| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7F8X | A | Peptide | Cholecystokinin | CCKA | Homo Sapiens | NN9056 | - | - | 3 | 2021-12-29 | doi.org/10.1038/s41589-021-00866-8 |
|

A 2D representation of the interactions of NH2 in 7F8X
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:Y360 | R:R:Y48 | 4.96 | 0 | Yes | No | 7 | 7 | 1 | 2 | | R:R:L90 | R:R:Y360 | 4.69 | 0 | No | Yes | 8 | 7 | 2 | 1 | | R:R:C94 | R:R:M121 | 8.1 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:M95 | R:R:Y360 | 15.57 | 0 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:L356 | R:R:N98 | 9.61 | 1 | No | No | 6 | 7 | 2 | 2 | | R:R:N98 | R:R:Y360 | 15.12 | 0 | No | Yes | 7 | 7 | 2 | 1 | | L:L:?6 | R:R:M121 | 4.01 | 1 | Yes | Yes | 0 | 7 | 2 | 1 | | L:L:?8 | R:R:M121 | 8.5 | 1 | Yes | Yes | 0 | 7 | 1 | 1 | | L:L:?9 | R:R:M121 | 14.07 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:F198 | R:R:Y176 | 19.6 | 1 | Yes | Yes | 5 | 7 | 2 | 2 | | L:L:?7 | R:R:Y176 | 7.89 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?8 | R:R:Y176 | 10.07 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?7 | R:R:F198 | 4.55 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | R:R:I329 | R:R:N333 | 7.08 | 1 | No | Yes | 7 | 7 | 2 | 2 | | R:R:I329 | R:R:L356 | 4.28 | 1 | No | No | 7 | 6 | 2 | 2 | | L:L:?8 | R:R:I329 | 3.68 | 1 | Yes | No | 0 | 7 | 1 | 2 | | L:L:?8 | R:R:F330 | 10.46 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?7 | R:R:N333 | 7.18 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?8 | R:R:N333 | 5.9 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?8 | R:R:L356 | 4.76 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:S359 | R:R:Y360 | 5.09 | 0 | No | Yes | 7 | 7 | 2 | 1 | | L:L:?9 | R:R:Y360 | 5.83 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?6 | L:L:?7 | 23.48 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?6 | L:L:?8 | 11.25 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?7 | L:L:?8 | 34.66 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?7 | L:L:?9 | 5.15 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?8 | L:L:?9 | 20.71 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 11.44 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 27.00 | | Average Links Mediated by Hubs In Shell | 25.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7F9Z | A | Peptide | Ghrelin | Ghrelin | Homo Sapiens | GHRP-6 | - | chim(NtGi1-Gs-CtGq)/Beta1/Gamma2 | 3.2 | 2021-08-18 | doi.org/10.1038/s41467-021-25364-2 |
|
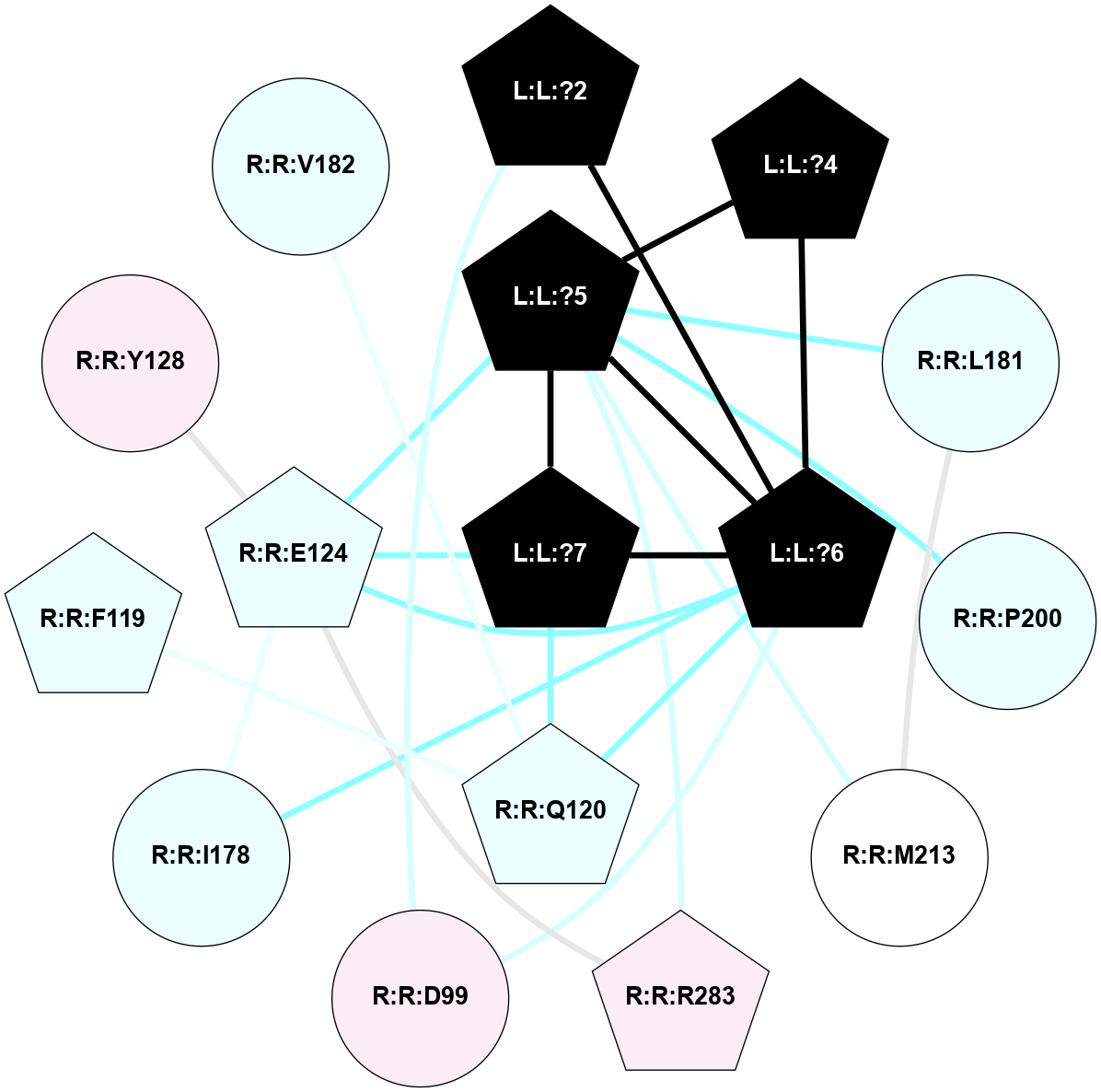
A 2D representation of the interactions of NH2 in 7F9Z
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?2 | L:L:?6 | 9.47 | 3 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?2 | R:R:D99 | 5.37 | 3 | Yes | No | 0 | 6 | 2 | 2 | | L:L:?4 | L:L:?5 | 13.89 | 3 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?4 | L:L:?6 | 9.47 | 3 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?5 | L:L:?6 | 28.35 | 3 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?5 | L:L:?7 | 5.82 | 3 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?5 | R:R:E124 | 5.6 | 3 | Yes | Yes | 0 | 4 | 1 | 1 | | L:L:?5 | R:R:L181 | 11.71 | 3 | Yes | No | 0 | 4 | 1 | 2 | | L:L:?5 | R:R:P200 | 6.94 | 3 | Yes | No | 0 | 4 | 1 | 2 | | L:L:?5 | R:R:M213 | 11.96 | 3 | Yes | No | 0 | 5 | 1 | 2 | | L:L:?5 | R:R:R283 | 27.74 | 3 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?6 | L:L:?7 | 23.83 | 3 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?6 | R:R:D99 | 4.69 | 3 | Yes | No | 0 | 6 | 1 | 2 | | L:L:?6 | R:R:Q120 | 21.87 | 3 | Yes | Yes | 0 | 4 | 1 | 1 | | L:L:?6 | R:R:E124 | 6.88 | 3 | Yes | Yes | 0 | 4 | 1 | 1 | | L:L:?6 | R:R:I178 | 4.94 | 3 | Yes | No | 0 | 4 | 1 | 2 | | L:L:?7 | R:R:Q120 | 9.93 | 3 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?7 | R:R:E124 | 13.19 | 3 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:F119 | R:R:Q120 | 4.68 | 0 | Yes | Yes | 4 | 4 | 2 | 1 | | R:R:E124 | R:R:Y128 | 4.49 | 3 | Yes | No | 4 | 6 | 1 | 2 | | R:R:E124 | R:R:I178 | 6.83 | 3 | Yes | No | 4 | 4 | 1 | 2 | | R:R:E124 | R:R:R283 | 5.82 | 3 | Yes | Yes | 4 | 6 | 1 | 2 | | R:R:L181 | R:R:M213 | 7.07 | 3 | No | No | 4 | 5 | 2 | 2 | | R:R:Q120 | R:R:V182 | 2.87 | 3 | Yes | No | 4 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 13.19 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 24.00 | | Average Links Mediated by Hubs In Shell | 23.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7M3G | C | Ion | Calcium Sensing | CaS | Homo Sapiens | - | Ca; Ca; Tryptophan; Evocalcet; Etelcalcetide; PO4 | - | 2.5 | 2021-06-30 | doi.org/10.1038/s41586-021-03691-0 |
|
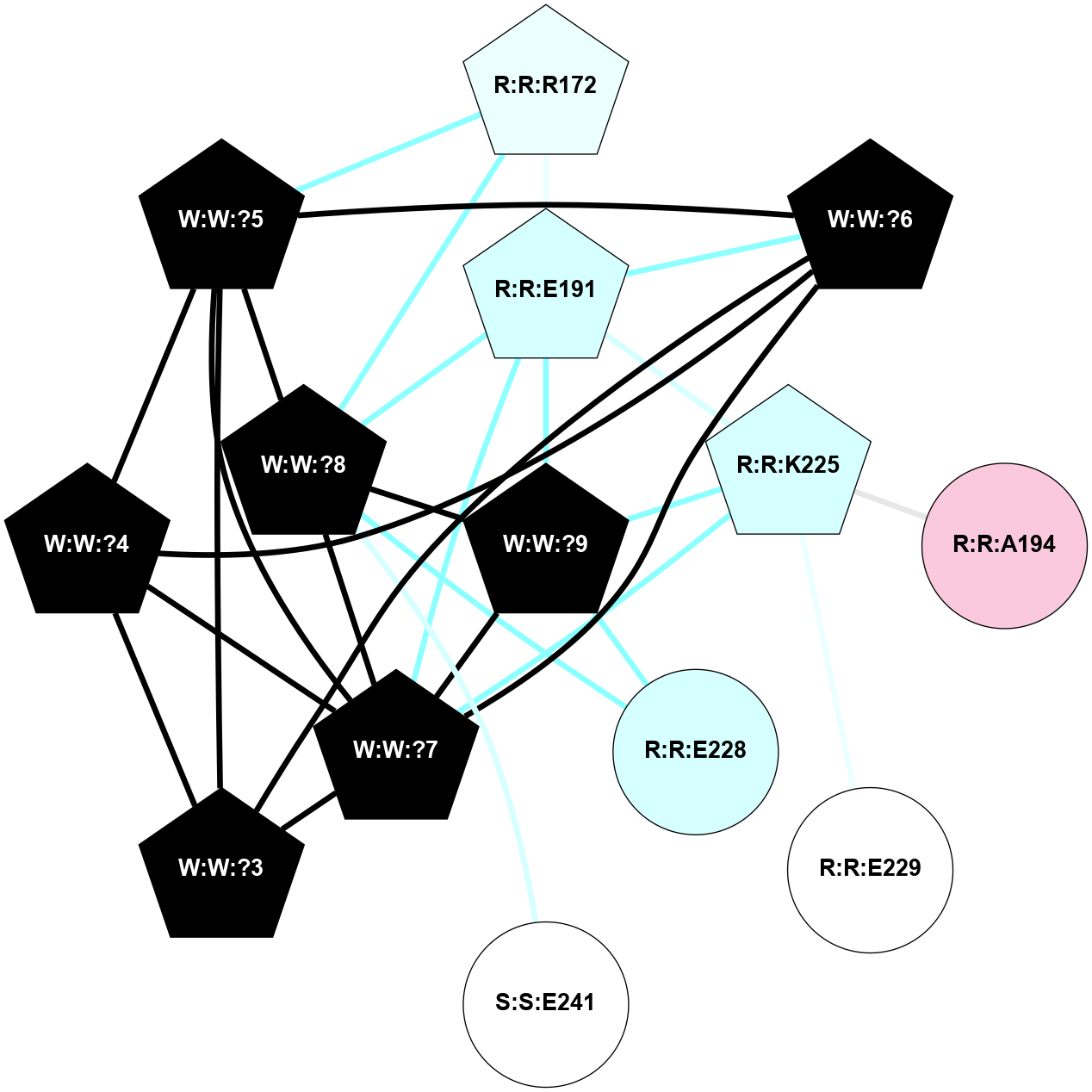
A 2D representation of the interactions of NH2 in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E191 | R:R:R172 | 9.3 | 2 | Yes | Yes | 3 | 4 | 1 | 2 | | R:R:R172 | W:W:?5 | 4.84 | 2 | Yes | Yes | 4 | 0 | 2 | 2 | | R:R:R172 | W:W:?8 | 5.81 | 2 | Yes | Yes | 4 | 0 | 2 | 1 | | R:R:E191 | R:R:K225 | 13.5 | 2 | Yes | Yes | 3 | 3 | 1 | 1 | | R:R:E191 | W:W:?6 | 6.34 | 2 | Yes | Yes | 3 | 0 | 1 | 2 | | R:R:E191 | W:W:?7 | 13.45 | 2 | Yes | Yes | 3 | 0 | 1 | 1 | | R:R:E191 | W:W:?8 | 10.57 | 2 | Yes | Yes | 3 | 0 | 1 | 1 | | R:R:E191 | W:W:?9 | 3.3 | 2 | Yes | Yes | 3 | 0 | 1 | 0 | | R:R:A194 | R:R:K225 | 3.21 | 0 | No | Yes | 7 | 3 | 2 | 1 | | R:R:E229 | R:R:K225 | 9.45 | 0 | No | Yes | 5 | 3 | 2 | 1 | | R:R:K225 | W:W:?7 | 10.02 | 2 | Yes | Yes | 3 | 0 | 1 | 1 | | R:R:K225 | W:W:?9 | 10.53 | 2 | Yes | Yes | 3 | 0 | 1 | 0 | | R:R:E228 | W:W:?8 | 21.13 | 2 | No | Yes | 3 | 0 | 1 | 1 | | R:R:E228 | W:W:?9 | 6.59 | 2 | No | Yes | 3 | 0 | 1 | 0 | | S:S:E241 | W:W:?8 | 8.45 | 0 | No | Yes | 5 | 0 | 2 | 1 | | W:W:?3 | W:W:?4 | 19.04 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | W:W:?3 | W:W:?5 | 3.36 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | W:W:?3 | W:W:?6 | 14.56 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | W:W:?3 | W:W:?7 | 2.85 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | W:W:?4 | W:W:?5 | 14.08 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | W:W:?4 | W:W:?6 | 6.16 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | W:W:?4 | W:W:?7 | 7.84 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | W:W:?5 | W:W:?6 | 14.96 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | W:W:?5 | W:W:?7 | 7.84 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | W:W:?5 | W:W:?8 | 6.16 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | W:W:?6 | W:W:?7 | 15.68 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | W:W:?7 | W:W:?8 | 23.52 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | W:W:?7 | W:W:?9 | 6.99 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | W:W:?8 | W:W:?9 | 13.73 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 8.23 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 29.00 | | Average Links Mediated by Hubs In Shell | 29.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
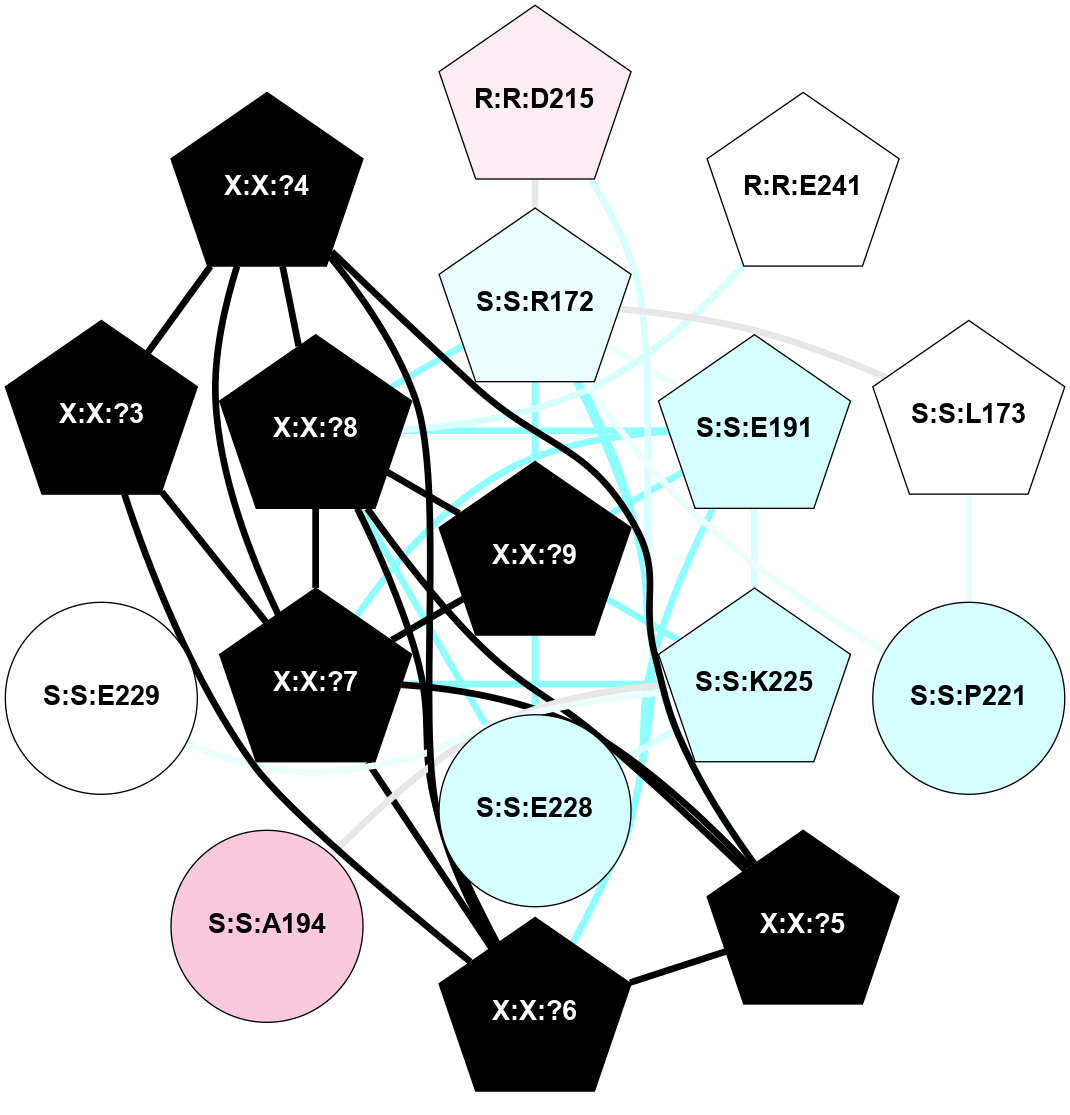
A 2D representation of the interactions of NH2 in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D215 | S:S:R172 | 9.53 | 1 | Yes | Yes | 6 | 4 | 2 | 1 | | R:R:D215 | X:X:?5 | 4.33 | 1 | Yes | Yes | 6 | 0 | 2 | 2 | | R:R:E241 | X:X:?8 | 7.4 | 1 | Yes | Yes | 5 | 0 | 2 | 1 | | S:S:L173 | S:S:R172 | 7.29 | 1 | Yes | Yes | 5 | 4 | 2 | 1 | | S:S:E191 | S:S:R172 | 8.14 | 1 | Yes | Yes | 3 | 4 | 1 | 1 | | S:S:P221 | S:S:R172 | 2.88 | 1 | No | Yes | 3 | 4 | 2 | 1 | | S:S:R172 | X:X:?5 | 4.84 | 1 | Yes | Yes | 4 | 0 | 1 | 2 | | S:S:R172 | X:X:?6 | 4.84 | 1 | Yes | Yes | 4 | 0 | 1 | 2 | | S:S:R172 | X:X:?8 | 7.75 | 1 | Yes | Yes | 4 | 0 | 1 | 1 | | S:S:R172 | X:X:?9 | 3.02 | 1 | Yes | Yes | 4 | 0 | 1 | 0 | | S:S:L173 | S:S:P221 | 3.28 | 1 | Yes | No | 5 | 3 | 2 | 2 | | S:S:E191 | S:S:K225 | 17.55 | 1 | Yes | Yes | 3 | 3 | 1 | 1 | | S:S:E191 | X:X:?6 | 8.45 | 1 | Yes | Yes | 3 | 0 | 1 | 2 | | S:S:E191 | X:X:?7 | 9.41 | 1 | Yes | Yes | 3 | 0 | 1 | 1 | | S:S:E191 | X:X:?8 | 3.17 | 1 | Yes | Yes | 3 | 0 | 1 | 1 | | S:S:E191 | X:X:?9 | 6.59 | 1 | Yes | Yes | 3 | 0 | 1 | 0 | | S:S:A194 | S:S:K225 | 3.21 | 0 | No | Yes | 7 | 3 | 2 | 1 | | S:S:E228 | S:S:K225 | 2.7 | 1 | No | Yes | 3 | 3 | 1 | 1 | | S:S:E229 | S:S:K225 | 9.45 | 0 | No | Yes | 5 | 3 | 2 | 1 | | S:S:K225 | X:X:?7 | 11.45 | 1 | Yes | Yes | 3 | 0 | 1 | 1 | | S:S:K225 | X:X:?9 | 10.53 | 1 | Yes | Yes | 3 | 0 | 1 | 0 | | S:S:E228 | X:X:?8 | 9.51 | 1 | No | Yes | 3 | 0 | 1 | 1 | | S:S:E228 | X:X:?9 | 3.3 | 1 | No | Yes | 3 | 0 | 1 | 0 | | X:X:?3 | X:X:?4 | 16.8 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | X:X:?3 | X:X:?6 | 16.8 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | X:X:?3 | X:X:?7 | 2.85 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | X:X:?4 | X:X:?5 | 15.84 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | X:X:?4 | X:X:?6 | 6.16 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | X:X:?4 | X:X:?7 | 5.6 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | X:X:?4 | X:X:?8 | 4.4 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | X:X:?5 | X:X:?6 | 15.84 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | X:X:?5 | X:X:?7 | 7.84 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | X:X:?5 | X:X:?8 | 8.8 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | X:X:?6 | X:X:?7 | 19.04 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | X:X:?6 | X:X:?8 | 4.4 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | X:X:?7 | X:X:?8 | 20.16 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | X:X:?7 | X:X:?9 | 6.99 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | X:X:?8 | X:X:?9 | 13.73 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 7.36 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 38.00 | | Average Links Mediated by Hubs In Shell | 38.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7P00 | A | Peptide | Tachykinin | NK1 | Homo Sapiens | Substance-P | - | chim(NtGi1-Gs-CtGq)/Beta1/Gamma2 | 2.71 | 2021-12-15 | doi.org/10.1126/sciadv.abk2872 |
|
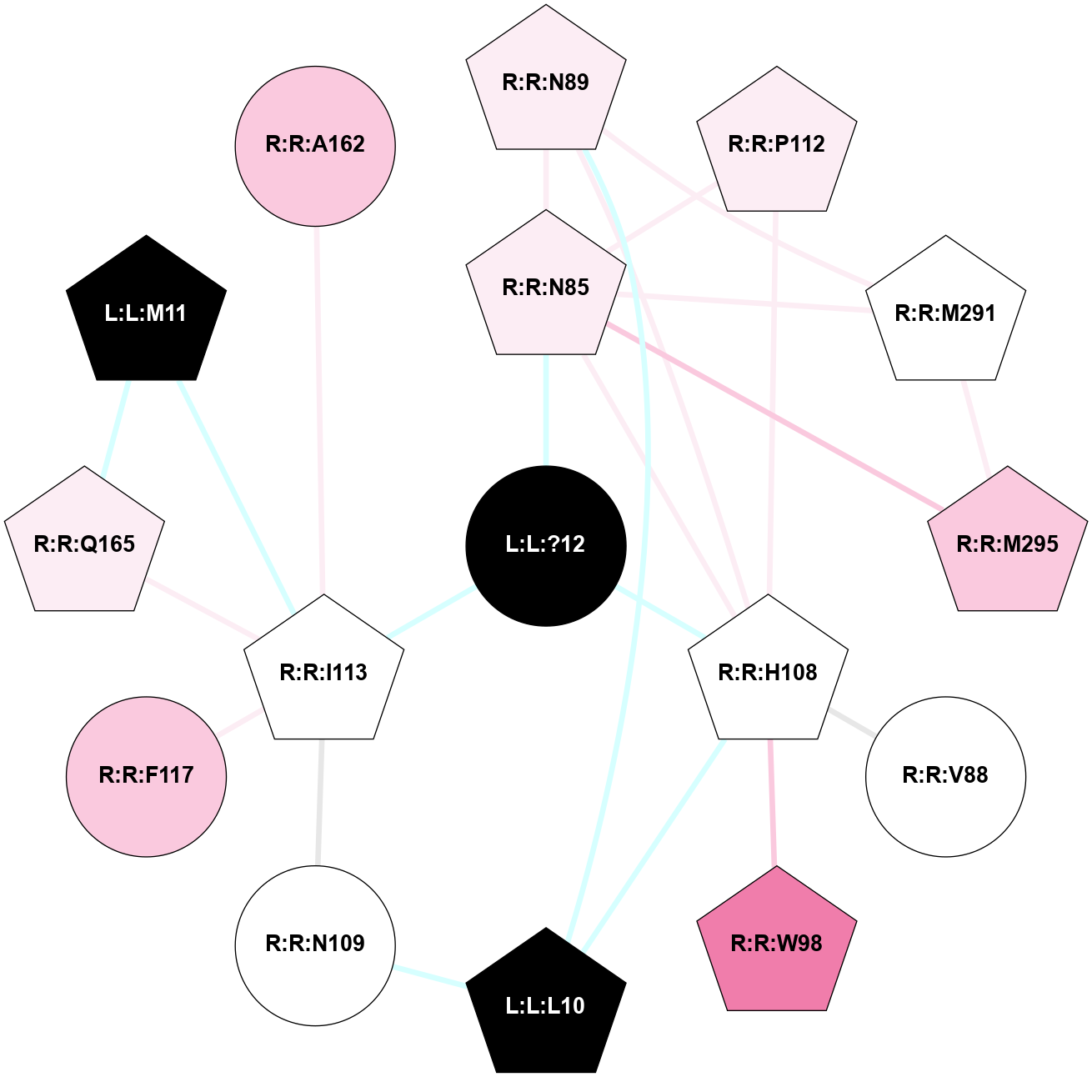
A 2D representation of the interactions of NH2 in 7P00
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N85 | R:R:N89 | 5.45 | 2 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:H108 | R:R:N85 | 8.93 | 2 | Yes | Yes | 5 | 6 | 1 | 1 | | R:R:N85 | R:R:P112 | 4.89 | 2 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:M291 | R:R:N85 | 4.21 | 2 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:M295 | R:R:N85 | 4.21 | 2 | Yes | Yes | 7 | 6 | 2 | 1 | | L:L:?12 | R:R:N85 | 10.25 | 2 | No | Yes | 0 | 6 | 0 | 1 | | R:R:H108 | R:R:V88 | 12.45 | 2 | Yes | No | 5 | 5 | 1 | 2 | | R:R:H108 | R:R:N89 | 12.75 | 2 | Yes | Yes | 5 | 6 | 1 | 2 | | R:R:M291 | R:R:N89 | 5.61 | 2 | Yes | Yes | 5 | 6 | 2 | 2 | | L:L:L10 | R:R:N89 | 6.87 | 2 | Yes | Yes | 0 | 6 | 2 | 2 | | R:R:H108 | R:R:W98 | 4.23 | 2 | Yes | Yes | 5 | 8 | 1 | 2 | | R:R:H108 | R:R:P112 | 10.68 | 2 | Yes | Yes | 5 | 6 | 1 | 2 | | L:L:L10 | R:R:H108 | 5.14 | 2 | Yes | Yes | 0 | 5 | 2 | 1 | | L:L:?12 | R:R:H108 | 6.4 | 2 | No | Yes | 0 | 5 | 0 | 1 | | R:R:I113 | R:R:N109 | 4.25 | 2 | Yes | No | 5 | 5 | 1 | 2 | | L:L:L10 | R:R:N109 | 8.24 | 2 | Yes | No | 0 | 5 | 2 | 2 | | R:R:F117 | R:R:I113 | 5.02 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:I113 | R:R:Q165 | 5.49 | 2 | Yes | Yes | 5 | 6 | 1 | 2 | | L:L:M11 | R:R:I113 | 4.37 | 2 | Yes | Yes | 0 | 5 | 2 | 1 | | L:L:?12 | R:R:I113 | 10.65 | 2 | No | Yes | 0 | 5 | 0 | 1 | | L:L:M11 | R:R:Q165 | 9.52 | 2 | Yes | Yes | 0 | 6 | 2 | 2 | | R:R:M291 | R:R:M295 | 5.78 | 2 | Yes | Yes | 5 | 7 | 2 | 2 | | R:R:A162 | R:R:I113 | 3.25 | 0 | No | Yes | 7 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 9.10 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 23.00 | | Average Links Mediated by Hubs In Shell | 23.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7P02 | A | Peptide | Tachykinin | NK1 | Homo Sapiens | Protachykinin-1 | - | chim(NtGi1-Gs)/Beta1/Gamma2 | 2.87 | 2021-12-15 | doi.org/10.1126/sciadv.abk2872 |
|
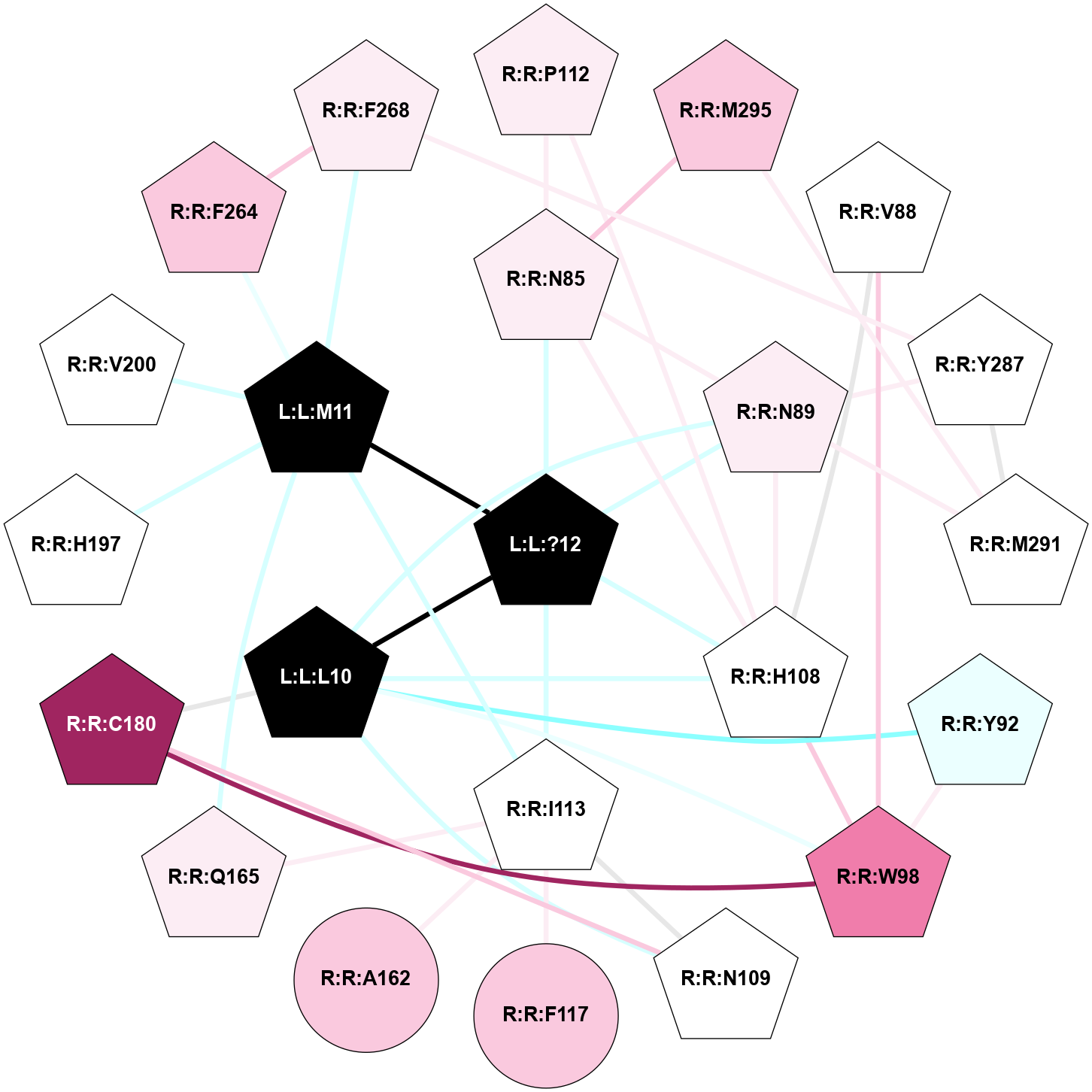
A 2D representation of the interactions of NH2 in 7P02
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N85 | R:R:N89 | 5.45 | 2 | Yes | Yes | 6 | 6 | 1 | 1 | | R:R:H108 | R:R:N85 | 7.65 | 2 | Yes | Yes | 5 | 6 | 1 | 1 | | R:R:N85 | R:R:P112 | 4.89 | 2 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:M295 | R:R:N85 | 7.01 | 0 | Yes | Yes | 7 | 6 | 2 | 1 | | L:L:?12 | R:R:N85 | 10.25 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:V88 | R:R:W98 | 3.68 | 2 | Yes | Yes | 5 | 8 | 2 | 2 | | R:R:H108 | R:R:V88 | 16.61 | 2 | Yes | Yes | 5 | 5 | 1 | 2 | | R:R:H108 | R:R:N89 | 12.75 | 2 | Yes | Yes | 5 | 6 | 1 | 1 | | R:R:N89 | R:R:Y287 | 8.14 | 2 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:M291 | R:R:N89 | 8.41 | 2 | Yes | Yes | 5 | 6 | 2 | 1 | | L:L:L10 | R:R:N89 | 6.87 | 2 | Yes | Yes | 0 | 6 | 1 | 1 | | L:L:?12 | R:R:N89 | 3.42 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:W98 | R:R:Y92 | 12.54 | 2 | Yes | Yes | 8 | 4 | 2 | 2 | | L:L:L10 | R:R:Y92 | 4.69 | 2 | Yes | Yes | 0 | 4 | 1 | 2 | | R:R:H108 | R:R:W98 | 4.23 | 2 | Yes | Yes | 5 | 8 | 1 | 2 | | R:R:C180 | R:R:W98 | 7.84 | 2 | Yes | Yes | 9 | 8 | 2 | 2 | | L:L:L10 | R:R:W98 | 3.42 | 2 | Yes | Yes | 0 | 8 | 1 | 2 | | R:R:H108 | R:R:P112 | 10.68 | 2 | Yes | Yes | 5 | 6 | 1 | 2 | | L:L:L10 | R:R:H108 | 3.86 | 2 | Yes | Yes | 0 | 5 | 1 | 1 | | L:L:?12 | R:R:H108 | 3.2 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:I113 | R:R:N109 | 4.25 | 2 | Yes | Yes | 5 | 5 | 1 | 2 | | R:R:C180 | R:R:N109 | 4.72 | 2 | Yes | Yes | 9 | 5 | 2 | 2 | | L:L:L10 | R:R:N109 | 9.61 | 2 | Yes | Yes | 0 | 5 | 1 | 2 | | R:R:F117 | R:R:I113 | 3.77 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:A162 | R:R:I113 | 3.25 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:I113 | R:R:Q165 | 8.23 | 2 | Yes | Yes | 5 | 6 | 1 | 2 | | L:L:M11 | R:R:I113 | 5.83 | 2 | Yes | Yes | 0 | 5 | 1 | 1 | | L:L:?12 | R:R:I113 | 10.65 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:M11 | R:R:Q165 | 8.16 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:L10 | R:R:C180 | 4.76 | 2 | Yes | Yes | 0 | 9 | 1 | 2 | | L:L:M11 | R:R:H197 | 3.94 | 2 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:M11 | R:R:V200 | 3.04 | 2 | Yes | Yes | 0 | 5 | 1 | 2 | | R:R:F264 | R:R:F268 | 12.86 | 2 | Yes | Yes | 7 | 6 | 2 | 2 | | L:L:M11 | R:R:F264 | 3.73 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | R:R:F268 | R:R:Y287 | 4.13 | 2 | Yes | Yes | 6 | 5 | 2 | 2 | | L:L:M11 | R:R:F268 | 9.95 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | R:R:M291 | R:R:Y287 | 10.78 | 2 | Yes | Yes | 5 | 5 | 2 | 2 | | R:R:M291 | R:R:M295 | 5.78 | 2 | Yes | Yes | 5 | 7 | 2 | 2 | | L:L:?12 | L:L:L10 | 3.44 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?12 | L:L:M11 | 3.52 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 5.75 | | Average Nodes In Shell | 23.00 | | Average Hubs In Shell | 21.00 | | Average Links In Shell | 40.00 | | Average Links Mediated by Hubs In Shell | 40.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7QVM | A | Peptide | Vasopressin and Oxytocin | OT | Homo Sapiens | Oxytocin | - | chim(NtGi1-Go-CtGq)/Beta1/Gamma2 | 3.25 | 2022-08-10 | doi.org/10.1038/s41467-022-31325-0 |
|
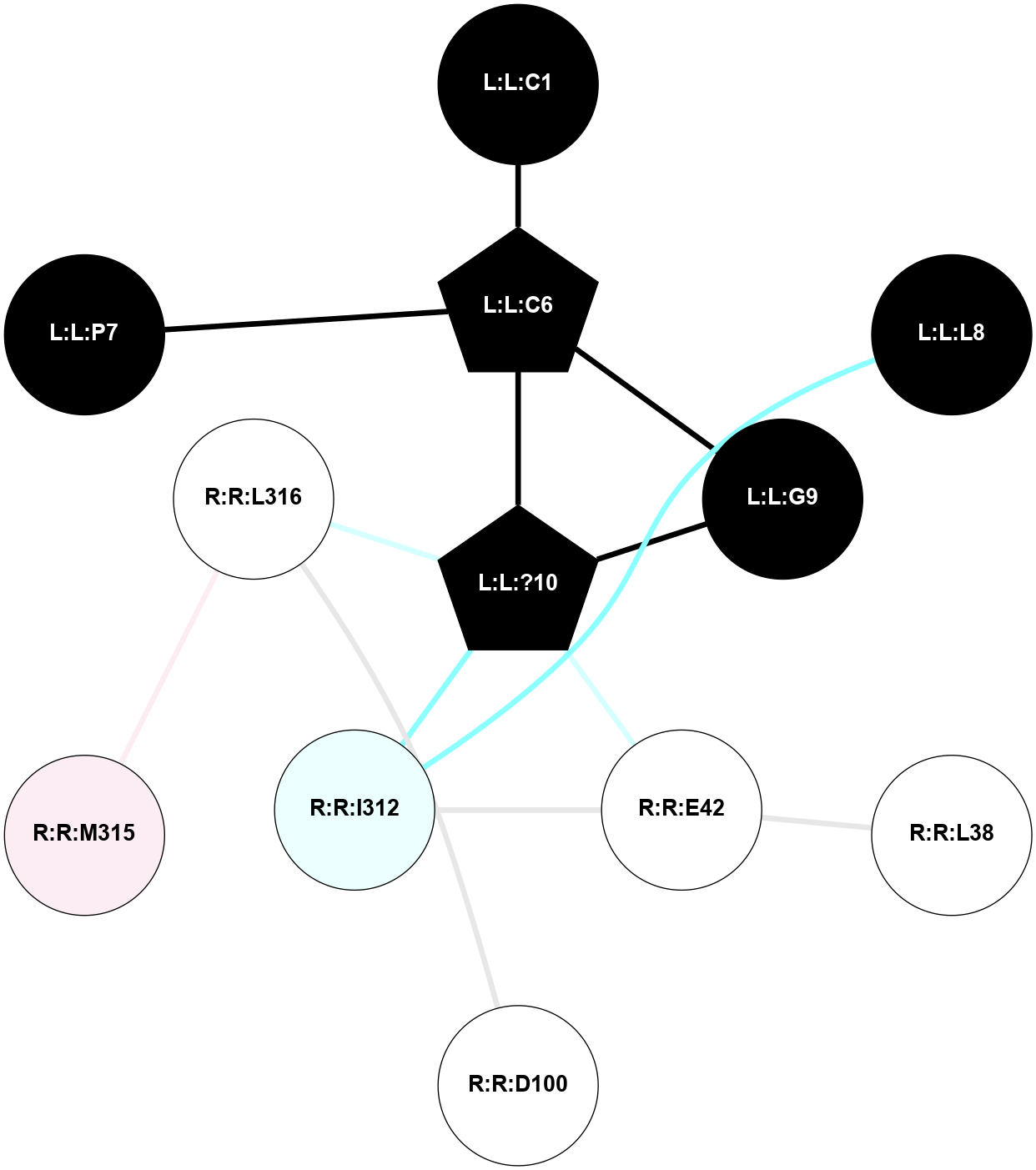
A 2D representation of the interactions of NH2 in 7QVM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:C1 | L:L:C6 | 7.28 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:C6 | L:L:G9 | 3.92 | 15 | Yes | No | 0 | 0 | 1 | 1 | | L:L:?10 | L:L:C6 | 3.95 | 15 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:L8 | R:R:I312 | 4.28 | 0 | No | No | 0 | 4 | 2 | 1 | | L:L:?10 | L:L:G9 | 4.26 | 15 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?10 | R:R:E42 | 9.89 | 15 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?10 | R:R:I312 | 3.55 | 15 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?10 | R:R:L316 | 3.44 | 15 | Yes | No | 0 | 5 | 0 | 1 | | R:R:E42 | R:R:L38 | 6.63 | 15 | No | No | 5 | 5 | 1 | 2 | | R:R:E42 | R:R:I312 | 4.1 | 15 | No | No | 5 | 4 | 1 | 1 | | R:R:D100 | R:R:L316 | 4.07 | 0 | No | No | 5 | 5 | 2 | 1 | | R:R:L316 | R:R:M315 | 2.83 | 0 | No | No | 5 | 6 | 1 | 2 | | L:L:C6 | L:L:P7 | 1.88 | 15 | Yes | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.02 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7R0C | A | Peptide | Vasopressin and Oxytocin | V2 | Homo Sapiens | AVP | - | Arrestin2 | 4.73 | 2022-09-14 | doi.org/10.1126/sciadv.abo7761 |
|
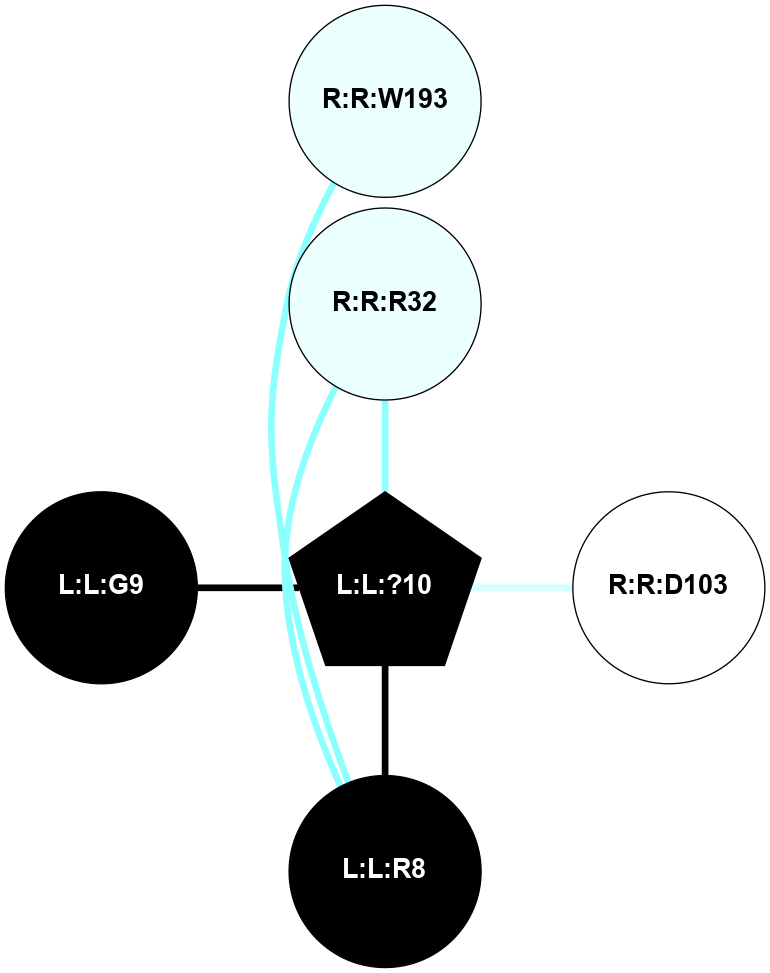
A 2D representation of the interactions of NH2 in 7R0C
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?10 | L:L:R8 | 3.02 | 16 | Yes | No | 0 | 0 | 0 | 1 | | L:L:R8 | R:R:R32 | 10.66 | 16 | No | No | 0 | 4 | 1 | 1 | | L:L:R8 | R:R:W193 | 12 | 16 | No | No | 0 | 4 | 1 | 2 | | L:L:?10 | L:L:G9 | 4.26 | 16 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?10 | R:R:R32 | 12.09 | 16 | Yes | No | 0 | 4 | 0 | 1 | | L:L:?10 | R:R:D103 | 3.38 | 16 | Yes | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 5.69 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7RMG | A | Peptide | Tachykinin | NK1 | Homo Sapiens | Substance-P | - | chim(Gs-CtGq)/Beta1/Gamma2 | 3 | 2021-11-03 | doi.org/10.1038/s41589-021-00890-8 |
|

A 2D representation of the interactions of NH2 in 7RMG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?12 | L:L:L10 | 3.44 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:L10 | R:R:Y92 | 4.69 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | L:L:L10 | R:R:N109 | 10.98 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:L10 | R:R:C180 | 3.17 | 1 | Yes | Yes | 0 | 9 | 1 | 2 | | L:L:?12 | L:L:M11 | 3.52 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:M11 | R:R:I113 | 5.83 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:M11 | R:R:Q165 | 8.16 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:M11 | R:R:F264 | 3.73 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:M11 | R:R:F268 | 12.44 | 1 | Yes | No | 0 | 6 | 1 | 2 | | L:L:?12 | R:R:N85 | 10.25 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?12 | R:R:N89 | 10.25 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?12 | R:R:H108 | 6.4 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:N85 | R:R:N89 | 6.81 | 1 | Yes | Yes | 6 | 6 | 1 | 1 | | R:R:H108 | R:R:N85 | 3.83 | 1 | Yes | Yes | 5 | 6 | 1 | 1 | | R:R:N85 | R:R:P112 | 6.52 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:H108 | R:R:V88 | 15.22 | 1 | Yes | No | 5 | 5 | 1 | 2 | | R:R:H108 | R:R:N89 | 8.93 | 1 | Yes | Yes | 5 | 6 | 1 | 1 | | R:R:N89 | R:R:Y287 | 4.65 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:M291 | R:R:N89 | 4.21 | 1 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:W98 | R:R:Y92 | 12.54 | 1 | Yes | Yes | 8 | 4 | 2 | 2 | | R:R:C180 | R:R:Y92 | 4.03 | 1 | Yes | Yes | 9 | 4 | 2 | 2 | | R:R:H108 | R:R:W98 | 3.17 | 1 | Yes | Yes | 5 | 8 | 1 | 2 | | R:R:C180 | R:R:W98 | 11.75 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:H108 | R:R:P112 | 6.1 | 1 | Yes | Yes | 5 | 6 | 1 | 2 | | R:R:C180 | R:R:N109 | 3.15 | 1 | Yes | Yes | 9 | 5 | 2 | 2 | | R:R:I113 | R:R:Q165 | 5.49 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:F264 | R:R:F268 | 13.93 | 1 | Yes | No | 7 | 6 | 2 | 2 | | R:R:M291 | R:R:Y287 | 4.79 | 1 | Yes | Yes | 5 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 6.77 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 16.00 | | Average Links In Shell | 28.00 | | Average Links Mediated by Hubs In Shell | 28.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7RMH | A | Peptide | Tachykinin | NK1 | Homo Sapiens | Substance-P | - | Gs/Beta1/Gamma2 | 3.1 | 2021-11-03 | doi.org/10.1038/s41589-021-00890-8 |
|
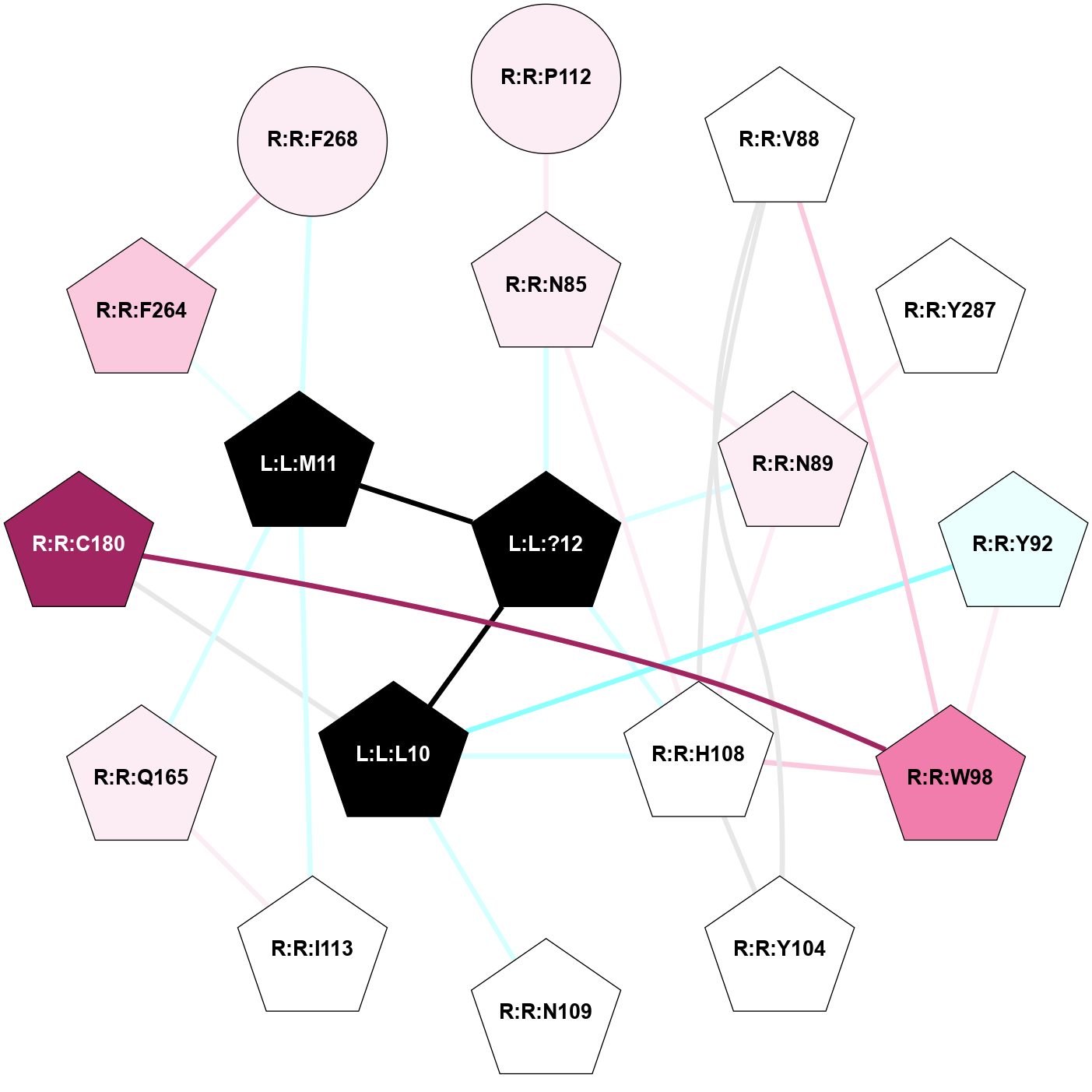
A 2D representation of the interactions of NH2 in 7RMH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?12 | L:L:L10 | 3.44 | 3 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:L10 | R:R:Y92 | 4.69 | 3 | Yes | Yes | 0 | 4 | 1 | 2 | | L:L:L10 | R:R:H108 | 2.57 | 3 | Yes | Yes | 0 | 5 | 1 | 1 | | L:L:L10 | R:R:N109 | 2.75 | 3 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:?12 | L:L:M11 | 3.52 | 3 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:M11 | R:R:I113 | 4.37 | 3 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:M11 | R:R:Q165 | 4.08 | 3 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:M11 | R:R:F264 | 2.49 | 3 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:M11 | R:R:F268 | 8.71 | 3 | Yes | No | 0 | 6 | 1 | 2 | | L:L:?12 | R:R:N85 | 10.25 | 3 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?12 | R:R:N89 | 10.25 | 3 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?12 | R:R:H108 | 6.4 | 3 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:N85 | R:R:N89 | 5.45 | 3 | Yes | Yes | 6 | 6 | 1 | 1 | | R:R:H108 | R:R:N85 | 2.55 | 3 | Yes | Yes | 5 | 6 | 1 | 1 | | R:R:N85 | R:R:P112 | 6.52 | 3 | Yes | No | 6 | 6 | 1 | 2 | | R:R:V88 | R:R:W98 | 2.45 | 3 | Yes | Yes | 5 | 8 | 2 | 2 | | R:R:V88 | R:R:Y104 | 5.05 | 3 | Yes | Yes | 5 | 5 | 2 | 2 | | R:R:H108 | R:R:V88 | 8.3 | 3 | Yes | Yes | 5 | 5 | 1 | 2 | | R:R:H108 | R:R:N89 | 10.2 | 3 | Yes | Yes | 5 | 6 | 1 | 1 | | R:R:N89 | R:R:Y287 | 4.65 | 3 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:W98 | R:R:Y92 | 8.68 | 3 | Yes | Yes | 8 | 4 | 2 | 2 | | R:R:H108 | R:R:W98 | 2.12 | 3 | Yes | Yes | 5 | 8 | 1 | 2 | | R:R:H108 | R:R:Y104 | 2.18 | 3 | Yes | Yes | 5 | 5 | 1 | 2 | | R:R:I113 | R:R:Q165 | 4.12 | 3 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:F264 | R:R:F268 | 12.86 | 3 | Yes | No | 7 | 6 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 6.77 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 15.00 | | Average Links In Shell | 25.00 | | Average Links Mediated by Hubs In Shell | 25.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7RMI | A | Peptide | Tachykinin | NK1 | Homo Sapiens | Substance-P | - | chim(Gs-CtGq)/Beta1/Gamma2 | 3.2 | 2021-11-03 | doi.org/10.1038/s41589-021-00890-8 |
|
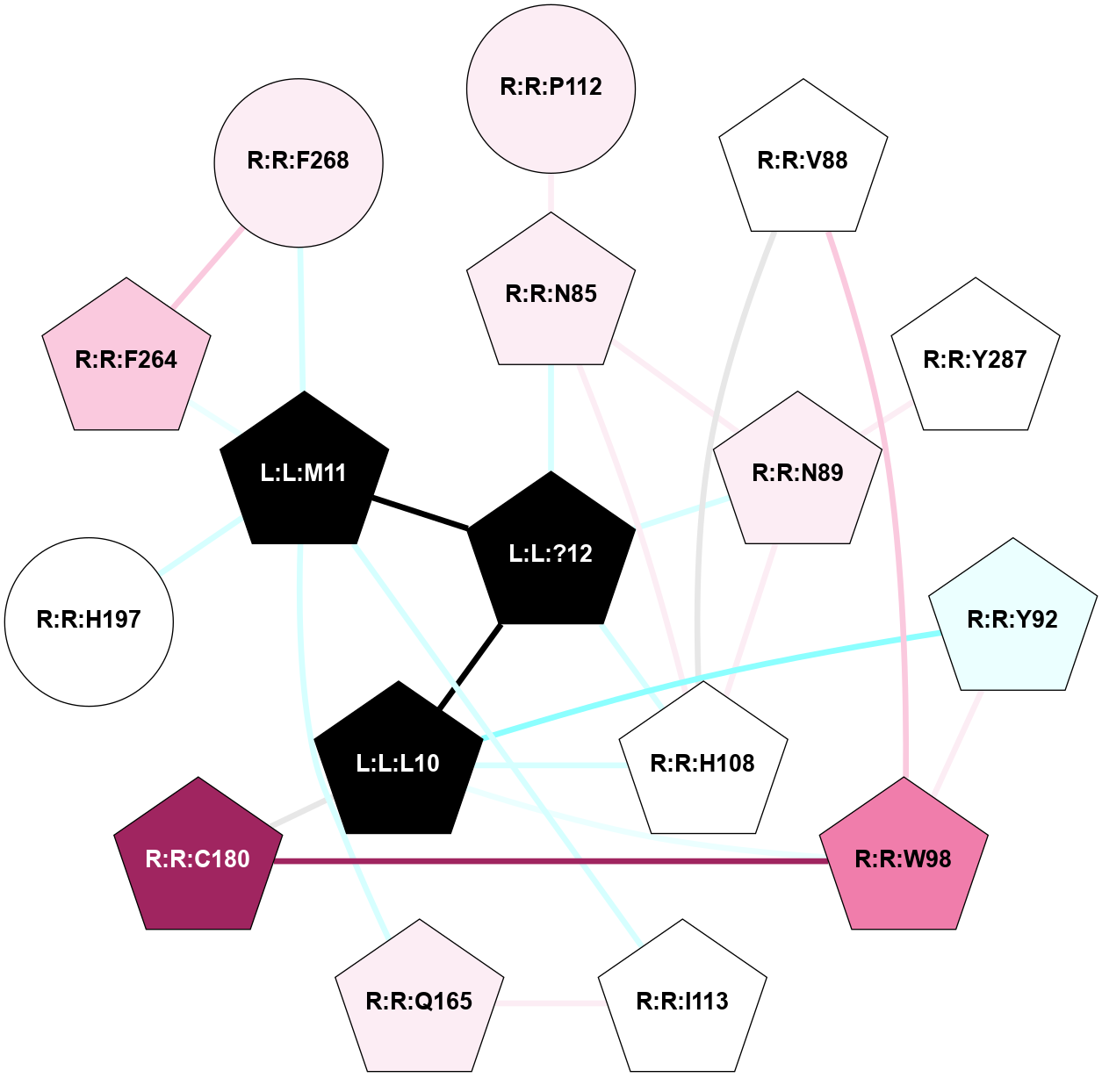
A 2D representation of the interactions of NH2 in 7RMI
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?12 | L:L:L10 | 3.44 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:L10 | R:R:Y92 | 4.69 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | L:L:L10 | R:R:W98 | 3.42 | 1 | Yes | Yes | 0 | 8 | 1 | 2 | | L:L:L10 | R:R:H108 | 5.14 | 1 | Yes | Yes | 0 | 5 | 1 | 1 | | L:L:L10 | R:R:C180 | 3.17 | 1 | Yes | Yes | 0 | 9 | 1 | 2 | | L:L:?12 | L:L:M11 | 3.52 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:M11 | R:R:I113 | 4.37 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:M11 | R:R:Q165 | 5.44 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:M11 | R:R:H197 | 2.63 | 1 | Yes | No | 0 | 5 | 1 | 2 | | L:L:M11 | R:R:F264 | 3.73 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:M11 | R:R:F268 | 11.2 | 1 | Yes | No | 0 | 6 | 1 | 2 | | L:L:?12 | R:R:N85 | 10.25 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?12 | R:R:N89 | 10.25 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?12 | R:R:H108 | 9.6 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:N85 | R:R:N89 | 9.54 | 1 | Yes | Yes | 6 | 6 | 1 | 1 | | R:R:H108 | R:R:N85 | 2.55 | 1 | Yes | Yes | 5 | 6 | 1 | 1 | | R:R:N85 | R:R:P112 | 4.89 | 1 | Yes | No | 6 | 6 | 1 | 2 | | R:R:V88 | R:R:W98 | 3.68 | 1 | Yes | Yes | 5 | 8 | 2 | 2 | | R:R:H108 | R:R:V88 | 9.69 | 1 | Yes | Yes | 5 | 5 | 1 | 2 | | R:R:H108 | R:R:N89 | 8.93 | 1 | Yes | Yes | 5 | 6 | 1 | 1 | | R:R:N89 | R:R:Y287 | 4.65 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:W98 | R:R:Y92 | 13.5 | 1 | Yes | Yes | 8 | 4 | 2 | 2 | | R:R:C180 | R:R:W98 | 14.37 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:I113 | R:R:Q165 | 8.23 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:F264 | R:R:F268 | 10.72 | 1 | Yes | No | 7 | 6 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 7.41 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 14.00 | | Average Links In Shell | 25.00 | | Average Links Mediated by Hubs In Shell | 25.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7RYC | A | Peptide | Vasopressin and Oxytocin | OT | Homo Sapiens | Oxytocin | - | chim(NtGi2L-Gs-CtGq)/Beta1/Gamma2 | 2.9 | 2022-03-09 | doi.org/10.1038/s41594-022-00728-4 |
|

A 2D representation of the interactions of NH2 in 7RYC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:G9 | L:L:L8 | 1.71 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:?10 | L:L:G9 | 4.26 | 22 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?10 | R:R:N35 | 3.42 | 22 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?10 | R:R:L38 | 10.33 | 22 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?10 | R:R:A39 | 3.92 | 22 | Yes | No | 0 | 5 | 0 | 1 | | R:R:A39 | R:R:R34 | 6.91 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:L38 | R:R:N35 | 9.61 | 22 | Yes | No | 5 | 5 | 1 | 1 | | R:R:A37 | R:R:L38 | 1.58 | 0 | No | Yes | 3 | 5 | 2 | 1 | | R:R:L38 | R:R:S309 | 1.5 | 22 | Yes | No | 5 | 2 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.48 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYF | B1 | Peptide | Calcitonin | CT (AMY1) | Homo Sapiens | Amylin | - | Gs/Beta1/Gamma2; RAMP1 | 2.2 | 2022-03-23 | doi.org/10.1126/science.abm9609 |
|

A 2D representation of the interactions of NH2 in 7TYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?38 | L:L:T36 | 7.33 | 3 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:T36 | R:R:W79 | 4.85 | 0 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?38 | R:R:W128 | 14.17 | 3 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?38 | R:R:S129 | 3.74 | 3 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?38 | R:R:Y131 | 5.83 | 3 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:L80 | 4.07 | 3 | Yes | Yes | 9 | 5 | 2 | 2 | | R:R:D77 | R:R:W82 | 8.93 | 3 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:D77 | R:R:S129 | 5.89 | 3 | Yes | Yes | 9 | 7 | 2 | 1 | | R:R:D77 | R:R:Y131 | 5.75 | 3 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:F102 | R:R:W79 | 4.01 | 3 | Yes | Yes | 4 | 6 | 2 | 2 | | R:R:W79 | R:R:Y131 | 23.15 | 3 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:L80 | R:R:V108 | 4.47 | 3 | Yes | Yes | 5 | 7 | 2 | 2 | | R:R:L80 | R:R:Y131 | 5.86 | 3 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:V108 | R:R:W82 | 3.68 | 3 | Yes | Yes | 7 | 9 | 2 | 2 | | R:R:K110 | R:R:W82 | 18.57 | 3 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:S129 | R:R:W82 | 4.94 | 3 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:F102 | R:R:Y131 | 3.09 | 3 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:V108 | R:R:Y131 | 12.62 | 3 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:K110 | R:R:S129 | 9.18 | 3 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:H121 | R:R:W128 | 6.35 | 3 | Yes | Yes | 4 | 5 | 2 | 1 | | H:H:?2 | R:R:H121 | 4.6 | 3 | Yes | Yes | 0 | 4 | 2 | 2 | | H:H:?2 | R:R:W128 | 10.19 | 3 | Yes | Yes | 0 | 5 | 2 | 1 | | R:R:S129 | R:R:Y131 | 6.36 | 3 | Yes | Yes | 7 | 5 | 1 | 1 | | L:L:G33 | R:R:W128 | 2.81 | 0 | No | Yes | 0 | 5 | 2 | 1 | | L:L:N31 | L:L:T36 | 1.46 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:N35 | L:L:T36 | 1.46 | 0 | No | Yes | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 7.77 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 14.00 | | Average Links In Shell | 26.00 | | Average Links Mediated by Hubs In Shell | 26.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYH | B1 | Peptide | Calcitonin | CT (AMY2) | Homo Sapiens | Calcitonin-1 | - | Gs/Beta1/Gamma2; RAMP2 | 3.3 | 2022-03-23 | doi.org/10.1126/science.abm9609 |
|
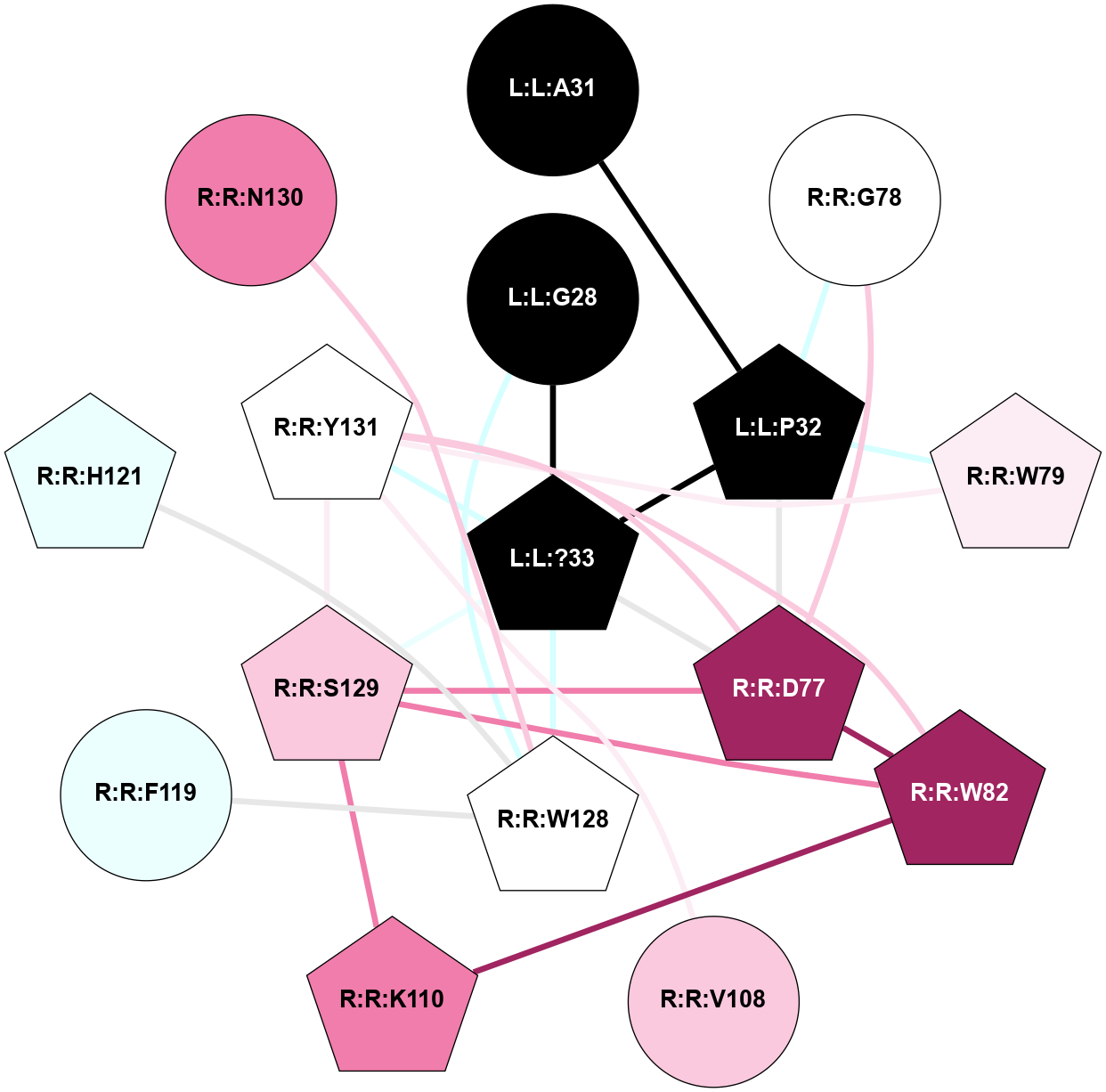
A 2D representation of the interactions of NH2 in 7TYH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?33 | L:L:G28 | 4.26 | 3 | Yes | No | 0 | 0 | 0 | 1 | | L:L:G28 | R:R:W128 | 4.22 | 0 | No | Yes | 0 | 5 | 1 | 2 | | L:L:A31 | L:L:P32 | 3.74 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?33 | L:L:P32 | 4.09 | 3 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:P32 | R:R:D77 | 4.83 | 3 | Yes | Yes | 0 | 9 | 1 | 1 | | L:L:P32 | R:R:G78 | 4.06 | 3 | Yes | No | 0 | 5 | 1 | 2 | | L:L:P32 | R:R:W79 | 5.4 | 3 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?33 | R:R:D77 | 3.38 | 3 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?33 | R:R:S129 | 7.47 | 3 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?33 | R:R:Y131 | 5.83 | 3 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:G78 | 3.35 | 3 | Yes | No | 9 | 5 | 1 | 2 | | R:R:D77 | R:R:W82 | 6.7 | 3 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D77 | R:R:K110 | 5.53 | 3 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:D77 | R:R:S129 | 2.94 | 3 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:D77 | R:R:Y131 | 8.05 | 3 | Yes | Yes | 9 | 5 | 1 | 1 | | R:R:W79 | R:R:Y131 | 19.29 | 0 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:K110 | R:R:W82 | 31.33 | 3 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:S129 | R:R:W82 | 3.71 | 3 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:W82 | R:R:Y131 | 2.89 | 3 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:K110 | R:R:S129 | 13.77 | 3 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:S129 | R:R:Y131 | 8.9 | 3 | Yes | Yes | 7 | 5 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 5.01 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 21.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
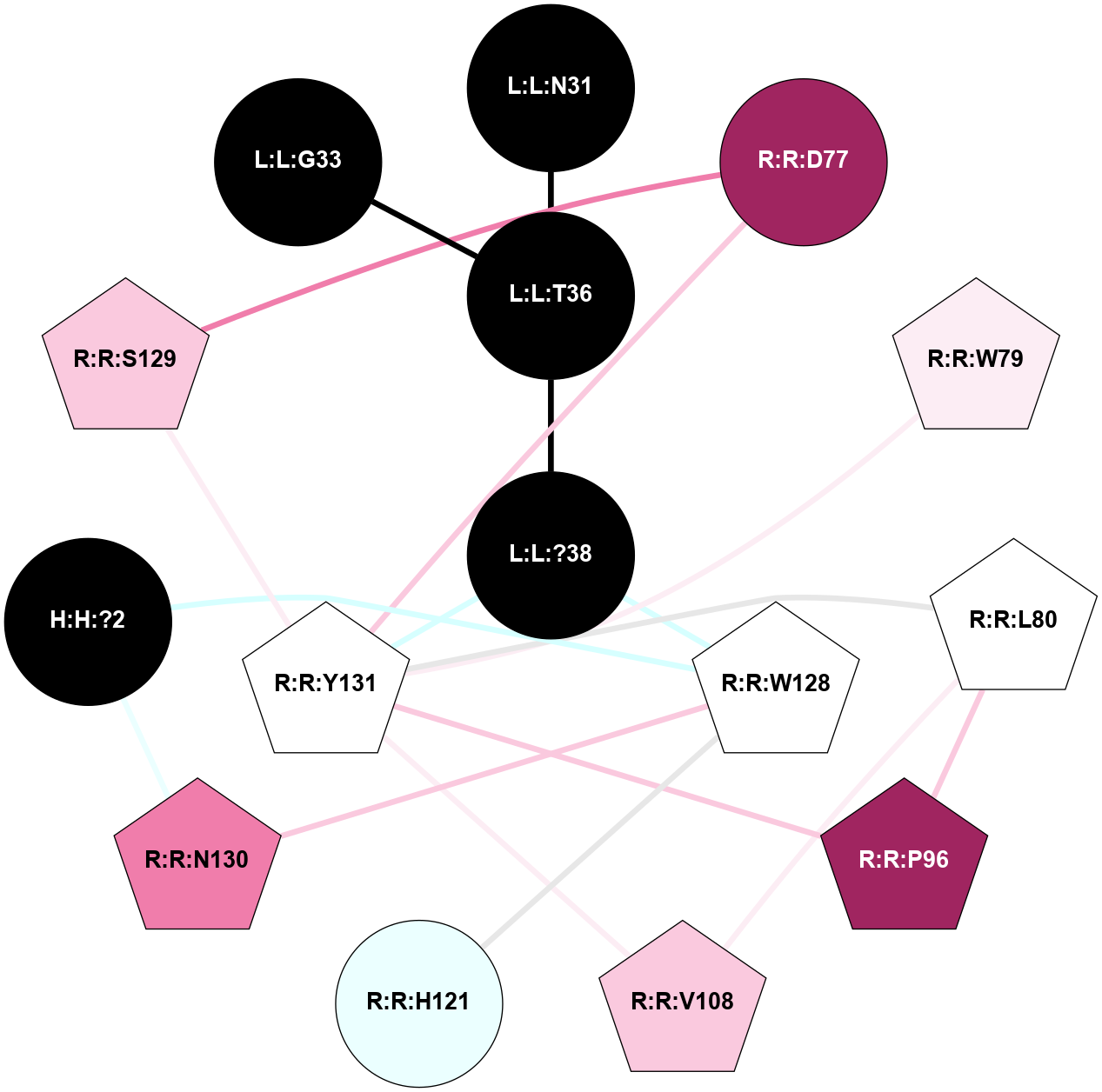
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYI | B1 | Peptide | Calcitonin | CT | Homo Sapiens | Amylin | - | Gs/Beta1/Gamma2 | 3.3 | 2022-03-30 | doi.org/10.1126/science.abm9609 |
|

A 2D representation of the interactions of NH2 in 7TYI
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:T36 | L:L:Y37 | 4.99 | 2 | No | Yes | 0 | 0 | 1 | 1 | | L:L:?38 | L:L:T36 | 7.33 | 2 | No | No | 0 | 0 | 0 | 1 | | L:L:?38 | L:L:Y37 | 8.75 | 2 | No | Yes | 0 | 0 | 0 | 1 | | L:L:Y37 | R:R:G78 | 8.69 | 2 | Yes | No | 0 | 5 | 1 | 2 | | L:L:Y37 | R:R:W79 | 12.54 | 2 | Yes | Yes | 0 | 6 | 1 | 1 | | L:L:?38 | R:R:W79 | 11.33 | 2 | No | Yes | 0 | 6 | 0 | 1 | | R:R:F99 | R:R:W79 | 9.02 | 2 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:F102 | R:R:W79 | 5.01 | 2 | Yes | Yes | 4 | 6 | 2 | 1 | | R:R:W79 | R:R:Y131 | 11.58 | 2 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F102 | R:R:F99 | 4.29 | 2 | Yes | Yes | 4 | 5 | 2 | 2 | | R:R:F102 | R:R:Y131 | 7.22 | 2 | Yes | Yes | 4 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.14 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYL | B1 | Peptide | Calcitonin | CT | Homo Sapiens | Amylin | - | Gs/Beta1/Gamma2 | 3.3 | 2022-03-23 | doi.org/10.1126/science.abm9609 |
|
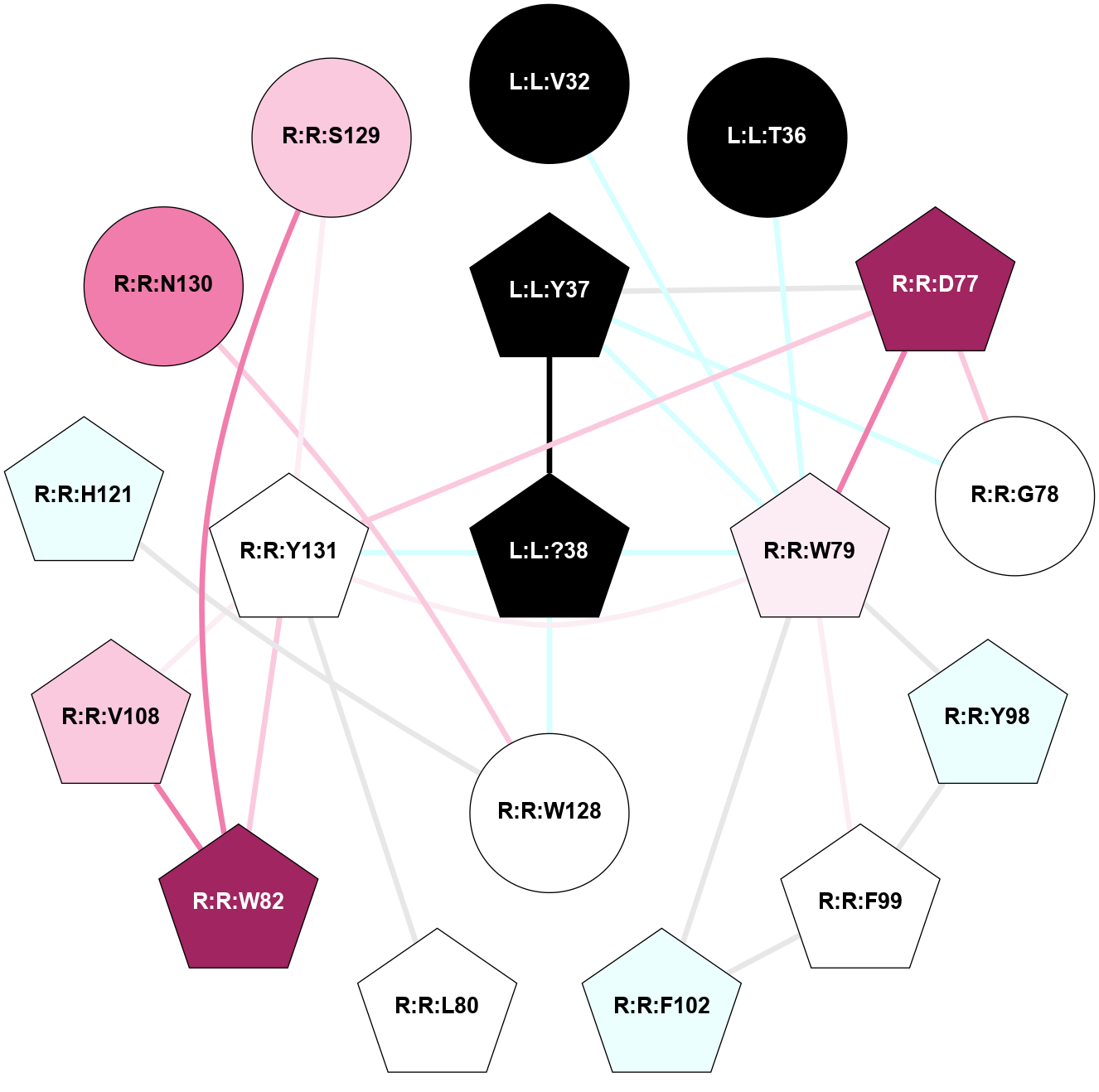
A 2D representation of the interactions of NH2 in 7TYL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:V32 | R:R:W79 | 4.9 | 0 | No | Yes | 0 | 6 | 2 | 1 | | L:L:T36 | R:R:W79 | 6.06 | 0 | No | Yes | 0 | 6 | 2 | 1 | | L:L:?38 | L:L:Y37 | 5.83 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:Y37 | R:R:D77 | 16.09 | 2 | Yes | Yes | 0 | 9 | 1 | 2 | | L:L:Y37 | R:R:G78 | 7.24 | 2 | Yes | No | 0 | 5 | 1 | 2 | | L:L:Y37 | R:R:W79 | 11.58 | 2 | Yes | Yes | 0 | 6 | 1 | 1 | | L:L:?38 | R:R:W79 | 2.83 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?38 | R:R:W128 | 8.5 | 2 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?38 | R:R:Y131 | 2.92 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:G78 | 6.71 | 2 | Yes | No | 9 | 5 | 2 | 2 | | R:R:D77 | R:R:W79 | 3.35 | 2 | Yes | Yes | 9 | 6 | 2 | 1 | | R:R:D77 | R:R:Y131 | 8.05 | 2 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:W79 | R:R:Y98 | 6.75 | 2 | Yes | Yes | 6 | 4 | 1 | 2 | | R:R:F99 | R:R:W79 | 6.01 | 2 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:F102 | R:R:W79 | 7.02 | 2 | Yes | Yes | 4 | 6 | 2 | 1 | | R:R:W79 | R:R:Y131 | 18.33 | 2 | Yes | Yes | 6 | 5 | 1 | 1 | | R:R:L80 | R:R:Y131 | 5.86 | 0 | No | Yes | 5 | 5 | 2 | 1 | | R:R:V108 | R:R:W82 | 6.13 | 2 | No | Yes | 7 | 9 | 2 | 2 | | R:R:S129 | R:R:W82 | 7.41 | 2 | No | Yes | 7 | 9 | 2 | 2 | | R:R:W82 | R:R:Y131 | 6.75 | 2 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:F99 | R:R:Y98 | 14.44 | 2 | Yes | Yes | 5 | 4 | 2 | 2 | | R:R:F102 | R:R:F99 | 7.5 | 2 | Yes | Yes | 4 | 5 | 2 | 2 | | R:R:V108 | R:R:Y131 | 12.62 | 2 | No | Yes | 7 | 5 | 2 | 1 | | R:R:H121 | R:R:W128 | 17.98 | 18 | Yes | No | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 7.91 | 0 | No | No | 8 | 5 | 2 | 1 | | R:R:S129 | R:R:Y131 | 6.36 | 2 | No | Yes | 7 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 5.02 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 26.00 | | Average Links Mediated by Hubs In Shell | 25.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYN | B1 | Peptide | Calcitonin | CT | Homo Sapiens | Calcitonin-1 | - | Gs/Beta1/Gamma2 | 2.6 | 2022-03-30 | doi.org/10.1126/science.abm9609 |
|
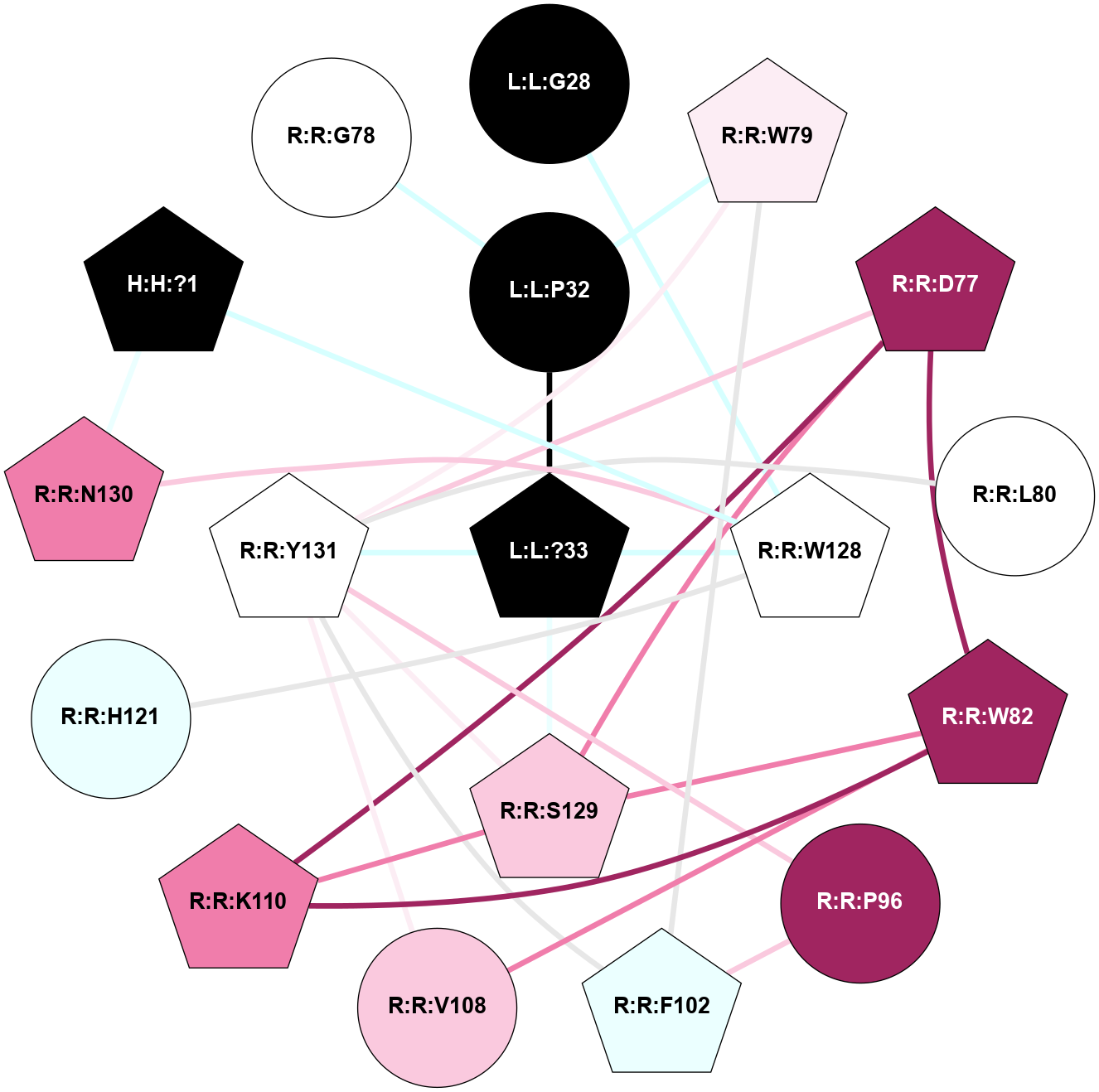
A 2D representation of the interactions of NH2 in 7TYN
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:W128 | 4.07 | 2 | Yes | Yes | 0 | 5 | 2 | 1 | | H:H:?1 | R:R:N130 | 24.56 | 2 | Yes | No | 0 | 8 | 2 | 2 | | L:L:G28 | R:R:W128 | 4.22 | 0 | No | Yes | 0 | 5 | 2 | 1 | | L:L:?33 | L:L:P32 | 12.26 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:P32 | R:R:D77 | 9.66 | 2 | No | Yes | 0 | 9 | 1 | 1 | | L:L:P32 | R:R:W79 | 21.62 | 2 | No | Yes | 0 | 6 | 1 | 2 | | L:L:?33 | R:R:D77 | 10.13 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?33 | R:R:W128 | 5.67 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?33 | R:R:S129 | 7.47 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?33 | R:R:Y131 | 5.83 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:S129 | 7.36 | 2 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:D77 | R:R:Y131 | 5.75 | 2 | Yes | Yes | 9 | 5 | 1 | 1 | | R:R:F102 | R:R:W79 | 5.01 | 2 | Yes | Yes | 4 | 6 | 2 | 2 | | R:R:W79 | R:R:Y131 | 15.43 | 2 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:L80 | R:R:P96 | 14.78 | 2 | No | Yes | 5 | 9 | 2 | 2 | | R:R:L80 | R:R:Y131 | 7.03 | 2 | No | Yes | 5 | 5 | 2 | 1 | | R:R:V108 | R:R:W82 | 3.68 | 0 | No | Yes | 7 | 9 | 2 | 2 | | R:R:S129 | R:R:W82 | 6.18 | 2 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:F102 | R:R:P96 | 5.78 | 2 | Yes | Yes | 4 | 9 | 2 | 2 | | R:R:P96 | R:R:Y131 | 4.17 | 2 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:F102 | R:R:Y131 | 6.19 | 2 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:V108 | R:R:Y131 | 12.62 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:H121 | R:R:W128 | 11.64 | 2 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 5.65 | 2 | No | Yes | 8 | 5 | 2 | 1 | | R:R:S129 | R:R:Y131 | 7.63 | 2 | Yes | Yes | 7 | 5 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 8.27 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 25.00 | | Average Links Mediated by Hubs In Shell | 25.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYO | B1 | Peptide | Calcitonin | CT | Homo Sapiens | Calcitonin | - | Gs/Beta1/Gamma2 | 2.7 | 2022-03-23 | doi.org/10.1126/science.abm9609 |
|
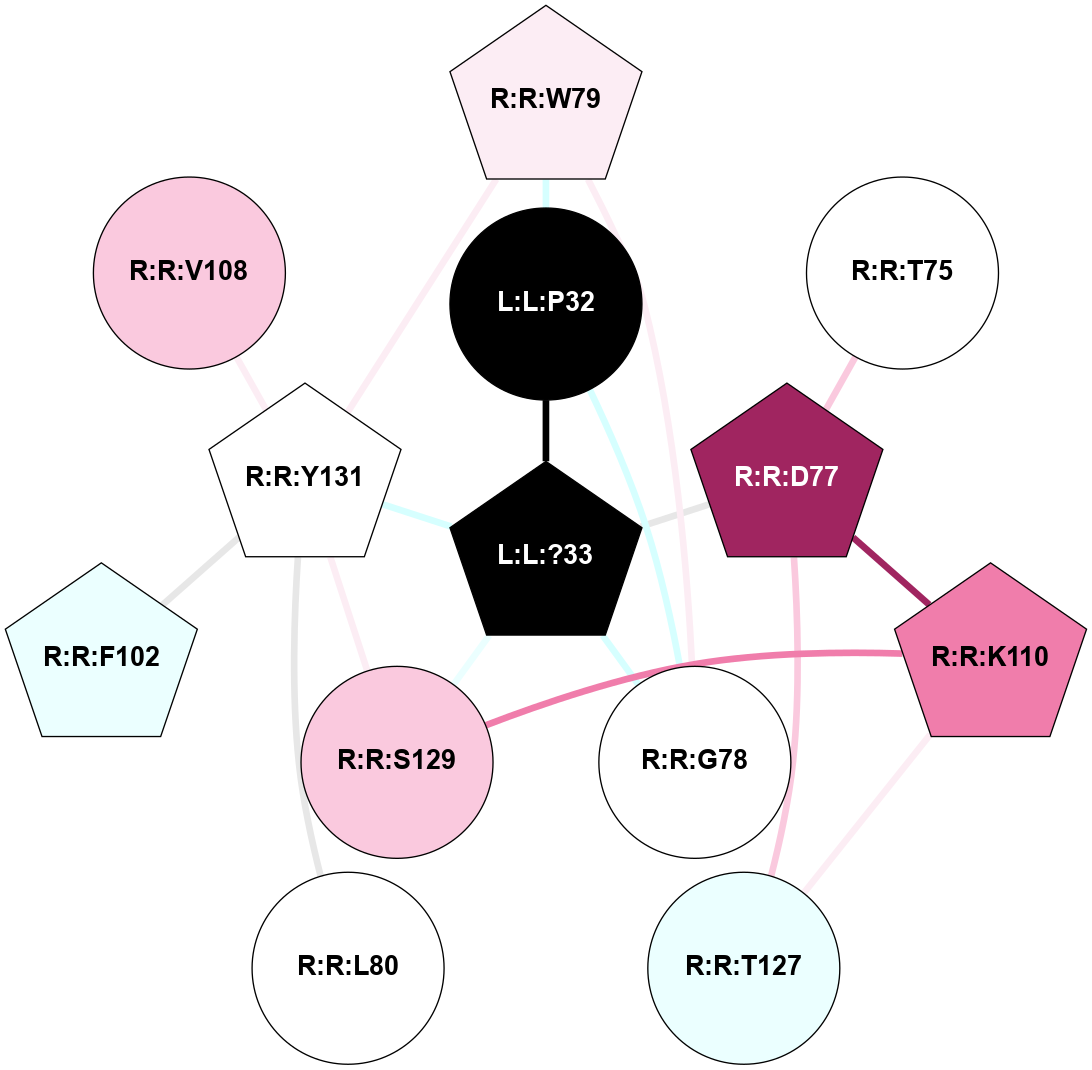
A 2D representation of the interactions of NH2 in 7TYO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?33 | L:L:P32 | 12.26 | 1 | Yes | No | 0 | 0 | 0 | 1 | | L:L:P32 | R:R:G78 | 4.06 | 1 | No | No | 0 | 5 | 1 | 1 | | L:L:P32 | R:R:W79 | 4.05 | 1 | No | Yes | 0 | 6 | 1 | 2 | | L:L:?33 | R:R:D77 | 3.38 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?33 | R:R:G78 | 4.26 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?33 | R:R:S129 | 3.74 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?33 | R:R:Y131 | 11.67 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:T75 | 5.78 | 1 | Yes | No | 9 | 5 | 1 | 2 | | R:R:D77 | R:R:K110 | 4.15 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:D77 | R:R:T127 | 8.67 | 1 | Yes | No | 9 | 4 | 1 | 2 | | R:R:G78 | R:R:W79 | 5.63 | 1 | No | Yes | 5 | 6 | 1 | 2 | | R:R:W79 | R:R:Y131 | 2.89 | 1 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:L80 | R:R:Y131 | 5.86 | 0 | No | Yes | 5 | 5 | 2 | 1 | | R:R:F102 | R:R:Y131 | 7.22 | 1 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:V108 | R:R:Y131 | 5.05 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:K110 | R:R:T127 | 3 | 1 | Yes | No | 8 | 4 | 2 | 2 | | R:R:K110 | R:R:S129 | 10.71 | 1 | Yes | No | 8 | 7 | 2 | 1 | | R:R:S129 | R:R:Y131 | 6.36 | 1 | No | Yes | 7 | 5 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 7.06 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYW | B1 | Peptide | Calcitonin | CT (AMY1) | Homo Sapiens | Calcitonin-1 | - | Gs/Beta1/Gamma2; RAMP1 | 3 | 2022-03-23 | doi.org/10.1126/science.abm9609 |
|
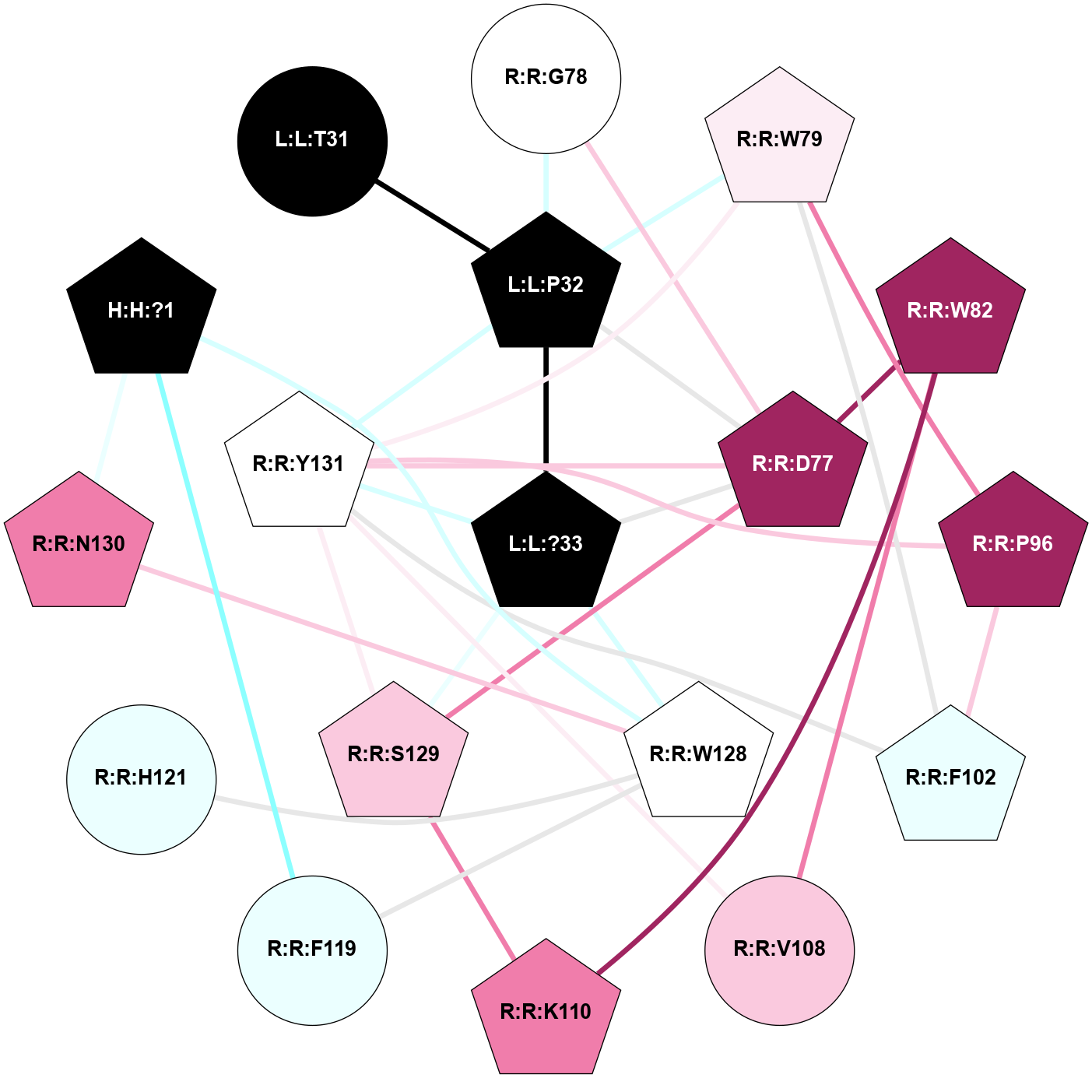
A 2D representation of the interactions of NH2 in 7TYW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:H121 | 4.6 | 2 | Yes | Yes | 0 | 4 | 2 | 2 | | H:H:?1 | R:R:W128 | 7.13 | 2 | Yes | Yes | 0 | 5 | 2 | 1 | | H:H:?1 | R:R:N130 | 24.56 | 2 | Yes | Yes | 0 | 8 | 2 | 2 | | L:L:?33 | L:L:P32 | 12.26 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:P32 | R:R:D77 | 8.05 | 2 | Yes | Yes | 0 | 9 | 1 | 1 | | L:L:P32 | R:R:G78 | 4.06 | 2 | Yes | No | 0 | 5 | 1 | 2 | | L:L:P32 | R:R:W79 | 20.27 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:P32 | R:R:Y131 | 5.56 | 2 | Yes | Yes | 0 | 5 | 1 | 1 | | L:L:?33 | R:R:D77 | 6.75 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?33 | R:R:W128 | 8.5 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?33 | R:R:S129 | 7.47 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?33 | R:R:Y131 | 8.75 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:G78 | 5.03 | 2 | Yes | No | 9 | 5 | 1 | 2 | | R:R:D77 | R:R:W82 | 8.93 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D77 | R:R:S129 | 8.83 | 2 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:D77 | R:R:Y131 | 6.9 | 2 | Yes | Yes | 9 | 5 | 1 | 1 | | R:R:P96 | R:R:W79 | 4.05 | 2 | Yes | Yes | 9 | 6 | 2 | 2 | | R:R:F102 | R:R:W79 | 5.01 | 2 | Yes | Yes | 4 | 6 | 2 | 2 | | R:R:W79 | R:R:Y131 | 19.29 | 2 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:V108 | R:R:W82 | 3.68 | 0 | No | Yes | 7 | 9 | 2 | 2 | | R:R:K110 | R:R:W82 | 20.89 | 2 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:F102 | R:R:P96 | 5.78 | 2 | Yes | Yes | 4 | 9 | 2 | 2 | | R:R:P96 | R:R:Y131 | 4.17 | 2 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:F102 | R:R:Y131 | 3.09 | 2 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:V108 | R:R:Y131 | 6.31 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:K110 | R:R:S129 | 6.12 | 2 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:H121 | R:R:W128 | 14.81 | 2 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 4.52 | 2 | Yes | Yes | 8 | 5 | 2 | 1 | | R:R:S129 | R:R:Y131 | 7.63 | 2 | Yes | Yes | 7 | 5 | 1 | 1 | | L:L:P32 | L:L:T31 | 1.75 | 2 | Yes | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 8.75 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 14.00 | | Average Links In Shell | 30.00 | | Average Links Mediated by Hubs In Shell | 30.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYX | B1 | Peptide | Calcitonin | CT (AMY2) | Homo Sapiens | Amylin | - | Gs/Beta1/Gamma2; RAMP2 | 2.55 | 2022-03-30 | doi.org/10.1126/science.abm9609 |
|
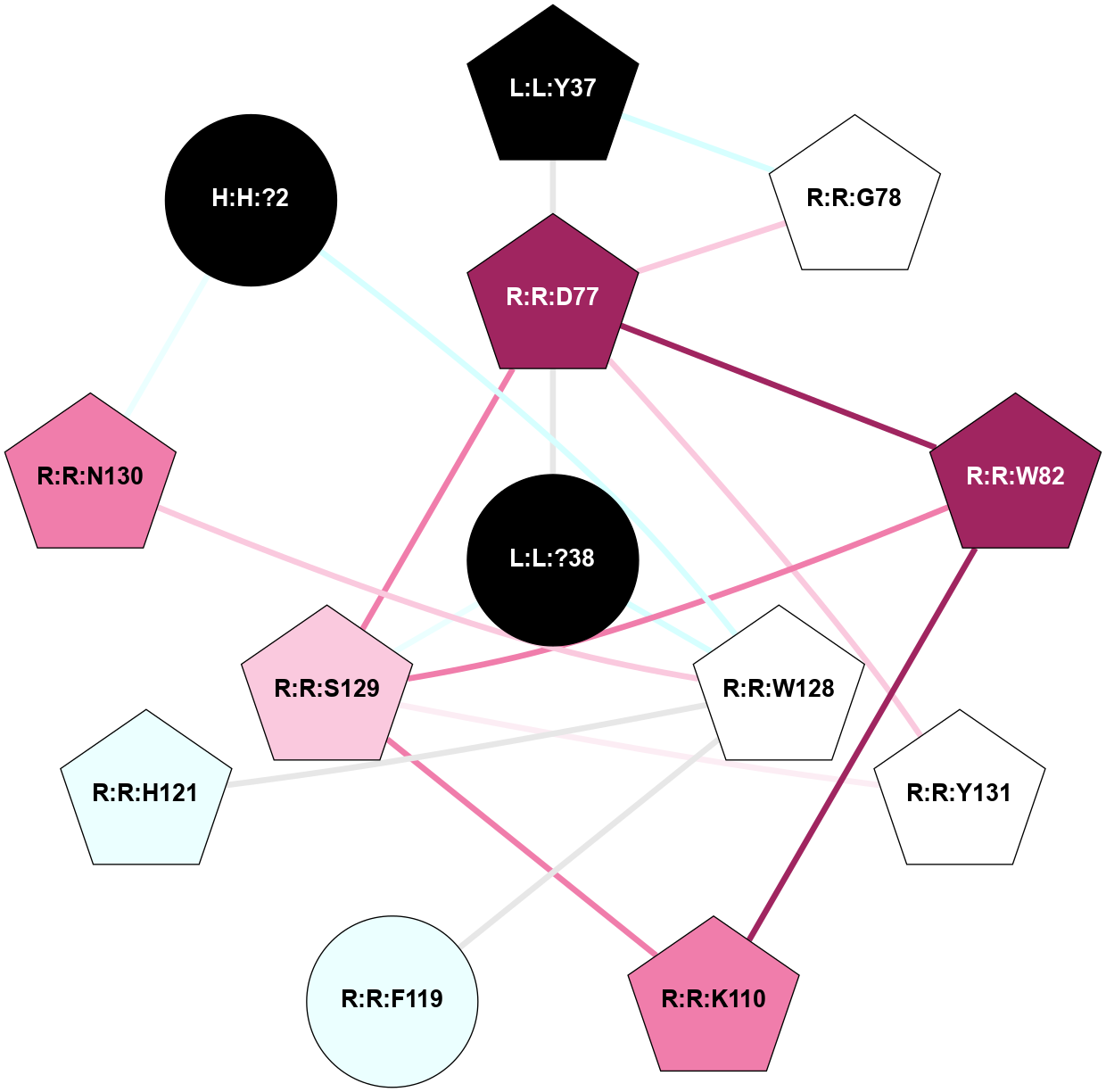
A 2D representation of the interactions of NH2 in 7TYX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:W128 | 6.11 | 1 | No | Yes | 0 | 5 | 2 | 1 | | H:H:?2 | R:R:N130 | 25.79 | 1 | No | Yes | 0 | 8 | 2 | 2 | | L:L:Y37 | R:R:D77 | 4.6 | 1 | Yes | Yes | 0 | 9 | 2 | 1 | | L:L:Y37 | R:R:G78 | 10.14 | 1 | Yes | Yes | 0 | 5 | 2 | 2 | | L:L:?38 | R:R:D77 | 3.38 | 1 | No | Yes | 0 | 9 | 0 | 1 | | L:L:?38 | R:R:W128 | 11.33 | 1 | No | Yes | 0 | 5 | 0 | 1 | | L:L:?38 | R:R:S129 | 3.74 | 1 | No | Yes | 0 | 7 | 0 | 1 | | R:R:D77 | R:R:G78 | 3.35 | 1 | Yes | Yes | 9 | 5 | 1 | 2 | | R:R:D77 | R:R:W82 | 4.47 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D77 | R:R:S129 | 8.83 | 1 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:D77 | R:R:Y131 | 10.34 | 1 | Yes | Yes | 9 | 5 | 1 | 2 | | R:R:K110 | R:R:W82 | 16.24 | 1 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:S129 | R:R:W82 | 3.71 | 1 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:K110 | R:R:S129 | 12.24 | 1 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:F119 | R:R:W128 | 4.01 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:H121 | R:R:W128 | 19.04 | 1 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 11.3 | 1 | Yes | Yes | 8 | 5 | 2 | 1 | | R:R:S129 | R:R:Y131 | 5.09 | 1 | Yes | Yes | 7 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 6.15 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 18.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYY | B1 | Peptide | Calcitonin | CT (AMY2) | Homo Sapiens | Calcitonin-1 | - | Gs/Beta1/Gamma2; RAMP2 | 3 | 2022-03-23 | doi.org/10.1126/science.abm9609 |
|
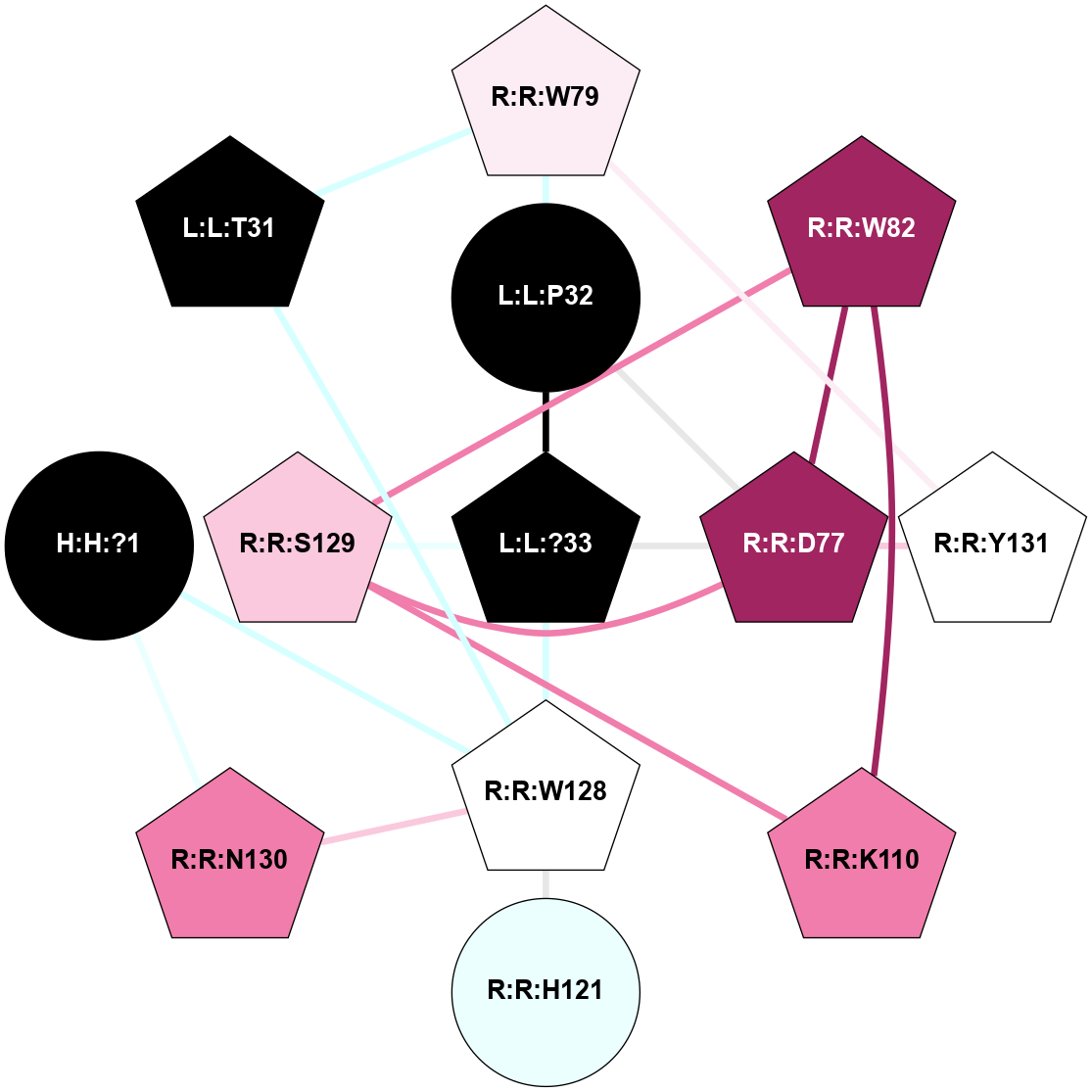
A 2D representation of the interactions of NH2 in 7TYY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:H121 | 4.6 | 1 | No | Yes | 0 | 4 | 2 | 2 | | H:H:?1 | R:R:W128 | 6.11 | 1 | No | Yes | 0 | 5 | 2 | 1 | | H:H:?1 | R:R:N130 | 24.56 | 1 | No | Yes | 0 | 8 | 2 | 2 | | L:L:?33 | L:L:P32 | 4.09 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:P32 | R:R:D77 | 4.83 | 1 | Yes | Yes | 0 | 9 | 1 | 1 | | L:L:P32 | R:R:W79 | 22.97 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?33 | R:R:D77 | 10.13 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?33 | R:R:W128 | 8.5 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?33 | R:R:S129 | 7.47 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:D77 | R:R:W82 | 6.7 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D77 | R:R:S129 | 7.36 | 1 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:D77 | R:R:Y131 | 8.05 | 1 | Yes | Yes | 9 | 5 | 1 | 2 | | R:R:W79 | R:R:Y131 | 13.5 | 1 | Yes | Yes | 6 | 5 | 2 | 2 | | R:R:K110 | R:R:W82 | 20.89 | 1 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:S129 | R:R:W82 | 7.41 | 1 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:K110 | R:R:S129 | 13.77 | 1 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:H121 | R:R:W128 | 20.1 | 1 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 11.3 | 1 | Yes | Yes | 8 | 5 | 2 | 1 | | E:E:P112 | L:L:P32 | 3.9 | 1 | No | Yes | 9 | 0 | 2 | 1 | | L:L:T31 | R:R:W79 | 2.43 | 0 | Yes | Yes | 0 | 6 | 2 | 2 | | L:L:T31 | R:R:W128 | 2.43 | 0 | Yes | Yes | 0 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 7.55 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 21.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TZF | B1 | Peptide | Calcitonin | CT (AMY3) | Homo Sapiens | Amylin | - | Gs/Beta1/Gamma2; RAMP3 | 2.4 | 2022-03-23 | doi.org/10.1126/science.abm9609 |
|

A 2D representation of the interactions of NH2 in 7TZF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:N31 | L:L:T36 | 5.85 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:?38 | L:L:T36 | 7.33 | 0 | No | No | 0 | 0 | 0 | 1 | | L:L:?38 | R:R:W128 | 11.33 | 0 | No | Yes | 0 | 5 | 0 | 1 | | L:L:?38 | R:R:Y131 | 5.83 | 0 | No | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:S129 | 5.89 | 1 | No | Yes | 9 | 7 | 2 | 2 | | R:R:D77 | R:R:Y131 | 8.05 | 1 | No | Yes | 9 | 5 | 2 | 1 | | R:R:W79 | R:R:Y131 | 12.54 | 1 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:L80 | R:R:P96 | 8.21 | 1 | Yes | Yes | 5 | 9 | 2 | 2 | | R:R:L80 | R:R:V108 | 4.47 | 1 | Yes | Yes | 5 | 7 | 2 | 2 | | R:R:L80 | R:R:Y131 | 8.21 | 1 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:P96 | R:R:Y131 | 4.17 | 1 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:V108 | R:R:Y131 | 13.88 | 1 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:F119 | R:R:W128 | 4.01 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:H121 | R:R:W128 | 17.98 | 7 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 12.43 | 7 | Yes | Yes | 8 | 5 | 2 | 1 | | H:H:?2 | R:R:W128 | 7.13 | 7 | Yes | Yes | 0 | 5 | 2 | 1 | | R:R:S129 | R:R:Y131 | 10.17 | 1 | Yes | Yes | 7 | 5 | 2 | 1 | | H:H:?2 | R:R:N130 | 22.1 | 7 | Yes | Yes | 0 | 8 | 2 | 2 | | R:R:C134 | R:R:Y131 | 4.03 | 0 | No | Yes | 9 | 5 | 2 | 1 | | L:L:G33 | L:L:T36 | 1.82 | 0 | No | No | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 8.16 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 20.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7W3Z | A | Peptide | Bombesin | BB2 | Homo Sapiens | Gastrin Releasing Peptide | - | chim(NtGi1L-Gq)/Beta1/Gamma2 | 3 | 2023-02-22 | doi.org/10.1073/pnas.2216230120 |
|
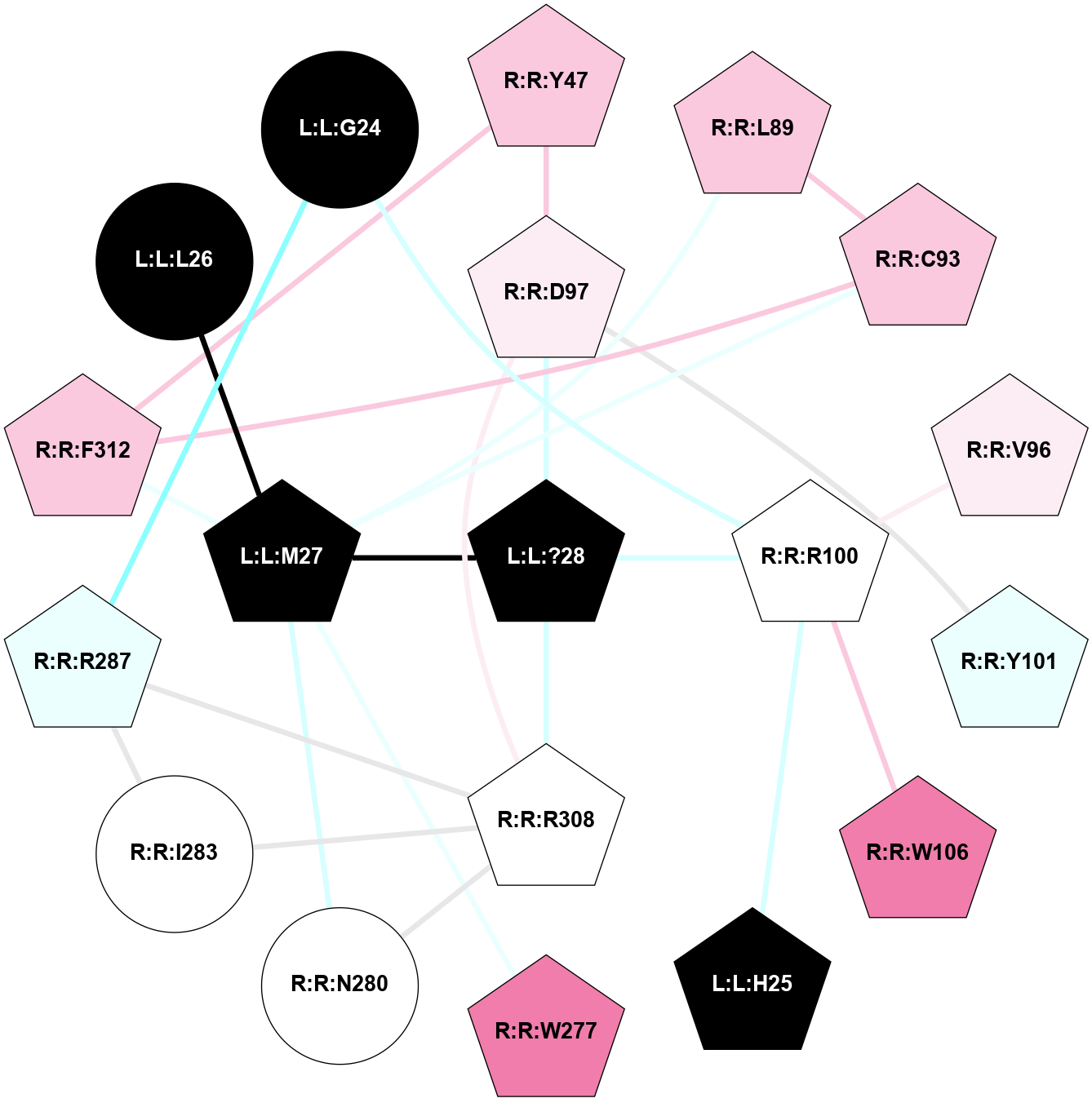
A 2D representation of the interactions of NH2 in 7W3Z
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D97 | R:R:Y47 | 6.9 | 1 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:F312 | R:R:Y47 | 4.13 | 1 | Yes | Yes | 7 | 7 | 2 | 2 | | R:R:C93 | R:R:L89 | 4.76 | 1 | Yes | Yes | 7 | 7 | 2 | 2 | | L:L:M27 | R:R:L89 | 7.07 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | R:R:C93 | R:R:F312 | 4.19 | 1 | Yes | Yes | 7 | 7 | 2 | 2 | | L:L:M27 | R:R:C93 | 4.86 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | R:R:R100 | R:R:V96 | 3.92 | 0 | Yes | Yes | 5 | 6 | 1 | 2 | | R:R:D97 | R:R:Y101 | 8.05 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | R:R:D97 | R:R:R308 | 4.76 | 1 | Yes | Yes | 6 | 5 | 1 | 1 | | L:L:?28 | R:R:D97 | 6.75 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:R100 | R:R:W106 | 9 | 0 | Yes | Yes | 5 | 8 | 1 | 2 | | L:L:H25 | R:R:R100 | 7.9 | 1 | Yes | Yes | 0 | 5 | 2 | 1 | | L:L:?28 | R:R:R100 | 3.02 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:M27 | R:R:W277 | 5.82 | 1 | Yes | Yes | 0 | 8 | 1 | 2 | | R:R:N280 | R:R:R308 | 9.64 | 0 | No | Yes | 5 | 5 | 2 | 1 | | L:L:M27 | R:R:N280 | 7.01 | 1 | Yes | No | 0 | 5 | 1 | 2 | | R:R:I283 | R:R:R287 | 6.26 | 1 | No | Yes | 5 | 4 | 2 | 2 | | R:R:I283 | R:R:R308 | 5.01 | 1 | No | Yes | 5 | 5 | 2 | 1 | | R:R:R287 | R:R:R308 | 7.46 | 1 | Yes | Yes | 4 | 5 | 2 | 1 | | L:L:?28 | R:R:R308 | 15.11 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:M27 | R:R:F312 | 4.98 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:L26 | L:L:M27 | 7.07 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?28 | L:L:M27 | 7.03 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:G24 | R:R:R100 | 1.5 | 0 | No | Yes | 0 | 5 | 2 | 1 | | L:L:G24 | R:R:R287 | 1.5 | 0 | No | Yes | 0 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 7.98 | | Average Nodes In Shell | 19.00 | | Average Hubs In Shell | 15.00 | | Average Links In Shell | 25.00 | | Average Links Mediated by Hubs In Shell | 25.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7X1T | A | Peptide | Thyrotropin Releasing Hormone | TRH | Homo Sapiens | Taltirelin | - | chim(NtGi2L-Gs-CtGq)/Beta1/Gamma2 | 3.26 | 2022-08-31 | doi.org/10.1038/s41422-022-00646-6 |
|

A 2D representation of the interactions of NH2 in 7X1T
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:H3 | L:L:P4 | 3.05 | 1 | No | Yes | 0 | 0 | 1 | 1 | | L:L:?5 | L:L:H3 | 3.2 | 1 | Yes | No | 0 | 0 | 0 | 1 | | L:L:H3 | R:R:Q105 | 2.47 | 1 | No | Yes | 0 | 7 | 1 | 2 | | L:L:?5 | L:L:P4 | 8.17 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:P4 | R:R:Q105 | 9.47 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:P4 | R:R:Y106 | 4.17 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:P4 | R:R:I109 | 3.39 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:P4 | R:R:Y282 | 4.17 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:P4 | R:R:R306 | 8.65 | 1 | Yes | Yes | 0 | 5 | 1 | 1 | | L:L:?5 | R:R:N82 | 3.42 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?5 | R:R:R306 | 21.16 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?5 | R:R:Y310 | 8.75 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:V32 | R:R:Y310 | 3.79 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:C36 | R:R:L75 | 6.35 | 1 | No | Yes | 6 | 7 | 2 | 2 | | R:R:C36 | R:R:G79 | 3.92 | 1 | No | No | 6 | 7 | 2 | 2 | | R:R:C36 | R:R:Y310 | 9.41 | 1 | No | Yes | 6 | 7 | 2 | 1 | | R:R:L75 | R:R:Y310 | 3.52 | 1 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:G79 | R:R:Y310 | 5.79 | 1 | No | Yes | 7 | 7 | 2 | 1 | | R:R:N82 | R:R:Y310 | 8.14 | 1 | No | Yes | 6 | 7 | 1 | 1 | | R:R:I109 | R:R:R306 | 5.01 | 1 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:I109 | R:R:I309 | 4.42 | 1 | Yes | Yes | 7 | 6 | 2 | 2 | | R:R:R306 | R:R:Y282 | 25.72 | 1 | Yes | Yes | 5 | 5 | 1 | 2 | | R:R:I309 | R:R:Y282 | 8.46 | 1 | Yes | Yes | 6 | 5 | 2 | 2 | | R:R:I309 | R:R:R306 | 20.04 | 1 | Yes | Yes | 6 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 8.94 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 24.00 | | Average Links Mediated by Hubs In Shell | 23.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7X1U | A | Peptide | Thyrotropin Releasing Hormone | TRH | Homo Sapiens | TRH | - | chim(NtGi2L-Gs-CtGq)/Beta1/Gamma2 | 3.19 | 2022-08-31 | doi.org/10.1038/s41422-022-00646-6 |
|
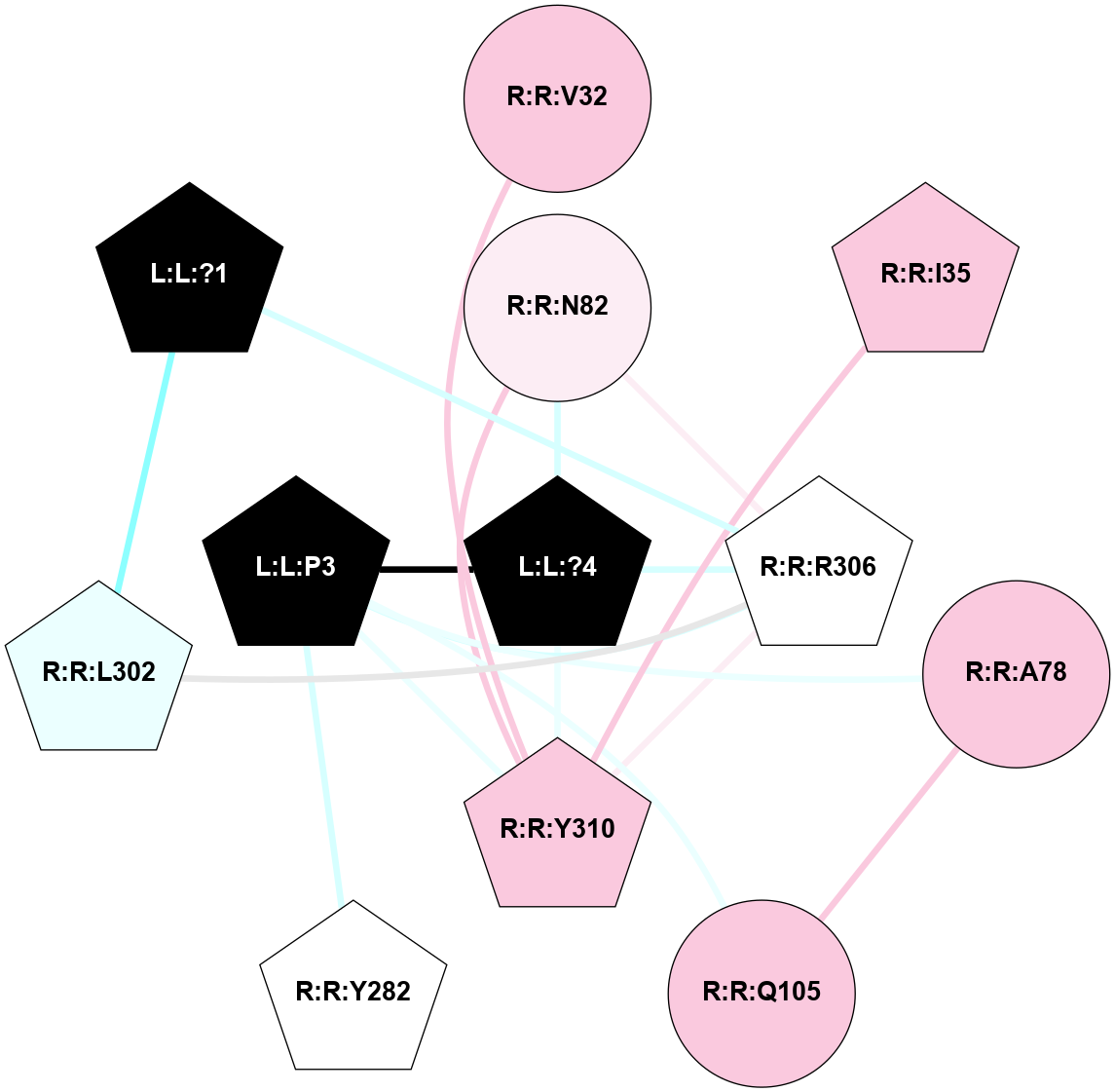
A 2D representation of the interactions of NH2 in 7X1U
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:L302 | 7.38 | 3 | Yes | No | 0 | 4 | 2 | 2 | | L:L:?1 | R:R:R306 | 6.48 | 3 | Yes | Yes | 0 | 5 | 2 | 1 | | L:L:?4 | L:L:P3 | 4.09 | 3 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:P3 | R:R:A78 | 3.74 | 3 | Yes | No | 0 | 7 | 1 | 2 | | L:L:P3 | R:R:Q105 | 11.05 | 3 | Yes | No | 0 | 7 | 1 | 2 | | L:L:P3 | R:R:Y282 | 4.17 | 3 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:P3 | R:R:R306 | 4.32 | 3 | Yes | Yes | 0 | 5 | 1 | 1 | | L:L:P3 | R:R:Y310 | 2.78 | 3 | Yes | Yes | 0 | 7 | 1 | 1 | | L:L:?4 | R:R:N82 | 10.25 | 3 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?4 | R:R:R306 | 18.13 | 3 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?4 | R:R:Y310 | 2.92 | 3 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:C36 | R:R:V32 | 5.12 | 3 | No | No | 6 | 7 | 2 | 2 | | R:R:V32 | R:R:Y310 | 7.57 | 3 | No | Yes | 7 | 7 | 2 | 1 | | R:R:C36 | R:R:Y310 | 8.06 | 3 | No | Yes | 6 | 7 | 2 | 1 | | R:R:A78 | R:R:Q105 | 4.55 | 3 | No | No | 7 | 7 | 2 | 2 | | R:R:N82 | R:R:R306 | 12.05 | 3 | No | Yes | 6 | 5 | 1 | 1 | | R:R:N82 | R:R:Y310 | 6.98 | 3 | No | Yes | 6 | 7 | 1 | 1 | | R:R:L302 | R:R:R306 | 4.86 | 3 | No | Yes | 4 | 5 | 2 | 1 | | R:R:R306 | R:R:Y310 | 8.23 | 3 | Yes | Yes | 5 | 7 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 8.85 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 19.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7XK8 | A | Peptide | Neuromedin U | NMUR2 | Homo Sapiens | Neuromedin-U-25 | - | Gi1/Beta1/Gamma2 | 3.3 | 2023-02-22 | doi.org/10.1038/s41467-022-34814-4 |
|
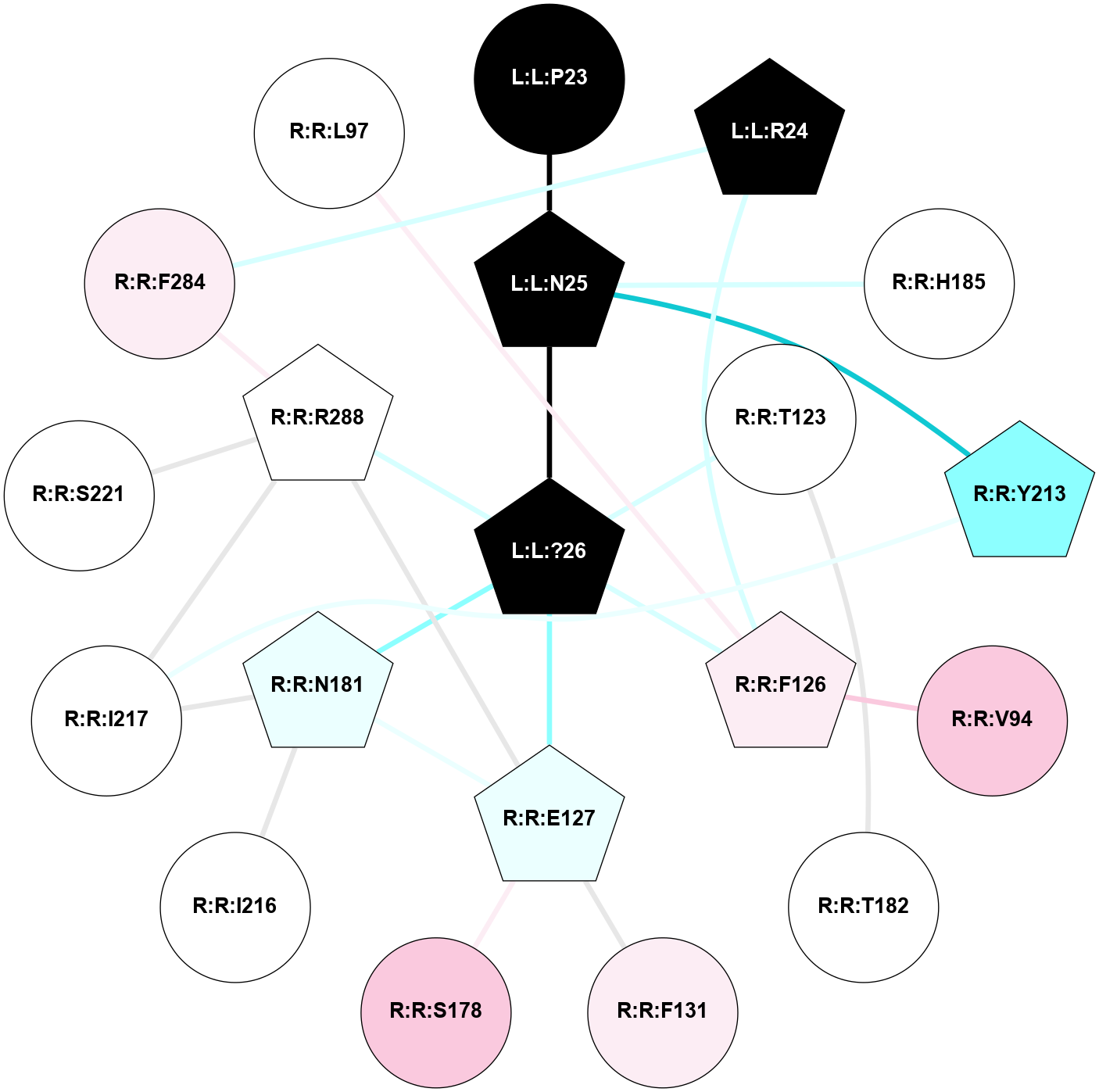
A 2D representation of the interactions of NH2 in 7XK8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:N25 | L:L:P23 | 8.15 | 0 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?26 | L:L:N25 | 6.83 | 27 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:N25 | R:R:H185 | 5.1 | 0 | Yes | No | 0 | 5 | 1 | 2 | | L:L:N25 | R:R:Y213 | 5.81 | 0 | Yes | No | 0 | 2 | 1 | 2 | | L:L:?26 | R:R:T123 | 3.67 | 27 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?26 | R:R:E127 | 13.19 | 27 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?26 | R:R:N181 | 3.42 | 27 | Yes | No | 0 | 4 | 0 | 1 | | R:R:T123 | R:R:T182 | 4.71 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:E127 | R:R:S178 | 5.75 | 27 | Yes | No | 4 | 7 | 1 | 2 | | R:R:E127 | R:R:N181 | 3.94 | 27 | Yes | No | 4 | 4 | 1 | 1 | | R:R:E127 | R:R:R288 | 3.49 | 27 | Yes | Yes | 4 | 5 | 1 | 2 | | R:R:I216 | R:R:N181 | 2.83 | 0 | No | No | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 6.78 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7XNA | A | Peptide | Somatostatin | SST2 | Homo Sapiens | CYN154806 | - | - | 2.65 | 2022-08-03 | doi.org/10.1038/s41422-022-00679-x |
|

A 2D representation of the interactions of NH2 in 7XNA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?9 | L:L:Y4 | 16.84 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?9 | L:L:T7 | 9.41 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:T7 | R:R:D295 | 4.34 | 1 | No | Yes | 0 | 2 | 2 | 2 | | L:L:?10 | L:L:?9 | 13.74 | 1 | No | Yes | 0 | 0 | 0 | 1 | | L:L:?9 | R:R:N43 | 6.58 | 1 | Yes | Yes | 0 | 3 | 1 | 1 | | L:L:?9 | R:R:V103 | 4.76 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | L:L:?9 | R:R:E106 | 10.58 | 1 | Yes | No | 0 | 4 | 1 | 2 | | L:L:?9 | R:R:D295 | 6.5 | 1 | Yes | Yes | 0 | 2 | 1 | 2 | | L:L:?10 | R:R:N43 | 10.25 | 1 | No | Yes | 0 | 3 | 0 | 1 | | R:R:N43 | R:R:T41 | 8.77 | 1 | Yes | No | 3 | 4 | 1 | 2 | | R:R:N43 | R:R:V103 | 4.43 | 1 | Yes | Yes | 3 | 4 | 1 | 2 | | R:R:E106 | R:R:N43 | 10.52 | 1 | No | Yes | 4 | 3 | 2 | 1 | | L:L:?10 | R:R:S42 | 3.74 | 1 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.24 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7XOU | A | Peptide | Cholecystokinin | CCKA | Homo Sapiens | Cholecystokinin-8 | - | Gs/Beta1/Gamma2 | 3.2 | 2022-07-20 | doi.org/10.1038/s41421-022-00420-3 |
|
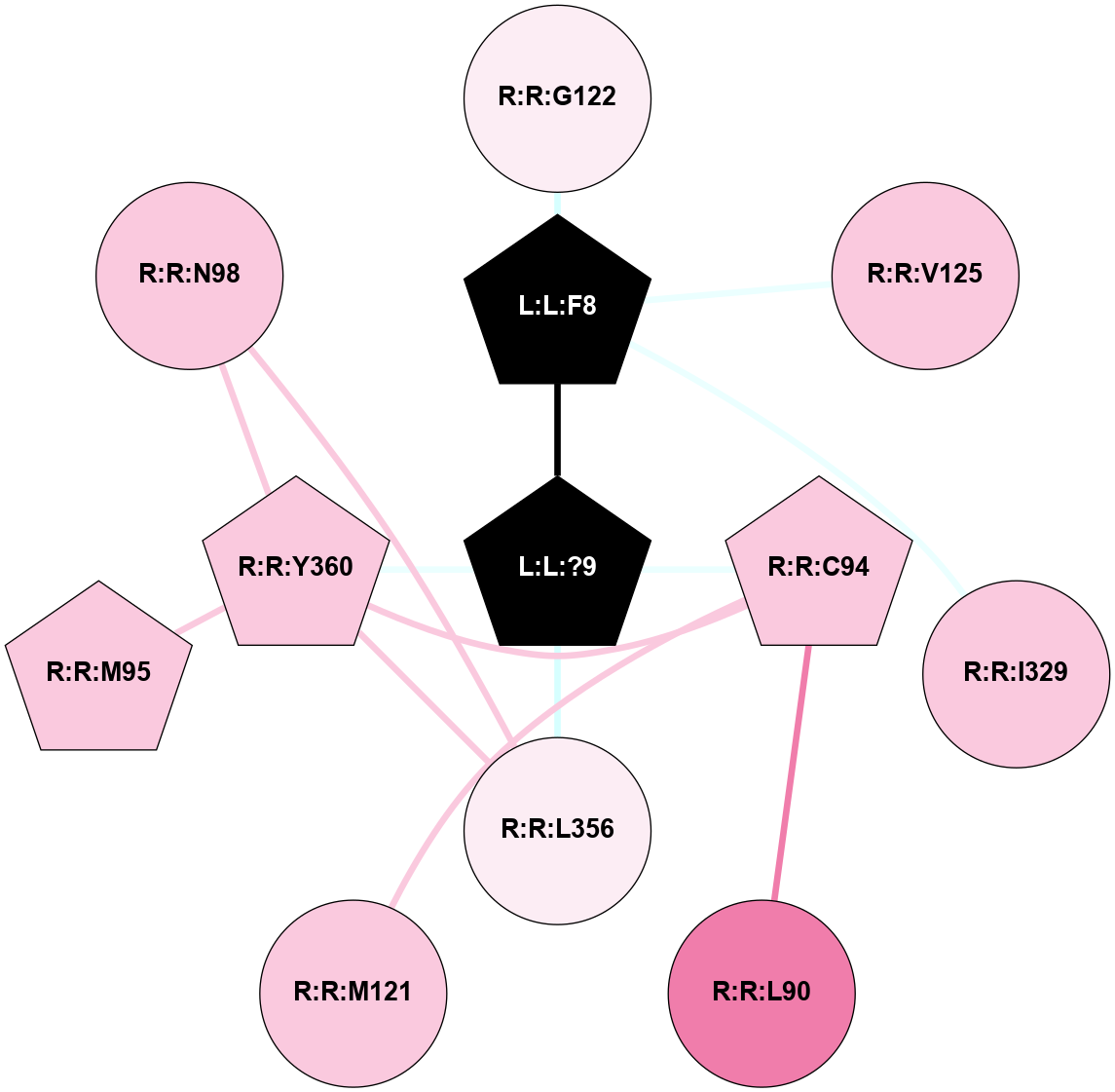
A 2D representation of the interactions of NH2 in 7XOU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?9 | L:L:F8 | 9.09 | 15 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:F8 | R:R:G122 | 3.01 | 0 | Yes | No | 0 | 6 | 1 | 2 | | L:L:F8 | R:R:V125 | 7.87 | 0 | Yes | No | 0 | 7 | 1 | 2 | | L:L:F8 | R:R:I329 | 5.02 | 0 | Yes | No | 0 | 7 | 1 | 2 | | L:L:?9 | R:R:C94 | 7.9 | 15 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?9 | R:R:L356 | 3.44 | 15 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?9 | R:R:Y360 | 8.75 | 15 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:C94 | R:R:L90 | 4.76 | 15 | Yes | No | 7 | 8 | 1 | 2 | | R:R:C94 | R:R:M121 | 4.86 | 15 | Yes | No | 7 | 7 | 1 | 2 | | R:R:C94 | R:R:Y360 | 8.06 | 15 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:M95 | R:R:Y360 | 4.79 | 0 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:L356 | R:R:N98 | 5.49 | 15 | No | No | 6 | 7 | 1 | 2 | | R:R:N98 | R:R:Y360 | 3.49 | 15 | No | Yes | 7 | 7 | 2 | 1 | | R:R:L356 | R:R:Y360 | 8.21 | 15 | No | Yes | 6 | 7 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 7.30 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7XW9 | A | Peptide | Thyrotropin Releasing Hormone | TRH | Homo Sapiens | TRH | - | chim(NtGi1L-Gs-CtGq)/Beta1/Gamma2 | 2.7 | 2022-12-28 | doi.org/10.1038/s41421-022-00477-0 |
|
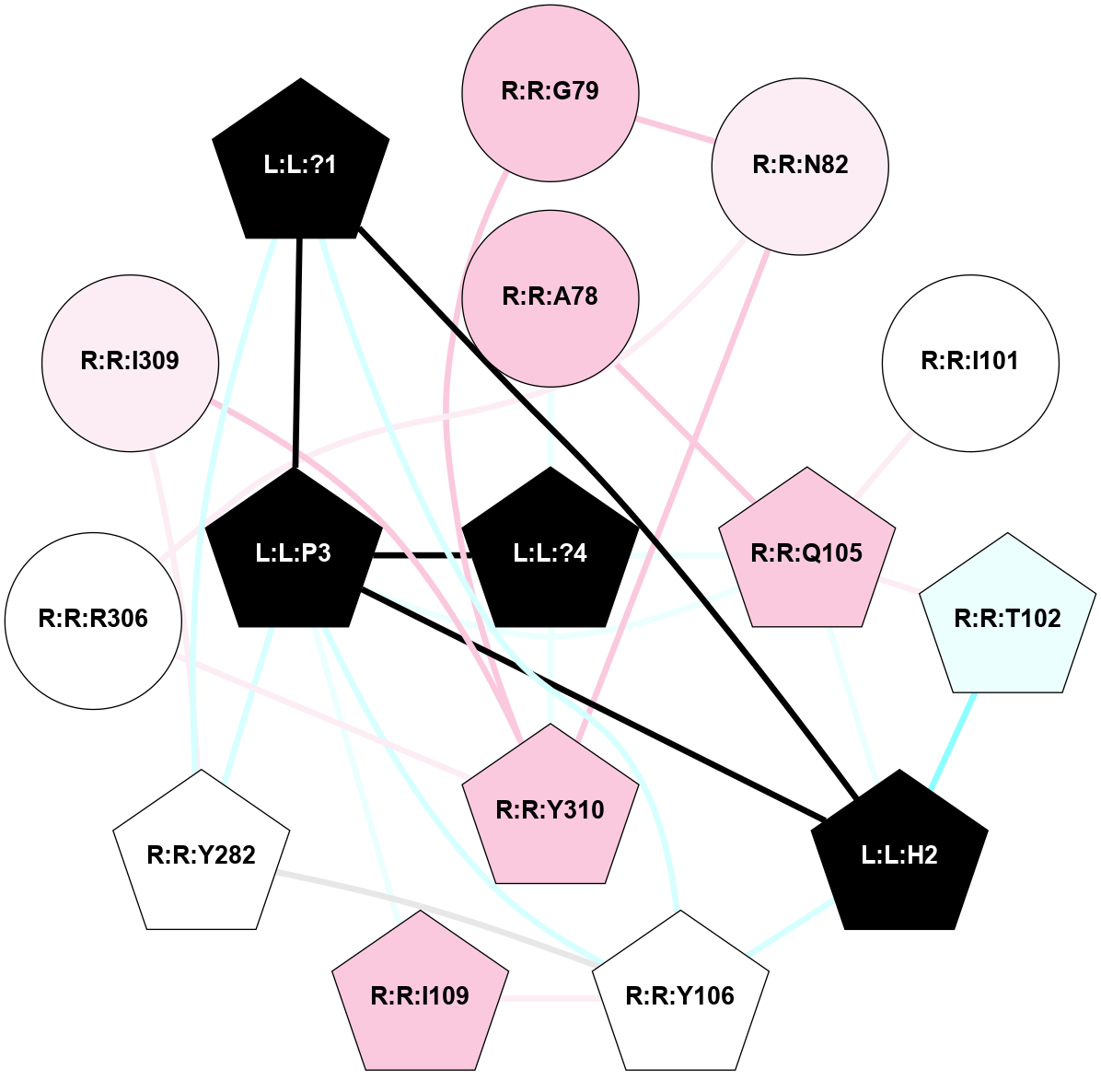
A 2D representation of the interactions of NH2 in 7XW9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | L:L:H2 | 9.63 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?1 | L:L:P3 | 3.51 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?1 | R:R:Y106 | 3.76 | 1 | Yes | Yes | 0 | 5 | 2 | 2 | | L:L:?1 | R:R:Y282 | 6.27 | 1 | Yes | Yes | 0 | 5 | 2 | 2 | | L:L:H2 | L:L:P3 | 4.58 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:H2 | R:R:T102 | 5.48 | 1 | Yes | Yes | 0 | 4 | 2 | 2 | | L:L:H2 | R:R:Q105 | 3.71 | 1 | Yes | Yes | 0 | 7 | 2 | 1 | | L:L:H2 | R:R:Y106 | 10.89 | 1 | Yes | Yes | 0 | 5 | 2 | 2 | | L:L:?4 | L:L:P3 | 12.19 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:P3 | R:R:Q105 | 12.63 | 1 | Yes | Yes | 0 | 7 | 1 | 1 | | L:L:P3 | R:R:Y106 | 11.13 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:P3 | R:R:I109 | 3.39 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:P3 | R:R:Y282 | 11.13 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:?4 | R:R:A78 | 3.9 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?4 | R:R:Q105 | 9.88 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?4 | R:R:Y310 | 8.7 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:A78 | R:R:Q105 | 4.55 | 1 | No | Yes | 7 | 7 | 1 | 1 | | R:R:G79 | R:R:N82 | 3.39 | 1 | No | No | 7 | 6 | 2 | 2 | | R:R:G79 | R:R:Y310 | 5.79 | 1 | No | Yes | 7 | 7 | 2 | 1 | | R:R:N82 | R:R:R306 | 3.62 | 1 | No | No | 6 | 5 | 2 | 2 | | R:R:N82 | R:R:Y310 | 8.14 | 1 | No | Yes | 6 | 7 | 2 | 1 | | R:R:I101 | R:R:Q105 | 4.12 | 0 | No | Yes | 5 | 7 | 2 | 1 | | R:R:Q105 | R:R:T102 | 5.67 | 1 | Yes | Yes | 7 | 4 | 1 | 2 | | R:R:I109 | R:R:Y106 | 3.63 | 1 | Yes | Yes | 7 | 5 | 2 | 2 | | R:R:Y106 | R:R:Y282 | 2.98 | 1 | Yes | Yes | 5 | 5 | 2 | 2 | | R:R:I309 | R:R:Y282 | 7.25 | 0 | No | Yes | 6 | 5 | 2 | 2 | | R:R:R306 | R:R:Y310 | 6.17 | 1 | No | Yes | 5 | 7 | 2 | 1 | | R:R:I309 | R:R:Y310 | 4.84 | 0 | No | Yes | 6 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 8.67 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 28.00 | | Average Links Mediated by Hubs In Shell | 26.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7Y35 | B1 | Peptide | Parathyroid Hormone | PTH1 | Homo Sapiens | Abaloparatide | - | Gs/Beta1/Gamma2 | 2.9 | 2023-07-05 | To be published |
|
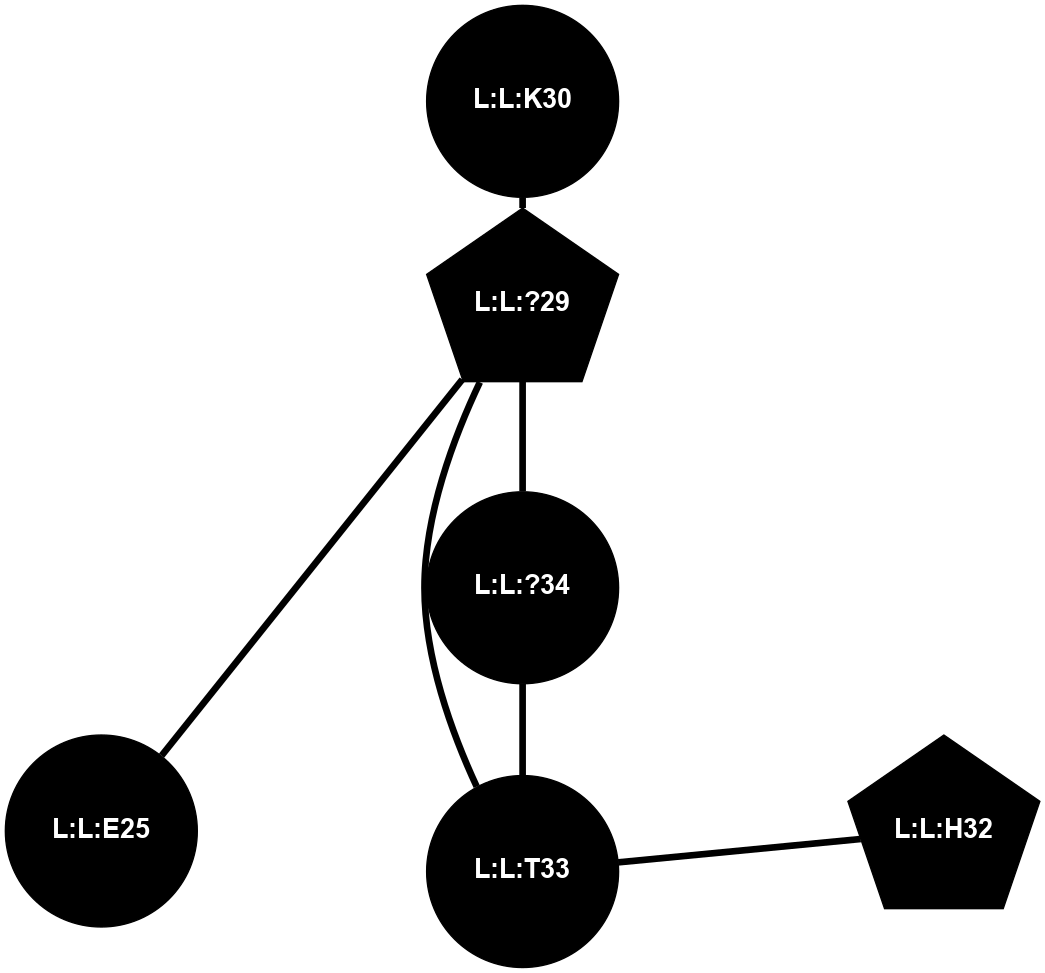
A 2D representation of the interactions of NH2 in 7Y35
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?29 | L:L:K30 | 4.05 | 4 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?29 | L:L:T33 | 8.47 | 4 | Yes | No | 0 | 0 | 1 | 1 | | L:L:?29 | L:L:?34 | 9.84 | 4 | Yes | No | 0 | 0 | 1 | 0 | | L:L:H32 | L:L:T33 | 2.74 | 4 | Yes | No | 0 | 0 | 2 | 1 | | L:L:?34 | L:L:T33 | 10.94 | 4 | No | No | 0 | 0 | 0 | 1 | | L:L:?29 | L:L:E25 | 1.27 | 4 | Yes | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 10.39 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8F0J | B1 | Peptide | Calcitonin | CT | Homo Sapiens | Pramlintide Analogue San45 | - | Gs/Beta1/Gamma2 | 2 | 2023-08-02 | doi.org/10.1038/s41589-023-01393-4 |
|
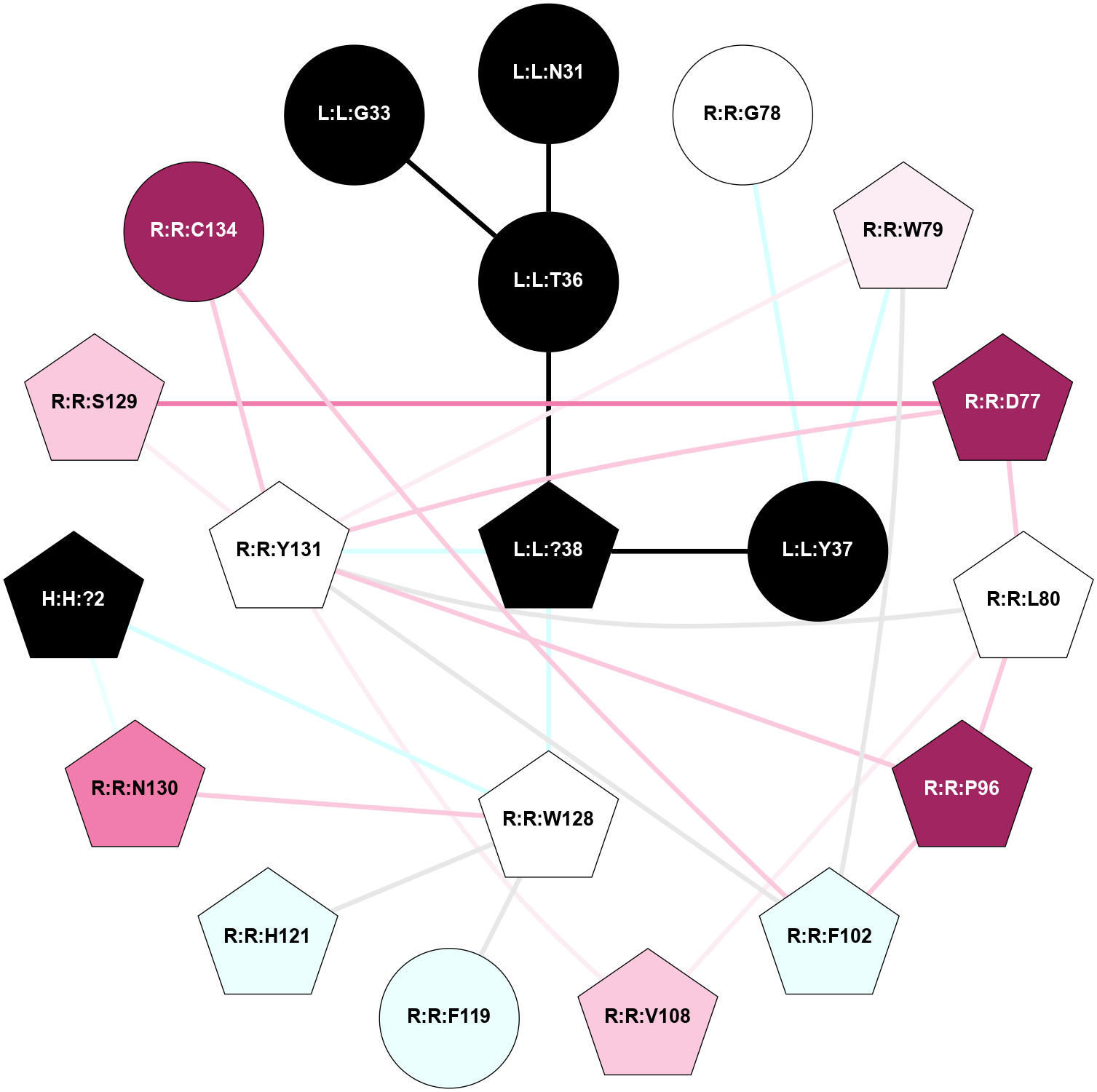
A 2D representation of the interactions of NH2 in 8F0J
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:W128 | 10.22 | 11 | Yes | Yes | 0 | 5 | 2 | 1 | | H:H:?2 | R:R:N130 | 25.87 | 11 | Yes | Yes | 0 | 8 | 2 | 2 | | L:L:N31 | L:L:T36 | 8.77 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:?38 | L:L:T36 | 7.3 | 0 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?38 | L:L:Y37 | 2.9 | 0 | Yes | No | 0 | 0 | 0 | 1 | | L:L:Y37 | R:R:G78 | 10.14 | 0 | No | No | 0 | 5 | 1 | 2 | | L:L:Y37 | R:R:W79 | 12.54 | 0 | No | Yes | 0 | 6 | 1 | 2 | | L:L:?38 | R:R:W128 | 8.46 | 0 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?38 | R:R:Y131 | 5.8 | 0 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:L80 | 4.07 | 6 | Yes | Yes | 9 | 5 | 2 | 2 | | R:R:D77 | R:R:S129 | 7.36 | 6 | Yes | Yes | 9 | 7 | 2 | 2 | | R:R:D77 | R:R:Y131 | 5.75 | 6 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:F102 | R:R:W79 | 6.01 | 6 | Yes | Yes | 4 | 6 | 2 | 2 | | R:R:W79 | R:R:Y131 | 18.33 | 6 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:L80 | R:R:P96 | 8.21 | 6 | Yes | Yes | 5 | 9 | 2 | 2 | | R:R:L80 | R:R:V108 | 4.47 | 6 | Yes | Yes | 5 | 7 | 2 | 2 | | R:R:L80 | R:R:Y131 | 8.21 | 6 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:F102 | R:R:P96 | 2.89 | 6 | Yes | Yes | 4 | 9 | 2 | 2 | | R:R:P96 | R:R:Y131 | 4.17 | 6 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:F102 | R:R:Y131 | 3.09 | 6 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:C134 | R:R:F102 | 8.38 | 6 | No | Yes | 9 | 4 | 2 | 2 | | R:R:V108 | R:R:Y131 | 15.14 | 6 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:F119 | R:R:W128 | 3.01 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:H121 | R:R:W128 | 23.27 | 11 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 9.04 | 11 | Yes | Yes | 8 | 5 | 2 | 1 | | R:R:S129 | R:R:Y131 | 6.36 | 6 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:C134 | R:R:Y131 | 4.03 | 6 | No | Yes | 9 | 5 | 2 | 1 | | L:L:G33 | L:L:T36 | 1.82 | 0 | No | No | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 6.12 | | Average Nodes In Shell | 20.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 28.00 | | Average Links Mediated by Hubs In Shell | 25.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8F0K | B1 | Peptide | Calcitonin | CT (AMY3) | Homo Sapiens | Pramlintide Analogue San385 | - | Gs/Beta1/Gamma2; RAMP3 | 1.9 | 2023-08-02 | doi.org/10.1038/s41589-023-01393-4 |
|
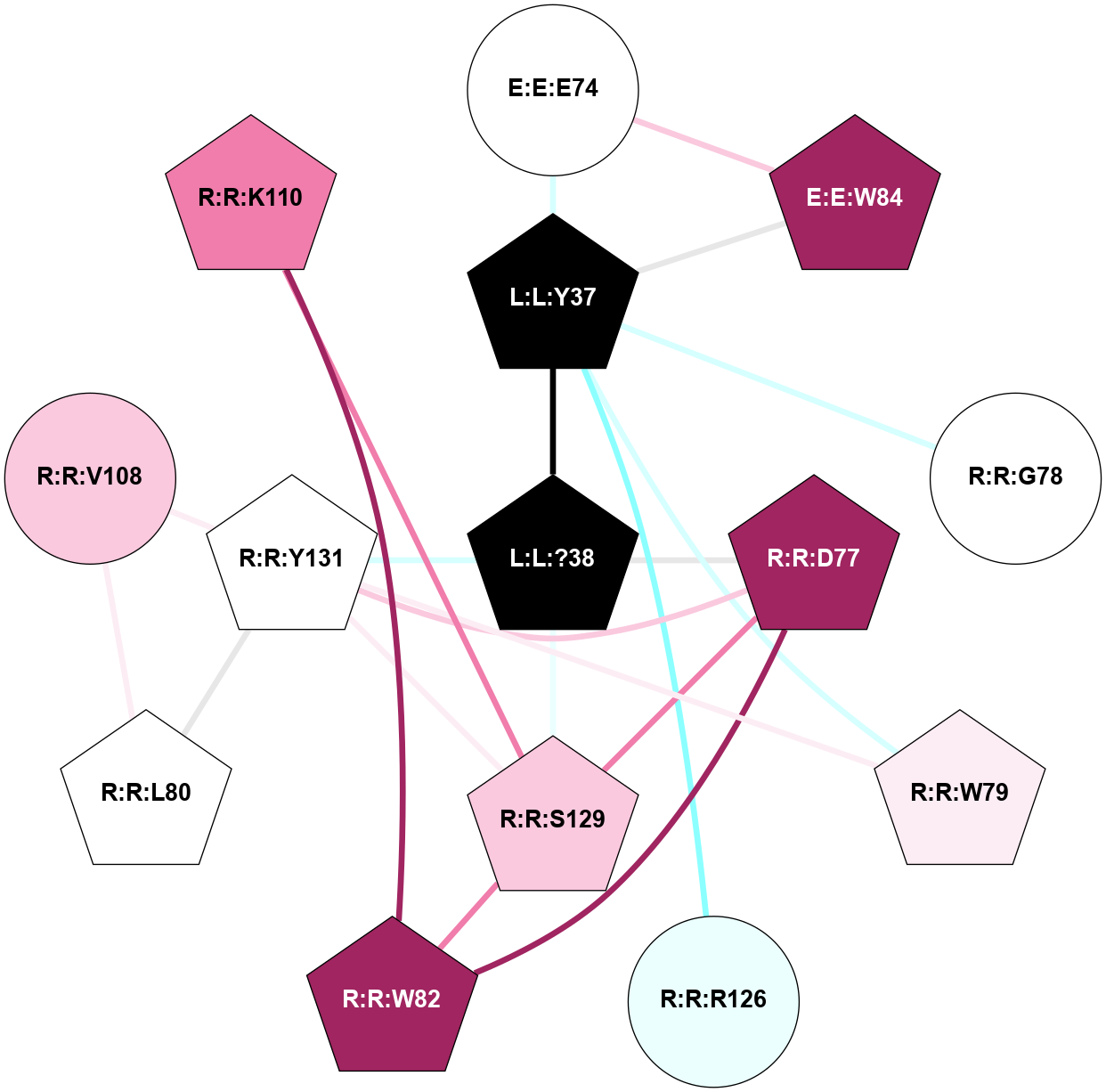
A 2D representation of the interactions of NH2 in 8F0K
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | E:E:E74 | E:E:W84 | 9.81 | 2 | No | Yes | 5 | 9 | 2 | 2 | | E:E:E74 | L:L:Y37 | 4.49 | 2 | No | Yes | 5 | 0 | 2 | 1 | | E:E:W84 | L:L:Y37 | 4.82 | 2 | Yes | Yes | 9 | 0 | 2 | 1 | | L:L:?38 | L:L:Y37 | 5.8 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:Y37 | R:R:G78 | 4.35 | 2 | Yes | No | 0 | 5 | 1 | 2 | | L:L:Y37 | R:R:W79 | 11.58 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:Y37 | R:R:R126 | 4.12 | 2 | Yes | No | 0 | 4 | 1 | 2 | | L:L:?38 | R:R:D77 | 10.08 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?38 | R:R:S129 | 7.43 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?38 | R:R:Y131 | 5.8 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:W82 | 7.82 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D77 | R:R:S129 | 7.36 | 2 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:D77 | R:R:Y131 | 6.9 | 2 | Yes | Yes | 9 | 5 | 1 | 1 | | R:R:W79 | R:R:Y131 | 17.36 | 2 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:L80 | R:R:V108 | 4.47 | 2 | Yes | No | 5 | 7 | 2 | 2 | | R:R:L80 | R:R:Y131 | 8.21 | 2 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:K110 | R:R:W82 | 23.21 | 2 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:S129 | R:R:W82 | 3.71 | 2 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:V108 | R:R:Y131 | 13.88 | 2 | No | Yes | 7 | 5 | 2 | 1 | | R:R:K110 | R:R:S129 | 10.71 | 2 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:S129 | R:R:Y131 | 5.09 | 2 | Yes | Yes | 7 | 5 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 7.28 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 21.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8F2A | B1 | Peptide | Calcitonin | CT (AMY3) | Homo Sapiens | Pramlintide Analogue San385 | - | Gs/Beta1/Gamma2; RAMP3 | 2.2 | 2023-08-02 | doi.org/10.1038/s41589-023-01393-4 |
|
A 2D representation of the interactions of NH2 in 8F2A
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:W128 | 7.15 | 12 | No | Yes | 0 | 5 | 2 | 1 | | H:H:?2 | R:R:N130 | 29.57 | 12 | No | Yes | 0 | 8 | 2 | 2 | | L:L:N31 | L:L:T36 | 4.39 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:?38 | L:L:T36 | 7.3 | 0 | No | No | 0 | 0 | 0 | 1 | | L:L:?38 | R:R:W128 | 11.28 | 0 | No | Yes | 0 | 5 | 0 | 1 | | L:L:?38 | R:R:Y131 | 5.8 | 0 | No | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:S129 | 7.36 | 2 | No | Yes | 9 | 7 | 2 | 2 | | R:R:D77 | R:R:Y131 | 11.49 | 2 | No | Yes | 9 | 5 | 2 | 1 | | R:R:W79 | R:R:Y131 | 17.36 | 2 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:L80 | R:R:P96 | 9.85 | 2 | Yes | Yes | 5 | 9 | 2 | 2 | | R:R:L80 | R:R:V108 | 4.47 | 2 | Yes | Yes | 5 | 7 | 2 | 2 | | R:R:L80 | R:R:Y131 | 8.21 | 2 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:P96 | R:R:Y131 | 4.17 | 2 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:V108 | R:R:Y131 | 13.88 | 2 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:H121 | R:R:W128 | 25.39 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 11.3 | 12 | Yes | Yes | 8 | 5 | 2 | 1 | | R:R:S129 | R:R:Y131 | 5.09 | 2 | Yes | Yes | 7 | 5 | 2 | 1 | | L:L:G33 | L:L:T36 | 1.82 | 0 | No | No | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 8.13 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8F2B | B1 | Peptide | Calcitonin | CT (AMY3) | Homo Sapiens | Pramlintide Analogue San45 | - | Gs/Beta1/Gamma2; RAMP3 | 2 | 2023-08-02 | doi.org/10.1038/s41589-023-01393-4 |
|
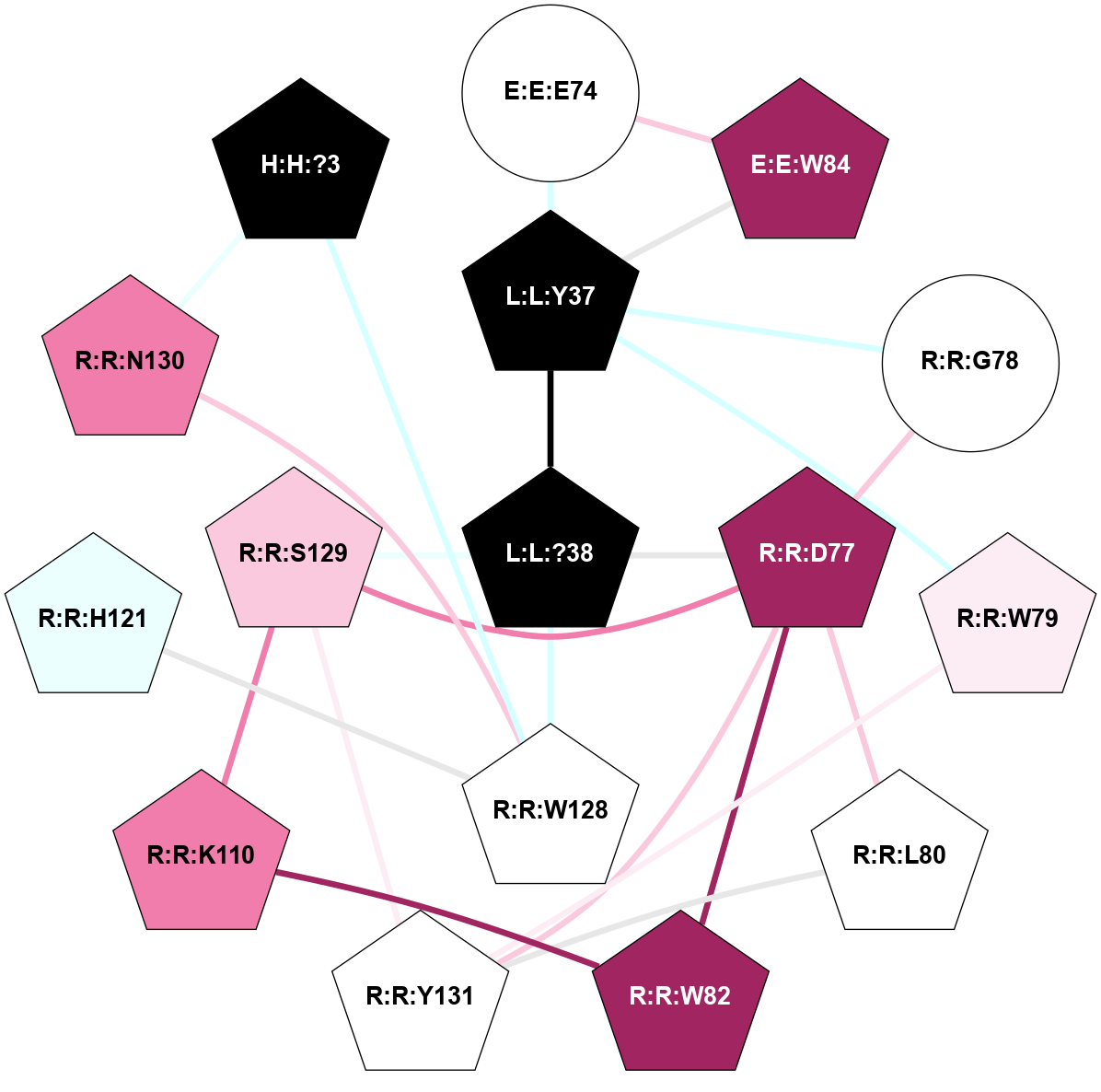
A 2D representation of the interactions of NH2 in 8F2B
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | E:E:E74 | E:E:W84 | 18.54 | 1 | No | Yes | 5 | 9 | 2 | 2 | | E:E:E74 | L:L:Y37 | 4.49 | 1 | No | Yes | 5 | 0 | 2 | 1 | | E:E:W84 | L:L:Y37 | 5.79 | 1 | Yes | Yes | 9 | 0 | 2 | 1 | | H:H:?3 | R:R:W128 | 5.11 | 1 | Yes | Yes | 0 | 5 | 2 | 1 | | H:H:?3 | R:R:N130 | 27.11 | 1 | Yes | Yes | 0 | 8 | 2 | 2 | | L:L:?38 | L:L:Y37 | 5.8 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:Y37 | R:R:G78 | 11.59 | 1 | Yes | No | 0 | 5 | 1 | 2 | | L:L:Y37 | R:R:W79 | 13.5 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?38 | R:R:D77 | 3.36 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?38 | R:R:W128 | 5.64 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?38 | R:R:S129 | 7.43 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:D77 | R:R:G78 | 3.35 | 1 | Yes | No | 9 | 5 | 1 | 2 | | R:R:D77 | R:R:L80 | 4.07 | 1 | Yes | Yes | 9 | 5 | 1 | 2 | | R:R:D77 | R:R:W82 | 10.05 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D77 | R:R:S129 | 5.89 | 1 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:D77 | R:R:Y131 | 8.05 | 1 | Yes | Yes | 9 | 5 | 1 | 2 | | R:R:W79 | R:R:Y131 | 14.47 | 1 | Yes | Yes | 6 | 5 | 2 | 2 | | R:R:L80 | R:R:Y131 | 9.38 | 1 | Yes | Yes | 5 | 5 | 2 | 2 | | R:R:K110 | R:R:W82 | 19.73 | 1 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:K110 | R:R:S129 | 6.12 | 1 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:H121 | R:R:W128 | 23.27 | 1 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 11.3 | 1 | Yes | Yes | 8 | 5 | 2 | 1 | | R:R:S129 | R:R:Y131 | 7.63 | 1 | Yes | Yes | 7 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 5.56 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 14.00 | | Average Links In Shell | 23.00 | | Average Links Mediated by Hubs In Shell | 23.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8F7R | A | Peptide | Opioid | MOP | Homo Sapiens | Endomorphin | - | Gi1/Beta1/Gamma2 | 3.28 | 2022-12-14 | doi.org/10.1016/j.cell.2022.12.026 |
|
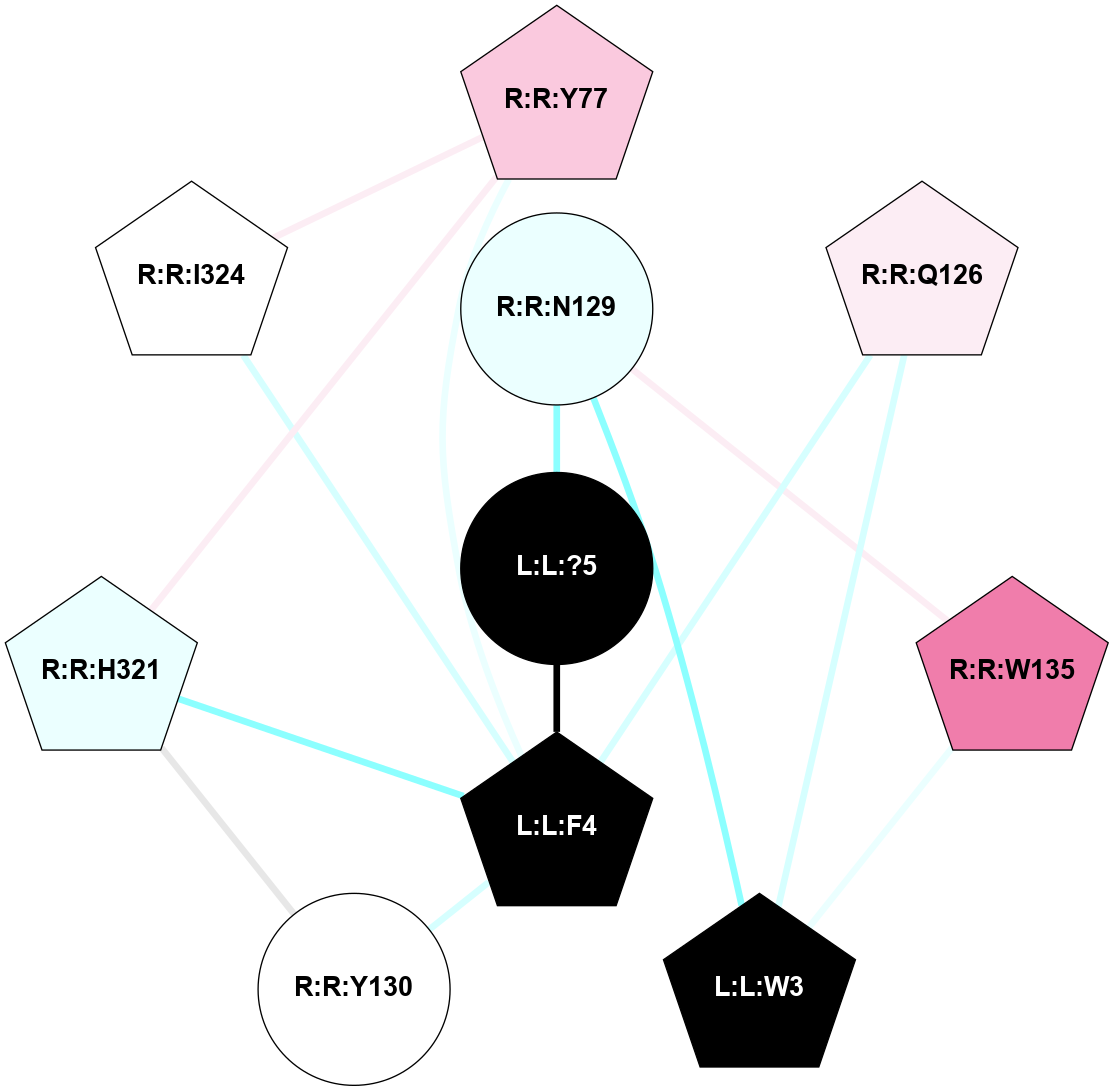
A 2D representation of the interactions of NH2 in 8F7R
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:W3 | R:R:Q126 | 5.48 | 1 | Yes | Yes | 0 | 6 | 2 | 2 | | L:L:W3 | R:R:N129 | 3.39 | 1 | Yes | No | 0 | 4 | 2 | 1 | | L:L:W3 | R:R:W135 | 4.69 | 1 | Yes | Yes | 0 | 8 | 2 | 2 | | L:L:?5 | L:L:F4 | 9.04 | 0 | No | Yes | 0 | 0 | 0 | 1 | | L:L:F4 | R:R:Y77 | 4.13 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:F4 | R:R:Q126 | 14.05 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:F4 | R:R:Y130 | 6.19 | 1 | Yes | No | 0 | 5 | 1 | 2 | | L:L:F4 | R:R:H321 | 6.79 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | L:L:F4 | R:R:I324 | 3.77 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:?5 | R:R:N129 | 13.59 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:H321 | R:R:Y77 | 10.89 | 1 | Yes | Yes | 4 | 7 | 2 | 2 | | R:R:I324 | R:R:Y77 | 8.46 | 1 | Yes | Yes | 5 | 7 | 2 | 2 | | R:R:N129 | R:R:W135 | 7.91 | 1 | No | Yes | 4 | 8 | 1 | 2 | | R:R:H321 | R:R:Y130 | 11.98 | 1 | Yes | No | 4 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 11.31 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8F7S | A | Peptide | Opioid | DOP | Homo Sapiens | Deltorphin | - | Gi1/Beta1/Gamma2 | 3 | 2022-12-14 | doi.org/10.1016/j.cell.2022.12.026 |
|
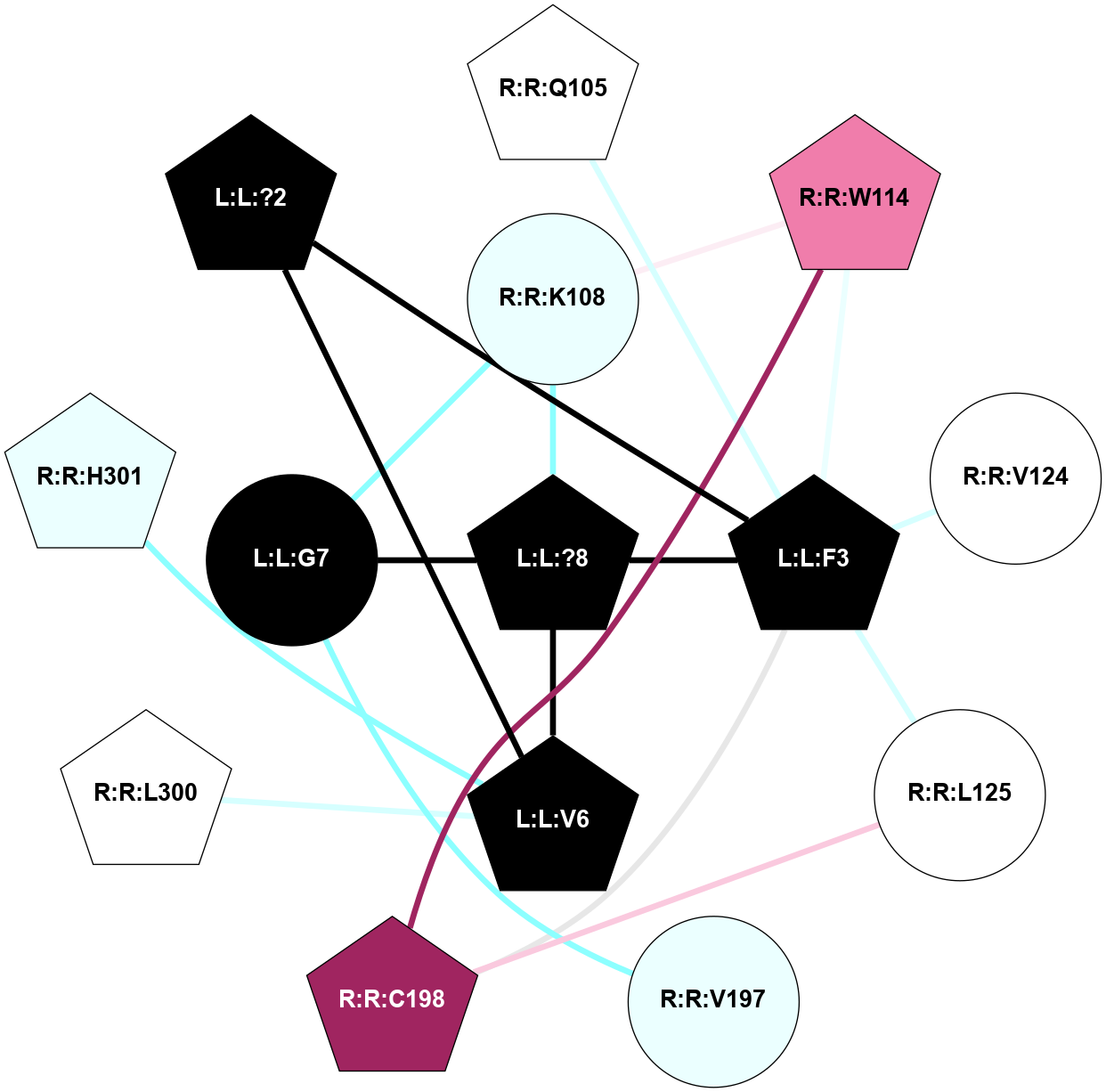
A 2D representation of the interactions of NH2 in 8F7S
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:F3 | R:R:Q105 | 10.54 | 5 | Yes | Yes | 0 | 5 | 1 | 2 | | R:R:K108 | R:R:W114 | 4.64 | 5 | No | Yes | 4 | 8 | 1 | 2 | | L:L:G7 | R:R:K108 | 3.49 | 5 | No | No | 0 | 4 | 1 | 1 | | L:L:?8 | R:R:K108 | 10.47 | 5 | Yes | No | 0 | 4 | 0 | 1 | | R:R:C198 | R:R:W114 | 6.53 | 5 | Yes | Yes | 9 | 8 | 2 | 2 | | L:L:F3 | R:R:W114 | 5.01 | 5 | Yes | Yes | 0 | 8 | 1 | 2 | | L:L:F3 | R:R:V124 | 2.62 | 5 | Yes | No | 0 | 5 | 1 | 2 | | R:R:C198 | R:R:L125 | 3.17 | 5 | Yes | No | 9 | 5 | 2 | 2 | | L:L:F3 | R:R:L125 | 4.87 | 5 | Yes | No | 0 | 5 | 1 | 2 | | L:L:G7 | R:R:V197 | 3.68 | 5 | No | No | 0 | 4 | 1 | 2 | | L:L:F3 | R:R:C198 | 5.59 | 5 | Yes | Yes | 0 | 9 | 1 | 2 | | L:L:V6 | R:R:L300 | 2.98 | 0 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:V6 | R:R:H301 | 2.77 | 0 | Yes | Yes | 0 | 4 | 1 | 2 | | L:L:?2 | L:L:F3 | 2.48 | 0 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?2 | L:L:V6 | 4.55 | 0 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?8 | L:L:F3 | 18.09 | 5 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?8 | L:L:V6 | 7.37 | 5 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?8 | L:L:G7 | 4.23 | 5 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 10.04 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8IA7 | A | Peptide | Cholecystokinin | CCKB | Homo Sapiens | Cholecystokinin-8 | - | chim(NtGi1L-Gq)/Beta1/Gamma2 | 3.1 | 2023-12-06 | doi.org/10.1038/s41421-022-00420-3 |
|

A 2D representation of the interactions of NH2 in 8IA7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:C107 | R:R:M134 | 6.48 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:C107 | R:R:Y380 | 8.06 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:M108 | R:R:Y380 | 3.59 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:T111 | R:R:Y380 | 3.75 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:M134 | R:R:V130 | 4.56 | 0 | Yes | No | 8 | 6 | 1 | 2 | | L:L:M6 | R:R:M134 | 2.89 | 0 | Yes | Yes | 0 | 8 | 2 | 1 | | L:L:?9 | R:R:M134 | 14 | 10 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:F8 | R:R:V138 | 6.55 | 0 | Yes | No | 0 | 7 | 1 | 2 | | L:L:F8 | R:R:Y189 | 4.13 | 0 | Yes | Yes | 0 | 7 | 1 | 2 | | R:R:S379 | R:R:V349 | 3.23 | 0 | No | No | 7 | 7 | 2 | 2 | | L:L:F8 | R:R:V349 | 3.93 | 0 | Yes | No | 0 | 7 | 1 | 2 | | R:R:H376 | R:R:I372 | 5.3 | 10 | No | Yes | 5 | 4 | 1 | 2 | | R:R:H376 | R:R:Y380 | 6.53 | 10 | No | Yes | 5 | 7 | 1 | 1 | | L:L:?9 | R:R:H376 | 3.18 | 10 | Yes | No | 0 | 5 | 0 | 1 | | R:R:S379 | R:R:Y380 | 3.82 | 0 | No | Yes | 7 | 7 | 2 | 1 | | L:L:?9 | R:R:Y380 | 2.9 | 10 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?9 | L:L:F8 | 9.04 | 10 | Yes | Yes | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 7.28 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8IOD | A | Peptide | Melanocortin | MC5 | Homo Sapiens | PG-901 | Ca | chim(NtGi1-Gs)/Beta1/Gamma2 | 2.59 | 2023-09-20 | doi.org/10.1038/s41421-023-00586-4 |
|
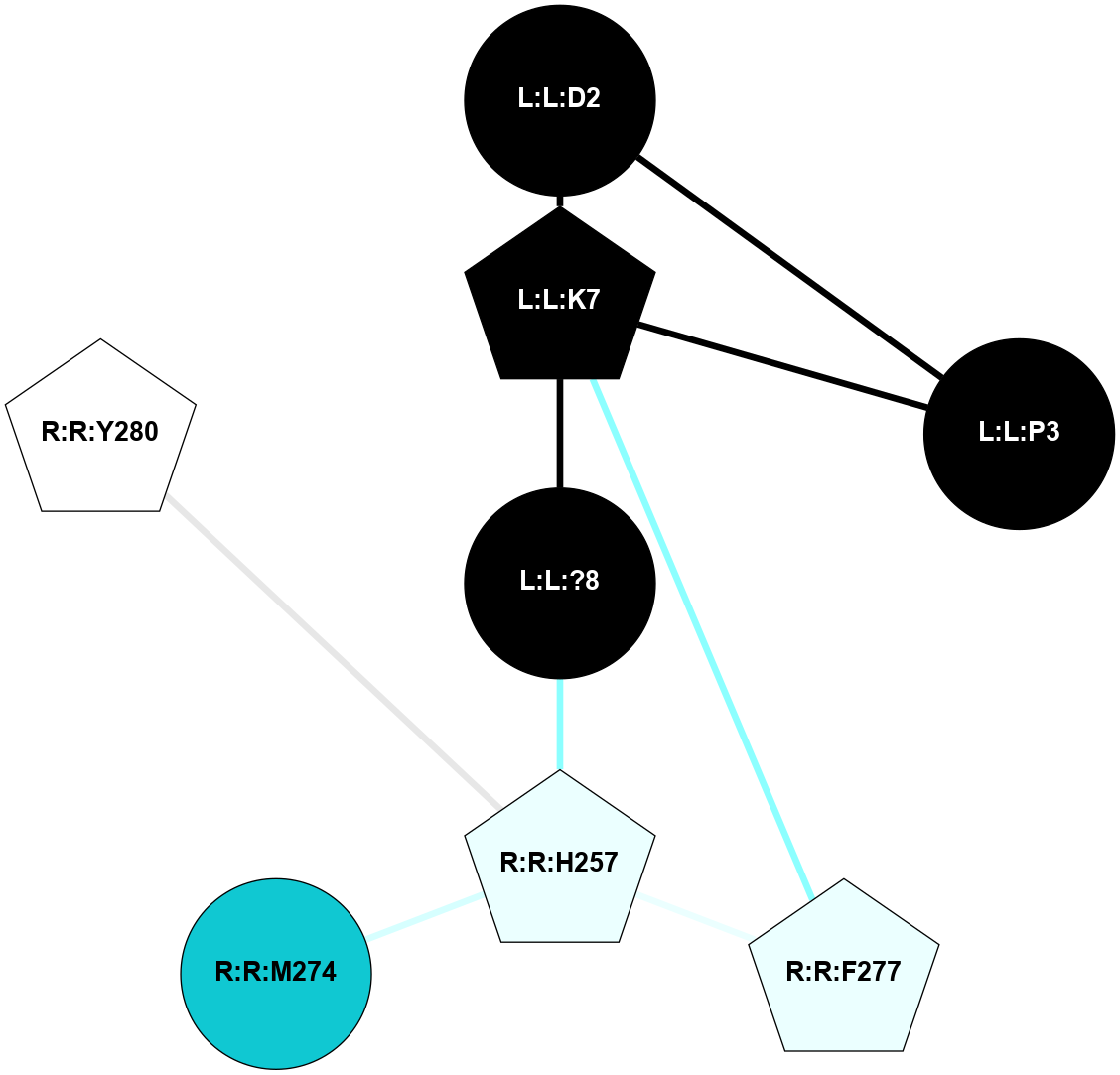
A 2D representation of the interactions of NH2 in 8IOD
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:D2 | L:L:P3 | 4.83 | 4 | No | No | 0 | 0 | 2 | 2 | | L:L:D2 | L:L:K7 | 9.68 | 4 | No | Yes | 0 | 0 | 2 | 1 | | L:L:K7 | L:L:P3 | 6.69 | 4 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?8 | L:L:K7 | 10.47 | 0 | No | Yes | 0 | 0 | 0 | 1 | | L:L:K7 | R:R:F277 | 19.85 | 4 | Yes | Yes | 0 | 4 | 1 | 2 | | L:L:?8 | R:R:H257 | 6.36 | 0 | No | Yes | 0 | 4 | 0 | 1 | | R:R:H257 | R:R:M274 | 7.88 | 0 | Yes | No | 4 | 1 | 1 | 2 | | R:R:F277 | R:R:H257 | 9.05 | 0 | Yes | Yes | 4 | 4 | 2 | 1 | | R:R:H257 | R:R:Y280 | 4.36 | 0 | Yes | Yes | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 8.42 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8WKY | A | Peptide | Melanocortin | MC4 | Homo sapiens | PG-934 | - | - | 2.9 | 2024-08-07 | doi.org/10.1021/acs.jmedchem.3c01822 |
|
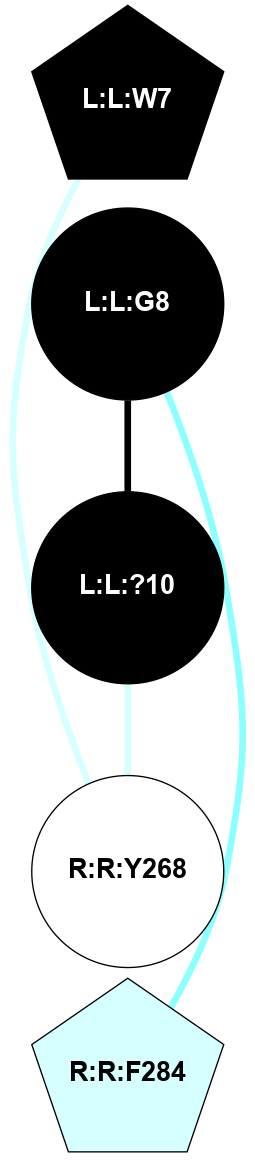
A 2D representation of the interactions of NH2 in 8WKY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:W7 | R:R:Y268 | 7.72 | 1 | Yes | No | 0 | 5 | 2 | 1 | | L:L:?10 | L:L:G8 | 4.23 | 0 | No | No | 0 | 0 | 0 | 1 | | L:L:G8 | R:R:F284 | 10.54 | 0 | No | Yes | 0 | 3 | 1 | 2 | | L:L:?10 | R:R:Y268 | 5.8 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 5.02 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8WKZ | A | Peptide | Melanocortin | MC4 | Homo sapiens | SBL-MC-31 | - | - | 3.3 | 2024-08-07 | doi.org/10.1021/acs.jmedchem.3c01822 |
|

A 2D representation of the interactions of NH2 in 8WKZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | L:L:?2 | 19.44 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?1 | L:L:K9 | 12.67 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?2 | L:L:D3 | 7.64 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?2 | L:L:K9 | 5.29 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:D3 | L:L:K9 | 9.68 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:W7 | R:R:Y268 | 9.65 | 1 | Yes | No | 0 | 5 | 2 | 1 | | L:L:?10 | L:L:K9 | 6.98 | 0 | No | Yes | 0 | 0 | 0 | 1 | | L:L:?10 | R:R:Y268 | 8.7 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 7.84 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8XGO | A | Peptide | Kisspeptin | Kiss1 | Homo Sapiens | TAK448 | - | chim(NtGi1-Gq)/Beta1/Gamma2 | 2.68 | 2024-10-30 | doi.org/10.1126/sciadv.adn7771 |
|

A 2D representation of the interactions of NH2 in 8XGO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:C95 | R:R:F91 | 6.98 | 6 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:C95 | R:R:Q122 | 7.63 | 6 | Yes | Yes | 7 | 6 | 1 | 1 | | R:R:C95 | R:R:Y313 | 5.38 | 6 | Yes | Yes | 7 | 7 | 1 | 1 | | L:L:?11 | R:R:C95 | 3.93 | 6 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:V96 | R:R:Y313 | 8.83 | 6 | No | Yes | 8 | 7 | 2 | 1 | | R:R:T99 | R:R:Y313 | 3.75 | 6 | No | Yes | 6 | 7 | 1 | 1 | | L:L:?11 | R:R:T99 | 10.94 | 6 | Yes | No | 0 | 6 | 0 | 1 | | R:R:Q122 | R:R:V118 | 5.73 | 6 | Yes | No | 6 | 5 | 1 | 2 | | L:L:L8 | R:R:Q122 | 5.32 | 0 | Yes | Yes | 0 | 6 | 2 | 1 | | L:L:W10 | R:R:Q122 | 6.57 | 0 | Yes | Yes | 0 | 6 | 2 | 1 | | L:L:?11 | R:R:Q122 | 9.88 | 6 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:H309 | R:R:I279 | 3.98 | 6 | Yes | Yes | 6 | 7 | 1 | 2 | | L:L:W10 | R:R:I279 | 3.52 | 0 | Yes | Yes | 0 | 7 | 2 | 2 | | R:R:H309 | R:R:K305 | 11.79 | 6 | Yes | No | 6 | 4 | 1 | 2 | | R:R:H309 | R:R:Y313 | 11.98 | 6 | Yes | Yes | 6 | 7 | 1 | 1 | | L:L:?11 | R:R:H309 | 9.55 | 6 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?11 | R:R:Y313 | 5.8 | 6 | Yes | Yes | 0 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 8.02 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8XGS | A | Peptide | Kisspeptin | Kiss1 | Homo Sapiens | Metastin | - | chim(NtGi1-Gq)/Beta1/Gamma2 | 2.95 | 2024-10-30 | doi.org/10.1126/sciadv.adn7771 |
|
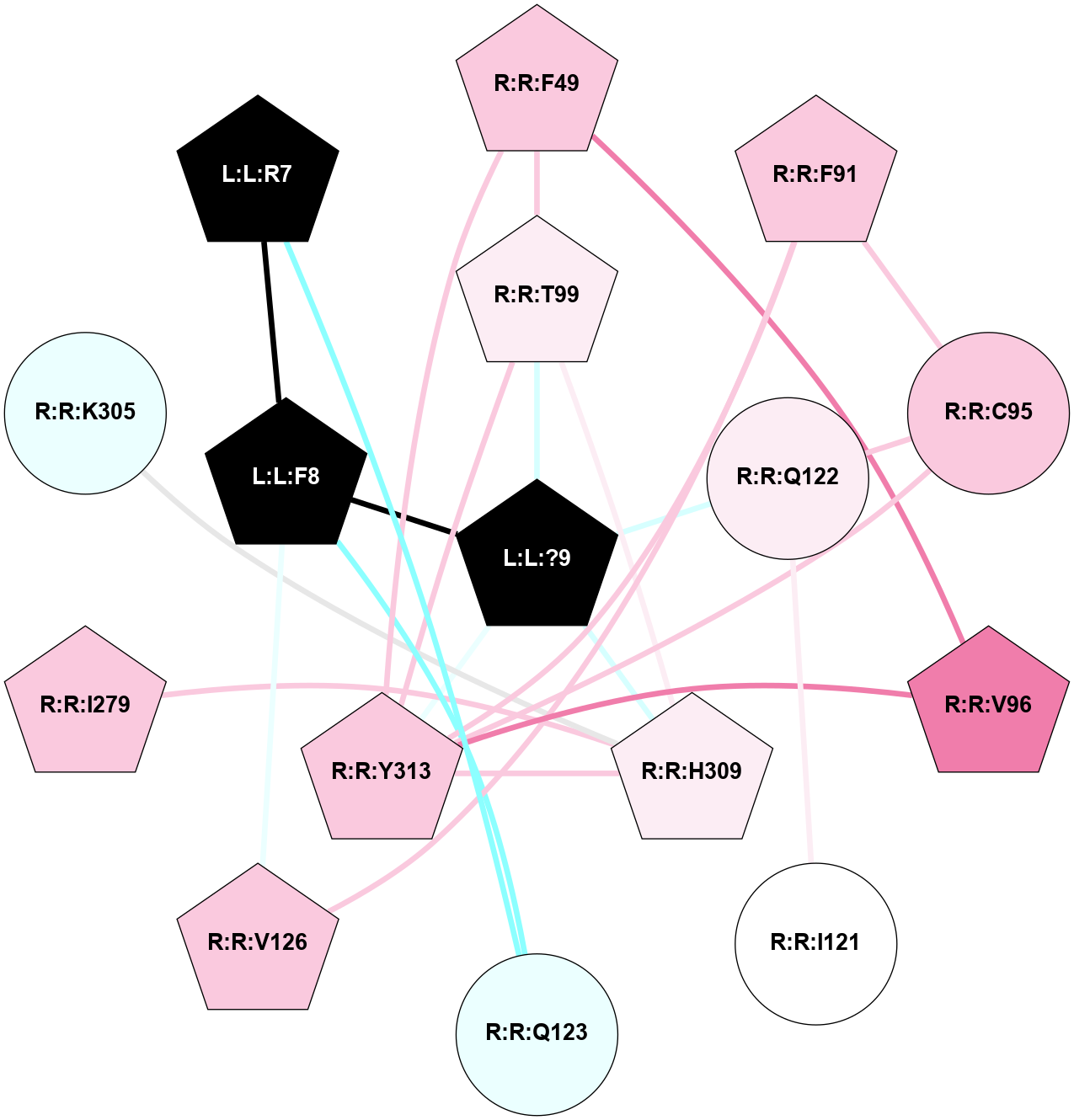
A 2D representation of the interactions of NH2 in 8XGS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F49 | R:R:V96 | 2.62 | 2 | Yes | Yes | 7 | 8 | 2 | 2 | | R:R:F49 | R:R:T99 | 2.59 | 2 | Yes | Yes | 7 | 6 | 2 | 1 | | R:R:F49 | R:R:Y313 | 7.22 | 2 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:C95 | R:R:F91 | 6.98 | 2 | No | Yes | 7 | 7 | 2 | 2 | | R:R:F91 | R:R:V126 | 2.62 | 2 | Yes | Yes | 7 | 7 | 2 | 2 | | R:R:F91 | R:R:Y313 | 4.13 | 2 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:C95 | R:R:Q122 | 12.21 | 2 | No | No | 7 | 6 | 2 | 1 | | R:R:C95 | R:R:Y313 | 6.72 | 2 | No | Yes | 7 | 7 | 2 | 1 | | R:R:V96 | R:R:Y313 | 5.05 | 2 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:H309 | R:R:T99 | 6.85 | 2 | Yes | Yes | 6 | 6 | 1 | 1 | | R:R:T99 | R:R:Y313 | 3.75 | 2 | Yes | Yes | 6 | 7 | 1 | 1 | | L:L:?9 | R:R:T99 | 10.94 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:I121 | R:R:Q122 | 2.74 | 0 | No | No | 5 | 6 | 2 | 1 | | L:L:?9 | R:R:Q122 | 3.29 | 2 | Yes | No | 0 | 6 | 0 | 1 | | L:L:R7 | R:R:Q123 | 4.67 | 2 | Yes | No | 0 | 4 | 2 | 2 | | L:L:F8 | R:R:Q123 | 7.03 | 2 | Yes | No | 0 | 4 | 1 | 2 | | L:L:F8 | R:R:V126 | 5.24 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | R:R:H309 | R:R:I279 | 2.65 | 2 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:H309 | R:R:K305 | 13.1 | 2 | Yes | No | 6 | 4 | 1 | 2 | | R:R:H309 | R:R:Y313 | 9.8 | 2 | Yes | Yes | 6 | 7 | 1 | 1 | | L:L:?9 | R:R:H309 | 9.55 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?9 | R:R:Y313 | 2.9 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:F8 | L:L:R7 | 7.48 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?9 | L:L:F8 | 3.01 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 5.94 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 24.00 | | Average Links Mediated by Hubs In Shell | 22.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8XGU | A | Peptide | Kisspeptin | Kiss1 | Homo Sapiens | TAK448 | - | Gi1/Beta1/Gamma2 | 3 | 2024-10-30 | doi.org/10.1126/sciadv.adn7771 |
|
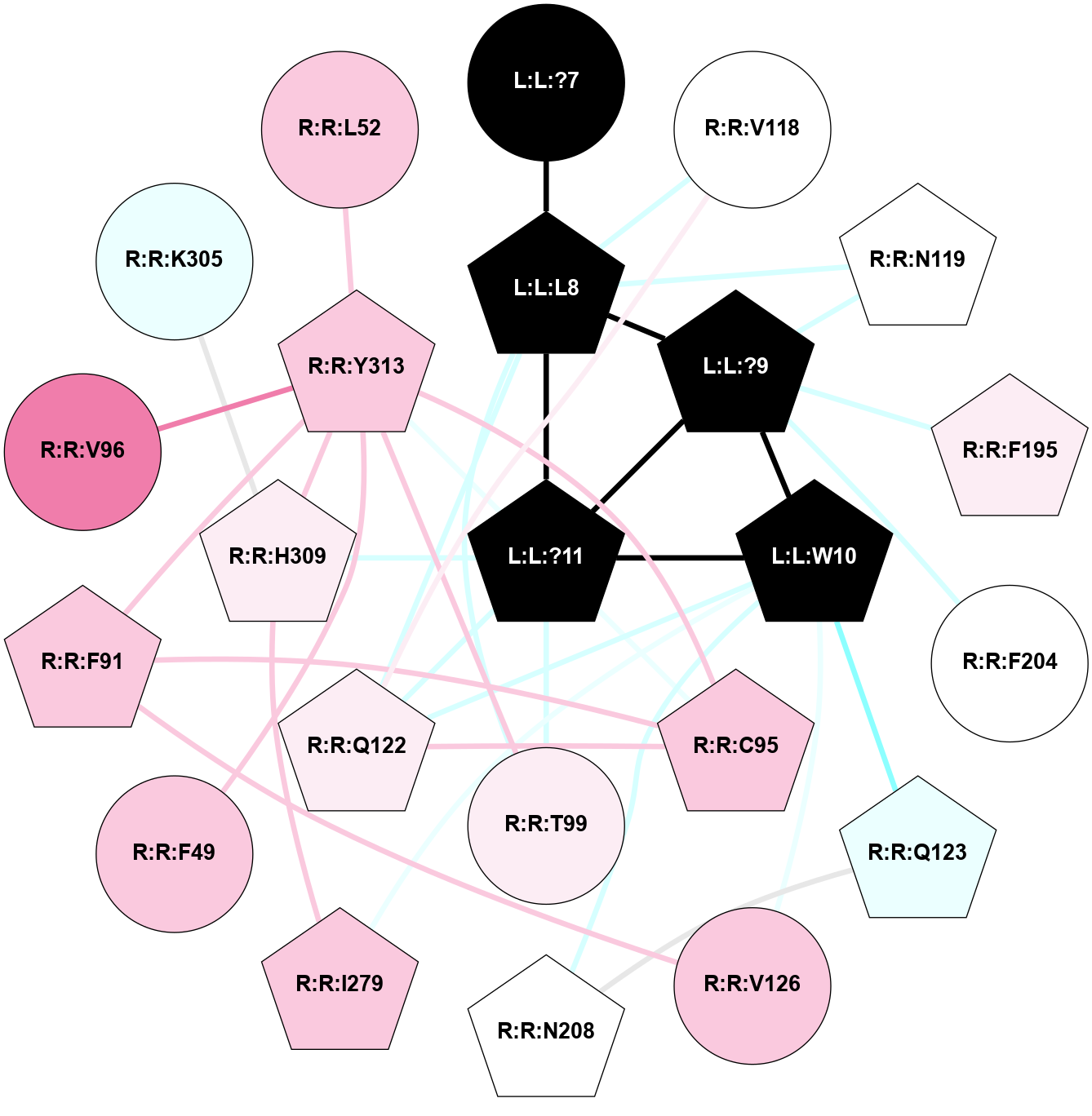
A 2D representation of the interactions of NH2 in 8XGU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?7 | L:L:L8 | 8.02 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:?9 | L:L:L8 | 3.29 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?11 | L:L:L8 | 10.28 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:L8 | R:R:T99 | 4.42 | 2 | Yes | No | 0 | 6 | 1 | 1 | | L:L:L8 | R:R:V118 | 2.98 | 2 | Yes | No | 0 | 5 | 1 | 2 | | L:L:L8 | R:R:N119 | 6.87 | 2 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:L8 | R:R:Q122 | 3.99 | 2 | Yes | Yes | 0 | 6 | 1 | 1 | | L:L:?9 | L:L:W10 | 17.16 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?11 | L:L:?9 | 8.15 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?9 | R:R:N119 | 3.27 | 2 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:?9 | R:R:F195 | 4.83 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?9 | R:R:F204 | 11.59 | 2 | Yes | No | 0 | 5 | 1 | 2 | | L:L:?11 | L:L:W10 | 2.82 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:W10 | R:R:Q122 | 3.29 | 2 | Yes | Yes | 0 | 6 | 1 | 1 | | L:L:W10 | R:R:Q123 | 8.76 | 2 | Yes | Yes | 0 | 4 | 1 | 2 | | L:L:W10 | R:R:V126 | 12.26 | 2 | Yes | No | 0 | 7 | 1 | 2 | | L:L:W10 | R:R:N208 | 7.91 | 2 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:W10 | R:R:I279 | 3.52 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?11 | R:R:C95 | 3.93 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?11 | R:R:T99 | 10.94 | 2 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?11 | R:R:Q122 | 6.59 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?11 | R:R:H309 | 9.55 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?11 | R:R:Y313 | 2.9 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:F49 | R:R:Y313 | 4.13 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:C95 | R:R:F91 | 6.98 | 2 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:F91 | R:R:V126 | 2.62 | 2 | Yes | No | 7 | 7 | 2 | 2 | | R:R:F91 | R:R:Y313 | 3.09 | 2 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:C95 | R:R:Q122 | 6.1 | 2 | Yes | Yes | 7 | 6 | 1 | 1 | | R:R:C95 | R:R:Y313 | 6.72 | 2 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:V96 | R:R:Y313 | 6.31 | 0 | No | Yes | 8 | 7 | 2 | 1 | | R:R:T99 | R:R:Y313 | 4.99 | 2 | No | Yes | 6 | 7 | 1 | 1 | | R:R:Q122 | R:R:V118 | 2.87 | 2 | Yes | No | 6 | 5 | 1 | 2 | | R:R:N208 | R:R:Q123 | 3.96 | 2 | Yes | Yes | 5 | 4 | 2 | 2 | | R:R:H309 | R:R:I279 | 2.65 | 2 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:H309 | R:R:K305 | 13.1 | 2 | Yes | No | 6 | 4 | 1 | 2 | | R:R:H309 | R:R:Y313 | 10.89 | 2 | Yes | Yes | 6 | 7 | 1 | 1 | | R:R:L52 | R:R:Y313 | 2.34 | 0 | No | Yes | 7 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 8.00 | | Average Number Of Links With An Hub | 7.00 | | Average Interaction Strength | 6.89 | | Average Nodes In Shell | 23.00 | | Average Hubs In Shell | 14.00 | | Average Links In Shell | 37.00 | | Average Links Mediated by Hubs In Shell | 37.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8ZH8 | A | Peptide | P518 | QRFP | Homo Sapiens | QRF-Amide | - | Gq/Beta1/Gamma1 | 3.19 | 2024-07-03 | doi.org/10.1038/s41467-024-49030-5 |
|
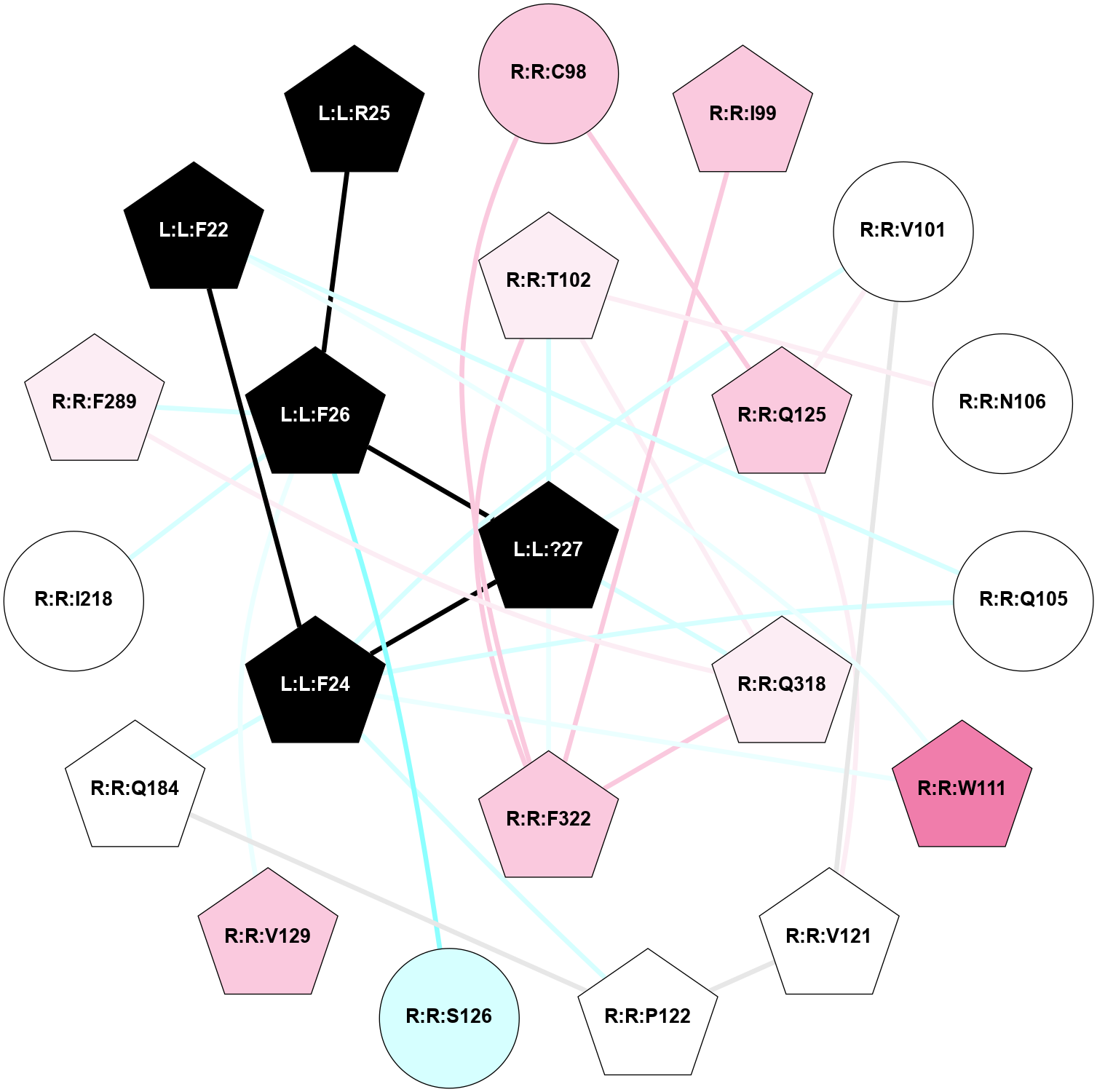
A 2D representation of the interactions of NH2 in 8ZH8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:C98 | R:R:Q125 | 6.1 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:C98 | R:R:F322 | 4.19 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:F322 | R:R:I99 | 5.02 | 1 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:V101 | R:R:V121 | 6.41 | 1 | No | Yes | 5 | 5 | 2 | 2 | | R:R:Q125 | R:R:V101 | 4.3 | 1 | Yes | No | 7 | 5 | 1 | 2 | | L:L:F24 | R:R:V101 | 6.55 | 1 | Yes | No | 0 | 5 | 1 | 2 | | R:R:N106 | R:R:T102 | 2.92 | 0 | No | Yes | 5 | 6 | 2 | 1 | | R:R:Q318 | R:R:T102 | 7.09 | 1 | Yes | Yes | 6 | 6 | 1 | 1 | | R:R:F322 | R:R:T102 | 6.49 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | L:L:?27 | R:R:T102 | 3.67 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:F22 | R:R:Q105 | 4.68 | 1 | Yes | No | 0 | 5 | 2 | 2 | | L:L:F24 | R:R:Q105 | 11.71 | 1 | Yes | No | 0 | 5 | 1 | 2 | | L:L:F22 | R:R:W111 | 15.03 | 1 | Yes | Yes | 0 | 8 | 2 | 2 | | L:L:F24 | R:R:W111 | 5.01 | 1 | Yes | Yes | 0 | 8 | 1 | 2 | | R:R:P122 | R:R:V121 | 3.53 | 1 | Yes | Yes | 5 | 5 | 2 | 2 | | R:R:Q125 | R:R:V121 | 4.3 | 1 | Yes | Yes | 7 | 5 | 1 | 2 | | R:R:P122 | R:R:Q184 | 12.63 | 1 | Yes | Yes | 5 | 5 | 2 | 2 | | L:L:F24 | R:R:P122 | 7.22 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:?27 | R:R:Q125 | 6.62 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:F26 | R:R:S126 | 6.61 | 0 | Yes | No | 0 | 3 | 1 | 2 | | L:L:F26 | R:R:V129 | 6.55 | 0 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:F24 | R:R:Q184 | 10.54 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:F26 | R:R:I218 | 8.79 | 0 | Yes | No | 0 | 5 | 1 | 2 | | R:R:F289 | R:R:Q318 | 3.51 | 1 | Yes | Yes | 6 | 6 | 2 | 1 | | L:L:F26 | R:R:F289 | 5.36 | 0 | Yes | Yes | 0 | 6 | 1 | 2 | | R:R:F322 | R:R:Q318 | 4.68 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | L:L:?27 | R:R:Q318 | 9.93 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?27 | R:R:F322 | 3.03 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:F22 | L:L:F24 | 4.29 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?27 | L:L:F24 | 3.03 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:F26 | L:L:R25 | 12.83 | 0 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?27 | L:L:F26 | 3.03 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 4.89 | | Average Nodes In Shell | 22.00 | | Average Hubs In Shell | 16.00 | | Average Links In Shell | 32.00 | | Average Links Mediated by Hubs In Shell | 32.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8ZJE | A | Peptide | Kisspeptin | Kiss1 | Homo Sapiens | TAK-448 | - | chim(NtGi1-Gq)/Beta1/Gamma1 | 3.07 | 2024-07-03 | doi.org/10.1016/j.celrep.2024.114389 |
|
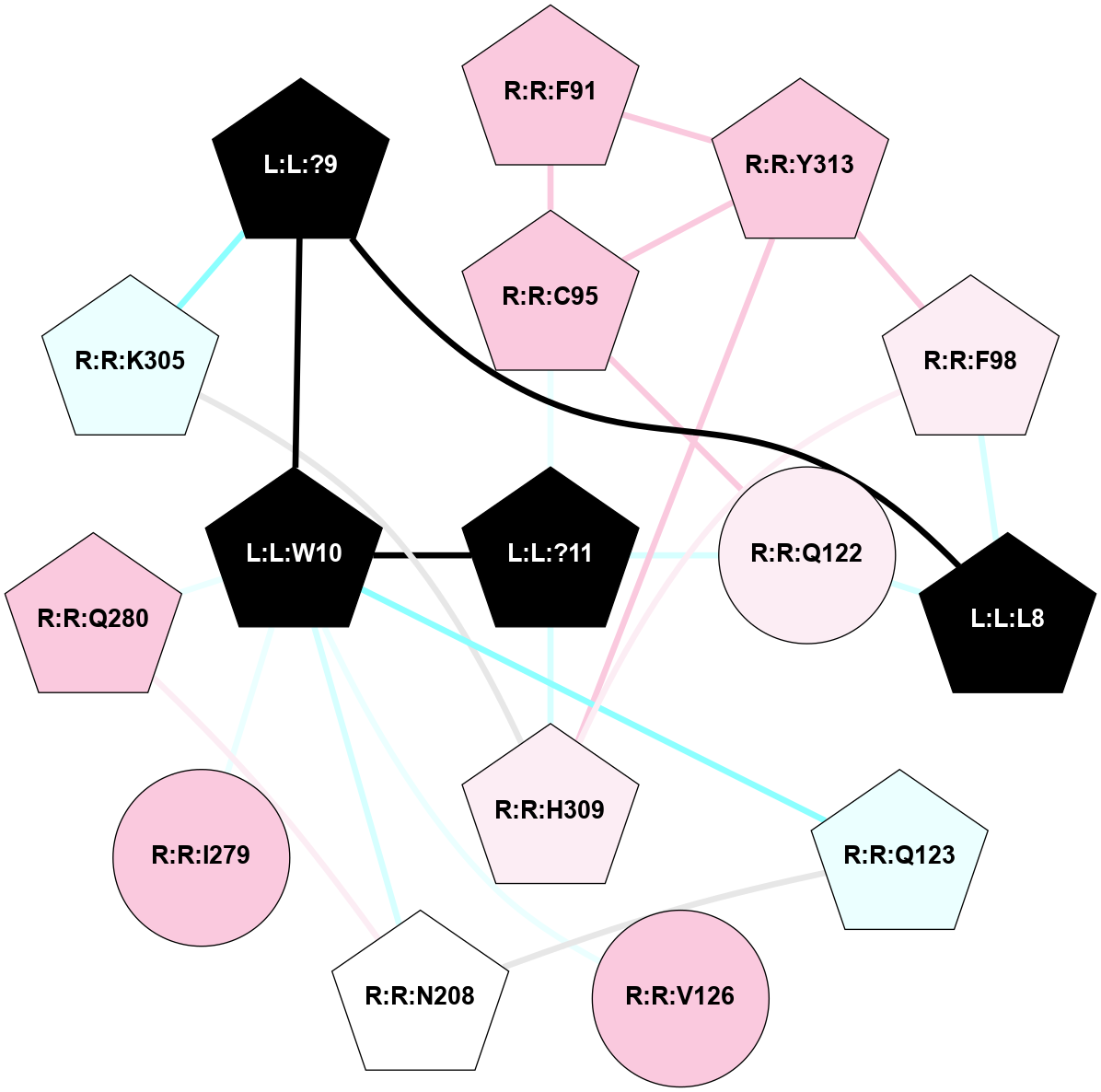
A 2D representation of the interactions of NH2 in 8ZJE
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:C95 | R:R:F91 | 4.19 | 1 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:F91 | R:R:Y313 | 3.09 | 1 | Yes | Yes | 7 | 7 | 2 | 2 | | R:R:C95 | R:R:Q122 | 6.1 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:C95 | R:R:Y313 | 6.72 | 1 | Yes | Yes | 7 | 7 | 1 | 2 | | L:L:?11 | R:R:C95 | 3.95 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:F98 | R:R:H309 | 14.71 | 1 | Yes | Yes | 6 | 6 | 2 | 1 | | R:R:F98 | R:R:Y313 | 4.13 | 1 | Yes | Yes | 6 | 7 | 2 | 2 | | L:L:L8 | R:R:F98 | 3.65 | 1 | Yes | Yes | 0 | 6 | 2 | 2 | | L:L:L8 | R:R:Q122 | 7.99 | 1 | Yes | No | 0 | 6 | 2 | 1 | | L:L:?11 | R:R:Q122 | 9.93 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:N208 | R:R:Q123 | 5.28 | 1 | Yes | Yes | 5 | 4 | 2 | 2 | | L:L:W10 | R:R:Q123 | 9.86 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | L:L:W10 | R:R:V126 | 15.94 | 1 | Yes | No | 0 | 7 | 1 | 2 | | R:R:N208 | R:R:Q280 | 6.6 | 1 | Yes | Yes | 5 | 7 | 2 | 2 | | L:L:W10 | R:R:N208 | 7.91 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:W10 | R:R:I279 | 8.22 | 1 | Yes | No | 0 | 7 | 1 | 2 | | L:L:W10 | R:R:Q280 | 4.38 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | R:R:H309 | R:R:K305 | 11.79 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | L:L:?9 | R:R:K305 | 3.29 | 1 | Yes | Yes | 0 | 4 | 2 | 2 | | R:R:H309 | R:R:Y313 | 5.44 | 1 | Yes | Yes | 6 | 7 | 1 | 2 | | L:L:?11 | R:R:H309 | 6.4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?9 | L:L:L8 | 3.23 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?9 | L:L:W10 | 12.39 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?11 | L:L:W10 | 8.5 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 7.20 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 24.00 | | Average Links Mediated by Hubs In Shell | 24.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8ZPS | A | Peptide | Prolactin-releasing peptide | PrRP | Homo sapiens | PrRP20 | - | chim(NtGi1-Gs-CtGq)/Beta1/Gamma2 | 2.97 | 2024-09-25 | doi.org/10.1038/s41421-024-00724-6 |
|

A 2D representation of the interactions of NH2 in 8ZPS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:F20 | L:L:R19 | 12.83 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:R19 | R:R:N298 | 2.41 | 2 | Yes | No | 0 | 6 | 2 | 2 | | L:L:?21 | L:L:F20 | 6.03 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:F20 | R:R:P142 | 2.89 | 2 | Yes | No | 0 | 5 | 1 | 2 | | L:L:F20 | R:R:V145 | 7.87 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:F20 | R:R:Y146 | 5.16 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:F20 | R:R:L294 | 2.44 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:F20 | R:R:N298 | 3.62 | 2 | Yes | No | 0 | 6 | 1 | 2 | | L:L:?21 | R:R:C113 | 7.86 | 2 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?21 | R:R:Q141 | 16.47 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?21 | R:R:H321 | 3.18 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:H321 | R:R:Y67 | 8.71 | 2 | Yes | Yes | 5 | 8 | 1 | 2 | | R:R:C113 | R:R:Q141 | 7.63 | 2 | No | Yes | 7 | 6 | 1 | 1 | | R:R:C113 | R:R:H321 | 2.95 | 2 | No | Yes | 7 | 5 | 1 | 1 | | R:R:L116 | R:R:T117 | 2.95 | 2 | No | No | 6 | 6 | 2 | 2 | | R:R:L116 | R:R:V137 | 5.96 | 2 | No | No | 6 | 6 | 2 | 2 | | R:R:L116 | R:R:Q141 | 3.99 | 2 | No | Yes | 6 | 6 | 2 | 1 | | R:R:H321 | R:R:T117 | 2.74 | 2 | Yes | No | 5 | 6 | 1 | 2 | | R:R:Q141 | R:R:V137 | 5.73 | 2 | Yes | No | 6 | 6 | 1 | 2 | | R:R:P142 | R:R:Y146 | 5.56 | 2 | No | Yes | 5 | 7 | 2 | 2 | | R:R:V145 | R:R:Y146 | 2.52 | 2 | Yes | Yes | 7 | 7 | 2 | 2 | | R:R:L294 | R:R:N298 | 5.49 | 2 | Yes | No | 7 | 6 | 2 | 2 | | R:R:H321 | R:R:M325 | 7.88 | 2 | Yes | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 8.38 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 23.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8ZPT | A | Peptide | Prolactin-releasing peptide | PrRP | Homo sapiens | PrRP20 | - | chim(NtGi1-Gs-CtGq)/Beta1/Gamma2 | 2.96 | 2024-09-25 | doi.org/10.1038/s41421-024-00724-6 |
|
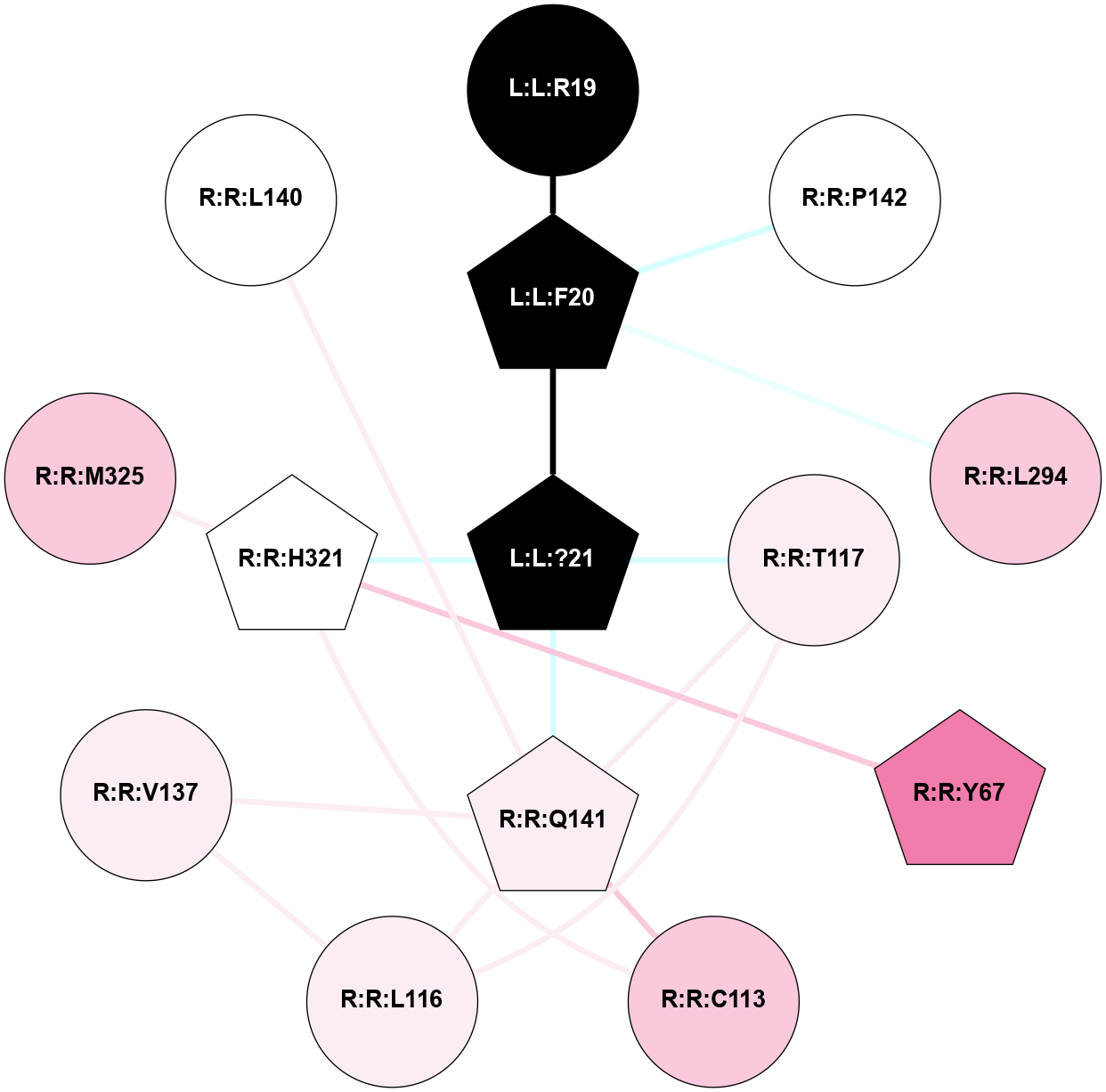
A 2D representation of the interactions of NH2 in 8ZPT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:F20 | L:L:R19 | 12.83 | 0 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?21 | L:L:F20 | 6.03 | 14 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:F20 | R:R:P142 | 5.78 | 0 | Yes | No | 0 | 5 | 1 | 2 | | L:L:F20 | R:R:L294 | 3.65 | 0 | Yes | No | 0 | 7 | 1 | 2 | | L:L:?21 | R:R:T117 | 10.94 | 14 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?21 | R:R:Q141 | 6.59 | 14 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?21 | R:R:H321 | 9.55 | 14 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:H321 | R:R:Y67 | 8.71 | 0 | Yes | Yes | 5 | 8 | 1 | 2 | | R:R:C113 | R:R:Q141 | 10.68 | 0 | No | Yes | 7 | 6 | 2 | 1 | | R:R:C113 | R:R:H321 | 2.95 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:L116 | R:R:T117 | 2.95 | 14 | No | No | 6 | 6 | 2 | 1 | | R:R:L116 | R:R:V137 | 7.45 | 14 | No | No | 6 | 6 | 2 | 2 | | R:R:L116 | R:R:Q141 | 3.99 | 14 | No | Yes | 6 | 6 | 2 | 1 | | R:R:Q141 | R:R:T117 | 4.25 | 14 | Yes | No | 6 | 6 | 1 | 1 | | R:R:Q141 | R:R:V137 | 5.73 | 14 | Yes | No | 6 | 6 | 1 | 2 | | R:R:H321 | R:R:M325 | 11.82 | 0 | Yes | No | 5 | 7 | 1 | 2 | | R:R:L140 | R:R:Q141 | 2.66 | 0 | No | Yes | 5 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 8.28 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9BKJ | A | Peptide | Cholecystokinin | CCKA | Homo Sapiens | Cholecystokinin-8 | - | chim(NtGi1-Gs-CtGq)/Beta1/Gamma2 | 2.59 | 2024-05-22 | doi.org/10.1371/journal.pbio.3002673 |
|

A 2D representation of the interactions of NH2 in 9BKJ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?2 | L:L:M6 | 5.07 | 3 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?101 | L:L:M6 | 3.5 | 3 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:M6 | R:R:N98 | 4.21 | 3 | Yes | Yes | 0 | 7 | 1 | 1 | | L:L:M6 | R:R:C196 | 3.24 | 3 | Yes | No | 0 | 9 | 1 | 2 | | L:L:?101 | L:L:F8 | 3.01 | 3 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:F8 | R:R:G122 | 3.01 | 0 | Yes | No | 0 | 6 | 1 | 2 | | L:L:F8 | R:R:Y176 | 6.19 | 0 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:F8 | R:R:F330 | 5.36 | 0 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?101 | R:R:N98 | 10.2 | 3 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?101 | R:R:M121 | 3.5 | 3 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?101 | R:R:L356 | 10.28 | 3 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:C94 | R:R:M121 | 8.1 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:C94 | R:R:Y360 | 6.72 | 0 | No | Yes | 7 | 7 | 2 | 2 | | R:R:M121 | R:R:N98 | 4.21 | 3 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:L356 | R:R:N98 | 4.12 | 3 | Yes | Yes | 6 | 7 | 1 | 1 | | R:R:N98 | R:R:Y360 | 9.3 | 3 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:M121 | R:R:T118 | 3.01 | 3 | Yes | No | 7 | 6 | 1 | 2 | | R:R:T118 | R:R:Y176 | 6.24 | 0 | No | Yes | 6 | 7 | 2 | 2 | | R:R:I329 | R:R:L356 | 4.28 | 3 | Yes | Yes | 7 | 6 | 2 | 1 | | R:R:I329 | R:R:S359 | 3.1 | 3 | Yes | Yes | 7 | 7 | 2 | 2 | | R:R:L356 | R:R:S359 | 3 | 3 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:L356 | R:R:Y360 | 8.21 | 3 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:S359 | R:R:Y360 | 6.36 | 3 | Yes | Yes | 7 | 7 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 6.10 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 23.00 | | Average Links Mediated by Hubs In Shell | 23.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9BKK | A | Peptide | Cholecystokinin | CCKA | Homo Sapiens | Cholecystokinin-8 | - | chim(NtGi1-Gs-CtGq)/Beta1/Gamma2 | 2.51 | 2024-05-22 | doi.org/10.1371/journal.pbio.3002673 |
|

A 2D representation of the interactions of NH2 in 9BKK
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?2 | L:L:M6 | 3.04 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?9 | L:L:M6 | 3.5 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:M6 | R:R:N98 | 7.01 | 2 | Yes | Yes | 0 | 7 | 1 | 1 | | L:L:M6 | R:R:M121 | 4.33 | 2 | Yes | Yes | 0 | 7 | 1 | 1 | | L:L:M6 | R:R:C196 | 4.86 | 2 | Yes | Yes | 0 | 9 | 1 | 2 | | L:L:D7 | L:L:F8 | 3.58 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?9 | L:L:F8 | 3.01 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:F8 | R:R:G122 | 3.01 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:F8 | R:R:V125 | 7.87 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:F8 | R:R:L213 | 3.65 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:F8 | R:R:F330 | 5.36 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?9 | R:R:N98 | 10.2 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?9 | R:R:M121 | 3.5 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?9 | R:R:L356 | 6.85 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?9 | R:R:Y360 | 2.9 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:Y360 | R:R:Y48 | 4.96 | 2 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:C94 | R:R:M121 | 8.1 | 2 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:C94 | R:R:Y360 | 8.06 | 2 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:M95 | R:R:Y360 | 8.38 | 2 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:M121 | R:R:N98 | 4.21 | 2 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:N98 | R:R:Y360 | 9.3 | 2 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:M121 | R:R:T118 | 4.52 | 2 | Yes | No | 7 | 6 | 1 | 2 | | R:R:G122 | R:R:M121 | 1.75 | 2 | Yes | Yes | 6 | 7 | 2 | 1 | | R:R:G122 | R:R:V125 | 1.84 | 2 | Yes | Yes | 6 | 7 | 2 | 2 | | R:R:F330 | R:R:I329 | 2.51 | 2 | Yes | Yes | 7 | 7 | 2 | 2 | | R:R:I329 | R:R:L356 | 4.28 | 2 | Yes | Yes | 7 | 6 | 2 | 1 | | R:R:I329 | R:R:S359 | 4.64 | 2 | Yes | No | 7 | 7 | 2 | 2 | | R:R:I352 | R:R:L356 | 2.85 | 0 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:L356 | R:R:Y360 | 8.21 | 2 | Yes | Yes | 6 | 7 | 1 | 1 | | R:R:S359 | R:R:Y360 | 3.82 | 2 | No | Yes | 7 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 6.00 | | Average Interaction Strength | 4.99 | | Average Nodes In Shell | 21.00 | | Average Hubs In Shell | 19.00 | | Average Links In Shell | 30.00 | | Average Links Mediated by Hubs In Shell | 30.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9BLB | B1 | Peptide | Calcitonin | CT | Homo Sapiens | Cagrilintide backbone (non-acylated) | - | Gs/Beta1/Gamma2 | 3.2 | 2025-04-16 | To be published |
|
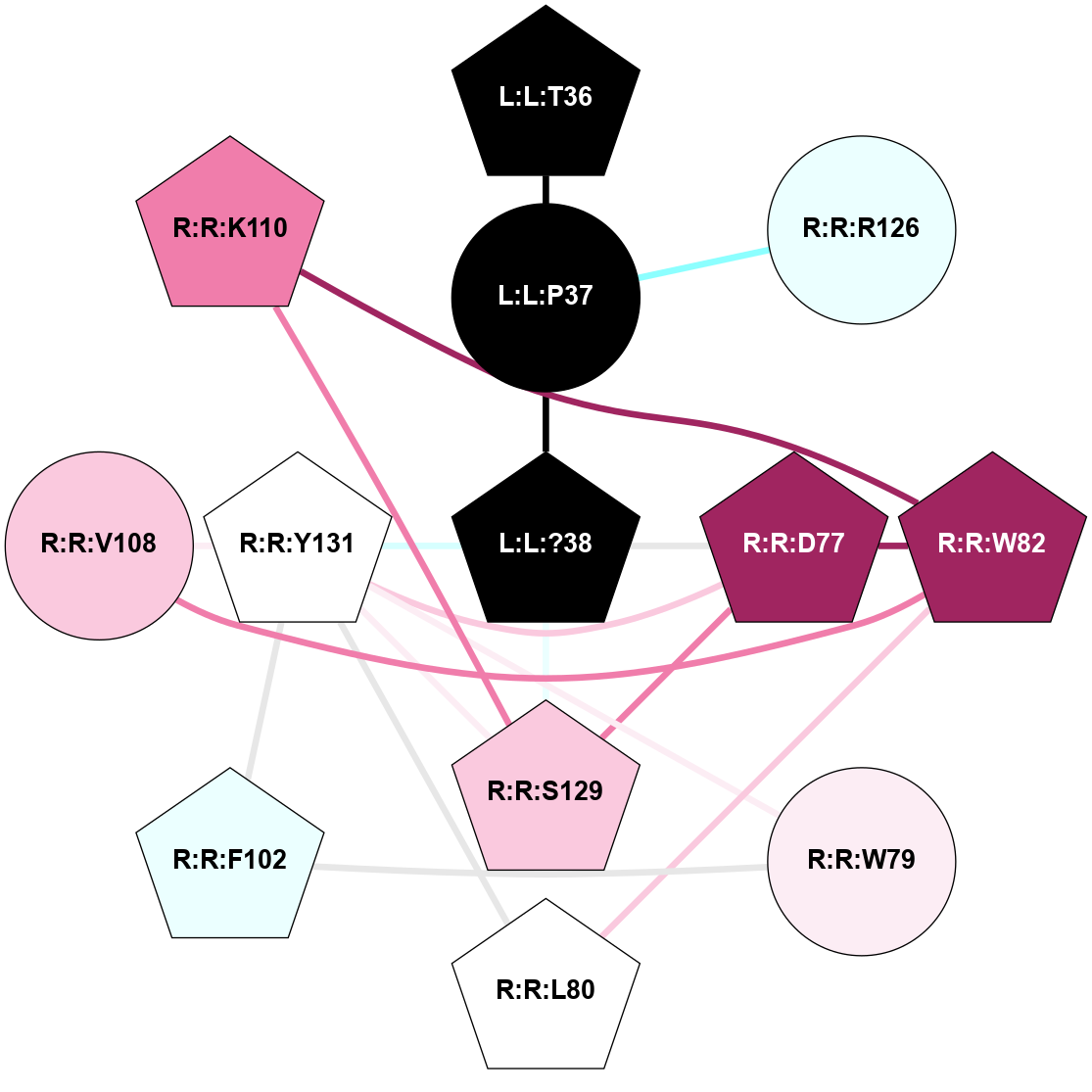
A 2D representation of the interactions of NH2 in 9BLB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:P37 | L:L:T36 | 3.5 | 0 | No | Yes | 0 | 0 | 1 | 2 | | L:L:?38 | L:L:P37 | 8.13 | 4 | Yes | No | 0 | 0 | 0 | 1 | | L:L:P37 | R:R:R126 | 2.88 | 0 | No | No | 0 | 4 | 1 | 2 | | L:L:?38 | R:R:D77 | 3.36 | 4 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?38 | R:R:S129 | 7.43 | 4 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?38 | R:R:Y131 | 5.8 | 4 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:W82 | 3.35 | 4 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D77 | R:R:S129 | 2.94 | 4 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:D77 | R:R:Y131 | 3.45 | 4 | Yes | Yes | 9 | 5 | 1 | 1 | | R:R:F102 | R:R:W79 | 5.01 | 4 | Yes | No | 4 | 6 | 2 | 2 | | R:R:W79 | R:R:Y131 | 15.43 | 4 | No | Yes | 6 | 5 | 2 | 1 | | R:R:L80 | R:R:W82 | 3.42 | 4 | Yes | Yes | 5 | 9 | 2 | 2 | | R:R:L80 | R:R:Y131 | 5.86 | 4 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:V108 | R:R:W82 | 3.68 | 0 | No | Yes | 7 | 9 | 2 | 2 | | R:R:K110 | R:R:W82 | 5.8 | 4 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:F102 | R:R:Y131 | 5.16 | 4 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:V108 | R:R:Y131 | 5.05 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:K110 | R:R:S129 | 9.18 | 4 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:S129 | R:R:Y131 | 3.82 | 4 | Yes | Yes | 7 | 5 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 6.18 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 19.00 | | Average Links Mediated by Hubs In Shell | 18.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9BLC | B1 | Peptide | Calcitonin | CT | Homo Sapiens | Cagrilintide backbone (non-acylated) | - | Gs/Beta1/Gamma2 | 3.3 | 2025-04-16 | To be published |
|
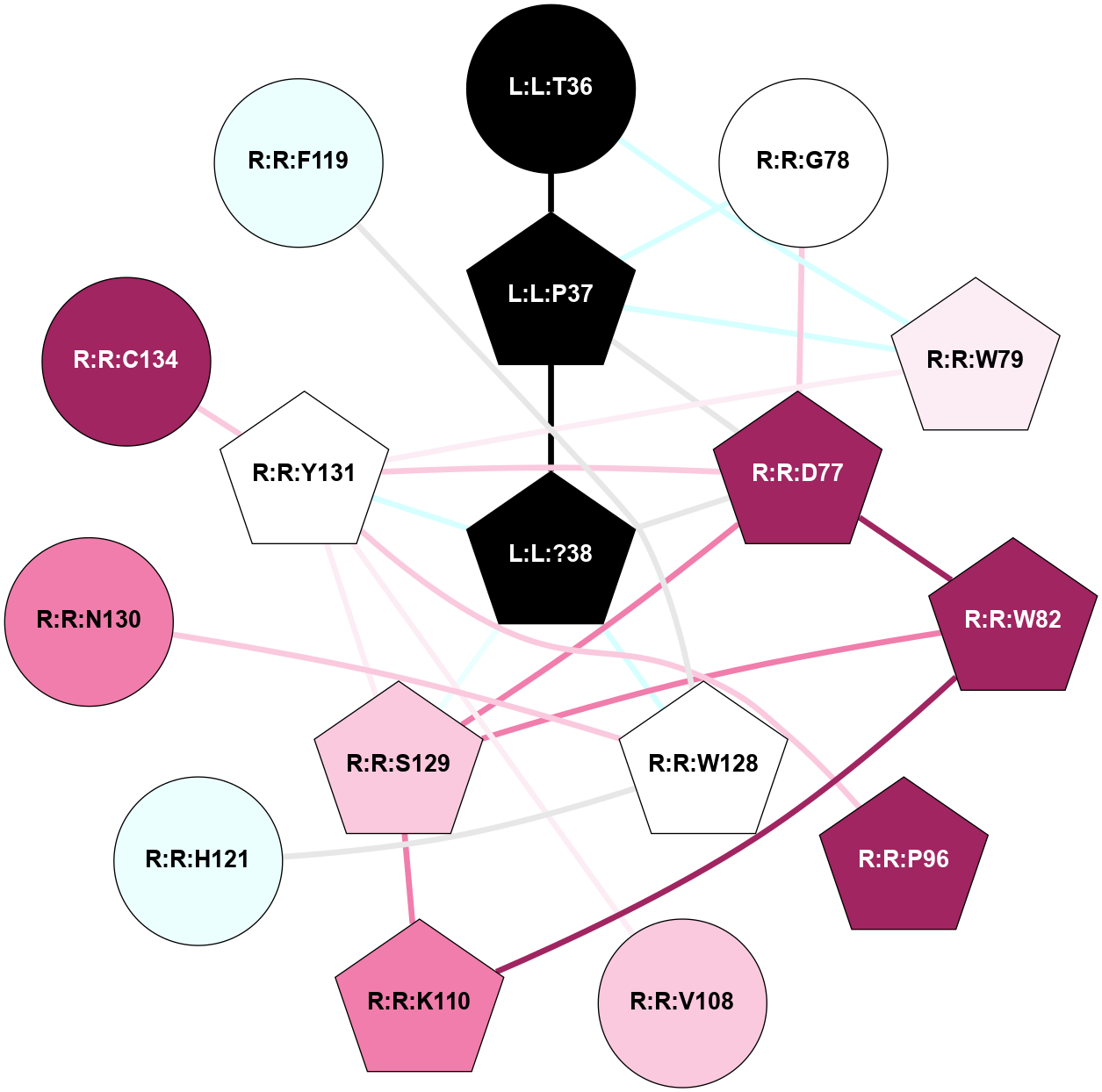
A 2D representation of the interactions of NH2 in 9BLC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:P37 | L:L:T36 | 3.5 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:T36 | R:R:W79 | 4.85 | 1 | No | Yes | 0 | 6 | 2 | 2 | | L:L:?38 | L:L:P37 | 4.06 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:P37 | R:R:D77 | 6.44 | 1 | Yes | Yes | 0 | 9 | 1 | 1 | | L:L:P37 | R:R:G78 | 4.06 | 1 | Yes | No | 0 | 5 | 1 | 2 | | L:L:P37 | R:R:W79 | 25.67 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?38 | R:R:D77 | 3.36 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?38 | R:R:W128 | 8.46 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?38 | R:R:S129 | 7.43 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?38 | R:R:Y131 | 5.8 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:G78 | 5.03 | 1 | Yes | No | 9 | 5 | 1 | 2 | | R:R:D77 | R:R:W82 | 5.58 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D77 | R:R:S129 | 2.94 | 1 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:D77 | R:R:Y131 | 3.45 | 1 | Yes | Yes | 9 | 5 | 1 | 1 | | R:R:W79 | R:R:Y131 | 13.5 | 1 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:K110 | R:R:W82 | 6.96 | 1 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:S129 | R:R:W82 | 4.94 | 1 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:P96 | R:R:Y131 | 4.17 | 1 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:V108 | R:R:Y131 | 13.88 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:K110 | R:R:S129 | 6.12 | 1 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:H121 | R:R:W128 | 19.04 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 7.91 | 0 | No | Yes | 8 | 5 | 2 | 1 | | R:R:S129 | R:R:Y131 | 7.63 | 1 | Yes | Yes | 7 | 5 | 1 | 1 | | R:R:C134 | R:R:Y131 | 4.03 | 1 | No | Yes | 9 | 5 | 2 | 1 | | R:R:F119 | R:R:W128 | 2 | 0 | No | Yes | 4 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 5.82 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 25.00 | | Average Links Mediated by Hubs In Shell | 25.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9BLW | B1 | Peptide | Calcitonin | CT (AMY1) | Homo Sapiens | Cagrilintide backbone (non-acylated) | - | Gs/Beta1/Gamma2; RAMP1 | 3.2 | 2025-04-16 | To be published |
|
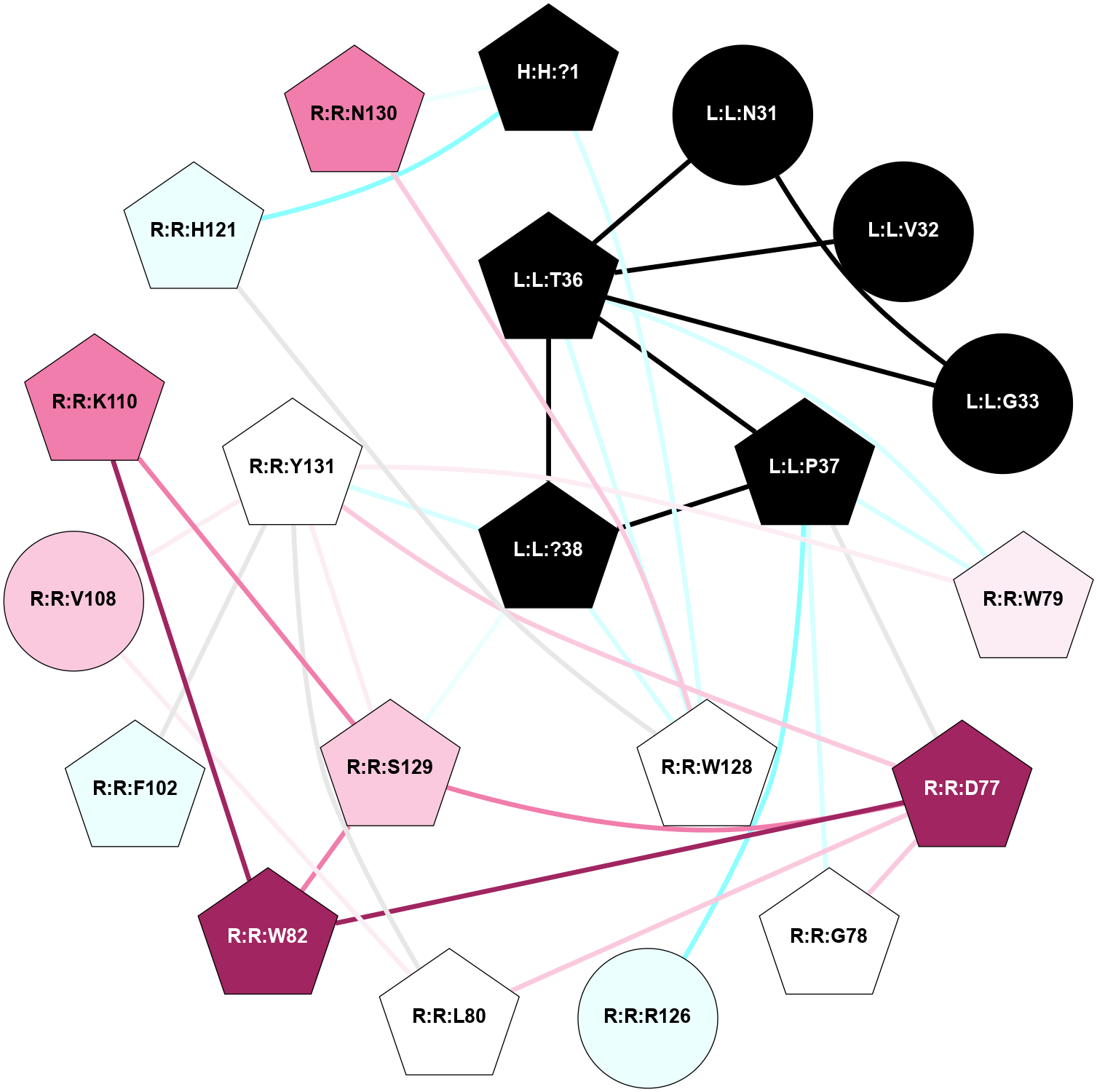
A 2D representation of the interactions of NH2 in 9BLW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:H121 | 5.77 | 2 | Yes | Yes | 0 | 4 | 2 | 2 | | H:H:?1 | R:R:W128 | 10.22 | 2 | Yes | Yes | 0 | 5 | 2 | 1 | | H:H:?1 | R:R:N130 | 30.8 | 2 | Yes | Yes | 0 | 8 | 2 | 2 | | L:L:G33 | L:L:N31 | 5.09 | 2 | No | No | 0 | 0 | 2 | 2 | | L:L:N31 | L:L:T36 | 4.39 | 2 | No | Yes | 0 | 0 | 2 | 1 | | L:L:T36 | L:L:V32 | 3.17 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:G33 | L:L:T36 | 3.64 | 2 | No | Yes | 0 | 0 | 2 | 1 | | L:L:P37 | L:L:T36 | 3.5 | 2 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?38 | L:L:T36 | 3.65 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:T36 | R:R:W79 | 4.85 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:T36 | R:R:W128 | 4.85 | 2 | Yes | Yes | 0 | 5 | 1 | 1 | | L:L:?38 | L:L:P37 | 12.19 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:P37 | R:R:D77 | 4.83 | 2 | Yes | Yes | 0 | 9 | 1 | 2 | | L:L:P37 | R:R:G78 | 4.06 | 2 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:P37 | R:R:W79 | 21.62 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:P37 | R:R:R126 | 5.76 | 2 | Yes | No | 0 | 4 | 1 | 2 | | L:L:?38 | R:R:W128 | 11.28 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?38 | R:R:S129 | 7.43 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?38 | R:R:Y131 | 5.8 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:G78 | 5.03 | 2 | Yes | Yes | 9 | 5 | 2 | 2 | | R:R:D77 | R:R:L80 | 4.07 | 2 | Yes | Yes | 9 | 5 | 2 | 2 | | R:R:D77 | R:R:W82 | 11.17 | 2 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:D77 | R:R:S129 | 5.89 | 2 | Yes | Yes | 9 | 7 | 2 | 1 | | R:R:D77 | R:R:Y131 | 8.05 | 2 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:W79 | R:R:Y131 | 20.26 | 2 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:L80 | R:R:V108 | 4.47 | 2 | Yes | No | 5 | 7 | 2 | 2 | | R:R:L80 | R:R:Y131 | 8.21 | 2 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:K110 | R:R:W82 | 23.21 | 2 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:S129 | R:R:W82 | 4.94 | 2 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:F102 | R:R:Y131 | 3.09 | 2 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:V108 | R:R:Y131 | 15.14 | 2 | No | Yes | 7 | 5 | 2 | 1 | | R:R:K110 | R:R:S129 | 7.65 | 2 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:H121 | R:R:W128 | 16.93 | 2 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 10.17 | 2 | Yes | Yes | 8 | 5 | 2 | 1 | | R:R:S129 | R:R:Y131 | 6.36 | 2 | Yes | Yes | 7 | 5 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 8.07 | | Average Nodes In Shell | 21.00 | | Average Hubs In Shell | 16.00 | | Average Links In Shell | 35.00 | | Average Links Mediated by Hubs In Shell | 34.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9BUE | B1 | Peptide | Calcitonin | CT | Homo Sapiens | Cagrilintide | - | Gs/Beta1/Gamma2 | 3.6 | 2025-04-16 | To be published |
|
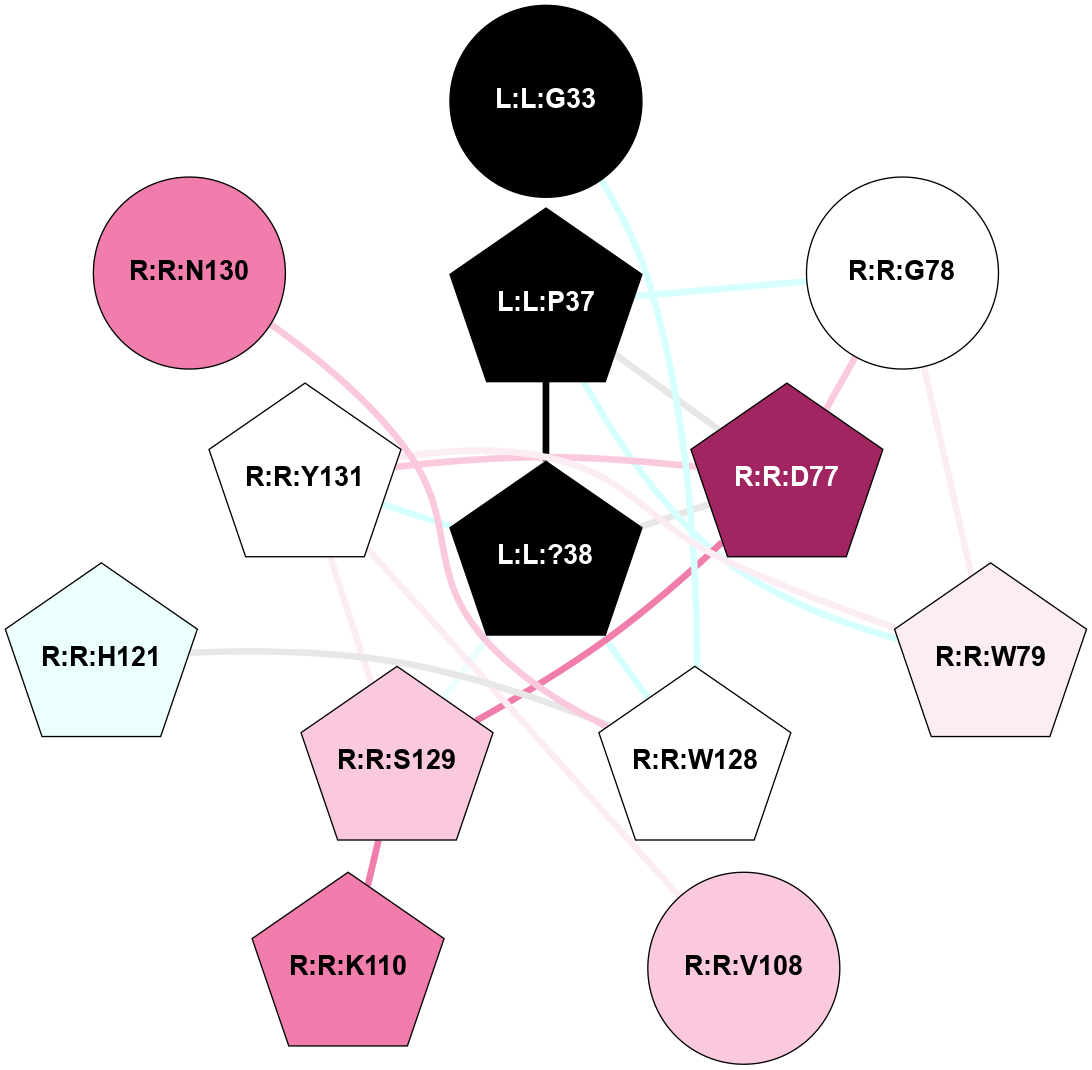
A 2D representation of the interactions of NH2 in 9BUE
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:G33 | R:R:W128 | 5.63 | 0 | No | Yes | 0 | 5 | 2 | 1 | | L:L:?38 | L:L:P37 | 8.13 | 5 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:P37 | R:R:D77 | 4.83 | 5 | Yes | Yes | 0 | 9 | 1 | 1 | | L:L:P37 | R:R:G78 | 4.06 | 5 | Yes | No | 0 | 5 | 1 | 2 | | L:L:P37 | R:R:W79 | 9.46 | 5 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?38 | R:R:D77 | 10.08 | 5 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?38 | R:R:W128 | 8.46 | 5 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?38 | R:R:S129 | 7.43 | 5 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?38 | R:R:Y131 | 11.61 | 5 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:G78 | 5.03 | 5 | Yes | No | 9 | 5 | 1 | 2 | | R:R:D77 | R:R:S129 | 5.89 | 5 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:D77 | R:R:Y131 | 9.2 | 5 | Yes | Yes | 9 | 5 | 1 | 1 | | R:R:G78 | R:R:W79 | 4.22 | 5 | No | Yes | 5 | 6 | 2 | 2 | | R:R:W79 | R:R:Y131 | 7.72 | 5 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:V108 | R:R:Y131 | 11.36 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:K110 | R:R:S129 | 4.59 | 5 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:H121 | R:R:W128 | 24.33 | 0 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 6.78 | 0 | No | Yes | 8 | 5 | 2 | 1 | | R:R:S129 | R:R:Y131 | 12.72 | 5 | Yes | Yes | 7 | 5 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 9.14 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 19.00 | | Average Links Mediated by Hubs In Shell | 19.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9IK9 | A | Peptide | Somatostatin | SST1 | Homo Sapiens | SST analogue | - | Gi1/Beta1/Gamma2 | 3.37 | 2025-04-30 | doi.org/10.1016/j.apsb.2025.03.043 |
|
A 2D representation of the interactions of NH2 in 9IK9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | L:L:?2 | 43.71 | 3 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?1 | L:L:?3 | 5.33 | 3 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?1 | L:L:?6 | 5.11 | 3 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?1 | L:L:?7 | 2.08 | 3 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?1 | L:L:?8 | 14.88 | 3 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?1 | L:L:?9 | 19.23 | 3 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?1 | R:R:D300 | 17.41 | 3 | Yes | No | 0 | 4 | 1 | 2 | | L:L:?2 | L:L:?3 | 16.61 | 3 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?2 | L:L:?4 | 3.09 | 3 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?2 | L:L:?6 | 5.97 | 3 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?2 | L:L:?7 | 18.26 | 3 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?2 | L:L:?8 | 3.73 | 3 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?2 | R:R:D300 | 15.27 | 3 | Yes | No | 0 | 4 | 2 | 2 | | L:L:?3 | L:L:?4 | 34.27 | 3 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:?3 | L:L:?6 | 11.21 | 3 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?3 | L:L:?7 | 2.08 | 3 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?4 | L:L:?6 | 13.15 | 3 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?4 | L:L:?7 | 6.18 | 3 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:?4 | R:R:S305 | 2.38 | 3 | Yes | No | 0 | 4 | 2 | 2 | | L:L:?6 | L:L:?7 | 22.7 | 3 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?6 | L:L:?8 | 15.87 | 3 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?6 | L:L:?9 | 3.15 | 3 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?6 | R:R:N209 | 12.64 | 3 | Yes | No | 0 | 5 | 1 | 2 | | L:L:?7 | L:L:?8 | 26.11 | 3 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:?7 | L:L:?9 | 3.21 | 3 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?7 | R:R:S305 | 8.45 | 3 | Yes | No | 0 | 4 | 1 | 2 | | L:L:?8 | L:L:?9 | 22.97 | 3 | Yes | Yes | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 12.14 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 27.00 | | Average Links Mediated by Hubs In Shell | 27.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9JFY | A | Peptide | Neuropeptide FF | NPFF2 | Homo Sapiens | hNPSF | - | Gi1/Beta1/Gamma2 | 3.21 | 2025-04-09 | doi.org/10.1038/s44319-025-00428-2 |
|
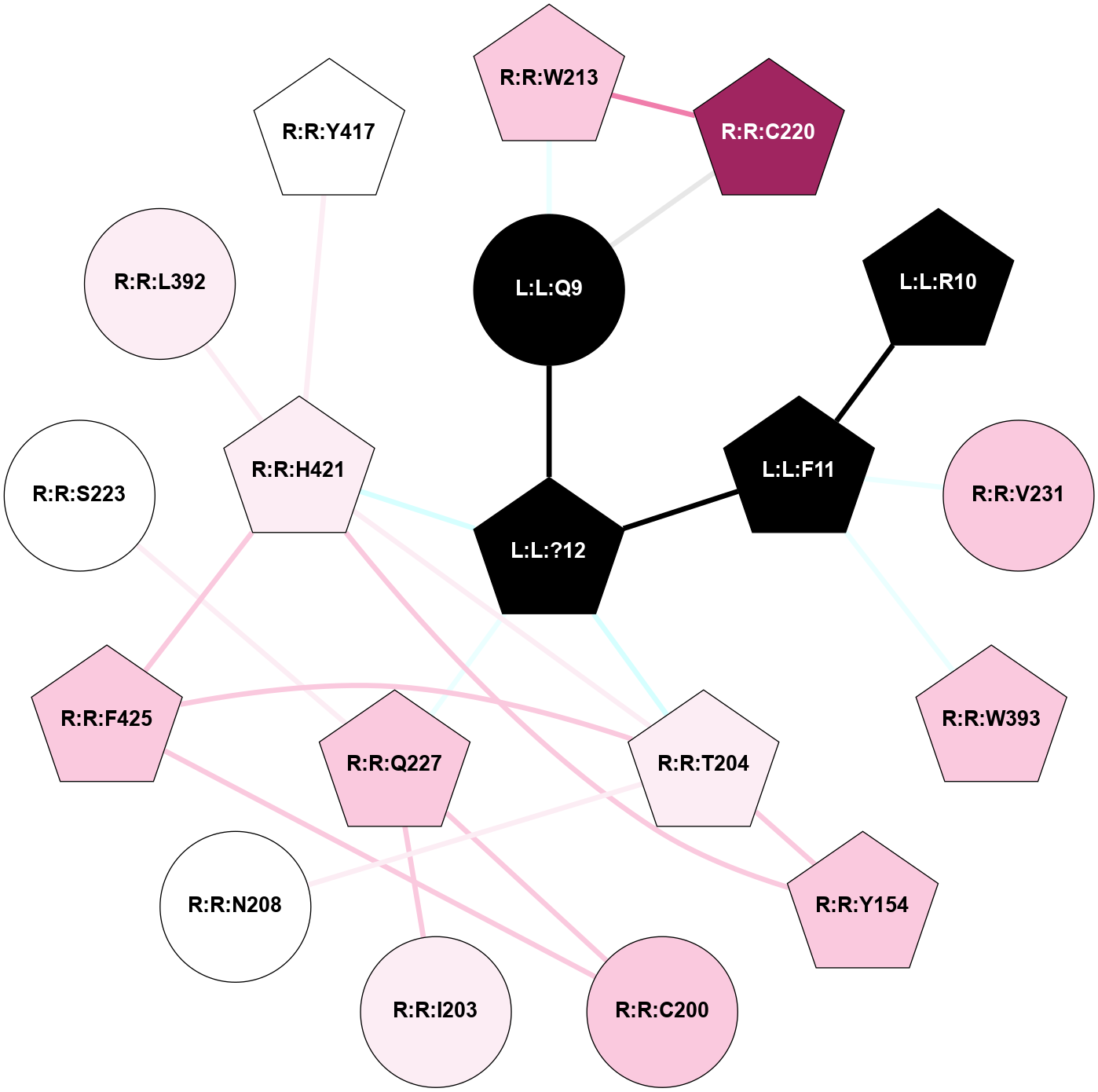
A 2D representation of the interactions of NH2 in 9JFY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?12 | L:L:Q9 | 3.29 | 2 | Yes | No | 0 | 0 | 0 | 1 | | L:L:Q9 | R:R:W213 | 4.38 | 2 | No | Yes | 0 | 7 | 1 | 2 | | L:L:Q9 | R:R:C220 | 4.58 | 2 | No | Yes | 0 | 9 | 1 | 2 | | L:L:F11 | L:L:R10 | 6.41 | 0 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?12 | L:L:F11 | 3.01 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:F11 | R:R:V231 | 9.18 | 0 | Yes | No | 0 | 7 | 1 | 2 | | L:L:F11 | R:R:W393 | 4.01 | 0 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?12 | R:R:T204 | 3.65 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?12 | R:R:Q227 | 3.29 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?12 | R:R:H421 | 9.55 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:T204 | R:R:Y154 | 2.5 | 2 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:H421 | R:R:Y154 | 3.27 | 2 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:C200 | R:R:Q227 | 6.1 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:C200 | R:R:F425 | 4.19 | 0 | No | Yes | 7 | 7 | 2 | 2 | | R:R:I203 | R:R:Q227 | 2.74 | 0 | No | Yes | 6 | 7 | 2 | 1 | | R:R:N208 | R:R:T204 | 2.92 | 0 | No | Yes | 5 | 6 | 2 | 1 | | R:R:H421 | R:R:T204 | 5.48 | 2 | Yes | Yes | 6 | 6 | 1 | 1 | | R:R:F425 | R:R:T204 | 2.59 | 2 | Yes | Yes | 7 | 6 | 2 | 1 | | R:R:C220 | R:R:W213 | 9.14 | 2 | Yes | Yes | 9 | 7 | 2 | 2 | | R:R:Q227 | R:R:S223 | 4.33 | 0 | Yes | No | 7 | 5 | 1 | 2 | | R:R:H421 | R:R:L392 | 2.57 | 2 | Yes | No | 6 | 6 | 1 | 2 | | R:R:H421 | R:R:Y417 | 3.27 | 2 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F425 | R:R:H421 | 7.92 | 2 | Yes | Yes | 7 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 4.56 | | Average Nodes In Shell | 19.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 23.00 | | Average Links Mediated by Hubs In Shell | 23.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9JR2 | B1 | Peptide | Parathyroid Hormone | PTH1 | Homo Sapiens | PTH | - | chim(NtGi1L-Gs-CtGq)/Beta1/Gamma2 | 2.8 | 2024-10-16 | To be published |
|
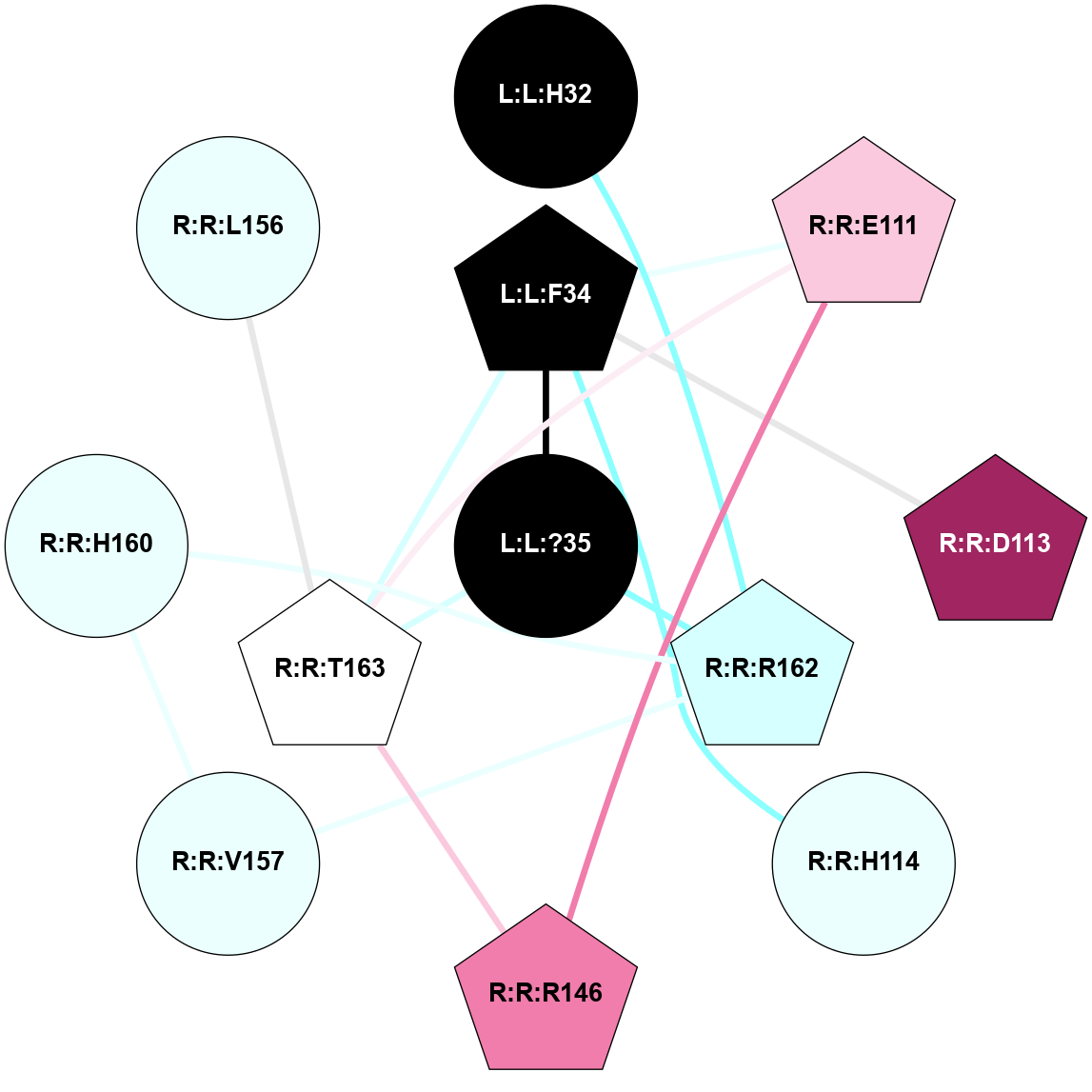
A 2D representation of the interactions of NH2 in 9JR2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:H32 | R:R:R162 | 5.64 | 0 | No | Yes | 0 | 3 | 2 | 1 | | L:L:?35 | L:L:F34 | 9.04 | 3 | No | Yes | 0 | 0 | 0 | 1 | | L:L:F34 | R:R:E111 | 9.33 | 3 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:F34 | R:R:D113 | 3.58 | 3 | Yes | Yes | 0 | 9 | 1 | 2 | | L:L:F34 | R:R:H114 | 6.79 | 3 | Yes | No | 0 | 4 | 1 | 2 | | L:L:F34 | R:R:T163 | 3.89 | 3 | Yes | Yes | 0 | 5 | 1 | 1 | | L:L:?35 | R:R:R162 | 3.01 | 3 | No | Yes | 0 | 3 | 0 | 1 | | L:L:?35 | R:R:T163 | 3.65 | 3 | No | Yes | 0 | 5 | 0 | 1 | | R:R:E111 | R:R:R146 | 11.63 | 3 | Yes | Yes | 7 | 8 | 2 | 2 | | R:R:E111 | R:R:T163 | 4.23 | 3 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:R146 | R:R:T163 | 5.17 | 3 | Yes | Yes | 8 | 5 | 2 | 1 | | R:R:H160 | R:R:V157 | 9.69 | 3 | No | No | 4 | 4 | 2 | 2 | | R:R:R162 | R:R:V157 | 18.31 | 3 | Yes | No | 3 | 4 | 1 | 2 | | R:R:H160 | R:R:R162 | 21.44 | 3 | No | Yes | 4 | 3 | 2 | 1 | | R:R:L156 | R:R:T163 | 1.47 | 0 | No | Yes | 4 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 5.23 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9JR3 | B1 | Peptide | Parathyroid Hormone | PTH1 | Homo Sapiens | PTH | - | chim(NtGi1L-Gs-CtGq)/Beta1/Gamma2 | 2.8 | 2024-10-16 | To be published |
|
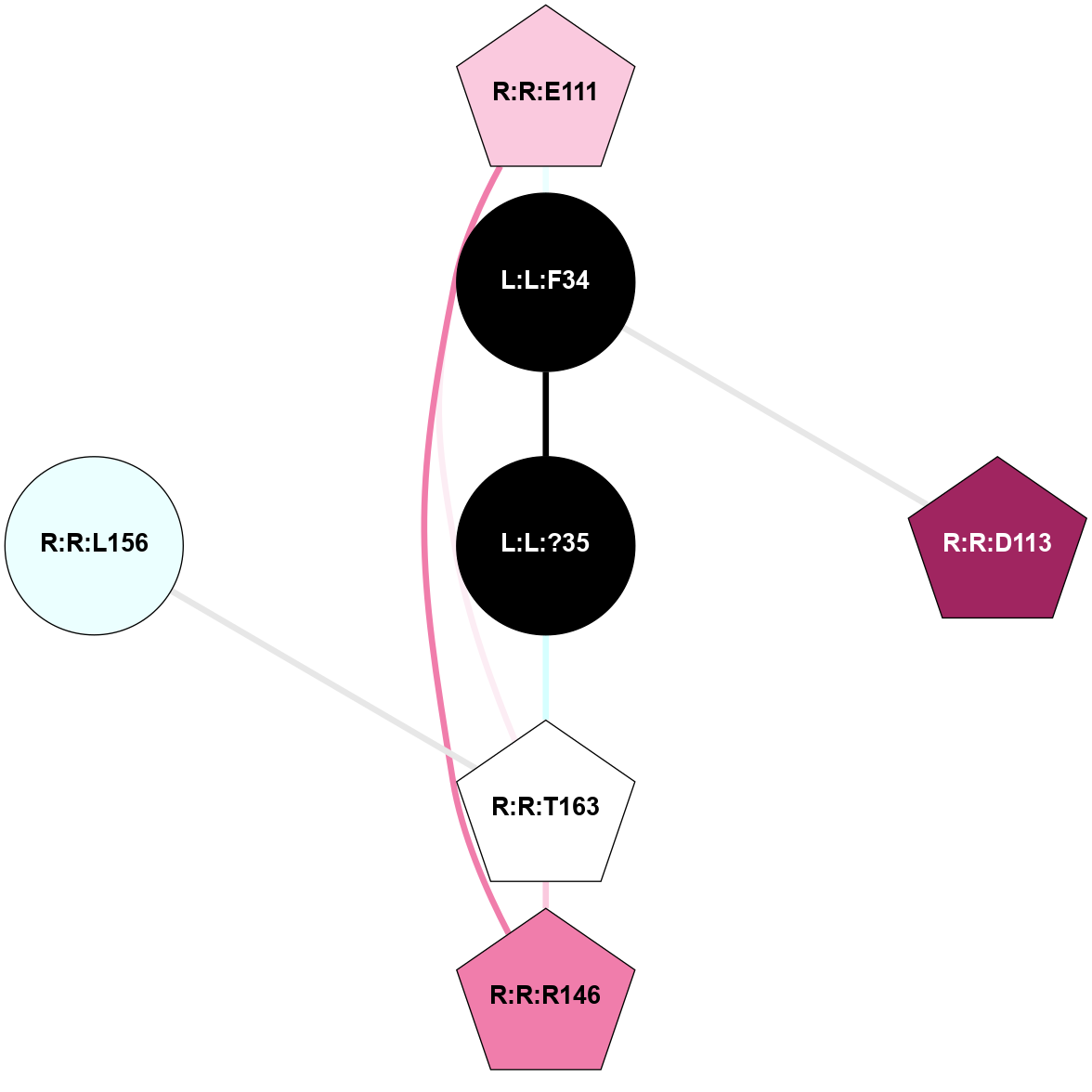
A 2D representation of the interactions of NH2 in 9JR3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?35 | L:L:F34 | 9.04 | 0 | No | No | 0 | 0 | 0 | 1 | | L:L:F34 | R:R:E111 | 10.49 | 0 | No | Yes | 0 | 7 | 1 | 2 | | L:L:F34 | R:R:D113 | 4.78 | 0 | No | Yes | 0 | 9 | 1 | 2 | | L:L:?35 | R:R:T163 | 3.65 | 0 | No | Yes | 0 | 5 | 0 | 1 | | R:R:E111 | R:R:R146 | 6.98 | 7 | Yes | Yes | 7 | 8 | 2 | 2 | | R:R:E111 | R:R:T163 | 2.82 | 7 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:R146 | R:R:T163 | 9.06 | 7 | Yes | Yes | 8 | 5 | 2 | 1 | | R:R:L156 | R:R:T163 | 2.95 | 0 | No | Yes | 4 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.34 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9K26 | A | Peptide | Prolactin-releasing peptide | PrRP | Homo sapiens | PrRP31 | - | Gi1/Beta1/Gamma2 | 3 | 2025-01-29 | To be published |
|

A 2D representation of the interactions of NH2 in 9K26
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:V28 | R:R:T117 | 3.17 | 0 | Yes | Yes | 0 | 6 | 2 | 1 | | L:L:F31 | L:L:R30 | 3.21 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?33 | L:L:F31 | 6.03 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:F31 | R:R:P142 | 4.33 | 1 | Yes | No | 0 | 5 | 1 | 2 | | L:L:F31 | R:R:V145 | 19.66 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:F31 | R:R:Y146 | 6.19 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:F31 | R:R:L294 | 3.65 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:F31 | R:R:H295 | 4.53 | 1 | Yes | Yes | 0 | 8 | 1 | 2 | | L:L:?33 | R:R:C113 | 3.93 | 1 | Yes | No | 0 | 7 | 0 | 1 | | L:L:?33 | R:R:T117 | 7.3 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?33 | R:R:Q141 | 16.47 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?33 | R:R:H321 | 6.36 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:H321 | R:R:Y67 | 9.8 | 1 | Yes | Yes | 5 | 8 | 1 | 2 | | R:R:C113 | R:R:Q141 | 4.58 | 1 | No | Yes | 7 | 6 | 1 | 1 | | R:R:C113 | R:R:M325 | 4.86 | 1 | No | No | 7 | 7 | 1 | 2 | | R:R:Q141 | R:R:T117 | 5.67 | 1 | Yes | Yes | 6 | 6 | 1 | 1 | | R:R:H321 | R:R:T117 | 8.21 | 1 | Yes | Yes | 5 | 6 | 1 | 1 | | R:R:Q141 | R:R:V137 | 4.3 | 1 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:P142 | R:R:Y146 | 4.17 | 1 | No | Yes | 5 | 7 | 2 | 2 | | R:R:V145 | R:R:Y146 | 3.79 | 1 | Yes | Yes | 7 | 7 | 2 | 2 | | R:R:H321 | R:R:M325 | 6.57 | 1 | Yes | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 8.02 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 21.00 | | Average Links Mediated by Hubs In Shell | 20.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9K27 | A | Peptide | Prolactin-releasing peptide | PrRP | Homo sapiens | PrRP31 | - | chim(NtGi1-Gq)/Beta1/Gamma2 | 2.68 | 2025-01-29 | To be published |
|
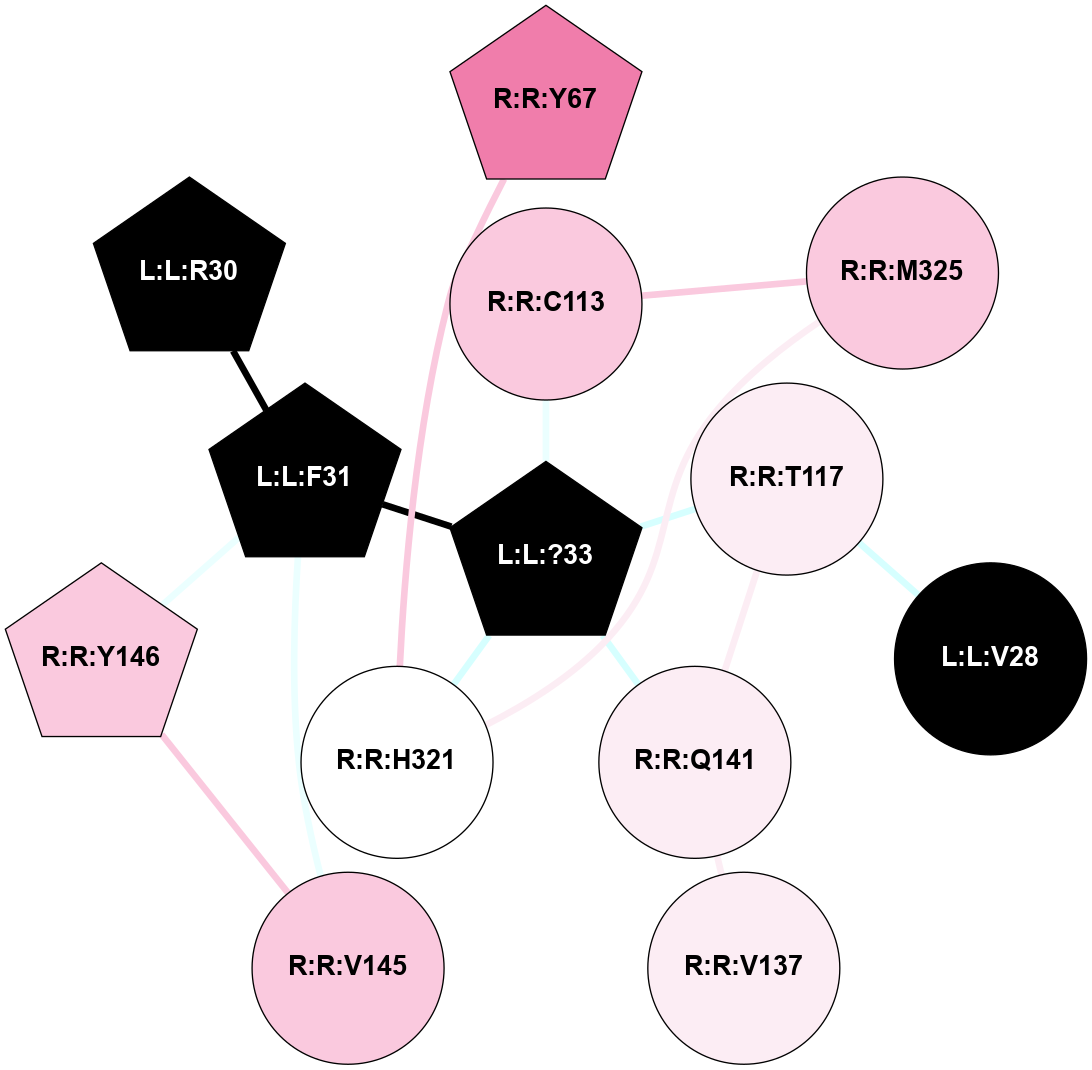
A 2D representation of the interactions of NH2 in 9K27
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:H321 | R:R:Y67 | 9.8 | 0 | No | Yes | 5 | 8 | 1 | 2 | | R:R:C113 | R:R:M325 | 3.24 | 0 | No | No | 7 | 7 | 1 | 2 | | L:L:?33 | R:R:C113 | 3.93 | 9 | Yes | No | 0 | 7 | 0 | 1 | | R:R:Q141 | R:R:T117 | 4.25 | 9 | No | No | 6 | 6 | 1 | 1 | | L:L:V28 | R:R:T117 | 3.17 | 0 | No | No | 0 | 6 | 2 | 1 | | L:L:?33 | R:R:T117 | 3.65 | 9 | Yes | No | 0 | 6 | 0 | 1 | | R:R:Q141 | R:R:V137 | 4.3 | 9 | No | No | 6 | 6 | 1 | 2 | | L:L:?33 | R:R:Q141 | 16.47 | 9 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V145 | R:R:Y146 | 5.05 | 9 | No | Yes | 7 | 7 | 2 | 2 | | L:L:F31 | R:R:V145 | 18.35 | 9 | Yes | No | 0 | 7 | 1 | 2 | | L:L:F31 | R:R:Y146 | 7.22 | 9 | Yes | Yes | 0 | 7 | 1 | 2 | | R:R:H321 | R:R:M325 | 10.51 | 0 | No | No | 5 | 7 | 1 | 2 | | L:L:?33 | R:R:H321 | 6.36 | 9 | Yes | No | 0 | 5 | 0 | 1 | | L:L:F31 | L:L:R30 | 3.21 | 9 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?33 | L:L:F31 | 6.03 | 9 | Yes | Yes | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 7.29 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9U4W | A | Peptide | Gonadotrophin Releasing Hormone | GnRH1 | Sus Scrofa | Gonadoliberin-1 | - | chim(NtGi1L-Gs-CtGq)/Beta1/Gamma2 | 3.18 | 2025-06-11 | doi.org/10.1073/pnas.2500112122 |
|
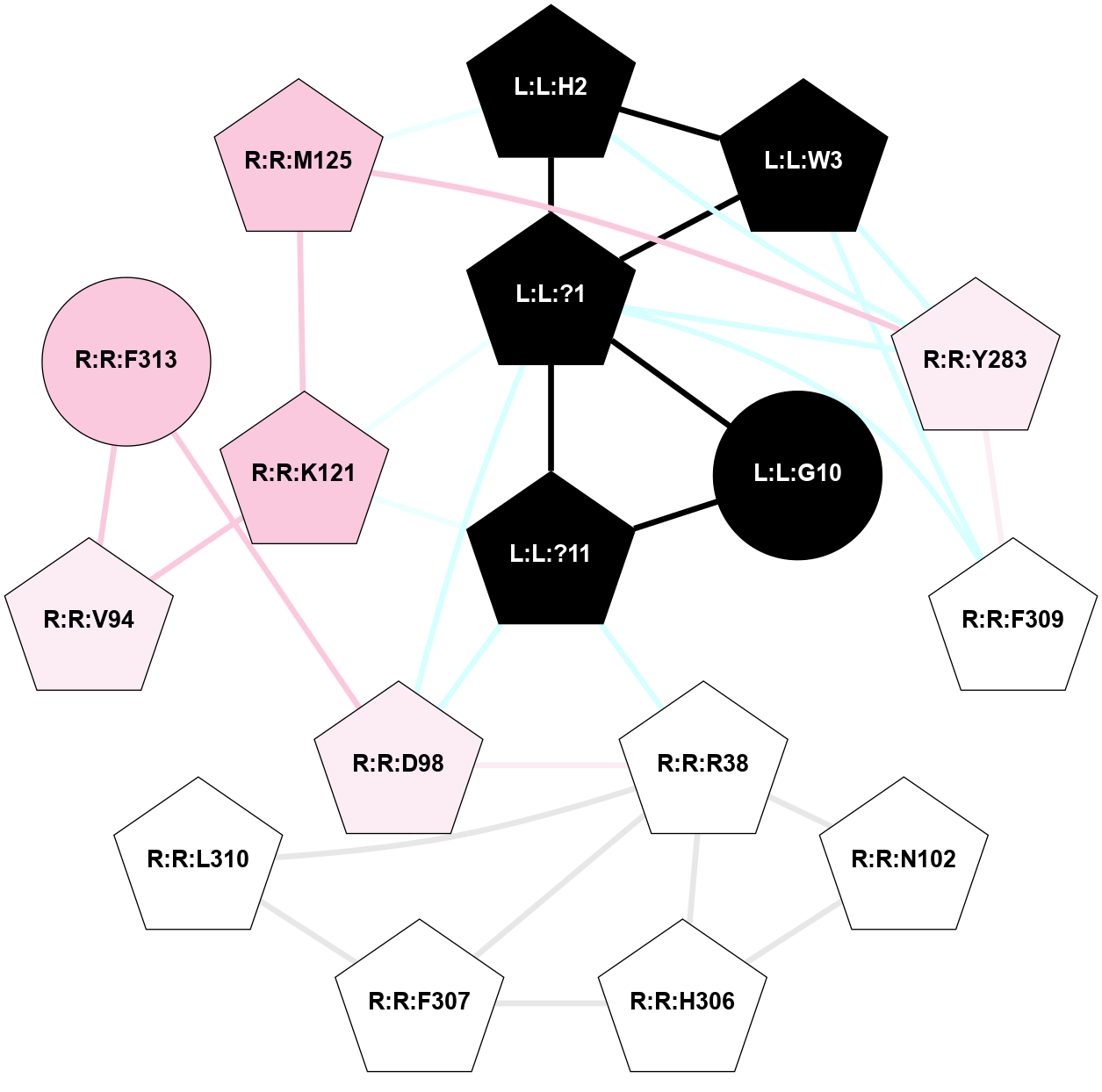
A 2D representation of the interactions of NH2 in 9U4W
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | L:L:H2 | 5.5 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?1 | L:L:W3 | 13.4 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?1 | L:L:G10 | 5.49 | 1 | Yes | No | 0 | 0 | 1 | 1 | | L:L:?1 | L:L:?11 | 21.99 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?1 | R:R:D98 | 4.36 | 1 | Yes | Yes | 0 | 6 | 1 | 1 | | L:L:?1 | R:R:K121 | 6.03 | 1 | Yes | Yes | 0 | 7 | 1 | 1 | | L:L:?1 | R:R:Y283 | 17.56 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?1 | R:R:F309 | 15.64 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:H2 | L:L:W3 | 9.52 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:H2 | R:R:M125 | 2.63 | 1 | Yes | Yes | 0 | 7 | 2 | 2 | | L:L:H2 | R:R:Y283 | 7.62 | 1 | Yes | Yes | 0 | 6 | 2 | 2 | | L:L:W3 | R:R:Y283 | 9.65 | 1 | Yes | Yes | 0 | 6 | 2 | 2 | | L:L:W3 | R:R:F309 | 4.01 | 1 | Yes | Yes | 0 | 5 | 2 | 2 | | L:L:?11 | L:L:G10 | 4.23 | 1 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?11 | R:R:R38 | 3.01 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?11 | R:R:D98 | 10.08 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?11 | R:R:K121 | 3.49 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:D98 | R:R:R38 | 5.96 | 1 | Yes | Yes | 6 | 5 | 1 | 1 | | R:R:N102 | R:R:R38 | 18.08 | 1 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:H306 | R:R:R38 | 11.28 | 1 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:F307 | R:R:R38 | 2.14 | 1 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:L310 | R:R:R38 | 4.86 | 1 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:K121 | R:R:V94 | 7.59 | 1 | Yes | Yes | 7 | 6 | 1 | 2 | | R:R:F313 | R:R:V94 | 2.62 | 0 | No | Yes | 7 | 6 | 2 | 2 | | R:R:D98 | R:R:F313 | 8.36 | 1 | Yes | No | 6 | 7 | 1 | 2 | | R:R:H306 | R:R:N102 | 6.38 | 1 | Yes | Yes | 5 | 5 | 2 | 2 | | R:R:K121 | R:R:M125 | 4.32 | 1 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:M125 | R:R:Y283 | 2.39 | 1 | Yes | Yes | 7 | 6 | 2 | 2 | | R:R:F309 | R:R:Y283 | 4.13 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:F307 | R:R:H306 | 4.53 | 1 | Yes | Yes | 5 | 5 | 2 | 2 | | R:R:F307 | R:R:L310 | 4.87 | 1 | Yes | Yes | 5 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 8.56 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 15.00 | | Average Links In Shell | 31.00 | | Average Links Mediated by Hubs In Shell | 31.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9U4Y | A | Peptide | Gonadotrophin Releasing Hormone | GnRH1 | Xenopus Laevis | Gonadoliberin-1 | - | chim(NtGi1L-Gs-CtGq)/Beta1/Gamma2 | 3.18 | 2025-06-11 | doi.org/10.1073/pnas.2500112122 |
|
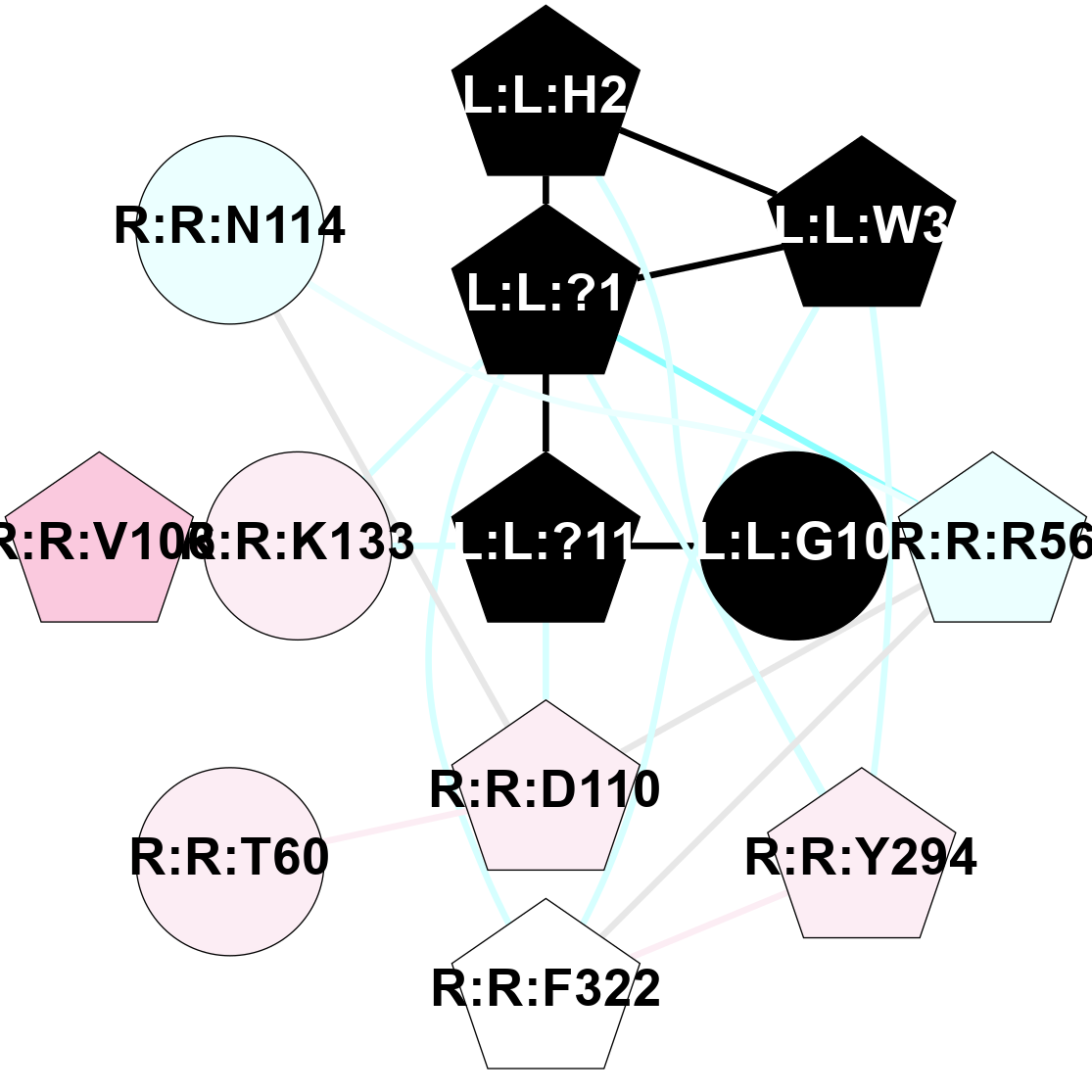
A 2D representation of the interactions of NH2 in 9U4Y
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | L:L:H2 | 5.5 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?1 | L:L:W3 | 25.59 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?1 | L:L:?11 | 14.66 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | L:L:?1 | R:R:R56 | 3.9 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | L:L:?1 | R:R:K133 | 4.53 | 1 | Yes | No | 0 | 6 | 1 | 1 | | L:L:?1 | R:R:Y294 | 17.56 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?1 | R:R:F322 | 20.85 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | L:L:H2 | L:L:W3 | 11.64 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:H2 | R:R:Y294 | 10.89 | 1 | Yes | Yes | 0 | 6 | 2 | 2 | | L:L:W3 | R:R:Y294 | 17.36 | 1 | Yes | Yes | 0 | 6 | 2 | 2 | | L:L:W3 | R:R:F322 | 4.01 | 1 | Yes | Yes | 0 | 5 | 2 | 2 | | L:L:?11 | L:L:G10 | 4.23 | 1 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?11 | R:R:D110 | 3.36 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?11 | R:R:K133 | 6.98 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:D110 | R:R:R56 | 10.72 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | R:R:N114 | R:R:R56 | 9.64 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:F322 | R:R:R56 | 5.34 | 1 | Yes | Yes | 5 | 4 | 2 | 2 | | R:R:D110 | R:R:T60 | 2.89 | 1 | Yes | No | 6 | 6 | 1 | 2 | | R:R:K133 | R:R:V106 | 4.55 | 1 | No | Yes | 6 | 7 | 1 | 2 | | R:R:D110 | R:R:N114 | 4.04 | 1 | Yes | No | 6 | 4 | 1 | 2 | | R:R:F322 | R:R:Y294 | 7.22 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 7.31 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 21.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9M0R | A | Peptide | Neuropeptide FF | NPFF1 | Homo Sapiens | Neuropeptide VF | - | Gi1/Beta1/Gamma2 | 2.47 | 2025-07-23 | doi.org/10.1016/j.celrep.2025.116160 |
|
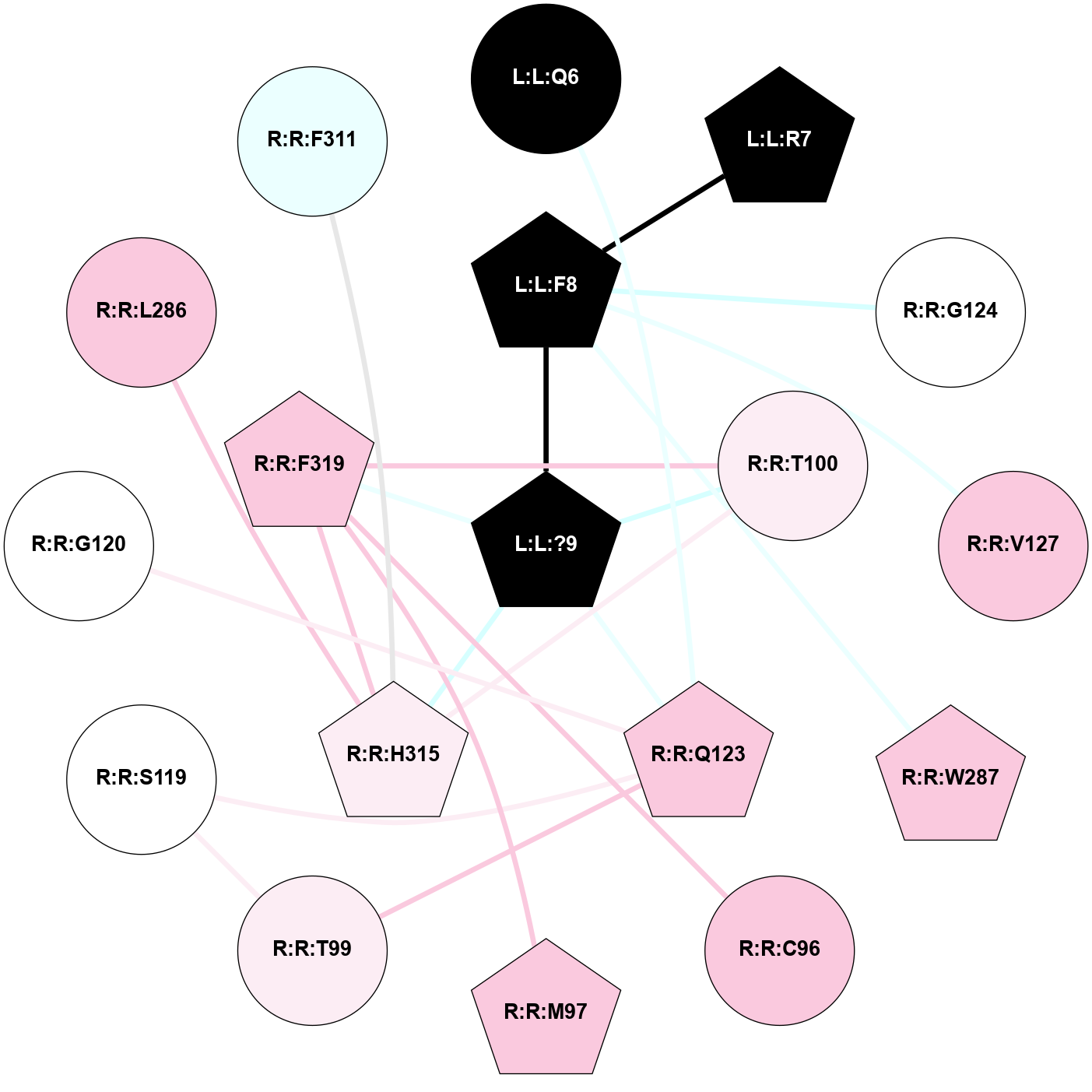
A 2D representation of the interactions of NH2 in 9M0R
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:Q6 | R:R:Q123 | 6.4 | 0 | No | Yes | 0 | 7 | 2 | 1 | | L:L:F8 | L:L:R7 | 11.76 | 0 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?9 | L:L:F8 | 3.01 | 9 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:F8 | R:R:G124 | 3.01 | 0 | Yes | No | 0 | 5 | 1 | 2 | | L:L:F8 | R:R:V127 | 9.18 | 0 | Yes | No | 0 | 7 | 1 | 2 | | L:L:F8 | R:R:W287 | 7.02 | 0 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?9 | R:R:T100 | 3.65 | 9 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?9 | R:R:Q123 | 9.88 | 9 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?9 | R:R:H315 | 6.36 | 9 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?9 | R:R:F319 | 6.03 | 9 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:C96 | R:R:F319 | 6.98 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:F319 | R:R:M97 | 8.71 | 9 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:S119 | R:R:T99 | 6.4 | 9 | No | No | 5 | 6 | 2 | 2 | | R:R:Q123 | R:R:T99 | 4.25 | 9 | Yes | No | 7 | 6 | 1 | 2 | | R:R:H315 | R:R:T100 | 8.21 | 9 | Yes | No | 6 | 6 | 1 | 1 | | R:R:F319 | R:R:T100 | 7.78 | 9 | Yes | No | 7 | 6 | 1 | 1 | | R:R:Q123 | R:R:S119 | 5.78 | 9 | Yes | No | 7 | 5 | 1 | 2 | | R:R:G120 | R:R:Q123 | 6.58 | 0 | No | Yes | 5 | 7 | 2 | 1 | | R:R:H315 | R:R:L286 | 12.86 | 9 | Yes | No | 6 | 7 | 1 | 2 | | R:R:F311 | R:R:H315 | 9.05 | 0 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F319 | R:R:H315 | 9.05 | 9 | Yes | Yes | 7 | 6 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 5.79 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 21.00 | | Average Links Mediated by Hubs In Shell | 20.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9M1O | A | Peptide | Neuropeptide FF | NPFF2 | Homo Sapiens | Morphine-Modulating Neuropeptide B | - | Gi1/Beta1/Gamma2 | 2.49 | 2025-07-23 | doi.org/10.1016/j.celrep.2025.116160 |
|
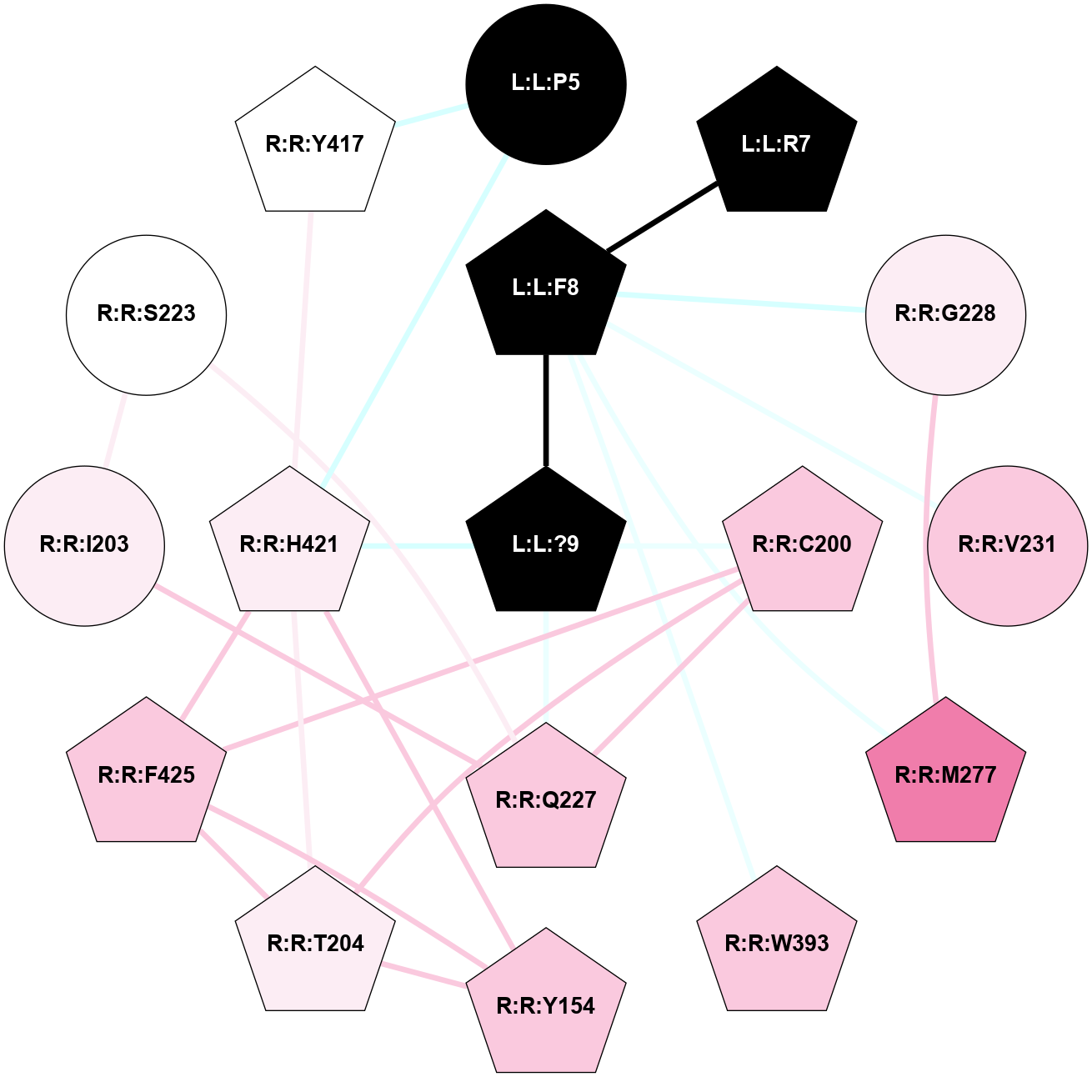
A 2D representation of the interactions of NH2 in 9M1O
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:P5 | R:R:Y417 | 19.47 | 2 | No | Yes | 0 | 5 | 2 | 2 | | L:L:P5 | R:R:H421 | 3.05 | 2 | No | Yes | 0 | 6 | 2 | 1 | | L:L:F8 | L:L:R7 | 4.28 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?9 | L:L:F8 | 9.04 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:F8 | R:R:G228 | 3.01 | 2 | Yes | No | 0 | 6 | 1 | 2 | | L:L:F8 | R:R:V231 | 10.49 | 2 | Yes | No | 0 | 7 | 1 | 2 | | L:L:F8 | R:R:M277 | 3.73 | 2 | Yes | Yes | 0 | 8 | 1 | 2 | | L:L:F8 | R:R:W393 | 4.01 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:?9 | R:R:C200 | 3.93 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?9 | R:R:Q227 | 16.47 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?9 | R:R:H421 | 3.18 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:T204 | R:R:Y154 | 3.75 | 2 | Yes | Yes | 6 | 7 | 2 | 2 | | R:R:H421 | R:R:Y154 | 3.27 | 2 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:F425 | R:R:Y154 | 6.19 | 2 | Yes | Yes | 7 | 7 | 2 | 2 | | R:R:C200 | R:R:T204 | 3.38 | 2 | Yes | Yes | 7 | 6 | 1 | 2 | | R:R:C200 | R:R:Q227 | 12.21 | 2 | Yes | Yes | 7 | 7 | 1 | 1 | | R:R:C200 | R:R:F425 | 5.59 | 2 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:I203 | R:R:S223 | 7.74 | 2 | No | No | 6 | 5 | 2 | 2 | | R:R:I203 | R:R:Q227 | 4.12 | 2 | No | Yes | 6 | 7 | 2 | 1 | | R:R:H421 | R:R:T204 | 5.48 | 2 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:F425 | R:R:T204 | 6.49 | 2 | Yes | Yes | 7 | 6 | 2 | 2 | | R:R:Q227 | R:R:S223 | 7.22 | 2 | Yes | No | 7 | 5 | 1 | 2 | | R:R:G228 | R:R:M277 | 3.49 | 2 | No | Yes | 6 | 8 | 2 | 2 | | R:R:H421 | R:R:Y417 | 5.44 | 2 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F425 | R:R:H421 | 9.05 | 2 | Yes | Yes | 7 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 8.15 | | Average Nodes In Shell | 17.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 25.00 | | Average Links Mediated by Hubs In Shell | 24.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9M2F | A | Peptide | Neuropeptide FF | NPFF1 | Homo Sapiens | Neuropeptide FF | - | Gi1/Beta1/Gamma2 | 2.93 | 2025-07-23 | doi.org/10.1016/j.celrep.2025.116160 |
|
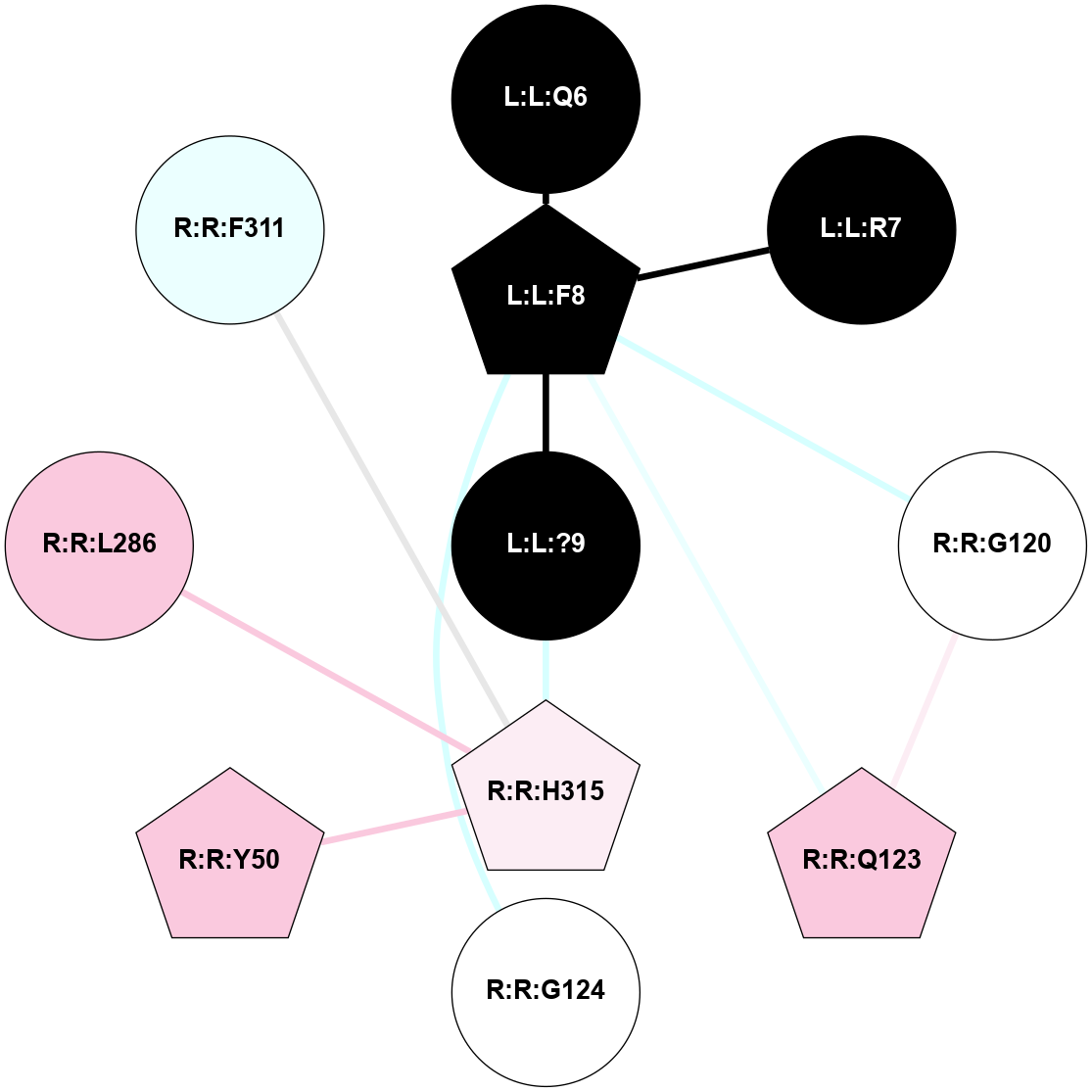
A 2D representation of the interactions of NH2 in 9M2F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:F8 | L:L:Q6 | 4.68 | 24 | Yes | No | 0 | 0 | 1 | 2 | | L:L:F8 | L:L:R7 | 14.97 | 24 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?9 | L:L:F8 | 6.03 | 0 | No | Yes | 0 | 0 | 0 | 1 | | L:L:F8 | R:R:G120 | 4.52 | 24 | Yes | No | 0 | 5 | 1 | 2 | | L:L:F8 | R:R:Q123 | 5.86 | 24 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:F8 | R:R:G124 | 4.52 | 24 | Yes | No | 0 | 5 | 1 | 2 | | L:L:?9 | R:R:H315 | 9.55 | 0 | No | Yes | 0 | 6 | 0 | 1 | | R:R:H315 | R:R:Y50 | 3.27 | 0 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:G120 | R:R:Q123 | 4.93 | 24 | No | Yes | 5 | 7 | 2 | 2 | | R:R:H315 | R:R:L286 | 5.14 | 0 | Yes | No | 6 | 7 | 1 | 2 | | R:R:F311 | R:R:H315 | 4.53 | 0 | No | Yes | 4 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 7.79 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9M54 | A | Peptide | Neuropeptide FF | NPFF2 | Homo Sapiens | Neuropeptide VF | - | Gi1/Beta1/Gamma2 | 3.24 | 2025-07-23 | doi.org/10.1016/j.celrep.2025.116160 |
|
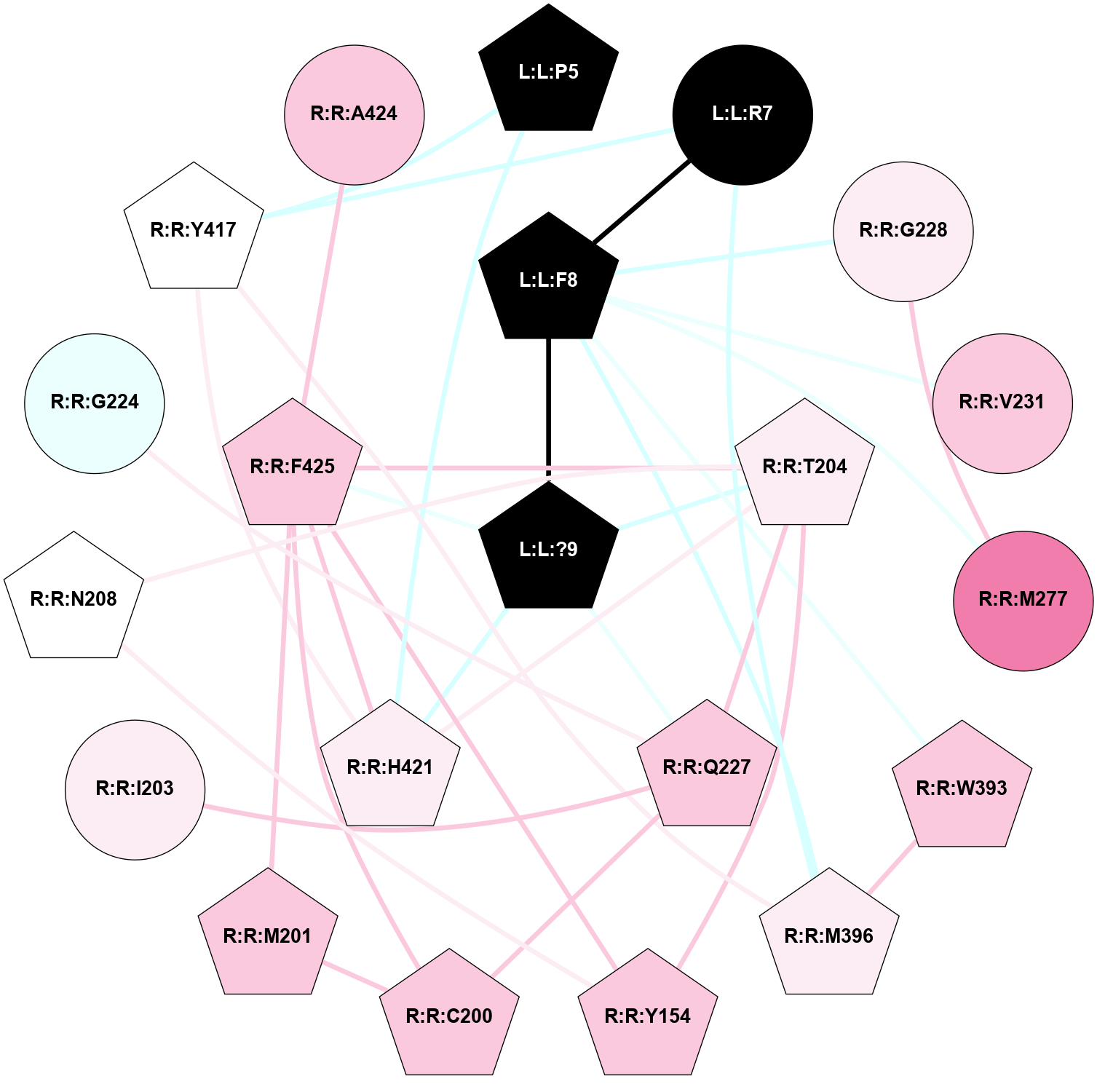
A 2D representation of the interactions of NH2 in 9M54
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:P5 | R:R:Y417 | 12.52 | 2 | Yes | Yes | 0 | 5 | 2 | 2 | | L:L:P5 | R:R:H421 | 3.05 | 2 | Yes | Yes | 0 | 6 | 2 | 1 | | L:L:F8 | L:L:R7 | 6.41 | 2 | Yes | No | 0 | 0 | 1 | 2 | | L:L:R7 | R:R:M396 | 3.72 | 2 | No | Yes | 0 | 6 | 2 | 2 | | L:L:R7 | R:R:Y417 | 6.17 | 2 | No | Yes | 0 | 5 | 2 | 2 | | L:L:?9 | L:L:F8 | 3.01 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:F8 | R:R:G228 | 3.01 | 2 | Yes | No | 0 | 6 | 1 | 2 | | L:L:F8 | R:R:V231 | 6.55 | 2 | Yes | No | 0 | 7 | 1 | 2 | | L:L:F8 | R:R:M277 | 7.46 | 2 | Yes | No | 0 | 8 | 1 | 2 | | L:L:F8 | R:R:W393 | 4.01 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | L:L:F8 | R:R:M396 | 3.73 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?9 | R:R:T204 | 3.65 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?9 | R:R:Q227 | 9.88 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?9 | R:R:H421 | 6.36 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | L:L:?9 | R:R:F425 | 6.03 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:T204 | R:R:Y154 | 7.49 | 2 | Yes | Yes | 6 | 7 | 1 | 2 | | R:R:N208 | R:R:Y154 | 3.49 | 2 | Yes | Yes | 5 | 7 | 2 | 2 | | R:R:F425 | R:R:Y154 | 7.22 | 2 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:C200 | R:R:M201 | 3.24 | 2 | Yes | Yes | 7 | 7 | 2 | 2 | | R:R:C200 | R:R:Q227 | 6.1 | 2 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:C200 | R:R:F425 | 8.38 | 2 | Yes | Yes | 7 | 7 | 2 | 1 | | R:R:F425 | R:R:M201 | 12.44 | 2 | Yes | Yes | 7 | 7 | 1 | 2 | | R:R:I203 | R:R:Q227 | 16.47 | 2 | No | Yes | 6 | 7 | 2 | 1 | | R:R:N208 | R:R:T204 | 4.39 | 2 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:Q227 | R:R:T204 | 4.25 | 2 | Yes | Yes | 7 | 6 | 1 | 1 | | R:R:H421 | R:R:T204 | 9.58 | 2 | Yes | Yes | 6 | 6 | 1 | 1 | | R:R:F425 | R:R:T204 | 7.78 | 2 | Yes | Yes | 7 | 6 | 1 | 1 | | R:R:G224 | R:R:Q227 | 3.29 | 0 | No | Yes | 4 | 7 | 2 | 1 | | R:R:G228 | R:R:M277 | 5.24 | 2 | No | No | 6 | 8 | 2 | 2 | | R:R:M396 | R:R:W393 | 9.31 | 2 | Yes | Yes | 6 | 7 | 2 | 2 | | R:R:M396 | R:R:Y417 | 3.59 | 2 | Yes | Yes | 6 | 5 | 2 | 2 | | R:R:H421 | R:R:Y417 | 15.24 | 2 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F425 | R:R:H421 | 9.05 | 2 | Yes | Yes | 7 | 6 | 1 | 1 | | R:R:A424 | R:R:F425 | 2.77 | 2 | No | Yes | 7 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 5.79 | | Average Nodes In Shell | 21.00 | | Average Hubs In Shell | 14.00 | | Average Links In Shell | 34.00 | | Average Links Mediated by Hubs In Shell | 33.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9BP3 | B1 | Peptide | Calcitonin | CT (AMY1) | Homo Sapiens | Cagrilintide | - | Gs/Beta1/Gamma2; RAMP1 | 2.2 | 2025-04-23 | To be published |
|
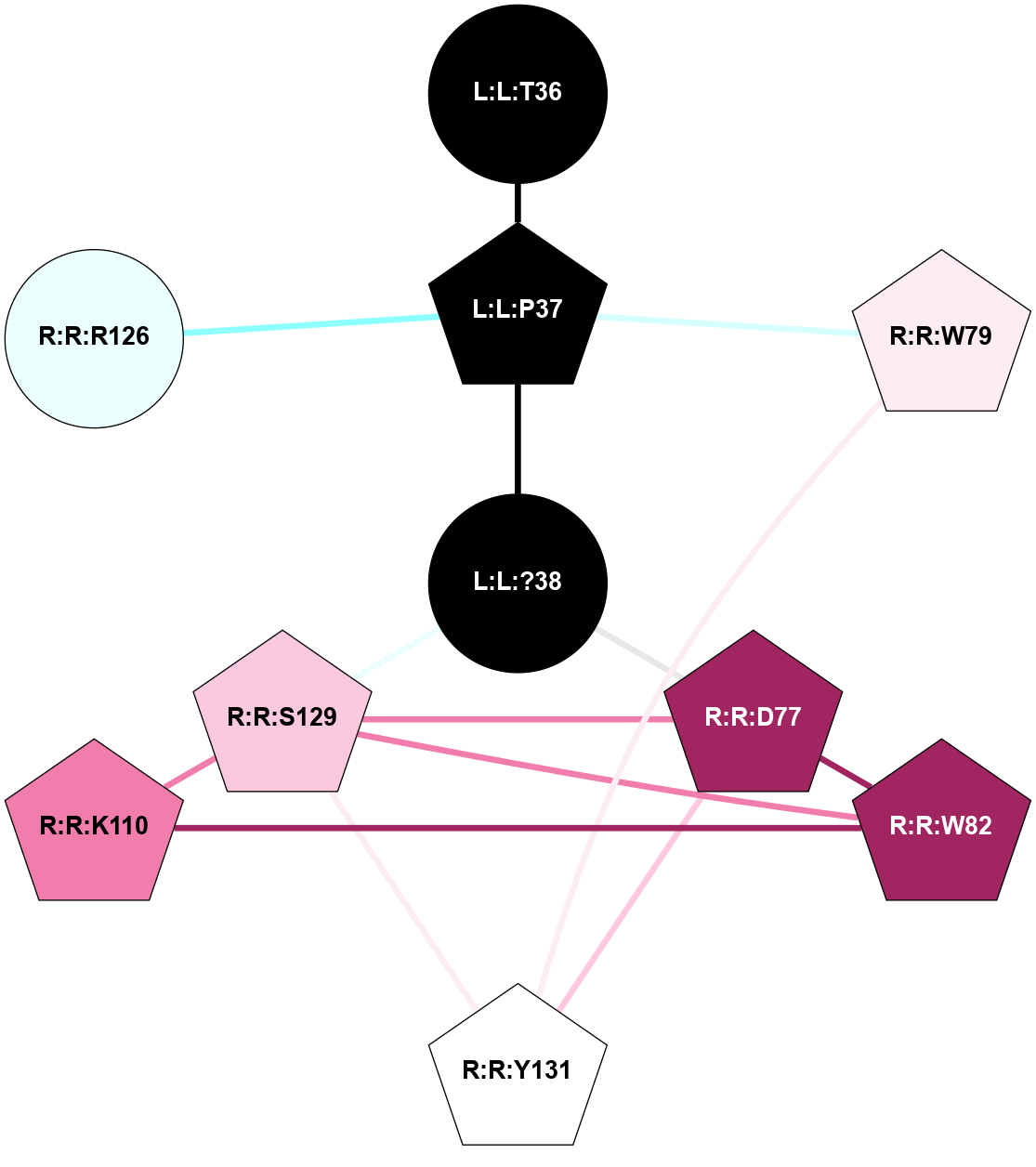
A 2D representation of the interactions of NH2 in 9BP3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:P37 | L:L:T36 | 3.5 | 0 | Yes | No | 0 | 0 | 1 | 2 | | L:L:?38 | L:L:P37 | 4.06 | 3 | No | Yes | 0 | 0 | 0 | 1 | | L:L:P37 | R:R:W79 | 17.57 | 0 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?38 | R:R:D77 | 10.08 | 3 | No | Yes | 0 | 9 | 0 | 1 | | L:L:?38 | R:R:S129 | 7.43 | 3 | No | Yes | 0 | 7 | 0 | 1 | | R:R:D77 | R:R:W82 | 10.05 | 3 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D77 | R:R:S129 | 5.89 | 3 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:D77 | R:R:Y131 | 5.75 | 3 | Yes | Yes | 9 | 5 | 1 | 2 | | R:R:W79 | R:R:Y131 | 15.43 | 3 | Yes | Yes | 6 | 5 | 2 | 2 | | R:R:K110 | R:R:W82 | 22.05 | 3 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:S129 | R:R:W82 | 3.71 | 3 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:K110 | R:R:S129 | 6.12 | 3 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:S129 | R:R:Y131 | 8.9 | 3 | Yes | Yes | 7 | 5 | 1 | 2 | | L:L:P37 | R:R:R126 | 2.88 | 0 | Yes | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 7.19 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9BQ3 | B1 | Peptide | Calcitonin | CT (AMY2) | Homo Sapiens | Cagrilintide | - | Gs/Beta1/Gamma2; RAMP2 | 2.8 | 2025-04-23 | To be published |
|
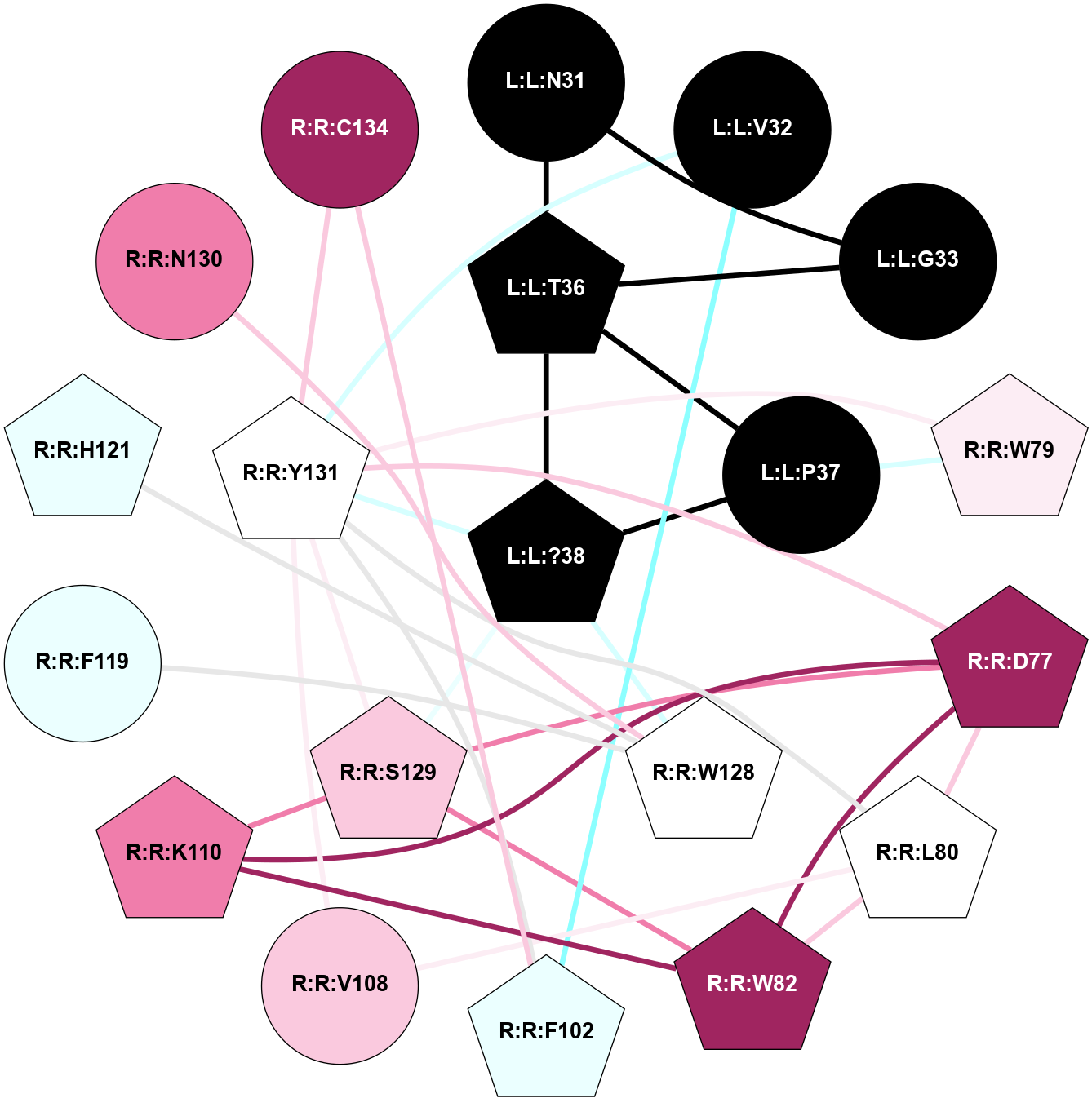
A 2D representation of the interactions of NH2 in 9BQ3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:G33 | L:L:N31 | 3.39 | 3 | No | No | 0 | 0 | 2 | 2 | | L:L:N31 | L:L:T36 | 5.85 | 3 | No | Yes | 0 | 0 | 2 | 1 | | L:L:V32 | R:R:F102 | 2.62 | 3 | No | Yes | 0 | 4 | 2 | 2 | | L:L:V32 | R:R:Y131 | 5.05 | 3 | No | Yes | 0 | 5 | 2 | 1 | | L:L:G33 | L:L:T36 | 3.64 | 3 | No | Yes | 0 | 0 | 2 | 1 | | L:L:P37 | L:L:T36 | 5.25 | 3 | No | Yes | 0 | 0 | 1 | 1 | | L:L:?38 | L:L:T36 | 3.65 | 3 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:?38 | L:L:P37 | 12.19 | 3 | Yes | No | 0 | 0 | 0 | 1 | | L:L:P37 | R:R:W79 | 25.67 | 3 | No | Yes | 0 | 6 | 1 | 2 | | L:L:?38 | R:R:W128 | 5.64 | 3 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?38 | R:R:S129 | 3.72 | 3 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?38 | R:R:Y131 | 5.8 | 3 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:L80 | 2.71 | 3 | Yes | Yes | 9 | 5 | 2 | 2 | | R:R:D77 | R:R:W82 | 11.17 | 3 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:D77 | R:R:K110 | 2.77 | 3 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:D77 | R:R:S129 | 5.89 | 3 | Yes | Yes | 9 | 7 | 2 | 1 | | R:R:D77 | R:R:Y131 | 6.9 | 3 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:W79 | R:R:Y131 | 16.4 | 3 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:L80 | R:R:W82 | 3.42 | 3 | Yes | Yes | 5 | 9 | 2 | 2 | | R:R:L80 | R:R:V108 | 2.98 | 3 | Yes | No | 5 | 7 | 2 | 2 | | R:R:L80 | R:R:Y131 | 7.03 | 3 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:K110 | R:R:W82 | 29.01 | 3 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:S129 | R:R:W82 | 3.71 | 3 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:F102 | R:R:Y131 | 3.09 | 3 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:C134 | R:R:F102 | 12.57 | 3 | No | Yes | 9 | 4 | 2 | 2 | | R:R:V108 | R:R:Y131 | 13.88 | 3 | No | Yes | 7 | 5 | 2 | 1 | | R:R:K110 | R:R:S129 | 7.65 | 3 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:F119 | R:R:W128 | 6.01 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:H121 | R:R:W128 | 11.64 | 35 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 9.04 | 0 | No | Yes | 8 | 5 | 2 | 1 | | R:R:S129 | R:R:Y131 | 6.36 | 3 | Yes | Yes | 7 | 5 | 1 | 1 | | R:R:C134 | R:R:Y131 | 2.69 | 3 | No | Yes | 9 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 6.20 | | Average Nodes In Shell | 20.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 32.00 | | Average Links Mediated by Hubs In Shell | 31.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9BUB | B1 | Peptide | Calcitonin | CT | Homo Sapiens | Cagrilintide | - | Gs/Beta1/Gamma2 | 2.3 | 2025-04-23 | doi.org/10.1038/s41467-025-58680-y |
|
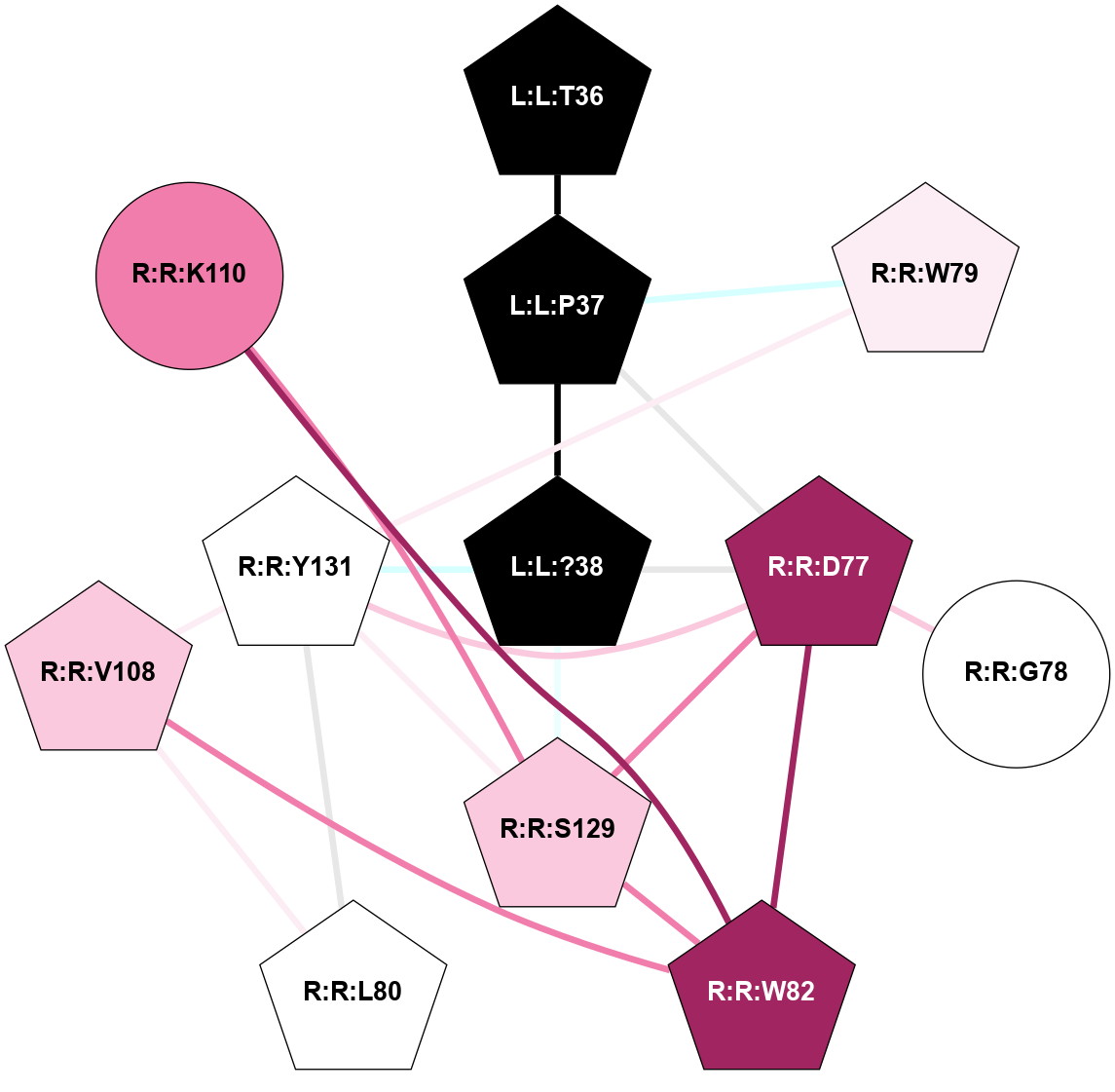
A 2D representation of the interactions of NH2 in 9BUB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:P37 | L:L:T36 | 3.5 | 4 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:?38 | L:L:P37 | 8.13 | 4 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:P37 | R:R:D77 | 4.83 | 4 | Yes | Yes | 0 | 9 | 1 | 1 | | L:L:P37 | R:R:W79 | 10.81 | 4 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?38 | R:R:D77 | 10.08 | 4 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?38 | R:R:S129 | 7.43 | 4 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?38 | R:R:Y131 | 8.7 | 4 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:G78 | 5.03 | 4 | Yes | No | 9 | 5 | 1 | 2 | | R:R:D77 | R:R:W82 | 5.58 | 4 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D77 | R:R:S129 | 5.89 | 4 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:D77 | R:R:Y131 | 9.2 | 4 | Yes | Yes | 9 | 5 | 1 | 1 | | R:R:W79 | R:R:Y131 | 13.5 | 4 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:L80 | R:R:V108 | 4.47 | 4 | Yes | Yes | 5 | 7 | 2 | 2 | | R:R:L80 | R:R:Y131 | 10.55 | 4 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:V108 | R:R:W82 | 3.68 | 4 | Yes | Yes | 7 | 9 | 2 | 2 | | R:R:K110 | R:R:W82 | 34.81 | 4 | No | Yes | 8 | 9 | 2 | 2 | | R:R:S129 | R:R:W82 | 4.94 | 4 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:V108 | R:R:Y131 | 12.62 | 4 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:K110 | R:R:S129 | 6.12 | 4 | No | Yes | 8 | 7 | 2 | 1 | | R:R:S129 | R:R:Y131 | 5.09 | 4 | Yes | Yes | 7 | 5 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 8.59 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 20.00 | | Average Links Mediated by Hubs In Shell | 20.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9BUD | B1 | Peptide | Calcitonin | CT | Homo Sapiens | Cagrilintide | - | Gs/Beta1/Gamma2 | 2.5 | 2025-04-23 | To be published |
|
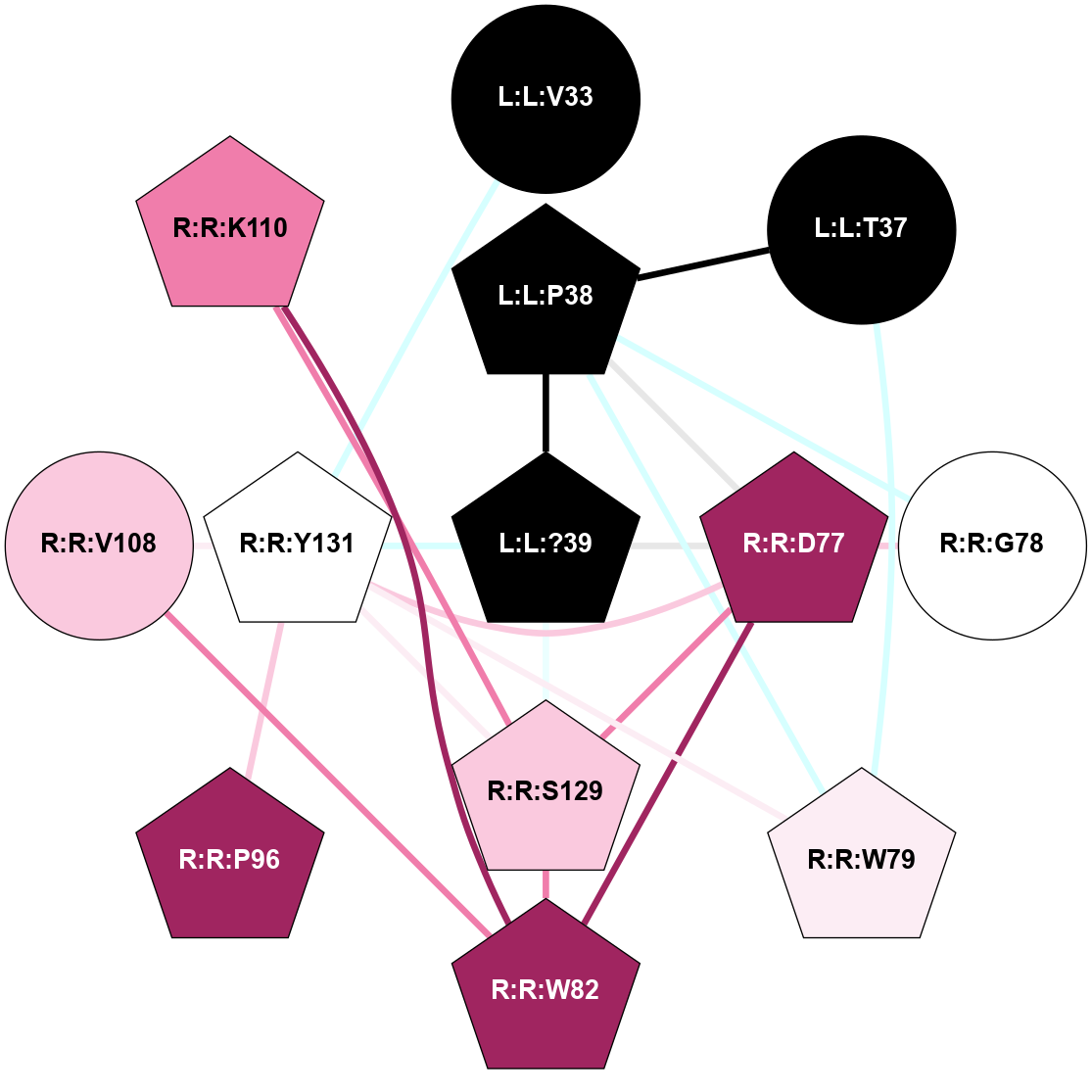
A 2D representation of the interactions of NH2 in 9BUD
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:V33 | R:R:Y131 | 2.52 | 0 | No | Yes | 0 | 5 | 2 | 1 | | L:L:P38 | L:L:T37 | 3.5 | 6 | Yes | No | 0 | 0 | 1 | 2 | | L:L:T37 | R:R:W79 | 3.64 | 6 | No | Yes | 0 | 6 | 2 | 2 | | L:L:?39 | L:L:P38 | 4.06 | 6 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:P38 | R:R:D77 | 4.83 | 6 | Yes | Yes | 0 | 9 | 1 | 1 | | L:L:P38 | R:R:G78 | 4.06 | 6 | Yes | No | 0 | 5 | 1 | 2 | | L:L:P38 | R:R:W79 | 32.43 | 6 | Yes | Yes | 0 | 6 | 1 | 2 | | L:L:?39 | R:R:D77 | 10.08 | 6 | Yes | Yes | 0 | 9 | 0 | 1 | | L:L:?39 | R:R:S129 | 7.43 | 6 | Yes | Yes | 0 | 7 | 0 | 1 | | L:L:?39 | R:R:Y131 | 5.8 | 6 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:D77 | R:R:G78 | 3.35 | 6 | Yes | No | 9 | 5 | 1 | 2 | | R:R:D77 | R:R:W82 | 3.35 | 6 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D77 | R:R:S129 | 7.36 | 6 | Yes | Yes | 9 | 7 | 1 | 1 | | R:R:D77 | R:R:Y131 | 6.9 | 6 | Yes | Yes | 9 | 5 | 1 | 1 | | R:R:W79 | R:R:Y131 | 19.29 | 6 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:V108 | R:R:W82 | 6.13 | 0 | No | Yes | 7 | 9 | 2 | 2 | | R:R:K110 | R:R:W82 | 17.4 | 6 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:S129 | R:R:W82 | 8.65 | 6 | Yes | Yes | 7 | 9 | 1 | 2 | | R:R:P96 | R:R:Y131 | 5.56 | 0 | Yes | Yes | 9 | 5 | 2 | 1 | | R:R:V108 | R:R:Y131 | 17.66 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:K110 | R:R:S129 | 9.18 | 6 | Yes | Yes | 8 | 7 | 2 | 1 | | R:R:S129 | R:R:Y131 | 7.63 | 6 | Yes | Yes | 7 | 5 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 6.84 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 22.00 | | Average Links Mediated by Hubs In Shell | 22.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|




































































