| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 1GZM | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.65 | 2003-11-20 | doi.org/10.1016/j.jmb.2004.08.090 |
|
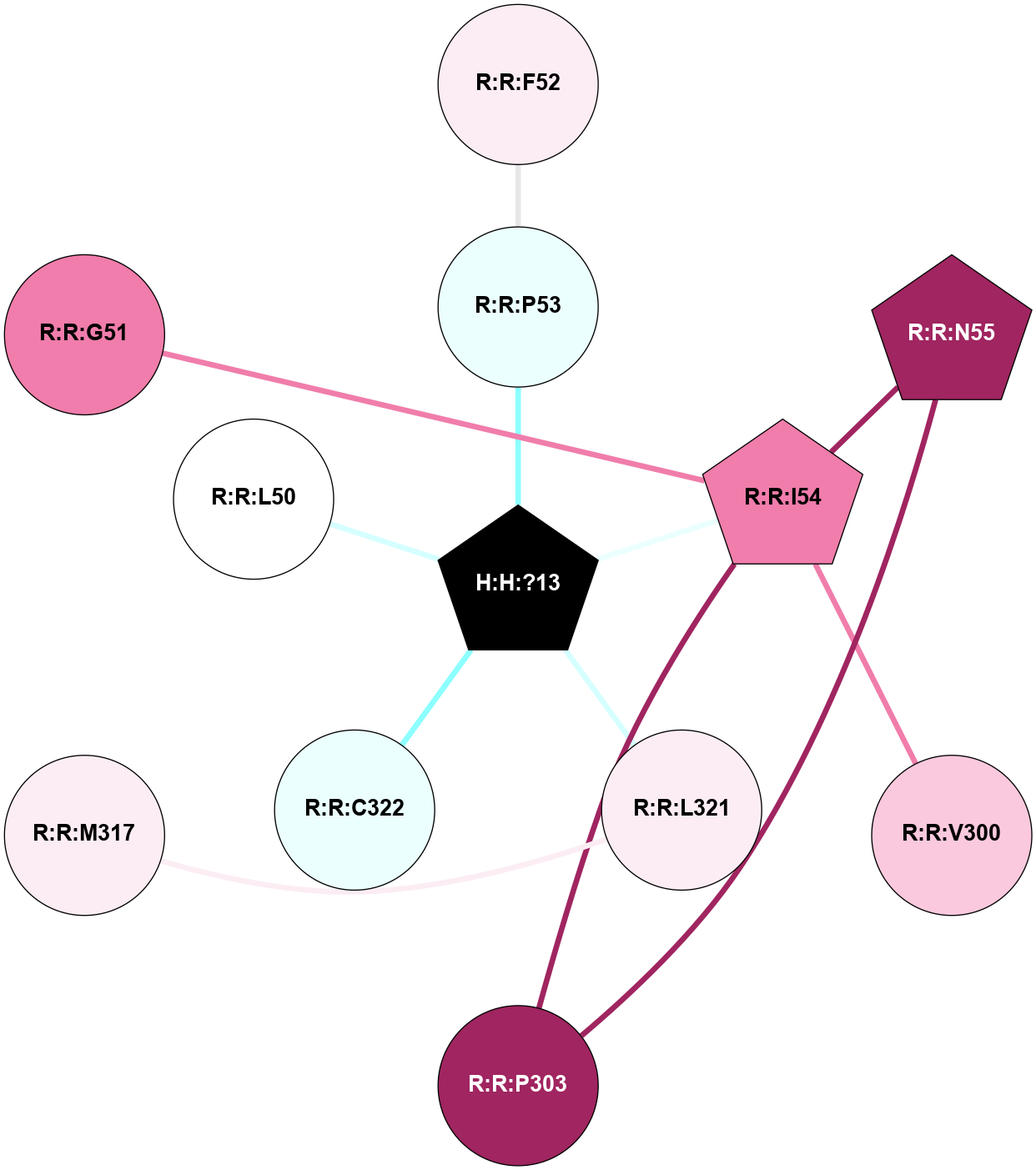
A 2D representation of the interactions of PLM in 1GZM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F52 | R:R:P53 | 4.33 | 0 | No | No | 6 | 4 | 2 | 1 | | H:H:?13 | R:R:P53 | 5.73 | 0 | Yes | No | 0 | 4 | 0 | 1 | | R:R:I54 | R:R:N55 | 4.25 | 6 | Yes | Yes | 8 | 9 | 1 | 2 | | R:R:I54 | R:R:V300 | 4.61 | 6 | Yes | No | 8 | 7 | 1 | 2 | | R:R:I54 | R:R:P303 | 6.77 | 6 | Yes | No | 8 | 9 | 1 | 2 | | H:H:?13 | R:R:I54 | 4.98 | 0 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:N55 | R:R:P303 | 6.52 | 6 | Yes | No | 9 | 9 | 2 | 2 | | R:R:L321 | R:R:M317 | 4.24 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?13 | R:R:L321 | 6.03 | 0 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?13 | R:R:C322 | 11.07 | 0 | Yes | No | 0 | 4 | 0 | 1 | | R:R:G51 | R:R:I54 | 3.53 | 0 | No | Yes | 8 | 8 | 2 | 1 | | H:H:?13 | R:R:L50 | 2.41 | 0 | Yes | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.04 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of PLM in 1GZM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F52 | R:R:P53 | 4.33 | 0 | No | No | 6 | 4 | 1 | 2 | | R:R:F52 | R:R:F88 | 6.43 | 0 | No | No | 6 | 4 | 1 | 2 | | H:H:?14 | R:R:F52 | 8.49 | 3 | Yes | No | 0 | 6 | 0 | 1 | | R:R:F56 | R:R:Y60 | 6.19 | 3 | No | No | 4 | 3 | 1 | 1 | | H:H:?14 | R:R:F56 | 9.55 | 3 | Yes | No | 0 | 4 | 0 | 1 | | R:R:L57 | R:R:V61 | 4.47 | 0 | No | No | 6 | 7 | 1 | 2 | | H:H:?14 | R:R:L57 | 6.03 | 3 | Yes | No | 0 | 6 | 0 | 1 | | R:R:Q64 | R:R:Y60 | 5.64 | 3 | No | No | 5 | 3 | 2 | 1 | | H:H:?14 | R:R:Y60 | 11.24 | 3 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?14 | R:R:C323 | 9.68 | 3 | Yes | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 9.00 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 1HZX | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.8 | 2001-07-04 | doi.org/10.1021/bi0155091 |
|
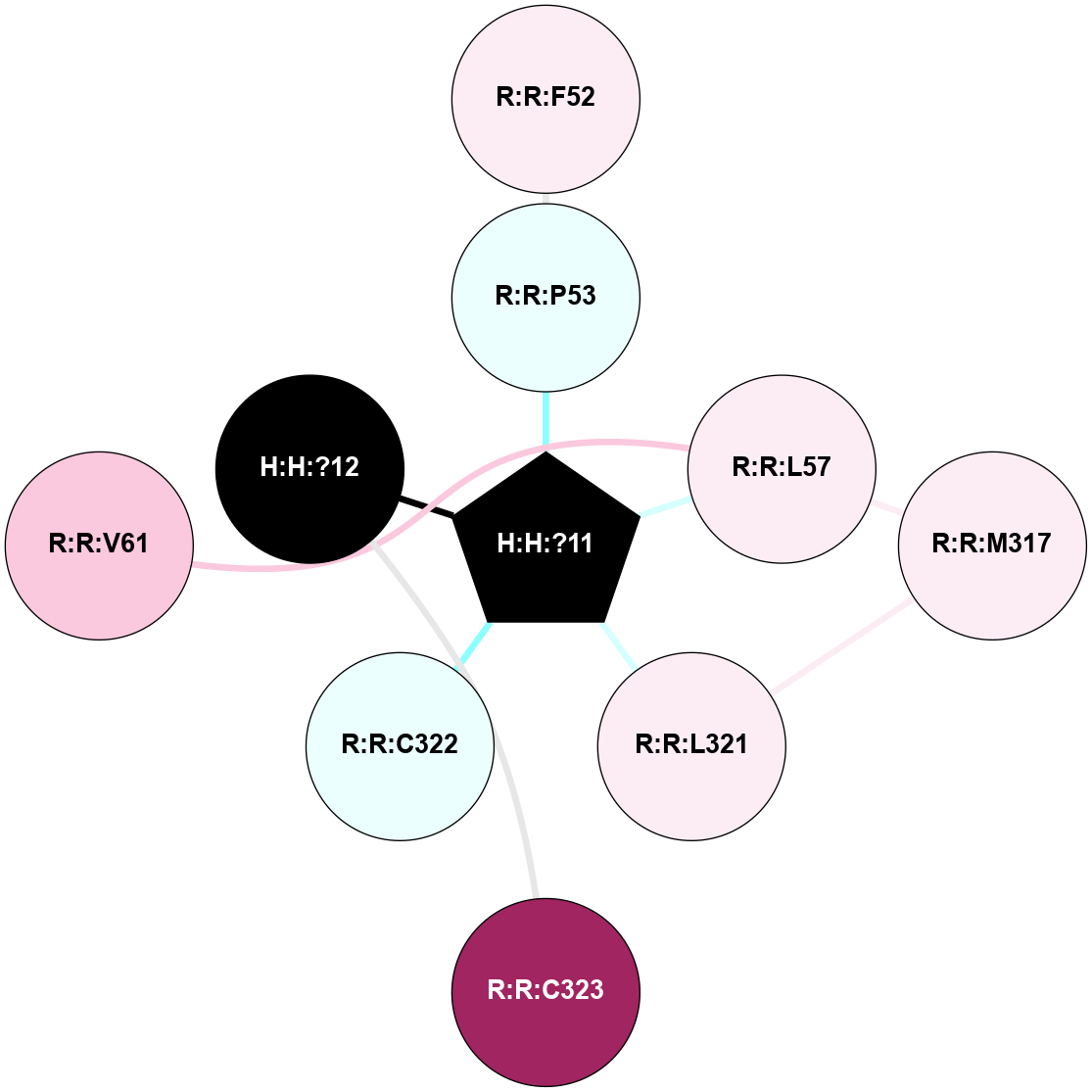
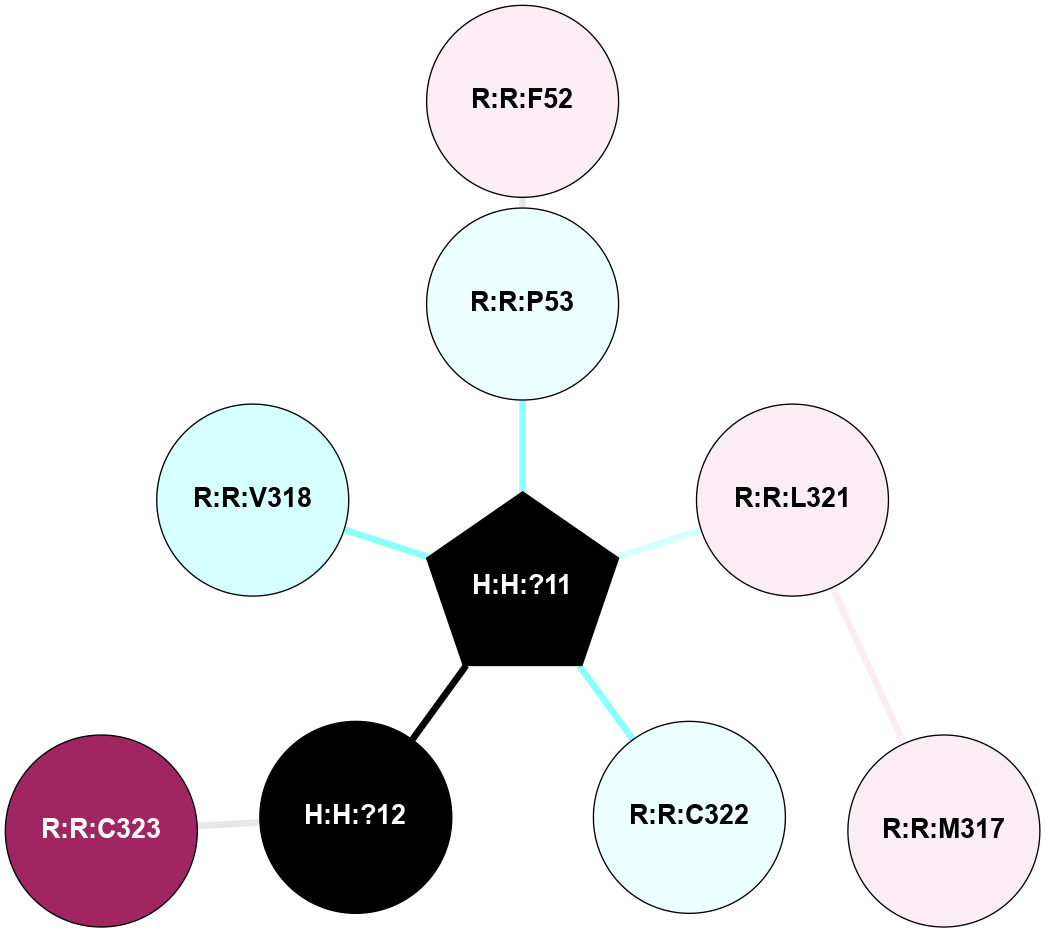
A 2D representation of the interactions of PLM in 1HZX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F52 | R:R:P53 | 4.33 | 0 | No | No | 6 | 4 | 2 | 1 | | H:H:?11 | R:R:P53 | 4.29 | 0 | Yes | No | 0 | 4 | 0 | 1 | | R:R:L57 | R:R:M317 | 5.65 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?11 | R:R:L57 | 3.62 | 0 | Yes | No | 0 | 6 | 0 | 1 | | R:R:L321 | R:R:M317 | 4.24 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?11 | R:R:L321 | 7.24 | 0 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?11 | R:R:C322 | 11.07 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?12 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 1 | 2 | | H:H:?11 | H:H:?12 | 4.21 | 0 | Yes | No | 0 | 0 | 0 | 1 | | R:R:L57 | R:R:V61 | 2.98 | 0 | No | No | 6 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.09 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
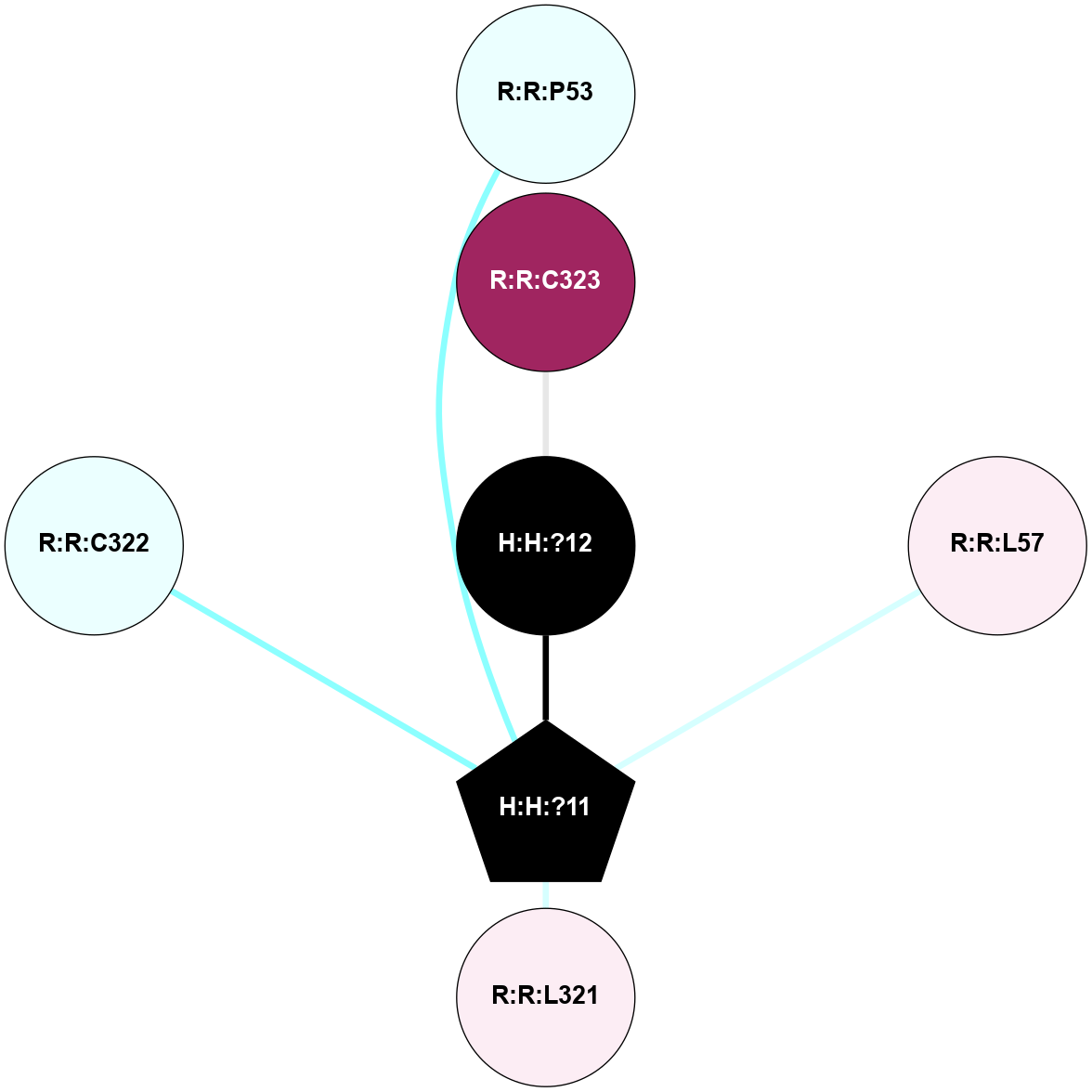
A 2D representation of the interactions of PLM in 1HZX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | R:R:P53 | 4.29 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?11 | R:R:L57 | 3.62 | 0 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?11 | R:R:L321 | 7.24 | 0 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?11 | R:R:C322 | 11.07 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?12 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?11 | H:H:?12 | 4.21 | 0 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.95 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 1L9H | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.6 | 2002-05-15 | doi.org/10.1073/pnas.082666399 |
|
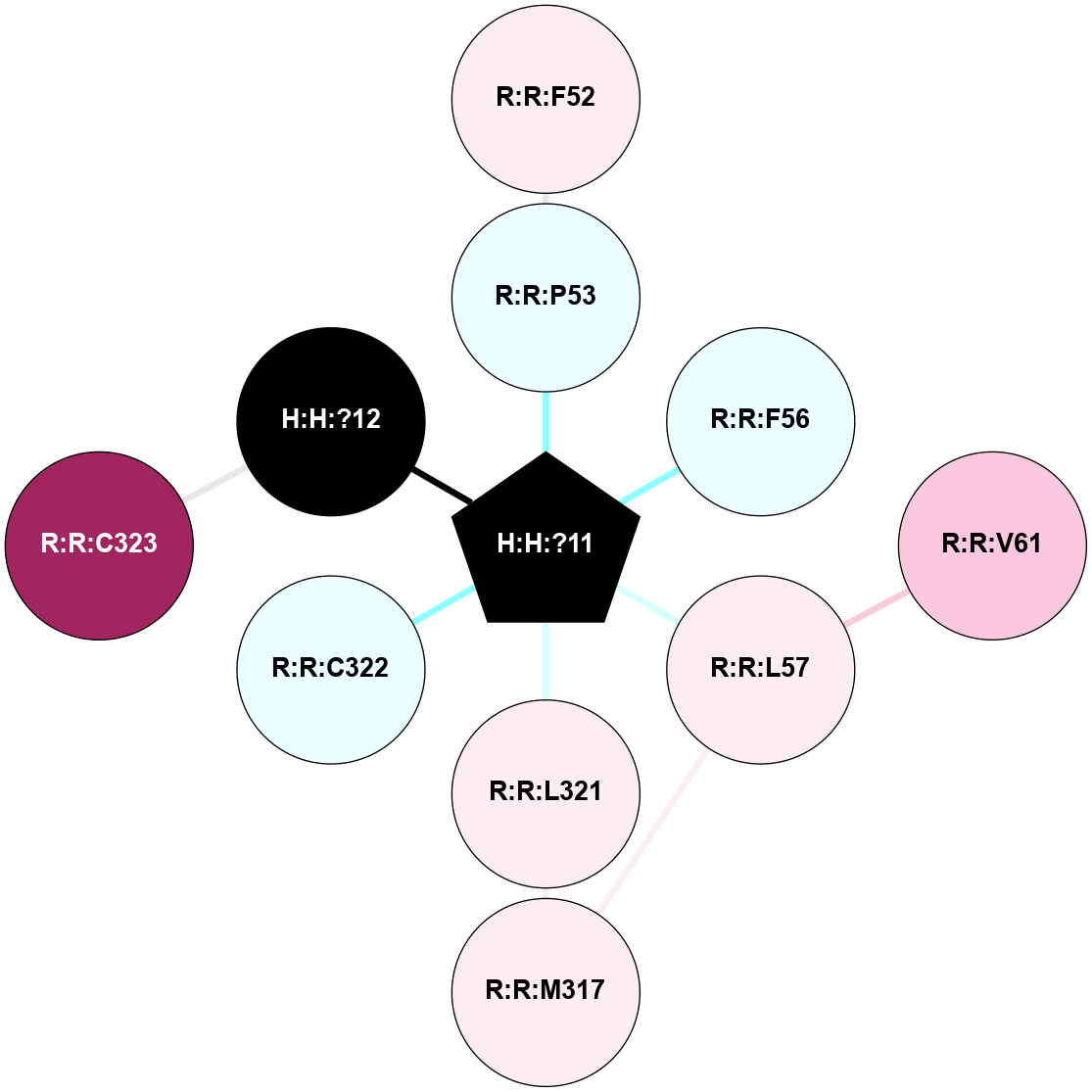
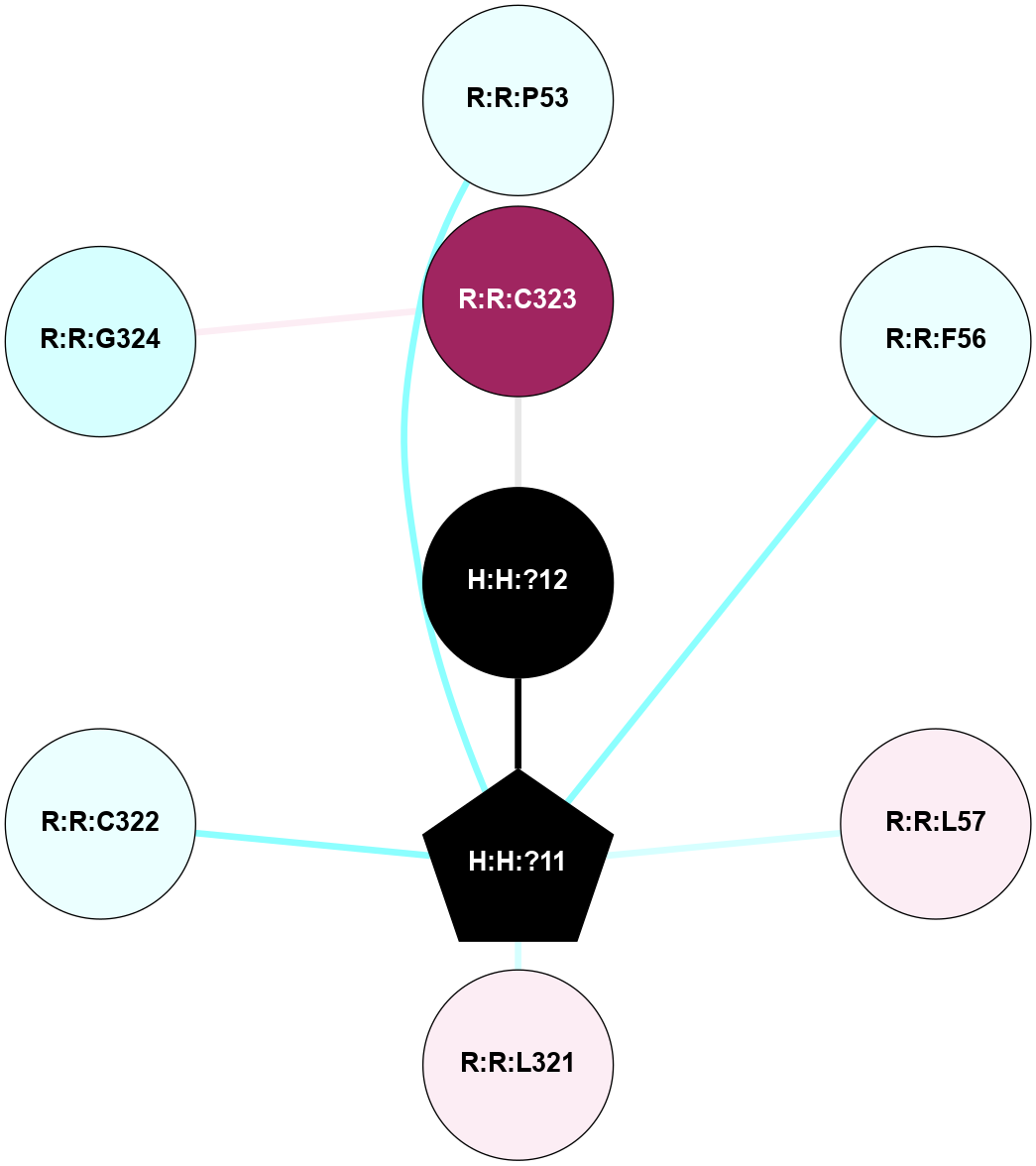
A 2D representation of the interactions of PLM in 1L9H
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F52 | R:R:P53 | 7.22 | 0 | No | No | 6 | 4 | 2 | 1 | | H:H:?11 | R:R:P53 | 4.29 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?11 | R:R:F56 | 4.25 | 0 | Yes | No | 0 | 4 | 0 | 1 | | R:R:L57 | R:R:V61 | 4.47 | 0 | No | No | 6 | 7 | 1 | 2 | | R:R:L57 | R:R:M317 | 5.65 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?11 | R:R:L57 | 4.83 | 0 | Yes | No | 0 | 6 | 0 | 1 | | R:R:L321 | R:R:M317 | 5.65 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?11 | R:R:L321 | 9.65 | 0 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?11 | R:R:C322 | 9.68 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?12 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 1 | 2 | | H:H:?11 | H:H:?12 | 6.31 | 0 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.50 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
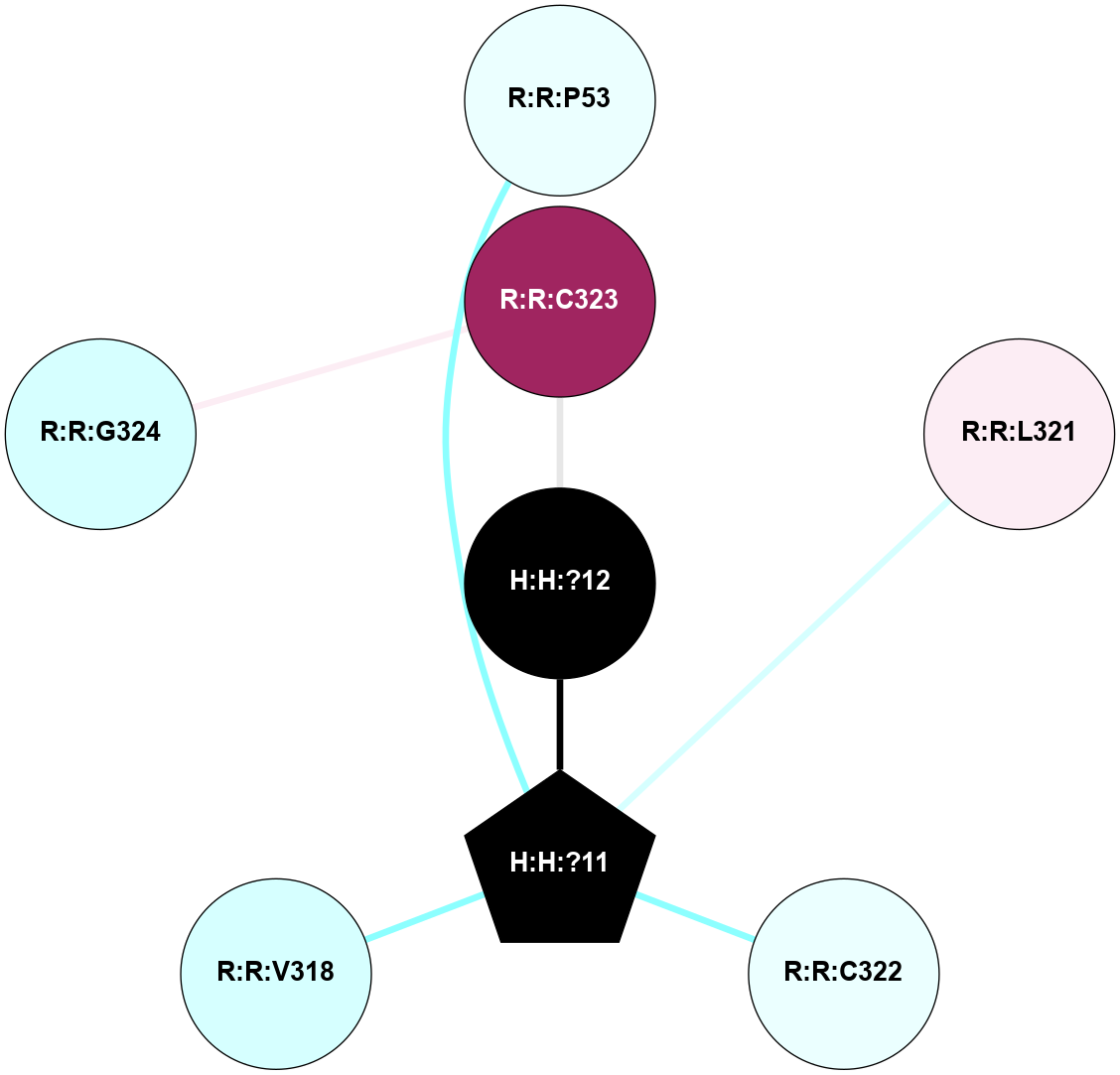
A 2D representation of the interactions of PLM in 1L9H
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | R:R:P53 | 4.29 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?11 | R:R:F56 | 4.25 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?11 | R:R:L57 | 4.83 | 0 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?11 | R:R:L321 | 9.65 | 0 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?11 | R:R:C322 | 9.68 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?12 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?11 | H:H:?12 | 6.31 | 0 | Yes | No | 0 | 0 | 1 | 0 | | R:R:C323 | R:R:G324 | 1.96 | 0 | No | No | 9 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 7.99 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 1U19 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.2 | 2004-10-12 | doi.org/10.1016/j.jmb.2004.07.044 |
|
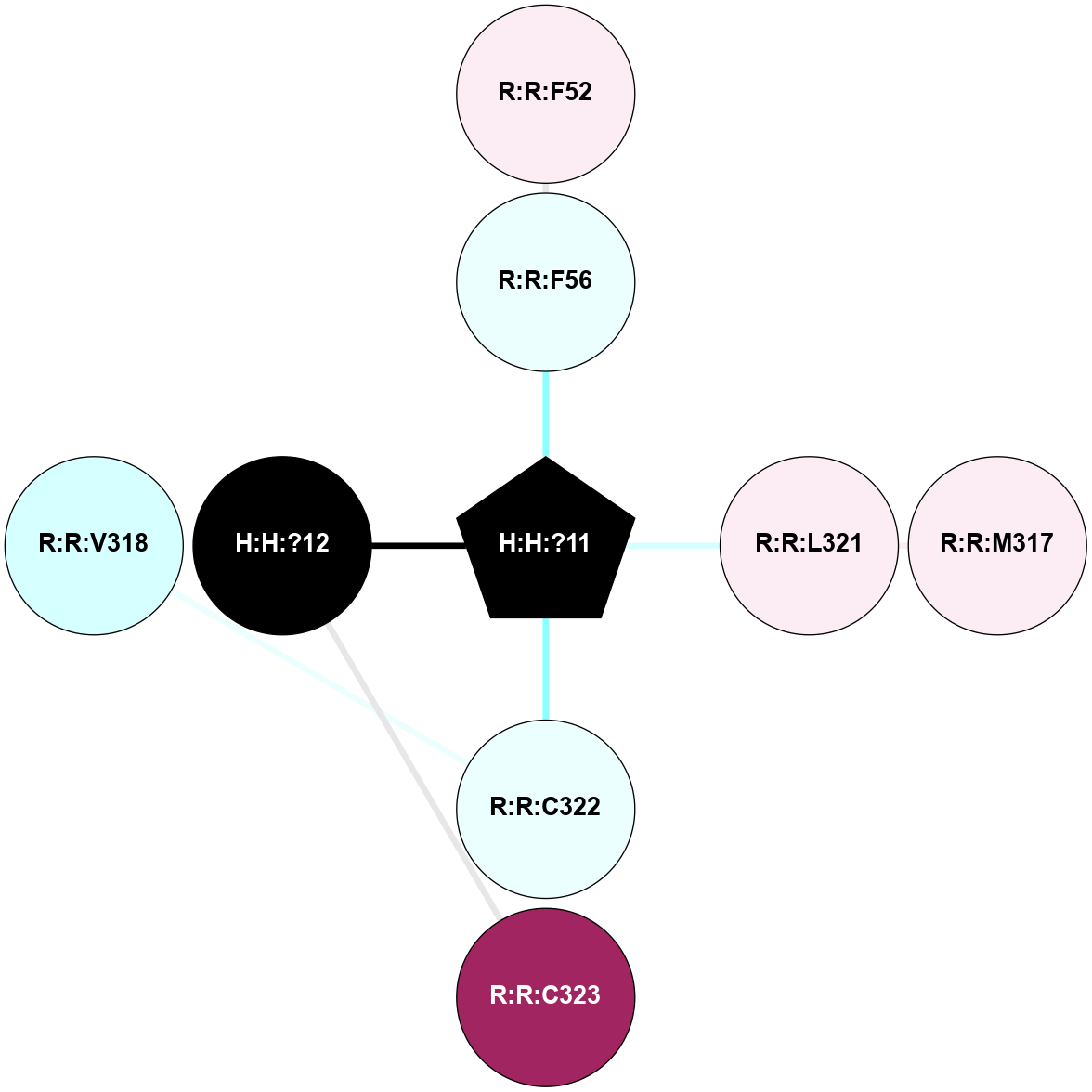
A 2D representation of the interactions of PLM in 1U19
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F52 | R:R:P53 | 4.33 | 0 | No | No | 6 | 4 | 2 | 1 | | H:H:?11 | R:R:P53 | 4.29 | 0 | Yes | No | 0 | 4 | 0 | 1 | | R:R:L321 | R:R:M317 | 4.24 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?11 | R:R:L321 | 14.48 | 0 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?11 | R:R:C322 | 9.68 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?12 | R:R:C323 | 11.07 | 0 | No | No | 0 | 9 | 1 | 2 | | H:H:?11 | H:H:?12 | 7.36 | 0 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?11 | R:R:V318 | 2.6 | 0 | Yes | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.68 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of PLM in 1U19
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | R:R:P53 | 4.29 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?11 | R:R:L321 | 14.48 | 0 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?11 | R:R:C322 | 9.68 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?12 | R:R:C323 | 11.07 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?11 | H:H:?12 | 7.36 | 0 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?11 | R:R:V318 | 2.6 | 0 | Yes | No | 0 | 3 | 1 | 2 | | R:R:C323 | R:R:G324 | 1.96 | 0 | No | No | 9 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 9.21 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2G87 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 2.6 | 2006-09-02 | doi.org/10.1002/anie.200600595 |
|
A 2D representation of the interactions of PLM in 2G87
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F52 | R:R:F56 | 4.29 | 0 | No | No | 6 | 4 | 2 | 1 | | H:H:?11 | R:R:F56 | 5.31 | 0 | Yes | No | 0 | 4 | 0 | 1 | | R:R:L321 | R:R:M317 | 4.24 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?11 | R:R:L321 | 12.06 | 0 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?11 | R:R:C322 | 9.68 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?12 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 1 | 2 | | H:H:?11 | H:H:?12 | 6.31 | 0 | Yes | No | 0 | 0 | 0 | 1 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 8.34 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of PLM in 2G87
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | R:R:F56 | 5.31 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?11 | R:R:L321 | 12.06 | 0 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?11 | R:R:C322 | 9.68 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?12 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?11 | H:H:?12 | 6.31 | 0 | Yes | No | 0 | 0 | 1 | 0 | | R:R:C323 | R:R:G324 | 1.96 | 0 | No | No | 9 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 7.99 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2HPY | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 2.8 | 2006-08-22 | doi.org/10.1073/pnas.0601765103 |
|
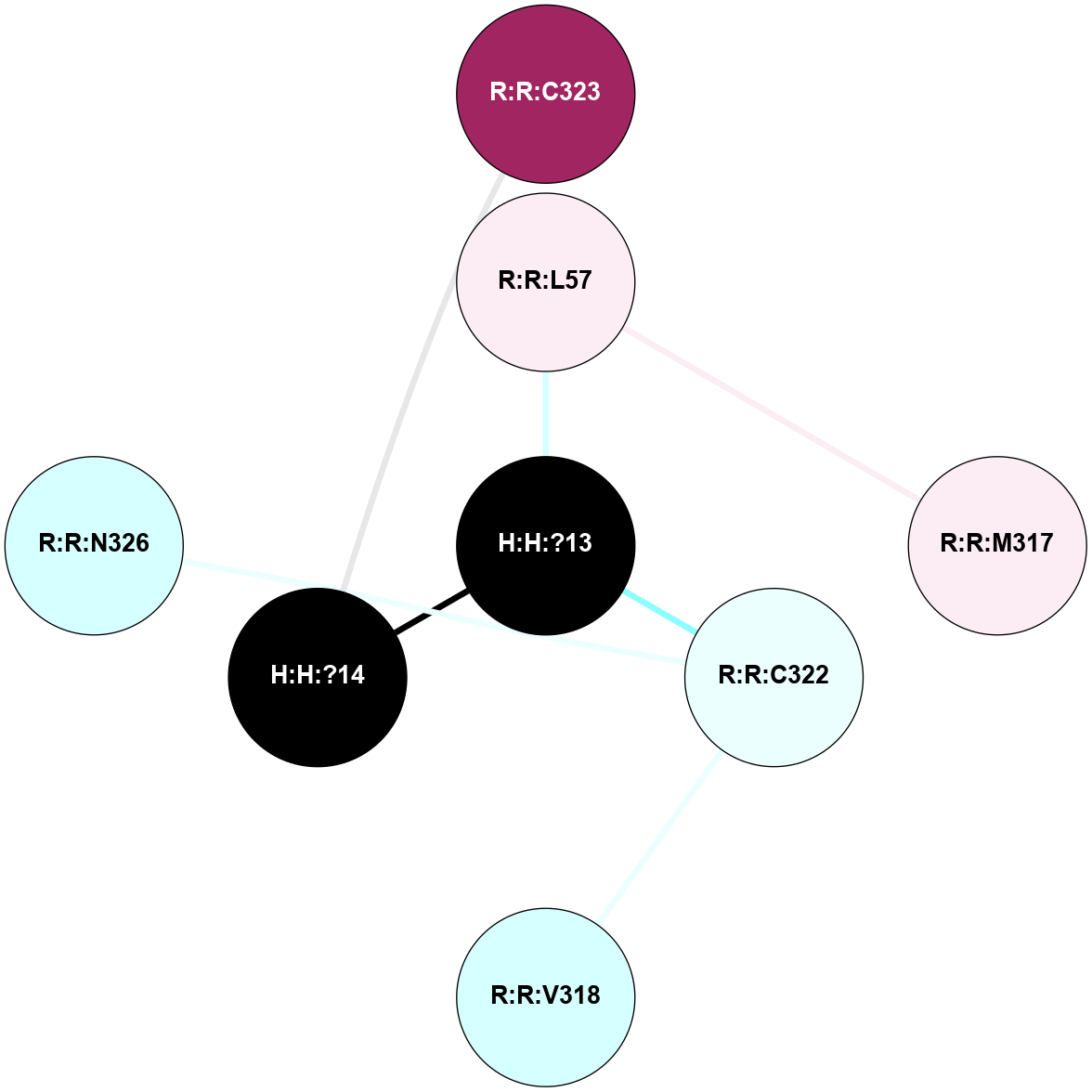
A 2D representation of the interactions of PLM in 2HPY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | R:R:L57 | 4.83 | 0 | No | No | 0 | 6 | 0 | 1 | | H:H:?13 | R:R:C322 | 11.07 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?14 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 1 | 2 | | H:H:?13 | H:H:?14 | 15.77 | 0 | No | No | 0 | 0 | 0 | 1 | | R:R:L57 | R:R:M317 | 2.83 | 0 | No | No | 6 | 6 | 1 | 2 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | | R:R:C322 | R:R:N326 | 1.57 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 10.56 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of PLM in 2HPY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | R:R:L57 | 4.83 | 0 | No | No | 0 | 6 | 1 | 2 | | H:H:?13 | R:R:C322 | 11.07 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?14 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?13 | H:H:?14 | 15.77 | 0 | No | No | 0 | 0 | 1 | 0 | | R:R:C323 | R:R:G324 | 1.96 | 0 | No | No | 9 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.72 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2I36 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | - | 4.1 | 2006-10-17 | doi.org/10.1073/pnas.0608022103 |
|
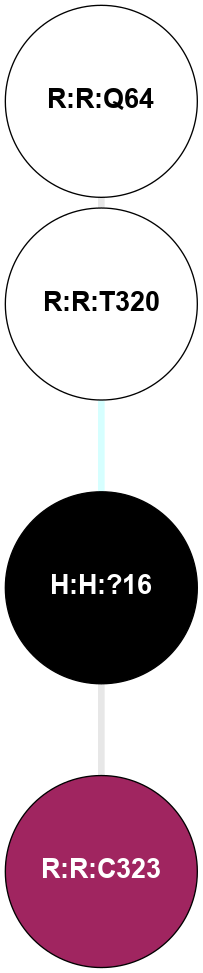
A 2D representation of the interactions of PLM in 2I36
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:Q64 | R:R:T320 | 8.5 | 0 | No | No | 5 | 5 | 2 | 1 | | H:H:?16 | R:R:T320 | 5.14 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?16 | R:R:C323 | 8.3 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.72 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2PED | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 9-cis-Retinal | - | - | 2.95 | 2007-10-30 | doi.org/10.1529/biophysj.107.108225 |
|
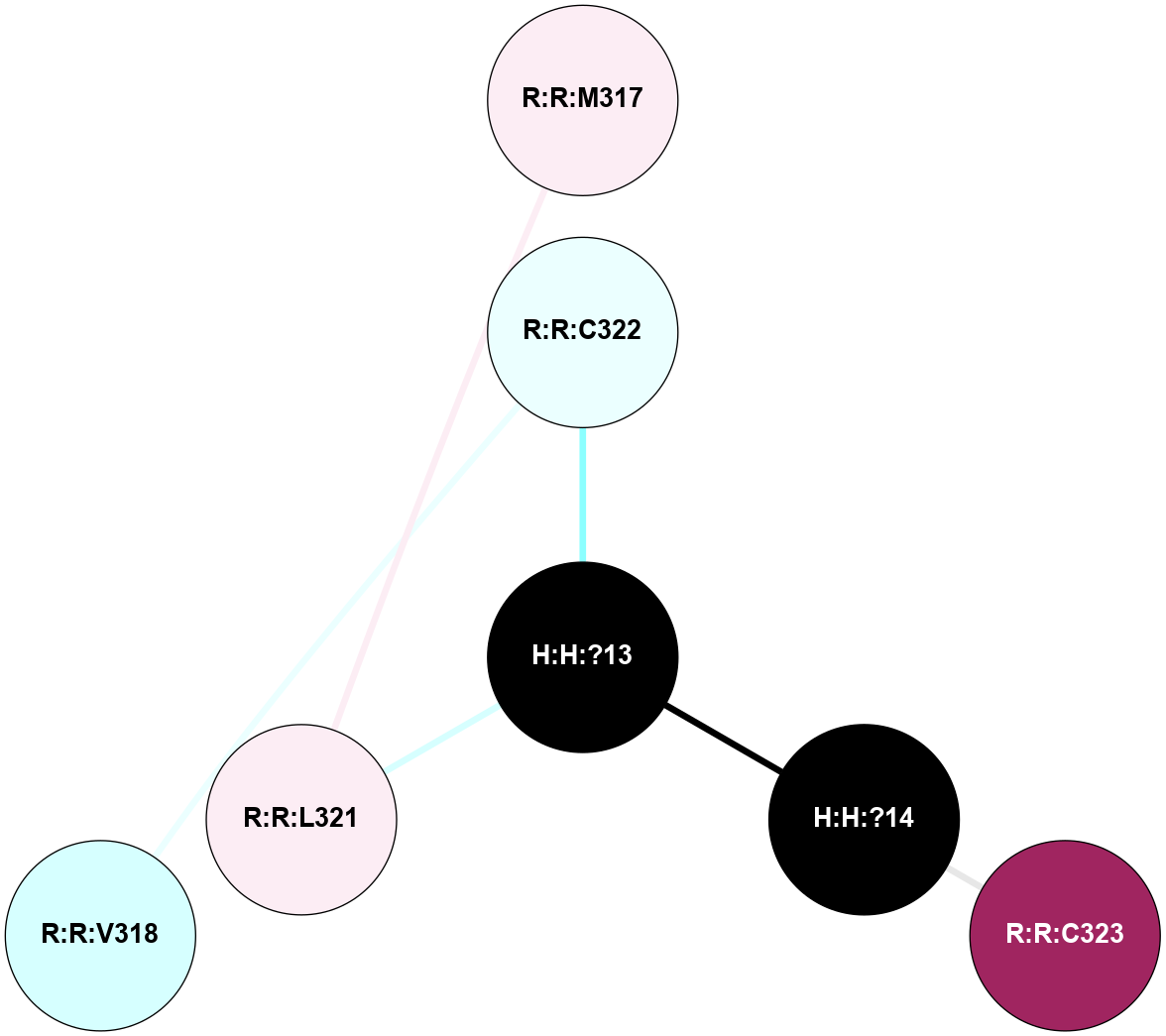
A 2D representation of the interactions of PLM in 2PED
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L321 | R:R:M317 | 5.65 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?13 | R:R:C322 | 11.07 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?14 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 1 | 2 | | H:H:?13 | H:H:?14 | 16.83 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?13 | R:R:L321 | 2.41 | 0 | No | No | 0 | 6 | 0 | 1 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 10.10 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
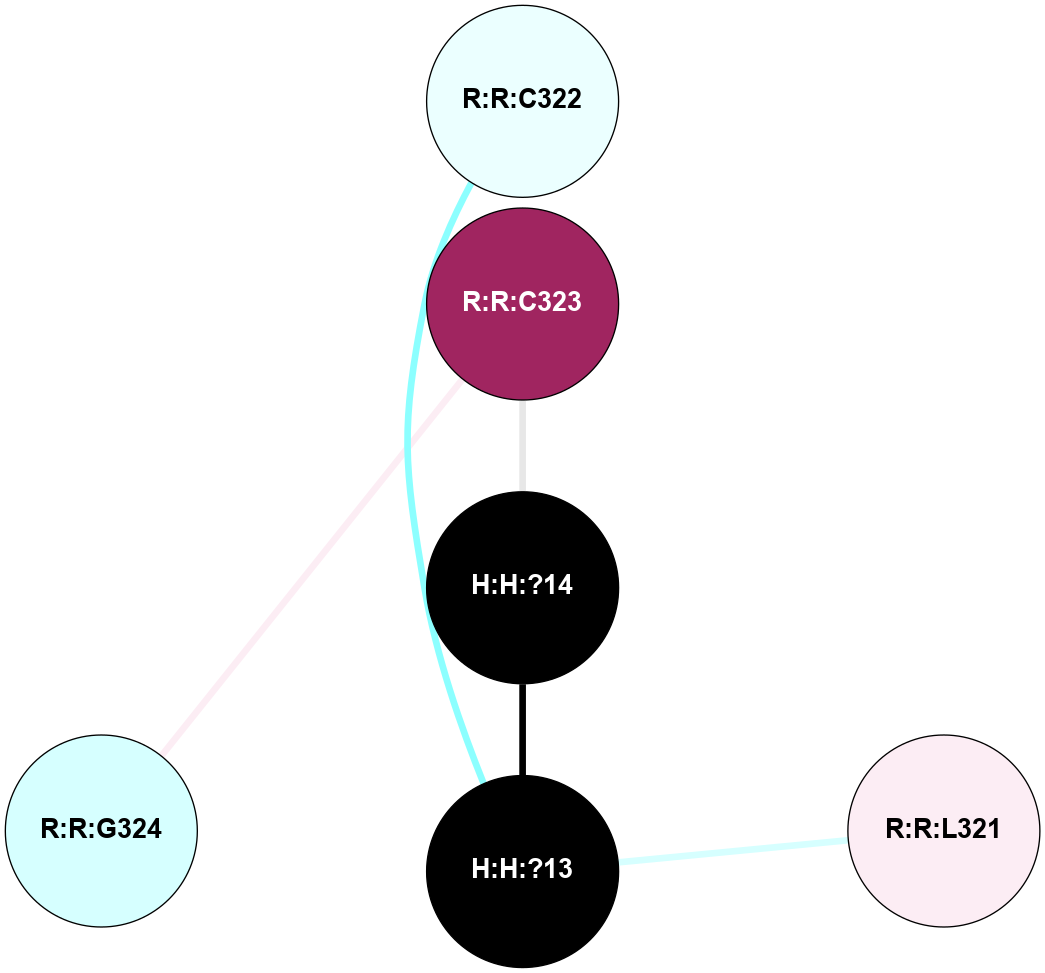
A 2D representation of the interactions of PLM in 2PED
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | R:R:C322 | 11.07 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?14 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?13 | H:H:?14 | 16.83 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?13 | R:R:L321 | 2.41 | 0 | No | No | 0 | 6 | 1 | 2 | | R:R:C323 | R:R:G324 | 1.96 | 0 | No | No | 9 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.25 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2RH1 | A | Amine | Adrenergic | Beta2 | Homo Sapiens | S-Carazolol | - | - | 2.4 | 2007-10-30 | doi.org/10.1126/science.1150577 |
|
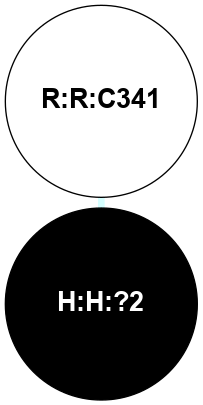
A 2D representation of the interactions of PLM in 2RH1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:C341 | 9.68 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 9.68 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2X72 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | Gt(CT) | 3 | 2011-03-16 | doi.org/10.1038/nature09795 |
|

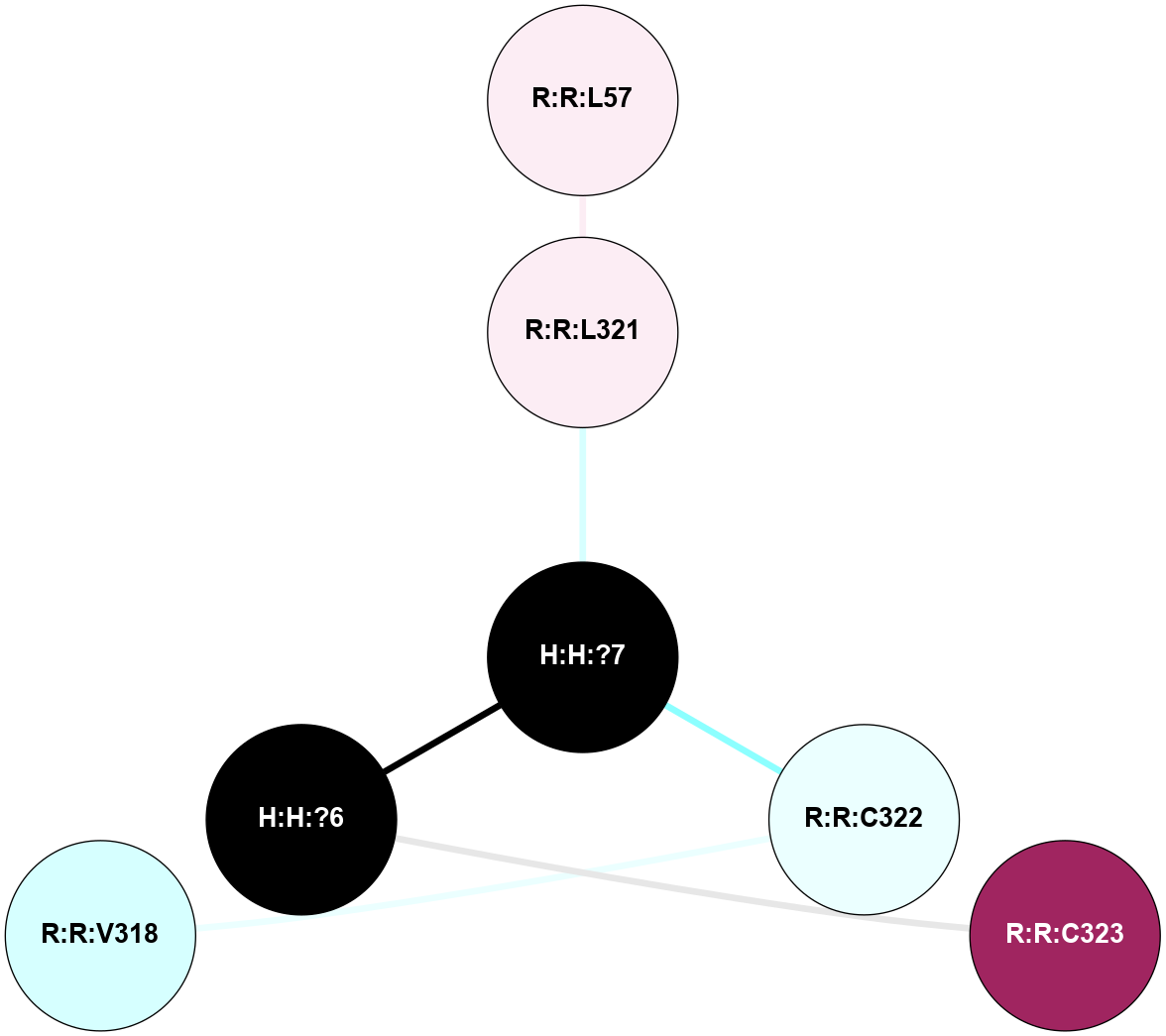
A 2D representation of the interactions of PLM in 2X72
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | R:R:L321 | 4.83 | 0 | No | No | 0 | 6 | 1 | 2 | | H:H:?7 | R:R:C322 | 9.68 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?6 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?6 | H:H:?7 | 19.98 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.83 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
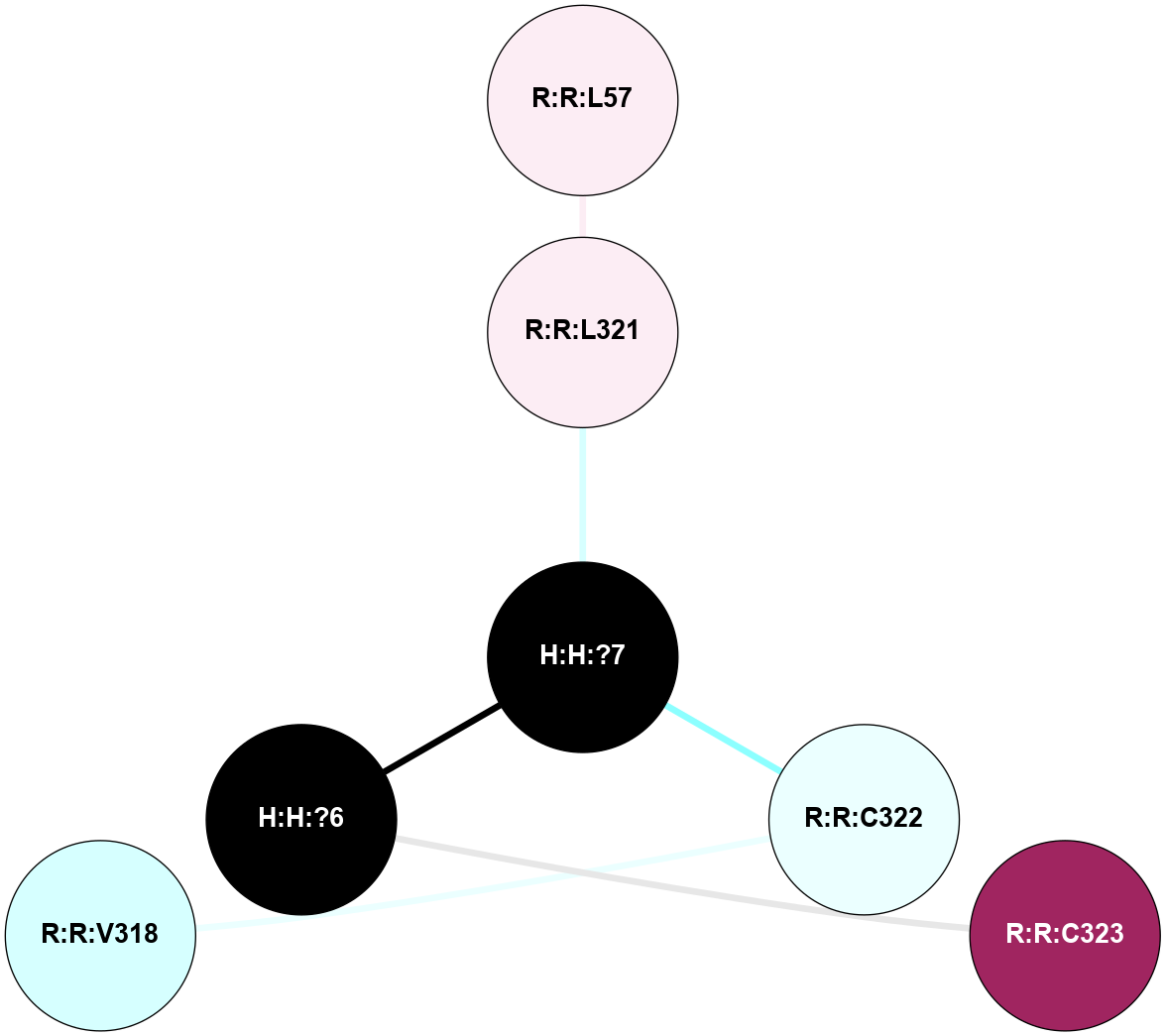
A 2D representation of the interactions of PLM in 2X72
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L321 | R:R:L57 | 4.15 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?7 | R:R:L321 | 4.83 | 0 | No | No | 0 | 6 | 0 | 1 | | H:H:?7 | R:R:C322 | 9.68 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?6 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 1 | 2 | | H:H:?6 | H:H:?7 | 19.98 | 0 | No | No | 0 | 0 | 1 | 0 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 11.50 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2Z73 | A | Sensory | Opsins | Rhodopsin | Todarodes Pacificus | 11-cis-Retinal | - | - | 2.5 | 2008-05-13 | doi.org/10.1038/nature06925 |
|

A 2D representation of the interactions of PLM in 2Z73
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I309 | R:R:I47 | 5.89 | 9 | Yes | No | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:M313 | 7.39 | 0 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:S316 | 6.54 | 0 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:V317 | 5.19 | 0 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:C337 | 8.3 | 0 | Yes | No | 0 | 9 | 0 | 1 | | R:R:I47 | R:R:M313 | 2.92 | 9 | No | No | 6 | 6 | 2 | 1 | | R:R:I309 | R:R:M313 | 2.92 | 9 | Yes | No | 7 | 6 | 2 | 1 | | R:R:C336 | R:R:C337 | 1.82 | 0 | No | No | 4 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.86 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2ZIY | A | Sensory | Opsins | Rhodopsin | Todarodes Pacificus | 11-cis-Retinal | - | - | 3.7 | 2008-05-06 | doi.org/10.1074/jbc.C800040200 |
|

A 2D representation of the interactions of PLM in 2ZIY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I43 | R:R:I47 | 4.42 | 0 | No | Yes | 4 | 6 | 1 | 2 | | H:H:?1 | R:R:I43 | 3.73 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:V317 | 3.9 | 0 | No | No | 0 | 5 | 1 | 2 | | H:H:?1 | R:R:C336 | 9.68 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:C337 | 9.68 | 0 | No | No | 0 | 9 | 1 | 2 | | H:H:?1 | H:H:?2 | 7.36 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.92 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of PLM in 2ZIY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:I43 | 3.73 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?2 | R:R:V317 | 3.9 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:C337 | R:R:L334 | 6.35 | 0 | No | No | 9 | 1 | 1 | 2 | | H:H:?1 | R:R:C336 | 9.68 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?2 | R:R:C337 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?1 | H:H:?2 | 7.36 | 0 | No | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.98 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3C9L | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.65 | 2008-08-05 | doi.org/10.1107/S0907444908017162 |
|
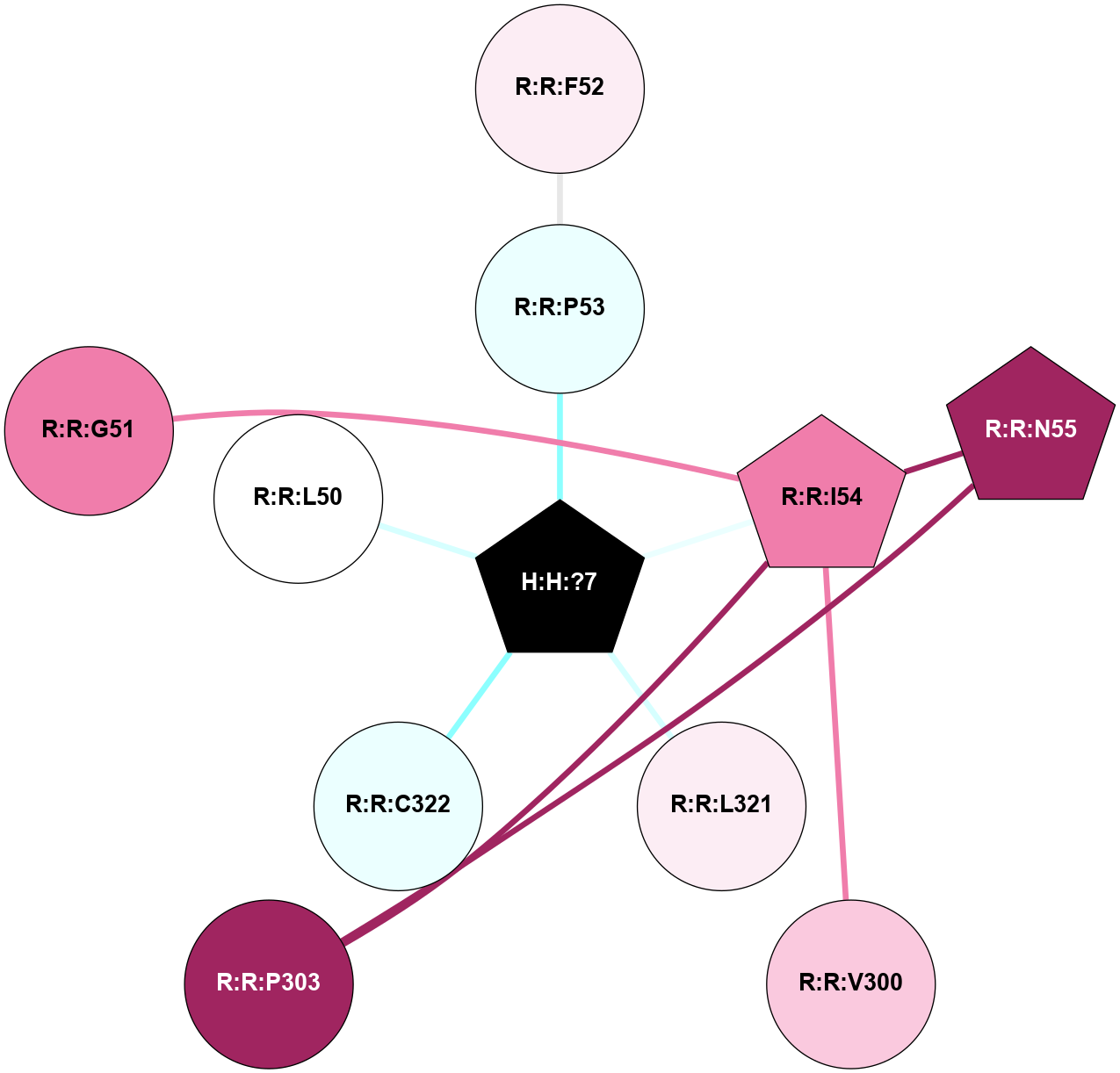
A 2D representation of the interactions of PLM in 3C9L
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | R:R:P53 | 5.73 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?7 | R:R:I54 | 4.98 | 0 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?7 | R:R:L321 | 6.03 | 0 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?7 | R:R:C322 | 11.07 | 0 | Yes | No | 0 | 4 | 0 | 1 | | R:R:F52 | R:R:P53 | 4.33 | 0 | No | No | 6 | 4 | 2 | 1 | | R:R:I54 | R:R:N55 | 4.25 | 5 | Yes | Yes | 8 | 9 | 1 | 2 | | R:R:I54 | R:R:V300 | 4.61 | 5 | Yes | No | 8 | 7 | 1 | 2 | | R:R:I54 | R:R:P303 | 8.47 | 5 | Yes | No | 8 | 9 | 1 | 2 | | R:R:N55 | R:R:P303 | 6.52 | 5 | Yes | No | 9 | 9 | 2 | 2 | | R:R:G51 | R:R:I54 | 3.53 | 0 | No | Yes | 8 | 8 | 2 | 1 | | H:H:?7 | R:R:L50 | 2.41 | 0 | Yes | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.04 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
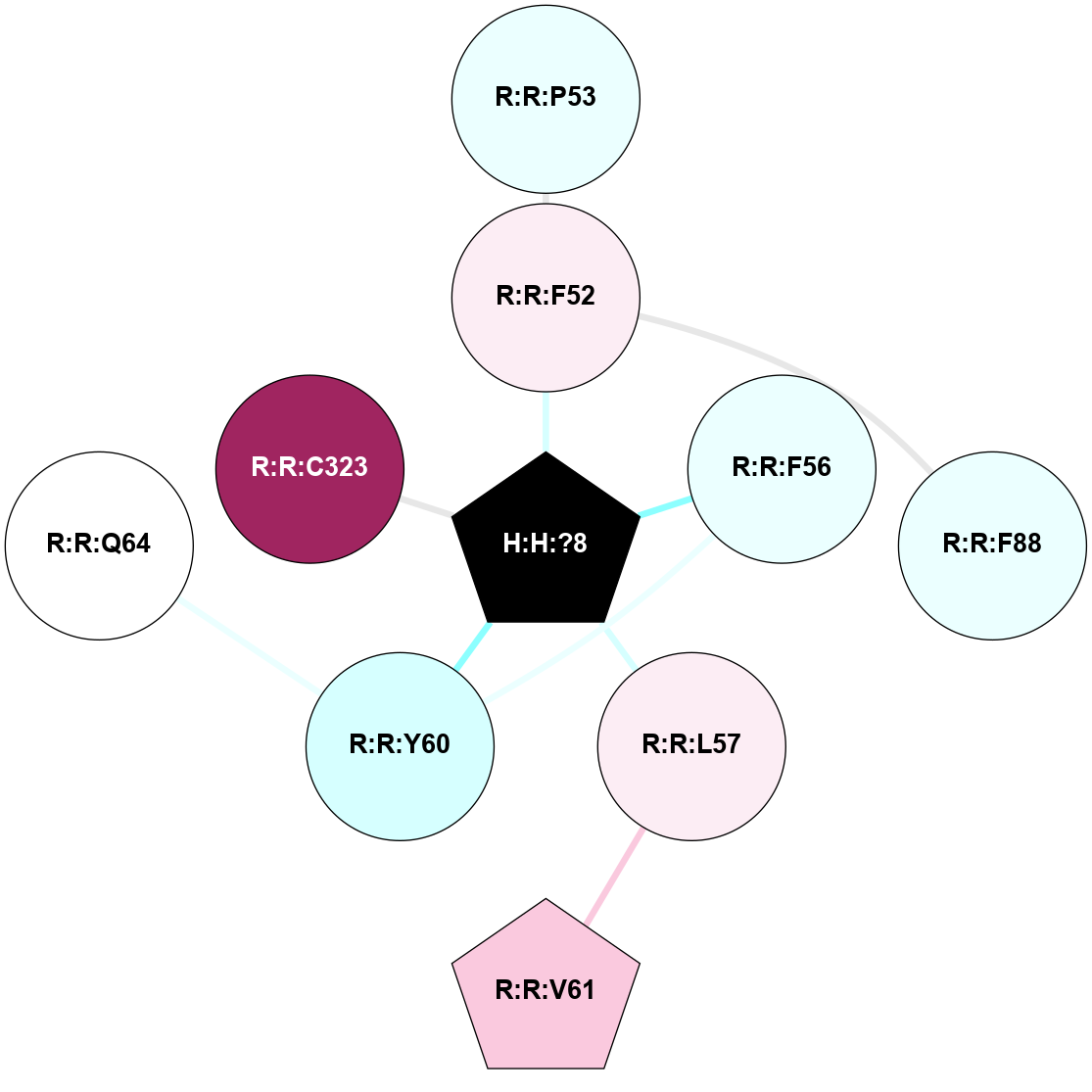
A 2D representation of the interactions of PLM in 3C9L
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | R:R:F52 | 7.43 | 6 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?8 | R:R:F56 | 7.43 | 6 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?8 | R:R:L57 | 6.03 | 6 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?8 | R:R:Y60 | 11.24 | 6 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?8 | R:R:C323 | 9.68 | 6 | Yes | No | 0 | 9 | 0 | 1 | | R:R:F52 | R:R:P53 | 4.33 | 0 | No | No | 6 | 4 | 1 | 2 | | R:R:F52 | R:R:F88 | 7.5 | 0 | No | No | 6 | 4 | 1 | 2 | | R:R:F56 | R:R:Y60 | 6.19 | 6 | No | No | 4 | 3 | 1 | 1 | | R:R:L57 | R:R:V61 | 4.47 | 0 | No | Yes | 6 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 8.36 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3CAP | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | - | 2.9 | 2008-06-24 | doi.org/10.1038/nature07063 |
|

A 2D representation of the interactions of PLM in 3CAP
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | R:R:C322 | 9.68 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:C322 | R:R:N326 | 3.15 | 0 | No | No | 4 | 3 | 1 | 2 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 9.68 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3DQB | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | Gt(CT) | 3.2 | 2008-09-23 | doi.org/10.1038/nature07330 |
|

A 2D representation of the interactions of PLM in 3DQB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | R:R:L321 | 3.62 | 0 | No | No | 0 | 6 | 1 | 2 | | H:H:?7 | R:R:C322 | 8.3 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?6 | R:R:C323 | 8.3 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?6 | H:H:?7 | 14.72 | 0 | No | No | 0 | 0 | 0 | 1 | | R:R:C323 | R:R:G324 | 1.96 | 0 | No | No | 9 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 11.51 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of PLM in 3DQB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L321 | R:R:L57 | 5.54 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?7 | R:R:L321 | 3.62 | 0 | No | No | 0 | 6 | 0 | 1 | | H:H:?7 | R:R:C322 | 8.3 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?6 | R:R:C323 | 8.3 | 0 | No | No | 0 | 9 | 1 | 2 | | H:H:?6 | H:H:?7 | 14.72 | 0 | No | No | 0 | 0 | 1 | 0 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 8.88 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3OAX | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.6 | 2011-01-19 | doi.org/10.1016/j.bpj.2010.08.003 |
|
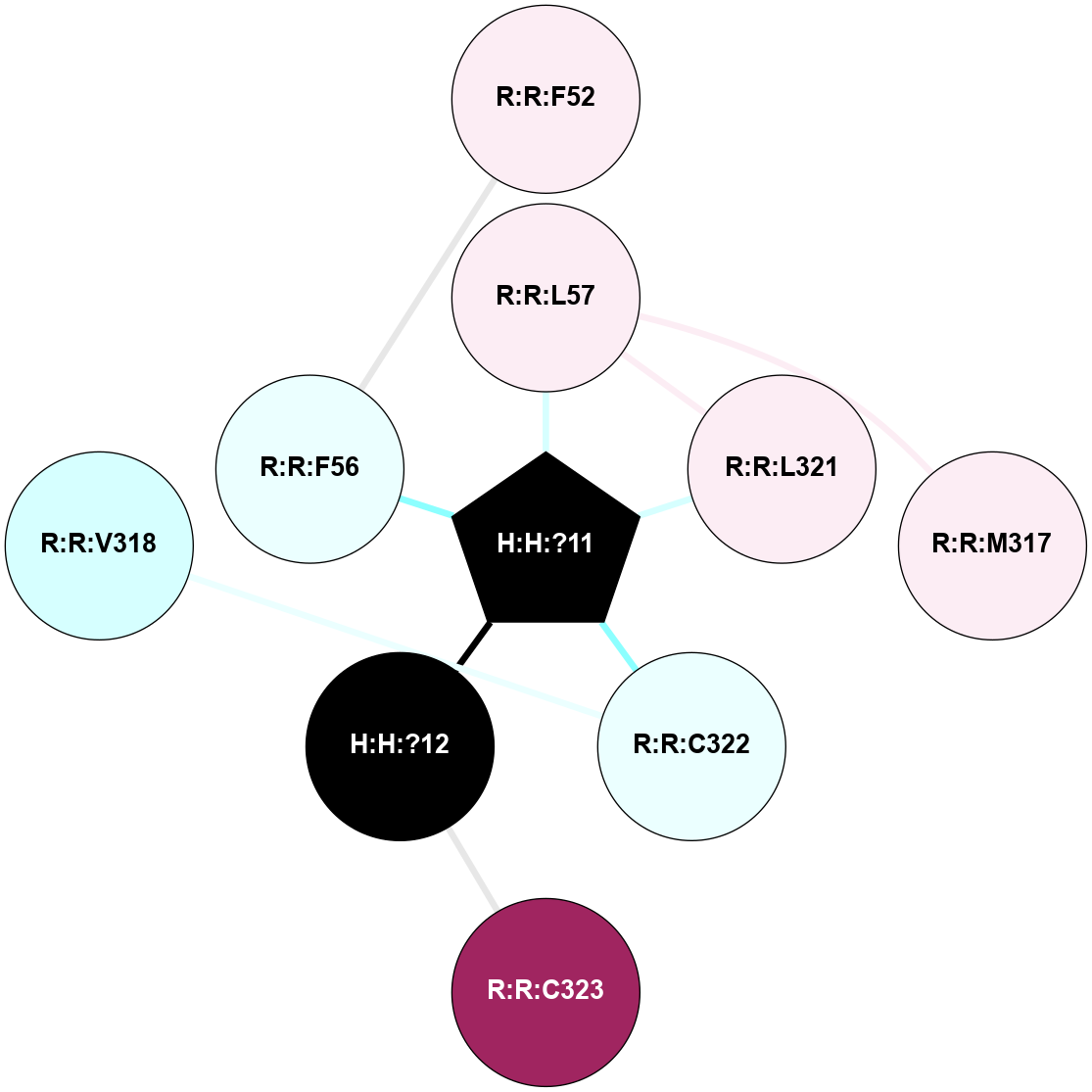
A 2D representation of the interactions of PLM in 3OAX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F52 | R:R:F56 | 6.43 | 0 | No | No | 6 | 4 | 2 | 1 | | R:R:L57 | R:R:M317 | 5.65 | 10 | No | No | 6 | 6 | 1 | 2 | | R:R:L321 | R:R:L57 | 5.54 | 10 | No | No | 6 | 6 | 1 | 1 | | H:H:?11 | R:R:L57 | 4.83 | 10 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?11 | R:R:L321 | 9.65 | 10 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?11 | R:R:C322 | 9.68 | 10 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?12 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 1 | 2 | | H:H:?11 | H:H:?12 | 5.26 | 10 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?11 | R:R:F56 | 3.18 | 10 | Yes | No | 0 | 4 | 0 | 1 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.52 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
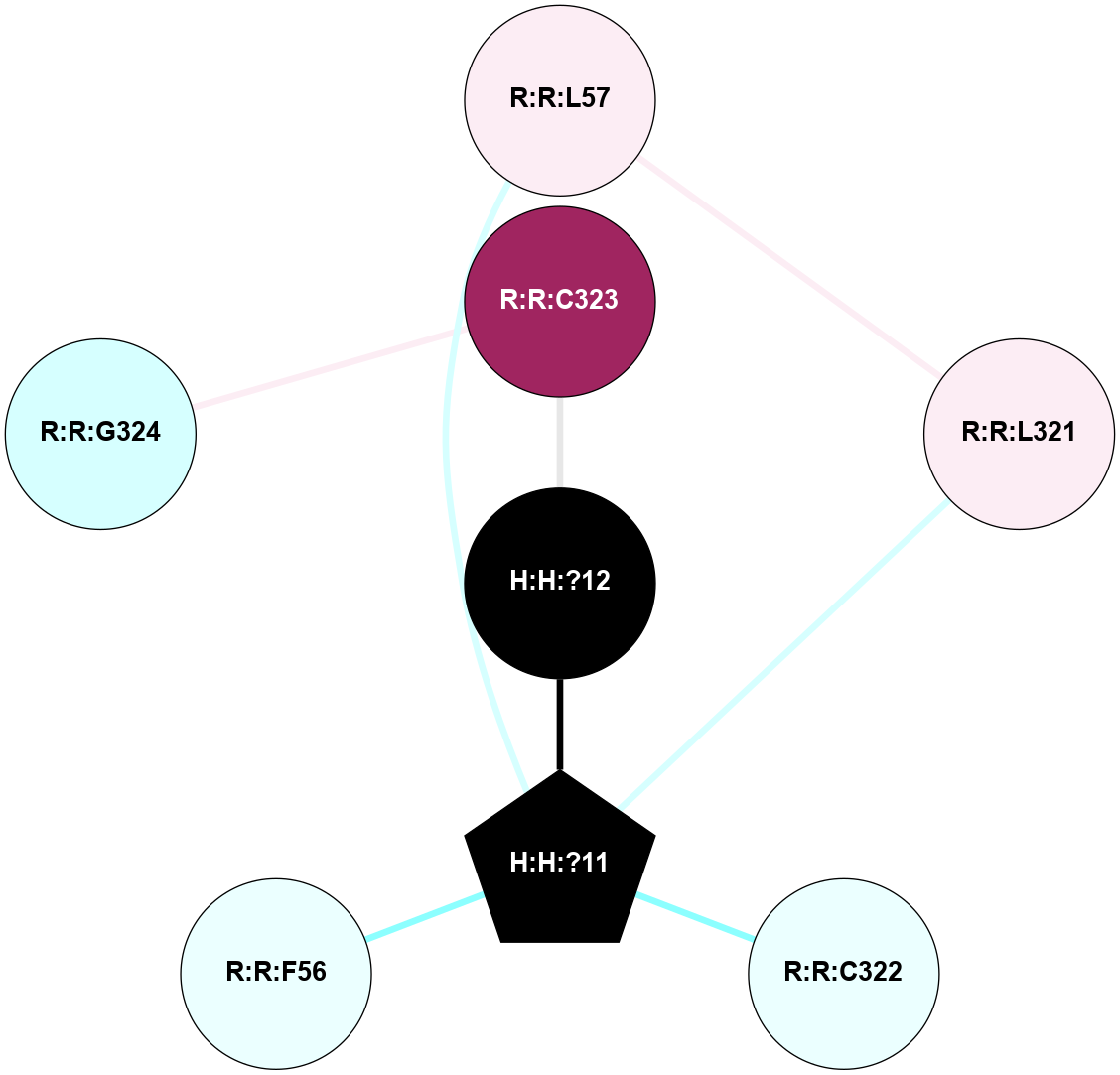
A 2D representation of the interactions of PLM in 3OAX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L321 | R:R:L57 | 5.54 | 10 | No | No | 6 | 6 | 2 | 2 | | H:H:?11 | R:R:L57 | 4.83 | 10 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?11 | R:R:L321 | 9.65 | 10 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?11 | R:R:C322 | 9.68 | 10 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?12 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?11 | H:H:?12 | 5.26 | 10 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?11 | R:R:F56 | 3.18 | 10 | Yes | No | 0 | 4 | 1 | 2 | | R:R:C323 | R:R:G324 | 1.96 | 0 | No | No | 9 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 7.47 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3PQR | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | Gt(CT) | 2.85 | 2011-03-09 | doi.org/10.1038/nature09789 |
|
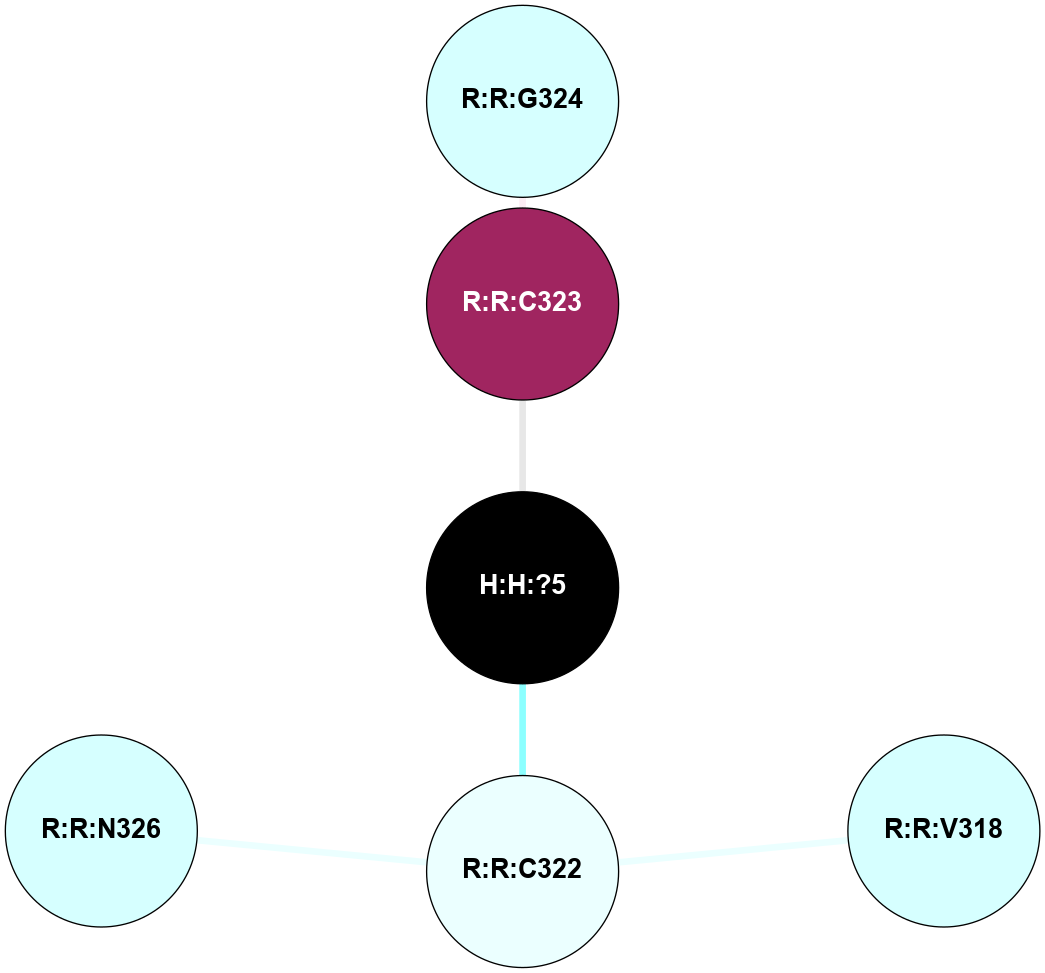
A 2D representation of the interactions of PLM in 3PQR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:C323 | R:R:G324 | 1.96 | 0 | No | No | 9 | 3 | 1 | 2 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | | R:R:C322 | R:R:N326 | 1.57 | 0 | No | No | 4 | 3 | 1 | 2 | | H:H:?5 | R:R:C322 | 1.38 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 5.53 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3PXO | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 3 | 2011-03-09 | doi.org/10.1038/nature09789 |
|

A 2D representation of the interactions of PLM in 3PXO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | R:R:C323 | 8.3 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:C323 | R:R:G324 | 1.96 | 0 | No | No | 9 | 3 | 1 | 2 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | | H:H:?6 | R:R:C322 | 1.38 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 4.84 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4A4M | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | Gt(CT) | 3.3 | 2012-01-25 | doi.org/10.1073/pnas.1114089108 |
|

A 2D representation of the interactions of PLM in 4A4M
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?3 | R:R:C322 | 2.77 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:C322 | R:R:N326 | 1.57 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.22 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4BEY | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | Gt(CT) | 2.9 | 2013-05-08 | doi.org/10.1038/embor.2013.44 |
|

A 2D representation of the interactions of PLM in 4BEY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 9.68 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4BEZ | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | - | 3.3 | 2013-04-24 | doi.org/10.1038/embor.2013.44 |
|

A 2D representation of the interactions of PLM in 4BEZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 9.68 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4IB4 | A | Amine | 5-Hydroxytryptamine | 5-HT2B | Homo Sapiens | Ergotamine | - | - | 2.7 | 2013-03-13 | doi.org/10.1126/science.1232808 |
|
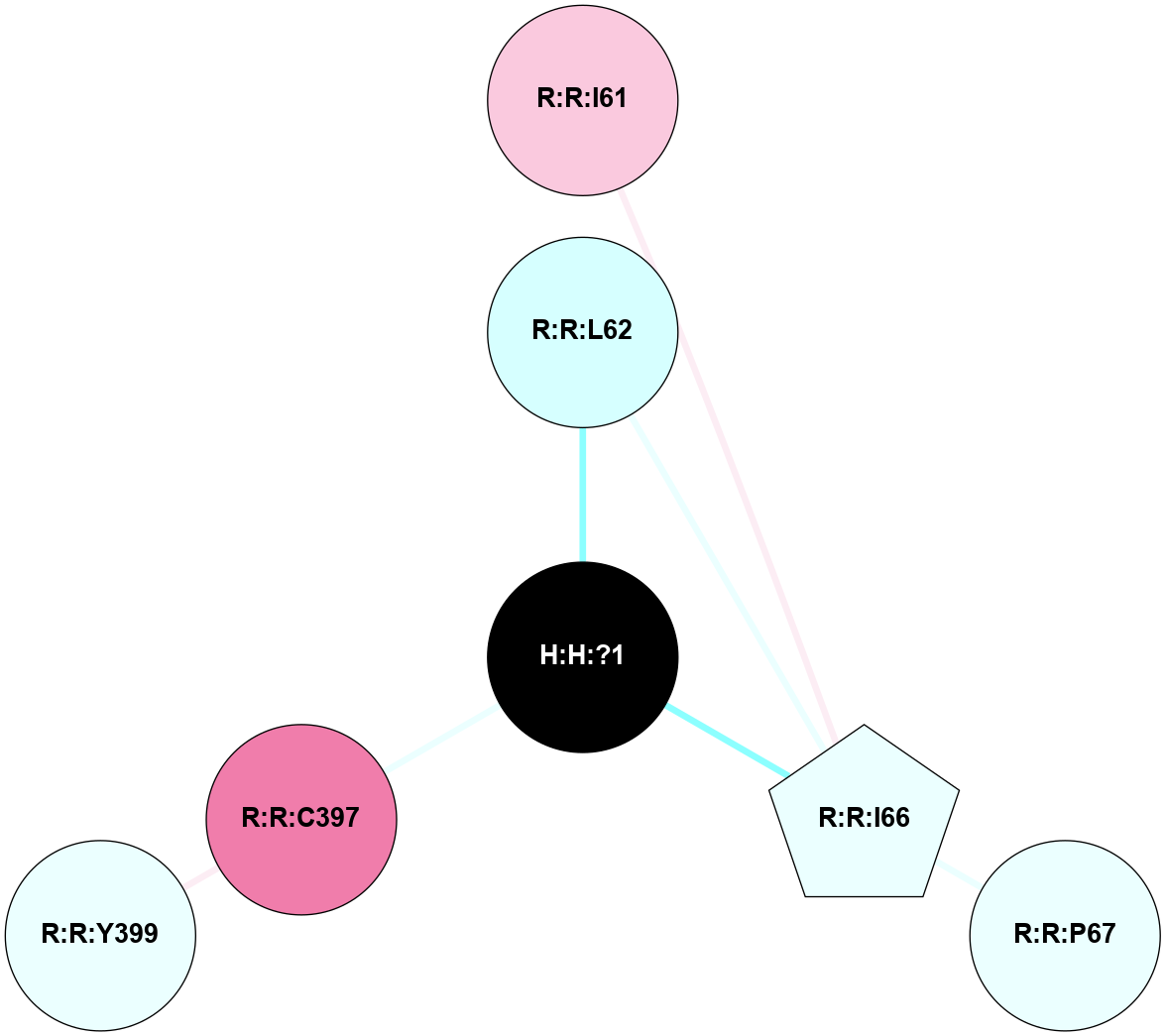
A 2D representation of the interactions of PLM in 4IB4
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:L62 | 7.24 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:I66 | 4.98 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:C397 | 9.68 | 0 | No | No | 0 | 8 | 0 | 1 | | R:R:I66 | R:R:P67 | 3.39 | 0 | No | No | 4 | 4 | 1 | 2 | | R:R:C397 | R:R:Y399 | 8.06 | 0 | No | No | 8 | 4 | 1 | 2 | | R:R:I61 | R:R:I66 | 2.94 | 0 | No | No | 7 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.30 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4J4Q | A | Sensory | Opsins | Rhodopsin | Bos Taurus | Octyl Beta-D-Glucopyranoside | - | Gt(CT) | 2.65 | 2013-10-30 | doi.org/10.1002/anie.201302374 |
|
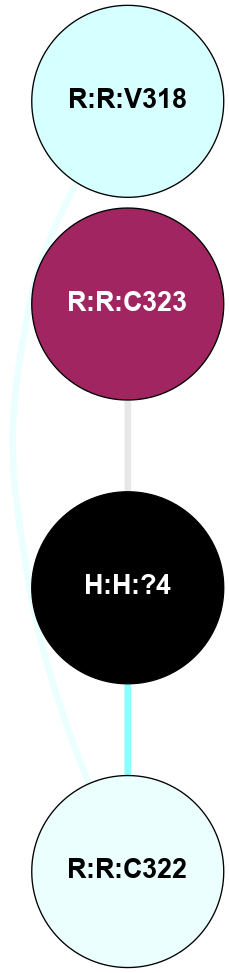
A 2D representation of the interactions of PLM in 4J4Q
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:C323 | 8.3 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | | H:H:?4 | R:R:C322 | 1.38 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 4.84 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4NC3 | A | Amine | 5-Hydroxytryptamine | 5-HT2B | Homo Sapiens | Ergotamine | - | - | 2.8 | 2013-12-18 | doi.org/10.1126/science.1244142 |
|
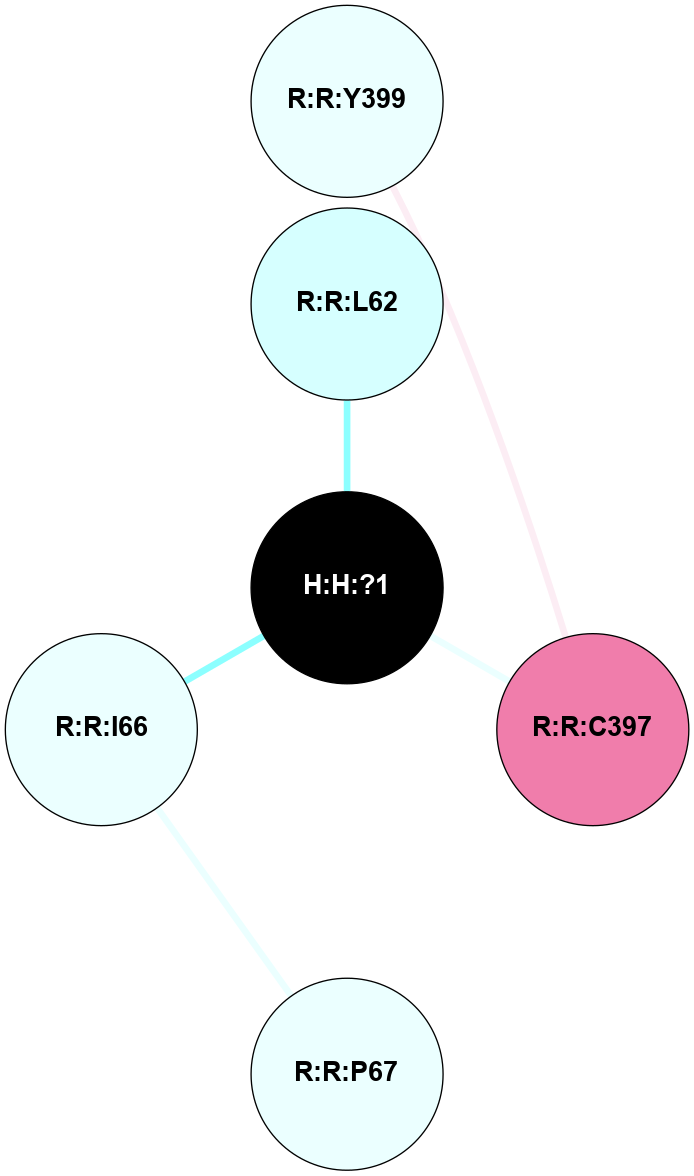
A 2D representation of the interactions of PLM in 4NC3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:L62 | 6.03 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:C397 | 9.68 | 0 | No | No | 0 | 8 | 0 | 1 | | R:R:I66 | R:R:P67 | 3.39 | 0 | No | No | 4 | 4 | 1 | 2 | | R:R:C397 | R:R:Y399 | 5.38 | 0 | No | No | 8 | 4 | 1 | 2 | | H:H:?1 | R:R:I66 | 2.49 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.07 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4PXF | A | Sensory | Opsins | Rhodopsin | Bos Taurus | Octyl-Beta-D-Glucopyranoside | - | Arrestin1 Finger Loop | 2.75 | 2014-09-17 | doi.org/10.1038/ncomms5801 |
|

A 2D representation of the interactions of PLM in 4PXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:C323 | 8.3 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?4 | R:R:C322 | 1.38 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 4.84 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4X1H | A | Sensory | Opsins | Rhodopsin | Bos Taurus | Nonyl-Beta-D-Glucopyranoside | - | Gt(CT) | 2.29 | 2015-11-04 | doi.org/10.1016/j.str.2015.09.015 |
|
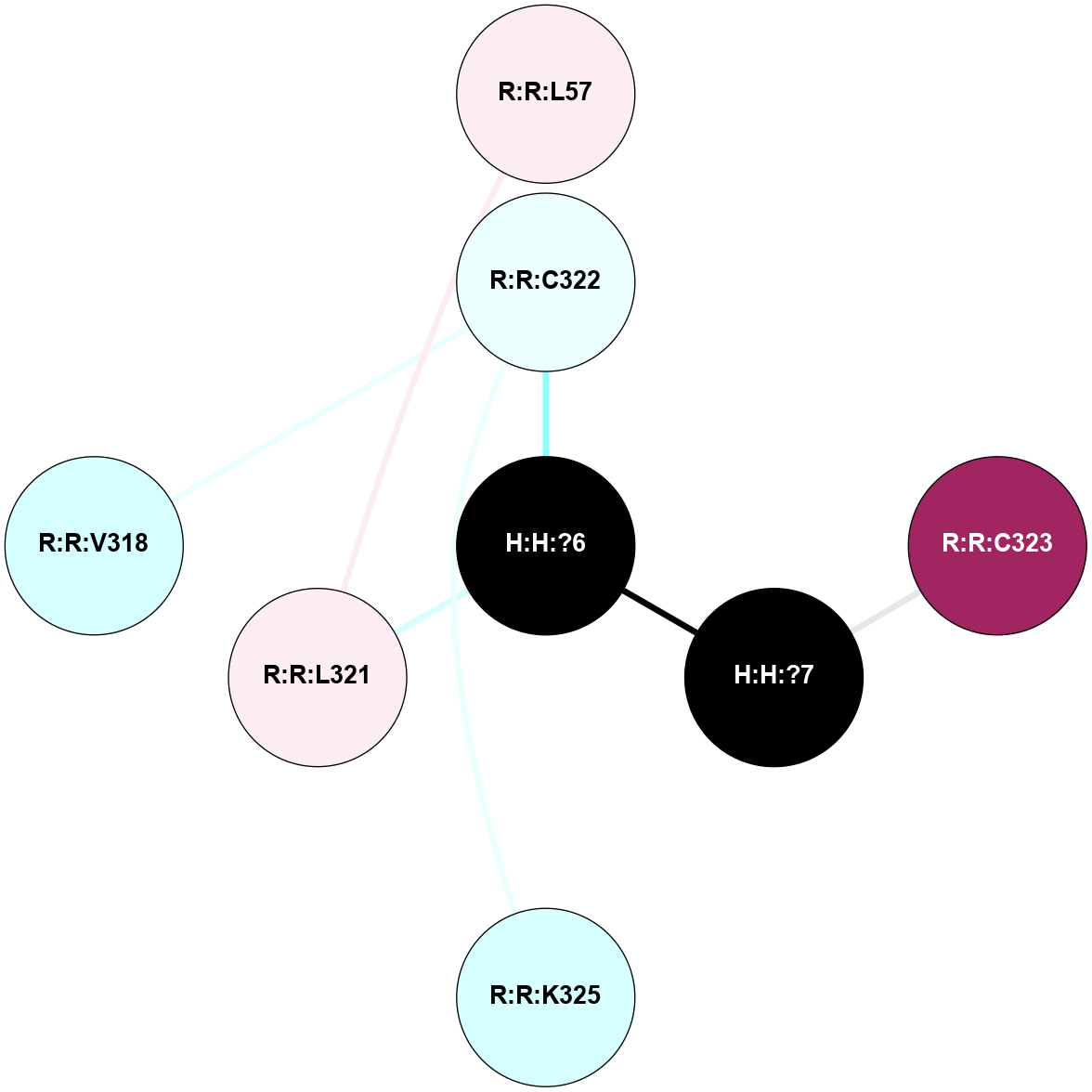
A 2D representation of the interactions of PLM in 4X1H
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L321 | R:R:L57 | 5.54 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?6 | R:R:C322 | 9.68 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?7 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 1 | 2 | | H:H:?6 | H:H:?7 | 12.62 | 0 | No | No | 0 | 0 | 0 | 1 | | R:R:C322 | R:R:K325 | 3.23 | 0 | No | No | 4 | 3 | 1 | 2 | | H:H:?6 | R:R:L321 | 2.41 | 0 | No | No | 0 | 6 | 0 | 1 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 8.24 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of PLM in 4X1H
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | R:R:C322 | 9.68 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?7 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?6 | H:H:?7 | 12.62 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?6 | R:R:L321 | 2.41 | 0 | No | No | 0 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 11.15 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5D5A | A | Amine | Adrenergic | Beta2 | Homo Sapiens | S-Carazolol | - | - | 2.48 | 2016-01-13 | doi.org/10.1107/S2059798315021683 |
|
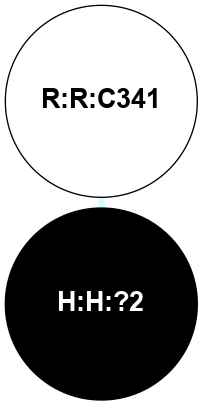
A 2D representation of the interactions of PLM in 5D5A
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:C341 | 9.68 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 9.68 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5D5B | A | Amine | Adrenergic | Beta2 | Homo Sapiens | S-Carazolol | - | - | 3.8 | 2016-01-13 | doi.org/10.1107/S2059798315021683 |
|
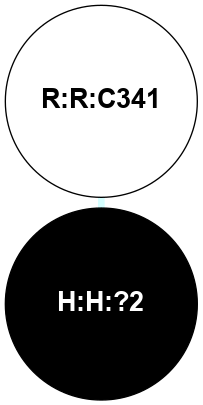
A 2D representation of the interactions of PLM in 5D5B
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:C341 | 12.45 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.45 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5D6L | A | Amine | Adrenergic | Beta2 | Homo Sapiens | S-Carazolol | - | - | 3.2 | 2016-08-17 | doi.org/10.1038/nprot.2017.057 |
|
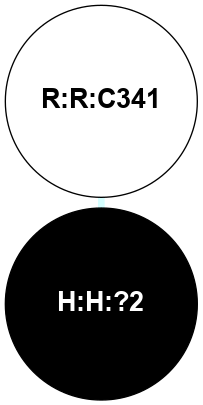
A 2D representation of the interactions of PLM in 5D6L
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:C341 | 9.68 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 9.68 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5DYS | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 2.3 | 2016-08-10 | doi.org/10.15252/embr.201642671 |
|

A 2D representation of the interactions of PLM in 5DYS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:C322 | 9.68 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?5 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 1 | 2 | | H:H:?4 | H:H:?5 | 10.52 | 0 | No | No | 0 | 0 | 0 | 1 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 10.10 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
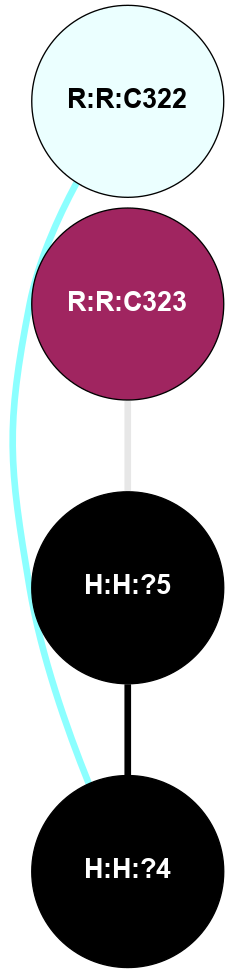
A 2D representation of the interactions of PLM in 5DYS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:C322 | 9.68 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?5 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?4 | H:H:?5 | 10.52 | 0 | No | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 10.10 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5EN0 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | Gt(CT) | 2.81 | 2016-08-10 | doi.org/10.15252/embr.201642671 |
|
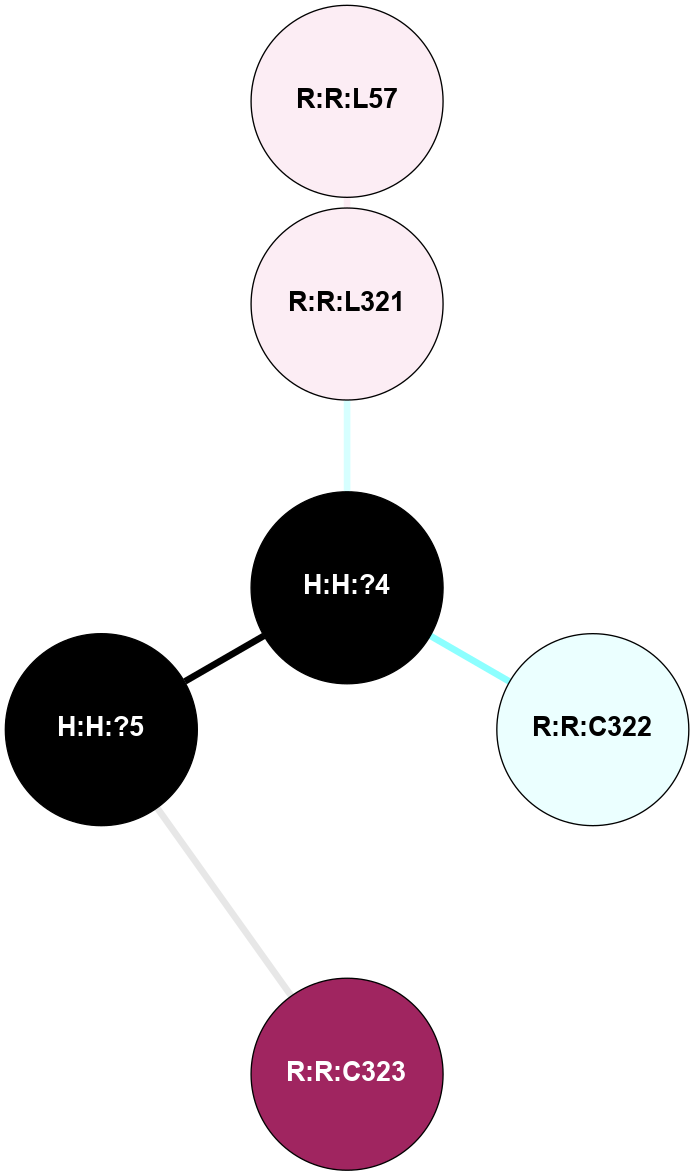
A 2D representation of the interactions of PLM in 5EN0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L321 | R:R:L57 | 5.54 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?4 | R:R:L321 | 4.85 | 0 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?4 | R:R:C322 | 9.74 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?5 | R:R:C323 | 9.74 | 0 | No | No | 0 | 9 | 1 | 2 | | H:H:?4 | H:H:?5 | 8.51 | 0 | Yes | No | 0 | 0 | 0 | 1 | | R:R:N326 | R:R:V318 | 1.48 | 0 | No | No | 3 | 3 | 2 | 1 | | R:R:R314 | R:R:V318 | 1.31 | 0 | No | No | 8 | 3 | 2 | 1 | | H:H:?4 | R:R:V318 | 1.31 | 0 | Yes | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.10 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of PLM in 5EN0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:L321 | 4.85 | 0 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?4 | R:R:C322 | 9.74 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?5 | R:R:C323 | 9.74 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?4 | H:H:?5 | 8.51 | 0 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?4 | R:R:V318 | 1.31 | 0 | Yes | No | 0 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 9.12 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5TE3 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | Octyl-Beta-D-Glucopyranoside | - | - | 2.7 | 2017-03-15 | doi.org/10.1073/pnas.1617446114 |
|
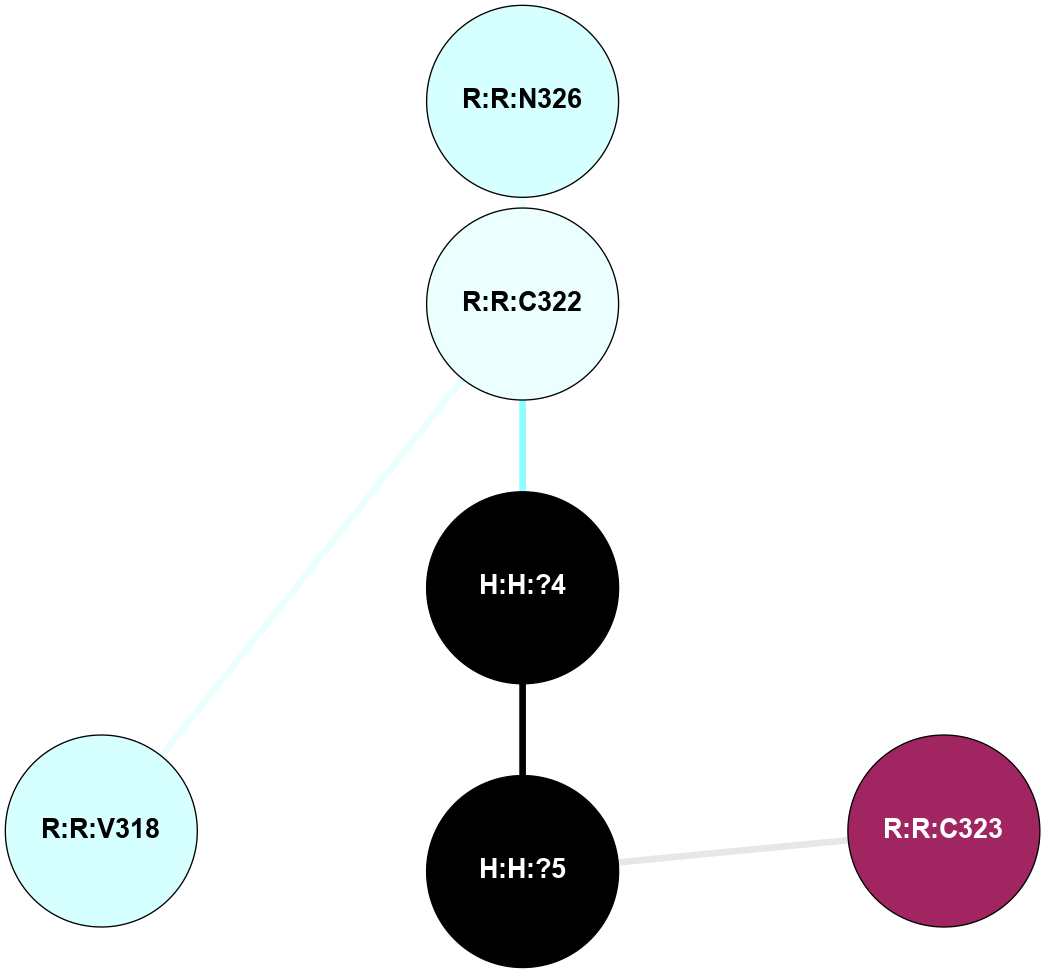
A 2D representation of the interactions of PLM in 5TE3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:C322 | R:R:N326 | 6.3 | 0 | No | No | 4 | 3 | 1 | 2 | | H:H:?4 | R:R:C322 | 9.68 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?5 | R:R:C323 | 11.07 | 0 | No | No | 0 | 9 | 1 | 2 | | H:H:?4 | H:H:?5 | 11.57 | 0 | No | No | 0 | 0 | 0 | 1 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 10.62 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
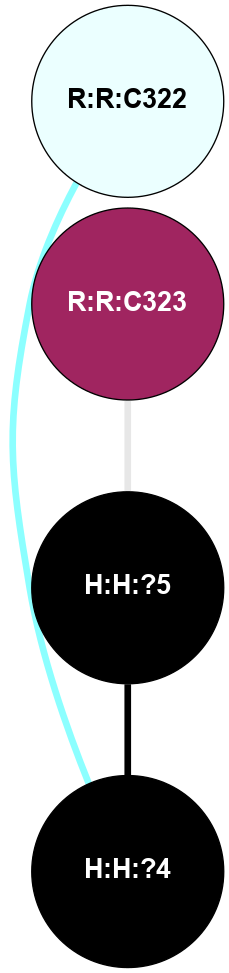
A 2D representation of the interactions of PLM in 5TE3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:C322 | 9.68 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?5 | R:R:C323 | 11.07 | 0 | No | No | 0 | 9 | 0 | 1 | | H:H:?4 | H:H:?5 | 11.57 | 0 | No | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 11.32 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FK6 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473033 | - | 2.36 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|

A 2D representation of the interactions of PLM in 6FK6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L321 | R:R:L57 | 5.54 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?4 | R:R:C322 | 11.07 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | | H:H:?4 | R:R:L321 | 1.21 | 0 | No | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.14 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FK7 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473034 | - | 2.62 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|

A 2D representation of the interactions of PLM in 6FK7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L321 | R:R:L57 | 5.54 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?6 | R:R:C322 | 12.45 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?6 | R:R:V318 | 2.6 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?6 | R:R:L321 | 1.21 | 0 | No | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 5.42 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FK8 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473035 | - | 2.87 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|
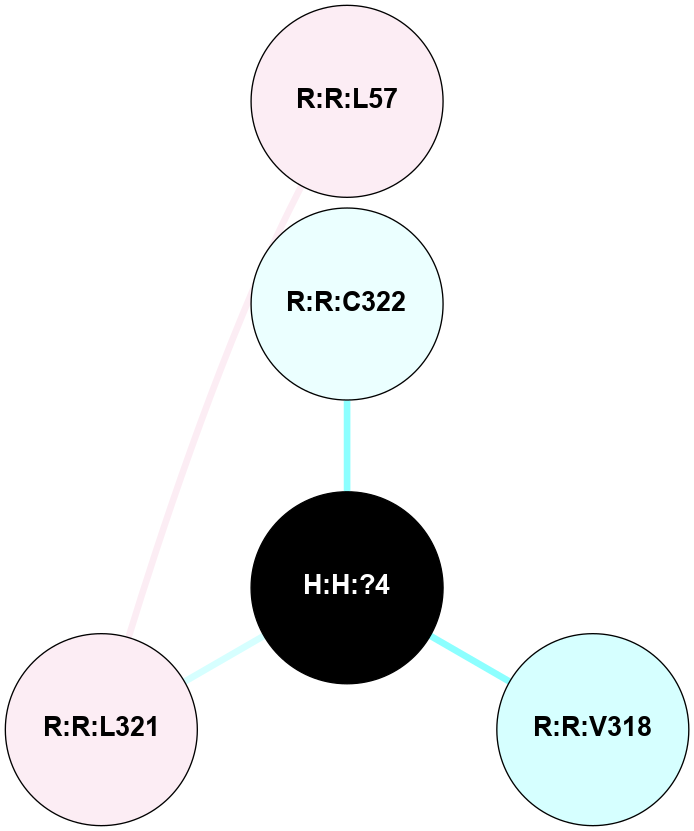
A 2D representation of the interactions of PLM in 6FK8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L321 | R:R:L57 | 5.54 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?4 | R:R:C322 | 12.45 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?4 | R:R:V318 | 1.3 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?4 | R:R:L321 | 1.21 | 0 | No | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 4.99 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FK9 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473036 | - | 2.63 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|
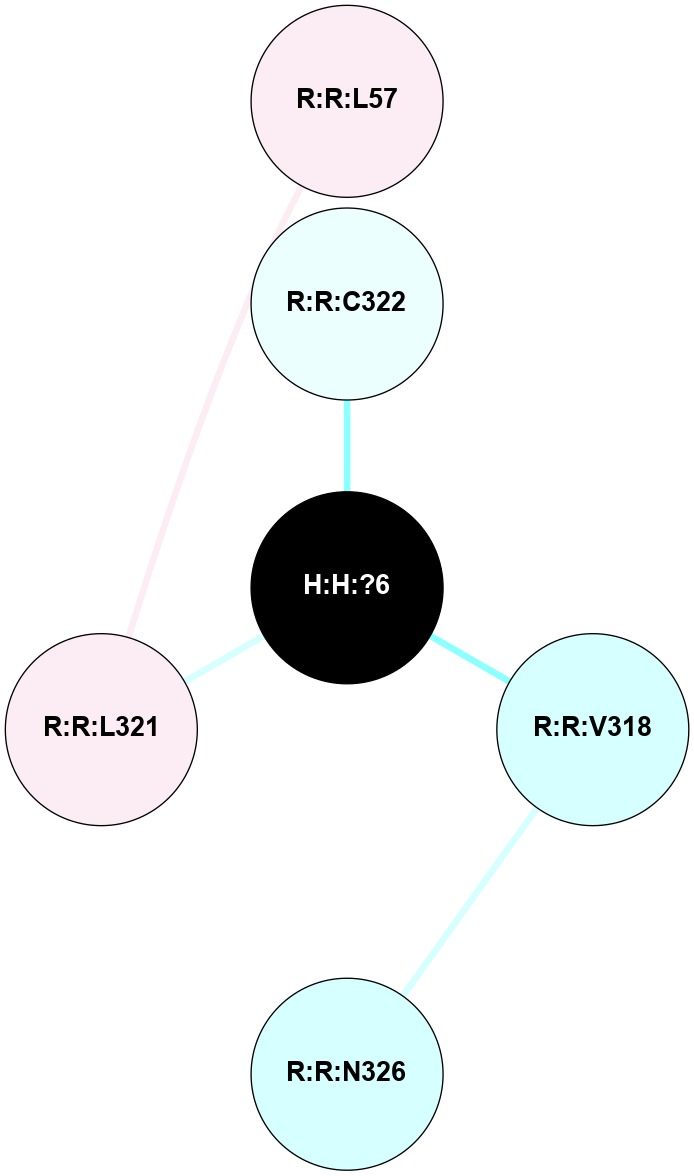
A 2D representation of the interactions of PLM in 6FK9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L321 | R:R:L57 | 5.54 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?6 | R:R:C322 | 12.45 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?6 | R:R:V318 | 2.6 | 0 | No | No | 0 | 3 | 0 | 1 | | R:R:N326 | R:R:V318 | 1.48 | 0 | No | No | 3 | 3 | 2 | 1 | | H:H:?6 | R:R:L321 | 1.21 | 0 | No | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 5.42 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FKA | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473037 | - | 2.7 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|

A 2D representation of the interactions of PLM in 6FKA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | R:R:C322 | 12.45 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:C322 | R:R:N326 | 3.15 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.45 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FKB | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473038 | - | 3.03 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|

A 2D representation of the interactions of PLM in 6FKB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L321 | R:R:L57 | 5.54 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?1 | R:R:C322 | 9.68 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:C322 | R:R:K325 | 1.62 | 0 | No | No | 4 | 3 | 1 | 2 | | H:H:?1 | R:R:L321 | 1.21 | 0 | No | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 5.45 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FKC | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473039 | - | 2.46 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|

A 2D representation of the interactions of PLM in 6FKC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L321 | R:R:L57 | 5.54 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?6 | R:R:C322 | 11.07 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:C322 | R:R:N326 | 3.15 | 0 | No | No | 4 | 3 | 1 | 2 | | H:H:?6 | R:R:L321 | 1.21 | 0 | No | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.14 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FKD | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 137349183 | - | 2.49 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|

A 2D representation of the interactions of PLM in 6FKD
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | R:R:C322 | 12.45 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?6 | R:R:V318 | 1.3 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.88 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6NWE | A | Sensory | Opsins | Rhodopsin | Bos Taurus | Octyl-Beta-D-Glucopyranoside | - | Gt(CT) | 2.71 | 2020-02-12 | To be published |
|

A 2D representation of the interactions of PLM in 6NWE
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:C323 | 8.3 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 8.30 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6PEL | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | Citronellol | Gt(CT) | 3.19 | 2020-06-24 | To be published |
|

A 2D representation of the interactions of PLM in 6PEL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 9.68 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6PGS | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | Geraniol | Gt(CT) | 2.9 | 2020-07-01 | To be published |
|

A 2D representation of the interactions of PLM in 6PGS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:C323 | 9.68 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 9.68 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7D76 | B2 | Adhesion | Adhesion | ADGRG3 | Homo Sapiens | Beclomethasone | - | Go/Beta1/Gamma2 | 3.1 | 2021-02-03 | doi.org/10.1038/s41586-020-03083-w |
|

A 2D representation of the interactions of PLM in 7D76
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | A:A:C351 | H:H:?1 | 11.13 | 0 | No | Yes | 4 | 0 | 1 | 0 | | A:A:L353 | R:R:L479 | 5.54 | 1 | Yes | Yes | 6 | 6 | 2 | 1 | | H:H:?1 | R:R:M356 | 6.2 | 1 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?1 | R:R:A360 | 5.52 | 1 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?1 | R:R:H362 | 4.51 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:L363 | 7.28 | 1 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?1 | R:R:F439 | 3.2 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:T442 | 3.88 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:F443 | 4.27 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G446 | 3 | 1 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?1 | R:R:L479 | 3.64 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:S483 | 11.85 | 1 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:G487 | 3 | 1 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?1 | R:R:W520 | 3.99 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:A305 | R:R:H309 | 2.93 | 0 | No | No | 7 | 8 | 2 | 2 | | R:R:A305 | R:R:H362 | 2.93 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:H362 | R:R:I308 | 6.63 | 0 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:H309 | R:R:W520 | 12.7 | 0 | No | Yes | 8 | 7 | 2 | 1 | | R:R:L312 | R:R:M356 | 2.83 | 0 | Yes | No | 8 | 9 | 2 | 1 | | R:R:F353 | R:R:Y438 | 3.09 | 1 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:F353 | R:R:F439 | 6.43 | 1 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:F353 | R:R:W490 | 8.02 | 1 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:F353 | R:R:I494 | 5.02 | 1 | Yes | Yes | 8 | 5 | 2 | 2 | | R:R:M356 | R:R:W490 | 8.14 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:A360 | R:R:F445 | 2.77 | 0 | No | No | 8 | 5 | 1 | 2 | | R:R:H362 | R:R:L366 | 9 | 0 | Yes | No | 7 | 6 | 1 | 2 | | R:R:H362 | R:R:Y378 | 7.62 | 0 | Yes | Yes | 7 | 6 | 1 | 2 | | R:R:L363 | R:R:L479 | 4.15 | 1 | No | Yes | 8 | 6 | 1 | 1 | | R:R:T442 | R:R:Y438 | 2.5 | 0 | No | Yes | 7 | 8 | 1 | 2 | | R:R:F439 | R:R:G491 | 4.52 | 1 | Yes | No | 6 | 6 | 1 | 2 | | R:R:F439 | R:R:I494 | 3.77 | 1 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:F443 | R:R:M447 | 8.71 | 1 | No | No | 7 | 3 | 1 | 2 | | R:R:F443 | R:R:G487 | 4.52 | 1 | No | No | 7 | 9 | 1 | 1 | | R:R:L450 | R:R:L479 | 2.77 | 0 | No | Yes | 7 | 6 | 2 | 1 | | R:R:L450 | R:R:S483 | 4.5 | 0 | No | No | 7 | 6 | 2 | 1 | | R:R:F516 | R:R:W520 | 4.01 | 0 | No | Yes | 6 | 7 | 2 | 1 | | R:R:L524 | R:R:W520 | 6.83 | 0 | No | Yes | 6 | 7 | 2 | 1 | | R:R:A360 | R:R:G357 | 1.95 | 0 | No | No | 8 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 13.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 5.50 | | Average Nodes In Shell | 32.00 | | Average Hubs In Shell | 13.00 | | Average Links In Shell | 38.00 | | Average Links Mediated by Hubs In Shell | 32.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7D77 | B2 | Adhesion | Adhesion | ADGRG3 | Homo Sapiens | Hydrocortisone | - | Go/Beta1/Gamma2 | 2.9 | 2021-02-03 | doi.org/10.1038/s41586-020-03083-w |
|
A 2D representation of the interactions of PLM in 7D77
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:H362 | R:R:I308 | 5.3 | 0 | Yes | No | 7 | 8 | 1 | 2 | | R:R:E359 | R:R:H309 | 9.85 | 0 | No | Yes | 9 | 8 | 2 | 1 | | R:R:H309 | R:R:W520 | 7.41 | 0 | Yes | No | 8 | 7 | 1 | 2 | | R:R:H309 | R:R:Y525 | 5.44 | 0 | Yes | Yes | 8 | 6 | 1 | 2 | | H:H:?1 | R:R:H309 | 5.64 | 0 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:F353 | R:R:Y438 | 4.13 | 15 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:F353 | R:R:F439 | 8.57 | 15 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:F353 | R:R:W490 | 6.01 | 15 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:F353 | R:R:I494 | 5.02 | 15 | Yes | Yes | 8 | 5 | 2 | 2 | | R:R:M356 | R:R:W490 | 5.82 | 0 | No | Yes | 9 | 9 | 1 | 2 | | H:H:?1 | R:R:M356 | 4.96 | 0 | Yes | No | 0 | 9 | 0 | 1 | | R:R:A360 | R:R:V449 | 3.39 | 0 | No | No | 8 | 7 | 1 | 2 | | H:H:?1 | R:R:A360 | 4.14 | 0 | Yes | No | 0 | 8 | 0 | 1 | | R:R:H362 | R:R:L366 | 9 | 0 | Yes | No | 7 | 6 | 1 | 2 | | R:R:H362 | R:R:Y378 | 7.62 | 0 | Yes | Yes | 7 | 6 | 1 | 2 | | H:H:?1 | R:R:H362 | 4.51 | 0 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:L363 | R:R:L479 | 5.54 | 0 | No | No | 8 | 6 | 1 | 2 | | H:H:?1 | R:R:L363 | 8.49 | 0 | Yes | No | 0 | 8 | 0 | 1 | | R:R:T442 | R:R:Y438 | 3.75 | 0 | No | Yes | 7 | 8 | 1 | 2 | | R:R:F439 | R:R:G491 | 4.52 | 15 | Yes | No | 6 | 6 | 1 | 2 | | R:R:F439 | R:R:I494 | 6.28 | 15 | Yes | Yes | 6 | 5 | 1 | 2 | | H:H:?1 | R:R:F439 | 6.41 | 0 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:T442 | 3.88 | 0 | Yes | No | 0 | 7 | 0 | 1 | | R:R:F443 | R:R:M447 | 8.71 | 0 | No | No | 7 | 3 | 1 | 2 | | R:R:F443 | R:R:G487 | 4.52 | 0 | No | No | 7 | 9 | 1 | 2 | | H:H:?1 | R:R:F443 | 3.2 | 0 | Yes | No | 0 | 7 | 0 | 1 | | R:R:L450 | R:R:S483 | 6.01 | 0 | No | No | 7 | 6 | 2 | 1 | | H:H:?1 | R:R:S483 | 7.9 | 0 | Yes | No | 0 | 6 | 0 | 1 | | A:A:C351 | H:H:?1 | 11.13 | 0 | No | Yes | 4 | 0 | 1 | 0 | | R:R:L363 | R:R:V453 | 2.98 | 0 | No | No | 8 | 7 | 1 | 2 | | R:R:A305 | R:R:H309 | 2.93 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:A305 | R:R:H362 | 2.93 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:G357 | R:R:T442 | 1.82 | 0 | No | No | 6 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 6.03 | | Average Nodes In Shell | 30.00 | | Average Hubs In Shell | 10.00 | | Average Links In Shell | 33.00 | | Average Links Mediated by Hubs In Shell | 26.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7ZBC | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 1.8 | 2023-03-29 | doi.org/10.1038/s41586-023-05863-6 |
|

A 2D representation of the interactions of PLM in 7ZBC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?410 | R:R:L50 | 3.64 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?410 | R:R:C322 | 9.74 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:L46 | R:R:L50 | 8.3 | 0 | No | No | 5 | 5 | 2 | 1 | | R:R:L321 | R:R:L57 | 2.77 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?410 | R:R:L321 | 2.43 | 0 | No | No | 0 | 6 | 0 | 1 | | R:R:G51 | R:R:L50 | 1.71 | 0 | No | No | 8 | 5 | 2 | 1 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 5.27 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7ZBE | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 1.8 | 2023-03-29 | doi.org/10.1038/s41586-023-05863-6 |
|
A 2D representation of the interactions of PLM in 7ZBE
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?410 | R:R:C322 | 8.35 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | | R:R:C322 | R:R:L321 | 1.59 | 0 | No | No | 4 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 8.35 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8A6C | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 1.8 | 2023-03-29 | doi.org/10.1038/s41586-023-05863-6 |
|

A 2D representation of the interactions of PLM in 8A6C
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?410 | R:R:C322 | 8.35 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 8.35 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8A6D | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 1.8 | 2023-03-29 | doi.org/10.1038/s41586-023-05863-6 |
|

A 2D representation of the interactions of PLM in 8A6D
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?410 | R:R:C322 | 8.35 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:C322 | R:R:L321 | 1.59 | 0 | No | No | 4 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 8.35 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8A6E | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 1.8 | 2023-03-29 | doi.org/10.1038/s41586-023-05863-6 |
|

A 2D representation of the interactions of PLM in 8A6E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?406 | R:R:L321 | 4.85 | 0 | No | No | 0 | 6 | 0 | 1 | | H:H:?406 | R:R:C322 | 8.35 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:L321 | R:R:L57 | 5.54 | 0 | No | No | 6 | 6 | 1 | 2 | | R:R:C322 | R:R:V318 | 1.71 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.60 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8U8F | A | Orphan | Orphan | GPR3 | Homo Sapiens | Palmitic Acid | - | Gs/Beta1/Gamma2 | 3.49 | 2024-03-06 | doi.org/10.1021/acs.biochem.3c00647 |
|
A 2D representation of the interactions of PLM in 8U8F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?1 | R:R:T121 | 5.14 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:I124 | 11.2 | 1 | Yes | No | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:G125 | 2.98 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:V187 | 5.19 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?1 | R:R:L190 | 6.03 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | L:L:?1 | R:R:L198 | 10.86 | 1 | Yes | No | 0 | 6 | 0 | 1 | | L:L:?1 | R:R:F202 | 6.37 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:V205 | 3.9 | 1 | Yes | No | 0 | 8 | 0 | 1 | | L:L:?1 | R:R:F263 | 4.25 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:P189 | R:R:V113 | 10.6 | 1 | Yes | No | 3 | 4 | 2 | 2 | | R:R:L190 | R:R:V113 | 4.47 | 1 | Yes | No | 3 | 4 | 1 | 2 | | R:R:A164 | R:R:T121 | 3.36 | 0 | No | No | 8 | 6 | 2 | 1 | | R:R:L167 | R:R:T121 | 5.9 | 0 | No | No | 6 | 6 | 2 | 1 | | R:R:I124 | R:R:V205 | 4.61 | 1 | No | No | 8 | 8 | 1 | 1 | | R:R:I124 | R:R:W260 | 9.4 | 1 | No | Yes | 8 | 8 | 1 | 2 | | R:R:G125 | R:R:V160 | 3.68 | 0 | No | No | 5 | 6 | 1 | 2 | | R:R:L128 | R:R:V205 | 5.96 | 0 | No | No | 7 | 8 | 2 | 1 | | R:R:L167 | R:R:L198 | 5.54 | 0 | No | No | 6 | 6 | 2 | 1 | | R:R:P189 | R:R:V187 | 3.53 | 1 | Yes | Yes | 3 | 5 | 2 | 1 | | R:R:L190 | R:R:V187 | 2.98 | 1 | Yes | Yes | 3 | 5 | 1 | 1 | | R:R:L195 | R:R:V187 | 2.98 | 0 | No | Yes | 5 | 5 | 2 | 1 | | R:R:L190 | R:R:P189 | 4.93 | 1 | Yes | Yes | 3 | 3 | 1 | 2 | | R:R:H194 | R:R:L198 | 12.86 | 0 | Yes | No | 7 | 6 | 2 | 1 | | R:R:C267 | R:R:L195 | 7.94 | 0 | No | No | 6 | 5 | 2 | 2 | | R:R:F202 | R:R:F203 | 5.36 | 1 | Yes | No | 8 | 4 | 1 | 2 | | R:R:F202 | R:R:F256 | 6.43 | 1 | Yes | Yes | 8 | 9 | 1 | 2 | | R:R:F202 | R:R:W260 | 9.02 | 1 | Yes | Yes | 8 | 8 | 1 | 2 | | R:R:F202 | R:R:T264 | 15.56 | 1 | Yes | No | 8 | 5 | 1 | 2 | | R:R:F203 | R:R:T264 | 7.78 | 1 | No | No | 4 | 5 | 2 | 2 | | R:R:F256 | R:R:W260 | 6.01 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:C267 | R:R:F263 | 8.38 | 0 | No | No | 6 | 6 | 2 | 1 | | L:L:?1 | R:R:A201 | 2.75 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:F202 | R:R:L261 | 2.44 | 1 | Yes | No | 8 | 4 | 1 | 2 | | R:R:F203 | R:R:L261 | 2.44 | 1 | No | No | 4 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 10.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 5.87 | | Average Nodes In Shell | 25.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 34.00 | | Average Links Mediated by Hubs In Shell | 24.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9AUC | B1 | Peptide | Calcitonin | CT (AMY1) | Homo Sapiens | Calcitonin gene-related peptide-1 | - | Gs/Beta1/Gamma2; RAMP1 | 2.4 | 2024-04-24 | doi.org/10.1021/acs.biochem.4c00114 |
|
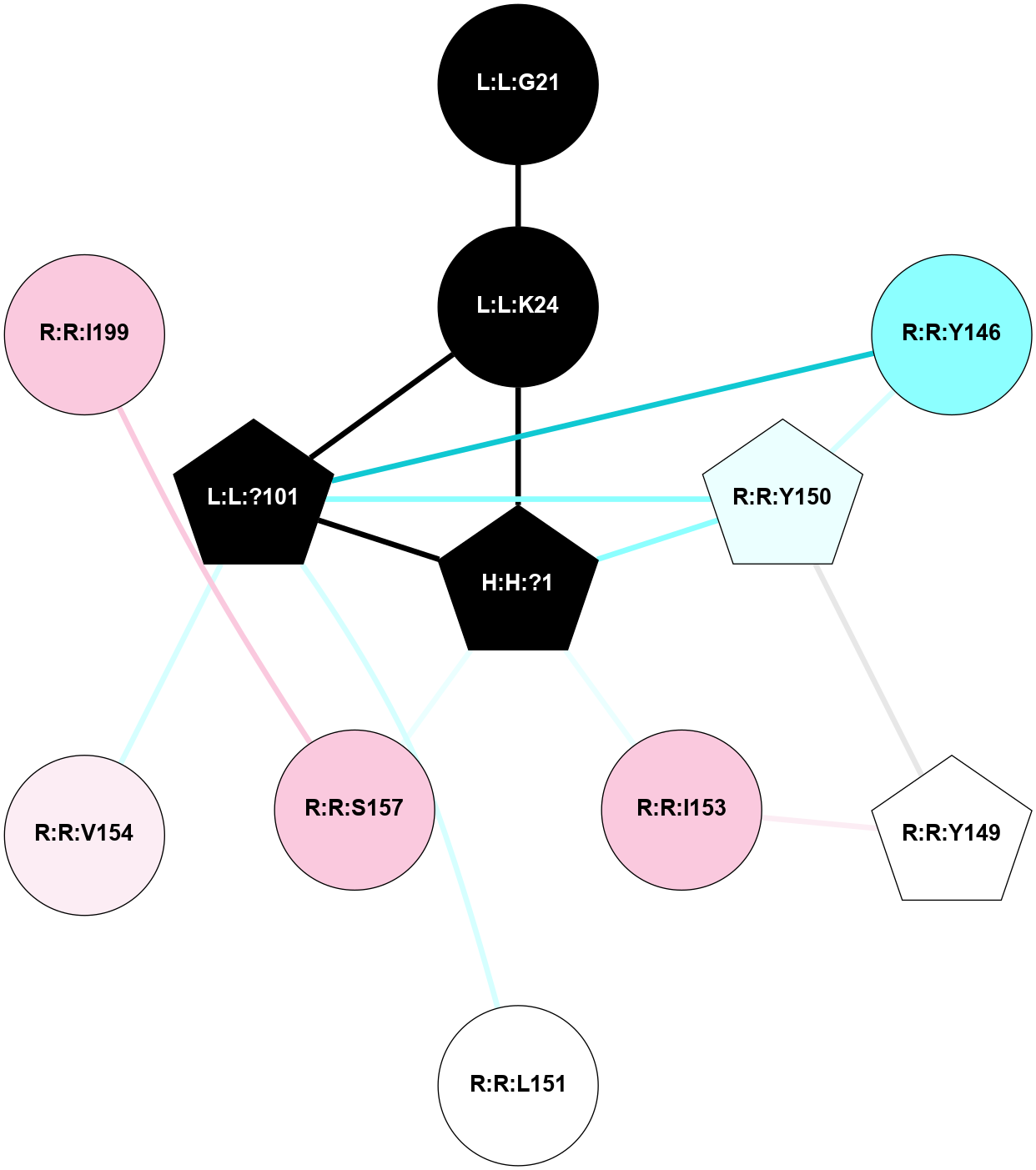
A 2D representation of the interactions of PLM in 9AUC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | L:L:K24 | 3.69 | 20 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | L:L:?101 | 5.26 | 20 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:Y150 | 7.15 | 20 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:I153 | 4.98 | 20 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:S157 | 9.16 | 20 | Yes | No | 0 | 7 | 0 | 1 | | L:L:G21 | L:L:K24 | 3.49 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:?101 | L:L:K24 | 12.29 | 20 | Yes | No | 0 | 0 | 1 | 1 | | L:L:?101 | R:R:Y146 | 11.24 | 20 | Yes | No | 0 | 2 | 1 | 2 | | L:L:?101 | R:R:Y150 | 10.22 | 20 | Yes | Yes | 0 | 4 | 1 | 1 | | L:L:?101 | R:R:L151 | 3.62 | 20 | Yes | No | 0 | 5 | 1 | 2 | | L:L:?101 | R:R:V154 | 3.9 | 20 | Yes | No | 0 | 6 | 1 | 2 | | R:R:Y146 | R:R:Y150 | 3.97 | 20 | No | Yes | 2 | 4 | 2 | 1 | | R:R:Y149 | R:R:Y150 | 11.91 | 0 | Yes | Yes | 5 | 4 | 2 | 1 | | R:R:I153 | R:R:Y149 | 8.46 | 0 | No | Yes | 7 | 5 | 1 | 2 | | R:R:I199 | R:R:S157 | 3.1 | 0 | No | No | 7 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 6.05 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of PLM in 9AUC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | L:L:K24 | 3.69 | 20 | Yes | No | 0 | 0 | 1 | 1 | | H:H:?1 | L:L:?101 | 5.26 | 20 | Yes | Yes | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:Y150 | 7.15 | 20 | Yes | Yes | 0 | 4 | 1 | 1 | | H:H:?1 | R:R:I153 | 4.98 | 20 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:S157 | 9.16 | 20 | Yes | No | 0 | 7 | 1 | 2 | | L:L:G21 | L:L:K24 | 3.49 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:V23 | R:R:Y146 | 10.09 | 0 | No | No | 0 | 2 | 2 | 1 | | L:L:?101 | L:L:K24 | 12.29 | 20 | Yes | No | 0 | 0 | 0 | 1 | | L:L:?101 | R:R:Y146 | 11.24 | 20 | Yes | No | 0 | 2 | 0 | 1 | | L:L:?101 | R:R:Y150 | 10.22 | 20 | Yes | Yes | 0 | 4 | 0 | 1 | | L:L:?101 | R:R:L151 | 3.62 | 20 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?101 | R:R:V154 | 3.9 | 20 | Yes | No | 0 | 6 | 0 | 1 | | R:R:Y146 | R:R:Y150 | 3.97 | 20 | No | Yes | 2 | 4 | 1 | 1 | | R:R:Y149 | R:R:Y150 | 11.91 | 0 | Yes | Yes | 5 | 4 | 2 | 1 | | R:R:I153 | R:R:Y149 | 8.46 | 0 | No | Yes | 7 | 5 | 2 | 2 | | R:R:L151 | R:R:S378 | 6.01 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:F382 | R:R:L151 | 3.65 | 0 | No | No | 7 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 7.75 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|




































































