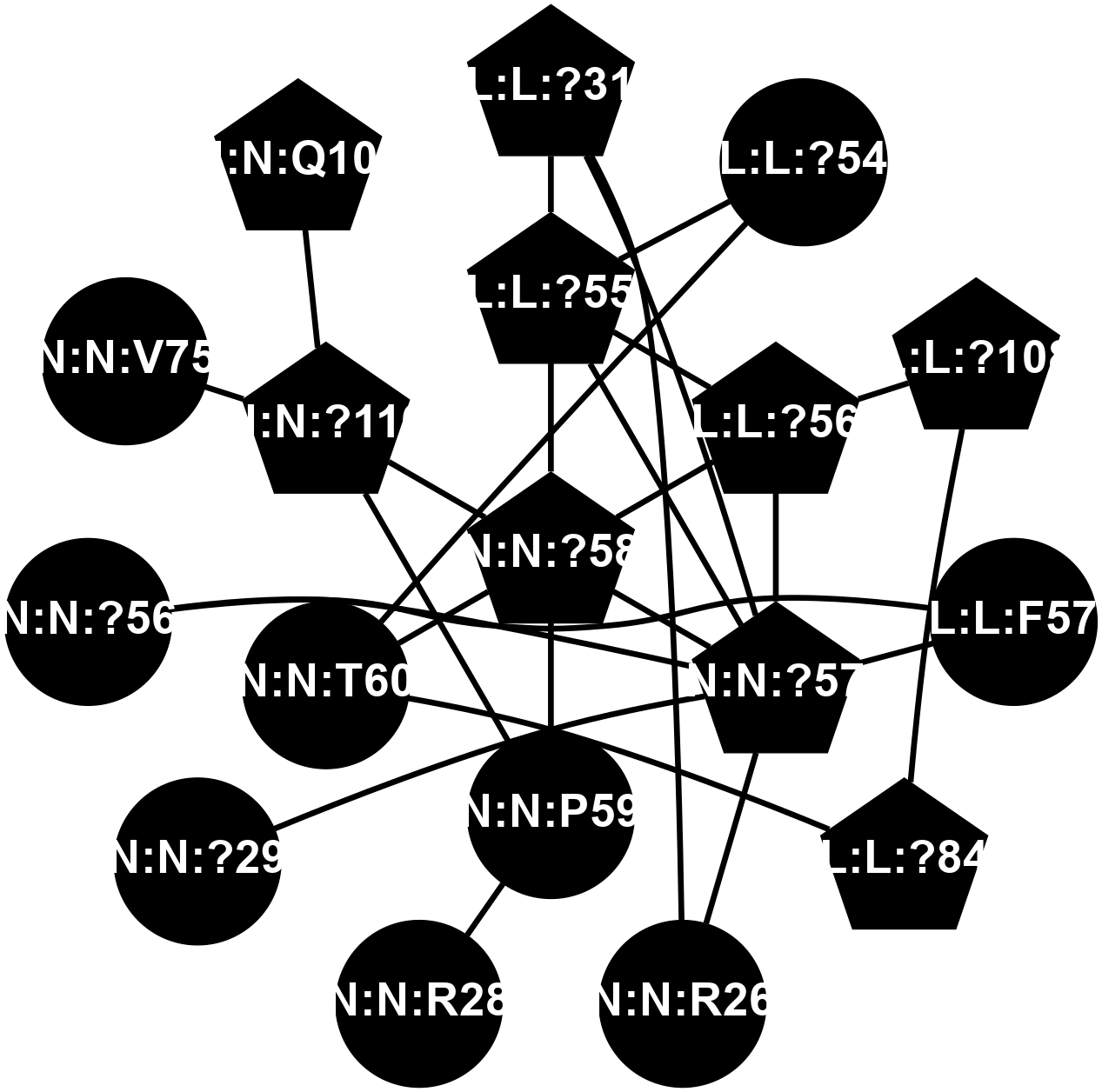
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?31 | L:L:?55 | 19.05 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?31 | N:N:R26 | 20.81 | 2 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?31 | N:N:?57 | 7.21 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?54 | L:L:?55 | 16.8 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?54 | N:N:T60 | 5.14 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:?55 | L:L:?56 | 26.98 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?55 | N:N:?57 | 11.53 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?55 | N:N:?58 | 14.29 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?108 | L:L:?56 | 9.52 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?56 | N:N:?57 | 27.39 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?56 | N:N:?58 | 15.87 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:F57 | N:N:?56 | 11.68 | 2 | No | No | 0 | 0 | 2 | 2 | | L:L:F57 | N:N:?57 | 18.95 | 2 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?108 | L:L:?84 | 14.29 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?84 | N:N:T60 | 6.31 | 2 | Yes | No | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:R26 | 16.54 | 2 | Yes | No | 0 | 0 | 1 | 2 | | N:N:P59 | N:N:R28 | 5.76 | 2 | No | No | 0 | 0 | 1 | 2 | | N:N:?29 | N:N:?57 | 12.97 | 0 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?56 | N:N:?57 | 21.13 | 2 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:?58 | 21.62 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:?58 | N:N:P59 | 19.34 | 2 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?58 | N:N:T60 | 4.74 | 2 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?110 | N:N:?58 | 12.7 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:?110 | N:N:P59 | 5.28 | 2 | Yes | No | 0 | 0 | 1 | 1 | | N:N:?110 | N:N:V75 | 14.36 | 2 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?110 | N:N:Q109 | 4.28 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 14.76 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 26.00 | | Average Links Mediated by Hubs In Shell | 23.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:S58 | N:N:?77 | 6.43 | 0 | No | Yes | 0 | 0 | 1 | 0 | | L:L:S81 | N:N:V76 | 4.85 | 0 | No | No | 0 | 0 | 2 | 1 | | N:N:?29 | N:N:R30 | 5.2 | 0 | No | No | 0 | 0 | 1 | 2 | | N:N:?29 | N:N:?57 | 12.97 | 0 | No | Yes | 0 | 0 | 1 | 2 | | N:N:?29 | N:N:?77 | 14.29 | 0 | No | Yes | 0 | 0 | 1 | 0 | | N:N:?77 | N:N:V76 | 7.98 | 0 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?77 | N:N:N78 | 4.41 | 0 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?79 | N:N:N78 | 18.7 | 9 | Yes | No | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 8.28 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
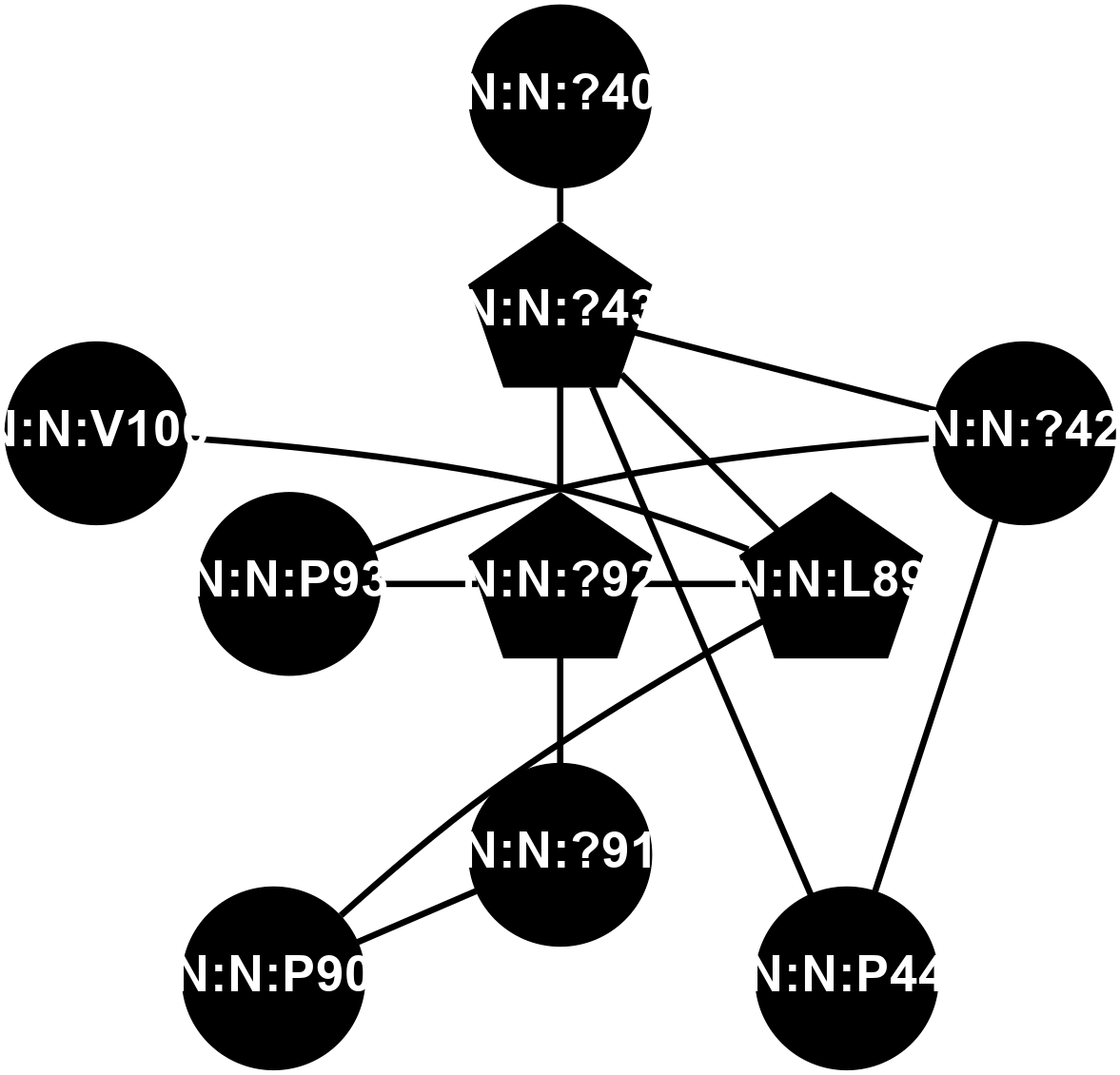
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | N:N:?40 | N:N:?43 | 10.61 | 0 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?42 | N:N:?43 | 19.39 | 16 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?42 | N:N:P44 | 4.3 | 16 | No | No | 0 | 0 | 2 | 2 | | N:N:?42 | N:N:P93 | 8.59 | 16 | No | No | 0 | 0 | 2 | 1 | | N:N:?43 | N:N:P44 | 17.58 | 16 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?43 | N:N:L89 | 4.45 | 16 | Yes | Yes | 0 | 0 | 1 | 1 | | N:N:?43 | N:N:?92 | 15.87 | 16 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:L89 | N:N:P90 | 4.93 | 16 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?92 | N:N:L89 | 11.86 | 16 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:L89 | N:N:V100 | 5.96 | 16 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?91 | N:N:P90 | 4.3 | 0 | No | No | 0 | 0 | 1 | 2 | | N:N:?91 | N:N:?92 | 21.97 | 0 | No | Yes | 0 | 0 | 1 | 0 | | N:N:?92 | N:N:P93 | 15.83 | 16 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 16.38 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
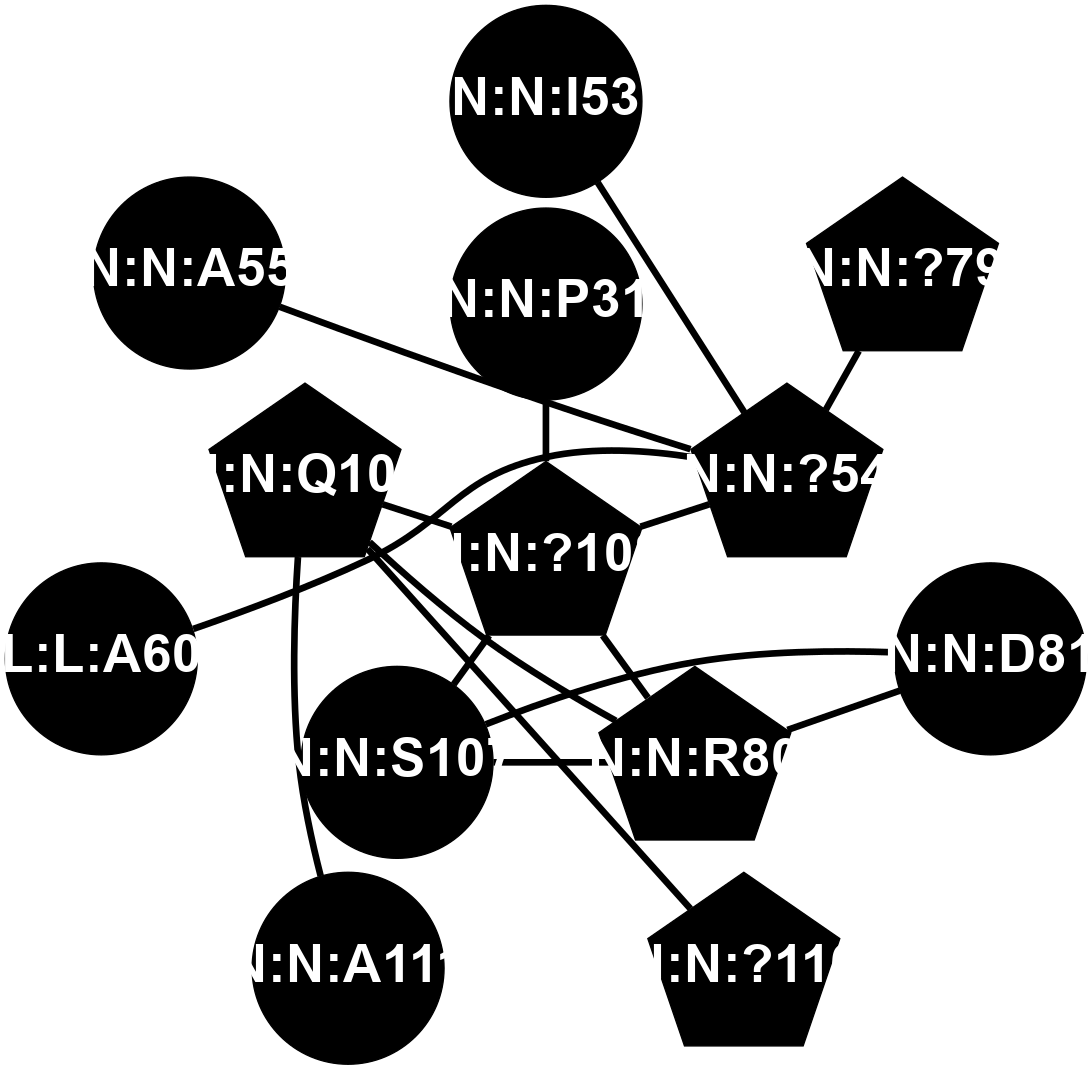
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | N:N:?108 | N:N:P31 | 8.79 | 2 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?54 | N:N:I53 | 9.17 | 0 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?54 | N:N:?79 | 23.07 | 0 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?108 | N:N:?54 | 14.29 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:D81 | N:N:R80 | 4.76 | 2 | No | Yes | 0 | 0 | 2 | 1 | | N:N:R80 | N:N:S107 | 11.86 | 2 | Yes | No | 0 | 0 | 1 | 1 | | N:N:?108 | N:N:R80 | 5.2 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:Q109 | N:N:R80 | 12.85 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | N:N:D81 | N:N:S107 | 10.31 | 2 | No | No | 0 | 0 | 2 | 1 | | N:N:?108 | N:N:S107 | 4.82 | 2 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?108 | N:N:Q109 | 5.7 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?110 | N:N:Q109 | 4.28 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:A111 | N:N:Q109 | 4.55 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:A60 | N:N:?54 | 3.37 | 0 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?54 | N:N:A55 | 3.37 | 0 | Yes | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 7.76 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
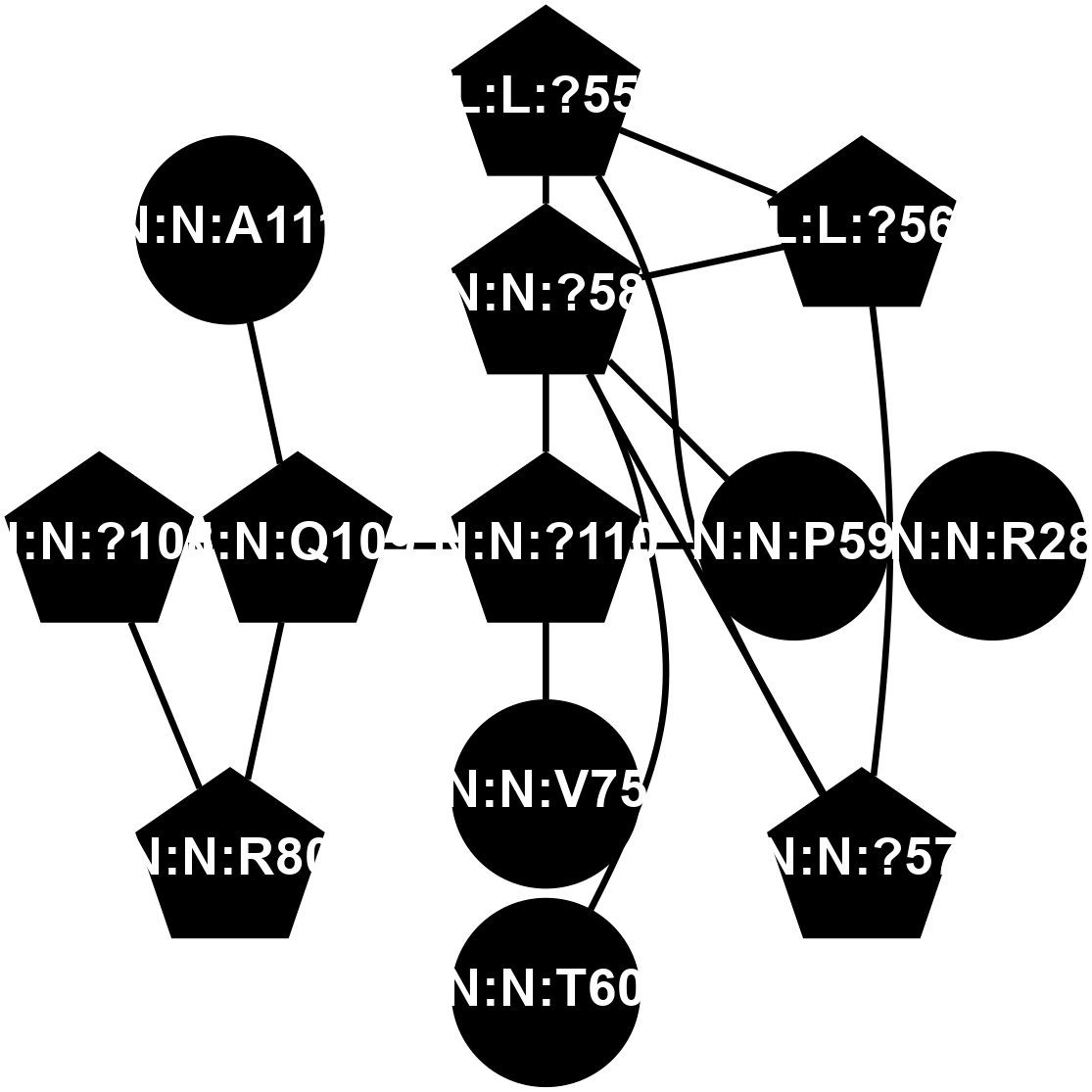
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?55 | L:L:?56 | 26.98 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?55 | N:N:?57 | 11.53 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?55 | N:N:?58 | 14.29 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?56 | N:N:?57 | 27.39 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?56 | N:N:?58 | 15.87 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:P59 | N:N:R28 | 5.76 | 2 | No | No | 0 | 0 | 1 | 2 | | N:N:?57 | N:N:?58 | 21.62 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?58 | N:N:P59 | 19.34 | 2 | Yes | No | 0 | 0 | 1 | 1 | | N:N:?58 | N:N:T60 | 4.74 | 2 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?110 | N:N:?58 | 12.7 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?110 | N:N:P59 | 5.28 | 2 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?110 | N:N:V75 | 14.36 | 2 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?108 | N:N:R80 | 5.2 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:Q109 | N:N:R80 | 12.85 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?108 | N:N:Q109 | 5.7 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?110 | N:N:Q109 | 4.28 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:A111 | N:N:Q109 | 4.55 | 0 | No | Yes | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.16 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
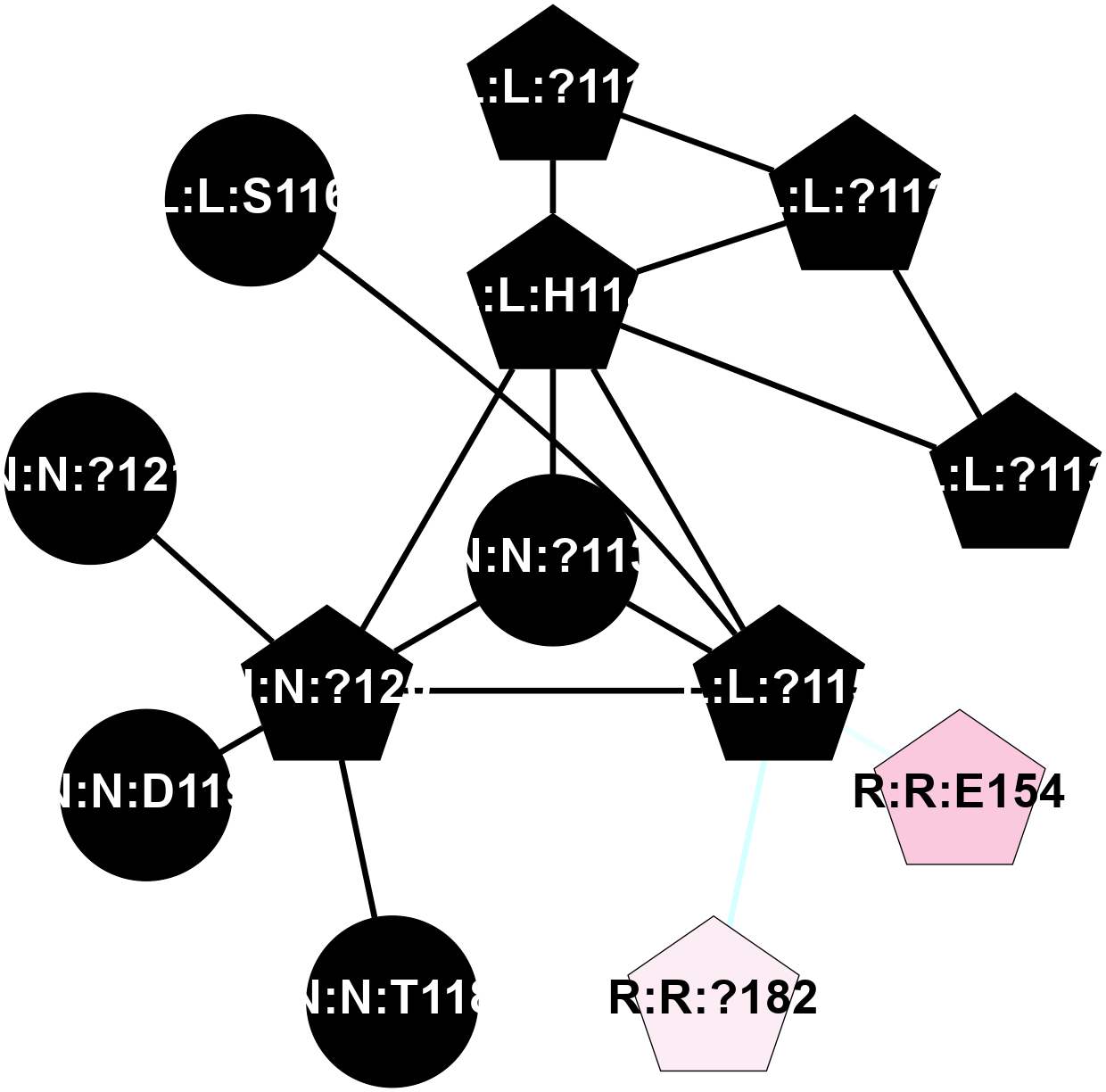
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?111 | L:L:?112 | 28.83 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?111 | L:L:H114 | 22.03 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?112 | L:L:?113 | 39.28 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?112 | L:L:H114 | 7.5 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?113 | L:L:H114 | 7.5 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?115 | L:L:H114 | 8.44 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:H114 | N:N:?113 | 5.51 | 2 | Yes | No | 0 | 0 | 1 | 0 | | L:L:H114 | N:N:?120 | 15.14 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?115 | N:N:?113 | 6.19 | 2 | Yes | No | 0 | 0 | 1 | 0 | | L:L:?115 | N:N:?120 | 6.19 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?115 | R:R:E154 | 7.11 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?115 | R:R:?182 | 5.62 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | N:N:?113 | N:N:?120 | 7.94 | 2 | No | Yes | 0 | 0 | 0 | 1 | | N:N:?120 | N:N:T118 | 4.74 | 2 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?120 | N:N:D119 | 4.36 | 2 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?120 | N:N:?121 | 14.22 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?115 | L:L:S116 | 3.58 | 2 | Yes | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 6.55 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:M71 | N:N:D119 | 4.16 | 2 | Yes | No | 0 | 0 | 2 | 1 | | L:L:V77 | N:N:T118 | 4.76 | 9 | Yes | No | 0 | 0 | 2 | 1 | | L:L:T78 | N:N:D119 | 13.01 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:S81 | N:N:?121 | 6.55 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:?111 | L:L:?112 | 28.83 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?111 | L:L:H114 | 22.03 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?112 | L:L:?113 | 39.28 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?112 | L:L:H114 | 7.5 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?113 | L:L:H114 | 7.5 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?115 | L:L:H114 | 8.44 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:H114 | N:N:?113 | 5.51 | 2 | Yes | No | 0 | 0 | 1 | 1 | | L:L:H114 | N:N:?120 | 15.14 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?115 | N:N:?113 | 6.19 | 2 | Yes | No | 0 | 0 | 1 | 1 | | L:L:?115 | N:N:?120 | 6.19 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?115 | R:R:E154 | 7.11 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?115 | R:R:?182 | 5.62 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | N:N:?113 | N:N:?120 | 7.94 | 2 | No | Yes | 0 | 0 | 1 | 0 | | N:N:?120 | N:N:T118 | 4.74 | 2 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?120 | N:N:D119 | 4.36 | 2 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?120 | N:N:?121 | 14.22 | 2 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?121 | N:N:?122 | 13.68 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:?115 | L:L:S116 | 3.58 | 2 | Yes | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 8.77 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 22.00 | | Average Links Mediated by Hubs In Shell | 19.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:R66 | N:N:E39 | 6.98 | 0 | No | No | 0 | 0 | 2 | 2 | | L:L:R66 | N:N:D131 | 7.15 | 0 | No | No | 0 | 0 | 2 | 1 | | N:N:?130 | N:N:A37 | 5.06 | 0 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?46 | N:N:E39 | 11.35 | 0 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?46 | N:N:V45 | 9.57 | 0 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?46 | N:N:I47 | 9.17 | 0 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?130 | N:N:?46 | 14.29 | 0 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?130 | N:N:T129 | 12.63 | 0 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?130 | N:N:D131 | 7.27 | 0 | Yes | No | 0 | 0 | 0 | 1 | | N:N:A37 | N:N:T48 | 1.68 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 9.81 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:M71 | R:R:N107 | 8.41 | 2 | Yes | Yes | 0 | 7 | 2 | 2 | | L:L:M71 | R:R:N132 | 7.01 | 2 | Yes | Yes | 0 | 8 | 2 | 1 | | L:L:L72 | R:R:N132 | 4.12 | 0 | No | Yes | 0 | 8 | 2 | 1 | | L:L:V73 | R:R:N132 | 4.43 | 0 | No | Yes | 0 | 8 | 2 | 1 | | N:N:R115 | R:R:D157 | 10.72 | 2 | Yes | No | 0 | 6 | 2 | 1 | | N:N:R115 | R:R:?182 | 8.27 | 2 | Yes | Yes | 0 | 6 | 2 | 2 | | R:R:N107 | R:R:N132 | 5.45 | 2 | Yes | Yes | 7 | 8 | 2 | 1 | | R:R:?131 | R:R:N132 | 5.88 | 2 | Yes | Yes | 7 | 8 | 0 | 1 | | R:R:?131 | R:R:T133 | 7.89 | 2 | Yes | Yes | 7 | 8 | 0 | 1 | | R:R:?131 | R:R:?156 | 30.16 | 2 | Yes | Yes | 7 | 7 | 0 | 1 | | R:R:?131 | R:R:D157 | 5.81 | 2 | Yes | No | 7 | 6 | 0 | 1 | | R:R:?131 | R:R:N158 | 5.88 | 2 | Yes | Yes | 7 | 9 | 0 | 1 | | R:R:?134 | R:R:T133 | 7.71 | 0 | Yes | Yes | 5 | 8 | 2 | 1 | | R:R:I135 | R:R:T133 | 7.6 | 0 | Yes | Yes | 7 | 8 | 2 | 1 | | R:R:N158 | R:R:T133 | 8.77 | 2 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:I135 | R:R:I161 | 4.42 | 0 | Yes | No | 7 | 7 | 2 | 2 | | R:R:?156 | R:R:E154 | 4.26 | 2 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:I155 | R:R:N158 | 4.25 | 2 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:I155 | R:R:N184 | 5.66 | 2 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:?156 | R:R:D157 | 17.44 | 2 | Yes | No | 7 | 6 | 1 | 1 | | R:R:?156 | R:R:N158 | 14.7 | 2 | Yes | Yes | 7 | 9 | 1 | 1 | | R:R:?156 | R:R:?182 | 15.86 | 2 | Yes | Yes | 7 | 6 | 1 | 2 | | R:R:?156 | R:R:N184 | 14.7 | 2 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:I161 | R:R:N158 | 5.66 | 0 | No | Yes | 7 | 9 | 2 | 1 | | R:R:N158 | R:R:N184 | 6.81 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:?182 | R:R:N184 | 10.68 | 2 | Yes | Yes | 6 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 11.12 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 14.00 | | Average Links In Shell | 26.00 | | Average Links Mediated by Hubs In Shell | 26.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?115 | R:R:E154 | 7.11 | 2 | Yes | Yes | 0 | 7 | 2 | 1 | | L:L:?115 | R:R:?182 | 5.62 | 2 | Yes | Yes | 0 | 6 | 2 | 1 | | N:N:R115 | R:R:D157 | 10.72 | 2 | Yes | No | 0 | 6 | 2 | 1 | | N:N:R115 | R:R:?182 | 8.27 | 2 | Yes | Yes | 0 | 6 | 2 | 1 | | N:N:R115 | R:R:?183 | 5.3 | 2 | Yes | Yes | 0 | 7 | 2 | 2 | | R:R:?127 | R:R:S129 | 10.22 | 2 | Yes | No | 6 | 7 | 2 | 2 | | R:R:?127 | R:R:E154 | 14.18 | 2 | Yes | Yes | 6 | 7 | 2 | 1 | | R:R:E154 | R:R:S129 | 10.06 | 2 | Yes | No | 7 | 7 | 1 | 2 | | R:R:?131 | R:R:N132 | 5.88 | 2 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:?131 | R:R:T133 | 7.89 | 2 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:?131 | R:R:?156 | 30.16 | 2 | Yes | Yes | 7 | 7 | 1 | 0 | | R:R:?131 | R:R:D157 | 5.81 | 2 | Yes | No | 7 | 6 | 1 | 1 | | R:R:?131 | R:R:N158 | 5.88 | 2 | Yes | Yes | 7 | 9 | 1 | 1 | | R:R:N158 | R:R:T133 | 8.77 | 2 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:?156 | R:R:E154 | 4.26 | 2 | Yes | Yes | 7 | 7 | 0 | 1 | | R:R:?180 | R:R:E154 | 8.7 | 0 | No | Yes | 5 | 7 | 2 | 1 | | R:R:I155 | R:R:N158 | 4.25 | 2 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:I155 | R:R:N184 | 5.66 | 2 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:?156 | R:R:D157 | 17.44 | 2 | Yes | No | 7 | 6 | 0 | 1 | | R:R:?156 | R:R:N158 | 14.7 | 2 | Yes | Yes | 7 | 9 | 0 | 1 | | R:R:?156 | R:R:?182 | 15.86 | 2 | Yes | Yes | 7 | 6 | 0 | 1 | | R:R:?156 | R:R:N184 | 14.7 | 2 | Yes | Yes | 7 | 9 | 0 | 1 | | R:R:I161 | R:R:N158 | 5.66 | 0 | No | Yes | 7 | 9 | 2 | 1 | | R:R:N158 | R:R:N184 | 6.81 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:F186 | R:R:I161 | 6.28 | 2 | Yes | No | 8 | 7 | 2 | 2 | | R:R:?180 | R:R:?182 | 16.87 | 0 | No | Yes | 5 | 6 | 2 | 1 | | R:R:?182 | R:R:?183 | 15.26 | 2 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:?182 | R:R:N184 | 10.68 | 2 | Yes | Yes | 6 | 9 | 1 | 1 | | R:R:?182 | R:R:E203 | 16.75 | 2 | Yes | No | 6 | 5 | 1 | 2 | | R:R:?182 | R:R:?205 | 8.03 | 2 | Yes | No | 6 | 7 | 1 | 2 | | R:R:?183 | R:R:N184 | 8.38 | 2 | Yes | Yes | 7 | 9 | 2 | 1 | | R:R:?183 | R:R:N207 | 5.99 | 2 | Yes | No | 7 | 9 | 2 | 2 | | R:R:F186 | R:R:N184 | 15.71 | 2 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:N184 | R:R:N207 | 5.45 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:F186 | R:R:N207 | 10.87 | 2 | Yes | No | 8 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 16.19 | | Average Nodes In Shell | 21.00 | | Average Hubs In Shell | 14.00 | | Average Links In Shell | 35.00 | | Average Links Mediated by Hubs In Shell | 35.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
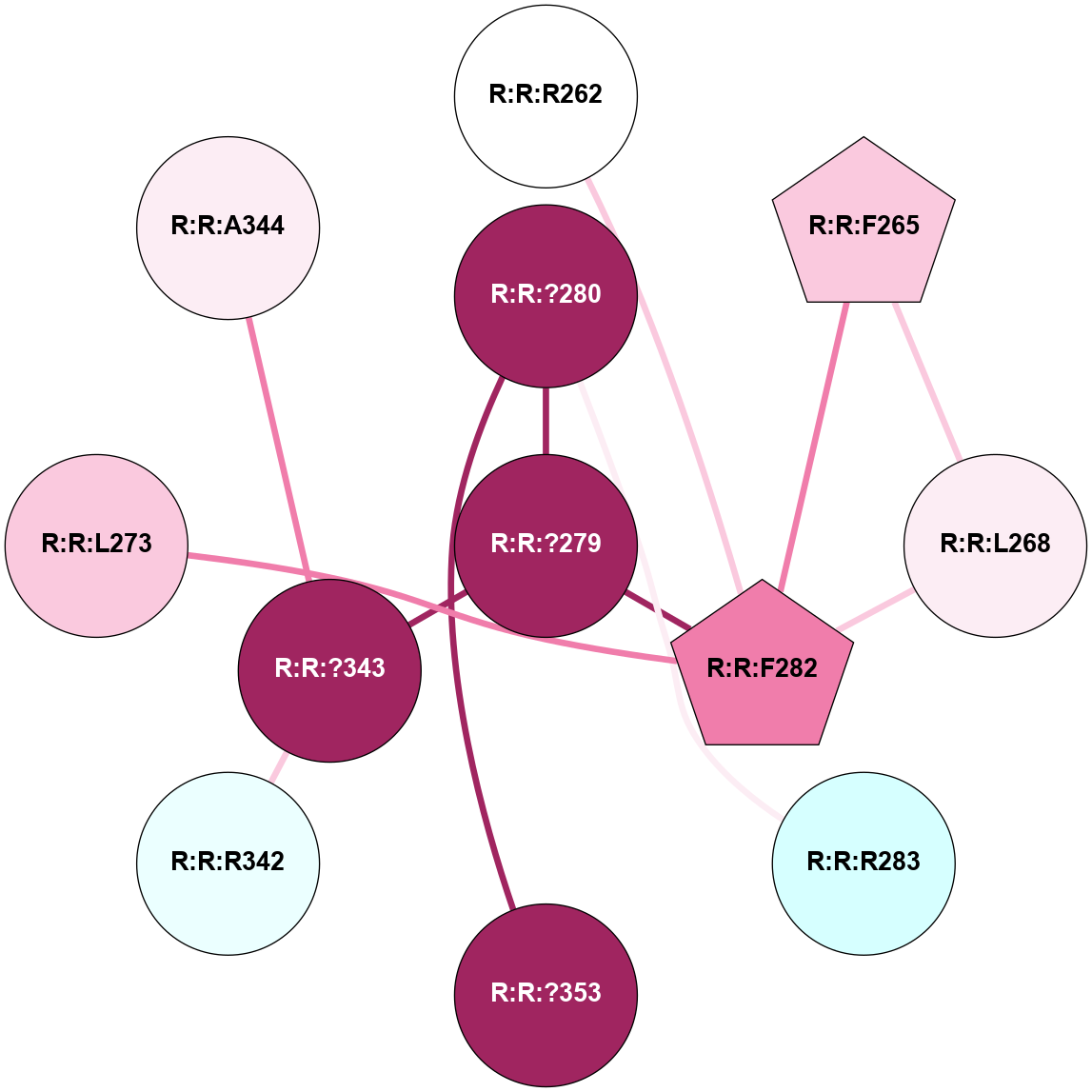
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F282 | R:R:R262 | 11.76 | 14 | Yes | No | 8 | 5 | 1 | 2 | | R:R:F265 | R:R:L268 | 9.74 | 14 | Yes | No | 7 | 6 | 2 | 2 | | R:R:F265 | R:R:F282 | 7.5 | 14 | Yes | Yes | 7 | 8 | 2 | 1 | | R:R:F282 | R:R:L268 | 7.31 | 14 | Yes | No | 8 | 6 | 1 | 2 | | R:R:?279 | R:R:?280 | 25.4 | 0 | No | No | 9 | 9 | 0 | 1 | | R:R:?279 | R:R:F282 | 7.83 | 0 | No | Yes | 9 | 8 | 0 | 1 | | R:R:?279 | R:R:?343 | 15.87 | 0 | No | No | 9 | 9 | 0 | 1 | | R:R:?280 | R:R:R283 | 10.41 | 0 | No | No | 9 | 3 | 1 | 2 | | R:R:?280 | R:R:?353 | 9.52 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:?343 | R:R:R342 | 7.81 | 0 | No | No | 9 | 4 | 1 | 2 | | R:R:F282 | R:R:L273 | 3.65 | 14 | Yes | No | 8 | 7 | 1 | 2 | | R:R:?343 | R:R:A344 | 3.37 | 0 | No | No | 9 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 16.37 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
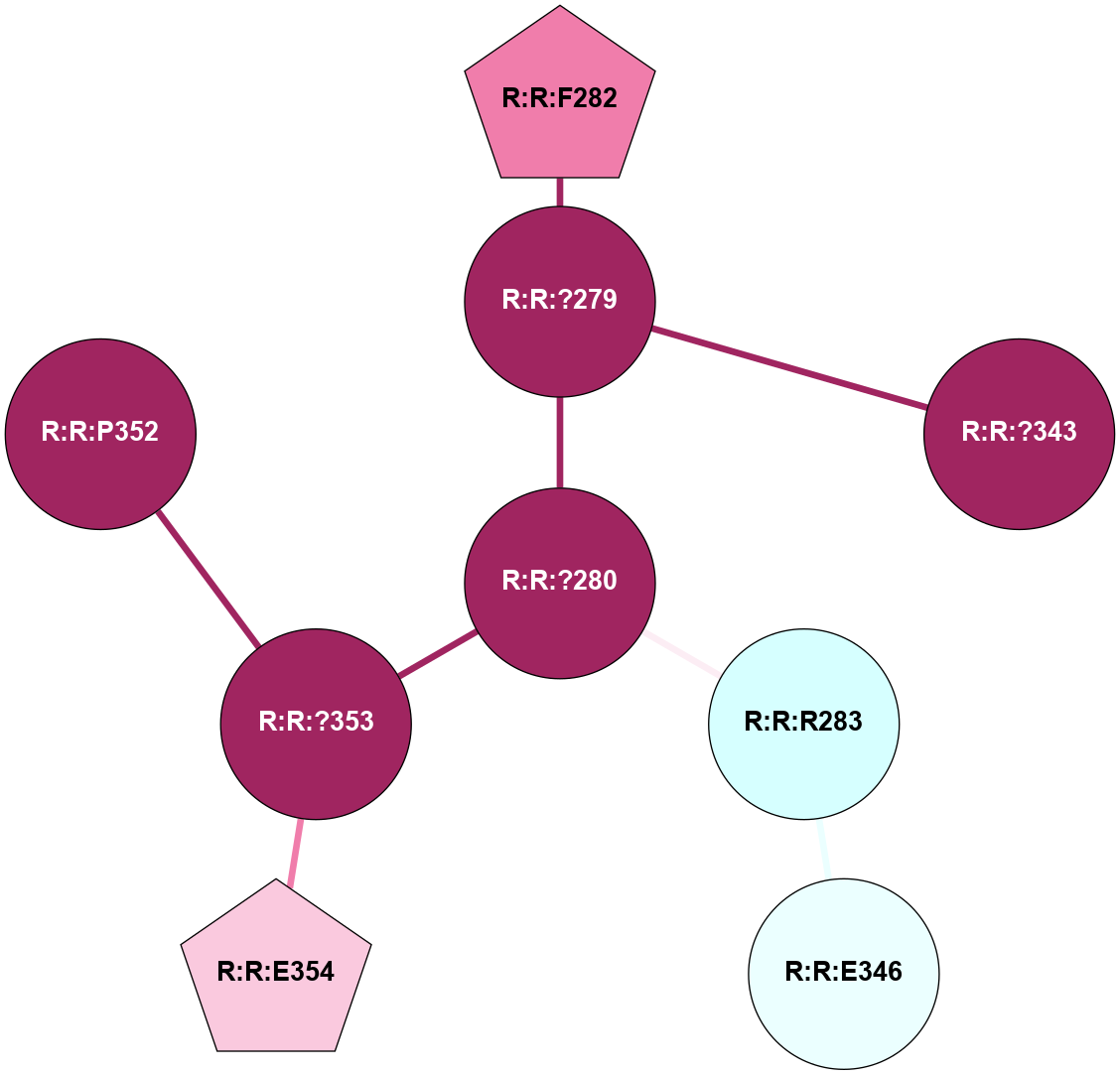
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?279 | R:R:?280 | 25.4 | 0 | No | No | 9 | 9 | 1 | 0 | | R:R:?279 | R:R:F282 | 7.83 | 0 | No | Yes | 9 | 8 | 1 | 2 | | R:R:?279 | R:R:?343 | 15.87 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:?280 | R:R:R283 | 10.41 | 0 | No | No | 9 | 3 | 0 | 1 | | R:R:?280 | R:R:?353 | 9.52 | 0 | No | No | 9 | 9 | 0 | 1 | | R:R:E346 | R:R:R283 | 10.47 | 0 | No | No | 4 | 3 | 2 | 1 | | R:R:?353 | R:R:E354 | 8.51 | 0 | No | Yes | 9 | 7 | 1 | 2 | | R:R:?353 | R:R:P352 | 1.76 | 0 | No | No | 9 | 9 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.11 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
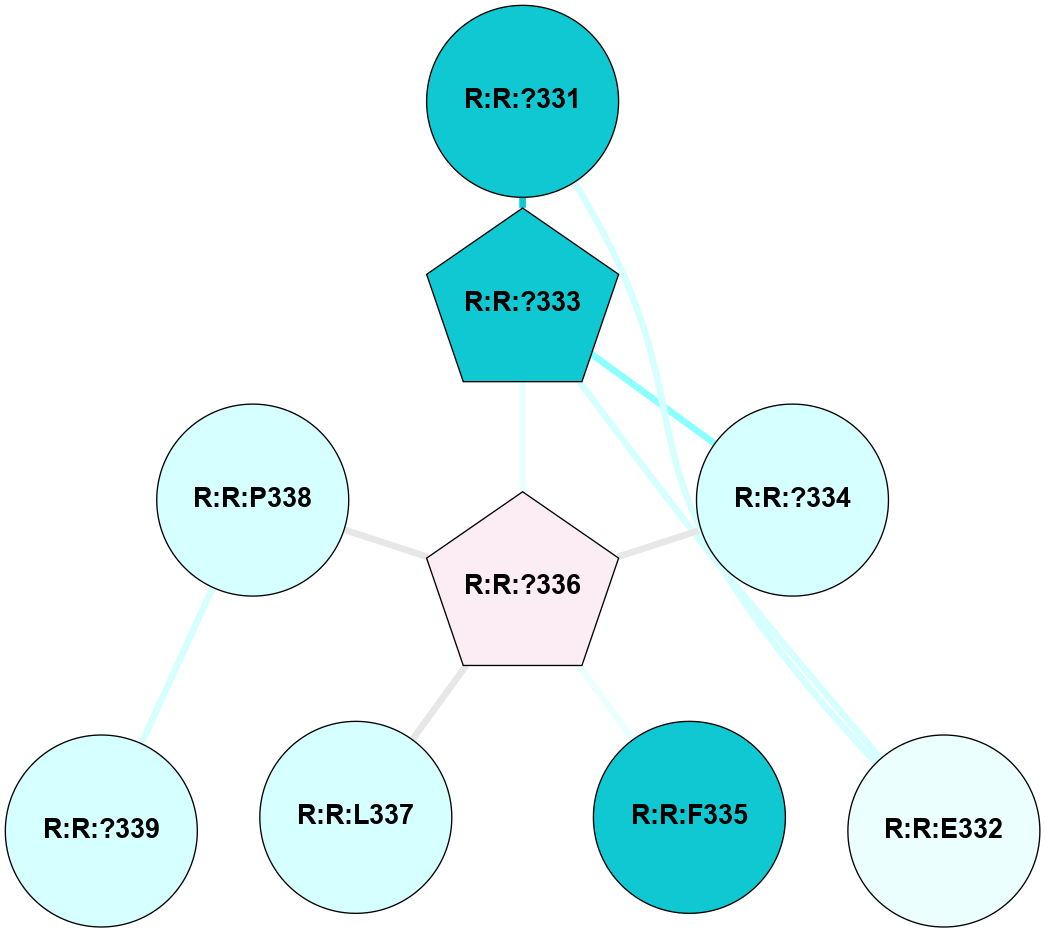
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?331 | R:R:?333 | 6.55 | 18 | No | Yes | 1 | 1 | 2 | 1 | | R:R:?333 | R:R:?334 | 18.78 | 18 | Yes | No | 1 | 3 | 1 | 1 | | R:R:?333 | R:R:?336 | 21.62 | 18 | Yes | Yes | 1 | 6 | 1 | 0 | | R:R:?334 | R:R:?336 | 6.46 | 18 | No | Yes | 3 | 6 | 1 | 0 | | R:R:F335 | R:R:P338 | 5.78 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:?336 | R:R:L337 | 7.41 | 18 | Yes | No | 6 | 3 | 0 | 1 | | R:R:?336 | R:R:P338 | 5.28 | 18 | Yes | No | 6 | 3 | 0 | 1 | | R:R:?339 | R:R:P338 | 2.94 | 0 | No | No | 3 | 3 | 2 | 1 | | R:R:?331 | R:R:E332 | 2.58 | 18 | No | No | 1 | 4 | 2 | 2 | | R:R:?333 | R:R:E332 | 2.58 | 18 | Yes | No | 1 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 10.19 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
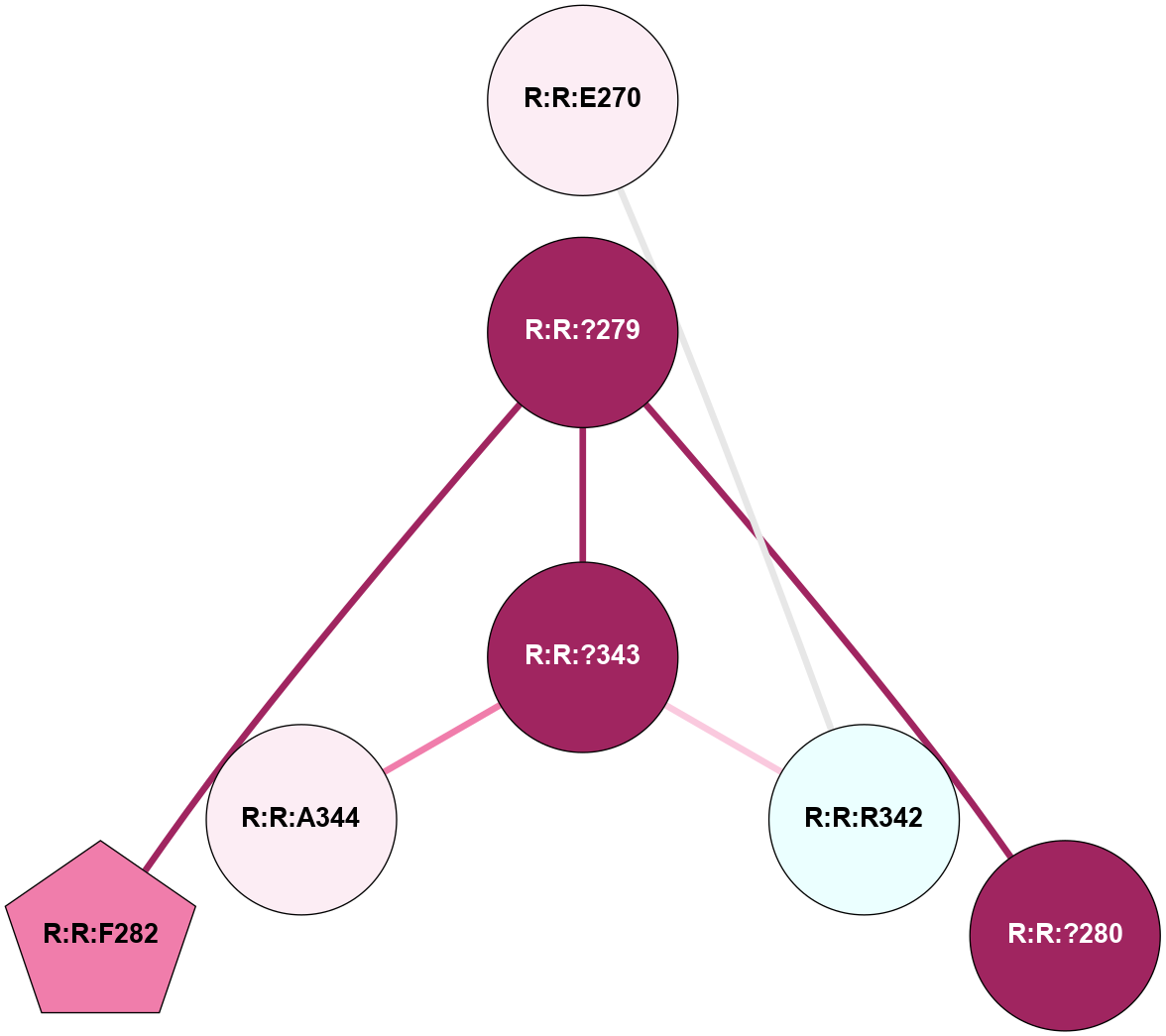
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E270 | R:R:R342 | 11.63 | 0 | No | No | 6 | 4 | 2 | 1 | | R:R:?279 | R:R:?280 | 25.4 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:?279 | R:R:F282 | 7.83 | 0 | No | Yes | 9 | 8 | 1 | 2 | | R:R:?279 | R:R:?343 | 15.87 | 0 | No | No | 9 | 9 | 1 | 0 | | R:R:?343 | R:R:R342 | 7.81 | 0 | No | No | 9 | 4 | 0 | 1 | | R:R:?343 | R:R:A344 | 3.37 | 0 | No | No | 9 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 9.02 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
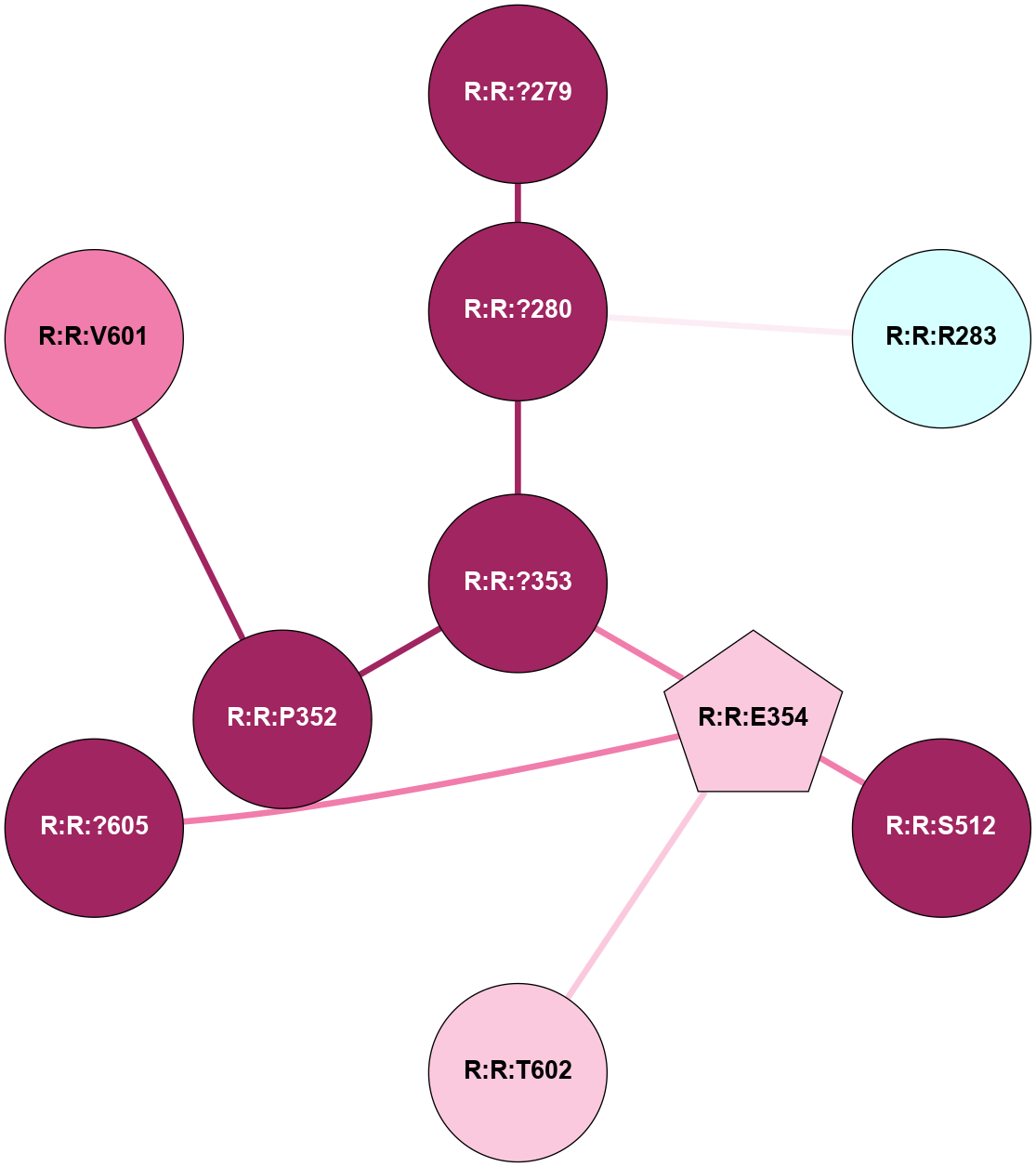
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?279 | R:R:?280 | 25.4 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:?280 | R:R:R283 | 10.41 | 0 | No | No | 9 | 3 | 1 | 2 | | R:R:?280 | R:R:?353 | 9.52 | 0 | No | No | 9 | 9 | 1 | 0 | | R:R:?353 | R:R:E354 | 8.51 | 0 | No | Yes | 9 | 7 | 0 | 1 | | R:R:E354 | R:R:S512 | 4.31 | 0 | Yes | No | 7 | 9 | 1 | 2 | | R:R:E354 | R:R:T602 | 12.7 | 0 | Yes | No | 7 | 7 | 1 | 2 | | R:R:?605 | R:R:E354 | 8.7 | 0 | No | Yes | 9 | 7 | 2 | 1 | | R:R:P352 | R:R:V601 | 3.53 | 0 | No | No | 9 | 8 | 1 | 2 | | R:R:?353 | R:R:P352 | 1.76 | 0 | No | No | 9 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.60 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
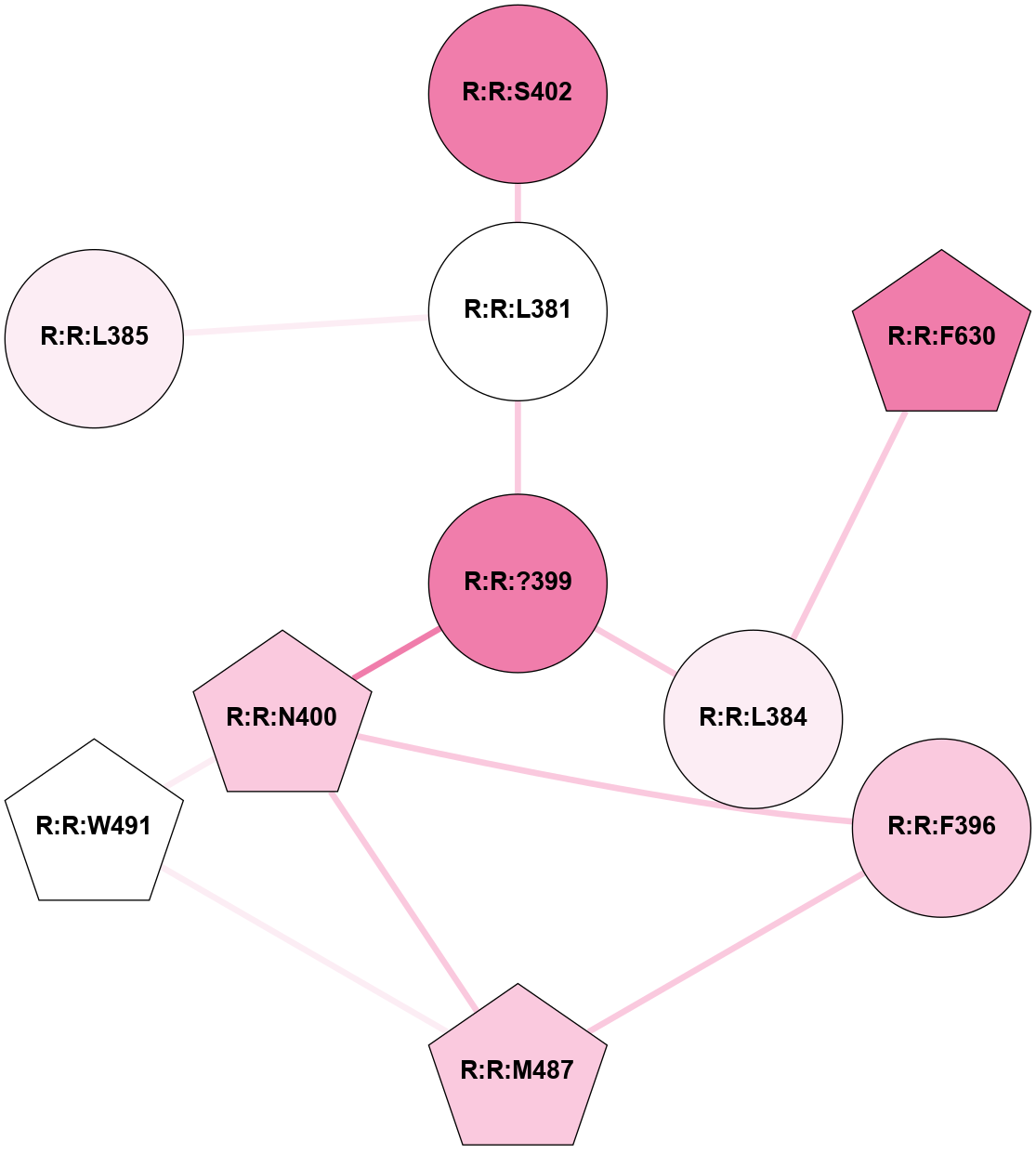
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?399 | R:R:L381 | 7.41 | 0 | No | No | 8 | 5 | 0 | 1 | | R:R:L381 | R:R:S402 | 4.5 | 0 | No | No | 5 | 8 | 1 | 2 | | R:R:?399 | R:R:L384 | 4.45 | 0 | No | No | 8 | 6 | 0 | 1 | | R:R:F630 | R:R:L384 | 4.87 | 0 | Yes | No | 8 | 6 | 2 | 1 | | R:R:F396 | R:R:N400 | 7.25 | 21 | No | Yes | 7 | 7 | 2 | 1 | | R:R:F396 | R:R:M487 | 4.98 | 21 | No | Yes | 7 | 7 | 2 | 2 | | R:R:?399 | R:R:N400 | 5.88 | 0 | No | Yes | 8 | 7 | 0 | 1 | | R:R:M487 | R:R:N400 | 16.83 | 21 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:N400 | R:R:W491 | 5.65 | 21 | Yes | Yes | 7 | 5 | 1 | 2 | | R:R:M487 | R:R:W491 | 4.65 | 21 | Yes | Yes | 7 | 5 | 2 | 2 | | R:R:L381 | R:R:L385 | 2.77 | 0 | No | No | 5 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.91 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
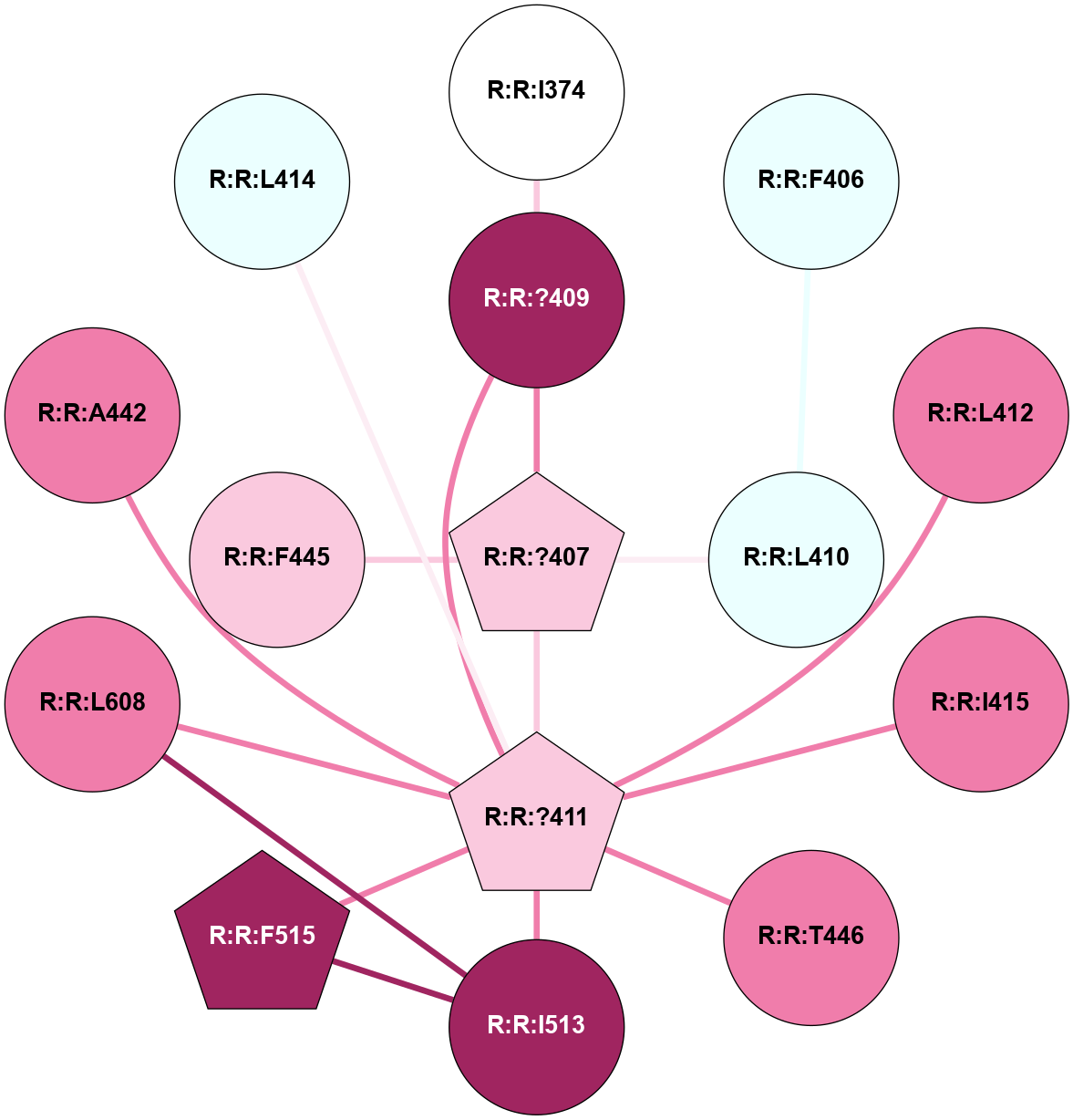
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?409 | R:R:I374 | 4.98 | 4 | No | No | 9 | 5 | 1 | 2 | | R:R:F406 | R:R:L410 | 10.96 | 0 | No | No | 4 | 4 | 2 | 1 | | R:R:?407 | R:R:?409 | 10.34 | 4 | Yes | No | 7 | 9 | 0 | 1 | | R:R:?407 | R:R:L410 | 8.89 | 4 | Yes | No | 7 | 4 | 0 | 1 | | R:R:?407 | R:R:?411 | 5.77 | 4 | Yes | Yes | 7 | 7 | 0 | 1 | | R:R:?409 | R:R:?411 | 9.39 | 4 | No | Yes | 9 | 7 | 1 | 1 | | R:R:?411 | R:R:L412 | 9.42 | 4 | Yes | No | 7 | 8 | 1 | 2 | | R:R:?411 | R:R:I415 | 18.05 | 4 | Yes | No | 7 | 8 | 1 | 2 | | R:R:?411 | R:R:T446 | 10.04 | 4 | Yes | No | 7 | 8 | 1 | 2 | | R:R:?411 | R:R:I513 | 4.16 | 4 | Yes | No | 7 | 9 | 1 | 2 | | R:R:?411 | R:R:F515 | 4.74 | 4 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:?411 | R:R:L608 | 6.73 | 4 | Yes | No | 7 | 8 | 1 | 2 | | R:R:F515 | R:R:I513 | 7.54 | 4 | Yes | No | 9 | 9 | 2 | 2 | | R:R:I513 | R:R:L608 | 5.71 | 4 | No | No | 9 | 8 | 2 | 2 | | R:R:?411 | R:R:A442 | 3.06 | 4 | Yes | No | 7 | 8 | 1 | 2 | | R:R:?411 | R:R:L414 | 2.69 | 4 | Yes | No | 7 | 4 | 1 | 2 | | R:R:?407 | R:R:F445 | 2.61 | 4 | Yes | No | 7 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.90 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
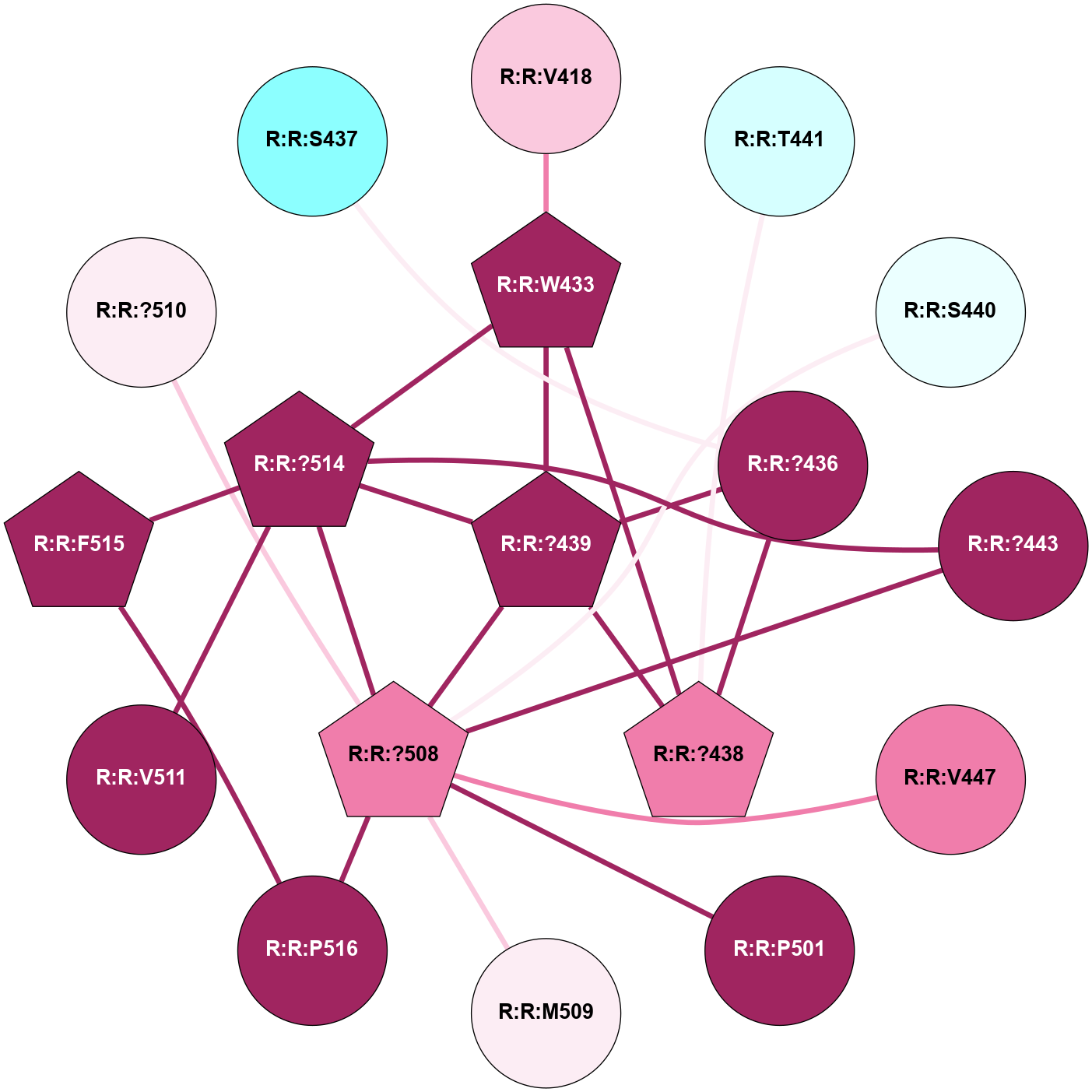
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:V418 | R:R:W433 | 13.48 | 0 | No | Yes | 7 | 9 | 2 | 1 | | R:R:?438 | R:R:W433 | 9.93 | 4 | Yes | Yes | 8 | 9 | 1 | 1 | | R:R:?439 | R:R:W433 | 29.27 | 4 | Yes | Yes | 9 | 9 | 0 | 1 | | R:R:?514 | R:R:W433 | 4.88 | 4 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:?436 | R:R:?438 | 6.32 | 4 | No | Yes | 9 | 8 | 1 | 1 | | R:R:?436 | R:R:?439 | 6.46 | 4 | No | Yes | 9 | 9 | 1 | 0 | | R:R:?438 | R:R:?439 | 19.39 | 4 | Yes | Yes | 8 | 9 | 1 | 0 | | R:R:?438 | R:R:T441 | 10.28 | 4 | Yes | No | 8 | 3 | 1 | 2 | | R:R:?439 | R:R:?508 | 4.32 | 4 | Yes | Yes | 9 | 8 | 0 | 1 | | R:R:?439 | R:R:?514 | 20.63 | 4 | Yes | Yes | 9 | 9 | 0 | 1 | | R:R:?508 | R:R:S440 | 7.3 | 4 | Yes | No | 8 | 4 | 1 | 2 | | R:R:?443 | R:R:?508 | 22.31 | 4 | No | Yes | 9 | 8 | 2 | 1 | | R:R:?443 | R:R:?514 | 7.76 | 4 | No | Yes | 9 | 9 | 2 | 1 | | R:R:?508 | R:R:V447 | 5.8 | 4 | Yes | No | 8 | 8 | 1 | 2 | | R:R:?508 | R:R:P501 | 19.17 | 4 | Yes | No | 8 | 9 | 1 | 2 | | R:R:?508 | R:R:M509 | 6.87 | 4 | Yes | No | 8 | 6 | 1 | 2 | | R:R:?508 | R:R:?514 | 18.74 | 4 | Yes | Yes | 8 | 9 | 1 | 1 | | R:R:?508 | R:R:P516 | 7.99 | 4 | Yes | No | 8 | 9 | 1 | 2 | | R:R:?514 | R:R:V511 | 4.79 | 4 | Yes | No | 9 | 9 | 1 | 2 | | R:R:?514 | R:R:F515 | 5.22 | 4 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:F515 | R:R:P516 | 4.33 | 4 | Yes | No | 9 | 9 | 2 | 2 | | R:R:?508 | R:R:?510 | 4.02 | 4 | Yes | No | 8 | 6 | 1 | 2 | | R:R:?436 | R:R:S437 | 3.93 | 4 | No | No | 9 | 2 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 16.01 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 23.00 | | Average Links Mediated by Hubs In Shell | 22.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
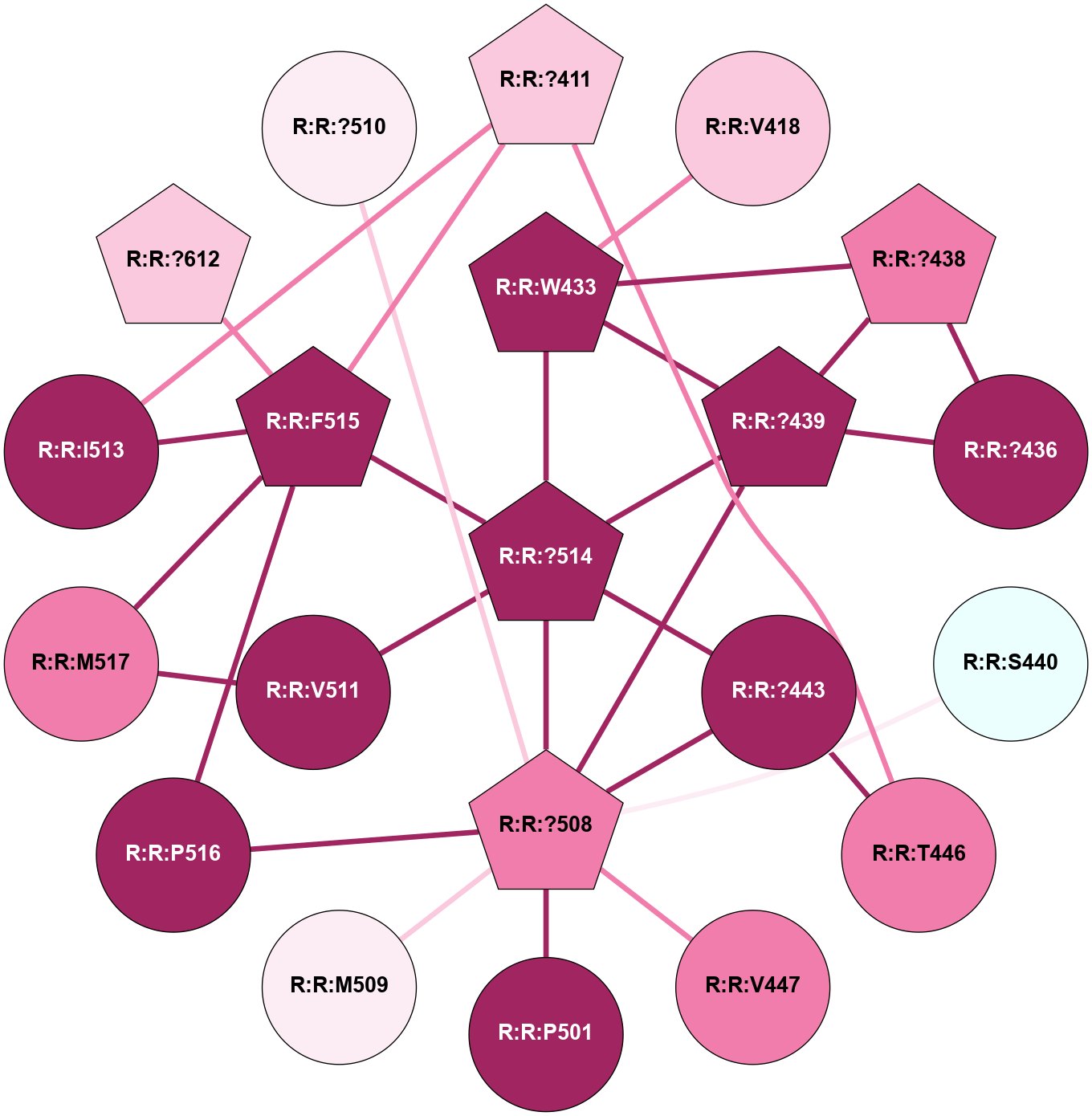
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?411 | R:R:T446 | 10.04 | 4 | Yes | No | 7 | 8 | 2 | 2 | | R:R:?411 | R:R:I513 | 4.16 | 4 | Yes | No | 7 | 9 | 2 | 2 | | R:R:?411 | R:R:F515 | 4.74 | 4 | Yes | Yes | 7 | 9 | 2 | 1 | | R:R:V418 | R:R:W433 | 13.48 | 0 | No | Yes | 7 | 9 | 2 | 1 | | R:R:?438 | R:R:W433 | 9.93 | 4 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:?439 | R:R:W433 | 29.27 | 4 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:?514 | R:R:W433 | 4.88 | 4 | Yes | Yes | 9 | 9 | 0 | 1 | | R:R:?436 | R:R:?438 | 6.32 | 4 | No | Yes | 9 | 8 | 2 | 2 | | R:R:?436 | R:R:?439 | 6.46 | 4 | No | Yes | 9 | 9 | 2 | 1 | | R:R:?438 | R:R:?439 | 19.39 | 4 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:?439 | R:R:?508 | 4.32 | 4 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:?439 | R:R:?514 | 20.63 | 4 | Yes | Yes | 9 | 9 | 1 | 0 | | R:R:?508 | R:R:S440 | 7.3 | 4 | Yes | No | 8 | 4 | 1 | 2 | | R:R:?443 | R:R:T446 | 9 | 4 | No | No | 9 | 8 | 1 | 2 | | R:R:?443 | R:R:?508 | 22.31 | 4 | No | Yes | 9 | 8 | 1 | 1 | | R:R:?443 | R:R:?514 | 7.76 | 4 | No | Yes | 9 | 9 | 1 | 0 | | R:R:?508 | R:R:V447 | 5.8 | 4 | Yes | No | 8 | 8 | 1 | 2 | | R:R:?508 | R:R:P501 | 19.17 | 4 | Yes | No | 8 | 9 | 1 | 2 | | R:R:?508 | R:R:M509 | 6.87 | 4 | Yes | No | 8 | 6 | 1 | 2 | | R:R:?508 | R:R:?514 | 18.74 | 4 | Yes | Yes | 8 | 9 | 1 | 0 | | R:R:?508 | R:R:P516 | 7.99 | 4 | Yes | No | 8 | 9 | 1 | 2 | | R:R:?514 | R:R:V511 | 4.79 | 4 | Yes | No | 9 | 9 | 0 | 1 | | R:R:M517 | R:R:V511 | 9.13 | 0 | No | No | 8 | 9 | 2 | 1 | | R:R:F515 | R:R:I513 | 7.54 | 4 | Yes | No | 9 | 9 | 1 | 2 | | R:R:?514 | R:R:F515 | 5.22 | 4 | Yes | Yes | 9 | 9 | 0 | 1 | | R:R:F515 | R:R:P516 | 4.33 | 4 | Yes | No | 9 | 9 | 1 | 2 | | R:R:F515 | R:R:M517 | 6.22 | 4 | Yes | No | 9 | 8 | 1 | 2 | | R:R:?612 | R:R:F515 | 11.85 | 4 | Yes | Yes | 7 | 9 | 2 | 1 | | R:R:?508 | R:R:?510 | 4.02 | 4 | Yes | No | 8 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 10.34 | | Average Nodes In Shell | 21.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 29.00 | | Average Links Mediated by Hubs In Shell | 27.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I542 | R:R:T461 | 9.12 | 0 | No | No | 8 | 8 | 1 | 2 | | R:R:?546 | R:R:T461 | 17.2 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:?546 | R:R:R464 | 9.45 | 6 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:?545 | R:R:W465 | 7.32 | 6 | Yes | No | 6 | 5 | 1 | 2 | | R:R:?543 | R:R:I542 | 4.59 | 6 | Yes | No | 7 | 8 | 0 | 1 | | R:R:D578 | R:R:I542 | 5.6 | 0 | No | No | 8 | 8 | 2 | 1 | | R:R:?543 | R:R:?545 | 12.7 | 6 | Yes | Yes | 7 | 6 | 0 | 1 | | R:R:?543 | R:R:?546 | 7.21 | 6 | Yes | Yes | 7 | 9 | 0 | 1 | | R:R:?543 | R:R:I547 | 10.7 | 6 | Yes | No | 7 | 5 | 0 | 1 | | R:R:?543 | R:R:I575 | 6.11 | 6 | Yes | No | 7 | 9 | 0 | 1 | | R:R:?545 | R:R:?546 | 25.95 | 6 | Yes | Yes | 6 | 9 | 1 | 1 | | R:R:?545 | R:R:?548 | 5.31 | 6 | Yes | No | 6 | 4 | 1 | 2 | | R:R:?545 | R:R:I549 | 6.11 | 6 | Yes | No | 6 | 9 | 1 | 2 | | R:R:?546 | R:R:I547 | 4.16 | 6 | Yes | No | 9 | 5 | 1 | 1 | | R:R:?546 | R:R:I549 | 11.11 | 6 | Yes | No | 9 | 9 | 1 | 2 | | R:R:?546 | R:R:?550 | 5.24 | 6 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:?546 | R:R:M571 | 16.5 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:?546 | R:R:L574 | 6.73 | 6 | Yes | No | 9 | 9 | 1 | 2 | | R:R:?546 | R:R:I575 | 4.16 | 6 | Yes | No | 9 | 9 | 1 | 1 | | R:R:?548 | R:R:?550 | 4.82 | 0 | No | Yes | 4 | 9 | 2 | 2 | | R:R:?550 | R:R:I549 | 5.55 | 6 | Yes | No | 9 | 9 | 2 | 2 | | R:R:?543 | R:R:A544 | 3.37 | 6 | Yes | No | 7 | 3 | 0 | 1 | | R:R:?545 | R:R:L462 | 2.96 | 6 | Yes | No | 6 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 7.45 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 23.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
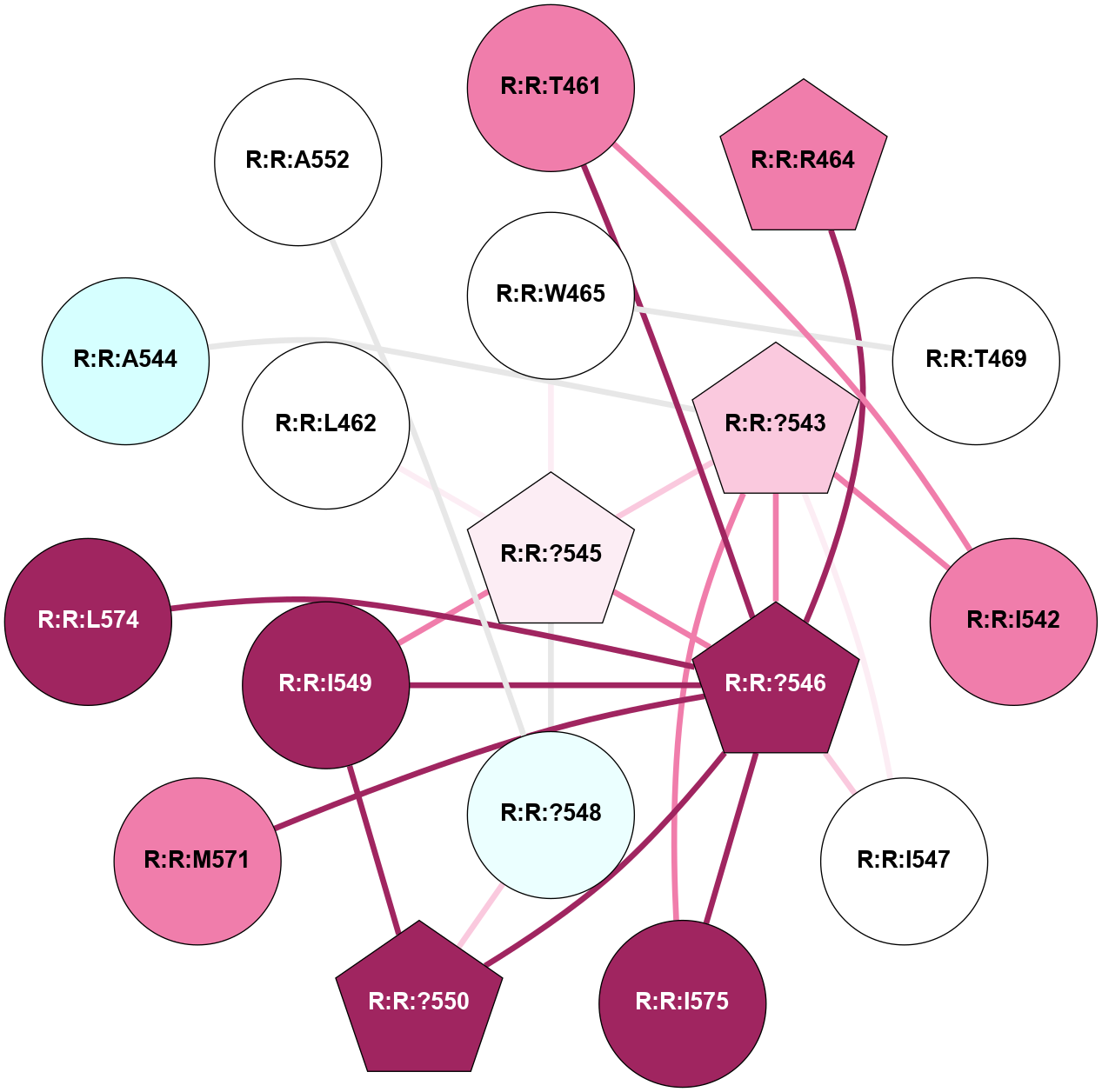
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I542 | R:R:T461 | 9.12 | 0 | No | No | 8 | 8 | 2 | 2 | | R:R:?546 | R:R:T461 | 17.2 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:?546 | R:R:R464 | 9.45 | 6 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:T469 | R:R:W465 | 16.98 | 0 | No | No | 5 | 5 | 2 | 1 | | R:R:?545 | R:R:W465 | 7.32 | 6 | Yes | No | 6 | 5 | 0 | 1 | | R:R:?543 | R:R:I542 | 4.59 | 6 | Yes | No | 7 | 8 | 1 | 2 | | R:R:?543 | R:R:?545 | 12.7 | 6 | Yes | Yes | 7 | 6 | 1 | 0 | | R:R:?543 | R:R:?546 | 7.21 | 6 | Yes | Yes | 7 | 9 | 1 | 1 | | R:R:?543 | R:R:I547 | 10.7 | 6 | Yes | No | 7 | 5 | 1 | 2 | | R:R:?543 | R:R:I575 | 6.11 | 6 | Yes | No | 7 | 9 | 1 | 2 | | R:R:?545 | R:R:?546 | 25.95 | 6 | Yes | Yes | 6 | 9 | 0 | 1 | | R:R:?545 | R:R:?548 | 5.31 | 6 | Yes | No | 6 | 4 | 0 | 1 | | R:R:?545 | R:R:I549 | 6.11 | 6 | Yes | No | 6 | 9 | 0 | 1 | | R:R:?546 | R:R:I547 | 4.16 | 6 | Yes | No | 9 | 5 | 1 | 2 | | R:R:?546 | R:R:I549 | 11.11 | 6 | Yes | No | 9 | 9 | 1 | 1 | | R:R:?546 | R:R:?550 | 5.24 | 6 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:?546 | R:R:M571 | 16.5 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:?546 | R:R:L574 | 6.73 | 6 | Yes | No | 9 | 9 | 1 | 2 | | R:R:?546 | R:R:I575 | 4.16 | 6 | Yes | No | 9 | 9 | 1 | 2 | | R:R:?548 | R:R:?550 | 4.82 | 0 | No | Yes | 4 | 9 | 1 | 2 | | R:R:?550 | R:R:I549 | 5.55 | 6 | Yes | No | 9 | 9 | 2 | 1 | | R:R:?543 | R:R:A544 | 3.37 | 6 | Yes | No | 7 | 3 | 1 | 2 | | R:R:?545 | R:R:L462 | 2.96 | 6 | Yes | No | 6 | 5 | 0 | 1 | | R:R:?548 | R:R:A552 | 1.88 | 0 | No | No | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 10.06 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 24.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?612 | R:R:M408 | 8.25 | 4 | Yes | No | 7 | 8 | 1 | 2 | | R:R:?612 | R:R:S450 | 4.38 | 4 | Yes | No | 7 | 8 | 1 | 2 | | R:R:?612 | R:R:F515 | 11.85 | 4 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:F576 | R:R:T580 | 5.19 | 0 | No | No | 8 | 6 | 2 | 1 | | R:R:D578 | R:R:N615 | 8.08 | 0 | No | No | 8 | 9 | 2 | 1 | | R:R:?581 | R:R:P584 | 5.28 | 4 | Yes | No | 9 | 9 | 0 | 1 | | R:R:?581 | R:R:I585 | 4.59 | 4 | Yes | No | 9 | 9 | 0 | 1 | | R:R:?581 | R:R:?612 | 4.32 | 4 | Yes | Yes | 9 | 7 | 0 | 1 | | R:R:?581 | R:R:I614 | 6.11 | 4 | Yes | No | 9 | 8 | 0 | 1 | | R:R:?581 | R:R:N615 | 13.23 | 4 | Yes | No | 9 | 9 | 0 | 1 | | R:R:F611 | R:R:P584 | 10.11 | 0 | No | No | 8 | 9 | 2 | 1 | | R:R:?612 | R:R:I585 | 6.94 | 4 | Yes | No | 7 | 9 | 1 | 1 | | R:R:?612 | R:R:P613 | 15.97 | 4 | Yes | No | 7 | 9 | 1 | 2 | | R:R:N615 | R:R:N619 | 5.45 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:?581 | R:R:T580 | 3.16 | 4 | Yes | No | 9 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.11 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
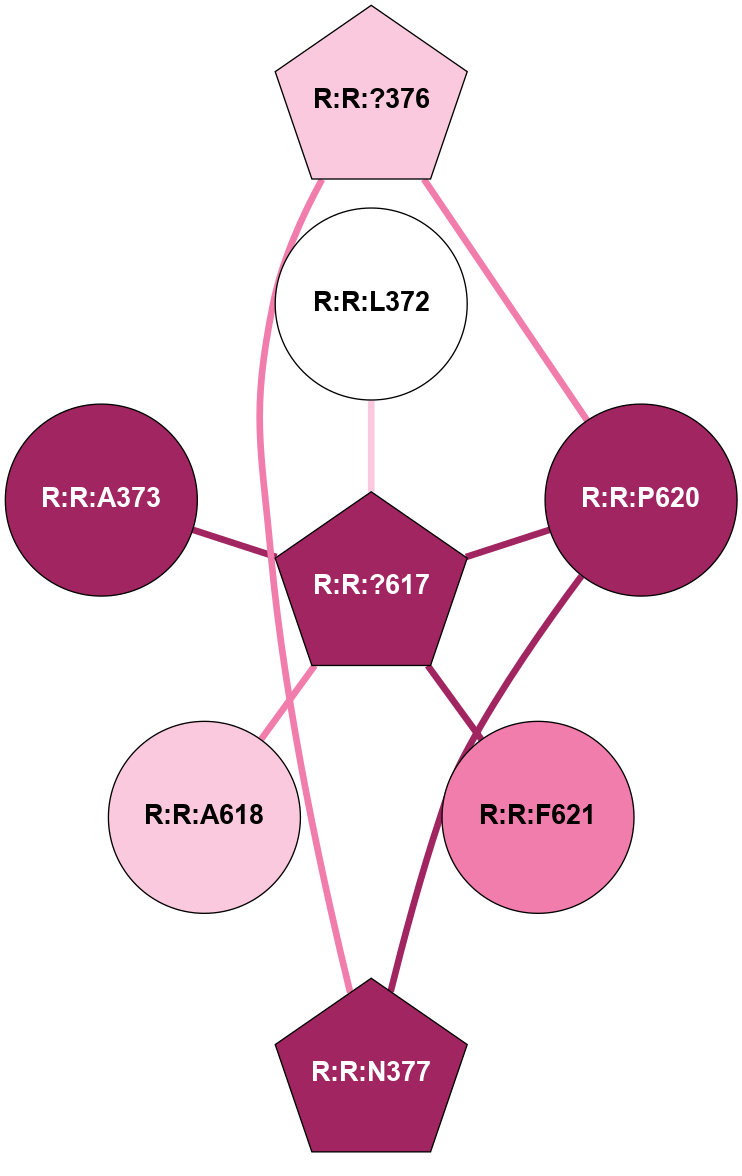
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?617 | R:R:L372 | 4.45 | 0 | Yes | No | 9 | 5 | 0 | 1 | | R:R:?376 | R:R:N377 | 4.79 | 6 | Yes | Yes | 7 | 9 | 2 | 2 | | R:R:?376 | R:R:P620 | 4.3 | 6 | Yes | No | 7 | 9 | 2 | 1 | | R:R:N377 | R:R:P620 | 11.4 | 6 | Yes | No | 9 | 9 | 2 | 1 | | R:R:?617 | R:R:P620 | 14.07 | 0 | Yes | No | 9 | 9 | 0 | 1 | | R:R:?617 | R:R:F621 | 6.52 | 0 | Yes | No | 9 | 8 | 0 | 1 | | R:R:?617 | R:R:A618 | 3.37 | 0 | Yes | No | 9 | 7 | 0 | 1 | | R:R:?617 | R:R:A373 | 1.69 | 0 | Yes | No | 9 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.02 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
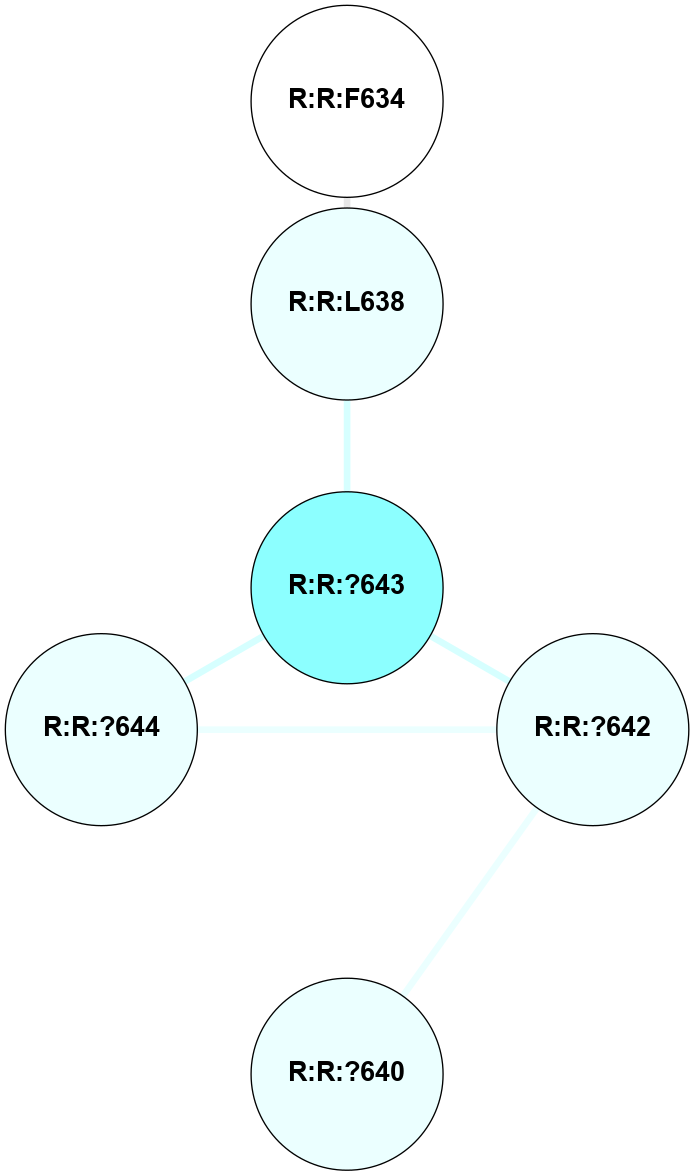
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F634 | R:R:L638 | 9.74 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:?643 | R:R:L638 | 10.38 | 34 | No | No | 2 | 4 | 0 | 1 | | R:R:?642 | R:R:?643 | 18.1 | 34 | No | No | 4 | 2 | 1 | 0 | | R:R:?642 | R:R:?644 | 7.76 | 34 | No | No | 4 | 4 | 1 | 1 | | R:R:?643 | R:R:?644 | 25.4 | 34 | No | No | 2 | 4 | 0 | 1 | | R:R:?640 | R:R:?642 | 2.88 | 0 | No | No | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.96 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of CMS in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?643 | R:R:L638 | 10.38 | 34 | No | No | 2 | 4 | 1 | 2 | | R:R:?642 | R:R:?643 | 18.1 | 34 | No | No | 4 | 2 | 1 | 1 | | R:R:?642 | R:R:?644 | 7.76 | 34 | No | No | 4 | 4 | 1 | 0 | | R:R:?643 | R:R:?644 | 25.4 | 34 | No | No | 2 | 4 | 1 | 0 | | R:R:?640 | R:R:?642 | 2.88 | 0 | No | No | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.58 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7FIH | A | Protein | Glycoprotein Hormone | LH | Homo Sapiens | hCG | Org43553 | chim(NtGi1-Gs)/Beta1/Gamma2 | 3.2 | 2021-09-29 | doi.org/10.1038/s41586-021-03924-2 |
|
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:?10 | A:A:T11 | 7.89 | 0 | No | No | 5 | 4 | 0 | 1 | | A:A:?10 | A:A:L12 | 4.45 | 0 | No | No | 5 | 2 | 0 | 1 | | A:A:D16 | A:A:L12 | 6.79 | 0 | No | No | 4 | 2 | 2 | 1 | | A:A:?17 | A:A:V20 | 4.44 | 0 | Yes | No | 3 | 3 | 1 | 2 | | A:A:?17 | A:A:A18 | 1.88 | 0 | Yes | No | 3 | 4 | 1 | 2 | | A:A:?10 | A:A:?17 | 0.88 | 0 | No | Yes | 5 | 3 | 0 | 1 | | A:A:?17 | A:A:E21 | 0.79 | 0 | Yes | No | 3 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 4.41 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:F238 | A:A:L44 | 15.83 | 9 | Yes | No | 9 | 9 | 1 | 2 | | A:A:F222 | B:B:W99 | 11.02 | 0 | No | Yes | 6 | 9 | 2 | 1 | | A:A:F238 | A:A:W234 | 4.01 | 9 | Yes | No | 9 | 9 | 1 | 2 | | A:A:?237 | A:A:F238 | 7.83 | 9 | No | Yes | 8 | 9 | 0 | 1 | | A:A:?237 | B:B:W99 | 10.98 | 9 | No | Yes | 8 | 9 | 0 | 1 | | A:A:F238 | A:A:V241 | 5.24 | 9 | Yes | No | 9 | 9 | 1 | 2 | | A:A:F238 | B:B:W99 | 13.03 | 9 | Yes | Yes | 9 | 9 | 1 | 1 | | B:B:Q75 | B:B:W99 | 5.48 | 0 | No | Yes | 9 | 9 | 2 | 1 | | B:B:L117 | B:B:W99 | 10.25 | 0 | No | Yes | 9 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.41 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:F273 | A:A:W277 | 10.02 | 3 | Yes | Yes | 9 | 6 | 2 | 1 | | A:A:F273 | A:A:V287 | 3.93 | 3 | Yes | No | 9 | 5 | 2 | 2 | | A:A:N278 | A:A:W277 | 4.52 | 0 | No | Yes | 8 | 6 | 2 | 1 | | A:A:V287 | A:A:W277 | 4.9 | 3 | No | Yes | 5 | 6 | 2 | 1 | | A:A:H357 | A:A:W277 | 26.45 | 0 | No | Yes | 7 | 6 | 2 | 1 | | A:A:?358 | A:A:W277 | 6.65 | 3 | Yes | Yes | 4 | 6 | 1 | 1 | | A:A:?359 | A:A:W277 | 7.32 | 3 | Yes | Yes | 6 | 6 | 0 | 1 | | A:A:?358 | A:A:S286 | 7.3 | 3 | Yes | No | 4 | 8 | 1 | 2 | | A:A:I382 | A:A:S286 | 9.29 | 0 | No | No | 7 | 8 | 2 | 2 | | A:A:L346 | A:A:S349 | 7.51 | 0 | No | No | 3 | 8 | 2 | 1 | | A:A:?359 | A:A:S349 | 6.43 | 3 | Yes | No | 6 | 8 | 0 | 1 | | A:A:?358 | A:A:?359 | 21.62 | 3 | Yes | Yes | 4 | 6 | 1 | 0 | | A:A:?358 | A:A:?360 | 10.47 | 3 | Yes | Yes | 4 | 8 | 1 | 1 | | A:A:?358 | A:A:R385 | 8.27 | 3 | Yes | Yes | 4 | 6 | 1 | 2 | | A:A:?358 | A:A:R389 | 7.09 | 3 | Yes | No | 4 | 5 | 1 | 2 | | A:A:?358 | A:A:L394 | 4.04 | 3 | Yes | Yes | 4 | 7 | 1 | 2 | | A:A:?359 | A:A:?360 | 27.39 | 3 | Yes | Yes | 6 | 8 | 0 | 1 | | A:A:?360 | A:A:P361 | 14.37 | 3 | Yes | No | 8 | 4 | 1 | 2 | | A:A:?360 | A:A:H362 | 6.25 | 3 | Yes | Yes | 8 | 9 | 1 | 2 | | A:A:?360 | A:A:D378 | 11.88 | 3 | Yes | No | 8 | 7 | 1 | 2 | | A:A:?360 | A:A:D381 | 13.2 | 3 | Yes | No | 8 | 8 | 1 | 2 | | A:A:?360 | A:A:I382 | 8.33 | 3 | Yes | No | 8 | 7 | 1 | 2 | | A:A:D378 | A:A:H362 | 13.87 | 3 | No | Yes | 7 | 9 | 2 | 2 | | A:A:D381 | A:A:R385 | 16.68 | 3 | No | Yes | 8 | 6 | 2 | 2 | | A:A:L394 | A:A:R385 | 3.64 | 3 | Yes | Yes | 7 | 6 | 2 | 2 | | A:A:L394 | A:A:R389 | 3.64 | 3 | Yes | No | 7 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 15.69 | | Average Nodes In Shell | 19.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 26.00 | | Average Links Mediated by Hubs In Shell | 24.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
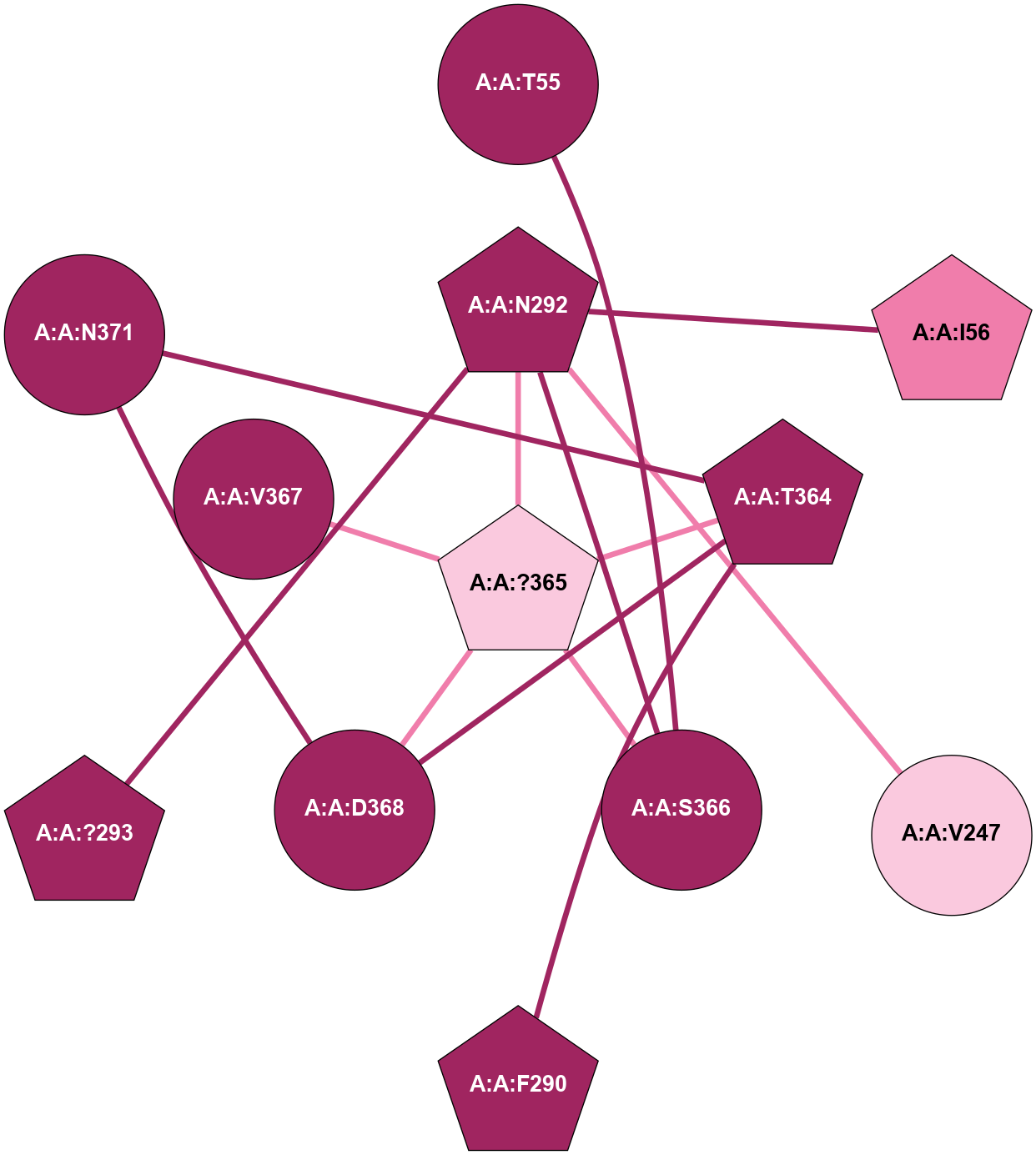
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:S366 | A:A:T55 | 4.8 | 5 | No | No | 9 | 9 | 1 | 2 | | A:A:I56 | A:A:N292 | 4.25 | 5 | Yes | Yes | 8 | 9 | 2 | 1 | | A:A:N292 | A:A:V247 | 4.43 | 5 | Yes | No | 9 | 7 | 1 | 2 | | A:A:F290 | A:A:T364 | 5.19 | 0 | Yes | Yes | 9 | 9 | 2 | 1 | | A:A:?293 | A:A:N292 | 8.19 | 0 | Yes | Yes | 9 | 9 | 2 | 1 | | A:A:?365 | A:A:N292 | 5.88 | 5 | Yes | Yes | 7 | 9 | 0 | 1 | | A:A:N292 | A:A:S366 | 5.96 | 5 | Yes | No | 9 | 9 | 1 | 1 | | A:A:?365 | A:A:T364 | 12.63 | 5 | Yes | Yes | 7 | 9 | 0 | 1 | | A:A:D368 | A:A:T364 | 5.78 | 5 | No | Yes | 9 | 9 | 1 | 1 | | A:A:N371 | A:A:T364 | 8.77 | 5 | No | Yes | 9 | 9 | 2 | 1 | | A:A:?365 | A:A:S366 | 4.82 | 5 | Yes | No | 7 | 9 | 0 | 1 | | A:A:?365 | A:A:D368 | 24.71 | 5 | Yes | No | 7 | 9 | 0 | 1 | | A:A:D368 | A:A:N371 | 4.04 | 5 | No | No | 9 | 9 | 1 | 2 | | A:A:?365 | A:A:V367 | 1.6 | 5 | Yes | No | 7 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.93 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
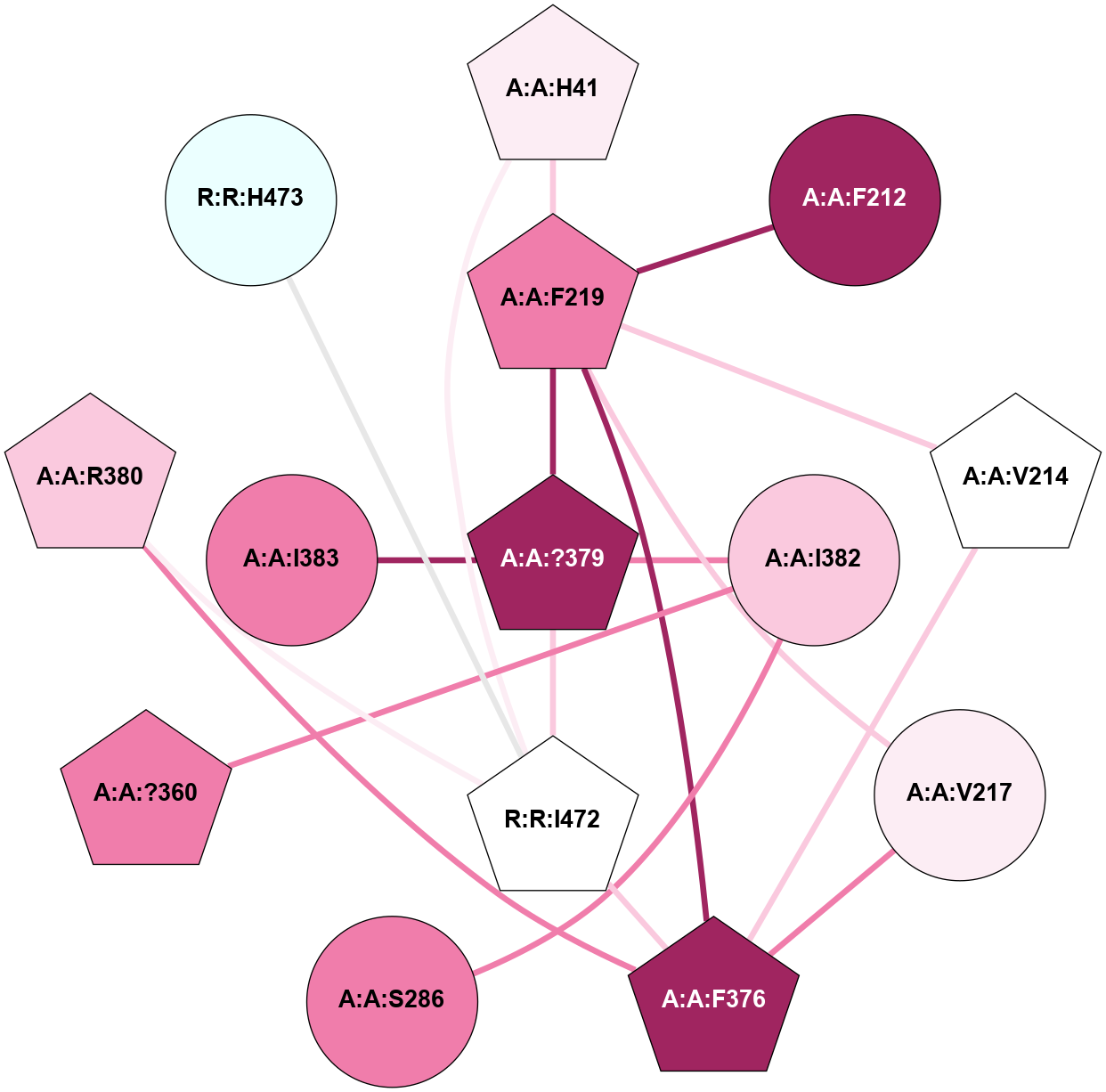
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:F219 | A:A:H41 | 7.92 | 5 | Yes | Yes | 8 | 6 | 1 | 2 | | A:A:H41 | R:R:I472 | 5.3 | 0 | Yes | Yes | 6 | 5 | 2 | 1 | | A:A:F212 | A:A:F219 | 11.79 | 5 | No | Yes | 9 | 8 | 2 | 1 | | A:A:F219 | A:A:V214 | 5.24 | 5 | Yes | Yes | 8 | 5 | 1 | 2 | | A:A:F376 | A:A:V214 | 6.55 | 5 | Yes | Yes | 9 | 5 | 2 | 2 | | A:A:F219 | A:A:V217 | 5.24 | 5 | Yes | No | 8 | 6 | 1 | 2 | | A:A:F376 | A:A:V217 | 3.93 | 5 | Yes | No | 9 | 6 | 2 | 2 | | A:A:F219 | A:A:F376 | 5.36 | 5 | Yes | Yes | 8 | 9 | 1 | 2 | | A:A:?379 | A:A:F219 | 5.22 | 0 | Yes | Yes | 9 | 8 | 0 | 1 | | A:A:I382 | A:A:S286 | 9.29 | 0 | No | No | 7 | 8 | 1 | 2 | | A:A:?360 | A:A:I382 | 8.33 | 3 | Yes | No | 8 | 7 | 2 | 1 | | A:A:F376 | A:A:R380 | 6.41 | 5 | Yes | Yes | 9 | 7 | 2 | 2 | | A:A:F376 | R:R:I472 | 8.79 | 5 | Yes | Yes | 9 | 5 | 2 | 1 | | A:A:?379 | A:A:I382 | 7.64 | 0 | Yes | No | 9 | 7 | 0 | 1 | | A:A:?379 | R:R:I472 | 4.59 | 0 | Yes | Yes | 9 | 5 | 0 | 1 | | A:A:R380 | R:R:I472 | 5.01 | 5 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:H473 | R:R:I472 | 6.63 | 0 | No | Yes | 4 | 5 | 2 | 1 | | A:A:?379 | A:A:I383 | 3.06 | 0 | Yes | No | 9 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 5.13 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
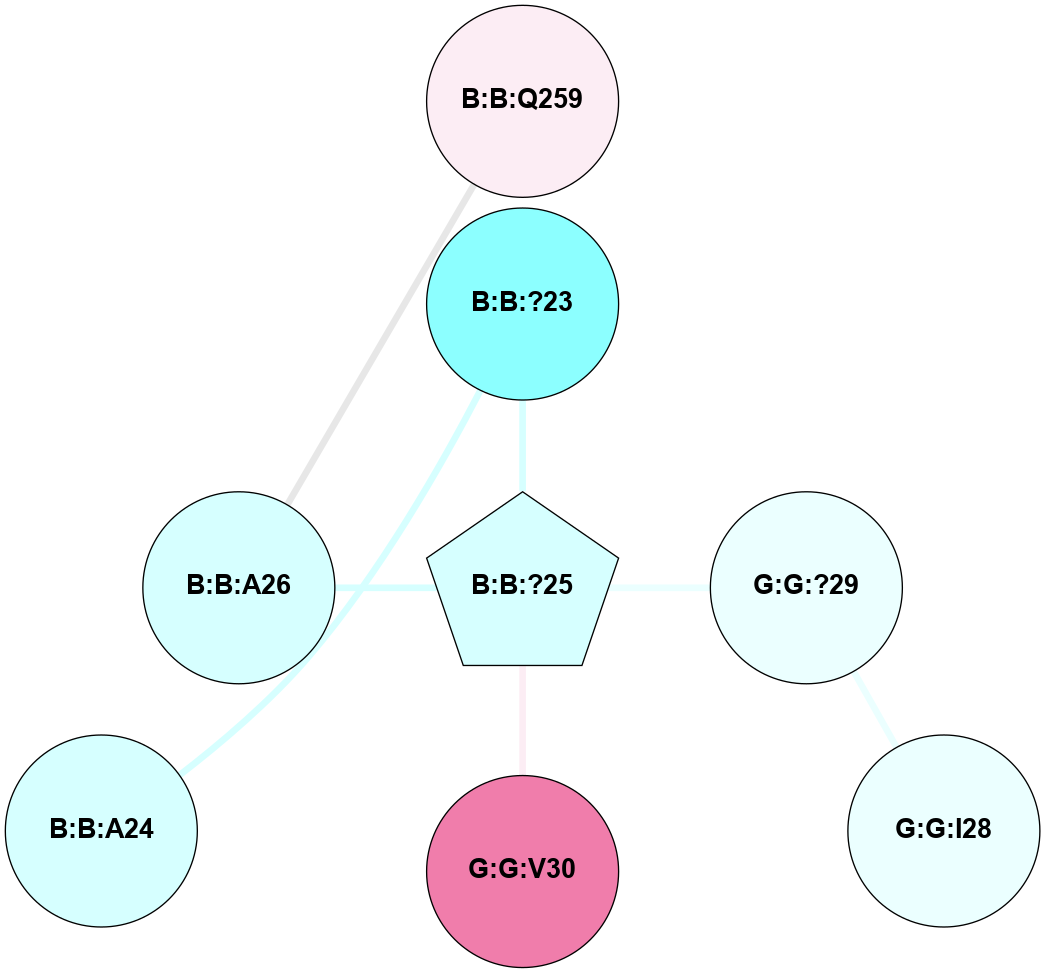
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:?23 | B:B:?25 | 7.07 | 0 | No | Yes | 2 | 3 | 1 | 0 | | B:B:?25 | G:G:?29 | 5.31 | 0 | Yes | No | 3 | 4 | 0 | 1 | | B:B:?25 | G:G:V30 | 4.79 | 0 | Yes | No | 3 | 8 | 0 | 1 | | B:B:A26 | B:B:Q259 | 4.55 | 0 | No | No | 3 | 6 | 1 | 2 | | G:G:?29 | G:G:I28 | 6.81 | 0 | No | No | 4 | 4 | 1 | 2 | | B:B:?25 | B:B:A26 | 3.37 | 0 | Yes | No | 3 | 3 | 0 | 1 | | B:B:?23 | B:B:A24 | 1.88 | 0 | No | No | 2 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 5.13 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:?103 | B:B:H62 | 5.51 | 4 | Yes | No | 8 | 6 | 0 | 1 | | B:B:?105 | B:B:H62 | 16.26 | 0 | Yes | No | 6 | 6 | 2 | 1 | | B:B:H62 | B:B:R150 | 6.77 | 4 | No | Yes | 6 | 6 | 1 | 1 | | B:B:L79 | B:B:V71 | 4.47 | 0 | No | Yes | 7 | 8 | 2 | 1 | | B:B:I81 | B:B:V71 | 6.14 | 0 | No | Yes | 7 | 8 | 2 | 1 | | B:B:?103 | B:B:V71 | 4.79 | 4 | Yes | Yes | 8 | 8 | 0 | 1 | | B:B:?105 | B:B:V71 | 13.04 | 0 | Yes | Yes | 6 | 8 | 2 | 1 | | B:B:?103 | B:B:T102 | 11.05 | 4 | Yes | Yes | 8 | 8 | 0 | 1 | | B:B:?115 | B:B:T102 | 3.86 | 0 | Yes | Yes | 9 | 8 | 2 | 1 | | B:B:?148 | B:B:T102 | 20.52 | 4 | Yes | Yes | 6 | 8 | 2 | 1 | | B:B:?149 | B:B:T102 | 15.78 | 4 | Yes | Yes | 8 | 8 | 2 | 1 | | B:B:?103 | B:B:A104 | 5.06 | 4 | Yes | No | 8 | 7 | 0 | 1 | | B:B:?103 | B:B:R150 | 3.9 | 4 | Yes | Yes | 8 | 6 | 0 | 1 | | B:B:?149 | B:B:A104 | 6.75 | 4 | Yes | No | 8 | 7 | 2 | 1 | | B:B:?148 | B:B:?149 | 28.57 | 4 | Yes | Yes | 6 | 8 | 2 | 2 | | B:B:?149 | B:B:R150 | 19.51 | 4 | Yes | Yes | 8 | 6 | 2 | 1 | | B:B:L192 | B:B:R150 | 3.64 | 0 | No | Yes | 5 | 6 | 2 | 1 | | B:B:?103 | B:B:A60 | 3.37 | 4 | Yes | No | 8 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 5.61 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 18.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
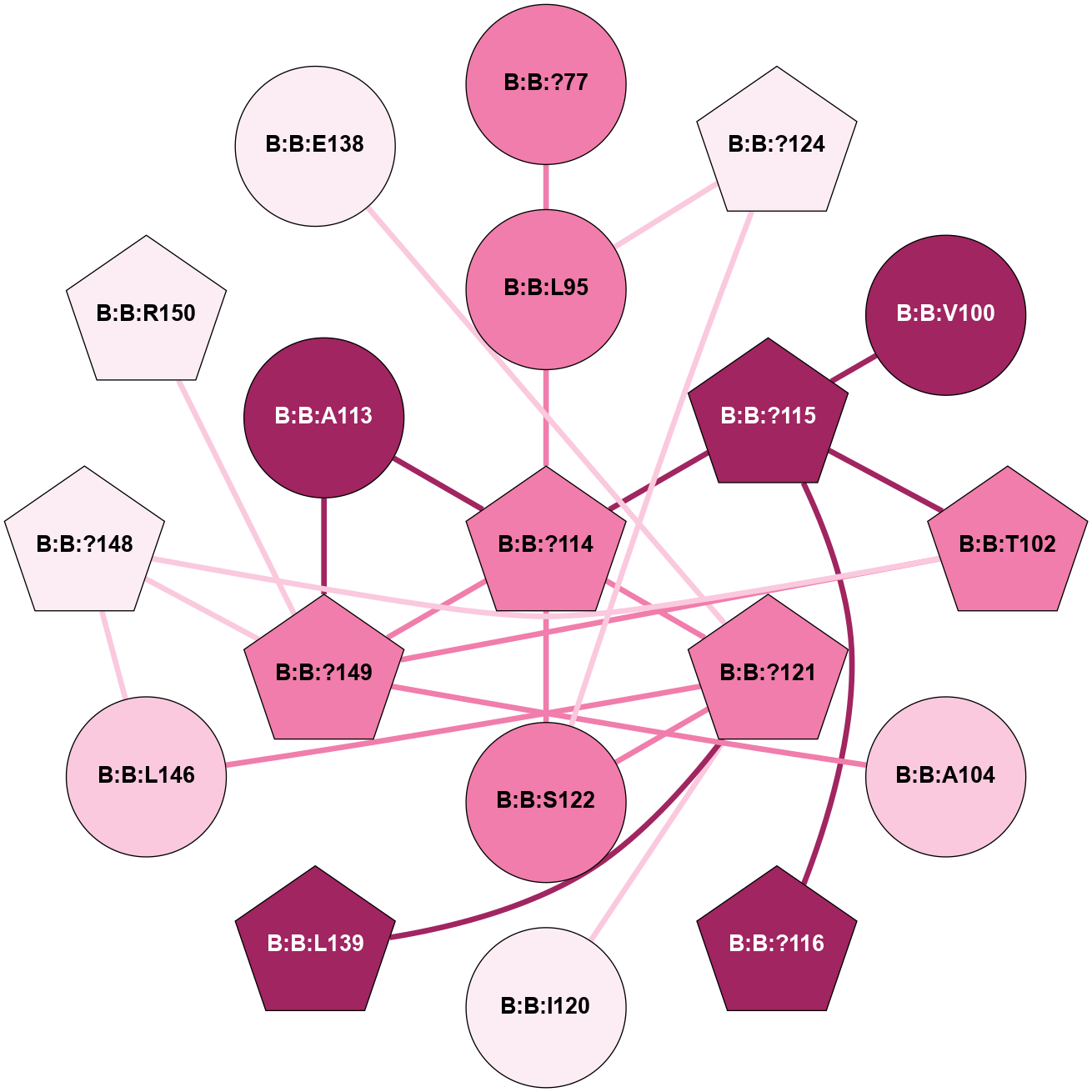
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:?77 | B:B:L95 | 9.66 | 10 | No | No | 8 | 8 | 2 | 1 | | B:B:?114 | B:B:L95 | 4.45 | 4 | Yes | No | 8 | 8 | 0 | 1 | | B:B:?124 | B:B:L95 | 5.38 | 4 | Yes | No | 6 | 8 | 2 | 1 | | B:B:?115 | B:B:V100 | 5.2 | 0 | Yes | No | 9 | 9 | 1 | 2 | | B:B:?115 | B:B:T102 | 3.86 | 0 | Yes | Yes | 9 | 8 | 1 | 2 | | B:B:?148 | B:B:T102 | 20.52 | 4 | Yes | Yes | 6 | 8 | 2 | 2 | | B:B:?149 | B:B:T102 | 15.78 | 4 | Yes | Yes | 8 | 8 | 1 | 2 | | B:B:?149 | B:B:A104 | 6.75 | 4 | Yes | No | 8 | 7 | 1 | 2 | | B:B:?114 | B:B:?115 | 16.8 | 4 | Yes | Yes | 8 | 9 | 0 | 1 | | B:B:?114 | B:B:?121 | 6.35 | 4 | Yes | Yes | 8 | 8 | 0 | 1 | | B:B:?114 | B:B:S122 | 12.86 | 4 | Yes | No | 8 | 8 | 0 | 1 | | B:B:?114 | B:B:?149 | 4.76 | 4 | Yes | Yes | 8 | 8 | 0 | 1 | | B:B:?115 | B:B:?116 | 10.53 | 0 | Yes | Yes | 9 | 9 | 1 | 2 | | B:B:?121 | B:B:I120 | 9.17 | 4 | Yes | No | 8 | 6 | 1 | 2 | | B:B:?121 | B:B:S122 | 4.82 | 4 | Yes | No | 8 | 8 | 1 | 1 | | B:B:?121 | B:B:L139 | 4.45 | 4 | Yes | Yes | 8 | 9 | 1 | 2 | | B:B:?121 | B:B:L146 | 4.45 | 4 | Yes | No | 8 | 7 | 1 | 2 | | B:B:?124 | B:B:S122 | 10.22 | 4 | Yes | No | 6 | 8 | 2 | 1 | | B:B:?148 | B:B:L146 | 5.93 | 4 | Yes | No | 6 | 7 | 2 | 2 | | B:B:?148 | B:B:?149 | 28.57 | 4 | Yes | Yes | 6 | 8 | 2 | 1 | | B:B:?149 | B:B:R150 | 19.51 | 4 | Yes | Yes | 8 | 6 | 1 | 2 | | B:B:?114 | B:B:A113 | 3.37 | 4 | Yes | No | 8 | 9 | 0 | 1 | | B:B:?149 | B:B:A113 | 3.37 | 4 | Yes | No | 8 | 9 | 1 | 1 | | B:B:?121 | B:B:E138 | 1.42 | 4 | Yes | No | 8 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 8.10 | | Average Nodes In Shell | 19.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 24.00 | | Average Links Mediated by Hubs In Shell | 23.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
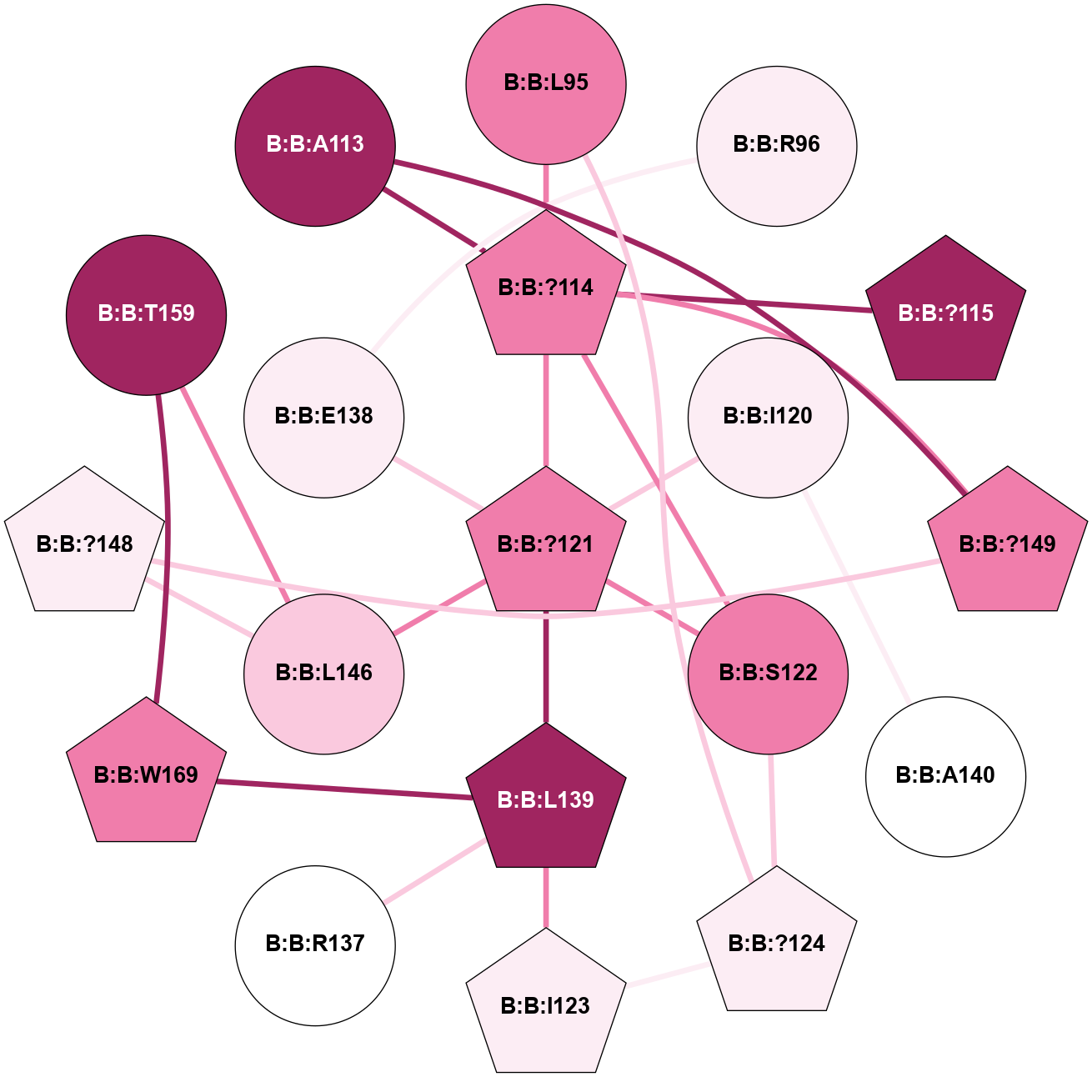
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:?114 | B:B:L95 | 4.45 | 4 | Yes | No | 8 | 8 | 1 | 2 | | B:B:?124 | B:B:L95 | 5.38 | 4 | Yes | No | 6 | 8 | 2 | 2 | | B:B:E138 | B:B:R96 | 8.14 | 0 | No | No | 6 | 6 | 1 | 2 | | B:B:?114 | B:B:?115 | 16.8 | 4 | Yes | Yes | 8 | 9 | 1 | 2 | | B:B:?114 | B:B:?121 | 6.35 | 4 | Yes | Yes | 8 | 8 | 1 | 0 | | B:B:?114 | B:B:S122 | 12.86 | 4 | Yes | No | 8 | 8 | 1 | 1 | | B:B:?114 | B:B:?149 | 4.76 | 4 | Yes | Yes | 8 | 8 | 1 | 2 | | B:B:?121 | B:B:I120 | 9.17 | 4 | Yes | No | 8 | 6 | 0 | 1 | | B:B:A140 | B:B:I120 | 6.5 | 0 | No | No | 5 | 6 | 2 | 1 | | B:B:?121 | B:B:S122 | 4.82 | 4 | Yes | No | 8 | 8 | 0 | 1 | | B:B:?121 | B:B:L139 | 4.45 | 4 | Yes | Yes | 8 | 9 | 0 | 1 | | B:B:?121 | B:B:L146 | 4.45 | 4 | Yes | No | 8 | 7 | 0 | 1 | | B:B:?124 | B:B:S122 | 10.22 | 4 | Yes | No | 6 | 8 | 2 | 1 | | B:B:?124 | B:B:I123 | 4.16 | 4 | Yes | Yes | 6 | 6 | 2 | 2 | | B:B:I123 | B:B:L139 | 4.28 | 0 | Yes | Yes | 6 | 9 | 2 | 1 | | B:B:L139 | B:B:R137 | 3.64 | 0 | Yes | No | 9 | 5 | 1 | 2 | | B:B:L139 | B:B:W169 | 7.97 | 0 | Yes | Yes | 9 | 8 | 1 | 2 | | B:B:?148 | B:B:L146 | 5.93 | 4 | Yes | No | 6 | 7 | 2 | 1 | | B:B:L146 | B:B:T159 | 8.84 | 0 | No | No | 7 | 9 | 1 | 2 | | B:B:?148 | B:B:?149 | 28.57 | 4 | Yes | Yes | 6 | 8 | 2 | 2 | | B:B:T159 | B:B:W169 | 3.64 | 0 | No | Yes | 9 | 8 | 2 | 2 | | B:B:?114 | B:B:A113 | 3.37 | 4 | Yes | No | 8 | 9 | 1 | 2 | | B:B:?149 | B:B:A113 | 3.37 | 4 | Yes | No | 8 | 9 | 2 | 2 | | B:B:?121 | B:B:E138 | 1.42 | 4 | Yes | No | 8 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 5.11 | | Average Nodes In Shell | 19.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 24.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
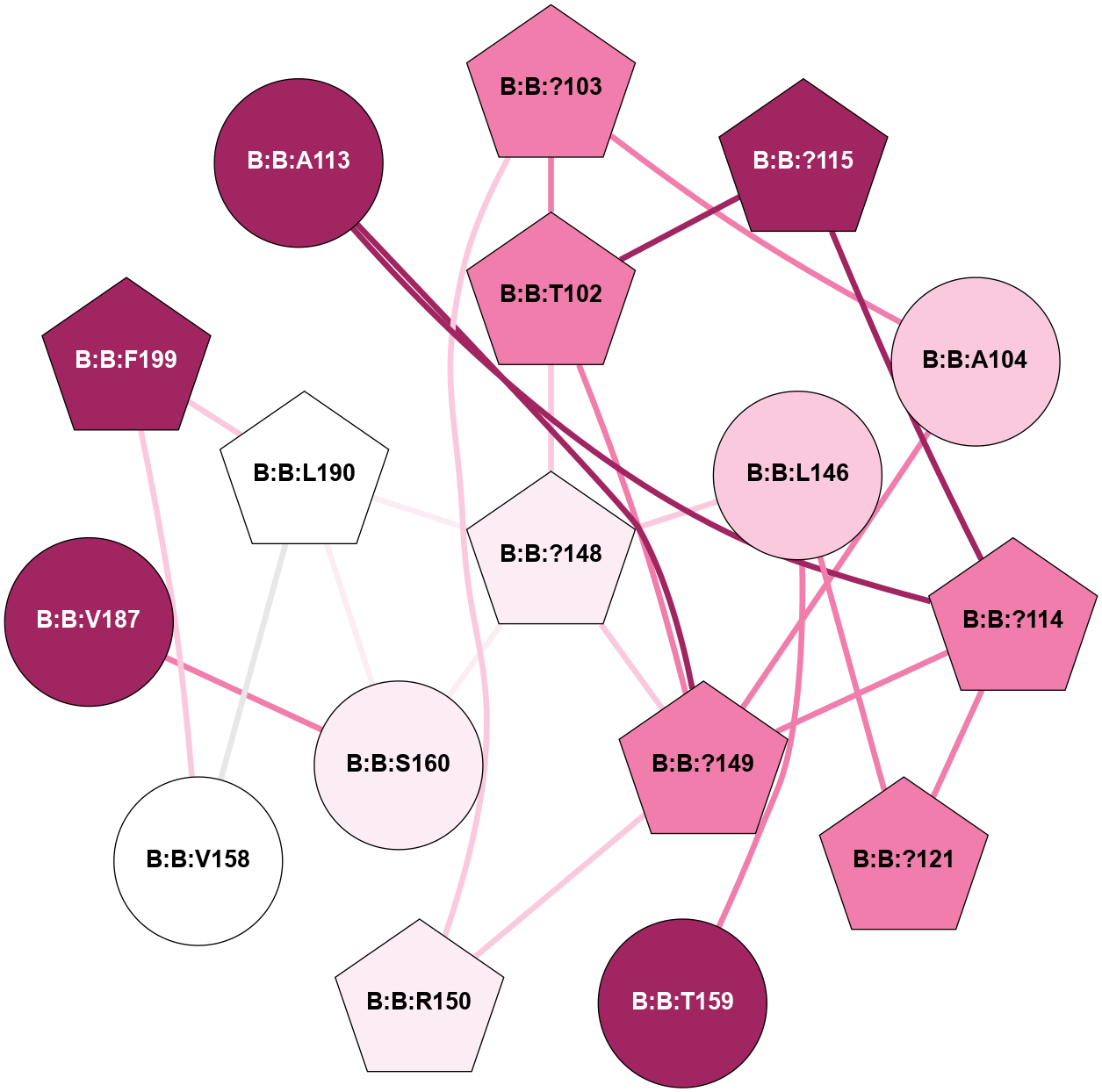
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:?103 | B:B:T102 | 11.05 | 4 | Yes | Yes | 8 | 8 | 2 | 1 | | B:B:?115 | B:B:T102 | 3.86 | 0 | Yes | Yes | 9 | 8 | 2 | 1 | | B:B:?148 | B:B:T102 | 20.52 | 4 | Yes | Yes | 6 | 8 | 0 | 1 | | B:B:?149 | B:B:T102 | 15.78 | 4 | Yes | Yes | 8 | 8 | 1 | 1 | | B:B:?103 | B:B:A104 | 5.06 | 4 | Yes | No | 8 | 7 | 2 | 2 | | B:B:?103 | B:B:R150 | 3.9 | 4 | Yes | Yes | 8 | 6 | 2 | 2 | | B:B:?149 | B:B:A104 | 6.75 | 4 | Yes | No | 8 | 7 | 1 | 2 | | B:B:?114 | B:B:?115 | 16.8 | 4 | Yes | Yes | 8 | 9 | 2 | 2 | | B:B:?114 | B:B:?121 | 6.35 | 4 | Yes | Yes | 8 | 8 | 2 | 2 | | B:B:?114 | B:B:?149 | 4.76 | 4 | Yes | Yes | 8 | 8 | 2 | 1 | | B:B:?121 | B:B:L146 | 4.45 | 4 | Yes | No | 8 | 7 | 2 | 1 | | B:B:?148 | B:B:L146 | 5.93 | 4 | Yes | No | 6 | 7 | 0 | 1 | | B:B:L146 | B:B:T159 | 8.84 | 0 | No | No | 7 | 9 | 1 | 2 | | B:B:?148 | B:B:?149 | 28.57 | 4 | Yes | Yes | 6 | 8 | 0 | 1 | | B:B:?148 | B:B:S160 | 11.26 | 4 | Yes | No | 6 | 6 | 0 | 1 | | B:B:?148 | B:B:L190 | 4.45 | 4 | Yes | Yes | 6 | 5 | 0 | 1 | | B:B:?149 | B:B:R150 | 19.51 | 4 | Yes | Yes | 8 | 6 | 1 | 2 | | B:B:L190 | B:B:V158 | 4.47 | 4 | Yes | No | 5 | 5 | 1 | 2 | | B:B:F199 | B:B:V158 | 3.93 | 4 | Yes | No | 9 | 5 | 2 | 2 | | B:B:S160 | B:B:V187 | 4.85 | 4 | No | No | 6 | 9 | 1 | 2 | | B:B:L190 | B:B:S160 | 9.01 | 4 | Yes | No | 5 | 6 | 1 | 1 | | B:B:F199 | B:B:L190 | 4.87 | 4 | Yes | Yes | 9 | 5 | 2 | 1 | | B:B:?114 | B:B:A113 | 3.37 | 4 | Yes | No | 8 | 9 | 2 | 2 | | B:B:?149 | B:B:A113 | 3.37 | 4 | Yes | No | 8 | 9 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 14.15 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 24.00 | | Average Links Mediated by Hubs In Shell | 22.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
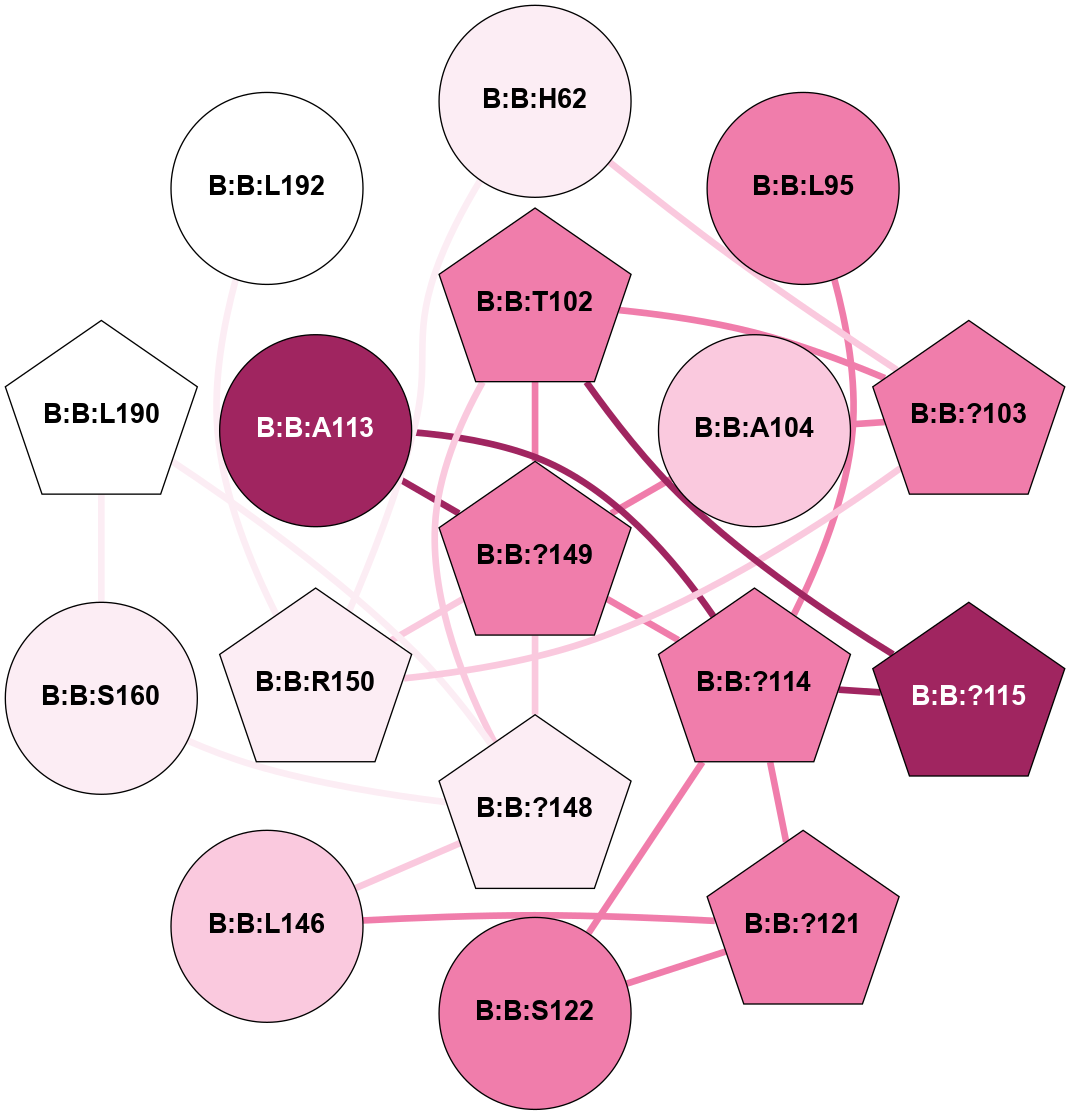
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:?103 | B:B:H62 | 5.51 | 4 | Yes | No | 8 | 6 | 2 | 2 | | B:B:H62 | B:B:R150 | 6.77 | 4 | No | Yes | 6 | 6 | 2 | 1 | | B:B:?114 | B:B:L95 | 4.45 | 4 | Yes | No | 8 | 8 | 1 | 2 | | B:B:?103 | B:B:T102 | 11.05 | 4 | Yes | Yes | 8 | 8 | 2 | 1 | | B:B:?115 | B:B:T102 | 3.86 | 0 | Yes | Yes | 9 | 8 | 2 | 1 | | B:B:?148 | B:B:T102 | 20.52 | 4 | Yes | Yes | 6 | 8 | 1 | 1 | | B:B:?149 | B:B:T102 | 15.78 | 4 | Yes | Yes | 8 | 8 | 0 | 1 | | B:B:?103 | B:B:A104 | 5.06 | 4 | Yes | No | 8 | 7 | 2 | 1 | | B:B:?103 | B:B:R150 | 3.9 | 4 | Yes | Yes | 8 | 6 | 2 | 1 | | B:B:?149 | B:B:A104 | 6.75 | 4 | Yes | No | 8 | 7 | 0 | 1 | | B:B:?114 | B:B:?115 | 16.8 | 4 | Yes | Yes | 8 | 9 | 1 | 2 | | B:B:?114 | B:B:?121 | 6.35 | 4 | Yes | Yes | 8 | 8 | 1 | 2 | | B:B:?114 | B:B:S122 | 12.86 | 4 | Yes | No | 8 | 8 | 1 | 2 | | B:B:?114 | B:B:?149 | 4.76 | 4 | Yes | Yes | 8 | 8 | 1 | 0 | | B:B:?121 | B:B:S122 | 4.82 | 4 | Yes | No | 8 | 8 | 2 | 2 | | B:B:?121 | B:B:L146 | 4.45 | 4 | Yes | No | 8 | 7 | 2 | 2 | | B:B:?148 | B:B:L146 | 5.93 | 4 | Yes | No | 6 | 7 | 1 | 2 | | B:B:?148 | B:B:?149 | 28.57 | 4 | Yes | Yes | 6 | 8 | 1 | 0 | | B:B:?148 | B:B:S160 | 11.26 | 4 | Yes | No | 6 | 6 | 1 | 2 | | B:B:?148 | B:B:L190 | 4.45 | 4 | Yes | Yes | 6 | 5 | 1 | 2 | | B:B:?149 | B:B:R150 | 19.51 | 4 | Yes | Yes | 8 | 6 | 0 | 1 | | B:B:L192 | B:B:R150 | 3.64 | 0 | No | Yes | 5 | 6 | 2 | 1 | | B:B:L190 | B:B:S160 | 9.01 | 4 | Yes | No | 5 | 6 | 2 | 2 | | B:B:?114 | B:B:A113 | 3.37 | 4 | Yes | No | 8 | 9 | 1 | 1 | | B:B:?149 | B:B:A113 | 3.37 | 4 | Yes | No | 8 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 13.12 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 25.00 | | Average Links Mediated by Hubs In Shell | 25.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
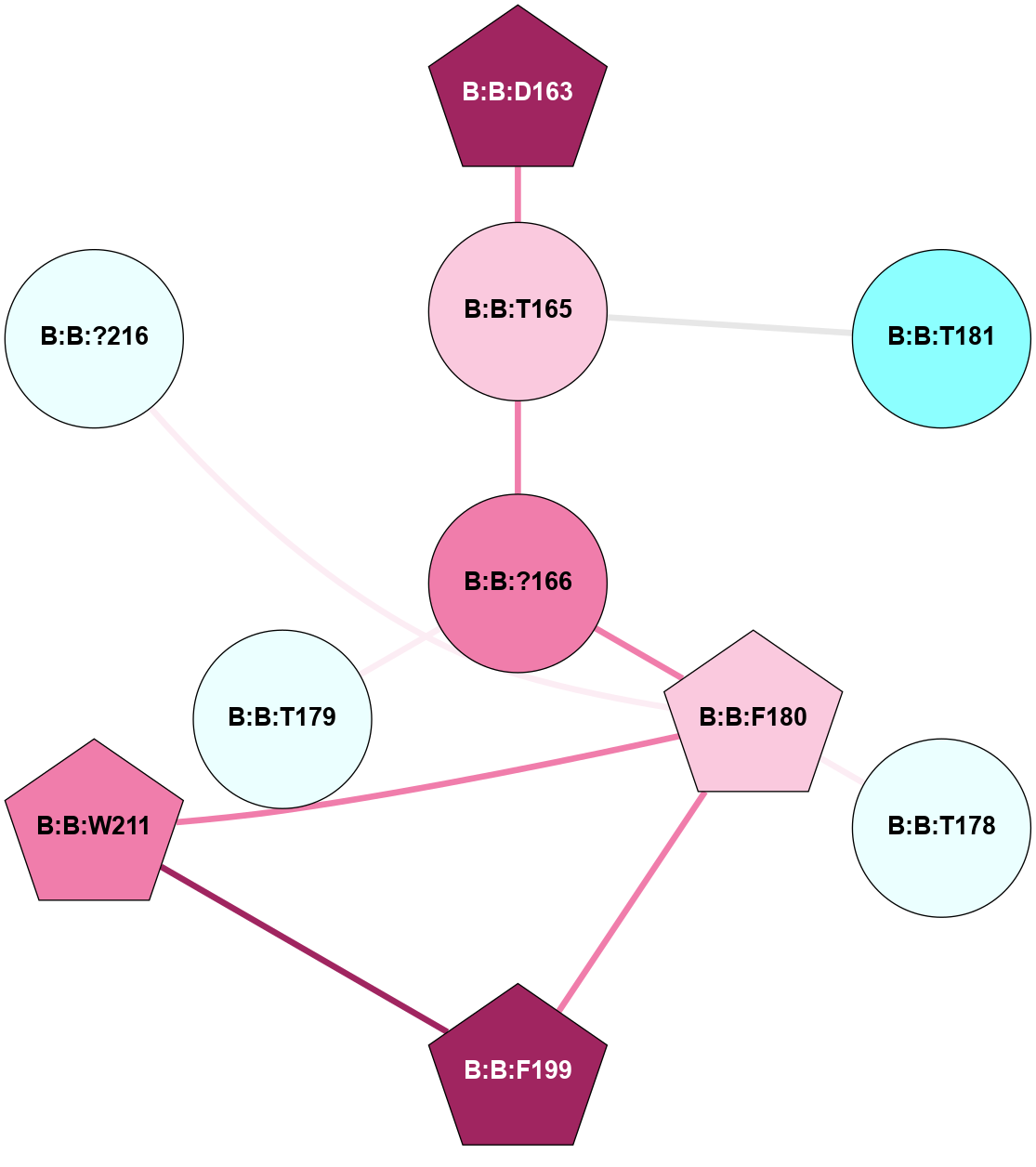
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:D163 | B:B:T165 | 11.56 | 7 | Yes | No | 9 | 7 | 2 | 1 | | B:B:?166 | B:B:T165 | 4.74 | 0 | No | No | 8 | 7 | 0 | 1 | | B:B:T165 | B:B:T181 | 4.71 | 0 | No | No | 7 | 2 | 1 | 2 | | B:B:?166 | B:B:F180 | 10.43 | 0 | No | Yes | 8 | 7 | 0 | 1 | | B:B:F180 | B:B:T178 | 6.49 | 4 | Yes | No | 7 | 4 | 1 | 2 | | B:B:F180 | B:B:F199 | 5.36 | 4 | Yes | Yes | 7 | 9 | 1 | 2 | | B:B:F180 | B:B:W211 | 7.02 | 4 | Yes | Yes | 7 | 8 | 1 | 2 | | B:B:?216 | B:B:F180 | 7.43 | 0 | No | Yes | 4 | 7 | 2 | 1 | | B:B:F199 | B:B:W211 | 10.02 | 4 | Yes | Yes | 9 | 8 | 2 | 2 | | B:B:?166 | B:B:T179 | 3.16 | 0 | No | No | 8 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.11 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
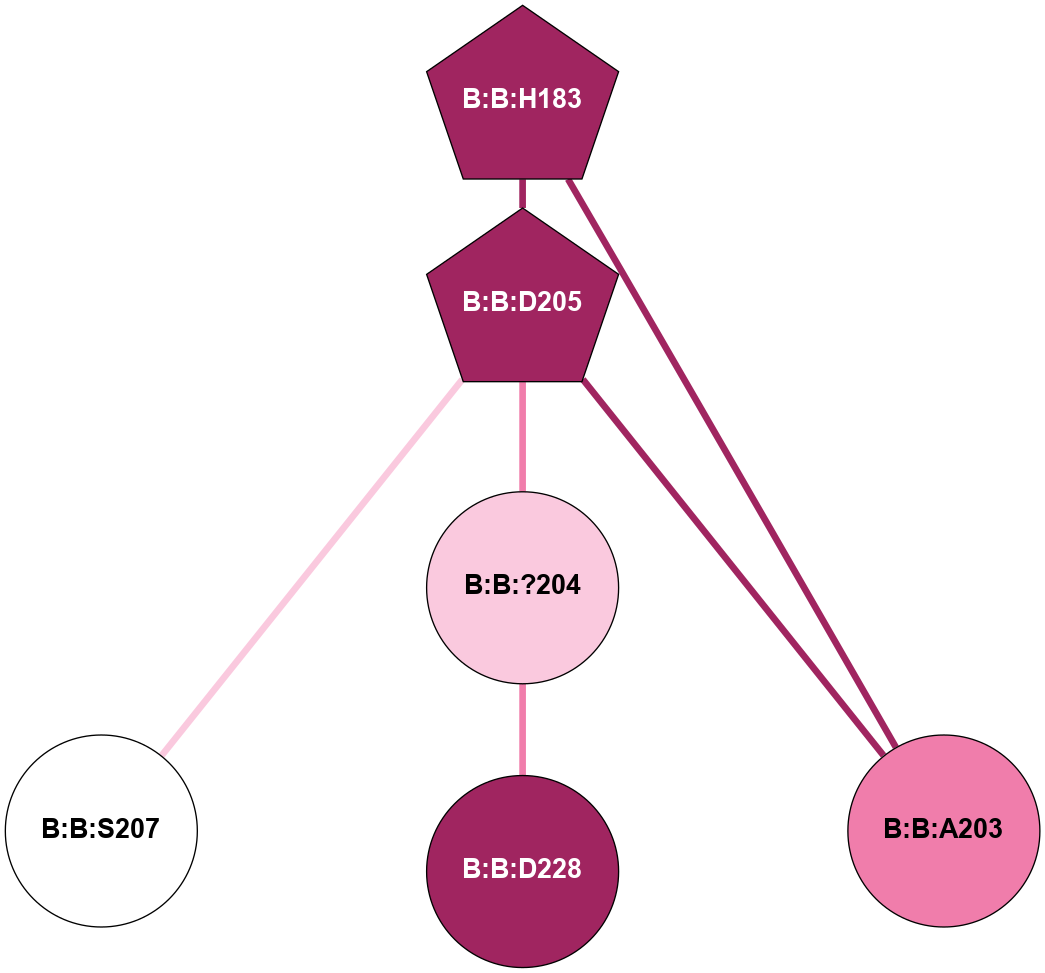
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:A203 | B:B:H183 | 4.39 | 4 | No | Yes | 8 | 9 | 2 | 2 | | B:B:D205 | B:B:H183 | 12.61 | 4 | Yes | Yes | 9 | 9 | 1 | 2 | | B:B:A203 | B:B:D205 | 4.63 | 4 | No | Yes | 8 | 9 | 2 | 1 | | B:B:?204 | B:B:D205 | 5.81 | 0 | No | Yes | 7 | 9 | 0 | 1 | | B:B:?204 | B:B:D228 | 21.8 | 0 | No | No | 7 | 9 | 0 | 1 | | B:B:D205 | B:B:S207 | 8.83 | 4 | Yes | No | 9 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 13.80 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:F180 | B:B:F199 | 5.36 | 4 | Yes | Yes | 7 | 9 | 2 | 2 | | B:B:F180 | B:B:W211 | 7.02 | 4 | Yes | Yes | 7 | 8 | 2 | 1 | | B:B:?216 | B:B:F180 | 7.43 | 0 | No | Yes | 4 | 7 | 2 | 2 | | B:B:?182 | B:B:H183 | 5.61 | 4 | No | Yes | 6 | 9 | 2 | 2 | | B:B:?182 | B:B:W211 | 9.93 | 4 | No | Yes | 6 | 8 | 2 | 1 | | B:B:?209 | B:B:H183 | 5.37 | 4 | No | Yes | 6 | 9 | 2 | 2 | | B:B:H183 | B:B:W211 | 4.23 | 4 | Yes | Yes | 9 | 8 | 2 | 1 | | B:B:F199 | B:B:W211 | 10.02 | 4 | Yes | Yes | 9 | 8 | 2 | 1 | | B:B:S201 | B:B:W211 | 7.41 | 0 | No | Yes | 9 | 8 | 2 | 1 | | B:B:?209 | B:B:W211 | 8.83 | 4 | No | Yes | 6 | 8 | 2 | 1 | | B:B:?218 | B:B:W211 | 19.51 | 0 | Yes | Yes | 5 | 8 | 0 | 1 | | B:B:D212 | B:B:E215 | 10.39 | 24 | No | No | 9 | 3 | 2 | 2 | | B:B:D212 | B:B:R219 | 11.91 | 24 | No | No | 9 | 5 | 2 | 1 | | B:B:E215 | B:B:M217 | 4.06 | 24 | No | No | 3 | 3 | 2 | 1 | | B:B:E215 | B:B:R219 | 11.63 | 24 | No | No | 3 | 5 | 2 | 1 | | B:B:?216 | B:B:M217 | 7.4 | 0 | No | No | 4 | 3 | 2 | 1 | | B:B:?218 | B:B:M217 | 7.57 | 0 | Yes | No | 5 | 3 | 0 | 1 | | B:B:?218 | B:B:R219 | 5.2 | 0 | Yes | No | 5 | 5 | 0 | 1 | | B:B:?218 | G:G:Q18 | 12.83 | 0 | Yes | No | 5 | 9 | 0 | 1 | | G:G:E22 | G:G:Q18 | 11.47 | 0 | No | No | 9 | 9 | 2 | 1 | | G:G:E17 | G:G:Q18 | 2.55 | 0 | No | No | 6 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 11.28 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 21.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
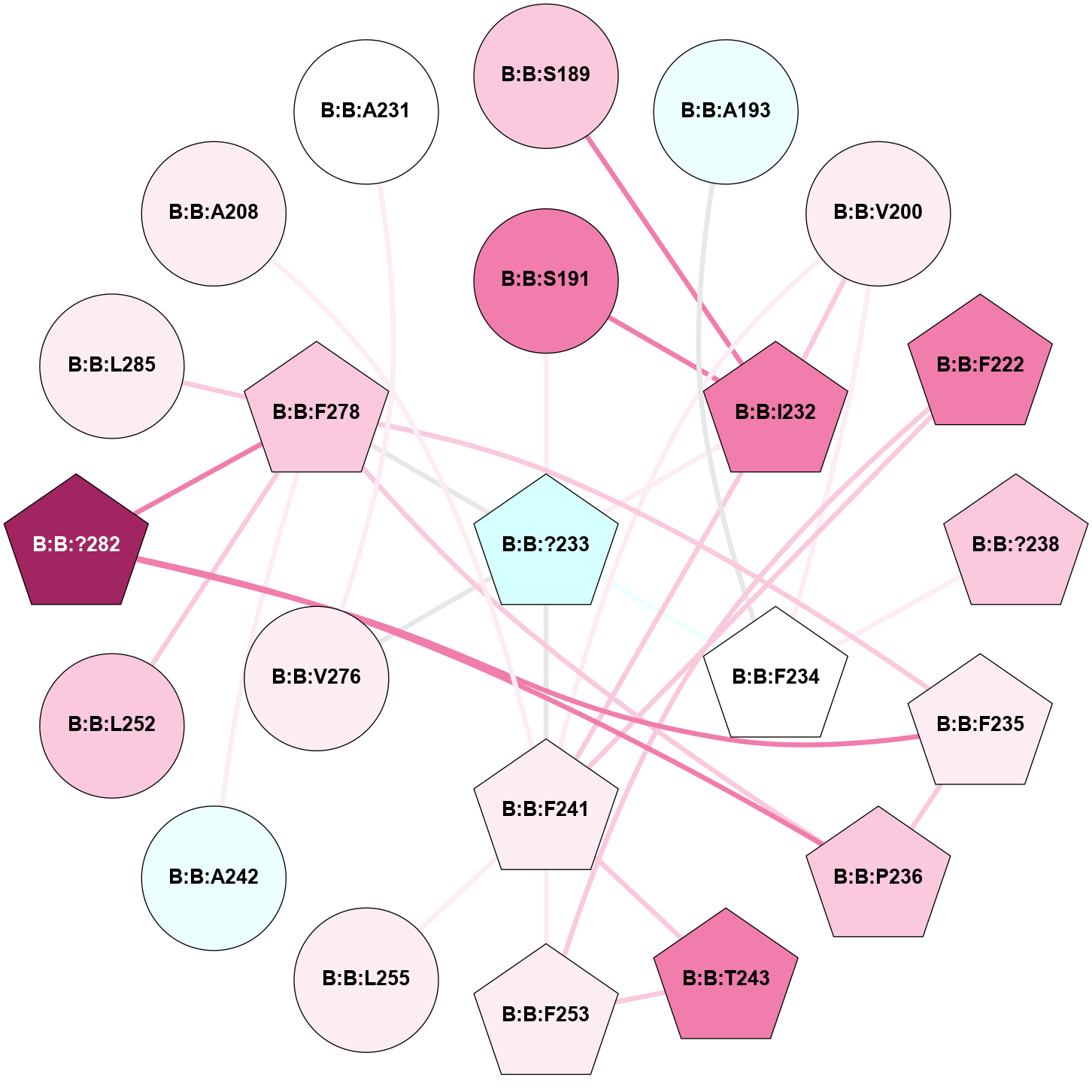
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:I232 | B:B:S189 | 6.19 | 2 | Yes | No | 8 | 7 | 1 | 2 | | B:B:I232 | B:B:S191 | 4.64 | 2 | Yes | No | 8 | 8 | 1 | 1 | | B:B:?233 | B:B:S191 | 11.26 | 2 | Yes | No | 3 | 8 | 0 | 1 | | B:B:A193 | B:B:F234 | 8.32 | 0 | No | Yes | 4 | 5 | 2 | 1 | | B:B:I232 | B:B:V200 | 9.22 | 2 | Yes | No | 8 | 6 | 1 | 2 | | B:B:F234 | B:B:V200 | 7.87 | 0 | Yes | No | 5 | 6 | 1 | 2 | | B:B:F241 | B:B:V200 | 6.55 | 2 | Yes | No | 6 | 6 | 1 | 2 | | B:B:F222 | B:B:F241 | 9.65 | 2 | Yes | Yes | 8 | 6 | 2 | 1 | | B:B:F222 | B:B:F253 | 4.29 | 2 | Yes | Yes | 8 | 6 | 2 | 2 | | B:B:?233 | B:B:I232 | 6.11 | 2 | Yes | Yes | 3 | 8 | 0 | 1 | | B:B:F241 | B:B:I232 | 6.28 | 2 | Yes | Yes | 6 | 8 | 1 | 1 | | B:B:?233 | B:B:F234 | 3.91 | 2 | Yes | Yes | 3 | 5 | 0 | 1 | | B:B:?233 | B:B:F241 | 3.91 | 2 | Yes | Yes | 3 | 6 | 0 | 1 | | B:B:?233 | B:B:V276 | 6.38 | 2 | Yes | No | 3 | 6 | 0 | 1 | | B:B:?233 | B:B:F278 | 6.52 | 2 | Yes | Yes | 3 | 7 | 0 | 1 | | B:B:?238 | B:B:F234 | 4.25 | 2 | Yes | Yes | 7 | 5 | 2 | 1 | | B:B:F235 | B:B:P236 | 10.11 | 2 | Yes | Yes | 6 | 7 | 2 | 2 | | B:B:F235 | B:B:F278 | 7.5 | 2 | Yes | Yes | 6 | 7 | 2 | 1 | | B:B:?282 | B:B:F235 | 5.31 | 2 | Yes | Yes | 9 | 6 | 2 | 2 | | B:B:F278 | B:B:P236 | 5.78 | 2 | Yes | Yes | 7 | 7 | 1 | 2 | | B:B:?282 | B:B:P236 | 4.3 | 2 | Yes | Yes | 9 | 7 | 2 | 2 | | B:B:F241 | B:B:T243 | 3.89 | 2 | Yes | Yes | 6 | 8 | 1 | 2 | | B:B:F241 | B:B:F253 | 11.79 | 2 | Yes | Yes | 6 | 6 | 1 | 2 | | B:B:F241 | B:B:L255 | 3.65 | 2 | Yes | No | 6 | 6 | 1 | 2 | | B:B:A242 | B:B:F278 | 4.16 | 0 | No | Yes | 4 | 7 | 2 | 1 | | B:B:F253 | B:B:T243 | 7.78 | 2 | Yes | Yes | 6 | 8 | 2 | 2 | | B:B:F278 | B:B:L252 | 3.65 | 2 | Yes | No | 7 | 7 | 1 | 2 | | B:B:?282 | B:B:F278 | 4.25 | 2 | Yes | Yes | 9 | 7 | 2 | 1 | | B:B:F278 | B:B:L285 | 6.09 | 2 | Yes | No | 7 | 6 | 1 | 2 | | B:B:A208 | B:B:F241 | 2.77 | 0 | No | Yes | 6 | 6 | 2 | 1 | | B:B:A231 | B:B:V276 | 1.7 | 0 | No | No | 5 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 6.35 | | Average Nodes In Shell | 23.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 31.00 | | Average Links Mediated by Hubs In Shell | 30.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:?224 | B:B:H225 | 4.48 | 2 | No | Yes | 6 | 9 | 2 | 2 | | B:B:?224 | B:B:R251 | 10.59 | 2 | No | Yes | 6 | 8 | 2 | 1 | | B:B:D247 | B:B:H225 | 5.04 | 2 | Yes | Yes | 9 | 9 | 2 | 2 | | B:B:H225 | B:B:R251 | 20.31 | 2 | Yes | Yes | 9 | 8 | 2 | 1 | | B:B:?244 | B:B:I273 | 7.47 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | B:B:D247 | B:B:T249 | 10.12 | 2 | Yes | Yes | 9 | 6 | 2 | 1 | | B:B:?250 | B:B:T249 | 4.74 | 2 | Yes | Yes | 8 | 6 | 0 | 1 | | B:B:R251 | B:B:T249 | 6.47 | 2 | Yes | Yes | 8 | 6 | 1 | 1 | | B:B:S265 | B:B:T249 | 4.8 | 0 | No | Yes | 2 | 6 | 2 | 1 | | B:B:?250 | B:B:R251 | 5.2 | 2 | Yes | Yes | 8 | 8 | 0 | 1 | | B:B:?250 | B:B:?264 | 25.95 | 2 | Yes | Yes | 8 | 5 | 0 | 1 | | B:B:?250 | B:B:I273 | 6.11 | 2 | Yes | Yes | 8 | 6 | 0 | 1 | | B:B:?250 | B:B:W297 | 4.88 | 2 | Yes | Yes | 8 | 8 | 0 | 1 | | B:B:F253 | B:B:R251 | 8.55 | 2 | Yes | Yes | 6 | 8 | 2 | 1 | | B:B:E260 | B:B:R251 | 16.28 | 2 | No | Yes | 6 | 8 | 2 | 1 | | B:B:R251 | B:B:T263 | 9.06 | 2 | Yes | No | 8 | 1 | 1 | 2 | | B:B:?264 | B:B:L252 | 9.42 | 2 | Yes | No | 5 | 7 | 1 | 2 | | B:B:E260 | B:B:F253 | 8.16 | 2 | No | Yes | 6 | 6 | 2 | 2 | | B:B:E260 | B:B:T263 | 5.64 | 2 | No | No | 6 | 1 | 2 | 2 | | B:B:?264 | B:B:M262 | 12.37 | 2 | Yes | No | 5 | 4 | 1 | 2 | | B:B:?264 | B:B:T263 | 7.17 | 2 | Yes | No | 5 | 1 | 1 | 2 | | B:B:?264 | B:B:S265 | 5.84 | 2 | Yes | No | 5 | 2 | 1 | 2 | | B:B:?264 | B:B:W297 | 14.4 | 2 | Yes | Yes | 5 | 8 | 1 | 1 | | B:B:?264 | B:B:A302 | 7.66 | 2 | Yes | No | 5 | 2 | 1 | 2 | | B:B:?272 | B:B:I273 | 4.98 | 2 | Yes | Yes | 4 | 6 | 2 | 1 | | B:B:?272 | B:B:?289 | 8.22 | 2 | Yes | Yes | 4 | 7 | 2 | 2 | | B:B:?288 | B:B:I273 | 4.98 | 2 | No | Yes | 7 | 6 | 2 | 1 | | B:B:?289 | B:B:I273 | 12.49 | 2 | Yes | Yes | 7 | 6 | 2 | 1 | | B:B:?288 | B:B:?289 | 25.83 | 2 | No | Yes | 7 | 7 | 2 | 2 | | B:B:?289 | B:B:N295 | 8.01 | 2 | Yes | Yes | 7 | 6 | 2 | 2 | | B:B:?289 | B:B:W297 | 7.75 | 2 | Yes | Yes | 7 | 8 | 2 | 1 | | B:B:N295 | B:B:W297 | 6.78 | 2 | Yes | Yes | 6 | 8 | 2 | 1 | | B:B:N295 | B:B:R304 | 3.62 | 2 | Yes | Yes | 6 | 5 | 2 | 2 | | B:B:R304 | B:B:W297 | 6 | 2 | Yes | Yes | 5 | 8 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 9.38 | | Average Nodes In Shell | 22.00 | | Average Hubs In Shell | 14.00 | | Average Links In Shell | 34.00 | | Average Links Mediated by Hubs In Shell | 33.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:W281 | B:B:D290 | 4.47 | 2 | Yes | Yes | 5 | 6 | 2 | 1 | | A:A:W281 | B:B:R314 | 7 | 2 | Yes | Yes | 5 | 8 | 2 | 2 | | B:B:?271 | B:B:A248 | 6.75 | 2 | Yes | No | 5 | 5 | 0 | 1 | | B:B:?271 | B:B:I269 | 4.59 | 2 | Yes | No | 5 | 4 | 0 | 1 | | B:B:?289 | B:B:I269 | 6.94 | 2 | Yes | No | 7 | 4 | 1 | 1 | | B:B:?271 | B:B:?272 | 18.1 | 2 | Yes | Yes | 5 | 4 | 0 | 1 | | B:B:?271 | B:B:?289 | 10.09 | 2 | Yes | Yes | 5 | 7 | 0 | 1 | | B:B:?271 | B:B:D290 | 8.72 | 2 | Yes | Yes | 5 | 6 | 0 | 1 | | B:B:?272 | B:B:I273 | 4.98 | 2 | Yes | Yes | 4 | 6 | 1 | 2 | | B:B:?272 | B:B:?289 | 8.22 | 2 | Yes | Yes | 4 | 7 | 1 | 1 | | B:B:?272 | B:B:D290 | 14.2 | 2 | Yes | Yes | 4 | 6 | 1 | 1 | | B:B:?288 | B:B:I273 | 4.98 | 2 | No | Yes | 7 | 6 | 2 | 2 | | B:B:?289 | B:B:I273 | 12.49 | 2 | Yes | Yes | 7 | 6 | 1 | 2 | | B:B:?288 | B:B:T274 | 5.14 | 2 | No | Yes | 7 | 8 | 2 | 2 | | B:B:?289 | B:B:T274 | 4.3 | 2 | Yes | Yes | 7 | 8 | 1 | 2 | | B:B:R314 | B:B:T274 | 9.06 | 2 | Yes | Yes | 8 | 8 | 2 | 2 | | B:B:?289 | B:B:A287 | 6.13 | 2 | Yes | No | 7 | 6 | 1 | 2 | | B:B:?288 | B:B:?289 | 25.83 | 2 | No | Yes | 7 | 7 | 2 | 1 | | B:B:?289 | B:B:D290 | 5.28 | 2 | Yes | Yes | 7 | 6 | 1 | 1 | | B:B:?289 | B:B:N295 | 8.01 | 2 | Yes | Yes | 7 | 6 | 1 | 2 | | B:B:?289 | B:B:W297 | 7.75 | 2 | Yes | Yes | 7 | 8 | 1 | 2 | | B:B:D290 | B:B:R314 | 7.15 | 2 | Yes | Yes | 6 | 8 | 1 | 2 | | B:B:N295 | B:B:W297 | 6.78 | 2 | Yes | Yes | 6 | 8 | 2 | 2 | | B:B:?272 | B:B:D246 | 3.55 | 2 | Yes | No | 4 | 9 | 1 | 2 | | B:B:?271 | B:B:I270 | 3.06 | 2 | Yes | No | 5 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 8.55 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 25.00 | | Average Links Mediated by Hubs In Shell | 25.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
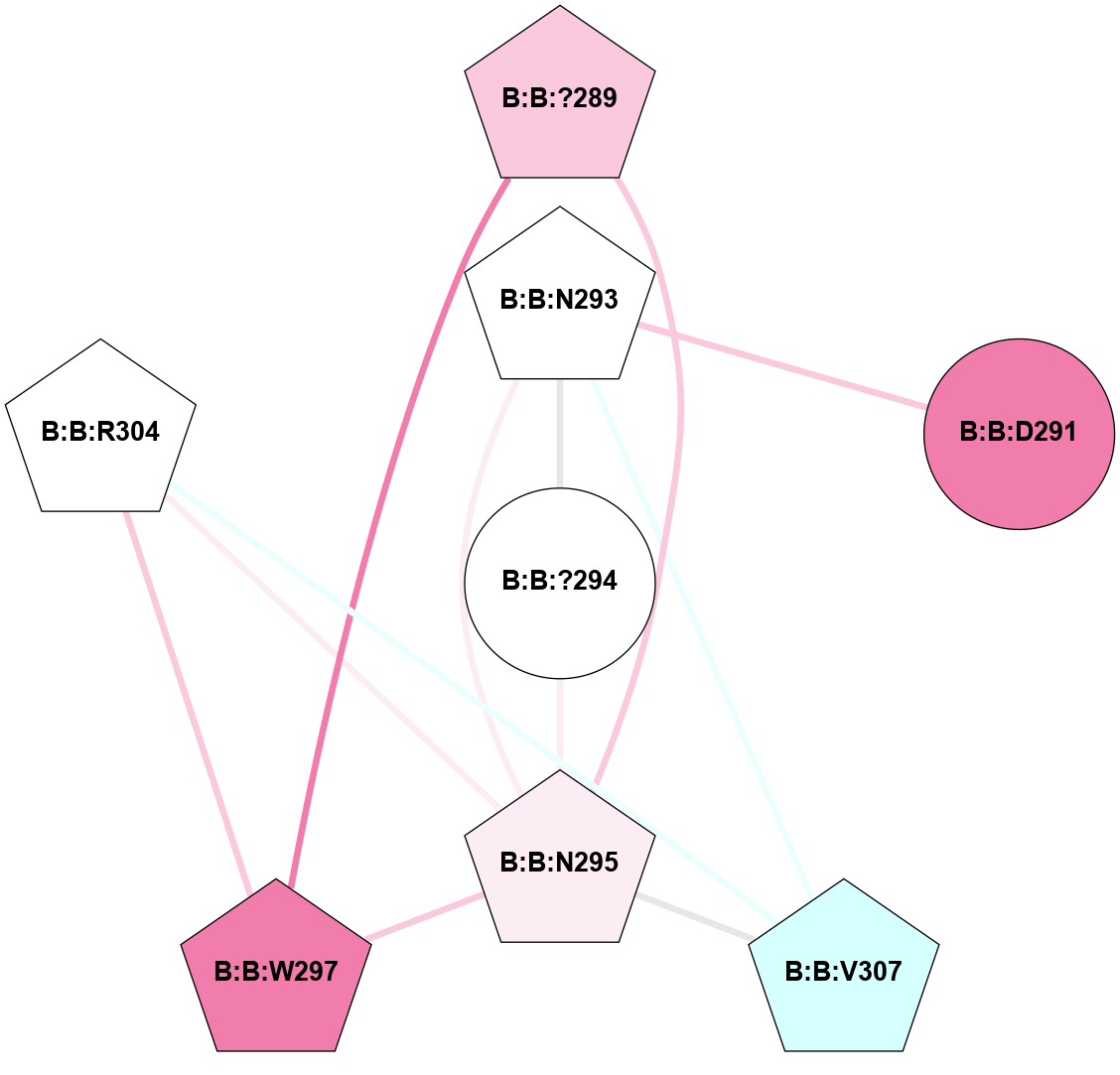
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:?289 | B:B:N295 | 8.01 | 2 | Yes | Yes | 7 | 6 | 2 | 1 | | B:B:?289 | B:B:W297 | 7.75 | 2 | Yes | Yes | 7 | 8 | 2 | 2 | | B:B:D291 | B:B:N293 | 14.81 | 0 | No | Yes | 8 | 5 | 2 | 1 | | B:B:?294 | B:B:N293 | 7.35 | 2 | No | Yes | 5 | 5 | 0 | 1 | | B:B:N293 | B:B:N295 | 5.45 | 2 | Yes | Yes | 5 | 6 | 1 | 1 | | B:B:N293 | B:B:V307 | 4.43 | 2 | Yes | Yes | 5 | 3 | 1 | 2 | | B:B:?294 | B:B:N295 | 11.76 | 2 | No | Yes | 5 | 6 | 0 | 1 | | B:B:N295 | B:B:W297 | 6.78 | 2 | Yes | Yes | 6 | 8 | 1 | 2 | | B:B:N295 | B:B:R304 | 3.62 | 2 | Yes | Yes | 6 | 5 | 1 | 2 | | B:B:N295 | B:B:V307 | 4.43 | 2 | Yes | Yes | 6 | 3 | 1 | 2 | | B:B:R304 | B:B:W297 | 6 | 2 | Yes | Yes | 5 | 8 | 2 | 2 | | B:B:R304 | B:B:V307 | 3.92 | 2 | Yes | Yes | 5 | 3 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.55 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
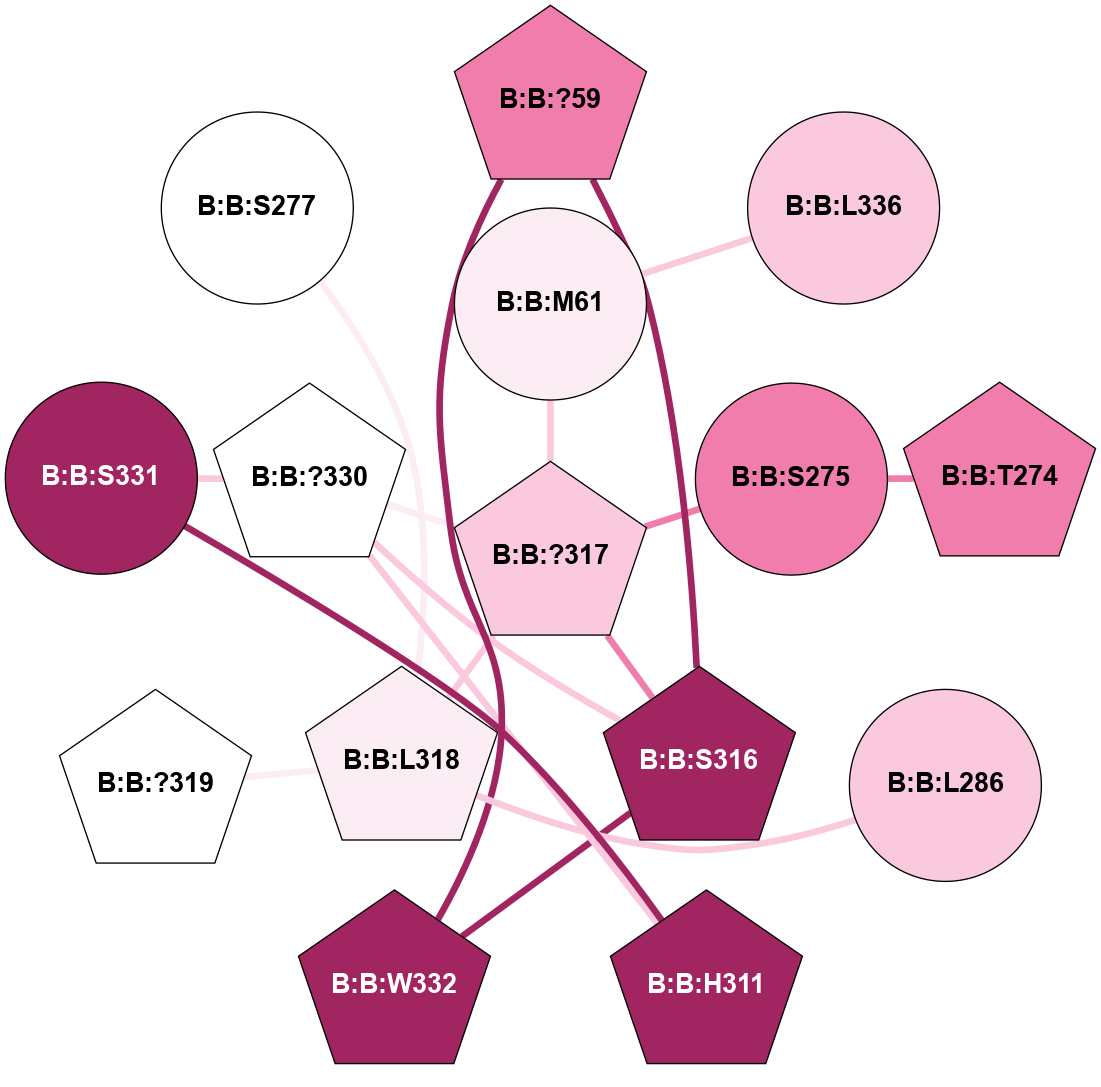
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:?59 | B:B:S316 | 11.68 | 2 | Yes | Yes | 8 | 9 | 2 | 1 | | B:B:?59 | B:B:W332 | 7.75 | 2 | Yes | Yes | 8 | 9 | 2 | 2 | | B:B:?317 | B:B:M61 | 9.08 | 2 | Yes | No | 7 | 6 | 0 | 1 | | B:B:L336 | B:B:M61 | 4.24 | 0 | No | No | 7 | 6 | 2 | 1 | | B:B:S275 | B:B:T274 | 4.8 | 0 | No | Yes | 8 | 8 | 1 | 2 | | B:B:?317 | B:B:S275 | 8.04 | 2 | Yes | No | 7 | 8 | 0 | 1 | | B:B:L286 | B:B:L318 | 4.15 | 0 | No | Yes | 7 | 6 | 2 | 1 | | B:B:?330 | B:B:H311 | 4.48 | 2 | Yes | Yes | 5 | 9 | 1 | 2 | | B:B:H311 | B:B:S331 | 13.95 | 2 | Yes | No | 9 | 9 | 2 | 2 | | B:B:?317 | B:B:S316 | 8.04 | 2 | Yes | Yes | 7 | 9 | 0 | 1 | | B:B:?330 | B:B:S316 | 3.93 | 2 | Yes | Yes | 5 | 9 | 1 | 1 | | B:B:S316 | B:B:W332 | 6.18 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | B:B:?317 | B:B:L318 | 4.45 | 2 | Yes | Yes | 7 | 6 | 0 | 1 | | B:B:?317 | B:B:?330 | 18.1 | 2 | Yes | Yes | 7 | 5 | 0 | 1 | | B:B:?319 | B:B:L318 | 7.24 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | B:B:?330 | B:B:S331 | 3.93 | 2 | Yes | No | 5 | 9 | 1 | 2 | | B:B:L318 | B:B:S277 | 3 | 0 | Yes | No | 6 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 9.54 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
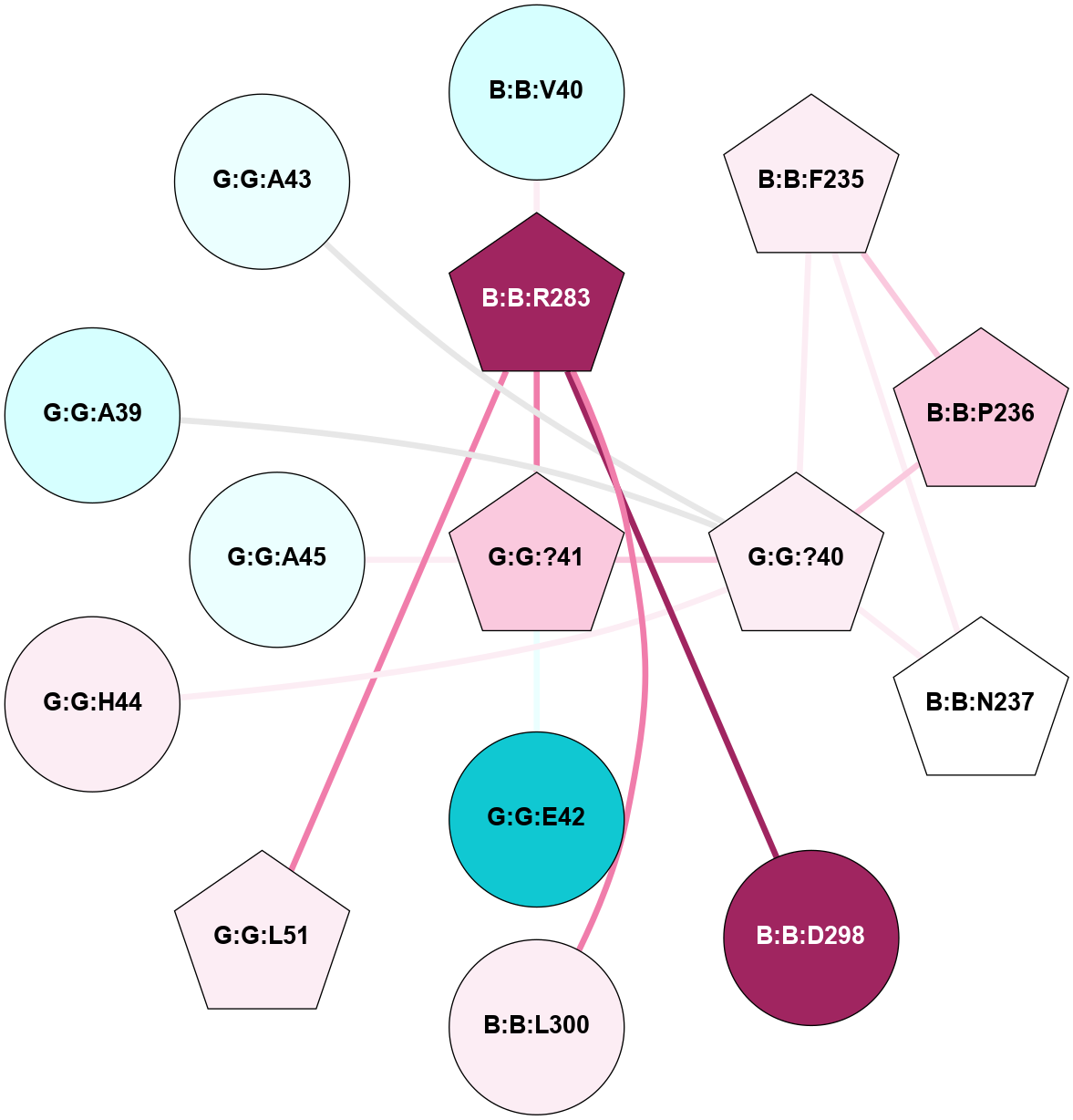
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | B:B:R283 | B:B:V40 | 5.23 | 0 | Yes | No | 9 | 3 | 1 | 2 | | B:B:F235 | B:B:P236 | 10.11 | 2 | Yes | Yes | 6 | 7 | 2 | 2 | | B:B:F235 | B:B:N237 | 10.87 | 2 | Yes | Yes | 6 | 5 | 2 | 2 | | B:B:F235 | G:G:?40 | 5.92 | 2 | Yes | Yes | 6 | 6 | 2 | 1 | | B:B:P236 | G:G:?40 | 6.39 | 2 | Yes | Yes | 7 | 6 | 2 | 1 | | B:B:N237 | G:G:?40 | 16.03 | 2 | Yes | Yes | 5 | 6 | 2 | 1 | | B:B:D298 | B:B:R283 | 14.29 | 0 | No | Yes | 9 | 9 | 2 | 1 | | B:B:L300 | B:B:R283 | 4.86 | 0 | No | Yes | 6 | 9 | 2 | 1 | | B:B:R283 | G:G:?41 | 3.9 | 0 | Yes | Yes | 9 | 7 | 1 | 0 | | B:B:R283 | G:G:L51 | 7.29 | 0 | Yes | Yes | 9 | 6 | 1 | 2 | | G:G:?40 | G:G:?41 | 27.39 | 2 | Yes | Yes | 6 | 7 | 1 | 0 | | G:G:?40 | G:G:H44 | 17.51 | 2 | Yes | No | 6 | 6 | 1 | 2 | | G:G:?40 | G:G:A39 | 3.06 | 2 | Yes | No | 6 | 3 | 1 | 2 | | G:G:?41 | G:G:E42 | 2.84 | 0 | Yes | No | 7 | 1 | 0 | 1 | | G:G:?41 | G:G:A45 | 1.69 | 0 | Yes | No | 7 | 4 | 0 | 1 | | G:G:?40 | G:G:A43 | 1.53 | 2 | Yes | No | 6 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 8.95 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
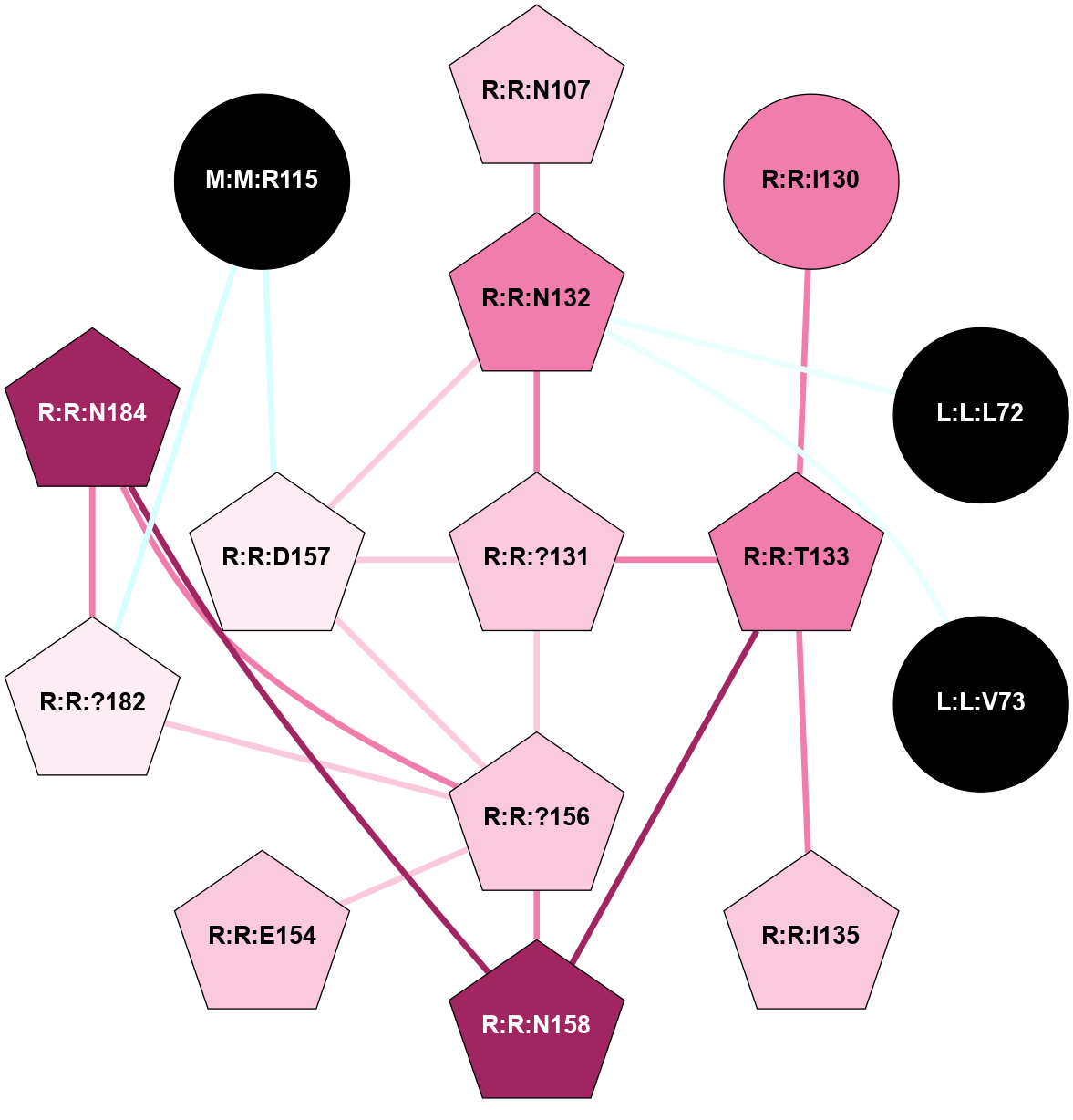
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N107 | R:R:N132 | 4.09 | 0 | Yes | Yes | 7 | 8 | 2 | 1 | | R:R:I130 | R:R:T133 | 4.56 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:?131 | R:R:N132 | 11.76 | 1 | Yes | Yes | 7 | 8 | 0 | 1 | | R:R:?131 | R:R:T133 | 6.31 | 1 | Yes | Yes | 7 | 8 | 0 | 1 | | R:R:?131 | R:R:?156 | 31.75 | 1 | Yes | Yes | 7 | 7 | 0 | 1 | | R:R:?131 | R:R:D157 | 4.36 | 1 | Yes | Yes | 7 | 6 | 0 | 1 | | R:R:D157 | R:R:N132 | 12.12 | 1 | Yes | Yes | 6 | 8 | 1 | 1 | | L:L:L72 | R:R:N132 | 5.49 | 1 | No | Yes | 0 | 8 | 2 | 1 | | L:L:V73 | R:R:N132 | 7.39 | 0 | No | Yes | 0 | 8 | 2 | 1 | | R:R:I135 | R:R:T133 | 7.6 | 1 | Yes | Yes | 7 | 8 | 2 | 1 | | R:R:N158 | R:R:T133 | 10.24 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:?156 | R:R:E154 | 4.26 | 1 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:?156 | R:R:D157 | 14.53 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | R:R:?156 | R:R:N158 | 13.23 | 1 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:?156 | R:R:?182 | 4.32 | 1 | Yes | Yes | 7 | 6 | 1 | 2 | | R:R:?156 | R:R:N184 | 13.23 | 1 | Yes | Yes | 7 | 9 | 1 | 2 | | N:N:R115 | R:R:D157 | 8.34 | 1 | No | Yes | 0 | 6 | 2 | 1 | | R:R:N158 | R:R:N184 | 5.45 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:?182 | R:R:N184 | 9.35 | 1 | Yes | Yes | 6 | 9 | 2 | 2 | | N:N:R115 | R:R:?182 | 11.82 | 1 | No | Yes | 0 | 6 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 13.54 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 20.00 | | Average Links Mediated by Hubs In Shell | 20.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
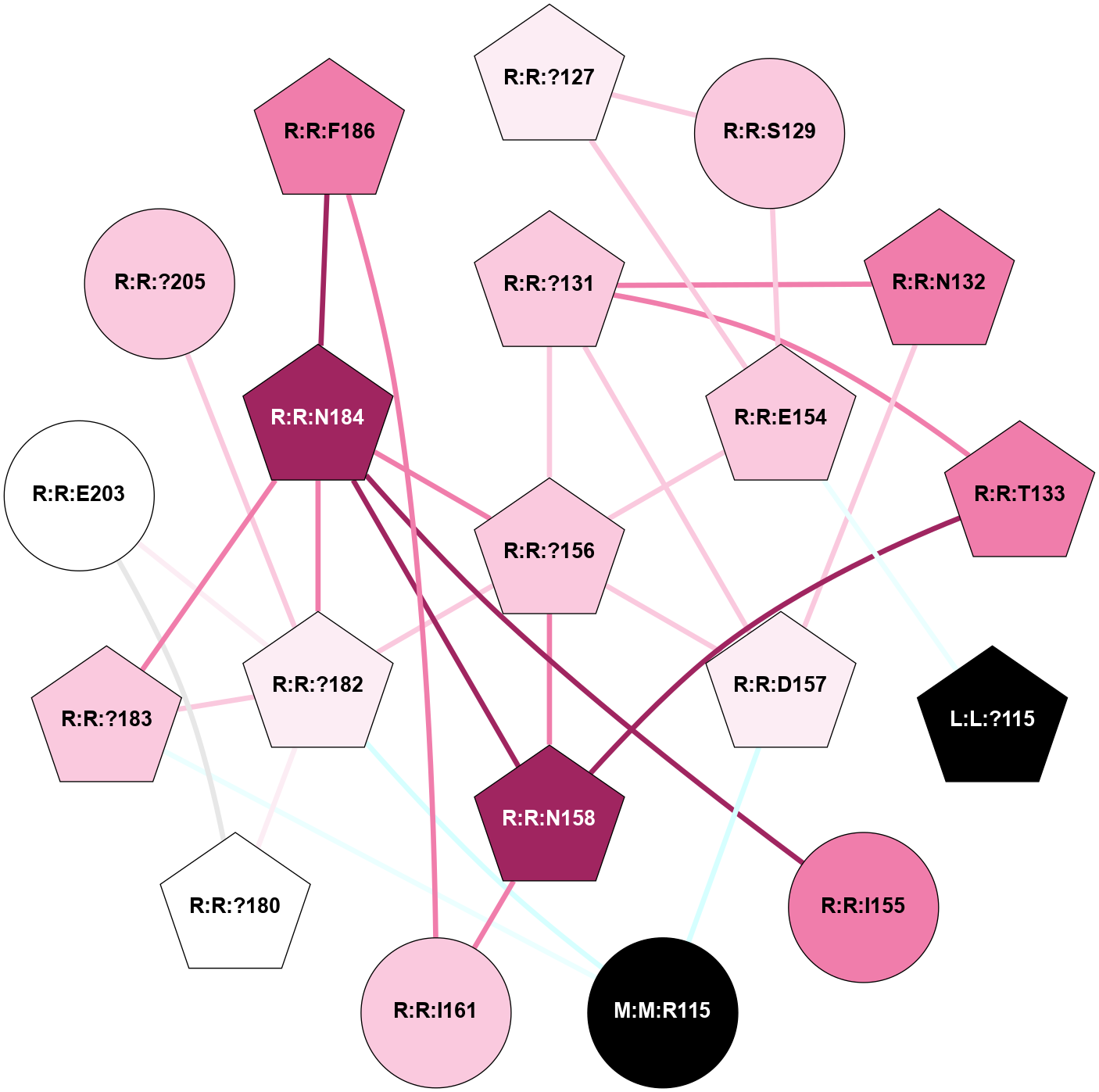
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?127 | R:R:S129 | 8.76 | 1 | Yes | No | 6 | 7 | 2 | 2 | | R:R:?127 | R:R:E154 | 9.02 | 1 | Yes | Yes | 6 | 7 | 2 | 1 | | R:R:E154 | R:R:S129 | 14.37 | 1 | Yes | No | 7 | 7 | 1 | 2 | | R:R:?131 | R:R:N132 | 11.76 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:?131 | R:R:T133 | 6.31 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:?131 | R:R:?156 | 31.75 | 1 | Yes | Yes | 7 | 7 | 1 | 0 | | R:R:?131 | R:R:D157 | 4.36 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | R:R:D157 | R:R:N132 | 12.12 | 1 | Yes | Yes | 6 | 8 | 1 | 2 | | R:R:N158 | R:R:T133 | 10.24 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:?156 | R:R:E154 | 4.26 | 1 | Yes | Yes | 7 | 7 | 0 | 1 | | L:L:?115 | R:R:E154 | 6.32 | 1 | Yes | Yes | 0 | 7 | 2 | 1 | | R:R:I155 | R:R:N184 | 7.08 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:?156 | R:R:D157 | 14.53 | 1 | Yes | Yes | 7 | 6 | 0 | 1 | | R:R:?156 | R:R:N158 | 13.23 | 1 | Yes | Yes | 7 | 9 | 0 | 1 | | R:R:?156 | R:R:?182 | 4.32 | 1 | Yes | Yes | 7 | 6 | 0 | 1 | | R:R:?156 | R:R:N184 | 13.23 | 1 | Yes | Yes | 7 | 9 | 0 | 1 | | N:N:R115 | R:R:D157 | 8.34 | 1 | No | Yes | 0 | 6 | 2 | 1 | | R:R:I161 | R:R:N158 | 8.5 | 0 | No | Yes | 7 | 9 | 2 | 1 | | R:R:N158 | R:R:N184 | 5.45 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:F186 | R:R:I161 | 5.02 | 0 | Yes | No | 8 | 7 | 2 | 2 | | R:R:?180 | R:R:?182 | 4.02 | 1 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:?180 | R:R:E203 | 5.53 | 1 | Yes | No | 5 | 5 | 2 | 2 | | R:R:?182 | R:R:?183 | 15.26 | 1 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:?182 | R:R:N184 | 9.35 | 1 | Yes | Yes | 6 | 9 | 1 | 1 | | R:R:?182 | R:R:E203 | 19.33 | 1 | Yes | No | 6 | 5 | 1 | 2 | | R:R:?182 | R:R:?205 | 13.65 | 1 | Yes | No | 6 | 7 | 1 | 2 | | N:N:R115 | R:R:?182 | 11.82 | 1 | No | Yes | 0 | 6 | 2 | 1 | | R:R:?183 | R:R:N184 | 4.79 | 1 | Yes | Yes | 7 | 9 | 2 | 1 | | N:N:R115 | R:R:?183 | 5.3 | 1 | No | Yes | 0 | 7 | 2 | 2 | | R:R:F186 | R:R:N184 | 14.5 | 0 | Yes | Yes | 8 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 13.55 | | Average Nodes In Shell | 20.00 | | Average Hubs In Shell | 14.00 | | Average Links In Shell | 30.00 | | Average Links Mediated by Hubs In Shell | 30.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
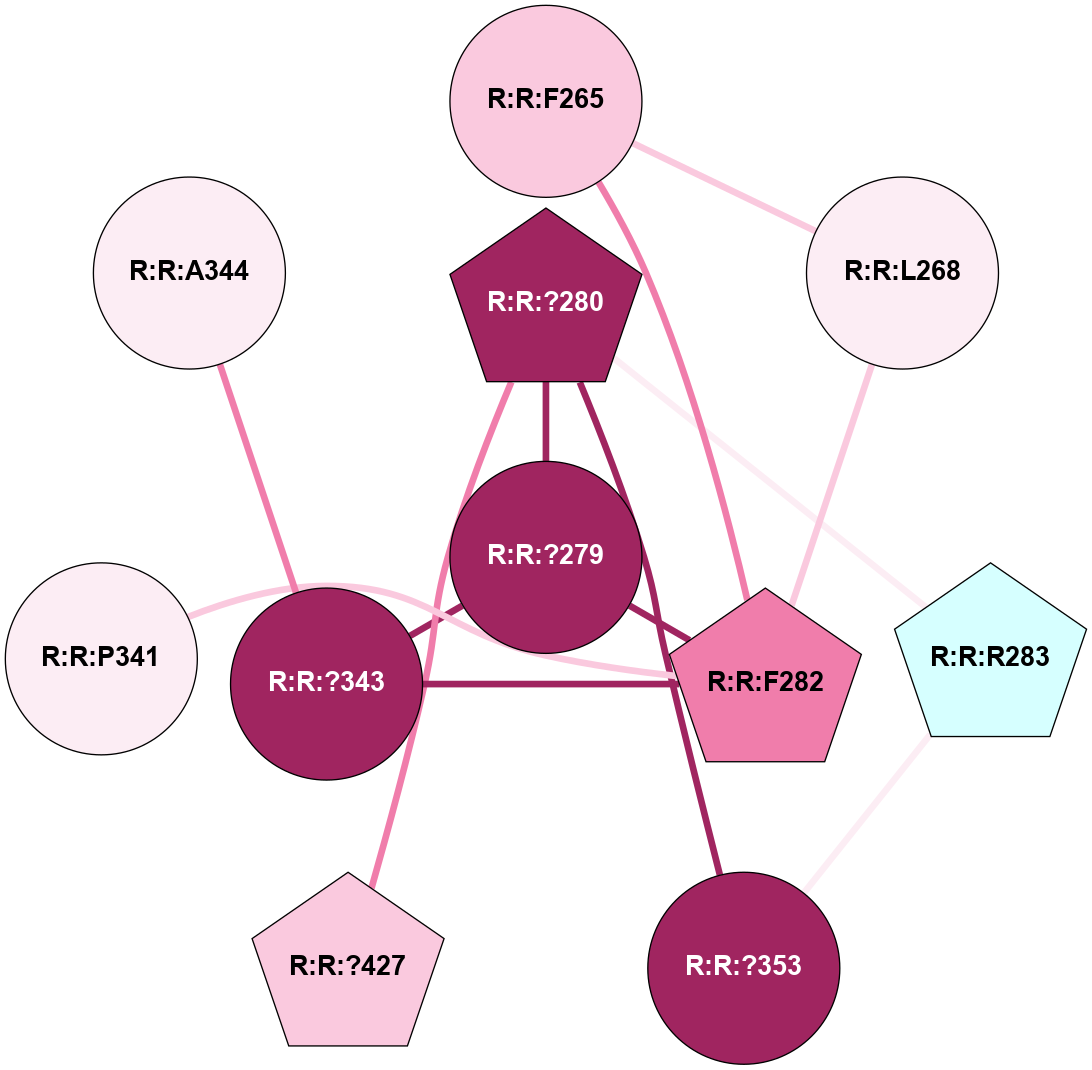
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F265 | R:R:L268 | 12.18 | 8 | No | No | 7 | 6 | 2 | 2 | | R:R:F265 | R:R:F282 | 4.29 | 8 | No | Yes | 7 | 8 | 2 | 1 | | R:R:F282 | R:R:L268 | 6.09 | 8 | Yes | No | 8 | 6 | 1 | 2 | | R:R:?279 | R:R:?280 | 26.98 | 8 | No | Yes | 9 | 9 | 0 | 1 | | R:R:?279 | R:R:F282 | 6.52 | 8 | No | Yes | 9 | 8 | 0 | 1 | | R:R:?279 | R:R:?343 | 17.46 | 8 | No | No | 9 | 9 | 0 | 1 | | R:R:?280 | R:R:R283 | 5.2 | 8 | Yes | Yes | 9 | 3 | 1 | 2 | | R:R:?280 | R:R:?353 | 15.87 | 8 | Yes | No | 9 | 9 | 1 | 2 | | R:R:?280 | R:R:?427 | 4.32 | 8 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F282 | R:R:P341 | 5.78 | 8 | Yes | No | 8 | 6 | 1 | 2 | | R:R:?343 | R:R:F282 | 3.91 | 8 | No | Yes | 9 | 8 | 1 | 1 | | R:R:?353 | R:R:R283 | 5.2 | 8 | No | Yes | 9 | 3 | 2 | 2 | | R:R:?343 | R:R:A344 | 3.37 | 8 | No | No | 9 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 16.99 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
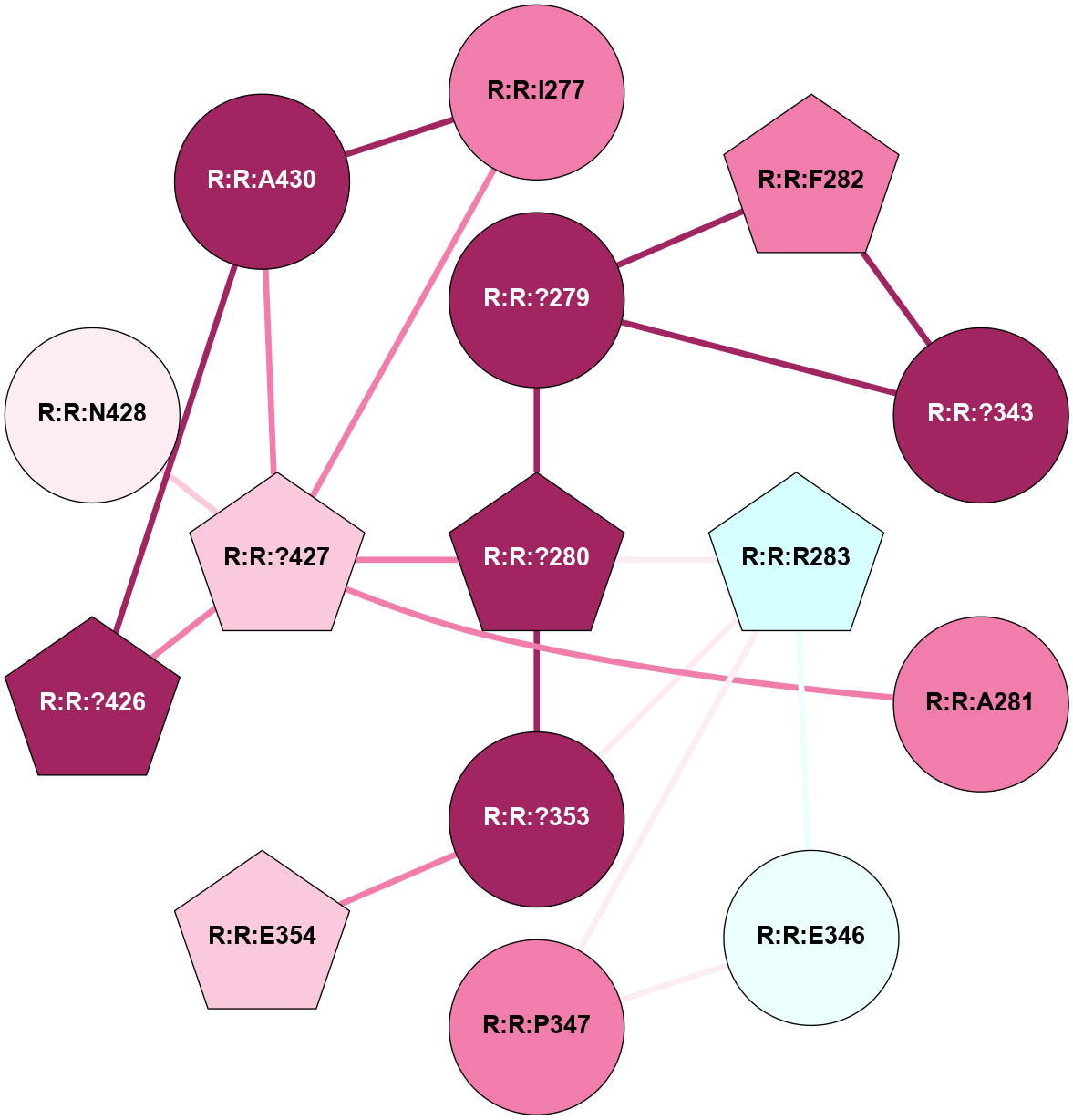
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?427 | R:R:I277 | 6.94 | 8 | Yes | No | 7 | 8 | 1 | 2 | | R:R:A430 | R:R:I277 | 4.87 | 8 | No | No | 9 | 8 | 2 | 2 | | R:R:?279 | R:R:?280 | 26.98 | 8 | No | Yes | 9 | 9 | 1 | 0 | | R:R:?279 | R:R:F282 | 6.52 | 8 | No | Yes | 9 | 8 | 1 | 2 | | R:R:?279 | R:R:?343 | 17.46 | 8 | No | No | 9 | 9 | 1 | 2 | | R:R:?280 | R:R:R283 | 5.2 | 8 | Yes | Yes | 9 | 3 | 0 | 1 | | R:R:?280 | R:R:?353 | 15.87 | 8 | Yes | No | 9 | 9 | 0 | 1 | | R:R:?280 | R:R:?427 | 4.32 | 8 | Yes | Yes | 9 | 7 | 0 | 1 | | R:R:?427 | R:R:A281 | 9.19 | 8 | Yes | No | 7 | 8 | 1 | 2 | | R:R:?343 | R:R:F282 | 3.91 | 8 | No | Yes | 9 | 8 | 2 | 2 | | R:R:E346 | R:R:R283 | 11.63 | 8 | No | Yes | 4 | 3 | 2 | 1 | | R:R:P347 | R:R:R283 | 4.32 | 8 | No | Yes | 8 | 3 | 2 | 1 | | R:R:?353 | R:R:R283 | 5.2 | 8 | No | Yes | 9 | 3 | 1 | 1 | | R:R:E346 | R:R:P347 | 7.86 | 8 | No | No | 4 | 8 | 2 | 2 | | R:R:?353 | R:R:E354 | 4.26 | 8 | No | Yes | 9 | 7 | 1 | 2 | | R:R:?426 | R:R:?427 | 27.5 | 8 | Yes | Yes | 9 | 7 | 2 | 1 | | R:R:?426 | R:R:A430 | 12.26 | 8 | Yes | No | 9 | 9 | 2 | 2 | | R:R:?427 | R:R:N428 | 18.7 | 8 | Yes | No | 7 | 6 | 1 | 2 | | R:R:?427 | R:R:A430 | 4.6 | 8 | Yes | No | 7 | 9 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 13.09 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 19.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L269 | R:R:L337 | 8.3 | 0 | No | No | 4 | 3 | 2 | 1 | | R:R:?334 | R:R:F335 | 11.68 | 0 | No | No | 3 | 1 | 2 | 1 | | R:R:?336 | R:R:L337 | 5.93 | 0 | No | No | 6 | 3 | 0 | 1 | | R:R:?336 | R:R:F335 | 1.3 | 0 | No | No | 6 | 1 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 3.61 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
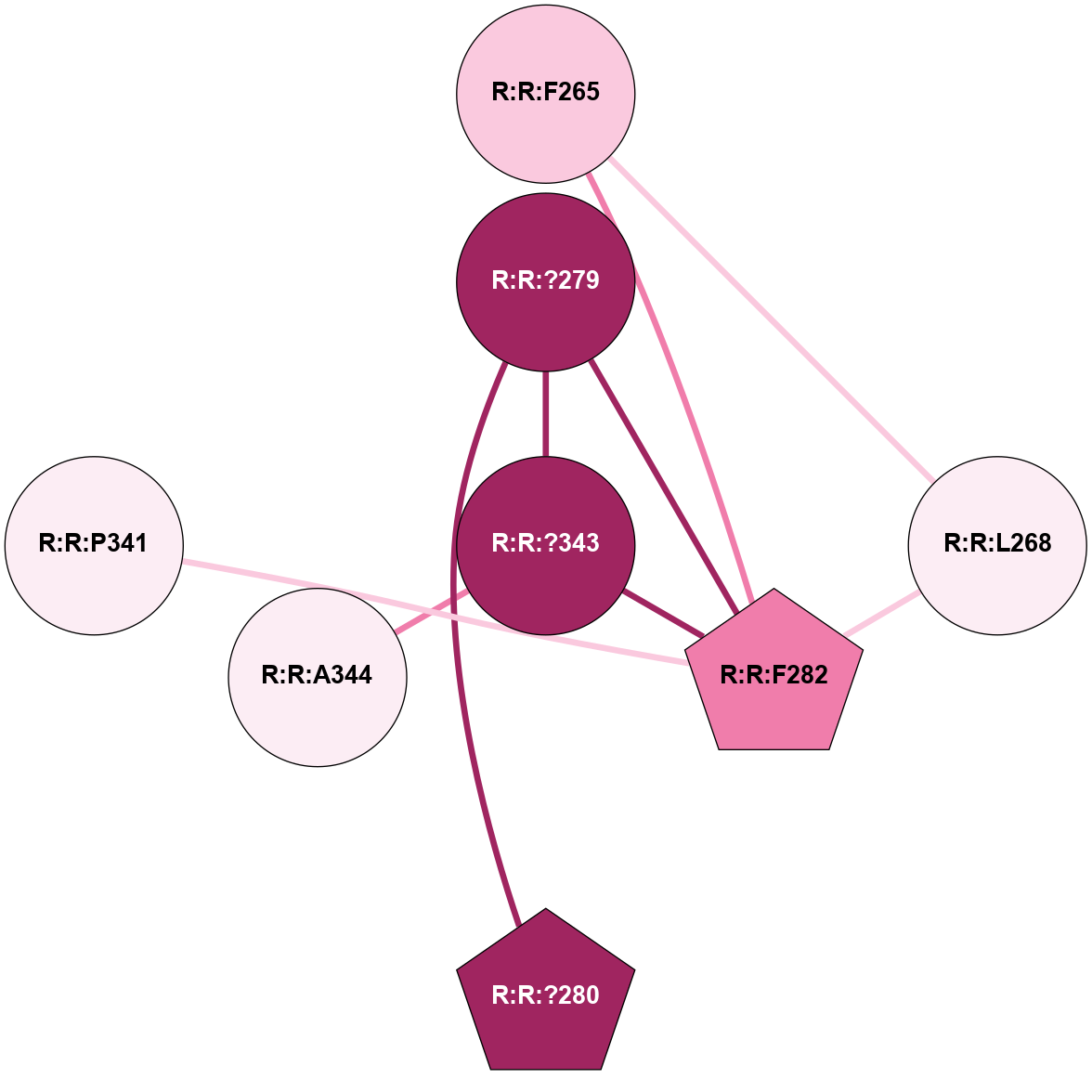
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F265 | R:R:L268 | 12.18 | 8 | No | No | 7 | 6 | 2 | 2 | | R:R:F265 | R:R:F282 | 4.29 | 8 | No | Yes | 7 | 8 | 2 | 1 | | R:R:F282 | R:R:L268 | 6.09 | 8 | Yes | No | 8 | 6 | 1 | 2 | | R:R:?279 | R:R:?280 | 26.98 | 8 | No | Yes | 9 | 9 | 1 | 2 | | R:R:?279 | R:R:F282 | 6.52 | 8 | No | Yes | 9 | 8 | 1 | 1 | | R:R:?279 | R:R:?343 | 17.46 | 8 | No | No | 9 | 9 | 1 | 0 | | R:R:F282 | R:R:P341 | 5.78 | 8 | Yes | No | 8 | 6 | 1 | 2 | | R:R:?343 | R:R:F282 | 3.91 | 8 | No | Yes | 9 | 8 | 0 | 1 | | R:R:?343 | R:R:A344 | 3.37 | 8 | No | No | 9 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 8.25 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
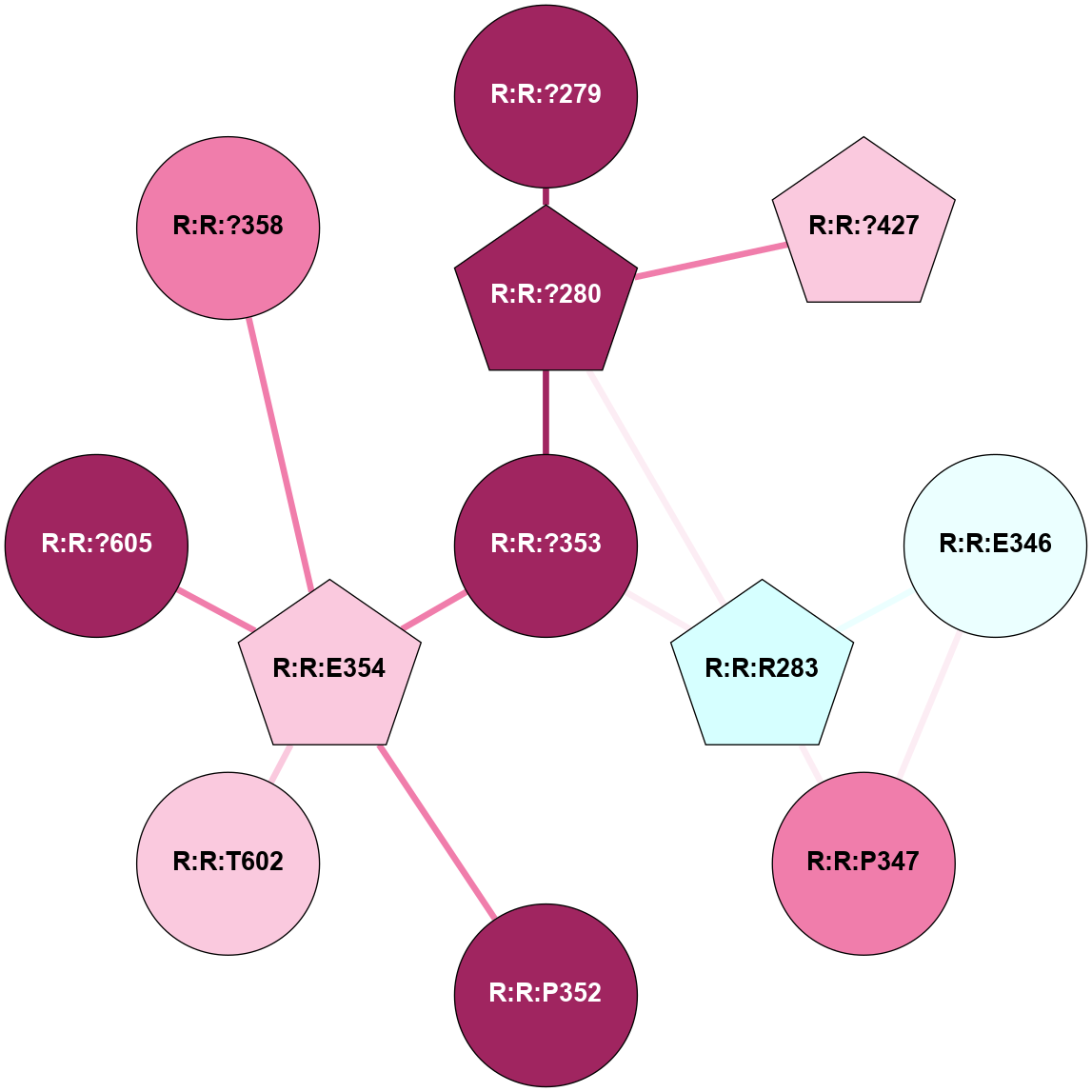
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?279 | R:R:?280 | 26.98 | 8 | No | Yes | 9 | 9 | 2 | 1 | | R:R:?280 | R:R:R283 | 5.2 | 8 | Yes | Yes | 9 | 3 | 1 | 1 | | R:R:?280 | R:R:?353 | 15.87 | 8 | Yes | No | 9 | 9 | 1 | 0 | | R:R:?280 | R:R:?427 | 4.32 | 8 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:E346 | R:R:R283 | 11.63 | 8 | No | Yes | 4 | 3 | 2 | 1 | | R:R:P347 | R:R:R283 | 4.32 | 8 | No | Yes | 8 | 3 | 2 | 1 | | R:R:?353 | R:R:R283 | 5.2 | 8 | No | Yes | 9 | 3 | 0 | 1 | | R:R:E346 | R:R:P347 | 7.86 | 8 | No | No | 4 | 8 | 2 | 2 | | R:R:E354 | R:R:P352 | 11 | 0 | Yes | No | 7 | 9 | 1 | 2 | | R:R:?353 | R:R:E354 | 4.26 | 8 | No | Yes | 9 | 7 | 0 | 1 | | R:R:E354 | R:R:T602 | 8.47 | 0 | Yes | No | 7 | 7 | 1 | 2 | | R:R:?605 | R:R:E354 | 4.74 | 0 | No | Yes | 9 | 7 | 2 | 1 | | R:R:?358 | R:R:E354 | 3.47 | 0 | No | Yes | 8 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 8.44 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
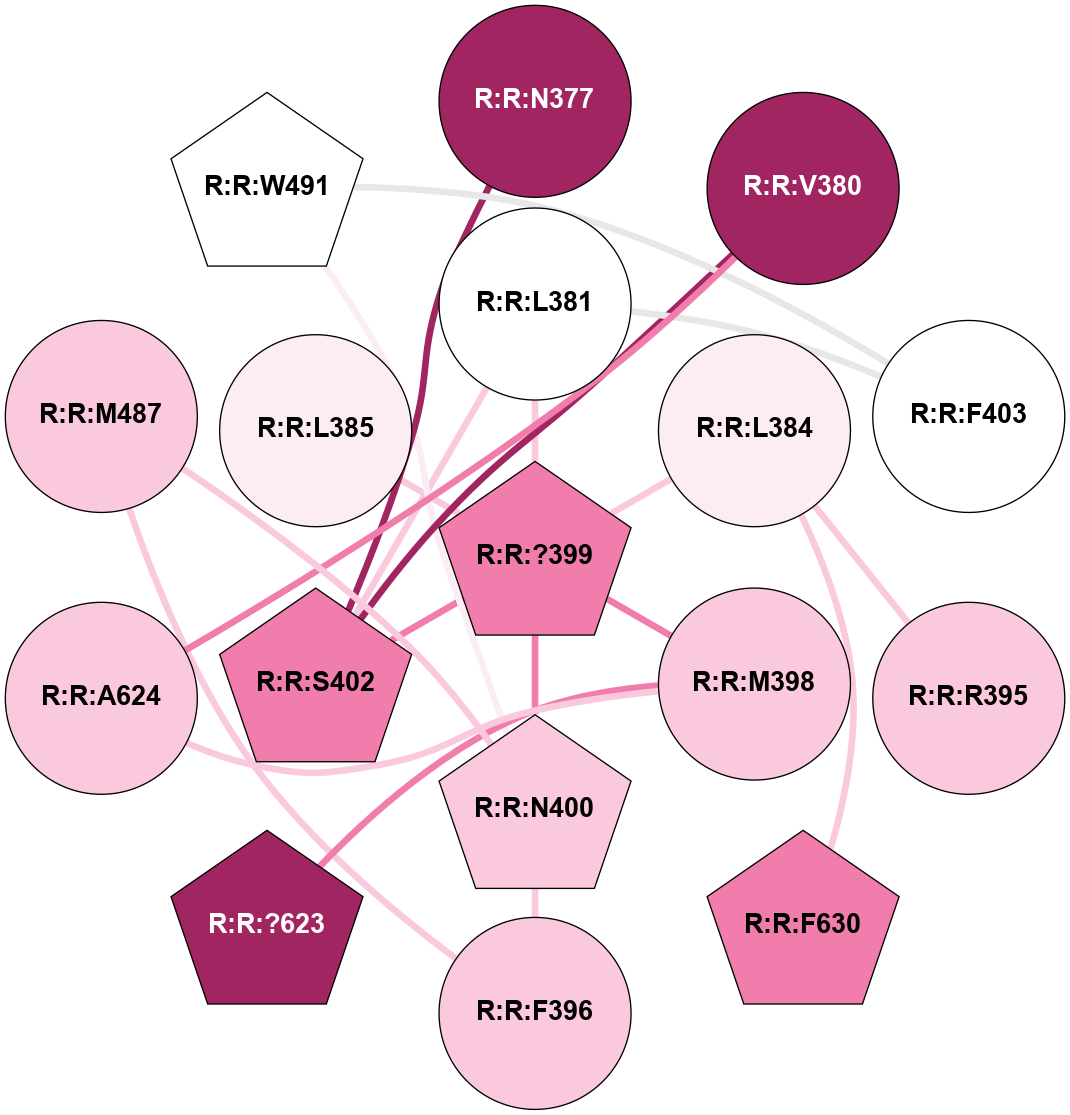
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N377 | R:R:S402 | 8.94 | 0 | No | Yes | 9 | 8 | 2 | 1 | | R:R:S402 | R:R:V380 | 6.46 | 15 | Yes | No | 8 | 9 | 1 | 2 | | R:R:A624 | R:R:V380 | 5.09 | 0 | No | No | 7 | 9 | 2 | 2 | | R:R:?399 | R:R:L381 | 7.41 | 15 | Yes | No | 8 | 5 | 0 | 1 | | R:R:L381 | R:R:S402 | 9.01 | 15 | No | Yes | 5 | 8 | 1 | 1 | | R:R:F403 | R:R:L381 | 3.65 | 0 | No | No | 5 | 5 | 2 | 1 | | R:R:L384 | R:R:R395 | 3.64 | 0 | No | No | 6 | 7 | 1 | 2 | | R:R:?399 | R:R:L384 | 5.93 | 15 | Yes | No | 8 | 6 | 0 | 1 | | R:R:F630 | R:R:L384 | 4.87 | 0 | Yes | No | 8 | 6 | 2 | 1 | | R:R:F396 | R:R:N400 | 13.29 | 15 | No | Yes | 7 | 7 | 2 | 1 | | R:R:F396 | R:R:M487 | 4.98 | 15 | No | No | 7 | 7 | 2 | 2 | | R:R:?399 | R:R:M398 | 4.54 | 15 | Yes | No | 8 | 7 | 0 | 1 | | R:R:?623 | R:R:M398 | 8.25 | 3 | Yes | No | 9 | 7 | 2 | 1 | | R:R:A624 | R:R:M398 | 4.83 | 0 | No | No | 7 | 7 | 2 | 1 | | R:R:?399 | R:R:N400 | 10.29 | 15 | Yes | Yes | 8 | 7 | 0 | 1 | | R:R:?399 | R:R:S402 | 4.82 | 15 | Yes | Yes | 8 | 8 | 0 | 1 | | R:R:M487 | R:R:N400 | 8.41 | 15 | No | Yes | 7 | 7 | 2 | 1 | | R:R:N400 | R:R:W491 | 4.52 | 15 | Yes | Yes | 7 | 5 | 1 | 2 | | R:R:F403 | R:R:W491 | 6.01 | 0 | No | Yes | 5 | 5 | 2 | 2 | | R:R:?399 | R:R:L385 | 1.48 | 15 | Yes | No | 8 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 5.74 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 20.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
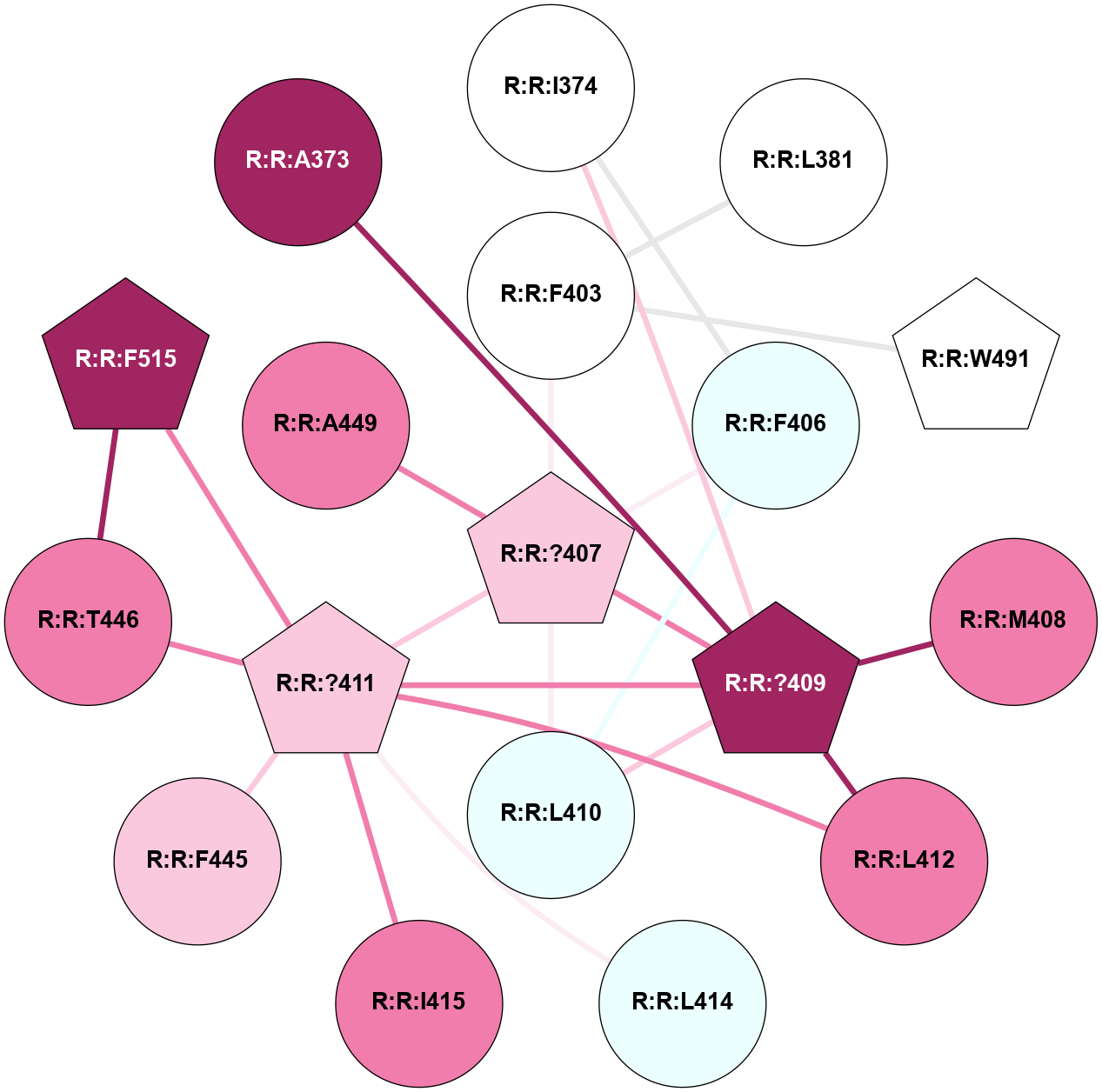
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F406 | R:R:I374 | 3.77 | 6 | No | No | 4 | 5 | 1 | 2 | | R:R:?409 | R:R:I374 | 4.98 | 6 | Yes | No | 9 | 5 | 1 | 2 | | R:R:F403 | R:R:L381 | 3.65 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:?407 | R:R:F403 | 5.22 | 6 | Yes | No | 7 | 5 | 0 | 1 | | R:R:F403 | R:R:W491 | 6.01 | 0 | No | Yes | 5 | 5 | 1 | 2 | | R:R:?407 | R:R:F406 | 3.91 | 6 | Yes | No | 7 | 4 | 0 | 1 | | R:R:F406 | R:R:L410 | 14.61 | 6 | No | No | 4 | 4 | 1 | 1 | | R:R:?407 | R:R:?409 | 10.34 | 6 | Yes | Yes | 7 | 9 | 0 | 1 | | R:R:?407 | R:R:L410 | 8.89 | 6 | Yes | No | 7 | 4 | 0 | 1 | | R:R:?407 | R:R:?411 | 5.77 | 6 | Yes | Yes | 7 | 7 | 0 | 1 | | R:R:?407 | R:R:A449 | 5.06 | 6 | Yes | No | 7 | 8 | 0 | 1 | | R:R:?409 | R:R:M408 | 3.7 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:?409 | R:R:L410 | 3.62 | 6 | Yes | No | 9 | 4 | 1 | 1 | | R:R:?409 | R:R:?411 | 9.39 | 6 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:?409 | R:R:L412 | 3.62 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:?411 | R:R:L412 | 9.42 | 6 | Yes | No | 7 | 8 | 1 | 2 | | R:R:?411 | R:R:L414 | 4.04 | 6 | Yes | No | 7 | 4 | 1 | 2 | | R:R:?411 | R:R:I415 | 18.05 | 6 | Yes | No | 7 | 8 | 1 | 2 | | R:R:?411 | R:R:F445 | 4.74 | 6 | Yes | No | 7 | 7 | 1 | 2 | | R:R:?411 | R:R:T446 | 11.47 | 6 | Yes | No | 7 | 8 | 1 | 2 | | R:R:?411 | R:R:F515 | 5.92 | 6 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:F515 | R:R:T446 | 5.19 | 6 | Yes | No | 9 | 8 | 2 | 2 | | R:R:?409 | R:R:A373 | 1.37 | 6 | Yes | No | 9 | 9 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 6.53 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 23.00 | | Average Links Mediated by Hubs In Shell | 20.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?436 | R:R:S437 | 3.93 | 6 | No | No | 9 | 2 | 1 | 2 | | R:R:?436 | R:R:?438 | 6.32 | 6 | No | Yes | 9 | 8 | 1 | 1 | | R:R:?436 | R:R:?439 | 6.46 | 6 | No | Yes | 9 | 9 | 1 | 0 | | R:R:?438 | R:R:S437 | 3.93 | 6 | Yes | No | 8 | 2 | 1 | 2 | | R:R:?438 | R:R:?439 | 19.39 | 6 | Yes | Yes | 8 | 9 | 1 | 0 | | R:R:?438 | R:R:T441 | 9 | 6 | Yes | No | 8 | 3 | 1 | 2 | | R:R:?439 | R:R:S440 | 4.82 | 6 | Yes | No | 9 | 4 | 0 | 1 | | R:R:?439 | R:R:A442 | 5.06 | 6 | Yes | No | 9 | 8 | 0 | 1 | | R:R:?439 | R:R:?443 | 3.88 | 6 | Yes | Yes | 9 | 9 | 0 | 1 | | R:R:?439 | R:R:?508 | 4.32 | 6 | Yes | Yes | 9 | 8 | 0 | 1 | | R:R:?439 | R:R:?514 | 19.05 | 6 | Yes | Yes | 9 | 9 | 0 | 1 | | R:R:?443 | R:R:T446 | 10.28 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:?443 | R:R:V447 | 3.9 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:?443 | R:R:?508 | 22.31 | 6 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:?443 | R:R:?514 | 5.17 | 6 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:F515 | R:R:T446 | 5.19 | 6 | Yes | No | 9 | 8 | 2 | 2 | | R:R:?508 | R:R:V447 | 7.24 | 6 | Yes | No | 8 | 8 | 1 | 2 | | R:R:?508 | R:R:P501 | 20.76 | 6 | Yes | No | 8 | 9 | 1 | 2 | | R:R:?508 | R:R:L502 | 4.04 | 6 | Yes | No | 8 | 7 | 1 | 2 | | R:R:?508 | R:R:M509 | 4.12 | 6 | Yes | No | 8 | 6 | 1 | 2 | | R:R:?508 | R:R:?514 | 23.07 | 6 | Yes | Yes | 8 | 9 | 1 | 1 | | R:R:?508 | R:R:P516 | 7.99 | 6 | Yes | No | 8 | 9 | 1 | 2 | | R:R:?514 | R:R:V511 | 4.79 | 6 | Yes | No | 9 | 9 | 1 | 2 | | R:R:?514 | R:R:F515 | 3.91 | 6 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:F515 | R:R:P516 | 4.33 | 6 | Yes | No | 9 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 9.00 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 25.00 | | Average Links Mediated by Hubs In Shell | 24.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
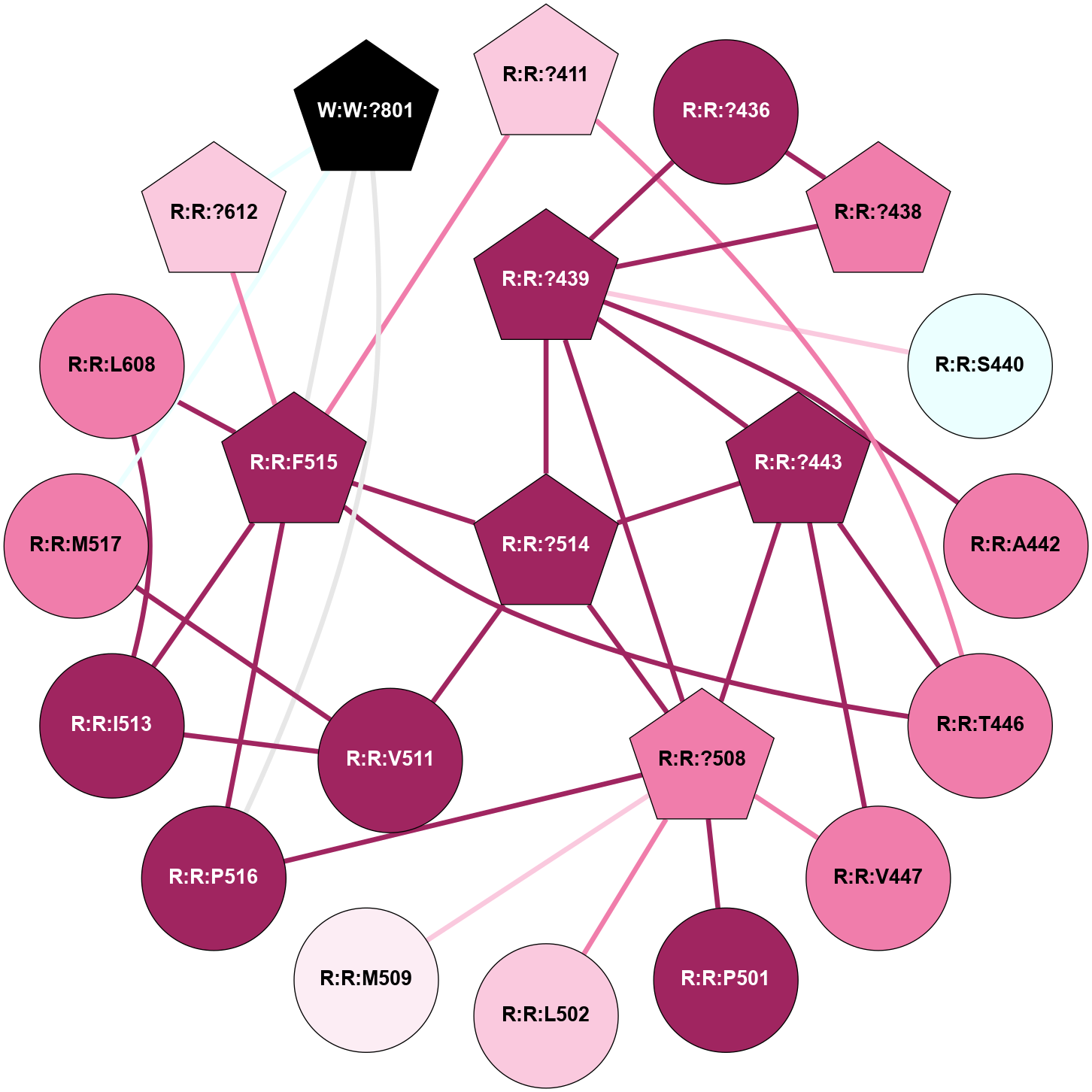
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?411 | R:R:T446 | 11.47 | 6 | Yes | No | 7 | 8 | 2 | 2 | | R:R:?411 | R:R:F515 | 5.92 | 6 | Yes | Yes | 7 | 9 | 2 | 1 | | R:R:?436 | R:R:?438 | 6.32 | 6 | No | Yes | 9 | 8 | 2 | 2 | | R:R:?436 | R:R:?439 | 6.46 | 6 | No | Yes | 9 | 9 | 2 | 1 | | R:R:?438 | R:R:?439 | 19.39 | 6 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:?439 | R:R:S440 | 4.82 | 6 | Yes | No | 9 | 4 | 1 | 2 | | R:R:?439 | R:R:A442 | 5.06 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:?439 | R:R:?443 | 3.88 | 6 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:?439 | R:R:?508 | 4.32 | 6 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:?439 | R:R:?514 | 19.05 | 6 | Yes | Yes | 9 | 9 | 1 | 0 | | R:R:?443 | R:R:T446 | 10.28 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:?443 | R:R:V447 | 3.9 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:?443 | R:R:?508 | 22.31 | 6 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:?443 | R:R:?514 | 5.17 | 6 | Yes | Yes | 9 | 9 | 1 | 0 | | R:R:F515 | R:R:T446 | 5.19 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:?508 | R:R:V447 | 7.24 | 6 | Yes | No | 8 | 8 | 1 | 2 | | R:R:?508 | R:R:P501 | 20.76 | 6 | Yes | No | 8 | 9 | 1 | 2 | | R:R:?508 | R:R:L502 | 4.04 | 6 | Yes | No | 8 | 7 | 1 | 2 | | R:R:?508 | R:R:M509 | 4.12 | 6 | Yes | No | 8 | 6 | 1 | 2 | | R:R:?508 | R:R:?514 | 23.07 | 6 | Yes | Yes | 8 | 9 | 1 | 0 | | R:R:?508 | R:R:P516 | 7.99 | 6 | Yes | No | 8 | 9 | 1 | 2 | | R:R:I513 | R:R:V511 | 4.61 | 6 | No | No | 9 | 9 | 2 | 1 | | R:R:?514 | R:R:V511 | 4.79 | 6 | Yes | No | 9 | 9 | 0 | 1 | | R:R:M517 | R:R:V511 | 6.09 | 0 | No | No | 8 | 9 | 2 | 1 | | R:R:F515 | R:R:I513 | 8.79 | 6 | Yes | No | 9 | 9 | 1 | 2 | | R:R:I513 | R:R:L608 | 8.56 | 6 | No | No | 9 | 8 | 2 | 2 | | R:R:?514 | R:R:F515 | 3.91 | 6 | Yes | Yes | 9 | 9 | 0 | 1 | | R:R:F515 | R:R:P516 | 4.33 | 6 | Yes | No | 9 | 9 | 1 | 2 | | R:R:F515 | R:R:L608 | 3.65 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:?612 | R:R:F515 | 10.66 | 6 | Yes | Yes | 7 | 9 | 2 | 1 | | R:R:F515 | W:W:?801 | 12.52 | 6 | Yes | Yes | 9 | 0 | 1 | 2 | | R:R:P516 | W:W:?801 | 6.03 | 6 | No | Yes | 9 | 0 | 2 | 2 | | R:R:M517 | W:W:?801 | 21.8 | 0 | No | Yes | 8 | 0 | 2 | 2 | | R:R:?612 | W:W:?801 | 3.95 | 6 | Yes | Yes | 7 | 0 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 11.20 | | Average Nodes In Shell | 22.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 34.00 | | Average Links Mediated by Hubs In Shell | 31.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
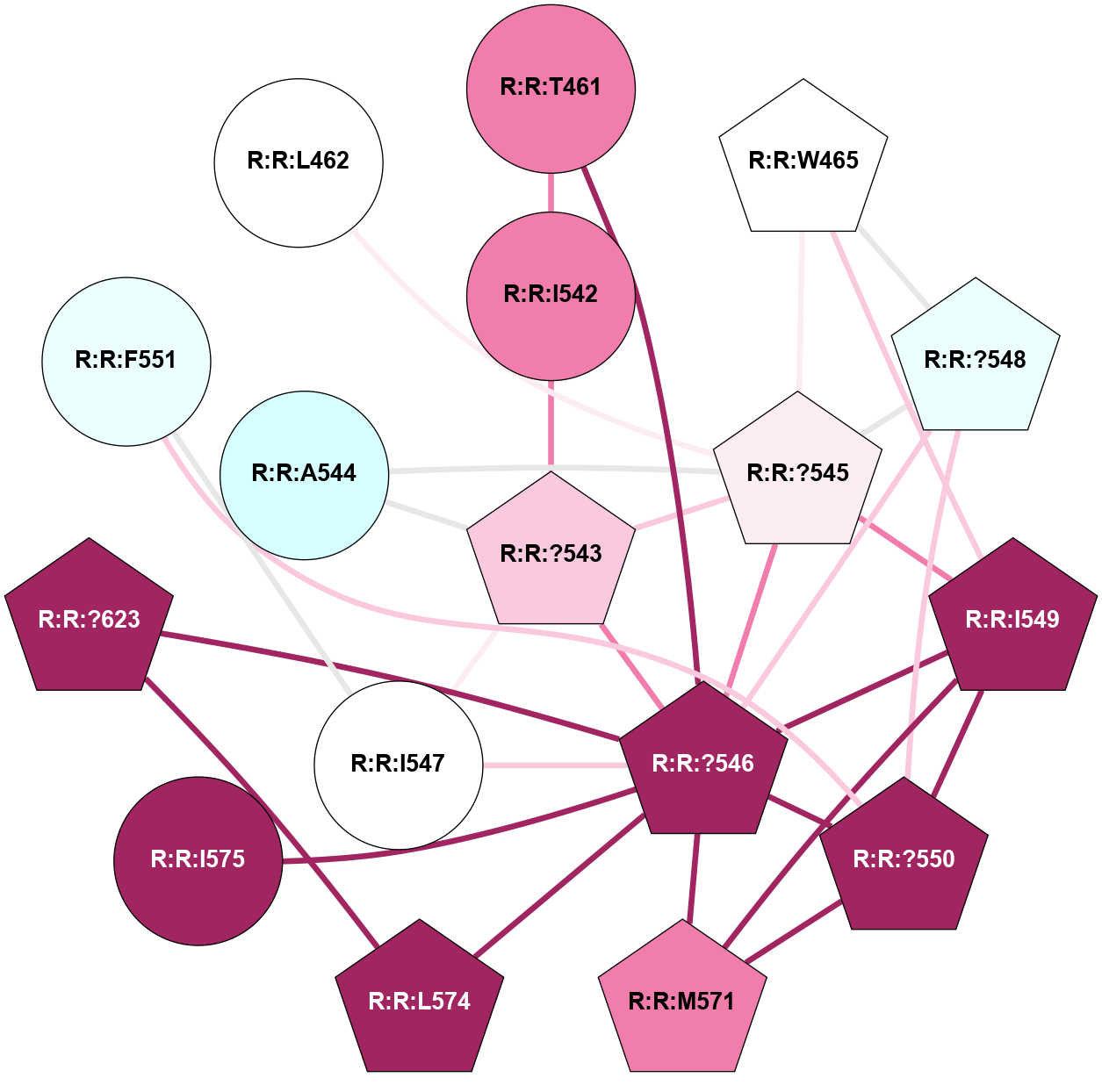
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I542 | R:R:T461 | 13.68 | 0 | No | No | 8 | 8 | 1 | 2 | | R:R:?546 | R:R:T461 | 14.34 | 3 | Yes | No | 9 | 8 | 1 | 2 | | R:R:?545 | R:R:W465 | 8.54 | 3 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:?548 | R:R:W465 | 4.08 | 3 | Yes | Yes | 4 | 5 | 2 | 2 | | R:R:I549 | R:R:W465 | 4.7 | 3 | Yes | Yes | 9 | 5 | 2 | 2 | | R:R:?543 | R:R:I542 | 4.59 | 3 | Yes | No | 7 | 8 | 0 | 1 | | R:R:?543 | R:R:?545 | 9.52 | 3 | Yes | Yes | 7 | 6 | 0 | 1 | | R:R:?543 | R:R:?546 | 8.65 | 3 | Yes | Yes | 7 | 9 | 0 | 1 | | R:R:?543 | R:R:I547 | 7.64 | 3 | Yes | No | 7 | 5 | 0 | 1 | | R:R:?545 | R:R:?546 | 25.95 | 3 | Yes | Yes | 6 | 9 | 1 | 1 | | R:R:?545 | R:R:?548 | 7.96 | 3 | Yes | Yes | 6 | 4 | 1 | 2 | | R:R:?545 | R:R:I549 | 6.11 | 3 | Yes | Yes | 6 | 9 | 1 | 2 | | R:R:?546 | R:R:I547 | 4.16 | 3 | Yes | No | 9 | 5 | 1 | 1 | | R:R:?546 | R:R:?548 | 6.42 | 3 | Yes | Yes | 9 | 4 | 1 | 2 | | R:R:?546 | R:R:I549 | 6.94 | 3 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:?546 | R:R:?550 | 5.24 | 3 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:?546 | R:R:M571 | 12.37 | 3 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:?546 | R:R:L574 | 4.04 | 3 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:?546 | R:R:I575 | 6.94 | 3 | Yes | No | 9 | 9 | 1 | 2 | | R:R:?546 | R:R:?623 | 6.55 | 3 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:F551 | R:R:I547 | 6.28 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:?548 | R:R:?550 | 5.62 | 3 | Yes | Yes | 4 | 9 | 2 | 2 | | R:R:?550 | R:R:I549 | 5.55 | 3 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:I549 | R:R:M571 | 4.37 | 3 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:?550 | R:R:F551 | 13.03 | 3 | Yes | No | 9 | 4 | 2 | 2 | | R:R:?550 | R:R:M571 | 4.12 | 3 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:?623 | R:R:L574 | 9.42 | 3 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:?543 | R:R:A544 | 3.37 | 3 | Yes | No | 7 | 3 | 0 | 1 | | R:R:?545 | R:R:A544 | 3.37 | 3 | Yes | No | 6 | 3 | 1 | 1 | | R:R:?545 | R:R:L462 | 2.96 | 3 | Yes | No | 6 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 6.75 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 30.00 | | Average Links Mediated by Hubs In Shell | 28.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
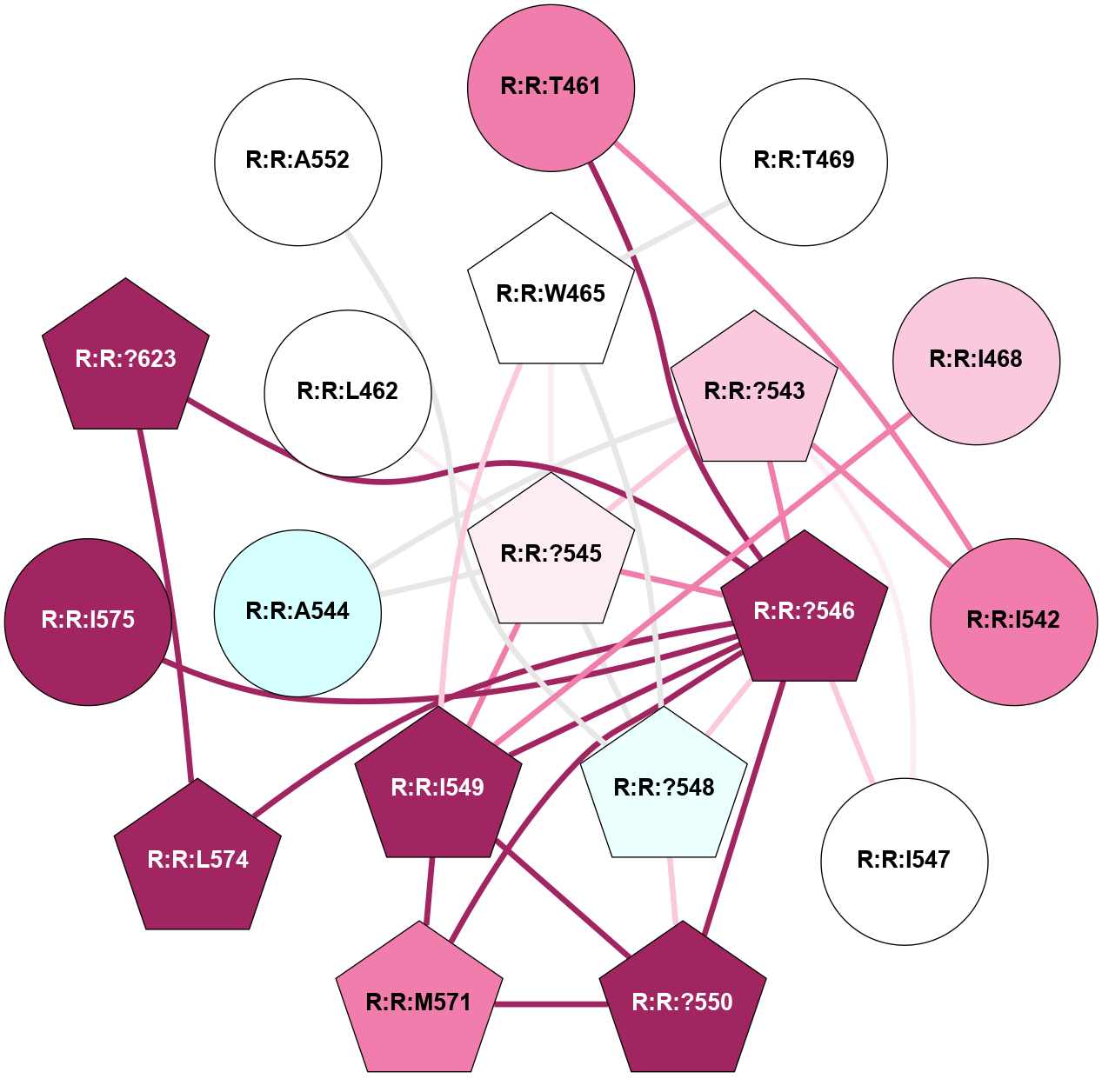
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I542 | R:R:T461 | 13.68 | 0 | No | No | 8 | 8 | 2 | 2 | | R:R:?546 | R:R:T461 | 14.34 | 3 | Yes | No | 9 | 8 | 1 | 2 | | R:R:T469 | R:R:W465 | 9.7 | 0 | No | Yes | 5 | 5 | 2 | 1 | | R:R:?545 | R:R:W465 | 8.54 | 3 | Yes | Yes | 6 | 5 | 0 | 1 | | R:R:?548 | R:R:W465 | 4.08 | 3 | Yes | Yes | 4 | 5 | 1 | 1 | | R:R:I549 | R:R:W465 | 4.7 | 3 | Yes | Yes | 9 | 5 | 1 | 1 | | R:R:I468 | R:R:I549 | 5.89 | 0 | No | Yes | 7 | 9 | 2 | 1 | | R:R:?543 | R:R:I542 | 4.59 | 3 | Yes | No | 7 | 8 | 1 | 2 | | R:R:?543 | R:R:?545 | 9.52 | 3 | Yes | Yes | 7 | 6 | 1 | 0 | | R:R:?543 | R:R:?546 | 8.65 | 3 | Yes | Yes | 7 | 9 | 1 | 1 | | R:R:?543 | R:R:I547 | 7.64 | 3 | Yes | No | 7 | 5 | 1 | 2 | | R:R:?545 | R:R:?546 | 25.95 | 3 | Yes | Yes | 6 | 9 | 0 | 1 | | R:R:?545 | R:R:?548 | 7.96 | 3 | Yes | Yes | 6 | 4 | 0 | 1 | | R:R:?545 | R:R:I549 | 6.11 | 3 | Yes | Yes | 6 | 9 | 0 | 1 | | R:R:?546 | R:R:I547 | 4.16 | 3 | Yes | No | 9 | 5 | 1 | 2 | | R:R:?546 | R:R:?548 | 6.42 | 3 | Yes | Yes | 9 | 4 | 1 | 1 | | R:R:?546 | R:R:I549 | 6.94 | 3 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:?546 | R:R:?550 | 5.24 | 3 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:?546 | R:R:M571 | 12.37 | 3 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:?546 | R:R:L574 | 4.04 | 3 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:?546 | R:R:I575 | 6.94 | 3 | Yes | No | 9 | 9 | 1 | 2 | | R:R:?546 | R:R:?623 | 6.55 | 3 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:?548 | R:R:?550 | 5.62 | 3 | Yes | Yes | 4 | 9 | 1 | 2 | | R:R:?550 | R:R:I549 | 5.55 | 3 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:I549 | R:R:M571 | 4.37 | 3 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:?550 | R:R:M571 | 4.12 | 3 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:?623 | R:R:L574 | 9.42 | 3 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:?543 | R:R:A544 | 3.37 | 3 | Yes | No | 7 | 3 | 1 | 1 | | R:R:?545 | R:R:A544 | 3.37 | 3 | Yes | No | 6 | 3 | 0 | 1 | | R:R:?545 | R:R:L462 | 2.96 | 3 | Yes | No | 6 | 5 | 0 | 1 | | R:R:?548 | R:R:A552 | 1.88 | 3 | Yes | No | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 9.20 | | Average Nodes In Shell | 19.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 31.00 | | Average Links Mediated by Hubs In Shell | 30.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D578 | R:R:N615 | 5.39 | 0 | No | No | 8 | 9 | 2 | 1 | | R:R:?581 | R:R:T580 | 6.31 | 0 | Yes | No | 9 | 6 | 0 | 1 | | R:R:?581 | R:R:I614 | 6.11 | 0 | Yes | No | 9 | 8 | 0 | 1 | | R:R:?581 | R:R:N615 | 7.35 | 0 | Yes | No | 9 | 9 | 0 | 1 | | R:R:N615 | R:R:N619 | 9.54 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:?581 | R:R:I585 | 3.06 | 0 | Yes | No | 9 | 9 | 0 | 1 | | R:R:I585 | R:R:L607 | 2.85 | 0 | No | No | 9 | 8 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 5.71 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
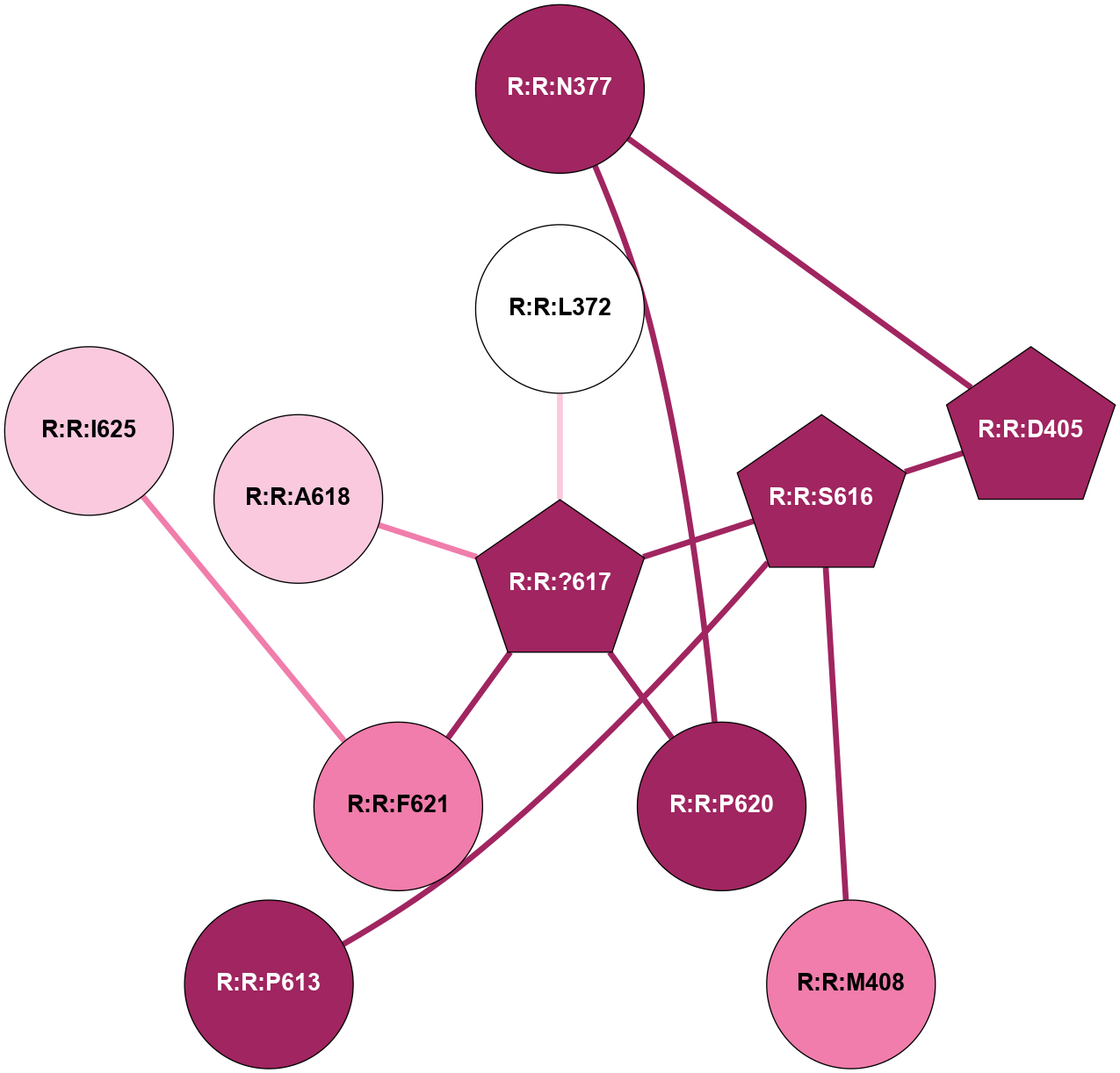
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?617 | R:R:L372 | 4.45 | 0 | Yes | No | 9 | 5 | 0 | 1 | | R:R:D405 | R:R:N377 | 9.42 | 0 | Yes | No | 9 | 9 | 2 | 2 | | R:R:N377 | R:R:P620 | 11.4 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D405 | R:R:S616 | 4.42 | 0 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:M408 | R:R:S616 | 4.6 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:P613 | R:R:S616 | 5.34 | 0 | No | Yes | 9 | 9 | 2 | 1 | | R:R:?617 | R:R:S616 | 6.43 | 0 | Yes | Yes | 9 | 9 | 0 | 1 | | R:R:?617 | R:R:P620 | 7.03 | 0 | Yes | No | 9 | 9 | 0 | 1 | | R:R:?617 | R:R:F621 | 3.91 | 0 | Yes | No | 9 | 8 | 0 | 1 | | R:R:F621 | R:R:I625 | 5.02 | 0 | No | No | 8 | 7 | 1 | 2 | | R:R:?617 | R:R:A618 | 3.37 | 0 | Yes | No | 9 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.04 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F634 | R:R:L638 | 7.31 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:?643 | R:R:L638 | 5.93 | 21 | No | No | 2 | 4 | 0 | 1 | | R:R:?644 | R:R:L638 | 8.89 | 21 | No | No | 4 | 4 | 1 | 1 | | R:R:?642 | R:R:?643 | 19.39 | 21 | No | No | 4 | 2 | 1 | 0 | | R:R:?642 | R:R:?644 | 5.17 | 21 | No | No | 4 | 4 | 1 | 1 | | R:R:?643 | R:R:?644 | 26.98 | 21 | No | No | 2 | 4 | 0 | 1 | | R:R:?640 | R:R:?642 | 2.88 | 0 | No | No | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.43 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F634 | R:R:L638 | 7.31 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:?643 | R:R:L638 | 5.93 | 21 | No | No | 2 | 4 | 1 | 1 | | R:R:?644 | R:R:L638 | 8.89 | 21 | No | No | 4 | 4 | 0 | 1 | | R:R:?642 | R:R:?643 | 19.39 | 21 | No | No | 4 | 2 | 1 | 1 | | R:R:?642 | R:R:?644 | 5.17 | 21 | No | No | 4 | 4 | 1 | 0 | | R:R:?643 | R:R:?644 | 26.98 | 21 | No | No | 2 | 4 | 1 | 0 | | R:R:?640 | R:R:?642 | 2.88 | 0 | No | No | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.68 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
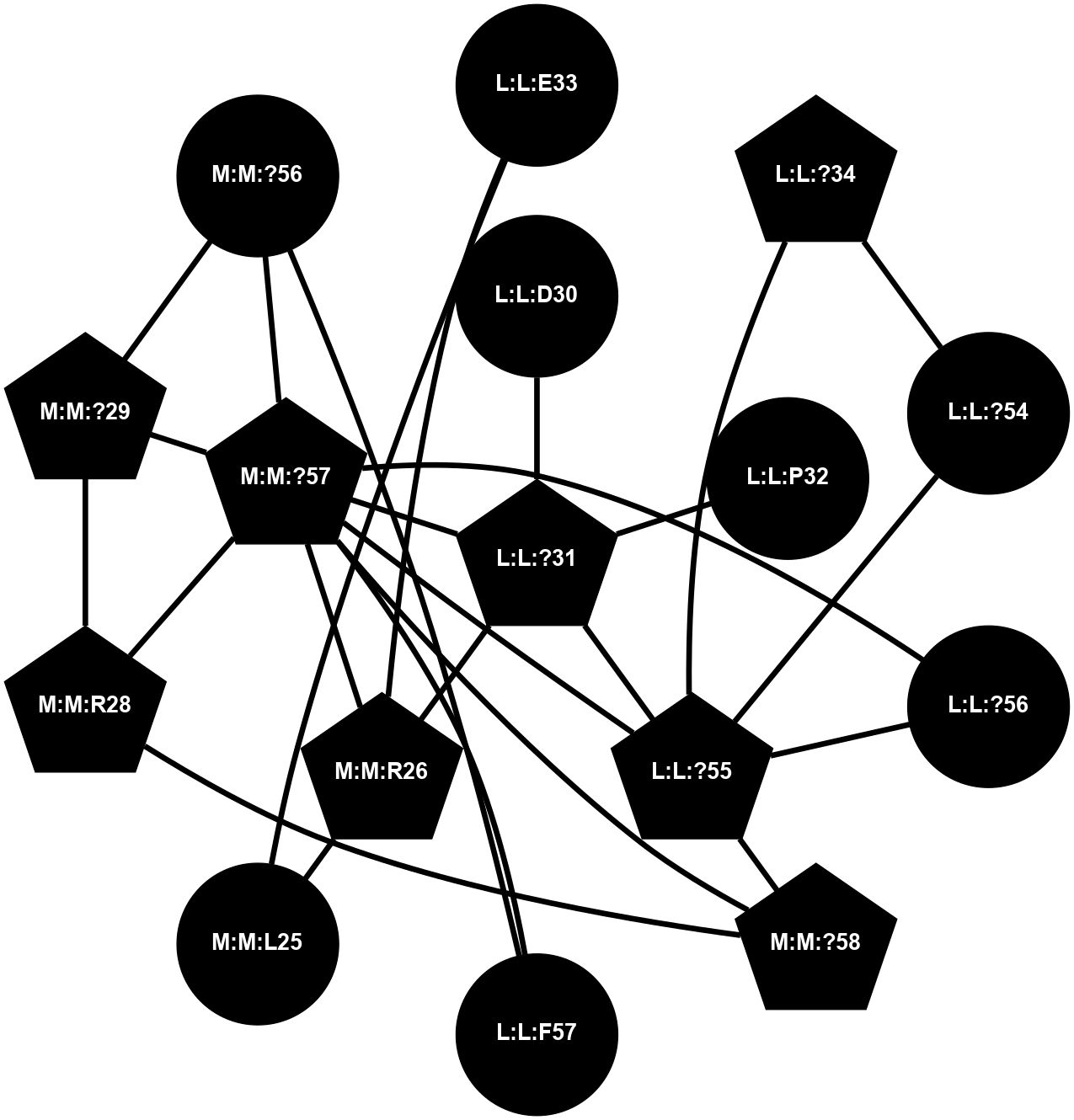
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?31 | L:L:D30 | 11.63 | 1 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?31 | L:L:P32 | 15.83 | 1 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?31 | L:L:?55 | 12.7 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?31 | N:N:R26 | 13.01 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?31 | N:N:?57 | 7.21 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:E33 | N:N:L25 | 3.98 | 1 | No | No | 0 | 0 | 2 | 2 | | L:L:E33 | N:N:R26 | 22.1 | 1 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?34 | L:L:?54 | 12.93 | 1 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?34 | L:L:?55 | 6.35 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?54 | L:L:?55 | 18.1 | 1 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?55 | L:L:?56 | 28.57 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?55 | N:N:?57 | 8.65 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?55 | N:N:?58 | 9.52 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?56 | N:N:?57 | 34.6 | 1 | No | Yes | 0 | 0 | 2 | 1 | | L:L:F57 | N:N:?56 | 5.31 | 1 | No | No | 0 | 0 | 2 | 2 | | L:L:F57 | N:N:?57 | 7.11 | 1 | No | Yes | 0 | 0 | 2 | 1 | | N:N:L25 | N:N:R26 | 4.86 | 1 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:R26 | 7.09 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | N:N:?29 | N:N:R28 | 6.5 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?57 | N:N:R28 | 5.91 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?58 | N:N:R28 | 5.2 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?29 | N:N:?56 | 7.76 | 1 | Yes | No | 0 | 0 | 2 | 2 | | N:N:?29 | N:N:?57 | 10.09 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?56 | N:N:?57 | 23.48 | 1 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:?58 | 24.51 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 12.08 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 25.00 | | Average Links Mediated by Hubs In Shell | 23.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
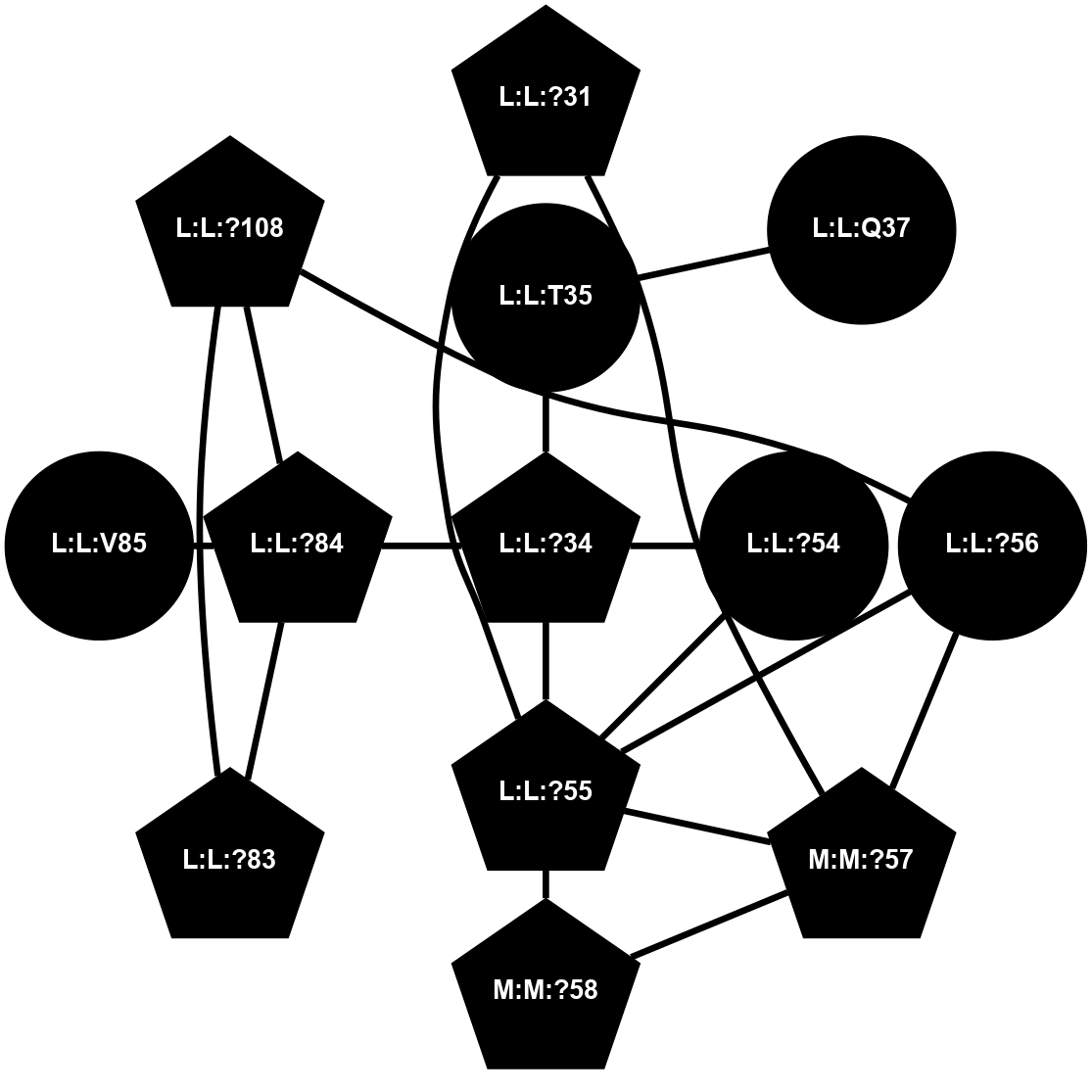
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?31 | L:L:?55 | 12.7 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?31 | N:N:?57 | 7.21 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?34 | L:L:T35 | 4.74 | 1 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?34 | L:L:?54 | 12.93 | 1 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?34 | L:L:?55 | 6.35 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?34 | L:L:?84 | 9.52 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:Q37 | L:L:T35 | 4.25 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:?54 | L:L:?55 | 18.1 | 1 | No | Yes | 0 | 0 | 1 | 1 | | L:L:?55 | L:L:?56 | 28.57 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?55 | N:N:?57 | 8.65 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?55 | N:N:?58 | 9.52 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:?56 | 9.52 | 1 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?56 | N:N:?57 | 34.6 | 1 | No | Yes | 0 | 0 | 2 | 2 | | L:L:?83 | L:L:?84 | 22.22 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?108 | L:L:?83 | 9.52 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?84 | L:L:V85 | 4.79 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:?84 | 12.7 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:?58 | 24.51 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 8.39 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
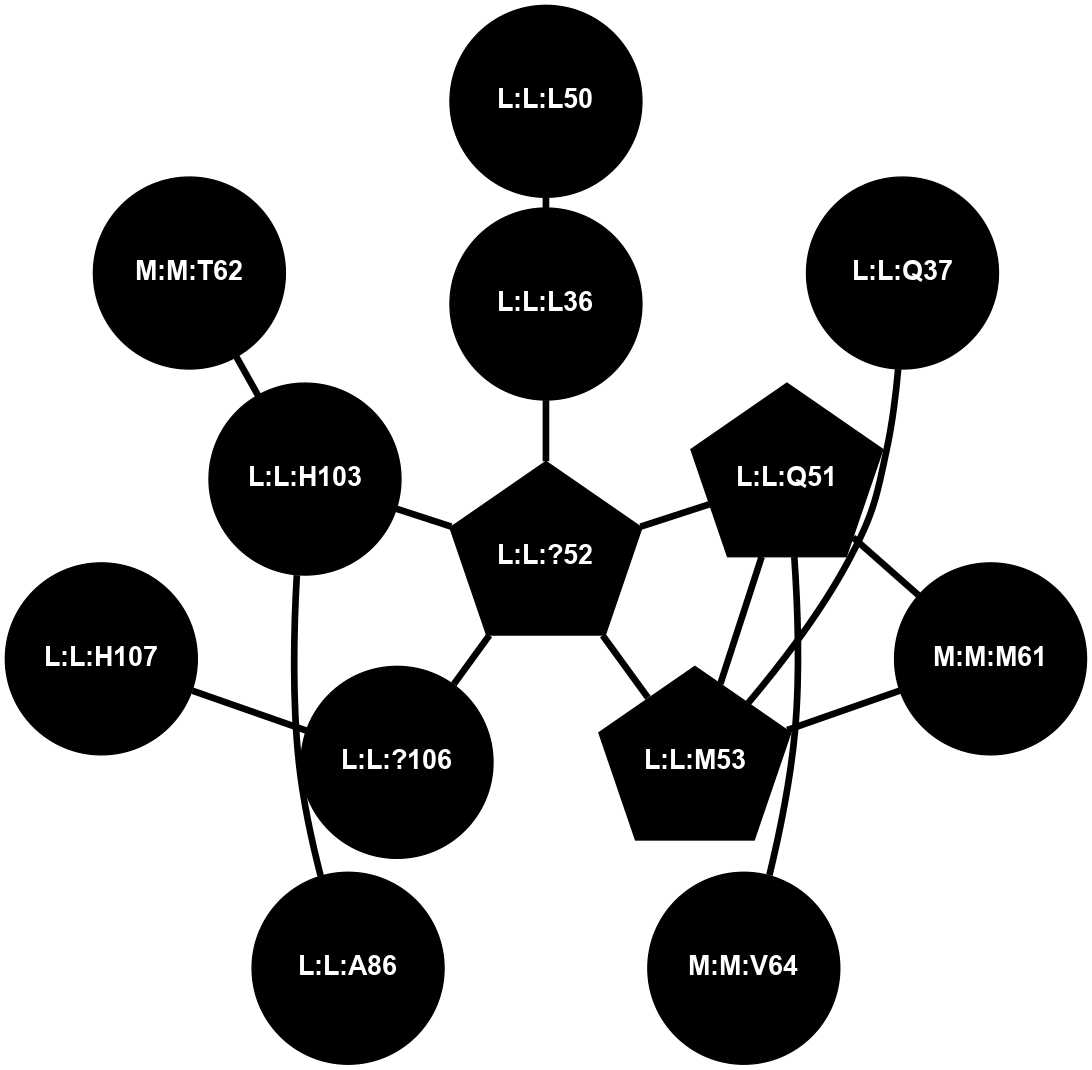
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:L36 | L:L:L50 | 12.46 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:?52 | L:L:L36 | 4.45 | 22 | Yes | No | 0 | 0 | 0 | 1 | | L:L:M53 | L:L:Q37 | 9.52 | 22 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?52 | L:L:Q51 | 7.13 | 22 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:M53 | L:L:Q51 | 4.08 | 22 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:Q51 | N:N:M61 | 5.44 | 22 | Yes | No | 0 | 0 | 1 | 2 | | L:L:Q51 | N:N:V64 | 4.3 | 22 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?52 | L:L:M53 | 9.08 | 22 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?106 | L:L:?52 | 9.52 | 0 | No | Yes | 0 | 0 | 1 | 0 | | L:L:M53 | N:N:M61 | 7.22 | 22 | Yes | No | 0 | 0 | 1 | 2 | | L:L:A86 | L:L:H103 | 8.78 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:?106 | L:L:H107 | 4.13 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:?52 | L:L:H103 | 2.75 | 22 | Yes | No | 0 | 0 | 0 | 1 | | L:L:H103 | N:N:T62 | 2.74 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 6.59 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
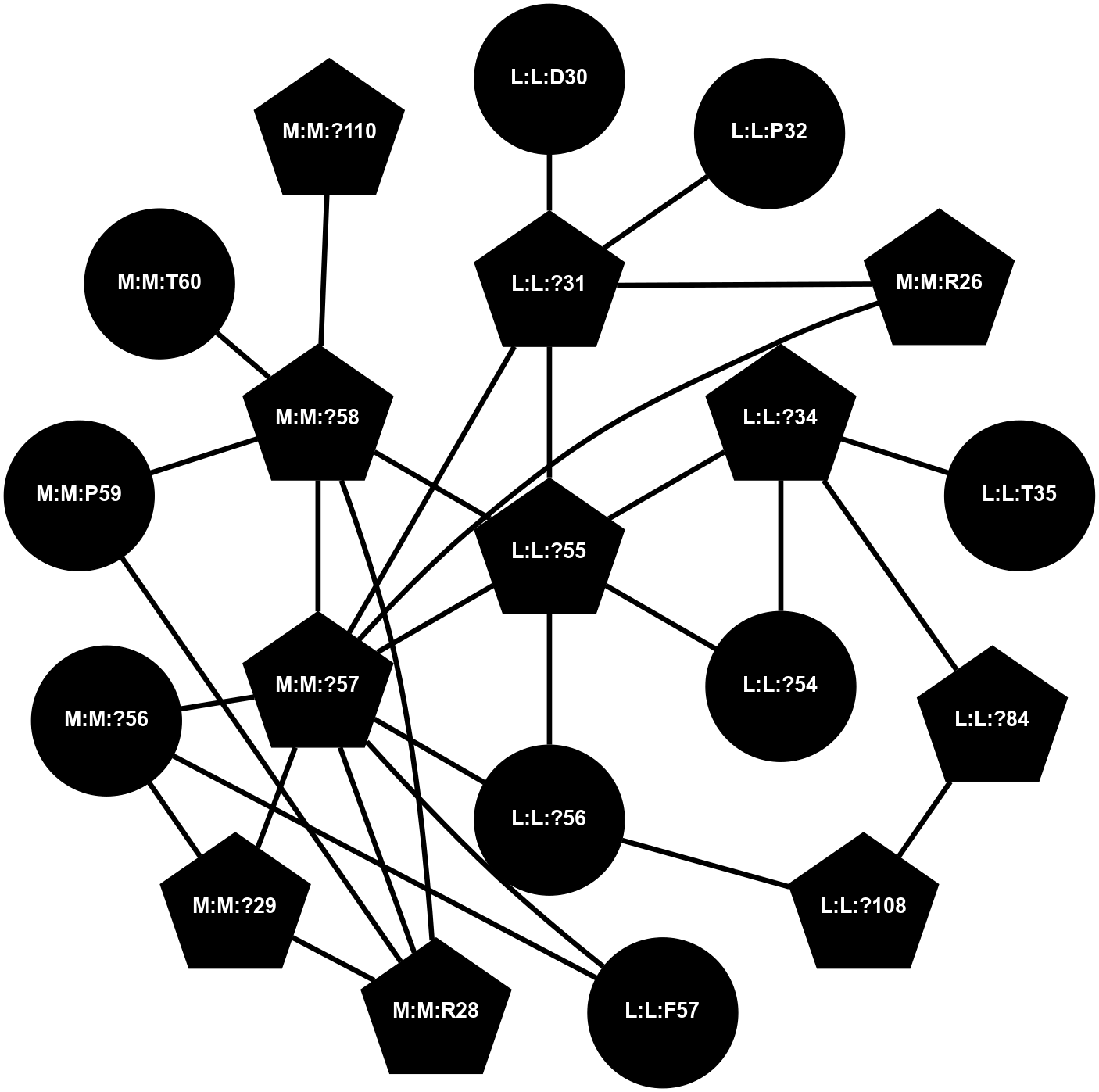
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?31 | L:L:D30 | 11.63 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?31 | L:L:P32 | 15.83 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?31 | L:L:?55 | 12.7 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?31 | N:N:R26 | 13.01 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?31 | N:N:?57 | 7.21 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?34 | L:L:T35 | 4.74 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?34 | L:L:?54 | 12.93 | 1 | Yes | No | 0 | 0 | 1 | 1 | | L:L:?34 | L:L:?55 | 6.35 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?34 | L:L:?84 | 9.52 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?54 | L:L:?55 | 18.1 | 1 | No | Yes | 0 | 0 | 1 | 0 | | L:L:?55 | L:L:?56 | 28.57 | 1 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?55 | N:N:?57 | 8.65 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?55 | N:N:?58 | 9.52 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?108 | L:L:?56 | 9.52 | 1 | Yes | No | 0 | 0 | 2 | 1 | | L:L:?56 | N:N:?57 | 34.6 | 1 | No | Yes | 0 | 0 | 1 | 1 | | L:L:F57 | N:N:?56 | 5.31 | 1 | No | No | 0 | 0 | 2 | 2 | | L:L:F57 | N:N:?57 | 7.11 | 1 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?108 | L:L:?84 | 12.7 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?57 | N:N:R26 | 7.09 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?29 | N:N:R28 | 6.5 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?57 | N:N:R28 | 5.91 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?58 | N:N:R28 | 5.2 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:P59 | N:N:R28 | 5.76 | 1 | No | Yes | 0 | 0 | 2 | 2 | | N:N:?29 | N:N:?56 | 7.76 | 1 | Yes | No | 0 | 0 | 2 | 2 | | N:N:?29 | N:N:?57 | 10.09 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?56 | N:N:?57 | 23.48 | 1 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:?58 | 24.51 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | N:N:?58 | N:N:P59 | 19.34 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?58 | N:N:T60 | 4.74 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?110 | N:N:?58 | 9.52 | 0 | Yes | Yes | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 13.98 | | Average Nodes In Shell | 20.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 30.00 | | Average Links Mediated by Hubs In Shell | 29.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
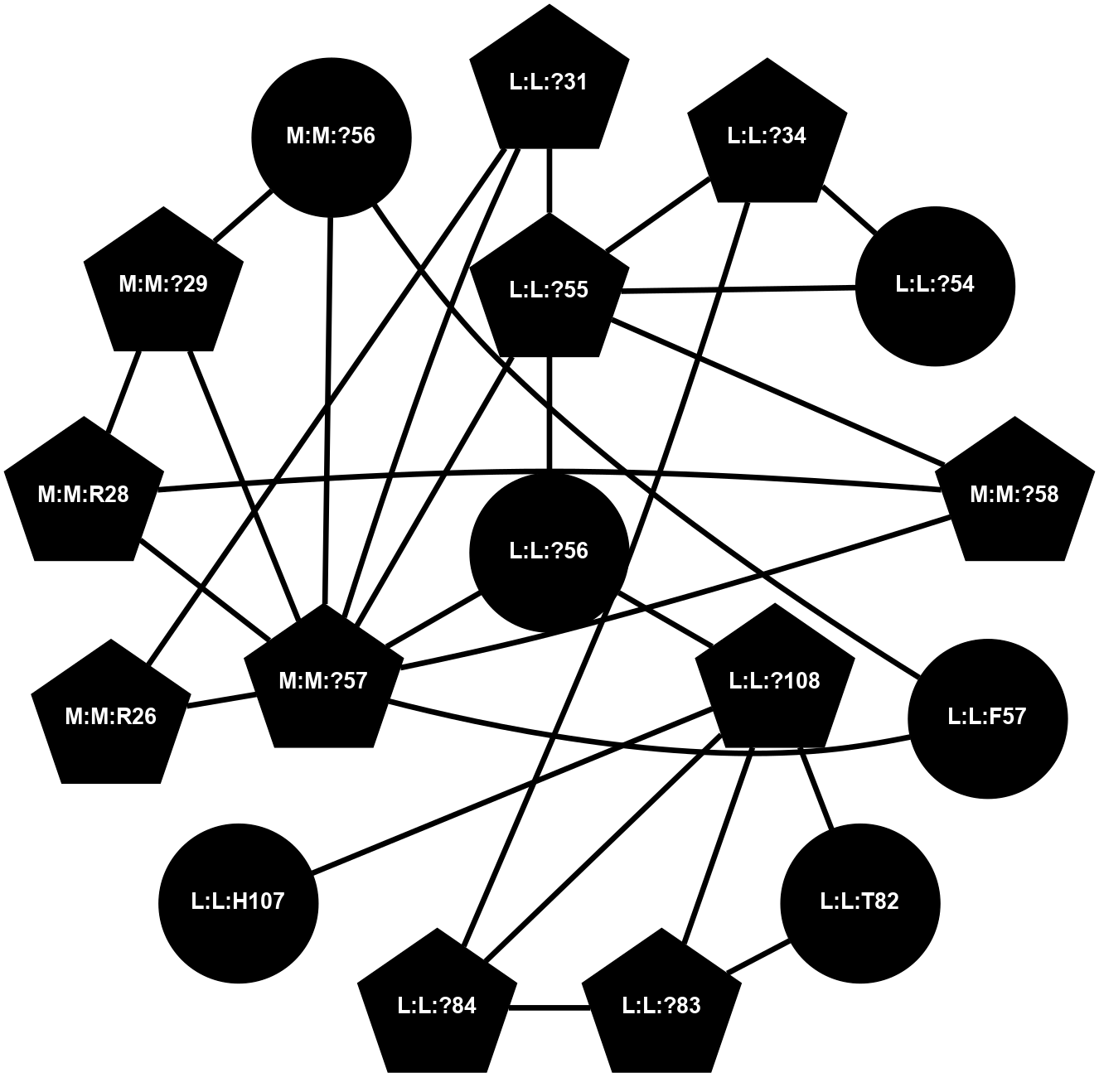
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?31 | L:L:?55 | 12.7 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?31 | N:N:R26 | 13.01 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?31 | N:N:?57 | 7.21 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?34 | L:L:?54 | 12.93 | 1 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?34 | L:L:?55 | 6.35 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?34 | L:L:?84 | 9.52 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?54 | L:L:?55 | 18.1 | 1 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?55 | L:L:?56 | 28.57 | 1 | Yes | No | 0 | 0 | 1 | 0 | | L:L:?55 | N:N:?57 | 8.65 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?55 | N:N:?58 | 9.52 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:?56 | 9.52 | 1 | Yes | No | 0 | 0 | 1 | 0 | | L:L:?56 | N:N:?57 | 34.6 | 1 | No | Yes | 0 | 0 | 0 | 1 | | L:L:F57 | N:N:?56 | 5.31 | 1 | No | No | 0 | 0 | 2 | 2 | | L:L:F57 | N:N:?57 | 7.11 | 1 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?83 | L:L:T82 | 4.74 | 1 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?108 | L:L:T82 | 7.89 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?83 | L:L:?84 | 22.22 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?108 | L:L:?83 | 9.52 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:?84 | 12.7 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:H107 | 15.14 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?57 | N:N:R26 | 7.09 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?29 | N:N:R28 | 6.5 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?57 | N:N:R28 | 5.91 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?58 | N:N:R28 | 5.2 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?29 | N:N:?56 | 7.76 | 1 | Yes | No | 0 | 0 | 2 | 2 | | N:N:?29 | N:N:?57 | 10.09 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?56 | N:N:?57 | 23.48 | 1 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:?58 | 24.51 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 24.23 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 28.00 | | Average Links Mediated by Hubs In Shell | 27.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?34 | L:L:?84 | 9.52 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?108 | L:L:?56 | 9.52 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?83 | L:L:T82 | 4.74 | 1 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?108 | L:L:T82 | 7.89 | 1 | Yes | No | 0 | 0 | 1 | 1 | | L:L:?83 | L:L:?84 | 22.22 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?108 | L:L:?83 | 9.52 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?111 | L:L:?83 | 15.87 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?83 | N:N:T60 | 4.74 | 1 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?83 | N:N:Q74 | 9.98 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?84 | L:L:V85 | 4.79 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:?84 | 12.7 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?108 | L:L:H107 | 15.14 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?111 | L:L:T110 | 7.89 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?112 | L:L:T110 | 7.17 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?111 | L:L:?112 | 30.27 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?111 | L:L:H114 | 22.03 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?111 | N:N:Q74 | 12.83 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?112 | L:L:?113 | 24.88 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?112 | L:L:H114 | 12.5 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?112 | N:N:Q74 | 5.18 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?113 | L:L:H114 | 7.5 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?113 | N:N:Q74 | 19.42 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?58 | N:N:T60 | 4.74 | 1 | Yes | No | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 11.18 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 23.00 | | Average Links Mediated by Hubs In Shell | 23.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?34 | L:L:T35 | 4.74 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?34 | L:L:?54 | 12.93 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?34 | L:L:?55 | 6.35 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?34 | L:L:?84 | 9.52 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?54 | L:L:?55 | 18.1 | 1 | No | Yes | 0 | 0 | 2 | 2 | | L:L:?55 | L:L:?56 | 28.57 | 1 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?108 | L:L:?56 | 9.52 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?83 | L:L:T82 | 4.74 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:T82 | 7.89 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?83 | L:L:?84 | 22.22 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?108 | L:L:?83 | 9.52 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?111 | L:L:?83 | 15.87 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?83 | N:N:T60 | 4.74 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?83 | N:N:Q74 | 9.98 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?84 | L:L:V85 | 4.79 | 1 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?108 | L:L:?84 | 12.7 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?108 | L:L:H107 | 15.14 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?111 | N:N:Q74 | 12.83 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 12.31 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 18.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?52 | L:L:L36 | 4.45 | 22 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?52 | L:L:Q51 | 7.13 | 22 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:M53 | L:L:Q51 | 4.08 | 22 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?52 | L:L:M53 | 9.08 | 22 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?106 | L:L:?52 | 9.52 | 0 | No | Yes | 0 | 0 | 0 | 1 | | L:L:?87 | L:L:H107 | 4.6 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:?106 | L:L:H107 | 4.13 | 0 | No | No | 0 | 0 | 0 | 1 | | L:L:?108 | L:L:H107 | 15.14 | 1 | Yes | No | 0 | 0 | 2 | 1 | | L:L:?52 | L:L:H103 | 2.75 | 22 | Yes | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.82 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
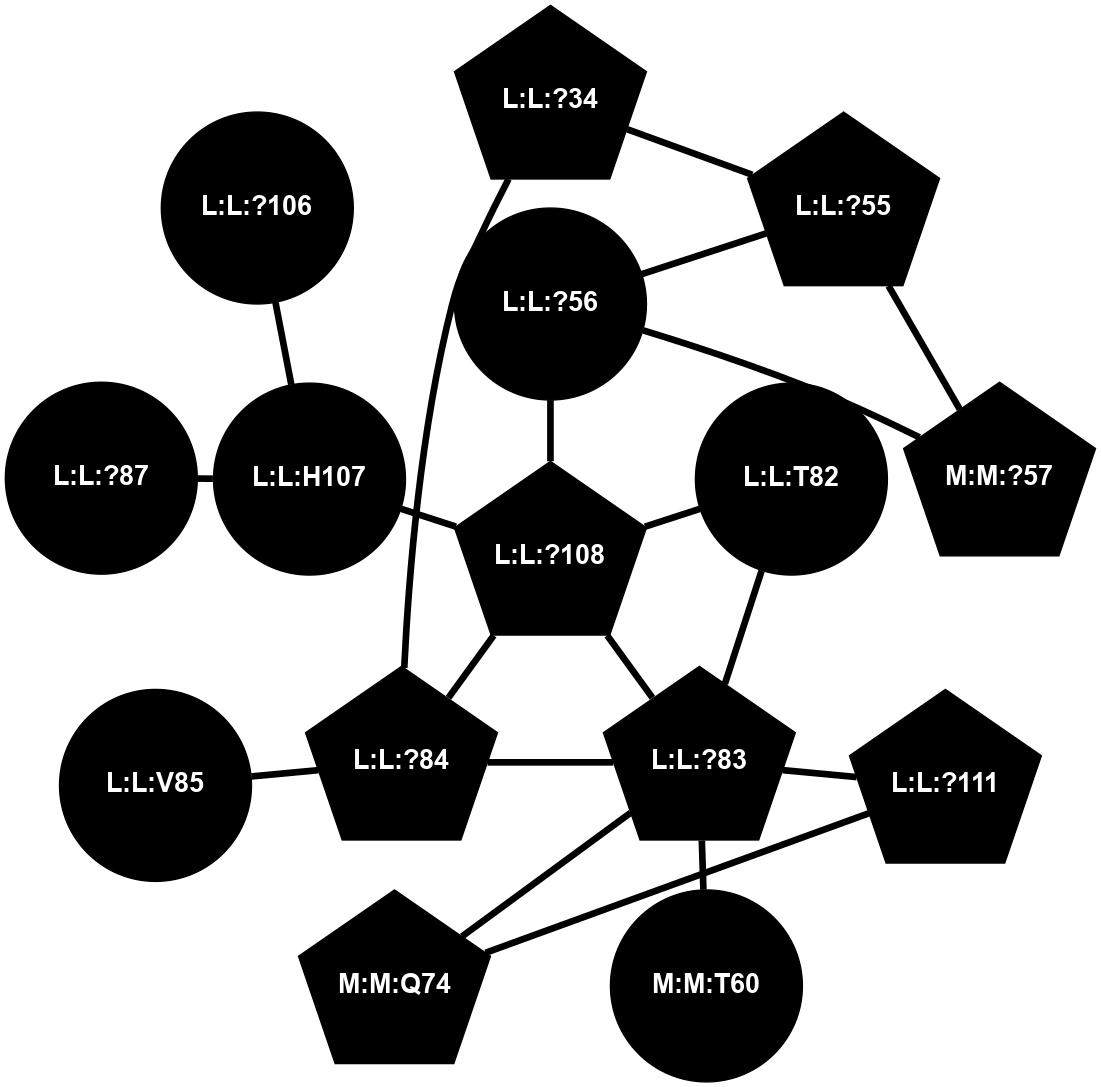
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?34 | L:L:?55 | 6.35 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?34 | L:L:?84 | 9.52 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?55 | L:L:?56 | 28.57 | 1 | Yes | No | 0 | 0 | 2 | 1 | | L:L:?55 | N:N:?57 | 8.65 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?108 | L:L:?56 | 9.52 | 1 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?56 | N:N:?57 | 34.6 | 1 | No | Yes | 0 | 0 | 1 | 2 | | L:L:?83 | L:L:T82 | 4.74 | 1 | Yes | No | 0 | 0 | 1 | 1 | | L:L:?108 | L:L:T82 | 7.89 | 1 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?83 | L:L:?84 | 22.22 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?108 | L:L:?83 | 9.52 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?111 | L:L:?83 | 15.87 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?83 | N:N:T60 | 4.74 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?83 | N:N:Q74 | 9.98 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?84 | L:L:V85 | 4.79 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:?84 | 12.7 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?87 | L:L:H107 | 4.6 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:?106 | L:L:H107 | 4.13 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:?108 | L:L:H107 | 15.14 | 1 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?111 | N:N:Q74 | 12.83 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 10.95 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 19.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
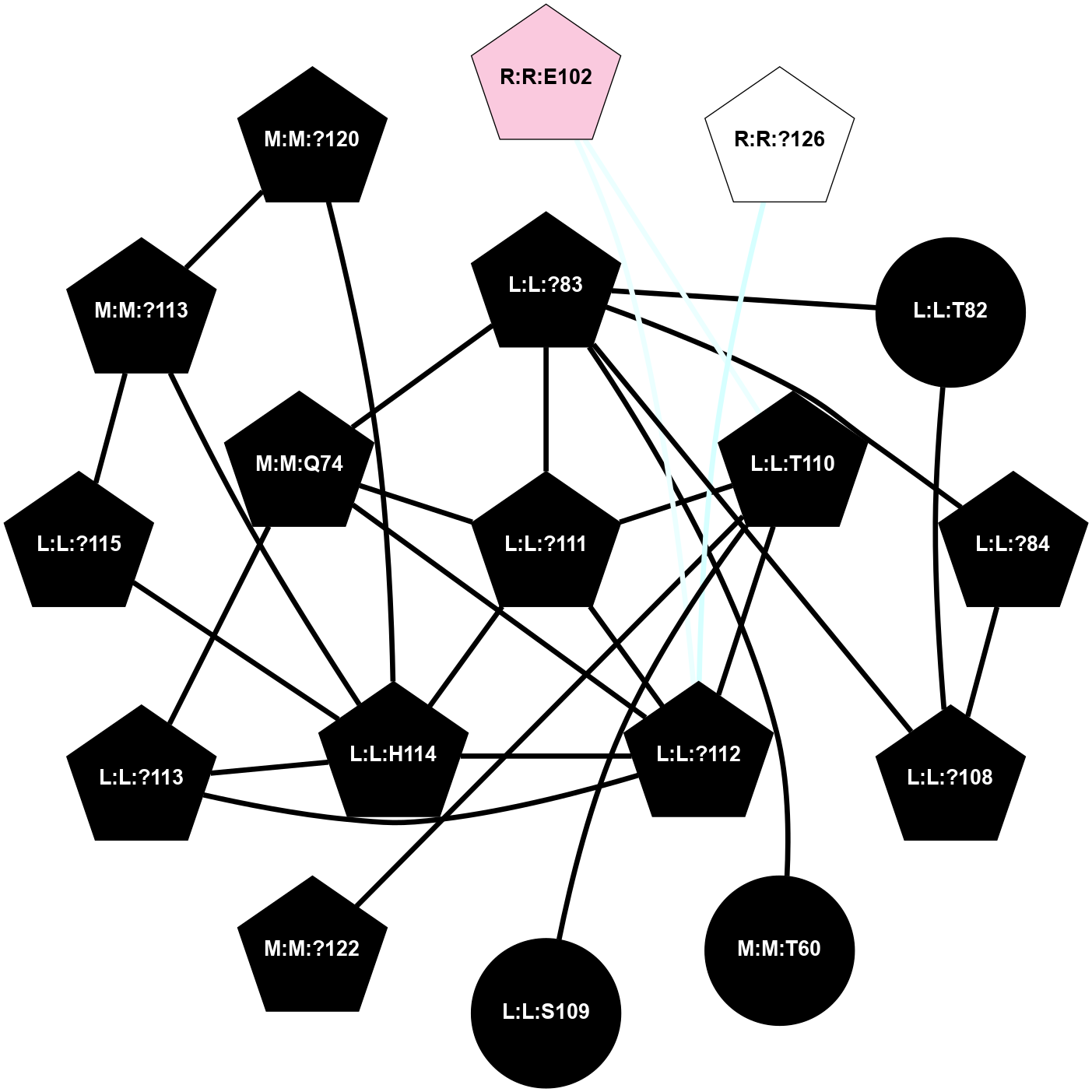
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:T110 | R:R:E102 | 7.06 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?112 | R:R:E102 | 9.02 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?112 | R:R:?126 | 4.82 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:?83 | L:L:T82 | 4.74 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:T82 | 7.89 | 1 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?83 | L:L:?84 | 22.22 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?108 | L:L:?83 | 9.52 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?111 | L:L:?83 | 15.87 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?83 | N:N:T60 | 4.74 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?83 | N:N:Q74 | 9.98 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?108 | L:L:?84 | 12.7 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:S109 | L:L:T110 | 4.8 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?111 | L:L:T110 | 7.89 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?112 | L:L:T110 | 7.17 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:T110 | N:N:?122 | 5.14 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?111 | L:L:?112 | 30.27 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?111 | L:L:H114 | 22.03 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?111 | N:N:Q74 | 12.83 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?112 | L:L:?113 | 24.88 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?112 | L:L:H114 | 12.5 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?112 | N:N:Q74 | 5.18 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?113 | L:L:H114 | 7.5 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?113 | N:N:Q74 | 19.42 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?115 | L:L:H114 | 12.27 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:H114 | N:N:?113 | 5.51 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:H114 | N:N:?120 | 12.39 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?115 | N:N:?113 | 7.96 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?113 | N:N:?120 | 11.11 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 17.78 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 15.00 | | Average Links In Shell | 28.00 | | Average Links Mediated by Hubs In Shell | 28.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:Q29 | N:N:R28 | 5.84 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?31 | L:L:?55 | 12.7 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?31 | N:N:R26 | 13.01 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?31 | N:N:?57 | 7.21 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?55 | L:L:?56 | 28.57 | 1 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?55 | N:N:?57 | 8.65 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?55 | N:N:?58 | 9.52 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?56 | N:N:?57 | 34.6 | 1 | No | Yes | 0 | 0 | 2 | 1 | | L:L:F57 | N:N:?56 | 5.31 | 1 | No | No | 0 | 0 | 2 | 1 | | L:L:F57 | N:N:?57 | 7.11 | 1 | No | Yes | 0 | 0 | 2 | 1 | | L:L:S58 | N:N:V76 | 6.46 | 1 | No | Yes | 0 | 0 | 2 | 2 | | L:L:S58 | N:N:?77 | 6.43 | 1 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:R26 | 7.09 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:P27 | N:N:R30 | 11.53 | 0 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?29 | N:N:R28 | 6.5 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?57 | N:N:R28 | 5.91 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | N:N:?58 | N:N:R28 | 5.2 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:P59 | N:N:R28 | 5.76 | 1 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?29 | N:N:R30 | 5.2 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?29 | N:N:?56 | 7.76 | 1 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?29 | N:N:?57 | 10.09 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?29 | N:N:?77 | 15.87 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?108 | N:N:?29 | 7.94 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:I32 | N:N:R30 | 12.53 | 0 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?54 | N:N:?77 | 6.35 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?54 | N:N:?79 | 4.32 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?108 | N:N:?54 | 9.52 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?56 | N:N:?57 | 23.48 | 1 | No | Yes | 0 | 0 | 1 | 1 | | N:N:?57 | N:N:?58 | 24.51 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?58 | N:N:P59 | 19.34 | 1 | Yes | No | 0 | 0 | 2 | 2 | | N:N:?110 | N:N:?58 | 9.52 | 0 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?77 | N:N:V76 | 12.76 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:N78 | N:N:V76 | 4.43 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?77 | N:N:N78 | 11.76 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?110 | N:N:?77 | 11.11 | 0 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?79 | N:N:N78 | 6.68 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?108 | N:N:?79 | 14.42 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?108 | N:N:R80 | 11.71 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:Q109 | N:N:R80 | 4.67 | 1 | Yes | No | 0 | 0 | 2 | 2 | | N:N:?108 | N:N:Q109 | 5.7 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?110 | N:N:Q109 | 17.1 | 0 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:P31 | N:N:R30 | 2.88 | 0 | No | Yes | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 8.89 | | Average Nodes In Shell | 26.00 | | Average Hubs In Shell | 16.00 | | Average Links In Shell | 42.00 | | Average Links Mediated by Hubs In Shell | 41.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | N:N:?40 | N:N:?43 | 7.07 | 0 | No | Yes | 0 | 0 | 1 | 0 | | N:N:?42 | N:N:?43 | 19.39 | 16 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:?42 | N:N:P44 | 4.3 | 16 | Yes | No | 0 | 0 | 1 | 1 | | N:N:?42 | N:N:P93 | 7.16 | 16 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?43 | N:N:P44 | 17.58 | 16 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?43 | N:N:L89 | 4.45 | 16 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?43 | N:N:?92 | 9.52 | 16 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:P44 | N:N:P93 | 3.9 | 16 | No | No | 0 | 0 | 1 | 2 | | N:N:I47 | N:N:L89 | 7.14 | 0 | No | No | 0 | 0 | 2 | 1 | | N:N:?92 | N:N:R88 | 3.9 | 16 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?92 | N:N:L89 | 4.45 | 16 | Yes | No | 0 | 0 | 1 | 1 | | N:N:?91 | N:N:?92 | 20.68 | 0 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?92 | N:N:P93 | 17.58 | 16 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?42 | N:N:E41 | 3.47 | 16 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?40 | N:N:V96 | 2.67 | 0 | No | No | 0 | 0 | 1 | 2 | | N:N:?40 | N:N:V100 | 1.78 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 11.60 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
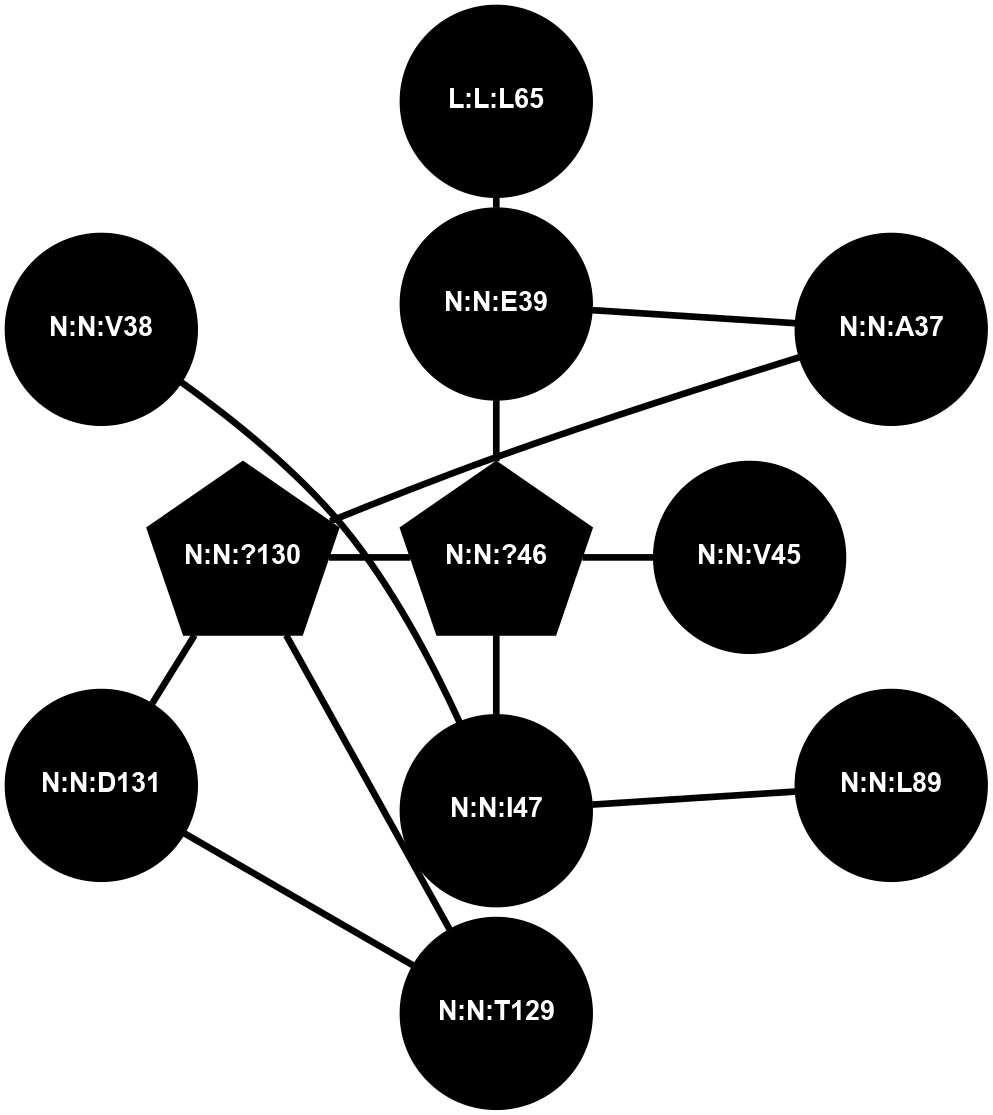
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:L65 | N:N:E39 | 7.95 | 0 | No | No | 0 | 0 | 2 | 1 | | N:N:A37 | N:N:E39 | 4.53 | 0 | No | No | 0 | 0 | 2 | 1 | | N:N:?130 | N:N:A37 | 6.75 | 34 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?46 | N:N:E39 | 8.51 | 0 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?46 | N:N:V45 | 9.57 | 0 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?46 | N:N:I47 | 10.7 | 0 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?130 | N:N:?46 | 9.52 | 34 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:I47 | N:N:L89 | 7.14 | 0 | No | No | 0 | 0 | 1 | 2 | | N:N:?130 | N:N:T129 | 9.47 | 34 | Yes | No | 0 | 0 | 1 | 2 | | N:N:D131 | N:N:T129 | 4.34 | 34 | No | No | 0 | 0 | 2 | 2 | | N:N:?130 | N:N:D131 | 18.89 | 34 | Yes | No | 0 | 0 | 1 | 2 | | N:N:I47 | N:N:V38 | 3.07 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 9.57 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
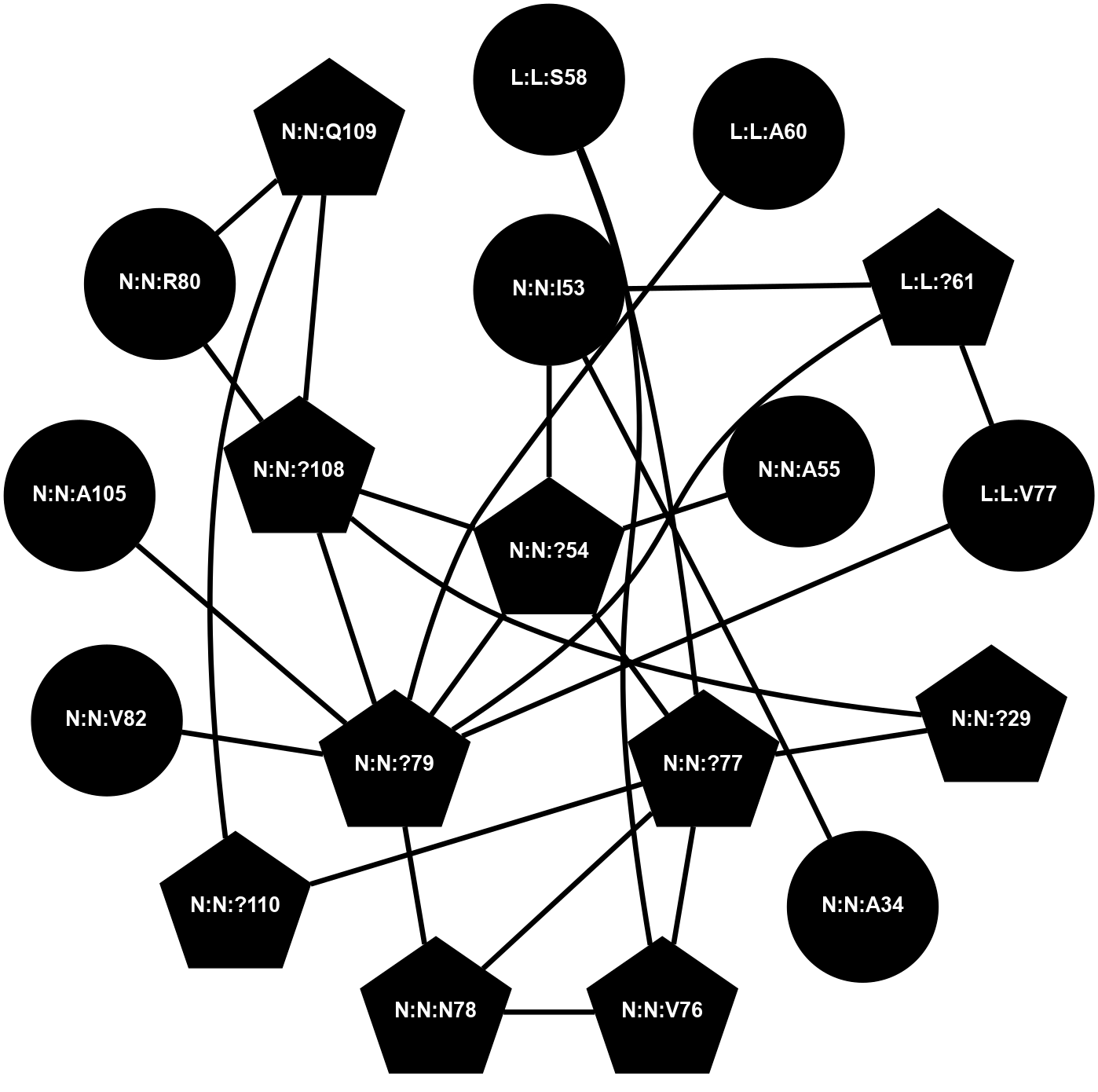
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:S58 | N:N:V76 | 6.46 | 1 | No | Yes | 0 | 0 | 2 | 2 | | L:L:S58 | N:N:?77 | 6.43 | 1 | No | Yes | 0 | 0 | 2 | 1 | | L:L:A60 | N:N:?79 | 4.6 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?61 | L:L:V77 | 7.24 | 1 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?61 | N:N:I53 | 4.16 | 1 | Yes | No | 0 | 0 | 2 | 1 | | L:L:?61 | N:N:?79 | 3.93 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:V77 | N:N:?79 | 7.24 | 1 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?29 | N:N:?77 | 15.87 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?108 | N:N:?29 | 7.94 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:A34 | N:N:I53 | 4.87 | 0 | No | No | 0 | 0 | 2 | 1 | | N:N:?54 | N:N:I53 | 9.17 | 1 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?54 | N:N:A55 | 5.06 | 1 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?54 | N:N:?77 | 6.35 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?54 | N:N:?79 | 4.32 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?108 | N:N:?54 | 9.52 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:?77 | N:N:V76 | 12.76 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:N78 | N:N:V76 | 4.43 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?77 | N:N:N78 | 11.76 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?110 | N:N:?77 | 11.11 | 0 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?79 | N:N:N78 | 6.68 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?79 | N:N:V82 | 5.8 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?79 | N:N:A105 | 4.6 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?108 | N:N:?79 | 14.42 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | N:N:?108 | N:N:R80 | 11.71 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:Q109 | N:N:R80 | 4.67 | 1 | Yes | No | 0 | 0 | 2 | 2 | | N:N:?108 | N:N:Q109 | 5.7 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?110 | N:N:Q109 | 17.1 | 0 | Yes | Yes | 0 | 0 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 6.88 | | Average Nodes In Shell | 19.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 27.00 | | Average Links Mediated by Hubs In Shell | 26.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:Q29 | N:N:R28 | 5.84 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?31 | L:L:?55 | 12.7 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?31 | N:N:R26 | 13.01 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?31 | N:N:?57 | 7.21 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?34 | L:L:?54 | 12.93 | 1 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?34 | L:L:?55 | 6.35 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?54 | L:L:?55 | 18.1 | 1 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?55 | L:L:?56 | 28.57 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?55 | N:N:?57 | 8.65 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?55 | N:N:?58 | 9.52 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?56 | N:N:?57 | 34.6 | 1 | No | Yes | 0 | 0 | 2 | 1 | | L:L:F57 | N:N:?56 | 5.31 | 1 | No | No | 0 | 0 | 2 | 2 | | L:L:F57 | N:N:?57 | 7.11 | 1 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?83 | N:N:T60 | 4.74 | 1 | Yes | No | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:R26 | 7.09 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?29 | N:N:R28 | 6.5 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:R28 | 5.91 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | N:N:?58 | N:N:R28 | 5.2 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:P59 | N:N:R28 | 5.76 | 1 | No | Yes | 0 | 0 | 1 | 1 | | N:N:?29 | N:N:?56 | 7.76 | 1 | Yes | No | 0 | 0 | 2 | 2 | | N:N:?29 | N:N:?57 | 10.09 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?29 | N:N:?77 | 15.87 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?56 | N:N:?57 | 23.48 | 1 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?57 | N:N:?58 | 24.51 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:?58 | N:N:P59 | 19.34 | 1 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?58 | N:N:T60 | 4.74 | 1 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?110 | N:N:?58 | 9.52 | 0 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:?110 | N:N:V75 | 19.14 | 0 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?110 | N:N:?77 | 11.11 | 0 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?110 | N:N:Q109 | 17.1 | 0 | Yes | Yes | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 12.14 | | Average Nodes In Shell | 20.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 30.00 | | Average Links Mediated by Hubs In Shell | 29.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
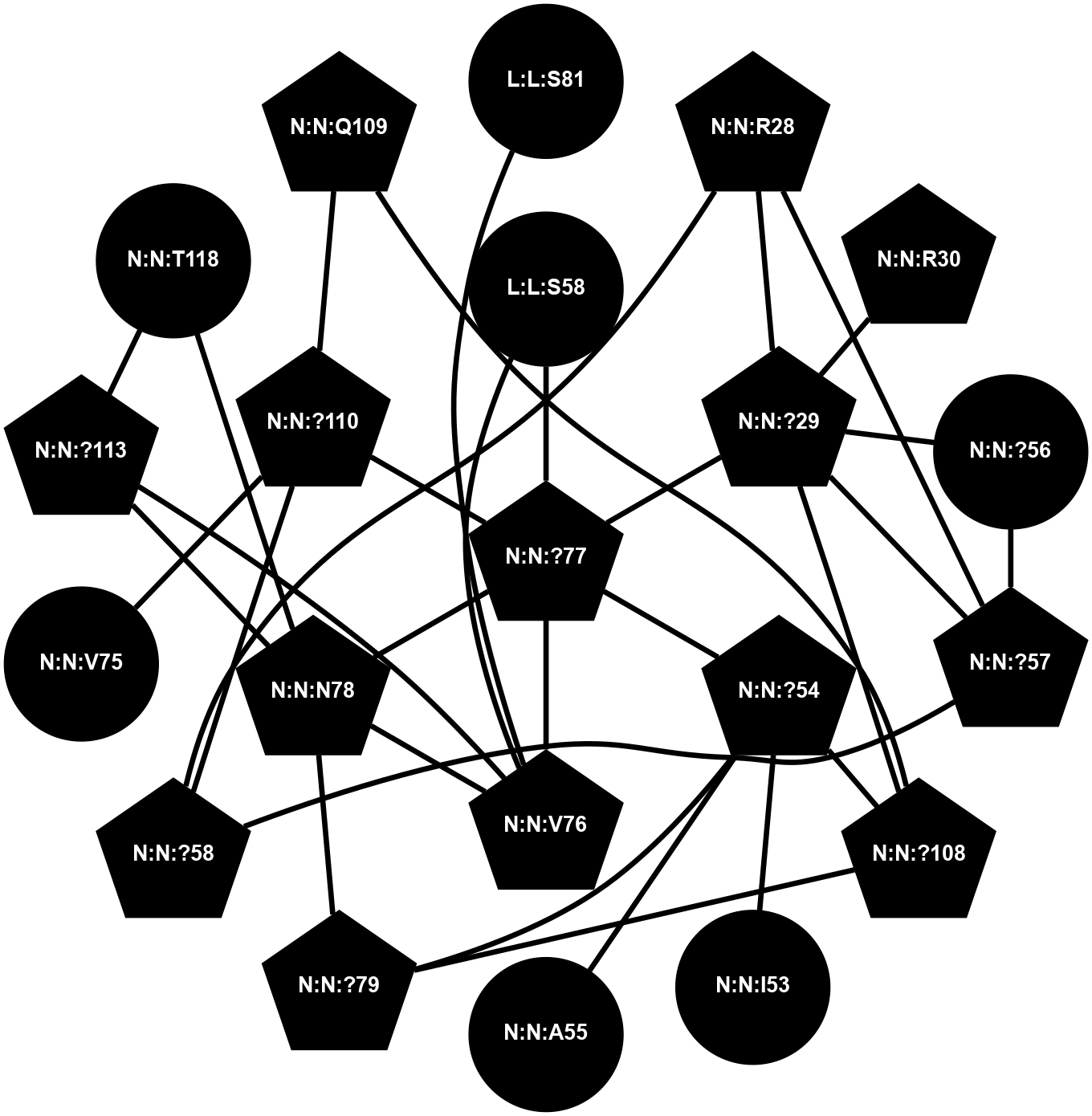
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:S58 | N:N:V76 | 6.46 | 1 | No | Yes | 0 | 0 | 1 | 1 | | L:L:S58 | N:N:?77 | 6.43 | 1 | No | Yes | 0 | 0 | 1 | 0 | | L:L:S81 | N:N:V76 | 8.08 | 0 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?29 | N:N:R28 | 6.5 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?57 | N:N:R28 | 5.91 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?58 | N:N:R28 | 5.2 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?29 | N:N:R30 | 5.2 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?29 | N:N:?56 | 7.76 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?29 | N:N:?57 | 10.09 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?29 | N:N:?77 | 15.87 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:?108 | N:N:?29 | 7.94 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?54 | N:N:I53 | 9.17 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?54 | N:N:A55 | 5.06 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?54 | N:N:?77 | 6.35 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:?54 | N:N:?79 | 4.32 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?108 | N:N:?54 | 9.52 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?56 | N:N:?57 | 23.48 | 1 | No | Yes | 0 | 0 | 2 | 2 | | N:N:?57 | N:N:?58 | 24.51 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?110 | N:N:?58 | 9.52 | 0 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?110 | N:N:V75 | 19.14 | 0 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?77 | N:N:V76 | 12.76 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:N78 | N:N:V76 | 4.43 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | N:N:?113 | N:N:V76 | 4.79 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?77 | N:N:N78 | 11.76 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?110 | N:N:?77 | 11.11 | 0 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:?79 | N:N:N78 | 6.68 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?113 | N:N:N78 | 4.41 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:N78 | N:N:T118 | 4.39 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?108 | N:N:?79 | 14.42 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?108 | N:N:Q109 | 5.7 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?110 | N:N:Q109 | 17.1 | 0 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?113 | N:N:T118 | 6.31 | 1 | Yes | No | 0 | 0 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 10.71 | | Average Nodes In Shell | 21.00 | | Average Hubs In Shell | 14.00 | | Average Links In Shell | 32.00 | | Average Links Mediated by Hubs In Shell | 32.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | N:N:?40 | N:N:?43 | 7.07 | 0 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?42 | N:N:?43 | 19.39 | 16 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?42 | N:N:P44 | 4.3 | 16 | Yes | No | 0 | 0 | 2 | 2 | | N:N:?42 | N:N:P93 | 7.16 | 16 | Yes | No | 0 | 0 | 2 | 1 | | N:N:?43 | N:N:P44 | 17.58 | 16 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?43 | N:N:L89 | 4.45 | 16 | Yes | No | 0 | 0 | 1 | 1 | | N:N:?43 | N:N:?92 | 9.52 | 16 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:P44 | N:N:P93 | 3.9 | 16 | No | No | 0 | 0 | 2 | 1 | | N:N:I47 | N:N:L89 | 7.14 | 0 | No | No | 0 | 0 | 2 | 1 | | N:N:?92 | N:N:R88 | 3.9 | 16 | Yes | No | 0 | 0 | 0 | 1 | | N:N:P98 | N:N:R88 | 7.21 | 0 | No | No | 0 | 0 | 2 | 1 | | N:N:?92 | N:N:L89 | 4.45 | 16 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?91 | N:N:P90 | 4.3 | 0 | No | No | 0 | 0 | 1 | 2 | | N:N:?91 | N:N:?92 | 20.68 | 0 | No | Yes | 0 | 0 | 1 | 0 | | N:N:?92 | N:N:P93 | 17.58 | 16 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 11.23 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:A60 | N:N:?79 | 4.6 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?61 | L:L:V77 | 7.24 | 1 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?61 | N:N:I53 | 4.16 | 1 | Yes | No | 0 | 0 | 2 | 2 | | L:L:?61 | N:N:?79 | 3.93 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:V77 | N:N:?79 | 7.24 | 1 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?29 | N:N:R28 | 6.5 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?57 | N:N:R28 | 5.91 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?29 | N:N:R30 | 5.2 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?29 | N:N:?56 | 7.76 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?29 | N:N:?57 | 10.09 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?29 | N:N:?77 | 15.87 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?108 | N:N:?29 | 7.94 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?54 | N:N:I53 | 9.17 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?54 | N:N:A55 | 5.06 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?54 | N:N:?77 | 6.35 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?54 | N:N:?79 | 4.32 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | N:N:?108 | N:N:?54 | 9.52 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?56 | N:N:?57 | 23.48 | 1 | No | Yes | 0 | 0 | 2 | 2 | | N:N:?77 | N:N:N78 | 11.76 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?110 | N:N:?77 | 11.11 | 0 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?79 | N:N:N78 | 6.68 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?79 | N:N:V82 | 5.8 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?79 | N:N:A105 | 4.6 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?108 | N:N:?79 | 14.42 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:R80 | N:N:S107 | 11.86 | 1 | No | No | 0 | 0 | 1 | 2 | | N:N:?108 | N:N:R80 | 11.71 | 1 | Yes | No | 0 | 0 | 0 | 1 | | N:N:Q109 | N:N:R80 | 4.67 | 1 | Yes | No | 0 | 0 | 1 | 1 | | N:N:?108 | N:N:Q109 | 5.7 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?110 | N:N:Q109 | 17.1 | 0 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:A111 | N:N:Q109 | 4.55 | 0 | No | Yes | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 9.86 | | Average Nodes In Shell | 22.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 30.00 | | Average Links Mediated by Hubs In Shell | 29.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
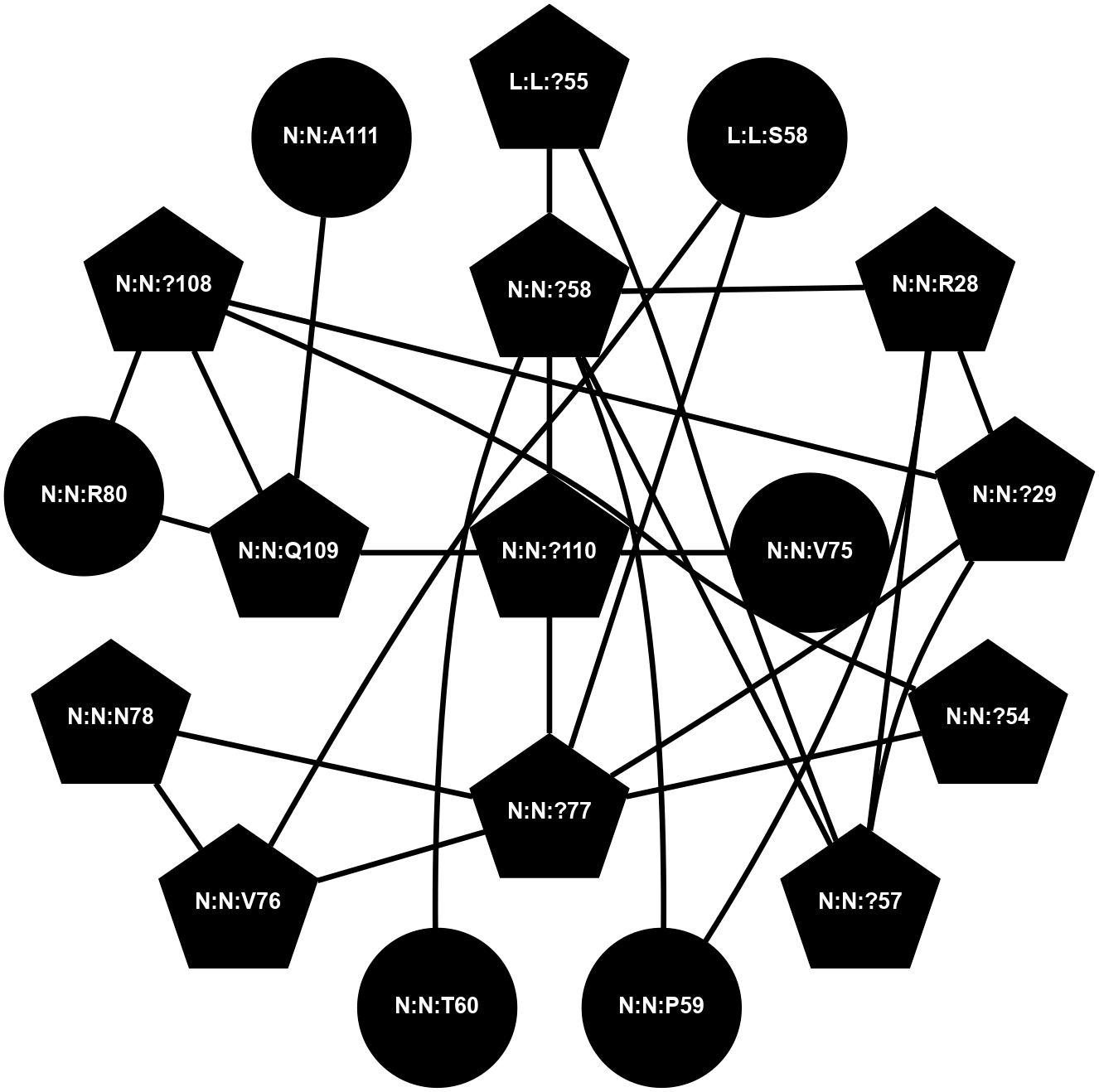
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?55 | N:N:?57 | 8.65 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?55 | N:N:?58 | 9.52 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:S58 | N:N:V76 | 6.46 | 1 | No | Yes | 0 | 0 | 2 | 2 | | L:L:S58 | N:N:?77 | 6.43 | 1 | No | Yes | 0 | 0 | 2 | 1 | | N:N:?29 | N:N:R28 | 6.5 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?57 | N:N:R28 | 5.91 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?58 | N:N:R28 | 5.2 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:P59 | N:N:R28 | 5.76 | 1 | No | Yes | 0 | 0 | 2 | 2 | | N:N:?29 | N:N:?57 | 10.09 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?29 | N:N:?77 | 15.87 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?108 | N:N:?29 | 7.94 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?54 | N:N:?77 | 6.35 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?108 | N:N:?54 | 9.52 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?57 | N:N:?58 | 24.51 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?58 | N:N:P59 | 19.34 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?58 | N:N:T60 | 4.74 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?110 | N:N:?58 | 9.52 | 0 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?110 | N:N:V75 | 19.14 | 0 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?77 | N:N:V76 | 12.76 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:N78 | N:N:V76 | 4.43 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?77 | N:N:N78 | 11.76 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?110 | N:N:?77 | 11.11 | 0 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?108 | N:N:R80 | 11.71 | 1 | Yes | No | 0 | 0 | 2 | 2 | | N:N:Q109 | N:N:R80 | 4.67 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?108 | N:N:Q109 | 5.7 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?110 | N:N:Q109 | 17.1 | 0 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:A111 | N:N:Q109 | 4.55 | 0 | No | Yes | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 14.22 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 27.00 | | Average Links Mediated by Hubs In Shell | 27.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
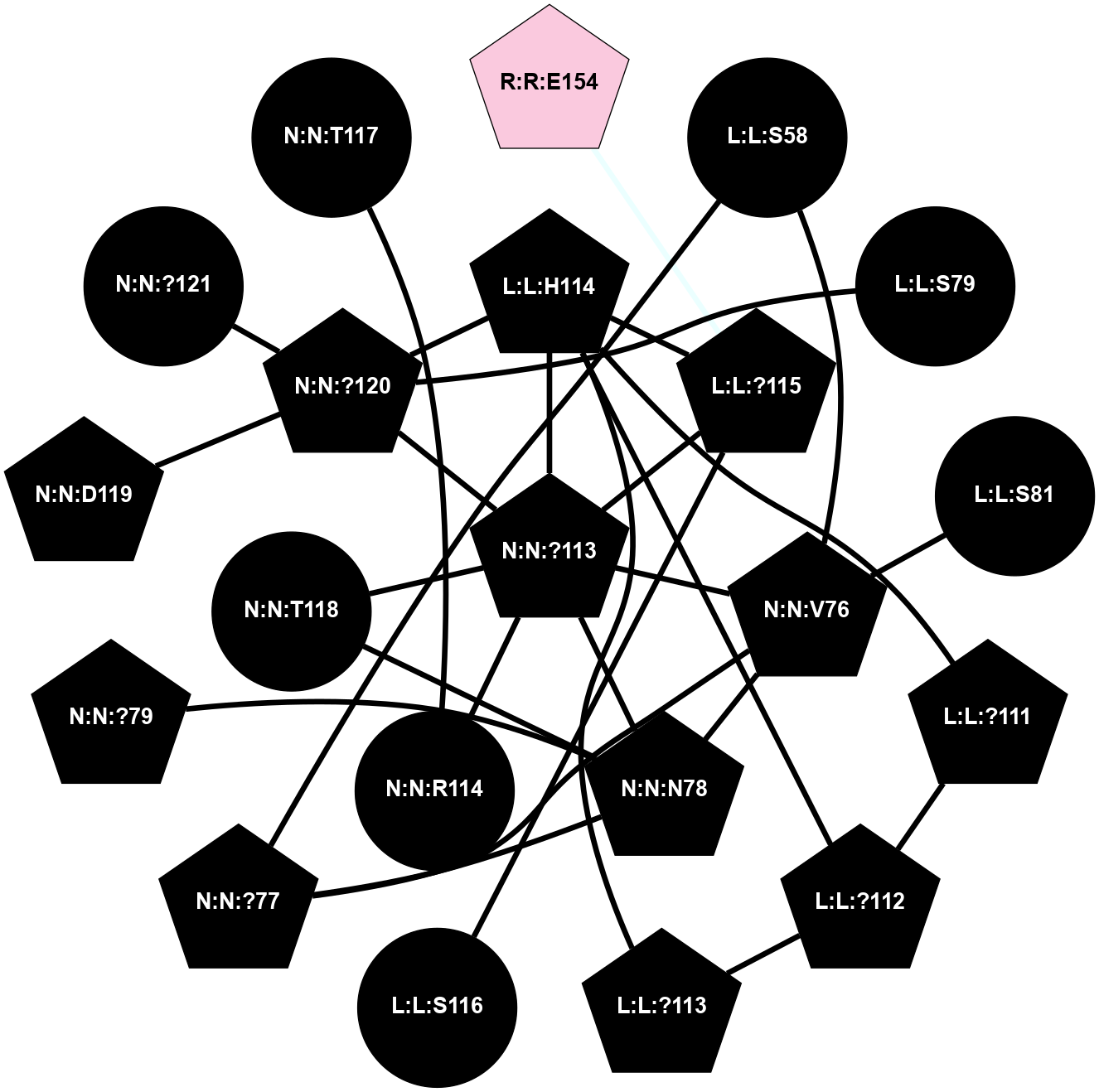
A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?115 | R:R:E154 | 6.32 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:S58 | N:N:V76 | 6.46 | 1 | No | Yes | 0 | 0 | 2 | 1 | | L:L:S58 | N:N:?77 | 6.43 | 1 | No | Yes | 0 | 0 | 2 | 2 | | L:L:S79 | N:N:?120 | 9.65 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:S81 | N:N:V76 | 8.08 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?111 | L:L:?112 | 30.27 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?111 | L:L:H114 | 22.03 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?112 | L:L:?113 | 24.88 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?112 | L:L:H114 | 12.5 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?113 | L:L:H114 | 7.5 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?115 | L:L:H114 | 12.27 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:H114 | N:N:?113 | 5.51 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:H114 | N:N:?120 | 12.39 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?115 | L:L:S116 | 5.37 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?115 | N:N:?113 | 7.96 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:?77 | N:N:V76 | 12.76 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:N78 | N:N:V76 | 4.43 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | N:N:?113 | N:N:V76 | 4.79 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?77 | N:N:N78 | 11.76 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?79 | N:N:N78 | 6.68 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:?113 | N:N:N78 | 4.41 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:N78 | N:N:T118 | 4.39 | 1 | Yes | No | 0 | 0 | 1 | 1 | | N:N:?113 | N:N:R114 | 3.9 | 1 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?113 | N:N:T118 | 6.31 | 1 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?113 | N:N:?120 | 11.11 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?120 | N:N:D119 | 5.81 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?120 | N:N:?121 | 15.51 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:R114 | N:N:T117 | 2.59 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 6.28 | | Average Nodes In Shell | 21.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 28.00 | | Average Links Mediated by Hubs In Shell | 27.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | N:N:D119 | R:R:Q106 | 6.53 | 0 | Yes | No | 0 | 7 | 1 | 2 | | N:N:?121 | R:R:Q106 | 4.64 | 0 | No | No | 0 | 7 | 1 | 2 | | L:L:M71 | N:N:D119 | 8.32 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:T78 | N:N:D119 | 5.78 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:S79 | N:N:?120 | 9.65 | 0 | No | Yes | 0 | 0 | 1 | 0 | | L:L:?111 | L:L:?112 | 30.27 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?111 | L:L:H114 | 22.03 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?112 | L:L:?113 | 24.88 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?112 | L:L:H114 | 12.5 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?113 | L:L:H114 | 7.5 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?115 | L:L:H114 | 12.27 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:H114 | N:N:?113 | 5.51 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:H114 | N:N:?120 | 12.39 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?115 | N:N:?113 | 7.96 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:N78 | N:N:V76 | 4.43 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | N:N:?113 | N:N:V76 | 4.79 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:?113 | N:N:N78 | 4.41 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:N78 | N:N:T118 | 4.39 | 1 | Yes | No | 0 | 0 | 2 | 2 | | N:N:?113 | N:N:R114 | 3.9 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?113 | N:N:T118 | 6.31 | 1 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?113 | N:N:?120 | 11.11 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | N:N:?120 | N:N:D119 | 5.81 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?120 | N:N:?121 | 15.51 | 1 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?121 | N:N:?122 | 12.63 | 0 | No | Yes | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 10.89 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 24.00 | | Average Links Mediated by Hubs In Shell | 23.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of CMS in 7FIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | N:N:A37 | N:N:E39 | 4.53 | 0 | No | No | 0 | 0 | 1 | 2 | | N:N:?130 | N:N:A37 | 6.75 | 34 | Yes | No | 0 | 0 | 0 | 1 | | N:N:?46 | N:N:E39 | 8.51 | 0 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?46 | N:N:V45 | 9.57 | 0 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?46 | N:N:I47 | 10.7 | 0 | Yes | No | 0 | 0 | 1 | 2 | | N:N:?130 | N:N:?46 | 9.52 | 34 | Yes | Yes | 0 | 0 | 0 | 1 | | N:N:?130 | N:N:T129 | 9.47 | 34 | Yes | No | 0 | 0 | 0 | 1 | | N:N:D131 | N:N:T129 | 4.34 | 34 | No | No | 0 | 0 | 1 | 1 | | N:N:?130 | N:N:D131 | 18.89 | 34 | Yes | No | 0 | 0 | 0 | 1 | | N:N:A37 | N:N:T48 | 1.68 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 11.16 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|





















































































































