| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3EML | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | - | - | 2.6 | 2008-10-14 | doi.org/10.1126/science.1164772 |
|
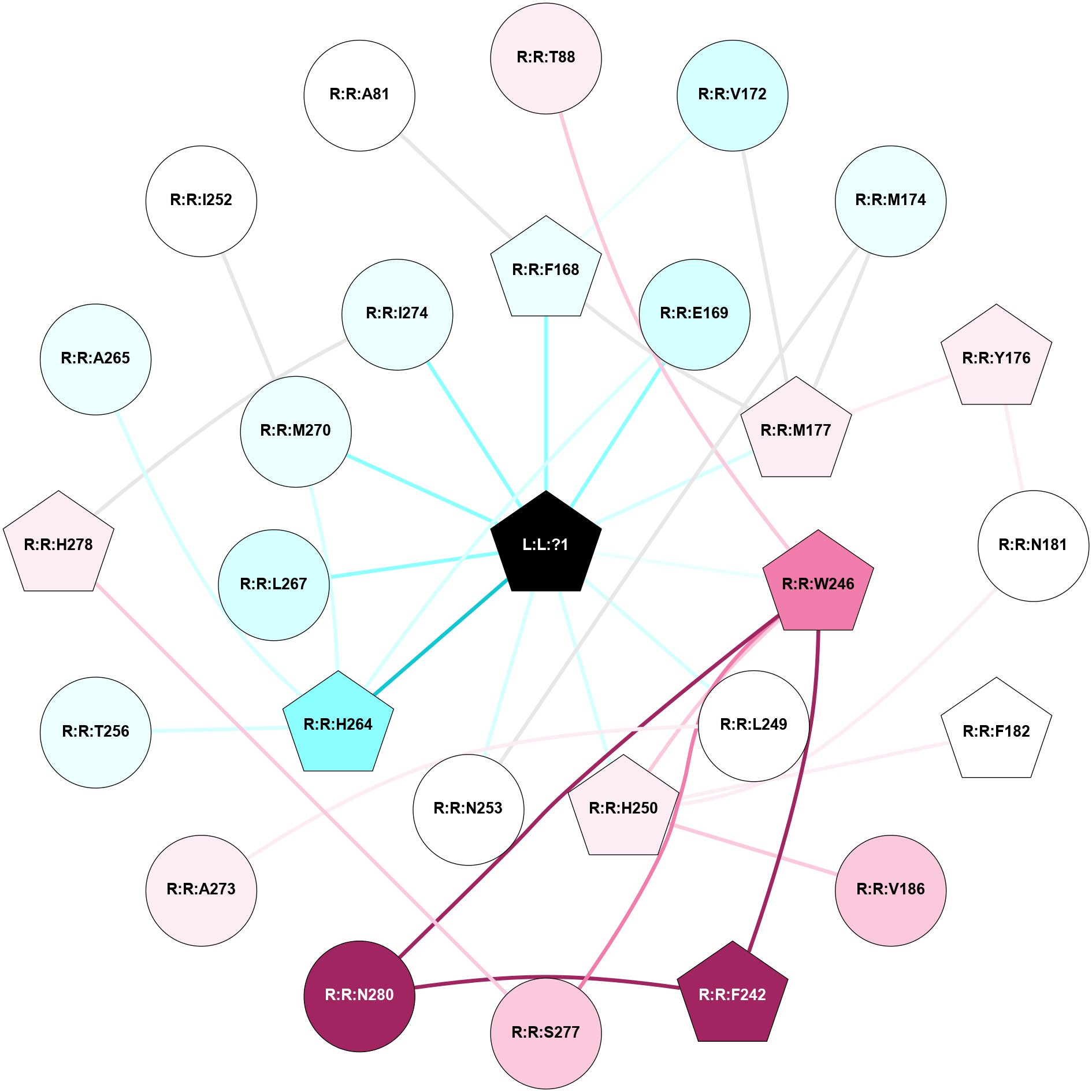
A 2D representation of the interactions of ZMA in 3EML
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:T88 | R:R:W246 | 10.92 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F168 | R:R:V172 | 13.11 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 6.22 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | L:L:?1 | R:R:F168 | 40.16 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:E169 | R:R:H264 | 11.08 | 1 | No | Yes | 3 | 2 | 1 | 1 | | L:L:?1 | R:R:E169 | 7.71 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:M177 | 4.33 | 0 | No | Yes | 4 | 6 | 2 | 1 | | R:R:M174 | R:R:N253 | 8.41 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 4.65 | 1 | No | Yes | 5 | 6 | 2 | 2 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:H250 | R:R:N181 | 3.83 | 1 | Yes | No | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 21.5 | 1 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 1 | Yes | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 12.03 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F242 | R:R:N280 | 9.67 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:H250 | R:R:W246 | 3.17 | 1 | Yes | Yes | 6 | 8 | 1 | 1 | | R:R:S277 | R:R:W246 | 13.59 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N280 | R:R:W246 | 6.78 | 1 | No | Yes | 9 | 8 | 2 | 1 | | L:L:?1 | R:R:W246 | 3.68 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | L:L:?1 | R:R:L249 | 16.11 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 4.99 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 10.65 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:H264 | R:R:T256 | 9.58 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 6.57 | 1 | Yes | No | 2 | 4 | 1 | 1 | | L:L:?1 | R:R:H264 | 5.82 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:L267 | 3.58 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M270 | 10.05 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 4 | Yes | No | 6 | 4 | 2 | 1 | | L:L:?1 | R:R:I274 | 4.61 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 4 | Yes | No | 6 | 7 | 2 | 2 | | R:R:A265 | R:R:H264 | 2.93 | 0 | No | Yes | 4 | 2 | 2 | 1 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 11.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 10.34 | | Average Nodes In Shell | 28.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 36.00 | | Average Links Mediated by Hubs In Shell | 33.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3PWH | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | - | - | 3.3 | 2011-09-07 | doi.org/10.1016/j.str.2011.06.014 |
|

A 2D representation of the interactions of ZMA in 3PWH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:Y271 | R:R:Y9 | 14.89 | 1 | Yes | No | 3 | 5 | 1 | 2 | | R:R:E13 | R:R:Y271 | 6.73 | 1 | Yes | Yes | 6 | 3 | 2 | 1 | | R:R:E13 | R:R:V275 | 5.7 | 1 | Yes | No | 6 | 5 | 2 | 2 | | R:R:E13 | R:R:H278 | 16 | 1 | Yes | Yes | 6 | 6 | 2 | 2 | | L:L:?1 | R:R:S67 | 13.59 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:E169 | R:R:L167 | 5.3 | 1 | No | No | 3 | 4 | 1 | 2 | | R:R:D170 | R:R:L167 | 5.43 | 1 | No | No | 4 | 4 | 2 | 2 | | R:R:F168 | R:R:V172 | 5.24 | 1 | No | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 6.22 | 1 | No | No | 4 | 6 | 1 | 1 | | L:L:?1 | R:R:F168 | 36.22 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:D170 | R:R:E169 | 9.09 | 1 | No | No | 4 | 3 | 2 | 1 | | L:L:?1 | R:R:E169 | 3.43 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | No | Yes | 6 | 6 | 1 | 2 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:I274 | R:R:L249 | 5.71 | 1 | No | No | 4 | 5 | 1 | 1 | | L:L:?1 | R:R:L249 | 17 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:N253 | 10.65 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:M270 | 6.4 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:V275 | R:R:Y271 | 8.83 | 1 | No | Yes | 5 | 3 | 2 | 1 | | L:L:?1 | R:R:Y271 | 6.82 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 1 | Yes | No | 6 | 4 | 2 | 1 | | L:L:?1 | R:R:I274 | 10.15 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 9.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 12.30 | | Average Nodes In Shell | 20.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 24.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3VG9 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Antibody | - | 2.7 | 2012-02-01 | doi.org/10.1038/nature10750 |
|
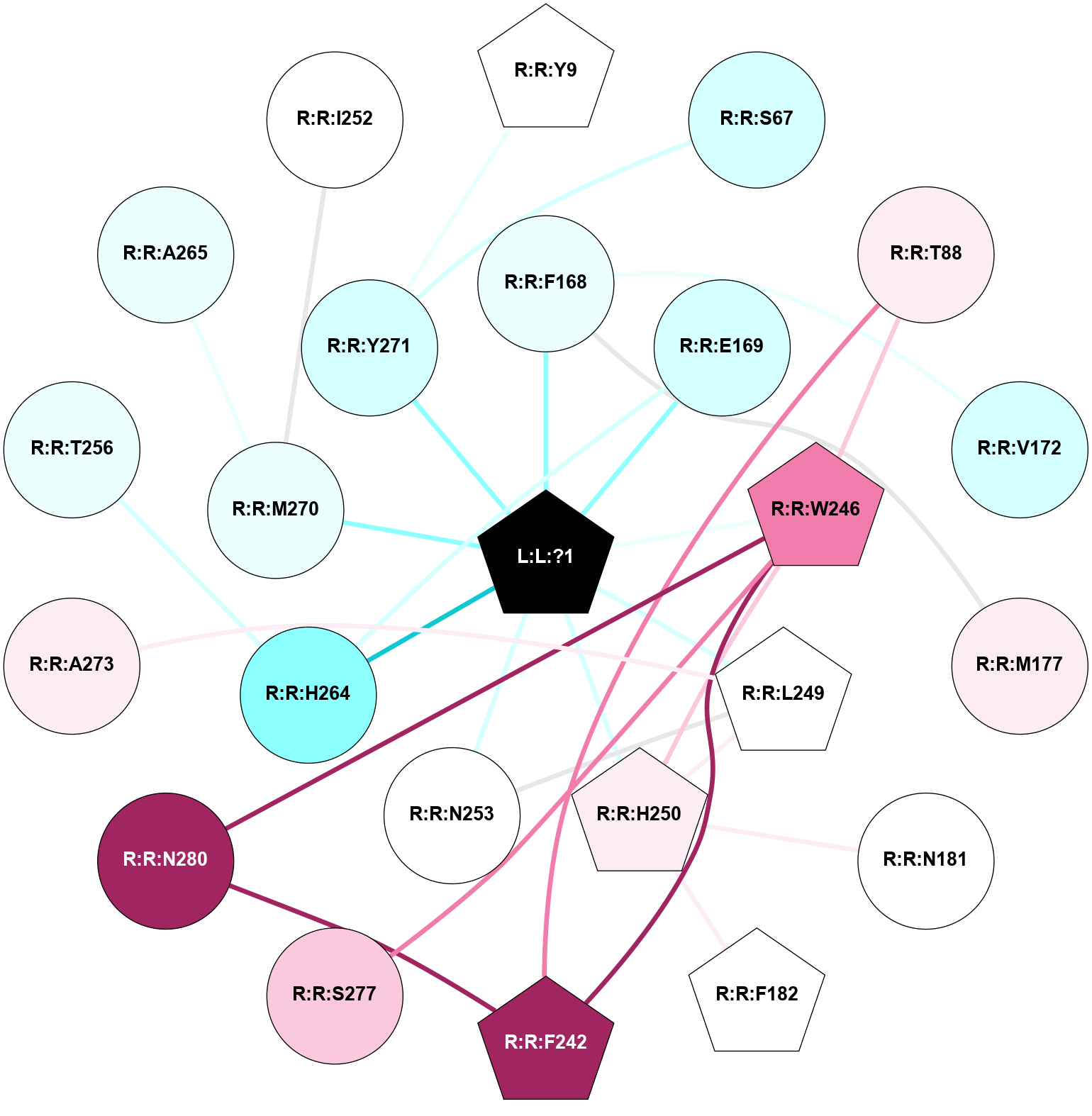
A 2D representation of the interactions of ZMA in 3VG9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:Y271 | R:R:Y9 | 11.91 | 0 | No | Yes | 3 | 5 | 1 | 2 | | R:R:S67 | R:R:Y271 | 5.09 | 0 | No | No | 3 | 3 | 2 | 1 | | R:R:F242 | R:R:T88 | 3.89 | 6 | Yes | No | 9 | 6 | 2 | 2 | | R:R:T88 | R:R:W246 | 16.98 | 6 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F168 | R:R:V172 | 26.22 | 0 | No | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 3.73 | 0 | No | No | 4 | 6 | 1 | 2 | | L:L:?1 | R:R:F168 | 37.01 | 6 | Yes | No | 0 | 4 | 0 | 1 | | R:R:E169 | R:R:H264 | 12.31 | 6 | No | No | 3 | 2 | 1 | 1 | | L:L:?1 | R:R:E169 | 11.14 | 6 | Yes | No | 0 | 3 | 0 | 1 | | R:R:H250 | R:R:N181 | 5.1 | 6 | Yes | No | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 26.02 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:F242 | R:R:W246 | 10.02 | 6 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F242 | R:R:N280 | 9.67 | 6 | Yes | No | 9 | 9 | 2 | 2 | | R:R:H250 | R:R:W246 | 3.17 | 6 | Yes | Yes | 6 | 8 | 1 | 1 | | R:R:S277 | R:R:W246 | 18.53 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N280 | R:R:W246 | 5.65 | 6 | No | Yes | 9 | 8 | 2 | 1 | | L:L:?1 | R:R:W246 | 5.89 | 6 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:H250 | R:R:L249 | 3.86 | 6 | Yes | Yes | 6 | 5 | 1 | 1 | | R:R:L249 | R:R:N253 | 5.49 | 6 | Yes | No | 5 | 5 | 1 | 1 | | R:R:A273 | R:R:L249 | 4.73 | 0 | No | Yes | 6 | 5 | 2 | 1 | | L:L:?1 | R:R:L249 | 13.42 | 6 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 4.16 | 6 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 9.76 | 6 | Yes | No | 0 | 5 | 0 | 1 | | R:R:H264 | R:R:T256 | 6.85 | 6 | No | No | 2 | 4 | 1 | 2 | | L:L:?1 | R:R:H264 | 3.32 | 6 | Yes | No | 0 | 2 | 0 | 1 | | R:R:A265 | R:R:M270 | 3.22 | 0 | No | No | 4 | 4 | 2 | 1 | | L:L:?1 | R:R:M270 | 9.14 | 6 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:Y271 | 3.79 | 6 | Yes | No | 0 | 3 | 0 | 1 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 9.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 10.85 | | Average Nodes In Shell | 24.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 29.00 | | Average Links Mediated by Hubs In Shell | 22.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3VGA | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Antibody | - | 3.1 | 2012-02-01 | doi.org/10.1038/nature10750 |
|
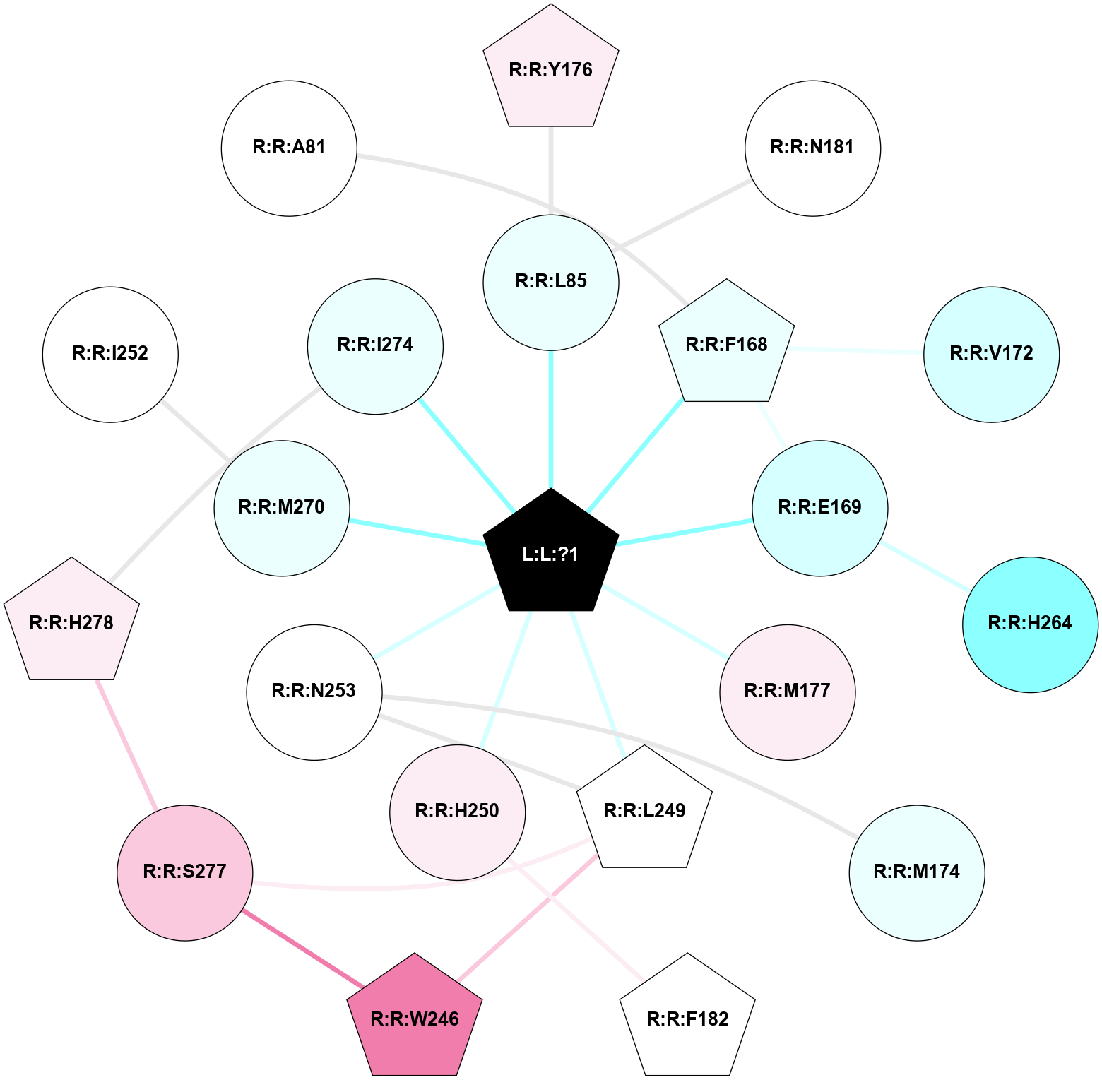
A 2D representation of the interactions of ZMA in 3VGA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:L85 | 3.58 | 5 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:F168 | 32.28 | 5 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:E169 | 23.13 | 5 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M177 | 10.97 | 5 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:L249 | 13.42 | 5 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 3.32 | 5 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 6.21 | 5 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:M270 | 5.48 | 5 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:I274 | 3.69 | 5 | Yes | No | 0 | 4 | 0 | 1 | | R:R:L85 | R:R:Y176 | 3.52 | 0 | No | Yes | 4 | 6 | 1 | 2 | | R:R:L85 | R:R:N181 | 4.12 | 0 | No | No | 4 | 5 | 1 | 2 | | R:R:E169 | R:R:F168 | 3.5 | 5 | No | Yes | 3 | 4 | 1 | 1 | | R:R:F168 | R:R:V172 | 13.11 | 5 | Yes | No | 4 | 3 | 1 | 2 | | R:R:E169 | R:R:H264 | 3.69 | 5 | No | No | 3 | 2 | 1 | 2 | | R:R:M174 | R:R:N253 | 8.41 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:F182 | R:R:H250 | 31.68 | 0 | Yes | No | 5 | 6 | 2 | 1 | | R:R:L249 | R:R:W246 | 5.69 | 5 | Yes | Yes | 5 | 8 | 1 | 2 | | R:R:S277 | R:R:W246 | 13.59 | 5 | No | Yes | 7 | 8 | 2 | 2 | | R:R:L249 | R:R:N253 | 4.12 | 5 | Yes | No | 5 | 5 | 1 | 1 | | R:R:L249 | R:R:S277 | 3 | 5 | Yes | No | 5 | 7 | 1 | 2 | | R:R:H278 | R:R:I274 | 5.3 | 5 | Yes | No | 6 | 4 | 2 | 1 | | R:R:H278 | R:R:S277 | 5.58 | 5 | Yes | No | 6 | 7 | 2 | 2 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 9.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 11.34 | | Average Nodes In Shell | 21.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 24.00 | | Average Links Mediated by Hubs In Shell | 20.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4EIY | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.8 | 2012-07-25 | doi.org/10.1126/science.1219218 |
|
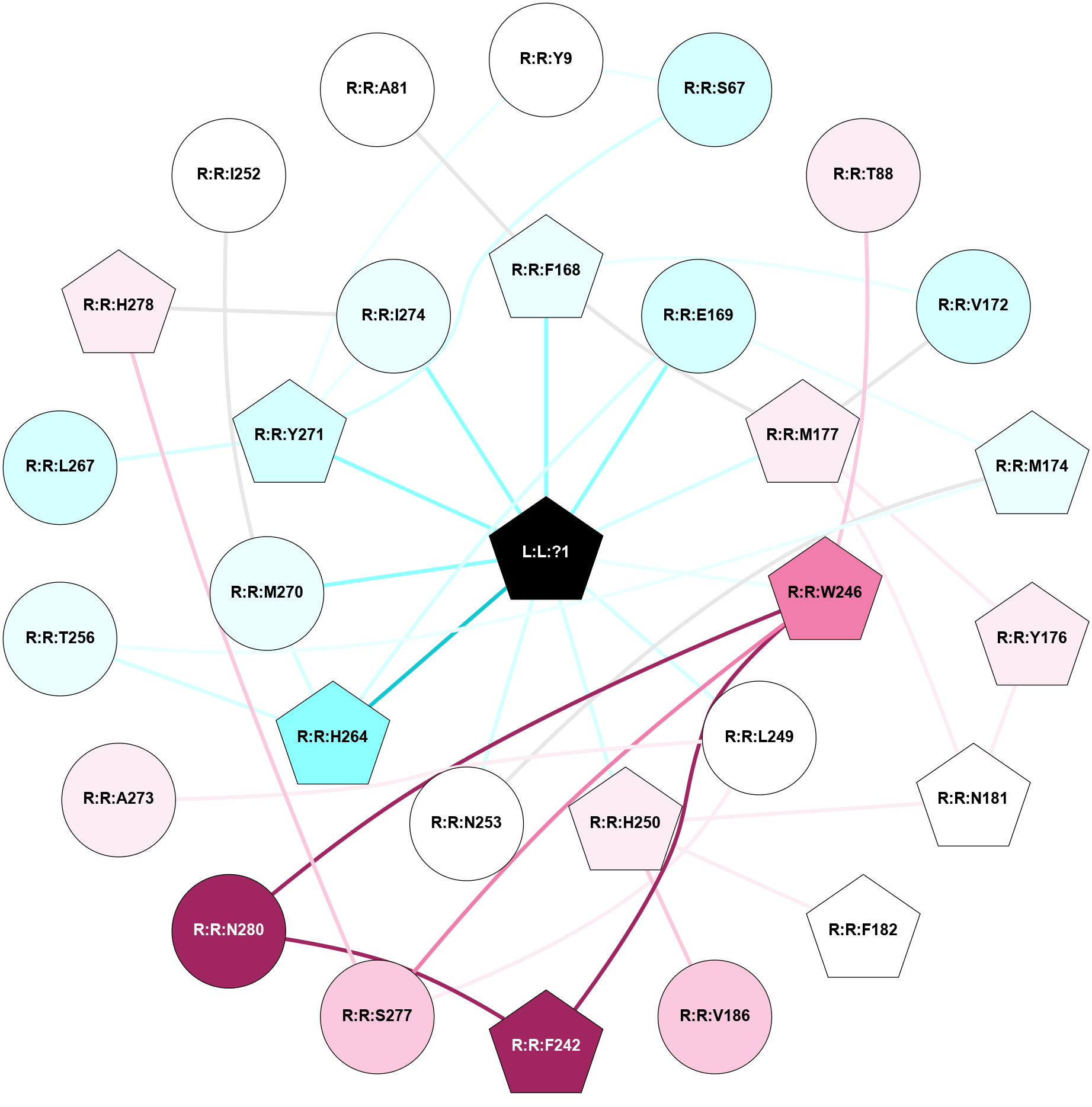
A 2D representation of the interactions of ZMA in 4EIY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:F168 | 38.58 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:E169 | 14.56 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:W246 | 3.68 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:L249 | 19.69 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 5.82 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H264 | 4.16 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:M270 | 9.14 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:Y271 | 4.55 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:I274 | 5.54 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:S67 | R:R:Y9 | 5.09 | 1 | No | No | 3 | 5 | 2 | 2 | | R:R:Y271 | R:R:Y9 | 15.89 | 1 | Yes | No | 3 | 5 | 1 | 2 | | R:R:S67 | R:R:Y271 | 3.82 | 1 | No | Yes | 3 | 3 | 2 | 1 | | R:R:T88 | R:R:W246 | 15.77 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F168 | R:R:V172 | 14.42 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 7.46 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | R:R:E169 | R:R:M174 | 5.41 | 1 | No | Yes | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 9.85 | 1 | No | Yes | 3 | 2 | 1 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 8.41 | 0 | Yes | No | 4 | 5 | 2 | 1 | | R:R:M174 | R:R:T256 | 3.01 | 0 | Yes | No | 4 | 4 | 2 | 2 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 6.98 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:M177 | R:R:N181 | 4.21 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:H250 | R:R:N181 | 3.83 | 0 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 27.15 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 0 | Yes | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 12.03 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F242 | R:R:N280 | 7.25 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:S277 | R:R:W246 | 12.36 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N280 | R:R:W246 | 3.39 | 1 | No | Yes | 9 | 8 | 2 | 1 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:L249 | R:R:S277 | 3 | 0 | No | No | 5 | 7 | 1 | 2 | | R:R:H264 | R:R:T256 | 12.32 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | R:R:L267 | R:R:Y271 | 4.69 | 0 | No | Yes | 3 | 3 | 2 | 1 | | R:R:I274 | R:R:Y271 | 3.63 | 1 | No | Yes | 4 | 3 | 1 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 1 | Yes | No | 6 | 4 | 2 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 1 | Yes | No | 6 | 7 | 2 | 2 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 11.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 11.32 | | Average Nodes In Shell | 30.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 42.00 | | Average Links Mediated by Hubs In Shell | 38.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5IU4 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.72 | 2016-06-29 | doi.org/10.1021/acs.jmedchem.6b00653 |
|

A 2D representation of the interactions of ZMA in 5IU4
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:S67 | R:R:Y9 | 5.09 | 1 | No | No | 3 | 5 | 2 | 2 | | R:R:Y271 | R:R:Y9 | 16.88 | 1 | Yes | No | 3 | 5 | 1 | 2 | | R:R:S67 | R:R:Y271 | 3.82 | 1 | No | Yes | 3 | 3 | 2 | 1 | | R:R:F168 | R:R:V172 | 14.42 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 7.46 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | L:L:?2 | R:R:F168 | 38.58 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:E169 | R:R:M174 | 5.41 | 1 | No | Yes | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 9.85 | 1 | No | Yes | 3 | 2 | 1 | 1 | | L:L:?2 | R:R:E169 | 14.56 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 9.82 | 0 | Yes | No | 4 | 5 | 2 | 1 | | R:R:M174 | R:R:T256 | 3.01 | 0 | Yes | No | 4 | 4 | 2 | 2 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 10.47 | 1 | No | Yes | 5 | 6 | 2 | 2 | | L:L:?2 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:H250 | R:R:N181 | 3.83 | 1 | Yes | No | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:V186 | 3.93 | 1 | Yes | No | 5 | 7 | 2 | 2 | | R:R:F182 | R:R:H250 | 24.89 | 1 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 1 | Yes | No | 6 | 7 | 1 | 2 | | R:R:H250 | R:R:W246 | 3.17 | 1 | Yes | Yes | 6 | 8 | 1 | 2 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | L:L:?2 | R:R:L249 | 19.69 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?2 | R:R:H250 | 7.48 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?2 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:H264 | R:R:T256 | 10.95 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | L:L:?2 | R:R:H264 | 4.16 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | R:R:L267 | R:R:Y271 | 7.03 | 0 | No | Yes | 3 | 3 | 2 | 1 | | L:L:?2 | R:R:M270 | 8.23 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?2 | R:R:Y271 | 4.55 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 1 | Yes | No | 6 | 4 | 2 | 1 | | L:L:?2 | R:R:I274 | 5.54 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 12.16 | | Average Nodes In Shell | 26.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 34.00 | | Average Links Mediated by Hubs In Shell | 31.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5JTB | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | - | - | 2.8 | 2017-05-31 | doi.org/10.1126/sciadv.1602952 |
|
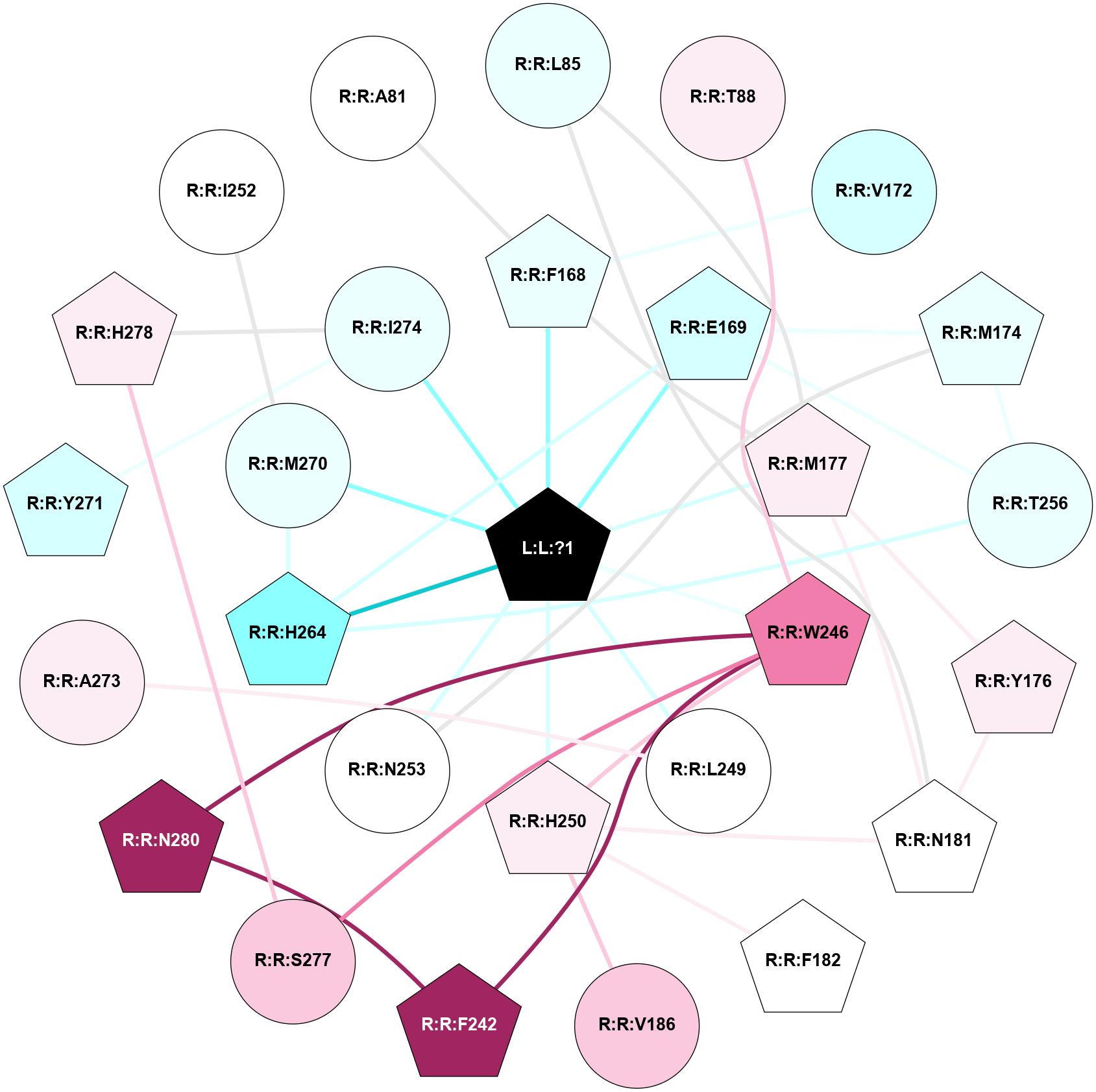
A 2D representation of the interactions of ZMA in 5JTB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L85 | R:R:M177 | 4.24 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:L85 | R:R:N181 | 8.24 | 1 | No | Yes | 4 | 5 | 2 | 2 | | R:R:T88 | R:R:W246 | 7.28 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F168 | R:R:V172 | 14.42 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 7.46 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | L:L:?1 | R:R:F168 | 37.01 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:E169 | R:R:M174 | 5.41 | 1 | Yes | Yes | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:T256 | 5.64 | 1 | Yes | No | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 9.85 | 1 | Yes | Yes | 3 | 2 | 1 | 1 | | L:L:?1 | R:R:E169 | 13.71 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | R:R:M174 | R:R:N253 | 8.41 | 1 | Yes | No | 4 | 5 | 2 | 1 | | R:R:M174 | R:R:T256 | 4.52 | 1 | Yes | No | 4 | 4 | 2 | 2 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 3.49 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:M177 | R:R:N181 | 5.61 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:H250 | R:R:N181 | 3.83 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 24.89 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 1 | Yes | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 12.03 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:H250 | R:R:W246 | 3.17 | 1 | Yes | Yes | 6 | 8 | 1 | 1 | | R:R:S277 | R:R:W246 | 16.06 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N280 | R:R:W246 | 4.52 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | L:L:?1 | R:R:W246 | 3.68 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | L:L:?1 | R:R:L249 | 20.58 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 3.32 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:H264 | R:R:T256 | 12.32 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 3.94 | 1 | Yes | No | 2 | 4 | 1 | 1 | | L:L:?1 | R:R:H264 | 5.82 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:M270 | 10.05 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:I274 | R:R:Y271 | 3.63 | 0 | No | Yes | 4 | 3 | 1 | 2 | | R:R:H278 | R:R:I274 | 3.98 | 3 | Yes | No | 6 | 4 | 2 | 1 | | L:L:?1 | R:R:I274 | 5.54 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 3 | Yes | No | 6 | 7 | 2 | 2 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 11.85 | | Average Nodes In Shell | 28.00 | | Average Hubs In Shell | 15.00 | | Average Links In Shell | 39.00 | | Average Links Mediated by Hubs In Shell | 37.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5K2A | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2.5 | 2016-09-21 | doi.org/10.1126/sciadv.1600292 |
|

A 2D representation of the interactions of ZMA in 5K2A
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:F168 | 33.07 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:E169 | 12.85 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M177 | 5.48 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:W246 | 3.68 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:L249 | 20.58 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 7.48 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H264 | 3.32 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:L267 | 5.37 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M270 | 6.4 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:I274 | 3.69 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:T88 | R:R:W246 | 12.13 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F168 | R:R:V172 | 11.8 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 6.22 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | R:R:E169 | R:R:M174 | 6.77 | 1 | No | No | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 11.08 | 1 | No | Yes | 3 | 2 | 1 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 7.01 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 4.65 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:M177 | R:R:N181 | 4.21 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:H250 | R:R:N181 | 3.83 | 0 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 23.76 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 0 | Yes | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 11.02 | 0 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:S277 | R:R:W246 | 14.83 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:L249 | R:R:S277 | 3 | 0 | No | No | 5 | 7 | 1 | 2 | | R:R:I252 | R:R:M270 | 4.37 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:H264 | R:R:T256 | 8.21 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | R:R:L267 | R:R:Y271 | 4.69 | 0 | No | Yes | 3 | 3 | 1 | 2 | | R:R:I274 | R:R:Y271 | 3.63 | 0 | No | Yes | 4 | 3 | 1 | 2 | | R:R:H278 | R:R:I274 | 3.98 | 4 | Yes | No | 6 | 4 | 2 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 4 | Yes | No | 6 | 7 | 2 | 2 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 11.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 10.40 | | Average Nodes In Shell | 27.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 36.00 | | Average Links Mediated by Hubs In Shell | 31.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5K2B | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2.5 | 2016-09-21 | doi.org/10.1126/sciadv.1600292 |
|
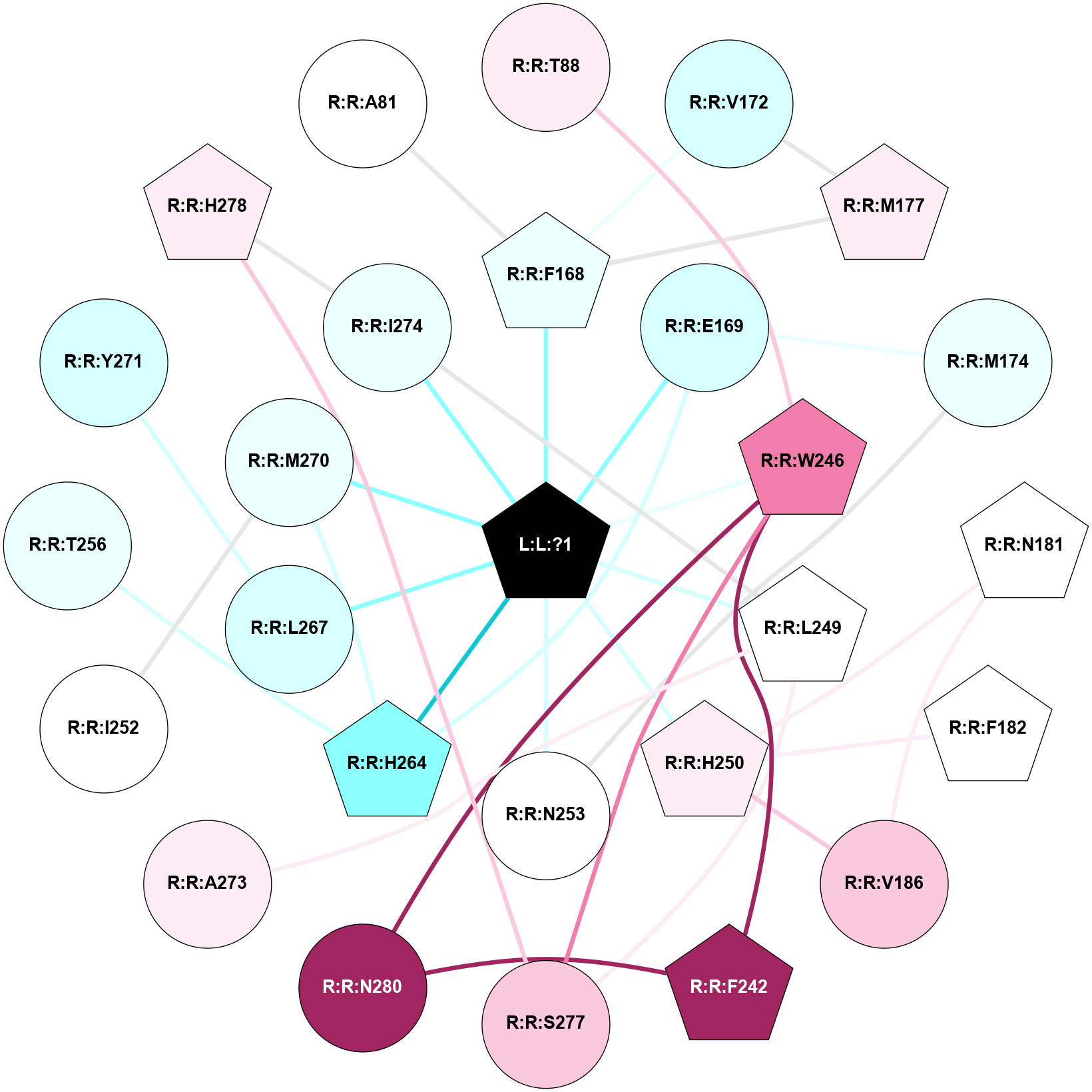
A 2D representation of the interactions of ZMA in 5K2B
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:F168 | 35.43 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:E169 | 12.85 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:W246 | 2.95 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:L249 | 19.69 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 6.65 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H264 | 3.32 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:L267 | 3.58 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M270 | 6.4 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:I274 | 4.61 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:T88 | R:R:W246 | 12.13 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F168 | R:R:V172 | 7.87 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 8.71 | 1 | Yes | Yes | 4 | 6 | 1 | 2 | | R:R:E169 | R:R:M174 | 6.77 | 1 | No | No | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 11.08 | 1 | No | Yes | 3 | 2 | 1 | 1 | | R:R:M177 | R:R:V172 | 4.56 | 1 | Yes | No | 6 | 3 | 2 | 2 | | R:R:M174 | R:R:N253 | 5.61 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:N181 | R:R:V186 | 2.96 | 1 | Yes | No | 5 | 7 | 2 | 2 | | R:R:H250 | R:R:N181 | 3.83 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 22.63 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 1 | Yes | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 11.02 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:S277 | R:R:W246 | 14.83 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N280 | R:R:W246 | 4.52 | 1 | No | Yes | 9 | 8 | 2 | 1 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:L249 | R:R:S277 | 3 | 0 | No | No | 5 | 7 | 1 | 2 | | R:R:H264 | R:R:T256 | 9.58 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | R:R:L267 | R:R:Y271 | 4.69 | 0 | No | No | 3 | 3 | 1 | 2 | | R:R:H278 | R:R:I274 | 3.98 | 3 | Yes | No | 6 | 4 | 2 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 3 | Yes | No | 6 | 7 | 2 | 2 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 10.79 | | Average Nodes In Shell | 27.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 34.00 | | Average Links Mediated by Hubs In Shell | 28.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5K2C | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.9 | 2016-09-21 | doi.org/10.1126/sciadv.1600292 |
|
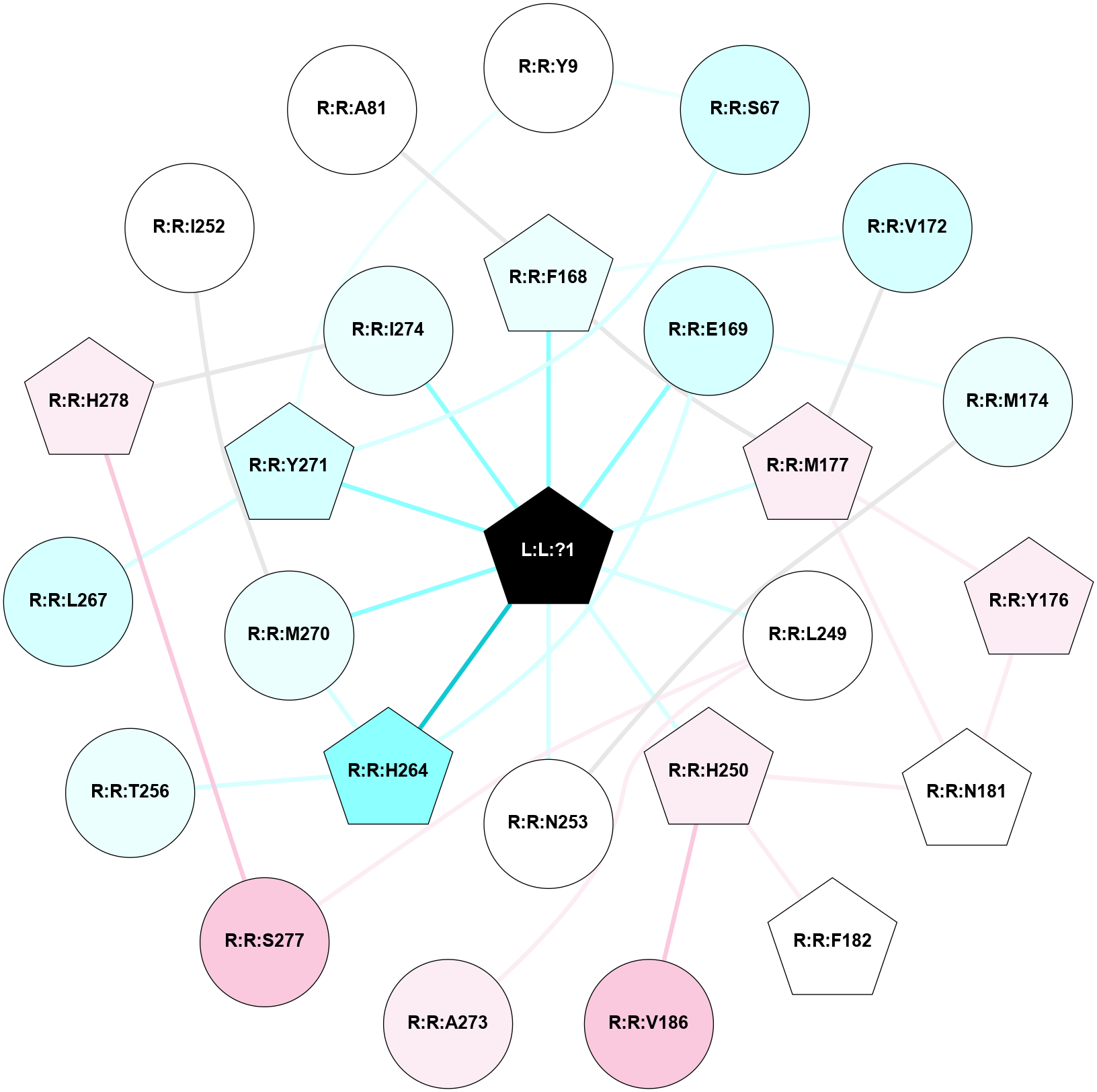
A 2D representation of the interactions of ZMA in 5K2C
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:F168 | 35.43 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:E169 | 13.71 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:L249 | 16.11 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 7.48 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H264 | 3.32 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:M270 | 9.14 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:Y271 | 3.03 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:I274 | 4.61 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:S67 | R:R:Y9 | 5.09 | 1 | No | No | 3 | 5 | 2 | 2 | | R:R:Y271 | R:R:Y9 | 15.89 | 1 | Yes | No | 3 | 5 | 1 | 2 | | R:R:S67 | R:R:Y271 | 3.82 | 1 | No | Yes | 3 | 3 | 2 | 1 | | R:R:F168 | R:R:V172 | 11.8 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 6.22 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | R:R:E169 | R:R:M174 | 6.77 | 1 | No | No | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 11.08 | 1 | No | Yes | 3 | 2 | 1 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 7.01 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 5.81 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:M177 | R:R:N181 | 4.21 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:H250 | R:R:N181 | 3.83 | 0 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 23.76 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 0 | Yes | No | 6 | 7 | 1 | 2 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:L249 | R:R:S277 | 3 | 0 | No | No | 5 | 7 | 1 | 2 | | R:R:H264 | R:R:T256 | 6.85 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | R:R:L267 | R:R:Y271 | 4.69 | 0 | No | Yes | 3 | 3 | 2 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 1 | Yes | No | 6 | 4 | 2 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 1 | Yes | No | 6 | 7 | 2 | 2 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 11.17 | | Average Nodes In Shell | 26.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 34.00 | | Average Links Mediated by Hubs In Shell | 28.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5K2D | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.9 | 2016-09-21 | doi.org/10.1126/sciadv.1600292 |
|

A 2D representation of the interactions of ZMA in 5K2D
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:F168 | 35.43 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:E169 | 13.71 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M177 | 5.48 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:L249 | 16.11 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 7.48 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H264 | 3.32 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:M270 | 8.23 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:I274 | 4.61 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:F168 | R:R:V172 | 13.11 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 6.22 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | R:R:E169 | R:R:M174 | 6.77 | 1 | No | No | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 11.08 | 1 | No | Yes | 3 | 2 | 1 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 7.01 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 5.81 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:M177 | R:R:N181 | 4.21 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:H250 | R:R:N181 | 3.83 | 0 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 22.63 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 0 | Yes | No | 6 | 7 | 1 | 2 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:L249 | R:R:S277 | 3 | 0 | No | No | 5 | 7 | 1 | 2 | | R:R:H264 | R:R:T256 | 6.85 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 2 | Yes | No | 6 | 4 | 2 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 2 | Yes | No | 6 | 7 | 2 | 2 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 9.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 11.87 | | Average Nodes In Shell | 22.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 29.00 | | Average Links Mediated by Hubs In Shell | 24.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5NLX | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2.14 | 2017-09-27 | doi.org/10.1038/s41467-017-00630-4 |
|
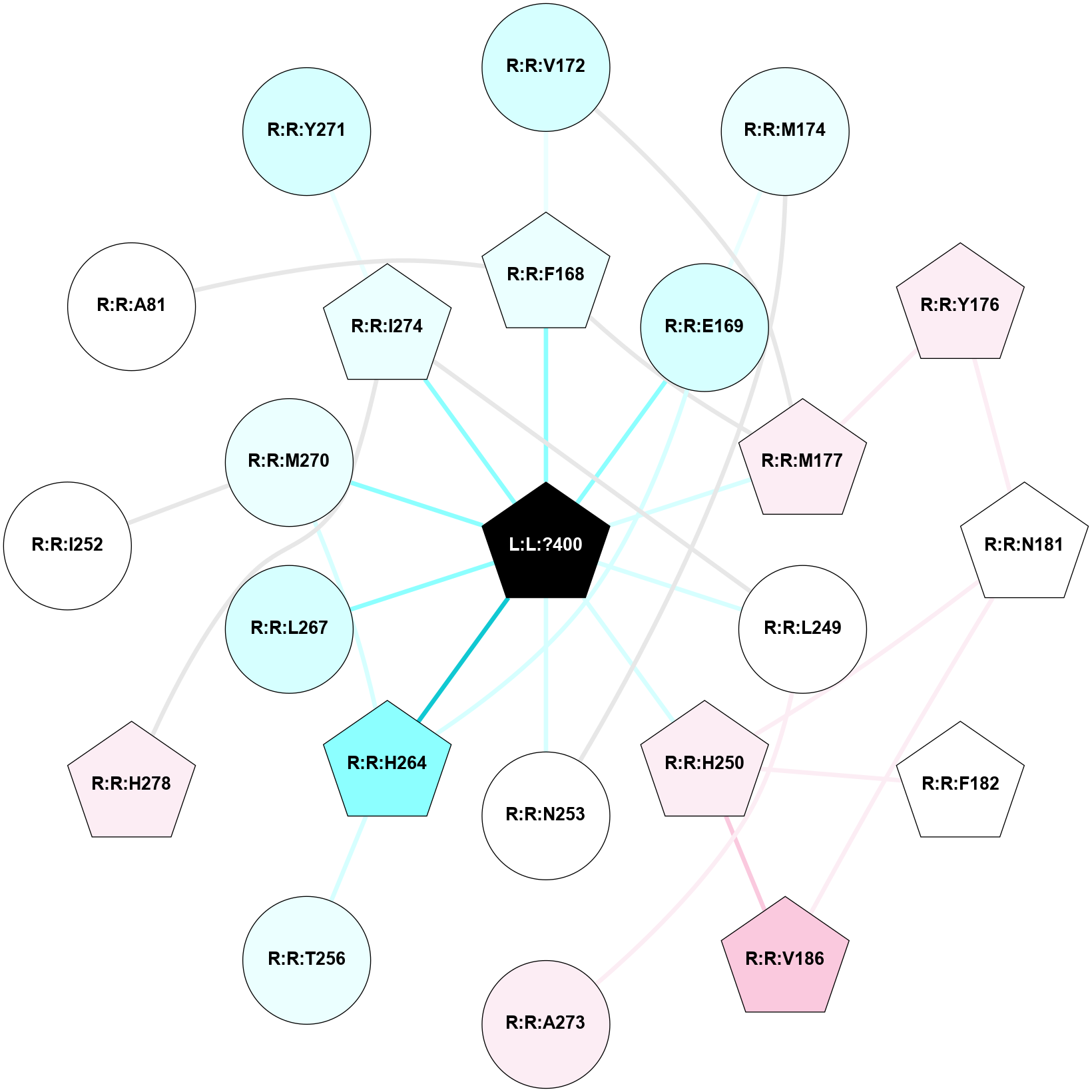
A 2D representation of the interactions of ZMA in 5NLX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?400 | R:R:F168 | 39.37 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?400 | R:R:E169 | 13.71 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?400 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?400 | R:R:L249 | 19.69 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?400 | R:R:H250 | 8.31 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?400 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?400 | R:R:H264 | 4.16 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?400 | R:R:L267 | 6.26 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?400 | R:R:M270 | 7.31 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?400 | R:R:I274 | 3.69 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:F168 | R:R:V172 | 11.8 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 7.46 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | R:R:E169 | R:R:M174 | 6.77 | 1 | No | No | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 9.85 | 1 | No | Yes | 3 | 2 | 1 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 5.61 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 8.14 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:N181 | R:R:V186 | 2.96 | 1 | Yes | Yes | 5 | 7 | 2 | 2 | | R:R:H250 | R:R:N181 | 5.1 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 22.63 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 5.54 | 1 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:A273 | R:R:L249 | 4.73 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:I274 | R:R:L249 | 4.28 | 1 | Yes | No | 4 | 5 | 1 | 1 | | R:R:H264 | R:R:T256 | 8.21 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 1 | Yes | Yes | 6 | 4 | 2 | 1 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | | R:R:I274 | R:R:Y271 | 2.42 | 1 | Yes | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 12.13 | | Average Nodes In Shell | 23.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 30.00 | | Average Links Mediated by Hubs In Shell | 26.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5NM2 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.95 | 2017-09-27 | doi.org/10.1038/s41467-017-00630-4 |
|
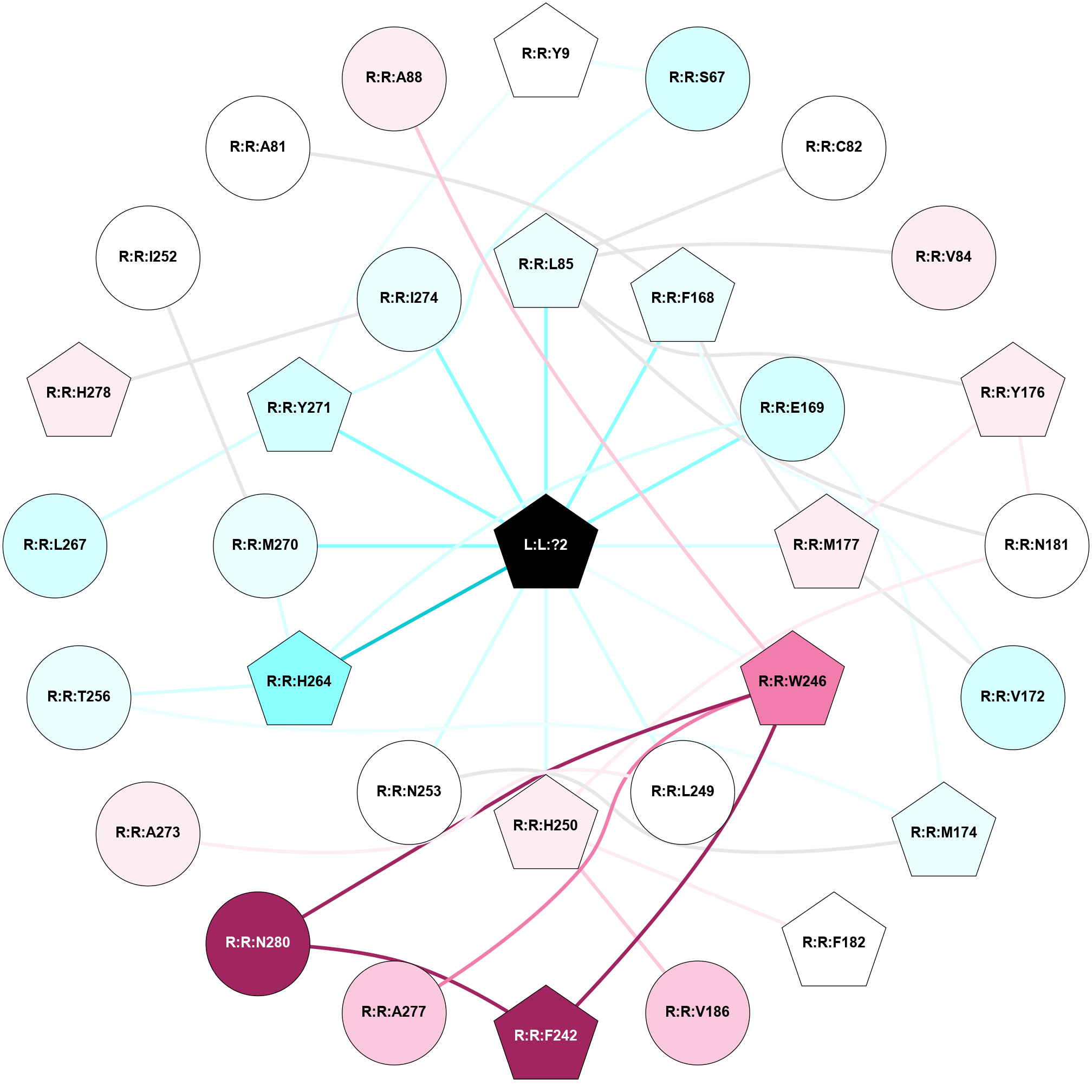
A 2D representation of the interactions of ZMA in 5NM2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:S67 | R:R:Y9 | 5.09 | 1 | No | Yes | 3 | 5 | 2 | 2 | | R:R:Y271 | R:R:Y9 | 16.88 | 1 | Yes | Yes | 3 | 5 | 1 | 2 | | R:R:S67 | R:R:Y271 | 3.82 | 1 | No | Yes | 3 | 3 | 2 | 1 | | R:R:C82 | R:R:L85 | 3.17 | 0 | No | Yes | 5 | 4 | 2 | 1 | | R:R:L85 | R:R:V84 | 2.98 | 1 | Yes | No | 4 | 6 | 1 | 2 | | R:R:L85 | R:R:Y176 | 3.52 | 1 | Yes | Yes | 4 | 6 | 1 | 2 | | R:R:L85 | R:R:N181 | 8.24 | 1 | Yes | No | 4 | 5 | 1 | 2 | | L:L:?2 | R:R:L85 | 3.58 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:F168 | R:R:V172 | 11.8 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 7.46 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | L:L:?2 | R:R:F168 | 38.58 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:E169 | R:R:M174 | 5.41 | 1 | No | Yes | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 11.08 | 1 | No | Yes | 3 | 2 | 1 | 1 | | L:L:?2 | R:R:E169 | 14.56 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 9.82 | 0 | Yes | No | 4 | 5 | 2 | 1 | | R:R:M174 | R:R:T256 | 3.01 | 0 | Yes | No | 4 | 4 | 2 | 2 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 9.3 | 1 | No | Yes | 5 | 6 | 2 | 2 | | L:L:?2 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:H250 | R:R:N181 | 3.83 | 0 | Yes | No | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 24.89 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 0 | Yes | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 17.04 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:A277 | R:R:W246 | 9.08 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N280 | R:R:W246 | 3.39 | 1 | No | Yes | 9 | 8 | 2 | 1 | | L:L:?2 | R:R:W246 | 3.68 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | L:L:?2 | R:R:L249 | 19.69 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?2 | R:R:H250 | 7.48 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?2 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:H264 | R:R:T256 | 12.32 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 6.57 | 1 | Yes | No | 2 | 4 | 1 | 1 | | L:L:?2 | R:R:H264 | 4.16 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | R:R:L267 | R:R:Y271 | 7.03 | 0 | No | Yes | 3 | 3 | 2 | 1 | | L:L:?2 | R:R:M270 | 9.14 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?2 | R:R:Y271 | 4.55 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 1 | Yes | No | 6 | 4 | 2 | 1 | | L:L:?2 | R:R:I274 | 5.54 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | | R:R:A88 | R:R:W246 | 2.59 | 0 | No | Yes | 6 | 8 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 12.00 | | Average Number Of Links With An Hub | 7.00 | | Average Interaction Strength | 10.82 | | Average Nodes In Shell | 33.00 | | Average Hubs In Shell | 14.00 | | Average Links In Shell | 43.00 | | Average Links Mediated by Hubs In Shell | 41.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5NM4 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.7 | 2017-09-27 | doi.org/10.1038/s41467-017-00630-4 |
|
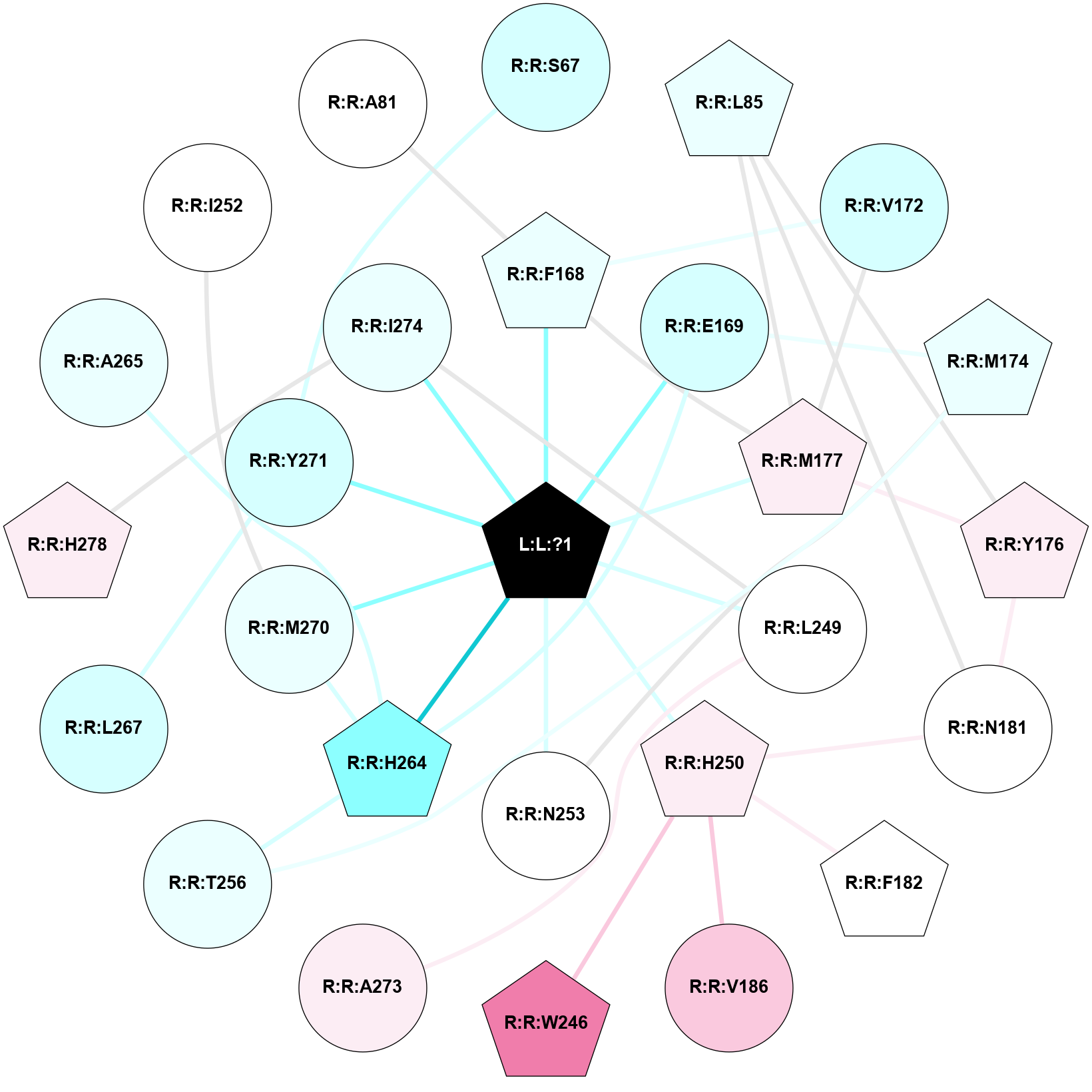
A 2D representation of the interactions of ZMA in 5NM4
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:F168 | 39.37 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:E169 | 13.71 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:L249 | 20.58 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 6.65 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H264 | 3.32 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:M270 | 8.23 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:Y271 | 3.03 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:I274 | 3.69 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:S67 | R:R:Y271 | 3.82 | 0 | No | No | 3 | 3 | 2 | 1 | | R:R:L85 | R:R:Y176 | 3.52 | 1 | Yes | Yes | 4 | 6 | 2 | 2 | | R:R:L85 | R:R:M177 | 4.24 | 1 | Yes | Yes | 4 | 6 | 2 | 1 | | R:R:L85 | R:R:N181 | 5.49 | 1 | Yes | No | 4 | 5 | 2 | 2 | | R:R:F168 | R:R:V172 | 11.8 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 7.46 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | R:R:E169 | R:R:M174 | 5.41 | 1 | No | Yes | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 11.08 | 1 | No | Yes | 3 | 2 | 1 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 9.82 | 0 | Yes | No | 4 | 5 | 2 | 1 | | R:R:M174 | R:R:T256 | 4.52 | 0 | Yes | No | 4 | 4 | 2 | 2 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 8.14 | 1 | No | Yes | 5 | 6 | 2 | 2 | | R:R:H250 | R:R:N181 | 5.1 | 0 | Yes | No | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 22.63 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 0 | Yes | No | 6 | 7 | 1 | 2 | | R:R:H250 | R:R:W246 | 3.17 | 0 | Yes | Yes | 6 | 8 | 1 | 2 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:I274 | R:R:L249 | 4.28 | 1 | No | No | 4 | 5 | 1 | 1 | | R:R:H264 | R:R:T256 | 10.95 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | R:R:L267 | R:R:Y271 | 5.86 | 0 | No | No | 3 | 3 | 2 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 1 | Yes | No | 6 | 4 | 2 | 1 | | R:R:A265 | R:R:H264 | 2.93 | 0 | No | Yes | 4 | 2 | 2 | 1 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 11.74 | | Average Nodes In Shell | 27.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 36.00 | | Average Links Mediated by Hubs In Shell | 31.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5OLG | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.87 | 2018-01-17 | doi.org/10.1038/s41598-017-18570-w |
|

A 2D representation of the interactions of ZMA in 5OLG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:S67 | R:R:Y9 | 5.09 | 1 | No | No | 3 | 5 | 2 | 2 | | R:R:Y271 | R:R:Y9 | 16.88 | 1 | Yes | No | 3 | 5 | 1 | 2 | | R:R:S67 | R:R:Y271 | 3.82 | 1 | No | Yes | 3 | 3 | 2 | 1 | | R:R:A88 | R:R:W246 | 3.89 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F168 | R:R:V172 | 11.8 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 7.46 | 1 | Yes | No | 4 | 6 | 1 | 1 | | L:L:?1 | R:R:F168 | 39.37 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:E169 | R:R:M174 | 5.41 | 1 | No | No | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 11.08 | 1 | No | Yes | 3 | 2 | 1 | 1 | | L:L:?1 | R:R:E169 | 15.42 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:M174 | R:R:N253 | 8.41 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | No | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 11.63 | 1 | No | Yes | 5 | 6 | 2 | 2 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:H250 | R:R:N181 | 3.83 | 1 | Yes | No | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:V186 | 3.93 | 1 | Yes | Yes | 5 | 7 | 2 | 2 | | R:R:F182 | R:R:H250 | 26.02 | 1 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 1 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 17.04 | 6 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:A277 | R:R:W246 | 7.78 | 0 | No | Yes | 7 | 8 | 2 | 1 | | L:L:?1 | R:R:W246 | 3.68 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:L249 | 18.79 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 6.65 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:H264 | R:R:T256 | 10.95 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | L:L:?1 | R:R:H264 | 4.99 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | R:R:L267 | R:R:Y271 | 7.03 | 0 | No | Yes | 3 | 3 | 2 | 1 | | L:L:?1 | R:R:M270 | 9.14 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:Y271 | 3.79 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 1 | Yes | No | 6 | 4 | 2 | 1 | | L:L:?1 | R:R:I274 | 5.54 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 11.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 11.47 | | Average Nodes In Shell | 29.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 35.00 | | Average Links Mediated by Hubs In Shell | 30.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5UVI | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | - | - | 3.2 | 2017-05-24 | doi.org/10.1107/S205225251700570X |
|

A 2D representation of the interactions of ZMA in 5UVI
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:F168 | 40.94 | 2 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:E169 | 6 | 2 | Yes | Yes | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M177 | 6.4 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:W246 | 3.68 | 2 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:L249 | 16.11 | 2 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 5.82 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 2 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H264 | 3.32 | 2 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:L267 | 6.26 | 2 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M270 | 7.31 | 2 | Yes | No | 0 | 4 | 0 | 1 | | R:R:L85 | R:R:Y176 | 3.52 | 2 | Yes | Yes | 4 | 6 | 2 | 2 | | R:R:L85 | R:R:M177 | 2.83 | 2 | Yes | Yes | 4 | 6 | 2 | 1 | | R:R:L85 | R:R:N181 | 8.24 | 2 | Yes | No | 4 | 5 | 2 | 2 | | R:R:T88 | R:R:W246 | 10.92 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F168 | R:R:V172 | 3.93 | 0 | No | No | 4 | 3 | 1 | 2 | | R:R:E169 | R:R:M174 | 6.77 | 2 | Yes | No | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:T256 | 5.64 | 2 | Yes | No | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 11.08 | 2 | Yes | Yes | 3 | 2 | 1 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 2 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 7.01 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:M177 | R:R:Y176 | 3.59 | 2 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:H250 | R:R:N181 | 3.83 | 0 | Yes | No | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 22.63 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 0 | Yes | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 11.02 | 0 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:S277 | R:R:W246 | 14.83 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:I274 | R:R:L249 | 2.85 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:L249 | R:R:S277 | 3 | 0 | No | No | 5 | 7 | 1 | 2 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:H264 | R:R:T256 | 9.58 | 2 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 3.94 | 2 | Yes | No | 2 | 4 | 1 | 1 | | R:R:L267 | R:R:Y271 | 4.69 | 0 | No | Yes | 3 | 3 | 1 | 2 | | R:R:I274 | R:R:Y271 | 3.63 | 0 | No | Yes | 4 | 3 | 2 | 2 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | No | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 10.83 | | Average Nodes In Shell | 26.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 34.00 | | Average Links Mediated by Hubs In Shell | 28.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5VRA | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2.35 | 2017-12-13 | doi.org/10.1038/nprot.2017.135 |
|
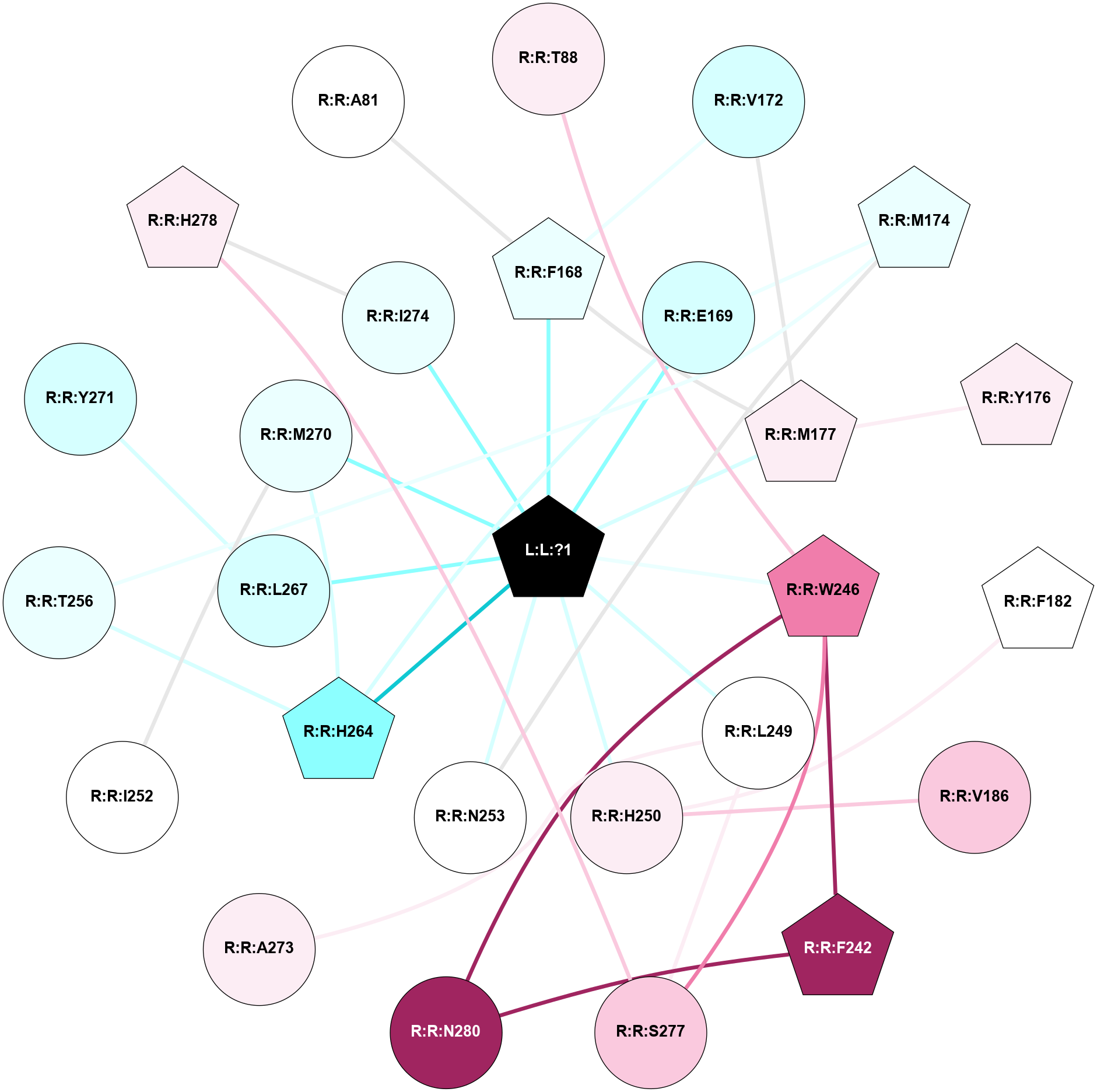
A 2D representation of the interactions of ZMA in 5VRA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:F168 | 37.79 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:E169 | 15.42 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:W246 | 5.15 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:L249 | 20.58 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 7.48 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H264 | 4.16 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:L267 | 3.58 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M270 | 8.23 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:I274 | 5.54 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:T88 | R:R:W246 | 10.92 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F168 | R:R:V172 | 14.42 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 7.46 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | R:R:E169 | R:R:M174 | 5.41 | 1 | No | Yes | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 8.62 | 1 | No | Yes | 3 | 2 | 1 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 8.41 | 0 | Yes | No | 4 | 5 | 2 | 1 | | R:R:M174 | R:R:T256 | 3.01 | 0 | Yes | No | 4 | 4 | 2 | 2 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:F182 | R:R:H250 | 27.15 | 0 | Yes | No | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 8.3 | 0 | No | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 11.02 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:S277 | R:R:W246 | 14.83 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N280 | R:R:W246 | 3.39 | 1 | No | Yes | 9 | 8 | 2 | 1 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:L249 | R:R:S277 | 3 | 0 | No | No | 5 | 7 | 1 | 2 | | R:R:I252 | R:R:M270 | 4.37 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:H264 | R:R:T256 | 9.58 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | R:R:L267 | R:R:Y271 | 5.86 | 0 | No | No | 3 | 3 | 1 | 2 | | R:R:H278 | R:R:I274 | 3.98 | 3 | Yes | No | 6 | 4 | 2 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 3 | Yes | No | 6 | 7 | 2 | 2 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 11.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 11.52 | | Average Nodes In Shell | 27.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 35.00 | | Average Links Mediated by Hubs In Shell | 30.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6AQF | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | - | - | 2.51 | 2018-01-10 | doi.org/10.1016/j.cell.2017.12.004 |
|

A 2D representation of the interactions of ZMA in 6AQF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:F168 | 37.01 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:E169 | 12.85 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:W246 | 4.42 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:L249 | 18.79 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 5.82 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:M270 | 4.57 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:Y271 | 3.03 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:I274 | 5.54 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:S67 | R:R:Y9 | 5.09 | 1 | No | No | 3 | 5 | 2 | 2 | | R:R:Y271 | R:R:Y9 | 14.89 | 1 | Yes | No | 3 | 5 | 1 | 2 | | R:R:S67 | R:R:Y271 | 3.82 | 1 | No | Yes | 3 | 3 | 2 | 1 | | R:R:L85 | R:R:M177 | 4.24 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:L85 | R:R:N181 | 8.24 | 1 | No | No | 4 | 5 | 2 | 2 | | R:R:F242 | R:R:T88 | 3.89 | 1 | Yes | No | 9 | 6 | 2 | 2 | | R:R:T88 | R:R:W246 | 12.13 | 1 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F168 | R:R:V172 | 14.42 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 6.22 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | R:R:E169 | R:R:M174 | 5.41 | 1 | Yes | Yes | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:T256 | 5.64 | 1 | Yes | No | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 11.08 | 1 | Yes | No | 3 | 2 | 1 | 2 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 7.01 | 1 | Yes | No | 4 | 5 | 2 | 1 | | R:R:M174 | R:R:T256 | 3.01 | 1 | Yes | No | 4 | 4 | 2 | 2 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 4.65 | 1 | No | Yes | 5 | 6 | 2 | 2 | | R:R:M177 | R:R:N181 | 4.21 | 1 | Yes | No | 6 | 5 | 1 | 2 | | R:R:M177 | R:R:N253 | 4.21 | 1 | Yes | No | 6 | 5 | 1 | 1 | | R:R:F182 | R:R:H250 | 19.23 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 1 | Yes | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 11.02 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:H250 | R:R:W246 | 3.17 | 1 | Yes | Yes | 6 | 8 | 1 | 1 | | R:R:S277 | R:R:W246 | 17.3 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:L249 | R:R:S277 | 3 | 0 | No | No | 5 | 7 | 1 | 2 | | R:R:H264 | R:R:T256 | 9.58 | 1 | No | No | 2 | 4 | 2 | 2 | | R:R:H264 | R:R:M270 | 3.94 | 1 | No | No | 2 | 4 | 2 | 1 | | R:R:A265 | R:R:M270 | 3.22 | 0 | No | No | 4 | 4 | 2 | 1 | | R:R:L267 | R:R:Y271 | 4.69 | 0 | No | Yes | 3 | 3 | 2 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 1 | Yes | No | 6 | 4 | 2 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 1 | Yes | No | 6 | 7 | 2 | 2 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 11.09 | | Average Nodes In Shell | 30.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 43.00 | | Average Links Mediated by Hubs In Shell | 36.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6JZH | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2.25 | 2019-10-30 | doi.org/10.1107/S1600576719012846 |
|

A 2D representation of the interactions of ZMA in 6JZH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?2 | R:R:F168 | 35.43 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?2 | R:R:E169 | 13.71 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?2 | R:R:M177 | 5.48 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?2 | R:R:L249 | 20.58 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?2 | R:R:H250 | 7.48 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?2 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?2 | R:R:H264 | 3.32 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?2 | R:R:M270 | 7.31 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?2 | R:R:I274 | 4.61 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:F168 | R:R:V172 | 11.8 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 6.22 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | R:R:E169 | R:R:H264 | 11.08 | 1 | No | Yes | 3 | 2 | 1 | 1 | | R:R:M174 | R:R:N253 | 7.01 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 4.65 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:M177 | R:R:N181 | 4.21 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:H250 | R:R:N181 | 3.83 | 0 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 26.02 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 8.3 | 0 | Yes | No | 6 | 7 | 1 | 2 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:H264 | R:R:T256 | 9.58 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 2 | Yes | No | 6 | 4 | 2 | 1 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 9.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 12.26 | | Average Nodes In Shell | 21.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 25.00 | | Average Links Mediated by Hubs In Shell | 22.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6LPJ | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.8 | 2020-11-25 | doi.org/10.1038/s41598-020-76277-x |
|

A 2D representation of the interactions of ZMA in 6LPJ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:S67 | R:R:Y9 | 5.09 | 1 | No | No | 3 | 5 | 2 | 2 | | R:R:Y271 | R:R:Y9 | 15.89 | 1 | Yes | No | 3 | 5 | 1 | 2 | | R:R:S67 | R:R:Y271 | 3.82 | 1 | No | Yes | 3 | 3 | 2 | 1 | | R:R:F168 | R:R:V172 | 14.42 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 6.22 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | L:L:?1 | R:R:F168 | 38.58 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:E169 | R:R:M174 | 6.77 | 1 | No | Yes | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 9.85 | 1 | No | Yes | 3 | 2 | 1 | 1 | | L:L:?1 | R:R:E169 | 13.71 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 7.01 | 0 | Yes | No | 4 | 5 | 2 | 1 | | R:R:M174 | R:R:T256 | 3.01 | 0 | Yes | No | 4 | 4 | 2 | 2 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 8.14 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:M177 | R:R:N181 | 5.61 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:H250 | R:R:N181 | 3.83 | 0 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 27.15 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 0 | Yes | No | 6 | 7 | 1 | 2 | | R:R:A273 | R:R:L249 | 4.73 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:L249 | R:R:S277 | 3 | 0 | No | No | 5 | 7 | 1 | 2 | | L:L:?1 | R:R:L249 | 20.58 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 7.48 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:H264 | R:R:T256 | 9.58 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | L:L:?1 | R:R:H264 | 3.32 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | R:R:L267 | R:R:Y271 | 3.52 | 1 | No | Yes | 3 | 3 | 1 | 1 | | L:L:?1 | R:R:L267 | 6.26 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M270 | 9.14 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:Y271 | 3.79 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 1 | Yes | No | 6 | 4 | 2 | 1 | | L:L:?1 | R:R:I274 | 6.46 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 1 | Yes | No | 6 | 7 | 2 | 2 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 11.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 11.65 | | Average Nodes In Shell | 26.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 36.00 | | Average Links Mediated by Hubs In Shell | 32.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6LPK | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.8 | 2020-11-25 | doi.org/10.1038/s41598-020-76277-x |
|
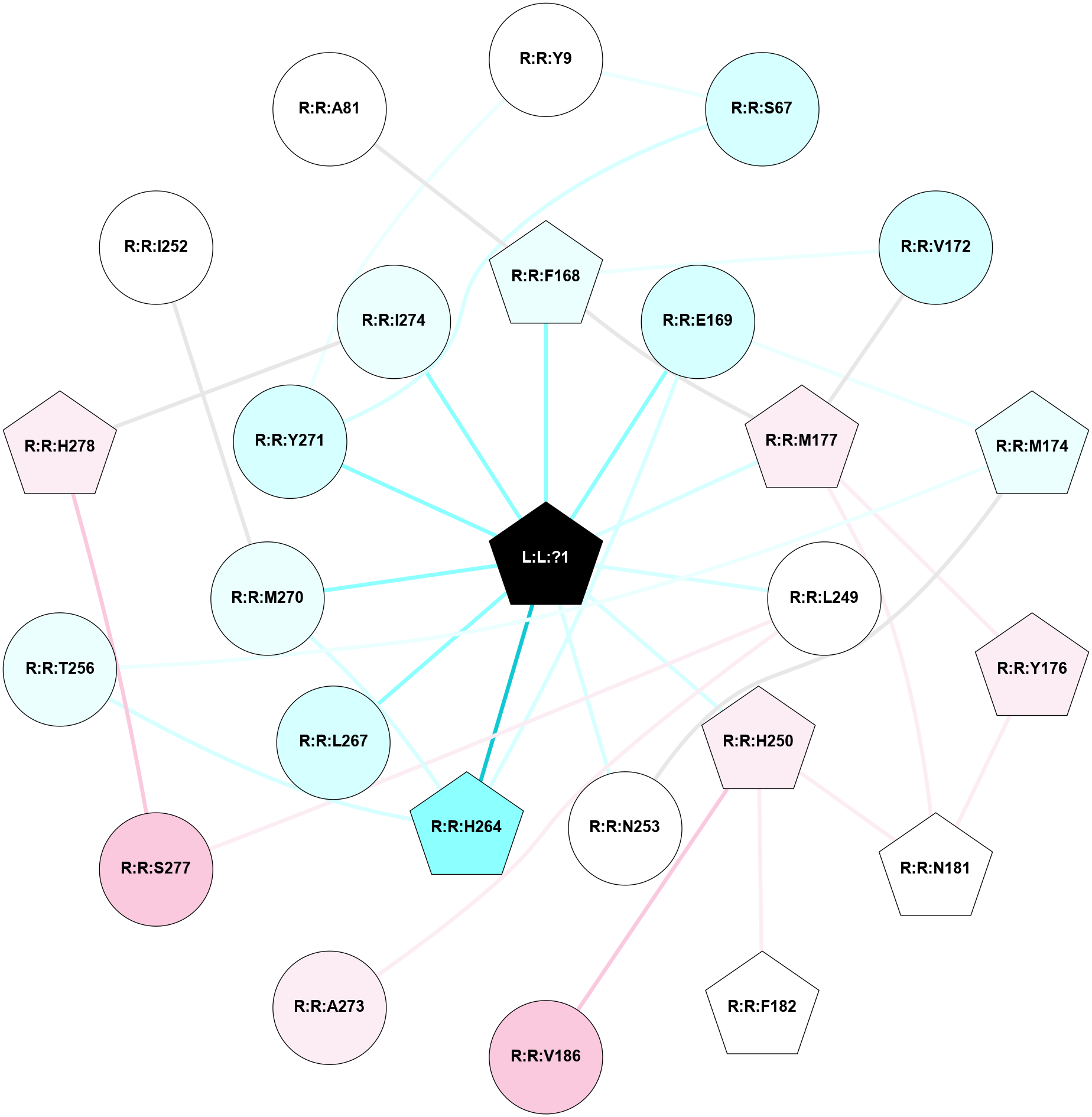
A 2D representation of the interactions of ZMA in 6LPK
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:S67 | R:R:Y9 | 5.09 | 1 | No | No | 3 | 5 | 2 | 2 | | R:R:Y271 | R:R:Y9 | 15.89 | 1 | No | No | 3 | 5 | 1 | 2 | | R:R:S67 | R:R:Y271 | 3.82 | 1 | No | No | 3 | 3 | 2 | 1 | | R:R:F168 | R:R:V172 | 15.73 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 6.22 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | L:L:?1 | R:R:F168 | 37.79 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:E169 | R:R:M174 | 6.77 | 1 | No | Yes | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 9.85 | 1 | No | Yes | 3 | 2 | 1 | 1 | | L:L:?1 | R:R:E169 | 13.71 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 7.01 | 0 | Yes | No | 4 | 5 | 2 | 1 | | R:R:M174 | R:R:T256 | 3.01 | 0 | Yes | No | 4 | 4 | 2 | 2 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 8.14 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:M177 | R:R:N181 | 5.61 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:H250 | R:R:N181 | 3.83 | 0 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 26.02 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 0 | Yes | No | 6 | 7 | 1 | 2 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:L249 | R:R:S277 | 3 | 0 | No | No | 5 | 7 | 1 | 2 | | L:L:?1 | R:R:L249 | 20.58 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 7.48 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:H264 | R:R:T256 | 10.95 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | L:L:?1 | R:R:H264 | 3.32 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:L267 | 7.16 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M270 | 9.14 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:Y271 | 3.03 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 1 | Yes | No | 6 | 4 | 2 | 1 | | L:L:?1 | R:R:I274 | 6.46 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 1 | Yes | No | 6 | 7 | 2 | 2 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 11.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 11.59 | | Average Nodes In Shell | 26.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 35.00 | | Average Links Mediated by Hubs In Shell | 29.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6LPL | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2 | 2020-11-25 | doi.org/10.1038/s41598-020-76277-x |
|
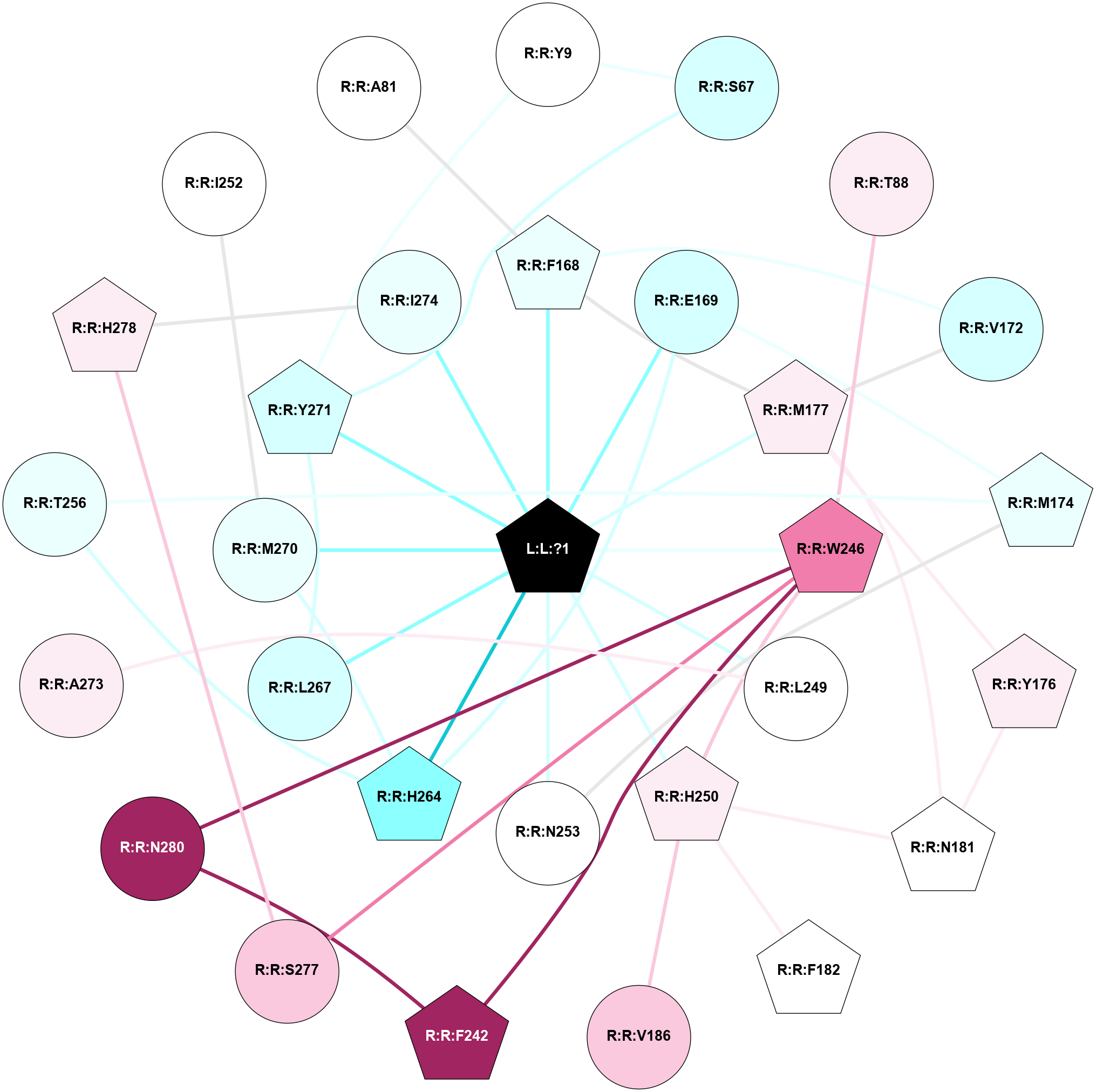
A 2D representation of the interactions of ZMA in 6LPL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:S67 | R:R:Y9 | 5.09 | 1 | No | No | 3 | 5 | 2 | 2 | | R:R:Y271 | R:R:Y9 | 15.89 | 1 | Yes | No | 3 | 5 | 1 | 2 | | R:R:S67 | R:R:Y271 | 3.82 | 1 | No | Yes | 3 | 3 | 2 | 1 | | R:R:T88 | R:R:W246 | 15.77 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F168 | R:R:V172 | 14.42 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 7.46 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | L:L:?1 | R:R:F168 | 39.37 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:E169 | R:R:M174 | 5.41 | 1 | No | Yes | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 11.08 | 1 | No | Yes | 3 | 2 | 1 | 1 | | L:L:?1 | R:R:E169 | 14.56 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 7.01 | 0 | Yes | No | 4 | 5 | 2 | 1 | | R:R:M174 | R:R:T256 | 4.52 | 0 | Yes | No | 4 | 4 | 2 | 2 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 9.3 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:M177 | R:R:N181 | 5.61 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:H250 | R:R:N181 | 3.83 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 28.28 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 1 | Yes | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 14.03 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F242 | R:R:N280 | 7.25 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:H250 | R:R:W246 | 3.17 | 1 | Yes | Yes | 6 | 8 | 1 | 1 | | R:R:S277 | R:R:W246 | 12.36 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N280 | R:R:W246 | 3.39 | 1 | No | Yes | 9 | 8 | 2 | 1 | | L:L:?1 | R:R:W246 | 3.68 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | L:L:?1 | R:R:L249 | 18.79 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 5.82 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:H264 | R:R:T256 | 12.32 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | L:L:?1 | R:R:H264 | 3.32 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | R:R:L267 | R:R:Y271 | 3.52 | 1 | No | Yes | 3 | 3 | 1 | 1 | | L:L:?1 | R:R:L267 | 6.26 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M270 | 9.14 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:Y271 | 4.55 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | R:R:H278 | R:R:I274 | 5.3 | 1 | Yes | No | 6 | 4 | 2 | 1 | | L:L:?1 | R:R:I274 | 6.46 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 1 | Yes | No | 6 | 7 | 2 | 2 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 12.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 10.90 | | Average Nodes In Shell | 30.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 42.00 | | Average Links Mediated by Hubs In Shell | 39.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6MH8 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | - | - | 4.2 | 2019-04-24 | doi.org/10.1107/S205225251900263X |
|
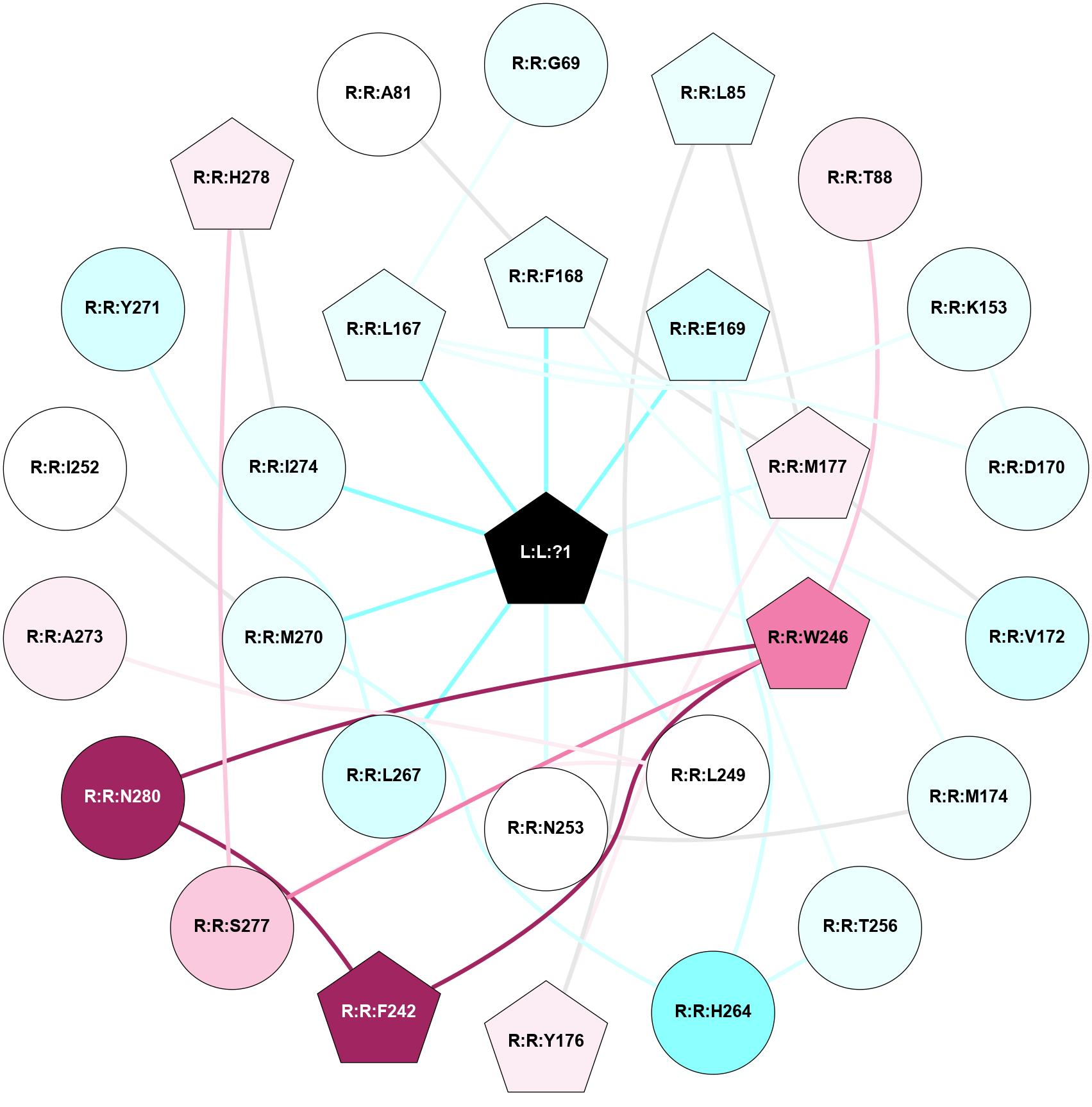
A 2D representation of the interactions of ZMA in 6MH8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:F168 | 31.5 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:E169 | 10.28 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M177 | 3.66 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:W246 | 2.95 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:L249 | 10.74 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:N253 | 10.65 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:L267 | 5.37 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M270 | 4.57 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:I274 | 6.46 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:G69 | R:R:L167 | 3.42 | 0 | No | Yes | 4 | 4 | 2 | 1 | | R:R:L85 | R:R:Y176 | 3.52 | 1 | Yes | Yes | 4 | 6 | 2 | 2 | | R:R:L85 | R:R:M177 | 4.24 | 1 | Yes | Yes | 4 | 6 | 2 | 1 | | R:R:T88 | R:R:W246 | 13.34 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:K153 | R:R:L167 | 4.23 | 1 | No | Yes | 4 | 4 | 2 | 1 | | R:R:D170 | R:R:K153 | 6.91 | 1 | No | No | 4 | 4 | 2 | 2 | | R:R:D170 | R:R:L167 | 8.14 | 1 | No | Yes | 4 | 4 | 2 | 1 | | R:R:F168 | R:R:V172 | 17.04 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 7.46 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | R:R:E169 | R:R:M174 | 5.41 | 1 | Yes | No | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:T256 | 7.06 | 1 | Yes | No | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 11.08 | 1 | Yes | No | 3 | 2 | 1 | 2 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 5.61 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:F242 | R:R:W246 | 18.04 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:S277 | R:R:W246 | 11.12 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N280 | R:R:W246 | 3.39 | 1 | No | Yes | 9 | 8 | 2 | 1 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:L249 | R:R:S277 | 3 | 0 | No | No | 5 | 7 | 1 | 2 | | R:R:I252 | R:R:M270 | 4.37 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:H264 | R:R:T256 | 9.58 | 1 | No | No | 2 | 4 | 2 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | No | No | 2 | 4 | 2 | 1 | | R:R:L267 | R:R:Y271 | 4.69 | 0 | No | No | 3 | 3 | 1 | 2 | | R:R:H278 | R:R:I274 | 6.63 | 2 | Yes | No | 6 | 4 | 2 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 2 | Yes | No | 6 | 7 | 2 | 2 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | | L:L:?1 | R:R:L167 | 2.68 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 8.89 | | Average Nodes In Shell | 29.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 38.00 | | Average Links Mediated by Hubs In Shell | 30.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6PS7 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.85 | 2019-11-13 | doi.org/10.1107/S2052252519013137 |
|
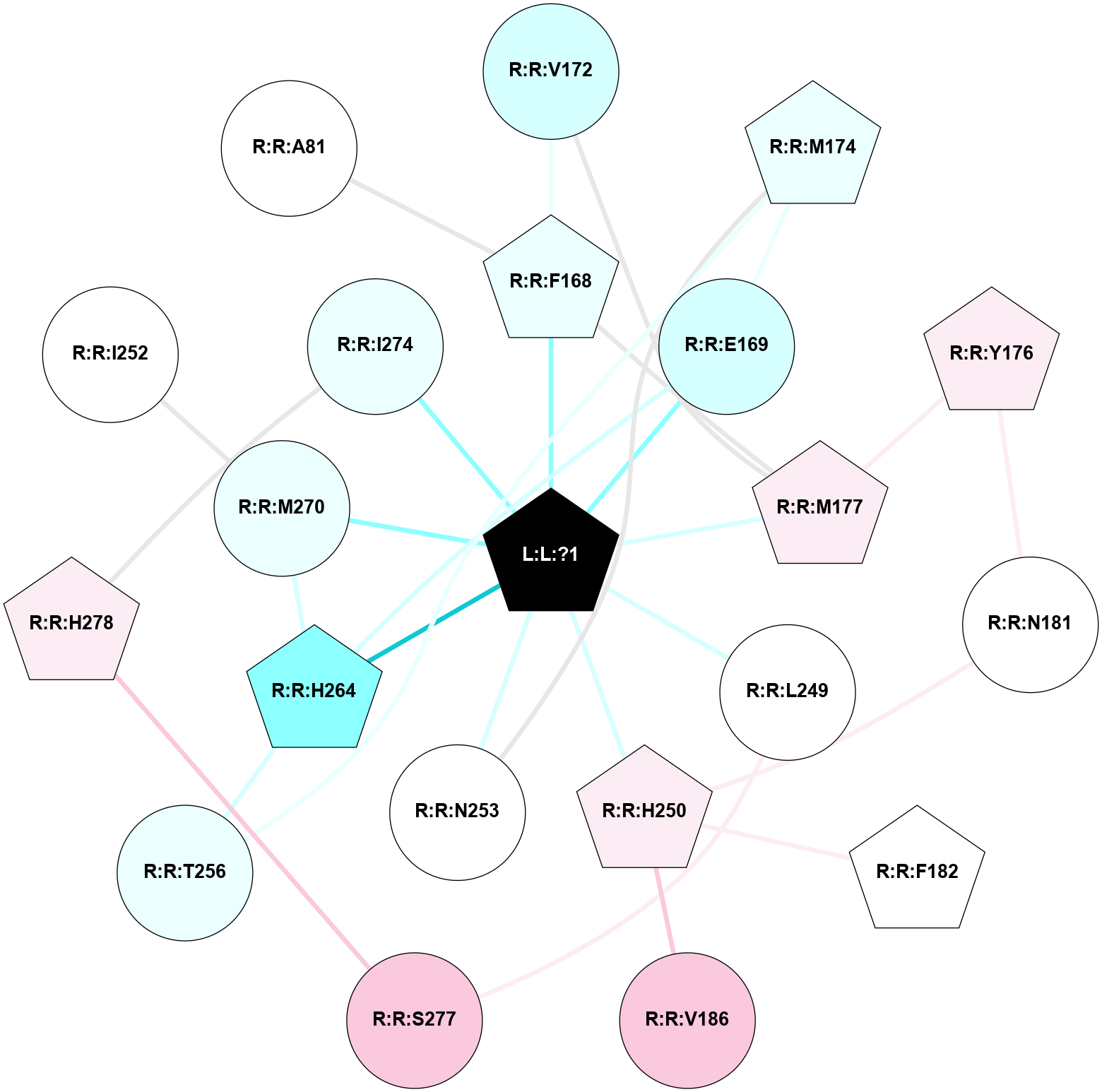
A 2D representation of the interactions of ZMA in 6PS7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:F168 | 37.01 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:E169 | 13.71 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:L249 | 20.58 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 4.99 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H264 | 3.32 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:M270 | 8.23 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:I274 | 4.61 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:F168 | R:R:V172 | 13.11 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 6.22 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | R:R:E169 | R:R:M174 | 5.41 | 1 | No | Yes | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 9.85 | 1 | No | Yes | 3 | 2 | 1 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 8.41 | 0 | Yes | No | 4 | 5 | 2 | 1 | | R:R:M174 | R:R:T256 | 3.01 | 0 | Yes | No | 4 | 4 | 2 | 2 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 5.81 | 1 | No | Yes | 5 | 6 | 2 | 2 | | R:R:H250 | R:R:N181 | 3.83 | 0 | Yes | No | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 23.76 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 0 | Yes | No | 6 | 7 | 1 | 2 | | R:R:L249 | R:R:S277 | 3 | 0 | No | No | 5 | 7 | 1 | 2 | | R:R:H264 | R:R:T256 | 10.95 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 2 | Yes | No | 6 | 4 | 2 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 2 | Yes | No | 6 | 7 | 2 | 2 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 9.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 12.36 | | Average Nodes In Shell | 21.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 28.00 | | Average Links Mediated by Hubs In Shell | 26.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6S0L | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | - | - | 2.65 | 2020-07-15 | doi.org/10.1107/S2052252520011379 |
|

A 2D representation of the interactions of ZMA in 6S0L
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F168 | R:R:V172 | 7.87 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 7.46 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | L:L:?1 | R:R:F168 | 40.94 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:E169 | R:R:M174 | 6.77 | 1 | Yes | Yes | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:T256 | 5.64 | 1 | Yes | No | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 11.08 | 1 | Yes | Yes | 3 | 2 | 1 | 1 | | L:L:?1 | R:R:E169 | 12.85 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 4.21 | 1 | Yes | No | 4 | 5 | 2 | 1 | | R:R:M174 | R:R:T256 | 3.01 | 1 | Yes | No | 4 | 4 | 2 | 2 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 10.47 | 0 | No | Yes | 5 | 6 | 2 | 2 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:H250 | R:R:N181 | 3.83 | 0 | Yes | No | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 21.5 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 0 | Yes | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 13.03 | 0 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:A277 | R:R:W246 | 9.08 | 0 | No | Yes | 7 | 8 | 2 | 1 | | L:L:?1 | R:R:W246 | 4.42 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:L249 | R:R:N253 | 4.12 | 1 | No | No | 5 | 5 | 1 | 1 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | L:L:?1 | R:R:L249 | 18.79 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 7.48 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 11.54 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:H264 | R:R:T256 | 10.95 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 6.57 | 1 | Yes | No | 2 | 4 | 1 | 1 | | L:L:?1 | R:R:H264 | 3.32 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | R:R:L267 | R:R:Y271 | 5.86 | 0 | No | No | 3 | 3 | 1 | 2 | | L:L:?1 | R:R:L267 | 5.37 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M270 | 9.14 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 3 | Yes | No | 6 | 4 | 2 | 1 | | L:L:?1 | R:R:I274 | 3.69 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | | R:R:A88 | R:R:W246 | 2.59 | 0 | No | Yes | 6 | 8 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 11.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 11.27 | | Average Nodes In Shell | 27.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 35.00 | | Average Links Mediated by Hubs In Shell | 31.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6S0Q | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | - | - | 2.65 | 2020-07-15 | doi.org/10.1107/S2052252520011379 |
|

A 2D representation of the interactions of ZMA in 6S0Q
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:S67 | R:R:Y9 | 3.82 | 1 | No | No | 3 | 5 | 2 | 2 | | R:R:Y271 | R:R:Y9 | 15.89 | 1 | Yes | No | 3 | 5 | 1 | 2 | | R:R:S67 | R:R:Y271 | 5.09 | 1 | No | Yes | 3 | 3 | 2 | 1 | | R:R:F168 | R:R:V172 | 11.8 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 6.22 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | L:L:?1 | R:R:F168 | 39.37 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:E169 | R:R:M174 | 6.77 | 1 | No | No | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 11.08 | 1 | No | Yes | 3 | 2 | 1 | 1 | | L:L:?1 | R:R:E169 | 13.71 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 4.21 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 9.3 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:M177 | R:R:N181 | 4.21 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:H250 | R:R:N181 | 3.83 | 0 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 22.63 | 5 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 0 | Yes | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 13.03 | 0 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:A277 | R:R:W246 | 7.78 | 0 | No | Yes | 7 | 8 | 2 | 1 | | L:L:?1 | R:R:W246 | 3.68 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:A273 | R:R:L249 | 4.73 | 0 | No | No | 6 | 5 | 2 | 1 | | L:L:?1 | R:R:L249 | 15.21 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 6.65 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 11.54 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:H264 | R:R:T256 | 12.32 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 5.25 | 1 | Yes | No | 2 | 4 | 1 | 1 | | L:L:?1 | R:R:H264 | 4.16 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | R:R:L267 | R:R:Y271 | 5.86 | 1 | No | Yes | 3 | 3 | 1 | 1 | | L:L:?1 | R:R:L267 | 5.37 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M270 | 10.97 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:Y271 | 3.03 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | R:R:H278 | R:R:I274 | 5.3 | 1 | Yes | No | 6 | 4 | 2 | 1 | | L:L:?1 | R:R:I274 | 3.69 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | | R:R:A88 | R:R:W246 | 2.59 | 0 | No | Yes | 6 | 8 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 12.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 10.31 | | Average Nodes In Shell | 29.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 37.00 | | Average Links Mediated by Hubs In Shell | 32.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6WQA | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2 | 2020-11-18 | doi.org/10.1107/S2052252520012701 |
|

A 2D representation of the interactions of ZMA in 6WQA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?2 | R:R:F168 | 35.43 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?2 | R:R:E169 | 12.85 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?2 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?2 | R:R:W246 | 2.95 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?2 | R:R:L249 | 19.69 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?2 | R:R:H250 | 7.48 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?2 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?2 | R:R:H264 | 3.32 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?2 | R:R:L267 | 3.58 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?2 | R:R:M270 | 8.23 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?2 | R:R:Y271 | 3.03 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | L:L:?2 | R:R:I274 | 3.69 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:S67 | R:R:Y9 | 5.09 | 1 | No | No | 3 | 5 | 2 | 2 | | R:R:Y271 | R:R:Y9 | 15.89 | 1 | Yes | No | 3 | 5 | 1 | 2 | | R:R:S67 | R:R:Y271 | 3.82 | 1 | No | Yes | 3 | 3 | 2 | 1 | | R:R:L85 | R:R:Y176 | 3.52 | 1 | Yes | Yes | 4 | 6 | 2 | 2 | | R:R:L85 | R:R:M177 | 4.24 | 1 | Yes | Yes | 4 | 6 | 2 | 1 | | R:R:L85 | R:R:N181 | 6.87 | 1 | Yes | Yes | 4 | 5 | 2 | 2 | | R:R:T88 | R:R:W246 | 14.55 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F168 | R:R:V172 | 13.11 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 6.22 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | R:R:E169 | R:R:M174 | 5.41 | 1 | No | Yes | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 11.08 | 1 | No | Yes | 3 | 2 | 1 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 7.01 | 0 | Yes | No | 4 | 5 | 2 | 1 | | R:R:M174 | R:R:T256 | 3.01 | 0 | Yes | No | 4 | 4 | 2 | 2 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 5.81 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:M177 | R:R:N181 | 4.21 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:M177 | R:R:N253 | 4.21 | 1 | Yes | No | 6 | 5 | 1 | 1 | | R:R:H250 | R:R:N181 | 5.1 | 0 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 23.76 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 8.3 | 0 | Yes | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 12.03 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F242 | R:R:N280 | 7.25 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:S277 | R:R:W246 | 12.36 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N280 | R:R:W246 | 4.52 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:L249 | R:R:S277 | 3 | 0 | No | No | 5 | 7 | 1 | 2 | | R:R:H264 | R:R:T256 | 9.58 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 3.94 | 1 | Yes | No | 2 | 4 | 1 | 1 | | R:R:L267 | R:R:Y271 | 4.69 | 1 | No | Yes | 3 | 3 | 1 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 1 | Yes | No | 6 | 4 | 2 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 1 | Yes | No | 6 | 7 | 2 | 2 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 12.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 9.92 | | Average Nodes In Shell | 30.00 | | Average Hubs In Shell | 15.00 | | Average Links In Shell | 45.00 | | Average Links Mediated by Hubs In Shell | 42.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7RM5 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2.79 | 2021-09-08 | doi.org/10.1073/pnas.2106041118 |
|

A 2D representation of the interactions of ZMA in 7RM5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:F168 | 37.79 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:E169 | 13.71 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:W246 | 4.42 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:L249 | 19.69 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 8.31 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 10.65 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H264 | 4.16 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:L267 | 7.16 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M270 | 8.23 | 1 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:Y271 | 3.79 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:I274 | 7.38 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:S67 | R:R:Y9 | 6.36 | 1 | No | No | 3 | 5 | 2 | 2 | | R:R:Y271 | R:R:Y9 | 14.89 | 1 | Yes | No | 3 | 5 | 1 | 2 | | R:R:S67 | R:R:Y271 | 8.9 | 1 | No | Yes | 3 | 3 | 2 | 1 | | R:R:T88 | R:R:W246 | 4.85 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F168 | R:R:V172 | 7.87 | 1 | Yes | Yes | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 7.46 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | R:R:E169 | R:R:H264 | 8.62 | 1 | No | Yes | 3 | 2 | 1 | 1 | | R:R:V172 | R:R:Y176 | 3.79 | 1 | Yes | Yes | 3 | 6 | 2 | 2 | | R:R:M174 | R:R:N253 | 4.21 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:M177 | R:R:N181 | 4.21 | 1 | Yes | No | 6 | 5 | 1 | 2 | | R:R:H250 | R:R:N181 | 5.1 | 1 | Yes | No | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:V186 | 3.93 | 1 | Yes | No | 5 | 7 | 2 | 2 | | R:R:F182 | R:R:H250 | 23.76 | 1 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 4.15 | 1 | Yes | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 11.02 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:S277 | R:R:W246 | 14.83 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N280 | R:R:W246 | 6.78 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:A273 | R:R:L249 | 4.73 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:H264 | R:R:T256 | 6.85 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 9.19 | 1 | Yes | No | 2 | 4 | 1 | 1 | | R:R:A265 | R:R:M270 | 3.22 | 0 | No | No | 4 | 4 | 2 | 1 | | R:R:I274 | R:R:Y271 | 3.63 | 1 | No | Yes | 4 | 3 | 1 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 1 | Yes | No | 6 | 4 | 2 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 1 | Yes | No | 6 | 7 | 2 | 2 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | | R:R:F168 | R:R:I66 | 2.51 | 1 | Yes | No | 4 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 12.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 10.97 | | Average Nodes In Shell | 31.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 40.00 | | Average Links Mediated by Hubs In Shell | 36.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7T32 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | - | - | 3.4 | 2022-08-10 | doi.org/10.1038/s41467-022-32125-2 |
|
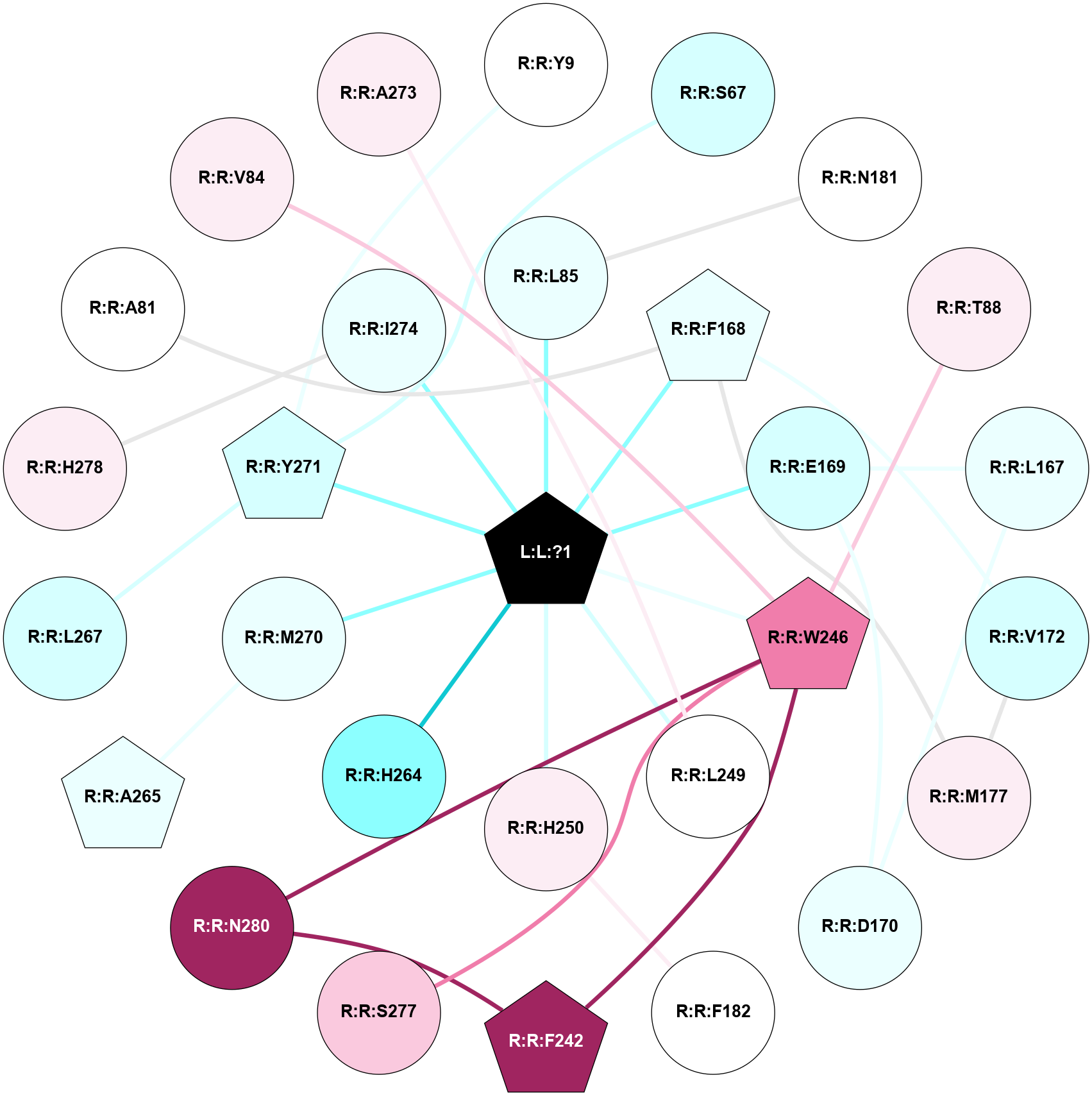
A 2D representation of the interactions of ZMA in 7T32
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:L85 | 5.37 | 0 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:F168 | 35.43 | 0 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:E169 | 15.42 | 0 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:W246 | 3.68 | 0 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:L249 | 8.95 | 0 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 5.82 | 0 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:H264 | 8.31 | 0 | Yes | No | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:M270 | 6.4 | 0 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:Y271 | 4.55 | 0 | Yes | Yes | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:I274 | 6.46 | 0 | Yes | No | 0 | 4 | 0 | 1 | | R:R:Y271 | R:R:Y9 | 17.87 | 0 | Yes | No | 3 | 5 | 1 | 2 | | R:R:S67 | R:R:Y271 | 3.82 | 0 | No | Yes | 3 | 3 | 2 | 1 | | R:R:L85 | R:R:N181 | 5.49 | 0 | No | No | 4 | 5 | 1 | 2 | | R:R:T88 | R:R:W246 | 19.41 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:E169 | R:R:L167 | 7.95 | 1 | No | No | 3 | 4 | 1 | 2 | | R:R:D170 | R:R:L167 | 6.79 | 1 | No | No | 4 | 4 | 2 | 2 | | R:R:F168 | R:R:V172 | 9.18 | 4 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 6.22 | 4 | Yes | No | 4 | 6 | 1 | 2 | | R:R:D170 | R:R:E169 | 3.9 | 1 | No | No | 4 | 3 | 2 | 1 | | R:R:M177 | R:R:V172 | 4.56 | 4 | No | No | 6 | 3 | 2 | 2 | | R:R:F182 | R:R:H250 | 11.31 | 8 | No | No | 5 | 6 | 2 | 1 | | R:R:F242 | R:R:W246 | 10.02 | 5 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F242 | R:R:N280 | 6.04 | 5 | Yes | No | 9 | 9 | 2 | 2 | | R:R:S277 | R:R:W246 | 18.53 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N280 | R:R:W246 | 3.39 | 5 | No | Yes | 9 | 8 | 2 | 1 | | R:R:A265 | R:R:M270 | 3.22 | 0 | Yes | No | 4 | 4 | 2 | 1 | | R:R:L267 | R:R:Y271 | 10.55 | 0 | No | Yes | 3 | 3 | 2 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 0 | No | No | 6 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | | R:R:V84 | R:R:W246 | 2.45 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:A273 | R:R:L249 | 1.58 | 0 | No | No | 6 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 10.04 | | Average Nodes In Shell | 29.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 31.00 | | Average Links Mediated by Hubs In Shell | 23.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8FYN | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2 | 2023-03-08 | doi.org/10.1038/s41467-023-36733-4 |
|

A 2D representation of the interactions of ZMA in 8FYN
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:F168 | 40.16 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:E169 | 14.56 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M177 | 5.48 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:W246 | 4.42 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:L249 | 22.37 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 5.82 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H264 | 4.16 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:L267 | 4.47 | 1 | Yes | No | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:M270 | 6.4 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?1 | R:R:Y271 | 3.79 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:I274 | 5.54 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:S67 | R:R:Y9 | 5.09 | 1 | No | Yes | 3 | 5 | 2 | 2 | | R:R:Y271 | R:R:Y9 | 15.89 | 1 | Yes | Yes | 3 | 5 | 1 | 2 | | R:R:I274 | R:R:Y9 | 3.63 | 1 | Yes | Yes | 4 | 5 | 1 | 2 | | R:R:S67 | R:R:Y271 | 3.82 | 1 | No | Yes | 3 | 3 | 2 | 1 | | R:R:L85 | R:R:M177 | 4.24 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:L85 | R:R:N181 | 12.36 | 1 | No | No | 4 | 5 | 2 | 2 | | R:R:F242 | R:R:T88 | 3.89 | 1 | Yes | No | 9 | 6 | 2 | 2 | | R:R:T88 | R:R:W246 | 13.34 | 1 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F168 | R:R:V172 | 14.42 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 6.22 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | R:R:E169 | R:R:M174 | 6.77 | 1 | No | Yes | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 11.08 | 1 | No | Yes | 3 | 2 | 1 | 1 | | R:R:M174 | R:R:N253 | 5.61 | 0 | Yes | No | 4 | 5 | 2 | 1 | | R:R:M174 | R:R:T256 | 4.52 | 0 | Yes | No | 4 | 4 | 2 | 2 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:N181 | R:R:Y176 | 11.63 | 1 | No | Yes | 5 | 6 | 2 | 2 | | R:R:M177 | R:R:N181 | 4.21 | 1 | Yes | No | 6 | 5 | 1 | 2 | | R:R:F182 | R:R:H250 | 27.15 | 0 | No | No | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 9.69 | 0 | No | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 14.03 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:S277 | R:R:W246 | 12.36 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N280 | R:R:W246 | 4.52 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:H264 | R:R:T256 | 10.95 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 6.57 | 1 | Yes | Yes | 2 | 4 | 1 | 1 | | R:R:L267 | R:R:Y271 | 5.86 | 1 | No | Yes | 3 | 3 | 1 | 1 | | R:R:I274 | R:R:Y271 | 3.63 | 1 | Yes | Yes | 4 | 3 | 1 | 1 | | R:R:H278 | R:R:I274 | 3.98 | 1 | Yes | Yes | 6 | 4 | 2 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 1 | Yes | No | 6 | 7 | 2 | 2 | | R:R:A265 | R:R:M270 | 3.22 | 0 | No | Yes | 4 | 4 | 2 | 1 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | Yes | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 12.00 | | Average Number Of Links With An Hub | 7.00 | | Average Interaction Strength | 10.80 | | Average Nodes In Shell | 32.00 | | Average Hubs In Shell | 14.00 | | Average Links In Shell | 45.00 | | Average Links Mediated by Hubs In Shell | 41.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8RQQ | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2.37 | 2024-04-03 | doi.org/10.1021/acs.cgd.4c00087 |
|

A 2D representation of the interactions of ZMA in 8RQQ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L85 | R:R:Y176 | 3.52 | 1 | Yes | Yes | 4 | 6 | 2 | 2 | | R:R:L85 | R:R:M177 | 4.24 | 1 | Yes | Yes | 4 | 6 | 2 | 1 | | R:R:T88 | R:R:W246 | 12.13 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F168 | R:R:V172 | 14.42 | 1 | Yes | No | 4 | 3 | 1 | 2 | | R:R:F168 | R:R:M177 | 7.46 | 1 | Yes | Yes | 4 | 6 | 1 | 1 | | L:L:?1 | R:R:F168 | 35.43 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:E169 | R:R:M174 | 5.41 | 1 | Yes | Yes | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:T256 | 4.23 | 1 | Yes | No | 3 | 4 | 1 | 2 | | R:R:E169 | R:R:H264 | 8.62 | 1 | Yes | Yes | 3 | 2 | 1 | 1 | | L:L:?1 | R:R:E169 | 12.85 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | R:R:M177 | R:R:V172 | 3.04 | 1 | Yes | No | 6 | 3 | 1 | 2 | | R:R:M174 | R:R:N253 | 7.01 | 1 | Yes | No | 4 | 5 | 2 | 1 | | R:R:M174 | R:R:T256 | 3.01 | 1 | Yes | No | 4 | 4 | 2 | 2 | | R:R:M177 | R:R:Y176 | 3.59 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:M177 | R:R:N253 | 4.21 | 1 | Yes | No | 6 | 5 | 1 | 1 | | L:L:?1 | R:R:M177 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:F182 | R:R:V186 | 3.93 | 1 | Yes | No | 5 | 7 | 2 | 2 | | R:R:F182 | R:R:H250 | 27.15 | 1 | Yes | No | 5 | 6 | 2 | 1 | | R:R:H250 | R:R:V186 | 6.92 | 1 | No | No | 6 | 7 | 1 | 2 | | R:R:F242 | R:R:W246 | 11.02 | 1 | Yes | Yes | 9 | 8 | 2 | 1 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:S277 | R:R:W246 | 12.36 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N280 | R:R:W246 | 3.39 | 1 | No | Yes | 9 | 8 | 2 | 1 | | L:L:?1 | R:R:W246 | 4.42 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:A273 | R:R:L249 | 3.15 | 0 | No | No | 6 | 5 | 2 | 1 | | L:L:?1 | R:R:L249 | 22.37 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:H250 | 4.99 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:N253 | 12.43 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:H264 | R:R:T256 | 10.95 | 1 | Yes | No | 2 | 4 | 1 | 2 | | R:R:H264 | R:R:M270 | 3.94 | 1 | Yes | No | 2 | 4 | 1 | 1 | | L:L:?1 | R:R:H264 | 3.32 | 1 | Yes | Yes | 0 | 2 | 0 | 1 | | L:L:?1 | R:R:M270 | 7.31 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:I274 | R:R:Y271 | 3.63 | 0 | No | Yes | 4 | 3 | 1 | 2 | | R:R:H278 | R:R:I274 | 3.98 | 0 | Yes | No | 6 | 4 | 2 | 1 | | L:L:?1 | R:R:I274 | 3.69 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:H278 | R:R:S277 | 4.18 | 0 | Yes | No | 6 | 7 | 2 | 2 | | R:R:I252 | R:R:M270 | 2.92 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:A81 | R:R:F168 | 2.77 | 0 | No | Yes | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 11.32 | | Average Nodes In Shell | 27.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 38.00 | | Average Links Mediated by Hubs In Shell | 35.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|






























