|
|
This ligand is also present in the following 41 networks: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1HZX | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.8 | 2001-07-04 | doi.org/10.1021/bi0155091 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
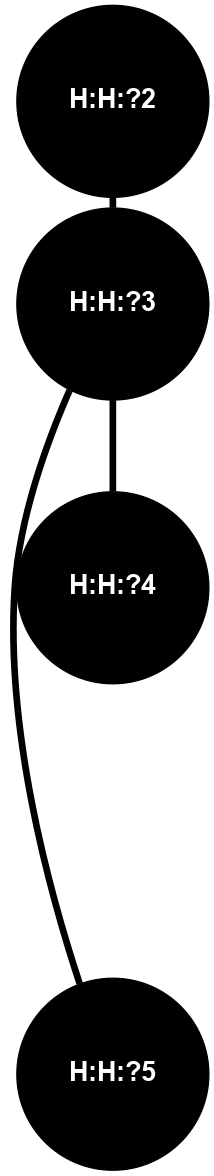
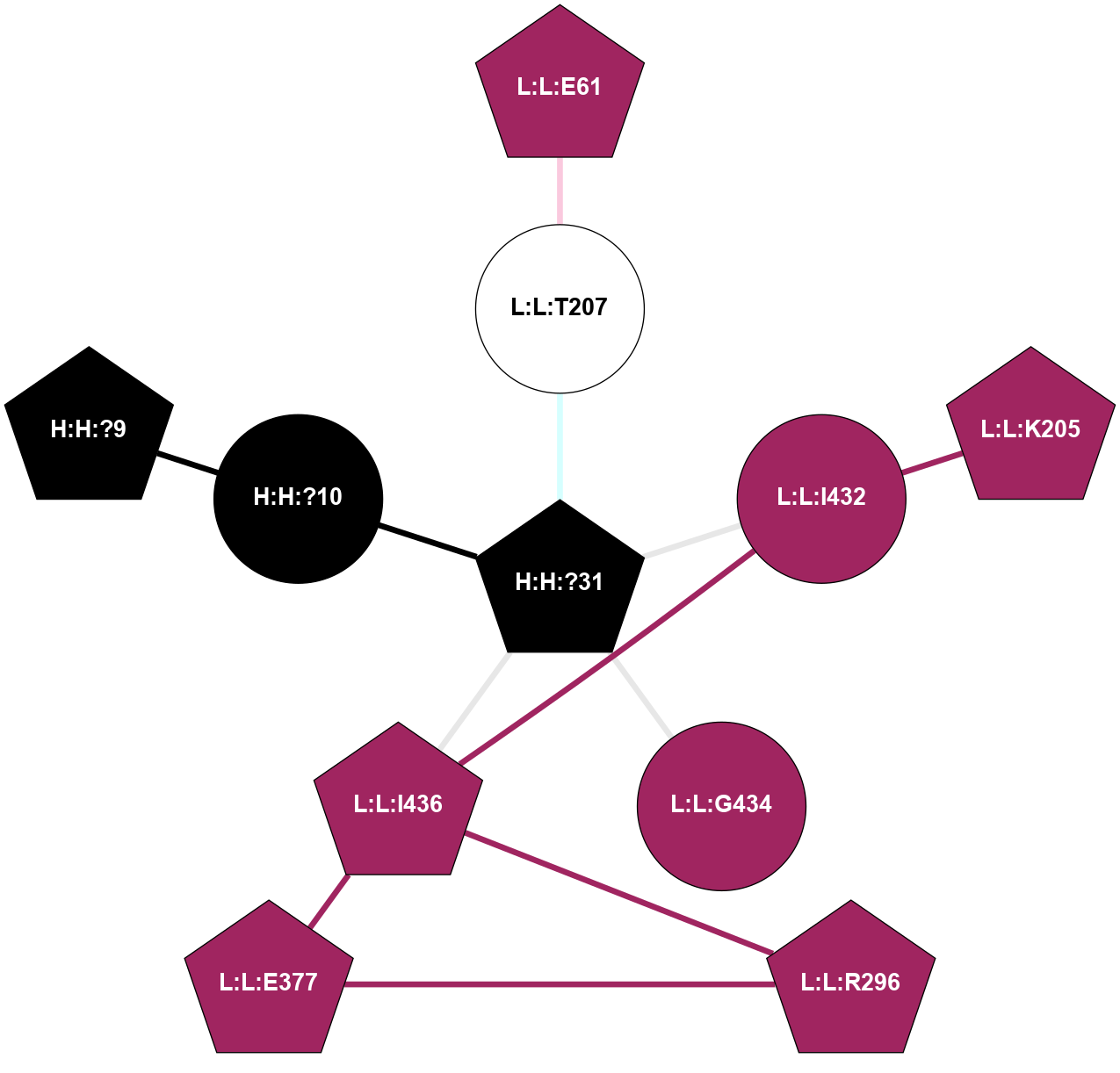
A 2D representation of the interactions of MAN in 1HZX n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1L9H | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.6 | 2002-05-15 | doi.org/10.1073/pnas.082666399 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
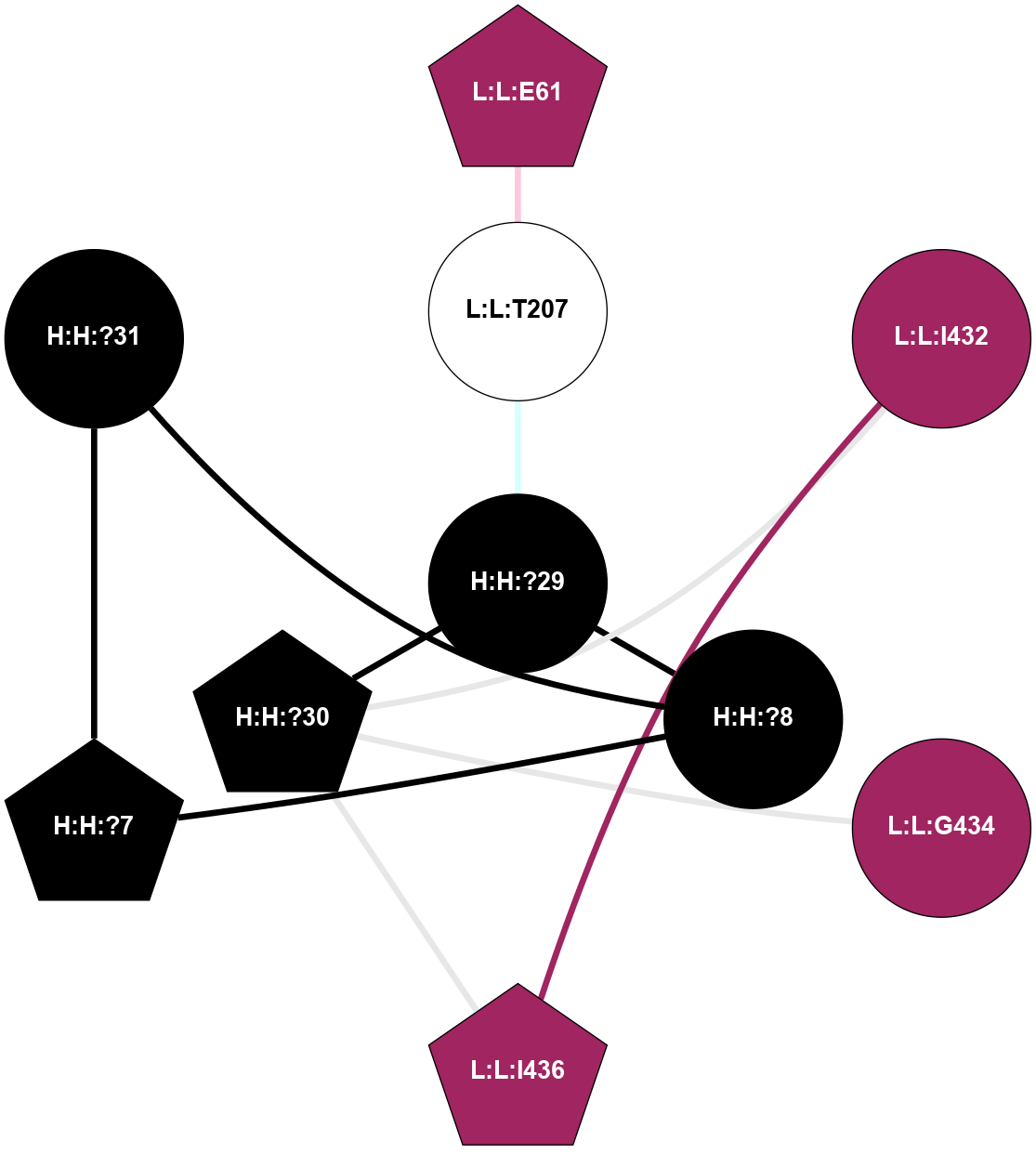
A 2D representation of the interactions of MAN in 1L9H n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1U19 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.2 | 2004-10-12 | doi.org/10.1016/j.jmb.2004.07.044 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
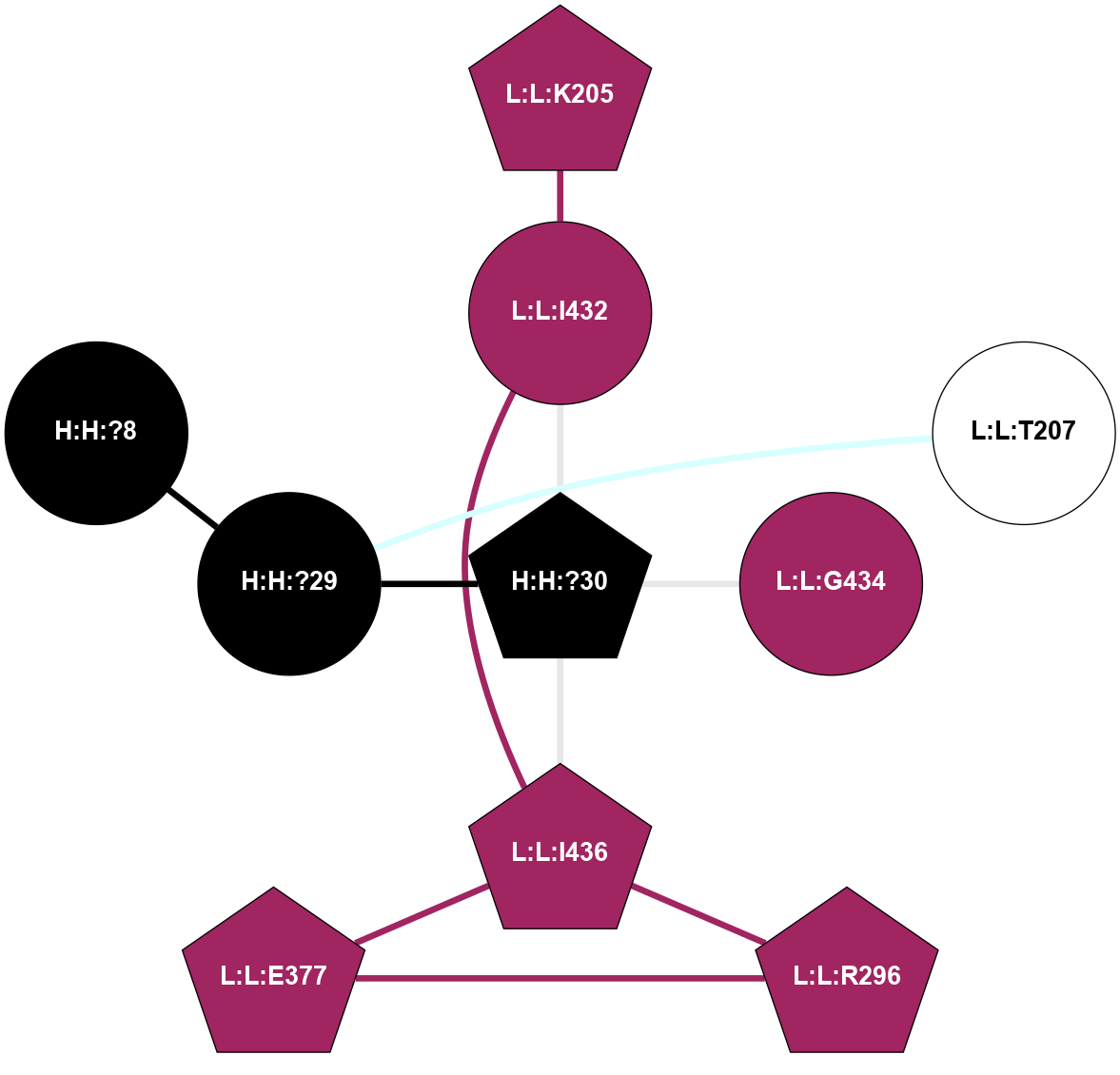
A 2D representation of the interactions of MAN in 1U19 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2G87 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 2.6 | 2006-09-02 | doi.org/10.1002/anie.200600595 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 2G87 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2I36 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | - | 4.1 | 2006-10-17 | doi.org/10.1073/pnas.0608022103 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 2I36 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2I37 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | - | 4.15 | 2006-10-17 | doi.org/10.1073/pnas.0608022103 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 2I37 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2X72 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | Gt(CT) | 3 | 2011-03-16 | doi.org/10.1038/nature09795 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 2X72 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 2X72 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3DQB | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | Gt(CT) | 3.2 | 2008-09-23 | doi.org/10.1038/nature07330 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 3DQB n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3PQR | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | Gt(CT) | 2.85 | 2011-03-09 | doi.org/10.1038/nature09789 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 3PQR n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4BEY | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | Gt(CT) | 2.9 | 2013-05-08 | doi.org/10.1038/embor.2013.44 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 4BEY n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4BEZ | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | - | 3.3 | 2013-04-24 | doi.org/10.1038/embor.2013.44 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 4BEZ n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4J4Q | A | Sensory | Opsins | Rhodopsin | Bos Taurus | Octyl Beta-D-Glucopyranoside | - | Gt(CT) | 2.65 | 2013-10-30 | doi.org/10.1002/anie.201302374 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 4J4Q n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4N4W | F | Protein | Frizzled | SMO | Homo Sapiens | - | SANT-1 | - | 2.8 | 2014-01-22 | doi.org/10.1038/ncomms5355 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 4N4W n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4PXF | A | Sensory | Opsins | Rhodopsin | Bos Taurus | Octyl-Beta-D-Glucopyranoside | - | Arrestin1 Finger Loop | 2.75 | 2014-09-17 | doi.org/10.1038/ncomms5801 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 4PXF n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5DYS | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 2.3 | 2016-08-10 | doi.org/10.15252/embr.201642671 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 5DYS n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5EN0 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | Gt(CT) | 2.81 | 2016-08-10 | doi.org/10.15252/embr.201642671 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 5EN0 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5TE3 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | Octyl-Beta-D-Glucopyranoside | - | - | 2.7 | 2017-03-15 | doi.org/10.1073/pnas.1617446114 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 5TE3 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5WKT | A | Sensory | Opsins | Rhodopsin | Bos Taurus | Octyl-Beta-D-Glucopyranoside | - | Gt(CT) | 3.2 | 2017-12-13 | doi.org/10.1038/nprot.2017.135 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 5WKT n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6FK6 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473033 | - | 2.36 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FK6 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FK6 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6FK7 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473034 | - | 2.62 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FK7 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FK7 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6FK8 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473035 | - | 2.87 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FK8 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FK8 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6FK9 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473036 | - | 2.63 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FK9 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FK9 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6FKA | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473037 | - | 2.7 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FKA n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FKA n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6FKB | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473038 | - | 3.03 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FKB n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FKB n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6FKC | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473039 | - | 2.46 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FKC n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FKC n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6FKD | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 137349183 | - | 2.49 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FKD n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6FKD n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6MEO | A | Protein | Chemokine | CCR5 | Homo Sapiens | Envelope Glycoprotein Gp160; T-Cell Surface Glycoprotein Cd4 | - | - | 3.9 | 2018-12-12 | doi.org/10.1038/s41586-018-0804-9 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6MEO n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6MET | A | Protein | Chemokine | CCR5 | Homo Sapiens | Envelope Glycoprotein Gp160; T-Cell Surface Glycoprotein Cd4 | - | - | 4.5 | 2018-12-12 | doi.org/10.1038/s41586-018-0804-9 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6MET n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6MET n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 6MET n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8FCZ | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 3.7 | 2023-08-30 | doi.org/10.1038/s41467-023-40911-9 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 8FCZ n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 8FCZ n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8FD0 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | - | 3.71 | 2023-08-30 | doi.org/10.1038/s41467-023-40911-9 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 8FD0 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 8FD0 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8FD1 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | - | 4.25 | 2023-08-30 | doi.org/10.1038/s41467-023-40911-9 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 8FD1 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A 2D representation of the interactions of MAN in 8FD1 n/a 1 2 3 4 5 6 7 8 9 (Click to enlarge 🔍) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | Location of the nodes interacting with this ligand (click to enlarge 🔍) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
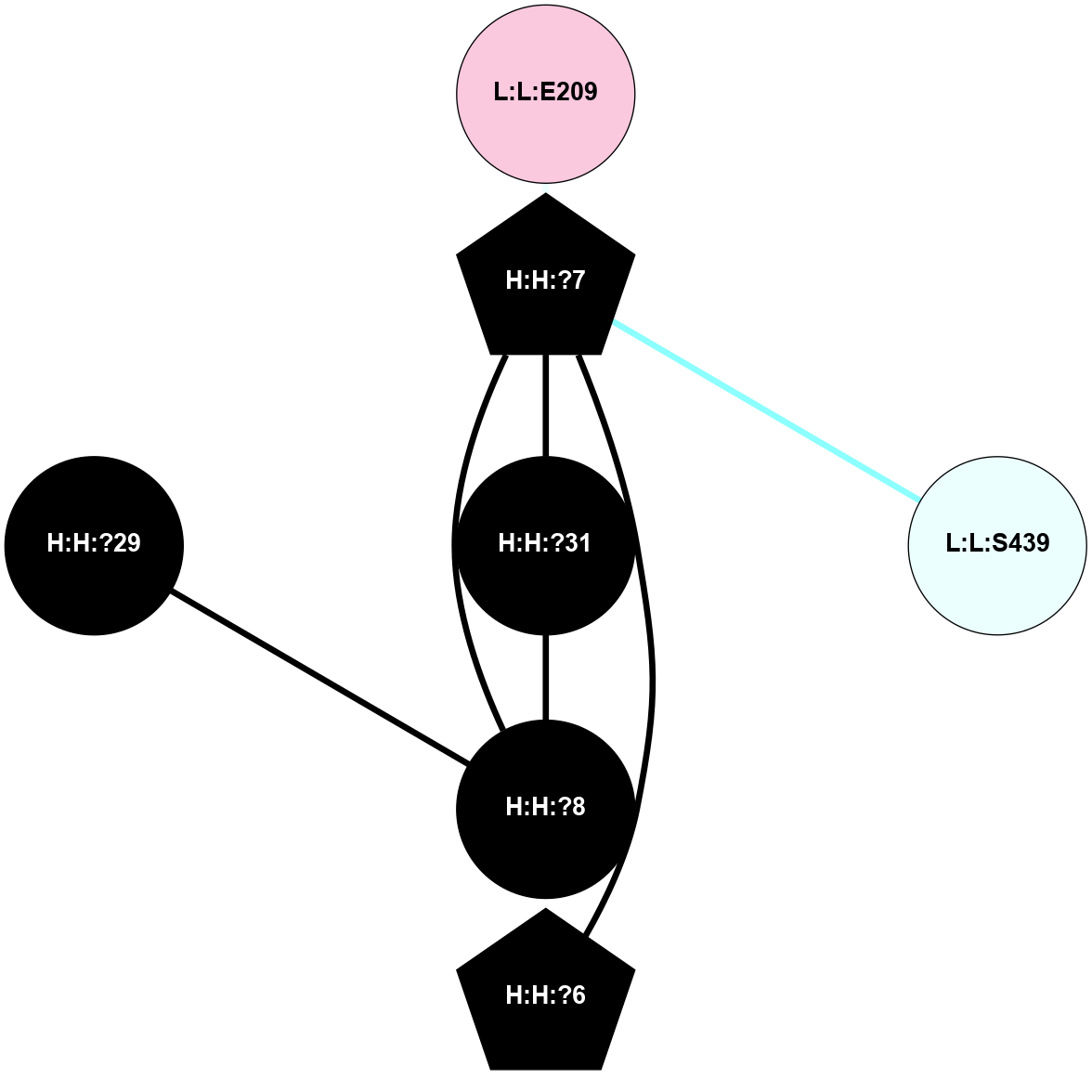
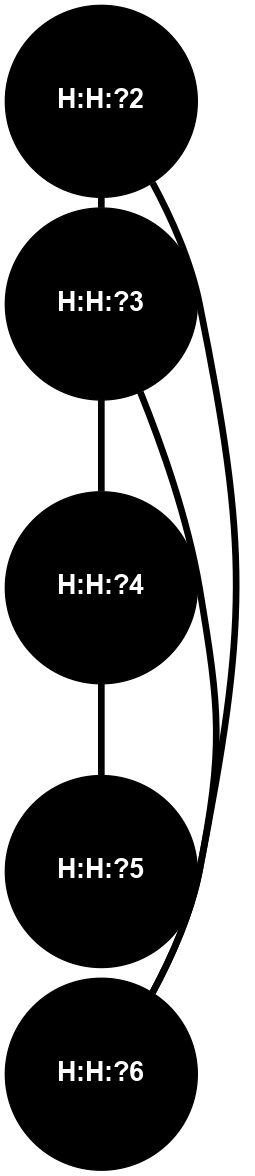
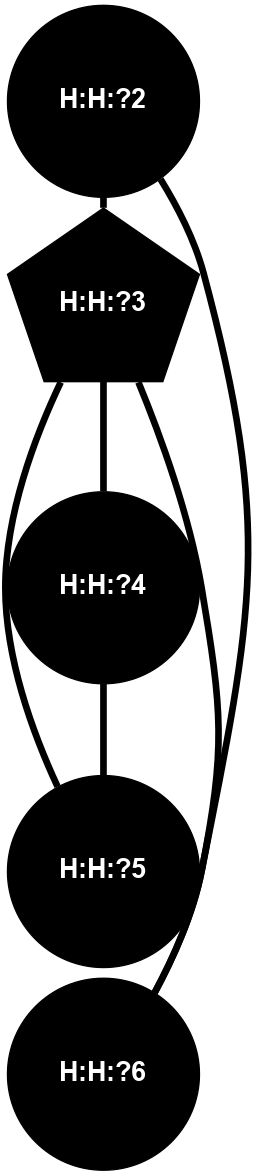
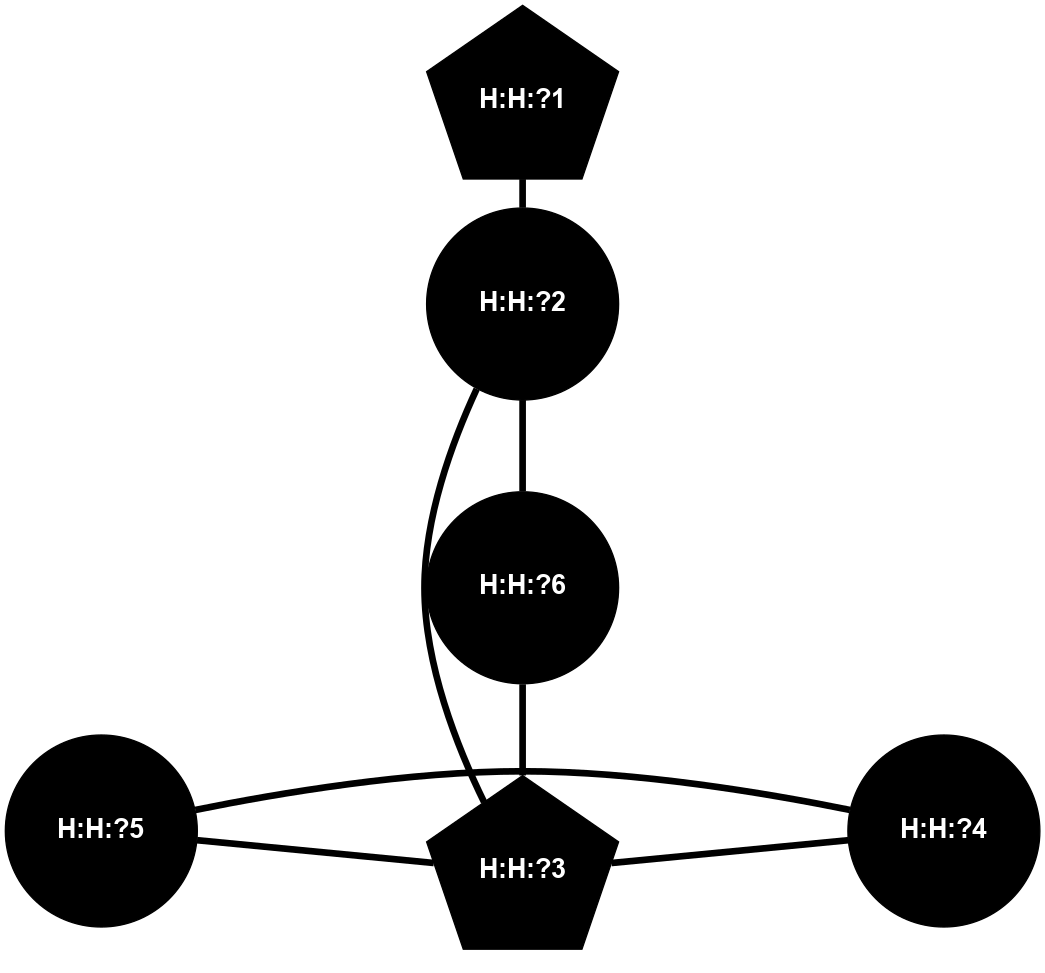
Physico-chemical properties of the nodes interacting with this ligand (Click to enlarge 🔍) |
Location of the nodes interacting with this ligand (Click to enlarge 🔍) |