| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3VW7 | A | Peptide | Proteinase Activated | PAR1 | Homo Sapiens | - | Vorapaxar; Na | - | 2.2 | 2012-12-12 | doi.org/10.1038/nature11701 |
|
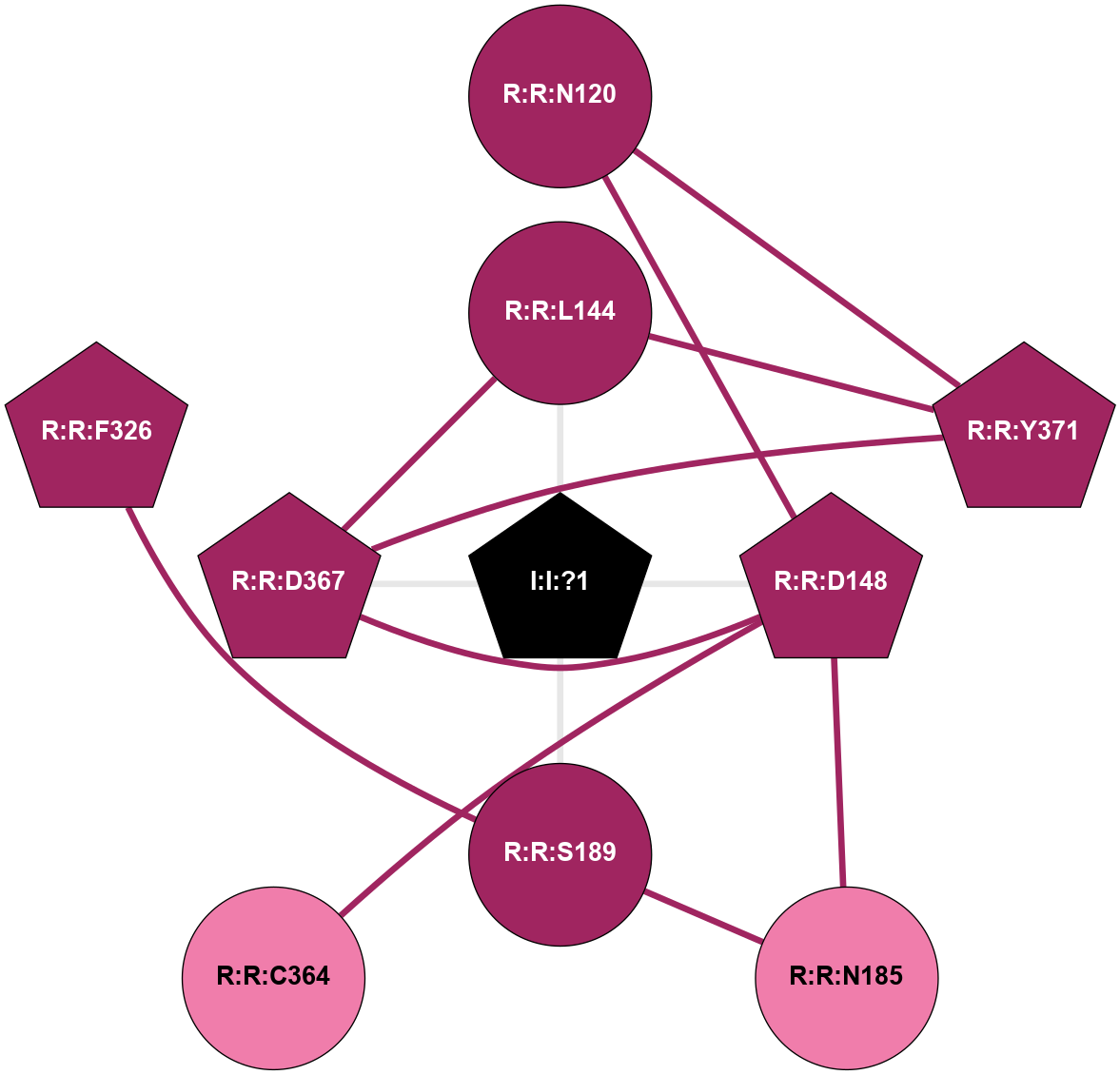
A 2D representation of the interactions of NA in 3VW7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D148 | R:R:N120 | 12.12 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:N120 | R:R:Y371 | 6.98 | 1 | No | Yes | 9 | 9 | 2 | 2 | | R:R:D367 | R:R:L144 | 10.86 | 1 | Yes | No | 9 | 9 | 1 | 1 | | R:R:L144 | R:R:Y371 | 5.86 | 1 | No | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:L144 | 5.48 | 1 | Yes | No | 0 | 9 | 0 | 1 | | R:R:D148 | R:R:N185 | 5.39 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:C364 | R:R:D148 | 10.89 | 1 | No | Yes | 8 | 9 | 2 | 1 | | R:R:D148 | R:R:D367 | 10.65 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:D148 | 8.06 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:N185 | R:R:S189 | 4.47 | 0 | No | No | 8 | 9 | 2 | 1 | | R:R:F326 | R:R:S189 | 3.96 | 0 | Yes | No | 9 | 9 | 2 | 1 | | I:I:?1 | R:R:S189 | 5.94 | 1 | Yes | No | 0 | 9 | 0 | 1 | | R:R:D367 | R:R:Y371 | 6.9 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D367 | 8.06 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 6.88 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3ZPQ | A | Amine | Adrenergic | Beta1 | Meleagris Gallopavo | 4-(piperazin-1- yl)-1H-indole | Na | - | 2.8 | 2013-04-03 | doi.org/10.1021/jm400140q |
|
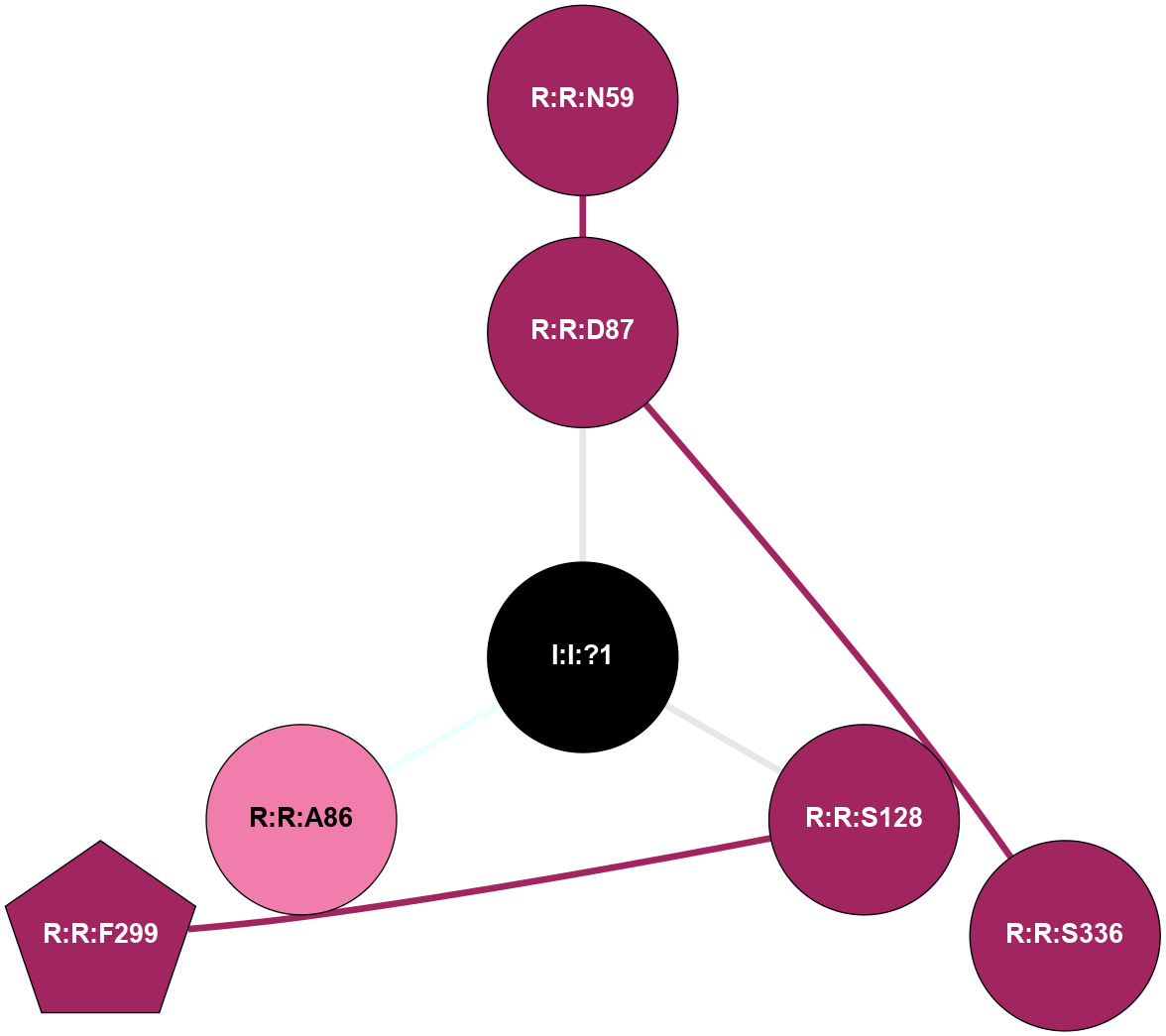
A 2D representation of the interactions of NA in 3ZPQ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D87 | R:R:N59 | 8.08 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:D87 | R:R:S336 | 8.83 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D87 | 10.74 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:F299 | R:R:S128 | 3.96 | 0 | Yes | No | 9 | 9 | 2 | 1 | | I:I:?1 | R:R:S128 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:A86 | 3.12 | 0 | No | No | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 6.60 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3ZPR | A | Amine | Adrenergic | Beta1 | Meleagris Gallopavo | 4-methyl-2-(piperazin-1-yl) | Na | - | 2.7 | 2013-04-03 | doi.org/10.1021/jm400140q |
|
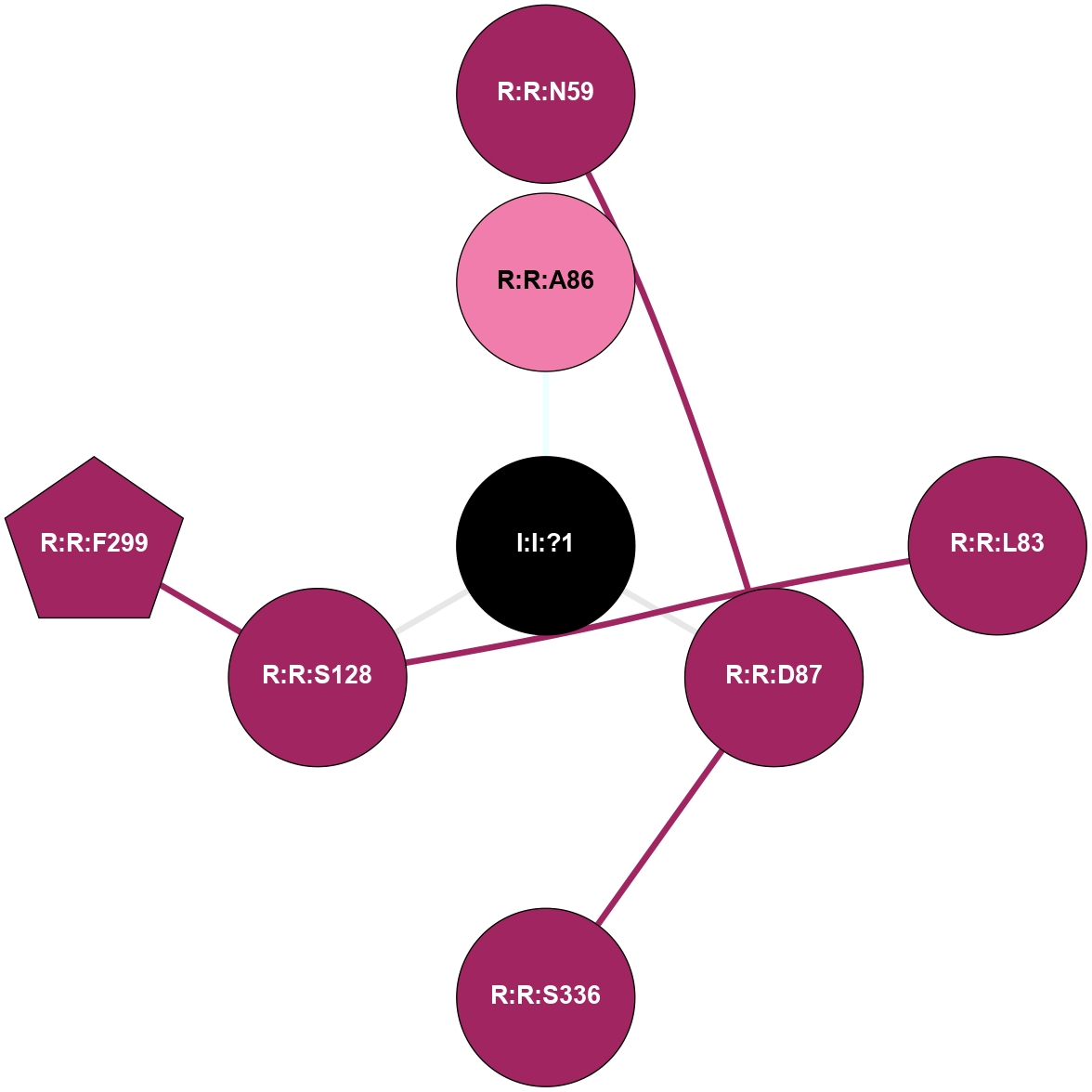
A 2D representation of the interactions of NA in 3ZPR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D87 | R:R:N59 | 10.77 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:L83 | R:R:S128 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | I:I:?1 | R:R:A86 | 3.12 | 0 | No | No | 0 | 8 | 0 | 1 | | R:R:D87 | R:R:S336 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D87 | 8.06 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:F299 | R:R:S128 | 3.96 | 0 | Yes | No | 9 | 9 | 2 | 1 | | I:I:?1 | R:R:S128 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 5.71 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4BVN | A | Amine | Adrenergic | Beta1 | Meleagris Gallopavo | Cyanopindolol | Na | - | 2.1 | 2014-04-02 | doi.org/10.1371/journal.pone.0092727 |
|
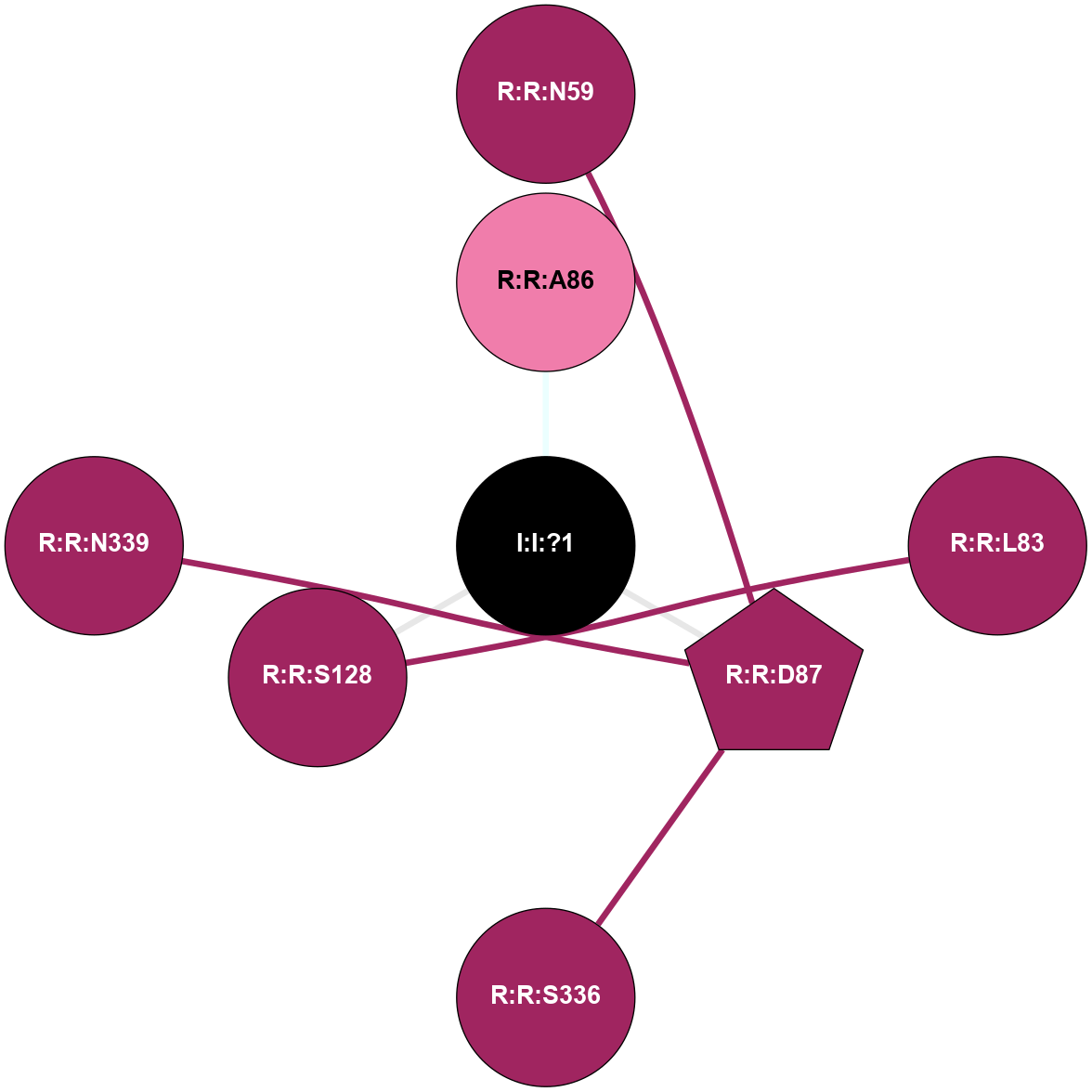
A 2D representation of the interactions of NA in 4BVN
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D87 | R:R:N59 | 13.46 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:L83 | R:R:S128 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | I:I:?1 | R:R:A86 | 3.12 | 0 | No | No | 0 | 8 | 0 | 1 | | R:R:D87 | R:R:S336 | 8.83 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D87 | R:R:N339 | 4.04 | 0 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D87 | 8.06 | 0 | No | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S128 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.71 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
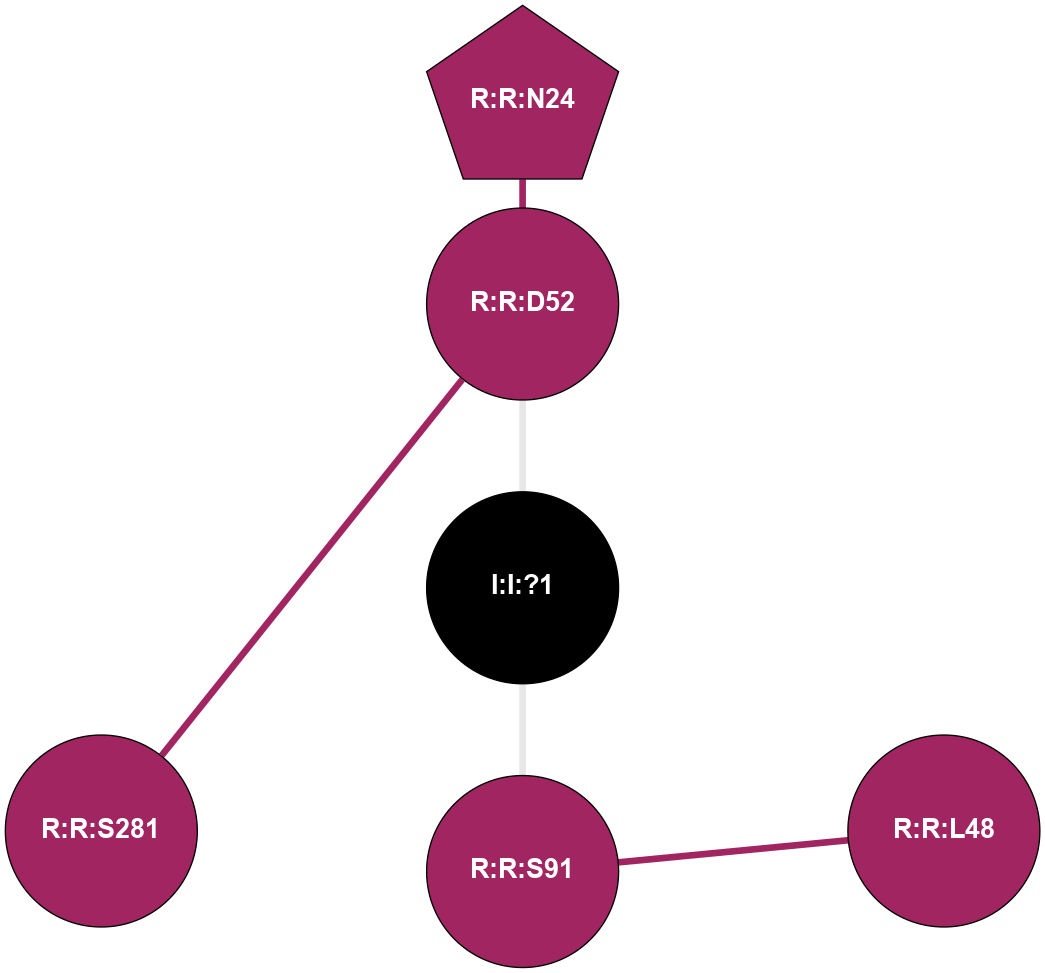
| 4EIY | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.8 | 2012-07-25 | doi.org/10.1126/science.1219218 |
|

A 2D representation of the interactions of NA in 4EIY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.06 | 0 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 8.08 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 10.31 | 0 | No | No | 9 | 9 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4N6H | A | Peptide | Opioid | DOP | Homo Sapiens | Naltrindole | Na | - | 1.8 | 2013-12-25 | doi.org/10.1038/nature12944 |
|
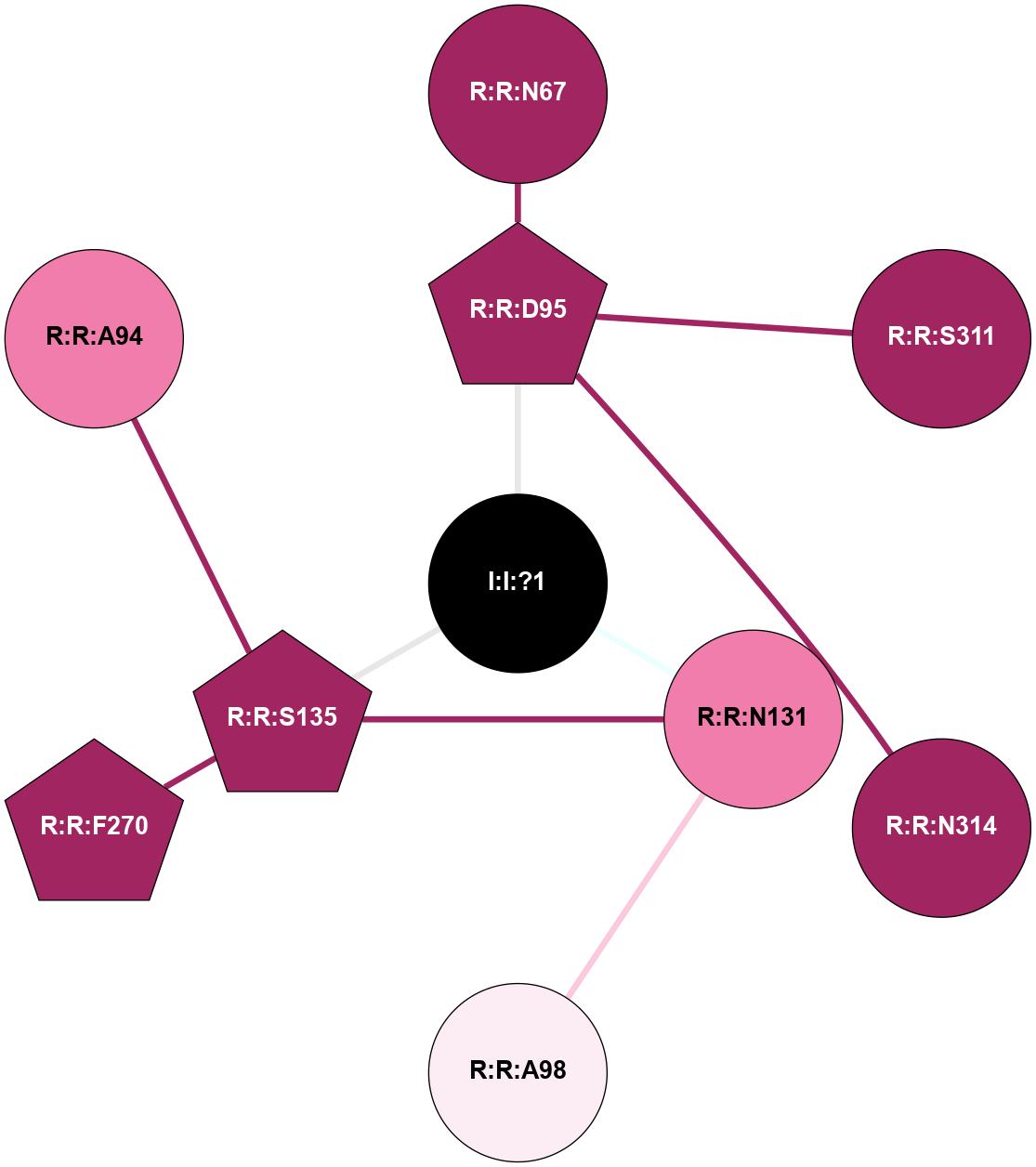
A 2D representation of the interactions of NA in 4N6H
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D95 | R:R:N67 | 9.42 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D95 | R:R:S311 | 11.78 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D95 | R:R:N314 | 5.39 | 0 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D95 | 10.74 | 1 | No | Yes | 0 | 9 | 0 | 1 | | R:R:A98 | R:R:N131 | 6.25 | 0 | No | No | 6 | 8 | 2 | 1 | | R:R:N131 | R:R:S135 | 4.47 | 1 | No | Yes | 8 | 9 | 1 | 1 | | I:I:?1 | R:R:N131 | 8.15 | 1 | No | No | 0 | 8 | 0 | 1 | | R:R:F270 | R:R:S135 | 3.96 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | | I:I:?1 | R:R:S135 | 5.94 | 1 | No | Yes | 0 | 9 | 0 | 1 | | R:R:A94 | R:R:S135 | 1.71 | 0 | No | Yes | 8 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 8.28 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4RWD | A | Peptide | Opioid | DOP | Homo Sapiens | DIPP-NH2 | Na | - | 2.7 | 2015-01-14 | doi.org/10.1038/nsmb.2965 |
|
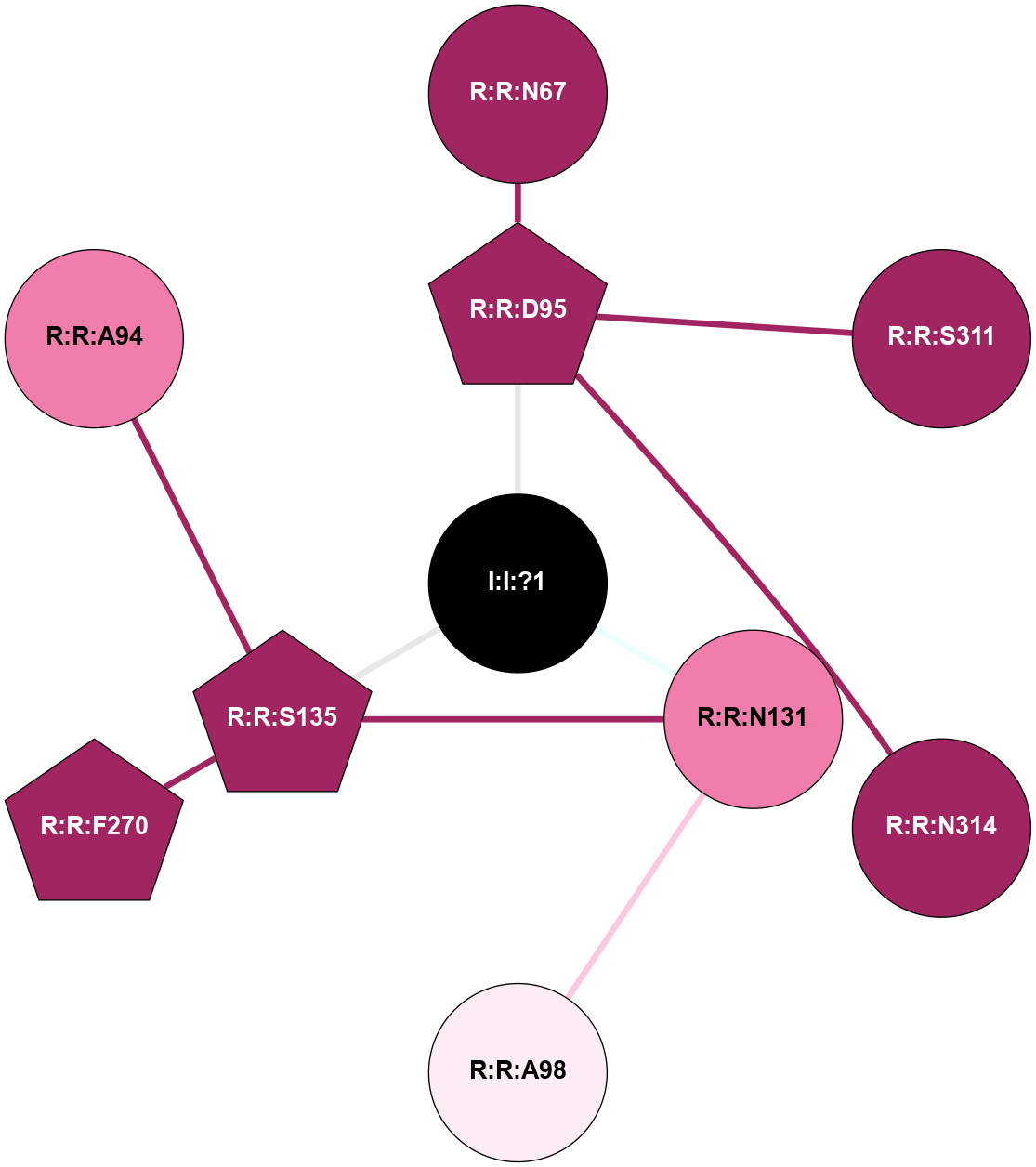
A 2D representation of the interactions of NA in 4RWD
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D95 | 8.06 | 4 | No | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:N131 | 8.15 | 4 | No | No | 0 | 8 | 0 | 1 | | I:I:?1 | R:R:S135 | 5.94 | 4 | No | Yes | 0 | 9 | 0 | 1 | | R:R:D95 | R:R:N67 | 8.08 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D95 | R:R:S311 | 11.78 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D95 | R:R:N314 | 6.73 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:A98 | R:R:N131 | 6.25 | 0 | No | No | 6 | 8 | 2 | 1 | | R:R:N131 | R:R:S135 | 4.47 | 4 | No | Yes | 8 | 9 | 1 | 1 | | R:R:F270 | R:R:S135 | 3.96 | 4 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:A94 | R:R:S135 | 1.71 | 0 | No | Yes | 8 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 7.38 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5A8E | A | Amine | Adrenergic | Beta1 | Meleagris Gallopavo | 7-Methylcyanopindolol | Na | - | 2.4 | 2015-09-30 | doi.org/10.1124/mol.115.101030 |
|
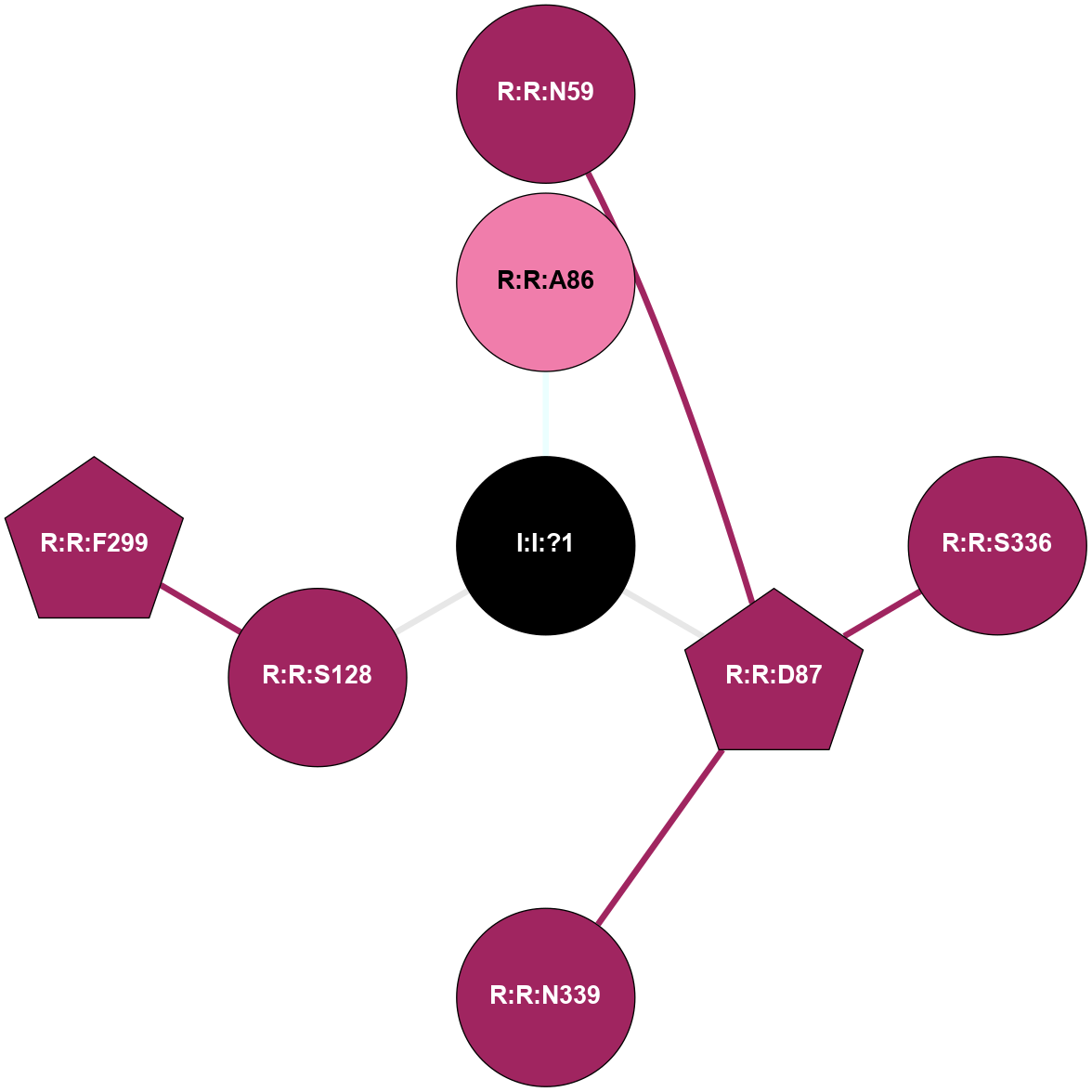
A 2D representation of the interactions of NA in 5A8E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D87 | R:R:N59 | 12.12 | 0 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:A86 | 3.12 | 0 | No | No | 0 | 8 | 0 | 1 | | R:R:D87 | R:R:S336 | 8.83 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D87 | R:R:N339 | 5.39 | 0 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D87 | 8.06 | 0 | No | Yes | 0 | 9 | 0 | 1 | | R:R:F299 | R:R:S128 | 3.96 | 1 | Yes | No | 9 | 9 | 2 | 1 | | I:I:?1 | R:R:S128 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.71 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5IU4 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.72 | 2016-06-29 | doi.org/10.1021/acs.jmedchem.6b00653 |
|
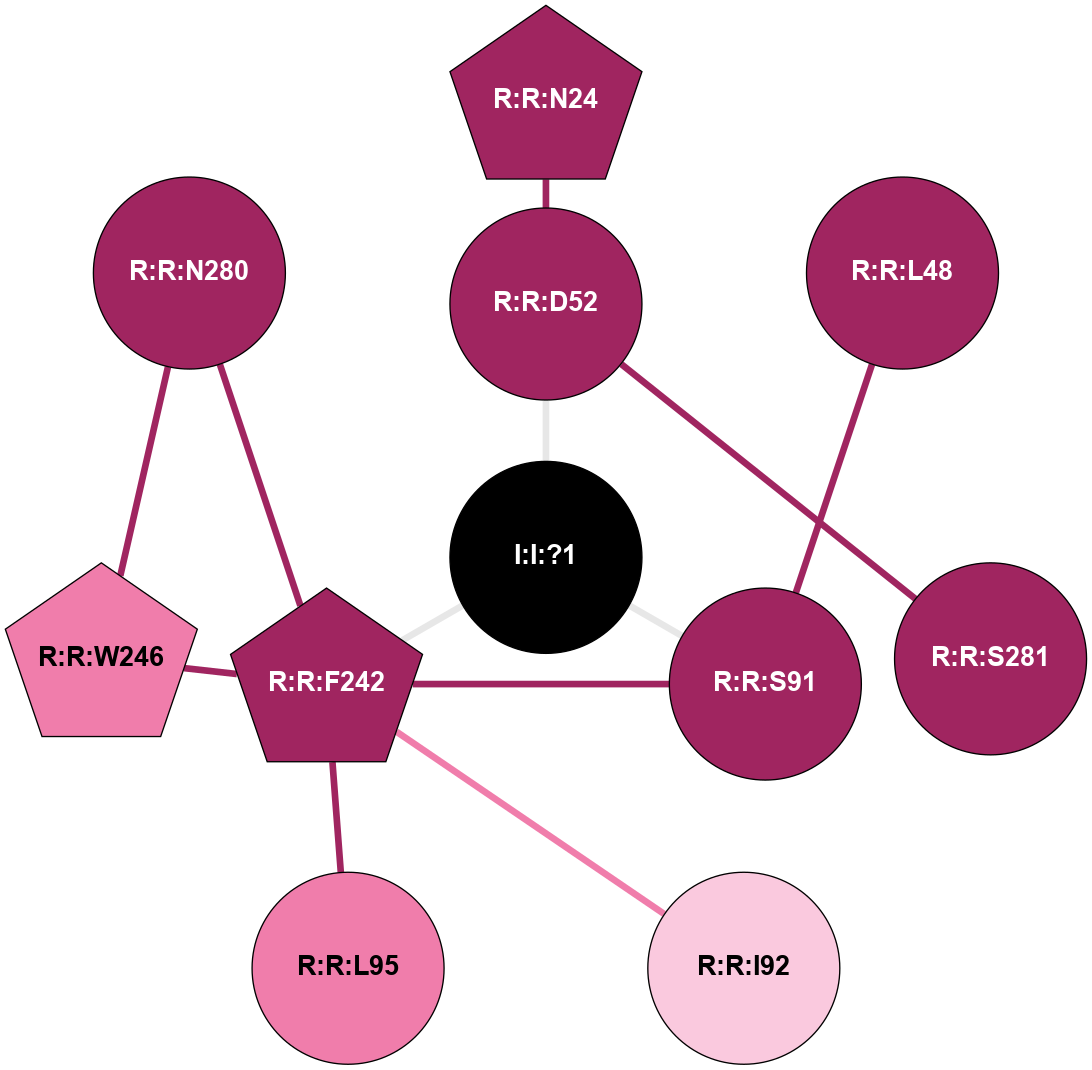
A 2D representation of the interactions of NA in 5IU4
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 12.12 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 1 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 1 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 1 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 6.28 | 1 | Yes | No | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 17.04 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 1 | No | Yes | 0 | 9 | 0 | 1 | | R:R:N280 | R:R:W246 | 3.39 | 1 | No | Yes | 9 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5IU7 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | Q27456346 | Na | - | 1.9 | 2016-06-29 | doi.org/10.1021/acs.jmedchem.6b00653 |
|
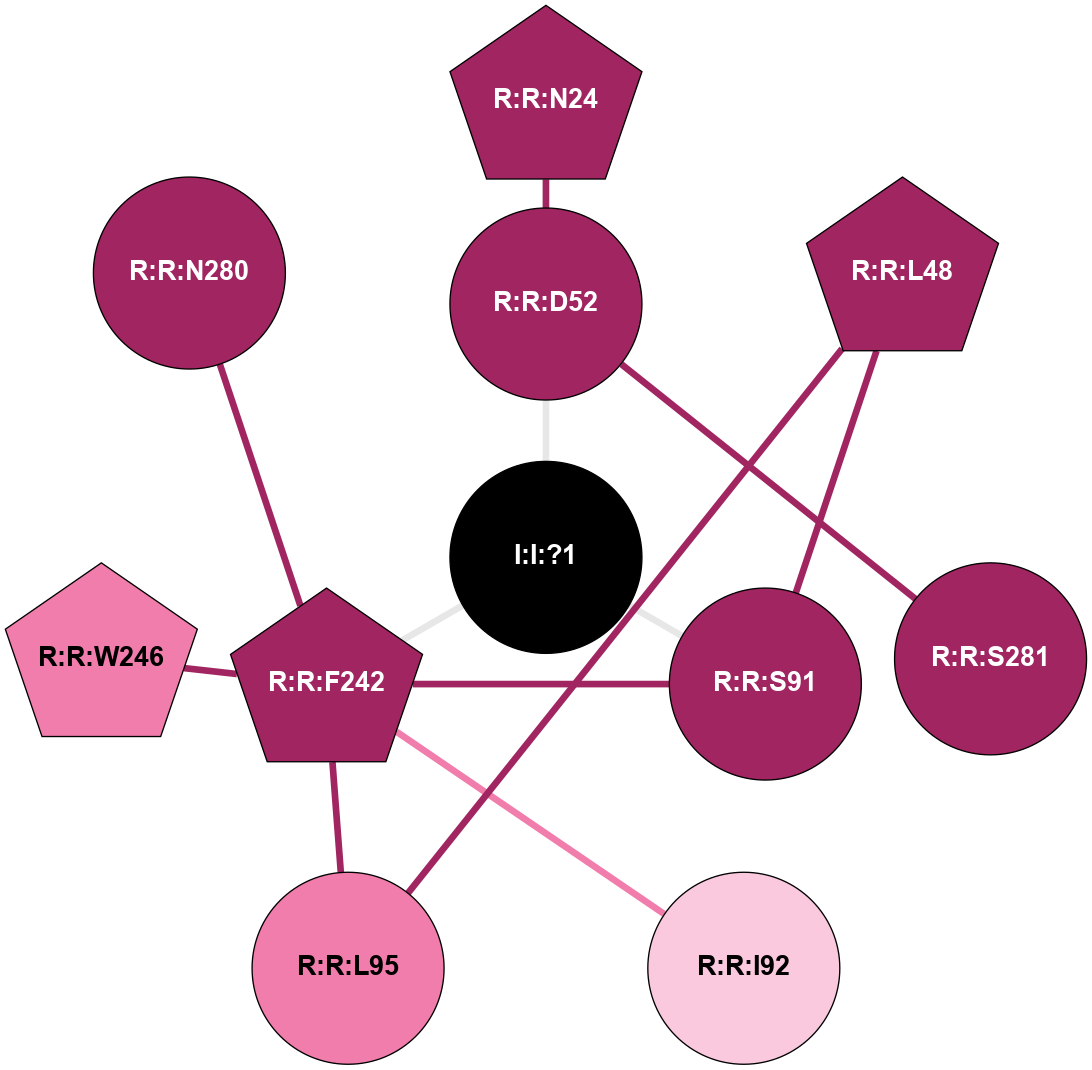
A 2D representation of the interactions of NA in 5IU7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 9.42 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | Yes | No | 9 | 9 | 2 | 1 | | R:R:L48 | R:R:L95 | 4.15 | 0 | Yes | No | 9 | 8 | 2 | 2 | | R:R:D52 | R:R:S281 | 10.31 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 1 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 1 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 1 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 6.28 | 1 | Yes | No | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 17.04 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 1 | No | Yes | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5IU8 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | Q27456347 | Na | - | 2 | 2016-06-29 | doi.org/10.1021/acs.jmedchem.6b00653 |
|
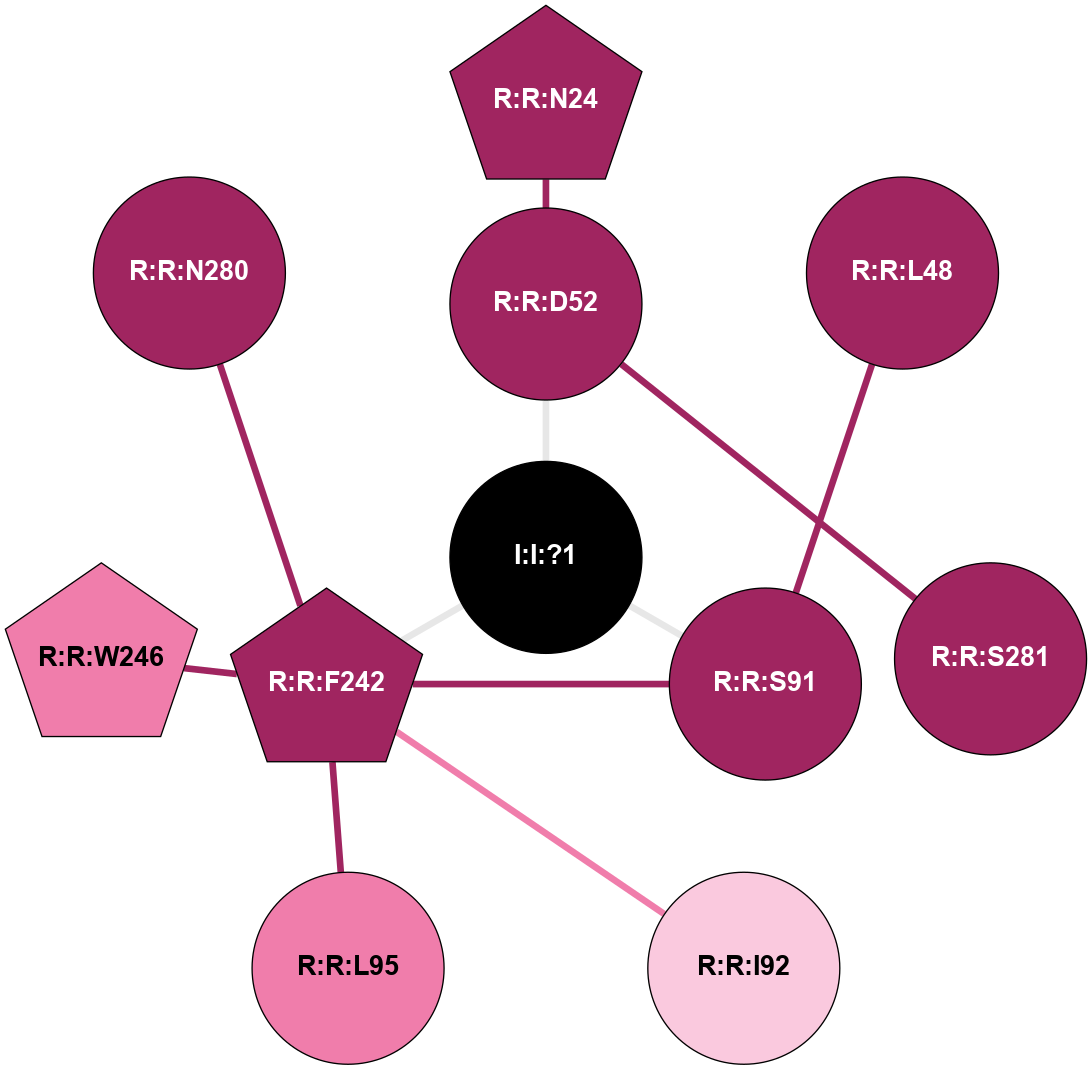
A 2D representation of the interactions of NA in 5IU8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 10.77 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 1 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 1 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 1 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 6.28 | 1 | Yes | No | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 17.04 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 1 | No | Yes | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5IUA | A | Nucleotide | Adenosine | A2A | Homo Sapiens | Q27456348 | Na | - | 2.2 | 2016-06-29 | doi.org/10.1021/acs.jmedchem.6b00653 |
|
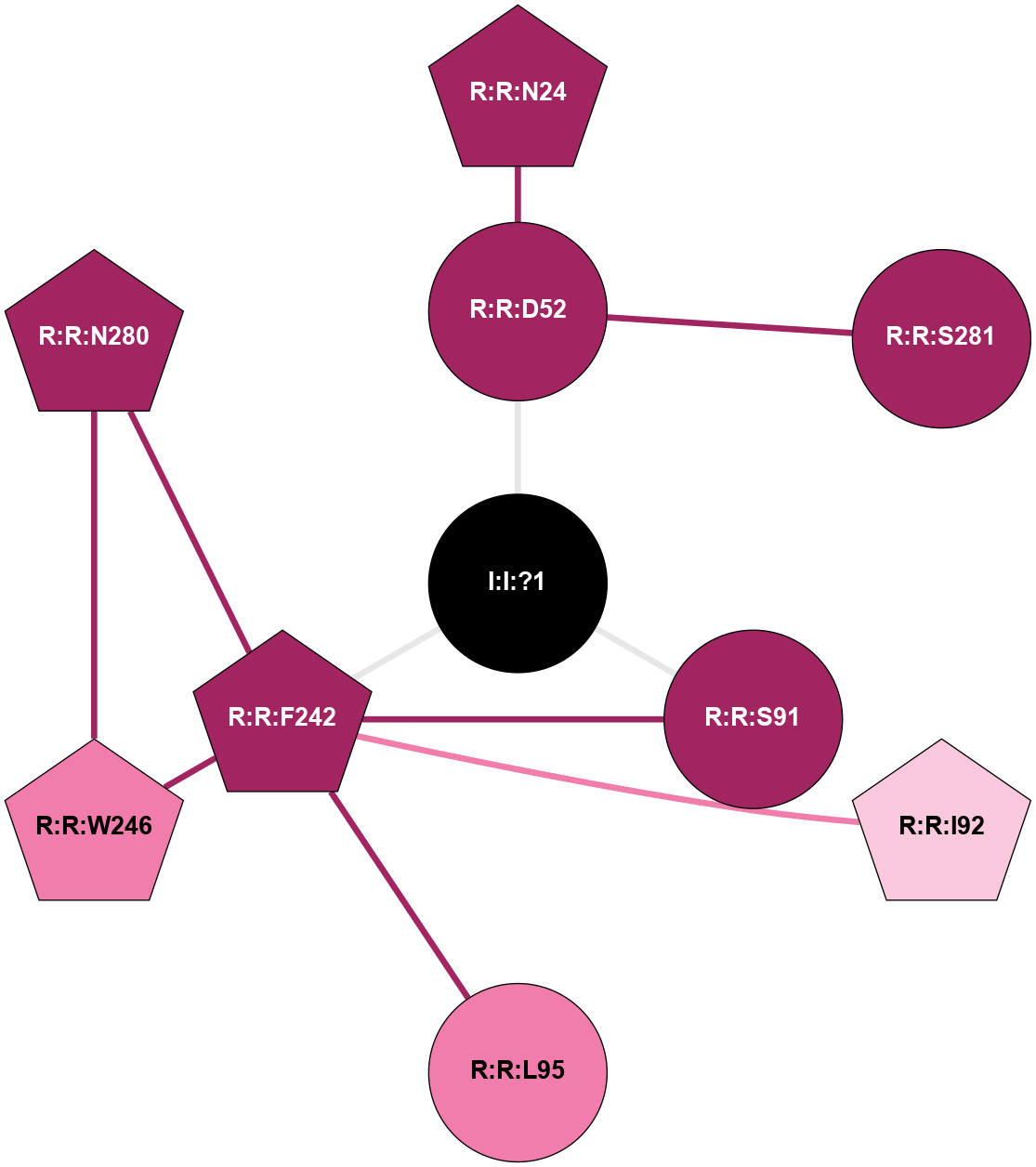
A 2D representation of the interactions of NA in 5IUA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 10.77 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 1 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 1 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 1 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 6.28 | 1 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 17.04 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 1 | No | Yes | 0 | 9 | 0 | 1 | | R:R:N280 | R:R:W246 | 3.39 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5IUB | A | Nucleotide | Adenosine | A2A | Homo Sapiens | Q27456344 | Na | - | 2.1 | 2016-06-29 | doi.org/10.1021/acs.jmedchem.6b00653 |
|
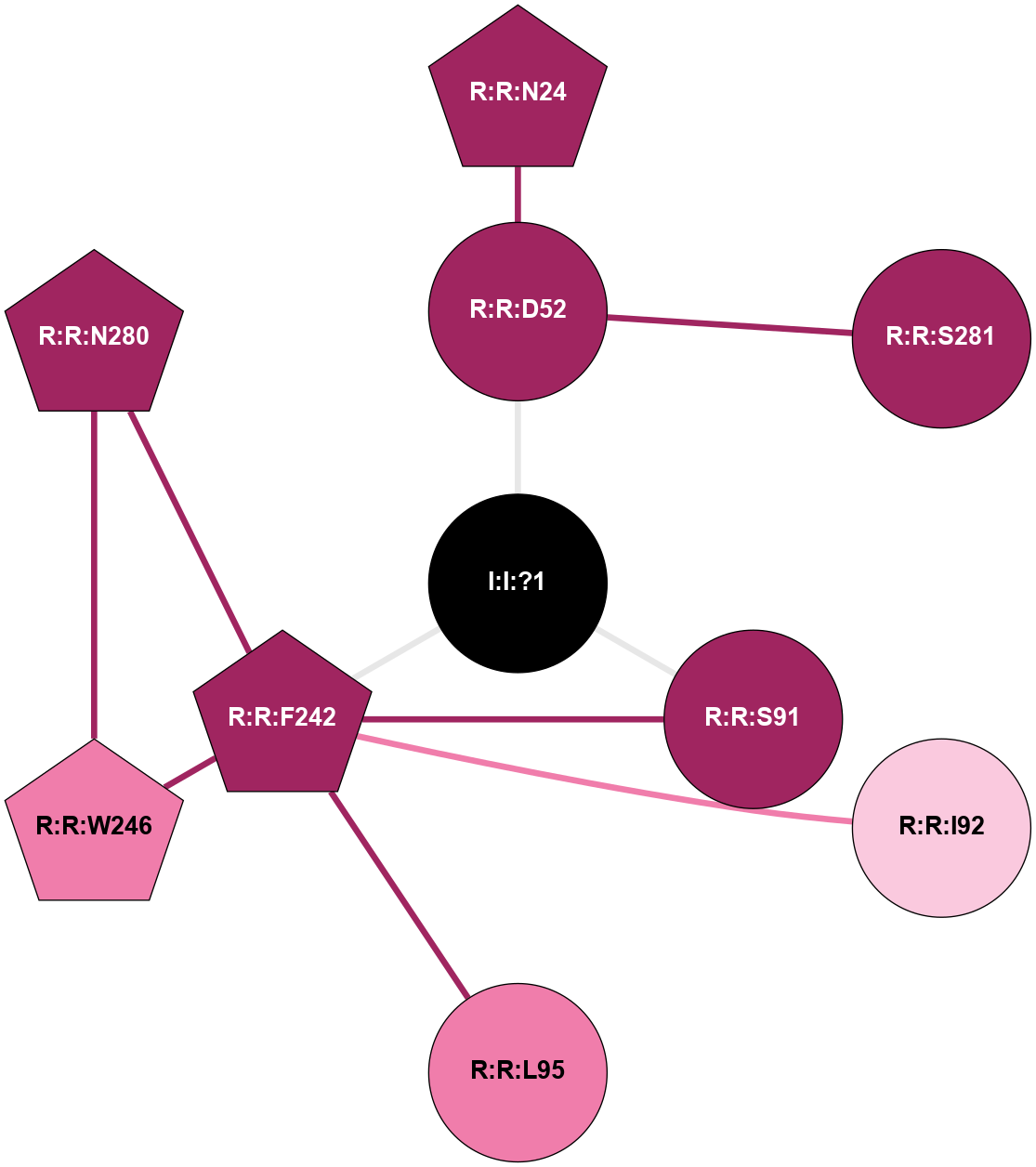
A 2D representation of the interactions of NA in 5IUB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 10.77 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 6 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 6 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 6 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 6.28 | 6 | Yes | No | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 16.04 | 6 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 6 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 6 | No | Yes | 0 | 9 | 0 | 1 | | R:R:N280 | R:R:W246 | 4.52 | 6 | Yes | Yes | 9 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5K2A | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2.5 | 2016-09-21 | doi.org/10.1126/sciadv.1600292 |
|
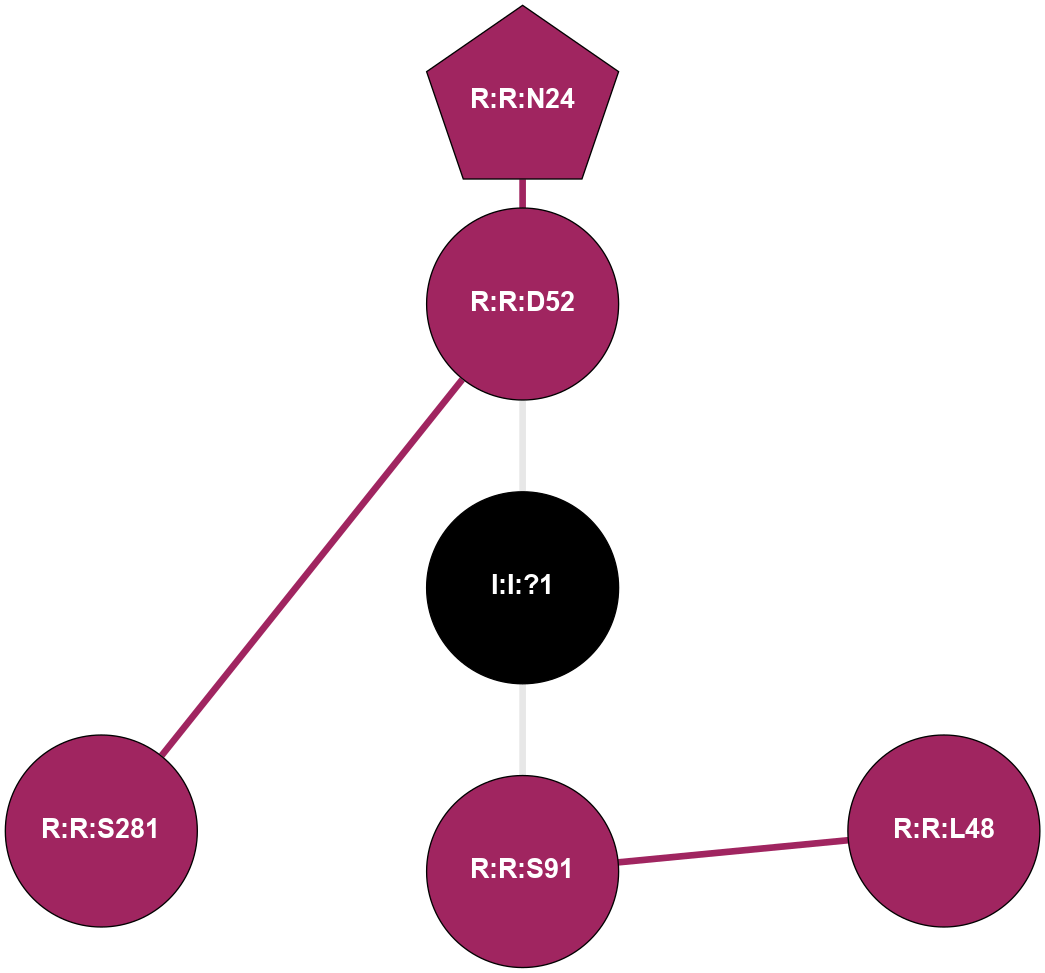
A 2D representation of the interactions of NA in 5K2A
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.06 | 0 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 9.42 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5K2B | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2.5 | 2016-09-21 | doi.org/10.1126/sciadv.1600292 |
|
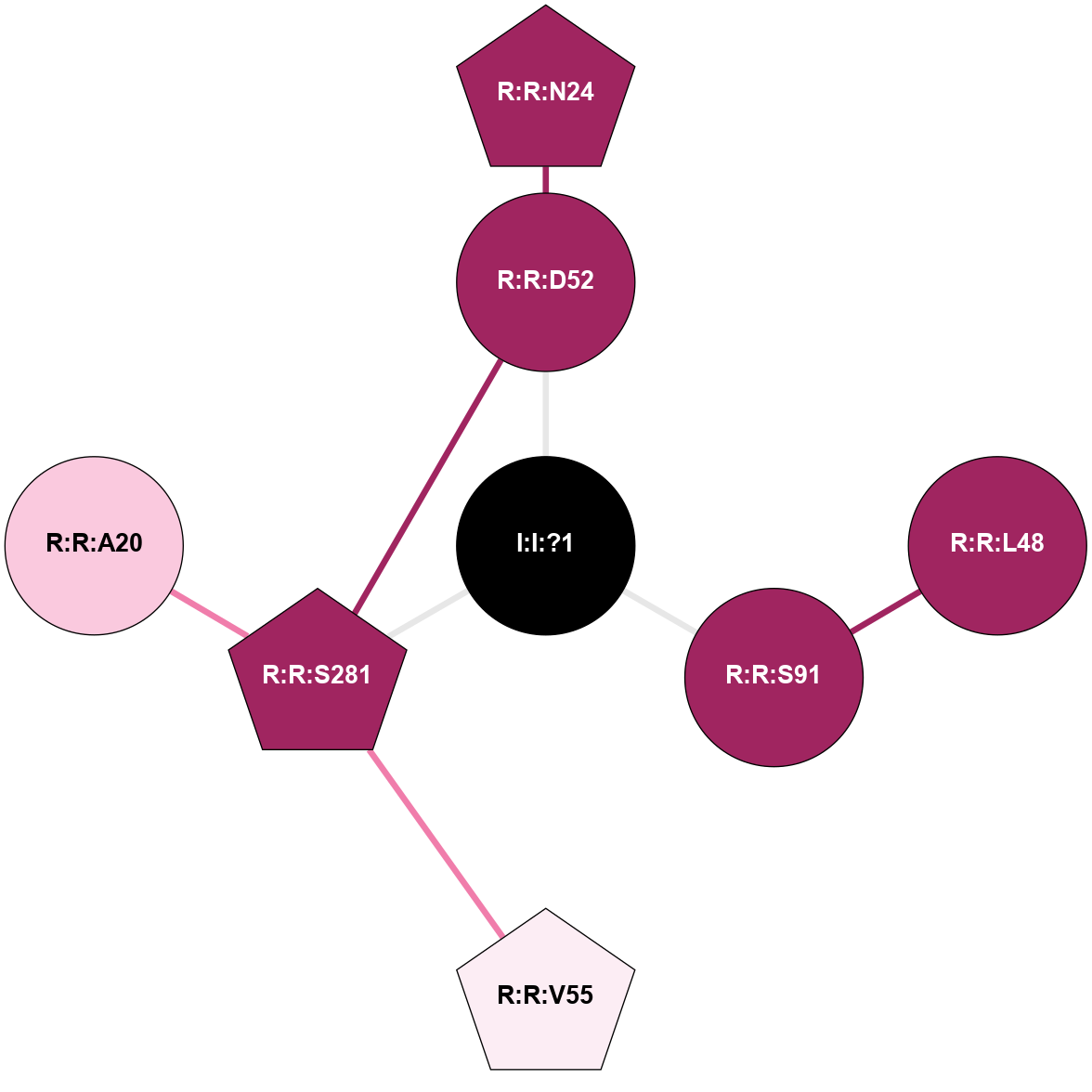
A 2D representation of the interactions of NA in 5K2B
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.06 | 7 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 7 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S281 | 2.97 | 7 | No | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 10.77 | 7 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 8.83 | 7 | No | Yes | 9 | 9 | 1 | 1 | | R:R:S281 | R:R:V55 | 8.08 | 7 | Yes | Yes | 9 | 6 | 1 | 2 | | R:R:A20 | R:R:S281 | 1.71 | 0 | No | Yes | 7 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.66 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5K2C | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.9 | 2016-09-21 | doi.org/10.1126/sciadv.1600292 |
|
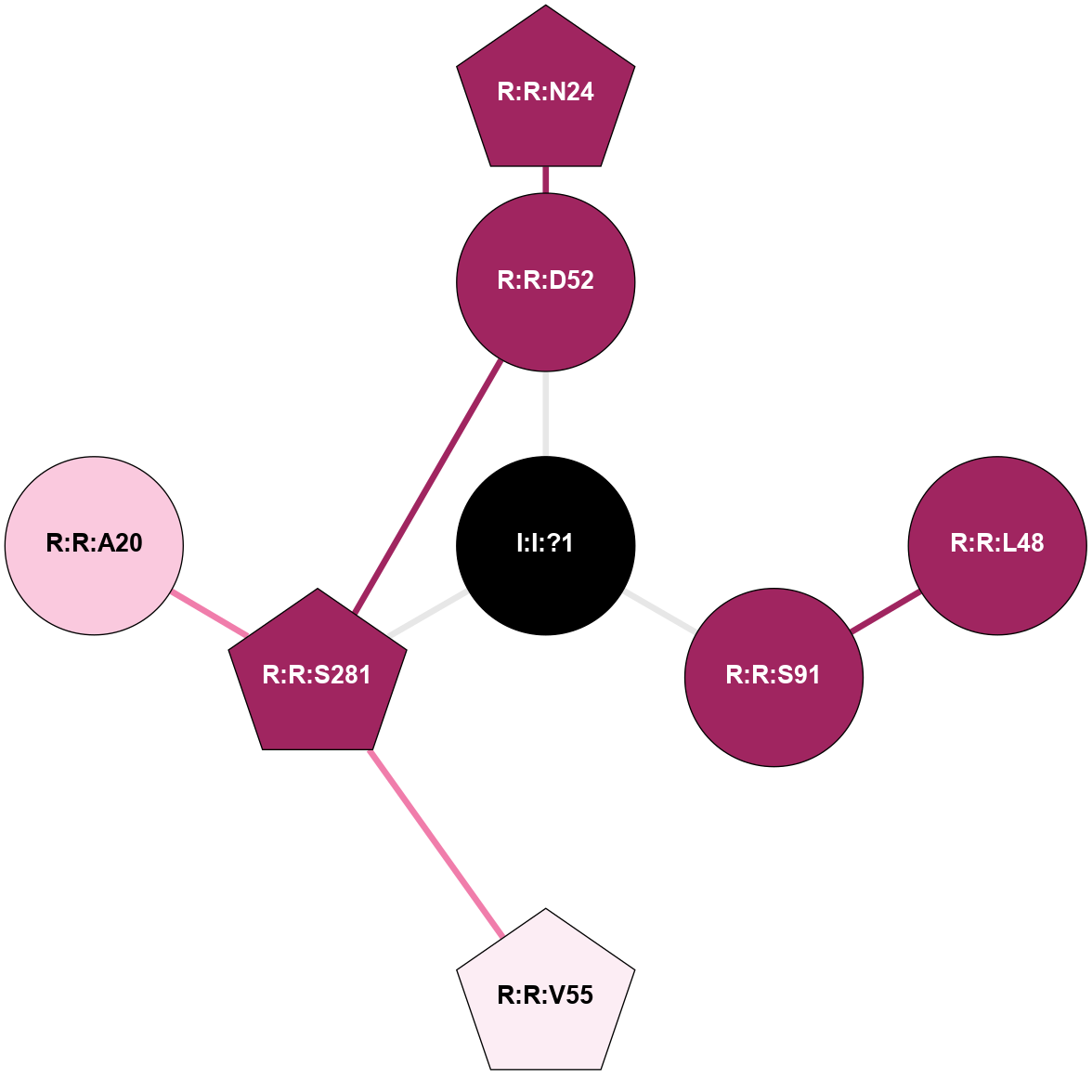
A 2D representation of the interactions of NA in 5K2C
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.06 | 7 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 7 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S281 | 2.97 | 7 | No | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 8.08 | 7 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 11.78 | 7 | No | Yes | 9 | 9 | 1 | 1 | | R:R:S281 | R:R:V55 | 9.7 | 7 | Yes | Yes | 9 | 6 | 1 | 2 | | R:R:A20 | R:R:S281 | 1.71 | 0 | No | Yes | 7 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.66 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5K2D | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.9 | 2016-09-21 | doi.org/10.1126/sciadv.1600292 |
|
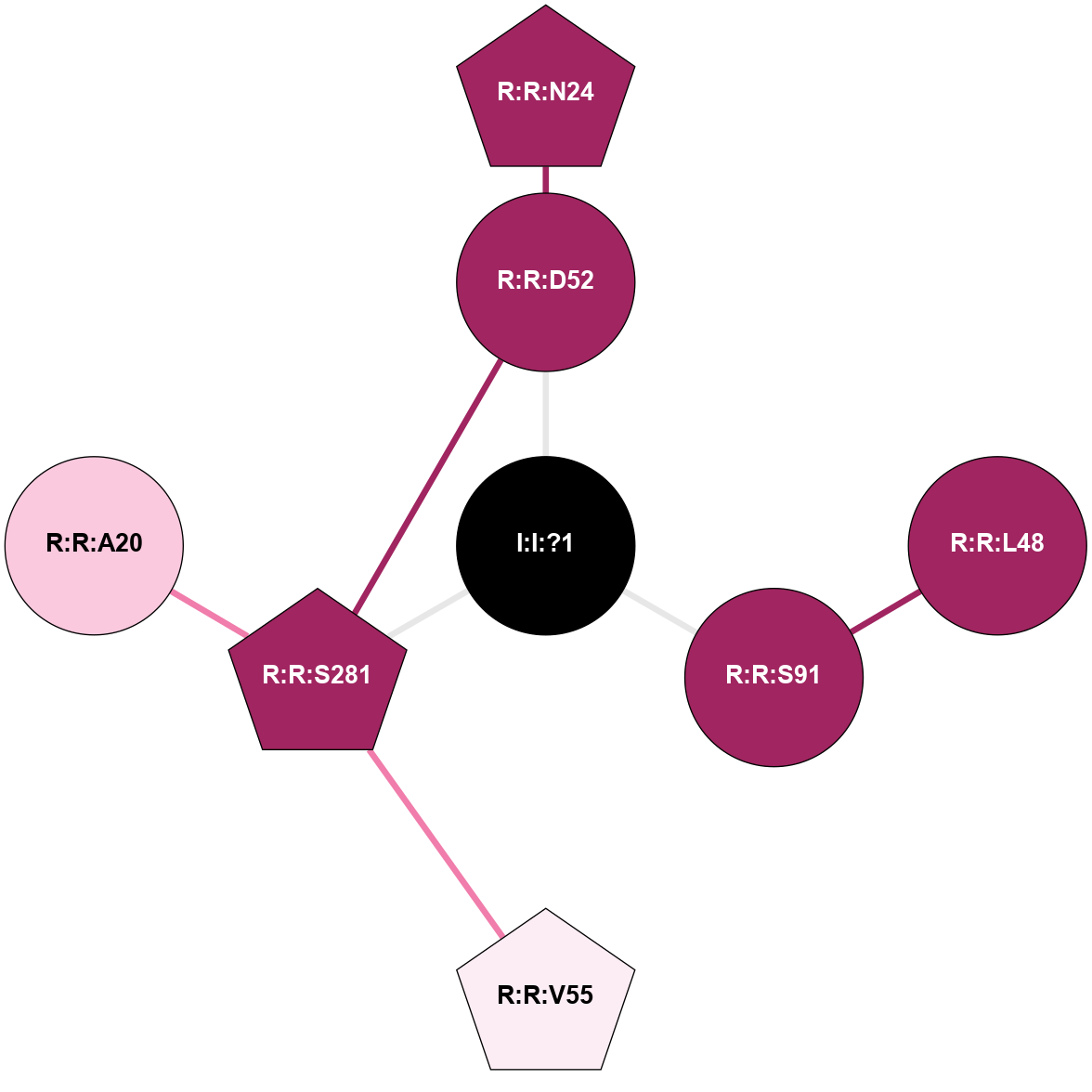
A 2D representation of the interactions of NA in 5K2D
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.05 | 9 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 9 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S281 | 2.97 | 9 | No | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 8.08 | 9 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 11.78 | 9 | No | Yes | 9 | 9 | 1 | 1 | | R:R:S281 | R:R:V55 | 9.7 | 9 | Yes | Yes | 9 | 6 | 1 | 2 | | R:R:A20 | R:R:S281 | 1.71 | 0 | No | Yes | 7 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.65 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5MZJ | A | Nucleotide | Adenosine | A2A | Homo Sapiens | Theophylline | Na | - | 2 | 2017-07-26 | doi.org/10.1016/j.str.2017.06.012 |
|
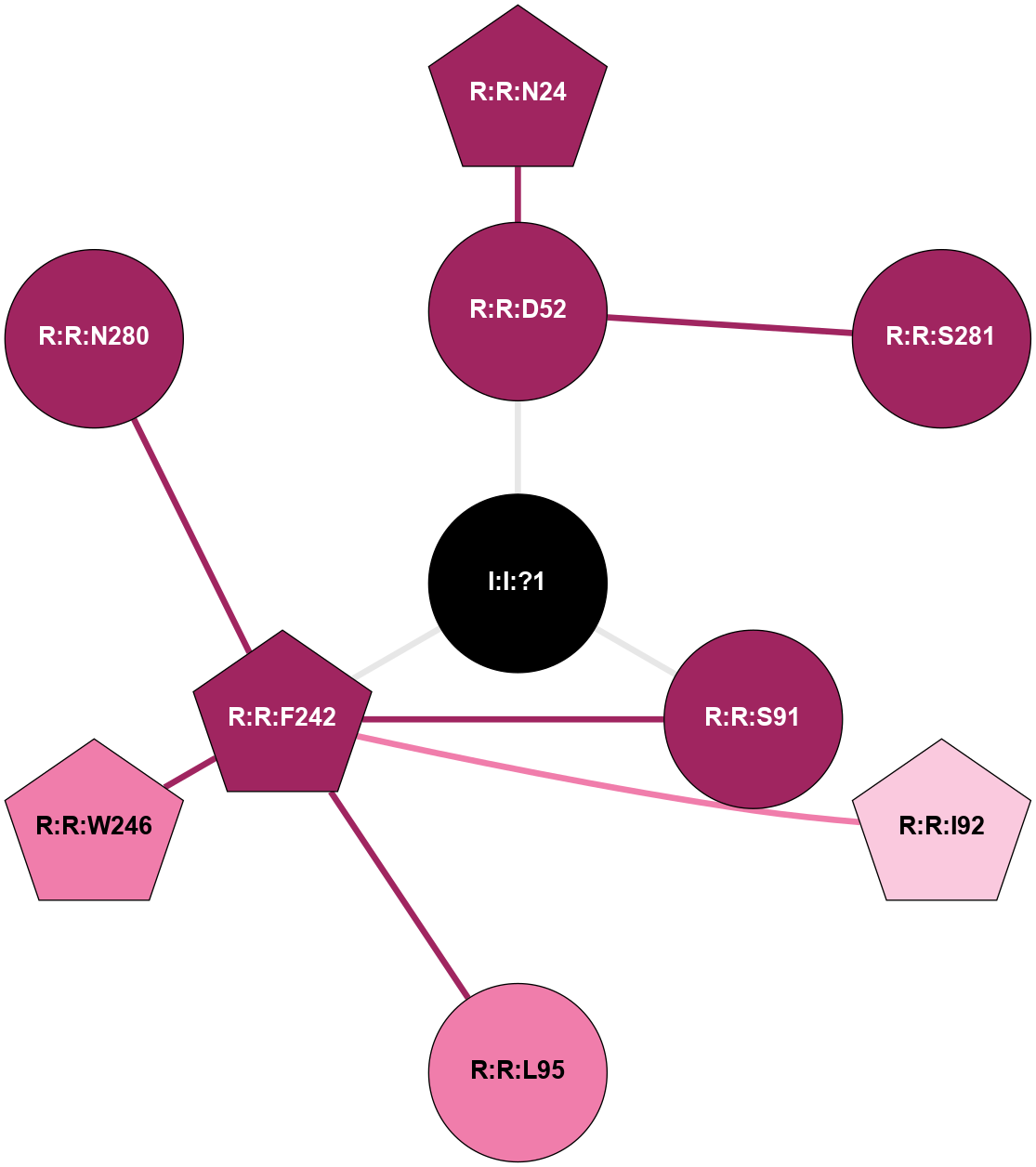
A 2D representation of the interactions of NA in 5MZJ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 12.12 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 9 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 3.96 | 9 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 9 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 9 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 9 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 16.04 | 9 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 9 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 9 | No | Yes | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5MZP | A | Nucleotide | Adenosine | A2A | Homo Sapiens | Caffeine | Na | - | 2.1 | 2017-07-26 | doi.org/10.1016/j.str.2017.06.012 |
|
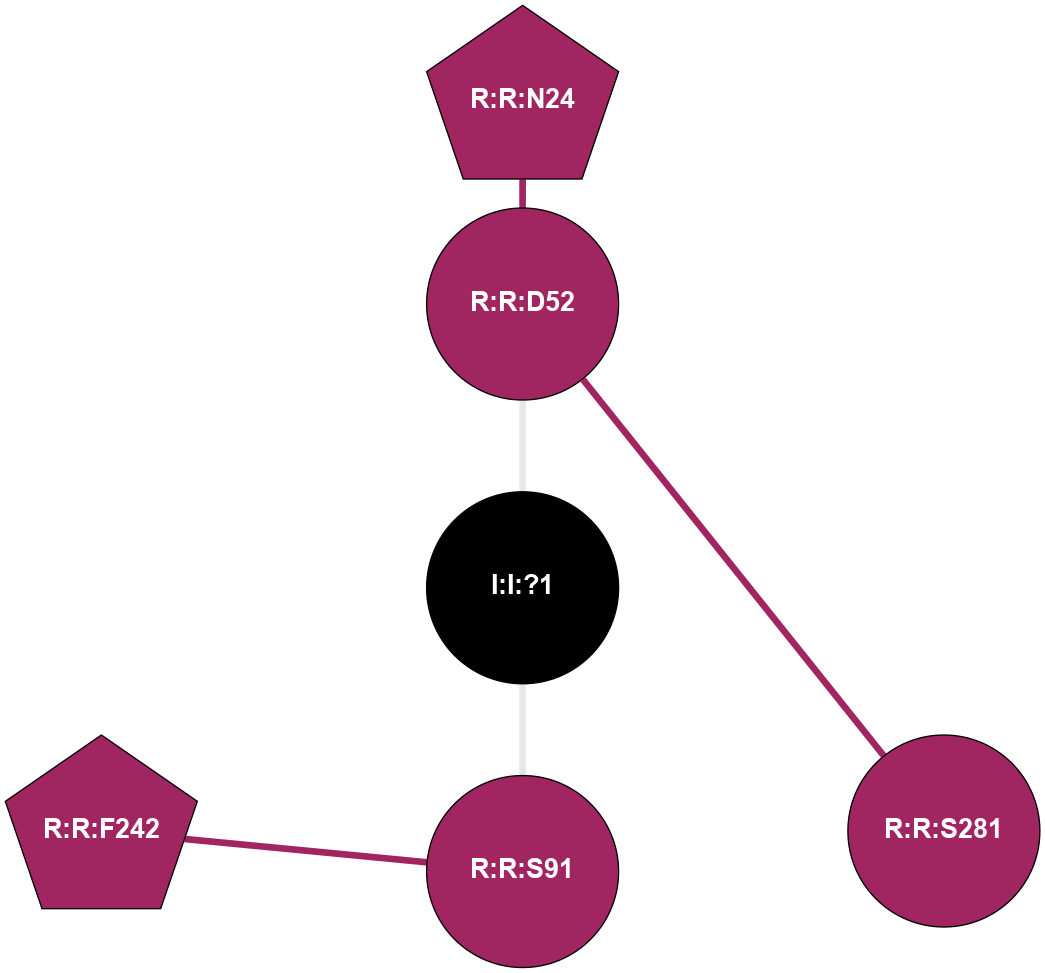
A 2D representation of the interactions of NA in 5MZP
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 13.46 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 10.31 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 0 | Yes | No | 9 | 9 | 2 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5N2R | A | Nucleotide | Adenosine | A2A | Homo Sapiens | PSB-36 | Na | - | 2.8 | 2017-07-26 | doi.org/10.1016/j.str.2017.06.012 |
|
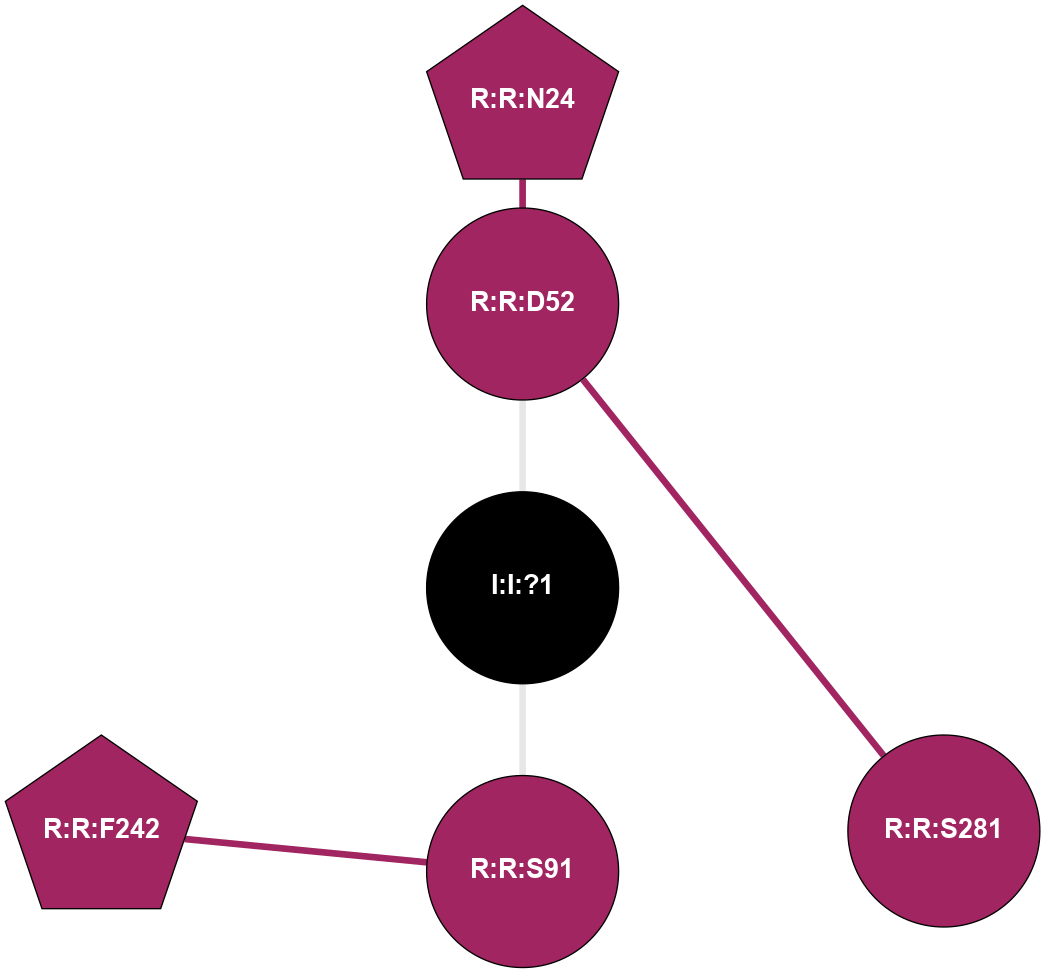
A 2D representation of the interactions of NA in 5N2R
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 9.42 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 8.83 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 0 | Yes | No | 9 | 9 | 2 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
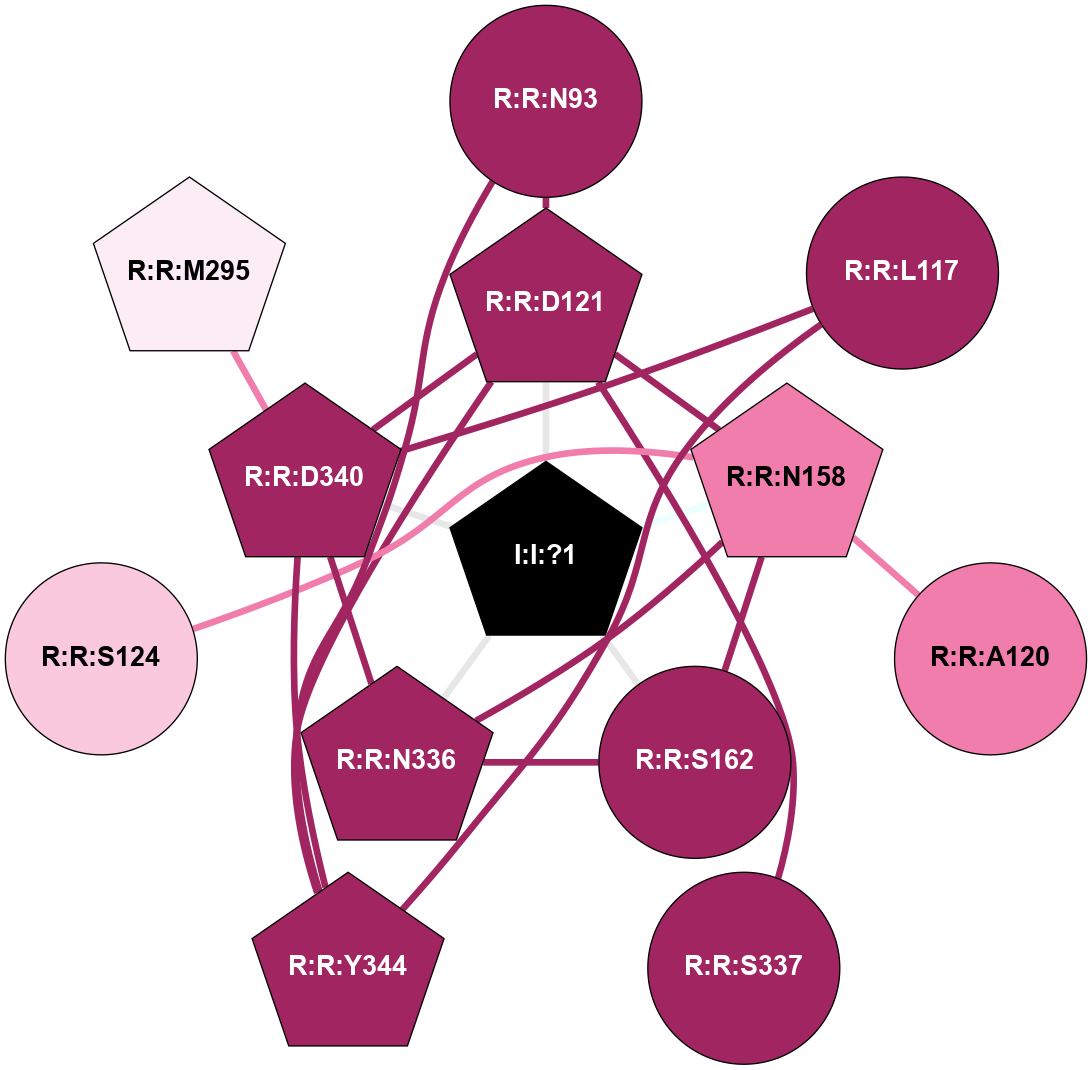
| 5NDD | A | Peptide | Proteinase Activated | PAR2 | Homo Sapiens | - | AZ8838; Na | - | 2.8 | 2017-05-03 | doi.org/10.1038/nature22309 |
|
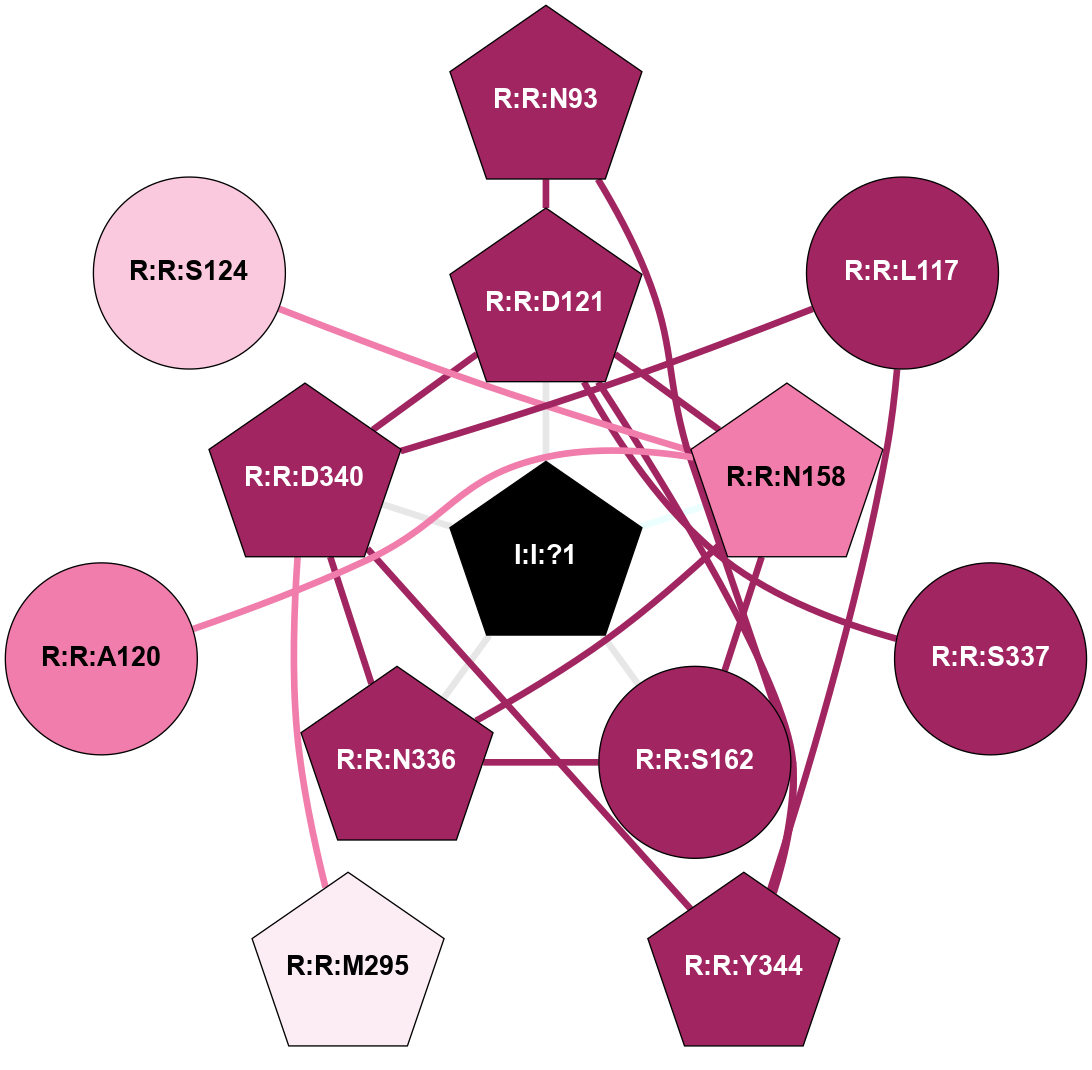
A 2D representation of the interactions of NA in 5NDD
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D121 | R:R:N93 | 14.81 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:N93 | R:R:Y344 | 6.98 | 2 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:D340 | R:R:L117 | 8.14 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:L117 | R:R:Y344 | 9.38 | 2 | No | Yes | 9 | 9 | 2 | 2 | | R:R:D121 | R:R:N158 | 4.04 | 2 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:D121 | R:R:S337 | 5.89 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D121 | R:R:D340 | 10.65 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:D121 | R:R:Y344 | 3.45 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D121 | 10.74 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:N158 | R:R:S162 | 5.96 | 2 | Yes | No | 8 | 9 | 1 | 1 | | R:R:N158 | R:R:N336 | 10.9 | 2 | Yes | Yes | 8 | 9 | 1 | 1 | | I:I:?1 | R:R:N158 | 8.15 | 2 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:N336 | R:R:S162 | 4.47 | 2 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S162 | 5.94 | 2 | Yes | No | 0 | 9 | 0 | 1 | | R:R:D340 | R:R:M295 | 4.16 | 2 | Yes | Yes | 9 | 6 | 1 | 2 | | R:R:D340 | R:R:N336 | 4.04 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:N336 | 10.87 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:D340 | R:R:Y344 | 6.9 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D340 | 8.06 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:A120 | R:R:N158 | 3.13 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:N158 | R:R:S124 | 1.49 | 2 | Yes | No | 8 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 8.75 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 21.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5NDZ | A | Peptide | Proteinase Activated | PAR2 | Homo Sapiens | - | AZ3451; Na | - | 3.6 | 2017-05-03 | doi.org/10.1038/nature22309 |
|
A 2D representation of the interactions of NA in 5NDZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D121 | R:R:N93 | 14.81 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:N93 | R:R:Y344 | 6.98 | 2 | No | Yes | 9 | 9 | 2 | 2 | | R:R:D340 | R:R:L117 | 12.21 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:L117 | R:R:Y344 | 10.55 | 2 | No | Yes | 9 | 9 | 2 | 2 | | R:R:A120 | R:R:N158 | 4.69 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:D121 | R:R:N158 | 5.39 | 2 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:D121 | R:R:S337 | 5.89 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D121 | R:R:D340 | 10.65 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:D121 | R:R:Y344 | 3.45 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D121 | 10.73 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:N158 | R:R:S124 | 4.47 | 2 | Yes | No | 8 | 7 | 1 | 2 | | R:R:N158 | R:R:S162 | 5.96 | 2 | Yes | No | 8 | 9 | 1 | 1 | | R:R:N158 | R:R:N336 | 8.17 | 2 | Yes | Yes | 8 | 9 | 1 | 1 | | I:I:?1 | R:R:N158 | 8.15 | 2 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:N336 | R:R:S162 | 4.47 | 2 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S162 | 5.94 | 2 | Yes | No | 0 | 9 | 0 | 1 | | R:R:D340 | R:R:M295 | 4.16 | 2 | Yes | Yes | 9 | 6 | 1 | 2 | | R:R:D340 | R:R:N336 | 4.04 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:N336 | 8.15 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:D340 | R:R:Y344 | 5.75 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D340 | 8.05 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 8.20 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 21.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5NLX | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2.14 | 2017-09-27 | doi.org/10.1038/s41467-017-00630-4 |
|
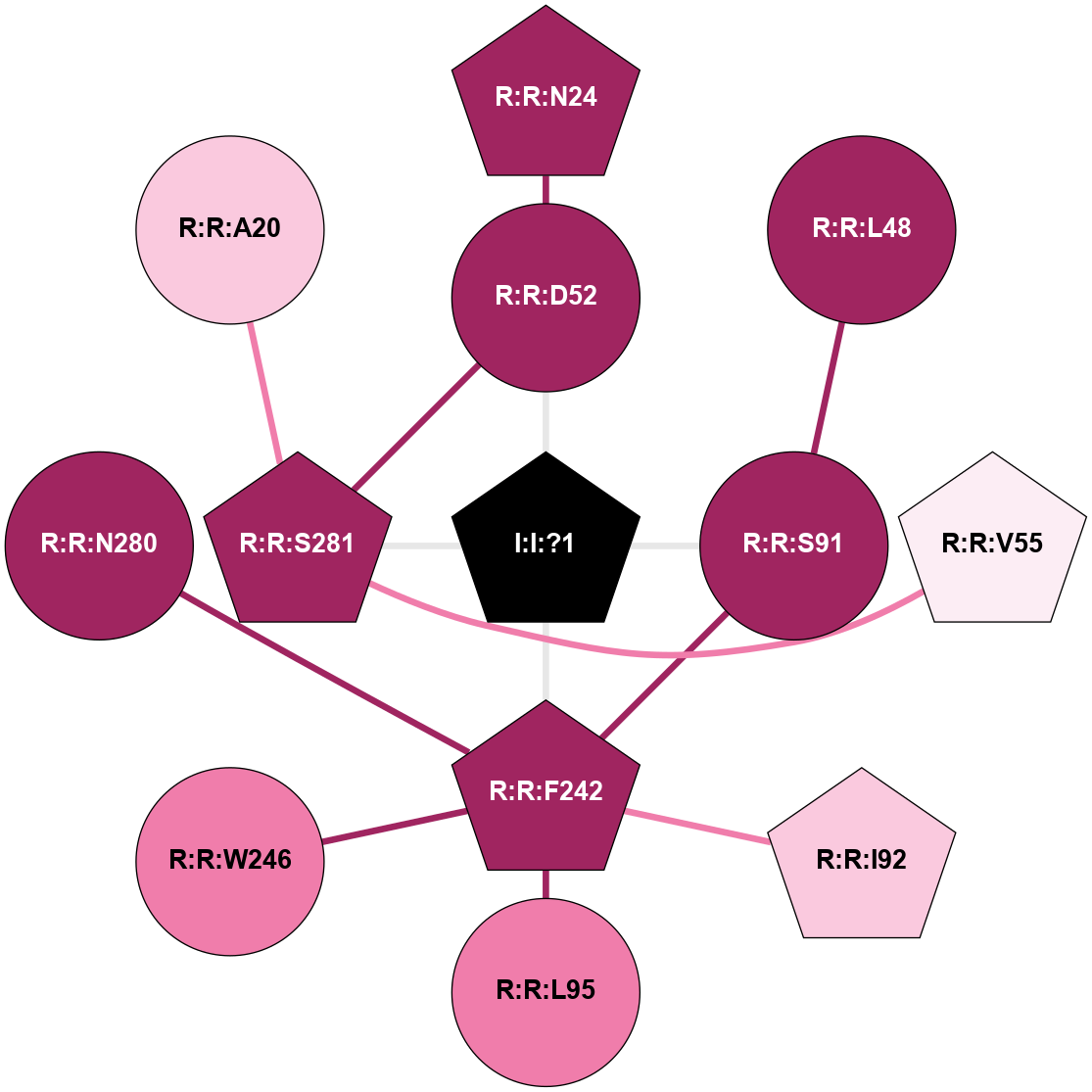
A 2D representation of the interactions of NA in 5NLX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.06 | 4 | Yes | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 4 | Yes | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:F242 | 4.82 | 4 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S281 | 2.97 | 4 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 10.77 | 4 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 11.78 | 4 | No | Yes | 9 | 9 | 1 | 1 | | R:R:S281 | R:R:V55 | 8.08 | 4 | Yes | Yes | 9 | 6 | 1 | 2 | | R:R:F242 | R:R:S91 | 5.28 | 4 | Yes | No | 9 | 9 | 1 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 4 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 4 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 15.03 | 4 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 7.25 | 4 | Yes | No | 9 | 9 | 1 | 2 | | R:R:A20 | R:R:S281 | 1.71 | 0 | No | Yes | 7 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 5.45 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5NM2 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.95 | 2017-09-27 | doi.org/10.1038/s41467-017-00630-4 |
|
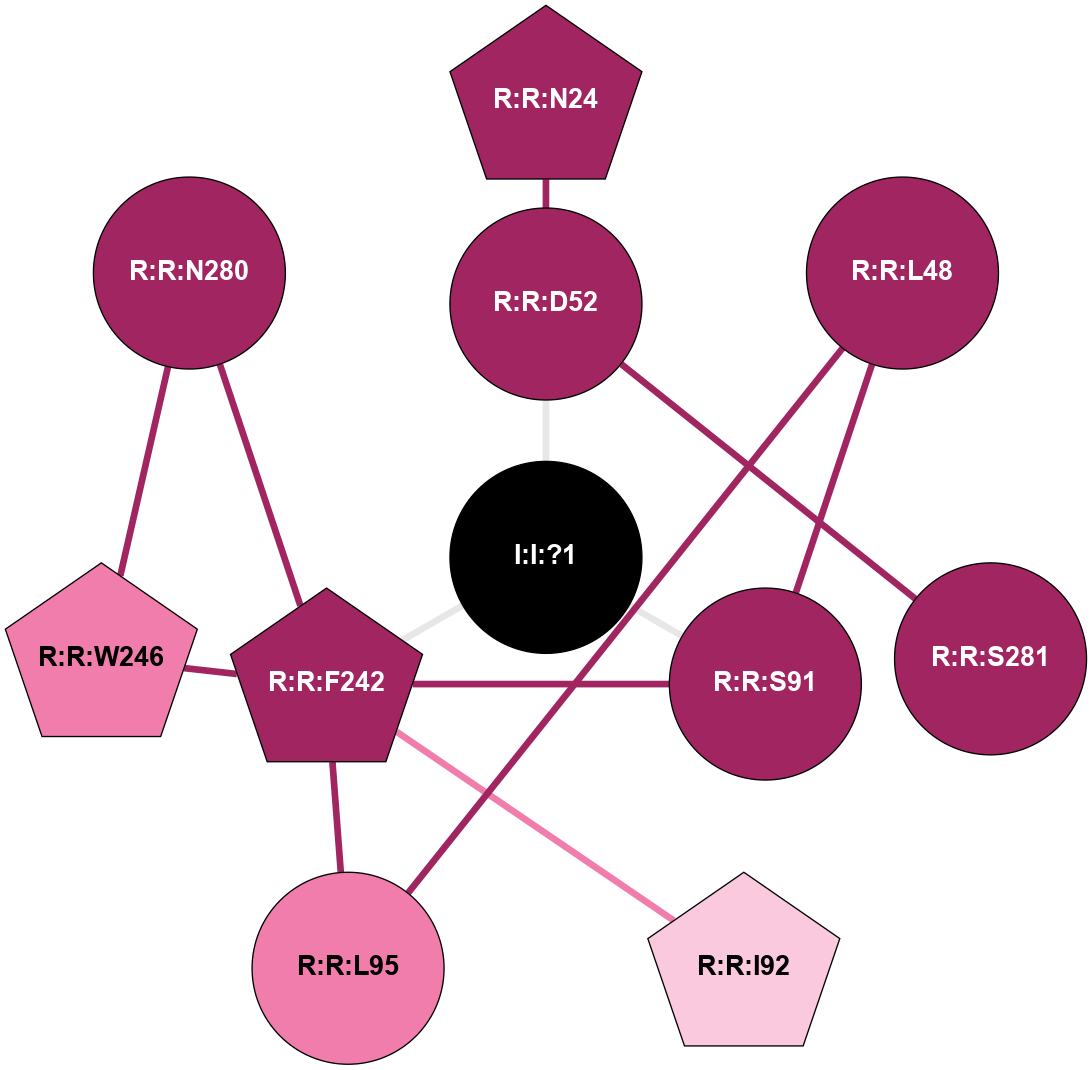
A 2D representation of the interactions of NA in 5NM2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 12.12 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:L48 | R:R:L95 | 4.15 | 0 | No | No | 9 | 8 | 2 | 2 | | R:R:D52 | R:R:S281 | 8.83 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 1 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 1 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 1 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 6.28 | 1 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 17.04 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 1 | No | Yes | 0 | 9 | 0 | 1 | | R:R:N280 | R:R:W246 | 3.39 | 1 | No | Yes | 9 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5NM4 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.7 | 2017-09-27 | doi.org/10.1038/s41467-017-00630-4 |
|
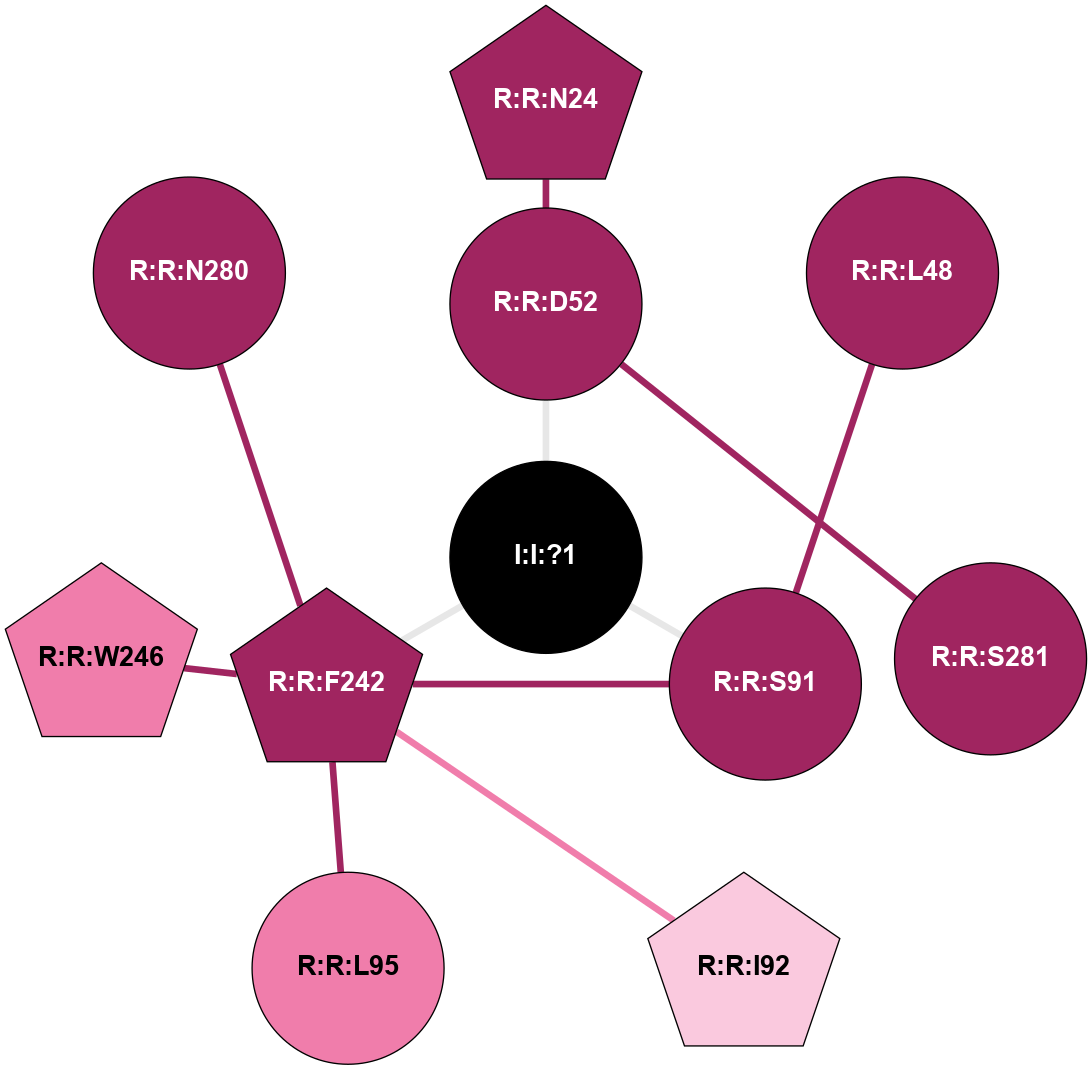
A 2D representation of the interactions of NA in 5NM4
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.06 | 8 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 8 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:F242 | 4.82 | 8 | No | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 10.77 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:F242 | R:R:S91 | 5.28 | 8 | Yes | No | 9 | 9 | 1 | 1 | | R:R:F242 | R:R:I92 | 6.28 | 8 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 8 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 16.04 | 8 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 8 | Yes | No | 9 | 9 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5OLG | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.87 | 2018-01-17 | doi.org/10.1038/s41598-017-18570-w |
|
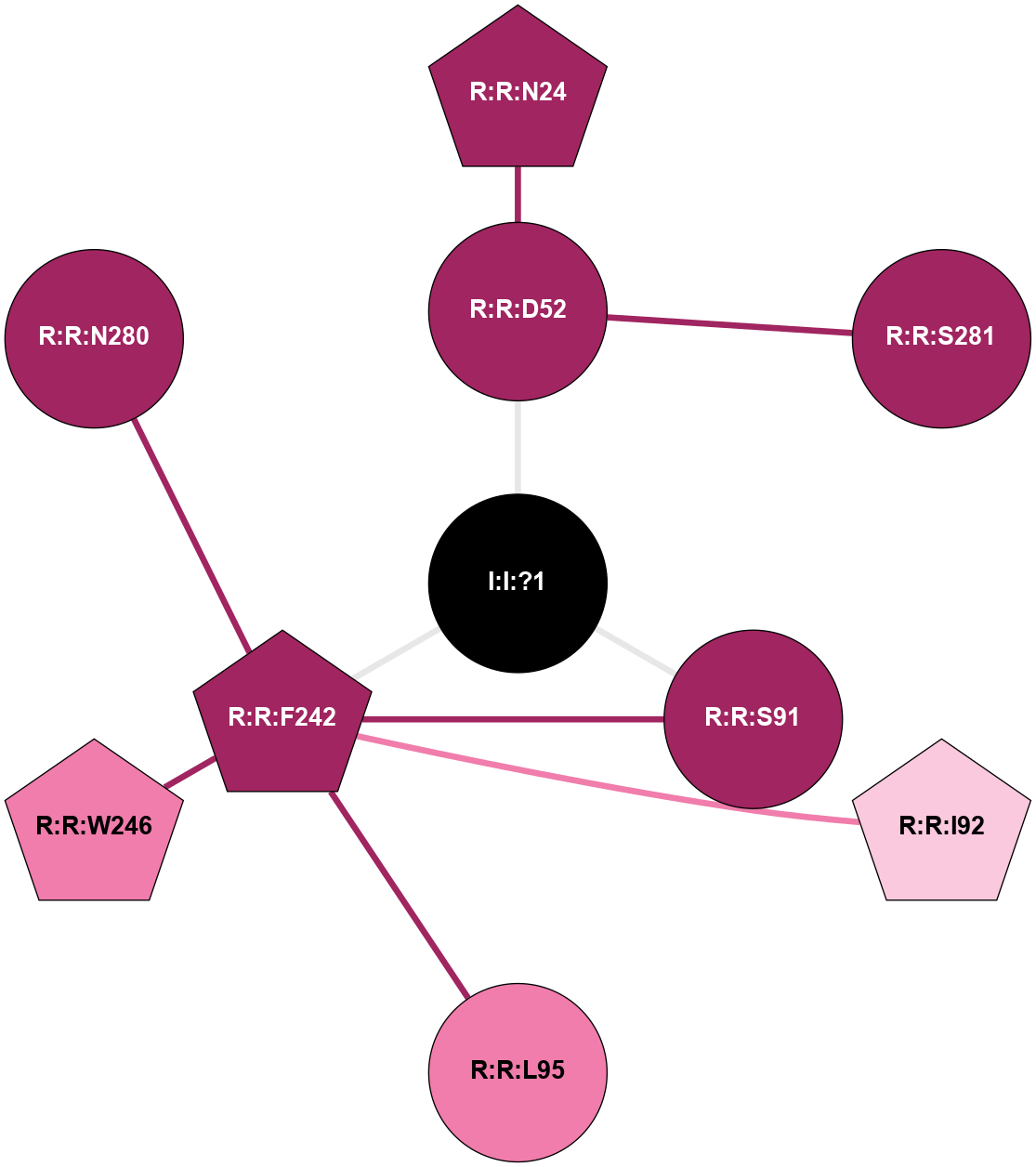
A 2D representation of the interactions of NA in 5OLG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 10.77 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 10.31 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 6 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 6 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 6 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 6 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 17.04 | 6 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 6 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 6 | No | Yes | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5OLH | A | Nucleotide | Adenosine | A2A | Homo Sapiens | Vipadenant | Na | - | 2.6 | 2018-01-17 | doi.org/10.1038/s41598-017-18570-w |
|
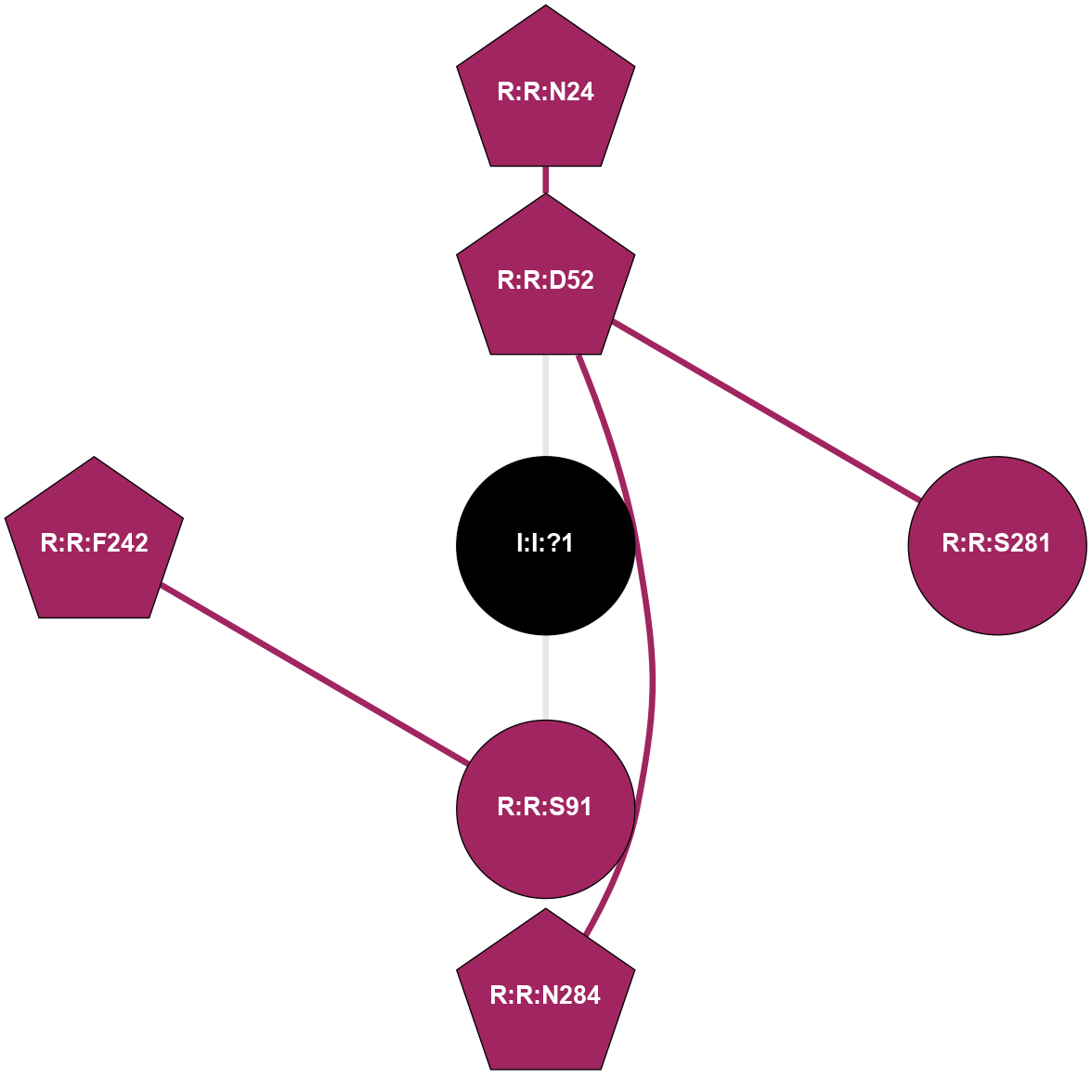
A 2D representation of the interactions of NA in 5OLH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 13.46 | 0 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 8.83 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:N284 | 4.04 | 0 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 0 | No | Yes | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 0 | Yes | No | 9 | 9 | 2 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5OLO | A | Nucleotide | Adenosine | A2A | Homo Sapiens | Tozadenant | Na | - | 3.1 | 2018-01-17 | doi.org/10.1038/s41598-017-18570-w |
|
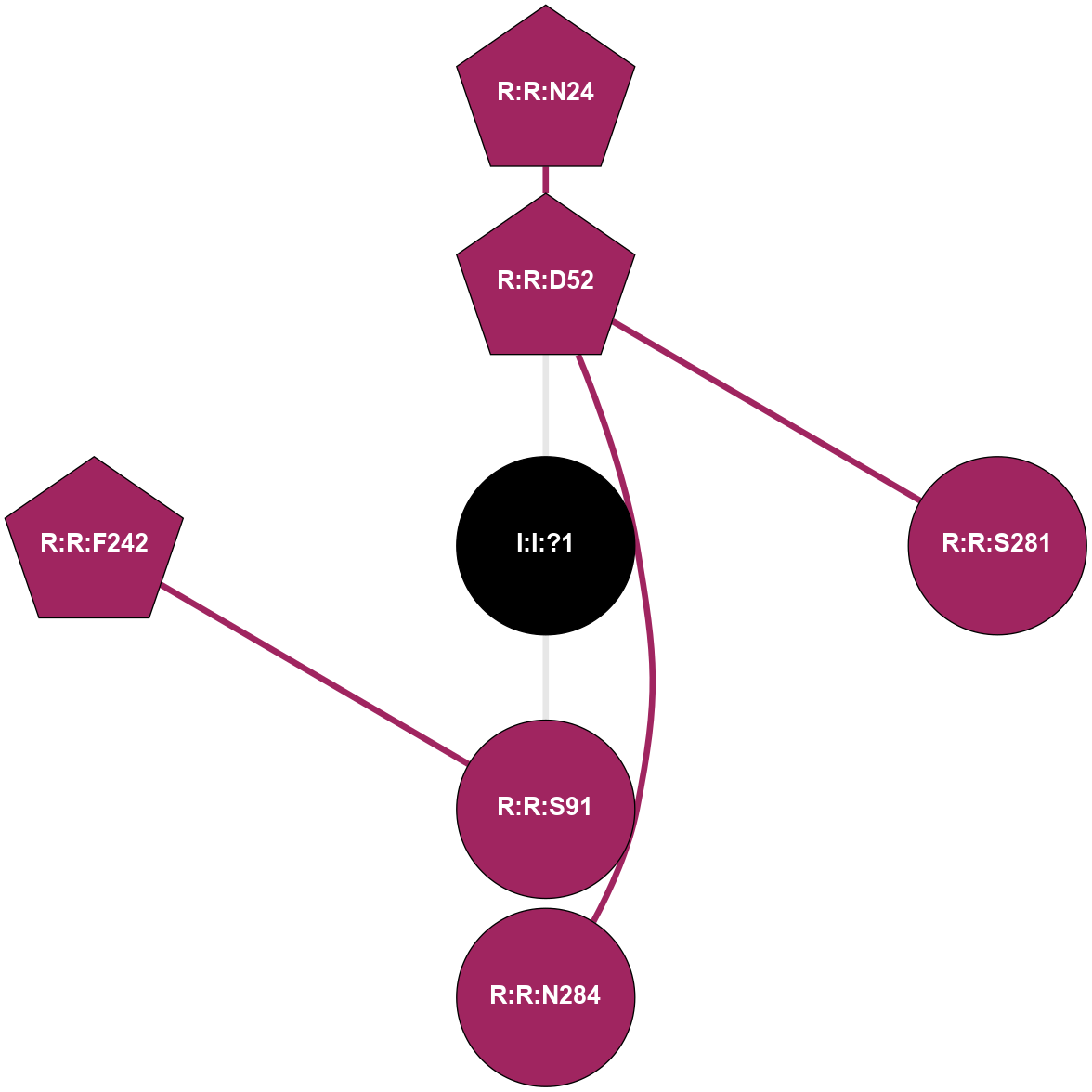
A 2D representation of the interactions of NA in 5OLO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 12.12 | 0 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 8.83 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:N284 | 6.73 | 0 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 5.37 | 0 | No | Yes | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 8 | Yes | No | 9 | 9 | 2 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.66 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5OLV | A | Nucleotide | Adenosine | A2A | Homo Sapiens | LUAA41063 | Na | - | 2 | 2018-01-17 | doi.org/10.1038/s41598-017-18570-w |
|
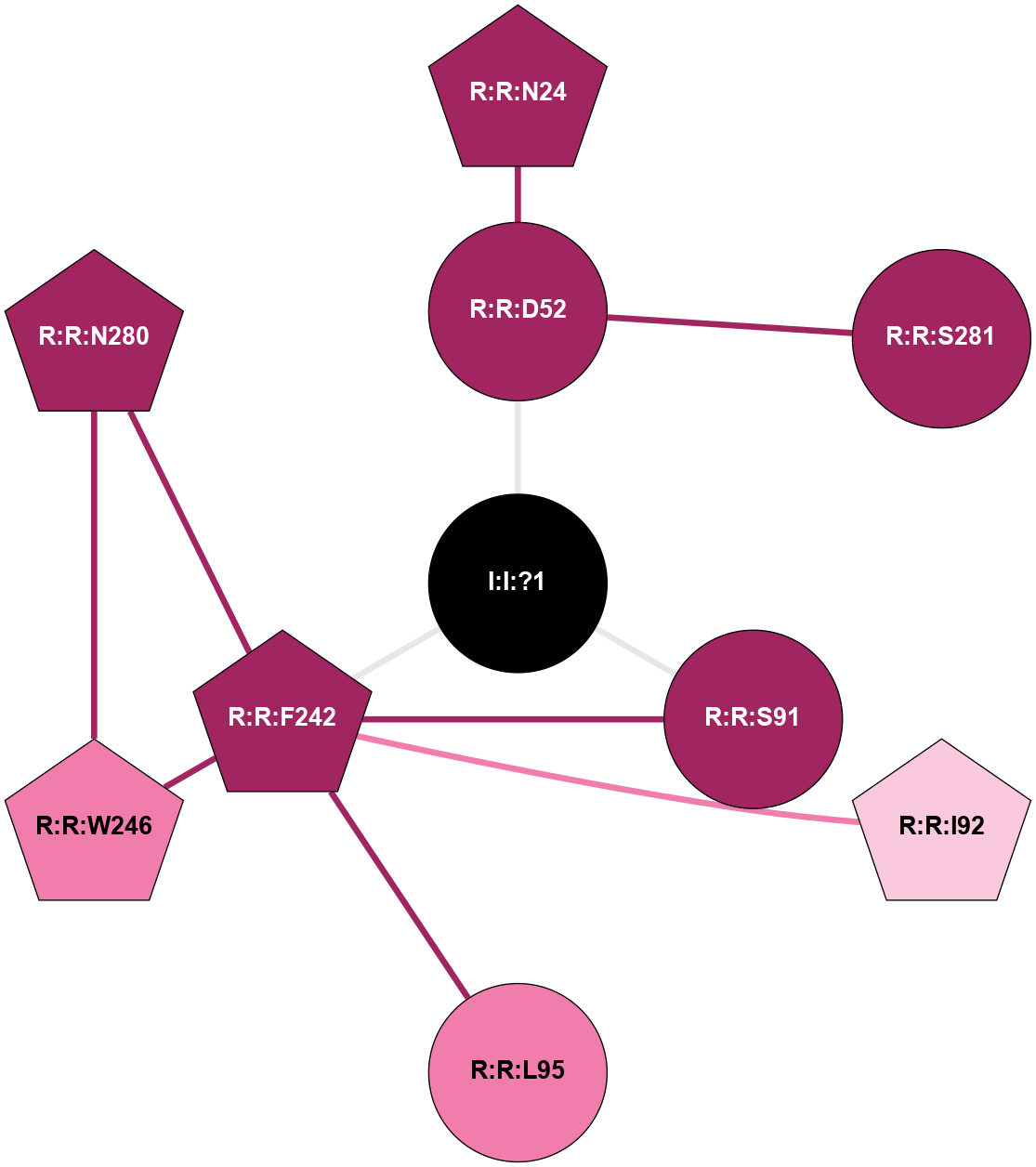
A 2D representation of the interactions of NA in 5OLV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 9.42 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 10.31 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 5 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 5 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 5 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 5 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 5 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 17.04 | 5 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 5 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 5 | No | Yes | 0 | 9 | 0 | 1 | | R:R:N280 | R:R:W246 | 3.39 | 5 | Yes | Yes | 9 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5OLZ | A | Nucleotide | Adenosine | A2A | Homo Sapiens | PubChem 135566609 | Na | - | 1.9 | 2018-01-17 | doi.org/10.1038/s41598-017-18570-w |
|
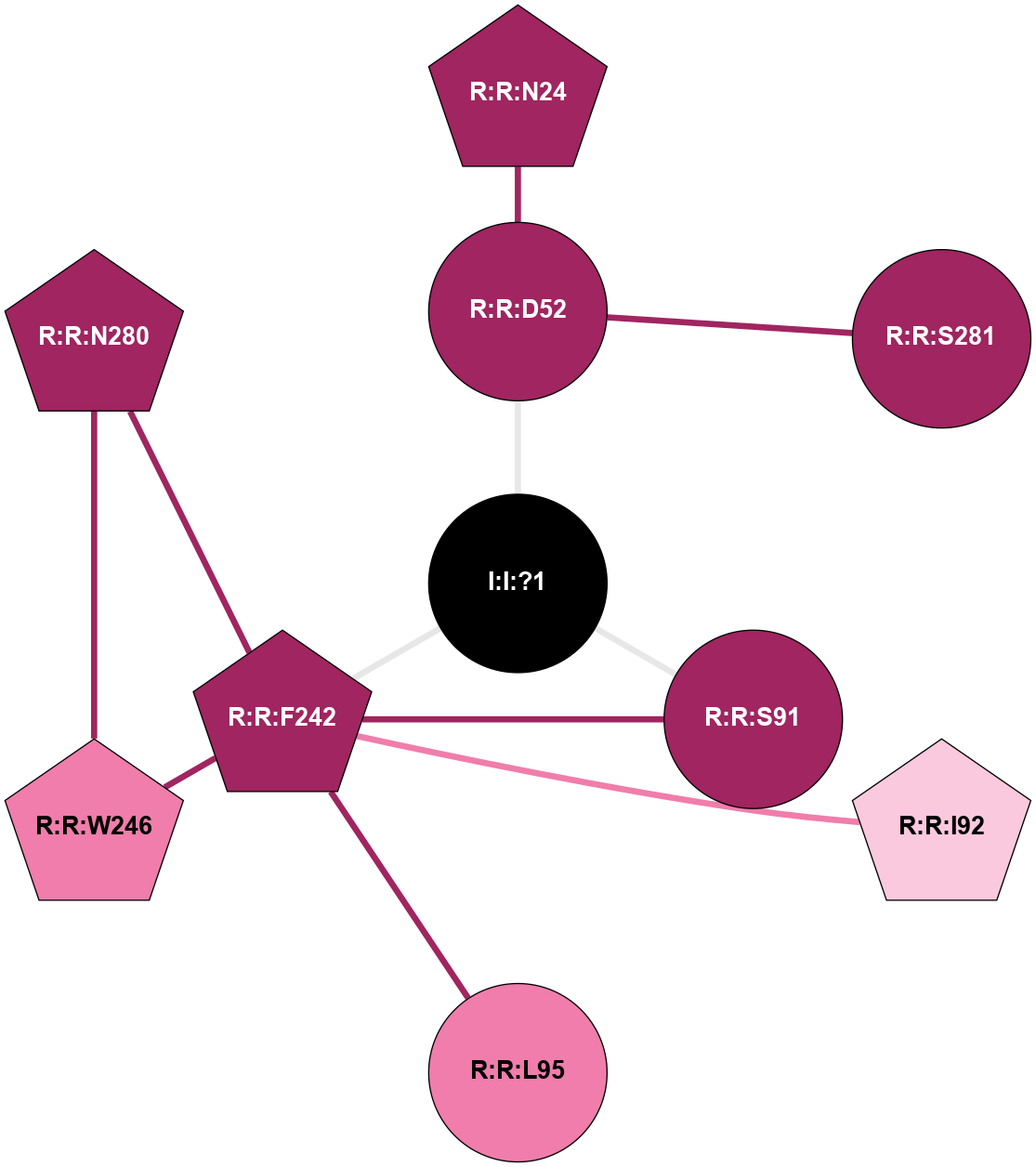
A 2D representation of the interactions of NA in 5OLZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 10.77 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 1 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 1 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 1 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 1 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 18.04 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 1 | No | Yes | 0 | 9 | 0 | 1 | | R:R:N280 | R:R:W246 | 3.39 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5OM1 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | PubChem 135566609 | Na | - | 2.1 | 2018-01-17 | doi.org/10.1038/s41598-017-18570-w |
|

A 2D representation of the interactions of NA in 5OM1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 10.77 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 7 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 7 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 7 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 7 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 7 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 16.04 | 7 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 7 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 7 | No | Yes | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5OM4 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | PubChem 135566609 | Na | - | 2 | 2018-01-17 | doi.org/10.1038/s41598-017-18570-w |
|
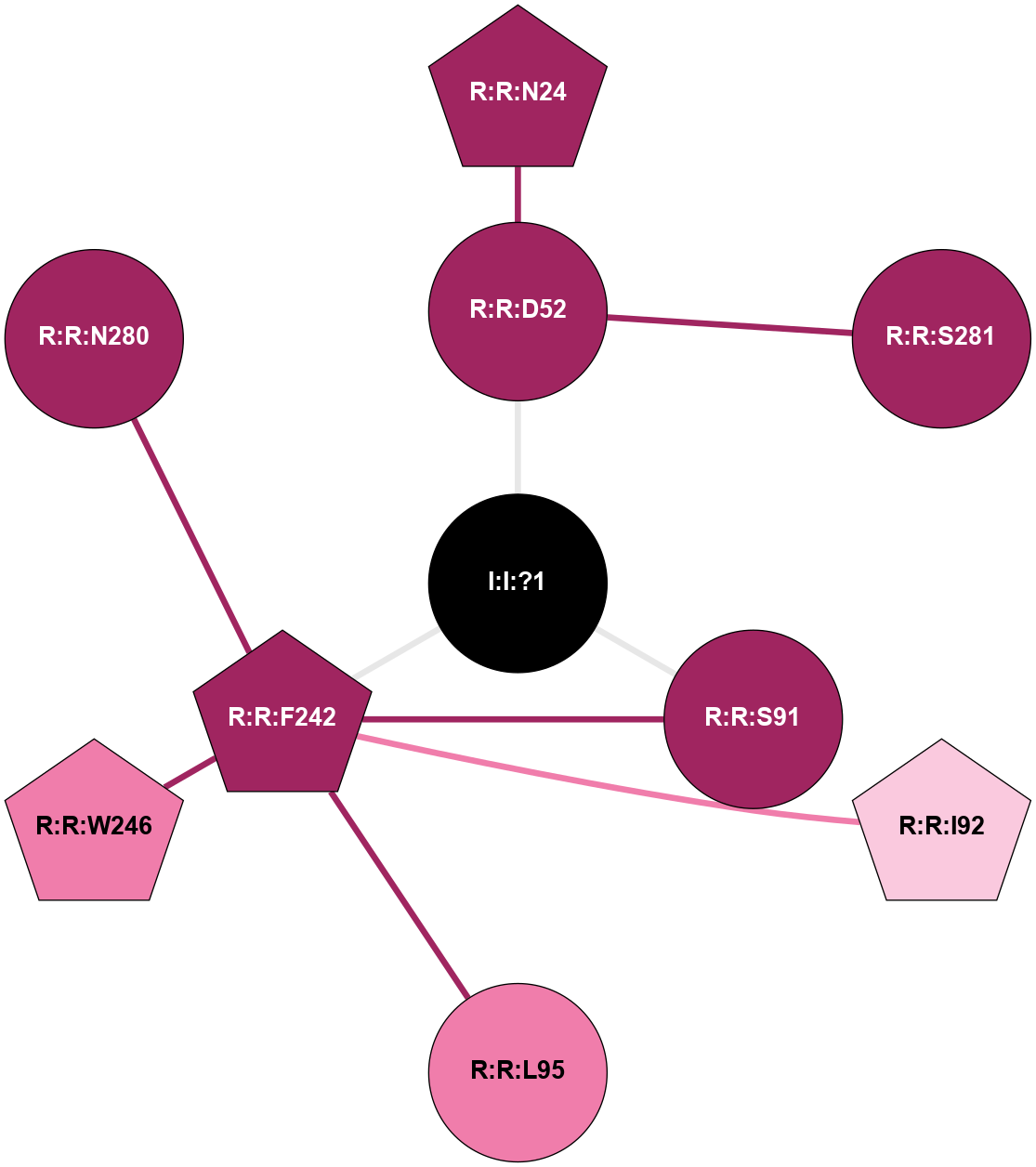
A 2D representation of the interactions of NA in 5OM4
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 12.12 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 6 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 6 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 6 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 6 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 16.04 | 6 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 6 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 6 | No | Yes | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5VRA | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2.35 | 2017-12-13 | doi.org/10.1038/nprot.2017.135 |
|
A 2D representation of the interactions of NA in 5VRA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.06 | 0 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 9.42 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5WIV | A | Amine | Dopamine | D4 | Homo Sapiens | Nemonapride | Na | - | 2.14 | 2017-10-18 | doi.org/10.1126/science.aan5468 |
|

A 2D representation of the interactions of NA in 5WIV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D80 | R:R:N52 | 10.77 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D80 | R:R:S393 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D80 | 8.06 | 0 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S122 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6C1R | A | Peptide | Complement Peptide | C5A1 | Homo Sapiens | PMX53 | Avacopan; Na | - | 2.2 | 2018-05-30 | doi.org/10.1038/s41594-018-0067-z |
|
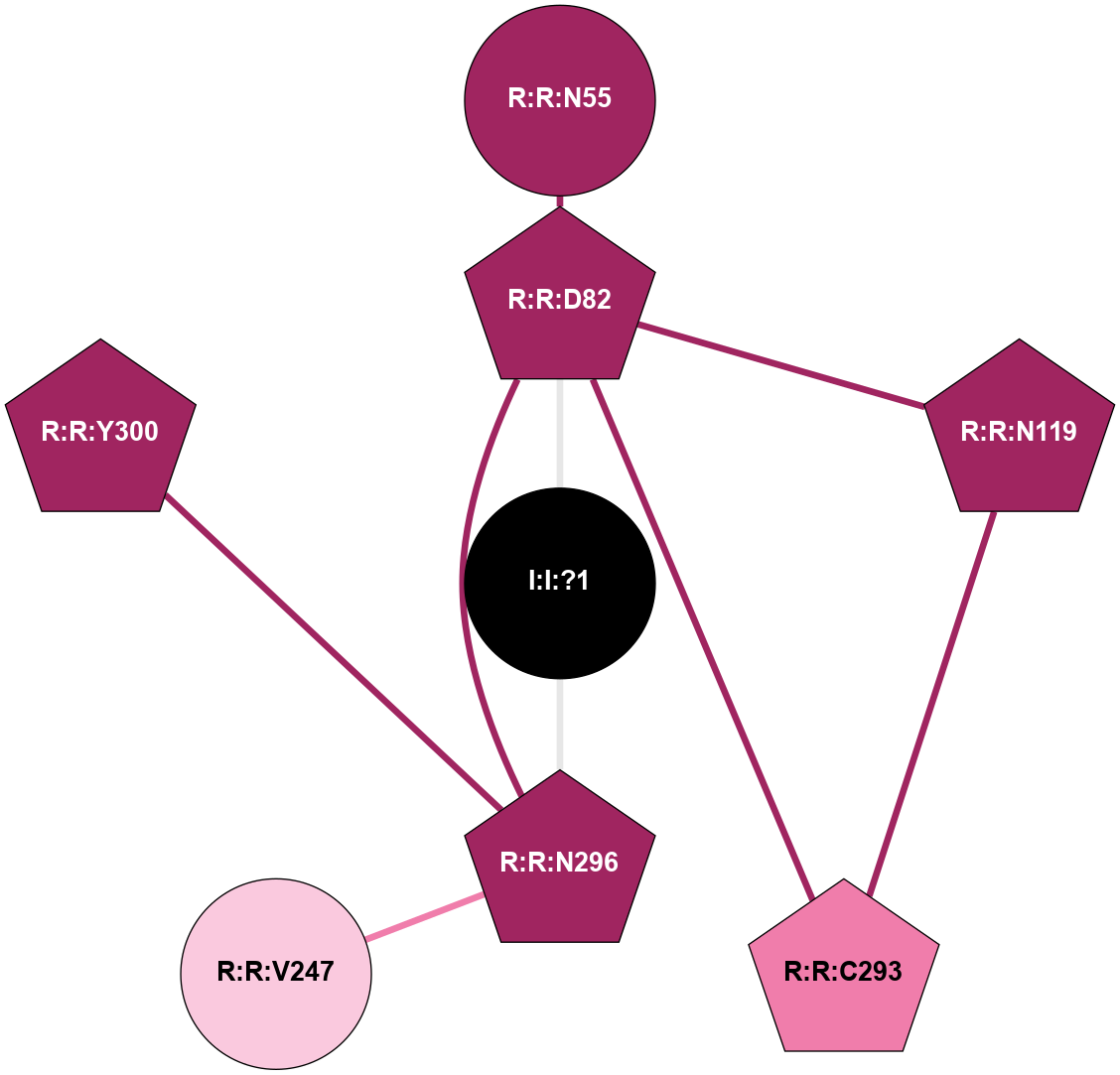
A 2D representation of the interactions of NA in 6C1R
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D82 | R:R:N55 | 9.42 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D82 | R:R:N119 | 6.73 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:C293 | R:R:D82 | 6.22 | 1 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:D82 | R:R:N296 | 5.39 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:D82 | 8.06 | 1 | No | Yes | 0 | 9 | 0 | 1 | | R:R:C293 | R:R:N119 | 6.3 | 1 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:N296 | R:R:V247 | 8.87 | 1 | Yes | No | 9 | 7 | 1 | 2 | | R:R:N296 | R:R:Y300 | 9.3 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:N296 | 10.87 | 1 | No | Yes | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.46 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6GT3 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | Imaradenant | Na | - | 2 | 2019-06-26 | doi.org/10.1136/jitc-2019-000417 |
|
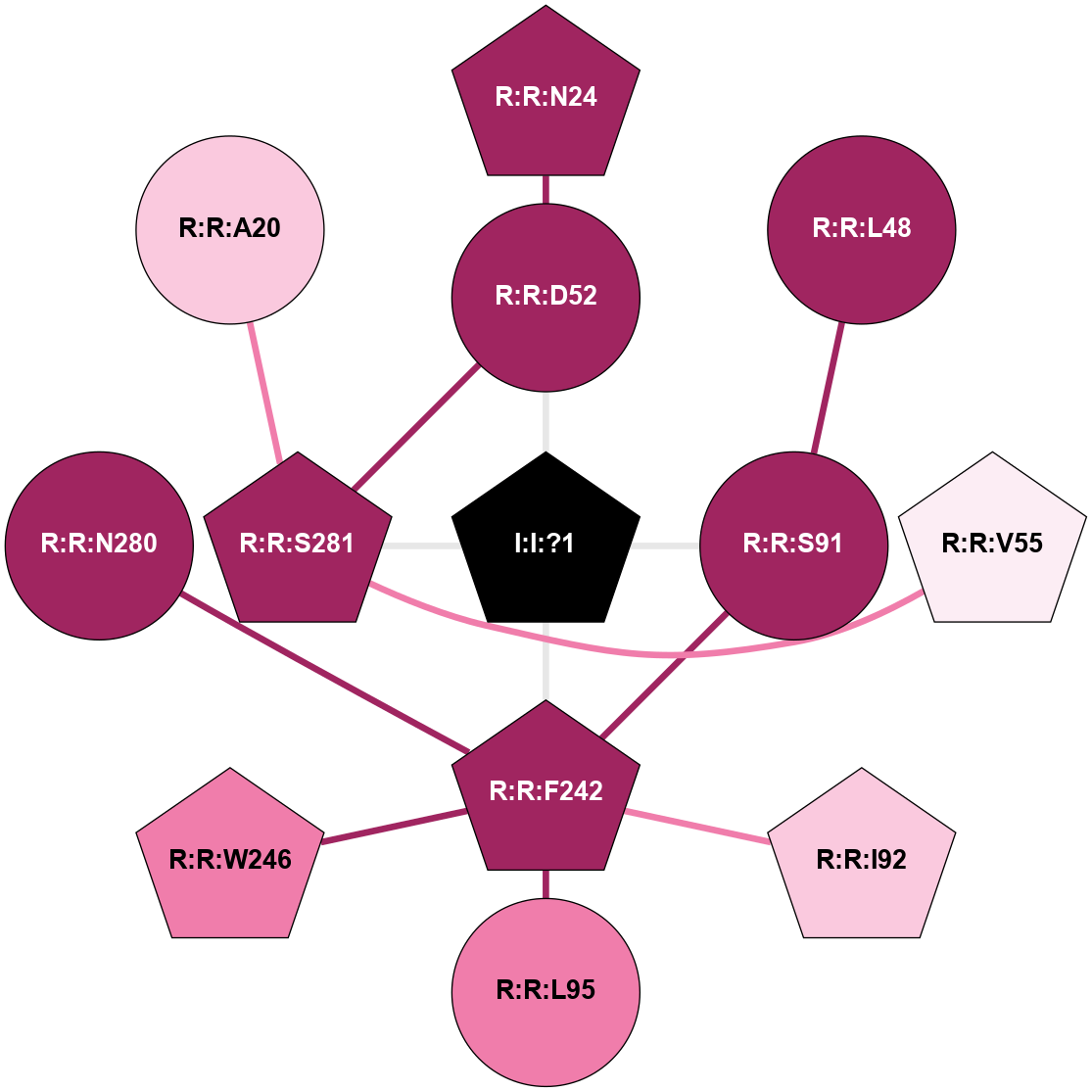
A 2D representation of the interactions of NA in 6GT3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 10.77 | 1 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 10.31 | 1 | No | Yes | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:D52 | 8.06 | 1 | Yes | No | 0 | 9 | 0 | 1 | | R:R:S281 | R:R:V55 | 8.08 | 1 | Yes | Yes | 9 | 6 | 1 | 2 | | R:R:F242 | R:R:S91 | 5.28 | 1 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 1 | Yes | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 1 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 16.04 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S281 | 2.97 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:A20 | R:R:S281 | 1.71 | 0 | No | Yes | 7 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 5.45 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6JZH | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2.25 | 2019-10-30 | doi.org/10.1107/S1600576719012846 |
|

A 2D representation of the interactions of NA in 6JZH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.06 | 0 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 9.42 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6LPJ | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.8 | 2020-11-25 | doi.org/10.1038/s41598-020-76277-x |
|
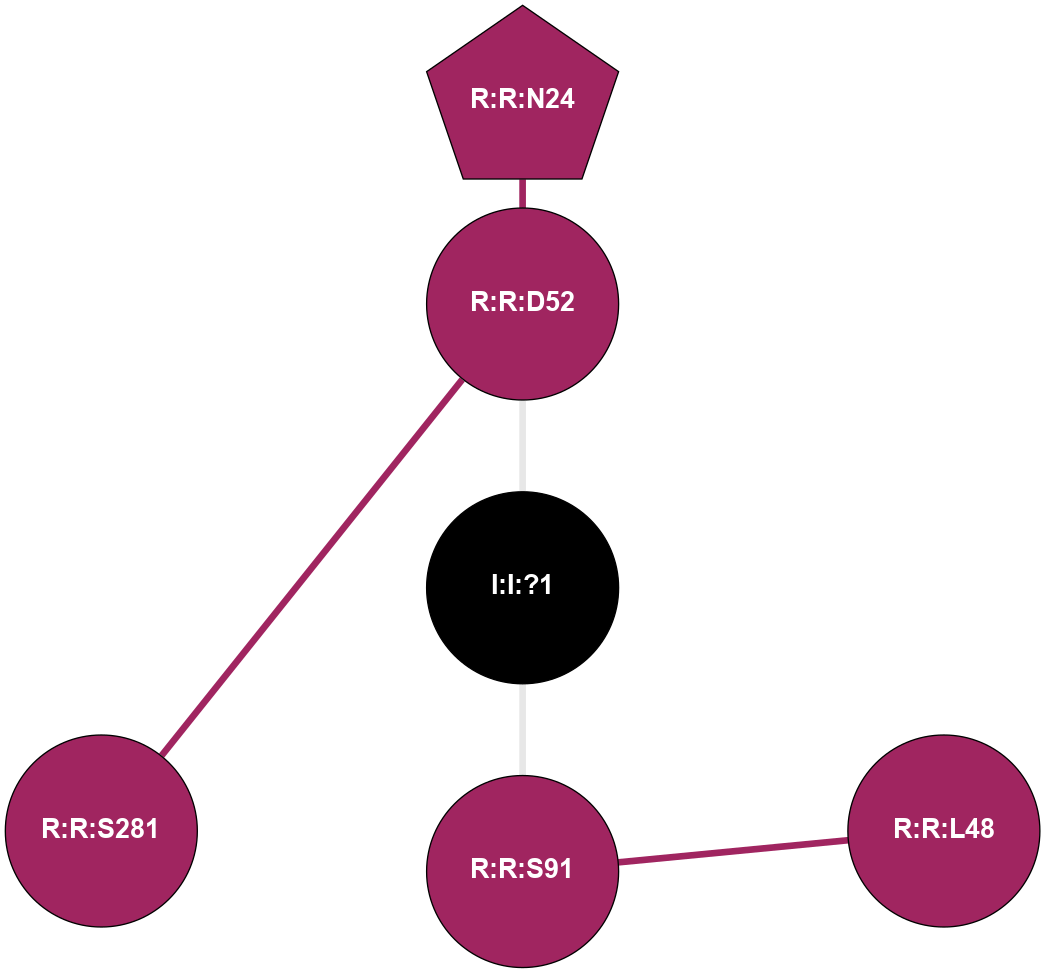
A 2D representation of the interactions of NA in 6LPJ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 9.42 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 0 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6LPK | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.8 | 2020-11-25 | doi.org/10.1038/s41598-020-76277-x |
|
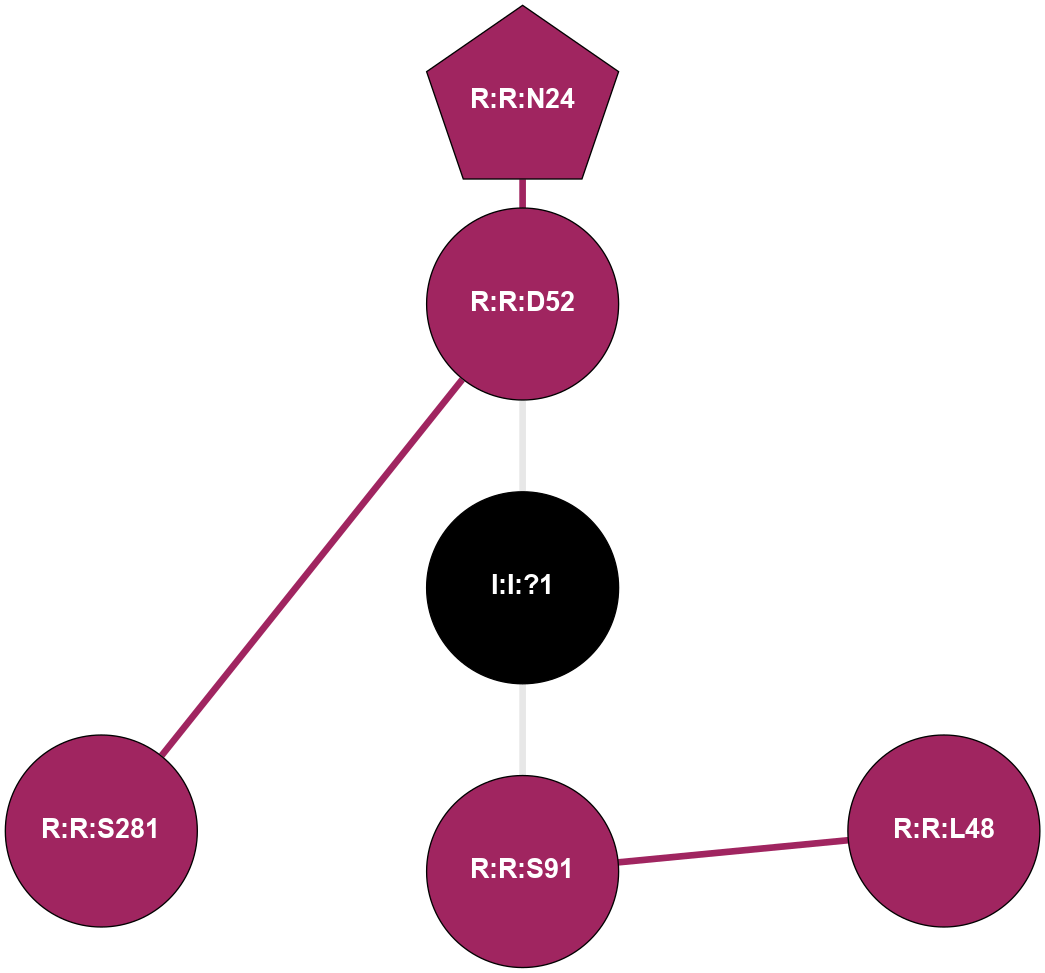
A 2D representation of the interactions of NA in 6LPK
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 10.77 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 0 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6LPL | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2 | 2020-11-25 | doi.org/10.1038/s41598-020-76277-x |
|

A 2D representation of the interactions of NA in 6LPL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 9.42 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 0 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6PS7 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 1.85 | 2019-11-13 | doi.org/10.1107/S2052252519013137 |
|
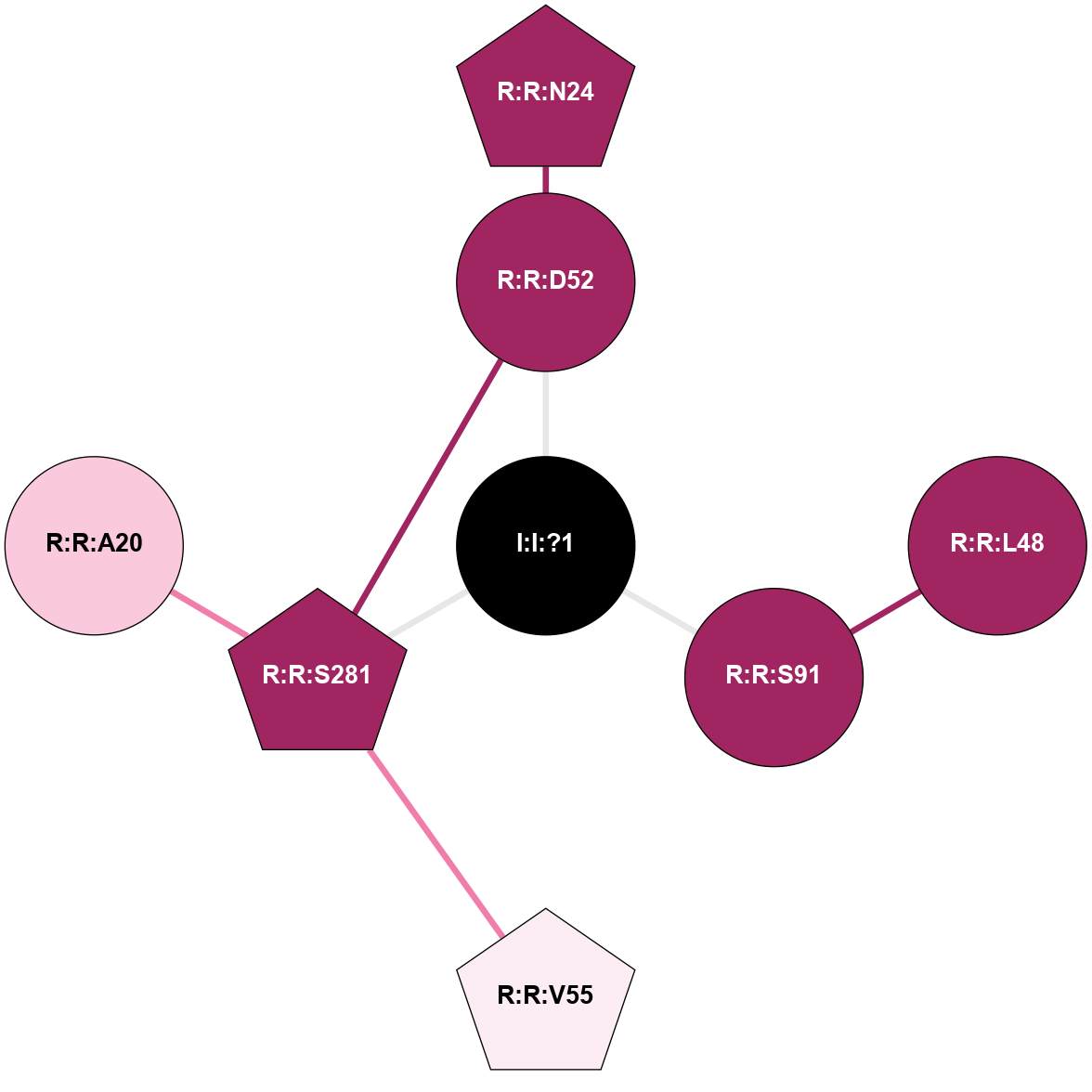
A 2D representation of the interactions of NA in 6PS7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.06 | 9 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 9 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S281 | 2.97 | 9 | No | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 8.08 | 9 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 11.78 | 9 | No | Yes | 9 | 9 | 1 | 1 | | R:R:S281 | R:R:V55 | 8.08 | 9 | Yes | Yes | 9 | 6 | 1 | 2 | | R:R:A20 | R:R:S281 | 1.71 | 0 | No | Yes | 7 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.66 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6QZH | A | Protein | Chemokine | CCR7 | Homo Sapiens | - | Cmp2105; Na | - | 2.1 | 2019-09-04 | doi.org/10.1016/j.cell.2019.07.028 |
|
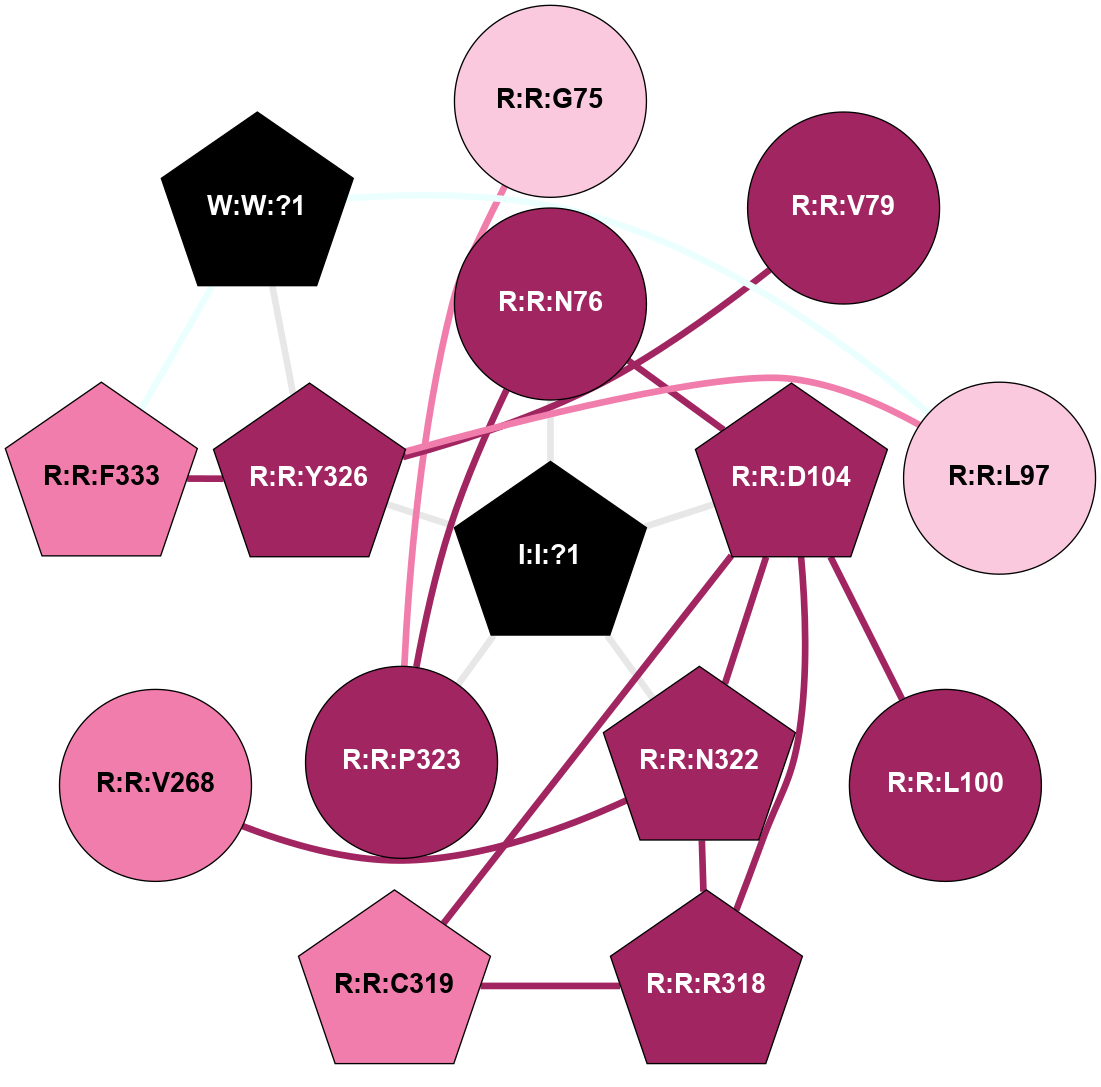
A 2D representation of the interactions of NA in 6QZH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:N76 | 8.15 | 1 | Yes | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:D104 | 8.06 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:N322 | 8.15 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:P323 | 6.5 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:Y326 | 4.64 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:G75 | R:R:P323 | 4.06 | 0 | No | Yes | 7 | 9 | 2 | 1 | | R:R:D104 | R:R:N76 | 10.77 | 1 | Yes | No | 9 | 9 | 1 | 1 | | R:R:N76 | R:R:P323 | 9.77 | 1 | No | Yes | 9 | 9 | 1 | 1 | | R:R:P323 | R:R:V79 | 3.53 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:V79 | R:R:Y326 | 5.05 | 0 | No | Yes | 9 | 9 | 2 | 1 | | R:R:L97 | R:R:Y326 | 11.72 | 1 | No | Yes | 7 | 9 | 2 | 1 | | R:R:L97 | W:W:?1 | 7.91 | 1 | No | Yes | 7 | 0 | 2 | 2 | | R:R:D104 | R:R:L100 | 4.07 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D104 | R:R:R318 | 10.72 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:C319 | R:R:D104 | 10.89 | 1 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:D104 | R:R:N322 | 4.04 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:N322 | R:R:V268 | 10.35 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:C319 | R:R:R318 | 5.57 | 1 | Yes | Yes | 8 | 9 | 2 | 2 | | R:R:N322 | R:R:R318 | 4.82 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:F333 | R:R:Y326 | 4.13 | 1 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:Y326 | W:W:?1 | 6.03 | 1 | Yes | Yes | 9 | 0 | 1 | 2 | | R:R:F333 | W:W:?1 | 4.18 | 1 | Yes | Yes | 8 | 0 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 7.10 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 22.00 | | Average Links Mediated by Hubs In Shell | 22.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6RZ4 | A | Lipid | Leukotriene | CysLT1 | Homo Sapiens | Pranlukast | Na | - | 2.7 | 2019-10-30 | doi.org/10.1126/sciadv.aax2518 |
|
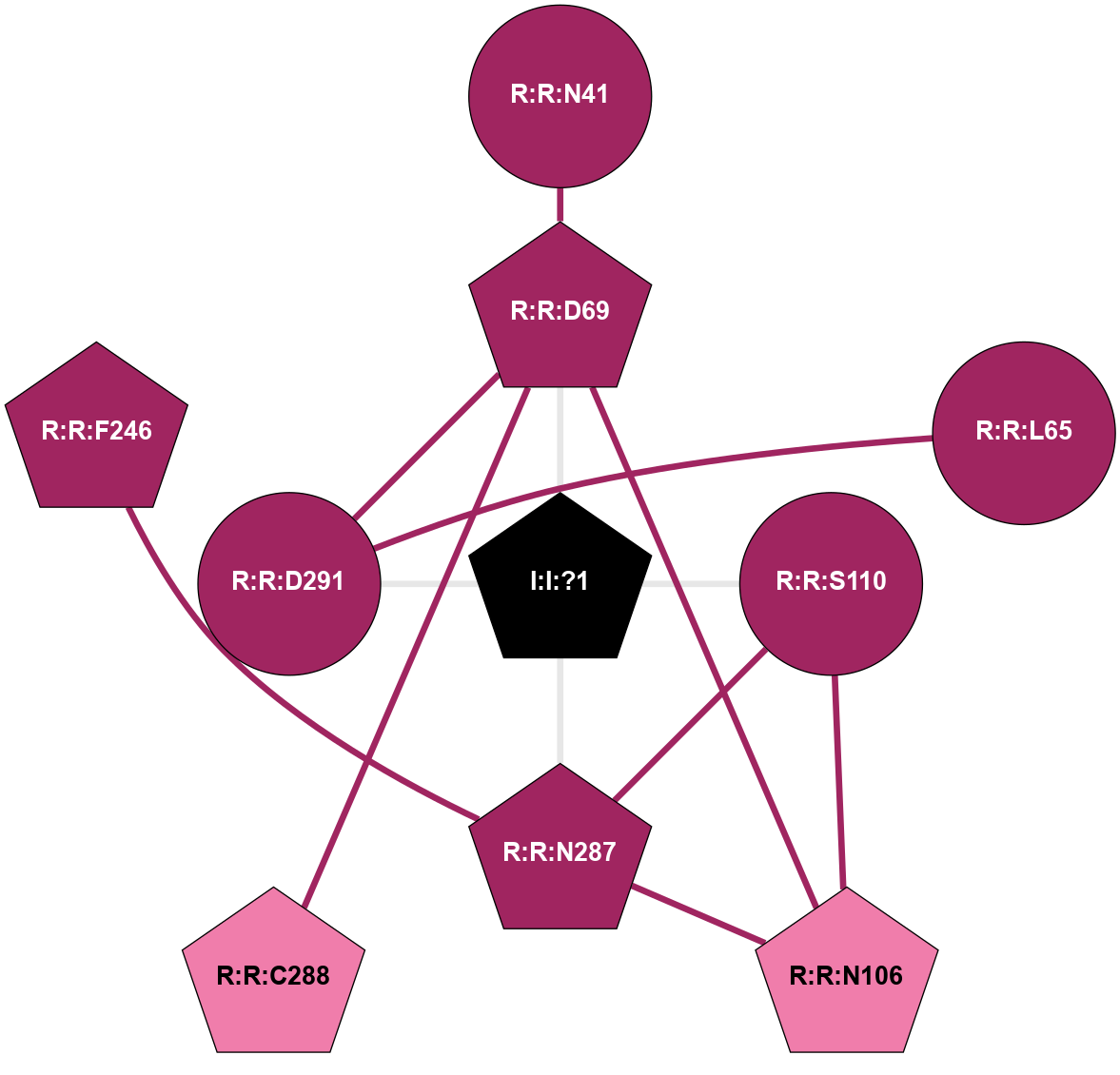
A 2D representation of the interactions of NA in 6RZ4
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D69 | 8.06 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S110 | 5.94 | 2 | Yes | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:N287 | 10.87 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:D291 | 8.06 | 2 | Yes | No | 0 | 9 | 0 | 1 | | R:R:D69 | R:R:N41 | 13.46 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D291 | R:R:L65 | 14.93 | 2 | No | No | 9 | 9 | 1 | 2 | | R:R:D69 | R:R:N106 | 6.73 | 2 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:C288 | R:R:D69 | 6.22 | 0 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:D291 | R:R:D69 | 7.98 | 2 | No | Yes | 9 | 9 | 1 | 1 | | R:R:N106 | R:R:S110 | 4.47 | 2 | Yes | No | 8 | 9 | 2 | 1 | | R:R:N106 | R:R:N287 | 9.54 | 2 | Yes | Yes | 8 | 9 | 2 | 1 | | R:R:N287 | R:R:S110 | 5.96 | 2 | Yes | No | 9 | 9 | 1 | 1 | | R:R:F246 | R:R:N287 | 3.62 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 8.23 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6RZ5 | A | Lipid | Leukotriene | CysLT1 | Homo Sapiens | Zafirlukast | Na | - | 2.53 | 2019-10-30 | doi.org/10.1126/sciadv.aax2518 |
|
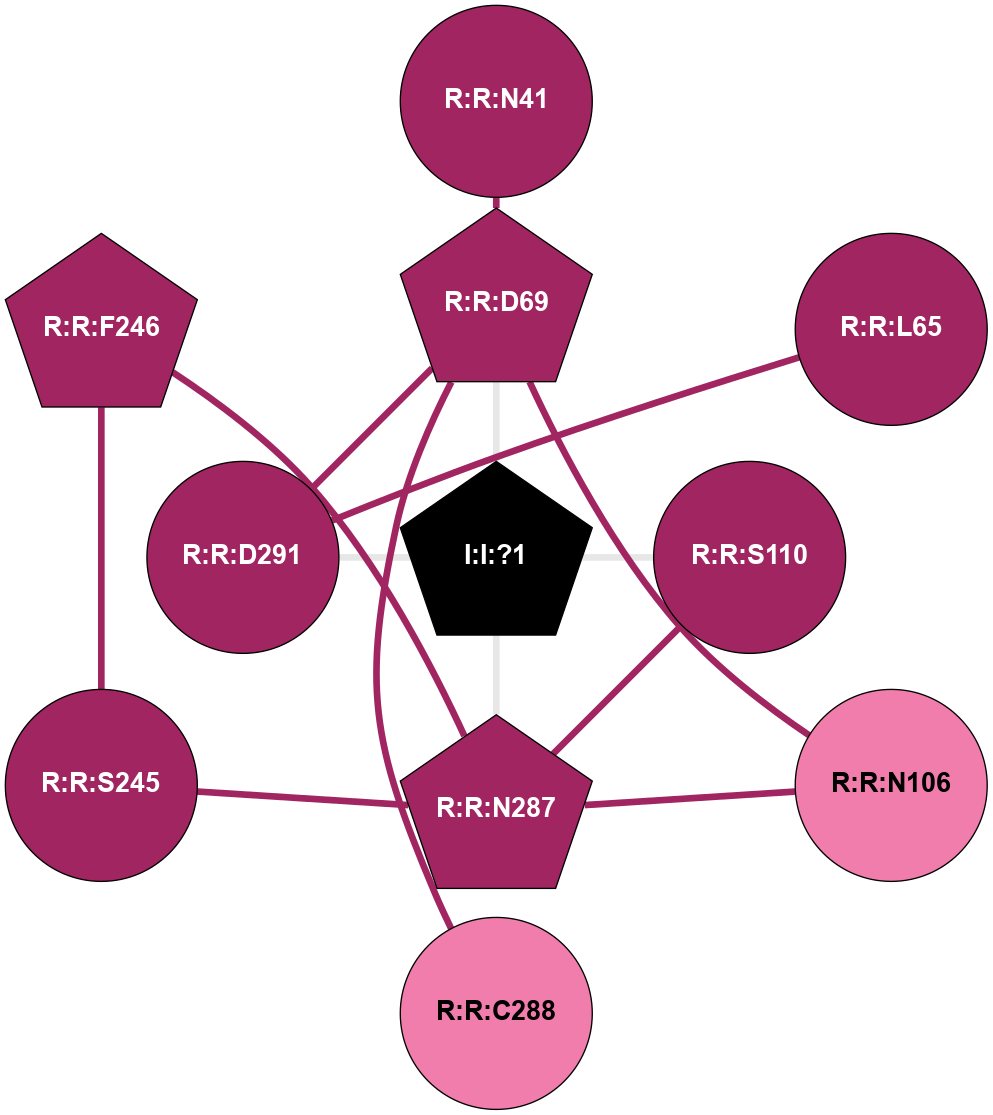
A 2D representation of the interactions of NA in 6RZ5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D69 | 8.06 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S110 | 5.94 | 1 | Yes | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:N287 | 8.15 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:D291 | 8.06 | 1 | Yes | No | 0 | 9 | 0 | 1 | | R:R:D69 | R:R:N41 | 13.46 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D291 | R:R:L65 | 12.21 | 1 | No | No | 9 | 9 | 1 | 2 | | R:R:D69 | R:R:N106 | 5.39 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:C288 | R:R:D69 | 6.22 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:D291 | R:R:D69 | 11.98 | 1 | No | Yes | 9 | 9 | 1 | 1 | | R:R:N106 | R:R:N287 | 12.26 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:N287 | R:R:S110 | 4.47 | 1 | Yes | No | 9 | 9 | 1 | 1 | | R:R:F246 | R:R:S245 | 3.96 | 1 | Yes | No | 9 | 9 | 2 | 2 | | R:R:N287 | R:R:S245 | 4.47 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:F246 | R:R:N287 | 3.62 | 1 | Yes | Yes | 9 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 7.55 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6TO7 | A | Peptide | Orexin | OX1 | Homo Sapiens | Suvorexant | Na | - | 2.26 | 2020-01-01 | doi.org/10.1021/acs.jmedchem.9b01787 |
|
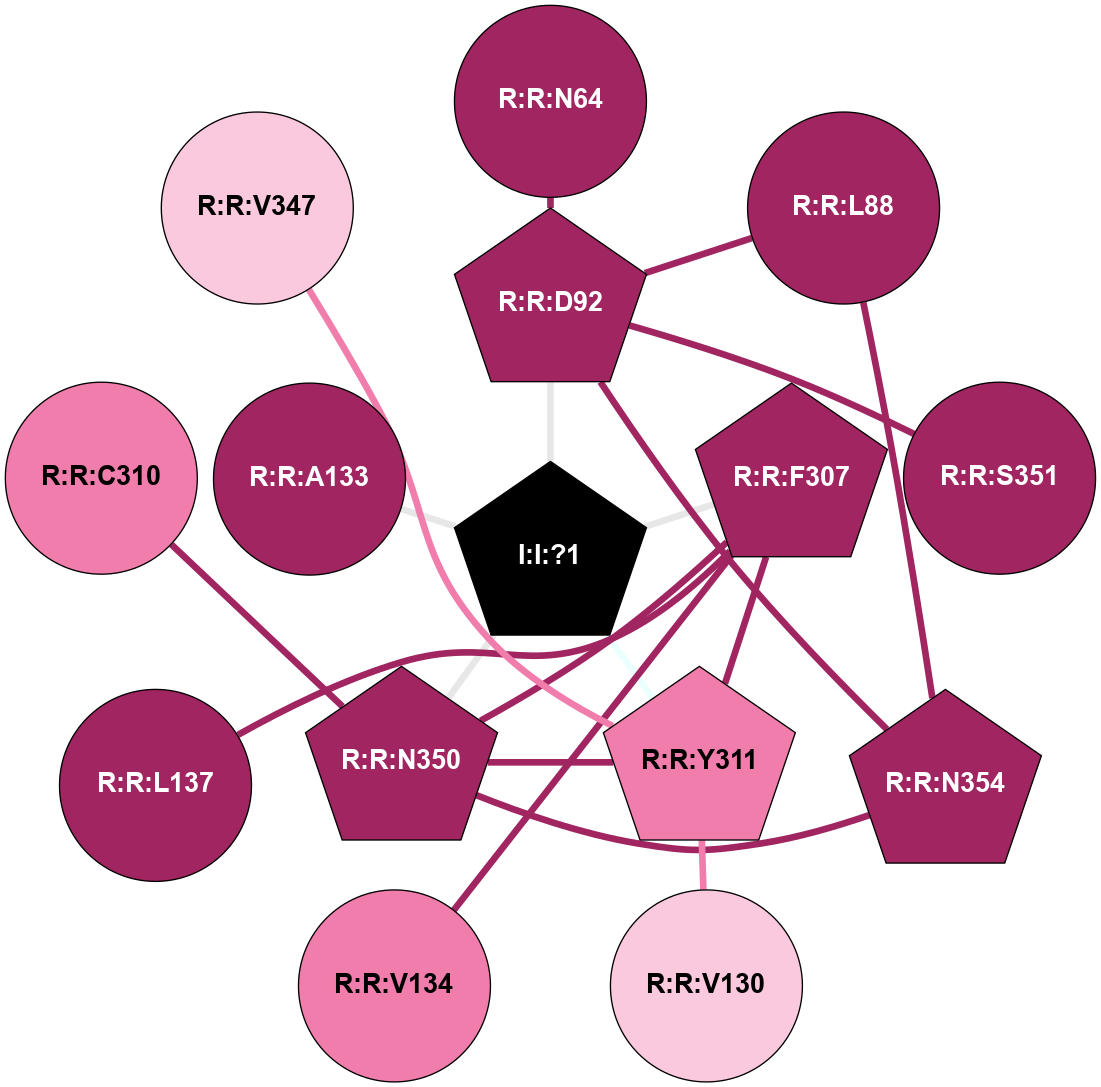
A 2D representation of the interactions of NA in 6TO7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D92 | R:R:N64 | 4.04 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D92 | R:R:L88 | 5.43 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:L88 | R:R:N354 | 5.49 | 2 | No | Yes | 9 | 9 | 2 | 2 | | R:R:D92 | R:R:S351 | 10.31 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D92 | R:R:N354 | 4.04 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D92 | 8.06 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:V130 | R:R:Y311 | 6.31 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:F307 | R:R:V134 | 5.24 | 2 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F307 | R:R:L137 | 6.09 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:F307 | R:R:Y311 | 17.54 | 2 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:F307 | R:R:N350 | 6.04 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:F307 | 4.82 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:C310 | R:R:N350 | 9.45 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:V347 | R:R:Y311 | 10.09 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N350 | R:R:Y311 | 3.49 | 2 | Yes | Yes | 9 | 8 | 1 | 1 | | I:I:?1 | R:R:Y311 | 6.96 | 2 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:N350 | R:R:N354 | 5.45 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:N350 | 8.15 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:A133 | 3.12 | 2 | Yes | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 6.22 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 19.00 | | Average Links Mediated by Hubs In Shell | 19.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6TOD | A | Peptide | Orexin | OX1 | Homo Sapiens | EMPA | Na | - | 2.11 | 2020-01-01 | doi.org/10.1021/acs.jmedchem.9b01787 |
|
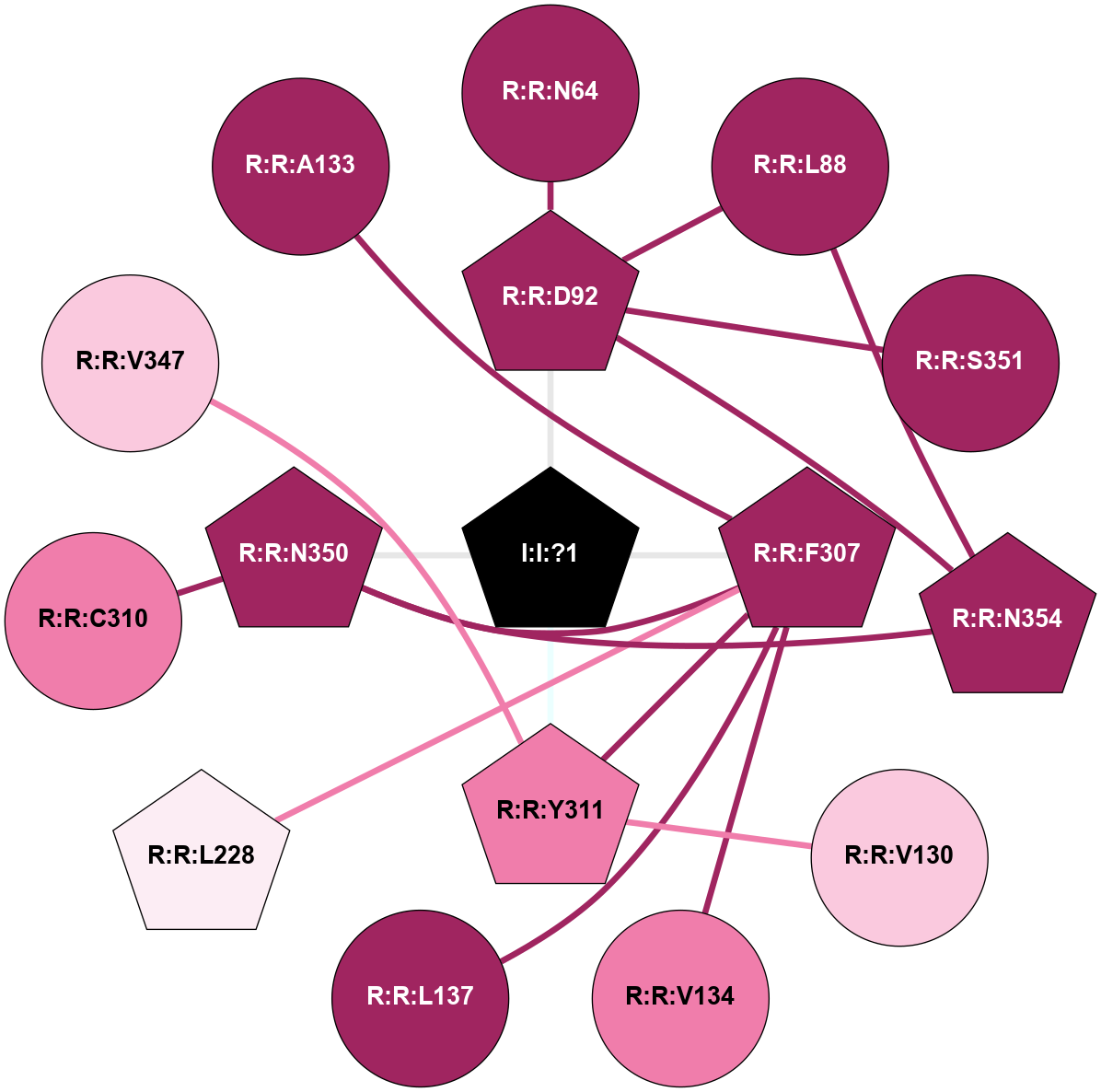
A 2D representation of the interactions of NA in 6TOD
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D92 | R:R:N64 | 4.04 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D92 | R:R:L88 | 5.43 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:L88 | R:R:N354 | 4.12 | 2 | No | Yes | 9 | 9 | 2 | 2 | | R:R:D92 | R:R:S351 | 11.78 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D92 | R:R:N354 | 4.04 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D92 | 8.06 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:V130 | R:R:Y311 | 6.31 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:F307 | R:R:V134 | 3.93 | 2 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F307 | R:R:L137 | 6.09 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:F307 | R:R:L228 | 3.65 | 2 | Yes | Yes | 9 | 6 | 1 | 2 | | R:R:F307 | R:R:Y311 | 22.69 | 2 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:F307 | R:R:N350 | 4.83 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:F307 | 4.82 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:C310 | R:R:N350 | 9.45 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:V347 | R:R:Y311 | 10.09 | 0 | No | Yes | 7 | 8 | 2 | 1 | | I:I:?1 | R:R:Y311 | 6.96 | 2 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:N350 | R:R:N354 | 6.81 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:N350 | 8.15 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:A133 | R:R:F307 | 2.77 | 0 | No | Yes | 9 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 19.00 | | Average Links Mediated by Hubs In Shell | 19.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6TOS | A | Peptide | Orexin | OX1 | Homo Sapiens | GSK1059865 | Na | - | 2.13 | 2020-01-15 | doi.org/10.1021/acs.jmedchem.9b01787 |
|

A 2D representation of the interactions of NA in 6TOS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D92 | R:R:N64 | 4.04 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D92 | R:R:L88 | 5.43 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:L88 | R:R:N354 | 4.12 | 2 | No | Yes | 9 | 9 | 2 | 2 | | R:R:D92 | R:R:S351 | 10.31 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D92 | R:R:N354 | 4.04 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D92 | 8.06 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:V130 | R:R:Y311 | 5.05 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:F307 | R:R:V134 | 5.24 | 2 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F307 | R:R:L137 | 6.09 | 2 | Yes | No | 9 | 9 | 1 | 2 | | R:R:F307 | R:R:Y311 | 22.69 | 2 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:F307 | R:R:N350 | 4.83 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:F307 | 4.82 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:C310 | R:R:N350 | 9.45 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:V347 | R:R:Y311 | 8.83 | 0 | No | Yes | 7 | 8 | 2 | 1 | | I:I:?1 | R:R:Y311 | 6.96 | 2 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:N350 | R:R:N354 | 8.17 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:N350 | 8.15 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:A133 | 3.12 | 2 | Yes | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 6.22 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 18.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6TQ4 | A | Peptide | Orexin | OX1 | Homo Sapiens | PubChem 16046844 | Na | - | 2.3 | 2020-01-01 | doi.org/10.1021/acs.jmedchem.9b01787 |
|
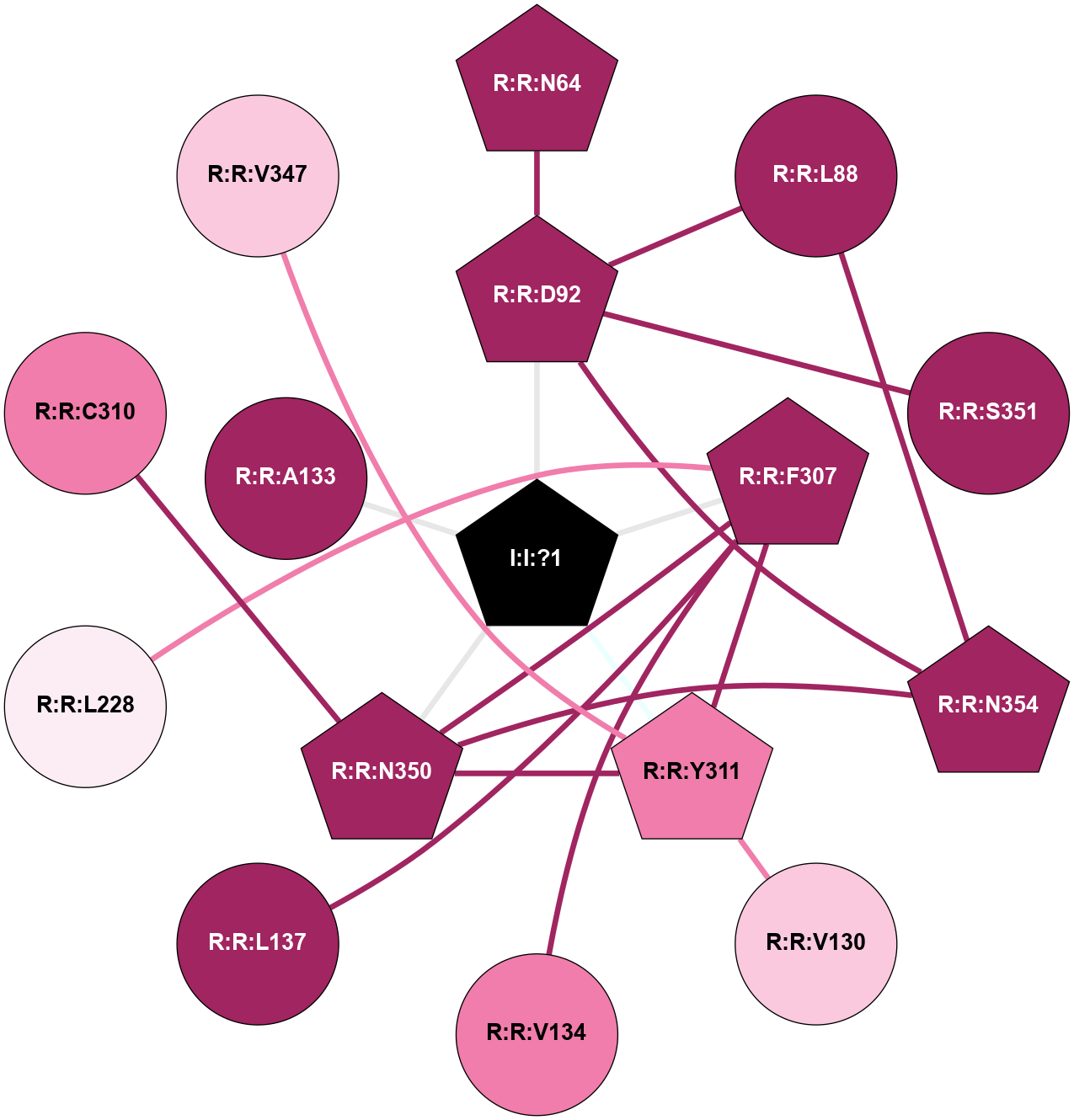
A 2D representation of the interactions of NA in 6TQ4
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D92 | R:R:N64 | 6.73 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D92 | R:R:L88 | 5.43 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:L88 | R:R:N354 | 5.49 | 1 | No | Yes | 9 | 9 | 2 | 2 | | R:R:D92 | R:R:S351 | 11.78 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D92 | R:R:N354 | 5.39 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D92 | 8.06 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:V130 | R:R:Y311 | 6.31 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:F307 | R:R:V134 | 3.93 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F307 | R:R:L137 | 7.31 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:F307 | R:R:L228 | 3.65 | 1 | Yes | No | 9 | 6 | 1 | 2 | | R:R:F307 | R:R:Y311 | 22.69 | 1 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:F307 | R:R:N350 | 7.25 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:F307 | 4.82 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:C310 | R:R:N350 | 9.45 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:V347 | R:R:Y311 | 10.09 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:N350 | R:R:Y311 | 3.49 | 1 | Yes | Yes | 9 | 8 | 1 | 1 | | I:I:?1 | R:R:Y311 | 6.96 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:N350 | R:R:N354 | 10.9 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:N350 | 8.15 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:A133 | 3.12 | 1 | Yes | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 6.22 | | Average Nodes In Shell | 16.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 20.00 | | Average Links Mediated by Hubs In Shell | 20.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6WQA | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2 | 2020-11-18 | doi.org/10.1107/S2052252520012701 |
|
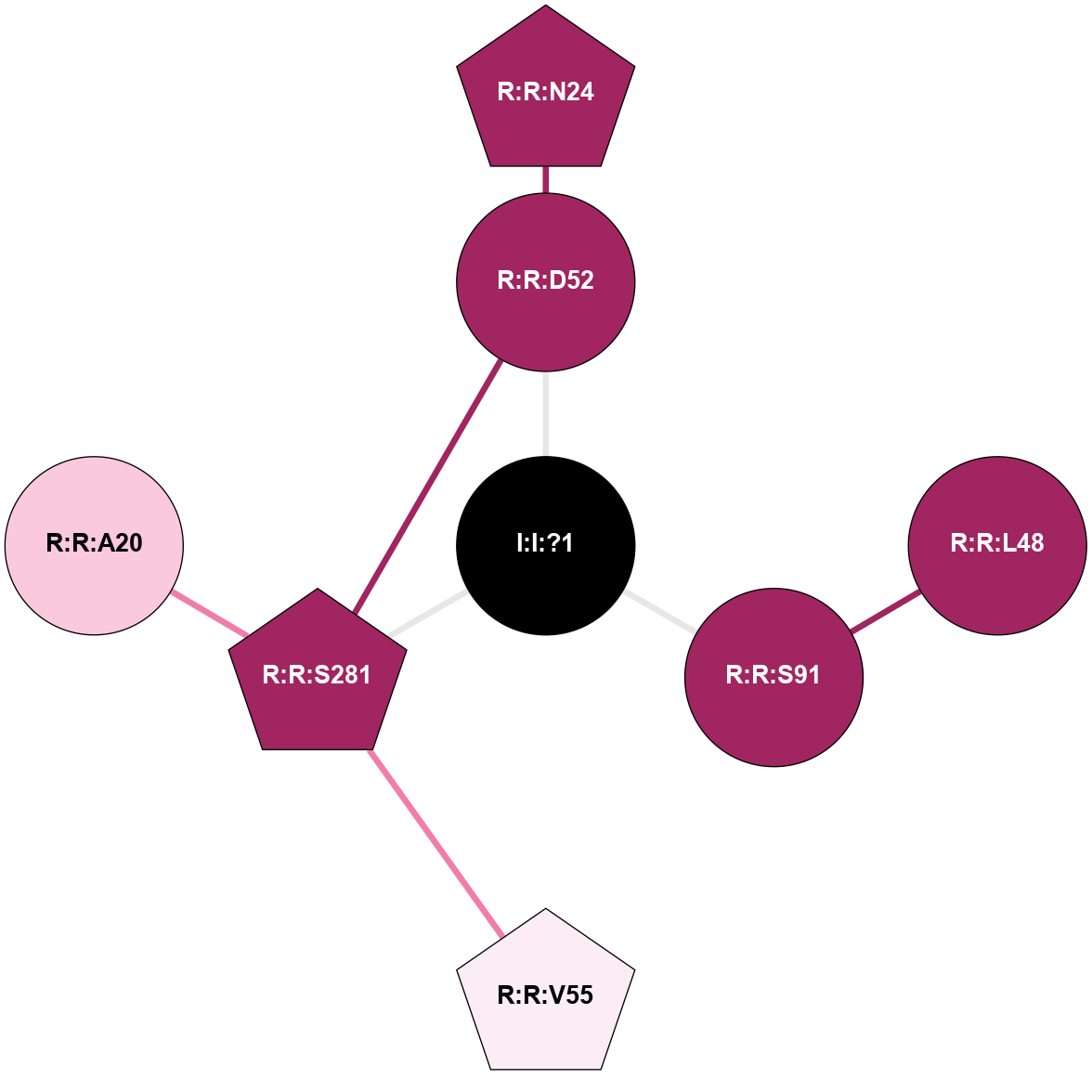
A 2D representation of the interactions of NA in 6WQA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.06 | 8 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 8 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S281 | 2.97 | 8 | No | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 8.08 | 8 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 8.83 | 8 | No | Yes | 9 | 9 | 1 | 1 | | R:R:S281 | R:R:V55 | 8.08 | 8 | Yes | Yes | 9 | 6 | 1 | 2 | | R:R:A20 | R:R:S281 | 1.71 | 0 | No | Yes | 7 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.66 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6ZDR | A | Nucleotide | Adenosine | A2A | Homo Sapiens | HMS1413f12 | Na | - | 1.92 | 2020-09-16 | doi.org/10.1002/anie.202003788 |
|

A 2D representation of the interactions of NA in 6ZDR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 8.08 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 0 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6ZDV | A | Nucleotide | Adenosine | A2A | Homo Sapiens | Chromone 5d | Na | - | 2.13 | 2020-09-16 | doi.org/10.1002/anie.202003788 |
|
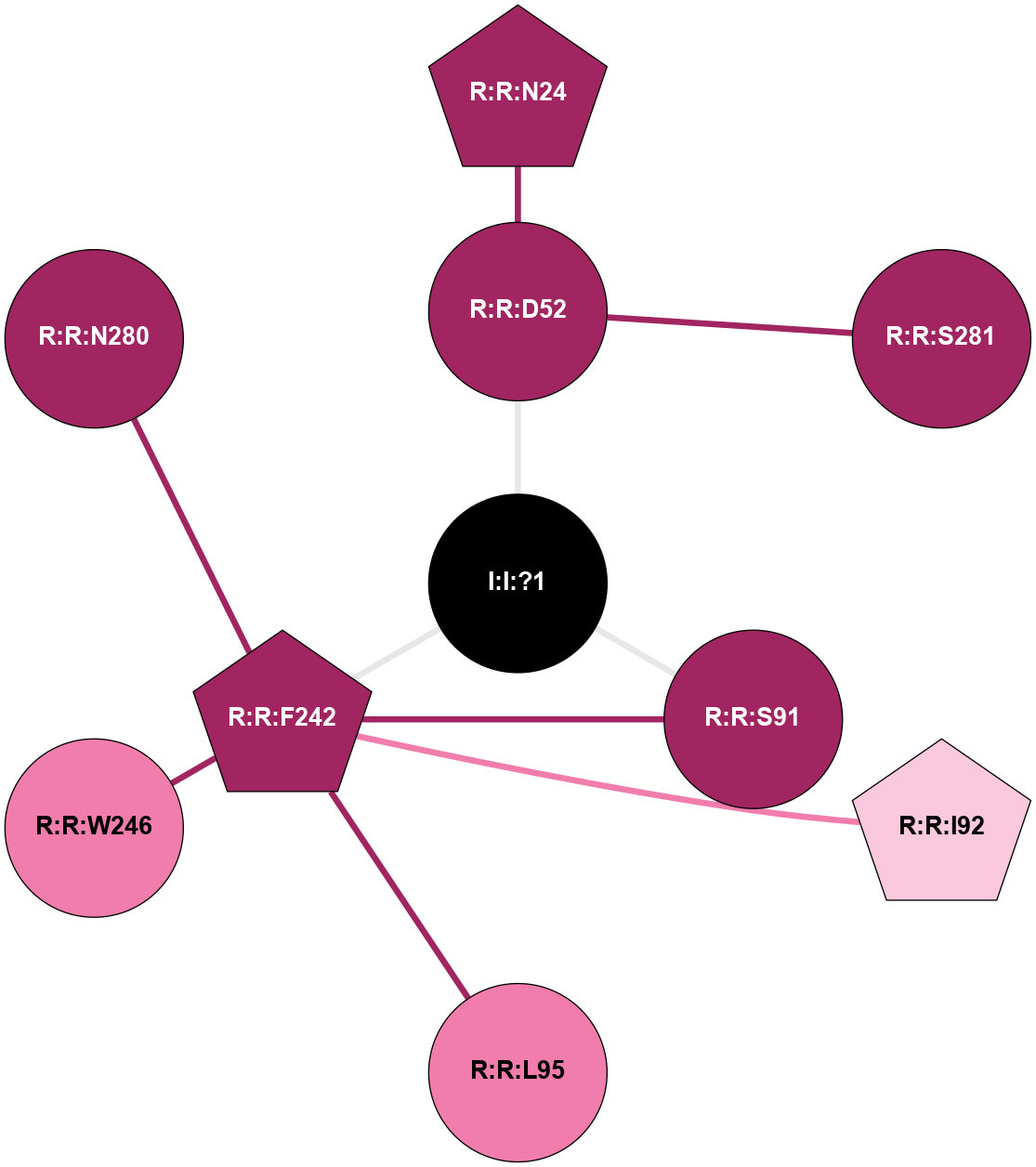
A 2D representation of the interactions of NA in 6ZDV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 9.42 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 6 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 3.96 | 6 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 6 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 6 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 17.04 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 6 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 6 | No | Yes | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7ARO | A | Nucleotide | Adenosine | A2A | Homo Sapiens | LUF5833 | Na | - | 3.12 | 2021-04-07 | doi.org/10.1021/acs.jmedchem.0c01856 |
|
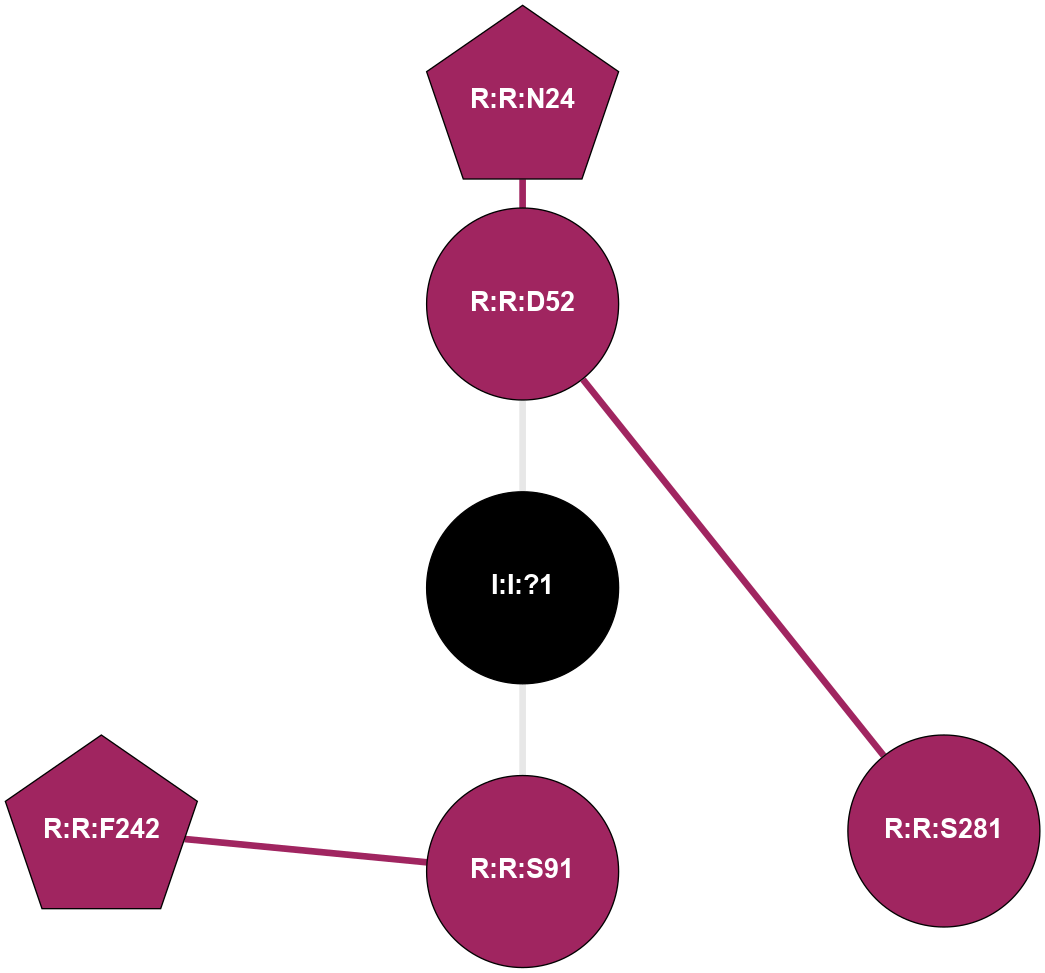
A 2D representation of the interactions of NA in 7ARO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 10.77 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.06 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 7 | Yes | No | 9 | 9 | 2 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7K15 | A | Lipid | Leukotriene | BLT1 | Homo Sapiens | MKD046 | Na | - | 2.88 | 2021-02-17 | doi.org/10.1038/s41467-021-23149-1 |
|
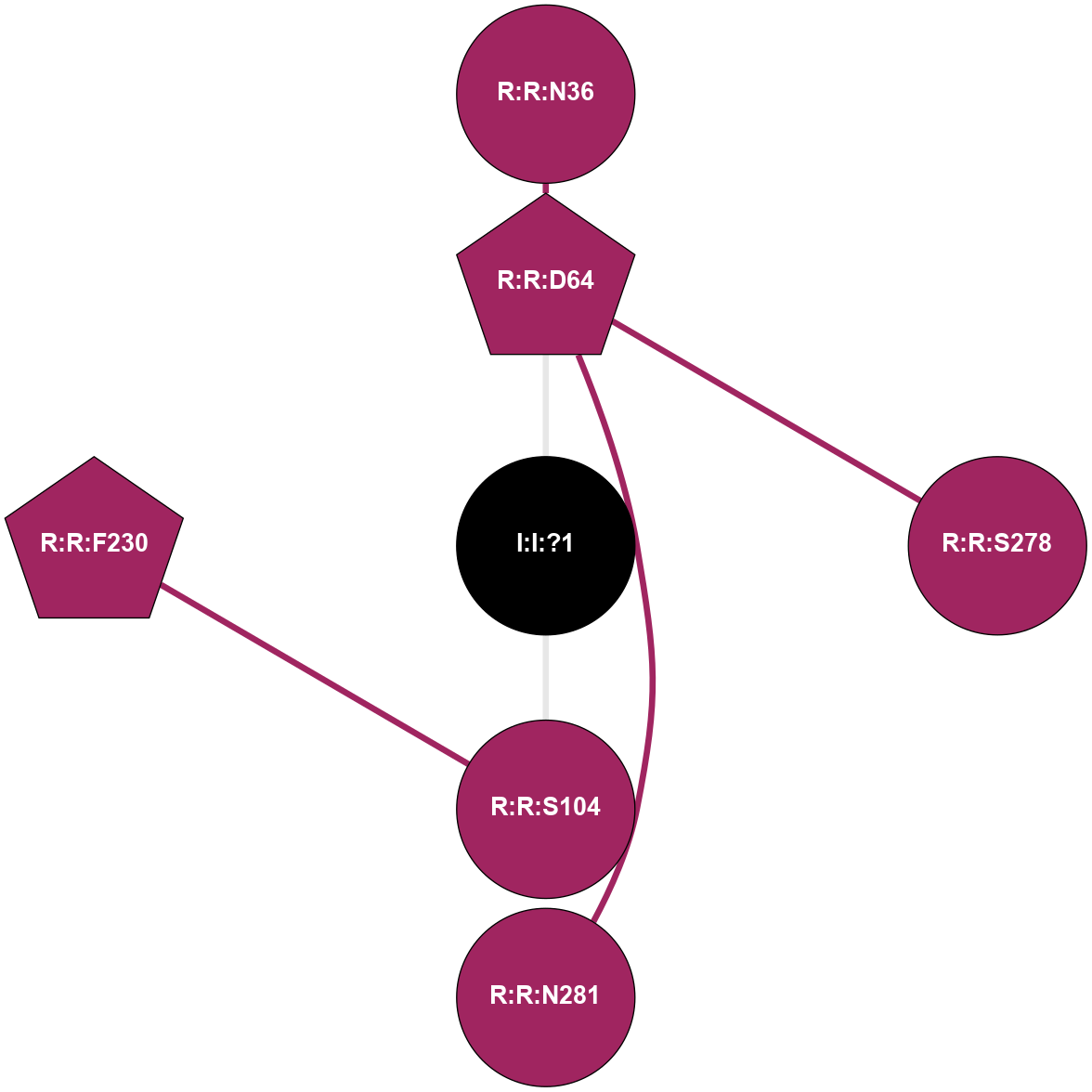
A 2D representation of the interactions of NA in 7K15
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D64 | 8.05 | 0 | No | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S104 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:D64 | R:R:N36 | 6.73 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:L60 | R:R:S104 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D64 | R:R:S278 | 8.83 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D64 | R:R:N281 | 6.73 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:F230 | R:R:S104 | 3.96 | 1 | Yes | No | 9 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7M8W | A | Lipid | Prostanoid | DP2 | Homo Sapiens | 15r-Methyl-Prostaglandin D2 | Na | - | 2.61 | 2021-08-25 | doi.org/10.1073/pnas.2102813118 |
|
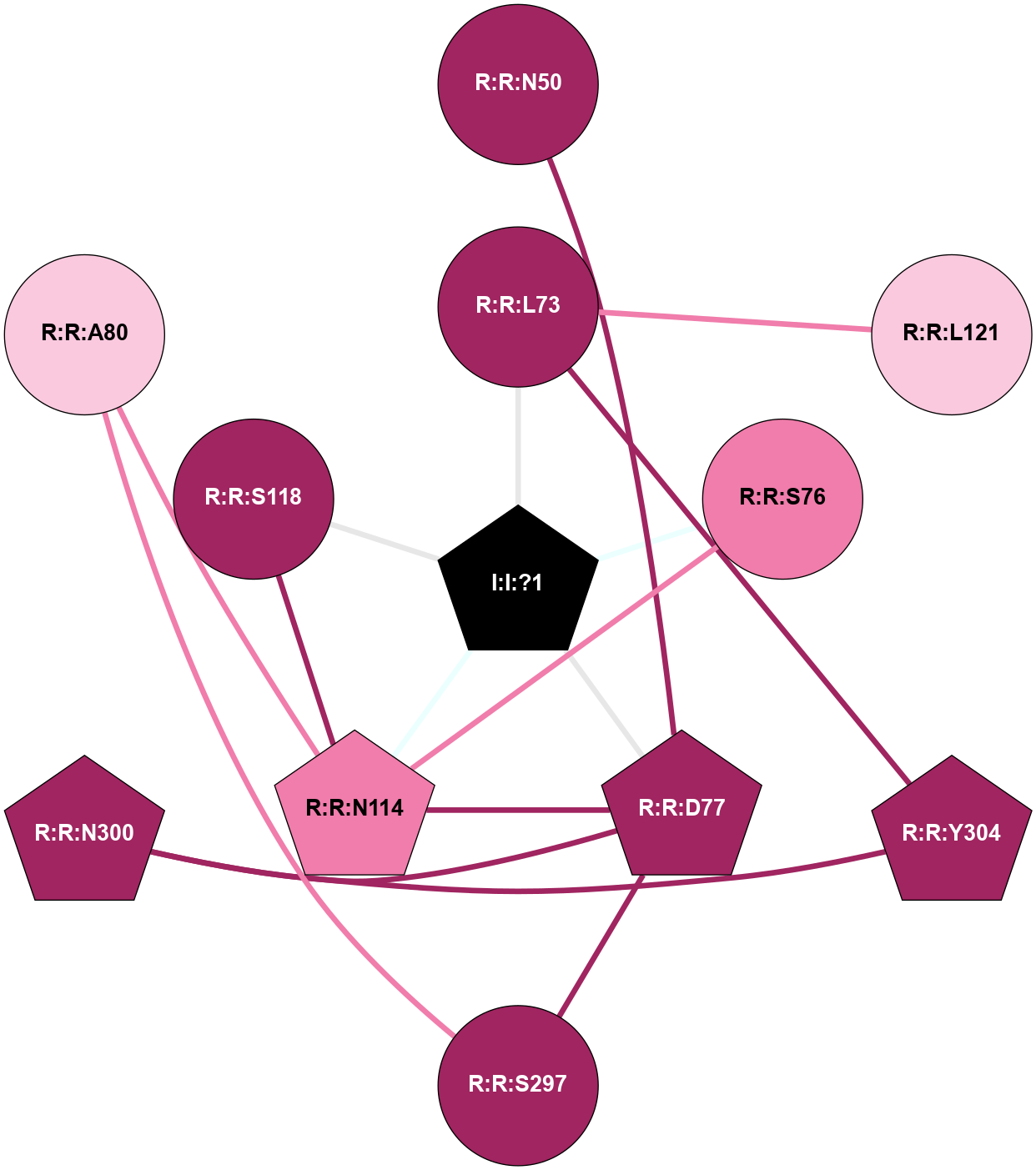
A 2D representation of the interactions of NA in 7M8W
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:L73 | 5.48 | 4 | Yes | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S76 | 5.94 | 4 | Yes | No | 0 | 8 | 0 | 1 | | I:I:?1 | R:R:D77 | 10.74 | 4 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:N114 | 8.15 | 4 | Yes | Yes | 0 | 8 | 0 | 1 | | I:I:?1 | R:R:S118 | 5.94 | 4 | Yes | No | 0 | 9 | 0 | 1 | | R:R:D77 | R:R:N50 | 6.73 | 4 | Yes | No | 9 | 9 | 1 | 2 | | R:R:L121 | R:R:L73 | 4.15 | 0 | No | No | 7 | 9 | 2 | 1 | | R:R:L73 | R:R:Y304 | 4.69 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:N114 | R:R:S76 | 7.45 | 4 | Yes | No | 8 | 8 | 1 | 1 | | R:R:D77 | R:R:N114 | 4.04 | 4 | Yes | Yes | 9 | 8 | 1 | 1 | | R:R:D77 | R:R:S297 | 8.83 | 4 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D77 | R:R:N300 | 8.08 | 4 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:A80 | R:R:N114 | 4.69 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:A80 | R:R:S297 | 3.42 | 0 | No | No | 7 | 9 | 2 | 2 | | R:R:N114 | R:R:S118 | 5.96 | 4 | Yes | No | 8 | 9 | 1 | 1 | | R:R:N300 | R:R:Y304 | 8.14 | 0 | Yes | Yes | 9 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 7.25 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7RM5 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2.79 | 2021-09-08 | doi.org/10.1073/pnas.2106041118 |
|
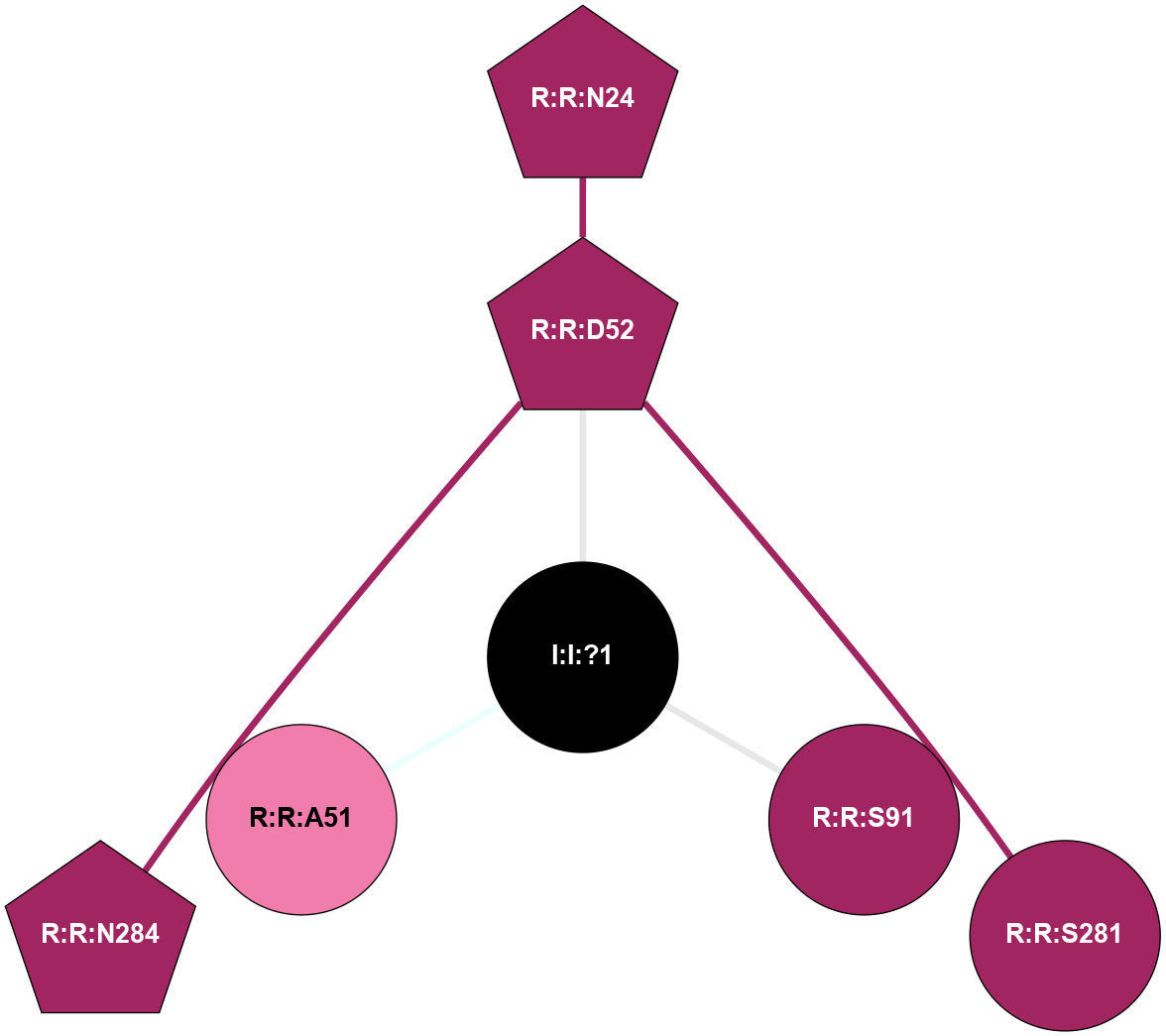
A 2D representation of the interactions of NA in 7RM5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.06 | 0 | No | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 6.73 | 0 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 8.83 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:N284 | 5.39 | 0 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:A51 | 3.12 | 0 | No | No | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.71 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7UL2 | A | Peptide | Neurotensin | NTS1 | Homo Sapiens | SR48692 | Na | - | 2.4 | 2022-06-29 | doi.org/10.1038/s41594-022-00859-8 |
|
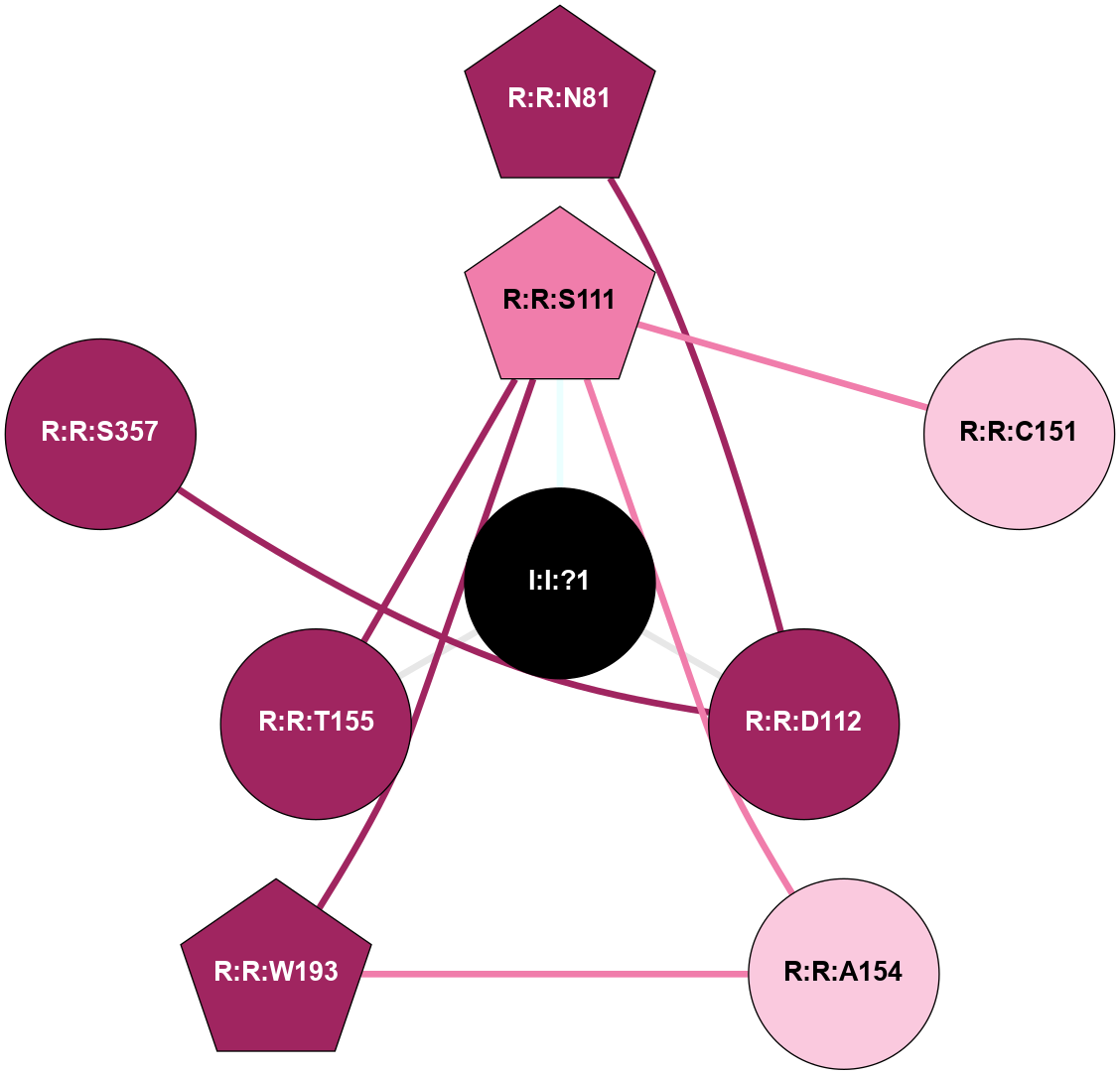
A 2D representation of the interactions of NA in 7UL2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D112 | R:R:N81 | 12.12 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:C151 | R:R:S111 | 3.44 | 0 | No | Yes | 7 | 8 | 2 | 1 | | R:R:A154 | R:R:S111 | 3.42 | 4 | No | Yes | 7 | 8 | 2 | 1 | | R:R:S111 | R:R:T155 | 4.8 | 4 | Yes | No | 8 | 9 | 1 | 1 | | R:R:S111 | R:R:W193 | 3.71 | 4 | Yes | Yes | 8 | 9 | 1 | 2 | | I:I:?1 | R:R:S111 | 5.94 | 4 | No | Yes | 0 | 8 | 0 | 1 | | R:R:D112 | R:R:S357 | 7.36 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D112 | 10.74 | 4 | No | No | 0 | 9 | 0 | 1 | | R:R:A154 | R:R:W193 | 3.89 | 4 | No | Yes | 7 | 9 | 2 | 2 | | I:I:?1 | R:R:T155 | 8.75 | 4 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 8.48 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7XK9 | A | Amine | Adrenergic | Beta2 | Homo Sapiens | Constrained Isoproterenol | Na | - | 3.4 | 2023-04-26 | doi.org/10.1038/s41467-023-37808-y |
|

A 2D representation of the interactions of NA in 7XK9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:N103 | 8.15 | 0 | No | No | 0 | 2 | 0 | 1 | | R:R:C106 | R:R:W99 | 5.22 | 1 | No | Yes | 9 | 7 | 1 | 2 | | R:R:C191 | R:R:W99 | 13.06 | 1 | No | Yes | 9 | 7 | 2 | 2 | | R:R:E188 | R:R:N103 | 13.15 | 0 | No | No | 3 | 2 | 2 | 1 | | R:R:C106 | R:R:C191 | 7.28 | 1 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:C106 | 3.14 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 5.65 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8A2O | A | Nucleotide | Adenosine | A2A | Homo Sapiens | Theophylline | Na | - | 3.45 | 2023-08-30 | doi.org/10.1107/S1600576723006428 |
|
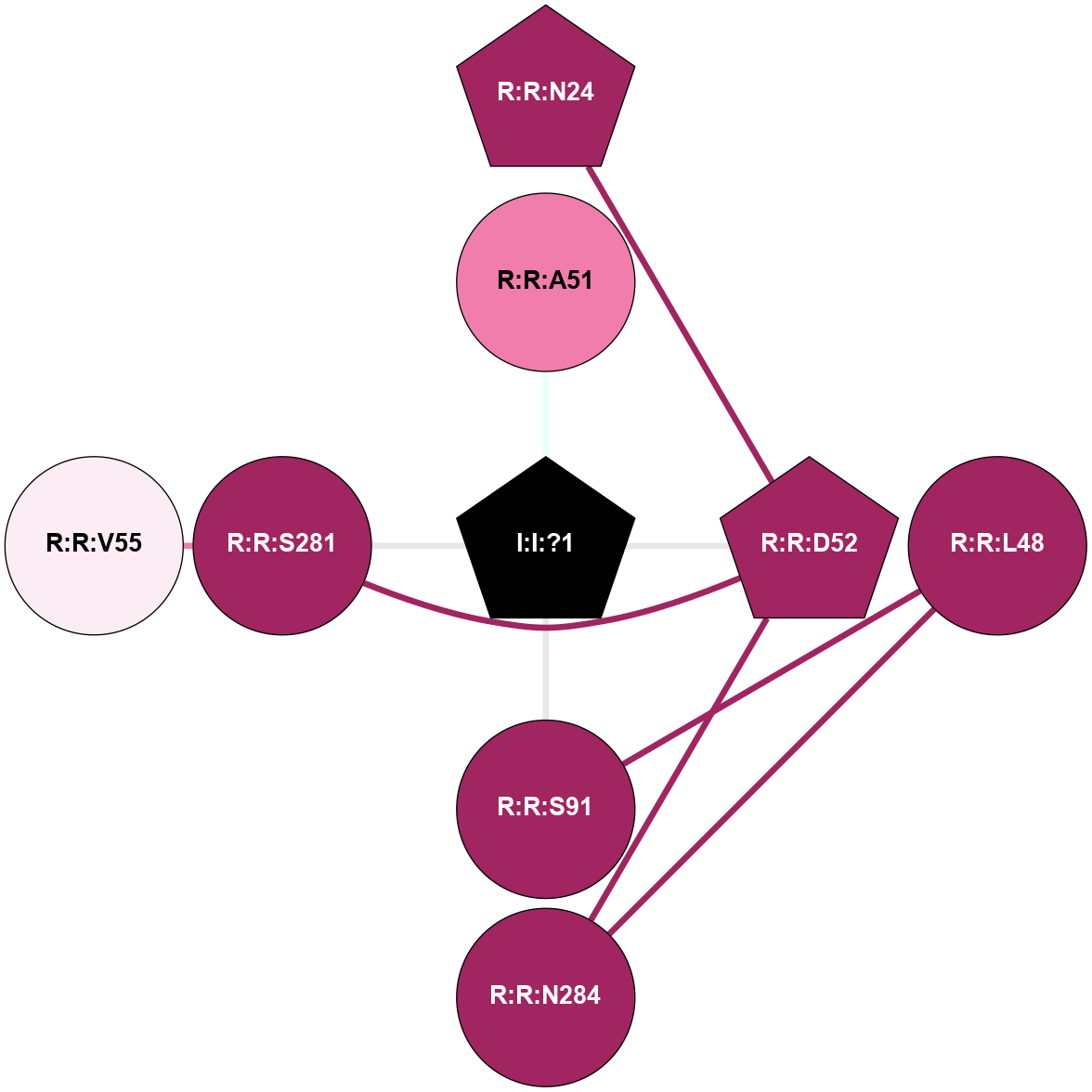
A 2D representation of the interactions of NA in 8A2O
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 13.46 | 9 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:L48 | R:R:N284 | 4.12 | 0 | No | No | 9 | 9 | 2 | 2 | | I:I:?1 | R:R:A51 | 3.12 | 9 | Yes | No | 0 | 8 | 0 | 1 | | R:R:D52 | R:R:S281 | 8.83 | 9 | Yes | No | 9 | 9 | 1 | 1 | | R:R:D52 | R:R:N284 | 6.73 | 9 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.05 | 9 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:S281 | R:R:V55 | 8.08 | 9 | No | No | 9 | 6 | 1 | 2 | | I:I:?1 | R:R:S91 | 5.94 | 9 | Yes | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S281 | 2.97 | 9 | Yes | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.02 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8A2P | A | Nucleotide | Adenosine | A2A | Homo Sapiens | LUAA47070 | Na | - | 3.5 | 2023-08-30 | doi.org/10.1107/S1600576723006428 |
|
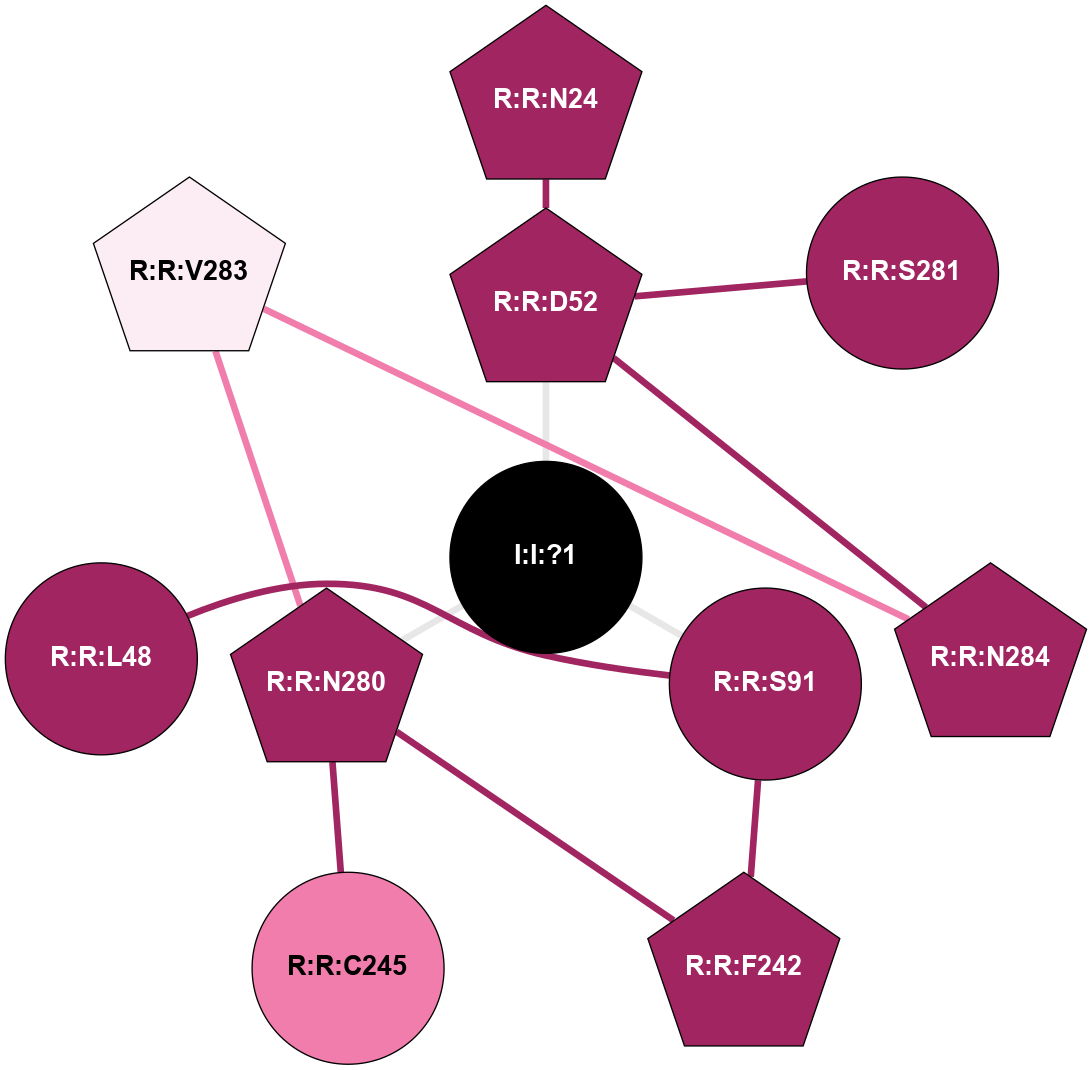
A 2D representation of the interactions of NA in 8A2P
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 13.46 | 0 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 8.83 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:N284 | 6.73 | 0 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.05 | 0 | No | Yes | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 3.96 | 0 | Yes | No | 9 | 9 | 2 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:N280 | 6.04 | 0 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:C245 | R:R:N280 | 4.72 | 0 | No | Yes | 8 | 9 | 2 | 1 | | I:I:?1 | R:R:N280 | 5.43 | 0 | No | Yes | 0 | 9 | 0 | 1 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:N280 | R:R:V283 | 2.96 | 0 | Yes | Yes | 9 | 6 | 1 | 2 | | R:R:N284 | R:R:V283 | 2.96 | 5 | Yes | Yes | 9 | 6 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 6.47 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8CIC | A | Nucleotide | Adenosine | A2A | Homo Sapiens | Etrumadenant | Na | - | 2.1 | 2023-05-31 | doi.org/10.1038/s42004-023-00894-6 |
|
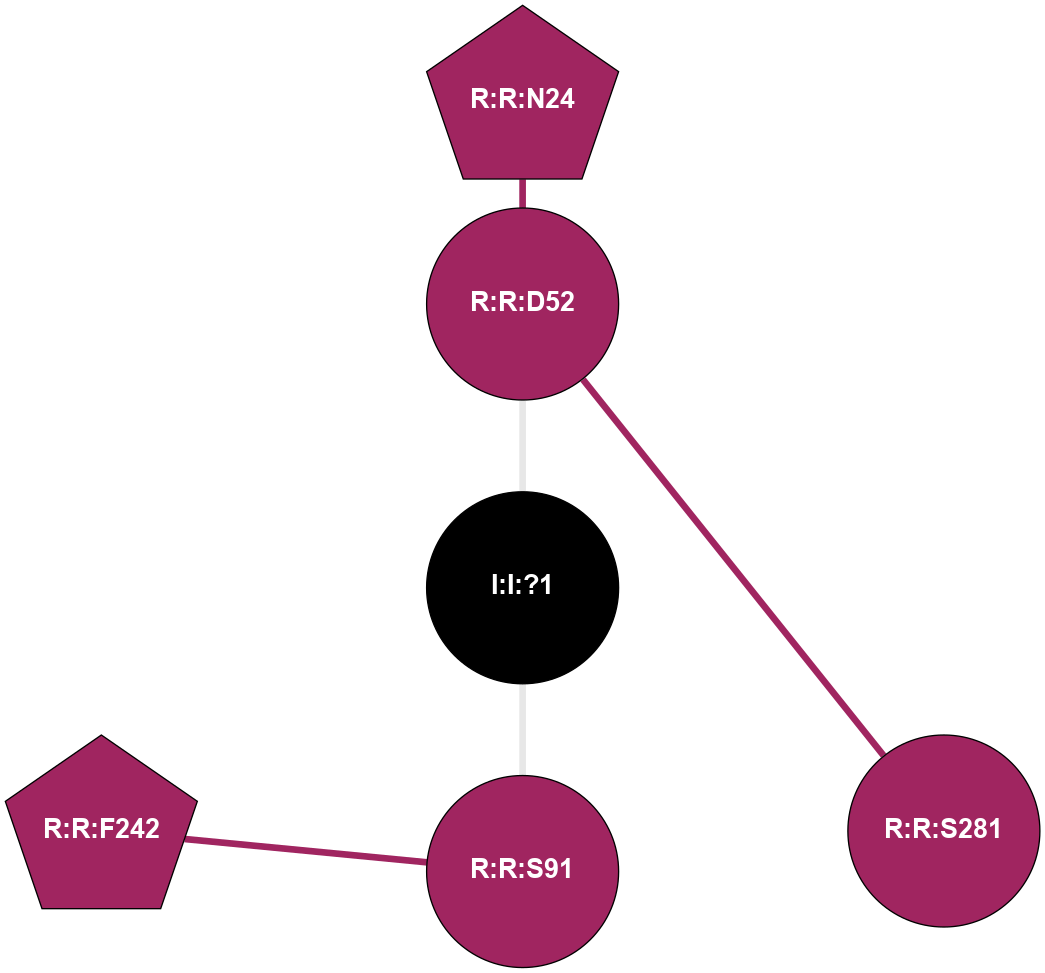
A 2D representation of the interactions of NA in 8CIC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 13.46 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 10.31 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.05 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 1 | Yes | No | 9 | 9 | 2 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8CU7 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | LJ-4517 | Na | - | 2.05 | 2022-08-31 | doi.org/10.1021/acs.jmedchem.2c00462 |
|
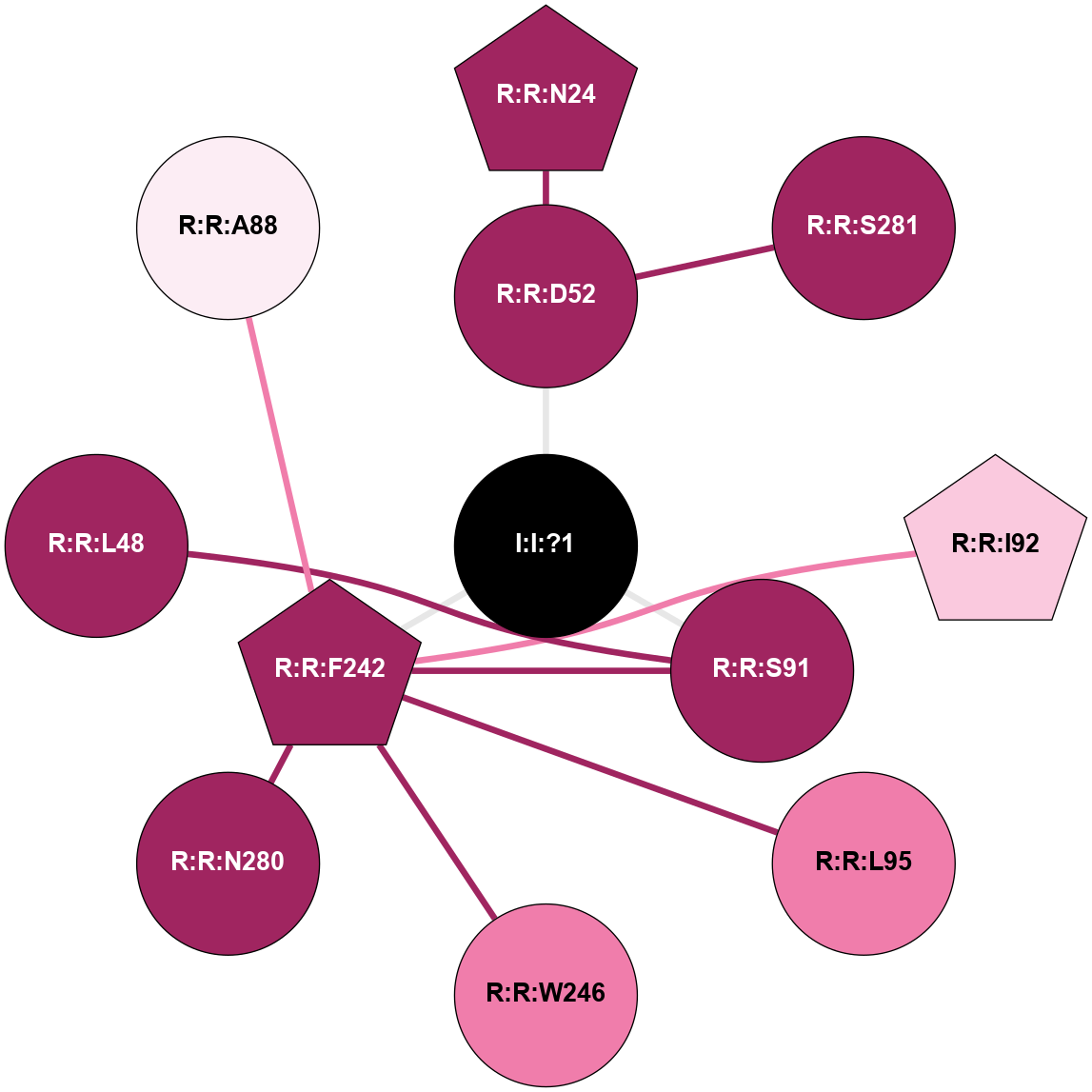
A 2D representation of the interactions of NA in 8CU7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 10.77 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 8.83 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.05 | 6 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 6 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 6 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 6 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 13.03 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 6 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 6 | No | Yes | 0 | 9 | 0 | 1 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:A88 | R:R:F242 | 1.39 | 0 | No | Yes | 6 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8FYN | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2 | 2023-03-08 | doi.org/10.1038/s41467-023-36733-4 |
|
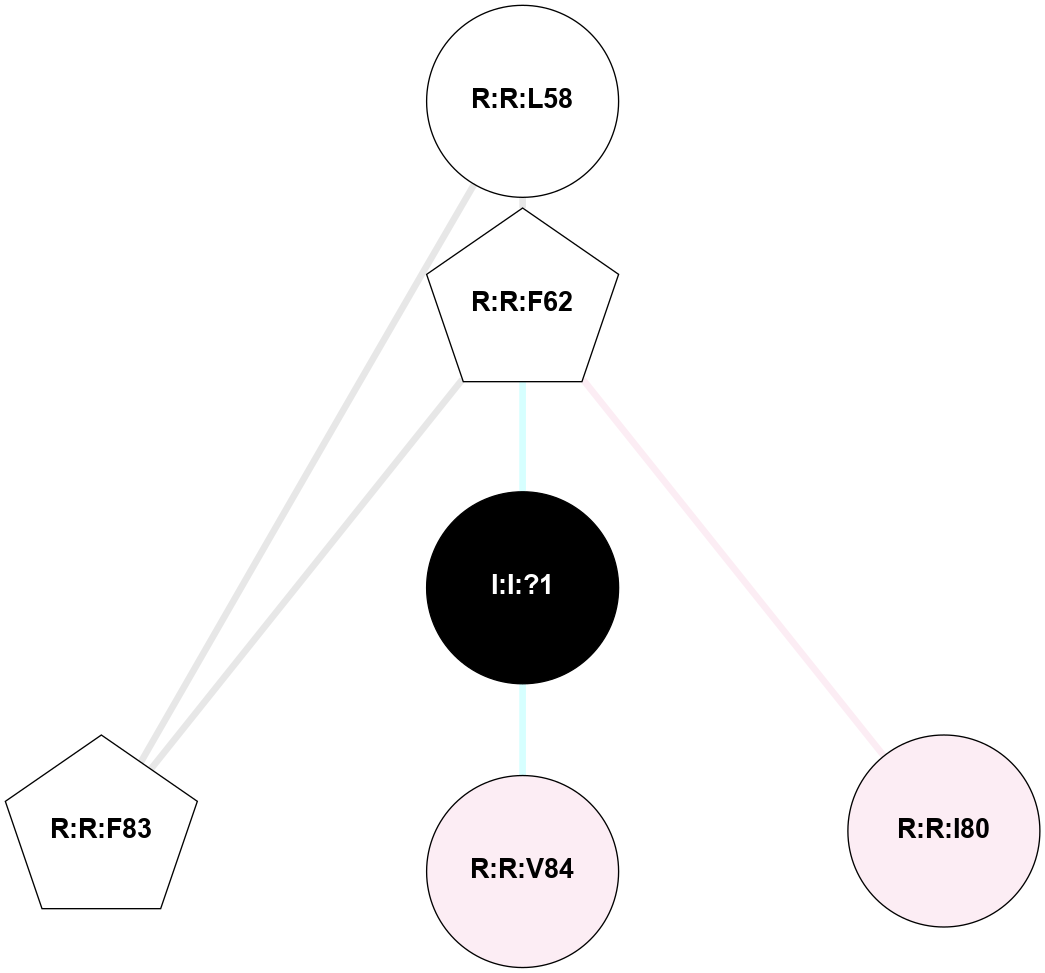
A 2D representation of the interactions of NA in 8FYN
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:F62 | 7.23 | 0 | No | Yes | 0 | 5 | 0 | 1 | | I:I:?1 | R:R:V84 | 5.89 | 0 | No | No | 0 | 6 | 0 | 1 | | R:R:F62 | R:R:L58 | 6.09 | 4 | Yes | No | 5 | 5 | 1 | 2 | | R:R:F83 | R:R:L58 | 8.53 | 4 | Yes | No | 5 | 5 | 2 | 2 | | R:R:F62 | R:R:I80 | 10.05 | 4 | Yes | No | 5 | 6 | 1 | 2 | | R:R:F62 | R:R:F83 | 6.43 | 4 | Yes | Yes | 5 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.56 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8GNE | A | Nucleotide | Adenosine | A2A | Homo Sapiens | KW-6356 | Na | - | 2.3 | 2023-03-22 | doi.org/10.1124/molpharm.122.000633 |
|
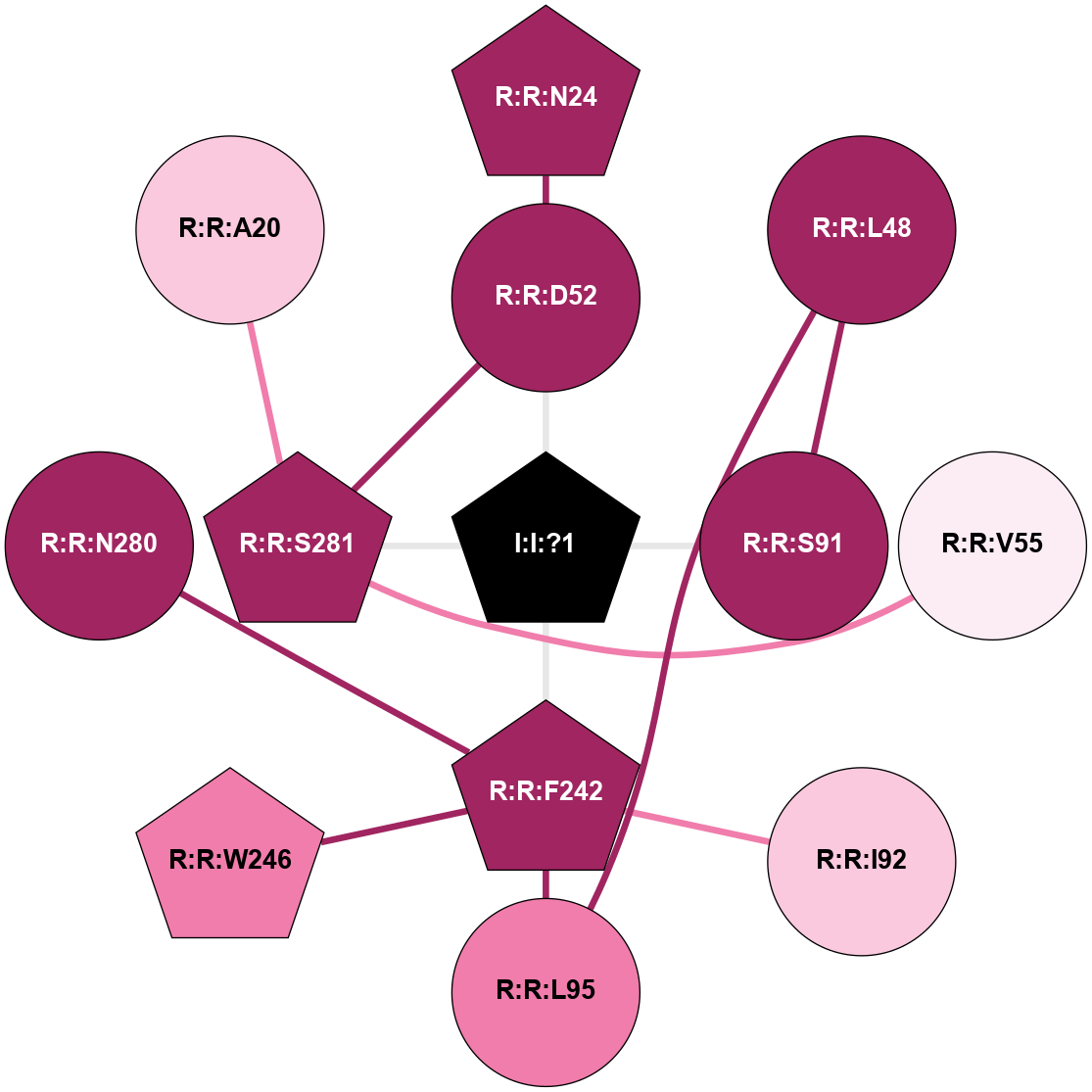
A 2D representation of the interactions of NA in 8GNE
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.05 | 8 | Yes | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 8 | Yes | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:F242 | 4.82 | 8 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S281 | 2.97 | 8 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 9.42 | 8 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:L48 | R:R:L95 | 4.15 | 0 | No | No | 9 | 8 | 2 | 2 | | R:R:D52 | R:R:S281 | 10.31 | 8 | No | Yes | 9 | 9 | 1 | 1 | | R:R:S281 | R:R:V55 | 9.7 | 8 | Yes | No | 9 | 6 | 1 | 2 | | R:R:F242 | R:R:I92 | 6.28 | 0 | Yes | No | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 3.65 | 0 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 11.02 | 0 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 7.25 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:A20 | R:R:S281 | 1.71 | 0 | No | Yes | 7 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 5.45 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8JWZ | A | Nucleotide | Adenosine | A2A | Homo Sapiens | AB928 | Na | - | 2.37 | 2023-08-16 | doi.org/10.1007/s11427-023-2459-8 |
|

A 2D representation of the interactions of NA in 8JWZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.06 | 0 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 12.12 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 10.31 | 0 | No | No | 9 | 9 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8RQQ | A | Nucleotide | Adenosine | A2A | Homo Sapiens | ZM-241385 | Na | - | 2.37 | 2024-04-03 | doi.org/10.1021/acs.cgd.4c00087 |
|

A 2D representation of the interactions of NA in 8RQQ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 13.46 | 0 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 8.83 | 0 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:N284 | 4.04 | 0 | Yes | Yes | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.05 | 0 | No | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8RW0 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | Istradefylline | Na | - | 1.94 | 2025-01-08 | doi.org/10.1038/s41467-024-55109-w |
|

A 2D representation of the interactions of NA in 8RW0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.05 | 0 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 9.42 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:F242 | R:R:S91 | 3.96 | 1 | Yes | No | 9 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8T1V | A | Orphan | Orphan | GPR6 | Homo Sapiens | PubChem 118310209 | Na | - | 2.6 | 2024-12-04 | doi.org/10.1126/scisignal.ado8741 |
|

A 2D representation of the interactions of NA in 8T1V
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D118 | 8.05 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S155 | 5.94 | 1 | Yes | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:F288 | 7.23 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:N321 | 8.15 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S322 | 2.97 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:N325 | 5.43 | 1 | Yes | Yes | 0 | 9 | 0 | 1 | | R:R:I86 | R:R:N90 | 2.83 | 0 | Yes | Yes | 7 | 9 | 2 | 2 | | R:R:I86 | R:R:S322 | 7.74 | 0 | Yes | Yes | 7 | 9 | 2 | 1 | | R:R:D118 | R:R:N90 | 6.73 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D118 | R:R:L114 | 8.14 | 1 | Yes | No | 9 | 9 | 1 | 2 | | R:R:L114 | R:R:N325 | 4.12 | 1 | No | Yes | 9 | 9 | 2 | 1 | | R:R:D118 | R:R:S322 | 11.78 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:D118 | R:R:N325 | 8.08 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:A121 | R:R:S322 | 3.42 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:F288 | R:R:S155 | 5.28 | 1 | Yes | No | 9 | 9 | 1 | 1 | | R:R:F288 | R:R:V156 | 5.24 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F288 | R:R:L159 | 8.53 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F234 | R:R:F238 | 6.43 | 1 | Yes | Yes | 7 | 6 | 2 | 2 | | R:R:F234 | R:R:F288 | 5.36 | 1 | Yes | Yes | 7 | 9 | 2 | 1 | | R:R:F238 | R:R:F288 | 3.22 | 1 | Yes | Yes | 6 | 9 | 2 | 1 | | R:R:N321 | R:R:T287 | 2.92 | 1 | Yes | No | 9 | 7 | 1 | 2 | | R:R:F288 | R:R:W292 | 10.02 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F288 | R:R:N321 | 6.04 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | | R:R:C291 | R:R:N321 | 7.87 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:N321 | R:R:W292 | 5.65 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:N321 | R:R:N325 | 12.26 | 1 | Yes | Yes | 9 | 9 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 6.30 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 11.00 | | Average Links In Shell | 26.00 | | Average Links Mediated by Hubs In Shell | 26.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9FUP | A | Nucleotide | Adenosine | A2A | Homo Sapiens | Istradefylline | Na | - | 2.5 | 2025-01-22 | doi.org/10.1038/s42004-024-01404-y |
|

A 2D representation of the interactions of NA in 9FUP
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.05 | 5 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 5 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:F242 | 4.82 | 5 | No | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 13.46 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 10.31 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:F242 | R:R:S91 | 5.28 | 5 | Yes | No | 9 | 9 | 1 | 1 | | R:R:F242 | R:R:I92 | 3.77 | 5 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 5 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 15.03 | 5 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 9.67 | 5 | Yes | No | 9 | 9 | 1 | 2 | | R:R:N280 | R:R:W246 | 6.78 | 5 | No | Yes | 9 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8RVW | A | Nucleotide | Adenosine | A2A | Homo Sapiens | StilSwitch3 | Na | - | 2.65 | 2025-01-08 | doi.org/10.1038/s41467-024-55109-w |
|

A 2D representation of the interactions of NA in 8RVW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 10.73 | 0 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 9.42 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 8.83 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:F242 | R:R:S91 | 3.96 | 9 | Yes | No | 9 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 8.34 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8RW4 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | AzoSwitch2 | Na | - | 2.2 | 2025-01-08 | doi.org/10.1038/s41467-024-55109-w |
|
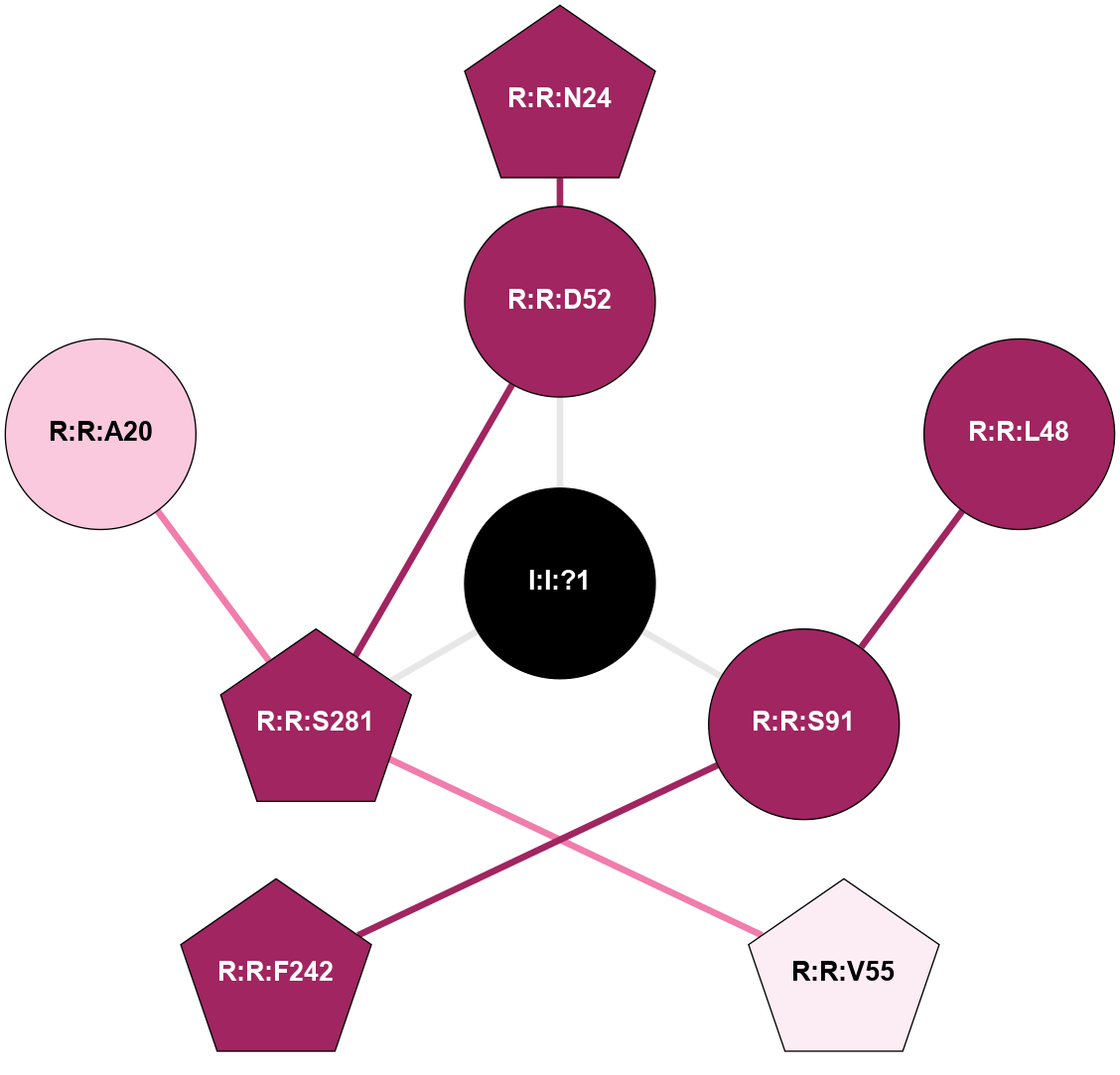
A 2D representation of the interactions of NA in 8RW4
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.05 | 6 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 6 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S281 | 2.97 | 6 | No | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 13.46 | 6 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 11.78 | 6 | No | Yes | 9 | 9 | 1 | 1 | | R:R:S281 | R:R:V55 | 8.08 | 6 | Yes | Yes | 9 | 6 | 1 | 2 | | R:R:F242 | R:R:S91 | 5.28 | 1 | Yes | No | 9 | 9 | 2 | 1 | | R:R:A20 | R:R:S281 | 1.71 | 0 | No | Yes | 7 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.65 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8RW7 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | StilSwitch1 | Na | - | 2.25 | 2025-01-08 | doi.org/10.1038/s41467-024-55109-w |
|

A 2D representation of the interactions of NA in 8RW7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.05 | 1 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 1 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:F242 | 4.82 | 1 | No | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 12.12 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:F242 | R:R:S91 | 5.28 | 1 | Yes | No | 9 | 9 | 1 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 1 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 17.04 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:N280 | R:R:W246 | 4.52 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8RWC | A | Nucleotide | Adenosine | A2A | Homo Sapiens | StilSwitch2 | Na | - | 2.31 | 2025-01-08 | doi.org/10.1038/s41467-024-55109-w |
|

A 2D representation of the interactions of NA in 8RWC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.05 | 1 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 1 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:F242 | 4.82 | 1 | No | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 9.42 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 10.31 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:F242 | R:R:S91 | 5.28 | 1 | Yes | No | 9 | 9 | 1 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 1 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 1 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 16.04 | 1 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 1 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:N280 | R:R:W246 | 4.52 | 1 | Yes | Yes | 9 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8RWD | A | Nucleotide | Adenosine | A2A | Homo Sapiens | StilSwitch3 | Na | - | 2.05 | 2025-01-08 | doi.org/10.1038/s41467-024-55109-w |
|

A 2D representation of the interactions of NA in 8RWD
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.05 | 6 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 6 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:F242 | 4.82 | 6 | No | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 13.46 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 11.78 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:F242 | R:R:S91 | 3.96 | 6 | Yes | No | 9 | 9 | 1 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 6 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 18.04 | 6 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 6 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:N280 | R:R:W246 | 4.52 | 6 | Yes | Yes | 9 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8RWE | A | Nucleotide | Adenosine | A2A | Homo Sapiens | StilSwitch4 | Na | - | 2.2 | 2025-01-08 | doi.org/10.1038/s41467-024-55109-w |
|

A 2D representation of the interactions of NA in 8RWE
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.05 | 6 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 6 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:F242 | 4.82 | 6 | No | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 13.46 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 8.83 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:F242 | R:R:S91 | 3.96 | 6 | Yes | No | 9 | 9 | 1 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 6 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 6 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 17.04 | 6 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 6 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:N280 | R:R:W246 | 3.39 | 6 | Yes | Yes | 9 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8RWH | A | Nucleotide | Adenosine | A2A | Homo Sapiens | StilSwitch2 | Na | - | 2.45 | 2025-01-08 | doi.org/10.1038/s41467-024-55109-w |
|

A 2D representation of the interactions of NA in 8RWH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.05 | 6 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 6 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S281 | 2.97 | 6 | No | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 8.08 | 6 | No | Yes | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:S91 | 3 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:D52 | R:R:S281 | 11.78 | 6 | No | Yes | 9 | 9 | 1 | 1 | | R:R:S281 | R:R:V55 | 8.08 | 6 | Yes | Yes | 9 | 6 | 1 | 2 | | R:R:F242 | R:R:S91 | 5.28 | 11 | Yes | No | 9 | 9 | 2 | 1 | | R:R:A20 | R:R:S281 | 1.71 | 0 | No | Yes | 7 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 5.65 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8RWI | A | Nucleotide | Adenosine | A2A | Homo Sapiens | StilSwitch2 | Na | - | 2.8 | 2025-01-08 | doi.org/10.1038/s41467-024-55109-w |
|

A 2D representation of the interactions of NA in 8RWI
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.05 | 0 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 13.46 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 8.83 | 0 | No | No | 9 | 9 | 1 | 2 | | R:R:F242 | R:R:S91 | 3.96 | 9 | Yes | No | 9 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.00 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8RWX | A | Nucleotide | Adenosine | A2A | Homo Sapiens | StilSwitch3 | Na | - | 3.05 | 2025-01-08 | doi.org/10.1038/s41467-024-55109-w |
|

A 2D representation of the interactions of NA in 8RWX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | I:I:?1 | R:R:D52 | 8.05 | 0 | No | Yes | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 0 | No | No | 0 | 9 | 0 | 1 | | I:I:?1 | R:R:N280 | 8.15 | 0 | No | Yes | 0 | 9 | 0 | 1 | | R:R:D52 | R:R:N24 | 13.46 | 4 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:L48 | 4.07 | 4 | Yes | No | 9 | 9 | 1 | 2 | | R:R:L48 | R:R:N284 | 5.49 | 4 | No | Yes | 9 | 9 | 2 | 2 | | R:R:D52 | R:R:S281 | 8.83 | 4 | Yes | No | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:N284 | 6.73 | 4 | Yes | Yes | 9 | 9 | 1 | 2 | | R:R:F242 | R:R:W246 | 15.03 | 5 | Yes | Yes | 9 | 8 | 2 | 2 | | R:R:F242 | R:R:N280 | 12.08 | 5 | Yes | Yes | 9 | 9 | 2 | 1 | | R:R:C245 | R:R:N280 | 3.15 | 0 | No | Yes | 8 | 9 | 2 | 1 | | R:R:N280 | R:R:W246 | 3.39 | 5 | Yes | Yes | 9 | 8 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 7.38 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9H2X | A | Nucleotide | Adenosine | A2A | Homo Sapiens | A1IR1 | Na | - | 1.75 | 2025-06-18 | To be published |
|
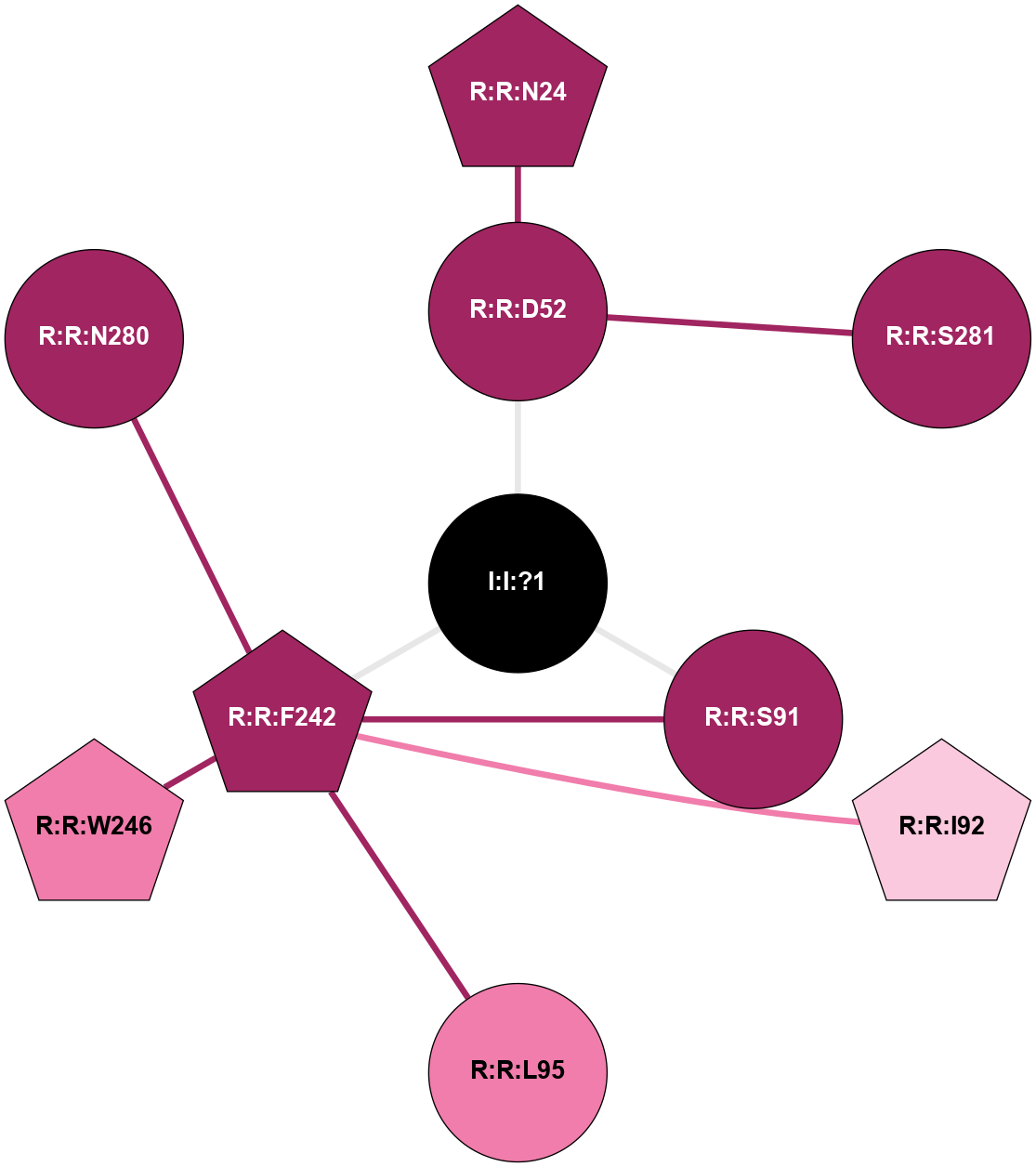
A 2D representation of the interactions of NA in 9H2X
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 10.77 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 10.31 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.05 | 7 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 7 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 7 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 7 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 7 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 15.03 | 7 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 7 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 7 | No | Yes | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9H37 | A | Nucleotide | Adenosine | A2A | Homo Sapiens | A1IR0 | Na | - | 1.71 | 2025-06-18 | To be published |
|
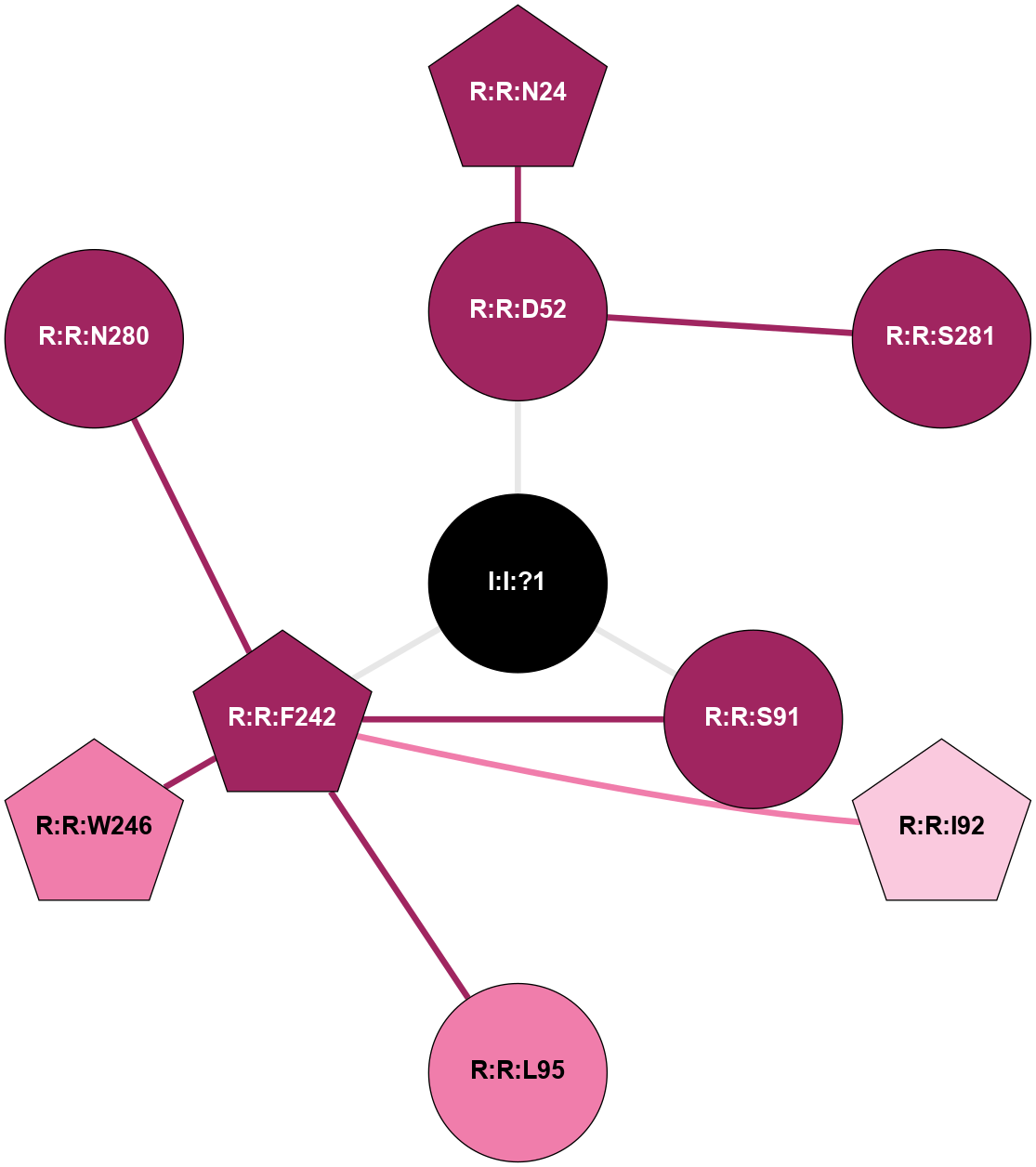
A 2D representation of the interactions of NA in 9H37
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D52 | R:R:N24 | 9.42 | 0 | No | Yes | 9 | 9 | 1 | 2 | | R:R:D52 | R:R:S281 | 10.31 | 0 | No | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:D52 | 8.05 | 8 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:S91 | 5.28 | 8 | Yes | No | 9 | 9 | 1 | 1 | | I:I:?1 | R:R:S91 | 5.94 | 8 | No | No | 0 | 9 | 0 | 1 | | R:R:F242 | R:R:I92 | 5.02 | 8 | Yes | Yes | 9 | 7 | 1 | 2 | | R:R:F242 | R:R:L95 | 4.87 | 8 | Yes | No | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:W246 | 16.04 | 8 | Yes | Yes | 9 | 8 | 1 | 2 | | R:R:F242 | R:R:N280 | 8.46 | 8 | Yes | No | 9 | 9 | 1 | 2 | | I:I:?1 | R:R:F242 | 4.82 | 8 | No | Yes | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 6.27 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|














































































