| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8YJP | C | Orphan | Orphan | GPR156 | Homo Sapiens | - | - | - | 3.09 | 2025-02-05 | doi.org/10.1038/s41467-024-54681-5 |
|
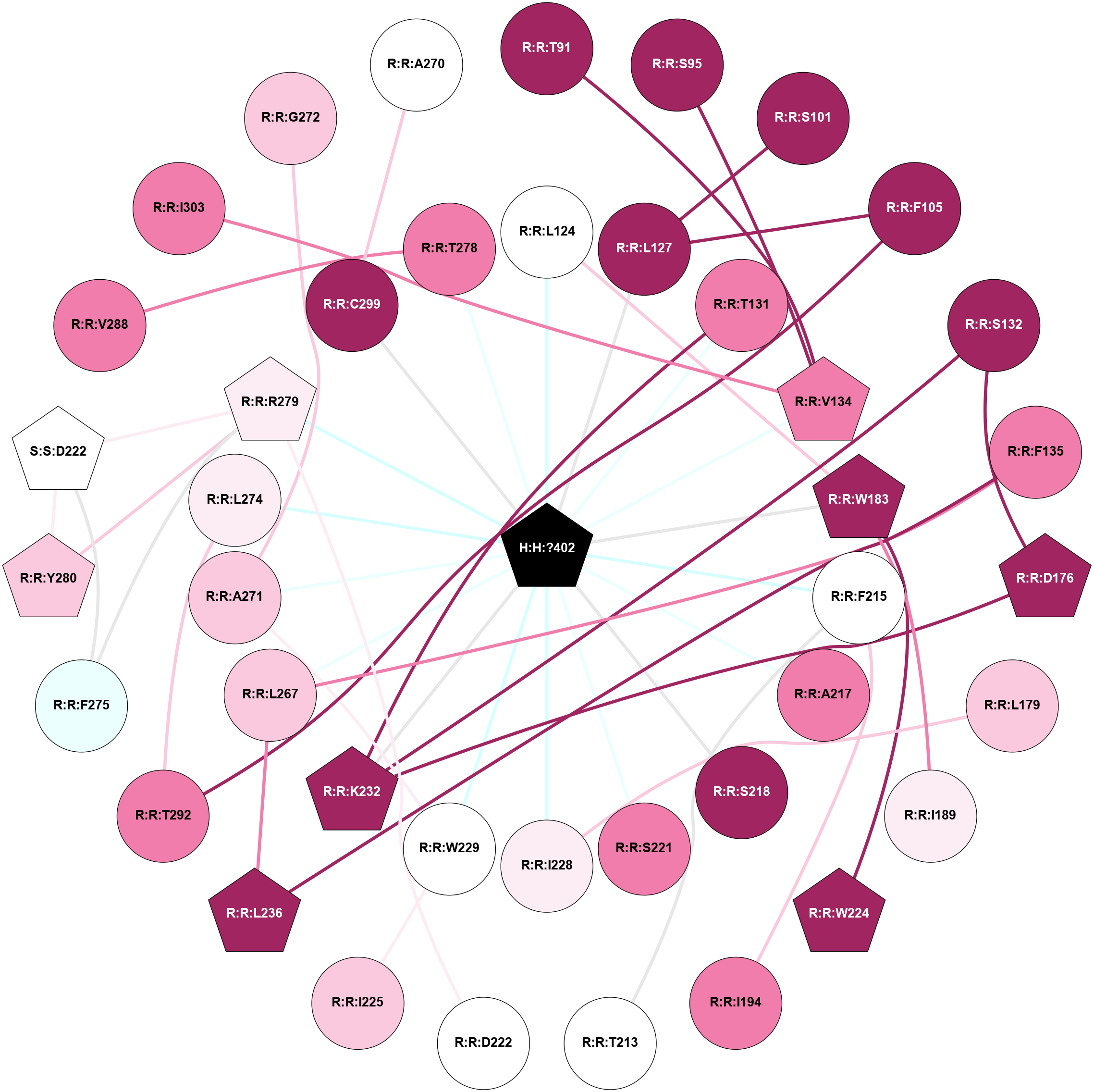
A 2D representation of the interactions of MW9 in 8YJP
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?402 | R:R:L124 | 7.74 | 1 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?402 | R:R:L127 | 3.22 | 1 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?402 | R:R:T131 | 8.93 | 1 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?402 | R:R:V134 | 3.47 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?402 | R:R:W183 | 5.84 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?402 | R:R:F215 | 3.97 | 1 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?402 | R:R:A217 | 7.34 | 1 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?402 | R:R:S218 | 4.2 | 1 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?402 | R:R:S221 | 4.9 | 1 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?402 | R:R:I228 | 3.99 | 1 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?402 | R:R:W229 | 4.24 | 1 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?402 | R:R:K232 | 3.94 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?402 | R:R:L267 | 7.09 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?402 | R:R:A271 | 3.67 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?402 | R:R:L274 | 3.87 | 1 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?402 | R:R:R279 | 7.92 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?402 | R:R:C299 | 3.7 | 1 | Yes | No | 0 | 9 | 0 | 1 | | R:R:T91 | R:R:V134 | 4.76 | 0 | No | Yes | 9 | 8 | 2 | 1 | | R:R:S95 | R:R:V134 | 3.23 | 2 | No | Yes | 9 | 8 | 2 | 1 | | R:R:L127 | R:R:S101 | 9.01 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:F105 | R:R:L127 | 4.87 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:F105 | R:R:T292 | 7.78 | 0 | No | No | 9 | 8 | 2 | 2 | | R:R:L124 | R:R:W183 | 5.69 | 1 | No | Yes | 5 | 9 | 1 | 1 | | R:R:K232 | R:R:T131 | 4.5 | 1 | Yes | No | 9 | 8 | 1 | 1 | | R:R:D176 | R:R:S132 | 8.83 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:K232 | R:R:S132 | 4.59 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:F135 | R:R:L236 | 3.65 | 1 | No | Yes | 8 | 9 | 2 | 2 | | R:R:F135 | R:R:L267 | 4.87 | 1 | No | No | 8 | 7 | 2 | 1 | | R:R:D176 | R:R:K232 | 6.91 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:I228 | R:R:L179 | 4.28 | 0 | No | No | 6 | 7 | 1 | 2 | | R:R:I189 | R:R:W183 | 5.87 | 0 | No | Yes | 6 | 9 | 2 | 1 | | R:R:W183 | R:R:W224 | 20.62 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:F215 | R:R:I194 | 3.77 | 0 | No | No | 5 | 8 | 1 | 2 | | R:R:F215 | R:R:T213 | 5.19 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:D222 | R:R:R279 | 10.72 | 1 | No | Yes | 5 | 6 | 2 | 1 | | R:R:I225 | R:R:W229 | 9.4 | 0 | No | No | 7 | 5 | 2 | 1 | | R:R:A271 | R:R:W229 | 5.19 | 1 | No | No | 7 | 5 | 1 | 1 | | R:R:L236 | R:R:L267 | 4.15 | 1 | Yes | No | 9 | 7 | 2 | 1 | | R:R:L274 | R:R:T292 | 5.9 | 0 | No | No | 6 | 8 | 1 | 2 | | R:R:F275 | R:R:R279 | 9.62 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F275 | S:S:D222 | 3.58 | 1 | No | Yes | 4 | 5 | 2 | 2 | | R:R:R279 | R:R:Y280 | 4.12 | 1 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:R279 | S:S:D222 | 13.1 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:Y280 | S:S:D222 | 11.49 | 1 | Yes | Yes | 7 | 5 | 2 | 2 | | R:R:T278 | R:R:V288 | 3.17 | 0 | No | No | 8 | 8 | 1 | 2 | | R:R:I303 | R:R:V134 | 3.07 | 0 | No | Yes | 8 | 8 | 2 | 1 | | H:H:?402 | R:R:T278 | 2.75 | 1 | Yes | No | 0 | 8 | 0 | 1 | | R:R:A271 | R:R:G272 | 1.95 | 1 | No | No | 7 | 7 | 1 | 2 | | R:R:A270 | R:R:C299 | 1.81 | 0 | No | No | 5 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 18.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 5.04 | | Average Nodes In Shell | 42.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 49.00 | | Average Links Mediated by Hubs In Shell | 36.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
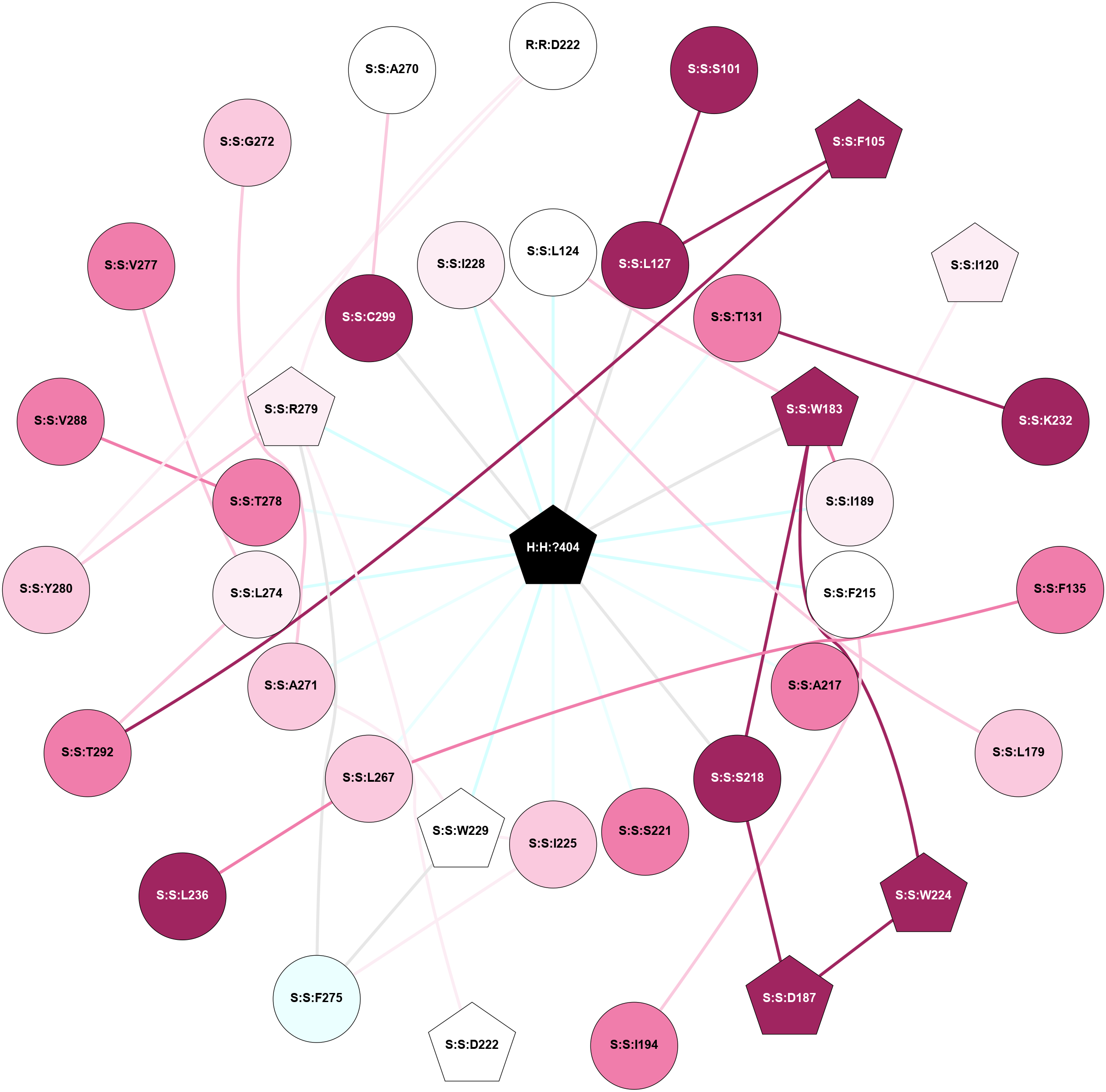
A 2D representation of the interactions of MW9 in 8YJP
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?404 | S:S:L124 | 7.74 | 1 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?404 | S:S:L127 | 3.87 | 1 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?404 | S:S:T131 | 5.49 | 1 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?404 | S:S:W183 | 8.49 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?404 | S:S:I189 | 3.32 | 1 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?404 | S:S:F215 | 6.24 | 1 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?404 | S:S:A217 | 3.67 | 1 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?404 | S:S:S218 | 3.5 | 1 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?404 | S:S:S221 | 4.9 | 1 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?404 | S:S:I225 | 4.65 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?404 | S:S:W229 | 14.32 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?404 | S:S:L267 | 7.09 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?404 | S:S:A271 | 3.67 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?404 | S:S:L274 | 4.51 | 1 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?404 | S:S:T278 | 4.12 | 1 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?404 | S:S:R279 | 6.79 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?404 | S:S:C299 | 3.7 | 1 | Yes | No | 0 | 9 | 0 | 1 | | R:R:D222 | S:S:R279 | 15.48 | 1 | No | Yes | 5 | 6 | 2 | 1 | | R:R:D222 | S:S:Y280 | 11.49 | 1 | No | No | 5 | 7 | 2 | 2 | | S:S:L127 | S:S:S101 | 6.01 | 0 | No | No | 9 | 9 | 1 | 2 | | S:S:F105 | S:S:L127 | 8.53 | 0 | Yes | No | 9 | 9 | 2 | 1 | | S:S:F105 | S:S:T292 | 9.08 | 0 | Yes | No | 9 | 8 | 2 | 2 | | S:S:I120 | S:S:I189 | 4.42 | 0 | Yes | No | 6 | 6 | 2 | 1 | | S:S:L124 | S:S:W183 | 5.69 | 1 | No | Yes | 5 | 9 | 1 | 1 | | S:S:K232 | S:S:T131 | 4.5 | 15 | No | No | 9 | 8 | 2 | 1 | | S:S:F135 | S:S:L267 | 4.87 | 0 | No | No | 8 | 7 | 2 | 1 | | S:S:I228 | S:S:L179 | 4.28 | 0 | No | No | 6 | 7 | 1 | 2 | | S:S:I189 | S:S:W183 | 7.05 | 1 | No | Yes | 6 | 9 | 1 | 1 | | S:S:S218 | S:S:W183 | 4.94 | 1 | No | Yes | 9 | 9 | 1 | 1 | | S:S:W183 | S:S:W224 | 18.74 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | S:S:D187 | S:S:S218 | 5.89 | 1 | Yes | No | 9 | 9 | 2 | 1 | | S:S:D187 | S:S:W224 | 7.82 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | S:S:F215 | S:S:I194 | 3.77 | 0 | No | No | 5 | 8 | 1 | 2 | | S:S:D222 | S:S:R279 | 10.72 | 1 | Yes | Yes | 5 | 6 | 2 | 1 | | S:S:I225 | S:S:W229 | 7.05 | 1 | No | Yes | 7 | 5 | 1 | 1 | | S:S:F275 | S:S:I225 | 6.28 | 1 | No | No | 4 | 7 | 2 | 1 | | S:S:A271 | S:S:W229 | 3.89 | 1 | No | Yes | 7 | 5 | 1 | 1 | | S:S:F275 | S:S:W229 | 4.01 | 1 | No | Yes | 4 | 5 | 2 | 1 | | S:S:L236 | S:S:L267 | 5.54 | 0 | No | No | 9 | 7 | 2 | 1 | | S:S:L274 | S:S:T292 | 5.9 | 0 | No | No | 6 | 8 | 1 | 2 | | S:S:F275 | S:S:R279 | 11.76 | 1 | No | Yes | 4 | 6 | 2 | 1 | | S:S:R279 | S:S:Y280 | 5.14 | 1 | Yes | No | 6 | 7 | 1 | 2 | | S:S:T278 | S:S:V288 | 3.17 | 0 | No | No | 8 | 8 | 1 | 2 | | S:S:L274 | S:S:V277 | 2.98 | 0 | No | No | 6 | 8 | 1 | 2 | | H:H:?404 | S:S:I228 | 2.66 | 1 | Yes | No | 0 | 6 | 0 | 1 | | S:S:A271 | S:S:G272 | 1.95 | 1 | No | No | 7 | 7 | 1 | 2 | | S:S:A270 | S:S:C299 | 1.81 | 0 | No | No | 5 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 18.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 5.49 | | Average Nodes In Shell | 38.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 47.00 | | Average Links Mediated by Hubs In Shell | 34.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8YK0 | C | Orphan | Orphan | GPR156 | Homo Sapiens | - | - | Gi3/Beta1/Gamma2 | 2.4 | 2025-02-05 | doi.org/10.1038/s41467-024-54681-5 |
|
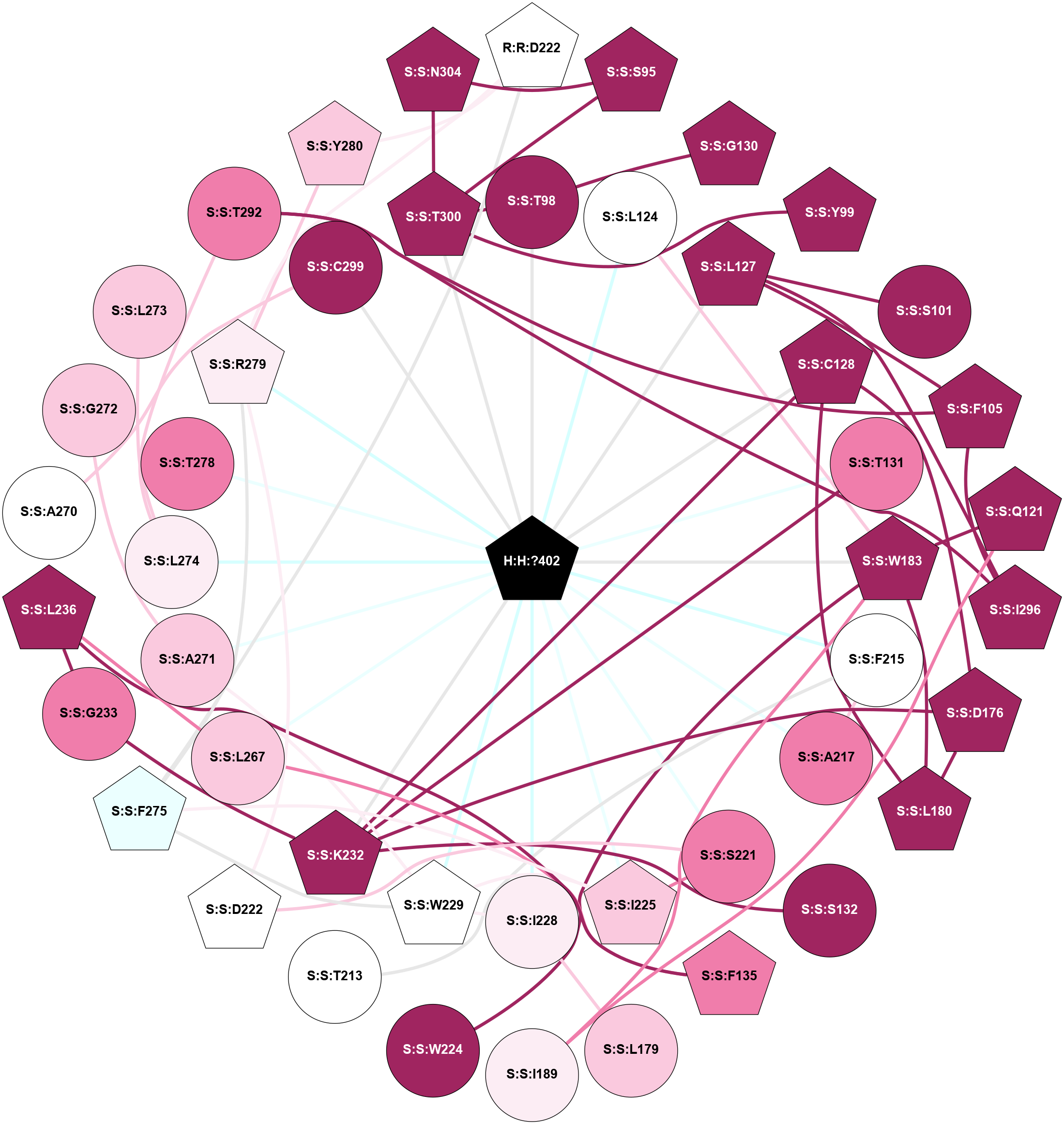
A 2D representation of the interactions of MW9 in 8YK0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?402 | S:S:T98 | 2.06 | 2 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?402 | S:S:L124 | 8.38 | 2 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?402 | S:S:L127 | 5.16 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?402 | S:S:C128 | 2.96 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?402 | S:S:T131 | 6.18 | 2 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?402 | S:S:W183 | 11.14 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?402 | S:S:F215 | 3.97 | 2 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?402 | S:S:A217 | 4.4 | 2 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?402 | S:S:S221 | 7.69 | 2 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?402 | S:S:I225 | 3.32 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?402 | S:S:I228 | 5.32 | 2 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?402 | S:S:W229 | 11.67 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?402 | S:S:K232 | 3.94 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?402 | S:S:L267 | 5.16 | 2 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?402 | S:S:A271 | 2.2 | 2 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?402 | S:S:L274 | 5.8 | 2 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?402 | S:S:T278 | 3.43 | 2 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?402 | S:S:R279 | 7.92 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?402 | S:S:C299 | 4.44 | 2 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?402 | S:S:T300 | 2.75 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:D222 | S:S:F275 | 4.78 | 2 | Yes | Yes | 5 | 4 | 2 | 2 | | R:R:D222 | S:S:R279 | 10.72 | 2 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:D222 | S:S:Y280 | 5.75 | 2 | Yes | Yes | 5 | 7 | 2 | 2 | | S:S:S95 | S:S:T300 | 3.2 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | | S:S:N304 | S:S:S95 | 10.43 | 2 | Yes | Yes | 9 | 9 | 2 | 2 | | S:S:G130 | S:S:T98 | 1.82 | 2 | Yes | No | 9 | 9 | 2 | 1 | | S:S:T300 | S:S:T98 | 7.85 | 2 | Yes | No | 9 | 9 | 1 | 1 | | S:S:T300 | S:S:Y99 | 12.48 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | S:S:L127 | S:S:S101 | 6.01 | 2 | Yes | No | 9 | 9 | 1 | 2 | | S:S:F105 | S:S:L127 | 2.44 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | | S:S:F105 | S:S:T292 | 9.08 | 2 | Yes | No | 9 | 8 | 2 | 2 | | S:S:F105 | S:S:I296 | 2.51 | 2 | Yes | Yes | 9 | 9 | 2 | 2 | | S:S:Q121 | S:S:W183 | 5.48 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | | S:S:I189 | S:S:Q121 | 6.86 | 2 | No | Yes | 6 | 9 | 2 | 2 | | S:S:L124 | S:S:W183 | 5.69 | 2 | No | Yes | 5 | 9 | 1 | 1 | | S:S:I296 | S:S:L127 | 2.85 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | | S:S:C128 | S:S:D176 | 10.89 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | S:S:C128 | S:S:L180 | 4.76 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | S:S:C128 | S:S:K232 | 3.23 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | S:S:K232 | S:S:T131 | 6.01 | 2 | Yes | No | 9 | 8 | 1 | 1 | | S:S:K232 | S:S:S132 | 3.06 | 2 | Yes | No | 9 | 9 | 1 | 2 | | S:S:F135 | S:S:L236 | 4.87 | 2 | Yes | Yes | 8 | 9 | 2 | 2 | | S:S:F135 | S:S:L267 | 3.65 | 2 | Yes | No | 8 | 7 | 2 | 1 | | S:S:D176 | S:S:L180 | 8.14 | 2 | Yes | Yes | 9 | 9 | 2 | 2 | | S:S:D176 | S:S:K232 | 2.77 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | | S:S:I228 | S:S:L179 | 2.85 | 2 | No | No | 6 | 7 | 1 | 2 | | S:S:L180 | S:S:W183 | 2.28 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | | S:S:I189 | S:S:W183 | 7.05 | 2 | No | Yes | 6 | 9 | 2 | 1 | | S:S:W183 | S:S:W224 | 16.87 | 2 | Yes | No | 9 | 9 | 1 | 2 | | S:S:F215 | S:S:T213 | 6.49 | 2 | No | No | 5 | 5 | 1 | 2 | | S:S:A217 | S:S:F215 | 2.77 | 2 | No | No | 8 | 5 | 1 | 1 | | S:S:D222 | S:S:S221 | 2.94 | 2 | Yes | No | 5 | 8 | 2 | 1 | | S:S:I225 | S:S:S221 | 3.1 | 2 | Yes | No | 7 | 8 | 1 | 1 | | S:S:D222 | S:S:R279 | 8.34 | 2 | Yes | Yes | 5 | 6 | 2 | 1 | | S:S:I225 | S:S:W229 | 9.4 | 2 | Yes | Yes | 7 | 5 | 1 | 1 | | S:S:F275 | S:S:I225 | 2.51 | 2 | Yes | Yes | 4 | 7 | 2 | 1 | | S:S:I228 | S:S:W229 | 3.52 | 2 | No | Yes | 6 | 5 | 1 | 1 | | S:S:A271 | S:S:W229 | 3.89 | 2 | No | Yes | 7 | 5 | 1 | 1 | | S:S:F275 | S:S:W229 | 4.01 | 2 | Yes | Yes | 4 | 5 | 2 | 1 | | S:S:G233 | S:S:K232 | 1.74 | 0 | No | Yes | 8 | 9 | 2 | 1 | | S:S:G233 | S:S:L236 | 3.42 | 0 | No | Yes | 8 | 9 | 2 | 2 | | S:S:L236 | S:S:L267 | 6.92 | 2 | Yes | No | 9 | 7 | 2 | 1 | | S:S:A270 | S:S:C299 | 1.81 | 0 | No | No | 5 | 9 | 2 | 1 | | S:S:A271 | S:S:G272 | 1.95 | 2 | No | No | 7 | 7 | 1 | 2 | | S:S:L273 | S:S:L274 | 4.15 | 0 | No | No | 7 | 6 | 2 | 1 | | S:S:L274 | S:S:T292 | 5.9 | 0 | No | No | 6 | 8 | 1 | 2 | | S:S:F275 | S:S:R279 | 11.76 | 2 | Yes | Yes | 4 | 6 | 2 | 1 | | S:S:R279 | S:S:Y280 | 8.23 | 2 | Yes | Yes | 6 | 7 | 1 | 2 | | S:S:I296 | S:S:T292 | 3.04 | 2 | Yes | No | 9 | 8 | 2 | 2 | | S:S:N304 | S:S:T300 | 2.92 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 20.00 | | Average Number Of Links With An Hub | 8.00 | | Average Interaction Strength | 5.39 | | Average Nodes In Shell | 47.00 | | Average Hubs In Shell | 24.00 | | Average Links In Shell | 70.00 | | Average Links Mediated by Hubs In Shell | 63.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
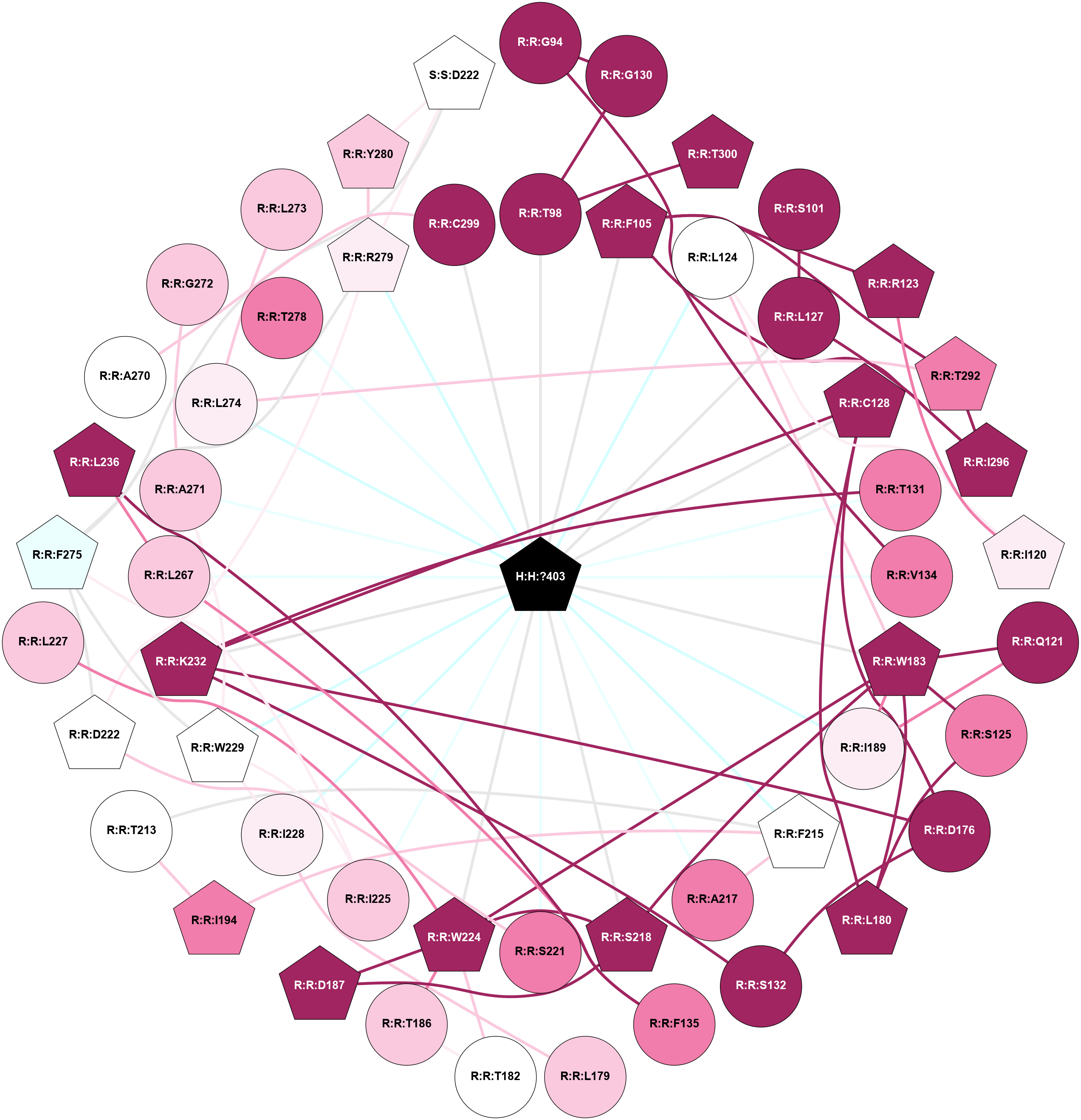
A 2D representation of the interactions of MW9 in 8YK0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?403 | R:R:T98 | 2.06 | 2 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?403 | R:R:F105 | 2.27 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?403 | R:R:L124 | 7.09 | 2 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?403 | R:R:L127 | 9.67 | 2 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?403 | R:R:C128 | 3.7 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?403 | R:R:T131 | 7.55 | 2 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?403 | R:R:V134 | 3.47 | 2 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?403 | R:R:W183 | 5.31 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?403 | R:R:I189 | 2.66 | 2 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?403 | R:R:F215 | 3.4 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?403 | R:R:A217 | 8.07 | 2 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?403 | R:R:S218 | 3.5 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?403 | R:R:S221 | 7.69 | 2 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?403 | R:R:W224 | 2.12 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?403 | R:R:I225 | 2.66 | 2 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?403 | R:R:I228 | 3.99 | 2 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?403 | R:R:W229 | 9.02 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?403 | R:R:K232 | 3.94 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?403 | R:R:L267 | 6.45 | 2 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?403 | R:R:A271 | 2.2 | 2 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?403 | R:R:L274 | 5.8 | 2 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?403 | R:R:T278 | 4.12 | 2 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?403 | R:R:R279 | 7.36 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?403 | R:R:C299 | 2.96 | 2 | Yes | No | 0 | 9 | 0 | 1 | | R:R:G130 | R:R:G94 | 2.11 | 0 | No | No | 9 | 9 | 2 | 2 | | R:R:G94 | R:R:V134 | 1.84 | 0 | No | No | 9 | 8 | 2 | 1 | | R:R:G130 | R:R:T98 | 1.82 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:T300 | R:R:T98 | 4.71 | 3 | Yes | No | 9 | 9 | 2 | 1 | | R:R:L127 | R:R:S101 | 6.01 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:F105 | R:R:R123 | 21.38 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:F105 | R:R:T292 | 10.38 | 2 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F105 | R:R:I296 | 2.51 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:I120 | R:R:R123 | 7.52 | 0 | Yes | Yes | 6 | 9 | 2 | 2 | | R:R:I120 | R:R:L124 | 2.85 | 0 | Yes | No | 6 | 5 | 2 | 1 | | R:R:Q121 | R:R:W183 | 3.29 | 2 | No | Yes | 9 | 9 | 2 | 1 | | R:R:I189 | R:R:Q121 | 5.49 | 2 | No | No | 6 | 9 | 1 | 2 | | R:R:L124 | R:R:W183 | 4.56 | 2 | No | Yes | 5 | 9 | 1 | 1 | | R:R:L180 | R:R:S125 | 10.51 | 2 | Yes | No | 9 | 8 | 2 | 2 | | R:R:S125 | R:R:W183 | 2.47 | 2 | No | Yes | 8 | 9 | 2 | 1 | | R:R:I296 | R:R:L127 | 5.71 | 2 | Yes | No | 9 | 9 | 2 | 1 | | R:R:C128 | R:R:D176 | 4.67 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:C128 | R:R:L180 | 6.35 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:C128 | R:R:K232 | 4.85 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:K232 | R:R:T131 | 6.01 | 2 | Yes | No | 9 | 8 | 1 | 1 | | R:R:D176 | R:R:S132 | 8.83 | 2 | No | No | 9 | 9 | 2 | 2 | | R:R:K232 | R:R:S132 | 4.59 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:F135 | R:R:L236 | 6.09 | 2 | No | Yes | 8 | 9 | 2 | 2 | | R:R:F135 | R:R:L267 | 6.09 | 2 | No | No | 8 | 7 | 2 | 1 | | R:R:D176 | R:R:K232 | 6.91 | 2 | No | Yes | 9 | 9 | 2 | 1 | | R:R:I228 | R:R:L179 | 4.28 | 2 | No | No | 6 | 7 | 1 | 2 | | R:R:L180 | R:R:W183 | 2.28 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:T182 | R:R:T186 | 6.28 | 2 | No | No | 5 | 7 | 2 | 2 | | R:R:T182 | R:R:W224 | 2.43 | 2 | No | Yes | 5 | 9 | 2 | 1 | | R:R:I189 | R:R:W183 | 7.05 | 2 | No | Yes | 6 | 9 | 1 | 1 | | R:R:S218 | R:R:W183 | 2.47 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:W183 | R:R:W224 | 19.68 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:T186 | R:R:W224 | 2.43 | 2 | No | Yes | 7 | 9 | 2 | 1 | | R:R:D187 | R:R:S218 | 8.83 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:D187 | R:R:W224 | 13.4 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:I194 | R:R:T213 | 3.04 | 2 | Yes | No | 8 | 5 | 2 | 2 | | R:R:F215 | R:R:I194 | 3.77 | 2 | Yes | Yes | 5 | 8 | 1 | 2 | | R:R:F215 | R:R:T213 | 7.78 | 2 | Yes | No | 5 | 5 | 1 | 2 | | R:R:A217 | R:R:F215 | 2.77 | 2 | No | Yes | 8 | 5 | 1 | 1 | | R:R:S218 | R:R:W224 | 2.47 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:D222 | R:R:S221 | 2.94 | 2 | Yes | No | 5 | 8 | 2 | 1 | | R:R:D222 | R:R:F275 | 2.39 | 2 | Yes | Yes | 5 | 4 | 2 | 2 | | R:R:D222 | R:R:R279 | 11.91 | 2 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:L227 | R:R:W224 | 2.28 | 0 | No | Yes | 7 | 9 | 2 | 1 | | R:R:I225 | R:R:W229 | 8.22 | 2 | No | Yes | 7 | 5 | 1 | 1 | | R:R:F275 | R:R:I225 | 8.79 | 2 | Yes | No | 4 | 7 | 2 | 1 | | R:R:I228 | R:R:W229 | 4.7 | 2 | No | Yes | 6 | 5 | 1 | 1 | | R:R:A271 | R:R:W229 | 3.89 | 2 | No | Yes | 7 | 5 | 1 | 1 | | R:R:F275 | R:R:W229 | 6.01 | 2 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:L236 | R:R:L267 | 6.92 | 2 | Yes | No | 9 | 7 | 2 | 1 | | R:R:A270 | R:R:C299 | 1.81 | 0 | No | No | 5 | 9 | 2 | 1 | | R:R:A271 | R:R:G272 | 1.95 | 2 | No | No | 7 | 7 | 1 | 2 | | R:R:L273 | R:R:L274 | 2.77 | 0 | No | No | 7 | 6 | 2 | 1 | | R:R:L274 | R:R:T292 | 5.9 | 0 | No | Yes | 6 | 8 | 1 | 2 | | R:R:F275 | R:R:R279 | 10.69 | 2 | Yes | Yes | 4 | 6 | 2 | 1 | | R:R:F275 | S:S:D222 | 3.58 | 2 | Yes | Yes | 4 | 5 | 2 | 2 | | R:R:R279 | R:R:Y280 | 7.2 | 2 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:R279 | S:S:D222 | 10.72 | 2 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:Y280 | S:S:D222 | 8.05 | 2 | Yes | Yes | 7 | 5 | 2 | 2 | | R:R:I296 | R:R:T292 | 3.04 | 2 | Yes | Yes | 9 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 24.00 | | Average Number Of Links With An Hub | 9.00 | | Average Interaction Strength | 4.88 | | Average Nodes In Shell | 54.00 | | Average Hubs In Shell | 23.00 | | Average Links In Shell | 84.00 | | Average Links Mediated by Hubs In Shell | 72.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|



