| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4OO9 | C | Aminoacid | Metabotropic Glutamate | mGlu5 | Homo Sapiens | - | Mavoglurant | - | 2.6 | 2014-07-02 | doi.org/10.1038/nature13396 |
|

A 2D representation of the interactions of YCM in 4OO9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L591 | R:R:L630 | 4.15 | 0 | No | No | 6 | 6 | 2 | 1 | | R:R:C626 | R:R:L630 | 3.17 | 0 | No | No | 4 | 6 | 2 | 1 | | R:R:?634 | R:R:L630 | 3.54 | 2 | Yes | No | 5 | 6 | 0 | 1 | | R:R:?634 | R:R:L635 | 3.54 | 2 | Yes | Yes | 5 | 7 | 0 | 1 | | R:R:?634 | R:R:A637 | 4.03 | 2 | Yes | No | 5 | 8 | 0 | 1 | | R:R:?634 | R:R:Y643 | 3 | 2 | Yes | Yes | 5 | 7 | 0 | 1 | | R:R:?634 | R:R:L646 | 9.43 | 2 | Yes | No | 5 | 5 | 0 | 1 | | R:R:?634 | R:R:Q647 | 9.07 | 2 | Yes | Yes | 5 | 8 | 0 | 1 | | R:R:?634 | R:R:L731 | 3.54 | 2 | Yes | No | 5 | 6 | 0 | 1 | | R:R:L635 | R:R:Q647 | 11.98 | 2 | Yes | Yes | 7 | 8 | 1 | 1 | | R:R:I651 | R:R:L635 | 4.28 | 0 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:C803 | R:R:L635 | 4.76 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:A637 | R:R:Y643 | 5.34 | 2 | No | Yes | 8 | 7 | 1 | 1 | | R:R:K640 | R:R:Y643 | 4.78 | 0 | No | Yes | 5 | 7 | 2 | 1 | | R:R:L646 | R:R:Y643 | 4.69 | 2 | No | Yes | 5 | 7 | 1 | 1 | | R:R:L731 | R:R:Q647 | 3.99 | 2 | No | Yes | 6 | 8 | 1 | 1 | | R:R:C733 | R:R:Q647 | 6.1 | 0 | No | Yes | 9 | 8 | 2 | 1 | | R:R:L731 | R:R:P639 | 1.64 | 2 | No | No | 6 | 9 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 5.16 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
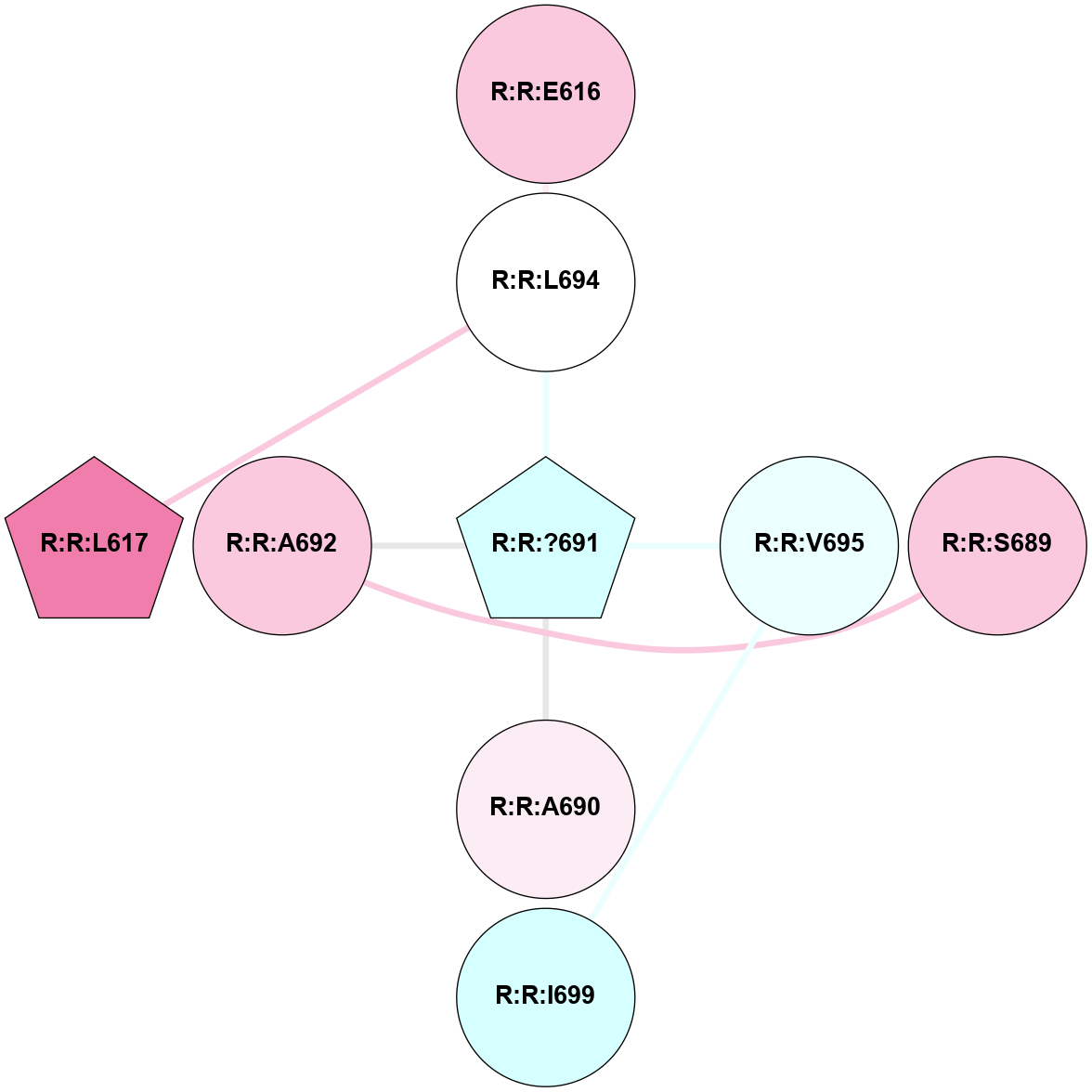
A 2D representation of the interactions of YCM in 4OO9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E616 | R:R:L694 | 7.95 | 0 | No | No | 7 | 5 | 2 | 1 | | R:R:A692 | R:R:S689 | 3.42 | 0 | No | No | 7 | 7 | 1 | 2 | | R:R:?691 | R:R:L694 | 5.89 | 0 | Yes | No | 3 | 5 | 0 | 1 | | R:R:?691 | R:R:V695 | 7.61 | 0 | Yes | No | 3 | 4 | 0 | 1 | | R:R:I699 | R:R:V695 | 3.07 | 0 | No | No | 3 | 4 | 2 | 1 | | R:R:L617 | R:R:L694 | 2.77 | 6 | Yes | No | 8 | 5 | 2 | 1 | | R:R:?691 | R:R:A690 | 2.68 | 0 | Yes | No | 3 | 6 | 0 | 1 | | R:R:?691 | R:R:A692 | 2.68 | 0 | Yes | No | 3 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 4.71 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5C1M | A | Peptide | Opioid | MOP | Mus Musculus | BU72 | - | - | 2.07 | 2015-08-05 | doi.org/10.1038/nature14886 |
|
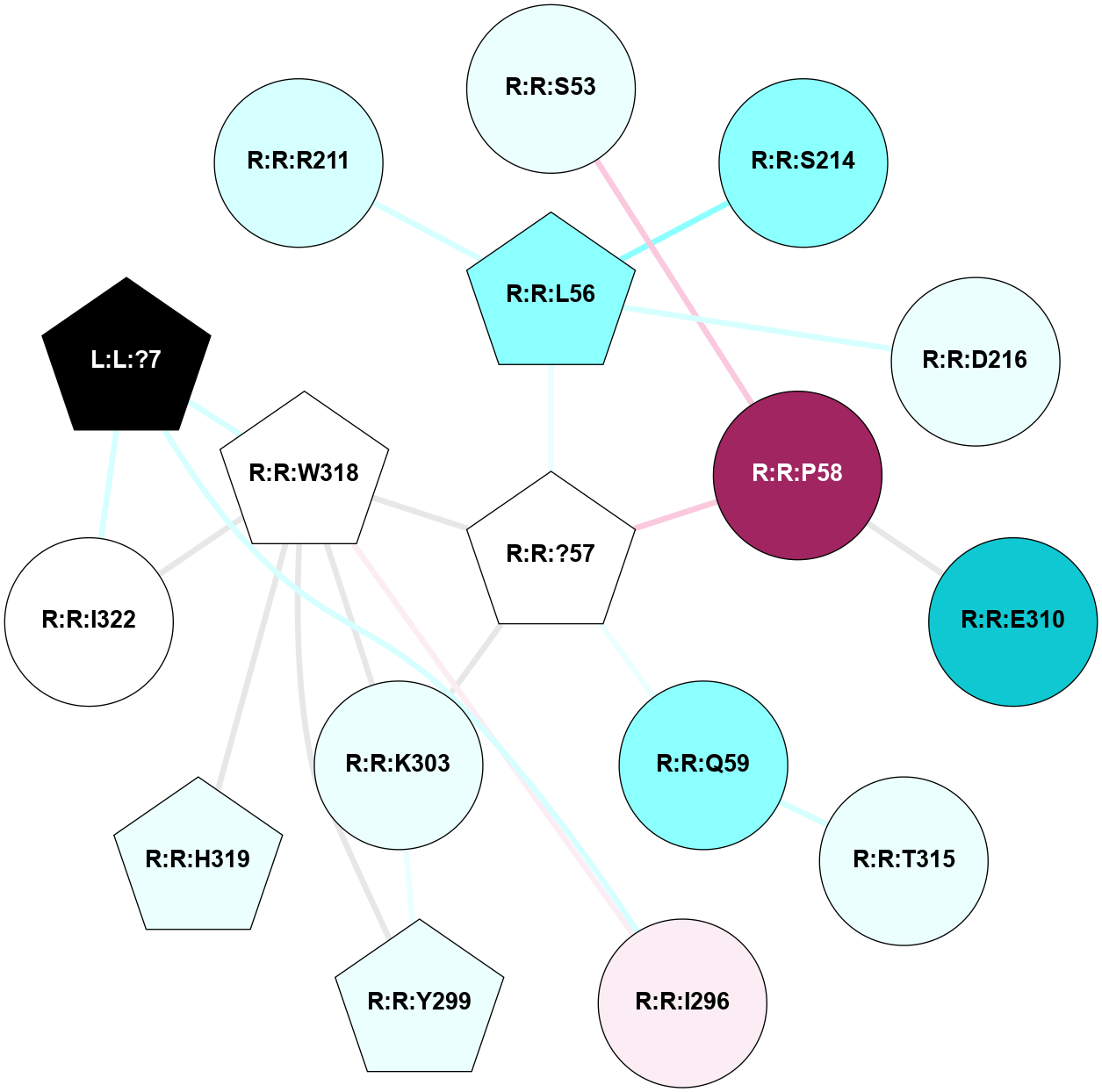
A 2D representation of the interactions of YCM in 5C1M
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?7 | R:R:I296 | 5.31 | 1 | Yes | No | 0 | 6 | 2 | 2 | | L:L:?7 | R:R:W318 | 3.53 | 1 | Yes | Yes | 0 | 5 | 2 | 1 | | L:L:?7 | R:R:I322 | 3.54 | 1 | Yes | No | 0 | 5 | 2 | 2 | | R:R:P58 | R:R:S53 | 3.56 | 0 | No | No | 9 | 4 | 1 | 2 | | R:R:?57 | R:R:L56 | 4.72 | 1 | Yes | Yes | 5 | 2 | 0 | 1 | | R:R:L56 | R:R:S214 | 7.51 | 0 | Yes | No | 2 | 2 | 1 | 2 | | R:R:D216 | R:R:L56 | 8.14 | 0 | No | Yes | 4 | 2 | 2 | 1 | | R:R:?57 | R:R:P58 | 18.18 | 1 | Yes | No | 5 | 9 | 0 | 1 | | R:R:?57 | R:R:Q59 | 13.6 | 1 | Yes | No | 5 | 2 | 0 | 1 | | R:R:?57 | R:R:K303 | 9.61 | 1 | Yes | No | 5 | 4 | 0 | 1 | | R:R:?57 | R:R:W318 | 21.34 | 1 | Yes | Yes | 5 | 5 | 0 | 1 | | R:R:E310 | R:R:P58 | 4.72 | 0 | No | No | 1 | 9 | 2 | 1 | | R:R:Q59 | R:R:T315 | 14.17 | 0 | No | No | 2 | 4 | 1 | 2 | | R:R:I296 | R:R:W318 | 3.52 | 1 | No | Yes | 6 | 5 | 2 | 1 | | R:R:K303 | R:R:Y299 | 4.78 | 1 | No | Yes | 4 | 4 | 1 | 2 | | R:R:W318 | R:R:Y299 | 13.5 | 1 | Yes | Yes | 5 | 4 | 1 | 2 | | R:R:K303 | R:R:W318 | 9.28 | 1 | No | Yes | 4 | 5 | 1 | 1 | | R:R:H319 | R:R:W318 | 6.35 | 1 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:I322 | R:R:W318 | 10.57 | 1 | No | Yes | 5 | 5 | 2 | 1 | | R:R:L56 | R:R:R211 | 2.43 | 0 | Yes | No | 2 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 13.49 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 20.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5T1A | A | Protein | Chemokine | CCR2 | Homo Sapiens | BMS-681 | PubChem 12093170 | - | 2.81 | 2016-12-14 | doi.org/10.1038/nature20605 |
|
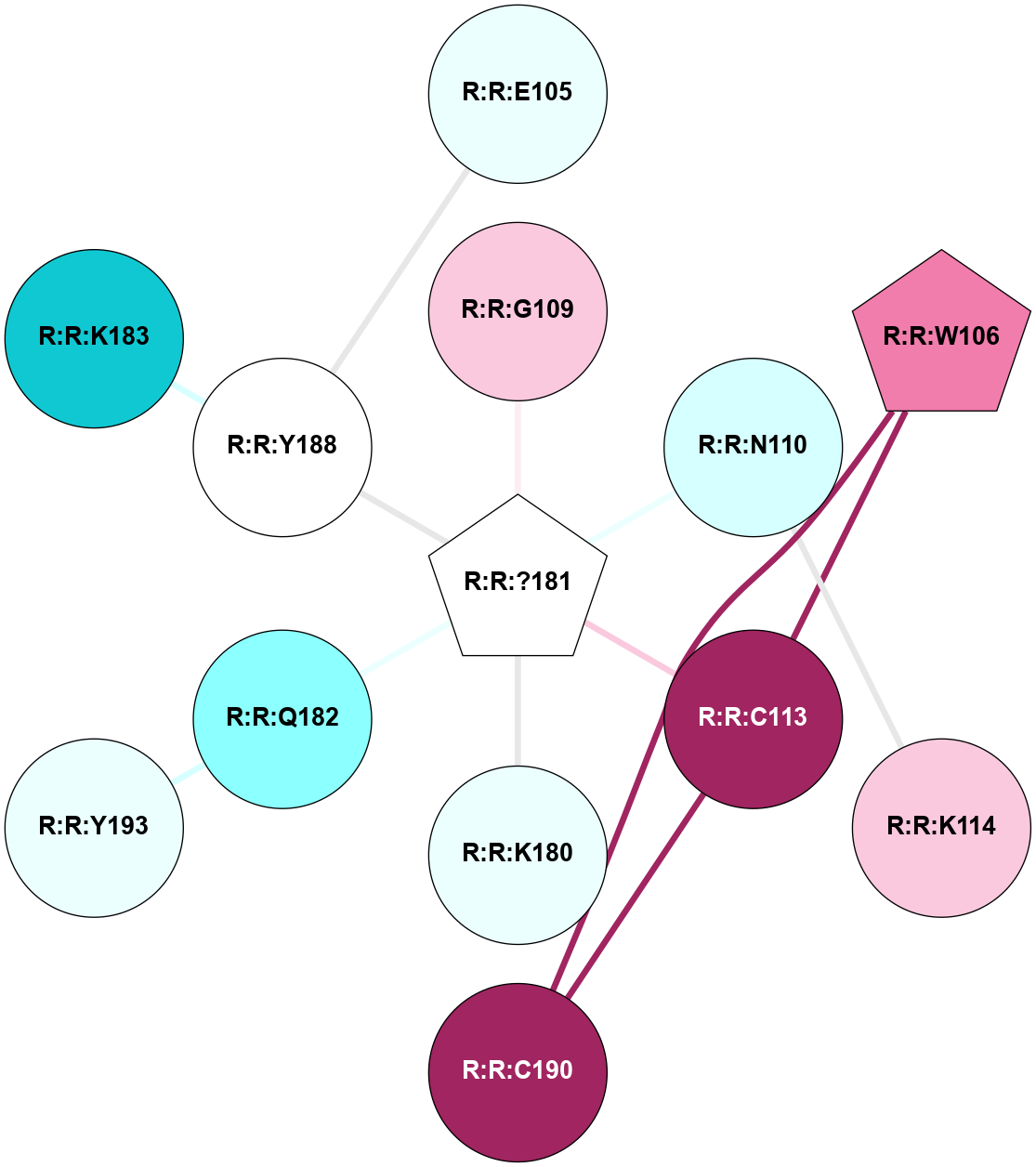
A 2D representation of the interactions of YCM in 5T1A
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E105 | R:R:Y188 | 15.71 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:C113 | R:R:W106 | 3.92 | 3 | No | Yes | 9 | 8 | 1 | 2 | | R:R:C190 | R:R:W106 | 13.06 | 3 | No | Yes | 9 | 8 | 2 | 2 | | R:R:?181 | R:R:G109 | 4.38 | 0 | Yes | No | 5 | 7 | 0 | 1 | | R:R:K114 | R:R:N110 | 5.6 | 0 | No | No | 7 | 3 | 2 | 1 | | R:R:?181 | R:R:N110 | 8.2 | 0 | Yes | No | 5 | 3 | 0 | 1 | | R:R:?181 | R:R:C113 | 4.06 | 0 | Yes | No | 5 | 9 | 0 | 1 | | R:R:C113 | R:R:C190 | 7.28 | 3 | No | No | 9 | 9 | 1 | 2 | | R:R:?181 | R:R:K180 | 4.81 | 0 | Yes | No | 5 | 4 | 0 | 1 | | R:R:?181 | R:R:Q182 | 4.54 | 0 | Yes | No | 5 | 2 | 0 | 1 | | R:R:?181 | R:R:Y188 | 4 | 0 | Yes | No | 5 | 5 | 0 | 1 | | R:R:Q182 | R:R:Y193 | 16.91 | 0 | No | No | 2 | 4 | 1 | 2 | | R:R:K183 | R:R:Y188 | 13.14 | 0 | No | No | 1 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 5.00 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5WB2 | A | Other | Unclassified | US28 | Human Cytomegalovirus (Strain Towne) | CX3CL1.35 | - | - | 3.5 | 2018-06-13 | doi.org/10.7554/eLife.35850 |
|
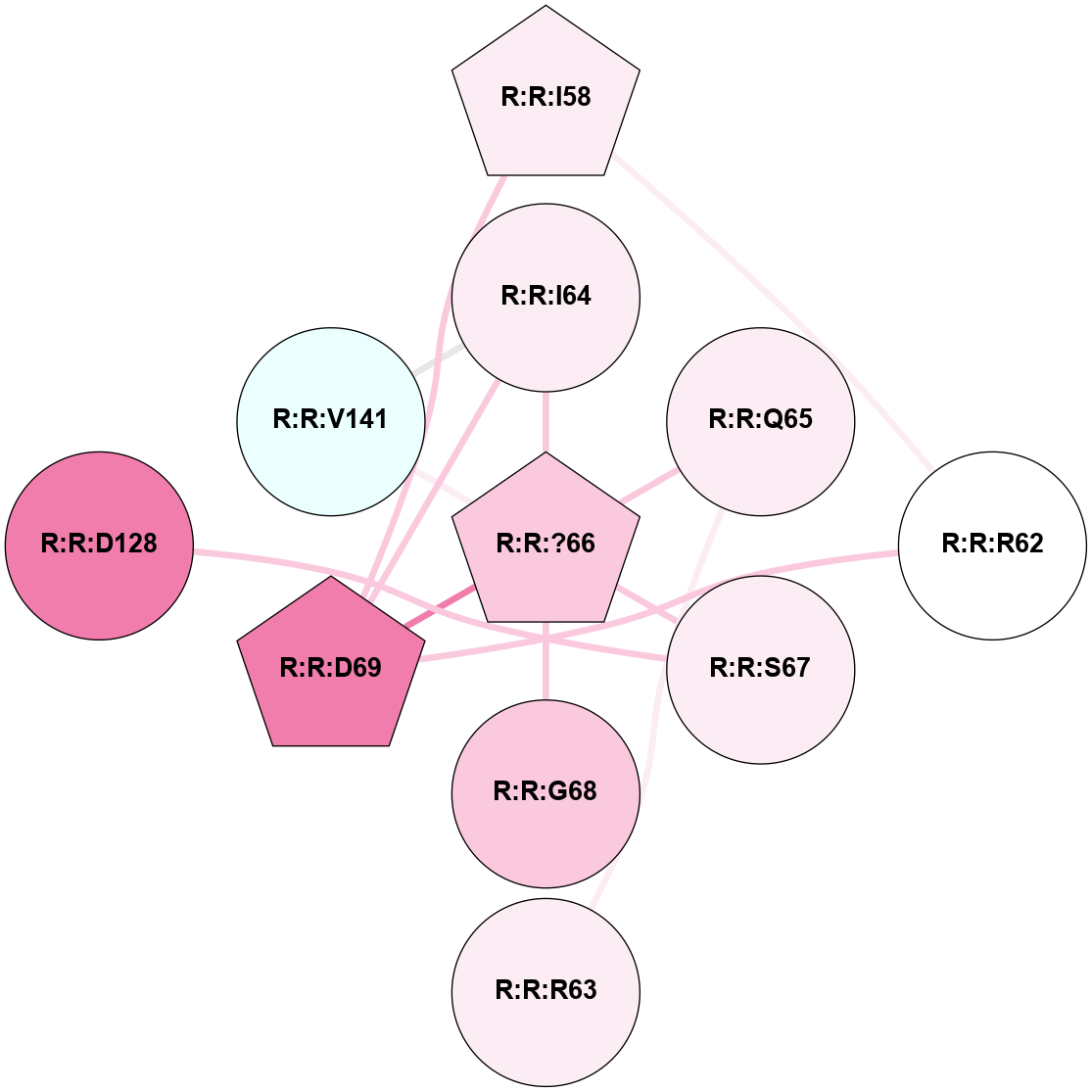
A 2D representation of the interactions of YCM in 5WB2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D69 | R:R:I58 | 5.6 | 5 | No | No | 8 | 6 | 1 | 2 | | R:R:?66 | R:R:I64 | 3.65 | 5 | Yes | No | 7 | 6 | 0 | 1 | | R:R:D69 | R:R:I64 | 7 | 5 | No | No | 8 | 6 | 1 | 1 | | R:R:I64 | R:R:V141 | 4.61 | 5 | No | No | 6 | 4 | 1 | 1 | | R:R:?66 | R:R:Q65 | 3.4 | 5 | Yes | No | 7 | 6 | 0 | 1 | | R:R:?66 | R:R:S67 | 5.12 | 5 | Yes | No | 7 | 6 | 0 | 1 | | R:R:?66 | R:R:G68 | 8.74 | 5 | Yes | No | 7 | 7 | 0 | 1 | | R:R:?66 | R:R:D69 | 15.03 | 5 | Yes | No | 7 | 8 | 0 | 1 | | R:R:?66 | R:R:V141 | 5.08 | 5 | Yes | No | 7 | 4 | 0 | 1 | | R:R:D128 | R:R:S67 | 8.83 | 0 | No | No | 8 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.84 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of YCM in 5WB2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L305 | R:R:W60 | 6.83 | 0 | No | No | 4 | 4 | 1 | 2 | | R:R:E301 | R:R:L305 | 6.63 | 0 | No | No | 4 | 4 | 2 | 1 | | R:R:?304 | R:R:H303 | 4.38 | 0 | No | No | 4 | 4 | 0 | 1 | | R:R:E308 | R:R:H303 | 3.69 | 0 | No | No | 4 | 4 | 2 | 1 | | R:R:?304 | R:R:L305 | 7.07 | 0 | No | No | 4 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 5.72 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6C1Q | A | Peptide | Complement Peptide | C5A1 | Homo Sapiens | PMX53 | NDT9513727 | - | 2.9 | 2018-05-30 | doi.org/10.1038/s41594-018-0067-z |
|
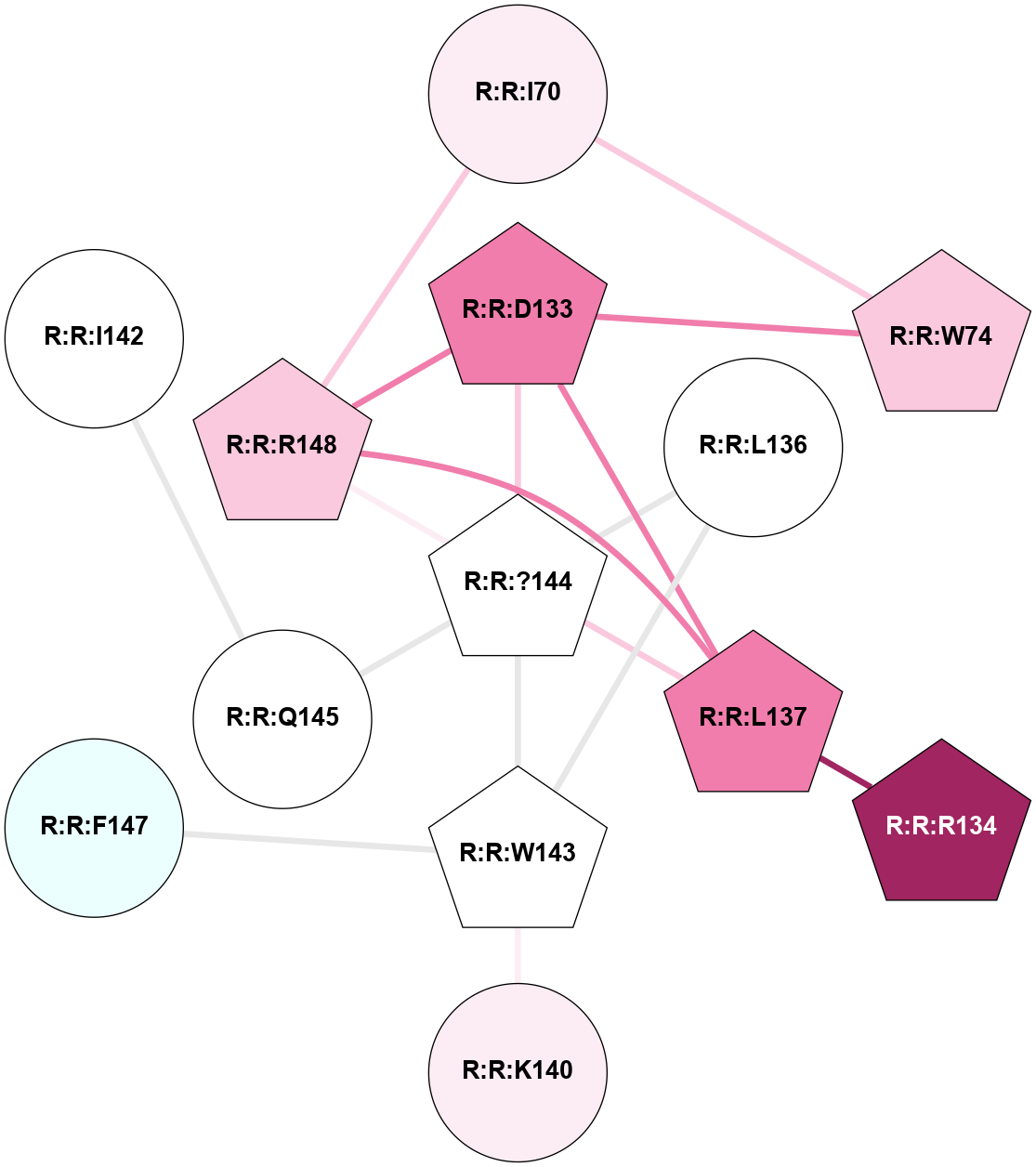
A 2D representation of the interactions of YCM in 6C1Q
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I70 | R:R:W74 | 7.05 | 2 | No | Yes | 6 | 7 | 2 | 2 | | R:R:I70 | R:R:R148 | 10.02 | 2 | No | Yes | 6 | 7 | 2 | 1 | | R:R:D133 | R:R:W74 | 8.93 | 2 | Yes | Yes | 8 | 7 | 1 | 2 | | R:R:D133 | R:R:L137 | 8.14 | 2 | Yes | Yes | 8 | 8 | 1 | 1 | | R:R:?144 | R:R:D133 | 6.94 | 2 | Yes | Yes | 5 | 8 | 0 | 1 | | R:R:D133 | R:R:R148 | 10.72 | 2 | Yes | Yes | 8 | 7 | 1 | 1 | | R:R:L137 | R:R:R134 | 6.07 | 2 | Yes | Yes | 8 | 9 | 1 | 2 | | R:R:L136 | R:R:W143 | 3.42 | 2 | No | Yes | 5 | 5 | 1 | 1 | | R:R:?144 | R:R:L136 | 12.97 | 2 | Yes | No | 5 | 5 | 0 | 1 | | R:R:?144 | R:R:L137 | 5.89 | 2 | Yes | Yes | 5 | 8 | 0 | 1 | | R:R:L137 | R:R:R148 | 7.29 | 2 | Yes | Yes | 8 | 7 | 1 | 1 | | R:R:K140 | R:R:W143 | 16.24 | 0 | No | Yes | 6 | 5 | 2 | 1 | | R:R:?144 | R:R:W143 | 3.88 | 2 | Yes | Yes | 5 | 5 | 0 | 1 | | R:R:F147 | R:R:W143 | 3.01 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:?144 | R:R:Q145 | 3.4 | 2 | Yes | No | 5 | 5 | 0 | 1 | | R:R:?144 | R:R:R148 | 11.38 | 2 | Yes | Yes | 5 | 7 | 0 | 1 | | R:R:I142 | R:R:Q145 | 2.74 | 0 | No | No | 5 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 7.41 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6C1R | A | Peptide | Complement Peptide | C5A1 | Homo Sapiens | PMX53 | Avacopan; Na | - | 2.2 | 2018-05-30 | doi.org/10.1038/s41594-018-0067-z |
|

A 2D representation of the interactions of YCM in 6C1R
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I70 | R:R:W74 | 5.87 | 0 | Yes | Yes | 6 | 7 | 2 | 2 | | R:R:I70 | R:R:R148 | 15.03 | 0 | Yes | Yes | 6 | 7 | 2 | 1 | | R:R:D133 | R:R:W74 | 11.17 | 4 | Yes | Yes | 8 | 7 | 1 | 2 | | R:R:D133 | R:R:L137 | 4.07 | 4 | Yes | Yes | 8 | 8 | 1 | 1 | | R:R:?144 | R:R:D133 | 6.94 | 4 | Yes | Yes | 5 | 8 | 0 | 1 | | R:R:D133 | R:R:R148 | 13.1 | 4 | Yes | Yes | 8 | 7 | 1 | 1 | | R:R:L137 | R:R:R134 | 6.07 | 4 | Yes | No | 8 | 9 | 1 | 2 | | R:R:?144 | R:R:L136 | 12.97 | 4 | Yes | No | 5 | 5 | 0 | 1 | | R:R:?144 | R:R:L137 | 5.89 | 4 | Yes | Yes | 5 | 8 | 0 | 1 | | R:R:L137 | R:R:R148 | 10.93 | 4 | Yes | Yes | 8 | 7 | 1 | 1 | | R:R:L137 | R:R:S327 | 7.51 | 4 | Yes | No | 8 | 4 | 1 | 2 | | R:R:?144 | R:R:R148 | 19.66 | 4 | Yes | Yes | 5 | 7 | 0 | 1 | | R:R:E326 | R:R:R148 | 3.49 | 0 | No | Yes | 5 | 7 | 2 | 1 | | R:R:L136 | R:R:W143 | 3.42 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:?144 | R:R:Q145 | 3.4 | 4 | Yes | No | 5 | 5 | 0 | 1 | | R:R:I142 | R:R:Q145 | 2.74 | 0 | No | No | 5 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 9.77 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6D26 | A | Lipid | Prostanoid | DP2 | Homo Sapiens | Fevipiprant | - | - | 2.8 | 2018-10-03 | doi.org/10.1016/j.molcel.2018.08.009 |
|

A 2D representation of the interactions of YCM in 6D26
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F244 | R:R:V66 | 9.18 | 0 | Yes | No | 7 | 7 | 1 | 2 | | R:R:F244 | R:R:T67 | 3.89 | 0 | Yes | No | 7 | 7 | 1 | 2 | | R:R:F244 | R:R:R243 | 8.55 | 0 | Yes | No | 7 | 7 | 1 | 2 | | R:R:F244 | R:R:L247 | 7.31 | 0 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:?308 | R:R:F244 | 7.26 | 6 | Yes | Yes | 8 | 7 | 0 | 1 | | R:R:?308 | R:R:R246 | 3.1 | 6 | Yes | No | 8 | 5 | 0 | 1 | | R:R:P309 | R:R:R246 | 8.65 | 6 | No | No | 5 | 5 | 1 | 1 | | R:R:L247 | R:R:Y304 | 7.03 | 1 | Yes | Yes | 7 | 9 | 2 | 2 | | R:R:L247 | R:R:T307 | 4.42 | 1 | Yes | No | 7 | 7 | 2 | 1 | | R:R:M311 | R:R:Y304 | 3.59 | 0 | No | Yes | 8 | 9 | 1 | 2 | | R:R:?308 | R:R:T307 | 3.77 | 6 | Yes | No | 8 | 7 | 0 | 1 | | R:R:?308 | R:R:P309 | 20.98 | 6 | Yes | No | 8 | 5 | 0 | 1 | | R:R:?308 | R:R:D310 | 9.25 | 6 | Yes | No | 8 | 5 | 0 | 1 | | R:R:?308 | R:R:M311 | 7.22 | 6 | Yes | No | 8 | 8 | 0 | 1 | | R:R:D310 | R:R:R313 | 13.1 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:M311 | R:R:M61 | 2.89 | 0 | No | No | 8 | 6 | 1 | 2 | | R:R:A250 | R:R:T307 | 1.68 | 0 | No | No | 6 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 8.60 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6D27 | A | Lipid | Prostanoid | DP2 | Homo Sapiens | CAY10471 | - | - | 2.74 | 2018-10-03 | doi.org/10.1016/j.molcel.2018.08.009 |
|

A 2D representation of the interactions of YCM in 6D27
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:M311 | R:R:V57 | 3.04 | 0 | No | No | 8 | 7 | 1 | 2 | | R:R:M311 | R:R:M61 | 2.89 | 0 | No | No | 8 | 6 | 1 | 2 | | R:R:F244 | R:R:V66 | 7.87 | 0 | Yes | No | 7 | 7 | 1 | 2 | | R:R:F244 | R:R:T67 | 2.59 | 0 | Yes | No | 7 | 7 | 1 | 2 | | R:R:F244 | R:R:R243 | 9.62 | 0 | Yes | No | 7 | 7 | 1 | 2 | | R:R:F244 | R:R:L247 | 7.31 | 0 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:?308 | R:R:F244 | 6.22 | 7 | Yes | Yes | 8 | 7 | 0 | 1 | | R:R:?308 | R:R:R246 | 3.1 | 7 | Yes | No | 8 | 5 | 0 | 1 | | R:R:P309 | R:R:R246 | 4.32 | 7 | No | No | 5 | 5 | 1 | 1 | | R:R:L312 | R:R:V305 | 5.96 | 0 | No | No | 7 | 7 | 1 | 2 | | R:R:?308 | R:R:P309 | 20.98 | 7 | Yes | No | 8 | 5 | 0 | 1 | | R:R:?308 | R:R:D310 | 10.4 | 7 | Yes | No | 8 | 5 | 0 | 1 | | R:R:?308 | R:R:M311 | 9.63 | 7 | Yes | No | 8 | 8 | 0 | 1 | | R:R:P309 | R:R:R313 | 4.32 | 7 | No | No | 5 | 5 | 1 | 2 | | R:R:D310 | R:R:R313 | 4.76 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:L312 | R:R:R316 | 3.64 | 0 | No | No | 7 | 5 | 1 | 2 | | R:R:?308 | R:R:L312 | 1.18 | 7 | Yes | No | 8 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 8.59 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FFH | C | Aminoacid | Metabotropic Glutamate | mGlu5 | Homo Sapiens | - | Fenobam | - | 2.65 | 2018-03-07 | doi.org/10.1021/acs.jmedchem.7b01722 |
|
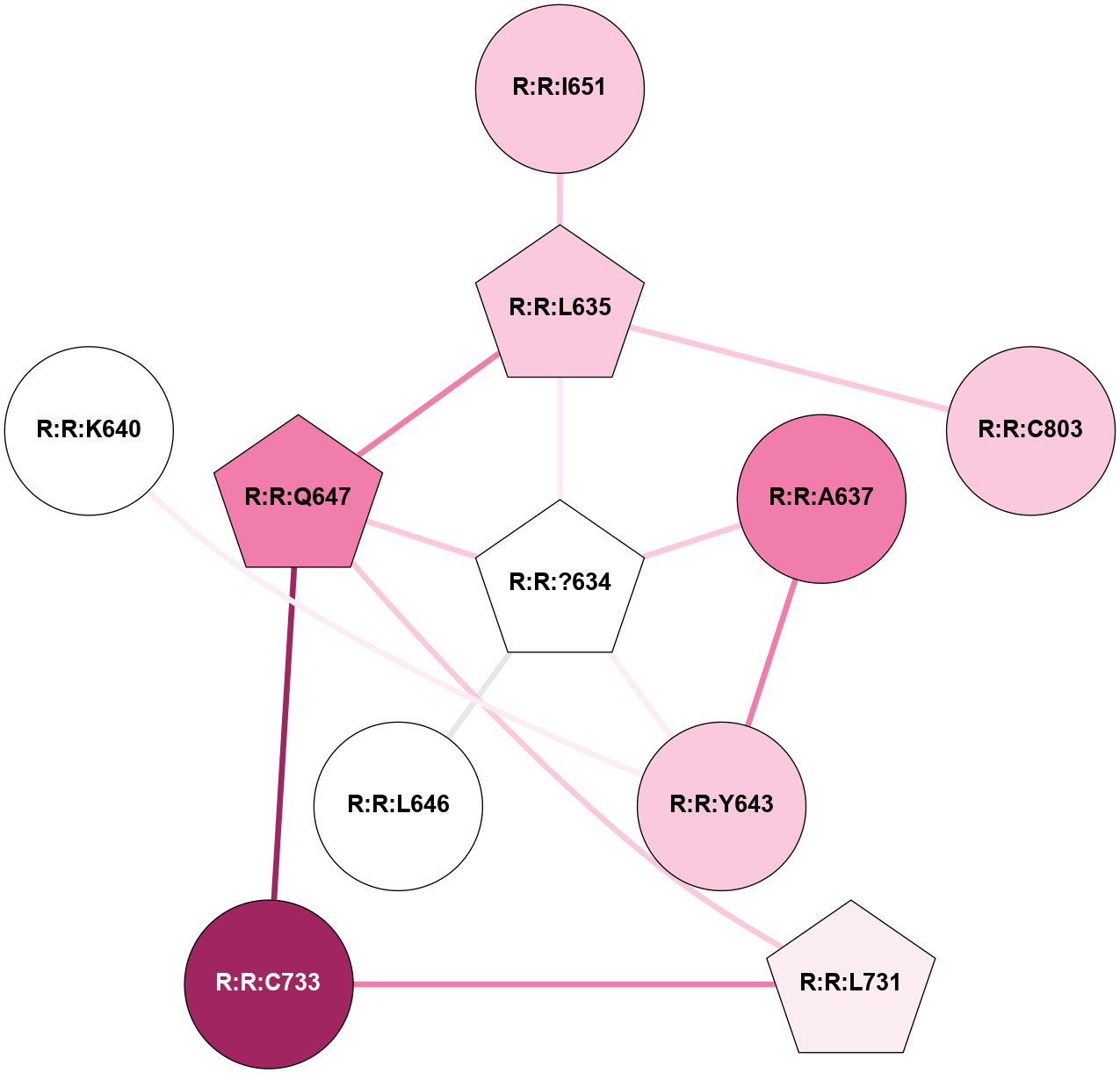
A 2D representation of the interactions of YCM in 6FFH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?634 | R:R:L635 | 3.54 | 2 | Yes | Yes | 5 | 7 | 0 | 1 | | R:R:?634 | R:R:A637 | 5.37 | 2 | Yes | No | 5 | 8 | 0 | 1 | | R:R:?634 | R:R:Y643 | 7.99 | 2 | Yes | No | 5 | 7 | 0 | 1 | | R:R:?634 | R:R:L646 | 10.61 | 2 | Yes | No | 5 | 5 | 0 | 1 | | R:R:?634 | R:R:Q647 | 7.94 | 2 | Yes | Yes | 5 | 8 | 0 | 1 | | R:R:L635 | R:R:Q647 | 10.65 | 2 | Yes | Yes | 7 | 8 | 1 | 1 | | R:R:I651 | R:R:L635 | 4.28 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:C803 | R:R:L635 | 4.76 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:A637 | R:R:Y643 | 5.34 | 2 | No | No | 8 | 7 | 1 | 1 | | R:R:L731 | R:R:Q647 | 3.99 | 2 | Yes | Yes | 6 | 8 | 2 | 1 | | R:R:C733 | R:R:Q647 | 6.1 | 2 | No | Yes | 9 | 8 | 2 | 1 | | R:R:C733 | R:R:L731 | 3.17 | 2 | No | Yes | 9 | 6 | 2 | 2 | | R:R:K640 | R:R:Y643 | 2.39 | 0 | No | No | 5 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 7.09 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FFI | C | Aminoacid | Metabotropic Glutamate | mGlu5 | Homo Sapiens | - | MMPEP | - | 2.2 | 2018-03-07 | doi.org/10.1021/acs.jmedchem.7b01722 |
|

A 2D representation of the interactions of YCM in 6FFI
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L591 | R:R:L630 | 5.54 | 0 | No | No | 6 | 6 | 2 | 1 | | R:R:C626 | R:R:L630 | 3.17 | 0 | No | No | 4 | 6 | 2 | 1 | | R:R:?634 | R:R:L630 | 3.54 | 5 | Yes | No | 5 | 6 | 0 | 1 | | R:R:?634 | R:R:L635 | 3.54 | 5 | Yes | Yes | 5 | 7 | 0 | 1 | | R:R:?634 | R:R:L646 | 8.25 | 5 | Yes | No | 5 | 5 | 0 | 1 | | R:R:?634 | R:R:Q647 | 9.07 | 5 | Yes | Yes | 5 | 8 | 0 | 1 | | R:R:L635 | R:R:Q647 | 10.65 | 5 | Yes | Yes | 7 | 8 | 1 | 1 | | R:R:I651 | R:R:L635 | 2.85 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:I799 | R:R:L635 | 2.85 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:C803 | R:R:L635 | 4.76 | 0 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:L731 | R:R:Q647 | 3.99 | 5 | Yes | Yes | 6 | 8 | 2 | 1 | | R:R:C733 | R:R:Q647 | 6.1 | 5 | No | Yes | 9 | 8 | 2 | 1 | | R:R:C733 | R:R:L731 | 3.17 | 5 | No | Yes | 9 | 6 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 6.10 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6HLO | A | Peptide | Tachykinin | NK1 | Homo Sapiens | Aprepitant | - | - | 2.4 | 2019-01-16 | doi.org/10.1038/s41467-018-07939-8 |
|

A 2D representation of the interactions of YCM in 6HLO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?322 | R:R:R321 | 4.14 | 0 | No | No | 9 | 5 | 0 | 1 | | R:R:?322 | R:R:C323 | 4.06 | 0 | No | No | 9 | 5 | 0 | 1 | | R:R:C323 | R:R:P324 | 7.53 | 0 | No | No | 5 | 3 | 1 | 2 | | R:R:H318 | R:R:R321 | 1.13 | 0 | No | No | 3 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 4.10 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6HLP | A | Peptide | Tachykinin | NK1 | Homo Sapiens | Netupitant | - | - | 2.2 | 2019-01-16 | doi.org/10.1038/s41467-018-07939-8 |
|
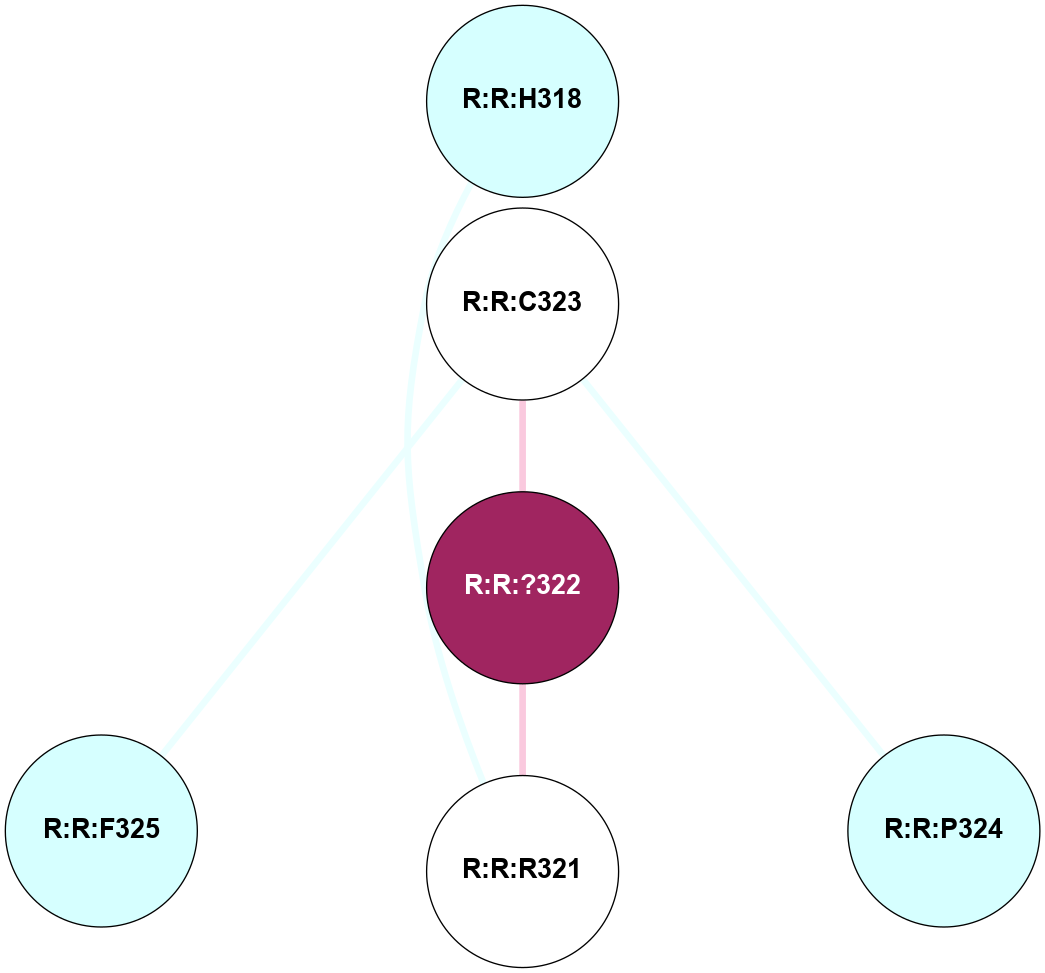
A 2D representation of the interactions of YCM in 6HLP
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:H318 | R:R:R321 | 11.28 | 0 | No | No | 3 | 5 | 2 | 1 | | R:R:?322 | R:R:C323 | 5.41 | 0 | No | No | 9 | 5 | 0 | 1 | | R:R:C323 | R:R:P324 | 3.77 | 0 | No | No | 5 | 3 | 1 | 2 | | R:R:C323 | R:R:F325 | 5.59 | 0 | No | No | 5 | 3 | 1 | 2 | | R:R:?322 | R:R:R321 | 3.1 | 0 | No | No | 9 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 4.25 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6ME2 | A | Melatonin | Melatonin | MT1 | Homo Sapiens | Ramelteon | - | - | 2.8 | 2019-04-24 | doi.org/10.1038/s41586-019-1141-3 |
|
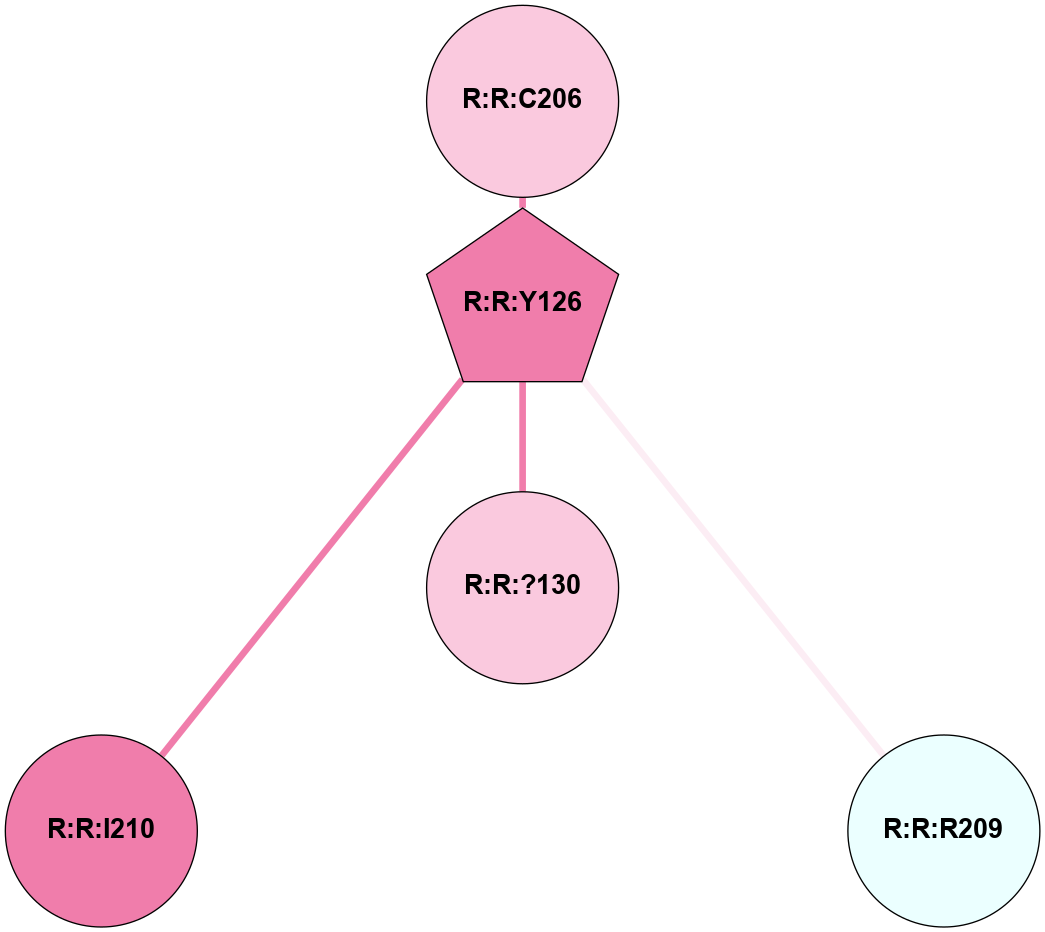
A 2D representation of the interactions of YCM in 6ME2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?130 | R:R:Y126 | 8.99 | 0 | No | Yes | 7 | 8 | 0 | 1 | | R:R:C206 | R:R:Y126 | 4.03 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:R209 | R:R:Y126 | 4.12 | 0 | No | Yes | 4 | 8 | 2 | 1 | | R:R:I210 | R:R:Y126 | 3.63 | 0 | No | Yes | 8 | 8 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 8.99 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6MXT | A | Amine | Adrenergic | Beta2 | Homo Sapiens | Salmeterol | - | - | 2.96 | 2018-11-14 | doi.org/10.1038/s41589-018-0145-x |
|

A 2D representation of the interactions of YCM in 6MXT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:K232 | R:R:Q229 | 8.14 | 0 | No | No | 4 | 7 | 2 | 1 | | R:R:?265 | R:R:Q229 | 7.94 | 2 | Yes | No | 3 | 7 | 0 | 1 | | R:R:E268 | R:R:Q229 | 8.92 | 2 | Yes | No | 7 | 7 | 1 | 1 | | R:R:?265 | R:R:L230 | 3.54 | 2 | Yes | No | 3 | 5 | 0 | 1 | | R:R:H269 | R:R:L230 | 3.86 | 2 | No | No | 5 | 5 | 1 | 1 | | R:R:?265 | R:R:I233 | 14.59 | 2 | Yes | No | 3 | 6 | 0 | 1 | | R:R:F264 | R:R:K263 | 19.85 | 0 | No | No | 4 | 2 | 1 | 2 | | R:R:?265 | R:R:E268 | 10.16 | 2 | Yes | Yes | 3 | 7 | 0 | 1 | | R:R:?265 | R:R:H269 | 10.95 | 2 | Yes | No | 3 | 5 | 0 | 1 | | R:R:K270 | R:R:L266 | 5.64 | 0 | No | No | 7 | 5 | 2 | 1 | | R:R:E268 | R:R:K267 | 9.45 | 2 | Yes | No | 7 | 6 | 1 | 2 | | R:R:A226 | R:R:E268 | 3.02 | 0 | No | Yes | 8 | 7 | 2 | 1 | | R:R:?265 | R:R:L266 | 2.36 | 2 | Yes | No | 3 | 5 | 0 | 1 | | R:R:?265 | R:R:F264 | 2.07 | 2 | Yes | No | 3 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 7.37 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of YCM in 6MXT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F49 | R:R:L340 | 3.65 | 0 | No | No | 4 | 6 | 2 | 1 | | R:R:L340 | R:R:L53 | 4.15 | 0 | No | No | 6 | 6 | 1 | 2 | | R:R:?341 | R:R:L342 | 3.54 | 0 | No | No | 5 | 8 | 0 | 1 | | R:R:?341 | R:R:Q337 | 3.4 | 0 | No | No | 5 | 4 | 0 | 1 | | R:R:?341 | R:R:L340 | 2.36 | 0 | No | No | 5 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 3.10 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6TPJ | A | Peptide | Orexin | OX2 | Homo Sapiens | Suvorexant | - | - | 2.74 | 2020-01-01 | doi.org/10.1021/acs.jmedchem.9b01787 |
|
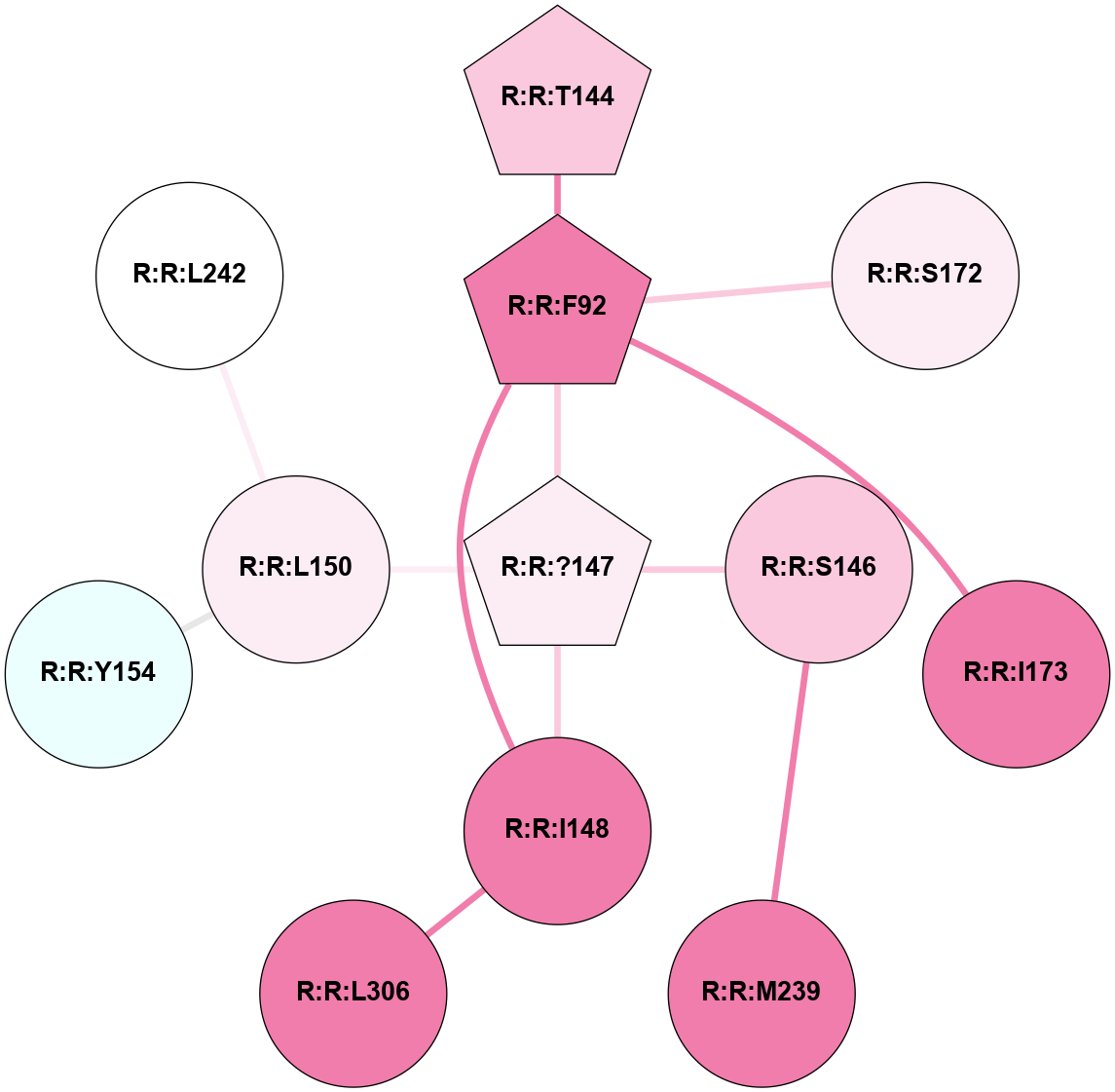
A 2D representation of the interactions of YCM in 6TPJ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F92 | R:R:T144 | 3.89 | 5 | Yes | Yes | 8 | 7 | 1 | 2 | | R:R:?147 | R:R:F92 | 6.22 | 5 | Yes | Yes | 6 | 8 | 0 | 1 | | R:R:F92 | R:R:I148 | 5.02 | 5 | Yes | No | 8 | 8 | 1 | 1 | | R:R:F92 | R:R:S172 | 3.96 | 5 | Yes | No | 8 | 6 | 1 | 2 | | R:R:F92 | R:R:I173 | 6.28 | 5 | Yes | No | 8 | 8 | 1 | 2 | | R:R:?147 | R:R:S146 | 3.84 | 5 | Yes | No | 6 | 7 | 0 | 1 | | R:R:M239 | R:R:S146 | 4.6 | 0 | No | No | 8 | 7 | 2 | 1 | | R:R:?147 | R:R:I148 | 3.65 | 5 | Yes | No | 6 | 8 | 0 | 1 | | R:R:?147 | R:R:L150 | 9.43 | 5 | Yes | No | 6 | 6 | 0 | 1 | | R:R:I148 | R:R:L306 | 4.28 | 5 | No | No | 8 | 8 | 1 | 2 | | R:R:L150 | R:R:Y154 | 4.69 | 0 | No | No | 6 | 4 | 1 | 2 | | R:R:L150 | R:R:L242 | 4.15 | 0 | No | No | 6 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.79 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7JNI | A | Peptide | Angiotensin | AT2 | Homo Sapiens | Olodanrigan | - | - | 3 | 2022-02-09 | doi.org/10.1073/pnas.2116289119 |
|
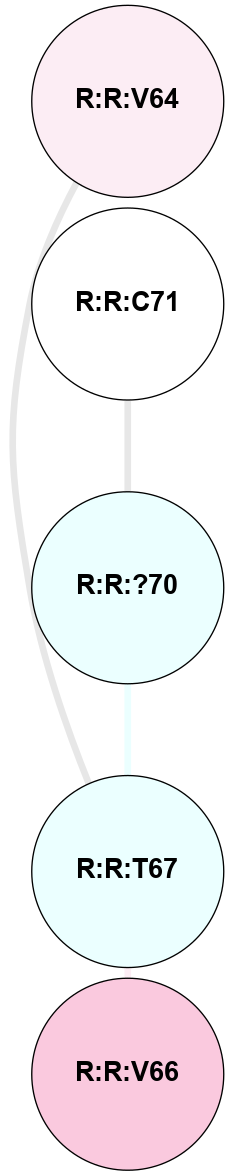
A 2D representation of the interactions of YCM in 7JNI
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?70 | R:R:C71 | 4.06 | 0 | No | No | 4 | 5 | 0 | 1 | | R:R:?70 | R:R:T67 | 2.51 | 0 | No | No | 4 | 4 | 0 | 1 | | R:R:T67 | R:R:V64 | 1.59 | 0 | No | No | 4 | 6 | 1 | 2 | | R:R:T67 | R:R:V66 | 1.59 | 0 | No | No | 4 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 3.28 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
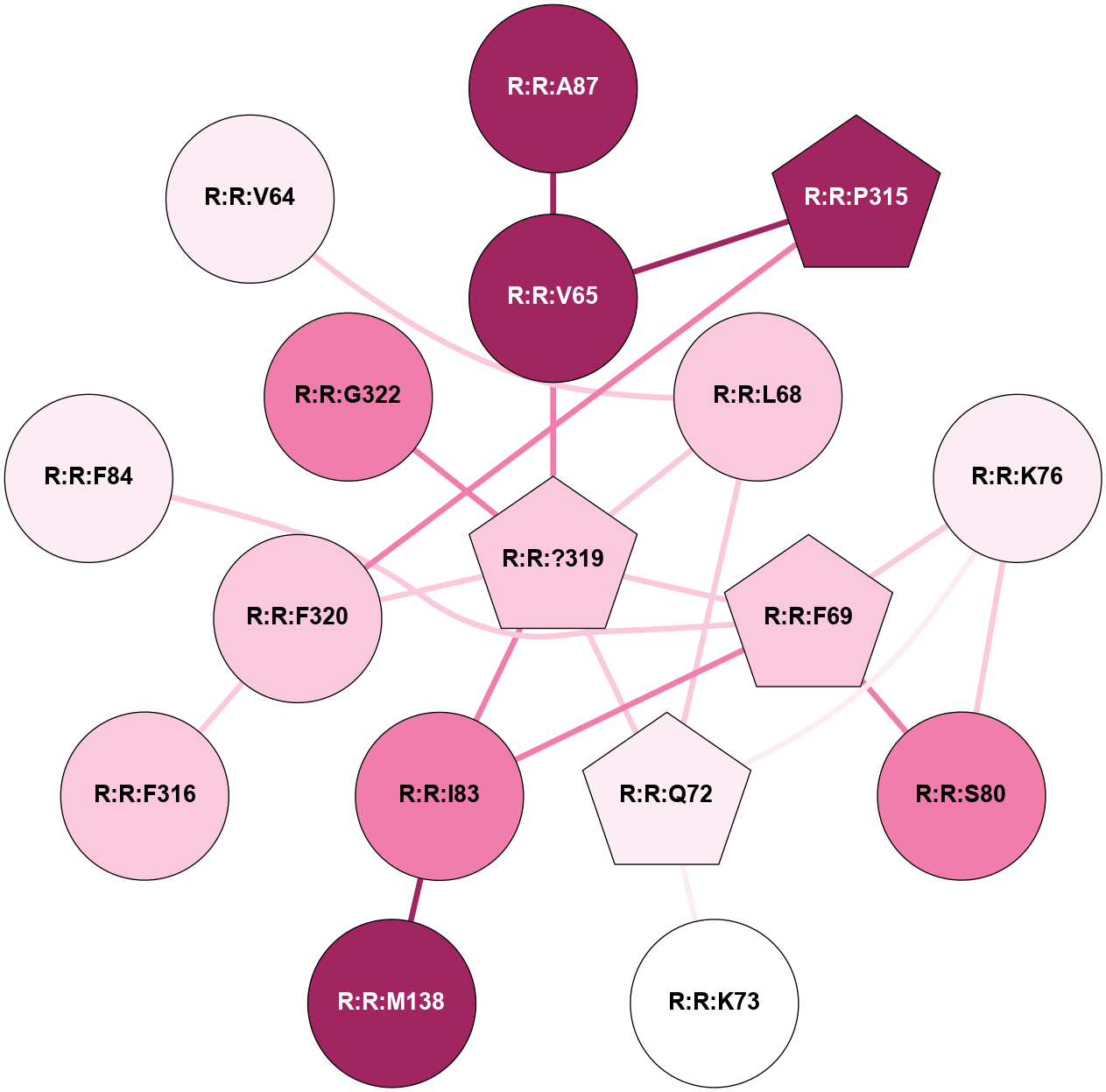
A 2D representation of the interactions of YCM in 7JNI
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:A87 | R:R:V65 | 3.39 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:P315 | R:R:V65 | 3.53 | 0 | Yes | No | 9 | 9 | 2 | 1 | | R:R:?319 | R:R:V65 | 8.88 | 4 | Yes | No | 7 | 9 | 0 | 1 | | R:R:L68 | R:R:Q72 | 3.99 | 4 | No | Yes | 7 | 6 | 1 | 1 | | R:R:?319 | R:R:L68 | 4.72 | 4 | Yes | No | 7 | 7 | 0 | 1 | | R:R:F69 | R:R:K76 | 4.96 | 4 | Yes | No | 7 | 6 | 1 | 2 | | R:R:F69 | R:R:S80 | 3.96 | 4 | Yes | No | 7 | 8 | 1 | 2 | | R:R:F69 | R:R:I83 | 3.77 | 4 | Yes | No | 7 | 8 | 1 | 1 | | R:R:?319 | R:R:F69 | 13.49 | 4 | Yes | Yes | 7 | 7 | 0 | 1 | | R:R:K73 | R:R:Q72 | 6.78 | 0 | No | Yes | 5 | 6 | 2 | 1 | | R:R:K76 | R:R:Q72 | 12.2 | 4 | No | Yes | 6 | 6 | 2 | 1 | | R:R:?319 | R:R:Q72 | 5.67 | 4 | Yes | Yes | 7 | 6 | 0 | 1 | | R:R:K76 | R:R:S80 | 10.71 | 4 | No | No | 6 | 8 | 2 | 2 | | R:R:I83 | R:R:M138 | 4.37 | 4 | No | No | 8 | 9 | 1 | 2 | | R:R:?319 | R:R:I83 | 4.86 | 4 | Yes | No | 7 | 8 | 0 | 1 | | R:R:F320 | R:R:P315 | 4.33 | 0 | No | Yes | 7 | 9 | 1 | 2 | | R:R:F316 | R:R:F320 | 11.79 | 0 | No | No | 7 | 7 | 2 | 1 | | R:R:?319 | R:R:F320 | 7.26 | 4 | Yes | No | 7 | 7 | 0 | 1 | | R:R:F69 | R:R:F84 | 3.22 | 4 | Yes | No | 7 | 6 | 1 | 2 | | R:R:L68 | R:R:V64 | 2.98 | 4 | No | No | 7 | 6 | 1 | 2 | | R:R:?319 | R:R:G322 | 2.91 | 4 | Yes | No | 7 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 6.83 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 21.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7M8W | A | Lipid | Prostanoid | DP2 | Homo Sapiens | 15r-Methyl-Prostaglandin D2 | Na | - | 2.61 | 2021-08-25 | doi.org/10.1073/pnas.2102813118 |
|

A 2D representation of the interactions of YCM in 7M8W
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F244 | R:R:V66 | 11.8 | 3 | Yes | No | 7 | 7 | 1 | 2 | | R:R:F244 | R:R:R243 | 11.76 | 3 | Yes | No | 7 | 7 | 1 | 1 | | R:R:?308 | R:R:R243 | 4.14 | 3 | Yes | No | 8 | 7 | 0 | 1 | | R:R:D310 | R:R:R243 | 5.96 | 3 | Yes | No | 5 | 7 | 1 | 1 | | R:R:F244 | R:R:L247 | 4.87 | 3 | Yes | No | 7 | 7 | 1 | 2 | | R:R:?308 | R:R:F244 | 6.22 | 3 | Yes | Yes | 8 | 7 | 0 | 1 | | R:R:P309 | R:R:R246 | 4.32 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:L247 | R:R:Y304 | 4.69 | 0 | No | Yes | 7 | 9 | 2 | 2 | | R:R:M311 | R:R:Y304 | 3.59 | 0 | No | Yes | 8 | 9 | 1 | 2 | | R:R:?308 | R:R:P309 | 18.18 | 3 | Yes | No | 8 | 5 | 0 | 1 | | R:R:?308 | R:R:D310 | 10.4 | 3 | Yes | Yes | 8 | 5 | 0 | 1 | | R:R:?308 | R:R:M311 | 7.22 | 3 | Yes | No | 8 | 8 | 0 | 1 | | R:R:D310 | R:R:R313 | 4.76 | 3 | Yes | No | 5 | 5 | 1 | 2 | | R:R:M311 | R:R:V305 | 3.04 | 0 | No | No | 8 | 7 | 1 | 2 | | R:R:D310 | R:R:K314 | 2.77 | 3 | Yes | No | 5 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.23 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7P2L | C | Aminoacid | Metabotropic Glutamate | mGlu5 | Homo Sapiens | - | 4YI | - | 2.54 | 2021-09-08 | doi.org/10.1016/j.celrep.2021.109648 |
|
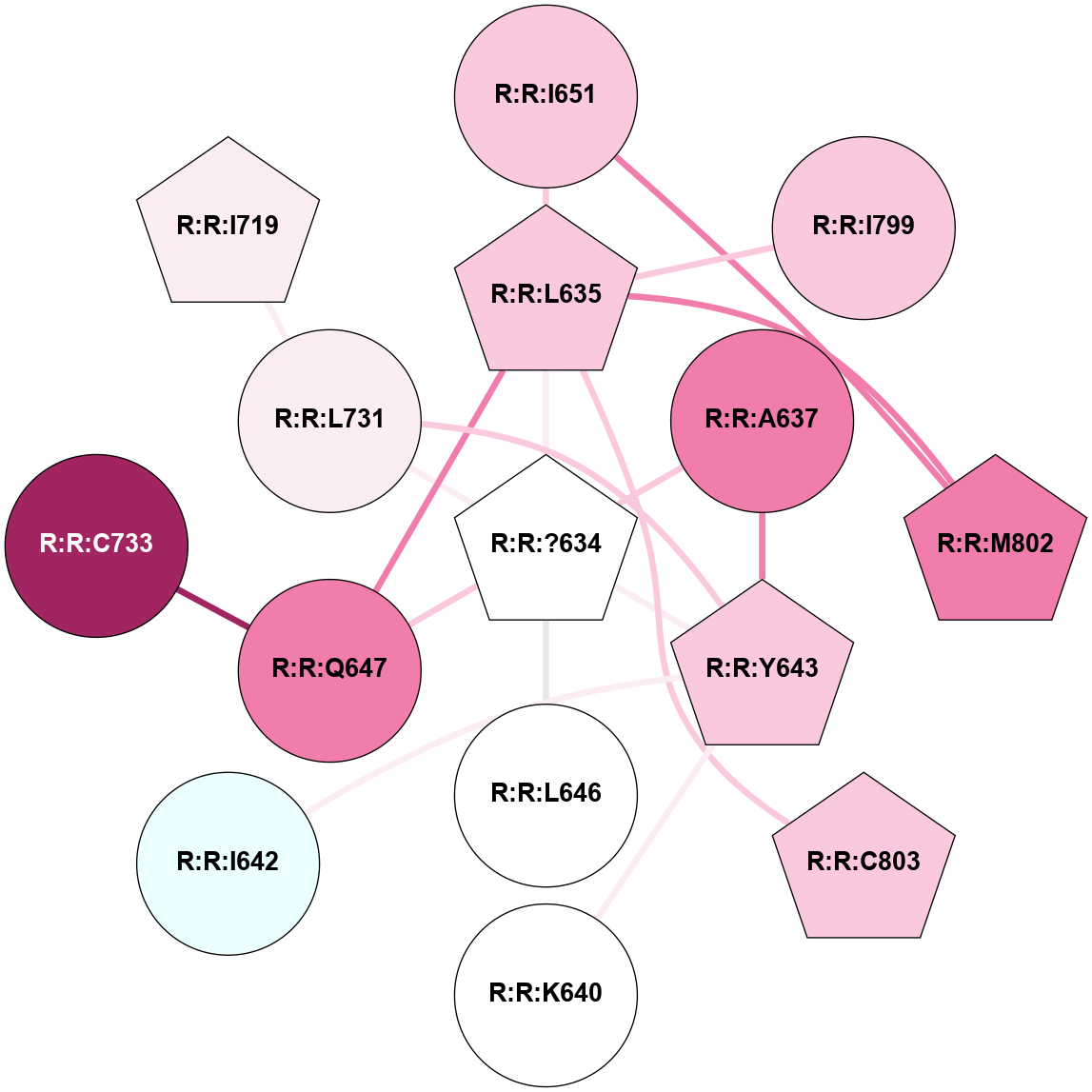
A 2D representation of the interactions of YCM in 7P2L
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?634 | R:R:L635 | 3.54 | 1 | Yes | Yes | 5 | 7 | 0 | 1 | | R:R:?634 | R:R:A637 | 4.03 | 1 | Yes | No | 5 | 8 | 0 | 1 | | R:R:?634 | R:R:Y643 | 3.99 | 1 | Yes | Yes | 5 | 7 | 0 | 1 | | R:R:?634 | R:R:L646 | 10.61 | 1 | Yes | No | 5 | 5 | 0 | 1 | | R:R:?634 | R:R:Q647 | 11.34 | 1 | Yes | No | 5 | 8 | 0 | 1 | | R:R:?634 | R:R:L731 | 3.54 | 1 | Yes | No | 5 | 6 | 0 | 1 | | R:R:L635 | R:R:Q647 | 9.32 | 1 | Yes | No | 7 | 8 | 1 | 1 | | R:R:I651 | R:R:L635 | 5.71 | 1 | No | Yes | 7 | 7 | 2 | 1 | | R:R:I799 | R:R:L635 | 4.28 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:L635 | R:R:M802 | 5.65 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:C803 | R:R:L635 | 4.76 | 0 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:A637 | R:R:Y643 | 5.34 | 1 | No | Yes | 8 | 7 | 1 | 1 | | R:R:K640 | R:R:Y643 | 3.58 | 0 | No | Yes | 5 | 7 | 2 | 1 | | R:R:I642 | R:R:Y643 | 8.46 | 0 | No | Yes | 4 | 7 | 2 | 1 | | R:R:L731 | R:R:Y643 | 3.52 | 1 | No | Yes | 6 | 7 | 1 | 1 | | R:R:C733 | R:R:Q647 | 6.1 | 0 | No | No | 9 | 8 | 2 | 1 | | R:R:I651 | R:R:M802 | 7.29 | 1 | No | Yes | 7 | 8 | 2 | 2 | | R:R:I719 | R:R:L731 | 2.85 | 0 | Yes | No | 6 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 6.18 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of YCM in 7P2L
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E616 | R:R:L694 | 3.98 | 0 | No | No | 7 | 5 | 2 | 1 | | R:R:A692 | R:R:S689 | 3.42 | 0 | No | No | 7 | 7 | 1 | 2 | | R:R:?691 | R:R:L694 | 3.54 | 0 | Yes | No | 3 | 5 | 0 | 1 | | R:R:?691 | R:R:V695 | 3.81 | 0 | Yes | No | 3 | 4 | 0 | 1 | | R:R:I699 | R:R:V695 | 3.07 | 0 | No | No | 3 | 4 | 2 | 1 | | R:R:?691 | R:R:A690 | 2.68 | 0 | Yes | No | 3 | 6 | 0 | 1 | | R:R:?691 | R:R:A692 | 2.68 | 0 | Yes | No | 3 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 3.18 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8E0G | A | Peptide | Opioid | MOP | Mus Musculus | BU72 | - | - | 2.1 | 2023-10-18 | doi.org/10.1186/s12915-023-01689-w |
|

A 2D representation of the interactions of YCM in 8E0G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:?54 | R:R:S53 | 4.53 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:?54 | R:R:S55 | 3.4 | 1 | Yes | Yes | 2 | 4 | 1 | 2 | | R:R:?54 | R:R:?57 | 3.56 | 1 | Yes | Yes | 2 | 5 | 1 | 0 | | R:R:?54 | R:R:I144 | 3.23 | 1 | Yes | Yes | 2 | 5 | 1 | 2 | | R:R:?54 | R:R:Y148 | 5.31 | 1 | Yes | Yes | 2 | 4 | 1 | 2 | | R:R:?54 | R:R:D216 | 7.17 | 1 | Yes | Yes | 2 | 4 | 1 | 2 | | R:R:?54 | R:R:T218 | 5.56 | 1 | Yes | No | 2 | 3 | 1 | 2 | | L:L:?1 | R:R:?54 | 29.77 | 1 | Yes | Yes | 0 | 2 | 2 | 1 | | R:R:D216 | R:R:S55 | 4.42 | 1 | Yes | Yes | 4 | 4 | 2 | 2 | | R:R:?57 | R:R:L56 | 3.54 | 1 | Yes | Yes | 5 | 2 | 0 | 1 | | R:R:L56 | R:R:S214 | 6.01 | 0 | Yes | No | 2 | 2 | 1 | 2 | | R:R:D216 | R:R:L56 | 6.79 | 1 | Yes | Yes | 4 | 2 | 2 | 1 | | R:R:?57 | R:R:P58 | 15.4 | 1 | Yes | No | 5 | 9 | 0 | 1 | | R:R:?57 | R:R:Q59 | 13.62 | 1 | Yes | No | 5 | 2 | 0 | 1 | | R:R:?57 | R:R:K303 | 6.01 | 1 | Yes | Yes | 5 | 4 | 0 | 1 | | R:R:?57 | R:R:W318 | 15.54 | 1 | Yes | Yes | 5 | 5 | 0 | 1 | | R:R:Q59 | R:R:T315 | 9.92 | 0 | No | No | 2 | 4 | 1 | 2 | | L:L:?1 | R:R:I144 | 5.31 | 1 | Yes | Yes | 0 | 5 | 2 | 2 | | L:L:?1 | R:R:Y148 | 8.72 | 1 | Yes | Yes | 0 | 4 | 2 | 2 | | R:R:D216 | R:R:T218 | 13.01 | 1 | Yes | No | 4 | 3 | 2 | 2 | | R:R:I296 | R:R:W318 | 4.7 | 1 | No | Yes | 6 | 5 | 2 | 1 | | L:L:?1 | R:R:I296 | 6.19 | 1 | Yes | No | 0 | 6 | 2 | 2 | | R:R:K303 | R:R:Y299 | 4.78 | 1 | Yes | Yes | 4 | 4 | 1 | 2 | | R:R:W318 | R:R:Y299 | 14.47 | 1 | Yes | Yes | 5 | 4 | 1 | 2 | | R:R:I308 | R:R:K303 | 4.36 | 0 | Yes | Yes | 1 | 4 | 2 | 1 | | R:R:K303 | R:R:W318 | 13.92 | 1 | Yes | Yes | 4 | 5 | 1 | 1 | | R:R:H319 | R:R:W318 | 5.29 | 1 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:I322 | R:R:W318 | 9.4 | 1 | No | Yes | 5 | 5 | 2 | 1 | | L:L:?1 | R:R:W318 | 3.53 | 1 | Yes | Yes | 0 | 5 | 2 | 1 | | L:L:?1 | R:R:I322 | 3.54 | 1 | Yes | No | 0 | 5 | 2 | 2 | | R:R:L56 | R:R:R211 | 2.43 | 0 | Yes | No | 2 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 9.61 | | Average Nodes In Shell | 22.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 31.00 | | Average Links Mediated by Hubs In Shell | 30.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8TF5 | A | Orphan | Orphan | GPR6 | Homo Sapiens | Oleic acid | - | - | 2.1 | 2024-12-04 | doi.org/10.2210/pdb8TF5/pdb |
|

A 2D representation of the interactions of YCM in 8TF5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I248 | R:R:L170 | 4.28 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:?249 | R:R:Y245 | 10.99 | 0 | Yes | Yes | 6 | 9 | 0 | 1 | | R:R:A282 | R:R:Y245 | 4 | 0 | No | Yes | 5 | 9 | 2 | 1 | | R:R:L285 | R:R:Y245 | 7.03 | 0 | No | Yes | 7 | 9 | 2 | 1 | | R:R:?249 | R:R:I248 | 4.87 | 0 | Yes | No | 6 | 9 | 0 | 1 | | R:R:?249 | R:R:W253 | 7.77 | 0 | Yes | No | 6 | 6 | 0 | 1 | | R:R:?249 | R:R:V278 | 5.08 | 0 | Yes | No | 6 | 7 | 0 | 1 | | R:R:I248 | R:R:L281 | 2.85 | 0 | No | No | 9 | 8 | 1 | 2 | | R:R:V252 | R:R:V278 | 1.6 | 0 | No | No | 8 | 7 | 2 | 1 | | R:R:V246 | R:R:Y245 | 1.26 | 0 | No | Yes | 4 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 7.18 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9HC0 | C | Aminoacid | Metabotropic Glutamate | mGlu5 | Homo sapiens | - | 4YI | - | 2.33 | 2025-06-25 | doi.org/10.1002/pro.70104 |
|
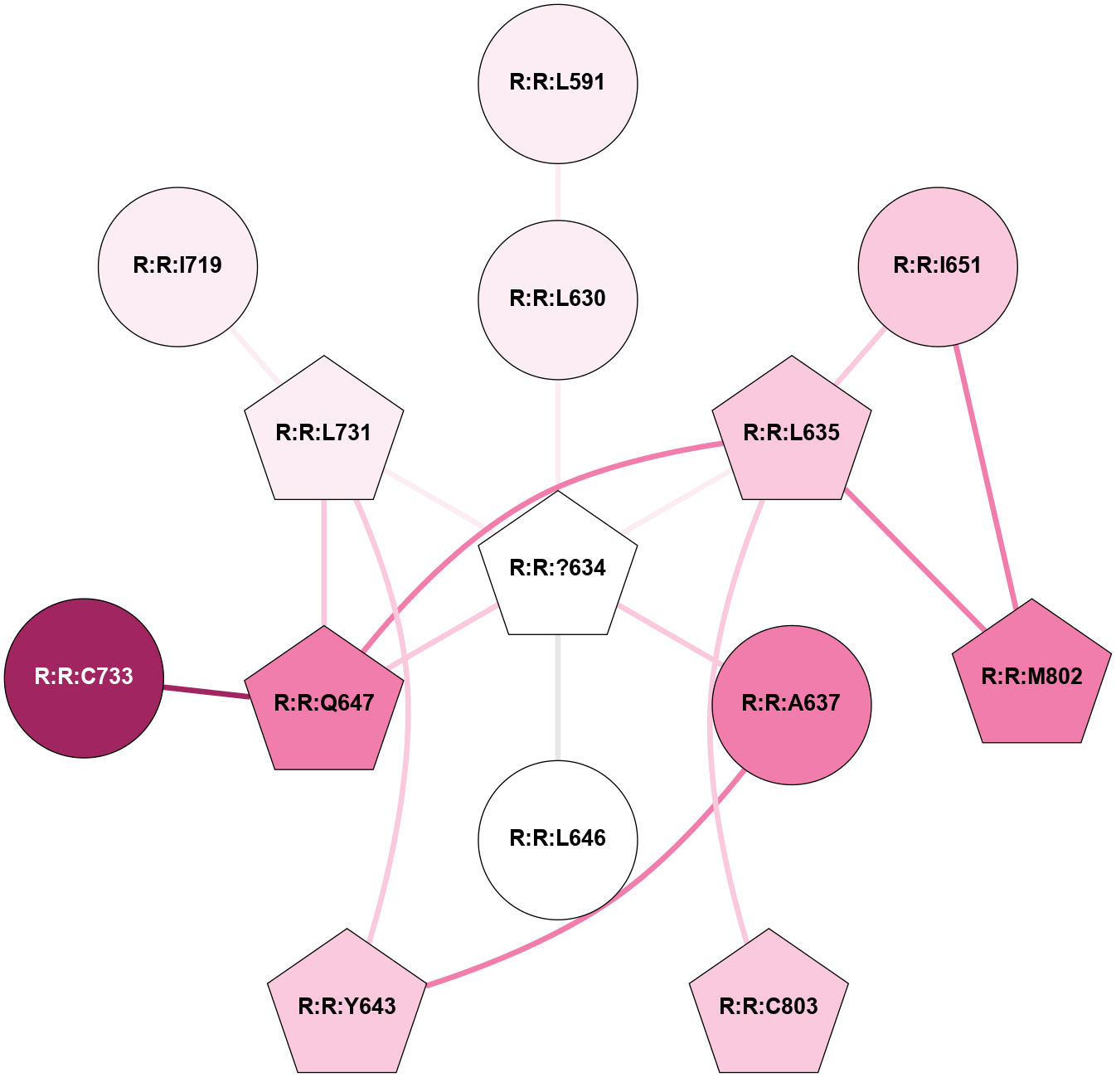
A 2D representation of the interactions of YCM in 9HC0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L591 | R:R:L630 | 5.54 | 0 | No | No | 6 | 6 | 2 | 1 | | R:R:?634 | R:R:L630 | 3.54 | 1 | Yes | No | 5 | 6 | 0 | 1 | | R:R:?634 | R:R:L635 | 3.54 | 1 | Yes | Yes | 5 | 7 | 0 | 1 | | R:R:?634 | R:R:A637 | 5.37 | 1 | Yes | No | 5 | 8 | 0 | 1 | | R:R:?634 | R:R:L646 | 10.62 | 1 | Yes | No | 5 | 5 | 0 | 1 | | R:R:?634 | R:R:Q647 | 10.21 | 1 | Yes | Yes | 5 | 8 | 0 | 1 | | R:R:?634 | R:R:L731 | 3.54 | 1 | Yes | Yes | 5 | 6 | 0 | 1 | | R:R:L635 | R:R:Q647 | 10.65 | 1 | Yes | Yes | 7 | 8 | 1 | 1 | | R:R:I651 | R:R:L635 | 4.28 | 1 | No | Yes | 7 | 7 | 2 | 1 | | R:R:L635 | R:R:M802 | 4.24 | 1 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:C803 | R:R:L635 | 4.76 | 0 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:A637 | R:R:Y643 | 5.34 | 0 | No | Yes | 8 | 7 | 1 | 2 | | R:R:L731 | R:R:Y643 | 3.52 | 1 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:L731 | R:R:Q647 | 3.99 | 1 | Yes | Yes | 6 | 8 | 1 | 1 | | R:R:C733 | R:R:Q647 | 6.1 | 0 | No | Yes | 9 | 8 | 2 | 1 | | R:R:I651 | R:R:M802 | 5.83 | 1 | No | Yes | 7 | 8 | 2 | 2 | | R:R:I719 | R:R:L731 | 4.28 | 0 | No | Yes | 6 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 6.14 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
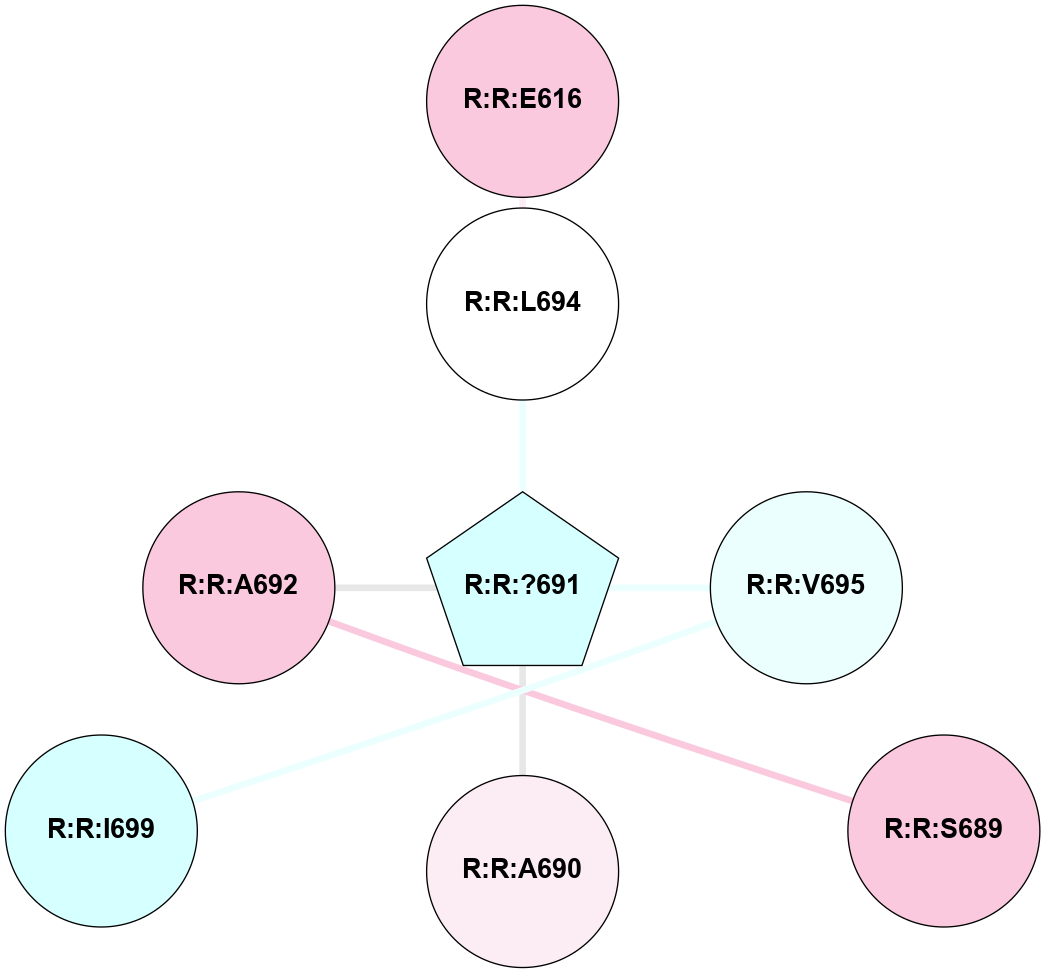
A 2D representation of the interactions of YCM in 9HC0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E616 | R:R:L694 | 5.3 | 0 | No | No | 7 | 5 | 2 | 1 | | R:R:A692 | R:R:S689 | 3.42 | 0 | No | No | 7 | 7 | 1 | 2 | | R:R:?691 | R:R:L694 | 3.54 | 0 | Yes | No | 3 | 5 | 0 | 1 | | R:R:?691 | R:R:V695 | 3.81 | 0 | Yes | No | 3 | 4 | 0 | 1 | | R:R:I699 | R:R:V695 | 3.07 | 0 | No | No | 3 | 4 | 2 | 1 | | R:R:?691 | R:R:A690 | 2.69 | 0 | Yes | No | 3 | 6 | 0 | 1 | | R:R:?691 | R:R:A692 | 2.69 | 0 | Yes | No | 3 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 3.18 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9HC3 | C | Aminoacid | Metabotropic Glutamate | mGlu5 | Homo sapiens | - | - | - | 2.9 | 2025-06-25 | doi.org/10.1002/pro.70104 |
|
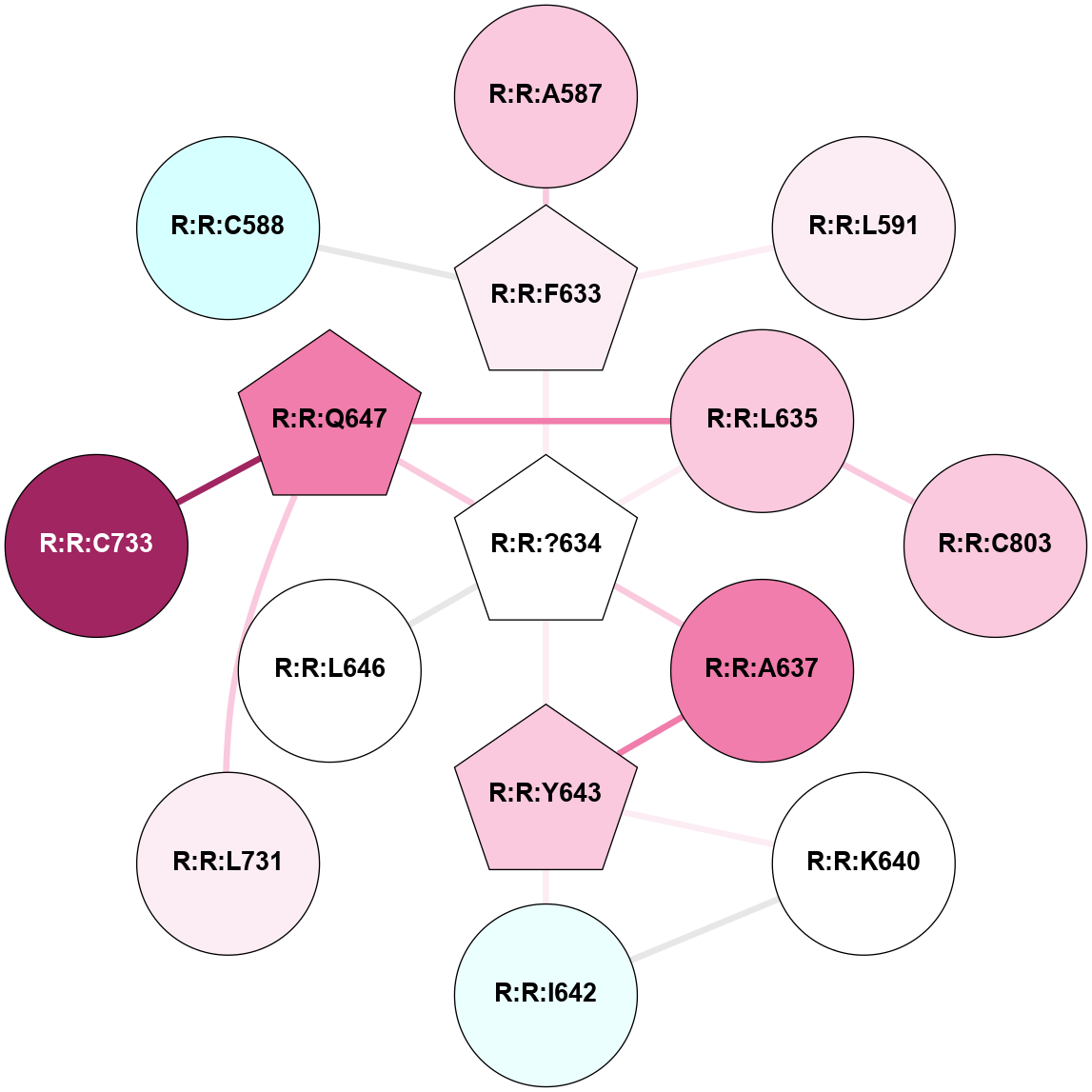
A 2D representation of the interactions of YCM in 9HC3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:A587 | R:R:F633 | 4.16 | 0 | No | Yes | 7 | 6 | 2 | 1 | | R:R:F633 | R:R:L591 | 3.65 | 0 | Yes | No | 6 | 6 | 1 | 2 | | R:R:?634 | R:R:F633 | 3.12 | 1 | Yes | Yes | 5 | 6 | 0 | 1 | | R:R:?634 | R:R:L635 | 3.54 | 1 | Yes | No | 5 | 7 | 0 | 1 | | R:R:?634 | R:R:A637 | 4.03 | 1 | Yes | No | 5 | 8 | 0 | 1 | | R:R:?634 | R:R:Y643 | 7 | 1 | Yes | Yes | 5 | 7 | 0 | 1 | | R:R:?634 | R:R:L646 | 3.54 | 1 | Yes | No | 5 | 5 | 0 | 1 | | R:R:?634 | R:R:Q647 | 9.08 | 1 | Yes | Yes | 5 | 8 | 0 | 1 | | R:R:L635 | R:R:Q647 | 10.65 | 1 | No | Yes | 7 | 8 | 1 | 1 | | R:R:C803 | R:R:L635 | 6.35 | 1 | No | No | 7 | 7 | 2 | 1 | | R:R:A637 | R:R:Y643 | 5.34 | 1 | No | Yes | 8 | 7 | 1 | 1 | | R:R:I642 | R:R:K640 | 4.36 | 1 | No | No | 4 | 5 | 2 | 2 | | R:R:K640 | R:R:Y643 | 4.78 | 1 | No | Yes | 5 | 7 | 2 | 1 | | R:R:I642 | R:R:Y643 | 3.63 | 1 | No | Yes | 4 | 7 | 2 | 1 | | R:R:L731 | R:R:Q647 | 3.99 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:C733 | R:R:Q647 | 6.1 | 0 | No | Yes | 9 | 8 | 2 | 1 | | R:R:C588 | R:R:F633 | 2.79 | 0 | No | Yes | 3 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 5.05 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
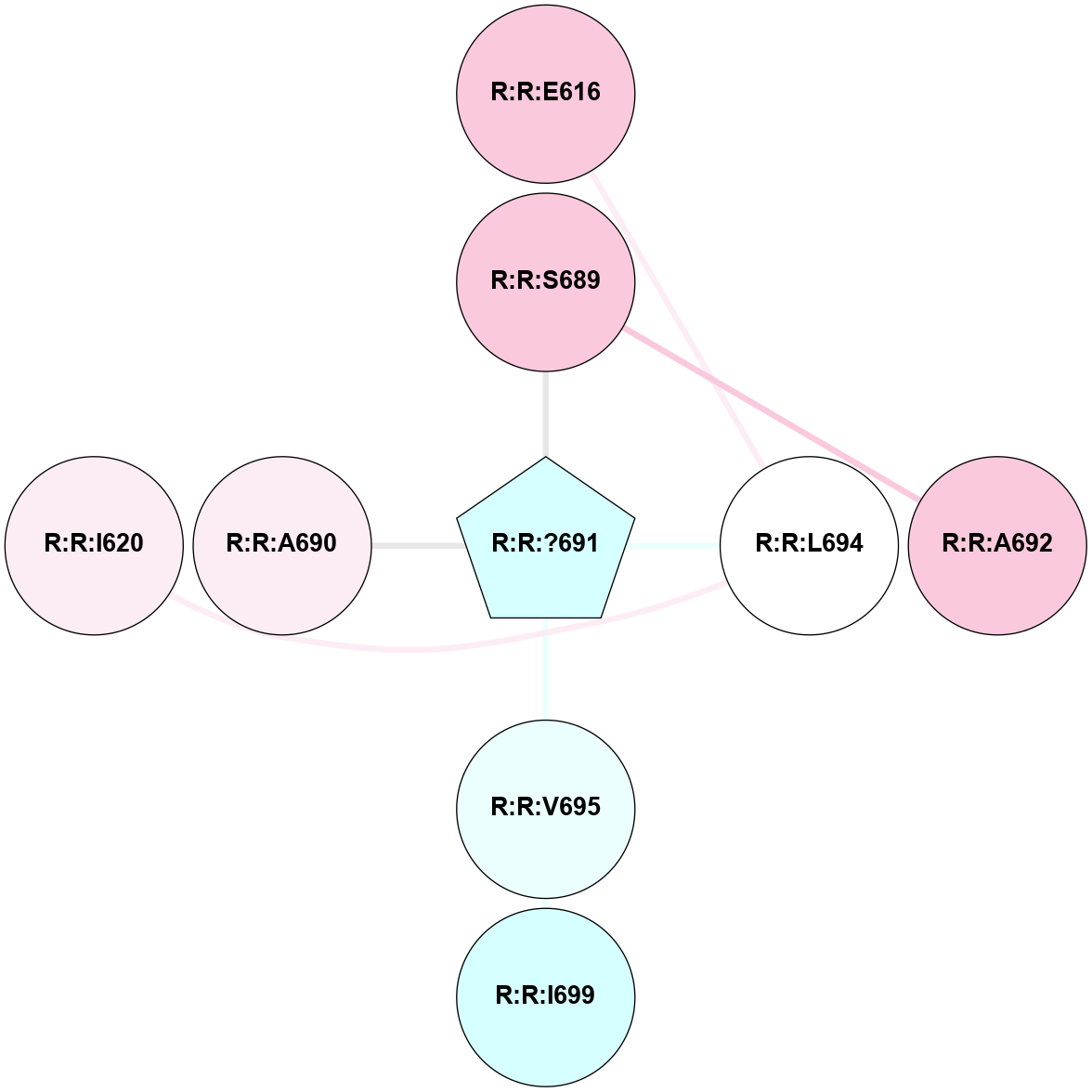
A 2D representation of the interactions of YCM in 9HC3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E616 | R:R:L694 | 5.3 | 0 | No | No | 7 | 5 | 2 | 1 | | R:R:?691 | R:R:S689 | 3.84 | 0 | Yes | No | 3 | 7 | 0 | 1 | | R:R:A692 | R:R:S689 | 3.42 | 0 | No | No | 7 | 7 | 2 | 1 | | R:R:?691 | R:R:L694 | 3.54 | 0 | Yes | No | 3 | 5 | 0 | 1 | | R:R:?691 | R:R:V695 | 3.81 | 0 | Yes | No | 3 | 4 | 0 | 1 | | R:R:I699 | R:R:V695 | 3.07 | 0 | No | No | 3 | 4 | 2 | 1 | | R:R:?691 | R:R:A690 | 2.69 | 0 | Yes | No | 3 | 6 | 0 | 1 | | R:R:I620 | R:R:L694 | 1.43 | 0 | No | No | 6 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 3.47 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|




























