| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9NS9 | A | Lipid | Free Fatty Acid | FFA2 | Homo Sapiens | TUG-1375 | Compound 187 | Gi1/Beta1/Gamma2 | 3.3 | 2025-06-25 | doi.org/10.1038/s41586-025-09186-6 |
|
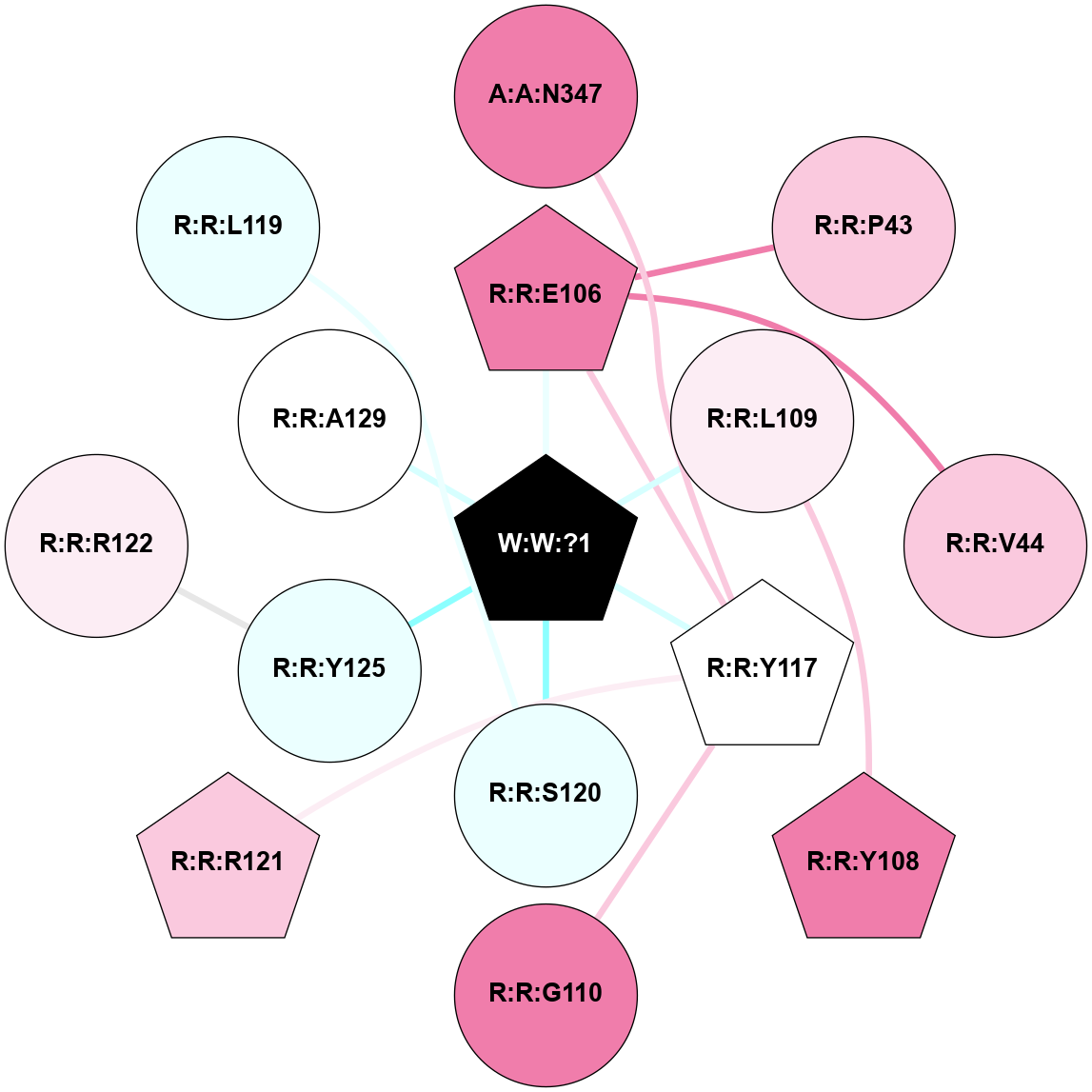
A 2D representation of the interactions of _W_ in 9NS9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:N347 | R:R:Y117 | 9.3 | 0 | No | Yes | 8 | 5 | 2 | 1 | | R:R:E106 | R:R:P43 | 11 | 22 | Yes | No | 8 | 7 | 1 | 2 | | R:R:E106 | R:R:V44 | 5.7 | 22 | Yes | No | 8 | 7 | 1 | 2 | | R:R:E106 | R:R:Y117 | 3.37 | 22 | Yes | Yes | 8 | 5 | 1 | 1 | | R:R:E106 | W:W:?1 | 12.66 | 22 | Yes | Yes | 8 | 0 | 1 | 0 | | R:R:L109 | R:R:Y108 | 3.52 | 0 | No | Yes | 6 | 8 | 1 | 2 | | R:R:L109 | W:W:?1 | 14.32 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:G110 | R:R:Y117 | 7.24 | 0 | No | Yes | 8 | 5 | 2 | 1 | | R:R:R121 | R:R:Y117 | 8.23 | 0 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:Y117 | W:W:?1 | 7.47 | 22 | Yes | Yes | 5 | 0 | 1 | 0 | | R:R:S120 | W:W:?1 | 3.59 | 0 | No | Yes | 4 | 0 | 1 | 0 | | R:R:R122 | R:R:Y125 | 10.29 | 0 | No | No | 6 | 4 | 2 | 1 | | R:R:Y125 | W:W:?1 | 15.87 | 0 | No | Yes | 4 | 0 | 1 | 0 | | R:R:A129 | W:W:?1 | 6.27 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:L119 | R:R:S120 | 1.5 | 0 | No | No | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 10.03 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8XXU | A | Lipid | Prostanoid | DP2 | Homo Sapiens | - | A1D5Q | Gi1/Beta1/Gamma2 | 2.54 | 2024-12-04 | doi.org/10.1073/pnas.2403304121 |
|
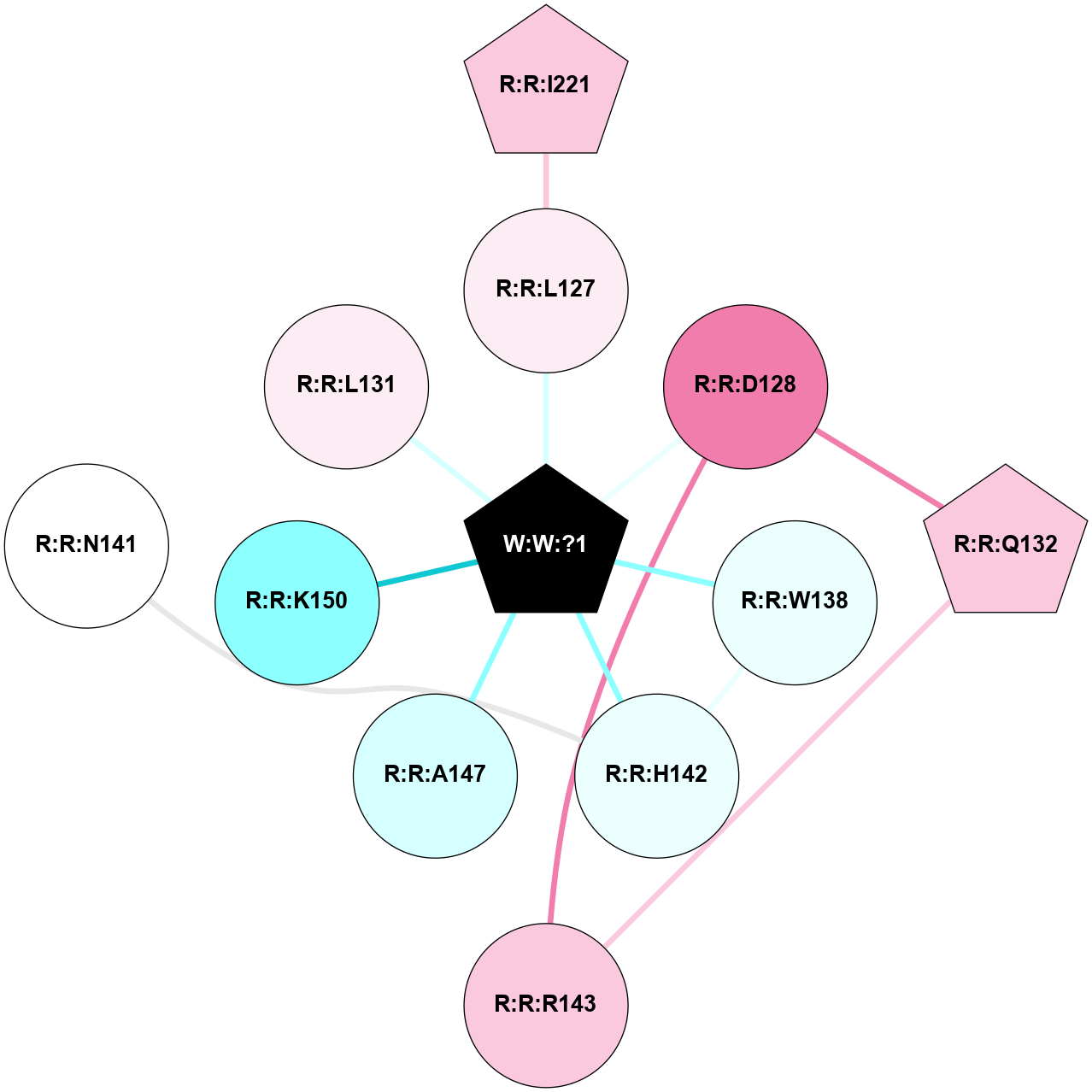
A 2D representation of the interactions of _W_ in 8XXU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I221 | R:R:L127 | 4.28 | 0 | Yes | No | 7 | 6 | 2 | 1 | | R:R:L127 | W:W:?1 | 6.2 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:D128 | R:R:Q132 | 10.44 | 11 | No | Yes | 8 | 7 | 1 | 2 | | R:R:D128 | R:R:R143 | 7.15 | 11 | No | No | 8 | 7 | 1 | 2 | | R:R:D128 | W:W:?1 | 6.08 | 11 | No | Yes | 8 | 0 | 1 | 0 | | R:R:Q132 | R:R:R143 | 11.68 | 11 | Yes | No | 7 | 7 | 2 | 2 | | R:R:H142 | R:R:W138 | 21.16 | 11 | No | No | 4 | 4 | 1 | 1 | | R:R:W138 | W:W:?1 | 8.5 | 11 | No | Yes | 4 | 0 | 1 | 0 | | R:R:H142 | R:R:N141 | 6.38 | 11 | No | No | 4 | 5 | 1 | 2 | | R:R:H142 | W:W:?1 | 27.83 | 11 | No | Yes | 4 | 0 | 1 | 0 | | R:R:A147 | W:W:?1 | 7.06 | 0 | No | Yes | 3 | 0 | 1 | 0 | | R:R:K150 | W:W:?1 | 7.37 | 0 | No | Yes | 2 | 0 | 1 | 0 | | R:R:L131 | W:W:?1 | 2.07 | 0 | No | Yes | 6 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 9.30 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8XXV | A | Lipid | Prostanoid | DP2 | Homo Sapiens | Indomethacin | A1D5Q | Gi1/Beta1/Gamma2 | 2.33 | 2024-12-04 | doi.org/10.1073/pnas.2403304121 |
|
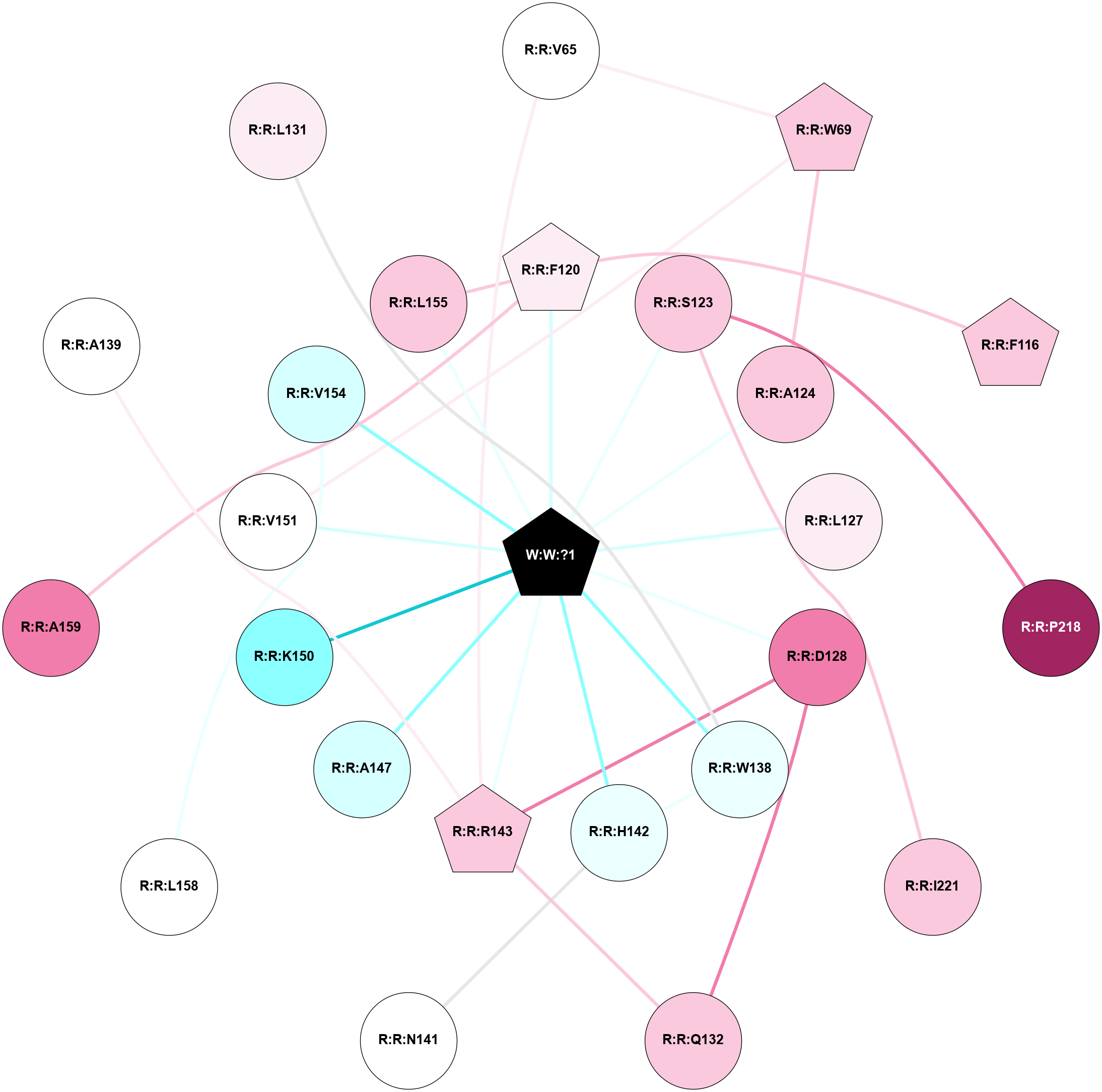
A 2D representation of the interactions of _W_ in 8XXV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:V65 | R:R:W69 | 3.68 | 3 | No | Yes | 5 | 7 | 2 | 2 | | R:R:R143 | R:R:V65 | 7.85 | 3 | Yes | No | 7 | 5 | 1 | 2 | | R:R:A124 | R:R:W69 | 5.19 | 0 | No | Yes | 7 | 7 | 1 | 2 | | R:R:V151 | R:R:W69 | 3.68 | 0 | No | Yes | 5 | 7 | 1 | 2 | | R:R:F116 | R:R:F120 | 8.57 | 0 | Yes | Yes | 7 | 6 | 2 | 1 | | R:R:F120 | R:R:L155 | 9.74 | 3 | Yes | No | 6 | 7 | 1 | 1 | | R:R:F120 | W:W:?1 | 3.64 | 3 | Yes | Yes | 6 | 0 | 1 | 0 | | R:R:P218 | R:R:S123 | 3.56 | 0 | No | No | 9 | 7 | 2 | 1 | | R:R:I221 | R:R:S123 | 4.64 | 0 | No | No | 7 | 7 | 2 | 1 | | R:R:S123 | W:W:?1 | 5.6 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:A124 | W:W:?1 | 5.88 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:L127 | W:W:?1 | 9.3 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:D128 | R:R:Q132 | 11.75 | 3 | No | No | 8 | 7 | 1 | 2 | | R:R:D128 | R:R:R143 | 8.34 | 3 | No | Yes | 8 | 7 | 1 | 1 | | R:R:D128 | W:W:?1 | 6.08 | 3 | No | Yes | 8 | 0 | 1 | 0 | | R:R:Q132 | R:R:R143 | 17.52 | 3 | No | Yes | 7 | 7 | 2 | 1 | | R:R:H142 | R:R:W138 | 17.98 | 3 | No | No | 4 | 4 | 1 | 1 | | R:R:W138 | W:W:?1 | 6.8 | 3 | No | Yes | 4 | 0 | 1 | 0 | | R:R:H142 | R:R:N141 | 6.38 | 3 | No | No | 4 | 5 | 1 | 2 | | R:R:H142 | W:W:?1 | 15.36 | 3 | No | Yes | 4 | 0 | 1 | 0 | | R:R:R143 | W:W:?1 | 4.53 | 3 | Yes | Yes | 7 | 0 | 1 | 0 | | R:R:A147 | W:W:?1 | 10.58 | 0 | No | Yes | 3 | 0 | 1 | 0 | | R:R:K150 | W:W:?1 | 9.47 | 0 | No | Yes | 2 | 0 | 1 | 0 | | R:R:V151 | W:W:?1 | 11.12 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:V154 | W:W:?1 | 8.9 | 0 | No | Yes | 3 | 0 | 1 | 0 | | R:R:L155 | W:W:?1 | 6.2 | 3 | No | Yes | 7 | 0 | 1 | 0 | | R:R:L158 | R:R:V154 | 2.98 | 0 | No | No | 5 | 3 | 2 | 1 | | R:R:A159 | R:R:F120 | 2.77 | 0 | No | Yes | 8 | 6 | 2 | 1 | | R:R:A139 | R:R:R143 | 2.77 | 0 | No | Yes | 5 | 7 | 2 | 1 | | R:R:L131 | R:R:W138 | 1.14 | 0 | No | No | 6 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 13.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 7.96 | | Average Nodes In Shell | 25.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 30.00 | | Average Links Mediated by Hubs In Shell | 23.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8Y6W | A | Lipid | Free Fatty Acid | FFA2 | Homo Sapiens | TUG-1375 | 4-CMTB | Gi1/Beta1/Gamma2 | 3.19 | 2025-04-02 | doi.org/10.1038/s41467-025-57983-4 |
|
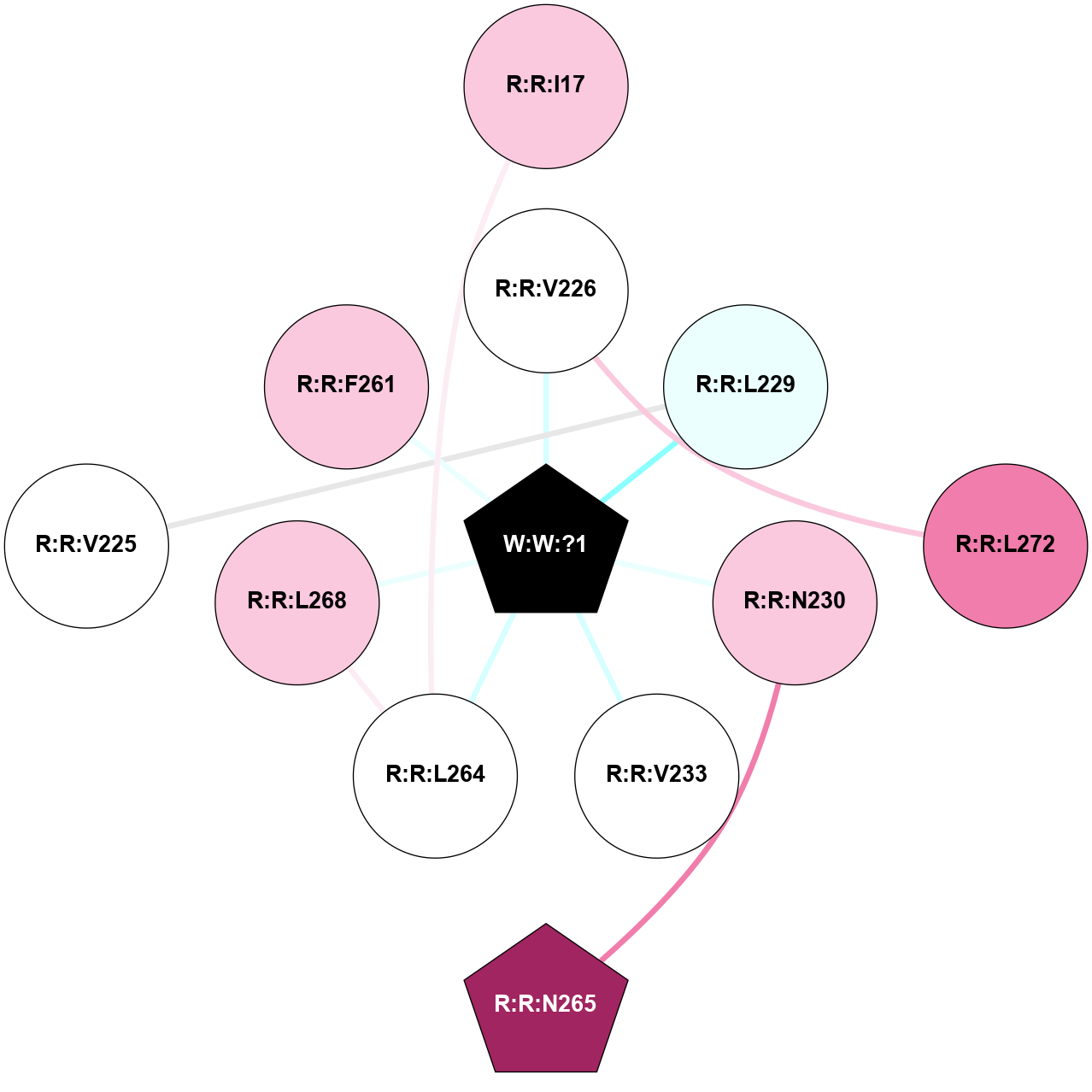
A 2D representation of the interactions of _W_ in 8Y6W
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I17 | R:R:L264 | 4.28 | 0 | No | No | 7 | 5 | 2 | 1 | | R:R:L272 | R:R:V226 | 4.47 | 0 | No | No | 8 | 5 | 2 | 1 | | R:R:V226 | W:W:?1 | 7.16 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:L229 | W:W:?1 | 8.32 | 0 | No | Yes | 4 | 0 | 1 | 0 | | R:R:N230 | R:R:N265 | 5.45 | 0 | No | Yes | 7 | 9 | 1 | 2 | | R:R:N230 | W:W:?1 | 23.11 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:V233 | W:W:?1 | 10.74 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:L264 | R:R:L268 | 6.92 | 28 | No | No | 5 | 7 | 1 | 1 | | R:R:L264 | W:W:?1 | 4.99 | 28 | No | Yes | 5 | 0 | 1 | 0 | | R:R:L268 | W:W:?1 | 16.64 | 28 | No | Yes | 7 | 0 | 1 | 0 | | R:R:L229 | R:R:V225 | 2.98 | 0 | No | No | 4 | 5 | 1 | 2 | | R:R:F261 | W:W:?1 | 2.93 | 0 | No | Yes | 7 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 10.56 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8Y73 | A | Peptide | Opioid | MOP | Homo Sapiens | Peptide | MPAM-15 | Gi1/Beta1/Gamma2 | 2.84 | 2025-08-06 | To be published |
|
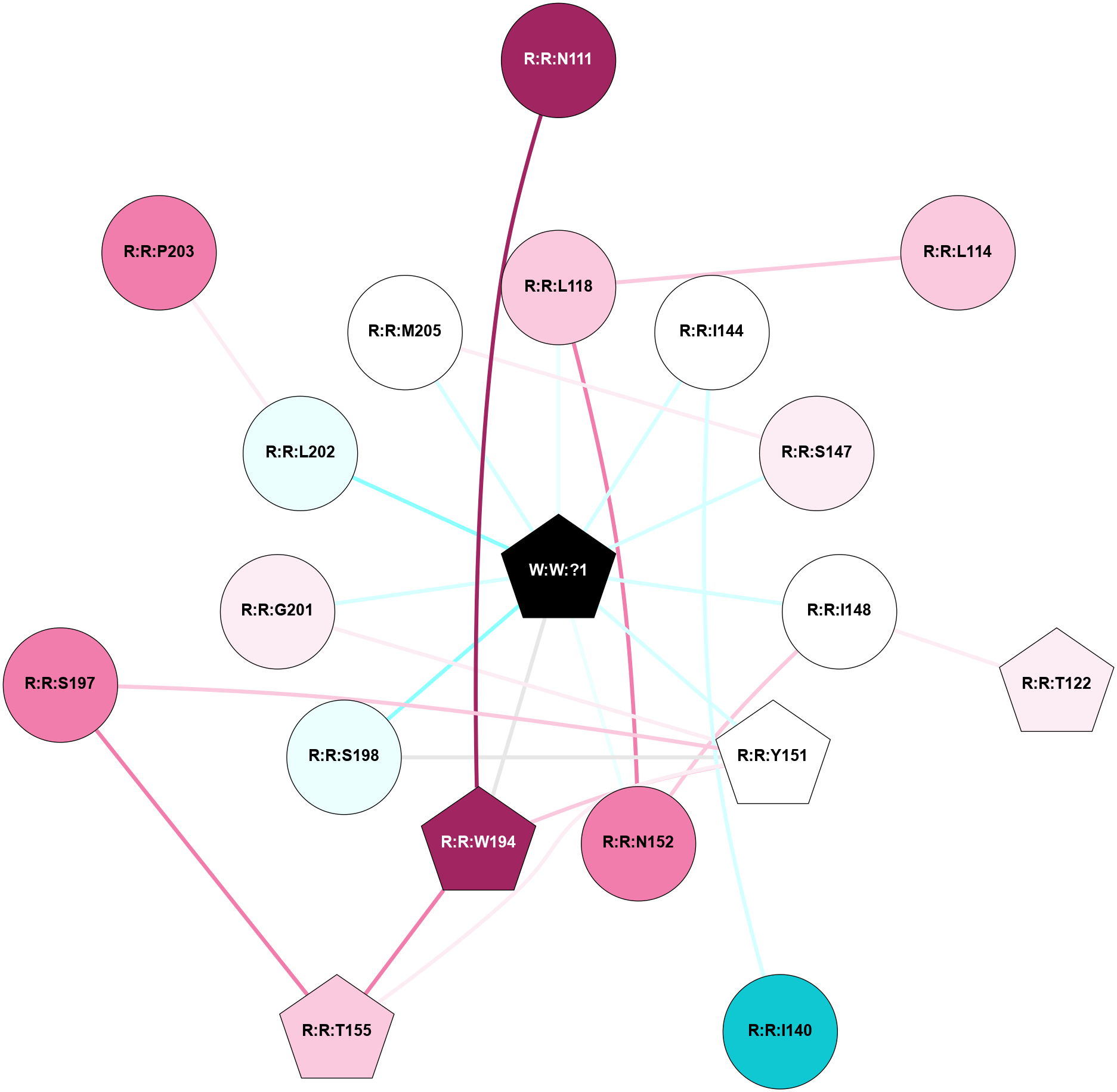
A 2D representation of the interactions of _W_ in 8Y73
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N111 | R:R:W194 | 14.69 | 0 | No | Yes | 9 | 9 | 2 | 1 | | R:R:L114 | R:R:L118 | 4.15 | 0 | No | No | 7 | 7 | 2 | 1 | | R:R:L118 | R:R:N152 | 9.61 | 1 | No | No | 7 | 8 | 1 | 1 | | R:R:L118 | W:W:?1 | 3.35 | 1 | No | Yes | 7 | 0 | 1 | 0 | | R:R:I148 | R:R:T122 | 3.04 | 1 | No | Yes | 5 | 6 | 1 | 2 | | R:R:I140 | R:R:I144 | 4.42 | 0 | No | No | 1 | 5 | 2 | 1 | | R:R:I144 | W:W:?1 | 3.45 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:M205 | R:R:S147 | 6.13 | 1 | No | No | 5 | 6 | 1 | 1 | | R:R:S147 | W:W:?1 | 7.27 | 1 | No | Yes | 6 | 0 | 1 | 0 | | R:R:I148 | R:R:N152 | 2.83 | 1 | No | No | 5 | 8 | 1 | 1 | | R:R:I148 | W:W:?1 | 12.67 | 1 | No | Yes | 5 | 0 | 1 | 0 | | R:R:T155 | R:R:Y151 | 3.75 | 1 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:W194 | R:R:Y151 | 10.61 | 1 | Yes | Yes | 9 | 5 | 1 | 1 | | R:R:S197 | R:R:Y151 | 6.36 | 1 | No | Yes | 8 | 5 | 2 | 1 | | R:R:S198 | R:R:Y151 | 5.09 | 1 | No | Yes | 4 | 5 | 1 | 1 | | R:R:G201 | R:R:Y151 | 2.9 | 1 | No | Yes | 6 | 5 | 1 | 1 | | R:R:Y151 | W:W:?1 | 42.56 | 1 | Yes | Yes | 5 | 0 | 1 | 0 | | R:R:N152 | W:W:?1 | 4.43 | 1 | No | Yes | 8 | 0 | 1 | 0 | | R:R:T155 | R:R:W194 | 15.77 | 1 | Yes | Yes | 7 | 9 | 2 | 1 | | R:R:S197 | R:R:T155 | 4.8 | 1 | No | Yes | 8 | 7 | 2 | 2 | | R:R:W194 | W:W:?1 | 6.43 | 1 | Yes | Yes | 9 | 0 | 1 | 0 | | R:R:S198 | W:W:?1 | 6.06 | 1 | No | Yes | 4 | 0 | 1 | 0 | | R:R:G201 | W:W:?1 | 4.14 | 1 | No | Yes | 6 | 0 | 1 | 0 | | R:R:L202 | W:W:?1 | 8.93 | 0 | No | Yes | 4 | 0 | 1 | 0 | | R:R:M205 | W:W:?1 | 6.84 | 1 | No | Yes | 5 | 0 | 1 | 0 | | R:R:L202 | R:R:P203 | 1.64 | 0 | No | No | 4 | 8 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 11.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.65 | | Average Nodes In Shell | 19.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 26.00 | | Average Links Mediated by Hubs In Shell | 20.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8YYX | A | Alicarboxylic acid | Oxoglutarate | Oxoglutarate | Homo Sapiens | - | Leukotriene e4 | chim(Gi1-CtGq)/Beta1/Gamma2 | 2.84 | 2025-04-09 | To be published |
|
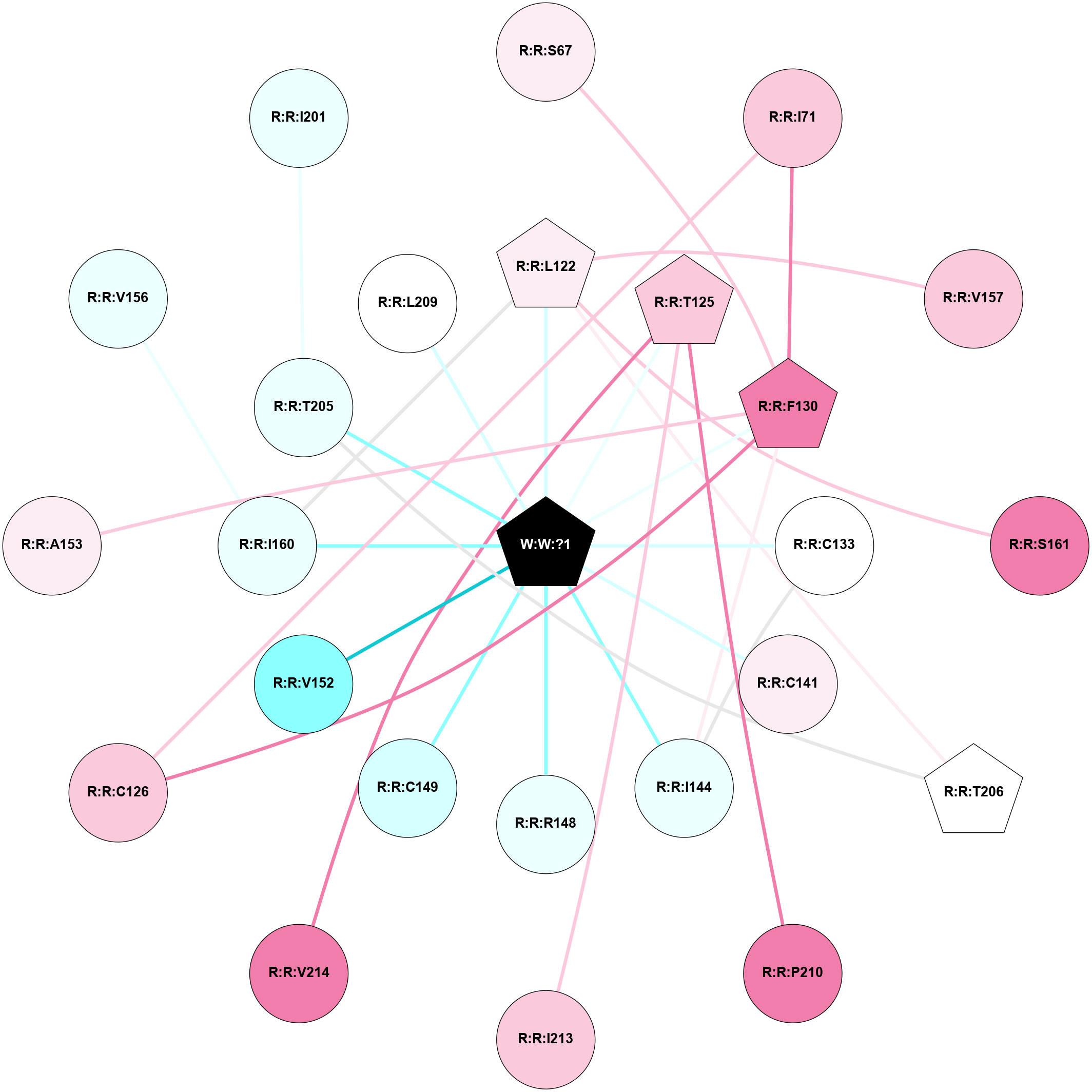
A 2D representation of the interactions of _W_ in 8YYX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F130 | R:R:S67 | 2.64 | 2 | Yes | No | 8 | 6 | 1 | 2 | | R:R:C126 | R:R:I71 | 3.27 | 2 | No | No | 7 | 7 | 2 | 2 | | R:R:F130 | R:R:I71 | 6.28 | 2 | Yes | No | 8 | 7 | 1 | 2 | | R:R:L122 | R:R:V157 | 4.47 | 2 | Yes | No | 6 | 7 | 1 | 2 | | R:R:I160 | R:R:L122 | 2.85 | 2 | No | Yes | 4 | 6 | 1 | 1 | | R:R:L122 | R:R:S161 | 3 | 2 | Yes | No | 6 | 8 | 1 | 2 | | R:R:L122 | R:R:T206 | 2.95 | 2 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:L122 | W:W:?1 | 5.26 | 2 | Yes | Yes | 6 | 0 | 1 | 0 | | R:R:P210 | R:R:T125 | 6.99 | 0 | No | Yes | 8 | 7 | 2 | 1 | | R:R:I213 | R:R:T125 | 3.04 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:T125 | R:R:V214 | 3.17 | 0 | Yes | No | 7 | 8 | 1 | 2 | | R:R:T125 | W:W:?1 | 1.87 | 0 | Yes | Yes | 7 | 0 | 1 | 0 | | R:R:C126 | R:R:F130 | 4.19 | 2 | No | Yes | 7 | 8 | 2 | 1 | | R:R:F130 | R:R:I144 | 10.05 | 2 | Yes | No | 8 | 4 | 1 | 1 | | R:R:A153 | R:R:F130 | 2.77 | 0 | No | Yes | 6 | 8 | 2 | 1 | | R:R:F130 | W:W:?1 | 3.09 | 2 | Yes | Yes | 8 | 0 | 1 | 0 | | R:R:C133 | R:R:I144 | 4.91 | 2 | No | No | 5 | 4 | 1 | 1 | | R:R:C133 | W:W:?1 | 2.01 | 2 | No | Yes | 5 | 0 | 1 | 0 | | R:R:C141 | W:W:?1 | 6.03 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:I144 | W:W:?1 | 5.43 | 2 | No | Yes | 4 | 0 | 1 | 0 | | R:R:R148 | W:W:?1 | 6.16 | 0 | No | Yes | 4 | 0 | 1 | 0 | | R:R:C149 | W:W:?1 | 16.09 | 0 | No | Yes | 3 | 0 | 1 | 0 | | R:R:V152 | W:W:?1 | 16.99 | 0 | No | Yes | 2 | 0 | 1 | 0 | | R:R:I160 | R:R:V156 | 3.07 | 2 | No | No | 4 | 4 | 1 | 2 | | R:R:I160 | W:W:?1 | 3.62 | 2 | No | Yes | 4 | 0 | 1 | 0 | | R:R:T205 | R:R:T206 | 4.71 | 0 | No | Yes | 4 | 5 | 1 | 2 | | R:R:T205 | W:W:?1 | 5.6 | 0 | No | Yes | 4 | 0 | 1 | 0 | | R:R:L209 | W:W:?1 | 3.51 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:I201 | R:R:T205 | 1.52 | 0 | No | No | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 12.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 6.30 | | Average Nodes In Shell | 25.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 29.00 | | Average Links Mediated by Hubs In Shell | 25.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9BJK | A | Peptide | Opioid | MOP | Mus Musculus | Naloxone | NAM | - | 3.26 | 2024-07-17 | doi.org/10.1038/s41586-024-07587-7 |
|
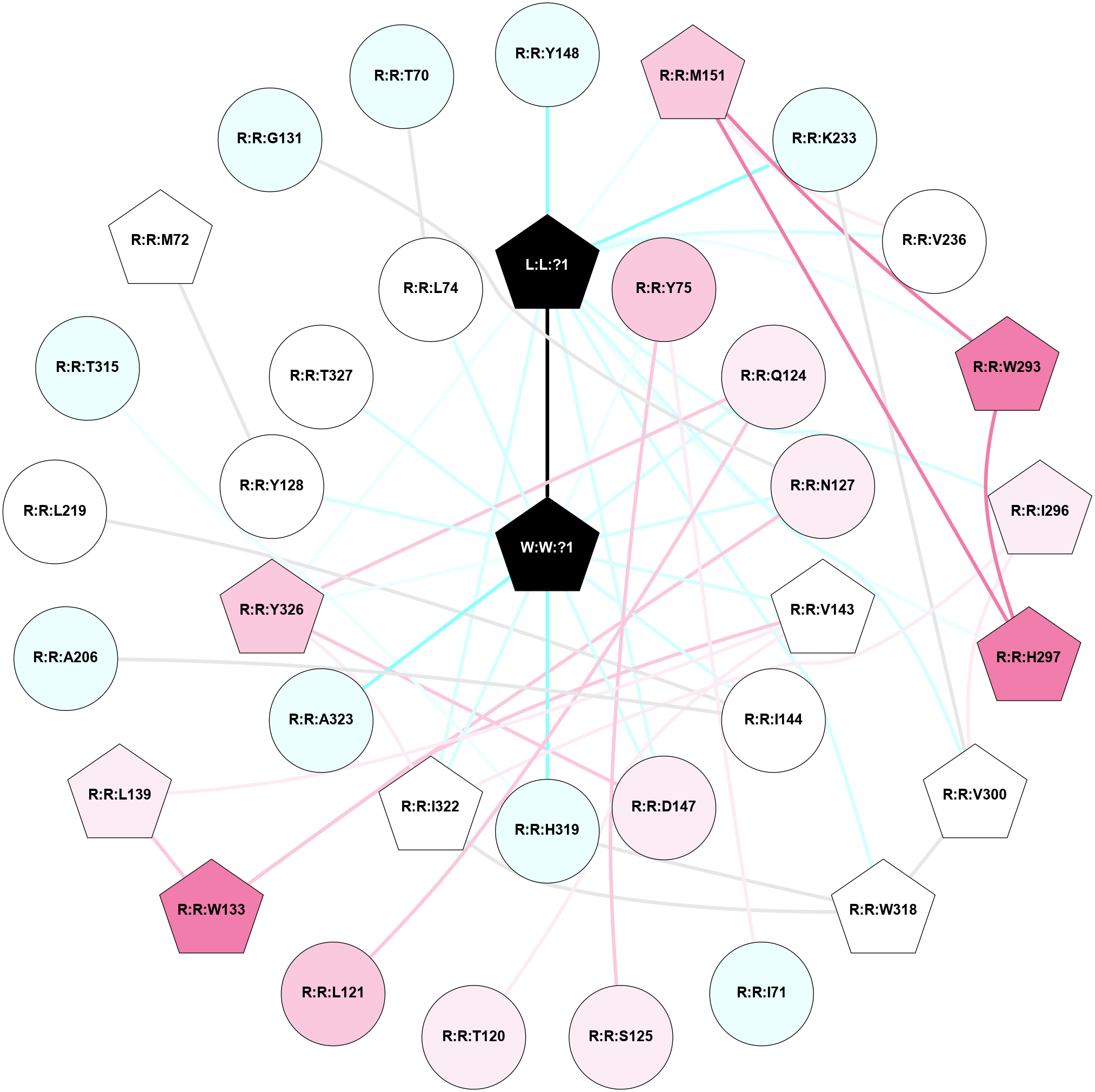
A 2D representation of the interactions of _W_ in 9BJK
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:D147 | 2.61 | 1 | Yes | No | 0 | 6 | 1 | 1 | | L:L:?1 | R:R:Y148 | 4.51 | 1 | Yes | No | 0 | 4 | 1 | 2 | | L:L:?1 | R:R:M151 | 5.44 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?1 | R:R:K233 | 2.71 | 1 | Yes | No | 0 | 4 | 1 | 2 | | L:L:?1 | R:R:V236 | 4.3 | 1 | Yes | No | 0 | 5 | 1 | 2 | | L:L:?1 | R:R:W293 | 9.87 | 1 | Yes | Yes | 0 | 8 | 1 | 2 | | L:L:?1 | R:R:I296 | 6.87 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?1 | R:R:H297 | 7.42 | 1 | Yes | Yes | 0 | 8 | 1 | 2 | | L:L:?1 | R:R:V300 | 20.07 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:?1 | R:R:W318 | 4.38 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:?1 | R:R:I322 | 2.75 | 1 | Yes | Yes | 0 | 5 | 1 | 1 | | L:L:?1 | R:R:Y326 | 11.28 | 1 | Yes | Yes | 0 | 7 | 1 | 1 | | L:L:?1 | W:W:?1 | 10.04 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | R:R:I71 | R:R:Y75 | 6.04 | 0 | No | No | 4 | 7 | 2 | 1 | | R:R:S125 | R:R:Y75 | 6.36 | 0 | No | No | 6 | 7 | 2 | 1 | | R:R:Y75 | W:W:?1 | 12.85 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:T120 | R:R:V143 | 6.35 | 0 | No | Yes | 6 | 5 | 2 | 1 | | R:R:L121 | R:R:Q124 | 2.66 | 0 | No | No | 7 | 6 | 2 | 1 | | R:R:Q124 | R:R:Y326 | 7.89 | 1 | No | Yes | 6 | 7 | 1 | 1 | | R:R:Q124 | W:W:?1 | 18.23 | 1 | No | Yes | 6 | 0 | 1 | 0 | | R:R:N127 | R:R:W133 | 6.78 | 0 | No | Yes | 6 | 8 | 1 | 2 | | R:R:N127 | W:W:?1 | 4.7 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:L139 | R:R:W133 | 4.56 | 1 | Yes | Yes | 6 | 8 | 2 | 2 | | R:R:V143 | R:R:W133 | 2.45 | 1 | Yes | Yes | 5 | 8 | 1 | 2 | | R:R:L139 | R:R:V143 | 2.98 | 1 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:V143 | W:W:?1 | 9.18 | 1 | Yes | Yes | 5 | 0 | 1 | 0 | | R:R:A206 | R:R:I144 | 4.87 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:I144 | R:R:L219 | 2.85 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:I144 | W:W:?1 | 9.78 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:D147 | R:R:Y326 | 6.9 | 1 | No | Yes | 6 | 7 | 1 | 1 | | R:R:D147 | W:W:?1 | 8.37 | 1 | No | Yes | 6 | 0 | 1 | 0 | | R:R:M151 | R:R:V236 | 3.04 | 1 | Yes | No | 7 | 5 | 2 | 2 | | R:R:M151 | R:R:W293 | 5.82 | 1 | Yes | Yes | 7 | 8 | 2 | 2 | | R:R:H297 | R:R:M151 | 5.25 | 1 | Yes | Yes | 8 | 7 | 2 | 2 | | R:R:K233 | R:R:V300 | 4.55 | 1 | No | Yes | 4 | 5 | 2 | 2 | | R:R:H297 | R:R:W293 | 3.17 | 1 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:I296 | R:R:V300 | 3.07 | 1 | Yes | Yes | 6 | 5 | 2 | 2 | | R:R:I296 | R:R:I322 | 5.89 | 1 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:V300 | R:R:W318 | 2.45 | 1 | Yes | Yes | 5 | 5 | 2 | 2 | | R:R:H319 | R:R:T315 | 6.85 | 0 | No | No | 4 | 4 | 1 | 2 | | R:R:H319 | R:R:W318 | 7.41 | 0 | No | Yes | 4 | 5 | 1 | 2 | | R:R:I322 | R:R:W318 | 4.7 | 1 | Yes | Yes | 5 | 5 | 1 | 2 | | R:R:H319 | W:W:?1 | 6.16 | 0 | No | Yes | 4 | 0 | 1 | 0 | | R:R:I322 | R:R:Y326 | 2.42 | 1 | Yes | Yes | 5 | 7 | 1 | 1 | | R:R:I322 | W:W:?1 | 10.75 | 1 | Yes | Yes | 5 | 0 | 1 | 0 | | R:R:A323 | W:W:?1 | 5.4 | 0 | No | Yes | 4 | 0 | 1 | 0 | | R:R:Y326 | W:W:?1 | 12.04 | 1 | Yes | Yes | 7 | 0 | 1 | 0 | | R:R:Y128 | W:W:?1 | 2.41 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:M72 | R:R:Y128 | 2.39 | 0 | Yes | No | 5 | 5 | 2 | 1 | | R:R:T327 | W:W:?1 | 2.02 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:G131 | R:R:N127 | 1.7 | 0 | No | No | 4 | 6 | 2 | 1 | | R:R:L74 | R:R:T70 | 1.47 | 0 | No | No | 5 | 4 | 1 | 2 | | R:R:L74 | W:W:?1 | 0.95 | 0 | No | Yes | 5 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 14.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 8.06 | | Average Nodes In Shell | 36.00 | | Average Hubs In Shell | 14.00 | | Average Links In Shell | 53.00 | | Average Links Mediated by Hubs In Shell | 45.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9C1P | C | Ion | Calcium Sensing | CaS | Homo Sapiens | Ca | Ca; Tryptophan; '6218; PO4 | - | 2.8 | 2024-10-02 | doi.org/10.1126/science.ado1868 |
|
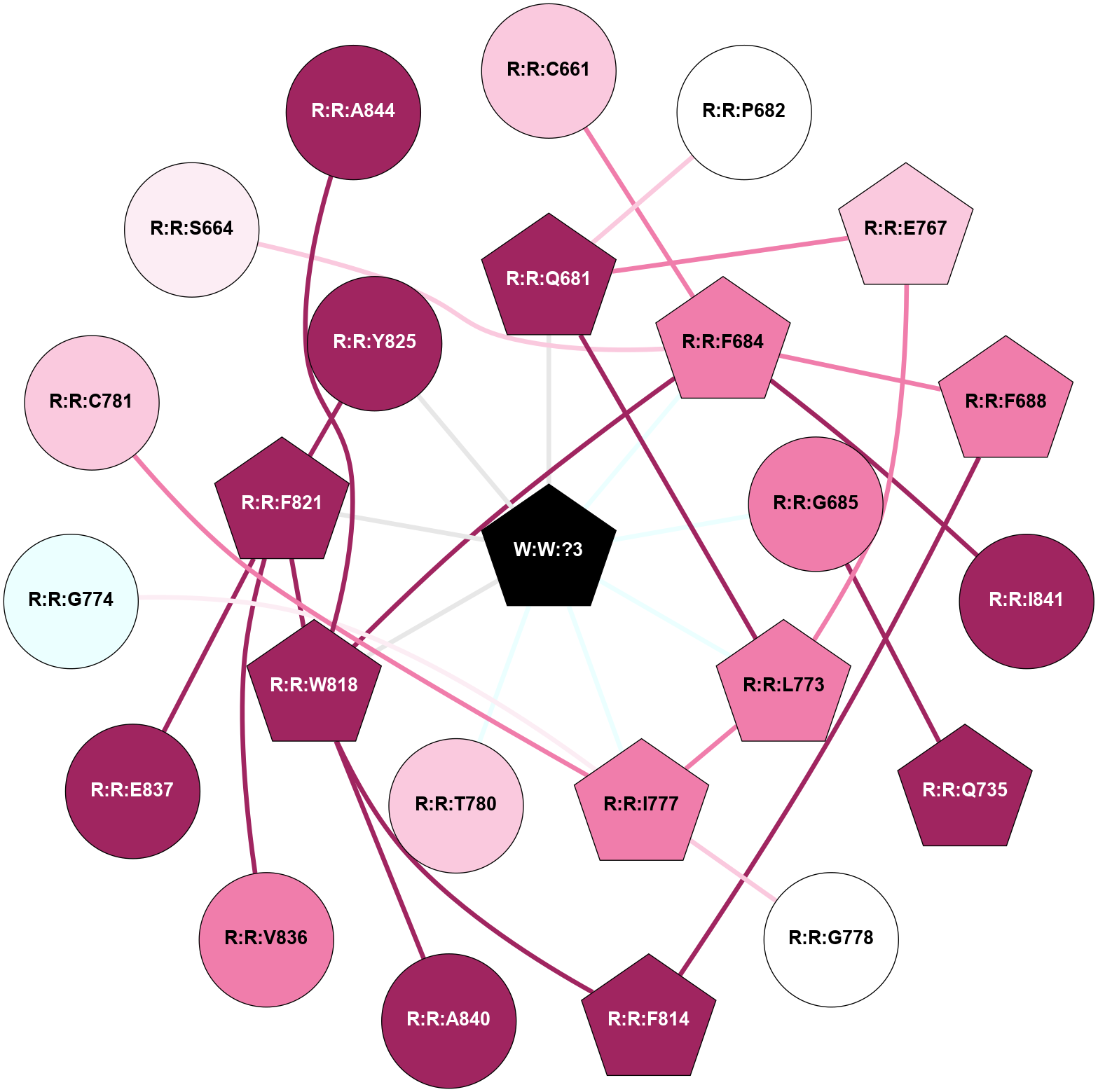
A 2D representation of the interactions of _W_ in 9C1P
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:C661 | R:R:F684 | 2.79 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:P682 | R:R:Q681 | 3.16 | 0 | No | Yes | 5 | 9 | 2 | 1 | | R:R:E767 | R:R:Q681 | 8.92 | 7 | Yes | Yes | 7 | 9 | 2 | 1 | | R:R:L773 | R:R:Q681 | 2.66 | 7 | Yes | Yes | 8 | 9 | 1 | 1 | | R:R:Q681 | W:W:?3 | 17.19 | 7 | Yes | Yes | 9 | 0 | 1 | 0 | | R:R:F684 | R:R:F688 | 4.29 | 7 | Yes | Yes | 8 | 8 | 1 | 2 | | R:R:F684 | R:R:W818 | 5.01 | 7 | Yes | Yes | 8 | 9 | 1 | 1 | | R:R:F684 | R:R:I841 | 11.3 | 7 | Yes | No | 8 | 9 | 1 | 2 | | R:R:F684 | W:W:?3 | 23.07 | 7 | Yes | Yes | 8 | 0 | 1 | 0 | | R:R:G685 | R:R:Q735 | 4.93 | 0 | No | Yes | 8 | 9 | 1 | 2 | | R:R:G685 | W:W:?3 | 2.94 | 0 | No | Yes | 8 | 0 | 1 | 0 | | R:R:F688 | R:R:F814 | 13.93 | 0 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:E767 | R:R:L773 | 2.65 | 7 | Yes | Yes | 7 | 8 | 2 | 1 | | R:R:I777 | R:R:L773 | 2.85 | 7 | Yes | Yes | 8 | 8 | 1 | 1 | | R:R:L773 | W:W:?3 | 3.57 | 7 | Yes | Yes | 8 | 0 | 1 | 0 | | R:R:G778 | R:R:I777 | 3.53 | 0 | No | Yes | 5 | 8 | 2 | 1 | | R:R:I777 | W:W:?3 | 15.97 | 7 | Yes | Yes | 8 | 0 | 1 | 0 | | R:R:T780 | W:W:?3 | 5.08 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:F814 | R:R:W818 | 9.02 | 7 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:F821 | R:R:W818 | 7.02 | 7 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:A840 | R:R:W818 | 6.48 | 0 | No | Yes | 9 | 9 | 2 | 1 | | R:R:W818 | W:W:?3 | 28.43 | 7 | Yes | Yes | 9 | 0 | 1 | 0 | | R:R:F821 | R:R:Y825 | 8.25 | 7 | Yes | No | 9 | 9 | 1 | 1 | | R:R:F821 | R:R:V836 | 6.55 | 7 | Yes | No | 9 | 8 | 1 | 2 | | R:R:E837 | R:R:F821 | 3.5 | 0 | No | Yes | 9 | 9 | 2 | 1 | | R:R:F821 | W:W:?3 | 9.44 | 7 | Yes | Yes | 9 | 0 | 1 | 0 | | R:R:Y825 | W:W:?3 | 8.07 | 7 | No | Yes | 9 | 0 | 1 | 0 | | R:R:G774 | R:R:I777 | 1.76 | 0 | No | Yes | 4 | 8 | 2 | 1 | | R:R:C781 | R:R:I777 | 1.64 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:F684 | R:R:S664 | 1.32 | 7 | Yes | No | 8 | 6 | 1 | 2 | | R:R:A844 | R:R:W818 | 1.3 | 0 | No | Yes | 9 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 9.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 12.64 | | Average Nodes In Shell | 25.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 31.00 | | Average Links Mediated by Hubs In Shell | 31.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
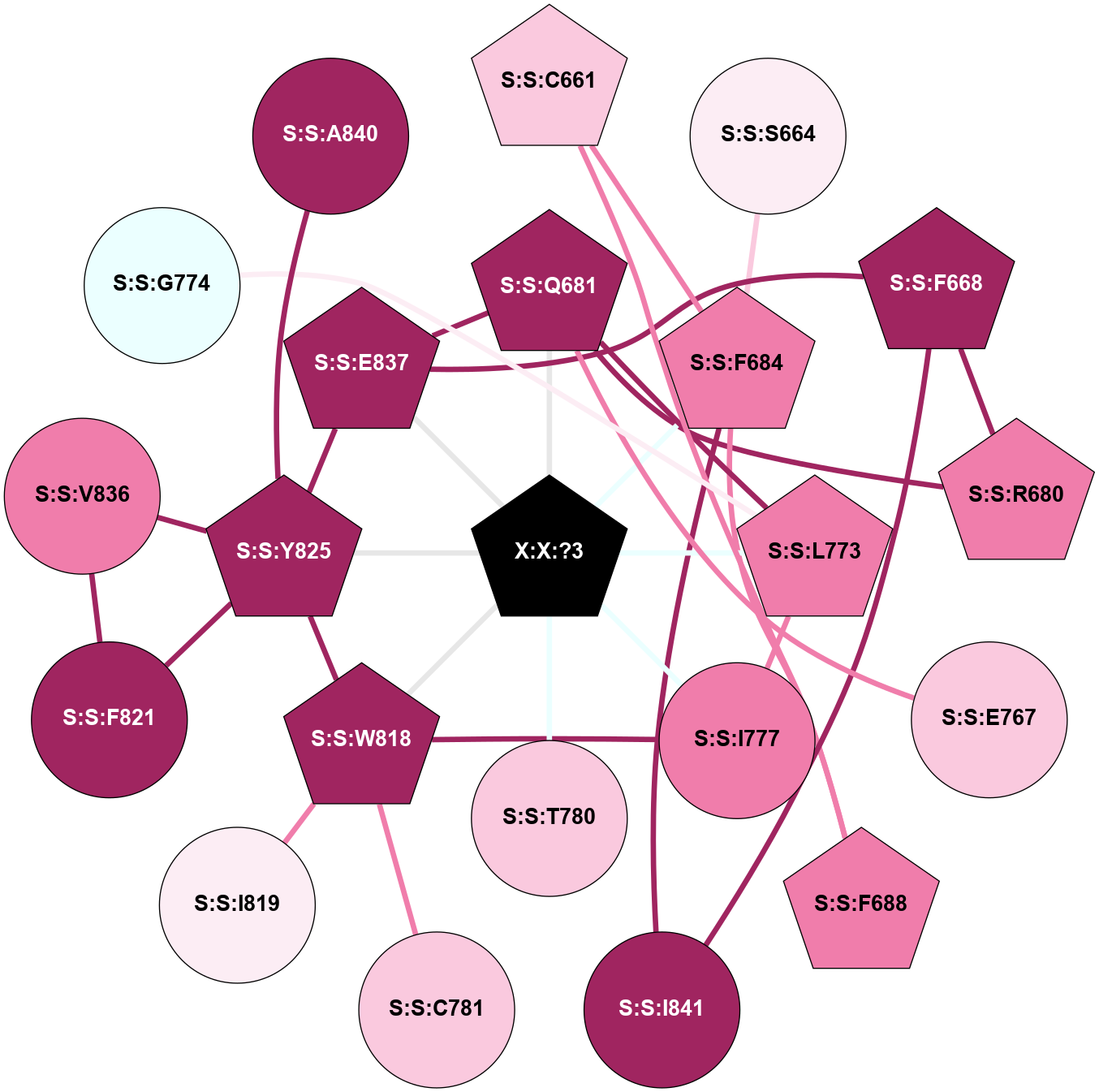
A 2D representation of the interactions of _W_ in 9C1P
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:C661 | S:S:F684 | 2.79 | 6 | Yes | Yes | 7 | 8 | 2 | 1 | | S:S:C661 | S:S:F688 | 2.79 | 6 | Yes | Yes | 7 | 8 | 2 | 2 | | S:S:F684 | S:S:S664 | 5.28 | 6 | Yes | No | 8 | 6 | 1 | 2 | | S:S:F668 | S:S:R680 | 11.76 | 0 | Yes | Yes | 9 | 8 | 2 | 2 | | S:S:E837 | S:S:F668 | 12.83 | 6 | Yes | Yes | 9 | 9 | 1 | 2 | | S:S:F668 | S:S:I841 | 3.77 | 0 | Yes | No | 9 | 9 | 2 | 2 | | S:S:Q681 | S:S:R680 | 4.67 | 6 | Yes | Yes | 9 | 8 | 1 | 2 | | S:S:E767 | S:S:Q681 | 6.37 | 0 | No | Yes | 7 | 9 | 2 | 1 | | S:S:L773 | S:S:Q681 | 2.66 | 6 | Yes | Yes | 8 | 9 | 1 | 1 | | S:S:E837 | S:S:Q681 | 6.37 | 6 | Yes | Yes | 9 | 9 | 1 | 1 | | S:S:Q681 | X:X:?3 | 8.02 | 6 | Yes | Yes | 9 | 0 | 1 | 0 | | S:S:F684 | S:S:F688 | 8.57 | 6 | Yes | Yes | 8 | 8 | 1 | 2 | | S:S:F684 | S:S:I841 | 20.09 | 6 | Yes | No | 8 | 9 | 1 | 2 | | S:S:F684 | X:X:?3 | 14.68 | 6 | Yes | Yes | 8 | 0 | 1 | 0 | | S:S:I777 | S:S:L773 | 5.71 | 6 | No | Yes | 8 | 8 | 1 | 1 | | S:S:L773 | X:X:?3 | 3.57 | 6 | Yes | Yes | 8 | 0 | 1 | 0 | | S:S:I777 | S:S:W818 | 4.7 | 6 | No | Yes | 8 | 9 | 1 | 1 | | S:S:I777 | X:X:?3 | 8.6 | 6 | No | Yes | 8 | 0 | 1 | 0 | | S:S:T780 | X:X:?3 | 7.61 | 0 | No | Yes | 7 | 0 | 1 | 0 | | S:S:C781 | S:S:W818 | 2.61 | 0 | No | Yes | 7 | 9 | 2 | 1 | | S:S:I819 | S:S:W818 | 3.52 | 0 | No | Yes | 6 | 9 | 2 | 1 | | S:S:W818 | S:S:Y825 | 2.89 | 6 | Yes | Yes | 9 | 9 | 1 | 1 | | S:S:W818 | X:X:?3 | 8.82 | 6 | Yes | Yes | 9 | 0 | 1 | 0 | | S:S:F821 | S:S:Y825 | 4.13 | 6 | No | Yes | 9 | 9 | 2 | 1 | | S:S:F821 | S:S:V836 | 7.87 | 6 | No | No | 9 | 8 | 2 | 2 | | S:S:V836 | S:S:Y825 | 3.79 | 6 | No | Yes | 8 | 9 | 2 | 1 | | S:S:E837 | S:S:Y825 | 6.73 | 6 | Yes | Yes | 9 | 9 | 1 | 1 | | S:S:Y825 | X:X:?3 | 11.1 | 6 | Yes | Yes | 9 | 0 | 1 | 0 | | S:S:E837 | X:X:?3 | 6.84 | 6 | Yes | Yes | 9 | 0 | 1 | 0 | | S:S:G774 | S:S:L773 | 1.71 | 0 | No | Yes | 4 | 8 | 2 | 1 | | S:S:A840 | S:S:Y825 | 1.33 | 0 | No | Yes | 9 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 8.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 8.65 | | Average Nodes In Shell | 22.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 31.00 | | Average Links Mediated by Hubs In Shell | 30.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9C2F | C | Ion | Calcium Sensing | CaS | Homo Sapiens | Ca | Ca; Ca; Tryptophan; '54149; PO4 | - | 2.8 | 2024-10-02 | doi.org/10.1126/science.ado1868 |
|
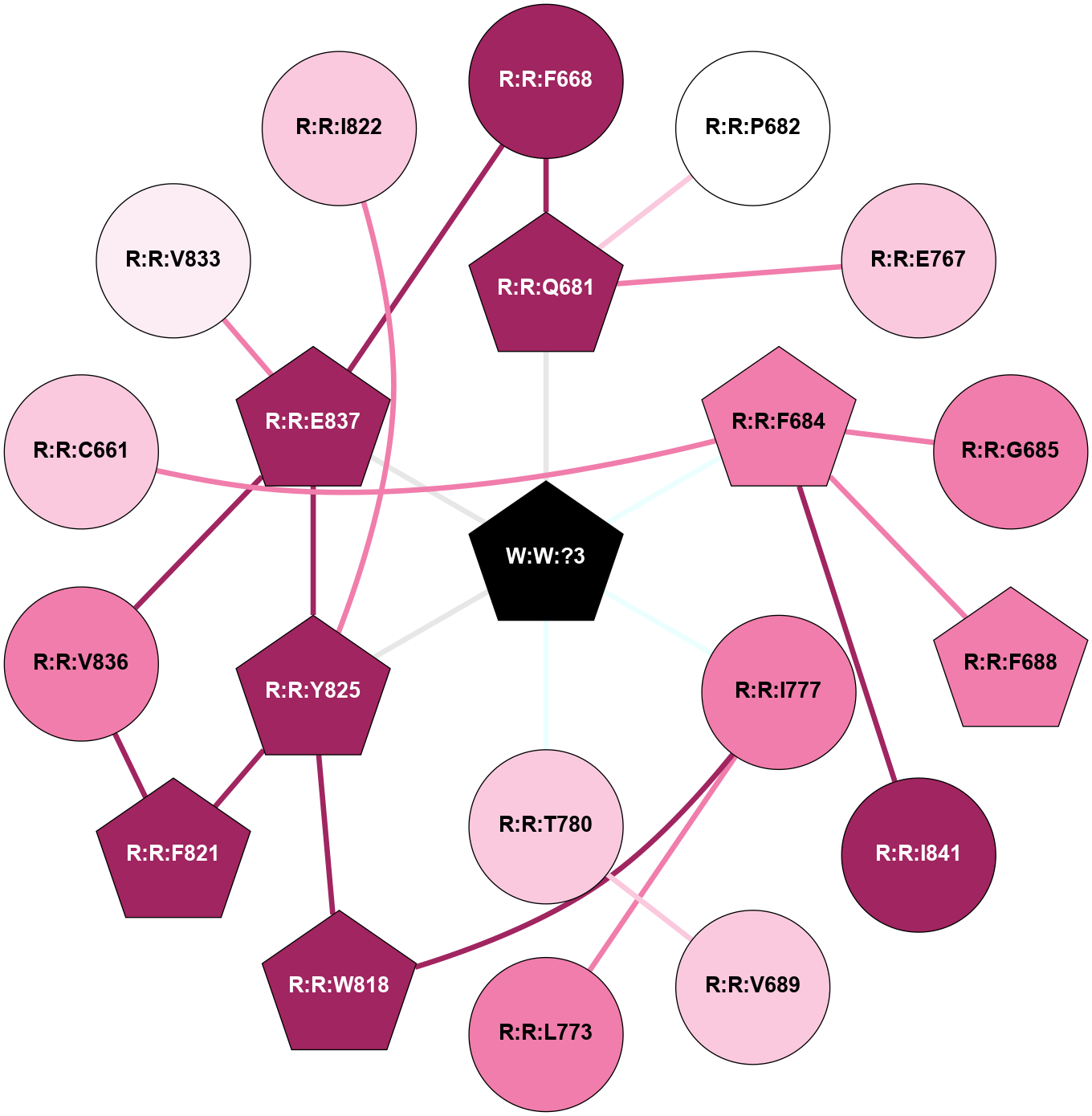
A 2D representation of the interactions of _W_ in 9C2F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F668 | R:R:Q681 | 4.68 | 0 | No | Yes | 9 | 9 | 2 | 1 | | R:R:E837 | R:R:F668 | 5.83 | 51 | Yes | No | 9 | 9 | 1 | 2 | | R:R:P682 | R:R:Q681 | 3.16 | 0 | No | Yes | 5 | 9 | 2 | 1 | | R:R:E767 | R:R:Q681 | 3.82 | 0 | No | Yes | 7 | 9 | 2 | 1 | | R:R:Q681 | W:W:?3 | 11.76 | 0 | Yes | Yes | 9 | 0 | 1 | 0 | | R:R:F684 | R:R:G685 | 3.01 | 0 | Yes | No | 8 | 8 | 1 | 2 | | R:R:F684 | R:R:F688 | 11.79 | 0 | Yes | Yes | 8 | 8 | 1 | 2 | | R:R:F684 | R:R:I841 | 11.3 | 0 | Yes | No | 8 | 9 | 1 | 2 | | R:R:F684 | W:W:?3 | 15.65 | 0 | Yes | Yes | 8 | 0 | 1 | 0 | | R:R:T780 | R:R:V689 | 3.17 | 0 | No | No | 7 | 7 | 1 | 2 | | R:R:I777 | R:R:L773 | 5.71 | 0 | No | No | 8 | 8 | 1 | 2 | | R:R:I777 | R:R:W818 | 3.52 | 0 | No | Yes | 8 | 9 | 1 | 2 | | R:R:I777 | W:W:?3 | 13.76 | 0 | No | Yes | 8 | 0 | 1 | 0 | | R:R:T780 | W:W:?3 | 5.92 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:W818 | R:R:Y825 | 2.89 | 0 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:F821 | R:R:Y825 | 7.22 | 0 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:F821 | R:R:V836 | 9.18 | 0 | Yes | No | 9 | 8 | 2 | 2 | | R:R:E837 | R:R:Y825 | 11.22 | 51 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:Y825 | W:W:?3 | 21.66 | 51 | Yes | Yes | 9 | 0 | 1 | 0 | | R:R:E837 | R:R:V836 | 4.28 | 51 | Yes | No | 9 | 8 | 1 | 2 | | R:R:E837 | W:W:?3 | 7.45 | 51 | Yes | Yes | 9 | 0 | 1 | 0 | | R:R:C661 | R:R:F684 | 2.79 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:E837 | R:R:V833 | 1.43 | 51 | Yes | No | 9 | 6 | 1 | 2 | | R:R:I822 | R:R:Y825 | 1.21 | 0 | No | Yes | 7 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 12.70 | | Average Nodes In Shell | 21.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 24.00 | | Average Links Mediated by Hubs In Shell | 22.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
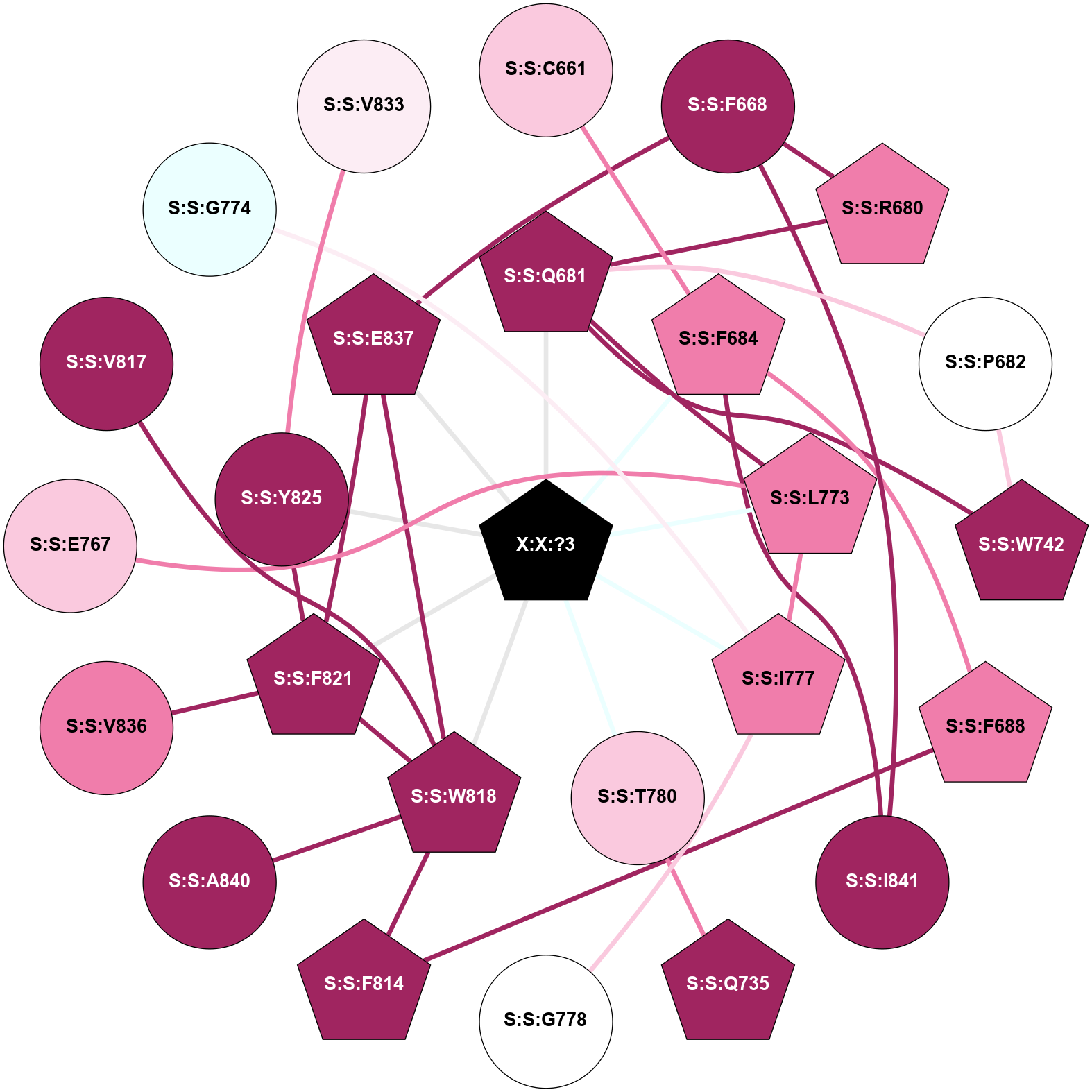
A 2D representation of the interactions of _W_ in 9C2F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:C661 | S:S:F684 | 4.19 | 0 | No | Yes | 7 | 8 | 2 | 1 | | S:S:F668 | S:S:R680 | 14.97 | 0 | No | Yes | 9 | 8 | 2 | 2 | | S:S:E837 | S:S:F668 | 8.16 | 5 | Yes | No | 9 | 9 | 1 | 2 | | S:S:F668 | S:S:I841 | 3.77 | 0 | No | No | 9 | 9 | 2 | 2 | | S:S:Q681 | S:S:R680 | 4.67 | 5 | Yes | Yes | 9 | 8 | 1 | 2 | | S:S:P682 | S:S:Q681 | 3.16 | 5 | No | Yes | 5 | 9 | 2 | 1 | | S:S:Q681 | S:S:W742 | 4.38 | 5 | Yes | Yes | 9 | 9 | 1 | 2 | | S:S:L773 | S:S:Q681 | 3.99 | 5 | Yes | Yes | 8 | 9 | 1 | 1 | | S:S:Q681 | X:X:?3 | 9.62 | 5 | Yes | Yes | 9 | 0 | 1 | 0 | | S:S:P682 | S:S:W742 | 4.05 | 5 | No | Yes | 5 | 9 | 2 | 2 | | S:S:F684 | S:S:F688 | 9.65 | 0 | Yes | Yes | 8 | 8 | 1 | 2 | | S:S:F684 | S:S:I841 | 13.82 | 0 | Yes | No | 8 | 9 | 1 | 2 | | S:S:F684 | X:X:?3 | 17.61 | 0 | Yes | Yes | 8 | 0 | 1 | 0 | | S:S:F688 | S:S:F814 | 11.79 | 5 | Yes | Yes | 8 | 9 | 2 | 2 | | S:S:Q735 | S:S:T780 | 4.25 | 65 | Yes | No | 9 | 7 | 2 | 1 | | S:S:I777 | S:S:L773 | 2.85 | 5 | Yes | Yes | 8 | 8 | 1 | 1 | | S:S:L773 | X:X:?3 | 5.56 | 5 | Yes | Yes | 8 | 0 | 1 | 0 | | S:S:G778 | S:S:I777 | 3.53 | 0 | No | Yes | 5 | 8 | 2 | 1 | | S:S:I777 | X:X:?3 | 16.05 | 5 | Yes | Yes | 8 | 0 | 1 | 0 | | S:S:T780 | X:X:?3 | 8.29 | 0 | No | Yes | 7 | 0 | 1 | 0 | | S:S:F814 | S:S:W818 | 8.02 | 5 | Yes | Yes | 9 | 9 | 2 | 1 | | S:S:F821 | S:S:W818 | 7.02 | 5 | Yes | Yes | 9 | 9 | 1 | 1 | | S:S:E837 | S:S:W818 | 3.27 | 5 | Yes | Yes | 9 | 9 | 1 | 1 | | S:S:A840 | S:S:W818 | 7.78 | 0 | No | Yes | 9 | 9 | 2 | 1 | | S:S:W818 | X:X:?3 | 19.21 | 5 | Yes | Yes | 9 | 0 | 1 | 0 | | S:S:F821 | S:S:Y825 | 5.16 | 5 | Yes | No | 9 | 9 | 1 | 1 | | S:S:F821 | S:S:V836 | 3.93 | 5 | Yes | No | 9 | 8 | 1 | 2 | | S:S:E837 | S:S:F821 | 4.66 | 5 | Yes | Yes | 9 | 9 | 1 | 1 | | S:S:F821 | X:X:?3 | 9.78 | 5 | Yes | Yes | 9 | 0 | 1 | 0 | | S:S:Y825 | X:X:?3 | 18.83 | 5 | No | Yes | 9 | 0 | 1 | 0 | | S:S:E837 | X:X:?3 | 15.96 | 5 | Yes | Yes | 9 | 0 | 1 | 0 | | S:S:E767 | S:S:L773 | 2.65 | 0 | No | Yes | 7 | 8 | 2 | 1 | | S:S:V817 | S:S:W818 | 2.45 | 0 | No | Yes | 9 | 9 | 2 | 1 | | S:S:G774 | S:S:I777 | 1.76 | 0 | No | Yes | 4 | 8 | 2 | 1 | | S:S:V833 | S:S:Y825 | 1.26 | 0 | No | No | 6 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 9.00 | | Average Number Of Links With An Hub | 7.00 | | Average Interaction Strength | 13.43 | | Average Nodes In Shell | 26.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 35.00 | | Average Links Mediated by Hubs In Shell | 33.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9CLW | A | Lipid | Free Fatty Acid | FFA2 | Homo Sapiens | TUG-1375 | 4-CMTB | chim(NtGi1L-Gs-CtGq)/Beta1/Gamma2 | 3.19 | 2025-06-25 | doi.org/10.1038/s41586-025-09186-6 |
|
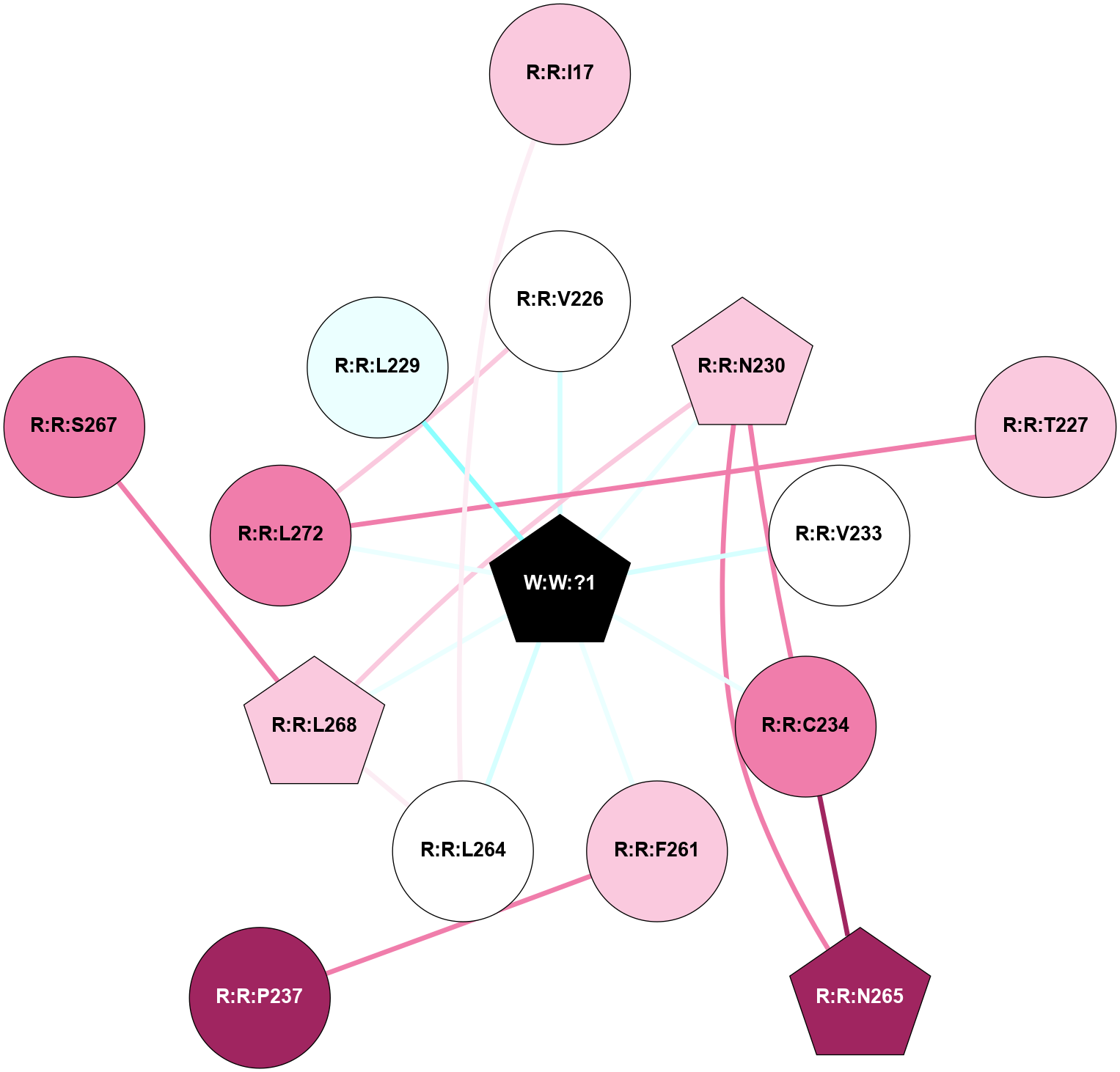
A 2D representation of the interactions of _W_ in 9CLW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I17 | R:R:L264 | 4.28 | 0 | No | No | 7 | 5 | 2 | 1 | | R:R:L272 | R:R:V226 | 5.96 | 1 | No | No | 8 | 5 | 1 | 1 | | R:R:V226 | W:W:?1 | 7.16 | 1 | No | Yes | 5 | 0 | 1 | 0 | | R:R:L272 | R:R:T227 | 5.9 | 1 | No | No | 8 | 7 | 1 | 2 | | R:R:C234 | R:R:N230 | 4.72 | 1 | No | Yes | 8 | 7 | 1 | 1 | | R:R:N230 | R:R:N265 | 5.45 | 1 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:L268 | R:R:N230 | 9.61 | 1 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:N230 | W:W:?1 | 21.46 | 1 | Yes | Yes | 7 | 0 | 1 | 0 | | R:R:V233 | W:W:?1 | 14.33 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:C234 | R:R:N265 | 9.45 | 1 | No | Yes | 8 | 9 | 1 | 2 | | R:R:C234 | W:W:?1 | 3.82 | 1 | No | Yes | 8 | 0 | 1 | 0 | | R:R:F261 | R:R:P237 | 5.78 | 0 | No | No | 7 | 9 | 1 | 2 | | R:R:F261 | W:W:?1 | 5.86 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:L264 | R:R:L268 | 6.92 | 1 | No | Yes | 5 | 7 | 1 | 1 | | R:R:L264 | W:W:?1 | 6.66 | 1 | No | Yes | 5 | 0 | 1 | 0 | | R:R:L268 | W:W:?1 | 16.64 | 1 | Yes | Yes | 7 | 0 | 1 | 0 | | R:R:L272 | W:W:?1 | 6.66 | 1 | No | Yes | 8 | 0 | 1 | 0 | | R:R:L268 | R:R:S267 | 3 | 1 | Yes | No | 7 | 8 | 1 | 2 | | R:R:L229 | W:W:?1 | 1.66 | 0 | No | Yes | 4 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 9.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.36 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 19.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9CM3 | A | Lipid | Free Fatty Acid | FFA2 | Homo Sapiens | TUG-1375 | Compound 187 | chim(NtGi1L-Gs-CtGq)/Beta1/Gamma2 | 3.06 | 2025-06-25 | doi.org/10.1038/s41586-025-09186-6 |
|
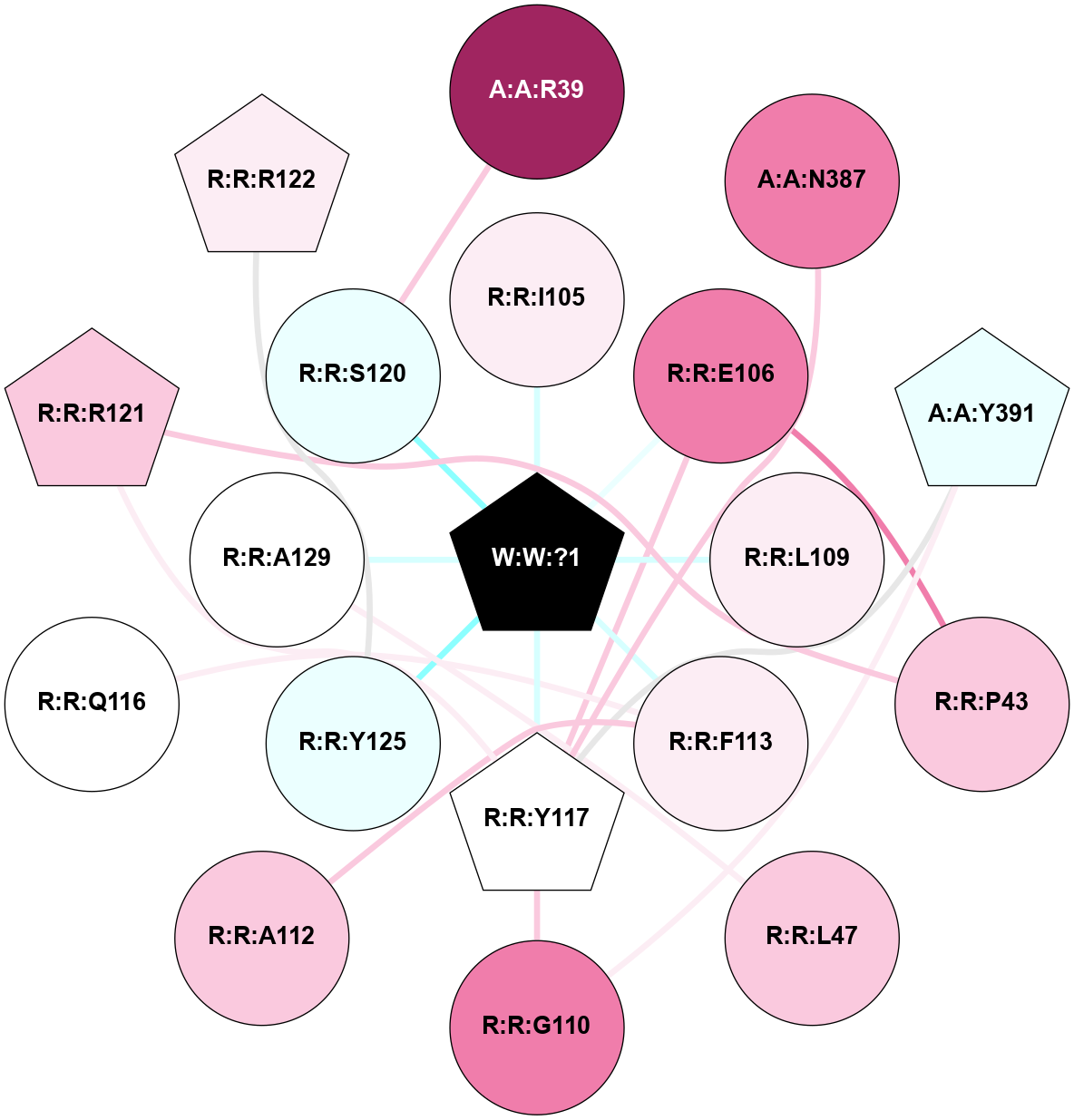
A 2D representation of the interactions of _W_ in 9CM3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:R39 | R:R:S120 | 5.27 | 0 | No | No | 9 | 4 | 2 | 1 | | A:A:N387 | R:R:Y117 | 10.47 | 0 | No | Yes | 8 | 5 | 2 | 1 | | A:A:Y391 | R:R:G110 | 4.35 | 5 | Yes | No | 4 | 8 | 2 | 2 | | A:A:Y391 | R:R:Y117 | 12.91 | 5 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:E106 | R:R:P43 | 12.58 | 5 | No | No | 8 | 7 | 1 | 2 | | R:R:P43 | R:R:R121 | 5.76 | 0 | No | Yes | 7 | 7 | 2 | 2 | | R:R:A129 | R:R:L47 | 4.73 | 0 | No | No | 5 | 7 | 1 | 2 | | R:R:I105 | W:W:?1 | 4.54 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:E106 | R:R:Y117 | 5.61 | 5 | No | Yes | 8 | 5 | 1 | 1 | | R:R:E106 | W:W:?1 | 16.88 | 5 | No | Yes | 8 | 0 | 1 | 0 | | R:R:L109 | W:W:?1 | 15.43 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:G110 | R:R:Y117 | 10.14 | 5 | No | Yes | 8 | 5 | 2 | 1 | | R:R:A112 | R:R:F113 | 8.32 | 0 | No | No | 7 | 6 | 2 | 1 | | R:R:F113 | R:R:Q116 | 7.03 | 0 | No | No | 6 | 5 | 1 | 2 | | R:R:F113 | W:W:?1 | 5.82 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:R121 | R:R:Y117 | 6.17 | 5 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:Y117 | W:W:?1 | 9.33 | 5 | Yes | Yes | 5 | 0 | 1 | 0 | | R:R:R122 | R:R:Y125 | 4.12 | 0 | Yes | No | 6 | 4 | 2 | 1 | | R:R:Y125 | W:W:?1 | 33.6 | 0 | No | Yes | 4 | 0 | 1 | 0 | | R:R:A129 | W:W:?1 | 6.27 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:S120 | W:W:?1 | 3.59 | 0 | No | Yes | 4 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 8.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 11.93 | | Average Nodes In Shell | 19.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 21.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9CM7 | A | Lipid | Free Fatty Acid | FFA2 | Homo Sapiens | TUG-1375 | AZ-1729 | Gi1/Beta1/Gamma2 | 3.29 | 2025-06-25 | doi.org/10.1038/s41586-025-09186-6 |
|
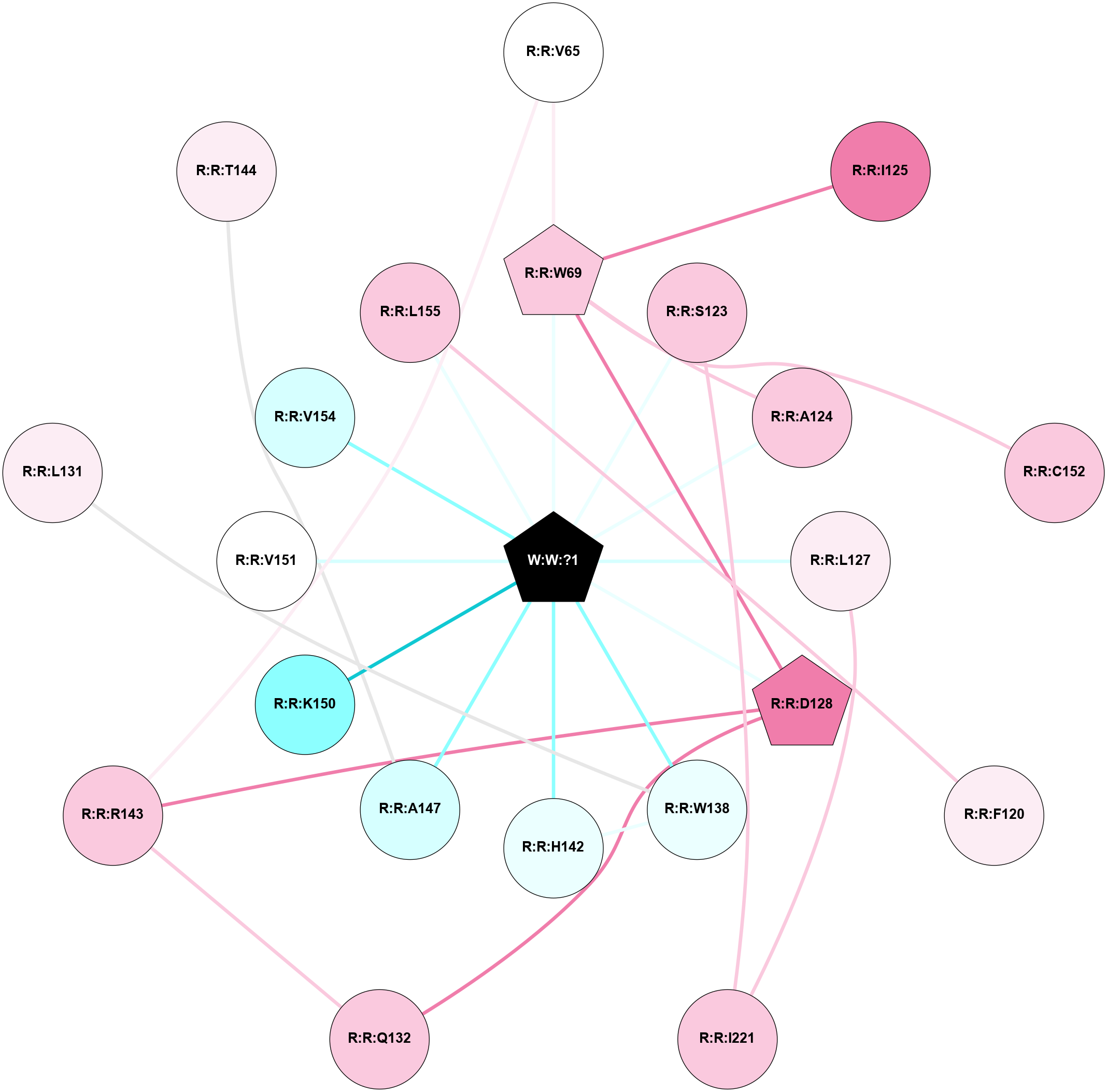
A 2D representation of the interactions of _W_ in 9CM7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:N347 | R:R:G110 | 5.09 | 3 | No | No | 8 | 8 | 2 | 2 | | A:A:N347 | R:R:Y117 | 12.79 | 3 | No | Yes | 8 | 5 | 2 | 1 | | R:R:E106 | R:R:P43 | 4.72 | 3 | Yes | No | 8 | 7 | 1 | 1 | | R:R:P43 | W:W:?1 | 7.71 | 3 | No | Yes | 7 | 0 | 1 | 0 | | R:R:E106 | R:R:V44 | 5.7 | 3 | Yes | No | 8 | 7 | 1 | 2 | | R:R:E106 | R:R:L47 | 3.98 | 3 | Yes | No | 8 | 7 | 1 | 2 | | R:R:P191 | R:R:T97 | 3.5 | 3 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:M136 | R:R:W98 | 12.8 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:L187 | R:R:W98 | 3.42 | 0 | No | Yes | 6 | 5 | 2 | 1 | | R:R:P191 | R:R:W98 | 16.21 | 3 | Yes | Yes | 9 | 5 | 1 | 1 | | R:R:W98 | W:W:?1 | 16.04 | 3 | Yes | Yes | 5 | 0 | 1 | 0 | | R:R:A101 | W:W:?1 | 4.93 | 0 | No | Yes | 8 | 0 | 1 | 0 | | R:R:I105 | W:W:?1 | 3.35 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:E106 | R:R:Y117 | 3.37 | 3 | Yes | Yes | 8 | 5 | 1 | 1 | | R:R:E106 | W:W:?1 | 14.52 | 3 | Yes | Yes | 8 | 0 | 1 | 0 | | R:R:G110 | R:R:Y117 | 5.79 | 3 | No | Yes | 8 | 5 | 2 | 1 | | R:R:Y117 | W:W:?1 | 4.59 | 3 | Yes | Yes | 5 | 0 | 1 | 0 | | R:R:S120 | W:W:?1 | 4.7 | 0 | No | Yes | 4 | 0 | 1 | 0 | | R:R:R122 | R:R:Y125 | 15.43 | 0 | No | No | 6 | 4 | 2 | 1 | | R:R:Y125 | W:W:?1 | 11.01 | 0 | No | Yes | 4 | 0 | 1 | 0 | | R:R:A129 | W:W:?1 | 7.4 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:V132 | W:W:?1 | 6.99 | 0 | No | Yes | 4 | 0 | 1 | 0 | | R:R:I190 | R:R:P191 | 3.39 | 0 | No | Yes | 5 | 9 | 2 | 1 | | R:R:P191 | W:W:?1 | 3.85 | 3 | Yes | Yes | 9 | 0 | 1 | 0 | | R:R:V194 | W:W:?1 | 6.99 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:G126 | R:R:P43 | 2.03 | 0 | No | No | 8 | 7 | 2 | 1 | | R:R:A101 | R:R:G102 | 1.95 | 0 | No | No | 8 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 12.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 7.67 | | Average Nodes In Shell | 24.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 27.00 | | Average Links Mediated by Hubs In Shell | 23.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9IVM | B1 | Peptide | Glucagon | GLP-1 | Homo Sapiens | GLP-1 (Truncated) | LSN3318839 | chim(NtGi1-Gs)/Beta2/Gamma2 | 3.22 | 2024-11-13 | doi.org/10.1016/j.apsb.2024.09.002 |
|
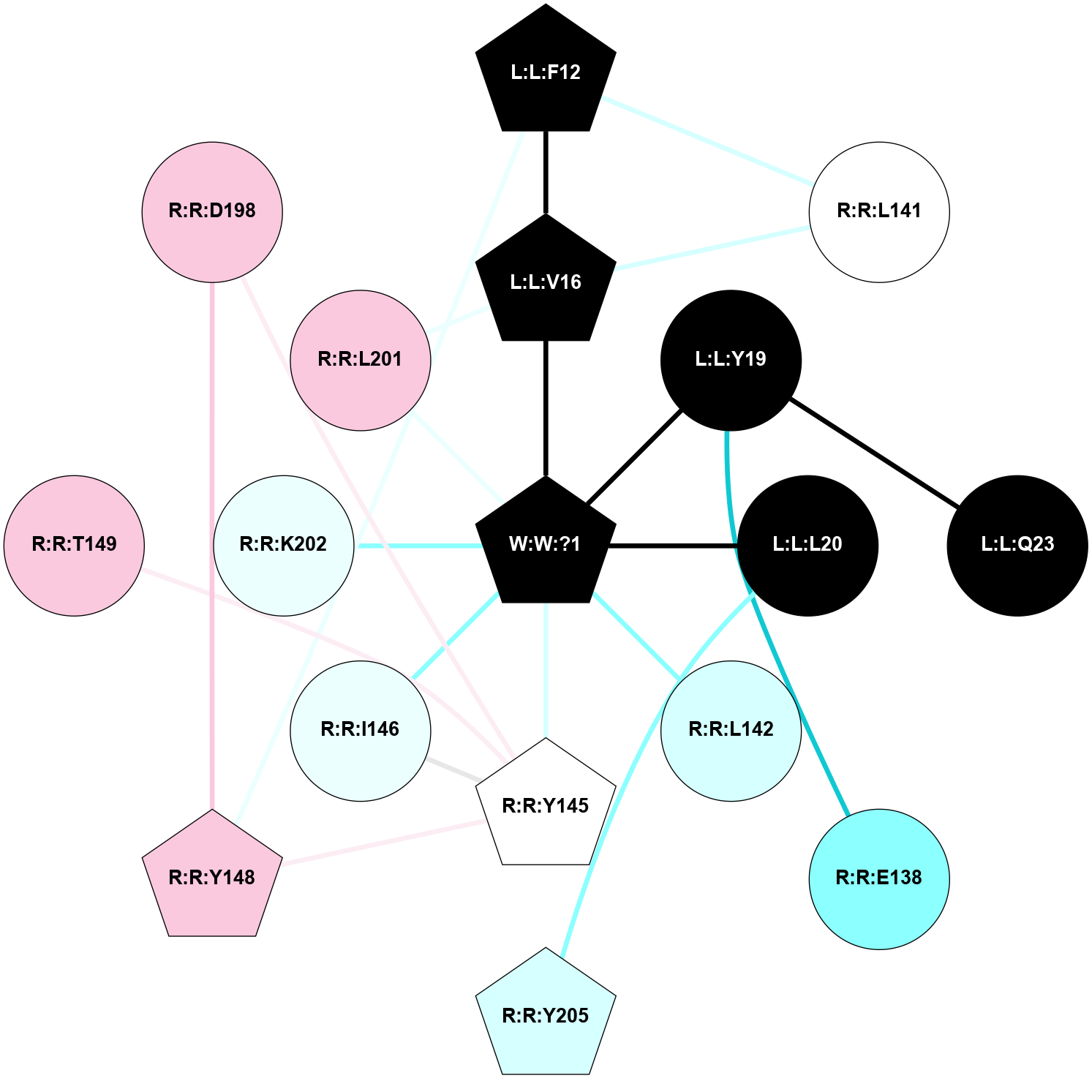
A 2D representation of the interactions of _W_ in 9IVM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:F12 | L:L:V16 | 5.24 | 3 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:F12 | R:R:L141 | 4.87 | 3 | Yes | No | 0 | 5 | 2 | 2 | | L:L:F12 | R:R:Y148 | 8.25 | 3 | Yes | Yes | 0 | 7 | 2 | 2 | | L:L:V16 | R:R:L141 | 4.47 | 3 | Yes | No | 0 | 5 | 1 | 2 | | L:L:V16 | W:W:?1 | 3.21 | 3 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:Q23 | L:L:Y19 | 7.89 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:Y19 | R:R:E138 | 7.86 | 0 | No | No | 0 | 2 | 1 | 2 | | L:L:Y19 | W:W:?1 | 4.21 | 0 | No | Yes | 0 | 0 | 1 | 0 | | L:L:L20 | R:R:Y205 | 10.55 | 0 | No | Yes | 0 | 3 | 1 | 2 | | L:L:L20 | W:W:?1 | 9.94 | 0 | No | Yes | 0 | 0 | 1 | 0 | | R:R:L142 | W:W:?1 | 17.9 | 0 | No | Yes | 3 | 0 | 1 | 0 | | R:R:I146 | R:R:Y145 | 3.63 | 3 | No | Yes | 4 | 5 | 1 | 1 | | R:R:Y145 | R:R:Y148 | 4.96 | 3 | Yes | Yes | 5 | 7 | 1 | 2 | | R:R:T149 | R:R:Y145 | 8.74 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:D198 | R:R:Y145 | 6.9 | 3 | No | Yes | 7 | 5 | 2 | 1 | | R:R:Y145 | W:W:?1 | 37.05 | 3 | Yes | Yes | 5 | 0 | 1 | 0 | | R:R:I146 | W:W:?1 | 3.08 | 3 | No | Yes | 4 | 0 | 1 | 0 | | R:R:D198 | R:R:Y148 | 9.2 | 3 | No | Yes | 7 | 7 | 2 | 2 | | R:R:K202 | W:W:?1 | 28.36 | 0 | No | Yes | 4 | 0 | 1 | 0 | | L:L:V16 | R:R:L201 | 2.98 | 3 | Yes | No | 0 | 7 | 1 | 1 | | R:R:L201 | W:W:?1 | 2.98 | 3 | No | Yes | 7 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 8.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 13.34 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 21.00 | | Average Links Mediated by Hubs In Shell | 19.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9IYB | A | Lipid | Prostanoid | DP2 | Homo Sapiens | Prostaglandin | A1D5Q | Gi1/Beta1/Gamma2 | 2.82 | 2024-12-04 | doi.org/10.1073/pnas.2403304121 |
|
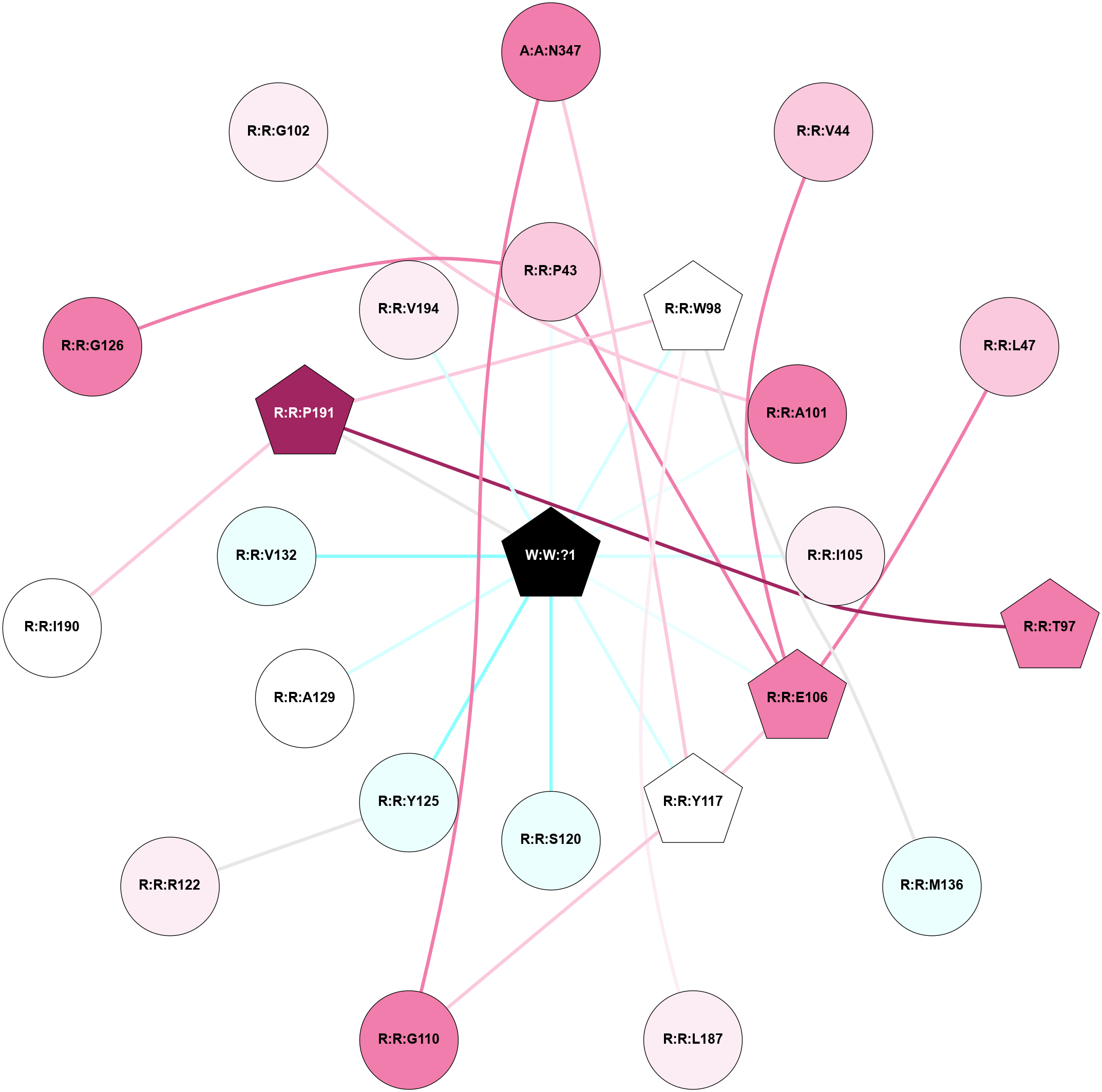
A 2D representation of the interactions of _W_ in 9IYB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:V65 | R:R:W69 | 4.9 | 0 | No | Yes | 5 | 7 | 2 | 1 | | R:R:R143 | R:R:V65 | 10.46 | 3 | No | No | 7 | 5 | 2 | 2 | | R:R:A124 | R:R:W69 | 5.19 | 3 | No | Yes | 7 | 7 | 1 | 1 | | R:R:I125 | R:R:W69 | 5.87 | 3 | No | Yes | 8 | 7 | 2 | 1 | | R:R:D128 | R:R:W69 | 5.58 | 3 | Yes | Yes | 8 | 7 | 1 | 1 | | R:R:C152 | R:R:W69 | 6.53 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:W69 | W:W:?1 | 6.8 | 3 | Yes | Yes | 7 | 0 | 1 | 0 | | R:R:F120 | R:R:L155 | 4.87 | 0 | No | No | 6 | 7 | 2 | 1 | | R:R:I221 | R:R:S123 | 4.64 | 0 | No | No | 7 | 7 | 2 | 1 | | R:R:S123 | W:W:?1 | 5.6 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:A124 | W:W:?1 | 3.53 | 3 | No | Yes | 7 | 0 | 1 | 0 | | R:R:I221 | R:R:L127 | 4.28 | 0 | No | No | 7 | 6 | 2 | 1 | | R:R:L127 | W:W:?1 | 8.27 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:D128 | R:R:Q132 | 3.92 | 3 | Yes | No | 8 | 7 | 1 | 2 | | R:R:D128 | R:R:R143 | 10.72 | 3 | Yes | No | 8 | 7 | 1 | 2 | | R:R:D128 | W:W:?1 | 4.05 | 3 | Yes | Yes | 8 | 0 | 1 | 0 | | R:R:L131 | R:R:W138 | 4.56 | 0 | No | No | 6 | 4 | 2 | 1 | | R:R:Q132 | R:R:R143 | 11.68 | 3 | No | No | 7 | 7 | 2 | 2 | | R:R:H142 | R:R:W138 | 20.1 | 3 | No | No | 4 | 4 | 1 | 1 | | R:R:W138 | W:W:?1 | 11.05 | 3 | No | Yes | 4 | 0 | 1 | 0 | | R:R:H142 | W:W:?1 | 26.87 | 3 | No | Yes | 4 | 0 | 1 | 0 | | R:R:A147 | W:W:?1 | 7.06 | 0 | No | Yes | 3 | 0 | 1 | 0 | | R:R:K150 | W:W:?1 | 8.42 | 0 | No | Yes | 2 | 0 | 1 | 0 | | R:R:V151 | W:W:?1 | 18.9 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:V154 | W:W:?1 | 6.67 | 0 | No | Yes | 3 | 0 | 1 | 0 | | R:R:L155 | W:W:?1 | 7.23 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:A147 | R:R:T144 | 1.68 | 0 | No | No | 3 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 12.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.54 | | Average Nodes In Shell | 22.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 27.00 | | Average Links Mediated by Hubs In Shell | 19.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9JCO | A | Orphan | Orphan | GPR4 | Homo sapiens | - | LPC | chim(NtGi1-Gs)/Beta1/Gamma2 | 2.36 | 2025-07-09 | doi.org/10.1038/s41421-025-00807-y |
|
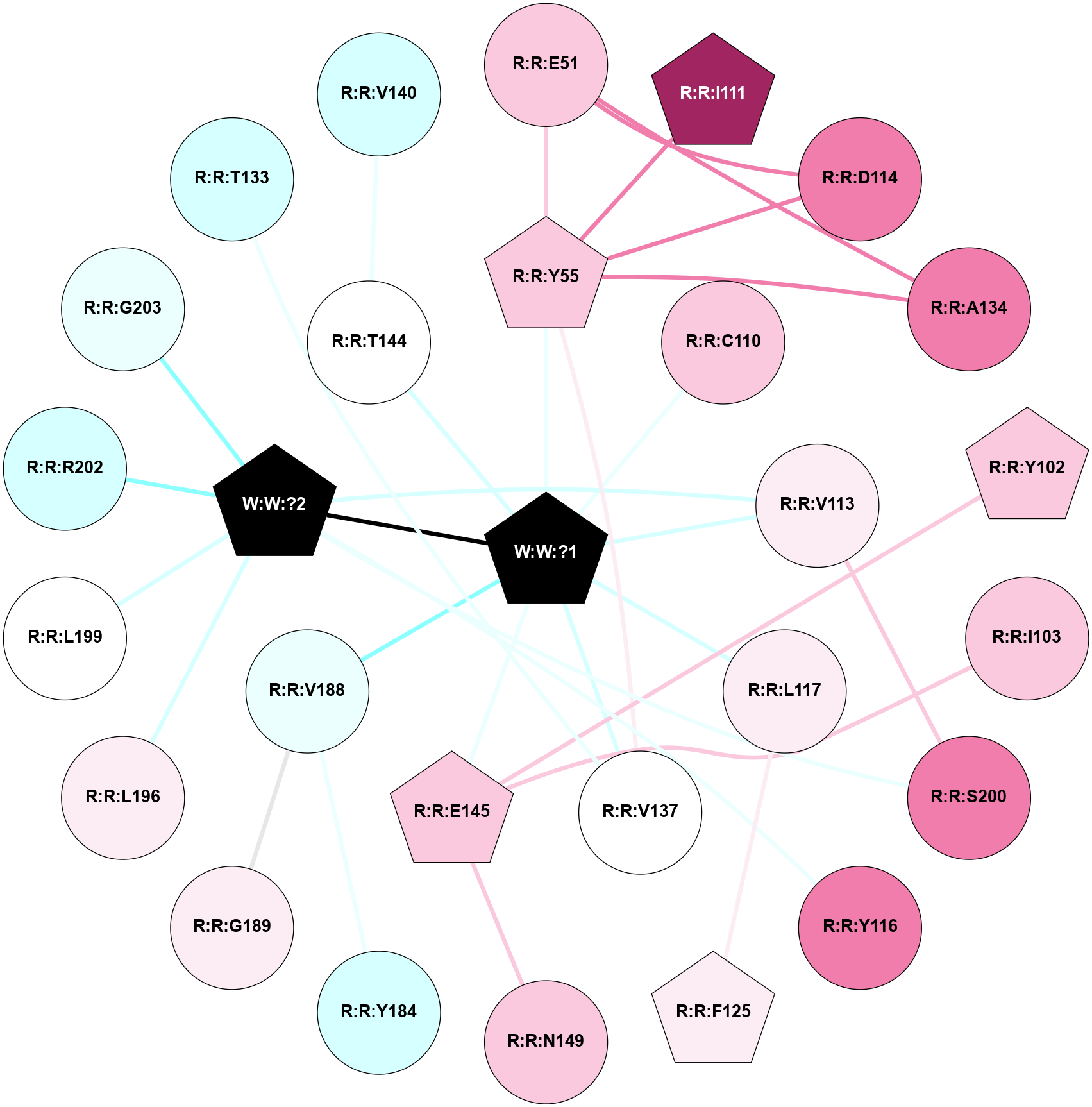
A 2D representation of the interactions of _W_ in 9JCO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E51 | R:R:Y55 | 7.86 | 2 | No | Yes | 7 | 7 | 2 | 1 | | R:R:D114 | R:R:E51 | 14.29 | 2 | No | No | 8 | 7 | 2 | 2 | | R:R:A134 | R:R:E51 | 4.53 | 2 | No | No | 8 | 7 | 2 | 2 | | R:R:I111 | R:R:Y55 | 3.63 | 2 | Yes | Yes | 9 | 7 | 2 | 1 | | R:R:D114 | R:R:Y55 | 4.6 | 2 | No | Yes | 8 | 7 | 2 | 1 | | R:R:A134 | R:R:Y55 | 5.34 | 2 | No | Yes | 8 | 7 | 2 | 1 | | R:R:V137 | R:R:Y55 | 6.31 | 2 | No | Yes | 5 | 7 | 1 | 1 | | R:R:Y55 | W:W:?1 | 3.74 | 2 | Yes | Yes | 7 | 0 | 1 | 0 | | R:R:E145 | R:R:Y102 | 3.37 | 0 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:E145 | R:R:I103 | 8.2 | 0 | Yes | No | 7 | 7 | 1 | 2 | | R:R:C110 | W:W:?1 | 6.74 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:S200 | R:R:V113 | 3.23 | 2 | No | No | 8 | 6 | 2 | 1 | | R:R:V113 | W:W:?1 | 9.5 | 2 | No | Yes | 6 | 0 | 1 | 0 | | R:R:V113 | W:W:?2 | 6.33 | 2 | No | Yes | 6 | 0 | 1 | 1 | | R:R:Y116 | W:W:?2 | 27.4 | 2 | No | Yes | 8 | 0 | 2 | 1 | | R:R:F125 | R:R:L117 | 3.65 | 2 | Yes | No | 6 | 6 | 2 | 1 | | R:R:L117 | W:W:?1 | 4.41 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:V137 | W:W:?1 | 9.5 | 2 | No | Yes | 5 | 0 | 1 | 0 | | R:R:E145 | R:R:N149 | 6.57 | 0 | Yes | No | 7 | 7 | 1 | 2 | | R:R:E145 | W:W:?1 | 5.63 | 0 | Yes | Yes | 7 | 0 | 1 | 0 | | R:R:V188 | R:R:Y184 | 12.62 | 0 | No | No | 4 | 3 | 1 | 2 | | R:R:G189 | R:R:V188 | 3.68 | 0 | No | No | 6 | 4 | 2 | 1 | | R:R:V188 | W:W:?1 | 3.17 | 0 | No | Yes | 4 | 0 | 1 | 0 | | R:R:L196 | W:W:?2 | 5.88 | 0 | No | Yes | 6 | 0 | 2 | 1 | | R:R:L199 | W:W:?2 | 11.76 | 0 | No | Yes | 5 | 0 | 2 | 1 | | R:R:S200 | W:W:?2 | 7.98 | 2 | No | Yes | 8 | 0 | 2 | 1 | | R:R:R202 | W:W:?2 | 7.74 | 0 | No | Yes | 3 | 0 | 2 | 1 | | R:R:G203 | W:W:?2 | 7.27 | 0 | No | Yes | 4 | 0 | 2 | 1 | | W:W:?1 | W:W:?2 | 7.81 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | R:R:T133 | R:R:V137 | 1.59 | 0 | No | No | 3 | 5 | 2 | 1 | | R:R:T144 | R:R:V140 | 1.59 | 0 | No | No | 5 | 3 | 1 | 2 | | R:R:T144 | W:W:?1 | 1.57 | 0 | No | Yes | 5 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 9.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 5.79 | | Average Nodes In Shell | 28.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 32.00 | | Average Links Mediated by Hubs In Shell | 25.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
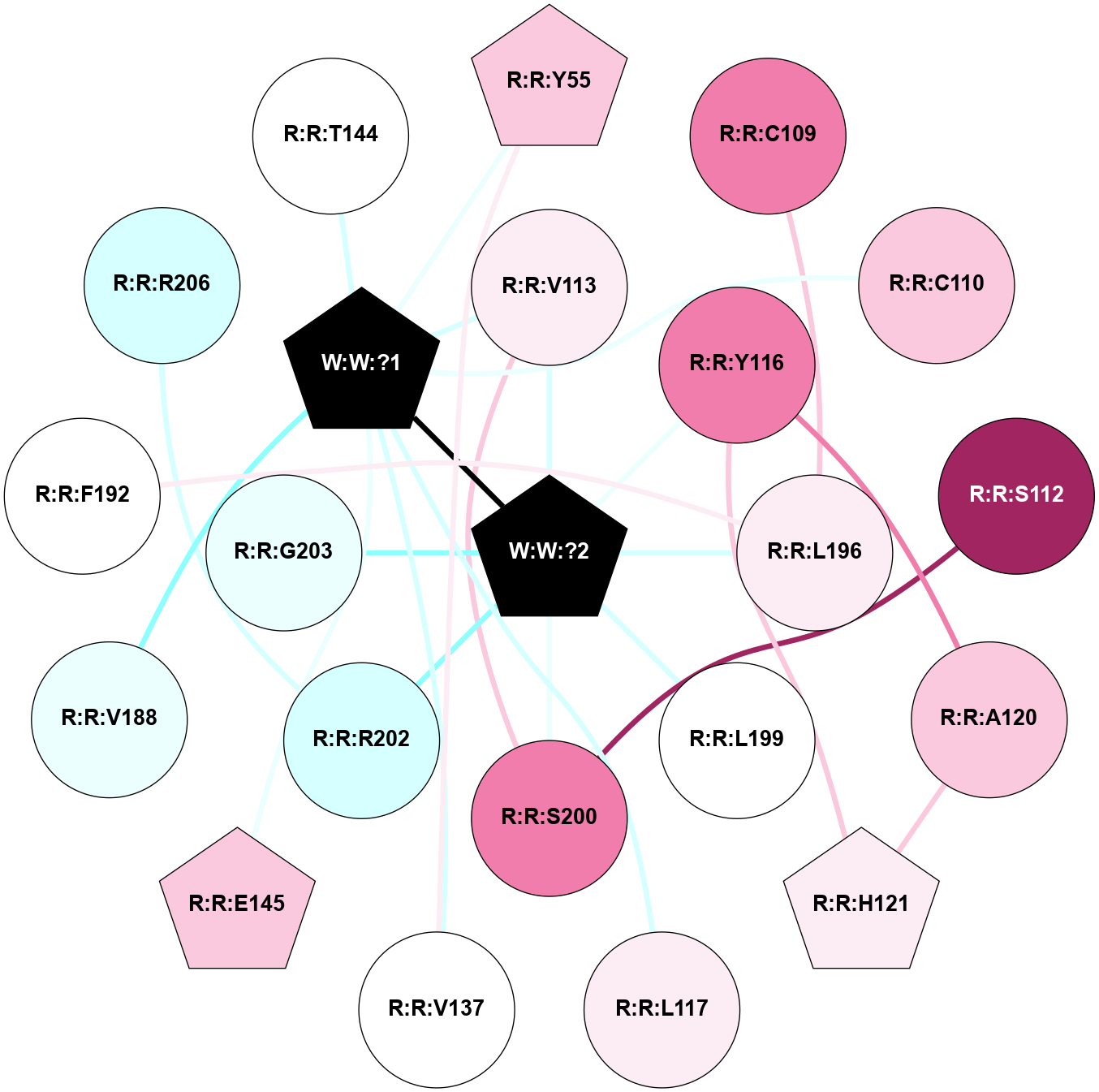
A 2D representation of the interactions of _W_ in 9JCO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:V137 | R:R:Y55 | 6.31 | 2 | No | Yes | 5 | 7 | 2 | 2 | | R:R:Y55 | W:W:?1 | 3.74 | 2 | Yes | Yes | 7 | 0 | 2 | 1 | | R:R:C109 | R:R:L196 | 3.17 | 0 | No | No | 8 | 6 | 2 | 1 | | R:R:C110 | W:W:?1 | 6.74 | 0 | No | Yes | 7 | 0 | 2 | 1 | | R:R:S112 | R:R:S200 | 6.52 | 2 | No | No | 9 | 8 | 2 | 1 | | R:R:S200 | R:R:V113 | 3.23 | 2 | No | No | 8 | 6 | 1 | 1 | | R:R:V113 | W:W:?1 | 9.5 | 2 | No | Yes | 6 | 0 | 1 | 1 | | R:R:V113 | W:W:?2 | 6.33 | 2 | No | Yes | 6 | 0 | 1 | 0 | | R:R:A120 | R:R:Y116 | 5.34 | 2 | No | No | 7 | 8 | 2 | 1 | | R:R:H121 | R:R:Y116 | 9.8 | 2 | Yes | No | 6 | 8 | 2 | 1 | | R:R:Y116 | W:W:?2 | 27.4 | 2 | No | Yes | 8 | 0 | 1 | 0 | | R:R:L117 | W:W:?1 | 4.41 | 0 | No | Yes | 6 | 0 | 2 | 1 | | R:R:A120 | R:R:H121 | 4.39 | 2 | No | Yes | 7 | 6 | 2 | 2 | | R:R:V137 | W:W:?1 | 9.5 | 2 | No | Yes | 5 | 0 | 2 | 1 | | R:R:E145 | W:W:?1 | 5.63 | 0 | Yes | Yes | 7 | 0 | 2 | 1 | | R:R:V188 | W:W:?1 | 3.17 | 0 | No | Yes | 4 | 0 | 2 | 1 | | R:R:F192 | R:R:L196 | 6.09 | 0 | No | No | 5 | 6 | 2 | 1 | | R:R:L196 | W:W:?2 | 5.88 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:L199 | W:W:?2 | 11.76 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:S200 | W:W:?2 | 7.98 | 2 | No | Yes | 8 | 0 | 1 | 0 | | R:R:R202 | R:R:R206 | 4.26 | 0 | No | No | 3 | 3 | 1 | 2 | | R:R:R202 | W:W:?2 | 7.74 | 0 | No | Yes | 3 | 0 | 1 | 0 | | R:R:G203 | W:W:?2 | 7.27 | 0 | No | Yes | 4 | 0 | 1 | 0 | | W:W:?1 | W:W:?2 | 7.81 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | R:R:T144 | W:W:?1 | 1.57 | 0 | No | Yes | 5 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 8.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 10.27 | | Average Nodes In Shell | 22.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 25.00 | | Average Links Mediated by Hubs In Shell | 19.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9JCP | A | Orphan | Orphan | GPR4 | Homo sapiens | - | LPC | chim(NtGi1-Gs-CtGq)/Beta1/Gamma2 | 2.55 | 2025-07-09 | doi.org/10.1038/s41421-025-00807-y |
|
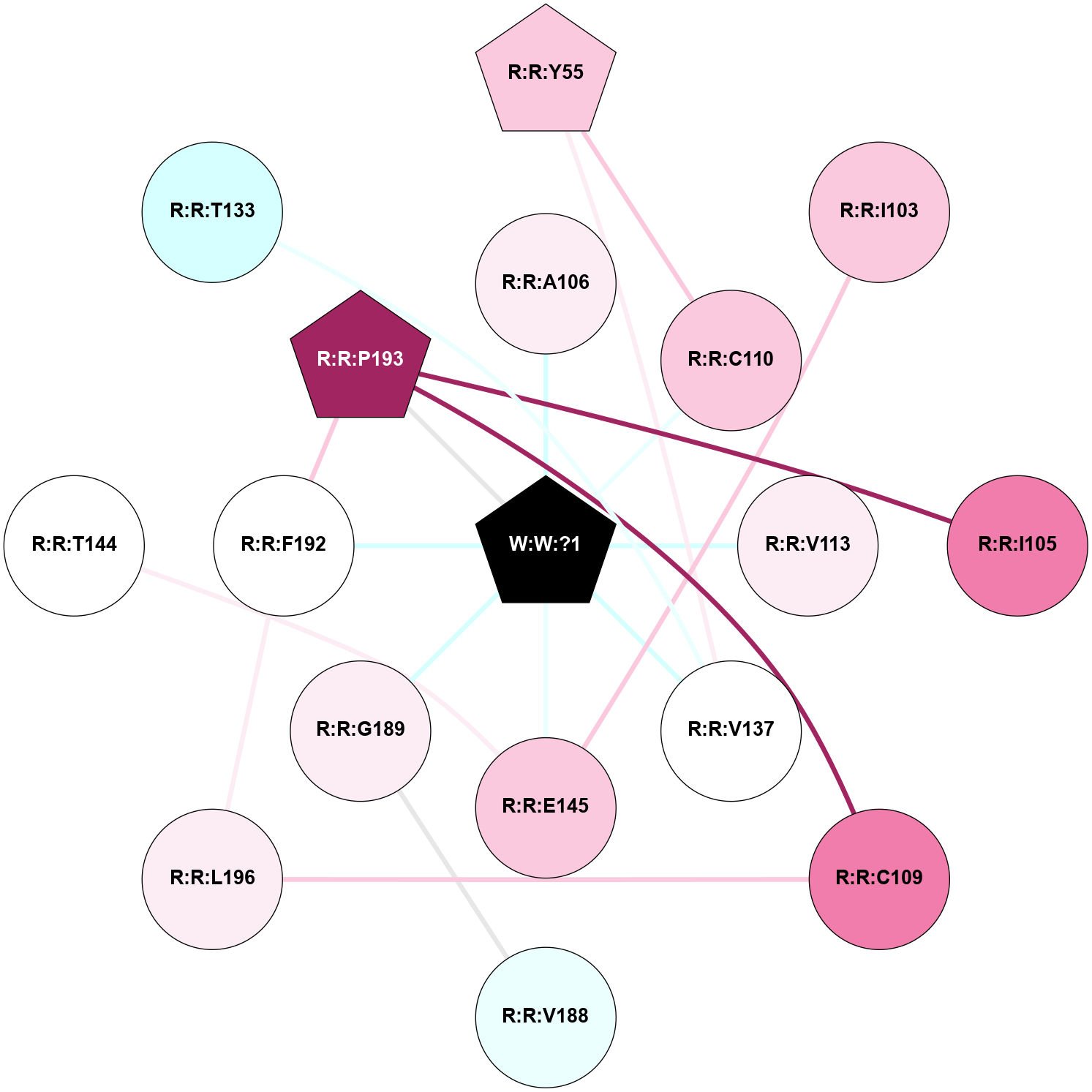
A 2D representation of the interactions of _W_ in 9JCP
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:C110 | R:R:Y55 | 5.38 | 0 | No | Yes | 7 | 7 | 1 | 2 | | R:R:V137 | R:R:Y55 | 6.31 | 0 | No | Yes | 5 | 7 | 1 | 2 | | R:R:E145 | R:R:I103 | 5.47 | 0 | No | No | 7 | 7 | 1 | 2 | | R:R:I105 | R:R:P193 | 3.39 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:A106 | W:W:?1 | 6.7 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:C109 | R:R:P193 | 3.77 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:C109 | R:R:L196 | 4.76 | 0 | No | No | 8 | 6 | 2 | 2 | | R:R:C110 | W:W:?1 | 10.12 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:V113 | W:W:?1 | 9.5 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:V137 | W:W:?1 | 6.33 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:E145 | W:W:?1 | 18.3 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:G189 | R:R:V188 | 3.68 | 0 | No | No | 6 | 4 | 1 | 2 | | R:R:G189 | W:W:?1 | 7.27 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:F192 | R:R:P193 | 4.33 | 30 | No | Yes | 5 | 9 | 1 | 1 | | R:R:F192 | R:R:L196 | 12.18 | 30 | No | No | 5 | 6 | 1 | 2 | | R:R:F192 | W:W:?1 | 3.88 | 30 | No | Yes | 5 | 0 | 1 | 0 | | R:R:P193 | W:W:?1 | 6.98 | 30 | Yes | Yes | 9 | 0 | 1 | 0 | | R:R:E145 | R:R:T144 | 2.82 | 0 | No | No | 7 | 5 | 1 | 2 | | R:R:T133 | R:R:V137 | 1.59 | 0 | No | No | 3 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 8.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 8.63 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 19.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
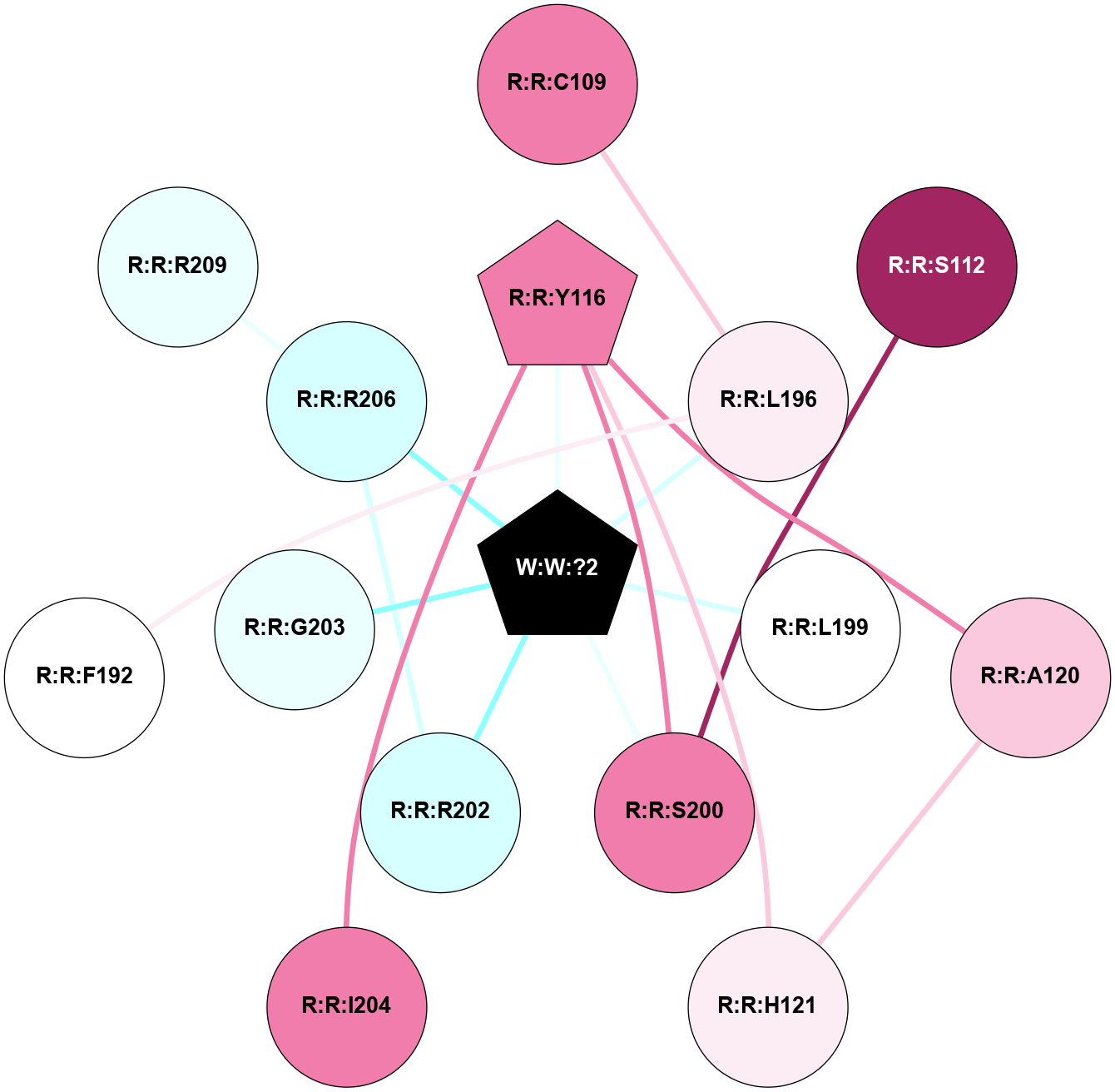
A 2D representation of the interactions of _W_ in 9JCP
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:C109 | R:R:L196 | 4.76 | 0 | No | No | 8 | 6 | 2 | 1 | | R:R:S112 | R:R:S200 | 6.52 | 1 | No | No | 9 | 8 | 2 | 1 | | R:R:A120 | R:R:Y116 | 5.34 | 1 | No | Yes | 7 | 8 | 2 | 1 | | R:R:H121 | R:R:Y116 | 5.44 | 1 | No | Yes | 6 | 8 | 2 | 1 | | R:R:S200 | R:R:Y116 | 3.82 | 1 | No | Yes | 8 | 8 | 1 | 1 | | R:R:I204 | R:R:Y116 | 3.63 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:Y116 | W:W:?2 | 28.65 | 1 | Yes | Yes | 8 | 0 | 1 | 0 | | R:R:A120 | R:R:H121 | 4.39 | 1 | No | No | 7 | 6 | 2 | 2 | | R:R:F192 | R:R:L196 | 12.18 | 30 | No | No | 5 | 6 | 2 | 1 | | R:R:L196 | W:W:?2 | 4.41 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:L199 | W:W:?2 | 7.35 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:S200 | W:W:?2 | 6.38 | 1 | No | Yes | 8 | 0 | 1 | 0 | | R:R:R202 | R:R:R206 | 4.26 | 1 | No | No | 3 | 3 | 1 | 1 | | R:R:R202 | W:W:?2 | 9.04 | 1 | No | Yes | 3 | 0 | 1 | 0 | | R:R:G203 | W:W:?2 | 5.45 | 0 | No | Yes | 4 | 0 | 1 | 0 | | R:R:R206 | W:W:?2 | 5.16 | 1 | No | Yes | 3 | 0 | 1 | 0 | | R:R:R206 | R:R:R209 | 3.2 | 1 | No | No | 3 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 9.49 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9JCQ | A | Orphan | Orphan | GPR4 | Homo sapiens | - | LPC | chim(NtGi1-Gs)/Beta1/Gamma2 | 2.59 | 2025-07-09 | doi.org/10.1038/s41421-025-00807-y |
|
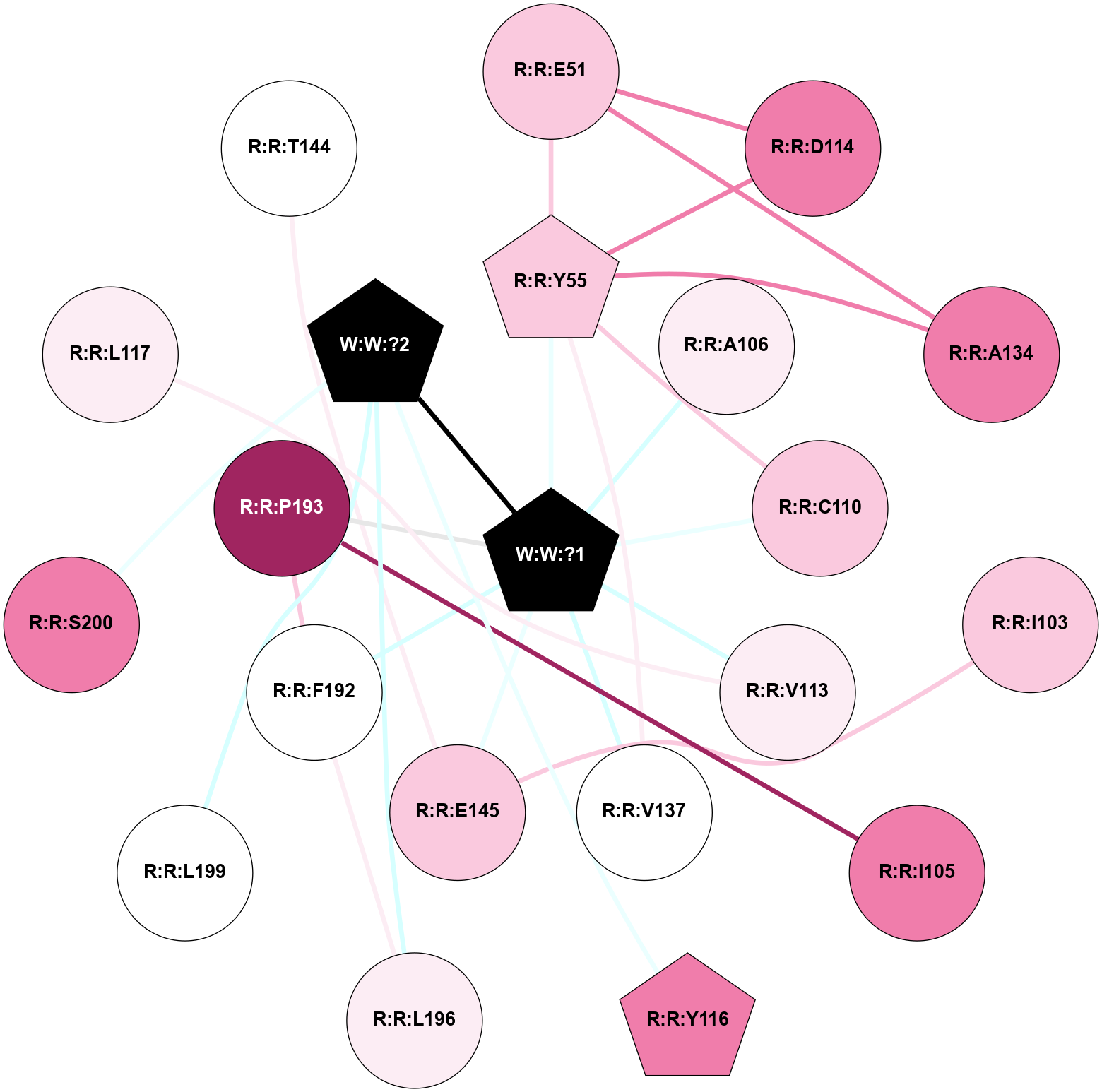
A 2D representation of the interactions of _W_ in 9JCQ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E51 | R:R:Y55 | 14.59 | 2 | No | Yes | 7 | 7 | 2 | 1 | | R:R:D114 | R:R:E51 | 14.29 | 2 | No | No | 8 | 7 | 2 | 2 | | R:R:A134 | R:R:E51 | 4.53 | 2 | No | No | 8 | 7 | 2 | 2 | | R:R:C110 | R:R:Y55 | 8.06 | 2 | No | Yes | 7 | 7 | 1 | 1 | | R:R:D114 | R:R:Y55 | 6.9 | 2 | No | Yes | 8 | 7 | 2 | 1 | | R:R:A134 | R:R:Y55 | 5.34 | 2 | No | Yes | 8 | 7 | 2 | 1 | | R:R:V137 | R:R:Y55 | 3.79 | 2 | No | Yes | 5 | 7 | 1 | 1 | | R:R:Y55 | W:W:?1 | 6.23 | 2 | Yes | Yes | 7 | 0 | 1 | 0 | | R:R:E145 | R:R:I103 | 6.83 | 0 | No | No | 7 | 7 | 1 | 2 | | R:R:I105 | R:R:P193 | 3.39 | 0 | No | No | 8 | 9 | 2 | 1 | | R:R:A106 | W:W:?1 | 3.35 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:C110 | W:W:?1 | 6.74 | 2 | No | Yes | 7 | 0 | 1 | 0 | | R:R:V113 | W:W:?1 | 15.83 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:Y116 | W:W:?2 | 27.4 | 0 | Yes | Yes | 8 | 0 | 2 | 1 | | R:R:V137 | W:W:?1 | 6.33 | 2 | No | Yes | 5 | 0 | 1 | 0 | | R:R:E145 | W:W:?1 | 9.86 | 0 | No | Yes | 7 | 0 | 1 | 0 | | R:R:F192 | R:R:P193 | 5.78 | 2 | No | No | 5 | 9 | 1 | 1 | | R:R:F192 | R:R:L196 | 13.4 | 2 | No | No | 5 | 6 | 1 | 2 | | R:R:F192 | W:W:?1 | 3.88 | 2 | No | Yes | 5 | 0 | 1 | 0 | | R:R:P193 | W:W:?1 | 5.23 | 2 | No | Yes | 9 | 0 | 1 | 0 | | R:R:L196 | W:W:?2 | 7.35 | 0 | No | Yes | 6 | 0 | 2 | 1 | | R:R:L199 | W:W:?2 | 5.88 | 0 | No | Yes | 5 | 0 | 2 | 1 | | R:R:S200 | W:W:?2 | 7.98 | 0 | No | Yes | 8 | 0 | 2 | 1 | | W:W:?1 | W:W:?2 | 6.25 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | R:R:L117 | R:R:V113 | 2.98 | 0 | No | No | 6 | 6 | 2 | 1 | | R:R:E145 | R:R:T144 | 1.41 | 0 | No | No | 7 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 9.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 7.08 | | Average Nodes In Shell | 21.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 26.00 | | Average Links Mediated by Hubs In Shell | 18.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
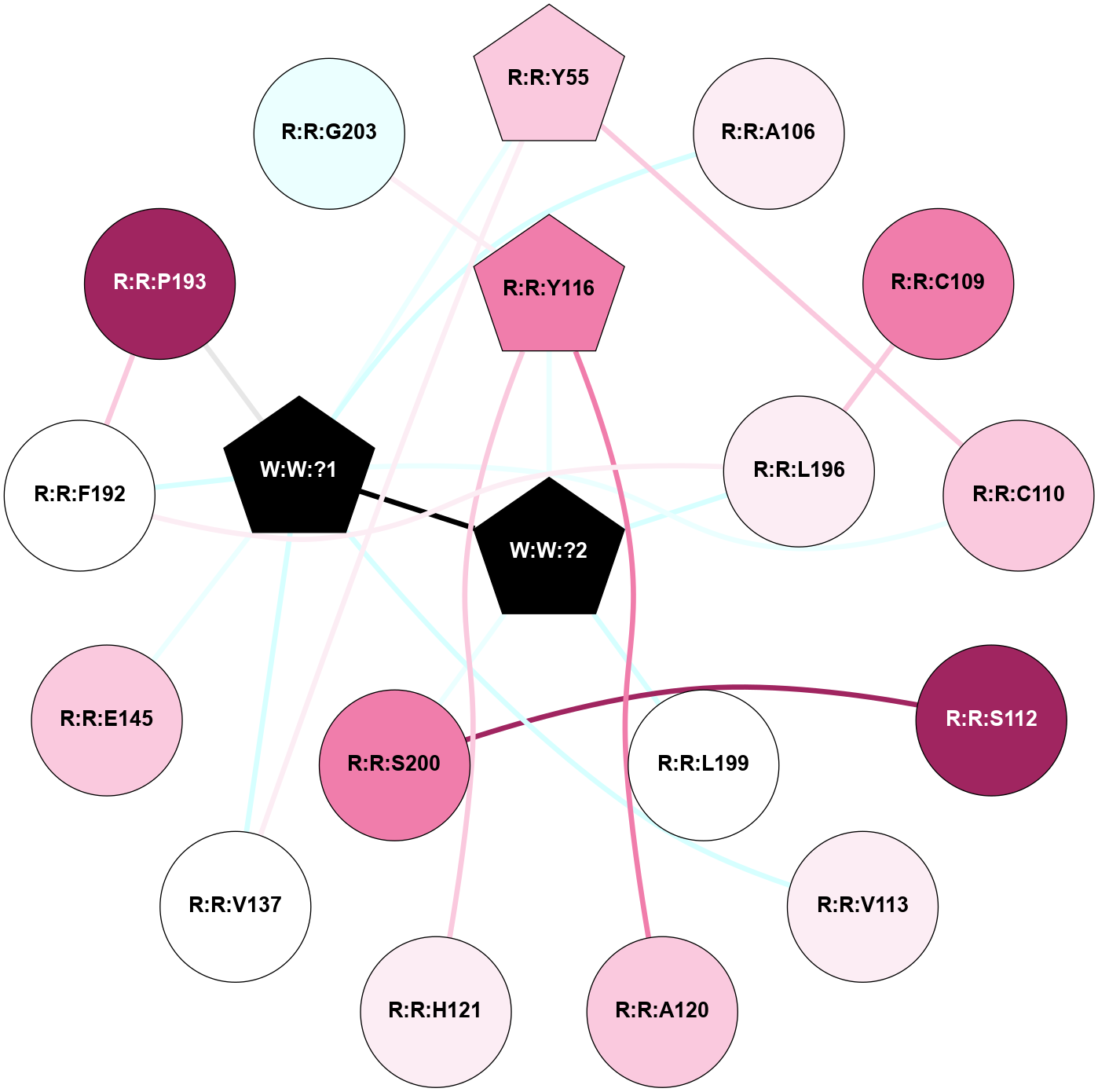
A 2D representation of the interactions of _W_ in 9JCQ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:C110 | R:R:Y55 | 8.06 | 2 | No | Yes | 7 | 7 | 2 | 2 | | R:R:V137 | R:R:Y55 | 3.79 | 2 | No | Yes | 5 | 7 | 2 | 2 | | R:R:Y55 | W:W:?1 | 6.23 | 2 | Yes | Yes | 7 | 0 | 2 | 1 | | R:R:A106 | W:W:?1 | 3.35 | 0 | No | Yes | 6 | 0 | 2 | 1 | | R:R:C109 | R:R:L196 | 4.76 | 0 | No | No | 8 | 6 | 2 | 1 | | R:R:C110 | W:W:?1 | 6.74 | 2 | No | Yes | 7 | 0 | 2 | 1 | | R:R:S112 | R:R:S200 | 6.52 | 0 | No | No | 9 | 8 | 2 | 1 | | R:R:V113 | W:W:?1 | 15.83 | 0 | No | Yes | 6 | 0 | 2 | 1 | | R:R:A120 | R:R:Y116 | 4 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:H121 | R:R:Y116 | 7.62 | 19 | No | Yes | 6 | 8 | 2 | 1 | | R:R:Y116 | W:W:?2 | 27.4 | 0 | Yes | Yes | 8 | 0 | 1 | 0 | | R:R:V137 | W:W:?1 | 6.33 | 2 | No | Yes | 5 | 0 | 2 | 1 | | R:R:E145 | W:W:?1 | 9.86 | 0 | No | Yes | 7 | 0 | 2 | 1 | | R:R:F192 | R:R:P193 | 5.78 | 2 | No | No | 5 | 9 | 2 | 2 | | R:R:F192 | R:R:L196 | 13.4 | 2 | No | No | 5 | 6 | 2 | 1 | | R:R:F192 | W:W:?1 | 3.88 | 2 | No | Yes | 5 | 0 | 2 | 1 | | R:R:P193 | W:W:?1 | 5.23 | 2 | No | Yes | 9 | 0 | 2 | 1 | | R:R:L196 | W:W:?2 | 7.35 | 0 | No | Yes | 6 | 0 | 1 | 0 | | R:R:L199 | W:W:?2 | 5.88 | 0 | No | Yes | 5 | 0 | 1 | 0 | | R:R:S200 | W:W:?2 | 7.98 | 0 | No | Yes | 8 | 0 | 1 | 0 | | W:W:?1 | W:W:?2 | 6.25 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | R:R:G203 | R:R:Y116 | 2.9 | 0 | No | Yes | 4 | 8 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 10.97 | | Average Nodes In Shell | 19.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 22.00 | | Average Links Mediated by Hubs In Shell | 18.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|





















