- Press and hold down the left mouse button and move the mouse to rotate.
- Press and hold down the central mouse button and move the mouse to translate.
- Use the mouse wheel to zoom in or out.
- Click on any atom on your molecule and the corresponding residue label will be placed beneath the viewer.
| Color | ConSurf Grade |
| No Conservation data available | |
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 | |
| 6 | |
| 7 | |
| 8 | |
| 9 |
Index: link id, click on each number to highlight the corresponding link in the 3D visualization.
Node1 Node2: the two nodes of the corresponding link.
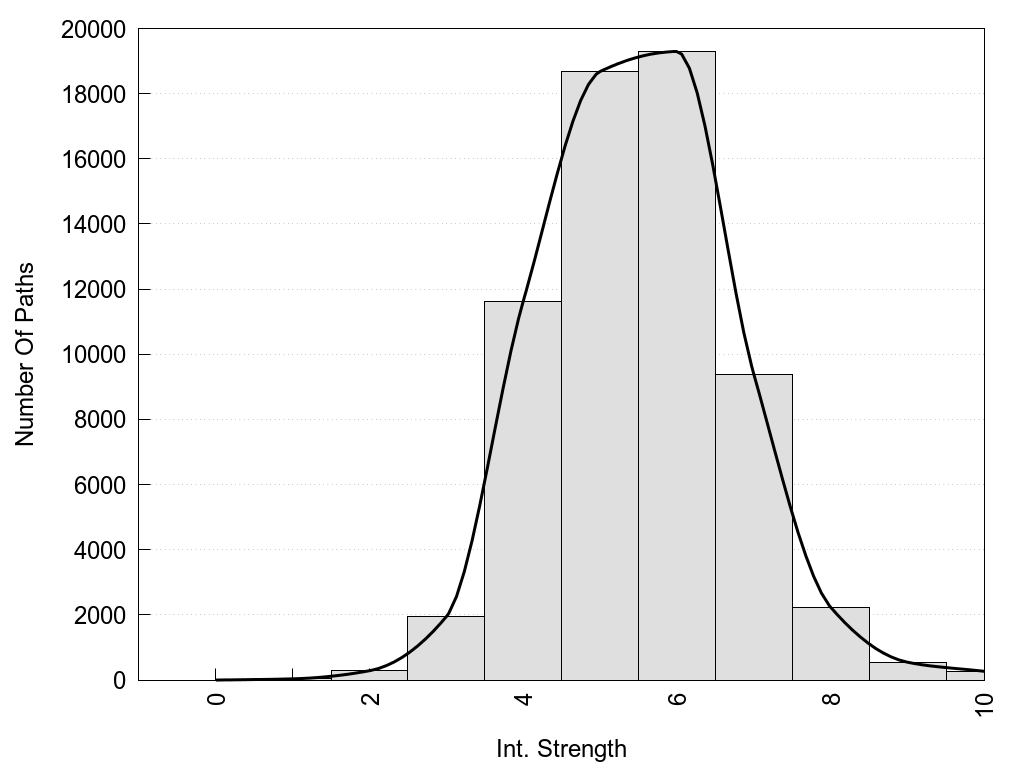
Int. Strength: the interaction strength between the two nodes.
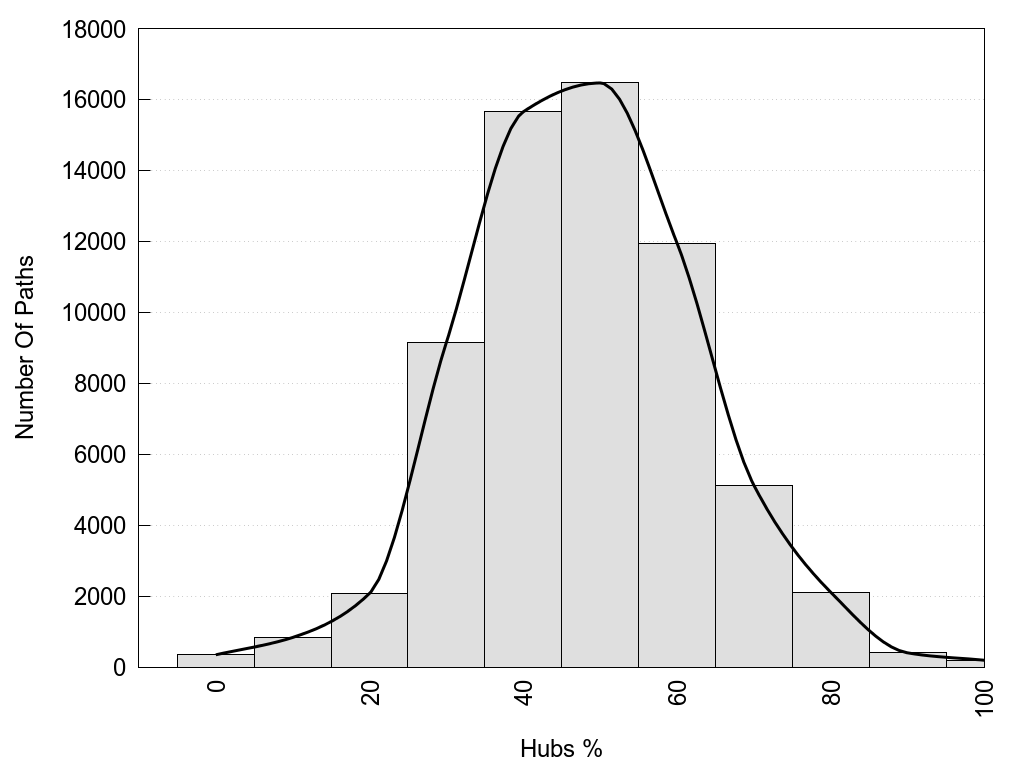
Hub1?, Hub2?: "Yes" if the corresponding node has more than 3 links, otherwise "No".
Community: the id of the community the link belong to, otherwise 0.
ConSurf1, ConSurf2: these columns report the ConSurf conservation grades of the two nodes involved in a link.
| Index | Node1 | Node2 | Int. Strength | Hub1? | Hub2? | Community | ConSurf1 | ConSurf2 |
|---|---|---|---|---|---|---|---|---|
| 1 | L:L:H1 | R:R:L267 | 3.86 | Yes | Yes | 1 | 0 | 6 |
| 2 | L:L:H1 | R:R:H268 | 15.53 | Yes | Yes | 1 | 0 | 7 |
| 3 | L:L:H1 | R:R:V271 | 13.84 | Yes | No | 0 | 0 | 6 |
| 4 | L:L:H1 | R:R:W340 | 8.46 | Yes | Yes | 1 | 0 | 6 |
| 5 | L:L:A2 | R:R:Q421 | 4.55 | No | No | 0 | 0 | 8 |
| 6 | L:L:D3 | R:R:Y186 | 6.9 | No | Yes | 1 | 0 | 7 |
| 7 | L:L:D3 | R:R:V228 | 8.76 | No | Yes | 1 | 0 | 7 |
| 8 | L:L:G4 | L:L:S7 | 3.71 | No | Yes | 0 | 0 | 0 |
| 9 | L:L:F6 | L:L:M10 | 4.98 | Yes | No | 1 | 0 | 0 |
| 10 | L:L:F6 | R:R:L175 | 3.65 | Yes | No | 0 | 0 | 5 |
| 11 | L:L:F6 | R:R:L178 | 4.87 | Yes | No | 1 | 0 | 5 |
| 12 | L:L:F6 | R:R:Q179 | 4.68 | Yes | Yes | 1 | 0 | 5 |
| 13 | L:L:F6 | R:R:Y182 | 12.38 | Yes | Yes | 1 | 0 | 7 |
| 14 | L:L:F6 | R:R:L422 | 3.65 | Yes | No | 1 | 0 | 7 |
| 15 | L:L:N11 | L:L:S7 | 2.98 | Yes | Yes | 1 | 0 | 0 |
| 16 | L:L:S7 | R:R:K231 | 4.59 | Yes | Yes | 1 | 0 | 7 |
| 17 | L:L:S7 | R:R:T332 | 3.2 | Yes | No | 1 | 0 | 6 |
| 18 | L:L:D8 | R:R:T333 | 2.89 | No | No | 0 | 0 | 4 |
| 19 | L:L:D8 | R:R:N334 | 4.04 | No | No | 0 | 0 | 7 |
| 20 | L:L:E9 | R:R:K414 | 6.75 | No | No | 0 | 0 | 5 |
| 21 | L:L:M10 | R:R:Y172 | 4.79 | No | Yes | 0 | 0 | 1 |
| 22 | L:L:M10 | R:R:Q179 | 8.16 | No | Yes | 1 | 0 | 5 |
| 23 | L:L:N11 | R:R:F235 | 13.29 | Yes | Yes | 1 | 0 | 8 |
| 24 | L:L:N11 | R:R:Y239 | 3.49 | Yes | Yes | 0 | 0 | 4 |
| 25 | L:L:N11 | R:R:T332 | 11.7 | Yes | No | 1 | 0 | 6 |
| 26 | L:L:I13 | R:R:R171 | 3.76 | No | No | 0 | 0 | 4 |
| 27 | L:L:I13 | R:R:Y172 | 3.63 | No | Yes | 0 | 0 | 1 |
| 28 | L:L:L14 | R:R:Y172 | 4.69 | No | Yes | 0 | 0 | 1 |
| 29 | L:L:L14 | R:R:Y239 | 7.03 | No | Yes | 0 | 0 | 4 |
| 30 | L:L:N16 | R:R:N168 | 8.17 | No | Yes | 0 | 0 | 3 |
| 31 | L:L:A18 | R:R:R242 | 4.15 | No | No | 0 | 0 | 2 |
| 32 | L:L:R20 | R:R:F165 | 3.21 | No | No | 0 | 0 | 1 |
| 33 | L:L:D21 | R:R:R242 | 19.06 | No | No | 0 | 0 | 2 |
| 34 | L:L:F22 | L:L:I23 | 3.77 | Yes | No | 0 | 0 | 0 |
| 35 | L:L:F22 | L:L:W25 | 4.01 | Yes | Yes | 3 | 0 | 0 |
| 36 | L:L:F22 | L:L:L26 | 4.87 | Yes | No | 3 | 0 | 0 |
| 37 | L:L:F22 | R:R:W249 | 20.04 | Yes | Yes | 3 | 0 | 3 |
| 38 | L:L:L26 | L:L:W25 | 5.69 | No | Yes | 3 | 0 | 0 |
| 39 | L:L:T29 | L:L:W25 | 6.06 | No | Yes | 0 | 0 | 0 |
| 40 | L:L:W25 | R:R:D244 | 5.58 | Yes | No | 0 | 0 | 1 |
| 41 | L:L:W25 | R:R:N247 | 3.39 | Yes | No | 0 | 0 | 4 |
| 42 | L:L:W25 | R:R:W249 | 15.93 | Yes | Yes | 3 | 0 | 3 |
| 43 | R:R:F165 | R:R:N168 | 4.83 | No | Yes | 0 | 1 | 3 |
| 44 | R:R:D170 | R:R:K166 | 6.91 | No | No | 0 | 1 | 3 |
| 45 | R:R:N168 | R:R:Q167 | 3.96 | Yes | No | 0 | 3 | 3 |
| 46 | R:R:L415 | R:R:R171 | 4.86 | Yes | No | 0 | 4 | 4 |
| 47 | R:R:Y172 | R:R:Y236 | 3.97 | Yes | No | 0 | 1 | 4 |
| 48 | R:R:L174 | R:R:L415 | 5.54 | No | Yes | 0 | 4 | 4 |
| 49 | R:R:F419 | R:R:L178 | 10.96 | No | No | 0 | 6 | 5 |
| 50 | R:R:L178 | R:R:L422 | 5.54 | No | No | 1 | 5 | 7 |
| 51 | R:R:Q179 | R:R:Y182 | 4.51 | Yes | Yes | 1 | 5 | 7 |
| 52 | R:R:D232 | R:R:Q179 | 6.53 | No | Yes | 1 | 8 | 5 |
| 53 | R:R:Y182 | R:R:Y186 | 5.96 | Yes | Yes | 1 | 7 | 7 |
| 54 | R:R:D232 | R:R:Y182 | 5.75 | No | Yes | 1 | 8 | 7 |
| 55 | R:R:L422 | R:R:Y182 | 3.52 | No | Yes | 1 | 7 | 7 |
| 56 | R:R:L229 | R:R:T183 | 2.95 | No | No | 0 | 7 | 7 |
| 57 | R:R:G185 | R:R:S426 | 3.71 | No | No | 0 | 8 | 9 |
| 58 | R:R:F427 | R:R:G185 | 4.52 | No | No | 0 | 8 | 8 |
| 59 | R:R:R224 | R:R:Y186 | 7.2 | Yes | Yes | 1 | 8 | 7 |
| 60 | R:R:T225 | R:R:Y186 | 8.74 | No | Yes | 0 | 8 | 7 |
| 61 | R:R:V228 | R:R:Y186 | 6.31 | Yes | Yes | 1 | 7 | 7 |
| 62 | R:R:S425 | R:R:Y186 | 3.82 | No | Yes | 1 | 6 | 7 |
| 63 | R:R:S426 | R:R:Y186 | 5.09 | No | Yes | 0 | 9 | 7 |
| 64 | R:R:F188 | R:R:F427 | 3.22 | No | No | 0 | 4 | 8 |
| 65 | R:R:F194 | R:R:L190 | 4.87 | No | No | 5 | 4 | 7 |
| 66 | R:R:I222 | R:R:L190 | 4.28 | Yes | No | 5 | 7 | 7 |
| 67 | R:R:L190 | R:R:T225 | 7.37 | No | No | 0 | 7 | 8 |
| 68 | R:R:F430 | R:R:S192 | 5.28 | No | No | 0 | 7 | 6 |
| 69 | R:R:F218 | R:R:L193 | 3.65 | Yes | Yes | 0 | 9 | 9 |
| 70 | R:R:F221 | R:R:L193 | 7.31 | Yes | Yes | 0 | 8 | 9 |
| 71 | R:R:I222 | R:R:L193 | 4.28 | Yes | Yes | 0 | 7 | 9 |
| 72 | R:R:A433 | R:R:L193 | 3.15 | No | Yes | 0 | 8 | 9 |
| 73 | R:R:F194 | R:R:I222 | 13.82 | No | Yes | 5 | 4 | 7 |
| 74 | R:R:F218 | R:R:L200 | 3.65 | Yes | No | 0 | 9 | 8 |
| 75 | R:R:L201 | R:R:M215 | 4.24 | No | No | 0 | 3 | 6 |
| 76 | R:R:F202 | R:R:L203 | 3.65 | No | No | 0 | 3 | 8 |
| 77 | R:R:L203 | R:R:L206 | 11.07 | No | Yes | 2 | 8 | 9 |
| 78 | R:R:H207 | R:R:R204 | 5.64 | No | No | 0 | 7 | 8 |
| 79 | R:R:K205 | R:R:L206 | 5.64 | No | Yes | 2 | 7 | 9 |
| 80 | R:R:E446 | R:R:K205 | 18.9 | No | No | 2 | 8 | 7 |
| 81 | R:R:E442 | R:R:L206 | 15.9 | No | Yes | 0 | 9 | 9 |
| 82 | R:R:E446 | R:R:L206 | 3.98 | No | Yes | 2 | 8 | 9 |
| 83 | R:R:H207 | R:R:Y212 | 13.07 | No | No | 0 | 7 | 6 |
| 84 | R:R:C208 | R:R:R210 | 4.18 | No | Yes | 2 | 8 | 9 |
| 85 | R:R:C208 | R:R:N211 | 3.15 | No | No | 2 | 8 | 9 |
| 86 | R:R:C208 | R:R:E296 | 3.04 | No | No | 0 | 8 | 7 |
| 87 | R:R:I213 | R:R:T209 | 3.04 | No | No | 2 | 9 | 7 |
| 88 | R:R:T209 | R:R:Y284 | 8.74 | No | Yes | 2 | 7 | 9 |
| 89 | R:R:N211 | R:R:R210 | 8.44 | No | Yes | 2 | 9 | 9 |
| 90 | R:R:H214 | R:R:R210 | 4.51 | No | Yes | 0 | 9 | 9 |
| 91 | R:R:R210 | R:R:Y284 | 5.14 | Yes | Yes | 2 | 9 | 9 |
| 92 | R:R:M215 | R:R:Y212 | 3.59 | No | No | 0 | 6 | 6 |
| 93 | R:R:L299 | R:R:Y212 | 3.52 | No | No | 0 | 4 | 6 |
| 94 | R:R:E281 | R:R:I213 | 6.83 | Yes | No | 0 | 9 | 9 |
| 95 | R:R:I213 | R:R:Y284 | 4.84 | No | Yes | 2 | 9 | 9 |
| 96 | R:R:E281 | R:R:H214 | 12.31 | Yes | No | 0 | 9 | 9 |
| 97 | R:R:H214 | R:R:Y436 | 8.71 | No | No | 0 | 9 | 8 |
| 98 | R:R:N216 | R:R:W277 | 18.08 | No | Yes | 6 | 9 | 9 |
| 99 | R:R:N216 | R:R:W308 | 6.78 | No | Yes | 6 | 9 | 9 |
| 100 | R:R:F221 | R:R:L217 | 4.87 | Yes | No | 0 | 8 | 9 |
| 101 | R:R:L217 | R:R:L278 | 5.54 | No | No | 0 | 9 | 9 |
| 102 | R:R:E281 | R:R:L217 | 5.3 | Yes | No | 0 | 9 | 9 |
| 103 | R:R:F218 | R:R:V432 | 3.93 | Yes | No | 0 | 9 | 9 |
| 104 | R:R:N274 | R:R:S220 | 5.96 | No | No | 0 | 9 | 9 |
| 105 | R:R:S220 | R:R:W308 | 3.71 | No | Yes | 0 | 9 | 9 |
| 106 | R:R:F221 | R:R:R224 | 3.21 | Yes | Yes | 1 | 8 | 8 |
| 107 | R:R:F221 | R:R:N274 | 15.71 | Yes | No | 1 | 8 | 9 |
| 108 | R:R:F221 | R:R:H428 | 9.05 | Yes | Yes | 0 | 8 | 9 |
| 109 | R:R:F221 | R:R:G429 | 9.03 | Yes | No | 0 | 8 | 9 |
| 110 | R:R:F221 | R:R:V432 | 3.93 | Yes | No | 0 | 8 | 9 |
| 111 | R:R:F270 | R:R:L223 | 9.74 | Yes | No | 0 | 5 | 7 |
| 112 | R:R:R224 | R:R:V271 | 3.92 | Yes | No | 0 | 8 | 6 |
| 113 | R:R:N274 | R:R:R224 | 7.23 | No | Yes | 1 | 9 | 8 |
| 114 | R:R:R224 | R:R:S425 | 3.95 | Yes | No | 1 | 8 | 6 |
| 115 | R:R:A227 | R:R:F270 | 5.55 | No | Yes | 0 | 6 | 5 |
| 116 | R:R:K231 | R:R:V228 | 3.04 | Yes | Yes | 1 | 7 | 7 |
| 117 | R:R:L267 | R:R:V228 | 2.98 | Yes | Yes | 1 | 6 | 7 |
| 118 | R:R:D232 | R:R:K231 | 8.3 | No | Yes | 1 | 8 | 7 |
| 119 | R:R:F235 | R:R:K231 | 6.2 | Yes | Yes | 1 | 8 | 7 |
| 120 | R:R:K231 | R:R:V263 | 3.04 | Yes | No | 1 | 7 | 5 |
| 121 | R:R:K231 | R:R:L267 | 4.23 | Yes | Yes | 1 | 7 | 6 |
| 122 | R:R:V234 | R:R:V263 | 3.21 | No | No | 1 | 4 | 5 |
| 123 | R:R:C260 | R:R:F235 | 4.19 | No | Yes | 1 | 9 | 8 |
| 124 | R:R:C330 | R:R:F235 | 4.19 | Yes | Yes | 1 | 9 | 8 |
| 125 | R:R:F235 | R:R:T332 | 3.89 | Yes | No | 1 | 8 | 6 |
| 126 | R:R:M256 | R:R:S238 | 4.6 | No | No | 0 | 6 | 4 |
| 127 | R:R:C330 | R:R:Y239 | 5.38 | Yes | Yes | 0 | 9 | 4 |
| 128 | R:R:K241 | R:R:Y252 | 16.72 | No | Yes | 0 | 3 | 3 |
| 129 | R:R:E255 | R:R:K241 | 10.8 | No | No | 0 | 4 | 3 |
| 130 | R:R:R242 | R:R:Y252 | 4.12 | No | Yes | 0 | 2 | 3 |
| 131 | R:R:P243 | R:R:W249 | 10.81 | No | Yes | 3 | 3 | 3 |
| 132 | R:R:P243 | R:R:Y252 | 4.17 | No | Yes | 3 | 3 | 3 |
| 133 | R:R:E246 | R:R:N245 | 15.77 | No | No | 0 | 5 | 4 |
| 134 | R:R:N245 | R:R:S251 | 4.47 | No | No | 0 | 4 | 2 |
| 135 | R:R:W249 | R:R:Y252 | 5.79 | Yes | Yes | 3 | 3 | 3 |
| 136 | R:R:C330 | R:R:M256 | 4.86 | Yes | No | 0 | 9 | 6 |
| 137 | R:R:C260 | R:R:C330 | 7.28 | No | Yes | 1 | 9 | 9 |
| 138 | R:R:N327 | R:R:R261 | 12.05 | No | No | 7 | 7 | 8 |
| 139 | R:R:R261 | R:R:W331 | 26.99 | No | Yes | 7 | 8 | 9 |
| 140 | R:R:L267 | R:R:Q264 | 5.32 | Yes | No | 0 | 6 | 6 |
| 141 | R:R:H268 | R:R:Q264 | 11.13 | Yes | No | 0 | 7 | 6 |
| 142 | R:R:F270 | R:R:L266 | 3.65 | Yes | No | 0 | 5 | 5 |
| 143 | R:R:H268 | R:R:W318 | 9.52 | Yes | Yes | 1 | 7 | 8 |
| 144 | R:R:H268 | R:R:W340 | 7.41 | Yes | Yes | 1 | 7 | 6 |
| 145 | R:R:H268 | R:R:I343 | 3.98 | Yes | No | 1 | 7 | 7 |
| 146 | R:R:F270 | R:R:Y269 | 8.25 | Yes | Yes | 0 | 5 | 8 |
| 147 | R:R:P311 | R:R:Y269 | 8.34 | No | Yes | 0 | 9 | 8 |
| 148 | R:R:V315 | R:R:Y269 | 3.79 | No | Yes | 0 | 6 | 8 |
| 149 | R:R:M347 | R:R:V271 | 4.56 | No | No | 0 | 7 | 6 |
| 150 | R:R:F314 | R:R:G272 | 3.01 | Yes | No | 0 | 6 | 5 |
| 151 | R:R:M347 | R:R:Y275 | 10.78 | No | No | 0 | 7 | 7 |
| 152 | R:R:F310 | R:R:L276 | 6.09 | No | No | 0 | 3 | 7 |
| 153 | R:R:L276 | R:R:P311 | 3.28 | No | No | 0 | 7 | 9 |
| 154 | R:R:V280 | R:R:W277 | 3.68 | No | Yes | 0 | 8 | 9 |
| 155 | R:R:L304 | R:R:W277 | 3.42 | No | Yes | 0 | 5 | 9 |
| 156 | R:R:G307 | R:R:W277 | 8.44 | No | Yes | 0 | 9 | 9 |
| 157 | R:R:W277 | R:R:W308 | 18.74 | Yes | Yes | 6 | 9 | 9 |
| 158 | R:R:L279 | R:R:L283 | 4.15 | No | No | 0 | 8 | 6 |
| 159 | R:R:C350 | R:R:L279 | 4.76 | No | No | 0 | 7 | 8 |
| 160 | R:R:L279 | R:R:V353 | 2.98 | No | No | 0 | 8 | 6 |
| 161 | R:R:L306 | R:R:V280 | 2.98 | No | No | 0 | 7 | 8 |
| 162 | R:R:E281 | R:R:L393 | 9.28 | Yes | No | 0 | 9 | 9 |
| 163 | R:R:L288 | R:R:Y284 | 5.86 | No | Yes | 0 | 8 | 9 |
| 164 | R:R:Y284 | R:R:Y303 | 2.98 | Yes | No | 0 | 9 | 7 |
| 165 | R:R:I361 | R:R:L285 | 5.71 | No | No | 0 | 9 | 9 |
| 166 | R:R:E290 | R:R:H286 | 4.92 | No | No | 0 | 6 | 8 |
| 167 | R:R:H286 | R:R:I357 | 6.63 | No | No | 0 | 8 | 8 |
| 168 | R:R:H286 | R:R:K360 | 9.17 | No | No | 0 | 8 | 8 |
| 169 | R:R:R297 | R:R:T287 | 5.17 | Yes | No | 0 | 5 | 7 |
| 170 | R:R:L288 | R:R:V293 | 4.47 | No | No | 0 | 8 | 7 |
| 171 | R:R:E290 | R:R:L289 | 5.3 | No | No | 0 | 6 | 8 |
| 172 | R:R:L289 | R:R:L364 | 4.15 | No | No | 0 | 8 | 8 |
| 173 | R:R:L294 | R:R:R297 | 10.93 | No | Yes | 0 | 6 | 5 |
| 174 | R:R:R297 | R:R:Y303 | 4.12 | Yes | No | 0 | 5 | 7 |
| 175 | R:R:R298 | R:R:W300 | 16.99 | No | No | 0 | 5 | 6 |
| 176 | R:R:L299 | R:R:Y303 | 4.69 | No | No | 0 | 4 | 7 |
| 177 | R:R:A309 | R:R:L305 | 3.15 | No | No | 0 | 5 | 3 |
| 178 | R:R:F310 | R:R:L306 | 4.87 | No | No | 0 | 3 | 7 |
| 179 | R:R:F310 | R:R:F314 | 4.29 | No | Yes | 0 | 3 | 6 |
| 180 | R:R:F314 | R:R:P311 | 4.33 | Yes | No | 0 | 6 | 9 |
| 181 | R:R:L313 | R:R:V312 | 2.98 | No | No | 0 | 4 | 3 |
| 182 | R:R:F314 | R:R:V315 | 3.93 | Yes | No | 0 | 6 | 6 |
| 183 | R:R:F314 | R:R:P346 | 11.56 | Yes | No | 0 | 6 | 9 |
| 184 | R:R:P317 | R:R:V316 | 3.53 | No | No | 0 | 4 | 3 |
| 185 | R:R:F320 | R:R:V316 | 5.24 | No | No | 0 | 4 | 3 |
| 186 | R:R:I339 | R:R:W318 | 5.87 | No | Yes | 0 | 4 | 8 |
| 187 | R:R:I343 | R:R:W318 | 10.57 | No | Yes | 1 | 7 | 8 |
| 188 | R:R:R322 | R:R:W331 | 11 | No | Yes | 0 | 8 | 9 |
| 189 | R:R:E326 | R:R:L325 | 9.28 | No | No | 8 | 6 | 3 |
| 190 | R:R:L325 | R:R:N336 | 4.12 | No | Yes | 8 | 3 | 6 |
| 191 | R:R:E326 | R:R:N336 | 3.94 | No | Yes | 8 | 6 | 6 |
| 192 | R:R:N327 | R:R:W331 | 6.78 | No | Yes | 7 | 7 | 9 |
| 193 | R:R:T328 | R:R:T333 | 4.71 | No | No | 0 | 4 | 4 |
| 194 | R:R:N334 | R:R:W340 | 11.3 | No | Yes | 0 | 7 | 6 |
| 195 | R:R:K338 | R:R:N336 | 5.6 | No | Yes | 0 | 2 | 6 |
| 196 | R:R:I339 | R:R:N336 | 5.66 | No | Yes | 0 | 4 | 6 |
| 197 | R:R:I343 | R:R:W340 | 3.52 | No | Yes | 1 | 7 | 6 |
| 198 | R:R:R344 | R:R:W340 | 14.99 | No | Yes | 1 | 6 | 6 |
| 199 | R:R:R344 | R:R:W341 | 15.99 | No | No | 1 | 6 | 7 |
| 200 | R:R:D406 | R:R:W341 | 6.7 | No | No | 1 | 5 | 7 |
| 201 | R:R:D406 | R:R:R344 | 11.91 | No | No | 1 | 5 | 6 |
| 202 | R:R:G345 | R:R:P346 | 4.06 | No | No | 0 | 4 | 9 |
| 203 | R:R:C350 | R:R:L349 | 4.76 | No | No | 0 | 7 | 3 |
| 204 | R:R:I399 | R:R:V351 | 3.07 | No | No | 0 | 6 | 7 |
| 205 | R:R:S402 | R:R:V351 | 4.85 | No | No | 0 | 6 | 7 |
| 206 | R:R:F356 | R:R:T352 | 10.38 | No | No | 0 | 3 | 4 |
| 207 | R:R:I357 | R:R:V353 | 3.07 | No | No | 0 | 8 | 6 |
| 208 | R:R:L394 | R:R:N354 | 6.87 | Yes | No | 0 | 9 | 9 |
| 209 | R:R:F355 | R:R:L359 | 8.53 | No | No | 0 | 6 | 5 |
| 210 | R:R:I357 | R:R:I361 | 2.94 | No | No | 0 | 8 | 9 |
| 211 | R:R:F358 | R:R:L390 | 6.09 | No | No | 0 | 9 | 9 |
| 212 | R:R:F358 | R:R:V396 | 3.93 | No | No | 0 | 9 | 7 |
| 213 | R:R:L362 | R:R:T387 | 4.42 | No | No | 0 | 7 | 9 |
| 214 | R:R:K368 | R:R:L364 | 7.05 | No | No | 0 | 9 | 8 |
| 215 | R:R:L365 | R:R:S386 | 3 | No | No | 0 | 9 | 9 |
| 216 | R:R:I366 | R:R:K370 | 5.82 | No | No | 9 | 6 | 6 |
| 217 | R:R:I366 | R:R:Y379 | 3.63 | No | Yes | 9 | 6 | 7 |
| 218 | R:R:I366 | R:R:L383 | 5.71 | No | No | 0 | 6 | 9 |
| 219 | R:R:L369 | R:R:Y379 | 7.03 | No | Yes | 0 | 8 | 7 |
| 220 | R:R:L369 | R:R:R382 | 3.64 | No | No | 0 | 8 | 8 |
| 221 | R:R:K370 | R:R:Y379 | 5.97 | No | Yes | 9 | 6 | 7 |
| 222 | R:R:M374 | R:R:Y379 | 3.59 | No | Yes | 0 | 5 | 7 |
| 223 | R:R:C375 | R:R:D378 | 4.67 | No | No | 0 | 5 | 5 |
| 224 | R:R:F376 | R:R:Y379 | 5.16 | No | Yes | 0 | 7 | 7 |
| 225 | R:R:F376 | R:R:K380 | 12.41 | No | No | 0 | 7 | 7 |
| 226 | R:R:K380 | R:R:R377 | 4.95 | No | No | 0 | 7 | 6 |
| 227 | R:R:K385 | R:R:Y381 | 3.58 | No | No | 0 | 8 | 7 |
| 228 | R:R:K385 | R:R:R382 | 6.19 | No | No | 0 | 8 | 8 |
| 229 | R:R:Q435 | R:R:V389 | 11.46 | No | No | 0 | 8 | 8 |
| 230 | R:R:I391 | R:R:P392 | 3.39 | No | No | 0 | 8 | 9 |
| 231 | R:R:H428 | R:R:P392 | 3.05 | Yes | No | 0 | 9 | 9 |
| 232 | R:R:L393 | R:R:L394 | 4.15 | No | Yes | 0 | 9 | 9 |
| 233 | R:R:L393 | R:R:Y436 | 19.93 | No | No | 0 | 9 | 8 |
| 234 | R:R:L394 | R:R:V396 | 2.98 | Yes | No | 0 | 9 | 7 |
| 235 | R:R:G395 | R:R:H428 | 3.18 | No | Yes | 0 | 9 | 9 |
| 236 | R:R:E398 | R:R:H397 | 19.69 | No | No | 0 | 8 | 8 |
| 237 | R:R:H397 | R:R:L424 | 5.14 | No | No | 0 | 8 | 6 |
| 238 | R:R:H397 | R:R:H428 | 10.75 | No | Yes | 0 | 8 | 9 |
| 239 | R:R:F401 | R:R:L400 | 8.53 | No | No | 0 | 6 | 6 |
| 240 | R:R:F401 | R:R:I420 | 3.77 | No | No | 0 | 6 | 5 |
| 241 | R:R:F401 | R:R:Q421 | 9.37 | No | No | 0 | 6 | 8 |
| 242 | R:R:I404 | R:R:Q408 | 8.23 | No | No | 0 | 5 | 3 |
| 243 | R:R:I404 | R:R:R417 | 5.01 | No | Yes | 0 | 5 | 8 |
| 244 | R:R:D407 | R:R:T405 | 2.89 | No | No | 0 | 4 | 8 |
| 245 | R:R:Q408 | R:R:T405 | 5.67 | No | No | 0 | 3 | 8 |
| 246 | R:R:D406 | R:R:D407 | 5.32 | No | No | 0 | 5 | 4 |
| 247 | R:R:F412 | R:R:L415 | 3.65 | No | Yes | 0 | 1 | 4 |
| 248 | R:R:K414 | R:R:R417 | 4.95 | No | Yes | 4 | 5 | 8 |
| 249 | R:R:K414 | R:R:L418 | 5.64 | No | No | 4 | 5 | 5 |
| 250 | R:R:F419 | R:R:L415 | 12.18 | No | Yes | 0 | 6 | 4 |
| 251 | R:R:F419 | R:R:I416 | 5.02 | No | No | 0 | 6 | 5 |
| 252 | R:R:L418 | R:R:R417 | 3.64 | No | Yes | 4 | 5 | 8 |
| 253 | R:R:Q421 | R:R:R417 | 4.67 | No | Yes | 0 | 8 | 8 |
| 254 | R:R:F427 | R:R:F430 | 4.29 | No | No | 0 | 8 | 7 |
| 255 | R:R:Q435 | R:R:Y436 | 5.64 | No | No | 0 | 8 | 8 |
| 256 | R:R:N440 | R:R:V443 | 5.91 | No | Yes | 0 | 9 | 9 |
| 257 | R:R:E446 | R:R:K449 | 5.4 | No | No | 2 | 8 | 7 |
| 258 | R:R:K449 | R:R:Y450 | 3.58 | No | No | 2 | 7 | 5 |
| 259 | R:R:K449 | R:R:R453 | 6.19 | No | No | 2 | 7 | 7 |
| 260 | R:R:R453 | R:R:Y450 | 6.17 | No | No | 2 | 7 | 5 |
| 261 | R:R:P301 | R:R:R298 | 2.88 | No | No | 0 | 3 | 5 |
| 262 | R:R:L197 | R:R:M215 | 2.83 | No | No | 0 | 6 | 6 |
| 263 | R:R:A196 | R:R:F218 | 2.77 | No | Yes | 0 | 9 | 9 |
| 264 | R:R:A196 | R:R:F438 | 2.77 | No | Yes | 0 | 9 | 9 |
| 265 | R:R:L206 | R:R:N211 | 2.75 | Yes | No | 2 | 9 | 9 |
| 266 | L:L:I27 | L:L:Q28 | 2.74 | No | No | 0 | 0 | 0 |
| 267 | L:L:S5 | R:R:R417 | 2.64 | No | Yes | 0 | 0 | 8 |
| 268 | R:R:R382 | R:R:S386 | 2.64 | No | No | 0 | 8 | 9 |
| 269 | R:R:F355 | R:R:V396 | 2.62 | No | No | 0 | 6 | 7 |
| 270 | R:R:R453 | R:R:V452 | 2.62 | No | No | 0 | 7 | 4 |
| 271 | R:R:A219 | R:R:W308 | 2.59 | No | Yes | 0 | 4 | 9 |
| 272 | R:R:A273 | R:R:W308 | 2.59 | No | Yes | 0 | 7 | 9 |
| 273 | R:R:R322 | R:R:T328 | 2.59 | No | No | 0 | 8 | 4 |
| 274 | R:R:A323 | R:R:W331 | 2.59 | No | Yes | 0 | 4 | 9 |
| 275 | R:R:S189 | R:R:Y186 | 2.54 | No | Yes | 0 | 9 | 7 |
| 276 | R:R:V312 | R:R:Y269 | 2.52 | No | Yes | 0 | 3 | 8 |
| 277 | R:R:V351 | R:R:Y275 | 2.52 | No | No | 0 | 7 | 7 |
| 278 | R:R:V265 | R:R:W331 | 2.45 | No | Yes | 0 | 6 | 9 |
| 279 | R:R:F438 | R:R:L195 | 2.44 | Yes | No | 0 | 9 | 6 |
| 280 | R:R:F438 | R:R:L199 | 2.44 | Yes | No | 0 | 9 | 8 |
| 281 | R:R:F358 | R:R:L362 | 2.44 | No | No | 0 | 9 | 7 |
| 282 | R:R:F430 | R:R:L431 | 2.44 | No | No | 0 | 7 | 6 |
| 283 | R:R:F438 | R:R:L434 | 2.44 | Yes | No | 0 | 9 | 5 |
| 284 | R:R:I342 | R:R:W318 | 2.35 | No | Yes | 0 | 8 | 8 |
| 285 | L:L:L17 | R:R:Y172 | 2.34 | No | Yes | 0 | 0 | 1 |
| 286 | L:L:D15 | R:R:Y239 | 2.3 | No | Yes | 0 | 0 | 4 |
| 287 | R:R:F355 | R:R:F356 | 2.14 | No | No | 0 | 6 | 3 |
| 288 | R:R:R448 | R:R:W451 | 2 | No | No | 0 | 5 | 5 |
| 289 | R:R:G185 | R:R:V184 | 1.84 | No | No | 0 | 8 | 6 |
| 290 | R:R:P295 | R:R:V293 | 1.77 | No | No | 0 | 6 | 7 |
| 291 | R:R:P291 | R:R:T287 | 1.75 | No | No | 0 | 5 | 7 |
| 292 | R:R:G437 | R:R:L200 | 1.71 | No | No | 0 | 9 | 8 |
| 293 | R:R:G282 | R:R:L285 | 1.71 | No | No | 0 | 9 | 9 |
| 294 | R:R:G282 | R:R:L394 | 1.71 | No | Yes | 0 | 9 | 9 |
| 295 | R:R:G335 | R:R:N336 | 1.7 | No | Yes | 0 | 3 | 6 |
| 296 | R:R:I342 | R:R:P317 | 1.69 | No | No | 0 | 8 | 4 |
| 297 | R:R:E410 | R:R:G411 | 1.64 | No | No | 0 | 4 | 4 |
| 298 | R:R:A321 | R:R:I339 | 1.62 | No | No | 0 | 4 | 4 |
| 299 | R:R:A321 | R:R:I342 | 1.62 | No | No | 0 | 4 | 8 |
| 300 | R:R:S187 | R:R:T183 | 1.6 | No | No | 0 | 6 | 7 |
| 301 | R:R:V230 | R:R:V234 | 1.6 | No | No | 1 | 6 | 4 |
| 302 | R:R:V230 | R:R:V263 | 1.6 | No | No | 1 | 6 | 5 |
| 303 | R:R:S257 | R:R:T258 | 1.6 | No | No | 0 | 4 | 3 |
| 304 | R:R:S259 | R:R:T258 | 1.6 | No | No | 0 | 4 | 3 |
| 305 | R:R:S386 | R:R:T387 | 1.6 | No | No | 0 | 9 | 9 |
| 306 | R:R:A439 | R:R:L434 | 1.58 | No | No | 0 | 6 | 5 |
| 307 | R:R:I191 | R:R:S192 | 1.55 | No | No | 0 | 3 | 6 |
| 308 | R:R:M181 | R:R:T423 | 1.51 | No | No | 0 | 6 | 5 |
| 309 | R:R:G411 | R:R:R171 | 1.5 | No | No | 0 | 4 | 4 |
| 310 | R:R:G329 | R:R:R261 | 1.5 | No | No | 0 | 4 | 8 |
| 311 | R:R:L203 | R:R:V443 | 1.49 | No | Yes | 2 | 8 | 9 |
| 312 | R:R:L206 | R:R:V443 | 1.49 | Yes | Yes | 2 | 9 | 9 |
| 313 | R:R:L447 | R:R:V443 | 1.49 | No | Yes | 0 | 5 | 9 |
| 314 | R:R:N237 | R:R:V233 | 1.48 | No | No | 0 | 3 | 3 |
| 315 | L:L:I23 | L:L:I27 | 1.47 | No | No | 0 | 0 | 0 |
| 316 | R:R:L174 | R:R:T177 | 1.47 | No | No | 0 | 4 | 3 |
| 317 | R:R:L199 | R:R:T198 | 1.47 | No | No | 0 | 8 | 1 |
| 318 | R:R:L388 | R:R:T387 | 1.47 | No | No | 0 | 8 | 9 |
| 319 | R:R:L424 | R:R:T423 | 1.47 | No | No | 0 | 6 | 5 |
| 320 | R:R:D170 | R:R:V169 | 1.46 | No | No | 0 | 1 | 3 |
| 321 | R:R:A371 | R:R:H372 | 1.46 | No | No | 0 | 6 | 5 |
| 322 | R:R:K241 | R:R:M256 | 1.44 | No | No | 0 | 3 | 6 |
| 323 | R:R:I222 | R:R:L226 | 1.43 | Yes | No | 0 | 7 | 3 |
| 324 | L:L:I23 | L:L:N24 | 1.42 | No | No | 0 | 0 | 0 |
| 325 | R:R:Q179 | R:R:T183 | 1.42 | Yes | No | 0 | 5 | 7 |
| 326 | R:R:G319 | R:R:W318 | 1.41 | No | Yes | 0 | 5 | 8 |
| 327 | R:R:G319 | R:R:W331 | 1.41 | No | Yes | 0 | 5 | 9 |
| 328 | R:R:D378 | R:R:M374 | 1.39 | No | No | 0 | 5 | 5 |
| 329 | R:R:D407 | R:R:K337 | 1.38 | No | No | 0 | 4 | 6 |
| 330 | L:L:L17 | R:R:N168 | 1.37 | No | Yes | 0 | 0 | 3 |
| 331 | R:R:D170 | R:R:Q167 | 1.31 | No | No | 0 | 1 | 3 |
| 332 | R:R:N237 | R:R:Y236 | 1.16 | No | No | 0 | 3 | 4 |
| 333 | R:R:N245 | R:R:W249 | 1.13 | No | Yes | 0 | 4 | 3 |
| 334 | R:R:R297 | R:R:R302 | 1.07 | Yes | No | 0 | 5 | 2 |
| Color | ConSurf Grade |
| No Conservation data available | |
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 | |
| 6 | |
| 7 | |
| 8 | |
| 9 |
Index: hub id, click on each number to highlight the corresponding hub in the 3D visualization.
Hub: the hub being considered.
Avg Int. Strength: the average interaction strength of all the links of the corresponding hub.
Num Of Links: the number of links of the corresponding hub.
Community: the id of the community the link belong to, otherwise 0.
ConSurf: this column reports the ConSurf conservation grades of each hub.
| Index | Hub | Avg Int. Strength | Num Of Links | Community | ConSurf |
|---|---|---|---|---|---|
| 1 | L:L:H1 | 10.4225 | 4 | 1 | 0 |
| 2 | L:L:F6 | 5.70167 | 6 | 1 | 0 |
| 3 | L:L:S7 | 3.62 | 4 | 1 | 0 |
| 4 | L:L:N11 | 7.865 | 4 | 1 | 0 |
| 5 | L:L:F22 | 8.1725 | 4 | 3 | 0 |
| 6 | L:L:W25 | 6.77667 | 6 | 3 | 0 |
| 7 | R:R:N168 | 4.5825 | 4 | 0 | 3 |
| 8 | R:R:Y172 | 3.884 | 5 | 0 | 1 |
| 9 | R:R:Q179 | 5.06 | 5 | 1 | 5 |
| 10 | R:R:Y182 | 6.424 | 5 | 1 | 7 |
| 11 | R:R:Y186 | 5.82 | 8 | 1 | 7 |
| 12 | R:R:L193 | 4.5975 | 4 | 0 | 9 |
| 13 | R:R:L206 | 6.805 | 6 | 2 | 9 |
| 14 | R:R:R210 | 5.5675 | 4 | 2 | 9 |
| 15 | R:R:F218 | 3.5 | 4 | 0 | 9 |
| 16 | R:R:F221 | 7.58714 | 7 | 1 | 8 |
| 17 | R:R:I222 | 5.9525 | 4 | 5 | 7 |
| 18 | R:R:R224 | 5.102 | 5 | 1 | 8 |
| 19 | R:R:V228 | 5.2725 | 4 | 1 | 7 |
| 20 | R:R:K231 | 4.9 | 6 | 1 | 7 |
| 21 | R:R:F235 | 6.352 | 5 | 1 | 8 |
| 22 | R:R:Y239 | 4.55 | 4 | 0 | 4 |
| 23 | R:R:W249 | 10.74 | 5 | 3 | 3 |
| 24 | R:R:Y252 | 7.7 | 4 | 3 | 3 |
| 25 | R:R:L267 | 4.0975 | 4 | 1 | 6 |
| 26 | R:R:H268 | 9.514 | 5 | 1 | 7 |
| 27 | R:R:Y269 | 5.725 | 4 | 0 | 8 |
| 28 | R:R:F270 | 6.7975 | 4 | 0 | 5 |
| 29 | R:R:W277 | 10.472 | 5 | 6 | 9 |
| 30 | R:R:E281 | 8.43 | 4 | 0 | 9 |
| 31 | R:R:Y284 | 5.512 | 5 | 2 | 9 |
| 32 | R:R:R297 | 5.3225 | 4 | 0 | 5 |
| 33 | R:R:W308 | 6.882 | 5 | 6 | 9 |
| 34 | R:R:F314 | 5.424 | 5 | 0 | 6 |
| 35 | R:R:W318 | 5.944 | 5 | 1 | 8 |
| 36 | R:R:C330 | 5.4275 | 4 | 1 | 9 |
| 37 | R:R:W331 | 8.53667 | 6 | 7 | 9 |
| 38 | R:R:N336 | 4.204 | 5 | 8 | 6 |
| 39 | R:R:W340 | 9.136 | 5 | 1 | 6 |
| 40 | R:R:Y379 | 5.076 | 5 | 9 | 7 |
| 41 | R:R:L394 | 3.9275 | 4 | 0 | 9 |
| 42 | R:R:L415 | 6.5575 | 4 | 0 | 4 |
| 43 | R:R:R417 | 4.182 | 5 | 4 | 8 |
| 44 | R:R:H428 | 6.5075 | 4 | 0 | 9 |
| 45 | R:R:F438 | 2.5225 | 4 | 0 | 9 |
| 46 | R:R:V443 | 2.595 | 4 | 2 | 9 |
| Color | ConSurf Grade |
| No Conservation data available | |
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 | |
| 6 | |
| 7 | |
| 8 | |
| 9 |
Index: link id, click on each number to highlight the corresponding link in the 3D visualization.
Node1 Node2: the two nodes of the corresponding link.
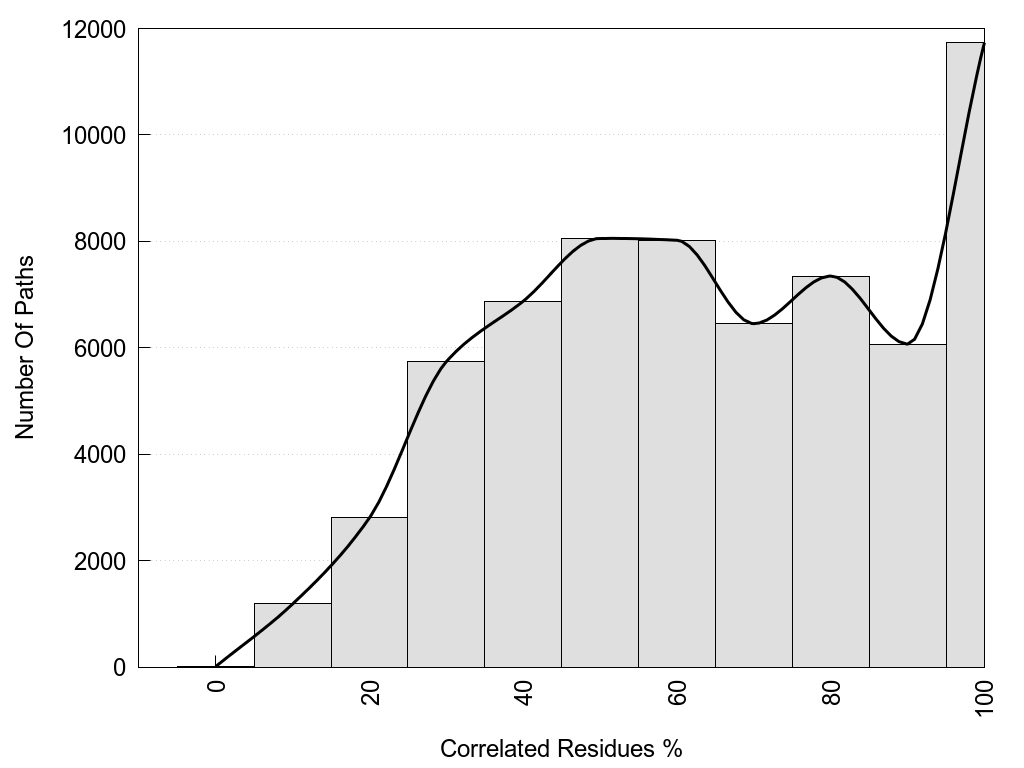
Recurrence: the relative Recurrence in the pool of shortest paths.
Int. Strength: the interaction strength between the two nodes.
Hub1?, Hub2?: "Yes" if the corresponding node has more than 3 links, otherwise "No".
Community: the id of the community the link belong to, otherwise 0.
ConSurf1, ConSurf2: these columns report the ConSurf conservation grades of the two nodes involved in a link.
| Index | Node1 | Node2 | Recurrence | Int. Strength | Hub1? | Hub2? | Community | ConSurf1 | ConSurf2 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | R:R:I404 | R:R:R417 | 16.8575 | 5.01 | No | Yes | 0 | 5 | 8 |
| 2 | R:R:I404 | R:R:Q408 | 18.4972 | 8.23 | No | No | 0 | 5 | 3 |
| 3 | R:R:Q408 | R:R:T405 | 20.1285 | 5.67 | No | No | 0 | 3 | 8 |
| 4 | R:R:D407 | R:R:T405 | 21.7513 | 2.89 | No | No | 0 | 4 | 8 |
| 5 | R:R:D406 | R:R:D407 | 24.9715 | 5.32 | No | No | 0 | 5 | 4 |
| 6 | R:R:D406 | R:R:R344 | 26.5013 | 11.91 | No | No | 1 | 5 | 6 |
| 7 | R:R:R344 | R:R:W340 | 29.7384 | 14.99 | No | Yes | 1 | 6 | 6 |
| 8 | L:L:H1 | R:R:W340 | 34.48 | 8.46 | Yes | Yes | 1 | 0 | 6 |
| 9 | R:R:L267 | R:R:V228 | 11.9385 | 2.98 | Yes | Yes | 1 | 6 | 7 |
| 10 | L:L:H1 | R:R:L267 | 29.244 | 3.86 | Yes | Yes | 1 | 0 | 6 |
| 11 | R:R:V228 | R:R:Y186 | 45.3239 | 6.31 | Yes | Yes | 1 | 7 | 7 |
| 12 | L:L:H1 | R:R:V271 | 45.5859 | 13.84 | Yes | No | 0 | 0 | 6 |
| 13 | R:R:R224 | R:R:V271 | 49.4908 | 3.92 | Yes | No | 0 | 8 | 6 |
| 14 | R:R:R224 | R:R:Y186 | 96.6065 | 7.2 | Yes | Yes | 1 | 8 | 7 |
| 15 | R:R:K231 | R:R:L267 | 30.8034 | 4.23 | Yes | Yes | 1 | 7 | 6 |
| 16 | R:R:D232 | R:R:Q179 | 11.6426 | 6.53 | No | Yes | 1 | 8 | 5 |
| 17 | R:R:D232 | R:R:K231 | 19.41 | 8.3 | No | Yes | 1 | 8 | 7 |
| 18 | L:L:F6 | R:R:Y182 | 33.3855 | 12.38 | Yes | Yes | 1 | 0 | 7 |
| 19 | R:R:Y182 | R:R:Y186 | 63.0985 | 5.96 | Yes | Yes | 1 | 7 | 7 |
| 20 | L:L:M10 | R:R:Q179 | 21.7259 | 8.16 | No | Yes | 1 | 0 | 5 |
| 21 | L:L:F6 | R:R:L178 | 18.0112 | 4.87 | Yes | No | 1 | 0 | 5 |
| 22 | R:R:L422 | R:R:Y182 | 16.2744 | 3.52 | No | Yes | 1 | 7 | 7 |
| 23 | R:R:L178 | R:R:L422 | 14.5079 | 5.54 | No | No | 1 | 5 | 7 |
| 24 | R:R:F235 | R:R:K231 | 40.6542 | 6.2 | Yes | Yes | 1 | 8 | 7 |
| 25 | L:L:M10 | R:R:Y172 | 36.2803 | 4.79 | No | Yes | 0 | 0 | 1 |
| 26 | R:R:C330 | R:R:F235 | 33.9475 | 4.19 | Yes | Yes | 1 | 9 | 8 |
| 27 | L:L:L17 | R:R:Y172 | 19.7988 | 2.34 | No | Yes | 0 | 0 | 1 |
| 28 | L:L:L17 | R:R:N168 | 17.7112 | 1.37 | No | Yes | 0 | 0 | 3 |
| 29 | R:R:K241 | R:R:Y252 | 28.441 | 16.72 | No | Yes | 0 | 3 | 3 |
| 30 | R:R:K241 | R:R:M256 | 31.0781 | 1.44 | No | No | 0 | 3 | 6 |
| 31 | R:R:C330 | R:R:M256 | 33.7235 | 4.86 | Yes | No | 0 | 9 | 6 |
| 32 | R:R:W249 | R:R:Y252 | 21.6329 | 5.79 | Yes | Yes | 3 | 3 | 3 |
| 33 | R:R:F419 | R:R:L178 | 28.2973 | 10.96 | No | No | 0 | 6 | 5 |
| 34 | R:R:F419 | R:R:L415 | 20.1834 | 12.18 | No | Yes | 0 | 6 | 4 |
| 35 | R:R:S426 | R:R:Y186 | 15.7038 | 5.09 | No | Yes | 0 | 9 | 7 |
| 36 | R:R:G185 | R:R:S426 | 13.9923 | 3.71 | No | No | 0 | 8 | 9 |
| 37 | R:R:F427 | R:R:G185 | 10.5439 | 4.52 | No | No | 0 | 8 | 8 |
| 38 | R:R:F221 | R:R:R224 | 97.0883 | 3.21 | Yes | Yes | 1 | 8 | 8 |
| 39 | R:R:F221 | R:R:L193 | 19.596 | 7.31 | Yes | Yes | 0 | 8 | 9 |
| 40 | R:R:F218 | R:R:L193 | 14.0303 | 3.65 | Yes | Yes | 0 | 9 | 9 |
| 41 | R:R:F221 | R:R:V432 | 15.1164 | 3.93 | Yes | No | 0 | 8 | 9 |
| 42 | R:R:F218 | R:R:V432 | 13.7768 | 3.93 | Yes | No | 0 | 9 | 9 |
| 43 | R:R:F221 | R:R:L217 | 100 | 4.87 | Yes | No | 0 | 8 | 9 |
| 44 | R:R:E281 | R:R:L217 | 98.5124 | 5.3 | Yes | No | 0 | 9 | 9 |
| 45 | R:R:E281 | R:R:I213 | 25.0264 | 6.83 | Yes | No | 0 | 9 | 9 |
| 46 | R:R:I213 | R:R:Y284 | 22.4824 | 4.84 | No | Yes | 2 | 9 | 9 |
| 47 | R:R:Y284 | R:R:Y303 | 18.5648 | 2.98 | Yes | No | 0 | 9 | 7 |
| 48 | R:R:L299 | R:R:Y303 | 10.172 | 4.69 | No | No | 0 | 4 | 7 |
| 49 | R:R:E281 | R:R:H214 | 26.8985 | 12.31 | Yes | No | 0 | 9 | 9 |
| 50 | R:R:H214 | R:R:R210 | 25.9223 | 4.51 | No | Yes | 0 | 9 | 9 |
| 51 | R:R:N211 | R:R:R210 | 22.2499 | 8.44 | No | Yes | 2 | 9 | 9 |
| 52 | R:R:L206 | R:R:N211 | 20.8131 | 2.75 | Yes | No | 2 | 9 | 9 |
| 53 | R:R:E281 | R:R:L393 | 59.6332 | 9.28 | Yes | No | 0 | 9 | 9 |
| 54 | R:R:N274 | R:R:R224 | 32.2233 | 7.23 | No | Yes | 1 | 9 | 8 |
| 55 | R:R:N274 | R:R:S220 | 53.2435 | 5.96 | No | No | 0 | 9 | 9 |
| 56 | R:R:S220 | R:R:W308 | 52.0771 | 3.71 | No | Yes | 0 | 9 | 9 |
| 57 | R:R:W277 | R:R:W308 | 47.1791 | 18.74 | Yes | Yes | 6 | 9 | 9 |
| 58 | R:R:F221 | R:R:H428 | 13.1175 | 9.05 | Yes | Yes | 0 | 8 | 9 |
| 59 | R:R:V280 | R:R:W277 | 43.6758 | 3.68 | No | Yes | 0 | 8 | 9 |
| 60 | R:R:L306 | R:R:V280 | 42.4418 | 2.98 | No | No | 0 | 7 | 8 |
| 61 | R:R:F310 | R:R:L306 | 41.1993 | 4.87 | No | No | 0 | 3 | 7 |
| 62 | R:R:F310 | R:R:L276 | 12.416 | 6.09 | No | No | 0 | 3 | 7 |
| 63 | R:R:L276 | R:R:P311 | 11.0764 | 3.28 | No | No | 0 | 7 | 9 |
| 64 | R:R:P311 | R:R:Y269 | 19.4692 | 8.34 | No | Yes | 0 | 9 | 8 |
| 65 | R:R:F270 | R:R:Y269 | 16.7646 | 8.25 | Yes | Yes | 0 | 5 | 8 |
| 66 | R:R:F310 | R:R:F314 | 27.5705 | 4.29 | No | Yes | 0 | 3 | 6 |
| 67 | R:R:F314 | R:R:P311 | 11.1778 | 4.33 | Yes | No | 0 | 6 | 9 |
| 68 | R:R:F314 | R:R:V315 | 11.1651 | 3.93 | Yes | No | 0 | 6 | 6 |
| 69 | L:L:H1 | R:R:H268 | 33.7742 | 15.53 | Yes | Yes | 1 | 0 | 7 |
| 70 | R:R:H268 | R:R:W318 | 43.866 | 9.52 | Yes | Yes | 1 | 7 | 8 |
| 71 | R:R:G319 | R:R:W318 | 17.9352 | 1.41 | No | Yes | 0 | 5 | 8 |
| 72 | R:R:G319 | R:R:W331 | 15.7419 | 1.41 | No | Yes | 0 | 5 | 9 |
| 73 | R:R:L267 | R:R:Q264 | 14.7276 | 5.32 | Yes | No | 0 | 6 | 6 |
| 74 | R:R:H268 | R:R:Q264 | 13.6585 | 11.13 | Yes | No | 0 | 7 | 6 |
| 75 | R:R:L393 | R:R:L394 | 58.9359 | 4.15 | No | Yes | 0 | 9 | 9 |
| 76 | R:R:G282 | R:R:L394 | 22.3556 | 1.71 | No | Yes | 0 | 9 | 9 |
| 77 | R:R:G282 | R:R:L285 | 20.9187 | 1.71 | No | No | 0 | 9 | 9 |
| 78 | R:R:I361 | R:R:L285 | 19.4692 | 5.71 | No | No | 0 | 9 | 9 |
| 79 | R:R:I357 | R:R:I361 | 18.0958 | 2.94 | No | No | 0 | 8 | 9 |
| 80 | R:R:I342 | R:R:W318 | 11.275 | 2.35 | No | Yes | 0 | 8 | 8 |
| 81 | R:R:I339 | R:R:W318 | 15.7461 | 5.87 | No | Yes | 0 | 4 | 8 |
| 82 | R:R:I339 | R:R:N336 | 11.3849 | 5.66 | No | Yes | 0 | 4 | 6 |
| 83 | R:R:L394 | R:R:V396 | 37.4551 | 2.98 | Yes | No | 0 | 9 | 7 |
| 84 | R:R:F358 | R:R:V396 | 30.6512 | 3.93 | No | No | 0 | 9 | 7 |
| 85 | R:R:F358 | R:R:L362 | 27.8621 | 2.44 | No | No | 0 | 9 | 7 |
| 86 | R:R:L362 | R:R:T387 | 26.5309 | 4.42 | No | No | 0 | 7 | 9 |
| 87 | R:R:S386 | R:R:T387 | 23.7797 | 1.6 | No | No | 0 | 9 | 9 |
| 88 | R:R:R382 | R:R:S386 | 20.9145 | 2.64 | No | No | 0 | 8 | 9 |
| 89 | R:R:L369 | R:R:R382 | 16.549 | 3.64 | No | No | 0 | 8 | 8 |
| 90 | R:R:L369 | R:R:Y379 | 15.1545 | 7.03 | No | Yes | 0 | 8 | 7 |
| 91 | R:R:A196 | R:R:F218 | 19.6087 | 2.77 | No | Yes | 0 | 9 | 9 |
| 92 | R:R:A196 | R:R:F438 | 16.8322 | 2.77 | No | Yes | 0 | 9 | 9 |
| 93 | L:L:F6 | L:L:M10 | 16.3716 | 4.98 | Yes | No | 1 | 0 | 0 |
| 94 | R:R:Q179 | R:R:Y182 | 18.5944 | 4.51 | Yes | Yes | 1 | 5 | 7 |
| 95 | R:R:K231 | R:R:V228 | 33.4446 | 3.04 | Yes | Yes | 1 | 7 | 7 |
| 96 | R:R:F221 | R:R:N274 | 22.1739 | 15.71 | Yes | No | 1 | 8 | 9 |
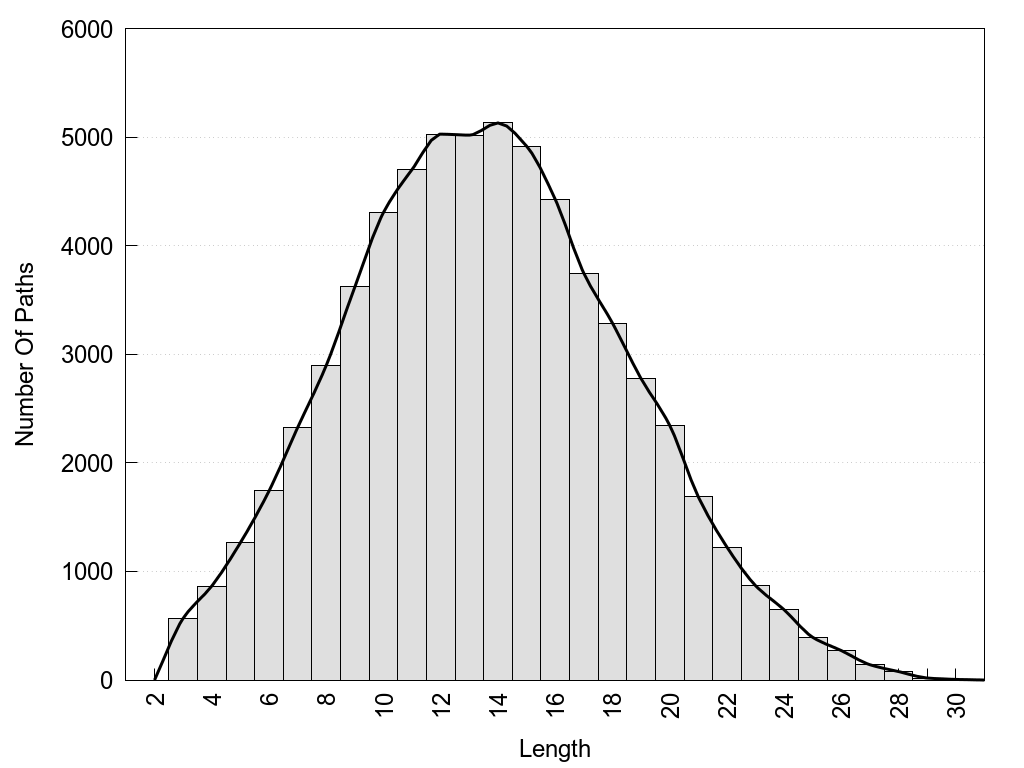
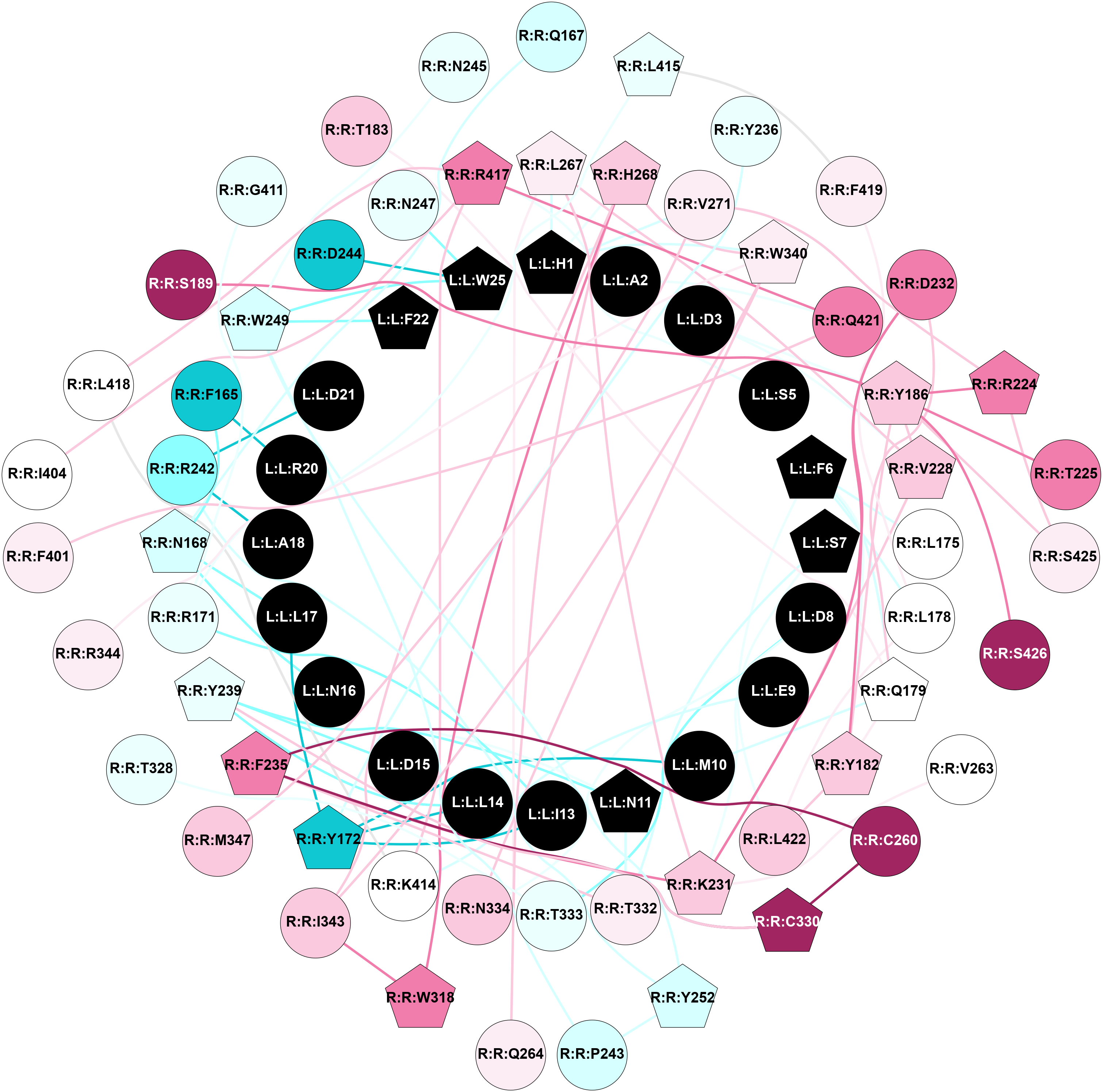
2D representation of the global metapath, ligand(s) interactions and
histograms of path distribution according to several parameters
(click on the image to enlarge it 🔍):

A 2D representation of the global communication in the network.
ConSurf Conservation Grade (See documentation):
n/a 1 2 3 4 5 6 7 8 9
2D representation of the interactions of this orthosteric/allosteric ligand. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Links and nodes colored according to ConSurf Conservation Grade (See documentation): n/a 1 2 3 4 5 6 7 8 9 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Location and physicochemical properties of the interaction partners of this ligand | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Interactions of this ligand | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Similarities between the interactions of this ligand and those of other networks | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|
Details about the values in these tables can be found in the corresponding documentation page .
| PDBsum | Open PDBsum Page |
| Chain | R |
| Protein | Receptor |
| UniProt | O95838 |
| Sequence | >7D68_nogp_Chain_R SFKQNVDRY ALLSTLQLM YTVGYSFSL ISLFLALTL LLFLRKLHC TRNYIHMNL FASFILRTL AVLVKDVVF YNSYSKRPD NENGWMSYL SEMSTSCRS VQVLLHYFV GANYLWLLV EGLYLHTLL EPTVLPERR LWPRYLLLG WAFPVLFVV PWGFARAHL ENTGCWTTN GNKKIWWII RGPMMLCVT VNFFIFLKI LKLLISKLK AHQMCFRDY KYRLAKSTL VLIPLLGVH EILFSFITD DQVEGFAKL IRLFIQLTL SSFHGFLVA LQYGFANGE VKAELRKYW VRFL Click on each residue to open a popup with some information about it. ConSurf Conservation Grade (See documentation): n/a 1 2 3 4 5 6 7 8 9 |
| This receptor, from the same or other species and bound to the same or other ligands, is also present in the following networks: | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7D68 | B1 | Peptide | Glucagon | GLP-2 | Homo sapiens | GLP-2 | - | Gs/β1/γ2 | 3 | 2020-12-16 | doi.org/10.1038/s41422-020-00442-0 | |
| 7D68 (No Gprot) | B1 | Peptide | Glucagon | GLP-2 | Homo sapiens | GLP-2 | - | 3 | 2020-12-16 | doi.org/10.1038/s41422-020-00442-0 | ||
You can download a compressed (zip) file with structure(s), 3D outputs (as PyMol and VMD scripts) and numerical data files (as csv and plain text files).
Download 7D68_nogp.zipYou can click to copy the link of this page to easily come back here later
or use this QR code to link and share this page.
You can also read or download a guide explaining the meaning of all files and numerical data.