| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 1F88 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.8 | 2000-08-04 | doi.org/10.1126/science.289.5480.739 |
|
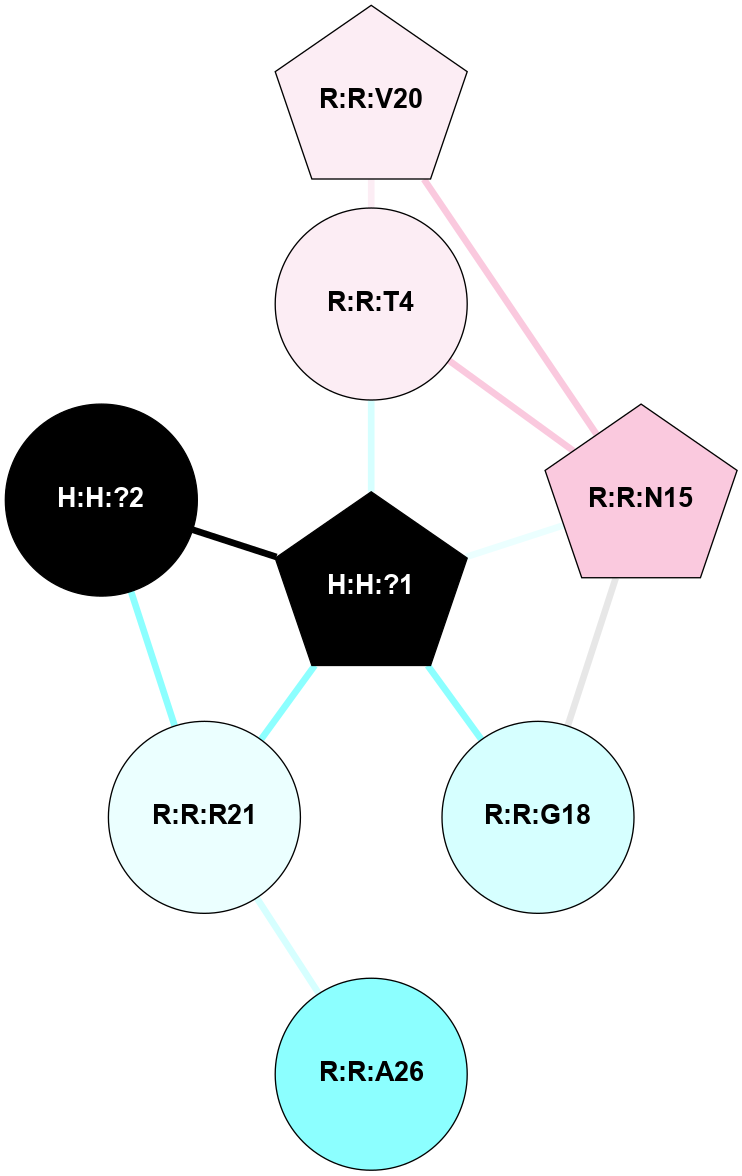
A 2D representation of the interactions of NAG in 1F88
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 35.42 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:N15 | 24.56 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:R21 | 14.12 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:R21 | 14.12 | 1 | No | No | 0 | 4 | 1 | 1 | | R:R:N15 | R:R:T4 | 7.31 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 8.87 | 1 | Yes | Yes | 7 | 6 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.10 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
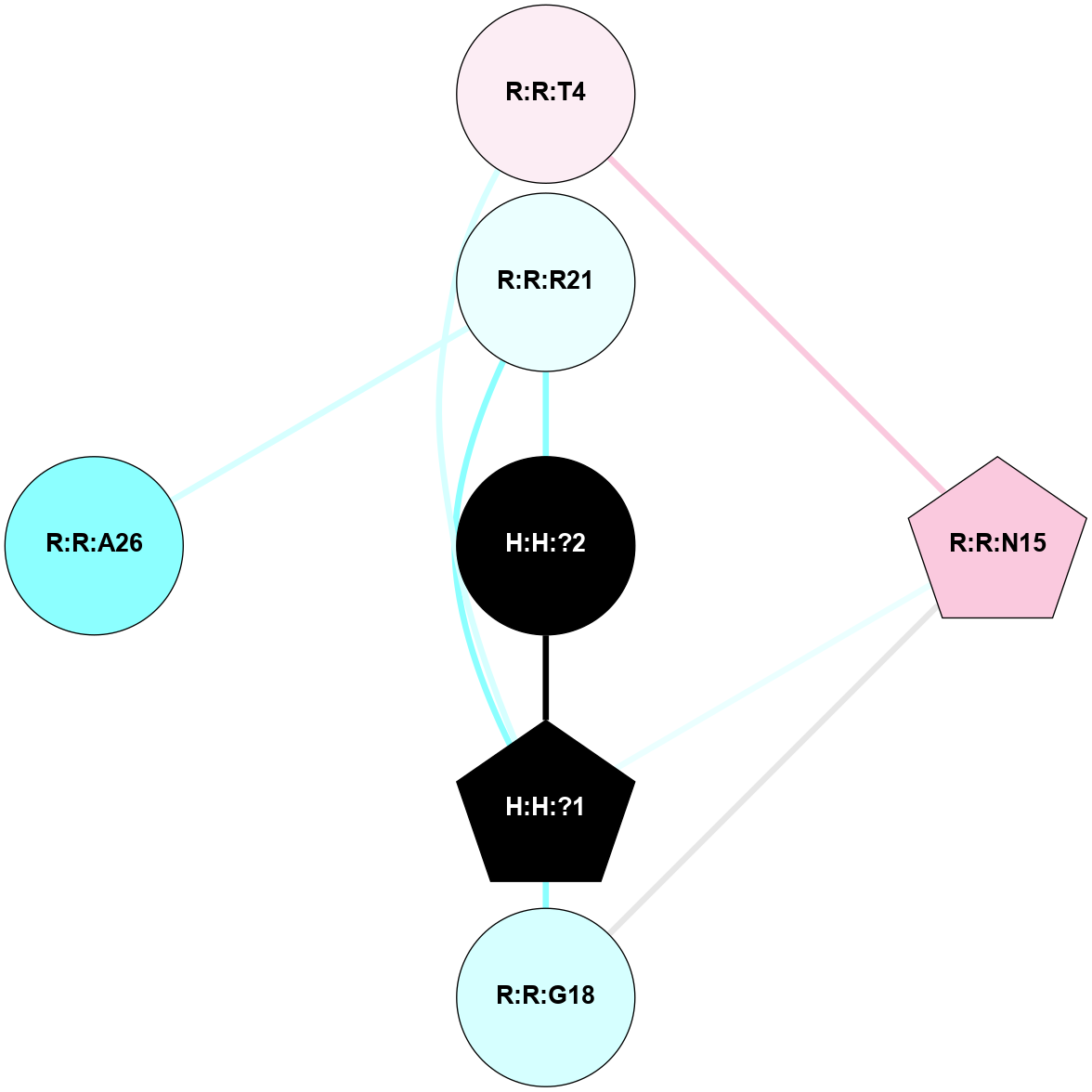
A 2D representation of the interactions of NAG in 1F88
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 35.42 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 24.56 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:R21 | 14.12 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?2 | R:R:R21 | 14.12 | 1 | No | No | 0 | 4 | 0 | 1 | | R:R:N15 | R:R:T4 | 7.31 | 1 | Yes | No | 7 | 6 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 2 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 24.77 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
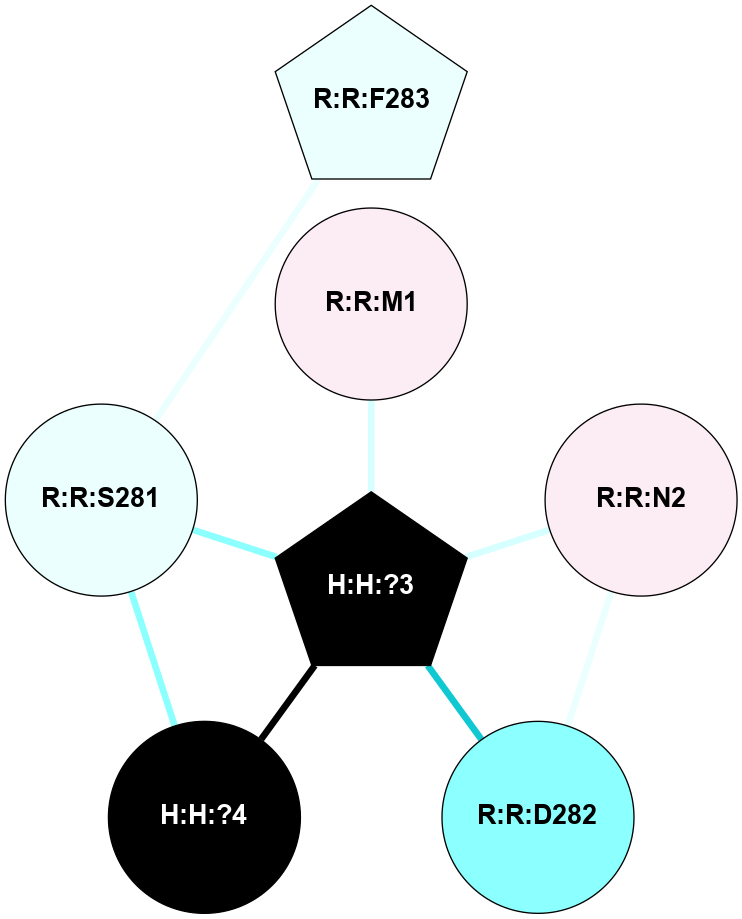
A 2D representation of the interactions of NAG in 1F88
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 33.21 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:M1 | 5.06 | 1 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?3 | R:R:N2 | 25.79 | 1 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?3 | R:R:D282 | 4.85 | 1 | Yes | No | 0 | 2 | 0 | 1 | | R:R:D282 | R:R:N2 | 13.46 | 1 | No | No | 2 | 6 | 1 | 1 | | R:R:F283 | R:R:S281 | 3.96 | 1 | Yes | No | 4 | 4 | 2 | 1 | | H:H:?3 | R:R:S281 | 2.69 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?4 | R:R:S281 | 2.69 | 1 | No | No | 0 | 4 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.32 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
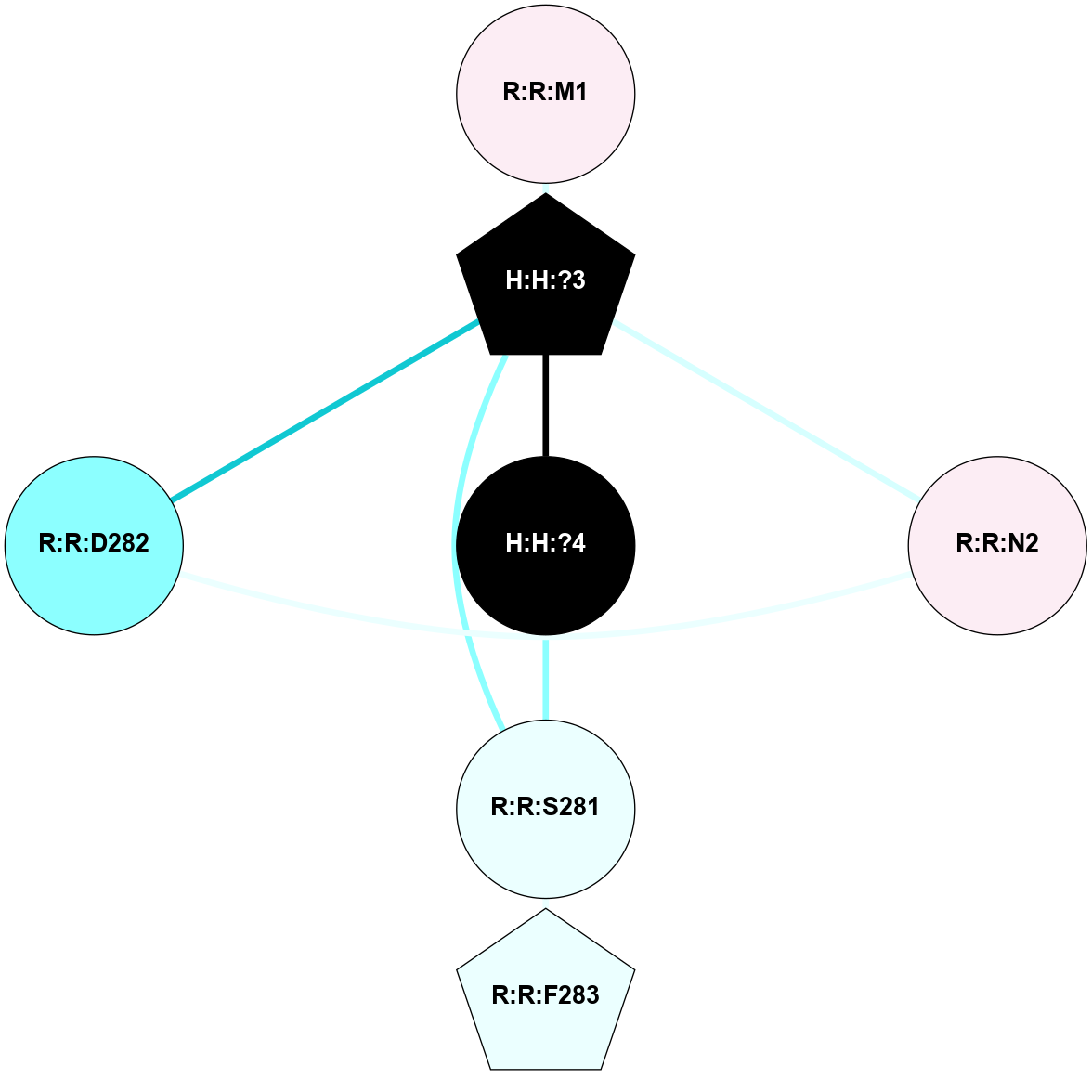
A 2D representation of the interactions of NAG in 1F88
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 33.21 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:M1 | 5.06 | 1 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?3 | R:R:N2 | 25.79 | 1 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?3 | R:R:D282 | 4.85 | 1 | Yes | No | 0 | 2 | 1 | 2 | | R:R:D282 | R:R:N2 | 13.46 | 1 | No | No | 2 | 6 | 2 | 2 | | R:R:F283 | R:R:S281 | 3.96 | 1 | Yes | No | 4 | 4 | 2 | 1 | | H:H:?3 | R:R:S281 | 2.69 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?4 | R:R:S281 | 2.69 | 1 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.95 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 1GZM | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.65 | 2003-11-20 | doi.org/10.1016/j.jmb.2004.08.090 |
|
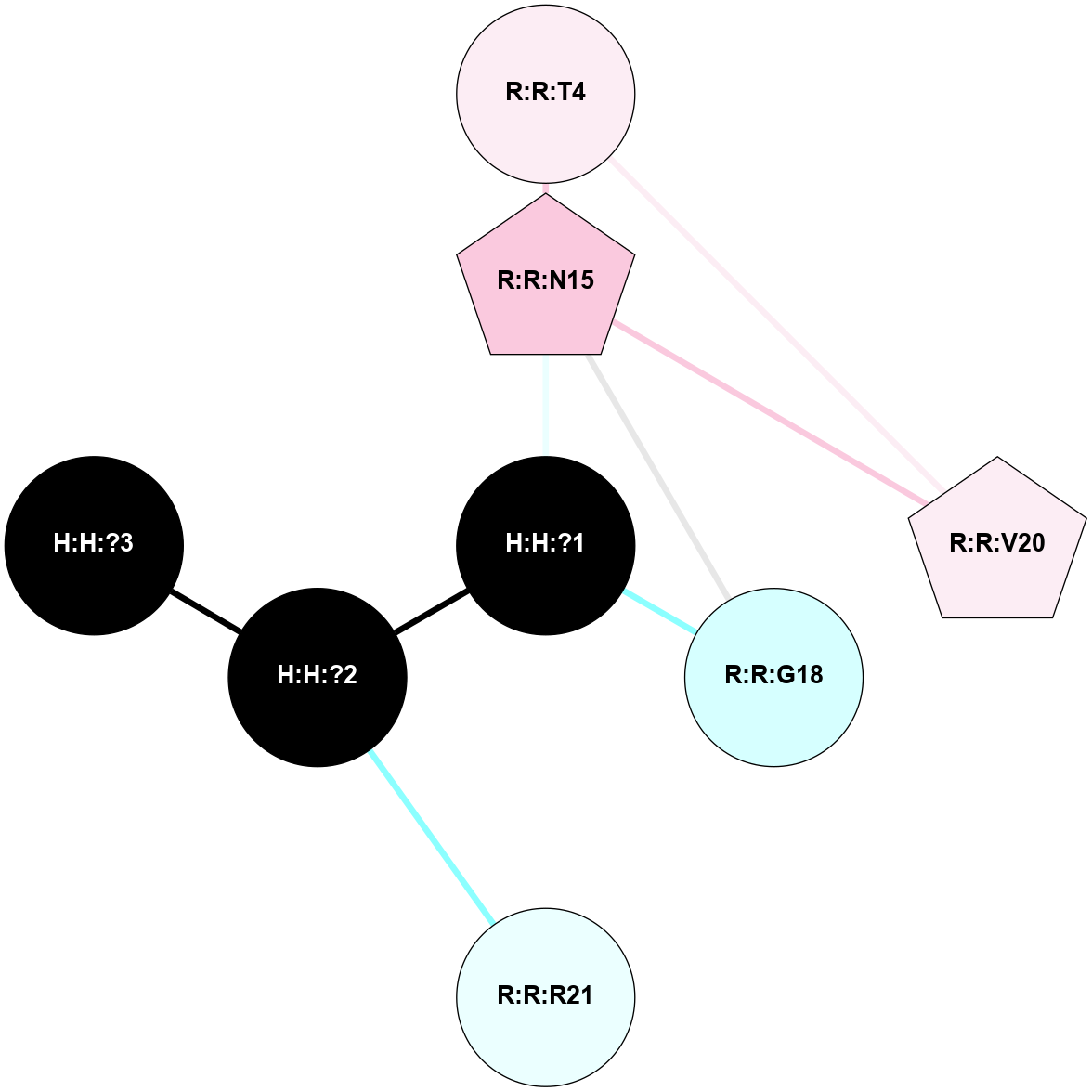
A 2D representation of the interactions of NAG in 1GZM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 5.85 | 2 | Yes | No | 7 | 6 | 1 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 2 | Yes | Yes | 7 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 22.1 | 2 | No | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 7.65 | 2 | No | No | 0 | 3 | 0 | 1 | | H:H:?2 | R:R:R21 | 5.43 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 28.78 | 2 | No | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 32.11 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.51 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
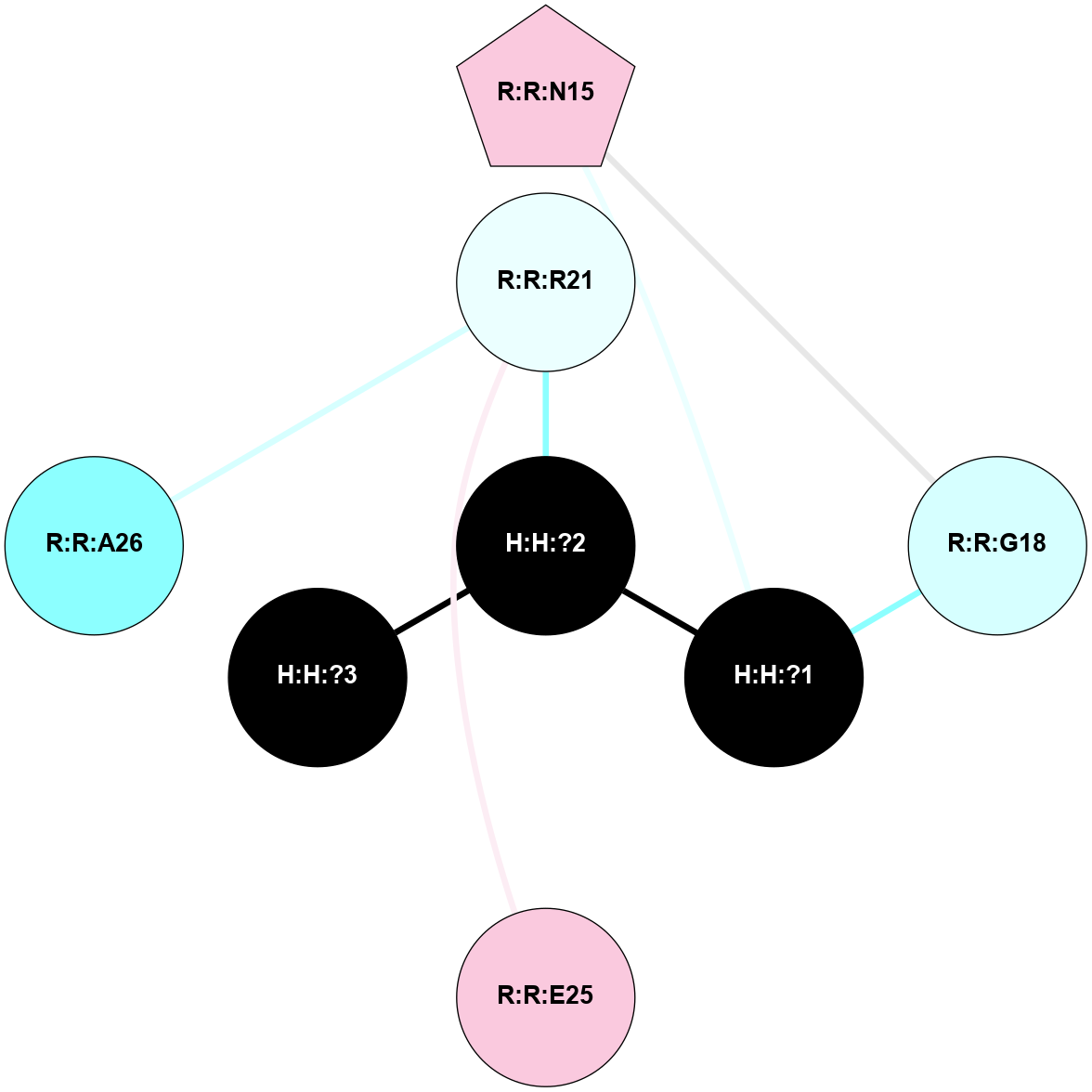
A 2D representation of the interactions of NAG in 1GZM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 2 | 2 | | H:H:?1 | R:R:N15 | 22.1 | 2 | No | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 7.65 | 2 | No | No | 0 | 3 | 1 | 2 | | R:R:E25 | R:R:R21 | 8.14 | 0 | No | No | 7 | 4 | 2 | 1 | | H:H:?2 | R:R:R21 | 5.43 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 28.78 | 2 | No | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 32.11 | 0 | No | No | 0 | 0 | 0 | 1 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 22.11 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
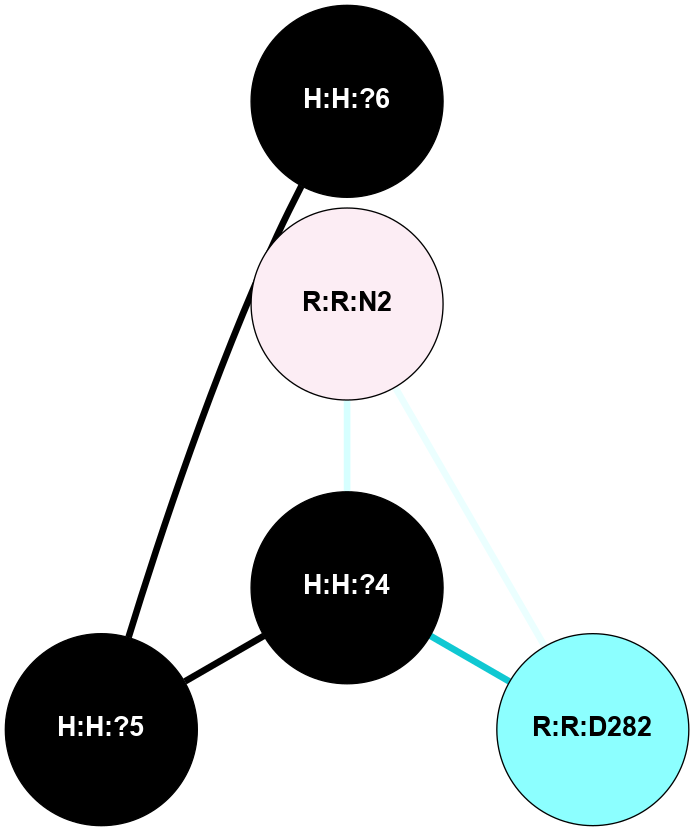
A 2D representation of the interactions of NAG in 1GZM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 16.16 | 4 | No | No | 2 | 6 | 1 | 1 | | H:H:?4 | R:R:N2 | 29.47 | 4 | No | No | 0 | 6 | 0 | 1 | | H:H:?4 | R:R:D282 | 9.71 | 4 | No | No | 0 | 2 | 0 | 1 | | H:H:?4 | H:H:?5 | 38.74 | 4 | No | No | 0 | 0 | 0 | 1 | | H:H:?5 | H:H:?6 | 28.25 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 25.97 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
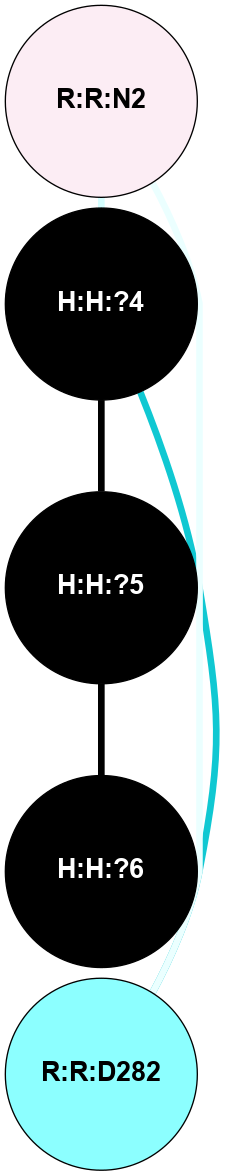
A 2D representation of the interactions of NAG in 1GZM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 16.16 | 4 | No | No | 2 | 6 | 2 | 2 | | H:H:?4 | R:R:N2 | 29.47 | 4 | No | No | 0 | 6 | 1 | 2 | | H:H:?4 | R:R:D282 | 9.71 | 4 | No | No | 0 | 2 | 1 | 2 | | H:H:?4 | H:H:?5 | 38.74 | 4 | No | No | 0 | 0 | 1 | 0 | | H:H:?5 | H:H:?6 | 28.25 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 33.50 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 1HZX | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.8 | 2001-07-04 | doi.org/10.1021/bi0155091 |
|
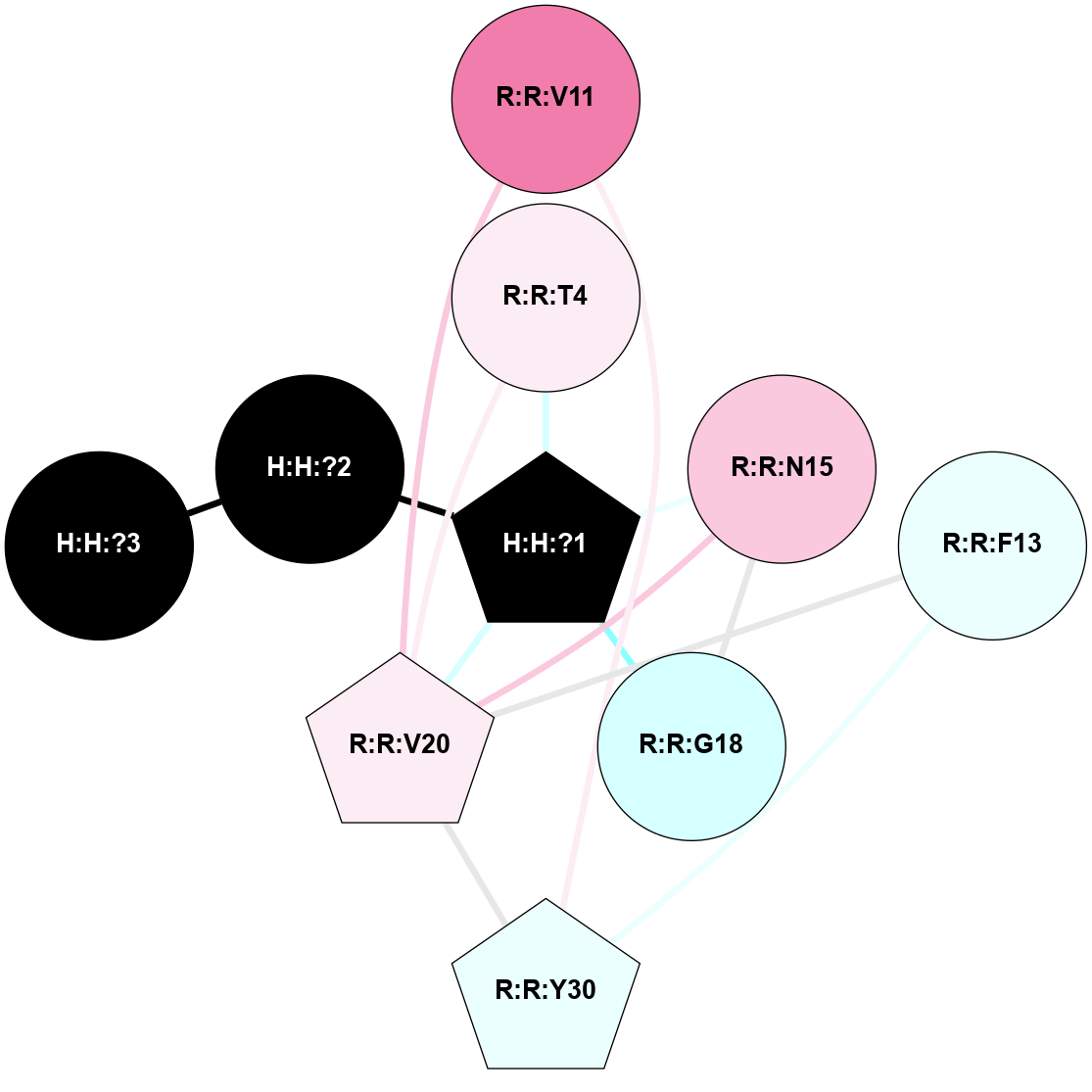
A 2D representation of the interactions of NAG in 1HZX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:T4 | R:R:V20 | 9.52 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | No | Yes | 8 | 6 | 2 | 1 | | R:R:V11 | R:R:Y30 | 3.79 | 1 | No | Yes | 8 | 4 | 2 | 2 | | R:R:F13 | R:R:V20 | 5.24 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 8.25 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | No | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 11.82 | 1 | No | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 28.24 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 7.65 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 10.09 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 6.66 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | H:H:?2 | 36.53 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 36.93 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 16.87 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
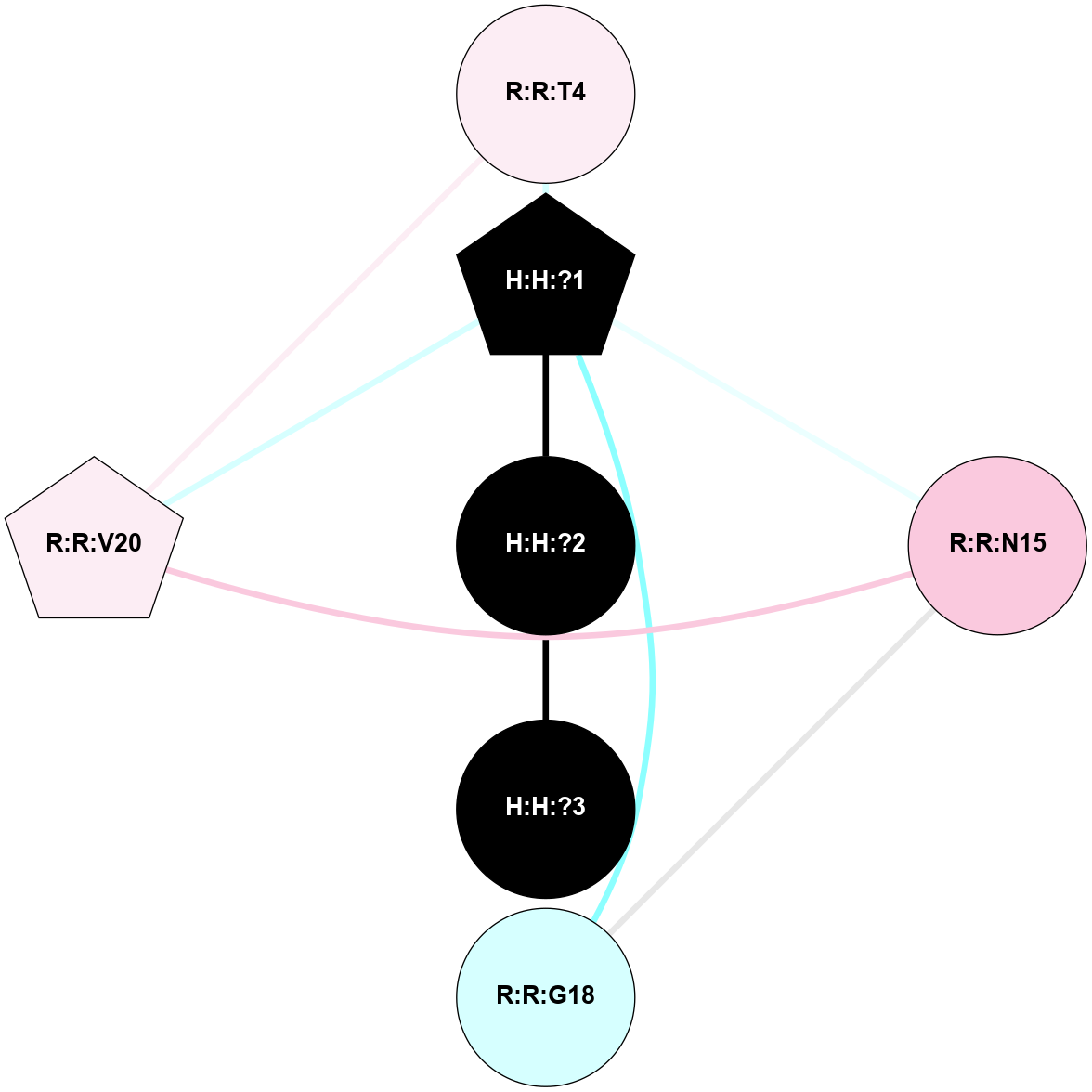
A 2D representation of the interactions of NAG in 1HZX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:T4 | R:R:V20 | 9.52 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | No | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 11.82 | 1 | No | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 28.24 | 1 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 7.65 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 6.66 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | H:H:?2 | 36.53 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 36.93 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 36.73 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
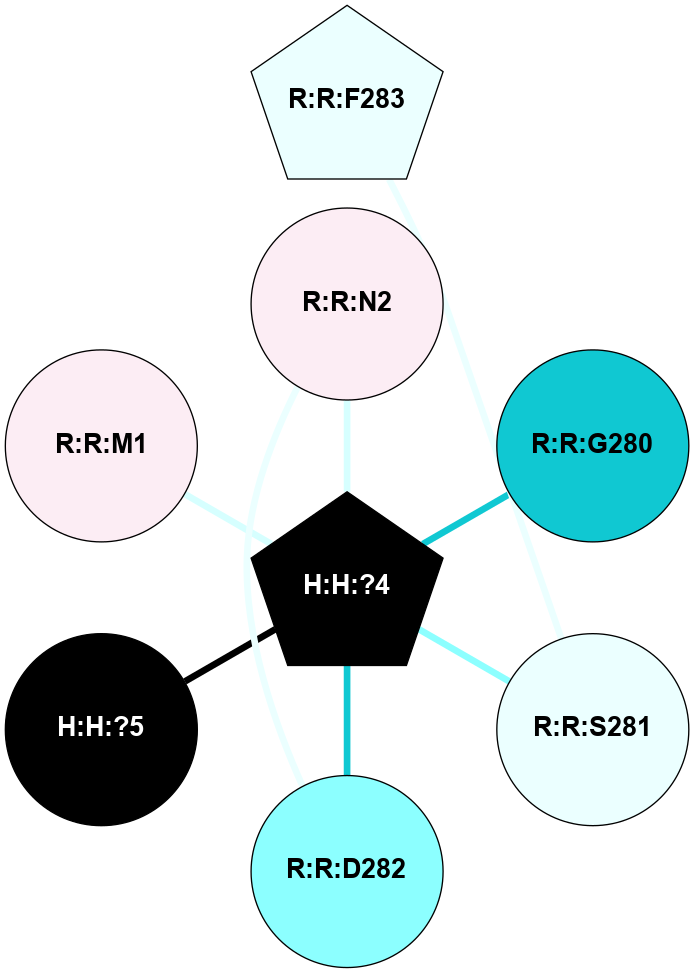
A 2D representation of the interactions of NAG in 1HZX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 10.77 | 4 | No | No | 2 | 6 | 1 | 1 | | H:H:?4 | R:R:N2 | 28.24 | 4 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?4 | R:R:G280 | 4.59 | 4 | Yes | No | 0 | 1 | 0 | 1 | | R:R:F283 | R:R:S281 | 3.96 | 1 | Yes | No | 4 | 4 | 2 | 1 | | H:H:?4 | R:R:S281 | 5.37 | 4 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?4 | R:R:D282 | 4.85 | 4 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?4 | H:H:?5 | 43.17 | 4 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?4 | R:R:M1 | 2.53 | 4 | Yes | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.79 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
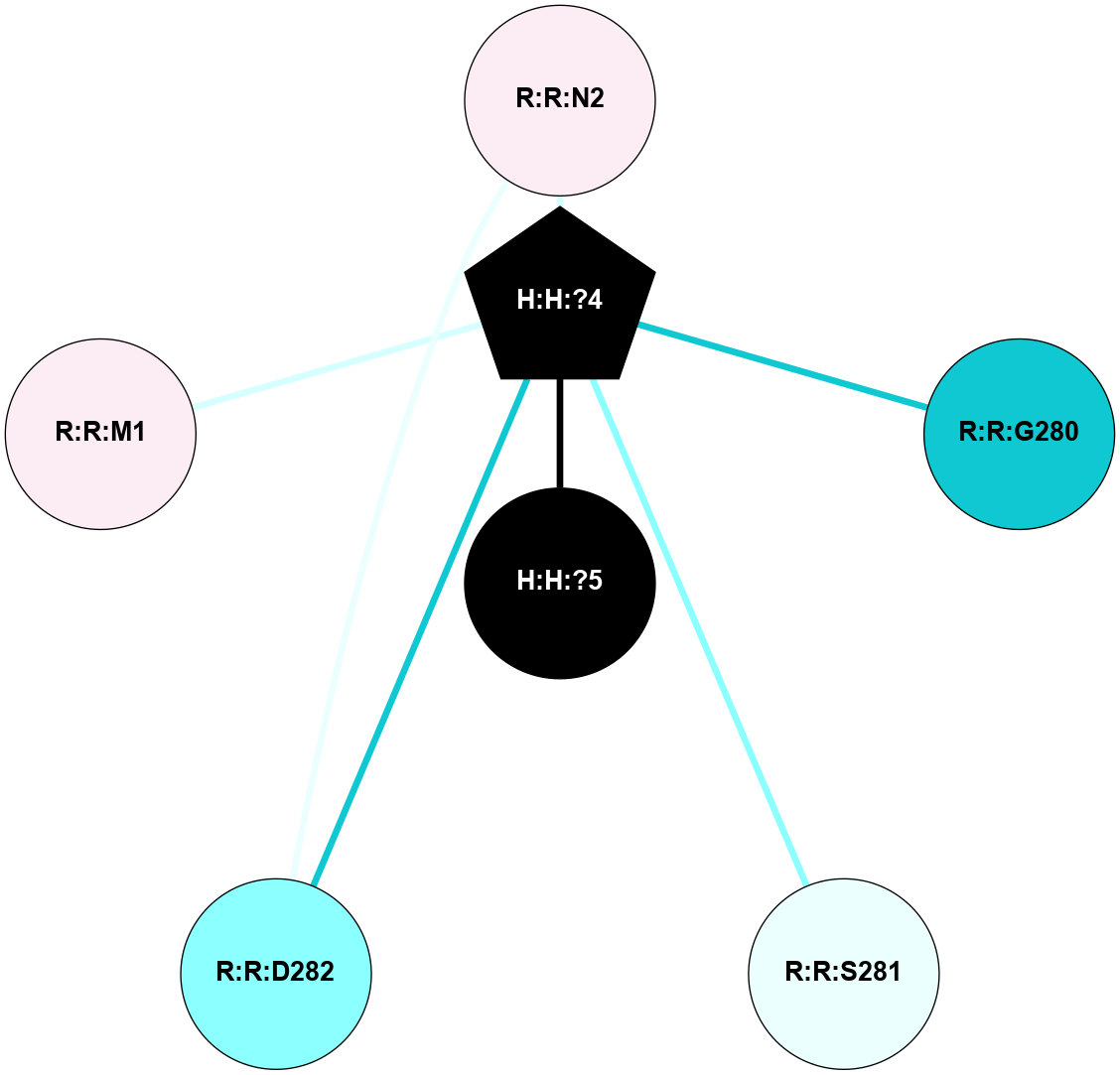
A 2D representation of the interactions of NAG in 1HZX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 10.77 | 4 | No | No | 2 | 6 | 2 | 2 | | H:H:?4 | R:R:N2 | 28.24 | 4 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?4 | R:R:G280 | 4.59 | 4 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?4 | R:R:S281 | 5.37 | 4 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?4 | R:R:D282 | 4.85 | 4 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?4 | H:H:?5 | 43.17 | 4 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?4 | R:R:M1 | 2.53 | 4 | Yes | No | 0 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 43.17 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 1L9H | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.6 | 2002-05-15 | doi.org/10.1073/pnas.082666399 |
|
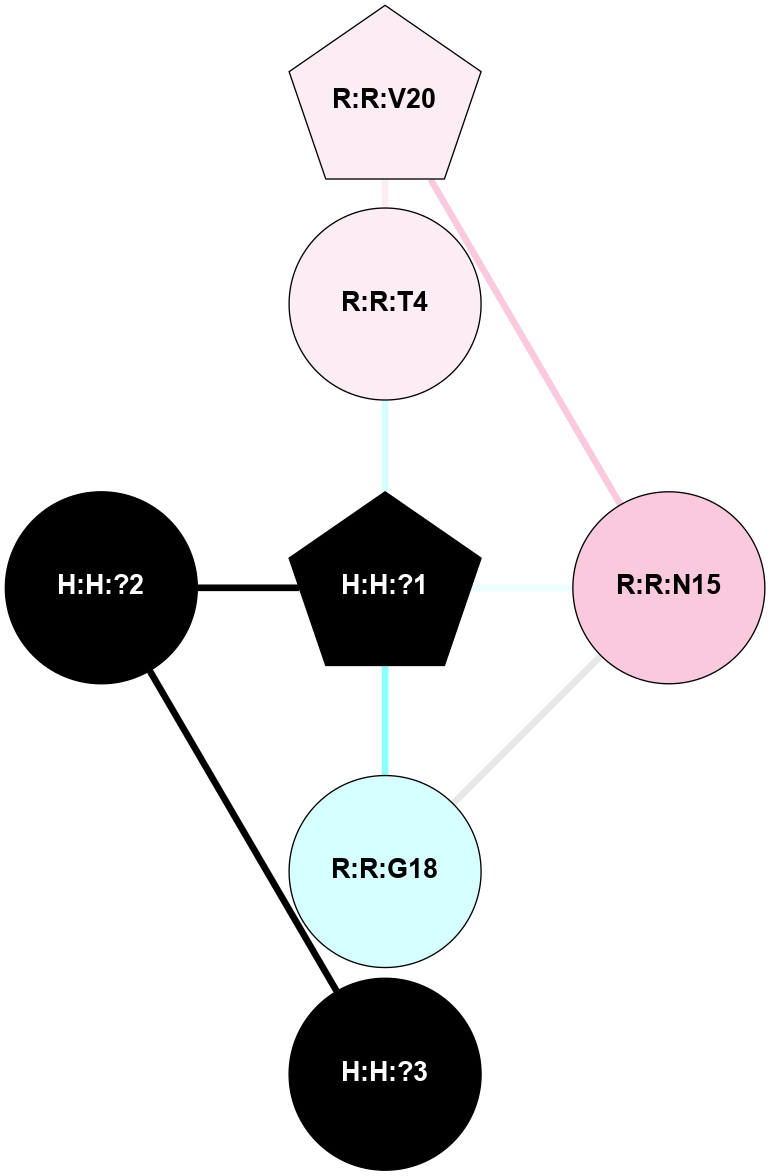
A 2D representation of the interactions of NAG in 1L9H
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:T4 | R:R:V20 | 9.52 | 0 | No | Yes | 6 | 6 | 1 | 2 | | H:H:?1 | R:R:T4 | 6.59 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | No | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 8.87 | 1 | No | Yes | 7 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 23.33 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | H:H:?2 | 43.17 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 47.11 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 19.80 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
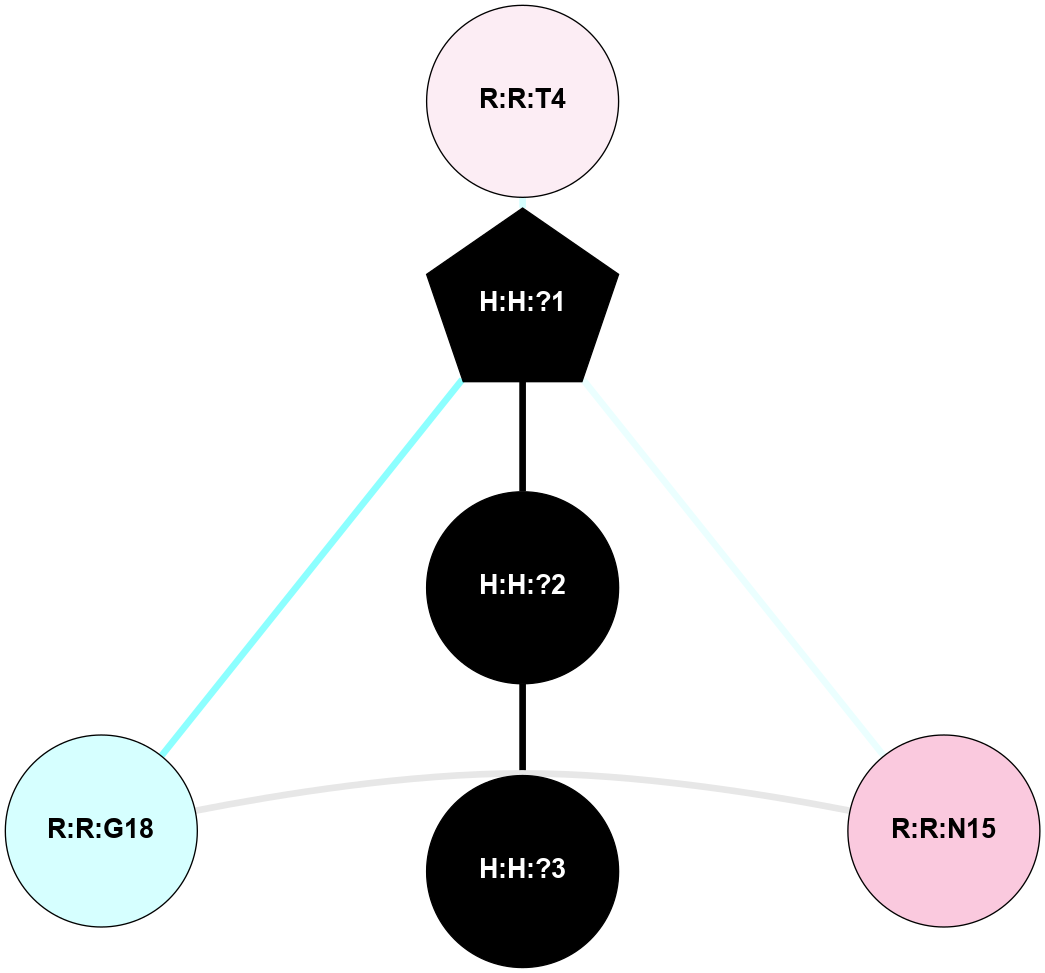
A 2D representation of the interactions of NAG in 1L9H
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:T4 | 6.59 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | No | 3 | 7 | 2 | 2 | | H:H:?1 | R:R:N15 | 23.33 | 1 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | H:H:?2 | 43.17 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 47.11 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 45.14 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
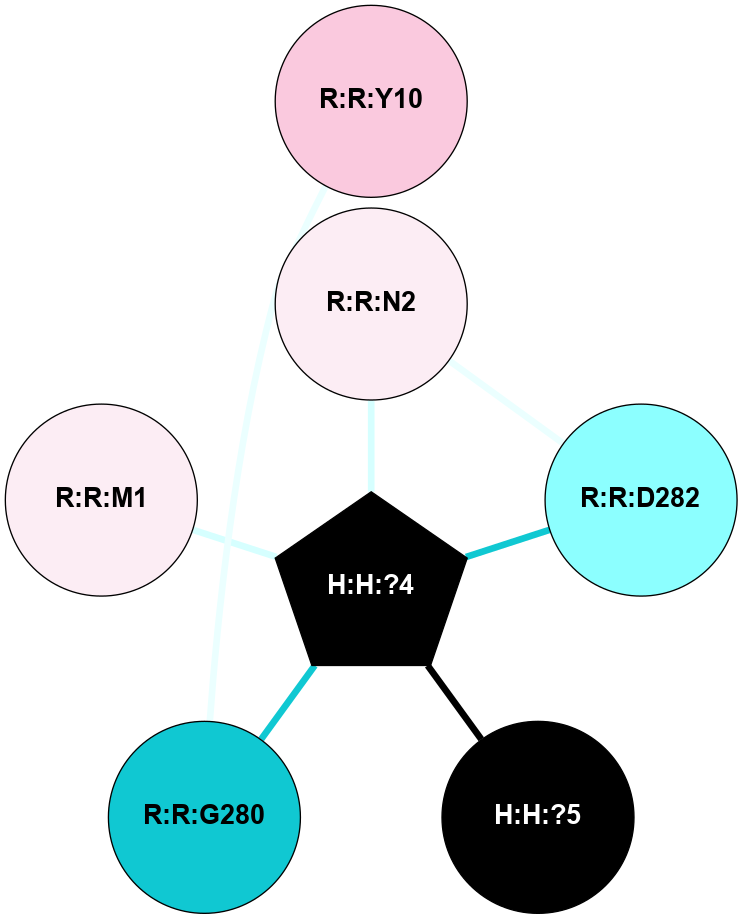
A 2D representation of the interactions of NAG in 1L9H
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 12.12 | 4 | No | No | 2 | 6 | 1 | 1 | | H:H:?4 | R:R:N2 | 24.56 | 4 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?4 | R:R:D282 | 6.07 | 4 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?4 | H:H:?5 | 50.92 | 4 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?4 | R:R:G280 | 3.06 | 4 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?4 | R:R:M1 | 2.53 | 4 | Yes | No | 0 | 6 | 0 | 1 | | R:R:G280 | R:R:Y10 | 1.45 | 0 | No | No | 1 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.43 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
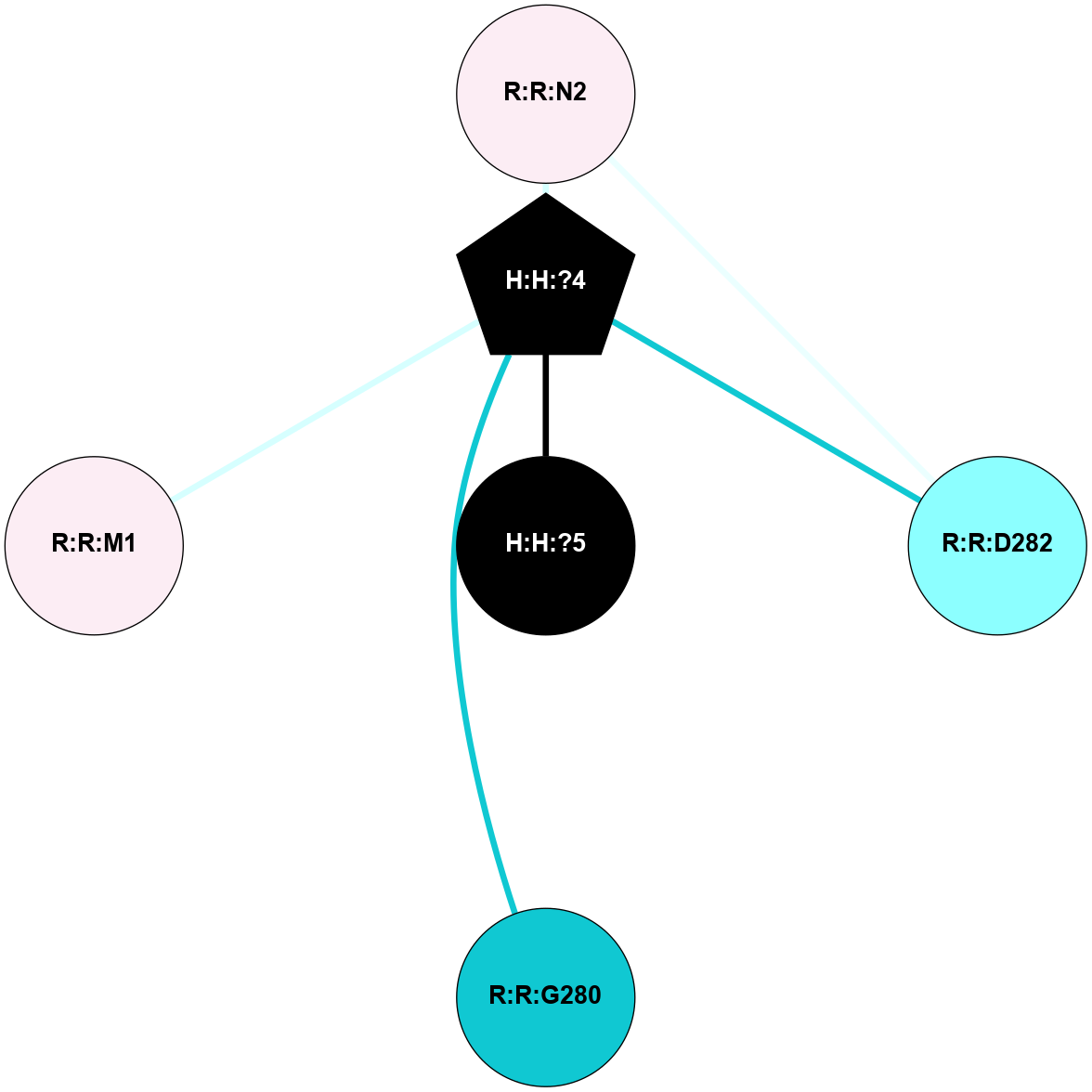
A 2D representation of the interactions of NAG in 1L9H
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 12.12 | 4 | No | No | 2 | 6 | 2 | 2 | | H:H:?4 | R:R:N2 | 24.56 | 4 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?4 | R:R:D282 | 6.07 | 4 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?4 | H:H:?5 | 50.92 | 4 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?4 | R:R:G280 | 3.06 | 4 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?4 | R:R:M1 | 2.53 | 4 | Yes | No | 0 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 50.92 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 1U19 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.2 | 2004-10-12 | doi.org/10.1016/j.jmb.2004.07.044 |
|
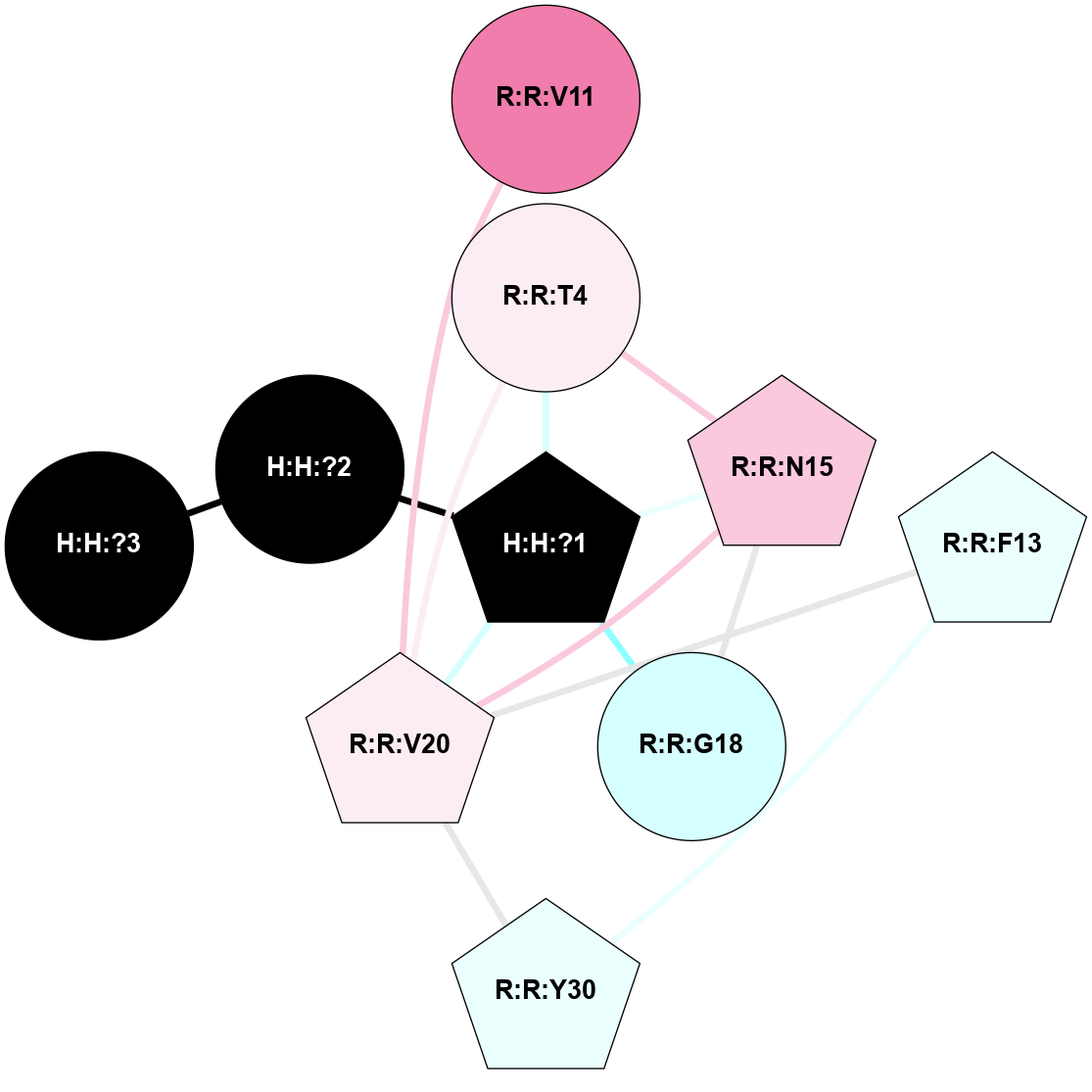
A 2D representation of the interactions of NAG in 1U19
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 4.39 | 2 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 0 | No | Yes | 8 | 6 | 2 | 1 | | R:R:F13 | R:R:V20 | 5.24 | 2 | Yes | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 8.25 | 2 | Yes | Yes | 4 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 6.79 | 2 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 2 | Yes | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 24.56 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 11.36 | 2 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | H:H:?2 | 42.06 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 31.83 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 16.40 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
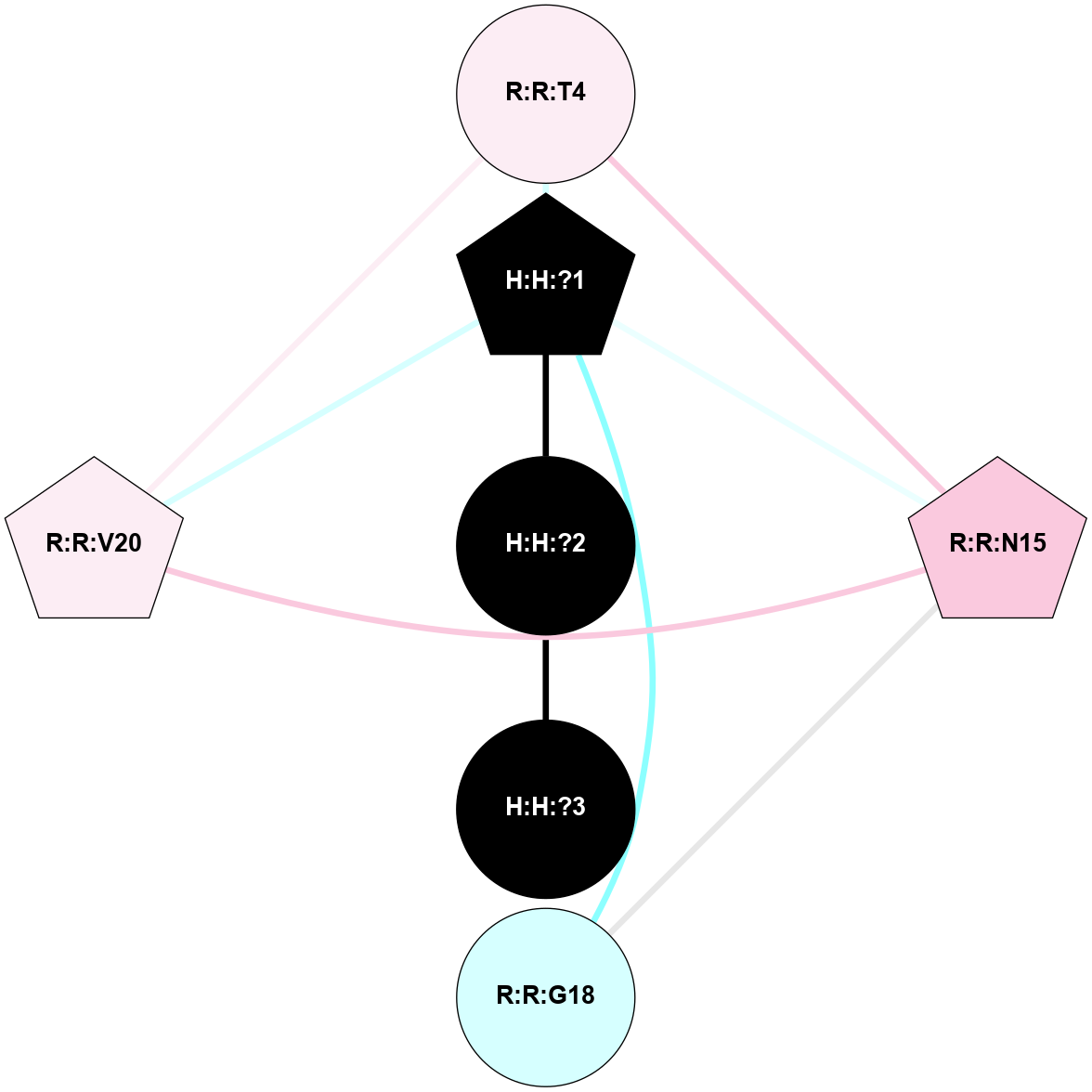
A 2D representation of the interactions of NAG in 1U19
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 4.39 | 2 | Yes | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 6.79 | 2 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 2 | Yes | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 24.56 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | H:H:?2 | 42.06 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 31.83 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 36.95 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
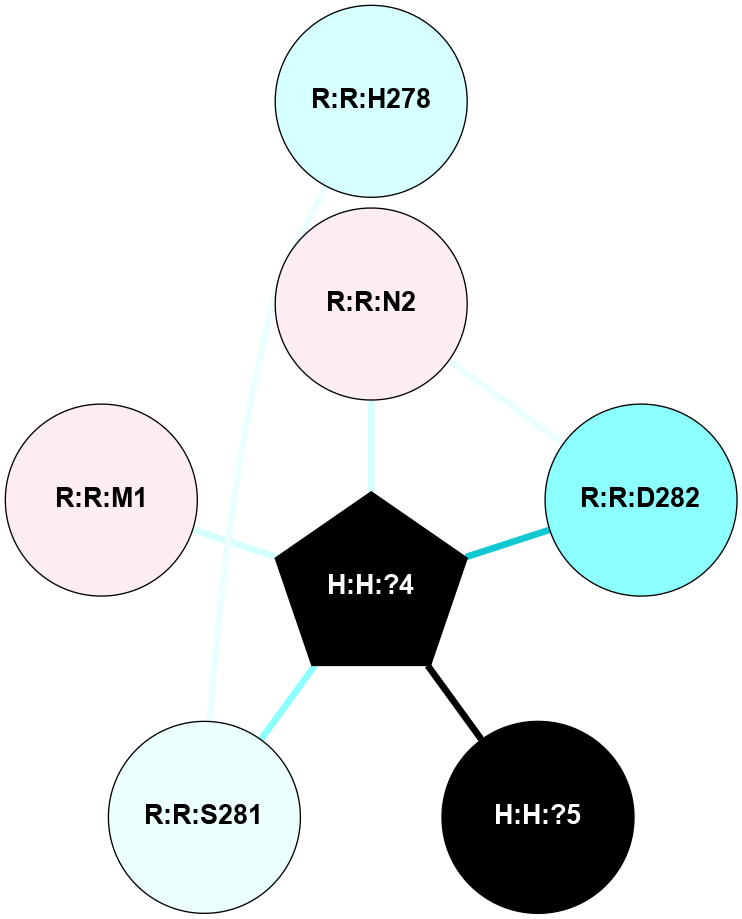
A 2D representation of the interactions of NAG in 1U19
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 13.46 | 8 | No | No | 2 | 6 | 1 | 1 | | H:H:?4 | R:R:N2 | 27.02 | 8 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?4 | R:R:D282 | 6.07 | 8 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?4 | H:H:?5 | 39.85 | 8 | Yes | No | 0 | 0 | 0 | 1 | | R:R:H278 | R:R:S281 | 2.79 | 0 | No | No | 3 | 4 | 2 | 1 | | H:H:?4 | R:R:S281 | 2.69 | 8 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?4 | R:R:M1 | 2.53 | 8 | Yes | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.63 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
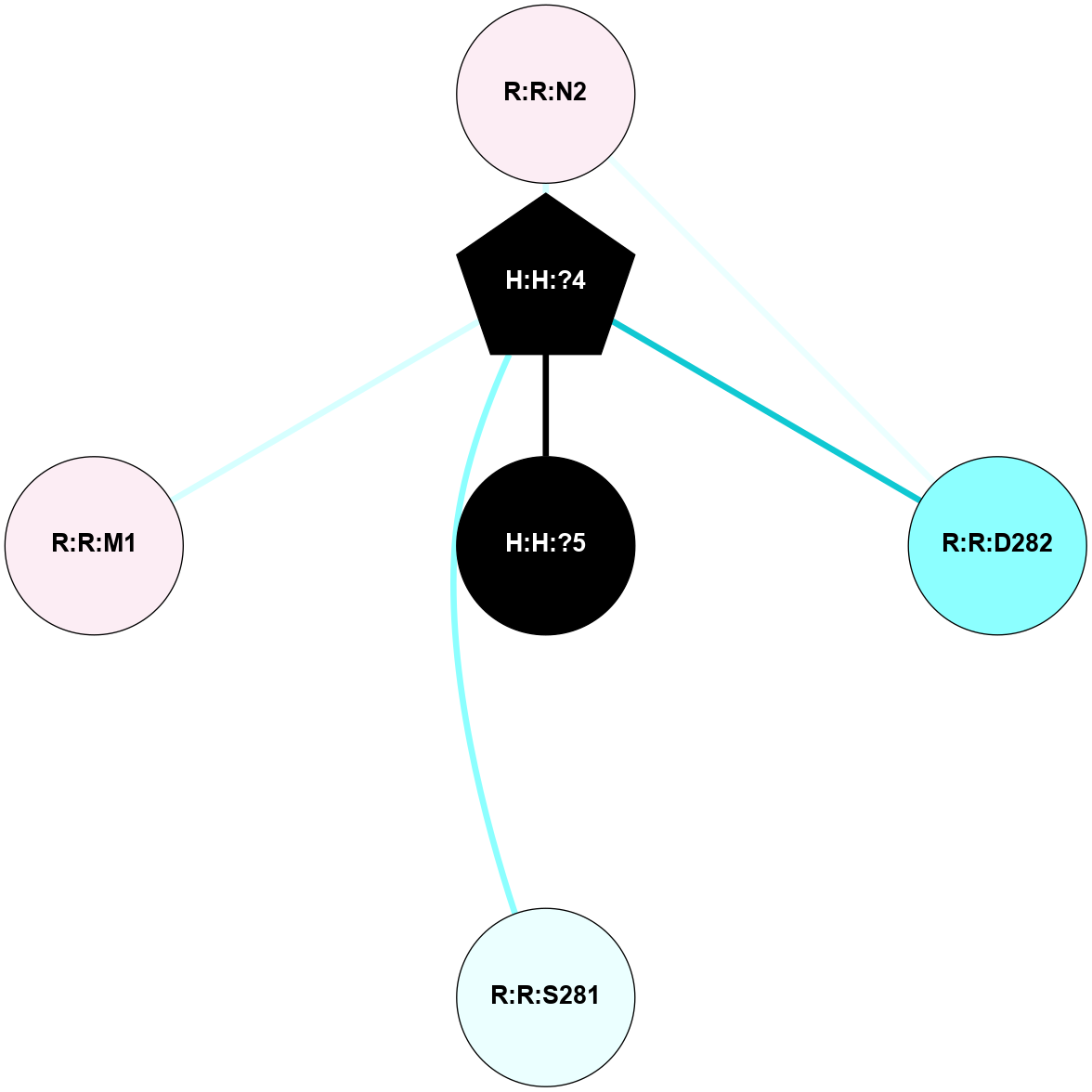
A 2D representation of the interactions of NAG in 1U19
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 13.46 | 8 | No | No | 2 | 6 | 2 | 2 | | H:H:?4 | R:R:N2 | 27.02 | 8 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?4 | R:R:D282 | 6.07 | 8 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?4 | H:H:?5 | 39.85 | 8 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?4 | R:R:S281 | 2.69 | 8 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?4 | R:R:M1 | 2.53 | 8 | Yes | No | 0 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 39.85 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2G87 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 2.6 | 2006-09-02 | doi.org/10.1002/anie.200600595 |
|
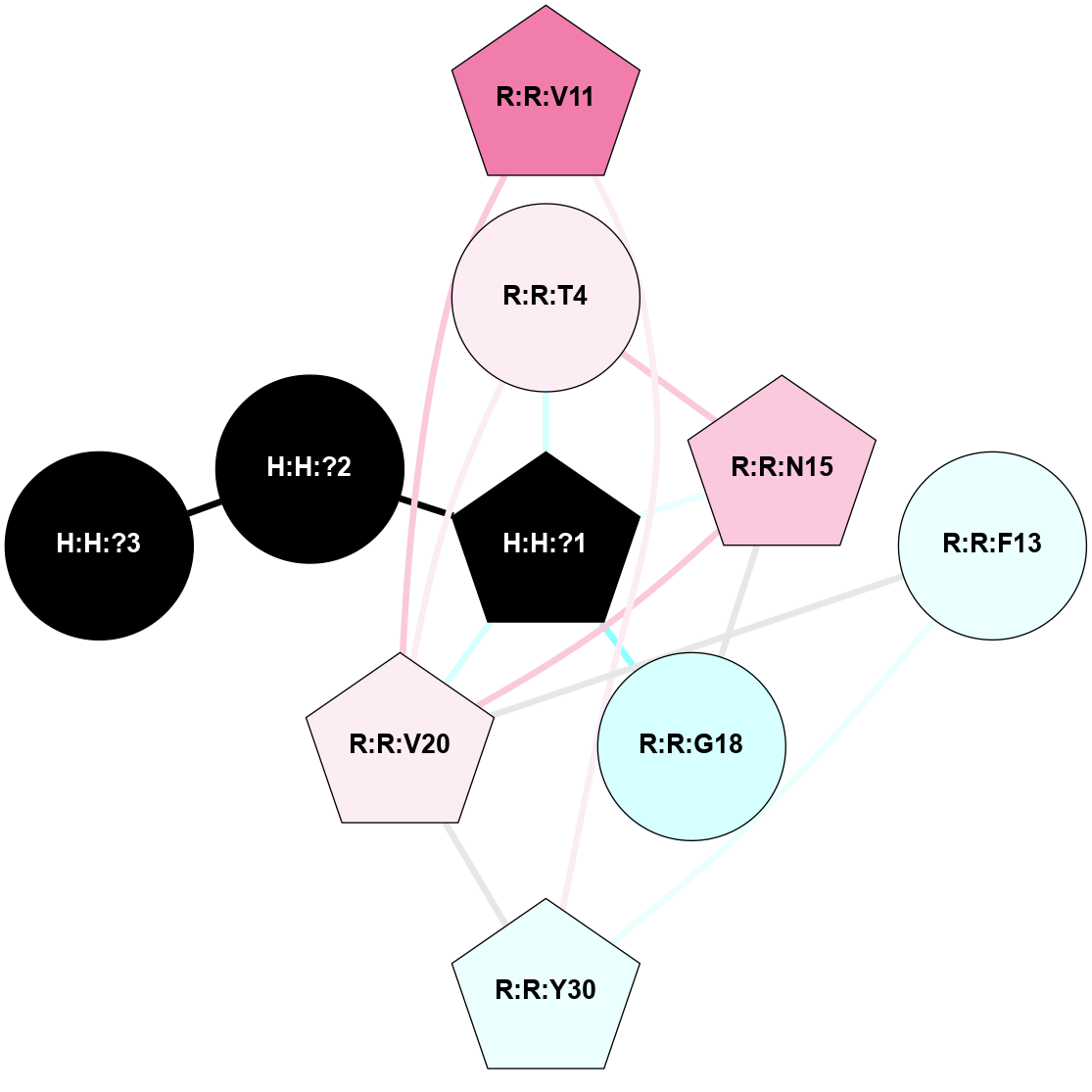
A 2D representation of the interactions of NAG in 2G87
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 5.85 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:V11 | R:R:Y30 | 3.79 | 1 | Yes | Yes | 8 | 4 | 2 | 2 | | R:R:F13 | R:R:V20 | 5.24 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 8.25 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 24.56 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 4.59 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 11.36 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 5.33 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | H:H:?2 | 40.96 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 31.83 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 16.14 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
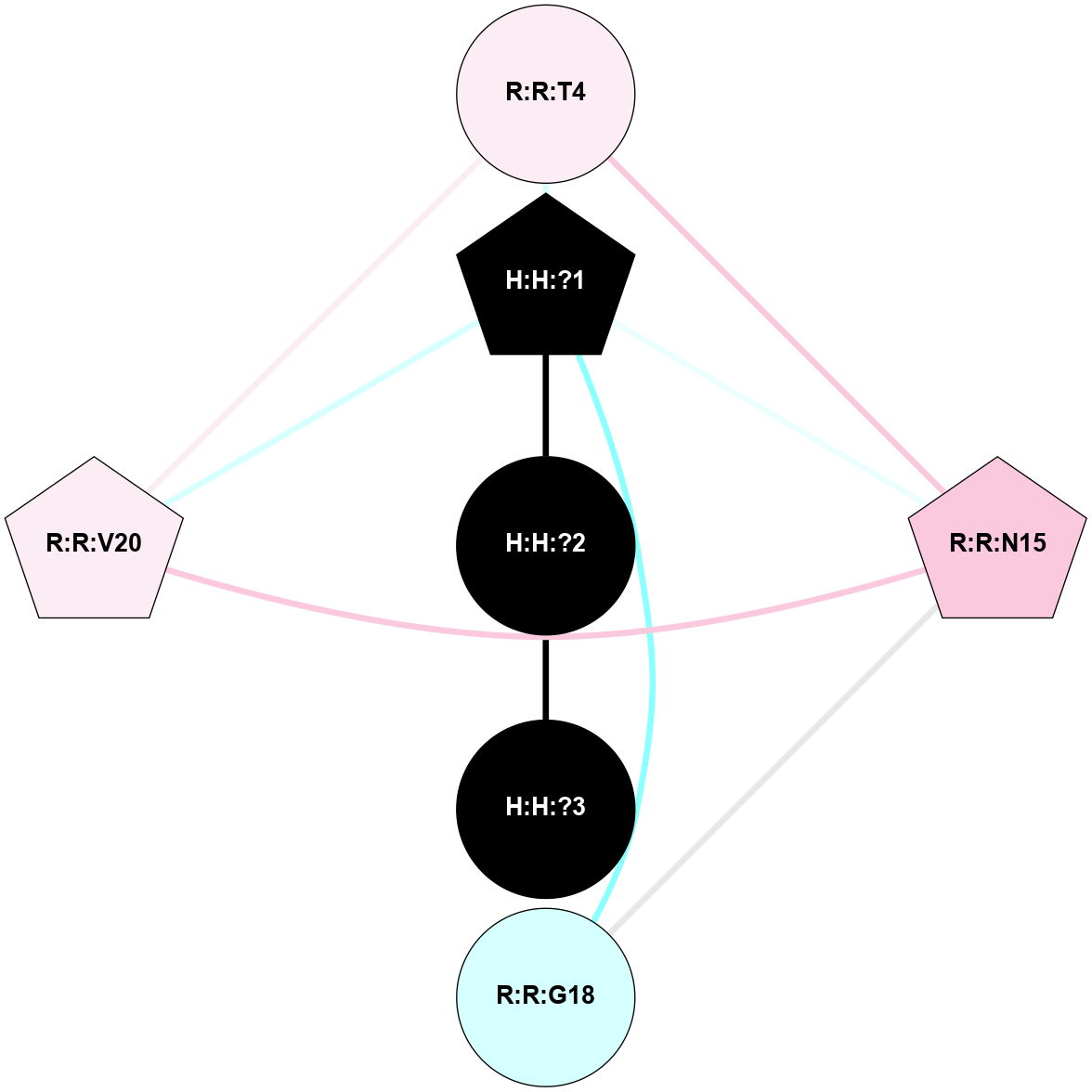
A 2D representation of the interactions of NAG in 2G87
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 5.85 | 1 | Yes | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 24.56 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 4.59 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 5.33 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | H:H:?2 | 40.96 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 31.83 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 36.39 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
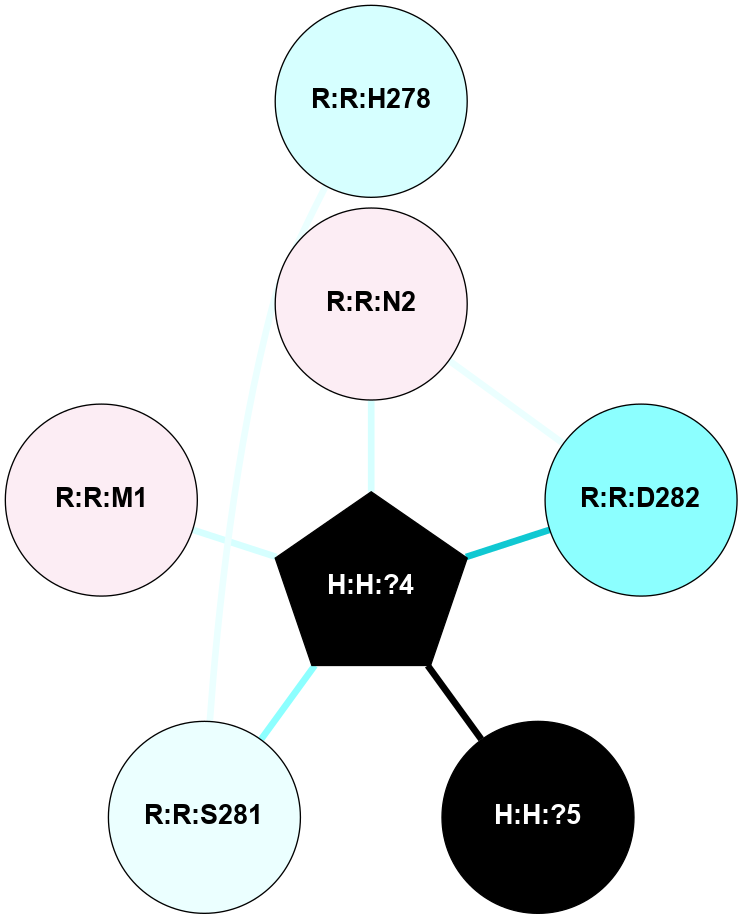
A 2D representation of the interactions of NAG in 2G87
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 13.46 | 9 | No | No | 2 | 6 | 1 | 1 | | H:H:?4 | R:R:N2 | 25.79 | 9 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?4 | R:R:D282 | 6.07 | 9 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?4 | H:H:?5 | 42.06 | 9 | Yes | No | 0 | 0 | 0 | 1 | | R:R:H278 | R:R:S281 | 2.79 | 0 | No | No | 3 | 4 | 2 | 1 | | H:H:?4 | R:R:S281 | 2.69 | 9 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?4 | R:R:M1 | 2.53 | 9 | Yes | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.83 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
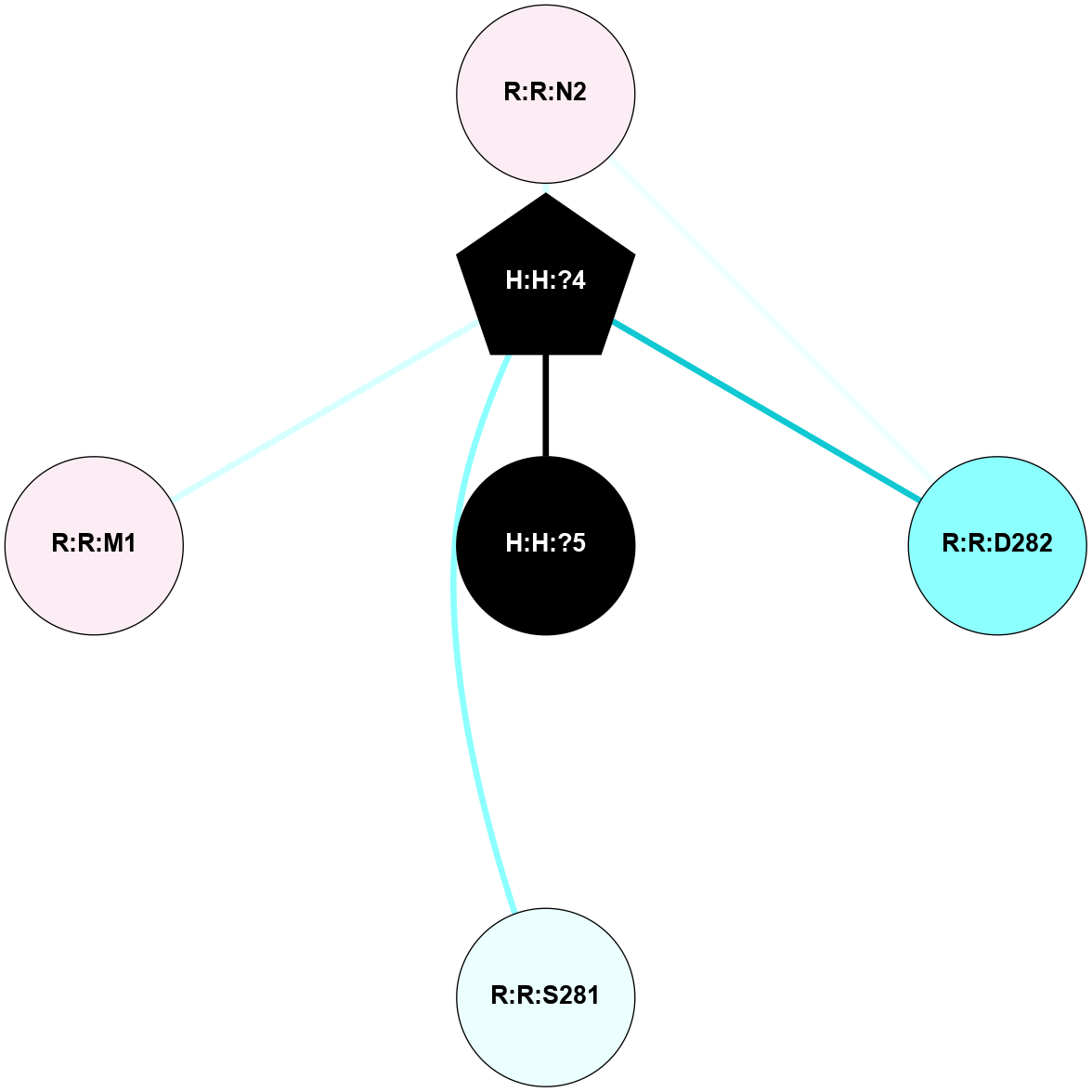
A 2D representation of the interactions of NAG in 2G87
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 13.46 | 9 | No | No | 2 | 6 | 2 | 2 | | H:H:?4 | R:R:N2 | 25.79 | 9 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?4 | R:R:D282 | 6.07 | 9 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?4 | H:H:?5 | 42.06 | 9 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?4 | R:R:S281 | 2.69 | 9 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?4 | R:R:M1 | 2.53 | 9 | Yes | No | 0 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 42.06 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2HPY | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 2.8 | 2006-08-22 | doi.org/10.1073/pnas.0601765103 |
|
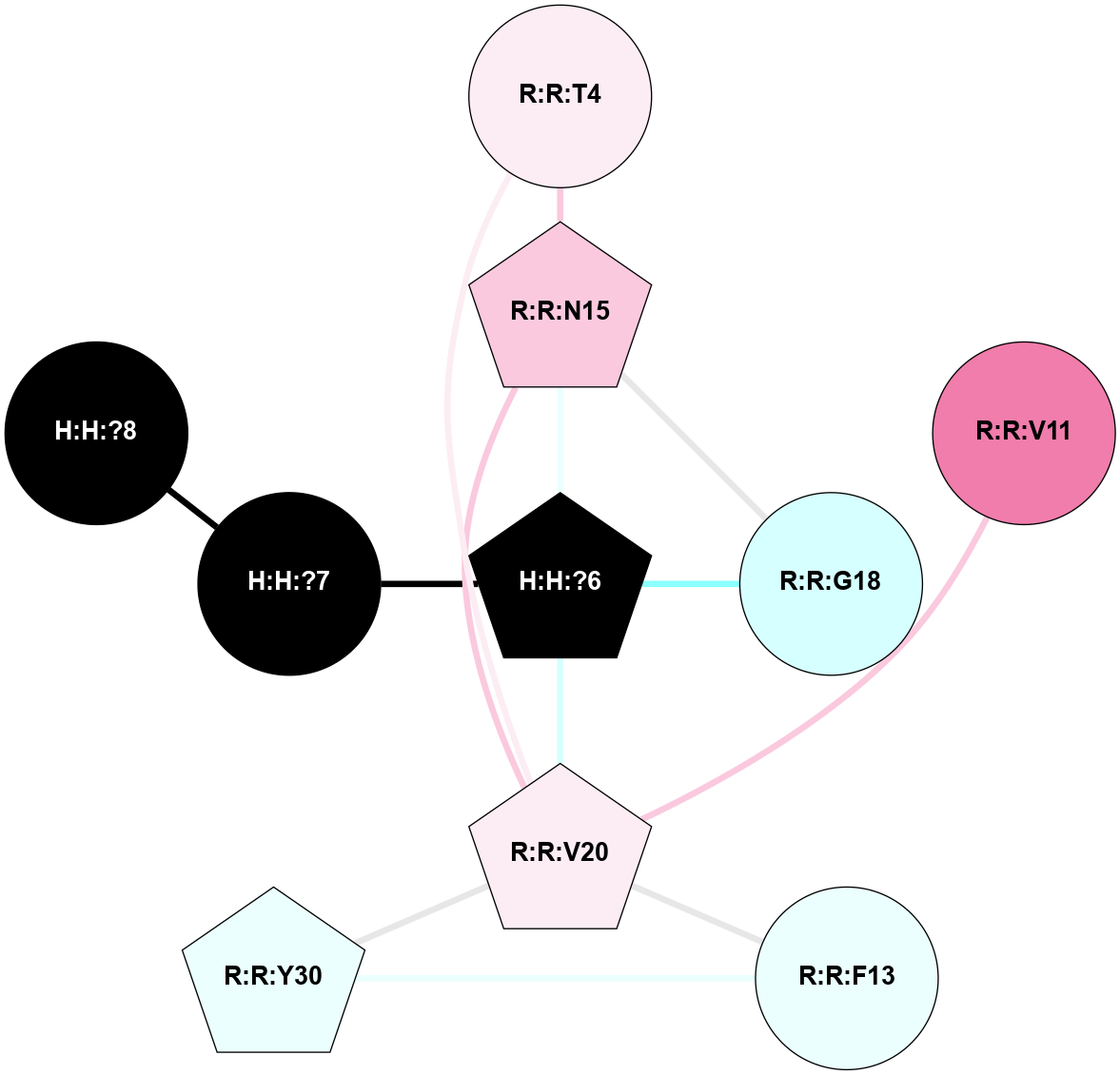
A 2D representation of the interactions of NAG in 2HPY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 2 | Yes | No | 7 | 6 | 1 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 2 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 0 | No | Yes | 8 | 6 | 2 | 1 | | R:R:F13 | R:R:V20 | 5.24 | 2 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 7.22 | 2 | No | Yes | 4 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 2 | Yes | Yes | 7 | 6 | 1 | 1 | | H:H:?6 | R:R:N15 | 29.47 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?6 | R:R:G18 | 7.65 | 2 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 10.09 | 2 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?6 | R:R:V20 | 7.99 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?6 | H:H:?7 | 43.17 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?7 | H:H:?8 | 26.97 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 22.07 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
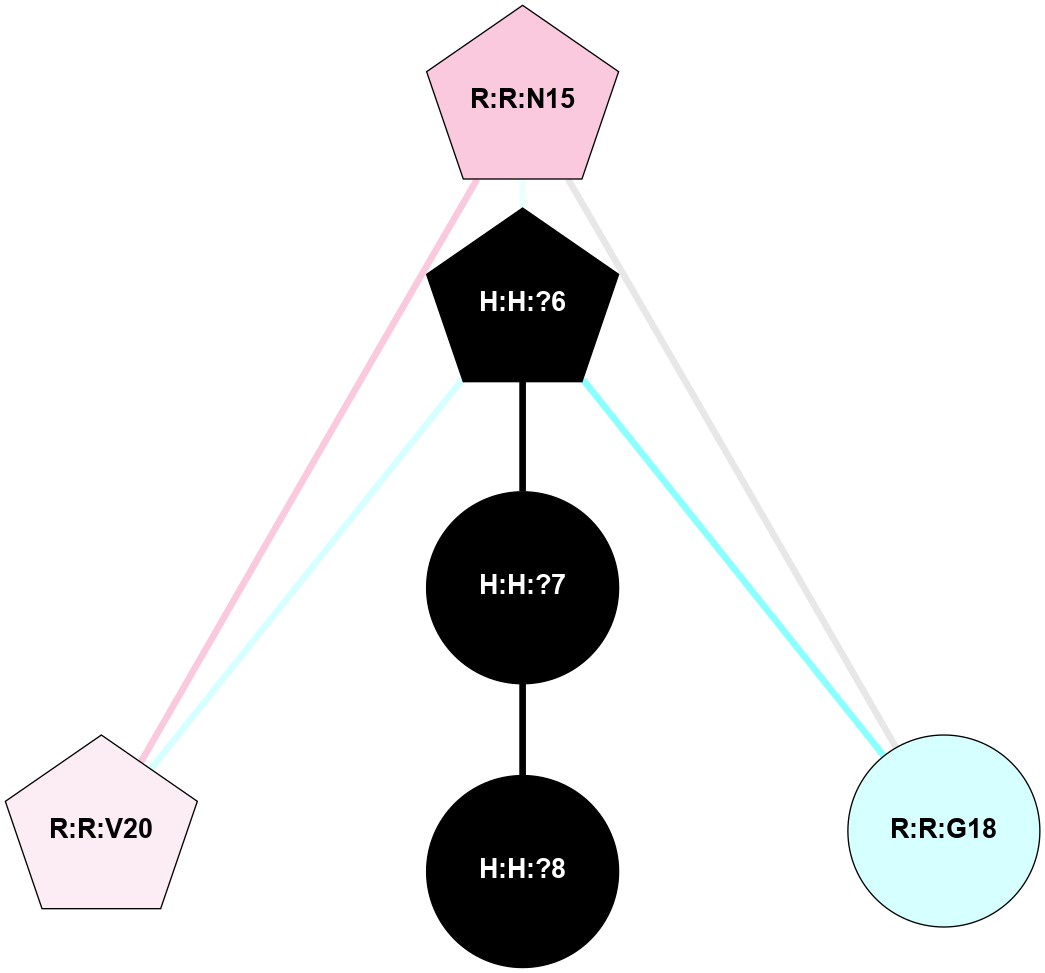
A 2D representation of the interactions of NAG in 2HPY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 2 | Yes | Yes | 7 | 6 | 2 | 2 | | H:H:?6 | R:R:N15 | 29.47 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?6 | R:R:G18 | 7.65 | 2 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?6 | R:R:V20 | 7.99 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?6 | H:H:?7 | 43.17 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?7 | H:H:?8 | 26.97 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 35.07 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
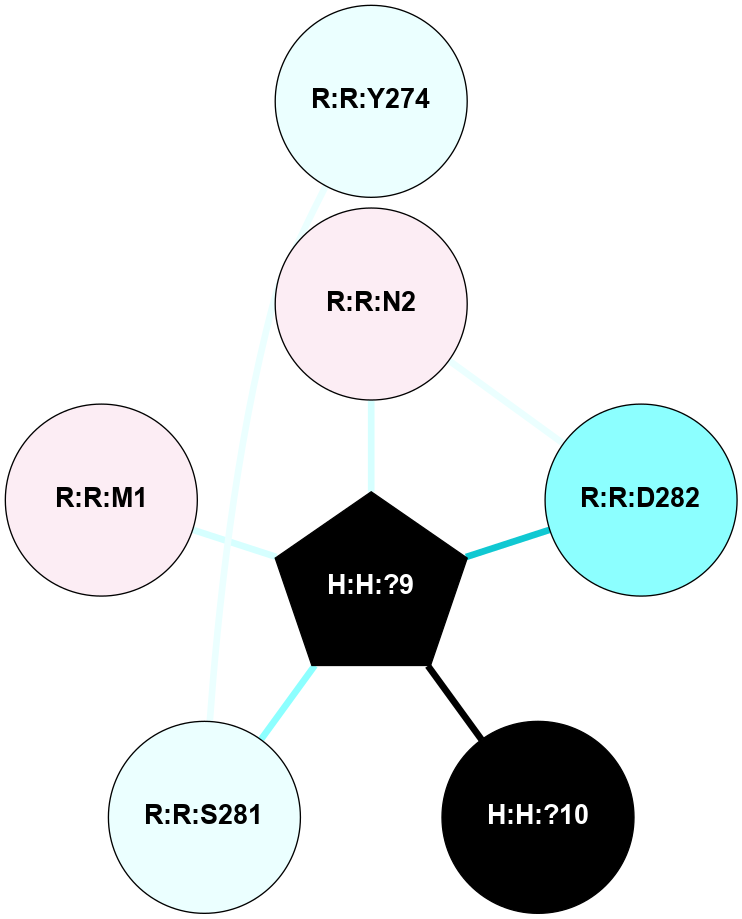
A 2D representation of the interactions of NAG in 2HPY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 6.73 | 10 | No | No | 2 | 6 | 1 | 1 | | H:H:?9 | R:R:N2 | 30.7 | 10 | Yes | No | 0 | 6 | 0 | 1 | | R:R:S281 | R:R:Y274 | 5.09 | 0 | No | No | 4 | 4 | 1 | 2 | | H:H:?9 | R:R:D282 | 6.07 | 10 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?10 | H:H:?9 | 36.53 | 0 | No | Yes | 0 | 0 | 1 | 0 | | H:H:?9 | R:R:S281 | 2.69 | 10 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?9 | R:R:M1 | 2.53 | 10 | Yes | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.70 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
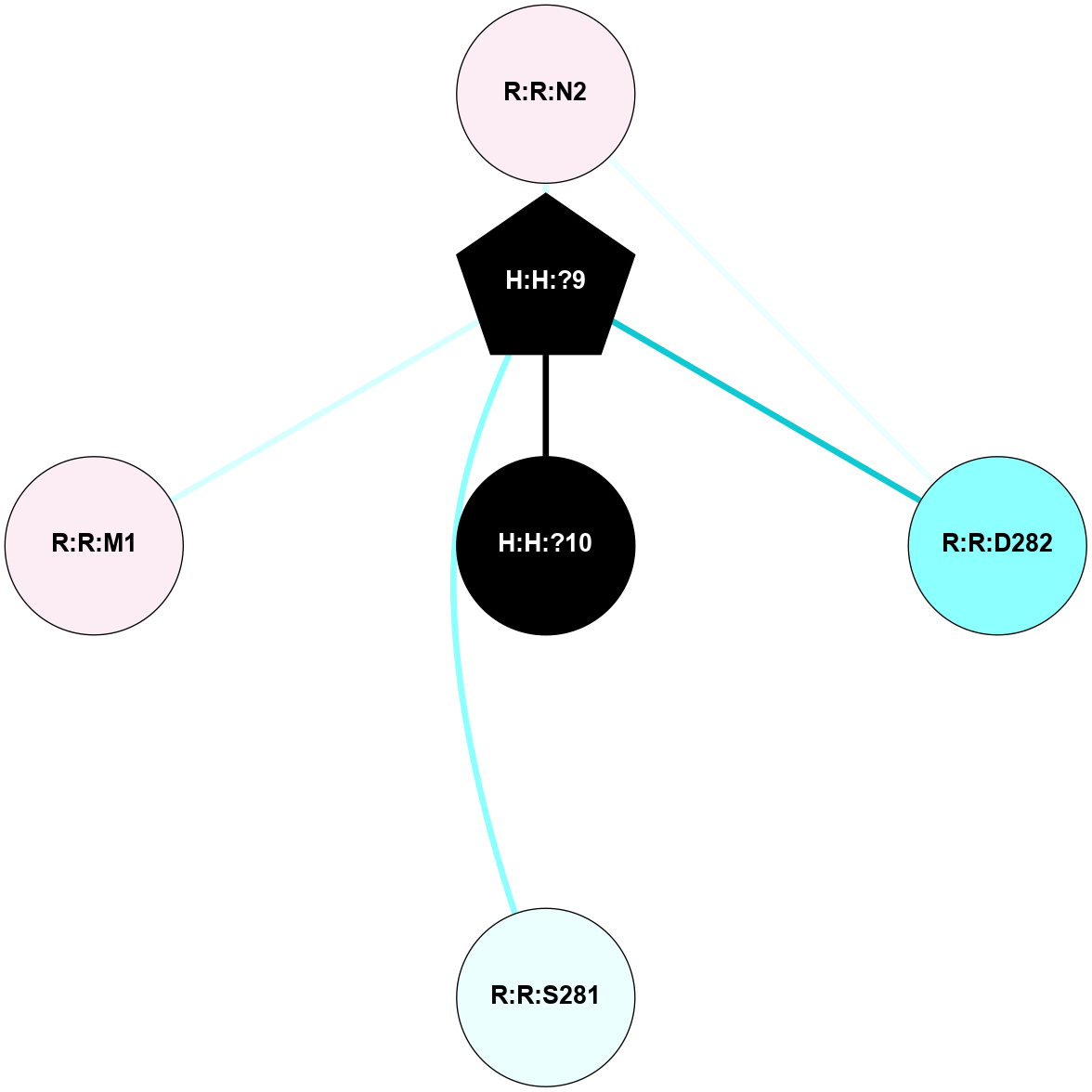
A 2D representation of the interactions of NAG in 2HPY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 6.73 | 10 | No | No | 2 | 6 | 2 | 2 | | H:H:?9 | R:R:N2 | 30.7 | 10 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?9 | R:R:D282 | 6.07 | 10 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?10 | H:H:?9 | 36.53 | 0 | No | Yes | 0 | 0 | 0 | 1 | | H:H:?9 | R:R:S281 | 2.69 | 10 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?9 | R:R:M1 | 2.53 | 10 | Yes | No | 0 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 36.53 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2I35 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 3.8 | 2006-10-17 | doi.org/10.1073/pnas.0608022103 |
|
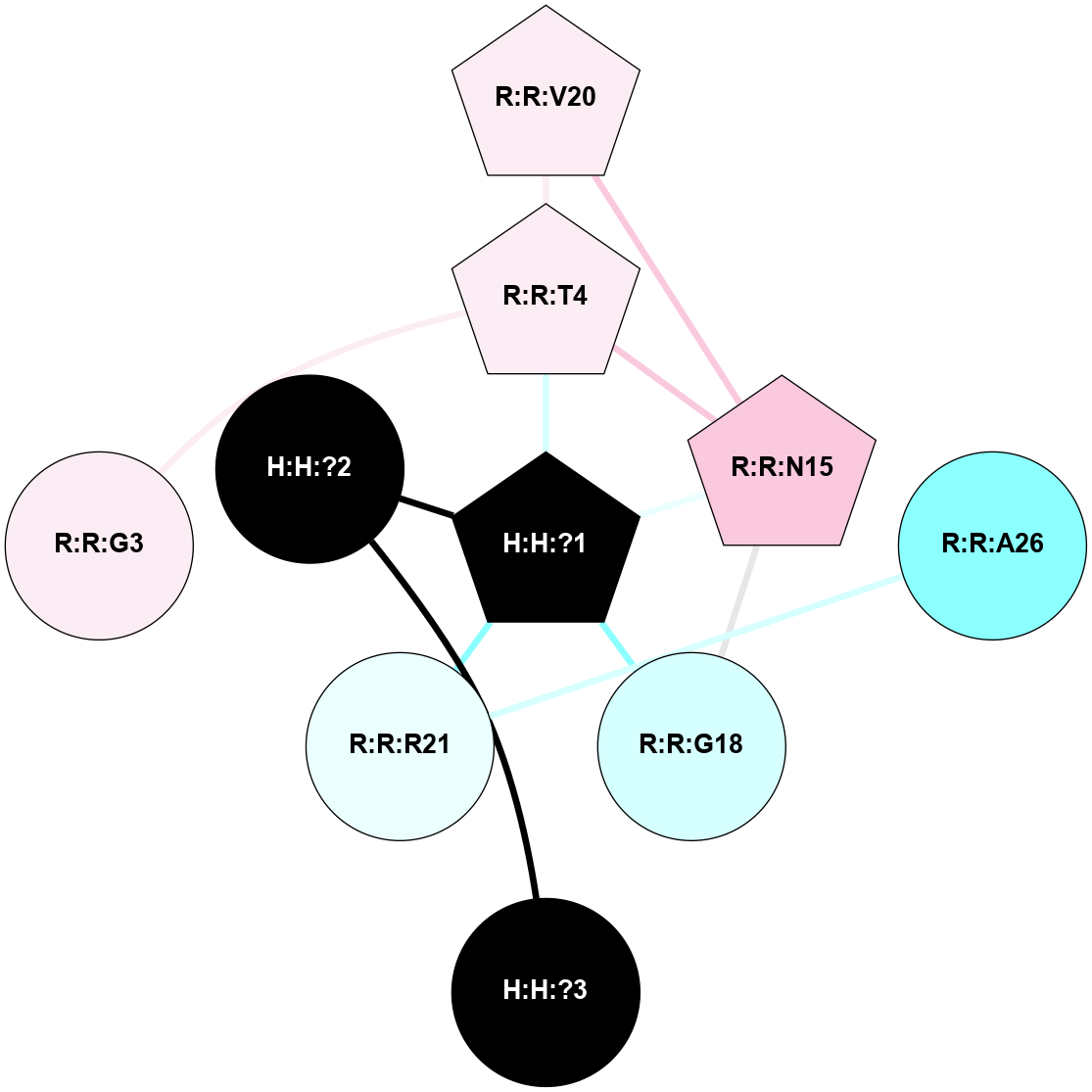
A 2D representation of the interactions of NAG in 2I35
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 33.21 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:T4 | 3.95 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:N15 | 24.56 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 7.65 | 2 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:R21 | 5.43 | 2 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | H:H:?3 | 30.82 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:N15 | R:R:T4 | 5.85 | 2 | Yes | Yes | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 7.93 | 2 | Yes | Yes | 6 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 11.82 | 2 | Yes | Yes | 7 | 6 | 1 | 2 | | R:R:A26 | R:R:R21 | 4.15 | 0 | No | No | 2 | 4 | 2 | 1 | | R:R:G3 | R:R:T4 | 1.82 | 0 | No | Yes | 6 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 14.96 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
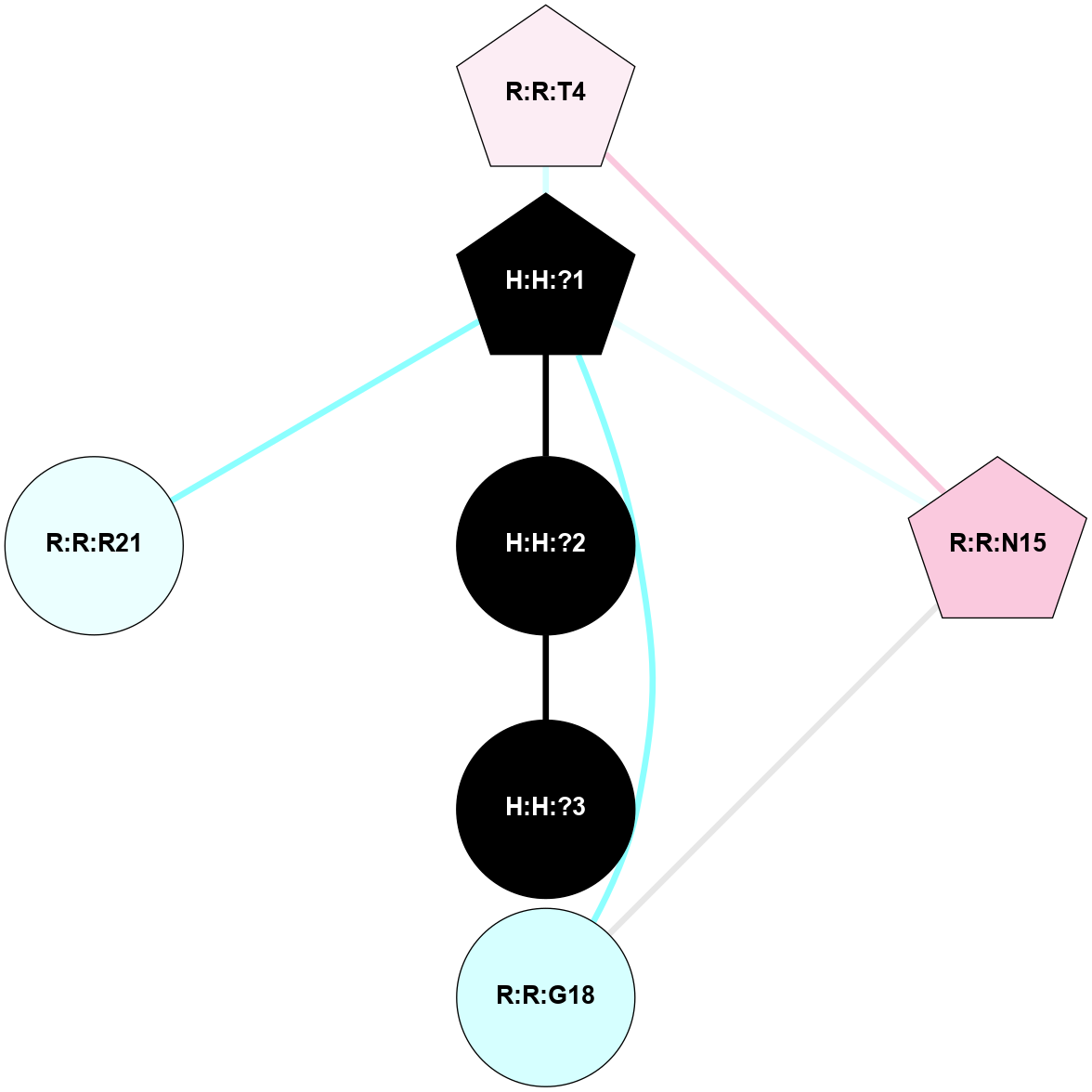
A 2D representation of the interactions of NAG in 2I35
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 33.21 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:T4 | 3.95 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 24.56 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 7.65 | 2 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:R21 | 5.43 | 2 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?2 | H:H:?3 | 30.82 | 0 | No | No | 0 | 0 | 0 | 1 | | R:R:N15 | R:R:T4 | 5.85 | 2 | Yes | Yes | 7 | 6 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 32.02 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
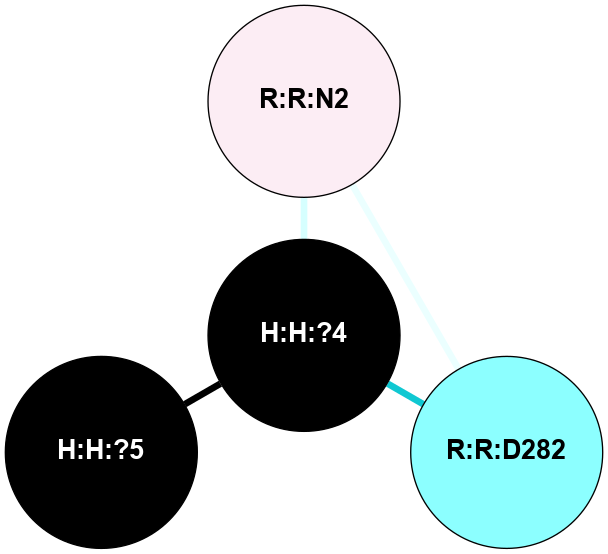
A 2D representation of the interactions of NAG in 2I35
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | H:H:?5 | 37.64 | 6 | No | No | 0 | 0 | 0 | 1 | | H:H:?4 | R:R:N2 | 25.79 | 6 | No | No | 0 | 6 | 0 | 1 | | H:H:?4 | R:R:D282 | 13.35 | 6 | No | No | 0 | 2 | 0 | 1 | | R:R:D282 | R:R:N2 | 8.08 | 6 | No | No | 2 | 6 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 25.59 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 2I35
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | H:H:?5 | 37.64 | 6 | No | No | 0 | 0 | 1 | 0 | | H:H:?4 | R:R:N2 | 25.79 | 6 | No | No | 0 | 6 | 1 | 2 | | H:H:?4 | R:R:D282 | 13.35 | 6 | No | No | 0 | 2 | 1 | 2 | | R:R:D282 | R:R:N2 | 8.08 | 6 | No | No | 2 | 6 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 37.64 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2I36 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | - | 4.1 | 2006-10-17 | doi.org/10.1073/pnas.0608022103 |
|
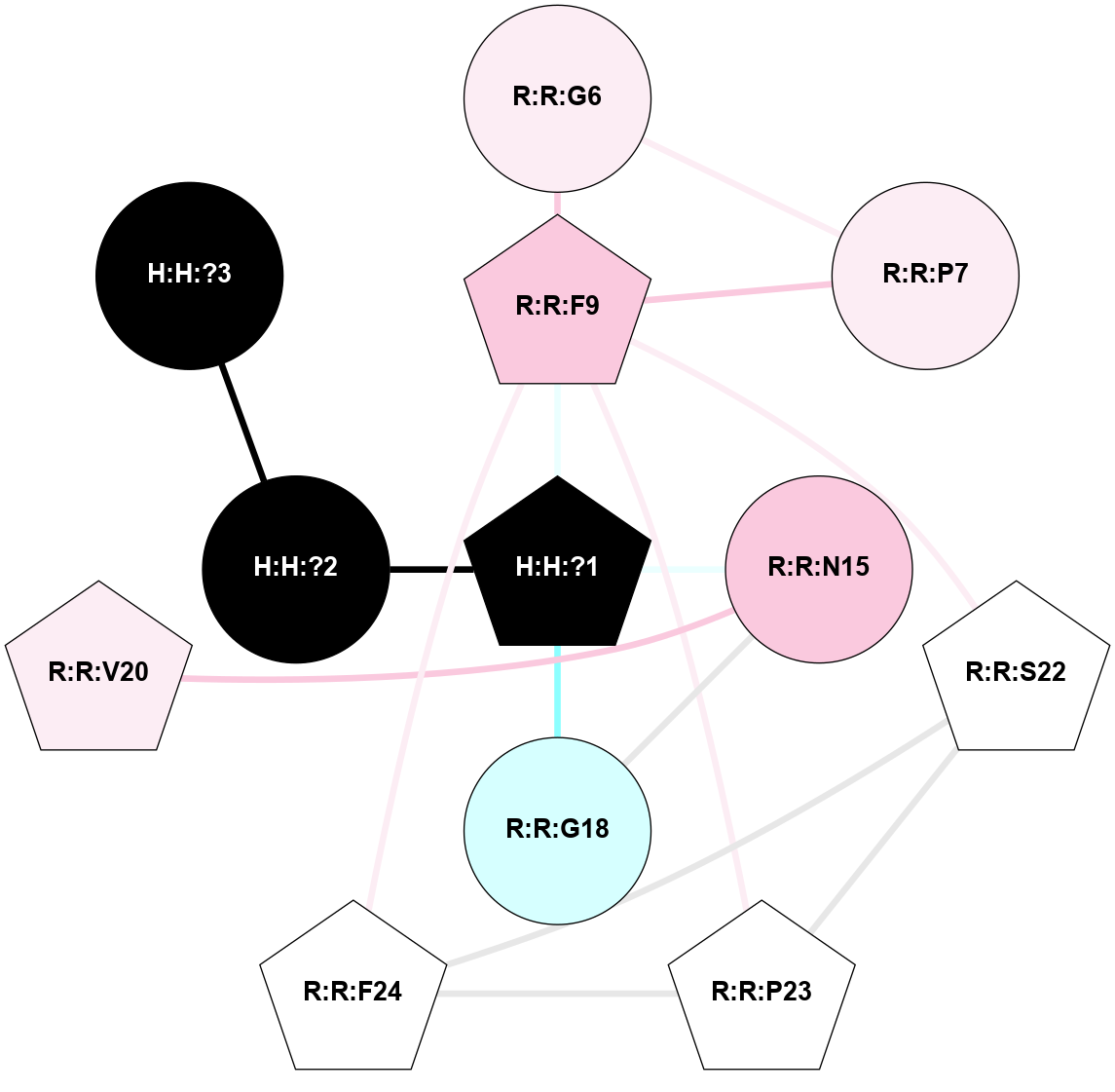
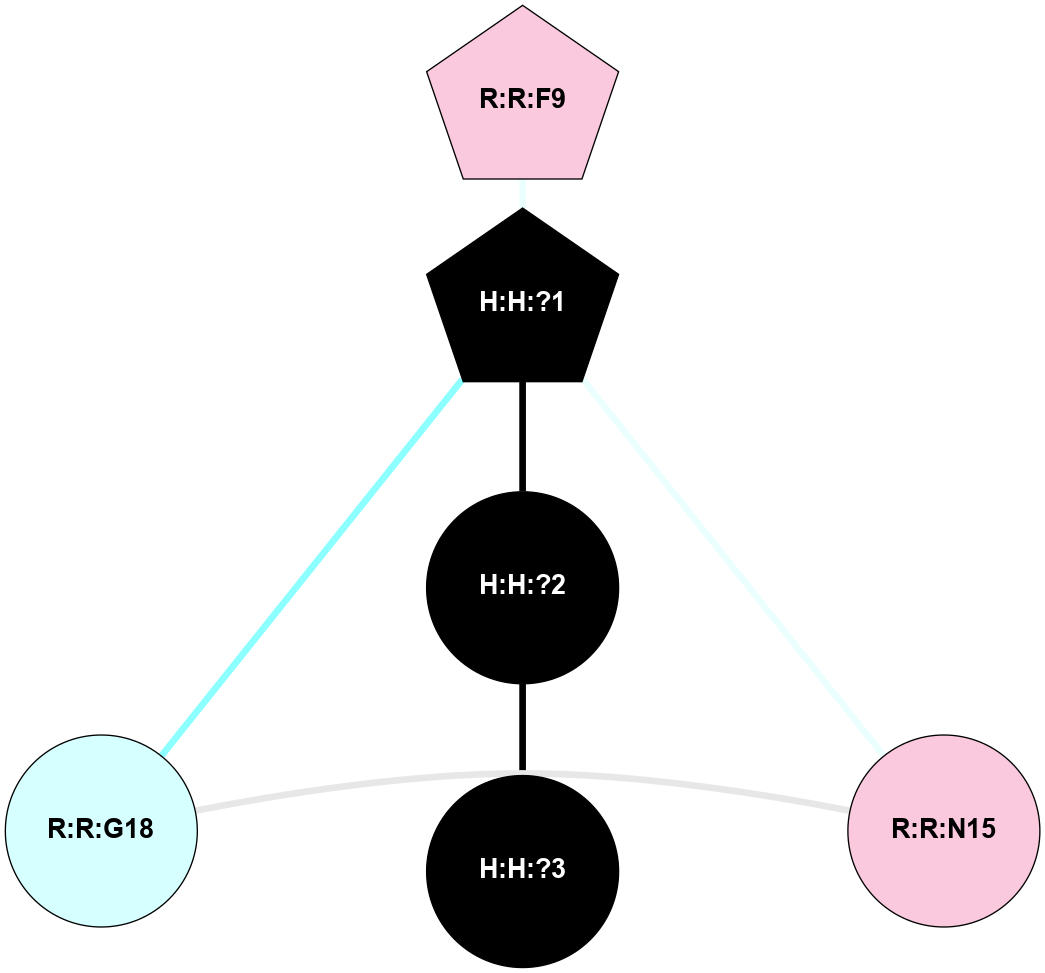
A 2D representation of the interactions of NAG in 2I36
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:G6 | R:R:P7 | 4.06 | 2 | No | No | 6 | 6 | 2 | 2 | | R:R:F9 | R:R:G6 | 9.03 | 2 | Yes | No | 7 | 6 | 1 | 2 | | R:R:F9 | R:R:P7 | 7.22 | 2 | Yes | No | 7 | 6 | 1 | 2 | | R:R:F9 | R:R:S22 | 6.61 | 2 | Yes | Yes | 7 | 5 | 1 | 2 | | R:R:F9 | R:R:P23 | 7.22 | 2 | Yes | Yes | 7 | 5 | 1 | 2 | | R:R:F24 | R:R:F9 | 5.36 | 2 | Yes | Yes | 5 | 7 | 2 | 1 | | H:H:?1 | R:R:F9 | 3.27 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:G18 | R:R:N15 | 3.39 | 2 | No | No | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 5.91 | 2 | No | Yes | 7 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 28.24 | 2 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 3.06 | 2 | Yes | No | 0 | 3 | 0 | 1 | | R:R:P23 | R:R:S22 | 5.34 | 2 | Yes | Yes | 5 | 5 | 2 | 2 | | R:R:F24 | R:R:S22 | 5.28 | 2 | Yes | Yes | 5 | 5 | 2 | 2 | | R:R:F24 | R:R:P23 | 8.67 | 2 | Yes | Yes | 5 | 5 | 2 | 2 | | H:H:?1 | H:H:?2 | 36.53 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 33.11 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.77 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 2I36
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:F9 | 3.27 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | R:R:G18 | R:R:N15 | 3.39 | 2 | No | No | 3 | 7 | 2 | 2 | | H:H:?1 | R:R:N15 | 28.24 | 2 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 3.06 | 2 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | H:H:?2 | 36.53 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 33.11 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 34.82 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
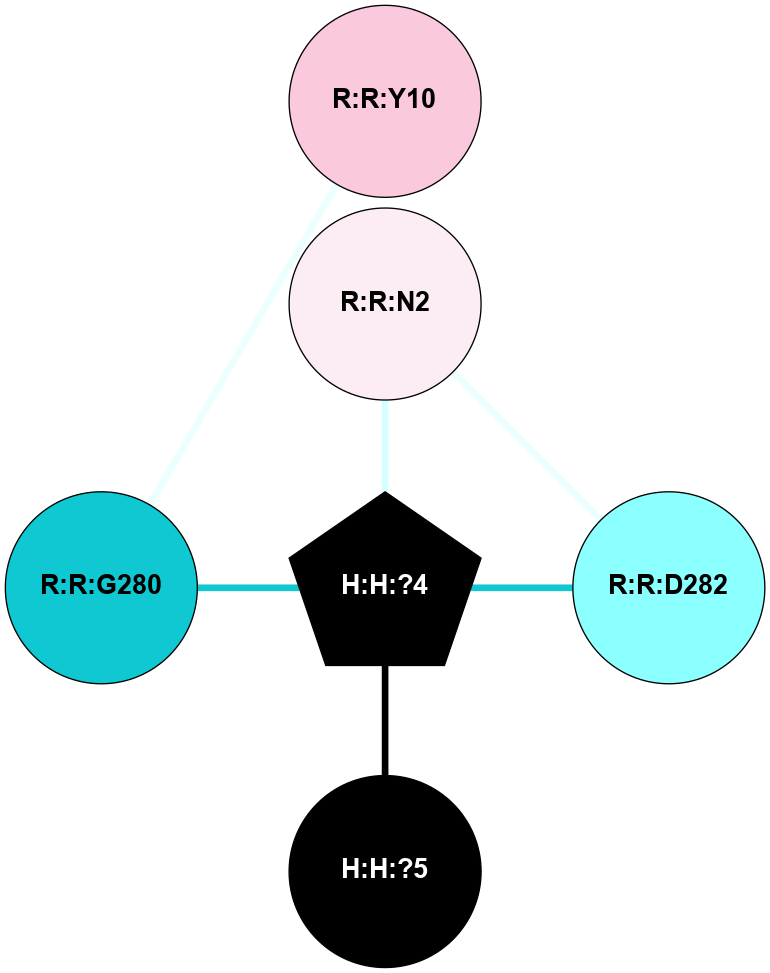
A 2D representation of the interactions of NAG in 2I36
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 10.77 | 6 | No | No | 2 | 6 | 1 | 1 | | H:H:?4 | R:R:N2 | 30.7 | 6 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?4 | R:R:D282 | 7.28 | 6 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?4 | H:H:?5 | 35.42 | 6 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?4 | R:R:G280 | 1.53 | 6 | Yes | No | 0 | 1 | 0 | 1 | | R:R:G280 | R:R:Y10 | 1.45 | 0 | No | No | 1 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.73 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
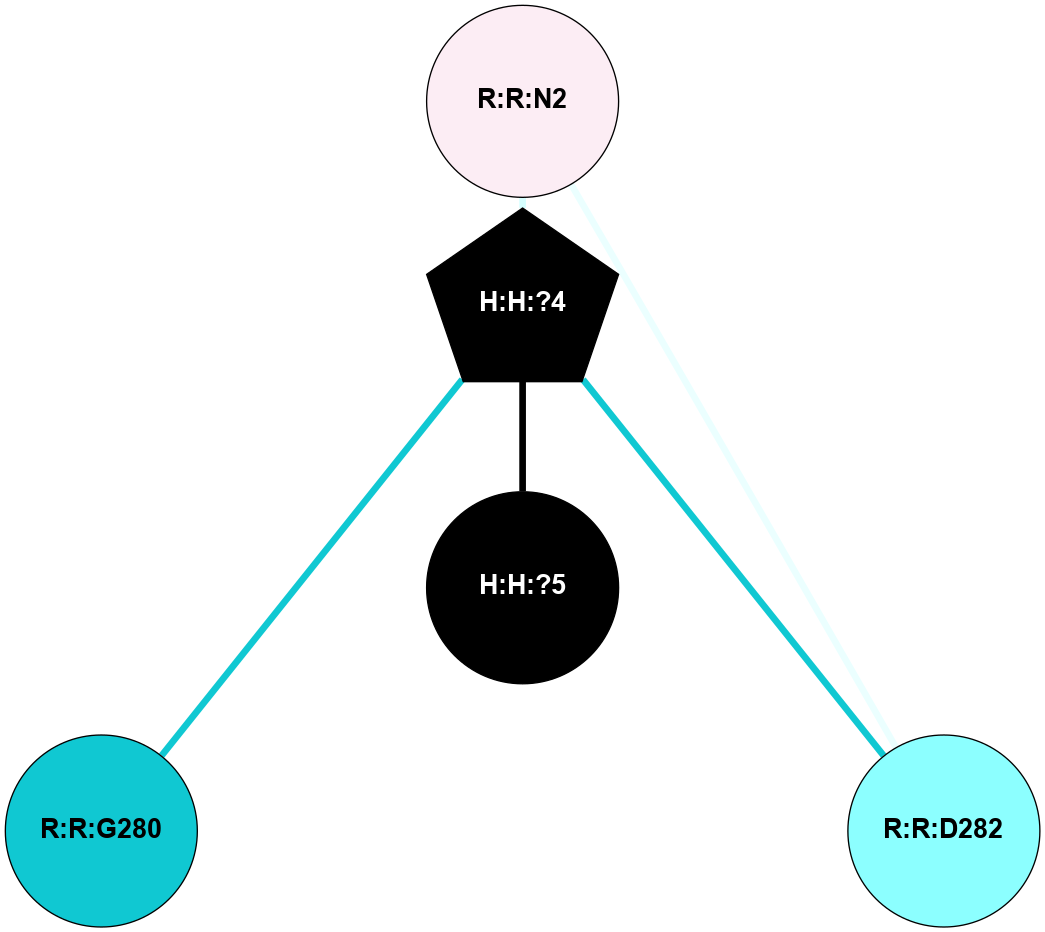
A 2D representation of the interactions of NAG in 2I36
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 10.77 | 6 | No | No | 2 | 6 | 2 | 2 | | H:H:?4 | R:R:N2 | 30.7 | 6 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?4 | R:R:D282 | 7.28 | 6 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?4 | H:H:?5 | 35.42 | 6 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?4 | R:R:G280 | 1.53 | 6 | Yes | No | 0 | 1 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 35.42 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2I37 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | - | 4.15 | 2006-10-17 | doi.org/10.1073/pnas.0608022103 |
|
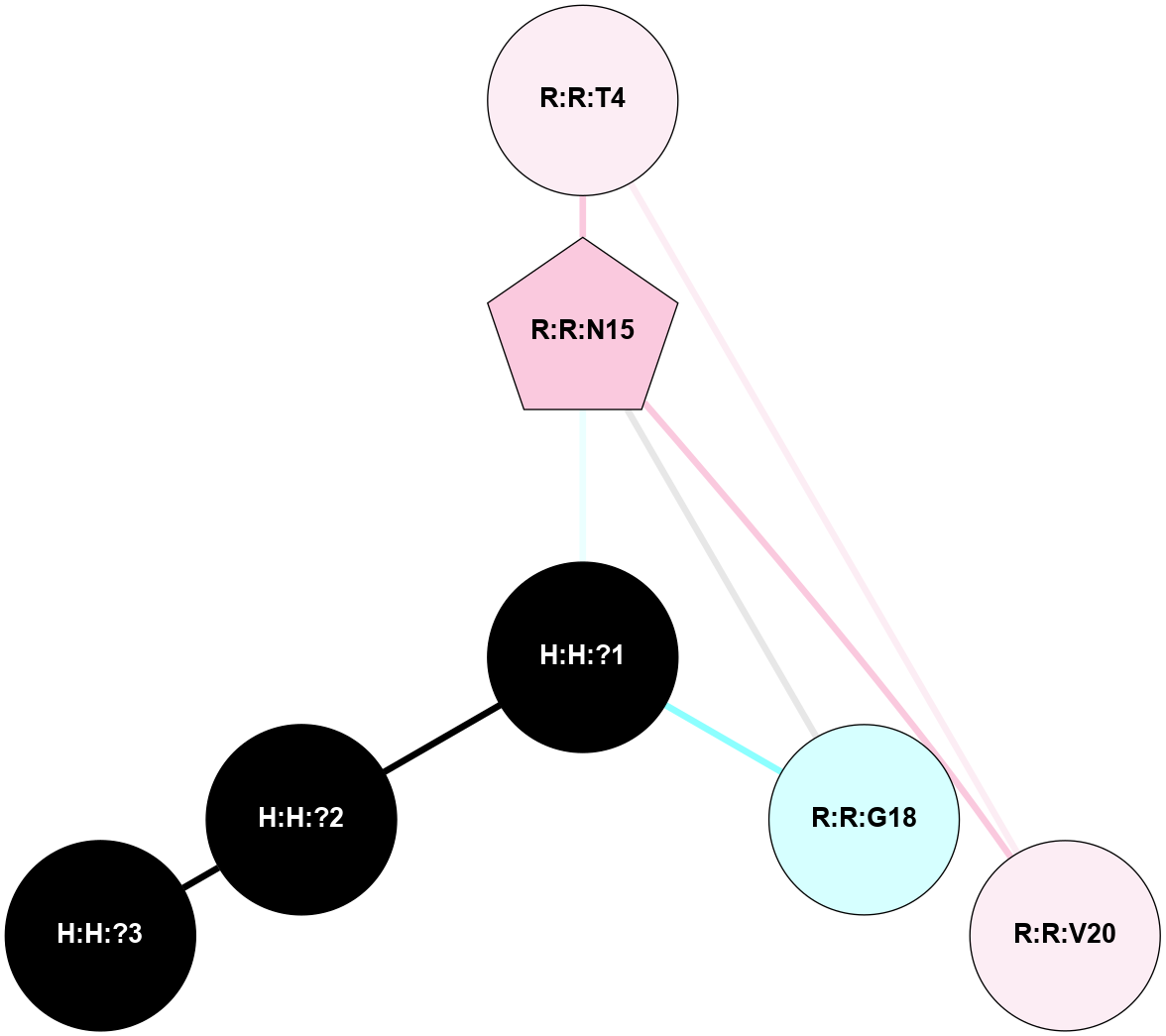
A 2D representation of the interactions of NAG in 2I37
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 2.92 | 2 | Yes | No | 7 | 6 | 1 | 2 | | R:R:T4 | R:R:V20 | 9.52 | 2 | No | No | 6 | 6 | 2 | 2 | | R:R:G18 | R:R:N15 | 3.39 | 2 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 8.87 | 2 | Yes | No | 7 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 29.47 | 2 | No | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 3.06 | 2 | No | No | 0 | 3 | 0 | 1 | | H:H:?1 | H:H:?2 | 34.31 | 2 | No | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 30.56 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 22.28 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 2I37
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:G18 | R:R:N15 | 3.39 | 2 | No | Yes | 3 | 7 | 2 | 2 | | H:H:?1 | R:R:N15 | 29.47 | 2 | No | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 3.06 | 2 | No | No | 0 | 3 | 1 | 2 | | H:H:?1 | H:H:?2 | 34.31 | 2 | No | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 30.56 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 32.44 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 2I37
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 13.46 | 4 | No | No | 2 | 6 | 1 | 1 | | H:H:?4 | R:R:N2 | 30.7 | 4 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?5 | R:R:H278 | 3.45 | 4 | No | No | 0 | 3 | 1 | 2 | | H:H:?4 | R:R:S281 | 4.03 | 4 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?5 | R:R:S281 | 4.03 | 4 | No | No | 0 | 4 | 1 | 1 | | H:H:?4 | R:R:D282 | 4.85 | 4 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?4 | H:H:?5 | 30.99 | 4 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.64 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 2I37
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 13.46 | 4 | No | No | 2 | 6 | 2 | 2 | | H:H:?4 | R:R:N2 | 30.7 | 4 | Yes | No | 0 | 6 | 1 | 2 | | R:R:H278 | R:R:Y274 | 25.04 | 0 | No | No | 3 | 4 | 1 | 2 | | R:R:H278 | R:R:T277 | 5.48 | 0 | No | No | 3 | 5 | 1 | 2 | | H:H:?5 | R:R:H278 | 3.45 | 4 | No | No | 0 | 3 | 0 | 1 | | H:H:?4 | R:R:S281 | 4.03 | 4 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?5 | R:R:S281 | 4.03 | 4 | No | No | 0 | 4 | 0 | 1 | | H:H:?4 | R:R:D282 | 4.85 | 4 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?4 | H:H:?5 | 30.99 | 4 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 12.82 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2J4Y | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 3.4 | 2007-09-25 | doi.org/10.1016/j.jmb.2007.03.007 |
|
A 2D representation of the interactions of NAG in 2J4Y
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:N15 | 22.1 | 0 | No | Yes | 0 | 7 | 0 | 1 | | R:R:N15 | R:R:T4 | 8.77 | 2 | Yes | No | 7 | 6 | 1 | 2 | | R:R:T4 | R:R:V20 | 14.28 | 2 | No | No | 6 | 6 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 0 | No | Yes | 3 | 7 | 2 | 1 | | R:R:N15 | R:R:V20 | 8.87 | 2 | Yes | No | 7 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 22.10 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2PED | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 9-cis-Retinal | - | - | 2.95 | 2007-10-30 | doi.org/10.1529/biophysj.107.108225 |
|
A 2D representation of the interactions of NAG in 2PED
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 11.11 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?6 | R:R:T4 | 3.95 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 0 | No | Yes | 8 | 6 | 2 | 1 | | R:R:F13 | R:R:V20 | 5.24 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 6.19 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 13.3 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | H:H:?6 | R:R:N15 | 29.47 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?6 | R:R:G18 | 7.65 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 10.09 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?6 | R:R:V20 | 9.33 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?6 | H:H:?7 | 46.49 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?7 | H:H:?8 | 26.97 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?7 | R:R:R21 | 3.26 | 0 | No | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 19.38 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
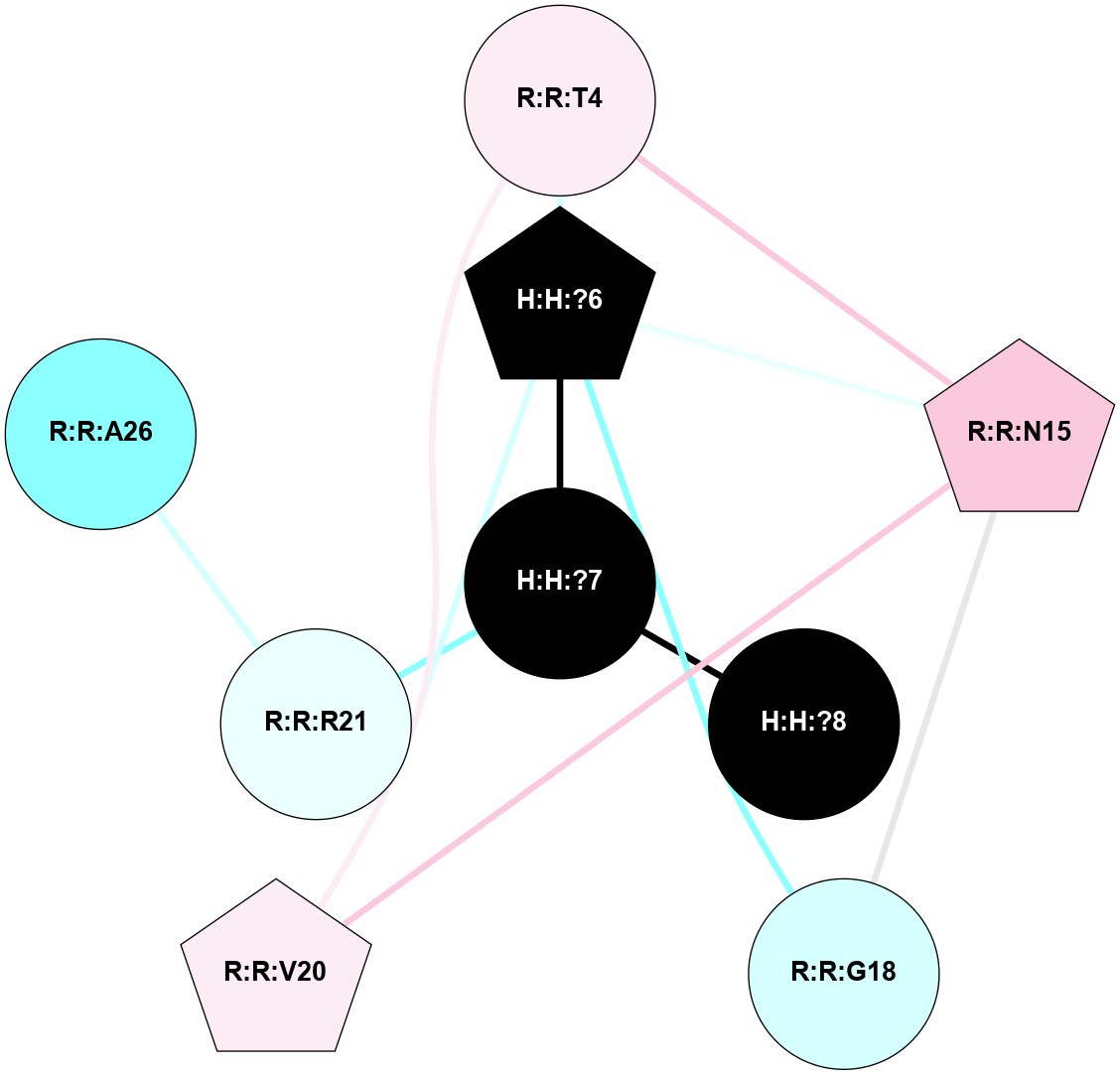
A 2D representation of the interactions of NAG in 2PED
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 1 | Yes | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 11.11 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?6 | R:R:T4 | 3.95 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 13.3 | 1 | Yes | Yes | 7 | 6 | 2 | 2 | | H:H:?6 | R:R:N15 | 29.47 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?6 | R:R:G18 | 7.65 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?6 | R:R:V20 | 9.33 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | R:R:A26 | R:R:R21 | 6.91 | 0 | No | No | 2 | 4 | 2 | 1 | | H:H:?6 | H:H:?7 | 46.49 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?7 | H:H:?8 | 26.97 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?7 | R:R:R21 | 3.26 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 25.57 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
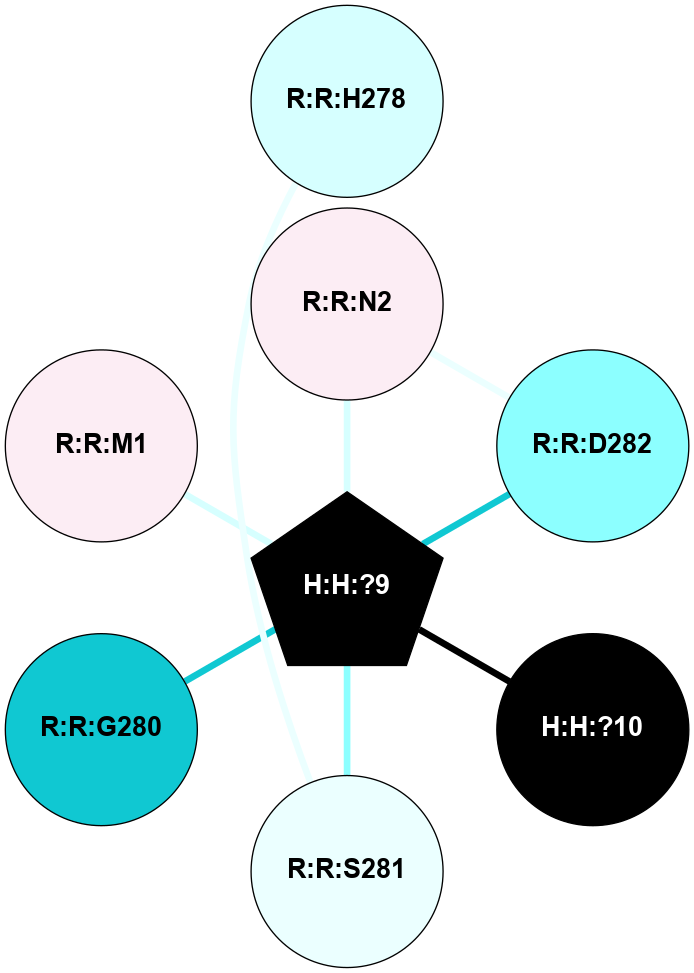
A 2D representation of the interactions of NAG in 2PED
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 14.81 | 10 | No | No | 2 | 6 | 1 | 1 | | H:H:?9 | R:R:N2 | 28.24 | 10 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?9 | R:R:D282 | 7.28 | 10 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?10 | H:H:?9 | 34.31 | 0 | No | Yes | 0 | 0 | 1 | 0 | | R:R:H278 | R:R:S281 | 2.79 | 0 | No | No | 3 | 4 | 2 | 1 | | H:H:?9 | R:R:S281 | 2.69 | 10 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?9 | R:R:G280 | 1.53 | 10 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?9 | R:R:M1 | 1.26 | 10 | Yes | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.55 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
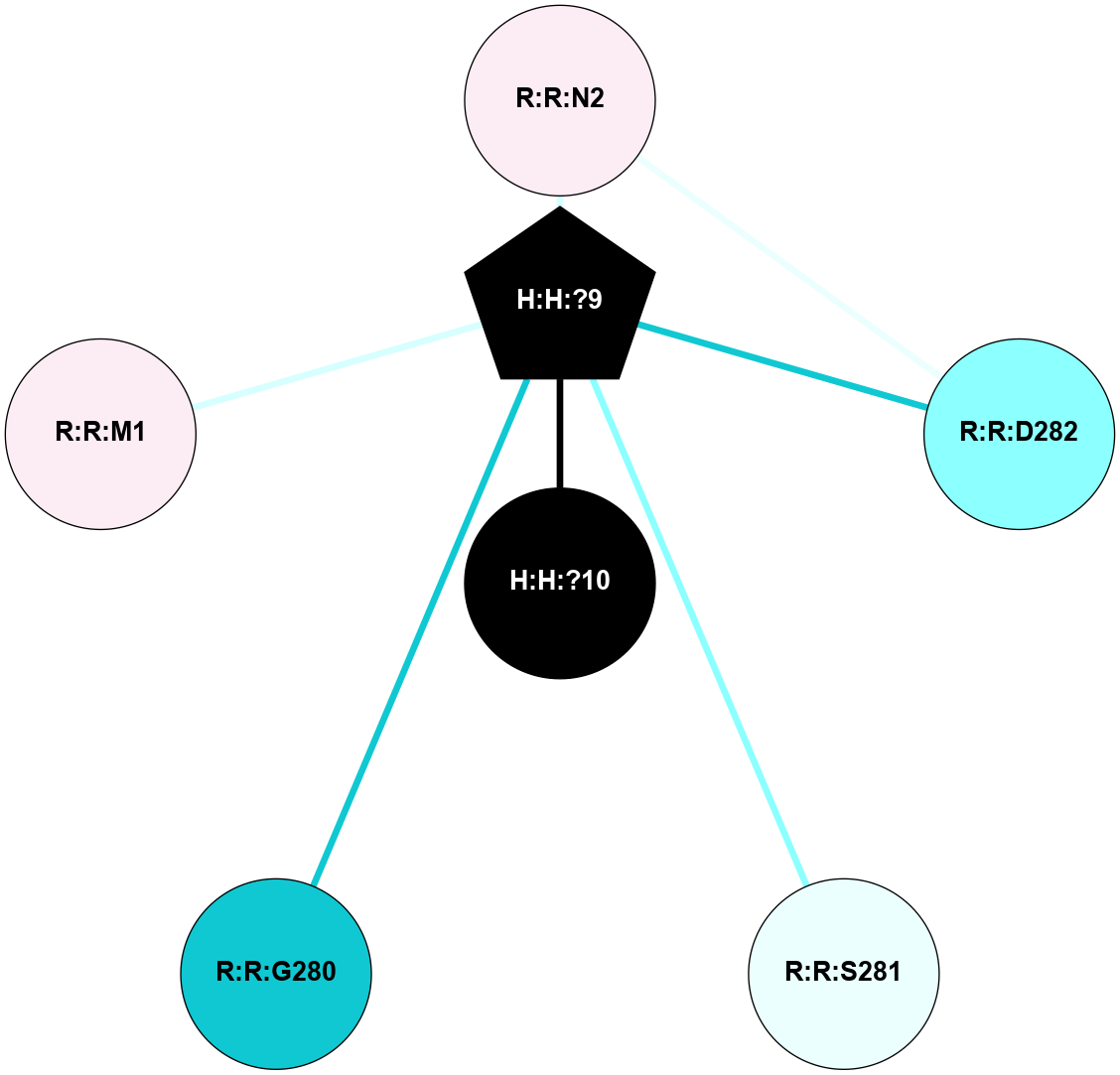
A 2D representation of the interactions of NAG in 2PED
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 14.81 | 10 | No | No | 2 | 6 | 2 | 2 | | H:H:?9 | R:R:N2 | 28.24 | 10 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?9 | R:R:D282 | 7.28 | 10 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?10 | H:H:?9 | 34.31 | 0 | No | Yes | 0 | 0 | 0 | 1 | | H:H:?9 | R:R:S281 | 2.69 | 10 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?9 | R:R:G280 | 1.53 | 10 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?9 | R:R:M1 | 1.26 | 10 | Yes | No | 0 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 34.31 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 2X72 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | Gt(CT) | 3 | 2011-03-16 | doi.org/10.1038/nature09795 |
|
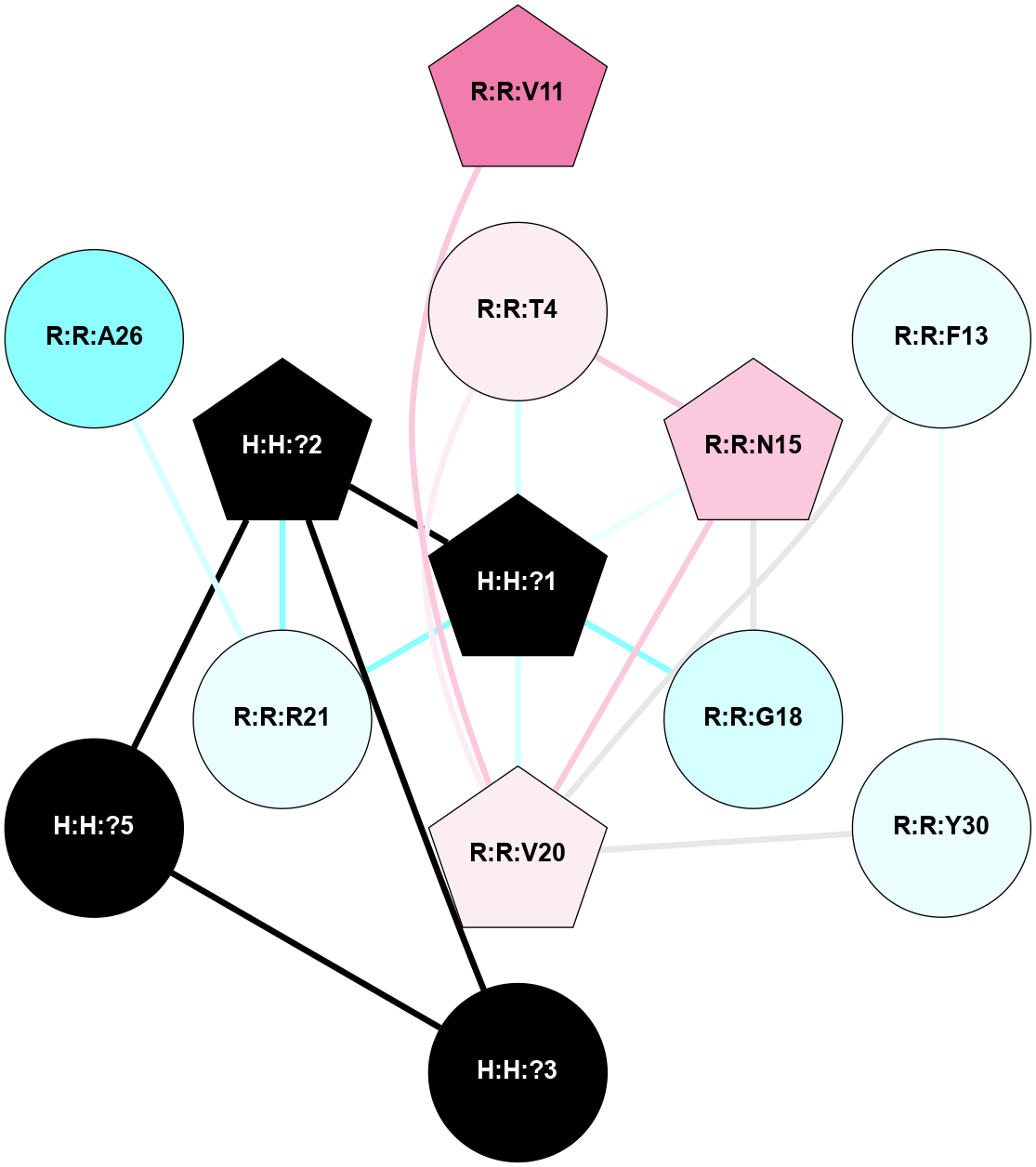
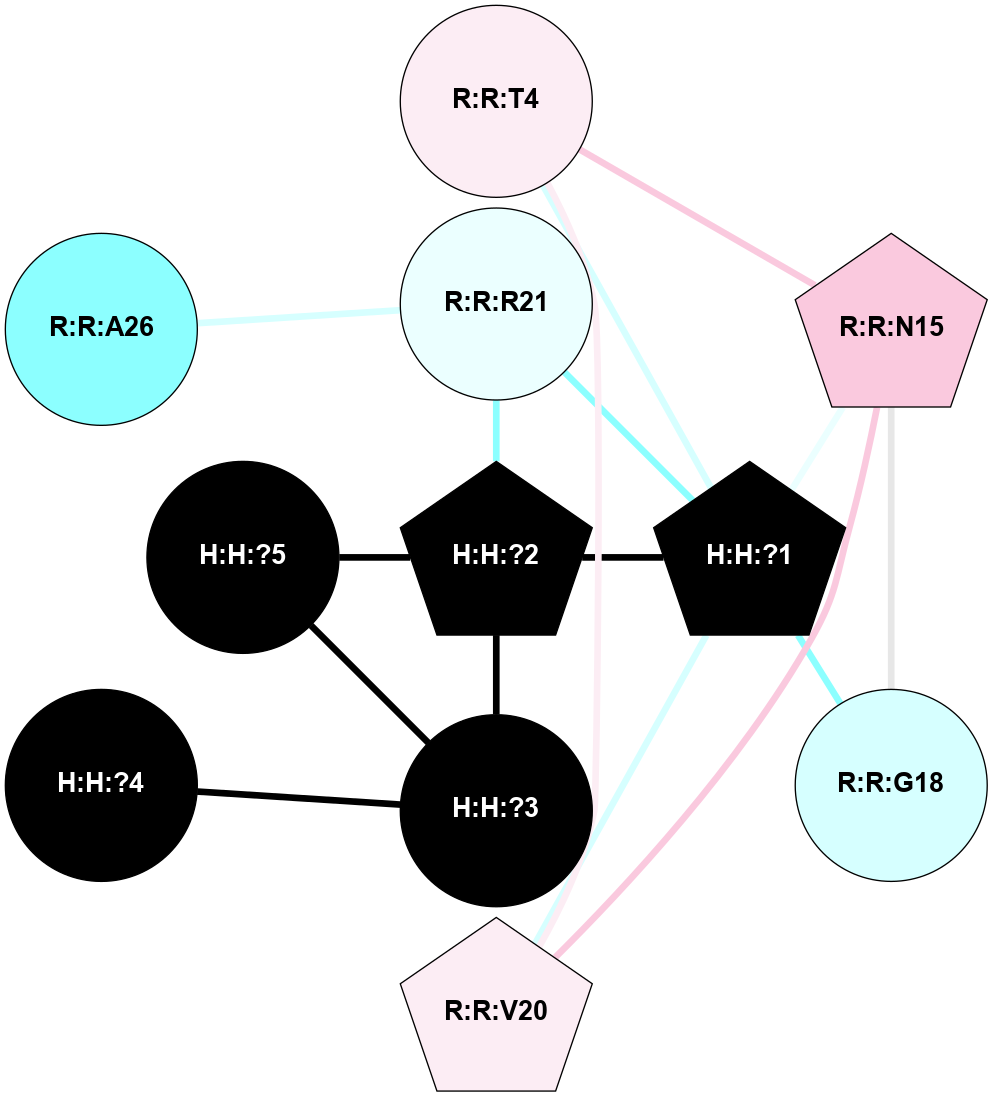
A 2D representation of the interactions of NAG in 2X72
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 6.59 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 0 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:F13 | R:R:V20 | 3.93 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 11.35 | 1 | No | No | 4 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 3.39 | 1 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 24.56 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 8.83 | 1 | Yes | No | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 6.66 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:R21 | 6.52 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:R21 | 8.69 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?1 | H:H:?2 | 39.85 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 30.82 | 1 | Yes | No | 0 | 0 | 1 | 2 | | H:H:?2 | H:H:?5 | 11.46 | 1 | Yes | No | 0 | 0 | 1 | 2 | | H:H:?3 | H:H:?5 | 28.07 | 1 | No | No | 0 | 0 | 2 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 15.05 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 19.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 2X72
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 1 | Yes | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 6.59 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 3.39 | 1 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 24.56 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 6.66 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:R21 | 6.52 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?2 | R:R:R21 | 8.69 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 39.85 | 1 | Yes | Yes | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 30.82 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?5 | 11.46 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?4 | 47.27 | 1 | No | No | 0 | 0 | 1 | 2 | | H:H:?3 | H:H:?5 | 28.07 | 1 | No | No | 0 | 0 | 1 | 1 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 22.70 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3C9L | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.65 | 2008-08-05 | doi.org/10.1107/S0907444908017162 |
|
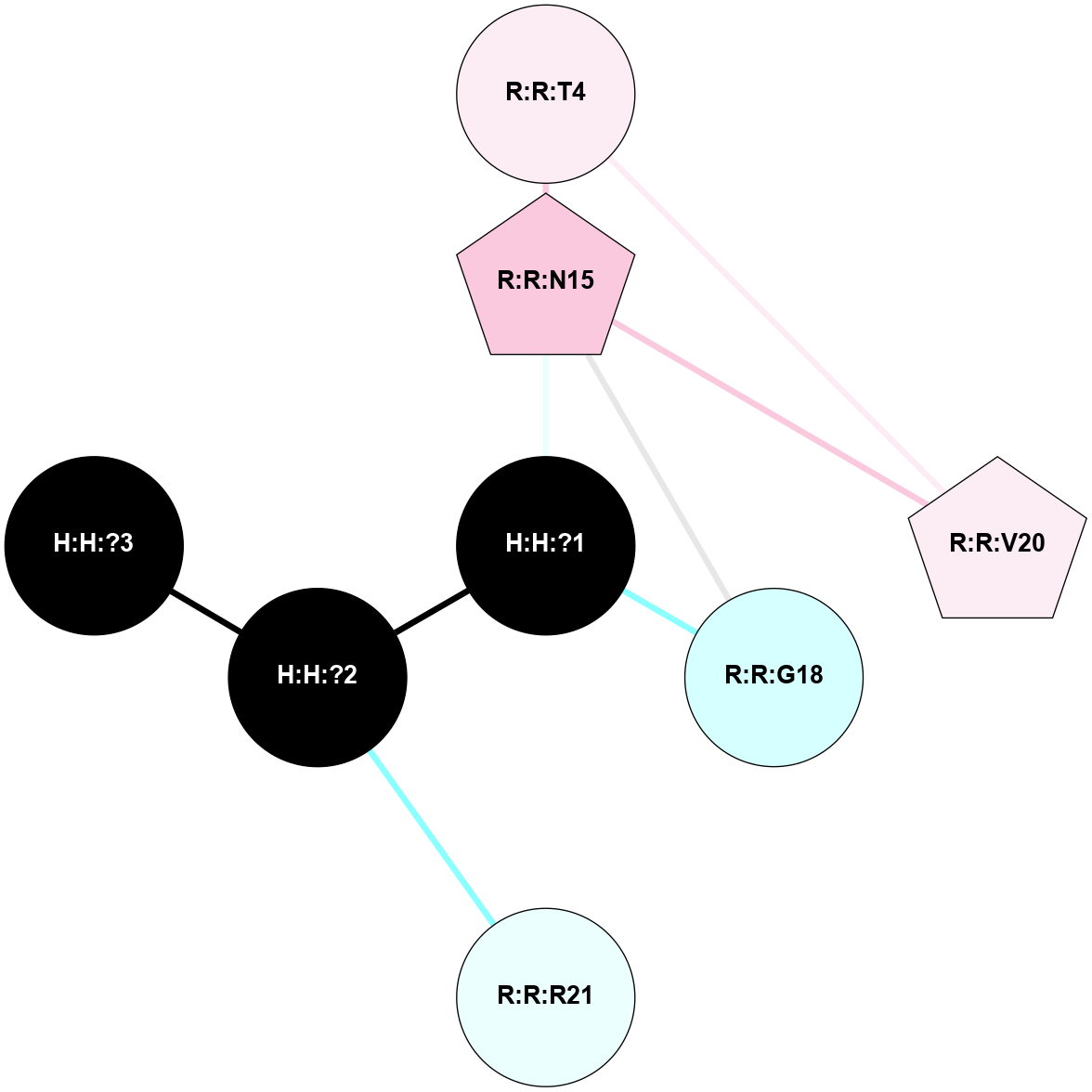
A 2D representation of the interactions of NAG in 3C9L
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 28.78 | 2 | No | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N15 | 24.56 | 2 | No | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 7.65 | 2 | No | No | 0 | 3 | 0 | 1 | | H:H:?2 | H:H:?3 | 32.11 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?2 | R:R:R21 | 5.43 | 0 | No | No | 0 | 4 | 1 | 2 | | R:R:N15 | R:R:T4 | 5.85 | 2 | Yes | No | 7 | 6 | 1 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 2 | Yes | Yes | 7 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 20.33 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
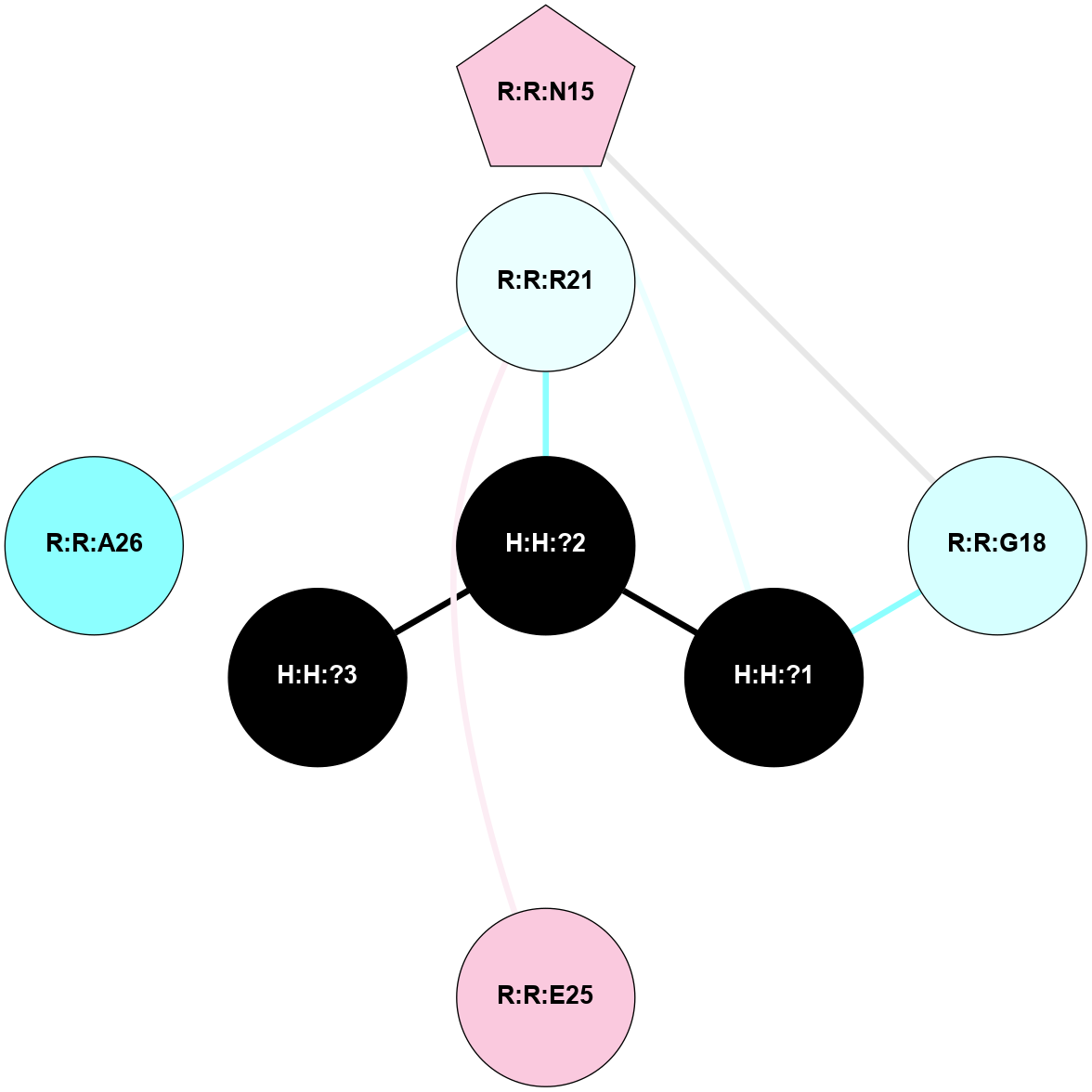
A 2D representation of the interactions of NAG in 3C9L
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 28.78 | 2 | No | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N15 | 24.56 | 2 | No | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 7.65 | 2 | No | No | 0 | 3 | 1 | 2 | | H:H:?2 | H:H:?3 | 32.11 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?2 | R:R:R21 | 5.43 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 2 | 2 | | R:R:E25 | R:R:R21 | 8.14 | 0 | No | No | 7 | 4 | 2 | 1 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 22.11 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
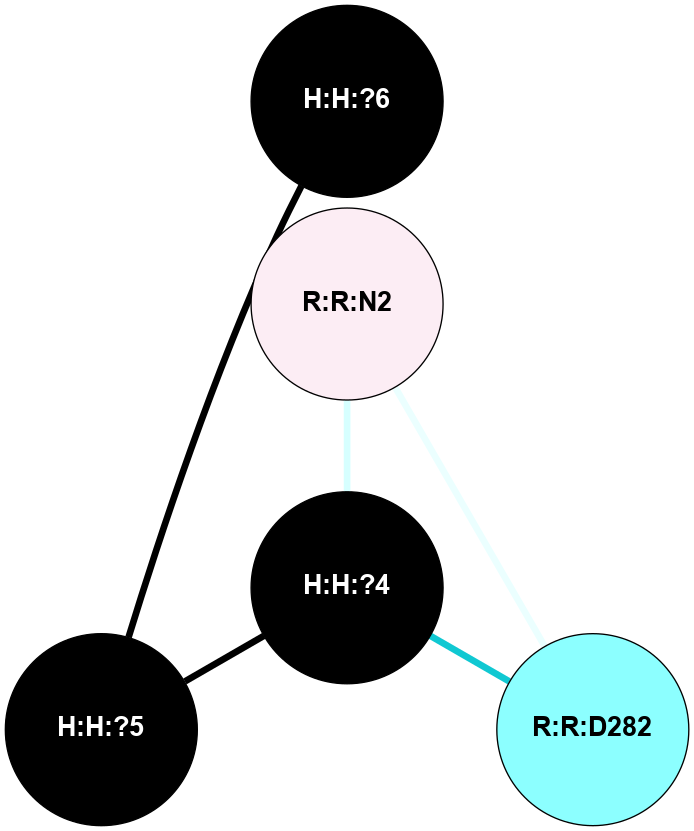
A 2D representation of the interactions of NAG in 3C9L
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | H:H:?5 | 38.74 | 4 | No | No | 0 | 0 | 0 | 1 | | H:H:?4 | R:R:N2 | 29.47 | 4 | No | No | 0 | 6 | 0 | 1 | | H:H:?4 | R:R:D282 | 9.71 | 4 | No | No | 0 | 2 | 0 | 1 | | H:H:?5 | H:H:?6 | 28.25 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:D282 | R:R:N2 | 16.16 | 4 | No | No | 2 | 6 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 25.97 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
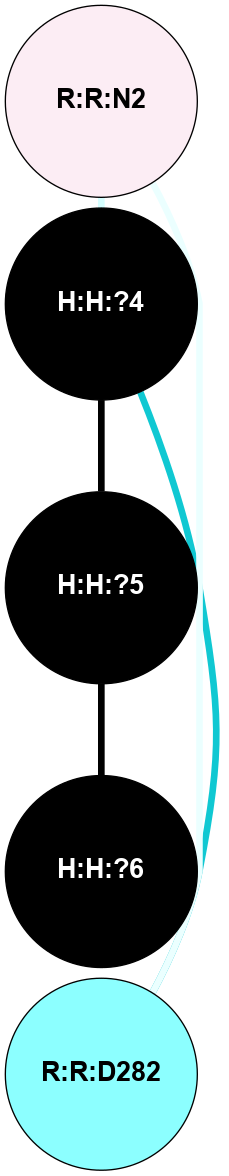
A 2D representation of the interactions of NAG in 3C9L
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | H:H:?5 | 38.74 | 4 | No | No | 0 | 0 | 1 | 0 | | H:H:?4 | R:R:N2 | 29.47 | 4 | No | No | 0 | 6 | 1 | 2 | | H:H:?4 | R:R:D282 | 9.71 | 4 | No | No | 0 | 2 | 1 | 2 | | H:H:?5 | H:H:?6 | 28.25 | 0 | No | No | 0 | 0 | 0 | 1 | | R:R:D282 | R:R:N2 | 16.16 | 4 | No | No | 2 | 6 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 33.50 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3C9M | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 3.4 | 2008-08-05 | doi.org/10.1107/S0907444908017162 |
|
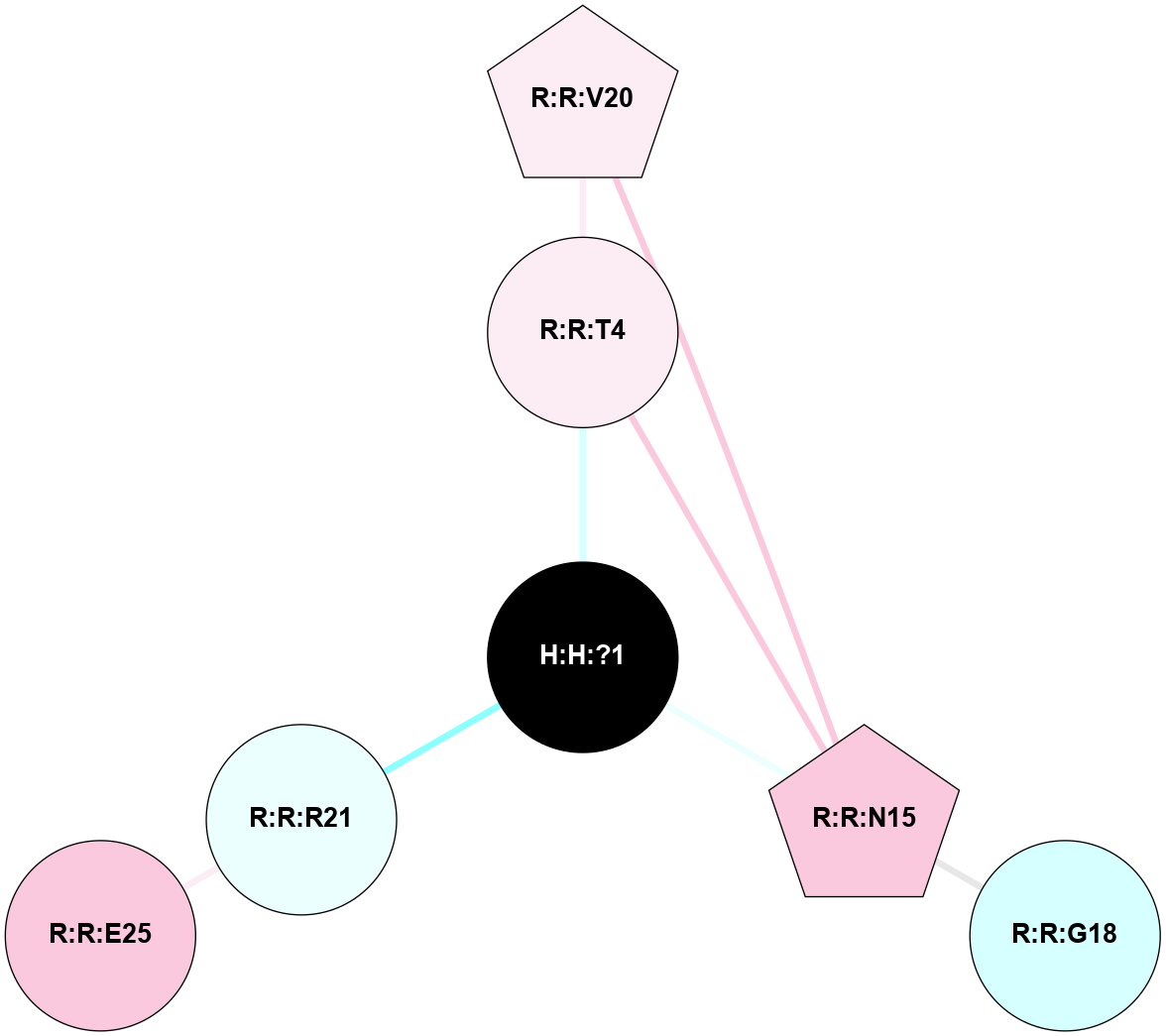
A 2D representation of the interactions of NAG in 3C9M
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:T4 | 3.95 | 1 | No | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:N15 | 24.56 | 1 | No | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:R21 | 4.35 | 1 | No | No | 0 | 4 | 0 | 1 | | R:R:N15 | R:R:T4 | 7.31 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 0 | No | Yes | 3 | 7 | 2 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 1 | 2 | | R:R:E25 | R:R:R21 | 8.14 | 0 | No | No | 7 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 10.95 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3CAP | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | - | 2.9 | 2008-06-24 | doi.org/10.1038/nature07063 |
|
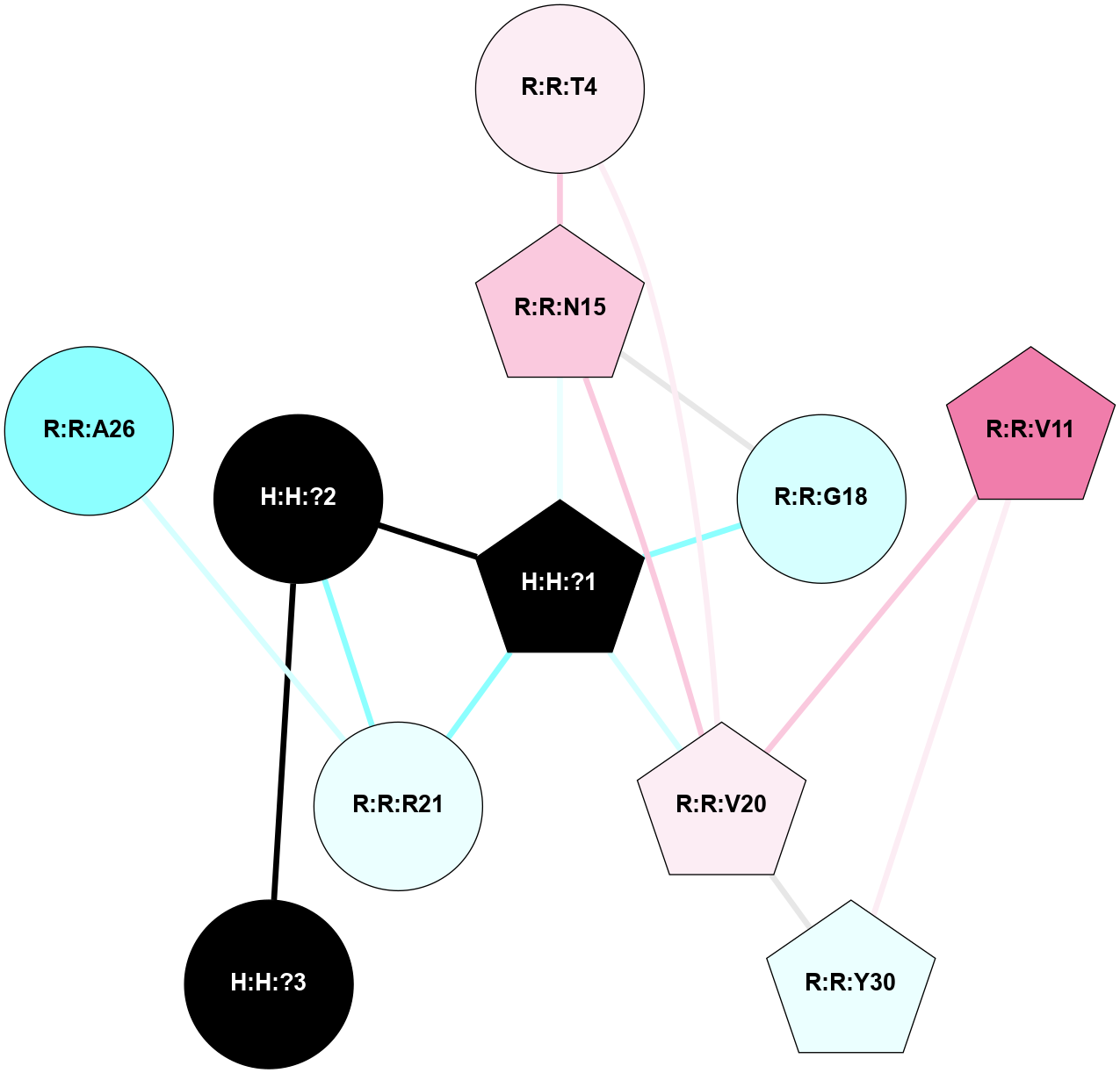
A 2D representation of the interactions of NAG in 3CAP
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 5.85 | 1 | Yes | No | 7 | 6 | 1 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 2 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:V11 | R:R:Y30 | 5.05 | 1 | Yes | Yes | 8 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 27.02 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 11.36 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 7.99 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:R21 | 4.35 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:R21 | 8.69 | 1 | No | No | 0 | 4 | 1 | 1 | | H:H:?1 | H:H:?2 | 37.64 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 32.11 | 1 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 16.62 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
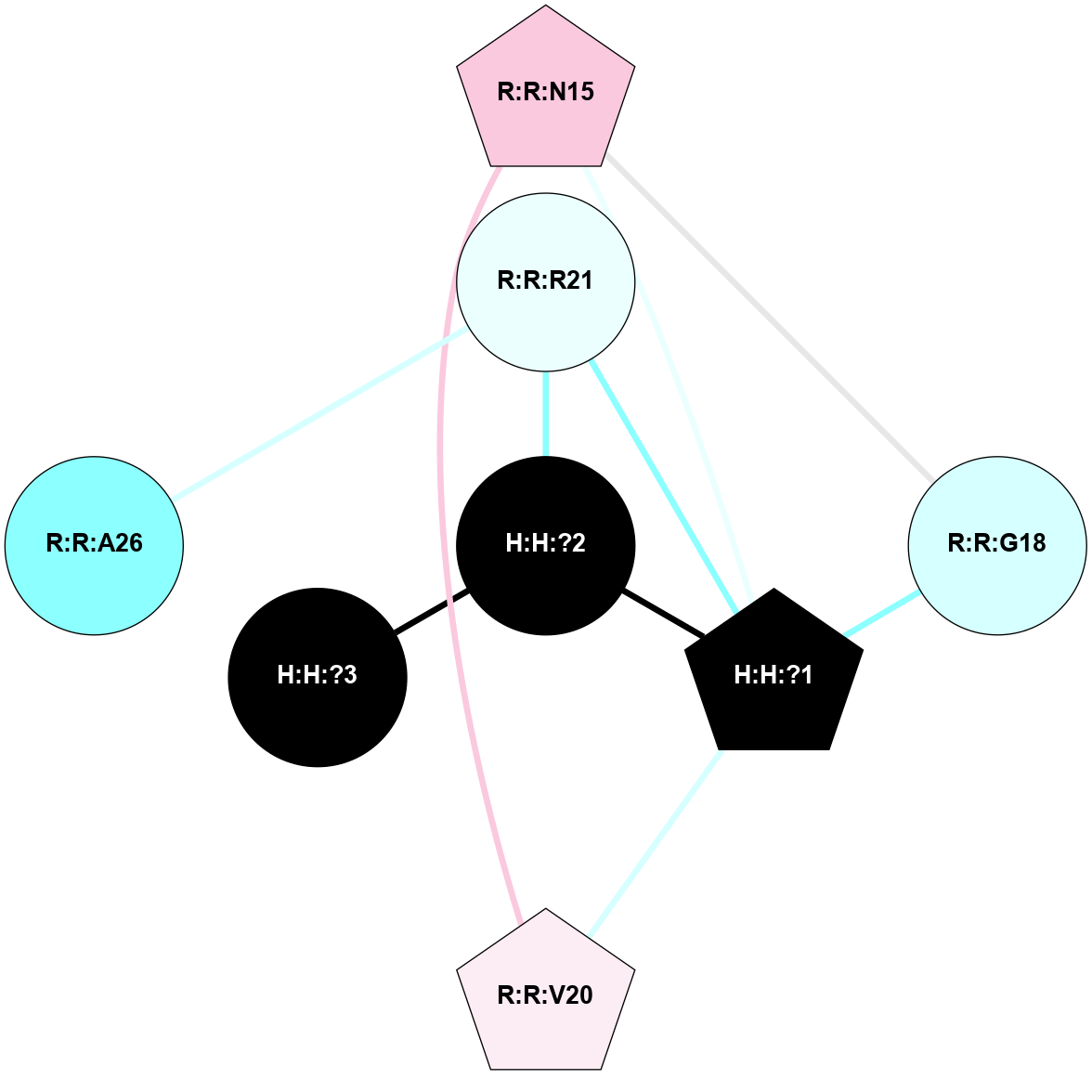
A 2D representation of the interactions of NAG in 3CAP
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 27.02 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 7.99 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:R21 | 4.35 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?2 | R:R:R21 | 8.69 | 1 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 37.64 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 32.11 | 1 | No | No | 0 | 0 | 0 | 1 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 26.15 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
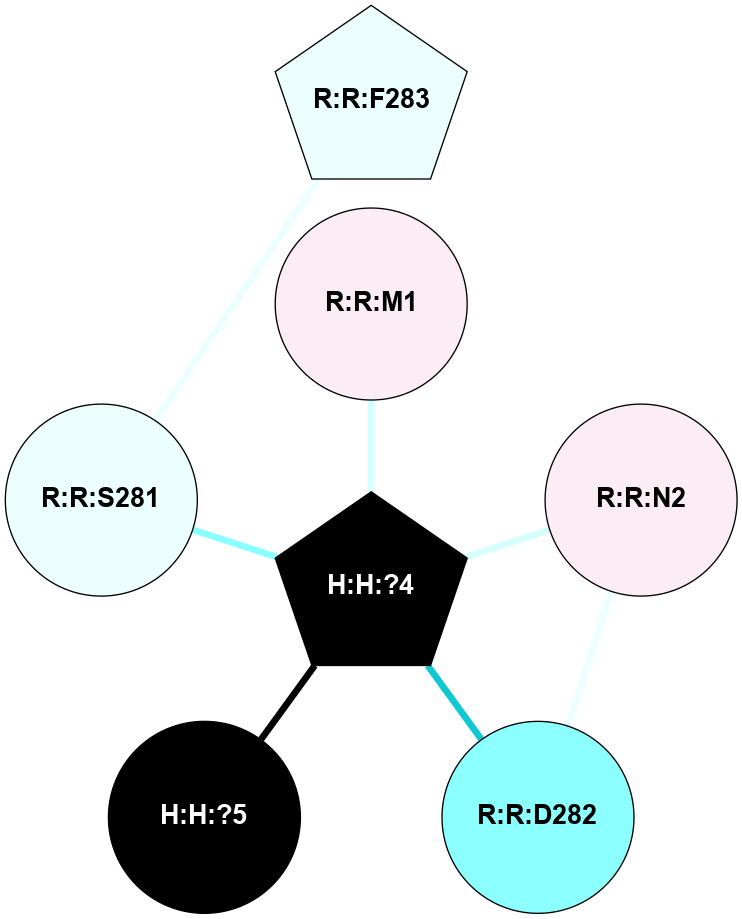
A 2D representation of the interactions of NAG in 3CAP
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:M1 | 3.79 | 7 | Yes | No | 0 | 6 | 0 | 1 | | R:R:D282 | R:R:N2 | 18.85 | 7 | No | No | 2 | 6 | 1 | 1 | | H:H:?4 | R:R:N2 | 36.84 | 7 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?4 | R:R:D282 | 12.14 | 7 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?4 | H:H:?5 | 36.53 | 7 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?4 | R:R:S281 | 1.34 | 7 | Yes | No | 0 | 4 | 0 | 1 | | R:R:F283 | R:R:S281 | 1.32 | 0 | Yes | No | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.13 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
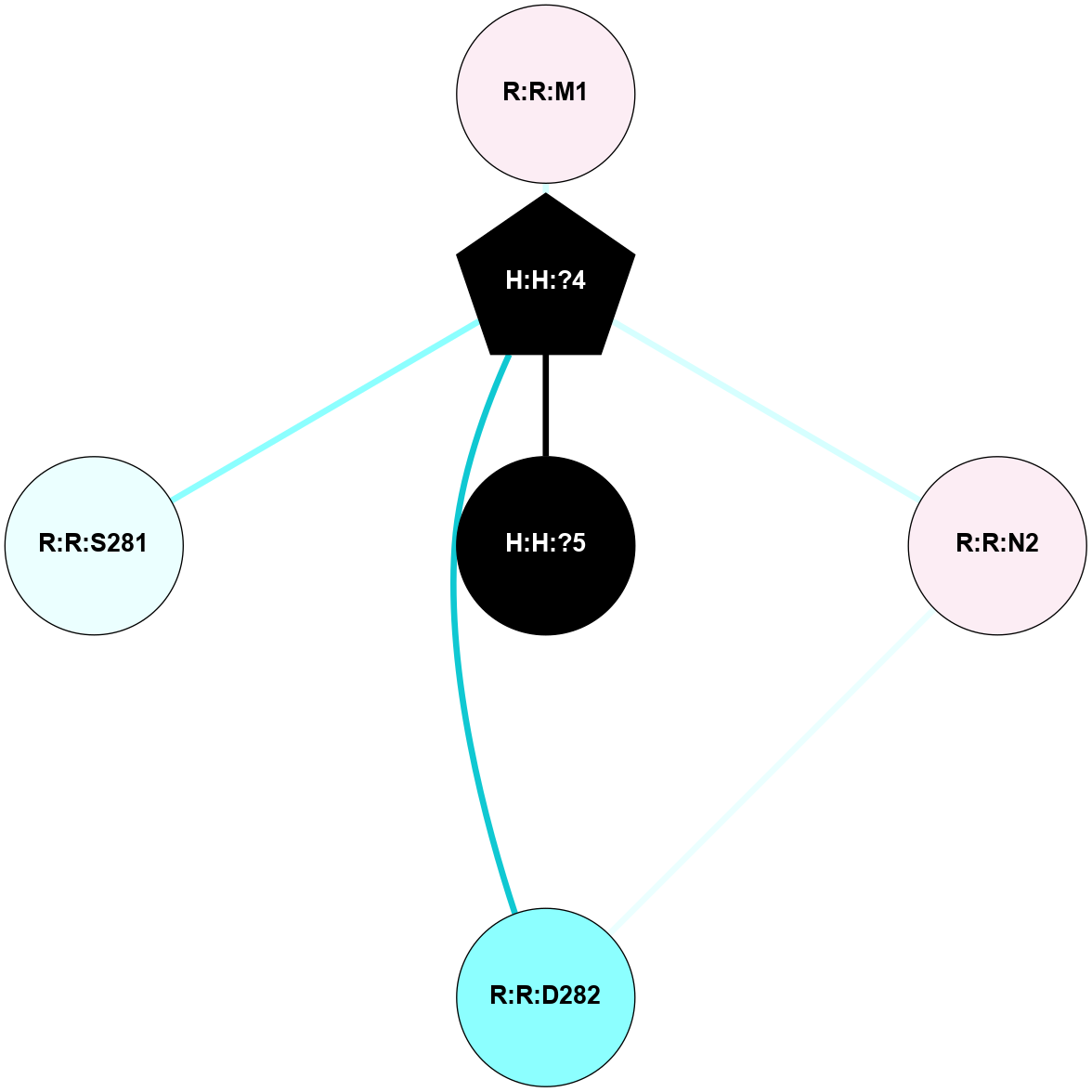
A 2D representation of the interactions of NAG in 3CAP
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:M1 | 3.79 | 7 | Yes | No | 0 | 6 | 1 | 2 | | R:R:D282 | R:R:N2 | 18.85 | 7 | No | No | 2 | 6 | 2 | 2 | | H:H:?4 | R:R:N2 | 36.84 | 7 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?4 | R:R:D282 | 12.14 | 7 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?4 | H:H:?5 | 36.53 | 7 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?4 | R:R:S281 | 1.34 | 7 | Yes | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 36.53 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3DQB | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | Gt(CT) | 3.2 | 2008-09-23 | doi.org/10.1038/nature07330 |
|
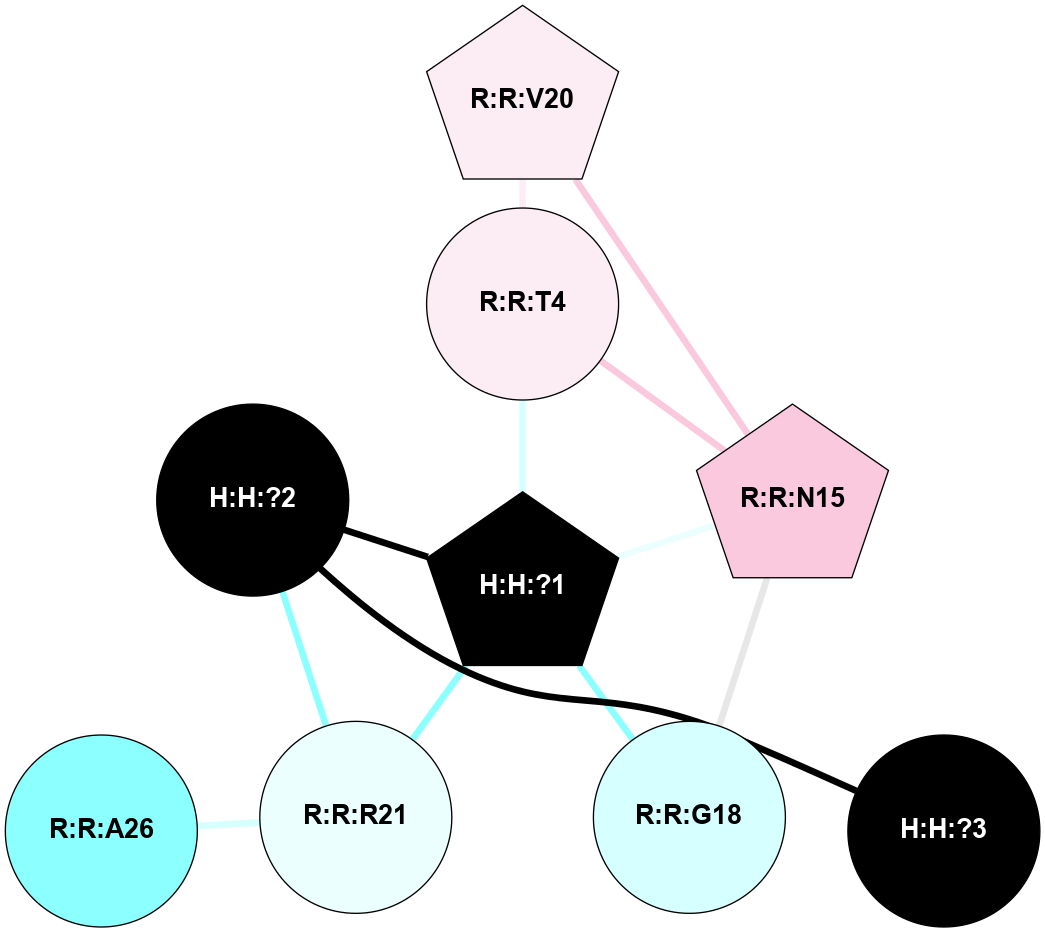
A 2D representation of the interactions of NAG in 3DQB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 5.85 | 2 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 1 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 0 | 1 | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 2 | Yes | Yes | 7 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 27.02 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:R21 | 4.35 | 2 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:R21 | 7.6 | 2 | No | No | 0 | 4 | 1 | 1 | | H:H:?1 | H:H:?2 | 35.42 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 33.39 | 2 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.64 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
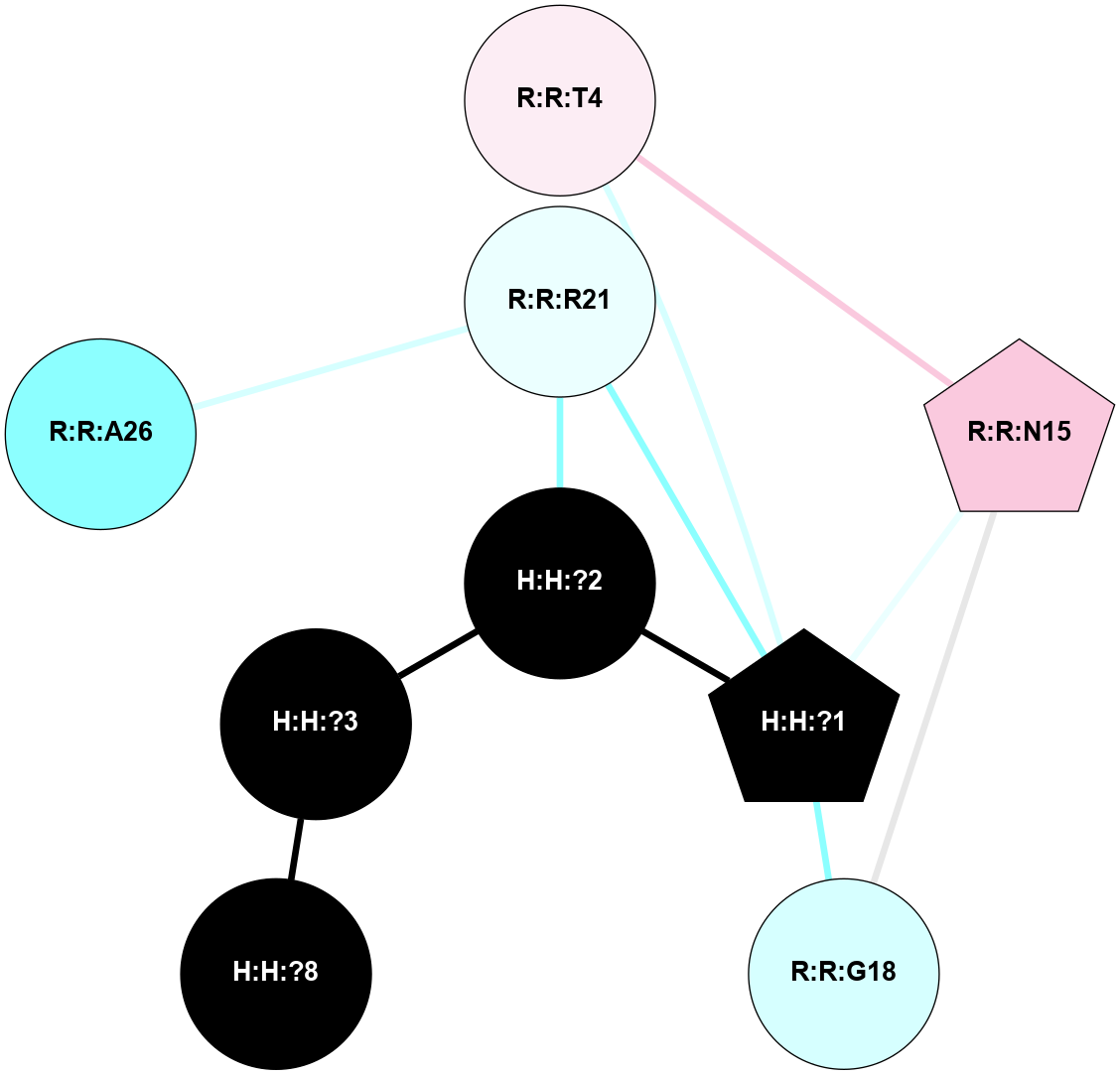
A 2D representation of the interactions of NAG in 3DQB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 5.85 | 2 | Yes | No | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 2 | 2 | | H:H:?1 | R:R:N15 | 27.02 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:R21 | 4.35 | 2 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?2 | R:R:R21 | 7.6 | 2 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 35.42 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 33.39 | 2 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?8 | 31.02 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 25.47 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
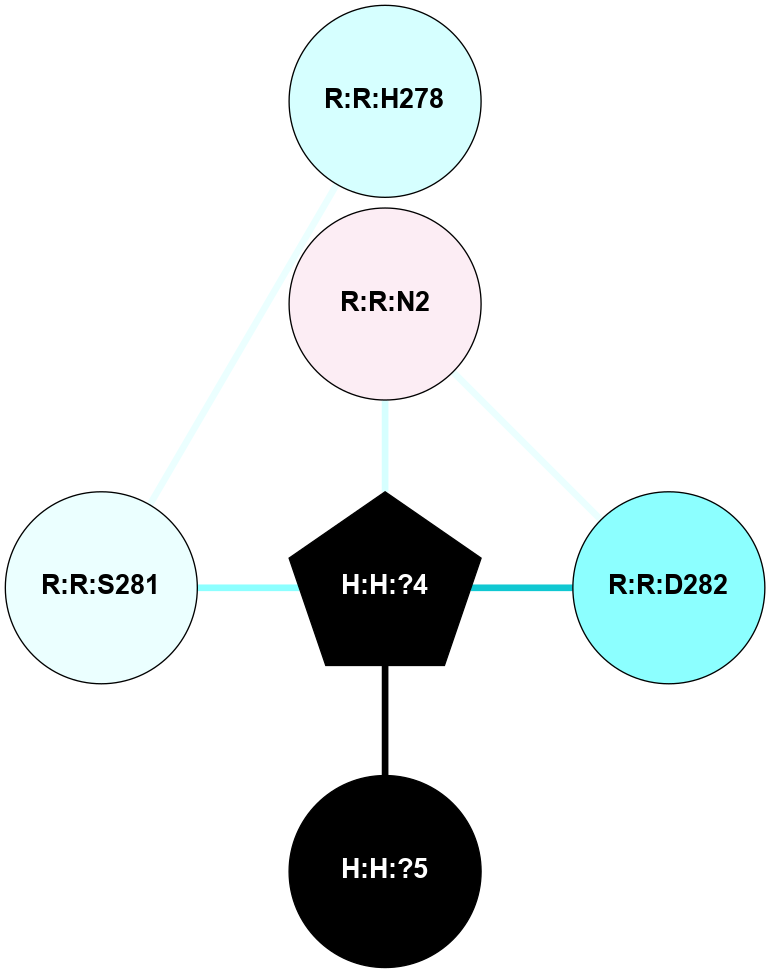
A 2D representation of the interactions of NAG in 3DQB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 20.2 | 6 | No | No | 2 | 6 | 1 | 1 | | H:H:?4 | R:R:N2 | 22.1 | 6 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?4 | R:R:D282 | 15.78 | 6 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?4 | H:H:?5 | 28.78 | 6 | Yes | No | 0 | 0 | 0 | 1 | | R:R:H278 | R:R:S281 | 2.79 | 0 | No | No | 3 | 4 | 2 | 1 | | H:H:?4 | R:R:S281 | 1.34 | 6 | Yes | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.00 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
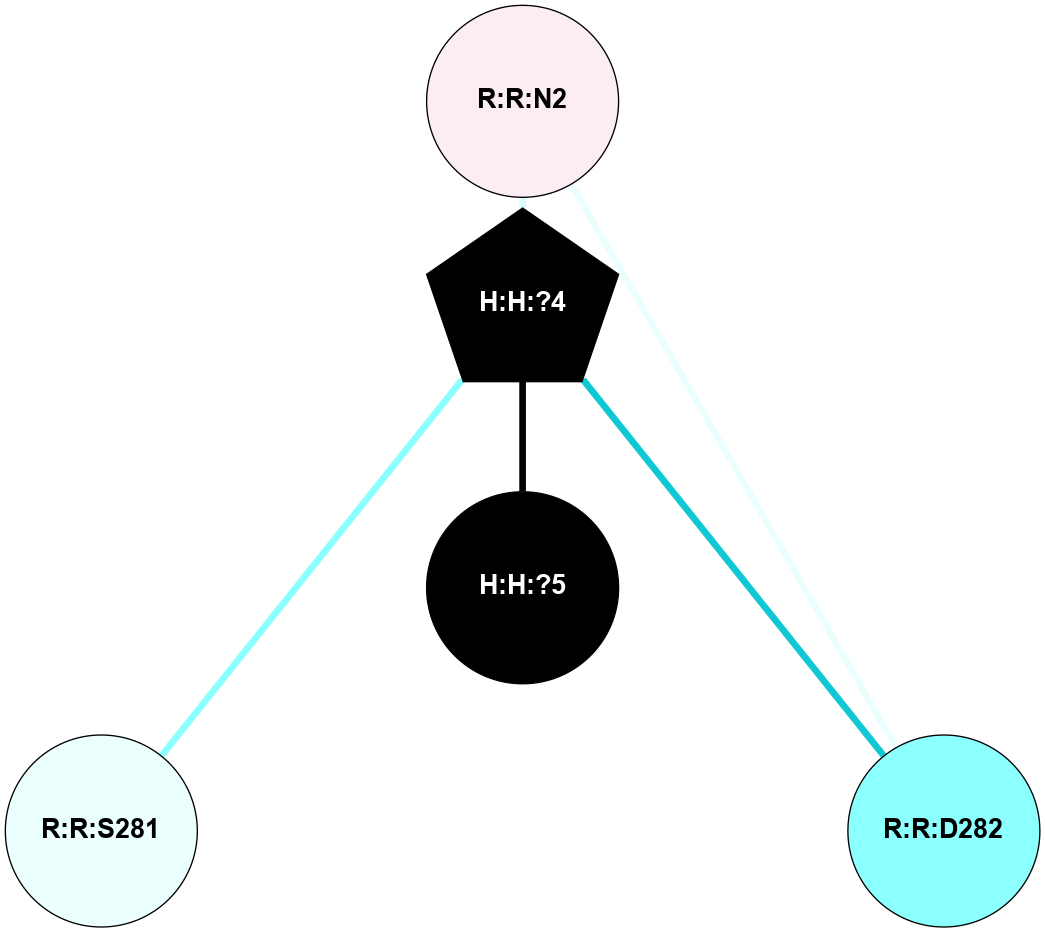
A 2D representation of the interactions of NAG in 3DQB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 20.2 | 6 | No | No | 2 | 6 | 2 | 2 | | H:H:?4 | R:R:N2 | 22.1 | 6 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?4 | R:R:D282 | 15.78 | 6 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?4 | H:H:?5 | 28.78 | 6 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?4 | R:R:S281 | 1.34 | 6 | Yes | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 28.78 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3OAX | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 2.6 | 2011-01-19 | doi.org/10.1016/j.bpj.2010.08.003 |
|
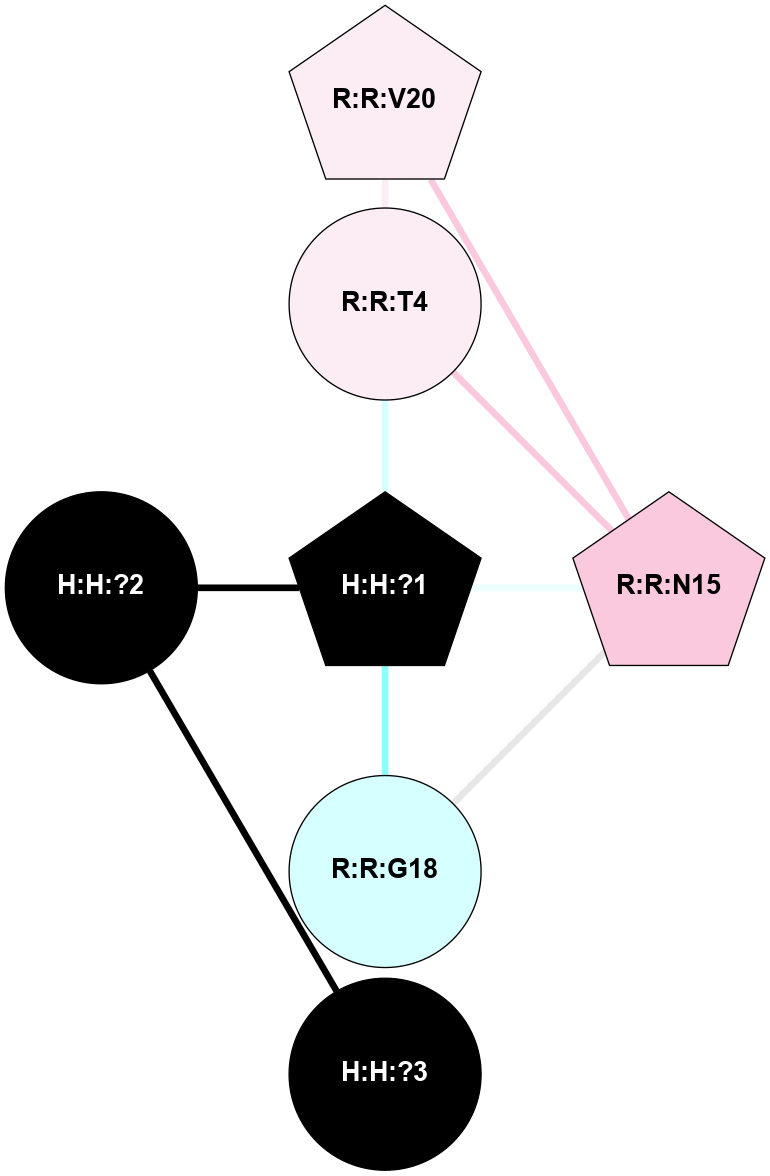
A 2D representation of the interactions of NAG in 3OAX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 5.85 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 1 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 23.33 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | H:H:?2 | 42.06 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 28.25 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.20 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
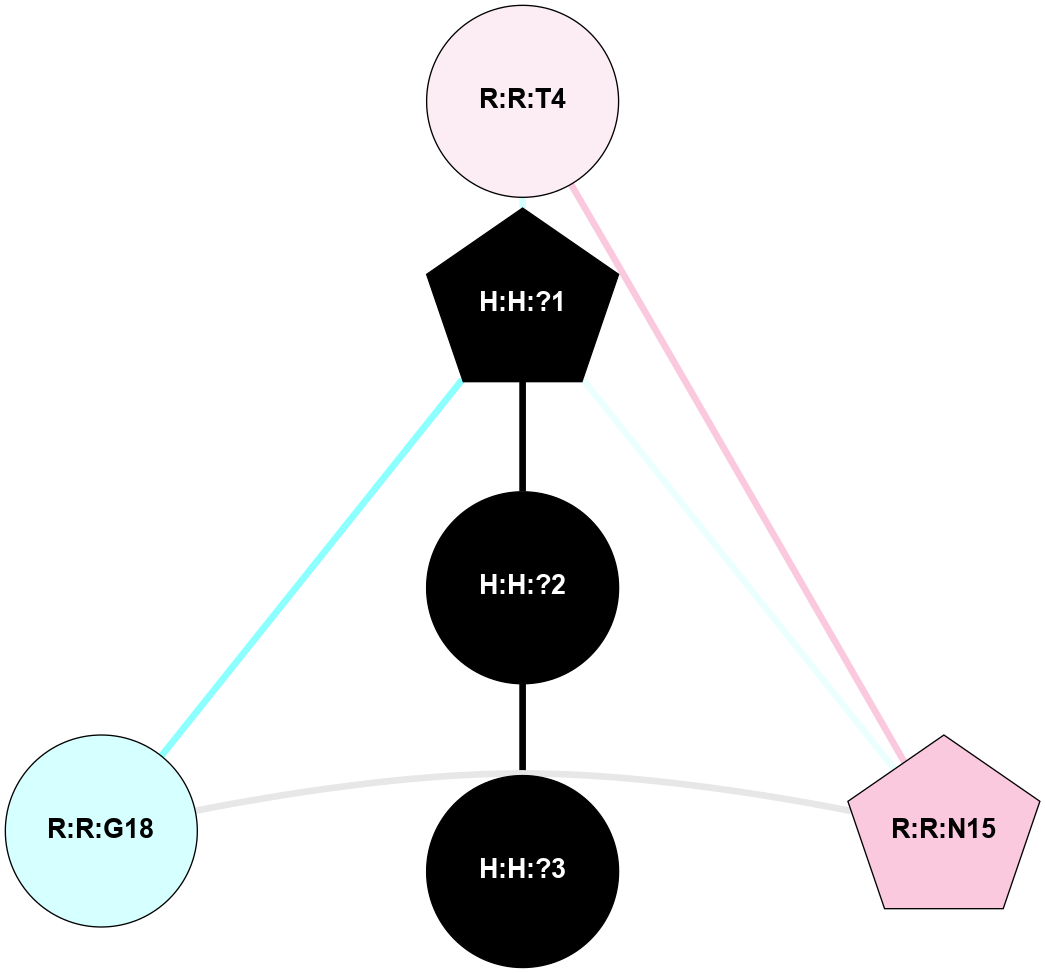
A 2D representation of the interactions of NAG in 3OAX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 5.85 | 1 | Yes | No | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 2 | 2 | | H:H:?1 | R:R:N15 | 23.33 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | H:H:?2 | 42.06 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 28.25 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 35.16 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
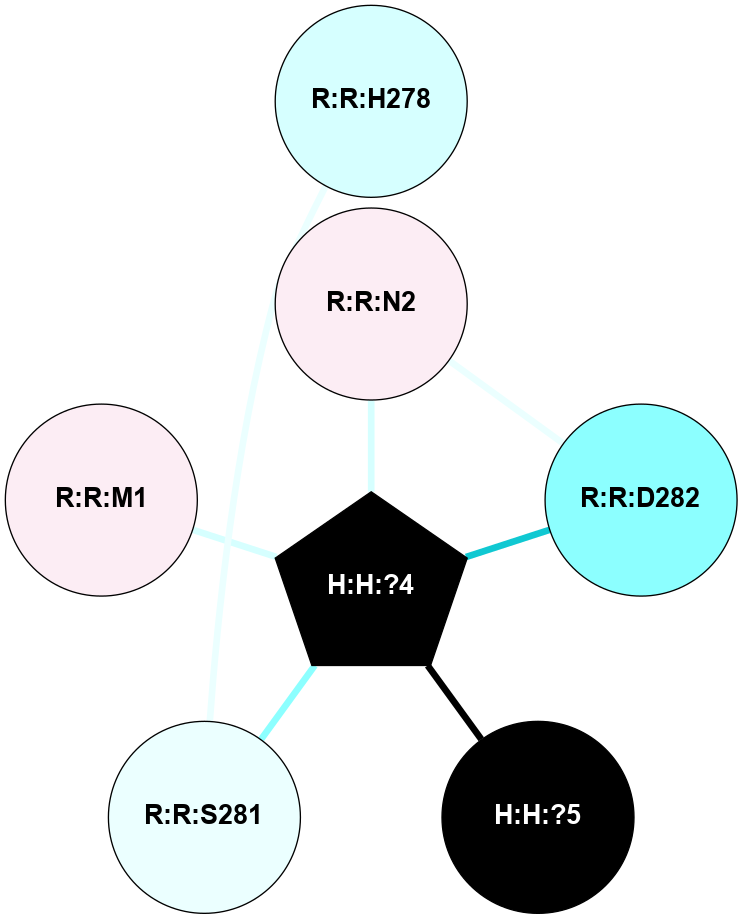
A 2D representation of the interactions of NAG in 3OAX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 14.81 | 7 | No | No | 2 | 6 | 1 | 1 | | H:H:?4 | R:R:N2 | 27.02 | 7 | Yes | No | 0 | 6 | 0 | 1 | | R:R:H278 | R:R:S281 | 4.18 | 0 | No | No | 3 | 4 | 2 | 1 | | H:H:?4 | R:R:D282 | 6.07 | 7 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?4 | H:H:?5 | 38.74 | 7 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?4 | R:R:S281 | 2.69 | 7 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?4 | R:R:M1 | 1.26 | 7 | Yes | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.16 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
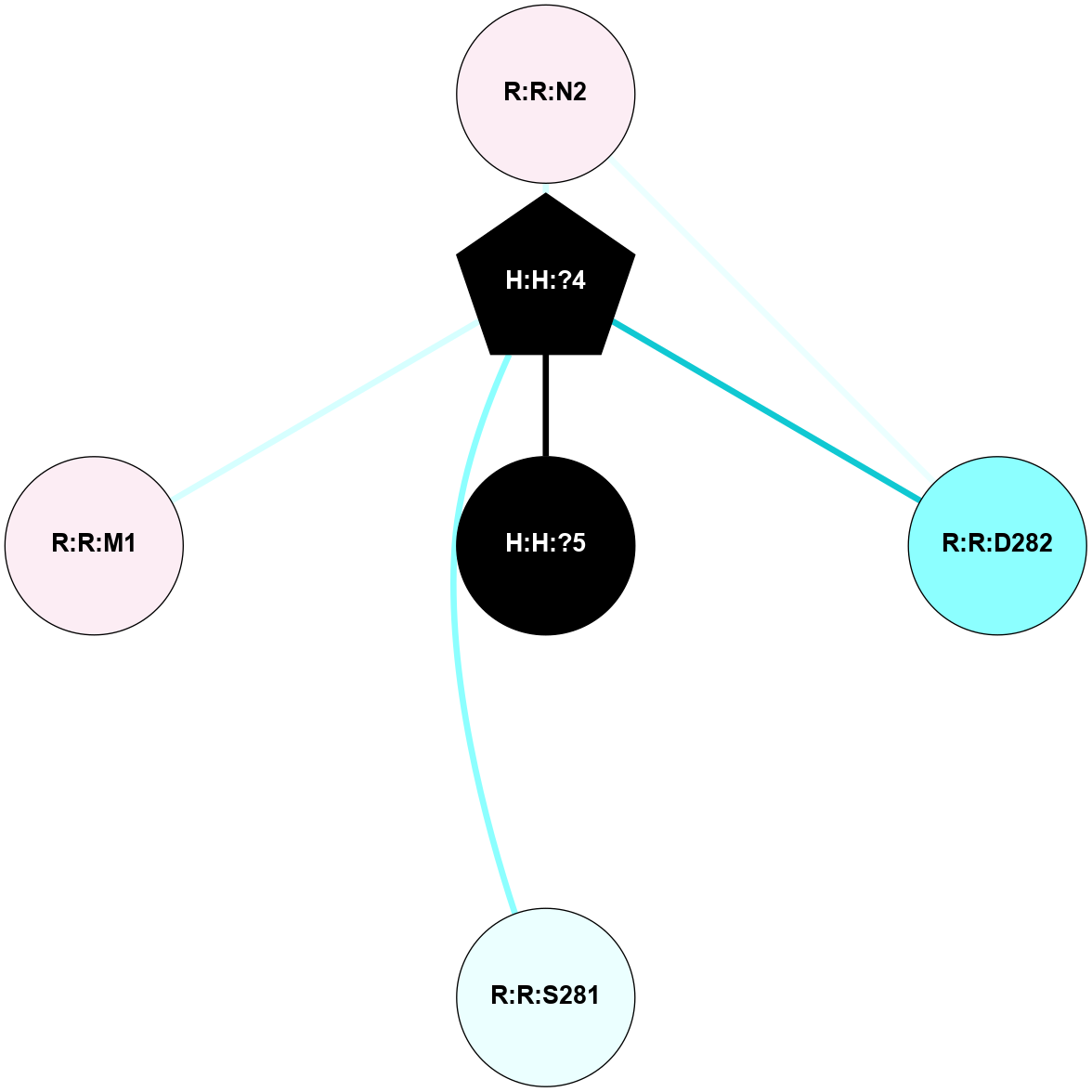
A 2D representation of the interactions of NAG in 3OAX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 14.81 | 7 | No | No | 2 | 6 | 2 | 2 | | H:H:?4 | R:R:N2 | 27.02 | 7 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?4 | R:R:D282 | 6.07 | 7 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?4 | H:H:?5 | 38.74 | 7 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?4 | R:R:S281 | 2.69 | 7 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?4 | R:R:M1 | 1.26 | 7 | Yes | No | 0 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 38.74 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3PQR | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | Gt(CT) | 2.85 | 2011-03-09 | doi.org/10.1038/nature09789 |
|
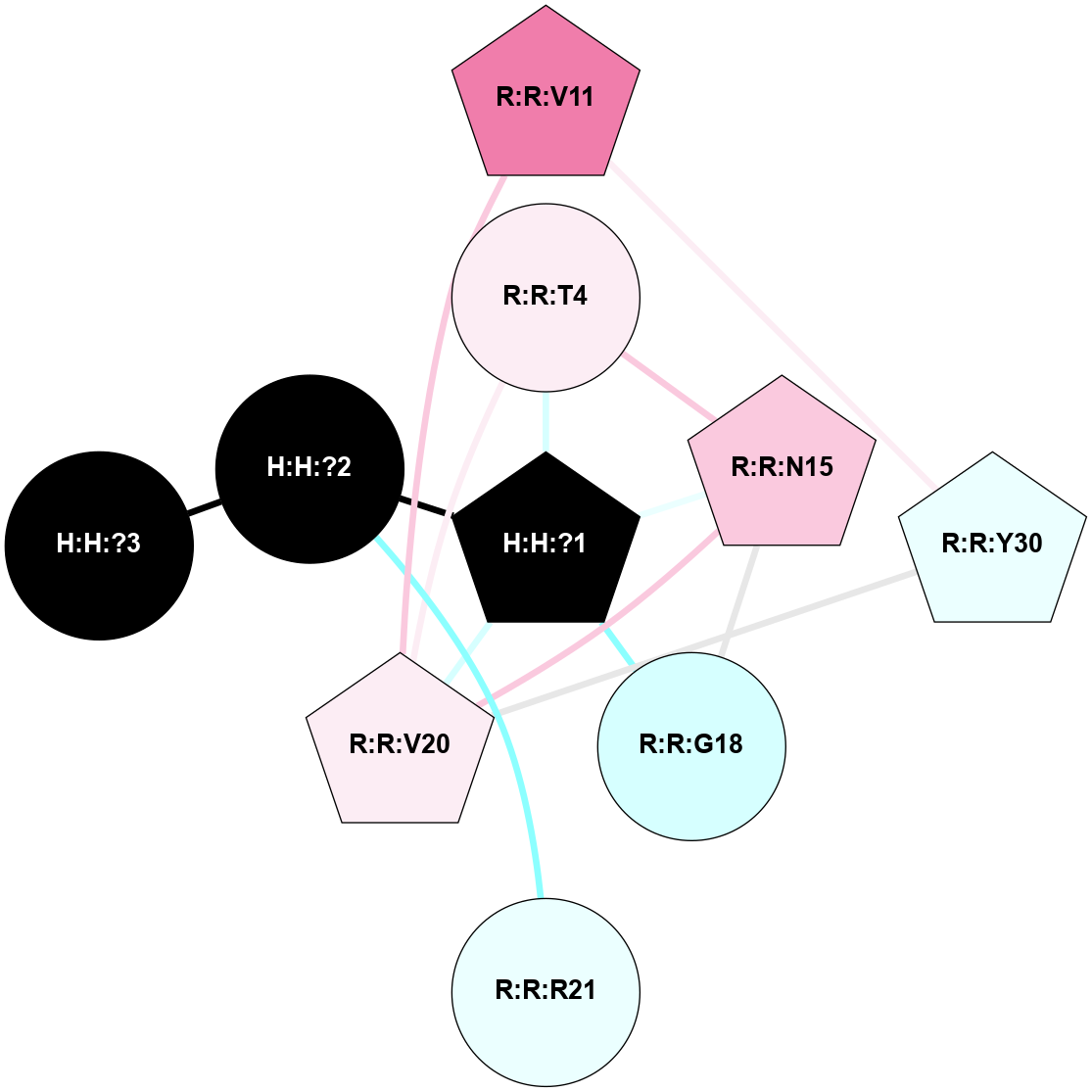
A 2D representation of the interactions of NAG in 3PQR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 2 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 2 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:V11 | R:R:Y30 | 5.05 | 2 | Yes | Yes | 8 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 2 | Yes | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 25.79 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 12.62 | 2 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 38.74 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 25.68 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 15.98 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
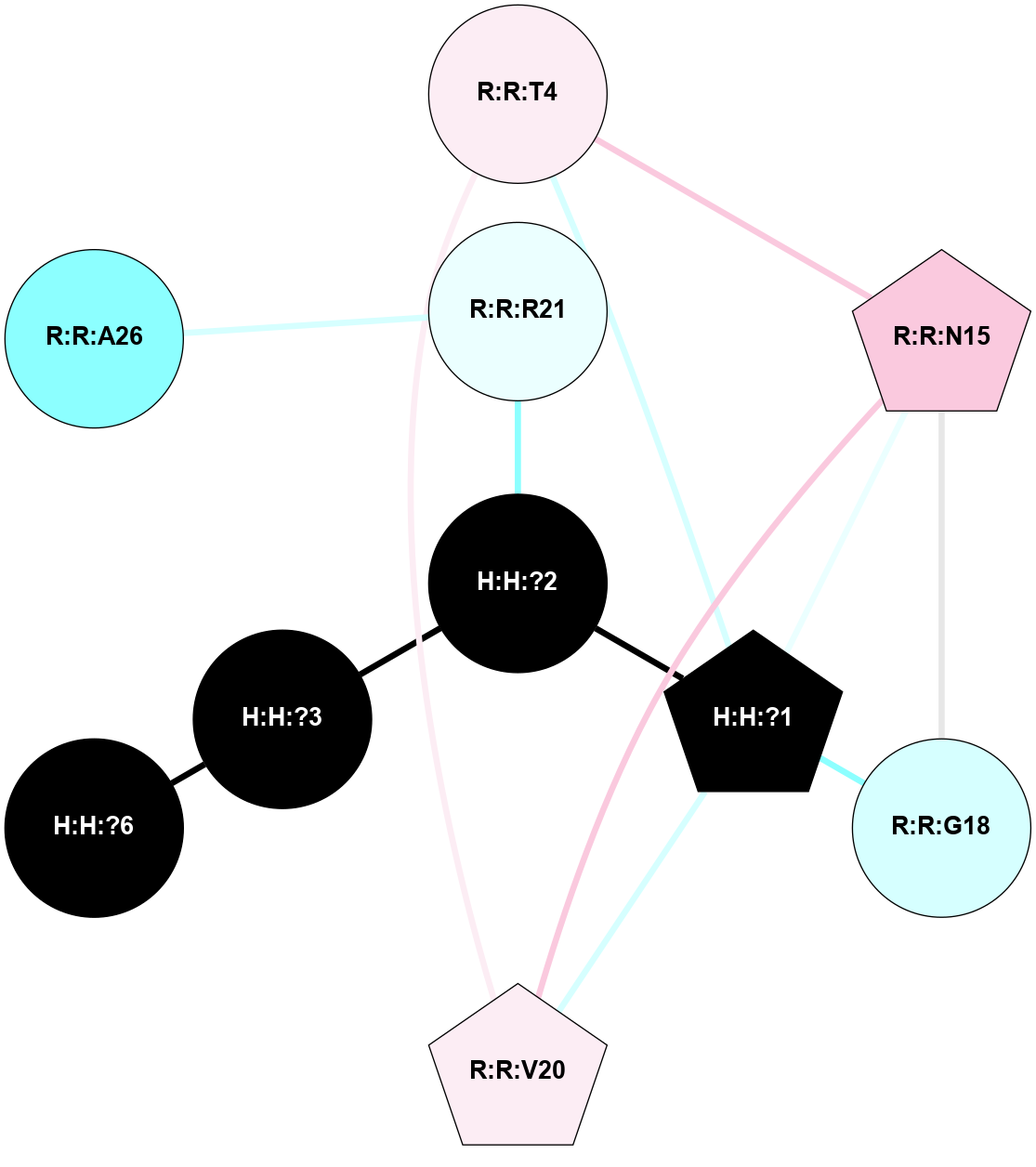
A 2D representation of the interactions of NAG in 3PQR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 2 | Yes | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 2 | Yes | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 25.79 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 38.74 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 25.68 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?6 | 39.89 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 23.65 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
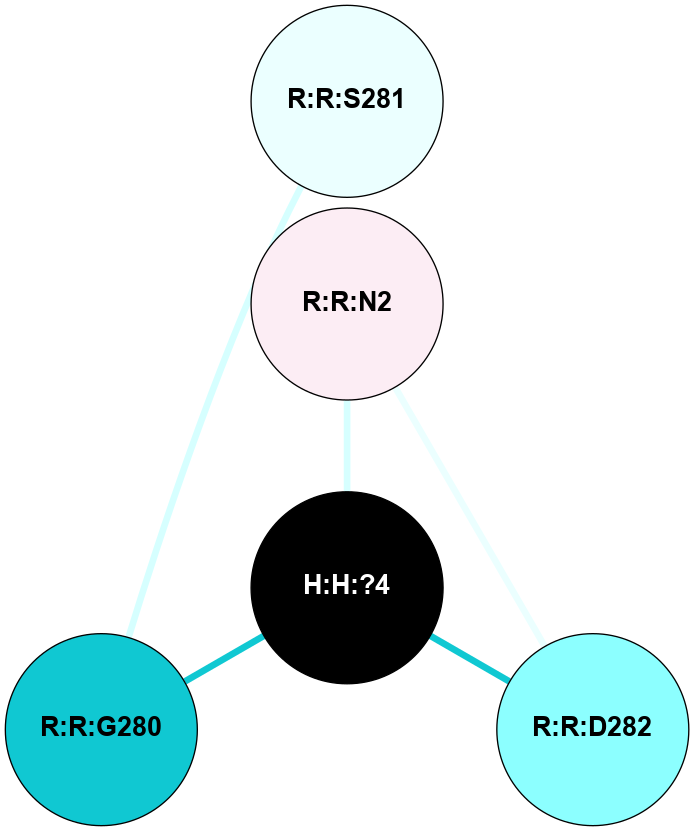
A 2D representation of the interactions of NAG in 3PQR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 13.46 | 5 | No | No | 2 | 6 | 1 | 1 | | H:H:?4 | R:R:N2 | 29.47 | 5 | No | No | 0 | 6 | 0 | 1 | | H:H:?4 | R:R:D282 | 21.85 | 5 | No | No | 0 | 2 | 0 | 1 | | R:R:G280 | R:R:S281 | 1.86 | 0 | No | No | 1 | 4 | 1 | 2 | | H:H:?4 | R:R:G280 | 1.53 | 5 | No | No | 0 | 1 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.62 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3PXO | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 3 | 2011-03-09 | doi.org/10.1038/nature09789 |
|
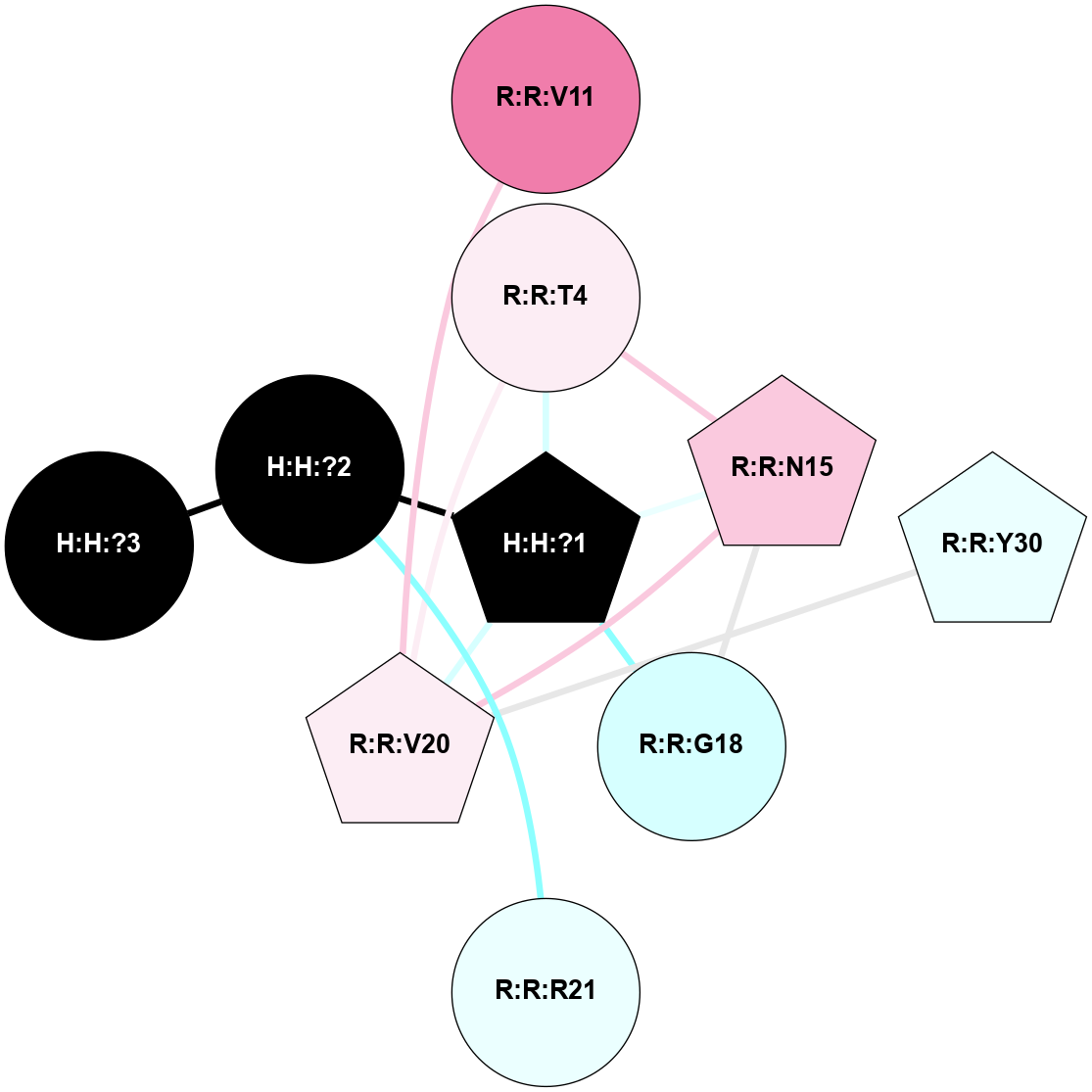
A 2D representation of the interactions of NAG in 3PXO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 11.11 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 0 | No | Yes | 8 | 6 | 2 | 1 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 27.02 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 12.62 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 36.53 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 28.25 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 15.79 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
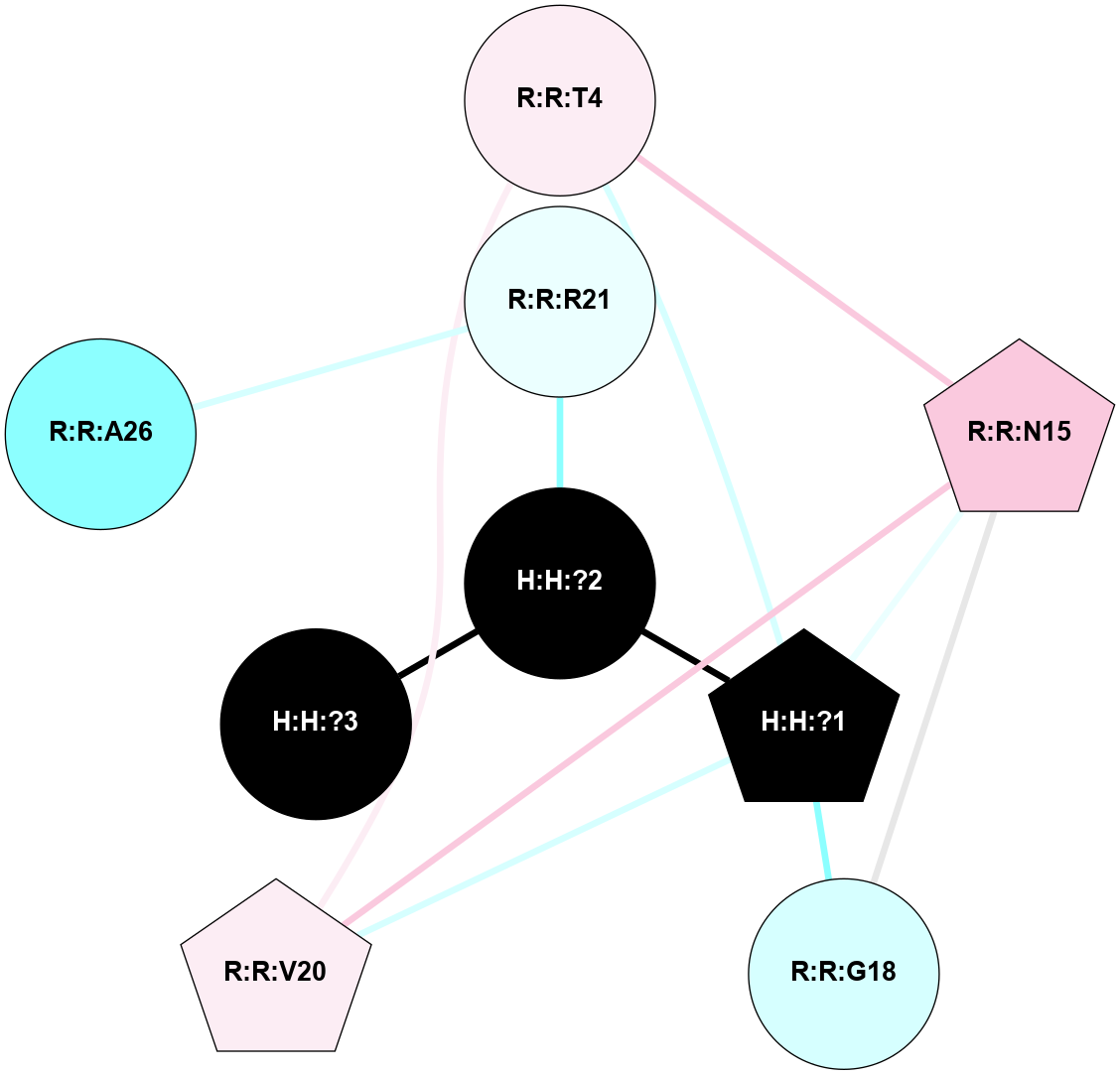
A 2D representation of the interactions of NAG in 3PXO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 1 | Yes | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 11.11 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 27.02 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 36.53 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 28.25 | 0 | No | No | 0 | 0 | 0 | 1 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 23.77 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
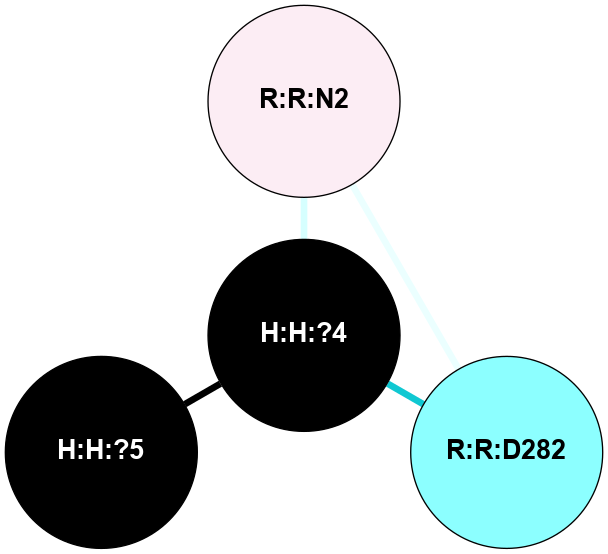
A 2D representation of the interactions of NAG in 3PXO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 18.85 | 6 | No | No | 2 | 6 | 1 | 1 | | H:H:?4 | R:R:N2 | 36.84 | 6 | No | No | 0 | 6 | 0 | 1 | | H:H:?4 | R:R:D282 | 12.14 | 6 | No | No | 0 | 2 | 0 | 1 | | H:H:?4 | H:H:?5 | 40.96 | 6 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 29.98 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 3PXO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 18.85 | 6 | No | No | 2 | 6 | 2 | 2 | | H:H:?4 | R:R:N2 | 36.84 | 6 | No | No | 0 | 6 | 1 | 2 | | H:H:?4 | R:R:D282 | 12.14 | 6 | No | No | 0 | 2 | 1 | 2 | | H:H:?4 | H:H:?5 | 40.96 | 6 | No | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 40.96 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3V2W | A | Lipid | Lysophospholipid | S1P1 | Homo Sapiens | PubChem 51892645 | - | - | 3.35 | 2012-02-15 | doi.org/10.1126/science.1215904 |
|

A 2D representation of the interactions of NAG in 3V2W
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N30 | 28.24 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:L35 | R:R:N30 | 5.49 | 0 | Yes | No | 6 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 28.24 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 3V2Y | A | Lipid | Lysophospholipid | S1P1 | Homo Sapiens | PubChem 51892645 | - | - | 2.8 | 2012-02-15 | doi.org/10.1126/science.1215904 |
|

A 2D representation of the interactions of NAG in 3V2Y
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N30 | 31.93 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:L35 | R:R:N30 | 6.87 | 0 | No | No | 6 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 31.93 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4A4M | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | Gt(CT) | 3.3 | 2012-01-25 | doi.org/10.1073/pnas.1114089108 |
|
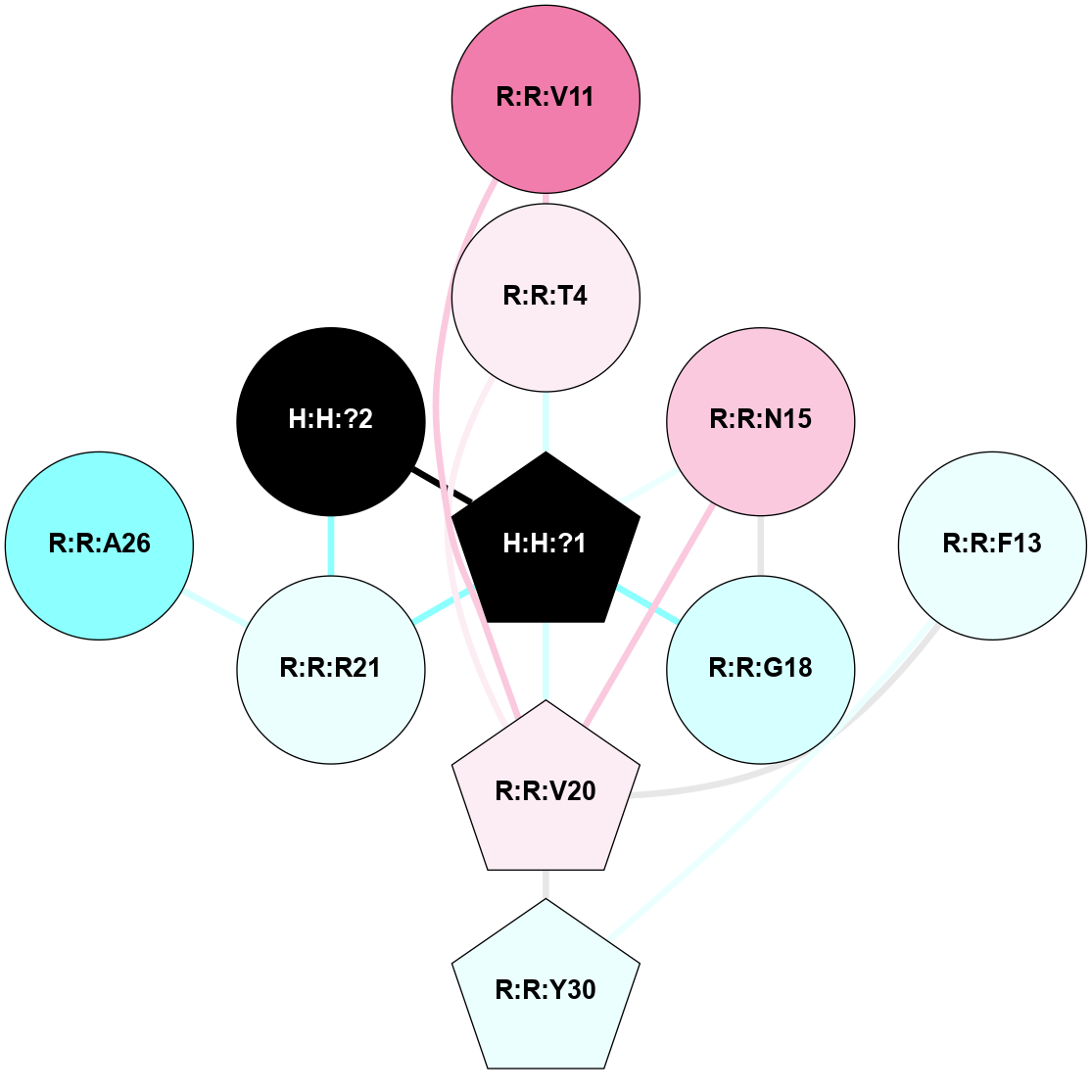
A 2D representation of the interactions of NAG in 4A4M
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:T4 | R:R:V11 | 4.76 | 1 | No | No | 6 | 8 | 1 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | No | Yes | 8 | 6 | 2 | 1 | | R:R:F13 | R:R:V20 | 3.93 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 12.38 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | No | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 13.3 | 1 | No | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 29.47 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 7.65 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 7.57 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 6.66 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:R21 | 7.6 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:R21 | 9.78 | 1 | No | No | 0 | 4 | 1 | 1 | | H:H:?1 | H:H:?2 | 35.42 | 1 | Yes | No | 0 | 0 | 0 | 1 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.34 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
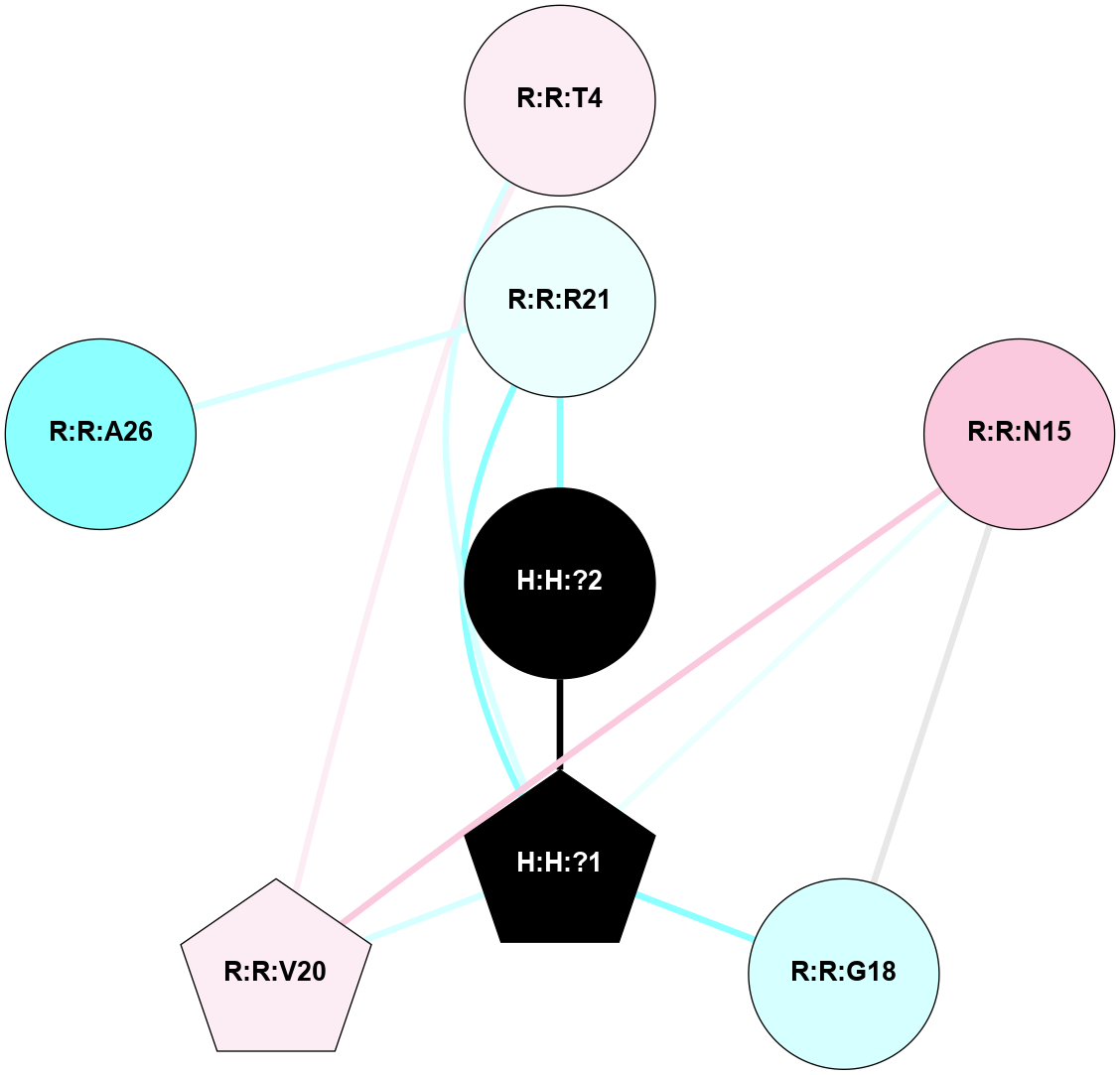
A 2D representation of the interactions of NAG in 4A4M
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | No | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 13.3 | 1 | No | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 29.47 | 1 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 7.65 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 6.66 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:R21 | 7.6 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?2 | R:R:R21 | 9.78 | 1 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 35.42 | 1 | Yes | No | 0 | 0 | 1 | 0 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 22.60 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4BEY | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | Gt(CT) | 2.9 | 2013-05-08 | doi.org/10.1038/embor.2013.44 |
|
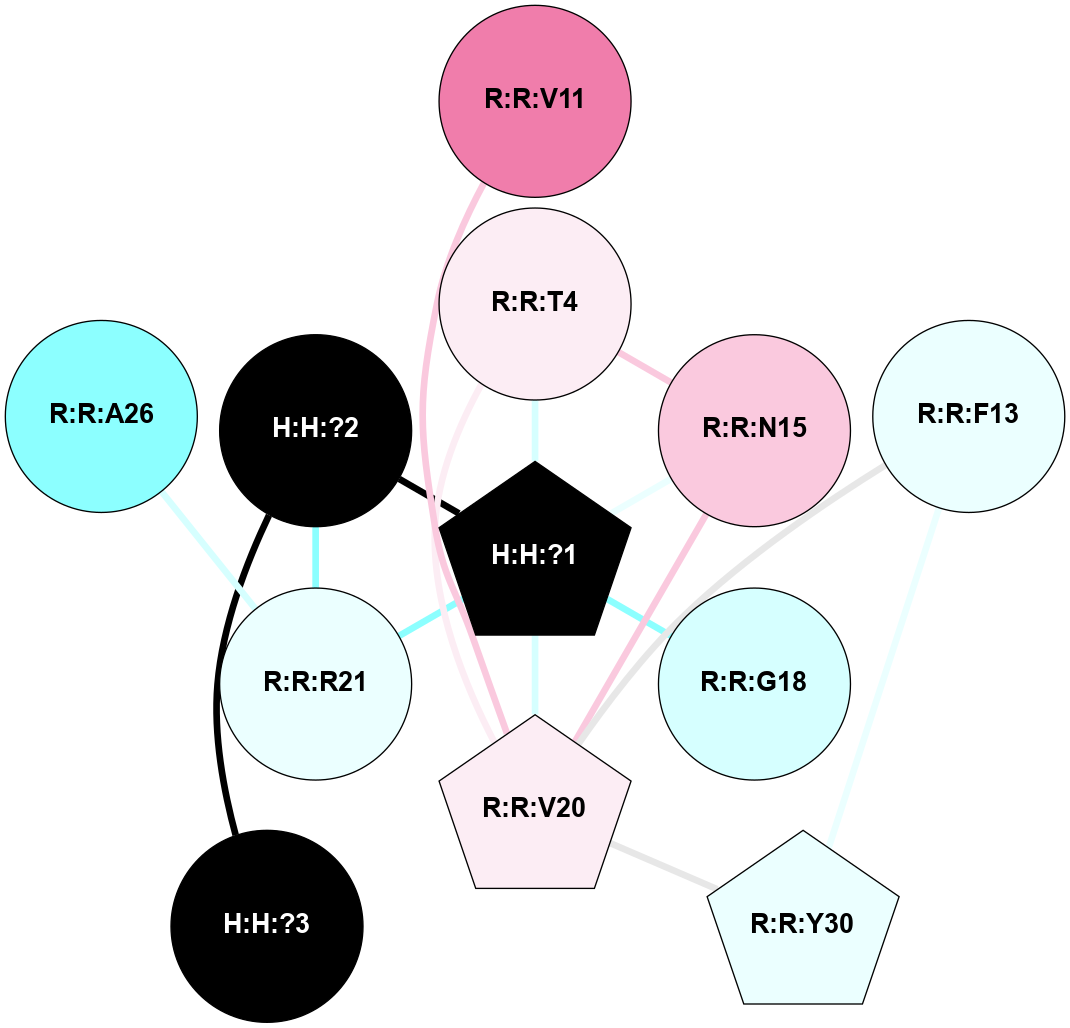
A 2D representation of the interactions of NAG in 4BEY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 5.85 | 1 | No | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 9.52 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | No | Yes | 8 | 6 | 2 | 1 | | R:R:F13 | R:R:V20 | 5.24 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 11.35 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | No | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 25.79 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 12.62 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 5.33 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:R21 | 5.43 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:R21 | 8.69 | 1 | No | No | 0 | 4 | 1 | 1 | | H:H:?1 | H:H:?2 | 37.64 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 30.82 | 1 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.26 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
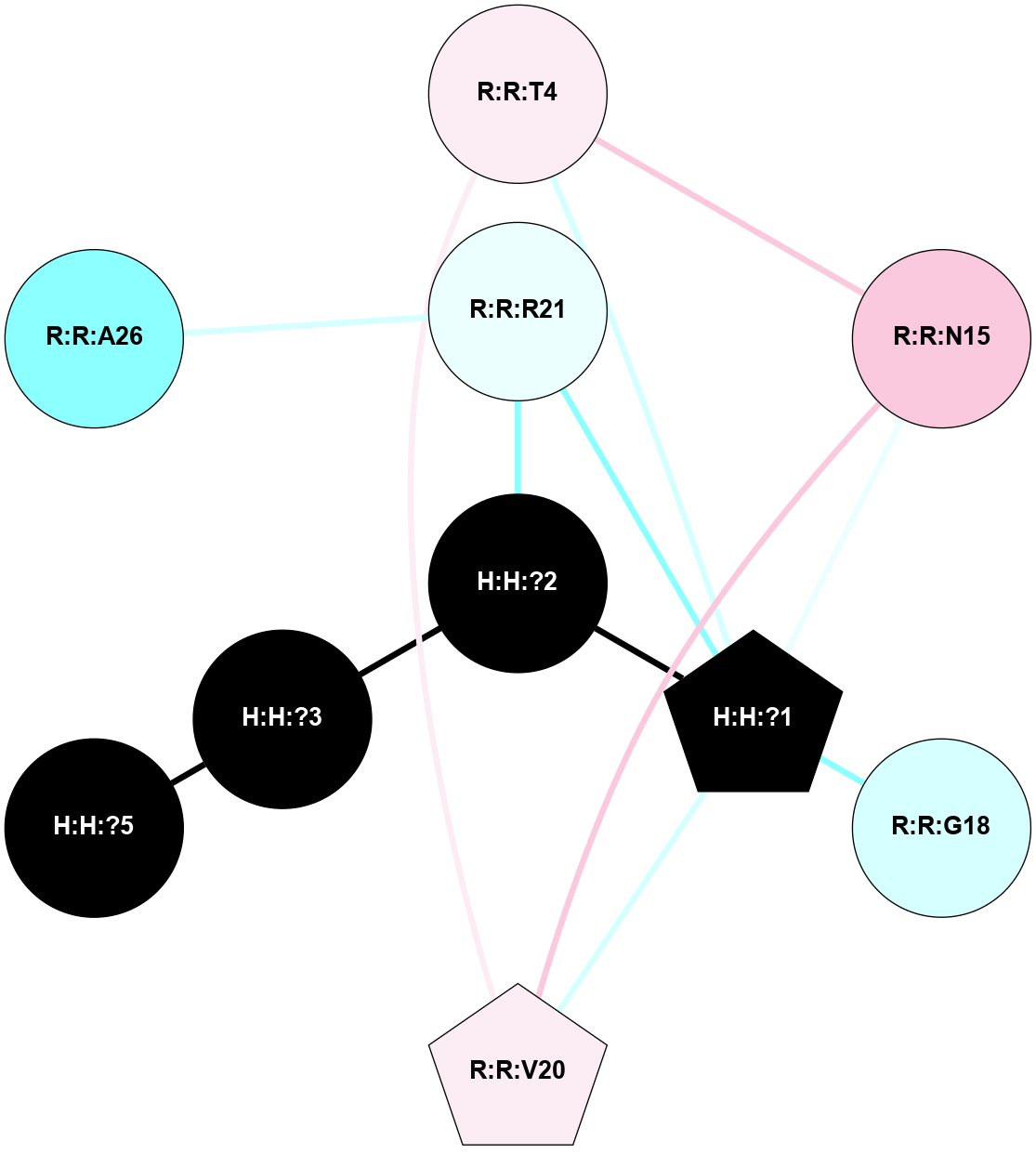
A 2D representation of the interactions of NAG in 4BEY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 5.85 | 1 | No | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 9.52 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | No | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 25.79 | 1 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 5.33 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:R21 | 5.43 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?2 | R:R:R21 | 8.69 | 1 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 37.64 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 30.82 | 1 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?5 | 48.75 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 25.72 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4BEZ | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | - | 3.3 | 2013-04-24 | doi.org/10.1038/embor.2013.44 |
|
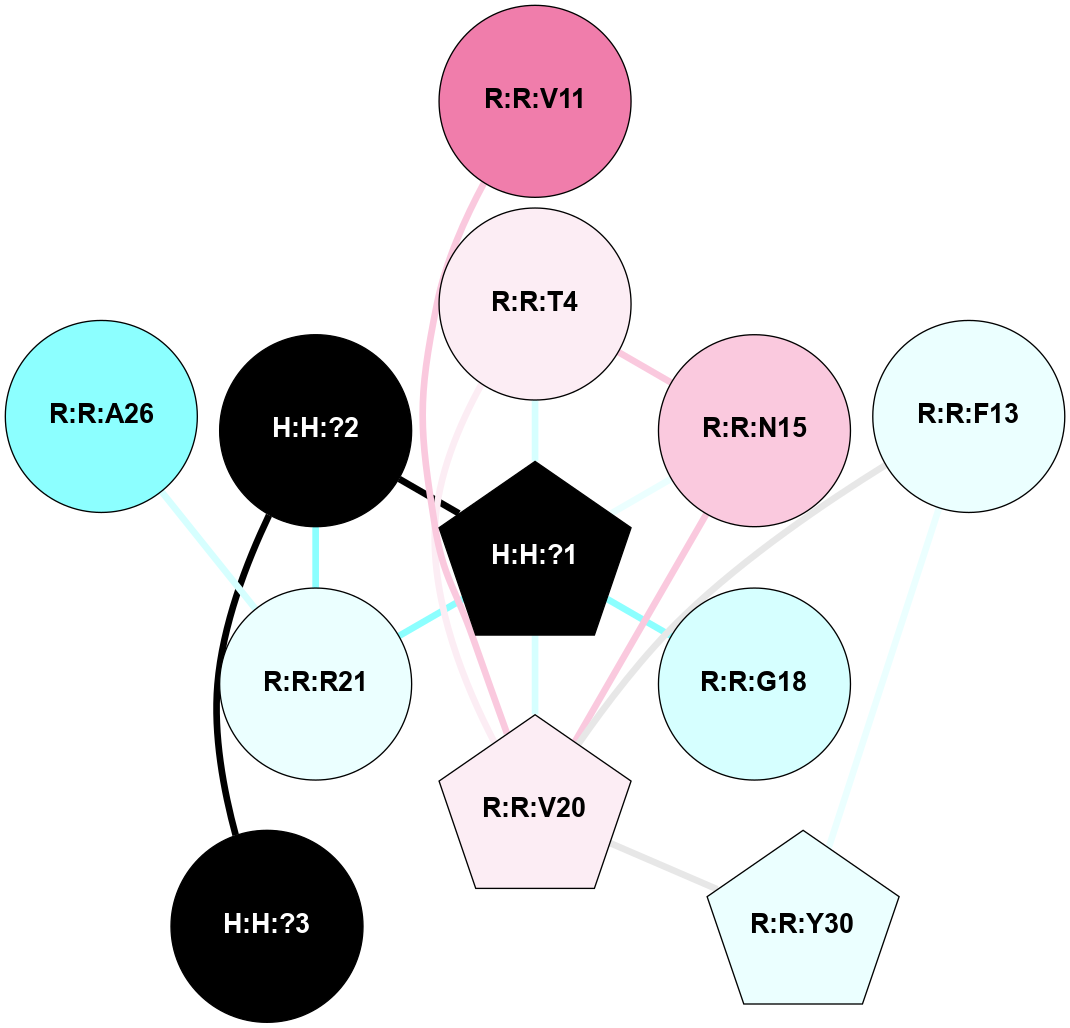
A 2D representation of the interactions of NAG in 4BEZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 4.39 | 1 | No | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 9.52 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | No | Yes | 8 | 6 | 2 | 1 | | R:R:F13 | R:R:V20 | 5.24 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 13.41 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | No | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 28.24 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 7.65 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 12.62 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 7.99 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:R21 | 6.52 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:R21 | 7.6 | 1 | No | No | 0 | 4 | 1 | 1 | | H:H:?1 | H:H:?2 | 37.64 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 34.67 | 1 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.55 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
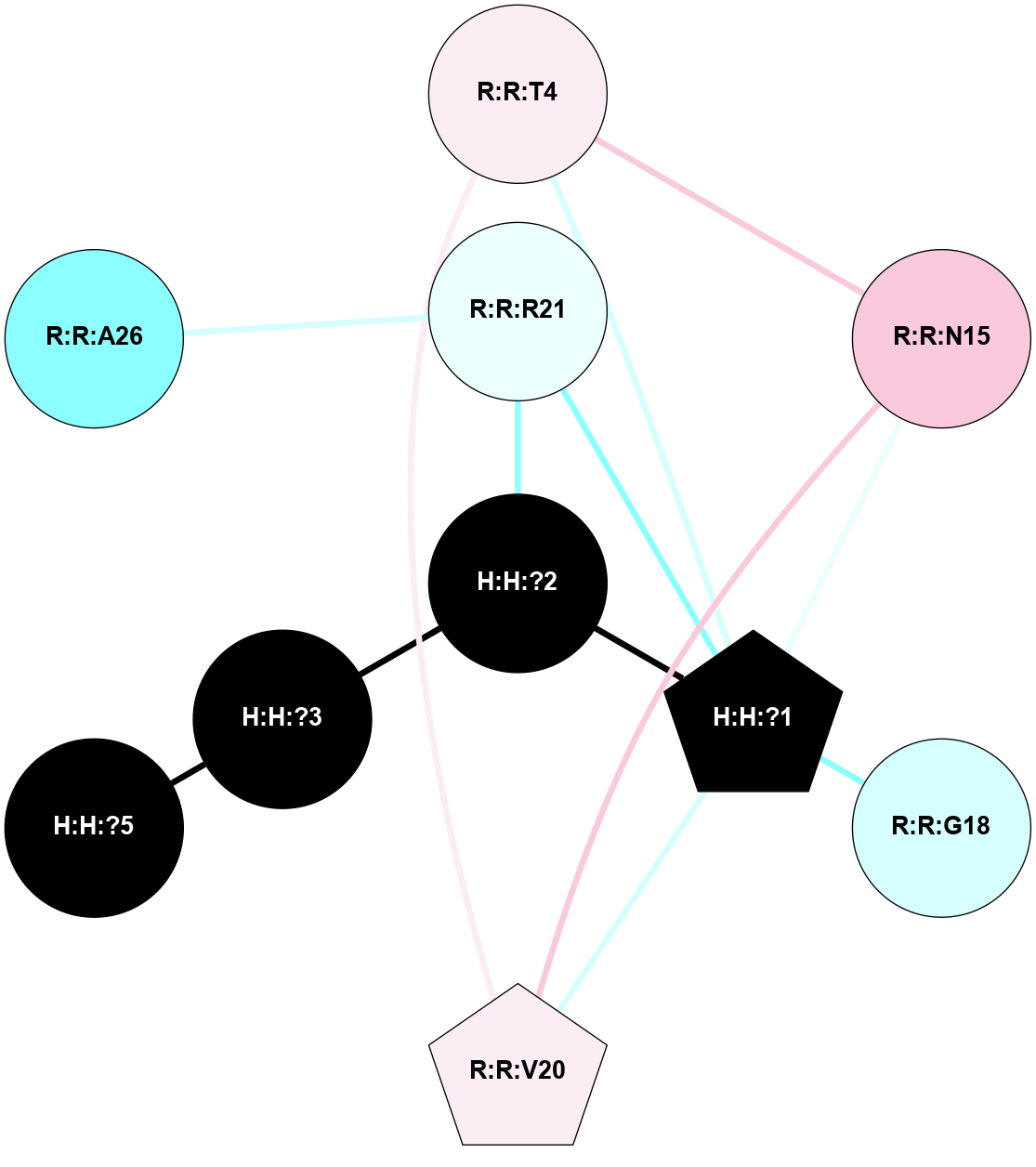
A 2D representation of the interactions of NAG in 4BEZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 4.39 | 1 | No | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 9.52 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | No | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 28.24 | 1 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 7.65 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 7.99 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:R21 | 6.52 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?2 | R:R:R21 | 7.6 | 1 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 37.64 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 34.67 | 1 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?5 | 44.32 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 26.64 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4J4Q | A | Sensory | Opsins | Rhodopsin | Bos Taurus | Octyl Beta-D-Glucopyranoside | - | Gt(CT) | 2.65 | 2013-10-30 | doi.org/10.1002/anie.201302374 |
|
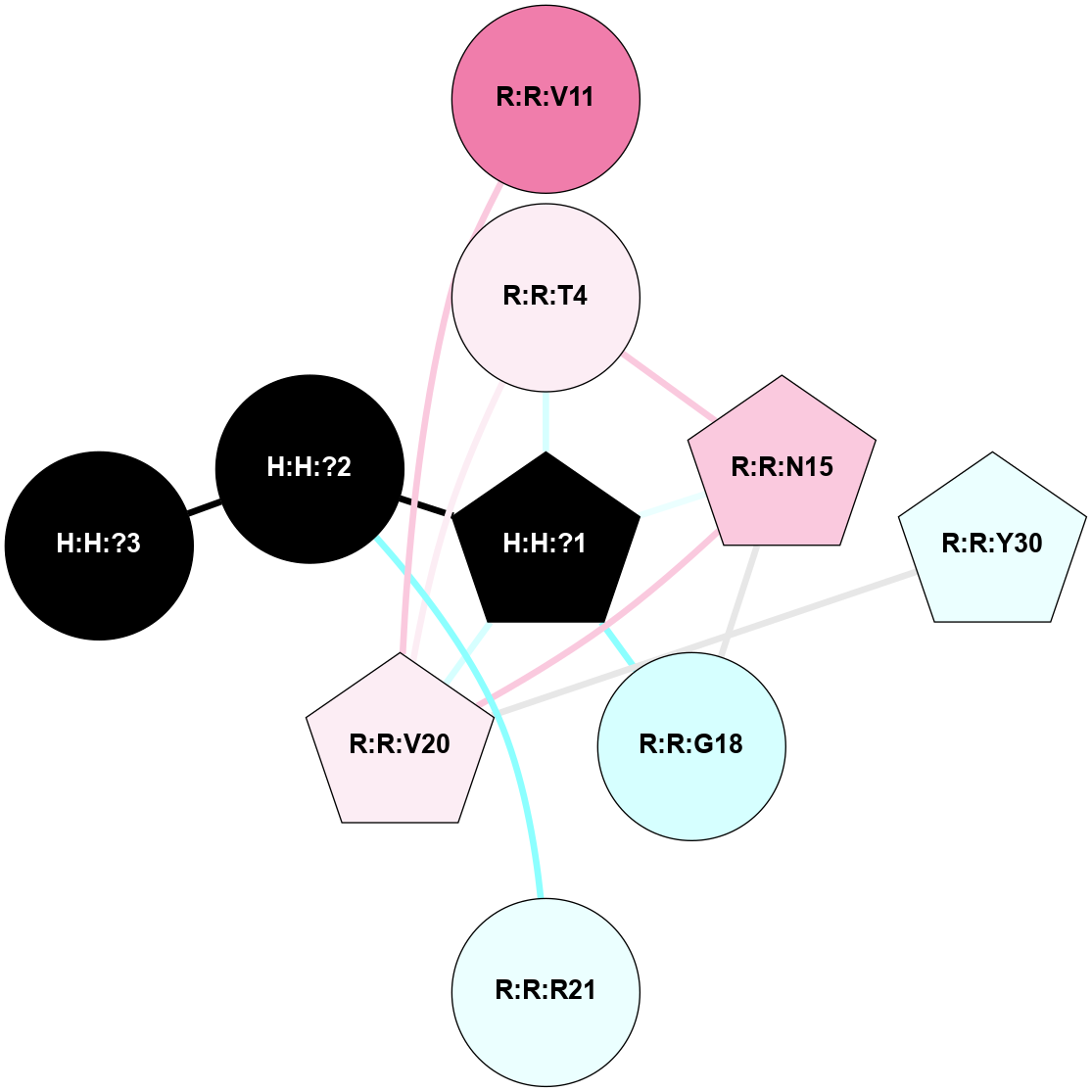
A 2D representation of the interactions of NAG in 4J4Q
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 4 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 9.52 | 4 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 4 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 0 | No | Yes | 8 | 6 | 2 | 1 | | R:R:G18 | R:R:N15 | 5.09 | 4 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 4 | Yes | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 24.56 | 4 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 4 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 12.62 | 4 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 5.33 | 4 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 33.21 | 4 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 28.25 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 14.90 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
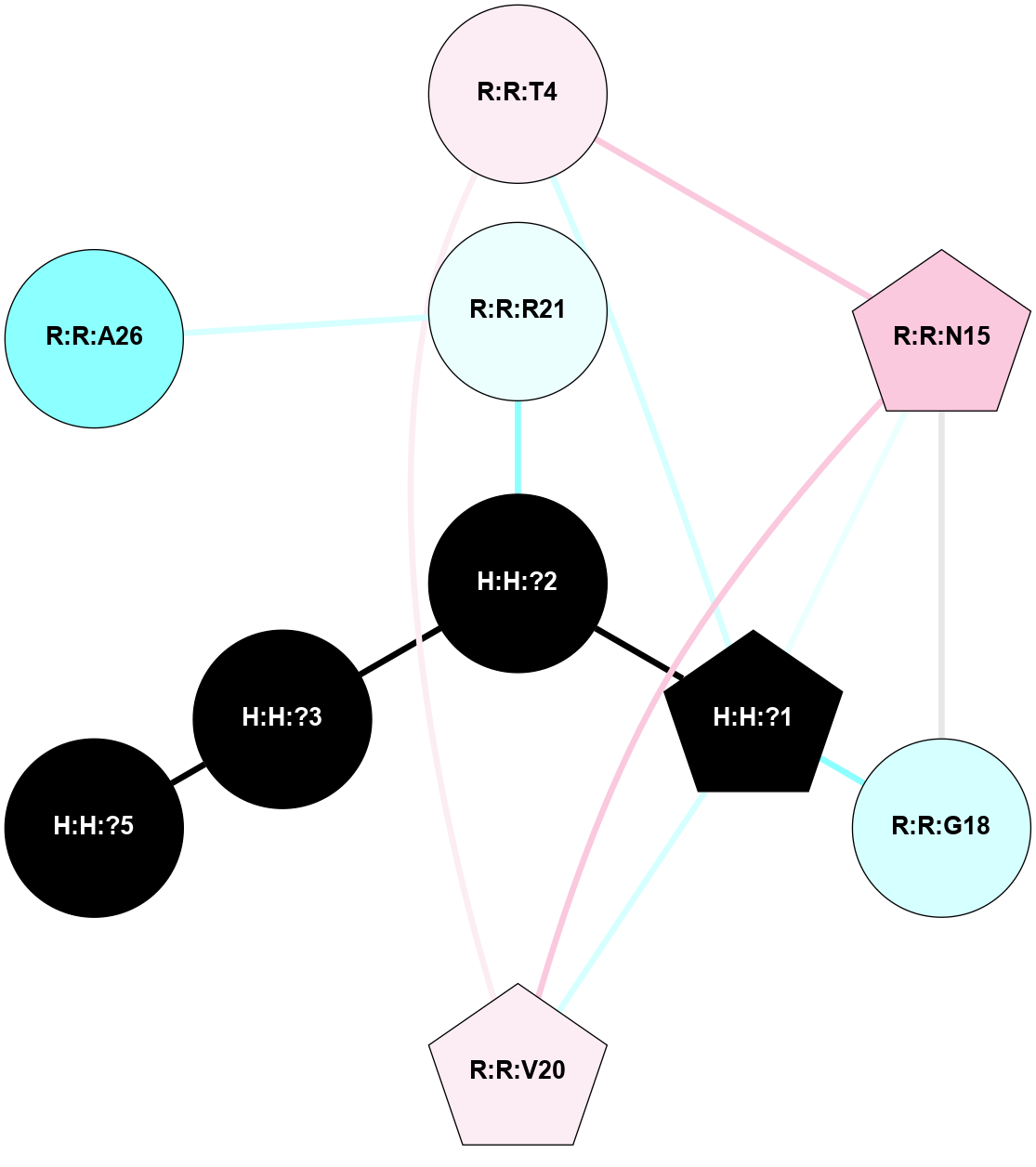
A 2D representation of the interactions of NAG in 4J4Q
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 4 | Yes | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 9.52 | 4 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 4 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 4 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 4 | Yes | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 24.56 | 4 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 4 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 5.33 | 4 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 33.21 | 4 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 28.25 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?5 | 29.55 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 22.66 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4N4W | F | Protein | Frizzled | SMO | Homo Sapiens | - | SANT-1 | - | 2.8 | 2014-01-22 | doi.org/10.1038/ncomms5355 |
|
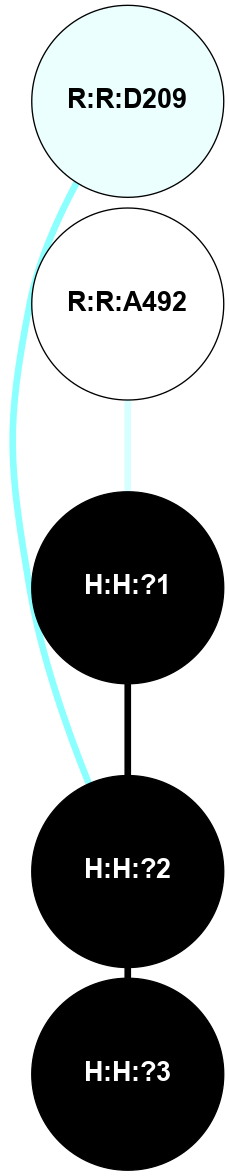
A 2D representation of the interactions of NAG in 4N4W
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 26.57 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:A492 | 4.23 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?2 | H:H:?3 | 28.25 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?2 | R:R:D209 | 7.28 | 0 | No | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.40 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
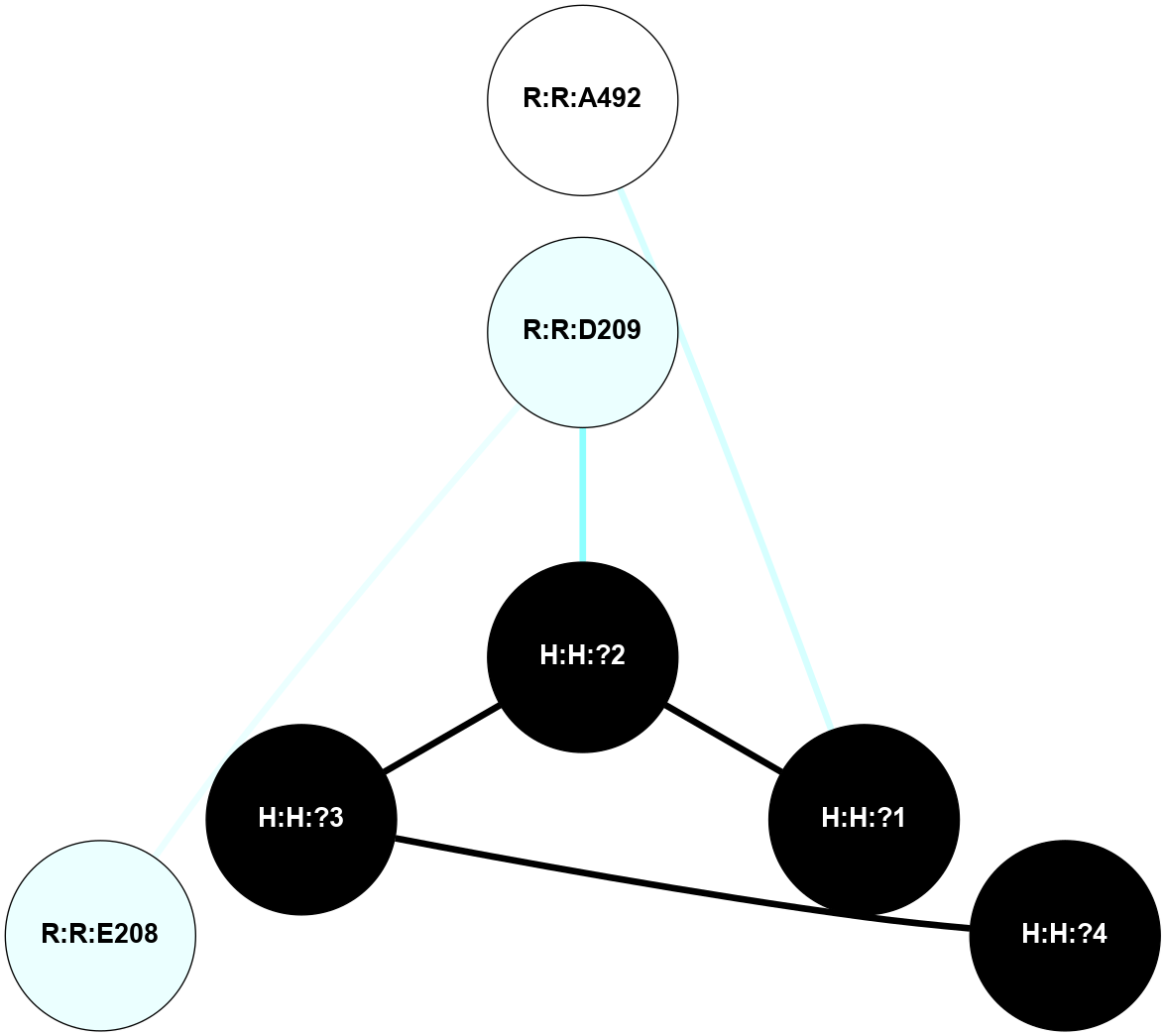
A 2D representation of the interactions of NAG in 4N4W
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 26.57 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:A492 | 4.23 | 0 | No | No | 0 | 5 | 1 | 2 | | H:H:?2 | H:H:?3 | 28.25 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?2 | R:R:D209 | 7.28 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?3 | H:H:?4 | 29.55 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:D209 | R:R:E208 | 2.6 | 0 | No | No | 4 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.70 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4PXF | A | Sensory | Opsins | Rhodopsin | Bos Taurus | Octyl-Beta-D-Glucopyranoside | - | Arrestin1 Finger Loop | 2.75 | 2014-09-17 | doi.org/10.1038/ncomms5801 |
|
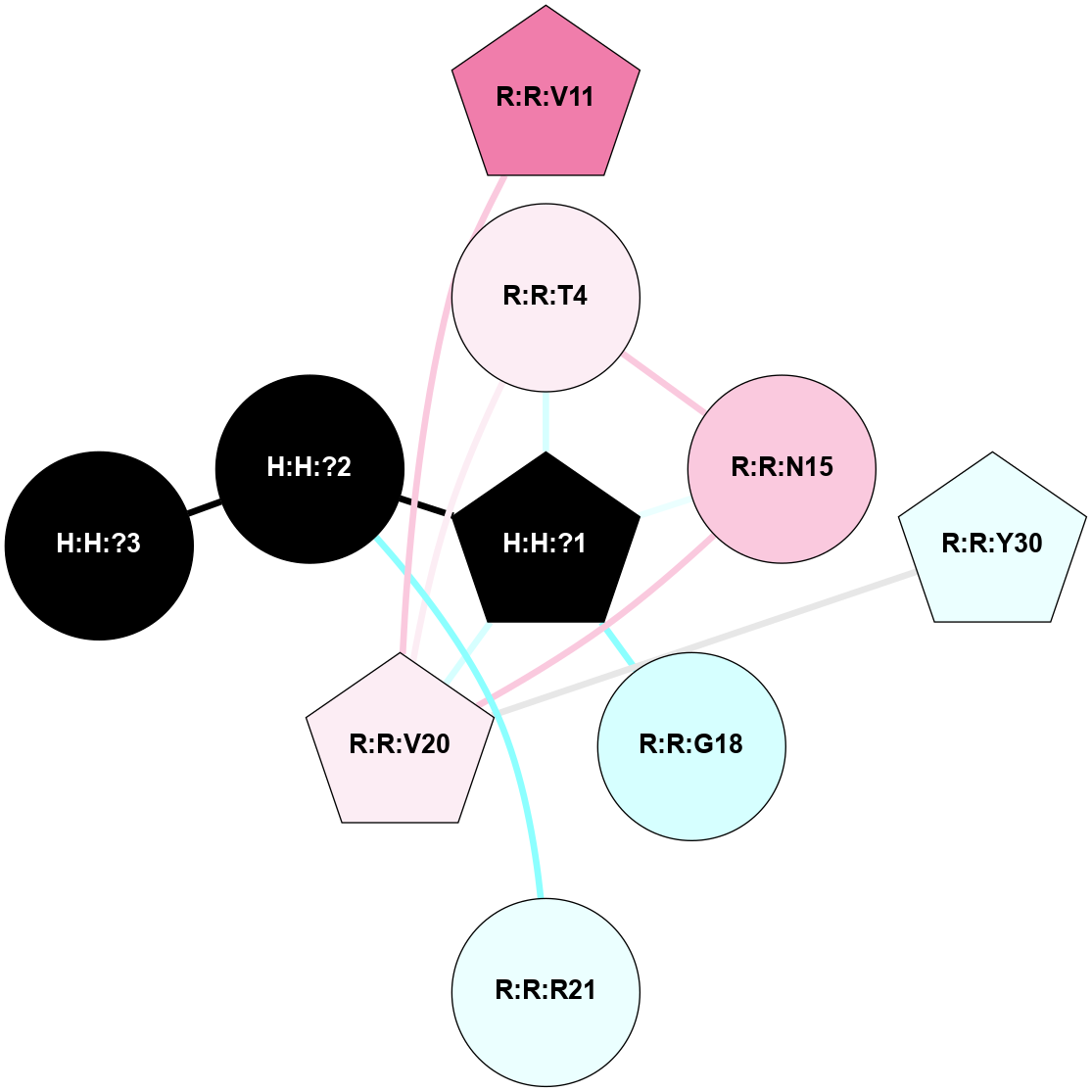
A 2D representation of the interactions of NAG in 4PXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 2 | No | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 2 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 2 | No | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 28.24 | 2 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 7.65 | 2 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 12.62 | 2 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 6.66 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:R21 | 5.43 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 37.64 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 29.54 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.09 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
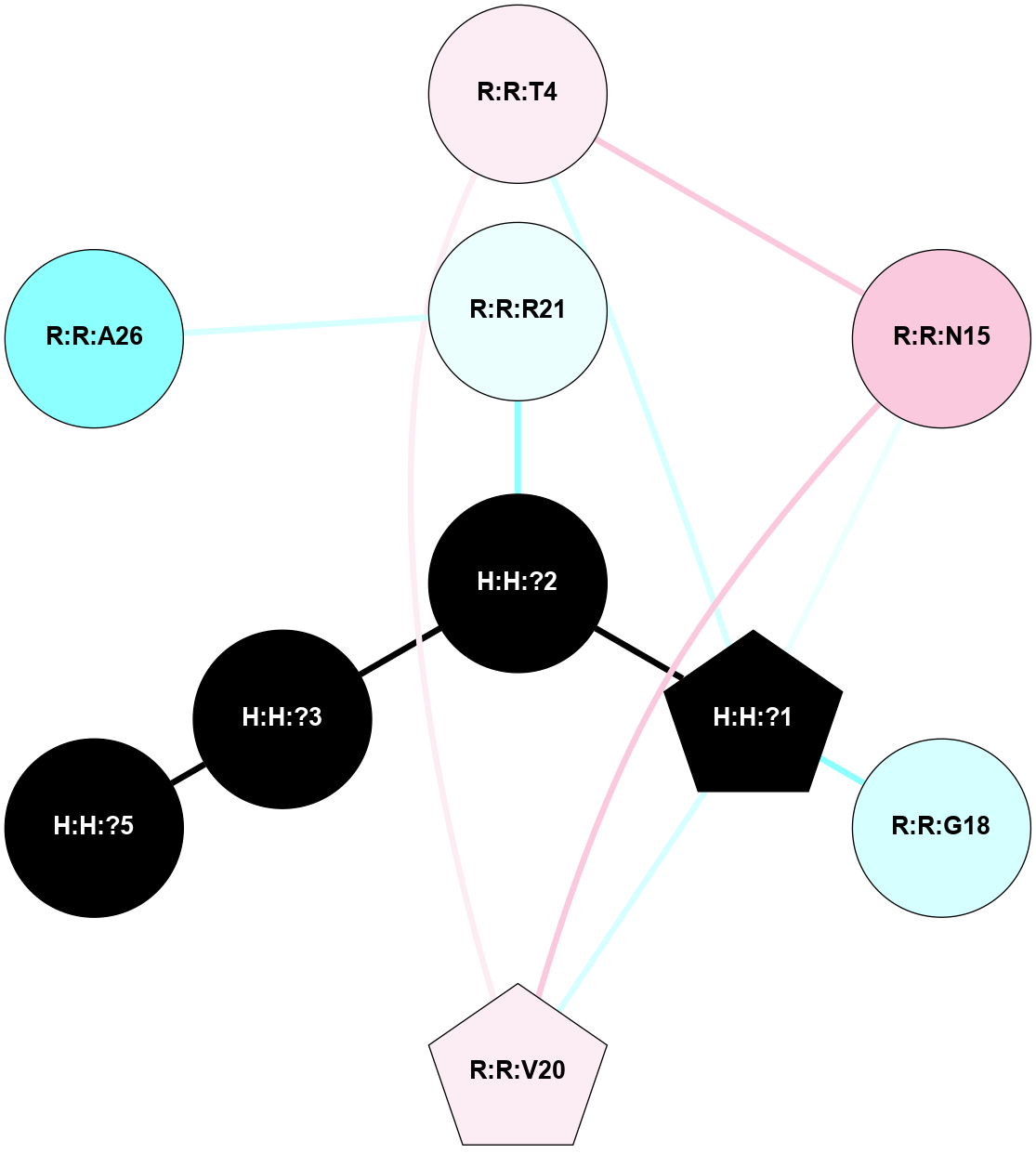
A 2D representation of the interactions of NAG in 4PXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 2 | No | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 1 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 2 | No | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 28.24 | 2 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 7.65 | 2 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 6.66 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?2 | R:R:R21 | 5.43 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 37.64 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 29.54 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?5 | 36.93 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 24.20 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4X1H | A | Sensory | Opsins | Rhodopsin | Bos Taurus | Nonyl-Beta-D-Glucopyranoside | - | Gt(CT) | 2.29 | 2015-11-04 | doi.org/10.1016/j.str.2015.09.015 |
|
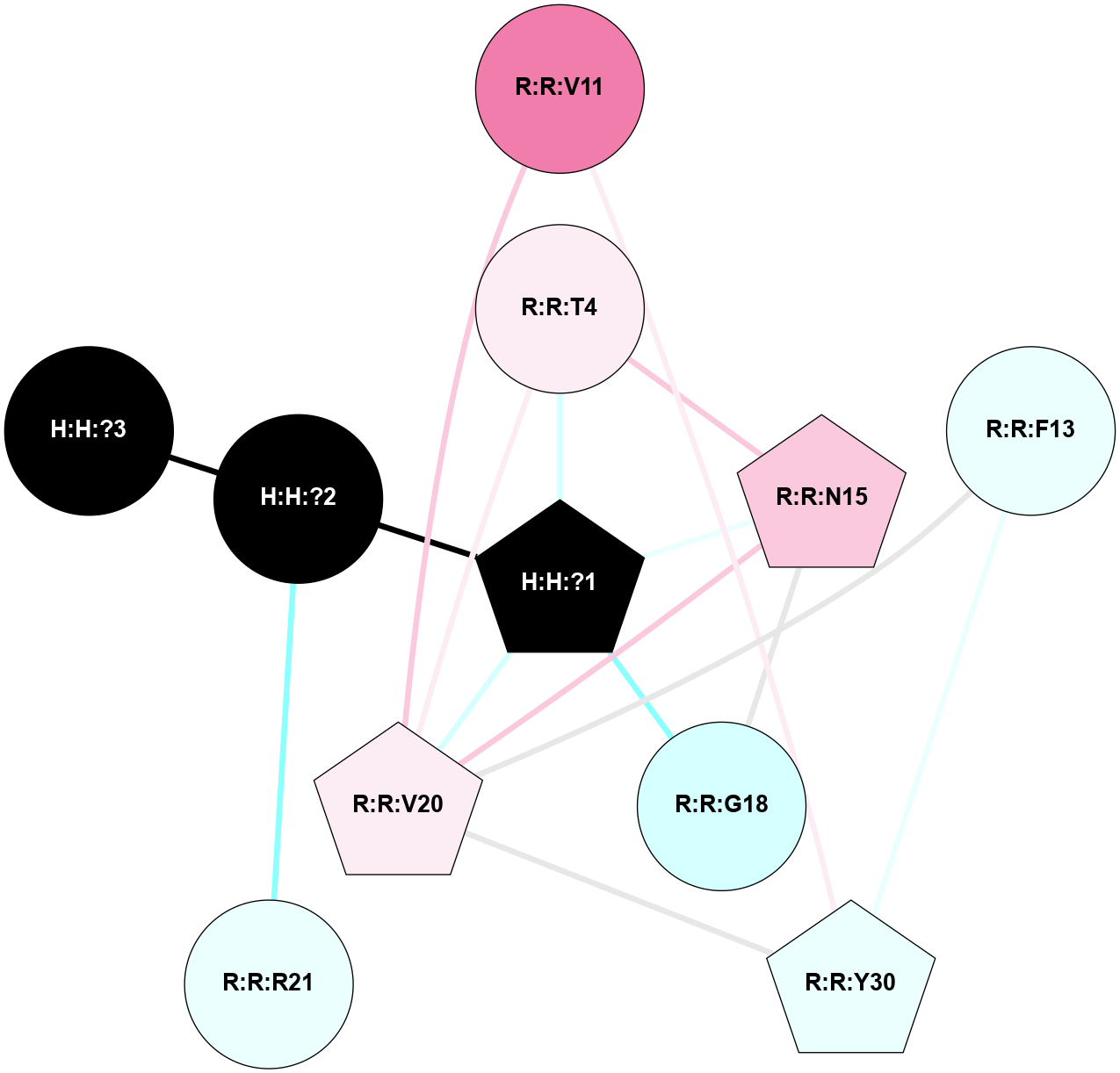
A 2D representation of the interactions of NAG in 4X1H
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 2 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 2 | No | Yes | 8 | 6 | 2 | 1 | | R:R:V11 | R:R:Y30 | 5.05 | 2 | No | Yes | 8 | 4 | 2 | 2 | | R:R:F13 | R:R:V20 | 3.93 | 2 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 7.22 | 2 | No | Yes | 4 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 2 | Yes | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 25.79 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 11.36 | 2 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 38.74 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 34.67 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 15.98 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
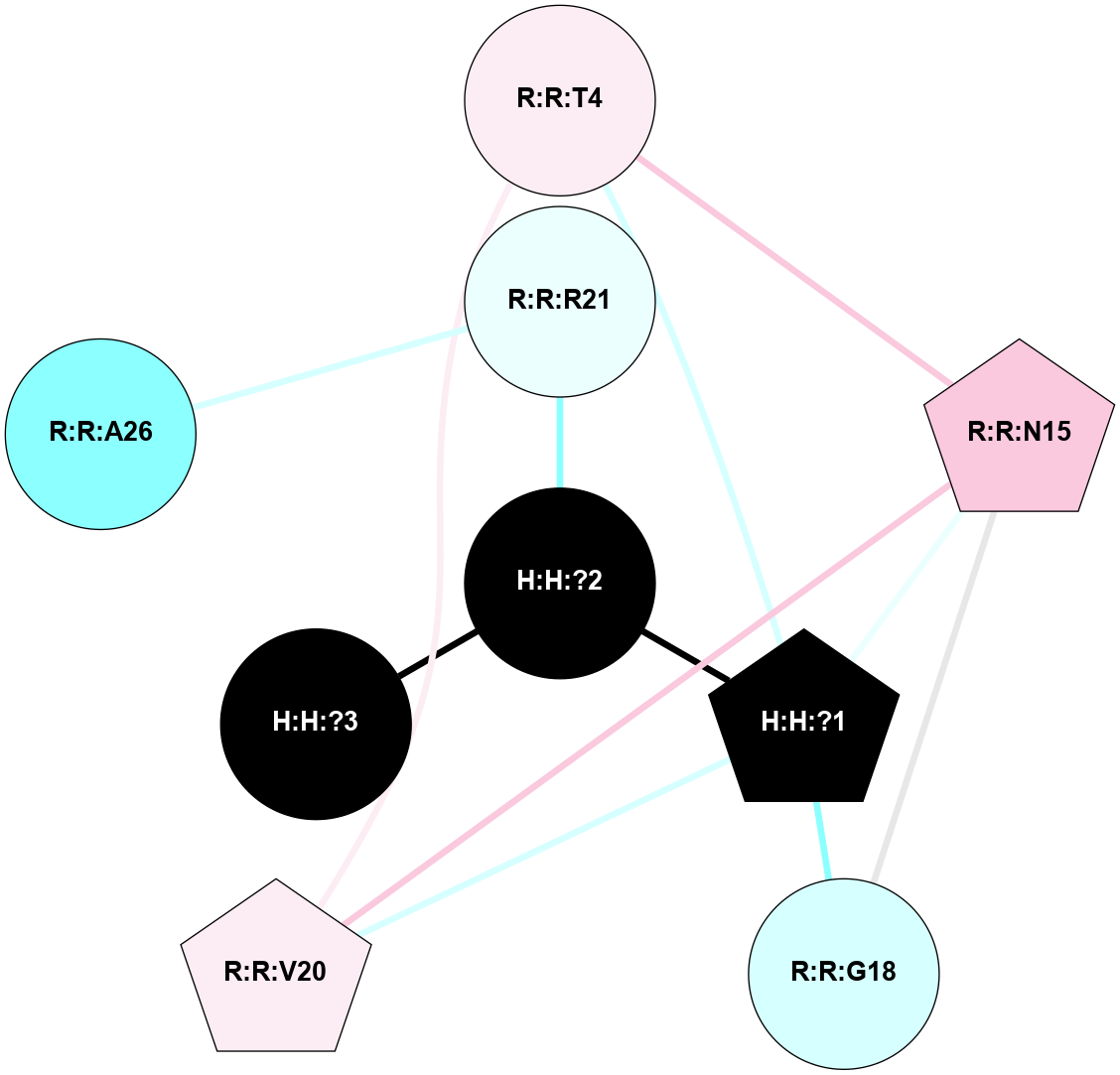
A 2D representation of the interactions of NAG in 4X1H
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 2 | Yes | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 2 | Yes | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 25.79 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 38.74 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 34.67 | 0 | No | No | 0 | 0 | 0 | 1 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 26.64 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
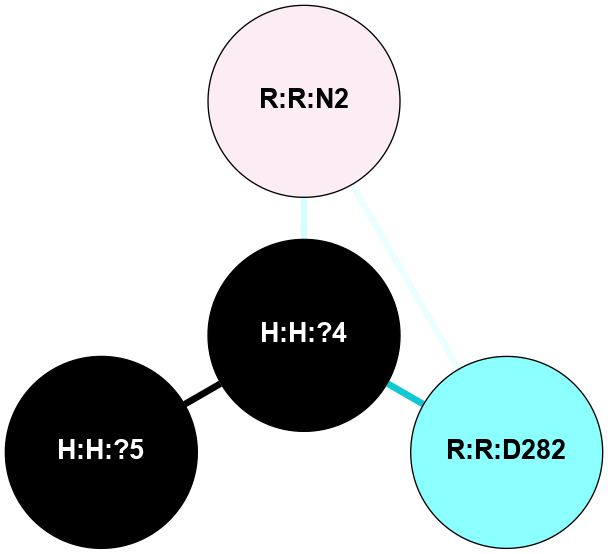
A 2D representation of the interactions of NAG in 4X1H
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 21.54 | 5 | No | No | 2 | 6 | 1 | 1 | | H:H:?4 | R:R:N2 | 30.7 | 5 | No | No | 0 | 6 | 0 | 1 | | H:H:?4 | R:R:D282 | 14.56 | 5 | No | No | 0 | 2 | 0 | 1 | | H:H:?4 | H:H:?5 | 38.74 | 5 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 28.00 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 4X1H
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D282 | R:R:N2 | 21.54 | 5 | No | No | 2 | 6 | 2 | 2 | | H:H:?4 | R:R:N2 | 30.7 | 5 | No | No | 0 | 6 | 1 | 2 | | H:H:?4 | R:R:D282 | 14.56 | 5 | No | No | 0 | 2 | 1 | 2 | | H:H:?4 | H:H:?5 | 38.74 | 5 | No | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 38.74 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 4XT3 | A | Other | Unclassified | US28 | Human Cytomegalovirus (Strain Ad169) | Fractalkine | - | - | 3.8 | 2015-03-04 | doi.org/10.1126/science.aaa5026 |
|
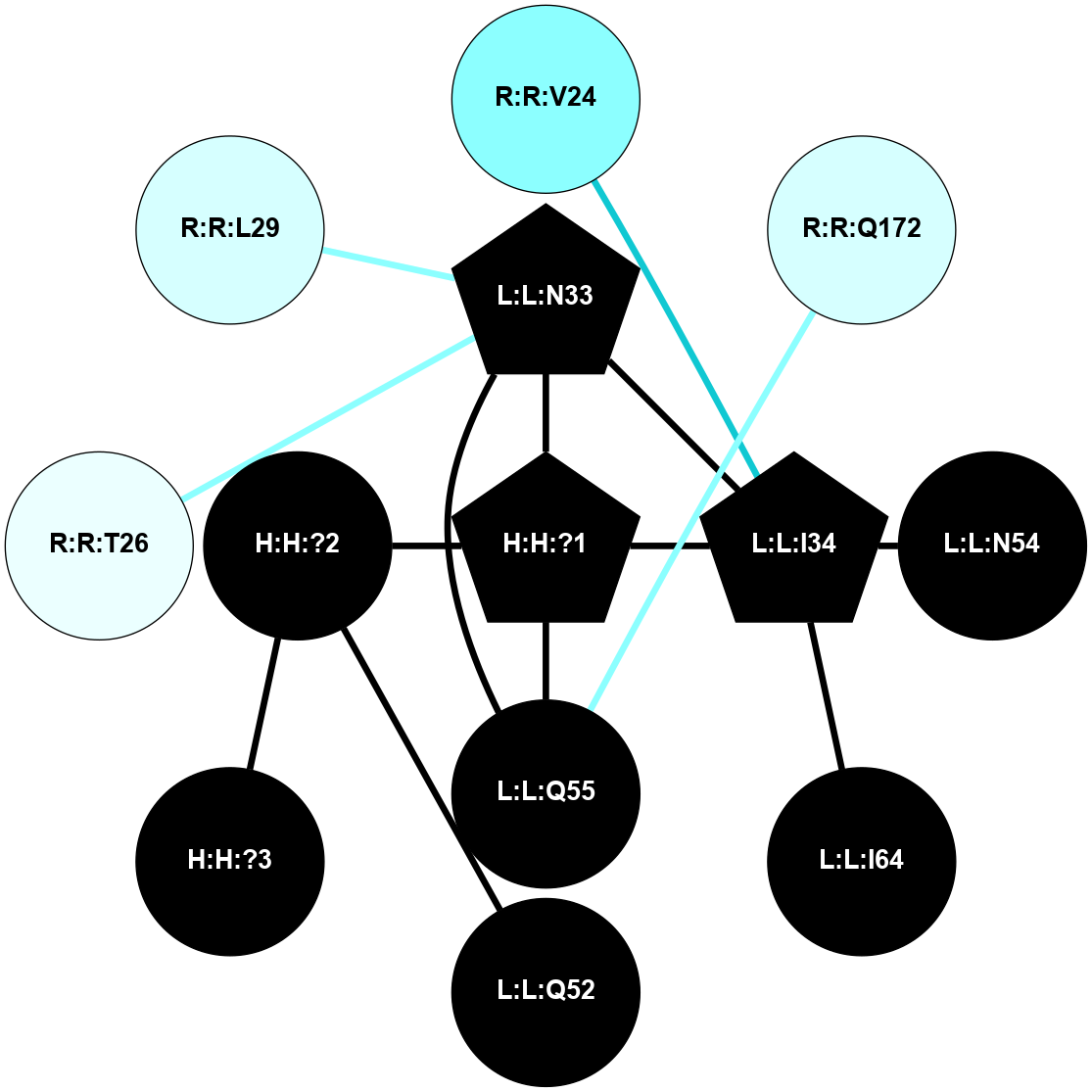
A 2D representation of the interactions of NAG in 4XT3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 48.71 | 7 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | L:L:N33 | 23.33 | 7 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?1 | L:L:I34 | 6.38 | 7 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?1 | L:L:Q55 | 7.14 | 7 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 38.74 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?2 | L:L:Q52 | 16.66 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:I34 | L:L:N33 | 4.25 | 7 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:N33 | L:L:Q55 | 6.6 | 7 | Yes | No | 0 | 0 | 1 | 1 | | L:L:I34 | L:L:N54 | 4.25 | 7 | Yes | No | 0 | 0 | 1 | 2 | | L:L:I34 | L:L:I64 | 7.36 | 7 | Yes | No | 0 | 0 | 1 | 2 | | L:L:I34 | R:R:V24 | 3.07 | 7 | Yes | No | 0 | 2 | 1 | 2 | | L:L:Q55 | R:R:Q172 | 6.4 | 7 | No | No | 0 | 3 | 1 | 2 | | L:L:N33 | R:R:T26 | 2.92 | 7 | Yes | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 21.39 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
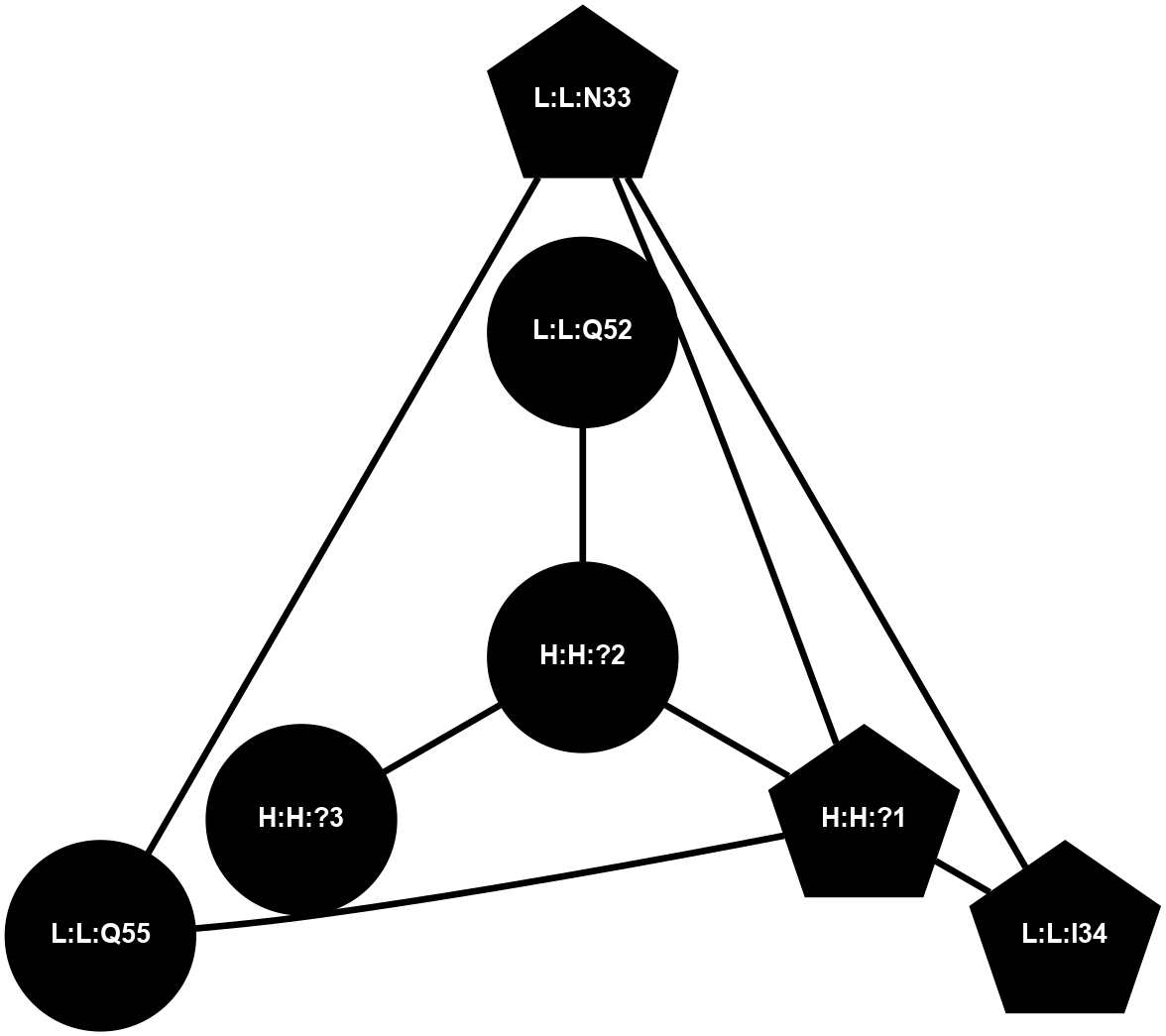
A 2D representation of the interactions of NAG in 4XT3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 48.71 | 7 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | L:L:N33 | 23.33 | 7 | Yes | Yes | 0 | 0 | 1 | 2 | | H:H:?1 | L:L:I34 | 6.38 | 7 | Yes | Yes | 0 | 0 | 1 | 2 | | H:H:?1 | L:L:Q55 | 7.14 | 7 | Yes | No | 0 | 0 | 1 | 2 | | H:H:?2 | H:H:?3 | 38.74 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?2 | L:L:Q52 | 16.66 | 0 | No | No | 0 | 0 | 0 | 1 | | L:L:I34 | L:L:N33 | 4.25 | 7 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:N33 | L:L:Q55 | 6.6 | 7 | Yes | No | 0 | 0 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 34.70 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
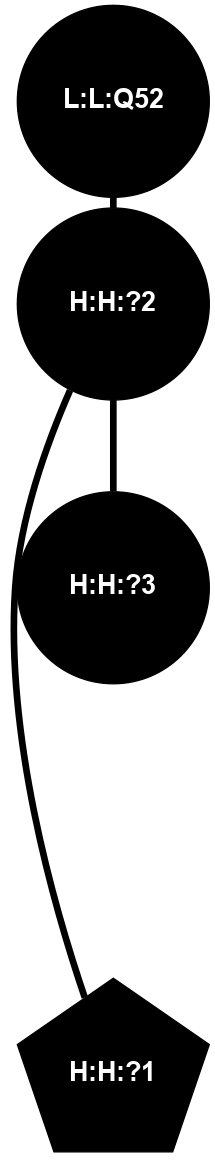
A 2D representation of the interactions of NAG in 4XT3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 48.71 | 7 | Yes | No | 0 | 0 | 2 | 1 | | H:H:?2 | H:H:?3 | 38.74 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?2 | L:L:Q52 | 16.66 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 38.74 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5DYS | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 2.3 | 2016-08-10 | doi.org/10.15252/embr.201642671 |
|
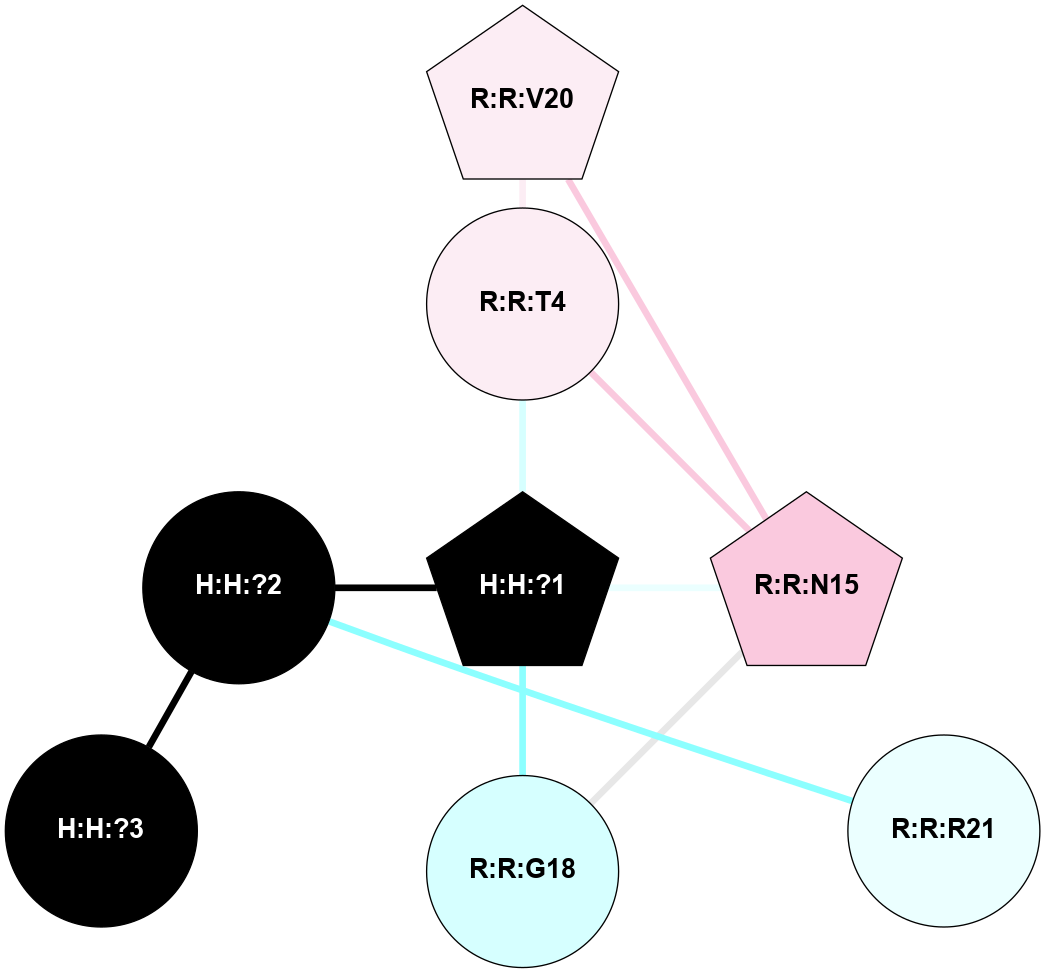
A 2D representation of the interactions of NAG in 5DYS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 1 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 24.56 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 37.64 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 28.25 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 18.40 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
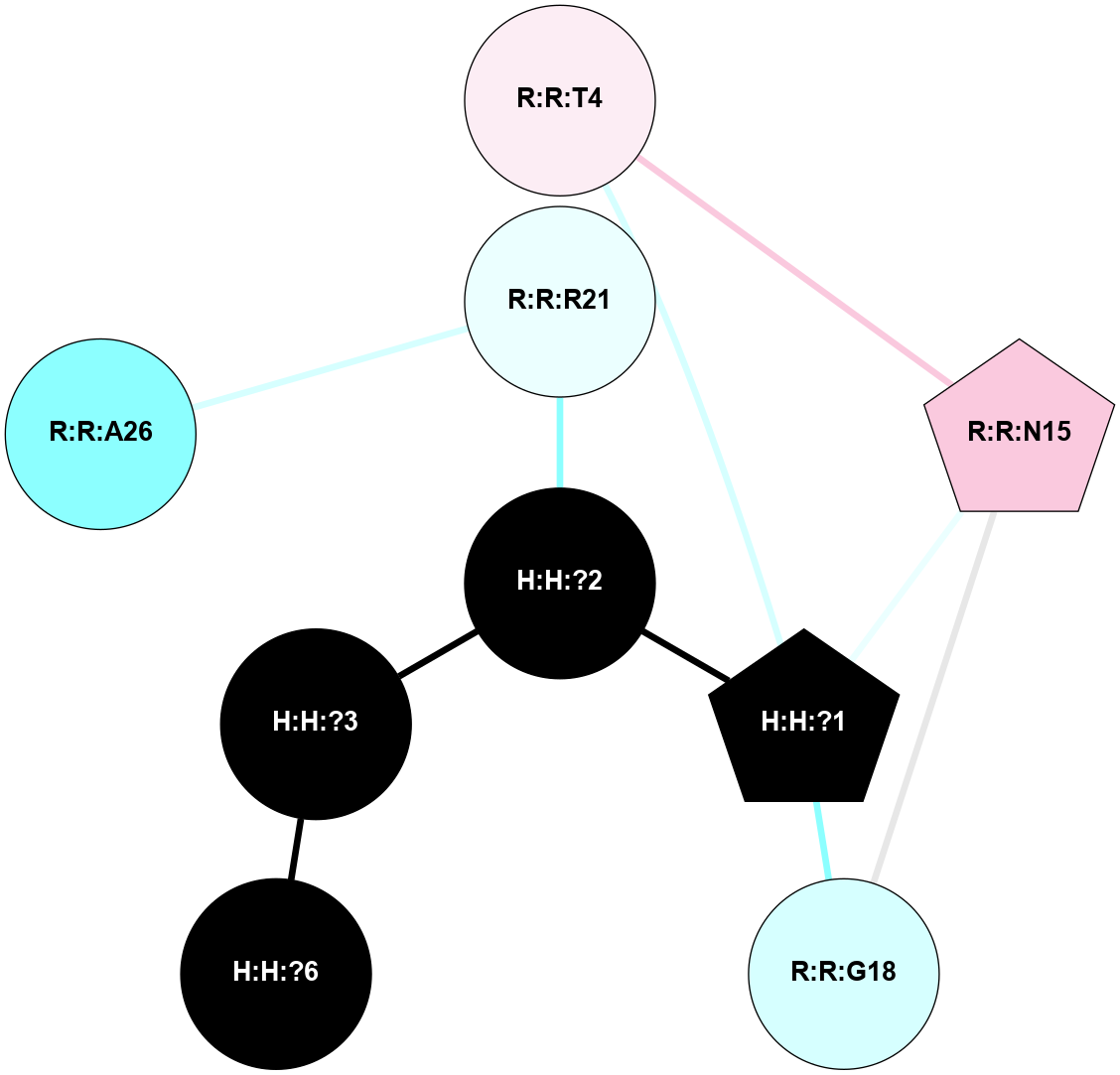
A 2D representation of the interactions of NAG in 5DYS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 1 | Yes | No | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 2 | 2 | | H:H:?1 | R:R:N15 | 24.56 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 37.64 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 28.25 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?6 | 39.89 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 24.14 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5EN0 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | Gt(CT) | 2.81 | 2016-08-10 | doi.org/10.15252/embr.201642671 |
|
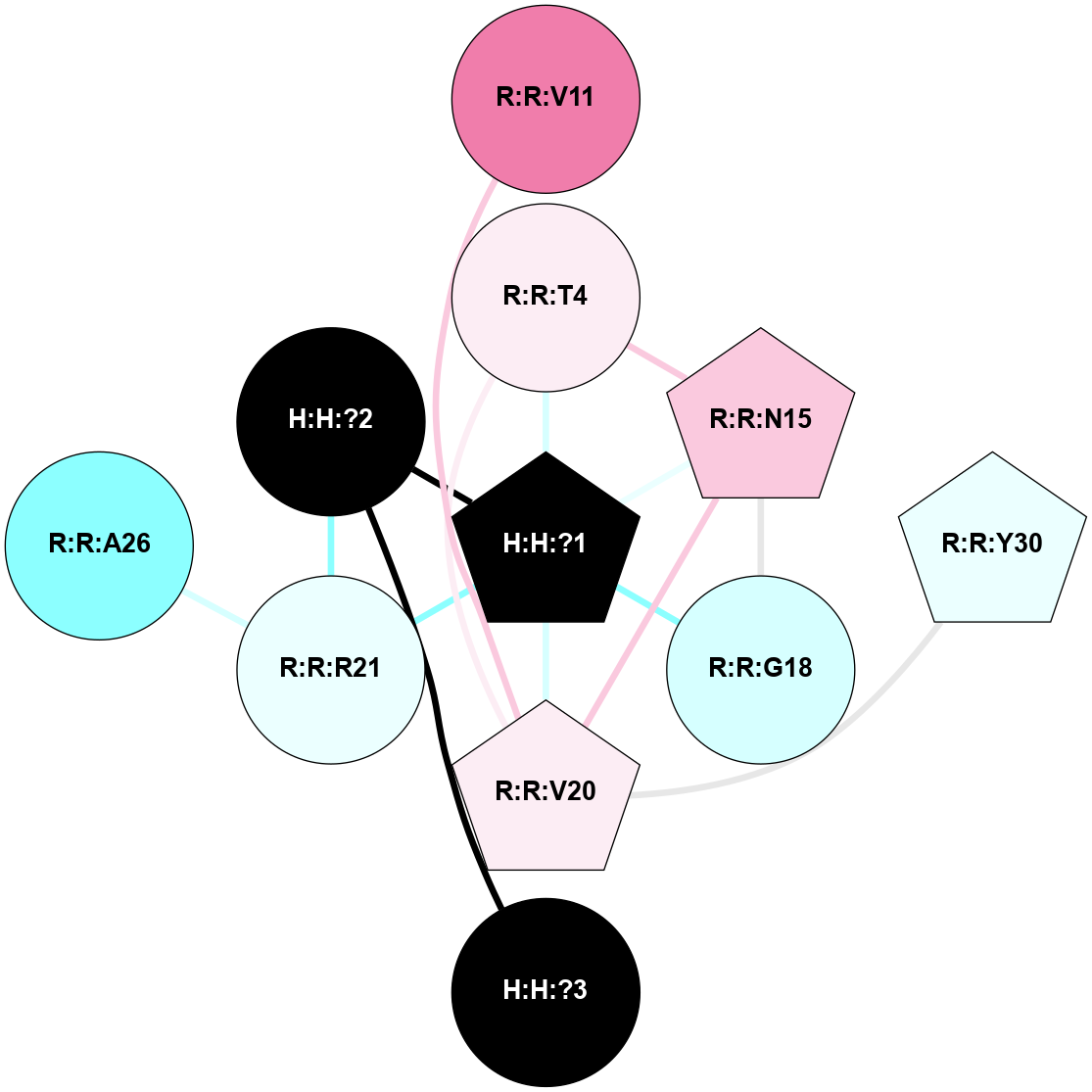
A 2D representation of the interactions of NAG in 5EN0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.29 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | No | Yes | 8 | 6 | 2 | 1 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 25.87 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.14 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 10.09 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 4.01 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:R21 | 5.45 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:R21 | 8.72 | 1 | No | No | 0 | 4 | 1 | 1 | | H:H:?1 | H:H:?2 | 37.89 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 25.98 | 1 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 14.11 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
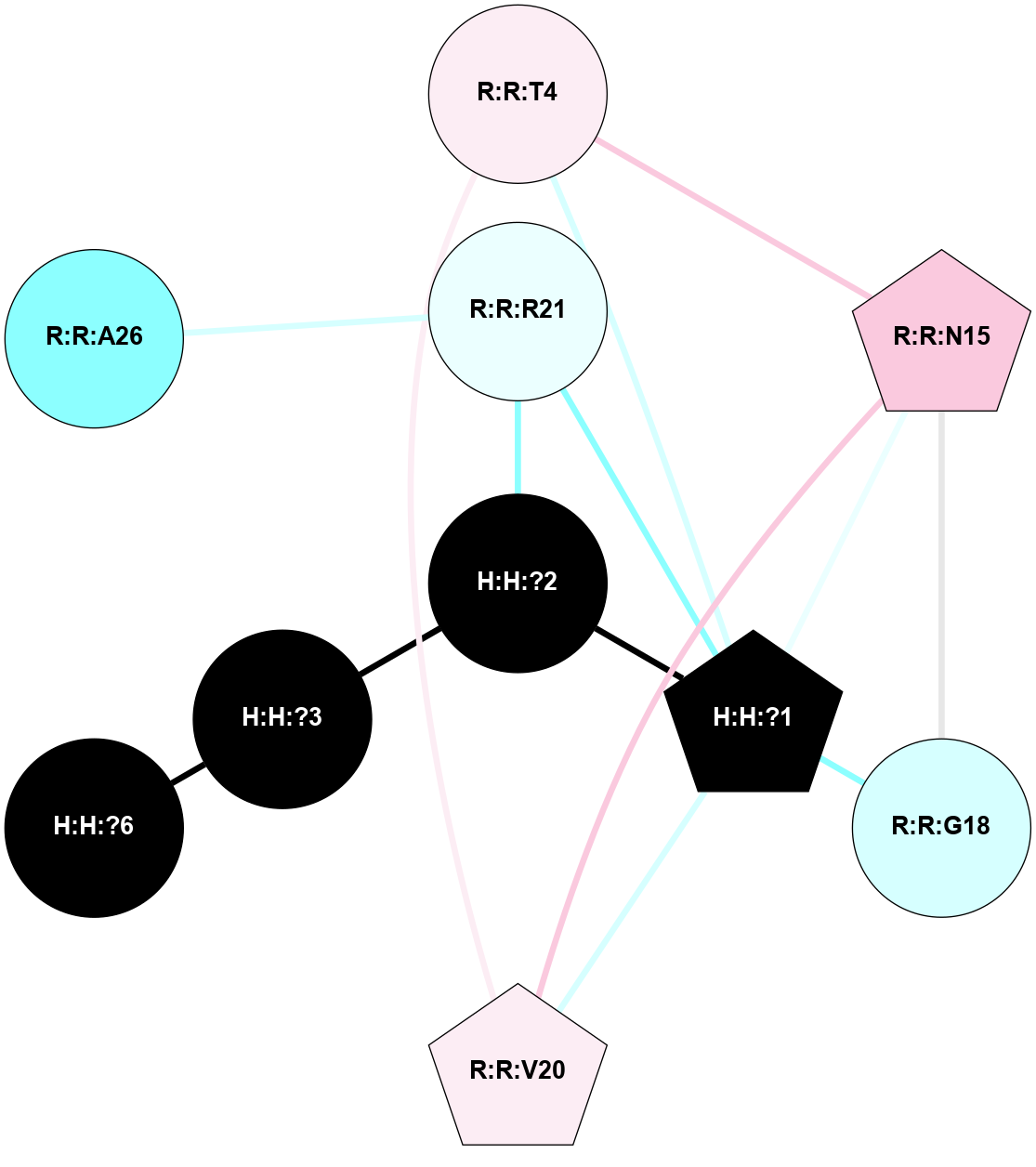
A 2D representation of the interactions of NAG in 5EN0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 1 | Yes | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.29 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 25.87 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.14 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 4.01 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:R21 | 5.45 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?2 | R:R:R21 | 8.72 | 1 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 37.89 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 25.98 | 1 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?6 | 44.81 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 24.20 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5L7D | F | Protein | Frizzled | SMO | Homo Sapiens | - | Cholesterol | - | 3.2 | 2016-07-20 | doi.org/10.1038/nature18934 |
|

A 2D representation of the interactions of NAG in 5L7D
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:K186 | R:R:N188 | 9.79 | 12 | No | No | 4 | 8 | 1 | 1 | | H:H:?1 | R:R:K186 | 10.09 | 12 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:N188 | 35.61 | 12 | No | No | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 22.85 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
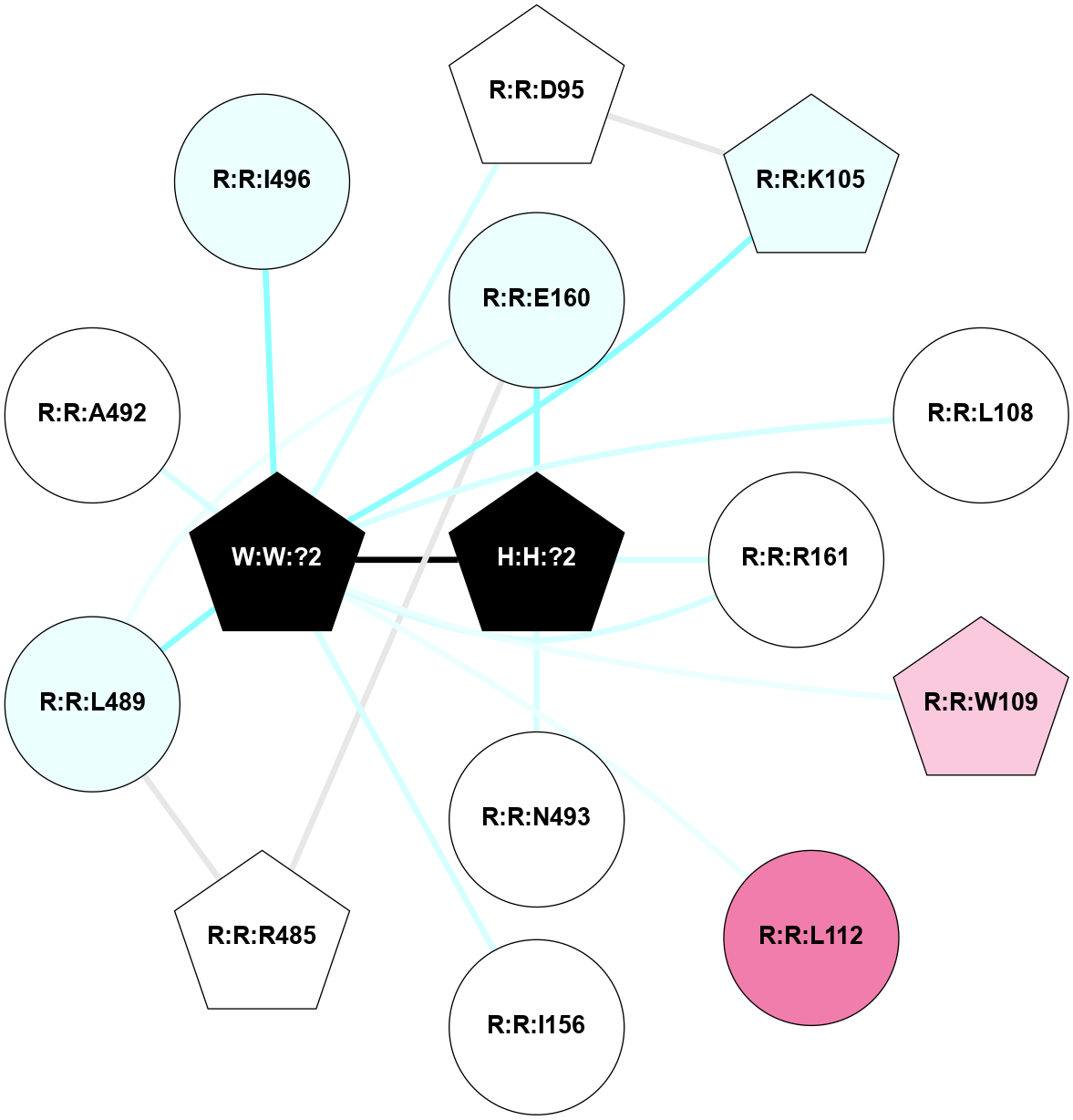
A 2D representation of the interactions of NAG in 5L7D
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D95 | R:R:K105 | 13.83 | 1 | Yes | Yes | 5 | 4 | 2 | 2 | | R:R:D95 | W:W:?2 | 8.19 | 1 | Yes | Yes | 5 | 0 | 2 | 1 | | R:R:K105 | W:W:?2 | 5.67 | 1 | Yes | Yes | 4 | 0 | 2 | 1 | | R:R:L108 | W:W:?2 | 5.57 | 0 | No | Yes | 5 | 0 | 2 | 1 | | R:R:W109 | W:W:?2 | 44.68 | 0 | Yes | Yes | 7 | 0 | 2 | 1 | | R:R:L112 | W:W:?2 | 12.53 | 0 | No | Yes | 8 | 0 | 2 | 1 | | R:R:I156 | W:W:?2 | 7.18 | 0 | No | Yes | 5 | 0 | 2 | 1 | | R:R:E160 | R:R:R485 | 4.65 | 1 | No | Yes | 4 | 5 | 1 | 2 | | R:R:E160 | R:R:L489 | 10.6 | 1 | No | No | 4 | 4 | 1 | 2 | | H:H:?2 | R:R:E160 | 14.22 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:R161 | W:W:?2 | 21.99 | 1 | No | Yes | 5 | 0 | 1 | 1 | | H:H:?2 | R:R:R161 | 6.52 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:L489 | R:R:R485 | 4.86 | 1 | No | Yes | 4 | 5 | 2 | 2 | | R:R:L489 | W:W:?2 | 5.57 | 1 | No | Yes | 4 | 0 | 2 | 1 | | R:R:A492 | W:W:?2 | 7.92 | 0 | No | Yes | 5 | 0 | 2 | 1 | | H:H:?2 | R:R:N493 | 28.24 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:I496 | W:W:?2 | 4.31 | 0 | No | Yes | 4 | 0 | 2 | 1 | | H:H:?2 | W:W:?2 | 7.47 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.11 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5NX2 | B1 | Peptide | Glucagon | GLP-1 | Homo Sapiens | GLP-1 (Truncated) | - | - | 3.7 | 2017-06-14 | doi.org/10.1038/nature22800 |
|
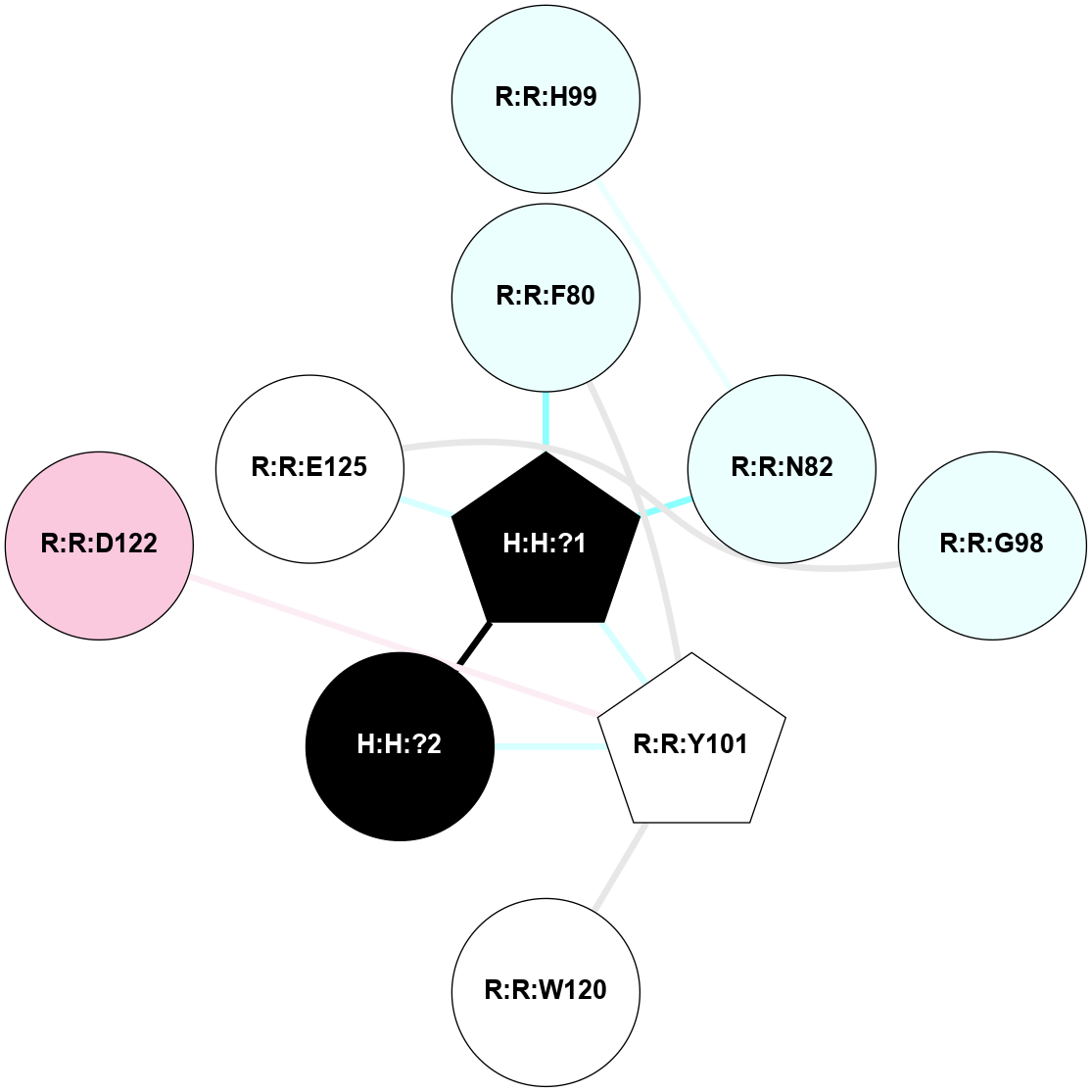
A 2D representation of the interactions of NAG in 5NX2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F80 | R:R:Y101 | 6.19 | 4 | No | Yes | 4 | 5 | 1 | 1 | | H:H:?1 | R:R:F80 | 7.62 | 4 | Yes | No | 0 | 4 | 0 | 1 | | R:R:H99 | R:R:N82 | 3.83 | 0 | No | No | 4 | 4 | 2 | 1 | | H:H:?1 | R:R:N82 | 38.07 | 4 | Yes | No | 0 | 4 | 0 | 1 | | R:R:E125 | R:R:G98 | 4.91 | 0 | No | No | 5 | 4 | 1 | 2 | | R:R:W120 | R:R:Y101 | 6.75 | 0 | No | Yes | 5 | 5 | 2 | 1 | | R:R:D122 | R:R:Y101 | 20.69 | 0 | No | Yes | 7 | 5 | 2 | 1 | | H:H:?1 | R:R:Y101 | 7.34 | 4 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:Y101 | 7.34 | 4 | No | Yes | 0 | 5 | 1 | 1 | | H:H:?1 | H:H:?2 | 32.1 | 4 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:E125 | 2.37 | 4 | Yes | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.50 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
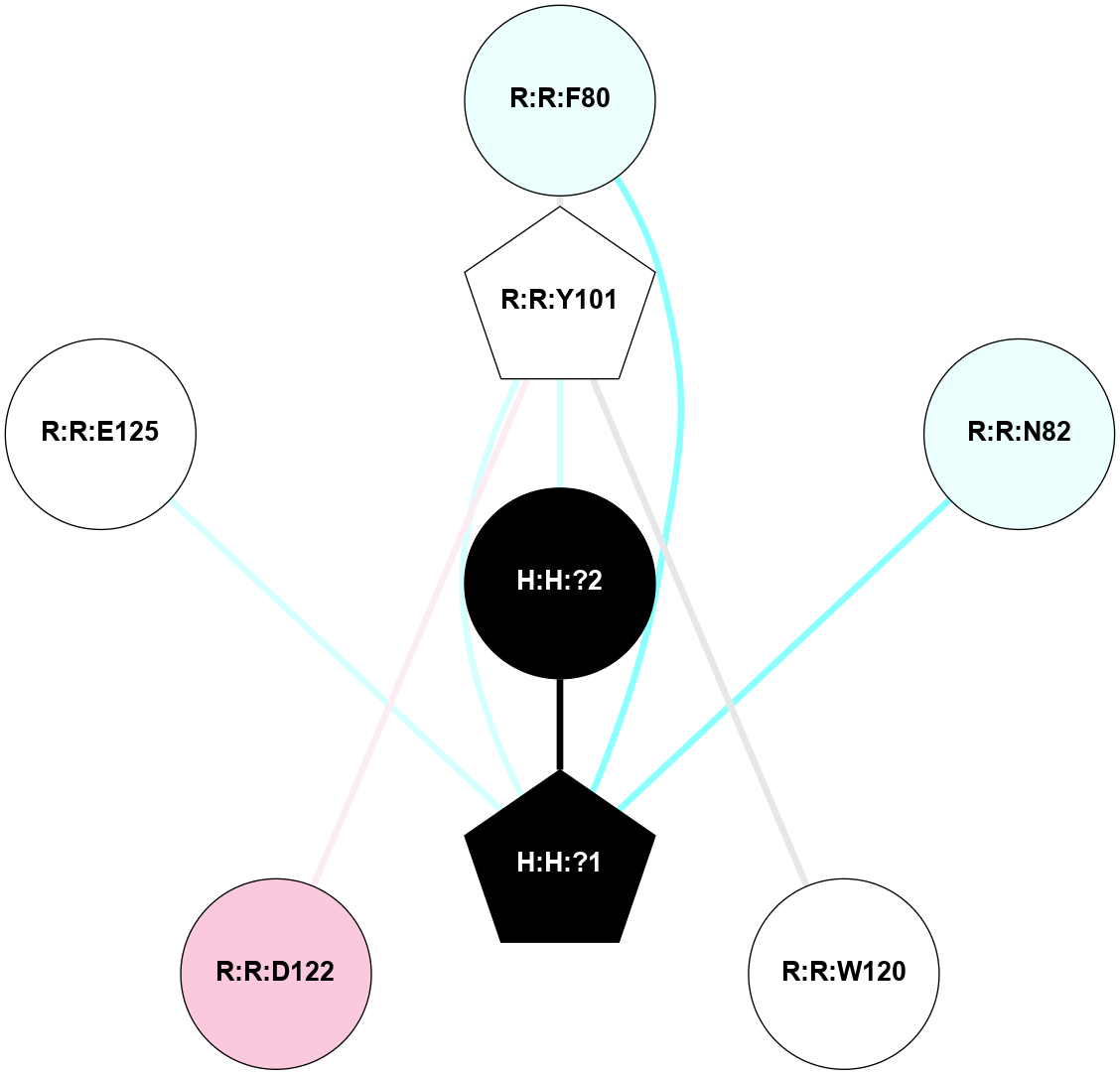
A 2D representation of the interactions of NAG in 5NX2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F80 | R:R:Y101 | 6.19 | 4 | No | Yes | 4 | 5 | 2 | 1 | | H:H:?1 | R:R:F80 | 7.62 | 4 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:N82 | 38.07 | 4 | Yes | No | 0 | 4 | 1 | 2 | | R:R:W120 | R:R:Y101 | 6.75 | 0 | No | Yes | 5 | 5 | 2 | 1 | | R:R:D122 | R:R:Y101 | 20.69 | 0 | No | Yes | 7 | 5 | 2 | 1 | | H:H:?1 | R:R:Y101 | 7.34 | 4 | Yes | Yes | 0 | 5 | 1 | 1 | | H:H:?2 | R:R:Y101 | 7.34 | 4 | No | Yes | 0 | 5 | 0 | 1 | | H:H:?1 | H:H:?2 | 32.1 | 4 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:E125 | 2.37 | 4 | Yes | No | 0 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 19.72 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
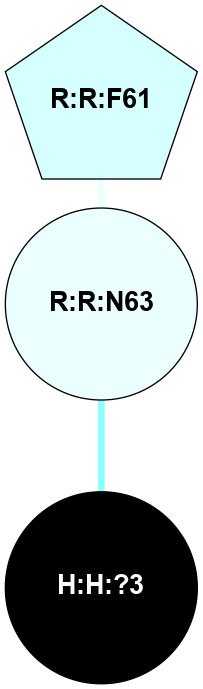
A 2D representation of the interactions of NAG in 5NX2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:N63 | 22.1 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:F61 | R:R:N63 | 2.42 | 6 | Yes | No | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 22.10 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5TE3 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | Octyl-Beta-D-Glucopyranoside | - | - | 2.7 | 2017-03-15 | doi.org/10.1073/pnas.1617446114 |
|
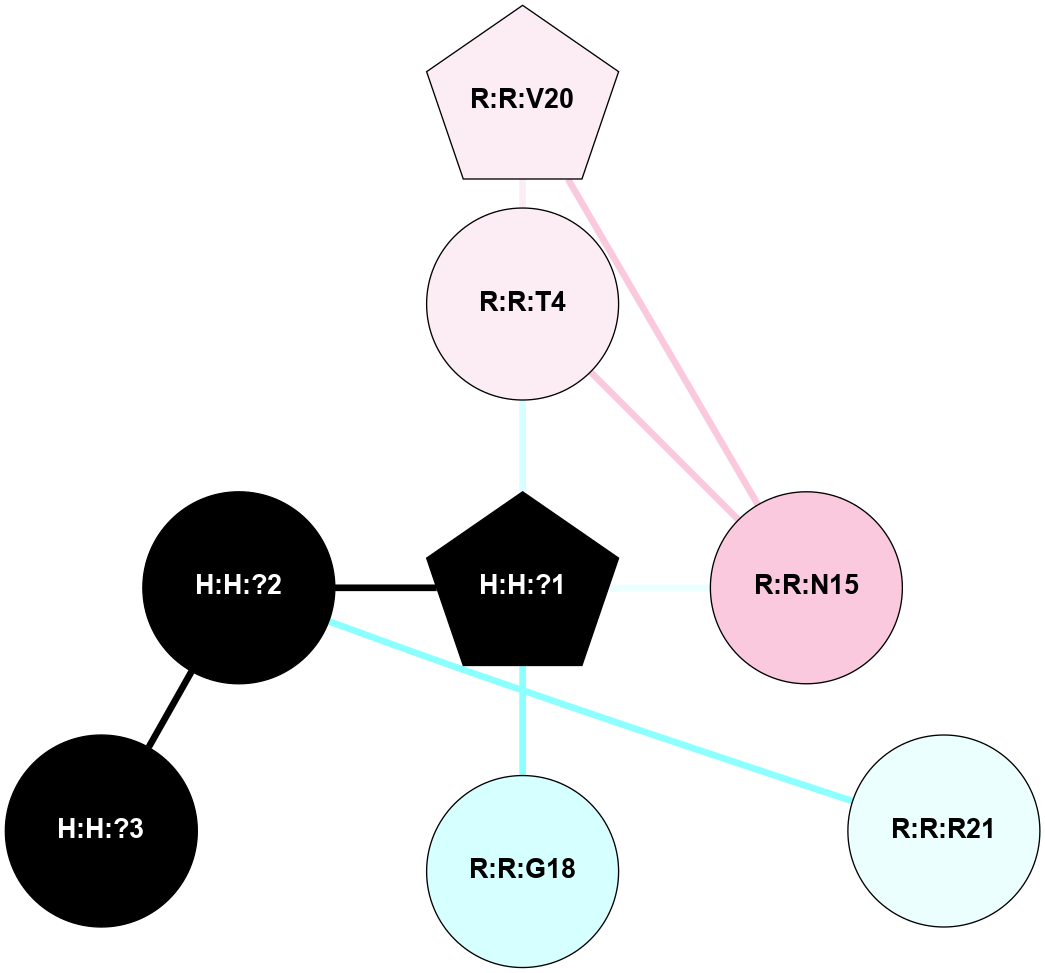
A 2D representation of the interactions of NAG in 5TE3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 2 | No | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 11.11 | 2 | No | Yes | 6 | 6 | 1 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 0 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 2 | No | Yes | 7 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 24.56 | 2 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?2 | R:R:R21 | 7.6 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 38.74 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 25.68 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.67 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
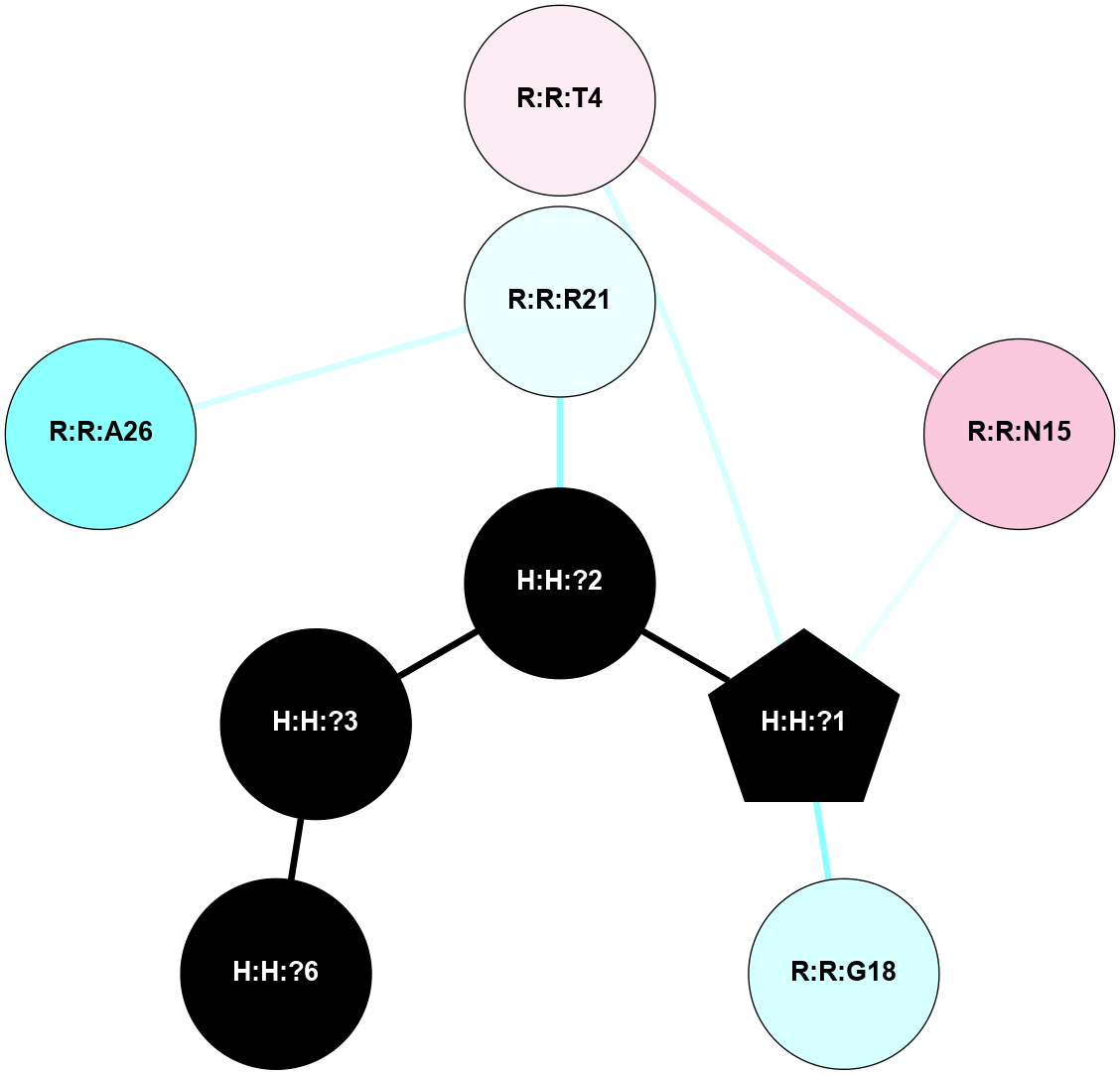
A 2D representation of the interactions of NAG in 5TE3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 2 | No | No | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 24.56 | 2 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?2 | R:R:R21 | 7.6 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 38.74 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 25.68 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?6 | 47.27 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 24.01 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5W0P | A | Sensory | Opsins | Rhodopsin | Homo Sapiens | - | - | Arrestin1 | 3.01 | 2017-08-09 | doi.org/10.1016/j.cell.2017.07.002 |
|
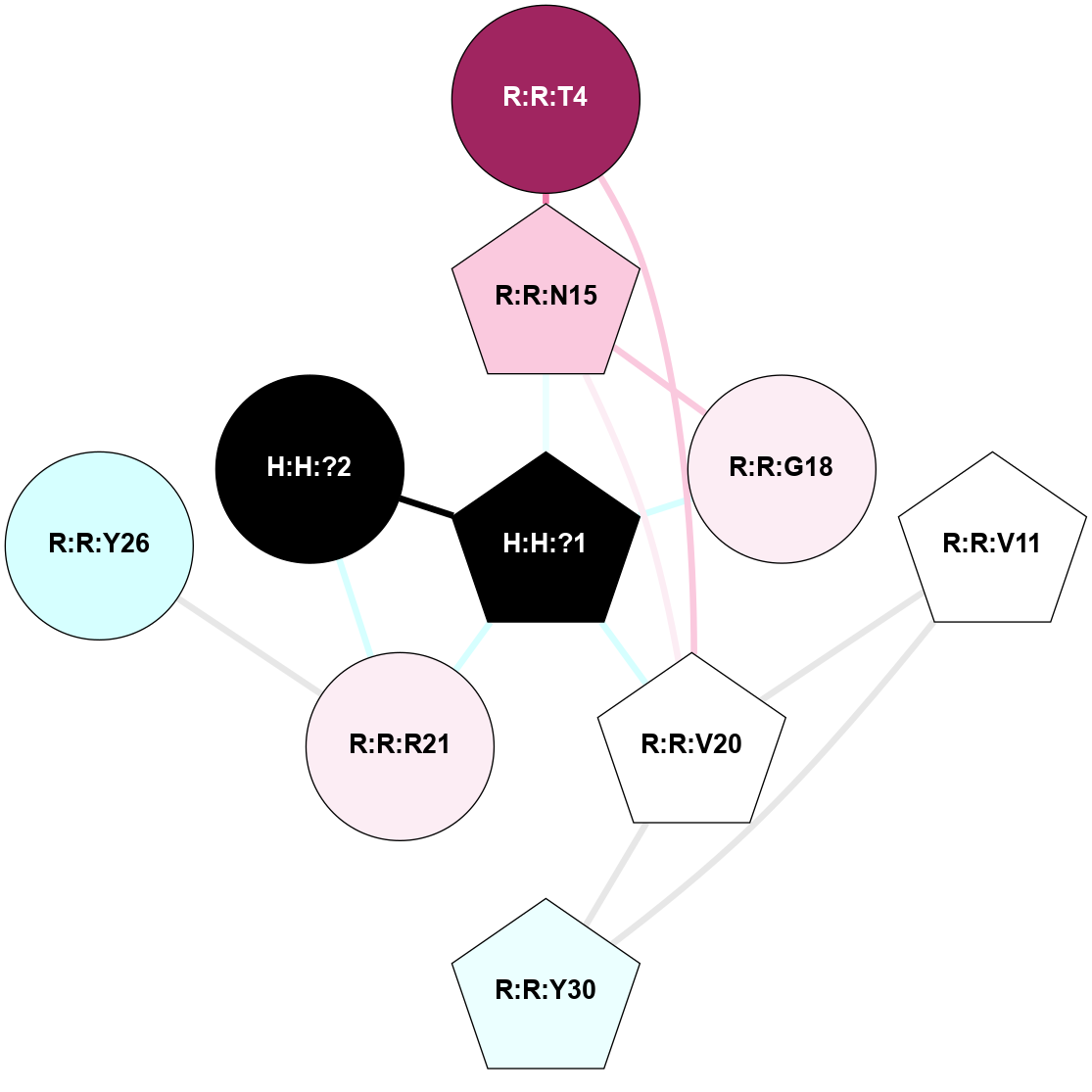
A 2D representation of the interactions of NAG in 5W0P
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 29.89 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N15 | 24.56 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 4.59 | 1 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:V20 | 5.33 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:R21 | 15.21 | 1 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:R21 | 9.78 | 1 | No | No | 0 | 6 | 1 | 1 | | R:R:N15 | R:R:T4 | 5.85 | 1 | Yes | No | 7 | 9 | 1 | 2 | | R:R:T4 | R:R:V20 | 7.93 | 1 | No | Yes | 9 | 5 | 2 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:V11 | R:R:Y30 | 3.79 | 1 | Yes | Yes | 5 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 3.39 | 1 | No | Yes | 6 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 5 | 1 | 1 | | R:R:V20 | R:R:Y30 | 8.83 | 1 | Yes | Yes | 5 | 4 | 1 | 2 | | R:R:R21 | R:R:Y26 | 8.23 | 1 | No | No | 6 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 15.92 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
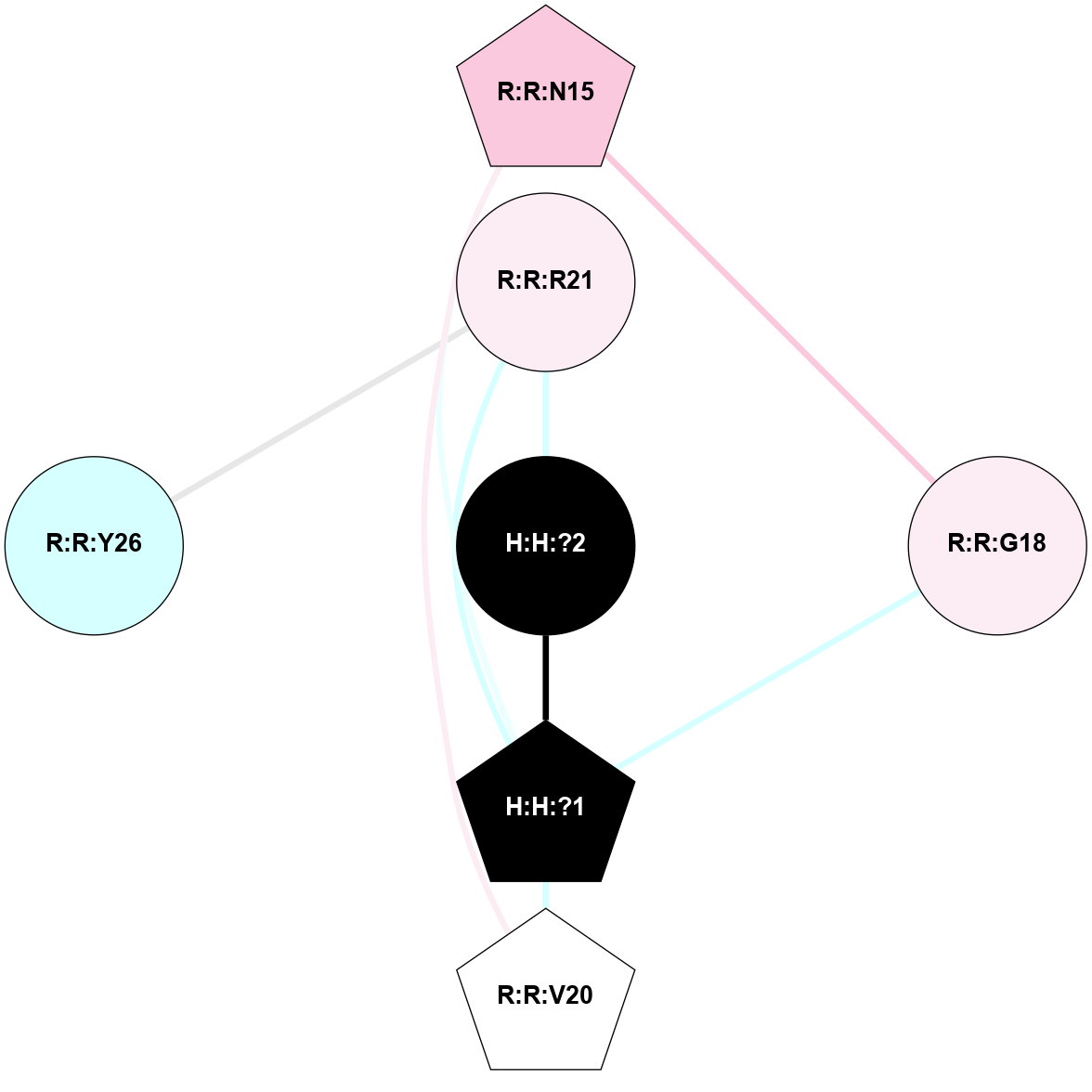
A 2D representation of the interactions of NAG in 5W0P
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 29.89 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N15 | 24.56 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 4.59 | 1 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:V20 | 5.33 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | H:H:?1 | R:R:R21 | 15.21 | 1 | Yes | No | 0 | 6 | 1 | 1 | | H:H:?2 | R:R:R21 | 9.78 | 1 | No | No | 0 | 6 | 0 | 1 | | R:R:G18 | R:R:N15 | 3.39 | 1 | No | Yes | 6 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 5 | 2 | 2 | | R:R:R21 | R:R:Y26 | 8.23 | 1 | No | No | 6 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.84 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5WKT | A | Sensory | Opsins | Rhodopsin | Bos Taurus | Octyl-Beta-D-Glucopyranoside | - | Gt(CT) | 3.2 | 2017-12-13 | doi.org/10.1038/nprot.2017.135 |
|
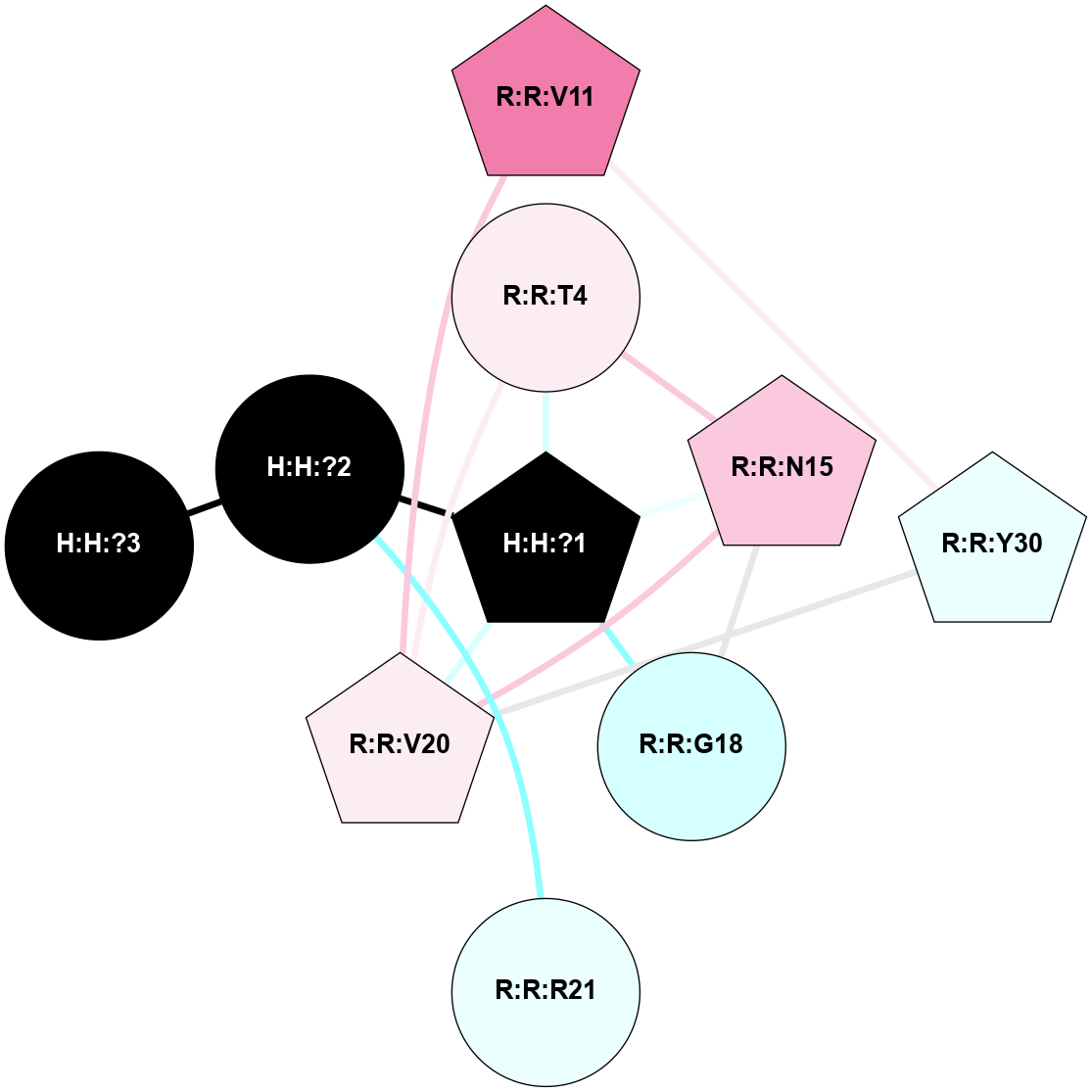
A 2D representation of the interactions of NAG in 5WKT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 14.28 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:V11 | R:R:Y30 | 5.05 | 1 | Yes | Yes | 8 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 3.39 | 1 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 24.56 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 7.65 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 11.36 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 5.33 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 34.31 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 30.82 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 15.42 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
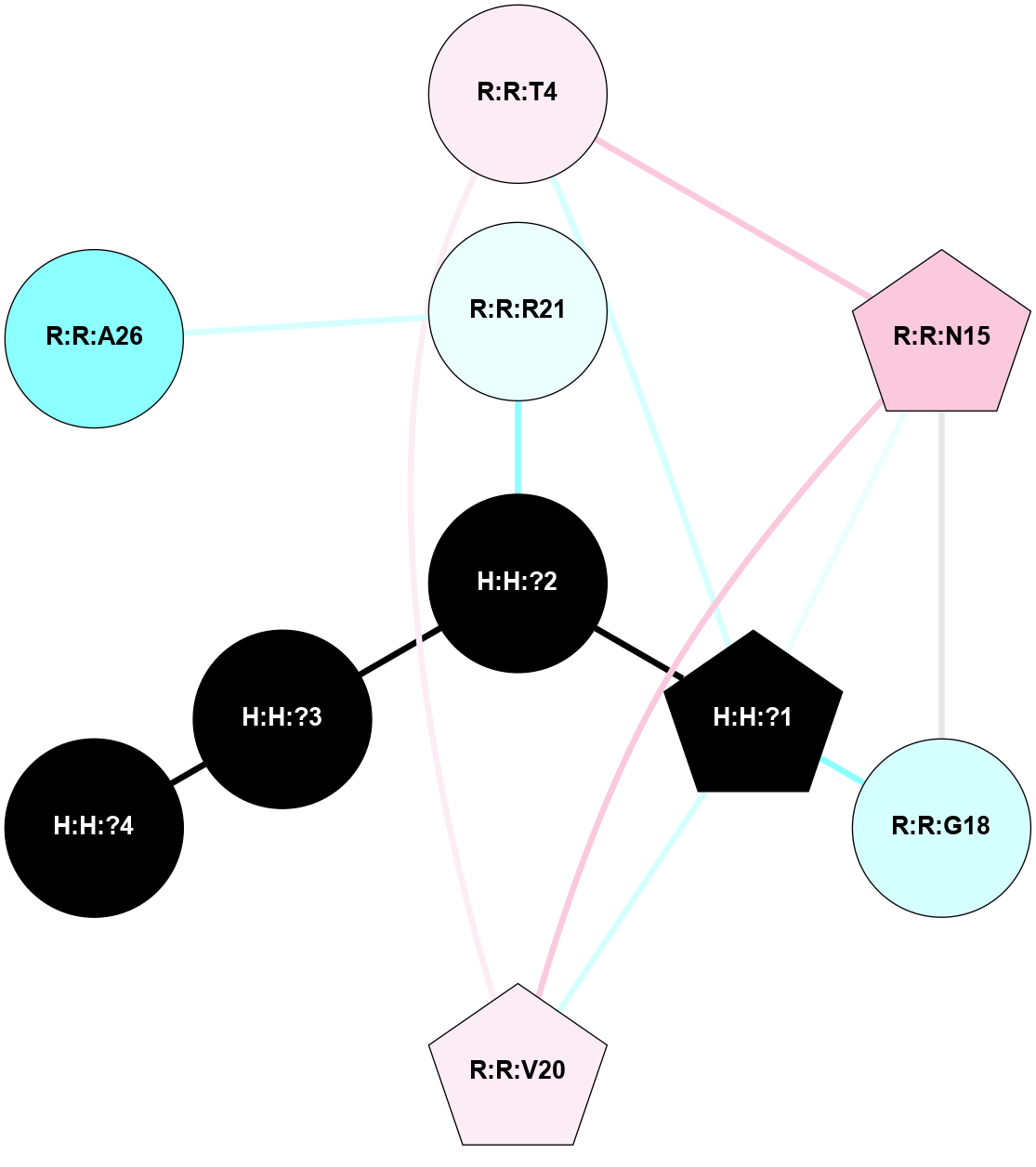
A 2D representation of the interactions of NAG in 5WKT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 1 | Yes | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 14.28 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 3.39 | 1 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 24.56 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 7.65 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 5.33 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 34.31 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 30.82 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?4 | 45.8 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 23.88 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5XEZ | B1 | Peptide | Glucagon | Glucagon | Homo Sapiens | - | NNC0640 | - | 3 | 2017-05-24 | doi.org/10.1038/nature22363 |
|
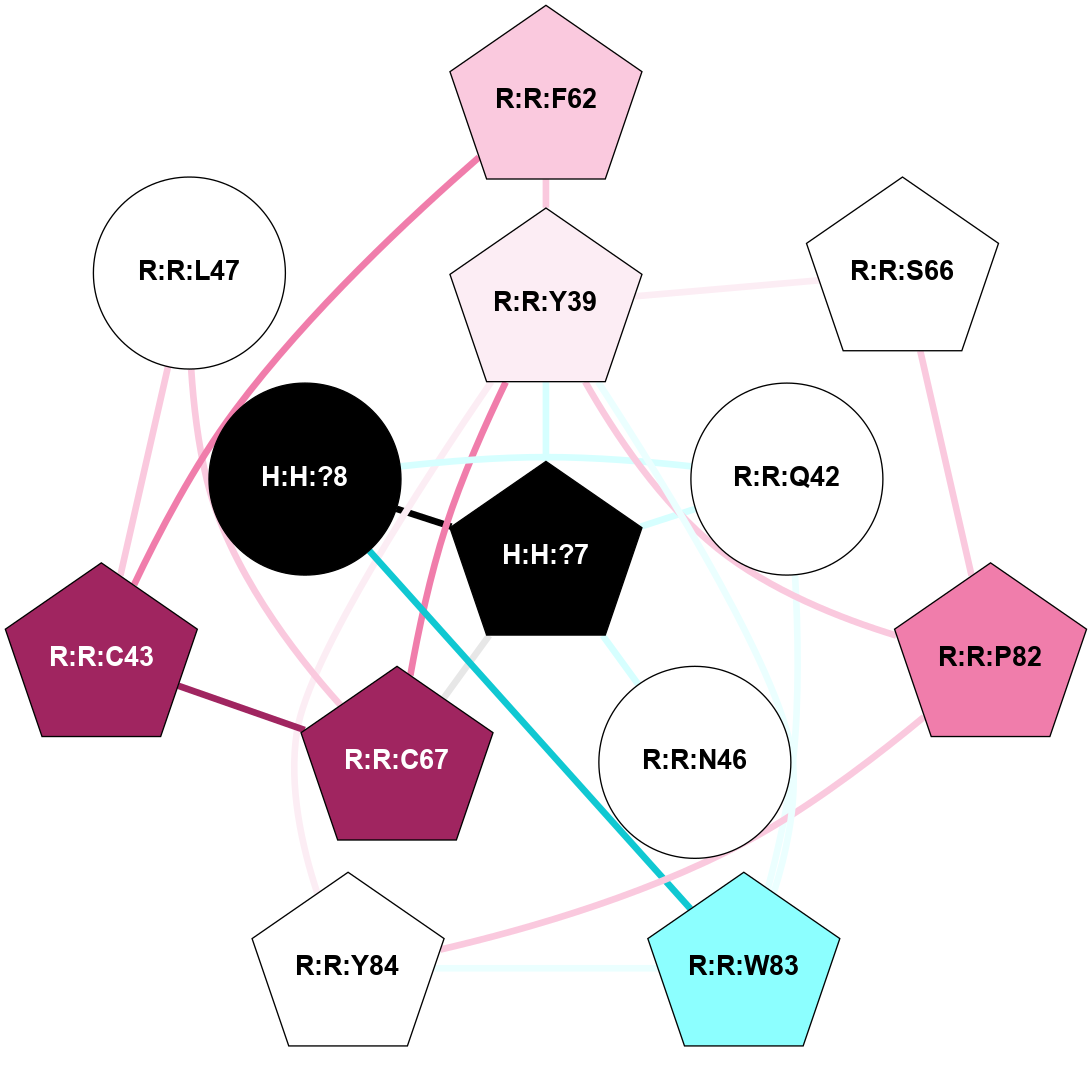
A 2D representation of the interactions of NAG in 5XEZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 36.53 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?7 | R:R:Y39 | 7.34 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?7 | R:R:Q42 | 15.47 | 2 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?7 | R:R:N46 | 31.93 | 2 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?7 | R:R:C67 | 2.84 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?8 | R:R:Q42 | 5.95 | 2 | No | No | 0 | 5 | 1 | 1 | | R:R:F62 | R:R:Y39 | 4.13 | 0 | Yes | Yes | 7 | 6 | 2 | 1 | | R:R:S66 | R:R:Y39 | 2.54 | 2 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:C67 | R:R:Y39 | 4.03 | 2 | Yes | Yes | 9 | 6 | 1 | 1 | | R:R:P82 | R:R:Y39 | 5.56 | 2 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:W83 | R:R:Y39 | 16.4 | 0 | Yes | Yes | 2 | 6 | 2 | 1 | | R:R:Y39 | R:R:Y84 | 8.94 | 2 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:Q42 | R:R:W83 | 6.57 | 2 | No | Yes | 5 | 2 | 1 | 2 | | R:R:C43 | R:R:L47 | 3.17 | 2 | No | No | 9 | 5 | 2 | 2 | | R:R:C43 | R:R:F62 | 8.38 | 2 | No | Yes | 9 | 7 | 2 | 2 | | R:R:C43 | R:R:C67 | 7.28 | 2 | No | Yes | 9 | 9 | 2 | 1 | | R:R:C67 | R:R:L47 | 4.76 | 2 | Yes | No | 9 | 5 | 1 | 2 | | R:R:P82 | R:R:S66 | 5.34 | 2 | Yes | Yes | 8 | 5 | 2 | 2 | | R:R:P82 | R:R:Y84 | 9.74 | 2 | Yes | Yes | 8 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 18.82 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 19.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
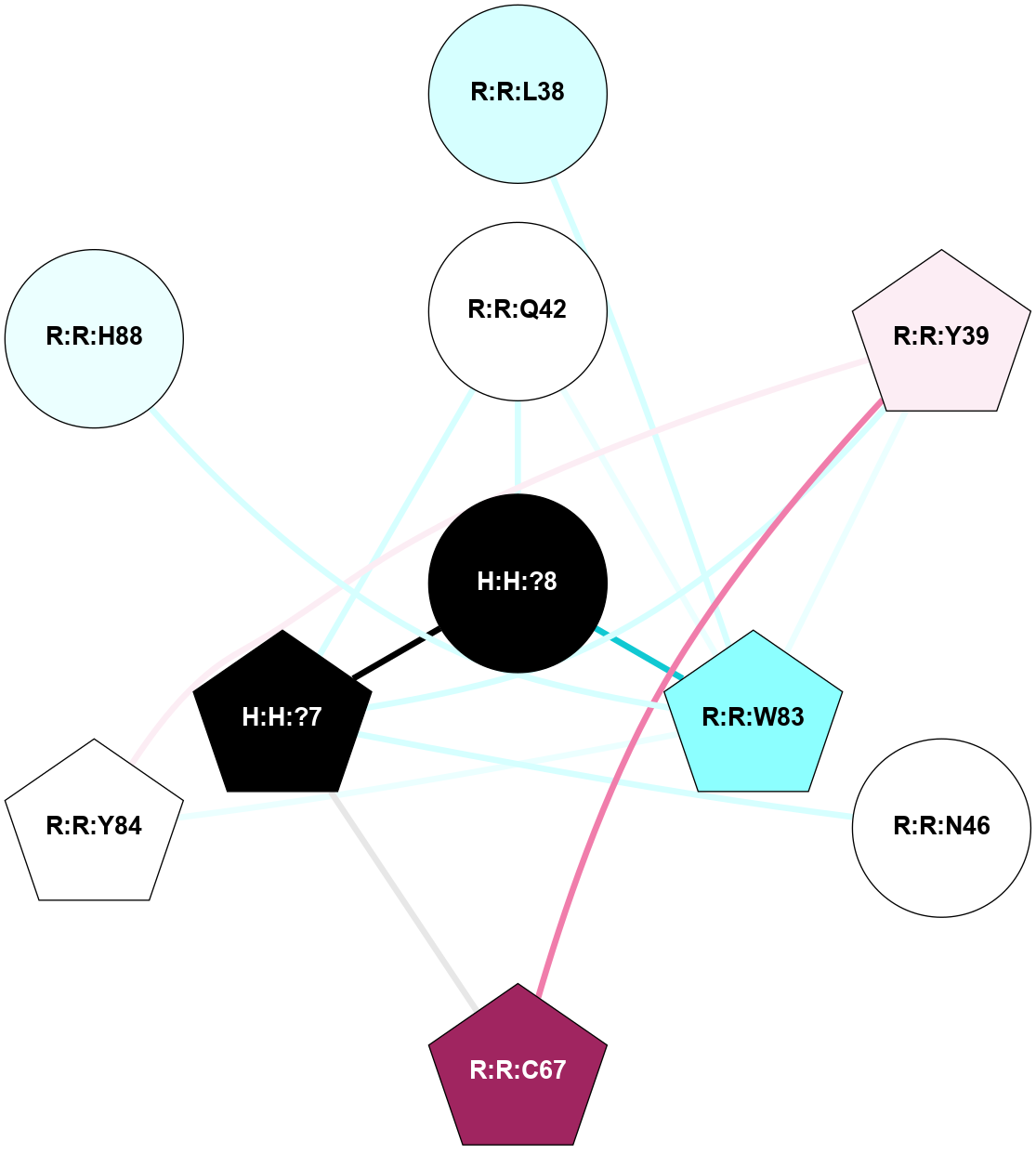
A 2D representation of the interactions of NAG in 5XEZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 36.53 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?7 | R:R:Y39 | 7.34 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?7 | R:R:Q42 | 15.47 | 2 | Yes | No | 0 | 5 | 1 | 1 | | H:H:?7 | R:R:N46 | 31.93 | 2 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?7 | R:R:C67 | 2.84 | 2 | Yes | Yes | 0 | 9 | 1 | 2 | | H:H:?8 | R:R:Q42 | 5.95 | 2 | No | No | 0 | 5 | 0 | 1 | | R:R:C67 | R:R:Y39 | 4.03 | 2 | Yes | Yes | 9 | 6 | 2 | 2 | | R:R:W83 | R:R:Y39 | 16.4 | 0 | Yes | Yes | 2 | 6 | 2 | 2 | | R:R:Q42 | R:R:W83 | 6.57 | 2 | No | Yes | 5 | 2 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 21.24 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
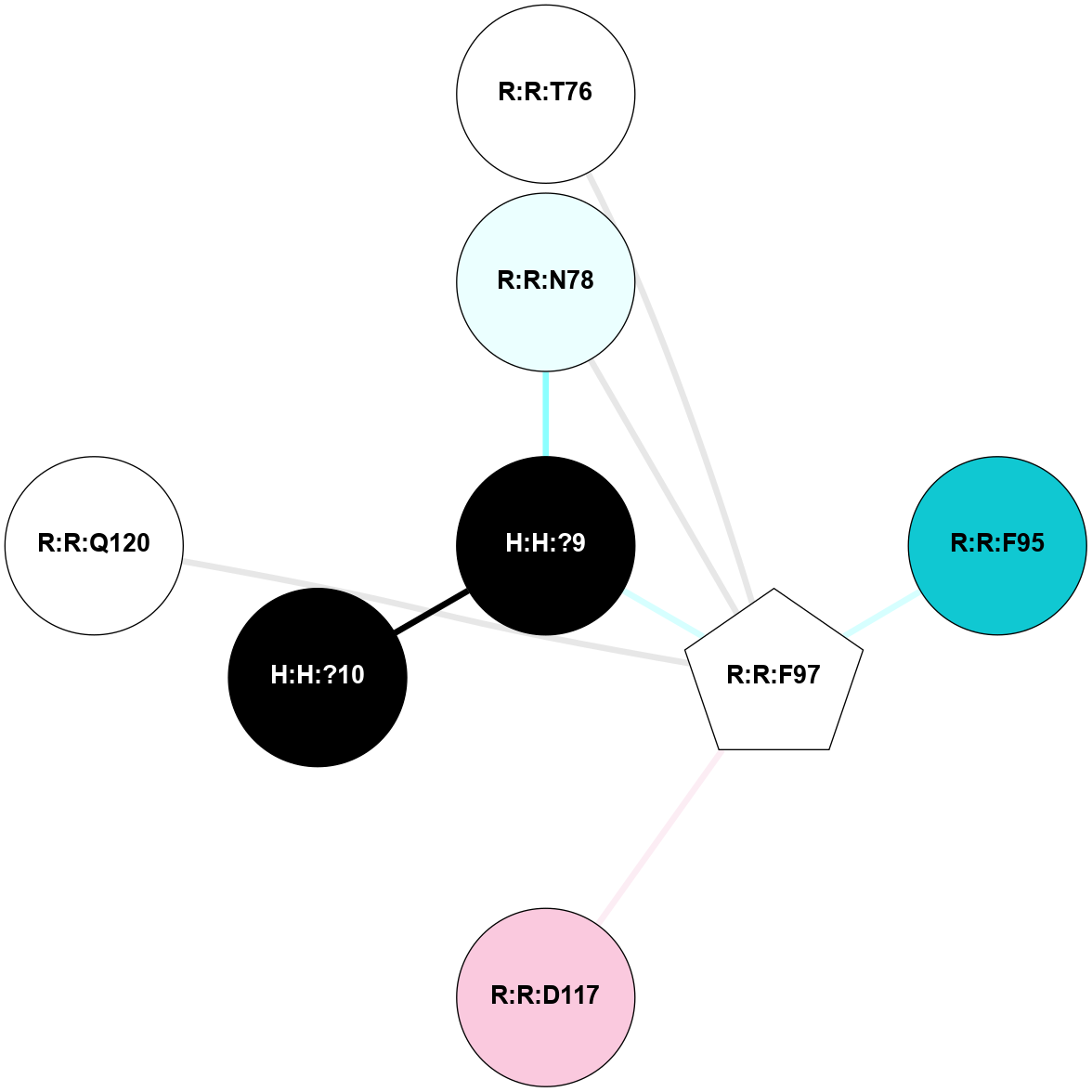
A 2D representation of the interactions of NAG in 5XEZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | H:H:?9 | 43.17 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?9 | R:R:N78 | 25.79 | 7 | No | No | 0 | 4 | 0 | 1 | | H:H:?9 | R:R:F97 | 6.54 | 7 | No | Yes | 0 | 5 | 0 | 1 | | R:R:F97 | R:R:T76 | 3.89 | 7 | Yes | No | 5 | 5 | 1 | 2 | | R:R:F97 | R:R:N78 | 4.83 | 7 | Yes | No | 5 | 4 | 1 | 1 | | R:R:F95 | R:R:F97 | 2.14 | 0 | No | Yes | 1 | 5 | 2 | 1 | | R:R:D117 | R:R:F97 | 2.39 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:F97 | R:R:Q120 | 3.51 | 7 | Yes | No | 5 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 25.17 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
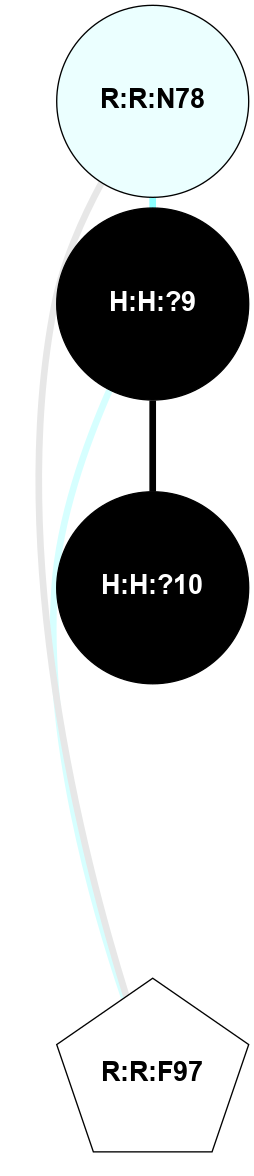
A 2D representation of the interactions of NAG in 5XEZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | H:H:?9 | 43.17 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?9 | R:R:N78 | 25.79 | 7 | No | No | 0 | 4 | 1 | 2 | | H:H:?9 | R:R:F97 | 6.54 | 7 | No | Yes | 0 | 5 | 1 | 2 | | R:R:F97 | R:R:N78 | 4.83 | 7 | Yes | No | 5 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 43.17 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
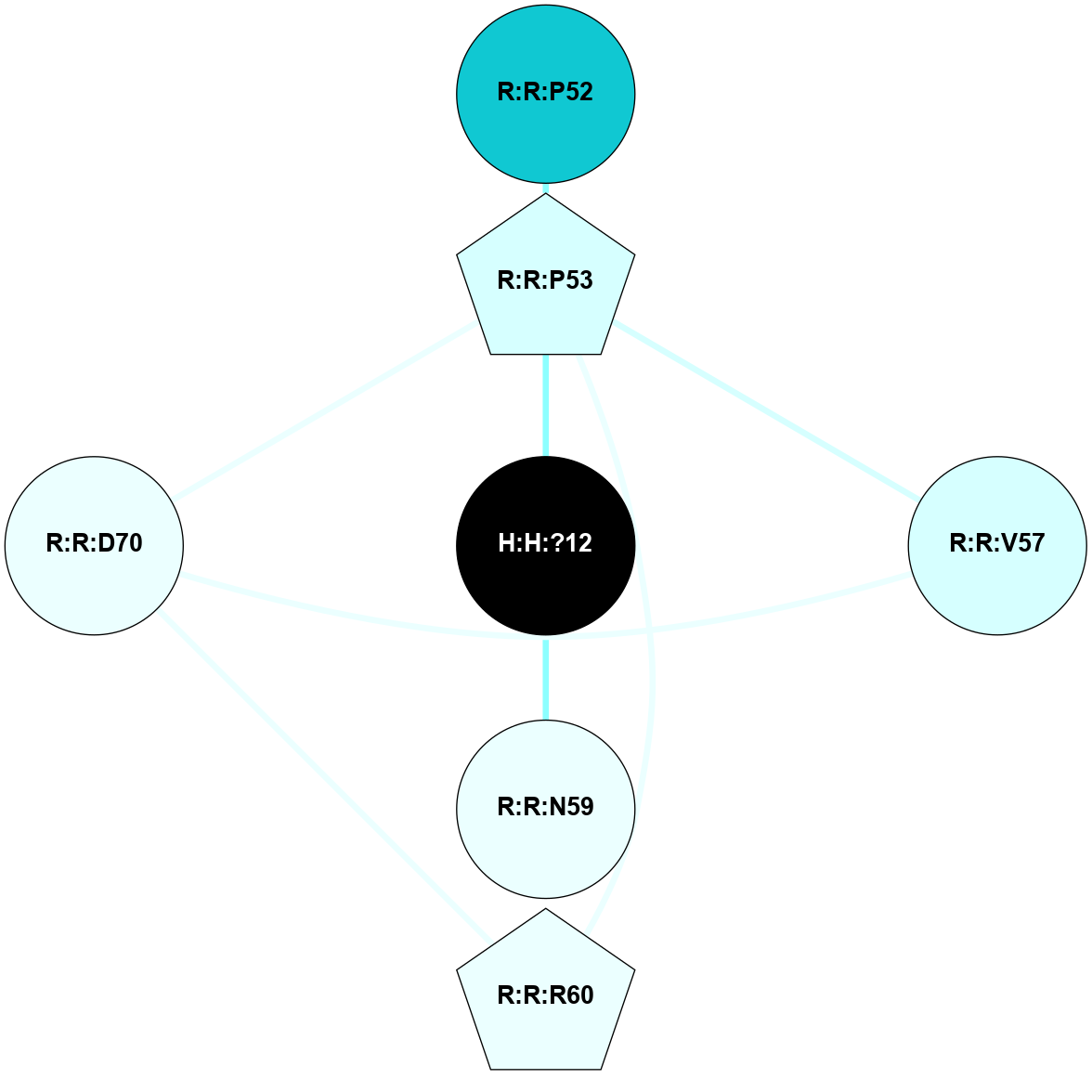
A 2D representation of the interactions of NAG in 5XEZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | R:R:P53 | 2.94 | 0 | No | Yes | 0 | 3 | 0 | 1 | | H:H:?12 | R:R:N59 | 25.79 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:P53 | R:R:V57 | 3.53 | 2 | Yes | No | 3 | 3 | 1 | 2 | | R:R:P53 | R:R:R60 | 2.88 | 2 | Yes | Yes | 3 | 4 | 1 | 2 | | R:R:D70 | R:R:P53 | 8.05 | 2 | No | Yes | 4 | 3 | 2 | 1 | | R:R:D70 | R:R:V57 | 5.84 | 2 | No | No | 4 | 3 | 2 | 2 | | R:R:D70 | R:R:R60 | 17.87 | 2 | No | Yes | 4 | 4 | 2 | 2 | | R:R:P52 | R:R:P53 | 1.95 | 0 | No | Yes | 1 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.37 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
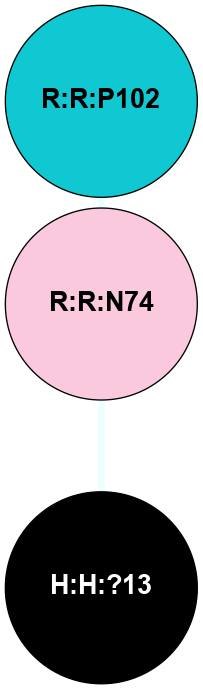
A 2D representation of the interactions of NAG in 5XEZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | R:R:N74 | 22.1 | 0 | No | No | 0 | 7 | 0 | 1 | | R:R:N74 | R:R:P102 | 4.89 | 0 | No | No | 7 | 1 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 22.10 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5XF1 | B1 | Peptide | Glucagon | Glucagon | Homo Sapiens | - | NNC0640 | - | 3.19 | 2017-05-24 | doi.org/10.1038/nature22363 |
|
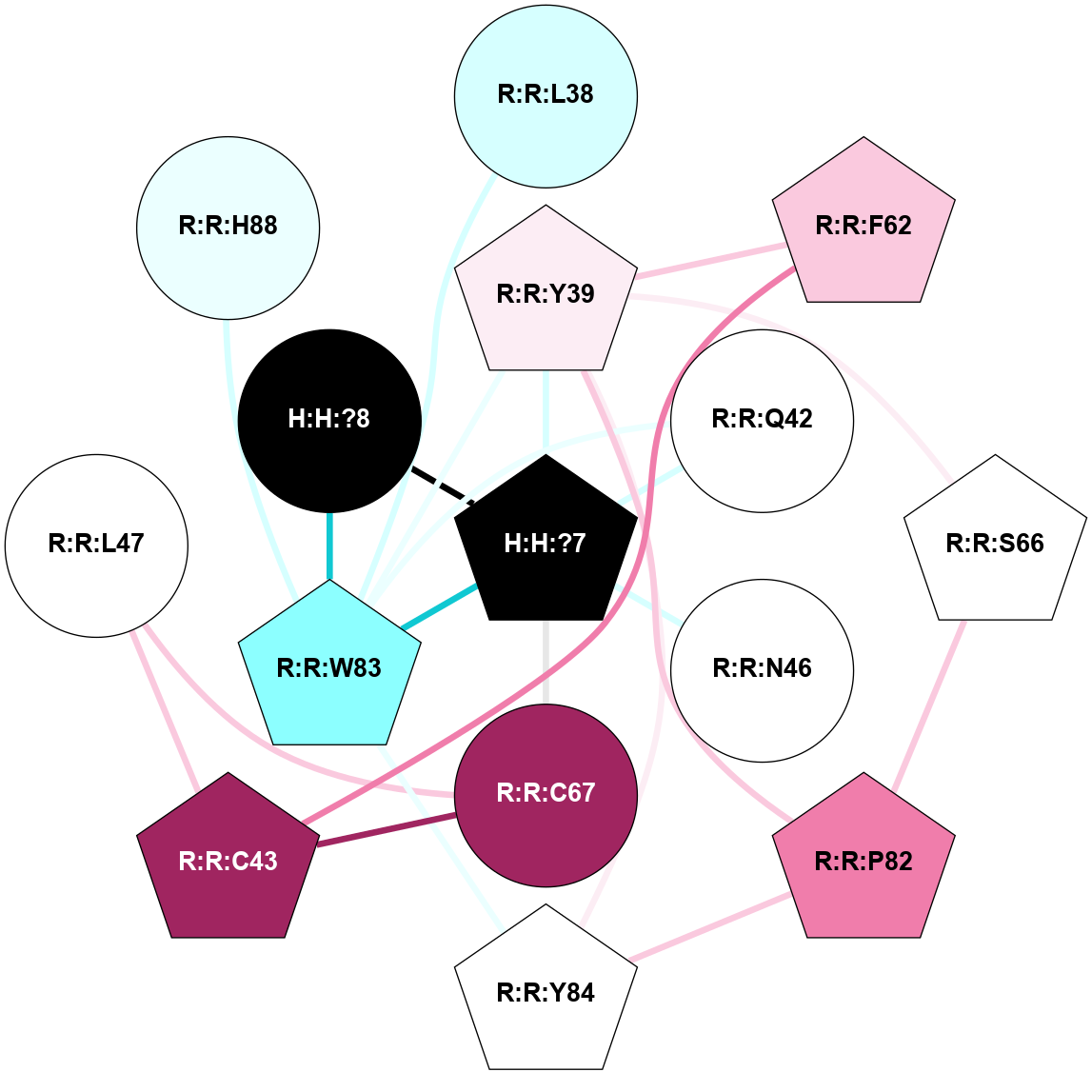
A 2D representation of the interactions of NAG in 5XF1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 39.85 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?7 | R:R:Y39 | 5.24 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?7 | R:R:Q42 | 15.47 | 2 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?7 | R:R:N46 | 24.56 | 2 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?7 | R:R:C67 | 2.84 | 2 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?7 | R:R:W83 | 2.04 | 2 | Yes | Yes | 0 | 2 | 0 | 1 | | H:H:?8 | R:R:W83 | 3.06 | 2 | No | Yes | 0 | 2 | 1 | 1 | | R:R:L38 | R:R:W83 | 2.28 | 0 | No | Yes | 3 | 2 | 2 | 1 | | R:R:F62 | R:R:Y39 | 4.13 | 2 | Yes | Yes | 7 | 6 | 2 | 1 | | R:R:S66 | R:R:Y39 | 2.54 | 2 | Yes | Yes | 5 | 6 | 2 | 1 | | R:R:P82 | R:R:Y39 | 5.56 | 2 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:W83 | R:R:Y39 | 13.5 | 2 | Yes | Yes | 2 | 6 | 1 | 1 | | R:R:Y39 | R:R:Y84 | 9.93 | 2 | Yes | Yes | 6 | 5 | 1 | 2 | | R:R:Q42 | R:R:W83 | 5.48 | 2 | No | Yes | 5 | 2 | 1 | 1 | | R:R:C43 | R:R:L47 | 3.17 | 2 | Yes | No | 9 | 5 | 2 | 2 | | R:R:C43 | R:R:F62 | 5.59 | 2 | Yes | Yes | 9 | 7 | 2 | 2 | | R:R:C43 | R:R:C67 | 7.28 | 2 | Yes | No | 9 | 9 | 2 | 1 | | R:R:C67 | R:R:L47 | 4.76 | 2 | No | No | 9 | 5 | 1 | 2 | | R:R:P82 | R:R:S66 | 5.34 | 2 | Yes | Yes | 8 | 5 | 2 | 2 | | R:R:P82 | R:R:Y84 | 9.74 | 2 | Yes | Yes | 8 | 5 | 2 | 2 | | R:R:W83 | R:R:Y84 | 1.93 | 2 | Yes | Yes | 2 | 5 | 1 | 2 | | R:R:H88 | R:R:W83 | 2.12 | 2 | No | Yes | 4 | 2 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 15.00 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 22.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
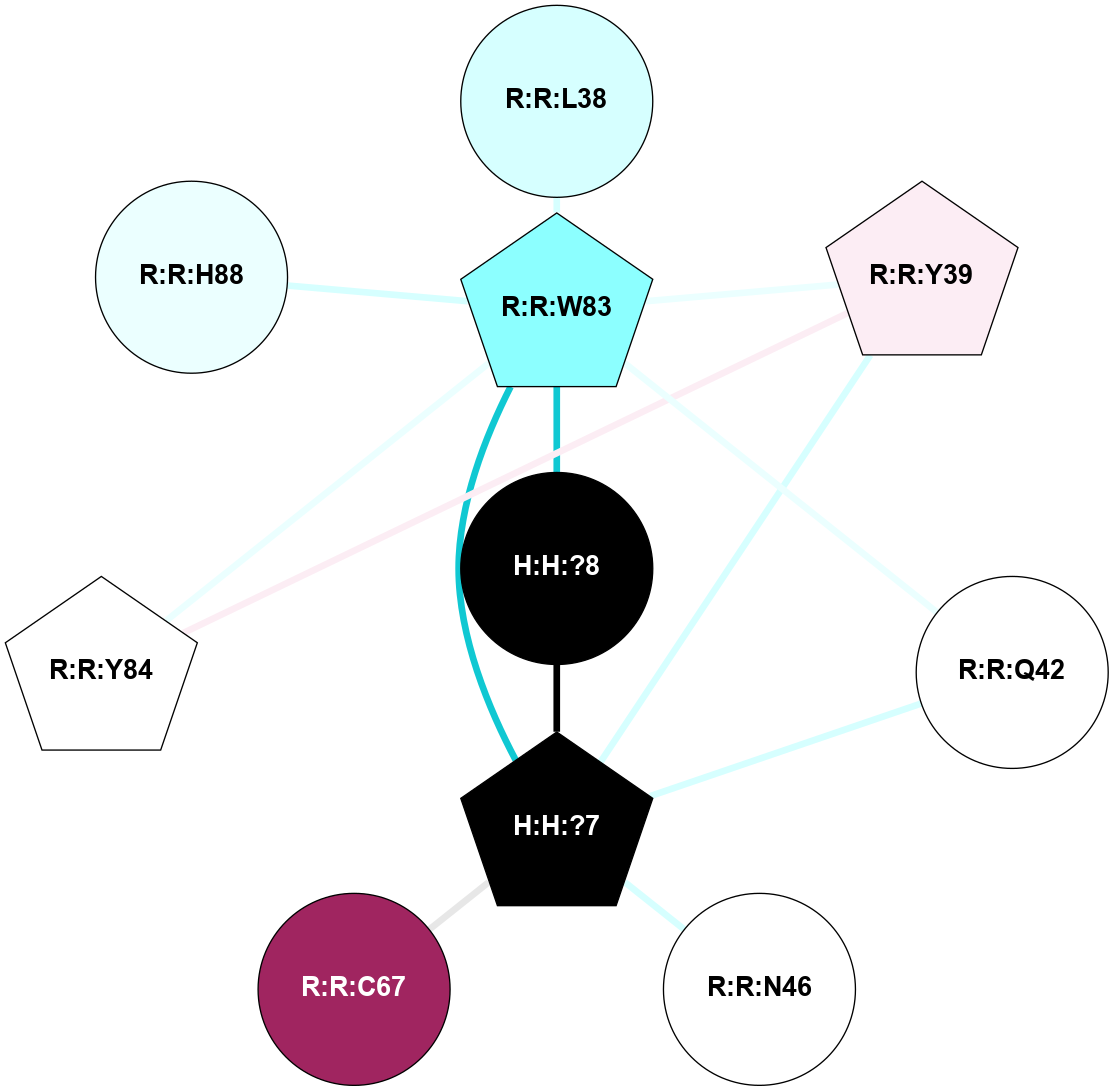
A 2D representation of the interactions of NAG in 5XF1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 39.85 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?7 | R:R:Y39 | 5.24 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?7 | R:R:Q42 | 15.47 | 2 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?7 | R:R:N46 | 24.56 | 2 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?7 | R:R:C67 | 2.84 | 2 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?7 | R:R:W83 | 2.04 | 2 | Yes | Yes | 0 | 2 | 1 | 1 | | H:H:?8 | R:R:W83 | 3.06 | 2 | No | Yes | 0 | 2 | 0 | 1 | | R:R:L38 | R:R:W83 | 2.28 | 0 | No | Yes | 3 | 2 | 2 | 1 | | R:R:W83 | R:R:Y39 | 13.5 | 2 | Yes | Yes | 2 | 6 | 1 | 2 | | R:R:Y39 | R:R:Y84 | 9.93 | 2 | Yes | Yes | 6 | 5 | 2 | 2 | | R:R:Q42 | R:R:W83 | 5.48 | 2 | No | Yes | 5 | 2 | 2 | 1 | | R:R:W83 | R:R:Y84 | 1.93 | 2 | Yes | Yes | 2 | 5 | 1 | 2 | | R:R:H88 | R:R:W83 | 2.12 | 2 | No | Yes | 4 | 2 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 21.46 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
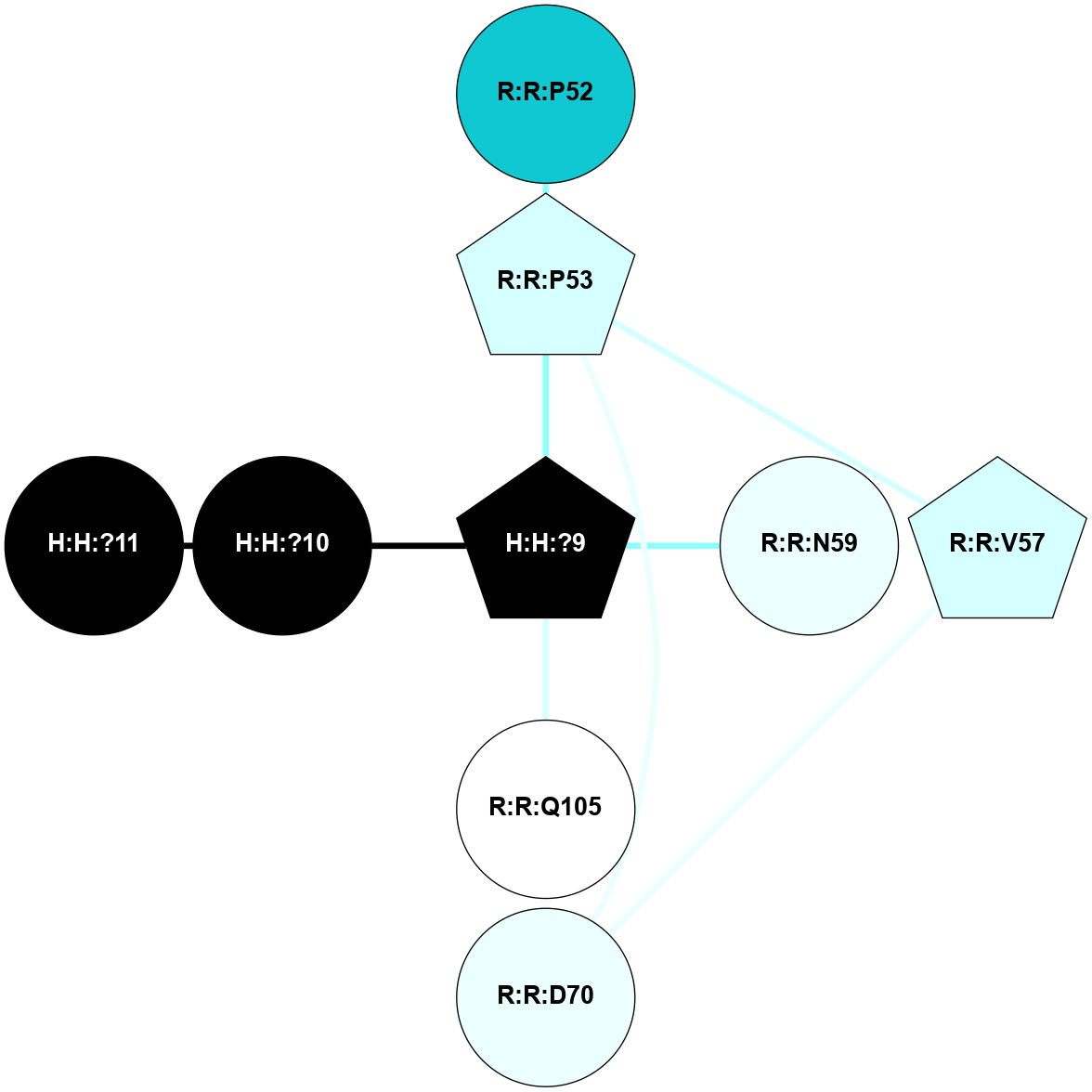
A 2D representation of the interactions of NAG in 5XF1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | H:H:?9 | 30.99 | 0 | No | Yes | 0 | 0 | 1 | 0 | | H:H:?9 | R:R:P53 | 2.94 | 0 | Yes | Yes | 0 | 3 | 0 | 1 | | H:H:?9 | R:R:N59 | 31.93 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?9 | R:R:Q105 | 7.14 | 0 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?10 | H:H:?11 | 26.57 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:P52 | R:R:P53 | 1.95 | 0 | No | Yes | 1 | 3 | 2 | 1 | | R:R:P53 | R:R:V57 | 1.77 | 7 | Yes | Yes | 3 | 3 | 1 | 2 | | R:R:D70 | R:R:P53 | 3.22 | 7 | No | Yes | 4 | 3 | 2 | 1 | | R:R:D70 | R:R:V57 | 4.38 | 7 | No | Yes | 4 | 3 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 18.25 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
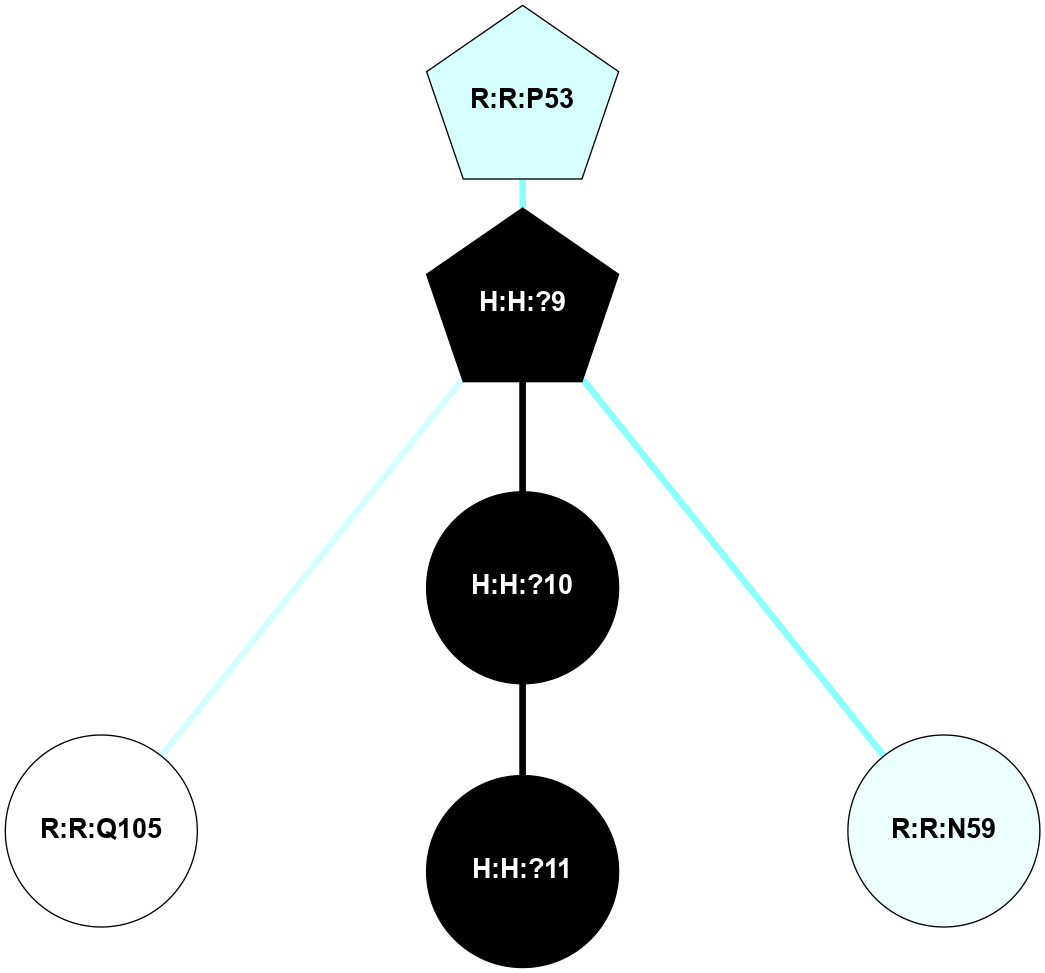
A 2D representation of the interactions of NAG in 5XF1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | H:H:?9 | 30.99 | 0 | No | Yes | 0 | 0 | 0 | 1 | | H:H:?9 | R:R:P53 | 2.94 | 0 | Yes | Yes | 0 | 3 | 1 | 2 | | H:H:?9 | R:R:N59 | 31.93 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?9 | R:R:Q105 | 7.14 | 0 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?10 | H:H:?11 | 26.57 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 28.78 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 5XF1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | H:H:?9 | 30.99 | 0 | No | Yes | 0 | 0 | 1 | 2 | | H:H:?10 | H:H:?11 | 26.57 | 0 | No | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 26.57 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
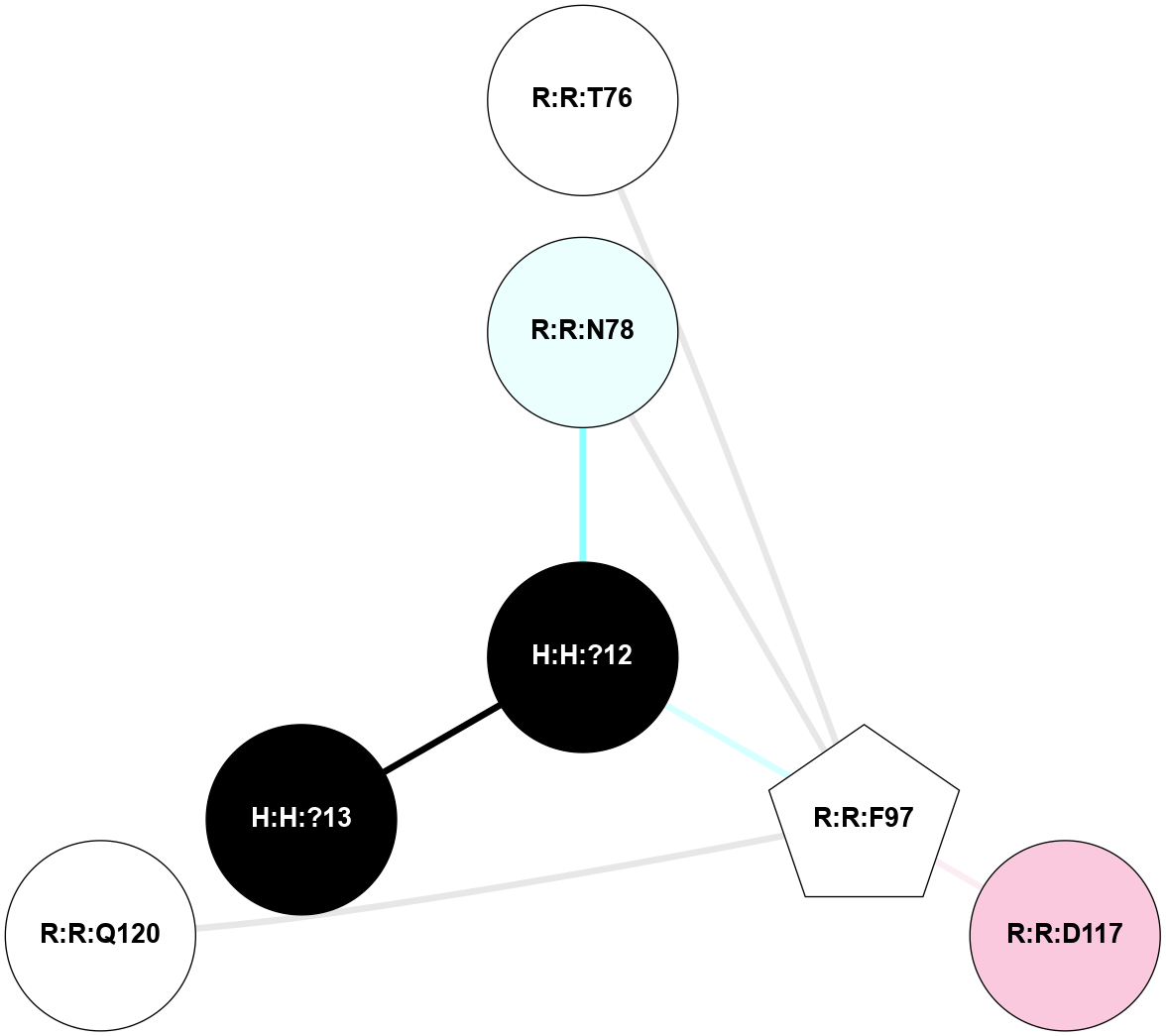
A 2D representation of the interactions of NAG in 5XF1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | H:H:?13 | 35.42 | 8 | No | No | 0 | 0 | 0 | 1 | | H:H:?12 | R:R:N78 | 24.56 | 8 | No | No | 0 | 4 | 0 | 1 | | H:H:?12 | R:R:F97 | 5.45 | 8 | No | Yes | 0 | 5 | 0 | 1 | | R:R:F97 | R:R:T76 | 2.59 | 8 | Yes | No | 5 | 5 | 1 | 2 | | R:R:F97 | R:R:N78 | 8.46 | 8 | Yes | No | 5 | 4 | 1 | 1 | | R:R:D117 | R:R:F97 | 4.78 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:F97 | R:R:Q120 | 3.51 | 8 | Yes | No | 5 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 21.81 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
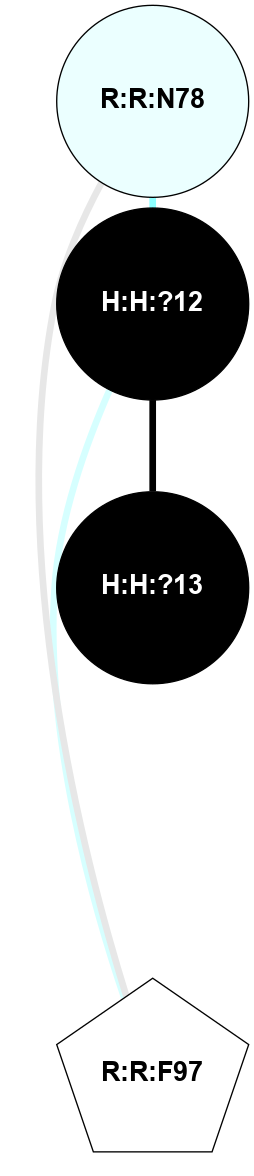
A 2D representation of the interactions of NAG in 5XF1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | H:H:?13 | 35.42 | 8 | No | No | 0 | 0 | 1 | 0 | | H:H:?12 | R:R:N78 | 24.56 | 8 | No | No | 0 | 4 | 1 | 2 | | H:H:?12 | R:R:F97 | 5.45 | 8 | No | Yes | 0 | 5 | 1 | 2 | | R:R:F97 | R:R:N78 | 8.46 | 8 | Yes | No | 5 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 35.42 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 5XF1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?14 | R:R:A73 | 4.23 | 0 | No | No | 0 | 6 | 0 | 1 | | H:H:?14 | R:R:N74 | 23.33 | 0 | No | No | 0 | 7 | 0 | 1 | | R:R:N74 | R:R:R99 | 2.41 | 0 | No | No | 7 | 3 | 1 | 2 | | R:R:N74 | R:R:P102 | 9.77 | 0 | No | No | 7 | 1 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.78 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 5YQZ | B1 | Peptide | Glucagon | Glucagon | Homo Sapiens | Glucagon analogue | - | - | 3 | 2018-01-17 | doi.org/10.1038/nature25153 |
|
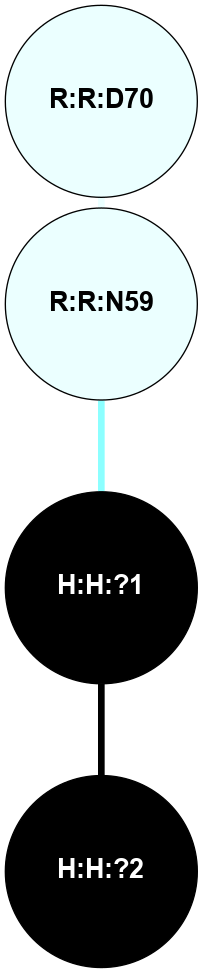
A 2D representation of the interactions of NAG in 5YQZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 27.67 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N59 | 31.93 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:D70 | R:R:N59 | 4.04 | 12 | Yes | No | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 29.80 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
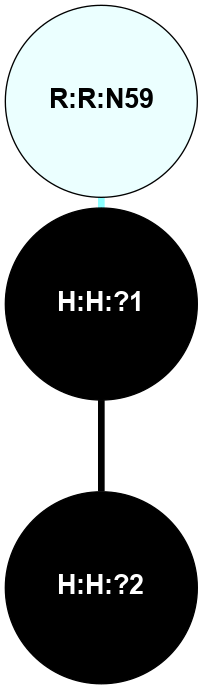
A 2D representation of the interactions of NAG in 5YQZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 27.67 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N59 | 31.93 | 0 | No | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.67 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
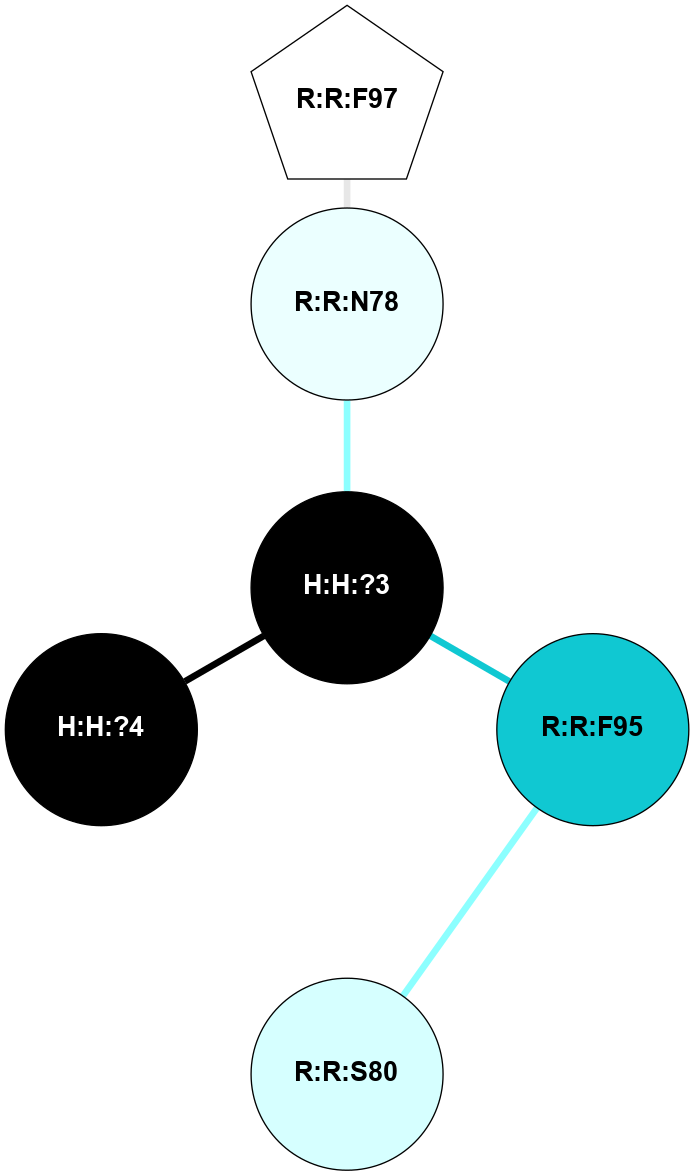
A 2D representation of the interactions of NAG in 5YQZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 27.67 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:N78 | 33.15 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?3 | R:R:F95 | 18.52 | 0 | No | No | 0 | 1 | 0 | 1 | | R:R:F97 | R:R:N78 | 7.25 | 0 | Yes | No | 5 | 4 | 2 | 1 | | R:R:F95 | R:R:S80 | 6.61 | 0 | No | No | 1 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 26.45 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
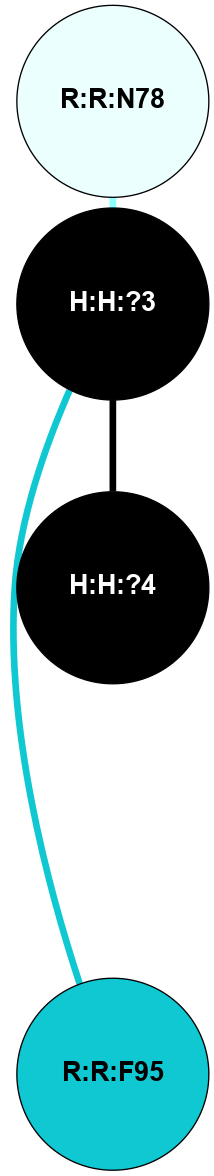
A 2D representation of the interactions of NAG in 5YQZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 27.67 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:N78 | 33.15 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?3 | R:R:F95 | 18.52 | 0 | No | No | 0 | 1 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.67 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6CMO | A | Sensory | Opsins | Rhodopsin | Homo Sapiens | - | - | Gi1/Beta1/Gamma2 | 4.5 | 2018-06-20 | doi.org/10.1038/s41586-018-0215-y |
|
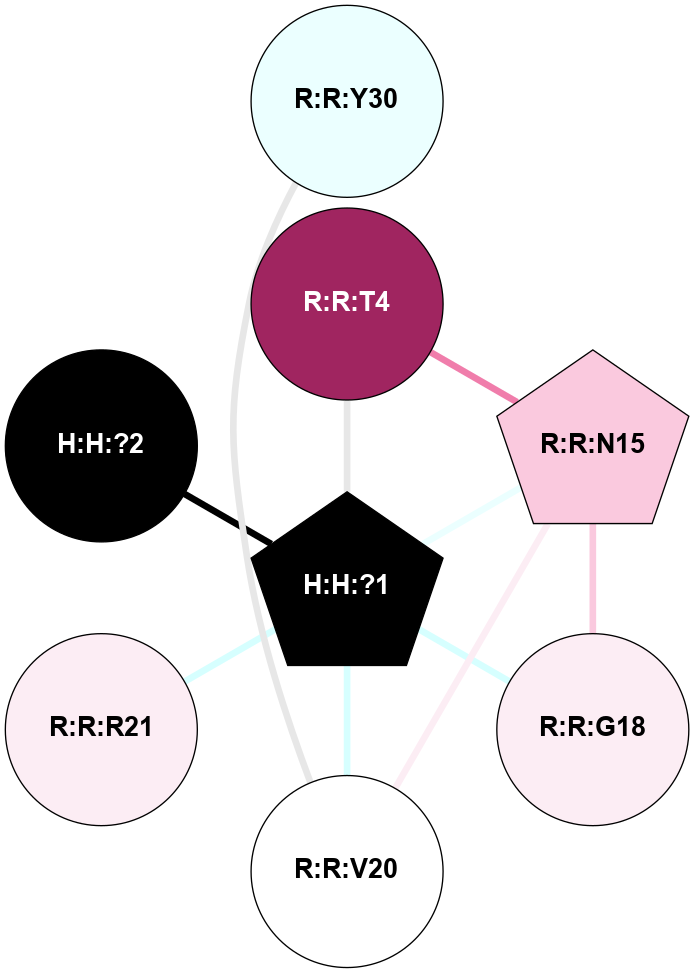
A 2D representation of the interactions of NAG in 6CMO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 2.92 | 17 | Yes | No | 7 | 9 | 1 | 1 | | H:H:?1 | R:R:T4 | 13.18 | 17 | Yes | No | 0 | 9 | 0 | 1 | | R:R:G18 | R:R:N15 | 3.39 | 17 | No | Yes | 6 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 11.82 | 17 | Yes | No | 7 | 5 | 1 | 1 | | H:H:?1 | R:R:N15 | 28.24 | 17 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 7.65 | 17 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V20 | R:R:Y30 | 2.52 | 17 | No | No | 5 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 6.66 | 17 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:R21 | 13.04 | 17 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | H:H:?2 | 38.74 | 17 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.92 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
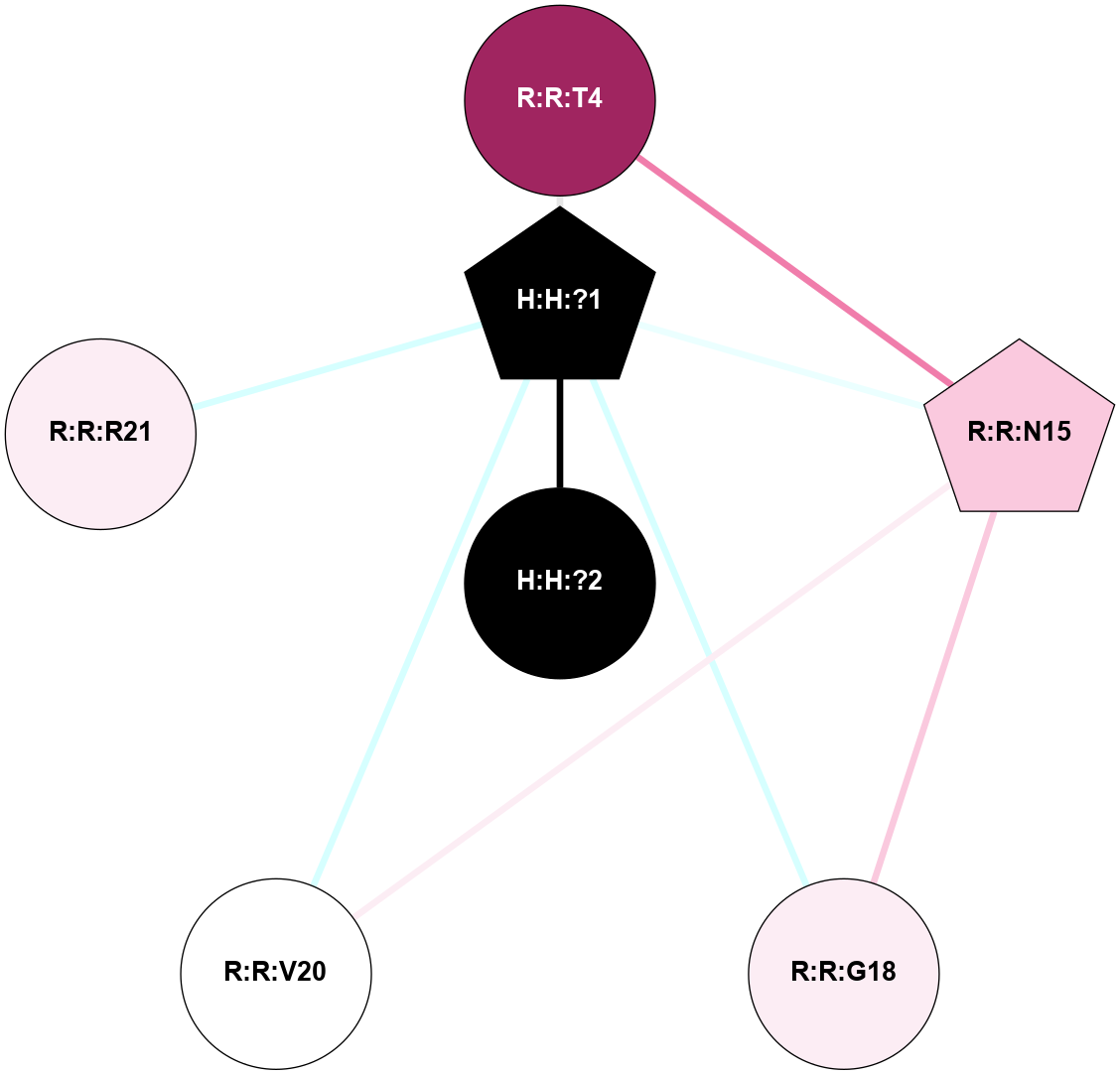
A 2D representation of the interactions of NAG in 6CMO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 2.92 | 17 | Yes | No | 7 | 9 | 2 | 2 | | H:H:?1 | R:R:T4 | 13.18 | 17 | Yes | No | 0 | 9 | 1 | 2 | | R:R:G18 | R:R:N15 | 3.39 | 17 | No | Yes | 6 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 11.82 | 17 | Yes | No | 7 | 5 | 2 | 2 | | H:H:?1 | R:R:N15 | 28.24 | 17 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 7.65 | 17 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:V20 | 6.66 | 17 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?1 | R:R:R21 | 13.04 | 17 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?1 | H:H:?2 | 38.74 | 17 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 38.74 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6DO1 | A | Peptide | Angiotensin | AT1 | Homo Sapiens | Angiotensin-like peptide S1I8 | - | - | 2.9 | 2019-01-30 | doi.org/10.1016/j.cell.2018.12.006 |
|
A 2D representation of the interactions of NAG in 6DO1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:N176 | 24.56 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:F171 | R:R:N176 | 4.83 | 6 | Yes | No | 6 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 24.56 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FJ3 | B1 | Peptide | Parathyroid Hormone | PTH1 | Homo Sapiens | PTH | - | - | 2.5 | 2018-11-21 | doi.org/10.1038/s41594-018-0151-4 |
|
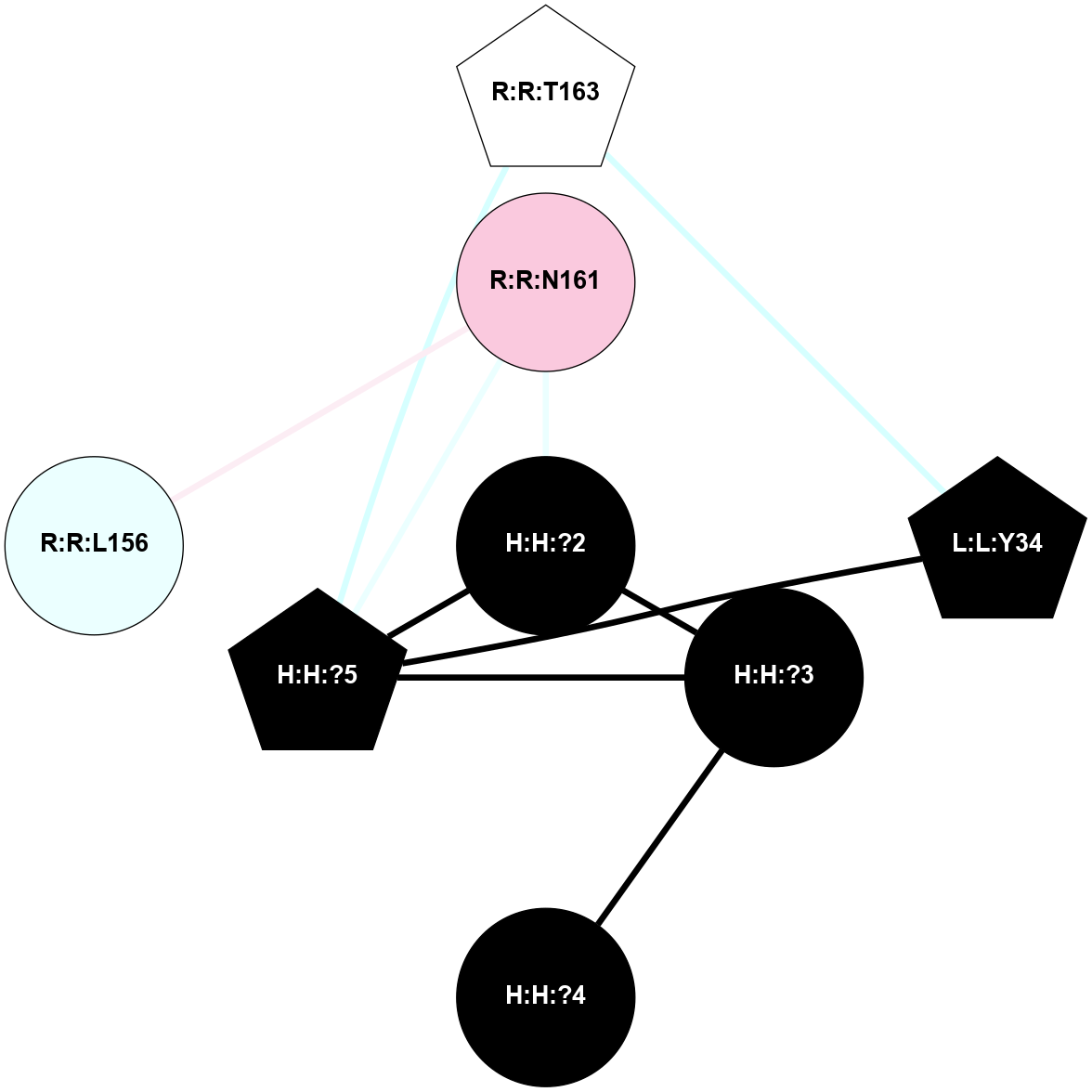
A 2D representation of the interactions of NAG in 6FJ3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:N161 | 29.47 | 2 | No | No | 0 | 7 | 0 | 1 | | H:H:?5 | R:R:N161 | 7.39 | 2 | Yes | No | 0 | 7 | 1 | 1 | | L:L:Y34 | R:R:T163 | 4.99 | 2 | Yes | Yes | 0 | 5 | 2 | 2 | | H:H:?5 | R:R:T163 | 9.52 | 2 | Yes | Yes | 0 | 5 | 1 | 2 | | H:H:?5 | L:L:Y34 | 17.67 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | H:H:?2 | H:H:?3 | 43.17 | 2 | No | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?5 | 27.98 | 2 | No | Yes | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?4 | 33.39 | 2 | No | No | 0 | 0 | 1 | 2 | | H:H:?3 | H:H:?5 | 5.33 | 2 | No | Yes | 0 | 0 | 1 | 1 | | R:R:L156 | R:R:N161 | 2.75 | 0 | No | No | 4 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 33.54 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
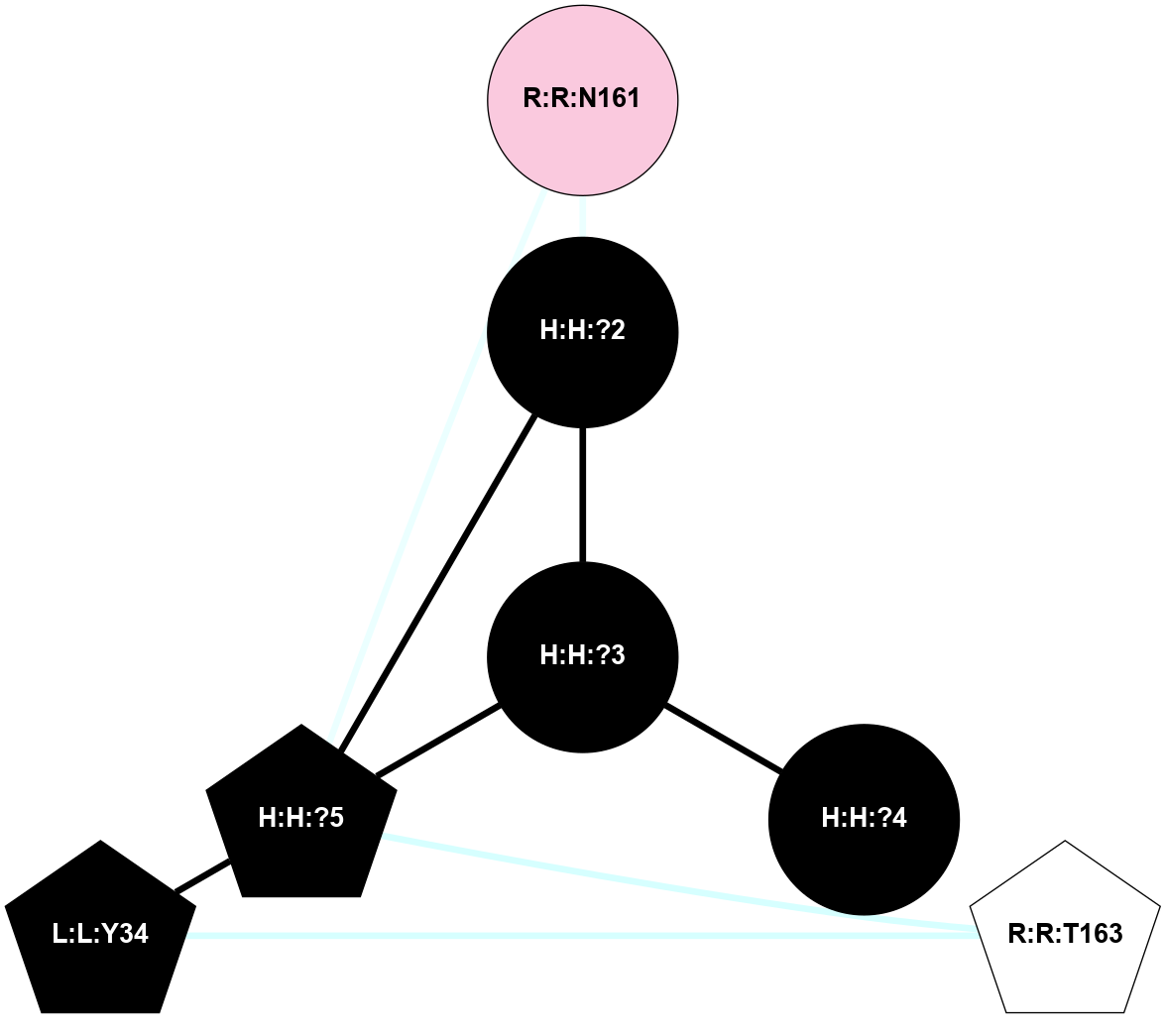
A 2D representation of the interactions of NAG in 6FJ3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:N161 | 29.47 | 2 | No | No | 0 | 7 | 1 | 2 | | H:H:?5 | R:R:N161 | 7.39 | 2 | Yes | No | 0 | 7 | 1 | 2 | | L:L:Y34 | R:R:T163 | 4.99 | 2 | Yes | Yes | 0 | 5 | 2 | 2 | | H:H:?5 | R:R:T163 | 9.52 | 2 | Yes | Yes | 0 | 5 | 1 | 2 | | H:H:?5 | L:L:Y34 | 17.67 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | H:H:?2 | H:H:?3 | 43.17 | 2 | No | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?5 | 27.98 | 2 | No | Yes | 0 | 0 | 1 | 1 | | H:H:?3 | H:H:?4 | 33.39 | 2 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?5 | 5.33 | 2 | No | Yes | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 27.30 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
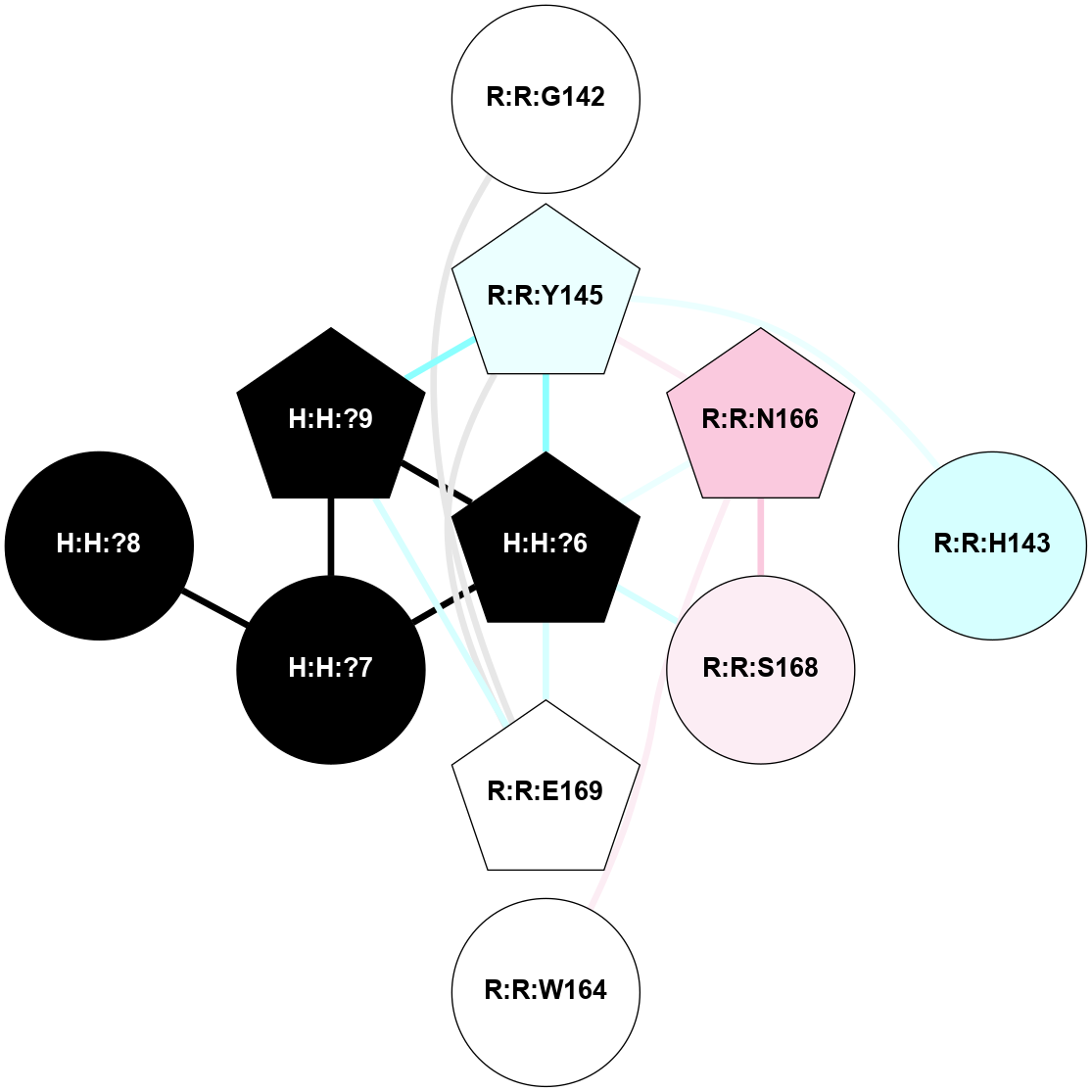
A 2D representation of the interactions of NAG in 6FJ3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E169 | R:R:G142 | 4.91 | 3 | Yes | No | 5 | 5 | 1 | 2 | | R:R:H143 | R:R:Y145 | 4.36 | 0 | No | Yes | 3 | 4 | 2 | 1 | | R:R:N166 | R:R:Y145 | 4.65 | 3 | Yes | Yes | 7 | 4 | 1 | 1 | | R:R:E169 | R:R:Y145 | 8.98 | 3 | Yes | Yes | 5 | 4 | 1 | 1 | | H:H:?6 | R:R:Y145 | 10.48 | 3 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?9 | R:R:Y145 | 22.72 | 3 | Yes | Yes | 0 | 4 | 1 | 1 | | R:R:N166 | R:R:W164 | 5.65 | 3 | Yes | No | 7 | 5 | 1 | 2 | | R:R:N166 | R:R:S168 | 5.96 | 3 | Yes | No | 7 | 6 | 1 | 1 | | H:H:?6 | R:R:N166 | 28.24 | 3 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?6 | R:R:S168 | 9.4 | 3 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?6 | R:R:E169 | 4.74 | 3 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?9 | R:R:E169 | 5.71 | 3 | Yes | Yes | 0 | 5 | 1 | 1 | | H:H:?6 | H:H:?7 | 29.89 | 3 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?6 | H:H:?9 | 22.65 | 3 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?7 | H:H:?8 | 30.82 | 3 | No | No | 0 | 0 | 1 | 2 | | H:H:?7 | H:H:?9 | 9.33 | 3 | No | Yes | 0 | 0 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 17.57 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
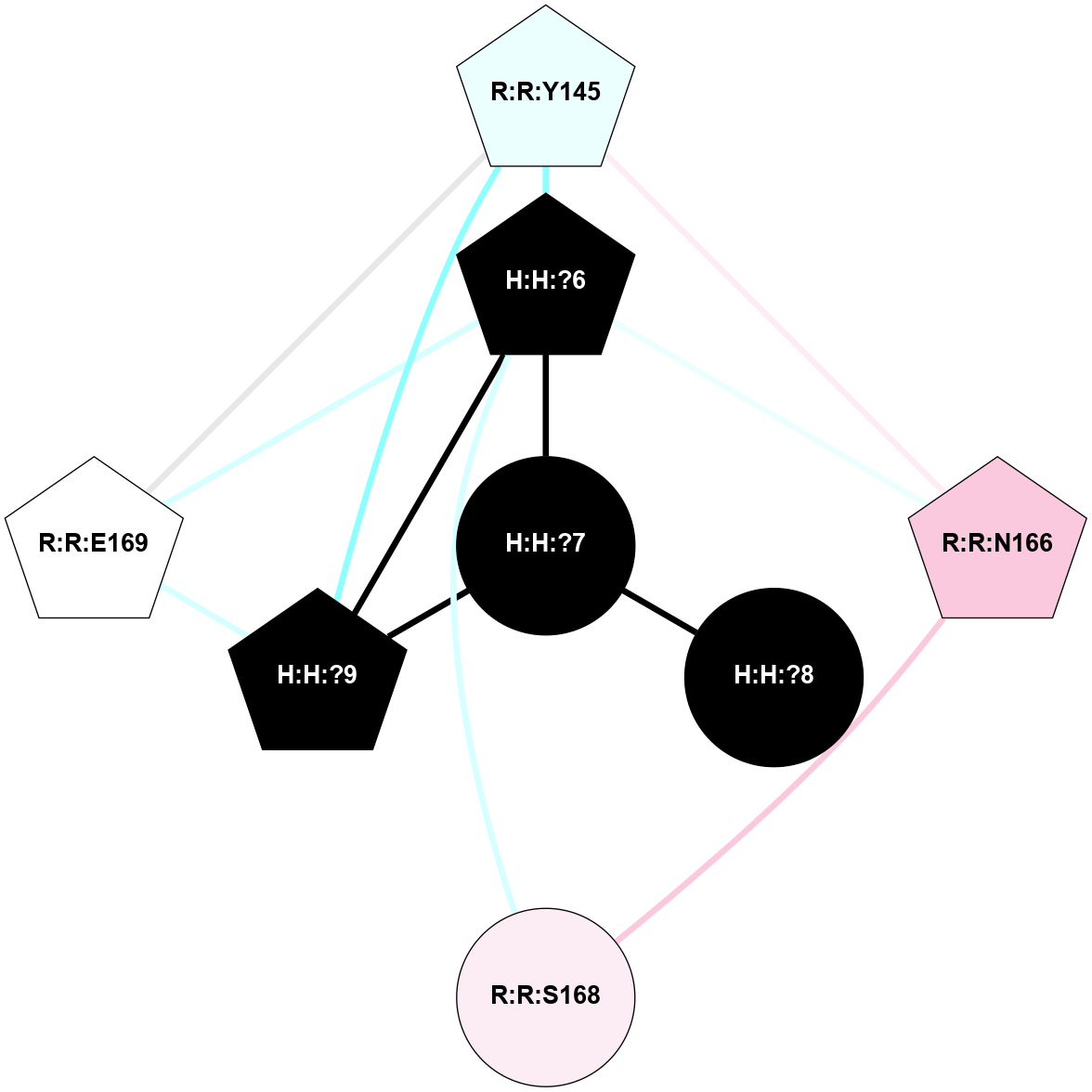
A 2D representation of the interactions of NAG in 6FJ3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N166 | R:R:Y145 | 4.65 | 3 | Yes | Yes | 7 | 4 | 2 | 2 | | R:R:E169 | R:R:Y145 | 8.98 | 3 | Yes | Yes | 5 | 4 | 2 | 2 | | H:H:?6 | R:R:Y145 | 10.48 | 3 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?9 | R:R:Y145 | 22.72 | 3 | Yes | Yes | 0 | 4 | 1 | 2 | | R:R:N166 | R:R:S168 | 5.96 | 3 | Yes | No | 7 | 6 | 2 | 2 | | H:H:?6 | R:R:N166 | 28.24 | 3 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?6 | R:R:S168 | 9.4 | 3 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?6 | R:R:E169 | 4.74 | 3 | Yes | Yes | 0 | 5 | 1 | 2 | | H:H:?9 | R:R:E169 | 5.71 | 3 | Yes | Yes | 0 | 5 | 1 | 2 | | H:H:?6 | H:H:?7 | 29.89 | 3 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?6 | H:H:?9 | 22.65 | 3 | Yes | Yes | 0 | 0 | 1 | 1 | | H:H:?7 | H:H:?8 | 30.82 | 3 | No | No | 0 | 0 | 0 | 1 | | H:H:?7 | H:H:?9 | 9.33 | 3 | No | Yes | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 23.35 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
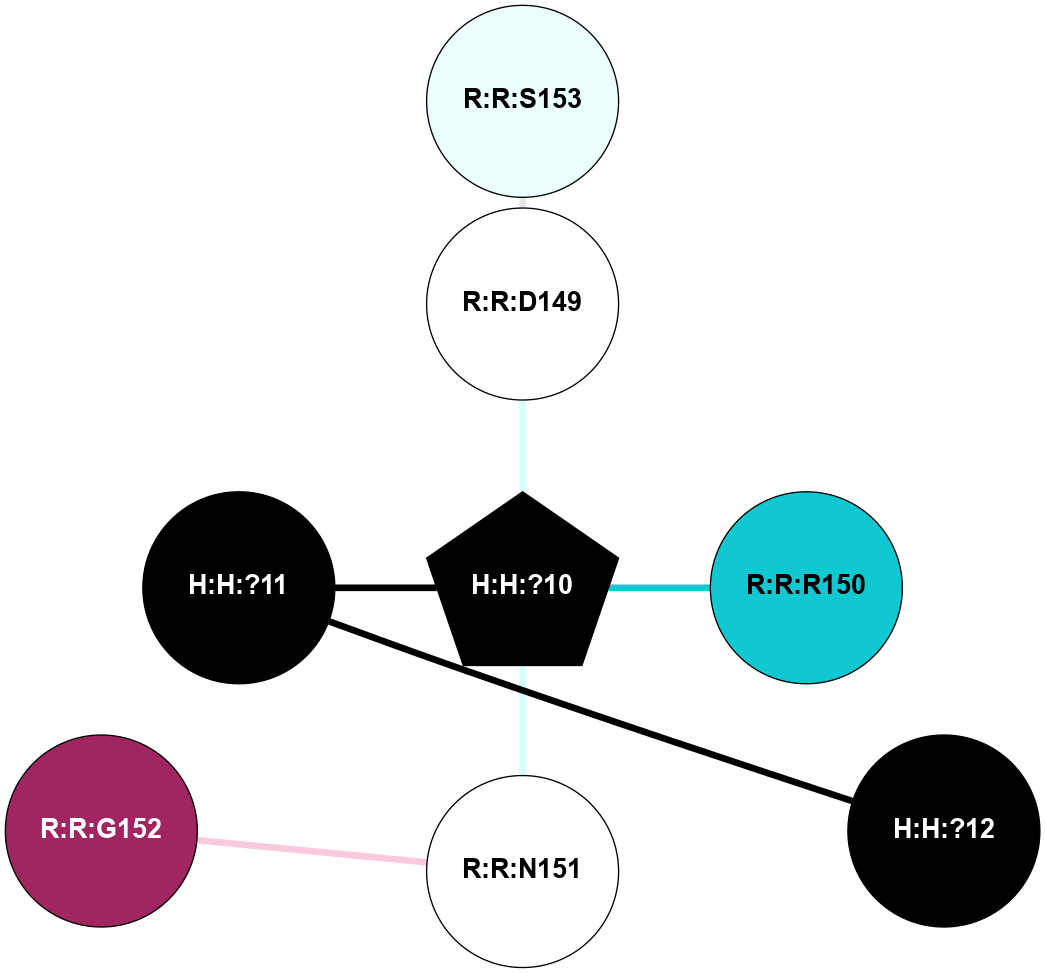
A 2D representation of the interactions of NAG in 6FJ3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D149 | R:R:S153 | 4.42 | 0 | No | No | 5 | 4 | 1 | 2 | | H:H:?10 | R:R:D149 | 6.07 | 0 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?10 | R:R:R150 | 9.78 | 0 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?10 | R:R:N151 | 24.56 | 0 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?10 | H:H:?11 | 42.06 | 0 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?11 | H:H:?12 | 34.67 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:G152 | R:R:N151 | 1.7 | 2 | No | No | 9 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.62 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
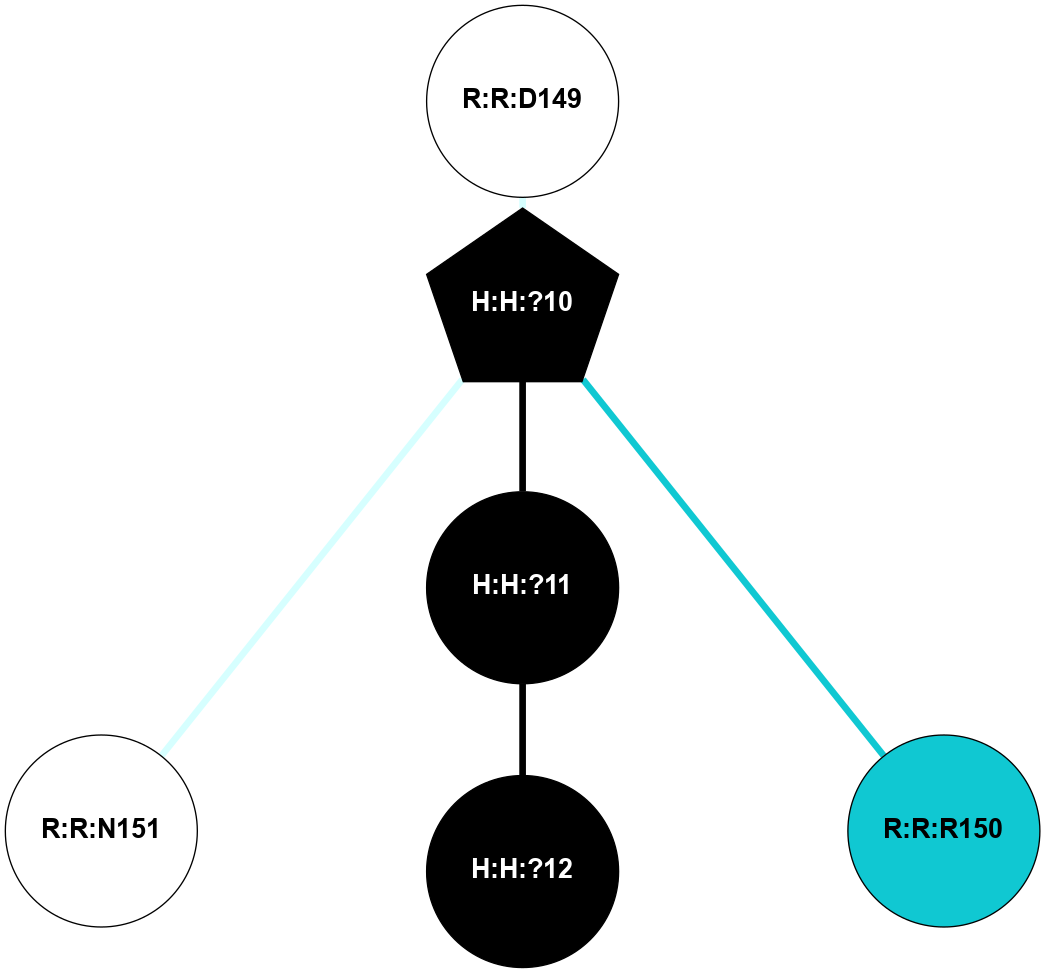
A 2D representation of the interactions of NAG in 6FJ3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | R:R:D149 | 6.07 | 0 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?10 | R:R:R150 | 9.78 | 0 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?10 | R:R:N151 | 24.56 | 0 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?10 | H:H:?11 | 42.06 | 0 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?11 | H:H:?12 | 34.67 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 38.37 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FK6 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473033 | - | 2.36 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|
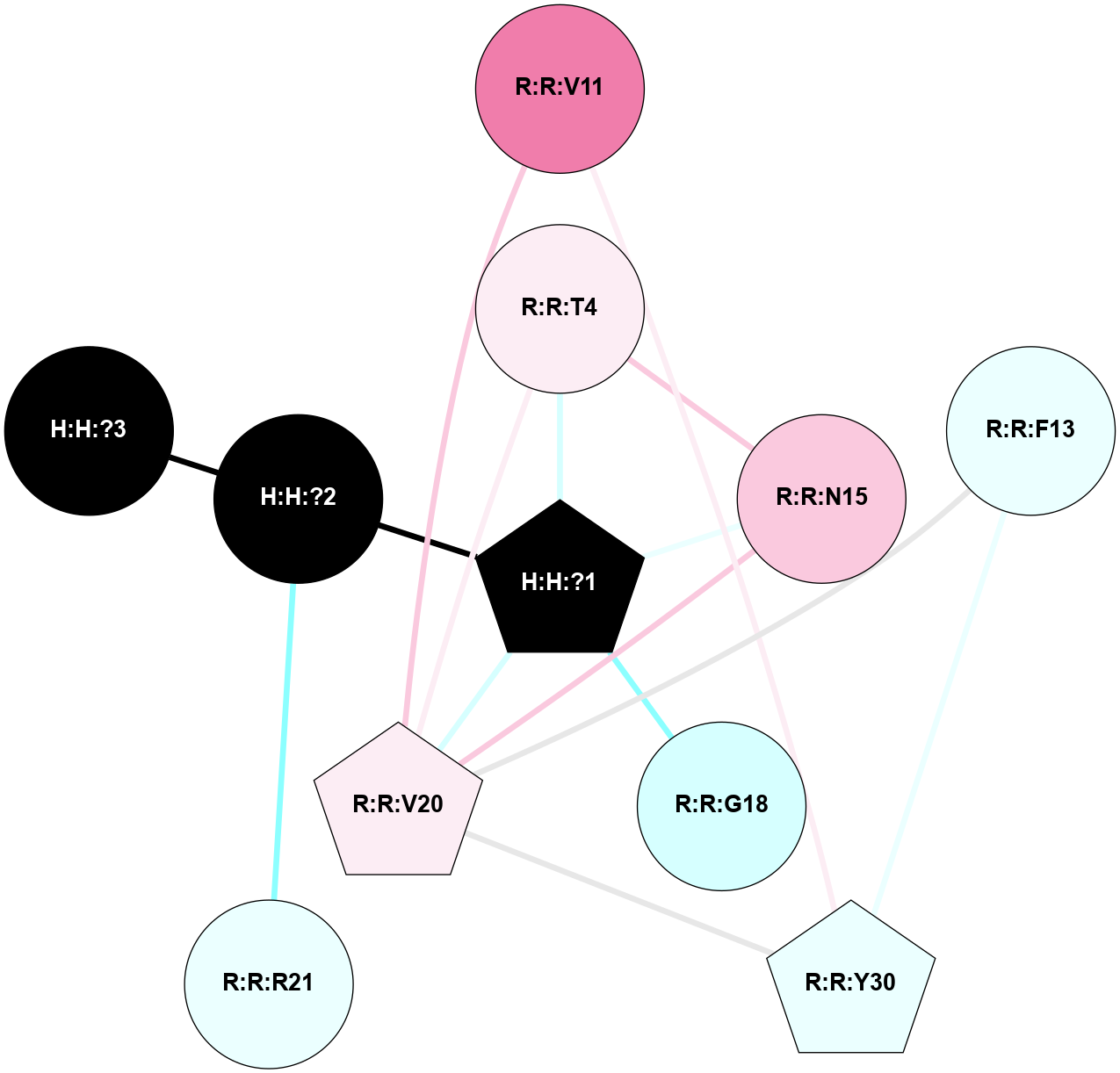
A 2D representation of the interactions of NAG in 6FK6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 2 | No | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 2 | No | Yes | 8 | 6 | 2 | 1 | | R:R:V11 | R:R:Y30 | 3.79 | 2 | No | Yes | 8 | 4 | 2 | 2 | | R:R:F13 | R:R:V20 | 5.24 | 2 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 13.41 | 2 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 2 | No | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 25.79 | 2 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 11.36 | 2 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 36.53 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 34.67 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.54 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
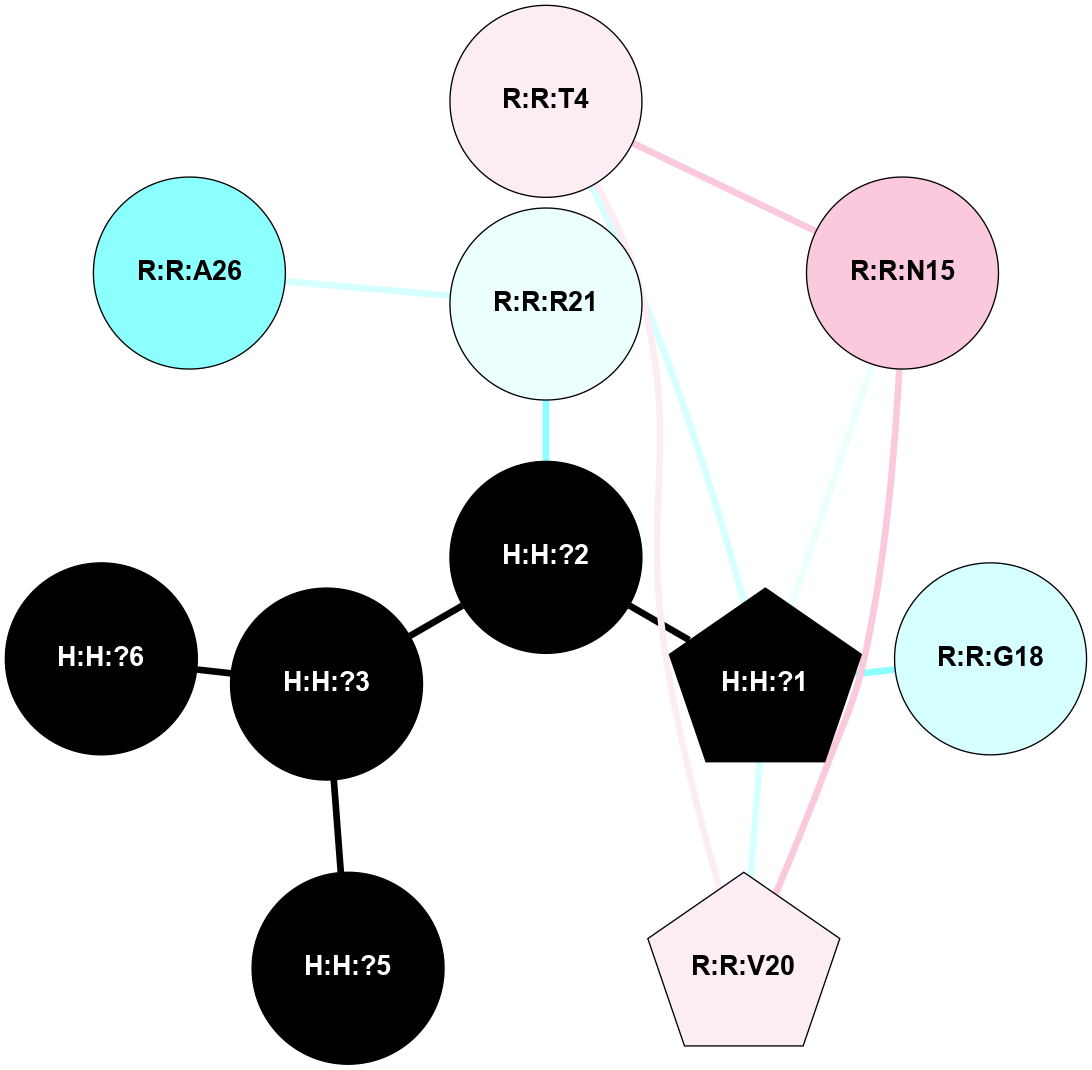
A 2D representation of the interactions of NAG in 6FK6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 2 | No | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 1 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 2 | No | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 25.79 | 2 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 36.53 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 34.67 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?5 | 50.23 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?3 | H:H:?6 | 35.45 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 25.91 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FK7 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473034 | - | 2.62 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|
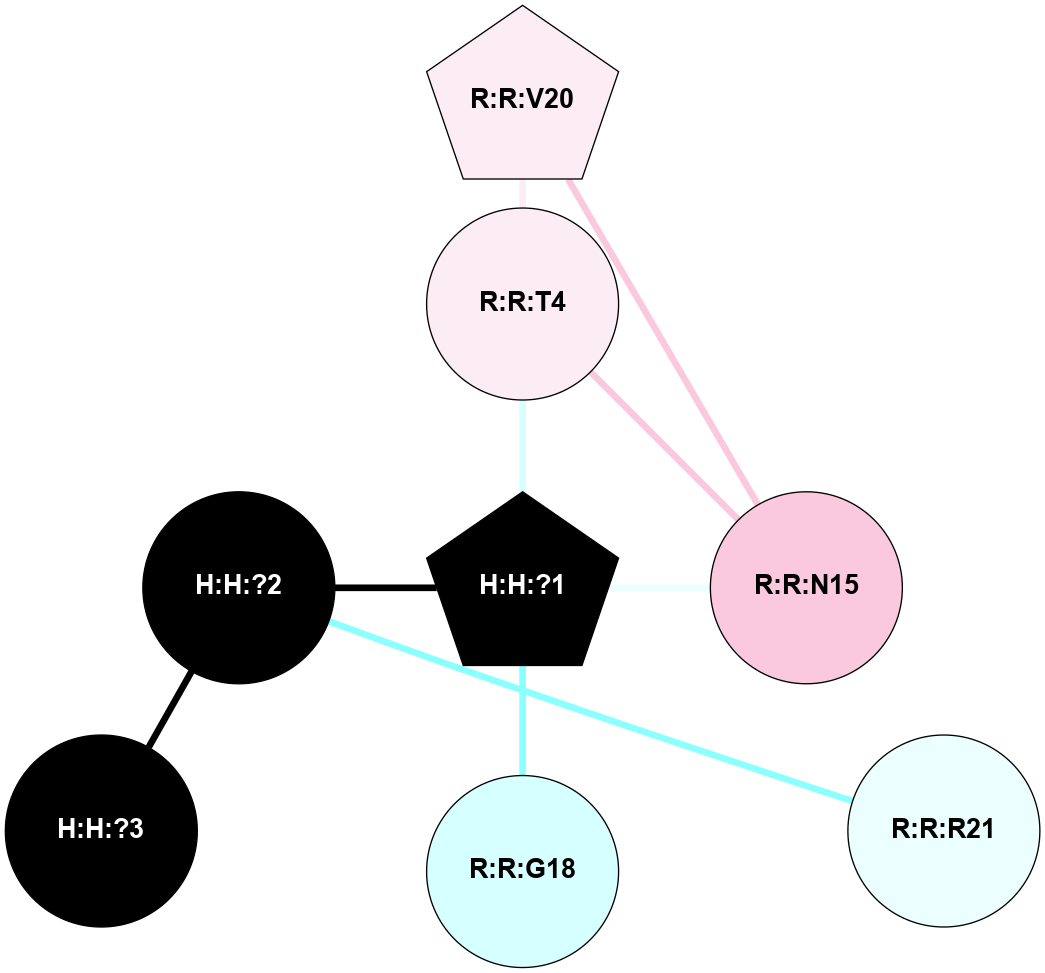
A 2D representation of the interactions of NAG in 6FK7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 10.24 | 2 | No | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 1 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 0 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 2 | No | Yes | 7 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 25.79 | 2 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?2 | R:R:R21 | 8.69 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 36.53 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 30.82 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.43 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
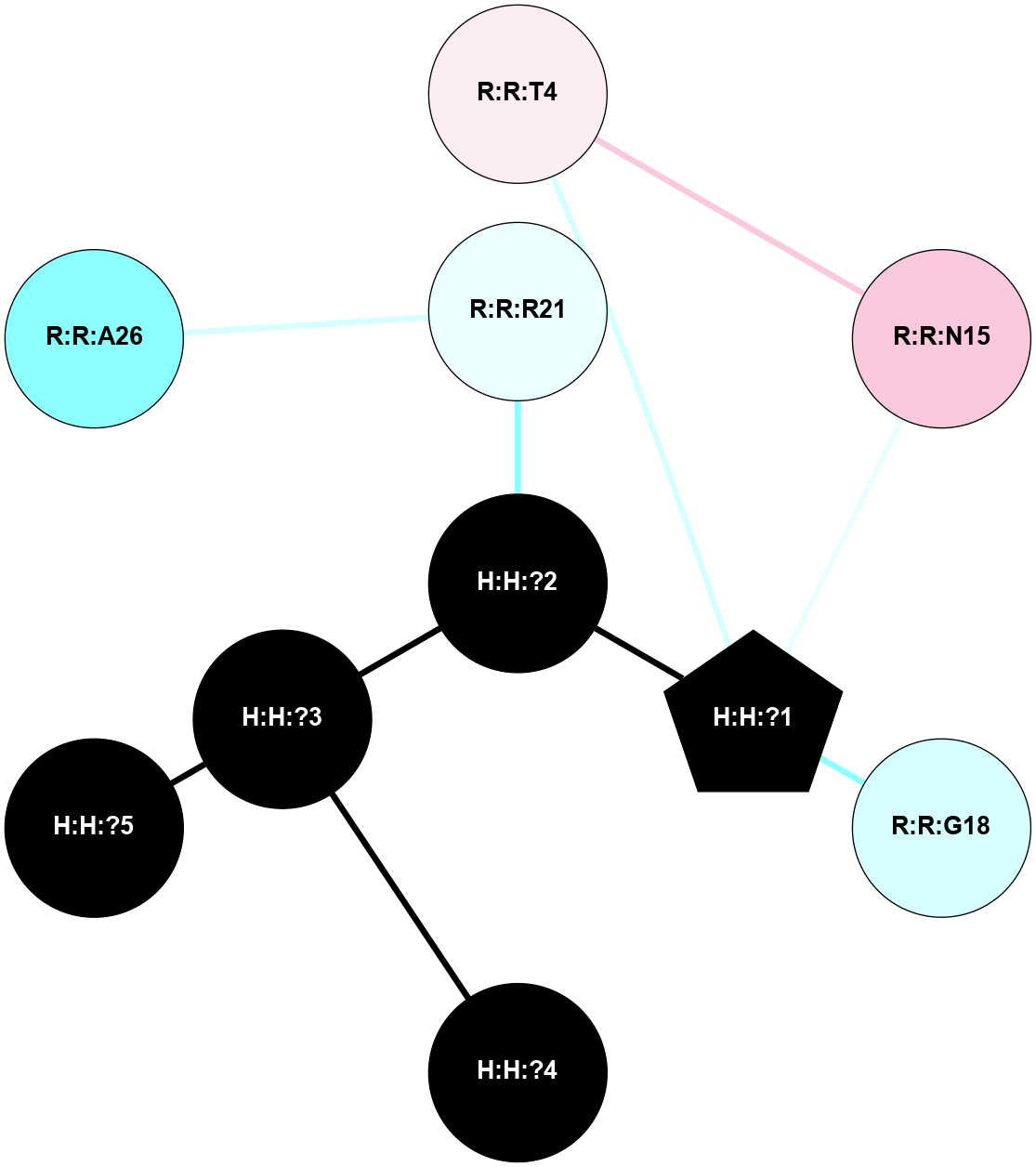
A 2D representation of the interactions of NAG in 6FK7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 10.24 | 2 | No | No | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 25.79 | 2 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?2 | R:R:R21 | 8.69 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 36.53 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 30.82 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?4 | 41.36 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?3 | H:H:?5 | 28.07 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 25.35 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FK8 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473035 | - | 2.87 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|
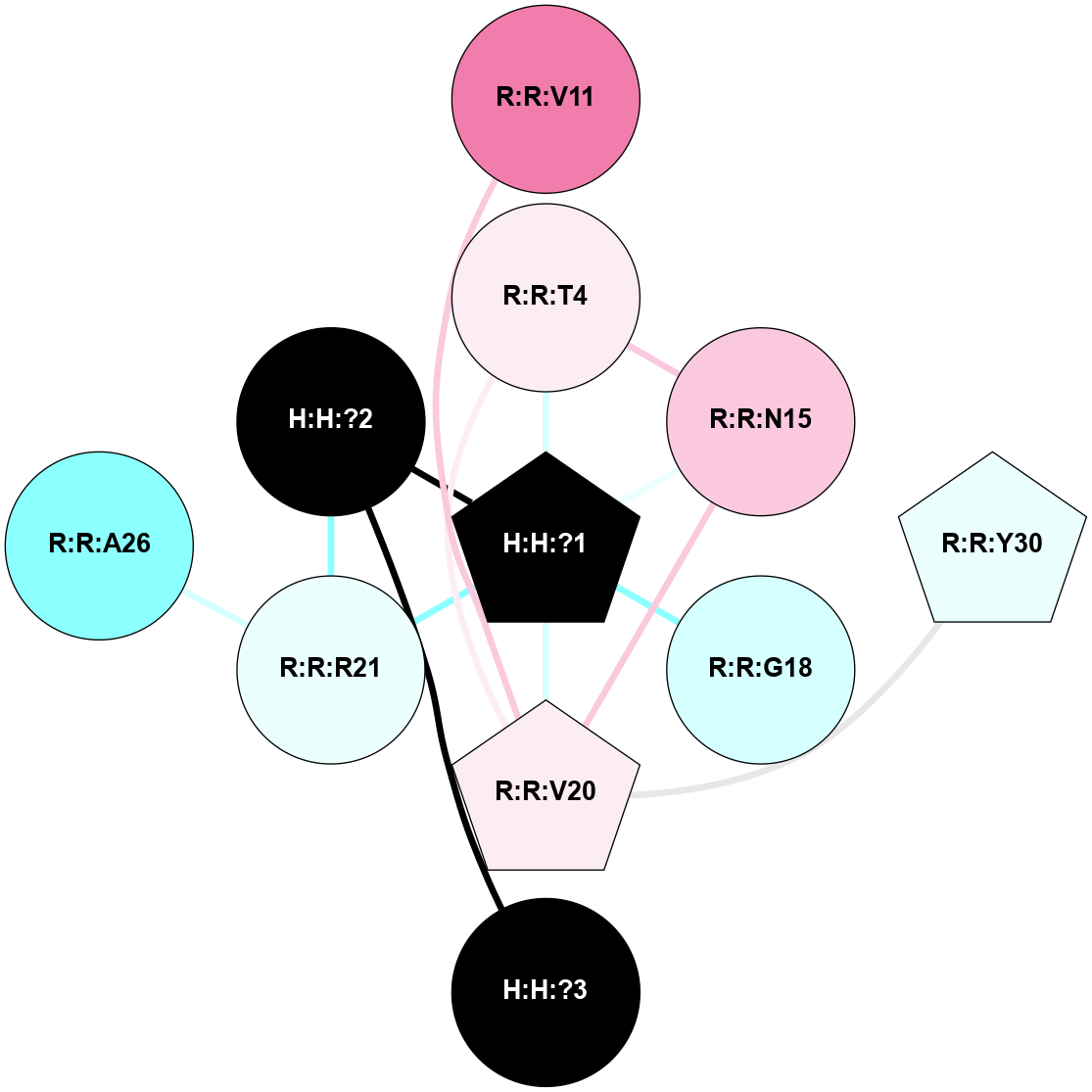
A 2D representation of the interactions of NAG in 6FK8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 1 | No | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 11.11 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | No | Yes | 8 | 6 | 2 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | No | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 25.79 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 11.36 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 7.99 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:R21 | 4.35 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:R21 | 8.69 | 1 | No | No | 0 | 4 | 1 | 1 | | H:H:?1 | H:H:?2 | 35.42 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 34.67 | 1 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.16 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
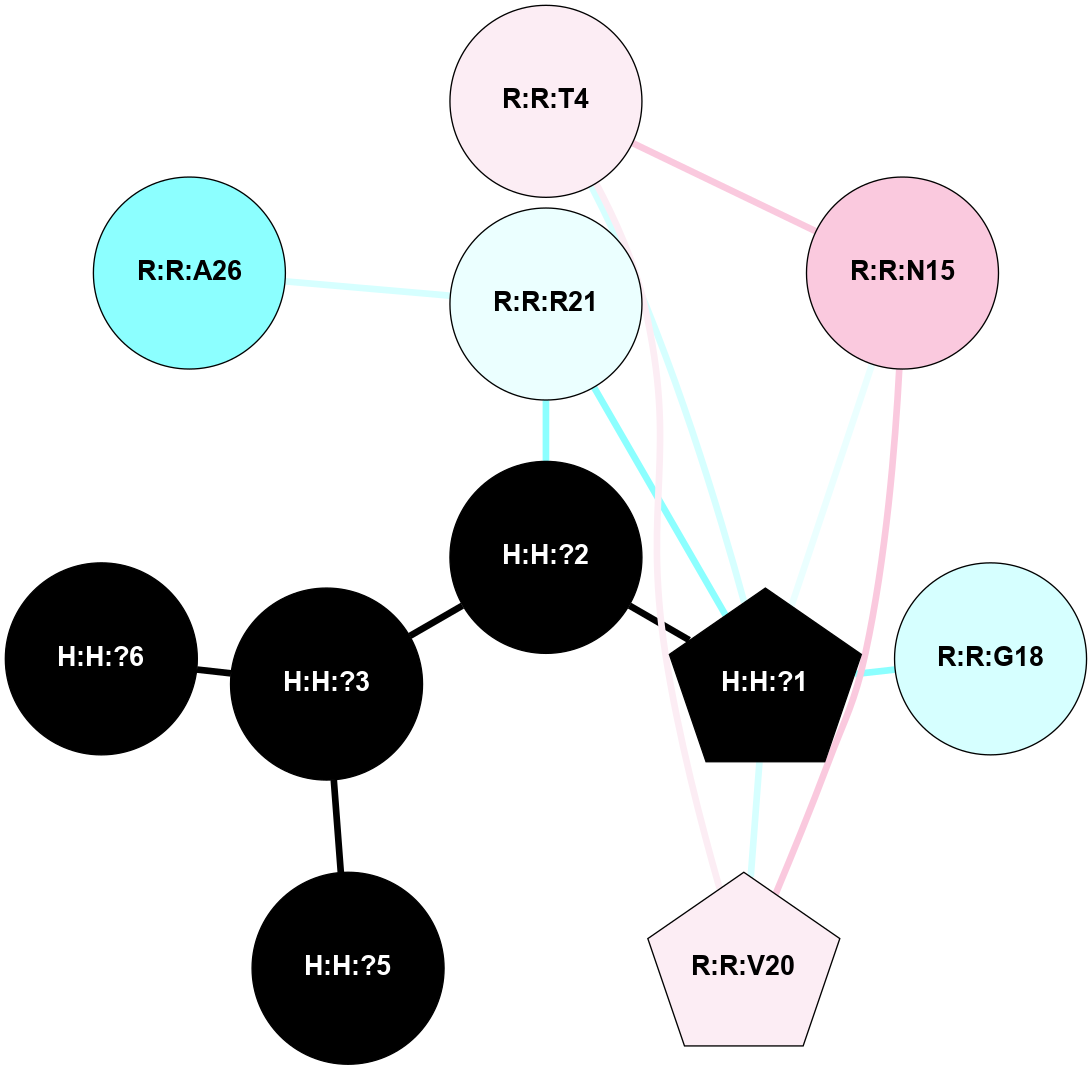
A 2D representation of the interactions of NAG in 6FK8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 1 | No | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 11.11 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | No | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 25.79 | 1 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 7.99 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:R21 | 4.35 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?2 | R:R:R21 | 8.69 | 1 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 35.42 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 34.67 | 1 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?5 | 45.8 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?3 | H:H:?6 | 26.59 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 26.26 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FK9 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473036 | - | 2.63 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|
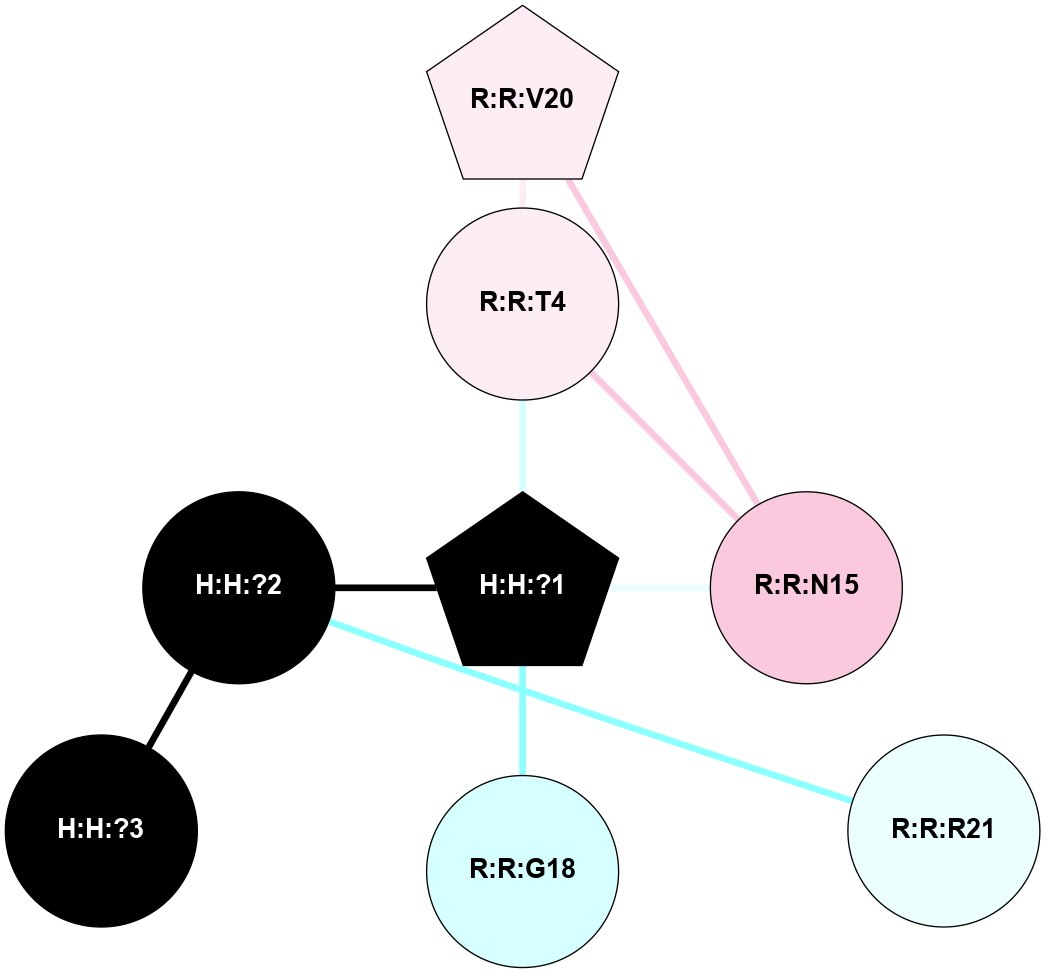
A 2D representation of the interactions of NAG in 6FK9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 10.24 | 1 | No | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | No | Yes | 8 | 6 | 2 | 1 | | R:R:F13 | R:R:V20 | 5.24 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 15.47 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | No | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 24.56 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 11.36 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:R21 | 7.6 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 38.74 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 30.82 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.74 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
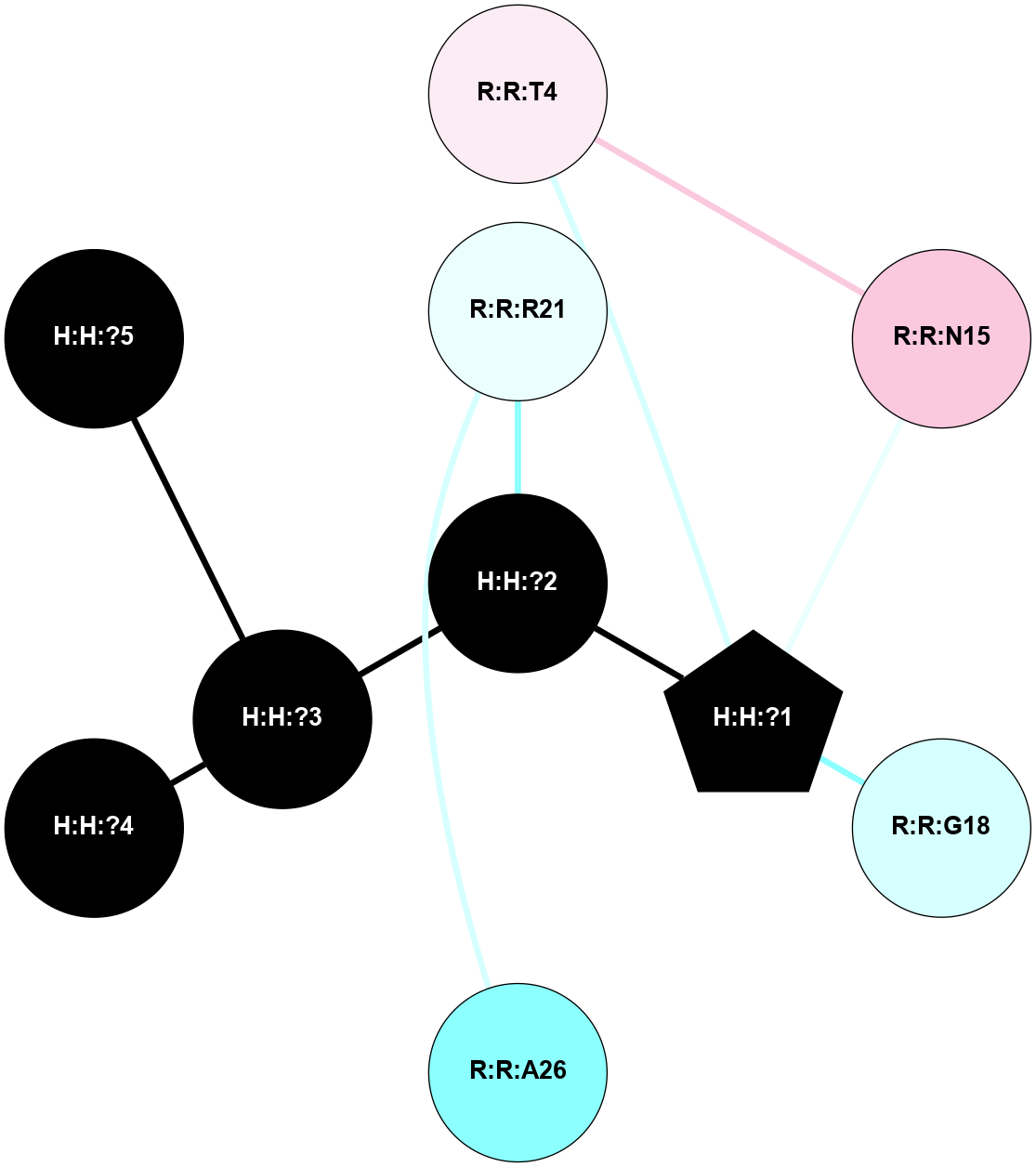
A 2D representation of the interactions of NAG in 6FK9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 10.24 | 1 | No | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | No | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 24.56 | 1 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | R:R:A26 | R:R:R21 | 4.15 | 0 | No | No | 2 | 4 | 2 | 1 | | H:H:?2 | R:R:R21 | 7.6 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 38.74 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 30.82 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?4 | 44.32 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?3 | H:H:?5 | 29.55 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 25.72 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FKA | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473037 | - | 2.7 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|
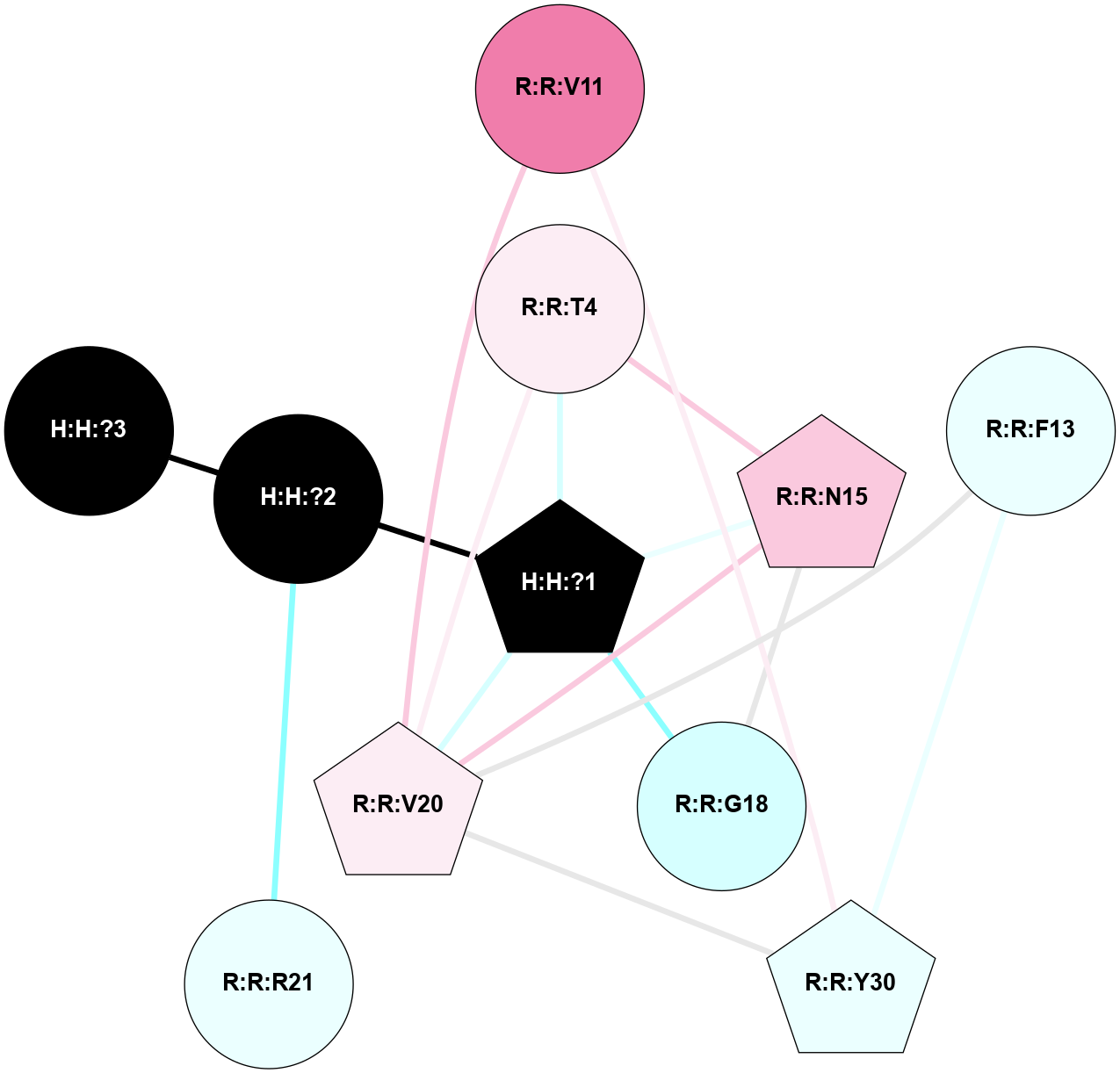
A 2D representation of the interactions of NAG in 6FKA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | No | Yes | 8 | 6 | 2 | 1 | | R:R:V11 | R:R:Y30 | 5.05 | 1 | No | Yes | 8 | 4 | 2 | 2 | | R:R:F13 | R:R:V20 | 5.24 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 14.44 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 25.79 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 11.36 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:R21 | 7.6 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 36.53 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 30.82 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 15.54 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
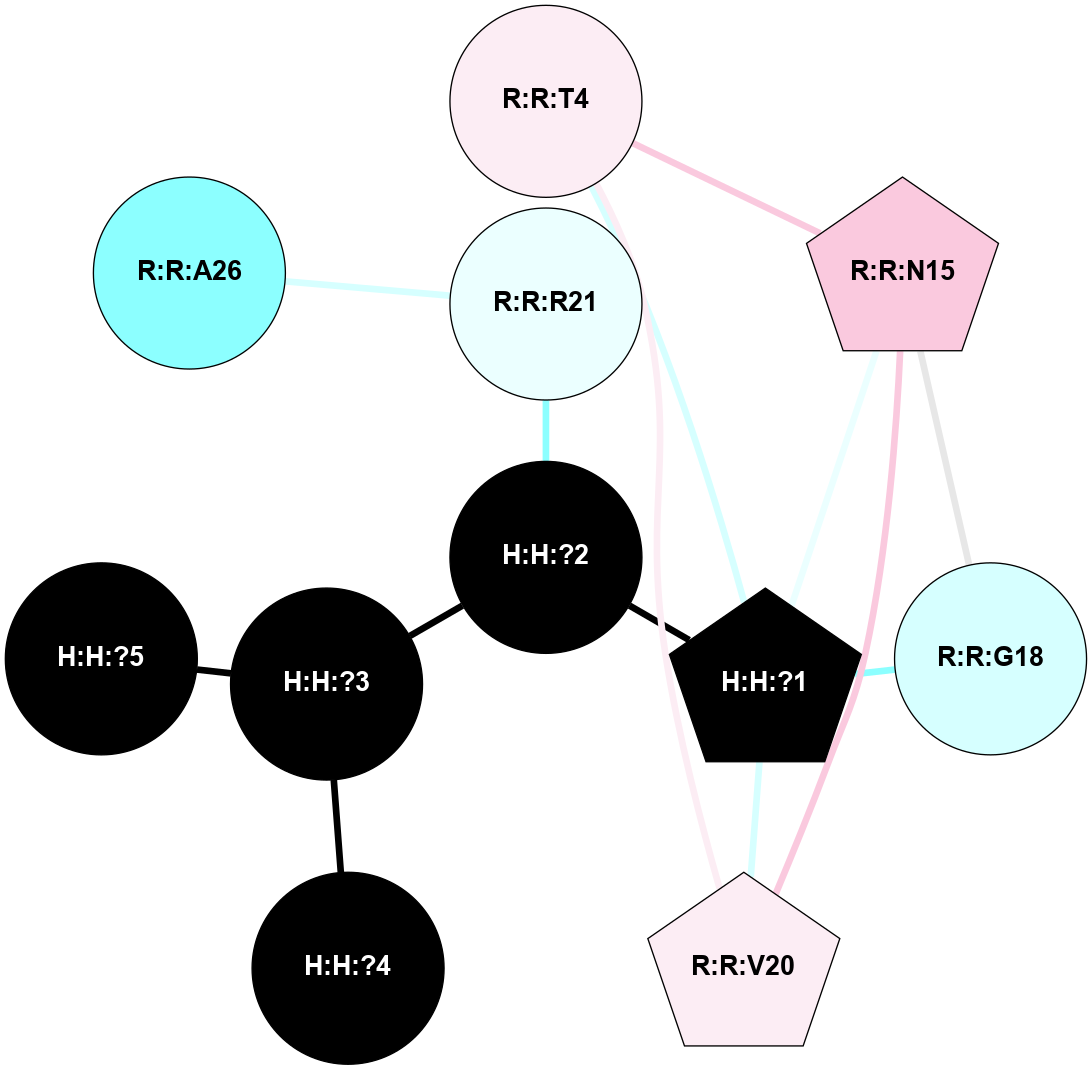
A 2D representation of the interactions of NAG in 6FKA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 1 | Yes | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 25.79 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?2 | R:R:R21 | 7.6 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 36.53 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 30.82 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?4 | 42.84 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?3 | H:H:?5 | 29.55 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 24.98 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FKB | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473038 | - | 3.03 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|
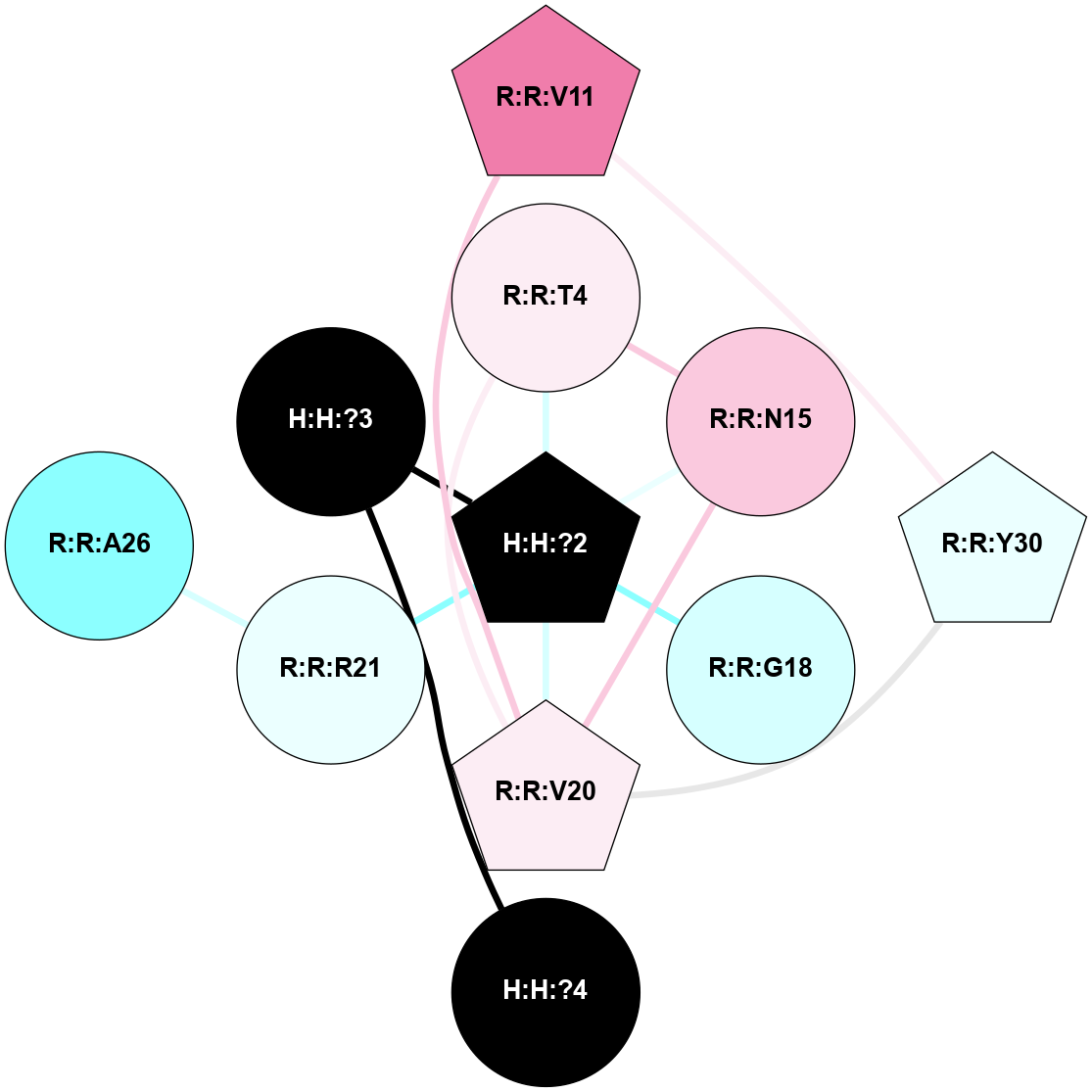
A 2D representation of the interactions of NAG in 6FKB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 11.7 | 1 | No | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?2 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:V11 | R:R:Y30 | 3.79 | 1 | Yes | Yes | 8 | 4 | 2 | 2 | | R:R:N15 | R:R:V20 | 7.39 | 1 | No | Yes | 7 | 6 | 1 | 1 | | H:H:?2 | R:R:N15 | 13.51 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?2 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 11.36 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?2 | R:R:V20 | 6.66 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:R21 | 8.69 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | H:H:?3 | 33.21 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?4 | 17.98 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 12.24 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
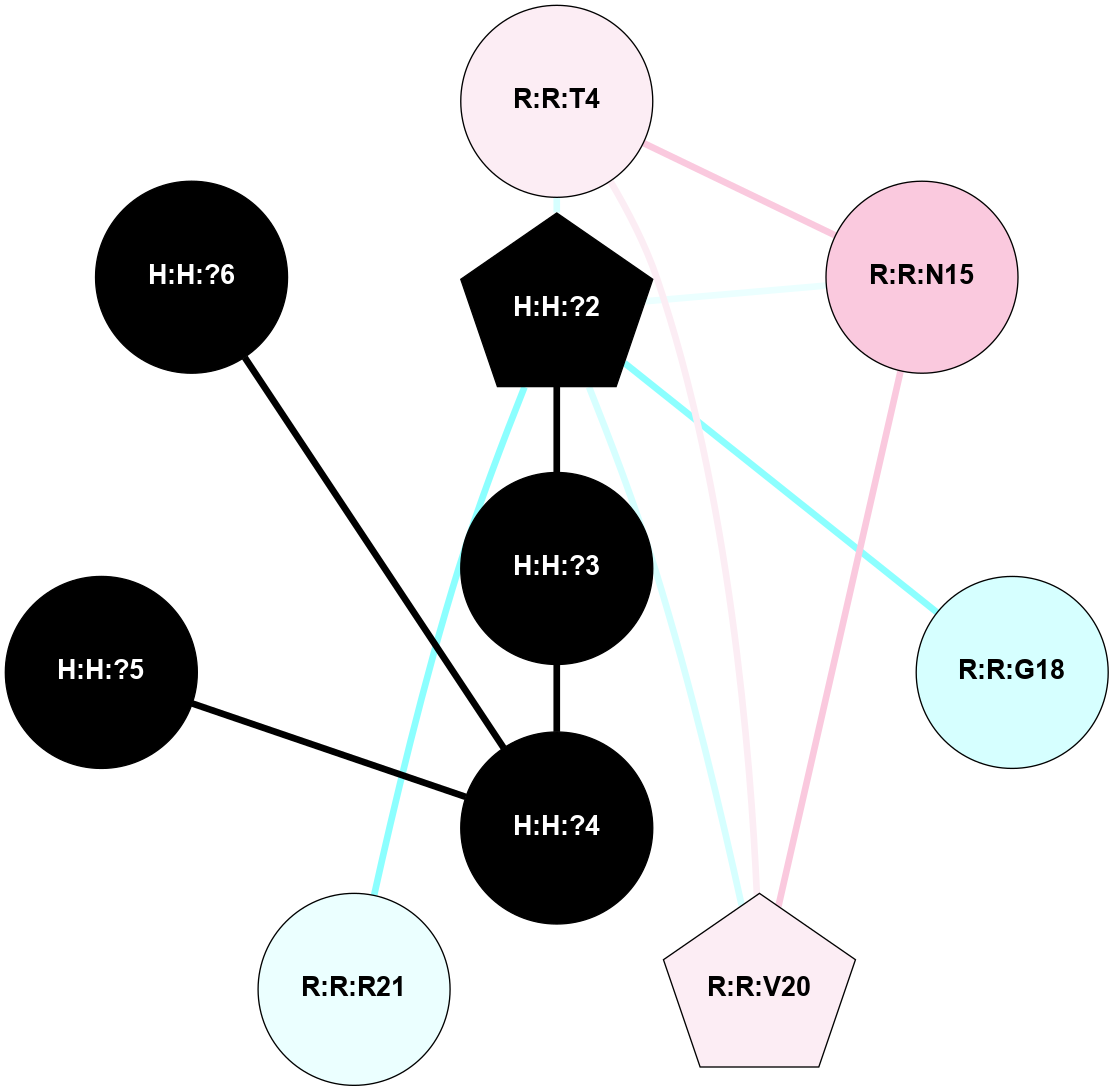
A 2D representation of the interactions of NAG in 6FKB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 11.7 | 1 | No | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?2 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:N15 | R:R:V20 | 7.39 | 1 | No | Yes | 7 | 6 | 2 | 2 | | H:H:?2 | R:R:N15 | 13.51 | 1 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?2 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?2 | R:R:V20 | 6.66 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?2 | R:R:R21 | 8.69 | 1 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?2 | H:H:?3 | 33.21 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?3 | H:H:?4 | 17.98 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?4 | H:H:?5 | 38.41 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?4 | H:H:?6 | 36.93 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 25.59 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FKC | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 132473039 | - | 2.46 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|
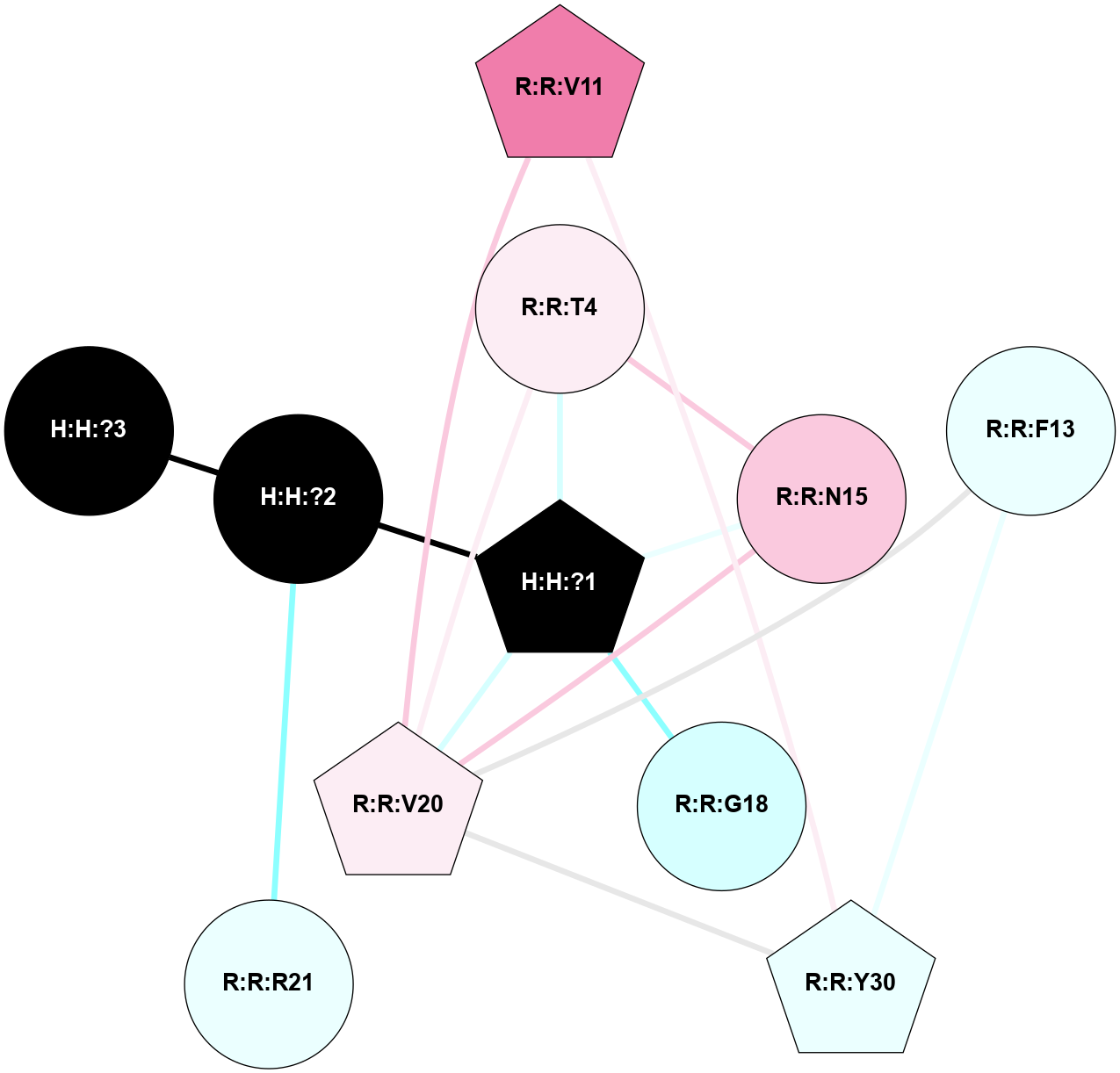
A 2D representation of the interactions of NAG in 6FKC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 1 | No | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:V11 | R:R:Y30 | 3.79 | 1 | Yes | Yes | 8 | 4 | 2 | 2 | | R:R:F13 | R:R:V20 | 5.24 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 13.41 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | No | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 25.79 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 10.09 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 36.53 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 30.82 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.54 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
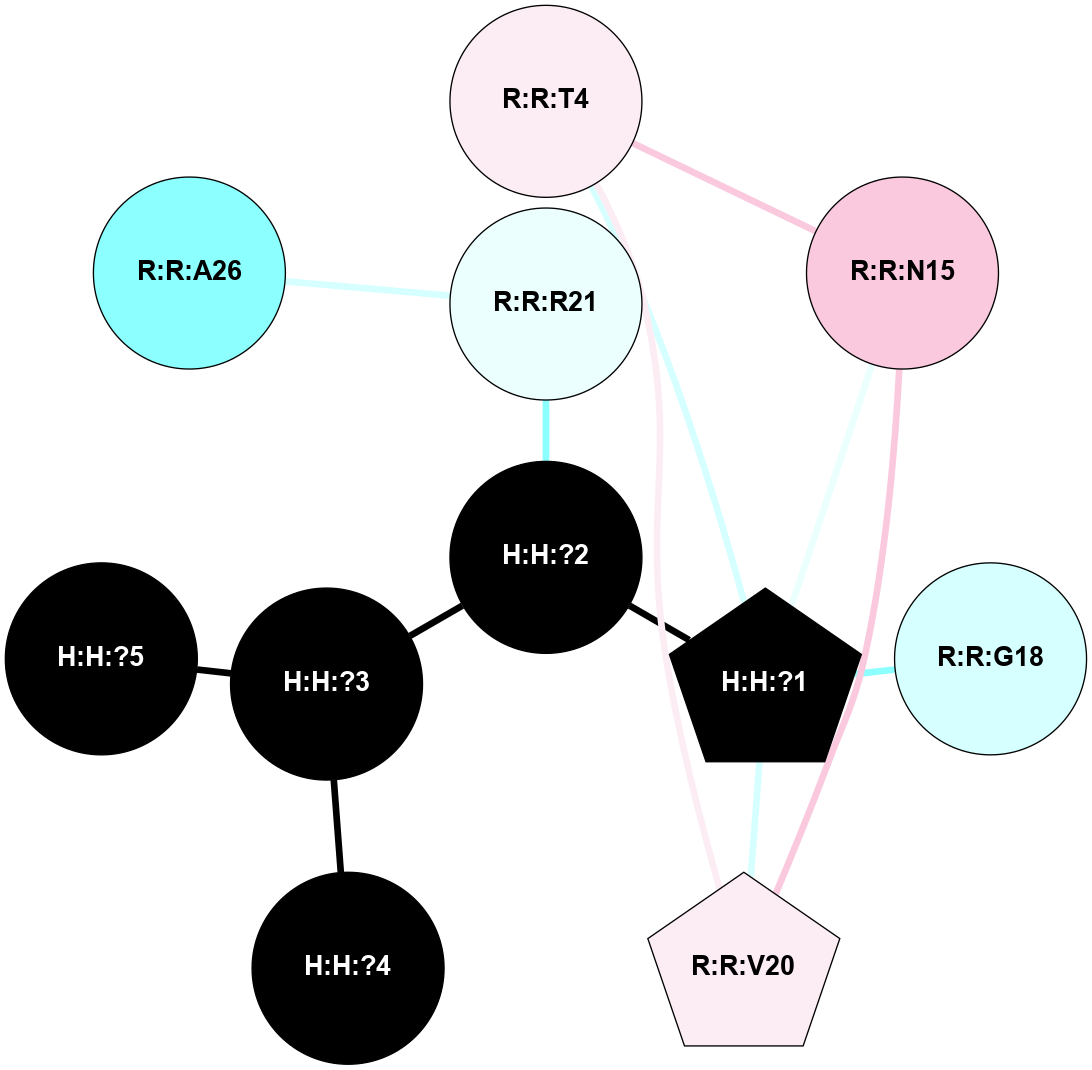
A 2D representation of the interactions of NAG in 6FKC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 1 | No | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | No | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 25.79 | 1 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 36.53 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 30.82 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?4 | 38.41 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?3 | H:H:?5 | 28.07 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 24.62 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FKD | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | PubChem 137349183 | - | 2.49 | 2018-04-04 | doi.org/10.1073/pnas.1718084115 |
|
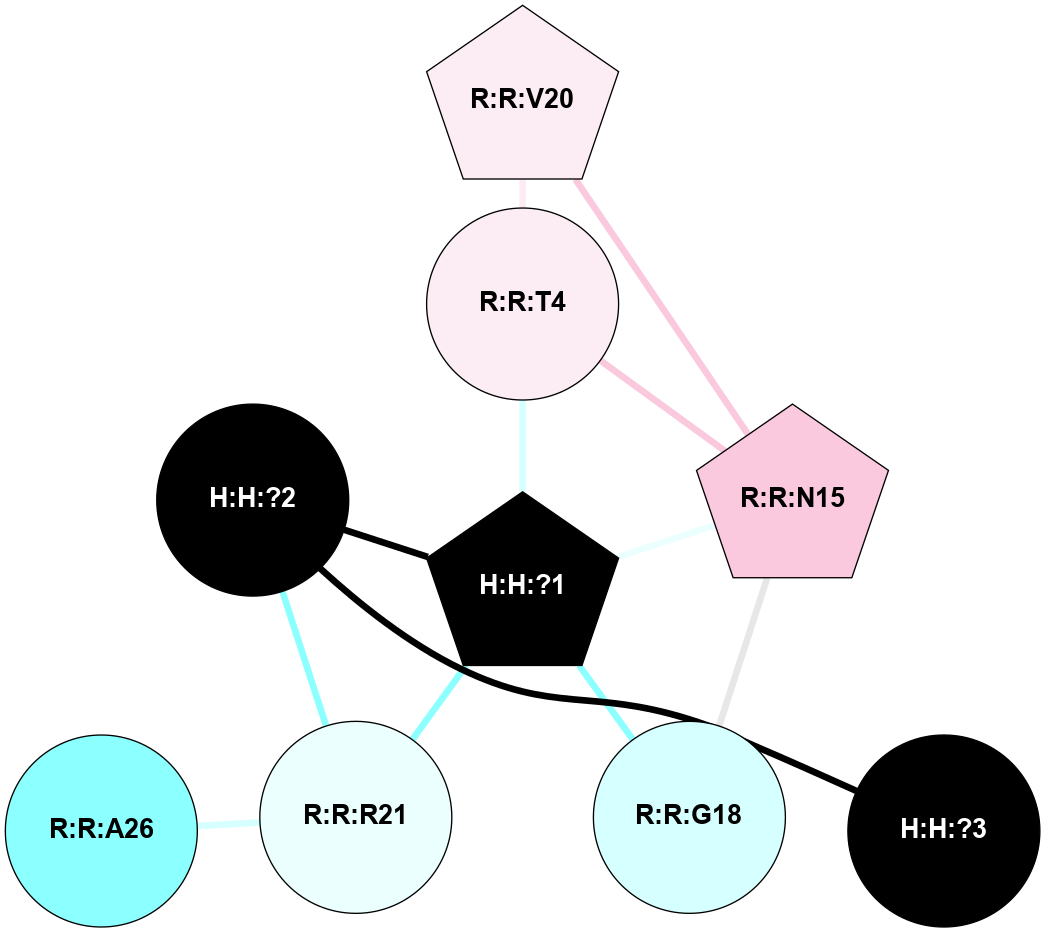
A 2D representation of the interactions of NAG in 6FKD
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 2 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 1 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 0 | 1 | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 2 | Yes | Yes | 7 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 25.79 | 2 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:R21 | 3.26 | 2 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:R21 | 6.52 | 2 | No | No | 0 | 4 | 1 | 1 | | H:H:?1 | H:H:?2 | 40.96 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 30.82 | 2 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 16.28 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
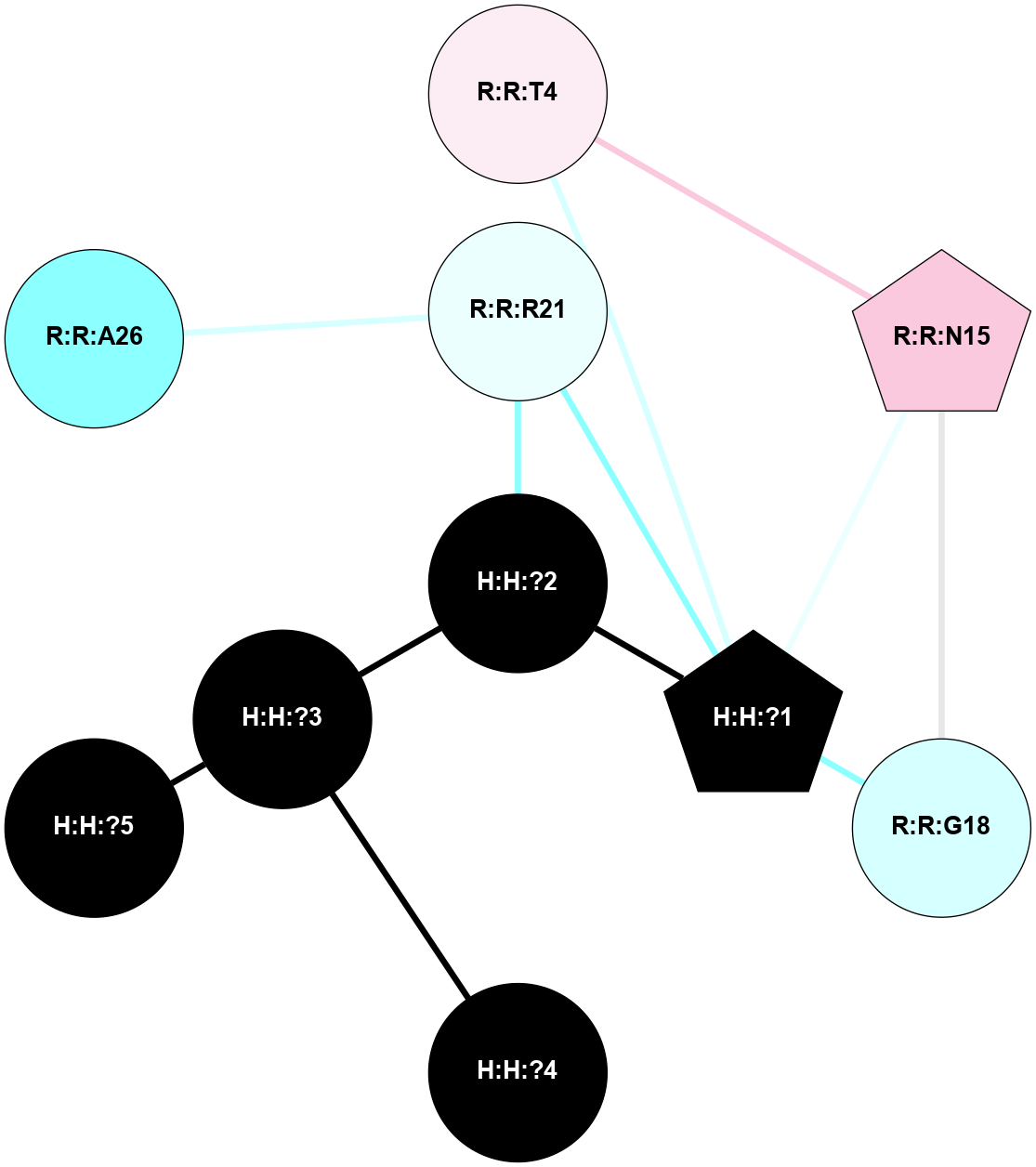
A 2D representation of the interactions of NAG in 6FKD
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 2 | Yes | No | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 2 | No | Yes | 3 | 7 | 2 | 2 | | H:H:?1 | R:R:N15 | 25.79 | 2 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:R21 | 3.26 | 2 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?2 | R:R:R21 | 6.52 | 2 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 40.96 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 30.82 | 2 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | H:H:?4 | 44.32 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?3 | H:H:?5 | 32.5 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 26.10 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6FUF | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | Go | 3.12 | 2018-10-03 | doi.org/10.1126/sciadv.aat7052 |
|
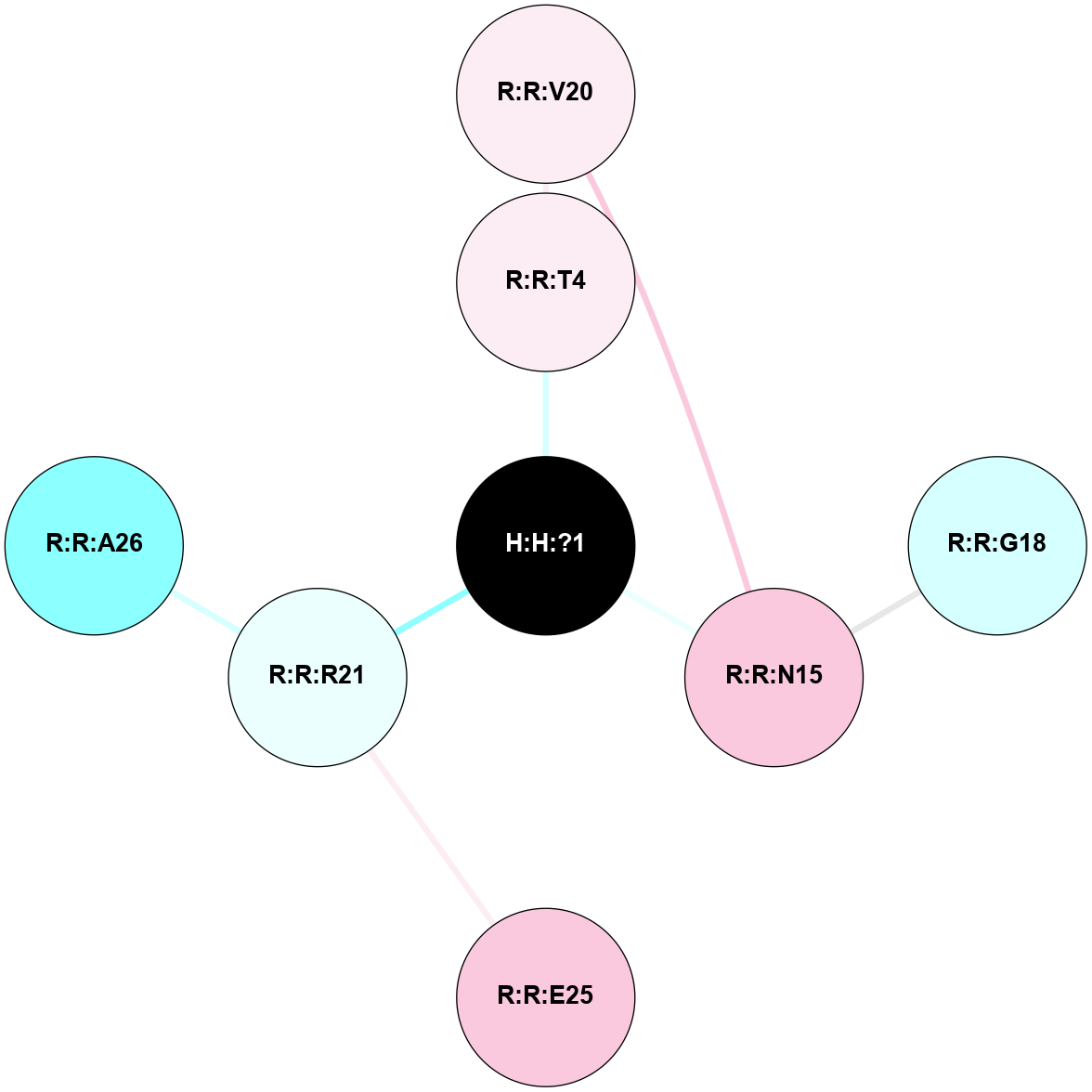
A 2D representation of the interactions of NAG in 6FUF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:T4 | 3.95 | 0 | No | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:N15 | 24.56 | 0 | No | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:R21 | 11.95 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:T4 | R:R:V20 | 11.11 | 0 | No | No | 6 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 3.39 | 0 | No | No | 3 | 7 | 2 | 1 | | R:R:N15 | R:R:V20 | 8.87 | 0 | No | No | 7 | 6 | 1 | 2 | | R:R:E25 | R:R:R21 | 8.14 | 0 | No | No | 7 | 4 | 2 | 1 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.49 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6LN2 | B1 | Peptide | Glucagon | GLP-1 | Homo Sapiens | - | PF-06372222 | - | 3.2 | 2020-03-18 | doi.org/10.1038/s41467-020-14934-5 |
|
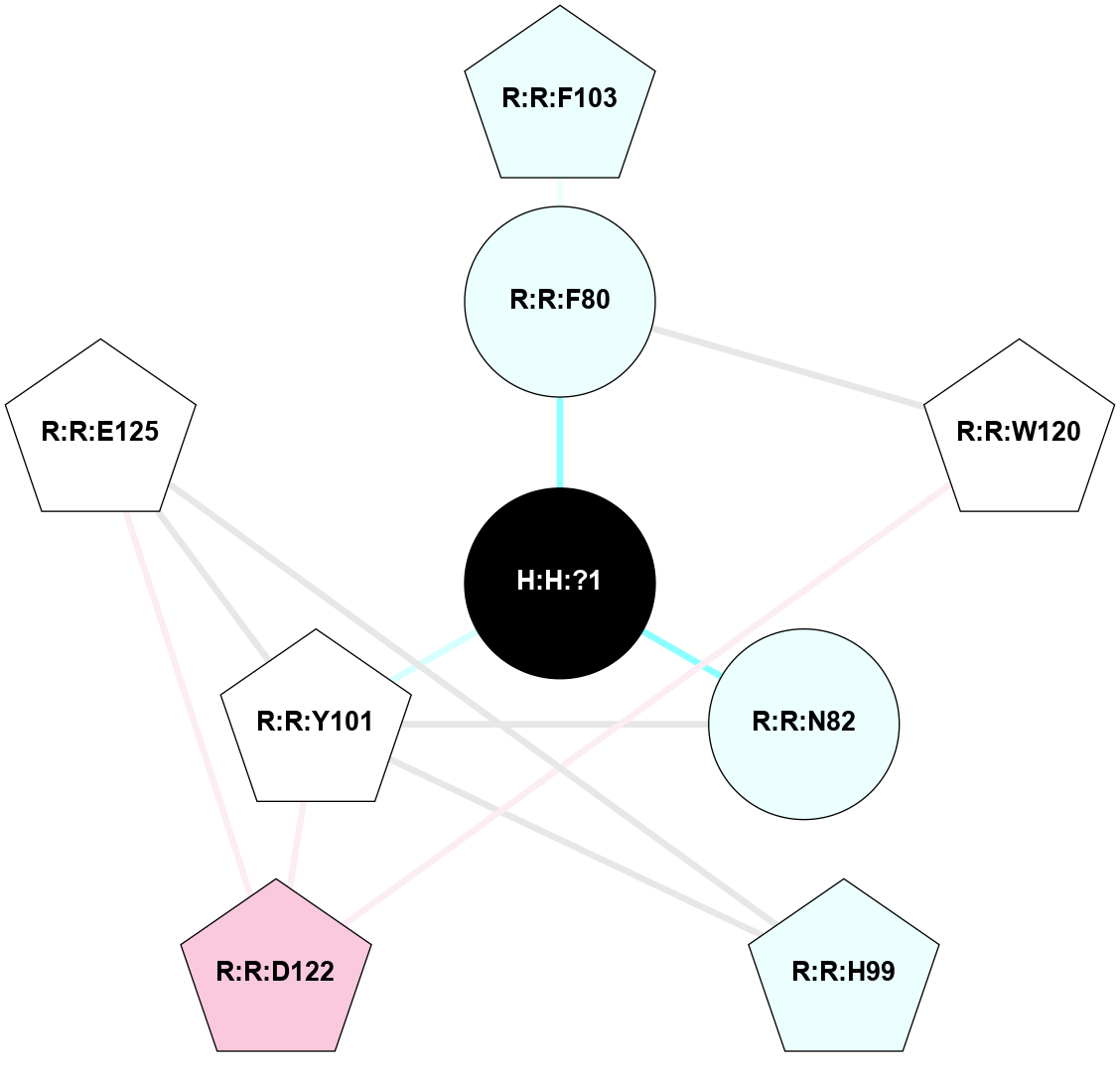
A 2D representation of the interactions of NAG in 6LN2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:F80 | 11.98 | 3 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:N82 | 25.79 | 3 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:Y101 | 6.29 | 3 | No | Yes | 0 | 5 | 0 | 1 | | R:R:F103 | R:R:F80 | 10.72 | 3 | Yes | No | 4 | 4 | 2 | 1 | | R:R:F80 | R:R:W120 | 5.01 | 3 | No | Yes | 4 | 5 | 1 | 2 | | R:R:N82 | R:R:Y101 | 17.44 | 3 | No | Yes | 4 | 5 | 1 | 1 | | R:R:H99 | R:R:Y101 | 10.89 | 3 | No | Yes | 4 | 5 | 2 | 1 | | R:R:E125 | R:R:H99 | 16 | 3 | No | No | 5 | 4 | 2 | 2 | | R:R:W120 | R:R:Y101 | 1.93 | 3 | Yes | Yes | 5 | 5 | 2 | 1 | | R:R:D122 | R:R:Y101 | 5.75 | 3 | No | Yes | 7 | 5 | 2 | 1 | | R:R:E125 | R:R:Y101 | 11.22 | 3 | No | Yes | 5 | 5 | 2 | 1 | | R:R:F103 | R:R:W120 | 2 | 3 | Yes | Yes | 4 | 5 | 2 | 2 | | R:R:D122 | R:R:W120 | 5.58 | 3 | No | Yes | 7 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.69 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6MEO | A | Protein | Chemokine | CCR5 | Homo Sapiens | Envelope Glycoprotein Gp160; T-Cell Surface Glycoprotein Cd4 | - | - | 3.9 | 2018-12-12 | doi.org/10.1038/s41586-018-0804-9 |
|
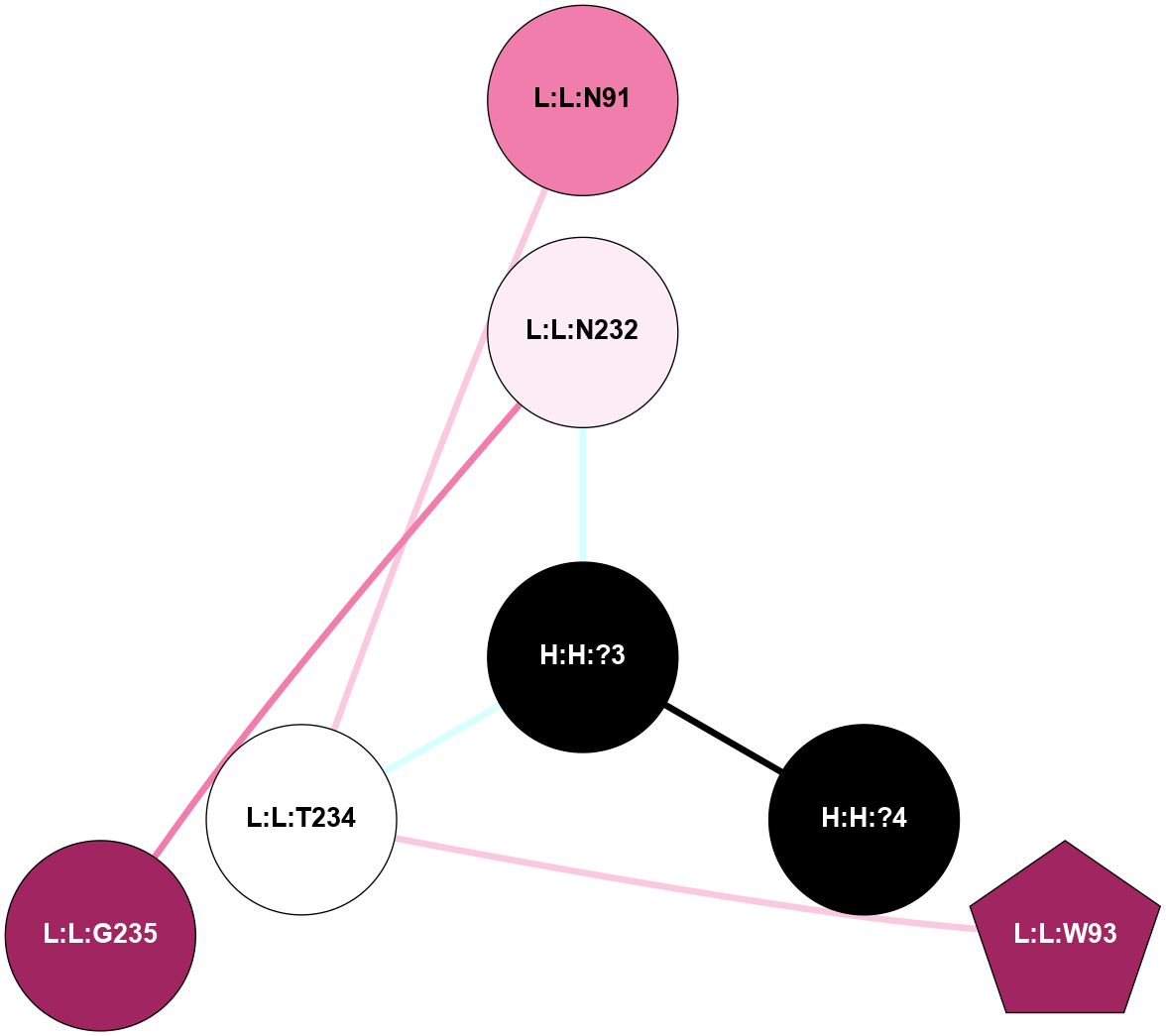
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 35.66 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | L:L:N232 | 25.87 | 0 | No | No | 0 | 6 | 0 | 1 | | L:L:N91 | L:L:T234 | 8.77 | 0 | No | No | 8 | 5 | 2 | 1 | | L:L:T234 | L:L:W93 | 3.64 | 0 | No | Yes | 5 | 9 | 1 | 2 | | L:L:G235 | L:L:N232 | 3.39 | 0 | No | No | 9 | 6 | 2 | 1 | | H:H:?3 | L:L:T234 | 1.32 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.95 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
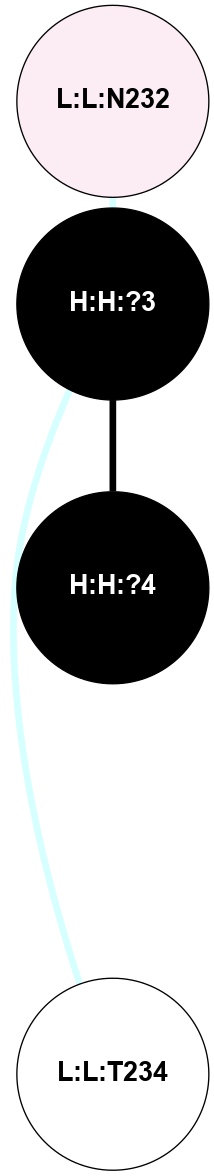
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 35.66 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?3 | L:L:N232 | 25.87 | 0 | No | No | 0 | 6 | 1 | 2 | | H:H:?3 | L:L:T234 | 1.32 | 0 | No | No | 0 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 35.66 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
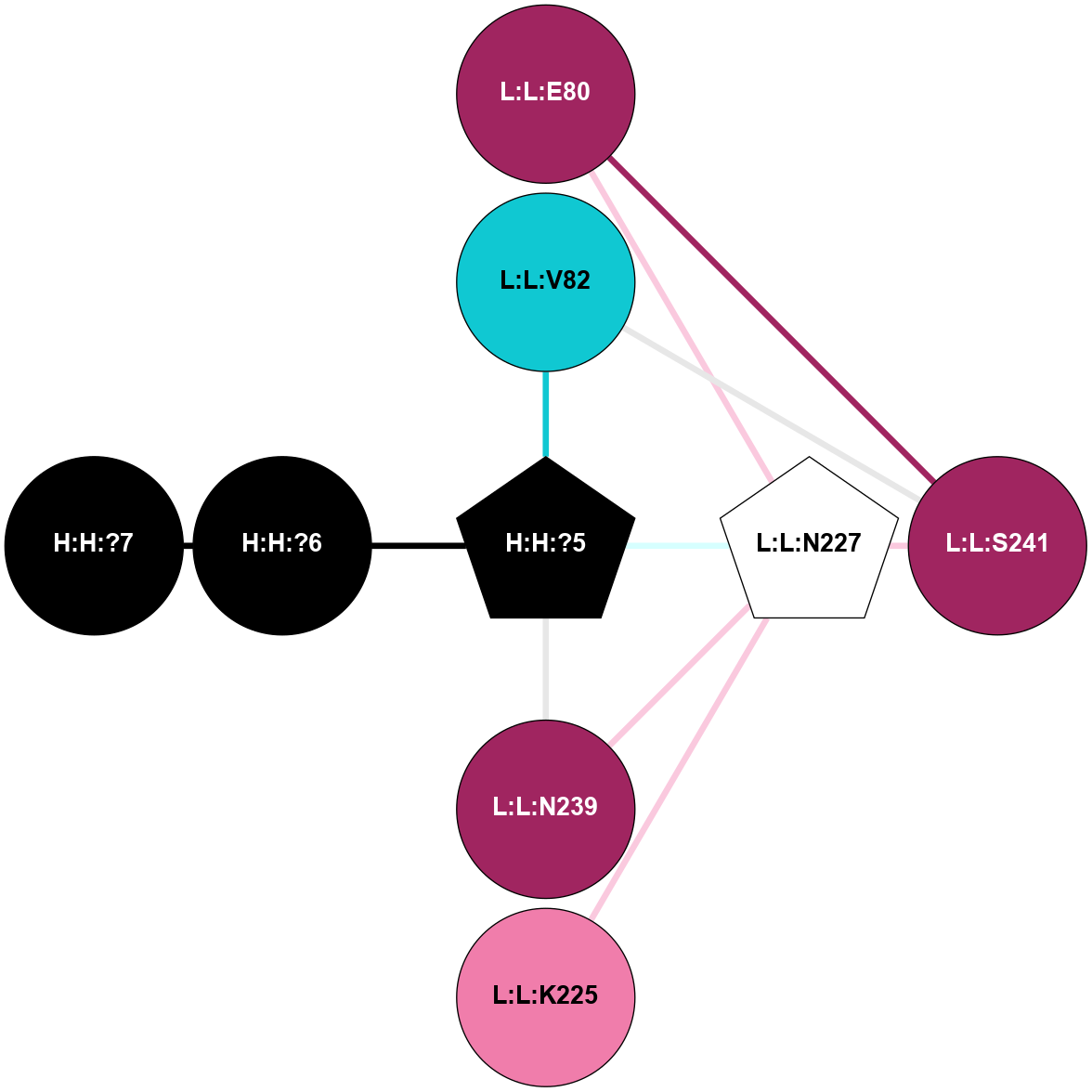
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 36.78 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?5 | L:L:V82 | 4.01 | 1 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?5 | L:L:N227 | 11.09 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?5 | L:L:N239 | 27.11 | 1 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?6 | H:H:?7 | 28.57 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:E80 | L:L:N227 | 3.94 | 1 | No | Yes | 9 | 5 | 2 | 1 | | L:L:E80 | L:L:S241 | 8.62 | 1 | No | No | 9 | 9 | 2 | 2 | | L:L:S241 | L:L:V82 | 3.23 | 1 | No | No | 9 | 1 | 2 | 1 | | L:L:K225 | L:L:N227 | 2.8 | 1 | No | Yes | 8 | 5 | 2 | 1 | | L:L:N227 | L:L:N239 | 12.26 | 1 | Yes | No | 5 | 9 | 1 | 1 | | L:L:N227 | L:L:S241 | 5.96 | 1 | Yes | No | 5 | 9 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.75 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
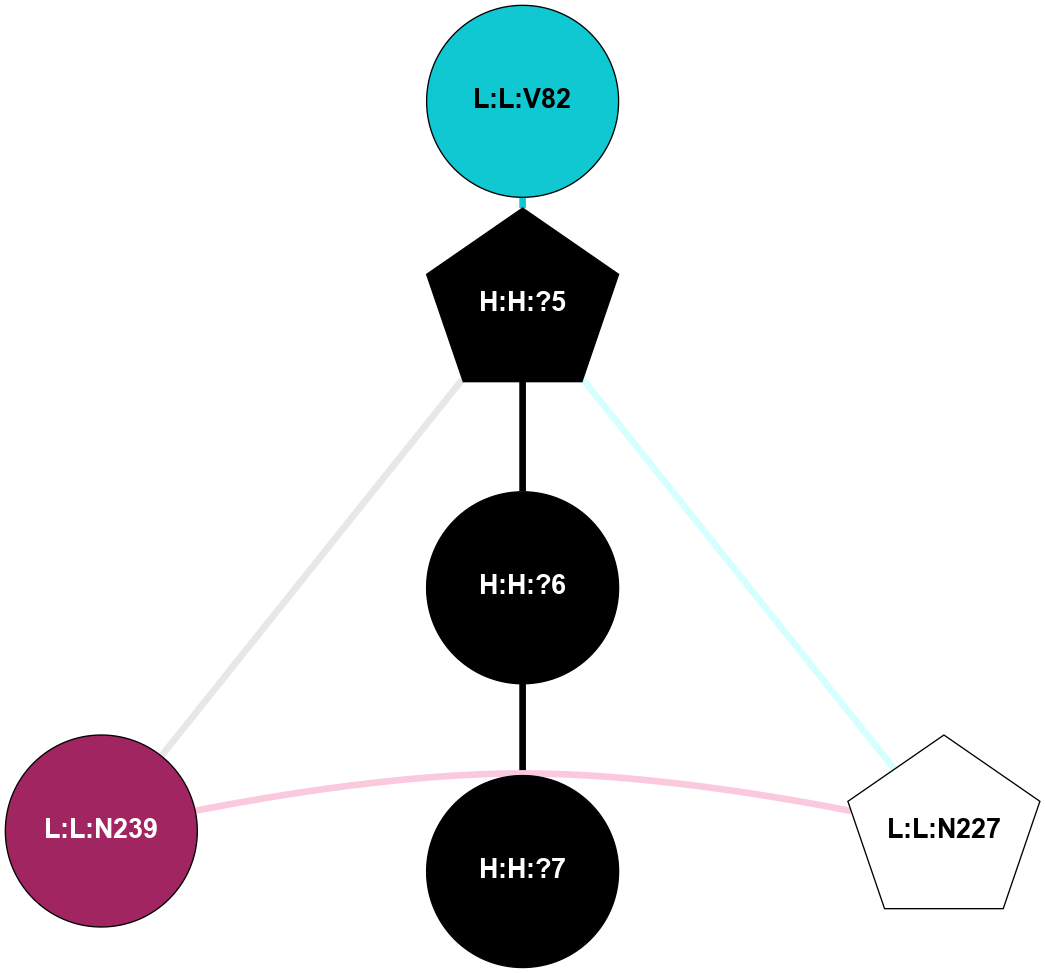
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 36.78 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?5 | L:L:V82 | 4.01 | 1 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?5 | L:L:N227 | 11.09 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | H:H:?5 | L:L:N239 | 27.11 | 1 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?6 | H:H:?7 | 28.57 | 0 | No | No | 0 | 0 | 0 | 1 | | L:L:N227 | L:L:N239 | 12.26 | 1 | Yes | No | 5 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 32.67 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
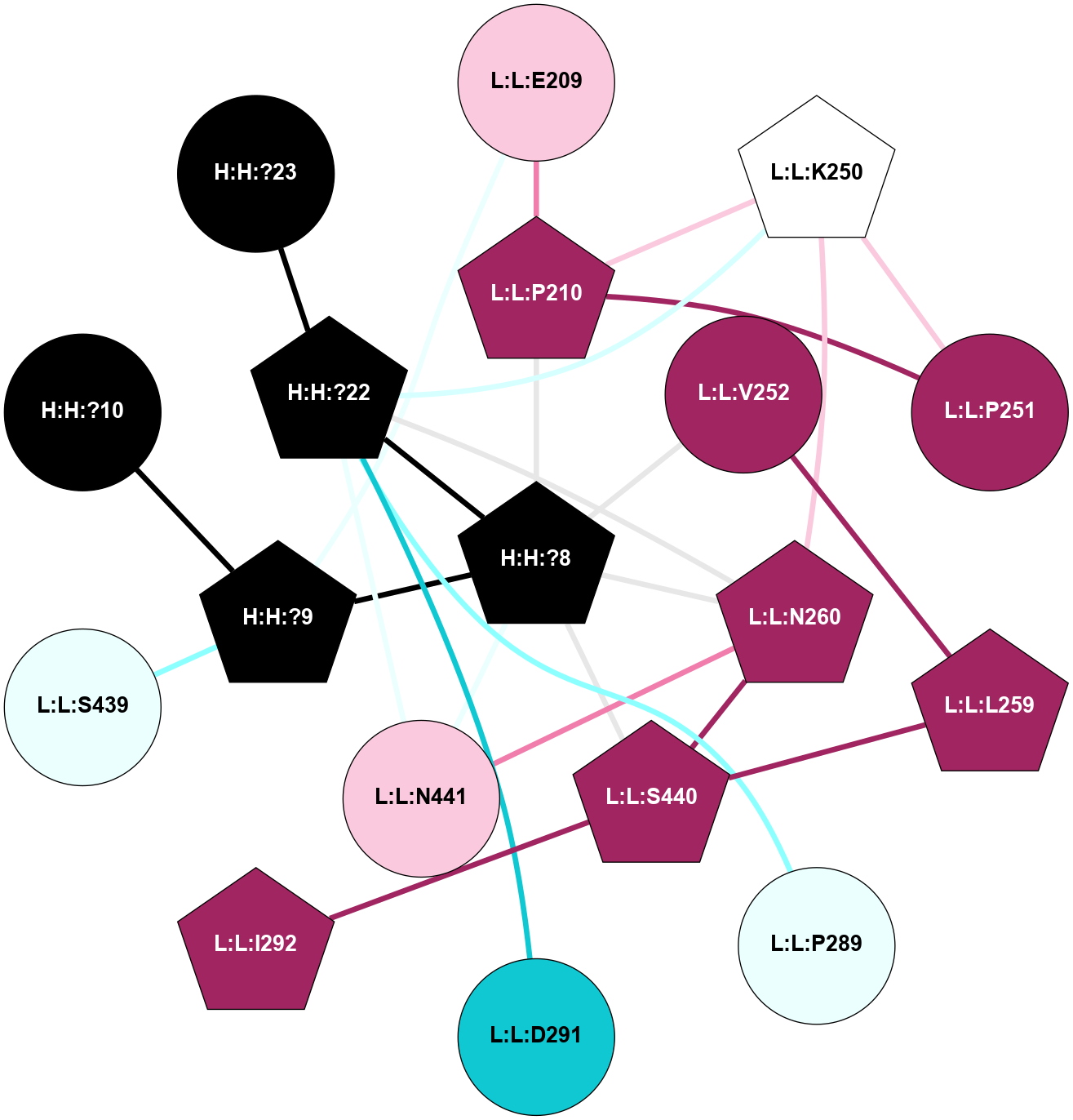
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | H:H:?9 | 31.2 | 3 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?22 | H:H:?8 | 3.34 | 3 | Yes | Yes | 0 | 0 | 1 | 0 | | H:H:?8 | L:L:P210 | 5.89 | 3 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?8 | L:L:V252 | 2.67 | 3 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?8 | L:L:N260 | 24.64 | 3 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?8 | L:L:S440 | 5.39 | 3 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?8 | L:L:N441 | 2.46 | 3 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?10 | H:H:?9 | 32.47 | 0 | No | Yes | 0 | 0 | 2 | 1 | | H:H:?9 | L:L:E209 | 10.7 | 0 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?9 | L:L:S439 | 8.08 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?22 | H:H:?23 | 33.43 | 3 | Yes | No | 0 | 0 | 1 | 2 | | H:H:?22 | L:L:K250 | 2.53 | 3 | Yes | Yes | 0 | 5 | 1 | 2 | | H:H:?22 | L:L:N260 | 3.7 | 3 | Yes | Yes | 0 | 9 | 1 | 1 | | H:H:?22 | L:L:P289 | 5.89 | 3 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?22 | L:L:D291 | 4.87 | 3 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?22 | L:L:N441 | 30.8 | 3 | Yes | No | 0 | 7 | 1 | 1 | | L:L:E209 | L:L:P210 | 3.14 | 0 | No | Yes | 7 | 9 | 2 | 1 | | L:L:K250 | L:L:P210 | 3.35 | 3 | Yes | Yes | 5 | 9 | 2 | 1 | | L:L:P210 | L:L:P251 | 3.9 | 3 | Yes | No | 9 | 9 | 1 | 2 | | L:L:P210 | L:L:V252 | 1.77 | 3 | Yes | No | 9 | 9 | 1 | 1 | | L:L:K250 | L:L:P251 | 3.35 | 3 | Yes | No | 5 | 9 | 2 | 2 | | L:L:K250 | L:L:N260 | 8.39 | 3 | Yes | Yes | 5 | 9 | 2 | 1 | | L:L:L259 | L:L:V252 | 5.96 | 0 | Yes | No | 9 | 9 | 2 | 1 | | L:L:L259 | L:L:S440 | 7.51 | 0 | Yes | Yes | 9 | 9 | 2 | 1 | | L:L:N260 | L:L:S440 | 4.47 | 3 | Yes | Yes | 9 | 9 | 1 | 1 | | L:L:N260 | L:L:N441 | 5.45 | 3 | Yes | No | 9 | 7 | 1 | 1 | | L:L:I292 | L:L:S440 | 3.1 | 3 | Yes | Yes | 9 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 10.80 | | Average Nodes In Shell | 18.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 27.00 | | Average Links Mediated by Hubs In Shell | 27.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
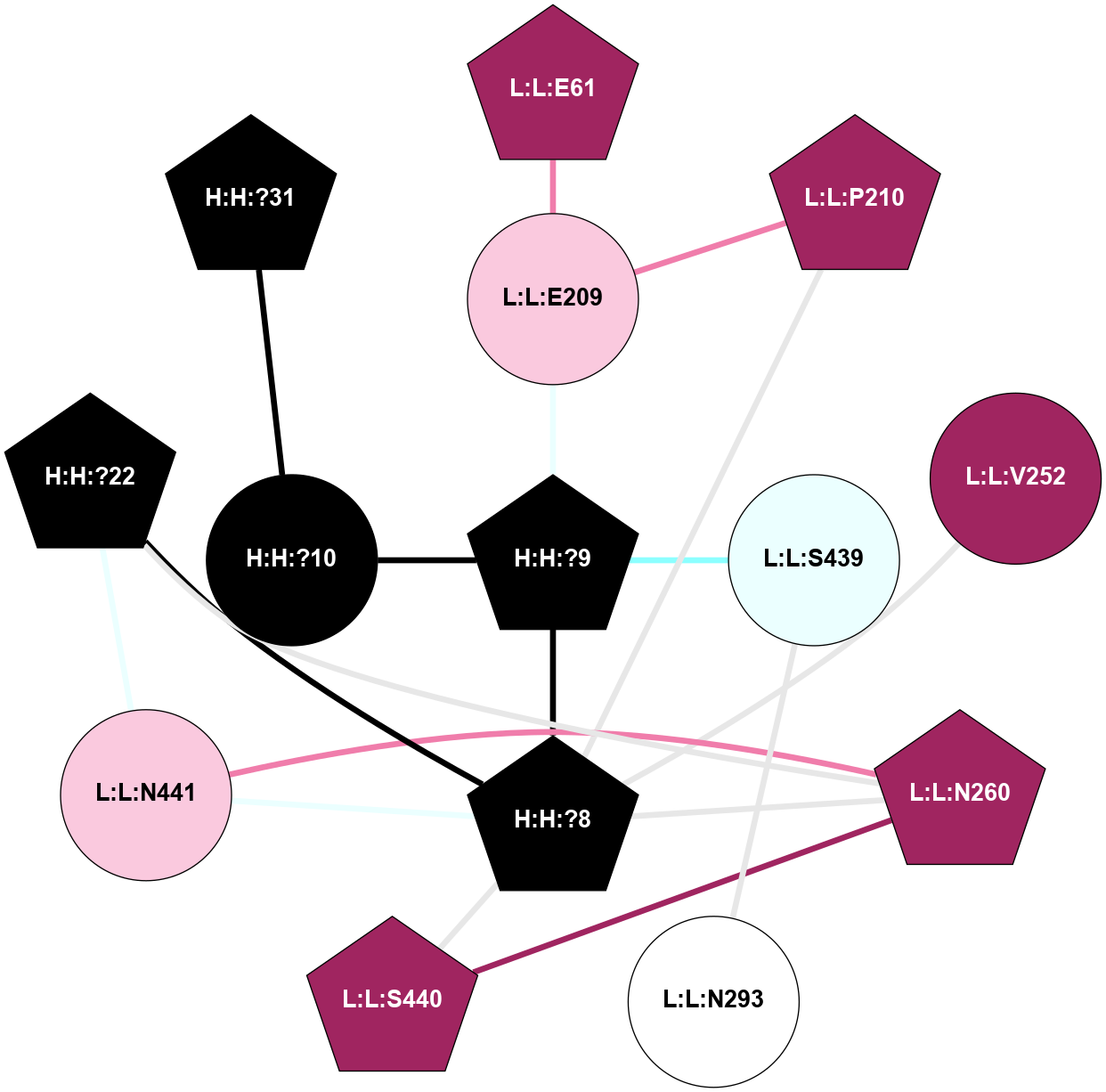
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | H:H:?9 | 31.2 | 3 | Yes | Yes | 0 | 0 | 1 | 0 | | H:H:?22 | H:H:?8 | 3.34 | 3 | Yes | Yes | 0 | 0 | 2 | 1 | | H:H:?8 | L:L:P210 | 5.89 | 3 | Yes | Yes | 0 | 9 | 1 | 2 | | H:H:?8 | L:L:V252 | 2.67 | 3 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?8 | L:L:N260 | 24.64 | 3 | Yes | Yes | 0 | 9 | 1 | 2 | | H:H:?8 | L:L:S440 | 5.39 | 3 | Yes | Yes | 0 | 9 | 1 | 2 | | H:H:?8 | L:L:N441 | 2.46 | 3 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?10 | H:H:?9 | 32.47 | 0 | No | Yes | 0 | 0 | 1 | 0 | | H:H:?9 | L:L:E209 | 10.7 | 0 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?9 | L:L:S439 | 8.08 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?10 | H:H:?31 | 38.83 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?22 | L:L:N260 | 3.7 | 3 | Yes | Yes | 0 | 9 | 2 | 2 | | H:H:?22 | L:L:N441 | 30.8 | 3 | Yes | No | 0 | 7 | 2 | 2 | | L:L:E209 | L:L:E61 | 3.81 | 0 | No | Yes | 7 | 9 | 1 | 2 | | L:L:E209 | L:L:P210 | 3.14 | 0 | No | Yes | 7 | 9 | 1 | 2 | | L:L:P210 | L:L:V252 | 1.77 | 3 | Yes | No | 9 | 9 | 2 | 2 | | L:L:N260 | L:L:S440 | 4.47 | 3 | Yes | Yes | 9 | 9 | 2 | 2 | | L:L:N260 | L:L:N441 | 5.45 | 3 | Yes | No | 9 | 7 | 2 | 2 | | L:L:N293 | L:L:S439 | 2.98 | 0 | No | No | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 20.61 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 19.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
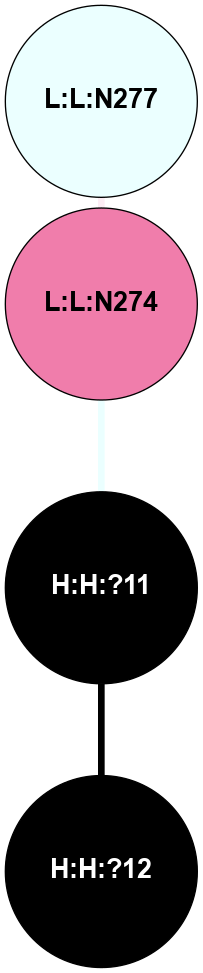
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | H:H:?12 | 40.12 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?11 | L:L:N274 | 33.27 | 0 | No | No | 0 | 8 | 0 | 1 | | L:L:N274 | L:L:N277 | 6.81 | 0 | No | No | 8 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 36.70 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | H:H:?12 | 40.12 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?11 | L:L:N274 | 33.27 | 0 | No | No | 0 | 8 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 40.12 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
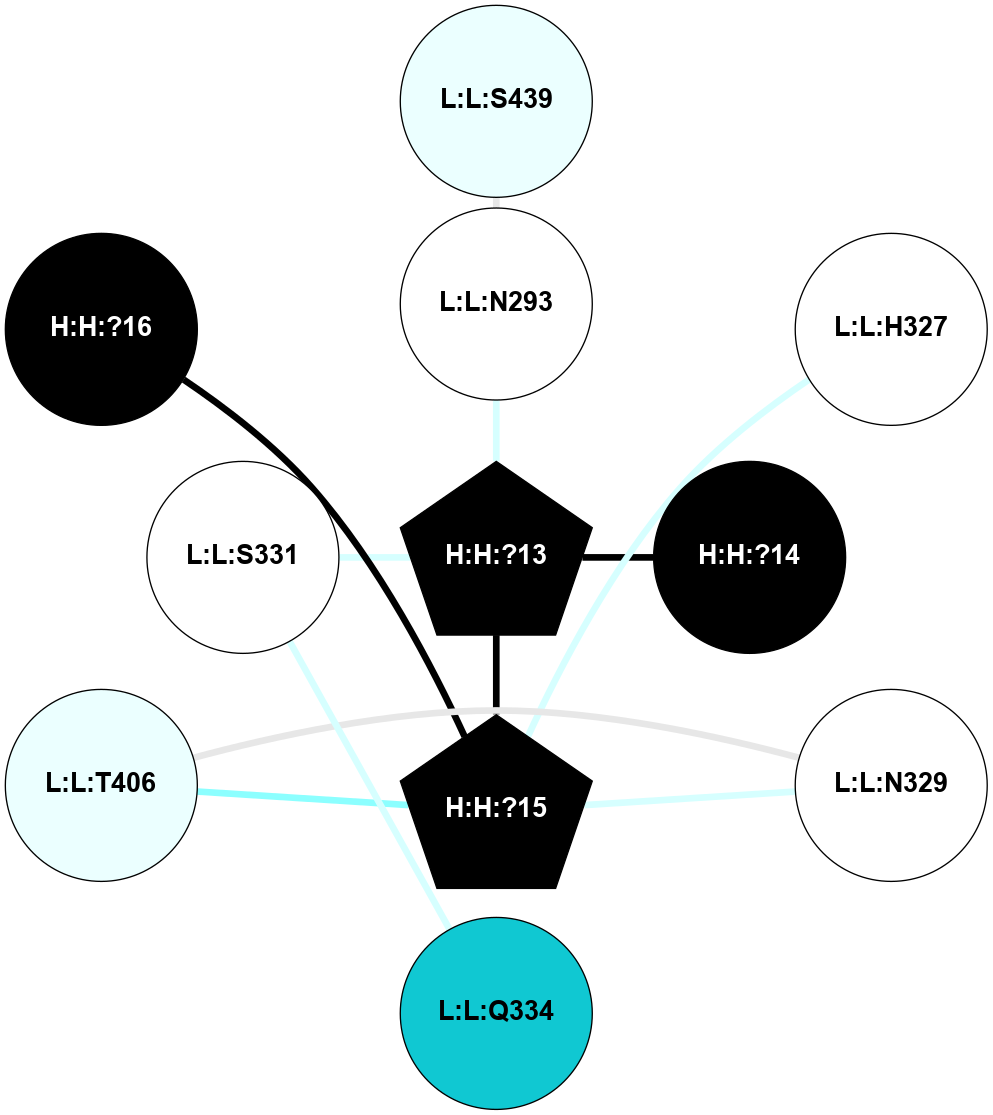
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | H:H:?14 | 35.66 | 0 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?13 | H:H:?15 | 4.46 | 0 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?13 | L:L:N293 | 25.87 | 0 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?15 | H:H:?16 | 34.55 | 15 | Yes | No | 0 | 0 | 1 | 2 | | H:H:?15 | L:L:H327 | 2.31 | 15 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?15 | L:L:N329 | 28.34 | 15 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?15 | L:L:T406 | 7.94 | 15 | Yes | No | 0 | 4 | 1 | 2 | | L:L:N293 | L:L:S439 | 2.98 | 0 | No | No | 5 | 4 | 1 | 2 | | L:L:N329 | L:L:T406 | 4.39 | 15 | No | No | 5 | 4 | 2 | 2 | | L:L:Q334 | L:L:S331 | 5.78 | 0 | No | No | 1 | 5 | 2 | 1 | | H:H:?13 | L:L:S331 | 1.35 | 0 | Yes | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 16.83 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
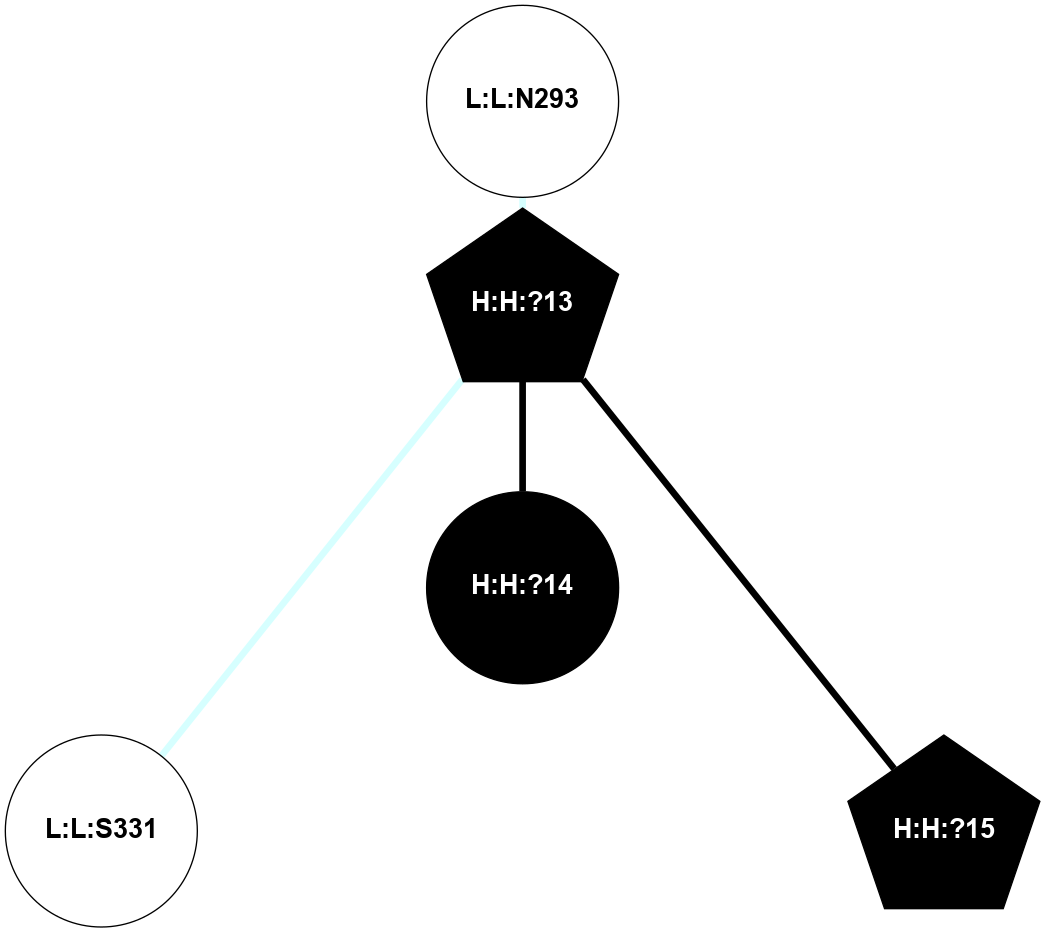
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | H:H:?14 | 35.66 | 0 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?13 | H:H:?15 | 4.46 | 0 | Yes | Yes | 0 | 0 | 1 | 2 | | H:H:?13 | L:L:N293 | 25.87 | 0 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?13 | L:L:S331 | 1.35 | 0 | Yes | No | 0 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 35.66 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
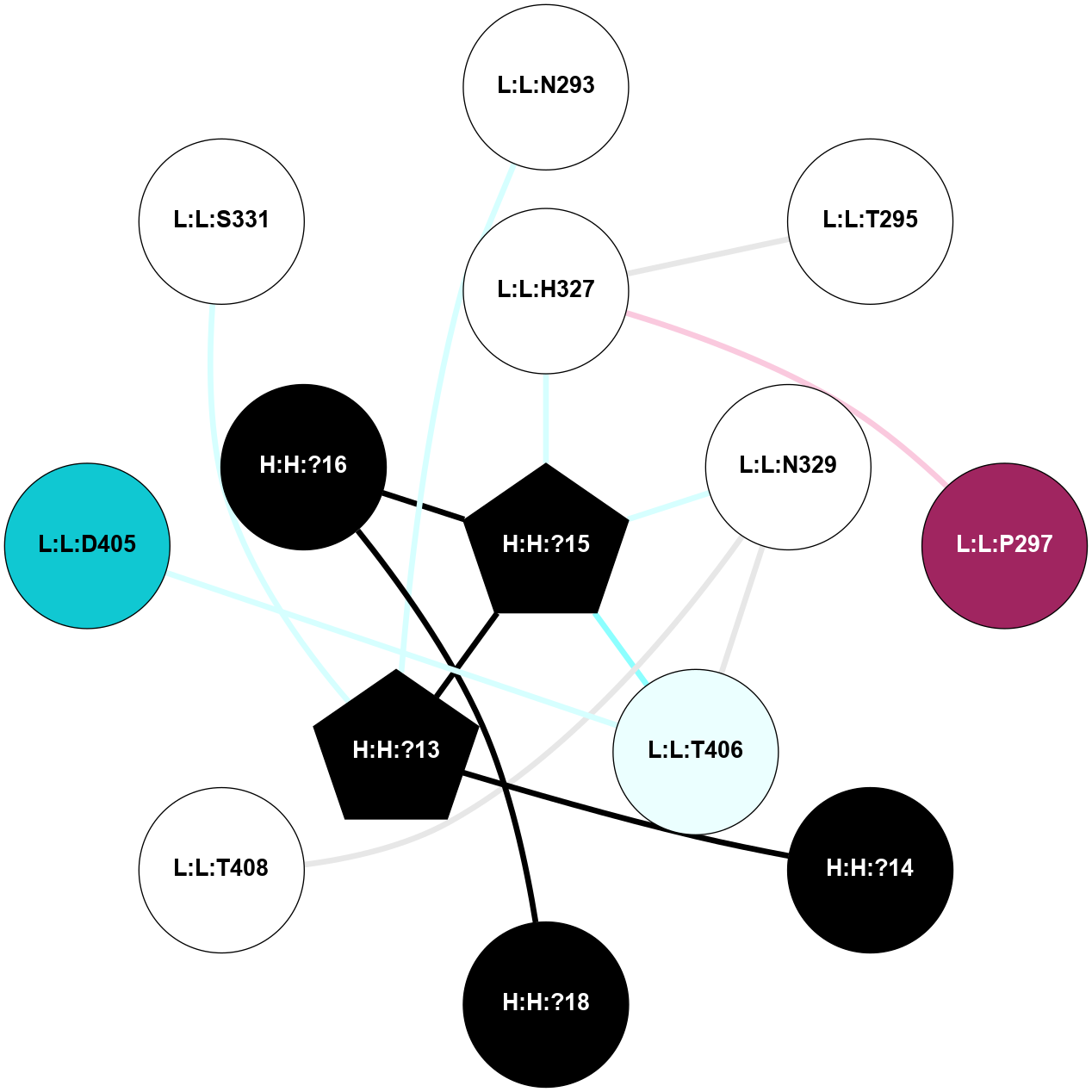
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | H:H:?14 | 35.66 | 0 | Yes | No | 0 | 0 | 1 | 2 | | H:H:?13 | H:H:?15 | 4.46 | 0 | Yes | Yes | 0 | 0 | 1 | 0 | | H:H:?13 | L:L:N293 | 25.87 | 0 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?15 | H:H:?16 | 34.55 | 15 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?15 | L:L:H327 | 2.31 | 15 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?15 | L:L:N329 | 28.34 | 15 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?15 | L:L:T406 | 7.94 | 15 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?16 | H:H:?18 | 4.46 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:H327 | L:L:T295 | 2.74 | 0 | No | No | 5 | 5 | 1 | 2 | | L:L:H327 | L:L:P297 | 6.1 | 0 | No | No | 5 | 9 | 1 | 2 | | L:L:N329 | L:L:T406 | 4.39 | 15 | No | No | 5 | 4 | 1 | 1 | | L:L:N329 | L:L:T408 | 1.46 | 15 | No | No | 5 | 5 | 1 | 2 | | L:L:D405 | L:L:T406 | 1.45 | 0 | No | No | 1 | 4 | 2 | 1 | | H:H:?13 | L:L:S331 | 1.35 | 0 | Yes | No | 0 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.52 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
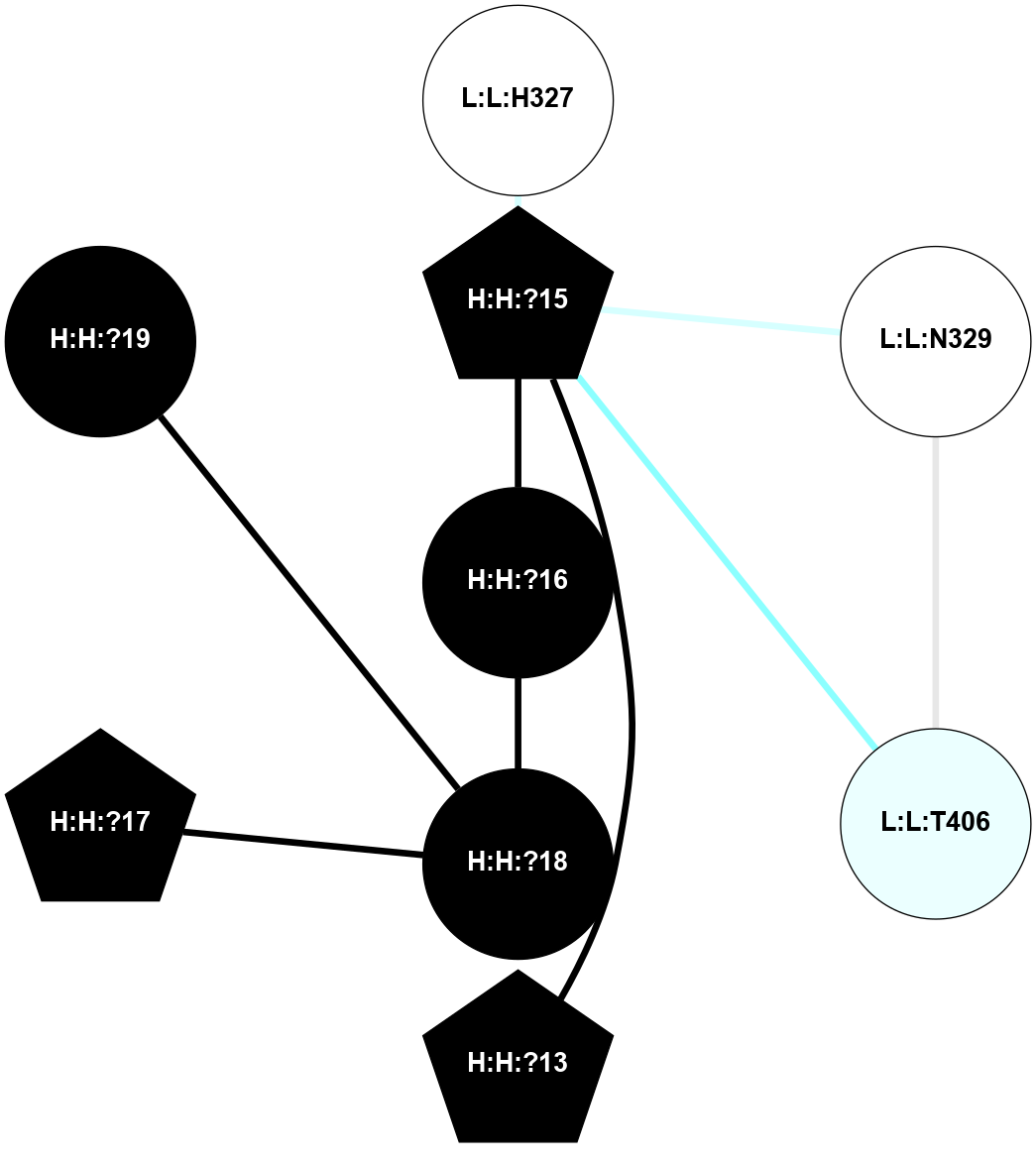
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | H:H:?15 | 4.46 | 0 | Yes | Yes | 0 | 0 | 2 | 1 | | H:H:?15 | H:H:?16 | 34.55 | 15 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?15 | L:L:H327 | 2.31 | 15 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?15 | L:L:N329 | 28.34 | 15 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?15 | L:L:T406 | 7.94 | 15 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?16 | H:H:?18 | 4.46 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?17 | H:H:?18 | 32.32 | 16 | Yes | No | 0 | 0 | 2 | 1 | | H:H:?18 | H:H:?19 | 29.87 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:N329 | L:L:T406 | 4.39 | 15 | No | No | 5 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.50 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
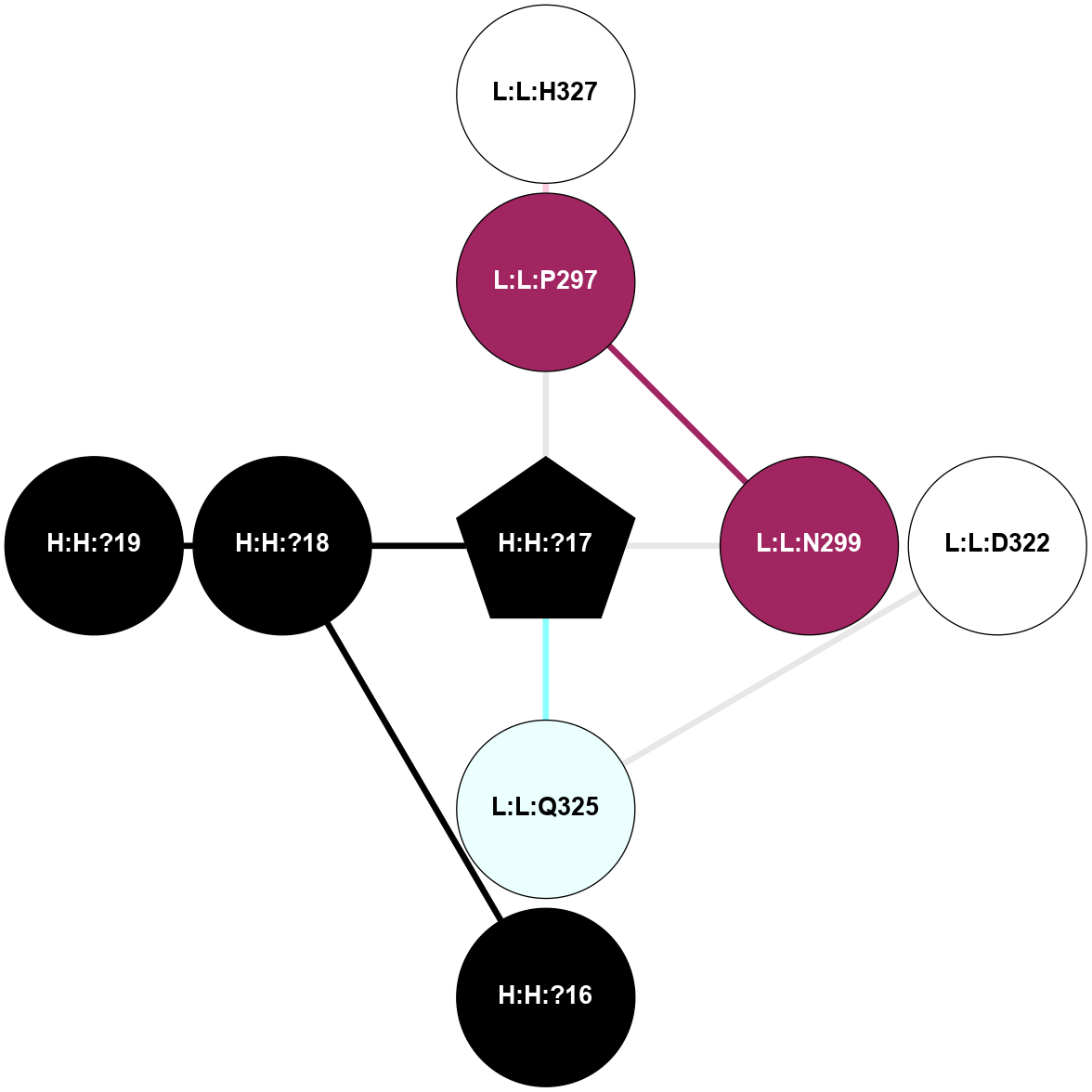
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?16 | H:H:?18 | 4.46 | 0 | No | No | 0 | 0 | 2 | 1 | | H:H:?17 | H:H:?18 | 32.32 | 16 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?17 | L:L:P297 | 14.73 | 16 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?17 | L:L:N299 | 23.41 | 16 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?17 | L:L:Q325 | 2.39 | 16 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?18 | H:H:?19 | 29.87 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:N299 | L:L:P297 | 4.89 | 16 | No | No | 9 | 9 | 1 | 1 | | L:L:H327 | L:L:P297 | 6.1 | 0 | No | No | 5 | 9 | 2 | 1 | | L:L:D322 | L:L:Q325 | 5.22 | 0 | No | No | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.21 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
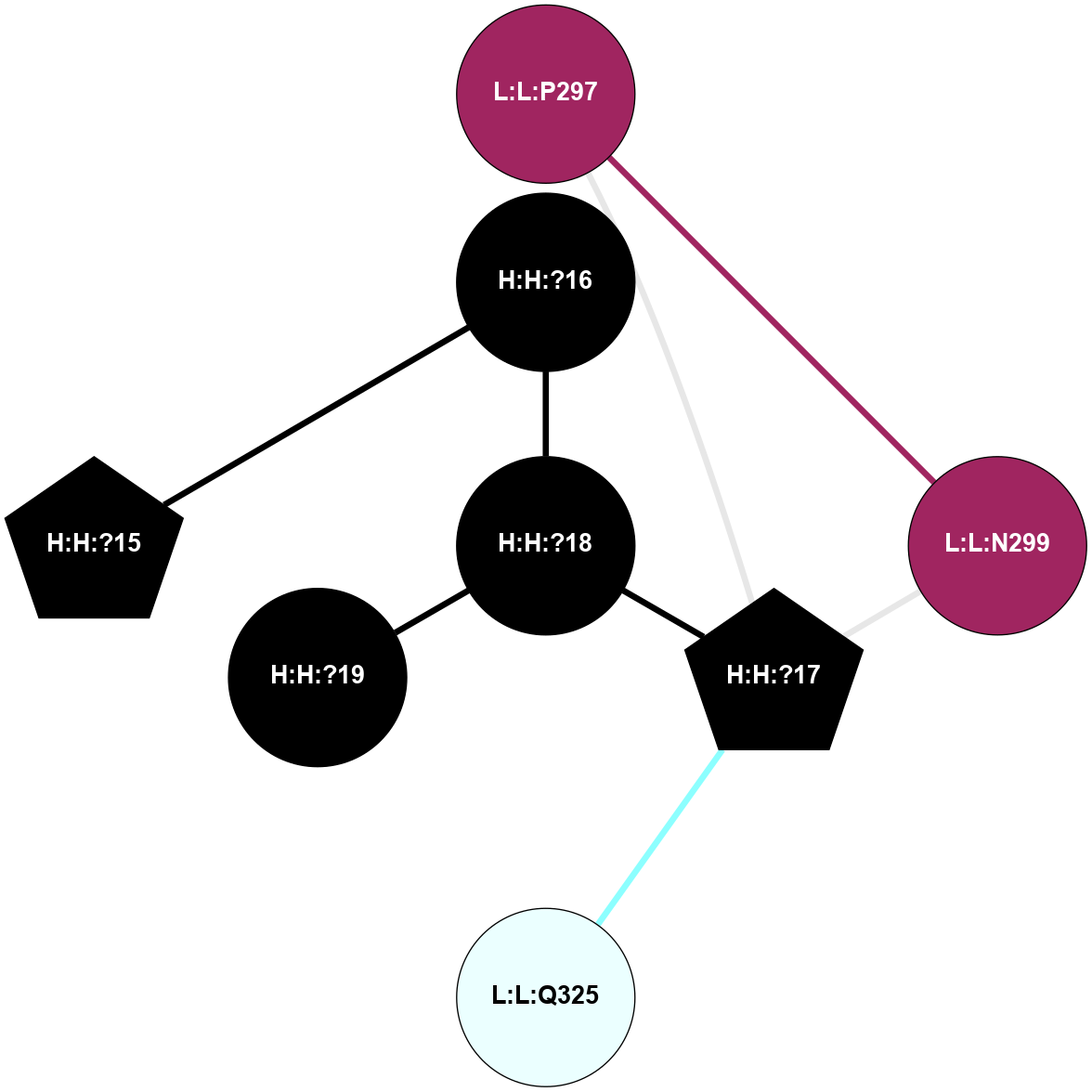
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?15 | H:H:?16 | 34.55 | 15 | Yes | No | 0 | 0 | 2 | 1 | | H:H:?16 | H:H:?18 | 4.46 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?17 | H:H:?18 | 32.32 | 16 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?17 | L:L:P297 | 14.73 | 16 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?17 | L:L:N299 | 23.41 | 16 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?17 | L:L:Q325 | 2.39 | 16 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?18 | H:H:?19 | 29.87 | 0 | No | No | 0 | 0 | 0 | 1 | | L:L:N299 | L:L:P297 | 4.89 | 16 | No | No | 9 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 22.22 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
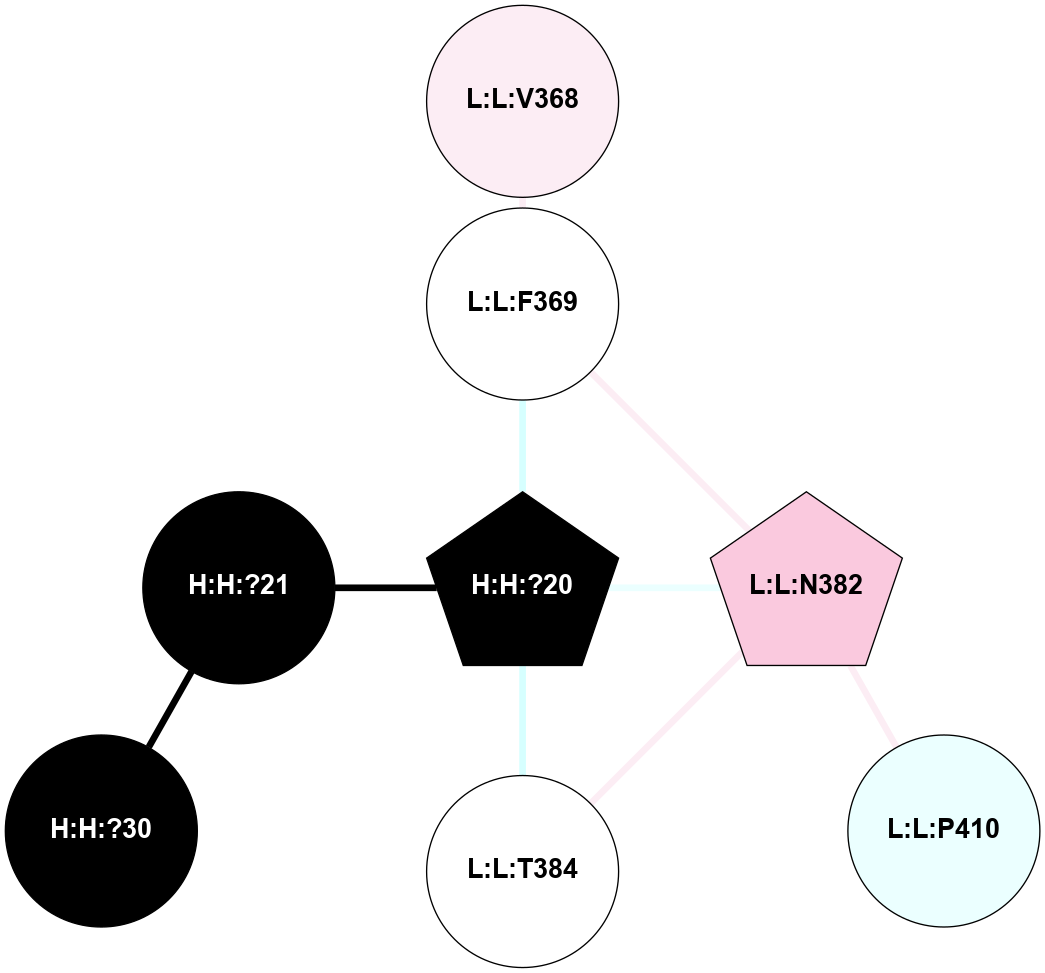
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?20 | H:H:?21 | 35.66 | 9 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?20 | L:L:F369 | 12.02 | 9 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?20 | L:L:N382 | 25.87 | 9 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?20 | L:L:T384 | 14.55 | 9 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?21 | H:H:?30 | 8.92 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:F369 | L:L:V368 | 3.93 | 9 | No | Yes | 5 | 6 | 1 | 2 | | L:L:F369 | L:L:N382 | 10.87 | 9 | No | Yes | 5 | 7 | 1 | 1 | | L:L:N382 | L:L:T384 | 7.31 | 9 | Yes | No | 7 | 5 | 1 | 1 | | L:L:N382 | L:L:P410 | 6.52 | 9 | Yes | No | 7 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 22.02 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
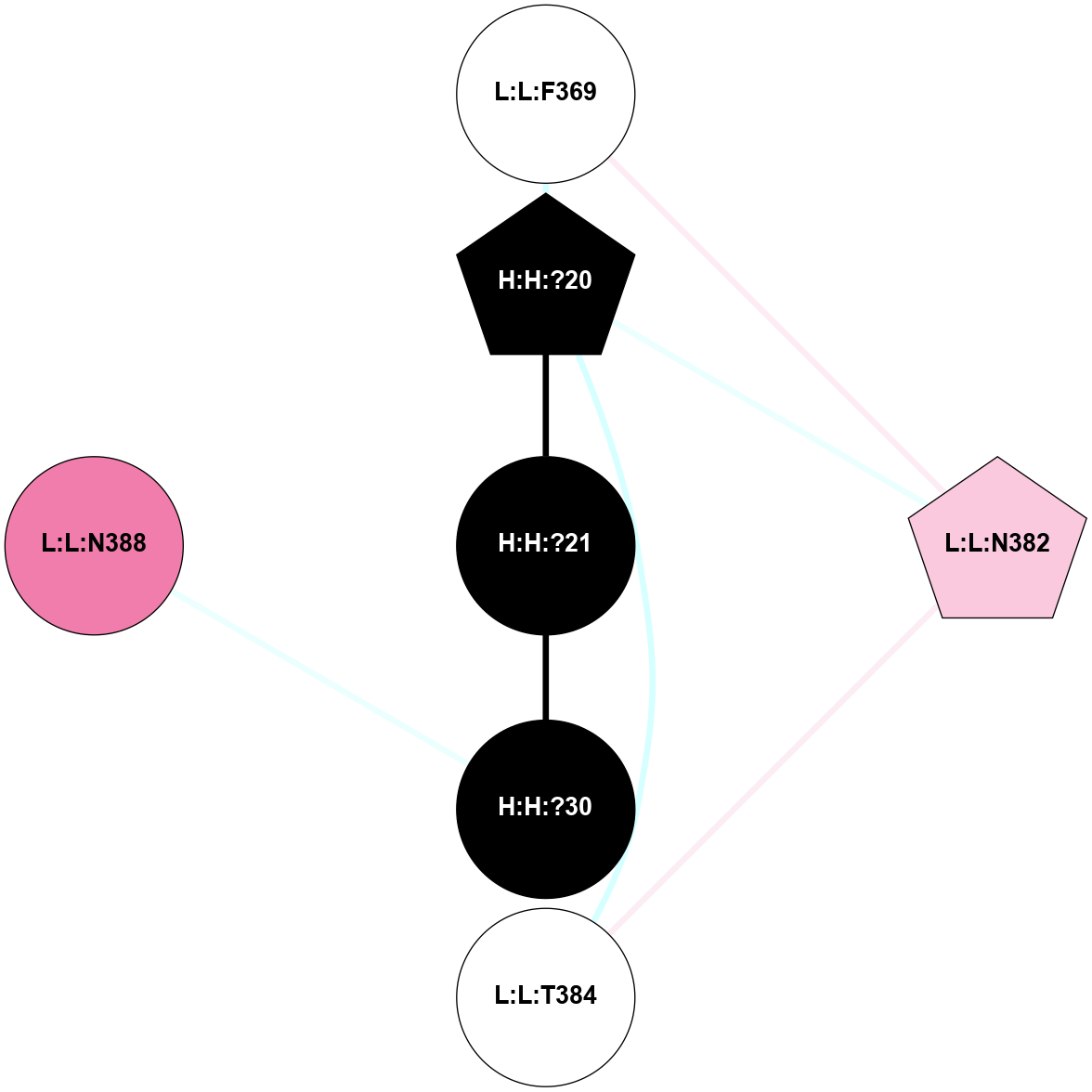
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?20 | H:H:?21 | 35.66 | 9 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?20 | L:L:F369 | 12.02 | 9 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?20 | L:L:N382 | 25.87 | 9 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?20 | L:L:T384 | 14.55 | 9 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?21 | H:H:?30 | 8.92 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?30 | L:L:N388 | 23.41 | 0 | No | No | 0 | 8 | 1 | 2 | | L:L:F369 | L:L:N382 | 10.87 | 9 | No | Yes | 5 | 7 | 2 | 2 | | L:L:N382 | L:L:T384 | 7.31 | 9 | Yes | No | 7 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 22.29 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
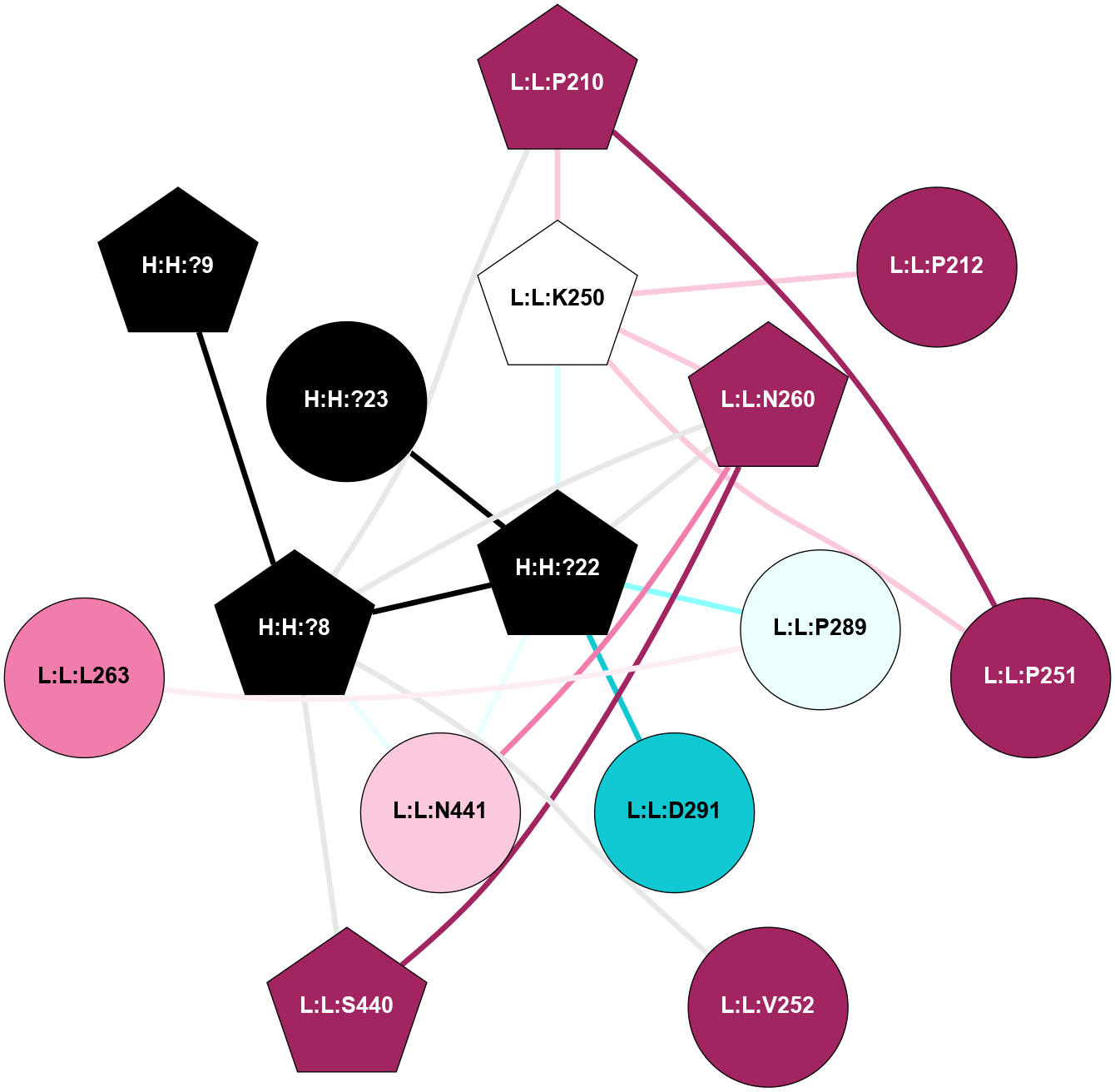
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | H:H:?9 | 31.2 | 3 | Yes | Yes | 0 | 0 | 1 | 2 | | H:H:?22 | H:H:?8 | 3.34 | 3 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?8 | L:L:P210 | 5.89 | 3 | Yes | Yes | 0 | 9 | 1 | 2 | | H:H:?8 | L:L:V252 | 2.67 | 3 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?8 | L:L:N260 | 24.64 | 3 | Yes | Yes | 0 | 9 | 1 | 1 | | H:H:?8 | L:L:S440 | 5.39 | 3 | Yes | Yes | 0 | 9 | 1 | 2 | | H:H:?8 | L:L:N441 | 2.46 | 3 | Yes | No | 0 | 7 | 1 | 1 | | H:H:?22 | H:H:?23 | 33.43 | 3 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?22 | L:L:K250 | 2.53 | 3 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?22 | L:L:N260 | 3.7 | 3 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?22 | L:L:P289 | 5.89 | 3 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?22 | L:L:D291 | 4.87 | 3 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?22 | L:L:N441 | 30.8 | 3 | Yes | No | 0 | 7 | 0 | 1 | | L:L:K250 | L:L:P210 | 3.35 | 3 | Yes | Yes | 5 | 9 | 1 | 2 | | L:L:P210 | L:L:P251 | 3.9 | 3 | Yes | No | 9 | 9 | 2 | 2 | | L:L:P210 | L:L:V252 | 1.77 | 3 | Yes | No | 9 | 9 | 2 | 2 | | L:L:K250 | L:L:P212 | 5.02 | 3 | Yes | No | 5 | 9 | 1 | 2 | | L:L:K250 | L:L:P251 | 3.35 | 3 | Yes | No | 5 | 9 | 1 | 2 | | L:L:K250 | L:L:N260 | 8.39 | 3 | Yes | Yes | 5 | 9 | 1 | 1 | | L:L:N260 | L:L:S440 | 4.47 | 3 | Yes | Yes | 9 | 9 | 1 | 2 | | L:L:N260 | L:L:N441 | 5.45 | 3 | Yes | No | 9 | 7 | 1 | 1 | | L:L:L263 | L:L:P289 | 6.57 | 0 | No | No | 8 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 12.08 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 22.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
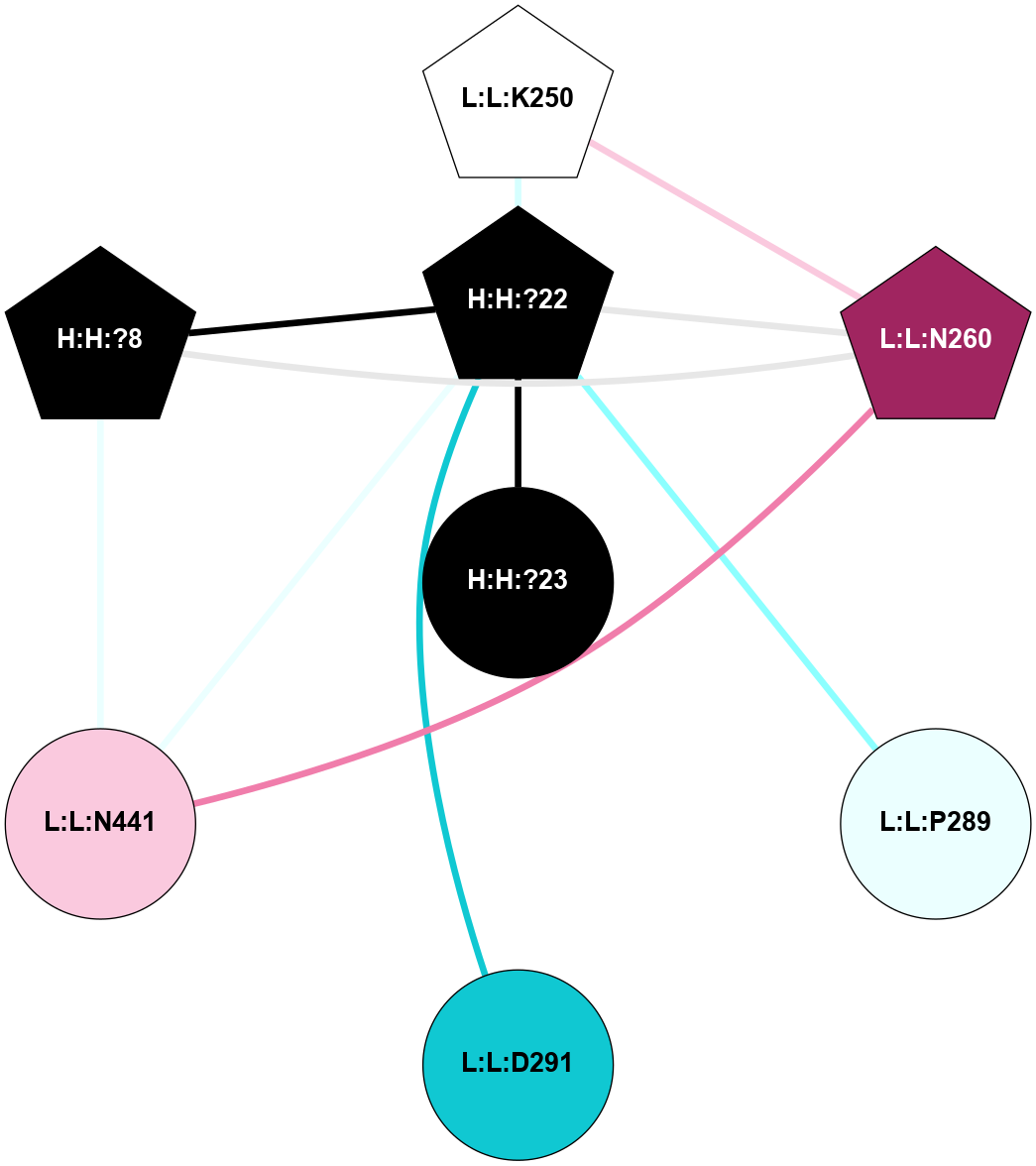
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?22 | H:H:?8 | 3.34 | 3 | Yes | Yes | 0 | 0 | 1 | 2 | | H:H:?8 | L:L:N260 | 24.64 | 3 | Yes | Yes | 0 | 9 | 2 | 2 | | H:H:?8 | L:L:N441 | 2.46 | 3 | Yes | No | 0 | 7 | 2 | 2 | | H:H:?22 | H:H:?23 | 33.43 | 3 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?22 | L:L:K250 | 2.53 | 3 | Yes | Yes | 0 | 5 | 1 | 2 | | H:H:?22 | L:L:N260 | 3.7 | 3 | Yes | Yes | 0 | 9 | 1 | 2 | | H:H:?22 | L:L:P289 | 5.89 | 3 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?22 | L:L:D291 | 4.87 | 3 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?22 | L:L:N441 | 30.8 | 3 | Yes | No | 0 | 7 | 1 | 2 | | L:L:K250 | L:L:N260 | 8.39 | 3 | Yes | Yes | 5 | 9 | 2 | 2 | | L:L:N260 | L:L:N441 | 5.45 | 3 | Yes | No | 9 | 7 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 33.43 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
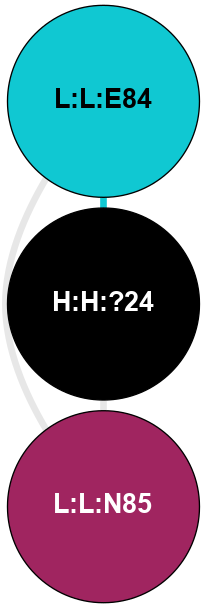
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?24 | L:L:E84 | 22.59 | 17 | No | No | 0 | 1 | 0 | 1 | | H:H:?24 | L:L:N85 | 24.64 | 17 | No | No | 0 | 9 | 0 | 1 | | L:L:E84 | L:L:N85 | 13.15 | 17 | No | No | 1 | 9 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 23.62 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
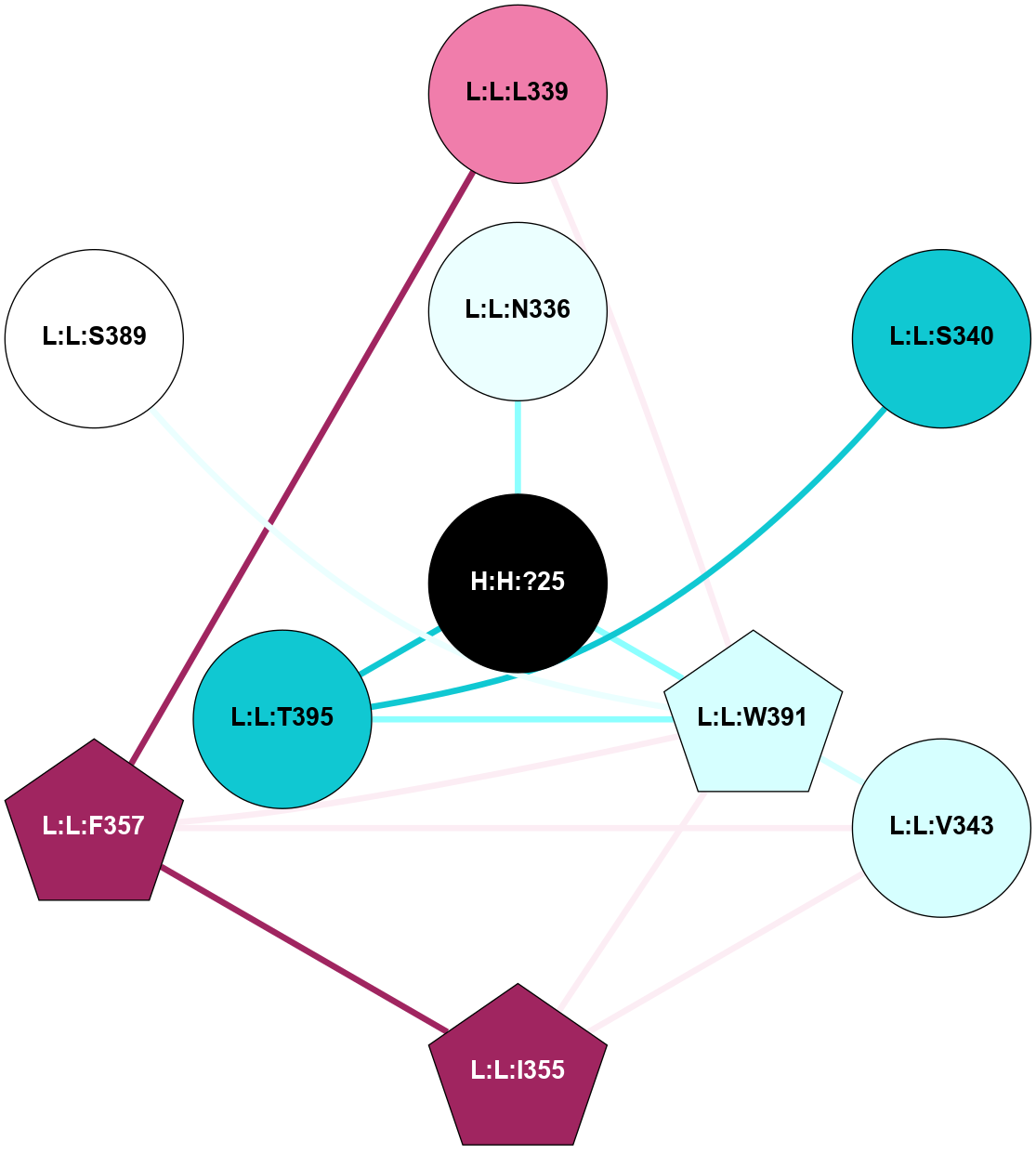
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?25 | L:L:N336 | 28.34 | 4 | No | No | 0 | 4 | 0 | 1 | | H:H:?25 | L:L:W391 | 5.11 | 4 | No | Yes | 0 | 3 | 0 | 1 | | H:H:?25 | L:L:T395 | 6.61 | 4 | No | No | 0 | 1 | 0 | 1 | | L:L:F357 | L:L:L339 | 2.44 | 4 | Yes | No | 9 | 8 | 2 | 2 | | L:L:L339 | L:L:W391 | 4.56 | 4 | No | Yes | 8 | 3 | 2 | 1 | | L:L:S340 | L:L:T395 | 3.2 | 0 | No | No | 1 | 1 | 2 | 1 | | L:L:I355 | L:L:V343 | 3.07 | 4 | Yes | No | 9 | 3 | 2 | 2 | | L:L:F357 | L:L:V343 | 2.62 | 4 | Yes | No | 9 | 3 | 2 | 2 | | L:L:V343 | L:L:W391 | 6.13 | 4 | No | Yes | 3 | 3 | 2 | 1 | | L:L:F357 | L:L:I355 | 3.77 | 4 | Yes | Yes | 9 | 9 | 2 | 2 | | L:L:I355 | L:L:W391 | 2.35 | 4 | Yes | Yes | 9 | 3 | 2 | 1 | | L:L:F357 | L:L:W391 | 4.01 | 4 | Yes | Yes | 9 | 3 | 2 | 1 | | L:L:S389 | L:L:W391 | 4.94 | 0 | No | Yes | 5 | 3 | 2 | 1 | | L:L:T395 | L:L:W391 | 4.85 | 4 | No | Yes | 1 | 3 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 13.35 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?26 | L:L:R347 | 2.18 | 18 | No | No | 0 | 3 | 0 | 1 | | H:H:?26 | L:L:N352 | 29.57 | 18 | No | No | 0 | 5 | 0 | 1 | | L:L:N352 | L:L:R347 | 7.23 | 18 | No | No | 5 | 3 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.88 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?27 | L:L:N393 | 24.64 | 19 | No | No | 0 | 1 | 0 | 1 | | H:H:?27 | L:L:N394 | 9.86 | 19 | No | No | 0 | 4 | 0 | 1 | | L:L:N393 | L:L:N394 | 8.17 | 19 | No | No | 1 | 4 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.25 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?28 | L:L:M456 | 7.61 | 0 | No | No | 0 | 1 | 0 | 1 | | H:H:?28 | L:L:N457 | 24.64 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.12 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
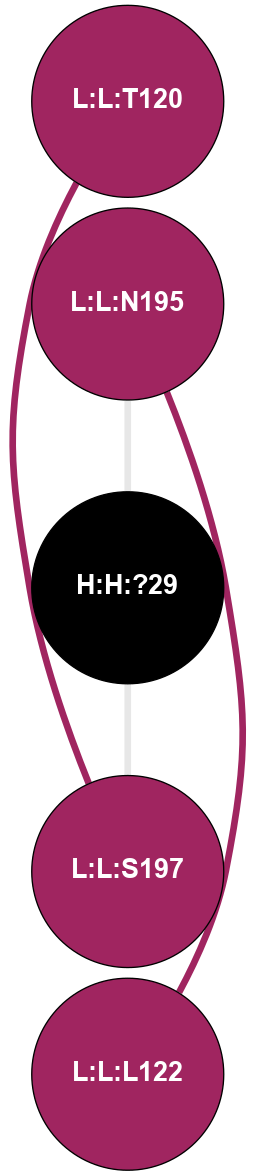
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?29 | L:L:N195 | 27.11 | 0 | No | No | 0 | 9 | 0 | 1 | | L:L:S197 | L:L:T120 | 1.6 | 0 | No | No | 9 | 9 | 1 | 2 | | L:L:L122 | L:L:N195 | 1.37 | 0 | No | No | 9 | 9 | 2 | 1 | | H:H:?29 | L:L:S197 | 1.35 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.23 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
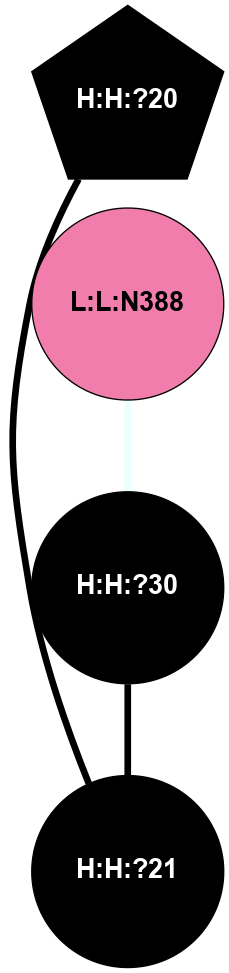
A 2D representation of the interactions of NAG in 6MEO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?20 | H:H:?21 | 35.66 | 9 | Yes | No | 0 | 0 | 2 | 1 | | H:H:?21 | H:H:?30 | 8.92 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?30 | L:L:N388 | 23.41 | 0 | No | No | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.16 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6MET | A | Protein | Chemokine | CCR5 | Homo Sapiens | Envelope Glycoprotein Gp160; T-Cell Surface Glycoprotein Cd4 | - | - | 4.5 | 2018-12-12 | doi.org/10.1038/s41586-018-0804-9 |
|
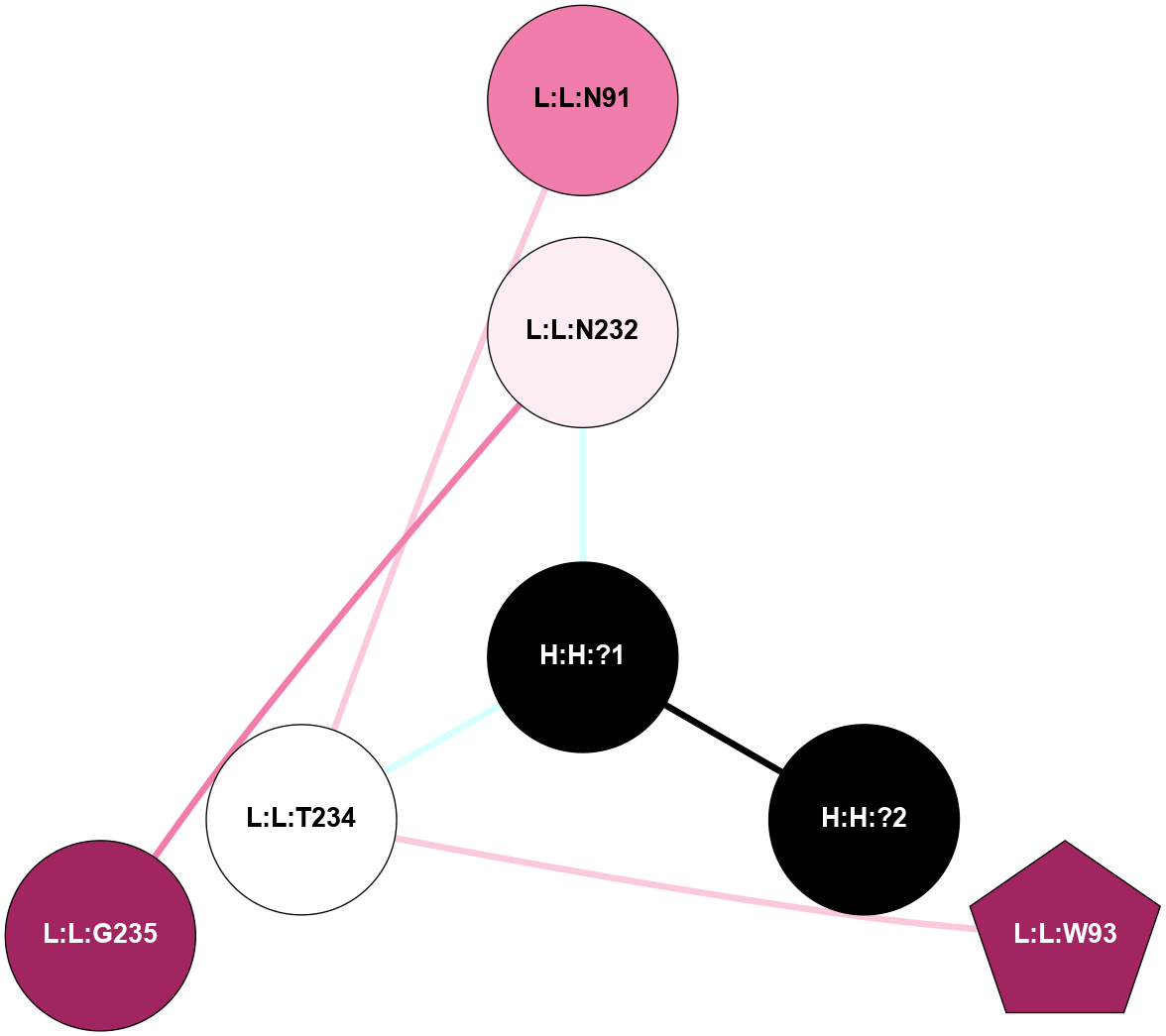
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 35.66 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?1 | L:L:N232 | 25.87 | 0 | No | No | 0 | 6 | 0 | 1 | | L:L:N91 | L:L:T234 | 5.85 | 0 | No | No | 8 | 5 | 2 | 1 | | L:L:T234 | L:L:W93 | 9.7 | 0 | No | Yes | 5 | 9 | 1 | 2 | | L:L:G235 | L:L:N232 | 3.39 | 0 | No | No | 9 | 6 | 2 | 1 | | H:H:?1 | L:L:T234 | 1.32 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.95 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 35.66 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?1 | L:L:N232 | 25.87 | 0 | No | No | 0 | 6 | 1 | 2 | | H:H:?1 | L:L:T234 | 1.32 | 0 | No | No | 0 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 35.66 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
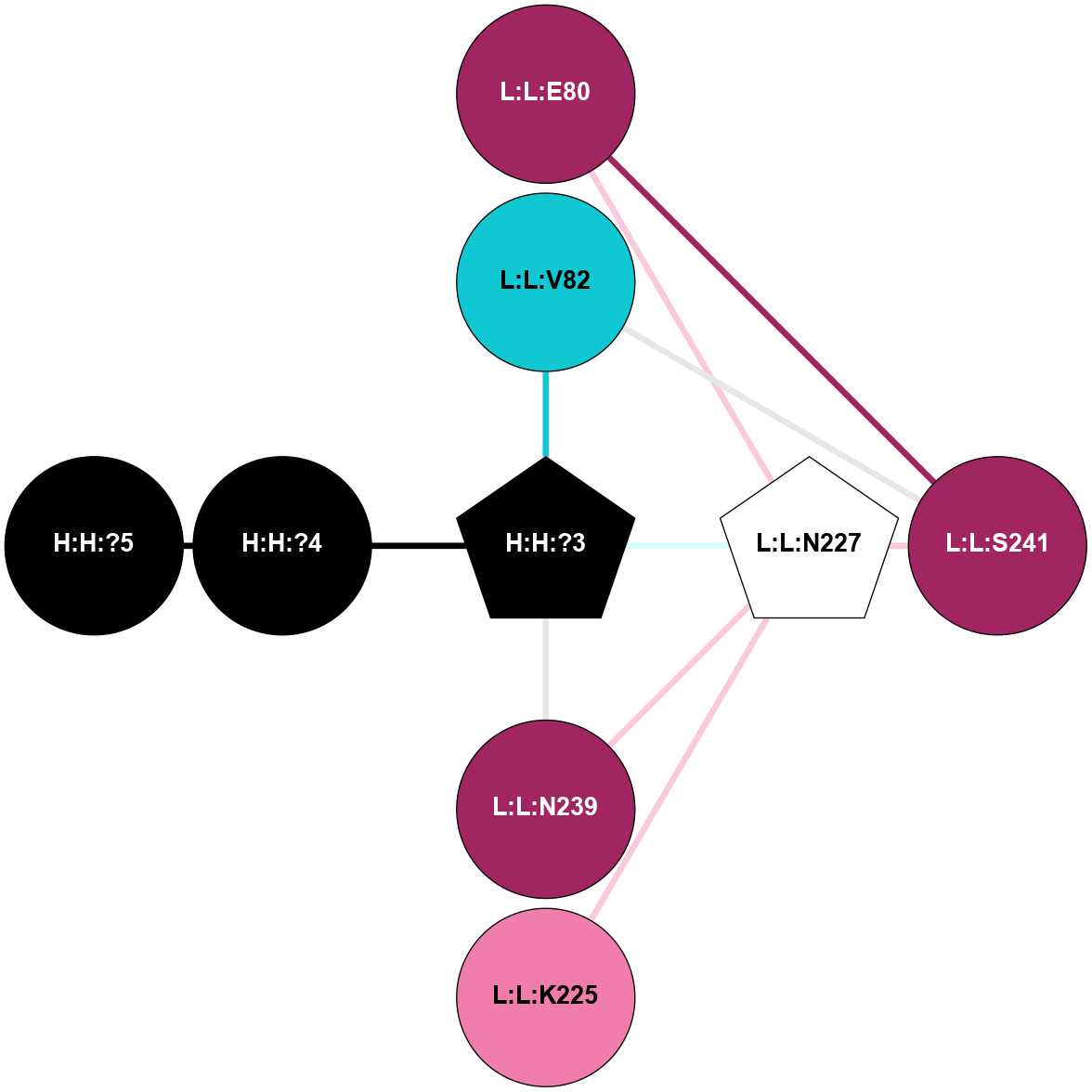
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 36.78 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | L:L:V82 | 4.01 | 1 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?3 | L:L:N227 | 11.09 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?3 | L:L:N239 | 27.11 | 1 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?4 | H:H:?5 | 28.57 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:E80 | L:L:N227 | 3.94 | 1 | No | Yes | 9 | 5 | 2 | 1 | | L:L:E80 | L:L:S241 | 8.62 | 1 | No | No | 9 | 9 | 2 | 2 | | L:L:S241 | L:L:V82 | 3.23 | 1 | No | No | 9 | 1 | 2 | 1 | | L:L:K225 | L:L:N227 | 2.8 | 1 | No | Yes | 8 | 5 | 2 | 1 | | L:L:N227 | L:L:N239 | 12.26 | 1 | Yes | No | 5 | 9 | 1 | 1 | | L:L:N227 | L:L:S241 | 5.96 | 1 | Yes | No | 5 | 9 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.75 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
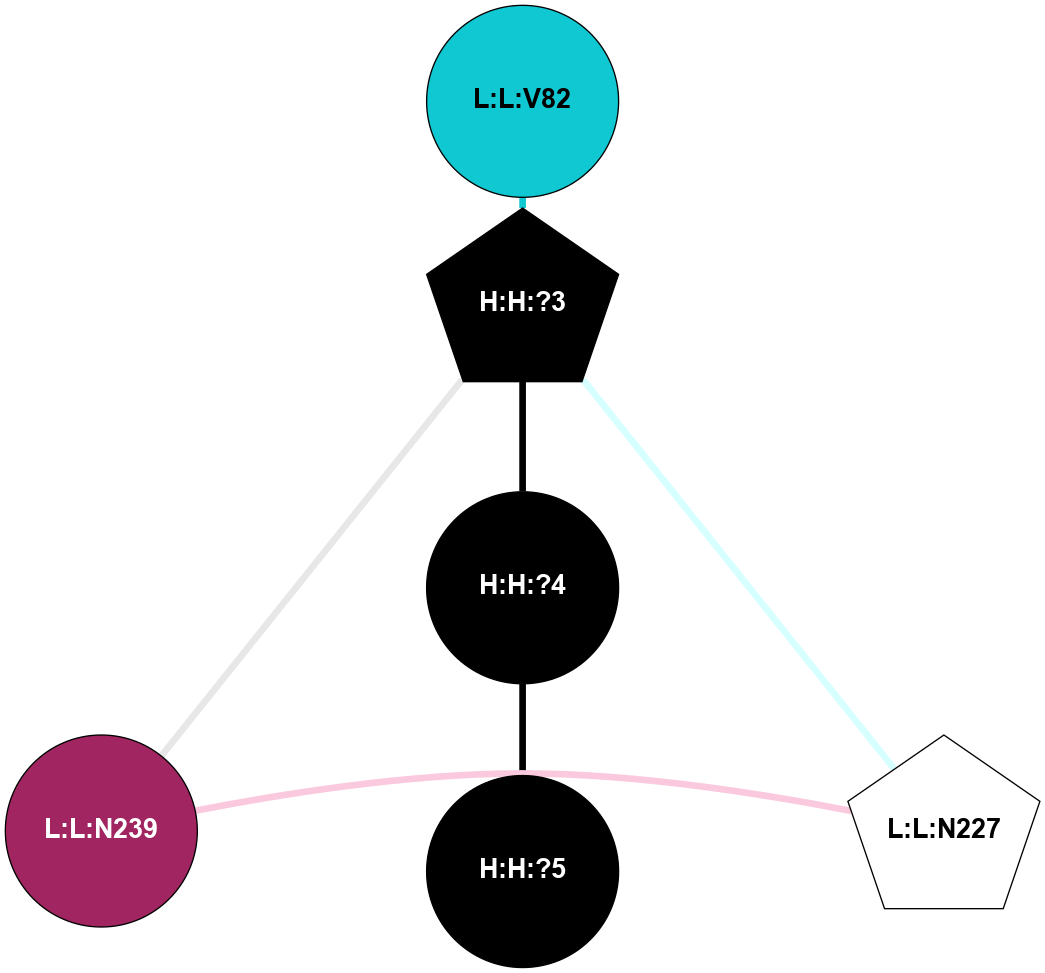
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 36.78 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?3 | L:L:V82 | 4.01 | 1 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?3 | L:L:N227 | 11.09 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | H:H:?3 | L:L:N239 | 27.11 | 1 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?4 | H:H:?5 | 28.57 | 0 | No | No | 0 | 0 | 0 | 1 | | L:L:N227 | L:L:N239 | 12.26 | 1 | Yes | No | 5 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 32.67 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
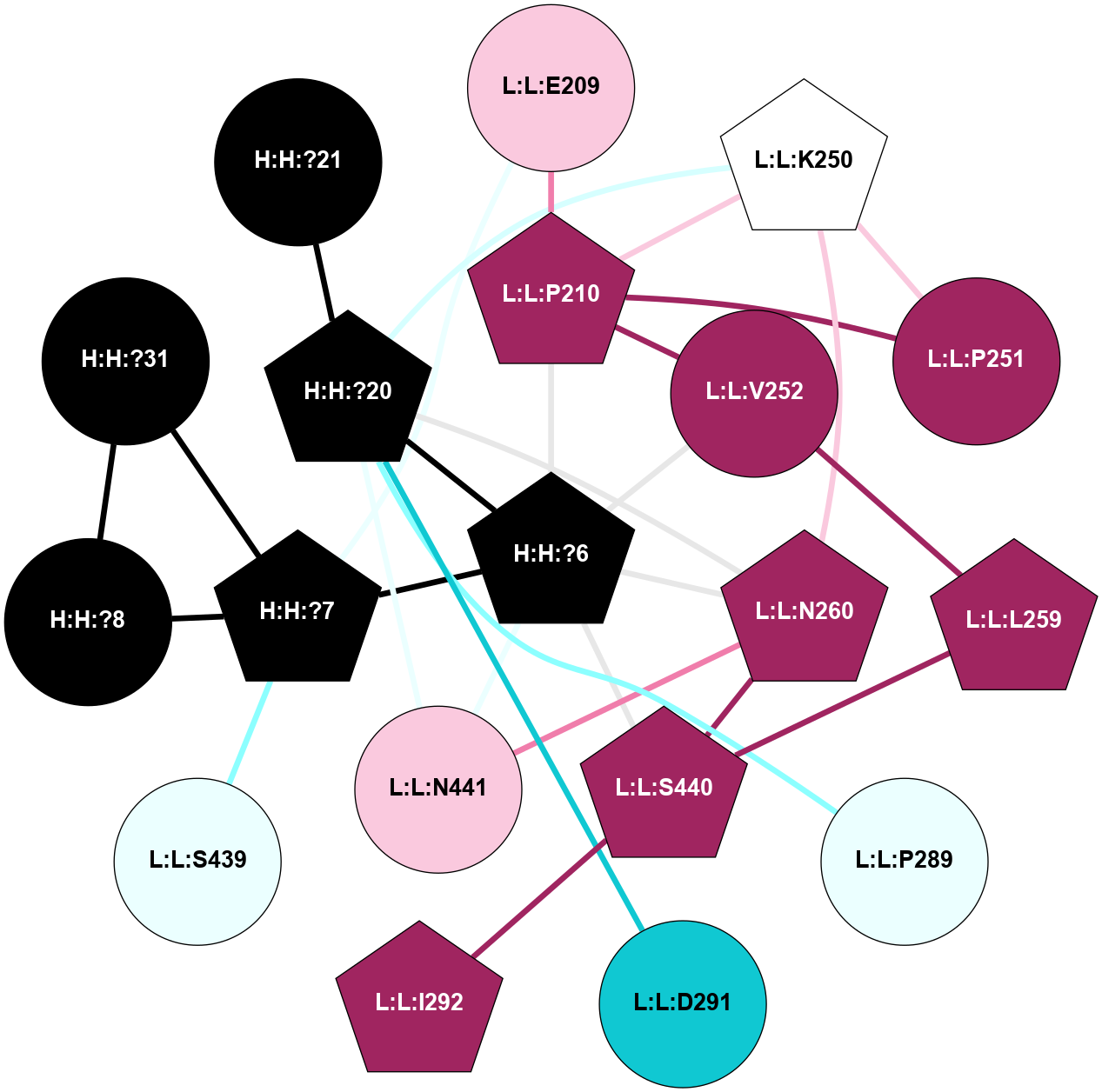
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | H:H:?7 | 31.2 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?20 | H:H:?6 | 3.34 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | H:H:?6 | L:L:P210 | 5.89 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?6 | L:L:V252 | 2.67 | 2 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?6 | L:L:N260 | 24.64 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?6 | L:L:S440 | 5.39 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?6 | L:L:N441 | 2.46 | 2 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?7 | H:H:?8 | 32.47 | 2 | Yes | No | 0 | 0 | 1 | 2 | | H:H:?31 | H:H:?7 | 3.84 | 2 | No | Yes | 0 | 0 | 2 | 1 | | H:H:?7 | L:L:E209 | 10.7 | 2 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?7 | L:L:S439 | 8.08 | 2 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?31 | H:H:?8 | 22.4 | 2 | No | No | 0 | 0 | 2 | 2 | | H:H:?20 | H:H:?21 | 33.43 | 2 | Yes | No | 0 | 0 | 1 | 2 | | H:H:?20 | L:L:K250 | 2.53 | 2 | Yes | Yes | 0 | 5 | 1 | 2 | | H:H:?20 | L:L:N260 | 3.7 | 2 | Yes | Yes | 0 | 9 | 1 | 1 | | H:H:?20 | L:L:P289 | 5.89 | 2 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?20 | L:L:D291 | 4.87 | 2 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?20 | L:L:N441 | 30.8 | 2 | Yes | No | 0 | 7 | 1 | 1 | | L:L:E209 | L:L:P210 | 3.14 | 0 | No | Yes | 7 | 9 | 2 | 1 | | L:L:K250 | L:L:P210 | 3.35 | 2 | Yes | Yes | 5 | 9 | 2 | 1 | | L:L:P210 | L:L:P251 | 3.9 | 2 | Yes | No | 9 | 9 | 1 | 2 | | L:L:P210 | L:L:V252 | 1.77 | 2 | Yes | No | 9 | 9 | 1 | 1 | | L:L:K250 | L:L:P251 | 3.35 | 2 | Yes | No | 5 | 9 | 2 | 2 | | L:L:K250 | L:L:N260 | 8.39 | 2 | Yes | Yes | 5 | 9 | 2 | 1 | | L:L:L259 | L:L:V252 | 5.96 | 0 | Yes | No | 9 | 9 | 2 | 1 | | L:L:L259 | L:L:S440 | 7.51 | 0 | Yes | Yes | 9 | 9 | 2 | 1 | | L:L:N260 | L:L:S440 | 4.47 | 2 | Yes | Yes | 9 | 9 | 1 | 1 | | L:L:N260 | L:L:N441 | 5.45 | 2 | Yes | No | 9 | 7 | 1 | 1 | | L:L:I292 | L:L:S440 | 3.1 | 2 | Yes | Yes | 9 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 10.80 | | Average Nodes In Shell | 19.00 | | Average Hubs In Shell | 9.00 | | Average Links In Shell | 29.00 | | Average Links Mediated by Hubs In Shell | 28.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
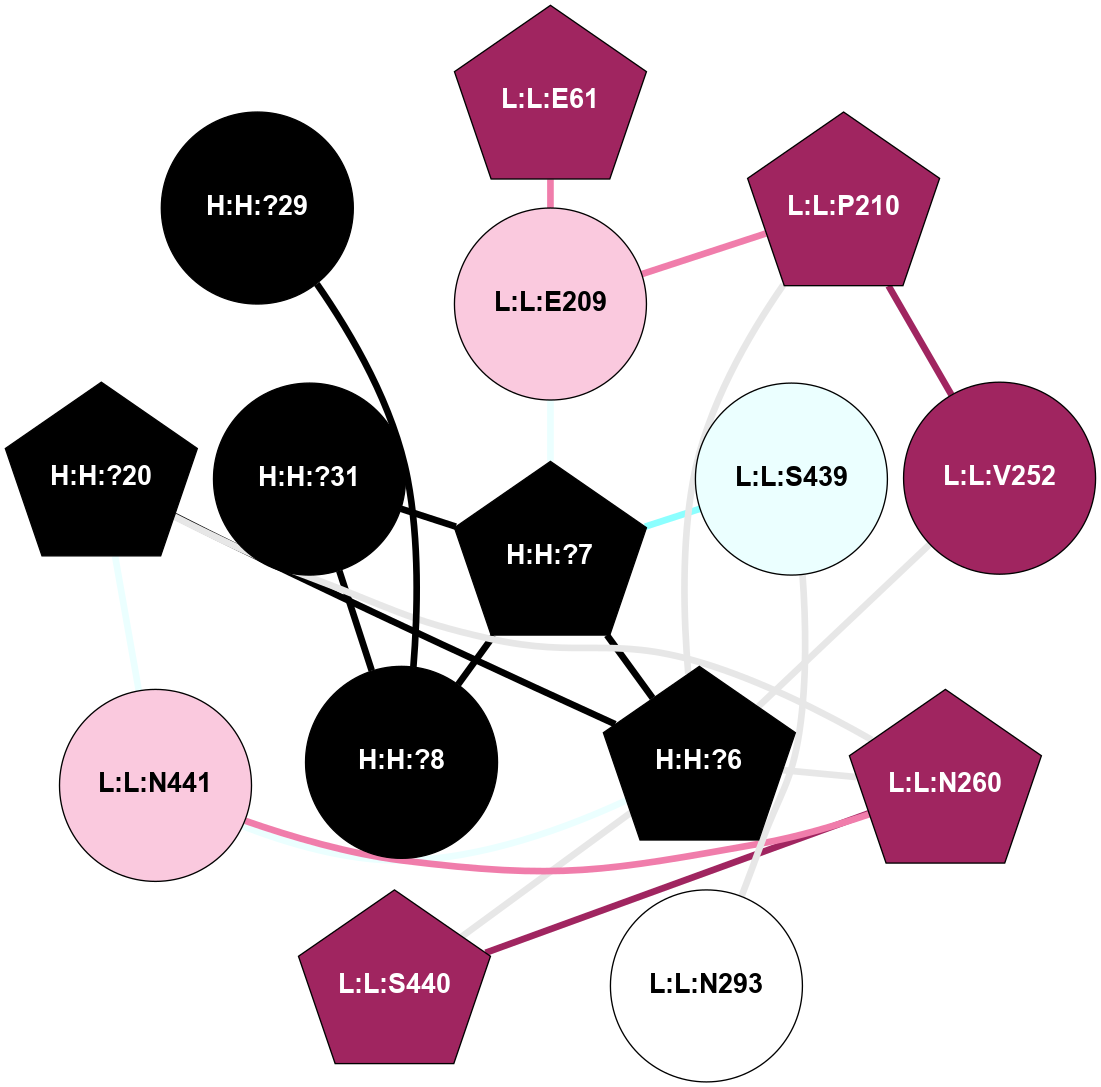
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | H:H:?7 | 31.2 | 2 | Yes | Yes | 0 | 0 | 1 | 0 | | H:H:?20 | H:H:?6 | 3.34 | 2 | Yes | Yes | 0 | 0 | 2 | 1 | | H:H:?6 | L:L:P210 | 5.89 | 2 | Yes | Yes | 0 | 9 | 1 | 2 | | H:H:?6 | L:L:V252 | 2.67 | 2 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?6 | L:L:N260 | 24.64 | 2 | Yes | Yes | 0 | 9 | 1 | 2 | | H:H:?6 | L:L:S440 | 5.39 | 2 | Yes | Yes | 0 | 9 | 1 | 2 | | H:H:?6 | L:L:N441 | 2.46 | 2 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?7 | H:H:?8 | 32.47 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?31 | H:H:?7 | 3.84 | 2 | No | Yes | 0 | 0 | 1 | 0 | | H:H:?7 | L:L:E209 | 10.7 | 2 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?7 | L:L:S439 | 8.08 | 2 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?29 | H:H:?8 | 38.83 | 0 | No | No | 0 | 0 | 2 | 1 | | H:H:?31 | H:H:?8 | 22.4 | 2 | No | No | 0 | 0 | 1 | 1 | | H:H:?20 | L:L:N260 | 3.7 | 2 | Yes | Yes | 0 | 9 | 2 | 2 | | H:H:?20 | L:L:N441 | 30.8 | 2 | Yes | No | 0 | 7 | 2 | 2 | | L:L:E209 | L:L:E61 | 3.81 | 0 | No | Yes | 7 | 9 | 1 | 2 | | L:L:E209 | L:L:P210 | 3.14 | 0 | No | Yes | 7 | 9 | 1 | 2 | | L:L:P210 | L:L:V252 | 1.77 | 2 | Yes | No | 9 | 9 | 2 | 2 | | L:L:N260 | L:L:S440 | 4.47 | 2 | Yes | Yes | 9 | 9 | 2 | 2 | | L:L:N260 | L:L:N441 | 5.45 | 2 | Yes | No | 9 | 7 | 2 | 2 | | L:L:N293 | L:L:S439 | 2.98 | 0 | No | No | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.26 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 21.00 | | Average Links Mediated by Hubs In Shell | 18.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
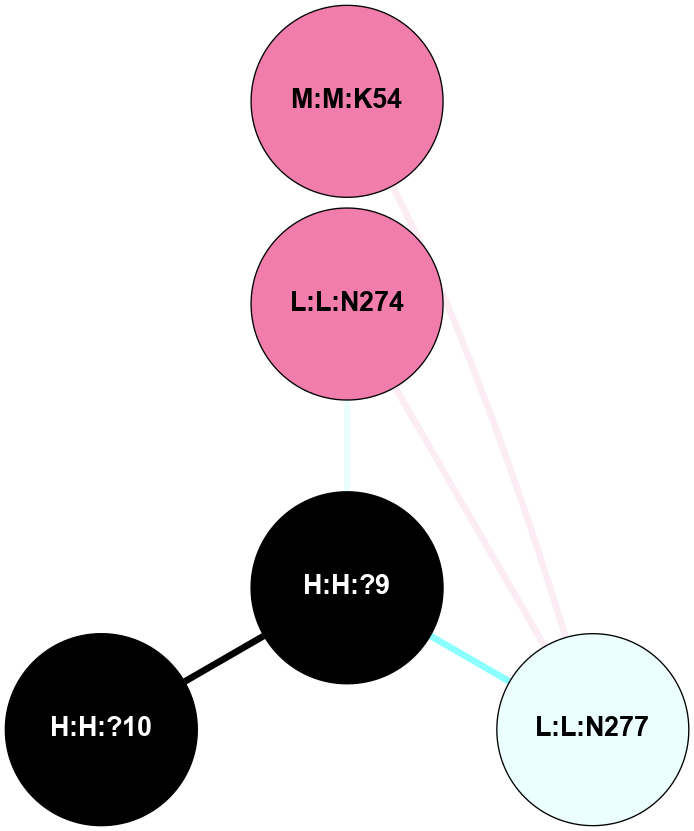
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | H:H:?9 | 40.12 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?9 | L:L:N274 | 33.27 | 20 | No | No | 0 | 8 | 0 | 1 | | H:H:?9 | L:L:N277 | 12.32 | 20 | No | No | 0 | 4 | 0 | 1 | | L:L:N274 | L:L:N277 | 5.45 | 20 | No | No | 8 | 4 | 1 | 1 | | L:L:N277 | N:N:K54 | 1.4 | 20 | No | No | 4 | 8 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 28.57 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
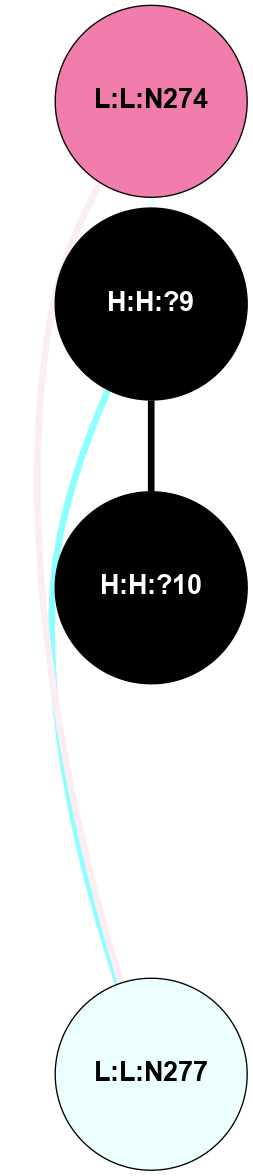
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | H:H:?9 | 40.12 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?9 | L:L:N274 | 33.27 | 20 | No | No | 0 | 8 | 1 | 2 | | H:H:?9 | L:L:N277 | 12.32 | 20 | No | No | 0 | 4 | 1 | 2 | | L:L:N274 | L:L:N277 | 5.45 | 20 | No | No | 8 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 40.12 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
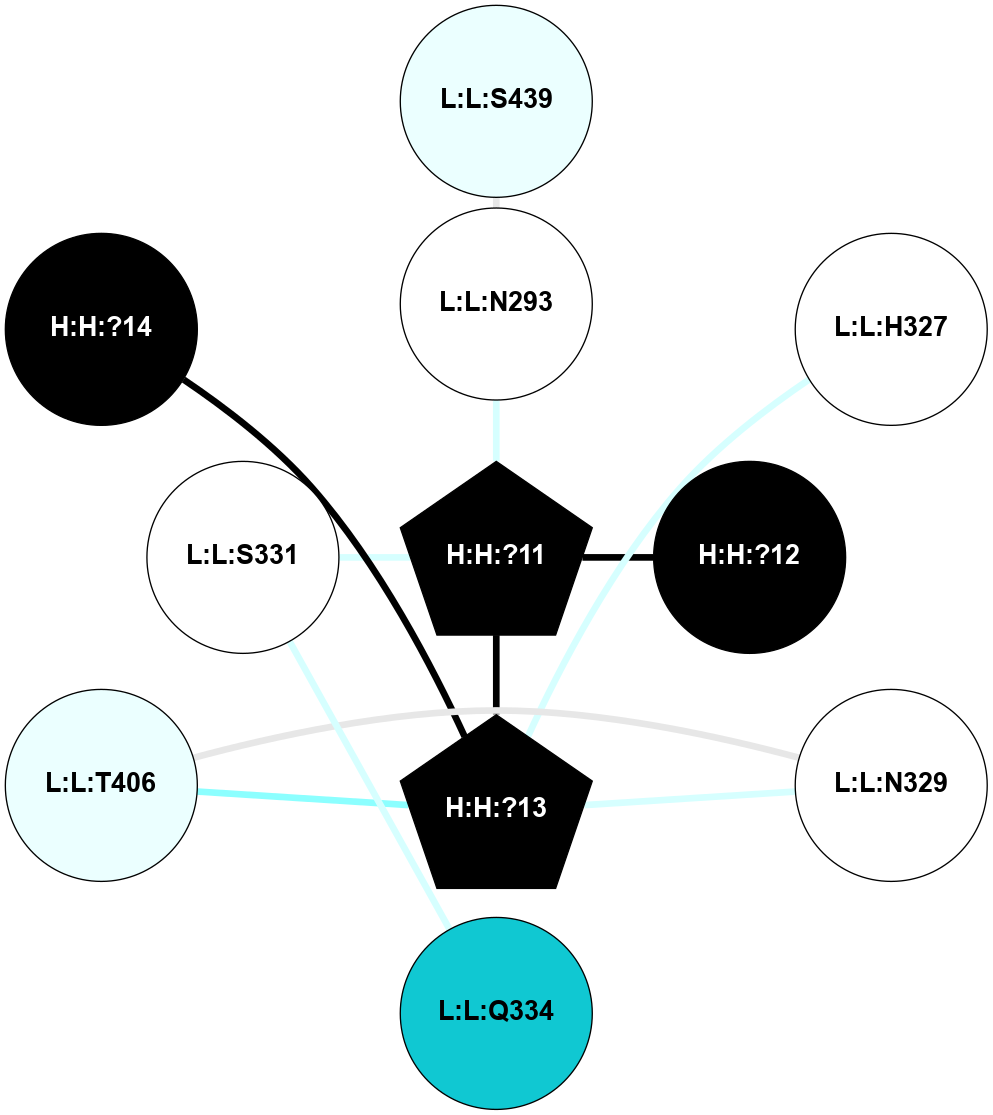
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | H:H:?12 | 35.66 | 0 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?11 | H:H:?13 | 4.46 | 0 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?11 | L:L:N293 | 25.87 | 0 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?13 | H:H:?14 | 34.55 | 21 | Yes | No | 0 | 0 | 1 | 2 | | H:H:?13 | L:L:H327 | 2.31 | 21 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?13 | L:L:N329 | 28.34 | 21 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?13 | L:L:T406 | 7.94 | 21 | Yes | No | 0 | 4 | 1 | 2 | | L:L:N293 | L:L:S439 | 2.98 | 0 | No | No | 5 | 4 | 1 | 2 | | L:L:N329 | L:L:T406 | 4.39 | 21 | No | No | 5 | 4 | 2 | 2 | | L:L:Q334 | L:L:S331 | 5.78 | 0 | No | No | 1 | 5 | 2 | 1 | | H:H:?11 | L:L:S331 | 1.35 | 0 | Yes | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 16.83 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
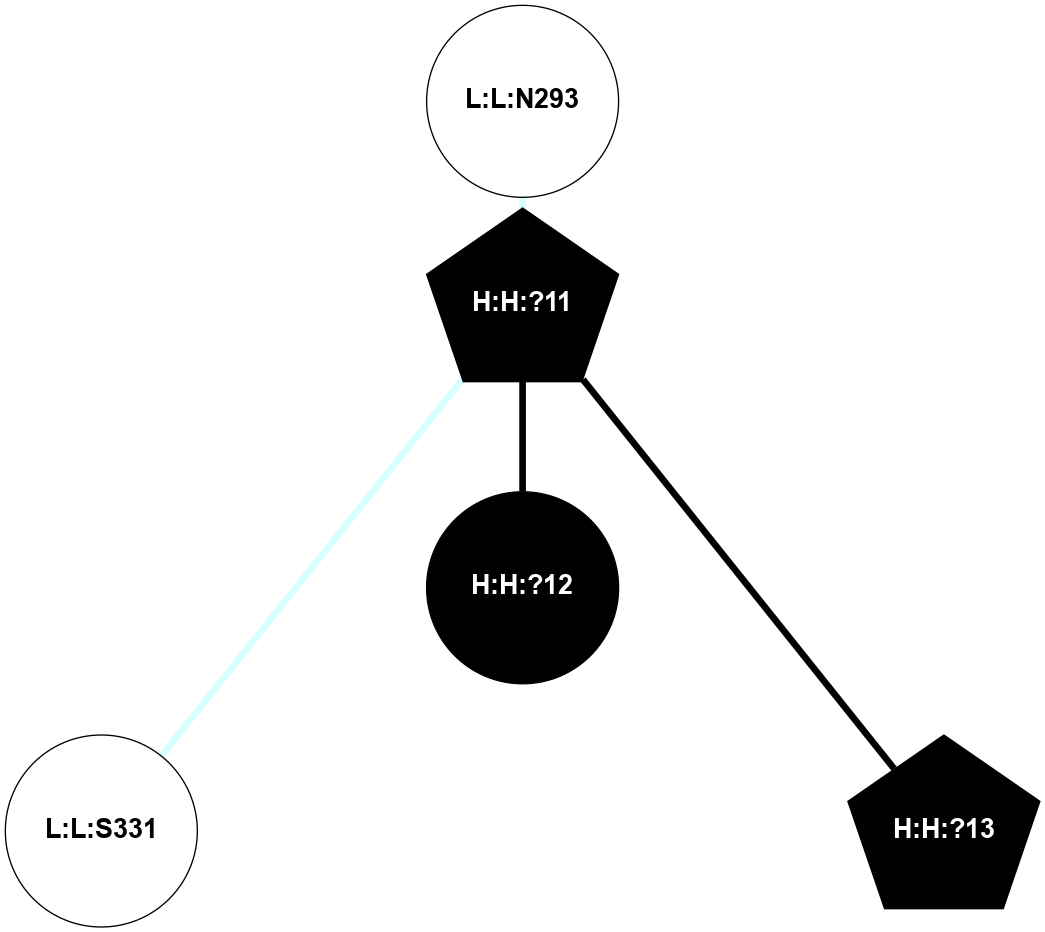
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | H:H:?12 | 35.66 | 0 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?11 | H:H:?13 | 4.46 | 0 | Yes | Yes | 0 | 0 | 1 | 2 | | H:H:?11 | L:L:N293 | 25.87 | 0 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?11 | L:L:S331 | 1.35 | 0 | Yes | No | 0 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 35.66 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
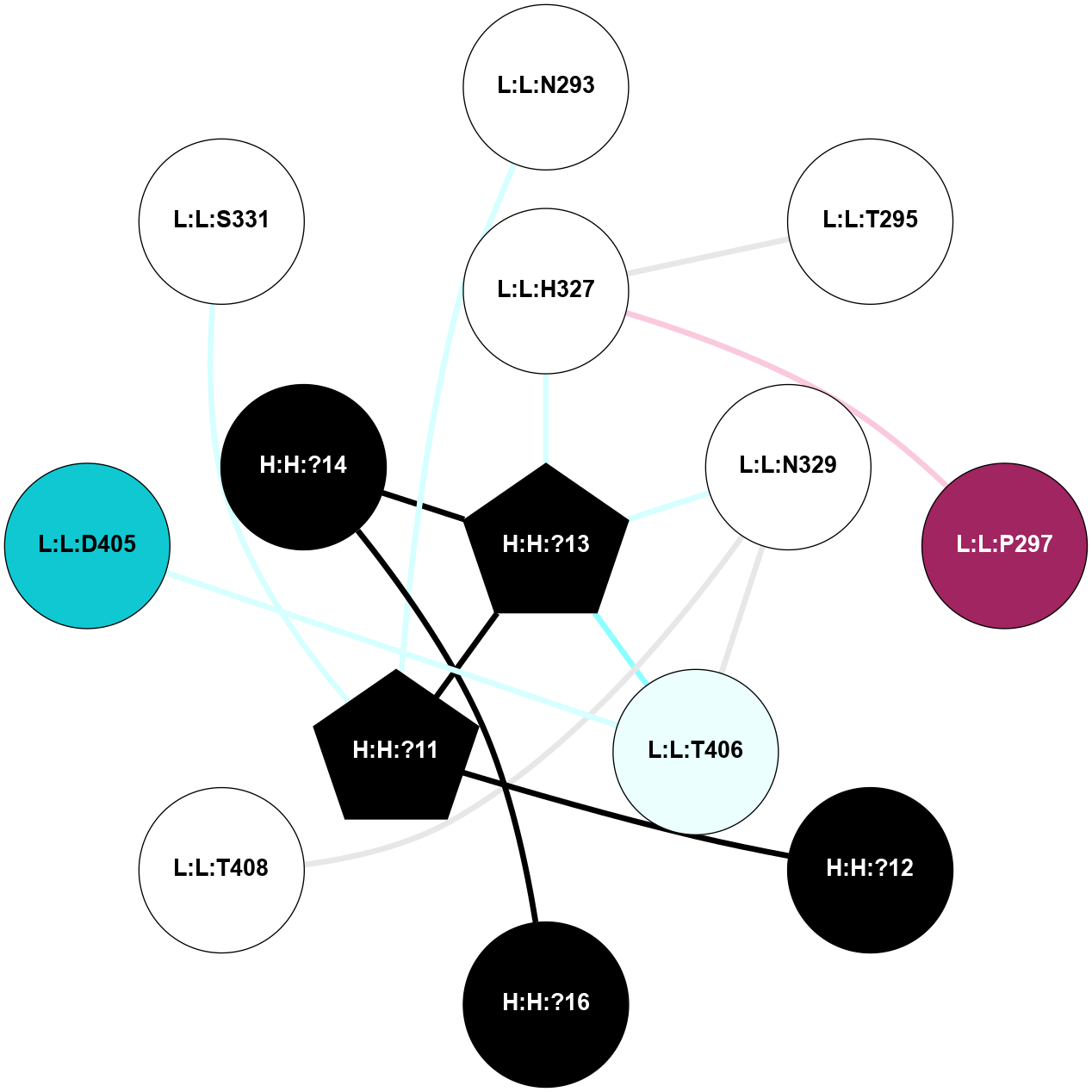
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | H:H:?12 | 35.66 | 0 | Yes | No | 0 | 0 | 1 | 2 | | H:H:?11 | H:H:?13 | 4.46 | 0 | Yes | Yes | 0 | 0 | 1 | 0 | | H:H:?11 | L:L:N293 | 25.87 | 0 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?13 | H:H:?14 | 34.55 | 21 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?13 | L:L:H327 | 2.31 | 21 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?13 | L:L:N329 | 28.34 | 21 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?13 | L:L:T406 | 7.94 | 21 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?14 | H:H:?16 | 4.46 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:H327 | L:L:T295 | 2.74 | 0 | No | No | 5 | 5 | 1 | 2 | | L:L:H327 | L:L:P297 | 6.1 | 0 | No | No | 5 | 9 | 1 | 2 | | L:L:N329 | L:L:T406 | 4.39 | 21 | No | No | 5 | 4 | 1 | 1 | | L:L:N329 | L:L:T408 | 1.46 | 21 | No | No | 5 | 5 | 1 | 2 | | L:L:D405 | L:L:T406 | 1.45 | 0 | No | No | 1 | 4 | 2 | 1 | | H:H:?11 | L:L:S331 | 1.35 | 0 | Yes | No | 0 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.52 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
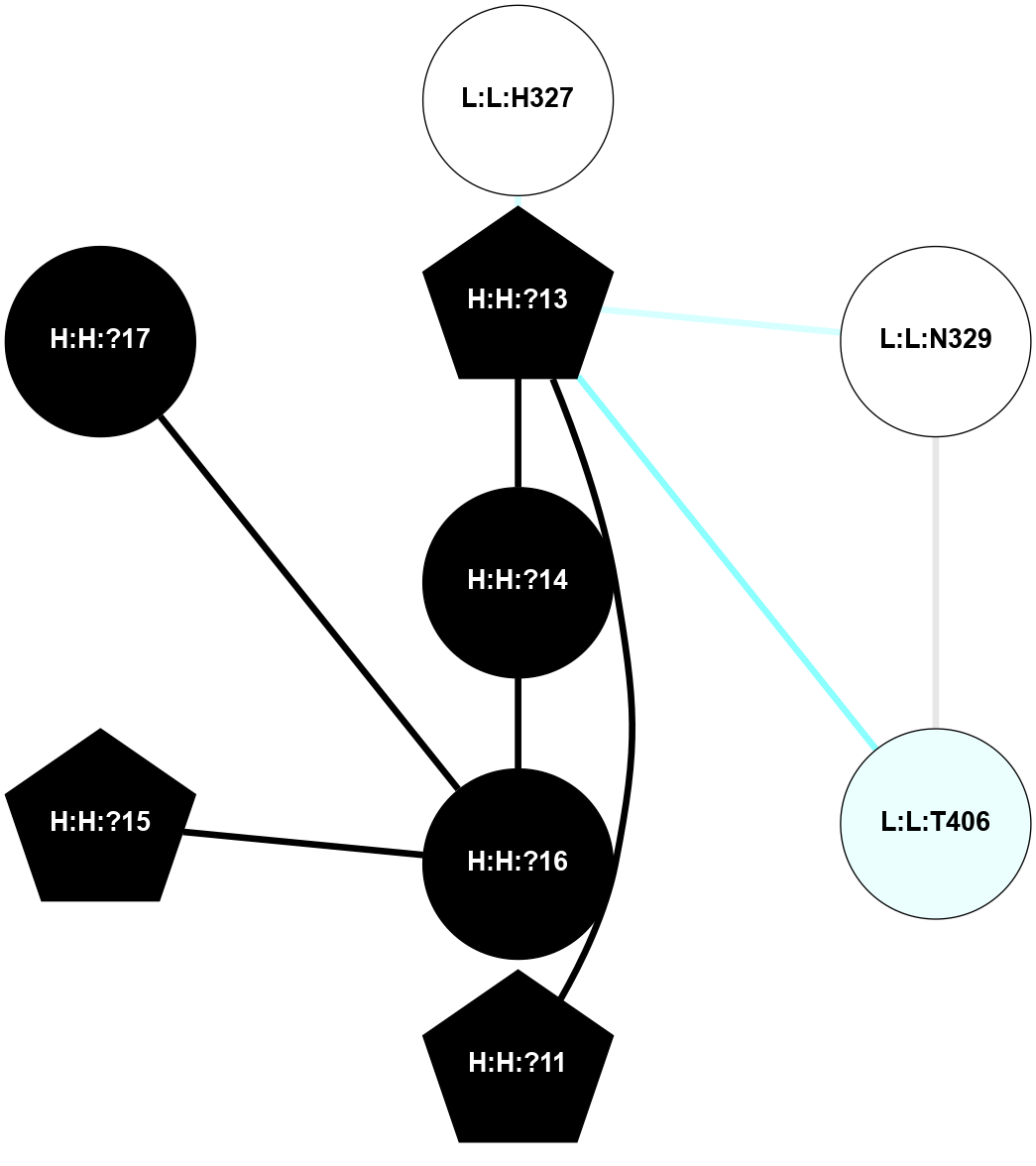
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | H:H:?13 | 4.46 | 0 | Yes | Yes | 0 | 0 | 2 | 1 | | H:H:?13 | H:H:?14 | 34.55 | 21 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?13 | L:L:H327 | 2.31 | 21 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?13 | L:L:N329 | 28.34 | 21 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?13 | L:L:T406 | 7.94 | 21 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?14 | H:H:?16 | 4.46 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?15 | H:H:?16 | 32.32 | 22 | Yes | No | 0 | 0 | 2 | 1 | | H:H:?16 | H:H:?17 | 29.87 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:N329 | L:L:T406 | 4.39 | 21 | No | No | 5 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.50 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
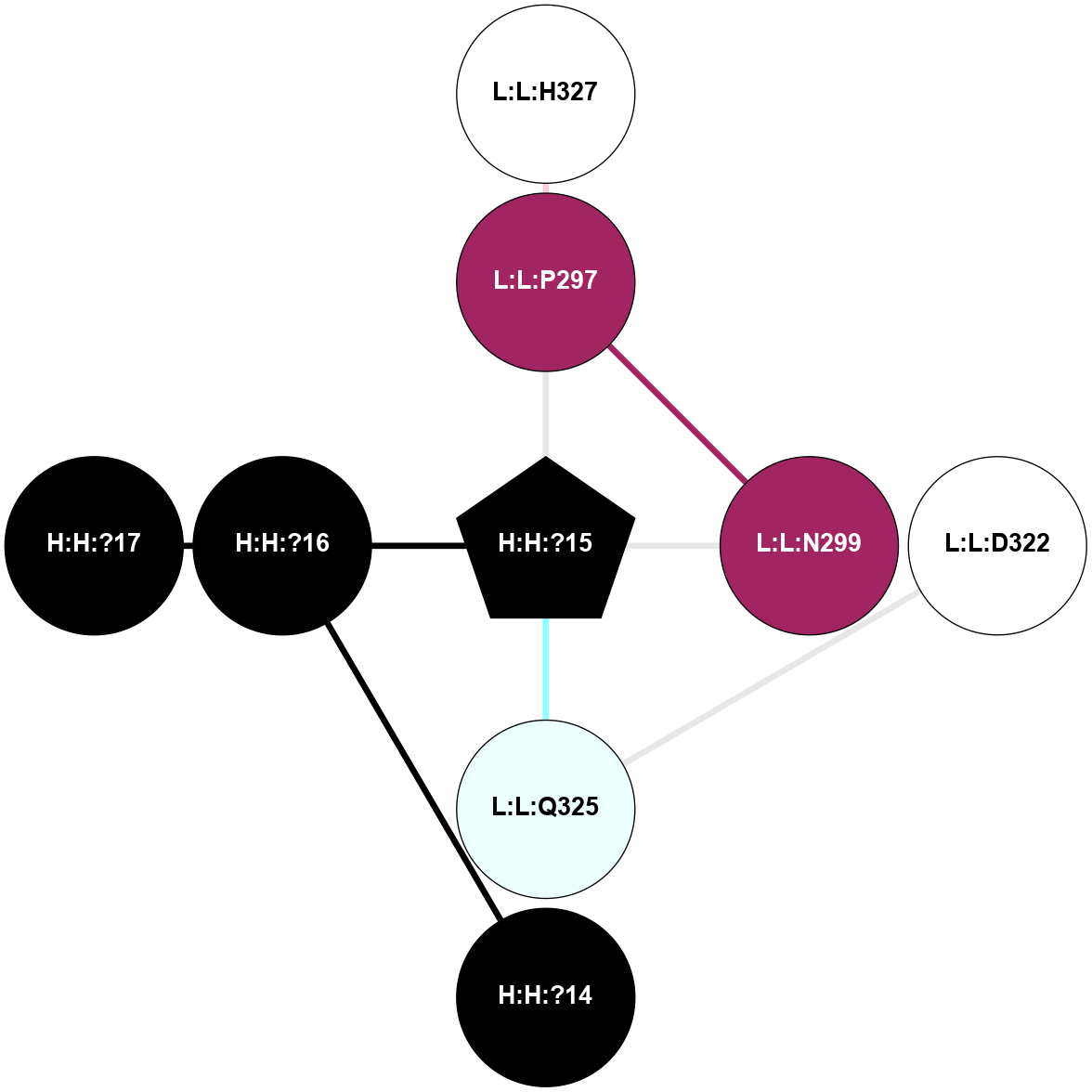
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?14 | H:H:?16 | 4.46 | 0 | No | No | 0 | 0 | 2 | 1 | | H:H:?15 | H:H:?16 | 32.32 | 22 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?15 | L:L:P297 | 14.73 | 22 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?15 | L:L:N299 | 23.41 | 22 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?15 | L:L:Q325 | 2.39 | 22 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?16 | H:H:?17 | 29.87 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:N299 | L:L:P297 | 4.89 | 22 | No | No | 9 | 9 | 1 | 1 | | L:L:H327 | L:L:P297 | 6.1 | 0 | No | No | 5 | 9 | 2 | 1 | | L:L:D322 | L:L:Q325 | 5.22 | 0 | No | No | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.21 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
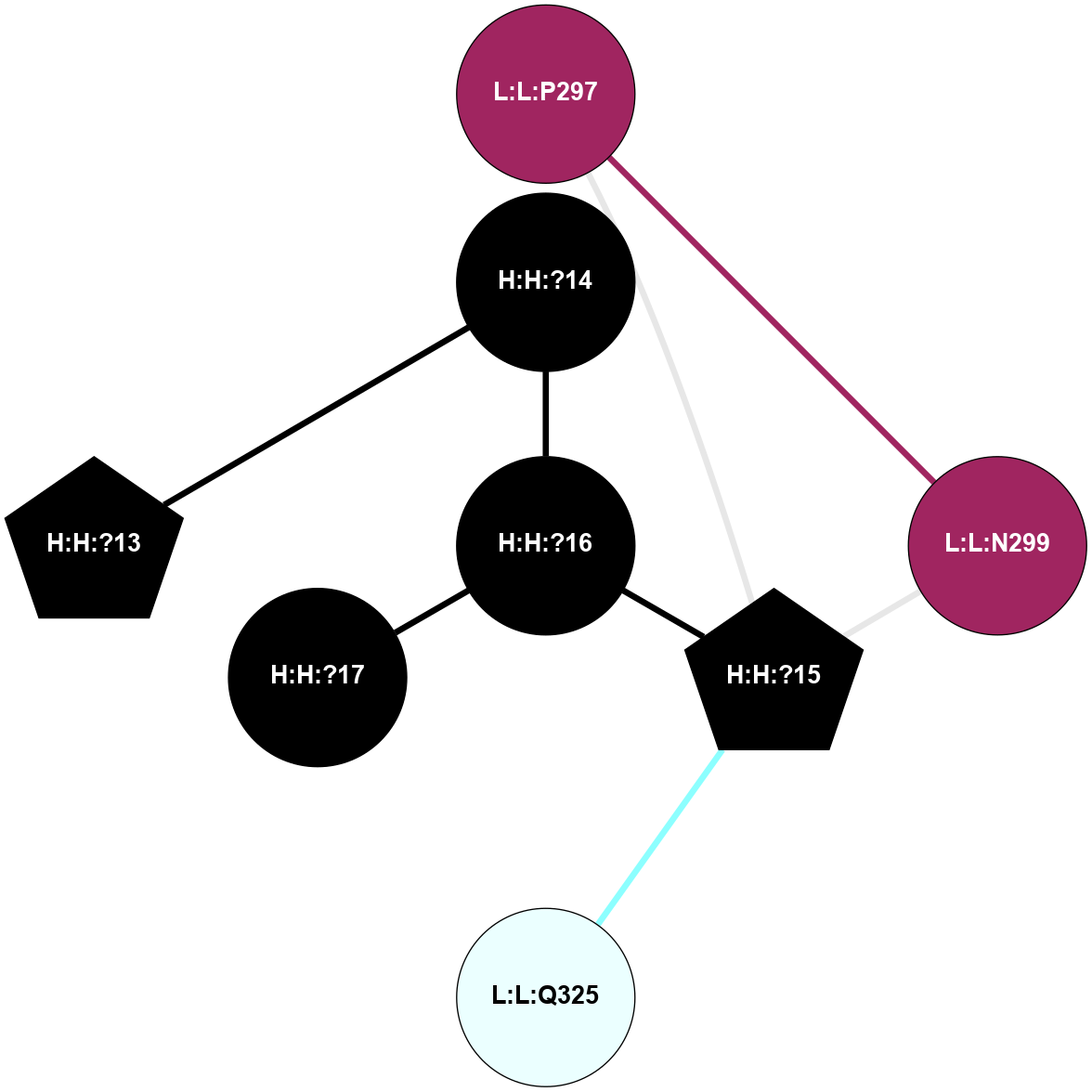
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | H:H:?14 | 34.55 | 21 | Yes | No | 0 | 0 | 2 | 1 | | H:H:?14 | H:H:?16 | 4.46 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?15 | H:H:?16 | 32.32 | 22 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?15 | L:L:P297 | 14.73 | 22 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?15 | L:L:N299 | 23.41 | 22 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?15 | L:L:Q325 | 2.39 | 22 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?16 | H:H:?17 | 29.87 | 0 | No | No | 0 | 0 | 0 | 1 | | L:L:N299 | L:L:P297 | 4.89 | 22 | No | No | 9 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 22.22 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
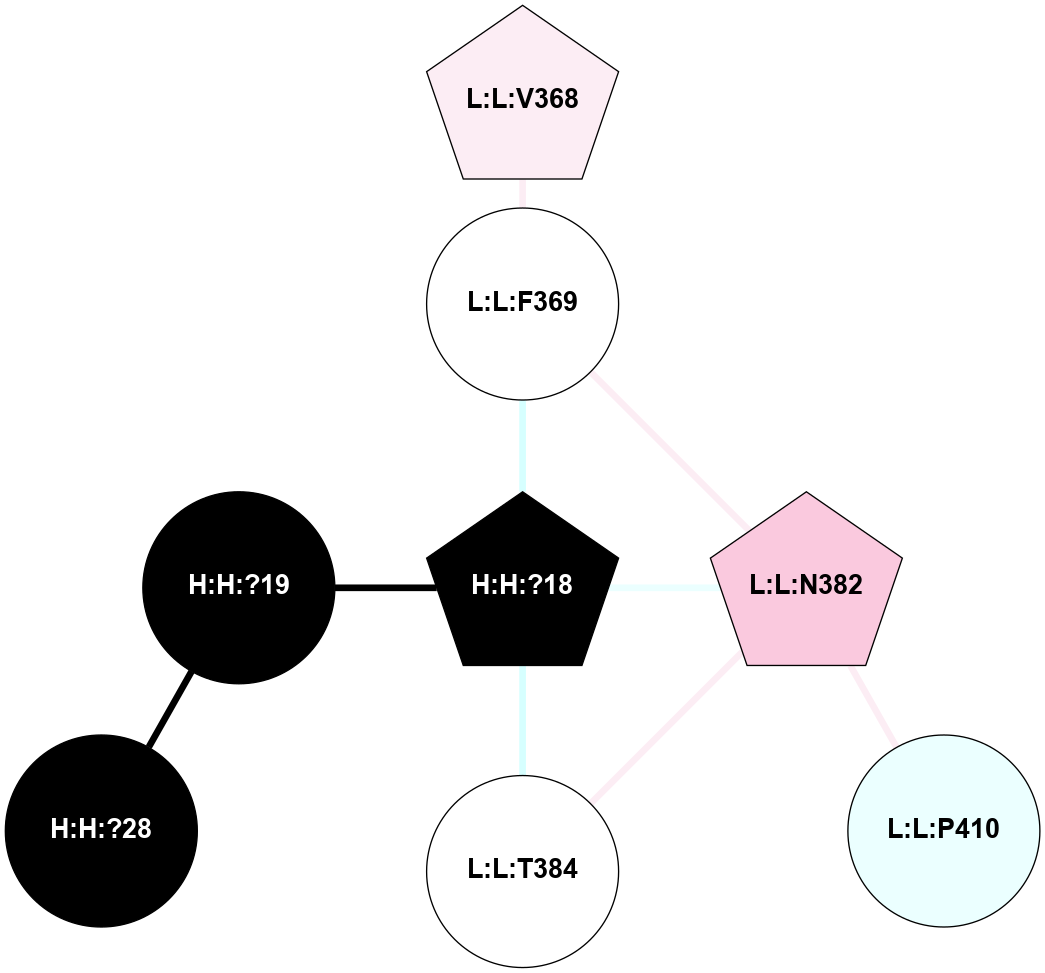
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?18 | H:H:?19 | 34.55 | 10 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?18 | L:L:F369 | 15.3 | 10 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?18 | L:L:N382 | 22.18 | 10 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?18 | L:L:T384 | 21.16 | 10 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?19 | H:H:?28 | 17.83 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:F369 | L:L:V368 | 3.93 | 10 | No | Yes | 5 | 6 | 1 | 2 | | L:L:F369 | L:L:N382 | 10.87 | 10 | No | Yes | 5 | 7 | 1 | 1 | | L:L:N382 | L:L:T384 | 10.24 | 10 | Yes | No | 7 | 5 | 1 | 1 | | L:L:N382 | L:L:P410 | 6.52 | 10 | Yes | No | 7 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 23.30 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
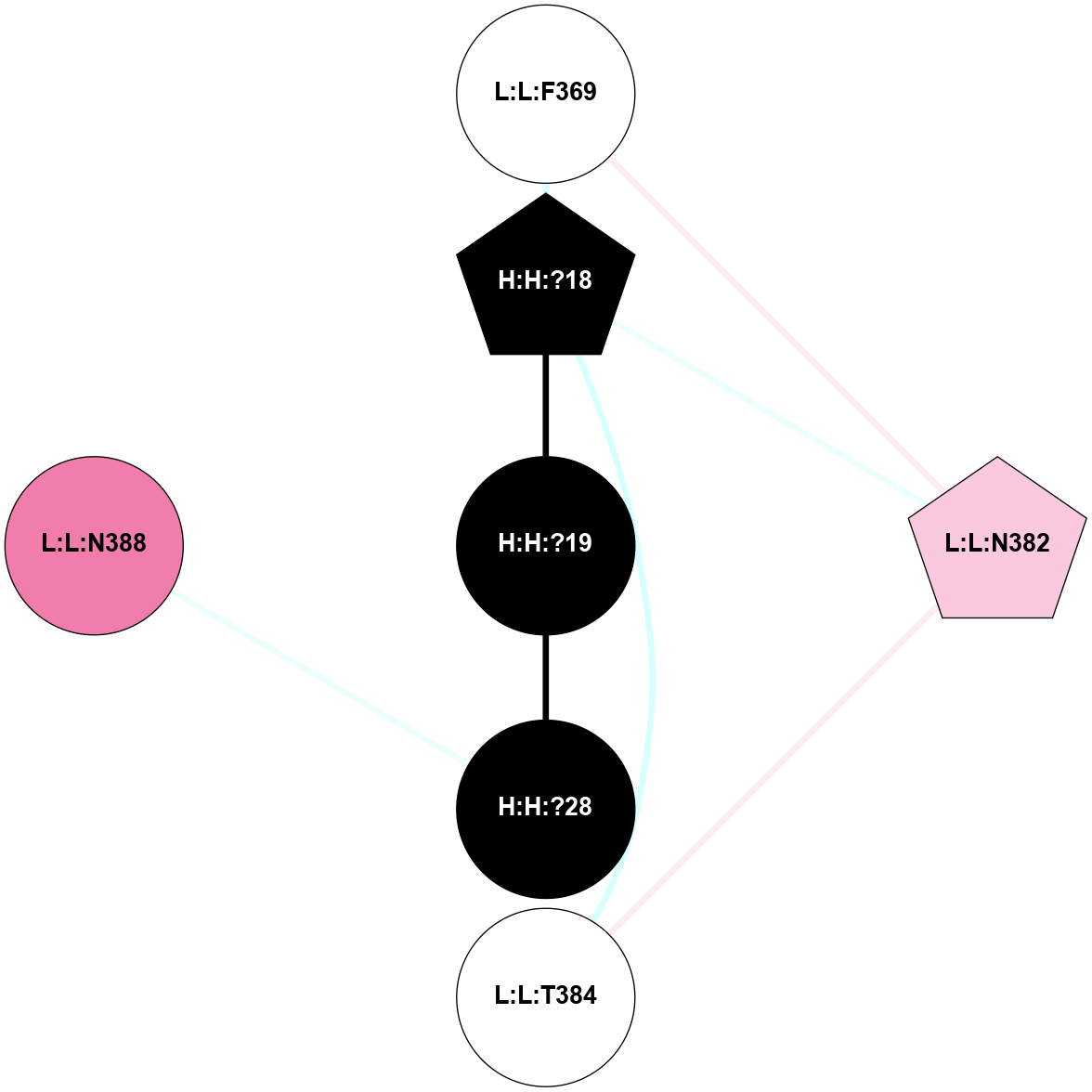
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?18 | H:H:?19 | 34.55 | 10 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?18 | L:L:F369 | 15.3 | 10 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?18 | L:L:N382 | 22.18 | 10 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?18 | L:L:T384 | 21.16 | 10 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?19 | H:H:?28 | 17.83 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?28 | L:L:N388 | 23.41 | 0 | No | No | 0 | 8 | 1 | 2 | | L:L:F369 | L:L:N382 | 10.87 | 10 | No | Yes | 5 | 7 | 2 | 2 | | L:L:N382 | L:L:T384 | 10.24 | 10 | Yes | No | 7 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 26.19 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
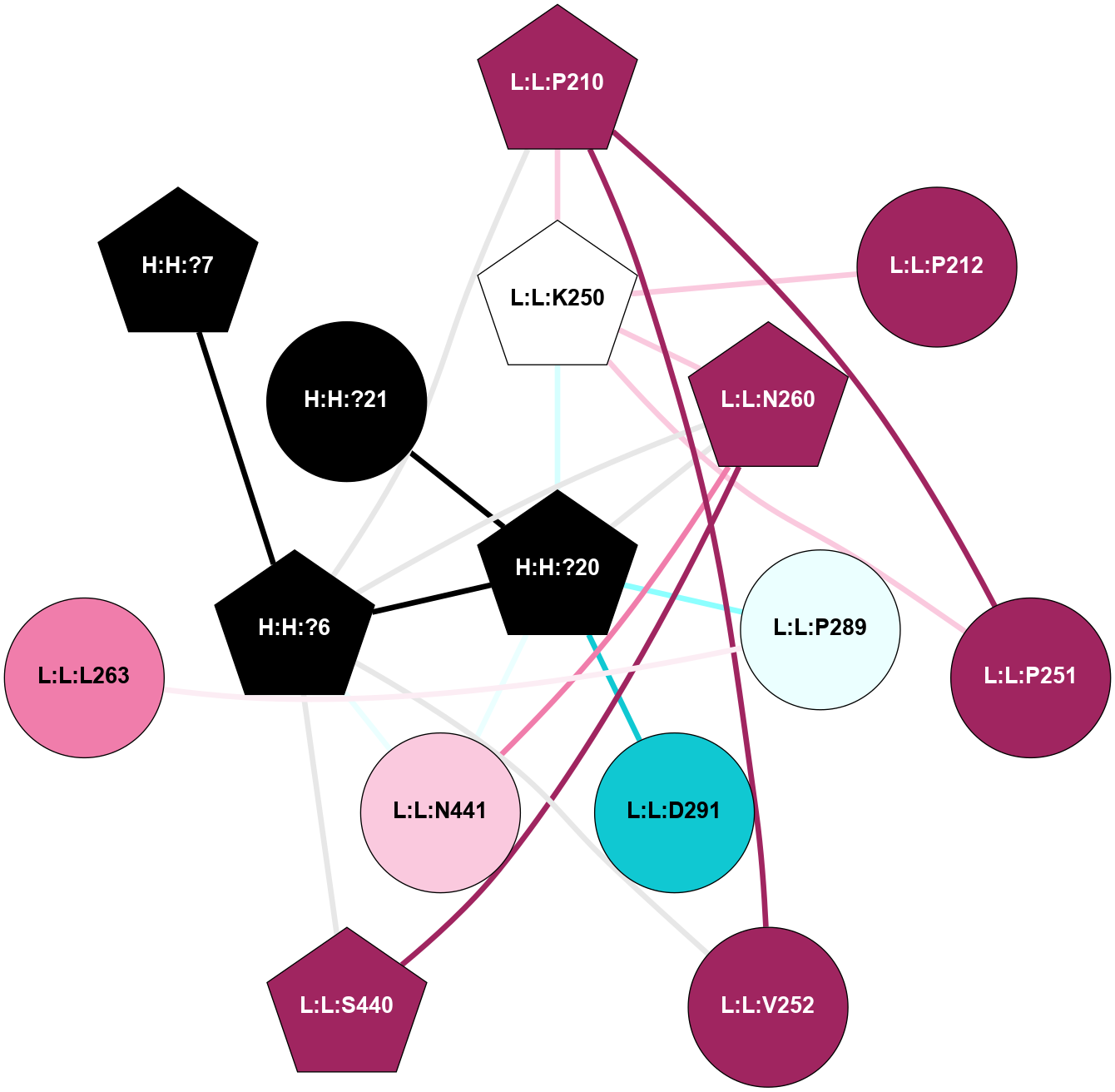
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | H:H:?7 | 31.2 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | H:H:?20 | H:H:?6 | 3.34 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?6 | L:L:P210 | 5.89 | 2 | Yes | Yes | 0 | 9 | 1 | 2 | | H:H:?6 | L:L:V252 | 2.67 | 2 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?6 | L:L:N260 | 24.64 | 2 | Yes | Yes | 0 | 9 | 1 | 1 | | H:H:?6 | L:L:S440 | 5.39 | 2 | Yes | Yes | 0 | 9 | 1 | 2 | | H:H:?6 | L:L:N441 | 2.46 | 2 | Yes | No | 0 | 7 | 1 | 1 | | H:H:?20 | H:H:?21 | 33.43 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?20 | L:L:K250 | 2.53 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?20 | L:L:N260 | 3.7 | 2 | Yes | Yes | 0 | 9 | 0 | 1 | | H:H:?20 | L:L:P289 | 5.89 | 2 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?20 | L:L:D291 | 4.87 | 2 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?20 | L:L:N441 | 30.8 | 2 | Yes | No | 0 | 7 | 0 | 1 | | L:L:K250 | L:L:P210 | 3.35 | 2 | Yes | Yes | 5 | 9 | 1 | 2 | | L:L:P210 | L:L:P251 | 3.9 | 2 | Yes | No | 9 | 9 | 2 | 2 | | L:L:P210 | L:L:V252 | 1.77 | 2 | Yes | No | 9 | 9 | 2 | 2 | | L:L:K250 | L:L:P212 | 5.02 | 2 | Yes | No | 5 | 9 | 1 | 2 | | L:L:K250 | L:L:P251 | 3.35 | 2 | Yes | No | 5 | 9 | 1 | 2 | | L:L:K250 | L:L:N260 | 8.39 | 2 | Yes | Yes | 5 | 9 | 1 | 1 | | L:L:N260 | L:L:S440 | 4.47 | 2 | Yes | Yes | 9 | 9 | 1 | 2 | | L:L:N260 | L:L:N441 | 5.45 | 2 | Yes | No | 9 | 7 | 1 | 1 | | L:L:L263 | L:L:P289 | 6.57 | 0 | No | No | 8 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 12.08 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 22.00 | | Average Links Mediated by Hubs In Shell | 21.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
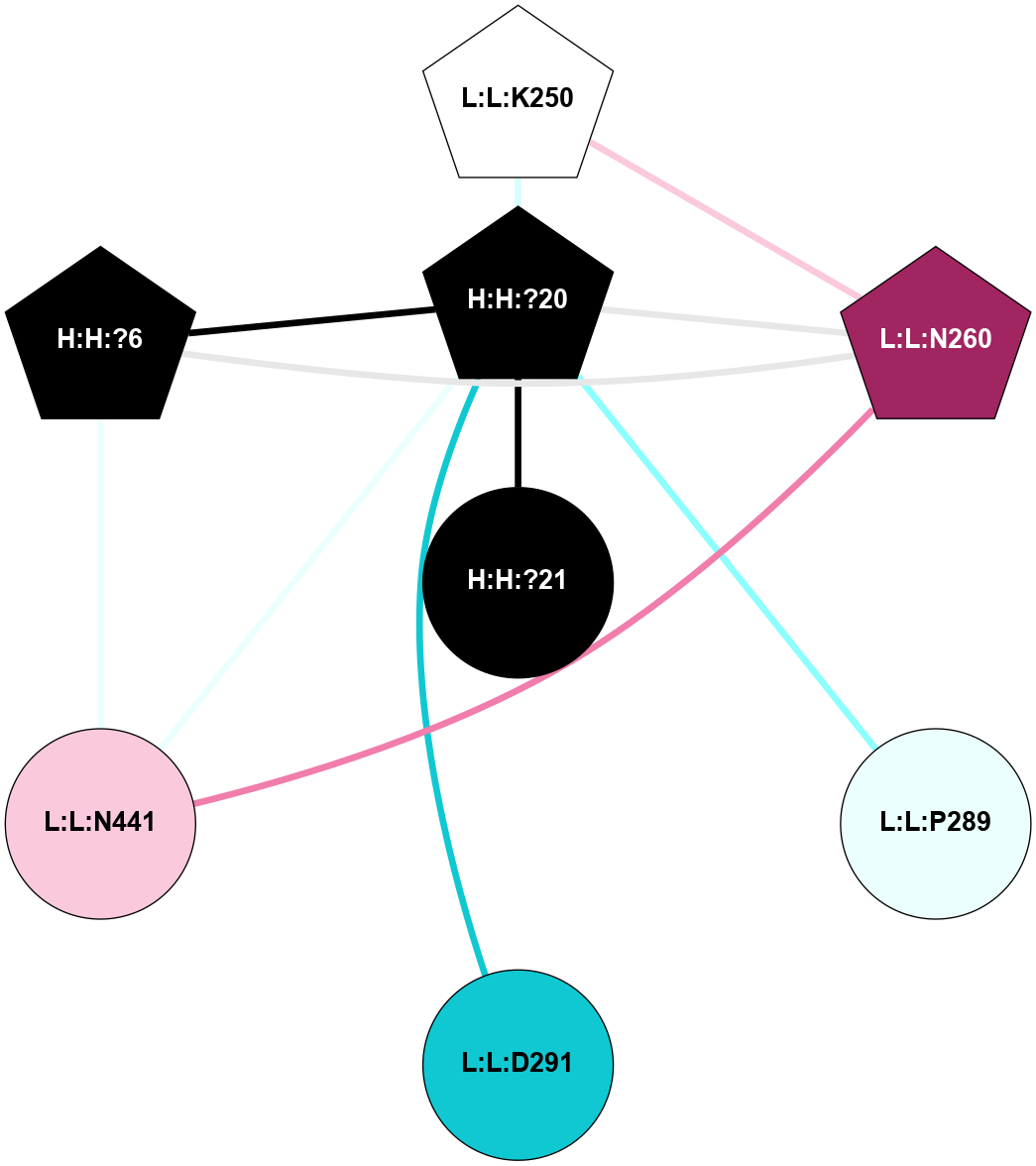
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?20 | H:H:?6 | 3.34 | 2 | Yes | Yes | 0 | 0 | 1 | 2 | | H:H:?6 | L:L:N260 | 24.64 | 2 | Yes | Yes | 0 | 9 | 2 | 2 | | H:H:?6 | L:L:N441 | 2.46 | 2 | Yes | No | 0 | 7 | 2 | 2 | | H:H:?20 | H:H:?21 | 33.43 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?20 | L:L:K250 | 2.53 | 2 | Yes | Yes | 0 | 5 | 1 | 2 | | H:H:?20 | L:L:N260 | 3.7 | 2 | Yes | Yes | 0 | 9 | 1 | 2 | | H:H:?20 | L:L:P289 | 5.89 | 2 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?20 | L:L:D291 | 4.87 | 2 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?20 | L:L:N441 | 30.8 | 2 | Yes | No | 0 | 7 | 1 | 2 | | L:L:K250 | L:L:N260 | 8.39 | 2 | Yes | Yes | 5 | 9 | 2 | 2 | | L:L:N260 | L:L:N441 | 5.45 | 2 | Yes | No | 9 | 7 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 33.43 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
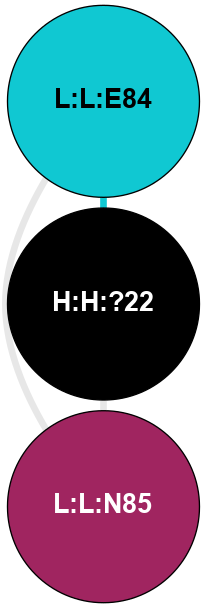
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?22 | L:L:E84 | 22.59 | 23 | No | No | 0 | 1 | 0 | 1 | | H:H:?22 | L:L:N85 | 24.64 | 23 | No | No | 0 | 9 | 0 | 1 | | L:L:E84 | L:L:N85 | 13.15 | 23 | No | No | 1 | 9 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 23.62 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
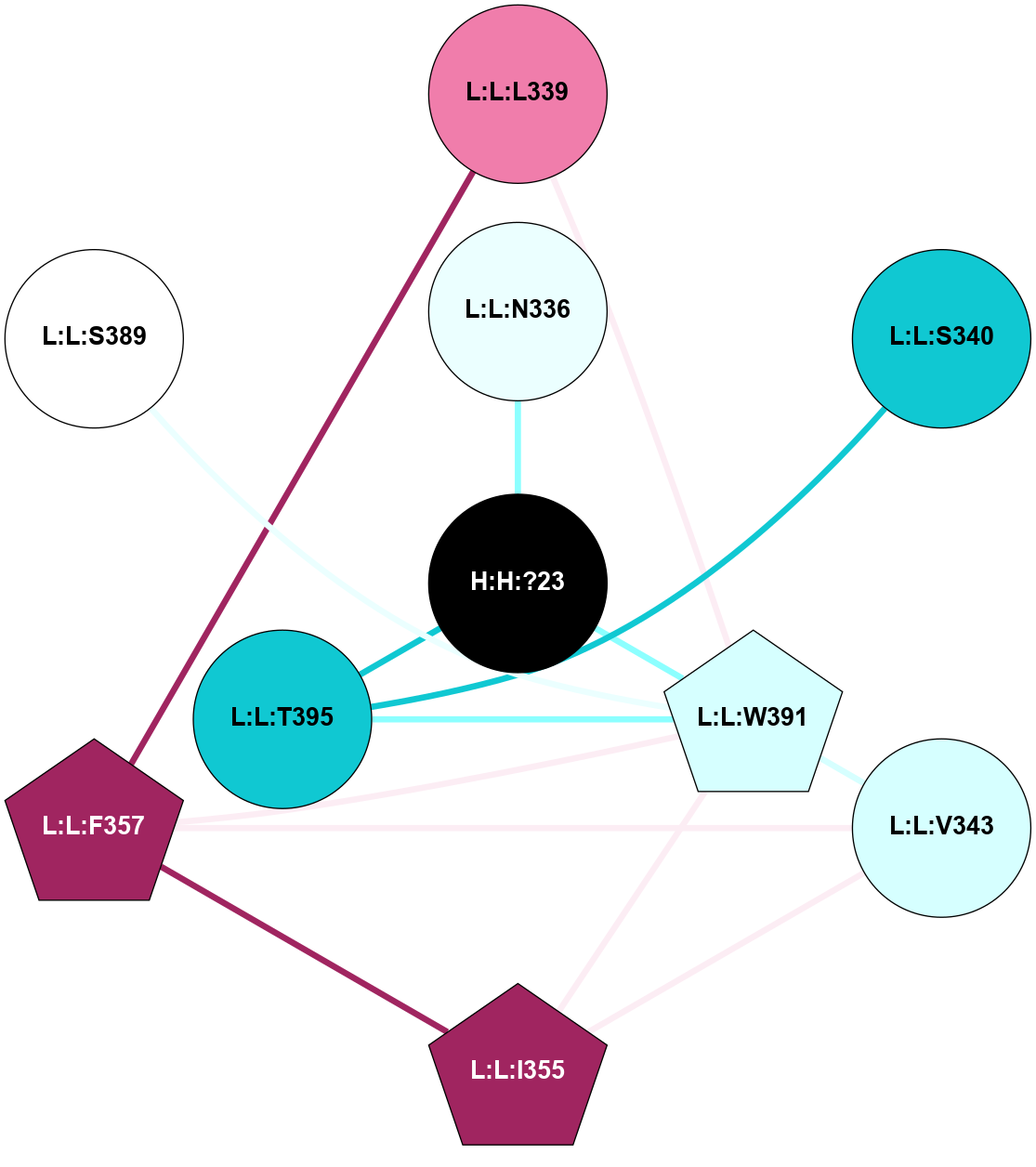
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?23 | L:L:N336 | 28.34 | 5 | No | No | 0 | 4 | 0 | 1 | | H:H:?23 | L:L:W391 | 5.11 | 5 | No | Yes | 0 | 3 | 0 | 1 | | H:H:?23 | L:L:T395 | 6.61 | 5 | No | No | 0 | 1 | 0 | 1 | | L:L:F357 | L:L:L339 | 2.44 | 5 | Yes | No | 9 | 8 | 2 | 2 | | L:L:L339 | L:L:W391 | 4.56 | 5 | No | Yes | 8 | 3 | 2 | 1 | | L:L:S340 | L:L:T395 | 3.2 | 0 | No | No | 1 | 1 | 2 | 1 | | L:L:I355 | L:L:V343 | 3.07 | 5 | Yes | No | 9 | 3 | 2 | 2 | | L:L:F357 | L:L:V343 | 2.62 | 5 | Yes | No | 9 | 3 | 2 | 2 | | L:L:V343 | L:L:W391 | 6.13 | 5 | No | Yes | 3 | 3 | 2 | 1 | | L:L:F357 | L:L:I355 | 3.77 | 5 | Yes | Yes | 9 | 9 | 2 | 2 | | L:L:I355 | L:L:W391 | 2.35 | 5 | Yes | Yes | 9 | 3 | 2 | 1 | | L:L:F357 | L:L:W391 | 4.01 | 5 | Yes | Yes | 9 | 3 | 2 | 1 | | L:L:S389 | L:L:W391 | 4.94 | 0 | No | Yes | 5 | 3 | 2 | 1 | | L:L:T395 | L:L:W391 | 4.85 | 5 | No | Yes | 1 | 3 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 13.35 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?24 | L:L:R347 | 2.18 | 24 | No | No | 0 | 3 | 0 | 1 | | H:H:?24 | L:L:N352 | 29.57 | 24 | No | No | 0 | 5 | 0 | 1 | | L:L:N352 | L:L:R347 | 7.23 | 24 | No | No | 5 | 3 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.88 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?25 | L:L:N393 | 24.64 | 25 | No | No | 0 | 1 | 0 | 1 | | H:H:?25 | L:L:N394 | 9.86 | 25 | No | No | 0 | 4 | 0 | 1 | | L:L:N393 | L:L:N394 | 8.17 | 25 | No | No | 1 | 4 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.25 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?26 | L:L:M456 | 7.61 | 0 | No | No | 0 | 1 | 0 | 1 | | H:H:?26 | L:L:N457 | 24.64 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.12 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?27 | L:L:N195 | 27.11 | 0 | No | No | 0 | 9 | 0 | 1 | | L:L:S197 | L:L:T120 | 1.6 | 0 | No | No | 9 | 9 | 1 | 2 | | L:L:L122 | L:L:N195 | 1.37 | 0 | No | No | 9 | 9 | 2 | 1 | | H:H:?27 | L:L:S197 | 1.35 | 0 | No | No | 0 | 9 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.23 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
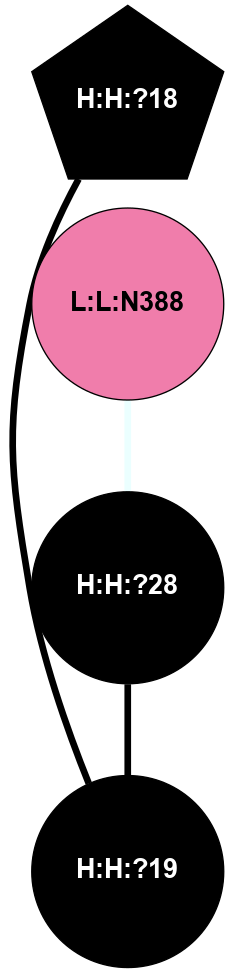
A 2D representation of the interactions of NAG in 6MET
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?18 | H:H:?19 | 34.55 | 10 | Yes | No | 0 | 0 | 2 | 1 | | H:H:?19 | H:H:?28 | 17.83 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?28 | L:L:N388 | 23.41 | 0 | No | No | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.62 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6N51 | C | Aminoacid | Metabotropic Glutamate | mGlu5; mGlu5 | Homo Sapiens | Quisqualate | - | - | 4 | 2019-01-23 | doi.org/10.1038/s41586-019-0881-4 |
|
A 2D representation of the interactions of NAG in 6N51
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | S:S:N210 | 25.79 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N210 | S:S:R508 | 1.21 | 0 | No | No | 4 | 1 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 25.79 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 6N51
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | S:S:N445 | 27.02 | 0 | No | No | 0 | 5 | 0 | 1 | | S:S:N445 | S:S:T453 | 4.39 | 0 | No | No | 5 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 6N51
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:N210 | 25.79 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N210 | R:R:R508 | 1.21 | 0 | No | No | 4 | 1 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 25.79 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
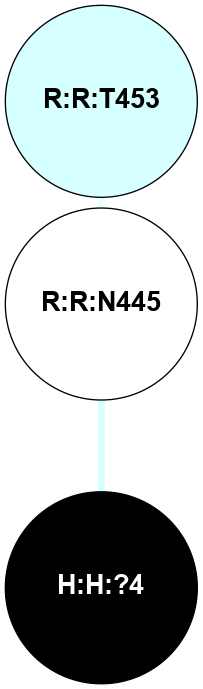
A 2D representation of the interactions of NAG in 6N51
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:N445 | 27.02 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:N445 | R:R:T453 | 4.39 | 0 | No | No | 5 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6N52 | C | Aminoacid | Metabotropic Glutamate | mGlu5; mGlu5 | Homo Sapiens | - | - | - | 4 | 2019-01-23 | doi.org/10.1038/s41586-019-0881-4 |
|

A 2D representation of the interactions of NAG in 6N52
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N210 | 23.33 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:N505 | 3.68 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N505 | R:R:W500 | 10.17 | 0 | No | No | 4 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.50 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6N52
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:N445 | 25.79 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:L455 | R:R:N445 | 5.49 | 0 | No | No | 3 | 5 | 2 | 1 | | R:R:N445 | R:R:T453 | 1.46 | 0 | No | No | 5 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 25.79 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
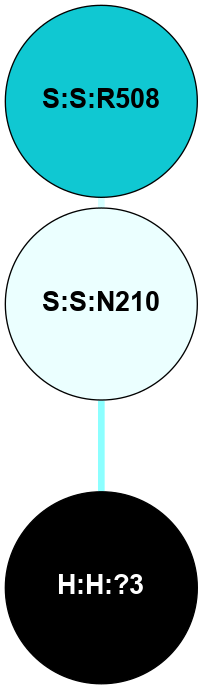
A 2D representation of the interactions of NAG in 6N52
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | S:S:N210 | 27.02 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:R508 | S:S:W211 | 6 | 0 | No | No | 1 | 9 | 1 | 2 | | H:H:?3 | S:S:R508 | 1.09 | 0 | No | No | 0 | 1 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.05 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6N52
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | S:S:N445 | 27.02 | 0 | No | No | 0 | 5 | 0 | 1 | | S:S:L455 | S:S:N445 | 5.49 | 0 | No | No | 3 | 5 | 2 | 1 | | S:S:N445 | S:S:T453 | 1.46 | 0 | No | No | 5 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6NWE | A | Sensory | Opsins | Rhodopsin | Bos Taurus | Octyl-Beta-D-Glucopyranoside | - | Gt(CT) | 2.71 | 2020-02-12 | To be published |
|
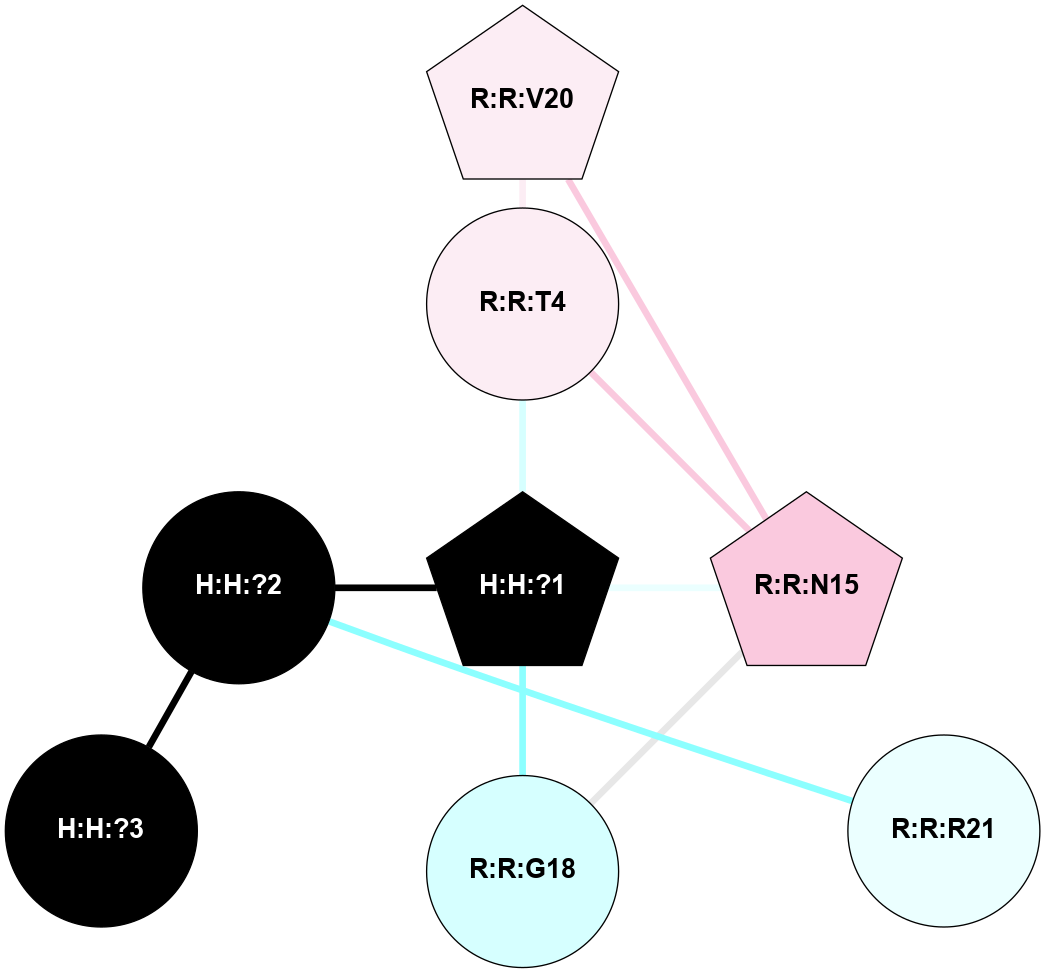
A 2D representation of the interactions of NAG in 6NWE
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 11.11 | 1 | No | Yes | 6 | 6 | 1 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 25.79 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 40.96 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 29.54 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.54 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
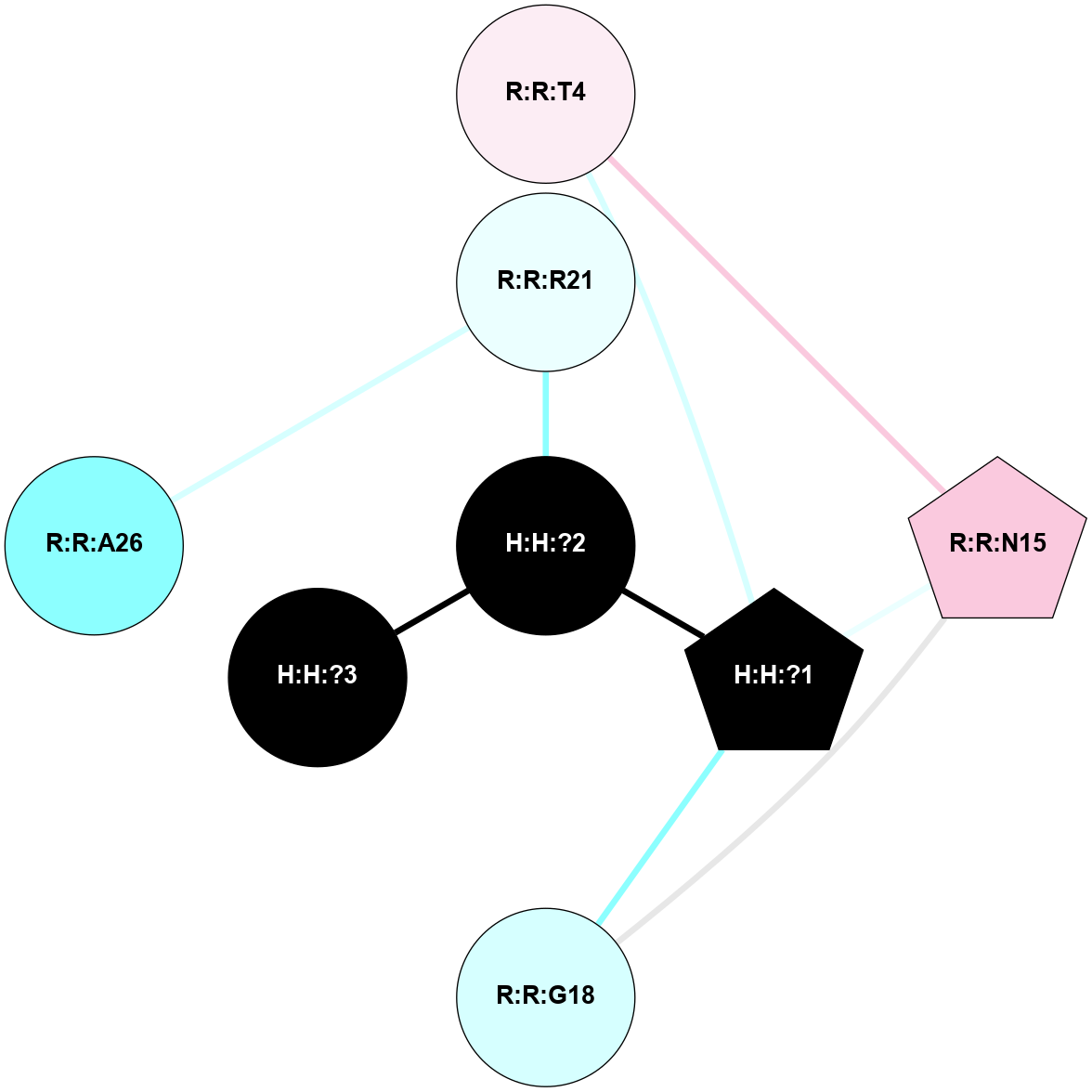
A 2D representation of the interactions of NAG in 6NWE
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 1 | Yes | No | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 2 | 2 | | H:H:?1 | R:R:N15 | 25.79 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?2 | R:R:R21 | 6.52 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 40.96 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 29.54 | 0 | No | No | 0 | 0 | 0 | 1 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 25.67 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6O3C | F | Protein | Frizzled | SMO | Mus Musculus | SAG21k | Cholesterol | - | 2.8 | 2019-07-03 | doi.org/10.1038/s41586-019-1355-4 |
|
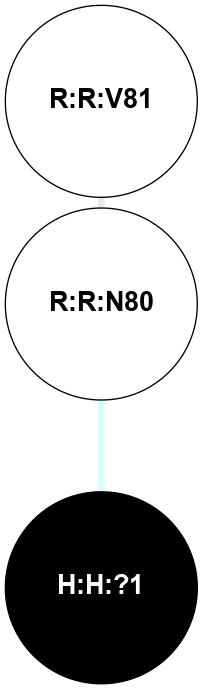
A 2D representation of the interactions of NAG in 6O3C
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N80 | 36.96 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:N80 | R:R:V81 | 4.43 | 0 | No | No | 5 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 36.96 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
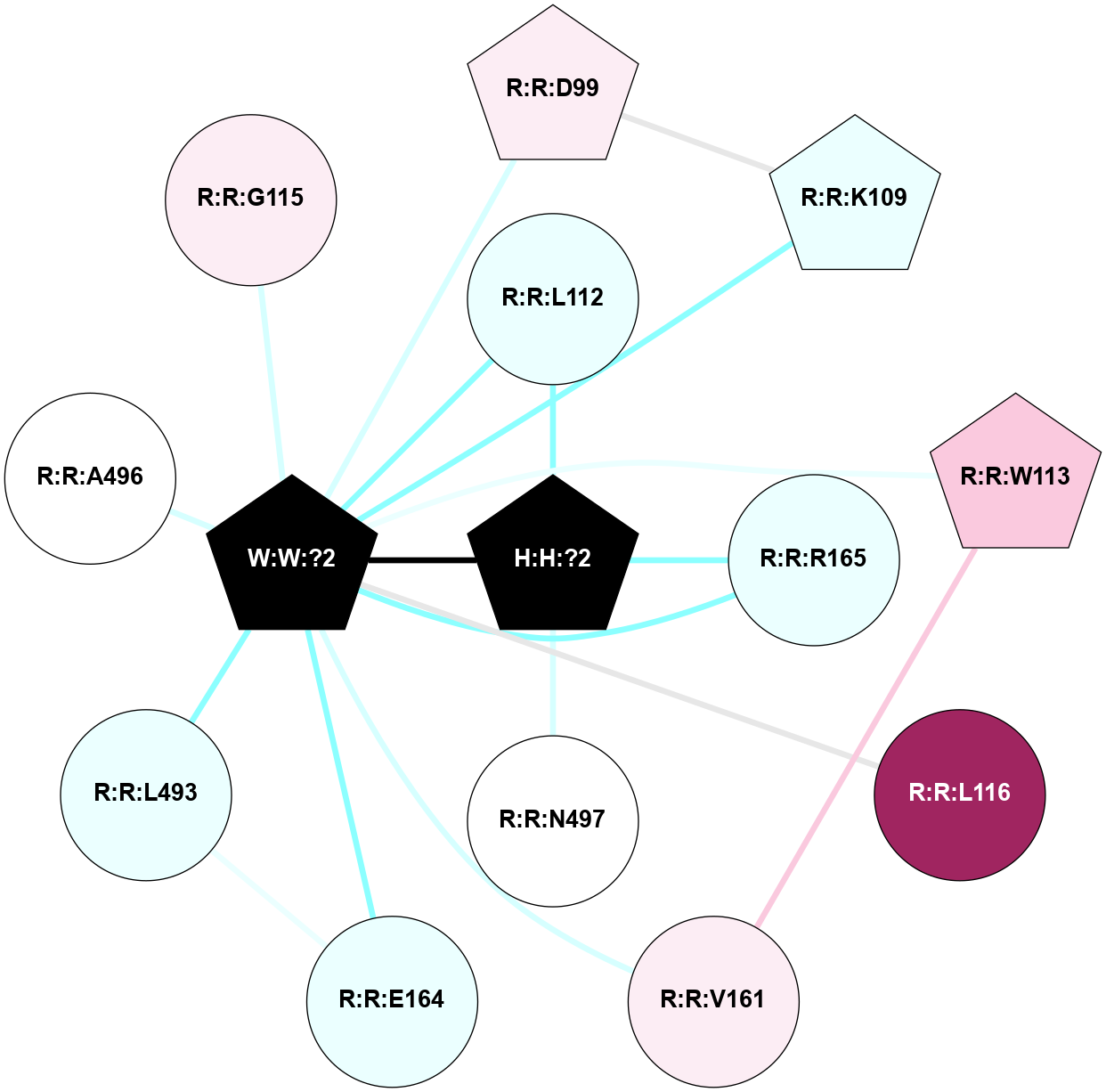
A 2D representation of the interactions of NAG in 6O3C
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:L112 | 4.97 | 2 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:R165 | 4.36 | 2 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:N497 | 29.57 | 2 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?2 | W:W:?2 | 6.19 | 2 | Yes | Yes | 0 | 0 | 0 | 1 | | R:R:D99 | R:R:K109 | 11.06 | 2 | Yes | Yes | 6 | 4 | 2 | 2 | | R:R:D99 | W:W:?2 | 6.77 | 2 | Yes | Yes | 6 | 0 | 2 | 1 | | R:R:K109 | W:W:?2 | 4.22 | 2 | Yes | Yes | 4 | 0 | 2 | 1 | | R:R:L112 | W:W:?2 | 9.66 | 2 | No | Yes | 4 | 0 | 1 | 1 | | R:R:V161 | R:R:W113 | 3.68 | 2 | No | Yes | 6 | 7 | 2 | 2 | | R:R:W113 | W:W:?2 | 34.07 | 2 | Yes | Yes | 7 | 0 | 2 | 1 | | R:R:L116 | W:W:?2 | 11.04 | 0 | No | Yes | 9 | 0 | 2 | 1 | | R:R:V161 | W:W:?2 | 4.46 | 2 | No | Yes | 6 | 0 | 2 | 1 | | R:R:E164 | R:R:L493 | 11.93 | 2 | No | No | 4 | 4 | 2 | 2 | | R:R:E164 | W:W:?2 | 3.96 | 2 | No | Yes | 4 | 0 | 2 | 1 | | R:R:R165 | W:W:?2 | 13.33 | 2 | No | Yes | 4 | 0 | 1 | 1 | | R:R:L493 | W:W:?2 | 4.14 | 2 | No | Yes | 4 | 0 | 2 | 1 | | R:R:A496 | W:W:?2 | 4.71 | 0 | No | Yes | 5 | 0 | 2 | 1 | | R:R:G115 | W:W:?2 | 1.71 | 0 | No | Yes | 6 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 11.27 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6OS0 | A | Peptide | Angiotensin | AT1 | Homo Sapiens | Angiotensinogen | - | - | 2.9 | 2020-02-19 | doi.org/10.1126/science.aay9813 |
|
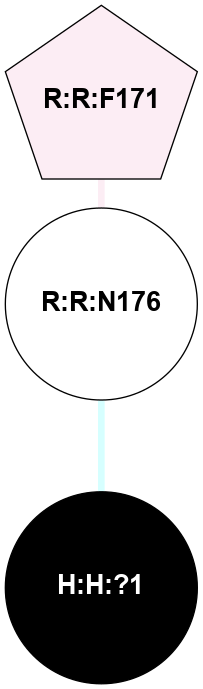
A 2D representation of the interactions of NAG in 6OS0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N176 | 30.7 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:F171 | R:R:N176 | 7.25 | 0 | Yes | No | 6 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 30.70 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6OS2 | A | Peptide | Angiotensin | AT1 | Homo Sapiens | TRV026 | - | - | 2.7 | 2020-02-19 | doi.org/10.1126/science.aay9813 |
|
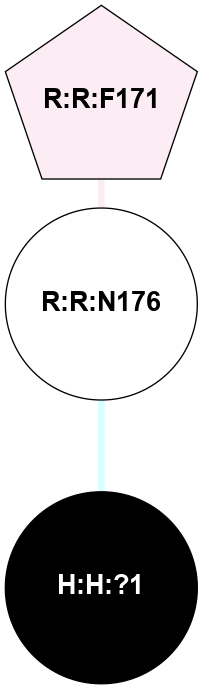
A 2D representation of the interactions of NAG in 6OS2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N176 | 28.24 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:F171 | R:R:N176 | 4.83 | 0 | Yes | No | 6 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 28.24 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6PEL | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | Citronellol | Gt(CT) | 3.19 | 2020-06-24 | To be published |
|
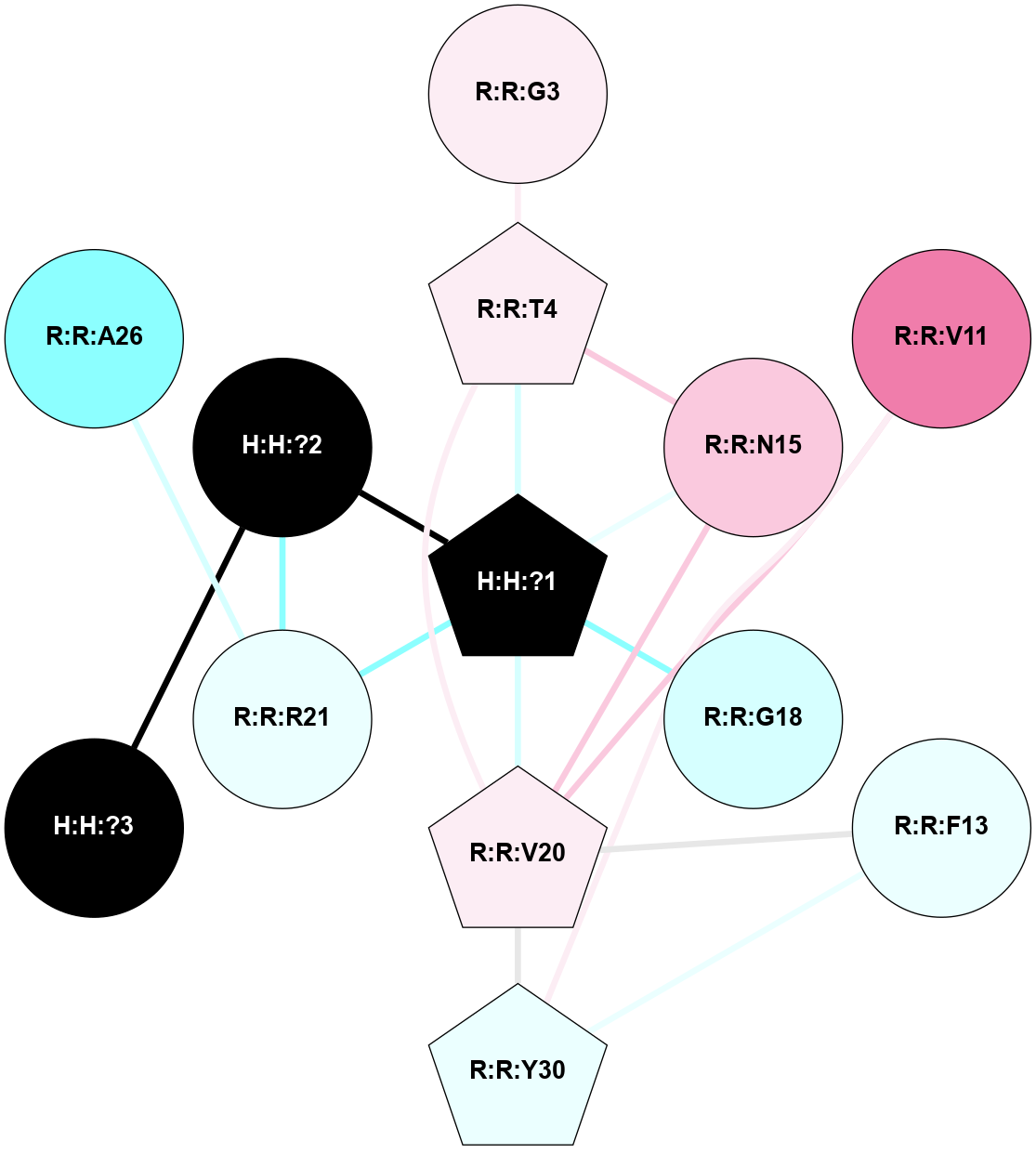
A 2D representation of the interactions of NAG in 6PEL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:G3 | R:R:T4 | 3.64 | 0 | No | Yes | 6 | 6 | 2 | 1 | | R:R:N15 | R:R:T4 | 8.77 | 2 | No | Yes | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 14.28 | 2 | Yes | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 2 | No | Yes | 8 | 6 | 2 | 1 | | R:R:V11 | R:R:Y30 | 5.05 | 2 | No | Yes | 8 | 4 | 2 | 2 | | R:R:F13 | R:R:V20 | 3.93 | 2 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 11.35 | 2 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 2 | No | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 24.56 | 2 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 7.65 | 2 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 8.83 | 2 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:R21 | 5.43 | 2 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:R21 | 7.6 | 2 | No | No | 0 | 4 | 1 | 1 | | H:H:?1 | H:H:?2 | 34.31 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 26.97 | 2 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 13.54 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
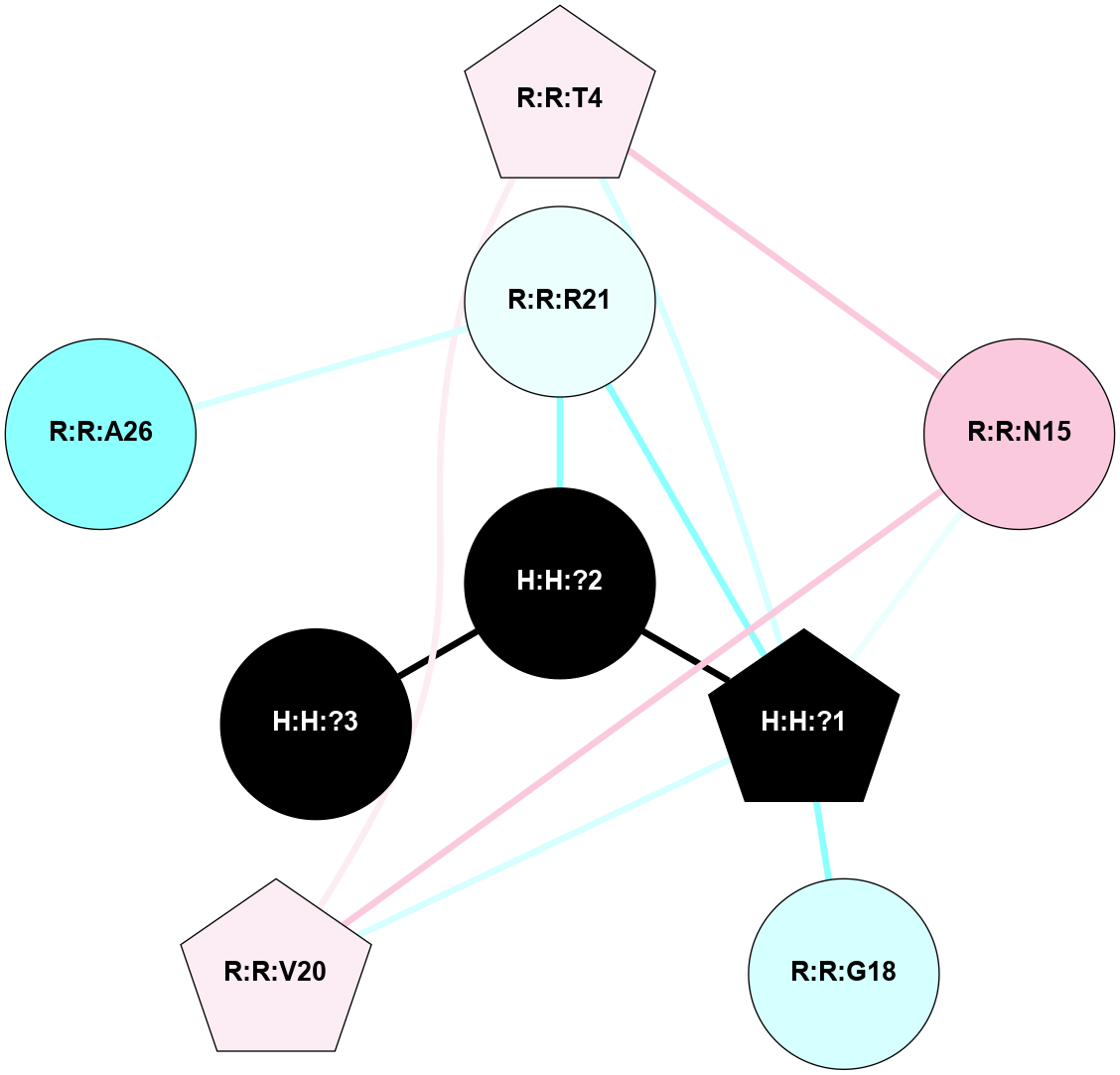
A 2D representation of the interactions of NAG in 6PEL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 2 | No | Yes | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 14.28 | 2 | Yes | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 2 | No | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 24.56 | 2 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 7.65 | 2 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 4 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:R21 | 5.43 | 2 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?2 | R:R:R21 | 7.6 | 2 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 34.31 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 26.97 | 2 | No | No | 0 | 0 | 0 | 1 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 22.96 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6PGS | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | Geraniol | Gt(CT) | 2.9 | 2020-07-01 | To be published |
|
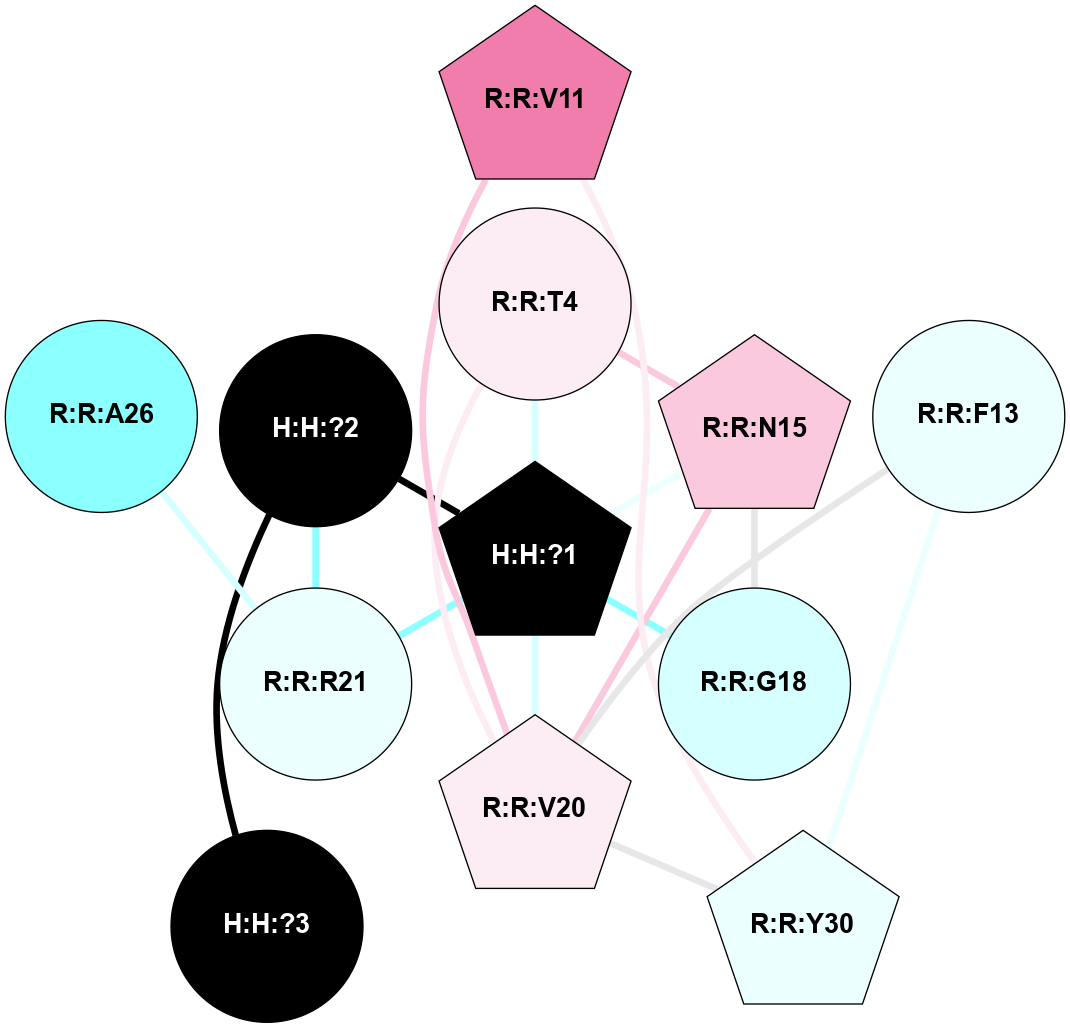
A 2D representation of the interactions of NAG in 6PGS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 11.11 | 1 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:V11 | R:R:Y30 | 5.05 | 1 | Yes | Yes | 8 | 4 | 2 | 2 | | R:R:F13 | R:R:V20 | 5.24 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 8.25 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 3.39 | 1 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 25.79 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 11.36 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 6.66 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:R21 | 5.43 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:R21 | 9.78 | 1 | No | No | 0 | 4 | 1 | 1 | | H:H:?1 | H:H:?2 | 40.96 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 29.54 | 1 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 15.04 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
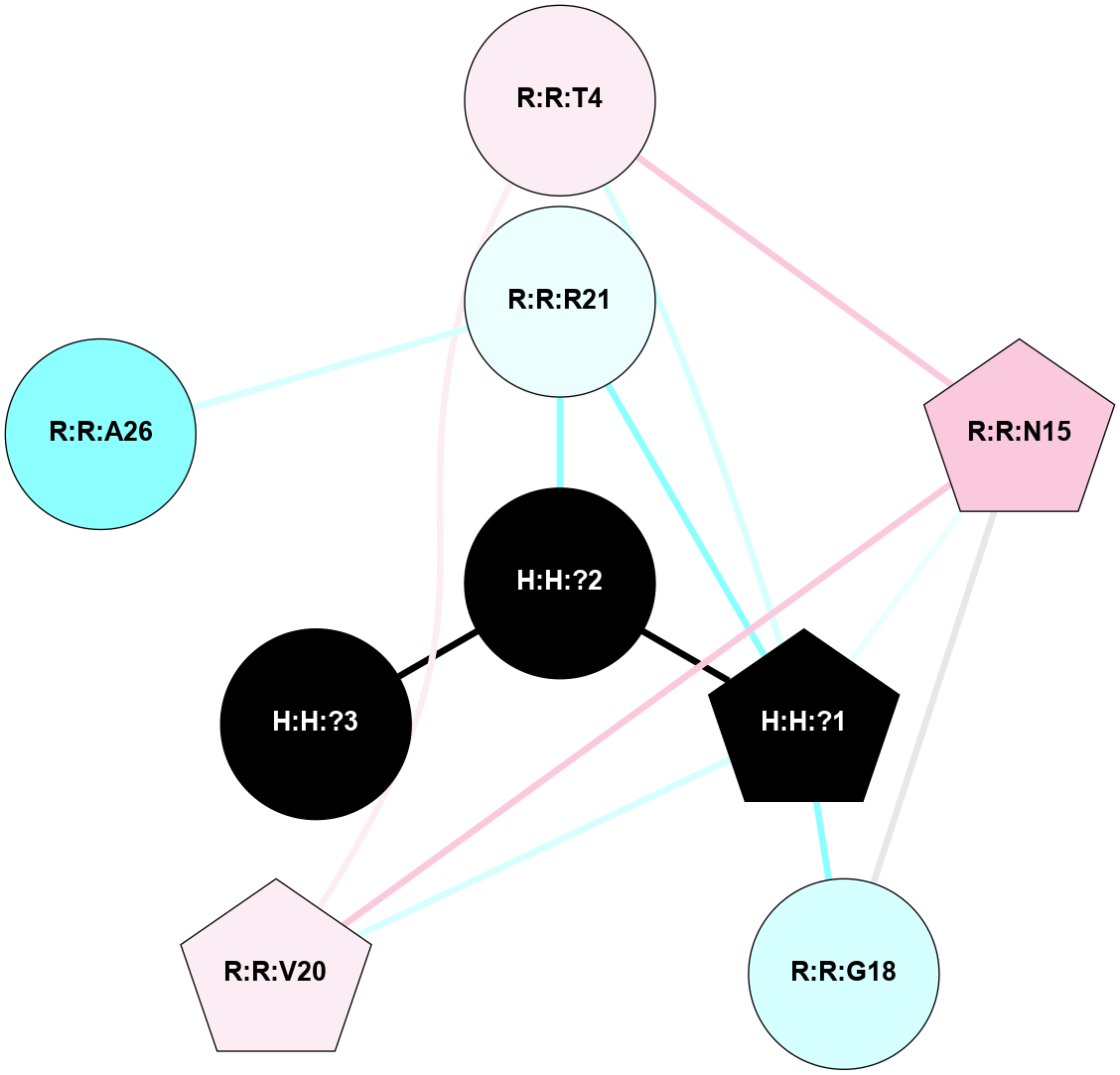
A 2D representation of the interactions of NAG in 6PGS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 1 | Yes | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 11.11 | 1 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:G18 | R:R:N15 | 3.39 | 1 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 25.79 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 6.66 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:R21 | 5.43 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?2 | R:R:R21 | 9.78 | 1 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 40.96 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 29.54 | 1 | No | No | 0 | 0 | 0 | 1 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 26.76 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6PH7 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | Nerol | Gt(CT) | 2.9 | 2020-07-01 | To be published |
|
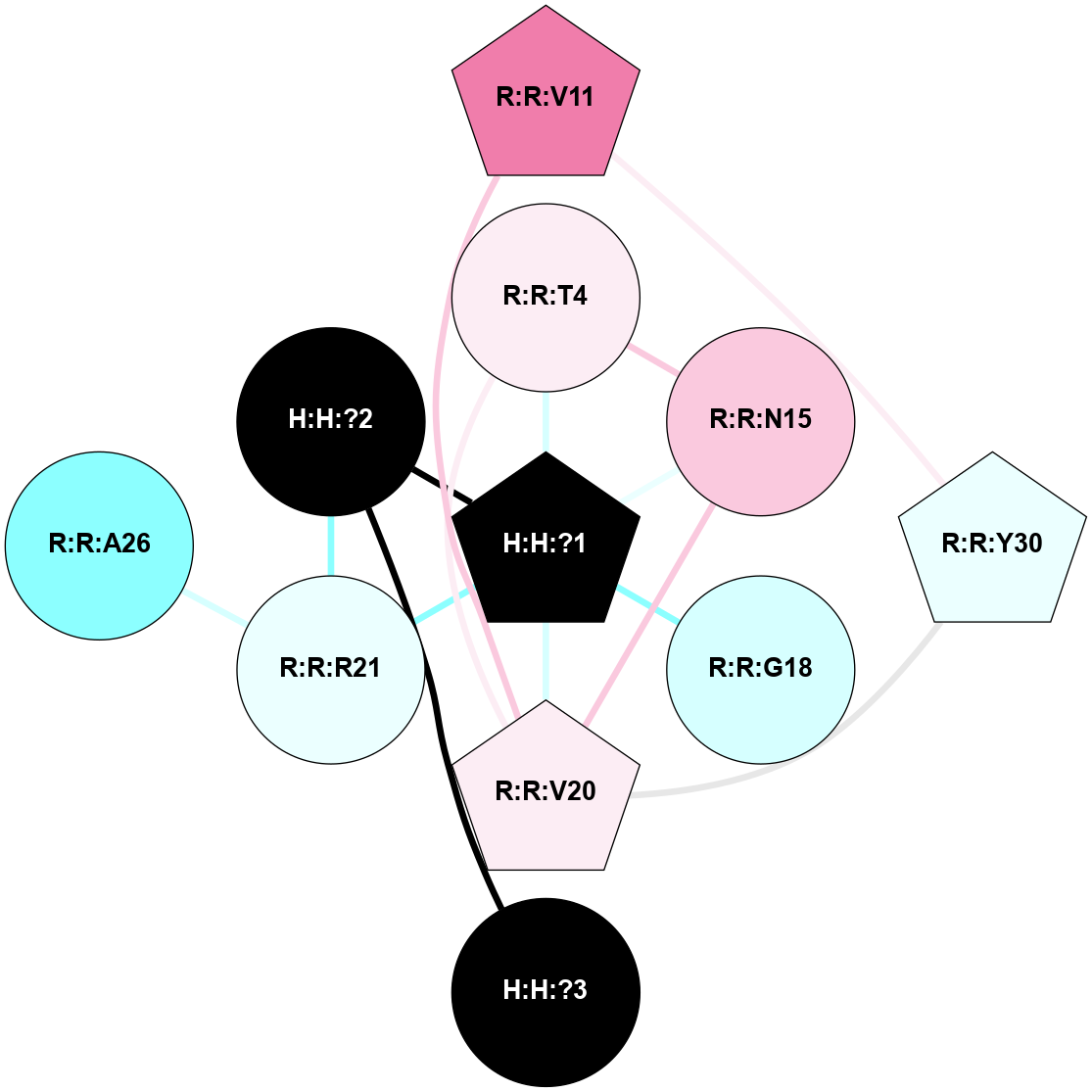
A 2D representation of the interactions of NAG in 6PH7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 2 | No | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 0 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 2 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:V11 | R:R:Y30 | 5.05 | 2 | Yes | Yes | 8 | 4 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 2 | No | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 29.47 | 2 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 0 | 1 | | R:R:V20 | R:R:Y30 | 11.36 | 2 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 5.33 | 2 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:R21 | 4.35 | 2 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:R21 | 8.69 | 2 | No | No | 0 | 4 | 1 | 1 | | H:H:?1 | H:H:?2 | 32.1 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?2 | H:H:?3 | 33.39 | 2 | No | No | 0 | 0 | 1 | 2 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 13.77 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
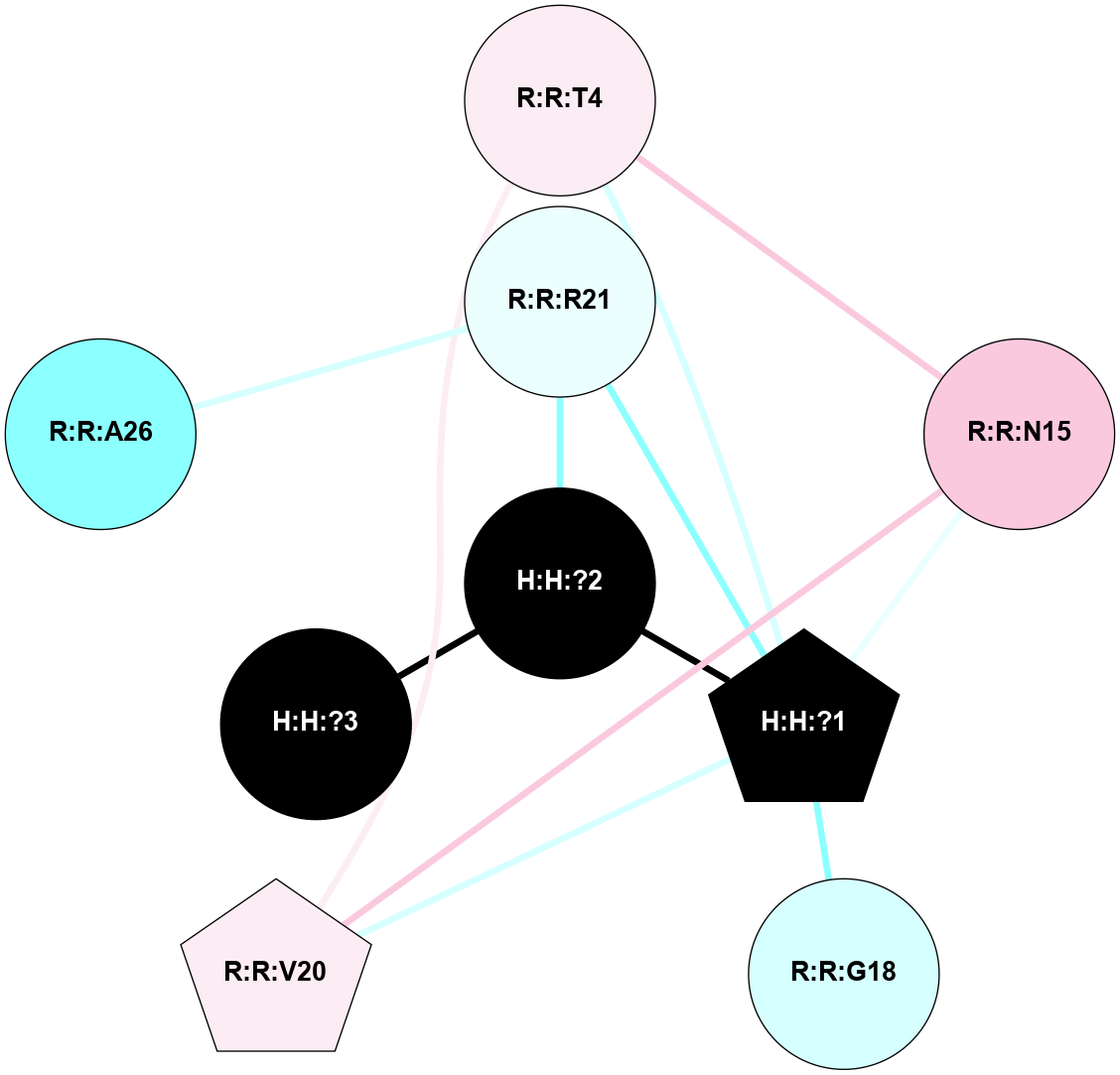
A 2D representation of the interactions of NAG in 6PH7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 8.77 | 2 | No | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 12.69 | 2 | No | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 5.27 | 2 | Yes | No | 0 | 6 | 1 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 2 | No | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 29.47 | 2 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.12 | 2 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 5.33 | 2 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:R21 | 4.35 | 2 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?2 | R:R:R21 | 8.69 | 2 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 32.1 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?2 | H:H:?3 | 33.39 | 2 | No | No | 0 | 0 | 0 | 1 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 24.73 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6QNO | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | Gi1/Beta1/Gamma1 | 4.38 | 2019-07-10 | doi.org/10.7554/eLife.46041 |
|
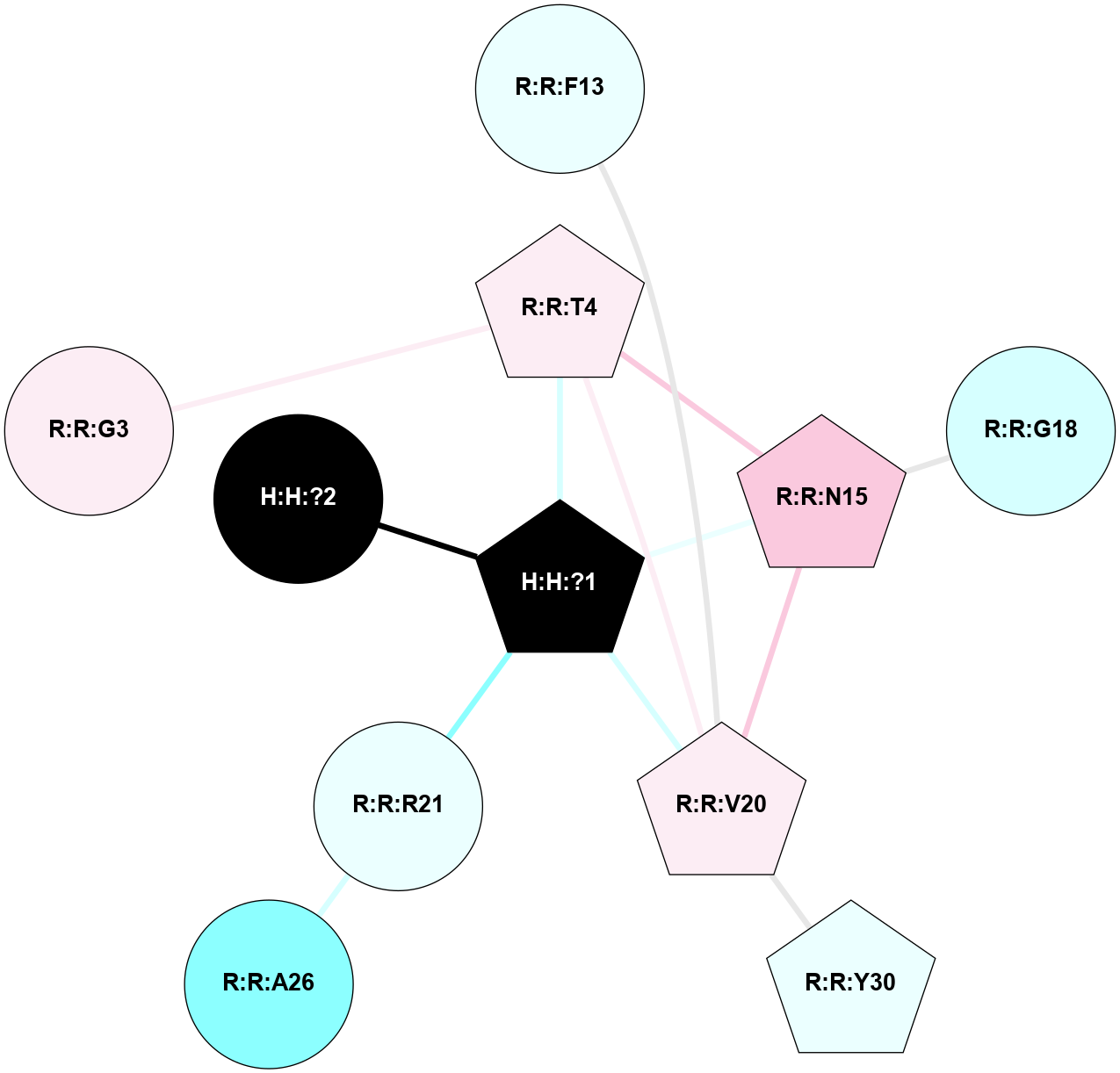
A 2D representation of the interactions of NAG in 6QNO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 11 | Yes | Yes | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 7.93 | 11 | Yes | Yes | 6 | 6 | 1 | 1 | | H:H:?1 | R:R:T4 | 3.95 | 11 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:F13 | R:R:V20 | 3.93 | 0 | No | Yes | 4 | 6 | 2 | 1 | | R:R:G18 | R:R:N15 | 3.39 | 0 | No | Yes | 3 | 7 | 2 | 1 | | R:R:N15 | R:R:V20 | 7.39 | 11 | Yes | Yes | 7 | 6 | 1 | 1 | | H:H:?1 | R:R:N15 | 25.79 | 11 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:V20 | R:R:Y30 | 5.05 | 11 | Yes | Yes | 6 | 4 | 1 | 2 | | H:H:?1 | R:R:V20 | 2.66 | 11 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:A26 | R:R:R21 | 2.77 | 0 | No | No | 2 | 4 | 2 | 1 | | H:H:?1 | R:R:R21 | 21.73 | 11 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 26.57 | 11 | Yes | No | 0 | 0 | 0 | 1 | | R:R:G3 | R:R:T4 | 1.82 | 0 | No | Yes | 6 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 16.14 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
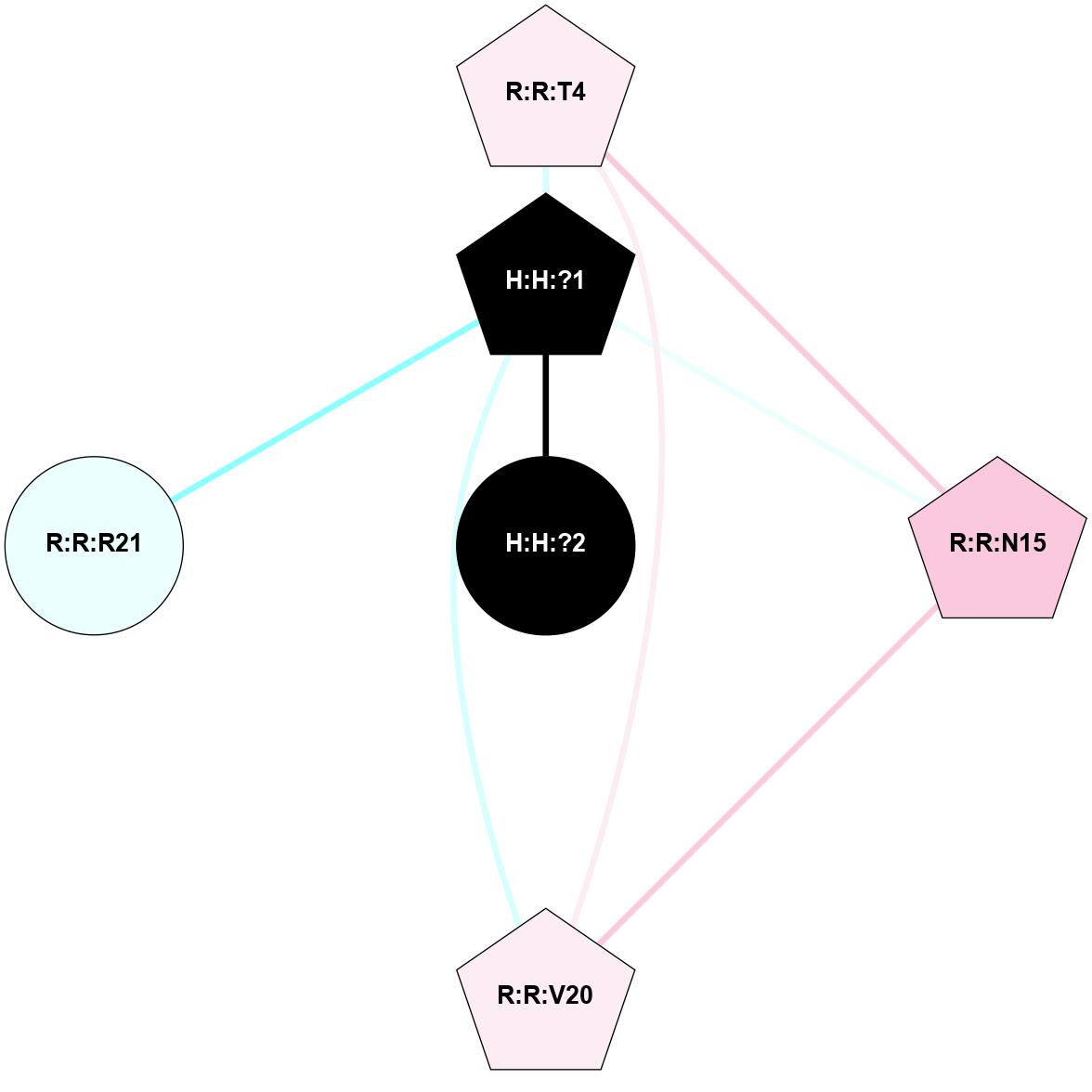
A 2D representation of the interactions of NAG in 6QNO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N15 | R:R:T4 | 7.31 | 11 | Yes | Yes | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 7.93 | 11 | Yes | Yes | 6 | 6 | 2 | 2 | | H:H:?1 | R:R:T4 | 3.95 | 11 | Yes | Yes | 0 | 6 | 1 | 2 | | R:R:N15 | R:R:V20 | 7.39 | 11 | Yes | Yes | 7 | 6 | 2 | 2 | | H:H:?1 | R:R:N15 | 25.79 | 11 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:V20 | 2.66 | 11 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:R21 | 21.73 | 11 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 26.57 | 11 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 26.57 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6UO8 | C | Aminoacid | GABAB | GABAB1; GABAB2 | Homo Sapiens | SKF97541 | GS39783 | - | 3.63 | 2020-06-10 | doi.org/10.1038/s41586-020-2408-4 |
|
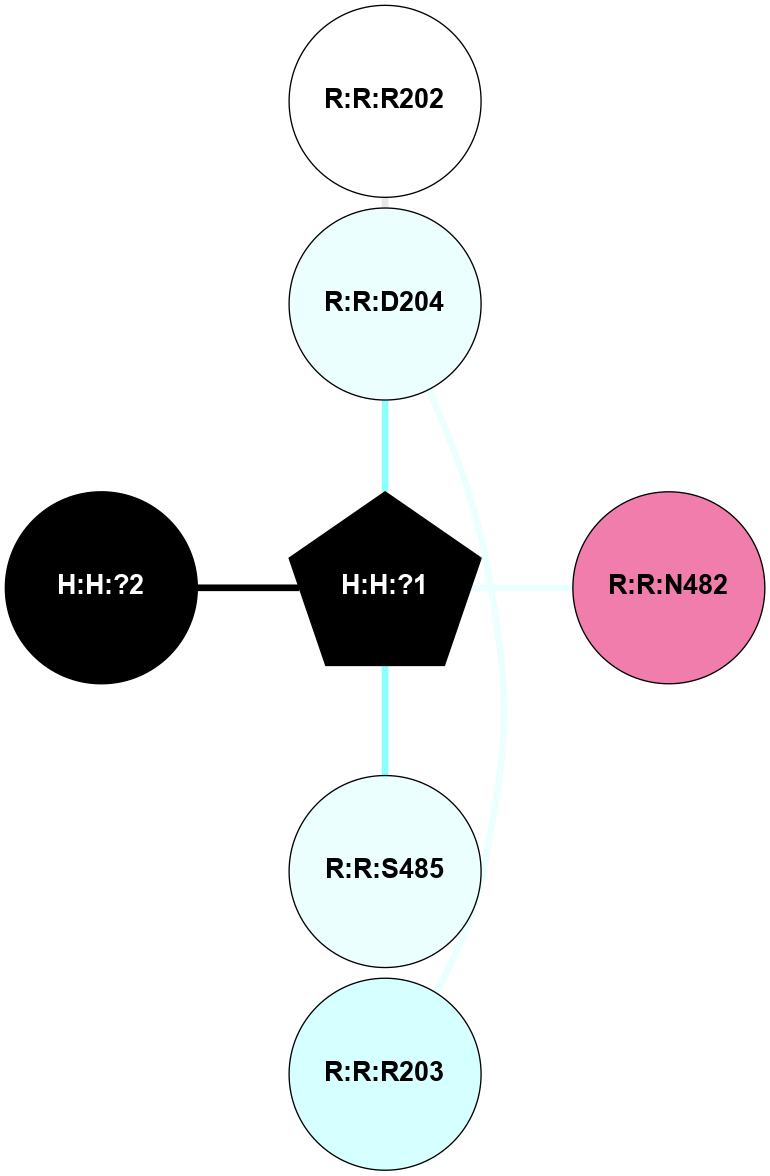
A 2D representation of the interactions of NAG in 6UO8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 34.31 | 0 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:D204 | 9.71 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:N482 | 25.79 | 0 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?1 | R:R:S485 | 2.69 | 0 | Yes | No | 0 | 4 | 0 | 1 | | R:R:D204 | R:R:R202 | 2.38 | 0 | No | No | 4 | 5 | 1 | 2 | | R:R:D204 | R:R:R203 | 2.38 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.12 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
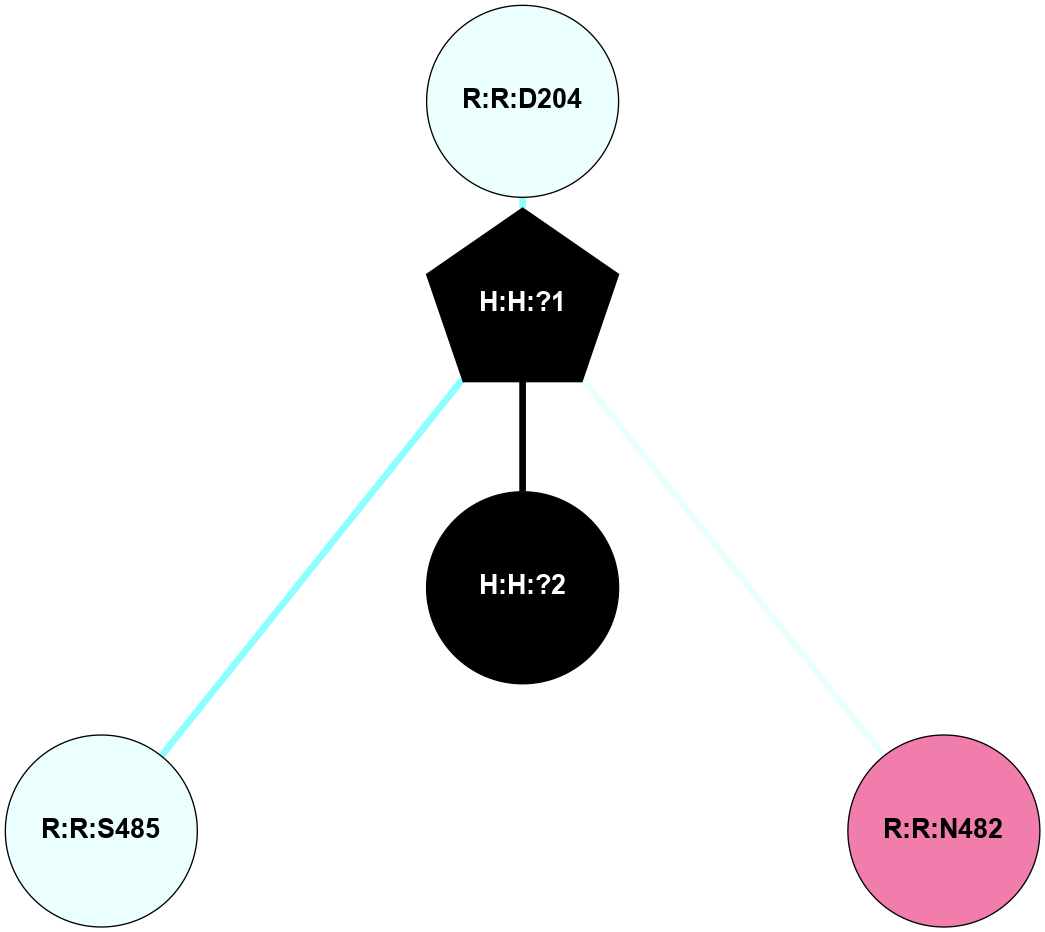
A 2D representation of the interactions of NAG in 6UO8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 34.31 | 0 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:D204 | 9.71 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:N482 | 25.79 | 0 | Yes | No | 0 | 8 | 1 | 2 | | H:H:?1 | R:R:S485 | 2.69 | 0 | Yes | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 34.31 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
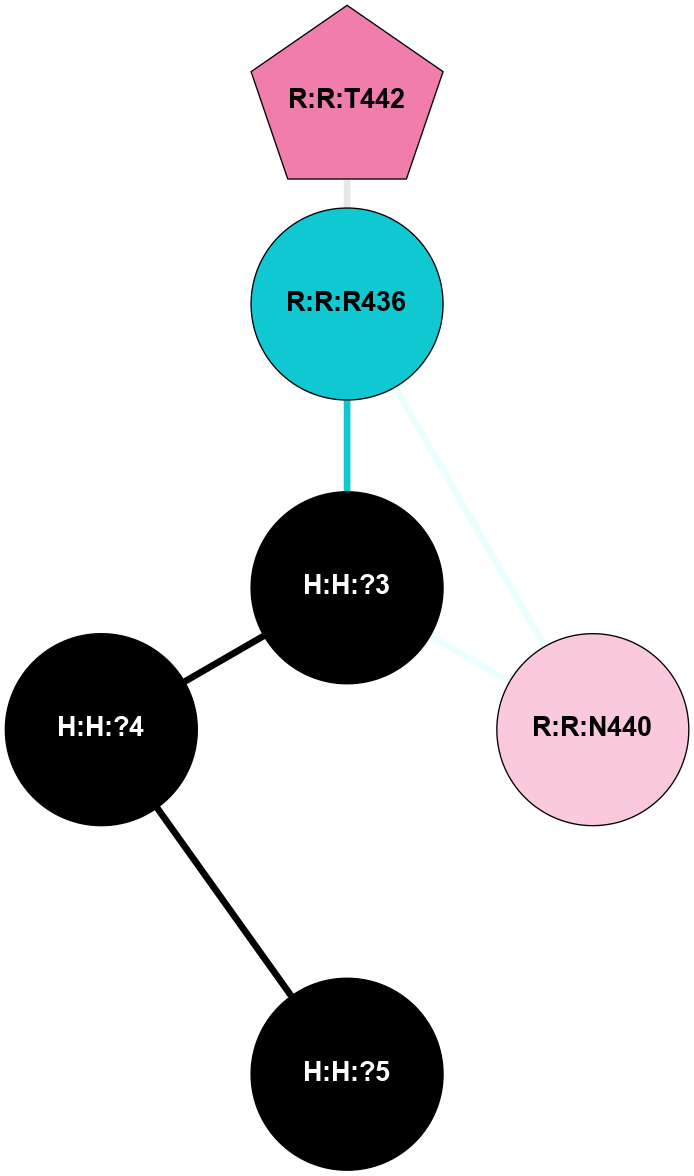
A 2D representation of the interactions of NAG in 6UO8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 30.99 | 11 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:R436 | 8.69 | 11 | No | No | 0 | 1 | 0 | 1 | | H:H:?3 | R:R:N440 | 29.47 | 11 | No | No | 0 | 7 | 0 | 1 | | H:H:?4 | H:H:?5 | 37.24 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:N440 | R:R:R436 | 4.82 | 11 | No | No | 7 | 1 | 1 | 1 | | R:R:R436 | R:R:T442 | 5.17 | 11 | No | Yes | 1 | 8 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 23.05 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6UO8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 30.99 | 11 | No | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:R436 | 8.69 | 11 | No | No | 0 | 1 | 1 | 2 | | H:H:?3 | R:R:N440 | 29.47 | 11 | No | No | 0 | 7 | 1 | 2 | | H:H:?4 | H:H:?5 | 37.24 | 0 | No | No | 0 | 0 | 0 | 1 | | R:R:N440 | R:R:R436 | 4.82 | 11 | No | No | 7 | 1 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 34.12 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
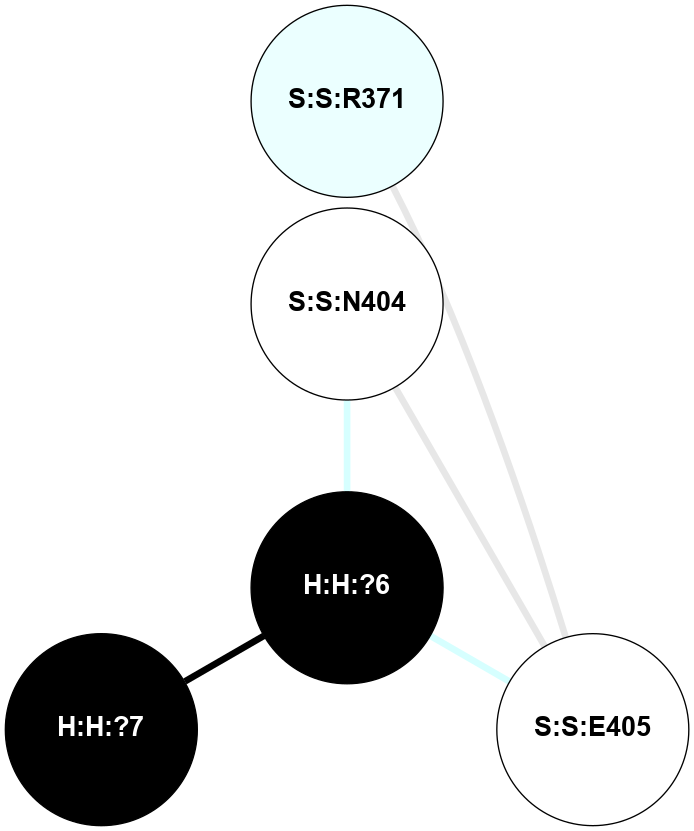
A 2D representation of the interactions of NAG in 6UO8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | H:H:?7 | 38.74 | 33 | No | No | 0 | 0 | 0 | 1 | | H:H:?6 | S:S:N404 | 23.33 | 33 | No | No | 0 | 5 | 0 | 1 | | H:H:?6 | S:S:E405 | 3.55 | 33 | No | No | 0 | 5 | 0 | 1 | | S:S:E405 | S:S:R371 | 2.33 | 33 | No | No | 5 | 4 | 1 | 2 | | S:S:E405 | S:S:N404 | 6.57 | 33 | No | No | 5 | 5 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 21.87 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6UO8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | H:H:?7 | 38.74 | 33 | No | No | 0 | 0 | 1 | 0 | | H:H:?6 | S:S:N404 | 23.33 | 33 | No | No | 0 | 5 | 1 | 2 | | H:H:?6 | S:S:E405 | 3.55 | 33 | No | No | 0 | 5 | 1 | 2 | | S:S:E405 | S:S:N404 | 6.57 | 33 | No | No | 5 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 38.74 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
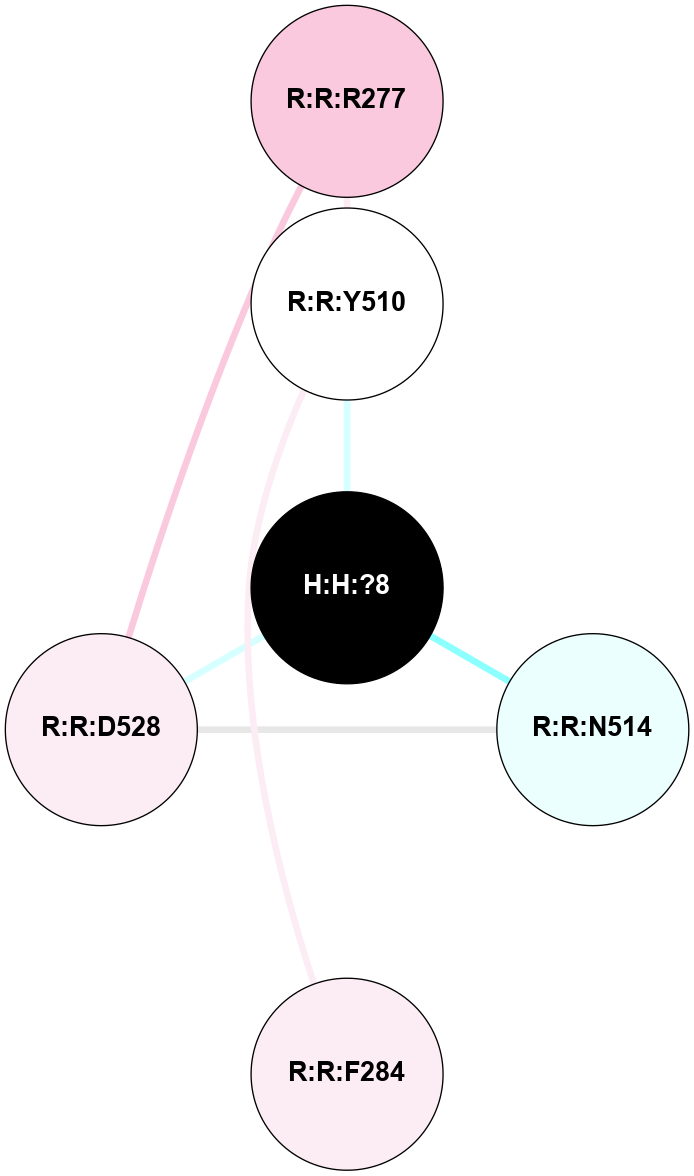
A 2D representation of the interactions of NAG in 6UO8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | R:R:Y510 | 2.1 | 34 | No | No | 0 | 5 | 0 | 1 | | H:H:?8 | R:R:N514 | 25.79 | 34 | No | No | 0 | 4 | 0 | 1 | | H:H:?8 | R:R:D528 | 3.64 | 34 | No | No | 0 | 6 | 0 | 1 | | R:R:R277 | R:R:Y510 | 4.12 | 0 | No | No | 7 | 5 | 2 | 1 | | R:R:D528 | R:R:R277 | 9.53 | 34 | No | No | 6 | 7 | 1 | 2 | | R:R:F284 | R:R:Y510 | 9.28 | 37 | No | No | 6 | 5 | 2 | 1 | | R:R:D528 | R:R:N514 | 9.42 | 34 | No | No | 6 | 4 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 10.51 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
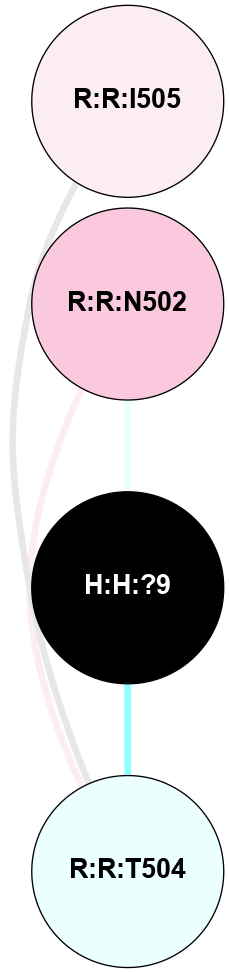
A 2D representation of the interactions of NAG in 6UO8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?9 | R:R:N502 | 29.47 | 35 | No | No | 0 | 7 | 0 | 1 | | H:H:?9 | R:R:T504 | 10.55 | 35 | No | No | 0 | 4 | 0 | 1 | | R:R:N502 | R:R:T504 | 8.77 | 35 | No | No | 7 | 4 | 1 | 1 | | R:R:I505 | R:R:T504 | 1.52 | 0 | No | No | 6 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.01 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
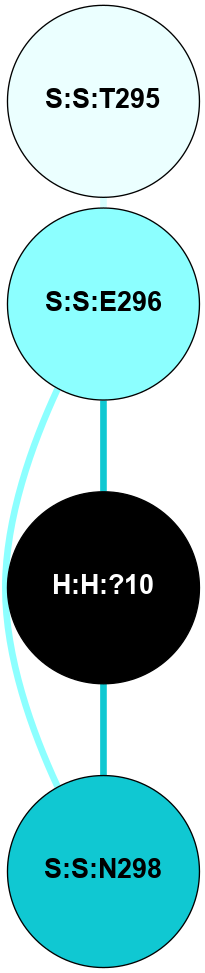
A 2D representation of the interactions of NAG in 6UO8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | S:S:H294 | 4.6 | 28 | No | Yes | 0 | 4 | 0 | 1 | | H:H:?10 | S:S:E296 | 14.22 | 28 | No | No | 0 | 2 | 0 | 1 | | H:H:?10 | S:S:N298 | 31.93 | 28 | No | No | 0 | 1 | 0 | 1 | | S:S:H294 | S:S:N298 | 7.65 | 28 | Yes | No | 4 | 1 | 1 | 1 | | S:S:H294 | S:S:S299 | 13.95 | 28 | Yes | No | 4 | 3 | 1 | 2 | | S:S:E296 | S:S:N298 | 6.57 | 28 | No | No | 2 | 1 | 1 | 1 | | S:S:E296 | S:S:T295 | 1.41 | 28 | No | No | 2 | 4 | 1 | 2 | | S:S:H294 | S:S:R304 | 1.13 | 28 | Yes | No | 4 | 1 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 16.92 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
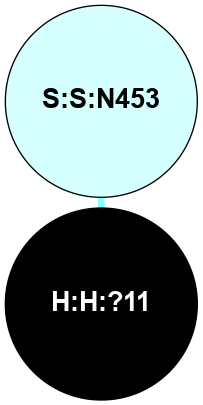
A 2D representation of the interactions of NAG in 6UO8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | S:S:N453 | 27.02 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
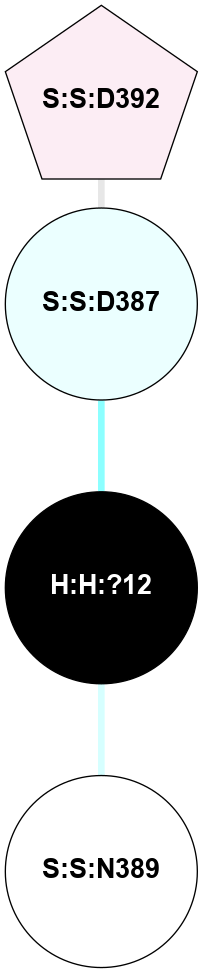
A 2D representation of the interactions of NAG in 6UO8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | S:S:D387 | 6.07 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?12 | S:S:N389 | 28.24 | 0 | No | No | 0 | 5 | 0 | 1 | | S:S:D387 | S:S:D392 | 3.99 | 0 | No | Yes | 4 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.16 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 6UO8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | S:S:N90 | 34.38 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 34.38 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6UO9 | C | Aminoacid | GABAB | GABAB1; GABAB2 | Homo Sapiens | SKF97541 | - | - | 4.8 | 2020-06-10 | doi.org/10.1038/s41586-020-2408-4 |
|
A 2D representation of the interactions of NAG in 6UO9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 44.28 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N409 | 29.47 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 36.88 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 6UO9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 44.28 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N409 | 29.47 | 0 | No | No | 0 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 44.28 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
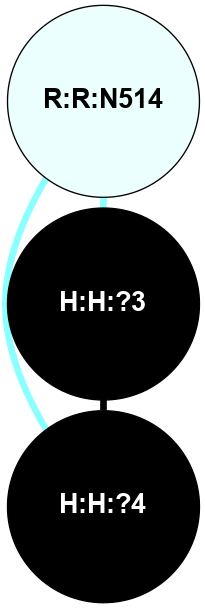
A 2D representation of the interactions of NAG in 6UO9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 49.81 | 24 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:N514 | 28.24 | 24 | No | No | 0 | 4 | 0 | 1 | | H:H:?4 | R:R:N514 | 2.46 | 24 | No | No | 0 | 4 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 39.02 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6UO9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 49.81 | 24 | No | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:N514 | 28.24 | 24 | No | No | 0 | 4 | 1 | 1 | | H:H:?4 | R:R:N514 | 2.46 | 24 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 26.14 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
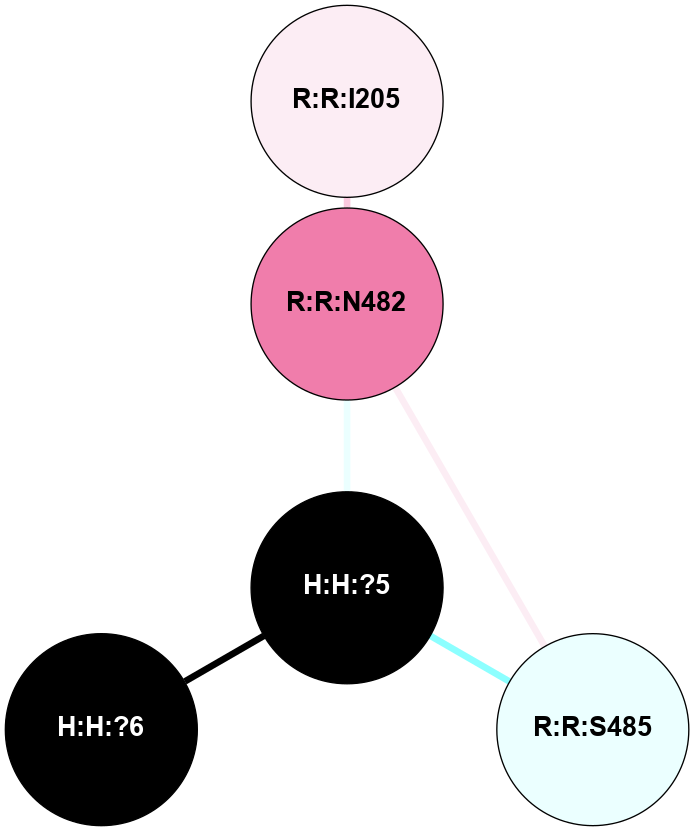
A 2D representation of the interactions of NAG in 6UO9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 32.1 | 25 | No | No | 0 | 0 | 0 | 1 | | H:H:?5 | R:R:N482 | 27.02 | 25 | No | No | 0 | 8 | 0 | 1 | | H:H:?5 | R:R:S485 | 5.37 | 25 | No | No | 0 | 4 | 0 | 1 | | R:R:N482 | R:R:S485 | 2.98 | 25 | No | No | 8 | 4 | 1 | 1 | | R:R:I205 | R:R:N482 | 1.42 | 0 | No | No | 6 | 8 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 21.50 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
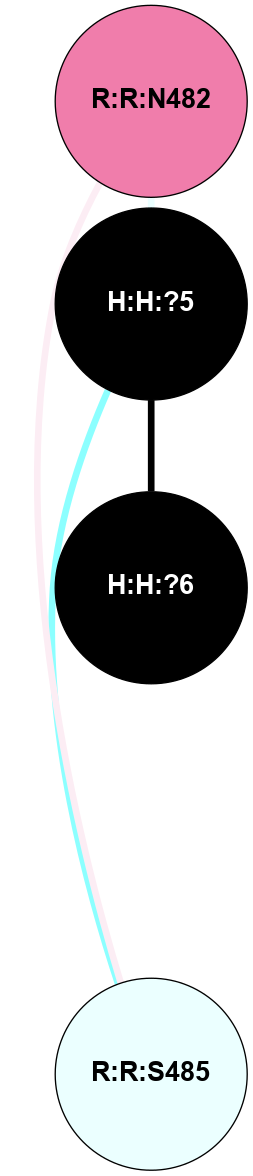
A 2D representation of the interactions of NAG in 6UO9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 32.1 | 25 | No | No | 0 | 0 | 1 | 0 | | H:H:?5 | R:R:N482 | 27.02 | 25 | No | No | 0 | 8 | 1 | 2 | | H:H:?5 | R:R:S485 | 5.37 | 25 | No | No | 0 | 4 | 1 | 2 | | R:R:N482 | R:R:S485 | 2.98 | 25 | No | No | 8 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 32.10 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
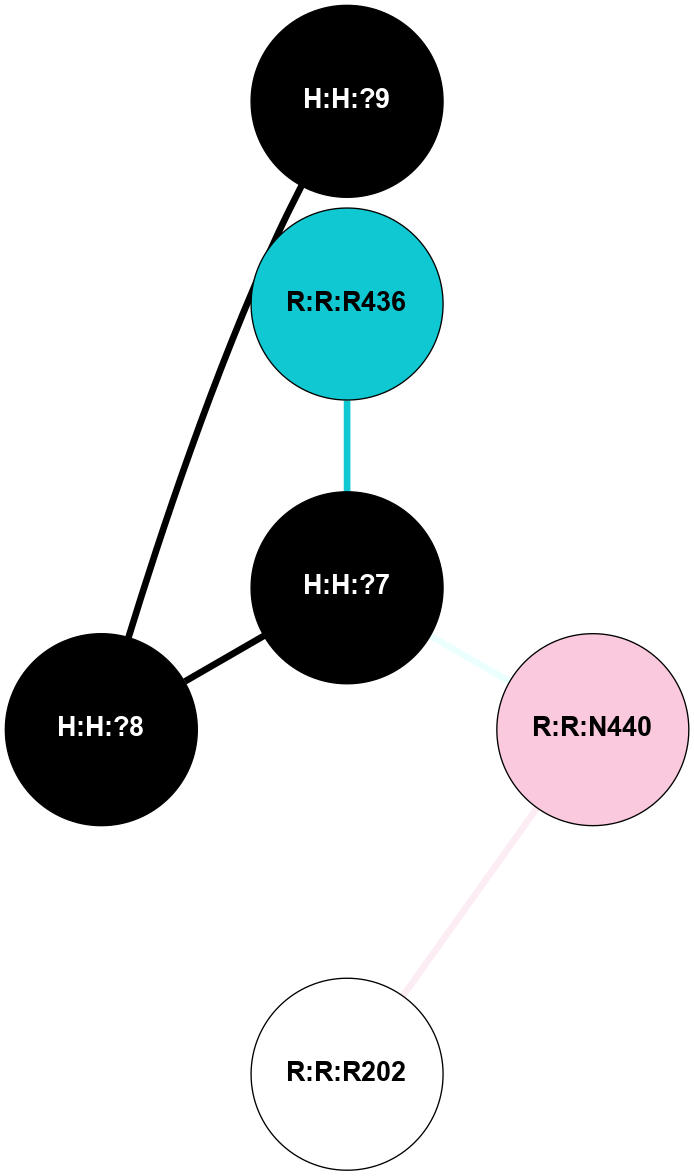
A 2D representation of the interactions of NAG in 6UO9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 34.31 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?7 | R:R:R436 | 4.35 | 0 | No | No | 0 | 1 | 0 | 1 | | H:H:?7 | R:R:N440 | 31.93 | 0 | No | No | 0 | 7 | 0 | 1 | | H:H:?8 | H:H:?9 | 33.39 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:N440 | R:R:R202 | 1.21 | 0 | No | No | 7 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 23.53 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
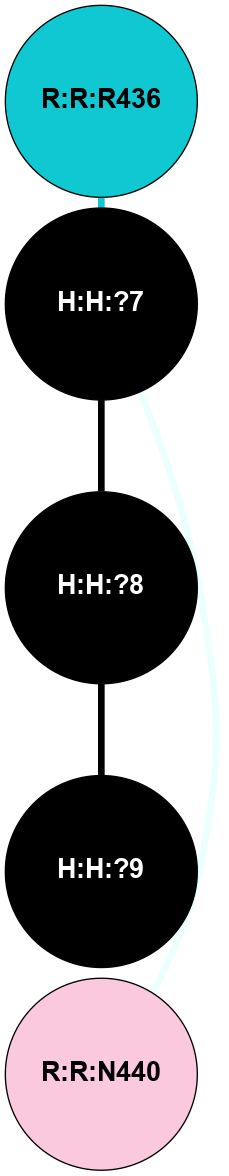
A 2D representation of the interactions of NAG in 6UO9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 34.31 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?7 | R:R:R436 | 4.35 | 0 | No | No | 0 | 1 | 1 | 2 | | H:H:?7 | R:R:N440 | 31.93 | 0 | No | No | 0 | 7 | 1 | 2 | | H:H:?8 | H:H:?9 | 33.39 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 33.85 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
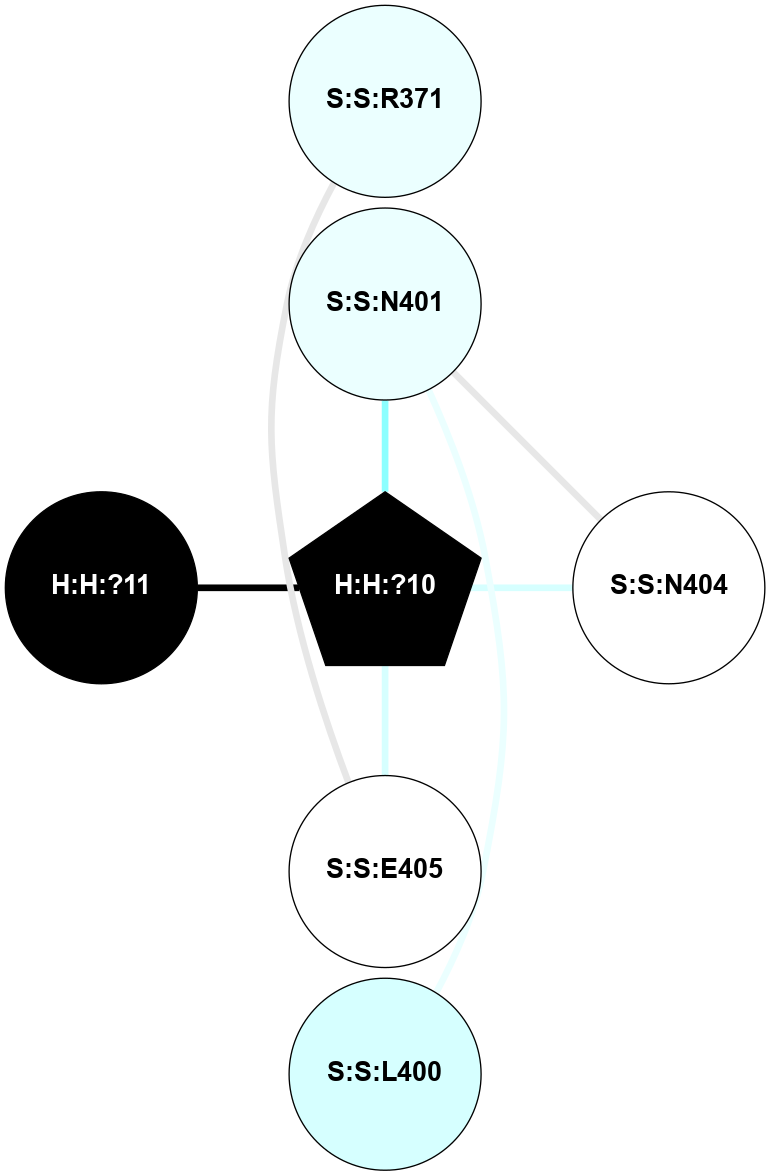
A 2D representation of the interactions of NAG in 6UO9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | H:H:?11 | 49.81 | 26 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?10 | S:S:N401 | 9.82 | 26 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?10 | S:S:N404 | 29.47 | 26 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?10 | S:S:E405 | 2.37 | 26 | Yes | No | 0 | 5 | 0 | 1 | | S:S:E405 | S:S:R371 | 12.79 | 0 | No | No | 5 | 4 | 1 | 2 | | S:S:N401 | S:S:N404 | 2.72 | 26 | No | No | 4 | 5 | 1 | 1 | | S:S:L400 | S:S:N401 | 1.37 | 0 | No | No | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 22.87 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
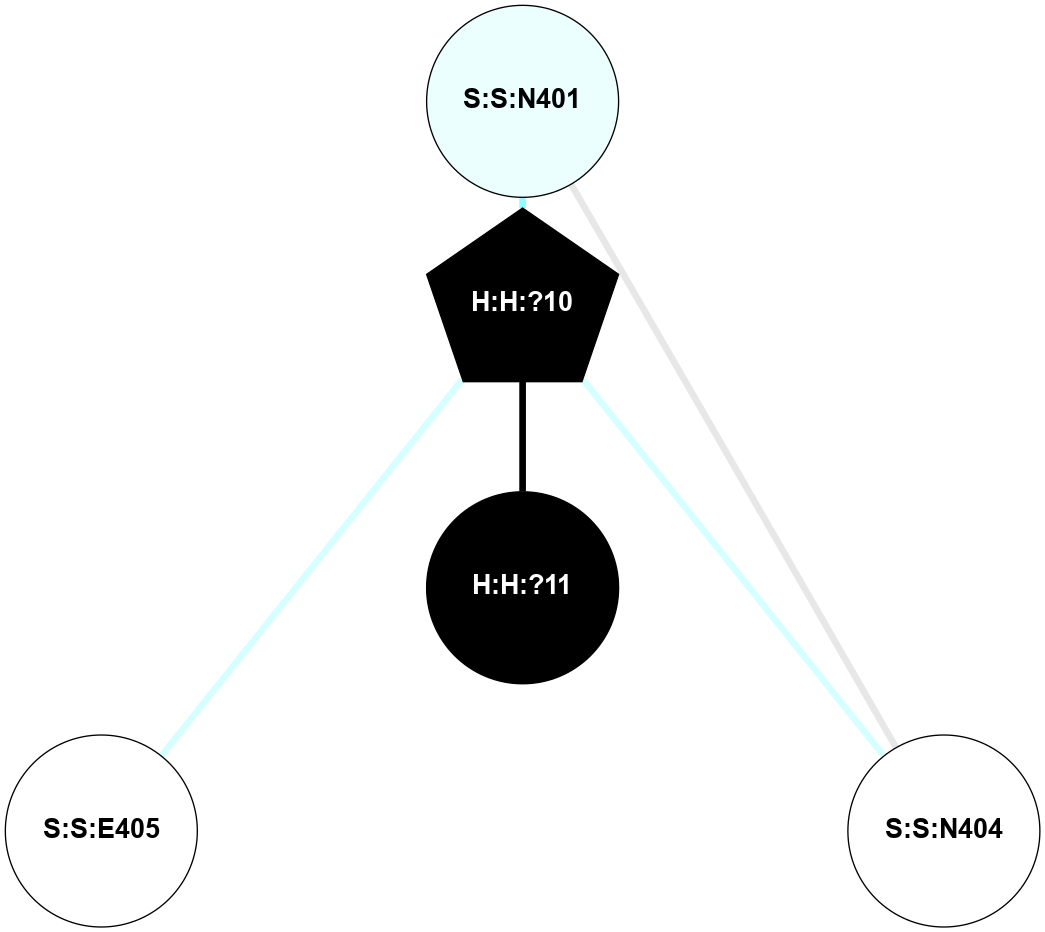
A 2D representation of the interactions of NAG in 6UO9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | H:H:?11 | 49.81 | 26 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?10 | S:S:N401 | 9.82 | 26 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?10 | S:S:N404 | 29.47 | 26 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?10 | S:S:E405 | 2.37 | 26 | Yes | No | 0 | 5 | 1 | 2 | | S:S:N401 | S:S:N404 | 2.72 | 26 | No | No | 4 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 49.81 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
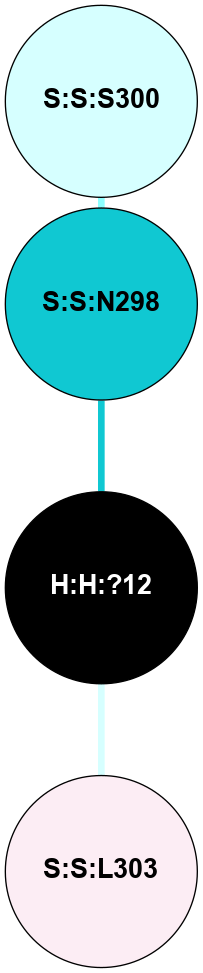
A 2D representation of the interactions of NAG in 6UO9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | S:S:N298 | 31.93 | 0 | No | No | 0 | 1 | 0 | 1 | | S:S:H294 | S:S:S299 | 2.79 | 0 | No | No | 4 | 3 | 1 | 2 | | S:S:H294 | S:S:R304 | 2.26 | 0 | No | No | 4 | 1 | 1 | 2 | | S:S:N298 | S:S:S300 | 4.47 | 0 | No | No | 1 | 3 | 1 | 2 | | H:H:?12 | S:S:L303 | 1.24 | 0 | No | No | 0 | 6 | 0 | 1 | | H:H:?12 | S:S:H294 | 1.15 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 11.44 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6UO9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | S:S:N453 | 33.15 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?13 | S:S:D454 | 1.21 | 0 | No | No | 0 | 2 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.18 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6UOA | C | Aminoacid | GABAB | GABAB1; GABAB2 | Homo Sapiens | - | - | - | 6.3 | 2020-06-10 | doi.org/10.1038/s41586-020-2408-4 |
|
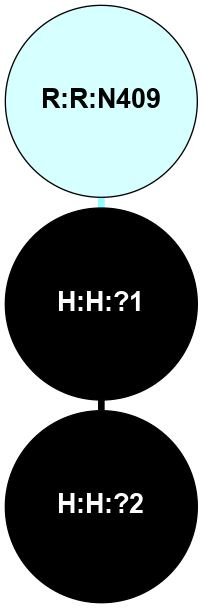
A 2D representation of the interactions of NAG in 6UOA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 52.03 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N409 | 31.93 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 41.98 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
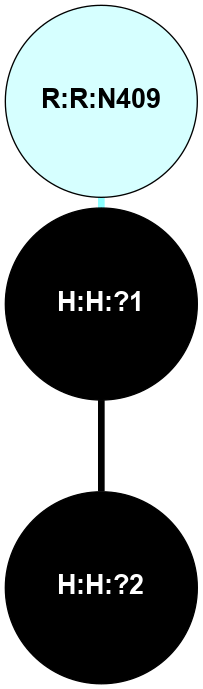
A 2D representation of the interactions of NAG in 6UOA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 52.03 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N409 | 31.93 | 0 | No | No | 0 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 52.03 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
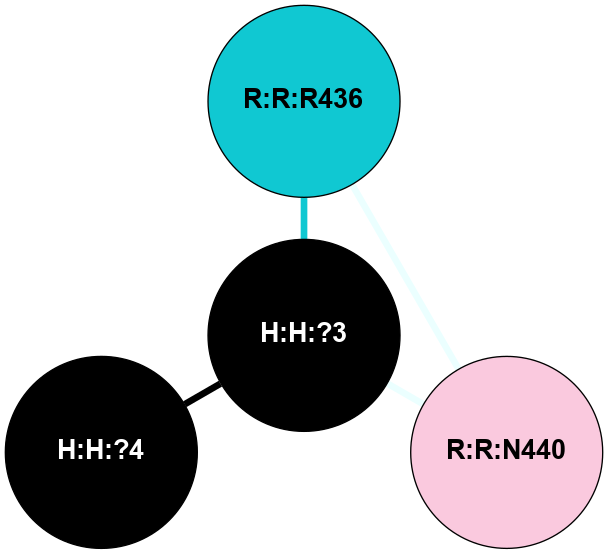
A 2D representation of the interactions of NAG in 6UOA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 44.28 | 34 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:R436 | 11.95 | 34 | No | No | 0 | 1 | 0 | 1 | | H:H:?3 | R:R:N440 | 31.93 | 34 | No | No | 0 | 7 | 0 | 1 | | R:R:N440 | R:R:R436 | 9.64 | 34 | No | No | 7 | 1 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 29.39 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6UOA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 44.28 | 34 | No | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:R436 | 11.95 | 34 | No | No | 0 | 1 | 1 | 2 | | H:H:?3 | R:R:N440 | 31.93 | 34 | No | No | 0 | 7 | 1 | 2 | | R:R:N440 | R:R:R436 | 9.64 | 34 | No | No | 7 | 1 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 44.28 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
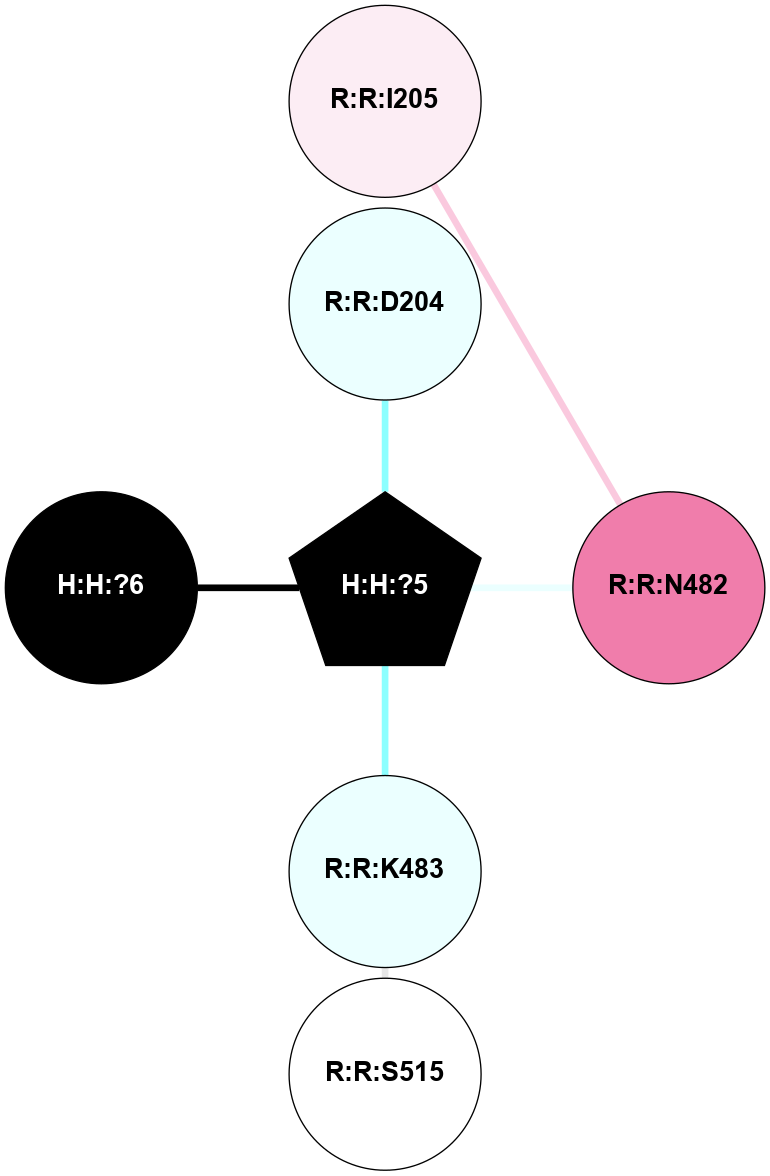
A 2D representation of the interactions of NAG in 6UOA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 46.49 | 0 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?5 | R:R:D204 | 4.85 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?5 | R:R:N482 | 28.24 | 0 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?5 | R:R:K483 | 6.31 | 0 | Yes | No | 0 | 4 | 0 | 1 | | R:R:I205 | R:R:N482 | 9.91 | 0 | No | No | 6 | 8 | 2 | 1 | | R:R:K483 | R:R:S515 | 4.59 | 0 | No | No | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 21.47 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
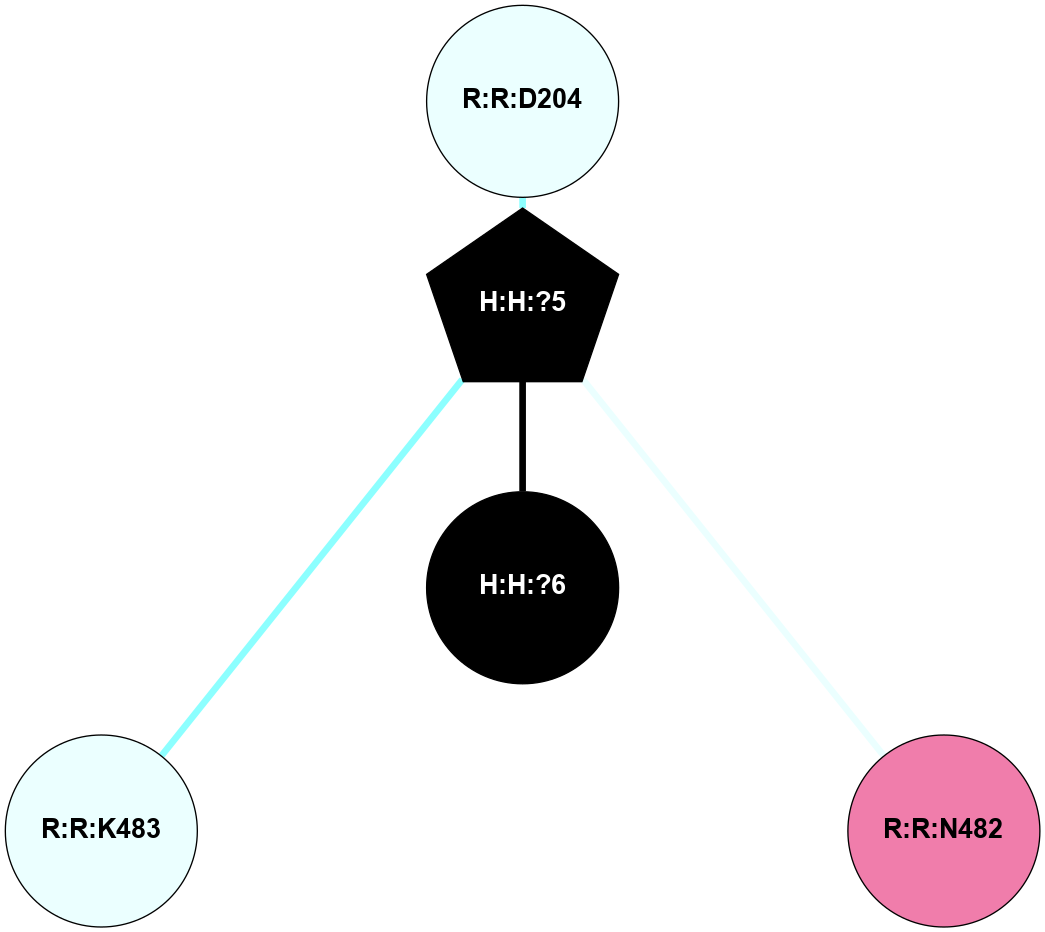
A 2D representation of the interactions of NAG in 6UOA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 46.49 | 0 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?5 | R:R:D204 | 4.85 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?5 | R:R:N482 | 28.24 | 0 | Yes | No | 0 | 8 | 1 | 2 | | H:H:?5 | R:R:K483 | 6.31 | 0 | Yes | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 46.49 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
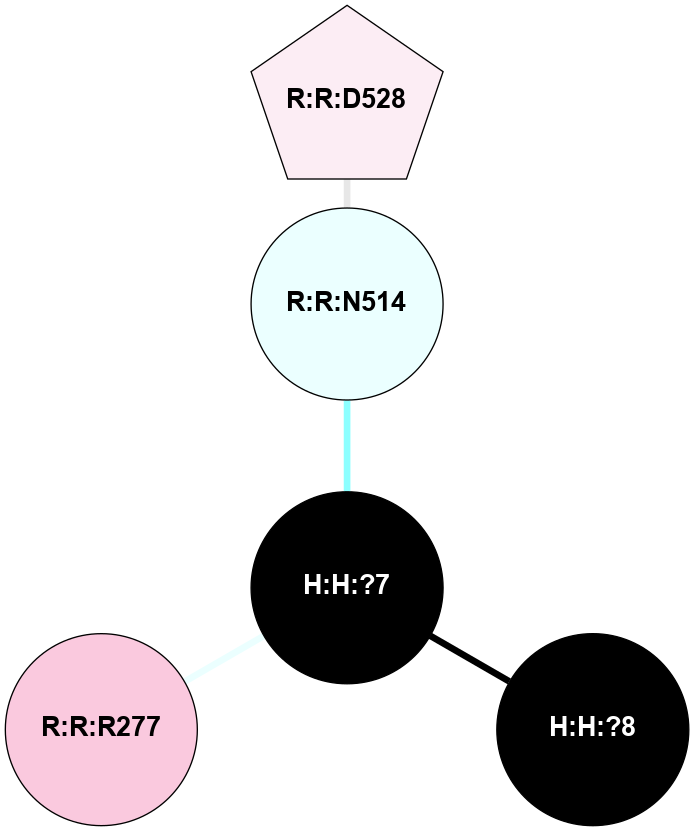
A 2D representation of the interactions of NAG in 6UOA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 47.6 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?7 | R:R:N514 | 28.24 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:D528 | R:R:N514 | 10.77 | 12 | Yes | No | 6 | 4 | 2 | 1 | | H:H:?7 | R:R:R277 | 1.09 | 0 | No | No | 0 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 25.64 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
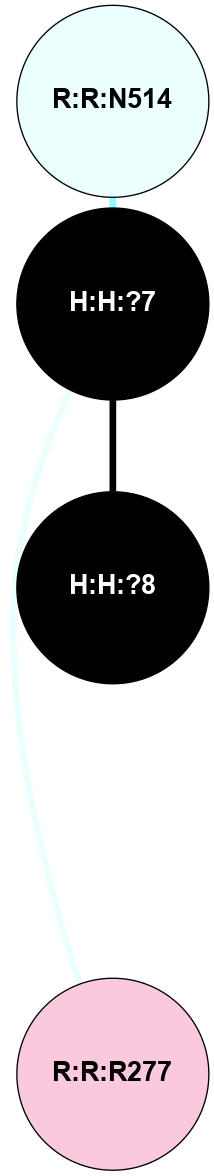
A 2D representation of the interactions of NAG in 6UOA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 47.6 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?7 | R:R:N514 | 28.24 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?7 | R:R:R277 | 1.09 | 0 | No | No | 0 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 47.60 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
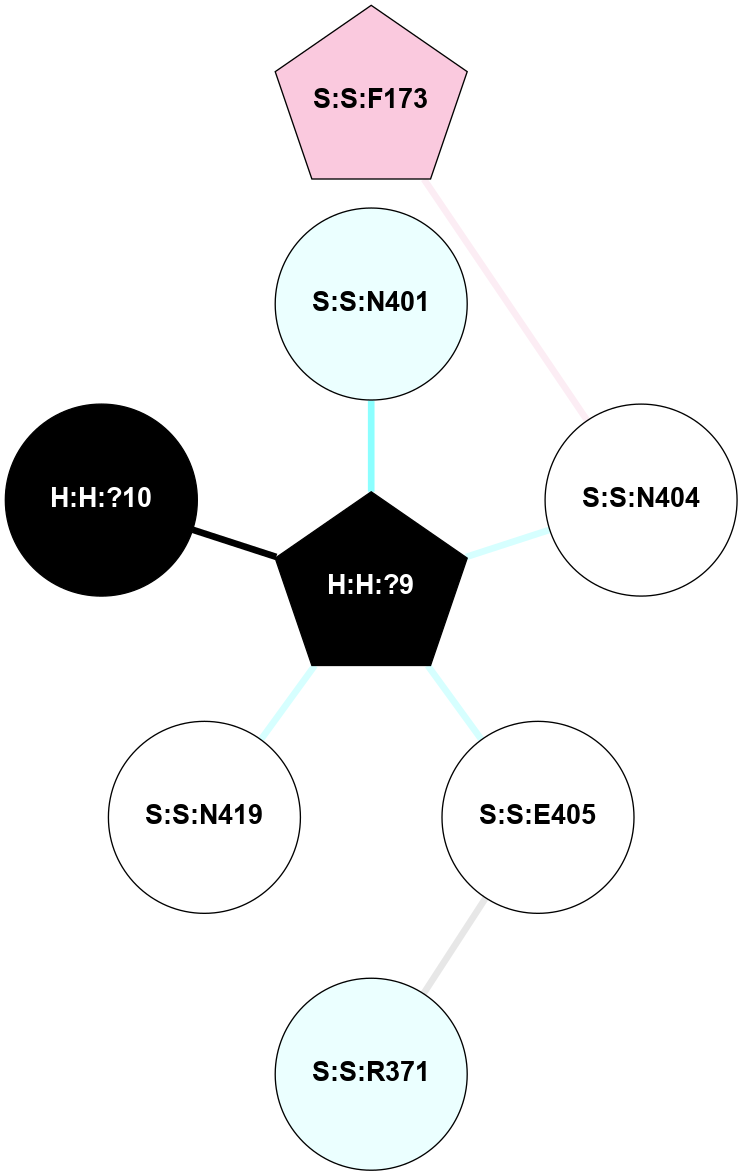
A 2D representation of the interactions of NAG in 6UOA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | H:H:?9 | 42.06 | 0 | No | Yes | 0 | 0 | 1 | 0 | | H:H:?9 | S:S:N401 | 3.68 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?9 | S:S:N404 | 33.15 | 0 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?9 | S:S:E405 | 2.37 | 0 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?9 | S:S:N419 | 9.82 | 0 | Yes | No | 0 | 5 | 0 | 1 | | S:S:F173 | S:S:N404 | 3.62 | 15 | Yes | No | 7 | 5 | 2 | 1 | | S:S:E405 | S:S:R371 | 11.63 | 0 | No | No | 5 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.22 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
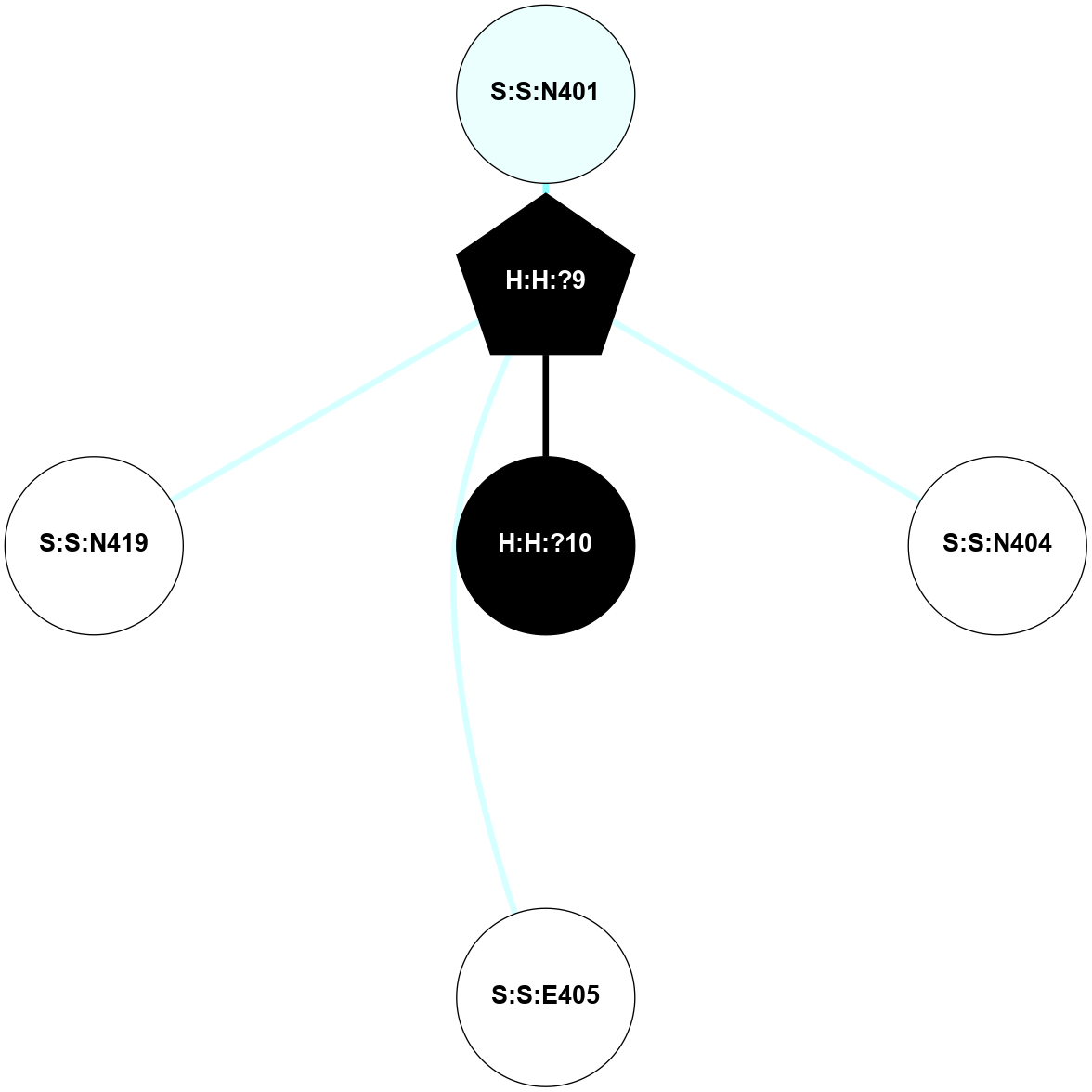
A 2D representation of the interactions of NAG in 6UOA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | H:H:?9 | 42.06 | 0 | No | Yes | 0 | 0 | 0 | 1 | | H:H:?9 | S:S:N401 | 3.68 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?9 | S:S:N404 | 33.15 | 0 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?9 | S:S:E405 | 2.37 | 0 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?9 | S:S:N419 | 9.82 | 0 | Yes | No | 0 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 42.06 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6UOA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | S:S:N453 | 28.24 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?11 | S:S:D454 | 6.07 | 0 | No | No | 0 | 2 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.16 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6VJM | C | Aminoacid | GABAB | GABAB1; GABAB2 | Homo Sapiens | - | - | - | 3.97 | 2020-06-10 | doi.org/10.1038/s41586-020-2408-4 |
|
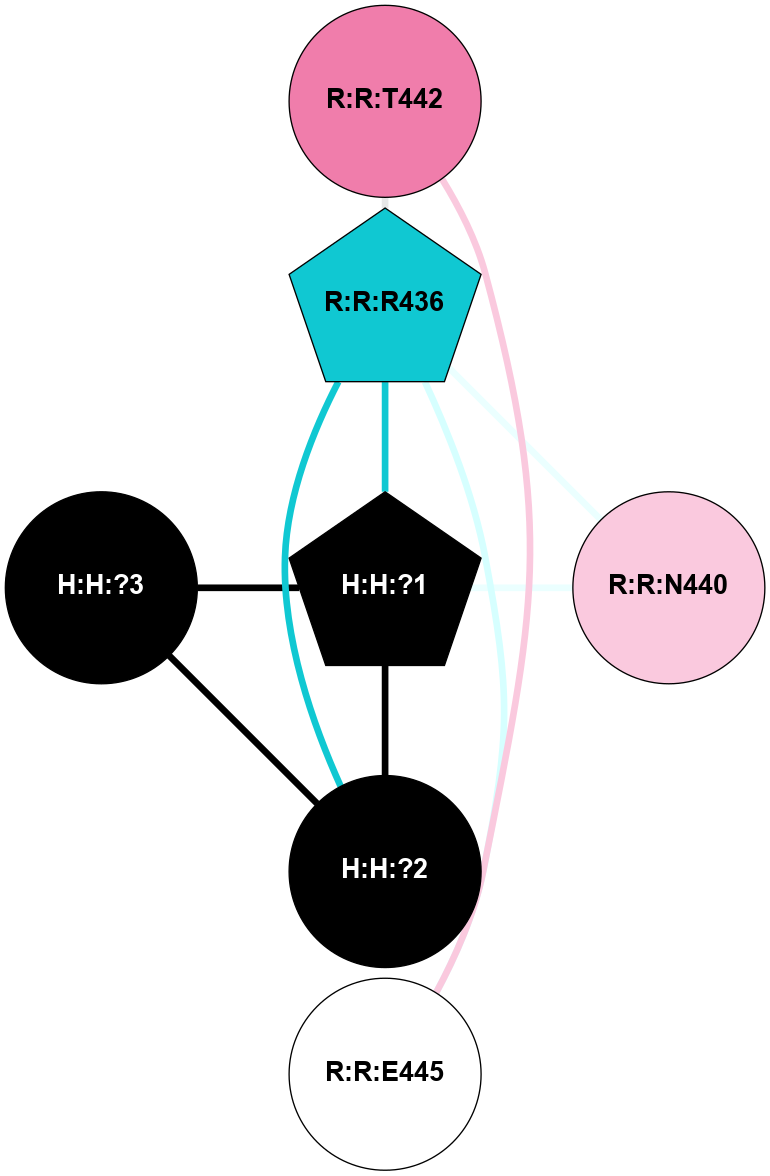
A 2D representation of the interactions of NAG in 6VJM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 46.49 | 6 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | H:H:?3 | 34.65 | 6 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:R436 | 13.04 | 6 | Yes | Yes | 0 | 1 | 0 | 1 | | H:H:?1 | R:R:N440 | 35.61 | 6 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?2 | H:H:?3 | 10.66 | 6 | No | No | 0 | 0 | 1 | 1 | | H:H:?2 | R:R:R436 | 2.17 | 6 | No | Yes | 0 | 1 | 1 | 1 | | R:R:N440 | R:R:R436 | 7.23 | 6 | No | Yes | 7 | 1 | 1 | 1 | | R:R:R436 | R:R:T442 | 2.59 | 6 | Yes | No | 1 | 8 | 1 | 2 | | R:R:E445 | R:R:R436 | 6.98 | 6 | No | Yes | 5 | 1 | 2 | 1 | | R:R:E445 | R:R:T442 | 12.7 | 6 | No | No | 5 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 32.45 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
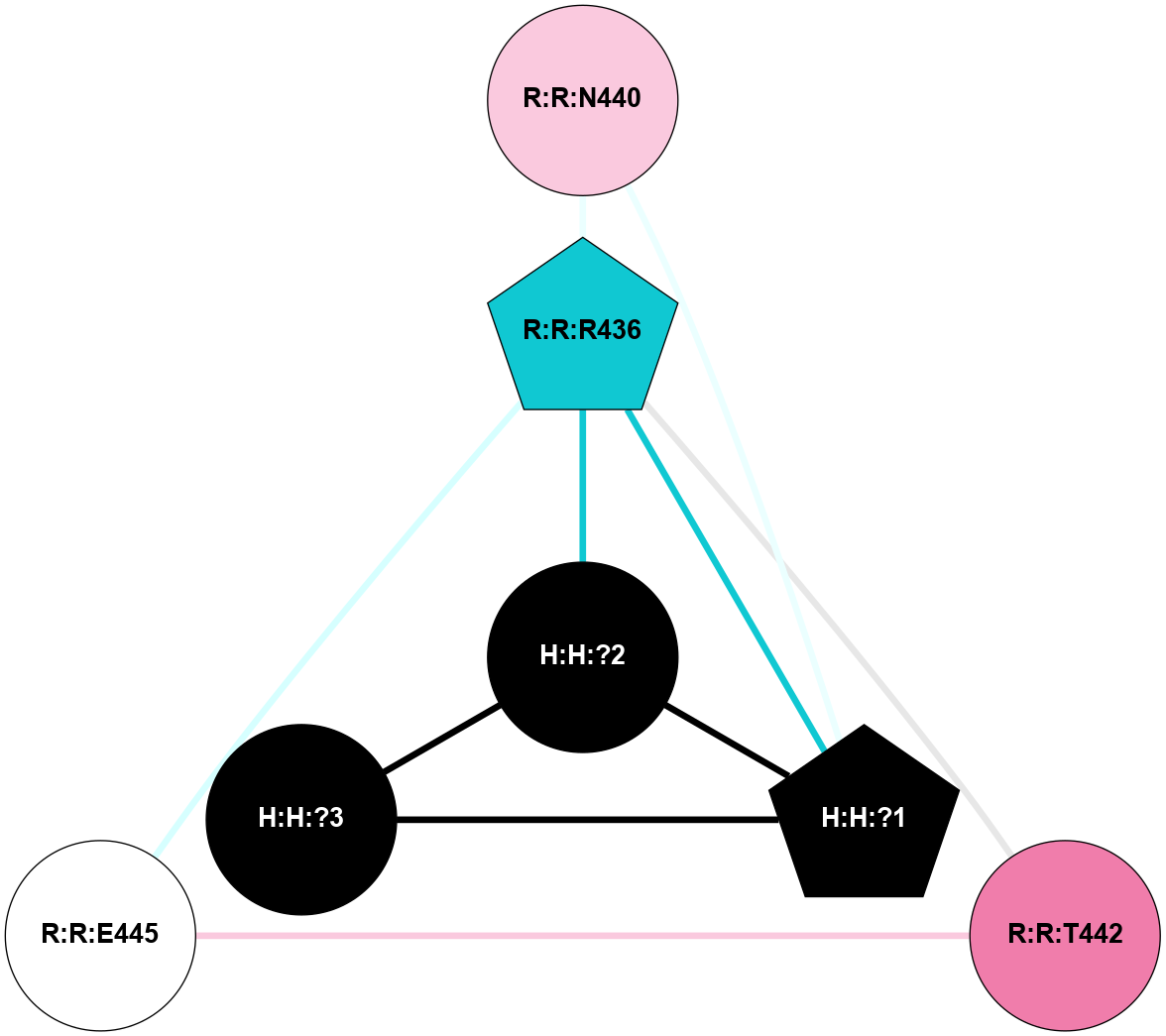
A 2D representation of the interactions of NAG in 6VJM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 46.49 | 6 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | H:H:?3 | 34.65 | 6 | Yes | No | 0 | 0 | 1 | 1 | | H:H:?1 | R:R:R436 | 13.04 | 6 | Yes | Yes | 0 | 1 | 1 | 1 | | H:H:?1 | R:R:N440 | 35.61 | 6 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?2 | H:H:?3 | 10.66 | 6 | No | No | 0 | 0 | 0 | 1 | | H:H:?2 | R:R:R436 | 2.17 | 6 | No | Yes | 0 | 1 | 0 | 1 | | R:R:N440 | R:R:R436 | 7.23 | 6 | No | Yes | 7 | 1 | 2 | 1 | | R:R:R436 | R:R:T442 | 2.59 | 6 | Yes | No | 1 | 8 | 1 | 2 | | R:R:E445 | R:R:R436 | 6.98 | 6 | No | Yes | 5 | 1 | 2 | 1 | | R:R:E445 | R:R:T442 | 12.7 | 6 | No | No | 5 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 19.77 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
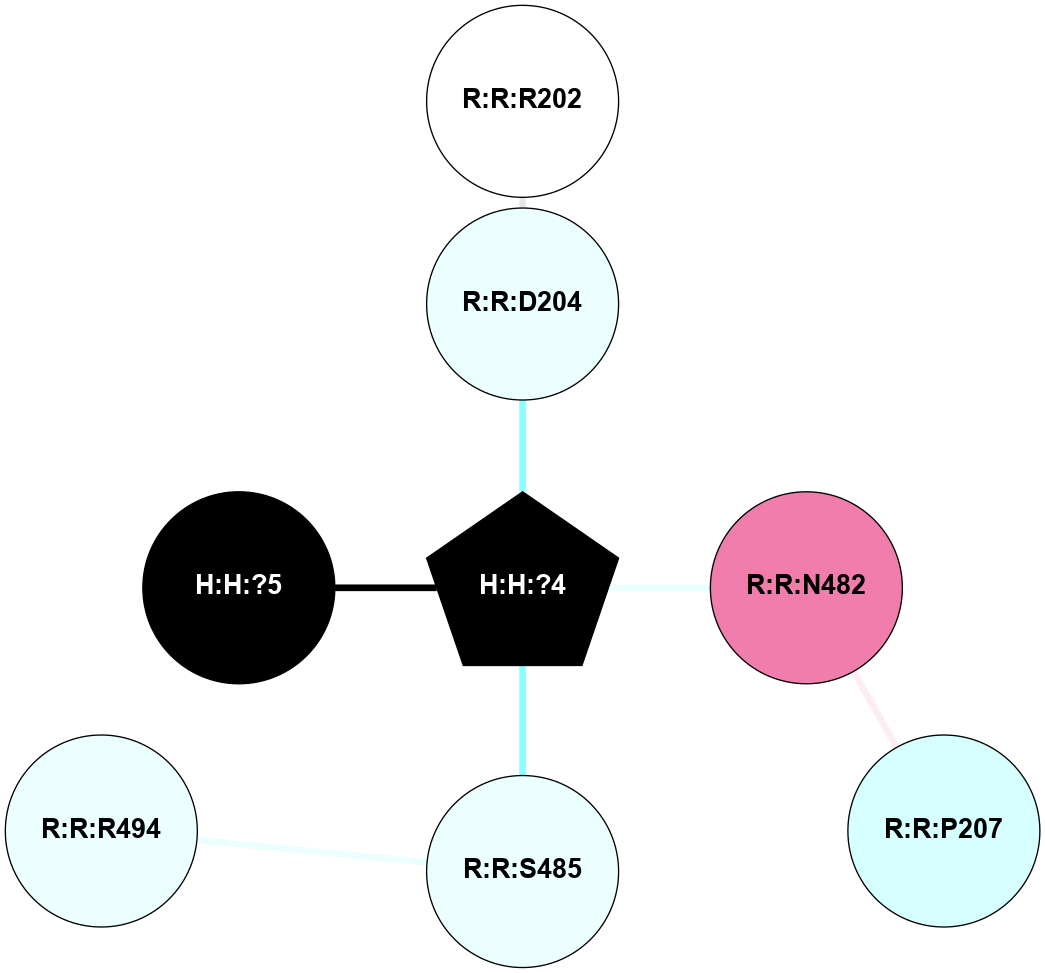
A 2D representation of the interactions of NAG in 6VJM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | H:H:?5 | 36.53 | 0 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?4 | R:R:D204 | 20.63 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?4 | R:R:N482 | 25.79 | 0 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?4 | R:R:S485 | 4.03 | 0 | Yes | No | 0 | 4 | 0 | 1 | | R:R:D204 | R:R:R202 | 2.38 | 0 | No | No | 4 | 5 | 1 | 2 | | R:R:N482 | R:R:P207 | 6.52 | 0 | No | No | 8 | 3 | 1 | 2 | | R:R:R494 | R:R:S485 | 3.95 | 0 | No | No | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 21.74 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
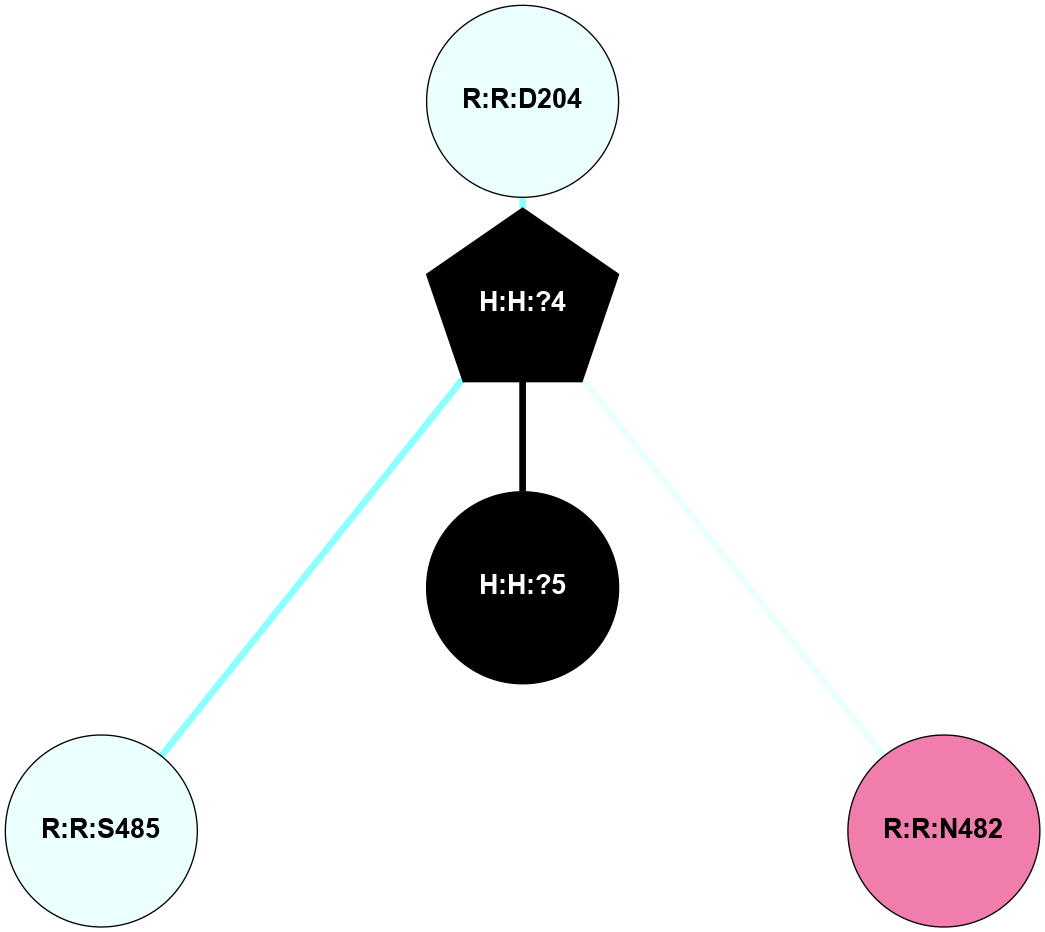
A 2D representation of the interactions of NAG in 6VJM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | H:H:?5 | 36.53 | 0 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?4 | R:R:D204 | 20.63 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?4 | R:R:N482 | 25.79 | 0 | Yes | No | 0 | 8 | 1 | 2 | | H:H:?4 | R:R:S485 | 4.03 | 0 | Yes | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 36.53 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
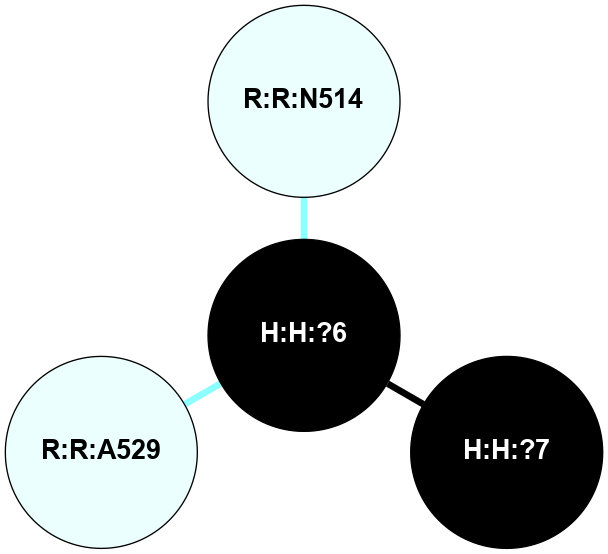
A 2D representation of the interactions of NAG in 6VJM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | H:H:?7 | 39.85 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?6 | R:R:N514 | 28.24 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?6 | R:R:A529 | 1.41 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 23.17 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6VJM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | H:H:?7 | 39.85 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?6 | R:R:N514 | 28.24 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?6 | R:R:A529 | 1.41 | 0 | No | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 39.85 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
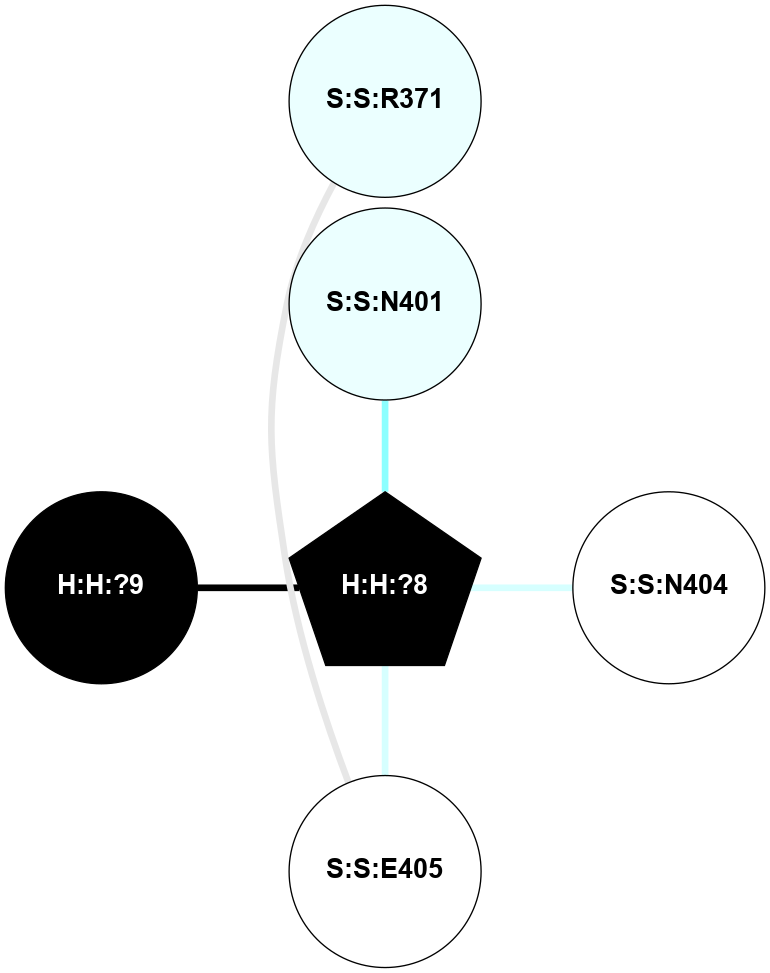
A 2D representation of the interactions of NAG in 6VJM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | H:H:?9 | 33.21 | 0 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?8 | S:S:N401 | 3.68 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?8 | S:S:N404 | 29.47 | 0 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?8 | S:S:E405 | 4.74 | 0 | Yes | No | 0 | 5 | 0 | 1 | | S:S:E405 | S:S:R371 | 9.3 | 0 | No | No | 5 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.77 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
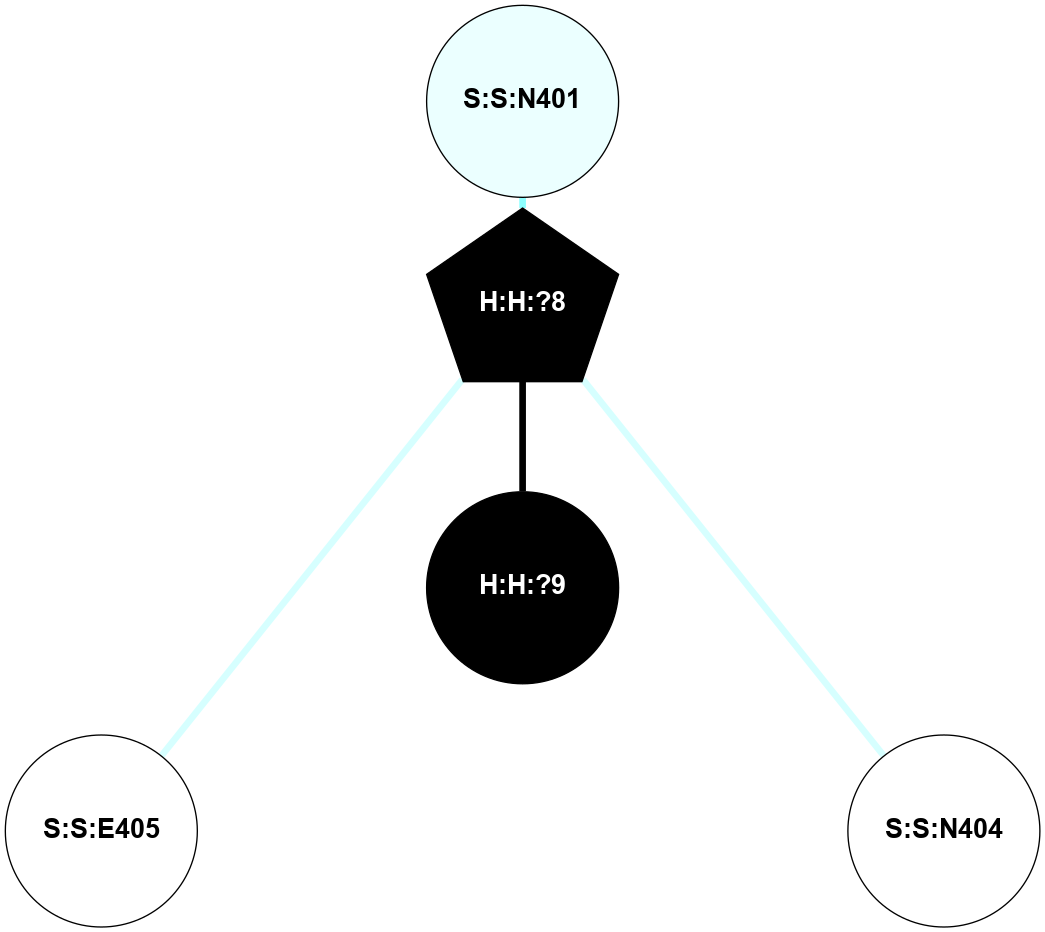
A 2D representation of the interactions of NAG in 6VJM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | H:H:?9 | 33.21 | 0 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?8 | S:S:N401 | 3.68 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?8 | S:S:N404 | 29.47 | 0 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?8 | S:S:E405 | 4.74 | 0 | Yes | No | 0 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 33.21 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
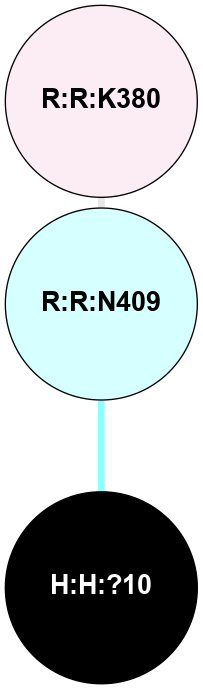
A 2D representation of the interactions of NAG in 6VJM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | R:R:N409 | 25.79 | 0 | No | No | 0 | 3 | 0 | 1 | | R:R:K380 | R:R:N409 | 2.8 | 0 | No | No | 6 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 25.79 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6VJM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | S:S:N453 | 29.47 | 28 | No | No | 0 | 3 | 0 | 1 | | H:H:?11 | S:S:D454 | 7.28 | 28 | No | No | 0 | 2 | 0 | 1 | | S:S:D454 | S:S:N453 | 9.42 | 28 | No | No | 2 | 3 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.38 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6W2X | C | Aminoacid | GABAB | GABAB1; GABAB2 | Homo Sapiens | PubChem 5311042 | - | - | 3.6 | 2020-07-01 | doi.org/10.1038/s41586-020-2469-4 |
|
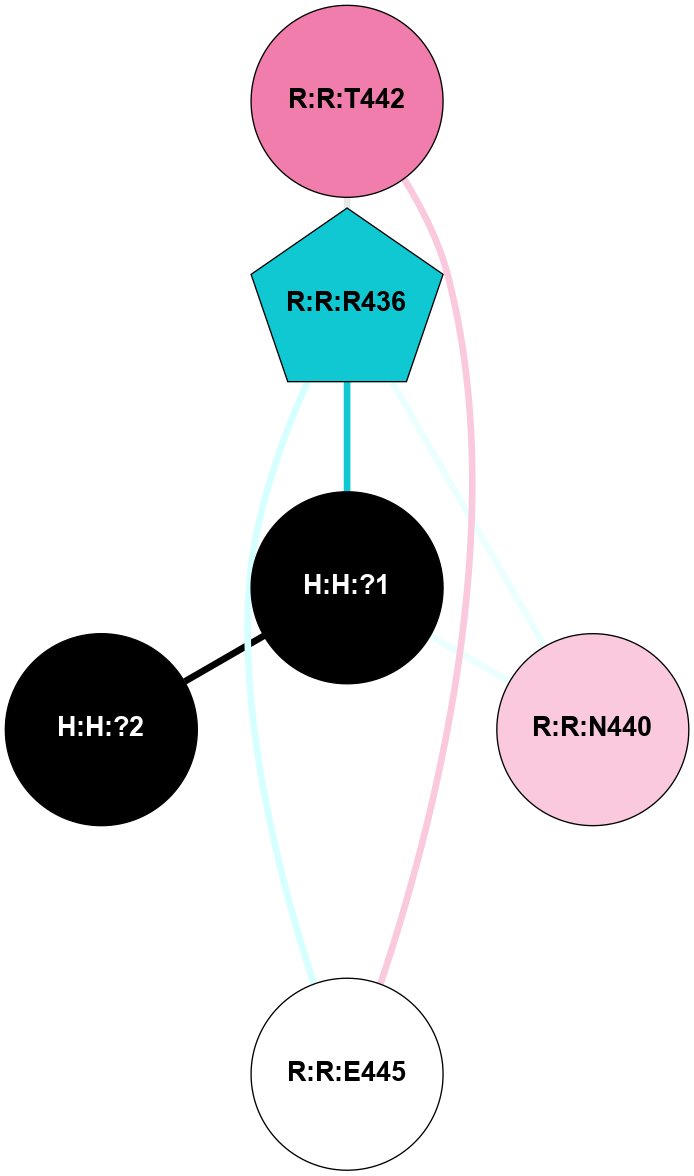
A 2D representation of the interactions of NAG in 6W2X
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 39.01 | 27 | No | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:R436 | 8.72 | 27 | No | No | 0 | 1 | 0 | 1 | | H:H:?1 | R:R:N440 | 27.11 | 27 | No | No | 0 | 7 | 0 | 1 | | R:R:N440 | R:R:R436 | 8.44 | 27 | No | No | 7 | 1 | 1 | 1 | | R:R:R436 | R:R:T442 | 3.88 | 27 | No | No | 1 | 8 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 24.95 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
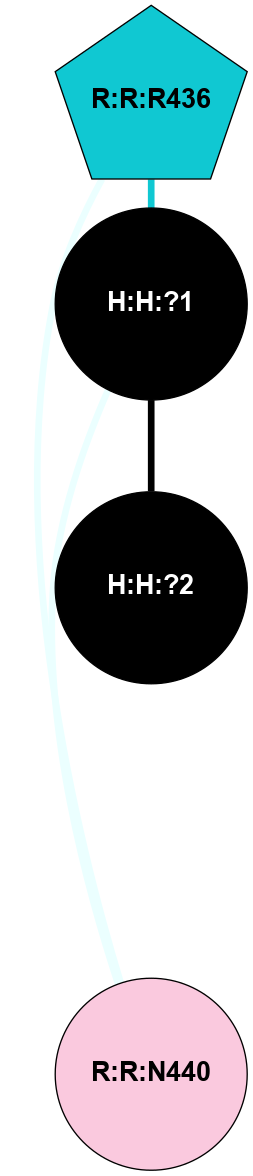
A 2D representation of the interactions of NAG in 6W2X
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 39.01 | 27 | No | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:R436 | 8.72 | 27 | No | No | 0 | 1 | 1 | 2 | | H:H:?1 | R:R:N440 | 27.11 | 27 | No | No | 0 | 7 | 1 | 2 | | R:R:N440 | R:R:R436 | 8.44 | 27 | No | No | 7 | 1 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 39.01 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
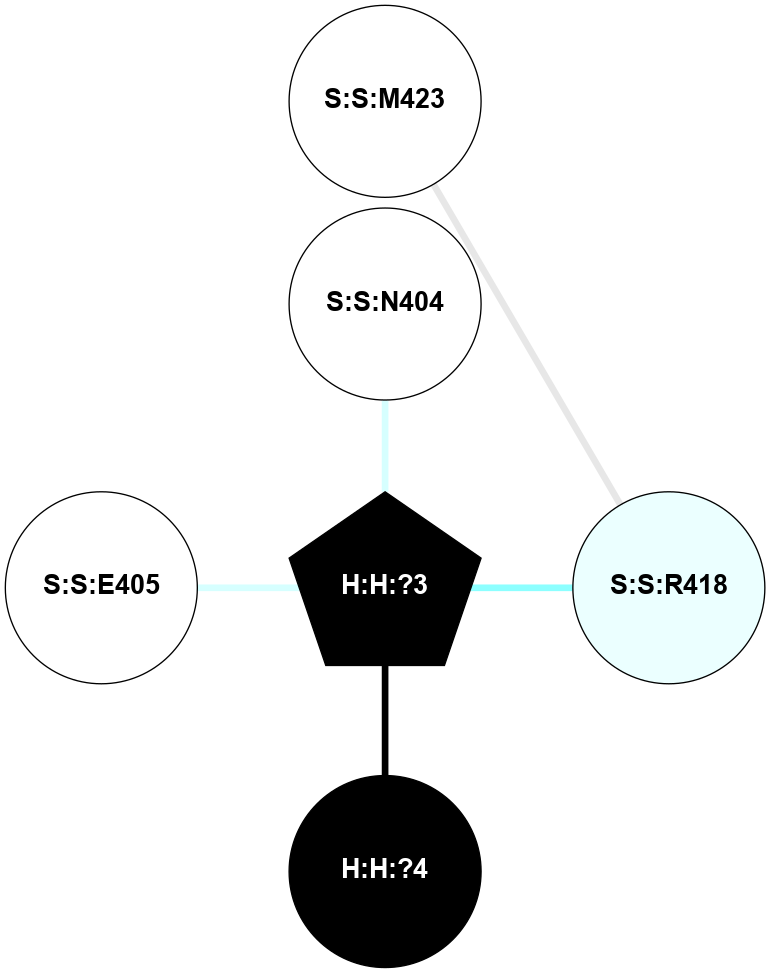
A 2D representation of the interactions of NAG in 6W2X
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 41.23 | 0 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | S:S:N404 | 29.57 | 0 | Yes | No | 0 | 5 | 0 | 1 | | S:S:N401 | S:S:R397 | 6.03 | 0 | No | No | 4 | 3 | 1 | 2 | | H:H:?3 | S:S:N401 | 1.23 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?3 | S:S:E405 | 1.19 | 0 | Yes | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.30 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
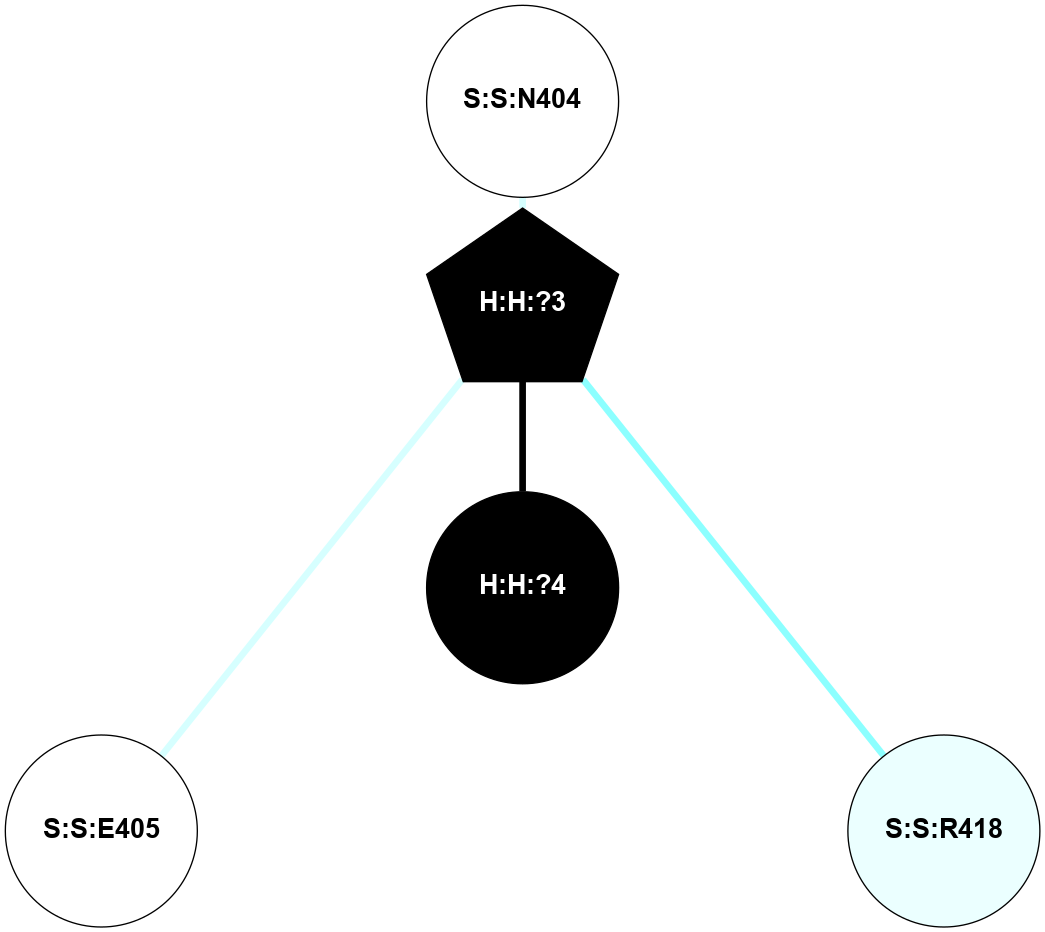
A 2D representation of the interactions of NAG in 6W2X
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 41.23 | 0 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?3 | S:S:N404 | 29.57 | 0 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | S:S:N401 | 1.23 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?3 | S:S:E405 | 1.19 | 0 | Yes | No | 0 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 41.23 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
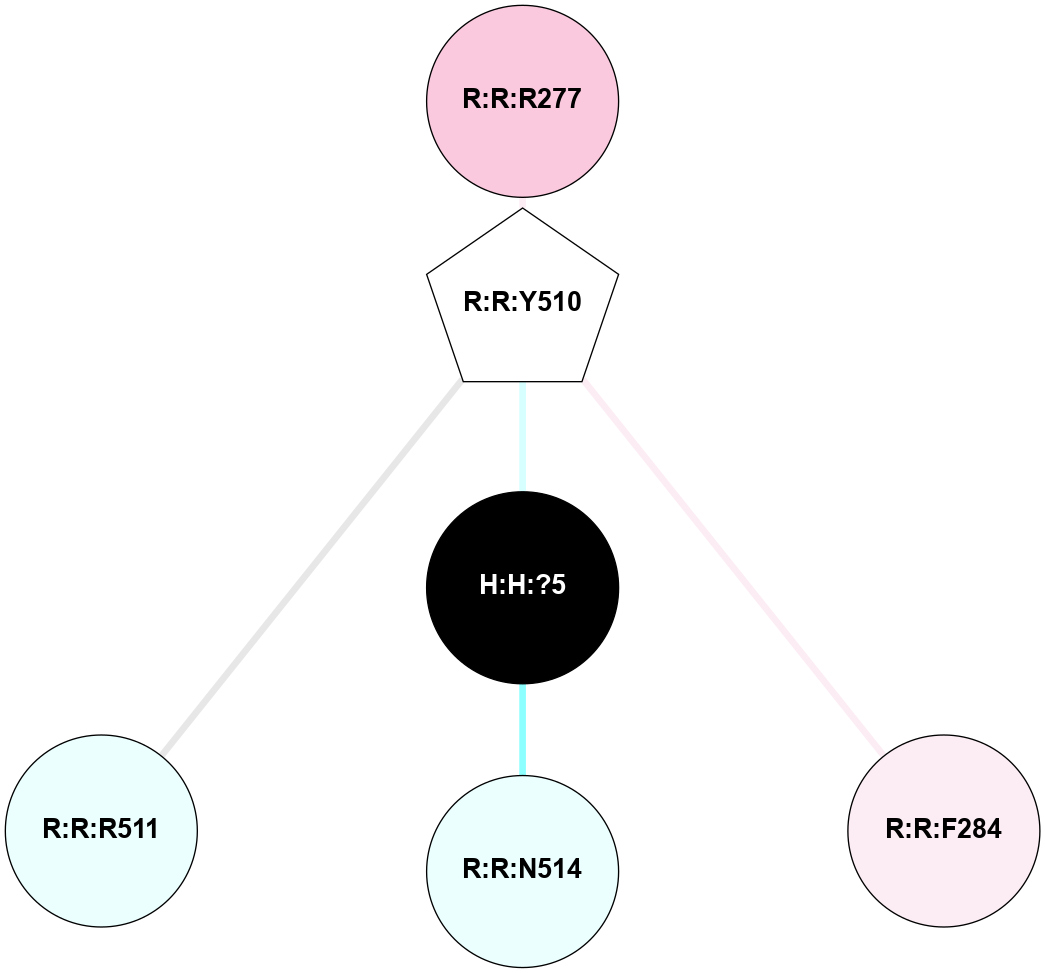
A 2D representation of the interactions of NAG in 6W2X
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | R:R:Y510 | 8.42 | 0 | No | Yes | 0 | 5 | 0 | 1 | | H:H:?5 | R:R:N514 | 27.11 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:R277 | R:R:Y510 | 4.12 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:F284 | R:R:Y510 | 3.09 | 0 | No | Yes | 6 | 5 | 2 | 1 | | R:R:R511 | R:R:Y510 | 2.06 | 0 | No | Yes | 4 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.77 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
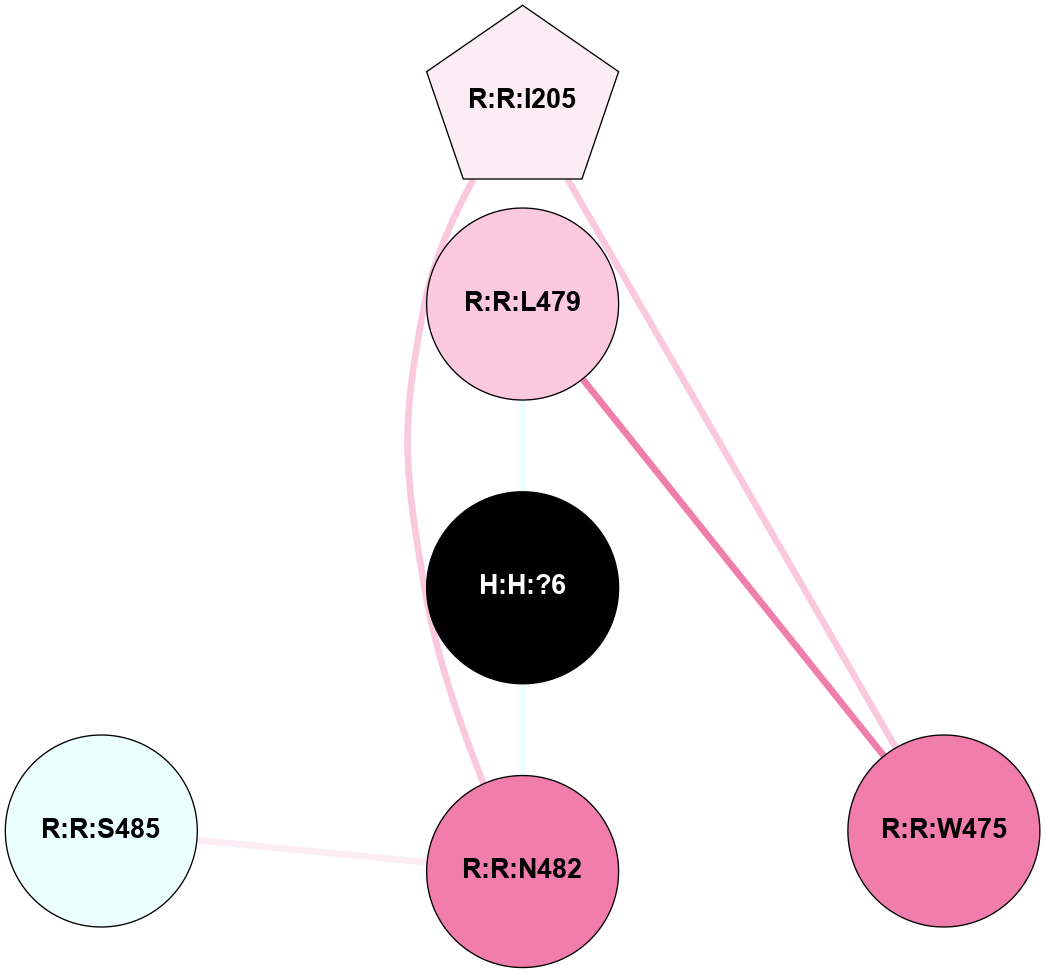
A 2D representation of the interactions of NAG in 6W2X
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | R:R:L479 | 2.48 | 0 | No | No | 0 | 7 | 0 | 1 | | H:H:?6 | R:R:N482 | 25.87 | 0 | No | No | 0 | 8 | 0 | 1 | | R:R:I205 | R:R:W475 | 3.52 | 15 | Yes | No | 6 | 8 | 2 | 2 | | R:R:I205 | R:R:N482 | 2.83 | 15 | Yes | No | 6 | 8 | 2 | 1 | | R:R:L479 | R:R:W475 | 2.28 | 0 | No | No | 7 | 8 | 1 | 2 | | R:R:N482 | R:R:S485 | 1.49 | 0 | No | No | 8 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.18 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6W2Y | C | Aminoacid | GABAB | GABAB1; GABAB1 | Homo Sapiens | PubChem 5311042 | - | - | 3.2 | 2020-07-01 | doi.org/10.1038/s41586-020-2469-4 |
|
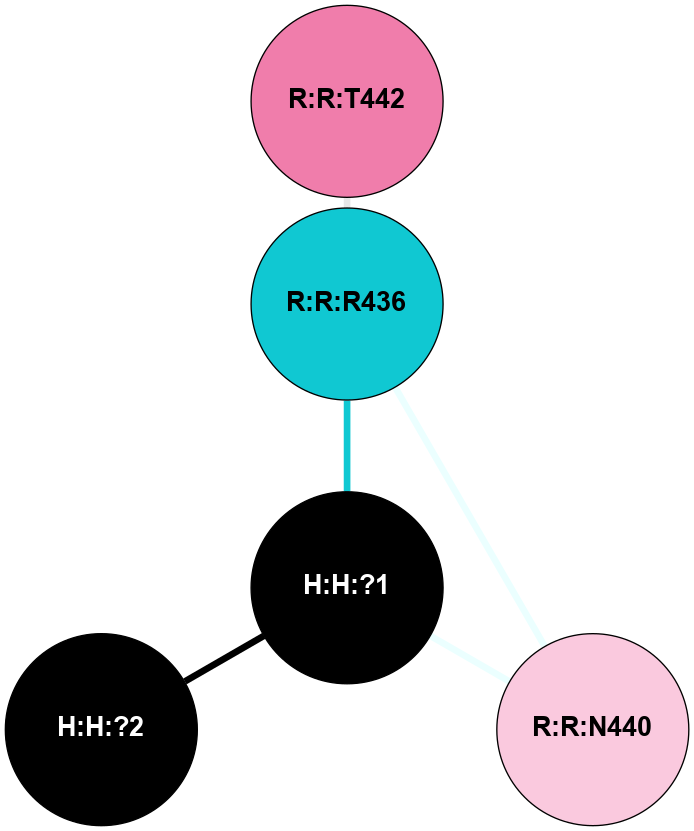
A 2D representation of the interactions of NAG in 6W2Y
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 32.32 | 25 | No | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:R436 | 15.26 | 25 | No | No | 0 | 1 | 0 | 1 | | H:H:?1 | R:R:N440 | 35.73 | 25 | No | No | 0 | 7 | 0 | 1 | | R:R:N440 | R:R:R436 | 7.23 | 25 | No | No | 7 | 1 | 1 | 1 | | R:R:R436 | R:R:T442 | 3.88 | 25 | No | No | 1 | 8 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.77 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
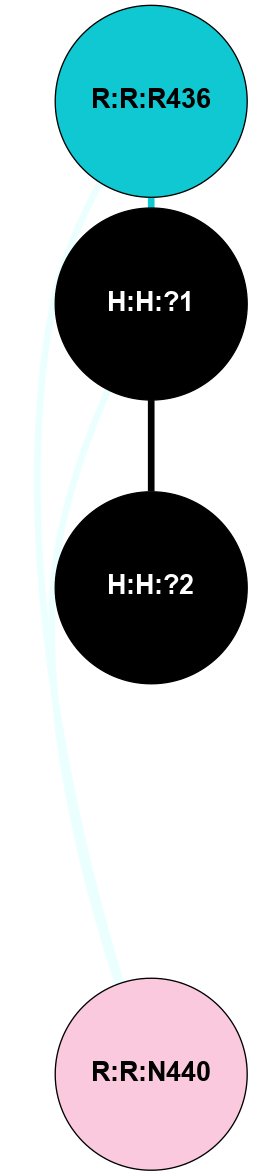
A 2D representation of the interactions of NAG in 6W2Y
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 32.32 | 25 | No | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:R436 | 15.26 | 25 | No | No | 0 | 1 | 1 | 2 | | H:H:?1 | R:R:N440 | 35.73 | 25 | No | No | 0 | 7 | 1 | 2 | | R:R:N440 | R:R:R436 | 7.23 | 25 | No | No | 7 | 1 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 32.32 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
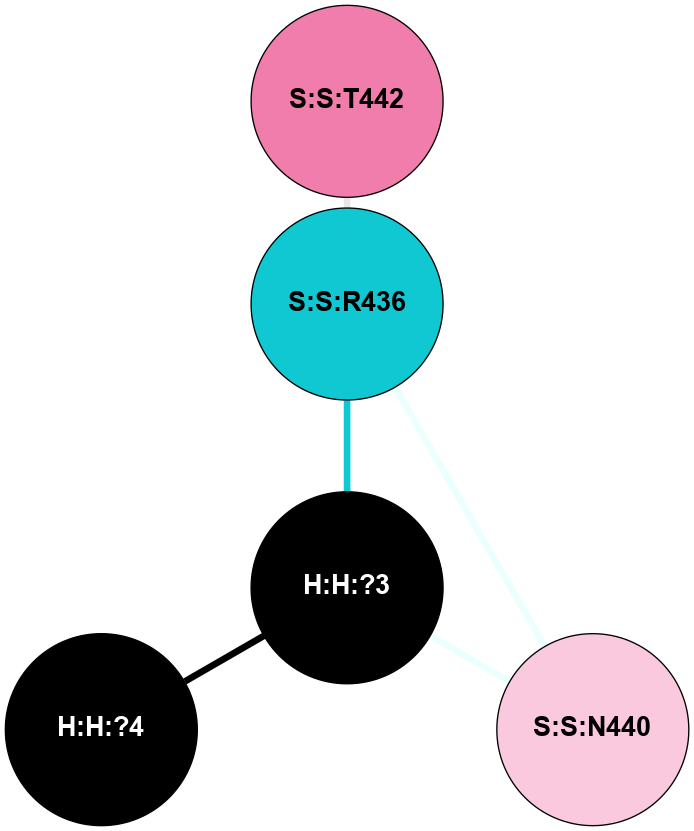
A 2D representation of the interactions of NAG in 6W2Y
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 37.89 | 26 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | S:S:R436 | 17.44 | 26 | No | No | 0 | 1 | 0 | 1 | | H:H:?3 | S:S:N440 | 28.34 | 26 | No | No | 0 | 7 | 0 | 1 | | S:S:N440 | S:S:R436 | 10.85 | 26 | No | No | 7 | 1 | 1 | 1 | | S:S:R436 | S:S:T442 | 2.59 | 26 | No | No | 1 | 8 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.89 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6W2Y
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 37.89 | 26 | No | No | 0 | 0 | 1 | 0 | | H:H:?3 | S:S:R436 | 17.44 | 26 | No | No | 0 | 1 | 1 | 2 | | H:H:?3 | S:S:N440 | 28.34 | 26 | No | No | 0 | 7 | 1 | 2 | | S:S:N440 | S:S:R436 | 10.85 | 26 | No | No | 7 | 1 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 37.89 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
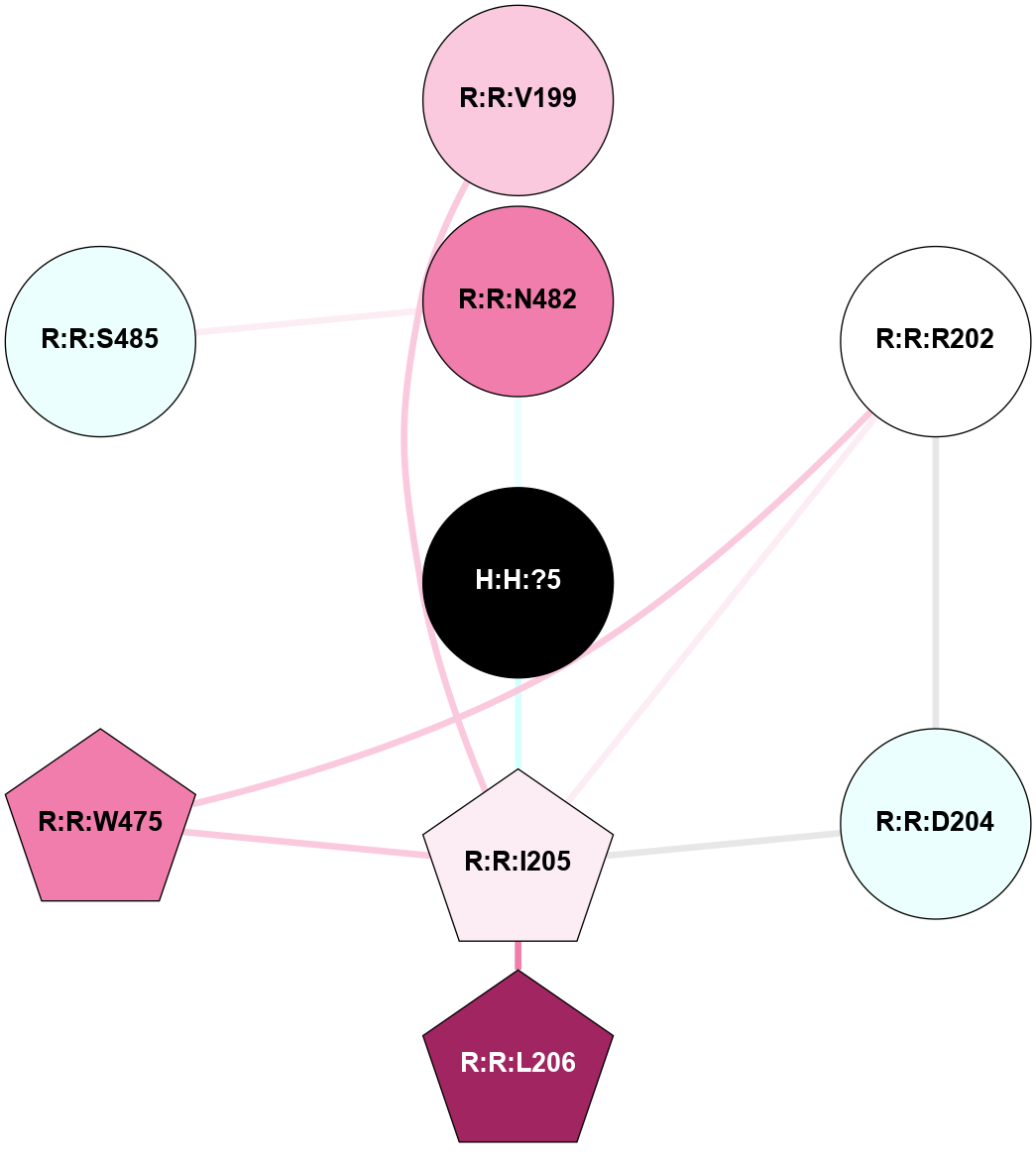
A 2D representation of the interactions of NAG in 6W2Y
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | R:R:N482 | 24.64 | 0 | No | No | 0 | 8 | 0 | 1 | | R:R:D204 | R:R:R202 | 9.53 | 8 | No | No | 4 | 5 | 2 | 2 | | R:R:I205 | R:R:R202 | 3.76 | 8 | Yes | No | 6 | 5 | 1 | 2 | | R:R:R202 | R:R:W475 | 5 | 8 | No | Yes | 5 | 8 | 2 | 2 | | R:R:D204 | R:R:I205 | 5.6 | 8 | No | Yes | 4 | 6 | 2 | 1 | | R:R:I205 | R:R:L206 | 4.28 | 8 | Yes | No | 6 | 9 | 1 | 2 | | R:R:I205 | R:R:W475 | 4.7 | 8 | Yes | Yes | 6 | 8 | 1 | 2 | | R:R:I205 | R:R:V199 | 3.07 | 8 | Yes | No | 6 | 7 | 1 | 2 | | H:H:?5 | R:R:I205 | 2.56 | 0 | No | Yes | 0 | 6 | 0 | 1 | | R:R:N482 | R:R:S485 | 1.49 | 0 | No | No | 8 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 13.60 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
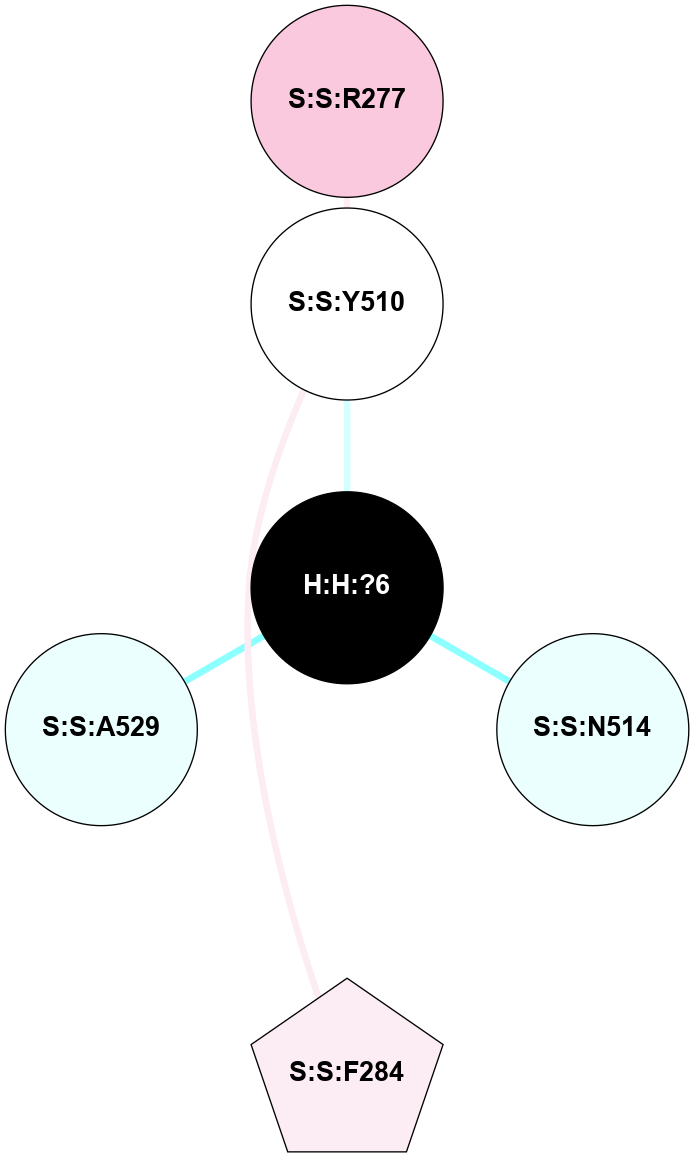
A 2D representation of the interactions of NAG in 6W2Y
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | S:S:Y510 | 10.52 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?6 | S:S:N514 | 25.87 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:R277 | S:S:Y510 | 7.2 | 0 | No | No | 7 | 5 | 2 | 1 | | S:S:F284 | S:S:Y510 | 8.25 | 0 | Yes | No | 6 | 5 | 2 | 1 | | H:H:?6 | S:S:A529 | 2.83 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.07 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6WIV | C | Aminoacid | GABAB | GABAB1; GABAB2 | Homo Sapiens | - | PubChem 52923113; PubChem 52924645 | - | 3.3 | 2020-07-01 | doi.org/10.1038/s41586-020-2452-0 |
|

A 2D representation of the interactions of NAG in 6WIV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I205 | R:R:L206 | 2.85 | 4 | Yes | Yes | 6 | 9 | 2 | 2 | | R:R:I205 | R:R:N482 | 2.83 | 4 | Yes | No | 6 | 8 | 2 | 1 | | R:R:L206 | R:R:N482 | 4.12 | 4 | Yes | No | 9 | 8 | 2 | 1 | | H:H:?1 | R:R:N482 | 25.87 | 0 | No | No | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 25.87 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 3.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 6WIV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F498 | R:R:I505 | 6.28 | 0 | Yes | Yes | 8 | 6 | 2 | 1 | | R:R:N502 | R:R:T504 | 13.16 | 23 | No | No | 7 | 4 | 1 | 2 | | R:R:I505 | R:R:N502 | 4.25 | 23 | Yes | No | 6 | 7 | 1 | 1 | | H:H:?2 | R:R:N502 | 24.64 | 23 | No | No | 0 | 7 | 0 | 1 | | R:R:I505 | R:R:T504 | 3.04 | 23 | Yes | No | 6 | 4 | 1 | 2 | | H:H:?2 | R:R:I505 | 3.84 | 23 | No | Yes | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.24 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
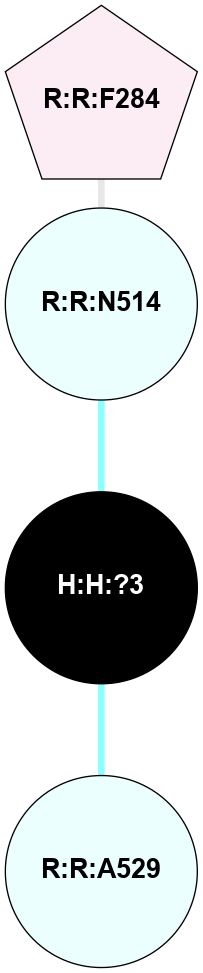
A 2D representation of the interactions of NAG in 6WIV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:N514 | 27.11 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:F284 | R:R:N514 | 2.42 | 4 | Yes | No | 6 | 4 | 2 | 1 | | H:H:?3 | R:R:A529 | 1.41 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.26 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
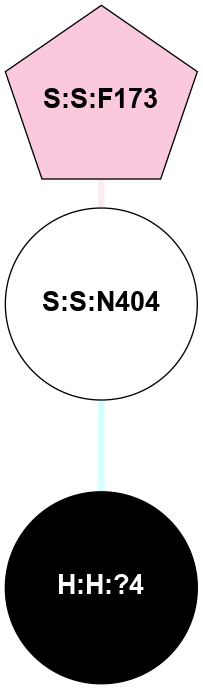
A 2D representation of the interactions of NAG in 6WIV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:F173 | S:S:N404 | 3.62 | 6 | Yes | No | 7 | 5 | 2 | 1 | | H:H:?4 | S:S:N404 | 27.11 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.11 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7AD3 | D1 | Ste2-like | STE2 | STE2 | Saccharomyces Cerevisiae | Alpha-factor mating pheromone | - | Gi1/STE4/Gamma2 | 3.3 | 2020-12-09 | doi.org/10.1038/s41586-020-2994-1 |
|
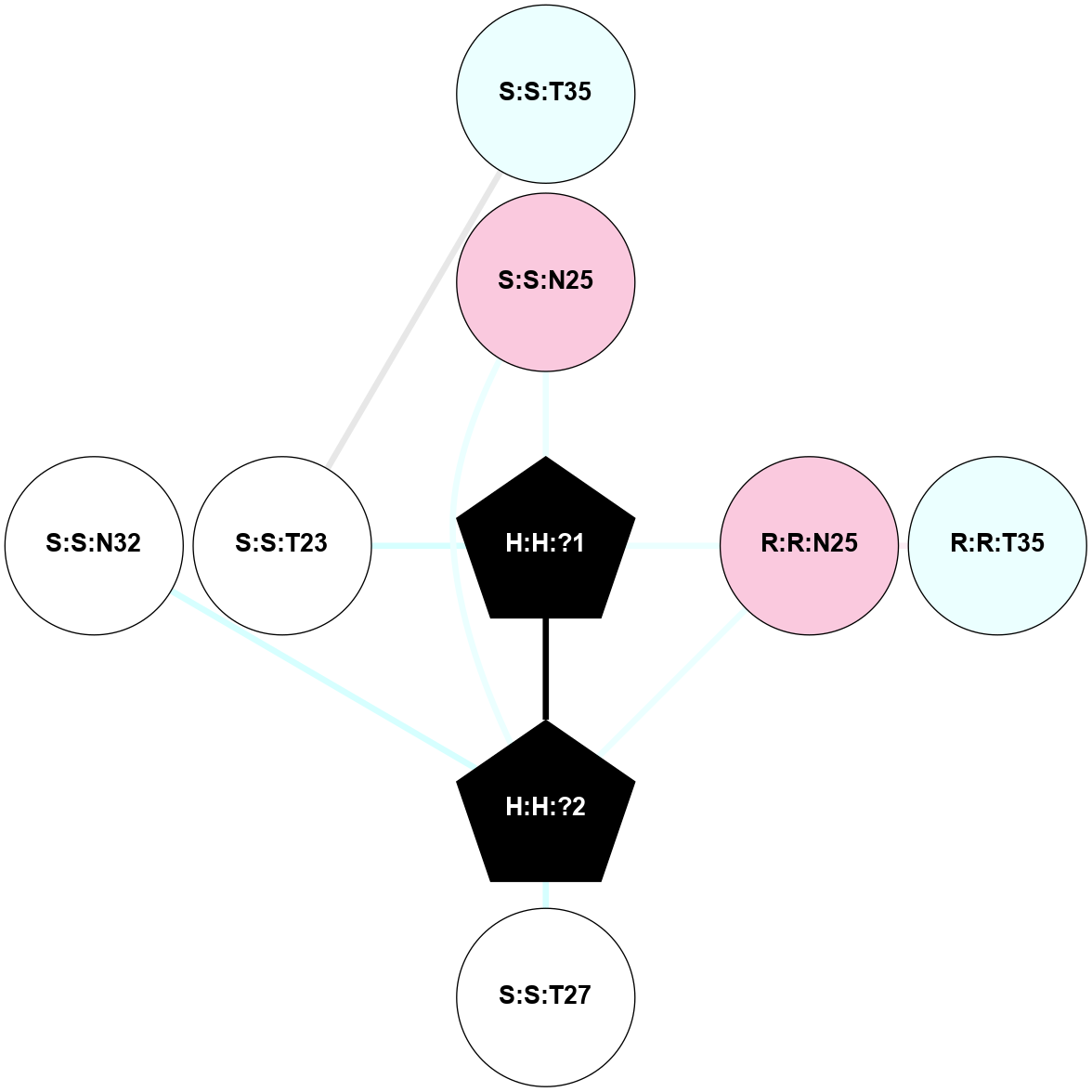
A 2D representation of the interactions of NAG in 7AD3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:T23 | S:S:T35 | 3.14 | 0 | No | No | 5 | 4 | 1 | 2 | | H:H:?1 | S:S:N25 | 25.87 | 16 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?2 | S:S:N25 | 4.93 | 16 | Yes | No | 0 | 7 | 1 | 1 | | H:H:?1 | R:R:N25 | 9.86 | 16 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?2 | R:R:N25 | 28.34 | 16 | Yes | No | 0 | 7 | 1 | 1 | | H:H:?1 | H:H:?2 | 12.26 | 16 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?1 | S:S:T23 | 2.65 | 16 | Yes | No | 0 | 5 | 0 | 1 | | R:R:N25 | R:R:T35 | 1.46 | 16 | No | No | 7 | 4 | 1 | 2 | | H:H:?2 | S:S:T27 | 1.32 | 16 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?2 | S:S:N32 | 1.23 | 16 | Yes | No | 0 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 12.66 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
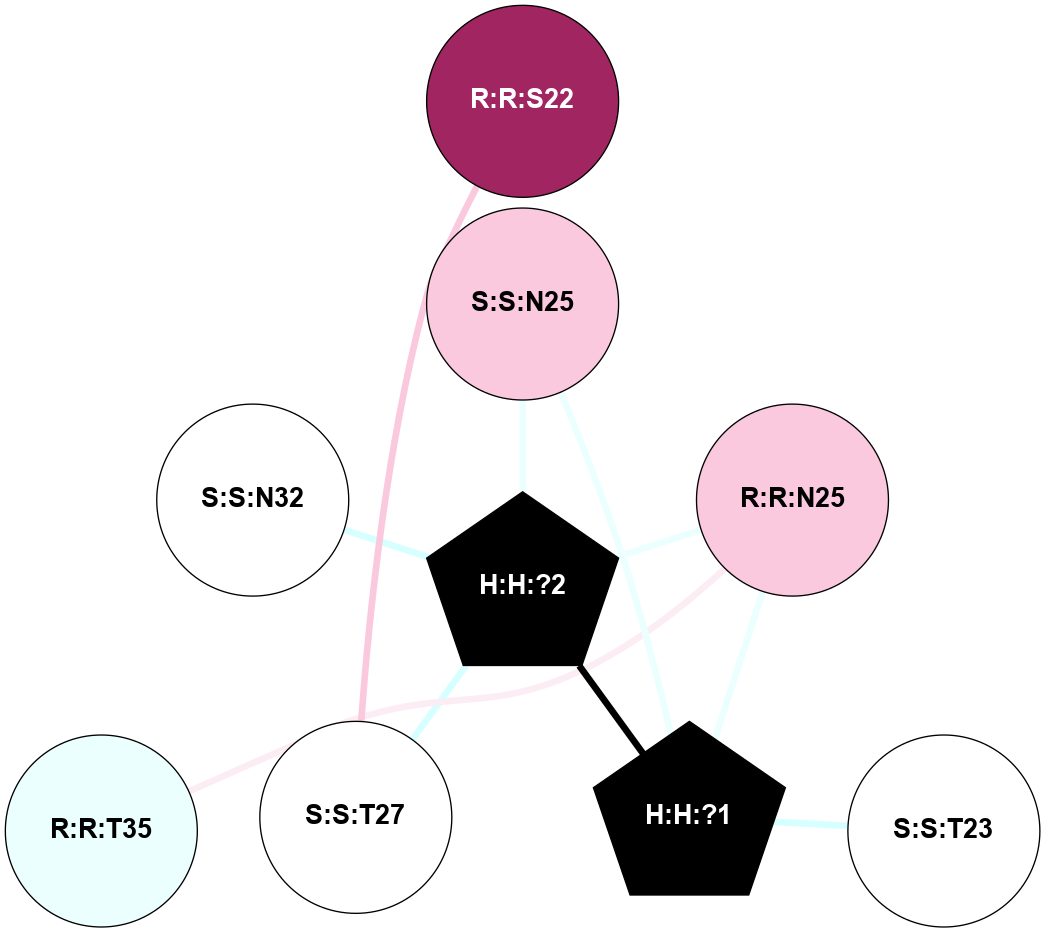
A 2D representation of the interactions of NAG in 7AD3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | S:S:N25 | 25.87 | 16 | Yes | No | 0 | 7 | 1 | 1 | | H:H:?2 | S:S:N25 | 4.93 | 16 | Yes | No | 0 | 7 | 0 | 1 | | R:R:S22 | S:S:T27 | 3.2 | 0 | No | No | 9 | 5 | 2 | 1 | | H:H:?1 | R:R:N25 | 9.86 | 16 | Yes | No | 0 | 7 | 1 | 1 | | H:H:?2 | R:R:N25 | 28.34 | 16 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?1 | H:H:?2 | 12.26 | 16 | Yes | Yes | 0 | 0 | 1 | 0 | | H:H:?1 | S:S:T23 | 2.65 | 16 | Yes | No | 0 | 5 | 1 | 2 | | R:R:N25 | R:R:T35 | 1.46 | 16 | No | No | 7 | 4 | 1 | 2 | | H:H:?2 | S:S:T27 | 1.32 | 16 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?2 | S:S:N32 | 1.23 | 16 | Yes | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 9.62 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7C7Q | C | Aminoacid | GABAB | GABAB1; GABAB2 | Homo Sapiens | Baclofen | BHFF | - | 3 | 2020-07-01 | doi.org/10.1038/s41422-020-0350-5 |
|
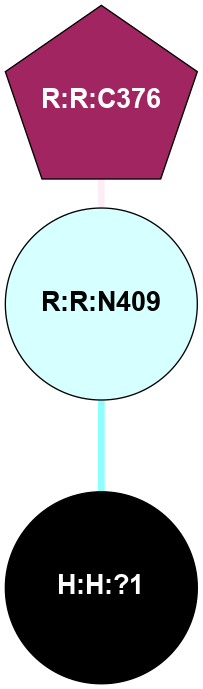
A 2D representation of the interactions of NAG in 7C7Q
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N409 | 25.79 | 0 | No | No | 0 | 3 | 0 | 1 | | R:R:C376 | R:R:N409 | 1.57 | 1 | Yes | No | 9 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 25.79 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
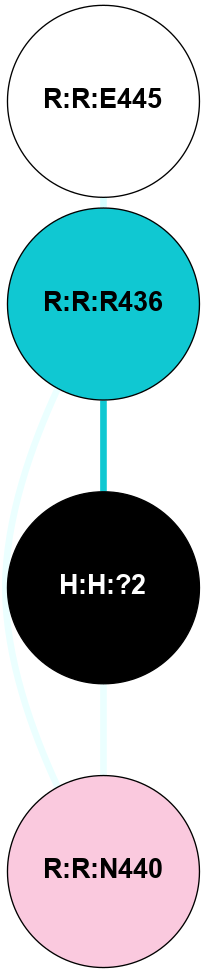
A 2D representation of the interactions of NAG in 7C7Q
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N440 | R:R:R436 | 8.44 | 29 | No | No | 7 | 1 | 1 | 1 | | R:R:E445 | R:R:R436 | 8.14 | 0 | No | No | 5 | 1 | 2 | 1 | | H:H:?2 | R:R:R436 | 6.52 | 29 | No | No | 0 | 1 | 0 | 1 | | H:H:?2 | R:R:N440 | 29.47 | 29 | No | No | 0 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.99 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
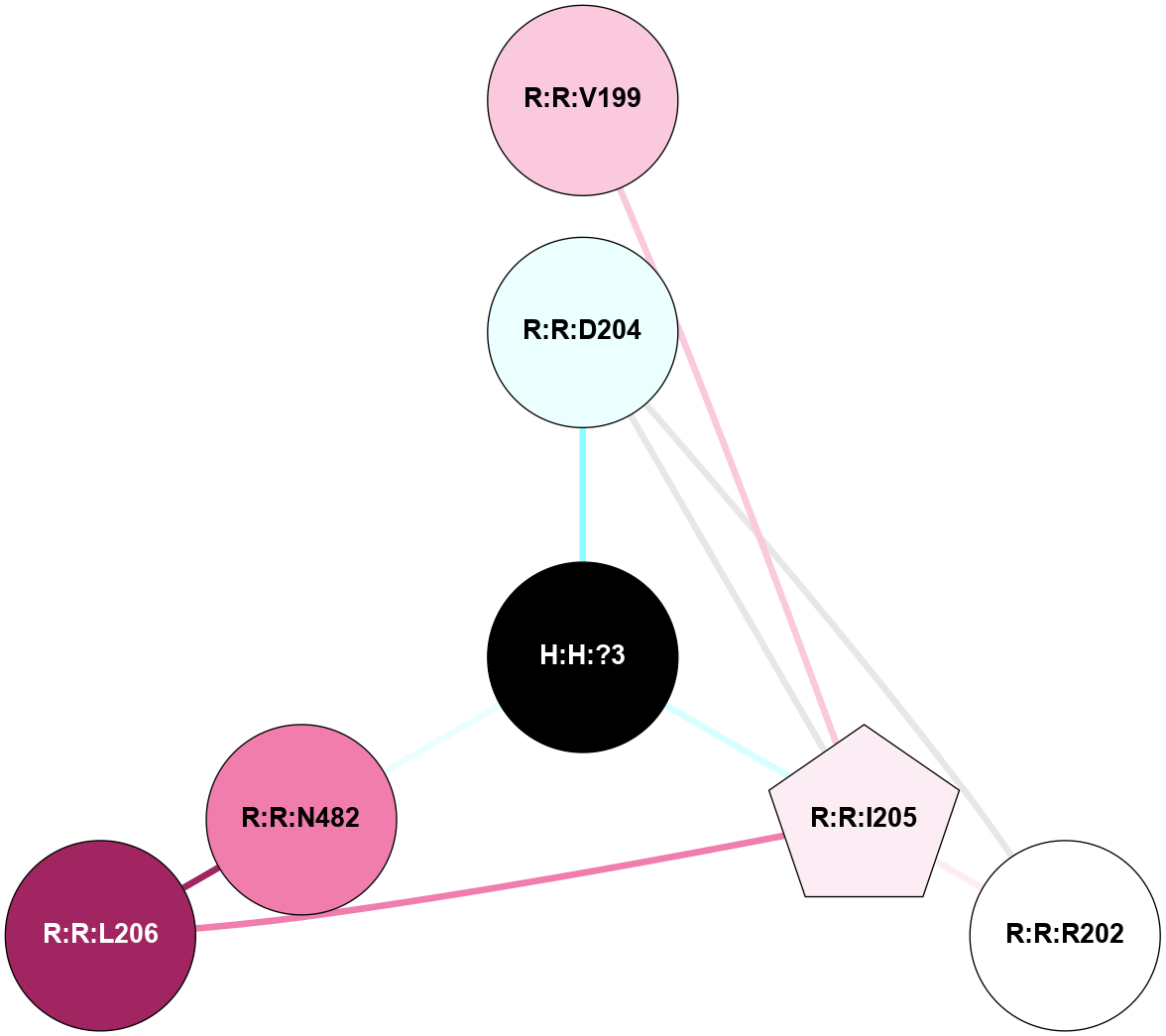
A 2D representation of the interactions of NAG in 7C7Q
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I205 | R:R:V199 | 3.07 | 15 | Yes | No | 6 | 7 | 1 | 2 | | R:R:D204 | R:R:R202 | 13.1 | 15 | No | No | 4 | 5 | 1 | 2 | | R:R:I205 | R:R:R202 | 3.76 | 15 | Yes | No | 6 | 5 | 1 | 2 | | R:R:D204 | R:R:I205 | 4.2 | 15 | No | Yes | 4 | 6 | 1 | 1 | | H:H:?3 | R:R:D204 | 7.28 | 15 | No | No | 0 | 4 | 0 | 1 | | R:R:I205 | R:R:L206 | 2.85 | 15 | Yes | No | 6 | 9 | 1 | 2 | | H:H:?3 | R:R:I205 | 3.83 | 15 | No | Yes | 0 | 6 | 0 | 1 | | R:R:L206 | R:R:N482 | 2.75 | 0 | No | No | 9 | 8 | 2 | 1 | | H:H:?3 | R:R:N482 | 24.56 | 15 | No | No | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 11.89 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
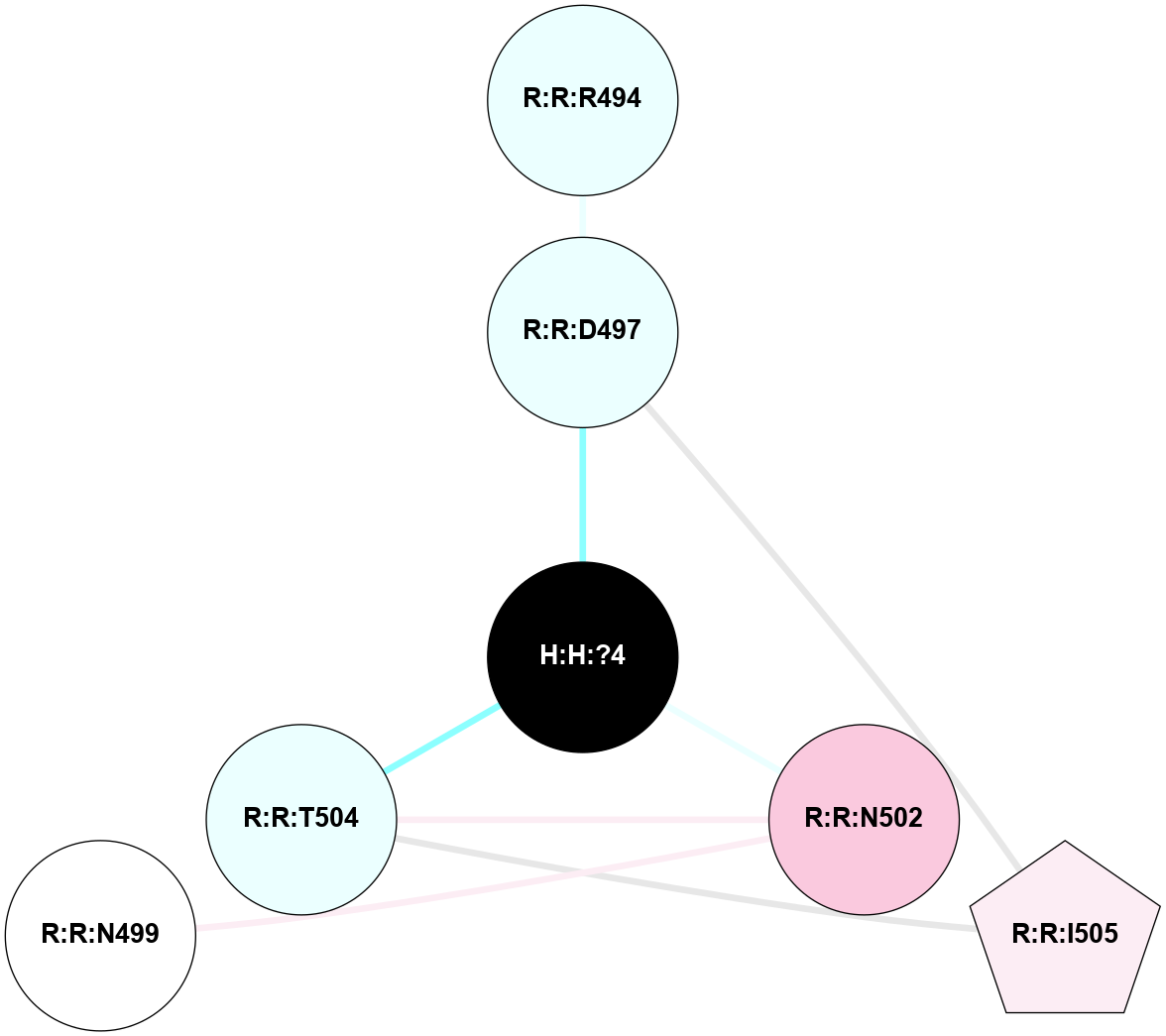
A 2D representation of the interactions of NAG in 7C7Q
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D497 | R:R:R494 | 8.34 | 0 | No | No | 4 | 4 | 1 | 2 | | R:R:D497 | R:R:I505 | 2.8 | 0 | No | Yes | 4 | 6 | 1 | 2 | | H:H:?4 | R:R:D497 | 13.35 | 30 | No | No | 0 | 4 | 0 | 1 | | R:R:N499 | R:R:N502 | 4.09 | 0 | No | No | 5 | 7 | 2 | 1 | | R:R:N502 | R:R:T504 | 8.77 | 30 | No | No | 7 | 4 | 1 | 1 | | H:H:?4 | R:R:N502 | 38.07 | 30 | No | No | 0 | 7 | 0 | 1 | | R:R:I505 | R:R:T504 | 3.04 | 0 | Yes | No | 6 | 4 | 2 | 1 | | H:H:?4 | R:R:T504 | 9.23 | 30 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.22 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
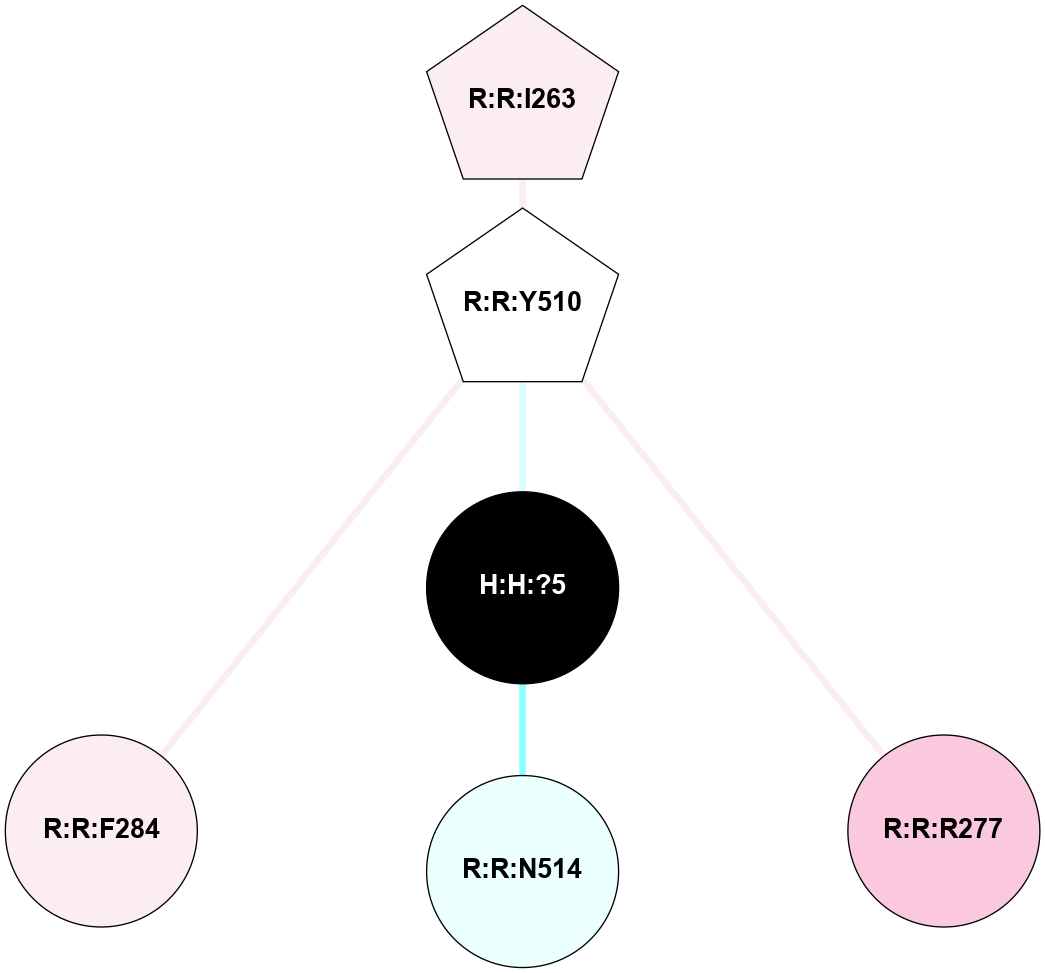
A 2D representation of the interactions of NAG in 7C7Q
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I263 | R:R:Y510 | 3.63 | 0 | Yes | Yes | 6 | 5 | 2 | 1 | | R:R:R277 | R:R:Y510 | 5.14 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:F284 | R:R:Y510 | 8.25 | 28 | No | Yes | 6 | 5 | 2 | 1 | | H:H:?5 | R:R:Y510 | 7.34 | 0 | No | Yes | 0 | 5 | 0 | 1 | | H:H:?5 | R:R:N514 | 20.88 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.11 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
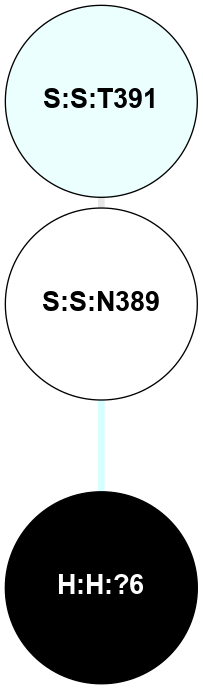
A 2D representation of the interactions of NAG in 7C7Q
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | S:S:N389 | 29.47 | 0 | No | No | 0 | 5 | 0 | 1 | | S:S:N389 | S:S:T391 | 1.46 | 0 | No | No | 5 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 29.47 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
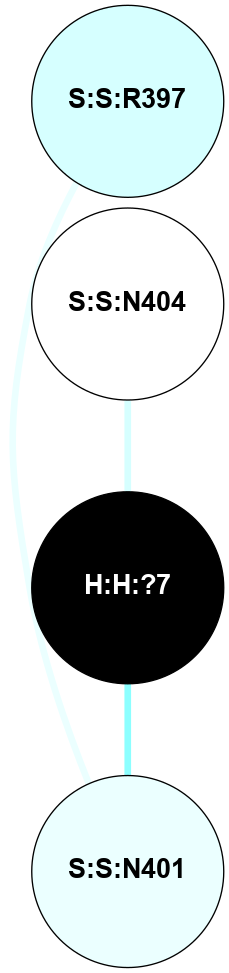
A 2D representation of the interactions of NAG in 7C7Q
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:N401 | S:S:R397 | 6.03 | 0 | No | No | 4 | 3 | 1 | 2 | | H:H:?7 | S:S:N404 | 34.38 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?7 | S:S:N401 | 1.23 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.80 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
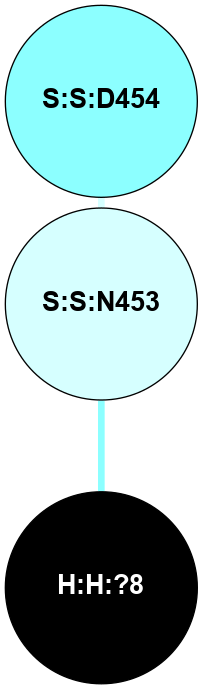
A 2D representation of the interactions of NAG in 7C7Q
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | S:S:N453 | 27.02 | 0 | No | No | 0 | 3 | 0 | 1 | | S:S:D454 | S:S:N453 | 1.35 | 0 | No | No | 2 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7C7S | C | Aminoacid | GABAB | GABAB1; GABAB2 | Homo Sapiens | CGP54626 | - | - | 2.9 | 2020-07-01 | doi.org/10.1038/s41422-020-0350-5 |
|
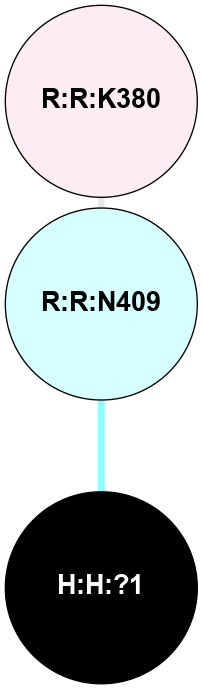
A 2D representation of the interactions of NAG in 7C7S
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:K380 | R:R:N409 | 2.8 | 0 | No | No | 6 | 3 | 2 | 1 | | H:H:?1 | R:R:N409 | 29.47 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 29.47 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
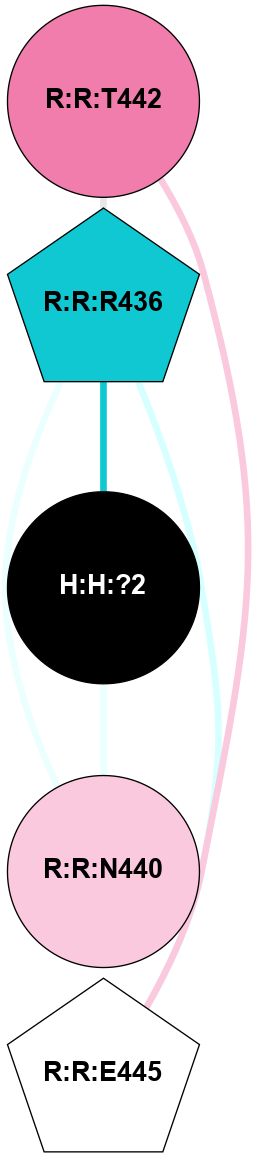
A 2D representation of the interactions of NAG in 7C7S
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N440 | R:R:R436 | 4.82 | 4 | No | Yes | 7 | 1 | 1 | 1 | | R:R:R436 | R:R:T442 | 6.47 | 4 | Yes | No | 1 | 8 | 1 | 2 | | R:R:E445 | R:R:R436 | 8.14 | 4 | Yes | Yes | 5 | 1 | 2 | 1 | | H:H:?2 | R:R:R436 | 3.26 | 4 | No | Yes | 0 | 1 | 0 | 1 | | H:H:?2 | R:R:N440 | 34.38 | 4 | No | No | 0 | 7 | 0 | 1 | | R:R:E445 | R:R:T442 | 11.29 | 4 | Yes | No | 5 | 8 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 18.82 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
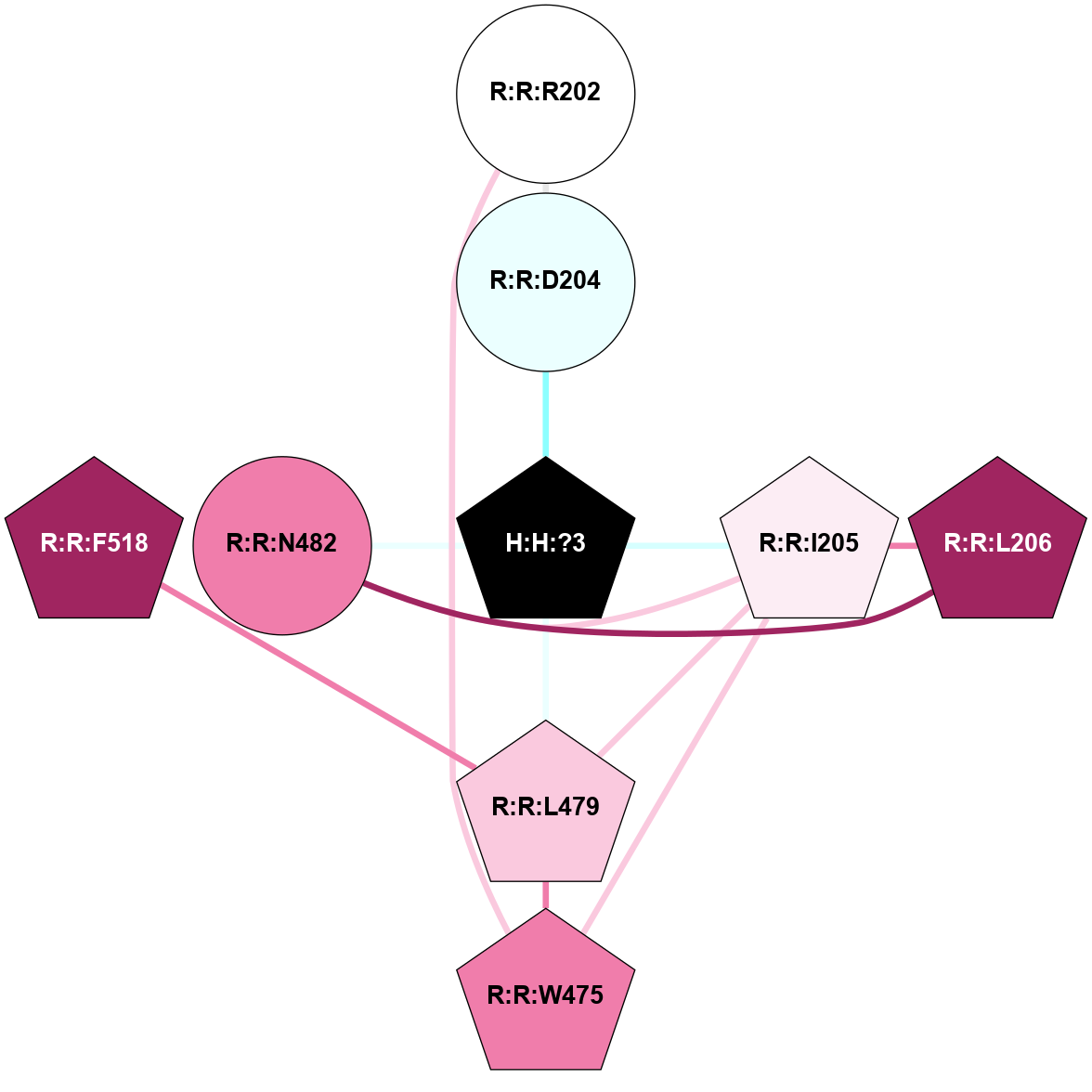
A 2D representation of the interactions of NAG in 7C7S
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D204 | R:R:R202 | 3.57 | 0 | No | No | 4 | 5 | 1 | 2 | | R:R:R202 | R:R:W475 | 4 | 0 | No | Yes | 5 | 8 | 2 | 2 | | H:H:?3 | R:R:D204 | 10.92 | 4 | Yes | No | 0 | 4 | 0 | 1 | | R:R:I205 | R:R:L206 | 4.28 | 4 | Yes | Yes | 6 | 9 | 1 | 2 | | R:R:I205 | R:R:W475 | 3.52 | 4 | Yes | Yes | 6 | 8 | 1 | 2 | | R:R:I205 | R:R:L479 | 2.85 | 4 | Yes | Yes | 6 | 7 | 1 | 1 | | R:R:I205 | R:R:N482 | 9.91 | 4 | Yes | No | 6 | 8 | 1 | 1 | | H:H:?3 | R:R:I205 | 6.38 | 4 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:L206 | R:R:N482 | 2.75 | 4 | Yes | No | 9 | 8 | 2 | 1 | | R:R:L479 | R:R:W475 | 3.42 | 4 | Yes | Yes | 7 | 8 | 1 | 2 | | R:R:F518 | R:R:L479 | 7.31 | 0 | Yes | Yes | 9 | 7 | 2 | 1 | | H:H:?3 | R:R:L479 | 3.71 | 4 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?3 | R:R:N482 | 24.56 | 4 | Yes | No | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 11.39 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
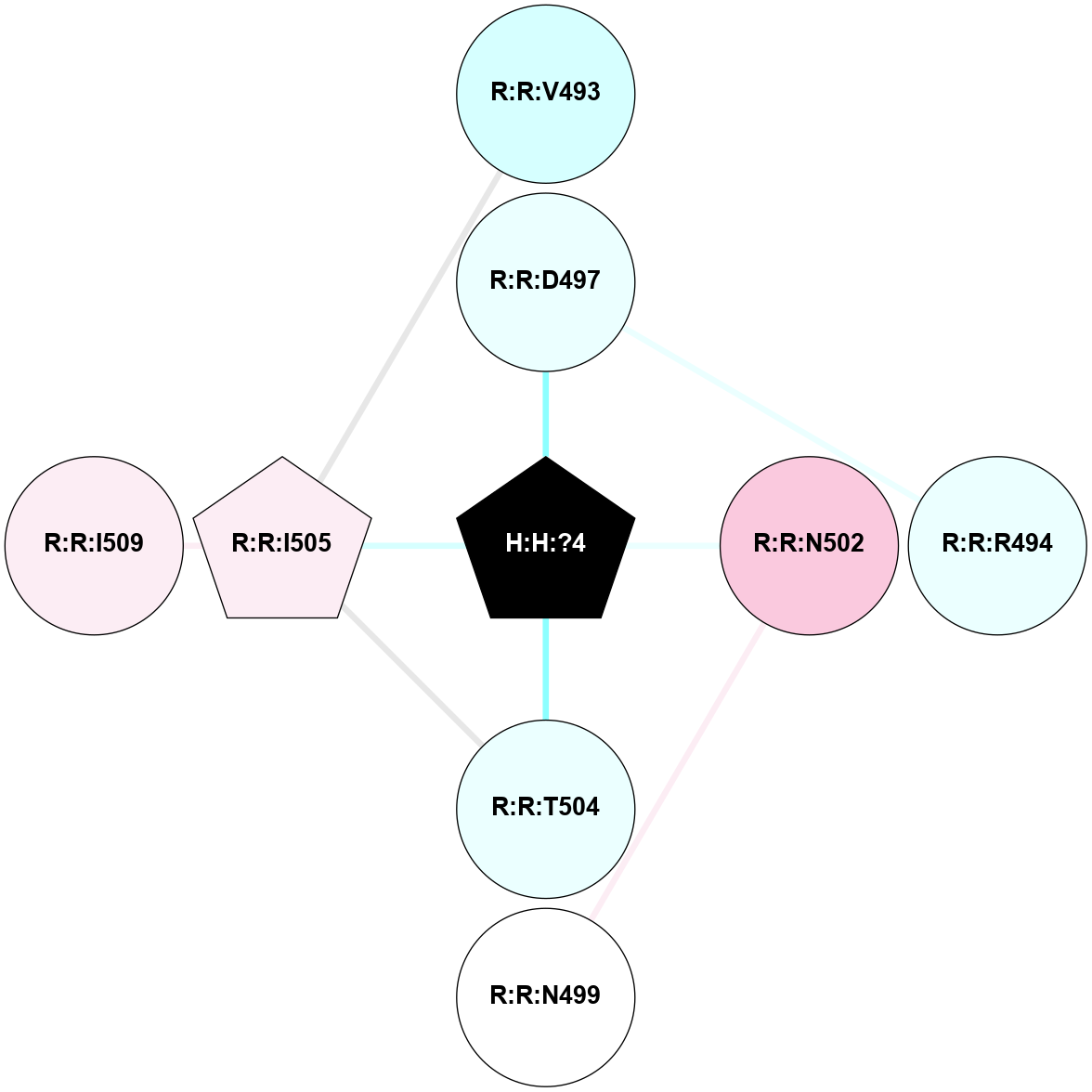
A 2D representation of the interactions of NAG in 7C7S
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:I505 | R:R:V493 | 6.14 | 32 | Yes | No | 6 | 3 | 1 | 2 | | R:R:D497 | R:R:R494 | 7.15 | 0 | No | No | 4 | 4 | 1 | 2 | | H:H:?4 | R:R:D497 | 7.28 | 32 | Yes | No | 0 | 4 | 0 | 1 | | R:R:N499 | R:R:N502 | 13.62 | 0 | No | No | 5 | 7 | 2 | 1 | | H:H:?4 | R:R:N502 | 27.02 | 32 | Yes | No | 0 | 7 | 0 | 1 | | R:R:I505 | R:R:T504 | 3.04 | 32 | Yes | No | 6 | 4 | 1 | 1 | | H:H:?4 | R:R:T504 | 3.95 | 32 | Yes | No | 0 | 4 | 0 | 1 | | R:R:I505 | R:R:I509 | 2.94 | 32 | Yes | No | 6 | 6 | 1 | 2 | | H:H:?4 | R:R:I505 | 8.93 | 32 | Yes | Yes | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 11.79 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
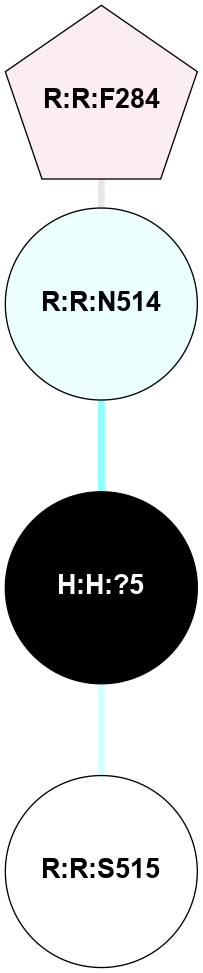
A 2D representation of the interactions of NAG in 7C7S
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | R:R:N514 | 24.56 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?5 | R:R:S515 | 2.69 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:F284 | R:R:N514 | 1.21 | 30 | Yes | No | 6 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.62 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
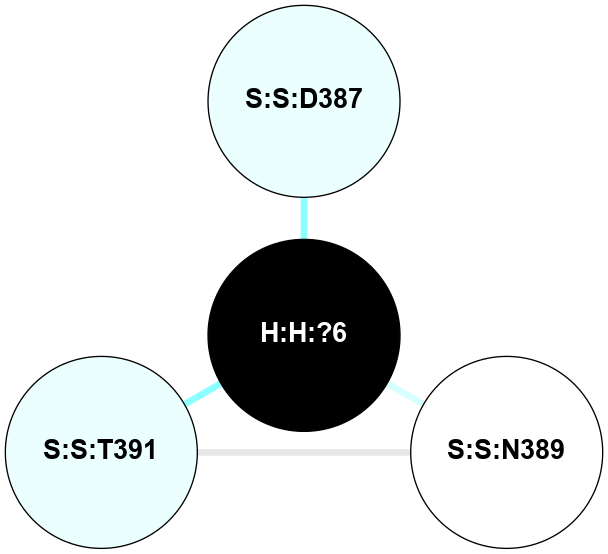
A 2D representation of the interactions of NAG in 7C7S
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | S:S:D387 | 14.56 | 41 | No | No | 0 | 4 | 0 | 1 | | S:S:N389 | S:S:T391 | 8.77 | 41 | No | No | 5 | 4 | 1 | 1 | | H:H:?6 | S:S:N389 | 25.79 | 41 | No | No | 0 | 5 | 0 | 1 | | H:H:?6 | S:S:T391 | 7.91 | 41 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.09 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
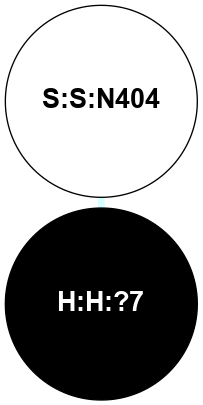
A 2D representation of the interactions of NAG in 7C7S
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | S:S:N404 | 23.33 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 23.33 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
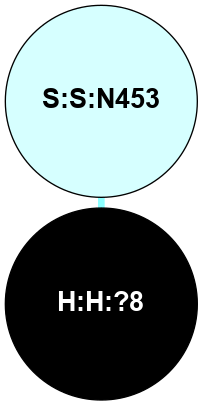
A 2D representation of the interactions of NAG in 7C7S
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | S:S:N453 | 24.56 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 24.56 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7DD5 | C | Ion | Calcium Sensing | CaS | Gallus Gallus | Ca | Ca; Ca; Tryptophan; NPS-2143 | - | 3.2 | 2021-06-16 | doi.org/10.1126/sciadv.abg1483 |
|
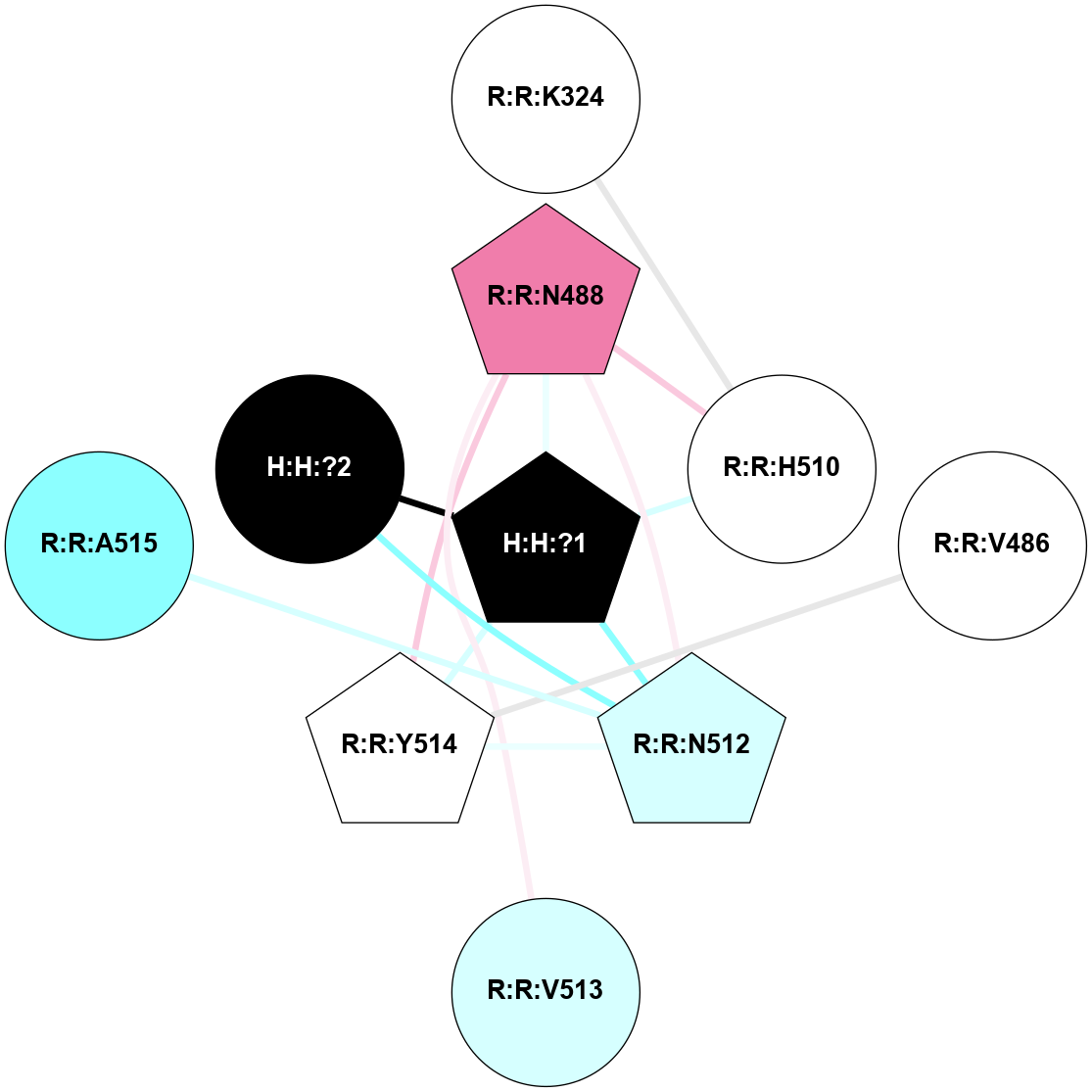
A 2D representation of the interactions of NAG in 7DD5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 26.57 | 17 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N488 | 23.33 | 17 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?1 | R:R:H510 | 13.8 | 17 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:N512 | 20.88 | 17 | Yes | Yes | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:Y514 | 13.63 | 17 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:N512 | 3.68 | 17 | No | Yes | 0 | 3 | 1 | 1 | | R:R:H510 | R:R:K324 | 9.17 | 17 | No | No | 5 | 5 | 1 | 2 | | R:R:V486 | R:R:Y514 | 8.83 | 0 | No | Yes | 5 | 5 | 2 | 1 | | R:R:H510 | R:R:N488 | 6.38 | 17 | No | Yes | 5 | 8 | 1 | 1 | | R:R:N488 | R:R:N512 | 13.62 | 17 | Yes | Yes | 8 | 3 | 1 | 1 | | R:R:N488 | R:R:V513 | 5.91 | 17 | Yes | No | 8 | 3 | 1 | 2 | | R:R:N488 | R:R:Y514 | 8.14 | 17 | Yes | Yes | 8 | 5 | 1 | 1 | | R:R:N512 | R:R:Y514 | 4.65 | 17 | Yes | Yes | 3 | 5 | 1 | 1 | | R:R:A515 | R:R:N512 | 3.13 | 0 | No | Yes | 2 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 19.64 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
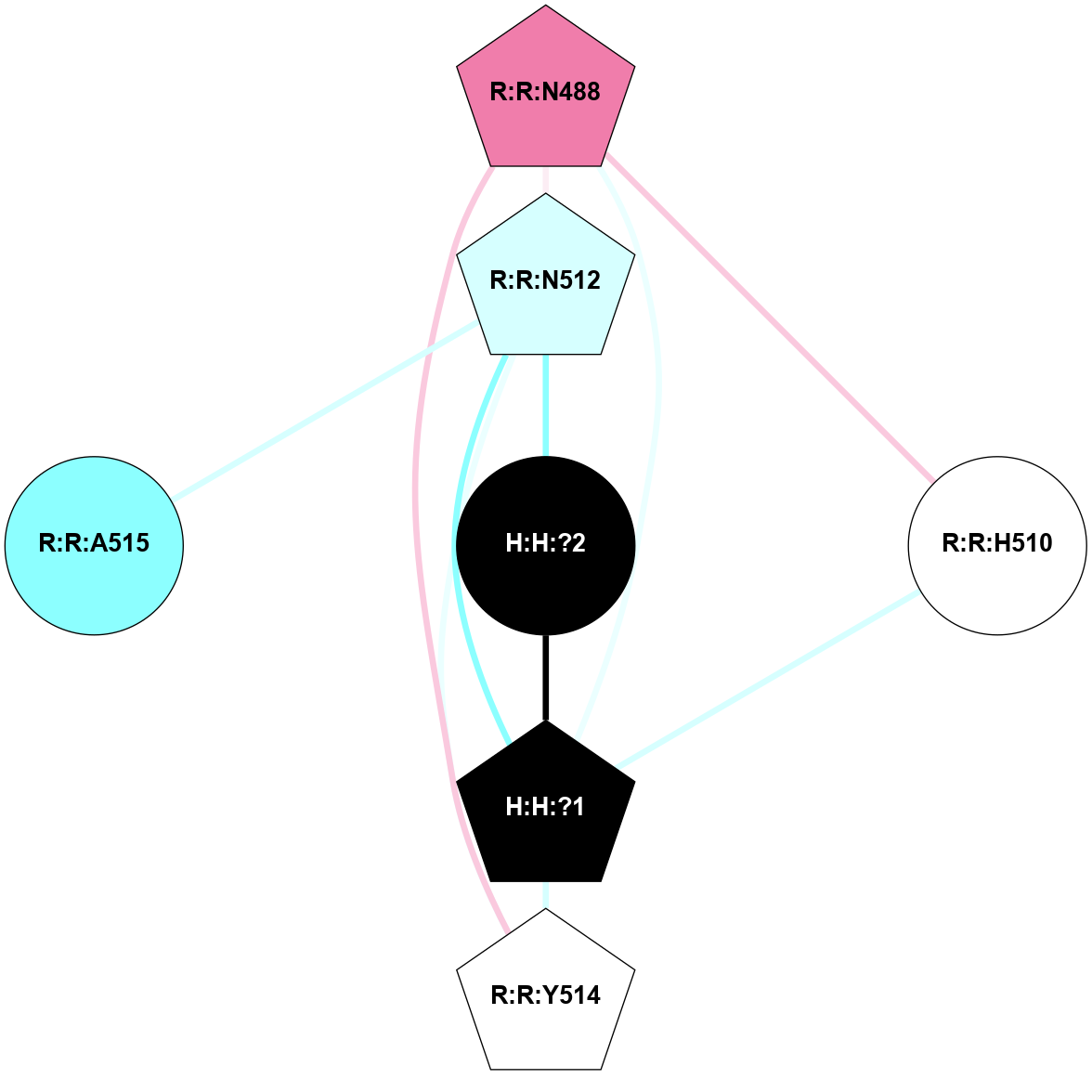
A 2D representation of the interactions of NAG in 7DD5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 26.57 | 17 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N488 | 23.33 | 17 | Yes | Yes | 0 | 8 | 1 | 2 | | H:H:?1 | R:R:H510 | 13.8 | 17 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?1 | R:R:N512 | 20.88 | 17 | Yes | Yes | 0 | 3 | 1 | 1 | | H:H:?1 | R:R:Y514 | 13.63 | 17 | Yes | Yes | 0 | 5 | 1 | 2 | | H:H:?2 | R:R:N512 | 3.68 | 17 | No | Yes | 0 | 3 | 0 | 1 | | R:R:H510 | R:R:N488 | 6.38 | 17 | No | Yes | 5 | 8 | 2 | 2 | | R:R:N488 | R:R:N512 | 13.62 | 17 | Yes | Yes | 8 | 3 | 2 | 1 | | R:R:N488 | R:R:Y514 | 8.14 | 17 | Yes | Yes | 8 | 5 | 2 | 2 | | R:R:N512 | R:R:Y514 | 4.65 | 17 | Yes | Yes | 3 | 5 | 1 | 2 | | R:R:A515 | R:R:N512 | 3.13 | 0 | No | Yes | 2 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 15.12 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
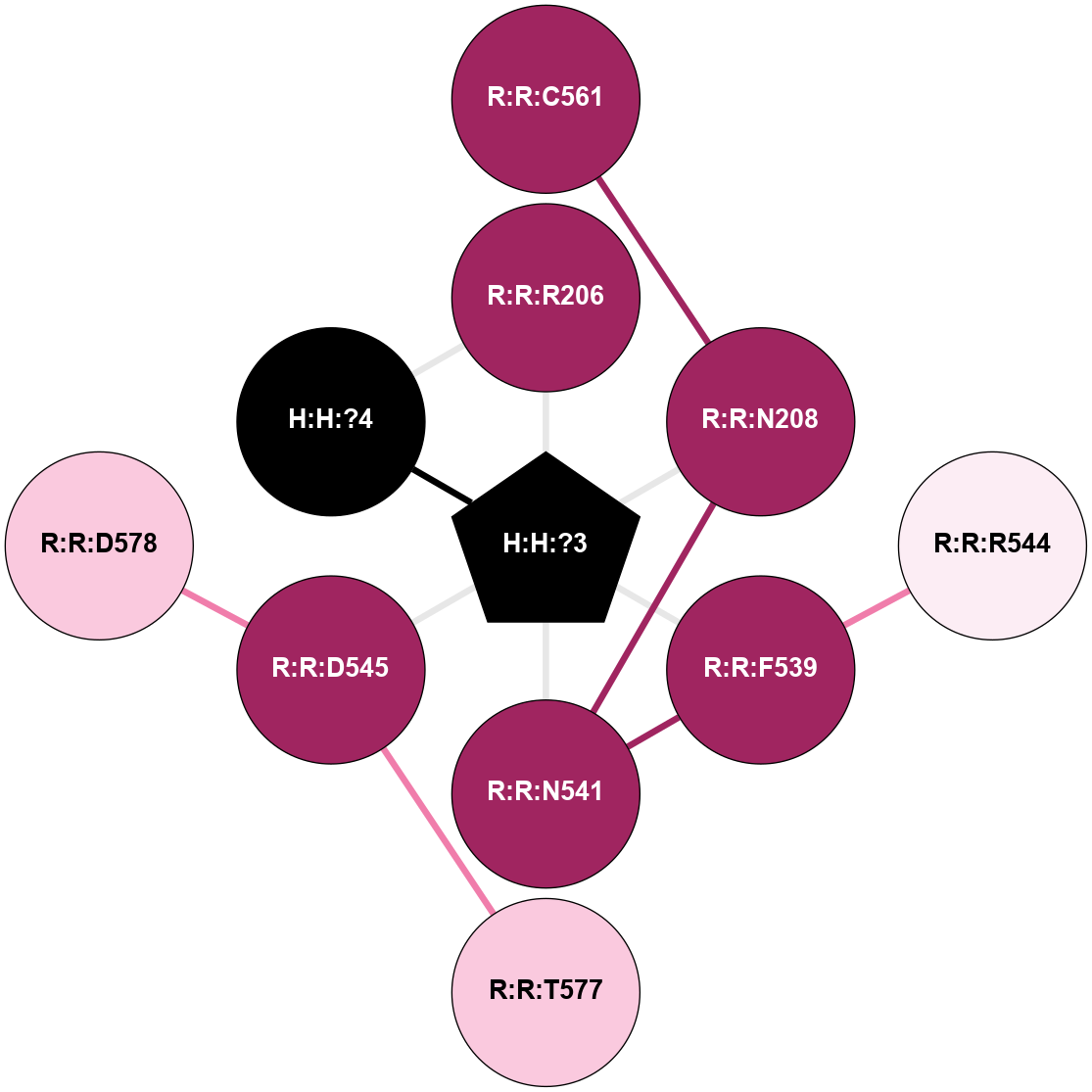
A 2D representation of the interactions of NAG in 7DD5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 35.42 | 18 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:R206 | 4.35 | 18 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?3 | R:R:N208 | 11.05 | 18 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?3 | R:R:F539 | 4.36 | 18 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?3 | R:R:N541 | 25.79 | 18 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?3 | R:R:D545 | 3.64 | 18 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?4 | R:R:R206 | 5.43 | 18 | No | No | 0 | 9 | 1 | 1 | | R:R:N208 | R:R:N541 | 12.26 | 18 | No | No | 9 | 9 | 1 | 1 | | R:R:C561 | R:R:N208 | 7.87 | 0 | No | No | 9 | 9 | 2 | 1 | | R:R:F539 | R:R:N541 | 6.04 | 18 | No | No | 9 | 9 | 1 | 1 | | R:R:F539 | R:R:R544 | 18.17 | 18 | No | No | 9 | 6 | 1 | 2 | | R:R:D545 | R:R:T577 | 2.89 | 0 | No | No | 9 | 7 | 1 | 2 | | R:R:D545 | R:R:D578 | 13.31 | 0 | No | No | 9 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.10 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
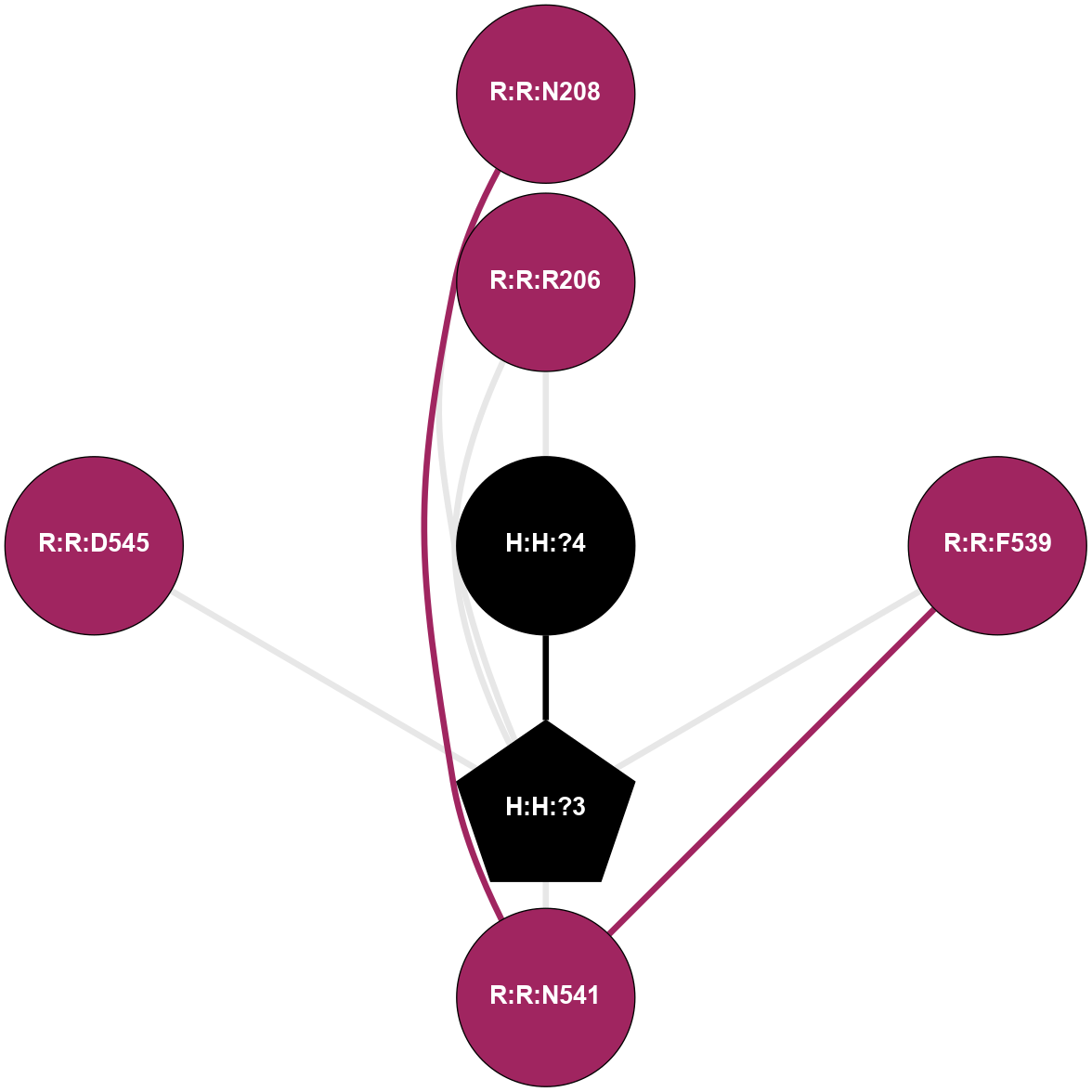
A 2D representation of the interactions of NAG in 7DD5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 35.42 | 18 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:R206 | 4.35 | 18 | Yes | No | 0 | 9 | 1 | 1 | | H:H:?3 | R:R:N208 | 11.05 | 18 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?3 | R:R:F539 | 4.36 | 18 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?3 | R:R:N541 | 25.79 | 18 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?3 | R:R:D545 | 3.64 | 18 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?4 | R:R:R206 | 5.43 | 18 | No | No | 0 | 9 | 0 | 1 | | R:R:N208 | R:R:N541 | 12.26 | 18 | No | No | 9 | 9 | 2 | 2 | | R:R:F539 | R:R:N541 | 6.04 | 18 | No | No | 9 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 20.43 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
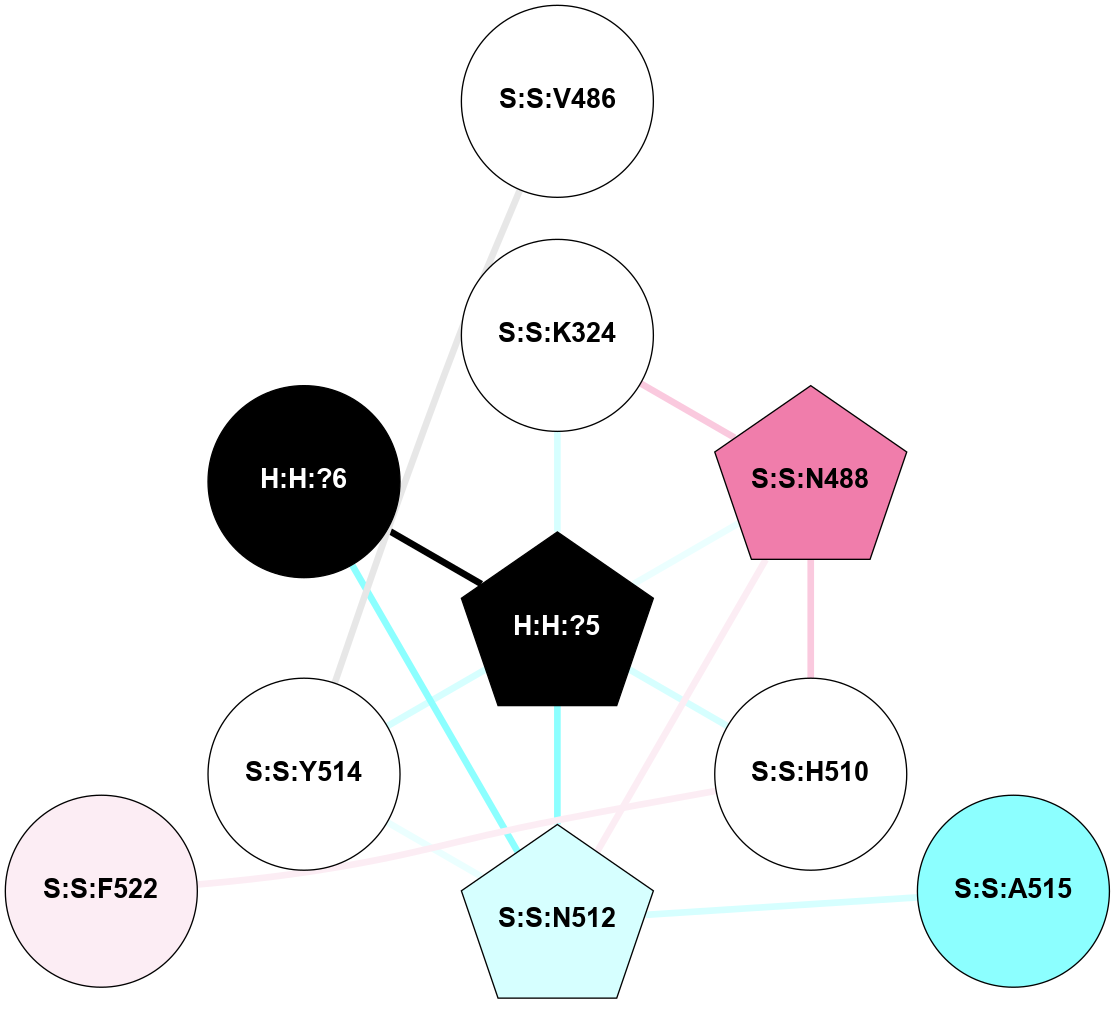
A 2D representation of the interactions of NAG in 7DD5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 34.31 | 14 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?5 | S:S:K324 | 13.87 | 14 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?5 | S:S:N488 | 27.02 | 14 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?5 | S:S:H510 | 11.5 | 14 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?5 | S:S:N512 | 19.65 | 14 | Yes | Yes | 0 | 3 | 0 | 1 | | H:H:?5 | S:S:Y514 | 12.58 | 14 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?6 | S:S:N512 | 3.68 | 14 | No | Yes | 0 | 3 | 1 | 1 | | S:S:K324 | S:S:N488 | 6.99 | 14 | No | No | 5 | 8 | 1 | 1 | | S:S:V486 | S:S:Y514 | 3.79 | 0 | No | No | 5 | 5 | 2 | 1 | | S:S:H510 | S:S:N488 | 6.38 | 14 | No | No | 5 | 8 | 1 | 1 | | S:S:N512 | S:S:Y514 | 4.65 | 14 | Yes | No | 3 | 5 | 1 | 1 | | S:S:A515 | S:S:N512 | 4.69 | 0 | No | Yes | 2 | 3 | 2 | 1 | | S:S:F522 | S:S:H510 | 1.13 | 0 | No | No | 6 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.82 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
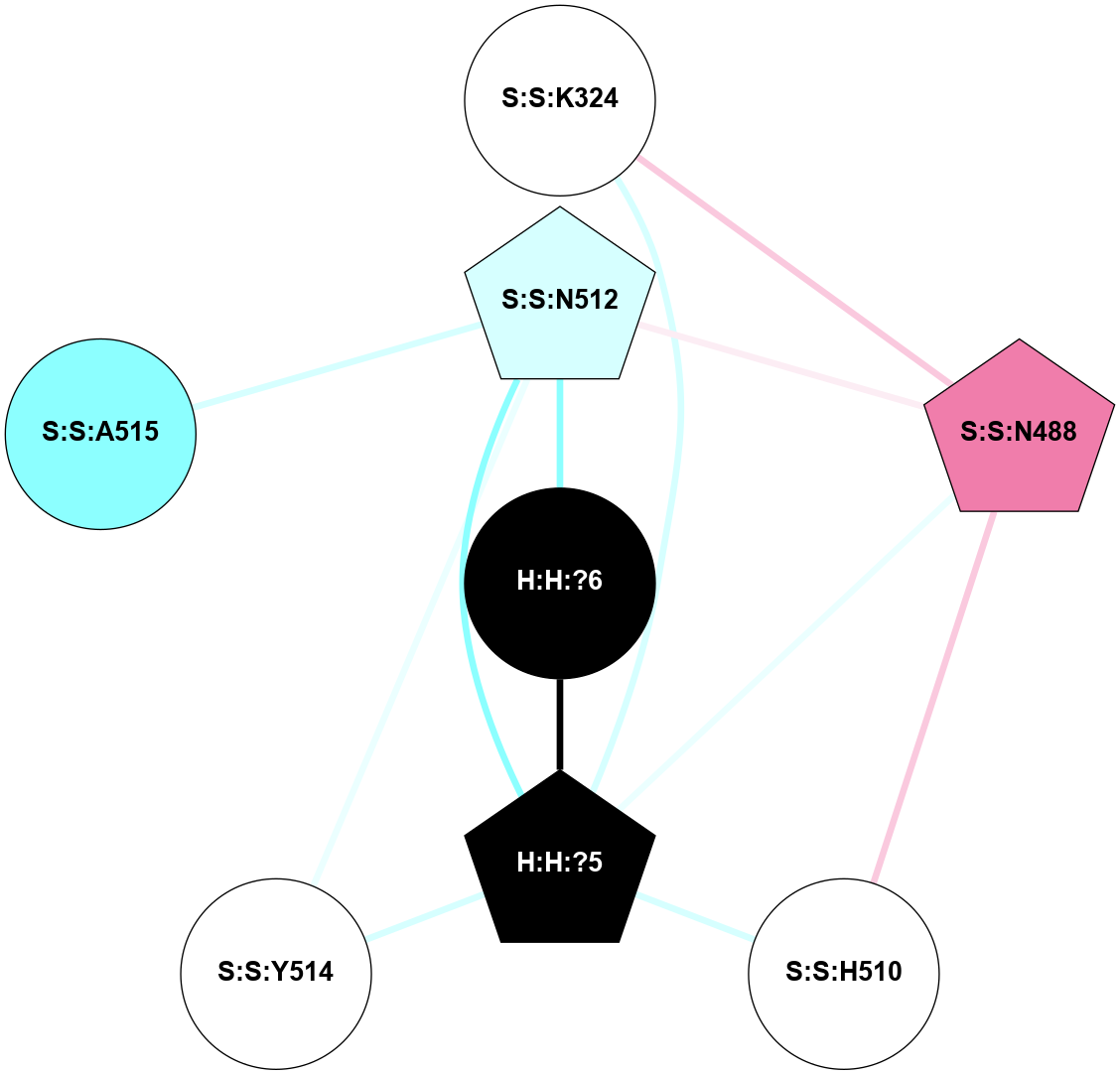
A 2D representation of the interactions of NAG in 7DD5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 34.31 | 14 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?5 | S:S:K324 | 13.87 | 14 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?5 | S:S:N488 | 27.02 | 14 | Yes | No | 0 | 8 | 1 | 2 | | H:H:?5 | S:S:H510 | 11.5 | 14 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?5 | S:S:N512 | 19.65 | 14 | Yes | Yes | 0 | 3 | 1 | 1 | | H:H:?5 | S:S:Y514 | 12.58 | 14 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?6 | S:S:N512 | 3.68 | 14 | No | Yes | 0 | 3 | 0 | 1 | | S:S:K324 | S:S:N488 | 6.99 | 14 | No | No | 5 | 8 | 2 | 2 | | S:S:H510 | S:S:N488 | 6.38 | 14 | No | No | 5 | 8 | 2 | 2 | | S:S:N512 | S:S:Y514 | 4.65 | 14 | Yes | No | 3 | 5 | 1 | 2 | | S:S:A515 | S:S:N512 | 4.69 | 0 | No | Yes | 2 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 19.00 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
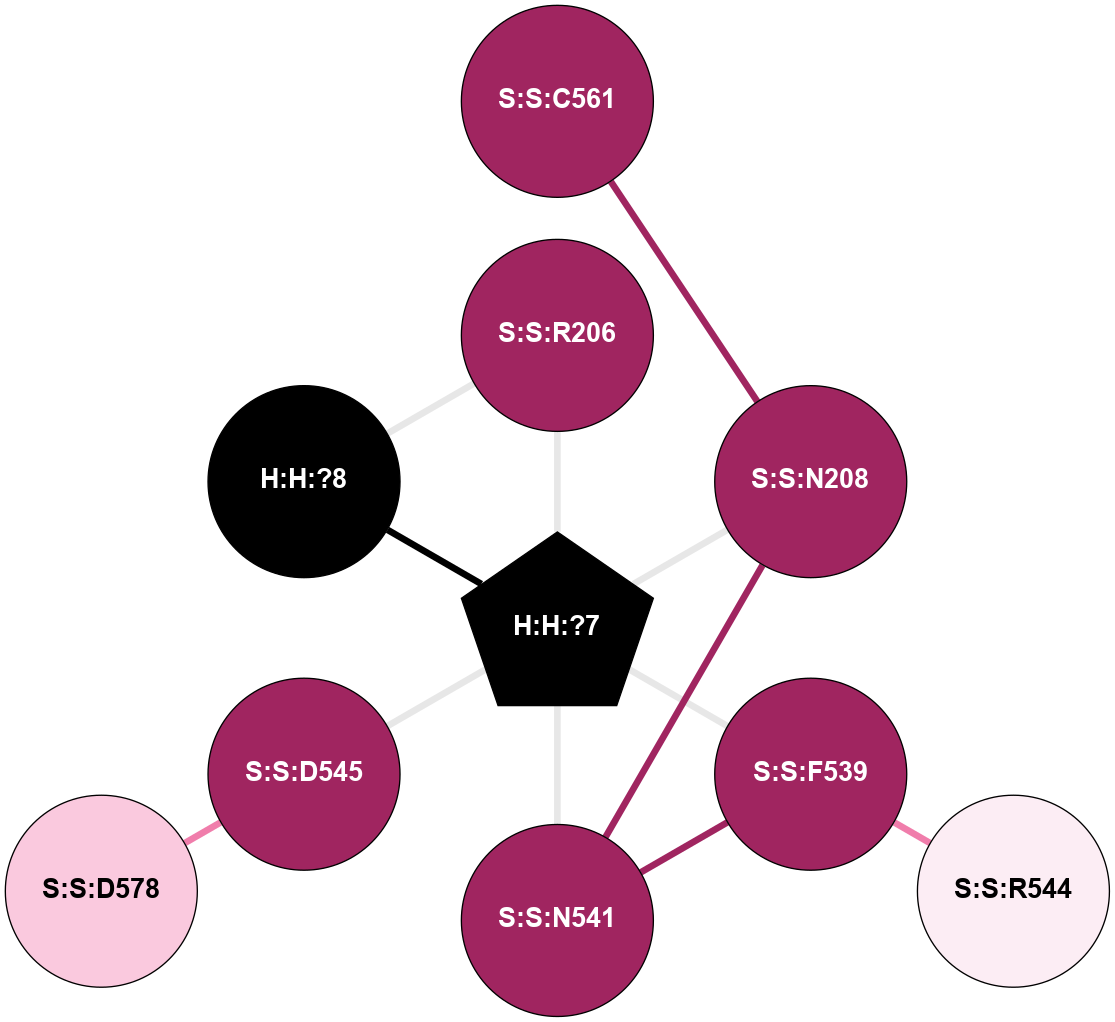
A 2D representation of the interactions of NAG in 7DD5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 37.64 | 11 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?7 | S:S:R206 | 5.43 | 11 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?7 | S:S:N208 | 7.37 | 11 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?7 | S:S:F539 | 4.36 | 11 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?7 | S:S:N541 | 23.33 | 11 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?7 | S:S:D545 | 6.07 | 11 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?8 | S:S:R206 | 15.21 | 11 | No | No | 0 | 9 | 1 | 1 | | S:S:N208 | S:S:N541 | 13.62 | 11 | No | No | 9 | 9 | 1 | 1 | | S:S:C561 | S:S:N208 | 4.72 | 11 | No | No | 9 | 9 | 2 | 1 | | S:S:F539 | S:S:N541 | 7.25 | 11 | No | No | 9 | 9 | 1 | 1 | | S:S:F539 | S:S:R544 | 3.21 | 11 | No | No | 9 | 6 | 1 | 2 | | S:S:D545 | S:S:D578 | 5.32 | 0 | No | No | 9 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.03 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
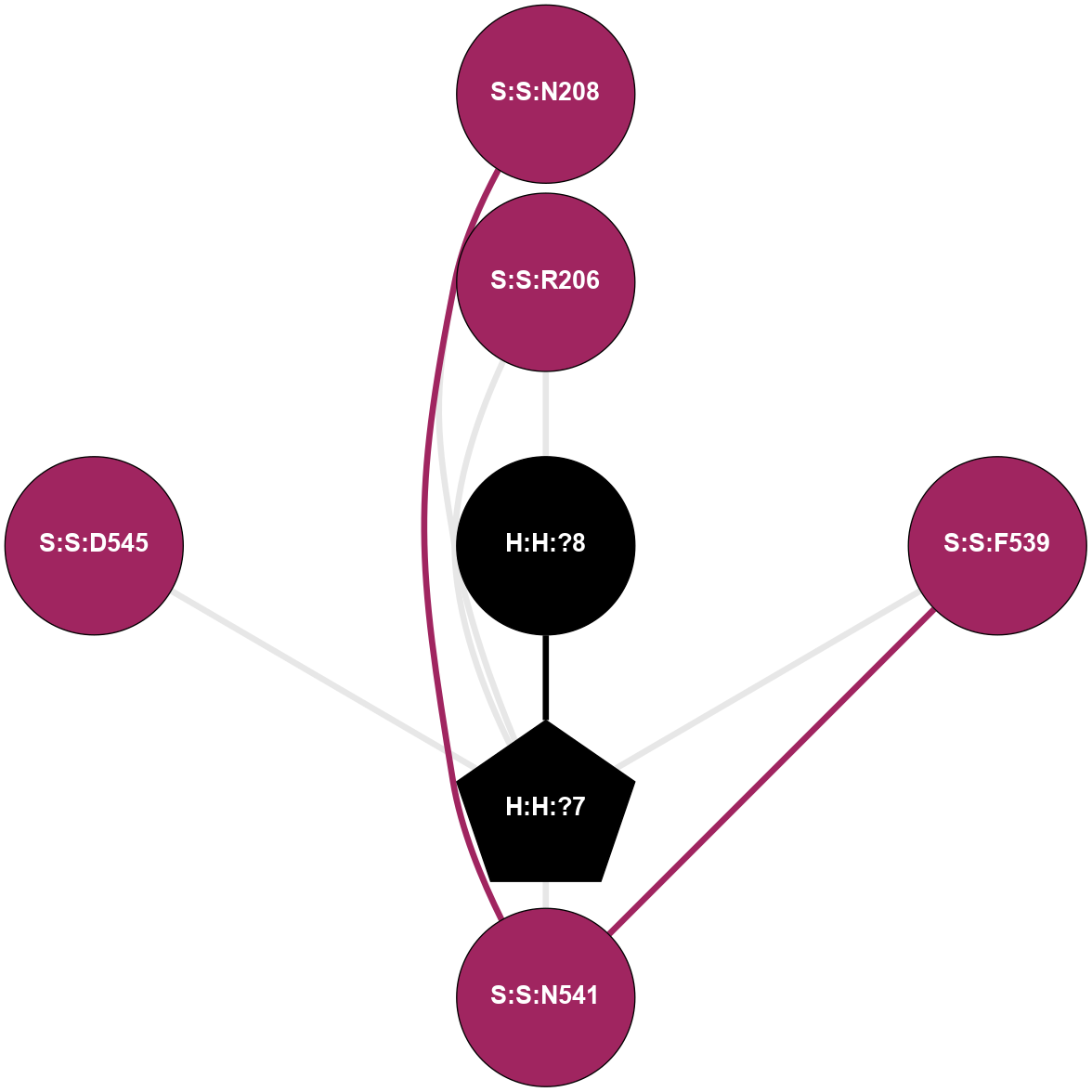
A 2D representation of the interactions of NAG in 7DD5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 37.64 | 11 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?7 | S:S:R206 | 5.43 | 11 | Yes | No | 0 | 9 | 1 | 1 | | H:H:?7 | S:S:N208 | 7.37 | 11 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?7 | S:S:F539 | 4.36 | 11 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?7 | S:S:N541 | 23.33 | 11 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?7 | S:S:D545 | 6.07 | 11 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?8 | S:S:R206 | 15.21 | 11 | No | No | 0 | 9 | 0 | 1 | | S:S:N208 | S:S:N541 | 13.62 | 11 | No | No | 9 | 9 | 2 | 2 | | S:S:F539 | S:S:N541 | 7.25 | 11 | No | No | 9 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 26.43 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
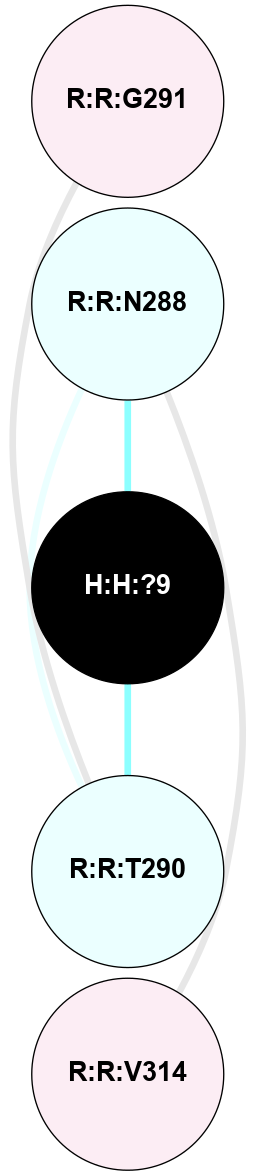
A 2D representation of the interactions of NAG in 7DD5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?9 | R:R:N288 | 28.24 | 25 | No | No | 0 | 4 | 0 | 1 | | H:H:?9 | R:R:T290 | 3.95 | 25 | No | No | 0 | 4 | 0 | 1 | | R:R:N288 | R:R:T290 | 2.92 | 25 | No | No | 4 | 4 | 1 | 1 | | R:R:G291 | R:R:T290 | 1.82 | 0 | No | No | 6 | 4 | 2 | 1 | | R:R:N288 | R:R:V314 | 1.48 | 25 | No | No | 4 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.09 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
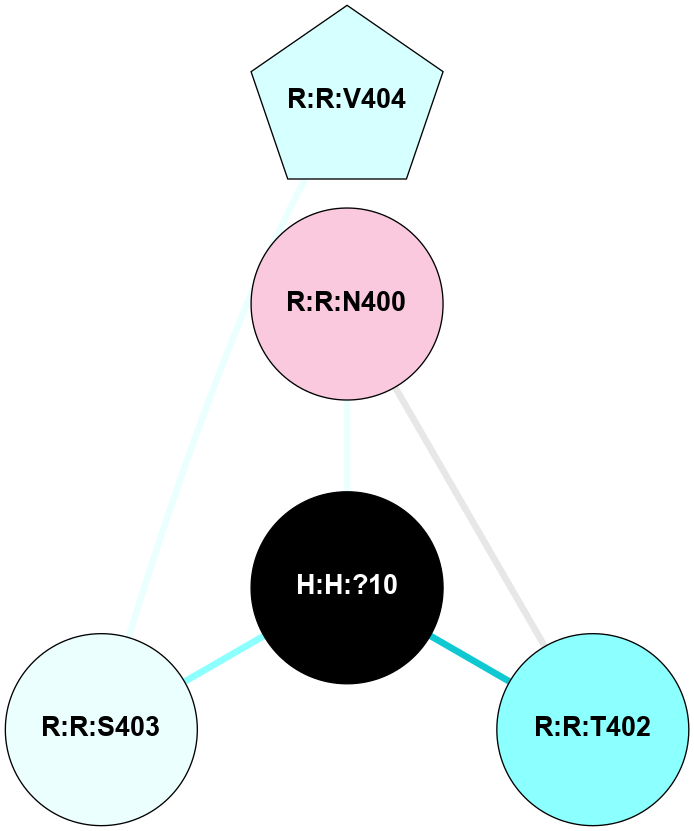
A 2D representation of the interactions of NAG in 7DD5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | R:R:N400 | 38.07 | 26 | No | No | 0 | 7 | 0 | 1 | | H:H:?10 | R:R:T402 | 10.55 | 26 | No | No | 0 | 2 | 0 | 1 | | H:H:?10 | R:R:S403 | 5.37 | 26 | No | No | 0 | 4 | 0 | 1 | | R:R:N400 | R:R:T402 | 14.62 | 26 | No | No | 7 | 2 | 1 | 1 | | R:R:S403 | R:R:V404 | 1.62 | 0 | No | Yes | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.00 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
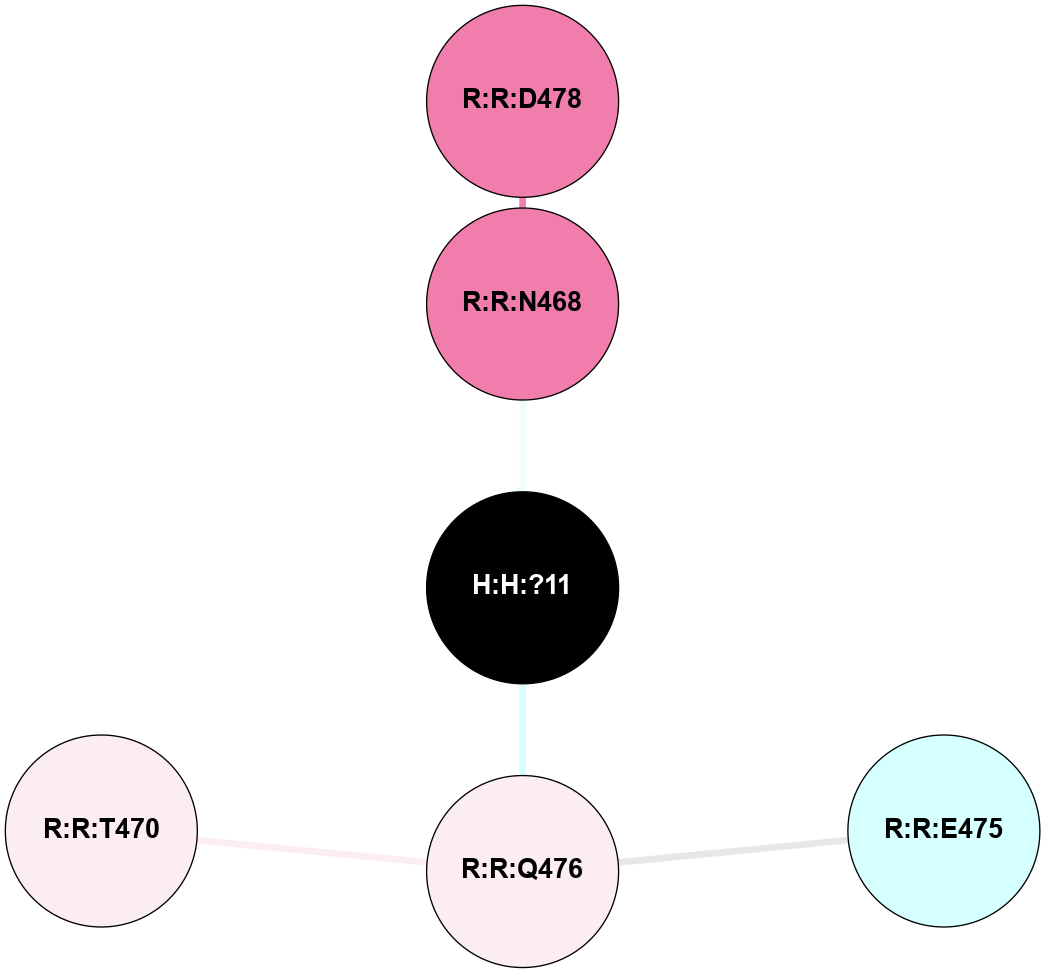
A 2D representation of the interactions of NAG in 7DD5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | R:R:N468 | 29.47 | 0 | No | No | 0 | 8 | 0 | 1 | | R:R:D478 | R:R:N468 | 5.39 | 0 | No | No | 8 | 8 | 2 | 1 | | R:R:E475 | R:R:Q476 | 2.55 | 0 | No | No | 3 | 6 | 2 | 1 | | R:R:Q476 | R:R:T470 | 1.42 | 0 | No | No | 6 | 6 | 1 | 2 | | H:H:?11 | R:R:Q476 | 1.19 | 0 | No | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.33 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
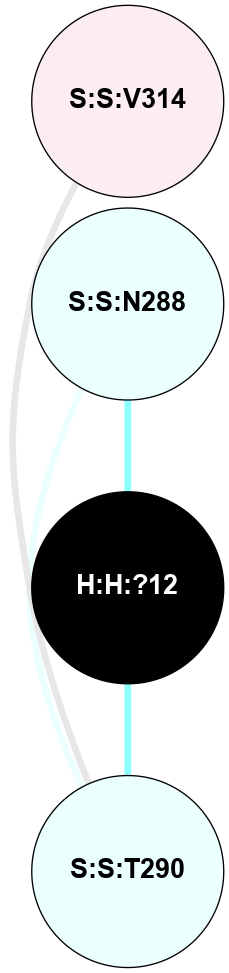
A 2D representation of the interactions of NAG in 7DD5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | S:S:N288 | 30.7 | 27 | No | No | 0 | 4 | 0 | 1 | | H:H:?12 | S:S:T290 | 3.95 | 27 | No | No | 0 | 4 | 0 | 1 | | S:S:N288 | S:S:T290 | 4.39 | 27 | No | No | 4 | 4 | 1 | 1 | | S:S:T290 | S:S:V314 | 1.59 | 27 | No | No | 4 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.32 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
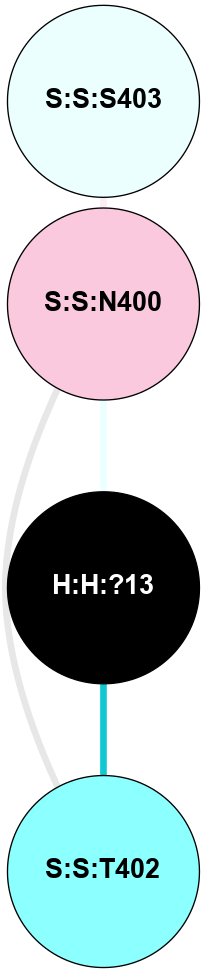
A 2D representation of the interactions of NAG in 7DD5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | S:S:N400 | 28.24 | 28 | No | No | 0 | 7 | 0 | 1 | | H:H:?13 | S:S:T402 | 15.82 | 28 | No | No | 0 | 2 | 0 | 1 | | S:S:N400 | S:S:T402 | 8.77 | 28 | No | No | 7 | 2 | 1 | 1 | | S:S:N400 | S:S:S403 | 2.98 | 28 | No | No | 7 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 22.03 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7DD5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?14 | S:S:N468 | 29.47 | 29 | No | No | 0 | 8 | 0 | 1 | | H:H:?14 | S:S:Q476 | 13.09 | 29 | No | No | 0 | 6 | 0 | 1 | | S:S:N468 | S:S:Q476 | 6.6 | 29 | No | No | 8 | 6 | 1 | 1 | | S:S:D478 | S:S:N468 | 9.42 | 0 | No | No | 8 | 8 | 2 | 1 | | S:S:Q476 | S:S:T470 | 5.67 | 29 | No | No | 6 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 21.28 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7DD6 | C | Ion | Calcium Sensing | CaS | Gallus Gallus | Ca | Ca; Ca; Tryptophan | - | 3.2 | 2021-06-16 | doi.org/10.1126/sciadv.abg1483 |
|
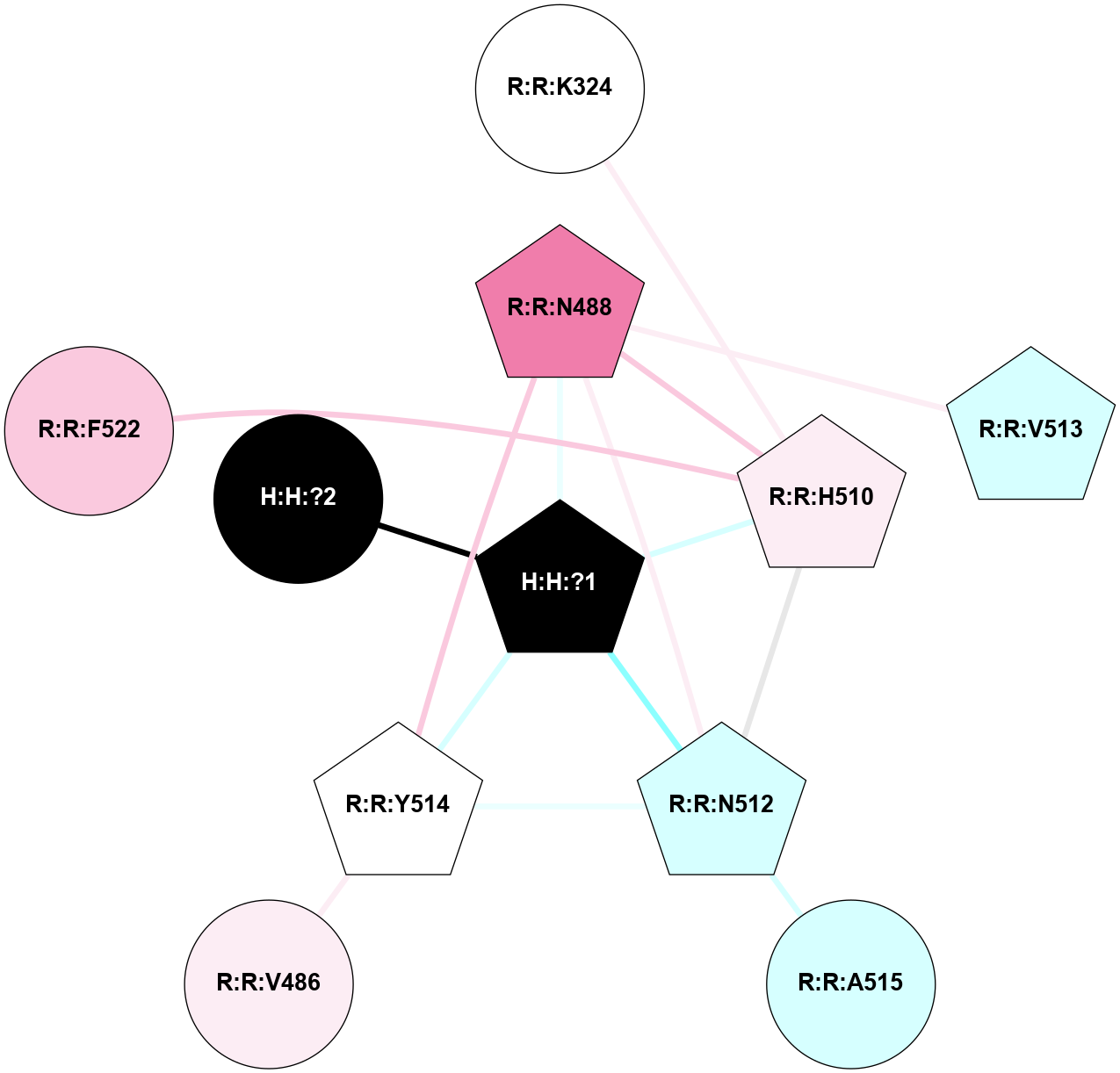
A 2D representation of the interactions of NAG in 7DD6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 30.99 | 3 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N488 | 24.56 | 3 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?1 | R:R:H510 | 13.8 | 3 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:N512 | 17.19 | 3 | Yes | Yes | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:Y514 | 13.63 | 3 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:H510 | R:R:K324 | 7.86 | 3 | Yes | No | 6 | 5 | 1 | 2 | | R:R:H510 | R:R:N488 | 3.83 | 3 | Yes | Yes | 6 | 8 | 1 | 1 | | R:R:N488 | R:R:N512 | 12.26 | 3 | Yes | Yes | 8 | 3 | 1 | 1 | | R:R:N488 | R:R:V513 | 4.43 | 3 | Yes | Yes | 8 | 3 | 1 | 2 | | R:R:N488 | R:R:Y514 | 8.14 | 3 | Yes | Yes | 8 | 5 | 1 | 1 | | R:R:H510 | R:R:N512 | 2.55 | 3 | Yes | Yes | 6 | 3 | 1 | 1 | | R:R:N512 | R:R:Y514 | 8.14 | 3 | Yes | Yes | 3 | 5 | 1 | 1 | | R:R:A515 | R:R:N512 | 1.56 | 0 | No | Yes | 3 | 3 | 2 | 1 | | R:R:V486 | R:R:Y514 | 1.26 | 0 | No | Yes | 6 | 5 | 2 | 1 | | R:R:F522 | R:R:H510 | 1.13 | 0 | No | Yes | 7 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 20.03 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
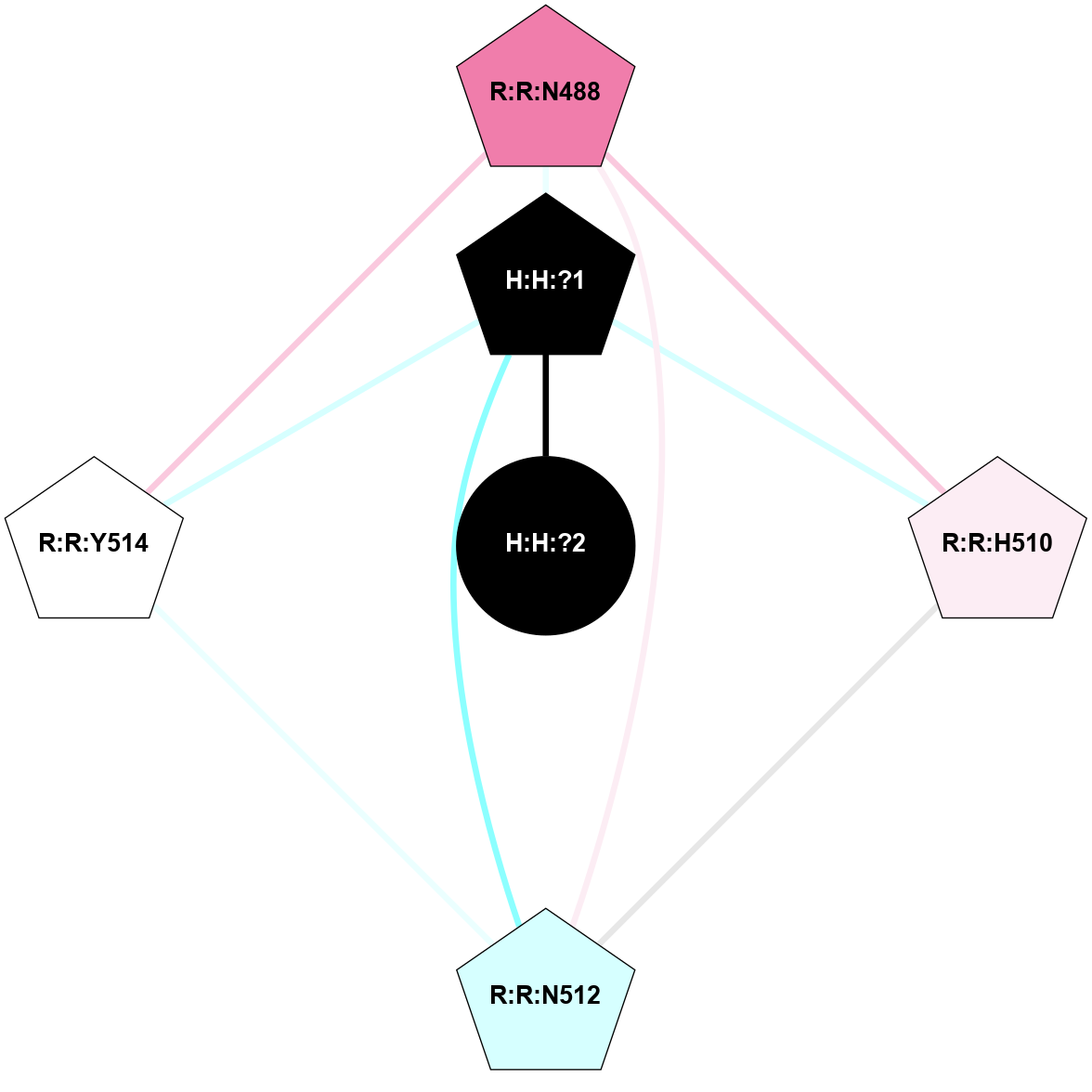
A 2D representation of the interactions of NAG in 7DD6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 30.99 | 3 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N488 | 24.56 | 3 | Yes | Yes | 0 | 8 | 1 | 2 | | H:H:?1 | R:R:H510 | 13.8 | 3 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:N512 | 17.19 | 3 | Yes | Yes | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:Y514 | 13.63 | 3 | Yes | Yes | 0 | 5 | 1 | 2 | | R:R:H510 | R:R:N488 | 3.83 | 3 | Yes | Yes | 6 | 8 | 2 | 2 | | R:R:N488 | R:R:N512 | 12.26 | 3 | Yes | Yes | 8 | 3 | 2 | 2 | | R:R:N488 | R:R:Y514 | 8.14 | 3 | Yes | Yes | 8 | 5 | 2 | 2 | | R:R:H510 | R:R:N512 | 2.55 | 3 | Yes | Yes | 6 | 3 | 2 | 2 | | R:R:N512 | R:R:Y514 | 8.14 | 3 | Yes | Yes | 3 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 30.99 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
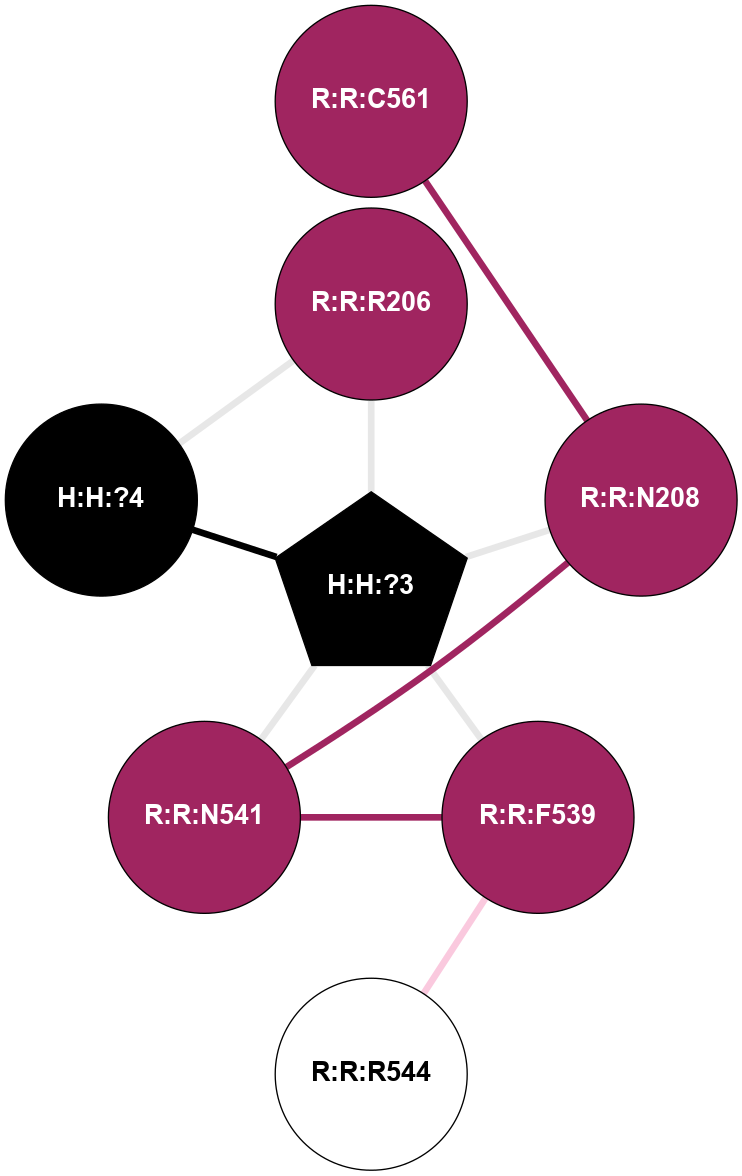
A 2D representation of the interactions of NAG in 7DD6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 33.21 | 8 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:R206 | 8.69 | 8 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?3 | R:R:N208 | 11.05 | 8 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?3 | R:R:F539 | 4.36 | 8 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?3 | R:R:N541 | 25.79 | 8 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?4 | R:R:R206 | 10.86 | 8 | No | No | 0 | 9 | 1 | 1 | | R:R:N208 | R:R:N541 | 12.26 | 8 | No | No | 9 | 9 | 1 | 1 | | R:R:C561 | R:R:N208 | 4.72 | 8 | No | No | 9 | 9 | 2 | 1 | | R:R:F539 | R:R:N541 | 6.04 | 8 | No | No | 9 | 9 | 1 | 1 | | R:R:F539 | R:R:R544 | 18.17 | 8 | No | No | 9 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.62 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
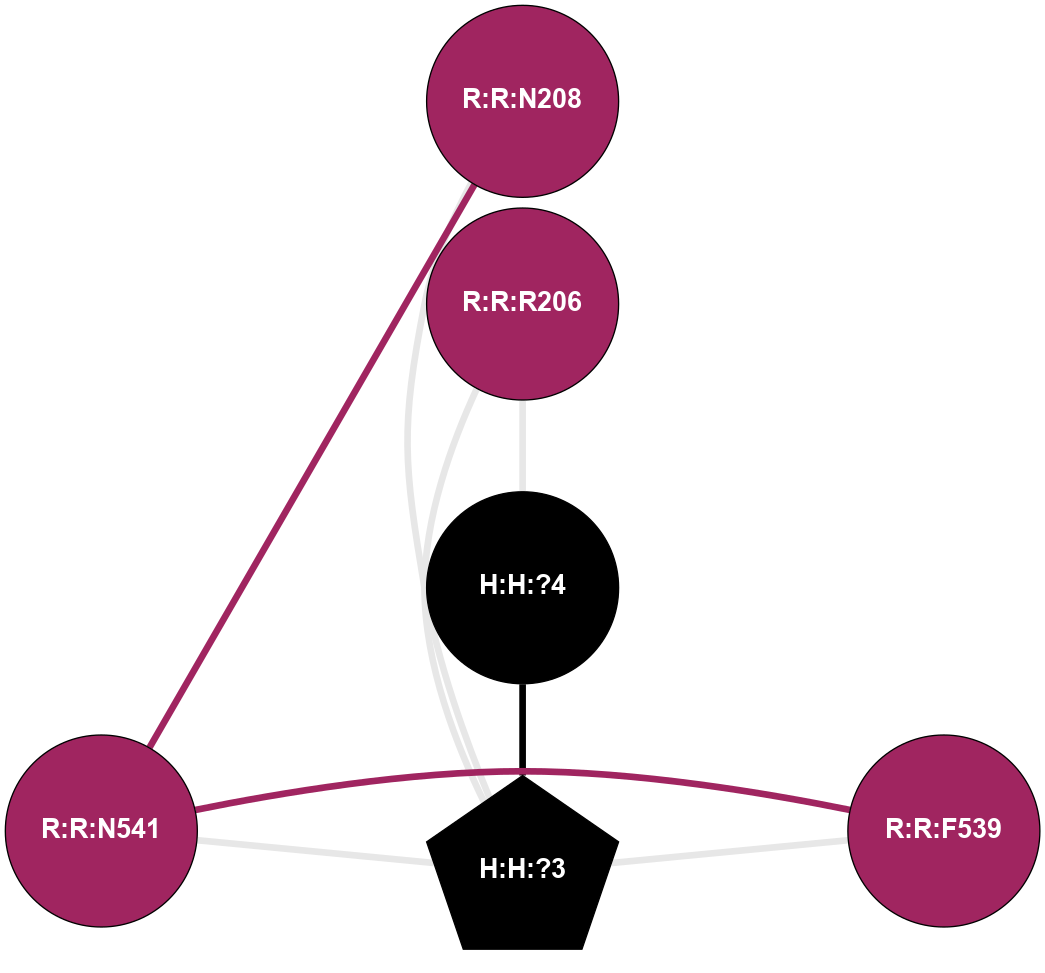
A 2D representation of the interactions of NAG in 7DD6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 33.21 | 8 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:R206 | 8.69 | 8 | Yes | No | 0 | 9 | 1 | 1 | | H:H:?3 | R:R:N208 | 11.05 | 8 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?3 | R:R:F539 | 4.36 | 8 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?3 | R:R:N541 | 25.79 | 8 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?4 | R:R:R206 | 10.86 | 8 | No | No | 0 | 9 | 0 | 1 | | R:R:N208 | R:R:N541 | 12.26 | 8 | No | No | 9 | 9 | 2 | 2 | | R:R:F539 | R:R:N541 | 6.04 | 8 | No | No | 9 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 22.04 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
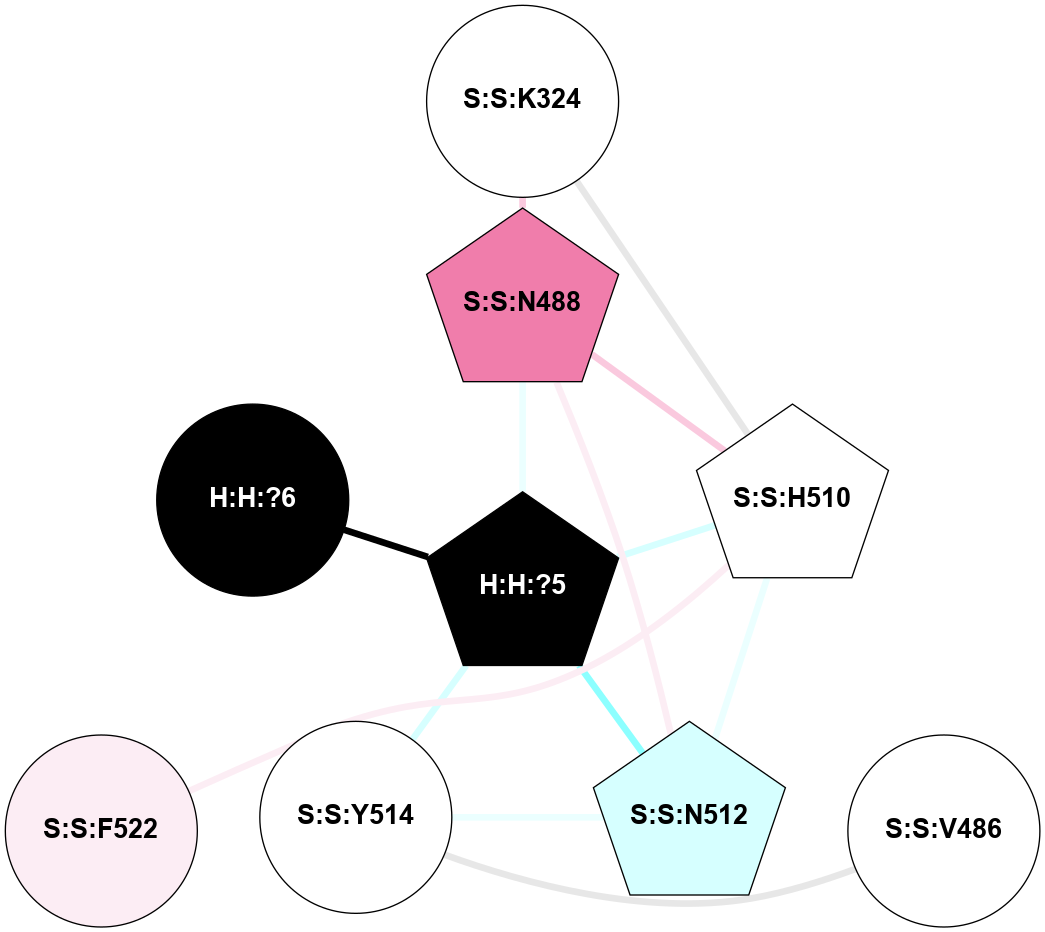
A 2D representation of the interactions of NAG in 7DD6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 29.89 | 16 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?5 | S:S:N488 | 27.02 | 16 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?5 | S:S:H510 | 12.65 | 16 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?5 | S:S:N512 | 19.65 | 16 | Yes | Yes | 0 | 3 | 0 | 1 | | H:H:?5 | S:S:Y514 | 11.53 | 16 | Yes | No | 0 | 5 | 0 | 1 | | S:S:K324 | S:S:N488 | 2.8 | 16 | No | Yes | 5 | 8 | 2 | 1 | | S:S:H510 | S:S:K324 | 6.55 | 16 | Yes | No | 5 | 5 | 1 | 2 | | S:S:V486 | S:S:Y514 | 3.79 | 0 | No | No | 5 | 5 | 2 | 1 | | S:S:H510 | S:S:N488 | 8.93 | 16 | Yes | Yes | 5 | 8 | 1 | 1 | | S:S:N488 | S:S:N512 | 6.81 | 16 | Yes | Yes | 8 | 3 | 1 | 1 | | S:S:H510 | S:S:N512 | 2.55 | 16 | Yes | Yes | 5 | 3 | 1 | 1 | | S:S:N512 | S:S:Y514 | 5.81 | 16 | Yes | No | 3 | 5 | 1 | 1 | | S:S:F522 | S:S:H510 | 1.13 | 0 | No | Yes | 6 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 20.15 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
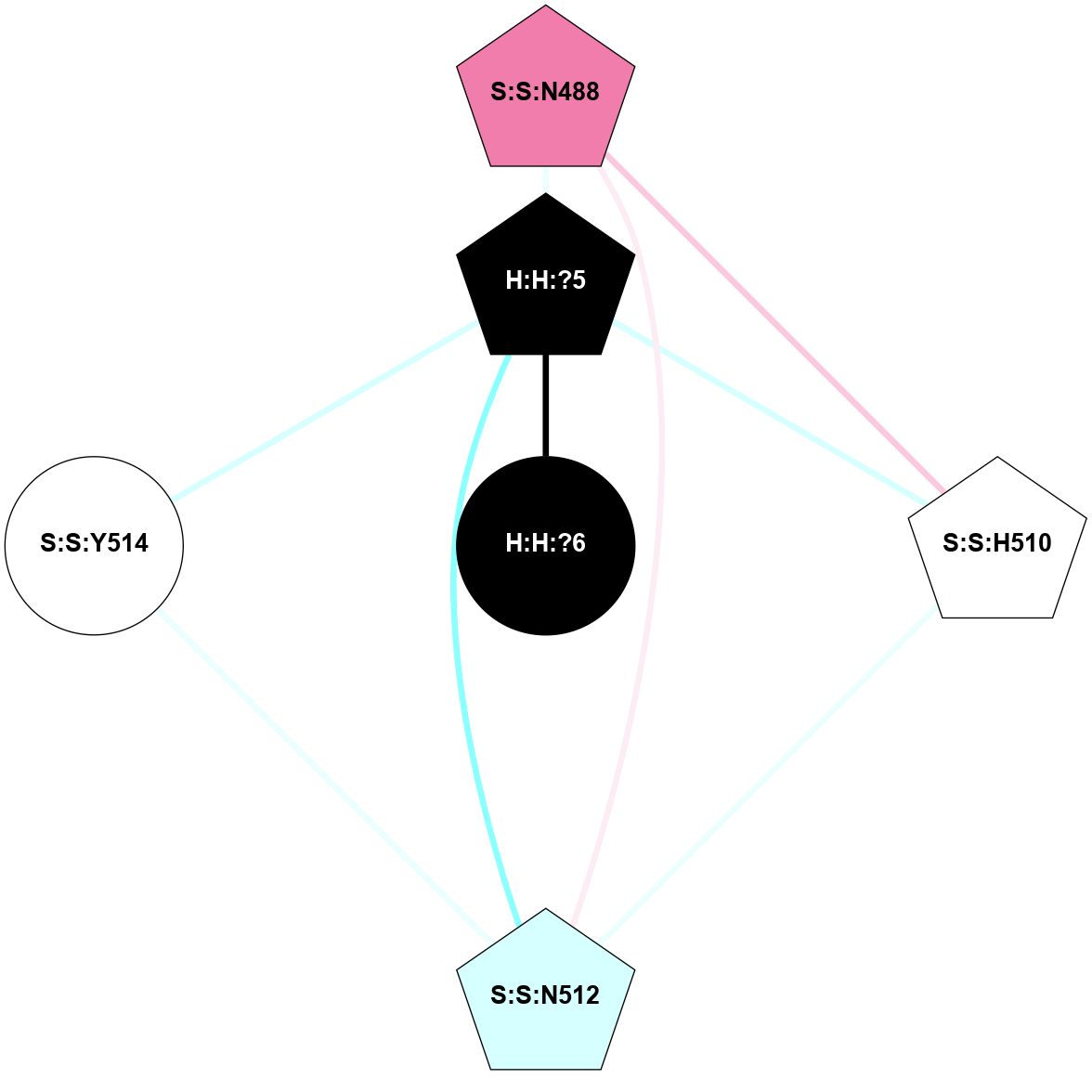
A 2D representation of the interactions of NAG in 7DD6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 29.89 | 16 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?5 | S:S:N488 | 27.02 | 16 | Yes | Yes | 0 | 8 | 1 | 2 | | H:H:?5 | S:S:H510 | 12.65 | 16 | Yes | Yes | 0 | 5 | 1 | 2 | | H:H:?5 | S:S:N512 | 19.65 | 16 | Yes | Yes | 0 | 3 | 1 | 2 | | H:H:?5 | S:S:Y514 | 11.53 | 16 | Yes | No | 0 | 5 | 1 | 2 | | S:S:H510 | S:S:N488 | 8.93 | 16 | Yes | Yes | 5 | 8 | 2 | 2 | | S:S:N488 | S:S:N512 | 6.81 | 16 | Yes | Yes | 8 | 3 | 2 | 2 | | S:S:H510 | S:S:N512 | 2.55 | 16 | Yes | Yes | 5 | 3 | 2 | 2 | | S:S:N512 | S:S:Y514 | 5.81 | 16 | Yes | No | 3 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 29.89 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
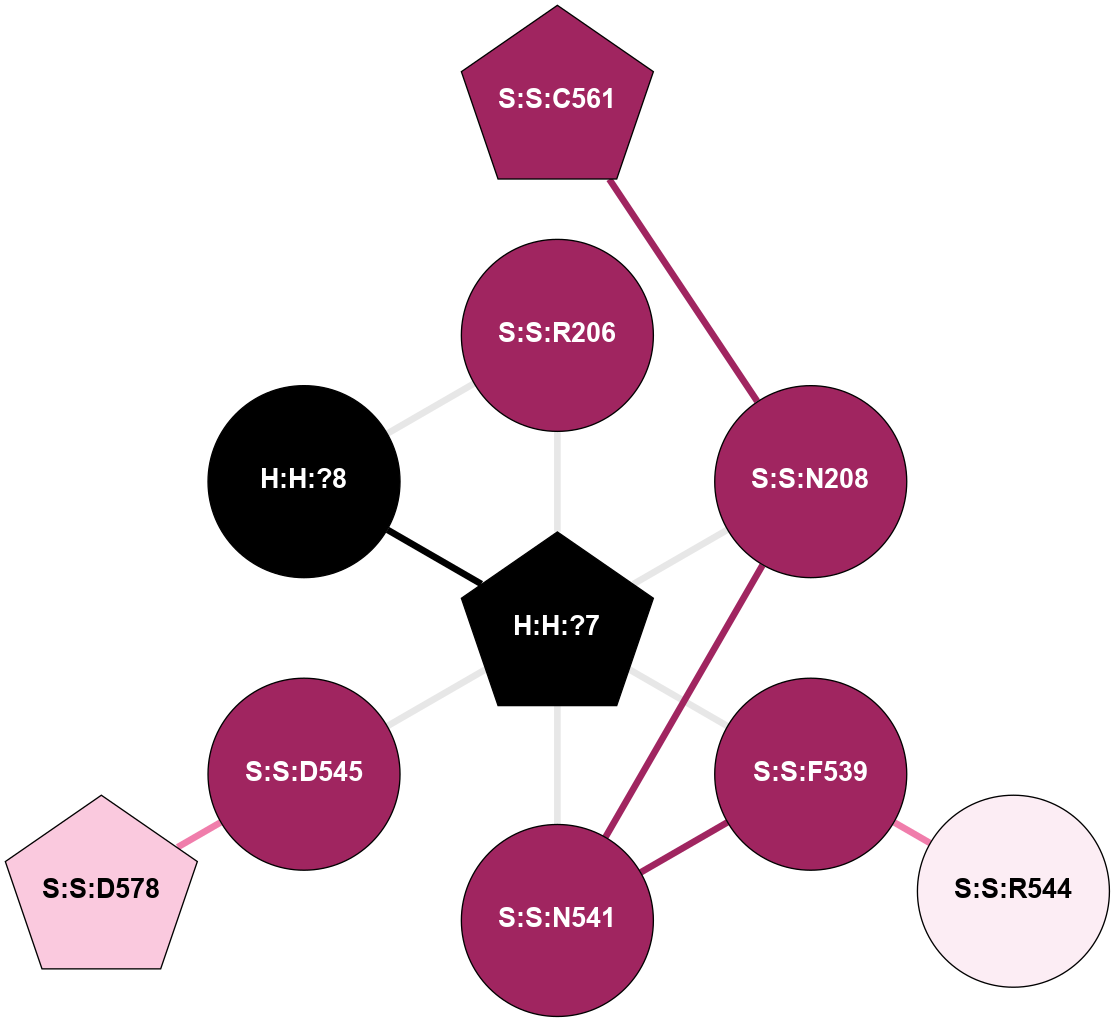
A 2D representation of the interactions of NAG in 7DD6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 39.85 | 6 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?7 | S:S:R206 | 4.35 | 6 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?7 | S:S:N208 | 7.37 | 6 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?7 | S:S:F539 | 5.45 | 6 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?7 | S:S:N541 | 25.79 | 6 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?7 | S:S:D545 | 6.07 | 6 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?8 | S:S:R206 | 15.21 | 6 | No | No | 0 | 9 | 1 | 1 | | S:S:N208 | S:S:N541 | 12.26 | 6 | No | No | 9 | 9 | 1 | 1 | | S:S:C561 | S:S:N208 | 4.72 | 6 | Yes | No | 9 | 9 | 2 | 1 | | S:S:F539 | S:S:N541 | 8.46 | 6 | No | No | 9 | 9 | 1 | 1 | | S:S:F539 | S:S:R544 | 14.97 | 6 | No | No | 9 | 6 | 1 | 2 | | S:S:D545 | S:S:D578 | 13.31 | 0 | No | Yes | 9 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.81 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
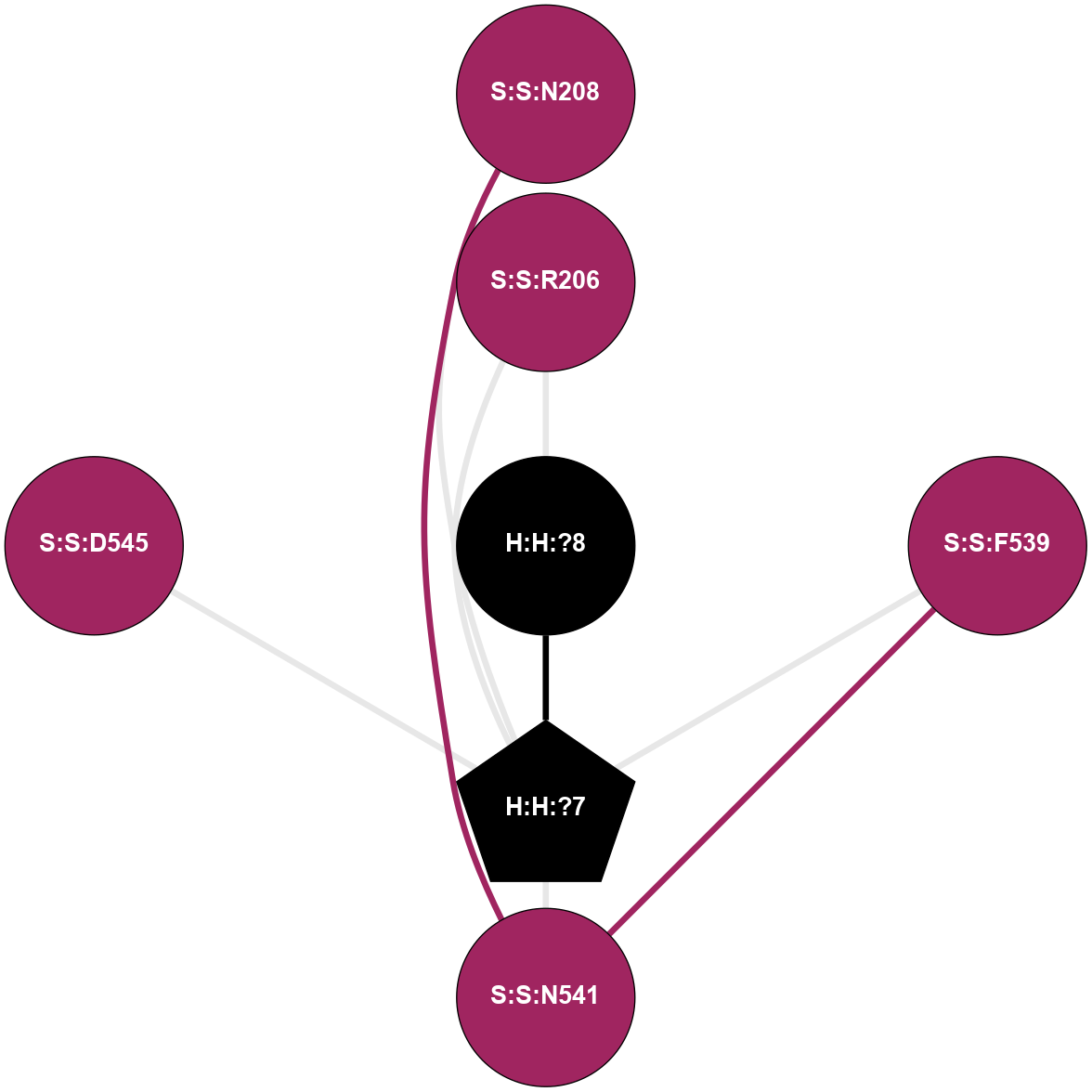
A 2D representation of the interactions of NAG in 7DD6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 39.85 | 6 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?7 | S:S:R206 | 4.35 | 6 | Yes | No | 0 | 9 | 1 | 1 | | H:H:?7 | S:S:N208 | 7.37 | 6 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?7 | S:S:F539 | 5.45 | 6 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?7 | S:S:N541 | 25.79 | 6 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?7 | S:S:D545 | 6.07 | 6 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?8 | S:S:R206 | 15.21 | 6 | No | No | 0 | 9 | 0 | 1 | | S:S:N208 | S:S:N541 | 12.26 | 6 | No | No | 9 | 9 | 2 | 2 | | S:S:F539 | S:S:N541 | 8.46 | 6 | No | No | 9 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 27.53 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
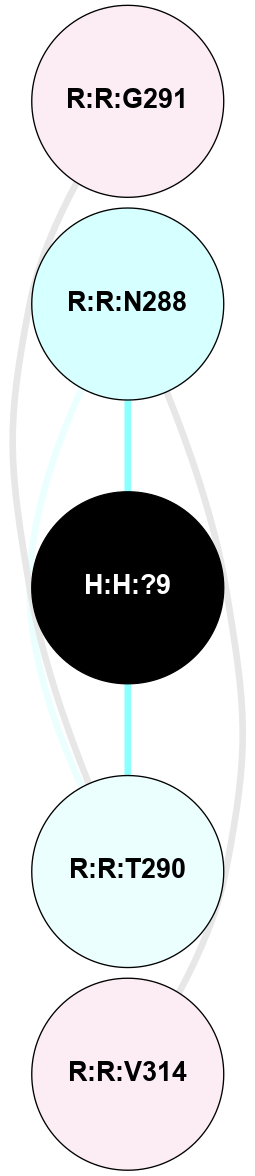
A 2D representation of the interactions of NAG in 7DD6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?9 | R:R:N288 | 30.7 | 30 | No | No | 0 | 3 | 0 | 1 | | H:H:?9 | R:R:T290 | 6.59 | 30 | No | No | 0 | 4 | 0 | 1 | | R:R:N288 | R:R:T290 | 4.39 | 30 | No | No | 3 | 4 | 1 | 1 | | R:R:G291 | R:R:T290 | 1.82 | 0 | No | No | 6 | 4 | 2 | 1 | | R:R:N288 | R:R:V314 | 1.48 | 30 | No | No | 3 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.64 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
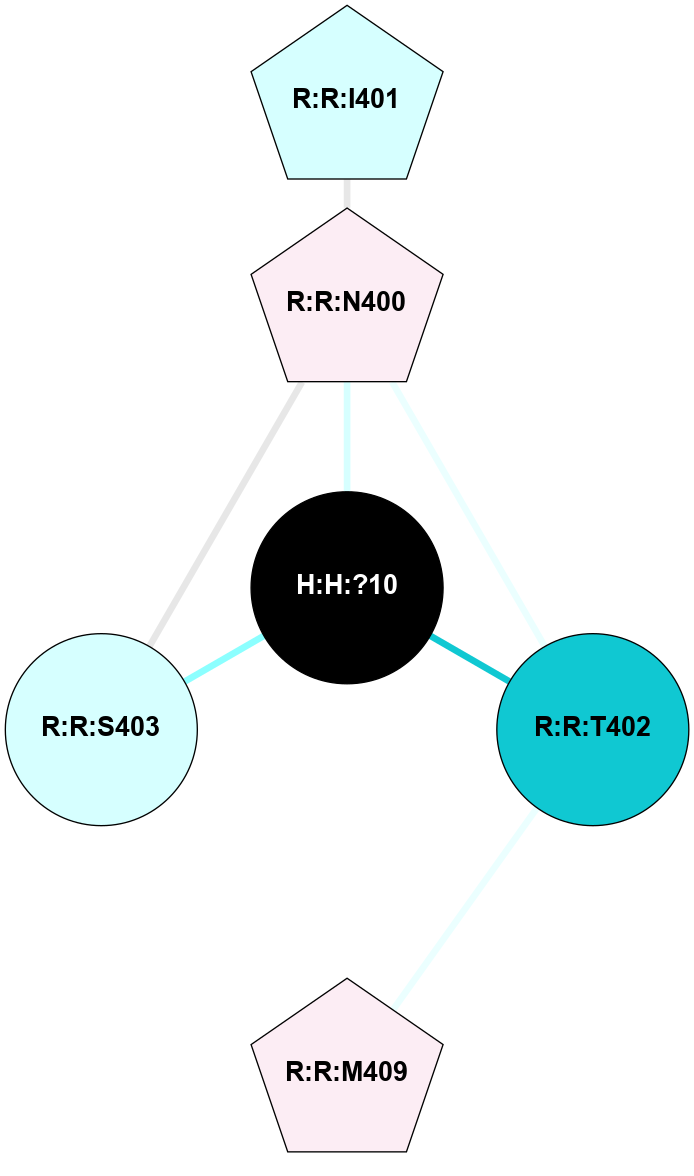
A 2D representation of the interactions of NAG in 7DD6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | R:R:N400 | 33.15 | 2 | No | Yes | 0 | 6 | 0 | 1 | | H:H:?10 | R:R:T402 | 9.23 | 2 | No | No | 0 | 1 | 0 | 1 | | H:H:?10 | R:R:S403 | 4.03 | 2 | No | No | 0 | 3 | 0 | 1 | | R:R:I401 | R:R:N400 | 2.83 | 2 | Yes | Yes | 3 | 6 | 2 | 1 | | R:R:N400 | R:R:T402 | 14.62 | 2 | Yes | No | 6 | 1 | 1 | 1 | | R:R:N400 | R:R:S403 | 4.47 | 2 | Yes | No | 6 | 3 | 1 | 1 | | R:R:M409 | R:R:T402 | 3.01 | 2 | Yes | No | 6 | 1 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.47 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7DD6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | R:R:N468 | 25.79 | 31 | No | No | 0 | 8 | 0 | 1 | | H:H:?11 | R:R:Q476 | 14.28 | 31 | No | No | 0 | 6 | 0 | 1 | | R:R:N468 | R:R:Q476 | 3.96 | 31 | No | No | 8 | 6 | 1 | 1 | | R:R:D478 | R:R:N468 | 5.39 | 0 | No | No | 9 | 8 | 2 | 1 | | R:R:Q476 | R:R:T470 | 5.67 | 31 | No | No | 6 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.04 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
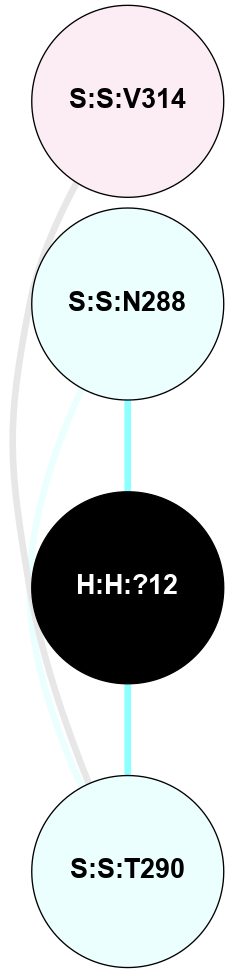
A 2D representation of the interactions of NAG in 7DD6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | S:S:N288 | 29.47 | 32 | No | No | 0 | 4 | 0 | 1 | | H:H:?12 | S:S:T290 | 5.27 | 32 | No | No | 0 | 4 | 0 | 1 | | S:S:N288 | S:S:T290 | 8.77 | 32 | No | No | 4 | 4 | 1 | 1 | | S:S:T290 | S:S:V314 | 7.93 | 32 | No | No | 4 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.37 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
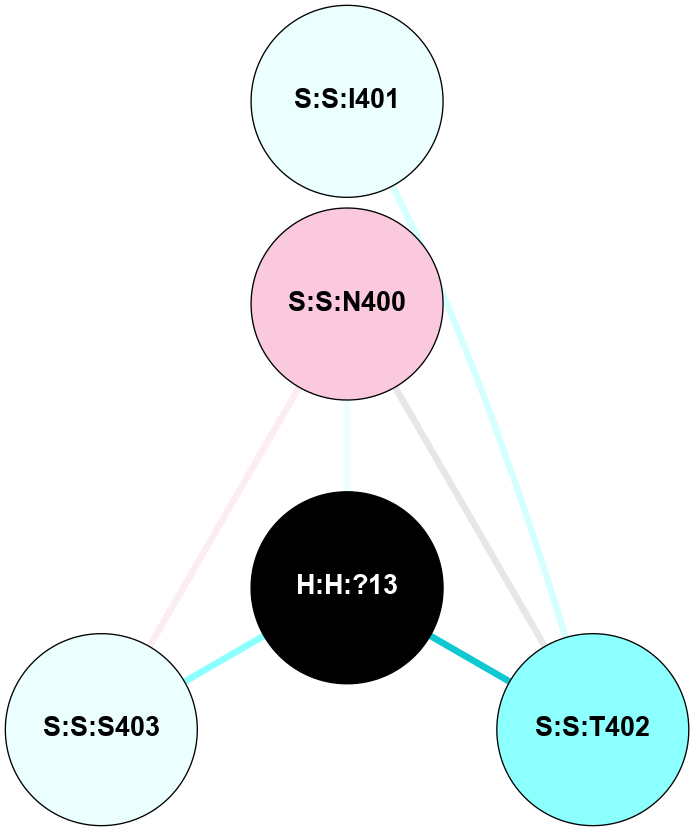
A 2D representation of the interactions of NAG in 7DD6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | S:S:N400 | 30.7 | 22 | No | No | 0 | 7 | 0 | 1 | | H:H:?13 | S:S:T402 | 14.5 | 22 | No | No | 0 | 2 | 0 | 1 | | H:H:?13 | S:S:S403 | 2.69 | 22 | No | No | 0 | 4 | 0 | 1 | | S:S:N400 | S:S:T402 | 10.24 | 22 | No | No | 7 | 2 | 1 | 1 | | S:S:N400 | S:S:S403 | 7.45 | 22 | No | No | 7 | 4 | 1 | 1 | | S:S:I401 | S:S:T402 | 1.52 | 0 | No | No | 4 | 2 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.96 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
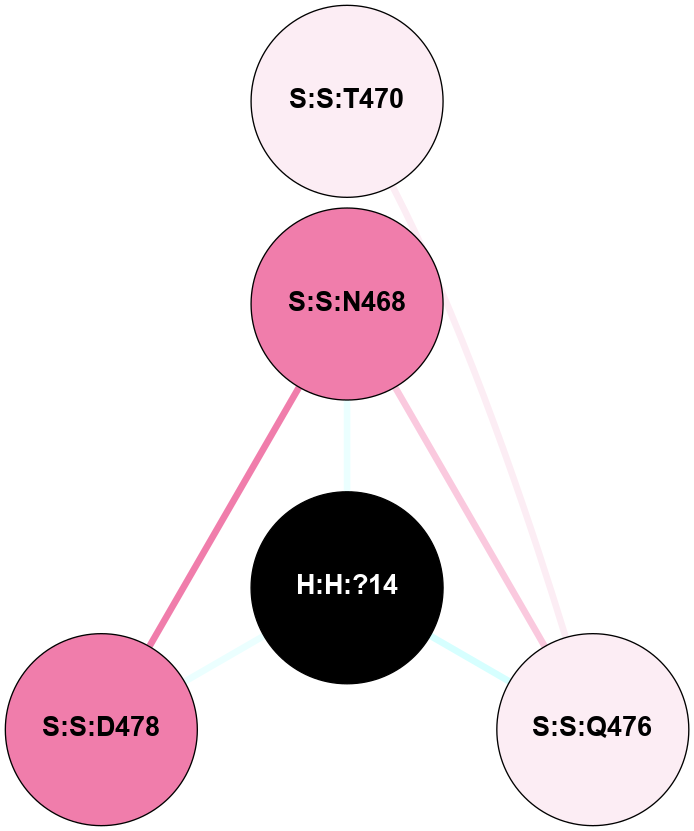
A 2D representation of the interactions of NAG in 7DD6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?14 | S:S:N468 | 29.47 | 23 | No | No | 0 | 8 | 0 | 1 | | H:H:?14 | S:S:Q476 | 14.28 | 23 | No | No | 0 | 6 | 0 | 1 | | H:H:?14 | S:S:D478 | 3.64 | 23 | No | No | 0 | 8 | 0 | 1 | | S:S:N468 | S:S:Q476 | 6.6 | 23 | No | No | 8 | 6 | 1 | 1 | | S:S:D478 | S:S:N468 | 10.77 | 23 | No | No | 8 | 8 | 1 | 1 | | S:S:Q476 | S:S:T470 | 5.67 | 23 | No | No | 6 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.80 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7DD7 | C | Ion | Calcium Sensing | CaS | Gallus Gallus | Ca | Ca; Ca; Tryptophan; Evocalcet | - | 3.2 | 2021-06-16 | doi.org/10.1126/sciadv.abg1483 |
|
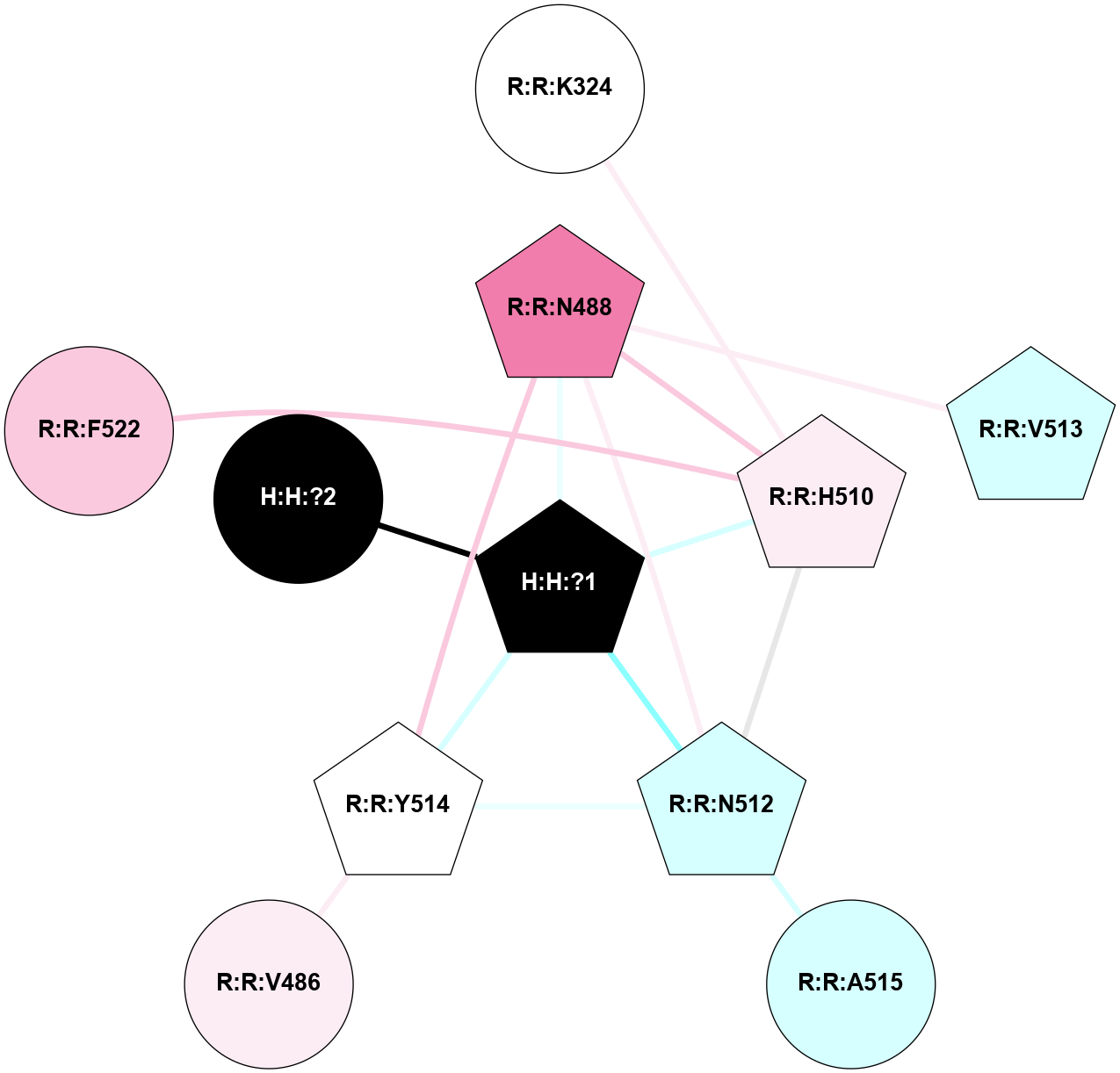
A 2D representation of the interactions of NAG in 7DD7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 30.99 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N488 | 24.56 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?1 | R:R:H510 | 13.8 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:N512 | 17.19 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:Y514 | 13.63 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:H510 | R:R:K324 | 7.86 | 1 | Yes | No | 6 | 5 | 1 | 2 | | R:R:H510 | R:R:N488 | 3.83 | 1 | Yes | Yes | 6 | 8 | 1 | 1 | | R:R:N488 | R:R:N512 | 12.26 | 1 | Yes | Yes | 8 | 3 | 1 | 1 | | R:R:N488 | R:R:V513 | 4.43 | 1 | Yes | Yes | 8 | 3 | 1 | 2 | | R:R:N488 | R:R:Y514 | 8.14 | 1 | Yes | Yes | 8 | 5 | 1 | 1 | | R:R:H510 | R:R:N512 | 2.55 | 1 | Yes | Yes | 6 | 3 | 1 | 1 | | R:R:N512 | R:R:Y514 | 8.14 | 1 | Yes | Yes | 3 | 5 | 1 | 1 | | R:R:A515 | R:R:N512 | 1.56 | 0 | No | Yes | 3 | 3 | 2 | 1 | | R:R:V486 | R:R:Y514 | 1.26 | 0 | No | Yes | 6 | 5 | 2 | 1 | | R:R:F522 | R:R:H510 | 1.13 | 0 | No | Yes | 7 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 20.03 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
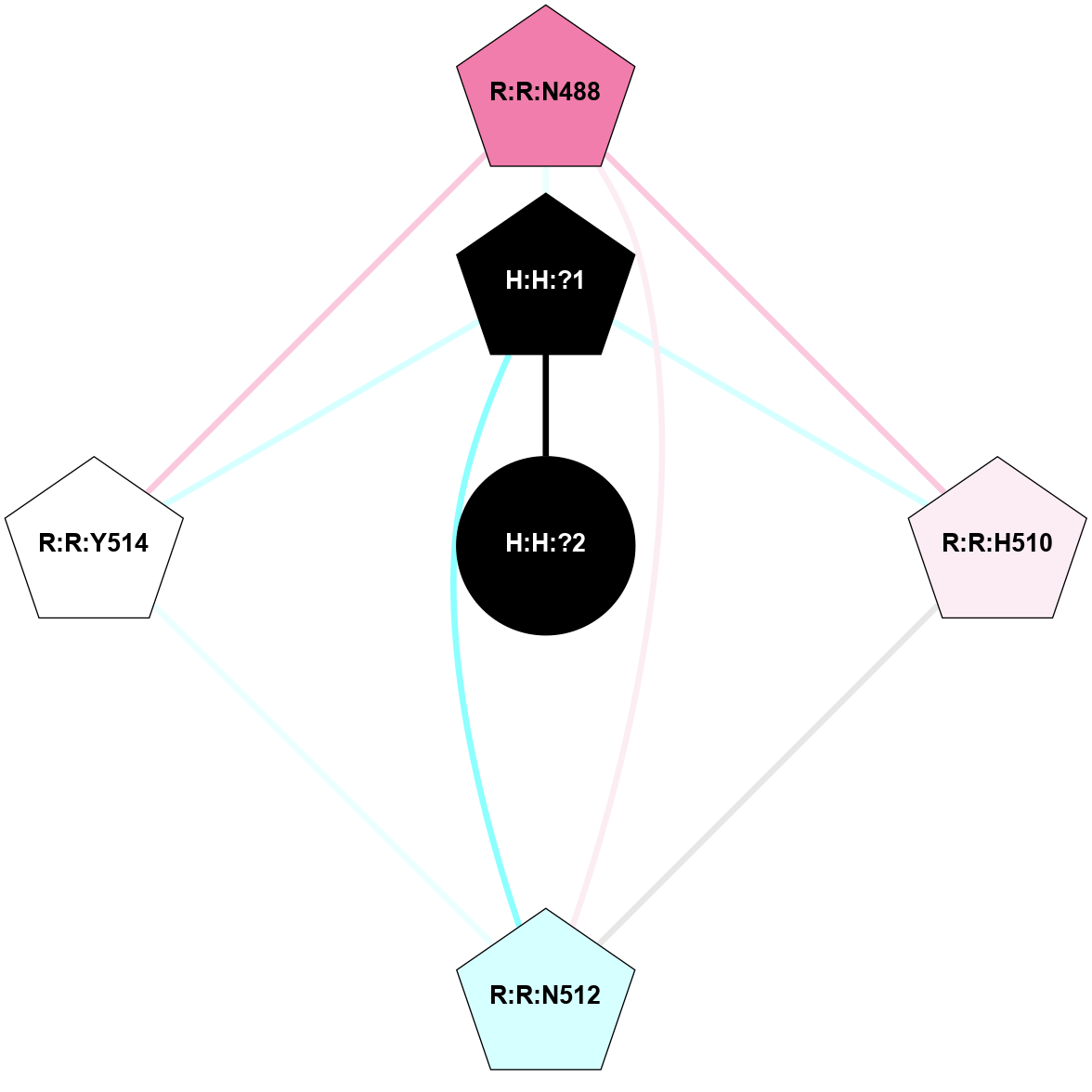
A 2D representation of the interactions of NAG in 7DD7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 30.99 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N488 | 24.56 | 1 | Yes | Yes | 0 | 8 | 1 | 2 | | H:H:?1 | R:R:H510 | 13.8 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:N512 | 17.19 | 1 | Yes | Yes | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:Y514 | 13.63 | 1 | Yes | Yes | 0 | 5 | 1 | 2 | | R:R:H510 | R:R:N488 | 3.83 | 1 | Yes | Yes | 6 | 8 | 2 | 2 | | R:R:N488 | R:R:N512 | 12.26 | 1 | Yes | Yes | 8 | 3 | 2 | 2 | | R:R:N488 | R:R:Y514 | 8.14 | 1 | Yes | Yes | 8 | 5 | 2 | 2 | | R:R:H510 | R:R:N512 | 2.55 | 1 | Yes | Yes | 6 | 3 | 2 | 2 | | R:R:N512 | R:R:Y514 | 8.14 | 1 | Yes | Yes | 3 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 30.99 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
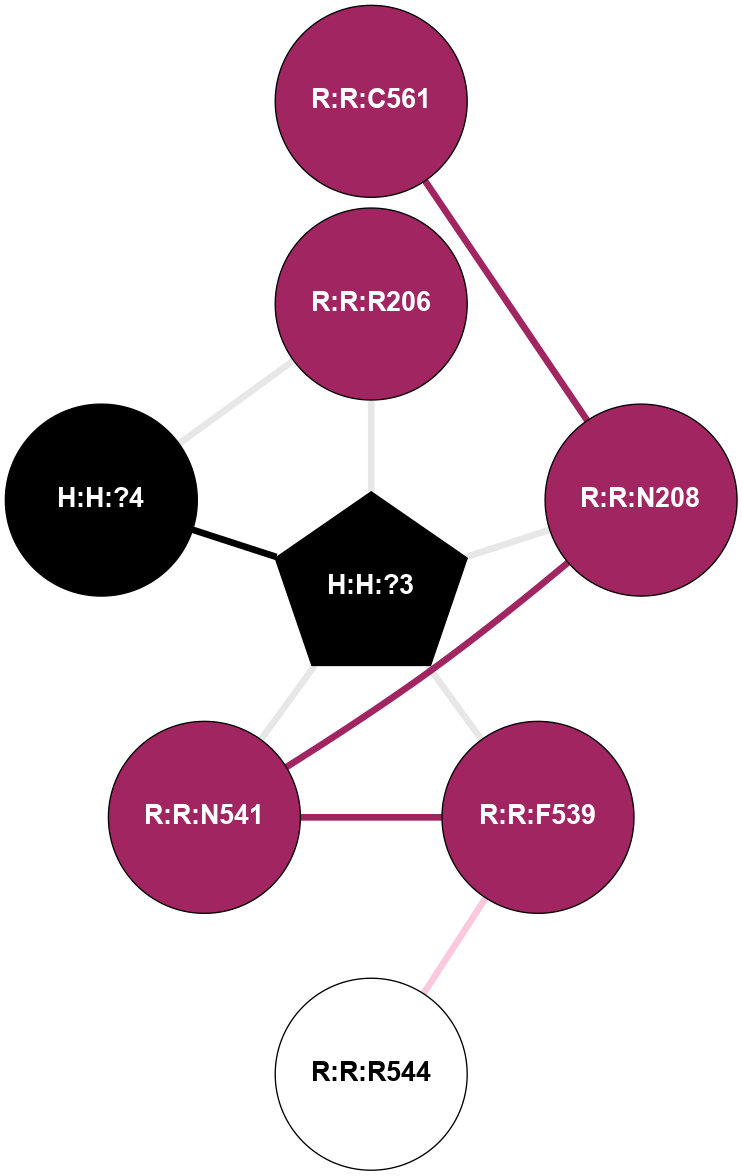
A 2D representation of the interactions of NAG in 7DD7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 33.21 | 7 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:R206 | 6.52 | 7 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?3 | R:R:N208 | 11.05 | 7 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?3 | R:R:F539 | 4.36 | 7 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?3 | R:R:N541 | 25.79 | 7 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?4 | R:R:R206 | 9.78 | 7 | No | No | 0 | 9 | 1 | 1 | | R:R:N208 | R:R:N541 | 12.26 | 7 | No | No | 9 | 9 | 1 | 1 | | R:R:C561 | R:R:N208 | 4.72 | 7 | No | No | 9 | 9 | 2 | 1 | | R:R:F539 | R:R:N541 | 6.04 | 7 | No | No | 9 | 9 | 1 | 1 | | R:R:F539 | R:R:R544 | 1.07 | 7 | No | No | 9 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.19 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
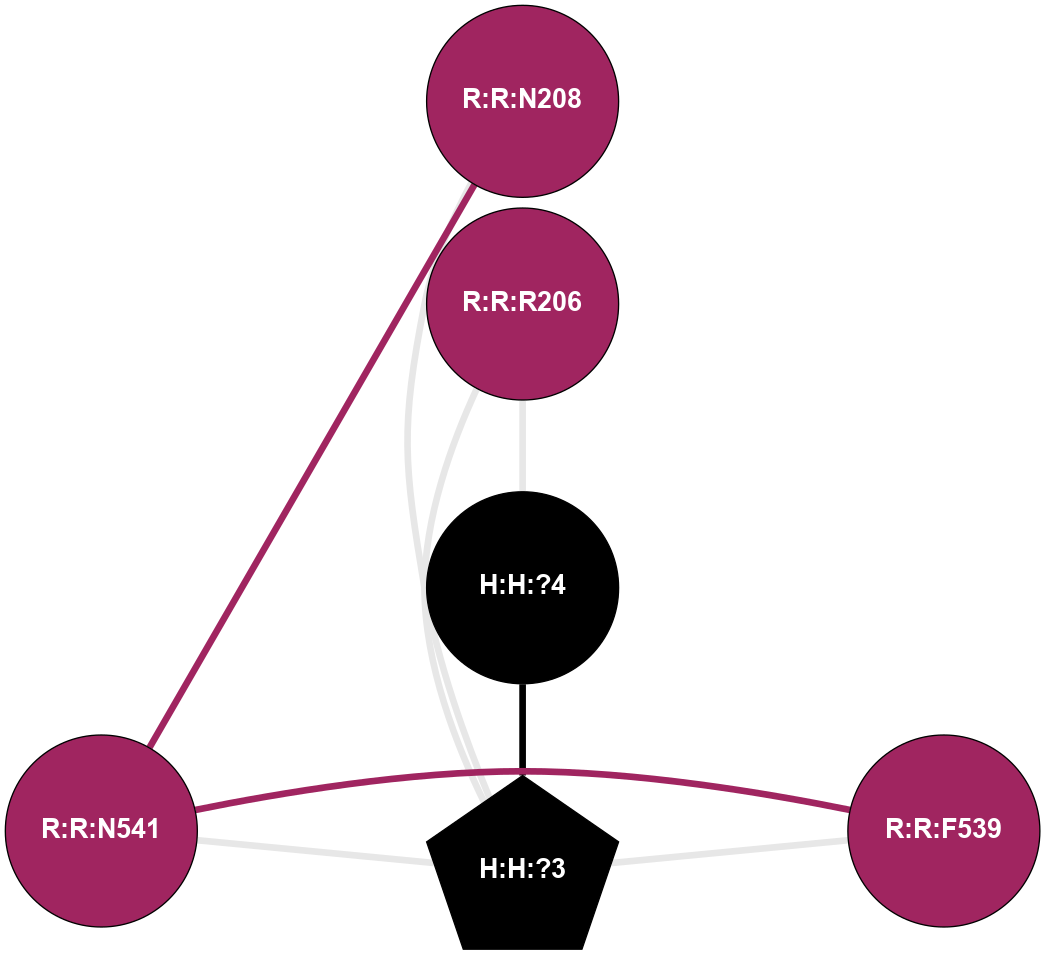
A 2D representation of the interactions of NAG in 7DD7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 33.21 | 7 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:R206 | 6.52 | 7 | Yes | No | 0 | 9 | 1 | 1 | | H:H:?3 | R:R:N208 | 11.05 | 7 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?3 | R:R:F539 | 4.36 | 7 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?3 | R:R:N541 | 25.79 | 7 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?4 | R:R:R206 | 9.78 | 7 | No | No | 0 | 9 | 0 | 1 | | R:R:N208 | R:R:N541 | 12.26 | 7 | No | No | 9 | 9 | 2 | 2 | | R:R:F539 | R:R:N541 | 6.04 | 7 | No | No | 9 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 21.50 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
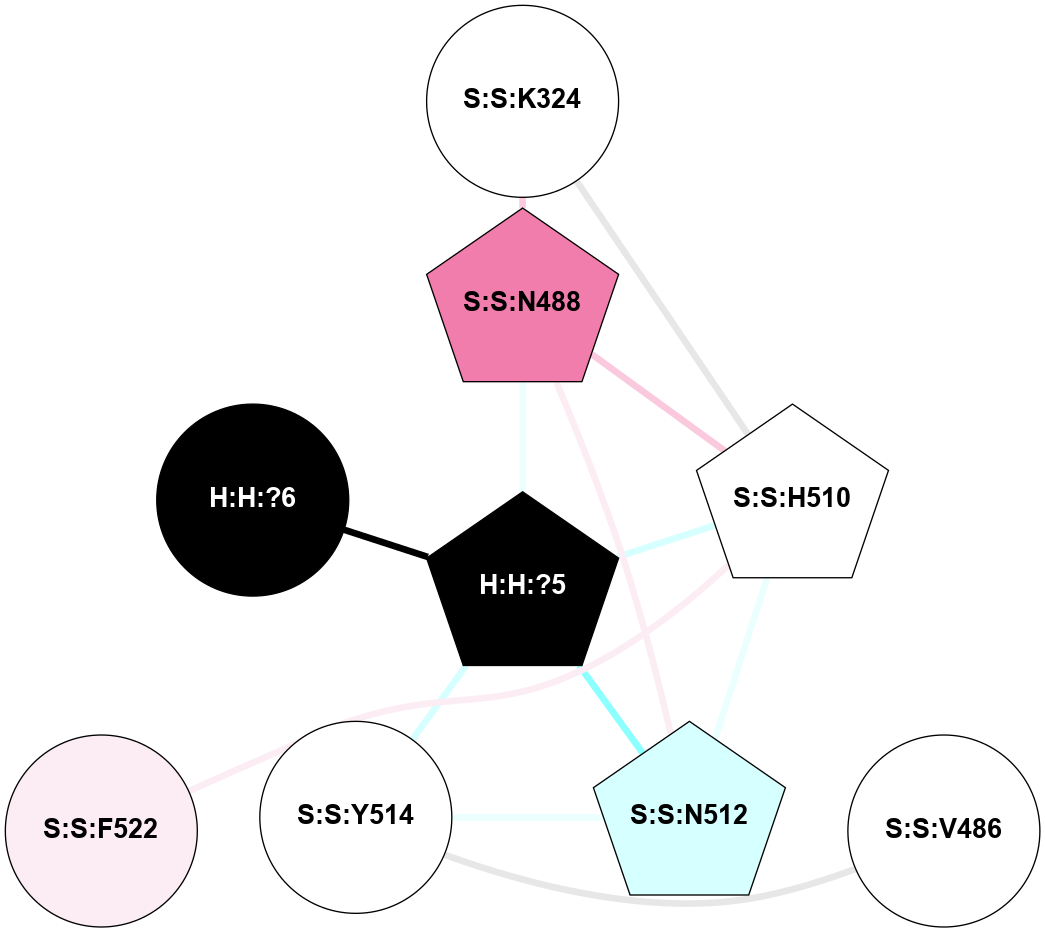
A 2D representation of the interactions of NAG in 7DD7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 29.89 | 15 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?5 | S:S:N488 | 27.02 | 15 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?5 | S:S:H510 | 12.65 | 15 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?5 | S:S:N512 | 19.65 | 15 | Yes | Yes | 0 | 3 | 0 | 1 | | H:H:?5 | S:S:Y514 | 11.53 | 15 | Yes | No | 0 | 5 | 0 | 1 | | S:S:K324 | S:S:N488 | 2.8 | 15 | No | Yes | 5 | 8 | 2 | 1 | | S:S:H510 | S:S:K324 | 6.55 | 15 | Yes | No | 5 | 5 | 1 | 2 | | S:S:V486 | S:S:Y514 | 3.79 | 0 | No | No | 5 | 5 | 2 | 1 | | S:S:H510 | S:S:N488 | 8.93 | 15 | Yes | Yes | 5 | 8 | 1 | 1 | | S:S:N488 | S:S:N512 | 6.81 | 15 | Yes | Yes | 8 | 3 | 1 | 1 | | S:S:H510 | S:S:N512 | 2.55 | 15 | Yes | Yes | 5 | 3 | 1 | 1 | | S:S:N512 | S:S:Y514 | 5.81 | 15 | Yes | No | 3 | 5 | 1 | 1 | | S:S:F522 | S:S:H510 | 1.13 | 0 | No | Yes | 6 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 20.15 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
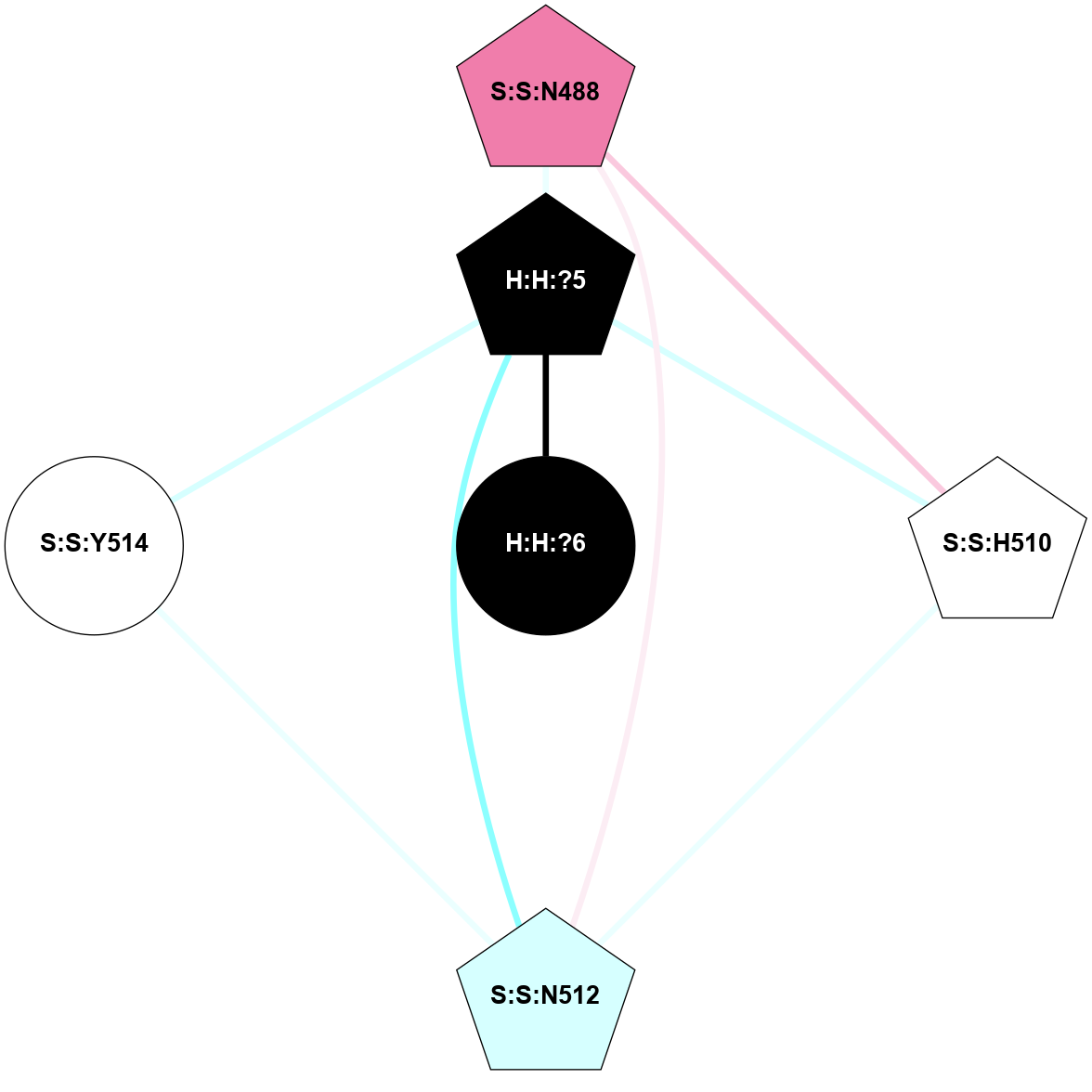
A 2D representation of the interactions of NAG in 7DD7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 29.89 | 15 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?5 | S:S:N488 | 27.02 | 15 | Yes | Yes | 0 | 8 | 1 | 2 | | H:H:?5 | S:S:H510 | 12.65 | 15 | Yes | Yes | 0 | 5 | 1 | 2 | | H:H:?5 | S:S:N512 | 19.65 | 15 | Yes | Yes | 0 | 3 | 1 | 2 | | H:H:?5 | S:S:Y514 | 11.53 | 15 | Yes | No | 0 | 5 | 1 | 2 | | S:S:H510 | S:S:N488 | 8.93 | 15 | Yes | Yes | 5 | 8 | 2 | 2 | | S:S:N488 | S:S:N512 | 6.81 | 15 | Yes | Yes | 8 | 3 | 2 | 2 | | S:S:H510 | S:S:N512 | 2.55 | 15 | Yes | Yes | 5 | 3 | 2 | 2 | | S:S:N512 | S:S:Y514 | 5.81 | 15 | Yes | No | 3 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 29.89 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
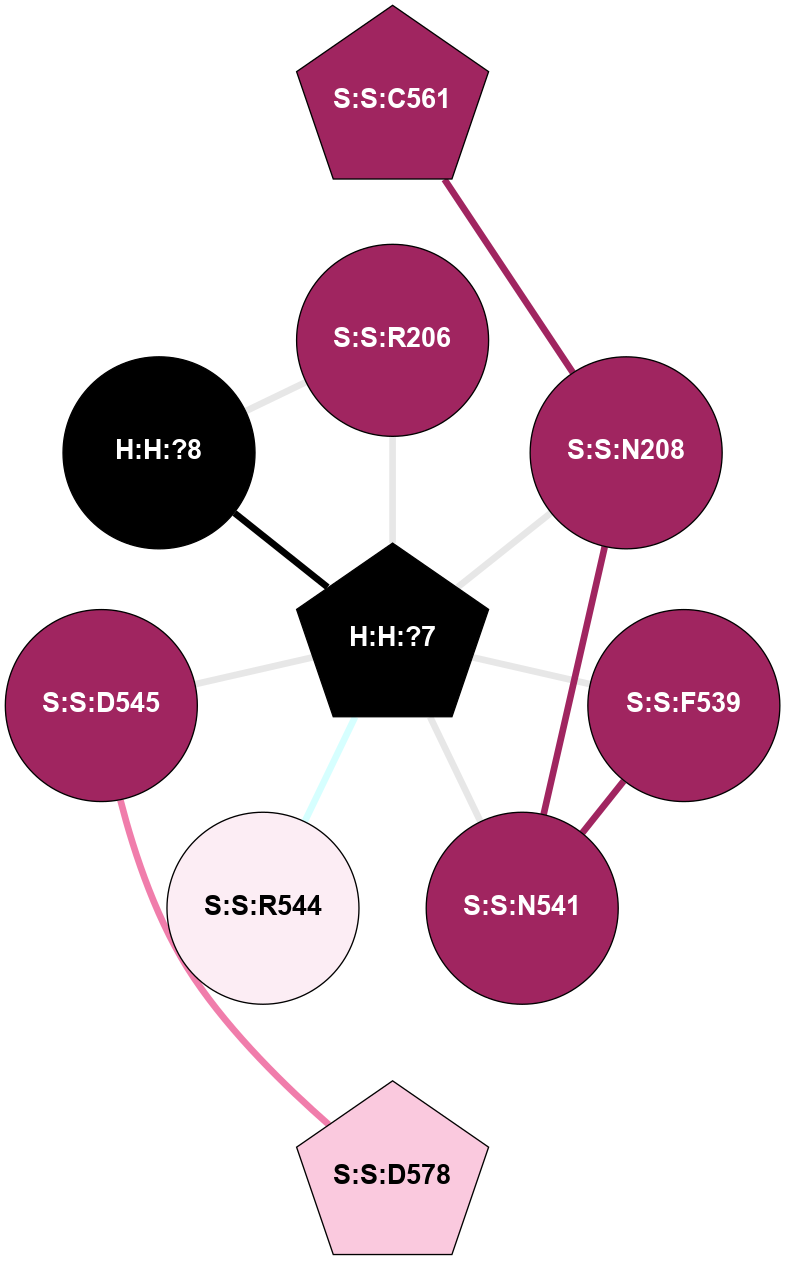
A 2D representation of the interactions of NAG in 7DD7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 39.85 | 5 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?7 | S:S:R206 | 4.35 | 5 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?7 | S:S:N208 | 7.37 | 5 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?7 | S:S:F539 | 5.45 | 5 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?7 | S:S:N541 | 25.79 | 5 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?7 | S:S:R544 | 3.26 | 5 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?7 | S:S:D545 | 6.07 | 5 | Yes | No | 0 | 9 | 0 | 1 | | H:H:?8 | S:S:R206 | 15.21 | 5 | No | No | 0 | 9 | 1 | 1 | | S:S:N208 | S:S:N541 | 12.26 | 5 | No | No | 9 | 9 | 1 | 1 | | S:S:C561 | S:S:N208 | 4.72 | 5 | Yes | No | 9 | 9 | 2 | 1 | | S:S:F539 | S:S:N541 | 8.46 | 5 | No | No | 9 | 9 | 1 | 1 | | S:S:D545 | S:S:D578 | 13.31 | 0 | No | Yes | 9 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.16 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
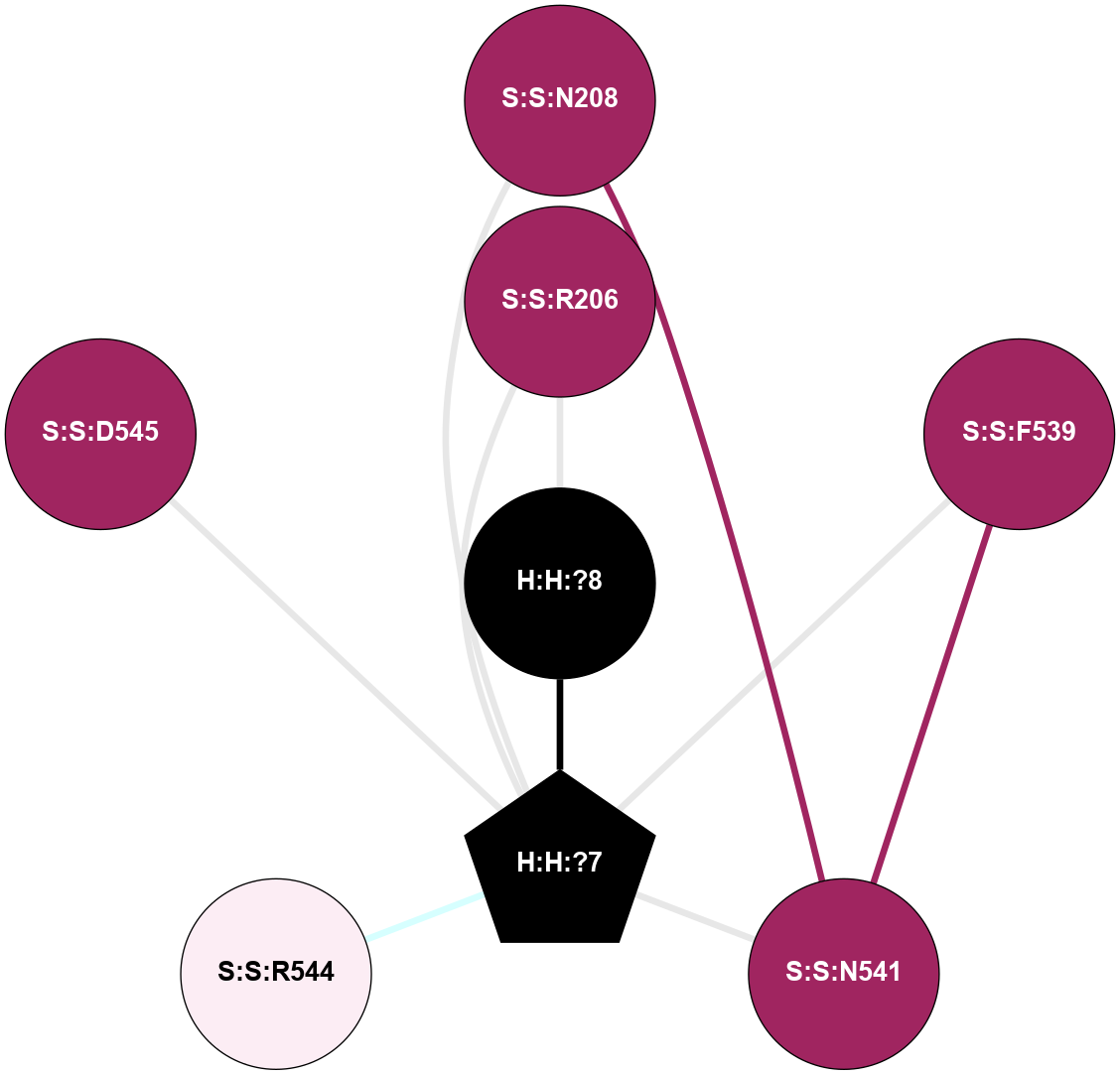
A 2D representation of the interactions of NAG in 7DD7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 39.85 | 5 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?7 | S:S:R206 | 4.35 | 5 | Yes | No | 0 | 9 | 1 | 1 | | H:H:?7 | S:S:N208 | 7.37 | 5 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?7 | S:S:F539 | 5.45 | 5 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?7 | S:S:N541 | 25.79 | 5 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?7 | S:S:R544 | 3.26 | 5 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?7 | S:S:D545 | 6.07 | 5 | Yes | No | 0 | 9 | 1 | 2 | | H:H:?8 | S:S:R206 | 15.21 | 5 | No | No | 0 | 9 | 0 | 1 | | S:S:N208 | S:S:N541 | 12.26 | 5 | No | No | 9 | 9 | 2 | 2 | | S:S:F539 | S:S:N541 | 8.46 | 5 | No | No | 9 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 27.53 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
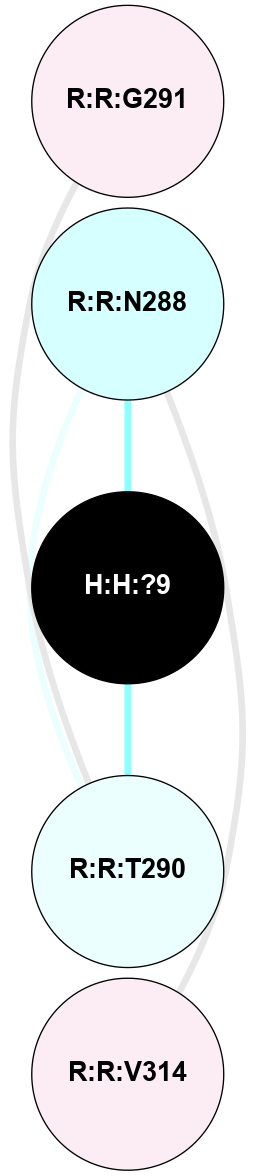
A 2D representation of the interactions of NAG in 7DD7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?9 | R:R:N288 | 30.7 | 36 | No | No | 0 | 3 | 0 | 1 | | H:H:?9 | R:R:T290 | 6.59 | 36 | No | No | 0 | 4 | 0 | 1 | | R:R:N288 | R:R:T290 | 4.39 | 36 | No | No | 3 | 4 | 1 | 1 | | R:R:G291 | R:R:T290 | 1.82 | 0 | No | No | 6 | 4 | 2 | 1 | | R:R:N288 | R:R:V314 | 1.48 | 36 | No | No | 3 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.64 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
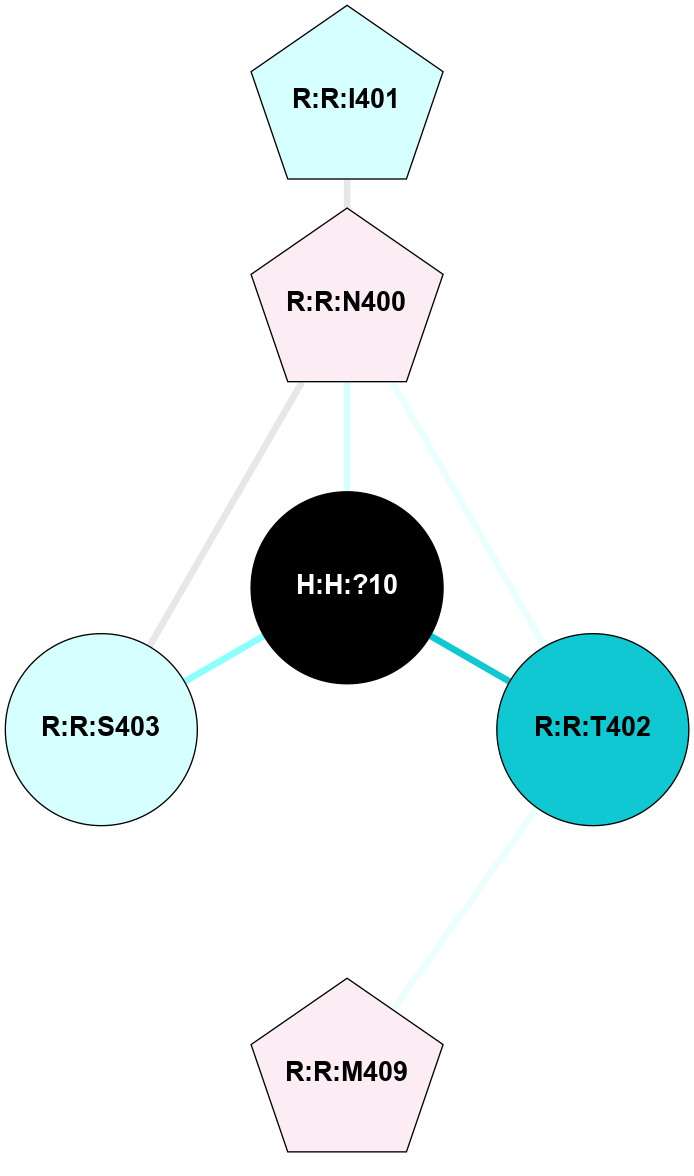
A 2D representation of the interactions of NAG in 7DD7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | R:R:N400 | 33.15 | 1 | No | Yes | 0 | 6 | 0 | 1 | | H:H:?10 | R:R:T402 | 9.23 | 1 | No | No | 0 | 1 | 0 | 1 | | H:H:?10 | R:R:S403 | 4.03 | 1 | No | No | 0 | 3 | 0 | 1 | | R:R:I401 | R:R:N400 | 2.83 | 1 | Yes | Yes | 3 | 6 | 2 | 1 | | R:R:N400 | R:R:T402 | 14.62 | 1 | Yes | No | 6 | 1 | 1 | 1 | | R:R:N400 | R:R:S403 | 4.47 | 1 | Yes | No | 6 | 3 | 1 | 1 | | R:R:M409 | R:R:T402 | 3.01 | 1 | Yes | No | 6 | 1 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.47 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
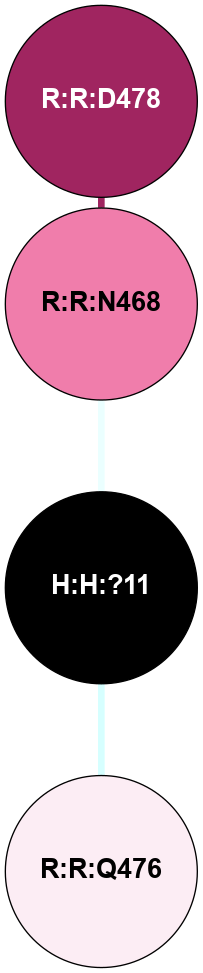
A 2D representation of the interactions of NAG in 7DD7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | R:R:N468 | 25.79 | 0 | No | No | 0 | 8 | 0 | 1 | | R:R:D478 | R:R:N468 | 5.39 | 0 | No | No | 9 | 8 | 2 | 1 | | H:H:?11 | R:R:Q476 | 2.38 | 0 | No | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.08 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
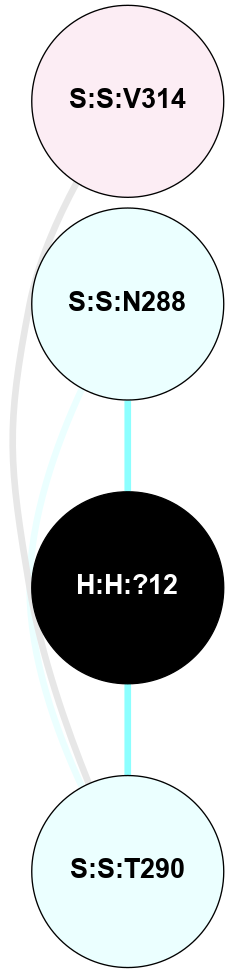
A 2D representation of the interactions of NAG in 7DD7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | S:S:N288 | 29.47 | 37 | No | No | 0 | 4 | 0 | 1 | | H:H:?12 | S:S:T290 | 5.27 | 37 | No | No | 0 | 4 | 0 | 1 | | S:S:N288 | S:S:T290 | 8.77 | 37 | No | No | 4 | 4 | 1 | 1 | | S:S:T290 | S:S:V314 | 7.93 | 37 | No | No | 4 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.37 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
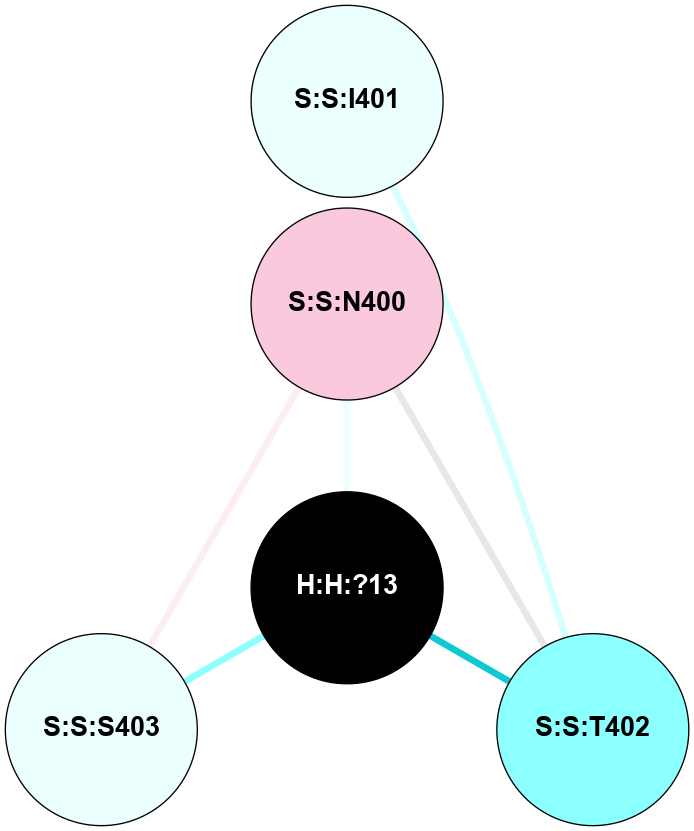
A 2D representation of the interactions of NAG in 7DD7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | S:S:N400 | 30.7 | 29 | No | No | 0 | 7 | 0 | 1 | | H:H:?13 | S:S:T402 | 14.5 | 29 | No | No | 0 | 2 | 0 | 1 | | H:H:?13 | S:S:S403 | 2.69 | 29 | No | No | 0 | 4 | 0 | 1 | | S:S:N400 | S:S:T402 | 10.24 | 29 | No | No | 7 | 2 | 1 | 1 | | S:S:N400 | S:S:S403 | 7.45 | 29 | No | No | 7 | 4 | 1 | 1 | | S:S:I401 | S:S:T402 | 1.52 | 0 | No | No | 4 | 2 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.96 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
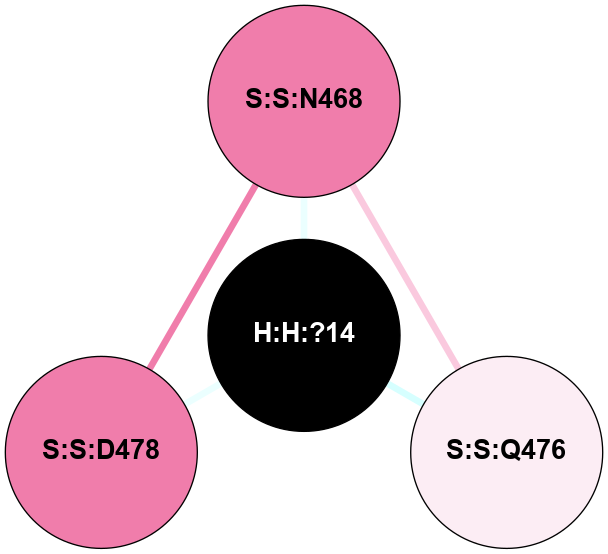
A 2D representation of the interactions of NAG in 7DD7
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?14 | S:S:N468 | 29.47 | 30 | No | No | 0 | 8 | 0 | 1 | | H:H:?14 | S:S:Q476 | 15.47 | 30 | No | No | 0 | 6 | 0 | 1 | | H:H:?14 | S:S:D478 | 3.64 | 30 | No | No | 0 | 8 | 0 | 1 | | S:S:N468 | S:S:Q476 | 6.6 | 30 | No | No | 8 | 6 | 1 | 1 | | S:S:D478 | S:S:N468 | 10.77 | 30 | No | No | 8 | 8 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.19 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7DTT | C | Ion | Calcium Sensing | CaS | Homo Sapiens | Ca | Ca | - | 3.8 | 2021-03-10 | doi.org/10.1038/s41422-021-00474-0 |
|
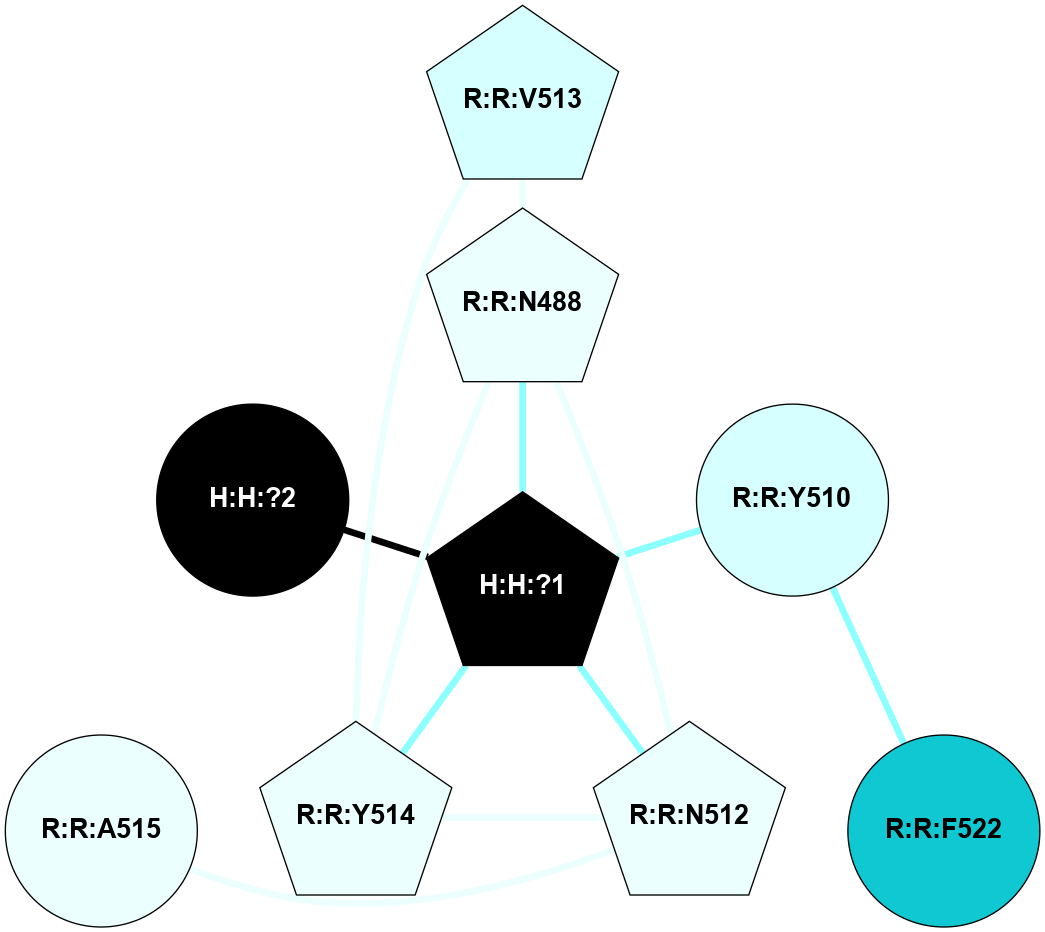
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 34.31 | 16 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N488 | 27.02 | 16 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:Y510 | 15.73 | 16 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:N512 | 17.19 | 16 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:Y514 | 13.63 | 16 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:N488 | R:R:N512 | 6.81 | 16 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:V513 | 5.91 | 16 | Yes | Yes | 4 | 3 | 1 | 2 | | R:R:N488 | R:R:Y514 | 6.98 | 16 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:F522 | R:R:Y510 | 15.47 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:N512 | R:R:Y514 | 5.81 | 16 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | | R:R:V513 | R:R:Y514 | 3.79 | 16 | Yes | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 21.58 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
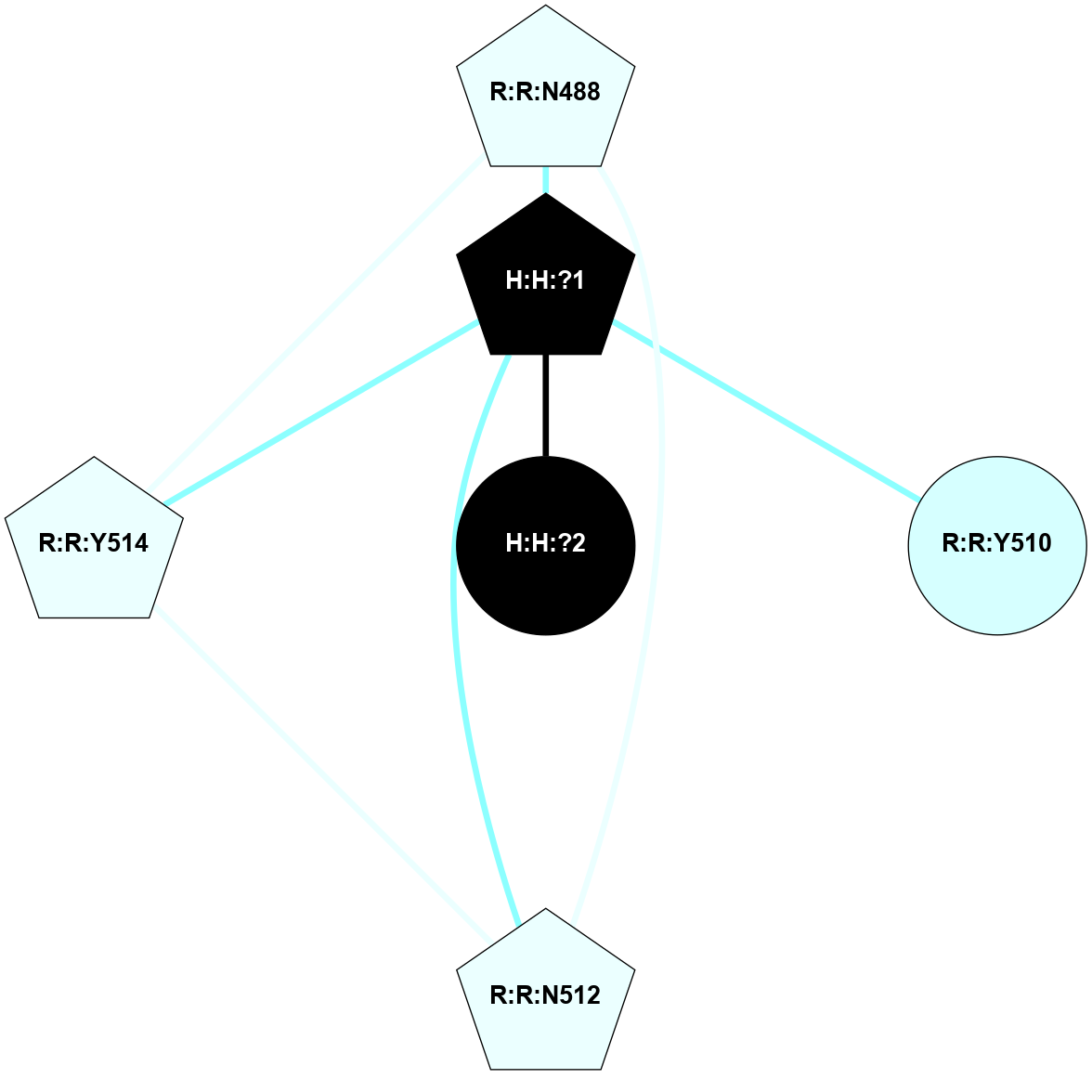
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 34.31 | 16 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N488 | 27.02 | 16 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:Y510 | 15.73 | 16 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:N512 | 17.19 | 16 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:Y514 | 13.63 | 16 | Yes | Yes | 0 | 4 | 1 | 2 | | R:R:N488 | R:R:N512 | 6.81 | 16 | Yes | Yes | 4 | 4 | 2 | 2 | | R:R:N488 | R:R:Y514 | 6.98 | 16 | Yes | Yes | 4 | 4 | 2 | 2 | | R:R:N512 | R:R:Y514 | 5.81 | 16 | Yes | Yes | 4 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 34.31 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
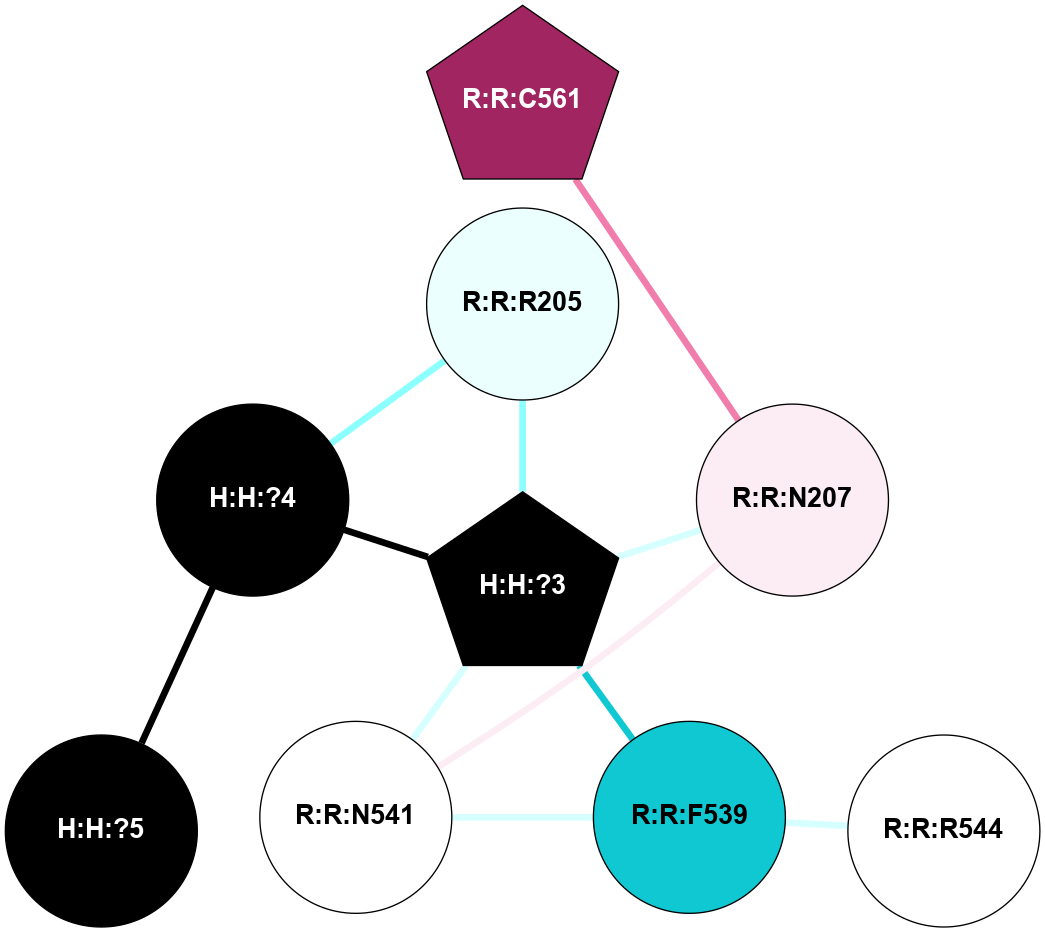
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 37.64 | 6 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:R205 | 6.52 | 6 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?3 | R:R:N207 | 15.96 | 6 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?3 | R:R:F539 | 6.54 | 6 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?3 | R:R:N541 | 28.24 | 6 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?4 | H:H:?5 | 33.21 | 6 | No | No | 0 | 0 | 1 | 2 | | H:H:?4 | R:R:R205 | 9.78 | 6 | No | No | 0 | 4 | 1 | 1 | | R:R:N207 | R:R:N541 | 13.62 | 6 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 11.02 | 6 | Yes | No | 9 | 6 | 2 | 1 | | R:R:F539 | R:R:N541 | 6.04 | 6 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 10.69 | 6 | No | No | 1 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.98 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
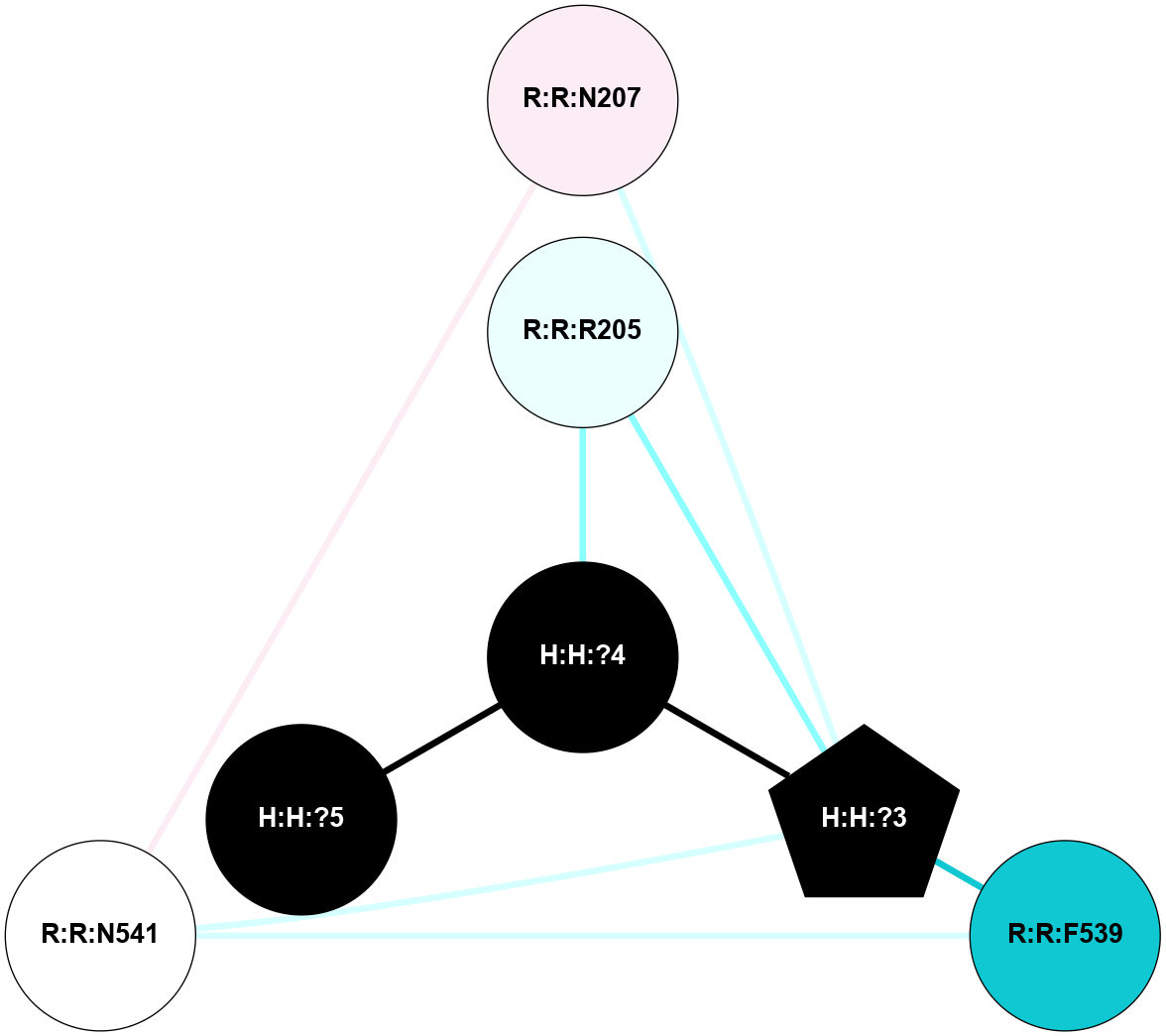
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 37.64 | 6 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:R205 | 6.52 | 6 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?3 | R:R:N207 | 15.96 | 6 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?3 | R:R:F539 | 6.54 | 6 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?3 | R:R:N541 | 28.24 | 6 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?4 | H:H:?5 | 33.21 | 6 | No | No | 0 | 0 | 0 | 1 | | H:H:?4 | R:R:R205 | 9.78 | 6 | No | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 13.62 | 6 | No | No | 6 | 5 | 2 | 2 | | R:R:F539 | R:R:N541 | 6.04 | 6 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 26.88 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
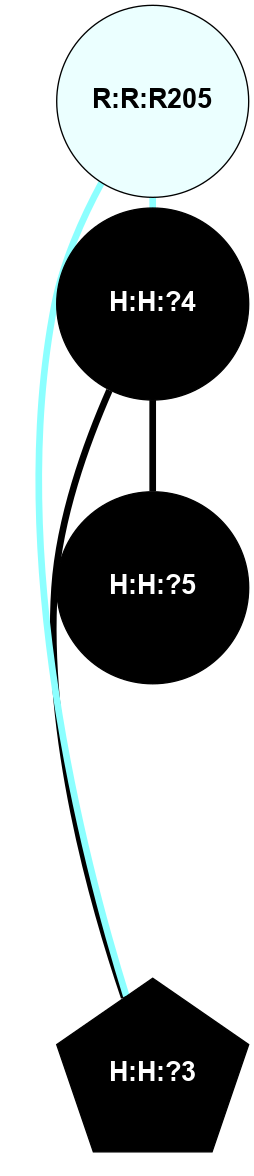
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 37.64 | 6 | Yes | No | 0 | 0 | 2 | 1 | | H:H:?3 | R:R:R205 | 6.52 | 6 | Yes | No | 0 | 4 | 2 | 2 | | H:H:?4 | H:H:?5 | 33.21 | 6 | No | No | 0 | 0 | 1 | 0 | | H:H:?4 | R:R:R205 | 9.78 | 6 | No | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 33.21 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
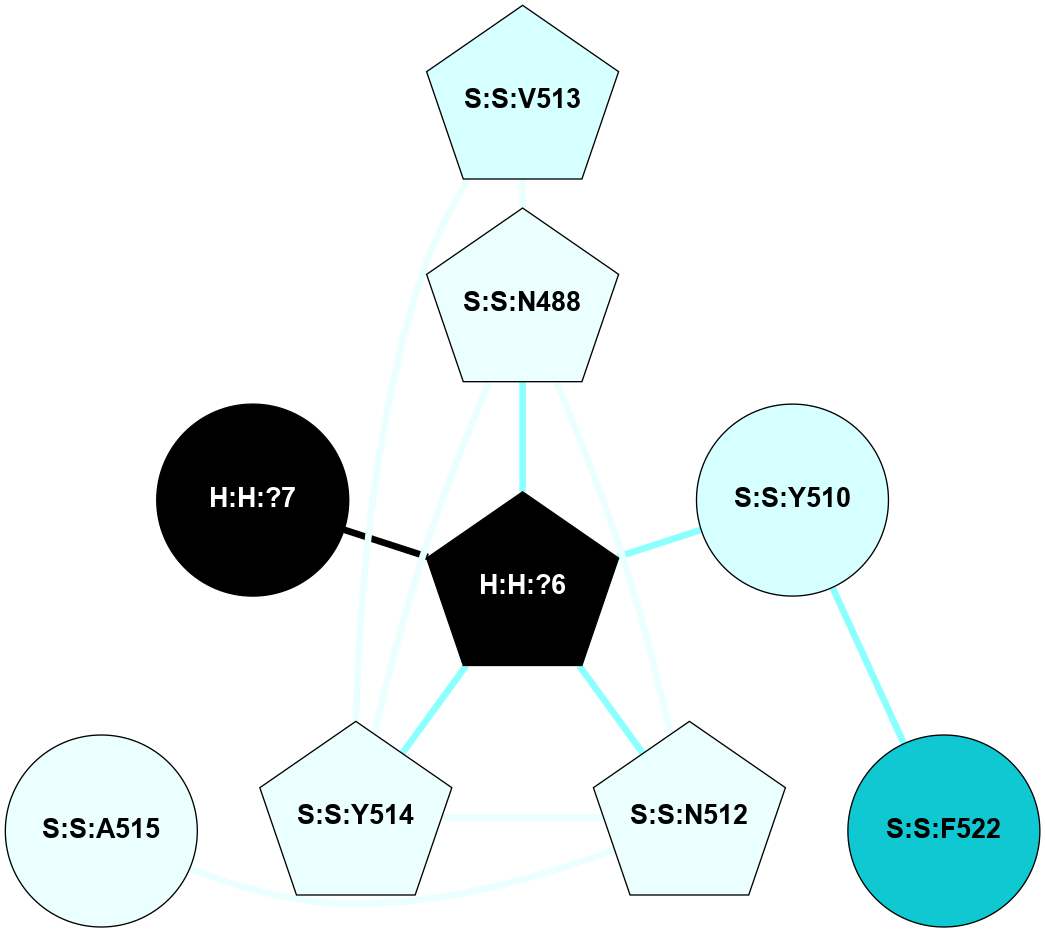
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | H:H:?7 | 34.31 | 17 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?6 | S:S:N488 | 27.02 | 17 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?6 | S:S:Y510 | 15.73 | 17 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?6 | S:S:N512 | 17.19 | 17 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?6 | S:S:Y514 | 13.63 | 17 | Yes | Yes | 0 | 4 | 0 | 1 | | S:S:N488 | S:S:N512 | 6.81 | 17 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:V513 | 5.91 | 17 | Yes | Yes | 4 | 3 | 1 | 2 | | S:S:N488 | S:S:Y514 | 6.98 | 17 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:F522 | S:S:Y510 | 15.47 | 0 | No | No | 1 | 3 | 2 | 1 | | S:S:N512 | S:S:Y514 | 5.81 | 17 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:A515 | S:S:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | | S:S:V513 | S:S:Y514 | 3.79 | 17 | Yes | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 21.58 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
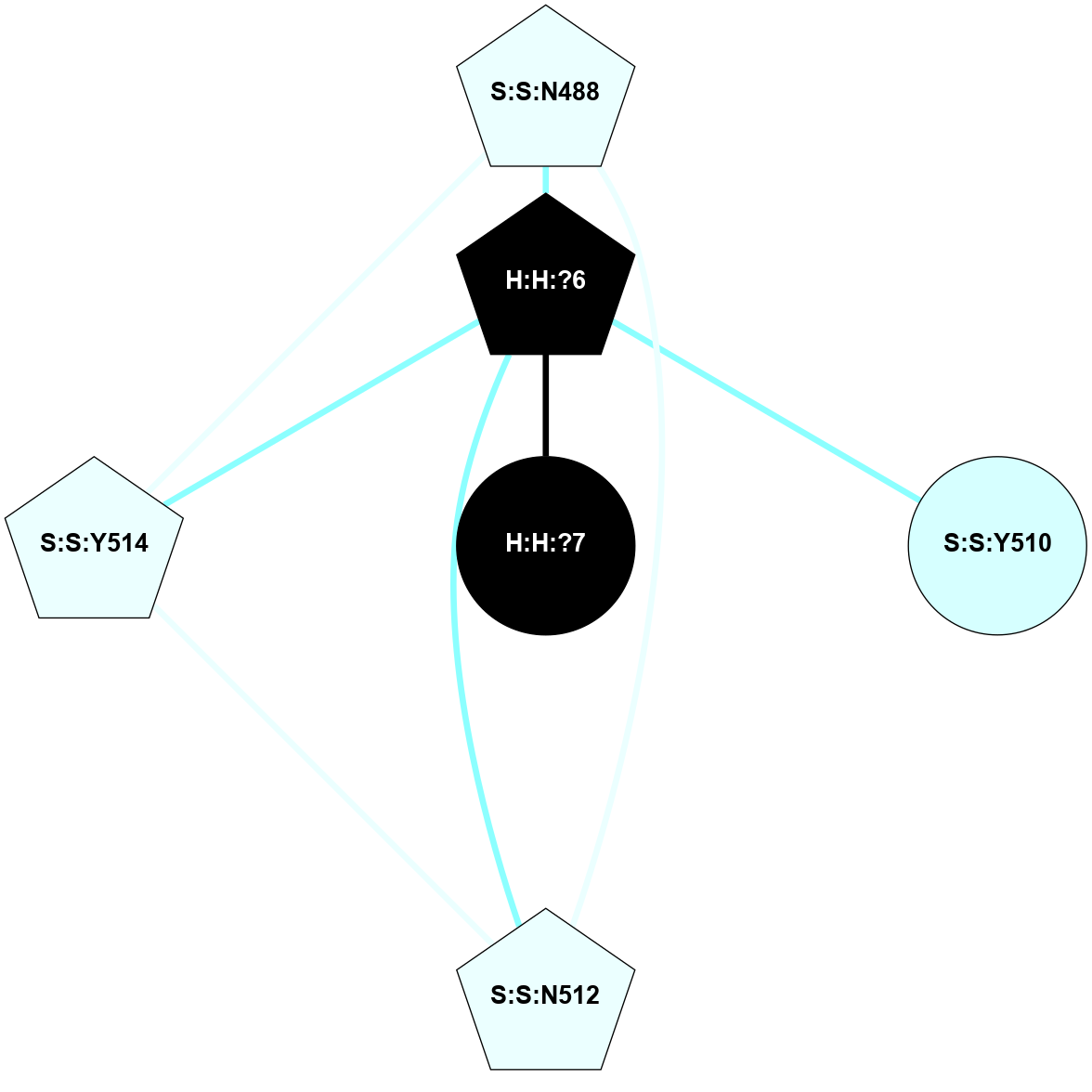
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | H:H:?7 | 34.31 | 17 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?6 | S:S:N488 | 27.02 | 17 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?6 | S:S:Y510 | 15.73 | 17 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?6 | S:S:N512 | 17.19 | 17 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?6 | S:S:Y514 | 13.63 | 17 | Yes | Yes | 0 | 4 | 1 | 2 | | S:S:N488 | S:S:N512 | 6.81 | 17 | Yes | Yes | 4 | 4 | 2 | 2 | | S:S:N488 | S:S:Y514 | 6.98 | 17 | Yes | Yes | 4 | 4 | 2 | 2 | | S:S:N512 | S:S:Y514 | 5.81 | 17 | Yes | Yes | 4 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 34.31 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
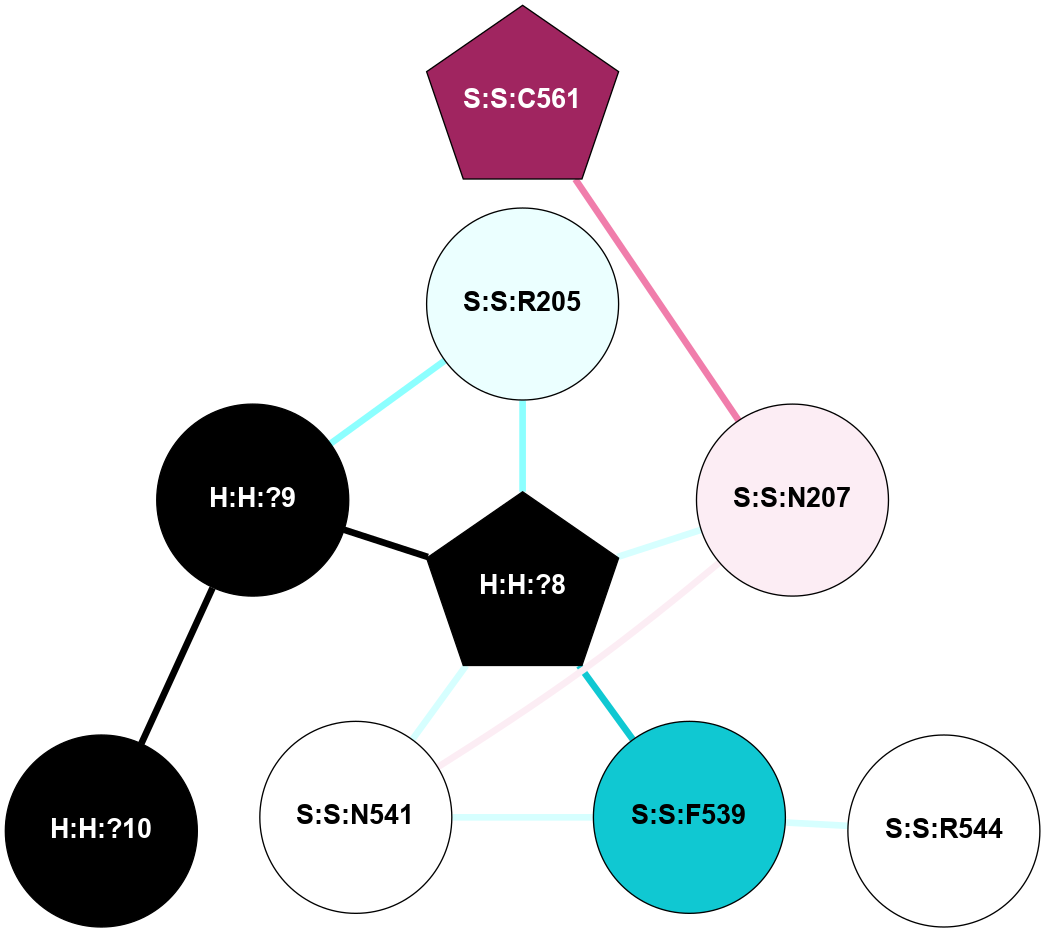
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | H:H:?9 | 37.64 | 7 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?8 | S:S:R205 | 6.52 | 7 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?8 | S:S:N207 | 15.96 | 7 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?8 | S:S:F539 | 6.54 | 7 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?8 | S:S:N541 | 28.24 | 7 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?10 | H:H:?9 | 33.21 | 0 | No | No | 0 | 0 | 2 | 1 | | H:H:?9 | S:S:R205 | 9.78 | 7 | No | No | 0 | 4 | 1 | 1 | | S:S:N207 | S:S:N541 | 13.62 | 7 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 11.02 | 7 | Yes | No | 9 | 6 | 2 | 1 | | S:S:F539 | S:S:N541 | 6.04 | 7 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 10.69 | 7 | No | No | 1 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.98 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
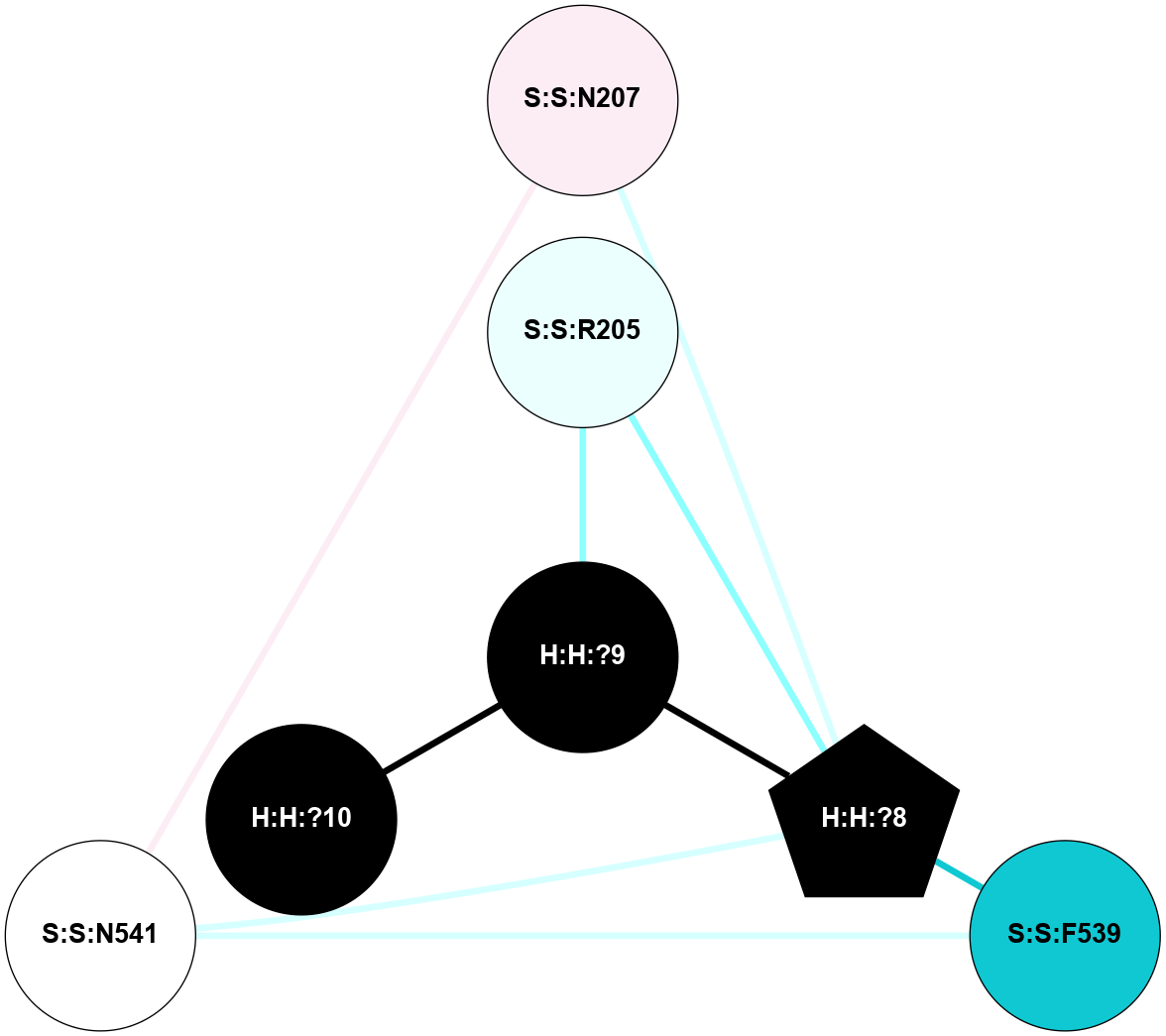
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | H:H:?9 | 37.64 | 7 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?8 | S:S:R205 | 6.52 | 7 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?8 | S:S:N207 | 15.96 | 7 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?8 | S:S:F539 | 6.54 | 7 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?8 | S:S:N541 | 28.24 | 7 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?10 | H:H:?9 | 33.21 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?9 | S:S:R205 | 9.78 | 7 | No | No | 0 | 4 | 0 | 1 | | S:S:N207 | S:S:N541 | 13.62 | 7 | No | No | 6 | 5 | 2 | 2 | | S:S:F539 | S:S:N541 | 6.04 | 7 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 26.88 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
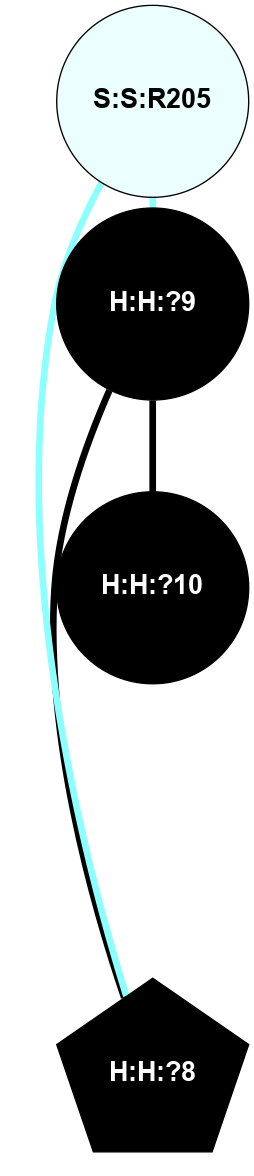
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | H:H:?9 | 37.64 | 7 | Yes | No | 0 | 0 | 2 | 1 | | H:H:?8 | S:S:R205 | 6.52 | 7 | Yes | No | 0 | 4 | 2 | 2 | | H:H:?10 | H:H:?9 | 33.21 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?9 | S:S:R205 | 9.78 | 7 | No | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 33.21 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
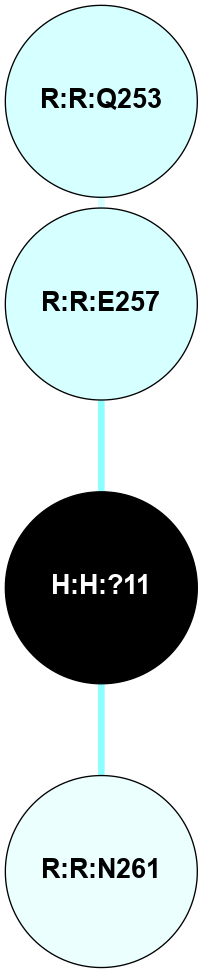
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | R:R:E257 | 5.92 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?11 | R:R:N261 | 25.79 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:E257 | R:R:Q253 | 6.37 | 0 | No | No | 3 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.86 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | R:R:N287 | 35.61 | 0 | No | No | 0 | 6 | 0 | 1 | | R:R:N287 | R:R:T289 | 2.92 | 0 | No | No | 6 | 5 | 1 | 2 | | R:R:H312 | R:R:N287 | 1.28 | 0 | No | No | 3 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 35.61 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
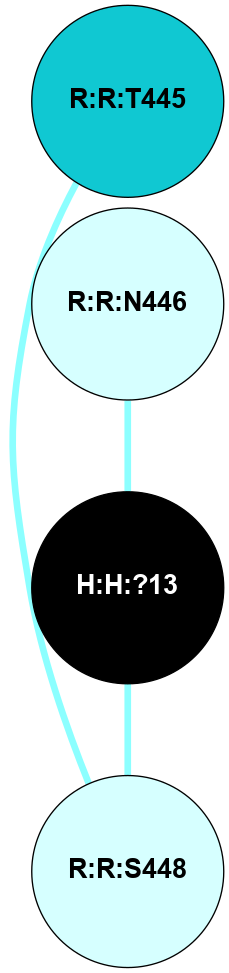
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | R:R:N446 | 35.61 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?13 | R:R:S448 | 2.69 | 0 | No | No | 0 | 3 | 0 | 1 | | R:R:S448 | R:R:T445 | 1.6 | 0 | No | No | 3 | 1 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 19.15 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
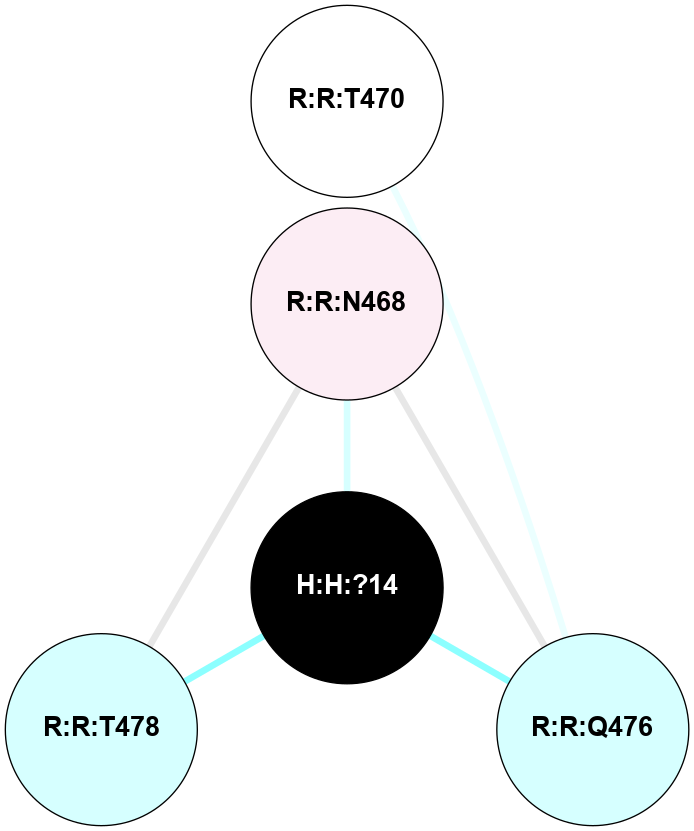
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?14 | R:R:N468 | 33.15 | 24 | No | No | 0 | 6 | 0 | 1 | | H:H:?14 | R:R:Q476 | 3.57 | 24 | No | No | 0 | 3 | 0 | 1 | | H:H:?14 | R:R:T478 | 5.27 | 24 | No | No | 0 | 3 | 0 | 1 | | R:R:N468 | R:R:Q476 | 6.6 | 24 | No | No | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 5.85 | 24 | No | No | 6 | 3 | 1 | 1 | | R:R:Q476 | R:R:T470 | 8.5 | 24 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.00 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?15 | S:S:E257 | 5.92 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?15 | S:S:N261 | 25.79 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:E257 | S:S:Q253 | 6.37 | 0 | No | No | 3 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.86 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?16 | S:S:N287 | 35.61 | 0 | No | No | 0 | 6 | 0 | 1 | | S:S:N287 | S:S:T289 | 2.92 | 0 | No | No | 6 | 5 | 1 | 2 | | S:S:H312 | S:S:N287 | 1.28 | 0 | No | No | 3 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 35.61 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?17 | S:S:N446 | 35.61 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?17 | S:S:S448 | 2.69 | 0 | No | No | 0 | 3 | 0 | 1 | | S:S:S448 | S:S:T445 | 1.6 | 0 | No | No | 3 | 1 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 19.15 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
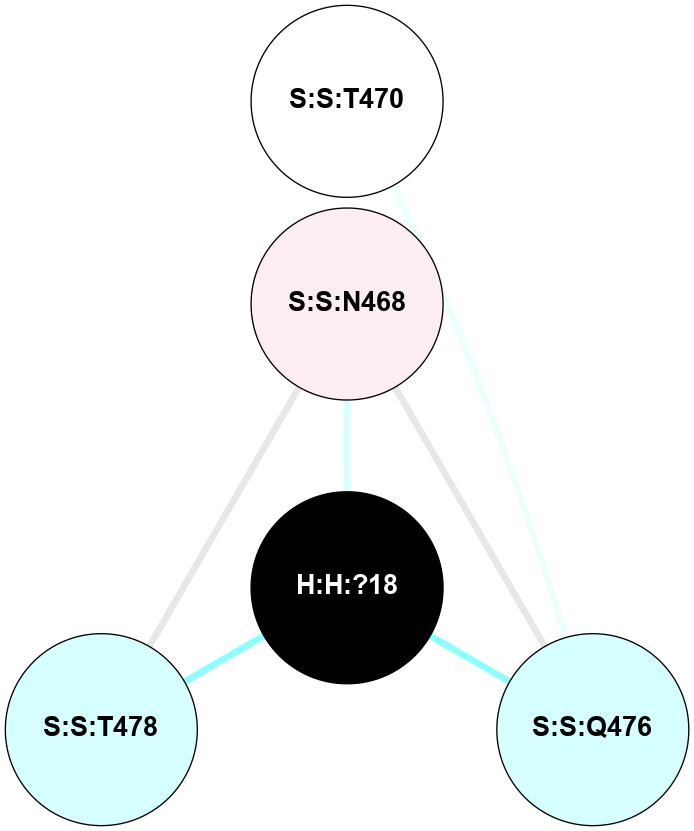
A 2D representation of the interactions of NAG in 7DTT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?18 | S:S:N468 | 33.15 | 25 | No | No | 0 | 6 | 0 | 1 | | H:H:?18 | S:S:Q476 | 3.57 | 25 | No | No | 0 | 3 | 0 | 1 | | H:H:?18 | S:S:T478 | 5.27 | 25 | No | No | 0 | 3 | 0 | 1 | | S:S:N468 | S:S:Q476 | 6.6 | 25 | No | No | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 5.85 | 25 | No | No | 6 | 3 | 1 | 1 | | S:S:Q476 | S:S:T470 | 8.5 | 25 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.00 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7DTU | C | Ion | Calcium Sensing | CaS | Homo Sapiens | - | Tryptophan | - | 4.4 | 2021-03-10 | doi.org/10.1038/s41422-021-00474-0 |
|
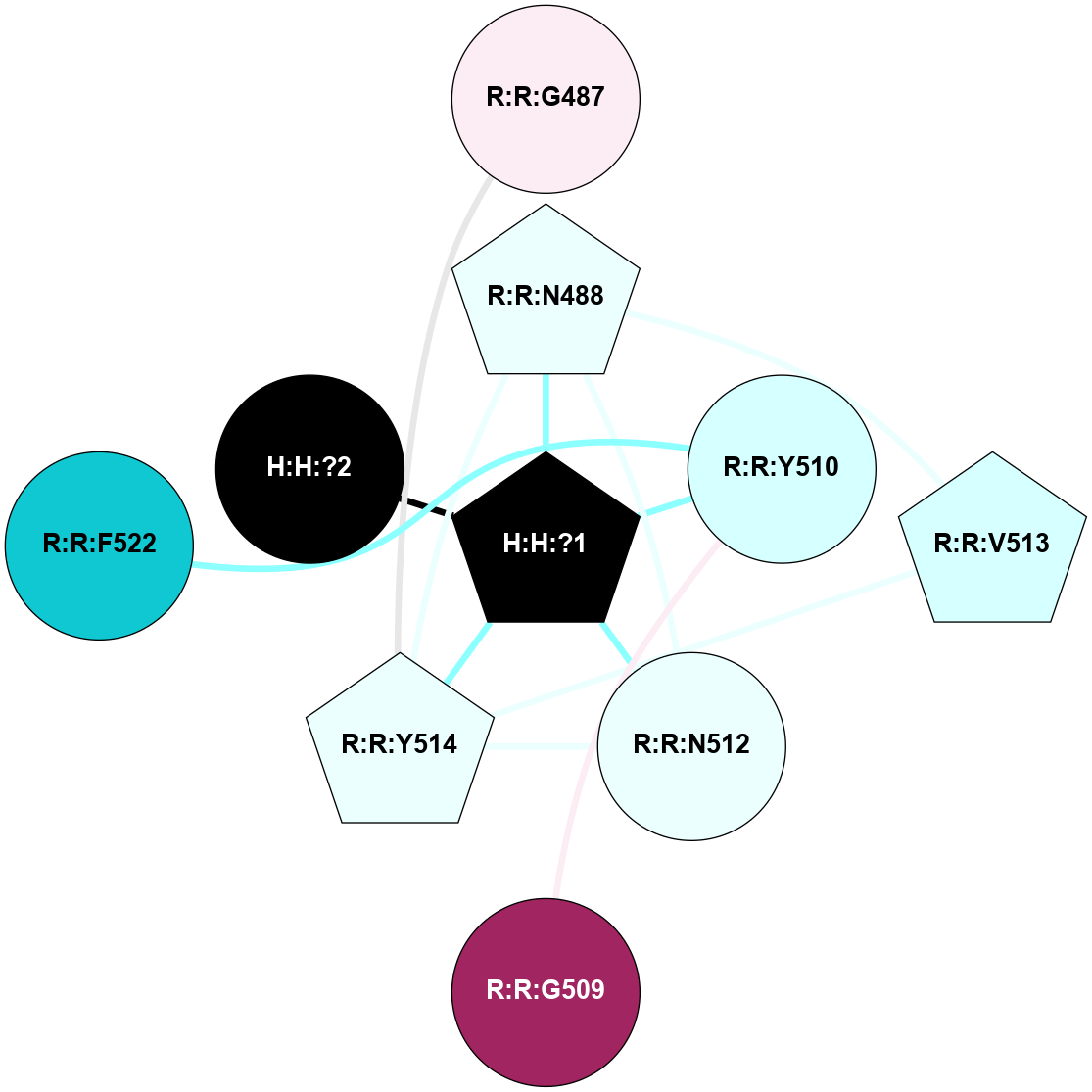
A 2D representation of the interactions of NAG in 7DTU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 38.74 | 22 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N488 | 23.33 | 22 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:Y510 | 6.29 | 22 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:N512 | 22.1 | 22 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:Y514 | 17.82 | 22 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:G487 | R:R:Y514 | 4.35 | 0 | No | Yes | 6 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 6.81 | 22 | Yes | No | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:V513 | 2.96 | 22 | Yes | Yes | 4 | 3 | 1 | 2 | | R:R:N488 | R:R:Y514 | 6.98 | 22 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:G509 | R:R:Y510 | 2.9 | 0 | No | No | 9 | 3 | 2 | 1 | | R:R:F522 | R:R:Y510 | 9.28 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:N512 | R:R:Y514 | 8.14 | 22 | No | Yes | 4 | 4 | 1 | 1 | | R:R:V513 | R:R:Y514 | 5.05 | 22 | Yes | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 21.66 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
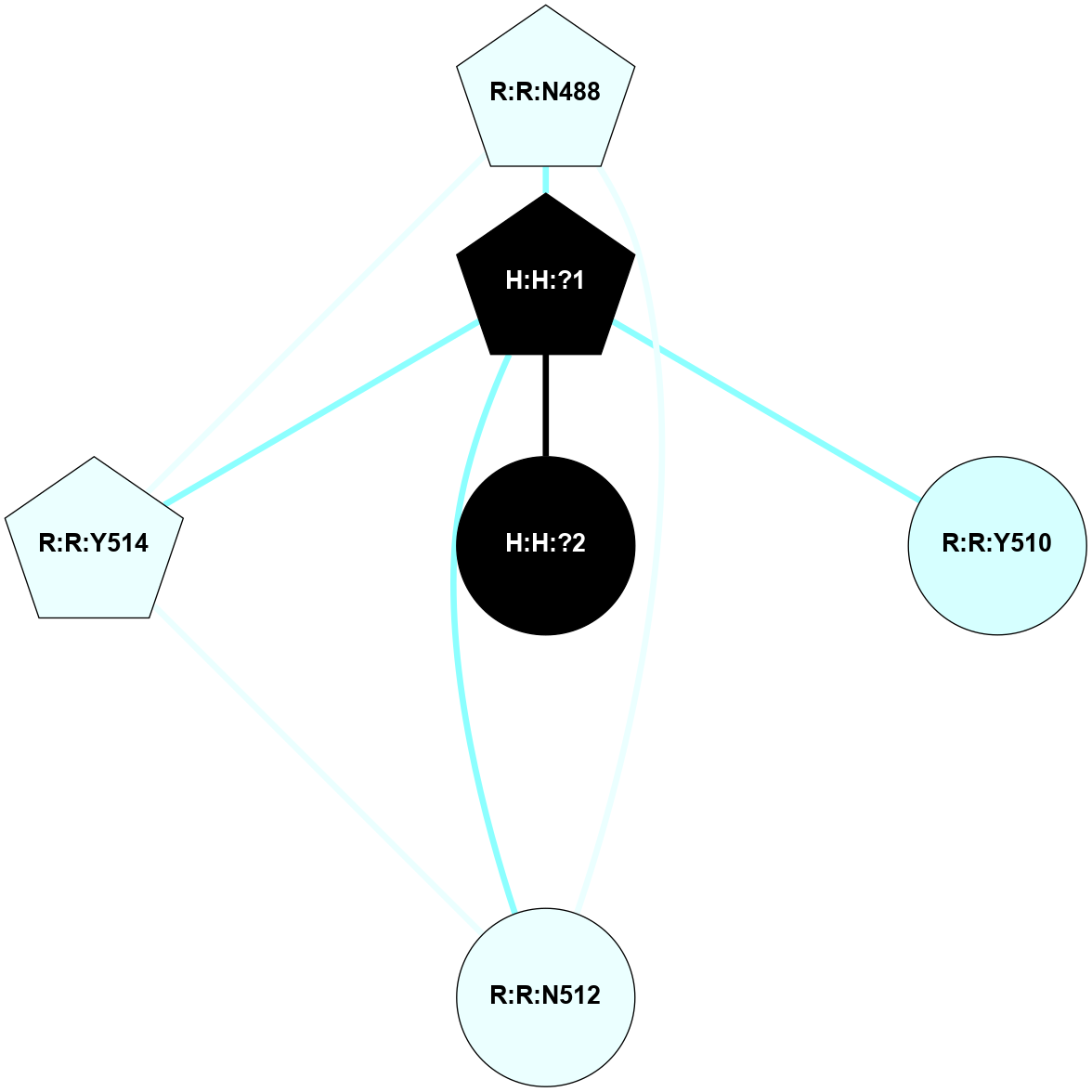
A 2D representation of the interactions of NAG in 7DTU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 38.74 | 22 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N488 | 23.33 | 22 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:Y510 | 6.29 | 22 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:N512 | 22.1 | 22 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:Y514 | 17.82 | 22 | Yes | Yes | 0 | 4 | 1 | 2 | | R:R:N488 | R:R:N512 | 6.81 | 22 | Yes | No | 4 | 4 | 2 | 2 | | R:R:N488 | R:R:Y514 | 6.98 | 22 | Yes | Yes | 4 | 4 | 2 | 2 | | R:R:N512 | R:R:Y514 | 8.14 | 22 | No | Yes | 4 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 38.74 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
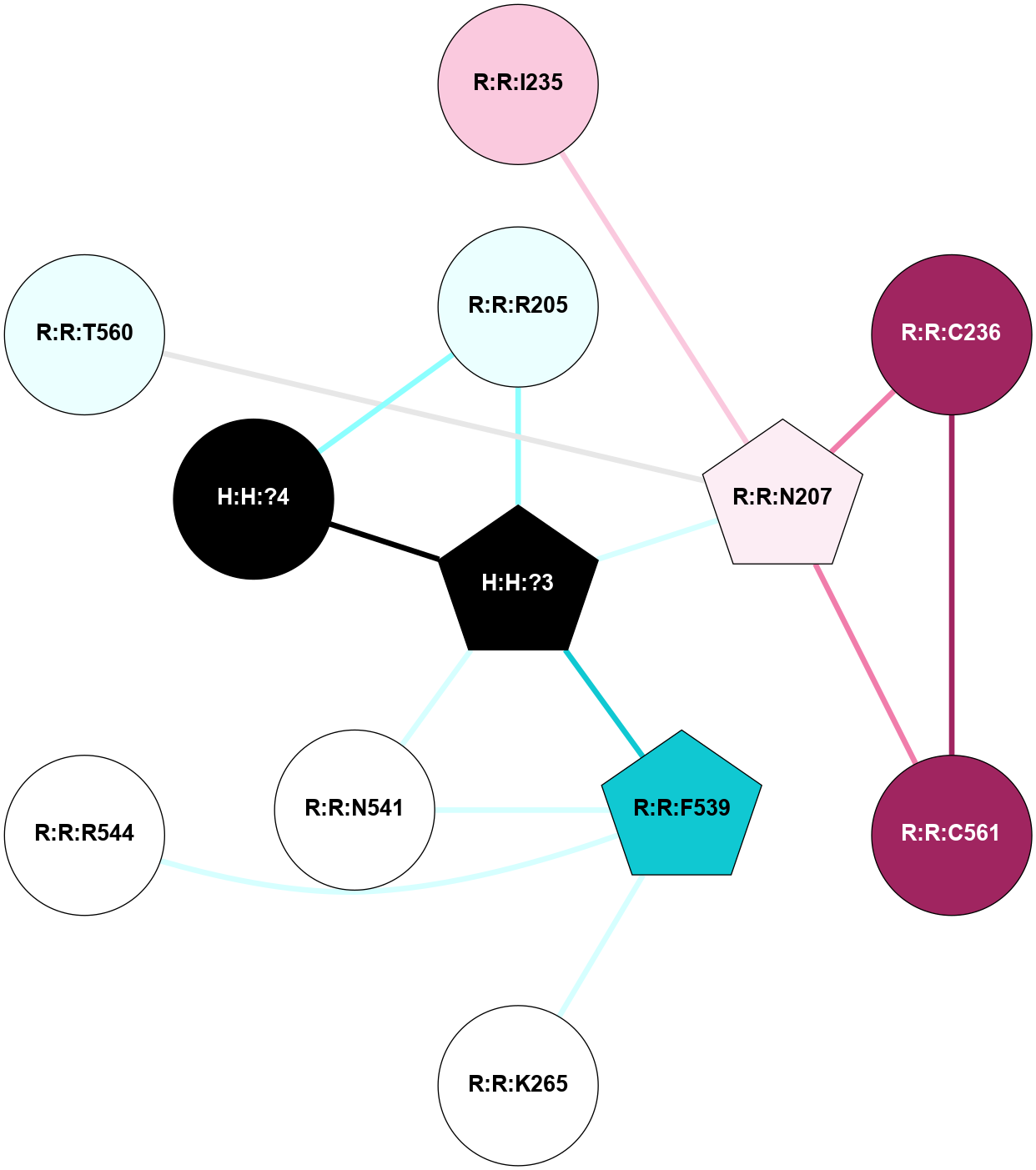
A 2D representation of the interactions of NAG in 7DTU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 33.21 | 11 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:R205 | 10.86 | 11 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?3 | R:R:N207 | 9.82 | 11 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?3 | R:R:F539 | 7.62 | 11 | Yes | Yes | 0 | 1 | 0 | 1 | | H:H:?3 | R:R:N541 | 30.7 | 11 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?3 | R:R:D545 | 9.71 | 11 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?4 | R:R:R205 | 15.21 | 11 | No | No | 0 | 4 | 1 | 1 | | R:R:C236 | R:R:N207 | 3.15 | 11 | No | Yes | 9 | 6 | 2 | 1 | | R:R:N207 | R:R:N541 | 2.72 | 11 | Yes | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 6.3 | 11 | No | Yes | 9 | 6 | 2 | 1 | | R:R:C236 | R:R:C561 | 7.28 | 11 | No | No | 9 | 9 | 2 | 2 | | R:R:F539 | R:R:K265 | 3.72 | 11 | Yes | No | 1 | 5 | 1 | 2 | | R:R:F539 | R:R:N541 | 7.25 | 11 | Yes | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 22.45 | 11 | Yes | No | 1 | 5 | 1 | 2 | | R:R:D545 | R:R:D578 | 3.99 | 0 | No | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 16.99 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
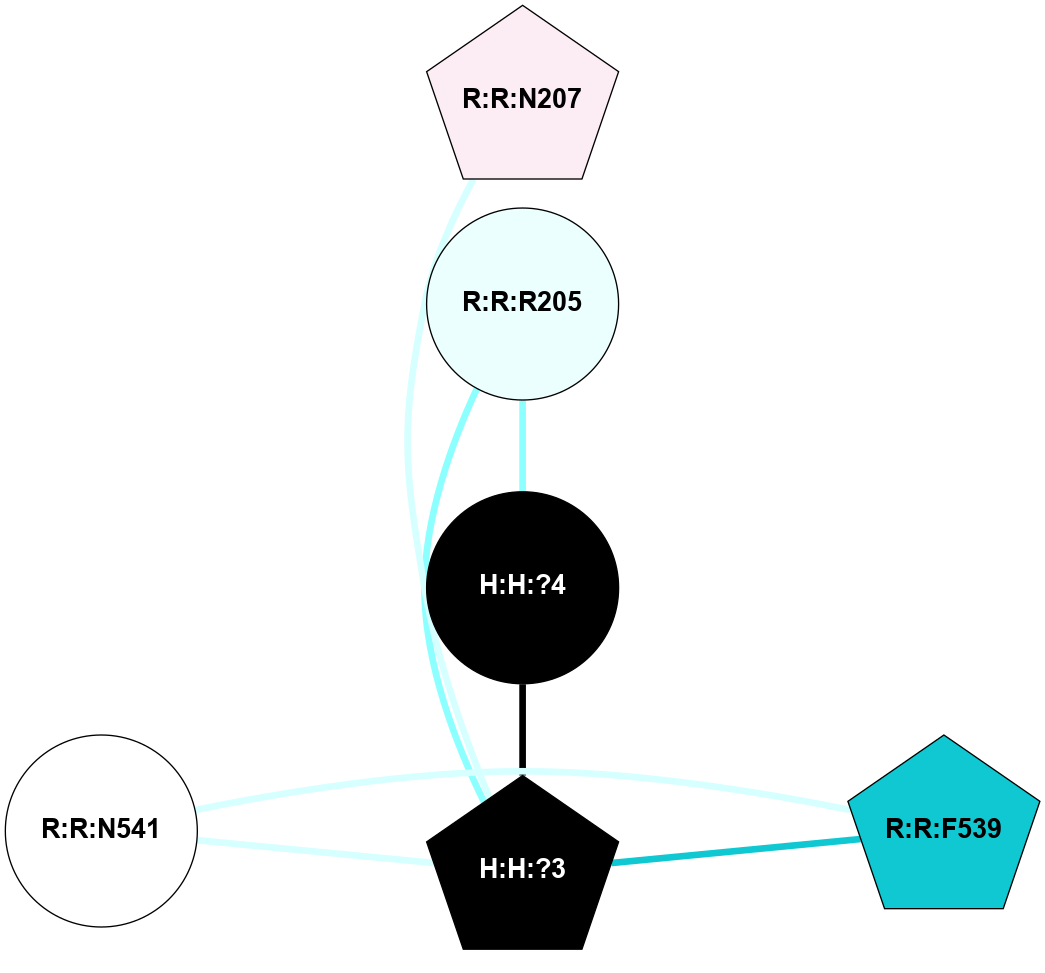
A 2D representation of the interactions of NAG in 7DTU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 33.21 | 11 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:R205 | 10.86 | 11 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?3 | R:R:N207 | 9.82 | 11 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?3 | R:R:F539 | 7.62 | 11 | Yes | Yes | 0 | 1 | 1 | 2 | | H:H:?3 | R:R:N541 | 30.7 | 11 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | R:R:D545 | 9.71 | 11 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?4 | R:R:R205 | 15.21 | 11 | No | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 2.72 | 11 | Yes | No | 6 | 5 | 2 | 2 | | R:R:F539 | R:R:N541 | 7.25 | 11 | Yes | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 24.21 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
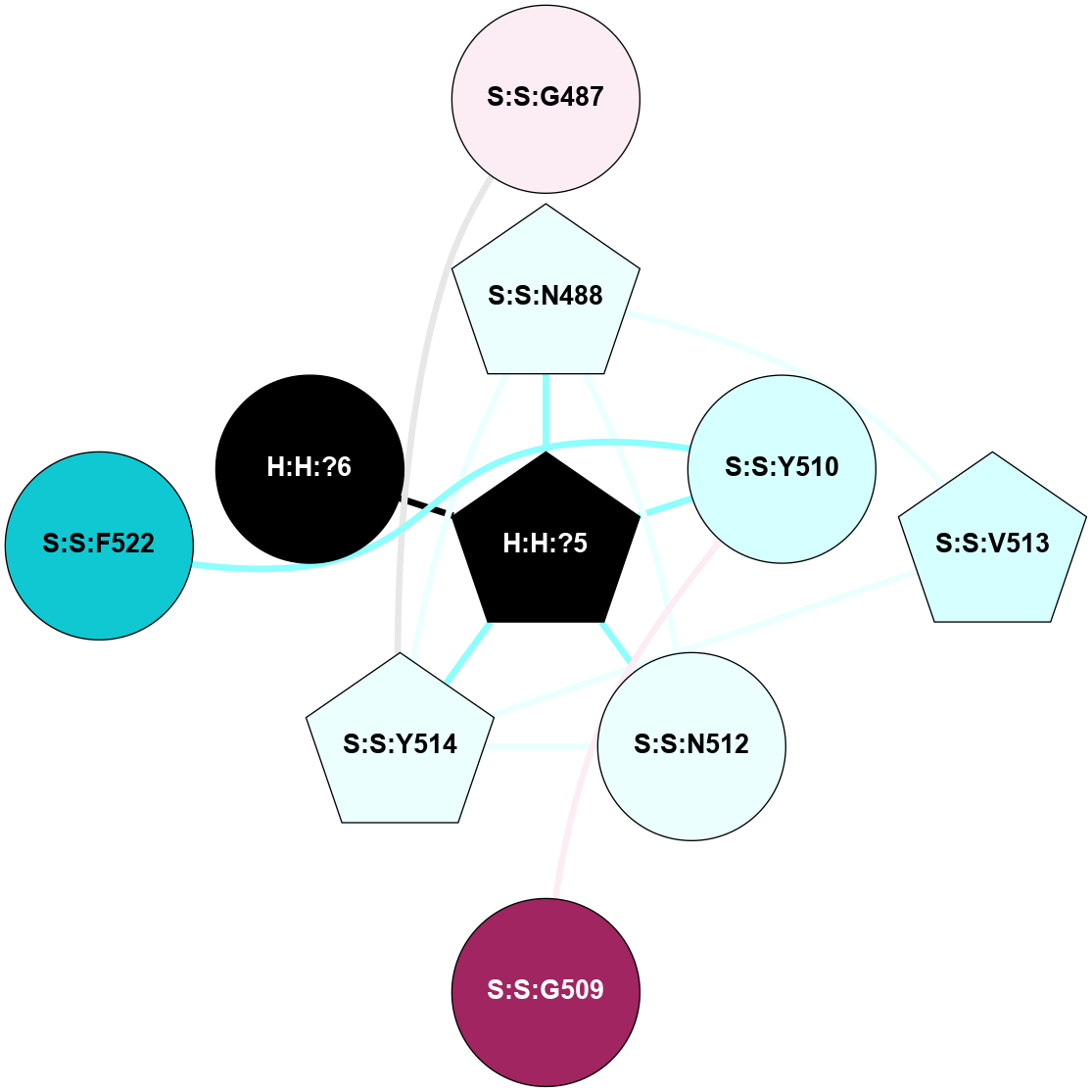
A 2D representation of the interactions of NAG in 7DTU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 38.74 | 26 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?5 | S:S:N488 | 23.33 | 26 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?5 | S:S:Y510 | 6.29 | 26 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?5 | S:S:N512 | 22.1 | 26 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?5 | S:S:Y514 | 17.82 | 26 | Yes | Yes | 0 | 4 | 0 | 1 | | S:S:G487 | S:S:Y514 | 4.35 | 0 | No | Yes | 6 | 4 | 2 | 1 | | S:S:N488 | S:S:N512 | 6.81 | 26 | Yes | No | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:V513 | 2.96 | 26 | Yes | Yes | 4 | 3 | 1 | 2 | | S:S:N488 | S:S:Y514 | 6.98 | 26 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:G509 | S:S:Y510 | 2.9 | 0 | No | No | 9 | 3 | 2 | 1 | | S:S:F522 | S:S:Y510 | 9.28 | 0 | No | No | 1 | 3 | 2 | 1 | | S:S:N512 | S:S:Y514 | 8.14 | 26 | No | Yes | 4 | 4 | 1 | 1 | | S:S:V513 | S:S:Y514 | 5.05 | 26 | Yes | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 21.66 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
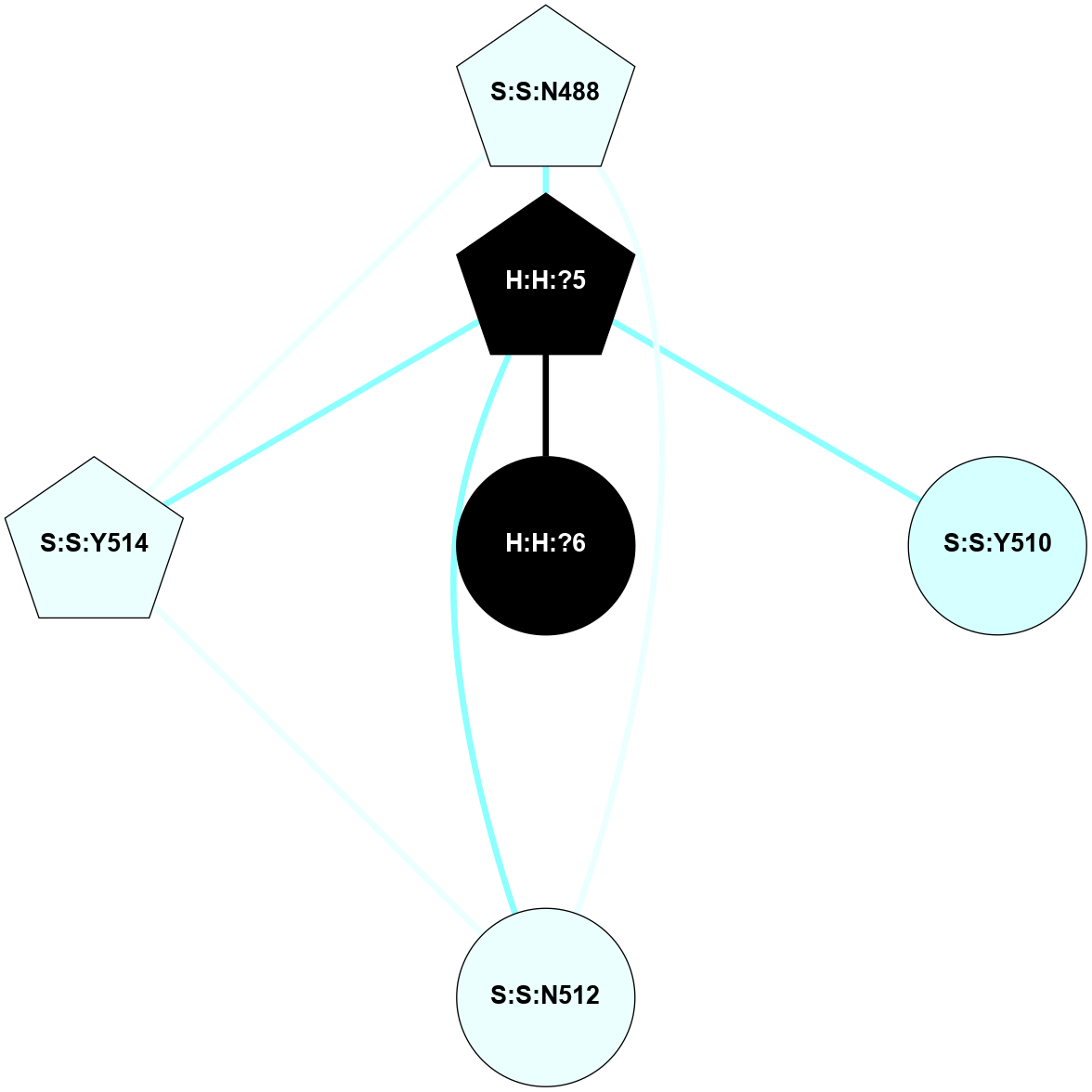
A 2D representation of the interactions of NAG in 7DTU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 38.74 | 26 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?5 | S:S:N488 | 23.33 | 26 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?5 | S:S:Y510 | 6.29 | 26 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?5 | S:S:N512 | 22.1 | 26 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?5 | S:S:Y514 | 17.82 | 26 | Yes | Yes | 0 | 4 | 1 | 2 | | S:S:N488 | S:S:N512 | 6.81 | 26 | Yes | No | 4 | 4 | 2 | 2 | | S:S:N488 | S:S:Y514 | 6.98 | 26 | Yes | Yes | 4 | 4 | 2 | 2 | | S:S:N512 | S:S:Y514 | 8.14 | 26 | No | Yes | 4 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 38.74 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
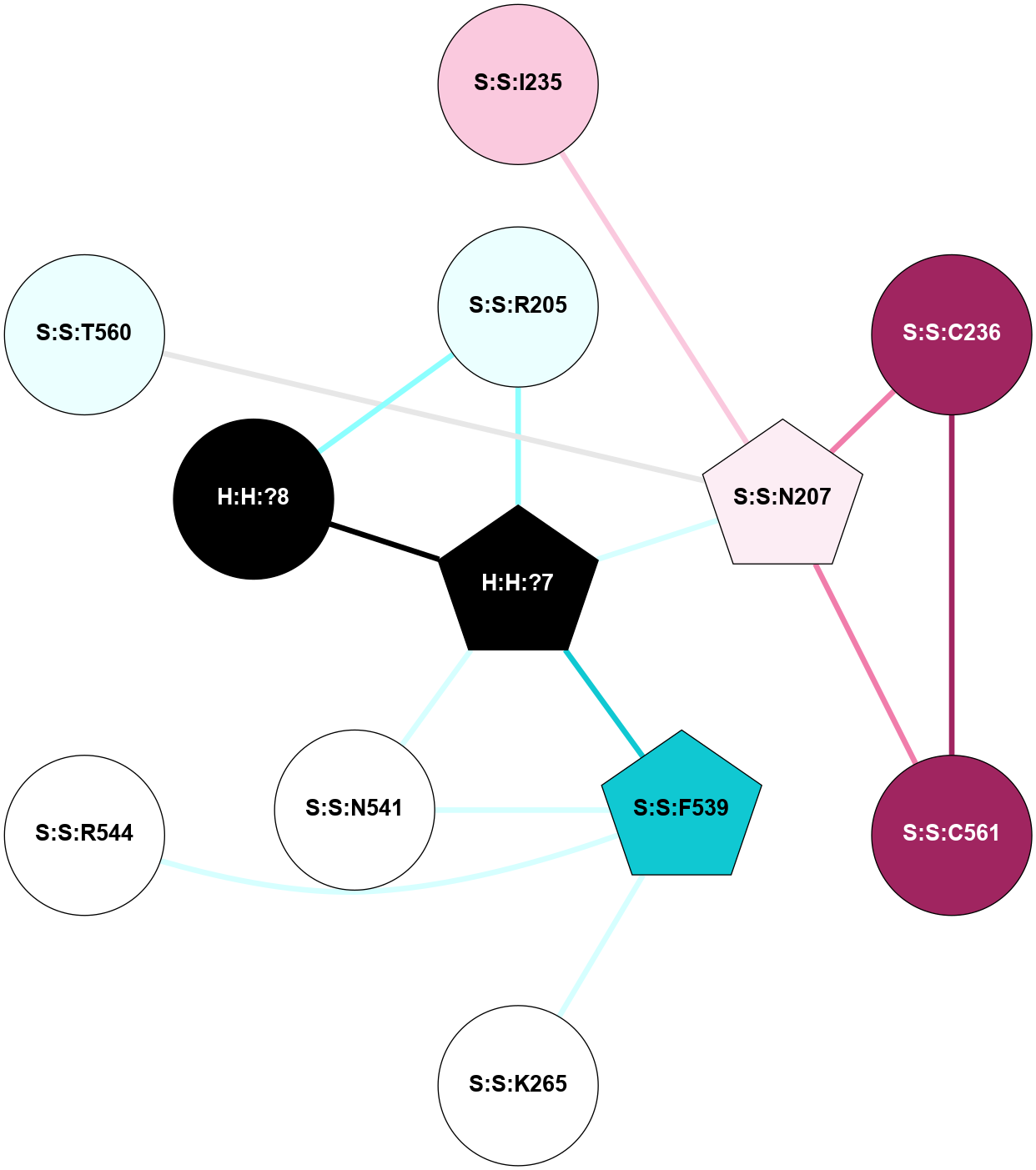
A 2D representation of the interactions of NAG in 7DTU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 33.21 | 12 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?7 | S:S:R205 | 10.86 | 12 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?7 | S:S:N207 | 7.37 | 12 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?7 | S:S:F539 | 7.62 | 12 | Yes | Yes | 0 | 1 | 0 | 1 | | H:H:?7 | S:S:N541 | 30.7 | 12 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?7 | S:S:D545 | 8.5 | 12 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?8 | S:S:R205 | 15.21 | 12 | No | No | 0 | 4 | 1 | 1 | | S:S:I235 | S:S:N207 | 4.25 | 0 | No | Yes | 7 | 6 | 2 | 1 | | S:S:C236 | S:S:N207 | 4.72 | 12 | No | Yes | 9 | 6 | 2 | 1 | | S:S:N207 | S:S:N541 | 2.72 | 12 | Yes | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 7.87 | 12 | No | Yes | 9 | 6 | 2 | 1 | | S:S:C236 | S:S:C561 | 7.28 | 12 | No | No | 9 | 9 | 2 | 2 | | S:S:F539 | S:S:K265 | 3.72 | 12 | Yes | No | 1 | 5 | 1 | 2 | | S:S:F539 | S:S:N541 | 7.25 | 12 | Yes | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 22.45 | 12 | Yes | No | 1 | 5 | 1 | 2 | | S:S:D545 | S:S:D578 | 9.31 | 0 | No | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 16.38 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
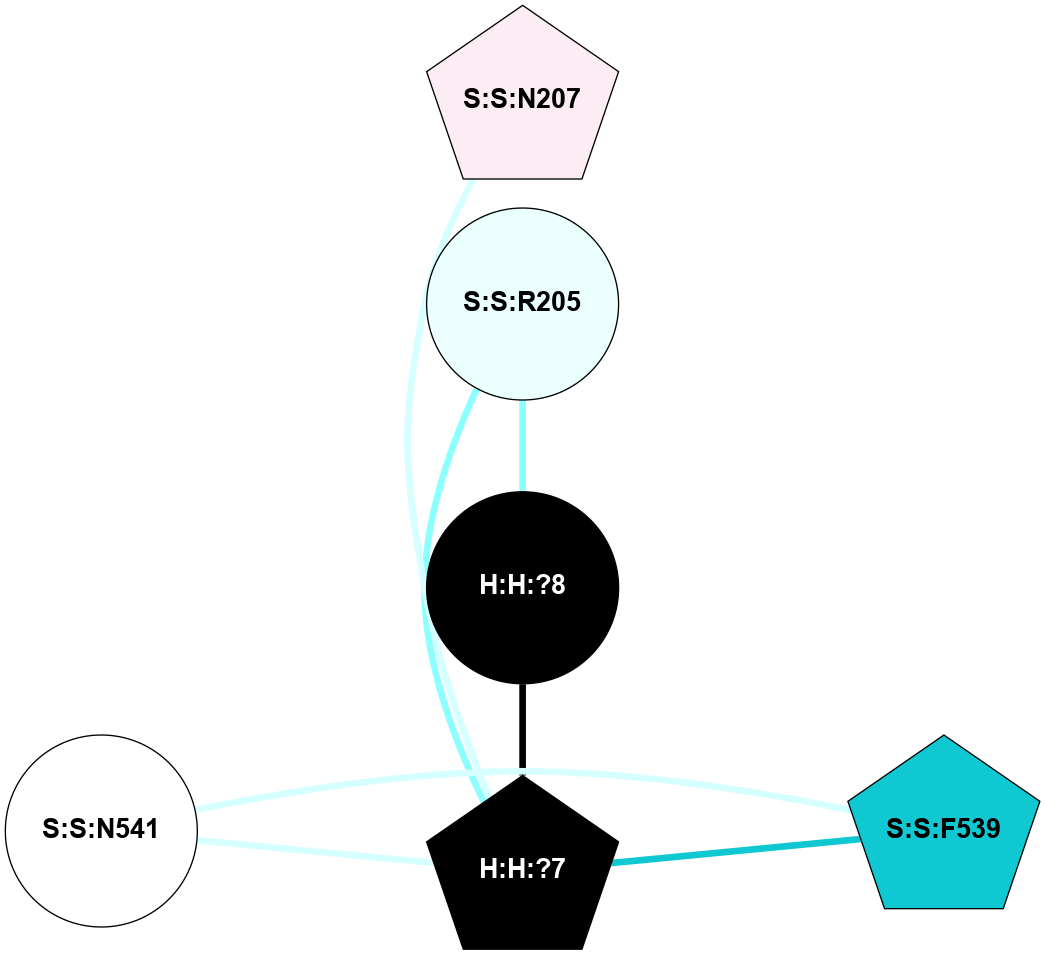
A 2D representation of the interactions of NAG in 7DTU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 33.21 | 12 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?7 | S:S:R205 | 10.86 | 12 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?7 | S:S:N207 | 7.37 | 12 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?7 | S:S:F539 | 7.62 | 12 | Yes | Yes | 0 | 1 | 1 | 2 | | H:H:?7 | S:S:N541 | 30.7 | 12 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?7 | S:S:D545 | 8.5 | 12 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?8 | S:S:R205 | 15.21 | 12 | No | No | 0 | 4 | 0 | 1 | | S:S:N207 | S:S:N541 | 2.72 | 12 | Yes | No | 6 | 5 | 2 | 2 | | S:S:F539 | S:S:N541 | 7.25 | 12 | Yes | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 24.21 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
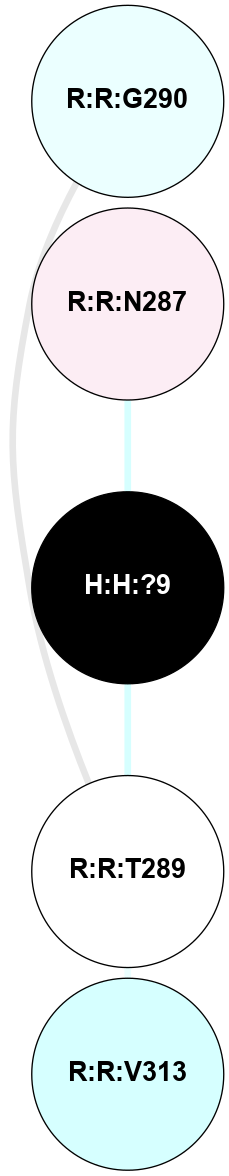
A 2D representation of the interactions of NAG in 7DTU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?9 | R:R:N287 | 34.38 | 0 | No | No | 0 | 6 | 0 | 1 | | H:H:?9 | R:R:T289 | 6.59 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:G290 | R:R:T289 | 1.82 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:T289 | R:R:V313 | 1.59 | 0 | No | No | 5 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.48 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
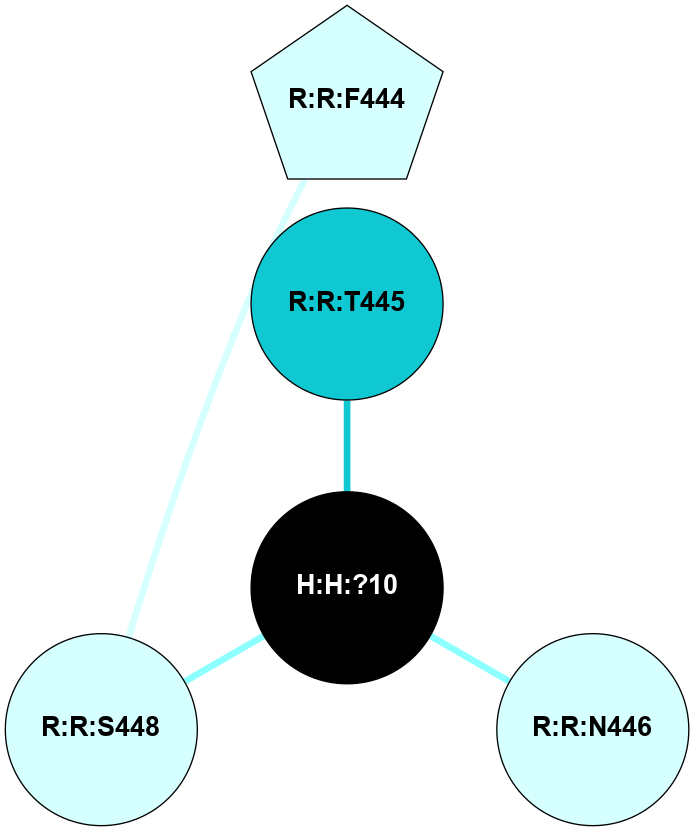
A 2D representation of the interactions of NAG in 7DTU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | R:R:T445 | 5.27 | 0 | No | No | 0 | 1 | 0 | 1 | | H:H:?10 | R:R:N446 | 35.61 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?10 | R:R:S448 | 2.69 | 0 | No | No | 0 | 3 | 0 | 1 | | R:R:F444 | R:R:S448 | 1.32 | 16 | Yes | No | 3 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.52 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7DTU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | R:R:N468 | 31.93 | 34 | No | No | 0 | 6 | 0 | 1 | | H:H:?11 | R:R:Q476 | 5.95 | 34 | No | Yes | 0 | 3 | 0 | 1 | | R:R:N468 | R:R:Q476 | 5.28 | 34 | No | Yes | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 5.85 | 34 | No | No | 6 | 3 | 1 | 2 | | R:R:Q476 | R:R:T470 | 7.09 | 34 | Yes | No | 3 | 5 | 1 | 2 | | R:R:Q476 | R:R:T478 | 4.25 | 34 | Yes | No | 3 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 18.94 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7DTU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | S:S:N287 | 34.38 | 0 | No | No | 0 | 6 | 0 | 1 | | H:H:?12 | S:S:T289 | 6.59 | 0 | No | No | 0 | 5 | 0 | 1 | | S:S:G290 | S:S:T289 | 1.82 | 0 | No | No | 4 | 5 | 2 | 1 | | S:S:T289 | S:S:V313 | 1.59 | 0 | No | No | 5 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.48 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
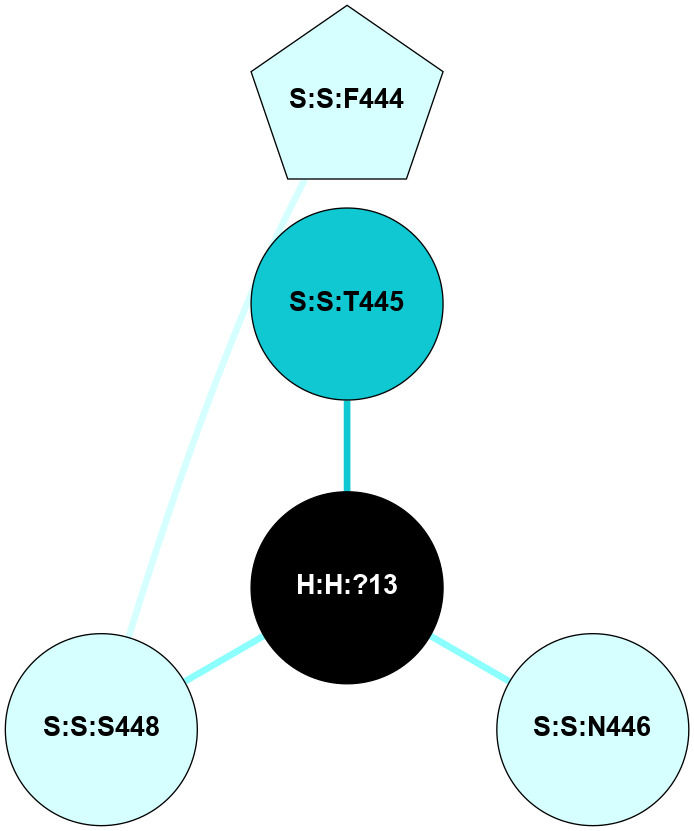
A 2D representation of the interactions of NAG in 7DTU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | S:S:T445 | 5.27 | 0 | No | No | 0 | 1 | 0 | 1 | | H:H:?13 | S:S:N446 | 35.61 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?13 | S:S:S448 | 2.69 | 0 | No | No | 0 | 3 | 0 | 1 | | S:S:F444 | S:S:S448 | 1.32 | 15 | Yes | No | 3 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.52 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7DTU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?14 | S:S:N468 | 31.93 | 35 | No | No | 0 | 6 | 0 | 1 | | H:H:?14 | S:S:Q476 | 5.95 | 35 | No | Yes | 0 | 3 | 0 | 1 | | S:S:N468 | S:S:Q476 | 5.28 | 35 | No | Yes | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 5.85 | 35 | No | No | 6 | 3 | 1 | 2 | | S:S:Q476 | S:S:T470 | 7.09 | 35 | Yes | No | 3 | 5 | 1 | 2 | | S:S:Q476 | S:S:T478 | 4.25 | 35 | Yes | No | 3 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 18.94 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7DTV | C | Ion | Calcium Sensing | CaS | Homo Sapiens | Ca | Ca; Tryptophan | - | 3.5 | 2021-03-10 | doi.org/10.1038/s41422-021-00474-0 |
|
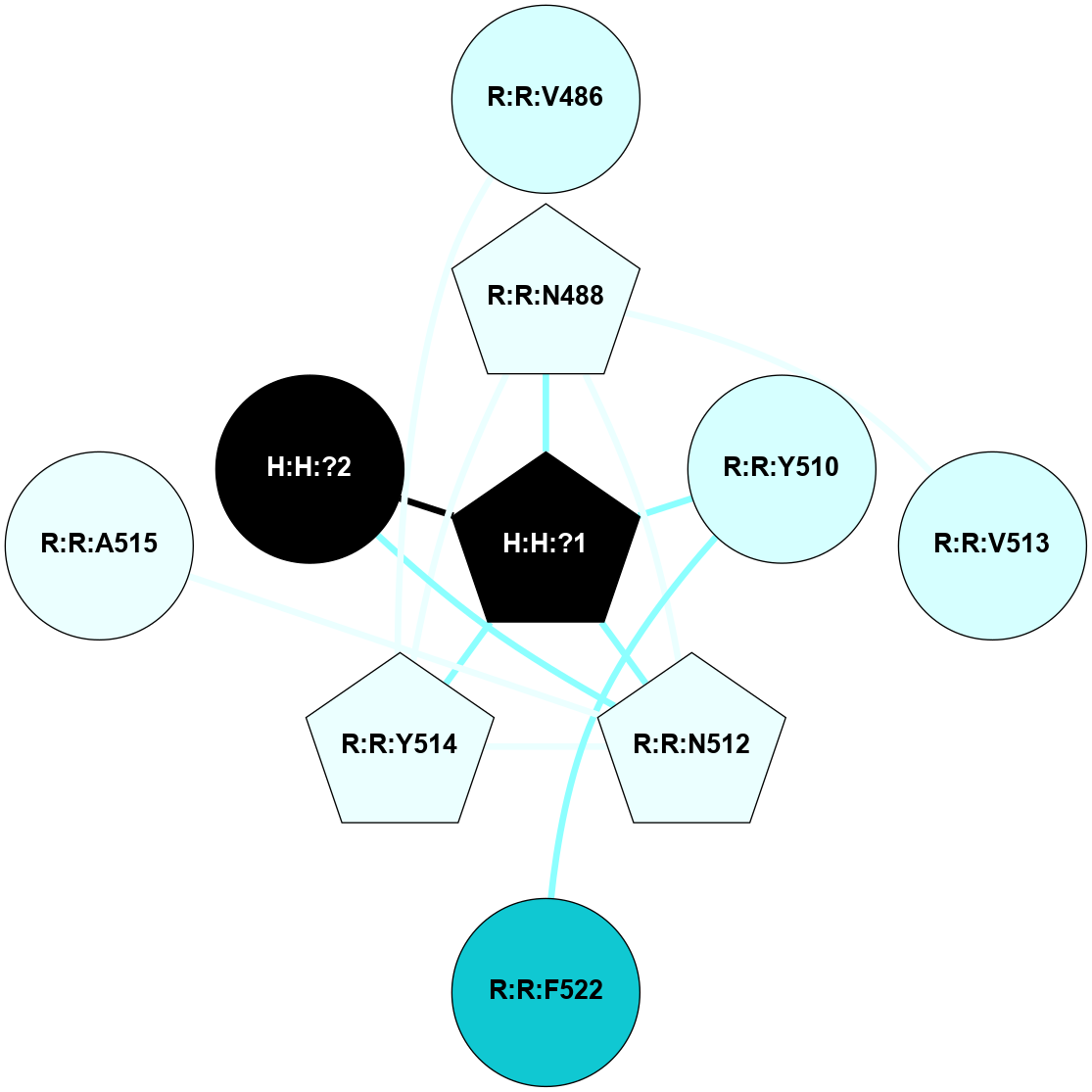
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 27.67 | 20 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N488 | 28.24 | 20 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:Y510 | 13.63 | 20 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:N512 | 18.42 | 20 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:Y514 | 11.53 | 20 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:N512 | 3.68 | 20 | No | Yes | 0 | 4 | 1 | 1 | | R:R:V486 | R:R:Y514 | 3.79 | 0 | No | Yes | 3 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 5.45 | 20 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:V513 | 5.91 | 20 | Yes | No | 4 | 3 | 1 | 2 | | R:R:N488 | R:R:Y514 | 6.98 | 20 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:F522 | R:R:Y510 | 14.44 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:N512 | R:R:Y514 | 3.49 | 20 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 19.90 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
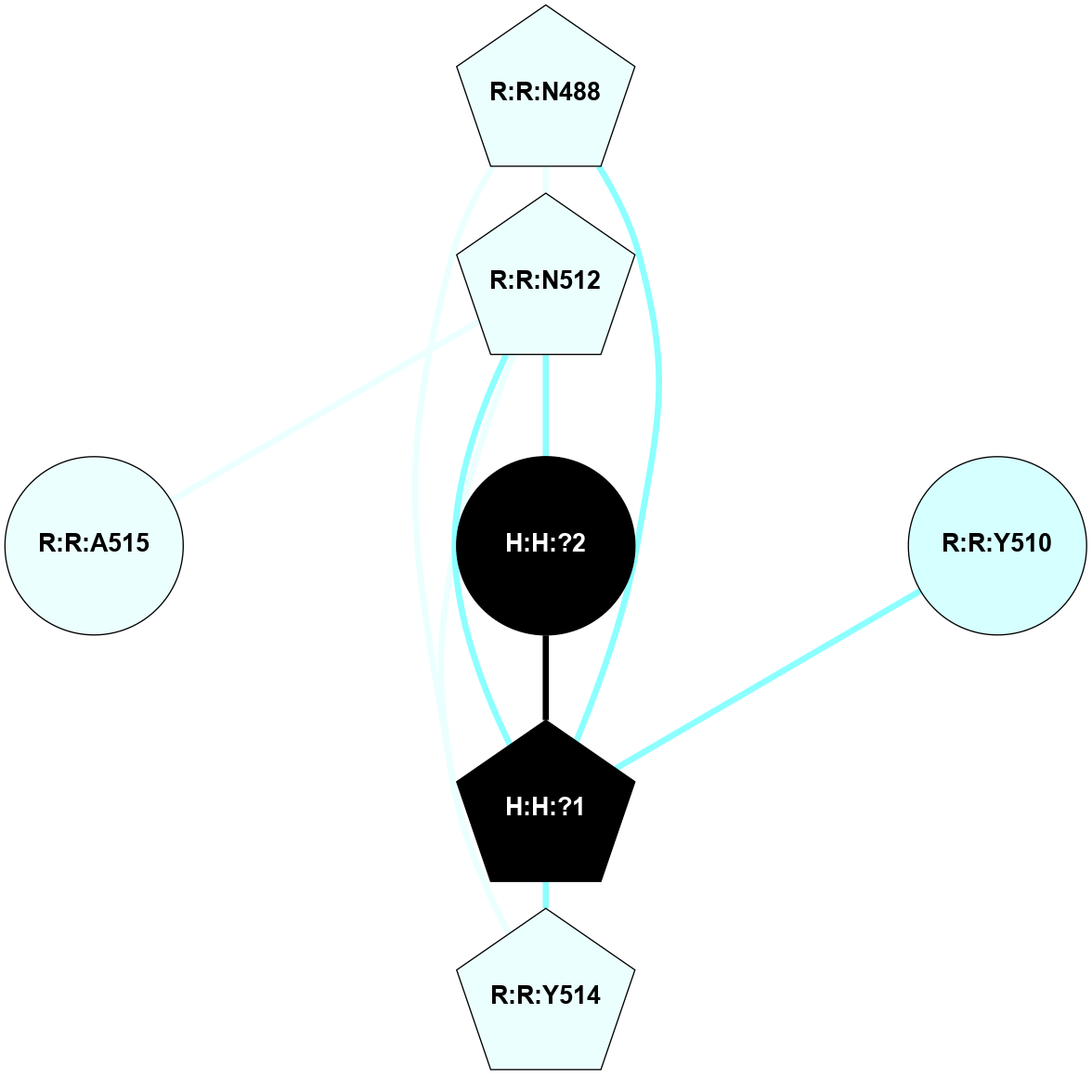
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 27.67 | 20 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N488 | 28.24 | 20 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:Y510 | 13.63 | 20 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:N512 | 18.42 | 20 | Yes | Yes | 0 | 4 | 1 | 1 | | H:H:?1 | R:R:Y514 | 11.53 | 20 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?2 | R:R:N512 | 3.68 | 20 | No | Yes | 0 | 4 | 0 | 1 | | R:R:N488 | R:R:N512 | 5.45 | 20 | Yes | Yes | 4 | 4 | 2 | 1 | | R:R:N488 | R:R:Y514 | 6.98 | 20 | Yes | Yes | 4 | 4 | 2 | 2 | | R:R:N512 | R:R:Y514 | 3.49 | 20 | Yes | Yes | 4 | 4 | 1 | 2 | | R:R:A515 | R:R:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 15.68 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
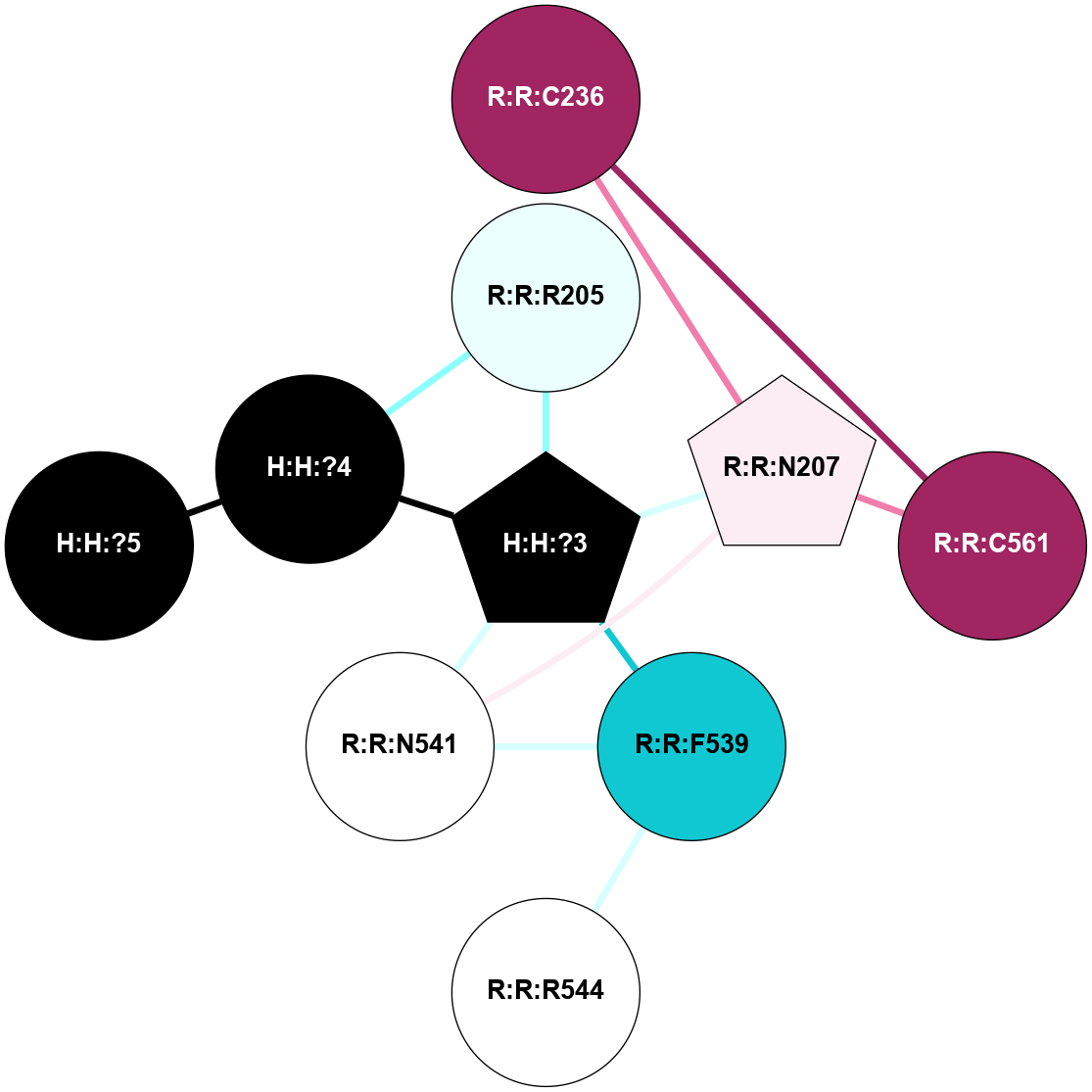
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 35.42 | 3 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:R205 | 4.35 | 3 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?3 | R:R:N207 | 15.96 | 3 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?3 | R:R:F539 | 4.36 | 3 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?3 | R:R:N541 | 25.79 | 3 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?4 | H:H:?5 | 34.31 | 3 | No | No | 0 | 0 | 1 | 2 | | H:H:?4 | R:R:R205 | 5.43 | 3 | No | No | 0 | 4 | 1 | 1 | | R:R:C236 | R:R:N207 | 3.15 | 3 | No | Yes | 9 | 6 | 2 | 1 | | R:R:N207 | R:R:N541 | 10.9 | 3 | Yes | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 4.72 | 3 | No | Yes | 9 | 6 | 2 | 1 | | R:R:C236 | R:R:C561 | 7.28 | 3 | No | No | 9 | 9 | 2 | 2 | | R:R:F539 | R:R:N541 | 7.25 | 3 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 27.79 | 3 | No | No | 1 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.18 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
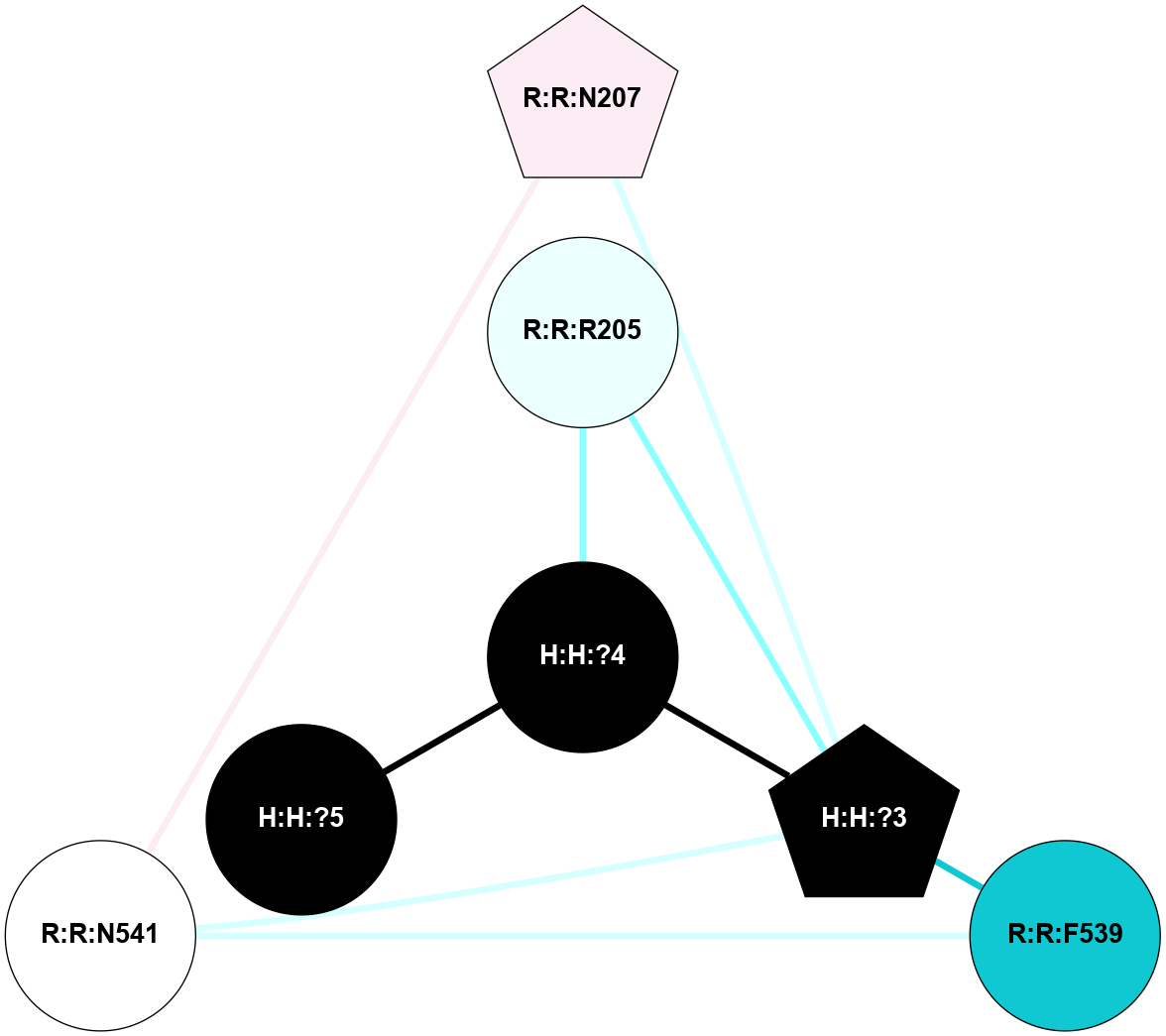
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 35.42 | 3 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:R205 | 4.35 | 3 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?3 | R:R:N207 | 15.96 | 3 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?3 | R:R:F539 | 4.36 | 3 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?3 | R:R:N541 | 25.79 | 3 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?4 | H:H:?5 | 34.31 | 3 | No | No | 0 | 0 | 0 | 1 | | H:H:?4 | R:R:R205 | 5.43 | 3 | No | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 10.9 | 3 | Yes | No | 6 | 5 | 2 | 2 | | R:R:F539 | R:R:N541 | 7.25 | 3 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 25.05 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 35.42 | 3 | Yes | No | 0 | 0 | 2 | 1 | | H:H:?3 | R:R:R205 | 4.35 | 3 | Yes | No | 0 | 4 | 2 | 2 | | H:H:?4 | H:H:?5 | 34.31 | 3 | No | No | 0 | 0 | 1 | 0 | | H:H:?4 | R:R:R205 | 5.43 | 3 | No | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 34.31 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | H:H:?7 | 27.67 | 21 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?6 | S:S:N488 | 28.24 | 21 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?6 | S:S:Y510 | 13.63 | 21 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?6 | S:S:N512 | 18.42 | 21 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?6 | S:S:Y514 | 11.53 | 21 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?7 | S:S:N512 | 3.68 | 21 | No | Yes | 0 | 4 | 1 | 1 | | S:S:V486 | S:S:Y514 | 3.79 | 0 | No | Yes | 3 | 4 | 2 | 1 | | S:S:N488 | S:S:N512 | 5.45 | 21 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:V513 | 5.91 | 21 | Yes | No | 4 | 3 | 1 | 2 | | S:S:N488 | S:S:Y514 | 6.98 | 21 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:F522 | S:S:Y510 | 14.44 | 0 | No | No | 1 | 3 | 2 | 1 | | S:S:N512 | S:S:Y514 | 3.49 | 21 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:A515 | S:S:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 19.90 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | H:H:?7 | 27.67 | 21 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?6 | S:S:N488 | 28.24 | 21 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?6 | S:S:Y510 | 13.63 | 21 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?6 | S:S:N512 | 18.42 | 21 | Yes | Yes | 0 | 4 | 1 | 1 | | H:H:?6 | S:S:Y514 | 11.53 | 21 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?7 | S:S:N512 | 3.68 | 21 | No | Yes | 0 | 4 | 0 | 1 | | S:S:N488 | S:S:N512 | 5.45 | 21 | Yes | Yes | 4 | 4 | 2 | 1 | | S:S:N488 | S:S:Y514 | 6.98 | 21 | Yes | Yes | 4 | 4 | 2 | 2 | | S:S:N512 | S:S:Y514 | 3.49 | 21 | Yes | Yes | 4 | 4 | 1 | 2 | | S:S:A515 | S:S:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 15.68 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | H:H:?9 | 35.42 | 4 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?8 | S:S:R205 | 4.35 | 4 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?8 | S:S:N207 | 15.96 | 4 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?8 | S:S:F539 | 4.36 | 4 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?8 | S:S:N541 | 25.79 | 4 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?10 | H:H:?9 | 34.31 | 0 | No | No | 0 | 0 | 2 | 1 | | H:H:?9 | S:S:R205 | 5.43 | 4 | No | No | 0 | 4 | 1 | 1 | | S:S:C236 | S:S:N207 | 3.15 | 4 | No | Yes | 9 | 6 | 2 | 1 | | S:S:N207 | S:S:N541 | 10.9 | 4 | Yes | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 4.72 | 4 | No | Yes | 9 | 6 | 2 | 1 | | S:S:C236 | S:S:C561 | 7.28 | 4 | No | No | 9 | 9 | 2 | 2 | | S:S:F539 | S:S:N541 | 7.25 | 4 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 27.79 | 4 | No | No | 1 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.18 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | H:H:?9 | 35.42 | 4 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?8 | S:S:R205 | 4.35 | 4 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?8 | S:S:N207 | 15.96 | 4 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?8 | S:S:F539 | 4.36 | 4 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?8 | S:S:N541 | 25.79 | 4 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?10 | H:H:?9 | 34.31 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?9 | S:S:R205 | 5.43 | 4 | No | No | 0 | 4 | 0 | 1 | | S:S:N207 | S:S:N541 | 10.9 | 4 | Yes | No | 6 | 5 | 2 | 2 | | S:S:F539 | S:S:N541 | 7.25 | 4 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 25.05 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
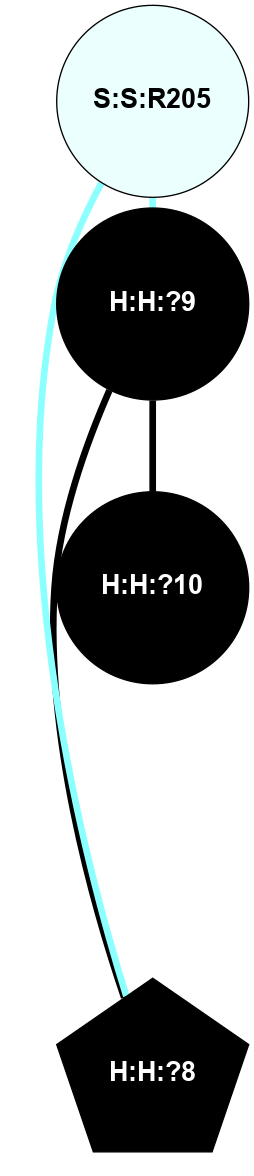
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | H:H:?9 | 35.42 | 4 | Yes | No | 0 | 0 | 2 | 1 | | H:H:?8 | S:S:R205 | 4.35 | 4 | Yes | No | 0 | 4 | 2 | 2 | | H:H:?10 | H:H:?9 | 34.31 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?9 | S:S:R205 | 5.43 | 4 | No | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 34.31 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
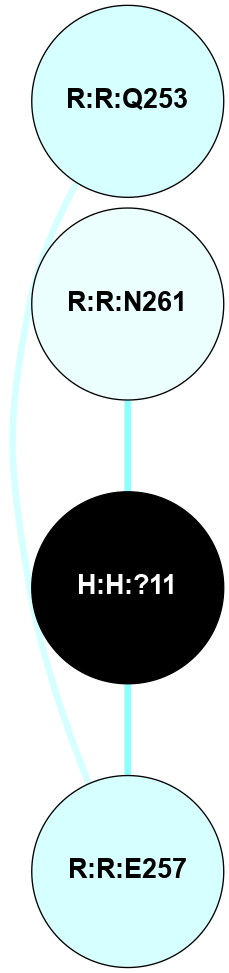
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | R:R:N261 | 30.7 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:E257 | R:R:Q253 | 6.37 | 0 | No | No | 3 | 3 | 1 | 2 | | H:H:?11 | R:R:E257 | 2.37 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.54 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
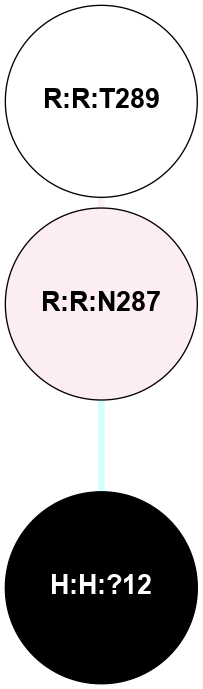
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | R:R:N287 | 29.47 | 0 | No | No | 0 | 6 | 0 | 1 | | R:R:N287 | R:R:T289 | 2.92 | 0 | No | No | 6 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 29.47 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | R:R:N446 | 35.61 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?13 | R:R:S448 | 2.69 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 19.15 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
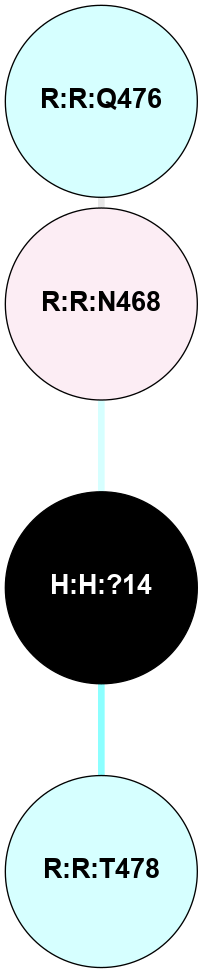
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?14 | R:R:N468 | 27.02 | 0 | No | No | 0 | 6 | 0 | 1 | | H:H:?14 | R:R:T478 | 3.95 | 0 | No | No | 0 | 3 | 0 | 1 | | R:R:N468 | R:R:Q476 | 3.96 | 0 | No | No | 6 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.48 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
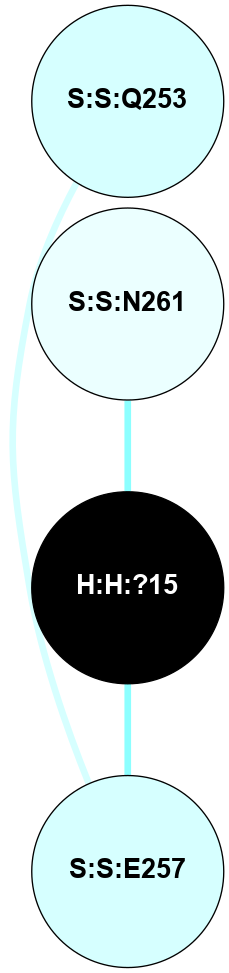
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?15 | S:S:N261 | 30.7 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:E257 | S:S:Q253 | 6.37 | 0 | No | No | 3 | 3 | 1 | 2 | | H:H:?15 | S:S:E257 | 2.37 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.54 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
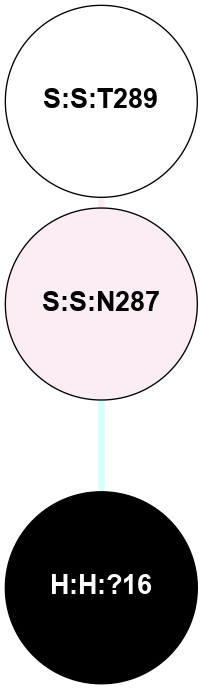
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?16 | S:S:N287 | 29.47 | 0 | No | No | 0 | 6 | 0 | 1 | | S:S:N287 | S:S:T289 | 2.92 | 0 | No | No | 6 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 29.47 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?17 | S:S:N446 | 35.61 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?17 | S:S:S448 | 2.69 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 19.15 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
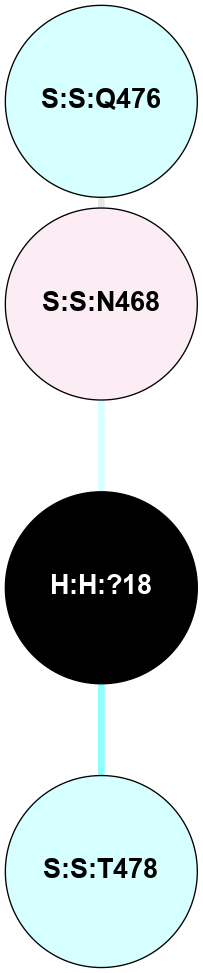
A 2D representation of the interactions of NAG in 7DTV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?18 | S:S:N468 | 27.02 | 0 | No | No | 0 | 6 | 0 | 1 | | H:H:?18 | S:S:T478 | 3.95 | 0 | No | No | 0 | 3 | 0 | 1 | | S:S:N468 | S:S:Q476 | 3.96 | 0 | No | No | 6 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.48 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7DTW | C | Ion | Calcium Sensing | CaS | Homo Sapiens | - | - | - | 4.5 | 2021-03-10 | doi.org/10.1038/s41422-021-00474-0 |
|
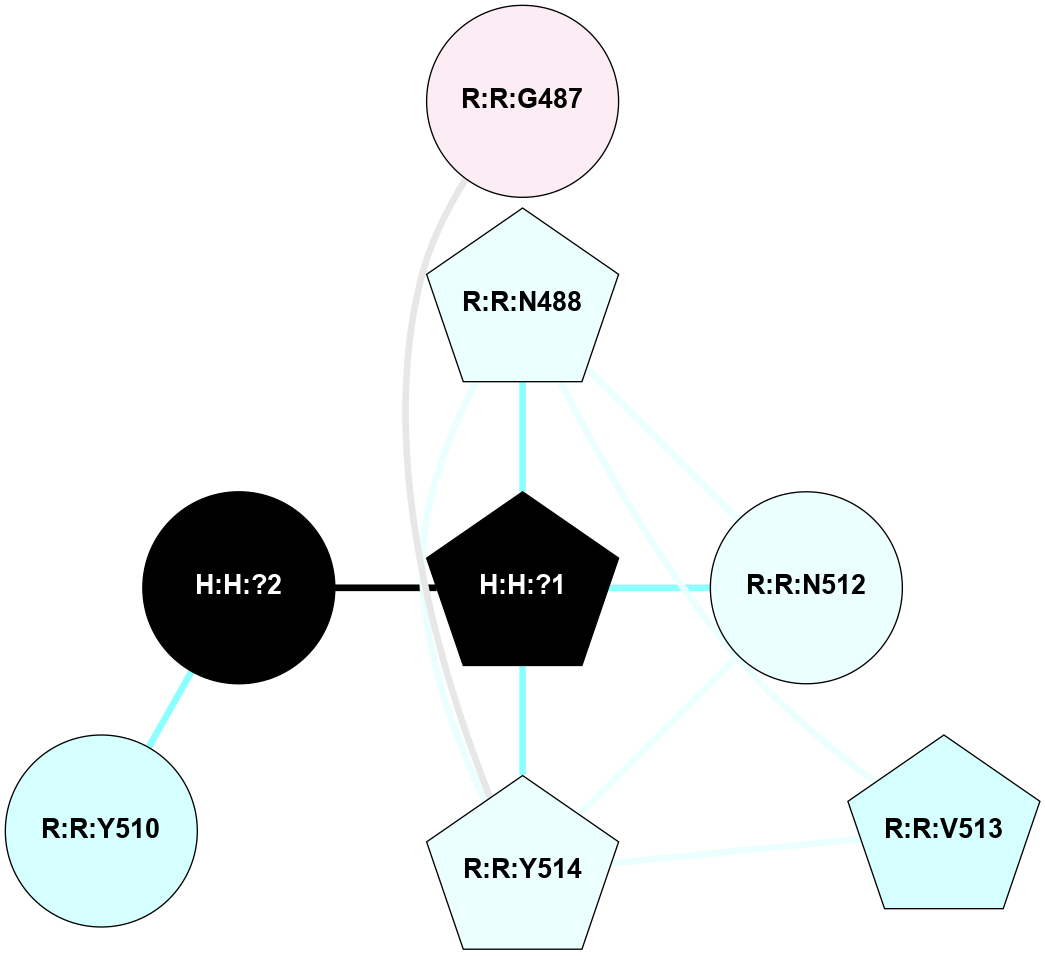
A 2D representation of the interactions of NAG in 7DTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 33.21 | 25 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N488 | 25.79 | 25 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:N512 | 17.19 | 25 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:Y514 | 15.73 | 25 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:Y510 | 5.24 | 0 | No | No | 0 | 3 | 1 | 2 | | R:R:G487 | R:R:Y514 | 2.9 | 0 | No | Yes | 6 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 9.54 | 25 | Yes | No | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:V513 | 5.91 | 25 | Yes | Yes | 4 | 3 | 1 | 2 | | R:R:N488 | R:R:Y514 | 6.98 | 25 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:N512 | R:R:Y514 | 8.14 | 25 | No | Yes | 4 | 4 | 1 | 1 | | R:R:V513 | R:R:Y514 | 8.83 | 25 | Yes | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 22.98 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
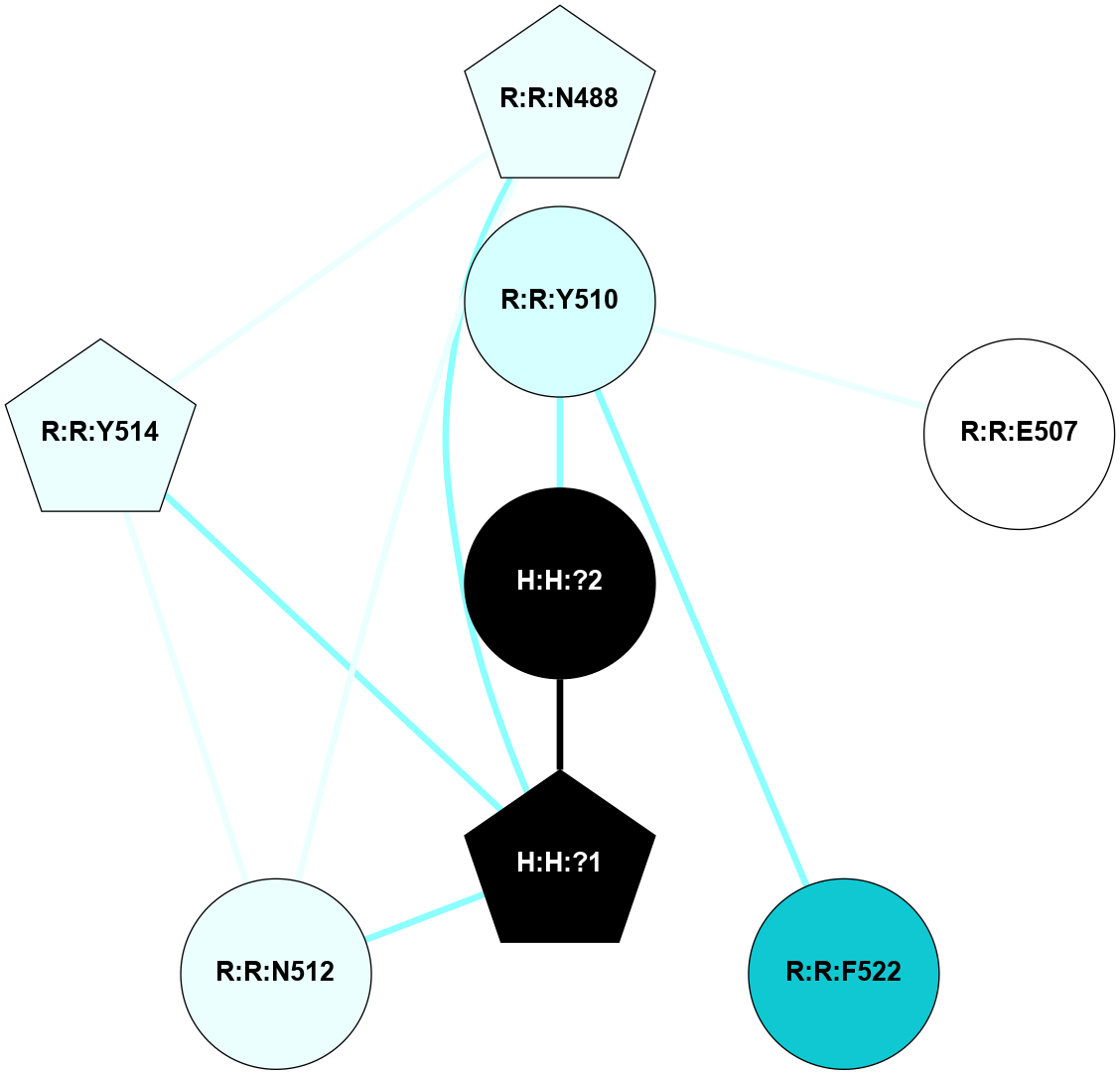
A 2D representation of the interactions of NAG in 7DTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 33.21 | 25 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N488 | 25.79 | 25 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:N512 | 17.19 | 25 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:Y514 | 15.73 | 25 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?2 | R:R:Y510 | 5.24 | 0 | No | No | 0 | 3 | 0 | 1 | | R:R:N488 | R:R:N512 | 9.54 | 25 | Yes | No | 4 | 4 | 2 | 2 | | R:R:N488 | R:R:Y514 | 6.98 | 25 | Yes | Yes | 4 | 4 | 2 | 2 | | R:R:E507 | R:R:Y510 | 4.49 | 0 | No | No | 5 | 3 | 2 | 1 | | R:R:F522 | R:R:Y510 | 14.44 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:N512 | R:R:Y514 | 8.14 | 25 | No | Yes | 4 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.23 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
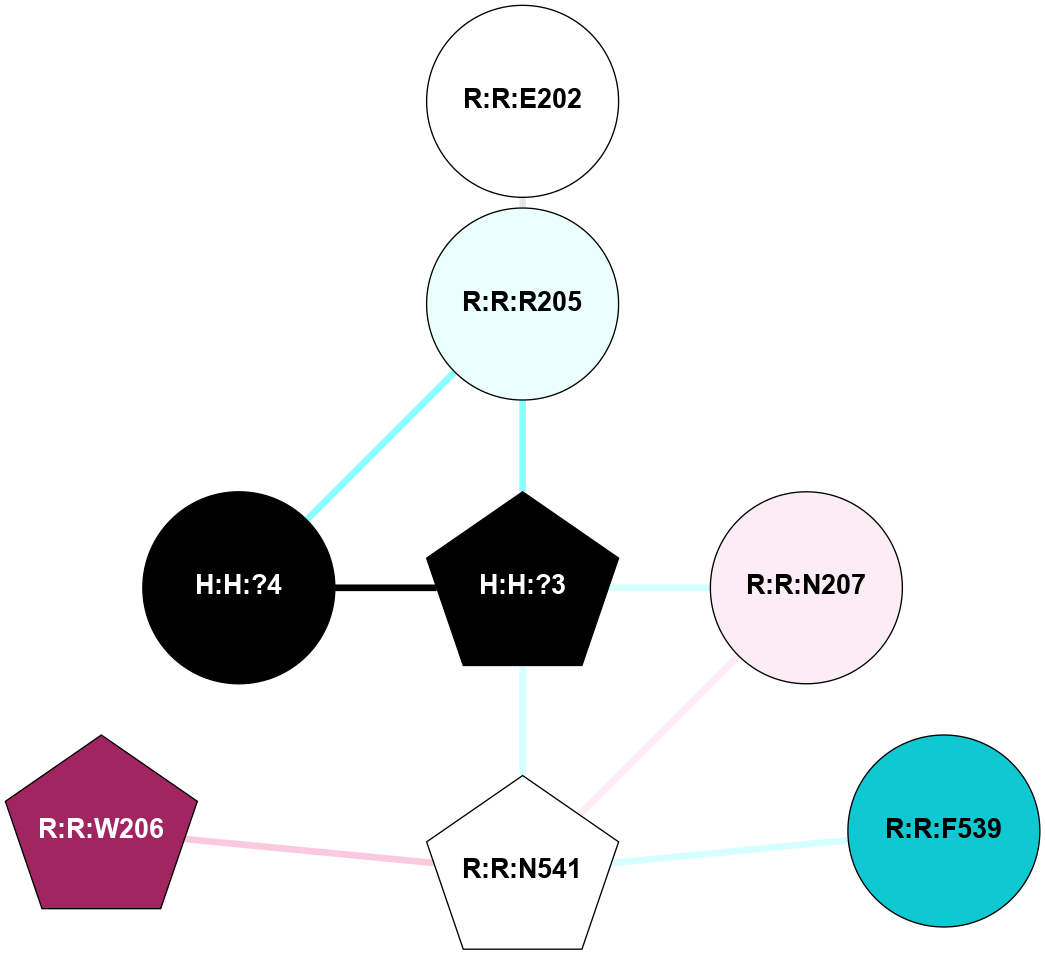
A 2D representation of the interactions of NAG in 7DTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 39.85 | 47 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:R205 | 15.21 | 47 | No | No | 0 | 4 | 0 | 1 | | H:H:?3 | R:R:N541 | 27.02 | 47 | No | No | 0 | 5 | 0 | 1 | | H:H:?4 | R:R:R205 | 21.73 | 47 | No | No | 0 | 4 | 1 | 1 | | R:R:E202 | R:R:R205 | 4.65 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:N207 | R:R:N541 | 4.09 | 0 | No | No | 6 | 5 | 2 | 1 | | R:R:F539 | R:R:N541 | 13.29 | 0 | No | No | 1 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.36 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
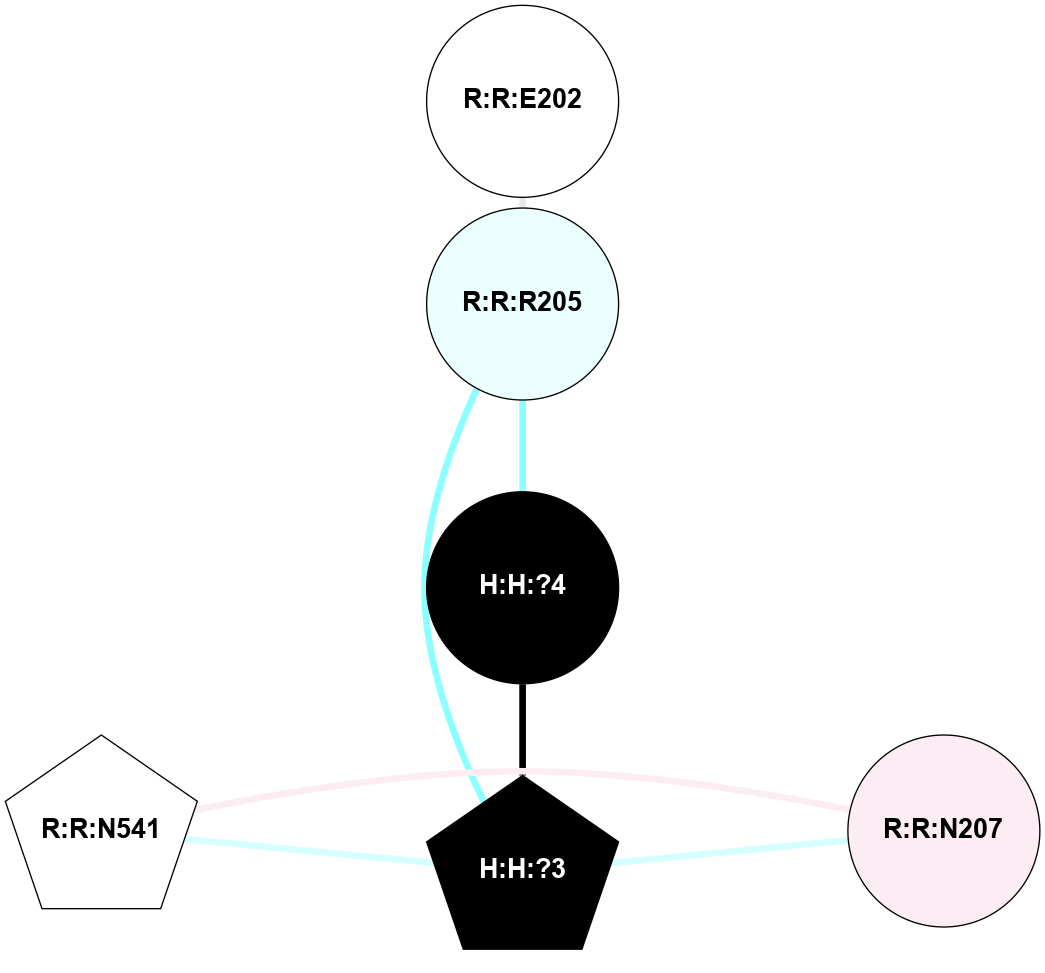
A 2D representation of the interactions of NAG in 7DTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 39.85 | 47 | No | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:R205 | 15.21 | 47 | No | No | 0 | 4 | 1 | 1 | | H:H:?3 | R:R:N541 | 27.02 | 47 | No | No | 0 | 5 | 1 | 2 | | H:H:?4 | R:R:R205 | 21.73 | 47 | No | No | 0 | 4 | 0 | 1 | | R:R:E202 | R:R:R205 | 4.65 | 0 | No | No | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 30.79 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
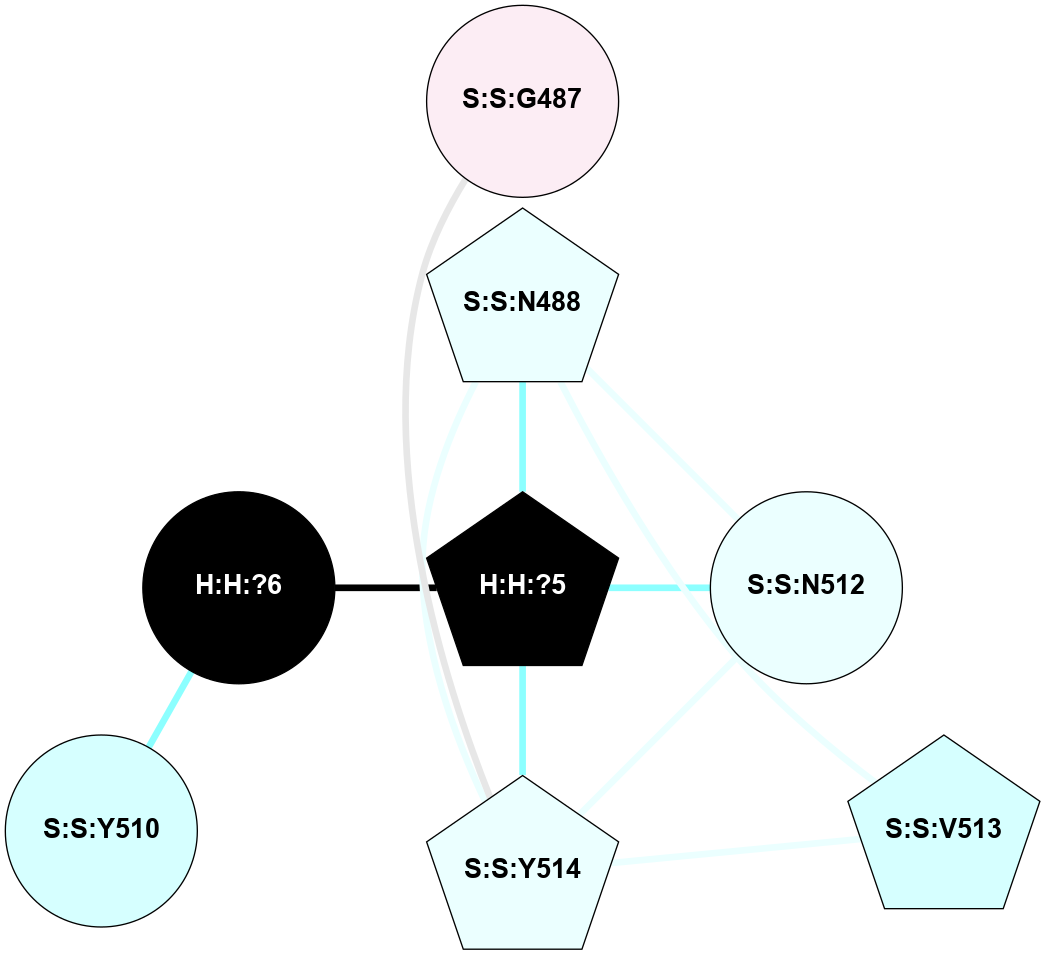
A 2D representation of the interactions of NAG in 7DTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 33.21 | 26 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?5 | S:S:N488 | 25.79 | 26 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?5 | S:S:N512 | 17.19 | 26 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?5 | S:S:Y514 | 15.73 | 26 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?6 | S:S:Y510 | 5.24 | 0 | No | No | 0 | 3 | 1 | 2 | | S:S:G487 | S:S:Y514 | 2.9 | 0 | No | Yes | 6 | 4 | 2 | 1 | | S:S:N488 | S:S:N512 | 9.54 | 26 | Yes | No | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:V513 | 5.91 | 26 | Yes | Yes | 4 | 3 | 1 | 2 | | S:S:N488 | S:S:Y514 | 6.98 | 26 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:N512 | S:S:Y514 | 8.14 | 26 | No | Yes | 4 | 4 | 1 | 1 | | S:S:V513 | S:S:Y514 | 8.83 | 26 | Yes | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 22.98 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
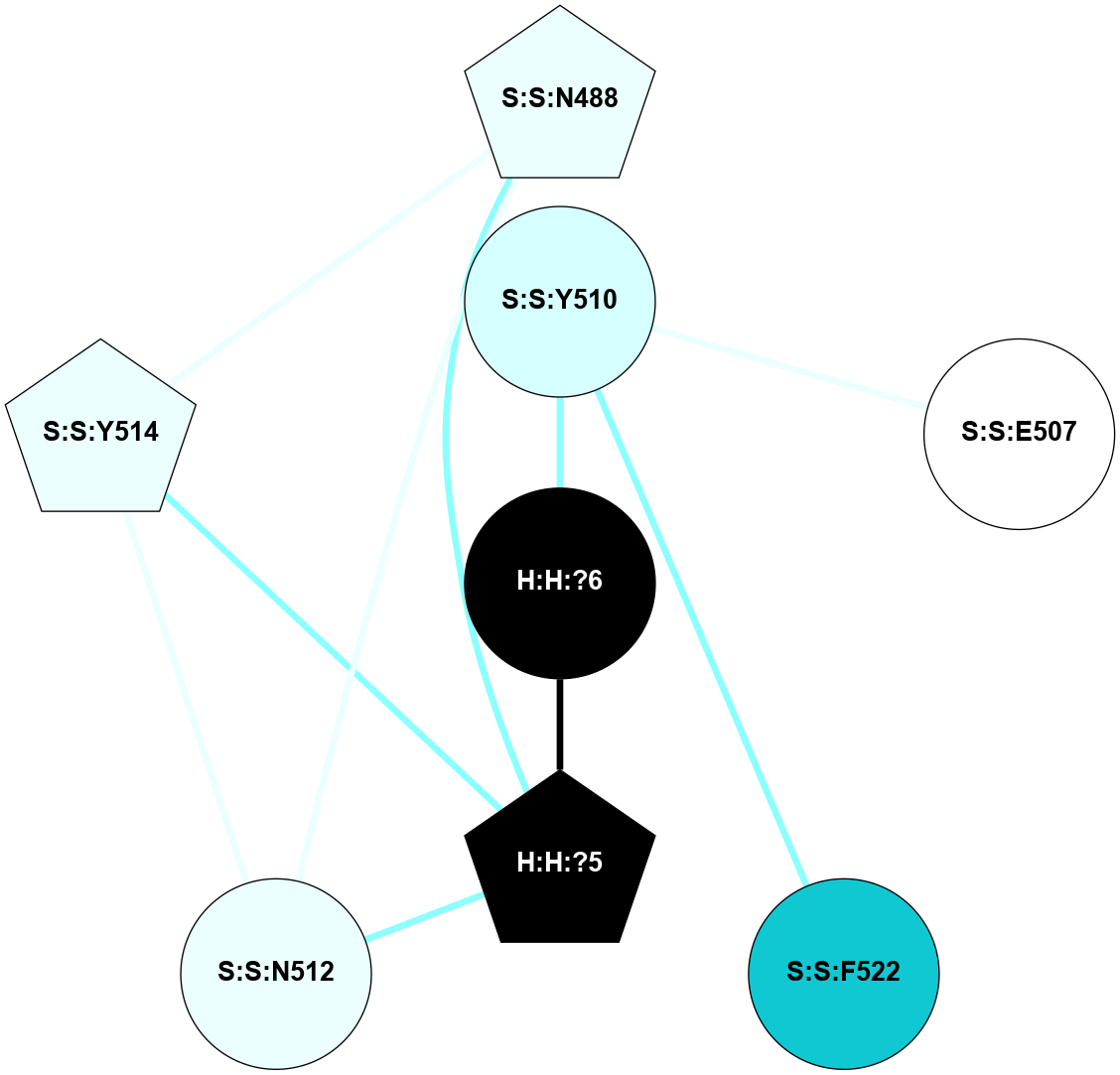
A 2D representation of the interactions of NAG in 7DTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 33.21 | 26 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?5 | S:S:N488 | 25.79 | 26 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?5 | S:S:N512 | 17.19 | 26 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?5 | S:S:Y514 | 15.73 | 26 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?6 | S:S:Y510 | 5.24 | 0 | No | No | 0 | 3 | 0 | 1 | | S:S:N488 | S:S:N512 | 9.54 | 26 | Yes | No | 4 | 4 | 2 | 2 | | S:S:N488 | S:S:Y514 | 6.98 | 26 | Yes | Yes | 4 | 4 | 2 | 2 | | S:S:E507 | S:S:Y510 | 4.49 | 0 | No | No | 5 | 3 | 2 | 1 | | S:S:F522 | S:S:Y510 | 14.44 | 0 | No | No | 1 | 3 | 2 | 1 | | S:S:N512 | S:S:Y514 | 8.14 | 26 | No | Yes | 4 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.23 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
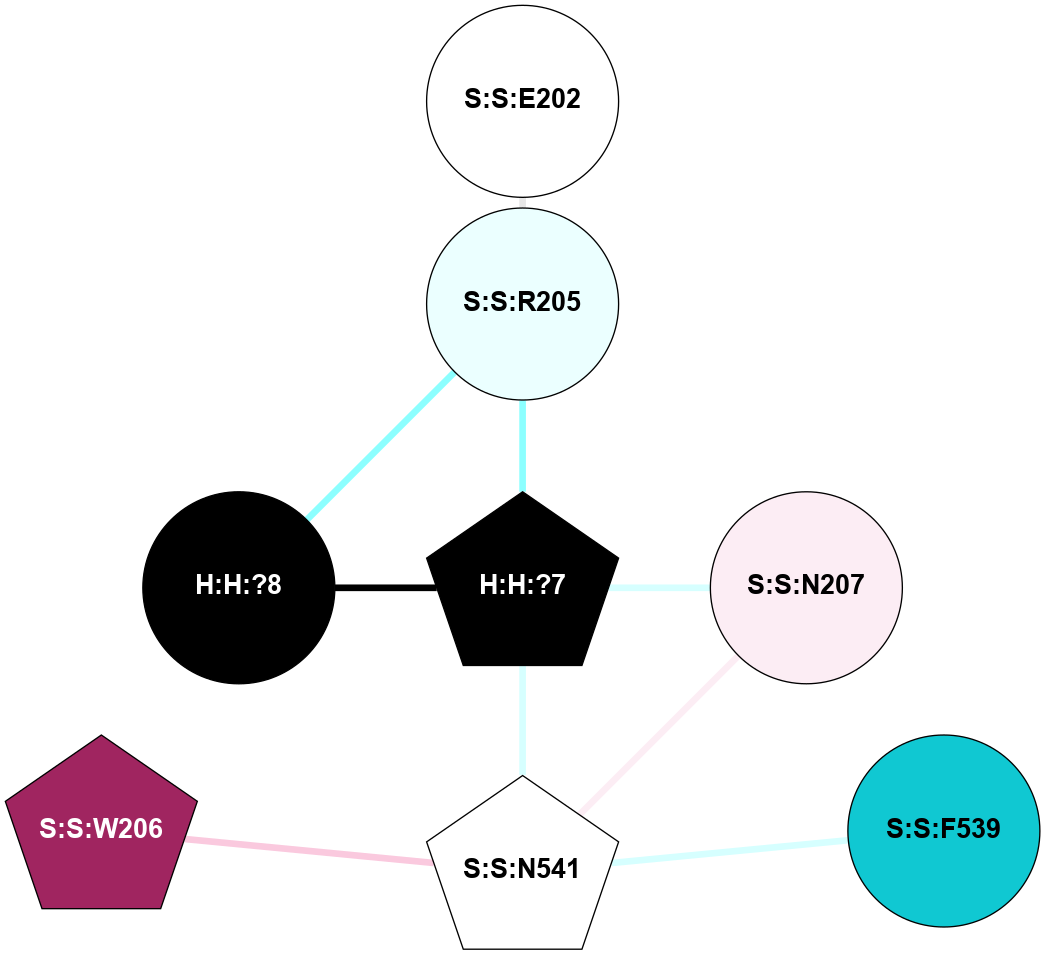
A 2D representation of the interactions of NAG in 7DTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 39.85 | 6 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?7 | S:S:R205 | 20.64 | 6 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?7 | S:S:N207 | 3.68 | 6 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?7 | S:S:N541 | 27.02 | 6 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?7 | S:S:D545 | 7.28 | 6 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?8 | S:S:R205 | 5.43 | 6 | No | No | 0 | 4 | 1 | 1 | | S:S:E202 | S:S:R205 | 10.47 | 0 | No | No | 5 | 4 | 2 | 1 | | S:S:I235 | S:S:N207 | 2.83 | 0 | No | Yes | 7 | 6 | 2 | 1 | | S:S:N207 | S:S:N541 | 4.09 | 6 | Yes | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 12.6 | 6 | No | Yes | 9 | 6 | 2 | 1 | | S:S:F539 | S:S:N541 | 13.29 | 0 | No | No | 1 | 5 | 2 | 1 | | S:S:D545 | S:S:D578 | 5.32 | 0 | No | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.69 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
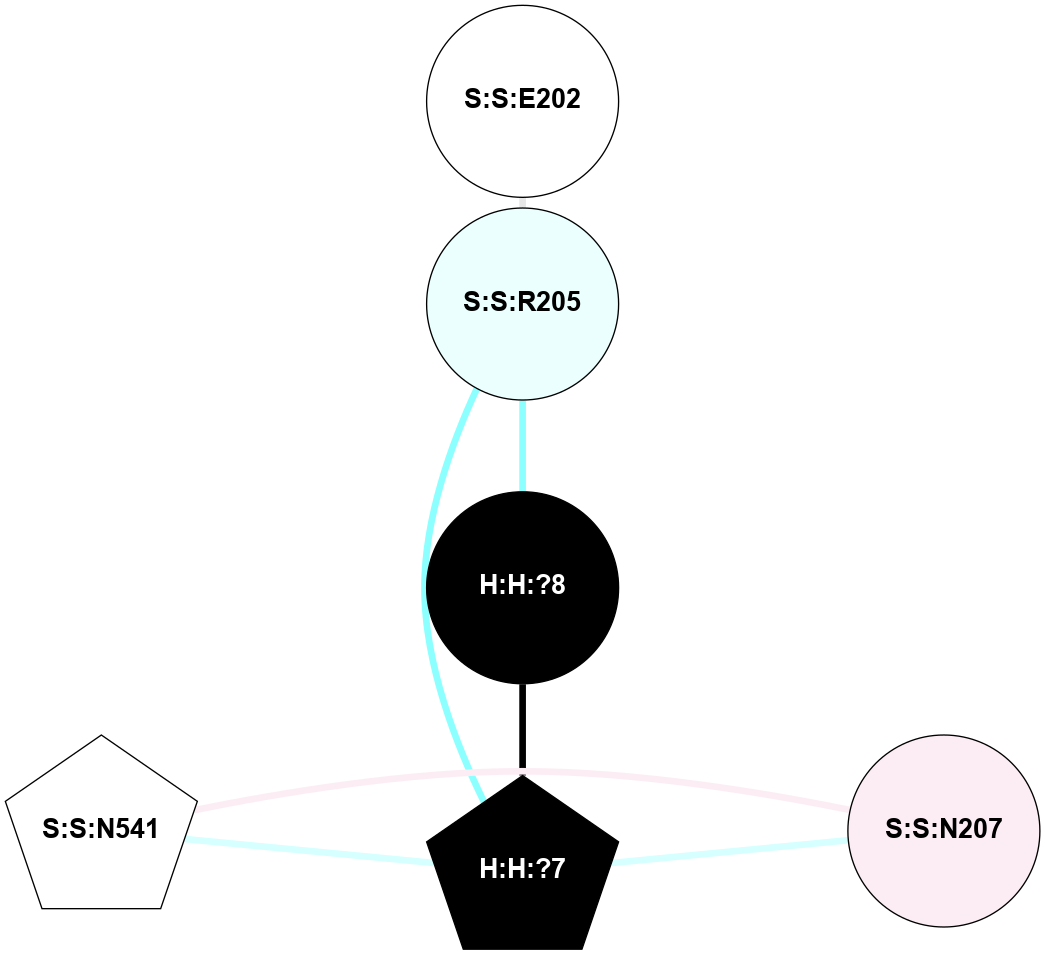
A 2D representation of the interactions of NAG in 7DTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 39.85 | 6 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?7 | S:S:R205 | 20.64 | 6 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?7 | S:S:N207 | 3.68 | 6 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?7 | S:S:N541 | 27.02 | 6 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?7 | S:S:D545 | 7.28 | 6 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?8 | S:S:R205 | 5.43 | 6 | No | No | 0 | 4 | 0 | 1 | | S:S:E202 | S:S:R205 | 10.47 | 0 | No | No | 5 | 4 | 2 | 1 | | S:S:N207 | S:S:N541 | 4.09 | 6 | Yes | No | 6 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 22.64 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
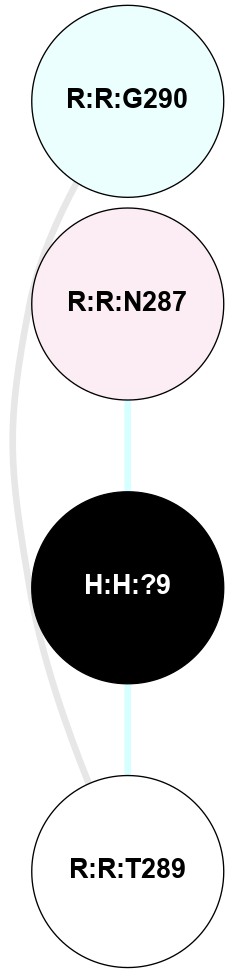
A 2D representation of the interactions of NAG in 7DTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?9 | R:R:N287 | 35.61 | 0 | No | No | 0 | 6 | 0 | 1 | | H:H:?9 | R:R:T289 | 13.18 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:G290 | R:R:T289 | 1.82 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:T289 | R:R:V313 | 1.59 | 0 | No | No | 5 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 24.39 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 7DTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | R:R:N446 | 27.02 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7DTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | R:R:N468 | 36.84 | 36 | No | No | 0 | 6 | 0 | 1 | | H:H:?11 | R:R:Q476 | 3.57 | 36 | No | No | 0 | 3 | 0 | 1 | | R:R:N468 | R:R:Q476 | 9.24 | 36 | No | No | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 4.39 | 36 | No | No | 6 | 3 | 1 | 2 | | R:R:Q476 | R:R:T478 | 4.25 | 36 | No | No | 3 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.21 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 7DTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | S:S:N287 | 35.61 | 0 | No | No | 0 | 6 | 0 | 1 | | H:H:?12 | S:S:T289 | 13.18 | 0 | No | No | 0 | 5 | 0 | 1 | | S:S:G290 | S:S:T289 | 1.82 | 0 | No | No | 4 | 5 | 2 | 1 | | S:S:T289 | S:S:V313 | 1.59 | 0 | No | No | 5 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 24.39 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 7DTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | S:S:N446 | 27.02 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7DTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?14 | S:S:N468 | 36.84 | 37 | No | No | 0 | 6 | 0 | 1 | | H:H:?14 | S:S:Q476 | 3.57 | 37 | No | Yes | 0 | 3 | 0 | 1 | | S:S:N468 | S:S:Q476 | 9.24 | 37 | No | Yes | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 2.92 | 37 | No | No | 6 | 3 | 1 | 2 | | S:S:Q476 | S:S:T478 | 4.25 | 37 | Yes | No | 3 | 3 | 1 | 2 | | S:S:Q476 | S:S:T470 | 1.42 | 37 | Yes | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 20.21 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7E6T | C | Ion | Calcium Sensing | CaS | Homo Sapiens | Ca | Ca; Ca; Cyclomethyltryptophan; PO4 | - | 3 | 2021-09-22 | doi.org/10.7554/eLife.68578 |
|
A 2D representation of the interactions of NAG in 7E6T
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | S:S:K323 | 3.78 | 51 | Yes | No | 0 | 5 | 0 | 1 | | S:S:E475 | S:S:N471 | 2.63 | 0 | No | Yes | 5 | 6 | 1 | 2 | | H:H:?1 | S:S:E475 | 3.55 | 51 | Yes | No | 0 | 5 | 0 | 1 | | S:S:V486 | S:S:Y514 | 5.05 | 0 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?1 | S:S:N488 | 28.24 | 51 | Yes | No | 0 | 4 | 0 | 1 | | S:S:N512 | S:S:Y514 | 3.49 | 51 | No | Yes | 4 | 4 | 1 | 1 | | S:S:A515 | S:S:N512 | 3.13 | 0 | No | No | 4 | 4 | 2 | 1 | | H:H:?1 | S:S:N512 | 11.05 | 51 | Yes | No | 0 | 4 | 0 | 1 | | S:S:V513 | S:S:Y514 | 7.57 | 0 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?1 | S:S:Y514 | 7.34 | 51 | Yes | Yes | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 10.79 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
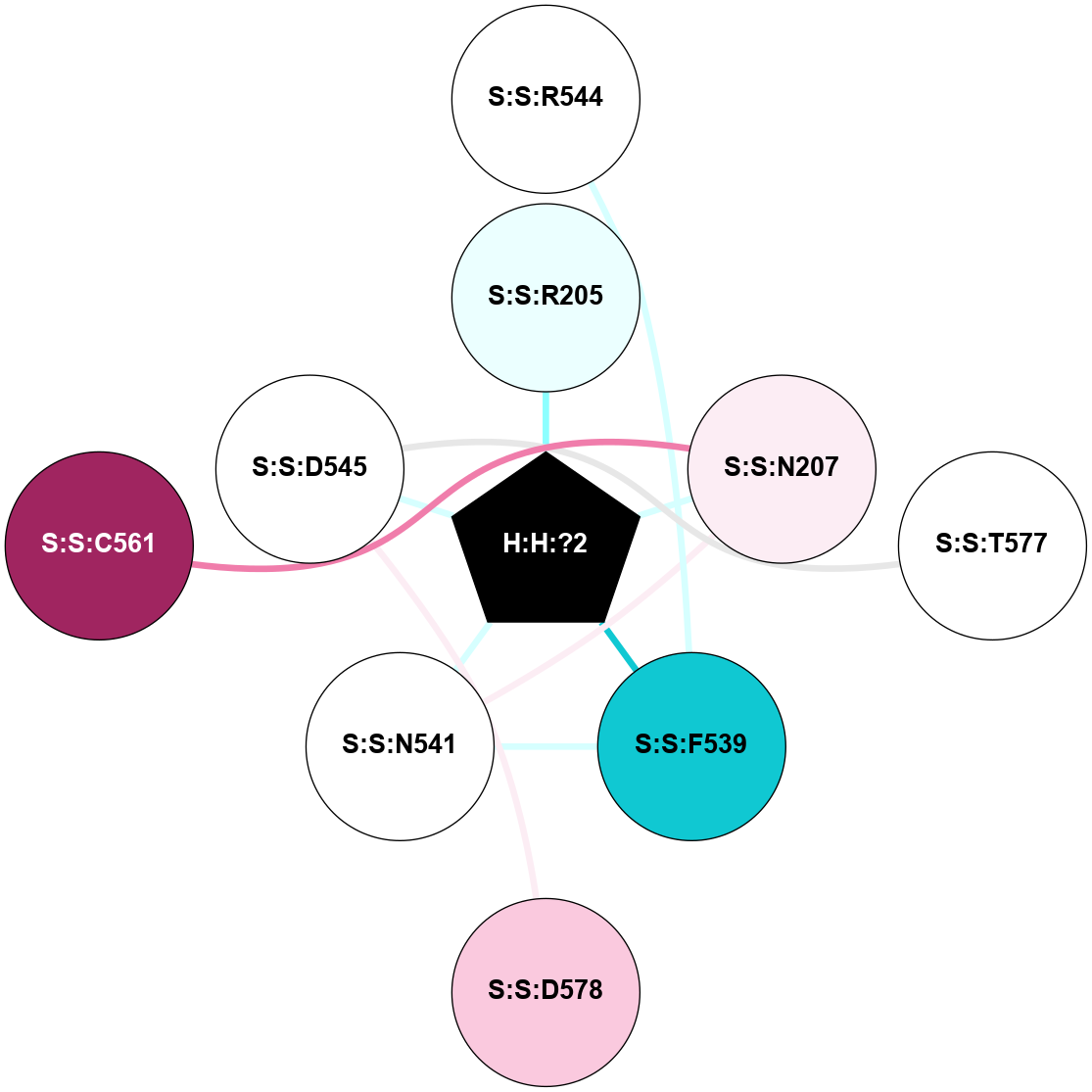
A 2D representation of the interactions of NAG in 7E6T
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | S:S:R205 | 6.52 | 34 | Yes | No | 0 | 4 | 0 | 1 | | S:S:N207 | S:S:N541 | 9.54 | 34 | No | No | 6 | 5 | 1 | 1 | | H:H:?2 | S:S:N207 | 7.37 | 34 | Yes | No | 0 | 6 | 0 | 1 | | S:S:F539 | S:S:N541 | 9.67 | 34 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 11.76 | 34 | No | No | 1 | 5 | 1 | 2 | | H:H:?2 | S:S:F539 | 6.54 | 34 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?2 | S:S:N541 | 22.1 | 34 | Yes | No | 0 | 5 | 0 | 1 | | S:S:D545 | S:S:T577 | 2.89 | 0 | No | No | 5 | 5 | 1 | 2 | | S:S:D545 | S:S:D578 | 2.66 | 0 | No | No | 5 | 7 | 1 | 2 | | H:H:?2 | S:S:D545 | 3.64 | 34 | Yes | No | 0 | 5 | 0 | 1 | | S:S:C561 | S:S:N207 | 1.57 | 0 | No | No | 9 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 9.23 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
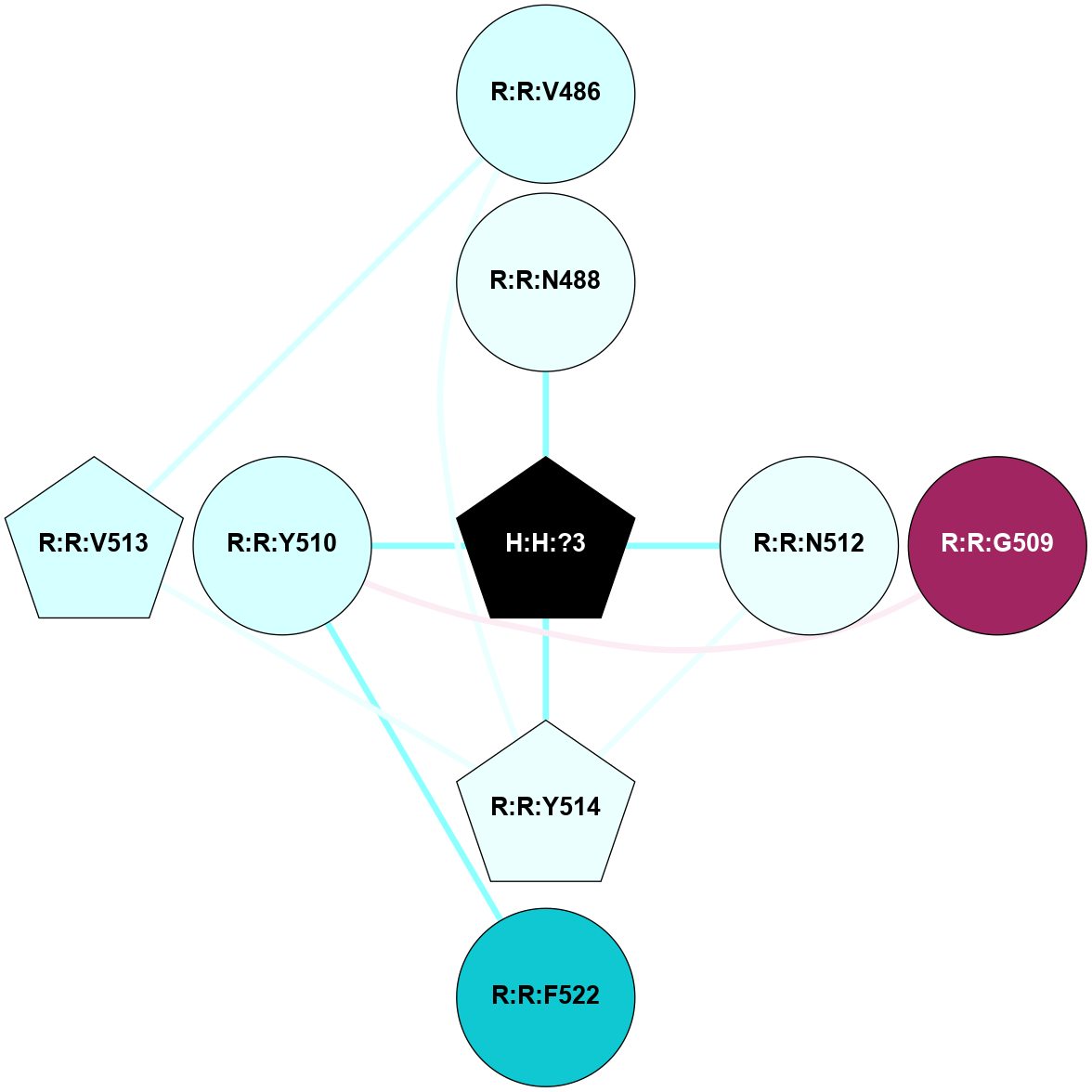
A 2D representation of the interactions of NAG in 7E6T
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:V486 | R:R:V513 | 3.21 | 1 | No | Yes | 3 | 3 | 2 | 2 | | R:R:V486 | R:R:Y514 | 3.79 | 1 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?3 | R:R:N488 | 29.47 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:G509 | R:R:Y510 | 2.9 | 0 | No | No | 9 | 3 | 2 | 1 | | R:R:F522 | R:R:Y510 | 12.38 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:N512 | R:R:Y514 | 3.49 | 1 | No | Yes | 4 | 4 | 1 | 1 | | H:H:?3 | R:R:N512 | 14.74 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:V513 | R:R:Y514 | 5.05 | 1 | Yes | Yes | 3 | 4 | 2 | 1 | | H:H:?3 | R:R:Y514 | 11.53 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?3 | R:R:Y510 | 1.05 | 1 | Yes | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.20 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
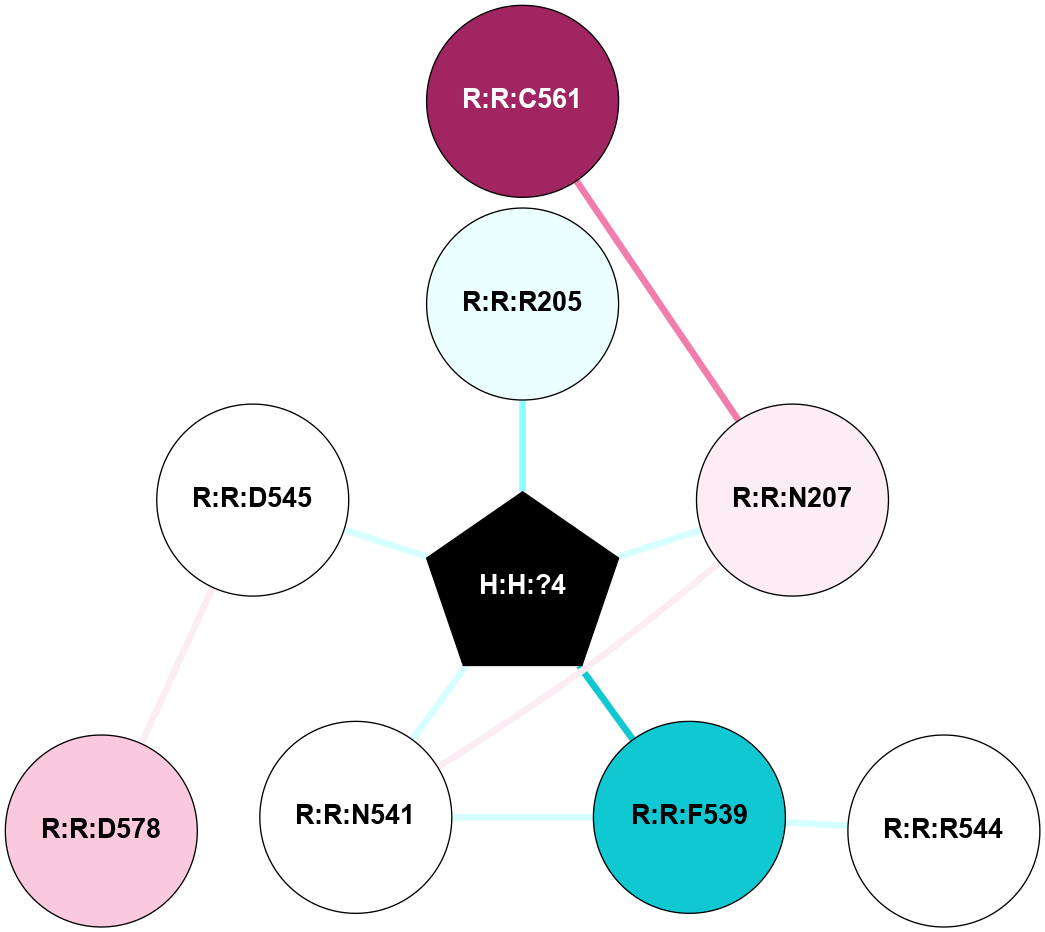
A 2D representation of the interactions of NAG in 7E6T
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:R205 | 3.26 | 30 | Yes | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 13.62 | 30 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 6.3 | 0 | No | No | 9 | 6 | 2 | 1 | | H:H:?4 | R:R:N207 | 11.05 | 30 | Yes | No | 0 | 6 | 0 | 1 | | R:R:F539 | R:R:N541 | 8.46 | 30 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 13.9 | 30 | No | No | 1 | 5 | 1 | 2 | | H:H:?4 | R:R:F539 | 5.45 | 30 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?4 | R:R:N541 | 23.33 | 30 | Yes | No | 0 | 5 | 0 | 1 | | R:R:D545 | R:R:D578 | 3.99 | 0 | No | No | 5 | 7 | 1 | 2 | | H:H:?4 | R:R:D545 | 16.99 | 30 | Yes | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.02 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7EVY | A | Lipid | Lysophospholipid | S1P1 | Homo Sapiens | Siponimod | - | Gi1/Beta1/Gamma2 | 2.98 | 2021-09-29 | doi.org/10.1038/s41422-021-00566-x |
|

A 2D representation of the interactions of NAG in 7EVY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N30 | 20.88 | 0 | No | No | 0 | 9 | 0 | 1 | | R:R:L35 | R:R:N30 | 2.75 | 0 | No | No | 6 | 9 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.88 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7F6G | A | Peptide | Angiotensin | AT1 | Homo Sapiens | SAR1-AngII | - | Gq/Beta1/Gamma2 | 2.9 | 2023-03-29 | doi.org/10.15252/embj.2022112940 |
|
A 2D representation of the interactions of NAG in 7F6G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F171 | R:R:N176 | 3.62 | 0 | No | No | 6 | 5 | 2 | 1 | | H:H:?401 | R:R:N176 | 30.7 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 30.70 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7FD8 | C | Aminoacid | Metabotropic Glutamate | mGlu5; mGlu5 | Homo Sapiens | Quisqualate | - | - | 3.8 | 2021-09-08 | doi.org/10.1016/j.celrep.2021.109648 |
|
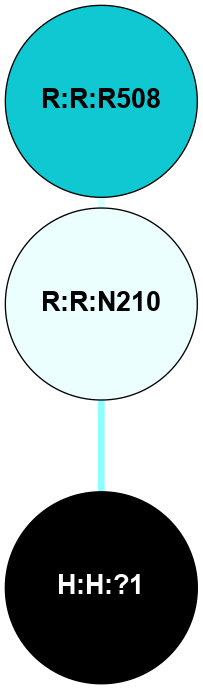
A 2D representation of the interactions of NAG in 7FD8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N210 | R:R:R508 | 6.03 | 0 | No | No | 4 | 1 | 1 | 2 | | H:H:?1 | R:R:N210 | 30.7 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 30.70 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
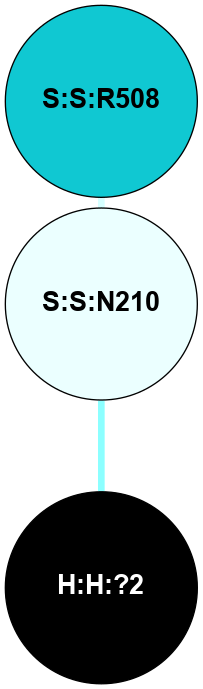
A 2D representation of the interactions of NAG in 7FD8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:N210 | S:S:R508 | 6.03 | 0 | No | No | 4 | 1 | 1 | 2 | | H:H:?2 | S:S:N210 | 30.7 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 30.70 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7FD9 | C | Aminoacid | Metabotropic Glutamate | mGlu5; mGlu5 | Homo Sapiens | LY341495 | - | - | 4 | 2021-09-08 | doi.org/10.1016/j.celrep.2021.109648 |
|
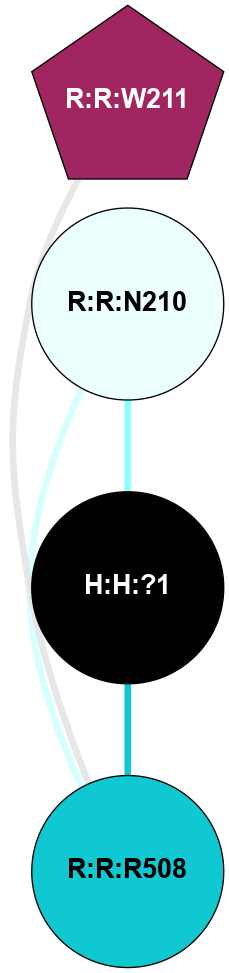
A 2D representation of the interactions of NAG in 7FD9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N210 | 24.56 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N505 | R:R:W500 | 10.17 | 0 | No | Yes | 4 | 7 | 1 | 2 | | H:H:?1 | R:R:N505 | 1.23 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.89 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
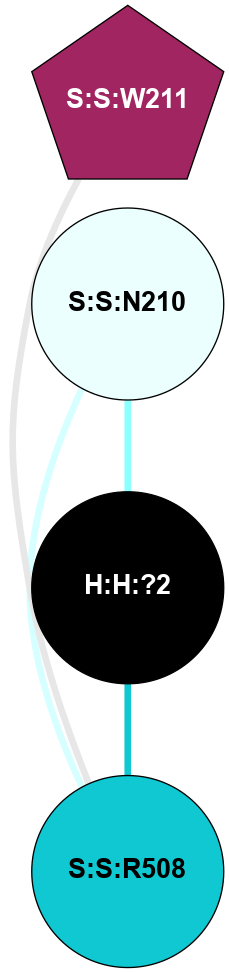
A 2D representation of the interactions of NAG in 7FD9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | S:S:N210 | 24.56 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?2 | S:S:R508 | 8.69 | 0 | No | No | 0 | 1 | 0 | 1 | | S:S:R508 | S:S:V510 | 1.31 | 0 | No | No | 1 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.62 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7FIG | A | Protein | Glycoprotein Hormone | LH | Homo Sapiens | hCG | - | chim(NtGi1L-Gs)/Beta1/Gamma2 | 3.9 | 2021-09-29 | doi.org/10.1038/s41586-021-03924-2 |
|
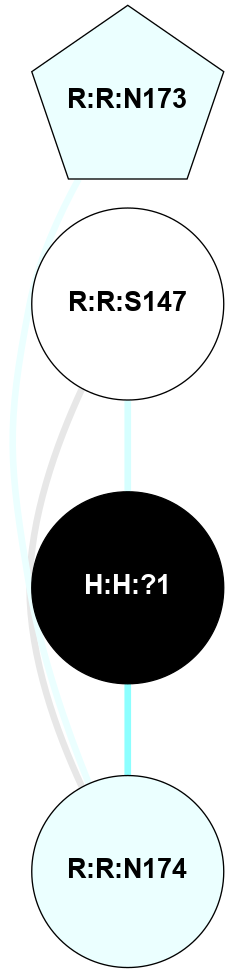
A 2D representation of the interactions of NAG in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:S147 | 12.13 | 25 | No | No | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:N174 | 29.57 | 25 | No | No | 0 | 4 | 0 | 1 | | R:R:N174 | R:R:S147 | 5.96 | 25 | No | No | 4 | 5 | 1 | 1 | | R:R:N173 | R:R:N174 | 1.36 | 0 | Yes | No | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.85 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
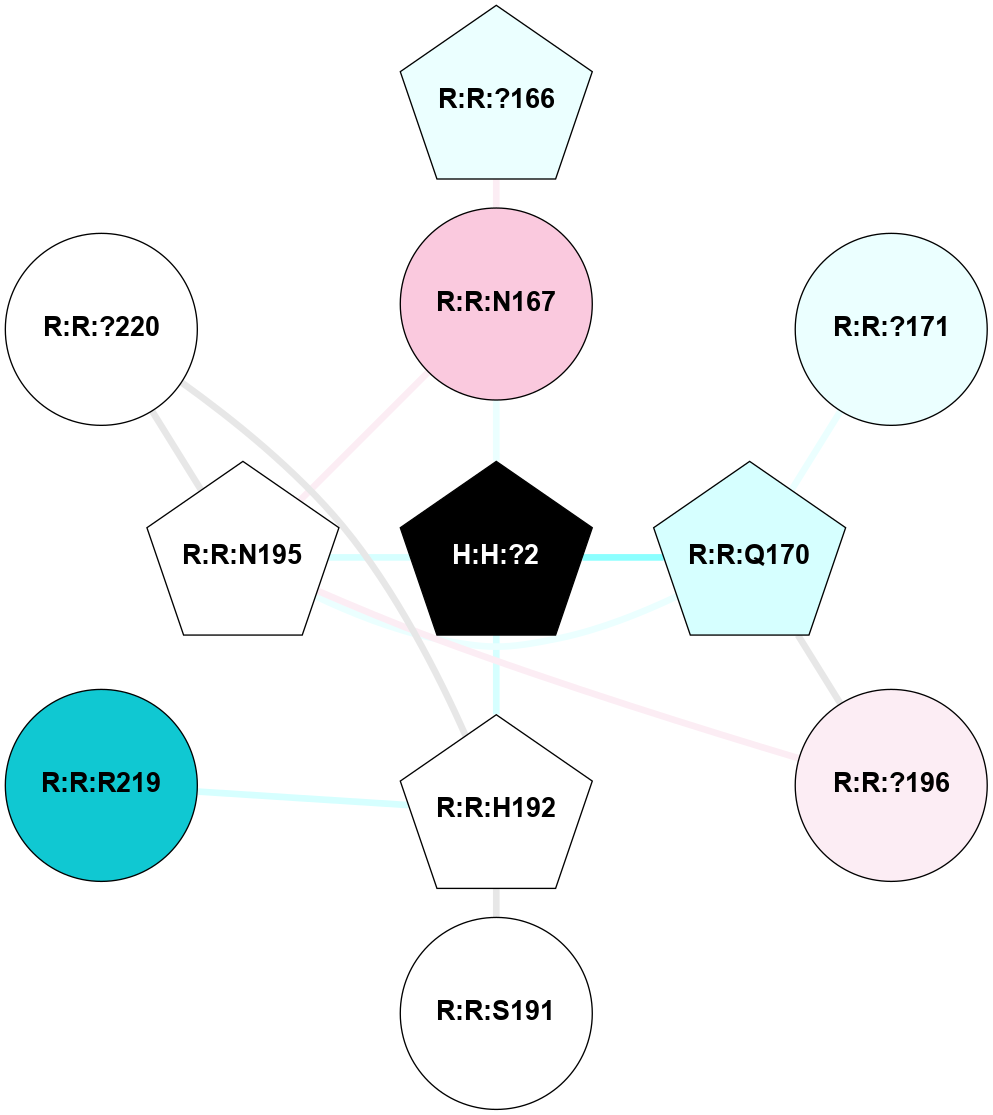
A 2D representation of the interactions of NAG in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:N167 | 19.71 | 15 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?2 | R:R:Q170 | 16.72 | 15 | Yes | Yes | 0 | 3 | 0 | 1 | | H:H:?2 | R:R:H192 | 8.08 | 15 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:N195 | 24.64 | 15 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:?166 | R:R:N167 | 9.58 | 0 | Yes | No | 4 | 7 | 2 | 1 | | R:R:N167 | R:R:N195 | 13.62 | 15 | No | Yes | 7 | 5 | 1 | 1 | | R:R:?171 | R:R:Q170 | 6.96 | 0 | No | Yes | 4 | 3 | 2 | 1 | | R:R:N195 | R:R:Q170 | 13.2 | 15 | Yes | Yes | 5 | 3 | 1 | 1 | | R:R:?196 | R:R:Q170 | 8.13 | 15 | No | Yes | 6 | 3 | 2 | 1 | | R:R:H192 | R:R:S191 | 4.18 | 0 | Yes | No | 5 | 5 | 1 | 2 | | R:R:H192 | R:R:R219 | 9.03 | 0 | Yes | No | 5 | 1 | 1 | 2 | | R:R:?220 | R:R:H192 | 4.48 | 0 | No | Yes | 5 | 5 | 2 | 1 | | R:R:?196 | R:R:N195 | 8.38 | 15 | No | Yes | 6 | 5 | 2 | 1 | | R:R:?220 | R:R:N195 | 10.78 | 0 | No | Yes | 5 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 17.29 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
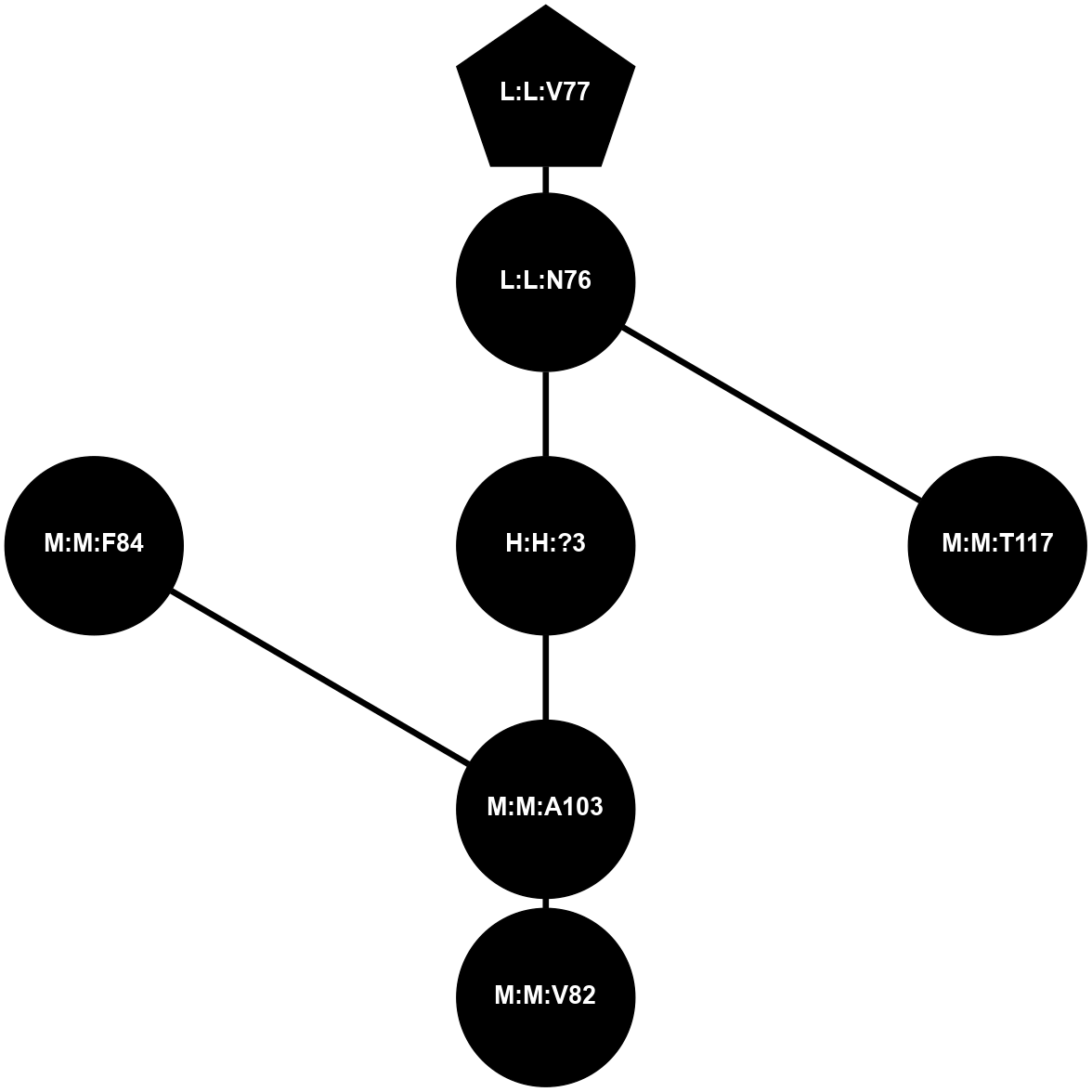
A 2D representation of the interactions of NAG in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | L:L:N76 | 25.87 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | N:N:A103 | 4.24 | 0 | No | No | 0 | 0 | 0 | 1 | | L:L:N76 | L:L:V77 | 4.43 | 0 | No | Yes | 0 | 0 | 1 | 2 | | L:L:N76 | N:N:T117 | 5.85 | 0 | No | No | 0 | 0 | 1 | 2 | | N:N:A103 | N:N:V82 | 5.09 | 0 | No | No | 0 | 0 | 1 | 2 | | N:N:A103 | N:N:F84 | 4.16 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.05 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
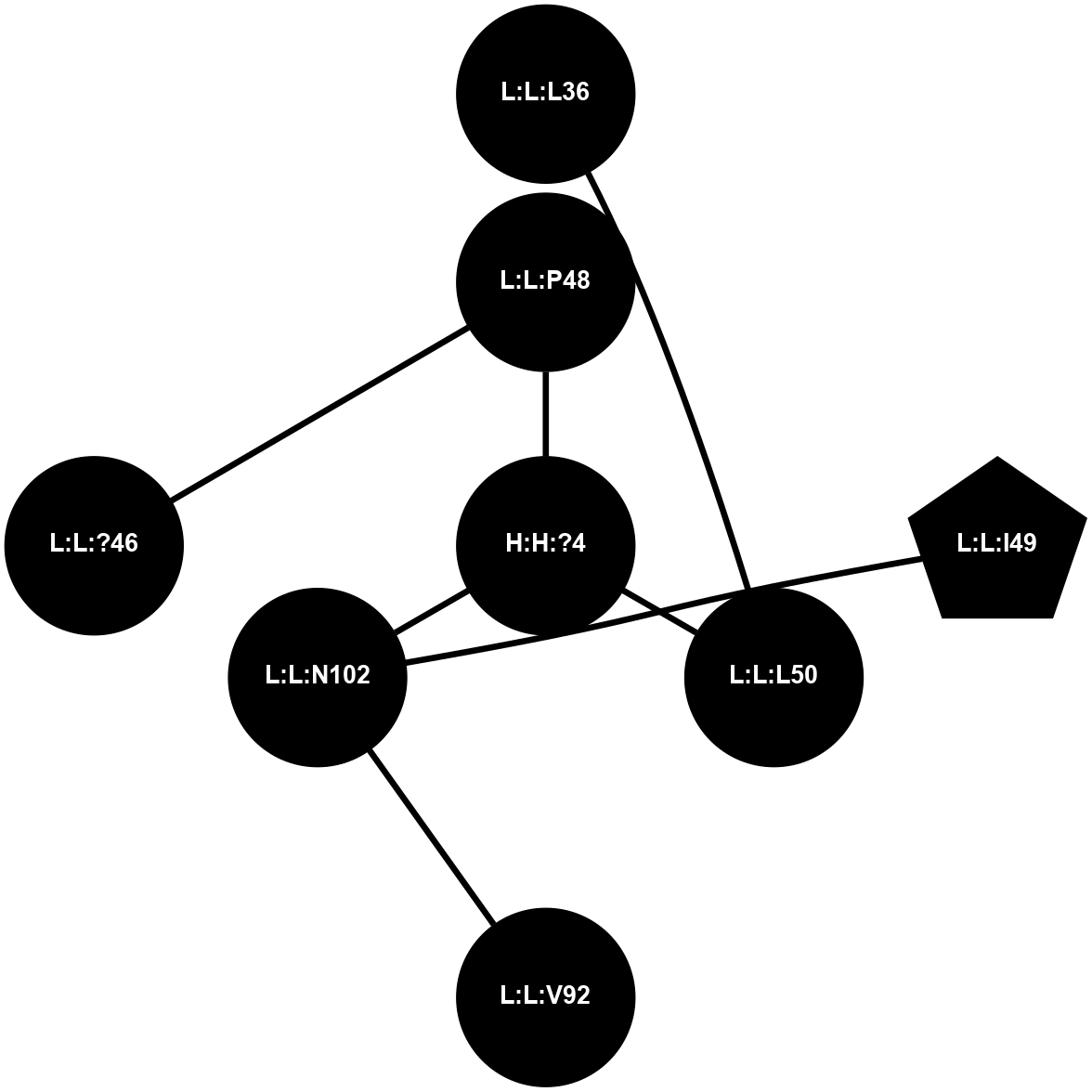
A 2D representation of the interactions of NAG in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | L:L:P48 | 7.37 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?4 | L:L:L50 | 7.45 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?4 | L:L:N102 | 23.41 | 0 | No | No | 0 | 0 | 0 | 1 | | L:L:L36 | L:L:L50 | 11.07 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:I49 | L:L:N102 | 4.25 | 0 | Yes | No | 0 | 0 | 2 | 1 | | L:L:N102 | L:L:V92 | 2.96 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:?46 | L:L:P48 | 2.86 | 0 | No | No | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.74 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 7FIG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | N:N:N33 | 27.11 | 0 | No | No | 0 | 0 | 0 | 1 | | N:N:N50 | N:N:T35 | 4.39 | 0 | No | No | 0 | 0 | 1 | 2 | | N:N:N33 | N:N:T52 | 1.46 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?5 | N:N:N50 | 1.23 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.17 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7M3E | C | Ion | Calcium Sensing | CaS | Homo Sapiens | - | Ca; Ca; Tryptophan; NPS-2143 | - | 3.2 | 2021-06-30 | doi.org/10.1038/s41586-021-03691-0 |
|
A 2D representation of the interactions of NAG in 7M3E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 30.99 | 5 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:R205 | 8.69 | 5 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:N207 | 9.82 | 5 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:F539 | 6.54 | 5 | Yes | Yes | 0 | 1 | 0 | 1 | | H:H:?1 | R:R:N541 | 28.24 | 5 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:D545 | 8.5 | 5 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:R205 | 16.3 | 5 | No | No | 0 | 4 | 1 | 1 | | R:R:N207 | R:R:N541 | 9.54 | 5 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 7.87 | 5 | No | No | 9 | 6 | 2 | 1 | | R:R:F539 | R:R:N541 | 6.04 | 5 | Yes | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 19.24 | 5 | Yes | No | 1 | 5 | 1 | 2 | | R:R:D545 | R:R:D578 | 6.65 | 0 | No | No | 5 | 7 | 1 | 2 | | R:R:F539 | R:R:K265 | 1.24 | 5 | Yes | No | 1 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.46 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 7M3E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 30.99 | 5 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:R205 | 8.69 | 5 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?1 | R:R:N207 | 9.82 | 5 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:F539 | 6.54 | 5 | Yes | Yes | 0 | 1 | 1 | 2 | | H:H:?1 | R:R:N541 | 28.24 | 5 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?1 | R:R:D545 | 8.5 | 5 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?2 | R:R:R205 | 16.3 | 5 | No | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 9.54 | 5 | No | No | 6 | 5 | 2 | 2 | | R:R:F539 | R:R:N541 | 6.04 | 5 | Yes | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 23.64 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 7M3E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:V258 | 2.66 | 0 | No | Yes | 0 | 4 | 0 | 1 | | H:H:?3 | R:R:N261 | 24.56 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:F239 | R:R:V258 | 3.93 | 5 | Yes | Yes | 6 | 4 | 2 | 1 | | R:R:E241 | R:R:H254 | 3.69 | 5 | No | No | 5 | 3 | 2 | 2 | | R:R:E241 | R:R:V258 | 11.41 | 5 | No | Yes | 5 | 4 | 2 | 1 | | R:R:H254 | R:R:V258 | 5.54 | 5 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?3 | R:R:E257 | 2.37 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 9.86 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
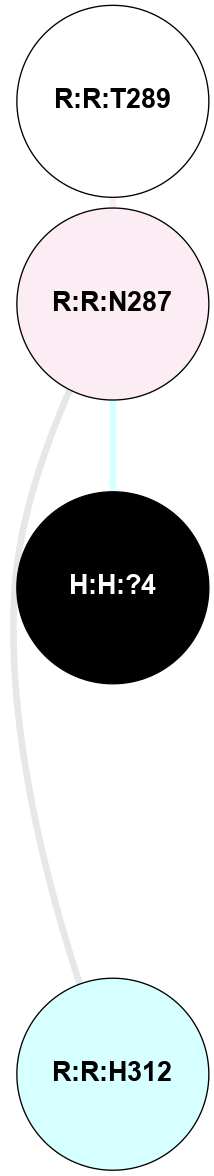
A 2D representation of the interactions of NAG in 7M3E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:N287 | 27.02 | 0 | No | No | 0 | 6 | 0 | 1 | | R:R:N287 | R:R:T289 | 2.92 | 0 | No | No | 6 | 5 | 1 | 2 | | R:R:H312 | R:R:N287 | 5.1 | 41 | No | No | 3 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
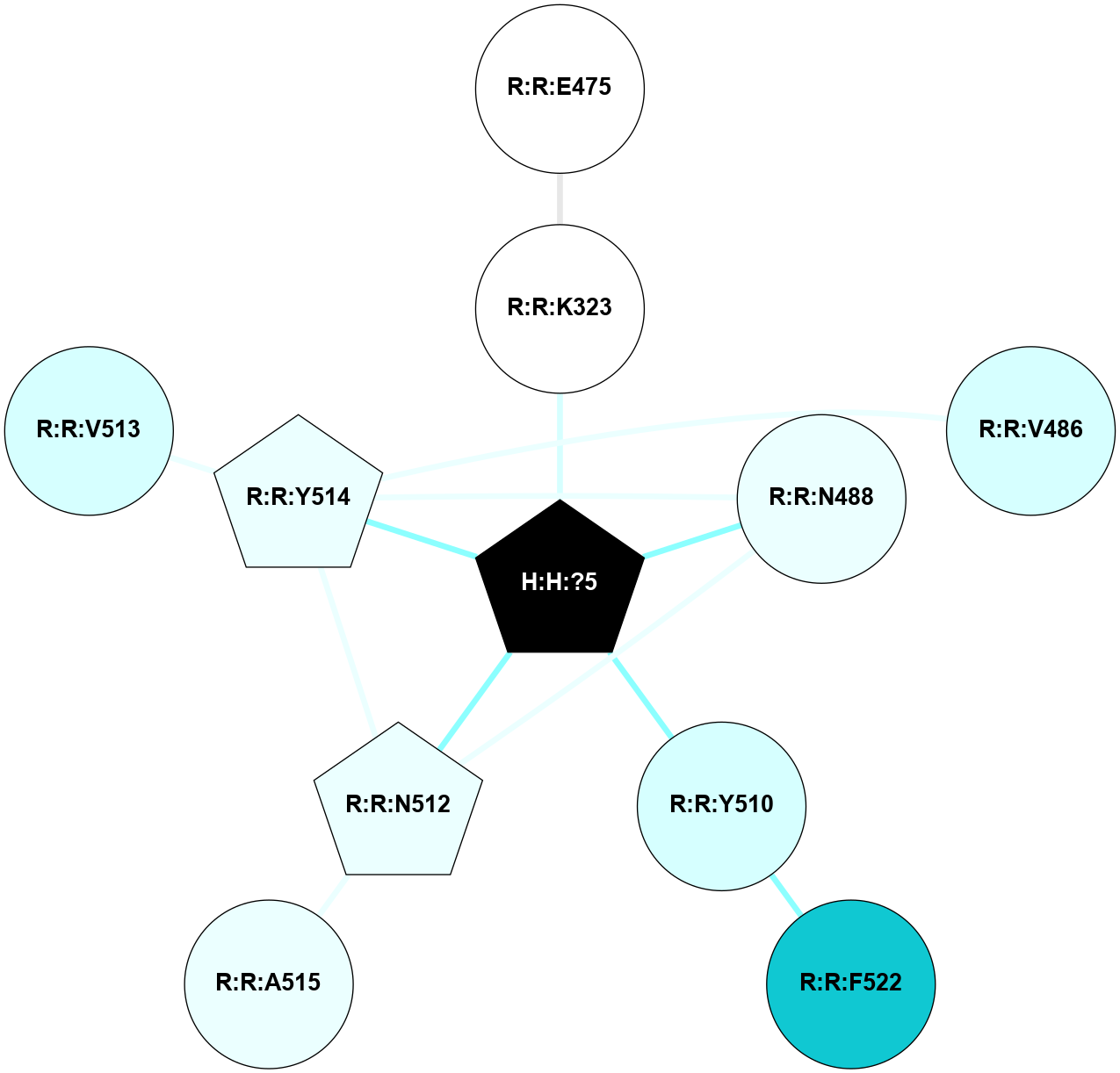
A 2D representation of the interactions of NAG in 7M3E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | R:R:K323 | 3.78 | 27 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?5 | R:R:N488 | 23.33 | 27 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?5 | R:R:Y510 | 8.39 | 27 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?5 | R:R:N512 | 15.96 | 27 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?5 | R:R:Y514 | 9.44 | 27 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:E475 | R:R:K323 | 5.4 | 0 | No | No | 5 | 5 | 2 | 1 | | R:R:V486 | R:R:Y514 | 5.05 | 0 | No | Yes | 3 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 6.81 | 27 | No | Yes | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:Y514 | 4.65 | 27 | No | Yes | 4 | 4 | 1 | 1 | | R:R:F522 | R:R:Y510 | 18.57 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:N512 | R:R:Y514 | 3.49 | 27 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | | R:R:V513 | R:R:Y514 | 6.31 | 0 | No | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 12.18 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7M3E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | R:R:N468 | 29.47 | 36 | No | No | 0 | 6 | 0 | 1 | | H:H:?6 | R:R:Q476 | 10.71 | 36 | No | No | 0 | 3 | 0 | 1 | | R:R:N468 | R:R:Q476 | 6.6 | 36 | No | No | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 7.31 | 36 | No | No | 6 | 3 | 1 | 2 | | R:R:Q476 | R:R:T470 | 5.67 | 36 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.09 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
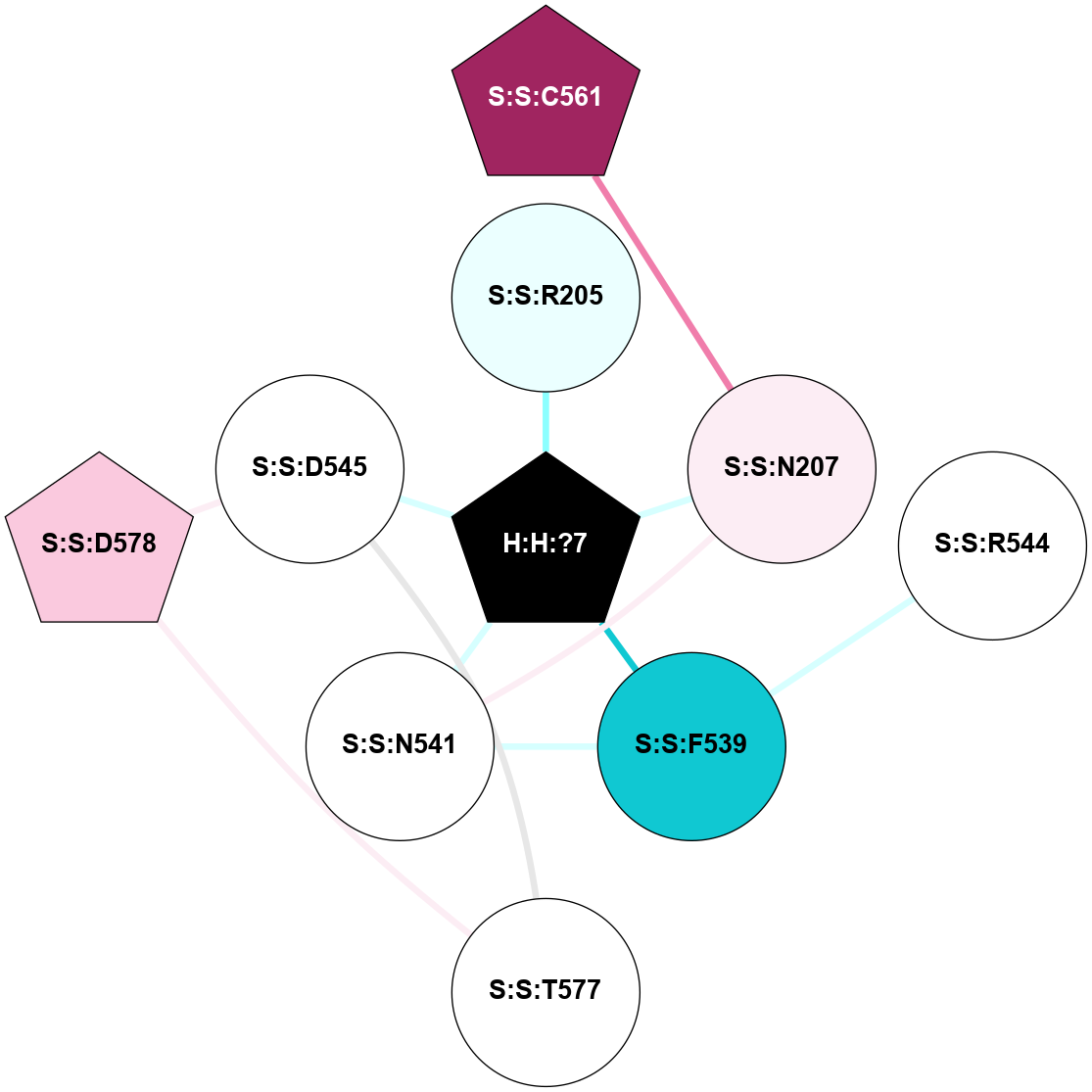
A 2D representation of the interactions of NAG in 7M3E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | S:S:R205 | 16.3 | 3 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?7 | S:S:N207 | 14.74 | 3 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?7 | S:S:F539 | 6.54 | 3 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?7 | S:S:N541 | 25.79 | 3 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?7 | S:S:D545 | 10.92 | 3 | Yes | No | 0 | 5 | 0 | 1 | | S:S:N207 | S:S:N541 | 8.17 | 3 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 4.72 | 3 | Yes | No | 9 | 6 | 2 | 1 | | S:S:F539 | S:S:N541 | 6.04 | 3 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 21.38 | 3 | No | No | 1 | 5 | 1 | 2 | | S:S:D545 | S:S:T577 | 4.34 | 3 | No | No | 5 | 5 | 1 | 2 | | S:S:D545 | S:S:D578 | 14.64 | 3 | No | Yes | 5 | 7 | 1 | 2 | | S:S:D578 | S:S:T577 | 4.34 | 3 | Yes | No | 7 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.86 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
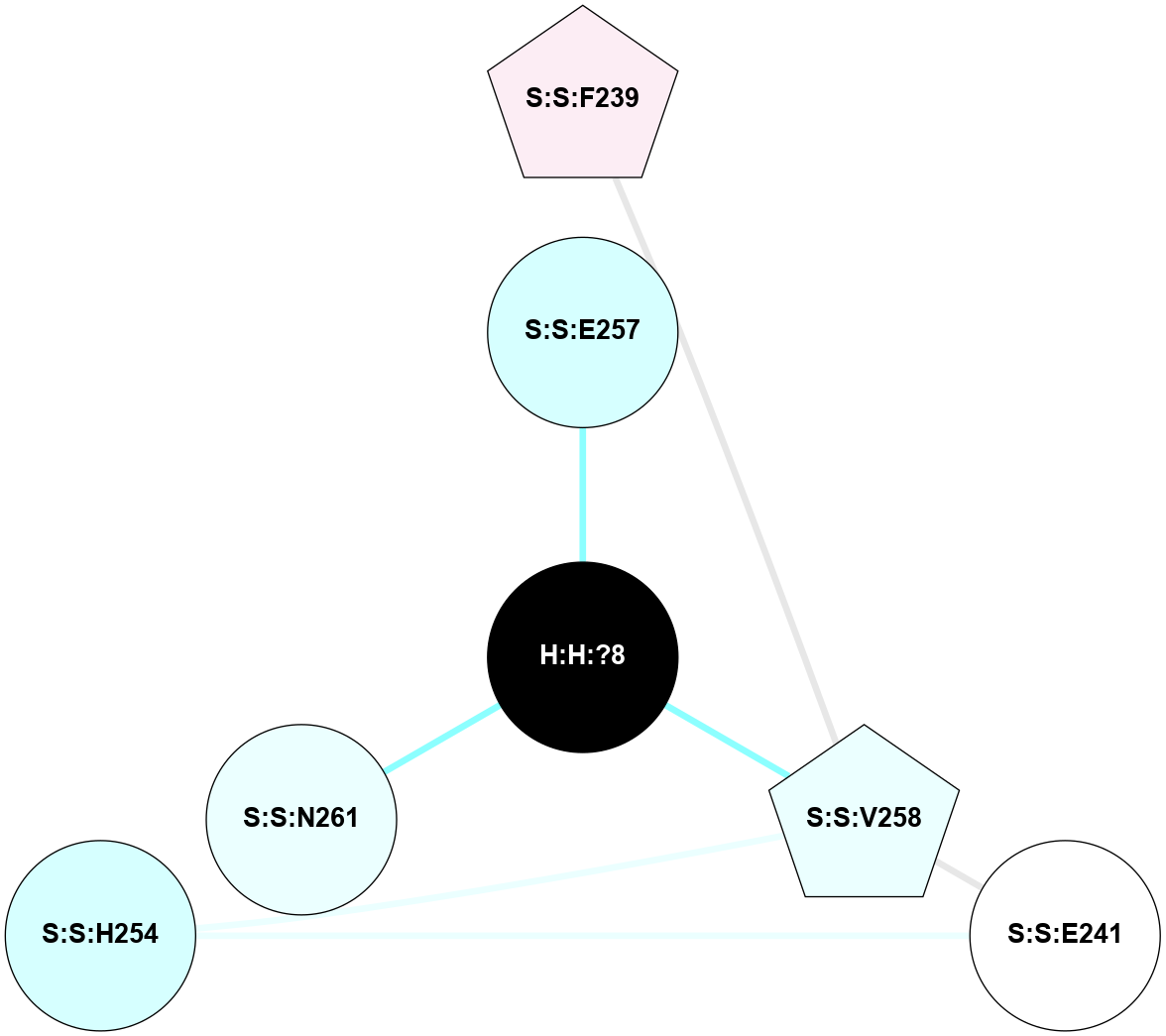
A 2D representation of the interactions of NAG in 7M3E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | S:S:E257 | 3.55 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?8 | S:S:V258 | 4 | 0 | No | Yes | 0 | 4 | 0 | 1 | | H:H:?8 | S:S:N261 | 25.79 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:F239 | S:S:V258 | 3.93 | 3 | Yes | Yes | 6 | 4 | 2 | 1 | | S:S:E241 | S:S:H254 | 3.69 | 3 | No | No | 5 | 3 | 2 | 2 | | S:S:E241 | S:S:V258 | 8.56 | 3 | No | Yes | 5 | 4 | 2 | 1 | | S:S:H254 | S:S:V258 | 4.15 | 3 | No | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 11.11 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7M3E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?9 | S:S:N287 | 25.79 | 4 | No | No | 0 | 6 | 0 | 1 | | H:H:?9 | S:S:T289 | 2.64 | 4 | No | No | 0 | 5 | 0 | 1 | | S:S:N287 | S:S:T289 | 2.92 | 4 | No | No | 6 | 5 | 1 | 1 | | S:S:H312 | S:S:N287 | 6.38 | 4 | No | No | 3 | 6 | 2 | 1 | | S:S:G290 | S:S:T289 | 1.82 | 0 | No | No | 4 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.21 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
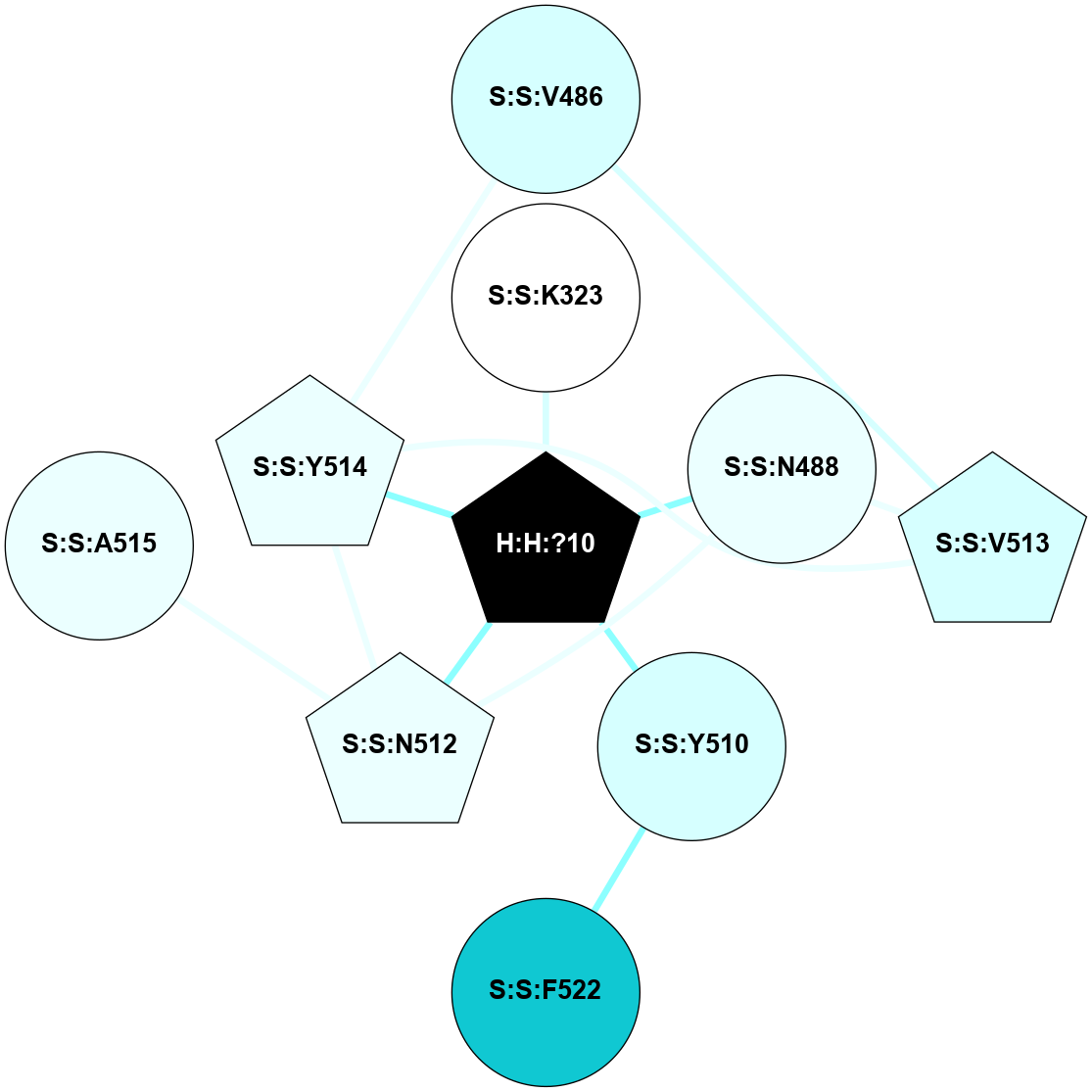
A 2D representation of the interactions of NAG in 7M3E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | S:S:K323 | 5.04 | 14 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?10 | S:S:N488 | 23.33 | 14 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?10 | S:S:Y510 | 7.34 | 14 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?10 | S:S:N512 | 14.74 | 14 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?10 | S:S:Y514 | 10.48 | 14 | Yes | Yes | 0 | 4 | 0 | 1 | | S:S:V486 | S:S:V513 | 3.21 | 14 | No | Yes | 3 | 3 | 2 | 2 | | S:S:V486 | S:S:Y514 | 3.79 | 14 | No | Yes | 3 | 4 | 2 | 1 | | S:S:N488 | S:S:N512 | 5.45 | 14 | No | Yes | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:V513 | 2.96 | 14 | No | Yes | 4 | 3 | 1 | 2 | | S:S:F522 | S:S:Y510 | 16.5 | 0 | No | No | 1 | 3 | 2 | 1 | | S:S:N512 | S:S:Y514 | 3.49 | 14 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:A515 | S:S:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | | S:S:V513 | S:S:Y514 | 5.05 | 14 | Yes | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 12.19 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
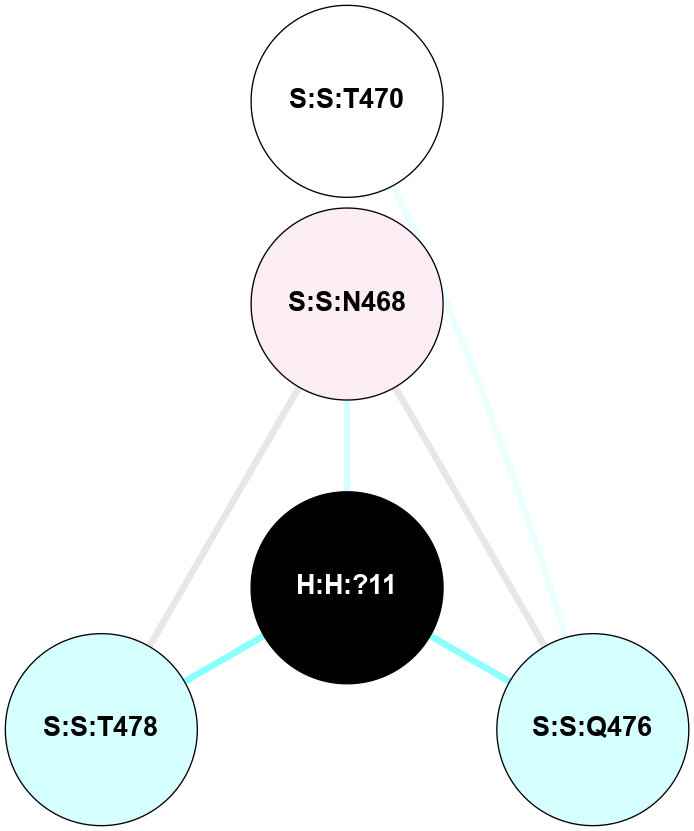
A 2D representation of the interactions of NAG in 7M3E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | S:S:N468 | 23.33 | 28 | No | No | 0 | 6 | 0 | 1 | | H:H:?11 | S:S:Q476 | 3.57 | 28 | No | No | 0 | 3 | 0 | 1 | | H:H:?11 | S:S:T478 | 6.59 | 28 | No | No | 0 | 3 | 0 | 1 | | S:S:N468 | S:S:Q476 | 6.6 | 28 | No | No | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 5.85 | 28 | No | No | 6 | 3 | 1 | 1 | | S:S:Q476 | S:S:T470 | 7.09 | 28 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 11.16 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7M3F | C | Ion | Calcium Sensing | CaS | Homo Sapiens | - | Ca; Ca; Tryptophan; Cinacalcet; PO4 | - | 2.8 | 2021-06-30 | doi.org/10.1038/s41586-021-03691-0 |
|
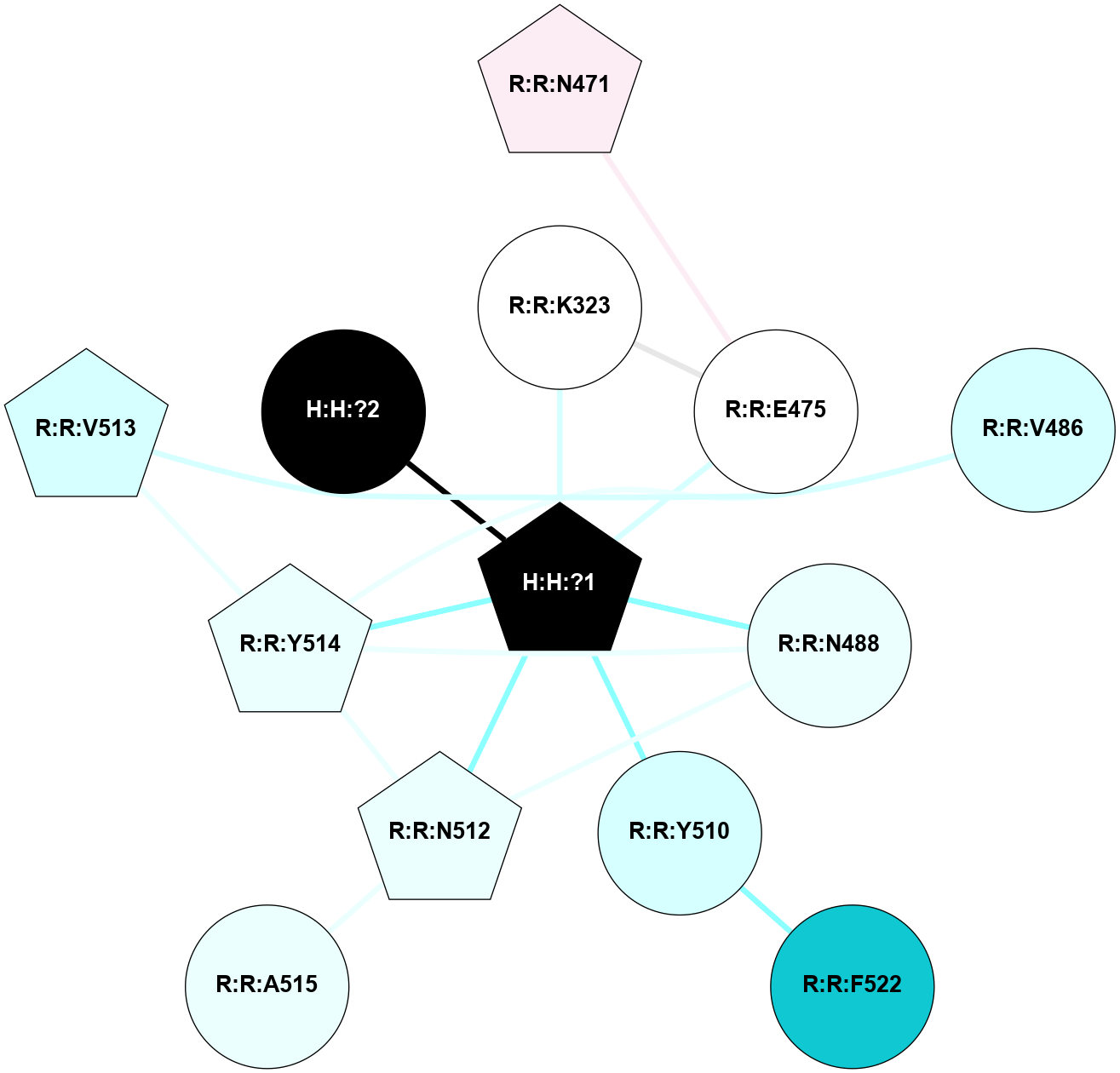
A 2D representation of the interactions of NAG in 7M3F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 34.31 | 2 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:K323 | 5.04 | 2 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:E475 | 3.55 | 2 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:N488 | 27.02 | 2 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:Y510 | 7.34 | 2 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:N512 | 17.19 | 2 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:Y514 | 8.39 | 2 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:E475 | R:R:K323 | 4.05 | 2 | No | No | 5 | 5 | 1 | 1 | | R:R:E475 | R:R:N471 | 3.94 | 2 | No | Yes | 5 | 6 | 1 | 2 | | R:R:V486 | R:R:V513 | 3.21 | 2 | No | Yes | 3 | 3 | 2 | 2 | | R:R:V486 | R:R:Y514 | 3.79 | 2 | No | Yes | 3 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 6.81 | 2 | No | Yes | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:Y514 | 4.65 | 2 | No | Yes | 4 | 4 | 1 | 1 | | R:R:F522 | R:R:Y510 | 16.5 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:N512 | R:R:Y514 | 3.49 | 2 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | | R:R:V513 | R:R:Y514 | 5.05 | 2 | Yes | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 14.69 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
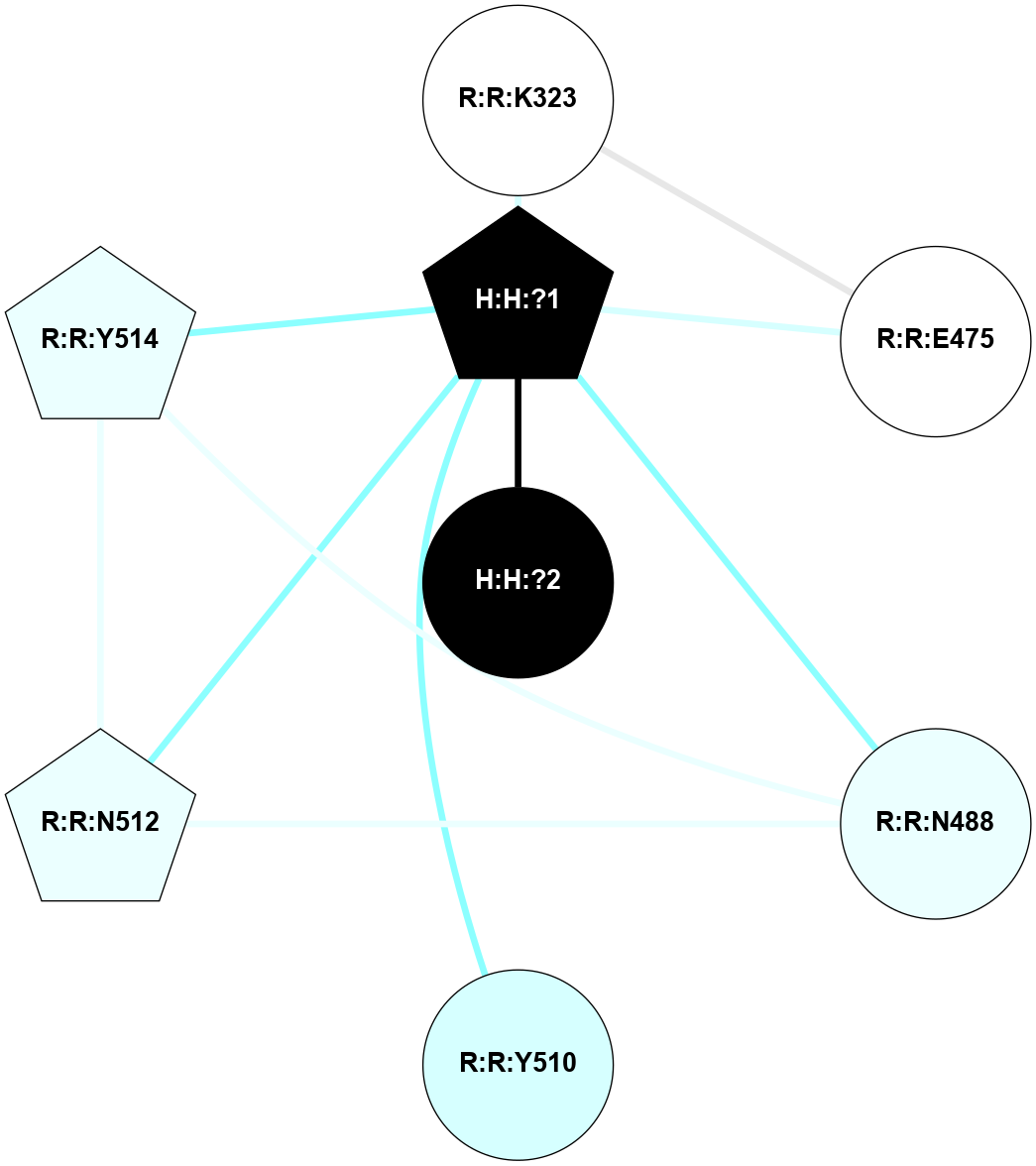
A 2D representation of the interactions of NAG in 7M3F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 34.31 | 2 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:K323 | 5.04 | 2 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?1 | R:R:E475 | 3.55 | 2 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?1 | R:R:N488 | 27.02 | 2 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:Y510 | 7.34 | 2 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:N512 | 17.19 | 2 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:Y514 | 8.39 | 2 | Yes | Yes | 0 | 4 | 1 | 2 | | R:R:E475 | R:R:K323 | 4.05 | 2 | No | No | 5 | 5 | 2 | 2 | | R:R:N488 | R:R:N512 | 6.81 | 2 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N488 | R:R:Y514 | 4.65 | 2 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N512 | R:R:Y514 | 3.49 | 2 | Yes | Yes | 4 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 34.31 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
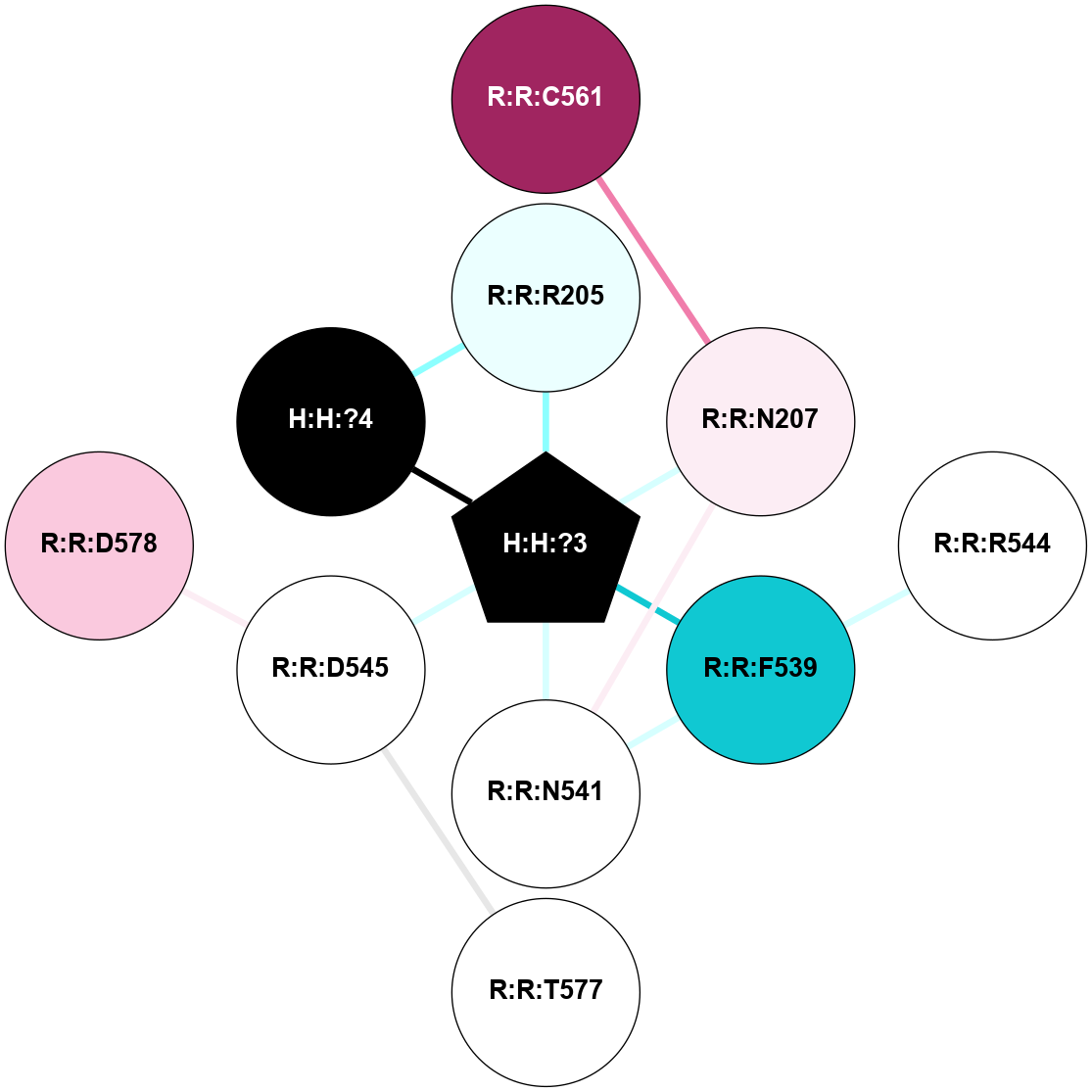
A 2D representation of the interactions of NAG in 7M3F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 34.31 | 3 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:R205 | 4.35 | 3 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?3 | R:R:N207 | 11.05 | 3 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?3 | R:R:F539 | 5.45 | 3 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?3 | R:R:N541 | 25.79 | 3 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?3 | R:R:D545 | 9.71 | 3 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?4 | R:R:R205 | 14.12 | 3 | No | No | 0 | 4 | 1 | 1 | | R:R:N207 | R:R:N541 | 16.35 | 3 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 7.87 | 3 | No | No | 9 | 6 | 2 | 1 | | R:R:F539 | R:R:N541 | 7.25 | 3 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 21.38 | 3 | No | No | 1 | 5 | 1 | 2 | | R:R:D545 | R:R:T577 | 4.34 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:D545 | R:R:D578 | 10.65 | 0 | No | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.11 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
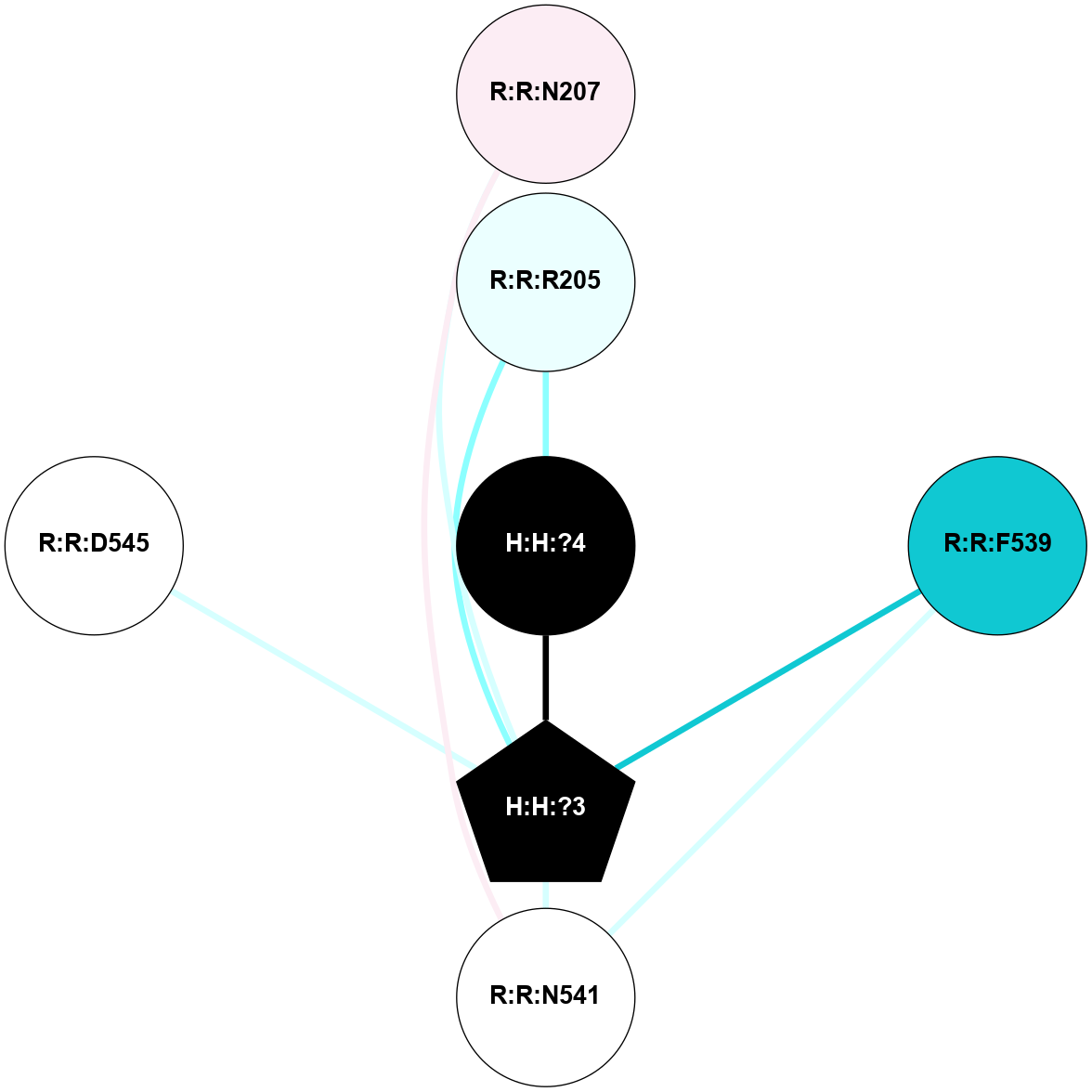
A 2D representation of the interactions of NAG in 7M3F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 34.31 | 3 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:R205 | 4.35 | 3 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?3 | R:R:N207 | 11.05 | 3 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?3 | R:R:F539 | 5.45 | 3 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?3 | R:R:N541 | 25.79 | 3 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | R:R:D545 | 9.71 | 3 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?4 | R:R:R205 | 14.12 | 3 | No | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 16.35 | 3 | No | No | 6 | 5 | 2 | 2 | | R:R:F539 | R:R:N541 | 7.25 | 3 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 24.21 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
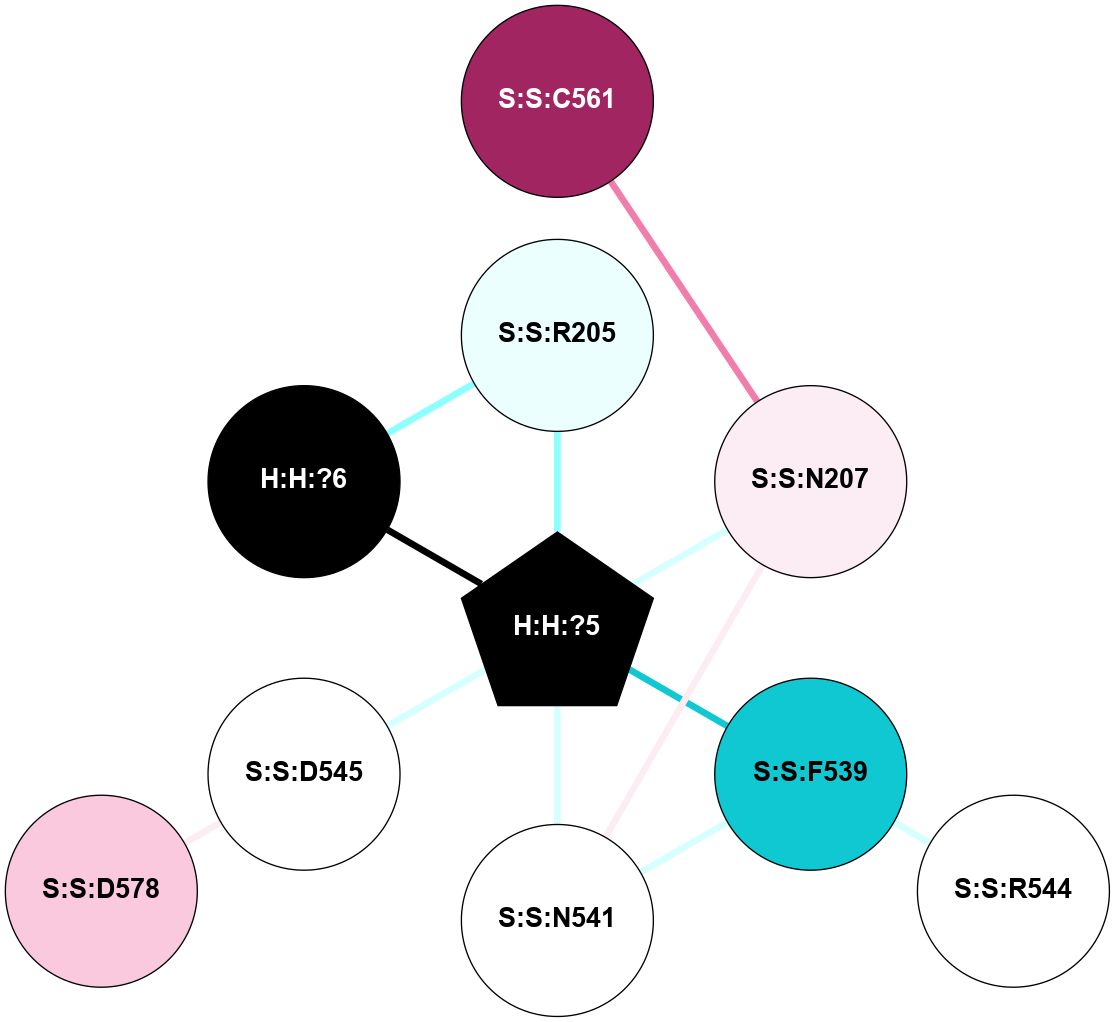
A 2D representation of the interactions of NAG in 7M3F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 30.99 | 14 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?5 | S:S:R205 | 14.12 | 14 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?5 | S:S:N207 | 8.6 | 14 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?5 | S:S:F539 | 5.45 | 14 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?5 | S:S:N541 | 27.02 | 14 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?5 | S:S:D545 | 8.5 | 14 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?6 | S:S:R205 | 18.47 | 14 | No | No | 0 | 4 | 1 | 1 | | S:S:N207 | S:S:N541 | 17.71 | 14 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 9.45 | 0 | No | No | 9 | 6 | 2 | 1 | | S:S:F539 | S:S:N541 | 4.83 | 14 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 23.52 | 14 | No | No | 1 | 5 | 1 | 2 | | S:S:D545 | S:S:D578 | 15.97 | 0 | No | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.78 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
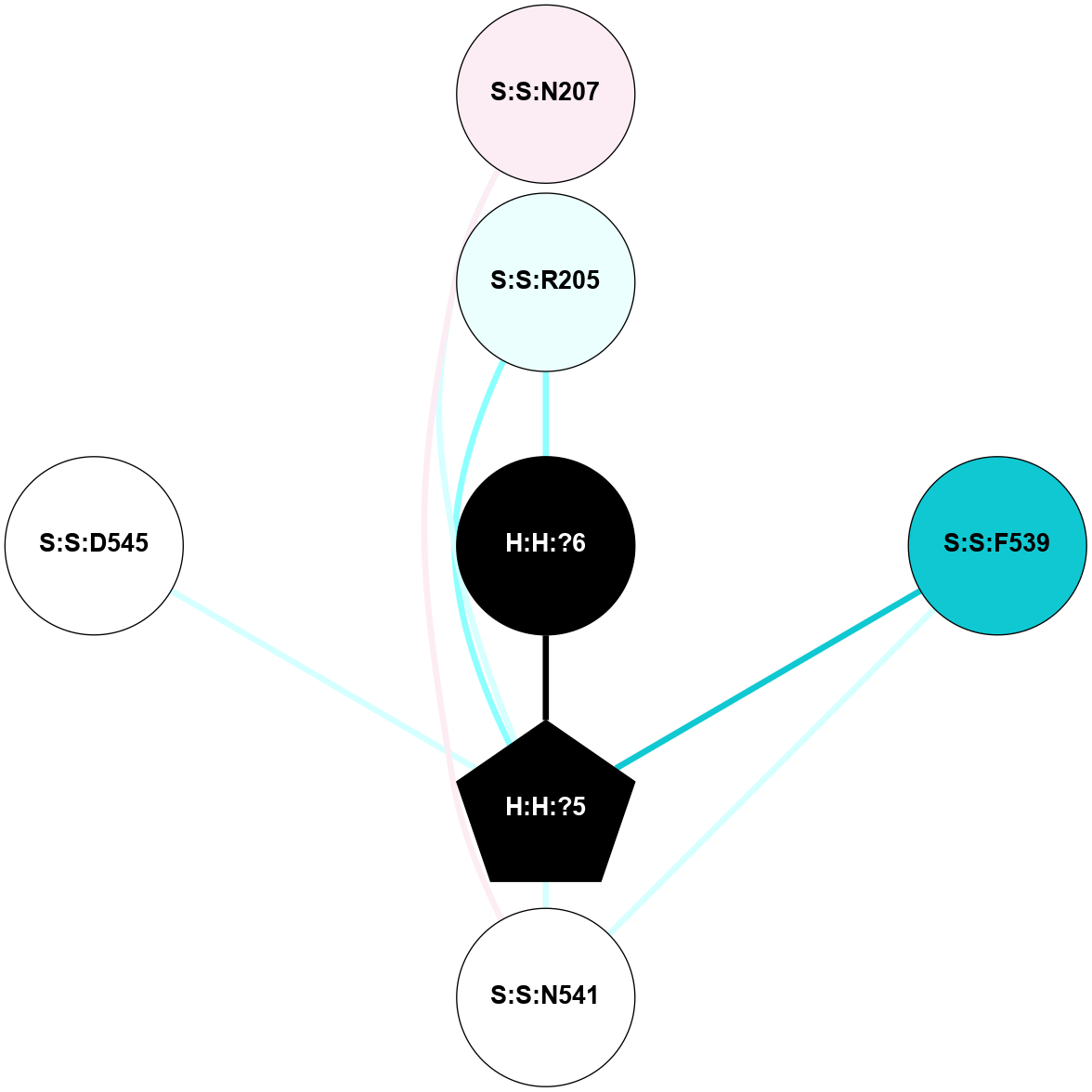
A 2D representation of the interactions of NAG in 7M3F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 30.99 | 14 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?5 | S:S:R205 | 14.12 | 14 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?5 | S:S:N207 | 8.6 | 14 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?5 | S:S:F539 | 5.45 | 14 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?5 | S:S:N541 | 27.02 | 14 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?5 | S:S:D545 | 8.5 | 14 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?6 | S:S:R205 | 18.47 | 14 | No | No | 0 | 4 | 0 | 1 | | S:S:N207 | S:S:N541 | 17.71 | 14 | No | No | 6 | 5 | 2 | 2 | | S:S:F539 | S:S:N541 | 4.83 | 14 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 24.73 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
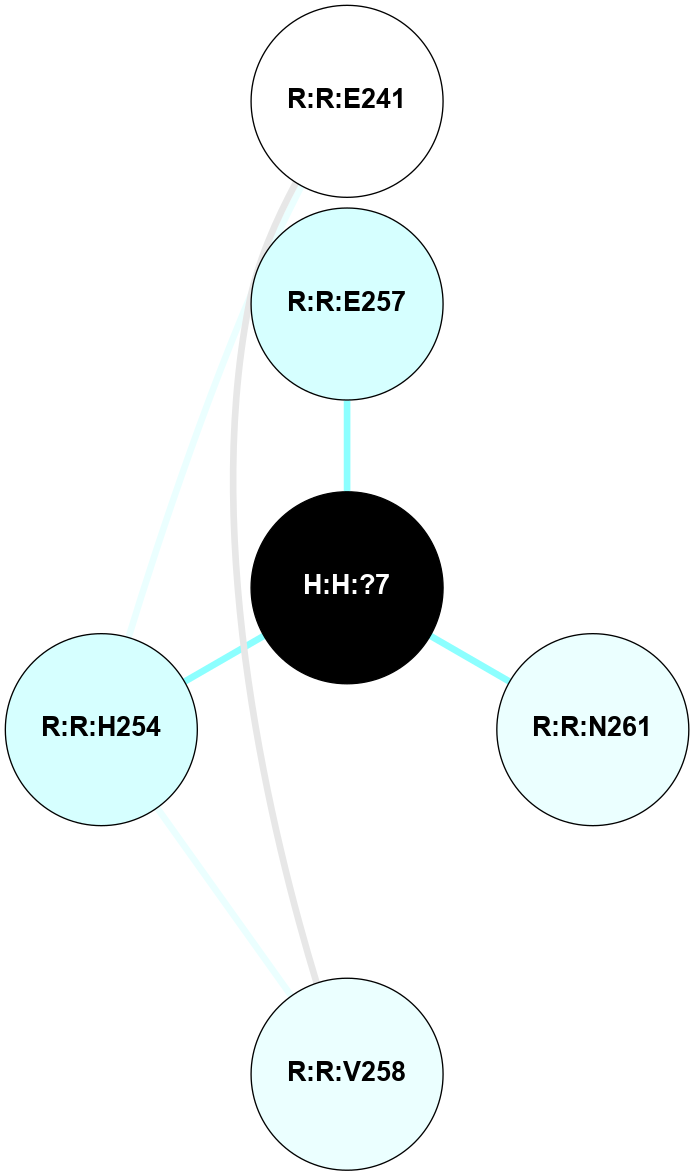
A 2D representation of the interactions of NAG in 7M3F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | R:R:E257 | 5.92 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?7 | R:R:N261 | 27.02 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:E241 | R:R:H254 | 3.69 | 3 | No | No | 5 | 3 | 2 | 1 | | R:R:E241 | R:R:V258 | 8.56 | 3 | No | No | 5 | 4 | 2 | 2 | | R:R:H254 | R:R:V258 | 5.54 | 3 | No | No | 3 | 4 | 1 | 2 | | H:H:?7 | R:R:H254 | 2.3 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 11.75 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
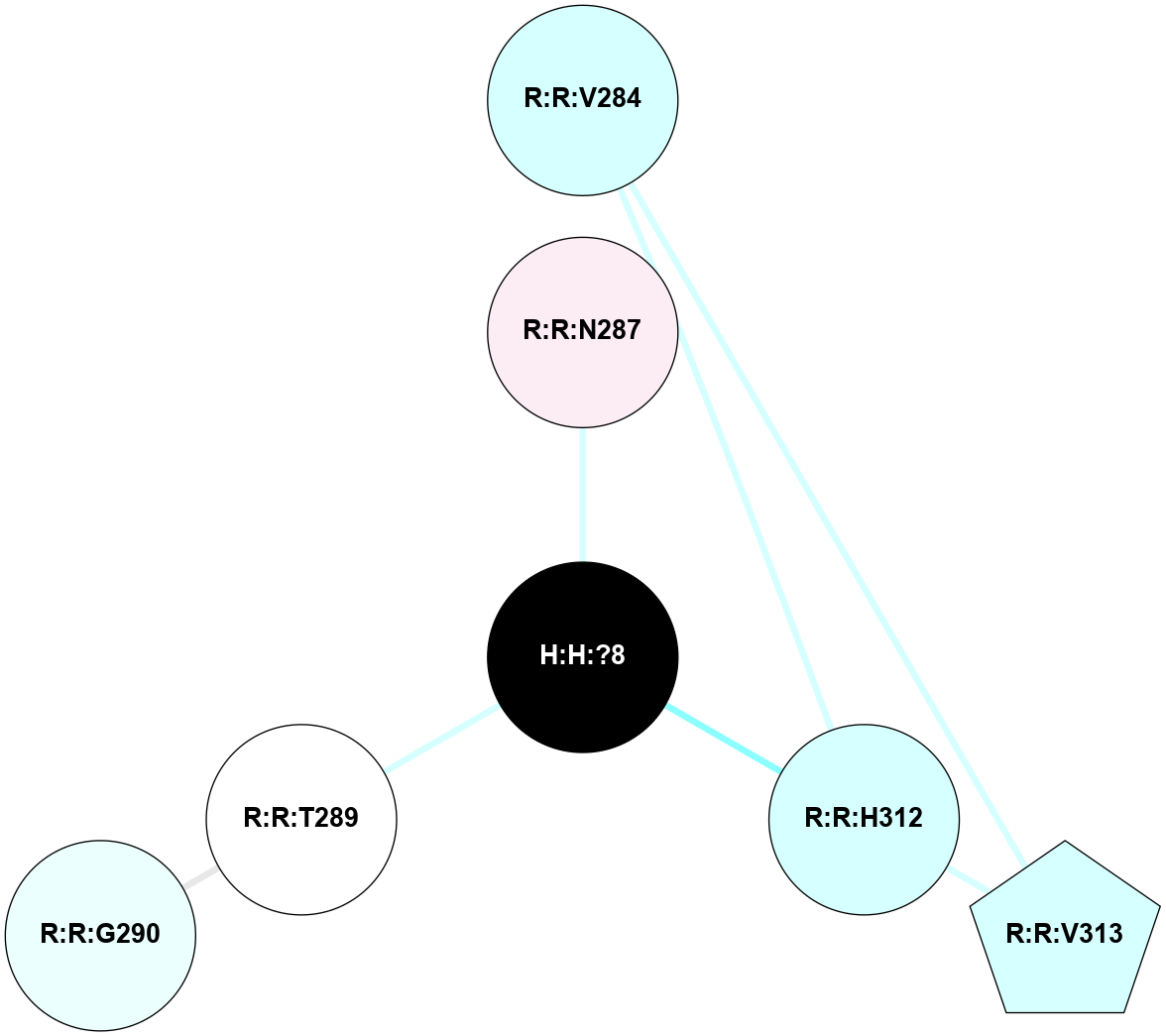
A 2D representation of the interactions of NAG in 7M3F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | R:R:N287 | 28.24 | 0 | No | No | 0 | 6 | 0 | 1 | | H:H:?8 | R:R:H312 | 4.6 | 0 | No | No | 0 | 3 | 0 | 1 | | R:R:H312 | R:R:V284 | 2.77 | 21 | No | No | 3 | 3 | 1 | 2 | | R:R:V284 | R:R:V313 | 6.41 | 21 | No | Yes | 3 | 3 | 2 | 2 | | R:R:H312 | R:R:V313 | 2.77 | 21 | No | Yes | 3 | 3 | 1 | 2 | | R:R:G290 | R:R:T289 | 1.82 | 0 | No | No | 4 | 5 | 2 | 1 | | H:H:?8 | R:R:T289 | 1.32 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 11.39 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7M3F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?9 | R:R:N468 | 25.79 | 22 | No | No | 0 | 6 | 0 | 1 | | H:H:?9 | R:R:Q476 | 7.14 | 22 | No | No | 0 | 3 | 0 | 1 | | R:R:N468 | R:R:Q476 | 7.92 | 22 | No | No | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 5.85 | 22 | No | No | 6 | 3 | 1 | 2 | | R:R:Q476 | R:R:T470 | 7.09 | 22 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.46 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
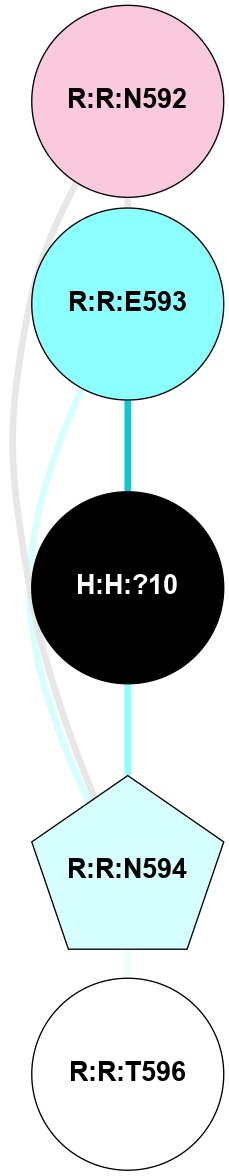
A 2D representation of the interactions of NAG in 7M3F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | R:R:N594 | 31.93 | 0 | No | No | 0 | 3 | 0 | 1 | | R:R:N594 | R:R:T596 | 2.92 | 0 | No | No | 3 | 5 | 1 | 2 | | R:R:N592 | R:R:N594 | 1.36 | 0 | No | No | 7 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 31.93 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
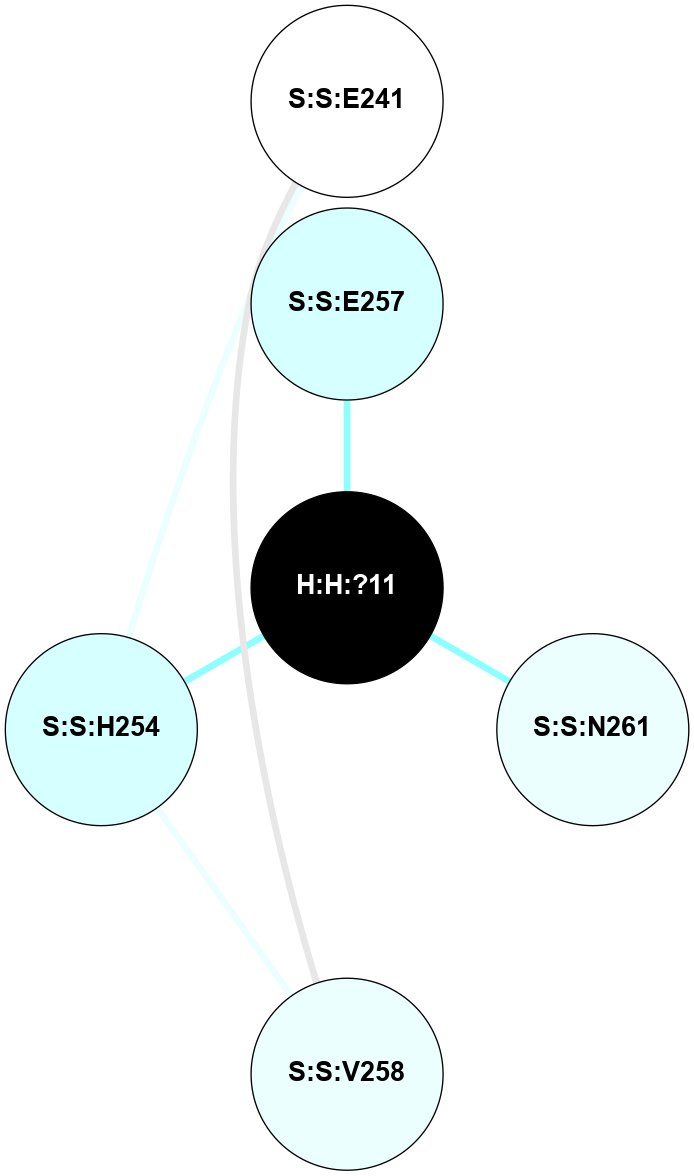
A 2D representation of the interactions of NAG in 7M3F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | S:S:E257 | 7.11 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?11 | S:S:N261 | 24.56 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:E241 | S:S:H254 | 4.92 | 1 | No | No | 5 | 3 | 2 | 1 | | S:S:E241 | S:S:V258 | 8.56 | 1 | No | No | 5 | 4 | 2 | 2 | | S:S:H254 | S:S:V258 | 5.54 | 1 | No | No | 3 | 4 | 1 | 2 | | H:H:?11 | S:S:H254 | 2.3 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 11.32 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7M3F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | S:S:N287 | 29.47 | 12 | No | No | 0 | 6 | 0 | 1 | | H:H:?12 | S:S:H312 | 3.45 | 12 | No | Yes | 0 | 3 | 0 | 1 | | S:S:H312 | S:S:V284 | 2.77 | 12 | Yes | No | 3 | 3 | 1 | 2 | | S:S:V284 | S:S:V313 | 6.41 | 12 | No | Yes | 3 | 3 | 2 | 2 | | S:S:H312 | S:S:N287 | 6.38 | 12 | Yes | No | 3 | 6 | 1 | 1 | | S:S:H312 | S:S:V313 | 2.77 | 12 | Yes | Yes | 3 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 16.46 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
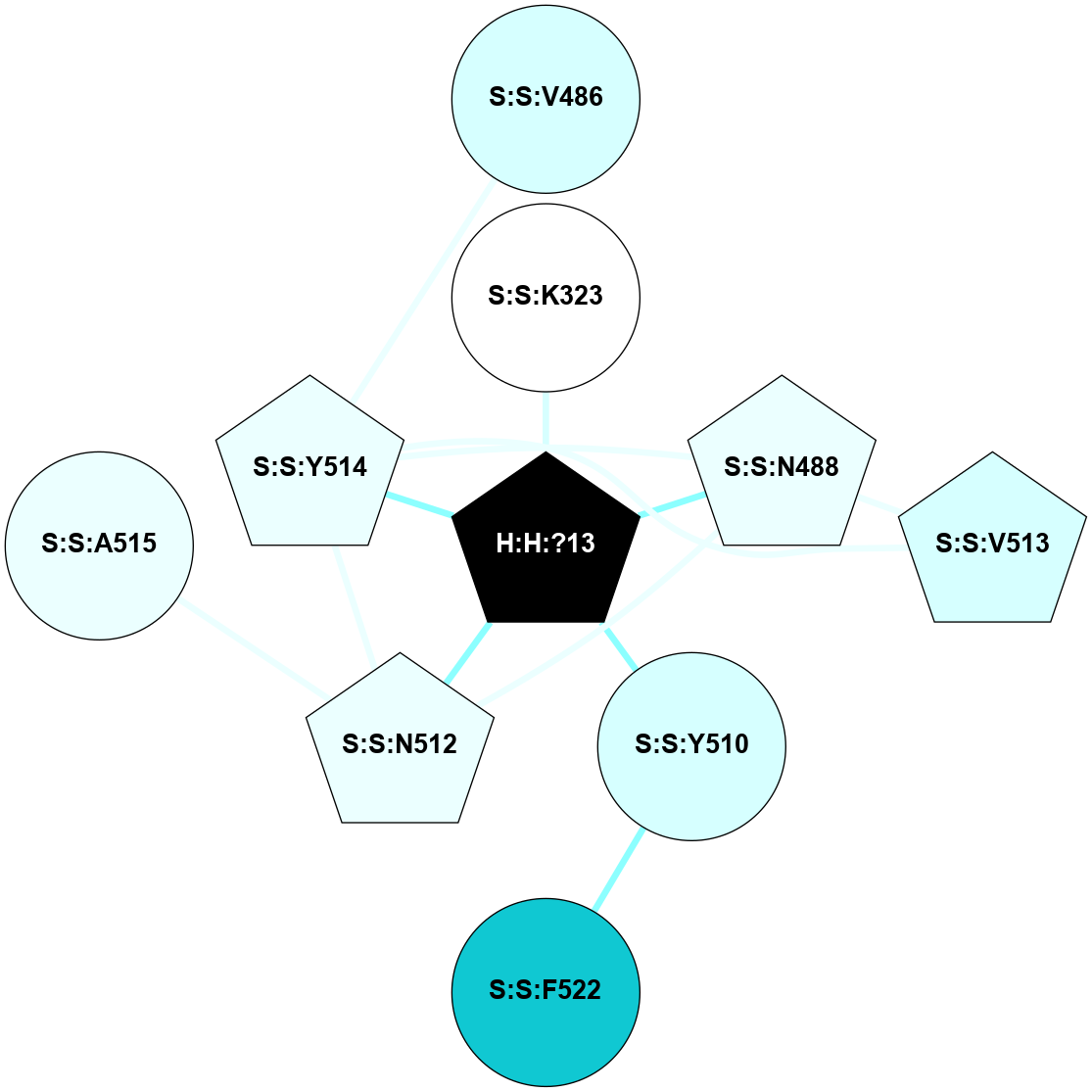
A 2D representation of the interactions of NAG in 7M3F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | S:S:K323 | 3.78 | 17 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?13 | S:S:N488 | 25.79 | 17 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?13 | S:S:Y510 | 8.39 | 17 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?13 | S:S:N512 | 13.51 | 17 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?13 | S:S:Y514 | 11.53 | 17 | Yes | Yes | 0 | 4 | 0 | 1 | | S:S:V486 | S:S:Y514 | 3.79 | 0 | No | Yes | 3 | 4 | 2 | 1 | | S:S:N488 | S:S:N512 | 5.45 | 17 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:V513 | 2.96 | 17 | Yes | No | 4 | 3 | 1 | 2 | | S:S:N488 | S:S:Y514 | 3.49 | 17 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:F522 | S:S:Y510 | 15.47 | 0 | No | No | 1 | 3 | 2 | 1 | | S:S:N512 | S:S:Y514 | 3.49 | 17 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:A515 | S:S:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 12.60 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
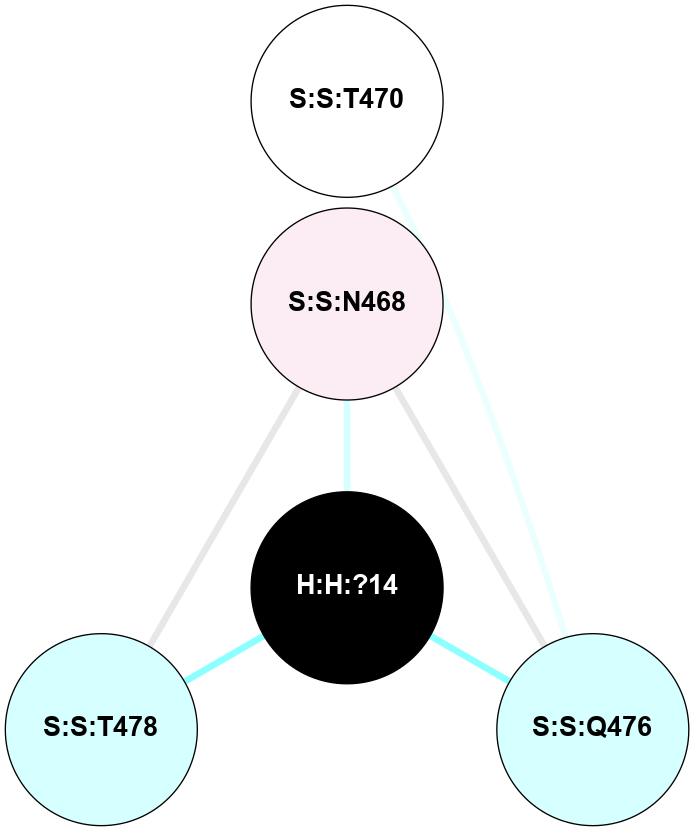
A 2D representation of the interactions of NAG in 7M3F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?14 | S:S:N468 | 23.33 | 18 | No | No | 0 | 6 | 0 | 1 | | H:H:?14 | S:S:Q476 | 8.33 | 18 | No | No | 0 | 3 | 0 | 1 | | H:H:?14 | S:S:T478 | 3.95 | 18 | No | No | 0 | 3 | 0 | 1 | | S:S:N468 | S:S:Q476 | 6.6 | 18 | No | No | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 8.77 | 18 | No | No | 6 | 3 | 1 | 1 | | S:S:Q476 | S:S:T470 | 8.5 | 18 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 11.87 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
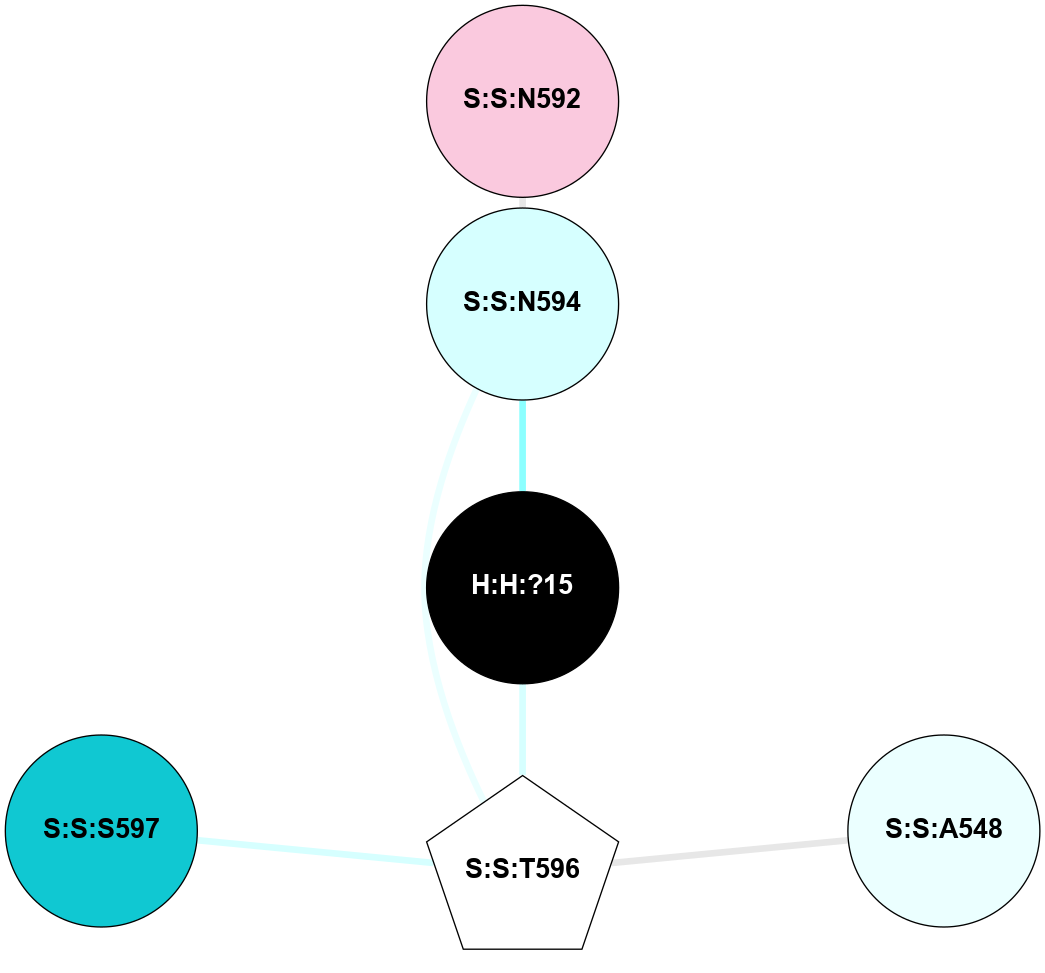
A 2D representation of the interactions of NAG in 7M3F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?15 | S:S:N594 | 25.79 | 23 | No | No | 0 | 3 | 0 | 1 | | H:H:?15 | S:S:T596 | 3.95 | 23 | No | Yes | 0 | 5 | 0 | 1 | | S:S:N592 | S:S:N594 | 2.72 | 0 | No | No | 7 | 3 | 2 | 1 | | S:S:N594 | S:S:T596 | 2.92 | 23 | No | Yes | 3 | 5 | 1 | 1 | | S:S:A548 | S:S:T596 | 1.68 | 0 | No | Yes | 4 | 5 | 2 | 1 | | S:S:S597 | S:S:T596 | 1.6 | 0 | No | Yes | 1 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.87 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7M3G | C | Ion | Calcium Sensing | CaS | Homo Sapiens | - | Ca; Ca; Tryptophan; Evocalcet; Etelcalcetide; PO4 | - | 2.5 | 2021-06-30 | doi.org/10.1038/s41586-021-03691-0 |
|
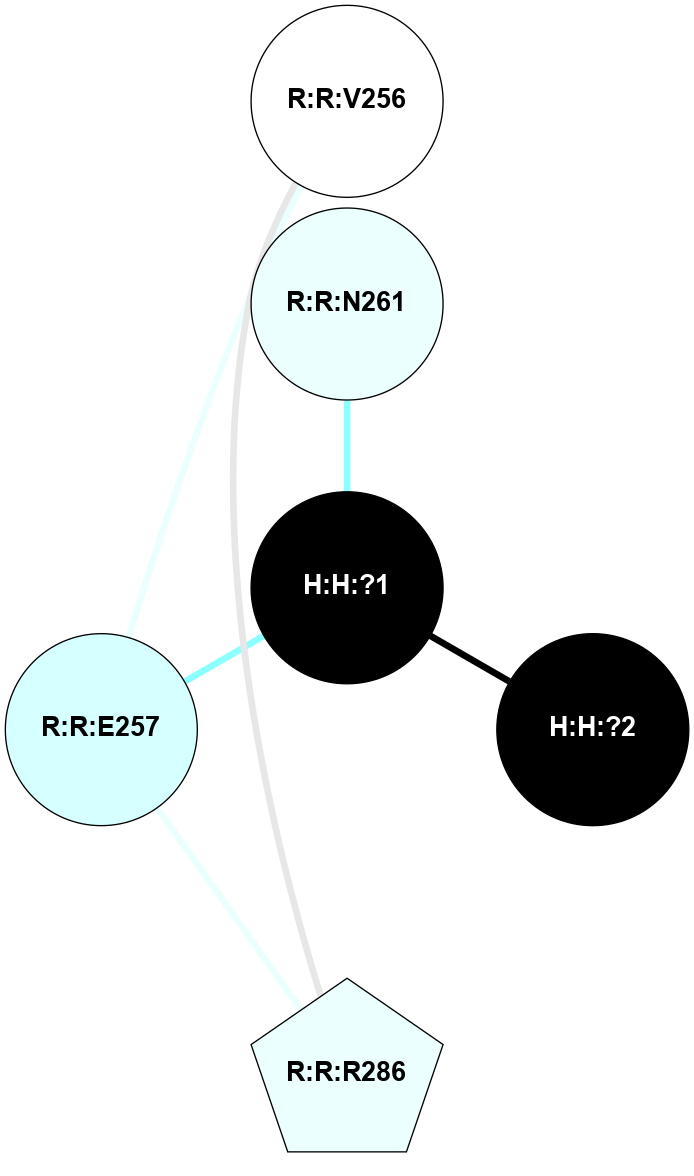
A 2D representation of the interactions of NAG in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 30.99 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N261 | 28.24 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:E257 | R:R:V256 | 2.85 | 25 | No | No | 3 | 5 | 1 | 2 | | R:R:R286 | R:R:V256 | 6.54 | 25 | Yes | No | 4 | 5 | 2 | 2 | | R:R:E257 | R:R:R286 | 10.47 | 25 | No | Yes | 3 | 4 | 1 | 2 | | H:H:?1 | R:R:E257 | 2.37 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.53 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 30.99 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N261 | 28.24 | 0 | No | No | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:E257 | 2.37 | 0 | No | No | 0 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 30.99 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
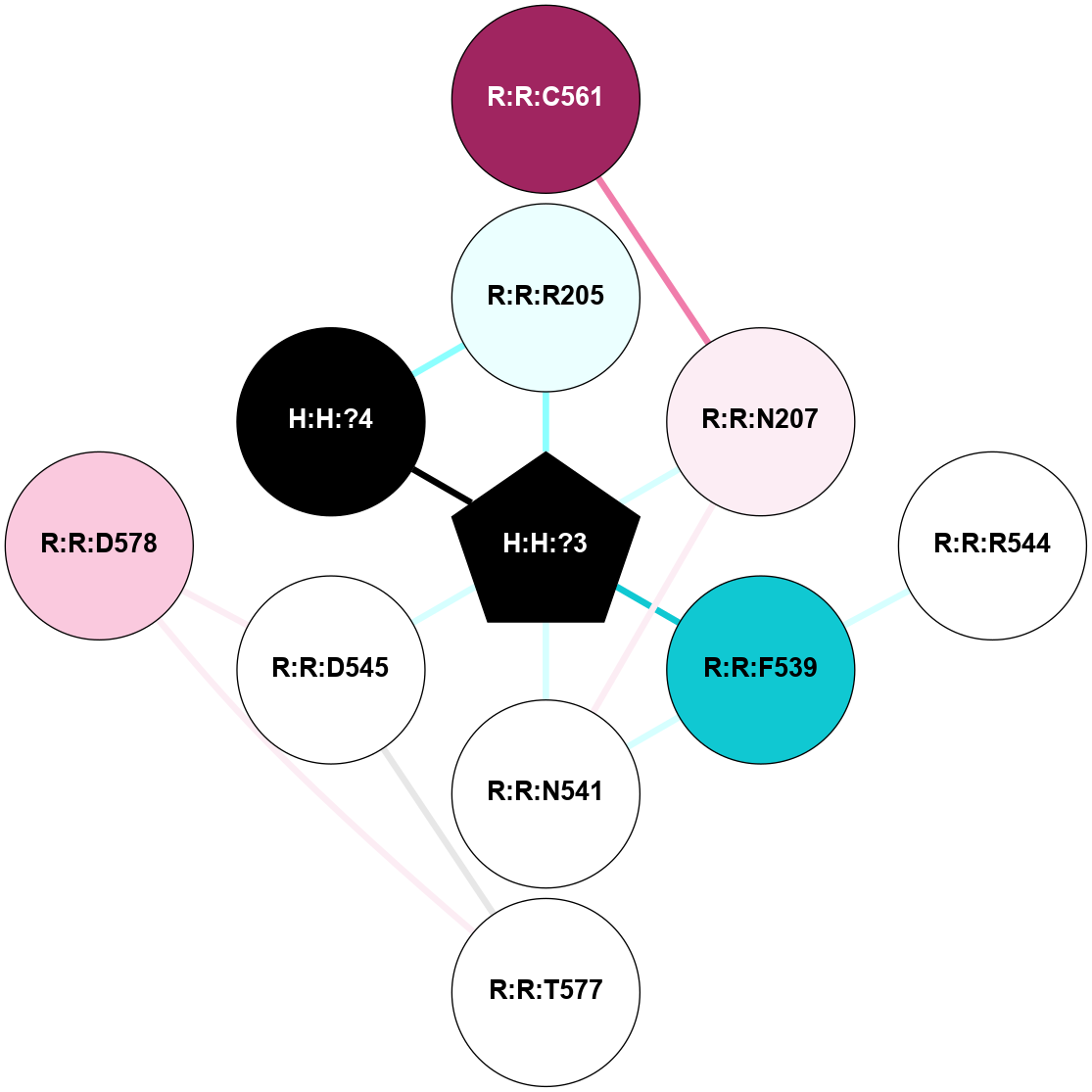
A 2D representation of the interactions of NAG in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 35.42 | 15 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:R205 | 6.52 | 15 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?3 | R:R:N207 | 14.74 | 15 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?3 | R:R:F539 | 5.45 | 15 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?3 | R:R:N541 | 27.02 | 15 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?3 | R:R:D545 | 8.5 | 15 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?4 | R:R:R205 | 16.3 | 15 | No | No | 0 | 4 | 1 | 1 | | R:R:N207 | R:R:N541 | 16.35 | 15 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 9.45 | 0 | No | No | 9 | 6 | 2 | 1 | | R:R:F539 | R:R:N541 | 4.83 | 15 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 24.59 | 15 | No | No | 1 | 5 | 1 | 2 | | R:R:D545 | R:R:T577 | 4.34 | 15 | No | No | 5 | 5 | 1 | 2 | | R:R:D545 | R:R:D578 | 13.31 | 15 | No | No | 5 | 7 | 1 | 2 | | R:R:D578 | R:R:T577 | 2.89 | 15 | No | No | 7 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.28 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
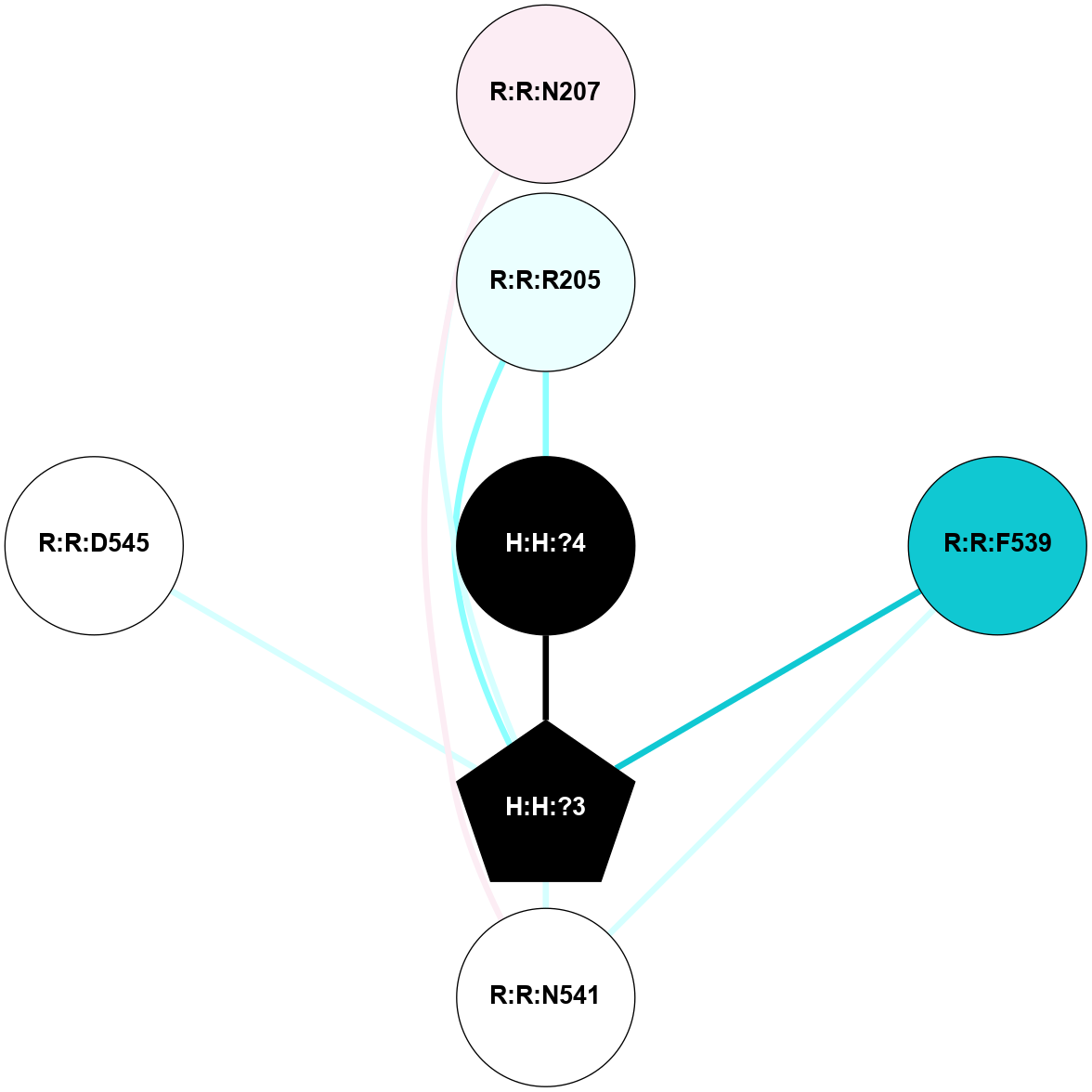
A 2D representation of the interactions of NAG in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 35.42 | 15 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:R205 | 6.52 | 15 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?3 | R:R:N207 | 14.74 | 15 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?3 | R:R:F539 | 5.45 | 15 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?3 | R:R:N541 | 27.02 | 15 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | R:R:D545 | 8.5 | 15 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?4 | R:R:R205 | 16.3 | 15 | No | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 16.35 | 15 | No | No | 6 | 5 | 2 | 2 | | R:R:F539 | R:R:N541 | 4.83 | 15 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 25.86 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
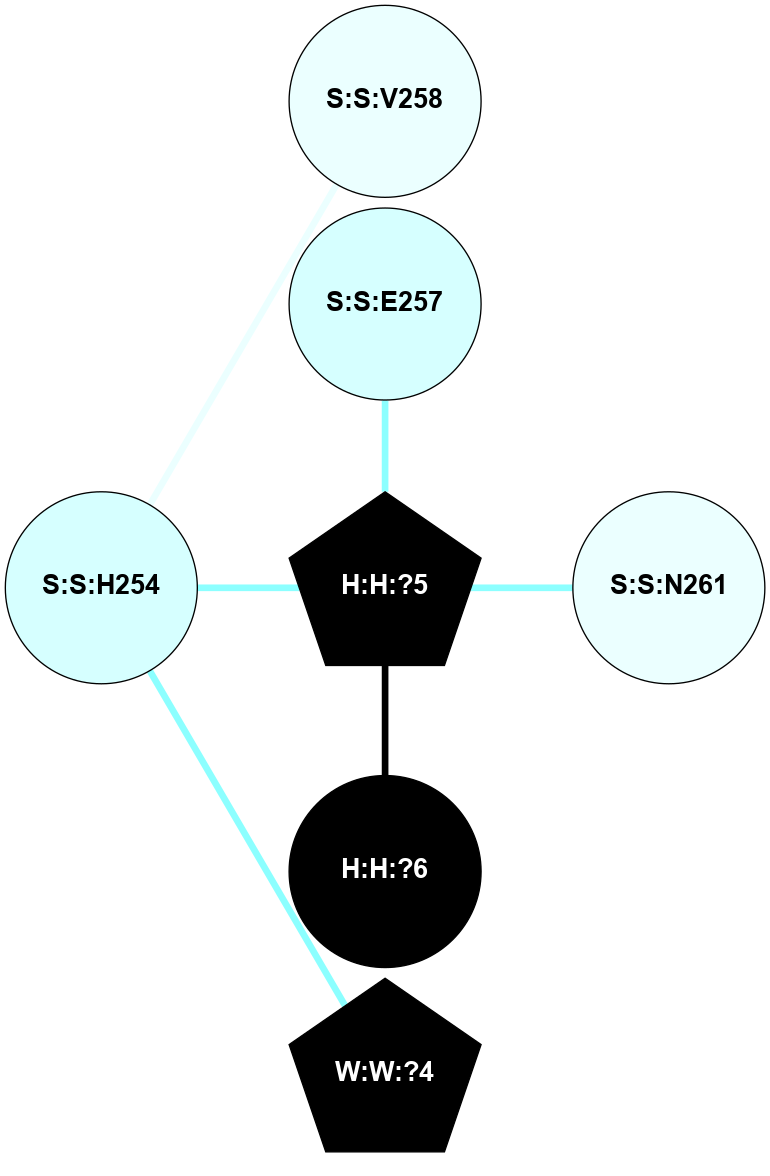
A 2D representation of the interactions of NAG in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 34.31 | 0 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?5 | S:S:E257 | 4.74 | 0 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?5 | S:S:N261 | 29.47 | 0 | Yes | No | 0 | 4 | 0 | 1 | | S:S:H254 | S:S:V258 | 4.15 | 0 | No | No | 3 | 4 | 1 | 2 | | S:S:H254 | W:W:?4 | 8.2 | 0 | No | Yes | 3 | 0 | 1 | 2 | | H:H:?5 | S:S:H254 | 2.3 | 0 | Yes | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.71 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
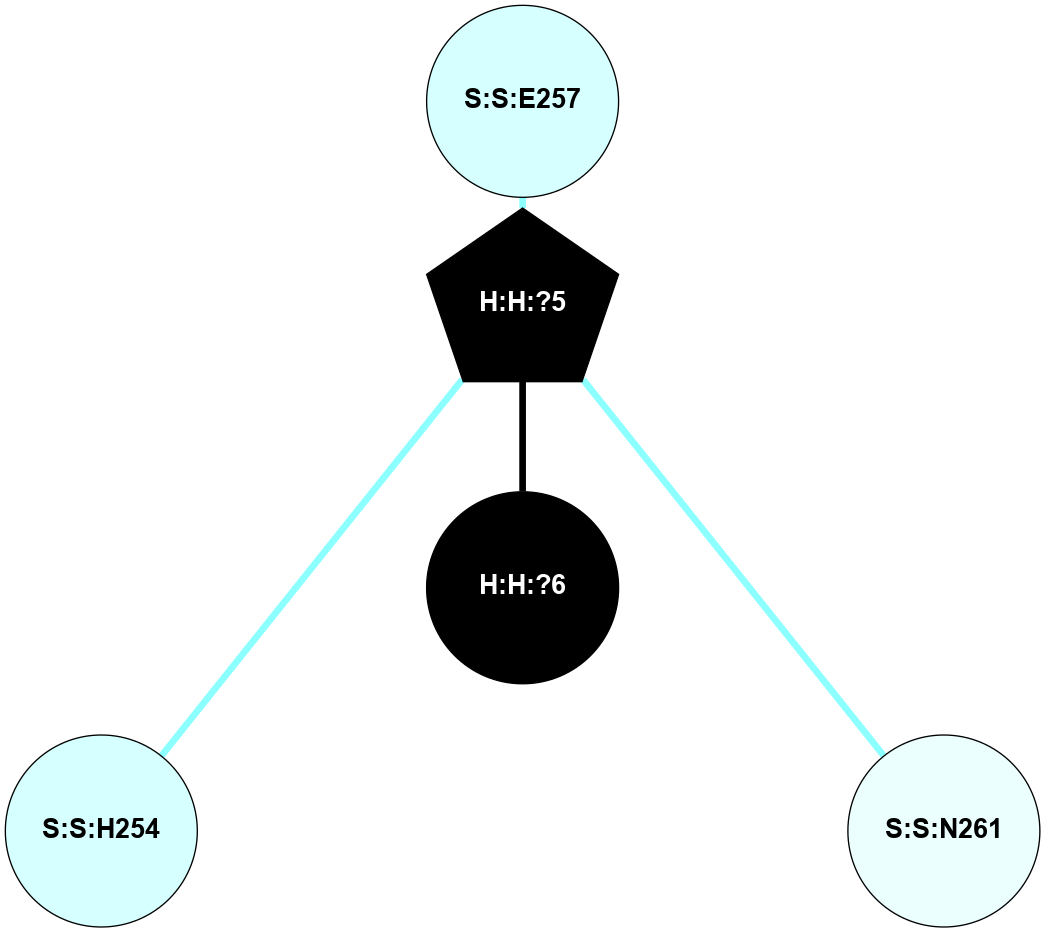
A 2D representation of the interactions of NAG in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 34.31 | 0 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?5 | S:S:E257 | 4.74 | 0 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?5 | S:S:N261 | 29.47 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?5 | S:S:H254 | 2.3 | 0 | Yes | No | 0 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 34.31 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
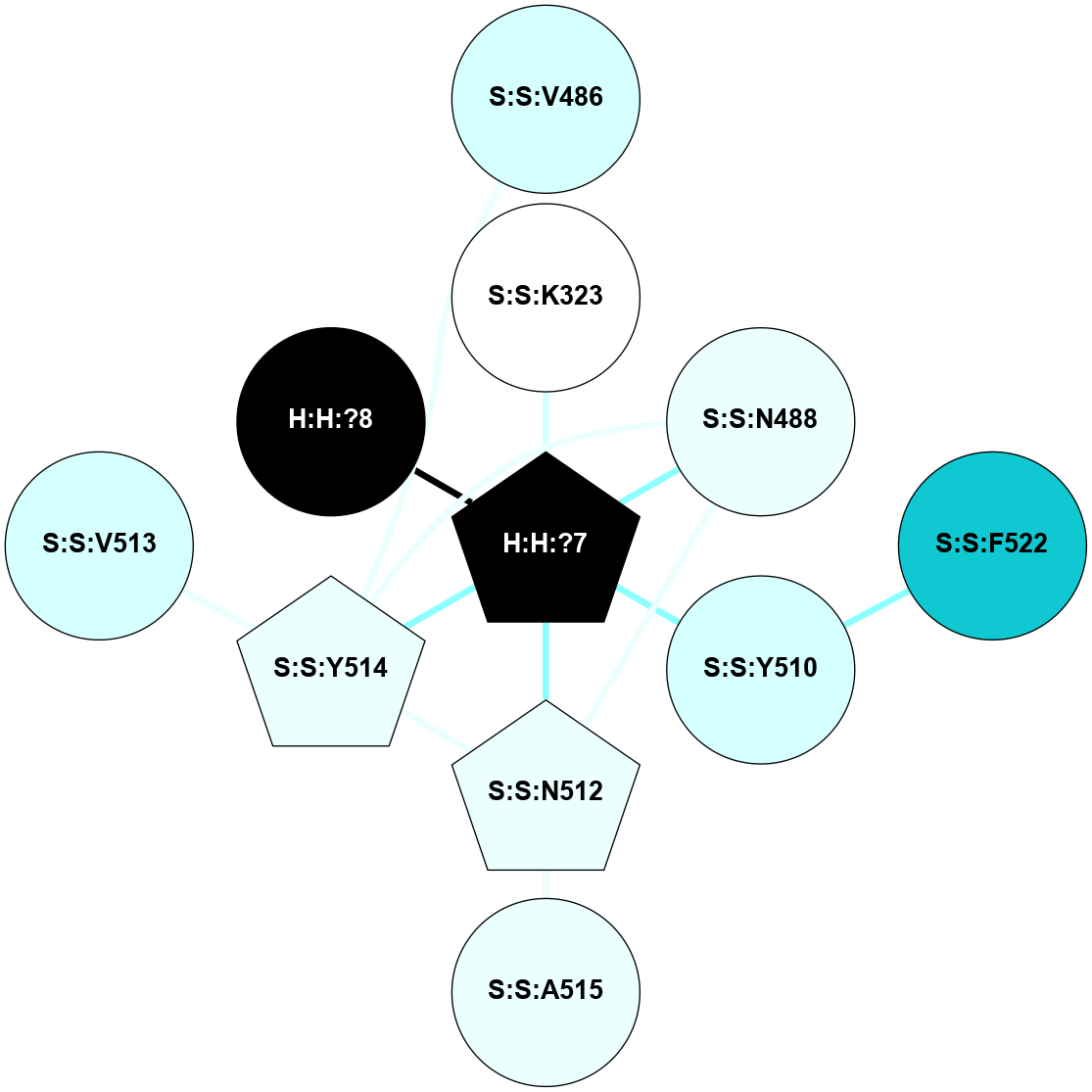
A 2D representation of the interactions of NAG in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 38.74 | 28 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?7 | S:S:K323 | 6.31 | 28 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?7 | S:S:N488 | 25.79 | 28 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?7 | S:S:Y510 | 11.53 | 28 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?7 | S:S:N512 | 17.19 | 28 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?7 | S:S:Y514 | 7.34 | 28 | Yes | Yes | 0 | 4 | 0 | 1 | | S:S:V486 | S:S:Y514 | 3.79 | 0 | No | Yes | 3 | 4 | 2 | 1 | | S:S:N488 | S:S:N512 | 6.81 | 28 | No | Yes | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:Y514 | 4.65 | 28 | No | Yes | 4 | 4 | 1 | 1 | | S:S:F522 | S:S:Y510 | 15.47 | 0 | No | No | 1 | 3 | 2 | 1 | | S:S:N512 | S:S:Y514 | 3.49 | 28 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:A515 | S:S:N512 | 4.69 | 0 | No | Yes | 4 | 4 | 2 | 1 | | S:S:V513 | S:S:Y514 | 5.05 | 0 | No | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 17.82 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
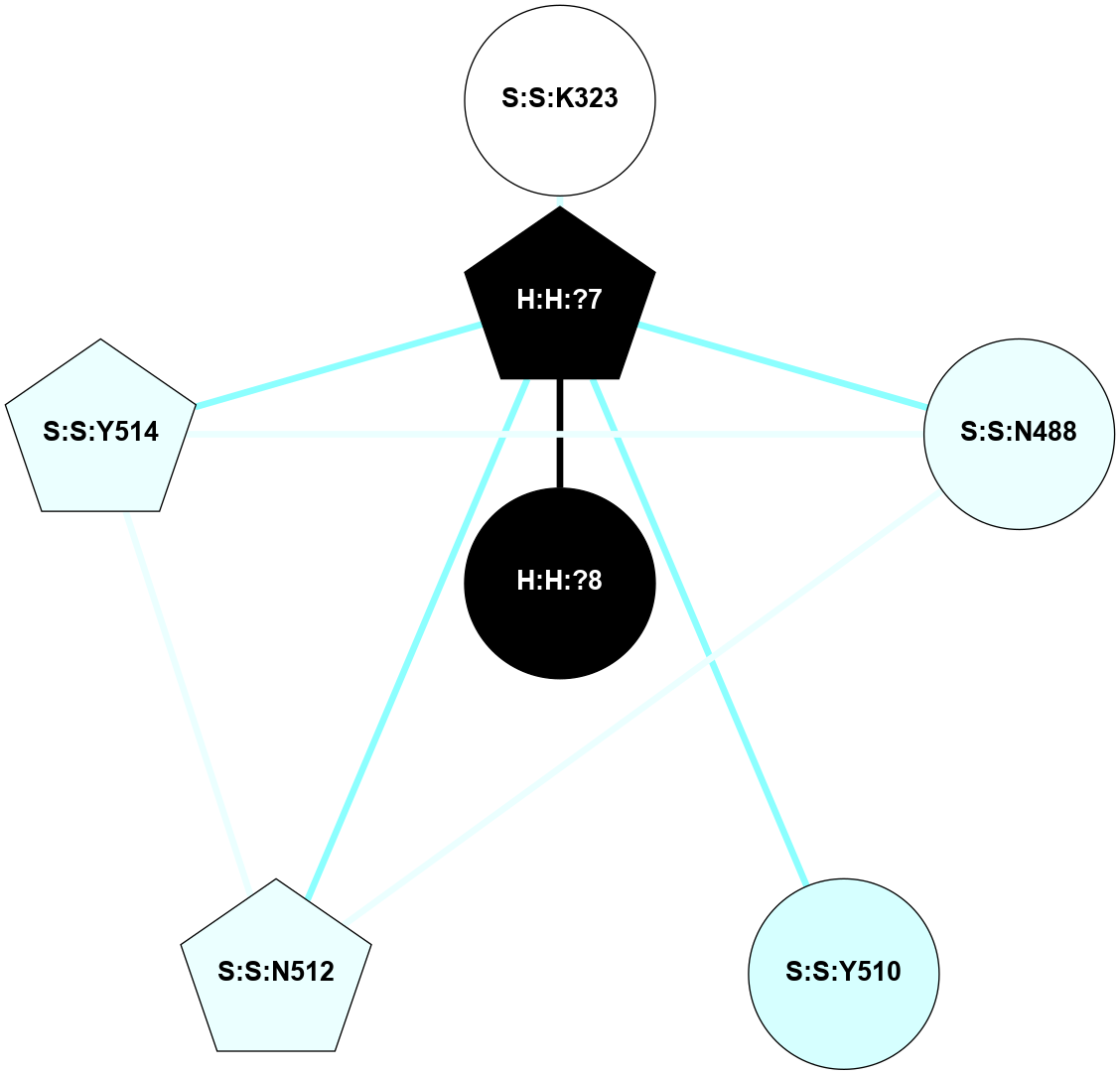
A 2D representation of the interactions of NAG in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 38.74 | 28 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?7 | S:S:K323 | 6.31 | 28 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?7 | S:S:N488 | 25.79 | 28 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?7 | S:S:Y510 | 11.53 | 28 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?7 | S:S:N512 | 17.19 | 28 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?7 | S:S:Y514 | 7.34 | 28 | Yes | Yes | 0 | 4 | 1 | 2 | | S:S:N488 | S:S:N512 | 6.81 | 28 | No | Yes | 4 | 4 | 2 | 2 | | S:S:N488 | S:S:Y514 | 4.65 | 28 | No | Yes | 4 | 4 | 2 | 2 | | S:S:N512 | S:S:Y514 | 3.49 | 28 | Yes | Yes | 4 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 38.74 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
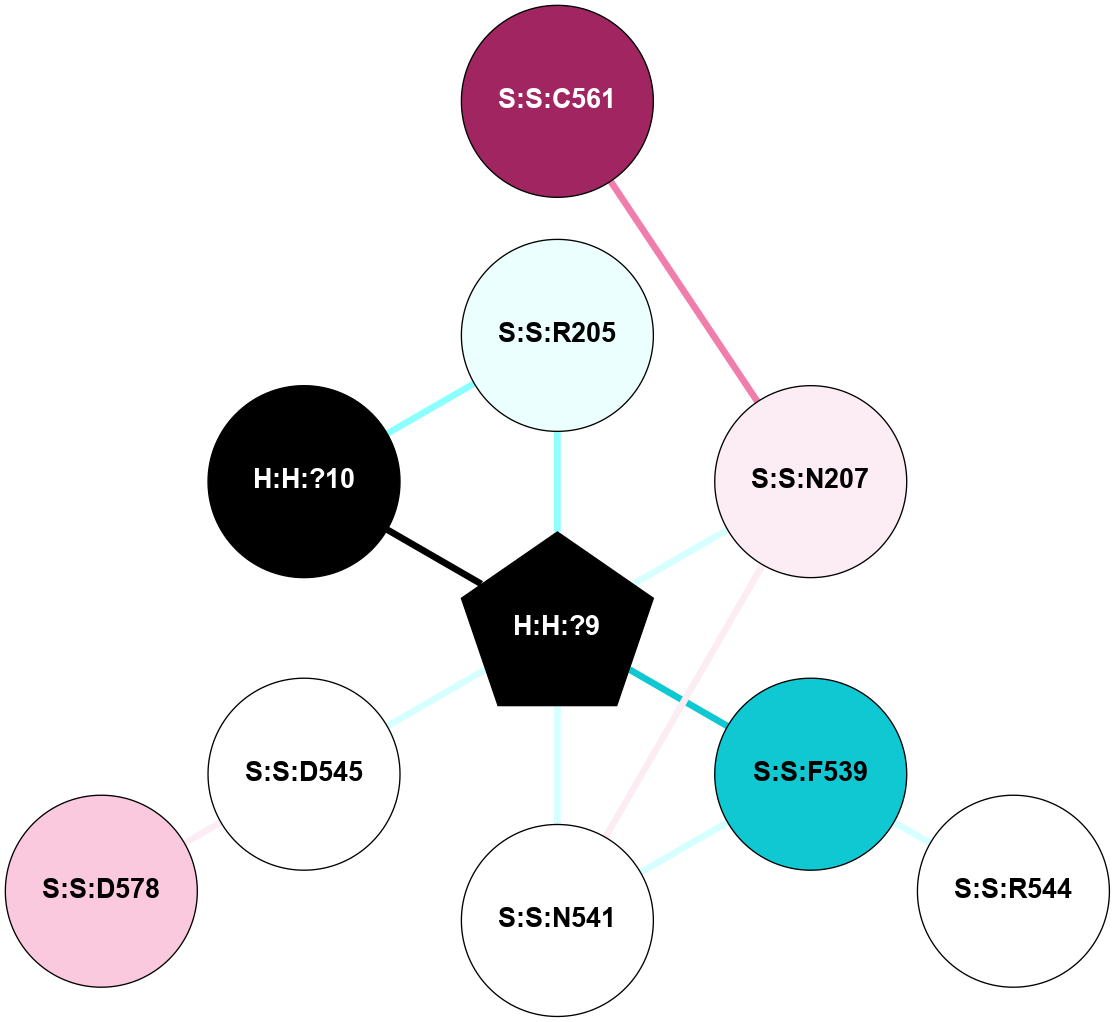
A 2D representation of the interactions of NAG in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | H:H:?9 | 35.42 | 20 | No | Yes | 0 | 0 | 1 | 0 | | H:H:?9 | S:S:R205 | 6.52 | 20 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?9 | S:S:N207 | 11.05 | 20 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?9 | S:S:F539 | 5.45 | 20 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?9 | S:S:N541 | 23.33 | 20 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?9 | S:S:D545 | 8.5 | 20 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?10 | S:S:R205 | 16.3 | 20 | No | No | 0 | 4 | 1 | 1 | | S:S:N207 | S:S:N541 | 14.98 | 20 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 7.87 | 0 | No | No | 9 | 6 | 2 | 1 | | S:S:F539 | S:S:N541 | 6.04 | 20 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 21.38 | 20 | No | No | 1 | 5 | 1 | 2 | | S:S:D545 | S:S:D578 | 10.65 | 0 | No | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.04 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
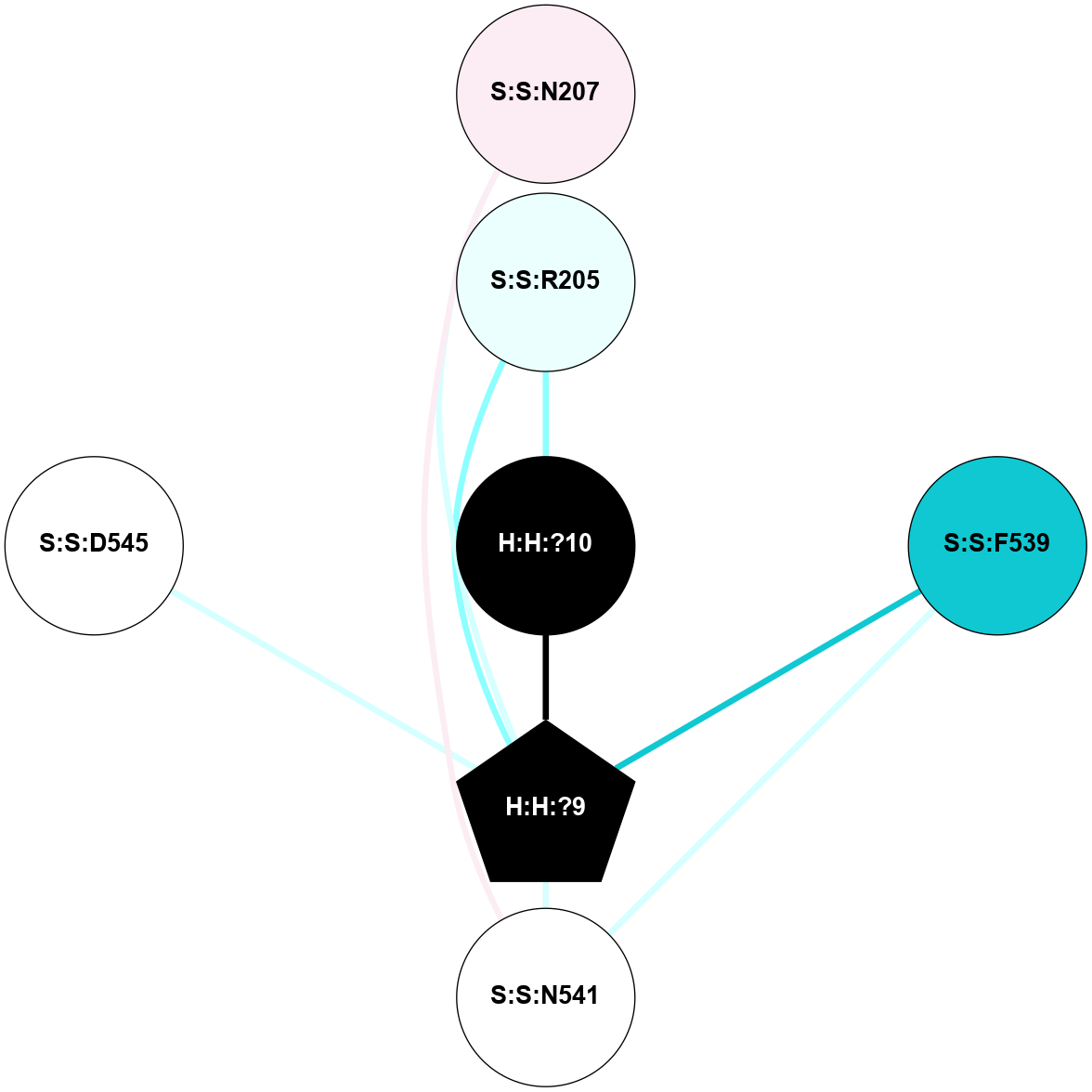
A 2D representation of the interactions of NAG in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | H:H:?9 | 35.42 | 20 | No | Yes | 0 | 0 | 0 | 1 | | H:H:?9 | S:S:R205 | 6.52 | 20 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?9 | S:S:N207 | 11.05 | 20 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?9 | S:S:F539 | 5.45 | 20 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?9 | S:S:N541 | 23.33 | 20 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?9 | S:S:D545 | 8.5 | 20 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?10 | S:S:R205 | 16.3 | 20 | No | No | 0 | 4 | 0 | 1 | | S:S:N207 | S:S:N541 | 14.98 | 20 | No | No | 6 | 5 | 2 | 2 | | S:S:F539 | S:S:N541 | 6.04 | 20 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 25.86 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
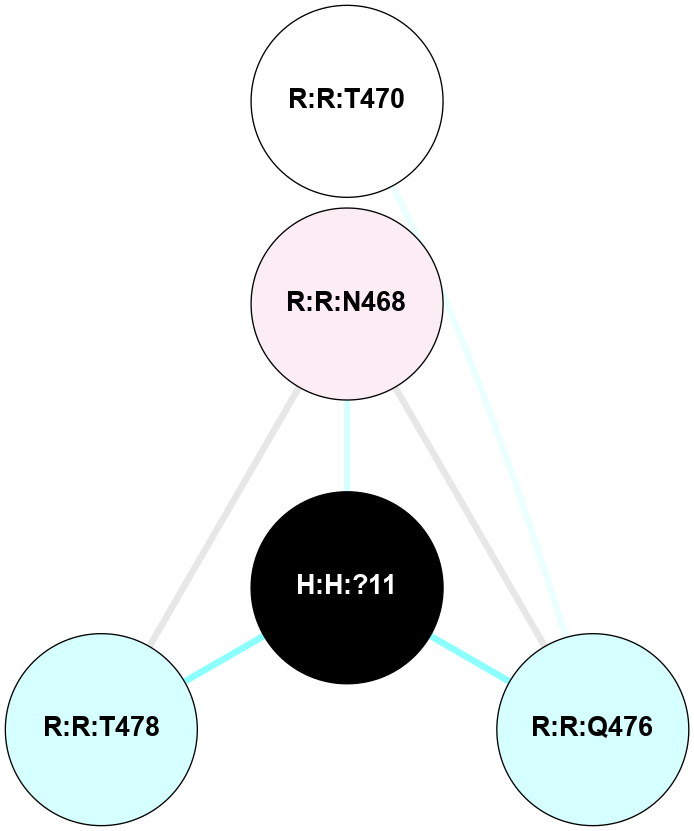
A 2D representation of the interactions of NAG in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | R:R:N468 | 23.33 | 29 | No | No | 0 | 6 | 0 | 1 | | H:H:?11 | R:R:Q476 | 3.57 | 29 | No | No | 0 | 3 | 0 | 1 | | H:H:?11 | R:R:T478 | 6.59 | 29 | No | No | 0 | 3 | 0 | 1 | | R:R:N468 | R:R:Q476 | 7.92 | 29 | No | No | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 5.85 | 29 | No | No | 6 | 3 | 1 | 1 | | R:R:Q476 | R:R:T470 | 8.5 | 29 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 11.16 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
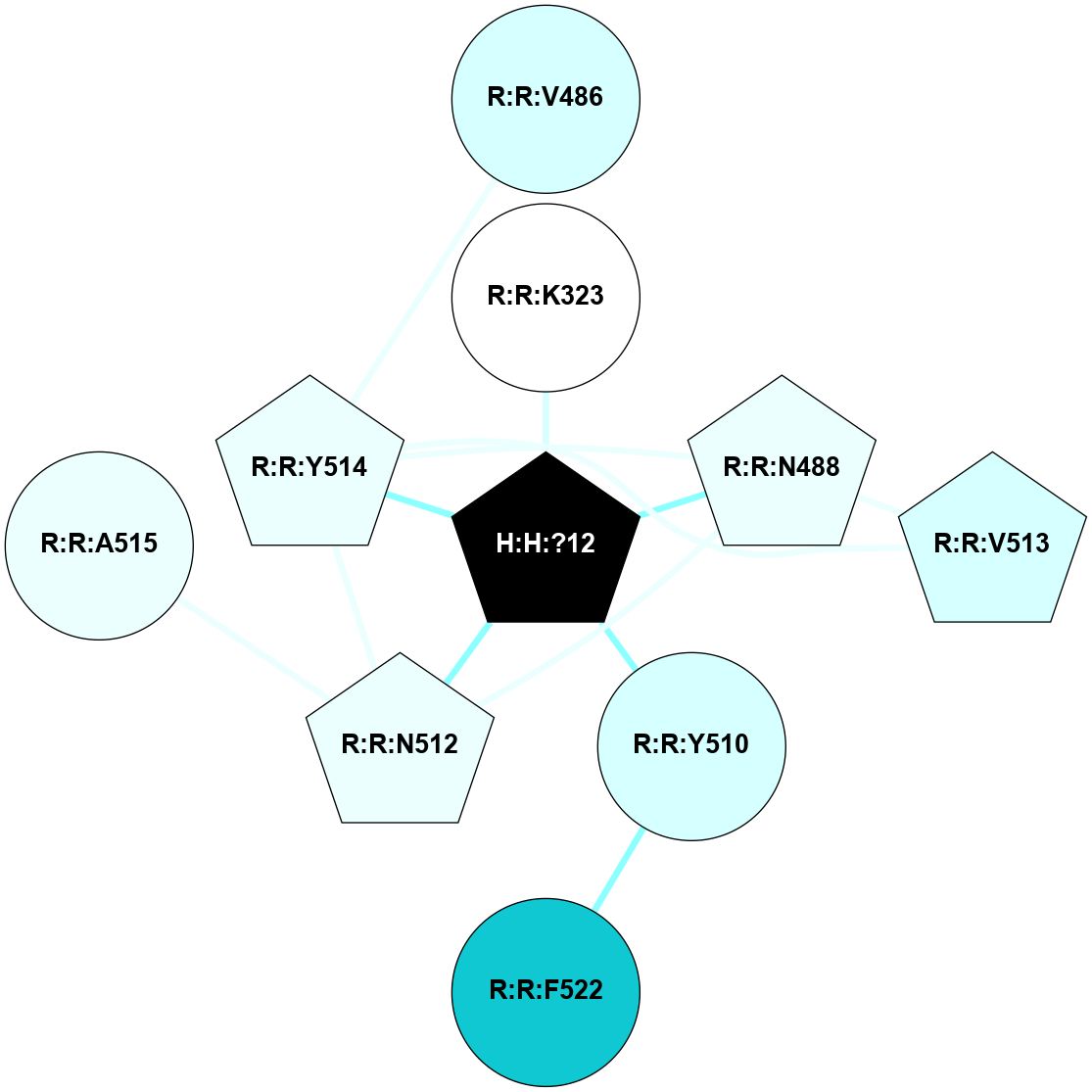
A 2D representation of the interactions of NAG in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | R:R:K323 | 5.04 | 24 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?12 | R:R:N488 | 24.56 | 24 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?12 | R:R:Y510 | 11.53 | 24 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?12 | R:R:N512 | 15.96 | 24 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?12 | R:R:Y514 | 12.58 | 24 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:V486 | R:R:Y514 | 3.79 | 0 | No | Yes | 3 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 6.81 | 24 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:V513 | 2.96 | 24 | Yes | Yes | 4 | 3 | 1 | 2 | | R:R:N488 | R:R:Y514 | 5.81 | 24 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:F522 | R:R:Y510 | 16.5 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:N512 | R:R:Y514 | 4.65 | 24 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | | R:R:V513 | R:R:Y514 | 5.05 | 24 | Yes | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 13.93 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
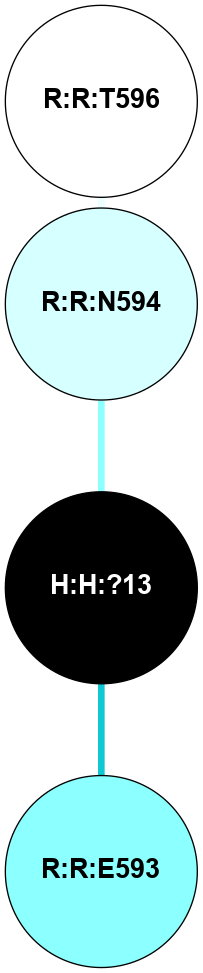
A 2D representation of the interactions of NAG in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | R:R:N594 | 27.02 | 0 | No | No | 0 | 3 | 0 | 1 | | R:R:N594 | R:R:T596 | 1.46 | 0 | No | No | 3 | 5 | 1 | 2 | | H:H:?13 | R:R:E593 | 1.18 | 0 | No | No | 0 | 2 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.10 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
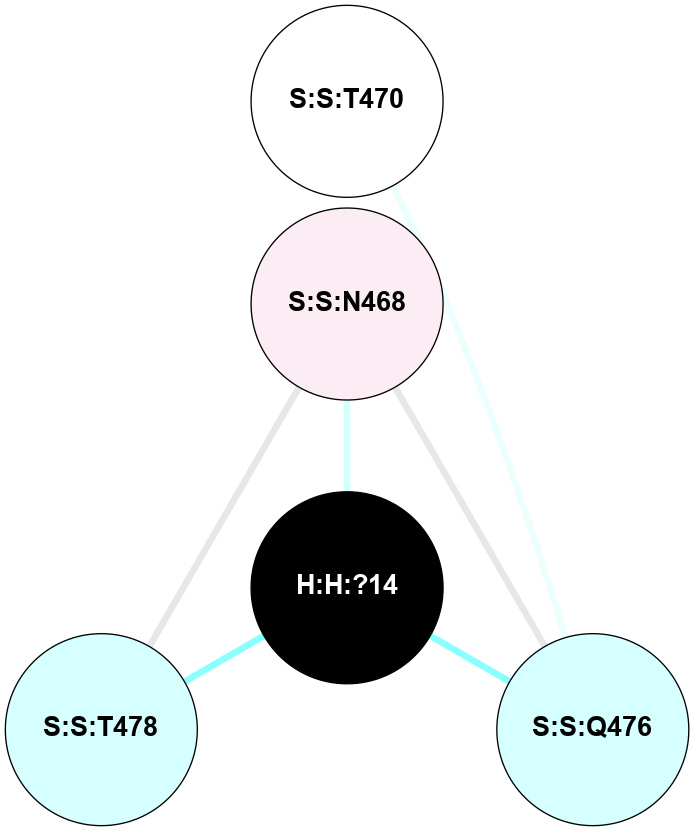
A 2D representation of the interactions of NAG in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?14 | S:S:N468 | 24.56 | 30 | No | No | 0 | 6 | 0 | 1 | | H:H:?14 | S:S:Q476 | 7.14 | 30 | No | No | 0 | 3 | 0 | 1 | | H:H:?14 | S:S:T478 | 6.59 | 30 | No | No | 0 | 3 | 0 | 1 | | S:S:N468 | S:S:Q476 | 6.6 | 30 | No | No | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 5.85 | 30 | No | No | 6 | 3 | 1 | 1 | | S:S:Q476 | S:S:T470 | 5.67 | 30 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.76 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
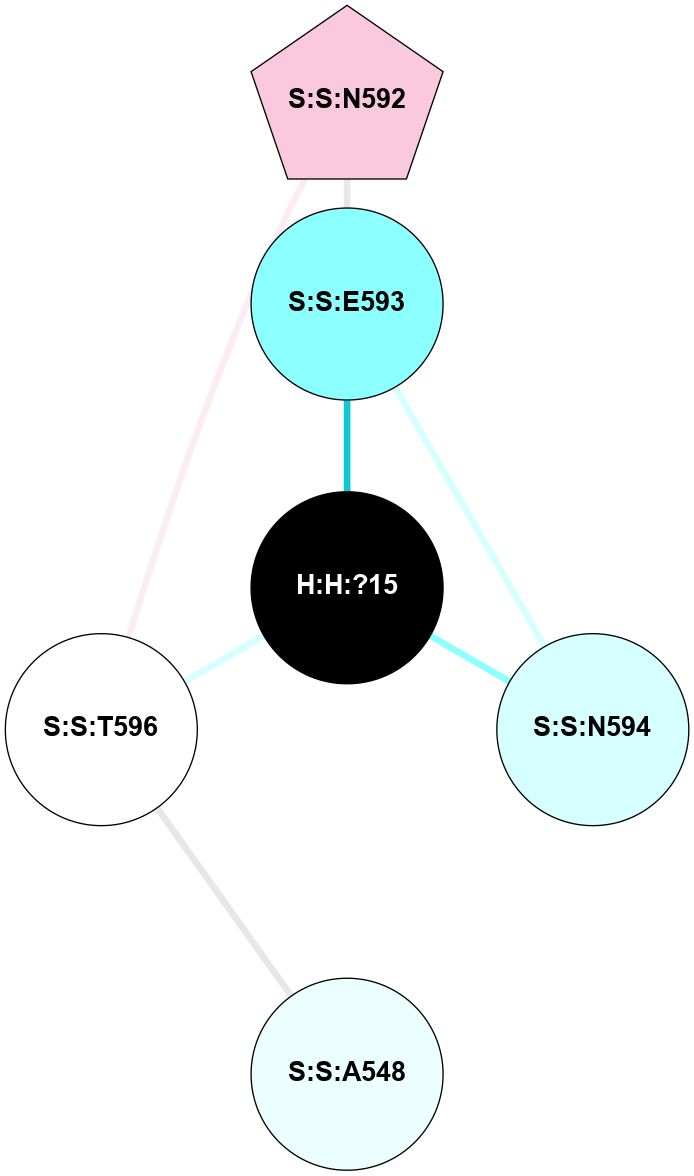
A 2D representation of the interactions of NAG in 7M3G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?15 | S:S:E593 | 7.11 | 36 | No | No | 0 | 2 | 0 | 1 | | H:H:?15 | S:S:N594 | 27.02 | 36 | No | No | 0 | 3 | 0 | 1 | | H:H:?15 | S:S:T596 | 5.27 | 36 | No | No | 0 | 5 | 0 | 1 | | S:S:E593 | S:S:N592 | 3.94 | 36 | No | Yes | 2 | 7 | 1 | 2 | | S:S:N592 | S:S:T596 | 2.92 | 0 | Yes | No | 7 | 5 | 2 | 1 | | S:S:E593 | S:S:N594 | 6.57 | 36 | No | No | 2 | 3 | 1 | 1 | | S:S:A548 | S:S:T596 | 1.68 | 0 | No | No | 4 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.13 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7M3J | C | Ion | Calcium Sensing | CaS | Homo Sapiens | - | NPS-2143; PO4 | - | 4.1 | 2021-06-30 | doi.org/10.1038/s41586-021-03691-0 |
|
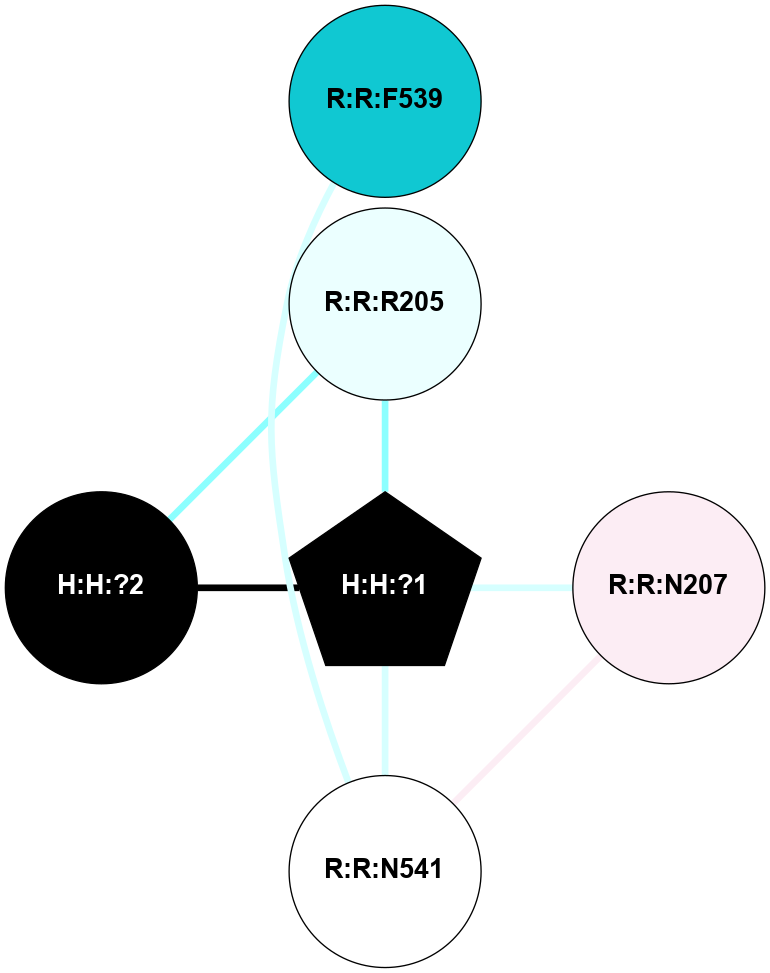
A 2D representation of the interactions of NAG in 7M3J
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 46.49 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:R205 | 2.17 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:N207 | 4.91 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:N541 | 33.15 | 1 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:R205 | 15.21 | 1 | No | No | 0 | 4 | 1 | 1 | | H:H:?2 | R:R:R233 | 5.43 | 1 | No | Yes | 0 | 4 | 1 | 2 | | R:R:R205 | R:R:R233 | 5.33 | 1 | No | Yes | 4 | 4 | 1 | 2 | | R:R:N207 | R:R:W208 | 3.39 | 1 | Yes | Yes | 6 | 9 | 1 | 2 | | R:R:C236 | R:R:N207 | 9.45 | 1 | Yes | Yes | 9 | 6 | 2 | 1 | | R:R:N207 | R:R:N541 | 4.09 | 1 | Yes | No | 6 | 5 | 1 | 1 | | R:R:C542 | R:R:N207 | 1.57 | 1 | Yes | Yes | 9 | 6 | 2 | 1 | | R:R:C561 | R:R:N207 | 11.02 | 1 | Yes | Yes | 9 | 6 | 2 | 1 | | R:R:C236 | R:R:W208 | 3.92 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:C542 | R:R:W208 | 14.37 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:C561 | R:R:W208 | 2.61 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:C236 | R:R:C561 | 7.28 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:F539 | R:R:N541 | 6.04 | 0 | No | No | 1 | 5 | 2 | 1 | | R:R:C542 | R:R:C561 | 1.82 | 1 | Yes | Yes | 9 | 9 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 21.68 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 7.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7M3J
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 46.49 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:R205 | 2.17 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?1 | R:R:N207 | 4.91 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:N541 | 33.15 | 1 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?2 | R:R:R205 | 15.21 | 1 | No | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:R233 | 5.43 | 1 | No | Yes | 0 | 4 | 0 | 1 | | R:R:E202 | R:R:R233 | 13.96 | 0 | No | Yes | 5 | 4 | 2 | 1 | | R:R:R205 | R:R:R233 | 5.33 | 1 | No | Yes | 4 | 4 | 1 | 1 | | R:R:N207 | R:R:N541 | 4.09 | 1 | Yes | No | 6 | 5 | 2 | 2 | | R:R:I235 | R:R:R233 | 11.27 | 0 | No | Yes | 7 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 22.38 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7M3J
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:E257 | 14.22 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?3 | R:R:N261 | 27.02 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:E257 | R:R:Q253 | 2.55 | 0 | No | No | 3 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.62 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
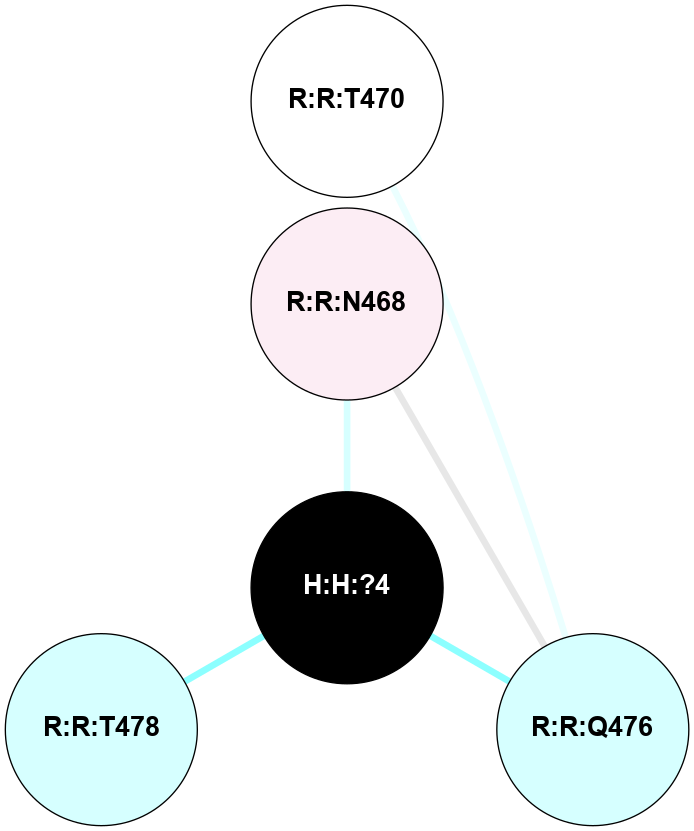
A 2D representation of the interactions of NAG in 7M3J
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:N468 | 30.7 | 26 | No | No | 0 | 6 | 0 | 1 | | H:H:?4 | R:R:Q476 | 8.33 | 26 | No | No | 0 | 3 | 0 | 1 | | H:H:?4 | R:R:T478 | 3.95 | 26 | No | No | 0 | 3 | 0 | 1 | | R:R:N468 | R:R:Q476 | 3.96 | 26 | No | No | 6 | 3 | 1 | 1 | | R:R:Q476 | R:R:T470 | 8.5 | 26 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.33 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
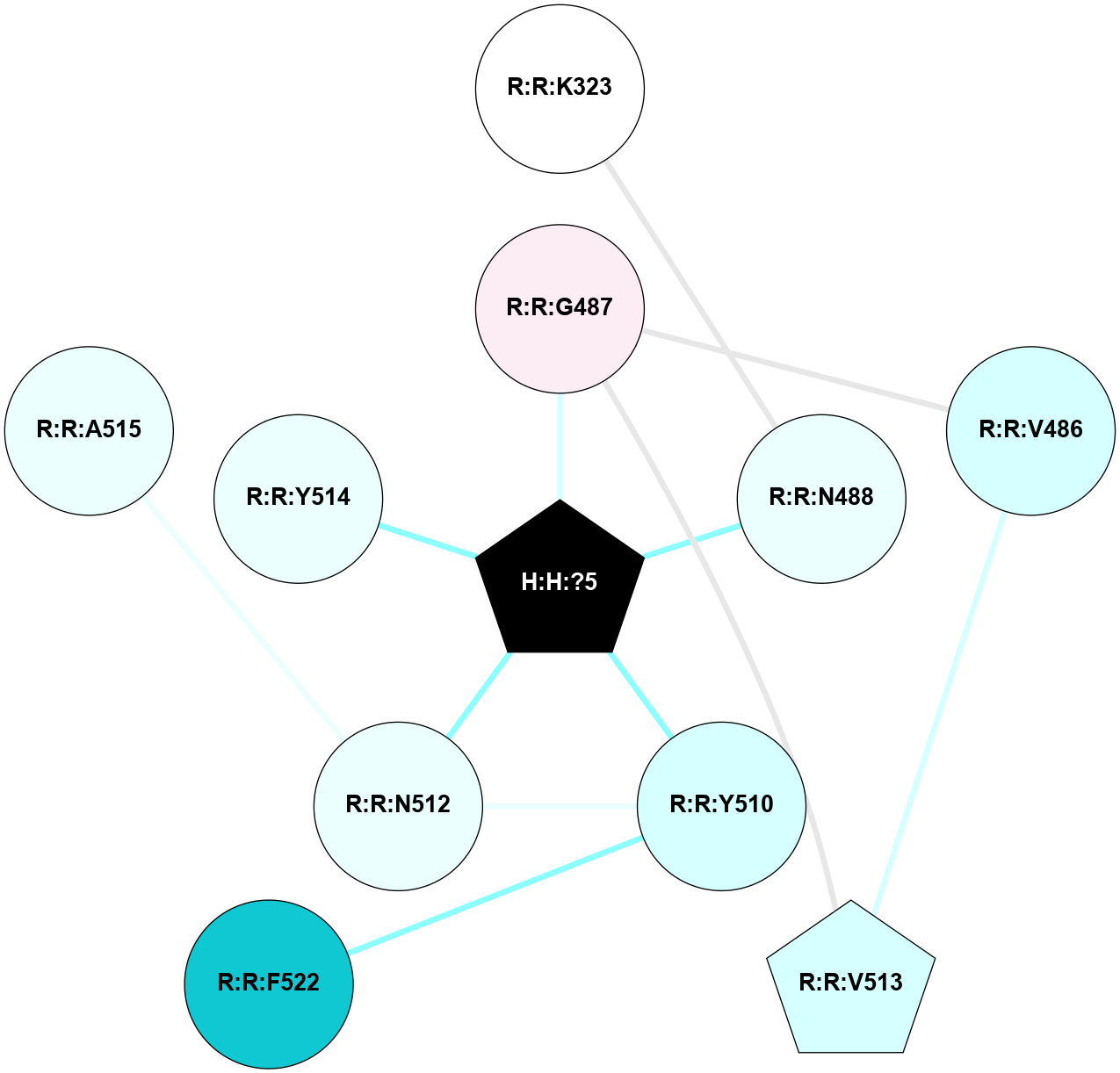
A 2D representation of the interactions of NAG in 7M3J
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | R:R:G487 | 1.53 | 17 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?5 | R:R:N488 | 28.24 | 17 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?5 | R:R:Y510 | 2.1 | 17 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?5 | R:R:N512 | 6.14 | 17 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?5 | R:R:Y514 | 3.15 | 17 | Yes | No | 0 | 4 | 0 | 1 | | R:R:K323 | R:R:N488 | 2.8 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:G487 | R:R:V486 | 1.84 | 17 | No | No | 6 | 3 | 1 | 2 | | R:R:V486 | R:R:V513 | 3.21 | 17 | No | Yes | 3 | 3 | 2 | 2 | | R:R:G487 | R:R:V513 | 1.84 | 17 | No | Yes | 6 | 3 | 1 | 2 | | R:R:N512 | R:R:Y510 | 8.14 | 17 | No | No | 4 | 3 | 1 | 1 | | R:R:F522 | R:R:Y510 | 12.38 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:A515 | R:R:N512 | 1.56 | 0 | No | No | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 8.23 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
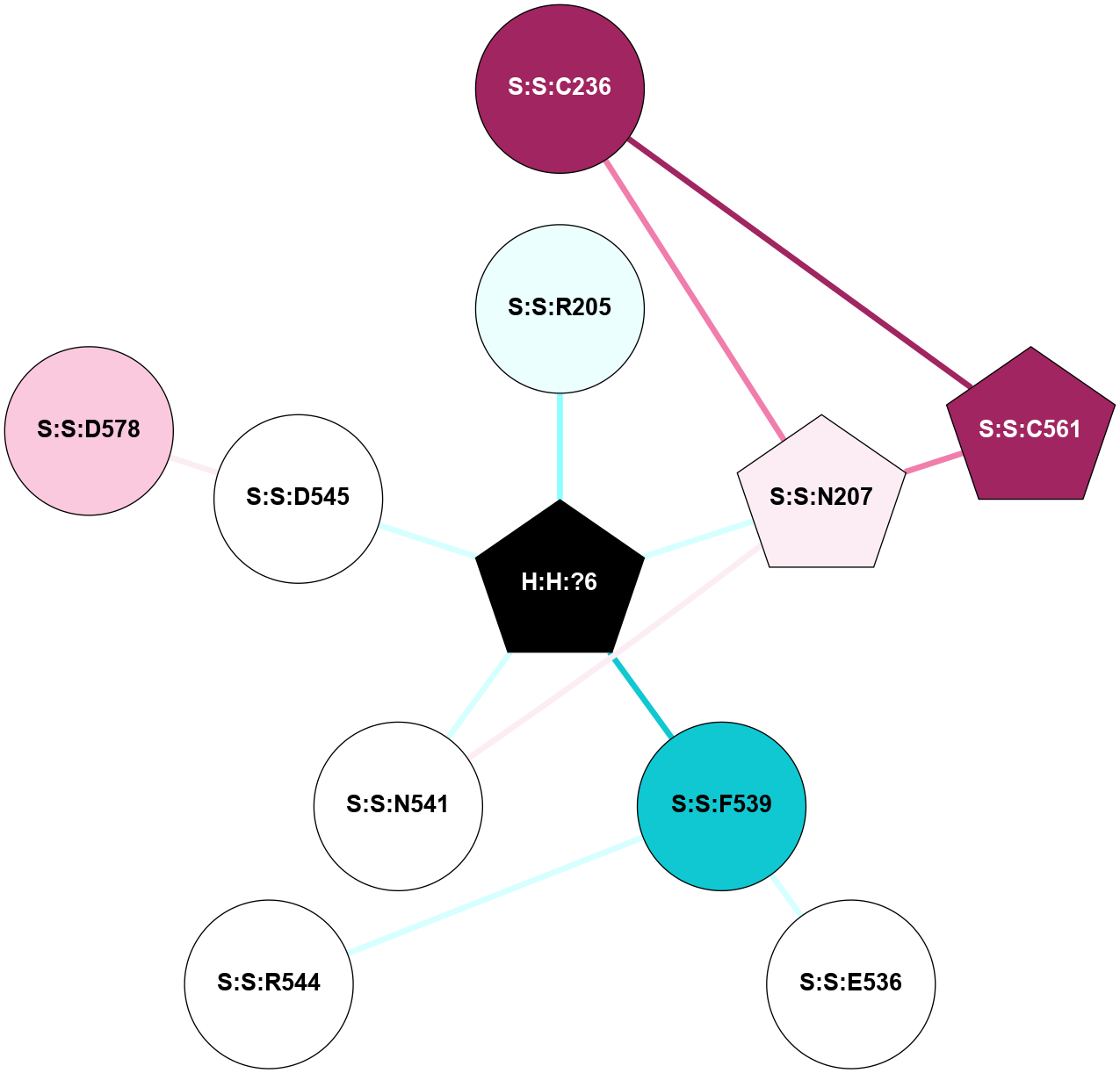
A 2D representation of the interactions of NAG in 7M3J
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | S:S:R205 | 2.17 | 5 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?6 | S:S:N207 | 9.82 | 5 | Yes | Yes | 0 | 6 | 0 | 1 | | H:H:?6 | S:S:F539 | 2.18 | 5 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?6 | S:S:N541 | 30.7 | 5 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?6 | S:S:D545 | 3.64 | 5 | Yes | No | 0 | 5 | 0 | 1 | | S:S:C236 | S:S:N207 | 1.57 | 5 | No | Yes | 9 | 6 | 2 | 1 | | S:S:N207 | S:S:N541 | 10.9 | 5 | Yes | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 1.57 | 5 | Yes | Yes | 9 | 6 | 2 | 1 | | S:S:C236 | S:S:C561 | 7.28 | 5 | No | Yes | 9 | 9 | 2 | 2 | | S:S:E536 | S:S:F539 | 4.66 | 0 | No | No | 5 | 1 | 2 | 1 | | S:S:F539 | S:S:R544 | 2.14 | 0 | No | No | 1 | 5 | 1 | 2 | | S:S:D545 | S:S:F563 | 1.19 | 0 | No | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 9.70 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
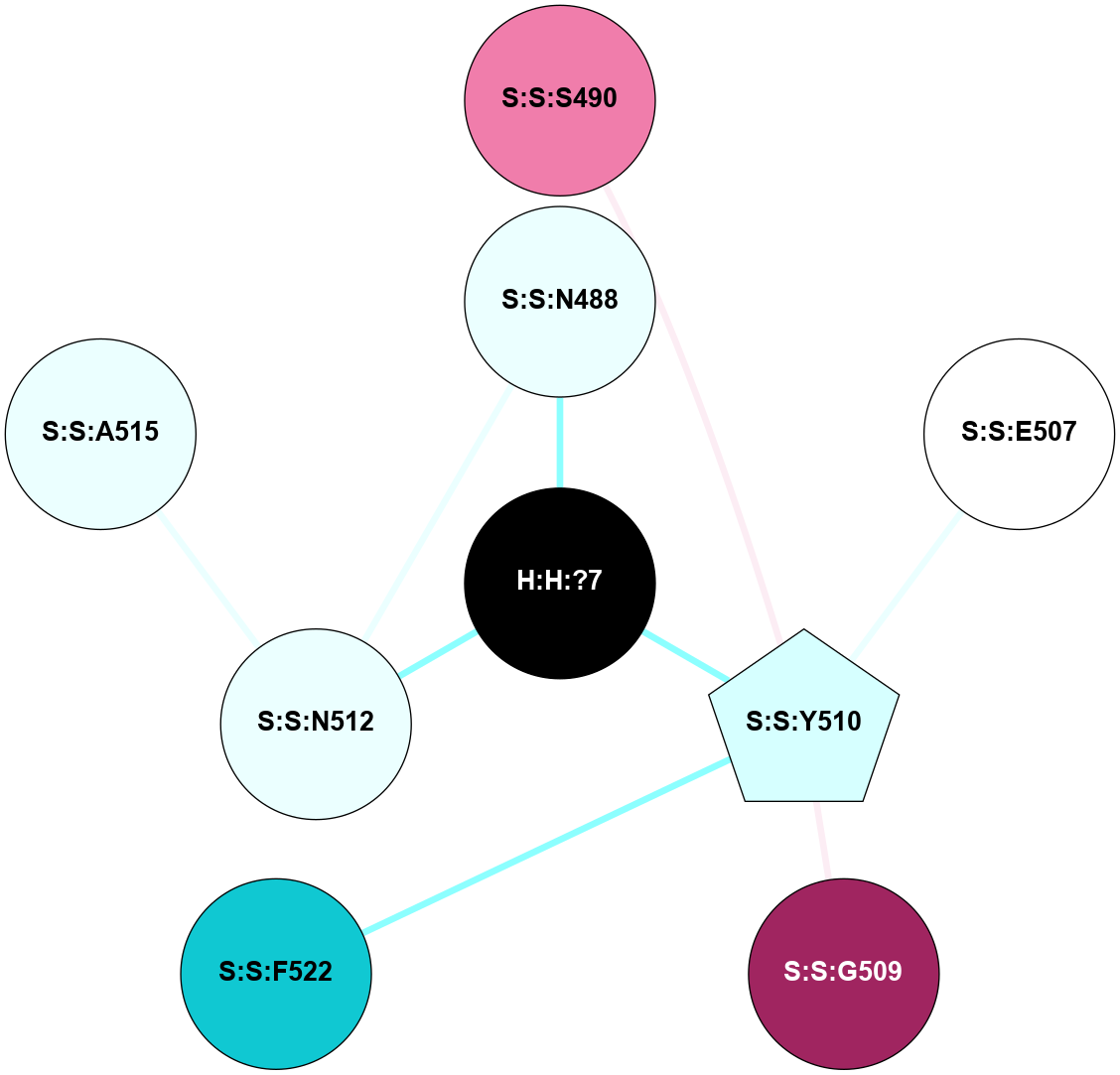
A 2D representation of the interactions of NAG in 7M3J
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | S:S:E475 | 5.92 | 27 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?7 | S:S:N488 | 31.93 | 27 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?7 | S:S:Y510 | 3.15 | 27 | Yes | Yes | 0 | 3 | 0 | 1 | | H:H:?7 | S:S:N512 | 11.05 | 27 | Yes | No | 0 | 4 | 0 | 1 | | S:S:N488 | S:S:N512 | 4.09 | 27 | No | No | 4 | 4 | 1 | 1 | | S:S:S490 | S:S:Y510 | 2.54 | 9 | No | Yes | 8 | 3 | 2 | 1 | | S:S:E507 | S:S:Y510 | 7.86 | 0 | No | Yes | 5 | 3 | 2 | 1 | | S:S:G509 | S:S:Y510 | 2.9 | 23 | No | Yes | 9 | 3 | 2 | 1 | | S:S:F522 | S:S:Y510 | 6.19 | 0 | No | Yes | 1 | 3 | 2 | 1 | | S:S:A515 | S:S:N512 | 3.13 | 0 | No | No | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 13.01 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7MTQ | C | Aminoacid | Metabotropic Glutamate | mGlu2; mGlu2 | Homo Sapiens | LY341495 | - | - | 3.65 | 2021-07-07 | doi.org/10.1038/s41586-021-03680-3 |
|

A 2D representation of the interactions of NAG in 7MTQ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N203 | 29.47 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:L495 | 3.71 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N203 | R:R:R200 | 3.62 | 0 | No | No | 4 | 4 | 1 | 2 | | R:R:L495 | R:R:P496 | 1.64 | 0 | No | No | 4 | 8 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.59 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7MTQ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | S:S:N203 | 29.47 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?2 | S:S:L495 | 3.71 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:L495 | S:S:P496 | 1.64 | 0 | No | No | 4 | 8 | 1 | 2 | | S:S:A488 | S:S:L495 | 1.58 | 0 | No | No | 1 | 4 | 2 | 1 | | S:S:N203 | S:S:R499 | 1.21 | 0 | No | No | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.59 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7MTR | C | Aminoacid | Metabotropic Glutamate | mGlu2; mGlu2 | Homo Sapiens | Glutamate | ADX55164 | - | 3.3 | 2021-07-07 | doi.org/10.1038/s41586-021-03680-3 |
|
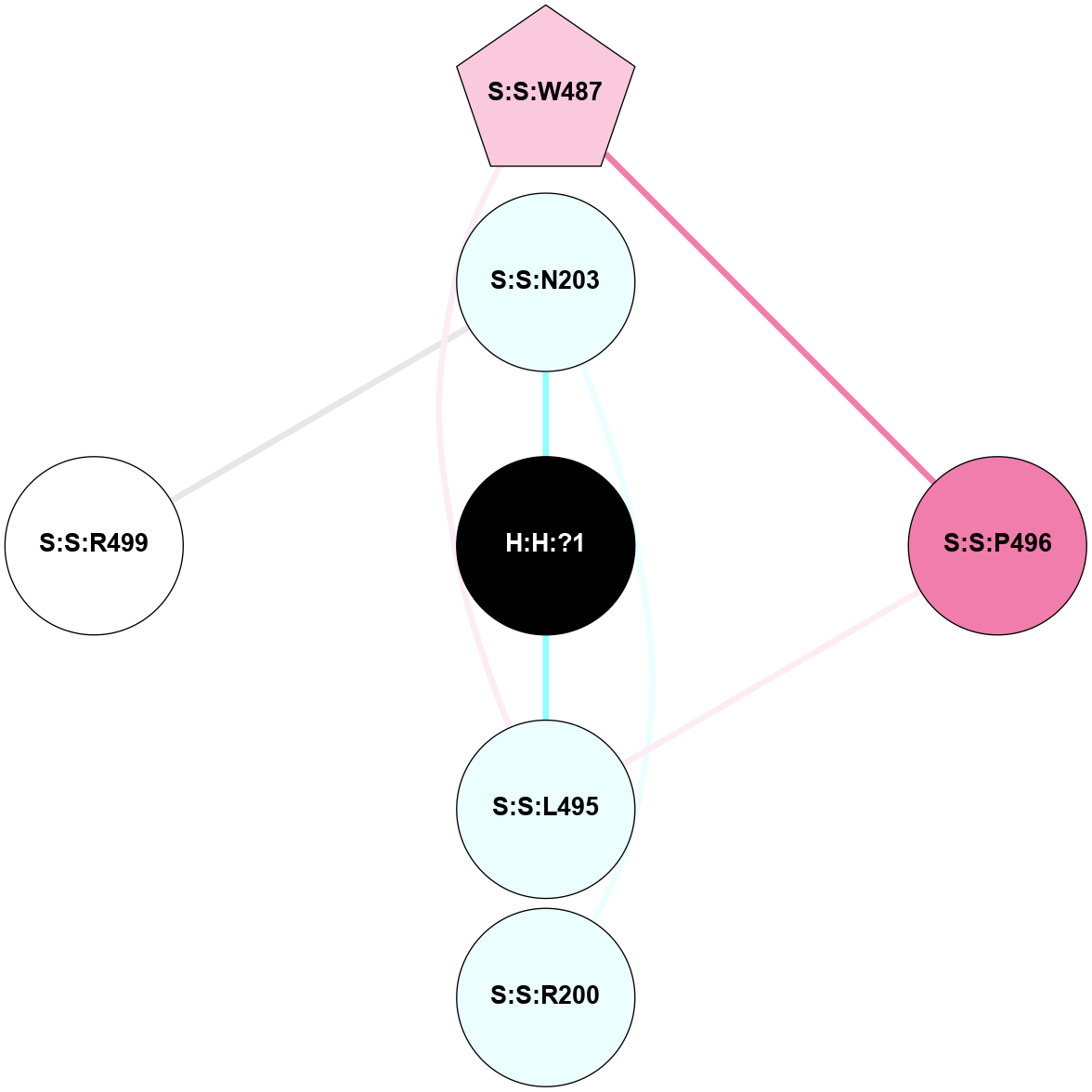
A 2D representation of the interactions of NAG in 7MTR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | S:S:N203 | 29.47 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:L495 | S:S:W487 | 2.28 | 20 | No | Yes | 4 | 7 | 1 | 2 | | S:S:P496 | S:S:W487 | 2.7 | 20 | No | Yes | 8 | 7 | 2 | 2 | | S:S:L495 | S:S:P496 | 3.28 | 20 | No | No | 4 | 8 | 1 | 2 | | H:H:?1 | S:S:L495 | 1.24 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N203 | S:S:R200 | 1.21 | 0 | No | No | 4 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.35 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
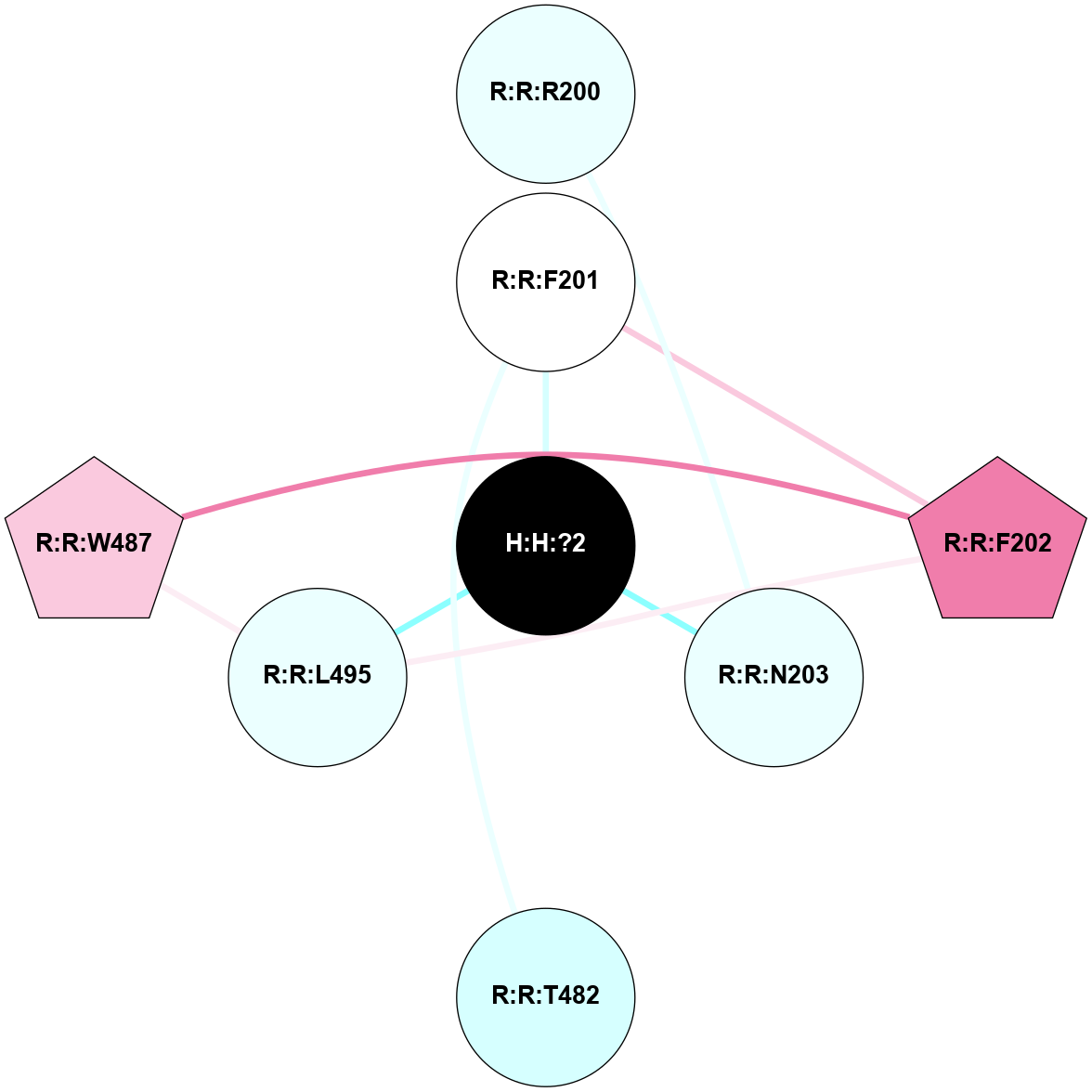
A 2D representation of the interactions of NAG in 7MTR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:F201 | 2.18 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:N203 | 33.15 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:L495 | 3.71 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N203 | R:R:R200 | 3.62 | 0 | No | No | 4 | 4 | 1 | 2 | | R:R:F201 | R:R:F202 | 9.65 | 0 | No | Yes | 5 | 8 | 1 | 2 | | R:R:F201 | R:R:T482 | 2.59 | 0 | No | No | 5 | 3 | 1 | 2 | | R:R:F202 | R:R:W487 | 11.02 | 1 | Yes | Yes | 8 | 7 | 2 | 2 | | R:R:F202 | R:R:L495 | 2.44 | 1 | Yes | No | 8 | 4 | 2 | 1 | | R:R:L495 | R:R:W487 | 3.42 | 1 | No | Yes | 4 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.01 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7MTS | C | Aminoacid | Metabotropic Glutamate | mGlu2; mGlu2 | Homo Sapiens | Glutamate | ADX55164 | Gi1/Beta1/Gamma2 | 3.2 | 2021-07-07 | doi.org/10.1038/s41586-021-03680-3 |
|
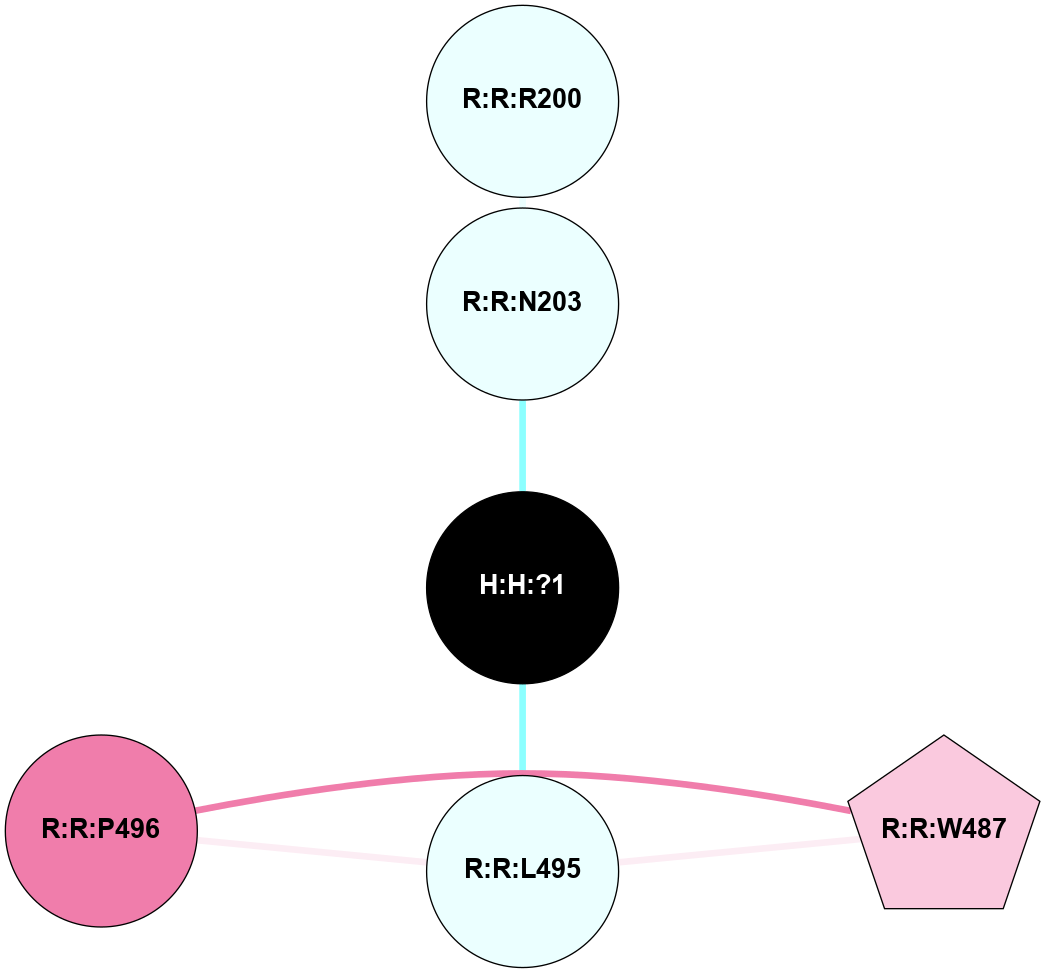
A 2D representation of the interactions of NAG in 7MTS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N203 | 27.02 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:L495 | 3.71 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N203 | R:R:R200 | 6.03 | 0 | No | No | 4 | 4 | 1 | 2 | | R:R:L495 | R:R:W487 | 2.28 | 1 | No | Yes | 4 | 7 | 1 | 2 | | R:R:P496 | R:R:W487 | 4.05 | 1 | No | Yes | 8 | 7 | 2 | 2 | | R:R:L495 | R:R:P496 | 3.28 | 1 | No | No | 4 | 8 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.37 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
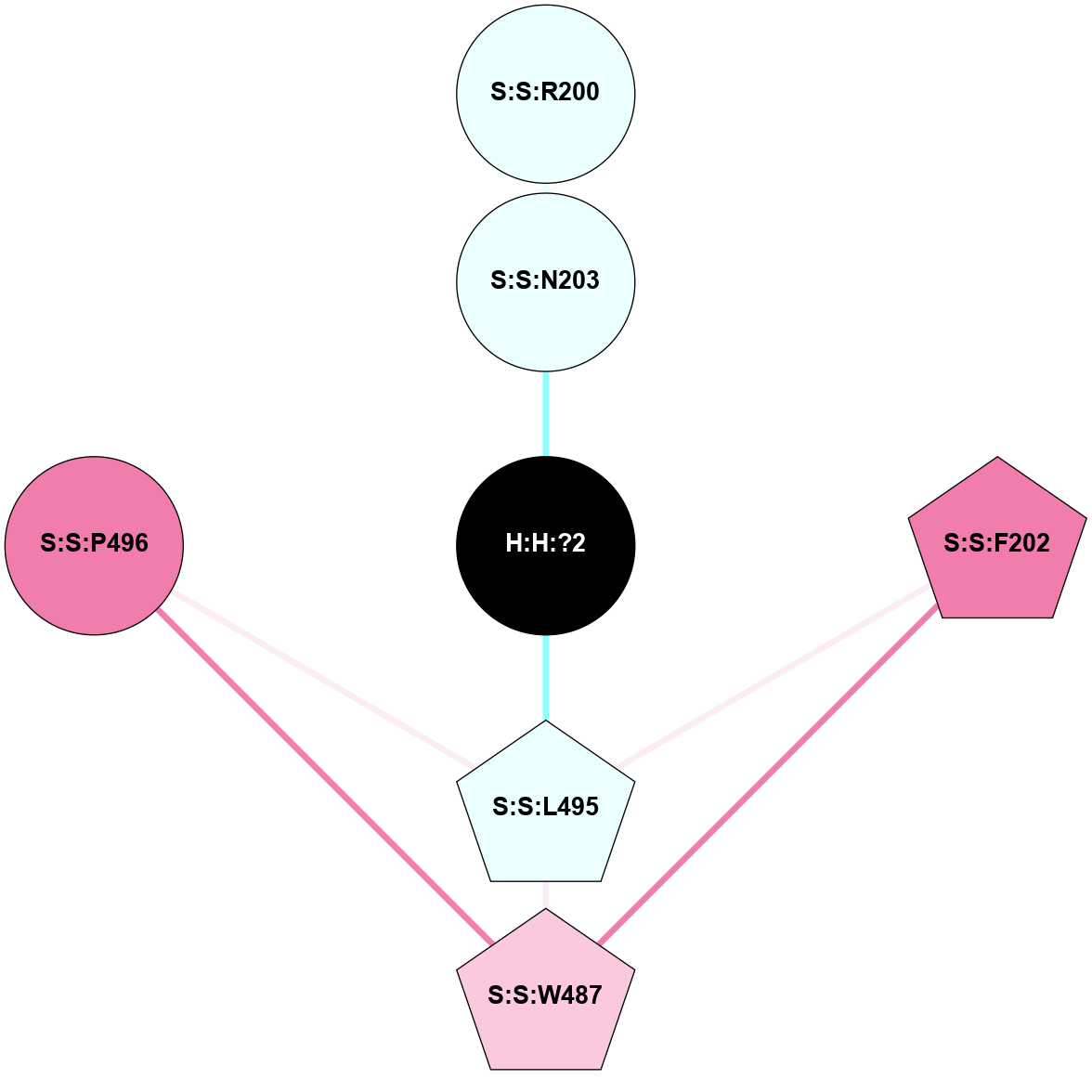
A 2D representation of the interactions of NAG in 7MTS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | S:S:N203 | 29.47 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?2 | S:S:L495 | 2.48 | 0 | No | Yes | 0 | 4 | 0 | 1 | | S:S:N203 | S:S:R200 | 4.82 | 0 | No | No | 4 | 4 | 1 | 2 | | S:S:F202 | S:S:W487 | 5.01 | 3 | Yes | Yes | 8 | 7 | 2 | 2 | | S:S:F202 | S:S:L495 | 2.44 | 3 | Yes | Yes | 8 | 4 | 2 | 1 | | S:S:L495 | S:S:W487 | 2.28 | 3 | Yes | Yes | 4 | 7 | 1 | 2 | | S:S:P496 | S:S:W487 | 6.76 | 3 | No | Yes | 8 | 7 | 2 | 2 | | S:S:L495 | S:S:P496 | 4.93 | 3 | Yes | No | 4 | 8 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.97 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7QA8 | D1 | Ste2-like | STE2 | STE2 | Saccharomyces Cerevisiae | Peptide | - | - | 2.7 | 2022-03-16 | doi.org/10.1038/s41586-022-04498-3 |
|

A 2D representation of the interactions of NAG in 7QA8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N25 | 29.47 | 0 | No | No | 0 | 7 | 0 | 1 | | H:H:?4 | R:R:N25 | 2.46 | 0 | Yes | No | 0 | 7 | 2 | 1 | | R:R:N25 | R:R:T23 | 2.92 | 0 | No | No | 7 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 29.47 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
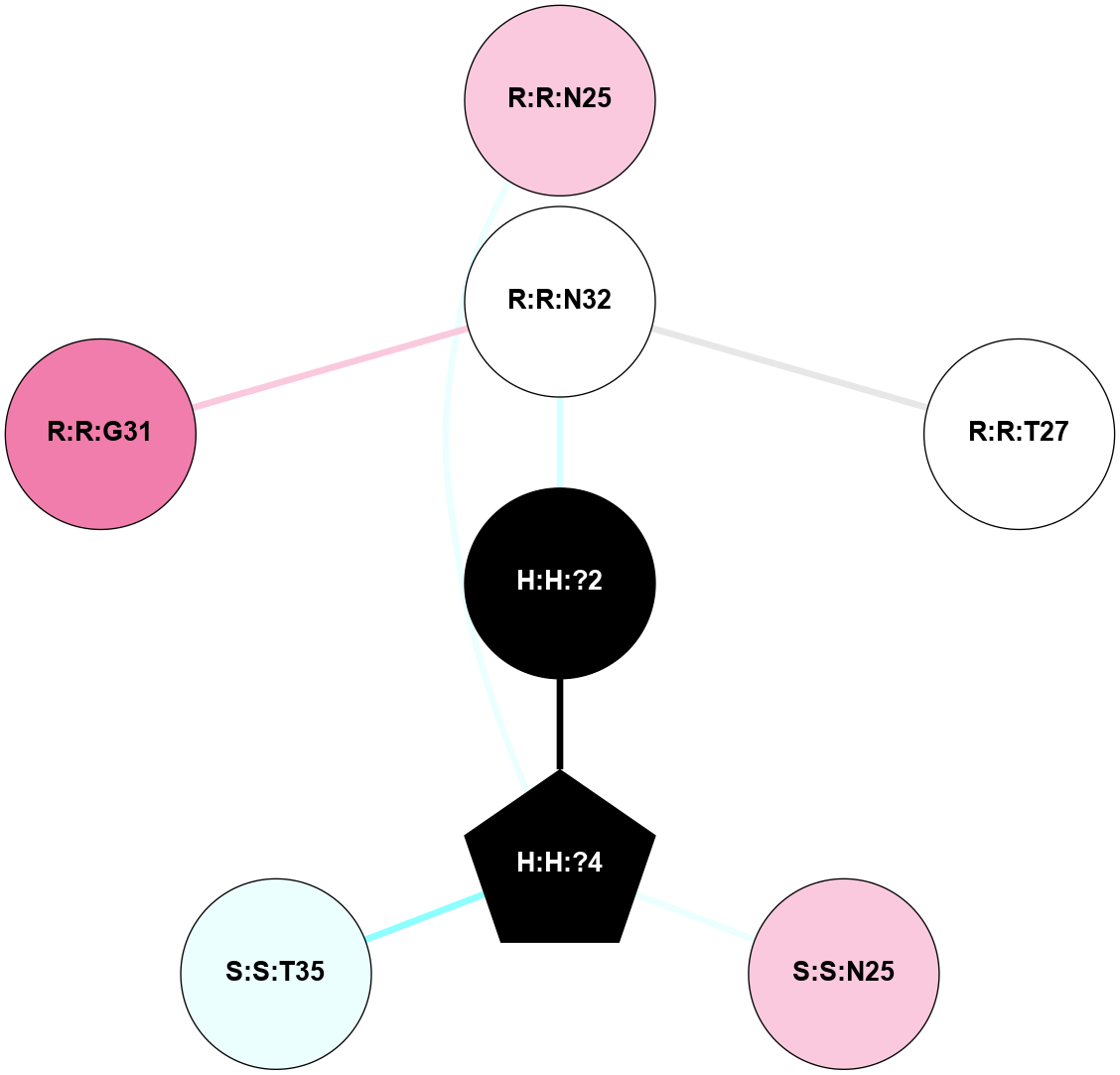
A 2D representation of the interactions of NAG in 7QA8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:N32 | 24.56 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?4 | R:R:N25 | 2.46 | 0 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?4 | S:S:N25 | 28.24 | 0 | Yes | No | 0 | 7 | 1 | 2 | | H:H:?4 | S:S:T35 | 2.64 | 0 | Yes | No | 0 | 4 | 1 | 2 | | R:R:N32 | R:R:T27 | 4.39 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:G31 | R:R:N32 | 1.7 | 0 | No | No | 8 | 5 | 2 | 1 | | H:H:?2 | H:H:?4 | 1.11 | 0 | No | Yes | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 12.83 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7QA8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | S:S:N32 | 24.56 | 0 | No | No | 0 | 5 | 0 | 1 | | S:S:N32 | S:S:T27 | 4.39 | 0 | No | No | 5 | 5 | 1 | 2 | | S:S:G31 | S:S:N32 | 1.7 | 0 | No | No | 8 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 24.56 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
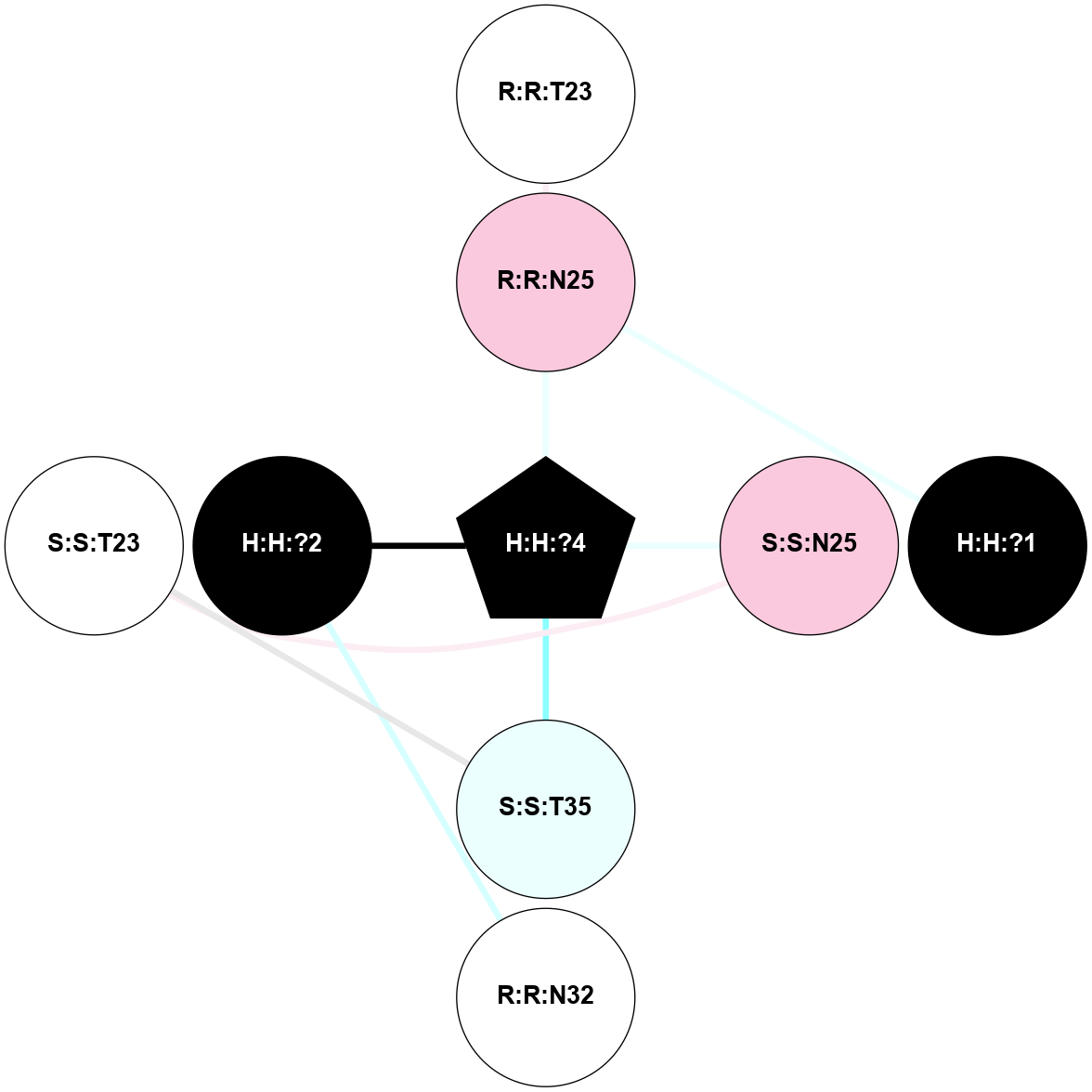
A 2D representation of the interactions of NAG in 7QA8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N25 | 29.47 | 0 | No | No | 0 | 7 | 2 | 1 | | H:H:?2 | R:R:N32 | 24.56 | 0 | No | No | 0 | 5 | 1 | 2 | | H:H:?4 | R:R:N25 | 2.46 | 0 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?4 | S:S:N25 | 28.24 | 0 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?4 | S:S:T35 | 2.64 | 0 | Yes | No | 0 | 4 | 0 | 1 | | R:R:N25 | R:R:T23 | 2.92 | 0 | No | No | 7 | 5 | 1 | 2 | | S:S:N25 | S:S:T23 | 2.92 | 0 | No | No | 7 | 5 | 1 | 2 | | S:S:T23 | S:S:T35 | 3.14 | 0 | No | No | 5 | 4 | 2 | 1 | | H:H:?2 | H:H:?4 | 1.11 | 0 | No | Yes | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 8.61 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7QB9 | D1 | Ste2-like | STE2 | STE2 | Saccharomyces Cerevisiae | - | - | - | 3.1 | 2022-03-16 | doi.org/10.1038/s41586-022-04498-3 |
|
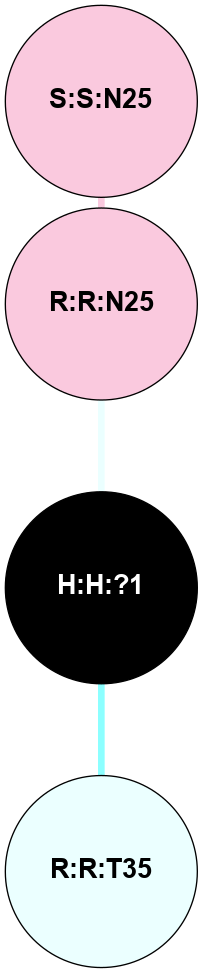
A 2D representation of the interactions of NAG in 7QB9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N25 | 28.24 | 0 | No | No | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:T35 | 2.64 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N25 | S:S:N25 | 6.81 | 0 | No | No | 7 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.44 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7QB9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:N32 | 29.47 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:N32 | R:R:T27 | 4.39 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:G31 | R:R:N32 | 1.7 | 0 | No | No | 8 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 29.47 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
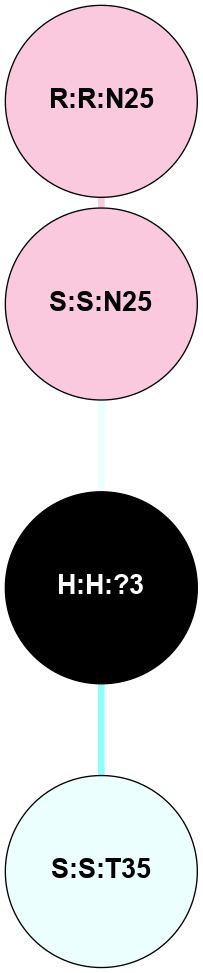
A 2D representation of the interactions of NAG in 7QB9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | S:S:N25 | 28.24 | 0 | No | No | 0 | 7 | 0 | 1 | | H:H:?3 | S:S:T35 | 2.64 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N25 | S:S:N25 | 6.81 | 0 | No | No | 7 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.44 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7QB9
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | S:S:N32 | 29.47 | 0 | No | No | 0 | 5 | 0 | 1 | | S:S:N32 | S:S:T27 | 4.39 | 0 | No | No | 5 | 5 | 1 | 2 | | S:S:G31 | S:S:N32 | 1.7 | 0 | No | No | 8 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 29.47 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7QBC | D1 | Ste2-like | STE2 | STE2 | Saccharomyces Cerevisiae | Alpha-factor mating pheromone | - | - | 3.53 | 2022-03-16 | doi.org/10.1038/s41586-022-04498-3 |
|
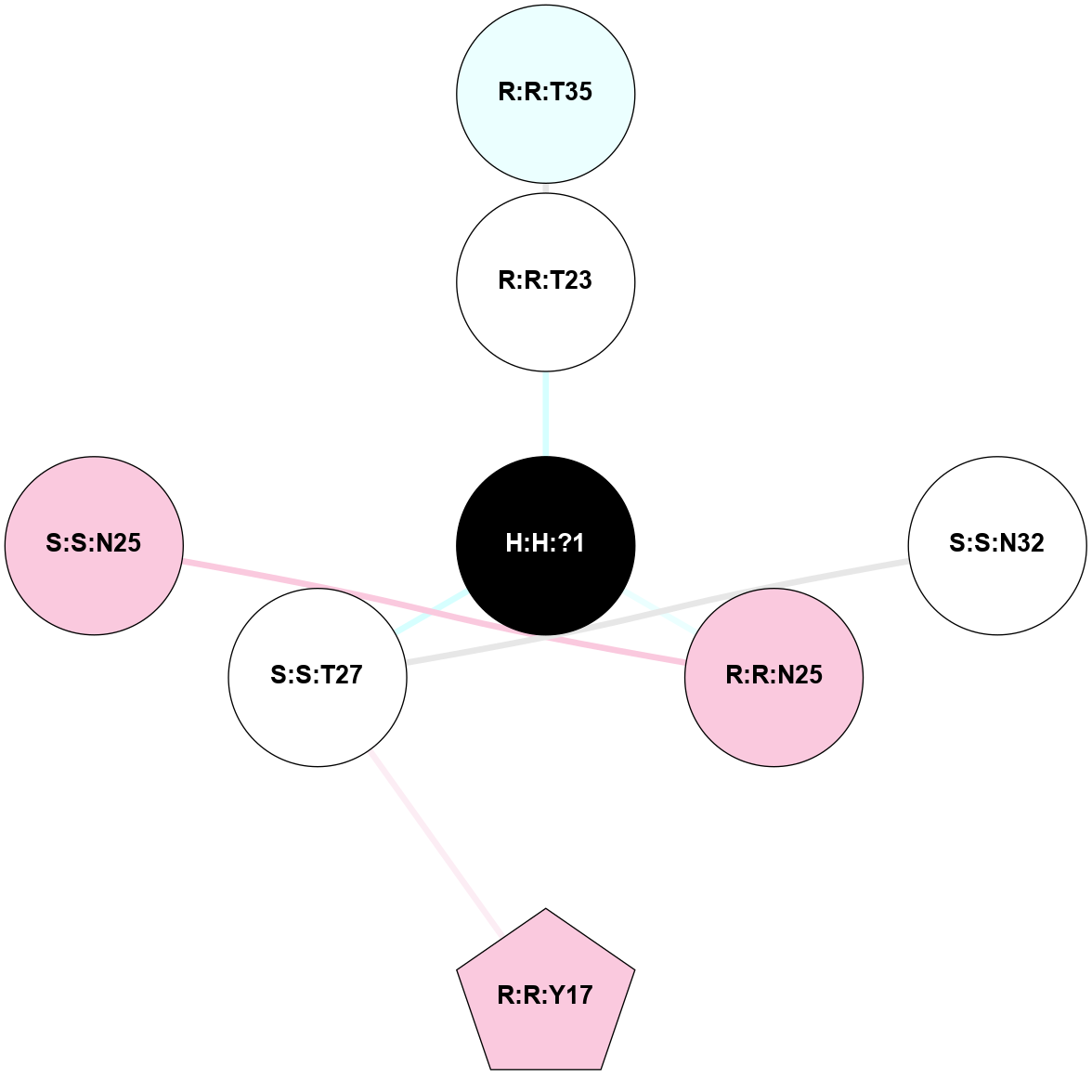
A 2D representation of the interactions of NAG in 7QBC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:T23 | 6.59 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:N25 | 30.7 | 0 | No | No | 0 | 7 | 0 | 1 | | R:R:T23 | R:R:T35 | 3.14 | 0 | No | No | 5 | 4 | 1 | 2 | | S:S:N32 | S:S:T27 | 2.92 | 0 | No | No | 5 | 5 | 2 | 1 | | R:R:Y17 | S:S:T27 | 2.5 | 0 | Yes | No | 7 | 5 | 2 | 1 | | R:R:N25 | S:S:N25 | 1.36 | 0 | No | No | 7 | 7 | 1 | 2 | | H:H:?1 | S:S:T27 | 1.32 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.87 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7QBC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:N32 | 34.38 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:N32 | R:R:T27 | 2.92 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:G33 | R:R:N32 | 1.7 | 0 | No | No | 2 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 34.38 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
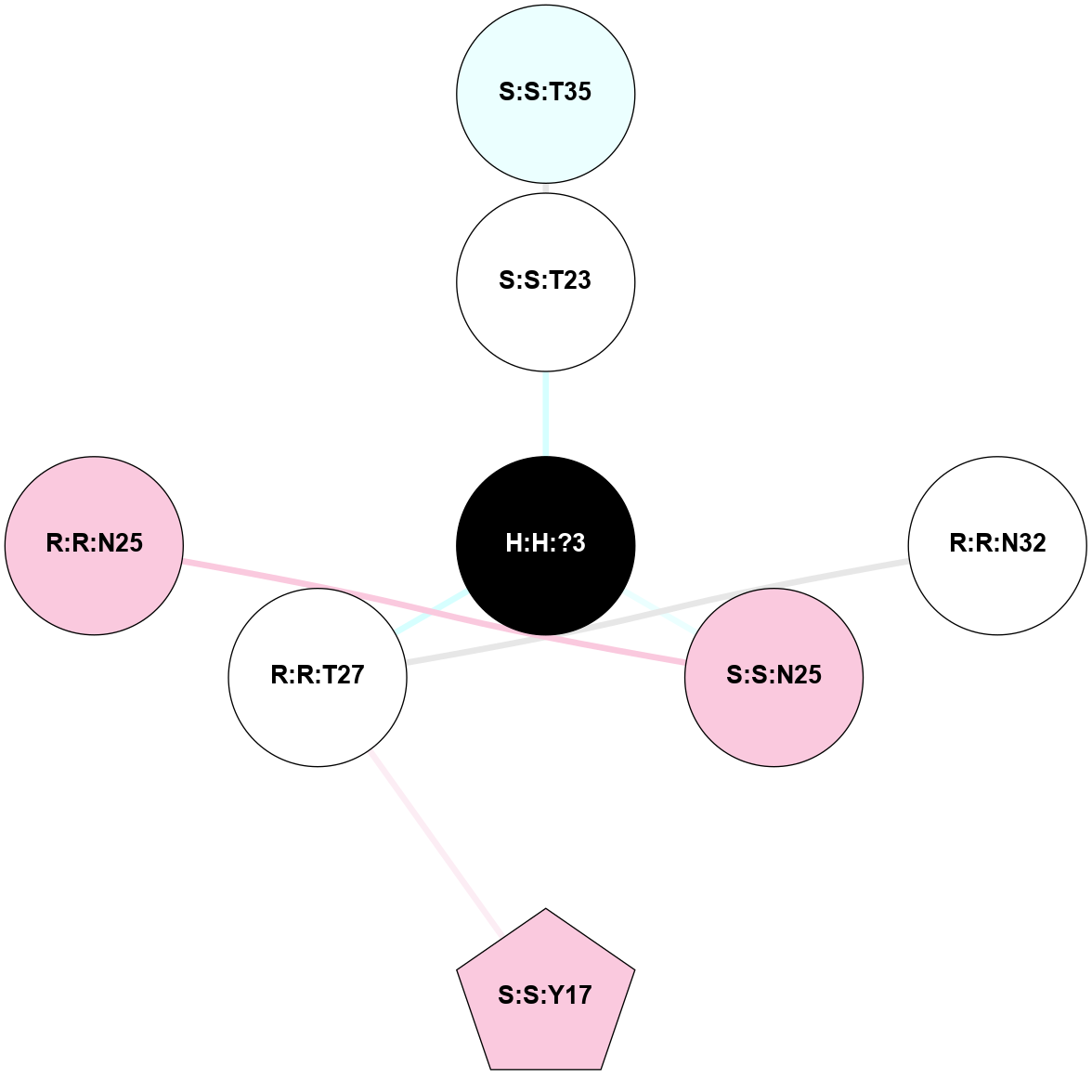
A 2D representation of the interactions of NAG in 7QBC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | S:S:T23 | 6.59 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?3 | S:S:N25 | 30.7 | 0 | No | No | 0 | 7 | 0 | 1 | | S:S:T23 | S:S:T35 | 3.14 | 0 | No | No | 5 | 4 | 1 | 2 | | R:R:N32 | R:R:T27 | 2.92 | 0 | No | No | 5 | 5 | 2 | 1 | | R:R:T27 | S:S:Y17 | 2.5 | 0 | No | Yes | 5 | 7 | 1 | 2 | | R:R:N25 | S:S:N25 | 1.36 | 0 | No | No | 7 | 7 | 2 | 1 | | H:H:?3 | R:R:T27 | 1.32 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.87 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7QBC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | S:S:N32 | 34.38 | 0 | No | No | 0 | 5 | 0 | 1 | | S:S:N32 | S:S:T27 | 2.92 | 0 | No | No | 5 | 5 | 1 | 2 | | S:S:G33 | S:S:N32 | 1.7 | 0 | No | No | 2 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 34.38 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7QBI | D1 | Ste2-like | STE2 | STE2 | Saccharomyces Cerevisiae | Alpha-factor mating pheromone | - | - | 3.46 | 2022-03-16 | doi.org/10.1038/s41586-022-04498-3 |
|
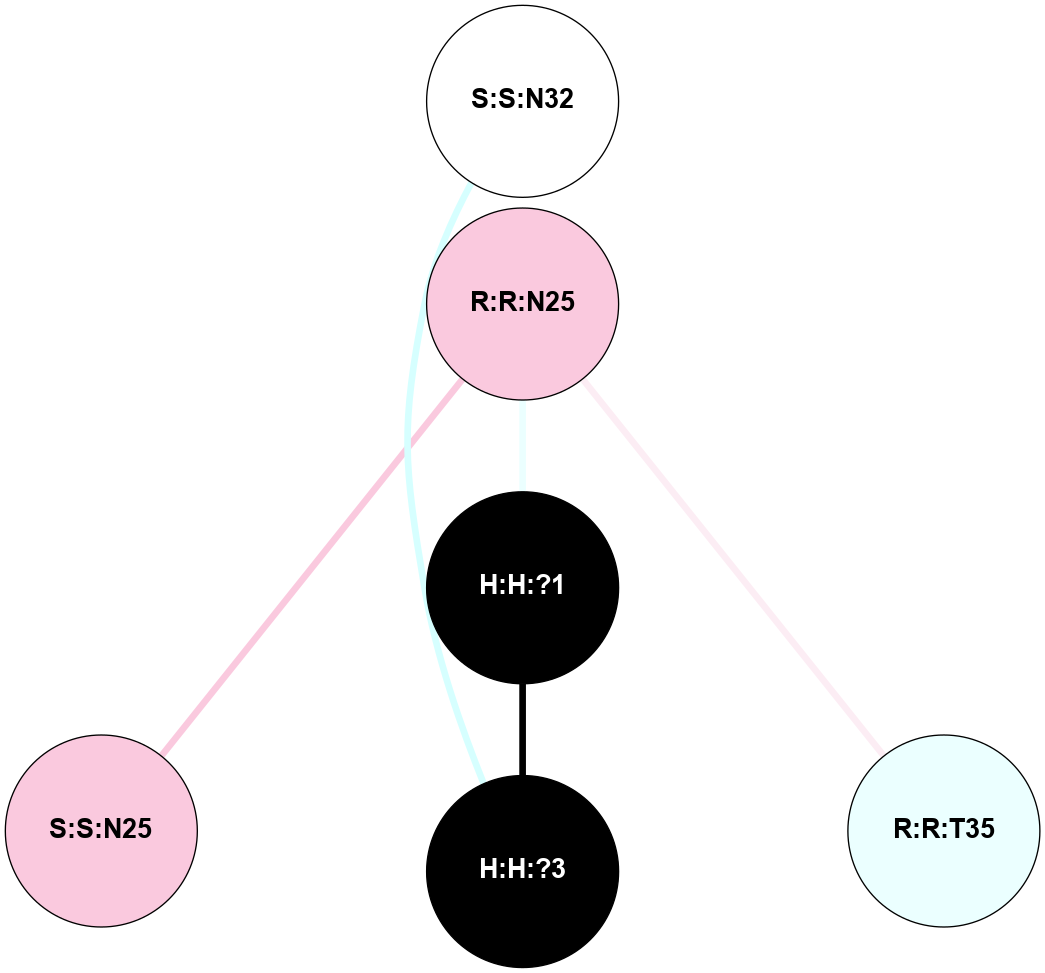
A 2D representation of the interactions of NAG in 7QBI
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?3 | 3.32 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N25 | 25.79 | 0 | No | No | 0 | 7 | 0 | 1 | | H:H:?3 | S:S:N32 | 23.33 | 0 | No | No | 0 | 5 | 1 | 2 | | R:R:N25 | R:R:T35 | 5.85 | 0 | No | No | 7 | 4 | 1 | 2 | | R:R:N25 | S:S:N25 | 1.36 | 0 | No | No | 7 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.55 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
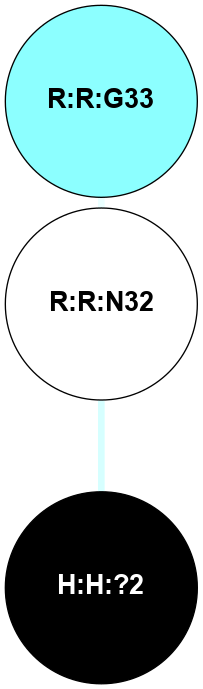
A 2D representation of the interactions of NAG in 7QBI
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:N32 | 23.33 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:G33 | R:R:N32 | 1.7 | 0 | No | No | 2 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 23.33 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
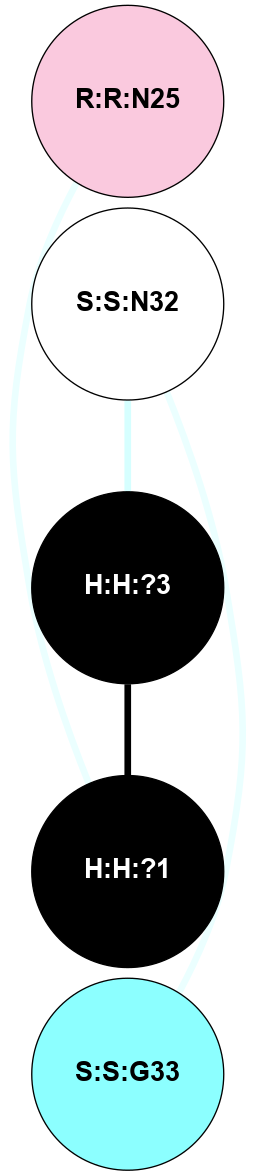
A 2D representation of the interactions of NAG in 7QBI
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?3 | 3.32 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N25 | 25.79 | 0 | No | No | 0 | 7 | 1 | 2 | | H:H:?3 | S:S:N32 | 23.33 | 0 | No | No | 0 | 5 | 0 | 1 | | S:S:G33 | S:S:N32 | 1.7 | 0 | No | No | 2 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.32 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
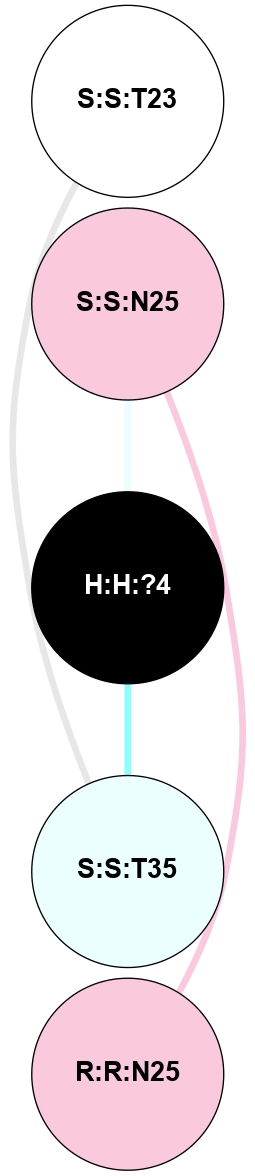
A 2D representation of the interactions of NAG in 7QBI
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | S:S:N25 | 27.02 | 8 | No | No | 0 | 7 | 0 | 1 | | H:H:?4 | S:S:T35 | 5.27 | 8 | No | No | 0 | 4 | 0 | 1 | | S:S:T23 | S:S:T35 | 7.85 | 0 | No | No | 5 | 4 | 2 | 1 | | S:S:N25 | S:S:T35 | 4.39 | 8 | No | No | 7 | 4 | 1 | 1 | | R:R:N25 | S:S:N25 | 1.36 | 0 | No | No | 7 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.14 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7RKF | A | Other | Unclassified | US28 | Human Betaherpesvirus 5 | Fractalkine | GDP | chim(NtGi1-G11)/Beta1/Gamma2 | 4 | 2022-01-26 | doi.org/10.1126/sciadv.abl5442 |
|
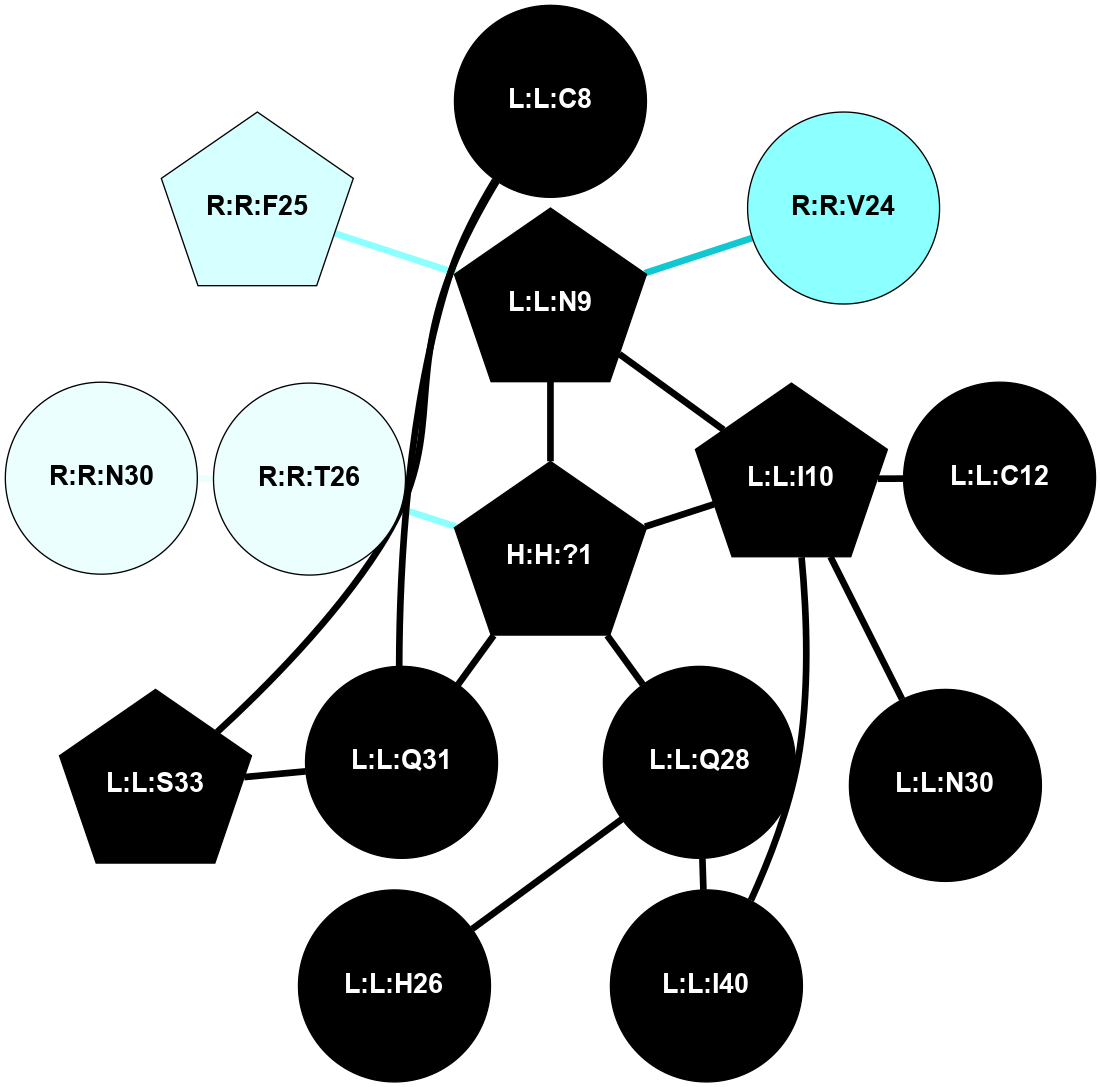
A 2D representation of the interactions of NAG in 7RKF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | L:L:N9 | 23.41 | 5 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?1 | L:L:I10 | 2.56 | 5 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?1 | L:L:Q28 | 4.78 | 5 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | L:L:Q31 | 5.97 | 5 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:T26 | 11.9 | 5 | Yes | No | 0 | 4 | 0 | 1 | | L:L:C8 | L:L:Q31 | 3.05 | 5 | No | No | 0 | 0 | 2 | 1 | | L:L:C8 | L:L:S33 | 3.44 | 5 | No | Yes | 0 | 0 | 2 | 2 | | L:L:I10 | L:L:N9 | 7.08 | 5 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:N9 | R:R:V24 | 5.91 | 5 | Yes | No | 0 | 2 | 1 | 2 | | L:L:C12 | L:L:I10 | 3.27 | 5 | No | Yes | 0 | 0 | 2 | 1 | | L:L:I10 | L:L:N30 | 2.83 | 5 | Yes | No | 0 | 0 | 1 | 2 | | L:L:I10 | L:L:I40 | 2.94 | 5 | Yes | No | 0 | 0 | 1 | 2 | | L:L:H26 | L:L:Q28 | 6.18 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:I40 | L:L:Q28 | 4.12 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:Q31 | L:L:S33 | 7.22 | 5 | No | Yes | 0 | 0 | 1 | 2 | | R:R:N30 | R:R:T26 | 5.85 | 0 | No | No | 4 | 4 | 2 | 1 | | L:L:N9 | R:R:F25 | 1.21 | 5 | Yes | Yes | 0 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.72 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7SIL | C | Ion | Calcium Sensing | CaS | Homo Sapiens | Ca | Ca; Ca; Ca; Cyclomethyltryptophan; 9IG; PO4 | - | 2.7 | 2022-01-19 | doi.org/10.1073/pnas.2115849118 |
|
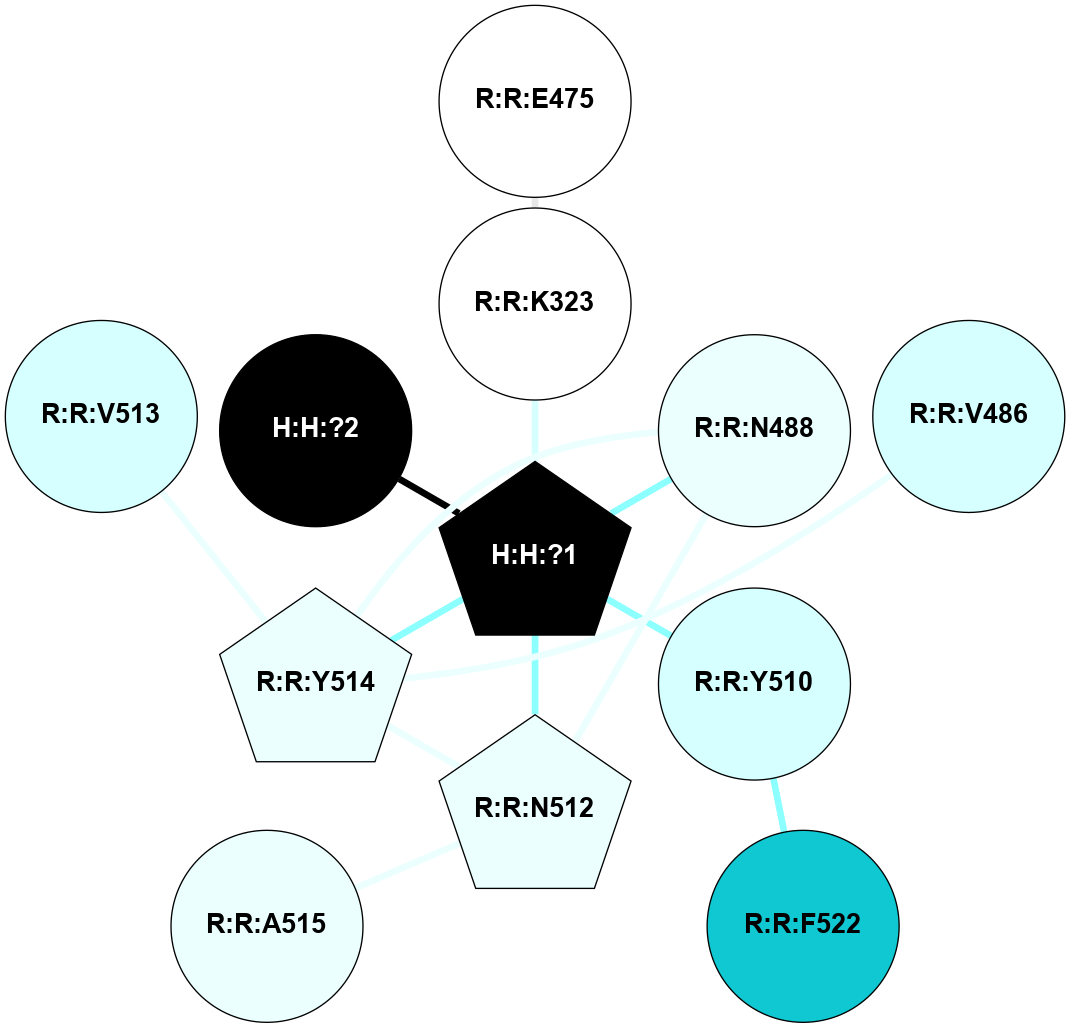
A 2D representation of the interactions of NAG in 7SIL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E475 | R:R:K323 | 6.75 | 0 | No | No | 5 | 5 | 2 | 1 | | H:H:?1 | R:R:K323 | 6.33 | 27 | Yes | No | 0 | 5 | 0 | 1 | | R:R:V486 | R:R:Y514 | 3.79 | 0 | No | Yes | 3 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 6.81 | 27 | No | Yes | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:Y514 | 4.65 | 27 | No | Yes | 4 | 4 | 1 | 1 | | H:H:?1 | R:R:N488 | 25.87 | 27 | Yes | No | 0 | 4 | 0 | 1 | | R:R:F522 | R:R:Y510 | 18.57 | 0 | No | No | 1 | 3 | 2 | 1 | | H:H:?1 | R:R:Y510 | 11.57 | 27 | Yes | No | 0 | 3 | 0 | 1 | | R:R:N512 | R:R:Y514 | 4.65 | 27 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 6.25 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?1 | R:R:N512 | 20.95 | 27 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:V513 | R:R:Y514 | 5.05 | 0 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?1 | R:R:Y514 | 10.52 | 27 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 37.89 | 27 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 18.86 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
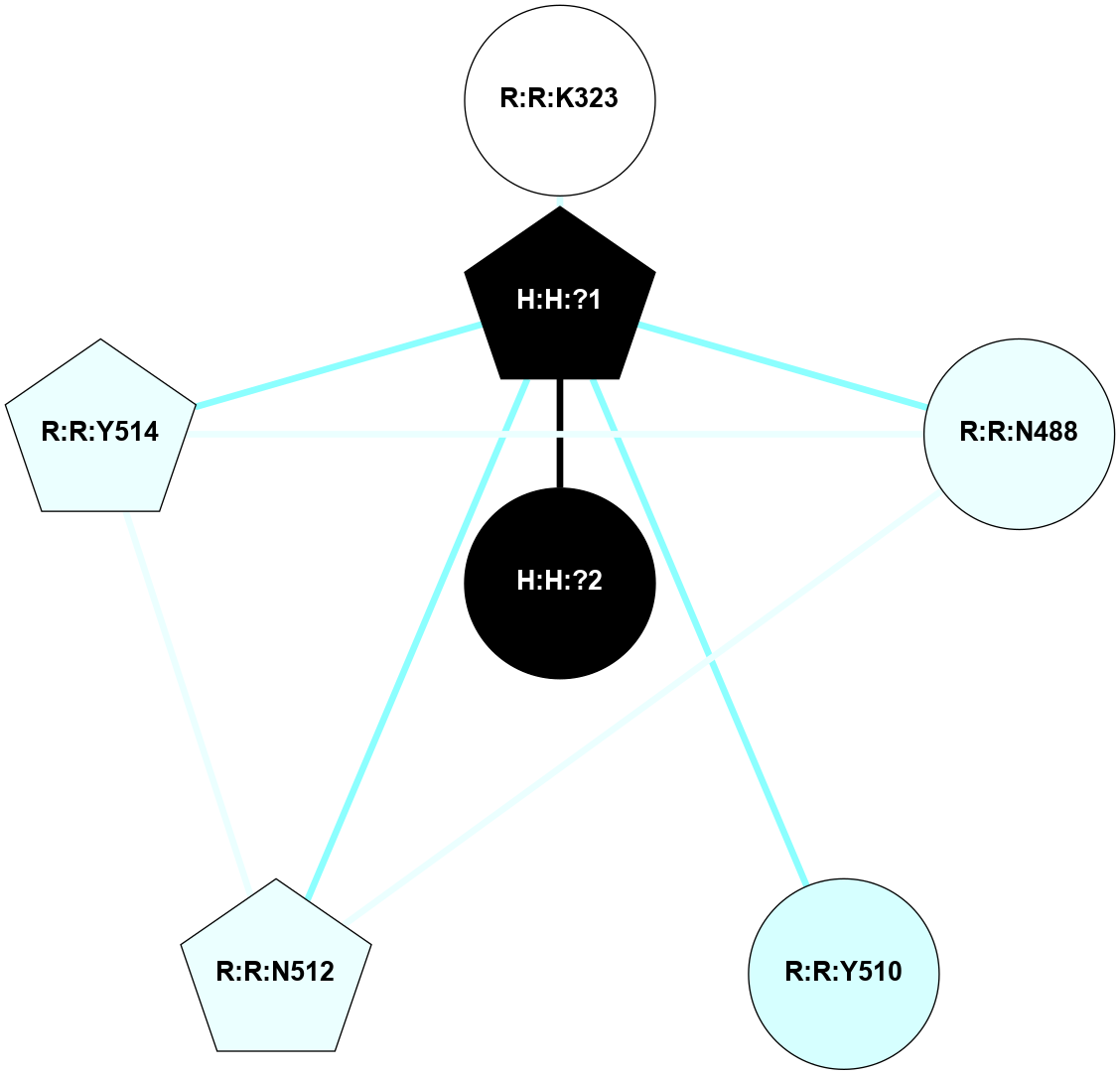
A 2D representation of the interactions of NAG in 7SIL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:K323 | 6.33 | 27 | Yes | No | 0 | 5 | 1 | 2 | | R:R:N488 | R:R:N512 | 6.81 | 27 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N488 | R:R:Y514 | 4.65 | 27 | No | Yes | 4 | 4 | 2 | 2 | | H:H:?1 | R:R:N488 | 25.87 | 27 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:Y510 | 11.57 | 27 | Yes | No | 0 | 3 | 1 | 2 | | R:R:N512 | R:R:Y514 | 4.65 | 27 | Yes | Yes | 4 | 4 | 2 | 2 | | H:H:?1 | R:R:N512 | 20.95 | 27 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:Y514 | 10.52 | 27 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 37.89 | 27 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 37.89 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
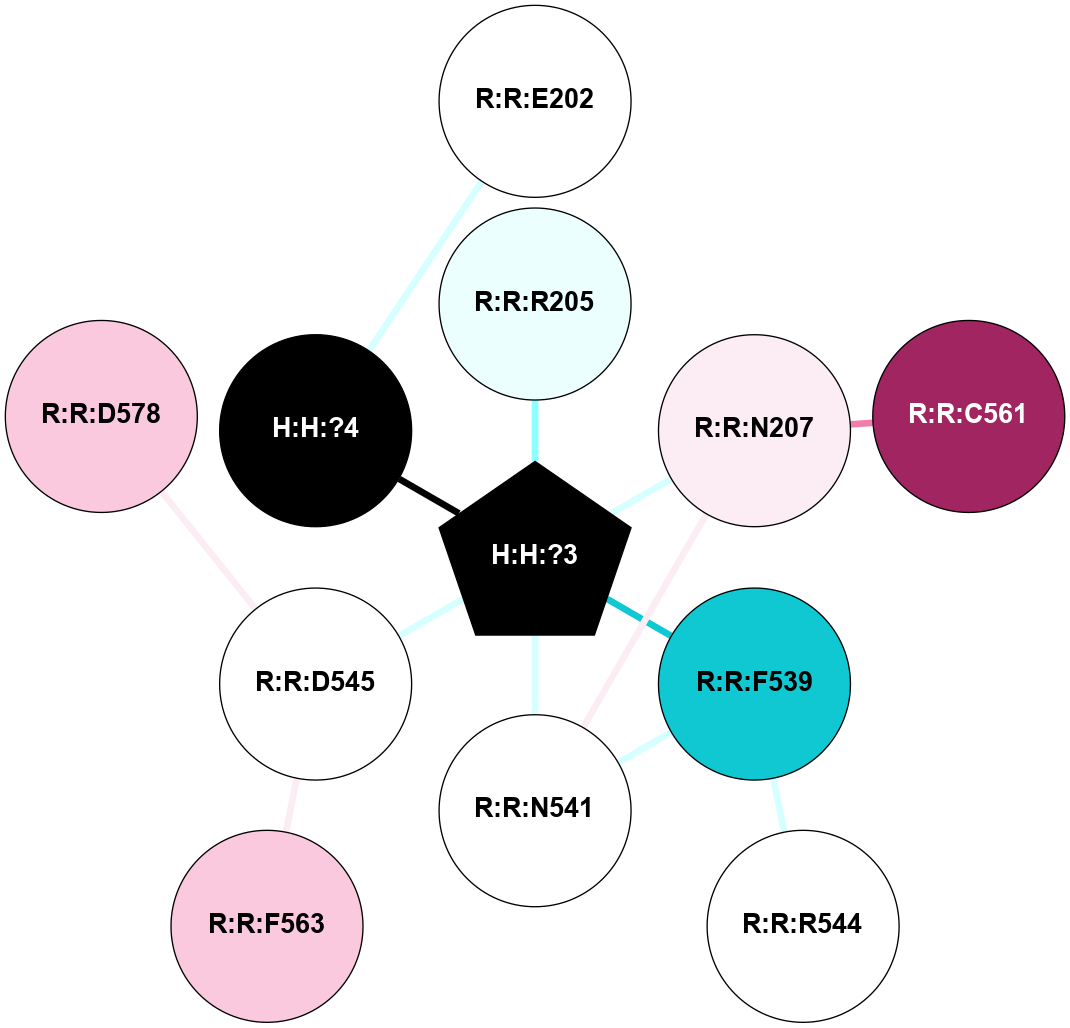
A 2D representation of the interactions of NAG in 7SIL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:E202 | 4.76 | 0 | No | No | 0 | 5 | 1 | 2 | | H:H:?3 | R:R:R205 | 8.72 | 25 | Yes | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 17.71 | 25 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 9.45 | 0 | No | No | 9 | 6 | 2 | 1 | | H:H:?3 | R:R:N207 | 16.02 | 25 | Yes | No | 0 | 6 | 0 | 1 | | R:R:F539 | R:R:N541 | 6.04 | 25 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 24.59 | 25 | No | No | 1 | 5 | 1 | 2 | | H:H:?3 | R:R:F539 | 5.46 | 25 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?3 | R:R:N541 | 25.87 | 25 | Yes | No | 0 | 5 | 0 | 1 | | R:R:D545 | R:R:F563 | 3.58 | 0 | No | No | 5 | 7 | 1 | 2 | | R:R:D545 | R:R:D578 | 15.97 | 0 | No | No | 5 | 7 | 1 | 2 | | H:H:?3 | R:R:D545 | 17.05 | 25 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?3 | H:H:?4 | 39.01 | 25 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.69 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
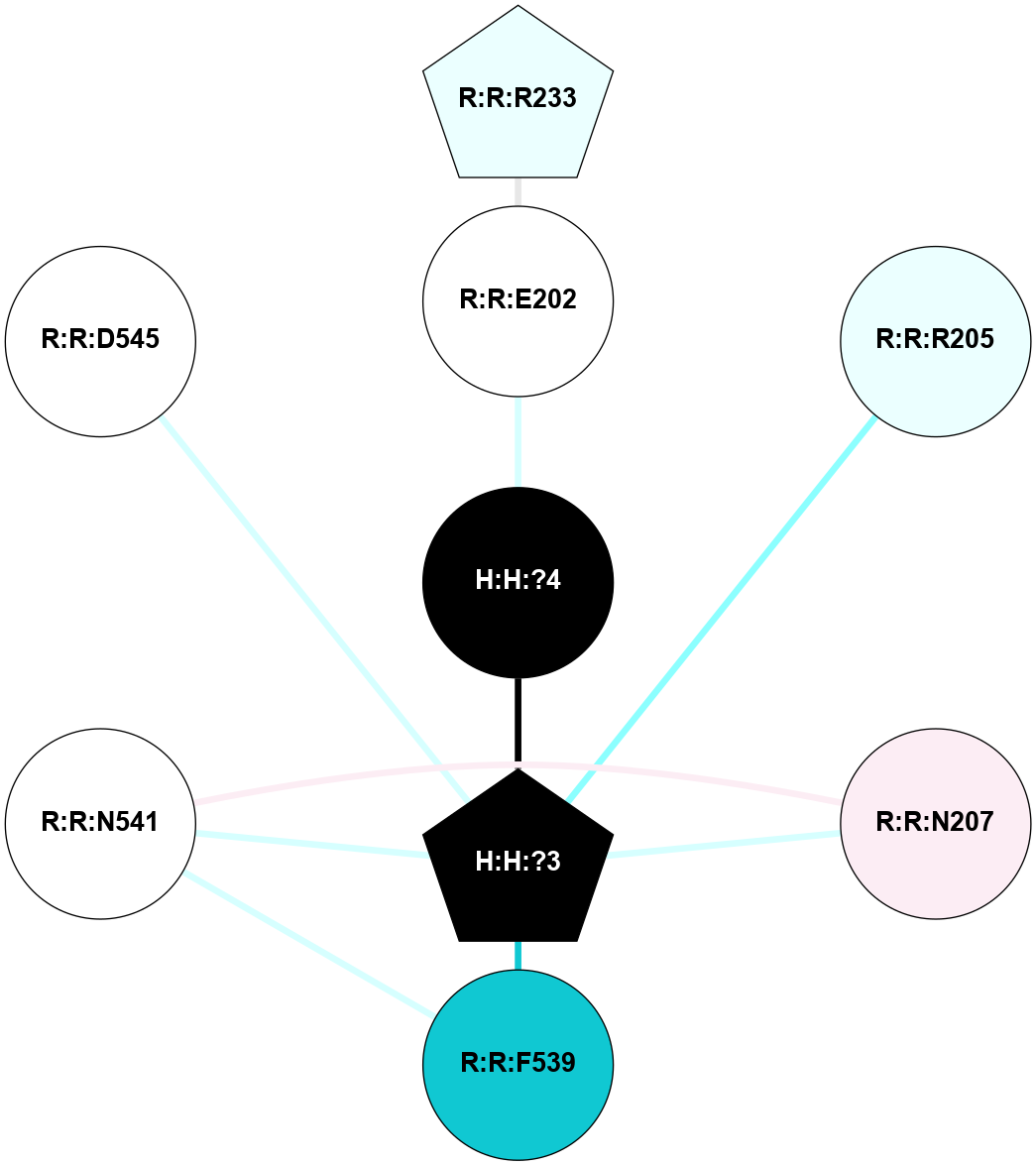
A 2D representation of the interactions of NAG in 7SIL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E202 | R:R:R233 | 13.96 | 0 | No | Yes | 5 | 4 | 1 | 2 | | H:H:?4 | R:R:E202 | 4.76 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?3 | R:R:R205 | 8.72 | 25 | Yes | No | 0 | 4 | 1 | 2 | | R:R:N207 | R:R:N541 | 17.71 | 25 | No | No | 6 | 5 | 2 | 2 | | H:H:?3 | R:R:N207 | 16.02 | 25 | Yes | No | 0 | 6 | 1 | 2 | | R:R:F539 | R:R:N541 | 6.04 | 25 | No | No | 1 | 5 | 2 | 2 | | H:H:?3 | R:R:F539 | 5.46 | 25 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?3 | R:R:N541 | 25.87 | 25 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | R:R:D545 | 17.05 | 25 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | H:H:?4 | 39.01 | 25 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 21.88 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
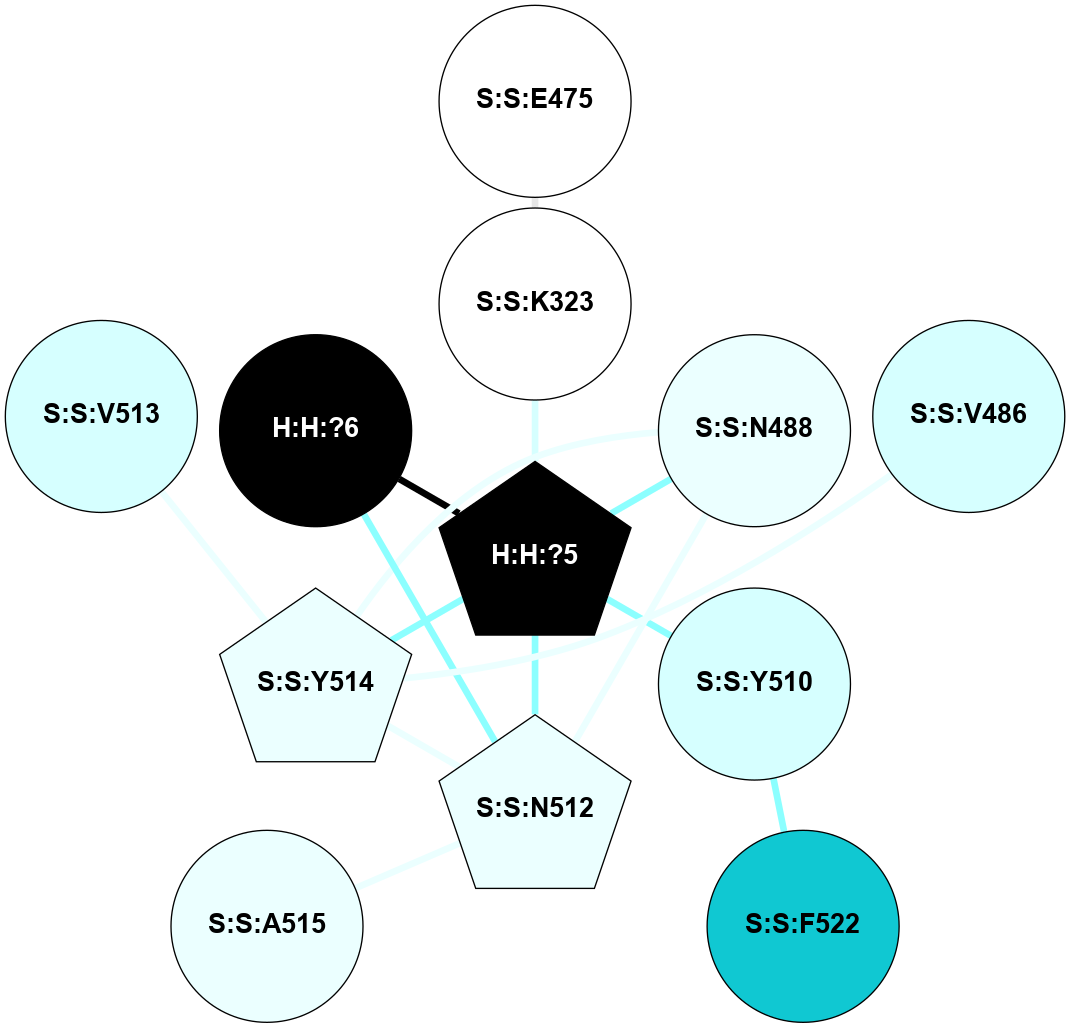
A 2D representation of the interactions of NAG in 7SIL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:E475 | S:S:K323 | 4.05 | 0 | No | No | 5 | 5 | 2 | 1 | | H:H:?5 | S:S:K323 | 7.59 | 22 | Yes | No | 0 | 5 | 0 | 1 | | S:S:V486 | S:S:Y514 | 3.79 | 0 | No | Yes | 3 | 4 | 2 | 1 | | S:S:N488 | S:S:N512 | 6.81 | 22 | No | Yes | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:Y514 | 4.65 | 22 | No | Yes | 4 | 4 | 1 | 1 | | H:H:?5 | S:S:N488 | 25.87 | 22 | Yes | No | 0 | 4 | 0 | 1 | | S:S:F522 | S:S:Y510 | 18.57 | 0 | No | No | 1 | 3 | 2 | 1 | | H:H:?5 | S:S:Y510 | 11.57 | 22 | Yes | No | 0 | 3 | 0 | 1 | | S:S:N512 | S:S:Y514 | 4.65 | 22 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:A515 | S:S:N512 | 6.25 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?5 | S:S:N512 | 19.71 | 22 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?6 | S:S:N512 | 3.7 | 22 | No | Yes | 0 | 4 | 1 | 1 | | S:S:V513 | S:S:Y514 | 5.05 | 0 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?5 | S:S:Y514 | 7.36 | 22 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?5 | H:H:?6 | 37.89 | 22 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 18.33 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
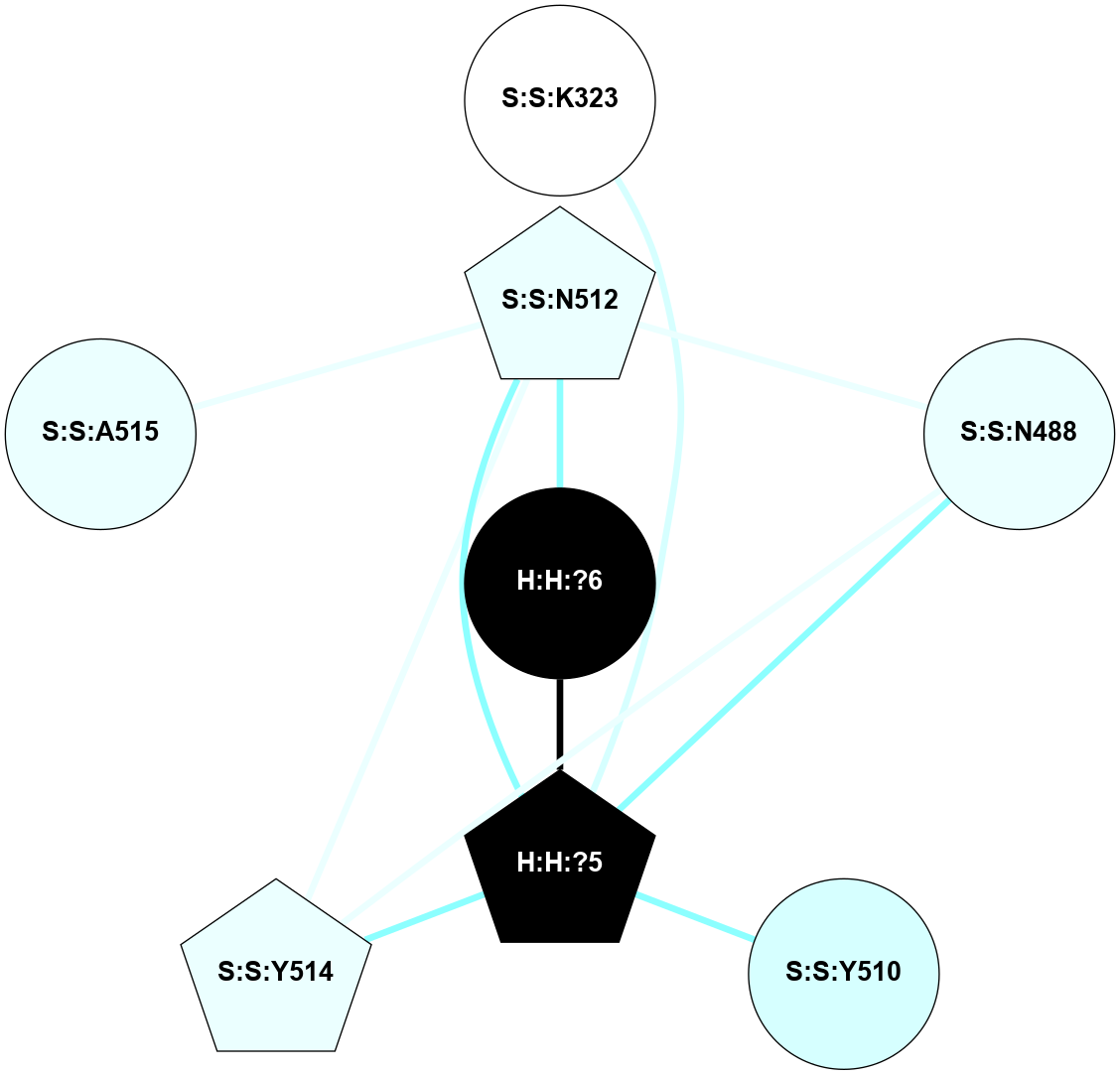
A 2D representation of the interactions of NAG in 7SIL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | S:S:K323 | 7.59 | 22 | Yes | No | 0 | 5 | 1 | 2 | | S:S:N488 | S:S:N512 | 6.81 | 22 | No | Yes | 4 | 4 | 2 | 1 | | S:S:N488 | S:S:Y514 | 4.65 | 22 | No | Yes | 4 | 4 | 2 | 2 | | H:H:?5 | S:S:N488 | 25.87 | 22 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?5 | S:S:Y510 | 11.57 | 22 | Yes | No | 0 | 3 | 1 | 2 | | S:S:N512 | S:S:Y514 | 4.65 | 22 | Yes | Yes | 4 | 4 | 1 | 2 | | S:S:A515 | S:S:N512 | 6.25 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?5 | S:S:N512 | 19.71 | 22 | Yes | Yes | 0 | 4 | 1 | 1 | | H:H:?6 | S:S:N512 | 3.7 | 22 | No | Yes | 0 | 4 | 0 | 1 | | H:H:?5 | S:S:Y514 | 7.36 | 22 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?5 | H:H:?6 | 37.89 | 22 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 20.80 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
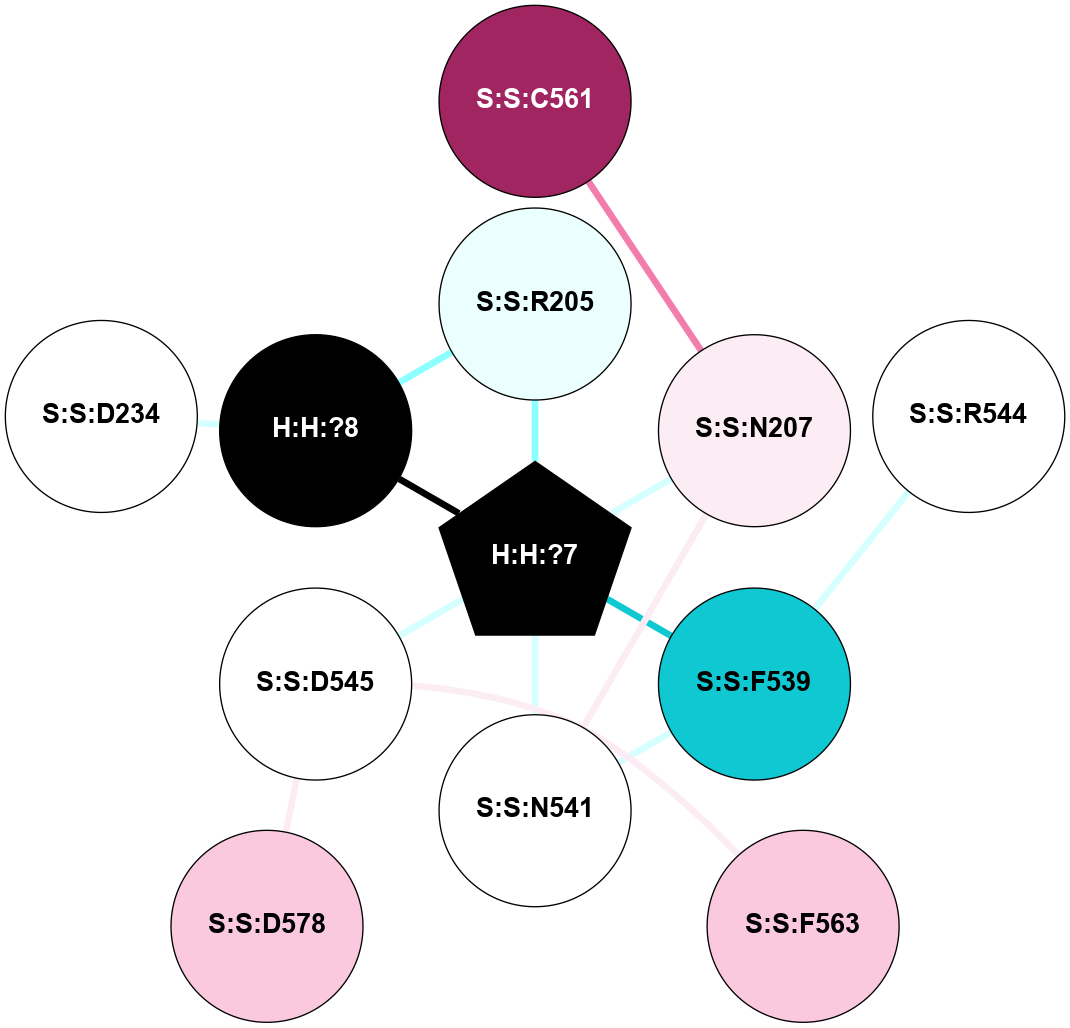
A 2D representation of the interactions of NAG in 7SIL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | S:S:R205 | 7.63 | 19 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?8 | S:S:R205 | 7.63 | 19 | No | No | 0 | 4 | 1 | 1 | | S:S:N207 | S:S:N541 | 17.71 | 19 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 9.45 | 0 | No | No | 9 | 6 | 2 | 1 | | H:H:?7 | S:S:N207 | 16.02 | 19 | Yes | No | 0 | 6 | 0 | 1 | | S:S:F539 | S:S:N541 | 6.04 | 19 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 25.66 | 19 | No | No | 1 | 5 | 1 | 2 | | H:H:?7 | S:S:F539 | 5.46 | 19 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?7 | S:S:N541 | 25.87 | 19 | Yes | No | 0 | 5 | 0 | 1 | | S:S:D545 | S:S:F563 | 3.58 | 0 | No | No | 5 | 7 | 1 | 2 | | S:S:D545 | S:S:D578 | 15.97 | 0 | No | No | 5 | 7 | 1 | 2 | | H:H:?7 | S:S:D545 | 18.27 | 19 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?7 | H:H:?8 | 41.23 | 19 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?8 | S:S:D234 | 1.22 | 19 | No | No | 0 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 19.08 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
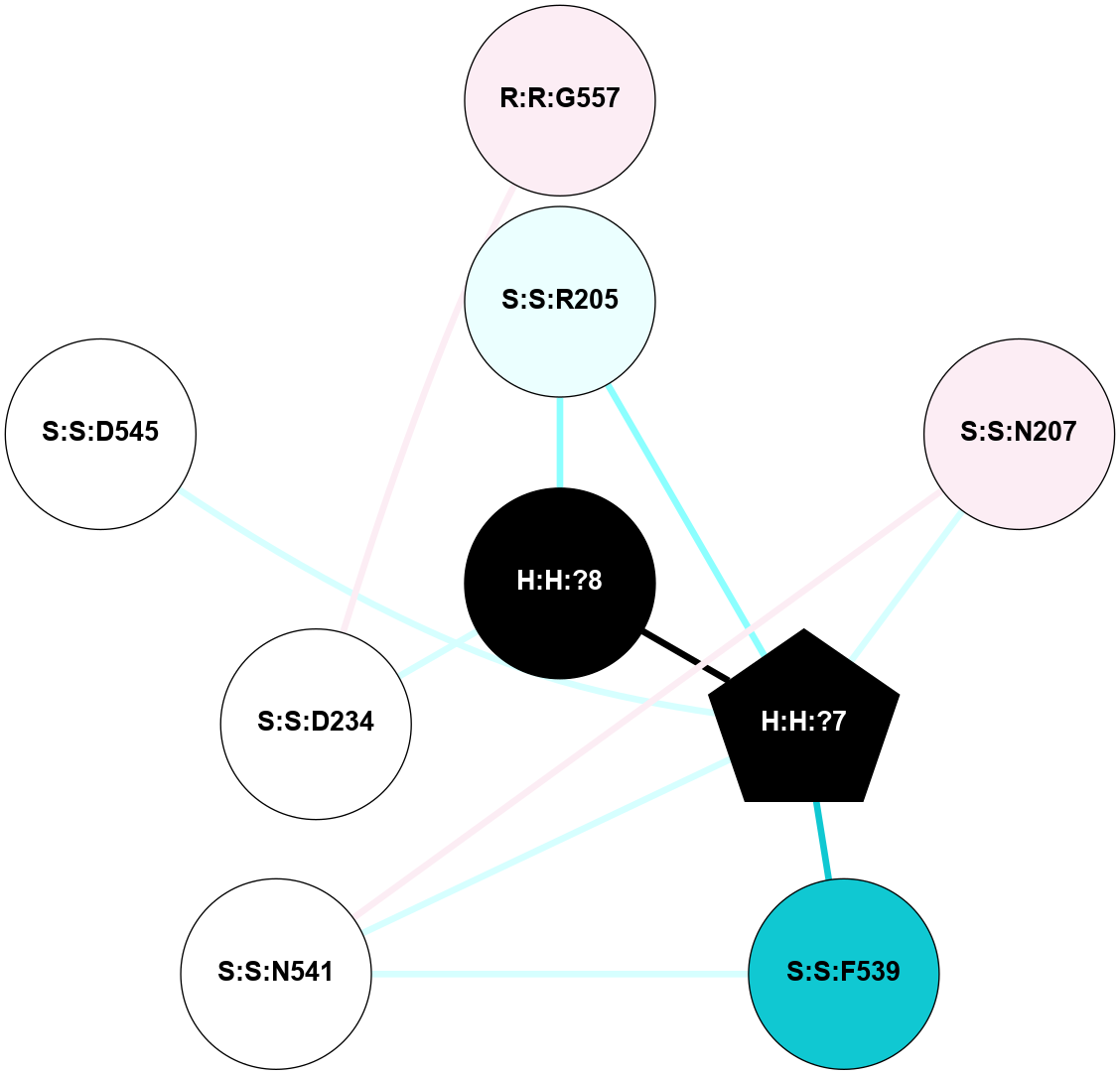
A 2D representation of the interactions of NAG in 7SIL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:G557 | S:S:D234 | 5.03 | 0 | No | No | 6 | 5 | 2 | 1 | | H:H:?7 | S:S:R205 | 7.63 | 19 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?8 | S:S:R205 | 7.63 | 19 | No | No | 0 | 4 | 0 | 1 | | S:S:N207 | S:S:N541 | 17.71 | 19 | No | No | 6 | 5 | 2 | 2 | | H:H:?7 | S:S:N207 | 16.02 | 19 | Yes | No | 0 | 6 | 1 | 2 | | S:S:F539 | S:S:N541 | 6.04 | 19 | No | No | 1 | 5 | 2 | 2 | | H:H:?7 | S:S:F539 | 5.46 | 19 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?7 | S:S:N541 | 25.87 | 19 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?7 | S:S:D545 | 18.27 | 19 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?7 | H:H:?8 | 41.23 | 19 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?8 | S:S:D234 | 1.22 | 19 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 16.69 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
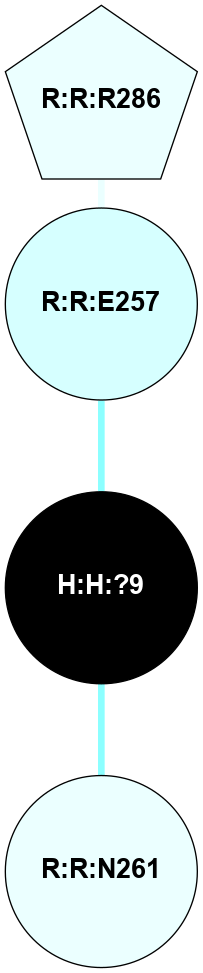
A 2D representation of the interactions of NAG in 7SIL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E257 | R:R:R286 | 6.98 | 0 | No | Yes | 3 | 4 | 1 | 2 | | H:H:?9 | R:R:E257 | 4.76 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?9 | R:R:N261 | 28.34 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.55 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
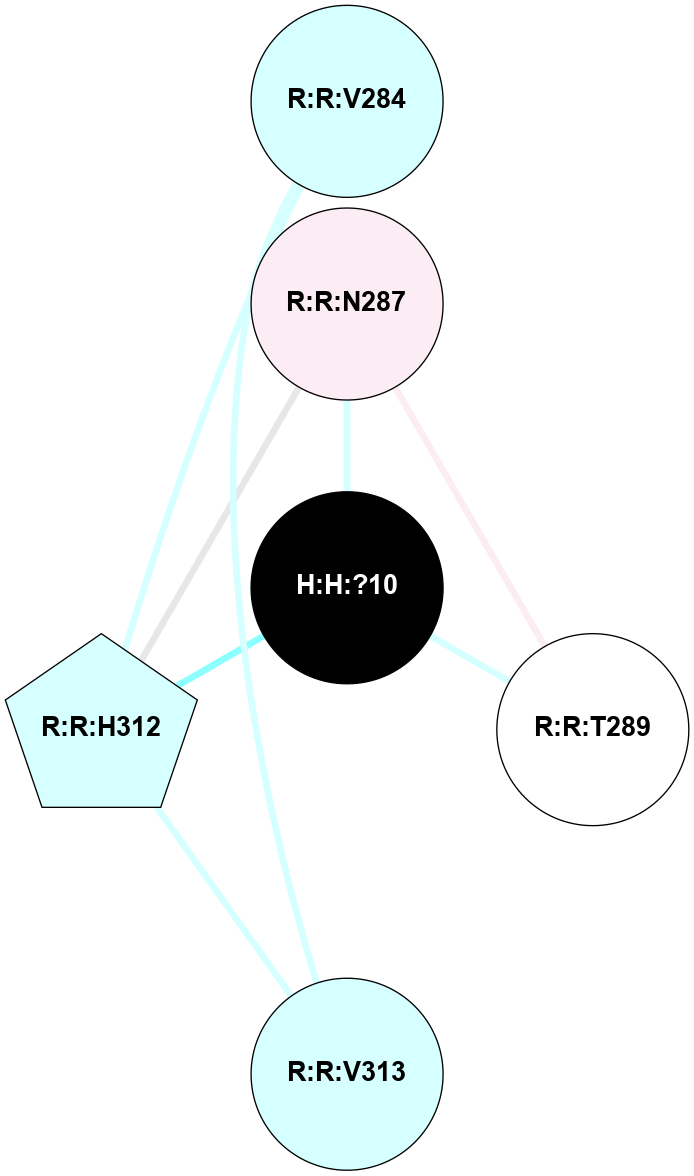
A 2D representation of the interactions of NAG in 7SIL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:H312 | R:R:V284 | 5.54 | 4 | Yes | No | 3 | 3 | 1 | 2 | | R:R:V284 | R:R:V313 | 6.41 | 4 | No | No | 3 | 3 | 2 | 2 | | R:R:N287 | R:R:T289 | 5.85 | 4 | No | No | 6 | 5 | 1 | 1 | | R:R:H312 | R:R:N287 | 6.38 | 4 | Yes | No | 3 | 6 | 1 | 1 | | H:H:?10 | R:R:N287 | 32.04 | 4 | No | No | 0 | 6 | 0 | 1 | | H:H:?10 | R:R:T289 | 10.58 | 4 | No | No | 0 | 5 | 0 | 1 | | R:R:H312 | R:R:V313 | 5.54 | 4 | Yes | No | 3 | 3 | 1 | 2 | | H:H:?10 | R:R:H312 | 3.46 | 4 | No | Yes | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.36 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
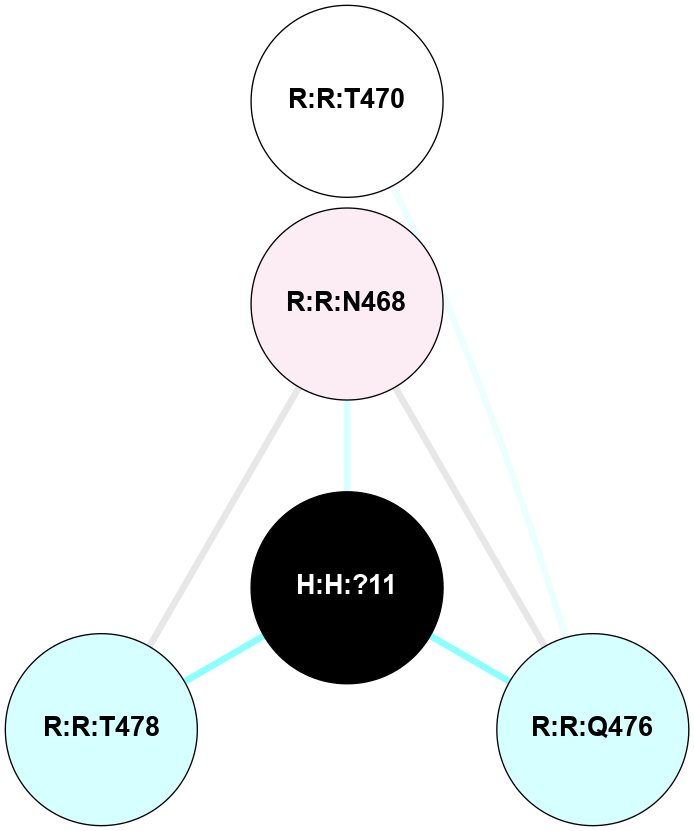
A 2D representation of the interactions of NAG in 7SIL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N468 | R:R:Q476 | 5.28 | 28 | No | No | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 5.85 | 28 | No | No | 6 | 3 | 1 | 1 | | H:H:?11 | R:R:N468 | 33.27 | 28 | No | No | 0 | 6 | 0 | 1 | | R:R:Q476 | R:R:T470 | 8.5 | 28 | No | No | 3 | 5 | 1 | 2 | | H:H:?11 | R:R:Q476 | 9.55 | 28 | No | No | 0 | 3 | 0 | 1 | | H:H:?11 | R:R:T478 | 3.97 | 28 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.60 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
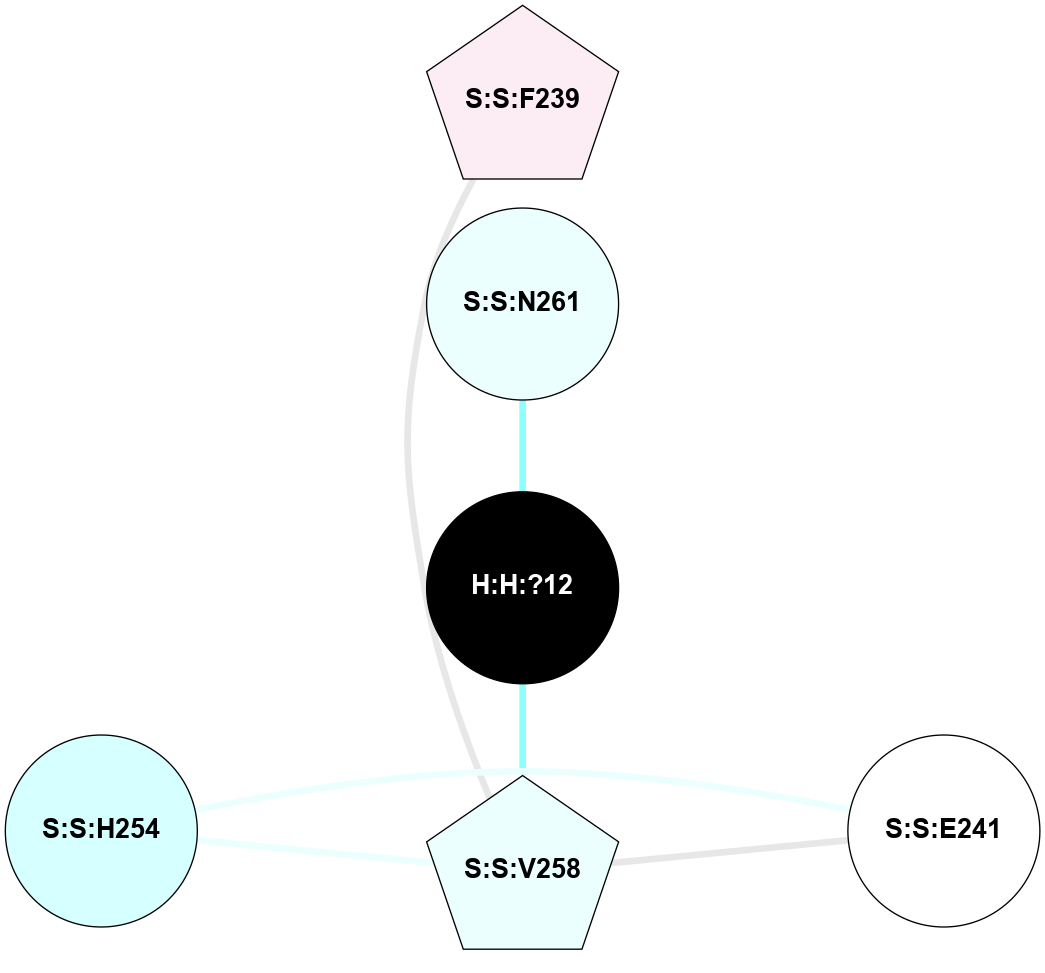
A 2D representation of the interactions of NAG in 7SIL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:F239 | S:S:V258 | 6.55 | 3 | Yes | Yes | 6 | 4 | 2 | 1 | | S:S:E241 | S:S:H254 | 4.92 | 3 | No | No | 5 | 3 | 2 | 2 | | S:S:E241 | S:S:V258 | 11.41 | 3 | No | Yes | 5 | 4 | 2 | 1 | | S:S:H254 | S:S:V258 | 5.54 | 3 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?12 | S:S:N261 | 28.34 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?12 | S:S:V258 | 1.34 | 0 | No | Yes | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.84 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
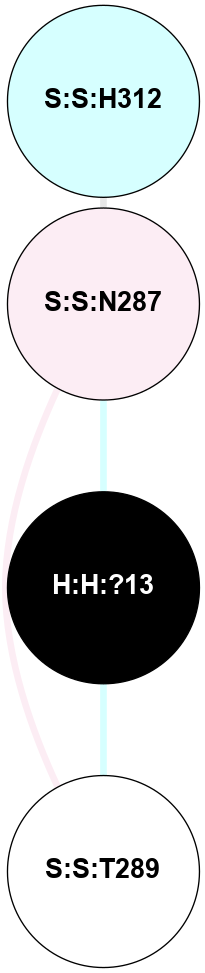
A 2D representation of the interactions of NAG in 7SIL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:N287 | S:S:T289 | 5.85 | 3 | No | No | 6 | 5 | 1 | 1 | | S:S:H312 | S:S:N287 | 3.83 | 3 | No | No | 3 | 6 | 2 | 1 | | H:H:?13 | S:S:N287 | 27.11 | 3 | No | No | 0 | 6 | 0 | 1 | | H:H:?13 | S:S:T289 | 6.61 | 3 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.86 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7SIL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:N468 | S:S:Q476 | 5.28 | 39 | No | No | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 5.85 | 39 | No | No | 6 | 3 | 1 | 2 | | H:H:?14 | S:S:N468 | 30.8 | 39 | No | No | 0 | 6 | 0 | 1 | | S:S:Q476 | S:S:T470 | 7.09 | 39 | No | No | 3 | 5 | 1 | 2 | | H:H:?14 | S:S:Q476 | 10.75 | 39 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.77 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7SIM | C | Ion | Calcium Sensing | CaS | Homo Sapiens | Ca | Ca; Ca; Cyclomethyltryptophan; PO4 | - | 2.7 | 2022-01-19 | doi.org/10.1073/pnas.2115849118 |
|
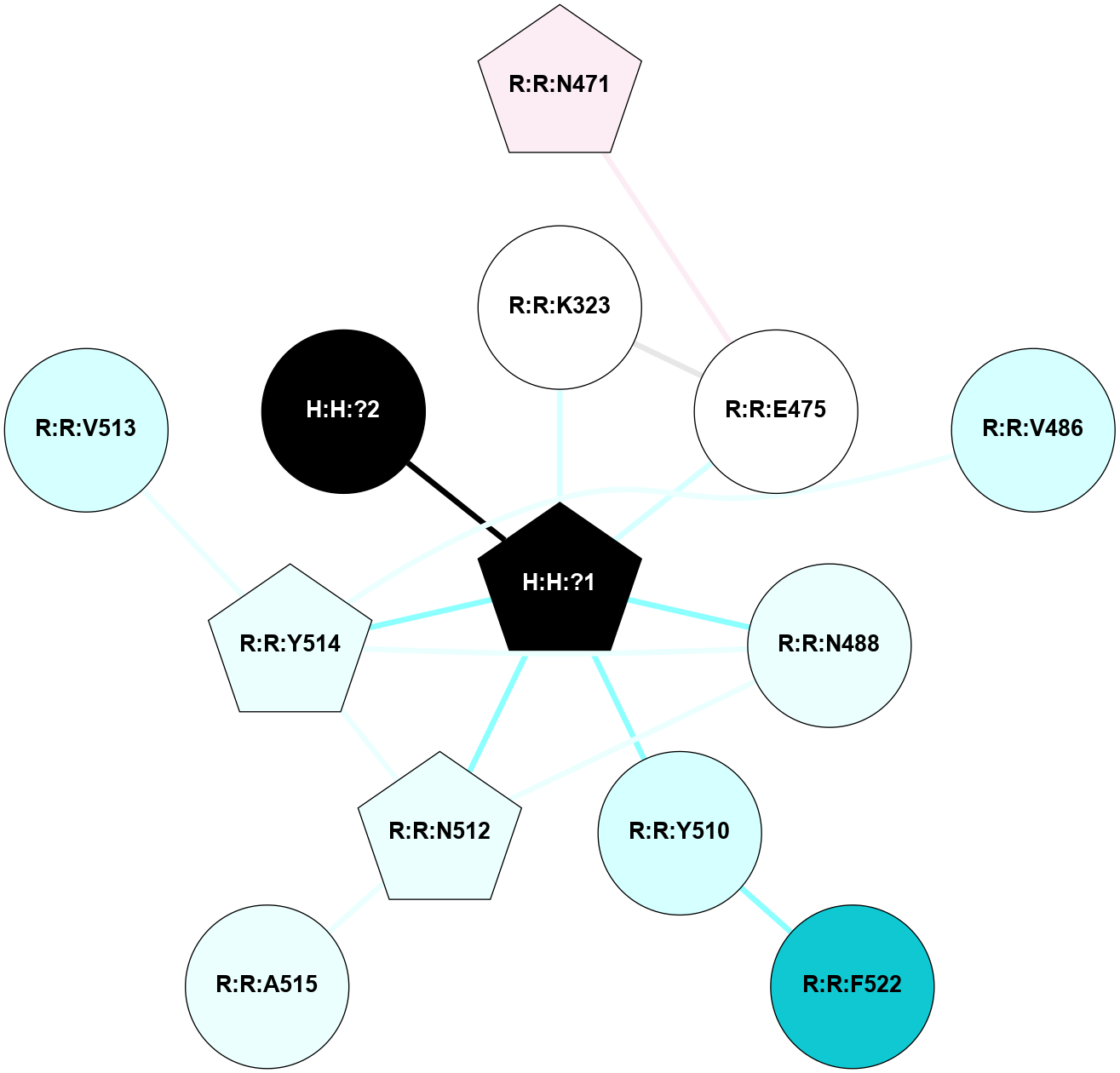
A 2D representation of the interactions of NAG in 7SIM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E475 | R:R:K323 | 5.4 | 1 | No | No | 5 | 5 | 1 | 1 | | H:H:?1 | R:R:K323 | 7.59 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:E475 | R:R:N471 | 5.26 | 1 | No | Yes | 5 | 6 | 1 | 2 | | H:H:?1 | R:R:E475 | 4.76 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:V486 | R:R:Y514 | 5.05 | 0 | No | Yes | 3 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 6.81 | 1 | No | Yes | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:Y514 | 4.65 | 1 | No | Yes | 4 | 4 | 1 | 1 | | H:H:?1 | R:R:N488 | 27.11 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:F522 | R:R:Y510 | 16.5 | 0 | No | No | 1 | 3 | 2 | 1 | | H:H:?1 | R:R:Y510 | 11.57 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:N512 | R:R:Y514 | 4.65 | 1 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 4.69 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?1 | R:R:N512 | 22.18 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:V513 | R:R:Y514 | 3.79 | 0 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?1 | R:R:Y514 | 9.47 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 43.46 | 1 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 18.02 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
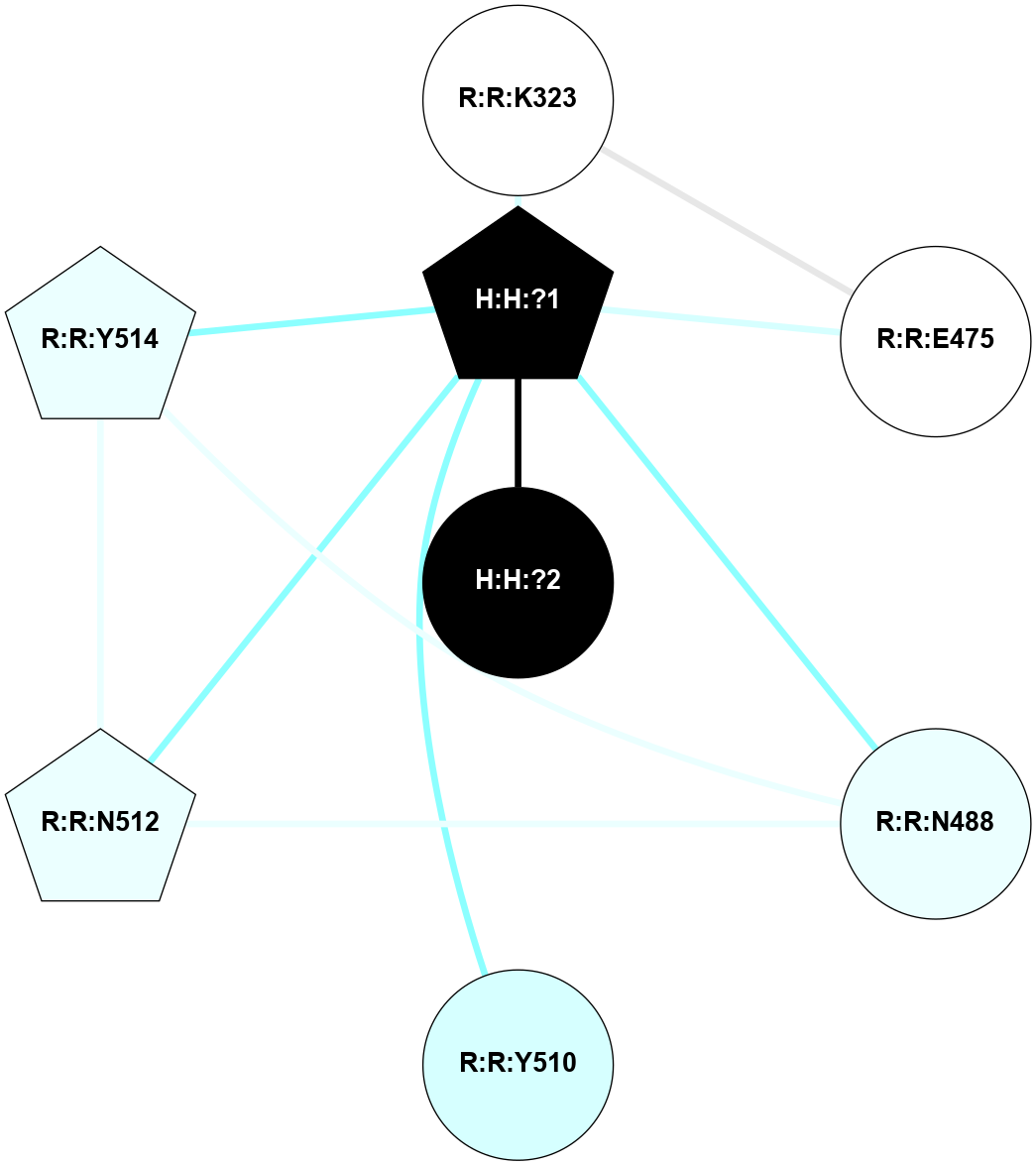
A 2D representation of the interactions of NAG in 7SIM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E475 | R:R:K323 | 5.4 | 1 | No | No | 5 | 5 | 2 | 2 | | H:H:?1 | R:R:K323 | 7.59 | 1 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?1 | R:R:E475 | 4.76 | 1 | Yes | No | 0 | 5 | 1 | 2 | | R:R:N488 | R:R:N512 | 6.81 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N488 | R:R:Y514 | 4.65 | 1 | No | Yes | 4 | 4 | 2 | 2 | | H:H:?1 | R:R:N488 | 27.11 | 1 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:Y510 | 11.57 | 1 | Yes | No | 0 | 3 | 1 | 2 | | R:R:N512 | R:R:Y514 | 4.65 | 1 | Yes | Yes | 4 | 4 | 2 | 2 | | H:H:?1 | R:R:N512 | 22.18 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:Y514 | 9.47 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 43.46 | 1 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 43.46 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
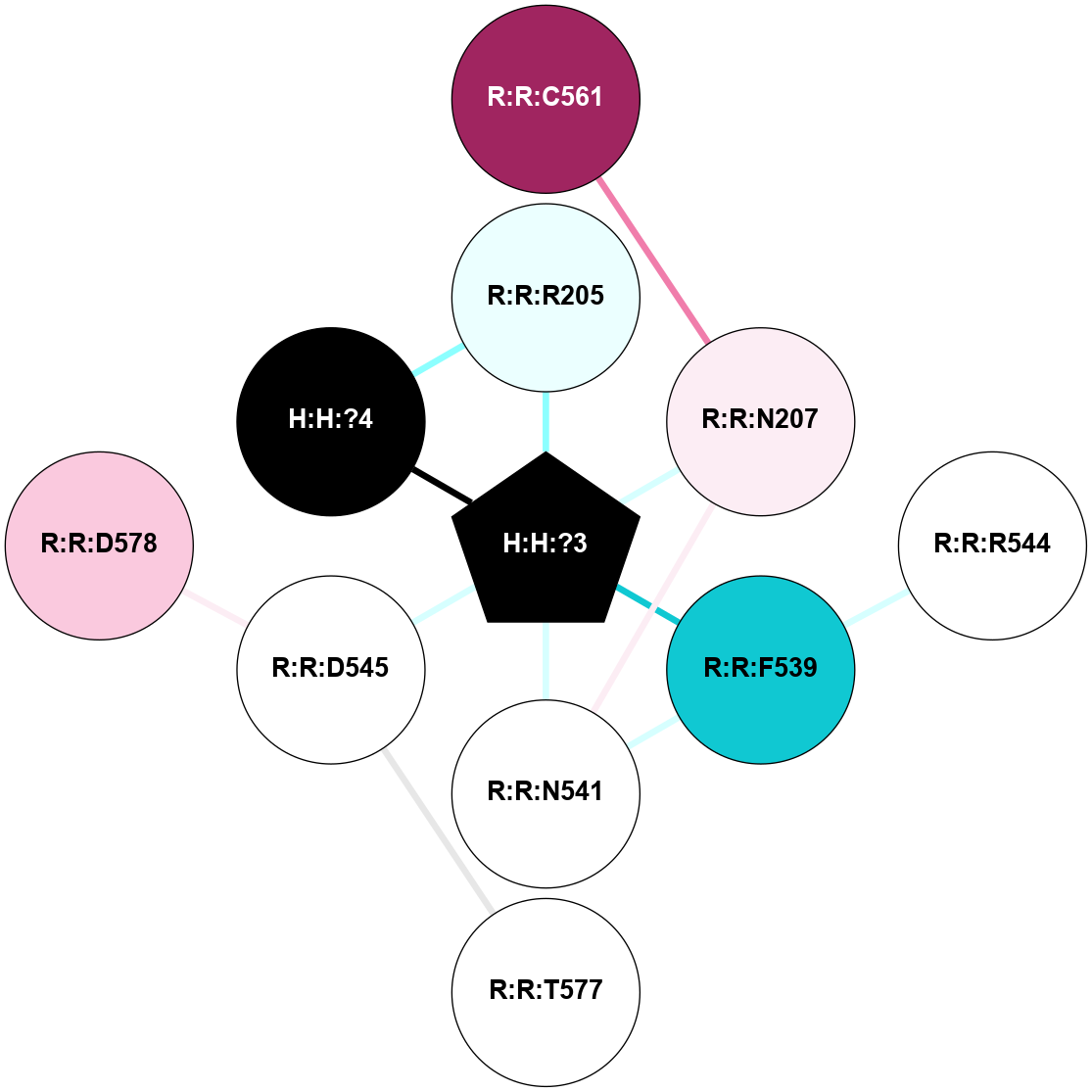
A 2D representation of the interactions of NAG in 7SIM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:R205 | 8.72 | 14 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?4 | R:R:R205 | 4.36 | 14 | No | No | 0 | 4 | 1 | 1 | | R:R:N207 | R:R:N541 | 17.71 | 14 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 7.87 | 0 | No | No | 9 | 6 | 2 | 1 | | H:H:?3 | R:R:N207 | 12.32 | 14 | Yes | No | 0 | 6 | 0 | 1 | | R:R:F539 | R:R:N541 | 7.25 | 14 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 26.72 | 14 | No | No | 1 | 5 | 1 | 2 | | H:H:?3 | R:R:F539 | 5.46 | 14 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?3 | R:R:N541 | 25.87 | 14 | Yes | No | 0 | 5 | 0 | 1 | | R:R:D545 | R:R:T577 | 5.78 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:D545 | R:R:D578 | 3.99 | 0 | No | No | 5 | 7 | 1 | 2 | | H:H:?3 | R:R:D545 | 3.65 | 14 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?3 | H:H:?4 | 36.78 | 14 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.47 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
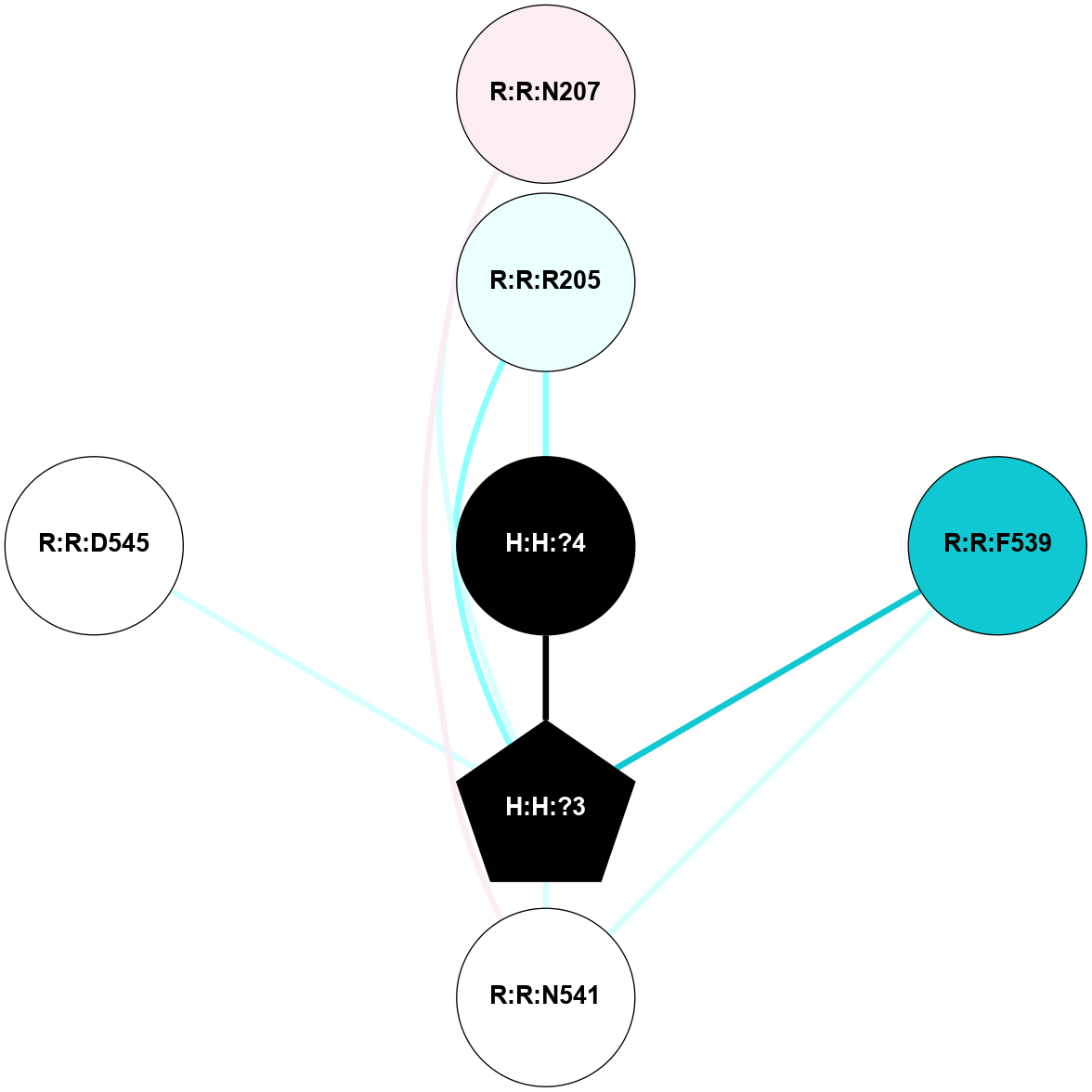
A 2D representation of the interactions of NAG in 7SIM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:R205 | 8.72 | 14 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?4 | R:R:R205 | 4.36 | 14 | No | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 17.71 | 14 | No | No | 6 | 5 | 2 | 2 | | H:H:?3 | R:R:N207 | 12.32 | 14 | Yes | No | 0 | 6 | 1 | 2 | | R:R:F539 | R:R:N541 | 7.25 | 14 | No | No | 1 | 5 | 2 | 2 | | H:H:?3 | R:R:F539 | 5.46 | 14 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?3 | R:R:N541 | 25.87 | 14 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | R:R:D545 | 3.65 | 14 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | H:H:?4 | 36.78 | 14 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 20.57 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
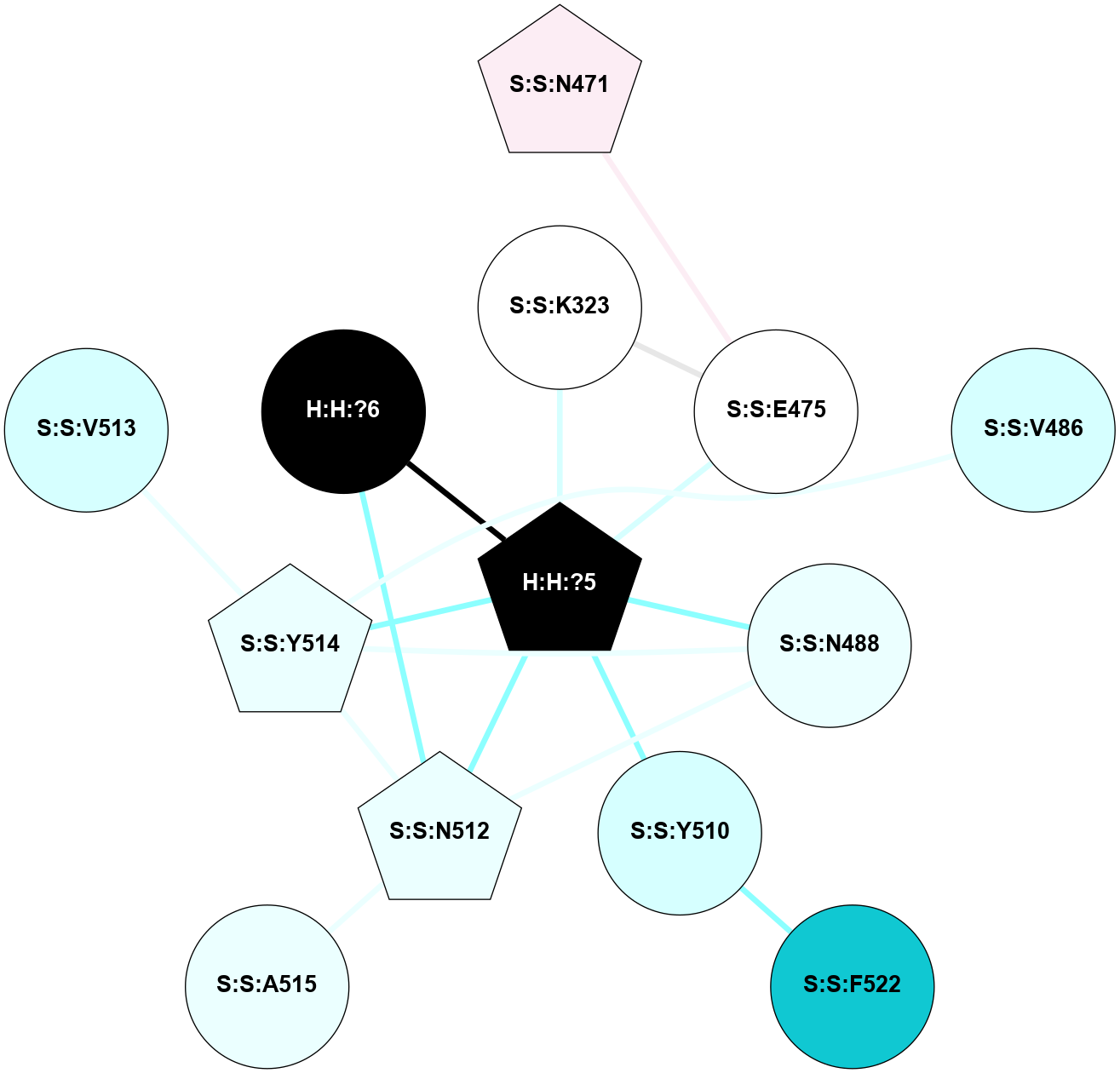
A 2D representation of the interactions of NAG in 7SIM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:E475 | S:S:K323 | 5.4 | 2 | No | No | 5 | 5 | 1 | 1 | | H:H:?5 | S:S:K323 | 7.59 | 2 | Yes | No | 0 | 5 | 0 | 1 | | S:S:E475 | S:S:N471 | 3.94 | 2 | No | Yes | 5 | 6 | 1 | 2 | | H:H:?5 | S:S:E475 | 3.57 | 2 | Yes | No | 0 | 5 | 0 | 1 | | S:S:V486 | S:S:Y514 | 5.05 | 0 | No | Yes | 3 | 4 | 2 | 1 | | S:S:N488 | S:S:N512 | 6.81 | 2 | No | Yes | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:Y514 | 4.65 | 2 | No | Yes | 4 | 4 | 1 | 1 | | H:H:?5 | S:S:N488 | 25.87 | 2 | Yes | No | 0 | 4 | 0 | 1 | | S:S:F522 | S:S:Y510 | 16.5 | 0 | No | No | 1 | 3 | 2 | 1 | | H:H:?5 | S:S:Y510 | 11.57 | 2 | Yes | No | 0 | 3 | 0 | 1 | | S:S:N512 | S:S:Y514 | 4.65 | 2 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:A515 | S:S:N512 | 4.69 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?5 | S:S:N512 | 23.41 | 2 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?6 | S:S:N512 | 3.7 | 2 | No | Yes | 0 | 4 | 1 | 1 | | S:S:V513 | S:S:Y514 | 3.79 | 0 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?5 | S:S:Y514 | 8.42 | 2 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?5 | H:H:?6 | 42.35 | 2 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 17.54 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
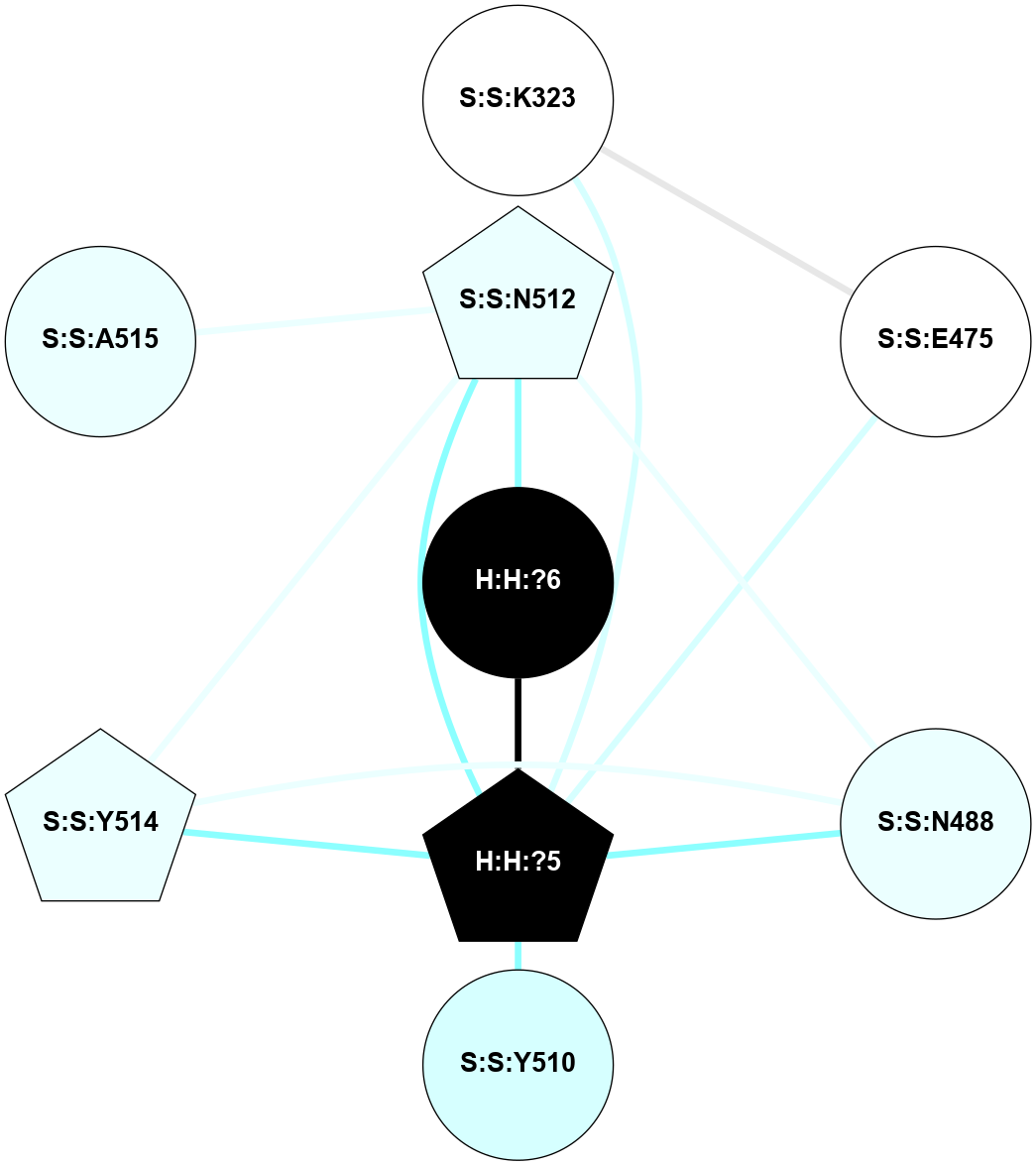
A 2D representation of the interactions of NAG in 7SIM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:E475 | S:S:K323 | 5.4 | 2 | No | No | 5 | 5 | 2 | 2 | | H:H:?5 | S:S:K323 | 7.59 | 2 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?5 | S:S:E475 | 3.57 | 2 | Yes | No | 0 | 5 | 1 | 2 | | S:S:N488 | S:S:N512 | 6.81 | 2 | No | Yes | 4 | 4 | 2 | 1 | | S:S:N488 | S:S:Y514 | 4.65 | 2 | No | Yes | 4 | 4 | 2 | 2 | | H:H:?5 | S:S:N488 | 25.87 | 2 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?5 | S:S:Y510 | 11.57 | 2 | Yes | No | 0 | 3 | 1 | 2 | | S:S:N512 | S:S:Y514 | 4.65 | 2 | Yes | Yes | 4 | 4 | 1 | 2 | | S:S:A515 | S:S:N512 | 4.69 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?5 | S:S:N512 | 23.41 | 2 | Yes | Yes | 0 | 4 | 1 | 1 | | H:H:?6 | S:S:N512 | 3.7 | 2 | No | Yes | 0 | 4 | 0 | 1 | | H:H:?5 | S:S:Y514 | 8.42 | 2 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?5 | H:H:?6 | 42.35 | 2 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 23.03 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
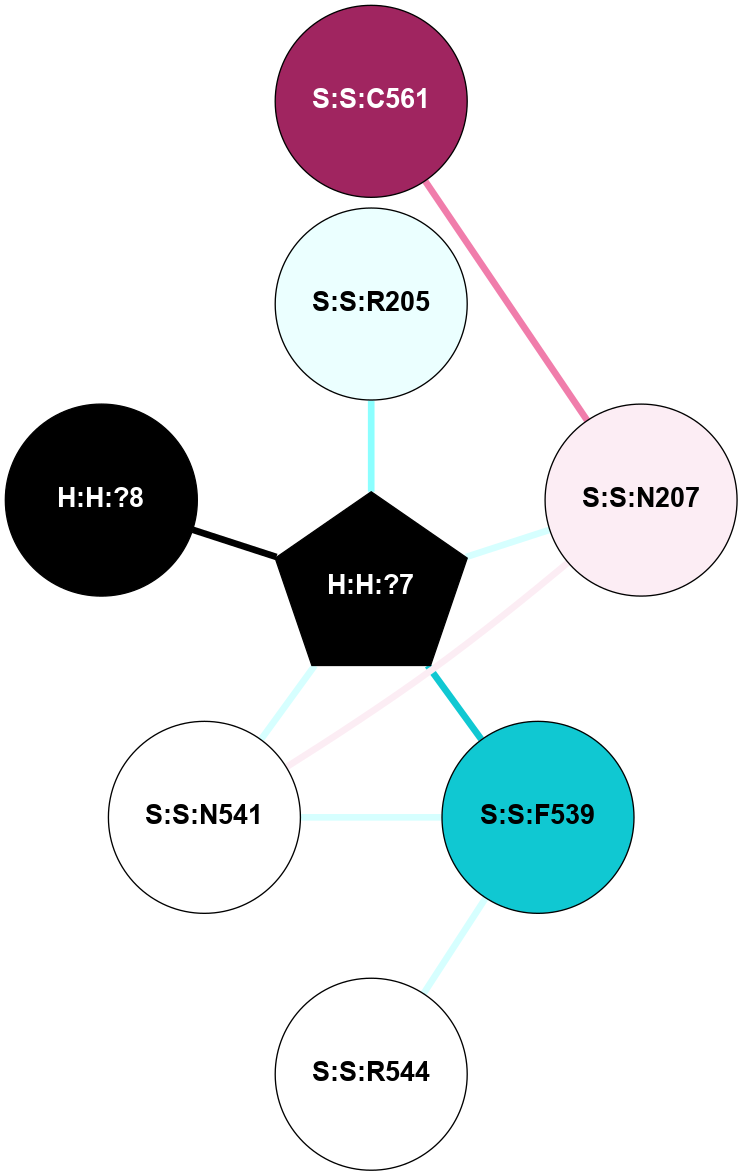
A 2D representation of the interactions of NAG in 7SIM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | S:S:R205 | 13.08 | 25 | Yes | No | 0 | 4 | 0 | 1 | | S:S:N207 | S:S:N541 | 17.71 | 25 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 7.87 | 0 | No | No | 9 | 6 | 2 | 1 | | H:H:?7 | S:S:N207 | 12.32 | 25 | Yes | No | 0 | 6 | 0 | 1 | | S:S:F539 | S:S:N541 | 7.25 | 25 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 32.07 | 25 | No | No | 1 | 5 | 1 | 2 | | H:H:?7 | S:S:F539 | 6.56 | 25 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?7 | S:S:N541 | 25.87 | 25 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?7 | H:H:?8 | 30.09 | 25 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.58 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
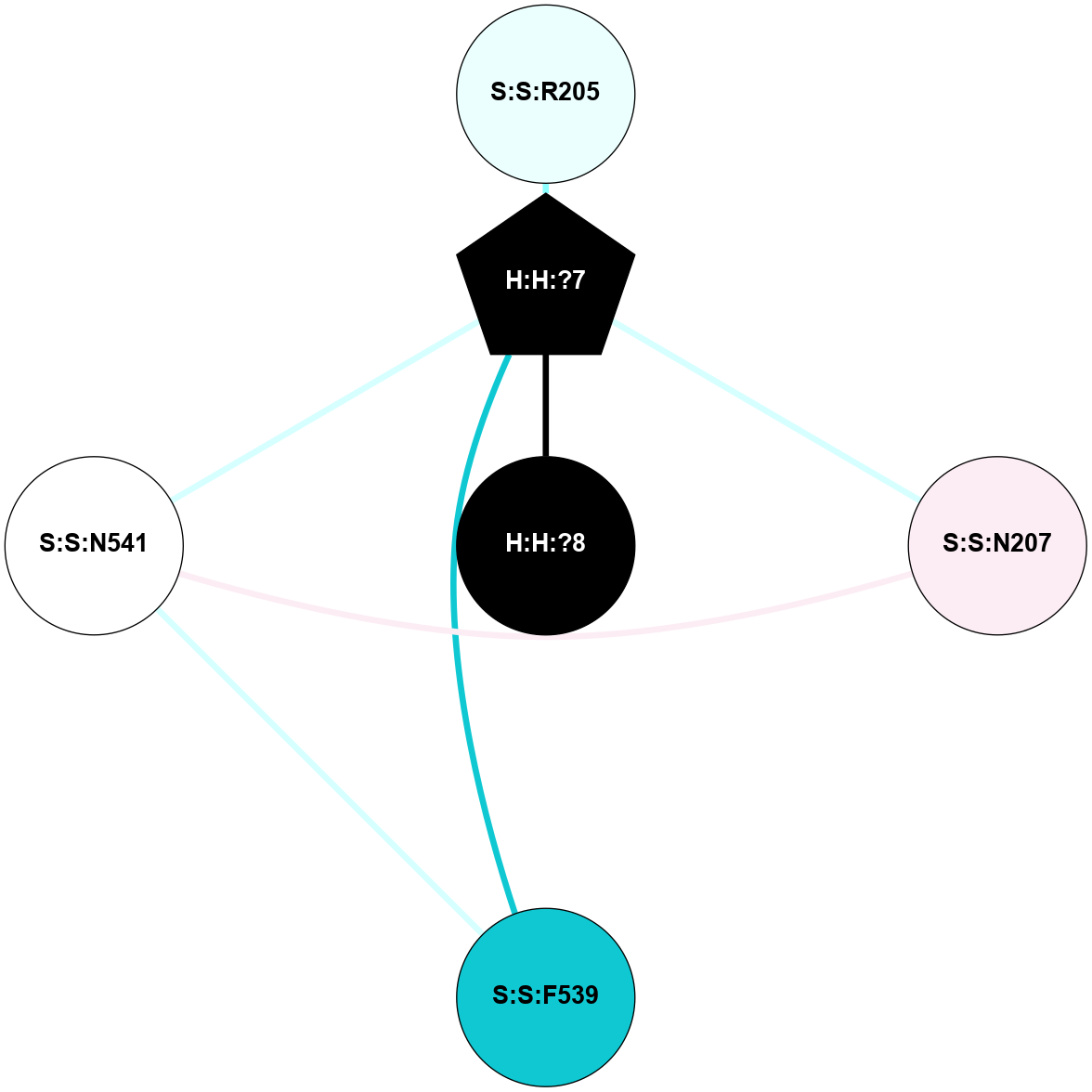
A 2D representation of the interactions of NAG in 7SIM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | S:S:R205 | 13.08 | 25 | Yes | No | 0 | 4 | 1 | 2 | | S:S:N207 | S:S:N541 | 17.71 | 25 | No | No | 6 | 5 | 2 | 2 | | H:H:?7 | S:S:N207 | 12.32 | 25 | Yes | No | 0 | 6 | 1 | 2 | | S:S:F539 | S:S:N541 | 7.25 | 25 | No | No | 1 | 5 | 2 | 2 | | H:H:?7 | S:S:F539 | 6.56 | 25 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?7 | S:S:N541 | 25.87 | 25 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?7 | H:H:?8 | 30.09 | 25 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 30.09 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7SIM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N468 | R:R:Q476 | 7.92 | 33 | No | No | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 8.77 | 33 | No | No | 6 | 3 | 1 | 2 | | H:H:?9 | R:R:N468 | 24.64 | 33 | No | No | 0 | 6 | 0 | 1 | | R:R:Q476 | R:R:T470 | 8.5 | 33 | No | No | 3 | 5 | 1 | 2 | | H:H:?9 | R:R:Q476 | 14.33 | 33 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 19.48 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7SIM
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:N468 | S:S:Q476 | 7.92 | 44 | No | No | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 10.24 | 44 | No | No | 6 | 3 | 1 | 2 | | H:H:?10 | S:S:N468 | 23.41 | 44 | No | No | 0 | 6 | 0 | 1 | | S:S:Q476 | S:S:T470 | 8.5 | 44 | No | No | 3 | 5 | 1 | 2 | | H:H:?10 | S:S:Q476 | 14.33 | 44 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.87 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7SRQ | A | Amine | 5-Hydroxytryptamine | 5-HT2B | Homo Sapiens | LSD | - | - | 2.7 | 2022-09-21 | doi.org/10.1016/j.neuron.2022.08.006 |
|
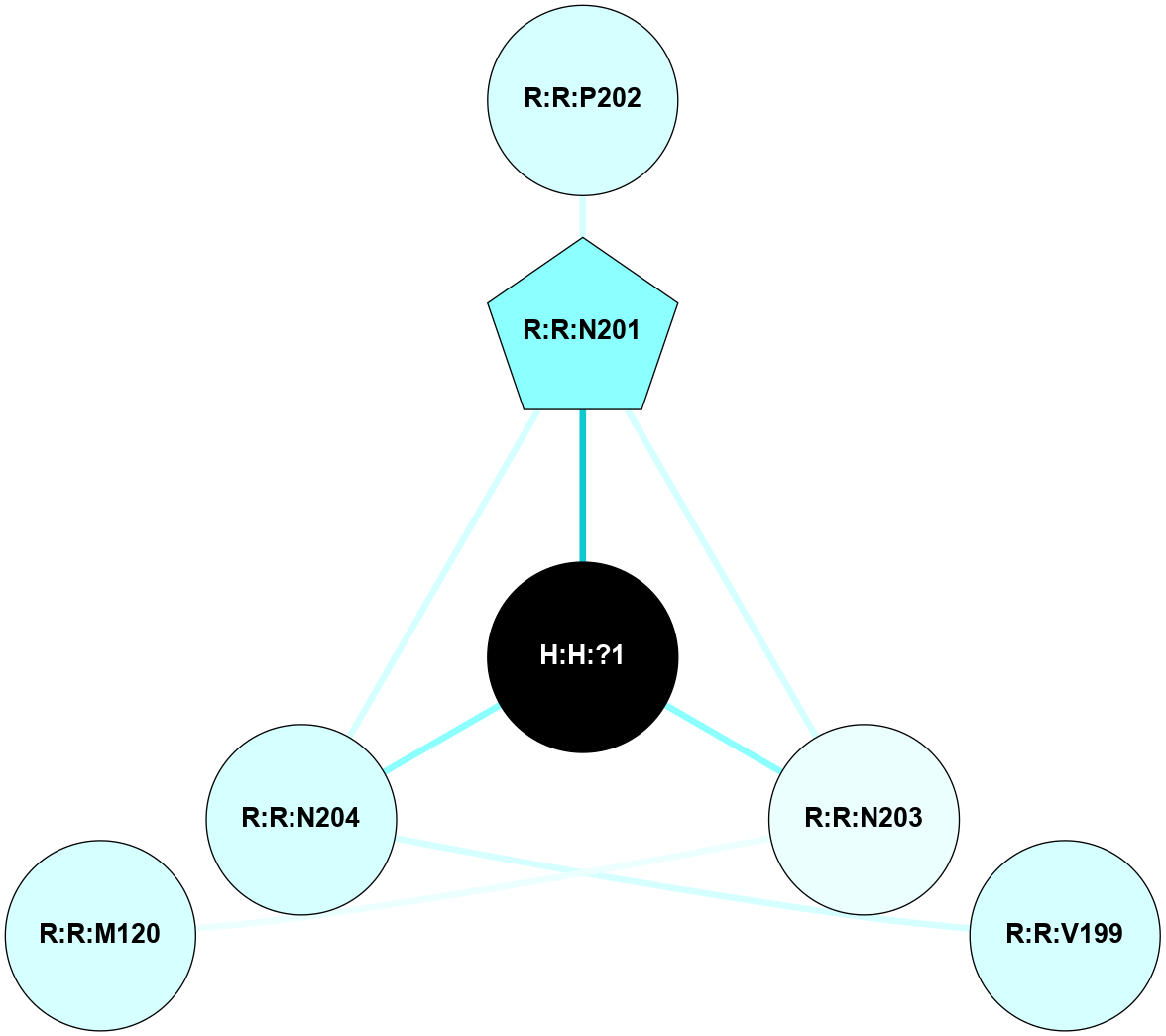
A 2D representation of the interactions of NAG in 7SRQ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N201 | 4.91 | 3 | No | Yes | 0 | 2 | 0 | 1 | | H:H:?1 | R:R:N203 | 7.37 | 3 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:N204 | 25.79 | 3 | No | No | 0 | 3 | 0 | 1 | | R:R:N201 | R:R:P202 | 4.89 | 3 | Yes | No | 2 | 3 | 1 | 2 | | R:R:N201 | R:R:N203 | 4.09 | 3 | Yes | No | 2 | 4 | 1 | 1 | | R:R:N201 | R:R:N204 | 12.26 | 3 | Yes | No | 2 | 3 | 1 | 1 | | R:R:N204 | R:R:V199 | 1.48 | 3 | No | No | 3 | 3 | 1 | 2 | | R:R:M120 | R:R:N203 | 1.4 | 0 | No | No | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 12.69 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7SRS | A | Amine | 5-Hydroxytryptamine | 5-HT2B | Homo Sapiens | LSD | - | Arrestin2 | 3.3 | 2022-09-21 | doi.org/10.1016/j.neuron.2022.08.006 |
|
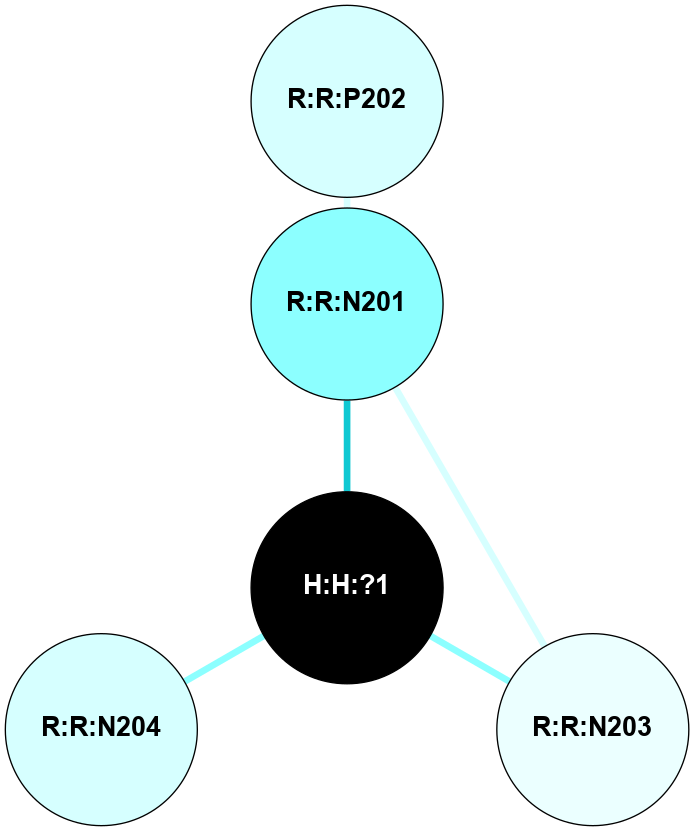
A 2D representation of the interactions of NAG in 7SRS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N201 | 4.91 | 12 | No | No | 0 | 2 | 0 | 1 | | H:H:?1 | R:R:N203 | 2.46 | 12 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:N204 | 29.47 | 12 | No | No | 0 | 3 | 0 | 1 | | R:R:N201 | R:R:P202 | 4.89 | 12 | No | No | 2 | 3 | 1 | 2 | | R:R:N201 | R:R:N203 | 2.72 | 12 | No | No | 2 | 4 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.28 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7T9I | A | Protein | Glycoprotein Hormone | TSH | Homo Sapiens | TSH | - | Gs/Beta1/Gamma2 | 2.9 | 2022-08-03 | doi.org/10.1038/s41586-022-05159-1 |
|
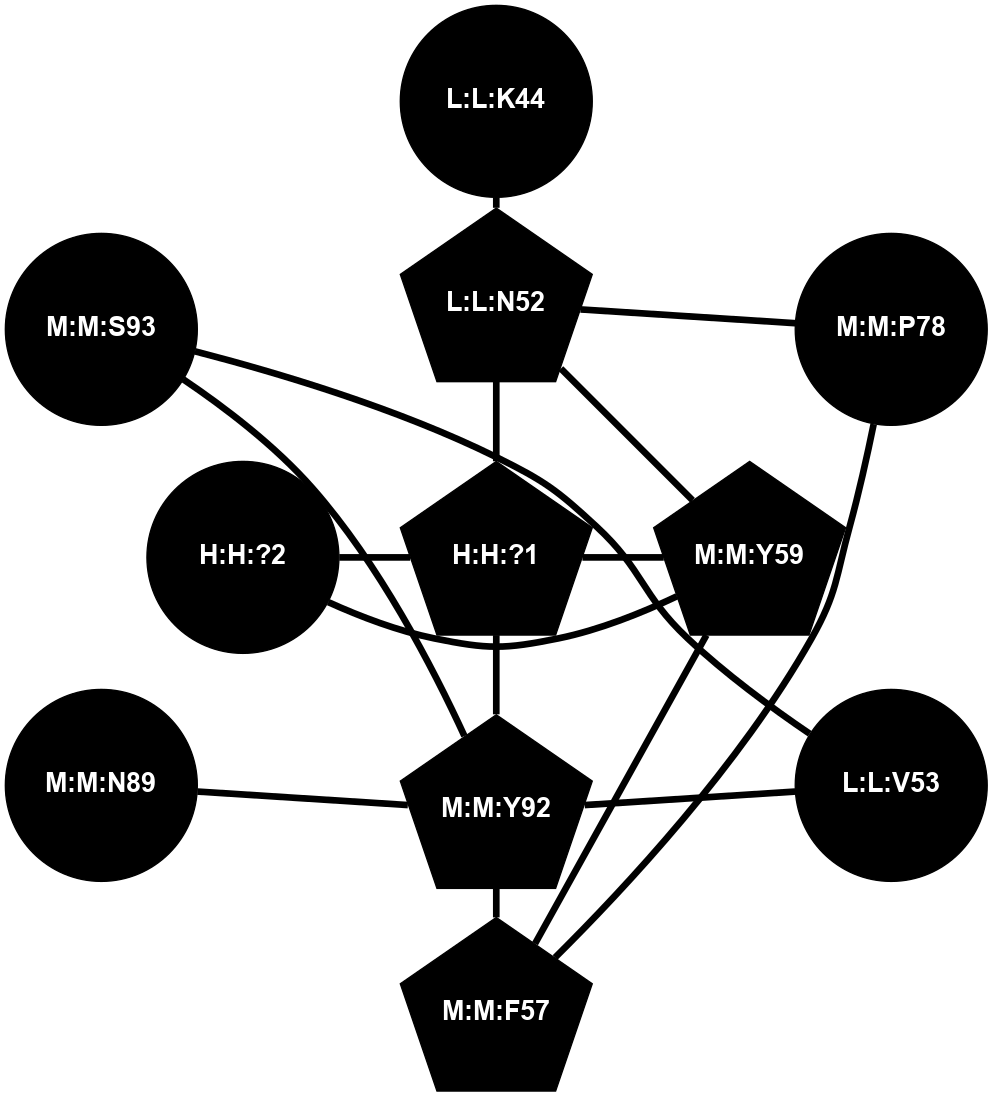
A 2D representation of the interactions of NAG in 7T9I
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:K44 | L:L:N52 | 4.2 | 0 | No | Yes | 0 | 0 | 2 | 1 | | L:L:N52 | N:N:Y59 | 5.81 | 15 | Yes | Yes | 0 | 0 | 1 | 1 | | L:L:N52 | N:N:P78 | 3.26 | 15 | Yes | No | 0 | 0 | 1 | 2 | | H:H:?1 | L:L:N52 | 34.5 | 15 | Yes | Yes | 0 | 0 | 0 | 1 | | L:L:V53 | N:N:Y92 | 3.79 | 15 | No | Yes | 0 | 0 | 2 | 1 | | L:L:V53 | N:N:S93 | 6.46 | 15 | No | No | 0 | 0 | 2 | 2 | | N:N:F57 | N:N:Y59 | 6.19 | 0 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:F57 | N:N:P78 | 5.78 | 0 | Yes | No | 0 | 0 | 2 | 2 | | N:N:F57 | N:N:Y92 | 8.25 | 0 | Yes | Yes | 0 | 0 | 2 | 1 | | H:H:?1 | N:N:Y59 | 16.83 | 15 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?2 | N:N:Y59 | 4.21 | 15 | No | Yes | 0 | 0 | 1 | 1 | | N:N:N89 | N:N:Y92 | 16.28 | 0 | No | Yes | 0 | 0 | 2 | 1 | | N:N:S93 | N:N:Y92 | 6.36 | 15 | No | Yes | 0 | 0 | 2 | 1 | | H:H:?1 | N:N:Y92 | 3.16 | 15 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?1 | H:H:?2 | 64.64 | 15 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 29.78 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
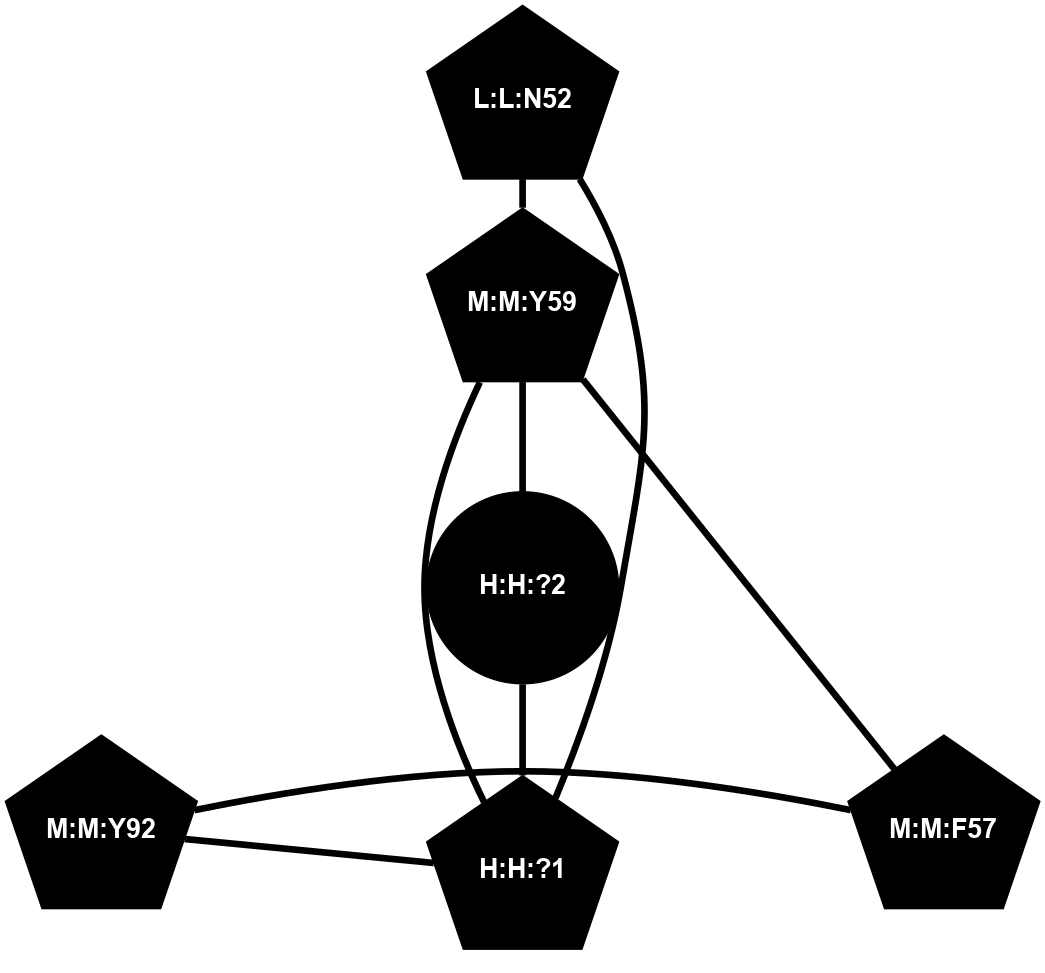
A 2D representation of the interactions of NAG in 7T9I
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:N52 | N:N:Y59 | 5.81 | 15 | Yes | Yes | 0 | 0 | 2 | 1 | | H:H:?1 | L:L:N52 | 34.5 | 15 | Yes | Yes | 0 | 0 | 1 | 2 | | N:N:F57 | N:N:Y59 | 6.19 | 0 | Yes | Yes | 0 | 0 | 2 | 1 | | N:N:F57 | N:N:Y92 | 8.25 | 0 | Yes | Yes | 0 | 0 | 2 | 2 | | H:H:?1 | N:N:Y59 | 16.83 | 15 | Yes | Yes | 0 | 0 | 1 | 1 | | H:H:?2 | N:N:Y59 | 4.21 | 15 | No | Yes | 0 | 0 | 0 | 1 | | H:H:?1 | N:N:Y92 | 3.16 | 15 | Yes | Yes | 0 | 0 | 1 | 2 | | H:H:?1 | H:H:?2 | 64.64 | 15 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 34.42 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
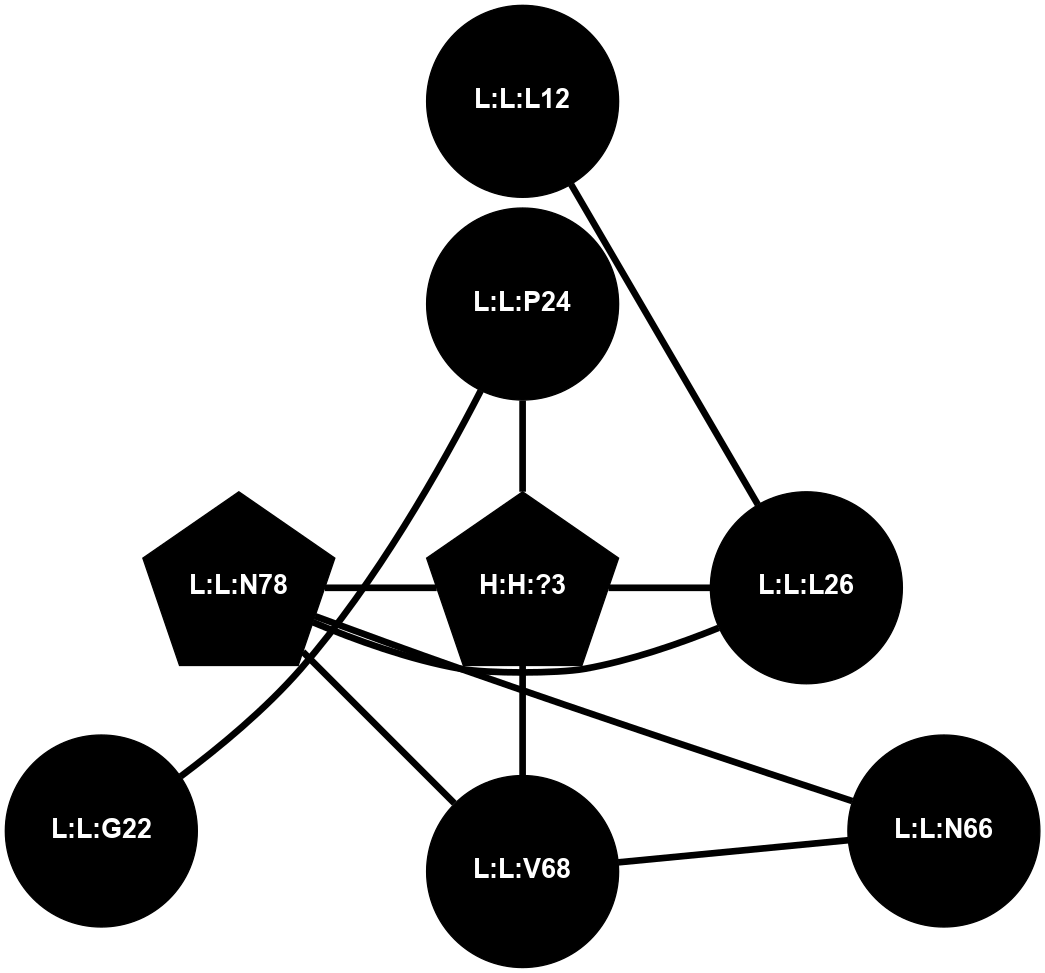
A 2D representation of the interactions of NAG in 7T9I
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:L12 | L:L:L26 | 8.3 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:I25 | L:L:S19 | 3.1 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:A23 | L:L:Q20 | 3.03 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?3 | L:L:A23 | 5.66 | 21 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | L:L:P24 | 10.31 | 21 | Yes | No | 0 | 0 | 0 | 1 | | L:L:I25 | L:L:V76 | 4.61 | 0 | No | Yes | 0 | 0 | 1 | 2 | | H:H:?3 | L:L:I25 | 5.12 | 21 | Yes | No | 0 | 0 | 0 | 1 | | L:L:L26 | L:L:N78 | 4.12 | 21 | No | Yes | 0 | 0 | 1 | 1 | | H:H:?3 | L:L:L26 | 8.69 | 21 | Yes | No | 0 | 0 | 0 | 1 | | L:L:S64 | L:L:T80 | 6.4 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:N66 | L:L:V68 | 4.43 | 21 | No | No | 0 | 0 | 2 | 1 | | L:L:N66 | L:L:N78 | 8.17 | 21 | No | Yes | 0 | 0 | 2 | 1 | | L:L:N66 | L:L:T80 | 8.77 | 21 | No | No | 0 | 0 | 2 | 1 | | L:L:N78 | L:L:V68 | 4.43 | 21 | Yes | No | 0 | 0 | 1 | 1 | | H:H:?3 | L:L:V68 | 8.02 | 21 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | L:L:N78 | 33.27 | 21 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?3 | L:L:T80 | 6.61 | 21 | Yes | No | 0 | 0 | 0 | 1 | | L:L:G22 | L:L:P24 | 2.03 | 0 | No | No | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 11.10 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7T9I
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | N:N:N23 | N:N:T8 | 7.31 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?4 | N:N:N23 | 33.27 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 33.27 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
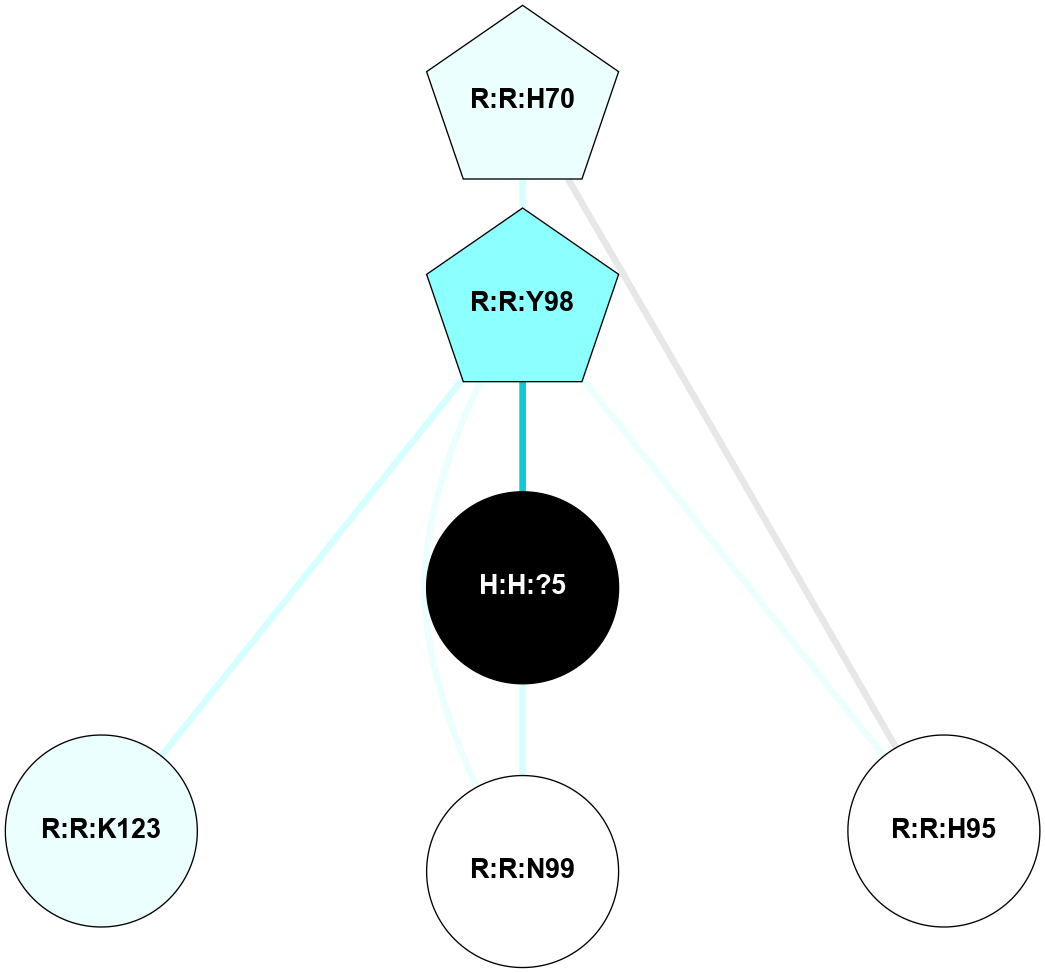
A 2D representation of the interactions of NAG in 7T9I
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:H70 | R:R:H95 | 8.36 | 22 | Yes | No | 4 | 5 | 2 | 2 | | R:R:H70 | R:R:Y98 | 4.36 | 22 | Yes | Yes | 4 | 2 | 2 | 1 | | R:R:H95 | R:R:Y98 | 11.98 | 22 | No | Yes | 5 | 2 | 2 | 1 | | R:R:N99 | R:R:Y98 | 6.98 | 22 | No | Yes | 5 | 2 | 1 | 1 | | R:R:K123 | R:R:Y98 | 3.58 | 0 | No | Yes | 4 | 2 | 2 | 1 | | H:H:?5 | R:R:Y98 | 8.42 | 22 | No | Yes | 0 | 2 | 0 | 1 | | H:H:?5 | R:R:N99 | 34.5 | 22 | No | No | 0 | 5 | 0 | 1 | | R:R:K102 | R:R:S101 | 3.06 | 0 | No | No | 4 | 4 | 2 | 1 | | H:H:?5 | R:R:S101 | 2.69 | 22 | No | No | 0 | 4 | 0 | 1 | | R:R:P126 | R:R:S101 | 1.78 | 0 | No | No | 5 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.20 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
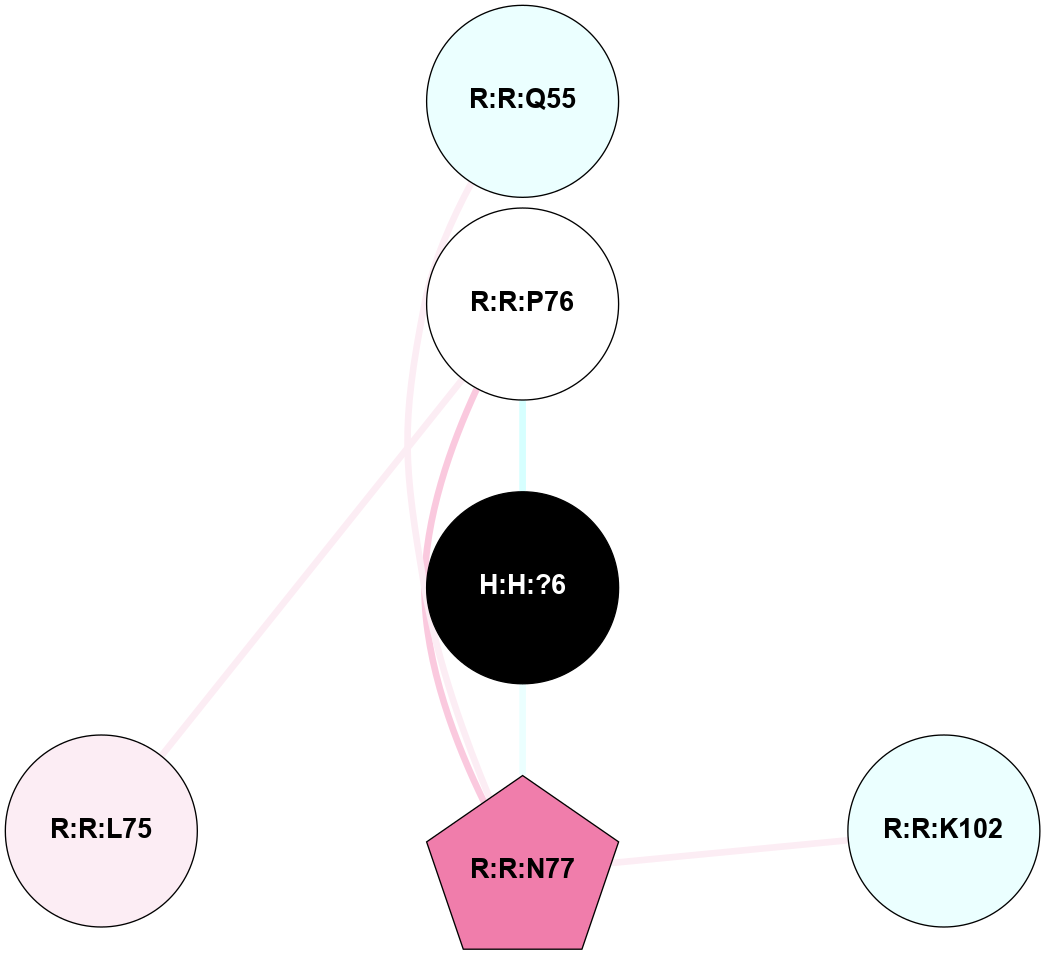
A 2D representation of the interactions of NAG in 7T9I
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:M22 | R:R:P52 | 5.03 | 0 | No | No | 2 | 3 | 2 | 1 | | R:R:M22 | R:R:S53 | 3.07 | 0 | No | No | 2 | 5 | 2 | 1 | | H:H:?6 | R:R:P52 | 2.95 | 35 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?6 | R:R:S53 | 4.04 | 35 | Yes | No | 0 | 5 | 0 | 1 | | R:R:N77 | R:R:Q55 | 6.6 | 35 | No | No | 8 | 4 | 1 | 2 | | R:R:N77 | R:R:P76 | 6.52 | 35 | No | No | 8 | 5 | 1 | 1 | | H:H:?6 | R:R:P76 | 8.84 | 35 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?6 | R:R:N77 | 29.57 | 35 | Yes | No | 0 | 8 | 0 | 1 | | R:R:K102 | R:R:S101 | 3.06 | 0 | No | No | 4 | 4 | 1 | 2 | | H:H:?6 | R:R:K102 | 3.8 | 35 | Yes | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 9.84 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
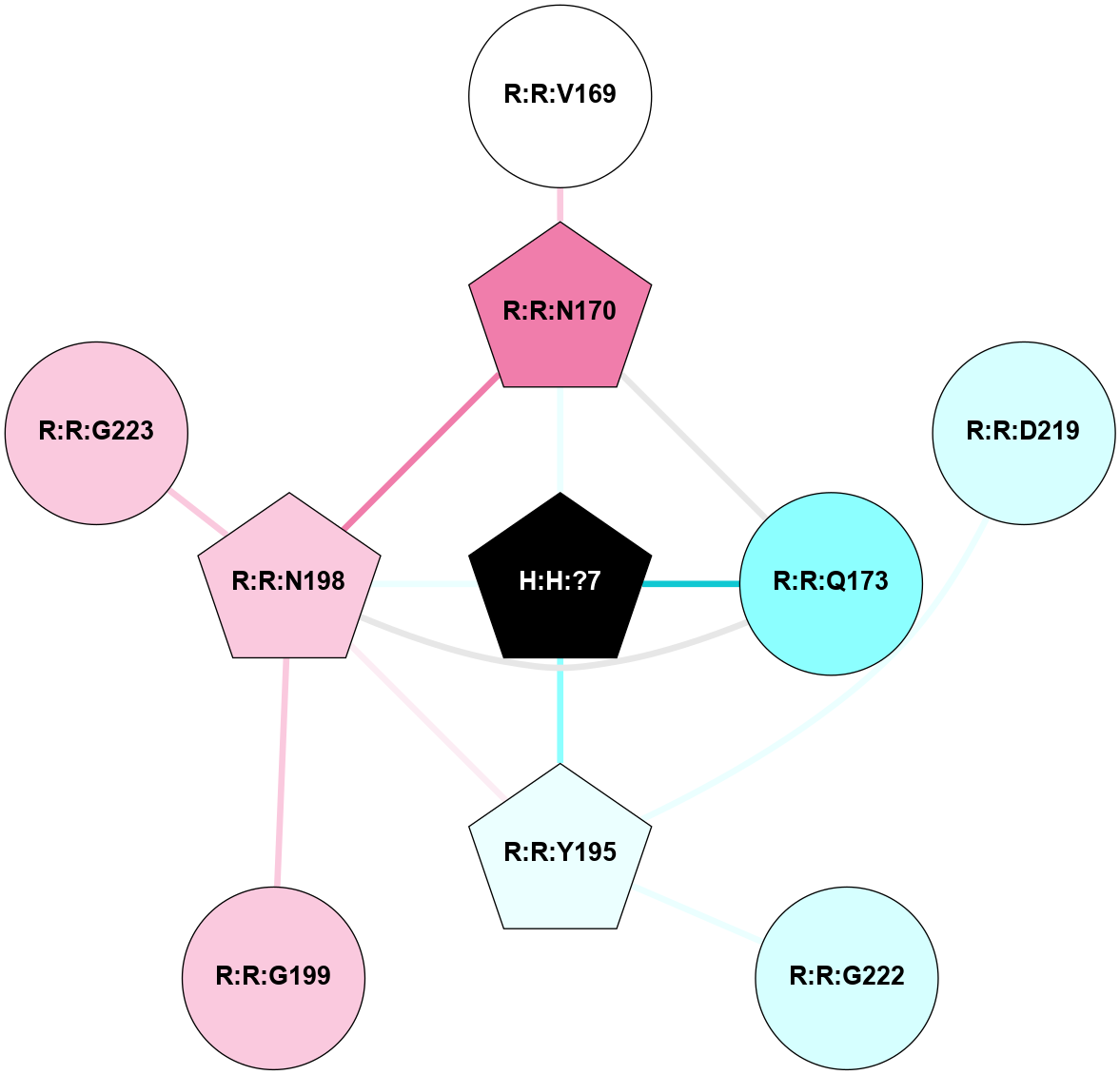
A 2D representation of the interactions of NAG in 7T9I
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N170 | R:R:V169 | 2.96 | 23 | Yes | No | 8 | 5 | 1 | 2 | | R:R:N170 | R:R:Q173 | 15.85 | 23 | Yes | No | 8 | 2 | 1 | 1 | | R:R:N170 | R:R:N198 | 5.45 | 23 | Yes | Yes | 8 | 7 | 1 | 1 | | H:H:?7 | R:R:N170 | 11.09 | 23 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:N198 | R:R:Q173 | 5.28 | 23 | Yes | No | 7 | 2 | 1 | 1 | | H:H:?7 | R:R:Q173 | 10.75 | 23 | Yes | No | 0 | 2 | 0 | 1 | | R:R:N198 | R:R:Y195 | 5.81 | 23 | Yes | Yes | 7 | 4 | 1 | 1 | | R:R:D219 | R:R:Y195 | 4.6 | 0 | No | Yes | 3 | 4 | 2 | 1 | | R:R:G222 | R:R:Y195 | 2.9 | 0 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?7 | R:R:Y195 | 12.62 | 23 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:G199 | R:R:N198 | 3.39 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:G223 | R:R:N198 | 3.39 | 0 | No | Yes | 7 | 7 | 2 | 1 | | H:H:?7 | R:R:N198 | 25.87 | 23 | Yes | Yes | 0 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 15.08 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
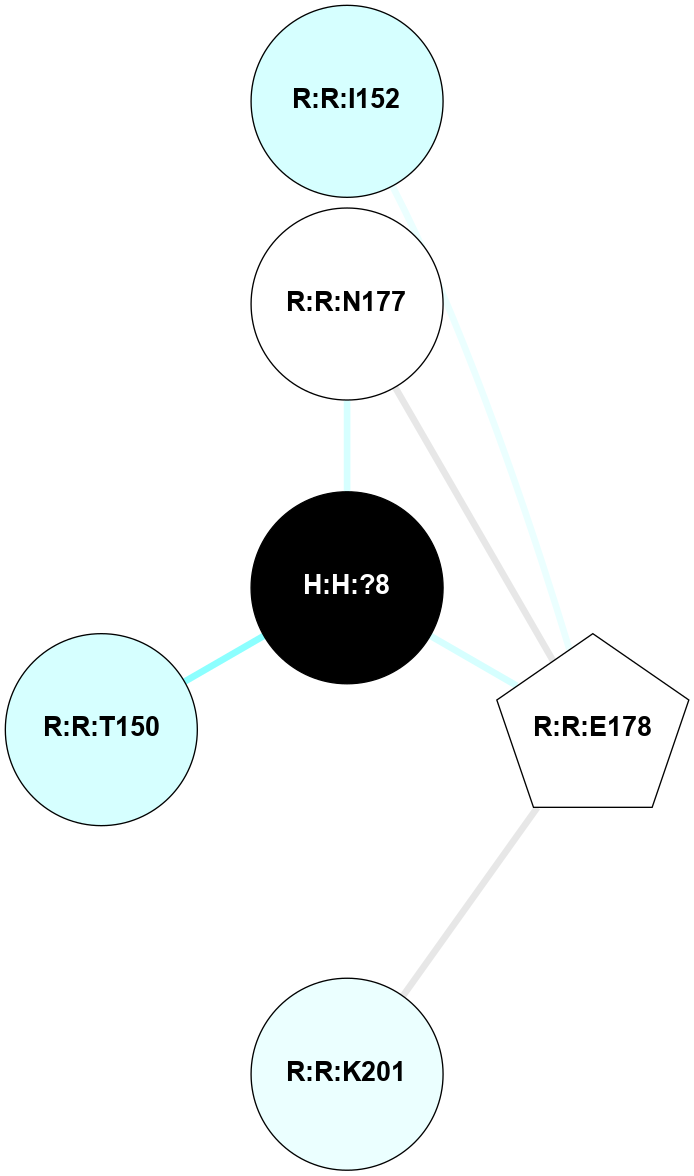
A 2D representation of the interactions of NAG in 7T9I
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | R:R:T150 | 5.29 | 38 | Yes | No | 0 | 3 | 0 | 1 | | R:R:E178 | R:R:I152 | 6.83 | 38 | No | No | 5 | 3 | 1 | 2 | | R:R:E178 | R:R:N177 | 3.94 | 38 | No | No | 5 | 5 | 1 | 1 | | H:H:?8 | R:R:N177 | 32.04 | 38 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?8 | R:R:E178 | 23.78 | 38 | Yes | No | 0 | 5 | 0 | 1 | | R:R:K201 | R:R:Y225 | 4.78 | 0 | No | No | 4 | 4 | 1 | 2 | | R:R:K201 | R:R:S226 | 4.59 | 0 | No | No | 4 | 4 | 1 | 2 | | H:H:?8 | R:R:K201 | 8.86 | 38 | Yes | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.49 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7T9M | A | Protein | Glycoprotein Hormone | TSH | Homo Sapiens | - | CS-17 Antibody | - | 3.1 | 2022-08-10 | doi.org/10.1038/s41586-022-05159-1 |
|
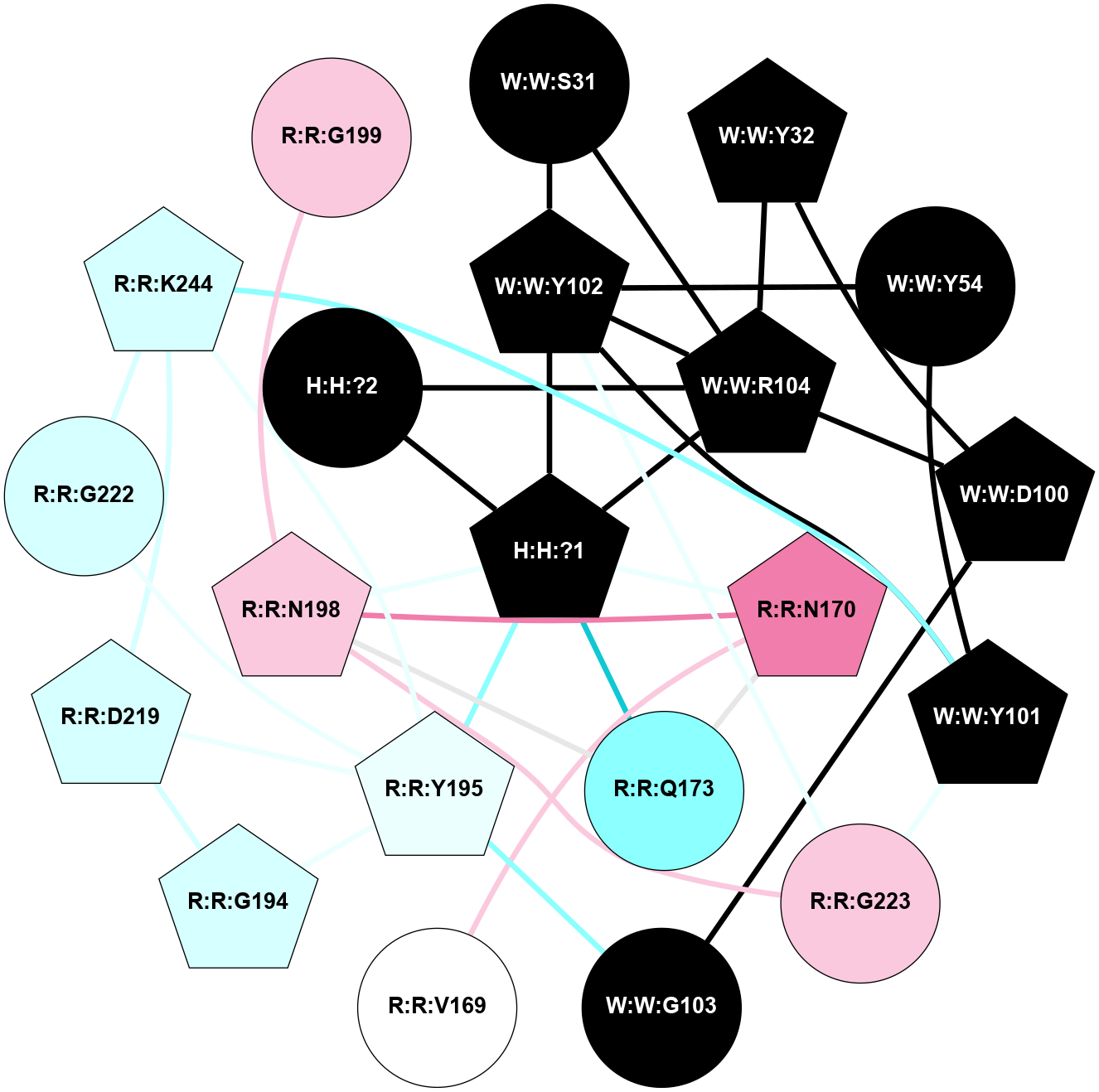
A 2D representation of the interactions of NAG in 7T9M
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 39.01 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N170 | 12.32 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?1 | R:R:Q173 | 10.75 | 1 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?1 | R:R:Y195 | 4.21 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:N198 | 24.64 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | W:W:Y102 | 3.16 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?1 | W:W:R104 | 4.36 | 1 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?2 | W:W:R104 | 8.72 | 1 | No | Yes | 0 | 0 | 1 | 1 | | R:R:N170 | R:R:V169 | 4.43 | 1 | Yes | No | 8 | 5 | 1 | 2 | | R:R:N170 | R:R:Q173 | 10.56 | 1 | Yes | No | 8 | 2 | 1 | 1 | | R:R:N170 | R:R:N198 | 8.17 | 1 | Yes | Yes | 8 | 7 | 1 | 1 | | R:R:N198 | R:R:Q173 | 2.64 | 1 | Yes | No | 7 | 2 | 1 | 1 | | R:R:G194 | R:R:Y195 | 2.9 | 1 | Yes | Yes | 3 | 4 | 2 | 1 | | R:R:D219 | R:R:G194 | 5.03 | 1 | Yes | Yes | 3 | 3 | 2 | 2 | | R:R:D219 | R:R:Y195 | 17.24 | 1 | Yes | Yes | 3 | 4 | 2 | 1 | | R:R:G222 | R:R:Y195 | 2.9 | 1 | No | Yes | 3 | 4 | 2 | 1 | | R:R:K244 | R:R:Y195 | 5.97 | 1 | Yes | Yes | 3 | 4 | 2 | 1 | | R:R:Y195 | W:W:G103 | 7.24 | 1 | Yes | No | 4 | 0 | 1 | 2 | | R:R:G199 | R:R:N198 | 3.39 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:G223 | R:R:N198 | 3.39 | 1 | No | Yes | 7 | 7 | 2 | 1 | | R:R:D219 | R:R:K244 | 4.15 | 1 | Yes | Yes | 3 | 3 | 2 | 2 | | R:R:G222 | R:R:K244 | 5.23 | 1 | No | Yes | 3 | 3 | 2 | 2 | | R:R:G223 | W:W:Y101 | 5.79 | 1 | No | Yes | 7 | 0 | 2 | 2 | | R:R:G223 | W:W:Y102 | 2.9 | 1 | No | Yes | 7 | 0 | 2 | 1 | | R:R:K244 | W:W:Y101 | 3.58 | 1 | Yes | Yes | 3 | 0 | 2 | 2 | | W:W:S31 | W:W:Y102 | 7.63 | 1 | No | Yes | 0 | 0 | 2 | 1 | | W:W:R104 | W:W:S31 | 3.95 | 1 | Yes | No | 0 | 0 | 1 | 2 | | W:W:D100 | W:W:Y32 | 17.24 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | W:W:R104 | W:W:Y32 | 5.14 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | W:W:Y101 | W:W:Y54 | 13.9 | 1 | Yes | No | 0 | 0 | 2 | 2 | | W:W:Y102 | W:W:Y54 | 6.95 | 1 | Yes | No | 0 | 0 | 1 | 2 | | W:W:D100 | W:W:G103 | 5.03 | 1 | Yes | No | 0 | 0 | 2 | 2 | | W:W:D100 | W:W:R104 | 15.48 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | W:W:Y101 | W:W:Y102 | 8.94 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | W:W:R104 | W:W:Y102 | 9.26 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 5.00 | | Average Interaction Strength | 14.06 | | Average Nodes In Shell | 21.00 | | Average Hubs In Shell | 12.00 | | Average Links In Shell | 35.00 | | Average Links Mediated by Hubs In Shell | 35.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
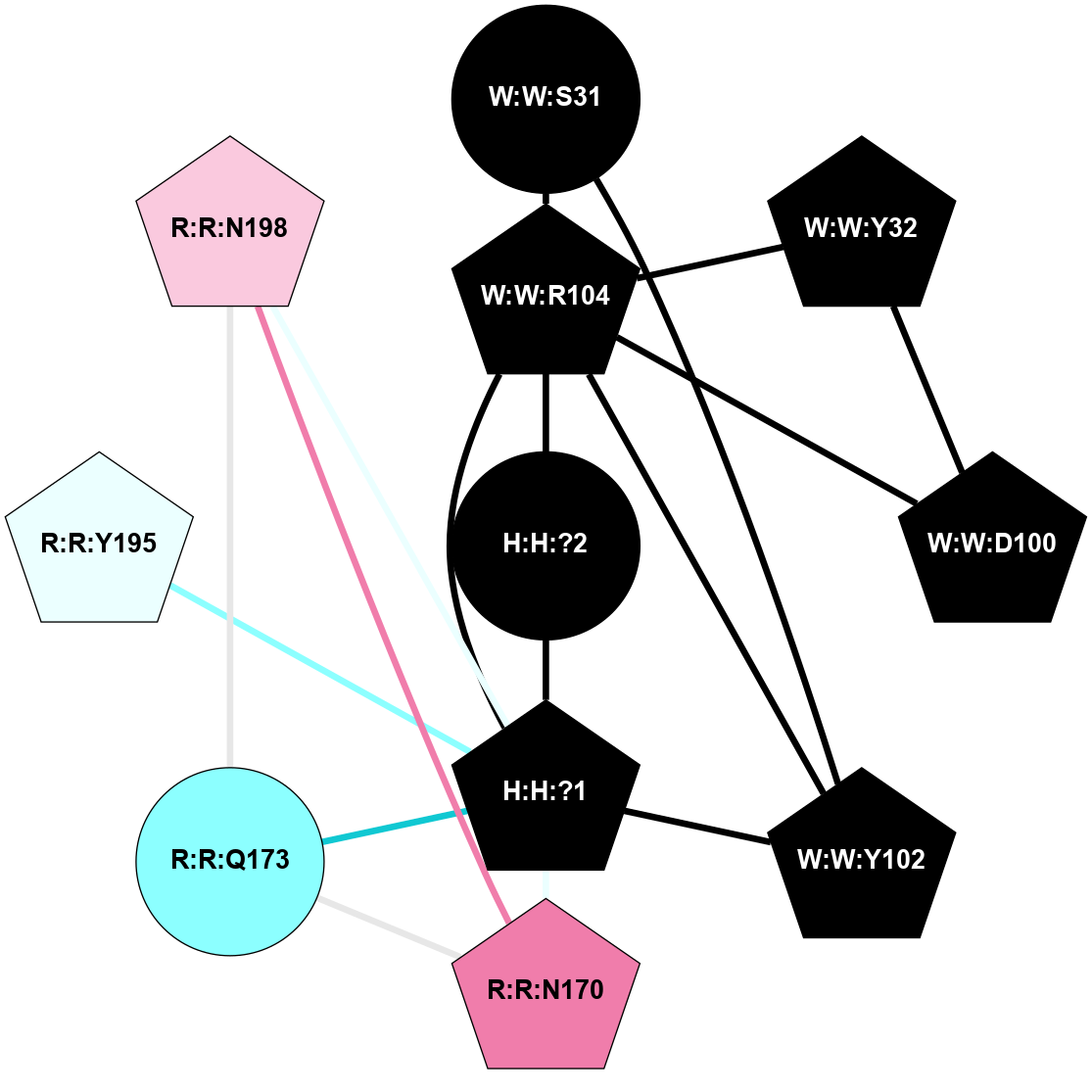
A 2D representation of the interactions of NAG in 7T9M
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 39.01 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N170 | 12.32 | 1 | Yes | Yes | 0 | 8 | 1 | 2 | | H:H:?1 | R:R:Q173 | 10.75 | 1 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?1 | R:R:Y195 | 4.21 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:N198 | 24.64 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | W:W:Y102 | 3.16 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | H:H:?1 | W:W:R104 | 4.36 | 1 | Yes | Yes | 0 | 0 | 1 | 1 | | H:H:?2 | W:W:R104 | 8.72 | 1 | No | Yes | 0 | 0 | 0 | 1 | | R:R:N170 | R:R:Q173 | 10.56 | 1 | Yes | No | 8 | 2 | 2 | 2 | | R:R:N170 | R:R:N198 | 8.17 | 1 | Yes | Yes | 8 | 7 | 2 | 2 | | R:R:N198 | R:R:Q173 | 2.64 | 1 | Yes | No | 7 | 2 | 2 | 2 | | W:W:S31 | W:W:Y102 | 7.63 | 1 | No | Yes | 0 | 0 | 2 | 2 | | W:W:R104 | W:W:S31 | 3.95 | 1 | Yes | No | 0 | 0 | 1 | 2 | | W:W:D100 | W:W:Y32 | 17.24 | 1 | Yes | Yes | 0 | 0 | 2 | 2 | | W:W:R104 | W:W:Y32 | 5.14 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | | W:W:D100 | W:W:R104 | 15.48 | 1 | Yes | Yes | 0 | 0 | 2 | 1 | | W:W:R104 | W:W:Y102 | 9.26 | 1 | Yes | Yes | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 23.86 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 8.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 17.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
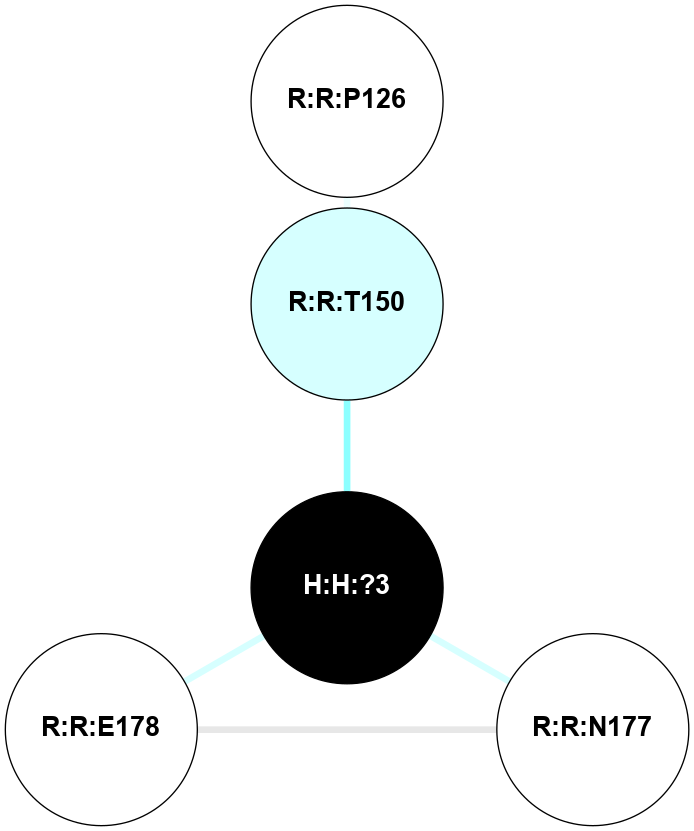
A 2D representation of the interactions of NAG in 7T9M
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:T150 | 5.29 | 20 | No | No | 0 | 3 | 0 | 1 | | H:H:?3 | R:R:N177 | 30.8 | 20 | No | No | 0 | 5 | 0 | 1 | | H:H:?3 | R:R:E178 | 10.7 | 20 | No | No | 0 | 5 | 0 | 1 | | R:R:P126 | R:R:T150 | 3.5 | 0 | No | No | 5 | 3 | 2 | 1 | | R:R:E178 | R:R:N177 | 3.94 | 20 | No | No | 5 | 5 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.60 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
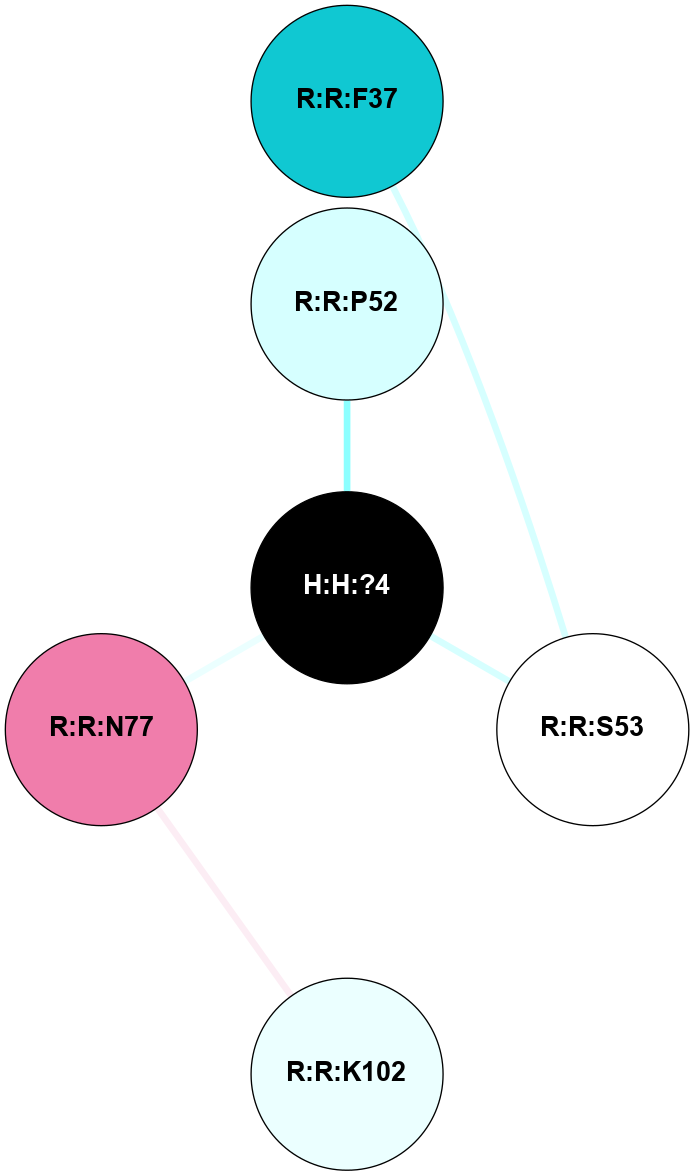
A 2D representation of the interactions of NAG in 7T9M
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:P52 | 2.95 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?4 | R:R:S53 | 4.04 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?4 | R:R:N77 | 20.95 | 0 | No | No | 0 | 8 | 0 | 1 | | R:R:F37 | R:R:S53 | 3.96 | 0 | No | No | 1 | 5 | 2 | 1 | | R:R:K102 | R:R:N77 | 4.2 | 0 | No | No | 4 | 8 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 9.31 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
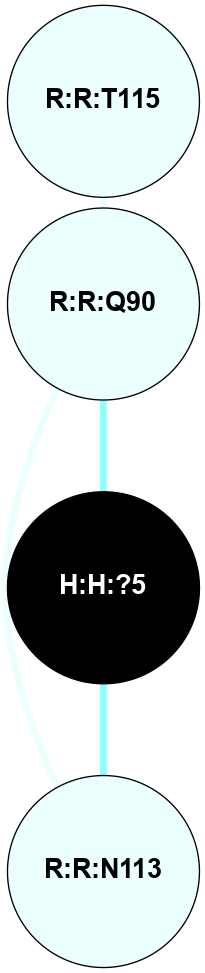
A 2D representation of the interactions of NAG in 7T9M
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | R:R:Q90 | 3.58 | 21 | No | No | 0 | 4 | 0 | 1 | | H:H:?5 | R:R:N113 | 25.87 | 21 | No | No | 0 | 4 | 0 | 1 | | R:R:N113 | R:R:Q90 | 3.96 | 21 | No | No | 4 | 4 | 1 | 1 | | R:R:Q90 | R:R:T115 | 4.25 | 21 | No | No | 4 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.73 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7T9N | A | Protein | Glycoprotein Hormone | TSH | Homo Sapiens | M22 Fab | DPPC | Gs/Beta1/Gamma2 | 2.9 | 2022-08-03 | doi.org/10.1038/s41586-022-05159-1 |
|
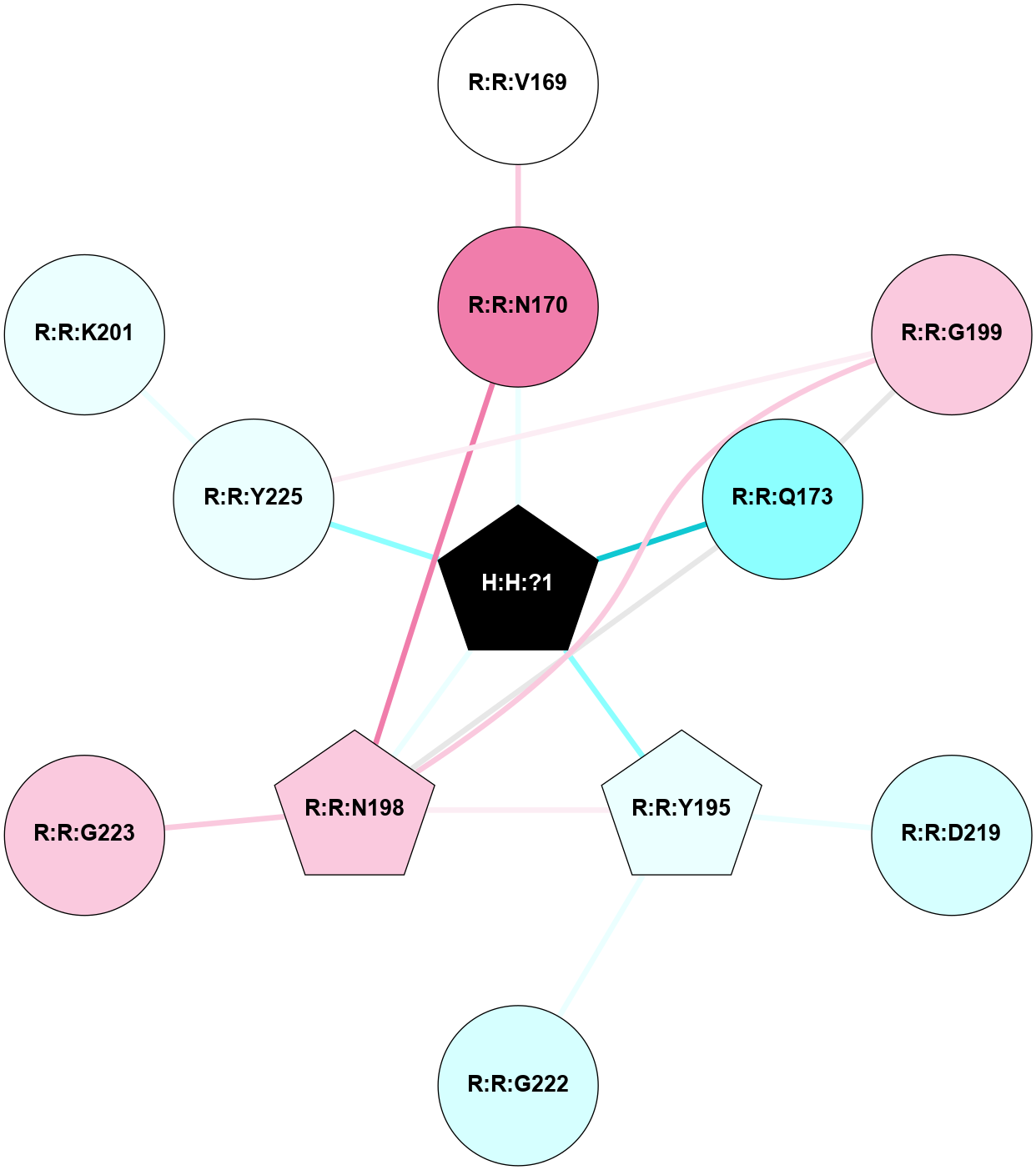
A 2D representation of the interactions of NAG in 7T9N
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N170 | R:R:V169 | 7.39 | 23 | No | No | 8 | 5 | 1 | 2 | | R:R:N170 | R:R:N198 | 9.54 | 23 | No | Yes | 8 | 7 | 1 | 1 | | H:H:?1 | R:R:N170 | 7.39 | 23 | Yes | No | 0 | 8 | 0 | 1 | | R:R:N198 | R:R:Q173 | 9.24 | 23 | Yes | No | 7 | 2 | 1 | 1 | | R:R:G199 | R:R:Q173 | 3.29 | 23 | No | No | 7 | 2 | 2 | 1 | | H:H:?1 | R:R:Q173 | 11.94 | 23 | Yes | No | 0 | 2 | 0 | 1 | | R:R:N198 | R:R:Y195 | 3.49 | 23 | Yes | Yes | 7 | 4 | 1 | 1 | | R:R:D219 | R:R:Y195 | 6.9 | 0 | No | Yes | 3 | 4 | 2 | 1 | | R:R:G222 | R:R:Y195 | 4.35 | 0 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?1 | R:R:Y195 | 5.26 | 23 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:G199 | R:R:N198 | 3.39 | 23 | No | Yes | 7 | 7 | 2 | 1 | | R:R:G223 | R:R:N198 | 3.39 | 0 | No | Yes | 7 | 7 | 2 | 1 | | H:H:?1 | R:R:N198 | 30.8 | 23 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:G199 | R:R:Y225 | 8.69 | 23 | No | No | 7 | 4 | 2 | 1 | | R:R:K201 | R:R:Y225 | 4.78 | 0 | No | No | 4 | 4 | 2 | 1 | | H:H:?1 | R:R:Y225 | 3.16 | 23 | Yes | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 11.71 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
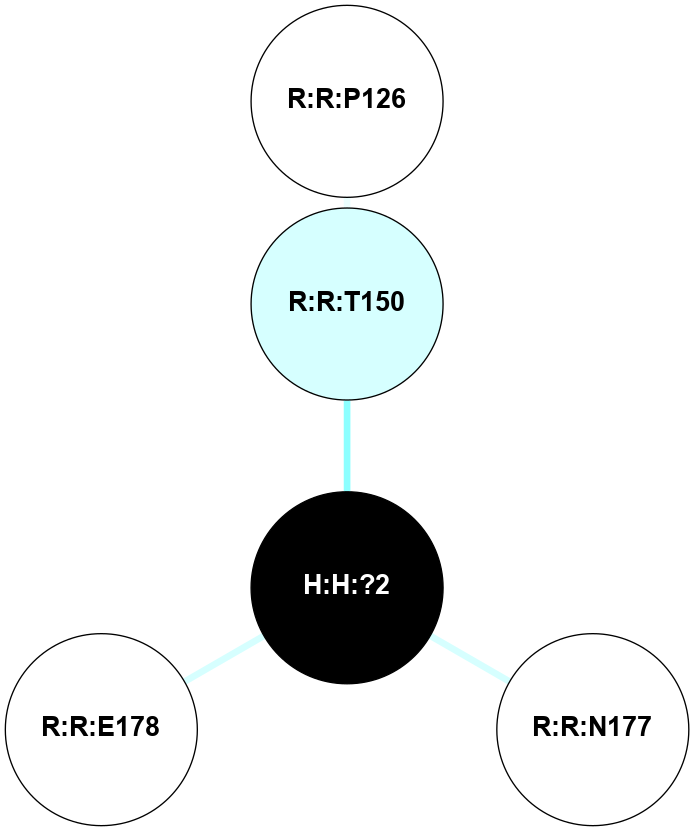
A 2D representation of the interactions of NAG in 7T9N
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:T150 | 10.58 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?2 | R:R:N177 | 33.27 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:E178 | 11.89 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:P126 | R:R:T150 | 1.75 | 0 | No | No | 5 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.58 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
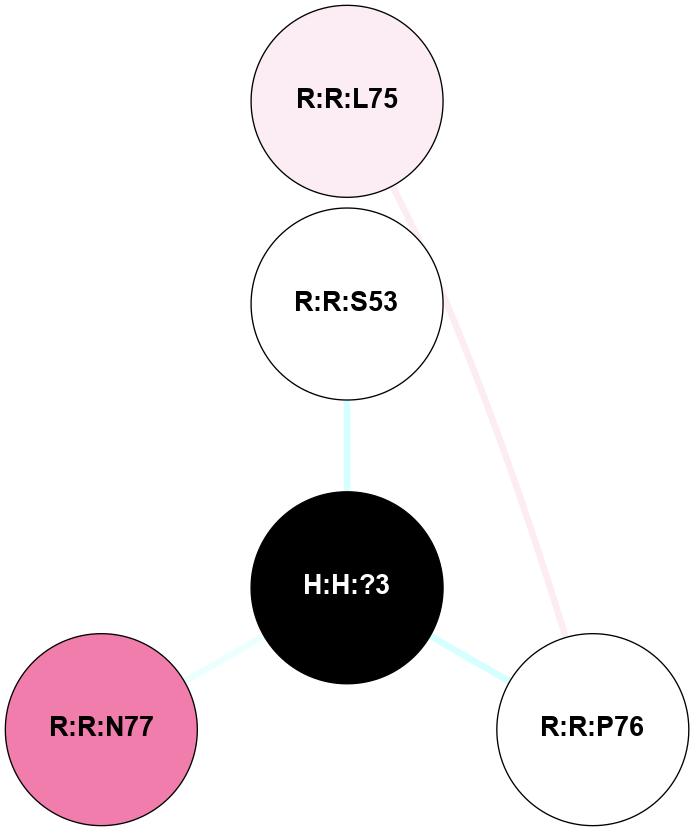
A 2D representation of the interactions of NAG in 7T9N
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:S53 | 16.17 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?3 | R:R:P76 | 10.31 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?3 | R:R:N77 | 39.43 | 0 | No | No | 0 | 8 | 0 | 1 | | R:R:L75 | R:R:P76 | 1.64 | 0 | No | No | 6 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 21.97 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TD3 | A | Lipid | Lysophospholipid | S1P1 | Homo Sapiens | S1P | - | Gi1/Beta1/Gamma2 | 3 | 2022-02-09 | doi.org/10.1038/s41467-022-28417-2 |
|

A 2D representation of the interactions of NAG in 7TD3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N30 | 28.24 | 25 | No | No | 0 | 9 | 0 | 1 | | H:H:?1 | R:R:L35 | 3.71 | 25 | No | Yes | 0 | 6 | 0 | 1 | | R:R:L35 | R:R:N30 | 5.49 | 25 | Yes | No | 6 | 9 | 1 | 1 | | R:R:I37 | R:R:L35 | 4.28 | 0 | No | Yes | 1 | 6 | 2 | 1 | | R:R:L35 | R:R:Y110 | 4.69 | 25 | Yes | Yes | 6 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.97 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TD4 | A | Lipid | Lysophospholipid | S1P1 | Homo Sapiens | Siponimod | - | Gi1/Beta1/Gamma2 | 2.6 | 2022-02-09 | doi.org/10.1038/s41467-022-28417-2 |
|
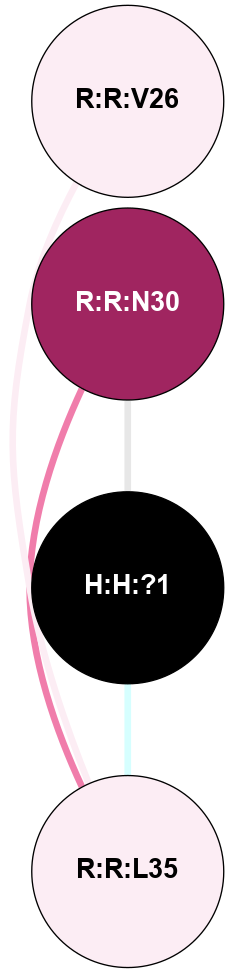
A 2D representation of the interactions of NAG in 7TD4
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:L35 | R:R:N30 | 5.49 | 27 | No | No | 6 | 9 | 1 | 1 | | H:H:?1 | R:R:N30 | 27.02 | 27 | No | No | 0 | 9 | 0 | 1 | | H:H:?1 | R:R:L35 | 4.95 | 27 | No | No | 0 | 6 | 0 | 1 | | R:R:L35 | R:R:V26 | 1.49 | 27 | No | No | 6 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.98 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYF | B1 | Peptide | Calcitonin | CT (AMY1) | Homo Sapiens | Amylin | - | Gs/Beta1/Gamma2; RAMP1 | 2.2 | 2022-03-23 | doi.org/10.1126/science.abm9609 |
|
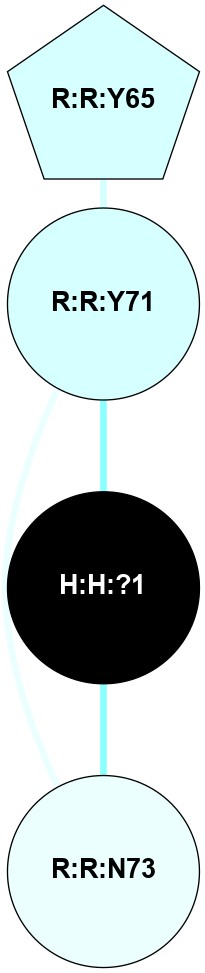
A 2D representation of the interactions of NAG in 7TYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:Y65 | R:R:Y71 | 29.79 | 0 | Yes | No | 3 | 3 | 2 | 1 | | R:R:N73 | R:R:Y71 | 8.14 | 32 | No | No | 4 | 3 | 1 | 1 | | H:H:?1 | R:R:Y71 | 4.19 | 32 | No | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:N73 | 29.47 | 32 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.83 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
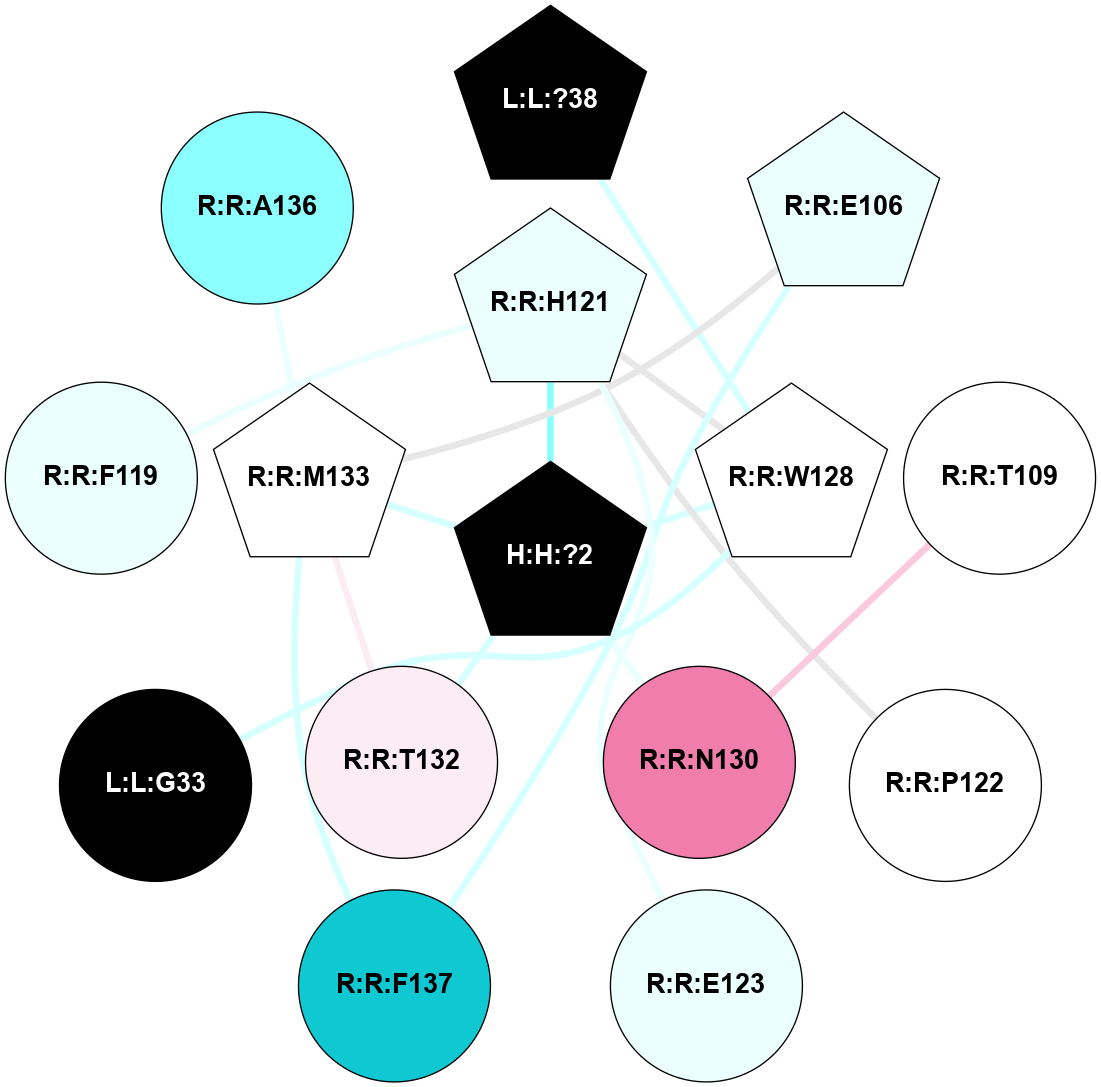
A 2D representation of the interactions of NAG in 7TYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?38 | R:R:W128 | 14.17 | 3 | Yes | Yes | 0 | 5 | 2 | 1 | | R:R:E106 | R:R:M133 | 5.41 | 3 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:E106 | R:R:F137 | 7 | 3 | Yes | No | 4 | 1 | 2 | 2 | | R:R:N130 | R:R:T109 | 11.7 | 0 | No | No | 8 | 5 | 1 | 2 | | R:R:H121 | R:R:P122 | 6.1 | 3 | Yes | No | 4 | 5 | 1 | 2 | | R:R:E123 | R:R:H121 | 18.46 | 0 | No | Yes | 4 | 4 | 2 | 1 | | R:R:H121 | R:R:W128 | 6.35 | 3 | Yes | Yes | 4 | 5 | 1 | 1 | | H:H:?2 | R:R:H121 | 4.6 | 3 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:W128 | 10.19 | 3 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:N130 | 25.79 | 3 | Yes | No | 0 | 8 | 0 | 1 | | R:R:M133 | R:R:T132 | 3.01 | 3 | Yes | No | 5 | 6 | 1 | 1 | | H:H:?2 | R:R:T132 | 9.23 | 3 | Yes | No | 0 | 6 | 0 | 1 | | R:R:F137 | R:R:M133 | 9.95 | 3 | No | Yes | 1 | 5 | 2 | 1 | | H:H:?2 | R:R:M133 | 7.59 | 3 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:G33 | R:R:W128 | 2.81 | 0 | No | Yes | 0 | 5 | 2 | 1 | | R:R:F119 | R:R:H121 | 2.26 | 0 | No | Yes | 4 | 4 | 2 | 1 | | R:R:A136 | R:R:M133 | 1.61 | 0 | No | Yes | 2 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 11.48 | | Average Nodes In Shell | 15.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 16.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYI | B1 | Peptide | Calcitonin | CT | Homo Sapiens | Amylin | - | Gs/Beta1/Gamma2 | 3.3 | 2022-03-30 | doi.org/10.1126/science.abm9609 |
|
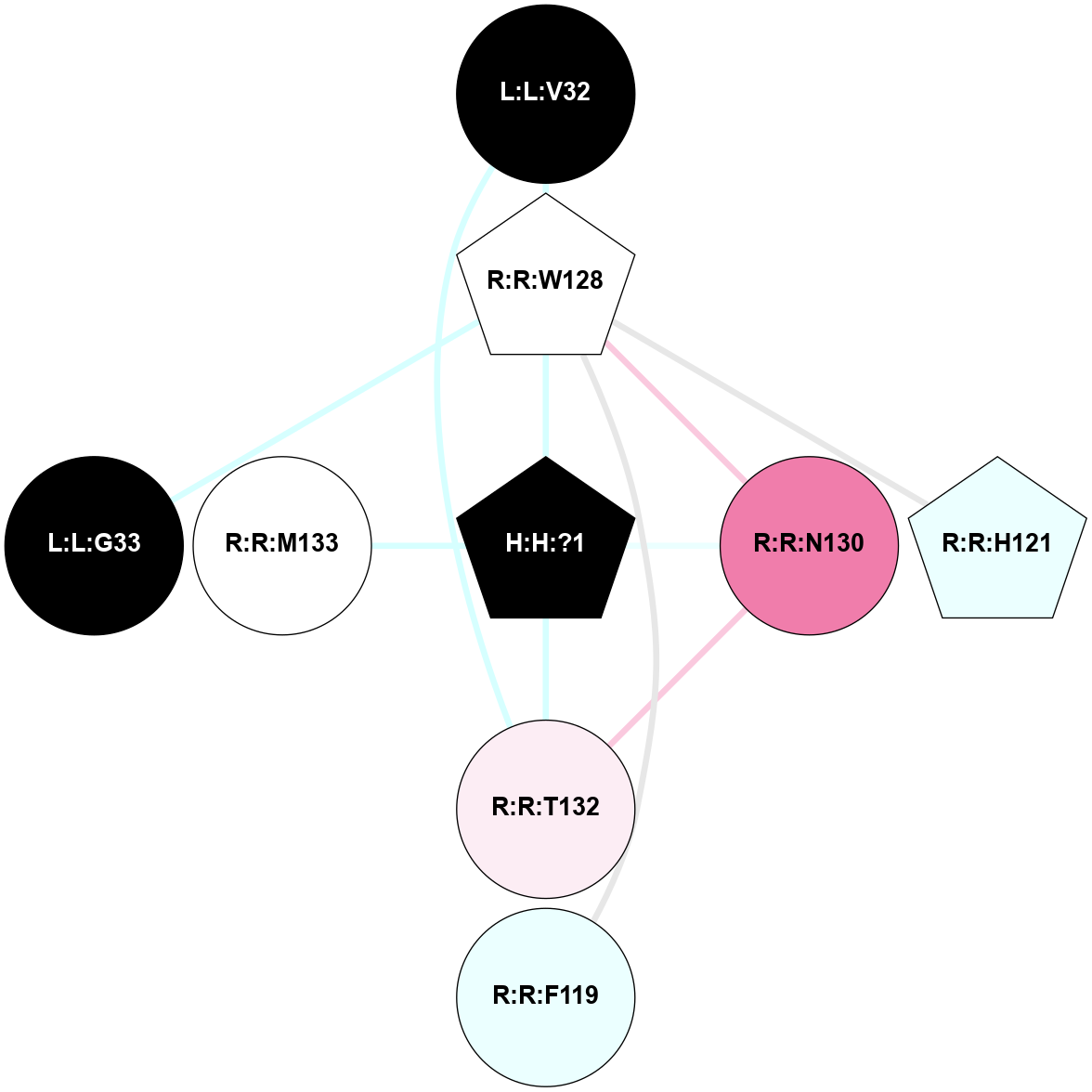
A 2D representation of the interactions of NAG in 7TYI
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:V32 | R:R:W128 | 3.68 | 0 | No | Yes | 0 | 5 | 2 | 1 | | L:L:V32 | R:R:T132 | 3.17 | 0 | No | No | 0 | 6 | 2 | 1 | | R:R:H121 | R:R:W128 | 7.41 | 0 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 5.65 | 17 | No | Yes | 8 | 5 | 1 | 1 | | H:H:?1 | R:R:W128 | 3.06 | 17 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:N130 | R:R:T132 | 5.85 | 17 | No | No | 8 | 6 | 1 | 1 | | H:H:?1 | R:R:N130 | 28.24 | 17 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?1 | R:R:T132 | 3.95 | 17 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:M133 | 3.79 | 17 | Yes | No | 0 | 5 | 0 | 1 | | R:R:F119 | R:R:W128 | 3.01 | 0 | No | Yes | 4 | 5 | 2 | 1 | | L:L:G33 | R:R:W128 | 2.81 | 0 | No | Yes | 0 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 9.76 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 7TYI
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:N73 | 34.38 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 34.38 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYN | B1 | Peptide | Calcitonin | CT | Homo Sapiens | Calcitonin-1 | - | Gs/Beta1/Gamma2 | 2.6 | 2022-03-30 | doi.org/10.1126/science.abm9609 |
|
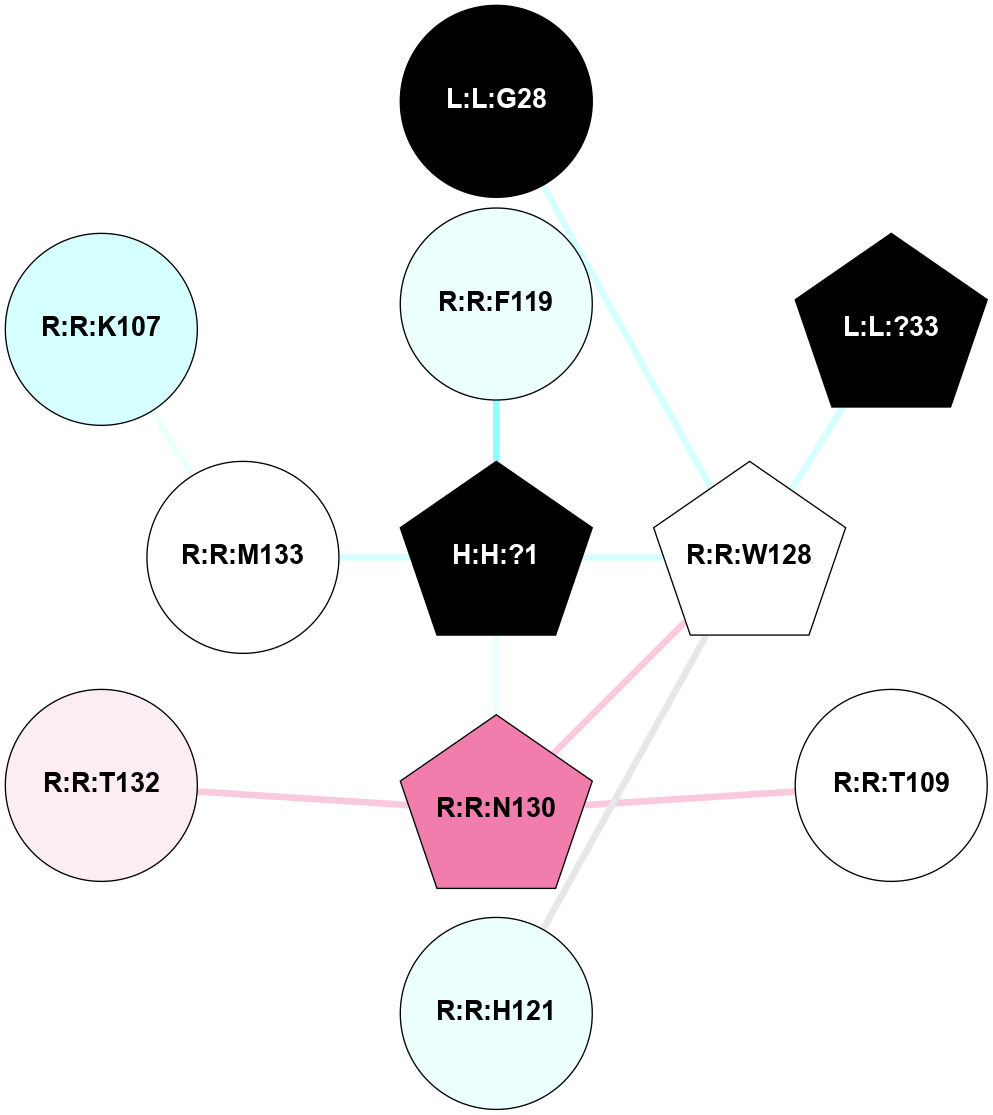
A 2D representation of the interactions of NAG in 7TYN
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:F119 | 8.71 | 2 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:W128 | 4.07 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:N130 | 24.56 | 2 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?1 | R:R:M133 | 3.79 | 2 | Yes | No | 0 | 5 | 0 | 1 | | L:L:G28 | R:R:W128 | 4.22 | 0 | No | Yes | 0 | 5 | 2 | 1 | | L:L:?33 | R:R:W128 | 5.67 | 2 | Yes | Yes | 0 | 5 | 2 | 1 | | R:R:E106 | R:R:M133 | 5.41 | 0 | Yes | No | 4 | 5 | 2 | 1 | | R:R:D113 | R:R:F119 | 3.58 | 0 | No | No | 5 | 4 | 2 | 1 | | R:R:H121 | R:R:W128 | 11.64 | 2 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 5.65 | 2 | No | Yes | 8 | 5 | 1 | 1 | | R:R:N130 | R:R:T132 | 8.77 | 2 | No | No | 8 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 10.28 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
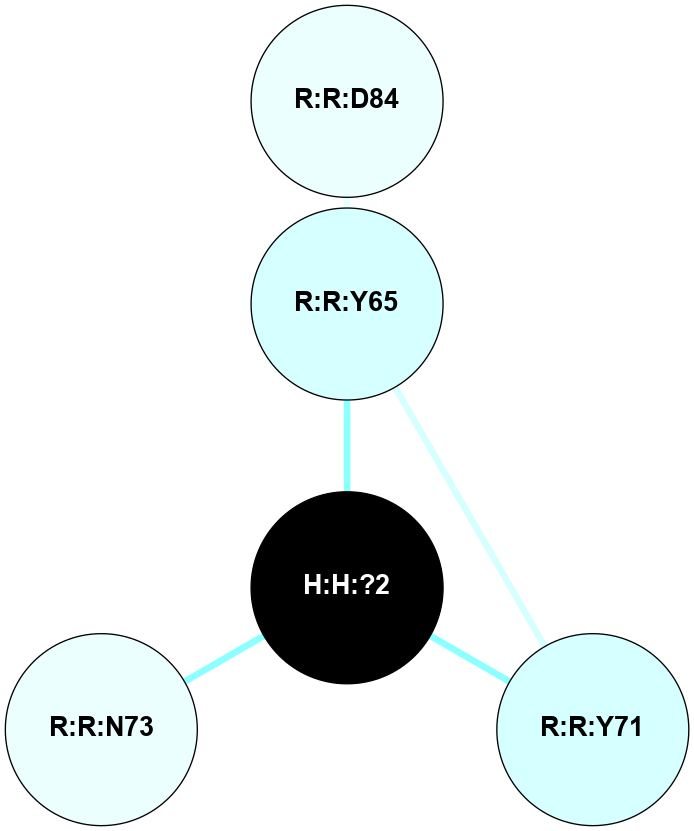
A 2D representation of the interactions of NAG in 7TYN
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:Y71 | 6.29 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?2 | R:R:N73 | 30.7 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:Y65 | R:R:Y71 | 3.97 | 0 | No | No | 3 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.50 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYO | B1 | Peptide | Calcitonin | CT | Homo Sapiens | Calcitonin | - | Gs/Beta1/Gamma2 | 2.7 | 2022-03-23 | doi.org/10.1126/science.abm9609 |
|
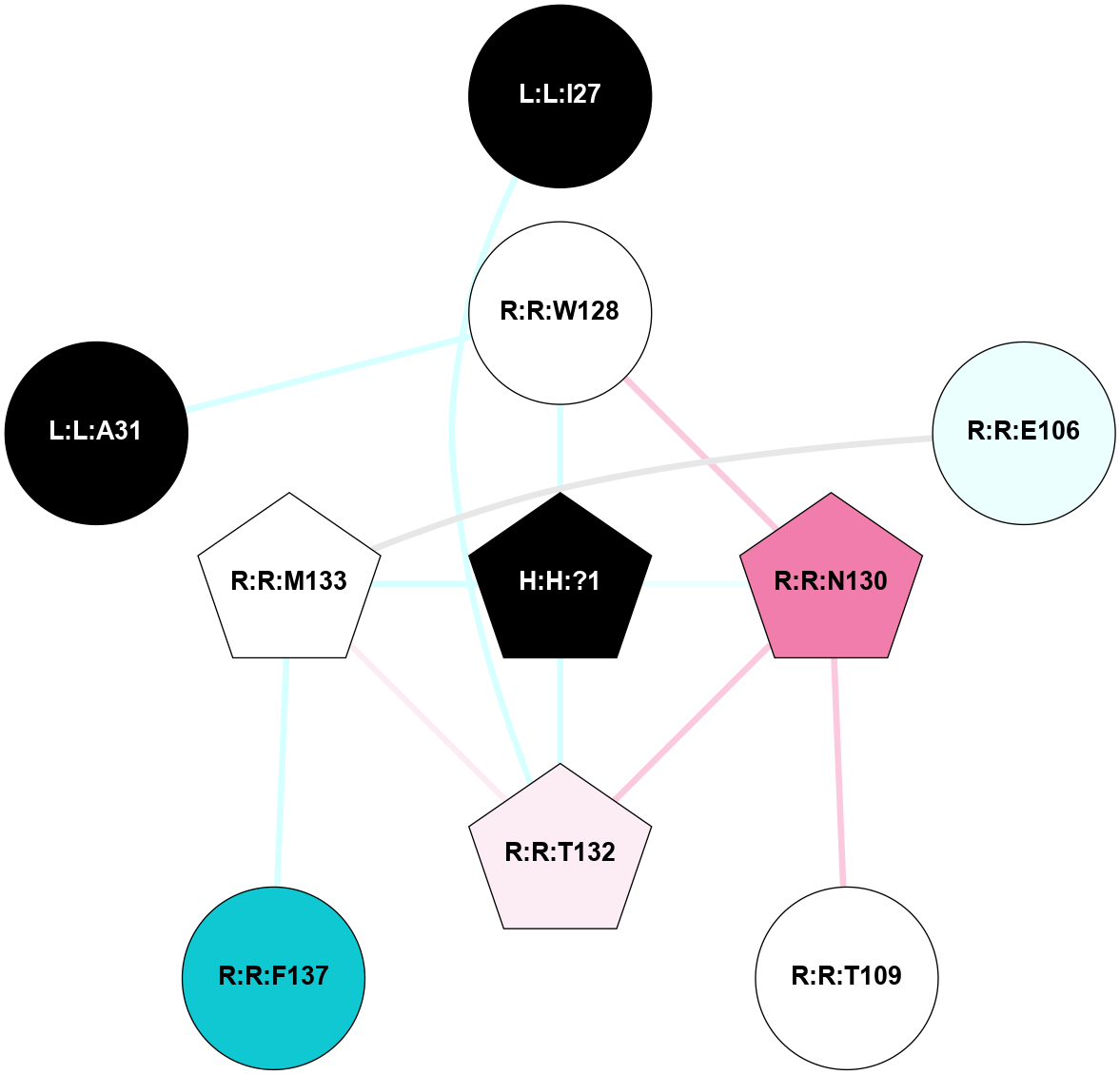
A 2D representation of the interactions of NAG in 7TYO
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:I27 | R:R:T132 | 3.04 | 0 | No | Yes | 0 | 6 | 2 | 1 | | R:R:E106 | R:R:M133 | 9.47 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:T109 | 8.77 | 14 | Yes | No | 8 | 5 | 1 | 2 | | R:R:N130 | R:R:W128 | 7.91 | 14 | Yes | No | 8 | 5 | 1 | 1 | | H:H:?1 | R:R:W128 | 4.07 | 14 | Yes | No | 0 | 5 | 0 | 1 | | R:R:N130 | R:R:T132 | 10.24 | 14 | Yes | Yes | 8 | 6 | 1 | 1 | | H:H:?1 | R:R:N130 | 25.79 | 14 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:M133 | R:R:T132 | 4.52 | 14 | Yes | Yes | 5 | 6 | 1 | 1 | | H:H:?1 | R:R:T132 | 17.14 | 14 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:F137 | R:R:M133 | 4.98 | 0 | No | Yes | 1 | 5 | 2 | 1 | | H:H:?1 | R:R:M133 | 3.79 | 14 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:A31 | R:R:W128 | 2.59 | 0 | No | No | 0 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 12.70 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYW | B1 | Peptide | Calcitonin | CT (AMY1) | Homo Sapiens | Calcitonin-1 | - | Gs/Beta1/Gamma2; RAMP1 | 3 | 2022-03-23 | doi.org/10.1126/science.abm9609 |
|
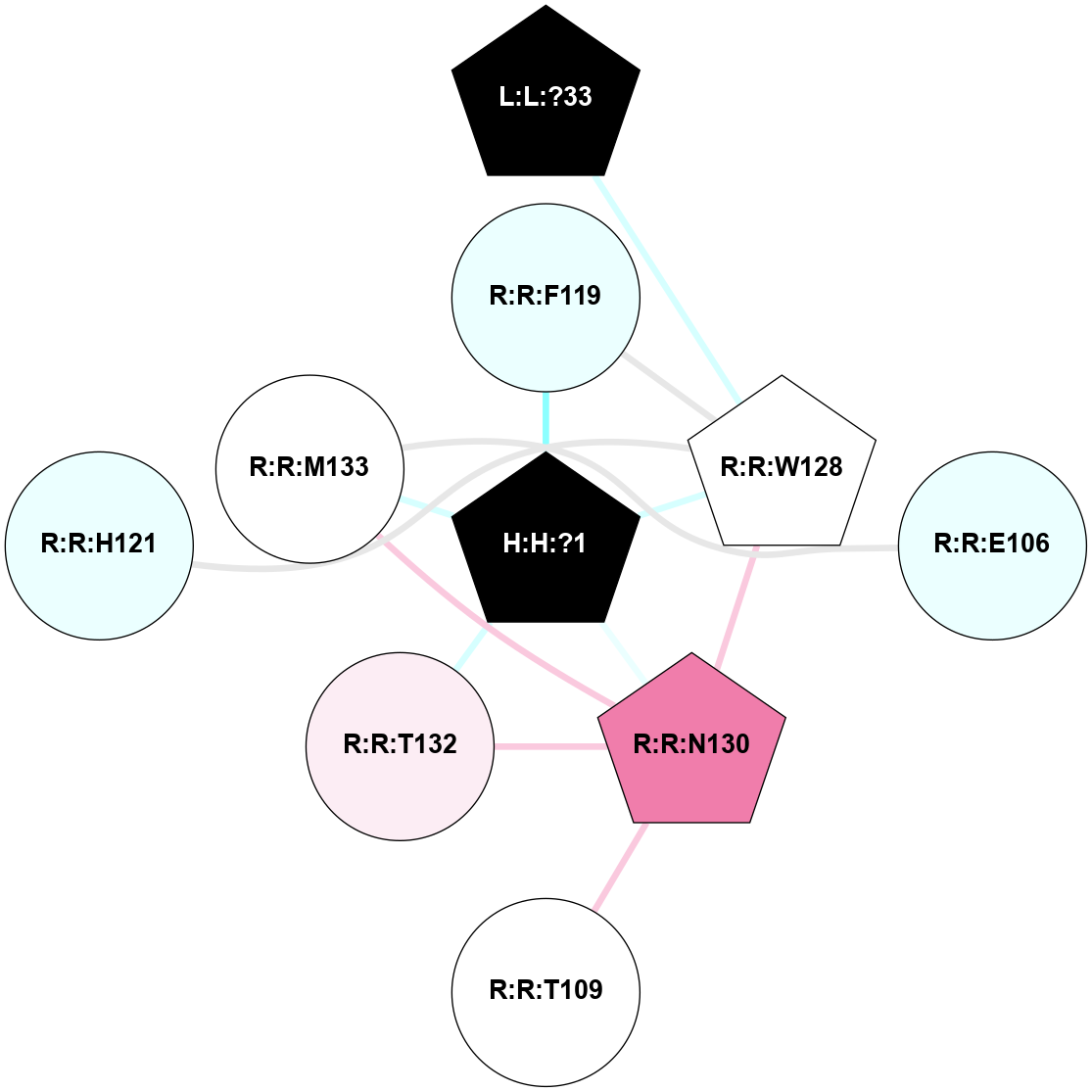
A 2D representation of the interactions of NAG in 7TYW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:H121 | 4.6 | 2 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:W128 | 7.13 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:N130 | 24.56 | 2 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?1 | R:R:T132 | 3.95 | 2 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:M133 | 10.11 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | L:L:?33 | R:R:W128 | 8.5 | 2 | Yes | Yes | 0 | 5 | 2 | 1 | | R:R:E106 | R:R:M133 | 8.12 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:K107 | R:R:M133 | 11.52 | 0 | No | Yes | 3 | 5 | 2 | 1 | | R:R:N130 | R:R:T109 | 5.85 | 2 | Yes | No | 8 | 5 | 1 | 2 | | R:R:H121 | R:R:P122 | 9.15 | 2 | Yes | No | 4 | 5 | 1 | 2 | | R:R:E123 | R:R:H121 | 7.39 | 0 | No | Yes | 4 | 4 | 2 | 1 | | R:R:H121 | R:R:W128 | 14.81 | 2 | Yes | Yes | 4 | 5 | 1 | 1 | | R:R:N130 | R:R:W128 | 4.52 | 2 | Yes | Yes | 8 | 5 | 1 | 1 | | R:R:N130 | R:R:T132 | 7.31 | 2 | Yes | No | 8 | 6 | 1 | 1 | | R:R:M133 | R:R:N130 | 4.21 | 2 | Yes | Yes | 5 | 8 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 10.07 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
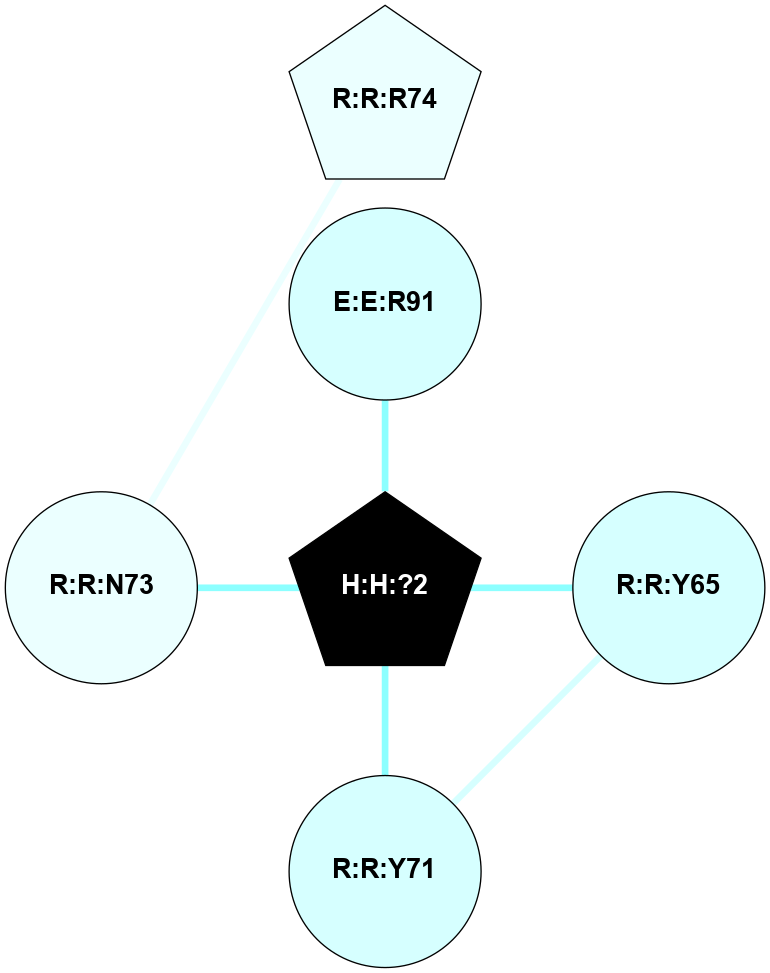
A 2D representation of the interactions of NAG in 7TYW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:Y65 | 4.19 | 31 | No | Yes | 0 | 3 | 0 | 1 | | H:H:?2 | R:R:Y71 | 6.29 | 31 | No | No | 0 | 3 | 0 | 1 | | H:H:?2 | R:R:N73 | 34.38 | 31 | No | No | 0 | 4 | 0 | 1 | | R:R:Q66 | R:R:Y65 | 3.38 | 0 | No | Yes | 1 | 3 | 2 | 1 | | R:R:G67 | R:R:Y65 | 5.79 | 0 | No | Yes | 2 | 3 | 2 | 1 | | R:R:Y65 | R:R:Y71 | 41.7 | 31 | Yes | No | 3 | 3 | 1 | 1 | | R:R:D84 | R:R:Y65 | 16.09 | 0 | No | Yes | 4 | 3 | 2 | 1 | | R:R:P86 | R:R:Y65 | 5.56 | 0 | No | Yes | 2 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.95 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYX | B1 | Peptide | Calcitonin | CT (AMY2) | Homo Sapiens | Amylin | - | Gs/Beta1/Gamma2; RAMP2 | 2.55 | 2022-03-30 | doi.org/10.1126/science.abm9609 |
|
A 2D representation of the interactions of NAG in 7TYX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N125 | 30.7 | 0 | No | No | 0 | 7 | 0 | 1 | | R:R:N125 | R:R:R126 | 4.82 | 0 | No | No | 7 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 30.70 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
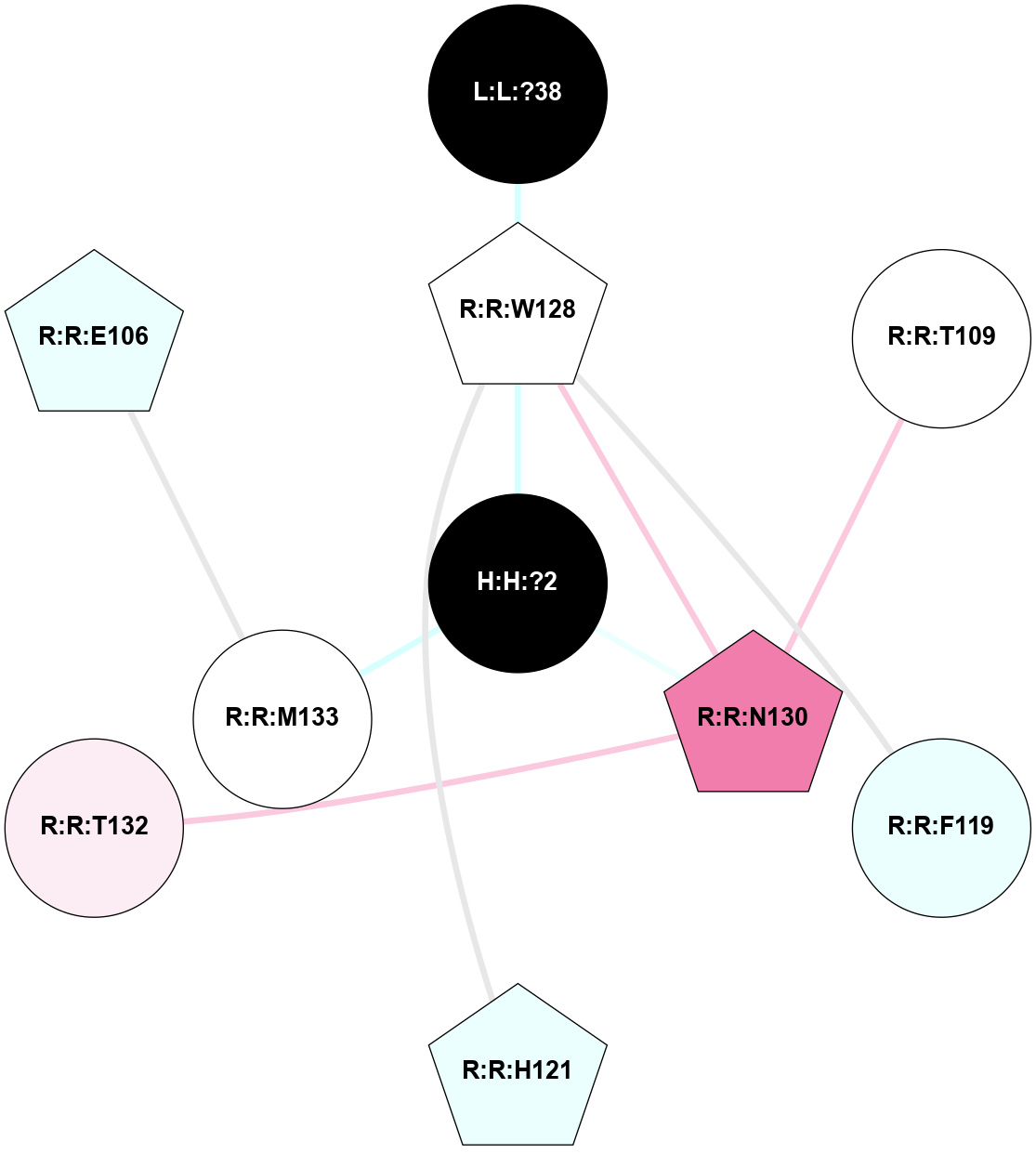
A 2D representation of the interactions of NAG in 7TYX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:W128 | 6.11 | 1 | No | Yes | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:N130 | 25.79 | 1 | No | Yes | 0 | 8 | 0 | 1 | | L:L:?38 | R:R:W128 | 11.33 | 1 | No | Yes | 0 | 5 | 2 | 1 | | R:R:N130 | R:R:T109 | 5.85 | 1 | Yes | No | 8 | 5 | 1 | 2 | | R:R:F119 | R:R:W128 | 4.01 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:H121 | R:R:W128 | 19.04 | 1 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 11.3 | 1 | Yes | Yes | 8 | 5 | 1 | 1 | | R:R:N130 | R:R:T132 | 7.31 | 1 | Yes | No | 8 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 15.95 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
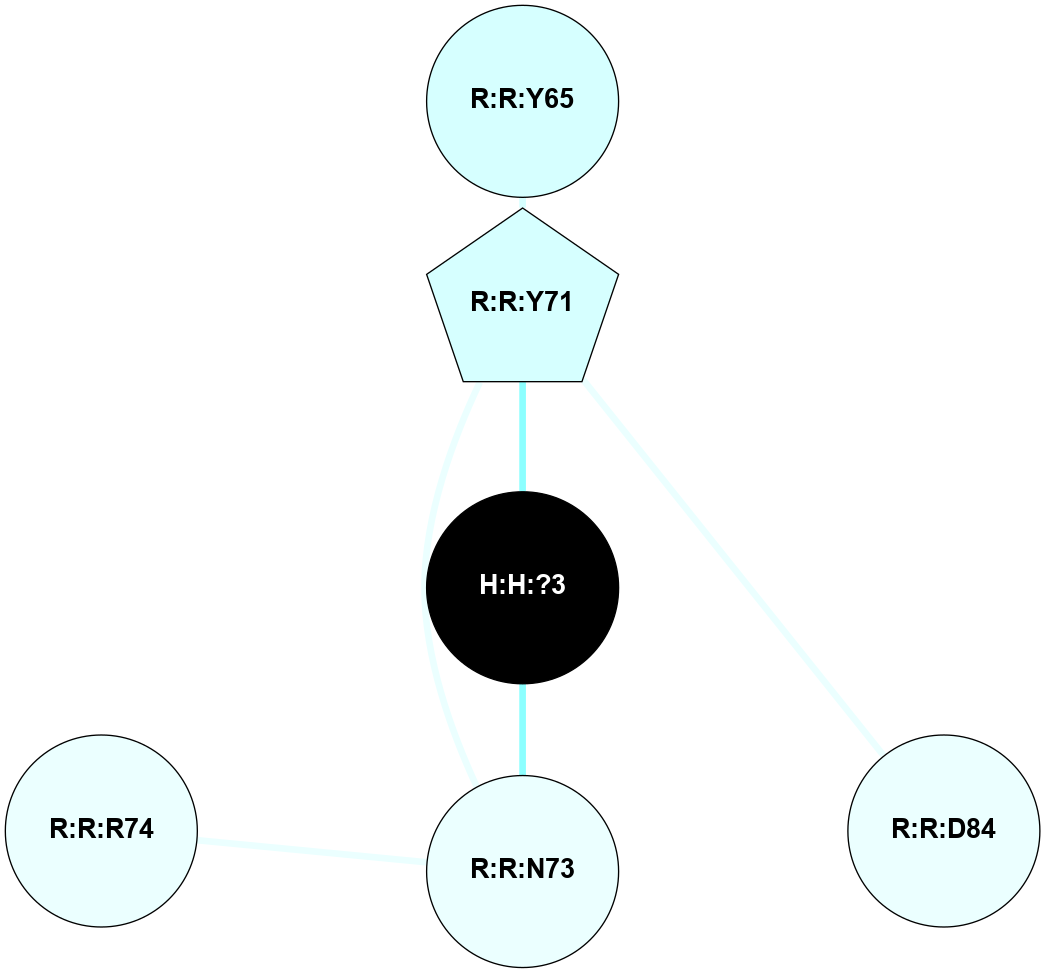
A 2D representation of the interactions of NAG in 7TYX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:Y71 | 12.58 | 21 | No | Yes | 0 | 3 | 0 | 1 | | H:H:?3 | R:R:N73 | 23.33 | 21 | No | No | 0 | 4 | 0 | 1 | | R:R:Y65 | R:R:Y71 | 42.69 | 21 | No | Yes | 3 | 3 | 2 | 1 | | R:R:D84 | R:R:Y65 | 11.49 | 21 | No | No | 4 | 3 | 2 | 2 | | R:R:N73 | R:R:Y71 | 6.98 | 21 | No | Yes | 4 | 3 | 1 | 1 | | R:R:D84 | R:R:Y71 | 12.64 | 21 | No | Yes | 4 | 3 | 2 | 1 | | R:R:N73 | R:R:R74 | 1.21 | 21 | No | No | 4 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.95 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TYY | B1 | Peptide | Calcitonin | CT (AMY2) | Homo Sapiens | Calcitonin-1 | - | Gs/Beta1/Gamma2; RAMP2 | 3 | 2022-03-23 | doi.org/10.1126/science.abm9609 |
|
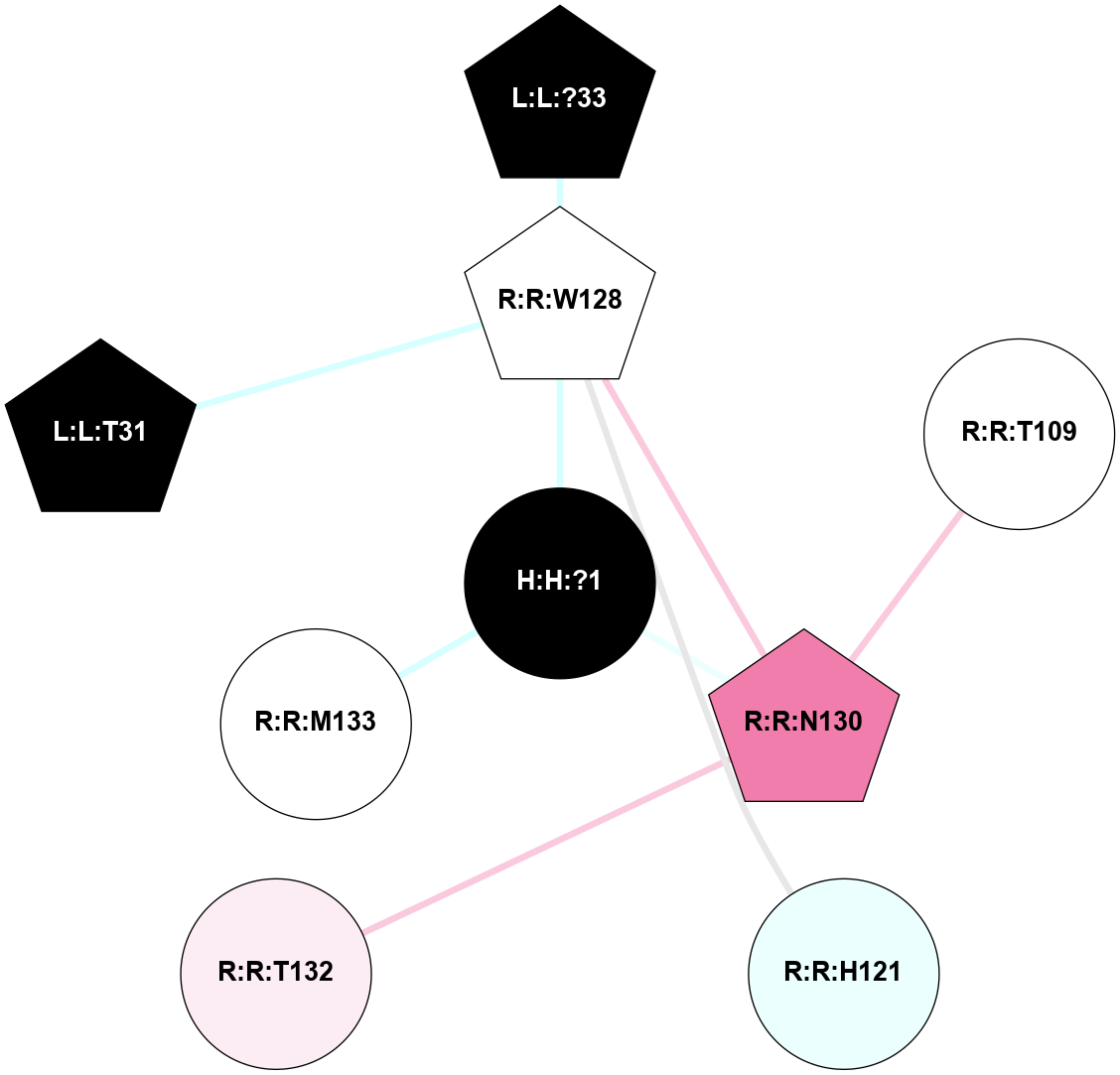
A 2D representation of the interactions of NAG in 7TYY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:H121 | 4.6 | 1 | No | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:W128 | 6.11 | 1 | No | Yes | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:N130 | 24.56 | 1 | No | Yes | 0 | 8 | 0 | 1 | | L:L:?33 | R:R:W128 | 8.5 | 1 | Yes | Yes | 0 | 5 | 2 | 1 | | R:R:H121 | R:R:P122 | 7.63 | 1 | Yes | No | 4 | 5 | 1 | 2 | | R:R:E123 | R:R:H121 | 6.15 | 0 | No | Yes | 4 | 4 | 2 | 1 | | R:R:H121 | R:R:W128 | 20.1 | 1 | Yes | Yes | 4 | 5 | 1 | 1 | | R:R:N130 | R:R:W128 | 11.3 | 1 | Yes | Yes | 8 | 5 | 1 | 1 | | R:R:N130 | R:R:T132 | 4.39 | 1 | Yes | No | 8 | 6 | 1 | 2 | | R:R:N130 | R:R:T109 | 2.92 | 1 | Yes | No | 8 | 5 | 1 | 2 | | L:L:T31 | R:R:W128 | 2.43 | 0 | Yes | Yes | 0 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 11.76 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7TZF | B1 | Peptide | Calcitonin | CT (AMY3) | Homo Sapiens | Amylin | - | Gs/Beta1/Gamma2; RAMP3 | 2.4 | 2022-03-23 | doi.org/10.1126/science.abm9609 |
|

A 2D representation of the interactions of NAG in 7TZF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | E:E:E67 | R:R:R45 | 16.28 | 0 | No | No | 4 | 7 | 1 | 2 | | E:E:E67 | H:H:?1 | 11.85 | 0 | No | No | 4 | 0 | 1 | 0 | | E:E:N71 | H:H:?1 | 29.47 | 0 | No | No | 4 | 0 | 1 | 0 | | E:E:M75 | E:E:N71 | 1.4 | 0 | No | No | 1 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.66 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
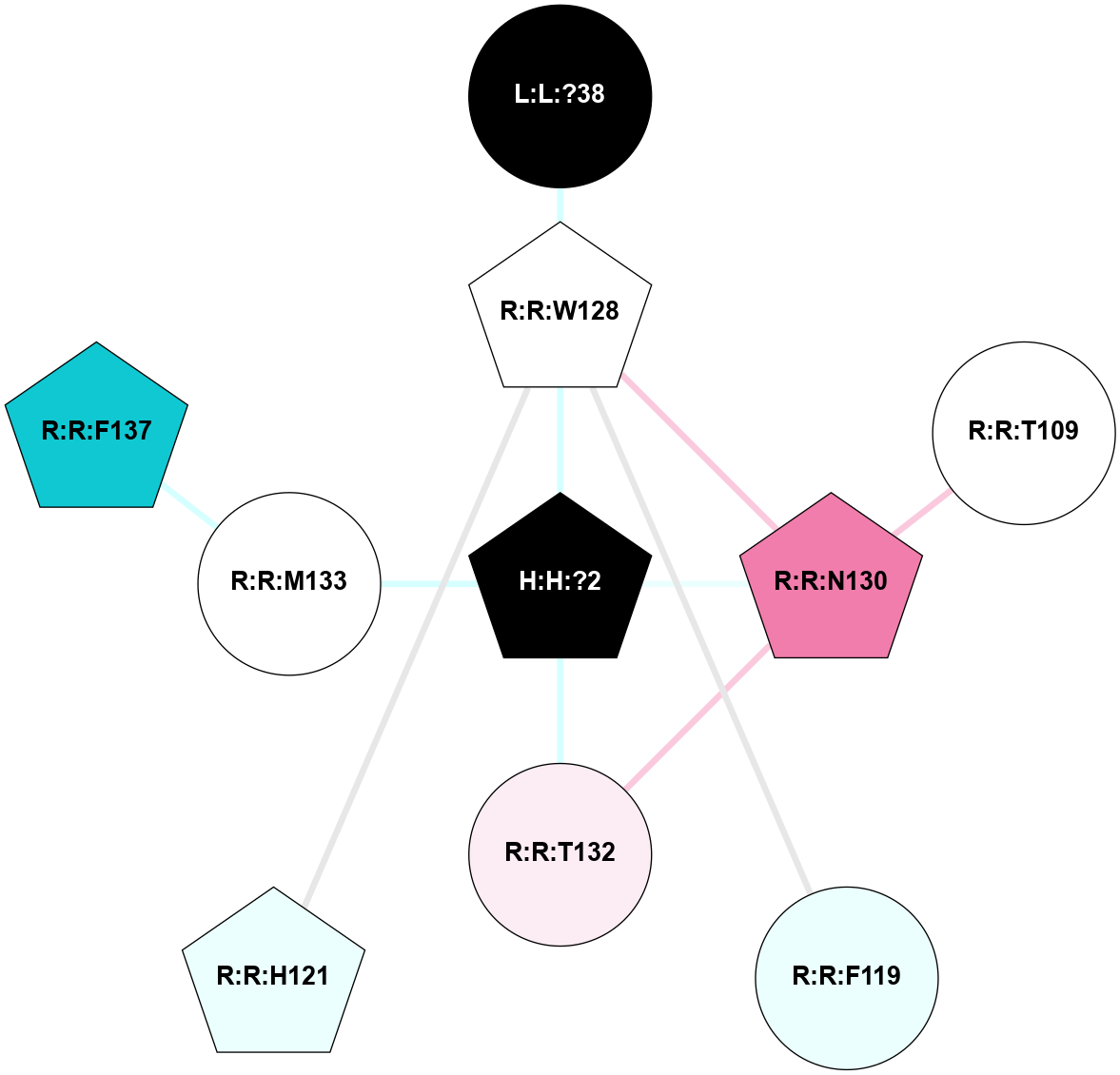
A 2D representation of the interactions of NAG in 7TZF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:?38 | R:R:W128 | 11.33 | 0 | No | Yes | 0 | 5 | 2 | 1 | | R:R:N130 | R:R:T109 | 8.77 | 7 | Yes | No | 8 | 5 | 1 | 2 | | R:R:F119 | R:R:W128 | 4.01 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:H121 | R:R:W128 | 17.98 | 7 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 12.43 | 7 | Yes | Yes | 8 | 5 | 1 | 1 | | H:H:?2 | R:R:W128 | 7.13 | 7 | Yes | Yes | 0 | 5 | 0 | 1 | | R:R:N130 | R:R:T132 | 8.77 | 7 | Yes | No | 8 | 6 | 1 | 1 | | H:H:?2 | R:R:N130 | 22.1 | 7 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?2 | R:R:T132 | 3.95 | 7 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:M133 | 7.59 | 7 | Yes | No | 0 | 5 | 0 | 1 | | R:R:F137 | R:R:M133 | 2.49 | 22 | Yes | No | 1 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 10.19 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
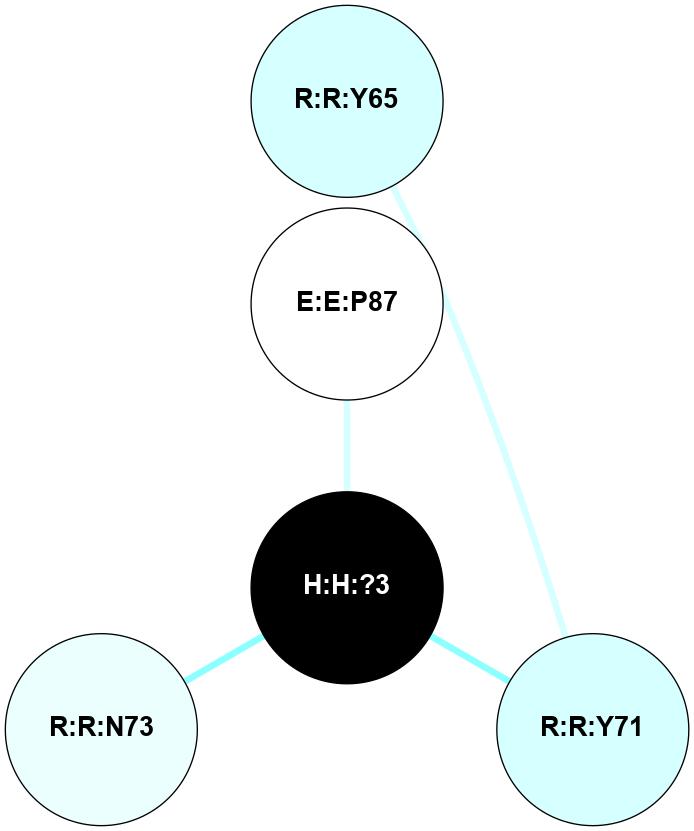
A 2D representation of the interactions of NAG in 7TZF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | E:E:P87 | H:H:?3 | 4.41 | 0 | No | No | 5 | 0 | 1 | 0 | | R:R:Y65 | R:R:Y71 | 19.86 | 0 | No | No | 3 | 3 | 2 | 1 | | H:H:?3 | R:R:Y71 | 7.34 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?3 | R:R:N73 | 29.47 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.74 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
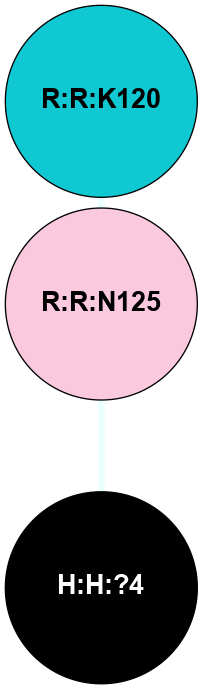
A 2D representation of the interactions of NAG in 7TZF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:K120 | R:R:N125 | 5.6 | 0 | No | No | 1 | 7 | 2 | 1 | | H:H:?4 | R:R:N125 | 30.7 | 0 | No | No | 0 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 30.70 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7UTZ | A | Protein | Glycoprotein Hormone | TSH | Homo Sapiens | TR1402 | DPPC | Gs/Beta1/Gamma2 | 2.4 | 2022-08-03 | doi.org/10.1038/s41586-022-05159-1 |
|
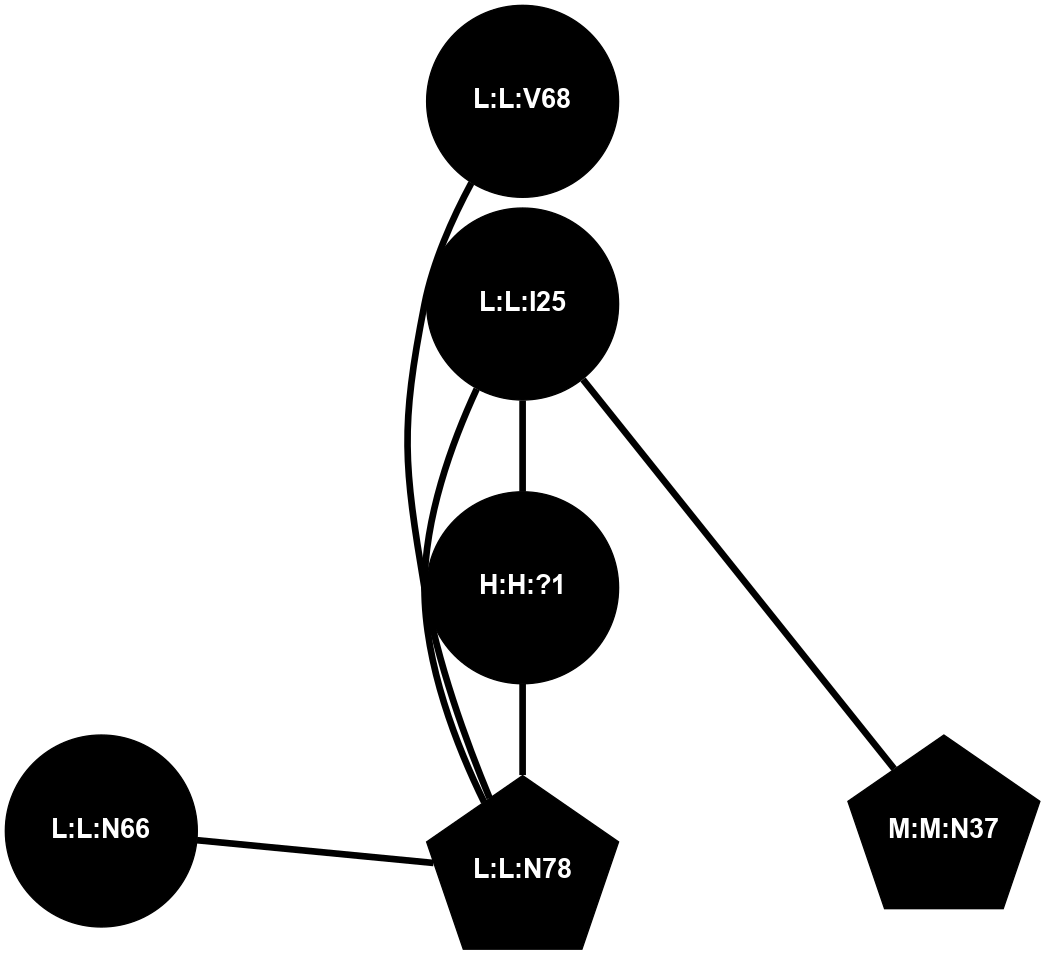
A 2D representation of the interactions of NAG in 7UTZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | L:L:I25 | 5.12 | 1 | No | No | 0 | 0 | 0 | 1 | | H:H:?1 | L:L:N78 | 25.87 | 1 | No | Yes | 0 | 0 | 0 | 1 | | L:L:I25 | L:L:N78 | 7.08 | 1 | No | Yes | 0 | 0 | 1 | 1 | | L:L:I25 | N:N:N37 | 2.83 | 1 | No | Yes | 0 | 0 | 1 | 2 | | L:L:N78 | L:L:V68 | 4.43 | 1 | Yes | No | 0 | 0 | 1 | 2 | | L:L:N66 | L:L:N78 | 2.72 | 0 | No | Yes | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.50 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
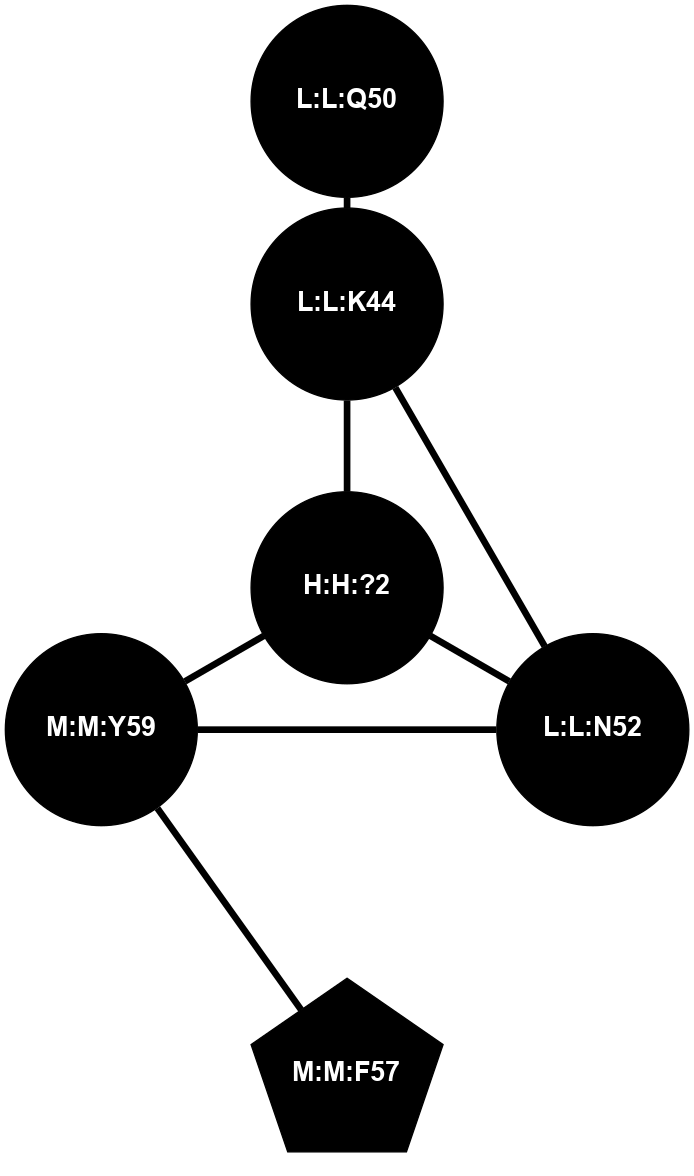
A 2D representation of the interactions of NAG in 7UTZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | L:L:K44 | 3.8 | 20 | No | No | 0 | 0 | 0 | 1 | | H:H:?2 | L:L:N52 | 28.34 | 20 | No | No | 0 | 0 | 0 | 1 | | H:H:?2 | N:N:Y59 | 14.73 | 20 | No | No | 0 | 0 | 0 | 1 | | L:L:K44 | L:L:Q50 | 9.49 | 20 | No | No | 0 | 0 | 1 | 2 | | L:L:K44 | L:L:N52 | 9.79 | 20 | No | No | 0 | 0 | 1 | 1 | | L:L:N52 | N:N:Y59 | 6.98 | 20 | No | No | 0 | 0 | 1 | 1 | | N:N:F57 | N:N:Y59 | 6.19 | 0 | Yes | No | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.62 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7UTZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | N:N:N23 | 30.8 | 0 | No | No | 0 | 0 | 0 | 1 | | N:N:E6 | N:N:N23 | 10.52 | 0 | No | No | 0 | 0 | 2 | 1 | | N:N:N23 | N:N:T8 | 4.39 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 30.80 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
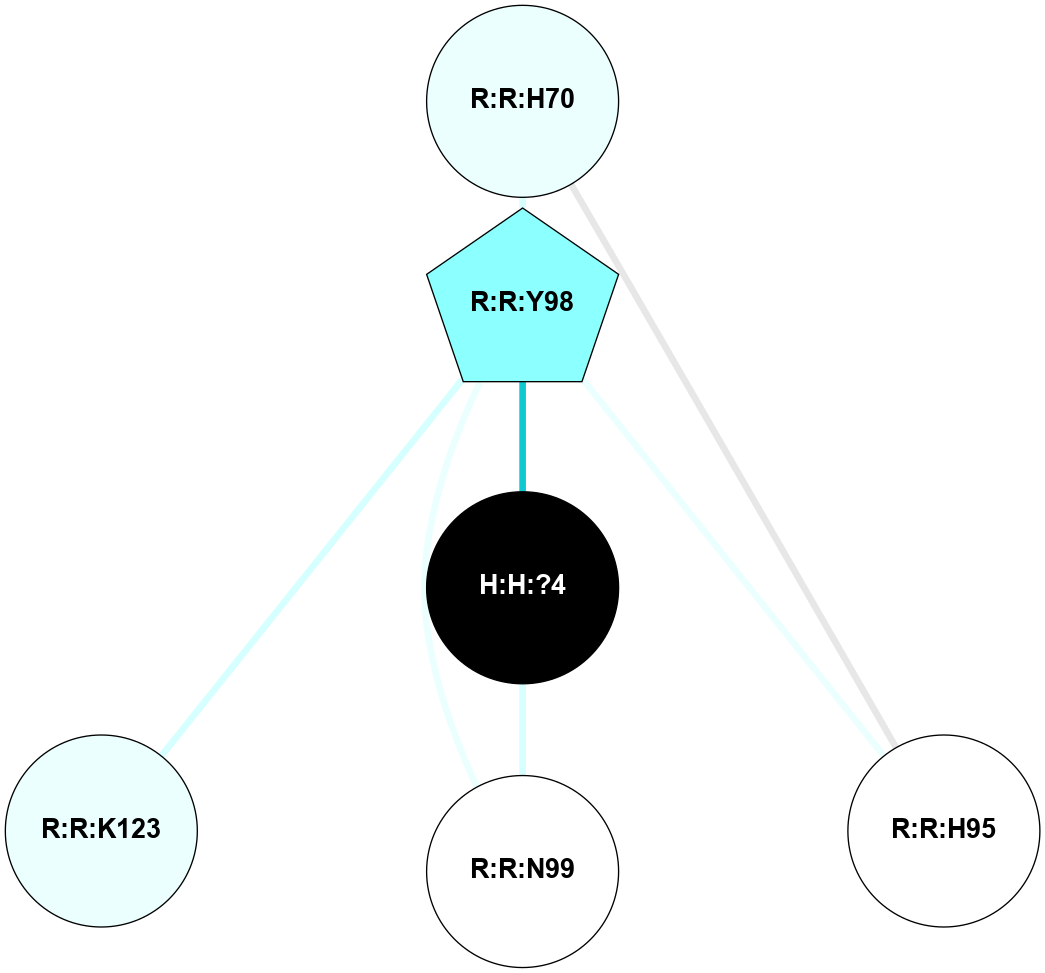
A 2D representation of the interactions of NAG in 7UTZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:Y98 | 4.21 | 18 | No | Yes | 0 | 2 | 0 | 1 | | H:H:?4 | R:R:N99 | 33.27 | 18 | No | No | 0 | 5 | 0 | 1 | | R:R:H70 | R:R:H95 | 8.36 | 18 | No | No | 4 | 5 | 2 | 2 | | R:R:H70 | R:R:Y98 | 3.27 | 18 | No | Yes | 4 | 2 | 2 | 1 | | R:R:H95 | R:R:Y98 | 13.07 | 18 | No | Yes | 5 | 2 | 2 | 1 | | R:R:N99 | R:R:Y98 | 6.98 | 18 | No | Yes | 5 | 2 | 1 | 1 | | R:R:K123 | R:R:Y98 | 3.58 | 0 | No | Yes | 4 | 2 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 18.74 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
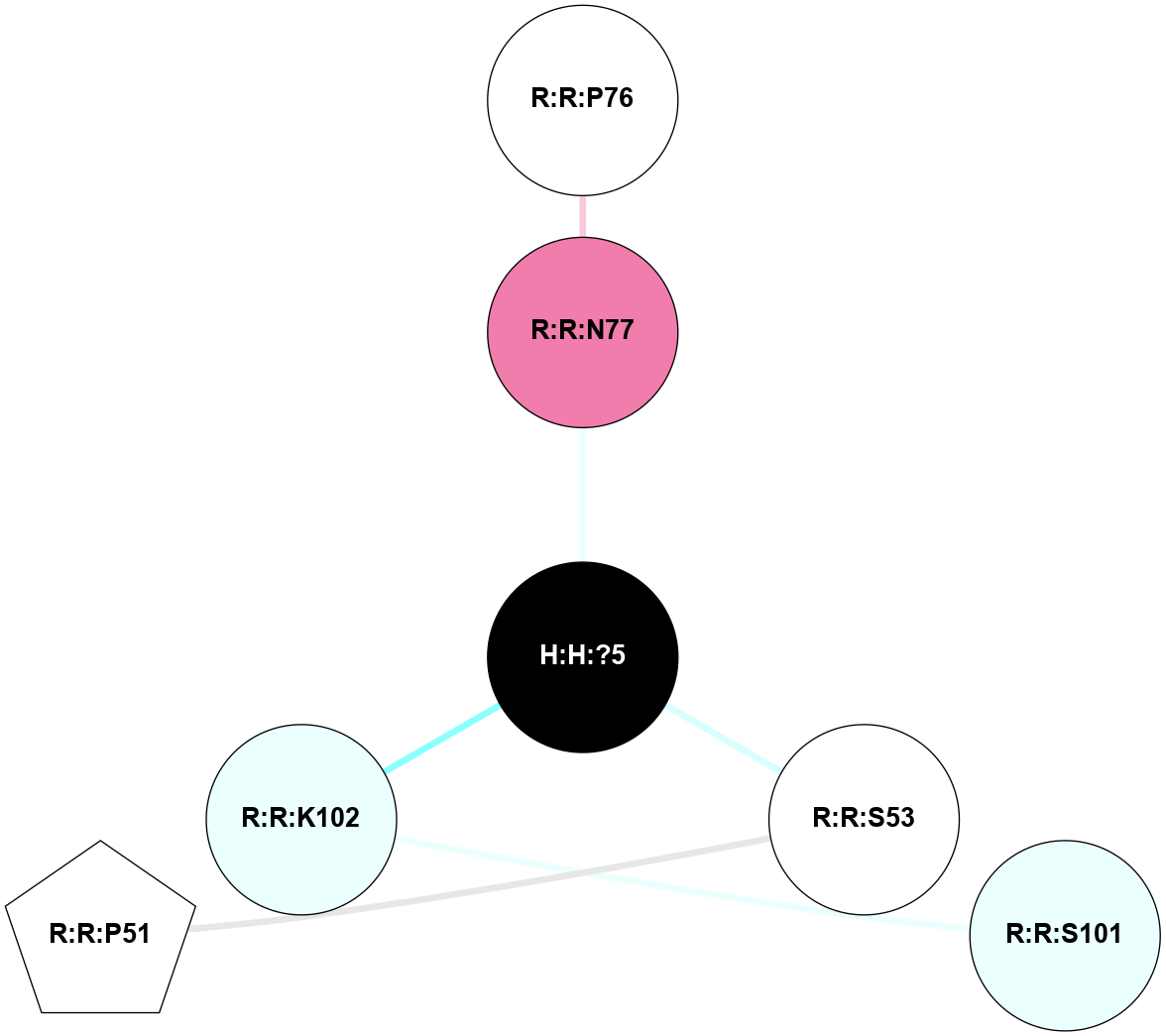
A 2D representation of the interactions of NAG in 7UTZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | R:R:P76 | 2.95 | 34 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?5 | R:R:N77 | 27.11 | 34 | Yes | No | 0 | 8 | 0 | 1 | | R:R:N77 | R:R:P76 | 6.52 | 34 | No | No | 8 | 5 | 1 | 1 | | R:R:K102 | R:R:S101 | 3.06 | 0 | No | No | 4 | 4 | 1 | 2 | | H:H:?5 | R:R:S53 | 2.69 | 34 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?5 | R:R:K102 | 2.53 | 34 | Yes | No | 0 | 4 | 0 | 1 | | R:R:P51 | R:R:S53 | 1.78 | 11 | Yes | No | 5 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 8.82 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
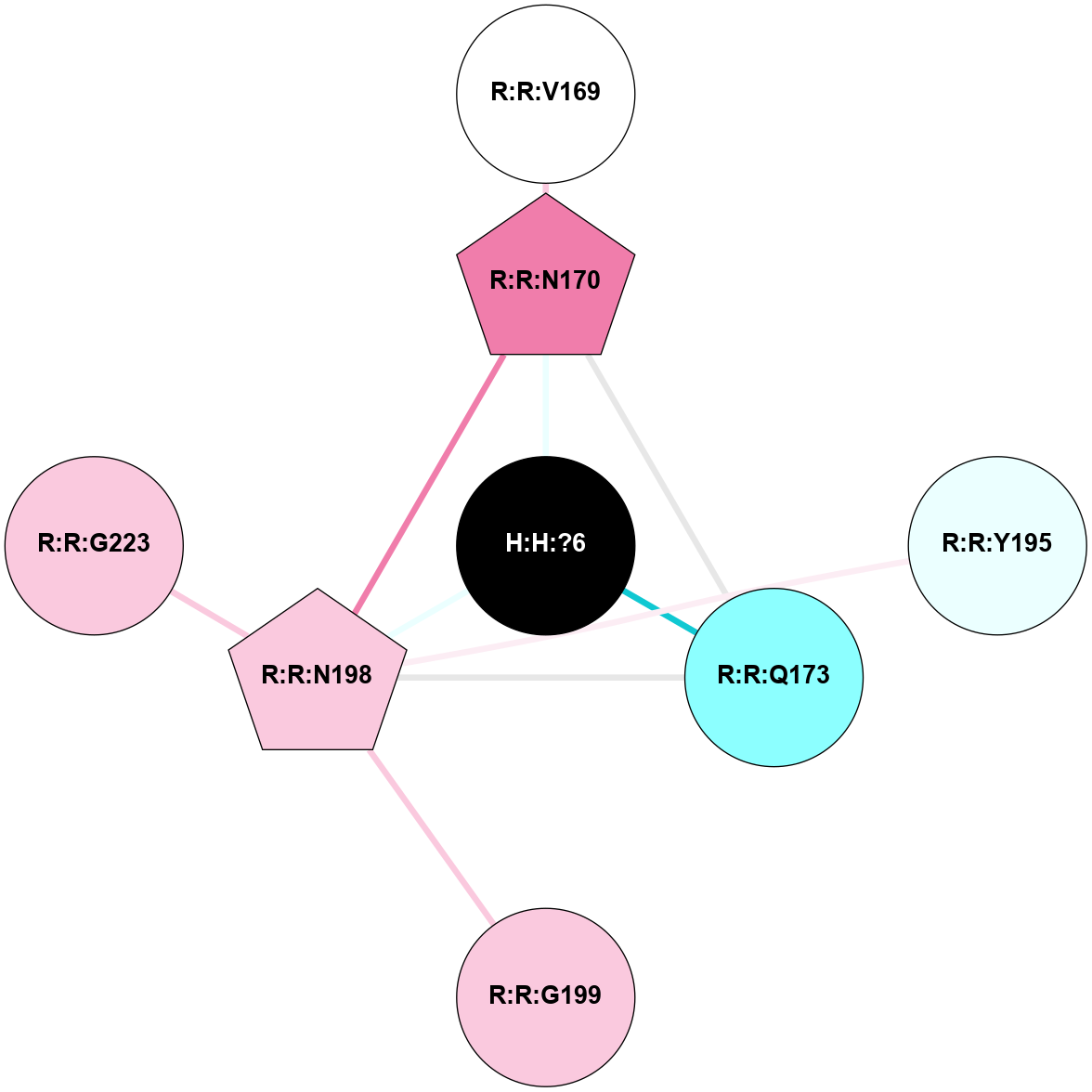
A 2D representation of the interactions of NAG in 7UTZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | R:R:N170 | 11.09 | 21 | No | Yes | 0 | 8 | 0 | 1 | | H:H:?6 | R:R:Q173 | 10.75 | 21 | No | Yes | 0 | 2 | 0 | 1 | | H:H:?6 | R:R:N198 | 24.64 | 21 | No | Yes | 0 | 7 | 0 | 1 | | R:R:N170 | R:R:V169 | 2.96 | 21 | Yes | No | 8 | 5 | 1 | 2 | | R:R:N170 | R:R:Q173 | 15.85 | 21 | Yes | Yes | 8 | 2 | 1 | 1 | | R:R:N170 | R:R:N198 | 5.45 | 21 | Yes | Yes | 8 | 7 | 1 | 1 | | R:R:N198 | R:R:Q173 | 6.6 | 21 | Yes | Yes | 7 | 2 | 1 | 1 | | R:R:N198 | R:R:Y195 | 3.49 | 21 | Yes | No | 7 | 4 | 1 | 2 | | R:R:G199 | R:R:N198 | 3.39 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:G223 | R:R:N198 | 5.09 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:G174 | R:R:Q173 | 1.64 | 0 | No | Yes | 9 | 2 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 15.49 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
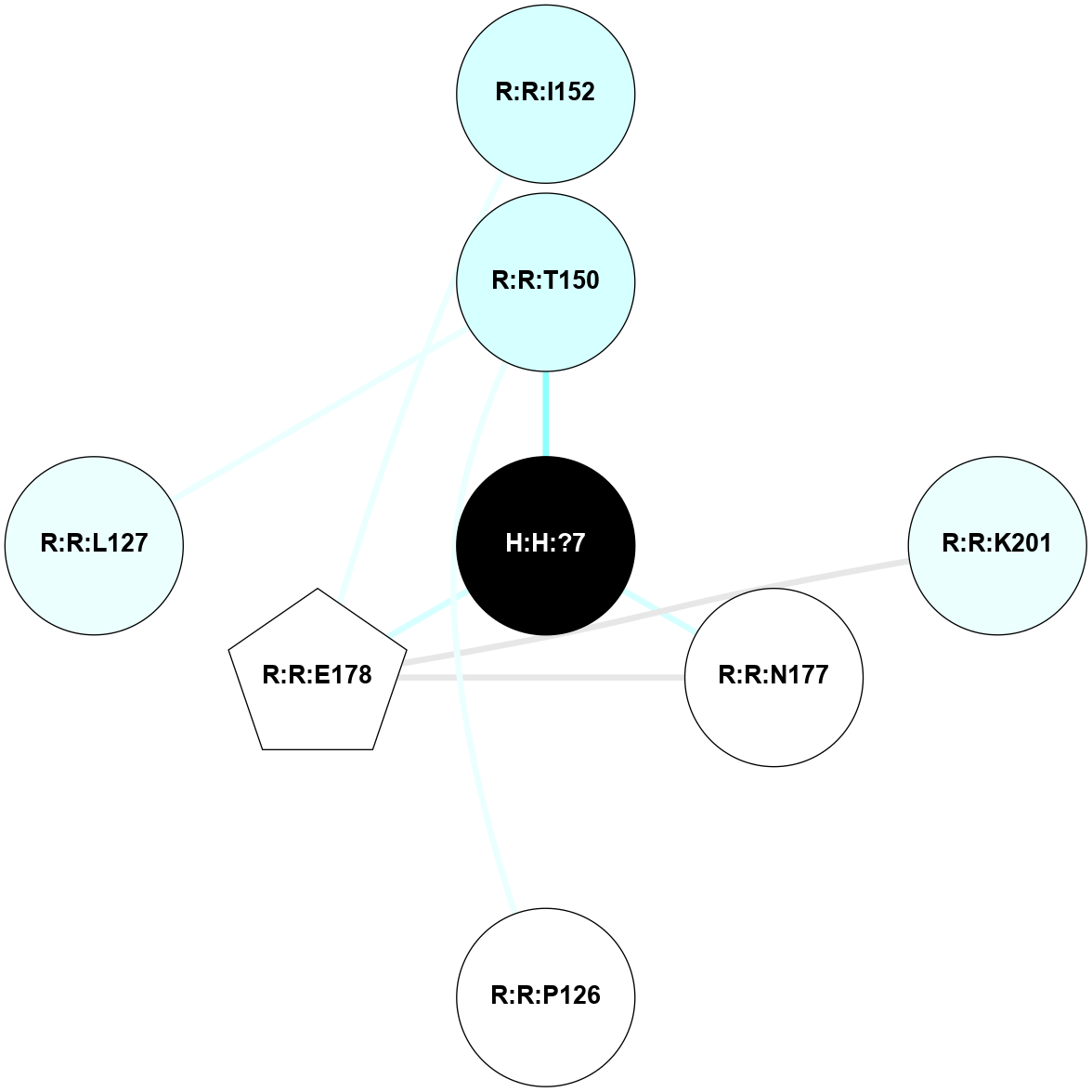
A 2D representation of the interactions of NAG in 7UTZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | R:R:T150 | 5.29 | 22 | No | Yes | 0 | 3 | 0 | 1 | | H:H:?7 | R:R:N177 | 29.57 | 22 | No | No | 0 | 5 | 0 | 1 | | H:H:?7 | R:R:E178 | 17.84 | 22 | No | Yes | 0 | 5 | 0 | 1 | | R:R:N177 | R:R:T150 | 2.92 | 22 | No | Yes | 5 | 3 | 1 | 1 | | R:R:E178 | R:R:I152 | 6.83 | 22 | Yes | No | 5 | 3 | 1 | 2 | | R:R:E178 | R:R:N177 | 3.94 | 22 | Yes | No | 5 | 5 | 1 | 1 | | R:R:E178 | R:R:K201 | 4.05 | 22 | Yes | No | 5 | 4 | 1 | 2 | | R:R:P126 | R:R:T150 | 1.75 | 0 | No | Yes | 5 | 3 | 2 | 1 | | R:R:L127 | R:R:T150 | 1.47 | 0 | No | Yes | 4 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 17.57 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7WI6 | C | Aminoacid | Metabotropic Glutamate | mGlu3; mGlu3 | Homo Sapiens | LY341495 | - | - | 3.71 | 2022-03-16 | doi.org/10.1038/s41422-022-00623-z |
|
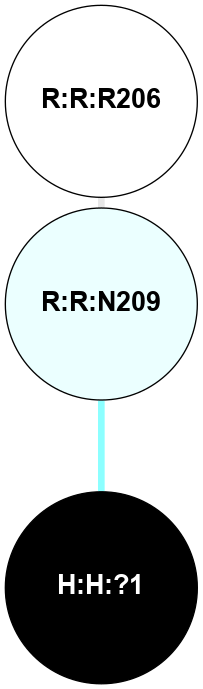
A 2D representation of the interactions of NAG in 7WI6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N209 | 23.33 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:F207 | R:R:F208 | 5.36 | 0 | No | Yes | 4 | 8 | 2 | 1 | | R:R:F208 | R:R:W210 | 14.03 | 18 | Yes | No | 8 | 9 | 1 | 2 | | R:R:F208 | R:R:W499 | 5.01 | 18 | Yes | Yes | 8 | 7 | 1 | 2 | | R:R:W210 | R:R:W499 | 12.18 | 18 | No | Yes | 9 | 7 | 2 | 2 | | R:R:F208 | R:R:V494 | 1.31 | 18 | Yes | No | 8 | 3 | 1 | 2 | | H:H:?1 | R:R:F208 | 1.09 | 0 | No | Yes | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 12.21 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
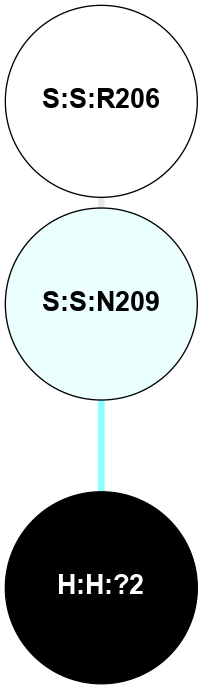
A 2D representation of the interactions of NAG in 7WI6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | S:S:N209 | 23.33 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:F207 | S:S:F208 | 5.36 | 0 | No | Yes | 4 | 8 | 2 | 1 | | S:S:F208 | S:S:W210 | 14.03 | 22 | Yes | No | 8 | 9 | 1 | 2 | | S:S:F208 | S:S:W499 | 5.01 | 22 | Yes | Yes | 8 | 7 | 1 | 2 | | S:S:W210 | S:S:W499 | 11.25 | 22 | No | Yes | 9 | 7 | 2 | 2 | | H:H:?2 | S:S:F208 | 1.09 | 0 | No | Yes | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 12.21 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7WI8 | C | Aminoacid | Metabotropic Glutamate | mGlu3; mGlu3 | Homo Sapiens | LY341495 | - | - | 4.17 | 2022-03-16 | doi.org/10.1038/s41422-022-00623-z |
|

A 2D representation of the interactions of NAG in 7WI8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:R206 | 2.17 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:N209 | 28.24 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:R206 | R:R:R237 | 13.86 | 0 | No | No | 5 | 4 | 1 | 2 | | R:R:N209 | R:R:T211 | 1.46 | 0 | No | No | 4 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.20 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7WI8
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | S:S:N209 | 28.24 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N209 | S:S:T211 | 1.46 | 0 | No | No | 4 | 7 | 1 | 2 | | S:S:N209 | S:S:R206 | 1.21 | 0 | No | No | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 28.24 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7WIH | C | Aminoacid | Metabotropic Glutamate | mGlu3; mGlu3 | Homo Sapiens | LY2794193 | - | - | 3.68 | 2022-03-16 | doi.org/10.1038/s41422-022-00623-z |
|
A 2D representation of the interactions of NAG in 7WIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N209 | 23.33 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N209 | R:R:R206 | 7.23 | 0 | No | No | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 23.33 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 7WIH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | S:S:N209 | 23.33 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N209 | S:S:R206 | 7.23 | 0 | No | No | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 23.33 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7XW5 | A | Protein | Glycoprotein Hormone | TSH | Homo Sapiens | TSH | ML109 | Gs/Beta1/Gamma2 | 2.96 | 2022-08-24 | doi.org/10.1038/s41586-022-05173-3 |
|
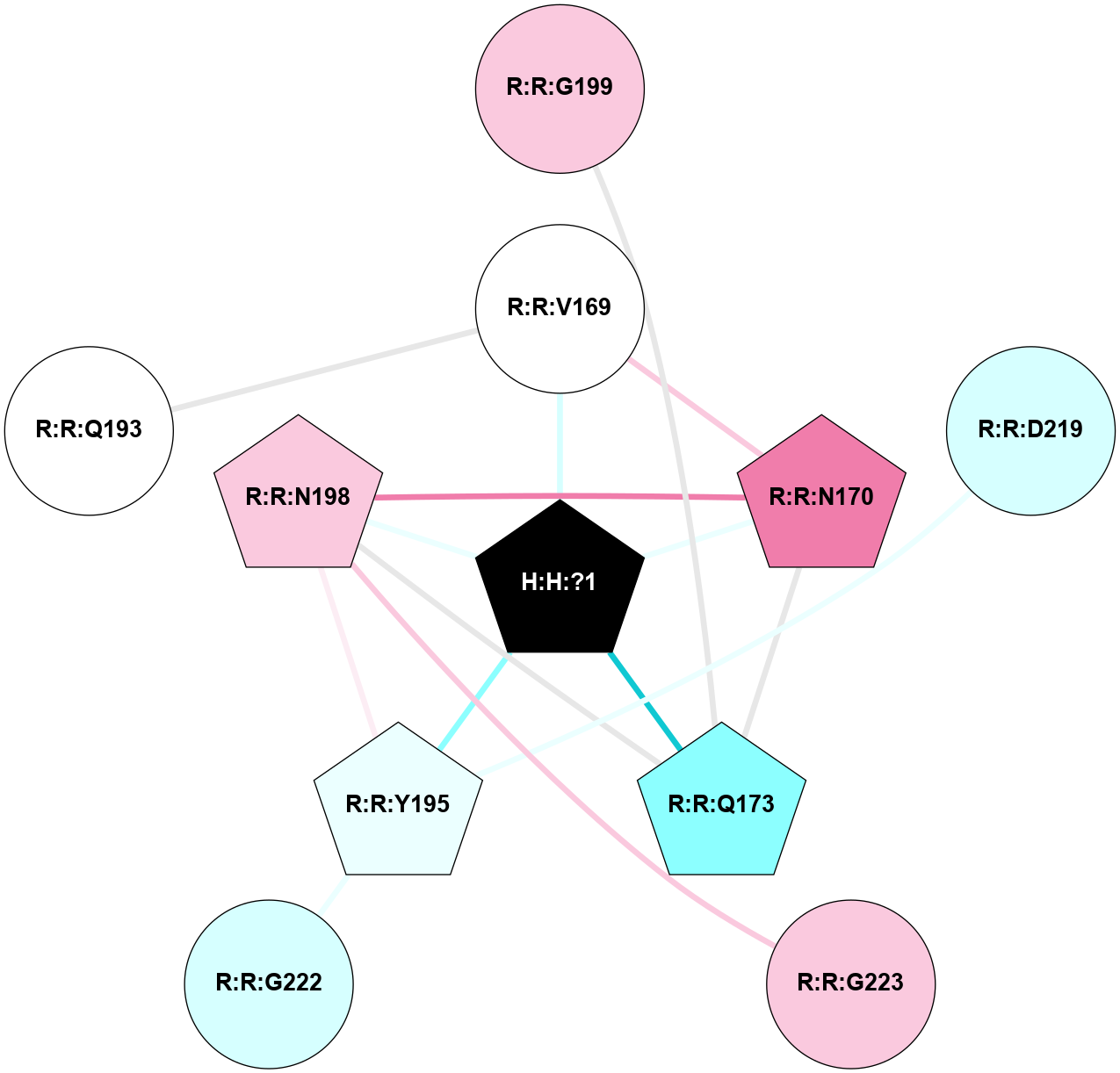
A 2D representation of the interactions of NAG in 7XW5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N170 | R:R:V169 | 5.91 | 21 | Yes | No | 8 | 5 | 1 | 1 | | H:H:?1 | R:R:V169 | 4.01 | 21 | Yes | No | 0 | 5 | 0 | 1 | | R:R:N170 | R:R:Q173 | 10.56 | 21 | Yes | Yes | 8 | 2 | 1 | 1 | | R:R:N170 | R:R:N198 | 8.17 | 21 | Yes | Yes | 8 | 7 | 1 | 1 | | H:H:?1 | R:R:N170 | 16.02 | 21 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:N198 | R:R:Q173 | 9.24 | 21 | Yes | Yes | 7 | 2 | 1 | 1 | | R:R:G199 | R:R:Q173 | 3.29 | 0 | No | Yes | 7 | 2 | 2 | 1 | | H:H:?1 | R:R:Q173 | 10.75 | 21 | Yes | Yes | 0 | 2 | 0 | 1 | | R:R:N198 | R:R:Y195 | 3.49 | 21 | Yes | Yes | 7 | 4 | 1 | 1 | | R:R:D219 | R:R:Y195 | 5.75 | 0 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?1 | R:R:Y195 | 9.47 | 21 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:G223 | R:R:N198 | 3.39 | 0 | No | Yes | 7 | 7 | 2 | 1 | | H:H:?1 | R:R:N198 | 24.64 | 21 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:G222 | R:R:Y195 | 2.9 | 0 | No | Yes | 3 | 4 | 2 | 1 | | R:R:Q193 | R:R:V169 | 2.87 | 0 | No | No | 5 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 4.00 | | Average Interaction Strength | 12.98 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
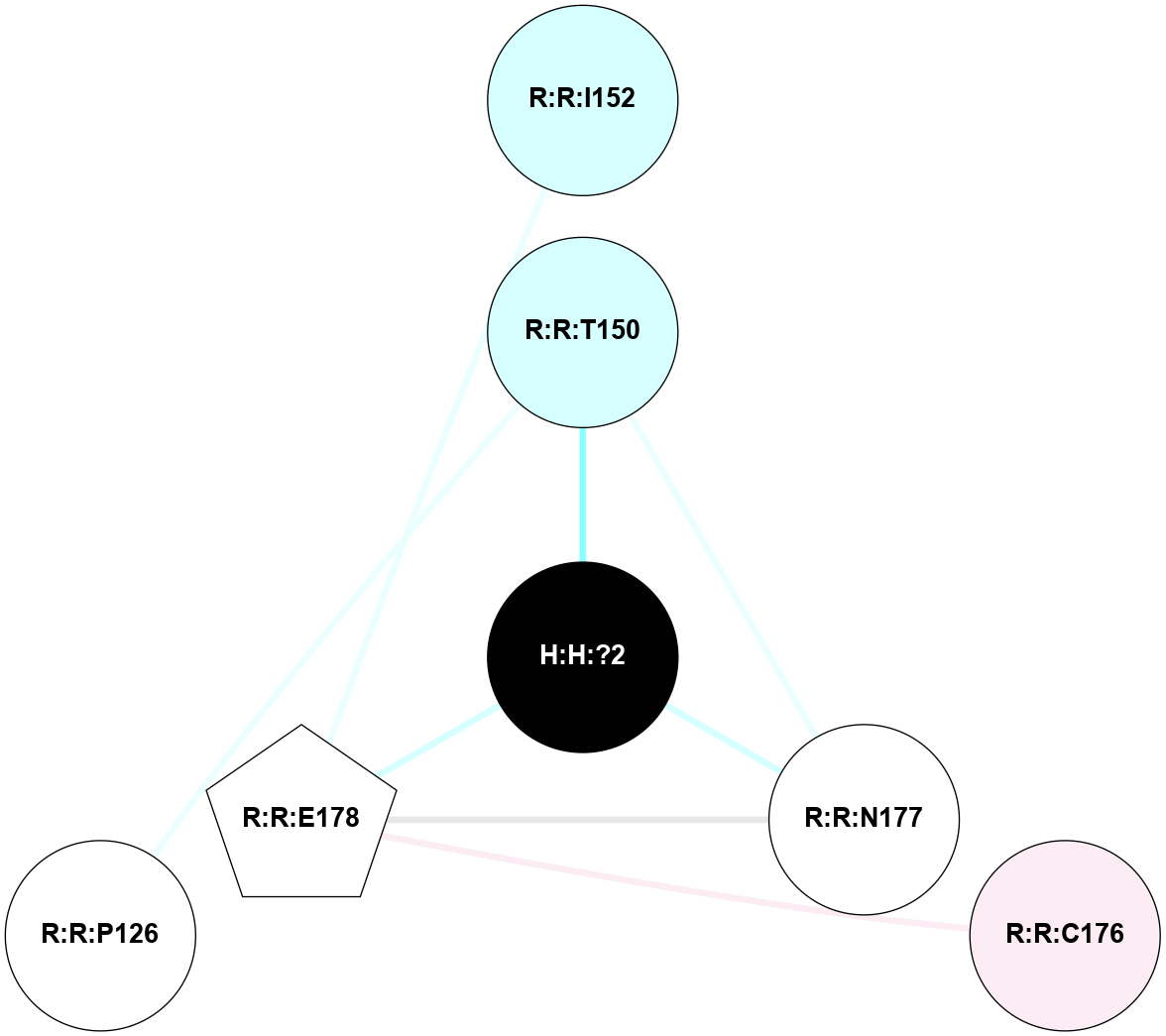
A 2D representation of the interactions of NAG in 7XW5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N177 | R:R:T150 | 4.39 | 4 | No | No | 5 | 3 | 1 | 1 | | H:H:?2 | R:R:T150 | 5.29 | 4 | No | No | 0 | 3 | 0 | 1 | | R:R:E178 | R:R:I152 | 9.56 | 4 | Yes | No | 5 | 3 | 1 | 2 | | R:R:C176 | R:R:E178 | 3.04 | 4 | No | Yes | 6 | 5 | 2 | 1 | | R:R:E178 | R:R:N177 | 3.94 | 4 | Yes | No | 5 | 5 | 1 | 1 | | H:H:?2 | R:R:N177 | 27.11 | 4 | No | No | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:E178 | 15.46 | 4 | No | Yes | 0 | 5 | 0 | 1 | | R:R:P126 | R:R:T150 | 1.75 | 0 | No | No | 5 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.95 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
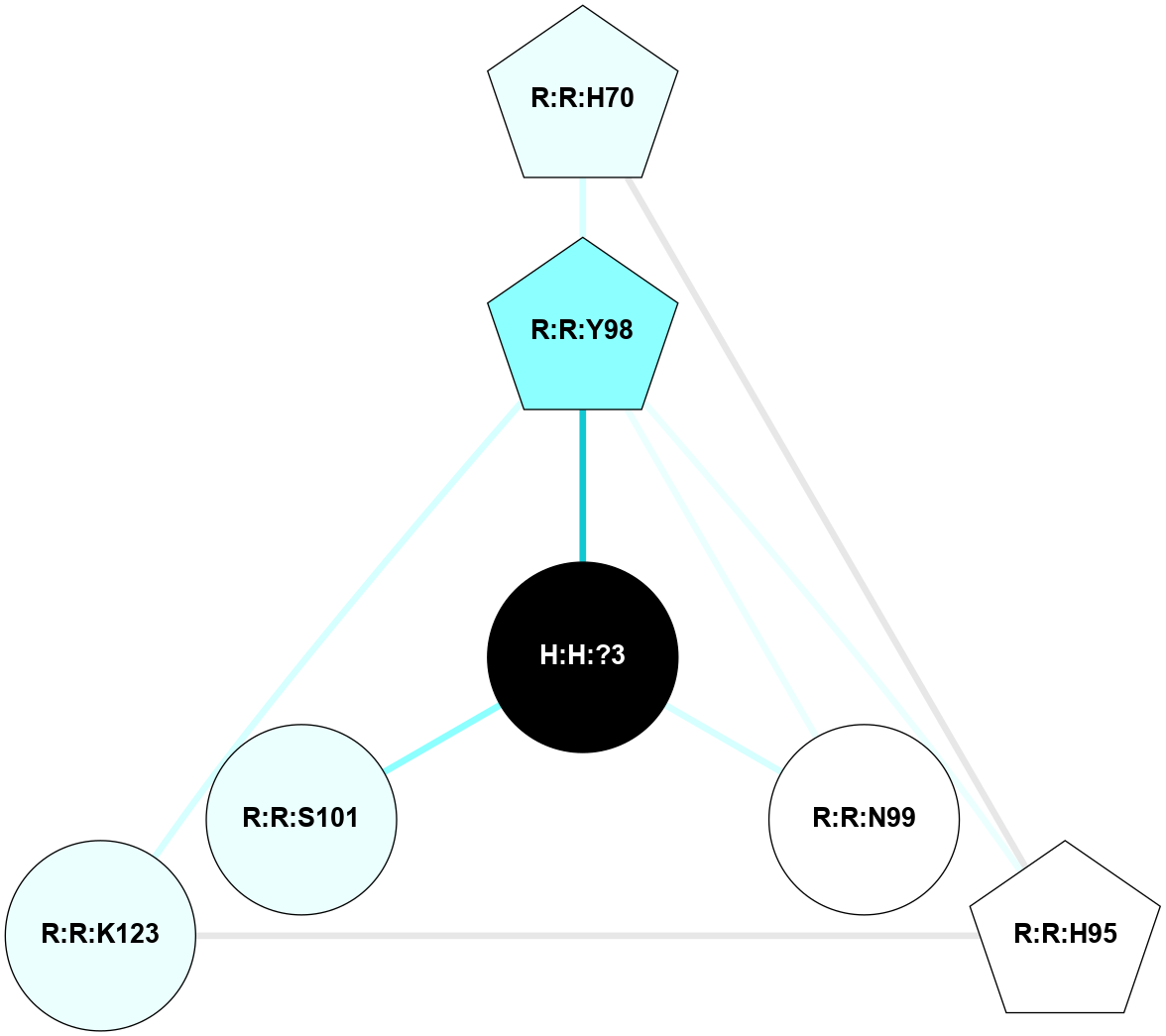
A 2D representation of the interactions of NAG in 7XW5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:H70 | R:R:H95 | 7.17 | 8 | Yes | Yes | 4 | 5 | 2 | 2 | | R:R:H70 | R:R:Y98 | 4.36 | 8 | Yes | Yes | 4 | 2 | 2 | 1 | | R:R:H95 | R:R:Y98 | 13.07 | 8 | Yes | Yes | 5 | 2 | 2 | 1 | | R:R:H95 | R:R:K123 | 5.24 | 8 | Yes | No | 5 | 4 | 2 | 2 | | R:R:N99 | R:R:Y98 | 8.14 | 8 | No | Yes | 5 | 2 | 1 | 1 | | R:R:K123 | R:R:Y98 | 7.17 | 8 | No | Yes | 4 | 2 | 2 | 1 | | H:H:?3 | R:R:Y98 | 10.52 | 8 | No | Yes | 0 | 2 | 0 | 1 | | H:H:?3 | R:R:N99 | 28.34 | 8 | No | No | 0 | 5 | 0 | 1 | | H:H:?3 | R:R:S101 | 2.69 | 8 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 13.85 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 7XW5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:N77 | 25.87 | 0 | No | No | 0 | 8 | 0 | 1 | | R:R:N77 | R:R:P76 | 1.63 | 0 | No | No | 8 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 25.87 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
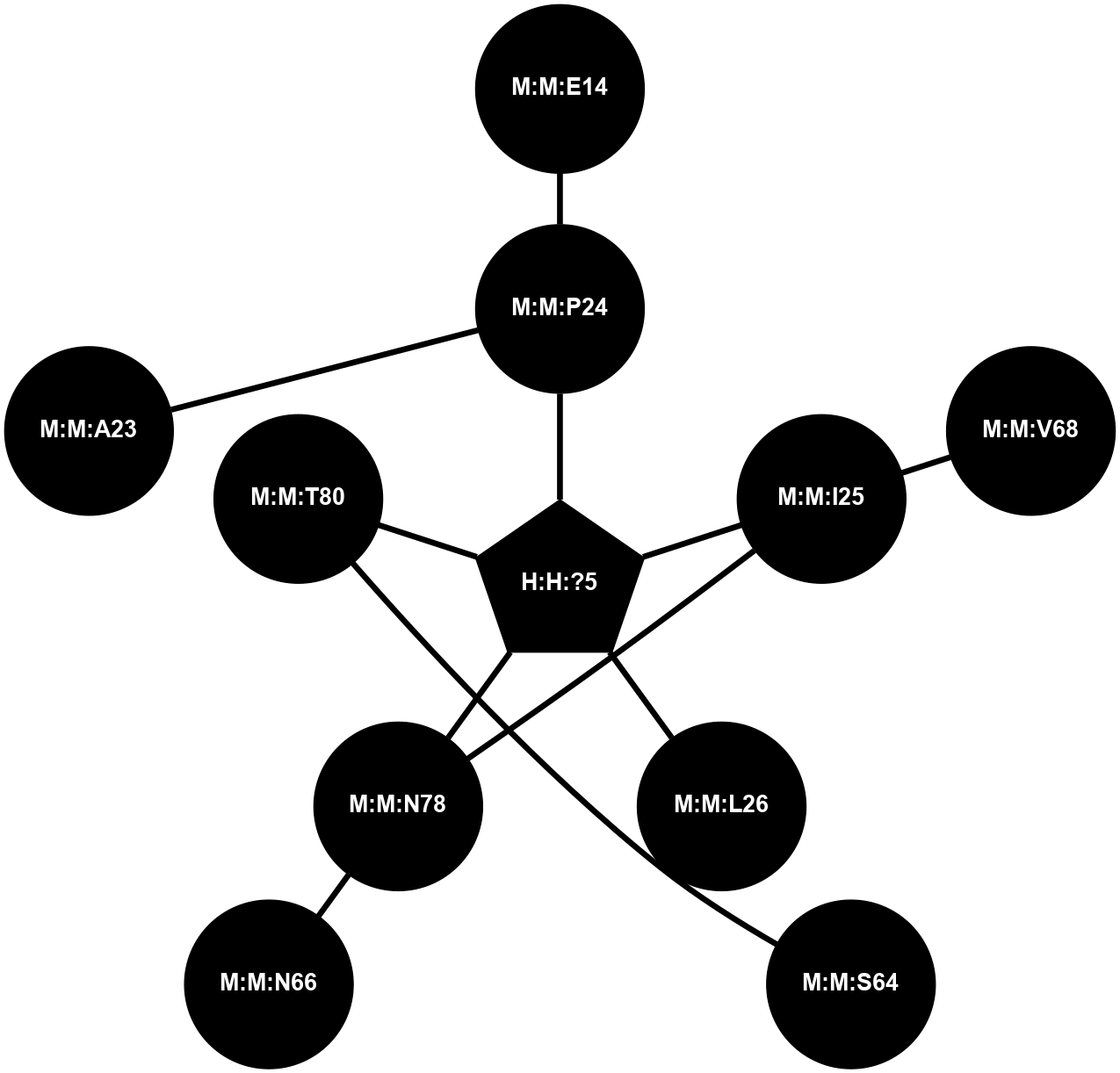
A 2D representation of the interactions of NAG in 7XW5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | N:N:E14 | N:N:P24 | 4.72 | 0 | No | No | 0 | 0 | 2 | 1 | | H:H:?5 | N:N:P24 | 4.42 | 45 | Yes | No | 0 | 0 | 0 | 1 | | N:N:I25 | N:N:V68 | 3.07 | 45 | No | No | 0 | 0 | 1 | 2 | | N:N:I25 | N:N:N78 | 5.66 | 45 | No | No | 0 | 0 | 1 | 1 | | H:H:?5 | N:N:I25 | 3.84 | 45 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?5 | N:N:L26 | 3.73 | 45 | Yes | No | 0 | 0 | 0 | 1 | | N:N:S64 | N:N:T80 | 4.8 | 0 | No | No | 0 | 0 | 2 | 1 | | H:H:?5 | N:N:N78 | 23.41 | 45 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?5 | N:N:T80 | 3.97 | 45 | Yes | No | 0 | 0 | 0 | 1 | | N:N:N66 | N:N:N78 | 2.72 | 0 | No | No | 0 | 0 | 2 | 1 | | N:N:A23 | N:N:P24 | 1.87 | 0 | No | No | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 7.87 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
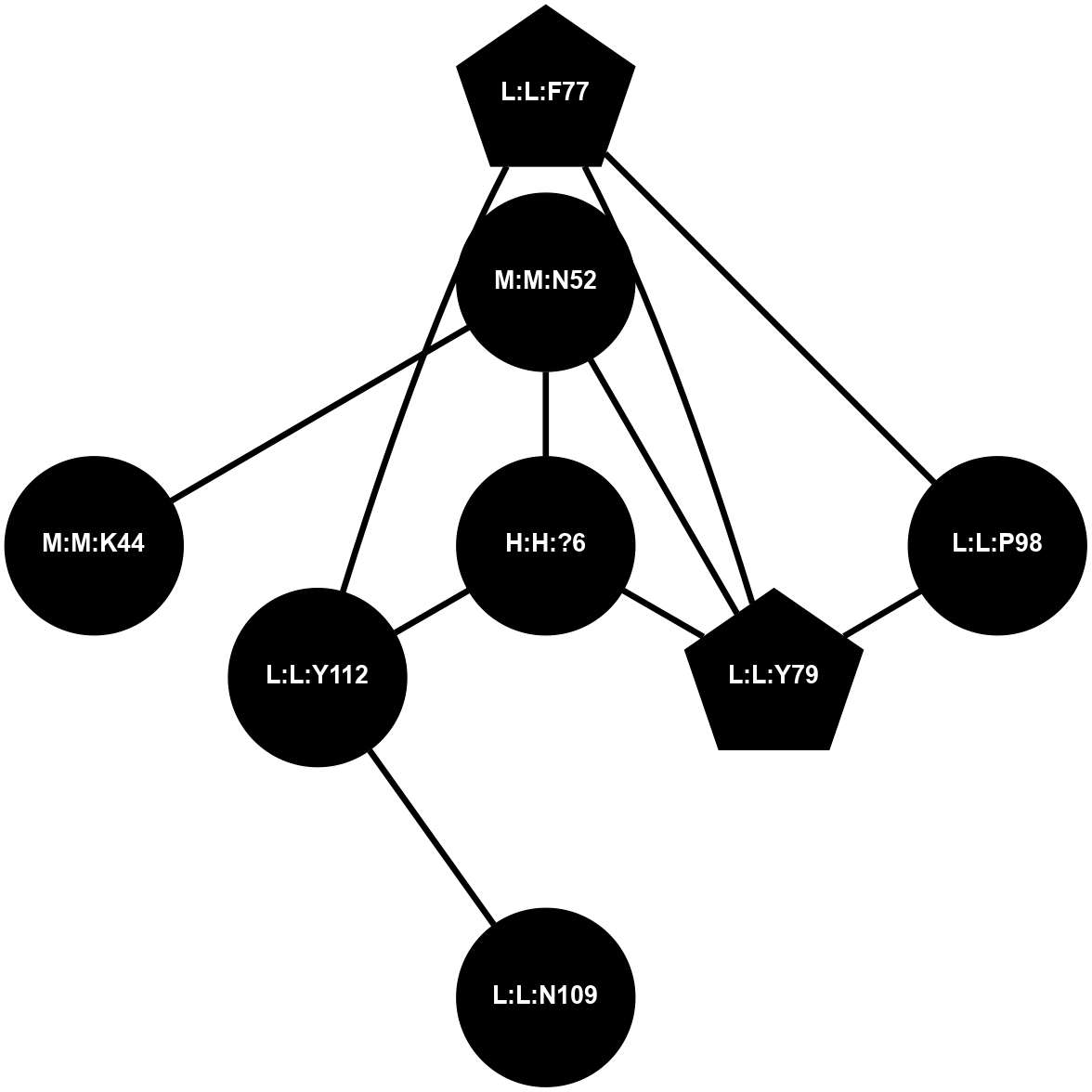
A 2D representation of the interactions of NAG in 7XW5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:Y79 | N:N:N52 | 4.65 | 26 | Yes | No | 0 | 0 | 1 | 1 | | H:H:?6 | N:N:N52 | 25.87 | 26 | No | No | 0 | 0 | 0 | 1 | | L:L:F77 | L:L:Y79 | 6.19 | 26 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:F77 | L:L:P98 | 4.33 | 26 | Yes | No | 0 | 0 | 2 | 2 | | L:L:F77 | L:L:Y112 | 9.28 | 26 | Yes | No | 0 | 0 | 2 | 1 | | L:L:P98 | L:L:Y79 | 5.56 | 26 | No | Yes | 0 | 0 | 2 | 1 | | H:H:?6 | L:L:Y79 | 30.5 | 26 | No | Yes | 0 | 0 | 0 | 1 | | L:L:N109 | L:L:Y112 | 15.12 | 0 | No | No | 0 | 0 | 2 | 1 | | H:H:?6 | L:L:Y112 | 3.16 | 26 | No | No | 0 | 0 | 0 | 1 | | N:N:K44 | N:N:N52 | 2.8 | 0 | No | No | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.84 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
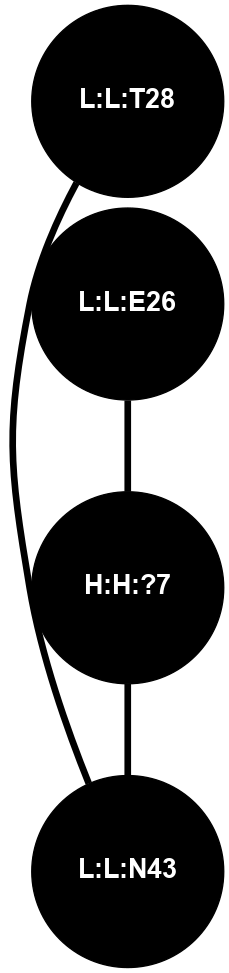
A 2D representation of the interactions of NAG in 7XW5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | L:L:E26 | 10.7 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?7 | L:L:N43 | 33.27 | 0 | No | No | 0 | 0 | 0 | 1 | | L:L:N43 | L:L:T28 | 1.46 | 0 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 21.98 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7XW6 | A | Protein | Glycoprotein Hormone | TSH | Homo Sapiens | M22 Fab | ML109 | Gs/Beta1/Gamma2 | 2.78 | 2022-08-17 | doi.org/10.1038/s41586-022-05173-3 |
|
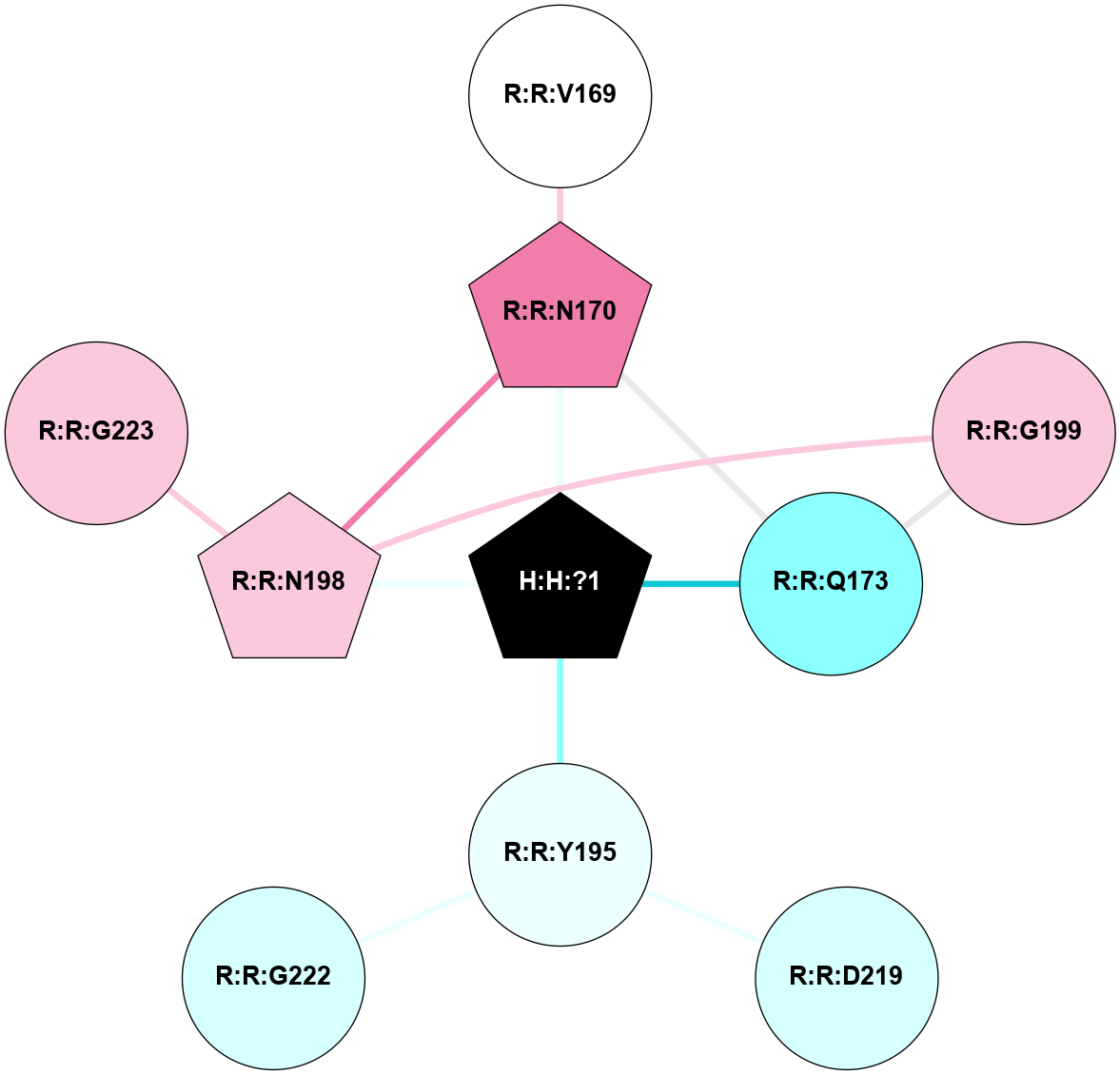
A 2D representation of the interactions of NAG in 7XW6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N170 | R:R:V169 | 5.91 | 27 | Yes | No | 8 | 5 | 1 | 2 | | R:R:N170 | R:R:Q173 | 10.56 | 27 | Yes | No | 8 | 2 | 1 | 1 | | R:R:N170 | R:R:N198 | 5.45 | 27 | Yes | Yes | 8 | 7 | 1 | 1 | | H:H:?1 | R:R:N170 | 12.32 | 27 | Yes | Yes | 0 | 8 | 0 | 1 | | R:R:G199 | R:R:Q173 | 3.29 | 0 | No | No | 7 | 2 | 2 | 1 | | H:H:?1 | R:R:Q173 | 8.36 | 27 | Yes | No | 0 | 2 | 0 | 1 | | R:R:D219 | R:R:Y195 | 12.64 | 0 | No | No | 3 | 4 | 2 | 1 | | R:R:G222 | R:R:Y195 | 2.9 | 0 | No | No | 3 | 4 | 2 | 1 | | H:H:?1 | R:R:Y195 | 7.36 | 27 | Yes | No | 0 | 4 | 0 | 1 | | R:R:G199 | R:R:N198 | 3.39 | 0 | No | Yes | 7 | 7 | 2 | 1 | | R:R:G223 | R:R:N198 | 3.39 | 0 | No | Yes | 7 | 7 | 2 | 1 | | H:H:?1 | R:R:N198 | 24.64 | 27 | Yes | Yes | 0 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 13.17 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
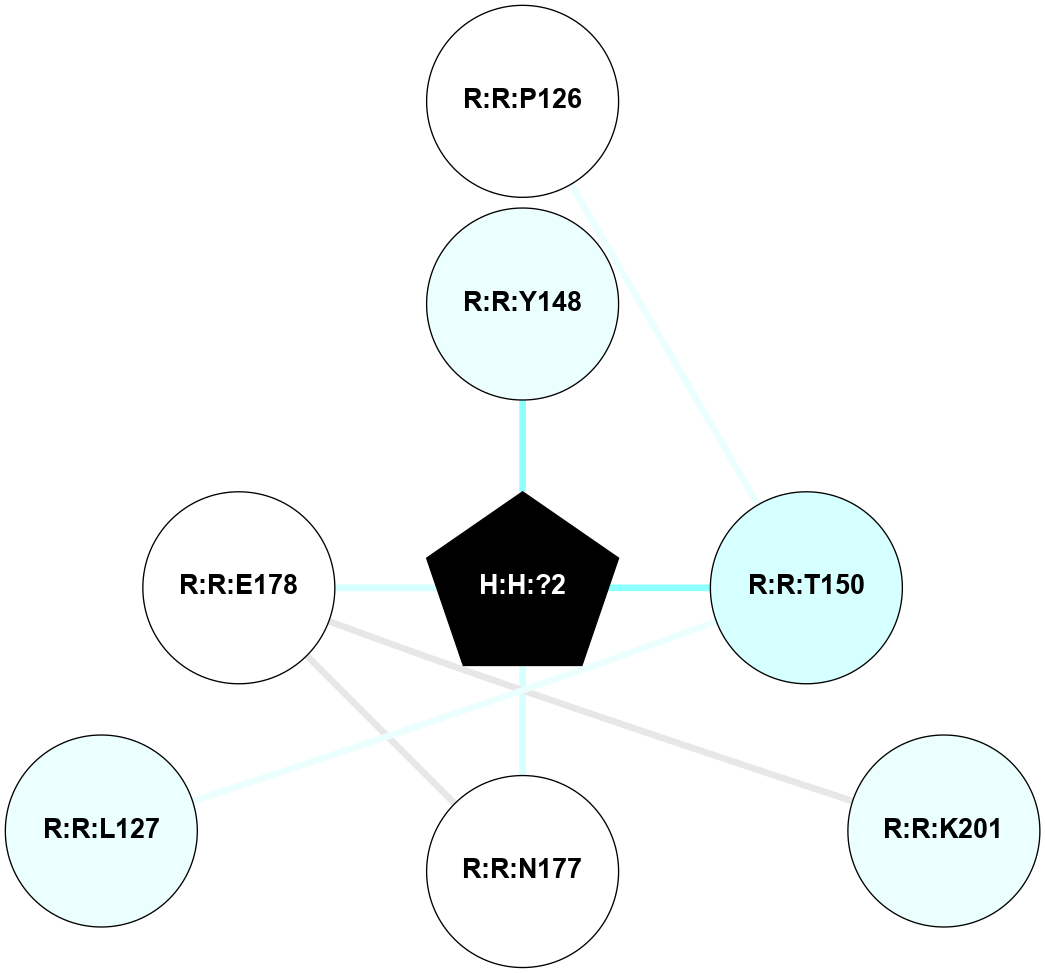
A 2D representation of the interactions of NAG in 7XW6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:P126 | R:R:T150 | 3.5 | 0 | No | No | 5 | 3 | 2 | 1 | | H:H:?2 | R:R:Y148 | 3.16 | 40 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:T150 | 6.61 | 40 | Yes | No | 0 | 3 | 0 | 1 | | R:R:E178 | R:R:N177 | 3.94 | 40 | No | No | 5 | 5 | 1 | 1 | | H:H:?2 | R:R:N177 | 29.57 | 40 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:E178 | 8.32 | 40 | Yes | No | 0 | 5 | 0 | 1 | | R:R:E178 | R:R:K201 | 2.7 | 40 | No | No | 5 | 4 | 1 | 2 | | R:R:L127 | R:R:T150 | 1.47 | 0 | No | No | 4 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 11.92 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
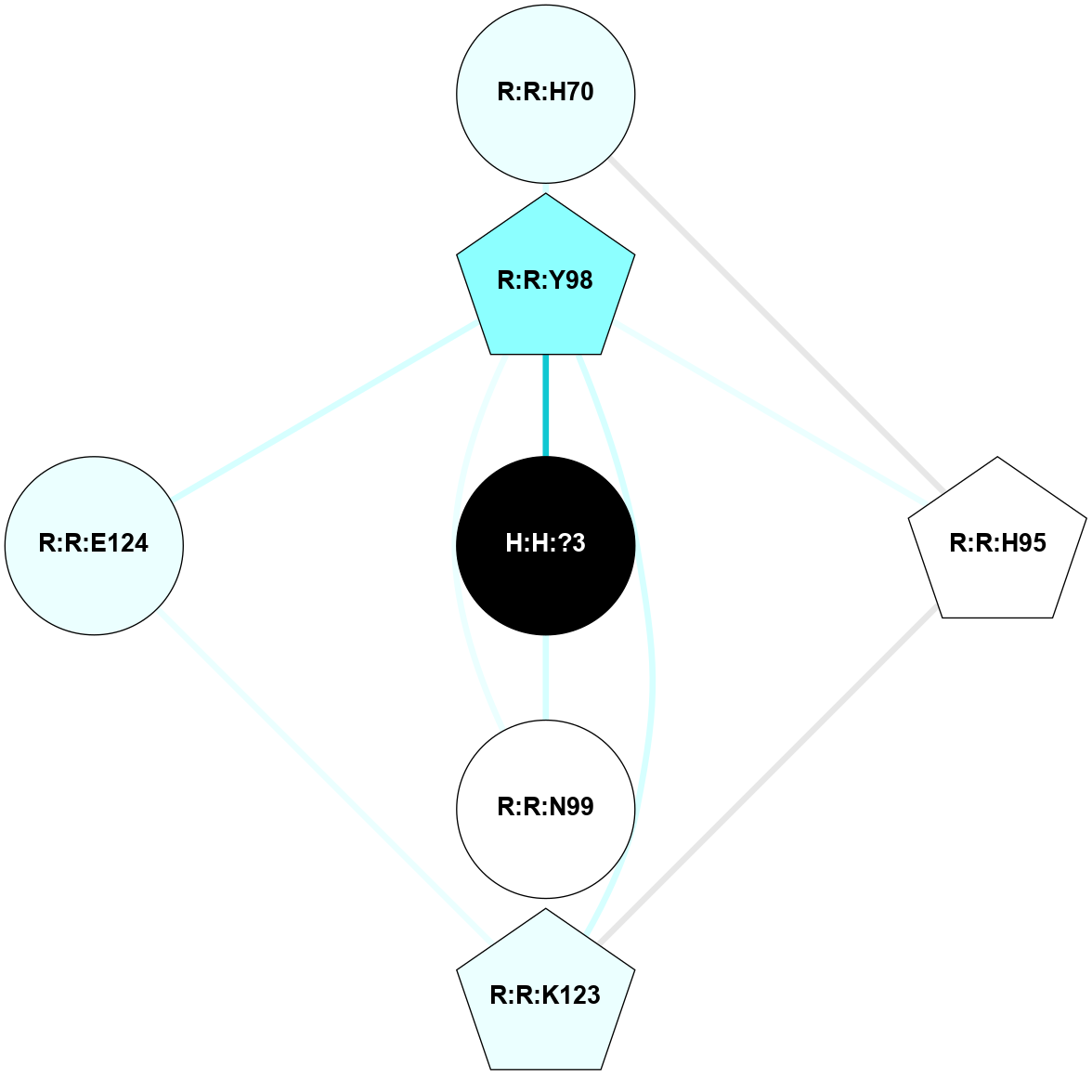
A 2D representation of the interactions of NAG in 7XW6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:H70 | R:R:H95 | 15.53 | 21 | No | Yes | 4 | 5 | 2 | 2 | | R:R:H70 | R:R:Y98 | 4.36 | 21 | No | Yes | 4 | 2 | 2 | 1 | | R:R:H95 | R:R:Y98 | 14.16 | 21 | Yes | Yes | 5 | 2 | 2 | 1 | | R:R:H95 | R:R:K123 | 3.93 | 21 | Yes | Yes | 5 | 4 | 2 | 2 | | R:R:N99 | R:R:Y98 | 10.47 | 21 | No | Yes | 5 | 2 | 1 | 1 | | R:R:K123 | R:R:Y98 | 9.55 | 21 | Yes | Yes | 4 | 2 | 2 | 1 | | R:R:E124 | R:R:Y98 | 13.47 | 21 | No | Yes | 4 | 2 | 2 | 1 | | H:H:?3 | R:R:Y98 | 10.52 | 21 | No | Yes | 0 | 2 | 0 | 1 | | H:H:?3 | R:R:N99 | 25.87 | 21 | No | No | 0 | 5 | 0 | 1 | | R:R:E124 | R:R:K123 | 16.2 | 21 | No | Yes | 4 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 18.20 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
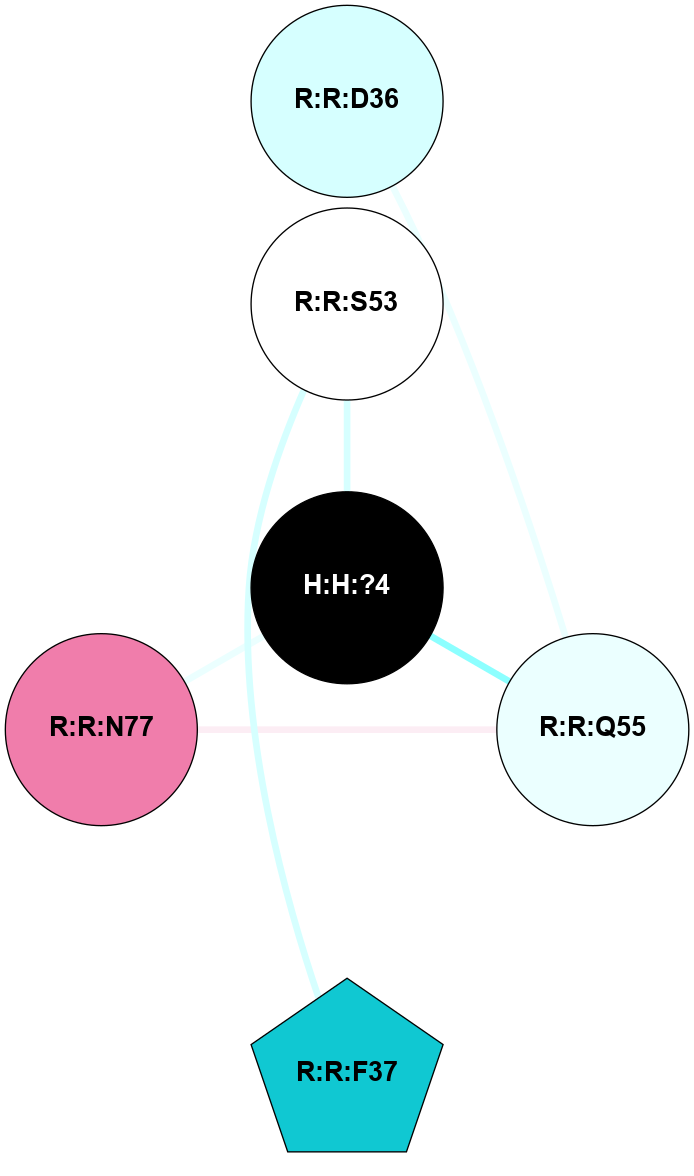
A 2D representation of the interactions of NAG in 7XW6
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D36 | R:R:Q55 | 15.66 | 0 | No | No | 3 | 4 | 2 | 1 | | R:R:F37 | R:R:S53 | 11.89 | 37 | Yes | No | 1 | 5 | 2 | 1 | | H:H:?4 | R:R:S53 | 4.04 | 38 | No | No | 0 | 5 | 0 | 1 | | R:R:N77 | R:R:Q55 | 13.2 | 38 | No | No | 8 | 4 | 1 | 1 | | H:H:?4 | R:R:Q55 | 5.97 | 38 | No | No | 0 | 4 | 0 | 1 | | H:H:?4 | R:R:N77 | 28.34 | 38 | No | No | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.78 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7YXA | A | Lipid | Lysophospholipid | S1P5 | Homo Sapiens | ONO-5430608 | - | - | 2.2 | 2022-08-10 | doi.org/10.1038/s41467-022-32447-1 |
|
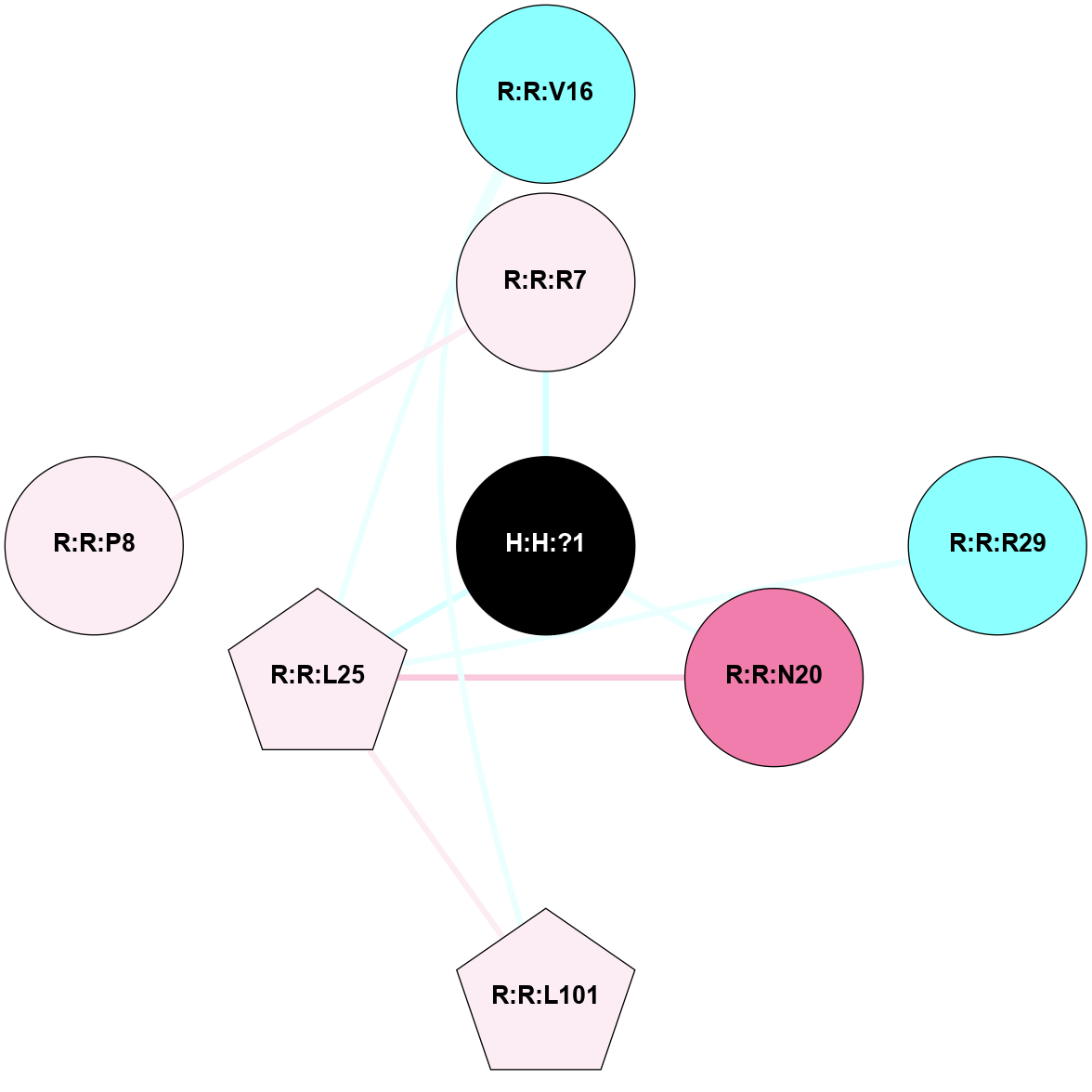
A 2D representation of the interactions of NAG in 7YXA
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:R7 | 4.36 | 1 | No | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:N20 | 22.18 | 1 | No | No | 0 | 8 | 0 | 1 | | H:H:?1 | R:R:L25 | 6.21 | 1 | No | Yes | 0 | 6 | 0 | 1 | | R:R:L25 | R:R:V16 | 5.96 | 1 | Yes | No | 6 | 2 | 1 | 2 | | R:R:L101 | R:R:V16 | 4.47 | 1 | Yes | No | 6 | 2 | 2 | 2 | | R:R:L25 | R:R:N20 | 5.49 | 1 | Yes | No | 6 | 8 | 1 | 1 | | R:R:L25 | R:R:R29 | 6.07 | 1 | Yes | No | 6 | 2 | 1 | 2 | | R:R:L101 | R:R:L25 | 4.15 | 1 | Yes | Yes | 6 | 6 | 2 | 1 | | R:R:P8 | R:R:R7 | 1.44 | 0 | No | No | 6 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 10.92 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7ZBC | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 1.8 | 2023-03-29 | doi.org/10.1038/s41586-023-05863-6 |
|
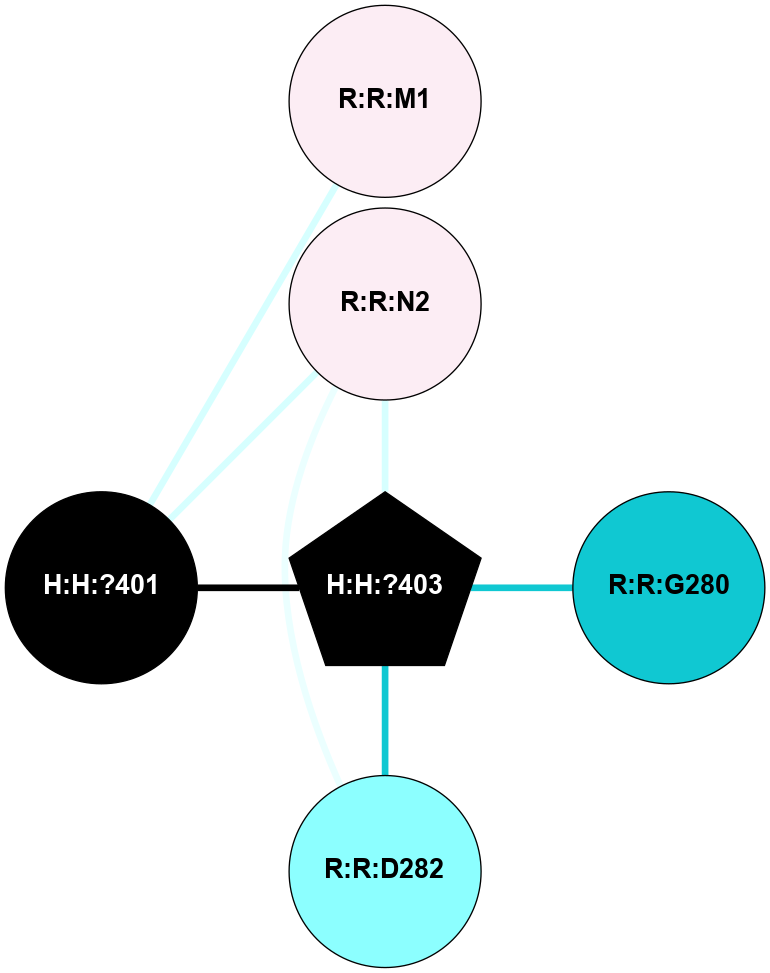
A 2D representation of the interactions of NAG in 7ZBC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?401 | H:H:?403 | 27.9 | 5 | No | Yes | 0 | 0 | 1 | 0 | | H:H:?401 | R:R:M1 | 6.35 | 5 | No | No | 0 | 6 | 1 | 2 | | H:H:?401 | R:R:N2 | 10.28 | 5 | No | No | 0 | 6 | 1 | 1 | | H:H:?403 | R:R:N2 | 24.64 | 5 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?403 | R:R:G280 | 4.6 | 5 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?403 | R:R:D282 | 6.09 | 5 | Yes | No | 0 | 2 | 0 | 1 | | R:R:D282 | R:R:N2 | 14.81 | 5 | No | No | 2 | 6 | 1 | 1 | | R:R:G280 | R:R:Y10 | 2.9 | 0 | No | Yes | 1 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.81 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7ZBE | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 1.8 | 2023-03-29 | doi.org/10.1038/s41586-023-05863-6 |
|
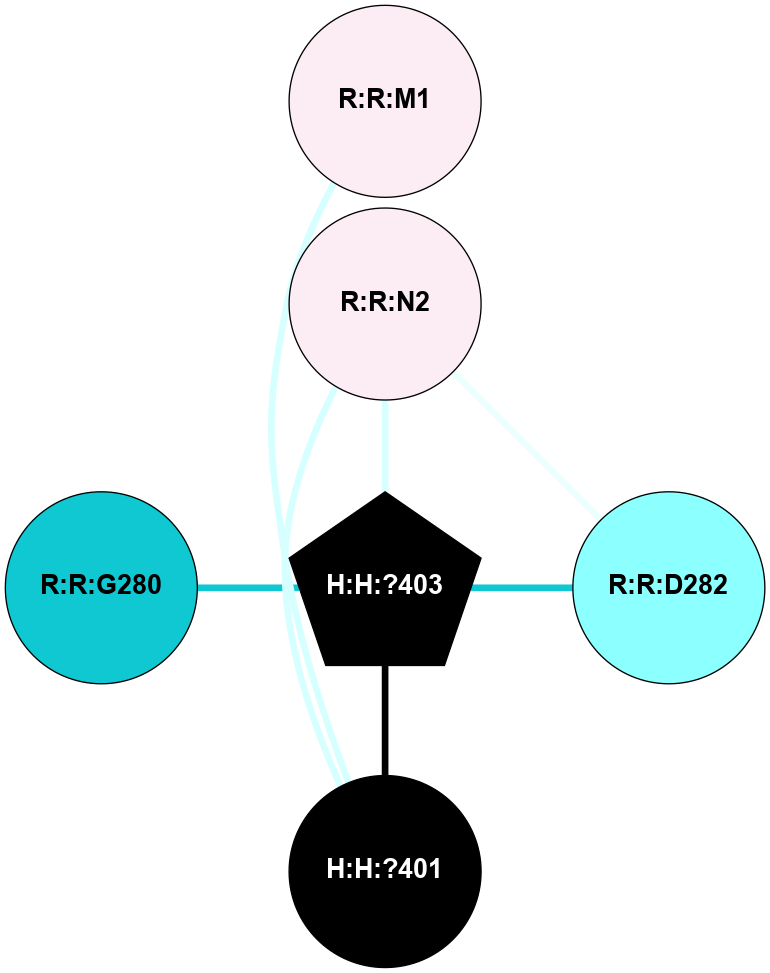
A 2D representation of the interactions of NAG in 7ZBE
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?401 | H:H:?403 | 26.04 | 4 | No | Yes | 0 | 0 | 1 | 0 | | H:H:?401 | R:R:M1 | 6.35 | 4 | No | No | 0 | 6 | 1 | 2 | | H:H:?401 | R:R:N2 | 10.28 | 4 | No | No | 0 | 6 | 1 | 1 | | H:H:?403 | R:R:N2 | 24.64 | 4 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?403 | R:R:D282 | 6.09 | 4 | Yes | No | 0 | 2 | 0 | 1 | | R:R:D282 | R:R:N2 | 14.81 | 4 | No | No | 2 | 6 | 1 | 1 | | H:H:?403 | R:R:G280 | 3.07 | 4 | Yes | No | 0 | 1 | 0 | 1 | | R:R:G280 | R:R:Y10 | 2.9 | 0 | No | Yes | 1 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.96 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 7ZI0 | F | Protein | Frizzled | SMO | Homo Sapiens | SAG | Cholesterol | - | 3 | 2022-06-15 | doi.org/10.1126/sciadv.abm5563 |
|

A 2D representation of the interactions of NAG in 7ZI0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:K186 | R:R:N188 | 5.6 | 9 | No | No | 4 | 8 | 1 | 1 | | H:H:?1 | R:R:K186 | 6.33 | 9 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:N188 | 30.8 | 9 | No | No | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.57 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8A6C | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 1.8 | 2023-03-29 | doi.org/10.1038/s41586-023-05863-6 |
|
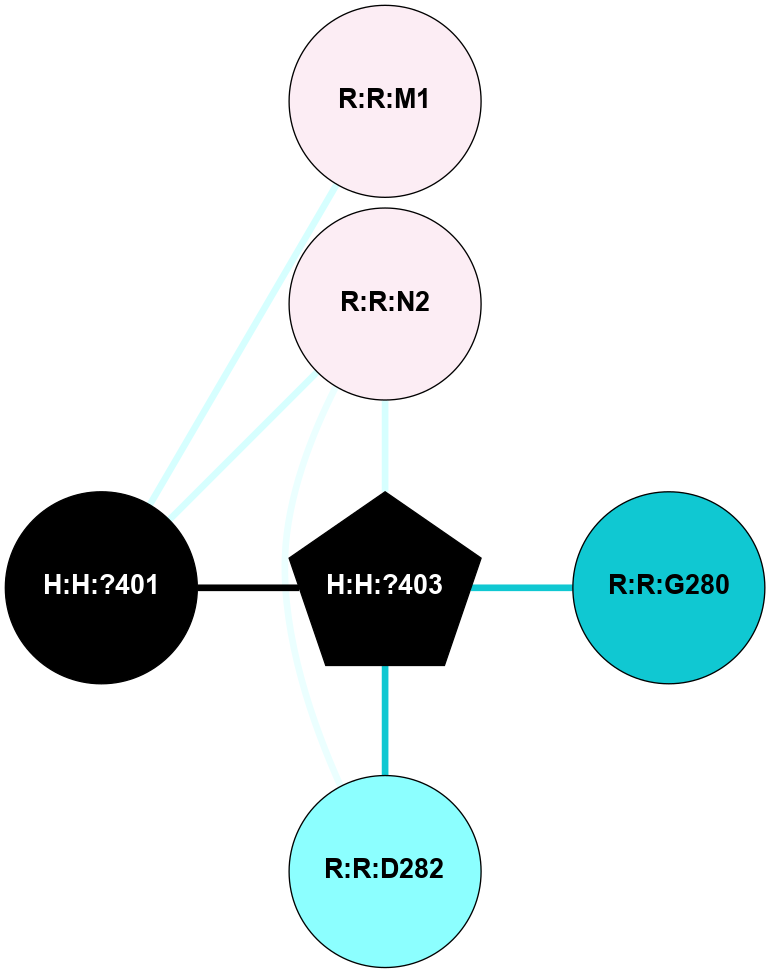
A 2D representation of the interactions of NAG in 8A6C
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?401 | H:H:?403 | 26.04 | 3 | No | Yes | 0 | 0 | 1 | 0 | | H:H:?401 | R:R:M1 | 6.35 | 3 | No | No | 0 | 6 | 1 | 2 | | H:H:?401 | R:R:N2 | 8.23 | 3 | No | No | 0 | 6 | 1 | 1 | | H:H:?403 | R:R:N2 | 23.41 | 3 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?403 | R:R:G280 | 3.07 | 3 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?403 | R:R:D282 | 6.09 | 3 | Yes | No | 0 | 2 | 0 | 1 | | R:R:D282 | R:R:N2 | 13.46 | 3 | No | No | 2 | 6 | 1 | 1 | | R:R:G280 | R:R:Y10 | 1.45 | 0 | No | Yes | 1 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.65 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8A6D | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 1.8 | 2023-03-29 | doi.org/10.1038/s41586-023-05863-6 |
|
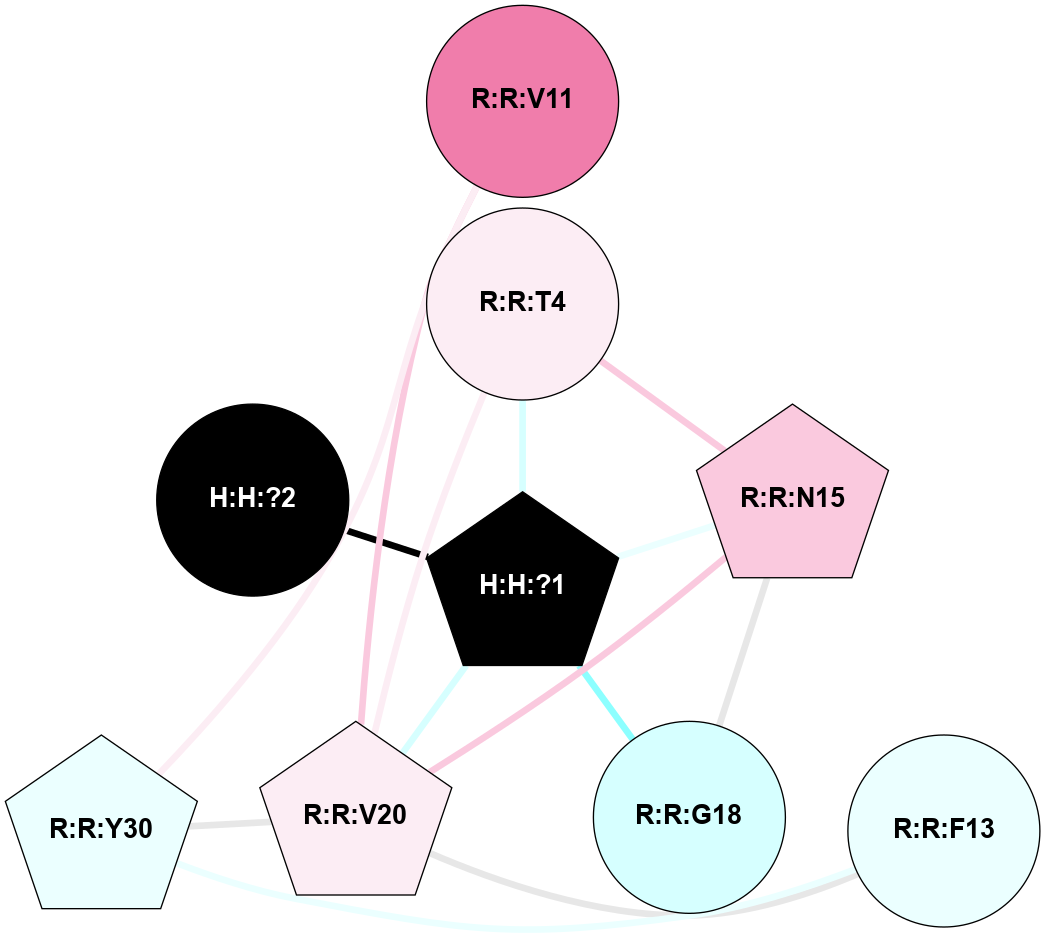
A 2D representation of the interactions of NAG in 8A6D
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 39.01 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:T4 | 3.97 | 1 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:N15 | 25.87 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.14 | 1 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:V20 | 4.01 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:N15 | R:R:T4 | 5.85 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 7.93 | 1 | No | Yes | 6 | 6 | 1 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | No | Yes | 8 | 6 | 2 | 1 | | R:R:V11 | R:R:Y30 | 6.31 | 1 | No | Yes | 8 | 4 | 2 | 2 | | R:R:F13 | R:R:V20 | 5.24 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 6.19 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | R:R:V20 | R:R:Y30 | 11.36 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 15.80 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
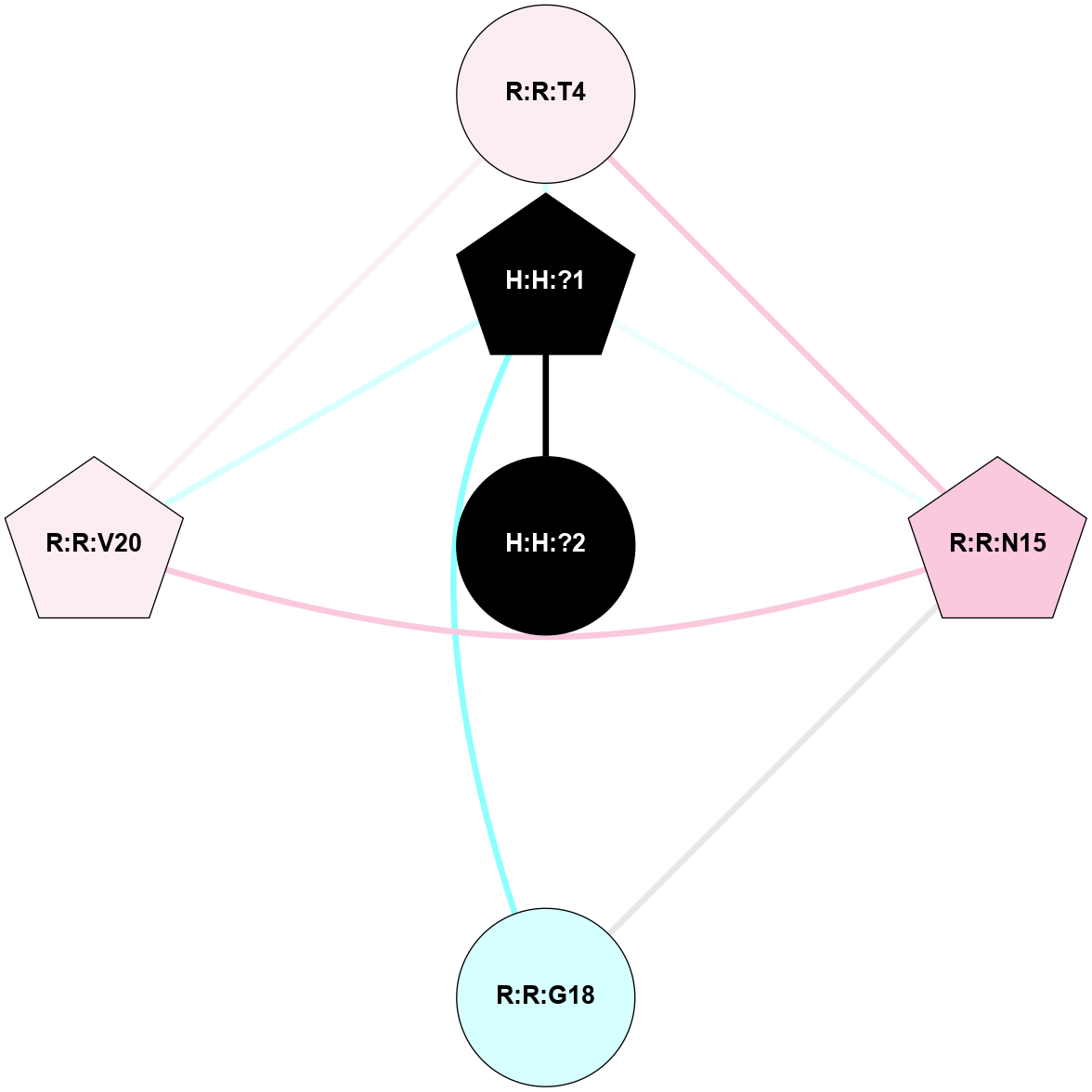
A 2D representation of the interactions of NAG in 8A6D
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 39.01 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:T4 | 3.97 | 1 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 25.87 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.14 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 4.01 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | R:R:N15 | R:R:T4 | 5.85 | 1 | Yes | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 7.93 | 1 | No | Yes | 6 | 6 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 39.01 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
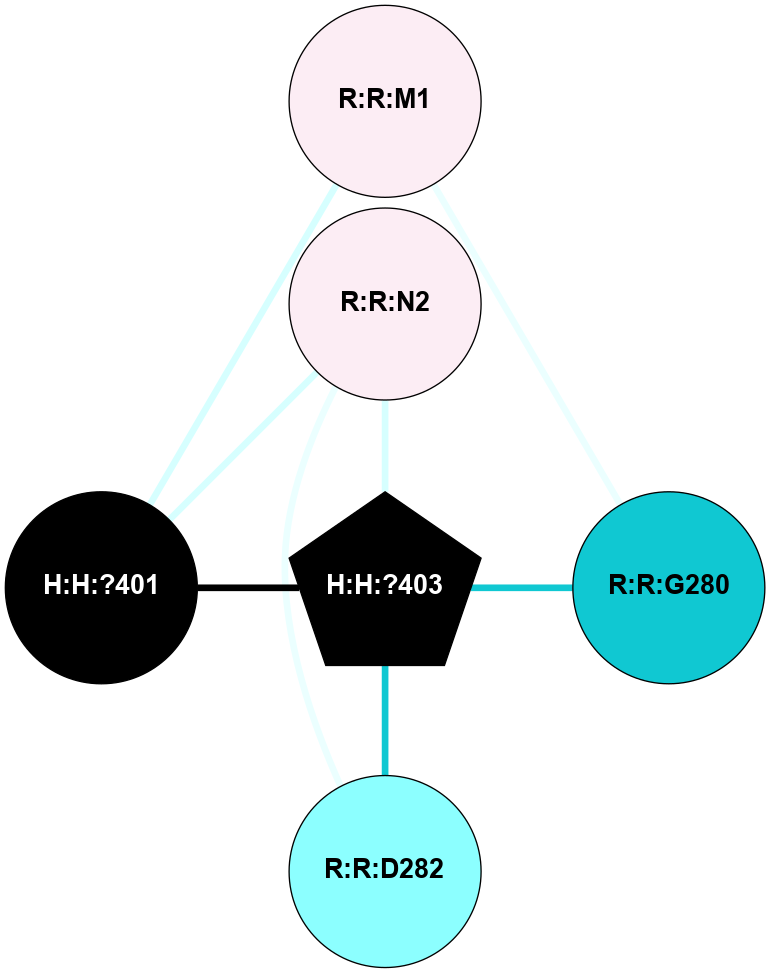
A 2D representation of the interactions of NAG in 8A6D
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?401 | H:H:?403 | 16.74 | 3 | No | Yes | 0 | 0 | 1 | 0 | | H:H:?401 | R:R:M1 | 6.35 | 3 | No | No | 0 | 6 | 1 | 2 | | H:H:?401 | R:R:N2 | 4.11 | 3 | No | No | 0 | 6 | 1 | 1 | | H:H:?403 | R:R:N2 | 24.64 | 3 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?403 | R:R:G280 | 4.6 | 3 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?403 | R:R:D282 | 8.52 | 3 | Yes | No | 0 | 2 | 0 | 1 | | R:R:D282 | R:R:N2 | 8.08 | 3 | No | No | 2 | 6 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.62 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8A6E | A | Sensory | Opsins | Rhodopsin | Bos Taurus | All-trans-Retinal | - | - | 1.8 | 2023-03-29 | doi.org/10.1038/s41586-023-05863-6 |
|
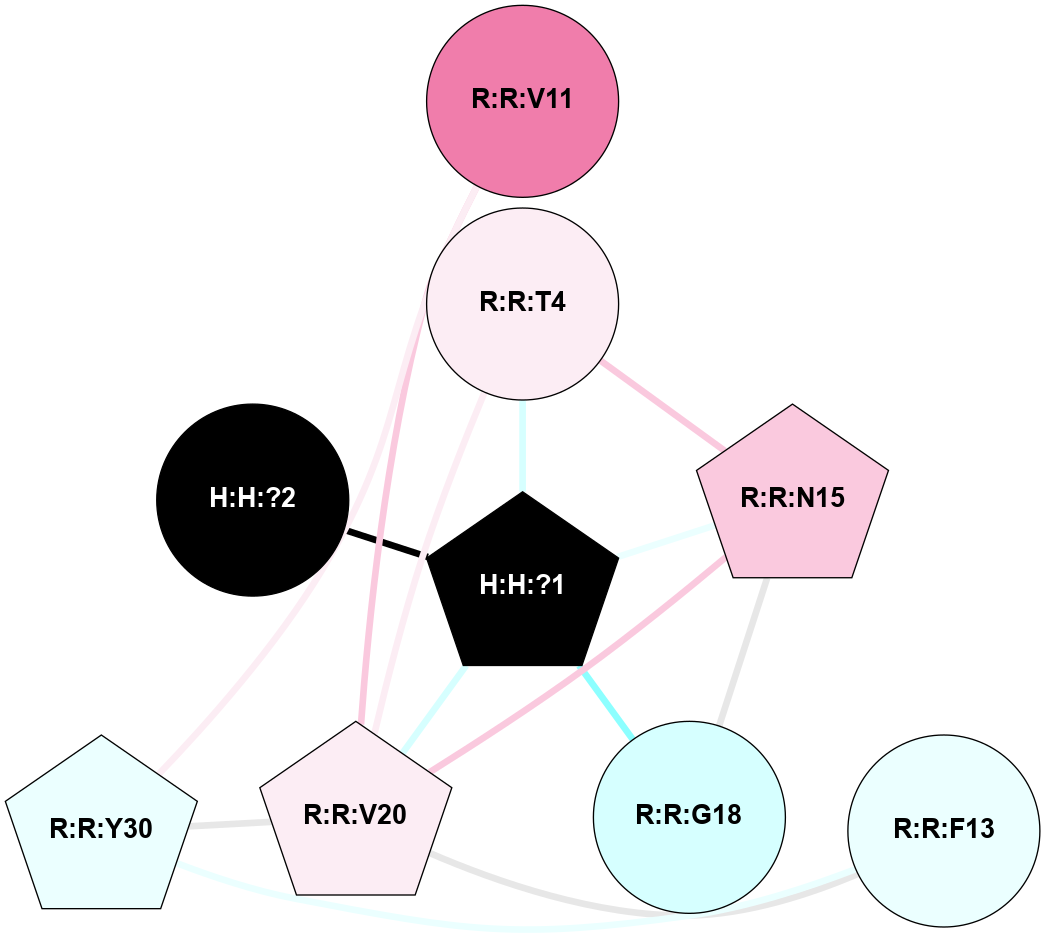
A 2D representation of the interactions of NAG in 8A6E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 35.66 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:T4 | 3.97 | 1 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:N15 | 27.11 | 1 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?1 | R:R:G18 | 6.14 | 1 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:V20 | 4.01 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:N15 | R:R:T4 | 5.85 | 1 | Yes | No | 7 | 6 | 1 | 1 | | R:R:T4 | R:R:V20 | 7.93 | 1 | No | Yes | 6 | 6 | 1 | 1 | | R:R:V11 | R:R:V20 | 4.81 | 1 | No | Yes | 8 | 6 | 2 | 1 | | R:R:V11 | R:R:Y30 | 6.31 | 1 | No | Yes | 8 | 4 | 2 | 2 | | R:R:F13 | R:R:V20 | 3.93 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:F13 | R:R:Y30 | 6.19 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 1 | 1 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 1 | 1 | | R:R:V20 | R:R:Y30 | 12.62 | 1 | Yes | Yes | 6 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 15.38 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
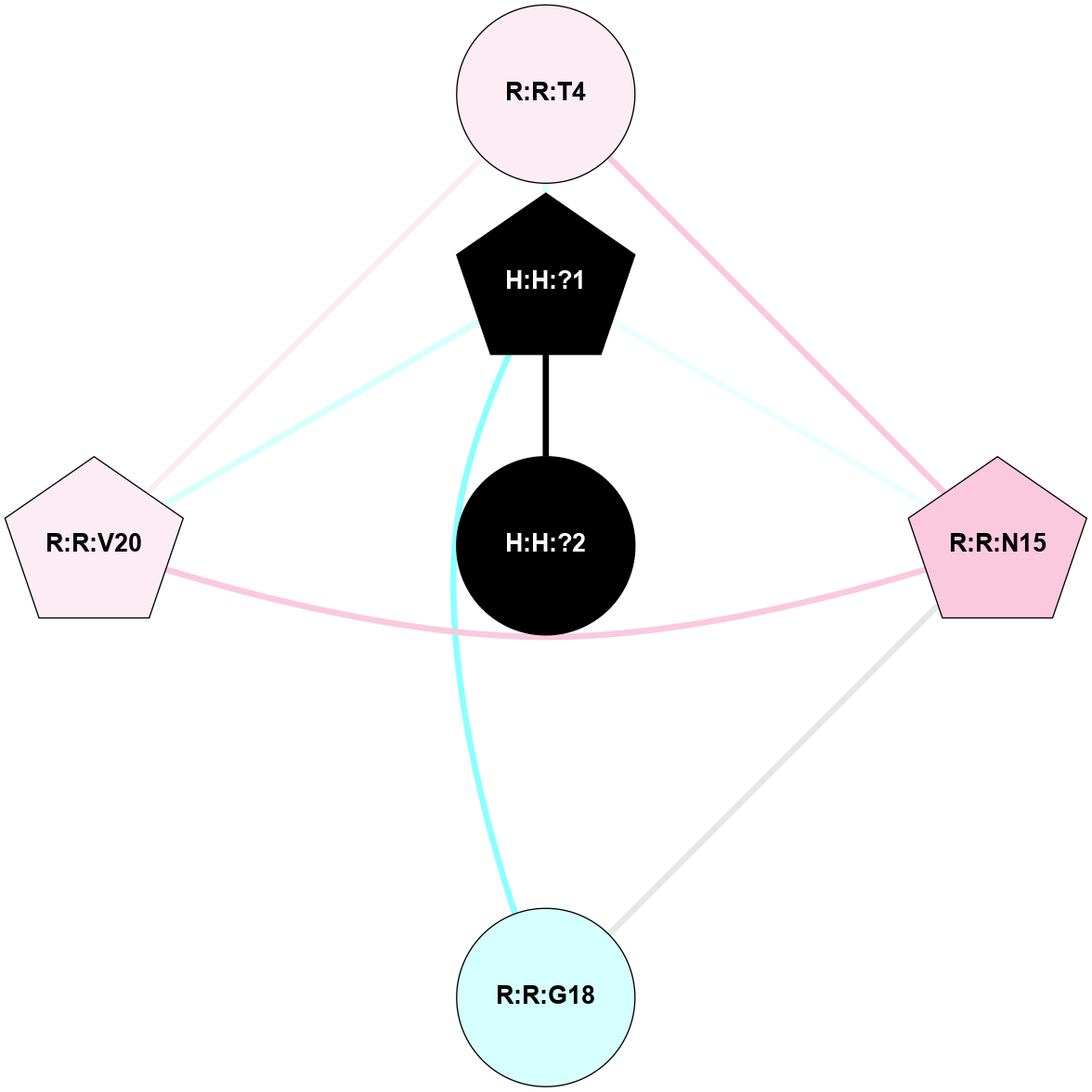
A 2D representation of the interactions of NAG in 8A6E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 35.66 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:T4 | 3.97 | 1 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:N15 | 27.11 | 1 | Yes | Yes | 0 | 7 | 1 | 2 | | H:H:?1 | R:R:G18 | 6.14 | 1 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:V20 | 4.01 | 1 | Yes | Yes | 0 | 6 | 1 | 2 | | R:R:N15 | R:R:T4 | 5.85 | 1 | Yes | No | 7 | 6 | 2 | 2 | | R:R:T4 | R:R:V20 | 7.93 | 1 | No | Yes | 6 | 6 | 2 | 2 | | R:R:G18 | R:R:N15 | 5.09 | 1 | No | Yes | 3 | 7 | 2 | 2 | | R:R:N15 | R:R:V20 | 10.35 | 1 | Yes | Yes | 7 | 6 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 35.66 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
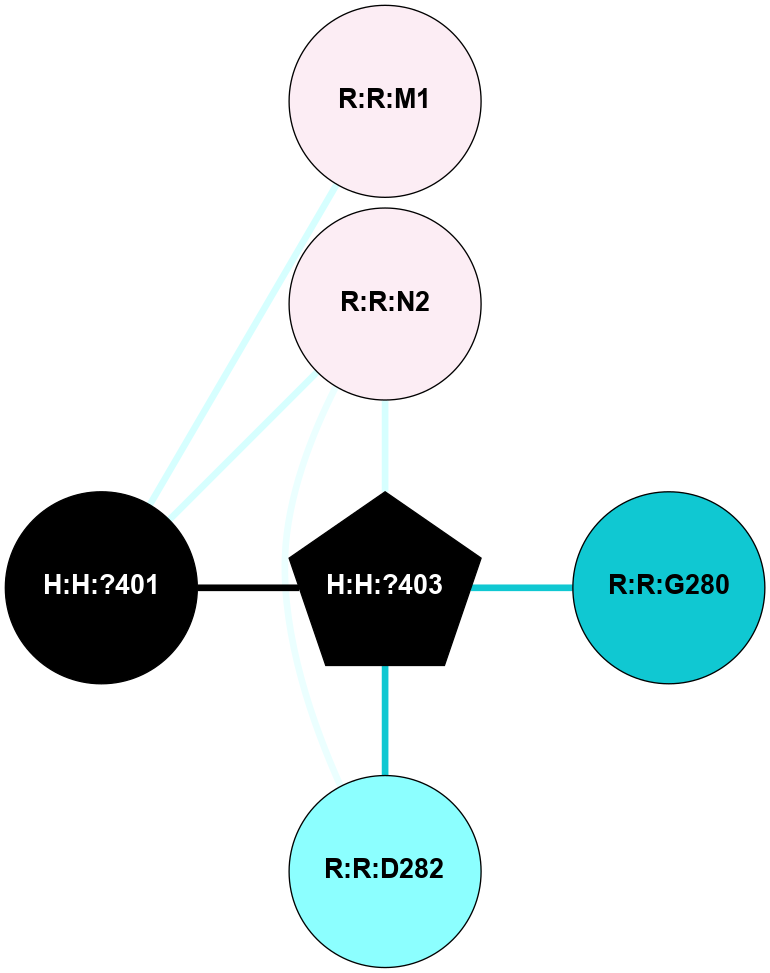
A 2D representation of the interactions of NAG in 8A6E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?401 | H:H:?403 | 16.74 | 6 | No | Yes | 0 | 0 | 1 | 0 | | H:H:?401 | R:R:M1 | 4.23 | 6 | No | No | 0 | 6 | 1 | 2 | | H:H:?401 | R:R:N2 | 4.11 | 6 | No | No | 0 | 6 | 1 | 1 | | H:H:?403 | R:R:N2 | 24.64 | 6 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?403 | R:R:G280 | 4.6 | 6 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?403 | R:R:D282 | 9.74 | 6 | Yes | No | 0 | 2 | 0 | 1 | | R:R:D282 | R:R:N2 | 10.77 | 6 | No | No | 2 | 6 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.93 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8F0J | B1 | Peptide | Calcitonin | CT | Homo Sapiens | Pramlintide Analogue San45 | - | Gs/Beta1/Gamma2 | 2 | 2023-08-02 | doi.org/10.1038/s41589-023-01393-4 |
|
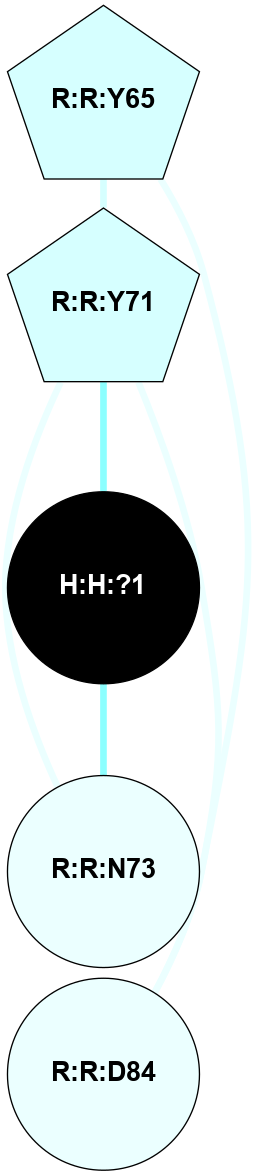
A 2D representation of the interactions of NAG in 8F0J
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:Y71 | 3.16 | 23 | No | Yes | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:N73 | 28.34 | 23 | No | No | 0 | 4 | 0 | 1 | | R:R:Y65 | R:R:Y71 | 10.92 | 23 | Yes | Yes | 3 | 3 | 2 | 1 | | R:R:D84 | R:R:Y65 | 6.9 | 23 | No | Yes | 4 | 3 | 2 | 2 | | R:R:N73 | R:R:Y71 | 5.81 | 23 | No | Yes | 4 | 3 | 1 | 1 | | R:R:D84 | R:R:Y71 | 10.34 | 23 | No | Yes | 4 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.75 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
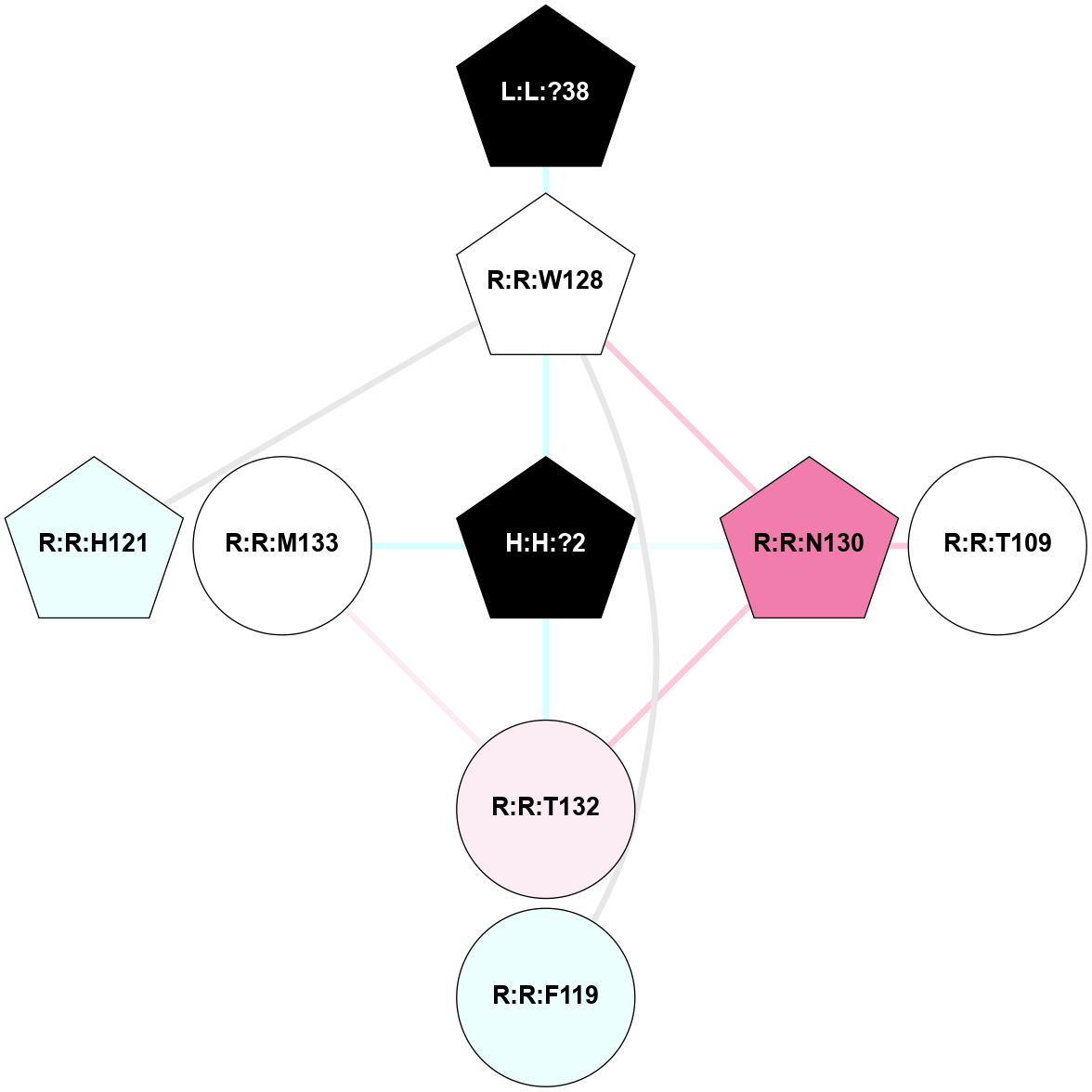
A 2D representation of the interactions of NAG in 8F0J
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:W128 | 10.22 | 11 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:N130 | 25.87 | 11 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?2 | R:R:T132 | 5.29 | 11 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:M133 | 5.07 | 11 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?38 | R:R:W128 | 8.46 | 0 | Yes | Yes | 0 | 5 | 2 | 1 | | R:R:N130 | R:R:T109 | 7.31 | 11 | Yes | No | 8 | 5 | 1 | 2 | | R:R:F119 | R:R:W128 | 3.01 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:H121 | R:R:W128 | 23.27 | 11 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 9.04 | 11 | Yes | Yes | 8 | 5 | 1 | 1 | | R:R:N130 | R:R:T132 | 8.77 | 11 | Yes | No | 8 | 6 | 1 | 1 | | R:R:M133 | R:R:T132 | 3.01 | 11 | No | No | 5 | 6 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 11.61 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8F0K | B1 | Peptide | Calcitonin | CT (AMY3) | Homo Sapiens | Pramlintide Analogue San385 | - | Gs/Beta1/Gamma2; RAMP3 | 1.9 | 2023-08-02 | doi.org/10.1038/s41589-023-01393-4 |
|
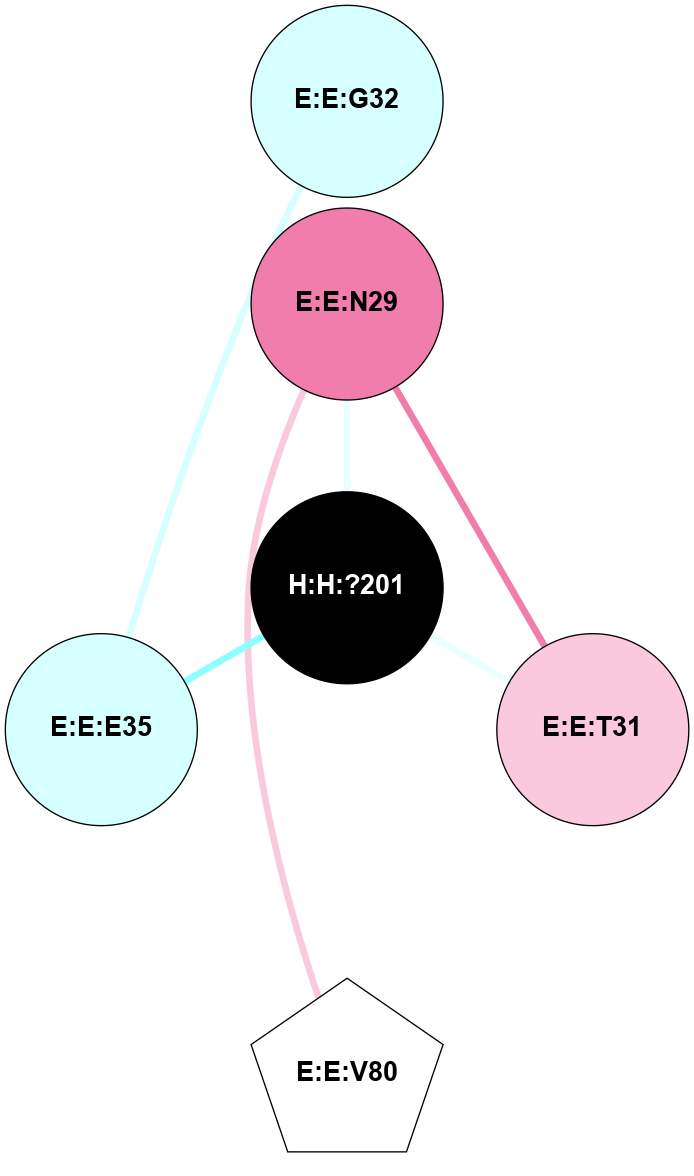
A 2D representation of the interactions of NAG in 8F0K
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | E:E:N29 | E:E:T31 | 5.85 | 26 | No | No | 8 | 7 | 1 | 1 | | E:E:N29 | H:H:?201 | 28.34 | 26 | No | No | 8 | 0 | 1 | 0 | | E:E:T31 | H:H:?201 | 5.29 | 26 | No | No | 7 | 0 | 1 | 0 | | E:E:E35 | H:H:?201 | 7.13 | 0 | No | No | 3 | 0 | 1 | 0 | | E:E:E35 | E:E:G32 | 3.27 | 0 | No | No | 3 | 3 | 1 | 2 | | E:E:N29 | E:E:V80 | 2.96 | 26 | No | Yes | 8 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.59 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
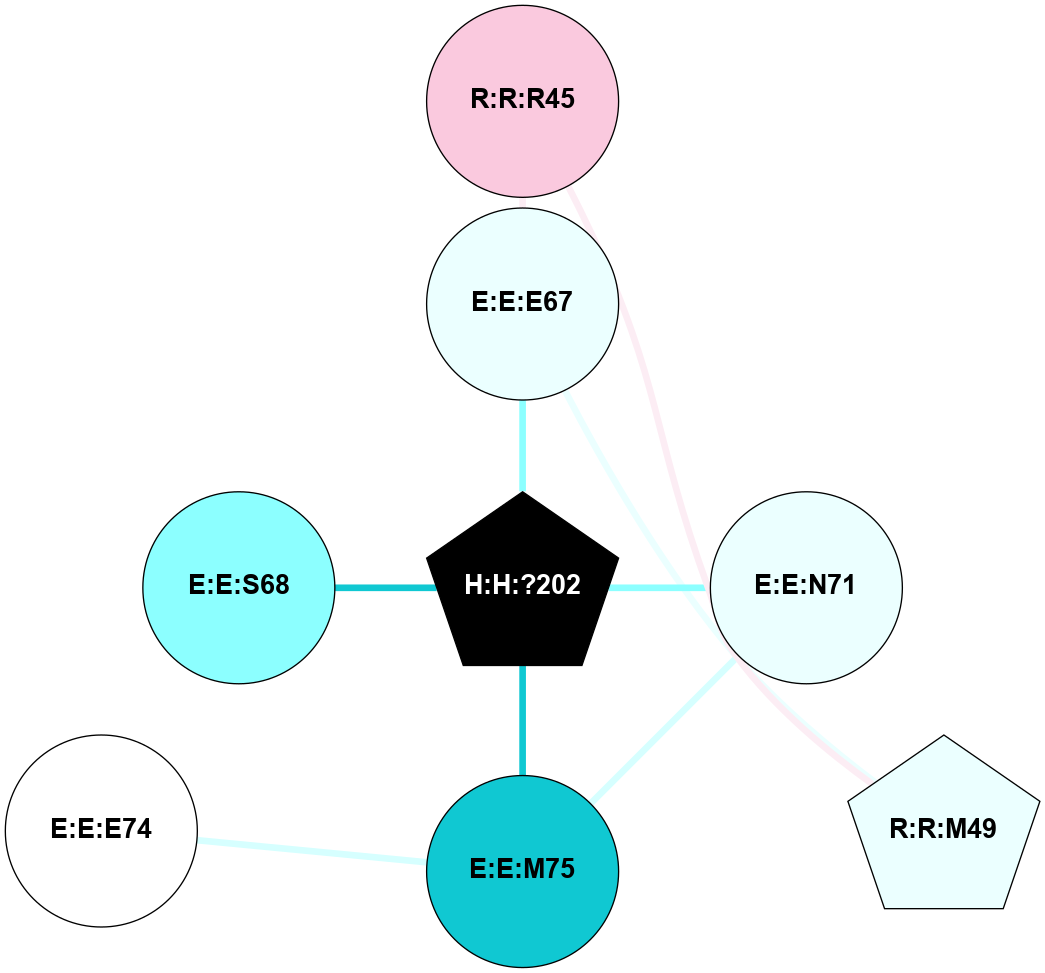
A 2D representation of the interactions of NAG in 8F0K
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | E:E:E67 | H:H:?202 | 14.27 | 2 | No | Yes | 4 | 0 | 1 | 0 | | E:E:E67 | R:R:R45 | 13.96 | 2 | No | No | 4 | 7 | 1 | 2 | | E:E:E67 | R:R:M49 | 6.77 | 2 | No | Yes | 4 | 4 | 1 | 2 | | E:E:M75 | E:E:N71 | 4.21 | 2 | No | No | 1 | 4 | 1 | 1 | | E:E:N71 | H:H:?202 | 24.64 | 2 | No | Yes | 4 | 0 | 1 | 0 | | E:E:E74 | E:E:M75 | 5.41 | 2 | No | No | 5 | 1 | 2 | 1 | | E:E:M75 | H:H:?202 | 7.61 | 2 | No | Yes | 1 | 0 | 1 | 0 | | R:R:M49 | R:R:R45 | 9.93 | 2 | Yes | No | 4 | 7 | 2 | 2 | | E:E:S68 | H:H:?202 | 2.69 | 0 | No | Yes | 2 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.30 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
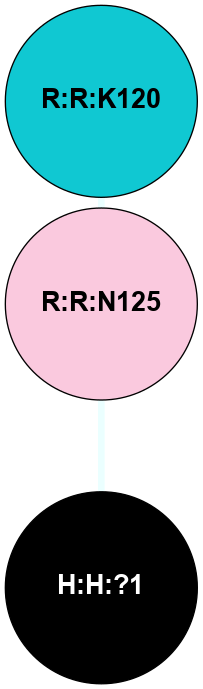
A 2D representation of the interactions of NAG in 8F0K
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N125 | 28.34 | 0 | No | No | 0 | 7 | 0 | 1 | | R:R:K120 | R:R:N125 | 1.4 | 0 | No | No | 1 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 28.34 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
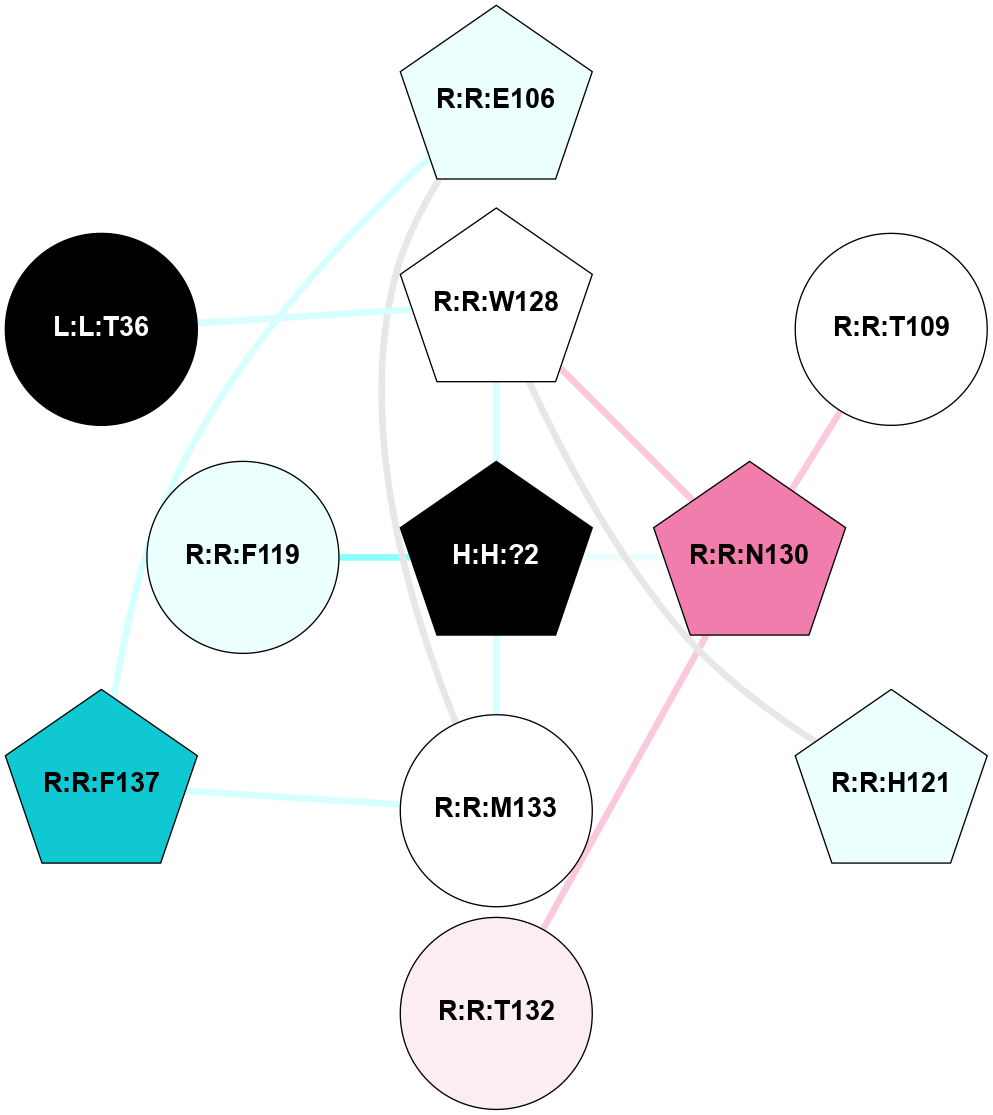
A 2D representation of the interactions of NAG in 8F0K
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:W128 | 6.13 | 7 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:N130 | 24.64 | 7 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?2 | R:R:M133 | 6.34 | 7 | Yes | No | 0 | 5 | 0 | 1 | | R:R:E106 | R:R:M133 | 4.06 | 7 | Yes | No | 4 | 5 | 2 | 1 | | R:R:E106 | R:R:F137 | 3.5 | 7 | Yes | Yes | 4 | 1 | 2 | 2 | | R:R:N130 | R:R:T109 | 5.85 | 7 | Yes | No | 8 | 5 | 1 | 2 | | R:R:H121 | R:R:W128 | 15.87 | 7 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 7.91 | 7 | Yes | Yes | 8 | 5 | 1 | 1 | | R:R:N130 | R:R:T132 | 5.85 | 7 | Yes | No | 8 | 6 | 1 | 2 | | R:R:F137 | R:R:M133 | 4.98 | 7 | Yes | No | 1 | 5 | 2 | 1 | | L:L:T36 | R:R:W128 | 2.43 | 0 | No | Yes | 0 | 5 | 2 | 1 | | H:H:?2 | R:R:F119 | 2.19 | 7 | Yes | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 9.82 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
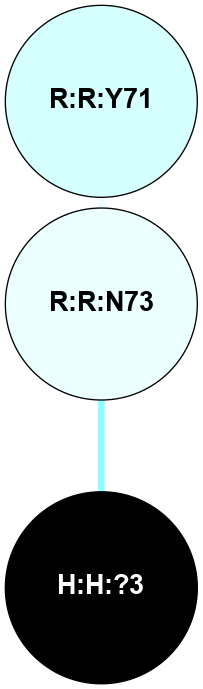
A 2D representation of the interactions of NAG in 8F0K
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:N73 | 25.87 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N73 | R:R:Y71 | 5.81 | 0 | No | No | 4 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 25.87 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8F2A | B1 | Peptide | Calcitonin | CT (AMY3) | Homo Sapiens | Pramlintide Analogue San385 | - | Gs/Beta1/Gamma2; RAMP3 | 2.2 | 2023-08-02 | doi.org/10.1038/s41589-023-01393-4 |
|

A 2D representation of the interactions of NAG in 8F2A
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | E:E:N29 | E:E:T31 | 4.39 | 0 | Yes | No | 8 | 7 | 1 | 2 | | E:E:G32 | E:E:N29 | 6.79 | 0 | No | Yes | 3 | 8 | 2 | 1 | | E:E:N29 | H:H:?201 | 29.57 | 0 | Yes | No | 8 | 0 | 1 | 0 | | E:E:N29 | E:E:V80 | 1.48 | 0 | Yes | No | 8 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 29.57 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
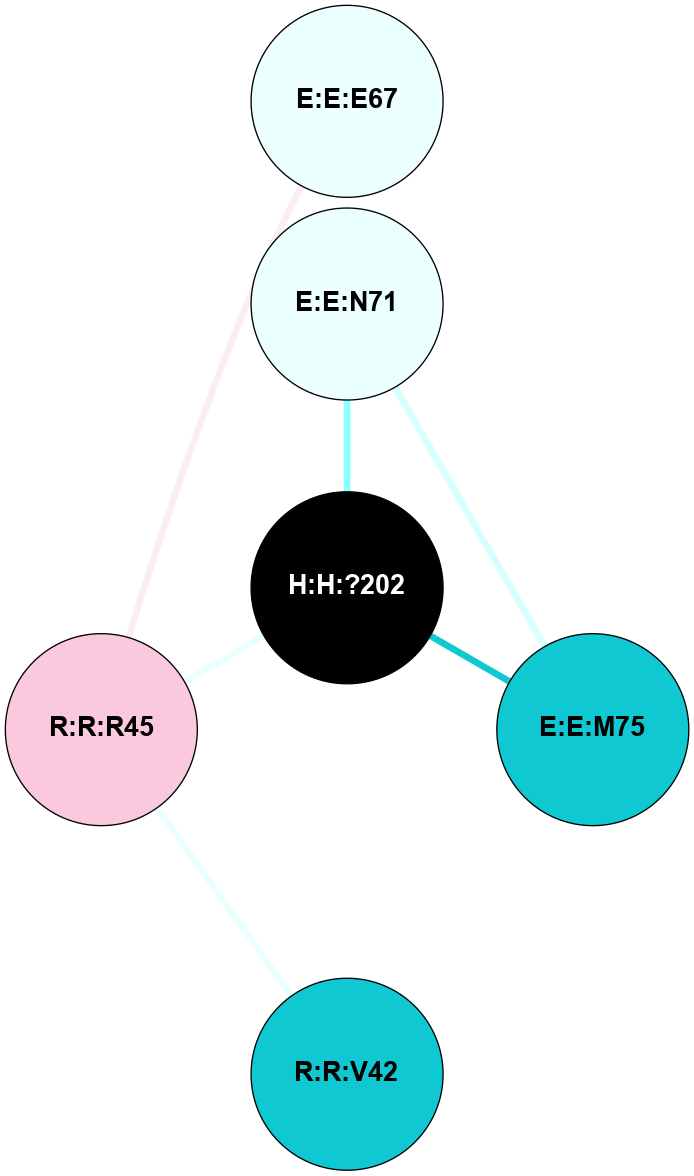
A 2D representation of the interactions of NAG in 8F2A
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | E:E:E67 | R:R:R45 | 12.79 | 0 | No | No | 4 | 7 | 2 | 1 | | E:E:M75 | E:E:N71 | 8.41 | 32 | No | No | 1 | 4 | 1 | 1 | | E:E:N71 | H:H:?202 | 30.8 | 32 | No | No | 4 | 0 | 1 | 0 | | E:E:M75 | H:H:?202 | 15.22 | 32 | No | No | 1 | 0 | 1 | 0 | | R:R:R45 | R:R:V42 | 1.31 | 0 | No | No | 7 | 1 | 1 | 2 | | H:H:?202 | R:R:R45 | 1.09 | 32 | No | No | 0 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.70 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 8F2A
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N73 | 29.57 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N73 | R:R:Y71 | 10.47 | 21 | No | No | 4 | 3 | 1 | 2 | | R:R:D84 | R:R:Y71 | 3.45 | 21 | Yes | No | 4 | 3 | 2 | 2 | | R:R:D84 | R:R:N73 | 4.04 | 21 | Yes | No | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 29.57 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
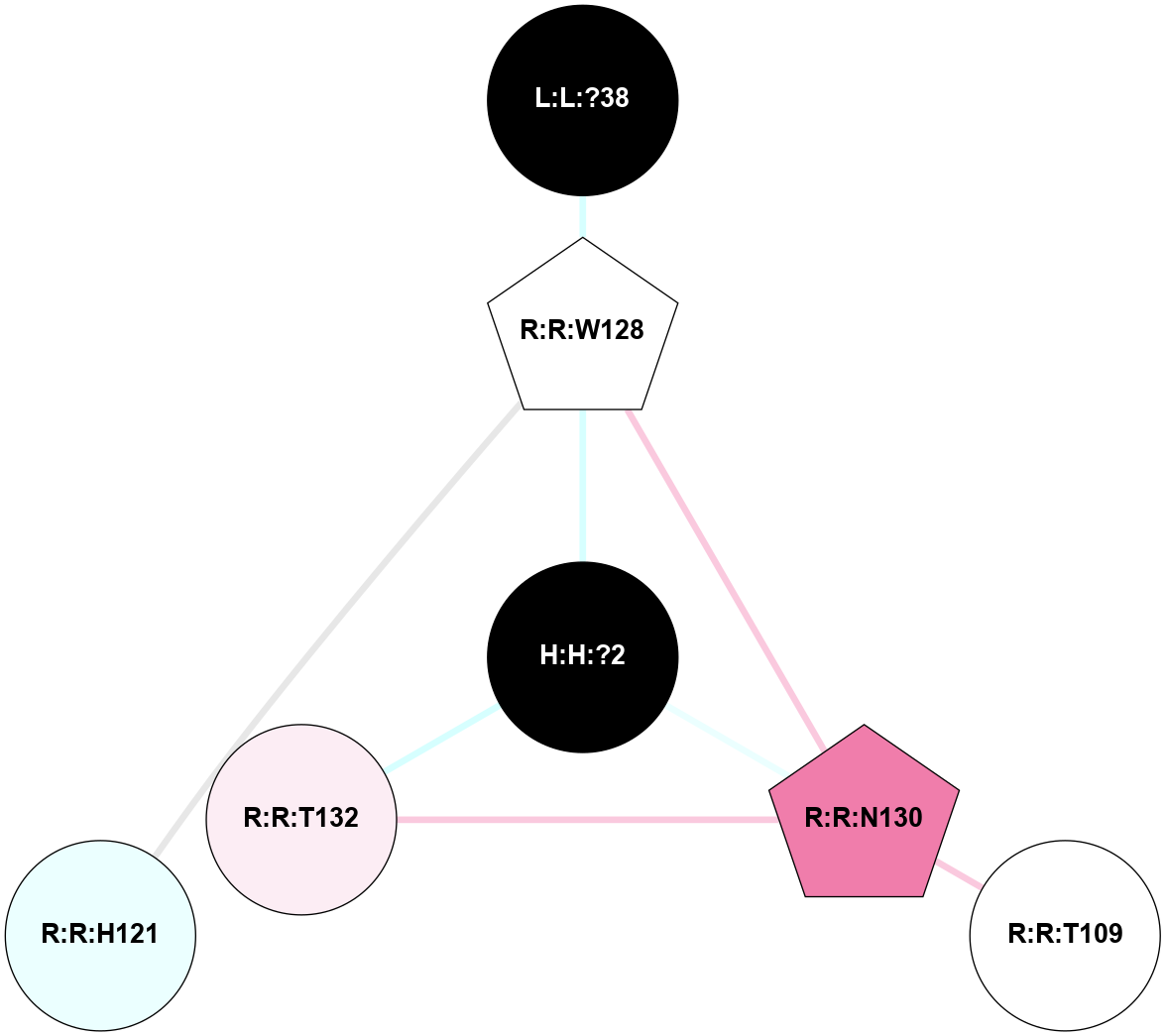
A 2D representation of the interactions of NAG in 8F2A
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:W128 | 7.15 | 12 | No | Yes | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:N130 | 29.57 | 12 | No | Yes | 0 | 8 | 0 | 1 | | H:H:?2 | R:R:T132 | 13.23 | 12 | No | No | 0 | 6 | 0 | 1 | | L:L:?38 | R:R:W128 | 11.28 | 0 | No | Yes | 0 | 5 | 2 | 1 | | R:R:N130 | R:R:T109 | 5.85 | 12 | Yes | No | 8 | 5 | 1 | 2 | | R:R:H121 | R:R:W128 | 25.39 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 11.3 | 12 | Yes | Yes | 8 | 5 | 1 | 1 | | R:R:N130 | R:R:T132 | 8.77 | 12 | Yes | No | 8 | 6 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 16.65 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8F2B | B1 | Peptide | Calcitonin | CT (AMY3) | Homo Sapiens | Pramlintide Analogue San45 | - | Gs/Beta1/Gamma2; RAMP3 | 2 | 2023-08-02 | doi.org/10.1038/s41589-023-01393-4 |
|
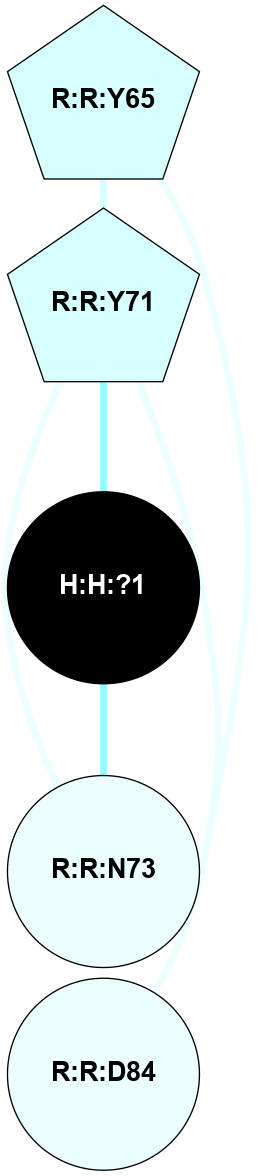
A 2D representation of the interactions of NAG in 8F2B
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:Y71 | 12.62 | 20 | No | Yes | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:N73 | 34.5 | 20 | No | No | 0 | 4 | 0 | 1 | | R:R:Y65 | R:R:Y71 | 19.86 | 20 | Yes | Yes | 3 | 3 | 2 | 1 | | R:R:D84 | R:R:Y65 | 5.75 | 20 | No | Yes | 4 | 3 | 2 | 2 | | R:R:N73 | R:R:Y71 | 3.49 | 20 | No | Yes | 4 | 3 | 1 | 1 | | R:R:D84 | R:R:Y71 | 5.75 | 20 | No | Yes | 4 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 23.56 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
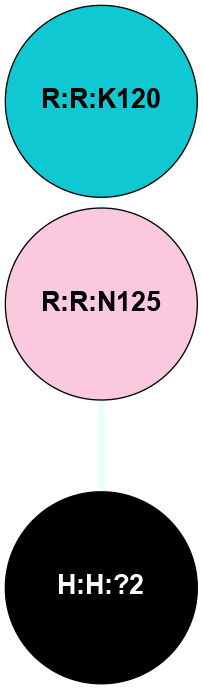
A 2D representation of the interactions of NAG in 8F2B
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:N125 | 27.11 | 0 | No | No | 0 | 7 | 0 | 1 | | R:R:K120 | R:R:N125 | 5.6 | 0 | No | No | 1 | 7 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.11 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
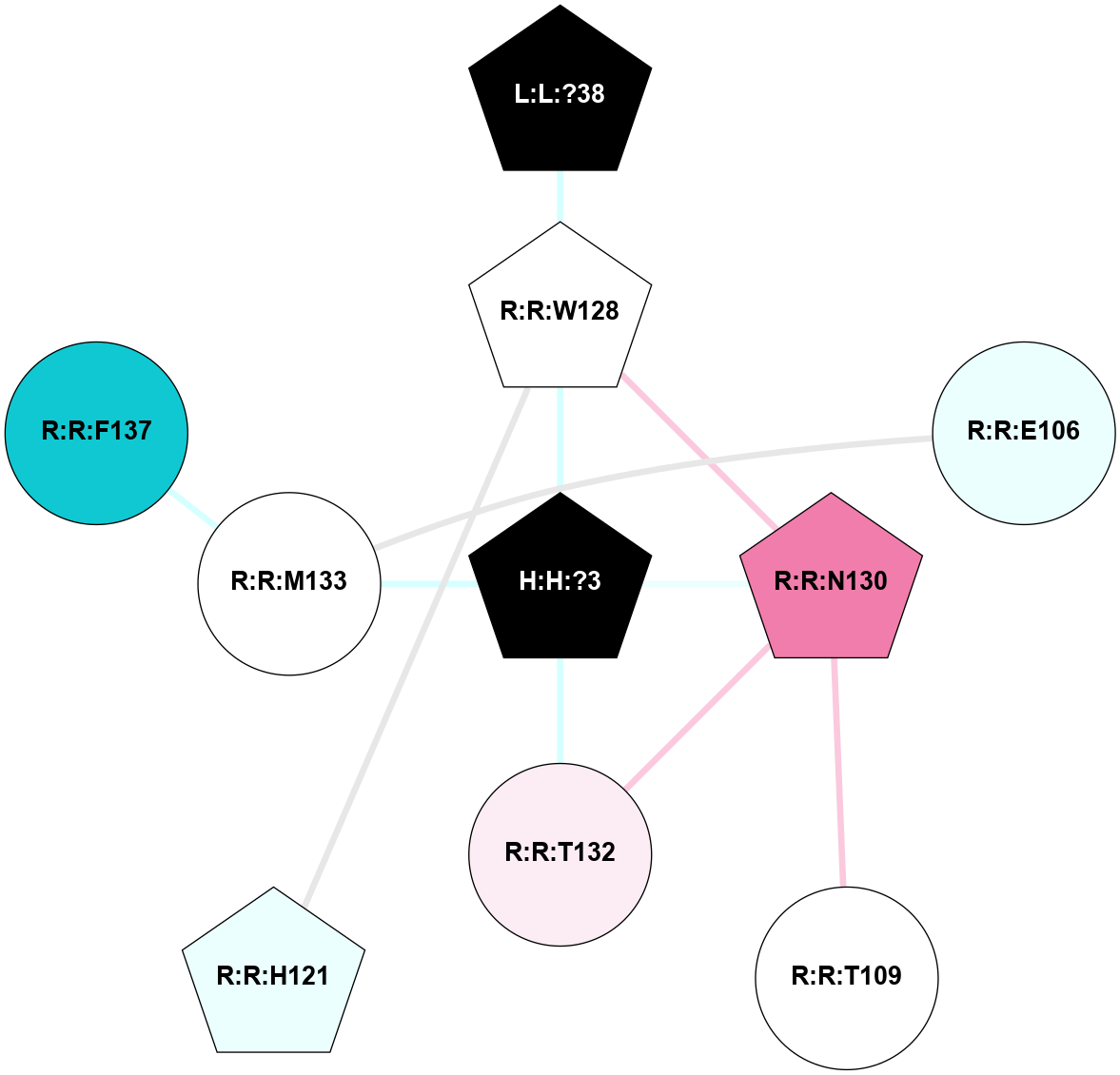
A 2D representation of the interactions of NAG in 8F2B
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:W128 | 5.11 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?3 | R:R:N130 | 27.11 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?3 | R:R:T132 | 6.61 | 1 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?3 | R:R:M133 | 5.07 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?38 | R:R:W128 | 5.64 | 1 | Yes | Yes | 0 | 5 | 2 | 1 | | R:R:E106 | R:R:M133 | 4.06 | 16 | No | No | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:T109 | 5.85 | 1 | Yes | No | 8 | 5 | 1 | 2 | | R:R:H121 | R:R:W128 | 23.27 | 1 | Yes | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 11.3 | 1 | Yes | Yes | 8 | 5 | 1 | 1 | | R:R:N130 | R:R:T132 | 7.31 | 1 | Yes | No | 8 | 6 | 1 | 1 | | R:R:F137 | R:R:M133 | 2.49 | 0 | No | No | 1 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 10.97 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8FCZ | A | Sensory | Opsins | Rhodopsin | Bos Taurus | 11-cis-Retinal | - | - | 3.7 | 2023-08-30 | doi.org/10.1038/s41467-023-40911-9 |
|
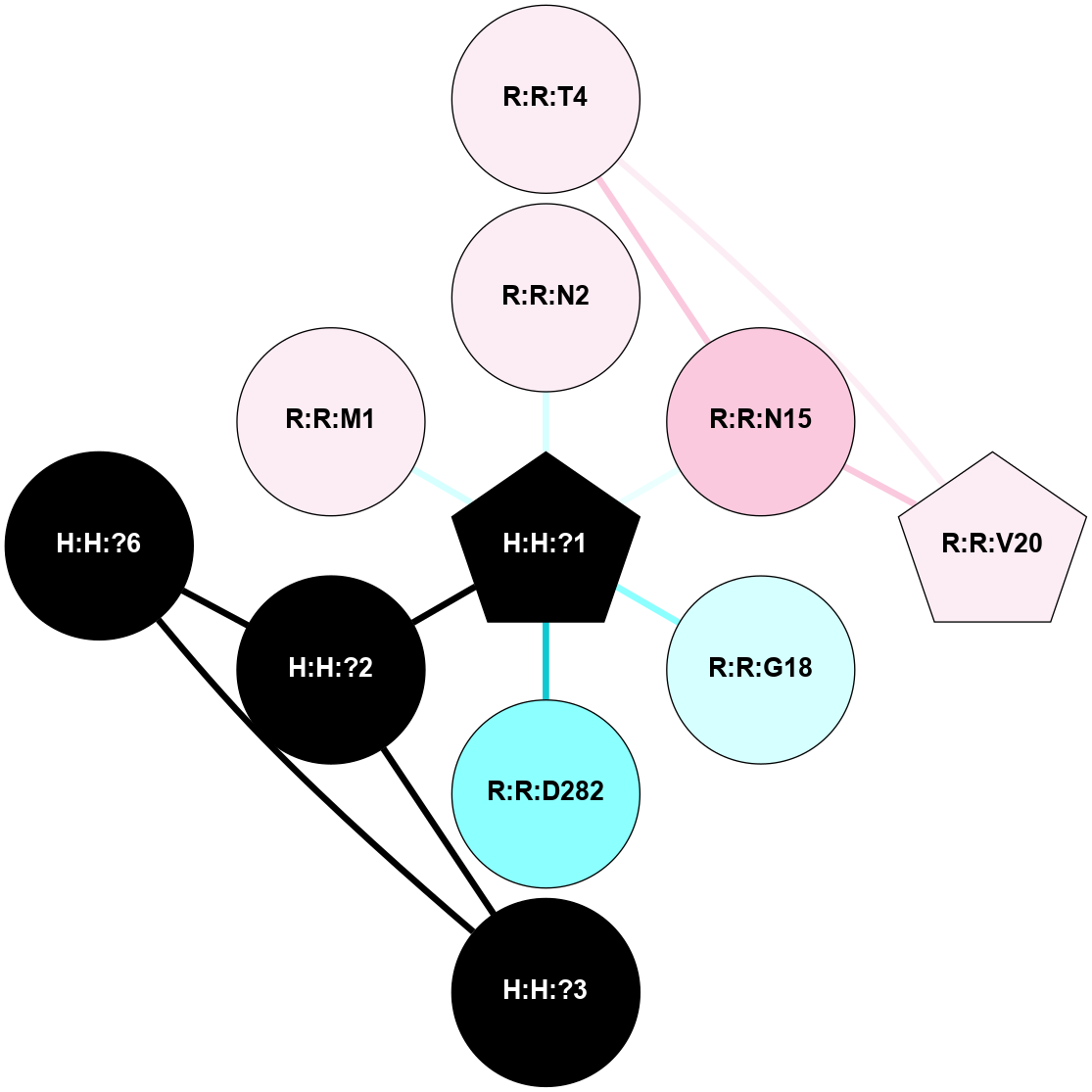
A 2D representation of the interactions of NAG in 8FCZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 36.78 | 0 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N2 | 25.87 | 0 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:D282 | 7.31 | 0 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?2 | H:H:?3 | 32.47 | 0 | No | No | 0 | 0 | 1 | 2 | | H:H:?1 | R:R:M1 | 2.54 | 0 | Yes | No | 0 | 6 | 0 | 1 | | R:R:G3 | R:R:N2 | 1.7 | 0 | No | No | 6 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.13 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
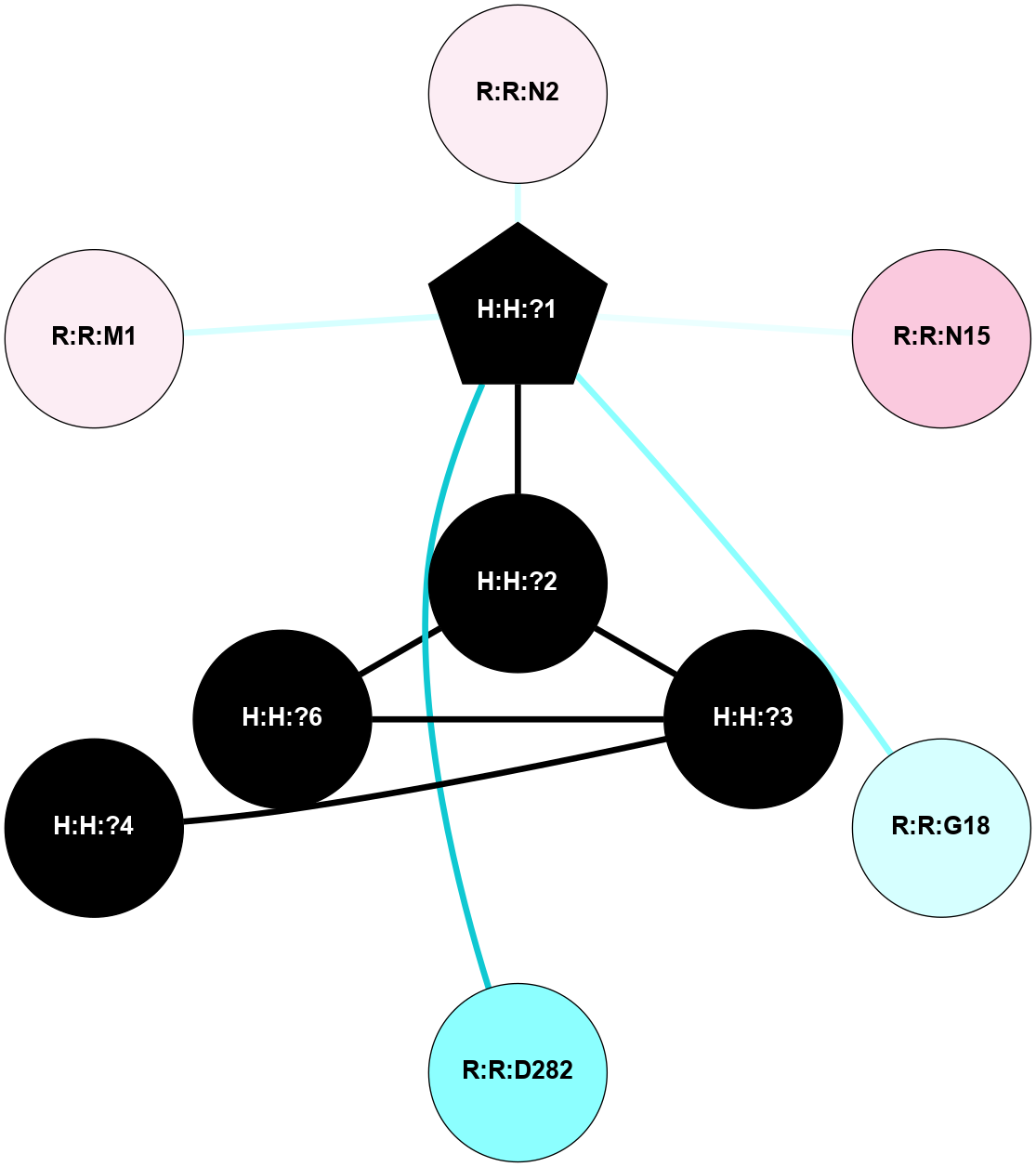
A 2D representation of the interactions of NAG in 8FCZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 36.78 | 0 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N2 | 25.87 | 0 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:D282 | 7.31 | 0 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?2 | H:H:?3 | 32.47 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:M1 | 2.54 | 0 | Yes | No | 0 | 6 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 34.62 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 8FCZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | H:H:?5 | 35.88 | 0 | No | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 35.88 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8FD0 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | - | 3.71 | 2023-08-30 | doi.org/10.1038/s41467-023-40911-9 |
|
A 2D representation of the interactions of NAG in 8FD0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 33.43 | 0 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N2 | 30.8 | 0 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:S281 | 4.04 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:D282 | 6.09 | 0 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?2 | H:H:?3 | 32.47 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:F283 | R:R:S281 | 5.28 | 1 | Yes | No | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.59 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 8FD0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 33.43 | 0 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N2 | 30.8 | 0 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:S281 | 4.04 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:D282 | 6.09 | 0 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?2 | H:H:?3 | 32.47 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 32.95 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 8FD0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | H:H:?5 | 32.04 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?5 | H:H:?6 | 1.28 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.66 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8FD1 | A | Sensory | Opsins | Rhodopsin | Bos Taurus | - | - | - | 4.25 | 2023-08-30 | doi.org/10.1038/s41467-023-40911-9 |
|
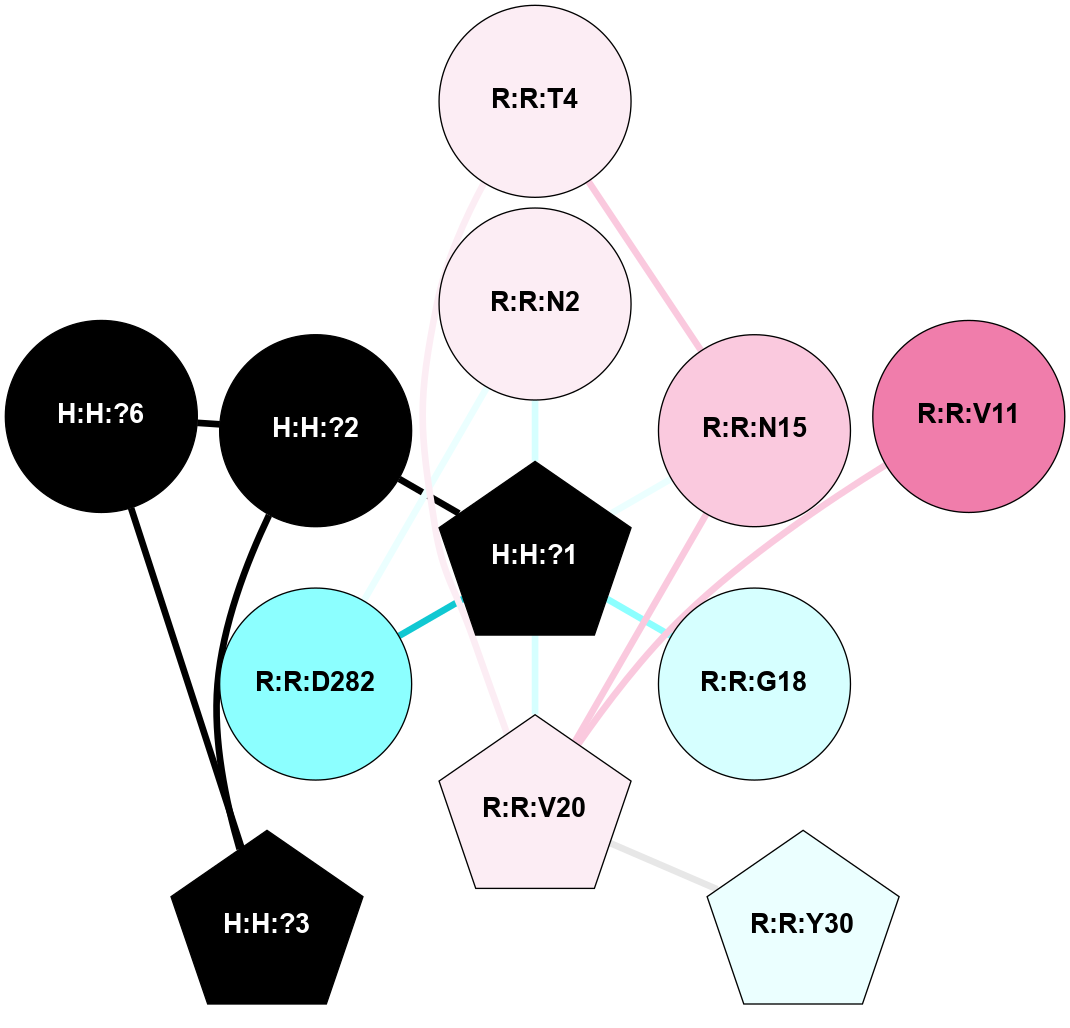
A 2D representation of the interactions of NAG in 8FD1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 36.78 | 6 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N2 | 28.34 | 6 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:D282 | 6.09 | 6 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?2 | H:H:?3 | 37.66 | 0 | No | No | 0 | 0 | 1 | 2 | | R:R:D282 | R:R:N2 | 10.77 | 6 | No | No | 2 | 6 | 1 | 1 | | R:R:F283 | R:R:S281 | 5.28 | 1 | Yes | No | 4 | 4 | 2 | 1 | | H:H:?1 | R:R:S281 | 4.04 | 6 | Yes | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.81 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
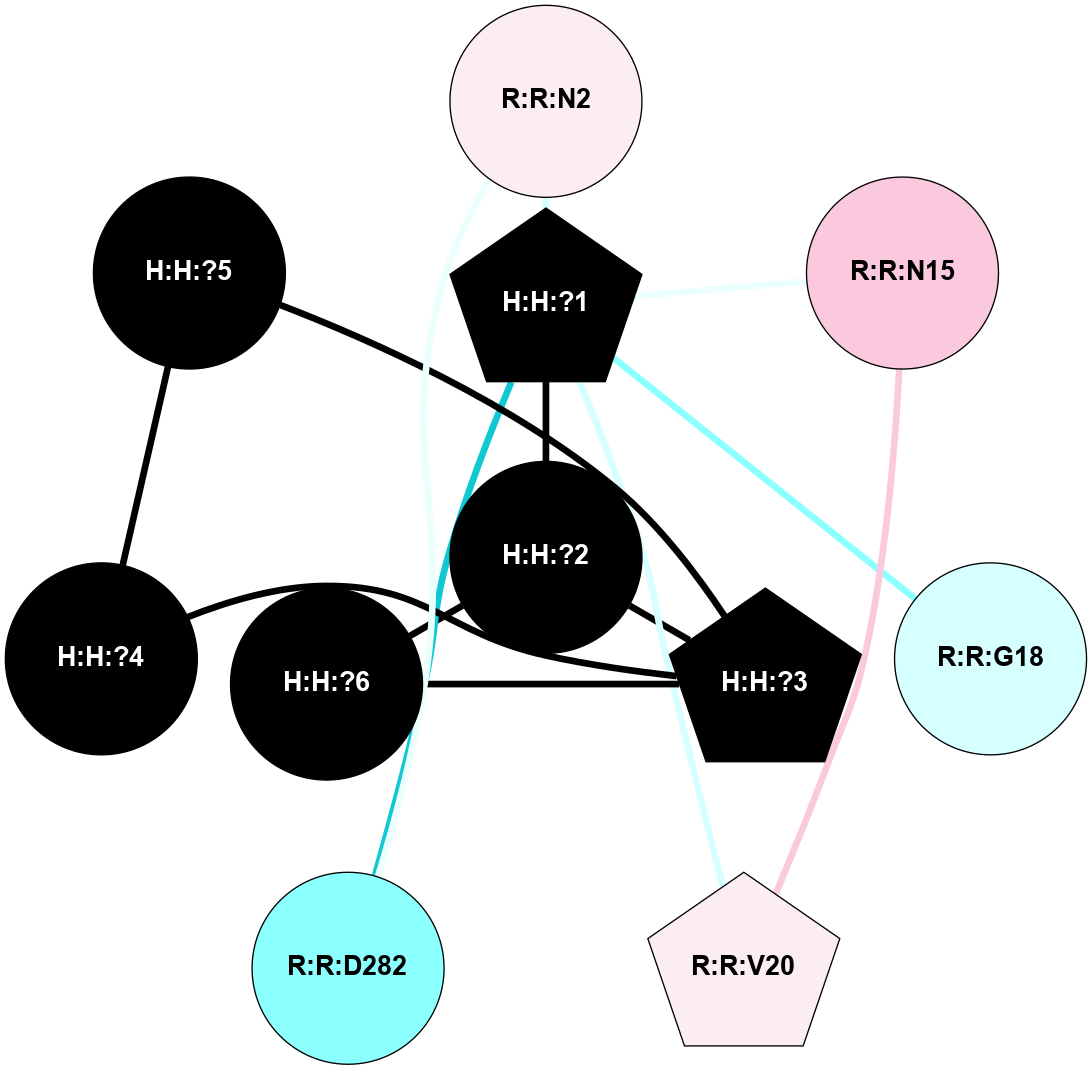
A 2D representation of the interactions of NAG in 8FD1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 36.78 | 6 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N2 | 28.34 | 6 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:D282 | 6.09 | 6 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?2 | H:H:?3 | 37.66 | 0 | No | No | 0 | 0 | 0 | 1 | | R:R:D282 | R:R:N2 | 10.77 | 6 | No | No | 2 | 6 | 2 | 2 | | H:H:?1 | R:R:S281 | 4.04 | 6 | Yes | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 37.22 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
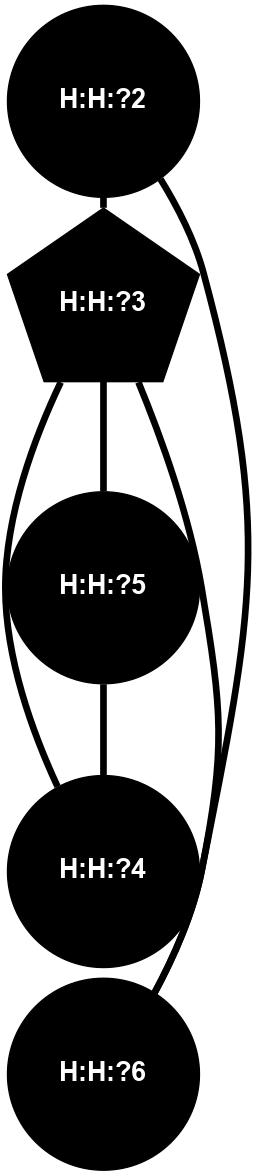
A 2D representation of the interactions of NAG in 8FD1
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | H:H:?5 | 29.48 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?5 | H:H:?6 | 3.84 | 0 | No | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.66 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8I2G | A | Protein | Glycoprotein Hormone | FSH | Homo Sapiens | Glycoprotein hormones alpha chain | Compound 716340 | Gs/Beta1/Gamma2 | 2.8 | 2023-03-29 | doi.org/10.1038/s41467-023-36170-3 |
|
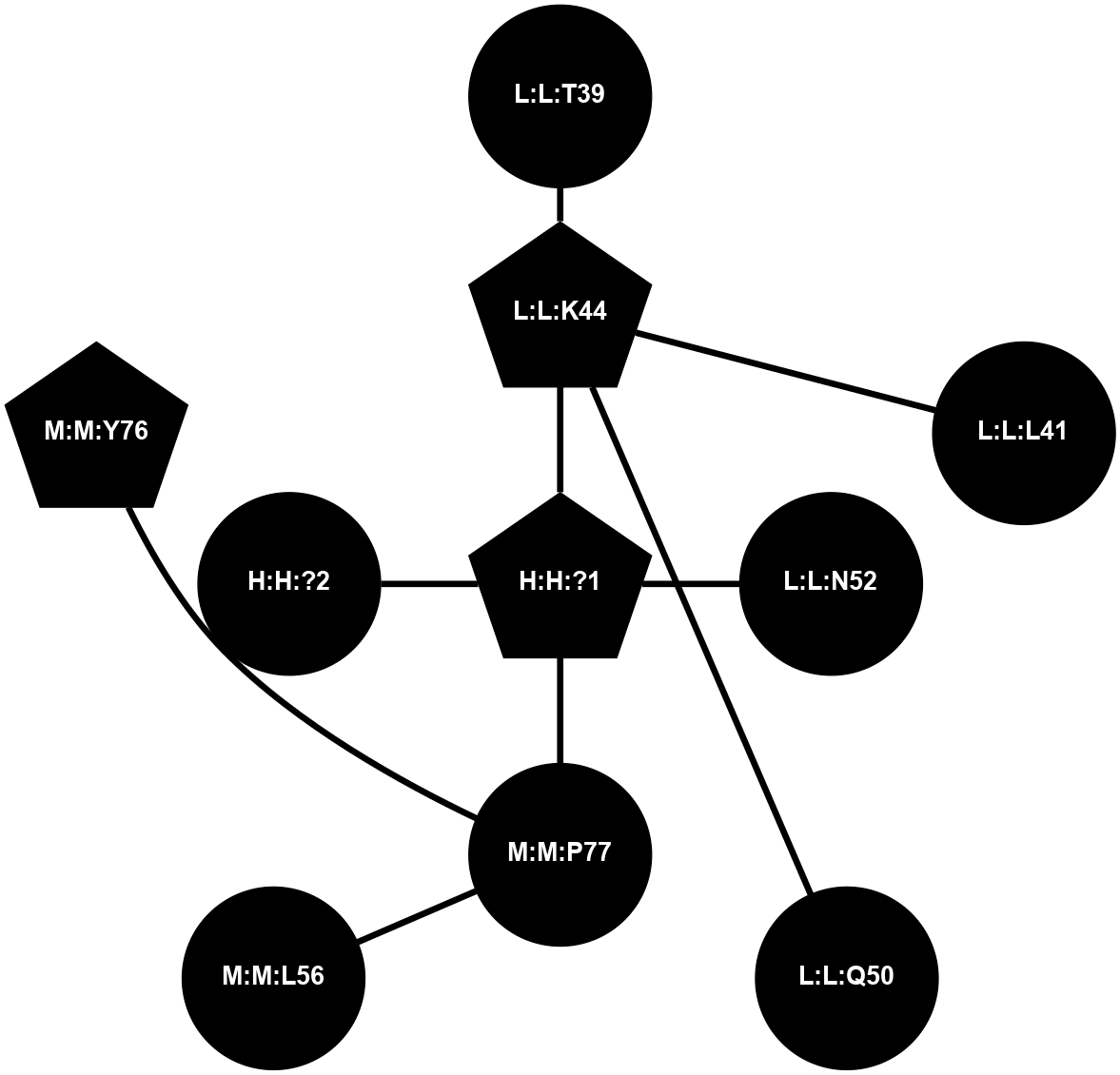
A 2D representation of the interactions of NAG in 8I2G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 33.43 | 0 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | L:L:K44 | 6.33 | 0 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?1 | L:L:N52 | 30.8 | 0 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | N:N:P77 | 2.95 | 0 | Yes | No | 0 | 0 | 0 | 1 | | L:L:K44 | L:L:T39 | 7.51 | 0 | Yes | No | 0 | 0 | 1 | 2 | | L:L:K44 | L:L:L41 | 2.82 | 0 | Yes | No | 0 | 0 | 1 | 2 | | L:L:K44 | L:L:Q50 | 8.14 | 0 | Yes | No | 0 | 0 | 1 | 2 | | N:N:L56 | N:N:P77 | 4.93 | 0 | No | No | 0 | 0 | 2 | 1 | | N:N:P77 | N:N:Y76 | 4.17 | 0 | No | Yes | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 18.38 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
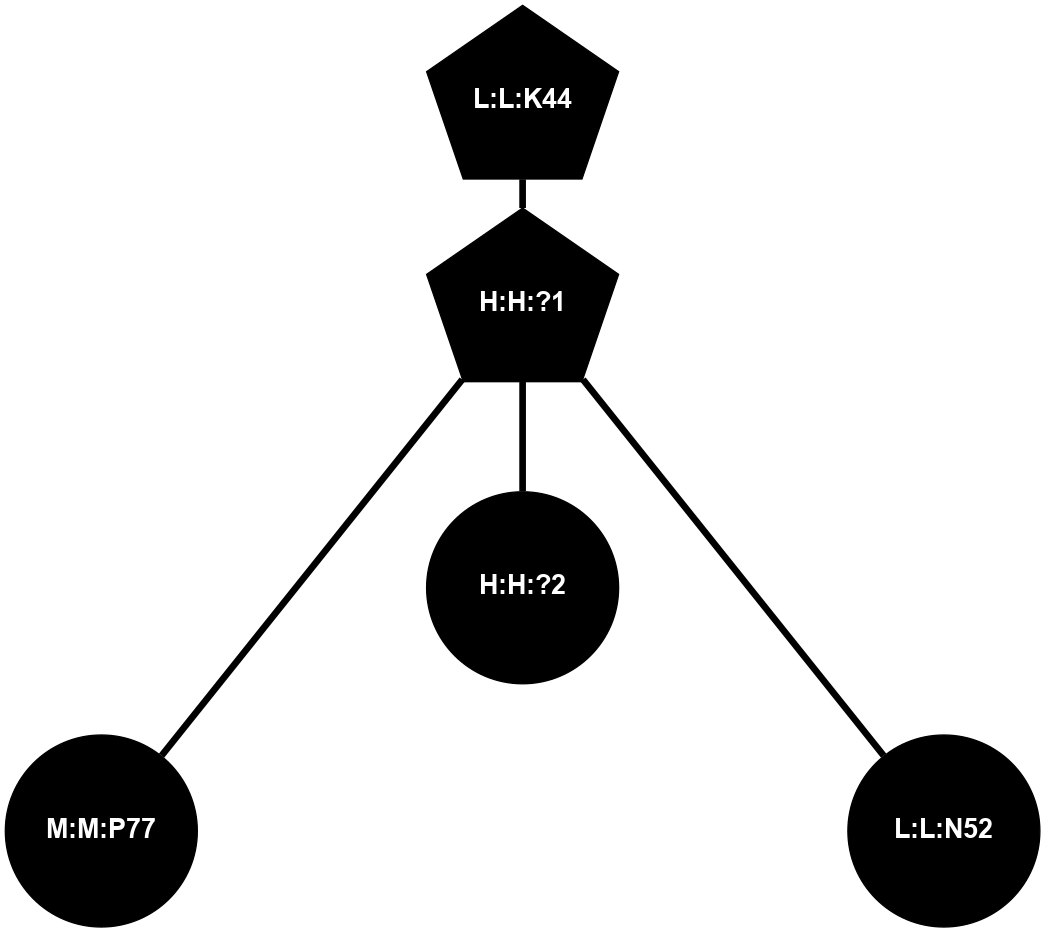
A 2D representation of the interactions of NAG in 8I2G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 33.43 | 0 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | L:L:K44 | 6.33 | 0 | Yes | Yes | 0 | 0 | 1 | 2 | | H:H:?1 | L:L:N52 | 30.8 | 0 | Yes | No | 0 | 0 | 1 | 2 | | H:H:?1 | N:N:P77 | 2.95 | 0 | Yes | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 33.43 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
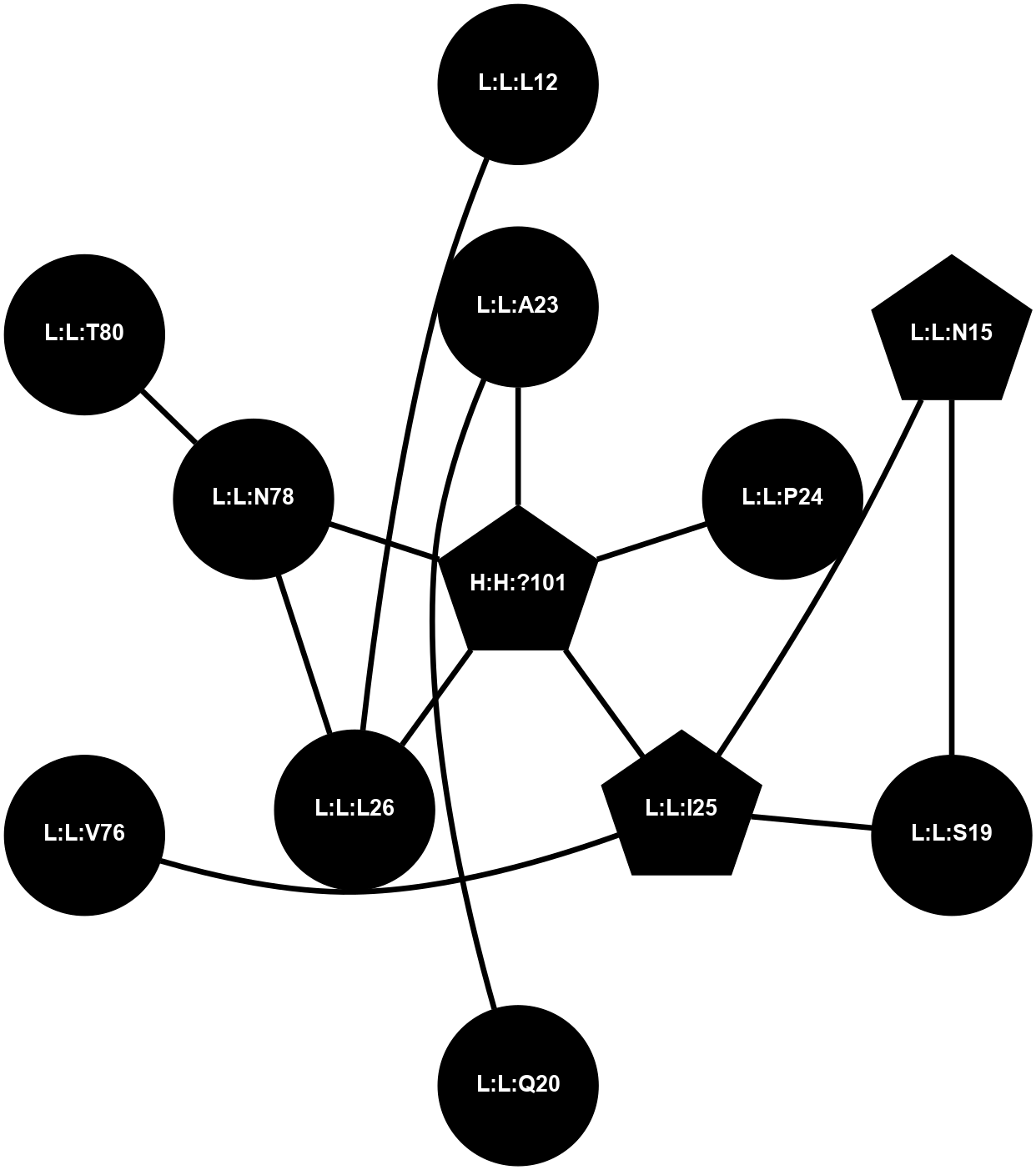
A 2D representation of the interactions of NAG in 8I2G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?101 | L:L:A23 | 2.83 | 10 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?101 | L:L:P24 | 10.31 | 10 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?101 | L:L:I25 | 3.84 | 10 | Yes | Yes | 0 | 0 | 0 | 1 | | H:H:?101 | L:L:L26 | 2.48 | 10 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?101 | L:L:N78 | 25.87 | 10 | Yes | No | 0 | 0 | 0 | 1 | | L:L:L12 | L:L:L26 | 12.46 | 0 | No | No | 0 | 0 | 2 | 1 | | L:L:N15 | L:L:S19 | 11.92 | 10 | Yes | No | 0 | 0 | 2 | 2 | | L:L:I25 | L:L:N15 | 2.83 | 10 | Yes | Yes | 0 | 0 | 1 | 2 | | L:L:I25 | L:L:S19 | 6.19 | 10 | Yes | No | 0 | 0 | 1 | 2 | | L:L:A23 | L:L:Q20 | 3.03 | 0 | No | No | 0 | 0 | 1 | 2 | | L:L:I25 | L:L:V76 | 4.61 | 10 | Yes | No | 0 | 0 | 1 | 2 | | L:L:L26 | L:L:N78 | 2.75 | 10 | No | No | 0 | 0 | 1 | 1 | | L:L:N78 | L:L:T80 | 4.39 | 10 | No | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 9.07 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
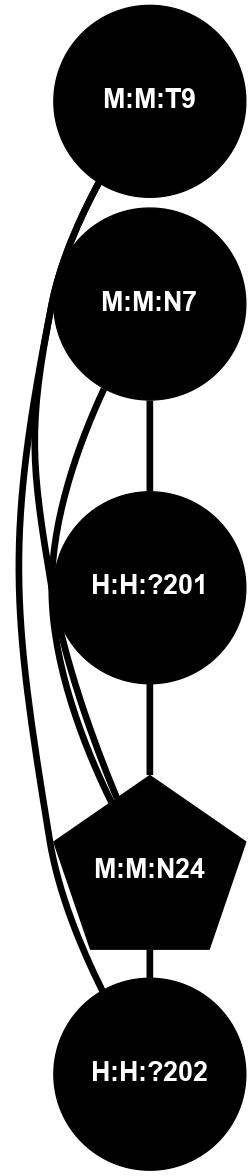
A 2D representation of the interactions of NAG in 8I2G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?201 | N:N:N7 | 30.8 | 19 | No | No | 0 | 0 | 0 | 1 | | H:H:?201 | N:N:N24 | 3.7 | 19 | No | Yes | 0 | 0 | 0 | 1 | | H:H:?202 | N:N:T9 | 13.23 | 19 | No | No | 0 | 0 | 2 | 2 | | H:H:?202 | N:N:N24 | 13.55 | 19 | No | Yes | 0 | 0 | 2 | 1 | | N:N:N24 | N:N:N7 | 2.72 | 19 | Yes | No | 0 | 0 | 1 | 1 | | N:N:N24 | N:N:T9 | 2.92 | 19 | Yes | No | 0 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.25 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
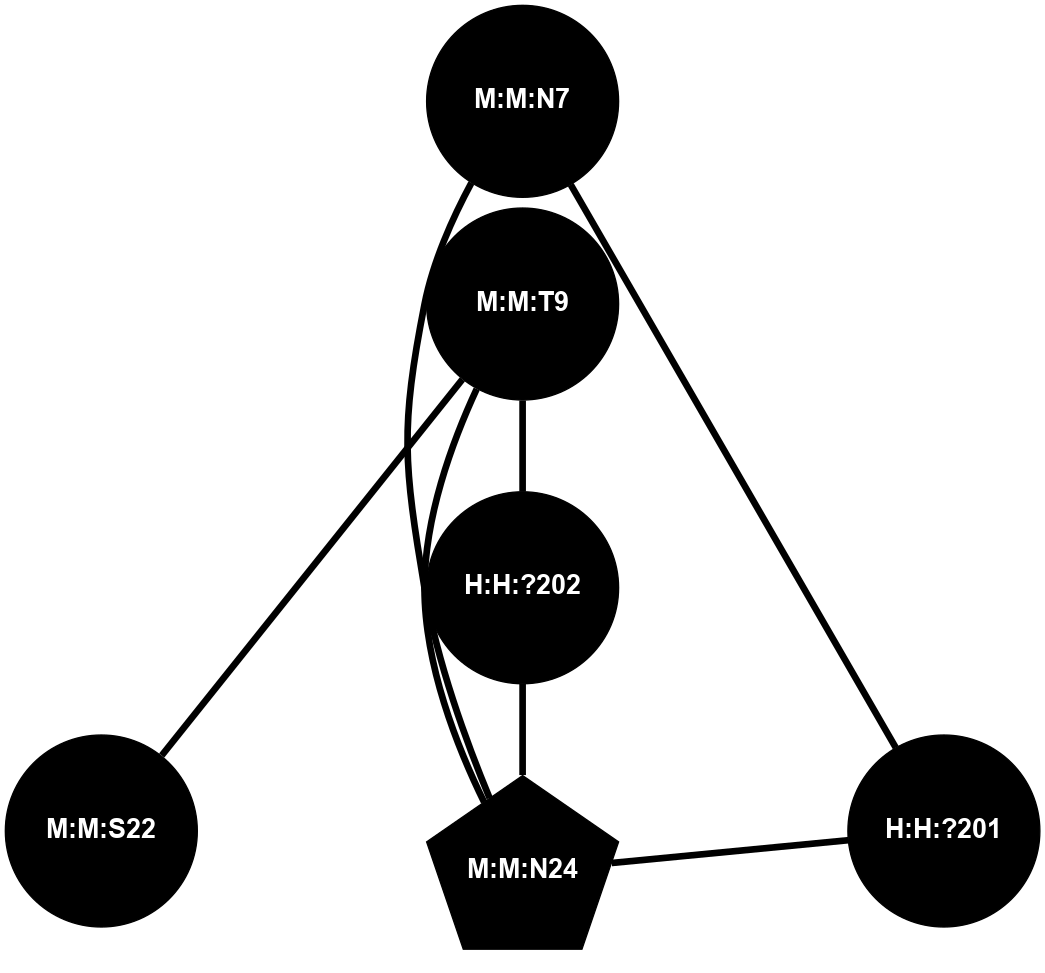
A 2D representation of the interactions of NAG in 8I2G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?201 | N:N:N7 | 30.8 | 19 | No | No | 0 | 0 | 2 | 2 | | H:H:?201 | N:N:N24 | 3.7 | 19 | No | Yes | 0 | 0 | 2 | 1 | | H:H:?202 | N:N:T9 | 13.23 | 19 | No | No | 0 | 0 | 0 | 1 | | H:H:?202 | N:N:N24 | 13.55 | 19 | No | Yes | 0 | 0 | 0 | 1 | | N:N:N24 | N:N:N7 | 2.72 | 19 | Yes | No | 0 | 0 | 1 | 2 | | N:N:N24 | N:N:T9 | 2.92 | 19 | Yes | No | 0 | 0 | 1 | 1 | | N:N:S22 | N:N:T9 | 1.6 | 0 | No | No | 0 | 0 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 13.39 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
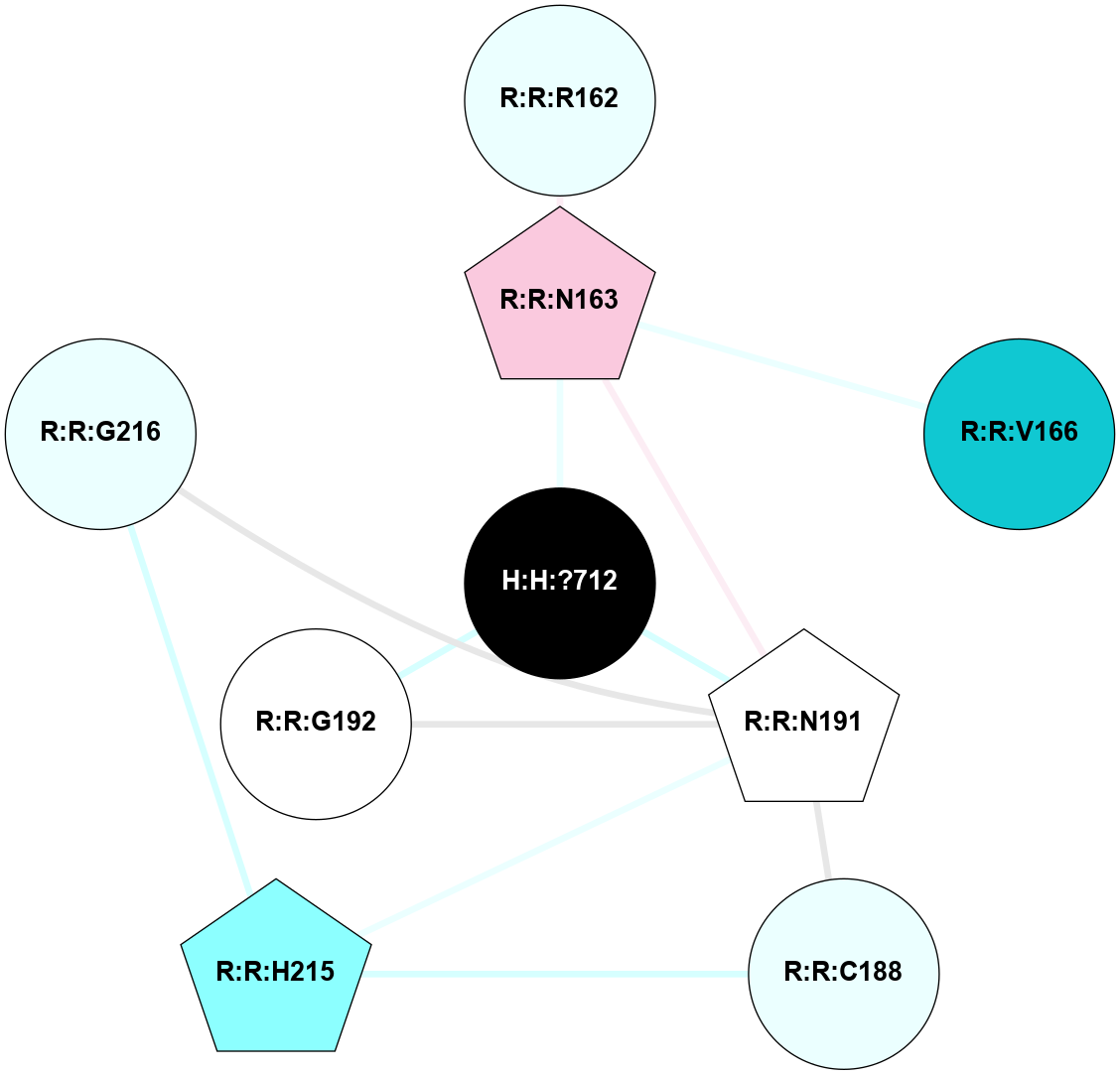
A 2D representation of the interactions of NAG in 8I2G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?712 | R:R:N163 | 12.32 | 14 | No | Yes | 0 | 7 | 0 | 1 | | H:H:?712 | R:R:N191 | 27.11 | 14 | No | Yes | 0 | 5 | 0 | 1 | | H:H:?712 | R:R:G192 | 3.07 | 14 | No | No | 0 | 5 | 0 | 1 | | R:R:N163 | R:R:R162 | 6.03 | 14 | Yes | No | 7 | 4 | 1 | 2 | | R:R:N163 | R:R:V166 | 5.91 | 14 | Yes | No | 7 | 1 | 1 | 2 | | R:R:N163 | R:R:N191 | 4.09 | 14 | Yes | Yes | 7 | 5 | 1 | 1 | | R:R:C188 | R:R:N191 | 4.72 | 14 | No | Yes | 4 | 5 | 2 | 1 | | R:R:C188 | R:R:H215 | 4.42 | 14 | No | Yes | 4 | 2 | 2 | 2 | | R:R:G192 | R:R:N191 | 5.09 | 14 | No | Yes | 5 | 5 | 1 | 1 | | R:R:H215 | R:R:N191 | 2.55 | 14 | Yes | Yes | 2 | 5 | 2 | 1 | | R:R:G216 | R:R:N191 | 3.39 | 14 | No | Yes | 4 | 5 | 2 | 1 | | R:R:G216 | R:R:H215 | 3.18 | 14 | No | Yes | 4 | 2 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 14.17 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8IHB | A | Alicarboxylic acid | Hydroxycarboxylic Acid | HCA2 | Homo Sapiens | GSK256073 | - | Gi1/Beta1/Gamma2 | 2.85 | 2023-09-13 | doi.org/10.1038/s41467-023-41650-7 |
|
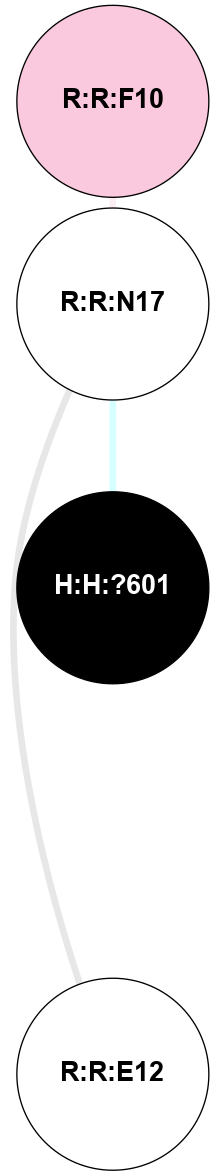
A 2D representation of the interactions of NAG in 8IHB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?601 | R:R:N17 | 30.8 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?601 | R:R:Q264 | 2.39 | 0 | No | No | 0 | 3 | 0 | 1 | | R:R:F10 | R:R:N17 | 7.25 | 0 | No | No | 7 | 5 | 2 | 1 | | R:R:E12 | R:R:N17 | 1.31 | 0 | No | No | 5 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.59 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8IHF | A | Alicarboxylic acid | Hydroxycarboxylic Acid | HCA2 | Homo Sapiens | MK6892 | - | Gi1/Beta1/Gamma2 | 2.97 | 2023-08-30 | doi.org/10.1038/s41467-023-41650-7 |
|
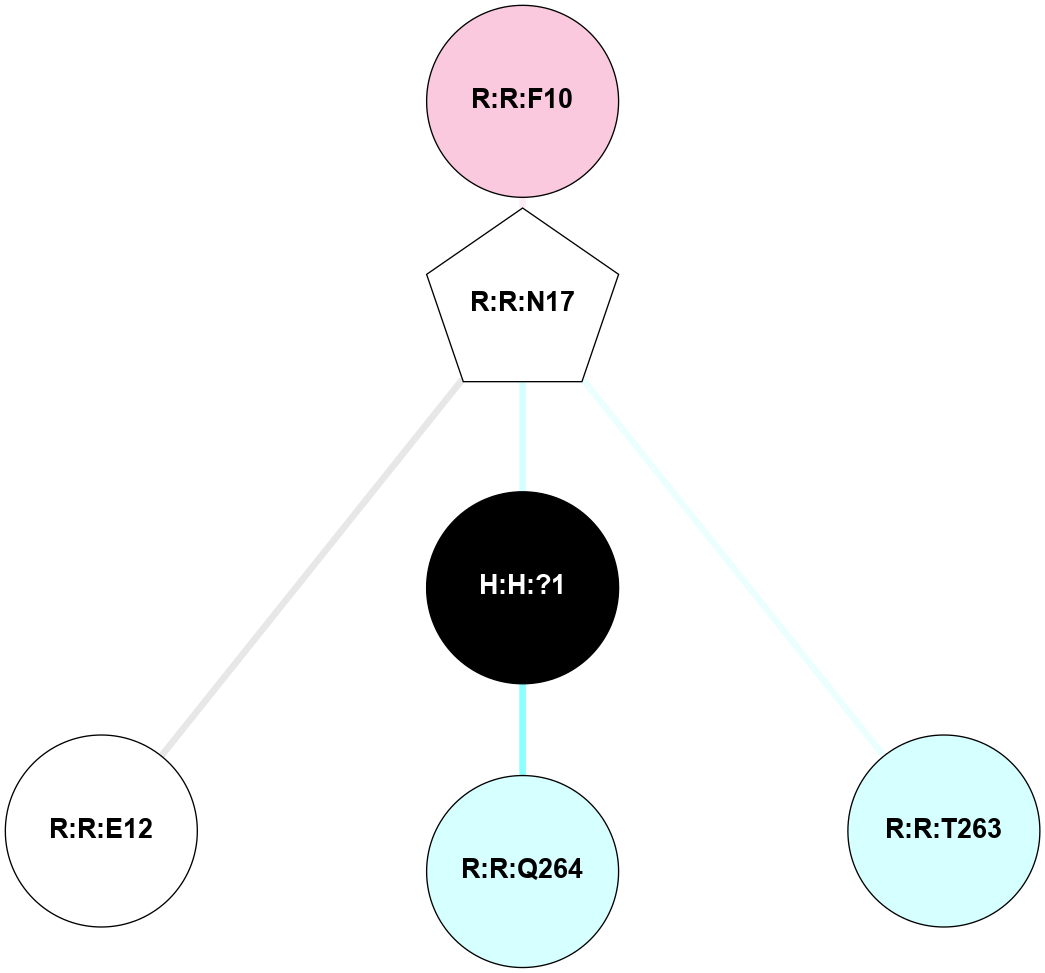
A 2D representation of the interactions of NAG in 8IHF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N17 | 25.87 | 0 | No | Yes | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:Q264 | 4.78 | 0 | No | No | 0 | 3 | 0 | 1 | | R:R:F10 | R:R:N17 | 8.46 | 0 | No | Yes | 7 | 5 | 2 | 1 | | R:R:N17 | R:R:T263 | 1.46 | 0 | Yes | No | 5 | 3 | 1 | 2 | | R:R:E12 | R:R:N17 | 1.31 | 0 | No | Yes | 5 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.33 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8IHH | A | Alicarboxylic acid | Hydroxycarboxylic Acid | HCA2 | Homo Sapiens | LUF6283 | - | Gi1/Beta1/Gamma2 | 3.06 | 2023-08-30 | doi.org/10.1038/s41467-023-41650-7 |
|

A 2D representation of the interactions of NAG in 8IHH
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F10 | R:R:N17 | 10.87 | 0 | No | No | 7 | 5 | 2 | 1 | | R:R:N17 | R:R:T263 | 7.31 | 0 | No | No | 5 | 3 | 1 | 2 | | H:H:?1 | R:R:N17 | 34.5 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:Q264 | 7.17 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.84 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8IHI | A | Alicarboxylic acid | Hydroxycarboxylic Acid | HCA2 | Homo Sapiens | Acifran | - | Gi1/Beta1/Gamma2 | 3.11 | 2023-08-30 | doi.org/10.1038/s41467-023-41650-7 |
|
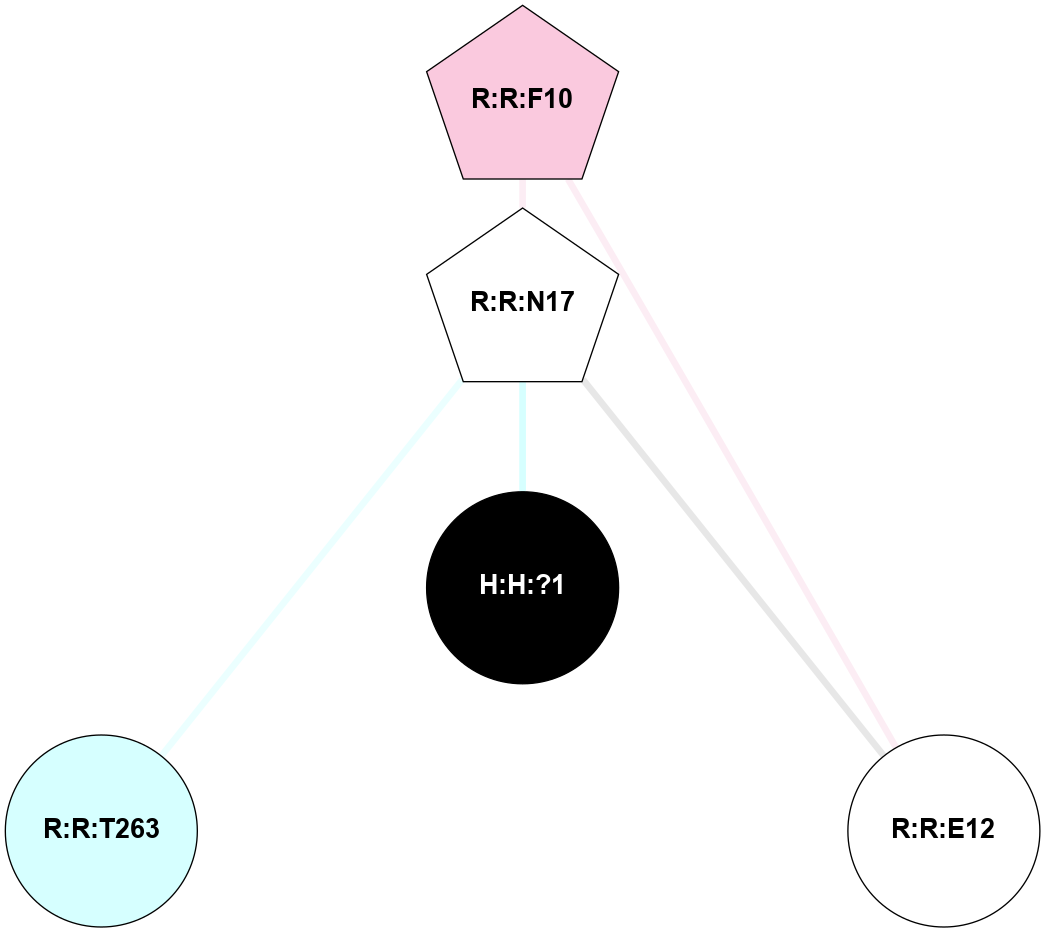
A 2D representation of the interactions of NAG in 8IHI
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E12 | R:R:F10 | 4.66 | 28 | No | Yes | 5 | 7 | 2 | 2 | | R:R:F10 | R:R:N17 | 9.67 | 28 | Yes | Yes | 7 | 5 | 2 | 1 | | R:R:E12 | R:R:N17 | 3.94 | 28 | No | Yes | 5 | 5 | 2 | 1 | | R:R:N17 | R:R:T263 | 4.39 | 28 | Yes | No | 5 | 3 | 1 | 2 | | H:H:?1 | R:R:N17 | 29.57 | 0 | No | Yes | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 29.57 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8IKJ | B2 | Adhesion | Adhesion | ADGRE5 | Homo Sapiens | - | - | - | 3.2 | 2024-02-14 | doi.org/10.1016/j.molcel.2023.12.020 |
|
A 2D representation of the interactions of NAG in 8IKJ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N413 | 36.96 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 36.96 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 8IKJ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:M309 | R:R:T409 | 1.51 | 1 | Yes | Yes | 8 | 4 | 2 | 1 | | R:R:A311 | R:R:P312 | 1.87 | 0 | No | Yes | 4 | 6 | 2 | 1 | | R:R:E316 | R:R:P312 | 6.29 | 1 | No | Yes | 4 | 6 | 2 | 1 | | R:R:P312 | R:R:T409 | 3.5 | 1 | Yes | Yes | 6 | 4 | 1 | 1 | | H:H:?2 | R:R:P312 | 2.95 | 1 | Yes | Yes | 0 | 6 | 0 | 1 | | R:R:E316 | R:R:T409 | 2.82 | 1 | No | Yes | 4 | 4 | 2 | 1 | | R:R:N406 | R:R:T408 | 5.85 | 1 | No | No | 5 | 4 | 1 | 2 | | R:R:N406 | R:R:T409 | 5.85 | 1 | No | Yes | 5 | 4 | 1 | 1 | | H:H:?2 | R:R:N406 | 30.8 | 1 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:T409 | 3.97 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:L469 | R:R:S471 | 1.5 | 0 | No | No | 3 | 4 | 2 | 1 | | H:H:?2 | R:R:S471 | 2.69 | 1 | Yes | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 10.10 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 8IKJ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:P480 | R:R:P481 | 1.95 | 0 | No | No | 7 | 4 | 2 | 1 | | H:H:?3 | R:R:P481 | 1.47 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?3 | R:R:K519 | 2.53 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?3 | R:R:N520 | 29.57 | 0 | No | No | 0 | 6 | 0 | 1 | | R:R:D484 | R:R:K519 | 1.38 | 0 | No | No | 2 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 11.19 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8J6P | A | Alicarboxylic acid | Hydroxycarboxylic Acid | HCA2 | Homo Sapiens | Niacin | Compound 9n | Gi1/Beta1/Gamma2 | 2.55 | 2023-12-06 | doi.org/10.1038/s41467-023-43537-z |
|

A 2D representation of the interactions of NAG in 8J6P
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F10 | R:R:N17 | 8.46 | 0 | No | No | 7 | 5 | 2 | 1 | | H:H:?405 | R:R:N17 | 30.7 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:N17 | R:R:T263 | 1.46 | 0 | No | No | 5 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 30.70 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8J6Q | A | Alicarboxylic acid | Hydroxycarboxylic Acid | HCA2 | Homo Sapiens | Beta-Hydroxybutyrate | Compound 9n | Gi1/Beta1/Gamma2 | 2.6 | 2023-12-06 | doi.org/10.1038/s41467-023-43537-z |
|

A 2D representation of the interactions of NAG in 8J6Q
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F10 | R:R:N17 | 8.46 | 0 | No | No | 7 | 5 | 2 | 1 | | H:H:?405 | R:R:N17 | 30.7 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:N17 | R:R:T263 | 1.46 | 0 | No | No | 5 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 30.70 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8J6R | A | Alicarboxylic acid | Hydroxycarboxylic Acid | HCA2 | Homo Sapiens | MK6892 | - | Gi1/Beta1/Gamma2 | 2.76 | 2023-12-06 | doi.org/10.1038/s41467-023-43537-z |
|

A 2D representation of the interactions of NAG in 8J6R
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F10 | R:R:N17 | 7.25 | 0 | No | No | 7 | 5 | 2 | 1 | | H:H:?402 | R:R:K15 | 2.53 | 0 | No | No | 0 | 3 | 0 | 1 | | R:R:N17 | R:R:T263 | 4.39 | 0 | No | No | 5 | 3 | 1 | 2 | | H:H:?402 | R:R:N17 | 24.64 | 0 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.59 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8JCU | C | Aminoacid | Metabotropic Glutamate | mGlu2; mGlu3 | Homo Sapiens | LY341495 | - | - | 2.8 | 2023-06-21 | doi.org/10.1038/s41422-023-00830-2 |
|
A 2D representation of the interactions of NAG in 8JCU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1102 | R:R:N203 | 25.79 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N203 | R:R:R200 | 1.21 | 0 | No | Yes | 4 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 25.79 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 8JCU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2001 | S:S:N209 | 27.02 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N209 | S:S:R206 | 4.82 | 0 | No | No | 4 | 5 | 1 | 2 | | S:S:P505 | S:S:S503 | 3.56 | 0 | No | No | 7 | 4 | 2 | 1 | | H:H:?2001 | S:S:S503 | 2.69 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.86 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8JCV | C | Aminoacid | Metabotropic Glutamate | mGlu2; mGlu3 | Homo Sapiens | LY341495 | - | - | 3.4 | 2023-06-21 | doi.org/10.1038/s41422-023-00830-2 |
|

A 2D representation of the interactions of NAG in 8JCV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2001 | R:R:N203 | 25.79 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:F201 | R:R:F202 | 4.29 | 0 | No | No | 5 | 8 | 2 | 1 | | R:R:F202 | R:R:W204 | 18.04 | 0 | No | No | 8 | 9 | 1 | 2 | | H:H:?2001 | R:R:F202 | 1.09 | 0 | No | No | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.44 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 8JCV
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2002 | S:S:N209 | 27.02 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N209 | S:S:R206 | 6.03 | 0 | No | No | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8JCW | C | Aminoacid | Metabotropic Glutamate | mGlu2; mGlu3 | Homo Sapiens | LY341495 | - | - | 3 | 2023-06-21 | doi.org/10.1038/s41422-023-00830-2 |
|
A 2D representation of the interactions of NAG in 8JCW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1002 | S:S:N209 | 27.02 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N209 | S:S:R206 | 1.21 | 0 | No | No | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 8JCW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1101 | R:R:N203 | 27.02 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N203 | R:R:R200 | 6.03 | 0 | No | No | 4 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8JCX | C | Aminoacid | Metabotropic Glutamate | mGlu2; mGlu3 | Homo Sapiens | LY341495 | - | - | 3 | 2023-06-21 | doi.org/10.1038/s41422-023-00830-2 |
|
A 2D representation of the interactions of NAG in 8JCX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1002 | S:S:N209 | 28.34 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N209 | S:S:R206 | 1.21 | 0 | No | Yes | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 28.34 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
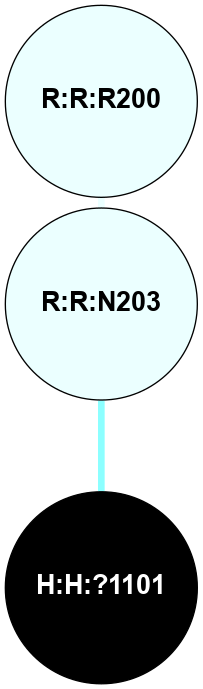
A 2D representation of the interactions of NAG in 8JCX
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1101 | R:R:N203 | 27.11 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N203 | R:R:R200 | 1.21 | 0 | No | No | 4 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.11 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8JCY | C | Aminoacid | Metabotropic Glutamate | mGlu2; mGlu3 | Homo Sapiens | LY341495 | - | - | 2.9 | 2023-06-21 | doi.org/10.1038/s41422-023-00830-2 |
|
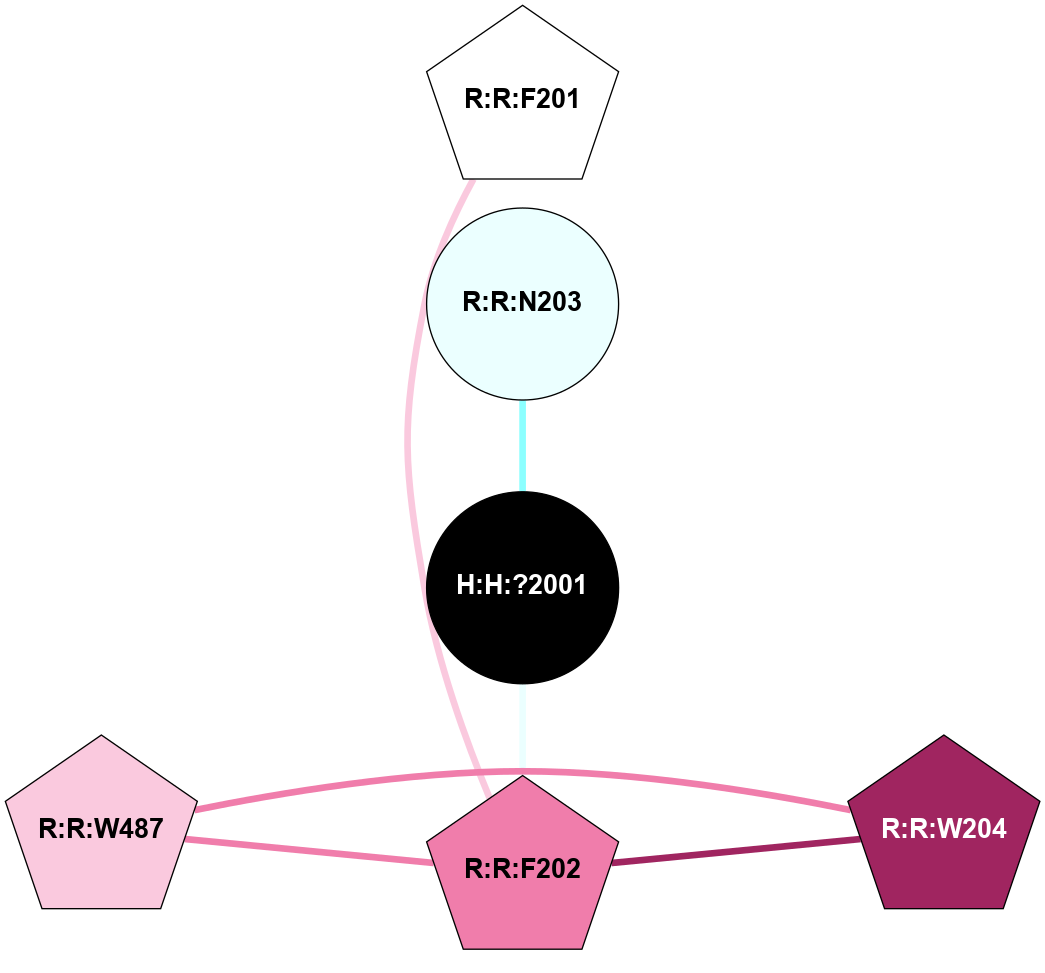
A 2D representation of the interactions of NAG in 8JCY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2001 | R:R:N203 | 27.02 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:F201 | R:R:F202 | 9.65 | 6 | Yes | Yes | 5 | 8 | 2 | 1 | | R:R:F202 | R:R:W204 | 12.03 | 6 | Yes | Yes | 8 | 9 | 1 | 2 | | R:R:F202 | R:R:W487 | 11.02 | 6 | Yes | Yes | 8 | 7 | 1 | 2 | | R:R:W204 | R:R:W487 | 5.62 | 6 | Yes | Yes | 9 | 7 | 2 | 2 | | H:H:?2001 | R:R:F202 | 1.09 | 0 | No | Yes | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.05 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
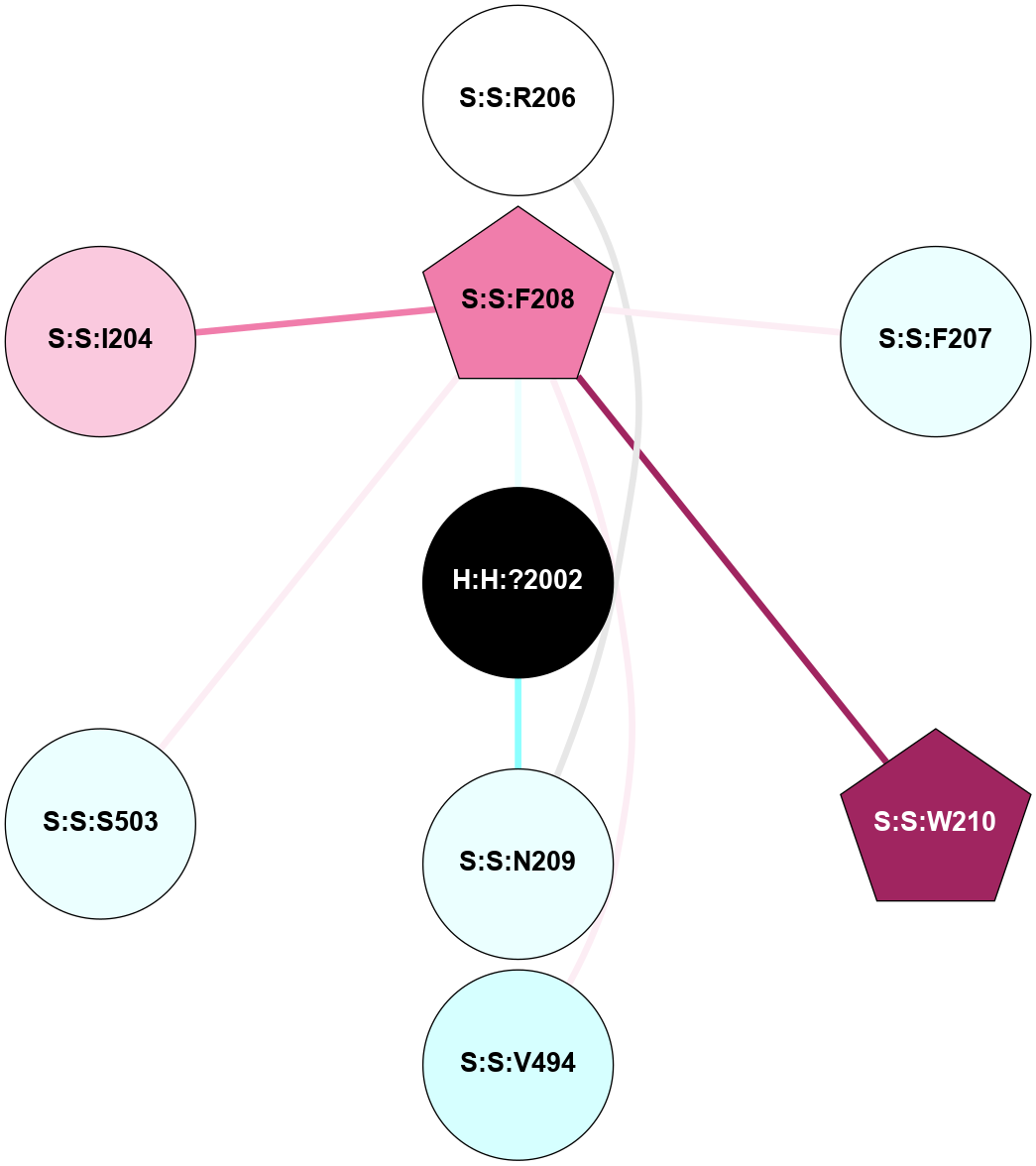
A 2D representation of the interactions of NAG in 8JCY
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2002 | S:S:F208 | 4.36 | 0 | No | Yes | 0 | 8 | 0 | 1 | | H:H:?2002 | S:S:N209 | 29.47 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N209 | S:S:R206 | 3.62 | 0 | No | No | 4 | 5 | 1 | 2 | | S:S:F207 | S:S:F208 | 11.79 | 0 | No | Yes | 4 | 8 | 2 | 1 | | S:S:F208 | S:S:W210 | 16.04 | 0 | Yes | Yes | 8 | 9 | 1 | 2 | | S:S:F208 | S:S:V494 | 3.93 | 0 | Yes | No | 8 | 3 | 1 | 2 | | S:S:F208 | S:S:S503 | 3.96 | 0 | Yes | No | 8 | 4 | 1 | 2 | | S:S:F208 | S:S:I204 | 2.51 | 0 | Yes | No | 8 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 16.91 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8JCZ | C | Aminoacid | Metabotropic Glutamate | mGlu2; mGlu3 | Homo Sapiens | LY341495 | - | - | 3 | 2023-06-21 | doi.org/10.1038/s41422-023-00830-2 |
|
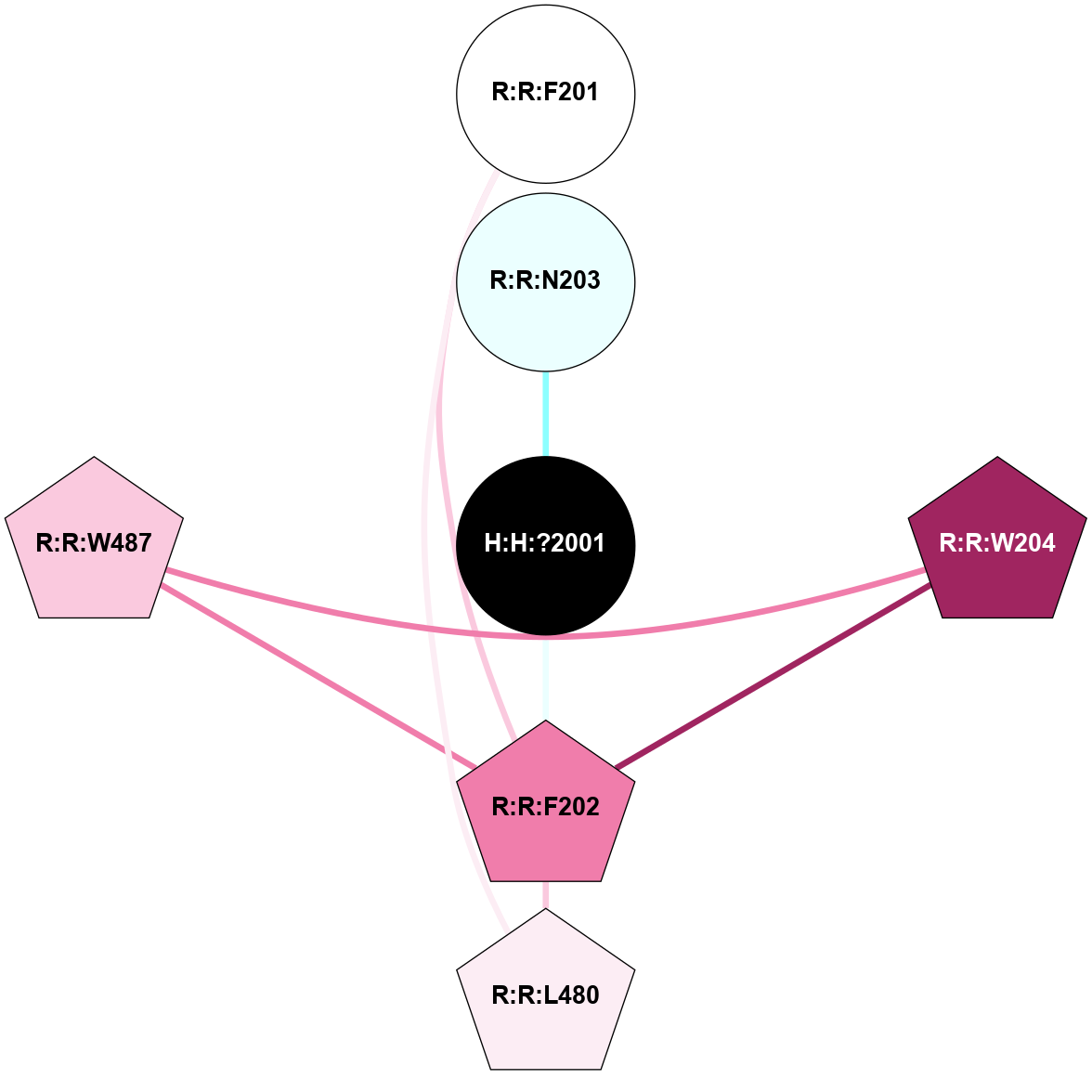
A 2D representation of the interactions of NAG in 8JCZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2001 | R:R:N203 | 28.24 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:F201 | R:R:F202 | 8.57 | 1 | No | Yes | 5 | 8 | 2 | 1 | | R:R:F201 | R:R:L480 | 4.87 | 1 | No | Yes | 5 | 6 | 2 | 2 | | R:R:F202 | R:R:W204 | 13.03 | 1 | Yes | Yes | 8 | 9 | 1 | 2 | | R:R:F202 | R:R:L480 | 3.65 | 1 | Yes | Yes | 8 | 6 | 1 | 2 | | R:R:F202 | R:R:W487 | 13.03 | 1 | Yes | Yes | 8 | 7 | 1 | 2 | | R:R:W204 | R:R:W487 | 7.5 | 1 | Yes | Yes | 9 | 7 | 2 | 2 | | H:H:?2001 | R:R:F202 | 1.09 | 0 | No | Yes | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.66 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 8JCZ
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2002 | S:S:N209 | 27.02 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N209 | S:S:R206 | 6.03 | 0 | No | No | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8JD0 | C | Aminoacid | Metabotropic Glutamate | mGlu2; mGlu3 | Homo Sapiens | Glutamate | NAM563 | - | 3.3 | 2023-06-21 | doi.org/10.1038/s41422-023-00830-2 |
|
A 2D representation of the interactions of NAG in 8JD0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1002 | S:S:N209 | 27.02 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N209 | S:S:R206 | 6.03 | 0 | No | No | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 8JD0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1101 | R:R:N203 | 27.02 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N203 | R:R:R200 | 1.21 | 0 | No | No | 4 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8JD2 | C | Aminoacid | Metabotropic Glutamate | mGlu2; mGlu3 | Homo Sapiens | Glutamate | - | - | 2.8 | 2023-06-21 | doi.org/10.1038/s41422-023-00830-2 |
|
A 2D representation of the interactions of NAG in 8JD2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1002 | S:S:N209 | 28.34 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N209 | S:S:R206 | 4.82 | 0 | No | No | 4 | 5 | 1 | 2 | | S:S:F207 | S:S:F208 | 5.36 | 0 | No | Yes | 4 | 8 | 1 | 2 | | S:S:F207 | S:S:L492 | 8.53 | 0 | No | No | 4 | 6 | 1 | 2 | | H:H:?1002 | S:S:F207 | 2.19 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.27 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
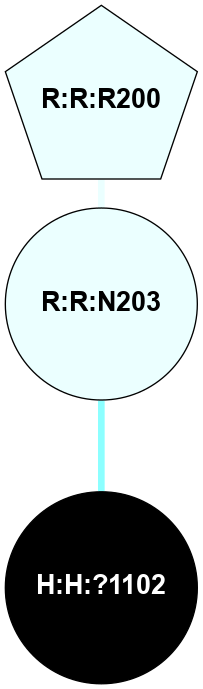
A 2D representation of the interactions of NAG in 8JD2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1102 | R:R:N203 | 30.8 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N203 | R:R:R200 | 3.62 | 0 | No | Yes | 4 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 30.80 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8JD4 | C | Aminoacid | Metabotropic Glutamate | mGlu2; mGlu4 | Homo Sapiens | Glutamate | - | - | 2.9 | 2023-06-21 | doi.org/10.1038/s41422-023-00830-2 |
|

A 2D representation of the interactions of NAG in 8JD4
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1102 | S:S:N454 | 28.24 | 23 | No | No | 0 | 5 | 0 | 1 | | H:H:?1102 | S:S:T464 | 9.23 | 23 | No | No | 0 | 3 | 0 | 1 | | S:S:N454 | S:S:T464 | 5.85 | 23 | No | No | 5 | 3 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.73 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
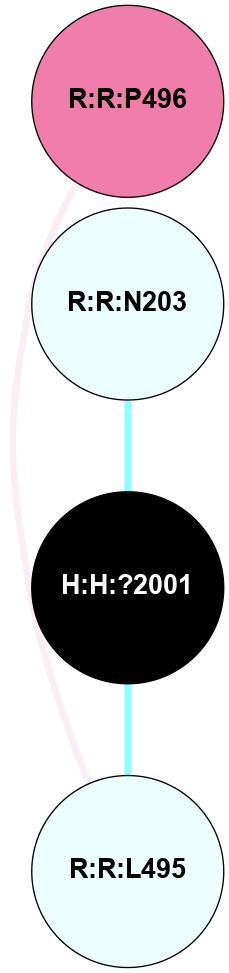
A 2D representation of the interactions of NAG in 8JD4
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2001 | R:R:N203 | 27.02 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N203 | R:R:R499 | 4.82 | 0 | No | No | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
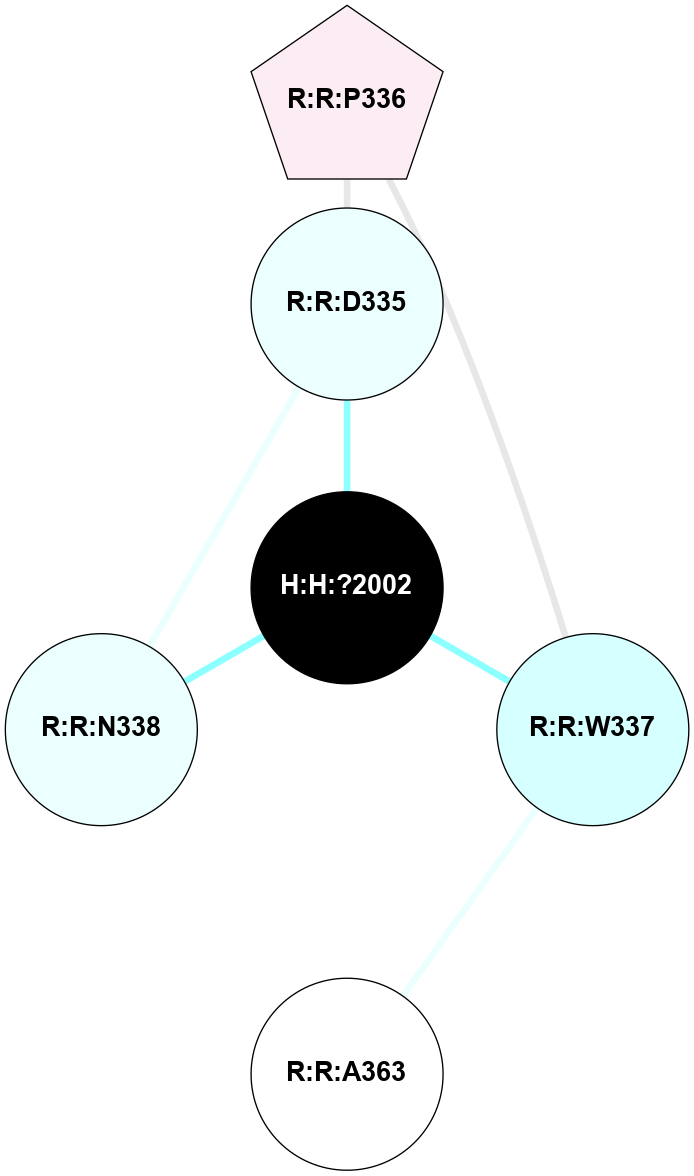
A 2D representation of the interactions of NAG in 8JD4
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2002 | R:R:D335 | 9.71 | 24 | No | No | 0 | 4 | 0 | 1 | | H:H:?2002 | R:R:W337 | 6.11 | 24 | No | No | 0 | 3 | 0 | 1 | | H:H:?2002 | R:R:N338 | 31.93 | 24 | No | No | 0 | 4 | 0 | 1 | | R:R:D335 | R:R:N338 | 12.12 | 24 | No | No | 4 | 4 | 1 | 1 | | R:R:P336 | R:R:W337 | 12.16 | 1 | Yes | No | 6 | 3 | 2 | 1 | | R:R:A363 | R:R:W337 | 2.59 | 0 | No | No | 5 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.92 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8JD5 | C | Aminoacid | Metabotropic Glutamate | mGlu2; mGlu4 | Homo Sapiens | Glutamate | JNJ-40411813; ADX88178 | Gi1/Beta1/Gamma2 | 3.6 | 2023-06-21 | doi.org/10.1038/s41422-023-00830-2 |
|

A 2D representation of the interactions of NAG in 8JD5
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | S:S:N454 | 25.87 | 36 | No | No | 0 | 5 | 0 | 1 | | H:H:?1 | S:S:T464 | 11.9 | 36 | No | No | 0 | 3 | 0 | 1 | | S:S:N454 | S:S:T464 | 2.92 | 36 | No | No | 5 | 3 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.89 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8SZF | C | Ion | Calcium Sensing | CaS | Homo Sapiens | - | Ca; Ca; Tryptophan; Cinacalcet; PO4; Spermine | - | 2.8 | 2024-02-07 | doi.org/10.1038/s41586-024-07055-2 |
|
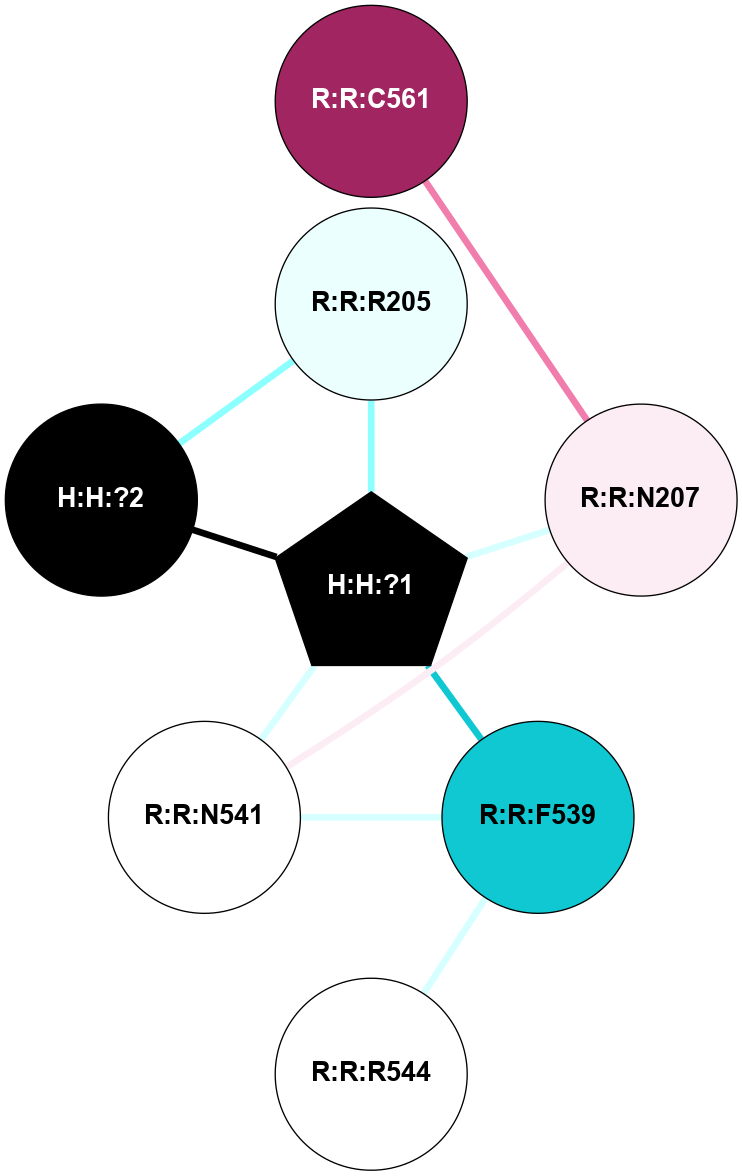
A 2D representation of the interactions of NAG in 8SZF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 33.43 | 4 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:R205 | 10.9 | 4 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:N207 | 12.32 | 4 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:F539 | 5.46 | 4 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?1 | R:R:N541 | 28.34 | 4 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:R205 | 5.45 | 4 | No | No | 0 | 4 | 1 | 1 | | R:R:N207 | R:R:N541 | 17.71 | 4 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 6.3 | 4 | No | No | 9 | 6 | 2 | 1 | | R:R:F539 | R:R:N541 | 6.04 | 4 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 18.17 | 4 | No | No | 1 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.09 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
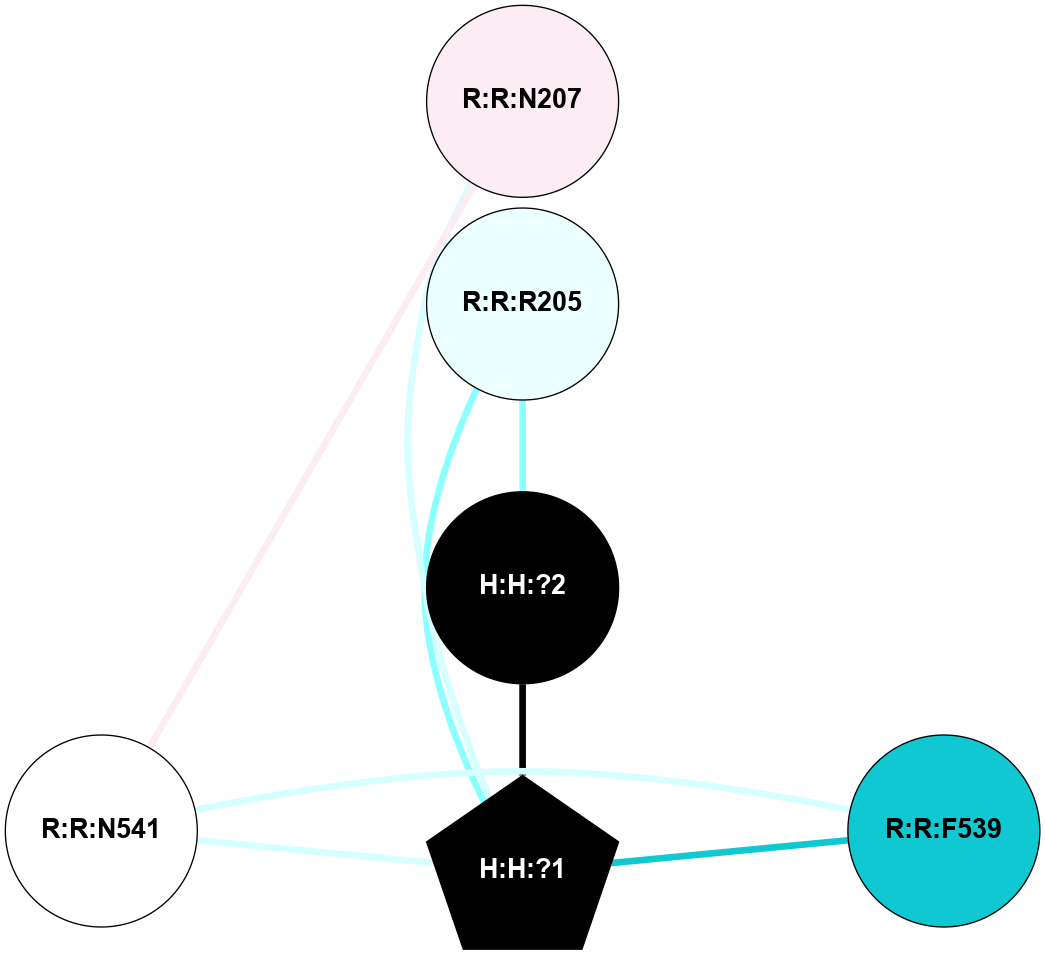
A 2D representation of the interactions of NAG in 8SZF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 33.43 | 4 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:R205 | 10.9 | 4 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?1 | R:R:N207 | 12.32 | 4 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:F539 | 5.46 | 4 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?1 | R:R:N541 | 28.34 | 4 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?2 | R:R:R205 | 5.45 | 4 | No | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 17.71 | 4 | No | No | 6 | 5 | 2 | 2 | | R:R:F539 | R:R:N541 | 6.04 | 4 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.44 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
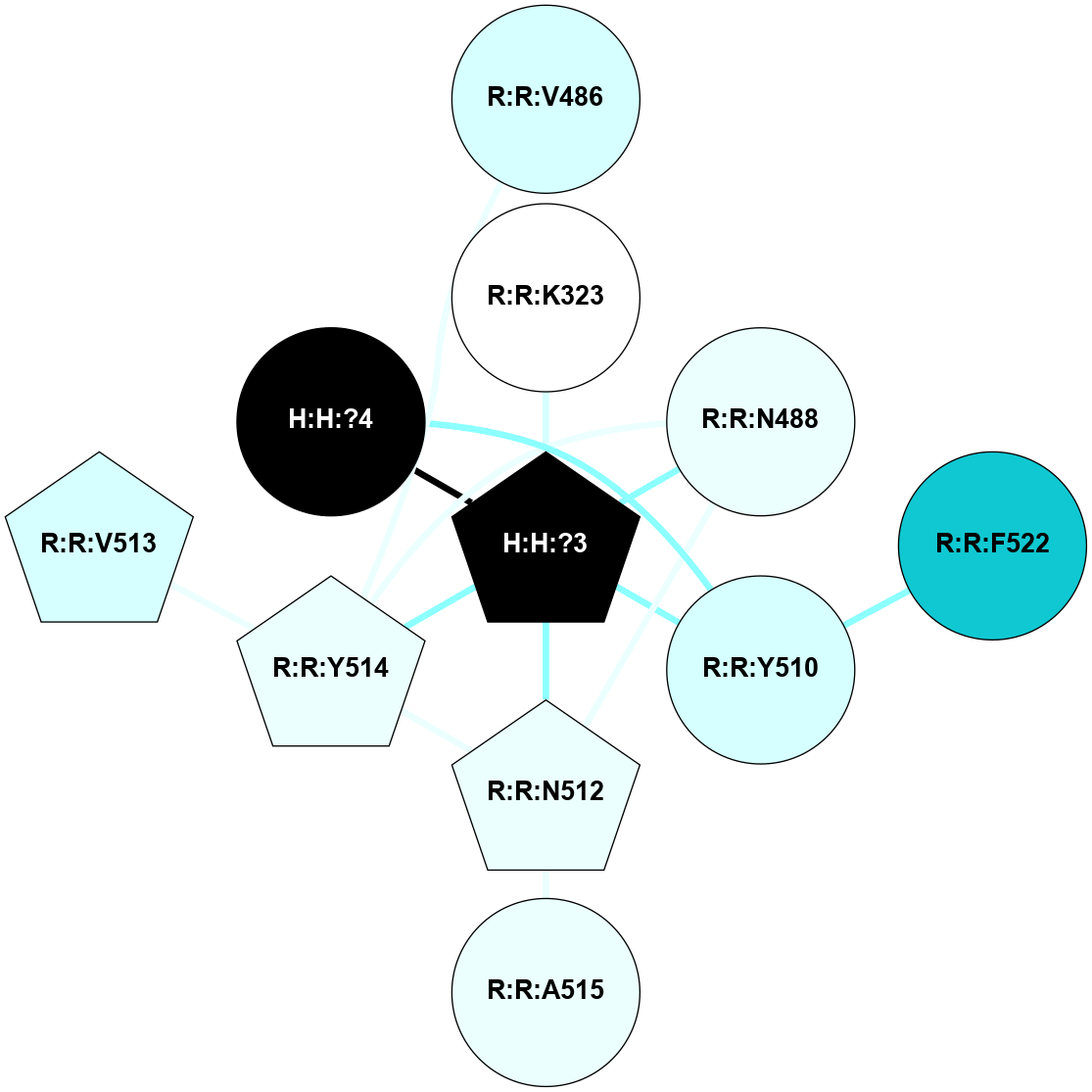
A 2D representation of the interactions of NAG in 8SZF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 41.23 | 16 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:K323 | 6.33 | 16 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?3 | R:R:N488 | 24.64 | 16 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?3 | R:R:Y510 | 9.47 | 16 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?3 | R:R:N512 | 14.79 | 16 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?3 | R:R:Y514 | 13.67 | 16 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?4 | R:R:Y510 | 3.16 | 16 | No | No | 0 | 3 | 1 | 1 | | R:R:V486 | R:R:Y514 | 3.79 | 0 | No | Yes | 3 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 5.45 | 16 | No | Yes | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:Y514 | 4.65 | 16 | No | Yes | 4 | 4 | 1 | 1 | | R:R:F522 | R:R:Y510 | 15.47 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:N512 | R:R:Y514 | 5.81 | 16 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 4.69 | 0 | No | Yes | 4 | 4 | 2 | 1 | | R:R:V513 | R:R:Y514 | 7.57 | 0 | Yes | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 18.35 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
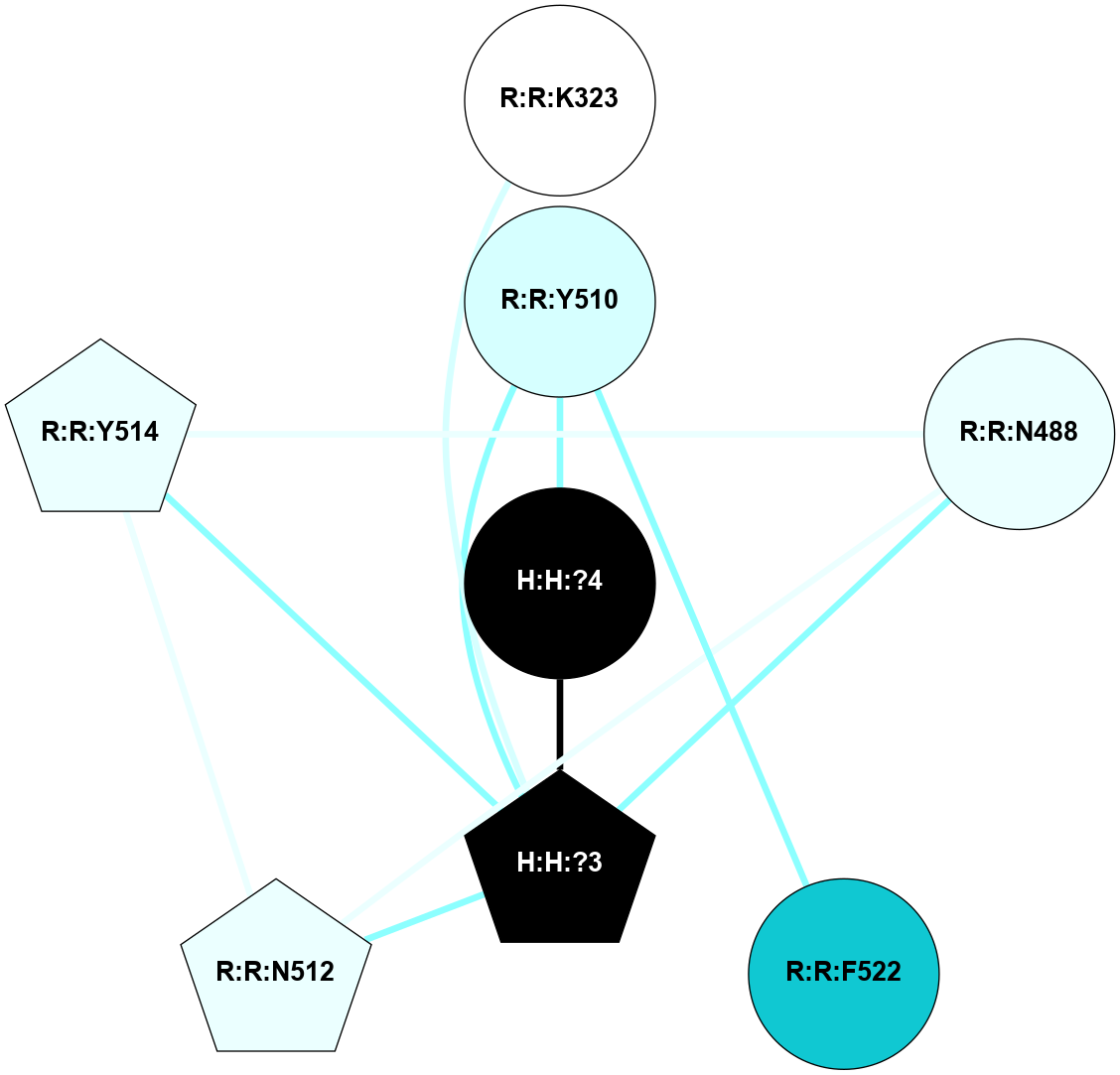
A 2D representation of the interactions of NAG in 8SZF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 41.23 | 16 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:K323 | 6.33 | 16 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | R:R:N488 | 24.64 | 16 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?3 | R:R:Y510 | 9.47 | 16 | Yes | No | 0 | 3 | 1 | 1 | | H:H:?3 | R:R:N512 | 14.79 | 16 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?3 | R:R:Y514 | 13.67 | 16 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?4 | R:R:Y510 | 3.16 | 16 | No | No | 0 | 3 | 0 | 1 | | R:R:N488 | R:R:N512 | 5.45 | 16 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N488 | R:R:Y514 | 4.65 | 16 | No | Yes | 4 | 4 | 2 | 2 | | R:R:F522 | R:R:Y510 | 15.47 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:N512 | R:R:Y514 | 5.81 | 16 | Yes | Yes | 4 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 22.20 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
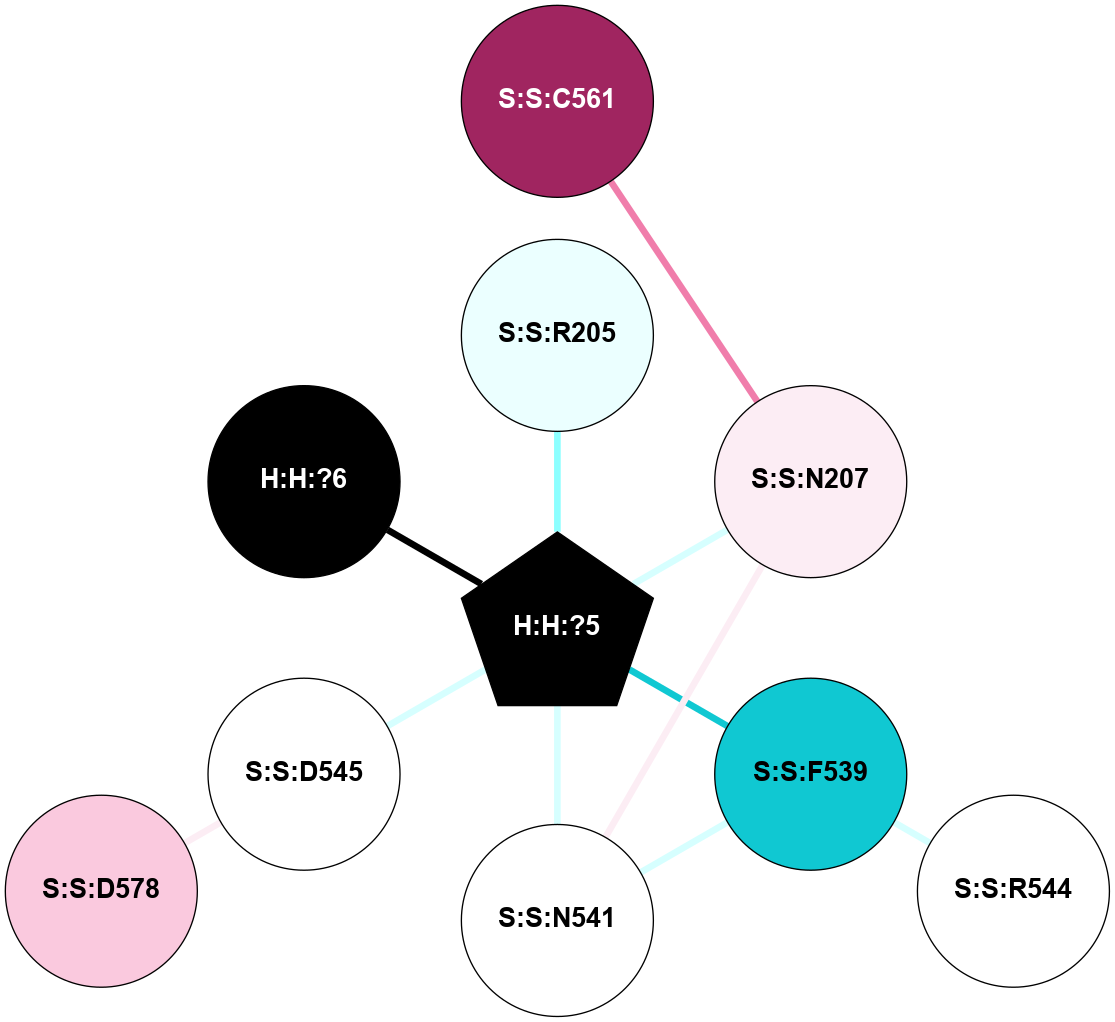
A 2D representation of the interactions of NAG in 8SZF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 37.89 | 3 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?5 | S:S:R205 | 6.54 | 3 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?5 | S:S:N207 | 14.79 | 3 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?5 | S:S:F539 | 5.46 | 3 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?5 | S:S:N541 | 29.57 | 3 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?5 | S:S:D545 | 7.31 | 3 | Yes | No | 0 | 5 | 0 | 1 | | S:S:N207 | S:S:N541 | 17.71 | 3 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 3.15 | 3 | No | No | 9 | 6 | 2 | 1 | | S:S:F539 | S:S:N541 | 6.04 | 3 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 25.66 | 3 | No | No | 1 | 5 | 1 | 2 | | S:S:D545 | S:S:D578 | 15.97 | 0 | No | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.93 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
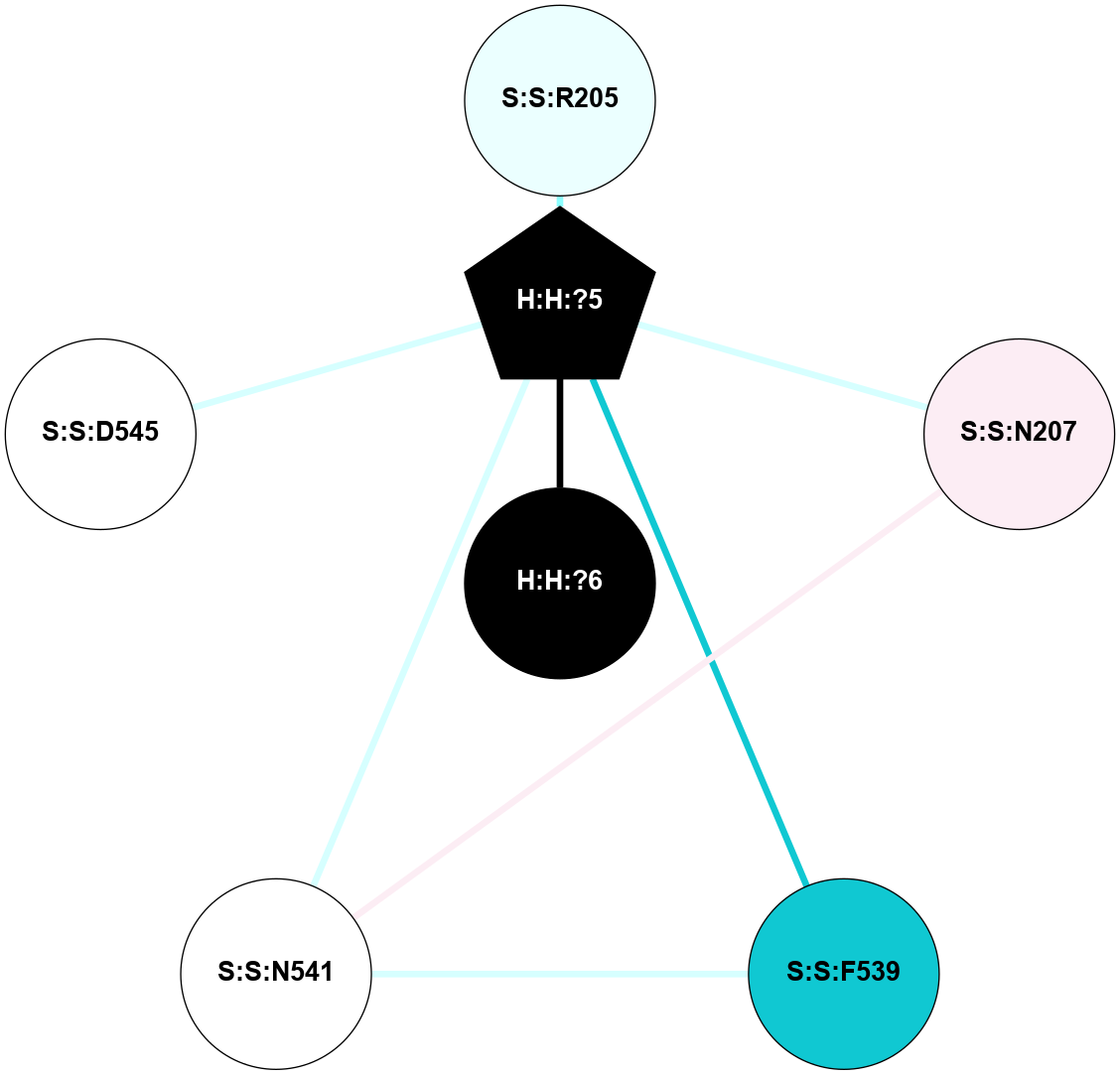
A 2D representation of the interactions of NAG in 8SZF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 37.89 | 3 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?5 | S:S:R205 | 6.54 | 3 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?5 | S:S:N207 | 14.79 | 3 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?5 | S:S:F539 | 5.46 | 3 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?5 | S:S:N541 | 29.57 | 3 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?5 | S:S:D545 | 7.31 | 3 | Yes | No | 0 | 5 | 1 | 2 | | S:S:N207 | S:S:N541 | 17.71 | 3 | No | No | 6 | 5 | 2 | 2 | | S:S:F539 | S:S:N541 | 6.04 | 3 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 37.89 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
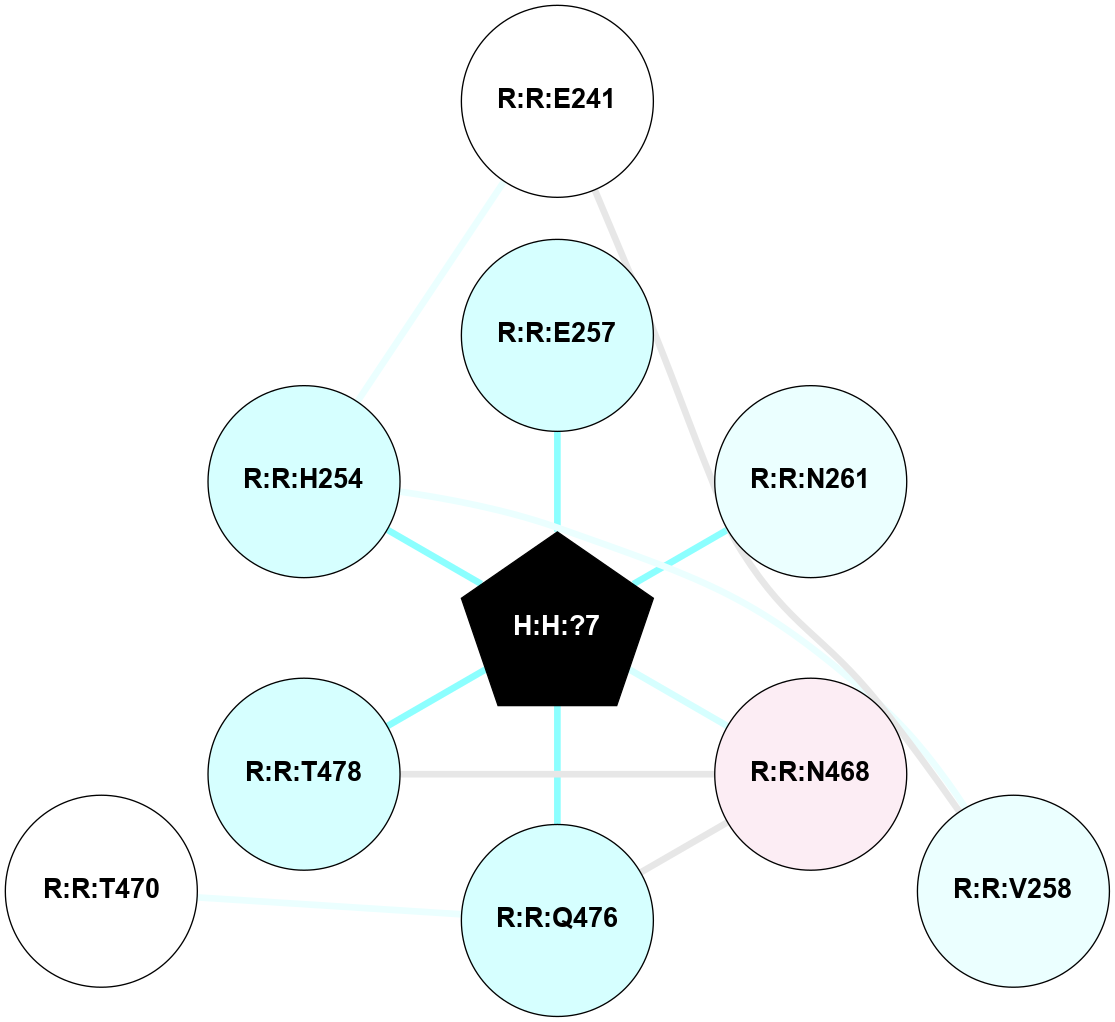
A 2D representation of the interactions of NAG in 8SZF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | R:R:E257 | 4.76 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?7 | R:R:N261 | 24.64 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:E241 | R:R:H254 | 3.69 | 4 | No | No | 5 | 3 | 2 | 1 | | R:R:E241 | R:R:V258 | 9.98 | 4 | No | No | 5 | 4 | 2 | 2 | | R:R:H254 | R:R:V258 | 5.54 | 4 | No | No | 3 | 4 | 1 | 2 | | H:H:?7 | R:R:H254 | 2.31 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 10.57 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
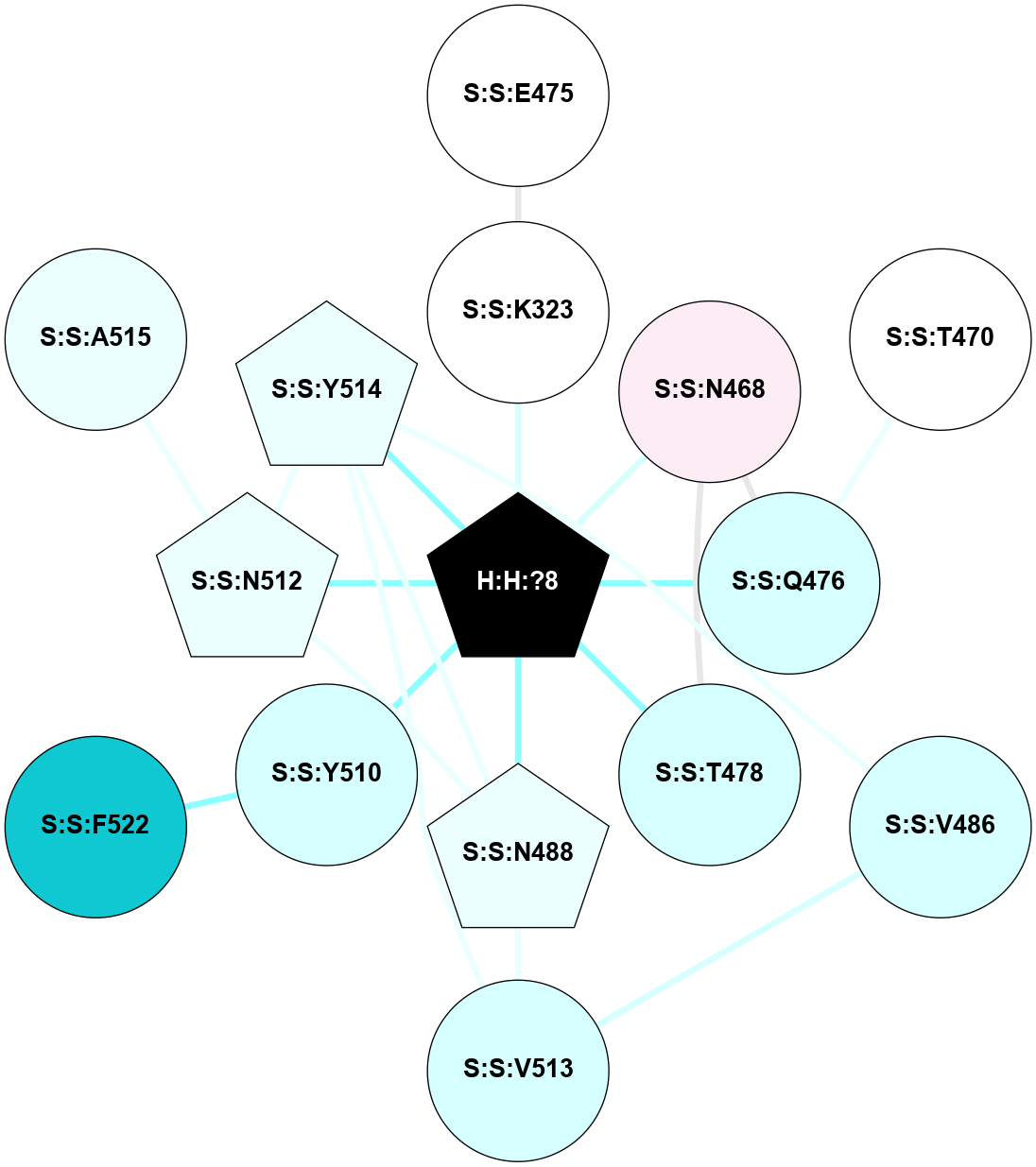
A 2D representation of the interactions of NAG in 8SZF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | S:S:K323 | 5.06 | 27 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?8 | S:S:N488 | 24.64 | 27 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?8 | S:S:Y510 | 7.36 | 27 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?8 | S:S:N512 | 16.02 | 27 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?8 | S:S:Y514 | 10.52 | 27 | Yes | Yes | 0 | 4 | 0 | 1 | | S:S:E475 | S:S:K323 | 4.05 | 0 | No | No | 5 | 5 | 2 | 1 | | S:S:V486 | S:S:V513 | 3.21 | 0 | No | No | 3 | 3 | 2 | 2 | | S:S:V486 | S:S:Y514 | 3.79 | 0 | No | Yes | 3 | 4 | 2 | 1 | | S:S:N488 | S:S:N512 | 5.45 | 27 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:V513 | 2.96 | 27 | Yes | No | 4 | 3 | 1 | 2 | | S:S:N488 | S:S:Y514 | 3.49 | 27 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:F522 | S:S:Y510 | 13.41 | 0 | No | No | 1 | 3 | 2 | 1 | | S:S:N512 | S:S:Y514 | 3.49 | 27 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:A515 | S:S:N512 | 4.69 | 0 | No | Yes | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 12.72 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
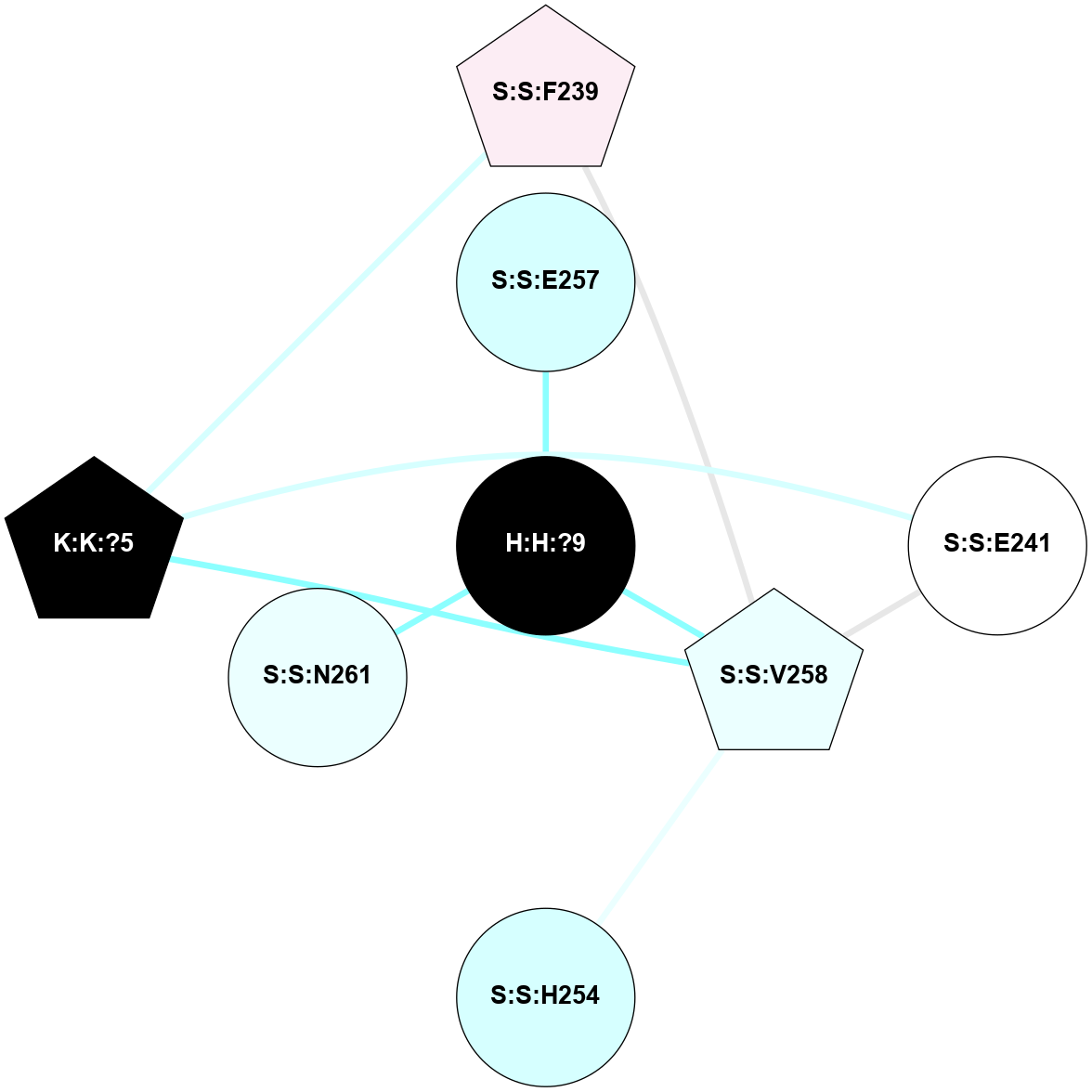
A 2D representation of the interactions of NAG in 8SZF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?9 | S:S:E257 | 4.76 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?9 | S:S:V258 | 4.01 | 0 | No | Yes | 0 | 4 | 0 | 1 | | H:H:?9 | S:S:N261 | 28.34 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:F239 | X:X:?5 | 5.84 | 3 | Yes | Yes | 6 | 0 | 2 | 2 | | S:S:E241 | X:X:?5 | 2.54 | 3 | Yes | Yes | 5 | 0 | 2 | 2 | | S:S:V258 | X:X:?5 | 2.86 | 3 | Yes | Yes | 4 | 0 | 1 | 2 | | X:X:?5 | X:X:?6 | 114.73 | 3 | Yes | Yes | 0 | 0 | 2 | 2 | | S:S:F239 | S:S:V258 | 3.93 | 3 | Yes | Yes | 6 | 4 | 2 | 1 | | S:S:F239 | X:X:?6 | 5.84 | 3 | Yes | Yes | 6 | 0 | 2 | 2 | | S:S:E241 | S:S:V258 | 9.98 | 3 | Yes | Yes | 5 | 4 | 2 | 1 | | S:S:E241 | X:X:?6 | 2.54 | 3 | Yes | Yes | 5 | 0 | 2 | 2 | | S:S:H254 | S:S:V258 | 4.15 | 0 | No | Yes | 3 | 4 | 2 | 1 | | S:S:V258 | X:X:?6 | 2.86 | 3 | Yes | Yes | 4 | 0 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 12.37 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8SZG | C | Ion | Calcium Sensing | CaS | Homo Sapiens | - | Ca; Ca; Tryptophan; Cinacalcet; PO4; Spermine | chim(NtGi1-Gq)/Beta1/Gamma2 | 3.6 | 2024-02-07 | doi.org/10.1038/s41586-024-07055-2 |
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8SZH | C | Ion | Calcium Sensing | CaS | Homo Sapiens | - | Ca; Ca; Tryptophan; Cinacalcet; PO4; Spermine | Gi1/Beta1/Gamma2 | 3.1 | 2024-02-07 | doi.org/10.1038/s41586-024-07055-2 |
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8SZI | C | Ion | Calcium Sensing | CaS | Homo Sapiens | - | Ca; Ca; Tryptophan; Cinacalcet; PO4 | Gi3/Beta1/Gamma2 | 3.5 | 2024-02-07 | doi.org/10.1038/s41586-024-07055-2 |
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8U1U | A | Protein | Chemokine | CCR8 | Homo Sapiens | CCL1 | - | Gi1/Beta1/Gamma2 | 3.1 | 2023-12-20 | doi.org/10.1038/s41467-023-43601-8 |
|
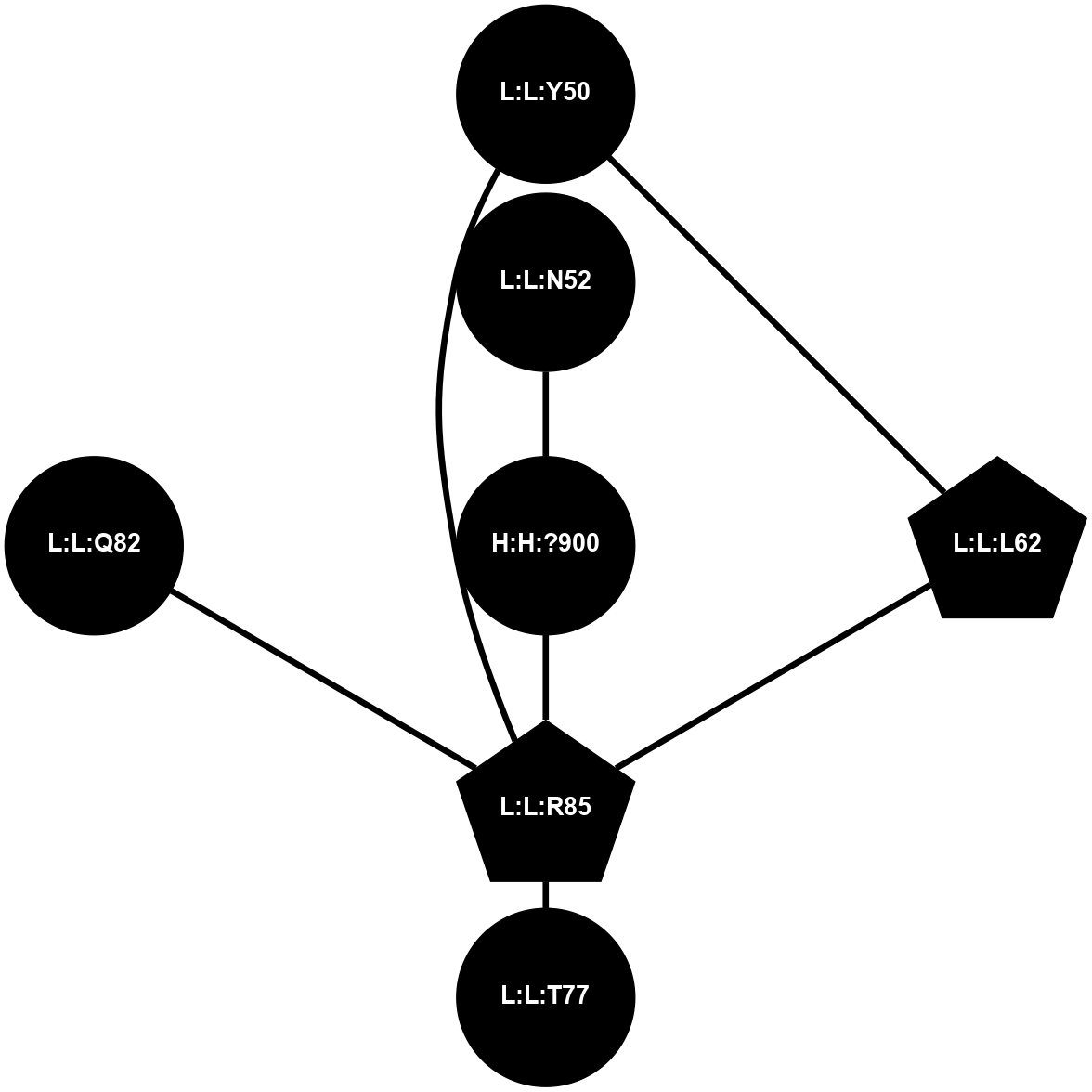
A 2D representation of the interactions of NAG in 8U1U
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | L:L:L62 | L:L:Y50 | 9.38 | 14 | Yes | No | 0 | 0 | 2 | 2 | | L:L:R85 | L:L:Y50 | 8.23 | 14 | Yes | No | 0 | 0 | 1 | 2 | | H:H:?900 | L:L:N52 | 28.24 | 0 | No | No | 0 | 0 | 0 | 1 | | L:L:L62 | L:L:R85 | 4.86 | 14 | Yes | Yes | 0 | 0 | 2 | 1 | | L:L:R85 | L:L:T77 | 5.17 | 14 | Yes | No | 0 | 0 | 1 | 2 | | L:L:Q82 | L:L:R85 | 7.01 | 0 | No | Yes | 0 | 0 | 2 | 1 | | H:H:?900 | L:L:R85 | 9.78 | 0 | No | Yes | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.01 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8WPG | C | Ion | Calcium Sensing | CaS | Homo Sapiens | - | Tryptophan; Cinacalcet; PO4; Ca | - | 2.7 | 2023-11-22 | doi.org/10.1038/s41422-023-00892-2 |
|
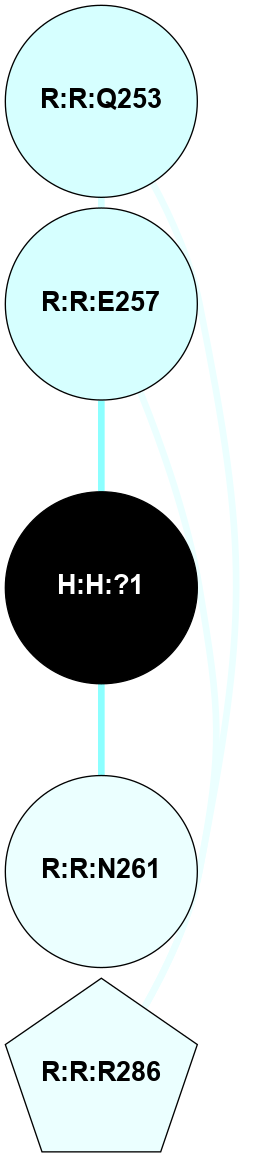
A 2D representation of the interactions of NAG in 8WPG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:E257 | 3.55 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:N261 | 27.02 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:E257 | R:R:Q253 | 5.1 | 18 | No | No | 3 | 3 | 1 | 2 | | R:R:Q253 | R:R:R286 | 8.18 | 18 | No | Yes | 3 | 4 | 2 | 2 | | R:R:E257 | R:R:R286 | 11.63 | 18 | No | Yes | 3 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.29 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 8WPG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:N287 | 29.47 | 0 | No | No | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:T289 | 6.59 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:H312 | R:R:N287 | 5.1 | 3 | No | No | 3 | 6 | 2 | 1 | | R:R:G290 | R:R:T289 | 3.64 | 0 | No | No | 4 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.03 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 8WPG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | S:S:E257 | 3.55 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?3 | S:S:N261 | 33.15 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:E257 | S:S:R286 | 11.63 | 0 | No | Yes | 3 | 4 | 1 | 2 | | S:S:E257 | S:S:Q253 | 1.27 | 0 | No | No | 3 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.35 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
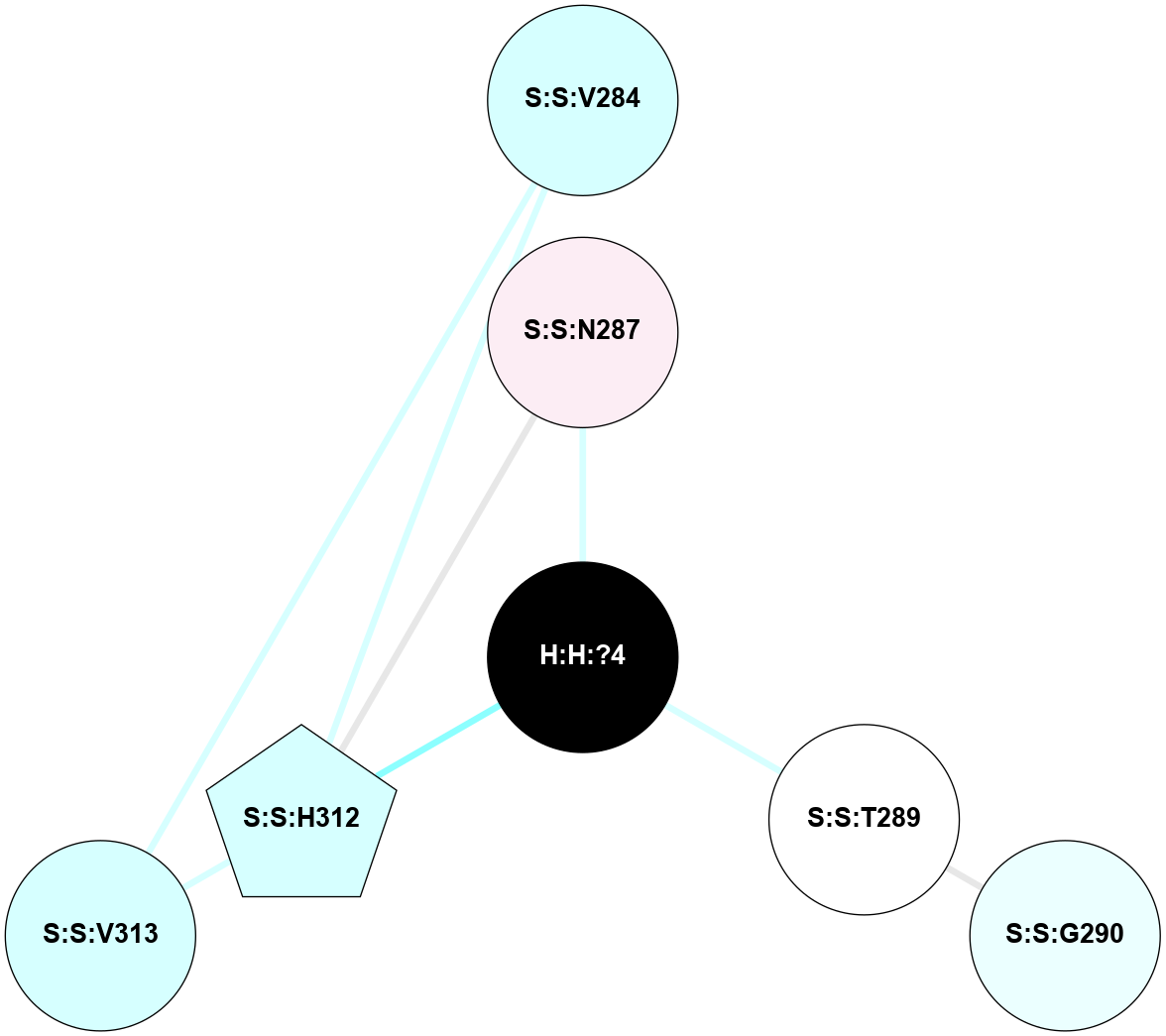
A 2D representation of the interactions of NAG in 8WPG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | S:S:N287 | 33.15 | 19 | No | No | 0 | 6 | 0 | 1 | | H:H:?4 | S:S:T289 | 6.59 | 19 | No | No | 0 | 5 | 0 | 1 | | H:H:?4 | S:S:H312 | 3.45 | 19 | No | Yes | 0 | 3 | 0 | 1 | | S:S:H312 | S:S:V284 | 5.54 | 19 | Yes | No | 3 | 3 | 1 | 2 | | S:S:V284 | S:S:V313 | 6.41 | 19 | No | No | 3 | 3 | 2 | 2 | | S:S:H312 | S:S:N287 | 7.65 | 19 | Yes | No | 3 | 6 | 1 | 1 | | S:S:G290 | S:S:T289 | 3.64 | 0 | No | No | 4 | 5 | 2 | 1 | | S:S:H312 | S:S:V313 | 6.92 | 19 | Yes | No | 3 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.40 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 8WPG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | S:S:N468 | 23.33 | 26 | No | No | 0 | 6 | 0 | 1 | | H:H:?5 | S:S:Q476 | 7.14 | 26 | No | No | 0 | 3 | 0 | 1 | | S:S:N468 | S:S:Q476 | 5.28 | 26 | No | No | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 10.24 | 26 | No | No | 6 | 3 | 1 | 2 | | S:S:Q476 | S:S:T470 | 5.67 | 26 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.23 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
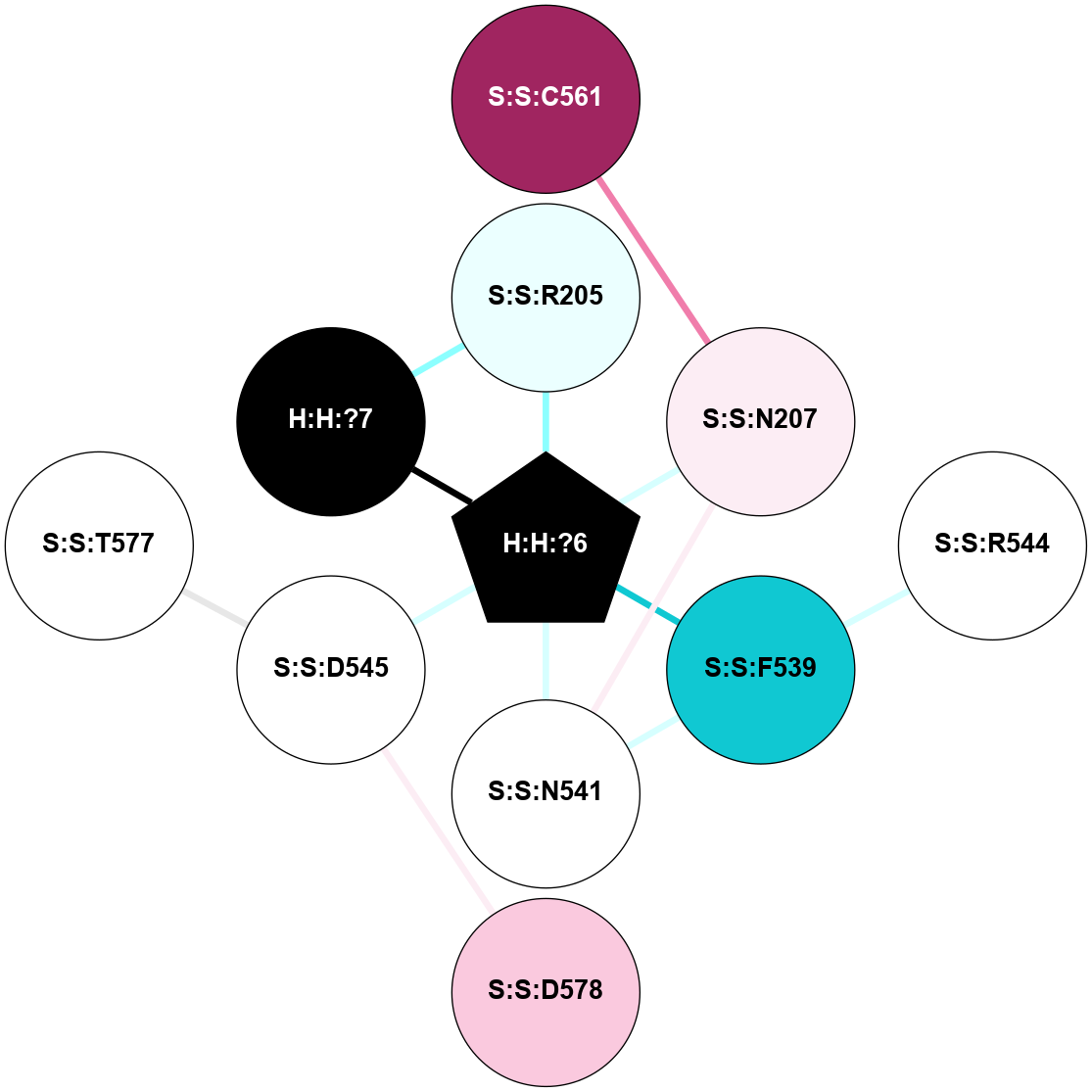
A 2D representation of the interactions of NAG in 8WPG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | H:H:?7 | 36.53 | 4 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?6 | S:S:R205 | 9.78 | 4 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?6 | S:S:N207 | 15.96 | 4 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?6 | S:S:F539 | 5.45 | 4 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?6 | S:S:N541 | 28.24 | 4 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?6 | S:S:D545 | 6.07 | 4 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?7 | S:S:R205 | 10.86 | 4 | No | No | 0 | 4 | 1 | 1 | | S:S:N207 | S:S:N541 | 16.35 | 4 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 9.45 | 4 | No | No | 9 | 6 | 2 | 1 | | S:S:F539 | S:S:N541 | 8.46 | 4 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 25.66 | 4 | No | No | 1 | 5 | 1 | 2 | | S:S:D545 | S:S:D578 | 7.98 | 0 | No | No | 5 | 7 | 1 | 2 | | S:S:D545 | S:S:T577 | 2.89 | 0 | No | No | 5 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.00 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
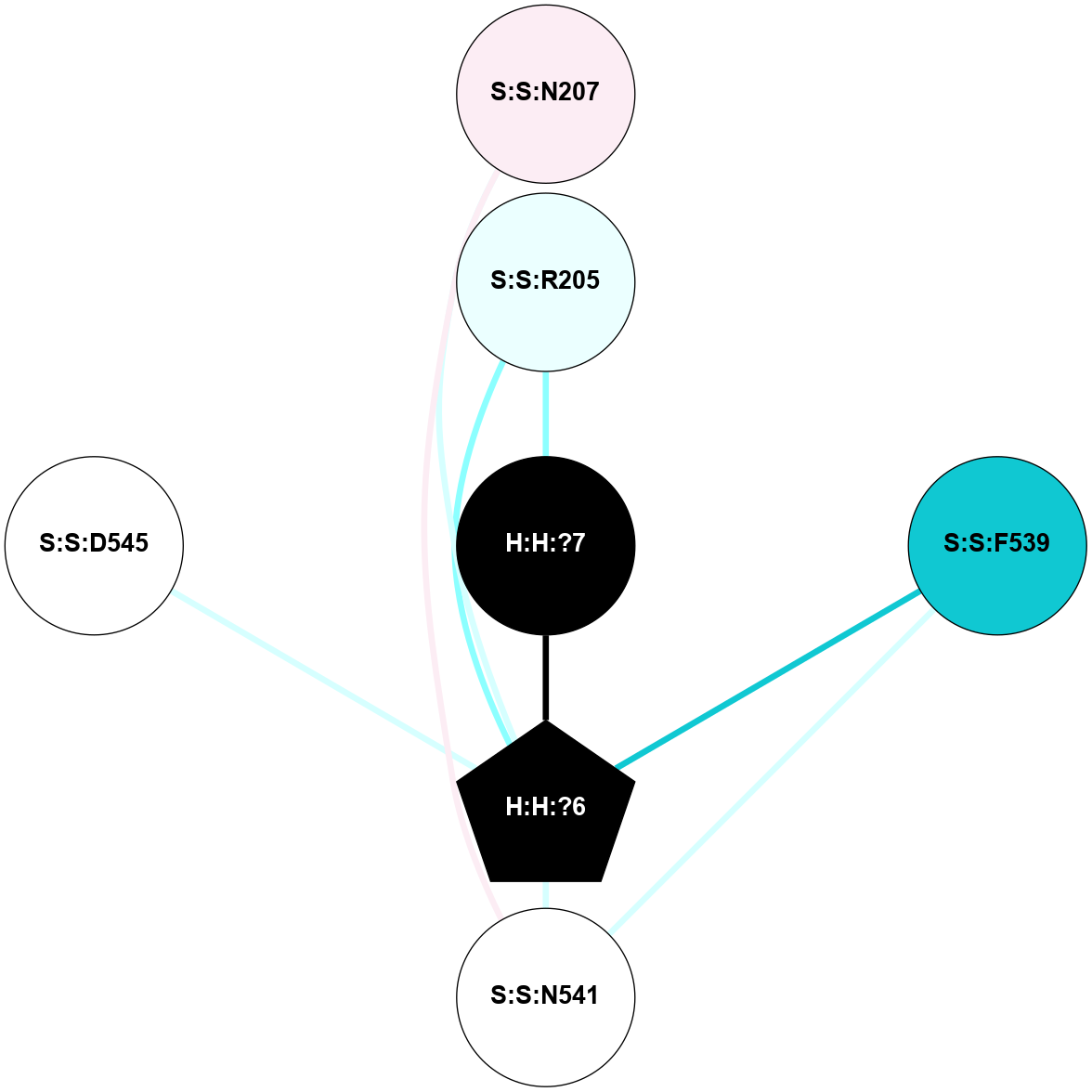
A 2D representation of the interactions of NAG in 8WPG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | H:H:?7 | 36.53 | 4 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?6 | S:S:R205 | 9.78 | 4 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?6 | S:S:N207 | 15.96 | 4 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?6 | S:S:F539 | 5.45 | 4 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?6 | S:S:N541 | 28.24 | 4 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?6 | S:S:D545 | 6.07 | 4 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?7 | S:S:R205 | 10.86 | 4 | No | No | 0 | 4 | 0 | 1 | | S:S:N207 | S:S:N541 | 16.35 | 4 | No | No | 6 | 5 | 2 | 2 | | S:S:F539 | S:S:N541 | 8.46 | 4 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 23.70 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
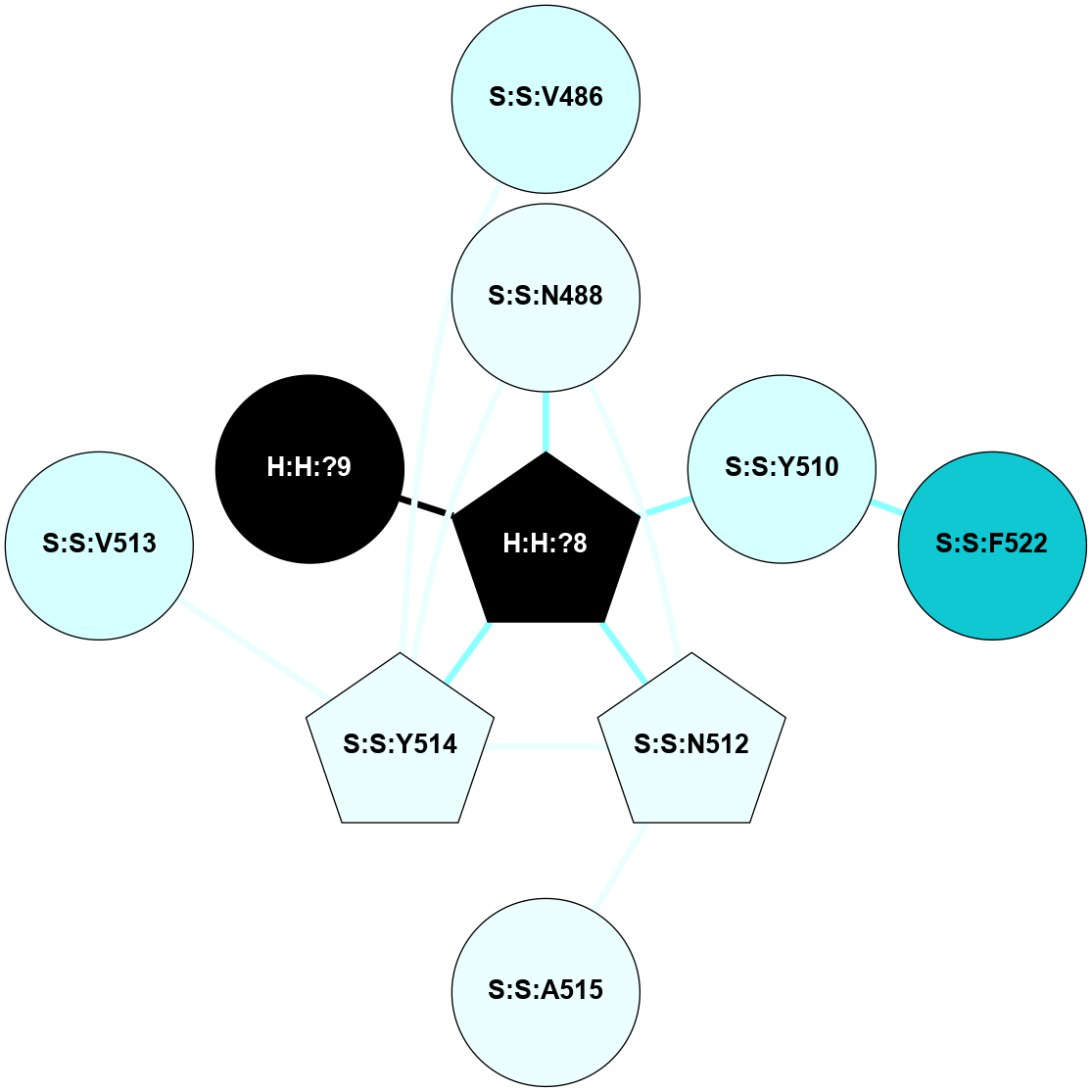
A 2D representation of the interactions of NAG in 8WPG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | H:H:?9 | 42.06 | 21 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?8 | S:S:N488 | 28.24 | 21 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?8 | S:S:Y510 | 10.48 | 21 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?8 | S:S:N512 | 17.19 | 21 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?8 | S:S:Y514 | 16.77 | 21 | Yes | Yes | 0 | 4 | 0 | 1 | | S:S:V486 | S:S:Y514 | 5.05 | 0 | No | Yes | 3 | 4 | 2 | 1 | | S:S:N488 | S:S:N512 | 4.09 | 21 | No | Yes | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:Y514 | 4.65 | 21 | No | Yes | 4 | 4 | 1 | 1 | | S:S:F522 | S:S:Y510 | 14.44 | 0 | No | No | 1 | 3 | 2 | 1 | | S:S:N512 | S:S:Y514 | 3.49 | 21 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:A515 | S:S:N512 | 4.69 | 0 | No | Yes | 4 | 4 | 2 | 1 | | S:S:V513 | S:S:Y514 | 5.05 | 0 | No | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 22.95 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
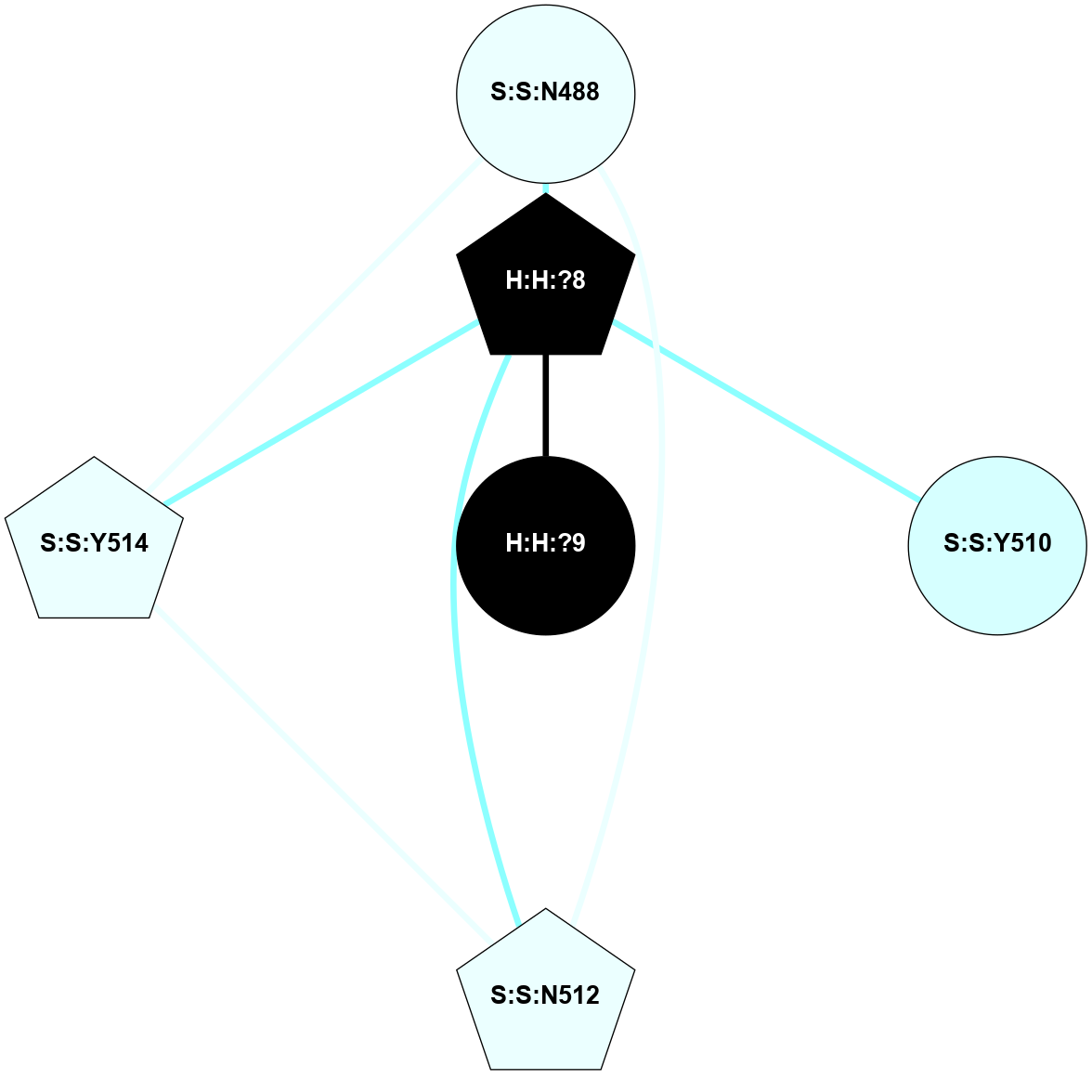
A 2D representation of the interactions of NAG in 8WPG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | H:H:?9 | 42.06 | 21 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?8 | S:S:N488 | 28.24 | 21 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?8 | S:S:Y510 | 10.48 | 21 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?8 | S:S:N512 | 17.19 | 21 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?8 | S:S:Y514 | 16.77 | 21 | Yes | Yes | 0 | 4 | 1 | 2 | | S:S:N488 | S:S:N512 | 4.09 | 21 | No | Yes | 4 | 4 | 2 | 2 | | S:S:N488 | S:S:Y514 | 4.65 | 21 | No | Yes | 4 | 4 | 2 | 2 | | S:S:N512 | S:S:Y514 | 3.49 | 21 | Yes | Yes | 4 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 42.06 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
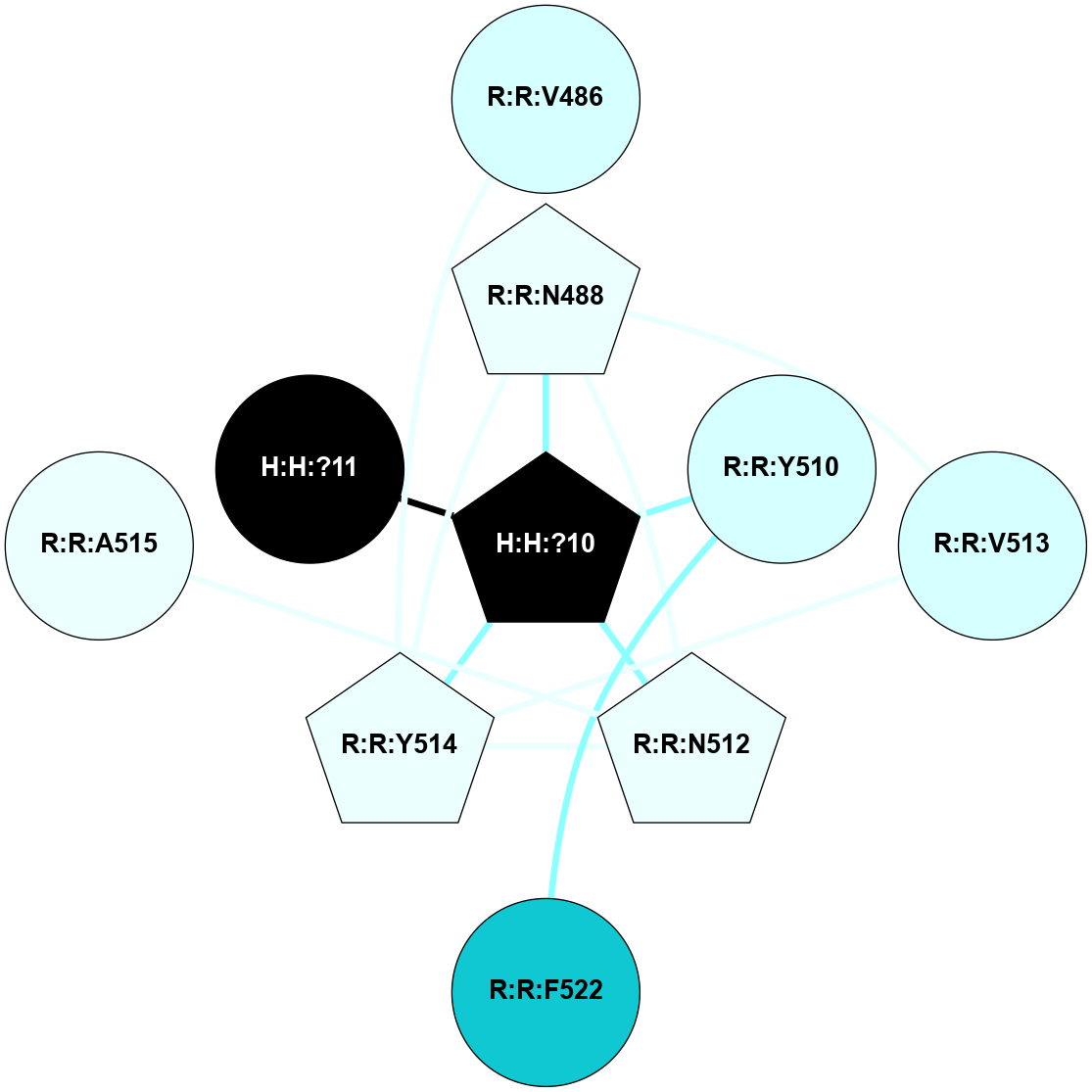
A 2D representation of the interactions of NAG in 8WPG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | H:H:?11 | 43.17 | 22 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?10 | R:R:N488 | 25.79 | 22 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?10 | R:R:Y510 | 8.39 | 22 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?10 | R:R:N512 | 17.19 | 22 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?10 | R:R:Y514 | 17.82 | 22 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:V486 | R:R:Y514 | 3.79 | 0 | No | Yes | 3 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 6.81 | 22 | No | Yes | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:Y514 | 8.14 | 22 | No | Yes | 4 | 4 | 1 | 1 | | R:R:F522 | R:R:Y510 | 13.41 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:N512 | R:R:Y514 | 6.98 | 22 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 4.69 | 0 | No | Yes | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 22.47 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
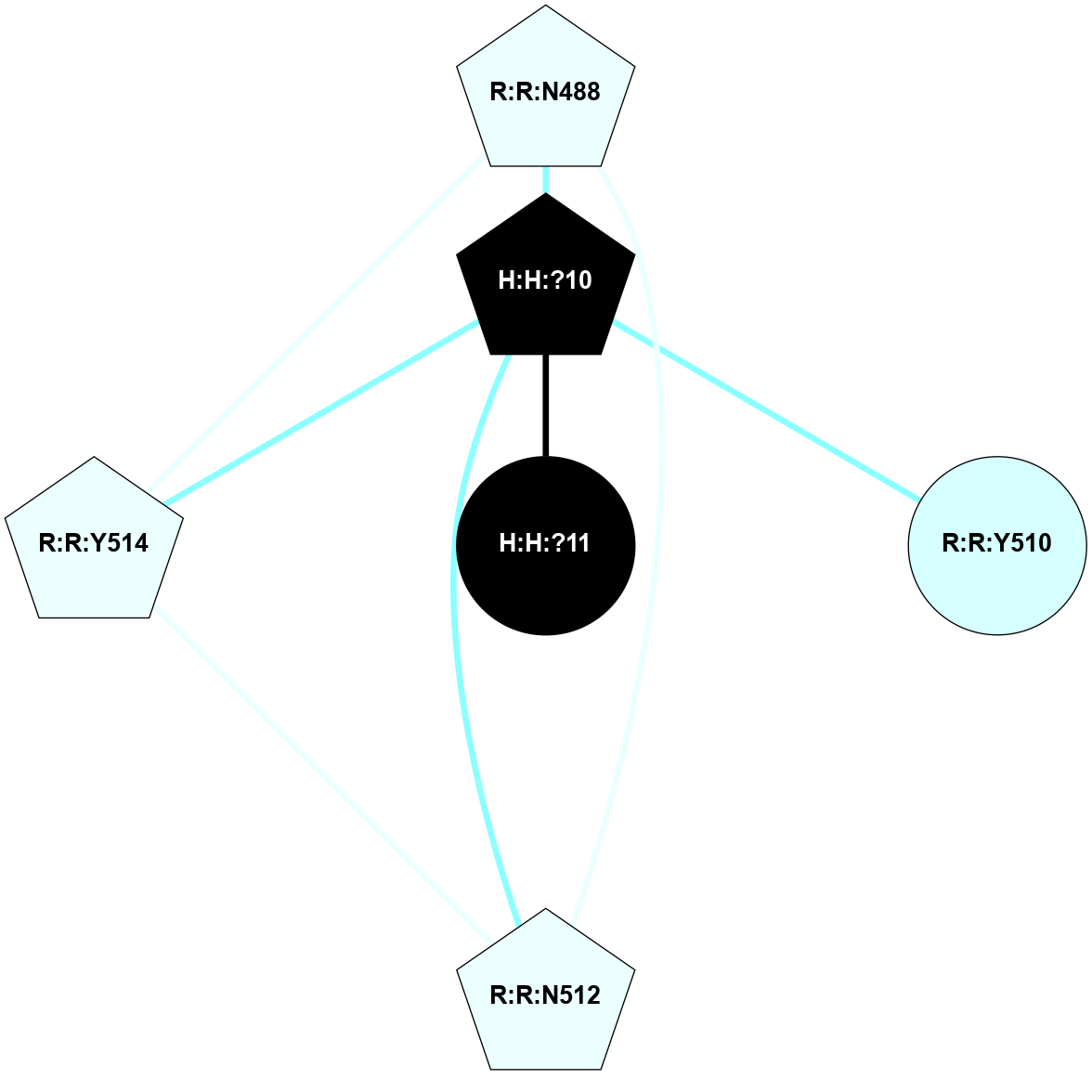
A 2D representation of the interactions of NAG in 8WPG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | H:H:?11 | 43.17 | 22 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?10 | R:R:N488 | 25.79 | 22 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?10 | R:R:Y510 | 8.39 | 22 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?10 | R:R:N512 | 17.19 | 22 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?10 | R:R:Y514 | 17.82 | 22 | Yes | Yes | 0 | 4 | 1 | 2 | | R:R:N488 | R:R:N512 | 6.81 | 22 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N488 | R:R:Y514 | 8.14 | 22 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N512 | R:R:Y514 | 6.98 | 22 | Yes | Yes | 4 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 43.17 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
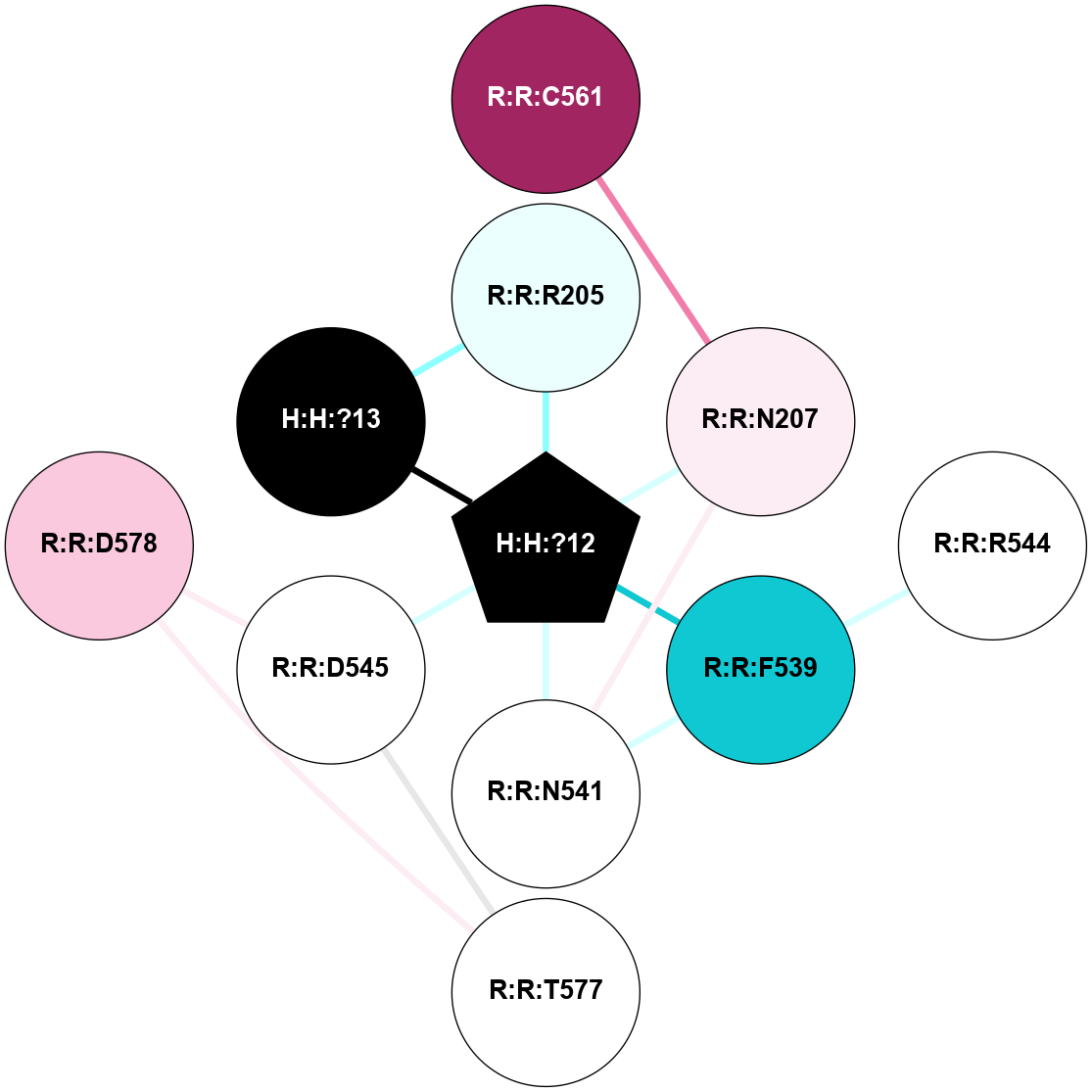
A 2D representation of the interactions of NAG in 8WPG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | H:H:?13 | 38.74 | 14 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?12 | R:R:R205 | 6.52 | 14 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?12 | R:R:N207 | 11.05 | 14 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?12 | R:R:F539 | 5.45 | 14 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?12 | R:R:N541 | 24.56 | 14 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?12 | R:R:D545 | 10.92 | 14 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?13 | R:R:R205 | 21.73 | 14 | No | No | 0 | 4 | 1 | 1 | | R:R:N207 | R:R:N541 | 16.35 | 14 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 7.87 | 14 | No | No | 9 | 6 | 2 | 1 | | R:R:F539 | R:R:N541 | 7.25 | 14 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 24.59 | 14 | No | No | 1 | 5 | 1 | 2 | | R:R:D545 | R:R:T577 | 4.34 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:D545 | R:R:D578 | 9.31 | 0 | No | Yes | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.21 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
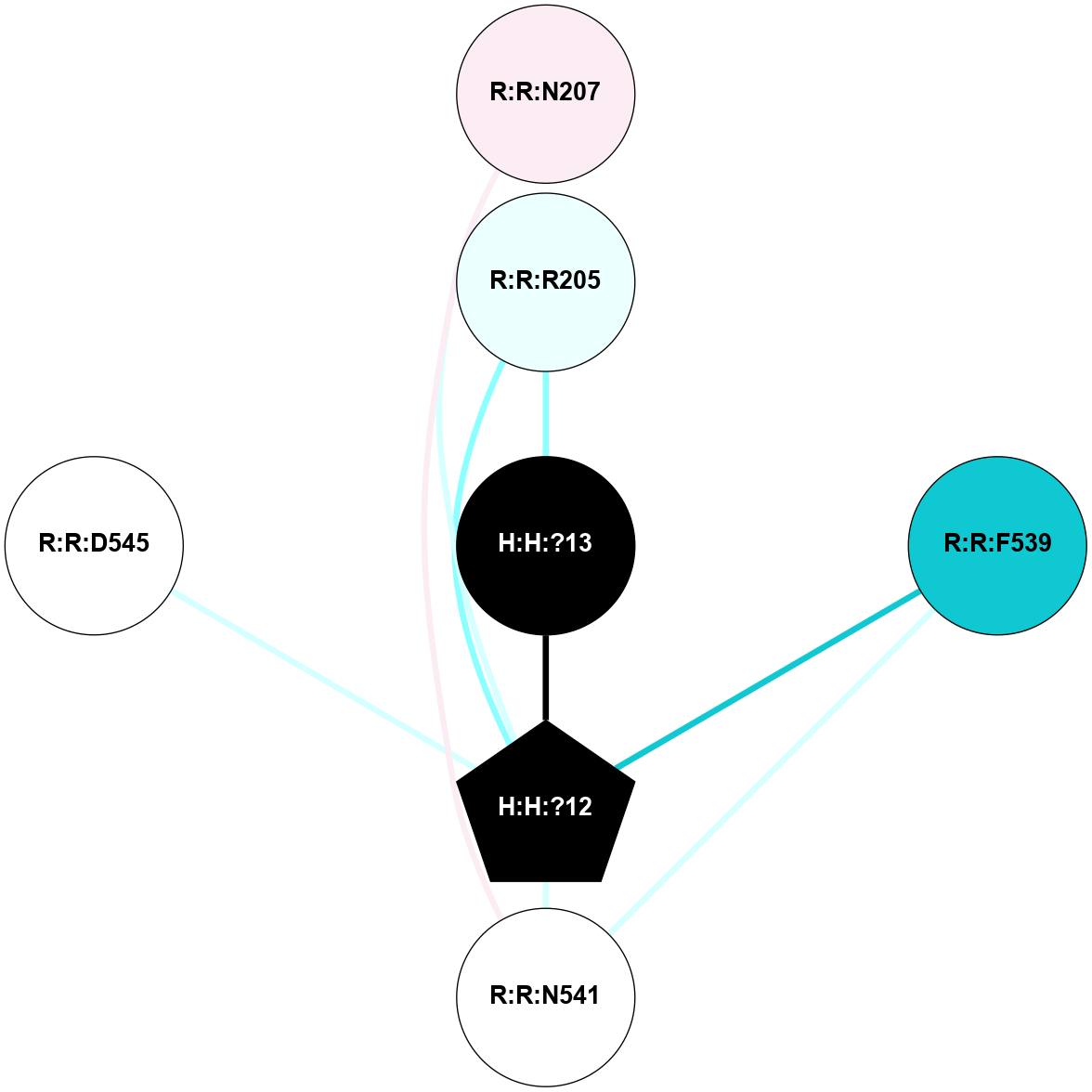
A 2D representation of the interactions of NAG in 8WPG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | H:H:?13 | 38.74 | 14 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?12 | R:R:R205 | 6.52 | 14 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?12 | R:R:N207 | 11.05 | 14 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?12 | R:R:F539 | 5.45 | 14 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?12 | R:R:N541 | 24.56 | 14 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?12 | R:R:D545 | 10.92 | 14 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?13 | R:R:R205 | 21.73 | 14 | No | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 16.35 | 14 | No | No | 6 | 5 | 2 | 2 | | R:R:F539 | R:R:N541 | 7.25 | 14 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 30.23 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8WPU | C | Ion | Calcium Sensing | CaS | Homo Sapiens | - | Tryptophan; Cinacalcet; PO4; Ca | chim(NtGi2L-Gs-CtGq)/Beta1/Gamma2 | 3.1 | 2023-11-22 | doi.org/10.1038/s41422-023-00892-2 |
|
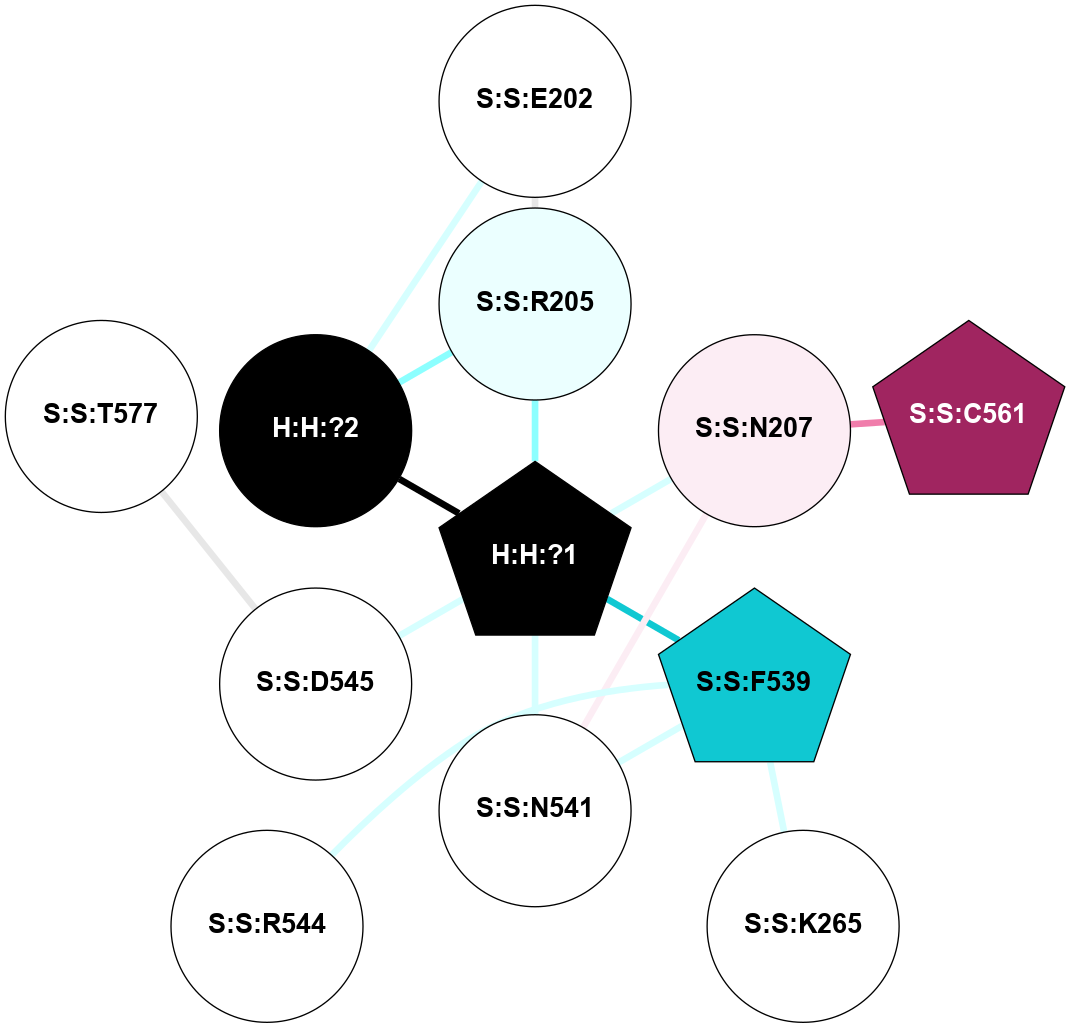
A 2D representation of the interactions of NAG in 8WPU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 33.21 | 6 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | S:S:R205 | 6.52 | 6 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?1 | S:S:N207 | 11.05 | 6 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | S:S:F539 | 7.62 | 6 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?1 | S:S:N541 | 28.24 | 6 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?1 | S:S:D545 | 6.07 | 6 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?2 | S:S:R205 | 5.43 | 6 | No | No | 0 | 4 | 1 | 1 | | S:S:N207 | S:S:N541 | 13.62 | 6 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 11.02 | 6 | Yes | No | 9 | 6 | 2 | 1 | | S:S:F539 | S:S:N541 | 8.46 | 6 | No | No | 1 | 5 | 1 | 1 | | S:S:D545 | S:S:T577 | 4.34 | 0 | No | No | 5 | 5 | 1 | 2 | | S:S:D545 | S:S:D578 | 13.31 | 0 | No | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.45 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
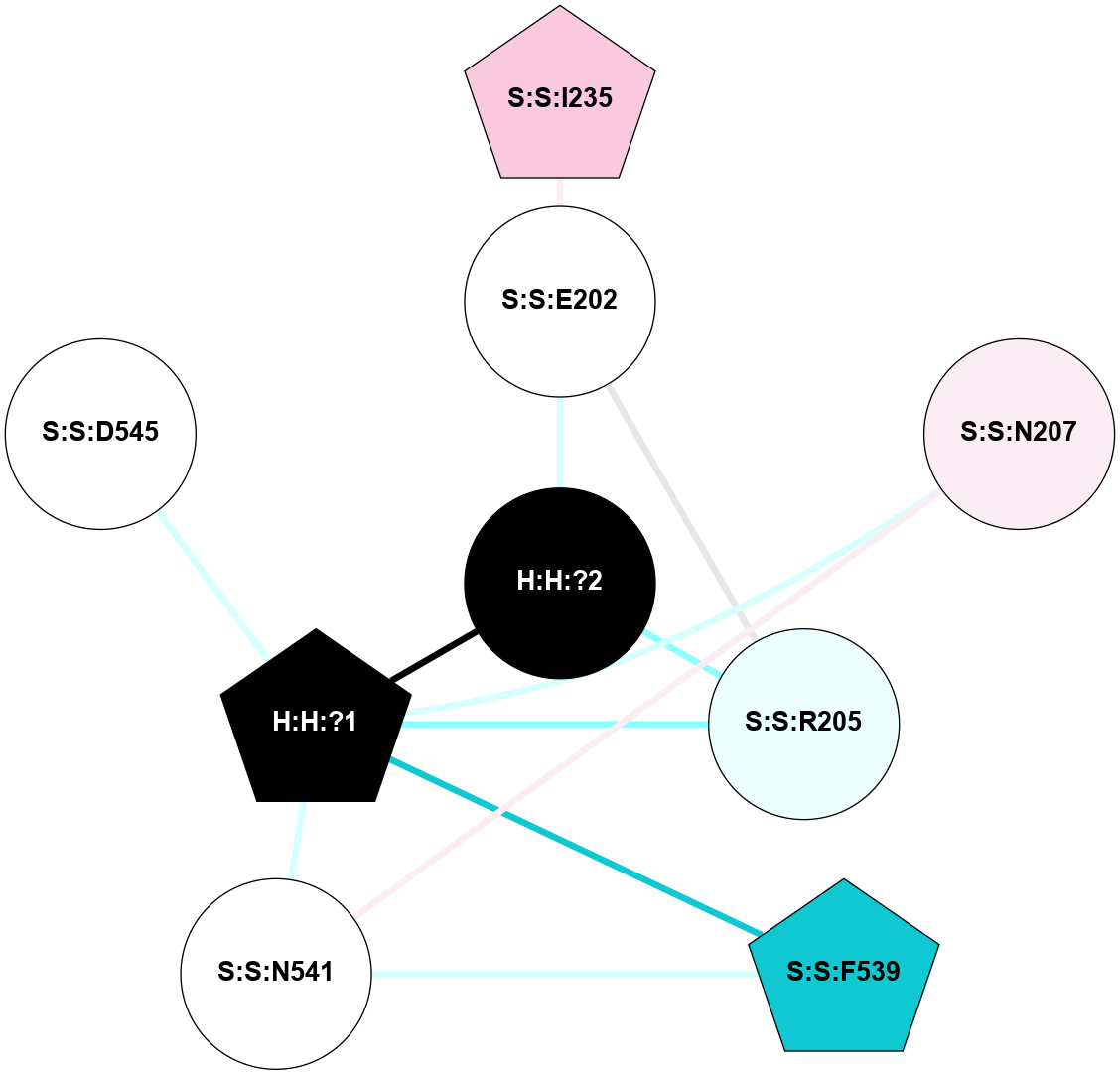
A 2D representation of the interactions of NAG in 8WPU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 33.21 | 6 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | S:S:R205 | 6.52 | 6 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?1 | S:S:N207 | 11.05 | 6 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?1 | S:S:F539 | 7.62 | 6 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?1 | S:S:N541 | 28.24 | 6 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?1 | S:S:D545 | 6.07 | 6 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?2 | S:S:R205 | 5.43 | 6 | No | No | 0 | 4 | 0 | 1 | | S:S:N207 | S:S:N541 | 13.62 | 6 | No | No | 6 | 5 | 2 | 2 | | S:S:F539 | S:S:N541 | 8.46 | 6 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 19.32 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
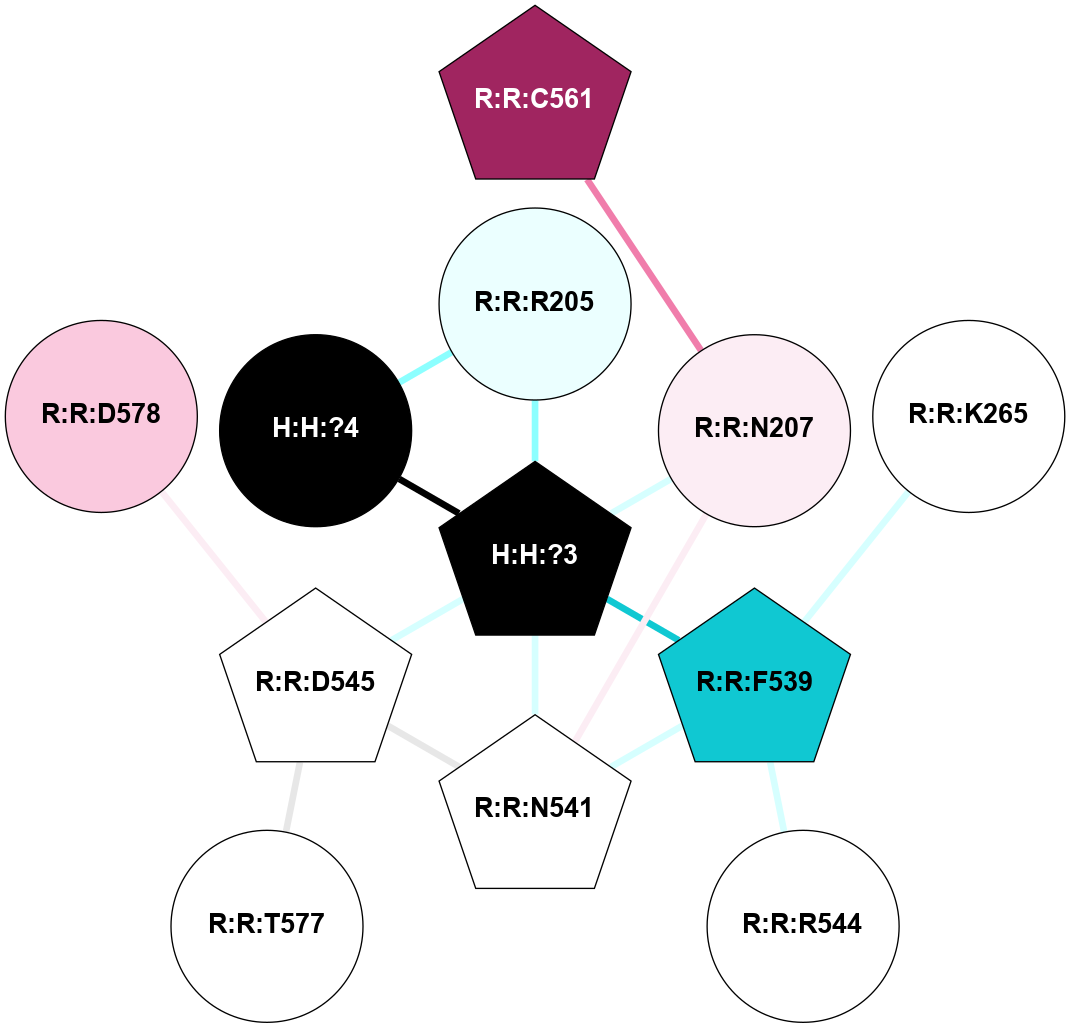
A 2D representation of the interactions of NAG in 8WPU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 35.42 | 16 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | R:R:R205 | 8.69 | 16 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?3 | R:R:N207 | 15.96 | 16 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?3 | R:R:F539 | 5.45 | 16 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?3 | R:R:N541 | 31.93 | 16 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?3 | R:R:D545 | 3.64 | 16 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?4 | R:R:R205 | 19.55 | 16 | No | No | 0 | 4 | 1 | 1 | | R:R:N207 | R:R:N541 | 12.26 | 16 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 7.87 | 16 | No | No | 9 | 6 | 2 | 1 | | R:R:F539 | R:R:N541 | 8.46 | 16 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 18.17 | 16 | No | No | 1 | 5 | 1 | 2 | | R:R:D545 | R:R:D578 | 14.64 | 0 | No | No | 5 | 7 | 1 | 2 | | R:R:D545 | R:R:T577 | 2.89 | 0 | No | No | 5 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.85 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
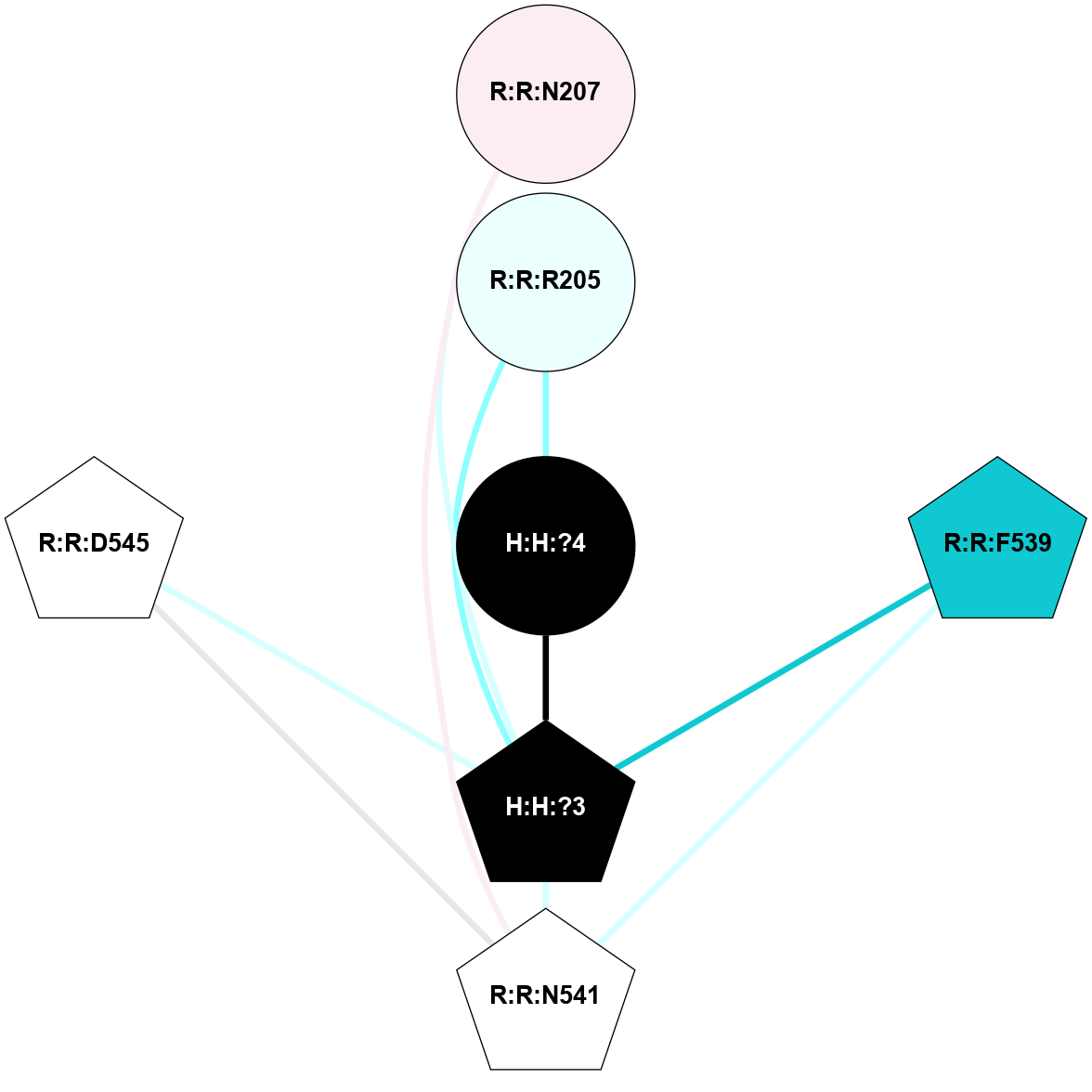
A 2D representation of the interactions of NAG in 8WPU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 35.42 | 16 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?3 | R:R:R205 | 8.69 | 16 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?3 | R:R:N207 | 15.96 | 16 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?3 | R:R:F539 | 5.45 | 16 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?3 | R:R:N541 | 31.93 | 16 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | R:R:D545 | 3.64 | 16 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?4 | R:R:R205 | 19.55 | 16 | No | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 12.26 | 16 | No | No | 6 | 5 | 2 | 2 | | R:R:F539 | R:R:N541 | 8.46 | 16 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 27.48 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
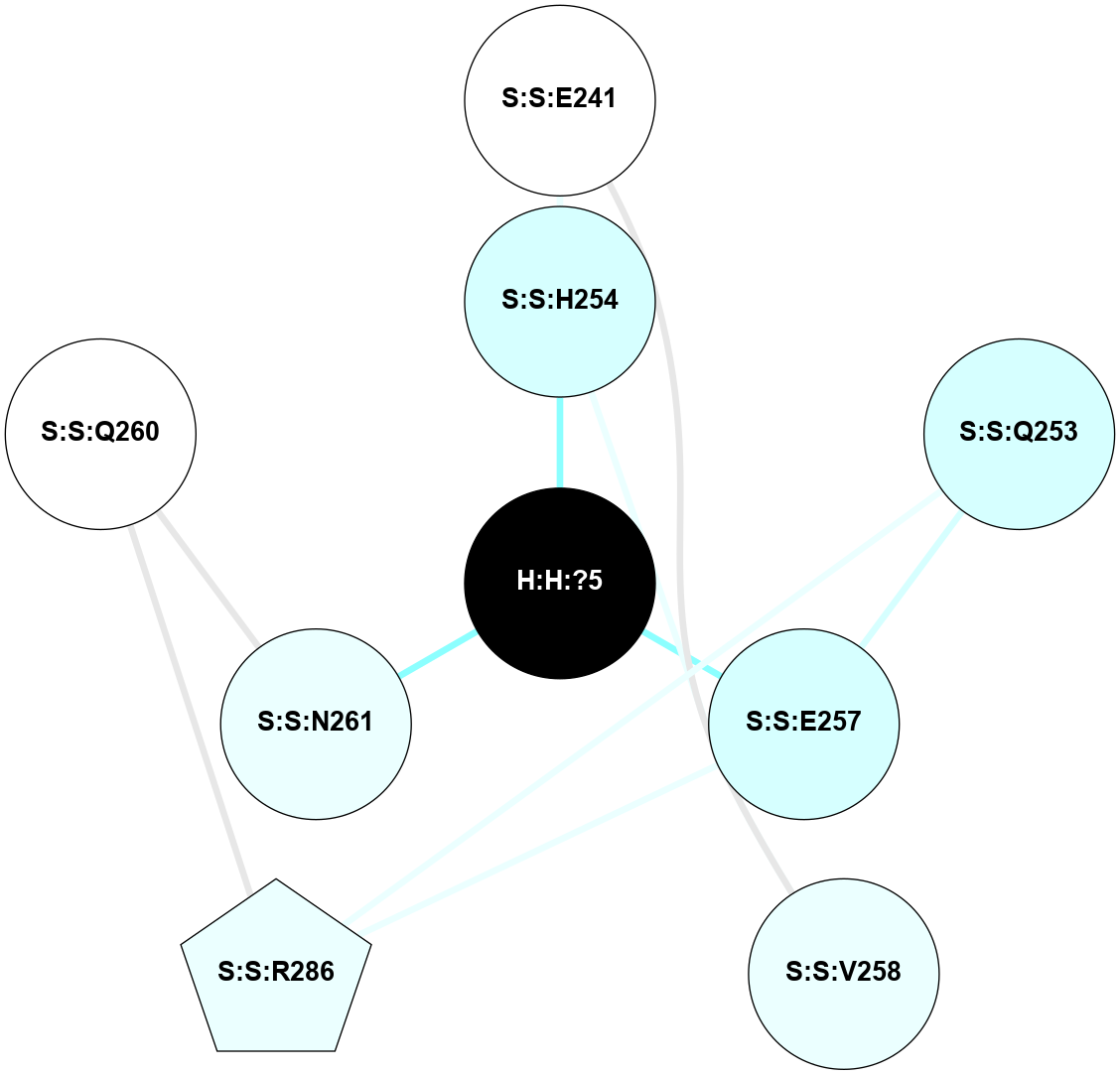
A 2D representation of the interactions of NAG in 8WPU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | S:S:E257 | 3.55 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?5 | S:S:N261 | 24.56 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:E257 | S:S:R286 | 3.49 | 0 | No | Yes | 3 | 4 | 1 | 2 | | S:S:E257 | S:S:Q253 | 2.55 | 0 | No | No | 3 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.05 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
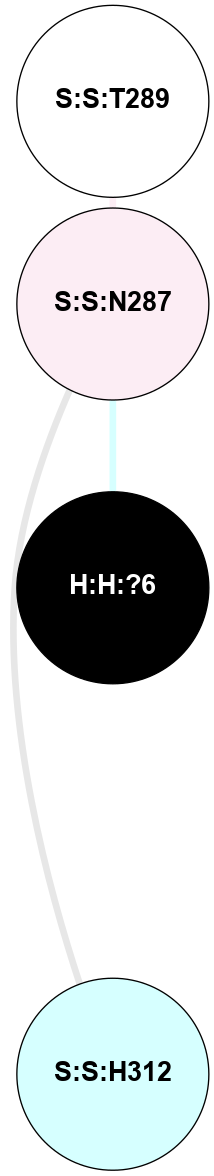
A 2D representation of the interactions of NAG in 8WPU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | S:S:N287 | 29.47 | 0 | No | No | 0 | 6 | 0 | 1 | | S:S:N287 | S:S:T289 | 4.39 | 0 | No | No | 6 | 5 | 1 | 2 | | S:S:H312 | S:S:N287 | 5.1 | 0 | No | No | 3 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 29.47 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
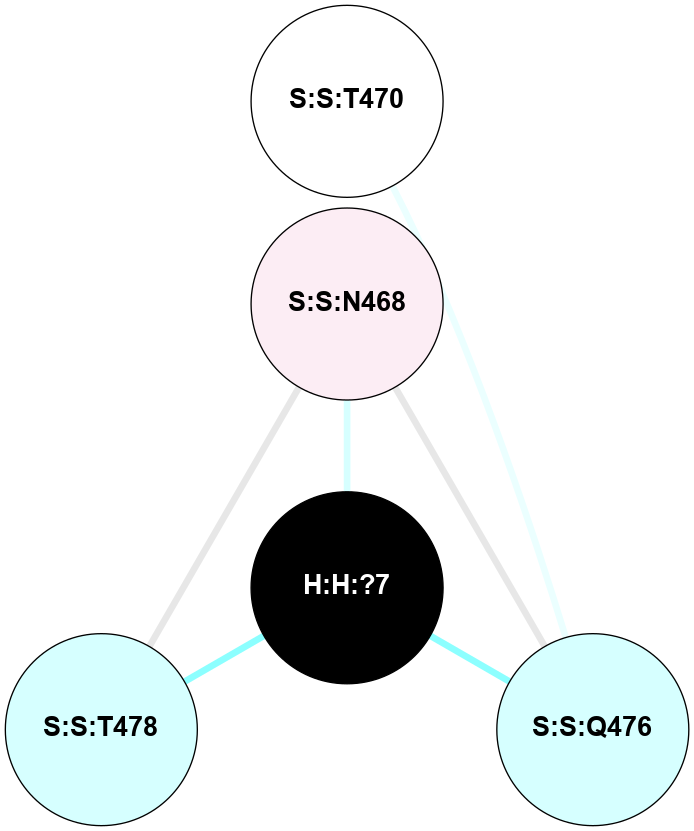
A 2D representation of the interactions of NAG in 8WPU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | S:S:N468 | 29.47 | 36 | No | No | 0 | 6 | 0 | 1 | | H:H:?7 | S:S:Q476 | 5.95 | 36 | No | No | 0 | 3 | 0 | 1 | | H:H:?7 | S:S:T478 | 3.95 | 36 | No | No | 0 | 3 | 0 | 1 | | S:S:N468 | S:S:Q476 | 6.6 | 36 | No | No | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 5.85 | 36 | No | No | 6 | 3 | 1 | 1 | | S:S:Q476 | S:S:T470 | 5.67 | 36 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.12 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
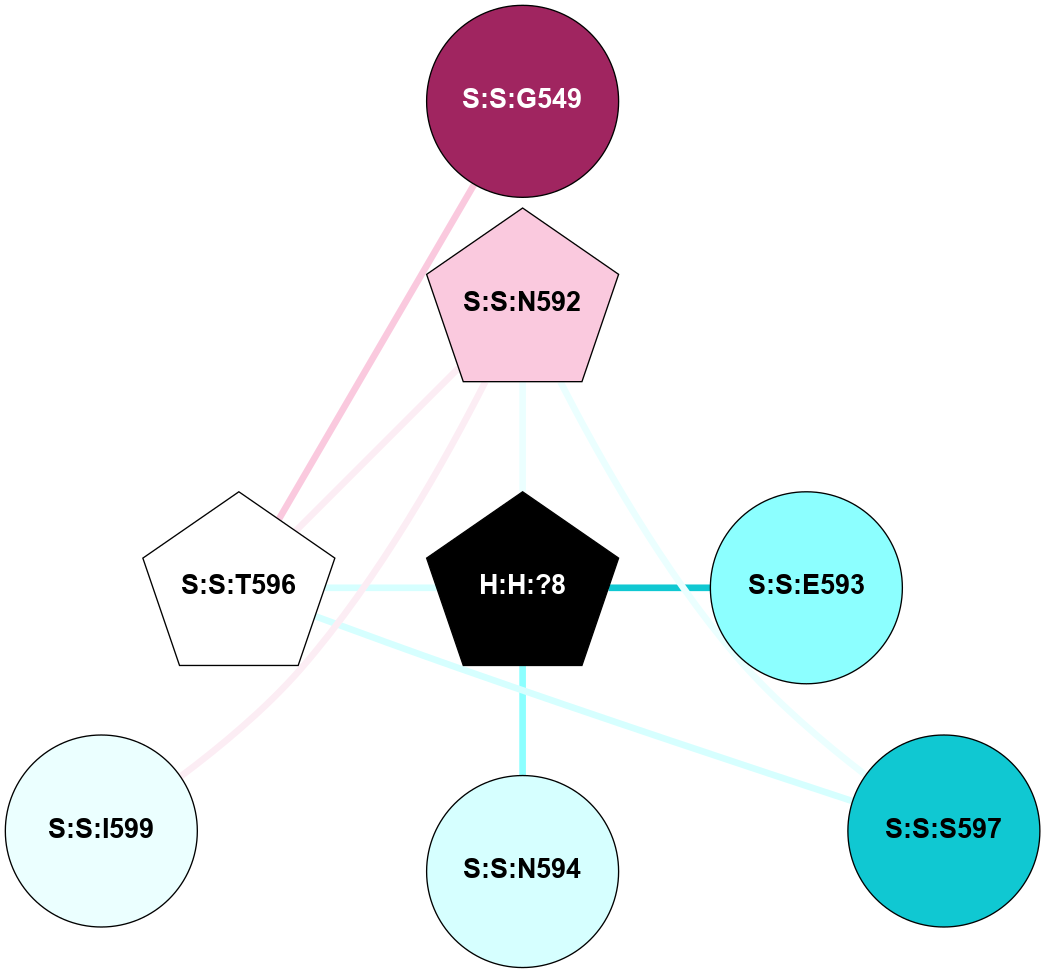
A 2D representation of the interactions of NAG in 8WPU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | S:S:N592 | 13.51 | 0 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?8 | S:S:E593 | 3.55 | 0 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?8 | S:S:N594 | 30.7 | 0 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?8 | S:S:T596 | 15.82 | 0 | Yes | No | 0 | 5 | 0 | 1 | | S:S:N592 | S:S:S597 | 8.94 | 0 | No | No | 7 | 1 | 1 | 2 | | S:S:I599 | S:S:N592 | 14.16 | 0 | No | No | 4 | 7 | 2 | 1 | | S:S:S597 | S:S:T596 | 3.2 | 0 | No | No | 1 | 5 | 2 | 1 | | S:S:G549 | S:S:T596 | 1.82 | 0 | No | No | 9 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.89 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
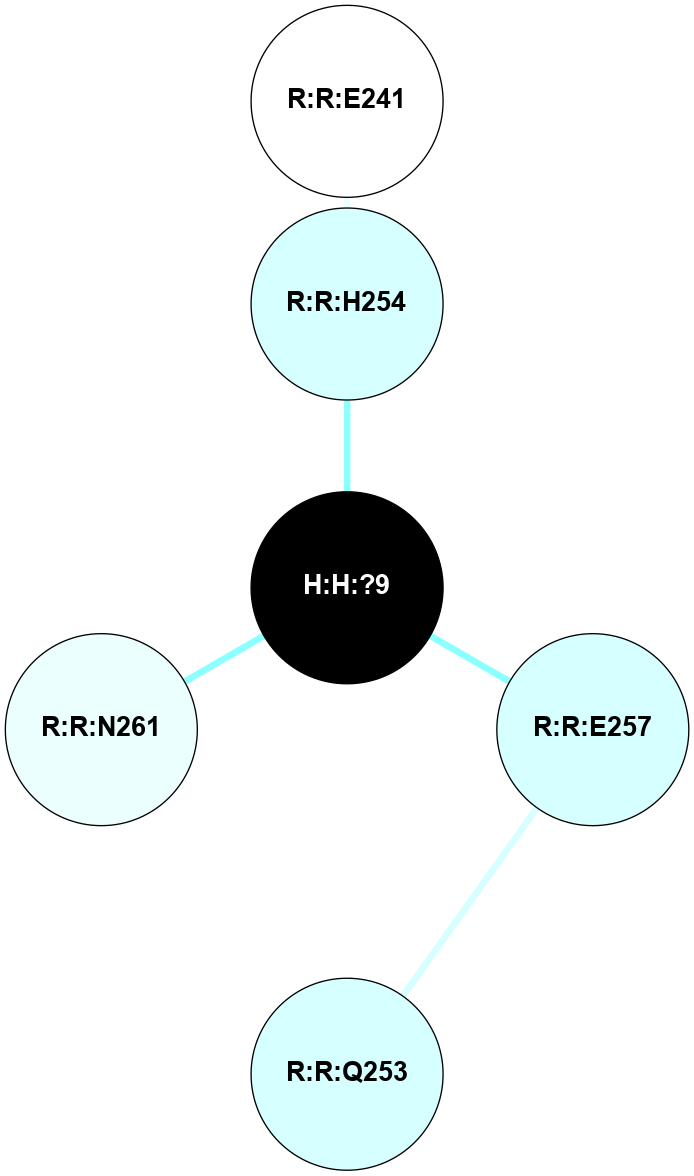
A 2D representation of the interactions of NAG in 8WPU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?9 | R:R:E257 | 5.92 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?9 | R:R:N261 | 24.56 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:E257 | R:R:Q253 | 2.55 | 0 | No | No | 3 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.24 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
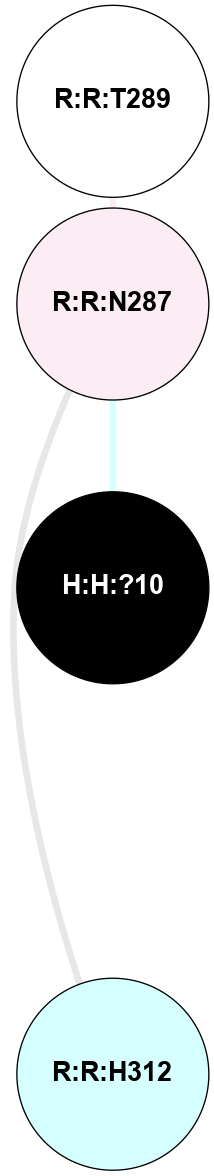
A 2D representation of the interactions of NAG in 8WPU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | R:R:N287 | 28.24 | 0 | No | No | 0 | 6 | 0 | 1 | | R:R:N287 | R:R:T289 | 4.39 | 0 | No | No | 6 | 5 | 1 | 2 | | R:R:H312 | R:R:N287 | 5.1 | 28 | No | No | 3 | 6 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 28.24 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
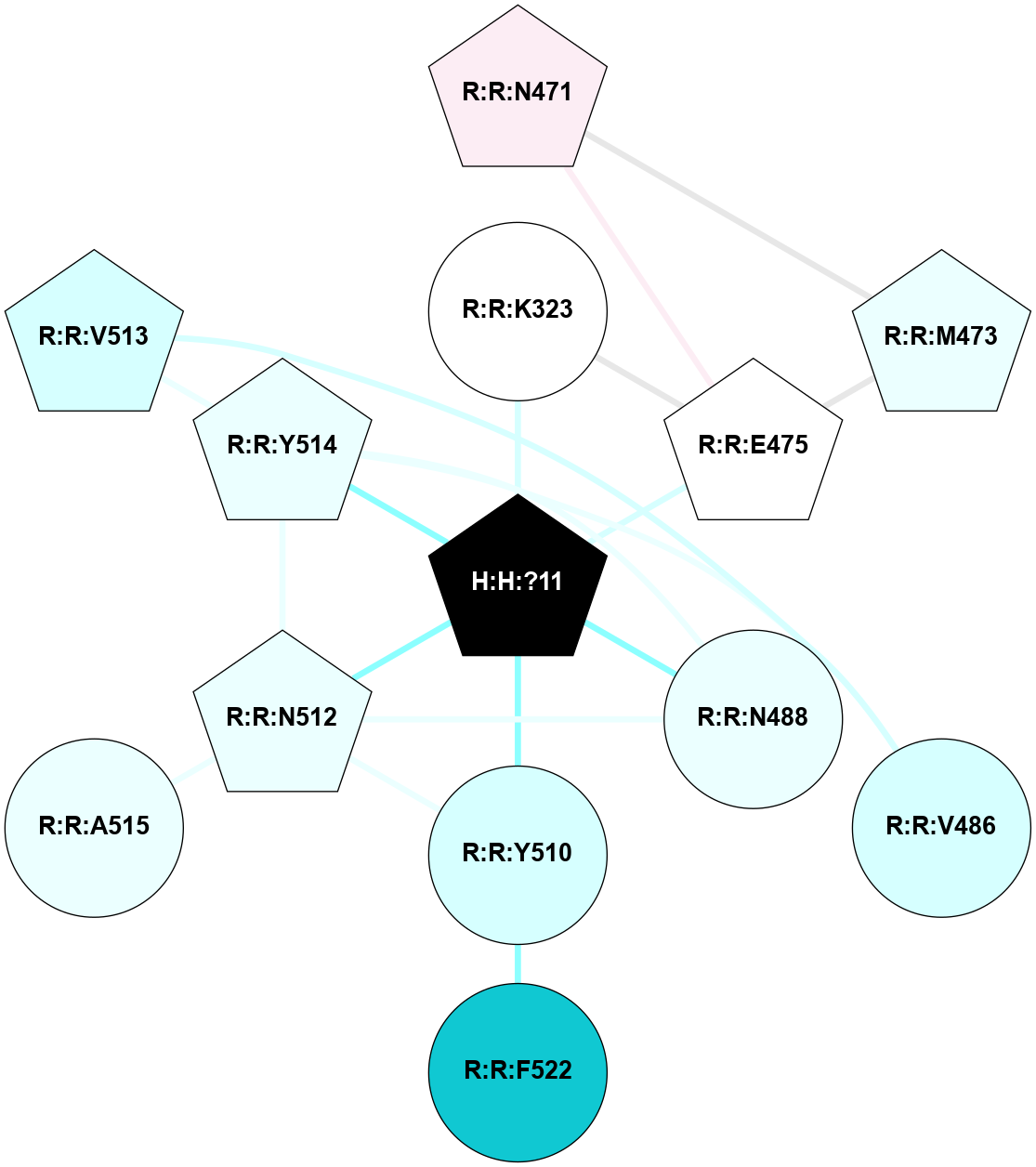
A 2D representation of the interactions of NAG in 8WPU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | R:R:E475 | 4.74 | 37 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?11 | R:R:N488 | 22.1 | 37 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?11 | R:R:Y510 | 8.39 | 37 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?11 | R:R:N512 | 17.19 | 37 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?11 | R:R:Y514 | 14.68 | 37 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:K323 | R:R:N488 | 4.2 | 0 | No | Yes | 5 | 4 | 2 | 1 | | R:R:E475 | R:R:N471 | 5.26 | 0 | No | Yes | 5 | 6 | 1 | 2 | | R:R:V486 | R:R:Y514 | 5.05 | 0 | No | Yes | 3 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 5.45 | 37 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:Y514 | 6.98 | 37 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:F522 | R:R:Y510 | 17.54 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:N512 | R:R:Y514 | 8.14 | 37 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 4.69 | 0 | No | Yes | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 13.42 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
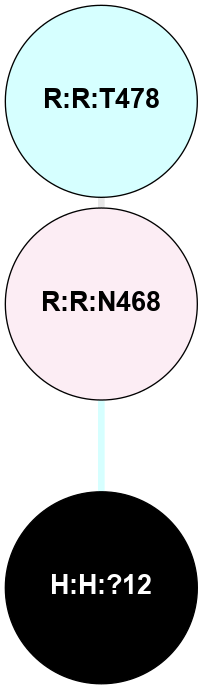
A 2D representation of the interactions of NAG in 8WPU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?12 | R:R:N468 | 25.79 | 49 | No | No | 0 | 6 | 0 | 1 | | H:H:?12 | R:R:Q476 | 17.85 | 49 | No | No | 0 | 3 | 0 | 1 | | R:R:N468 | R:R:Q476 | 6.6 | 49 | No | No | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 5.85 | 49 | No | No | 6 | 3 | 1 | 2 | | R:R:Q476 | R:R:T470 | 5.67 | 49 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 21.82 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
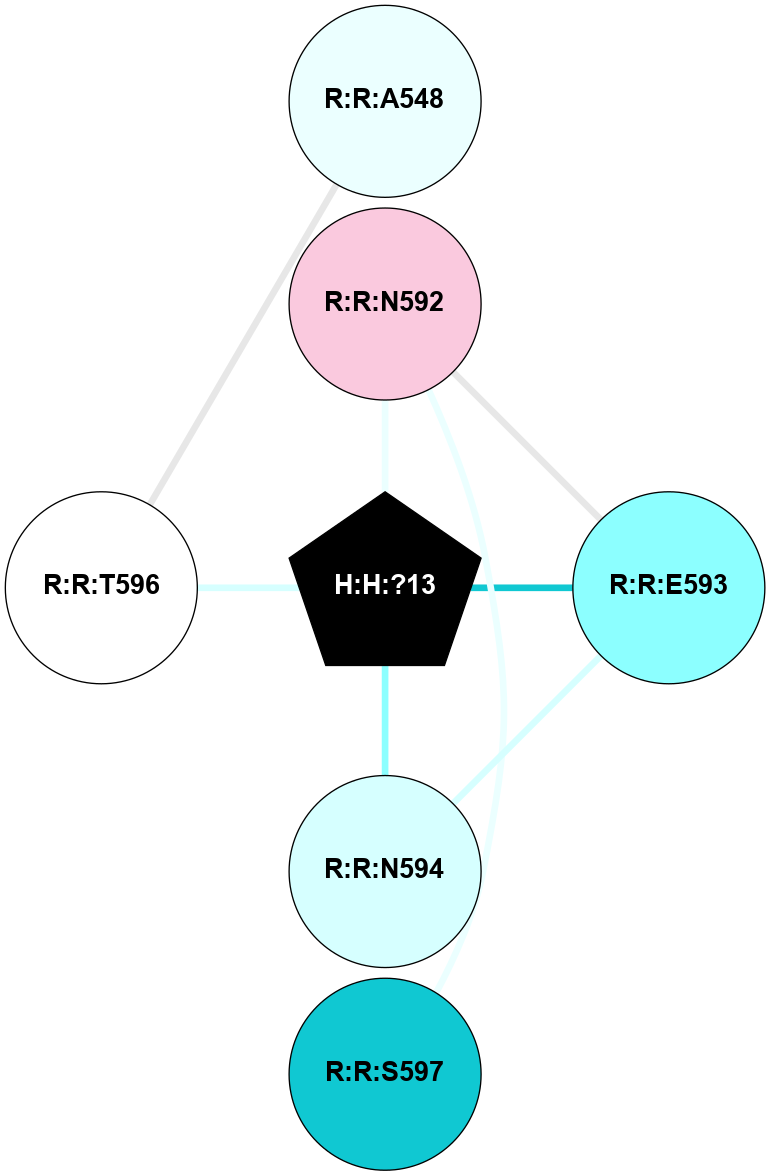
A 2D representation of the interactions of NAG in 8WPU
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?13 | R:R:N592 | 11.05 | 38 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?13 | R:R:E593 | 13.03 | 38 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?13 | R:R:N594 | 28.24 | 38 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?13 | R:R:T596 | 7.91 | 38 | Yes | No | 0 | 5 | 0 | 1 | | R:R:A548 | R:R:T596 | 3.36 | 0 | No | No | 4 | 5 | 2 | 1 | | R:R:E593 | R:R:N592 | 3.94 | 38 | No | No | 2 | 7 | 1 | 1 | | R:R:E593 | R:R:N594 | 6.57 | 38 | No | No | 2 | 3 | 1 | 1 | | R:R:N592 | R:R:S597 | 1.49 | 38 | No | No | 7 | 1 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.06 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8X0B | C | Aminoacid | Metabotropic Glutamate | mGlu5; mGlu5 | Homo Sapiens | Quisqualate | VU0424465 | - | 3.1 | 2024-11-06 | doi.org/10.1038/s41467-024-55439-9 |
|
A 2D representation of the interactions of NAG in 8X0B
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N210 | 29.57 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 29.57 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 8X0B
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:N210 | S:S:R508 | 6.03 | 0 | No | No | 4 | 1 | 1 | 2 | | H:H:?2 | S:S:N210 | 25.87 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 25.87 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8X0C | C | Aminoacid | Metabotropic Glutamate | mGlu5; mGlu5 | Homo Sapiens | Quisqualate | VU0424465 | - | 3.2 | 2024-11-06 | doi.org/10.1038/s41467-024-55439-9 |
|
A 2D representation of the interactions of NAG in 8X0C
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N210 | 28.34 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 28.34 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 8X0C
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:N210 | S:S:R508 | 6.03 | 0 | No | No | 4 | 1 | 1 | 2 | | H:H:?2 | S:S:N210 | 27.11 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.11 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8X0D | C | Aminoacid | Metabotropic Glutamate | mGlu5; mGlu5 | Homo Sapiens | Quisqualate | VU0424465 | - | 3.5 | 2024-11-06 | doi.org/10.1038/s41467-024-55439-9 |
|

A 2D representation of the interactions of NAG in 8X0D
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N210 | 28.34 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:D496 | R:R:V499 | 1.46 | 0 | No | No | 3 | 7 | 1 | 2 | | H:H:?1 | R:R:D496 | 1.22 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.78 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
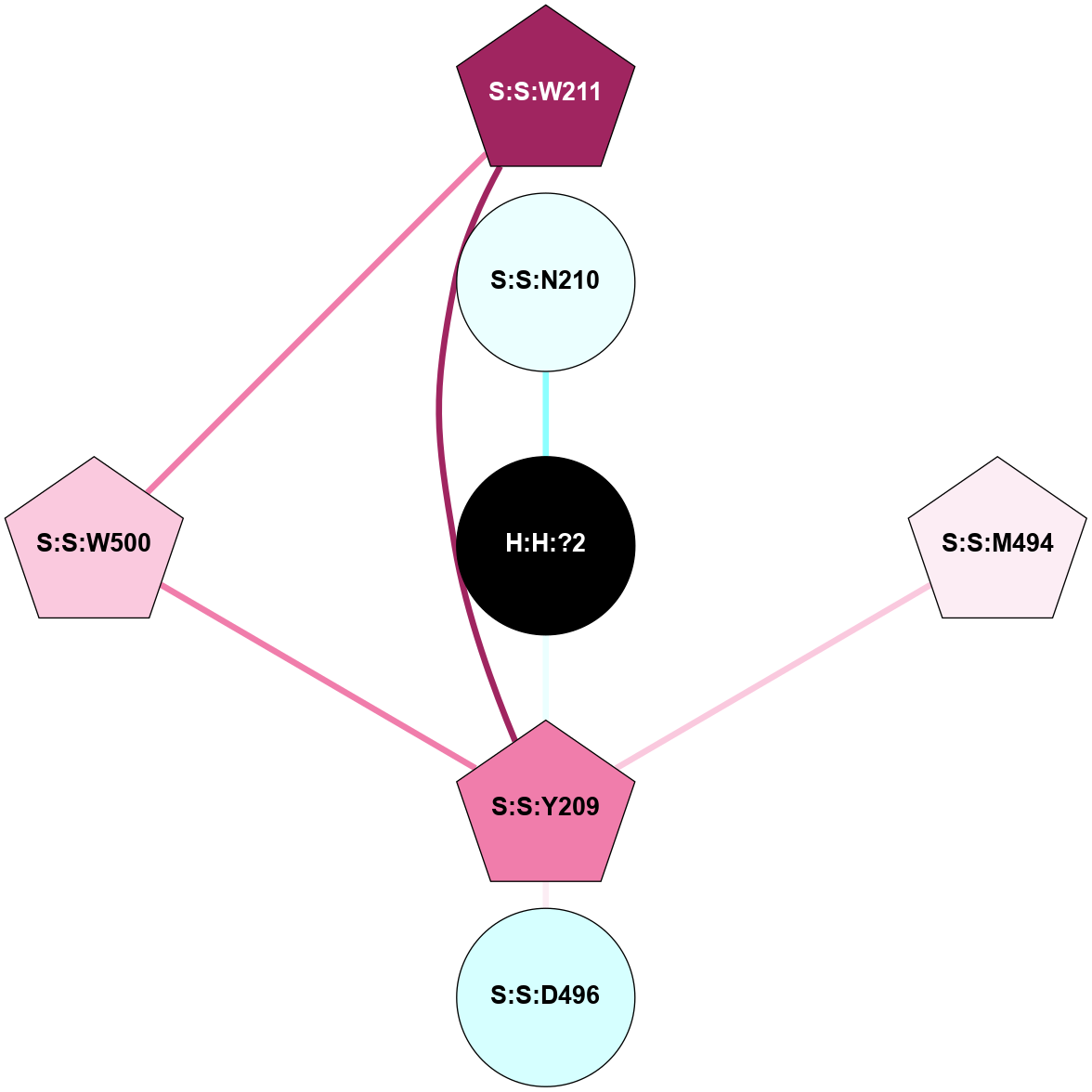
A 2D representation of the interactions of NAG in 8X0D
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:W211 | S:S:Y209 | 12.54 | 2 | Yes | Yes | 9 | 8 | 2 | 1 | | S:S:M494 | S:S:Y209 | 7.18 | 2 | Yes | Yes | 6 | 8 | 2 | 1 | | S:S:D496 | S:S:Y209 | 11.49 | 0 | No | Yes | 3 | 8 | 2 | 1 | | S:S:W500 | S:S:Y209 | 15.43 | 2 | Yes | Yes | 7 | 8 | 2 | 1 | | H:H:?2 | S:S:N210 | 32.04 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:W211 | S:S:W500 | 4.69 | 2 | Yes | Yes | 9 | 7 | 2 | 2 | | H:H:?2 | S:S:Y209 | 2.1 | 0 | No | Yes | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.07 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8X0E | C | Aminoacid | Metabotropic Glutamate | mGlu5; mGlu5 | Homo Sapiens | Quisqualate | - | - | 3.4 | 2024-11-06 | doi.org/10.1038/s41467-024-55439-9 |
|
A 2D representation of the interactions of NAG in 8X0E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N210 | 27.11 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N210 | R:R:R508 | 2.41 | 0 | No | Yes | 4 | 1 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.11 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 8X0E
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:N210 | S:S:R508 | 10.85 | 0 | No | No | 4 | 1 | 1 | 2 | | H:H:?2 | S:S:N210 | 28.34 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 28.34 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8X0F | C | Aminoacid | Metabotropic Glutamate | mGlu5; mGlu5 | Homo Sapiens | Quisqualate | VU0424465 | - | 3.3 | 2024-11-06 | doi.org/10.1038/s41467-024-55439-9 |
|
A 2D representation of the interactions of NAG in 8X0F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:D496 | R:R:Y209 | 11.49 | 0 | No | Yes | 3 | 8 | 1 | 2 | | H:H:?1 | R:R:N210 | 27.11 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:D496 | 1.22 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.16 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 8X0F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:R208 | S:S:Y209 | 3.09 | 1 | No | Yes | 5 | 8 | 2 | 2 | | S:S:D496 | S:S:R208 | 8.34 | 1 | No | No | 3 | 5 | 1 | 2 | | S:S:D496 | S:S:Y209 | 10.34 | 1 | No | Yes | 3 | 8 | 1 | 2 | | H:H:?2 | S:S:N210 | 28.34 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?2 | S:S:D496 | 1.22 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.78 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8X0G | C | Aminoacid | Metabotropic Glutamate | mGlu5; mGlu5 | Homo Sapiens | Quisqualate | - | - | 3 | 2024-11-06 | doi.org/10.1038/s41467-024-55439-9 |
|
A 2D representation of the interactions of NAG in 8X0G
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N210 | R:R:R508 | 7.23 | 0 | No | No | 4 | 1 | 1 | 2 | | H:H:?2 | R:R:N210 | 27.11 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?2 | S:S:N210 | 27.11 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.11 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 8X0H | C | Aminoacid | Metabotropic Glutamate | mGlu5; mGlu5 | Homo Sapiens | Quisqualate | - | - | 4.1 | 2024-11-06 | doi.org/10.1038/s41467-024-55439-9 |
|
A 2D representation of the interactions of NAG in 8X0H
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N210 | 30.8 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 30.80 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 8X0H
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | S:S:N210 | 27.11 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N505 | S:S:W500 | 3.39 | 0 | No | Yes | 4 | 7 | 1 | 2 | | S:S:N505 | S:S:R508 | 4.82 | 0 | No | No | 4 | 1 | 1 | 2 | | H:H:?2 | S:S:N505 | 2.46 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.79 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9ASB | C | Ion | Calcium Sensing | CaS | Homo Sapiens | Ca | Ca; Ca; Cyclomethyltryptophan; 9IG; PO4 | chim(NtGi1-Gs-CtGq)/Beta1/Gamma2 | 3.4 | 2024-04-17 | doi.org/10.1038/s41586-024-07331-1 |
|
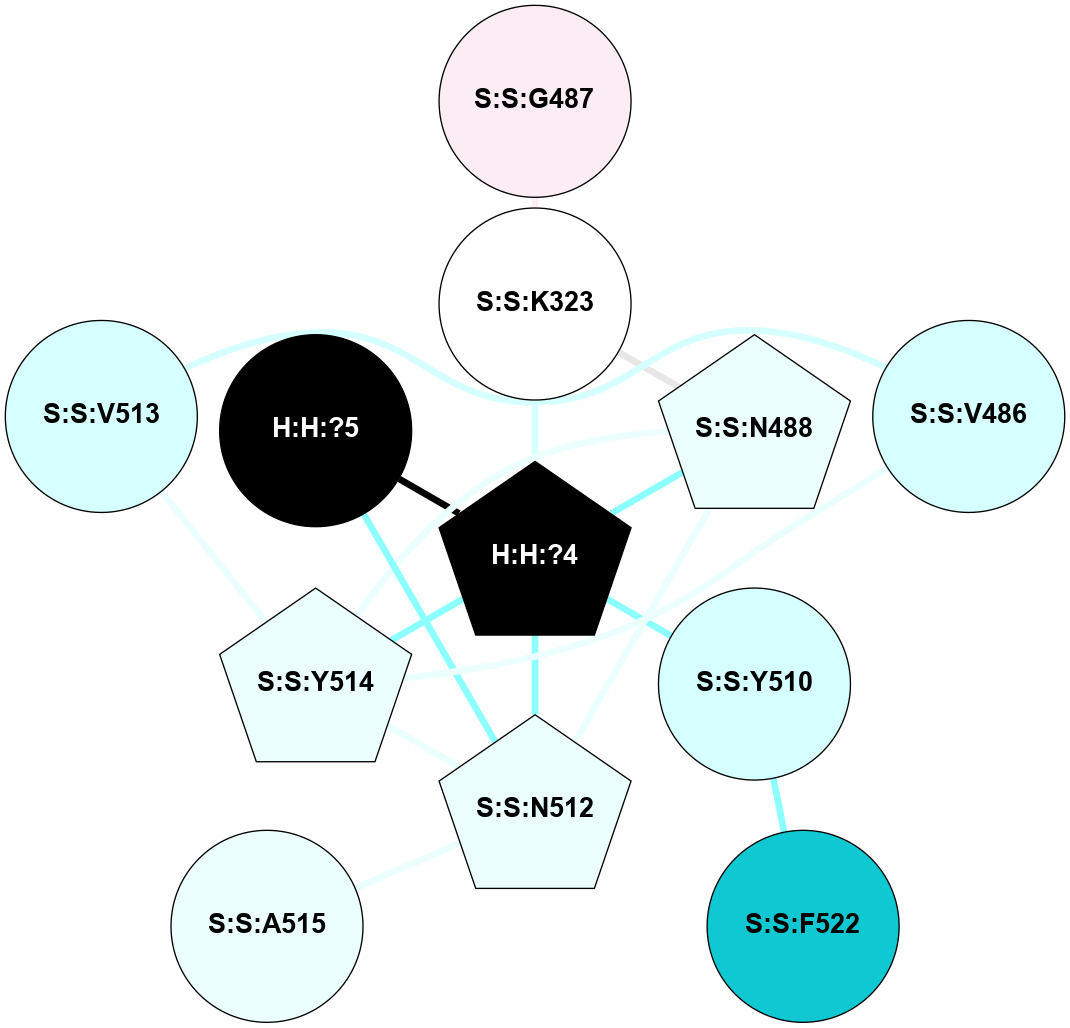
A 2D representation of the interactions of NAG in 9ASB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:G487 | S:S:K323 | 3.49 | 0 | No | No | 6 | 5 | 2 | 1 | | S:S:K323 | S:S:N488 | 4.2 | 23 | No | Yes | 5 | 4 | 1 | 1 | | H:H:?4 | S:S:K323 | 7.59 | 23 | Yes | No | 0 | 5 | 0 | 1 | | S:S:V486 | S:S:V513 | 3.21 | 23 | No | No | 3 | 3 | 2 | 2 | | S:S:V486 | S:S:Y514 | 10.09 | 23 | No | Yes | 3 | 4 | 2 | 1 | | S:S:N488 | S:S:N512 | 6.81 | 23 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:Y514 | 4.65 | 23 | Yes | Yes | 4 | 4 | 1 | 1 | | H:H:?4 | S:S:N488 | 22.18 | 23 | Yes | Yes | 0 | 4 | 0 | 1 | | S:S:F522 | S:S:Y510 | 20.63 | 0 | No | No | 1 | 3 | 2 | 1 | | H:H:?4 | S:S:Y510 | 11.57 | 23 | Yes | No | 0 | 3 | 0 | 1 | | S:S:N512 | S:S:Y514 | 3.49 | 23 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:A515 | S:S:N512 | 6.25 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?4 | S:S:N512 | 17.25 | 23 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?5 | S:S:N512 | 3.7 | 23 | No | Yes | 0 | 4 | 1 | 1 | | S:S:V513 | S:S:Y514 | 5.05 | 23 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?4 | S:S:Y514 | 8.42 | 23 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?4 | H:H:?5 | 37.89 | 23 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 17.48 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
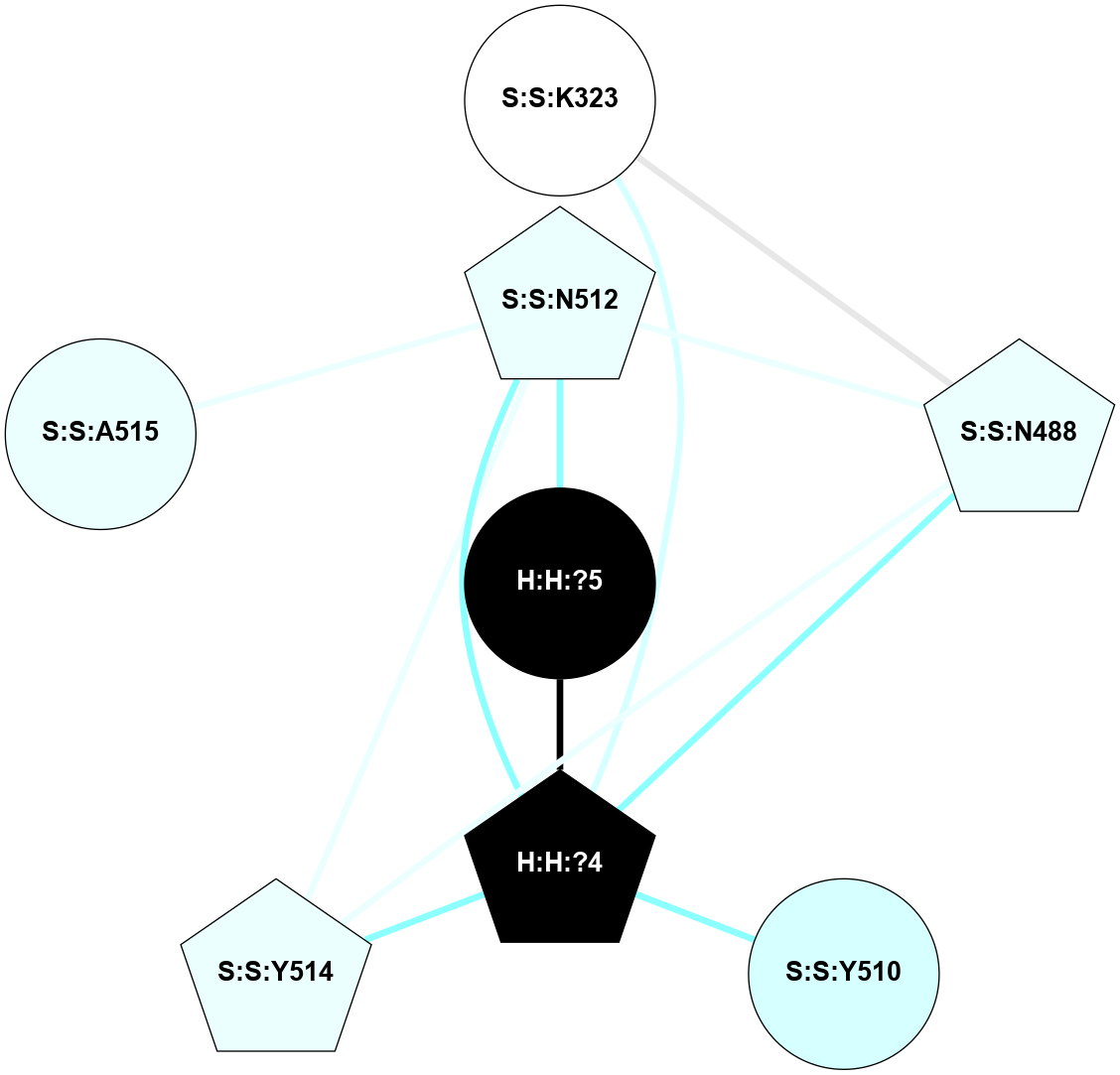
A 2D representation of the interactions of NAG in 9ASB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:K323 | S:S:N488 | 4.2 | 23 | No | Yes | 5 | 4 | 2 | 2 | | H:H:?4 | S:S:K323 | 7.59 | 23 | Yes | No | 0 | 5 | 1 | 2 | | S:S:N488 | S:S:N512 | 6.81 | 23 | Yes | Yes | 4 | 4 | 2 | 1 | | S:S:N488 | S:S:Y514 | 4.65 | 23 | Yes | Yes | 4 | 4 | 2 | 2 | | H:H:?4 | S:S:N488 | 22.18 | 23 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?4 | S:S:Y510 | 11.57 | 23 | Yes | No | 0 | 3 | 1 | 2 | | S:S:N512 | S:S:Y514 | 3.49 | 23 | Yes | Yes | 4 | 4 | 1 | 2 | | S:S:A515 | S:S:N512 | 6.25 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?4 | S:S:N512 | 17.25 | 23 | Yes | Yes | 0 | 4 | 1 | 1 | | H:H:?5 | S:S:N512 | 3.7 | 23 | No | Yes | 0 | 4 | 0 | 1 | | H:H:?4 | S:S:Y514 | 8.42 | 23 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?4 | H:H:?5 | 37.89 | 23 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 20.80 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
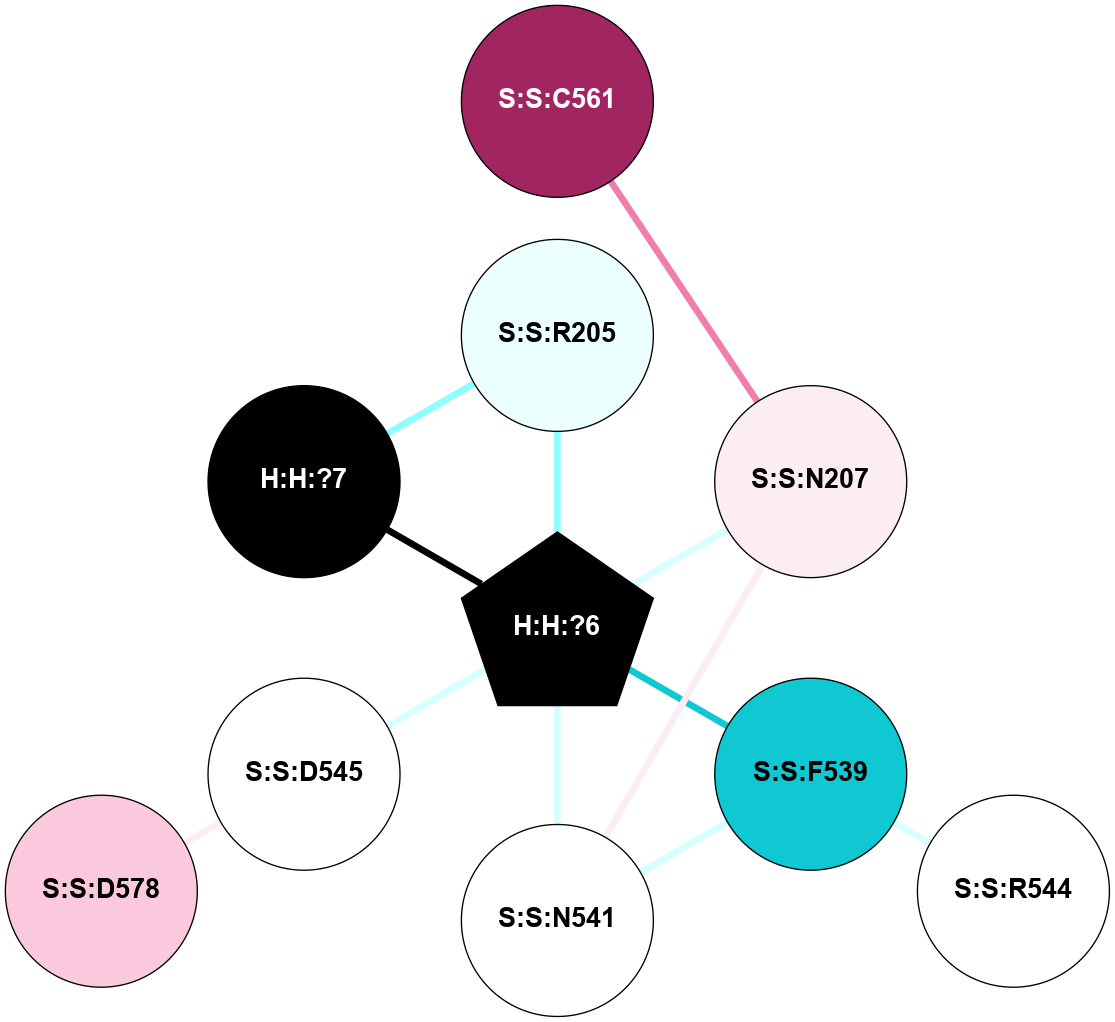
A 2D representation of the interactions of NAG in 9ASB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | S:S:R205 | 13.08 | 3 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?7 | S:S:R205 | 8.72 | 3 | No | No | 0 | 4 | 1 | 1 | | S:S:N207 | S:S:N541 | 16.35 | 3 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 11.02 | 3 | No | No | 9 | 6 | 2 | 1 | | H:H:?6 | S:S:N207 | 12.32 | 3 | Yes | No | 0 | 6 | 0 | 1 | | S:S:F539 | S:S:N541 | 8.46 | 3 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 29.93 | 3 | No | No | 1 | 5 | 1 | 2 | | H:H:?6 | S:S:F539 | 6.56 | 3 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?6 | S:S:N541 | 29.57 | 3 | Yes | No | 0 | 5 | 0 | 1 | | S:S:D545 | S:S:D578 | 13.31 | 0 | No | No | 5 | 7 | 1 | 2 | | H:H:?6 | S:S:D545 | 9.74 | 3 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?6 | H:H:?7 | 42.35 | 3 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.94 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9ASB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | S:S:R205 | 13.08 | 3 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?7 | S:S:R205 | 8.72 | 3 | No | No | 0 | 4 | 0 | 1 | | S:S:N207 | S:S:N541 | 16.35 | 3 | No | No | 6 | 5 | 2 | 2 | | H:H:?6 | S:S:N207 | 12.32 | 3 | Yes | No | 0 | 6 | 1 | 2 | | S:S:F539 | S:S:N541 | 8.46 | 3 | No | No | 1 | 5 | 2 | 2 | | H:H:?6 | S:S:F539 | 6.56 | 3 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?6 | S:S:N541 | 29.57 | 3 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?6 | S:S:D545 | 9.74 | 3 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?6 | H:H:?7 | 42.35 | 3 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 25.54 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
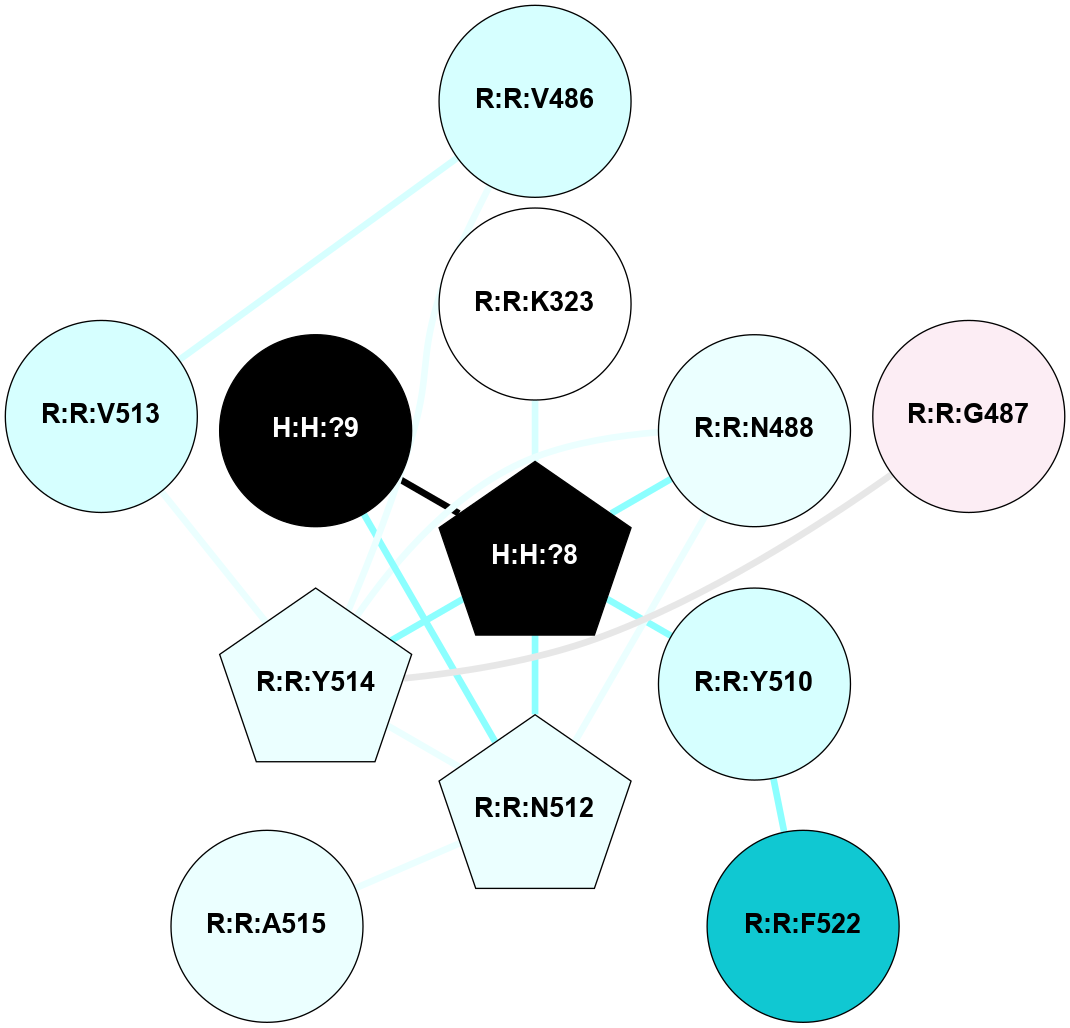
A 2D representation of the interactions of NAG in 9ASB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | R:R:K323 | 5.06 | 27 | Yes | No | 0 | 5 | 0 | 1 | | R:R:V486 | R:R:V513 | 3.21 | 27 | No | No | 3 | 3 | 2 | 2 | | R:R:V486 | R:R:Y514 | 3.79 | 27 | No | Yes | 3 | 4 | 2 | 1 | | R:R:G487 | R:R:Y514 | 2.9 | 0 | No | Yes | 6 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 4.09 | 27 | No | Yes | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:Y514 | 5.81 | 27 | No | Yes | 4 | 4 | 1 | 1 | | H:H:?8 | R:R:N488 | 27.11 | 27 | Yes | No | 0 | 4 | 0 | 1 | | R:R:F522 | R:R:Y510 | 18.57 | 0 | No | No | 1 | 3 | 2 | 1 | | H:H:?8 | R:R:Y510 | 11.57 | 27 | Yes | No | 0 | 3 | 0 | 1 | | R:R:N512 | R:R:Y514 | 4.65 | 27 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 4.69 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?8 | R:R:N512 | 20.95 | 27 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?9 | R:R:N512 | 3.7 | 27 | No | Yes | 0 | 4 | 1 | 1 | | R:R:V513 | R:R:Y514 | 7.57 | 27 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?8 | R:R:Y514 | 15.78 | 27 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?8 | H:H:?9 | 41.23 | 27 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 20.28 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9ASB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?8 | R:R:K323 | 5.06 | 27 | Yes | No | 0 | 5 | 1 | 2 | | R:R:N488 | R:R:N512 | 4.09 | 27 | No | Yes | 4 | 4 | 2 | 1 | | R:R:N488 | R:R:Y514 | 5.81 | 27 | No | Yes | 4 | 4 | 2 | 2 | | H:H:?8 | R:R:N488 | 27.11 | 27 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?8 | R:R:Y510 | 11.57 | 27 | Yes | No | 0 | 3 | 1 | 2 | | R:R:N512 | R:R:Y514 | 4.65 | 27 | Yes | Yes | 4 | 4 | 1 | 2 | | R:R:A515 | R:R:N512 | 4.69 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?8 | R:R:N512 | 20.95 | 27 | Yes | Yes | 0 | 4 | 1 | 1 | | H:H:?9 | R:R:N512 | 3.7 | 27 | No | Yes | 0 | 4 | 0 | 1 | | H:H:?8 | R:R:Y514 | 15.78 | 27 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?8 | H:H:?9 | 41.23 | 27 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 22.46 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
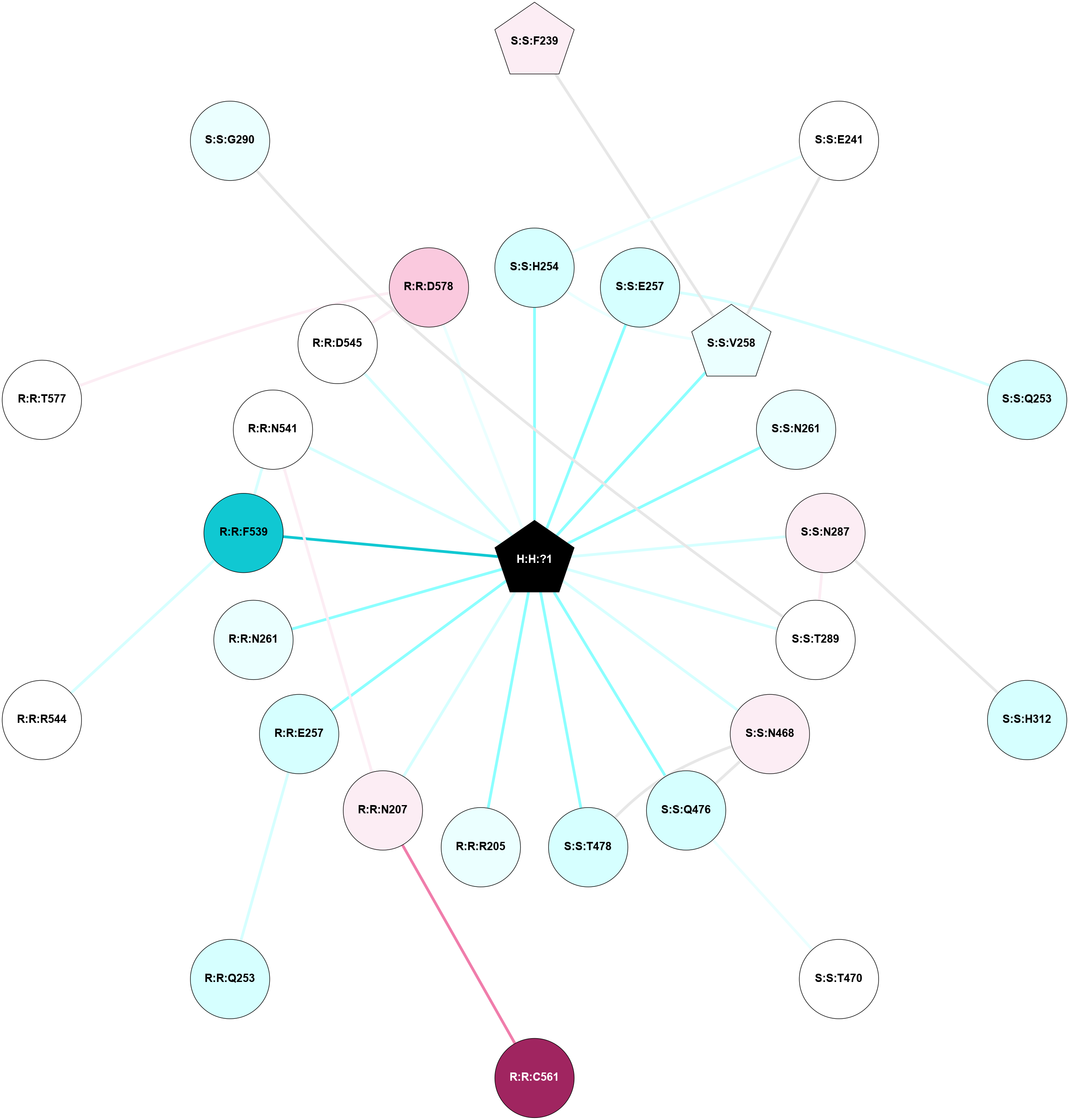
A 2D representation of the interactions of NAG in 9ASB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:F239 | S:S:V258 | 5.24 | 3 | Yes | Yes | 6 | 4 | 2 | 1 | | S:S:E241 | S:S:H254 | 7.39 | 3 | No | No | 5 | 3 | 2 | 1 | | S:S:E241 | S:S:V258 | 9.98 | 3 | No | Yes | 5 | 4 | 2 | 1 | | S:S:E257 | S:S:Q253 | 7.65 | 0 | No | No | 3 | 3 | 1 | 2 | | S:S:H254 | S:S:V258 | 6.92 | 3 | No | Yes | 3 | 4 | 1 | 1 | | H:H:?1 | S:S:H254 | 5.77 | 3 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | S:S:E257 | 10.7 | 3 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | S:S:V258 | 4.01 | 3 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | S:S:N261 | 25.87 | 3 | Yes | No | 0 | 4 | 0 | 1 | | S:S:N287 | S:S:T289 | 2.92 | 3 | No | No | 6 | 5 | 1 | 1 | | S:S:H312 | S:S:N287 | 6.38 | 0 | No | No | 3 | 6 | 2 | 1 | | H:H:?1 | S:S:N287 | 29.57 | 3 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | S:S:T289 | 3.97 | 3 | Yes | No | 0 | 5 | 0 | 1 | | S:S:N468 | S:S:Q476 | 10.56 | 3 | No | No | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 5.85 | 3 | No | No | 6 | 3 | 1 | 1 | | H:H:?1 | S:S:N468 | 28.34 | 3 | Yes | No | 0 | 6 | 0 | 1 | | S:S:Q476 | S:S:T470 | 4.25 | 3 | No | No | 3 | 5 | 1 | 2 | | H:H:?1 | S:S:Q476 | 13.14 | 3 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | S:S:T478 | 6.61 | 3 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:R205 | 40.33 | 3 | Yes | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 16.35 | 3 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 9.45 | 3 | No | No | 9 | 6 | 2 | 1 | | H:H:?1 | R:R:N207 | 12.32 | 3 | Yes | No | 0 | 6 | 0 | 1 | | R:R:E257 | R:R:Q253 | 10.19 | 0 | No | No | 3 | 3 | 1 | 2 | | H:H:?1 | R:R:E257 | 16.65 | 3 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:N261 | 30.8 | 3 | Yes | No | 0 | 4 | 0 | 1 | | R:R:F539 | R:R:N541 | 7.25 | 3 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 31 | 3 | No | No | 1 | 5 | 1 | 2 | | H:H:?1 | R:R:F539 | 6.56 | 3 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?1 | R:R:N541 | 30.8 | 3 | Yes | No | 0 | 5 | 0 | 1 | | R:R:D545 | R:R:D578 | 13.31 | 3 | No | No | 5 | 7 | 1 | 1 | | H:H:?1 | R:R:D545 | 18.27 | 3 | Yes | No | 0 | 5 | 0 | 1 | | R:R:D578 | R:R:T577 | 4.34 | 3 | No | No | 7 | 5 | 1 | 2 | | H:H:?1 | R:R:D578 | 6.09 | 3 | Yes | No | 0 | 7 | 0 | 1 | | S:S:G290 | S:S:T289 | 1.82 | 0 | No | No | 4 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 17.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.05 | | Average Nodes In Shell | 28.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 35.00 | | Average Links Mediated by Hubs In Shell | 20.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9ASB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N287 | R:R:T289 | 2.92 | 0 | No | No | 6 | 5 | 1 | 2 | | R:R:H312 | R:R:N287 | 5.1 | 4 | No | No | 3 | 6 | 2 | 1 | | H:H:?2 | R:R:N287 | 29.57 | 0 | No | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 29.57 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9ASB
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N468 | R:R:Q476 | 7.92 | 34 | No | No | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 5.85 | 34 | No | No | 6 | 3 | 1 | 1 | | H:H:?3 | R:R:N468 | 29.57 | 34 | No | No | 0 | 6 | 0 | 1 | | R:R:Q476 | R:R:T470 | 8.5 | 34 | No | No | 3 | 5 | 1 | 2 | | H:H:?3 | R:R:Q476 | 11.94 | 34 | No | No | 0 | 3 | 0 | 1 | | H:H:?3 | R:R:T478 | 3.97 | 34 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.16 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9AUC | B1 | Peptide | Calcitonin | CT (AMY1) | Homo Sapiens | Calcitonin gene-related peptide-1 | - | Gs/Beta1/Gamma2; RAMP1 | 2.4 | 2024-04-24 | doi.org/10.1021/acs.biochem.4c00114 |
|
A 2D representation of the interactions of NAG in 9AUC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:N73 | 31.93 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N73 | R:R:R74 | 3.62 | 0 | No | No | 4 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 31.93 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 9AUC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:N125 | 27.02 | 0 | No | No | 0 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.02 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
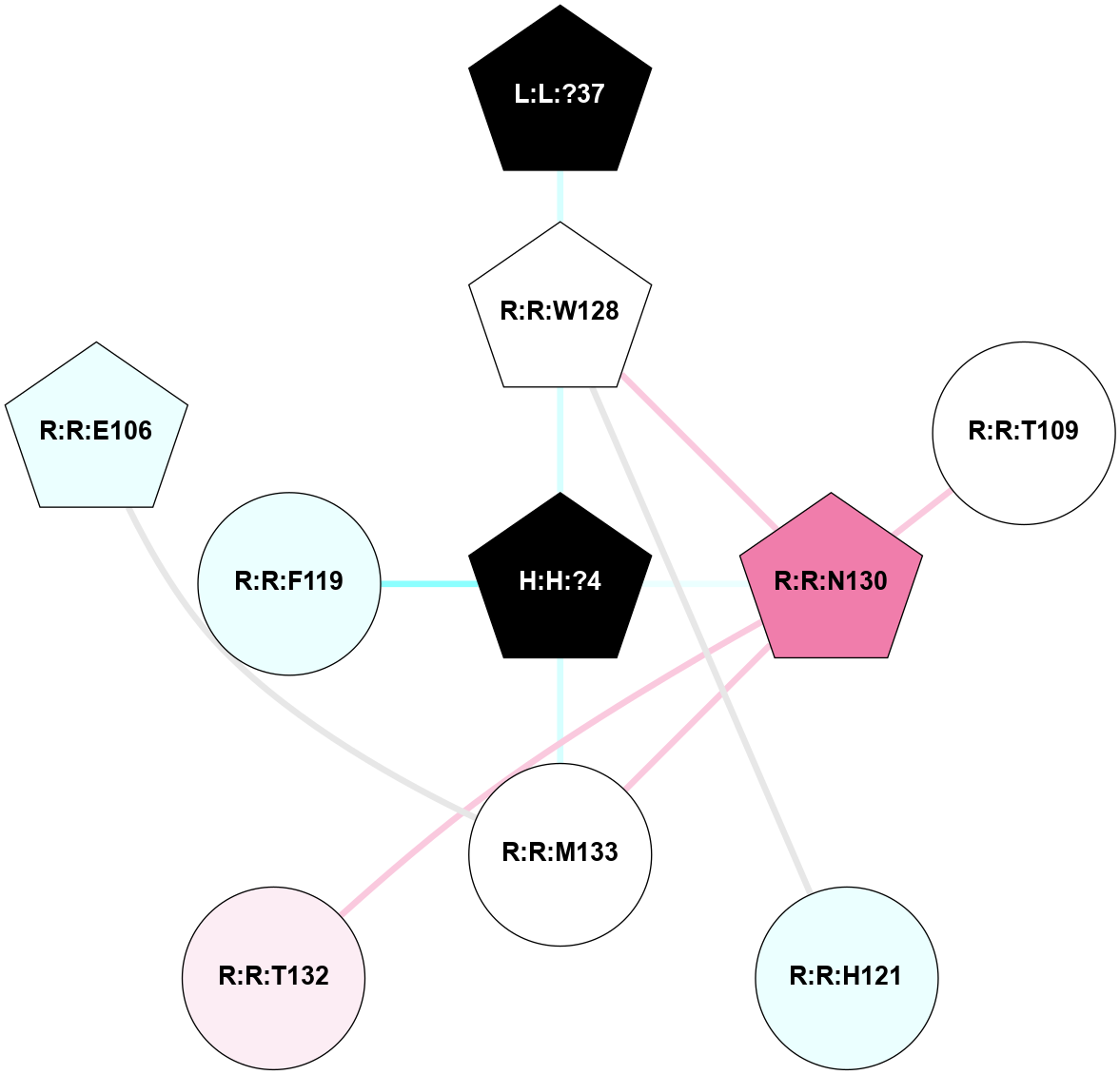
A 2D representation of the interactions of NAG in 9AUC
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | R:R:W128 | 7.13 | 1 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?4 | R:R:N130 | 28.24 | 1 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?4 | R:R:M133 | 7.59 | 1 | Yes | No | 0 | 5 | 0 | 1 | | L:L:?37 | R:R:W128 | 6.44 | 1 | Yes | Yes | 0 | 5 | 2 | 1 | | R:R:N130 | R:R:T109 | 4.39 | 1 | Yes | No | 8 | 5 | 1 | 2 | | R:R:H121 | R:R:W128 | 19.04 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 7.91 | 1 | Yes | Yes | 8 | 5 | 1 | 1 | | R:R:N130 | R:R:T132 | 4.39 | 1 | Yes | No | 8 | 6 | 1 | 2 | | R:R:M133 | R:R:N130 | 5.61 | 1 | No | Yes | 5 | 8 | 1 | 1 | | R:R:E106 | R:R:M133 | 2.71 | 1 | Yes | No | 4 | 5 | 2 | 1 | | H:H:?4 | R:R:F119 | 2.18 | 1 | Yes | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 11.28 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9AVG | C | Ion | Calcium Sensing | CaS | Homo Sapiens | Ca | Ca; Ca; Cyclomethyltryptophan; 9IG; PO4 | chim(NtGi1-Gs)/Beta1/Gamma2 | 3.6 | 2024-04-17 | doi.org/10.1038/s41586-024-07331-1 |
|

A 2D representation of the interactions of NAG in 9AVG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N287 | 29.57 | 1 | No | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:T289 | 9.26 | 1 | No | No | 0 | 5 | 0 | 1 | | R:R:N287 | R:R:T289 | 4.39 | 1 | No | No | 6 | 5 | 1 | 1 | | R:R:H312 | R:R:N287 | 6.38 | 1 | No | No | 3 | 6 | 2 | 1 | | R:R:G290 | R:R:T289 | 3.64 | 0 | No | No | 4 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 19.41 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9AVG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:N468 | 28.34 | 33 | No | No | 0 | 6 | 0 | 1 | | H:H:?2 | R:R:Q476 | 17.91 | 33 | No | No | 0 | 3 | 0 | 1 | | H:H:?2 | R:R:T478 | 7.94 | 33 | No | No | 0 | 3 | 0 | 1 | | R:R:N468 | R:R:Q476 | 9.24 | 33 | No | No | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 5.85 | 33 | No | No | 6 | 3 | 1 | 1 | | R:R:Q476 | R:R:T470 | 9.92 | 33 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.06 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
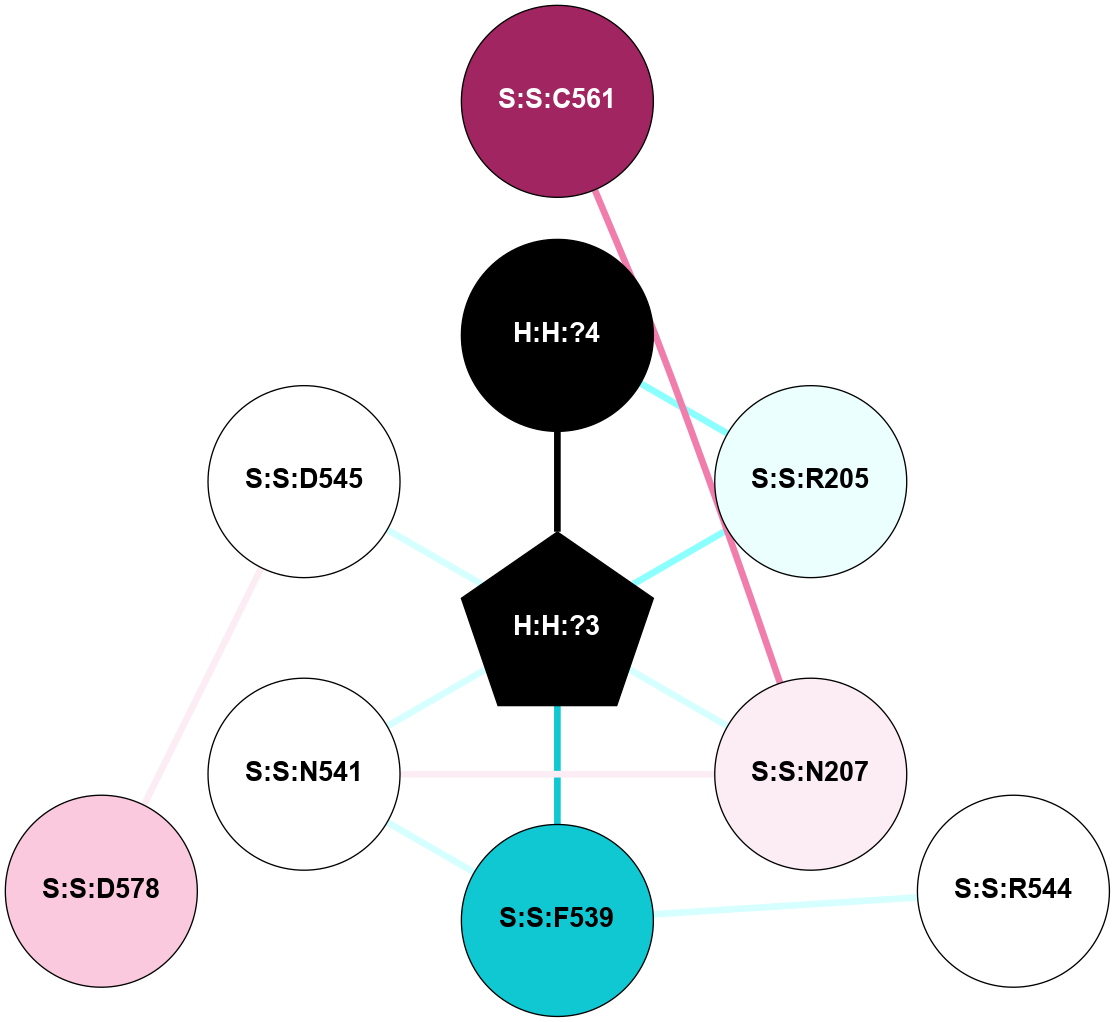
A 2D representation of the interactions of NAG in 9AVG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 40.12 | 21 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?3 | S:S:R205 | 9.81 | 21 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?3 | S:S:N207 | 9.86 | 21 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?3 | S:S:F539 | 8.74 | 21 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?3 | S:S:N541 | 27.11 | 21 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?3 | S:S:D545 | 13.4 | 21 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?4 | S:S:R205 | 9.81 | 21 | No | No | 0 | 4 | 1 | 1 | | S:S:N207 | S:S:N541 | 13.62 | 21 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 7.87 | 0 | No | No | 9 | 6 | 2 | 1 | | S:S:F539 | S:S:N541 | 8.46 | 21 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 33.14 | 21 | No | No | 1 | 5 | 1 | 2 | | S:S:D545 | S:S:D578 | 15.97 | 0 | No | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.17 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
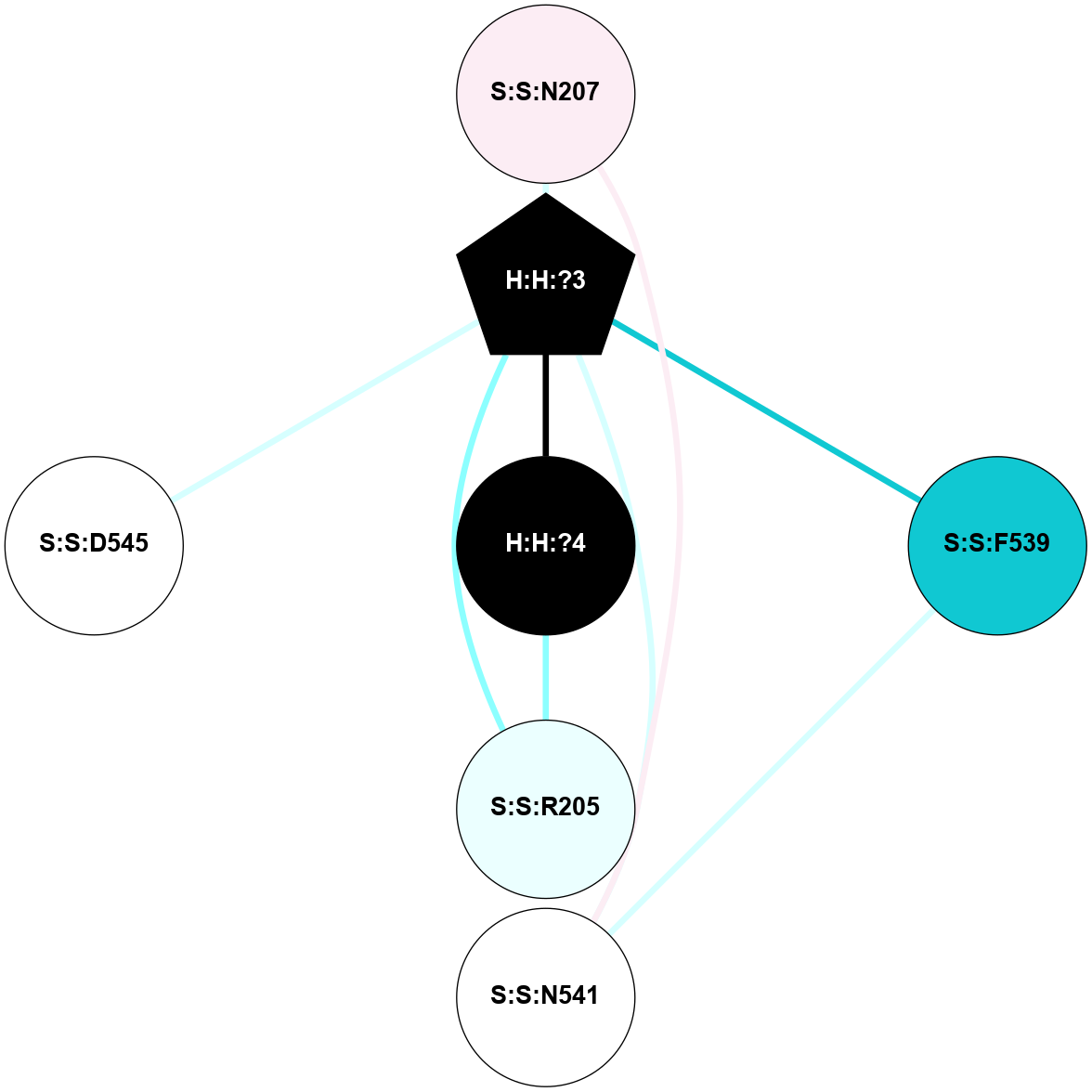
A 2D representation of the interactions of NAG in 9AVG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 40.12 | 21 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?3 | S:S:R205 | 9.81 | 21 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?3 | S:S:N207 | 9.86 | 21 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?3 | S:S:F539 | 8.74 | 21 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?3 | S:S:N541 | 27.11 | 21 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | S:S:D545 | 13.4 | 21 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?4 | S:S:R205 | 9.81 | 21 | No | No | 0 | 4 | 0 | 1 | | S:S:N207 | S:S:N541 | 13.62 | 21 | No | No | 6 | 5 | 2 | 2 | | S:S:F539 | S:S:N541 | 8.46 | 21 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 24.96 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
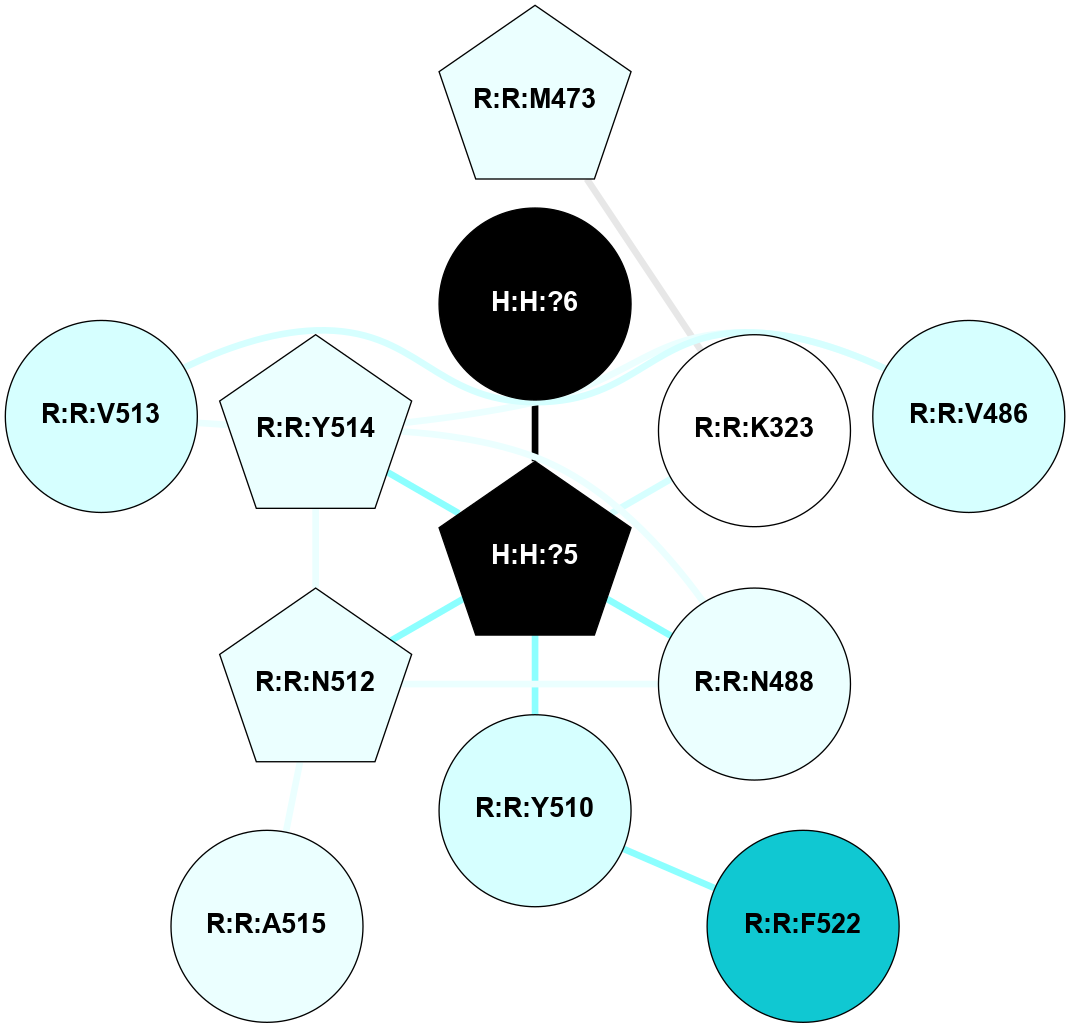
A 2D representation of the interactions of NAG in 9AVG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 37.89 | 22 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?5 | R:R:K323 | 3.8 | 22 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?5 | R:R:N488 | 25.87 | 22 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?5 | R:R:Y510 | 11.57 | 22 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?5 | R:R:N512 | 17.25 | 22 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?5 | R:R:Y514 | 16.83 | 22 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:K323 | R:R:M473 | 2.88 | 0 | No | Yes | 5 | 4 | 1 | 2 | | R:R:V486 | R:R:V513 | 3.21 | 22 | No | No | 3 | 3 | 2 | 2 | | R:R:V486 | R:R:Y514 | 11.36 | 22 | No | Yes | 3 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 5.45 | 22 | No | Yes | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:Y514 | 6.98 | 22 | No | Yes | 4 | 4 | 1 | 1 | | R:R:F522 | R:R:Y510 | 14.44 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:N512 | R:R:Y514 | 4.65 | 22 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 6.25 | 0 | No | Yes | 4 | 4 | 2 | 1 | | R:R:V513 | R:R:Y514 | 5.05 | 22 | No | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 18.87 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
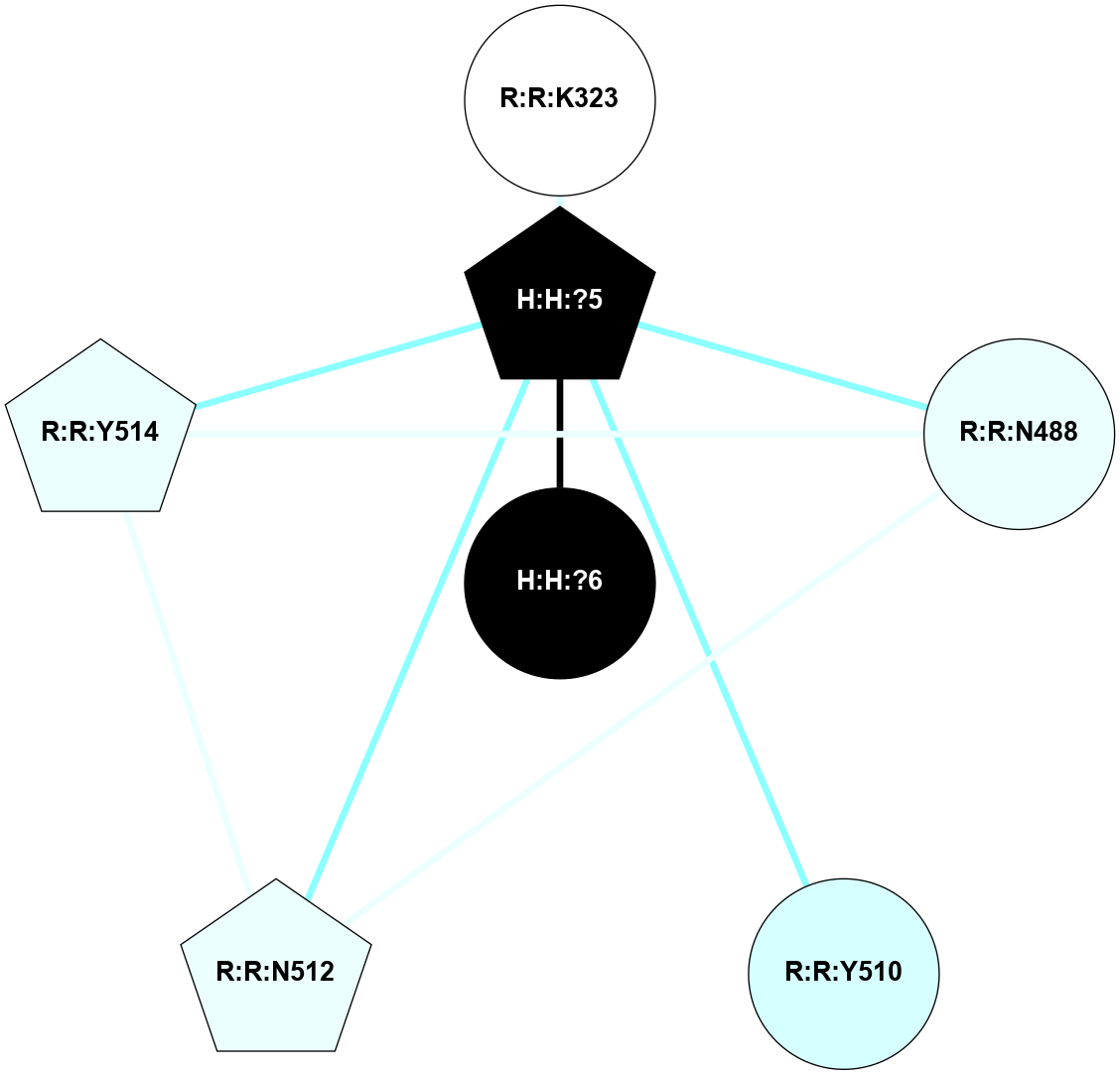
A 2D representation of the interactions of NAG in 9AVG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | H:H:?6 | 37.89 | 22 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?5 | R:R:K323 | 3.8 | 22 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?5 | R:R:N488 | 25.87 | 22 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?5 | R:R:Y510 | 11.57 | 22 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?5 | R:R:N512 | 17.25 | 22 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?5 | R:R:Y514 | 16.83 | 22 | Yes | Yes | 0 | 4 | 1 | 2 | | R:R:N488 | R:R:N512 | 5.45 | 22 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N488 | R:R:Y514 | 6.98 | 22 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N512 | R:R:Y514 | 4.65 | 22 | Yes | Yes | 4 | 4 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 37.89 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
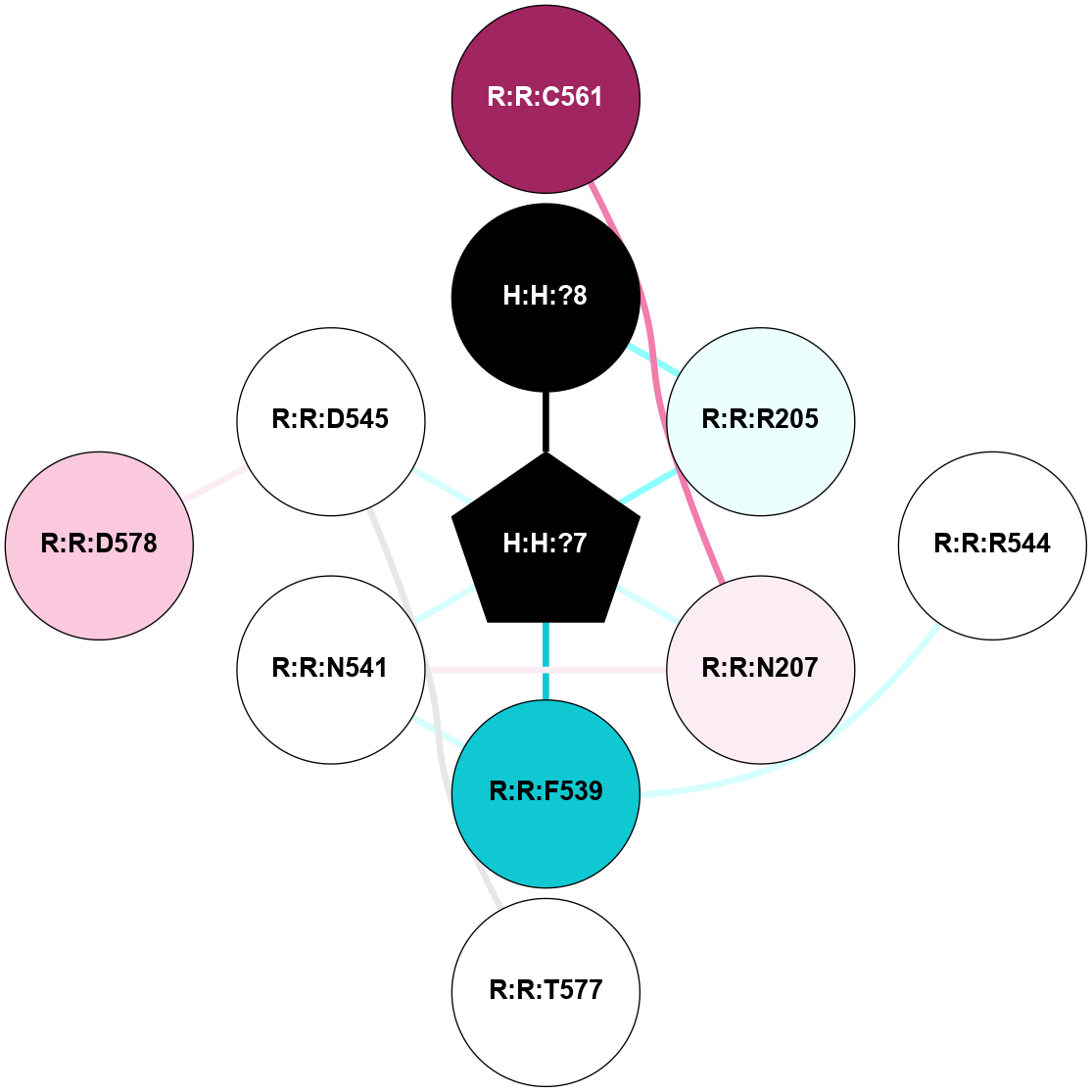
A 2D representation of the interactions of NAG in 9AVG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 36.78 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?7 | R:R:R205 | 7.63 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?7 | R:R:N207 | 12.32 | 1 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?7 | R:R:F539 | 7.65 | 1 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?7 | R:R:N541 | 30.8 | 1 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?7 | R:R:D545 | 14.61 | 1 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?8 | R:R:R205 | 10.9 | 1 | No | No | 0 | 4 | 1 | 1 | | R:R:N207 | R:R:N541 | 16.35 | 1 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 9.45 | 1 | No | No | 9 | 6 | 2 | 1 | | R:R:F539 | R:R:N541 | 7.25 | 1 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 32.07 | 1 | No | No | 1 | 5 | 1 | 2 | | R:R:D545 | R:R:T577 | 4.34 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:D545 | R:R:D578 | 11.98 | 0 | No | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.30 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
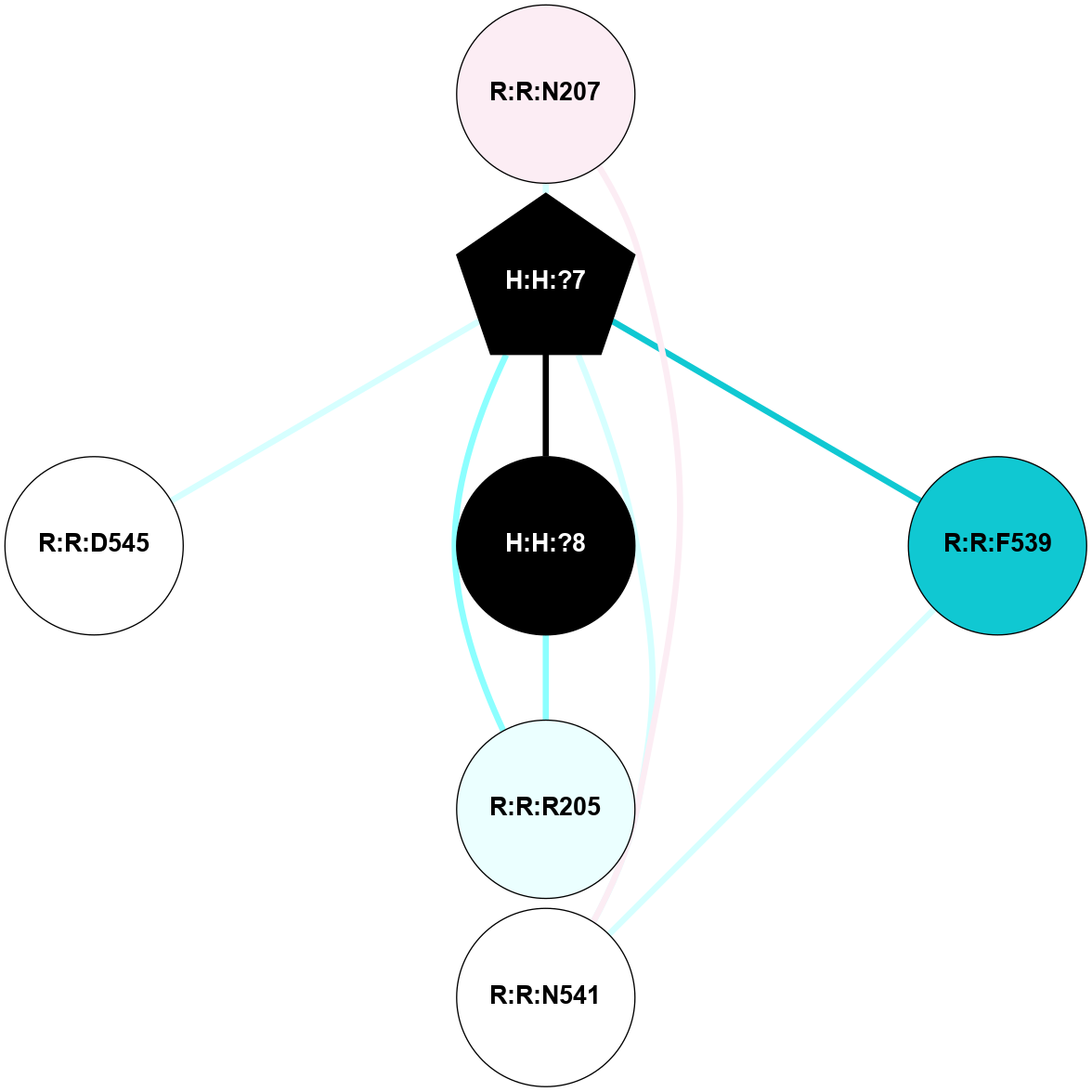
A 2D representation of the interactions of NAG in 9AVG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | H:H:?8 | 36.78 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?7 | R:R:R205 | 7.63 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?7 | R:R:N207 | 12.32 | 1 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?7 | R:R:F539 | 7.65 | 1 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?7 | R:R:N541 | 30.8 | 1 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?7 | R:R:D545 | 14.61 | 1 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?8 | R:R:R205 | 10.9 | 1 | No | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 16.35 | 1 | No | No | 6 | 5 | 2 | 2 | | R:R:F539 | R:R:N541 | 7.25 | 1 | No | No | 1 | 5 | 2 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 23.84 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
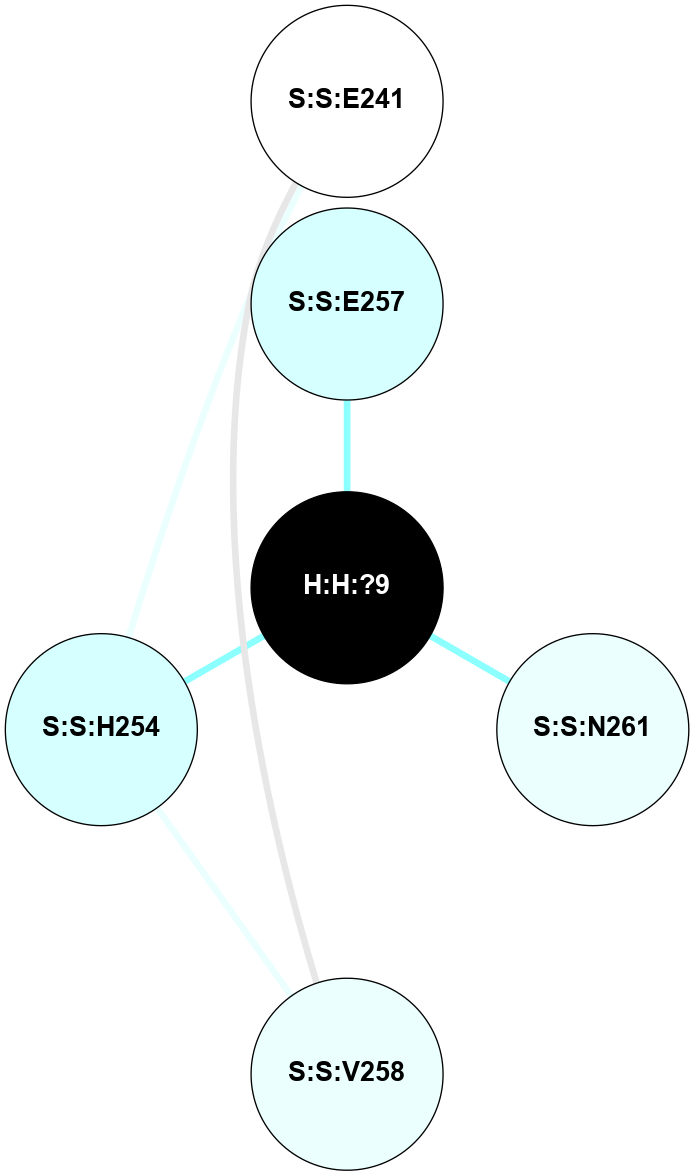
A 2D representation of the interactions of NAG in 9AVG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?9 | S:S:E257 | 4.76 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?9 | S:S:N261 | 33.27 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:E241 | S:S:H254 | 8.62 | 4 | No | No | 5 | 3 | 2 | 1 | | S:S:E241 | S:S:V258 | 11.41 | 4 | No | No | 5 | 4 | 2 | 2 | | S:S:H254 | S:S:V258 | 6.92 | 4 | No | No | 3 | 4 | 1 | 2 | | H:H:?9 | S:S:H254 | 2.31 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.45 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
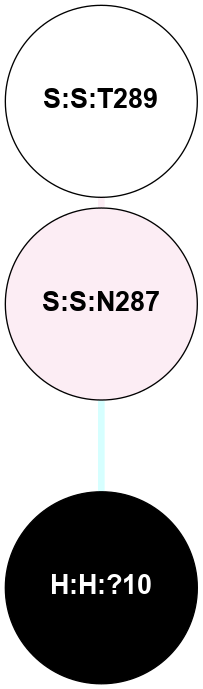
A 2D representation of the interactions of NAG in 9AVG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?10 | S:S:N287 | 28.34 | 0 | No | No | 0 | 6 | 0 | 1 | | S:S:N287 | S:S:T289 | 2.92 | 0 | No | No | 6 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 28.34 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
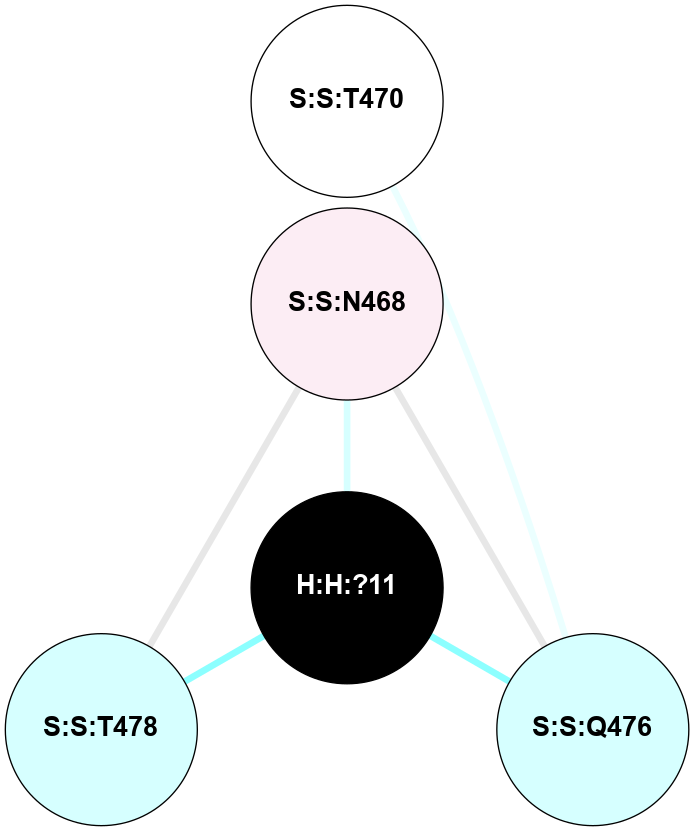
A 2D representation of the interactions of NAG in 9AVG
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?11 | S:S:N468 | 34.5 | 34 | No | No | 0 | 6 | 0 | 1 | | H:H:?11 | S:S:Q476 | 16.72 | 34 | No | No | 0 | 3 | 0 | 1 | | H:H:?11 | S:S:T478 | 5.29 | 34 | No | No | 0 | 3 | 0 | 1 | | S:S:N468 | S:S:Q476 | 14.52 | 34 | No | No | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 10.24 | 34 | No | No | 6 | 3 | 1 | 1 | | S:S:Q476 | S:S:T470 | 7.09 | 34 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.84 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9AVL | C | Ion | Calcium Sensing | CaS | Homo Sapiens | Ca | Ca; Ca; Cyclomethyltryptophan; 9IG; PO4 | Gi3/Beta2/Gamma2 | 3.8 | 2024-04-17 | doi.org/10.1038/s41586-024-07331-1 |
|
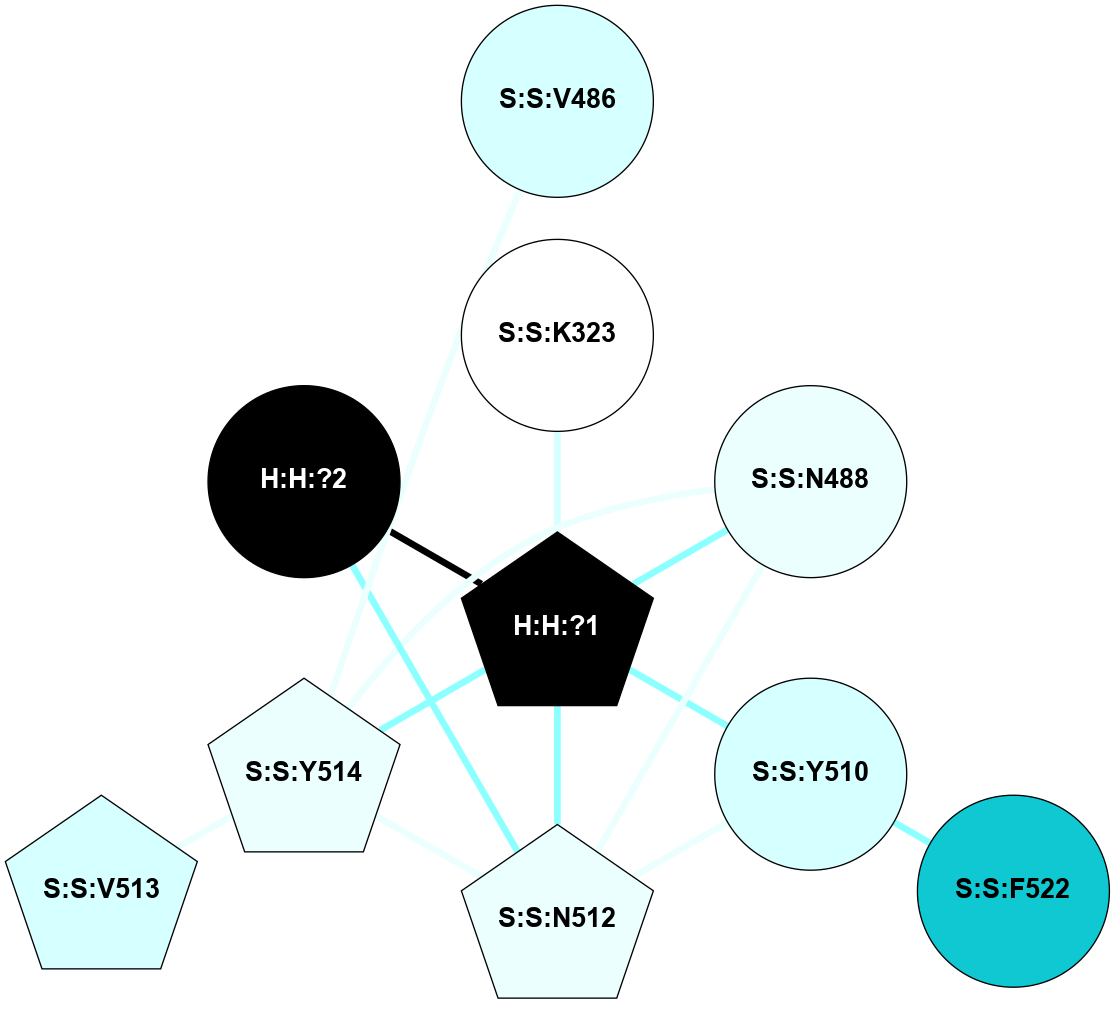
A 2D representation of the interactions of NAG in 9AVL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | S:S:K323 | 6.33 | 2 | Yes | No | 0 | 5 | 0 | 1 | | S:S:V486 | S:S:Y514 | 6.31 | 0 | No | Yes | 3 | 4 | 2 | 1 | | S:S:N488 | S:S:N512 | 6.81 | 2 | No | Yes | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:Y514 | 2.33 | 2 | No | Yes | 4 | 4 | 1 | 1 | | H:H:?1 | S:S:N488 | 24.64 | 2 | Yes | No | 0 | 4 | 0 | 1 | | S:S:N512 | S:S:Y510 | 2.33 | 2 | Yes | No | 4 | 3 | 1 | 1 | | S:S:F522 | S:S:Y510 | 17.54 | 0 | No | No | 1 | 3 | 2 | 1 | | H:H:?1 | S:S:Y510 | 10.52 | 2 | Yes | No | 0 | 3 | 0 | 1 | | S:S:N512 | S:S:Y514 | 4.65 | 2 | Yes | Yes | 4 | 4 | 1 | 1 | | H:H:?1 | S:S:N512 | 20.95 | 2 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?2 | S:S:N512 | 3.7 | 2 | No | Yes | 0 | 4 | 1 | 1 | | S:S:V513 | S:S:Y514 | 3.79 | 2 | Yes | Yes | 3 | 4 | 2 | 1 | | H:H:?1 | S:S:Y514 | 7.36 | 2 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 36.78 | 2 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 17.76 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
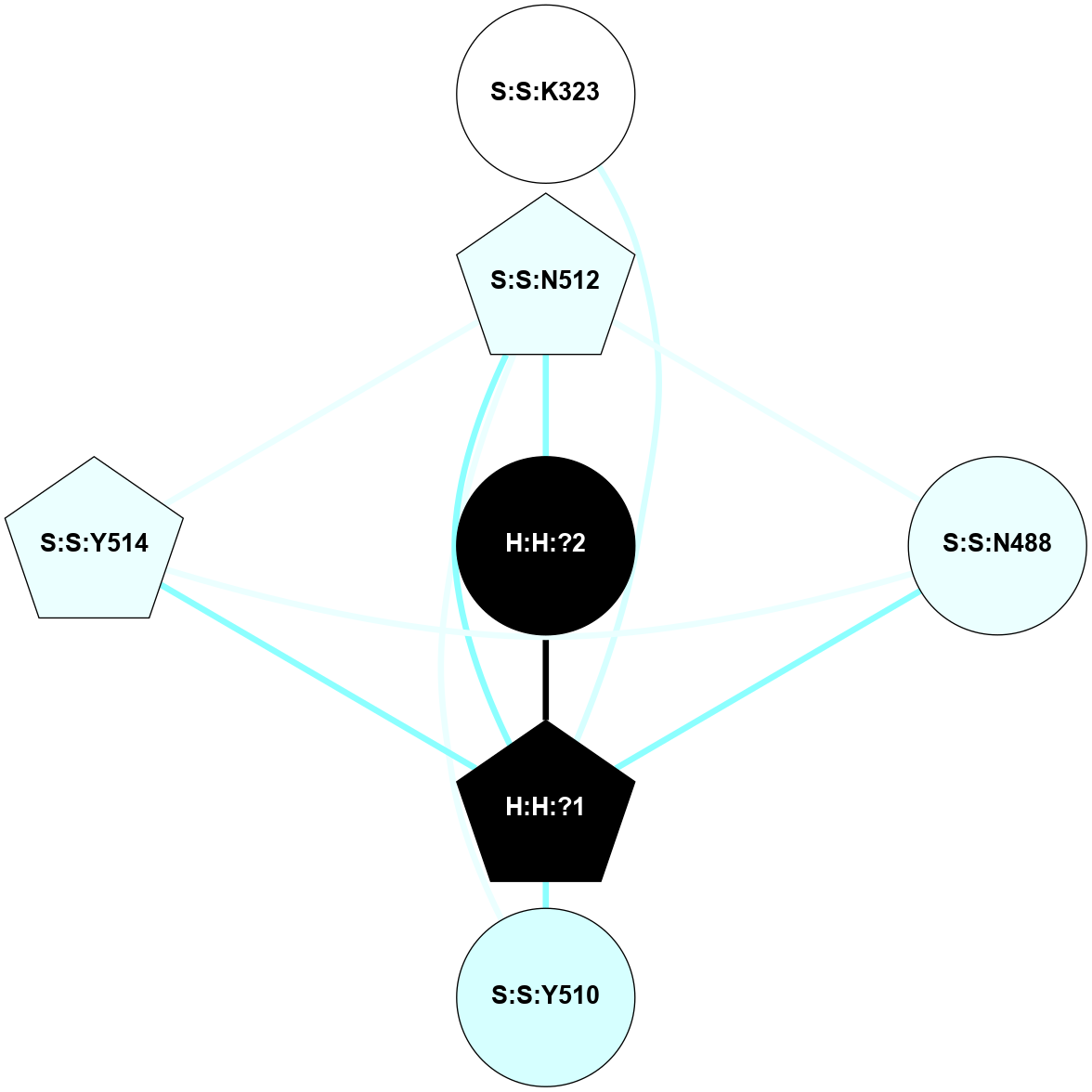
A 2D representation of the interactions of NAG in 9AVL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | S:S:K323 | 6.33 | 2 | Yes | No | 0 | 5 | 1 | 2 | | S:S:N488 | S:S:N512 | 6.81 | 2 | No | Yes | 4 | 4 | 2 | 1 | | S:S:N488 | S:S:Y514 | 2.33 | 2 | No | Yes | 4 | 4 | 2 | 2 | | H:H:?1 | S:S:N488 | 24.64 | 2 | Yes | No | 0 | 4 | 1 | 2 | | S:S:N512 | S:S:Y510 | 2.33 | 2 | Yes | No | 4 | 3 | 1 | 2 | | H:H:?1 | S:S:Y510 | 10.52 | 2 | Yes | No | 0 | 3 | 1 | 2 | | S:S:N512 | S:S:Y514 | 4.65 | 2 | Yes | Yes | 4 | 4 | 1 | 2 | | H:H:?1 | S:S:N512 | 20.95 | 2 | Yes | Yes | 0 | 4 | 1 | 1 | | H:H:?2 | S:S:N512 | 3.7 | 2 | No | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | S:S:Y514 | 7.36 | 2 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 36.78 | 2 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 20.24 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 11.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
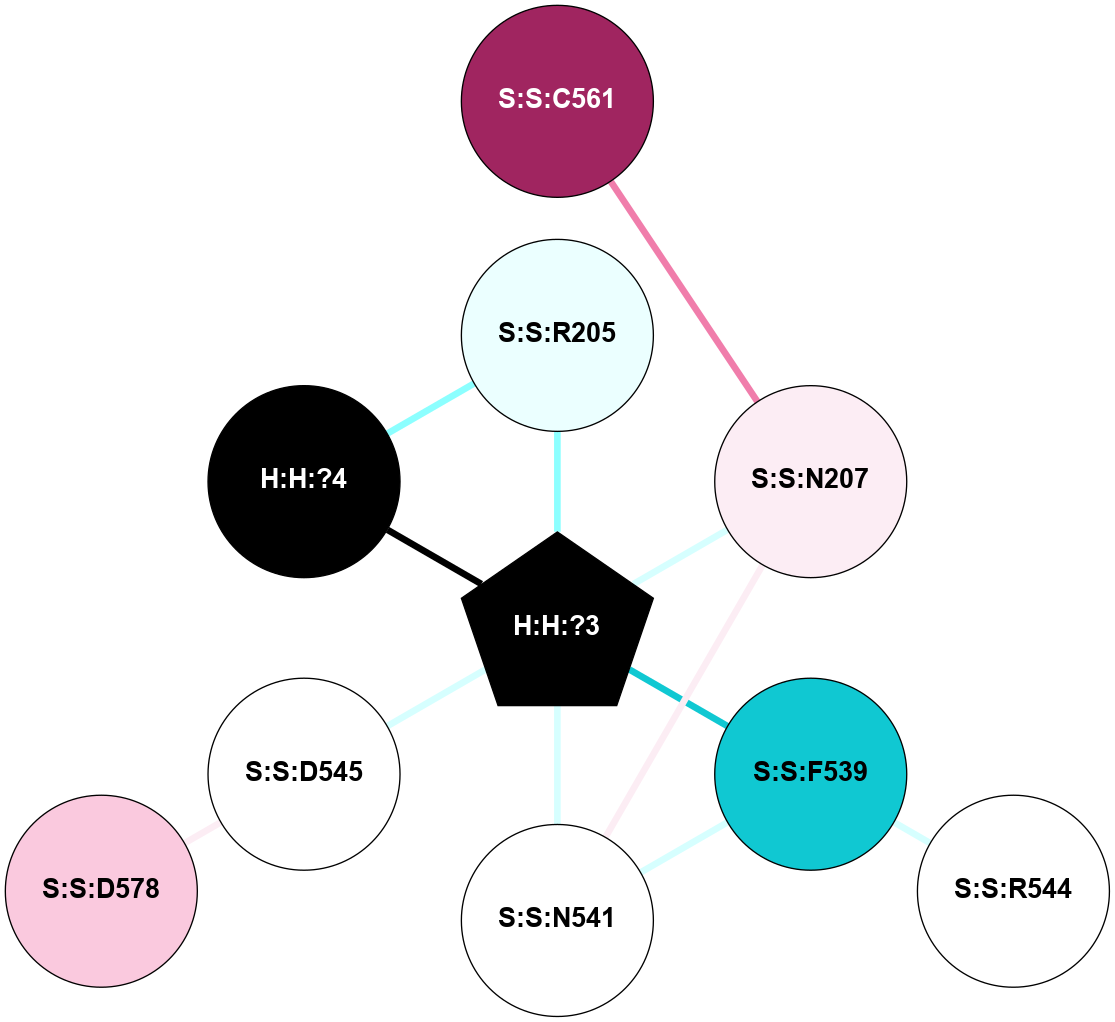
A 2D representation of the interactions of NAG in 9AVL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | S:S:R205 | 10.9 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?4 | S:S:R205 | 9.81 | 1 | No | No | 0 | 4 | 1 | 1 | | S:S:N207 | S:S:N541 | 16.35 | 1 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 7.87 | 1 | No | No | 9 | 6 | 2 | 1 | | H:H:?3 | S:S:N207 | 11.09 | 1 | Yes | No | 0 | 6 | 0 | 1 | | S:S:F539 | S:S:N541 | 8.46 | 1 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 26.72 | 1 | No | No | 1 | 5 | 1 | 2 | | H:H:?3 | S:S:F539 | 5.46 | 1 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?3 | S:S:N541 | 29.57 | 1 | Yes | No | 0 | 5 | 0 | 1 | | S:S:D545 | S:S:D578 | 15.97 | 0 | No | No | 5 | 7 | 1 | 2 | | H:H:?3 | S:S:D545 | 9.74 | 1 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?3 | H:H:?4 | 35.66 | 1 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.07 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
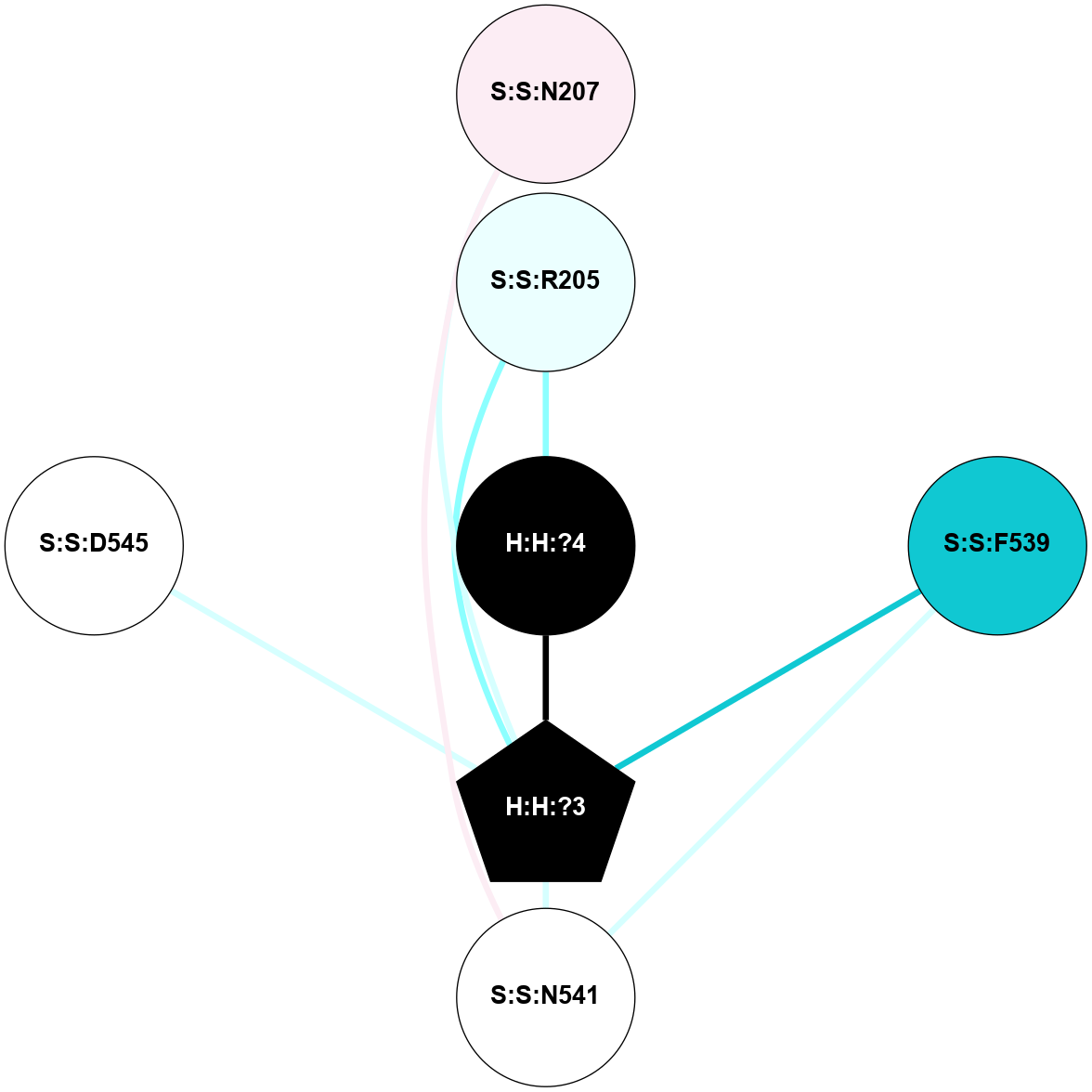
A 2D representation of the interactions of NAG in 9AVL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | S:S:R205 | 10.9 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?4 | S:S:R205 | 9.81 | 1 | No | No | 0 | 4 | 0 | 1 | | S:S:N207 | S:S:N541 | 16.35 | 1 | No | No | 6 | 5 | 2 | 2 | | H:H:?3 | S:S:N207 | 11.09 | 1 | Yes | No | 0 | 6 | 1 | 2 | | S:S:F539 | S:S:N541 | 8.46 | 1 | No | No | 1 | 5 | 2 | 2 | | H:H:?3 | S:S:F539 | 5.46 | 1 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?3 | S:S:N541 | 29.57 | 1 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | S:S:D545 | 9.74 | 1 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | H:H:?4 | 35.66 | 1 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 22.73 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
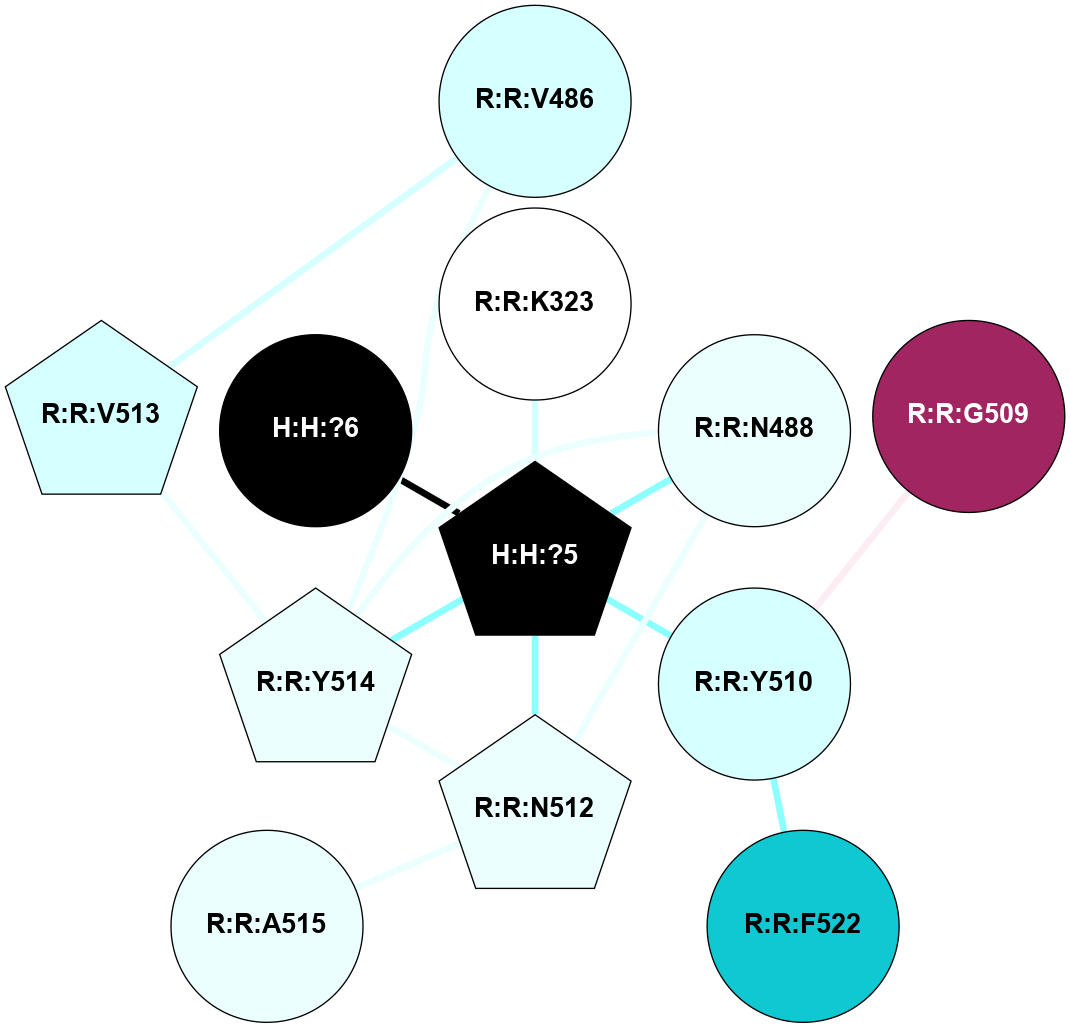
A 2D representation of the interactions of NAG in 9AVL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | R:R:K323 | 2.53 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:V486 | R:R:V513 | 3.21 | 1 | No | Yes | 3 | 3 | 2 | 2 | | R:R:V486 | R:R:Y514 | 7.57 | 1 | No | Yes | 3 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 5.45 | 1 | No | Yes | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:Y514 | 3.49 | 1 | No | Yes | 4 | 4 | 1 | 1 | | H:H:?5 | R:R:N488 | 24.64 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:G509 | R:R:Y510 | 2.9 | 0 | No | No | 9 | 3 | 2 | 1 | | R:R:F522 | R:R:Y510 | 17.54 | 0 | No | No | 1 | 3 | 2 | 1 | | H:H:?5 | R:R:Y510 | 5.26 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:N512 | R:R:Y514 | 2.33 | 1 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 4.69 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?5 | R:R:N512 | 14.79 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:V513 | R:R:Y514 | 7.57 | 1 | Yes | Yes | 3 | 4 | 2 | 1 | | H:H:?5 | R:R:Y514 | 12.62 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?5 | H:H:?6 | 39.01 | 1 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 16.47 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
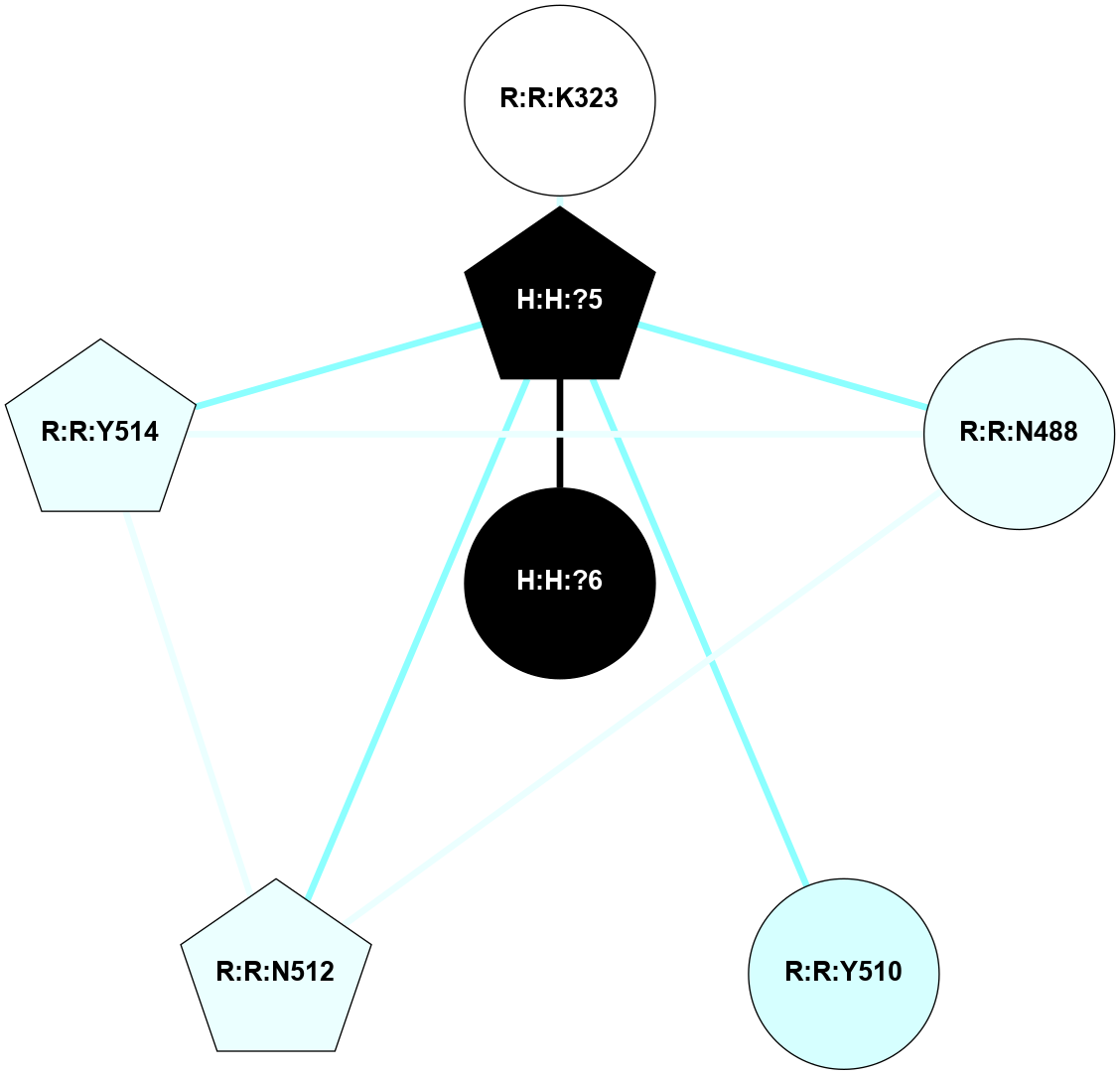
A 2D representation of the interactions of NAG in 9AVL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | R:R:K323 | 2.53 | 1 | Yes | No | 0 | 5 | 1 | 2 | | R:R:N488 | R:R:N512 | 5.45 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N488 | R:R:Y514 | 3.49 | 1 | No | Yes | 4 | 4 | 2 | 2 | | H:H:?5 | R:R:N488 | 24.64 | 1 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?5 | R:R:Y510 | 5.26 | 1 | Yes | No | 0 | 3 | 1 | 2 | | R:R:N512 | R:R:Y514 | 2.33 | 1 | Yes | Yes | 4 | 4 | 2 | 2 | | H:H:?5 | R:R:N512 | 14.79 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?5 | R:R:Y514 | 12.62 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?5 | H:H:?6 | 39.01 | 1 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 39.01 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9AVL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | R:R:R205 | 13.08 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?8 | R:R:R205 | 10.9 | 1 | No | No | 0 | 4 | 1 | 1 | | R:R:N207 | R:R:N541 | 16.35 | 1 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 11.02 | 1 | Yes | No | 9 | 6 | 2 | 1 | | H:H:?7 | R:R:N207 | 13.55 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:F539 | R:R:N541 | 8.46 | 1 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 26.72 | 1 | No | No | 1 | 5 | 1 | 2 | | H:H:?7 | R:R:F539 | 5.46 | 1 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?7 | R:R:N541 | 24.64 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:D545 | R:R:T577 | 4.34 | 1 | No | No | 5 | 5 | 1 | 2 | | R:R:D545 | R:R:D578 | 15.97 | 1 | No | No | 5 | 7 | 1 | 2 | | H:H:?7 | R:R:D545 | 12.18 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:D578 | R:R:T577 | 5.78 | 1 | No | No | 7 | 5 | 2 | 2 | | H:H:?7 | H:H:?8 | 39.01 | 1 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.99 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
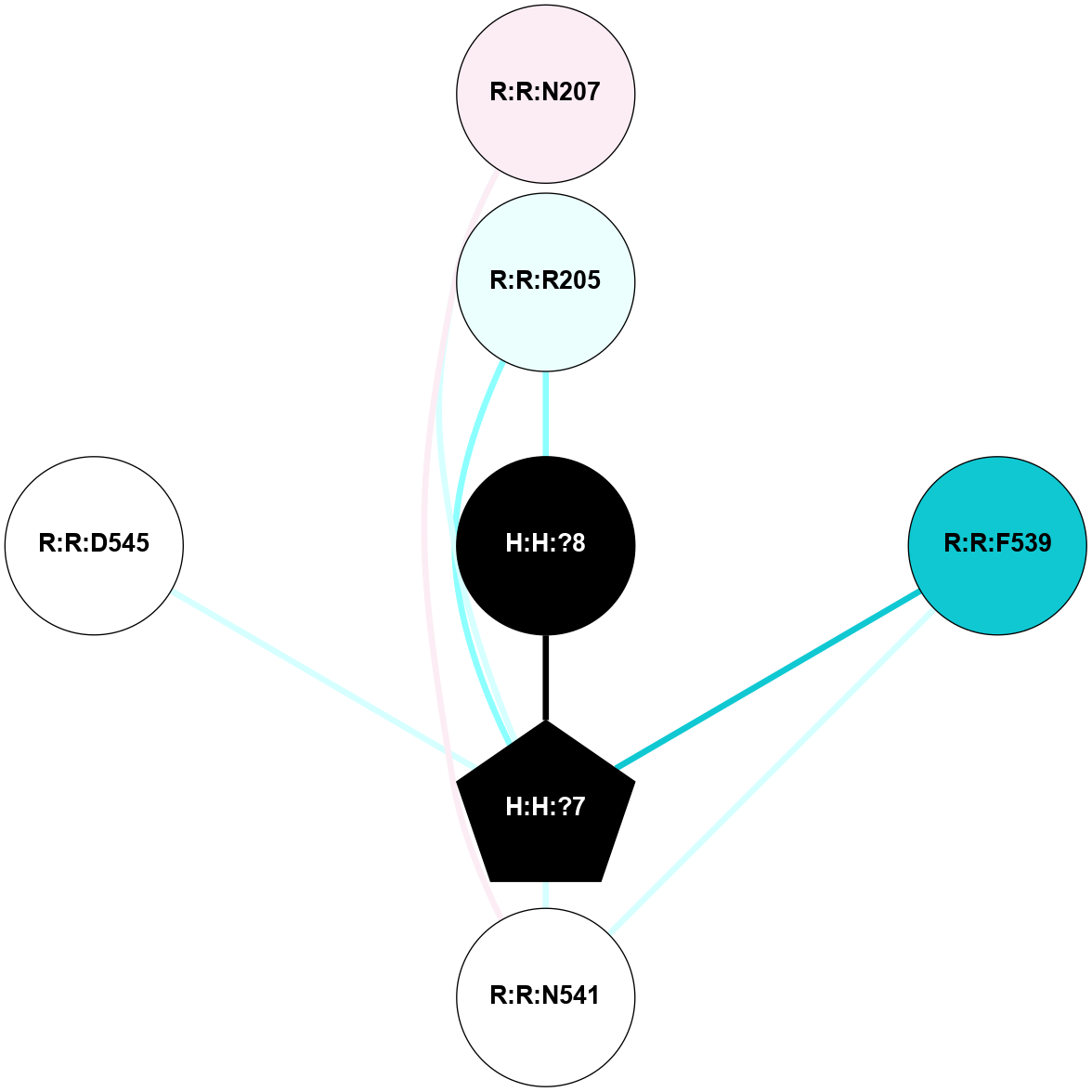
A 2D representation of the interactions of NAG in 9AVL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | R:R:R205 | 13.08 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?8 | R:R:R205 | 10.9 | 1 | No | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 16.35 | 1 | No | No | 6 | 5 | 2 | 2 | | H:H:?7 | R:R:N207 | 13.55 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:F539 | R:R:N541 | 8.46 | 1 | No | No | 1 | 5 | 2 | 2 | | H:H:?7 | R:R:F539 | 5.46 | 1 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?7 | R:R:N541 | 24.64 | 1 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?7 | R:R:D545 | 12.18 | 1 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?7 | H:H:?8 | 39.01 | 1 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 24.95 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
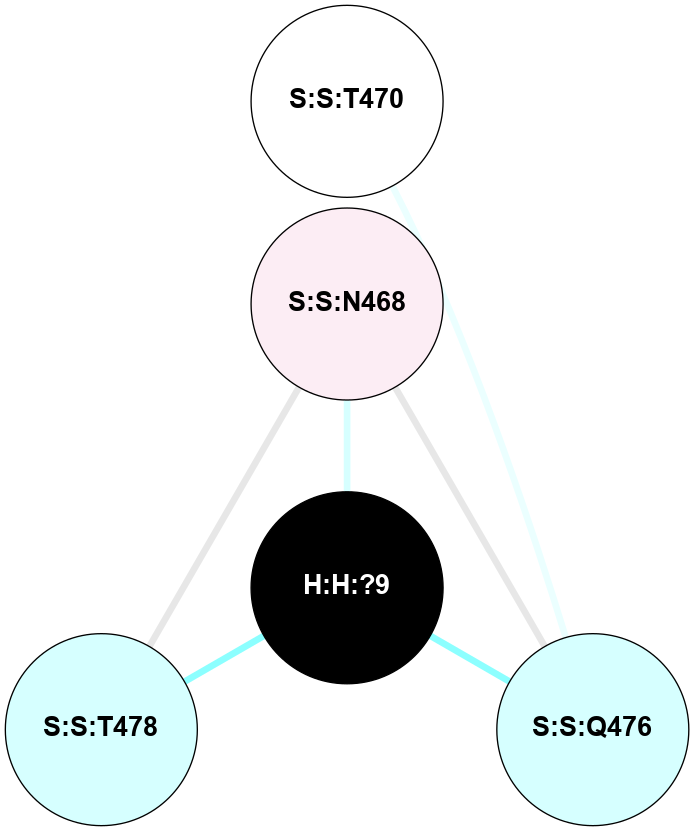
A 2D representation of the interactions of NAG in 9AVL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:N468 | S:S:Q476 | 6.6 | 22 | No | No | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 10.24 | 22 | No | No | 6 | 3 | 1 | 1 | | H:H:?9 | S:S:N468 | 27.11 | 22 | No | No | 0 | 6 | 0 | 1 | | S:S:Q476 | S:S:T470 | 11.34 | 22 | No | No | 3 | 5 | 1 | 2 | | H:H:?9 | S:S:Q476 | 15.53 | 22 | No | No | 0 | 3 | 0 | 1 | | H:H:?9 | S:S:T478 | 2.65 | 22 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 15.10 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
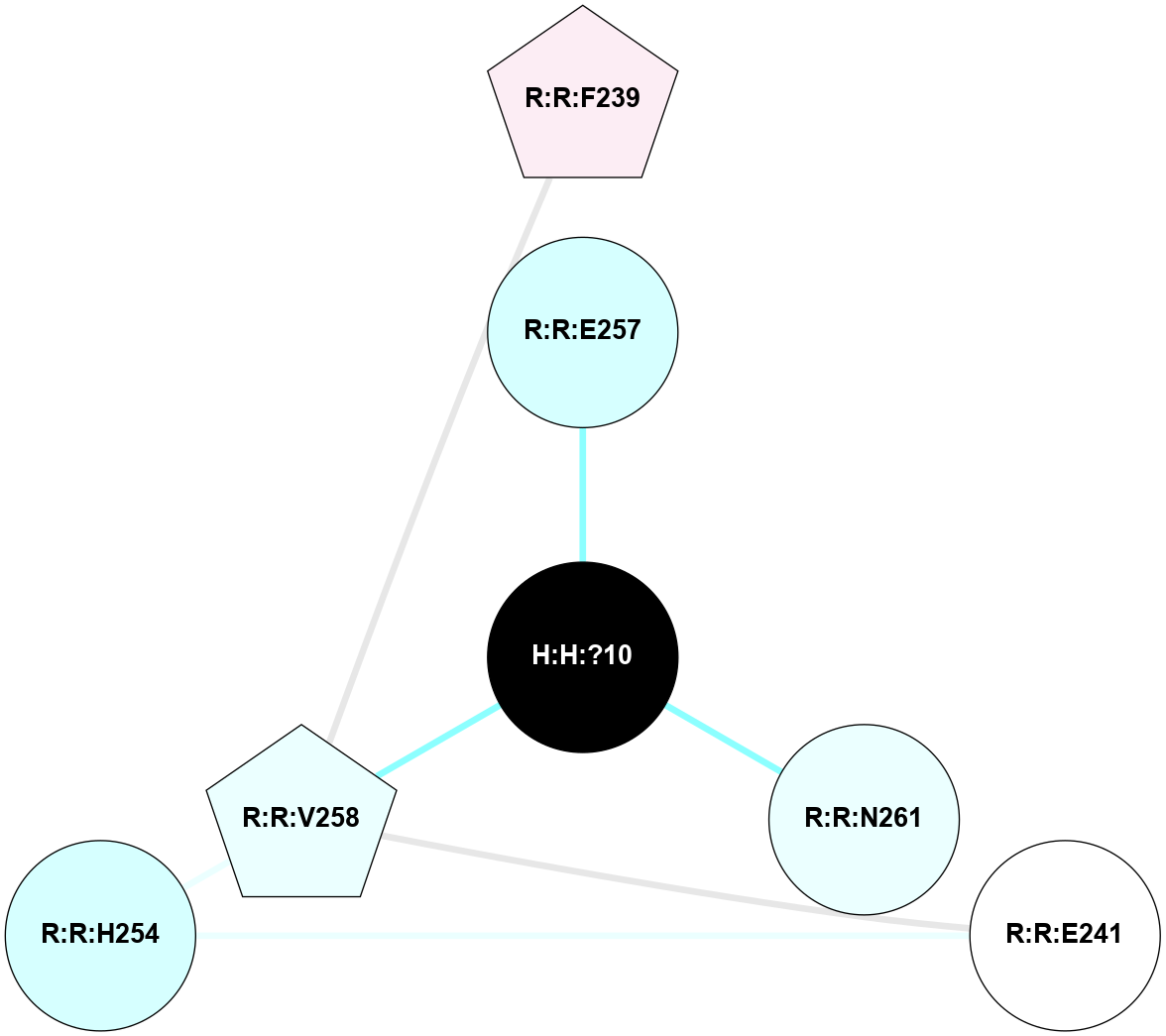
A 2D representation of the interactions of NAG in 9AVL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:F239 | R:R:V258 | 5.24 | 1 | Yes | Yes | 6 | 4 | 2 | 1 | | R:R:E241 | R:R:H254 | 7.39 | 1 | No | No | 5 | 3 | 2 | 2 | | R:R:E241 | R:R:V258 | 11.41 | 1 | No | Yes | 5 | 4 | 2 | 1 | | R:R:H254 | R:R:V258 | 4.15 | 1 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?10 | R:R:E257 | 8.32 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?10 | R:R:N261 | 25.87 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?10 | R:R:V258 | 1.34 | 0 | No | Yes | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 11.84 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9AVL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N287 | R:R:T289 | 4.39 | 0 | No | No | 6 | 5 | 1 | 2 | | H:H:?11 | R:R:N287 | 28.34 | 0 | No | No | 0 | 6 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 28.34 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9AVL
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N468 | R:R:Q476 | 7.92 | 26 | No | No | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 5.85 | 26 | No | No | 6 | 3 | 1 | 1 | | H:H:?12 | R:R:N468 | 29.57 | 26 | No | No | 0 | 6 | 0 | 1 | | R:R:Q476 | R:R:T470 | 7.09 | 26 | No | No | 3 | 5 | 1 | 2 | | H:H:?12 | R:R:Q476 | 17.91 | 26 | No | No | 0 | 3 | 0 | 1 | | H:H:?12 | R:R:T478 | 6.61 | 26 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.03 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9AXF | C | Ion | Calcium Sensing | CaS | Homo Sapiens | Ca | Ca; Ca; Ca; Cyclomethyltryptophan; 9IG; PO4 | chim(NtGi1-Gs-CtGq)/Beta1/Gamma2 | 3.5 | 2024-04-17 | doi.org/10.1038/s41586-024-07331-1 |
|
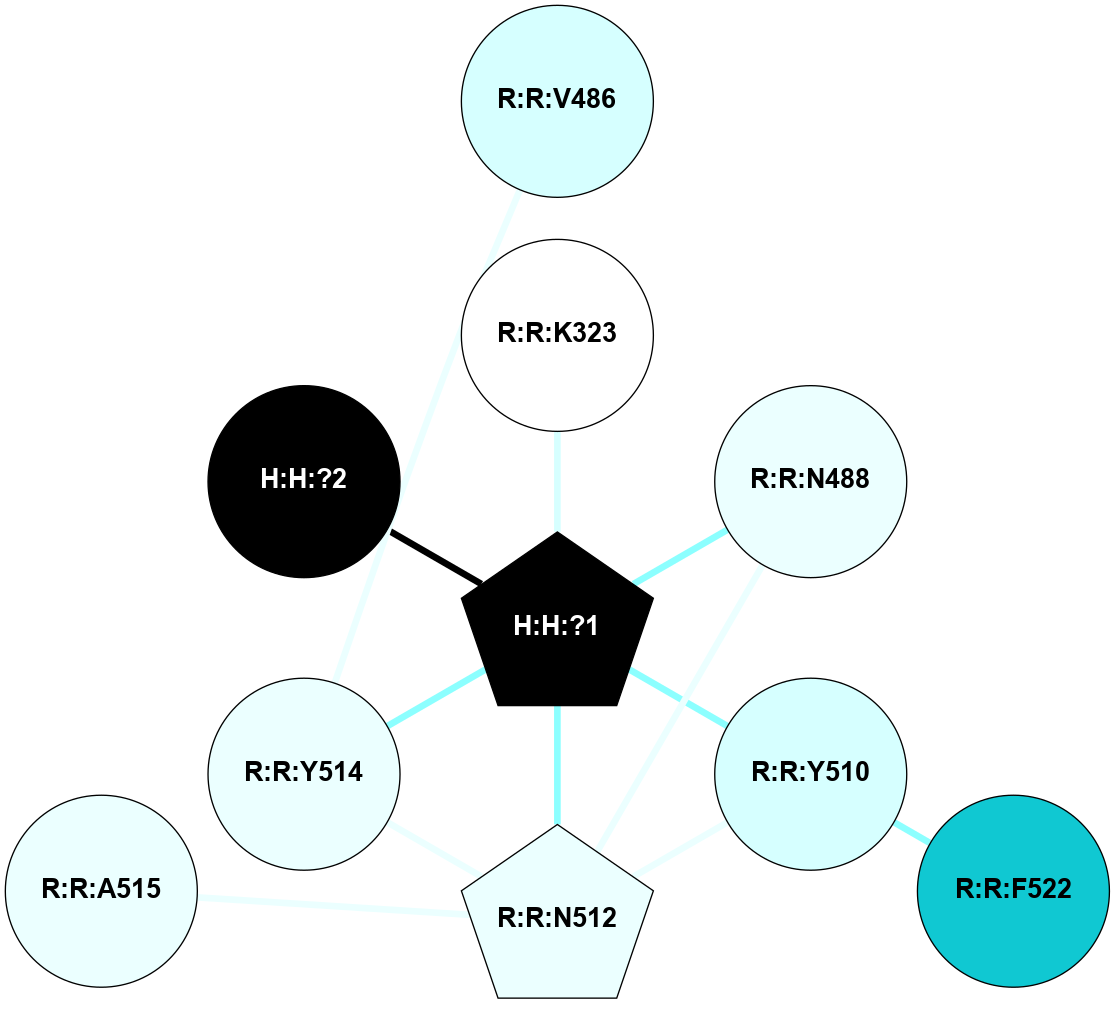
A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:K323 | 5.04 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:V486 | R:R:Y514 | 3.79 | 1 | No | No | 3 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 4.09 | 1 | No | Yes | 4 | 4 | 1 | 1 | | H:H:?1 | R:R:N488 | 23.33 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:N512 | R:R:Y510 | 2.33 | 1 | Yes | No | 4 | 3 | 1 | 1 | | R:R:F522 | R:R:Y510 | 16.5 | 0 | No | No | 1 | 3 | 2 | 1 | | H:H:?1 | R:R:Y510 | 10.48 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:N512 | R:R:Y514 | 3.49 | 1 | Yes | No | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 4.69 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?1 | R:R:N512 | 14.74 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:Y514 | 5.24 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 38.74 | 1 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 16.26 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
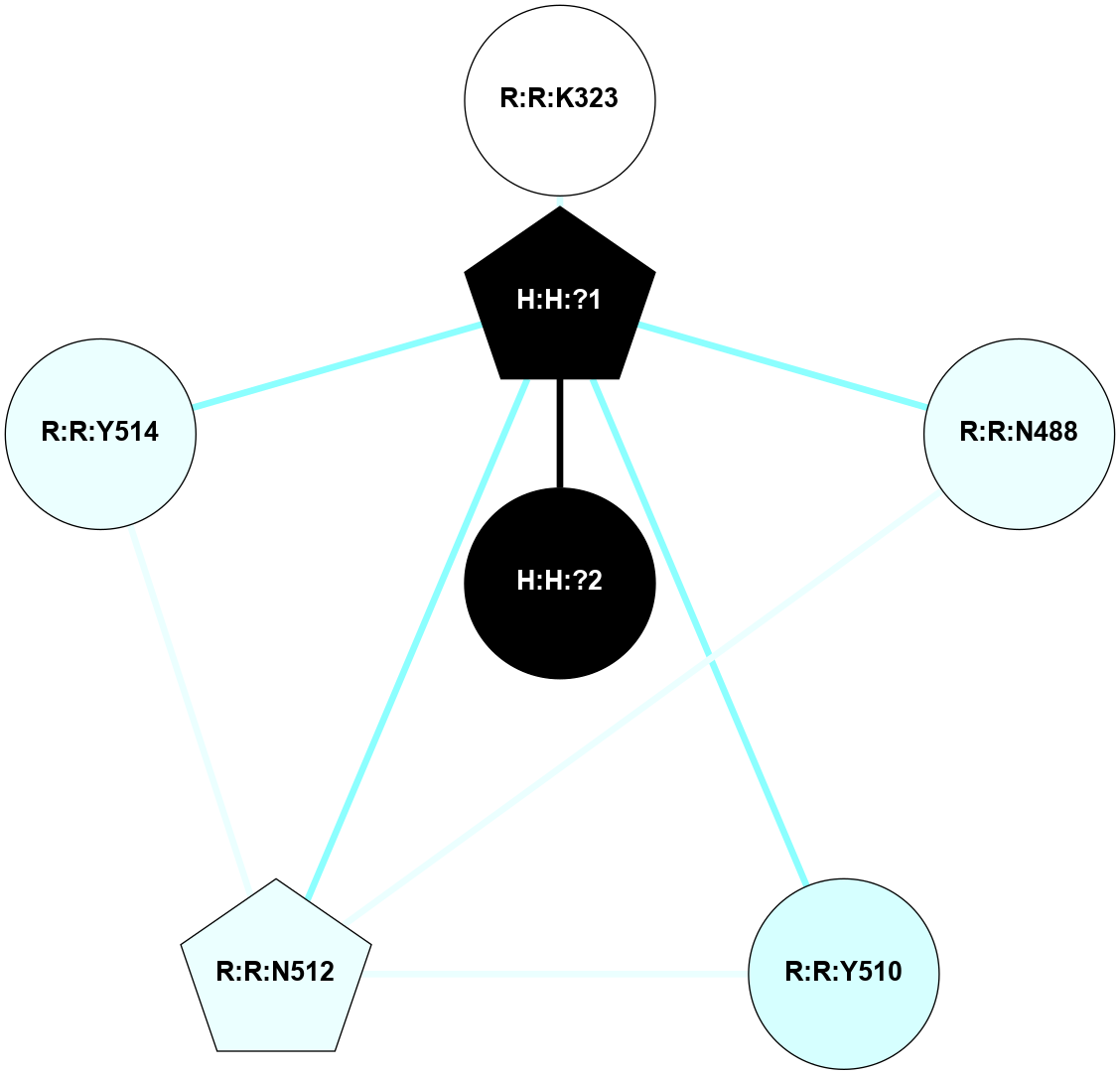
A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:K323 | 5.04 | 1 | Yes | No | 0 | 5 | 1 | 2 | | R:R:N488 | R:R:N512 | 4.09 | 1 | No | Yes | 4 | 4 | 2 | 2 | | H:H:?1 | R:R:N488 | 23.33 | 1 | Yes | No | 0 | 4 | 1 | 2 | | R:R:N512 | R:R:Y510 | 2.33 | 1 | Yes | No | 4 | 3 | 2 | 2 | | H:H:?1 | R:R:Y510 | 10.48 | 1 | Yes | No | 0 | 3 | 1 | 2 | | R:R:N512 | R:R:Y514 | 3.49 | 1 | Yes | No | 4 | 4 | 2 | 2 | | H:H:?1 | R:R:N512 | 14.74 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?1 | R:R:Y514 | 5.24 | 1 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 38.74 | 1 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 38.74 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
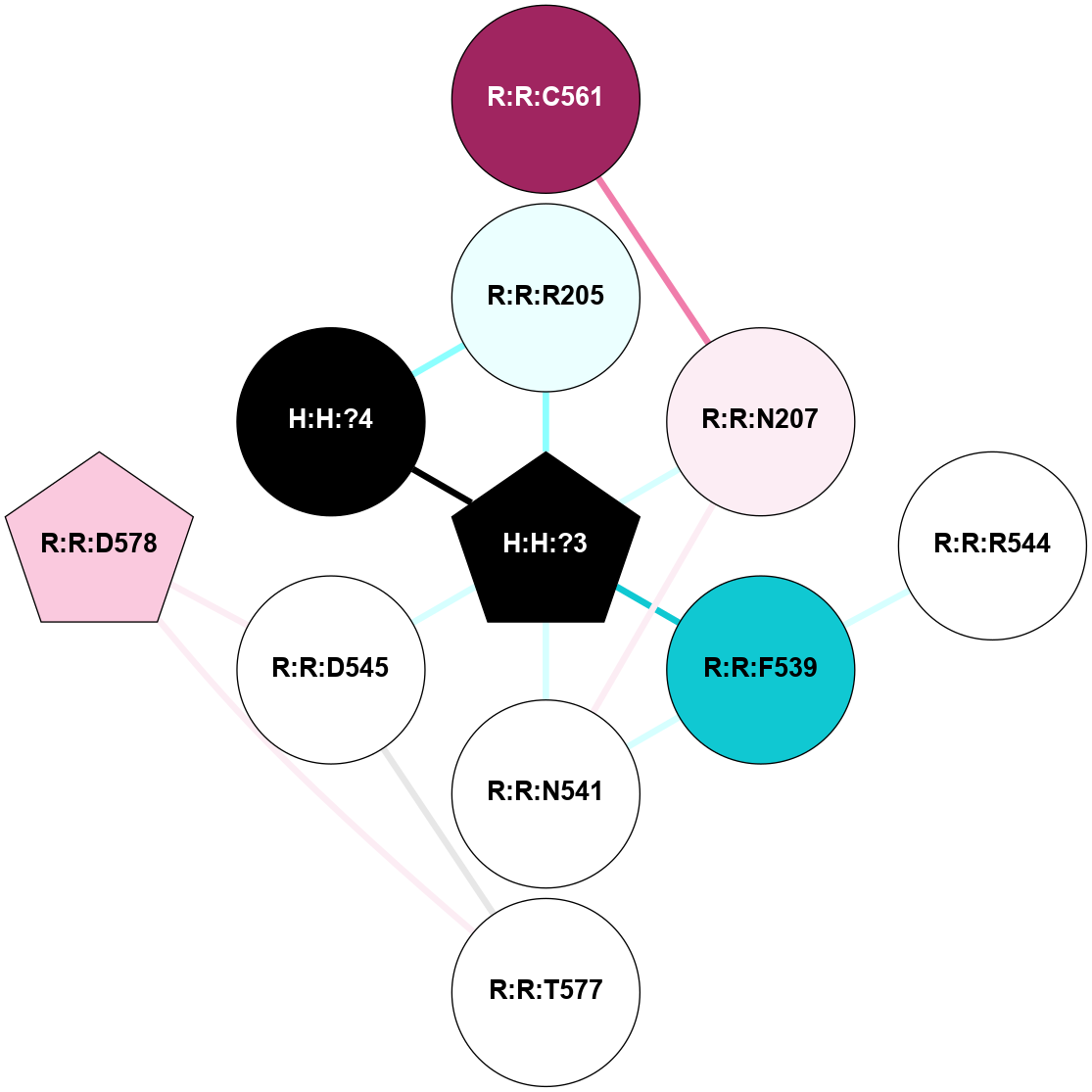
A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:R205 | 6.52 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?4 | R:R:R205 | 8.69 | 1 | No | No | 0 | 4 | 1 | 1 | | R:R:N207 | R:R:N541 | 13.62 | 1 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 11.02 | 0 | No | No | 9 | 6 | 2 | 1 | | H:H:?3 | R:R:N207 | 18.42 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:F539 | R:R:N541 | 10.87 | 1 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 20.31 | 1 | No | No | 1 | 5 | 1 | 2 | | H:H:?3 | R:R:F539 | 3.27 | 1 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?3 | R:R:N541 | 27.02 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:D545 | R:R:T577 | 2.89 | 1 | No | No | 5 | 5 | 1 | 2 | | R:R:D545 | R:R:D578 | 3.99 | 1 | No | Yes | 5 | 7 | 1 | 2 | | H:H:?3 | R:R:D545 | 3.64 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:D578 | R:R:T577 | 2.89 | 1 | Yes | No | 7 | 5 | 2 | 2 | | H:H:?3 | H:H:?4 | 38.74 | 1 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.27 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
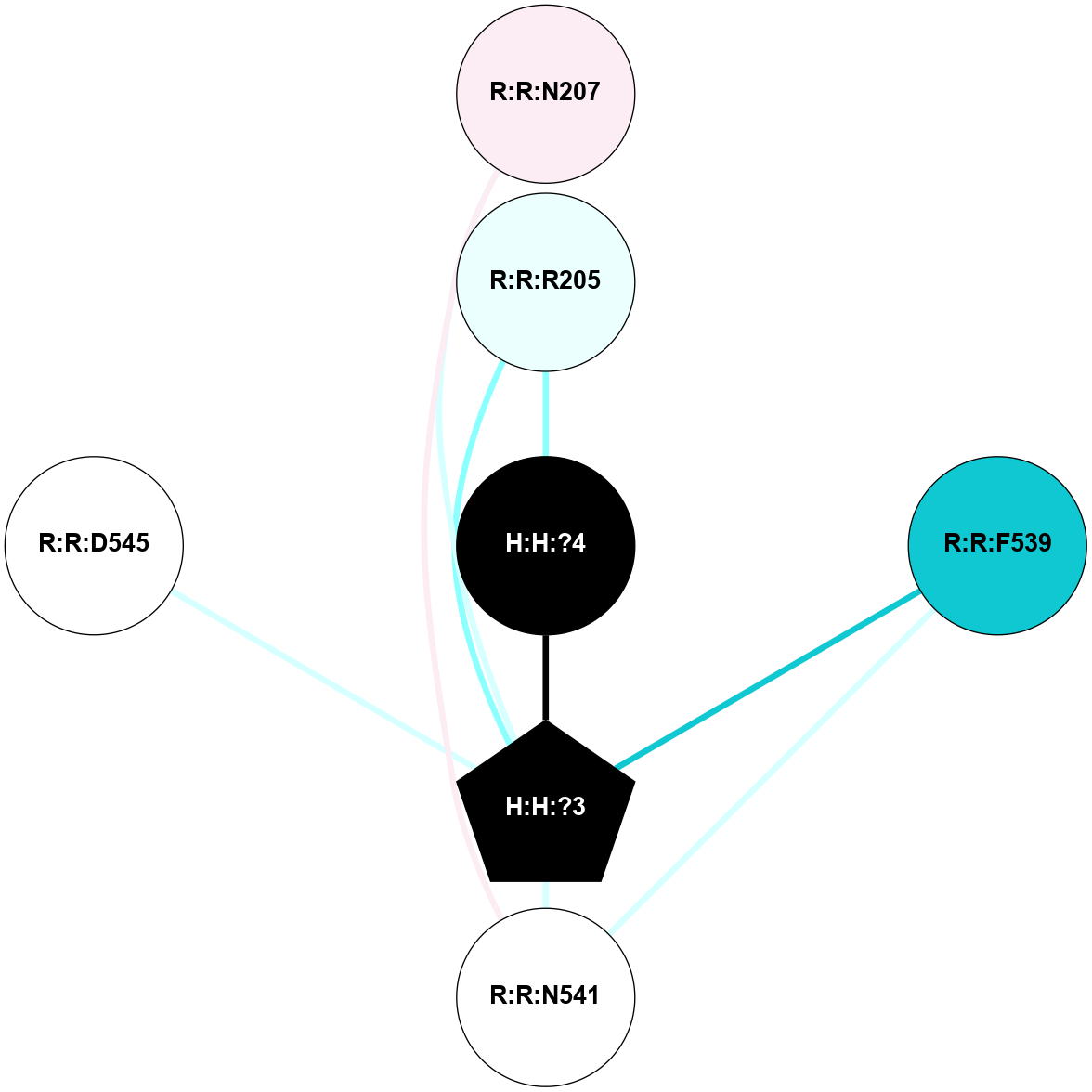
A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:R205 | 6.52 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?4 | R:R:R205 | 8.69 | 1 | No | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 13.62 | 1 | No | No | 6 | 5 | 2 | 2 | | H:H:?3 | R:R:N207 | 18.42 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:F539 | R:R:N541 | 10.87 | 1 | No | No | 1 | 5 | 2 | 2 | | H:H:?3 | R:R:F539 | 3.27 | 1 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?3 | R:R:N541 | 27.02 | 1 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | R:R:D545 | 3.64 | 1 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | H:H:?4 | 38.74 | 1 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 23.71 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
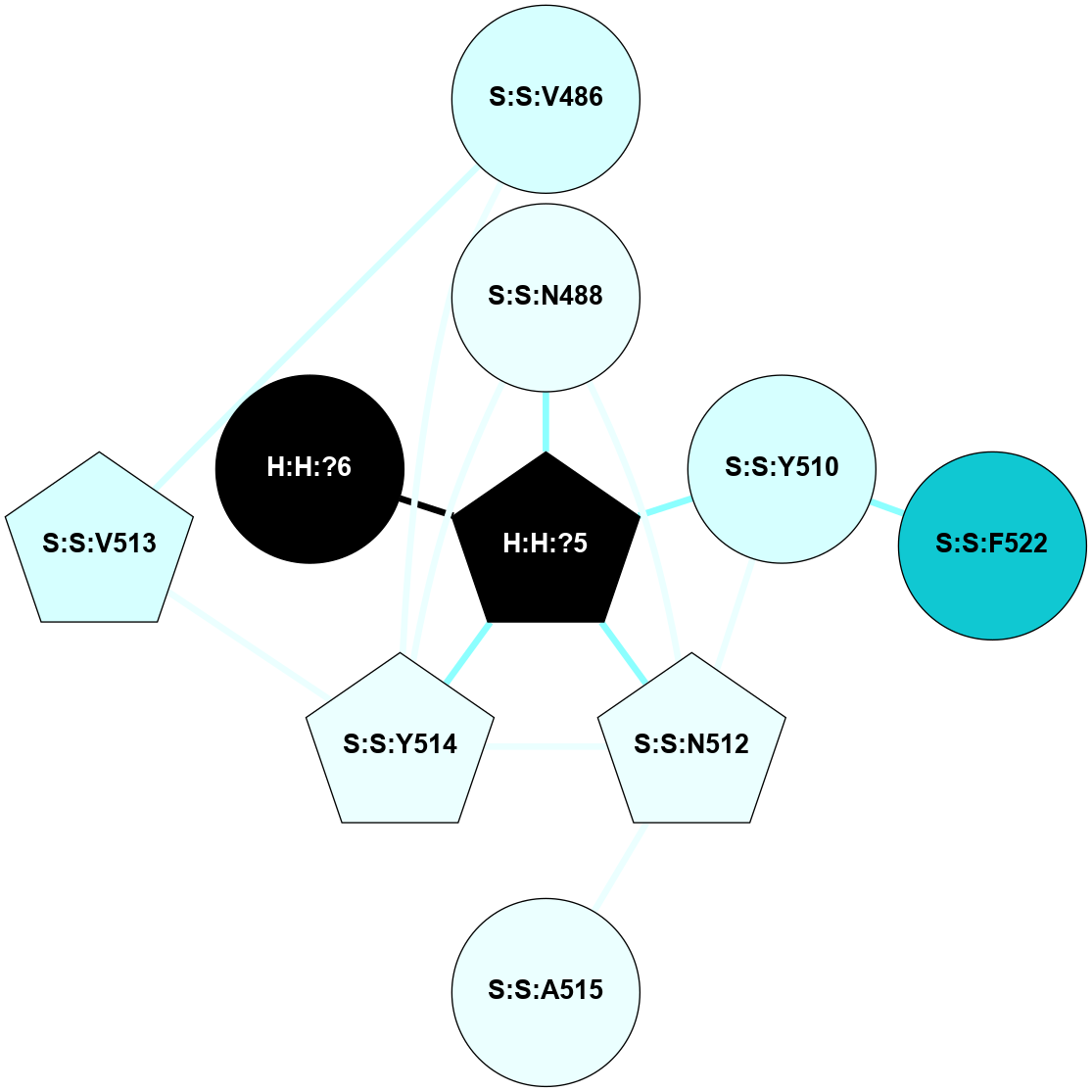
A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:V486 | S:S:V513 | 3.21 | 1 | No | Yes | 3 | 3 | 2 | 2 | | S:S:V486 | S:S:Y514 | 6.31 | 1 | No | Yes | 3 | 4 | 2 | 1 | | S:S:N488 | S:S:N512 | 6.81 | 1 | No | Yes | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:Y514 | 3.49 | 1 | No | Yes | 4 | 4 | 1 | 1 | | H:H:?5 | S:S:N488 | 27.02 | 1 | Yes | No | 0 | 4 | 0 | 1 | | S:S:N512 | S:S:Y510 | 2.33 | 1 | Yes | No | 4 | 3 | 1 | 1 | | S:S:F522 | S:S:Y510 | 10.32 | 0 | No | No | 1 | 3 | 2 | 1 | | H:H:?5 | S:S:Y510 | 10.48 | 1 | Yes | No | 0 | 3 | 0 | 1 | | S:S:N512 | S:S:Y514 | 6.98 | 1 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:A515 | S:S:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?5 | S:S:N512 | 17.19 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | S:S:V513 | S:S:Y514 | 7.57 | 1 | Yes | Yes | 3 | 4 | 2 | 1 | | H:H:?5 | S:S:Y514 | 11.53 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?5 | H:H:?6 | 40.96 | 1 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 21.44 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
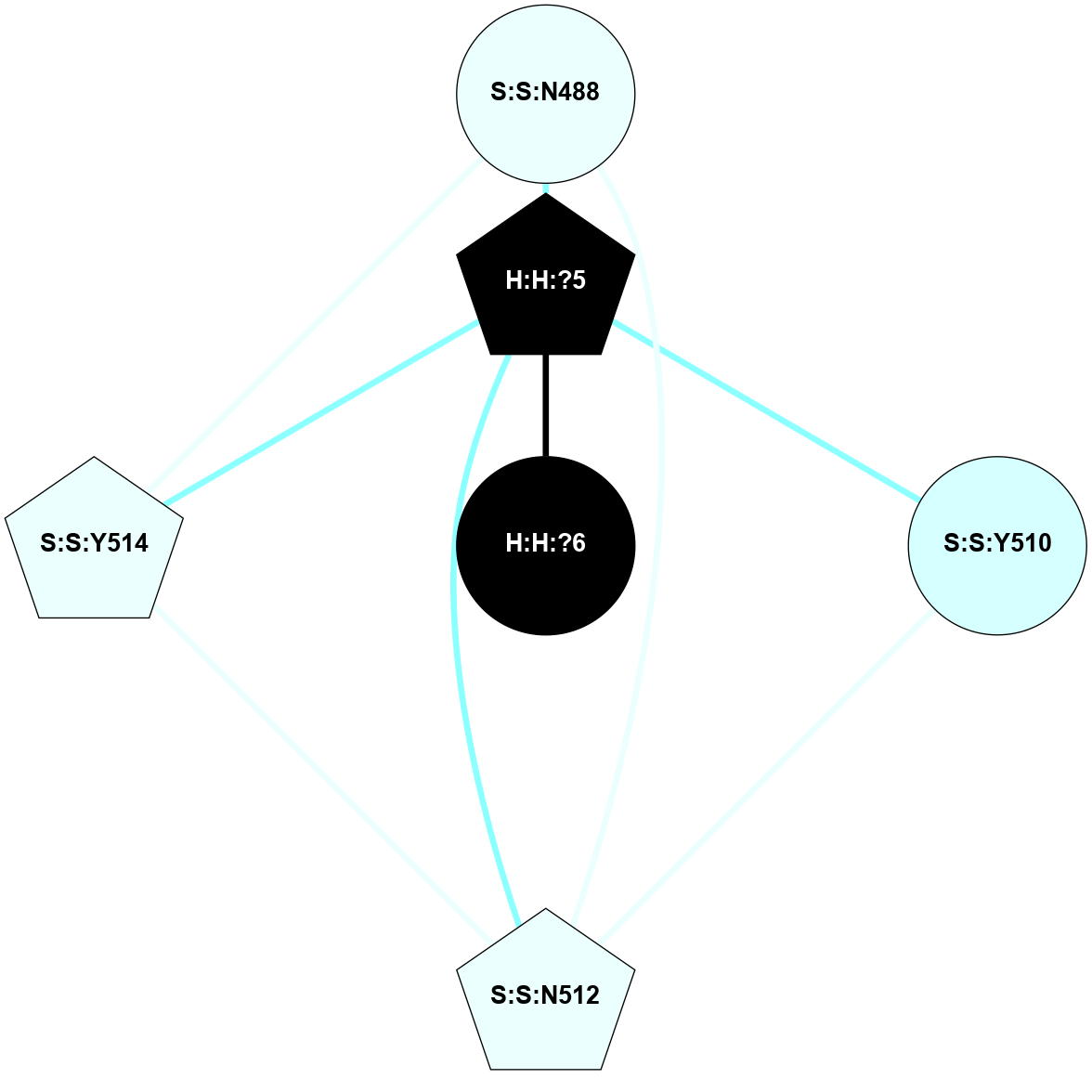
A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:N488 | S:S:N512 | 6.81 | 1 | No | Yes | 4 | 4 | 2 | 2 | | S:S:N488 | S:S:Y514 | 3.49 | 1 | No | Yes | 4 | 4 | 2 | 2 | | H:H:?5 | S:S:N488 | 27.02 | 1 | Yes | No | 0 | 4 | 1 | 2 | | S:S:N512 | S:S:Y510 | 2.33 | 1 | Yes | No | 4 | 3 | 2 | 2 | | H:H:?5 | S:S:Y510 | 10.48 | 1 | Yes | No | 0 | 3 | 1 | 2 | | S:S:N512 | S:S:Y514 | 6.98 | 1 | Yes | Yes | 4 | 4 | 2 | 2 | | H:H:?5 | S:S:N512 | 17.19 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?5 | S:S:Y514 | 11.53 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?5 | H:H:?6 | 40.96 | 1 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 40.96 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
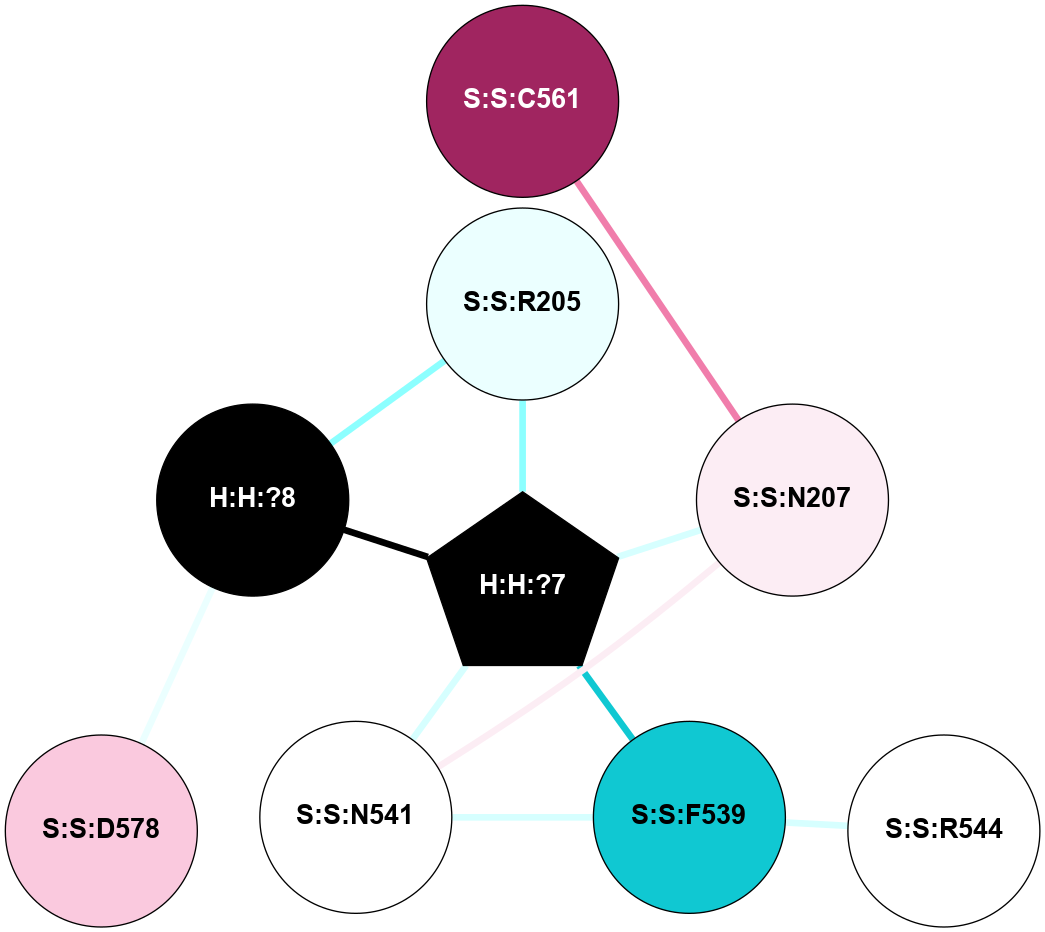
A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | S:S:R205 | 10.86 | 7 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?8 | S:S:R205 | 7.6 | 7 | No | No | 0 | 4 | 1 | 1 | | S:S:N207 | S:S:N541 | 17.71 | 7 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 4.72 | 0 | No | No | 9 | 6 | 2 | 1 | | H:H:?7 | S:S:N207 | 11.05 | 7 | Yes | No | 0 | 6 | 0 | 1 | | S:S:F539 | S:S:N541 | 7.25 | 7 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 26.72 | 7 | No | No | 1 | 5 | 1 | 2 | | H:H:?7 | S:S:F539 | 5.45 | 7 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?7 | S:S:N541 | 29.47 | 7 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?8 | S:S:D578 | 2.43 | 7 | No | No | 0 | 7 | 1 | 2 | | H:H:?7 | H:H:?8 | 40.96 | 7 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 19.56 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
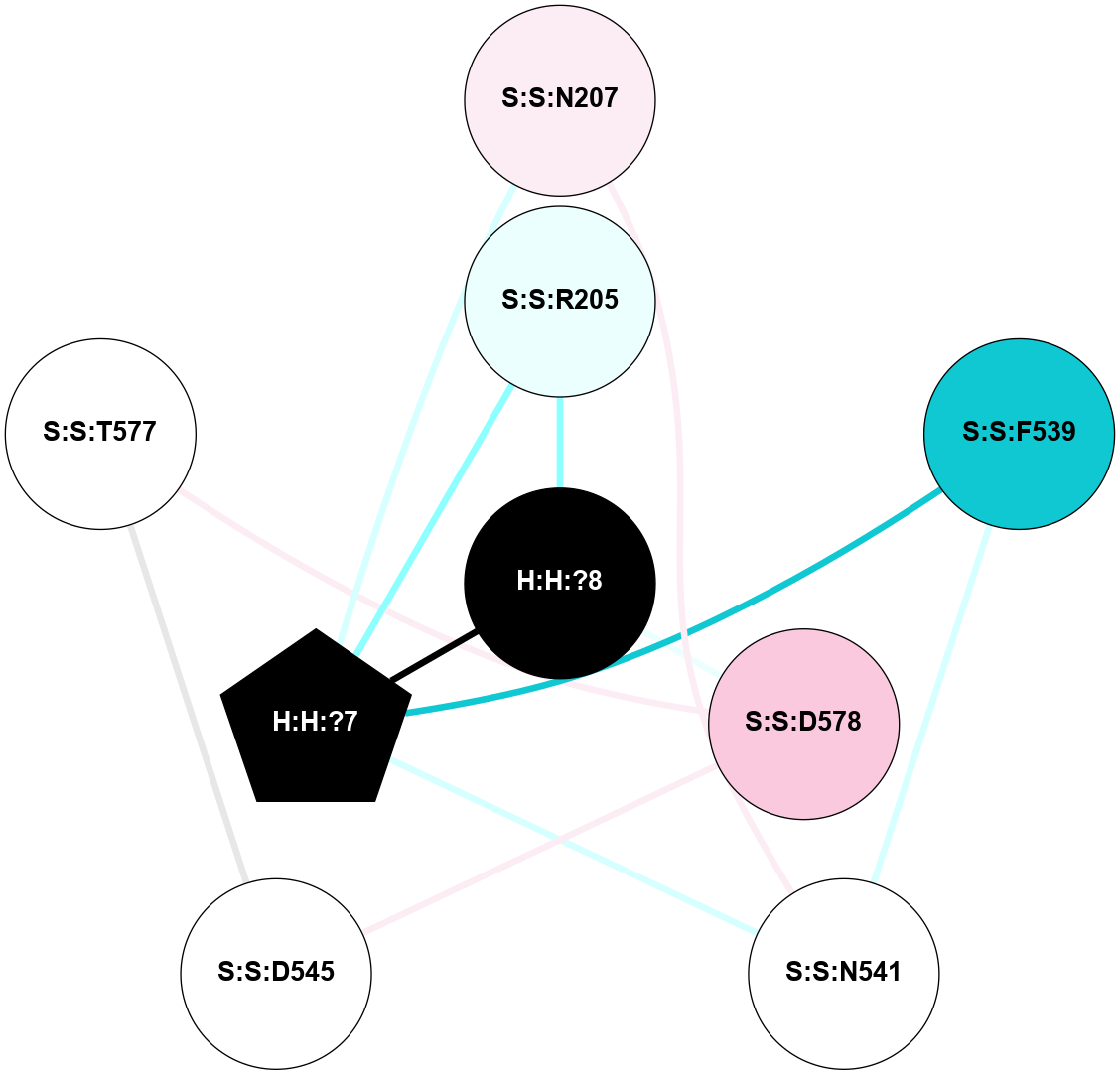
A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | S:S:R205 | 10.86 | 7 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?8 | S:S:R205 | 7.6 | 7 | No | No | 0 | 4 | 0 | 1 | | S:S:N207 | S:S:N541 | 17.71 | 7 | No | No | 6 | 5 | 2 | 2 | | H:H:?7 | S:S:N207 | 11.05 | 7 | Yes | No | 0 | 6 | 1 | 2 | | S:S:F539 | S:S:N541 | 7.25 | 7 | No | No | 1 | 5 | 2 | 2 | | H:H:?7 | S:S:F539 | 5.45 | 7 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?7 | S:S:N541 | 29.47 | 7 | Yes | No | 0 | 5 | 1 | 2 | | S:S:D545 | S:S:T577 | 2.89 | 7 | No | No | 5 | 5 | 2 | 2 | | S:S:D545 | S:S:D578 | 9.31 | 7 | No | No | 5 | 7 | 2 | 1 | | S:S:D578 | S:S:T577 | 2.89 | 7 | No | No | 7 | 5 | 1 | 2 | | H:H:?8 | S:S:D578 | 2.43 | 7 | No | No | 0 | 7 | 0 | 1 | | H:H:?7 | H:H:?8 | 40.96 | 7 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.00 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
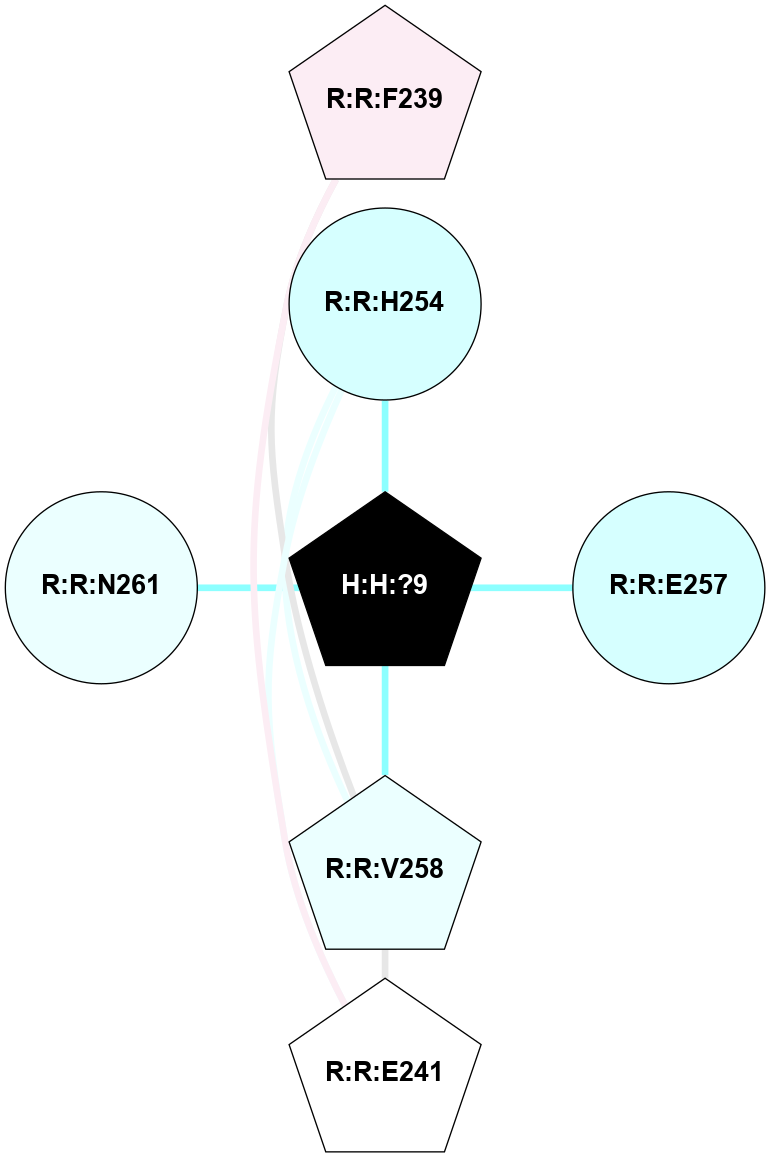
A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E241 | R:R:F239 | 2.33 | 1 | Yes | Yes | 5 | 6 | 2 | 2 | | R:R:F239 | R:R:V258 | 5.24 | 1 | Yes | Yes | 6 | 4 | 2 | 1 | | R:R:E241 | R:R:H254 | 6.15 | 1 | Yes | No | 5 | 3 | 2 | 1 | | R:R:E241 | R:R:V258 | 12.84 | 1 | Yes | Yes | 5 | 4 | 2 | 1 | | R:R:H254 | R:R:V258 | 4.15 | 1 | No | Yes | 3 | 4 | 1 | 1 | | H:H:?9 | R:R:H254 | 2.3 | 1 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?9 | R:R:E257 | 22.51 | 1 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?9 | R:R:V258 | 4 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?9 | R:R:N261 | 29.47 | 1 | Yes | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.57 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
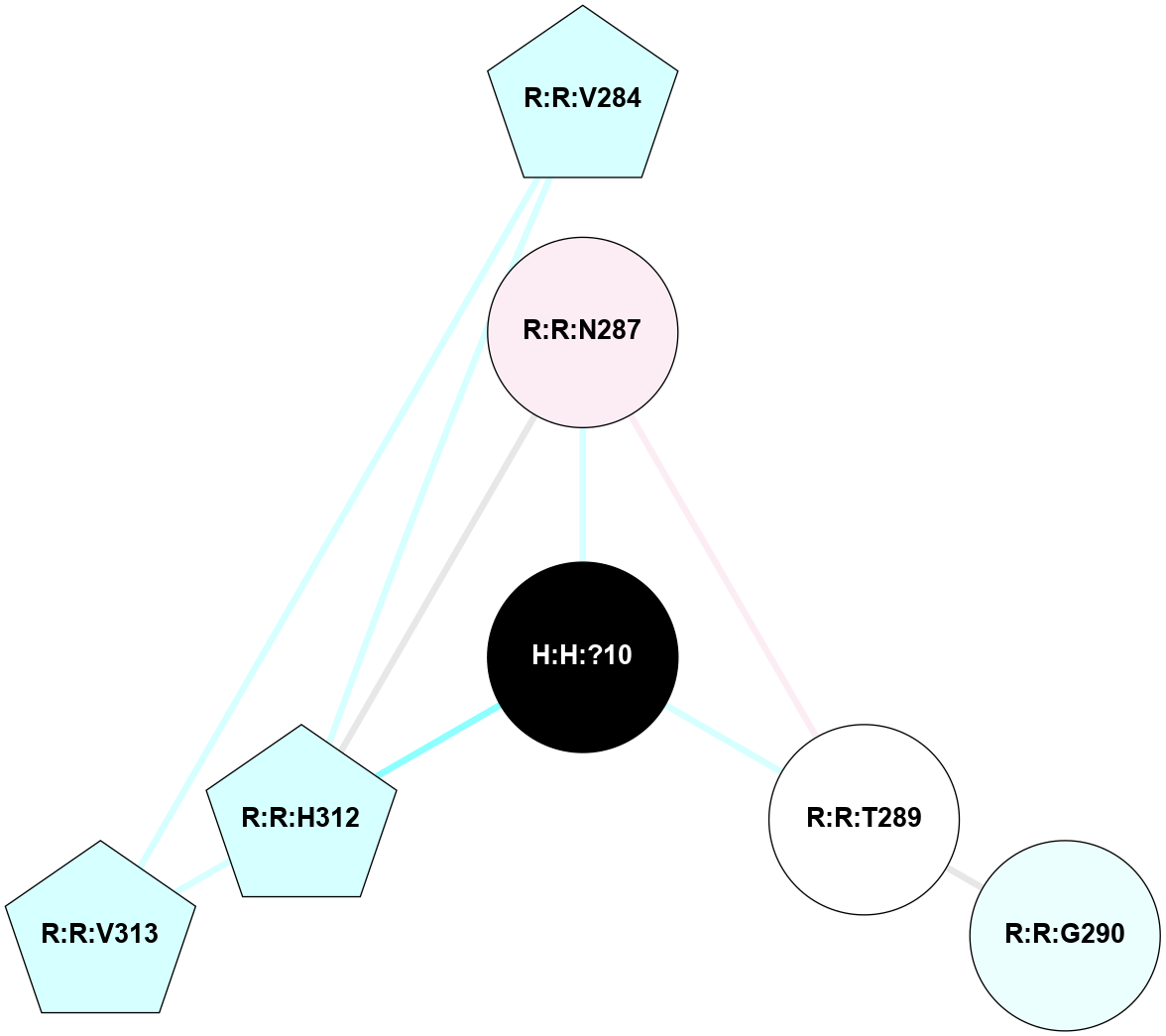
A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:H312 | R:R:V284 | 2.77 | 1 | Yes | Yes | 3 | 3 | 1 | 2 | | R:R:V284 | R:R:V313 | 6.41 | 1 | Yes | Yes | 3 | 3 | 2 | 2 | | R:R:N287 | R:R:T289 | 5.85 | 1 | No | No | 6 | 5 | 1 | 1 | | R:R:H312 | R:R:N287 | 7.65 | 1 | Yes | No | 3 | 6 | 1 | 1 | | H:H:?10 | R:R:N287 | 31.93 | 1 | No | No | 0 | 6 | 0 | 1 | | R:R:G290 | R:R:T289 | 1.82 | 0 | No | No | 4 | 5 | 2 | 1 | | H:H:?10 | R:R:T289 | 2.64 | 1 | No | No | 0 | 5 | 0 | 1 | | R:R:H312 | R:R:V313 | 5.54 | 1 | Yes | Yes | 3 | 3 | 1 | 2 | | H:H:?10 | R:R:H312 | 3.45 | 1 | No | Yes | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 12.67 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N468 | R:R:Q476 | 5.28 | 15 | No | No | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 5.85 | 15 | No | No | 6 | 3 | 1 | 2 | | H:H:?11 | R:R:N468 | 28.24 | 15 | No | No | 0 | 6 | 0 | 1 | | R:R:Q476 | R:R:T470 | 9.92 | 15 | No | No | 3 | 5 | 1 | 2 | | H:H:?11 | R:R:Q476 | 15.47 | 15 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 21.86 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N594 | R:R:T596 | 4.39 | 0 | No | No | 3 | 5 | 1 | 2 | | H:H:?12 | R:R:N594 | 25.79 | 0 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 25.79 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:F239 | S:S:V258 | 5.24 | 1 | Yes | Yes | 6 | 4 | 2 | 1 | | S:S:E241 | S:S:H254 | 7.39 | 1 | No | No | 5 | 3 | 2 | 2 | | S:S:E241 | S:S:V258 | 7.13 | 1 | No | Yes | 5 | 4 | 2 | 1 | | S:S:H254 | S:S:V258 | 5.54 | 1 | No | Yes | 3 | 4 | 2 | 1 | | H:H:?13 | S:S:N261 | 28.24 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?13 | S:S:V258 | 1.33 | 0 | No | Yes | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.79 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
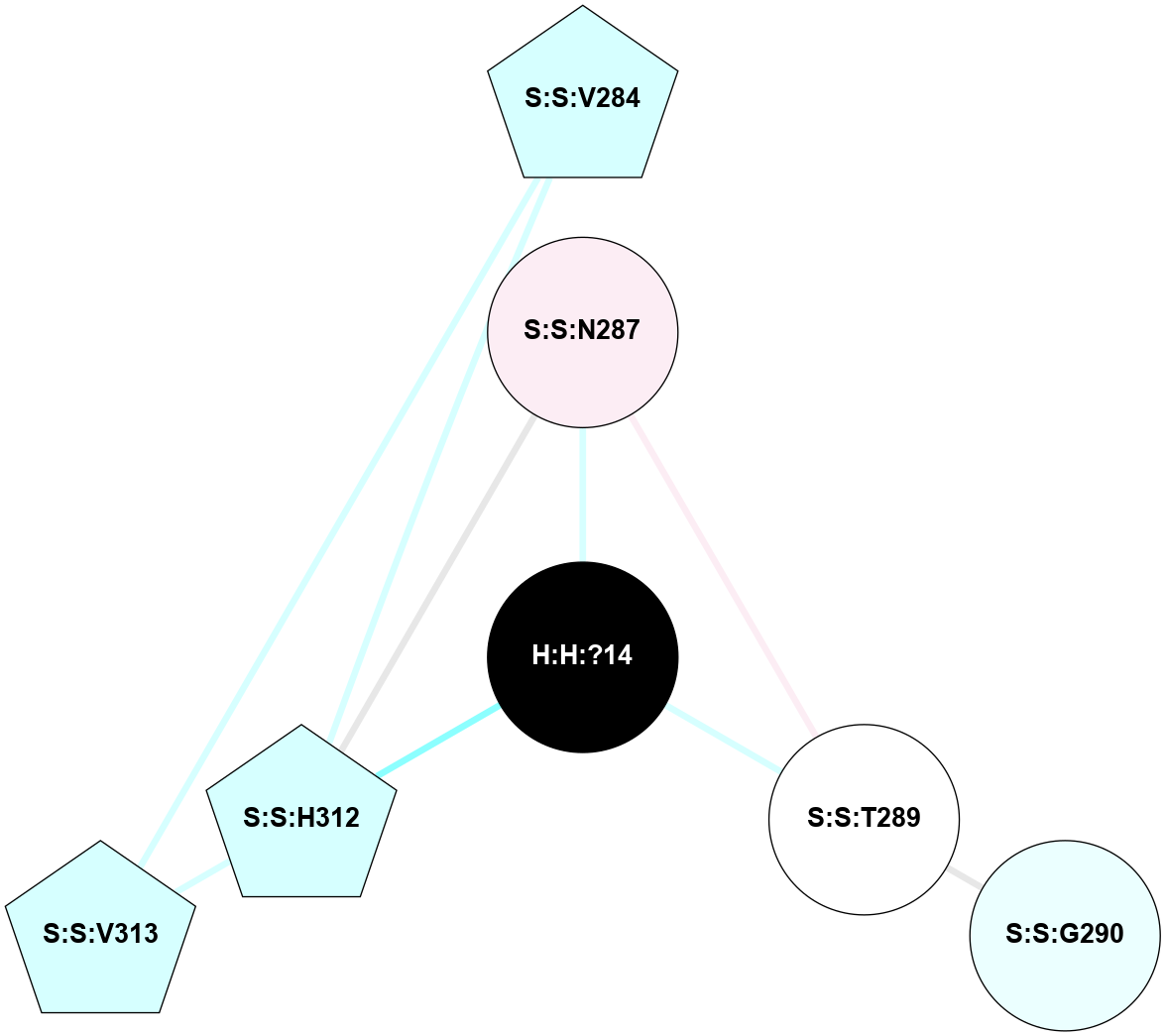
A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:H312 | S:S:V284 | 4.15 | 1 | Yes | Yes | 3 | 3 | 1 | 2 | | S:S:V284 | S:S:V313 | 6.41 | 1 | Yes | Yes | 3 | 3 | 2 | 2 | | S:S:N287 | S:S:T289 | 4.39 | 1 | No | No | 6 | 5 | 1 | 1 | | S:S:H312 | S:S:N287 | 7.65 | 1 | Yes | No | 3 | 6 | 1 | 1 | | H:H:?14 | S:S:N287 | 27.02 | 1 | No | No | 0 | 6 | 0 | 1 | | S:S:G290 | S:S:T289 | 1.82 | 0 | No | No | 4 | 5 | 2 | 1 | | H:H:?14 | S:S:T289 | 5.27 | 1 | No | No | 0 | 5 | 0 | 1 | | S:S:H312 | S:S:V313 | 5.54 | 1 | Yes | Yes | 3 | 3 | 1 | 2 | | H:H:?14 | S:S:H312 | 5.75 | 1 | No | Yes | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 12.68 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:N468 | S:S:Q476 | 7.92 | 19 | No | No | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 5.85 | 19 | No | No | 6 | 3 | 1 | 2 | | H:H:?15 | S:S:N468 | 25.79 | 19 | No | No | 0 | 6 | 0 | 1 | | S:S:Q476 | S:S:T470 | 7.09 | 19 | No | No | 3 | 5 | 1 | 2 | | H:H:?15 | S:S:Q476 | 11.9 | 19 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 18.84 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9AXF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:N592 | S:S:N594 | 5.45 | 0 | No | Yes | 7 | 3 | 2 | 1 | | S:S:E593 | S:S:N594 | 9.2 | 23 | No | Yes | 2 | 3 | 1 | 1 | | H:H:?16 | S:S:E593 | 14.22 | 23 | No | No | 0 | 2 | 0 | 1 | | S:S:N594 | S:S:T596 | 10.24 | 23 | Yes | No | 3 | 5 | 1 | 2 | | H:H:?16 | S:S:N594 | 29.47 | 23 | No | Yes | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 21.84 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9AYF | C | Ion | Calcium Sensing | CaS | Homo Sapiens | Ca | Ca; Ca; Ca; Cyclomethyltryptophan; 9IG; PO4 | Gi1/Beta1/Gamma2 | 3.6 | 2024-04-17 | doi.org/10.1038/s41586-024-07331-1 |
|
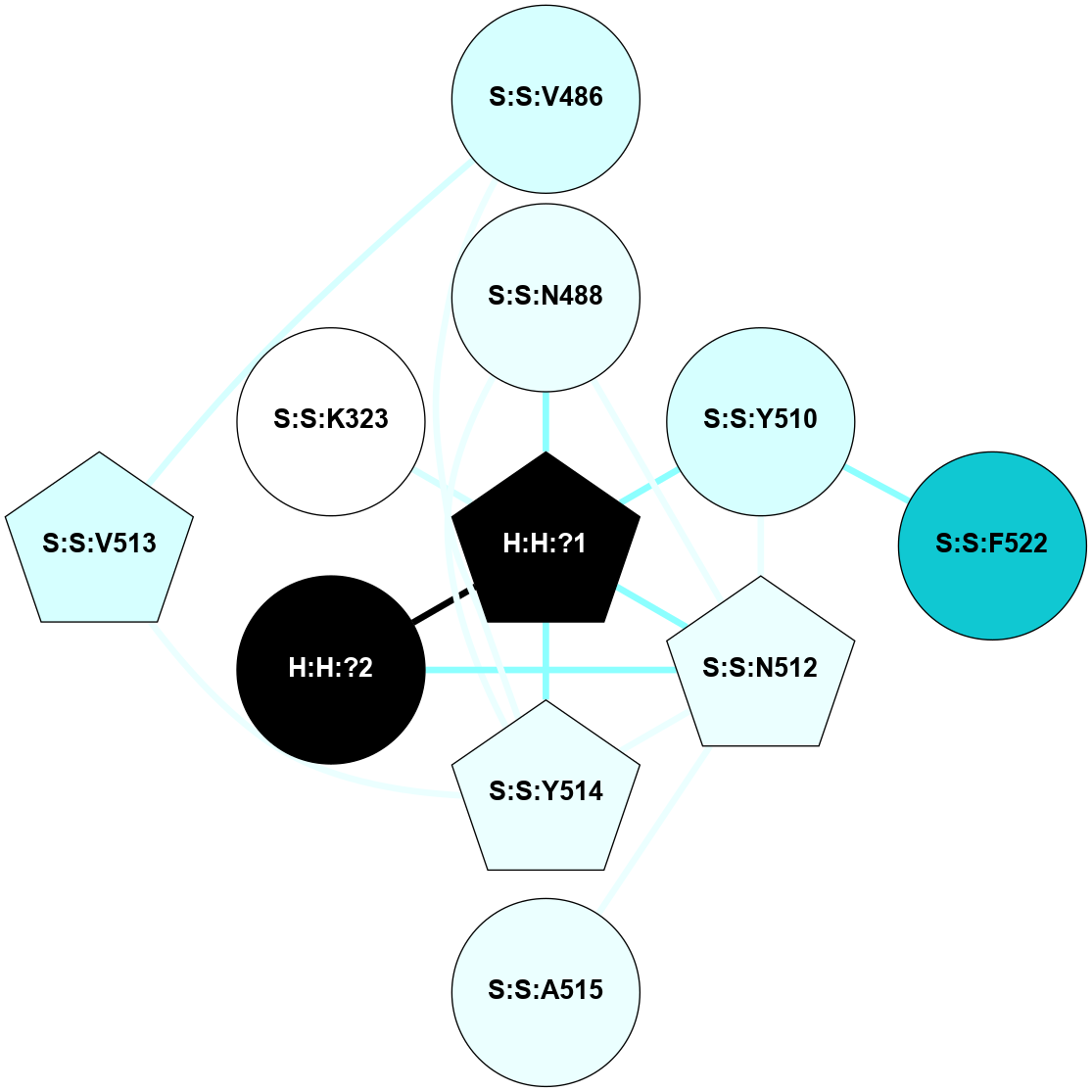
A 2D representation of the interactions of NAG in 9AYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:V486 | S:S:V513 | 3.21 | 1 | No | Yes | 3 | 3 | 2 | 2 | | S:S:V486 | S:S:Y514 | 3.79 | 1 | No | Yes | 3 | 4 | 2 | 1 | | S:S:N488 | S:S:N512 | 5.45 | 1 | No | Yes | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:Y514 | 4.65 | 1 | No | Yes | 4 | 4 | 1 | 1 | | H:H:?1 | S:S:N488 | 29.57 | 1 | Yes | No | 0 | 4 | 0 | 1 | | S:S:N512 | S:S:Y510 | 2.33 | 1 | Yes | No | 4 | 3 | 1 | 1 | | S:S:F522 | S:S:Y510 | 21.66 | 0 | No | No | 1 | 3 | 2 | 1 | | H:H:?1 | S:S:Y510 | 8.42 | 1 | Yes | No | 0 | 3 | 0 | 1 | | S:S:N512 | S:S:Y514 | 5.81 | 1 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:A515 | S:S:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?1 | S:S:N512 | 18.48 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?2 | S:S:N512 | 3.7 | 1 | No | Yes | 0 | 4 | 1 | 1 | | S:S:V513 | S:S:Y514 | 7.57 | 1 | Yes | Yes | 3 | 4 | 2 | 1 | | H:H:?1 | S:S:Y514 | 13.67 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 40.12 | 1 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | S:S:K323 | 1.27 | 1 | Yes | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 18.59 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 16.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
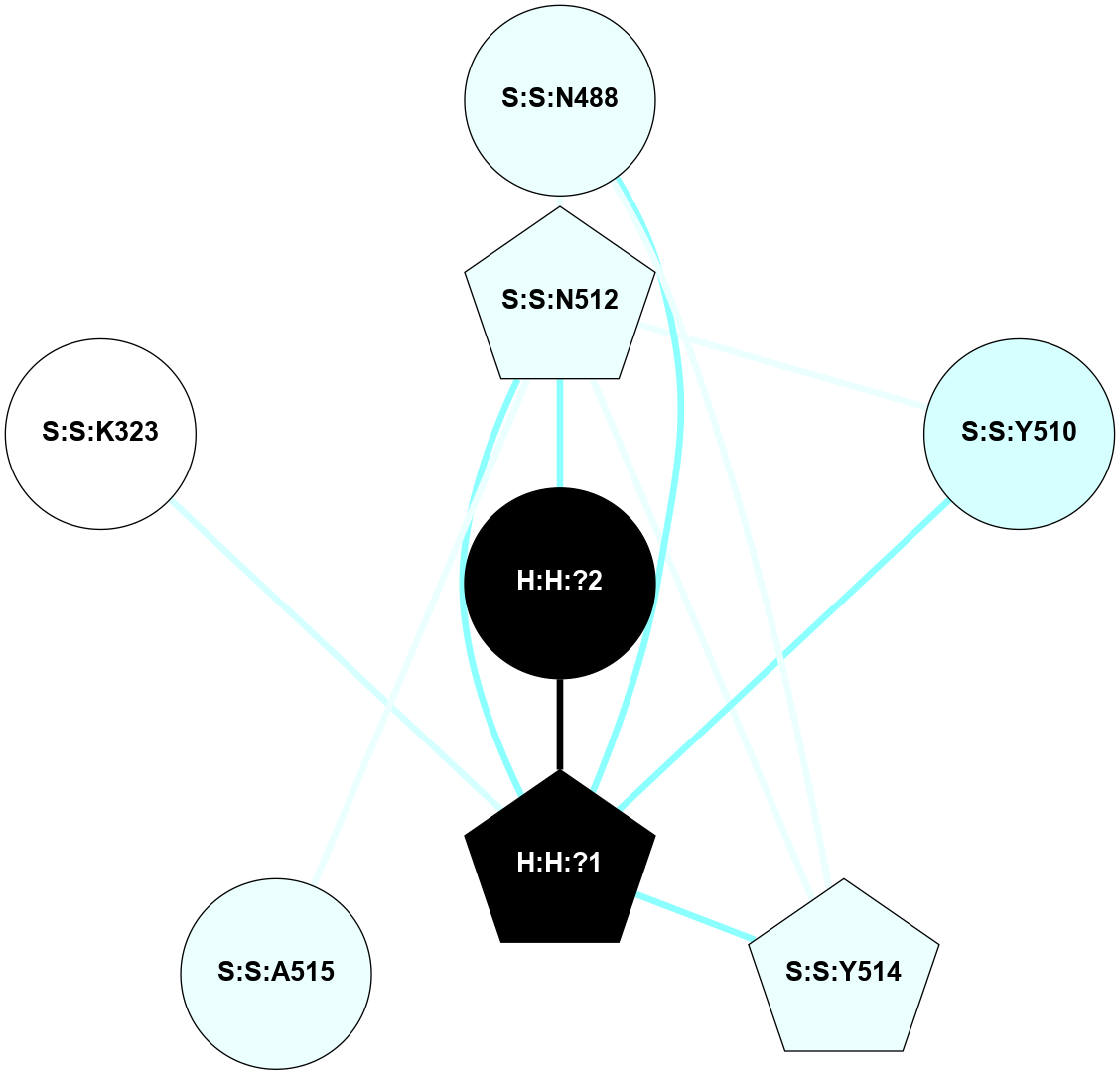
A 2D representation of the interactions of NAG in 9AYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:N488 | S:S:N512 | 5.45 | 1 | No | Yes | 4 | 4 | 2 | 1 | | S:S:N488 | S:S:Y514 | 4.65 | 1 | No | Yes | 4 | 4 | 2 | 2 | | H:H:?1 | S:S:N488 | 29.57 | 1 | Yes | No | 0 | 4 | 1 | 2 | | S:S:N512 | S:S:Y510 | 2.33 | 1 | Yes | No | 4 | 3 | 1 | 2 | | H:H:?1 | S:S:Y510 | 8.42 | 1 | Yes | No | 0 | 3 | 1 | 2 | | S:S:N512 | S:S:Y514 | 5.81 | 1 | Yes | Yes | 4 | 4 | 1 | 2 | | S:S:A515 | S:S:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?1 | S:S:N512 | 18.48 | 1 | Yes | Yes | 0 | 4 | 1 | 1 | | H:H:?2 | S:S:N512 | 3.7 | 1 | No | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | S:S:Y514 | 13.67 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 40.12 | 1 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | S:S:K323 | 1.27 | 1 | Yes | No | 0 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 21.91 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
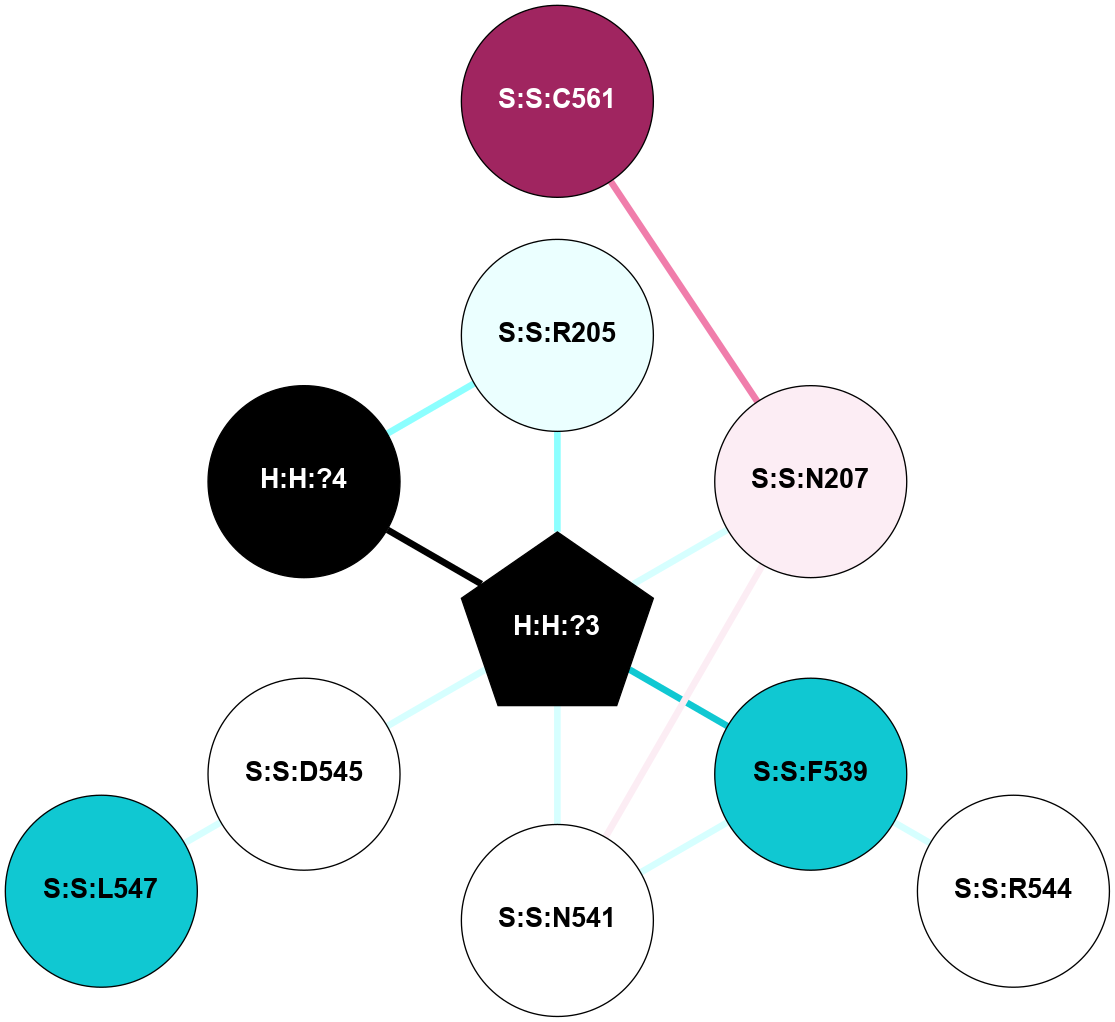
A 2D representation of the interactions of NAG in 9AYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | S:S:R205 | 3.27 | 10 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?4 | S:S:R205 | 18.53 | 10 | No | No | 0 | 4 | 1 | 1 | | S:S:N207 | S:S:N541 | 13.62 | 10 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 7.87 | 0 | No | No | 9 | 6 | 2 | 1 | | H:H:?3 | S:S:N207 | 7.39 | 10 | Yes | No | 0 | 6 | 0 | 1 | | S:S:F539 | S:S:N541 | 9.67 | 10 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 25.66 | 10 | No | No | 1 | 5 | 1 | 2 | | H:H:?3 | S:S:F539 | 8.74 | 10 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?3 | S:S:N541 | 24.64 | 10 | Yes | No | 0 | 5 | 0 | 1 | | S:S:D545 | S:S:L547 | 5.43 | 0 | No | No | 5 | 1 | 1 | 2 | | H:H:?3 | S:S:D545 | 2.44 | 10 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?3 | H:H:?4 | 37.89 | 10 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.06 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
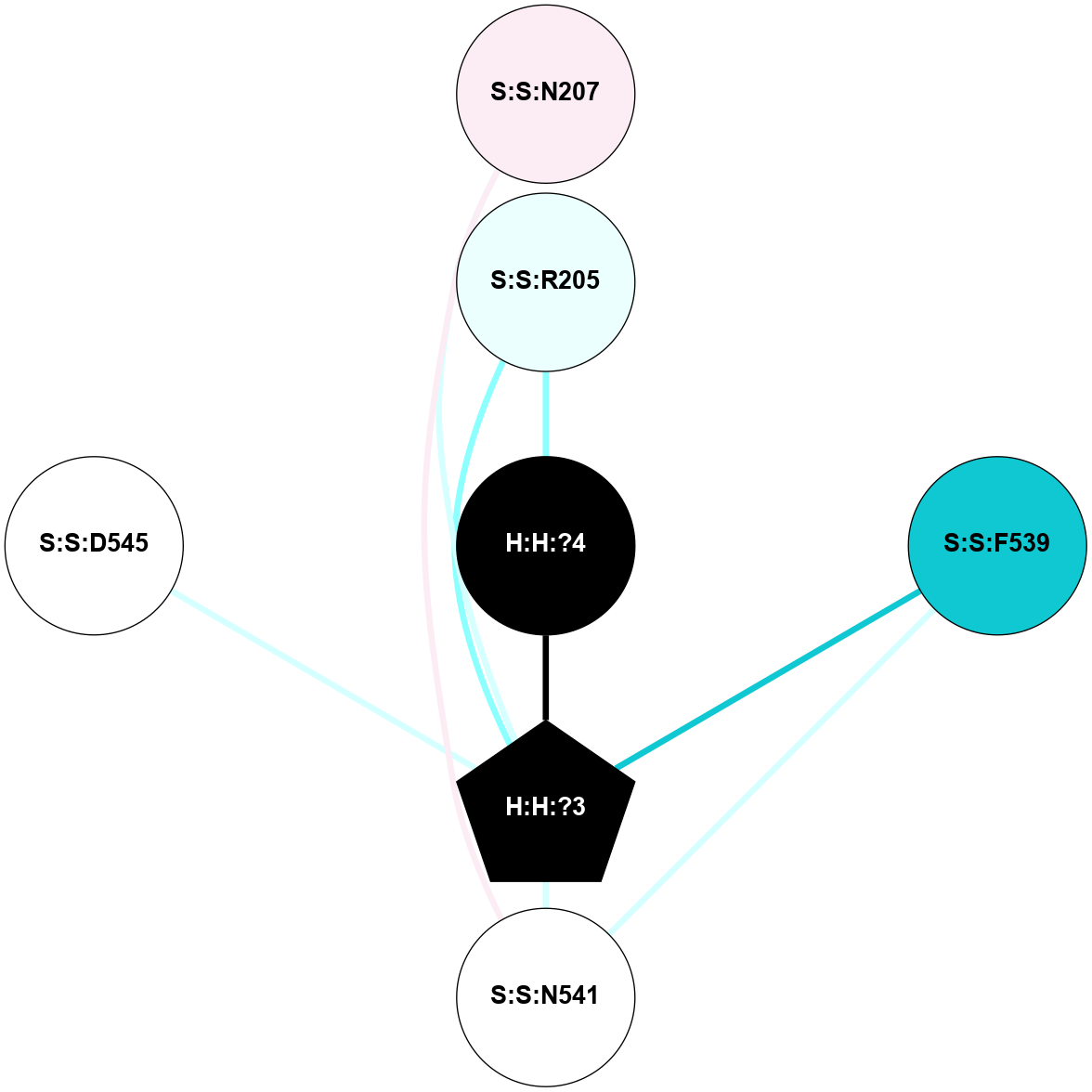
A 2D representation of the interactions of NAG in 9AYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | S:S:R205 | 3.27 | 10 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?4 | S:S:R205 | 18.53 | 10 | No | No | 0 | 4 | 0 | 1 | | S:S:N207 | S:S:N541 | 13.62 | 10 | No | No | 6 | 5 | 2 | 2 | | H:H:?3 | S:S:N207 | 7.39 | 10 | Yes | No | 0 | 6 | 1 | 2 | | S:S:F539 | S:S:N541 | 9.67 | 10 | No | No | 1 | 5 | 2 | 2 | | H:H:?3 | S:S:F539 | 8.74 | 10 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?3 | S:S:N541 | 24.64 | 10 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | S:S:D545 | 2.44 | 10 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?3 | H:H:?4 | 37.89 | 10 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 28.21 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
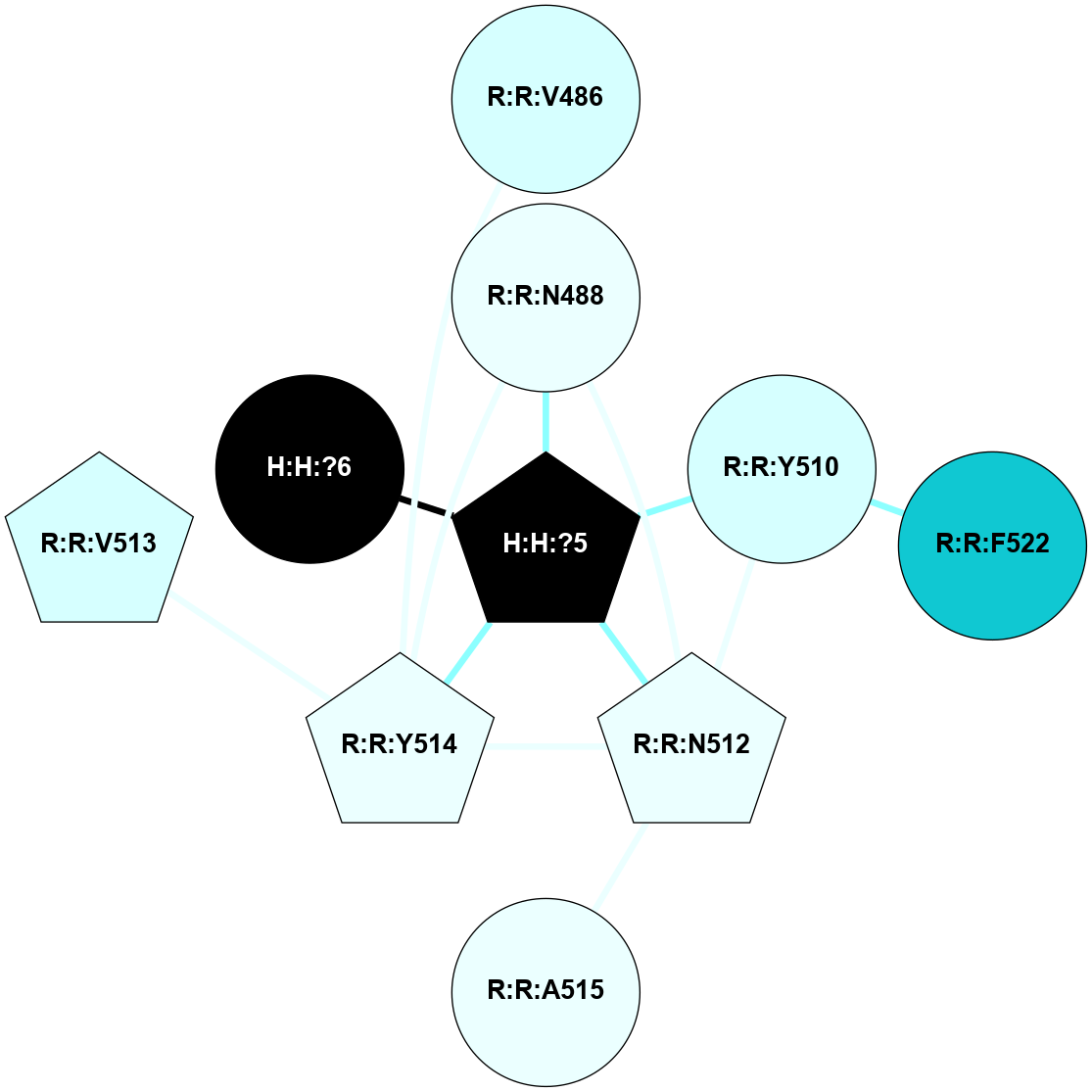
A 2D representation of the interactions of NAG in 9AYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:V486 | R:R:Y514 | 5.05 | 0 | No | Yes | 3 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 6.81 | 1 | No | Yes | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:Y514 | 4.65 | 1 | No | Yes | 4 | 4 | 1 | 1 | | H:H:?5 | R:R:N488 | 28.34 | 1 | Yes | No | 0 | 4 | 0 | 1 | | R:R:N512 | R:R:Y510 | 2.33 | 1 | Yes | No | 4 | 3 | 1 | 1 | | R:R:F522 | R:R:Y510 | 19.6 | 0 | No | No | 1 | 3 | 2 | 1 | | H:H:?5 | R:R:Y510 | 7.36 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:N512 | R:R:Y514 | 5.81 | 1 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | | H:H:?5 | R:R:N512 | 17.25 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:V513 | R:R:Y514 | 5.05 | 1 | Yes | Yes | 3 | 4 | 2 | 1 | | H:H:?5 | R:R:Y514 | 16.83 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?5 | H:H:?6 | 42.35 | 1 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 22.43 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
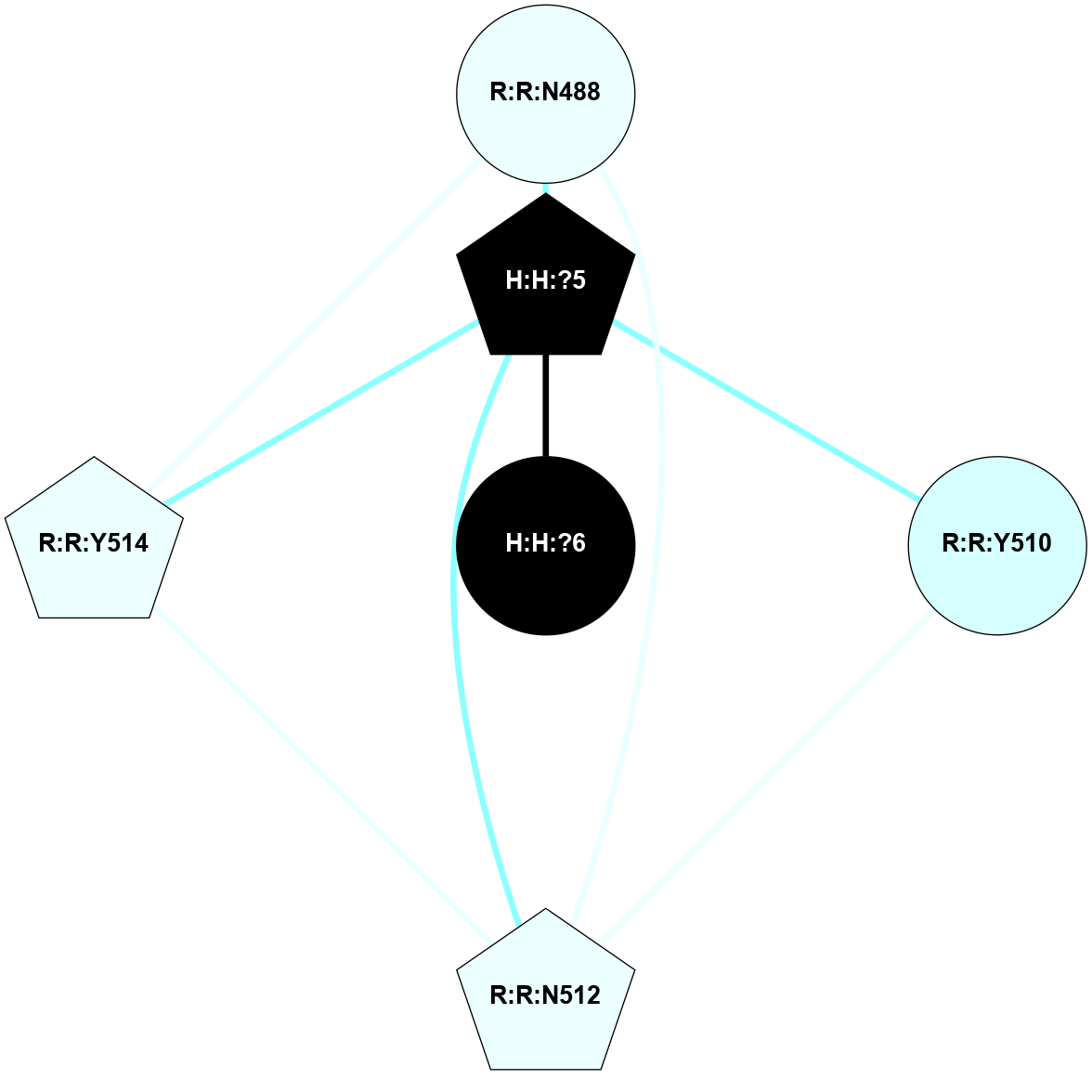
A 2D representation of the interactions of NAG in 9AYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N488 | R:R:N512 | 6.81 | 1 | No | Yes | 4 | 4 | 2 | 2 | | R:R:N488 | R:R:Y514 | 4.65 | 1 | No | Yes | 4 | 4 | 2 | 2 | | H:H:?5 | R:R:N488 | 28.34 | 1 | Yes | No | 0 | 4 | 1 | 2 | | R:R:N512 | R:R:Y510 | 2.33 | 1 | Yes | No | 4 | 3 | 2 | 2 | | H:H:?5 | R:R:Y510 | 7.36 | 1 | Yes | No | 0 | 3 | 1 | 2 | | R:R:N512 | R:R:Y514 | 5.81 | 1 | Yes | Yes | 4 | 4 | 2 | 2 | | H:H:?5 | R:R:N512 | 17.25 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?5 | R:R:Y514 | 16.83 | 1 | Yes | Yes | 0 | 4 | 1 | 2 | | H:H:?5 | H:H:?6 | 42.35 | 1 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 42.35 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
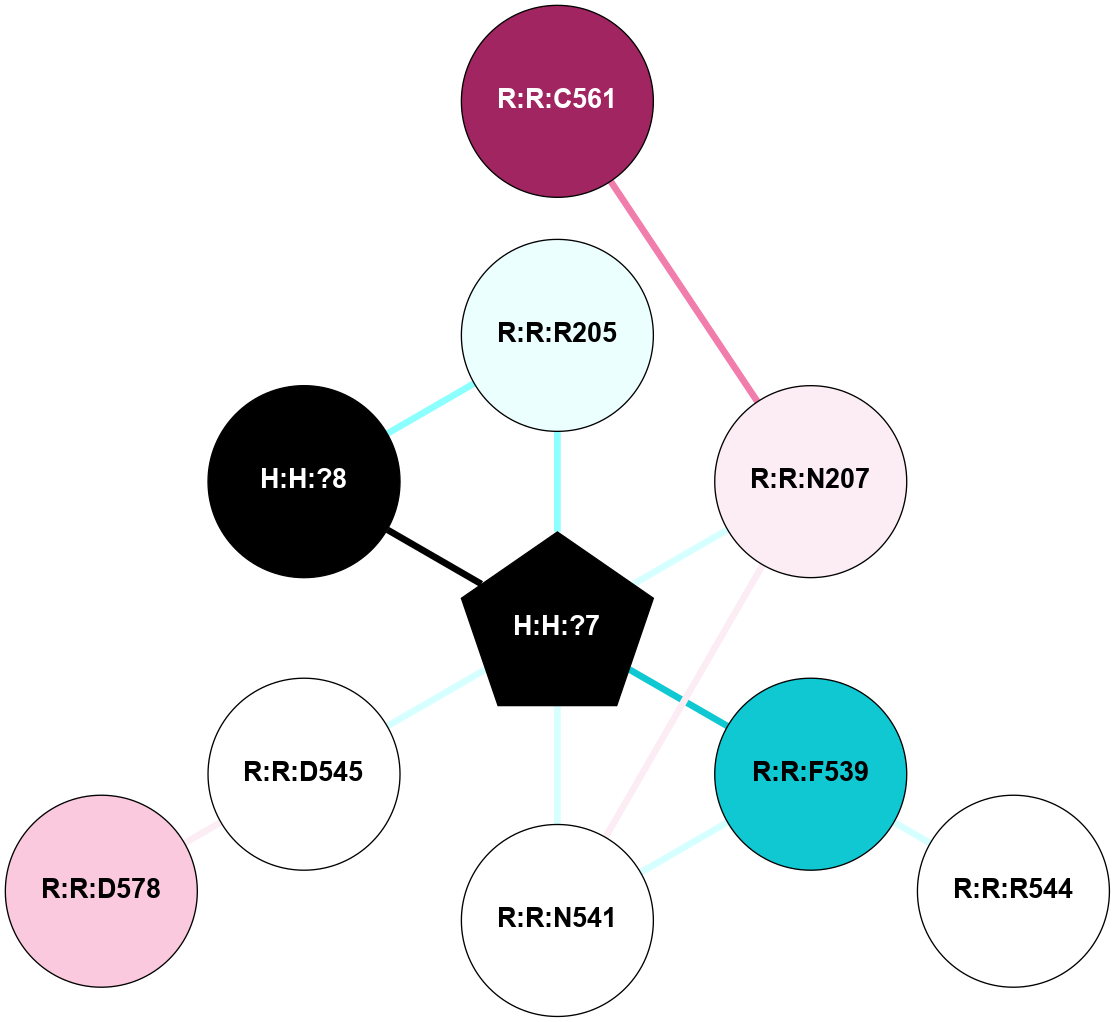
A 2D representation of the interactions of NAG in 9AYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | R:R:R205 | 3.27 | 1 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?8 | R:R:R205 | 2.18 | 1 | No | No | 0 | 4 | 1 | 1 | | R:R:N207 | R:R:N541 | 16.35 | 1 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 7.87 | 1 | No | No | 9 | 6 | 2 | 1 | | H:H:?7 | R:R:N207 | 13.55 | 1 | Yes | No | 0 | 6 | 0 | 1 | | R:R:F539 | R:R:N541 | 7.25 | 1 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 22.45 | 1 | No | No | 1 | 5 | 1 | 2 | | H:H:?7 | R:R:F539 | 6.56 | 1 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?7 | R:R:N541 | 30.8 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:D545 | R:R:D578 | 9.31 | 0 | No | No | 5 | 7 | 1 | 2 | | H:H:?7 | R:R:D545 | 12.18 | 1 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?7 | H:H:?8 | 39.01 | 1 | Yes | No | 0 | 0 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.56 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
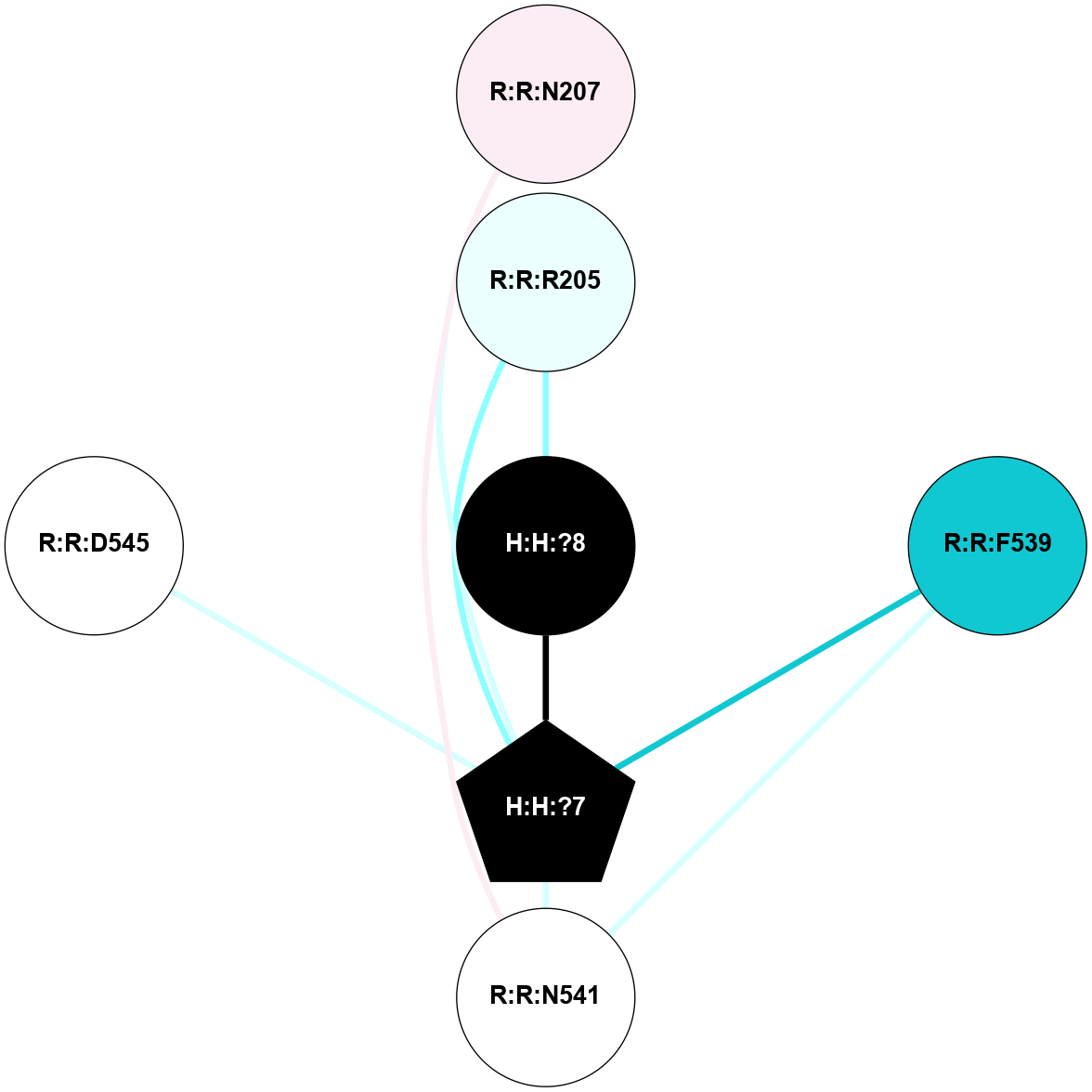
A 2D representation of the interactions of NAG in 9AYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | R:R:R205 | 3.27 | 1 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?8 | R:R:R205 | 2.18 | 1 | No | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 16.35 | 1 | No | No | 6 | 5 | 2 | 2 | | H:H:?7 | R:R:N207 | 13.55 | 1 | Yes | No | 0 | 6 | 1 | 2 | | R:R:F539 | R:R:N541 | 7.25 | 1 | No | No | 1 | 5 | 2 | 2 | | H:H:?7 | R:R:F539 | 6.56 | 1 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?7 | R:R:N541 | 30.8 | 1 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?7 | R:R:D545 | 12.18 | 1 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?7 | H:H:?8 | 39.01 | 1 | Yes | No | 0 | 0 | 1 | 0 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 20.59 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
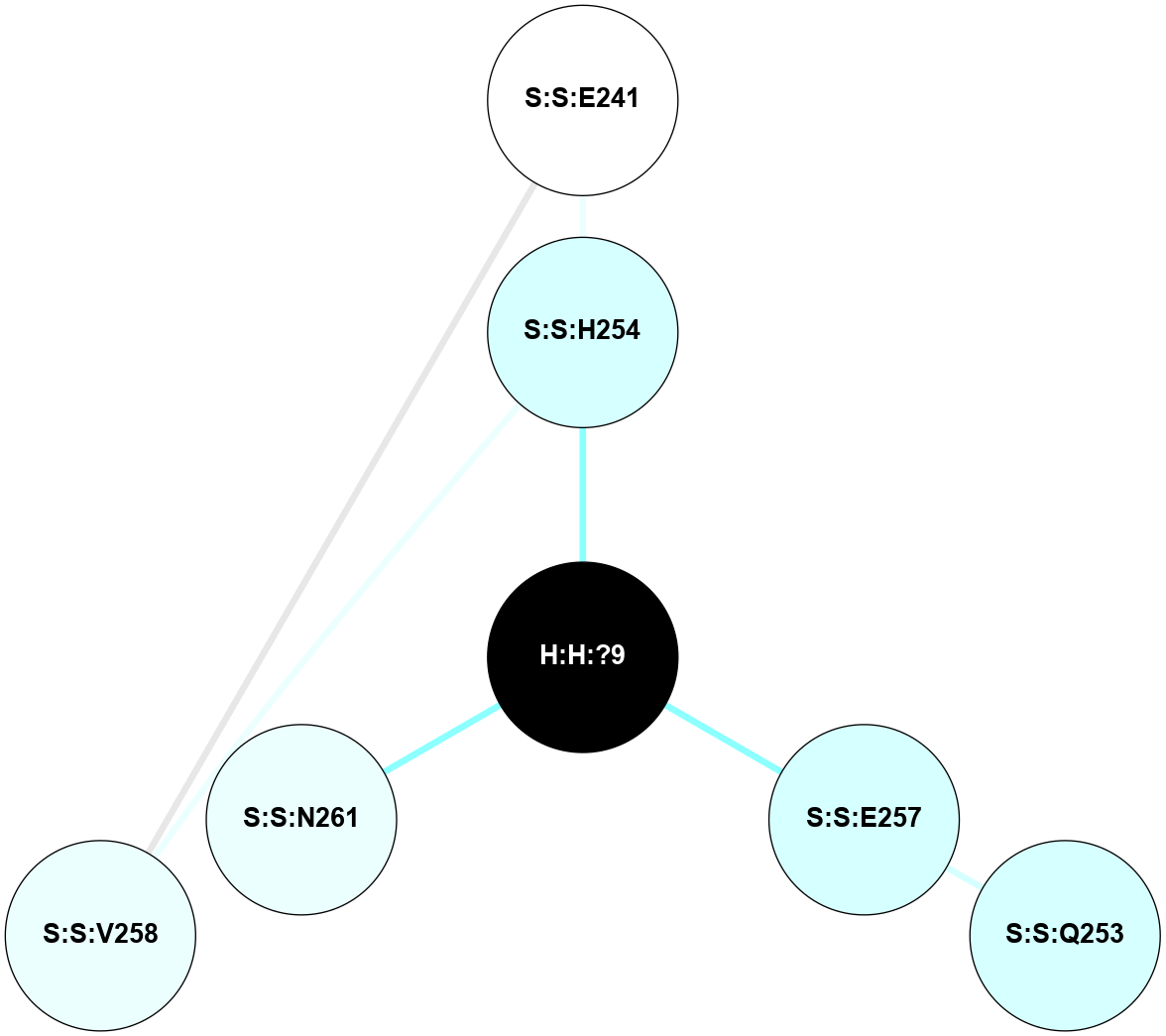
A 2D representation of the interactions of NAG in 9AYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:E241 | S:S:H254 | 7.39 | 1 | No | No | 5 | 3 | 2 | 1 | | S:S:E241 | S:S:V258 | 8.56 | 1 | No | No | 5 | 4 | 2 | 2 | | S:S:E257 | S:S:Q253 | 2.55 | 0 | No | No | 3 | 3 | 1 | 2 | | S:S:H254 | S:S:V258 | 6.92 | 1 | No | No | 3 | 4 | 1 | 2 | | H:H:?9 | S:S:H254 | 4.61 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?9 | S:S:E257 | 11.89 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?9 | S:S:N261 | 23.41 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.30 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9AYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:H312 | S:S:V284 | 2.77 | 1 | Yes | Yes | 3 | 3 | 1 | 2 | | S:S:V284 | S:S:V313 | 6.41 | 1 | Yes | Yes | 3 | 3 | 2 | 2 | | S:S:H312 | S:S:N287 | 8.93 | 1 | Yes | No | 3 | 6 | 1 | 1 | | H:H:?10 | S:S:N287 | 30.8 | 1 | No | No | 0 | 6 | 0 | 1 | | S:S:H312 | S:S:V313 | 6.92 | 1 | Yes | Yes | 3 | 3 | 1 | 2 | | H:H:?10 | S:S:H312 | 13.84 | 1 | No | Yes | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 22.32 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9AYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:N468 | S:S:Q476 | 5.28 | 22 | No | No | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 7.31 | 22 | No | No | 6 | 3 | 1 | 2 | | H:H:?11 | S:S:N468 | 29.57 | 22 | No | No | 0 | 6 | 0 | 1 | | S:S:Q476 | S:S:T470 | 8.5 | 22 | No | No | 3 | 5 | 1 | 2 | | H:H:?11 | S:S:Q476 | 11.94 | 22 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.75 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
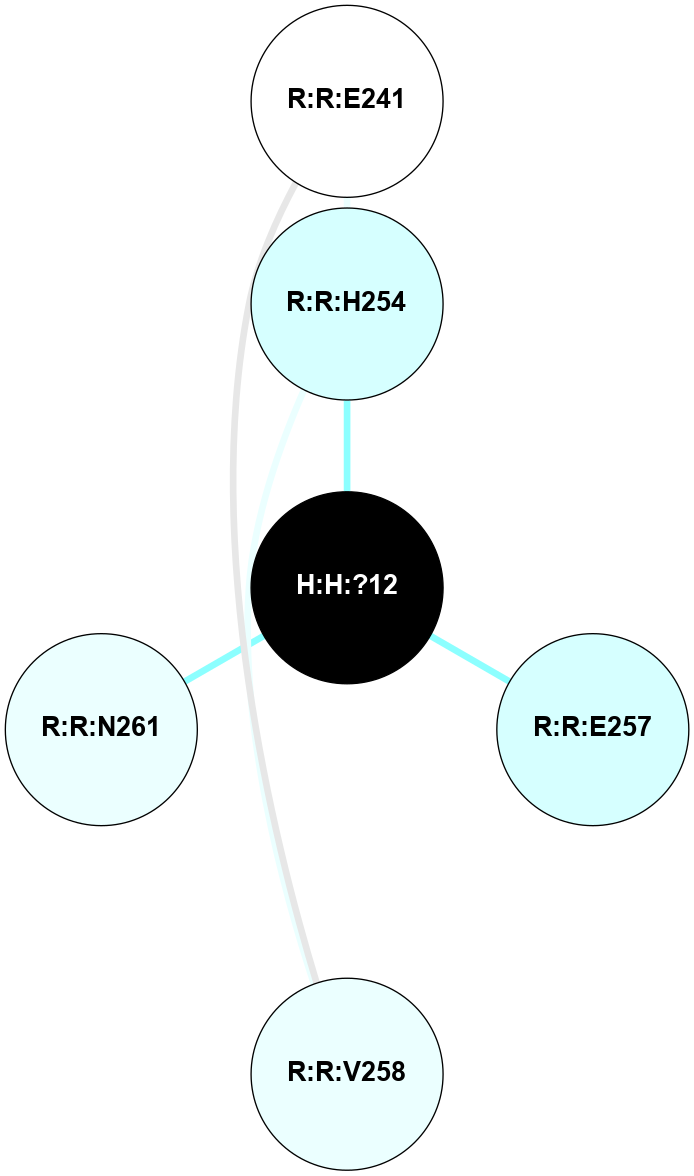
A 2D representation of the interactions of NAG in 9AYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E241 | R:R:H254 | 7.39 | 1 | No | No | 5 | 3 | 2 | 1 | | R:R:E241 | R:R:V258 | 9.98 | 1 | No | No | 5 | 4 | 2 | 2 | | R:R:H254 | R:R:V258 | 5.54 | 1 | No | No | 3 | 4 | 1 | 2 | | H:H:?12 | R:R:H254 | 2.31 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?12 | R:R:E257 | 7.13 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?12 | R:R:N261 | 28.34 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.59 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9AYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:H312 | R:R:V284 | 4.15 | 1 | Yes | No | 3 | 3 | 1 | 2 | | R:R:V284 | R:R:V313 | 4.81 | 1 | No | Yes | 3 | 3 | 2 | 2 | | R:R:H312 | R:R:N287 | 7.65 | 1 | Yes | No | 3 | 6 | 1 | 1 | | H:H:?13 | R:R:N287 | 29.57 | 1 | No | No | 0 | 6 | 0 | 1 | | R:R:H312 | R:R:V313 | 5.54 | 1 | Yes | Yes | 3 | 3 | 1 | 2 | | H:H:?13 | R:R:H312 | 2.31 | 1 | No | Yes | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.94 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
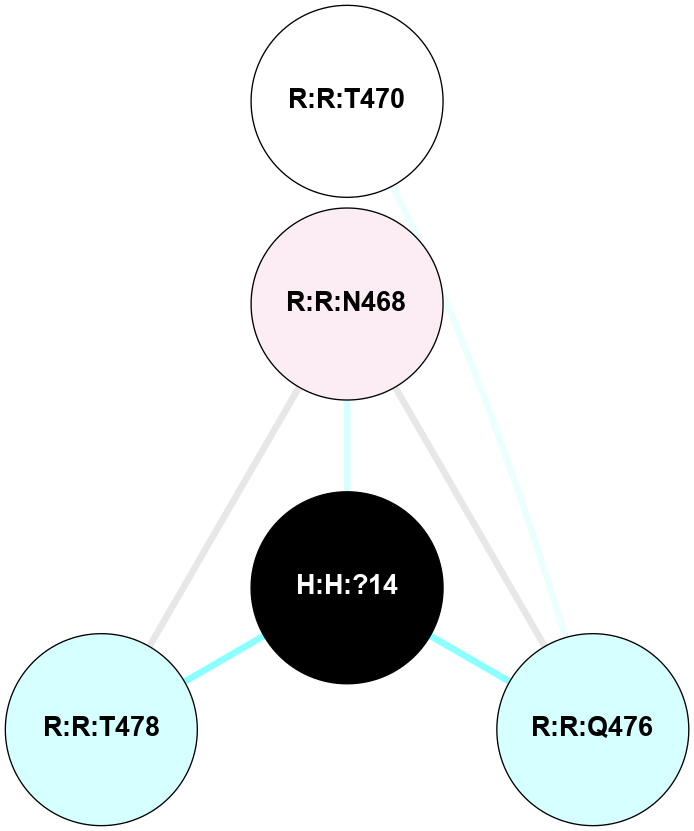
A 2D representation of the interactions of NAG in 9AYF
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:N468 | R:R:Q476 | 10.56 | 17 | No | No | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 5.85 | 17 | No | No | 6 | 3 | 1 | 1 | | H:H:?14 | R:R:N468 | 20.95 | 17 | No | No | 0 | 6 | 0 | 1 | | R:R:Q476 | R:R:T470 | 4.25 | 17 | No | No | 3 | 5 | 1 | 2 | | H:H:?14 | R:R:Q476 | 13.14 | 17 | No | No | 0 | 3 | 0 | 1 | | H:H:?14 | R:R:T478 | 3.97 | 17 | No | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.69 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9BLW | B1 | Peptide | Calcitonin | CT (AMY1) | Homo Sapiens | Cagrilintide backbone (non-acylated) | - | Gs/Beta1/Gamma2; RAMP1 | 3.2 | 2025-04-16 | To be published |
|
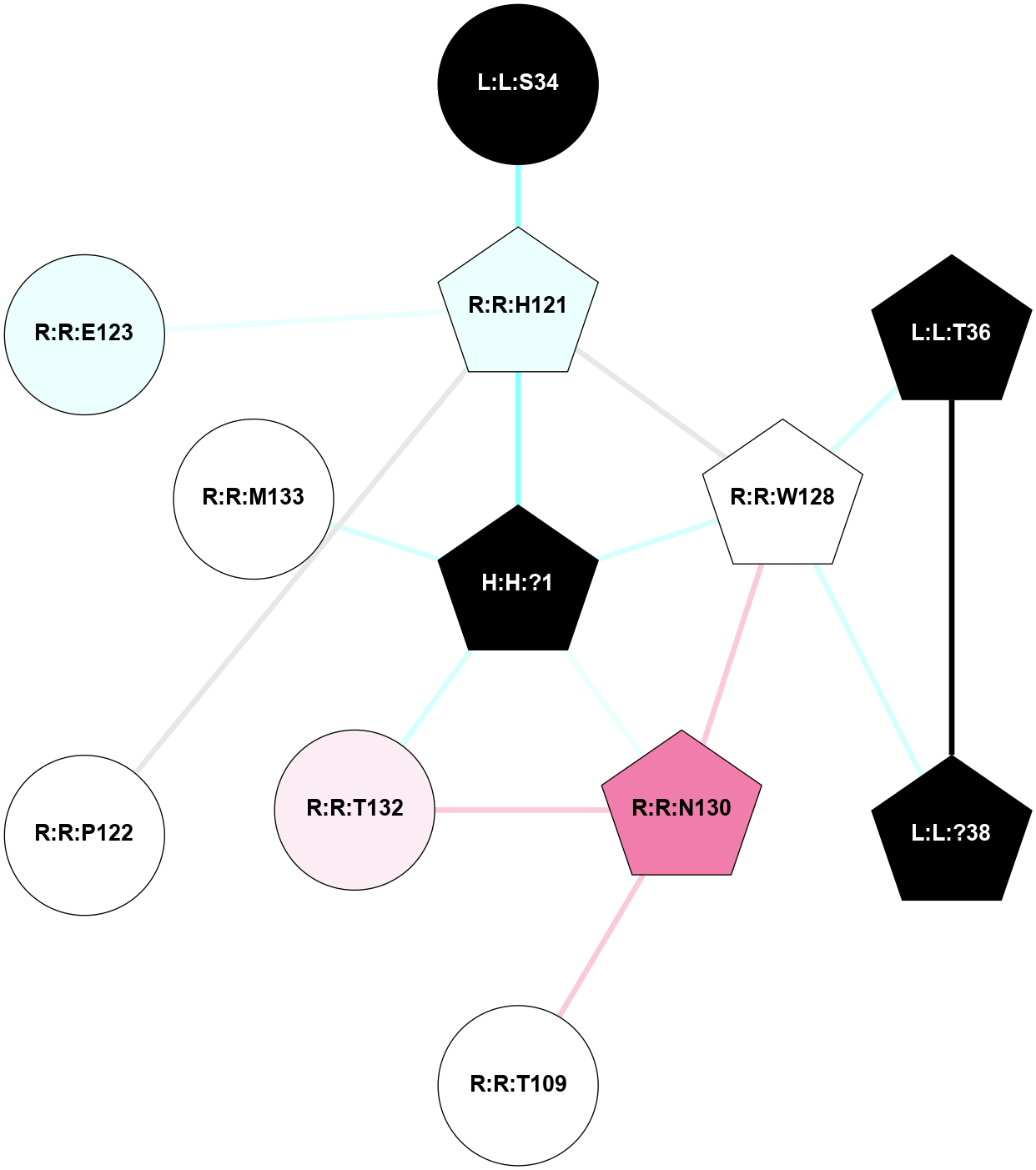
A 2D representation of the interactions of NAG in 9BLW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:H121 | 5.77 | 2 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:W128 | 10.22 | 2 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:N130 | 30.8 | 2 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?1 | R:R:T132 | 9.26 | 2 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:M133 | 3.81 | 2 | Yes | No | 0 | 5 | 0 | 1 | | L:L:S34 | R:R:H121 | 8.37 | 0 | No | Yes | 0 | 4 | 2 | 1 | | L:L:?38 | L:L:T36 | 3.65 | 2 | Yes | Yes | 0 | 0 | 2 | 2 | | L:L:T36 | R:R:W128 | 4.85 | 2 | Yes | Yes | 0 | 5 | 2 | 1 | | L:L:?38 | R:R:W128 | 11.28 | 2 | Yes | Yes | 0 | 5 | 2 | 1 | | R:R:N130 | R:R:T109 | 4.39 | 2 | Yes | No | 8 | 5 | 1 | 2 | | R:R:H121 | R:R:P122 | 7.63 | 2 | Yes | No | 4 | 5 | 1 | 2 | | R:R:E123 | R:R:H121 | 4.92 | 0 | No | Yes | 4 | 4 | 2 | 1 | | R:R:H121 | R:R:W128 | 16.93 | 2 | Yes | Yes | 4 | 5 | 1 | 1 | | R:R:N130 | R:R:W128 | 10.17 | 2 | Yes | Yes | 8 | 5 | 1 | 1 | | R:R:N130 | R:R:T132 | 5.85 | 2 | Yes | No | 8 | 6 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 3.00 | | Average Interaction Strength | 11.97 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9BTW | B1 | Peptide | Calcitonin | CT (AMY3) | Homo Sapiens | Cagrilintide | - | Gs/Beta1/Gamma2; RAMP3 | 3 | 2025-04-16 | To be published |
|
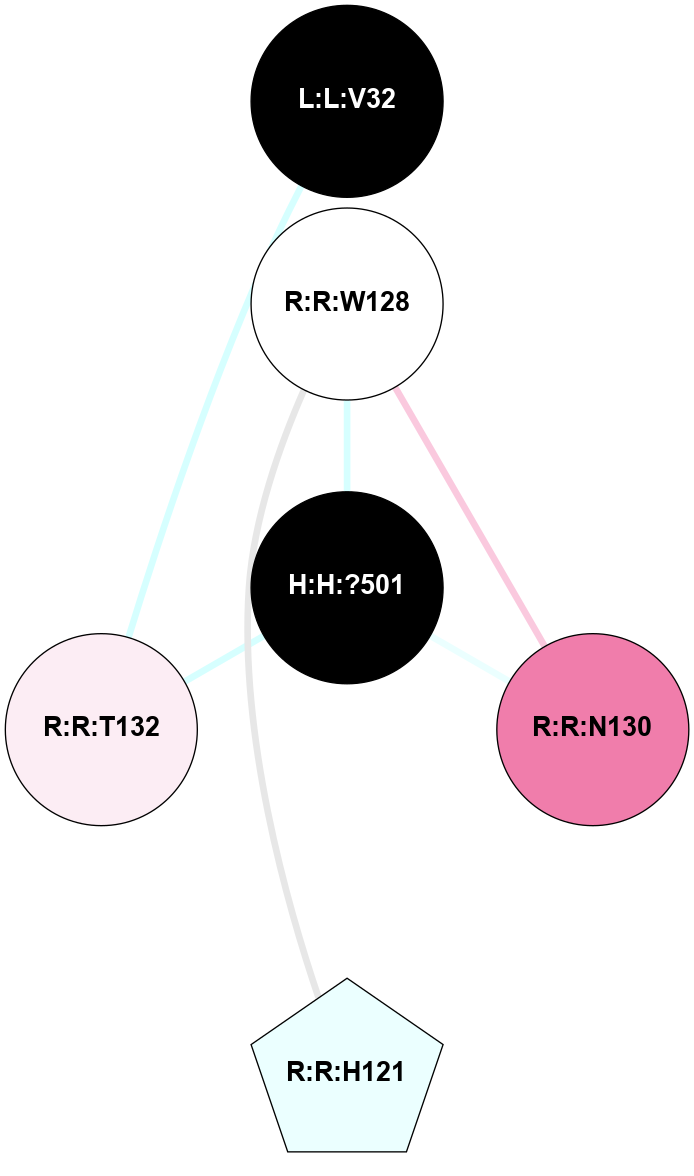
A 2D representation of the interactions of NAG in 9BTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?501 | R:R:W128 | 6.13 | 10 | No | No | 0 | 5 | 0 | 1 | | H:H:?501 | R:R:N130 | 24.64 | 10 | No | No | 0 | 8 | 0 | 1 | | H:H:?501 | R:R:T132 | 6.61 | 10 | No | No | 0 | 6 | 0 | 1 | | L:L:V32 | R:R:T132 | 6.35 | 0 | No | No | 0 | 6 | 2 | 1 | | R:R:H121 | R:R:W128 | 19.04 | 10 | Yes | No | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 4.52 | 10 | No | No | 8 | 5 | 1 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 12.46 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
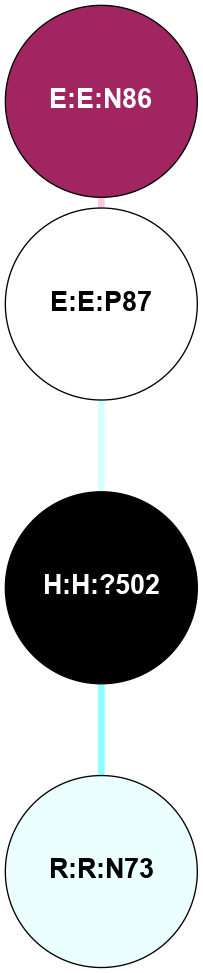
A 2D representation of the interactions of NAG in 9BTW
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | E:E:N86 | E:E:P87 | 6.52 | 0 | No | No | 9 | 5 | 2 | 1 | | E:E:P87 | H:H:?502 | 2.95 | 0 | No | No | 5 | 0 | 1 | 0 | | H:H:?502 | R:R:N73 | 29.57 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.26 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9II2 | C | Aminoacid | Metabotropic Glutamate | mGlu3; mGlu3 | Homo Sapiens | Glutamate | - | Arrestin2 | 3.7 | 2025-03-05 | doi.org/10.1038/s41589-025-01858-8 |
|
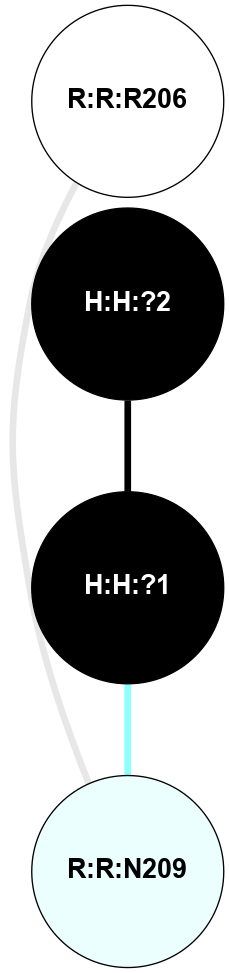
A 2D representation of the interactions of NAG in 9II2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 42.35 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N209 | 29.57 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N209 | R:R:R206 | 6.03 | 0 | No | No | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 35.96 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9II2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 42.35 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N209 | 29.57 | 0 | No | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 42.35 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9II2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 42.35 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | S:S:N209 | 29.57 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N209 | S:S:R206 | 6.03 | 0 | No | No | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 35.96 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
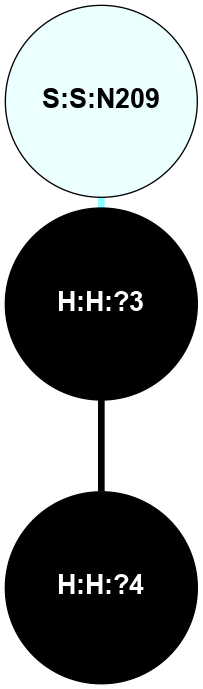
A 2D representation of the interactions of NAG in 9II2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 42.35 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?3 | S:S:N209 | 29.57 | 0 | No | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 42.35 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9II3 | C | Aminoacid | Metabotropic Glutamate | mGlu3; mGlu3 | Homo Sapiens | Glutamate | - | Arrestin2 | 3.9 | 2025-03-05 | doi.org/10.1038/s41589-025-01858-8 |
|
A 2D representation of the interactions of NAG in 9II3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 44.58 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N209 | 35.73 | 0 | No | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 40.16 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 9II3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 44.58 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N209 | 35.73 | 0 | No | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 44.58 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 9II3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 39.01 | 0 | No | No | 0 | 0 | 0 | 1 | | H:H:?3 | S:S:N209 | 24.64 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N209 | S:S:Q508 | 1.32 | 0 | No | No | 4 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 31.82 | | Average Nodes In Shell | 4.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 3.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
A 2D representation of the interactions of NAG in 9II3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | H:H:?4 | 39.01 | 0 | No | No | 0 | 0 | 1 | 0 | | H:H:?3 | S:S:N209 | 24.64 | 0 | No | No | 0 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 39.01 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9JR3 | B1 | Peptide | Parathyroid Hormone | PTH1 | Homo Sapiens | PTH | - | chim(NtGi1L-Gs-CtGq)/Beta1/Gamma2 | 2.8 | 2024-10-16 | To be published |
|

A 2D representation of the interactions of NAG in 9JR3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 30.09 | 15 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:N176 | 28.34 | 15 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:T178 | 15.87 | 15 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:R179 | 22.89 | 15 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:E182 | 4.76 | 15 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?2 | R:R:R179 | 4.36 | 15 | No | No | 0 | 3 | 1 | 1 | | R:R:N176 | R:R:T178 | 7.31 | 15 | No | No | 3 | 3 | 1 | 1 | | R:R:N176 | R:R:R179 | 8.44 | 15 | No | No | 3 | 3 | 1 | 1 | | R:R:E177 | R:R:T178 | 1.41 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:E182 | R:R:R186 | 1.16 | 0 | No | No | 2 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.39 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
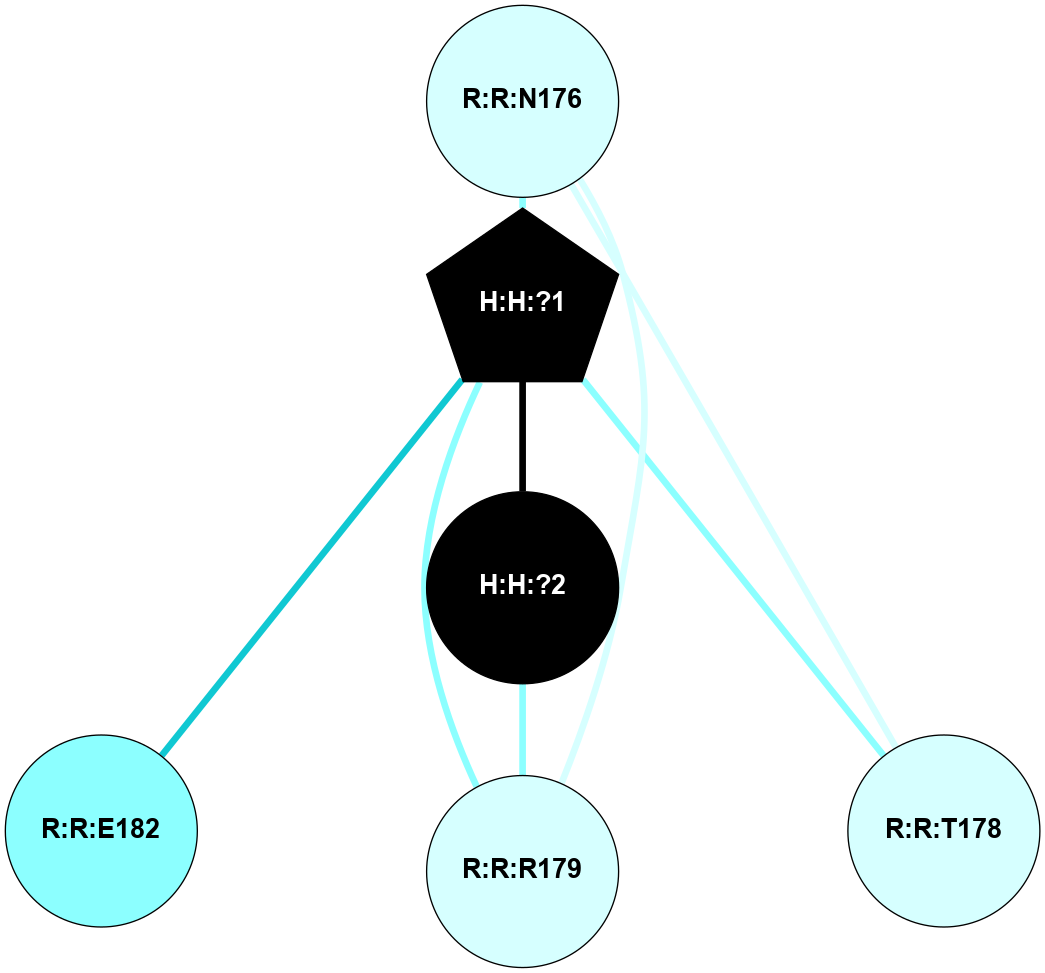
A 2D representation of the interactions of NAG in 9JR3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 30.09 | 15 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:N176 | 28.34 | 15 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:T178 | 15.87 | 15 | Yes | No | 0 | 3 | 1 | 2 | | H:H:?1 | R:R:R179 | 22.89 | 15 | Yes | No | 0 | 3 | 1 | 1 | | H:H:?1 | R:R:E182 | 4.76 | 15 | Yes | No | 0 | 2 | 1 | 2 | | H:H:?2 | R:R:R179 | 4.36 | 15 | No | No | 0 | 3 | 0 | 1 | | R:R:N176 | R:R:T178 | 7.31 | 15 | No | No | 3 | 3 | 2 | 2 | | R:R:N176 | R:R:R179 | 8.44 | 15 | No | No | 3 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.23 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9NOS | C | Sensory | Taste1 | TAS1R2; TAS1R3 | Homo Sapiens | - | - | - | 3.5 | 2025-04-09 | doi.org/10.1016/j.cell.2025.04.021 |
|
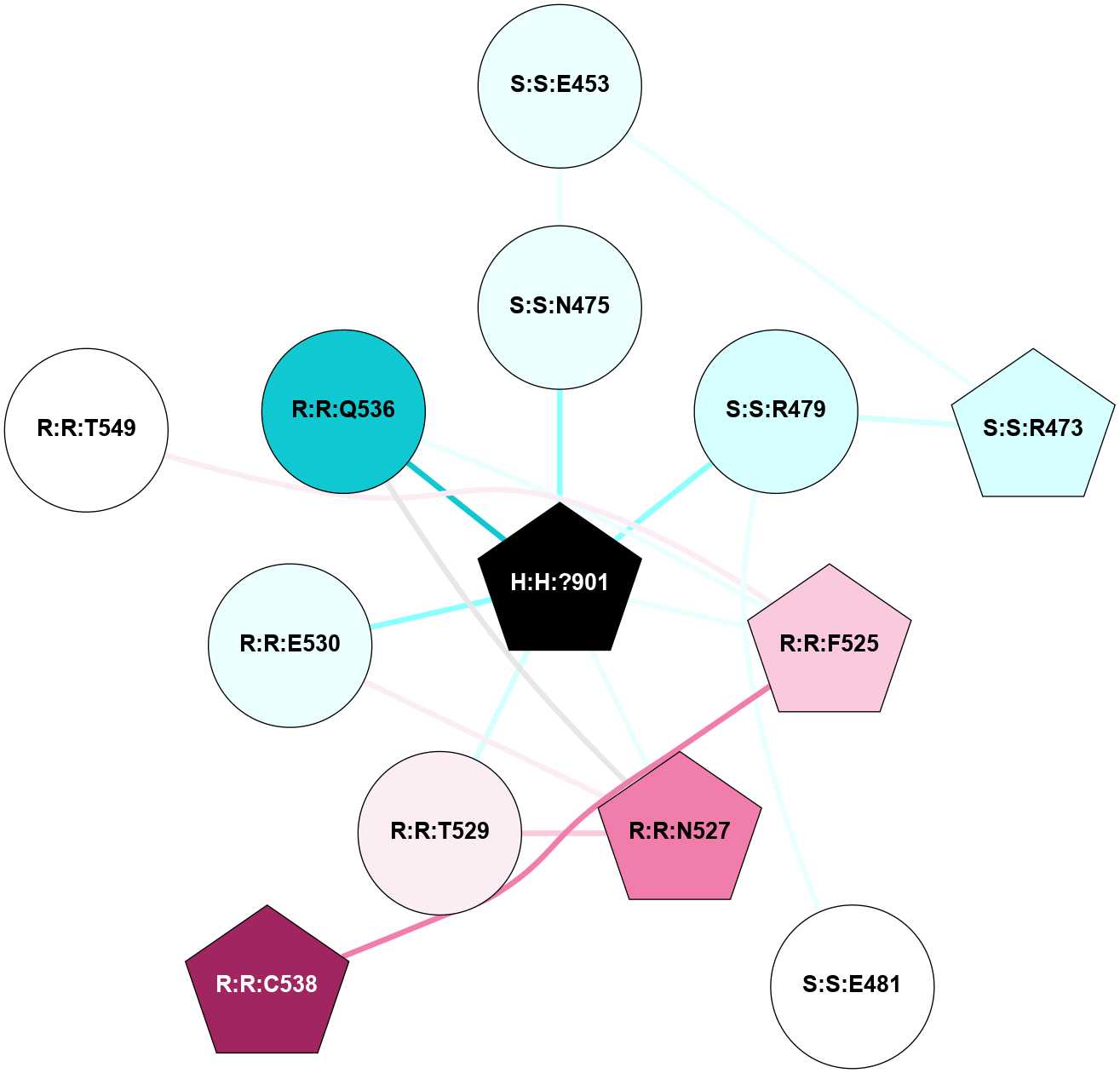
A 2D representation of the interactions of NAG in 9NOS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:E453 | S:S:R473 | 10.47 | 23 | No | Yes | 4 | 3 | 2 | 2 | | S:S:E453 | S:S:N475 | 9.2 | 23 | No | No | 4 | 4 | 2 | 1 | | S:S:R473 | S:S:R479 | 6.4 | 23 | Yes | No | 3 | 3 | 2 | 1 | | H:H:?901 | S:S:N475 | 30.8 | 10 | Yes | No | 0 | 4 | 0 | 1 | | S:S:E481 | S:S:R479 | 12.79 | 0 | No | No | 5 | 3 | 2 | 1 | | H:H:?901 | S:S:R479 | 3.27 | 10 | Yes | No | 0 | 3 | 0 | 1 | | R:R:F525 | R:R:Q536 | 5.86 | 10 | Yes | No | 7 | 1 | 1 | 1 | | H:H:?901 | R:R:F525 | 4.37 | 10 | Yes | Yes | 0 | 7 | 0 | 1 | | R:R:N527 | R:R:T529 | 7.31 | 10 | Yes | No | 8 | 6 | 1 | 1 | | R:R:E530 | R:R:N527 | 9.2 | 10 | No | Yes | 4 | 8 | 1 | 1 | | R:R:N527 | R:R:Q536 | 6.6 | 10 | Yes | No | 8 | 1 | 1 | 1 | | H:H:?901 | R:R:N527 | 29.57 | 10 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?901 | R:R:T529 | 9.26 | 10 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?901 | R:R:E530 | 13.08 | 10 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?901 | R:R:Q536 | 3.58 | 10 | Yes | No | 0 | 1 | 0 | 1 | | R:R:C538 | R:R:F525 | 2.79 | 10 | Yes | Yes | 9 | 7 | 2 | 1 | | R:R:F525 | R:R:T549 | 1.3 | 10 | Yes | No | 7 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 13.42 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
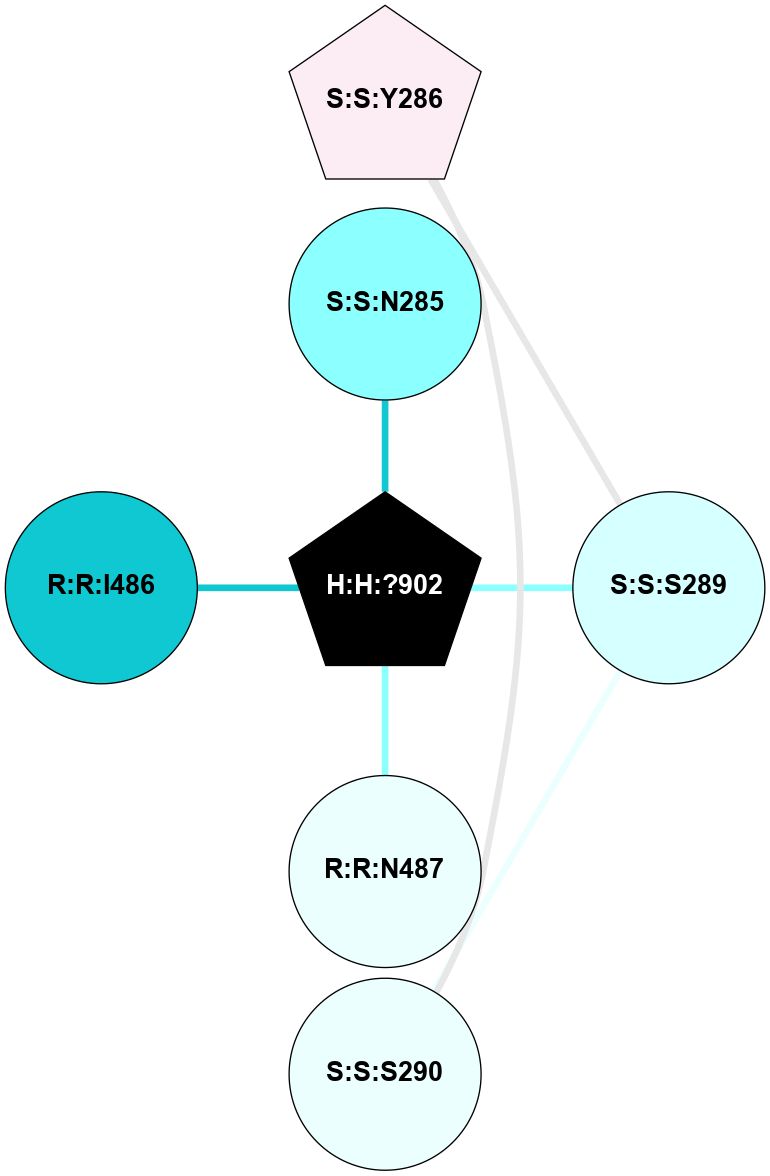
A 2D representation of the interactions of NAG in 9NOS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?902 | S:S:N285 | 32.03 | 0 | Yes | No | 0 | 2 | 0 | 1 | | S:S:S289 | S:S:Y286 | 3.82 | 37 | No | Yes | 3 | 6 | 1 | 2 | | S:S:S290 | S:S:Y286 | 11.45 | 37 | No | Yes | 4 | 6 | 2 | 2 | | S:S:S289 | S:S:S290 | 3.26 | 37 | No | No | 3 | 4 | 1 | 2 | | H:H:?902 | S:S:S289 | 8.08 | 0 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?902 | R:R:N487 | 28.34 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?902 | R:R:I486 | 2.56 | 0 | Yes | No | 0 | 1 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.75 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
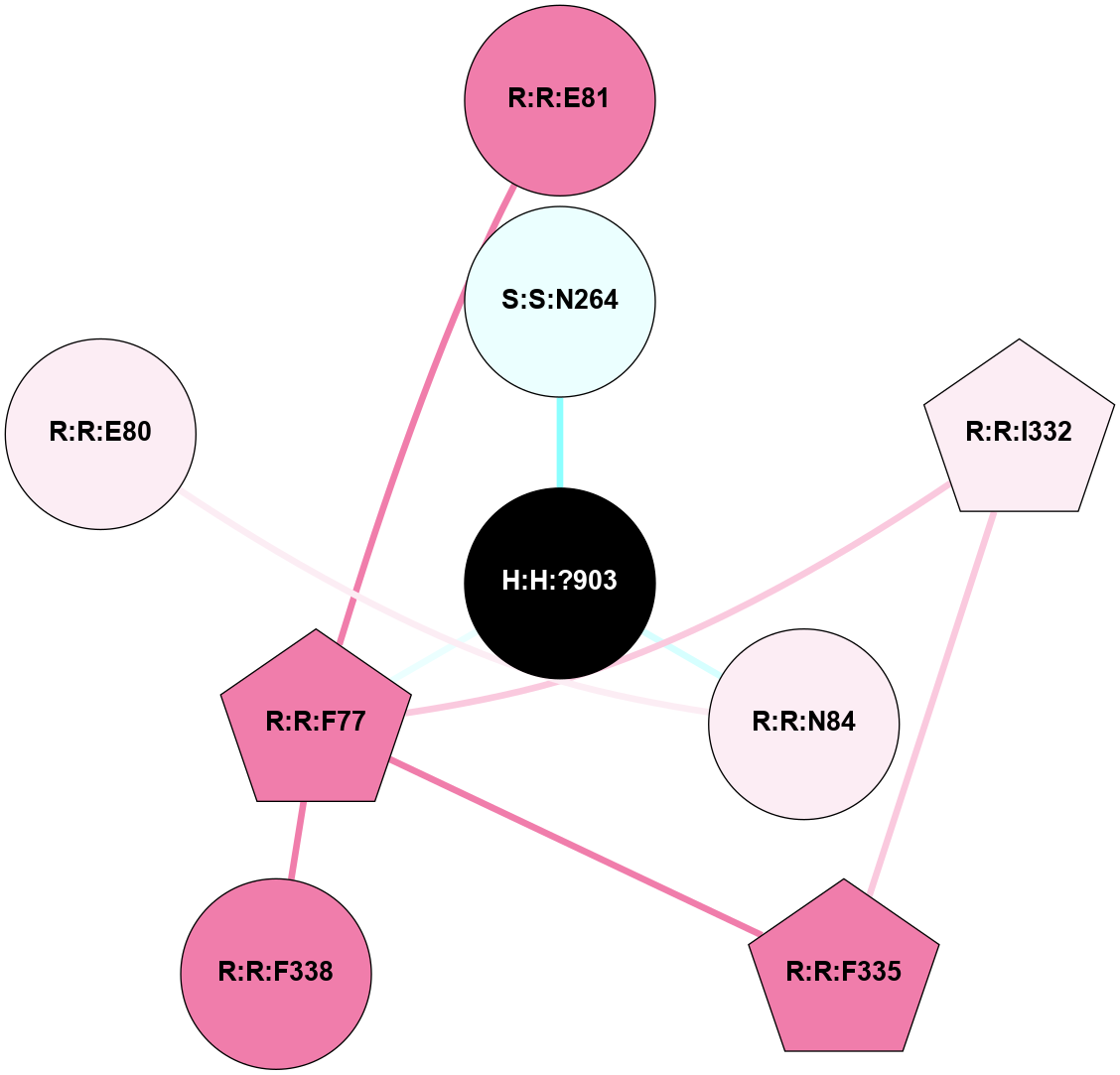
A 2D representation of the interactions of NAG in 9NOS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?903 | S:S:N264 | 25.87 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:E81 | R:R:F77 | 3.5 | 3 | No | Yes | 8 | 8 | 2 | 1 | | R:R:F77 | R:R:I332 | 3.77 | 3 | Yes | Yes | 8 | 6 | 1 | 2 | | R:R:F335 | R:R:F77 | 9.65 | 3 | Yes | Yes | 8 | 8 | 2 | 1 | | R:R:F338 | R:R:F77 | 6.43 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:E80 | R:R:N84 | 3.94 | 0 | No | No | 6 | 6 | 2 | 1 | | H:H:?903 | R:R:N84 | 25.87 | 0 | No | No | 0 | 6 | 0 | 1 | | R:R:F335 | R:R:I332 | 3.77 | 3 | Yes | Yes | 8 | 6 | 2 | 2 | | H:H:?903 | R:R:F77 | 2.19 | 0 | No | Yes | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.98 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
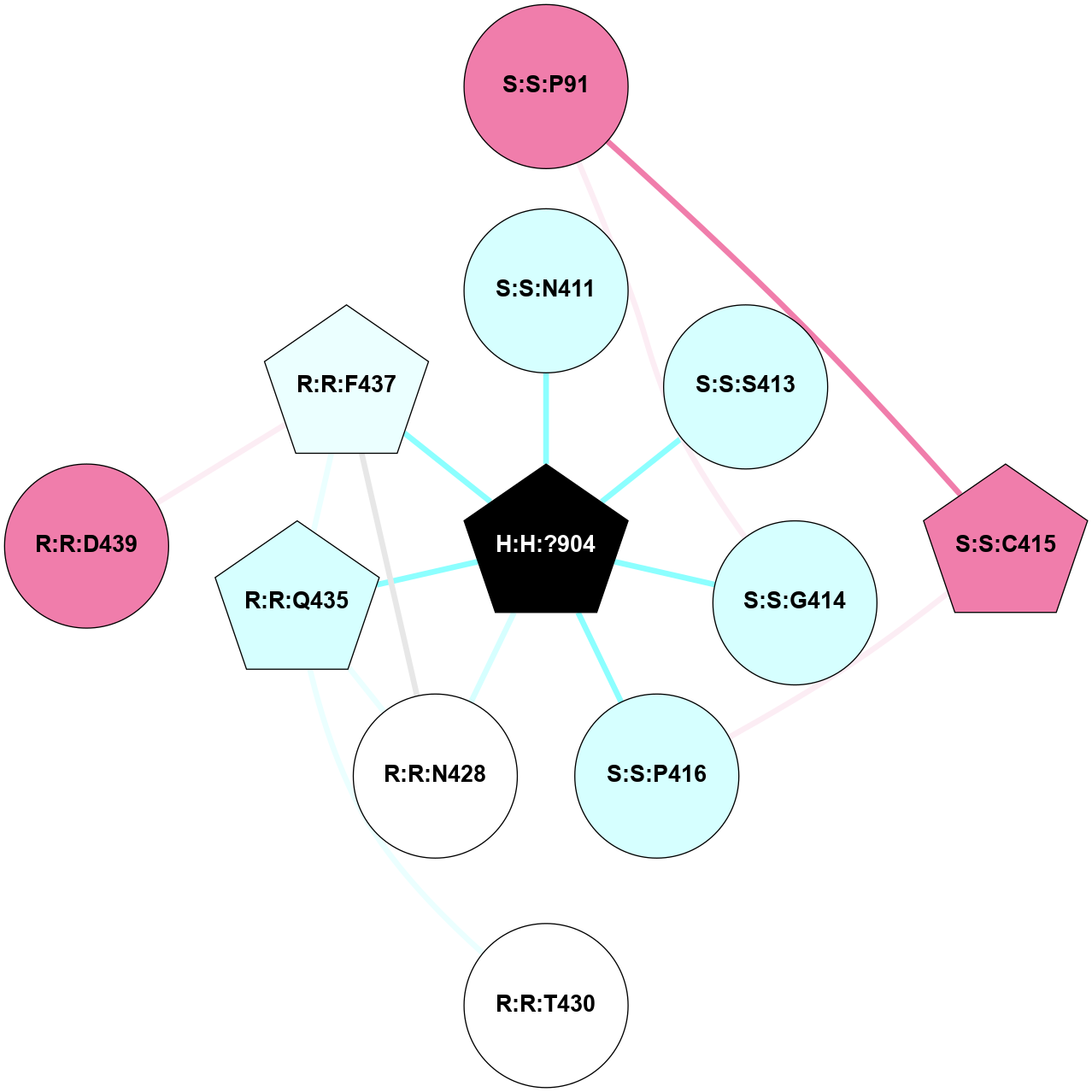
A 2D representation of the interactions of NAG in 9NOS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:G414 | S:S:P91 | 4.06 | 0 | No | No | 3 | 8 | 1 | 2 | | S:S:C415 | S:S:P91 | 3.77 | 0 | Yes | No | 8 | 8 | 2 | 2 | | H:H:?904 | S:S:N411 | 28.34 | 24 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?904 | S:S:S413 | 4.04 | 24 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?904 | S:S:G414 | 3.07 | 24 | Yes | No | 0 | 3 | 0 | 1 | | S:S:C415 | S:S:P416 | 3.77 | 0 | Yes | No | 8 | 3 | 2 | 1 | | H:H:?904 | S:S:P416 | 8.84 | 24 | Yes | No | 0 | 3 | 0 | 1 | | R:R:N428 | R:R:Q435 | 6.6 | 24 | No | Yes | 5 | 3 | 1 | 1 | | R:R:F437 | R:R:N428 | 6.04 | 24 | Yes | No | 4 | 5 | 1 | 1 | | H:H:?904 | R:R:N428 | 24.64 | 24 | Yes | No | 0 | 5 | 0 | 1 | | R:R:Q435 | R:R:T430 | 4.25 | 24 | Yes | No | 3 | 5 | 1 | 2 | | R:R:F437 | R:R:Q435 | 4.68 | 24 | Yes | Yes | 4 | 3 | 1 | 1 | | H:H:?904 | R:R:Q435 | 4.78 | 24 | Yes | Yes | 0 | 3 | 0 | 1 | | H:H:?904 | R:R:F437 | 4.37 | 24 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:D439 | R:R:F437 | 2.39 | 0 | No | Yes | 8 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 11.15 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
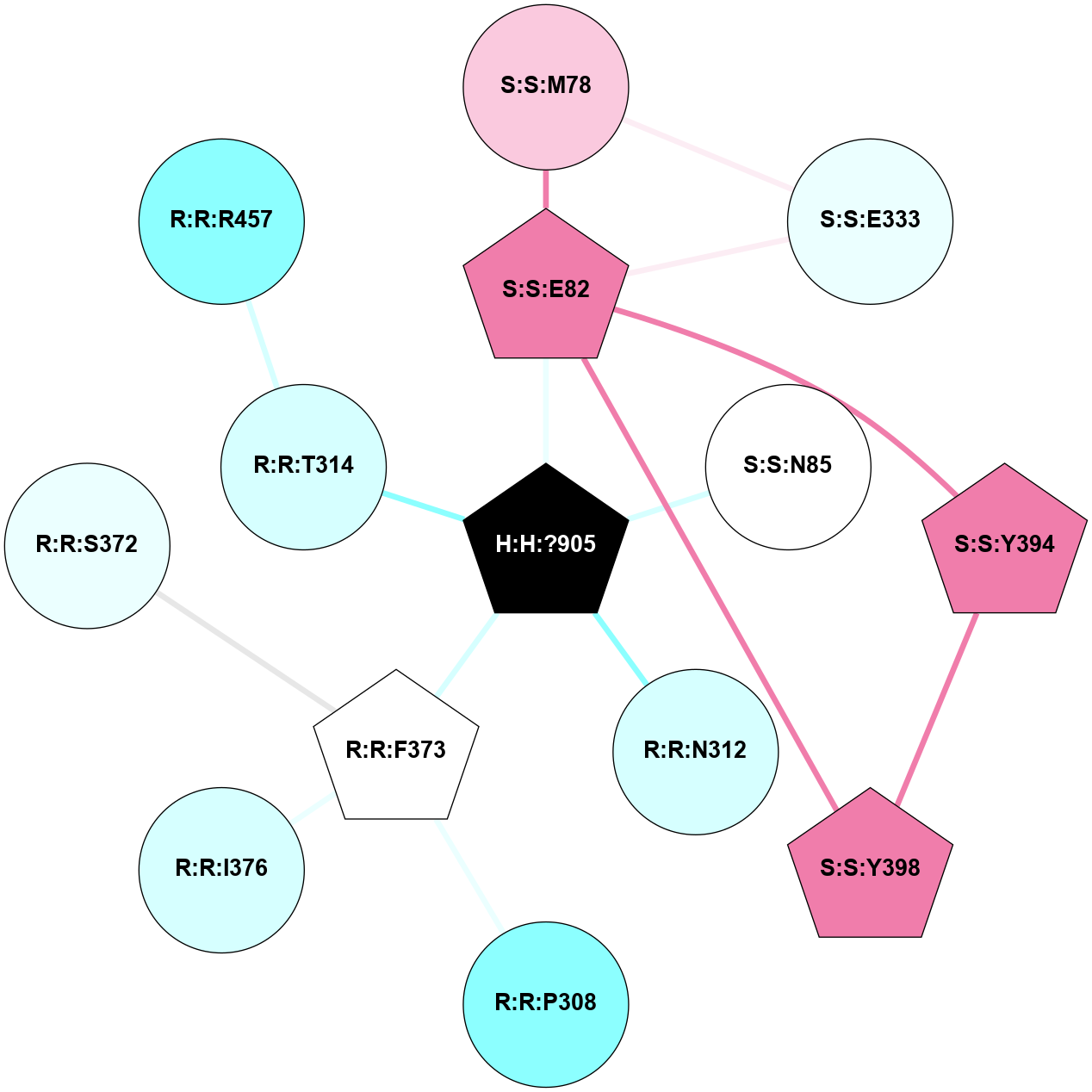
A 2D representation of the interactions of NAG in 9NOS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:E82 | S:S:M78 | 4.06 | 6 | Yes | No | 8 | 7 | 1 | 2 | | S:S:E333 | S:S:M78 | 5.41 | 6 | No | No | 4 | 7 | 2 | 2 | | S:S:E333 | S:S:E82 | 5.07 | 6 | No | Yes | 4 | 8 | 2 | 1 | | S:S:E82 | S:S:Y394 | 7.86 | 6 | Yes | Yes | 8 | 8 | 1 | 2 | | S:S:E82 | S:S:Y398 | 3.37 | 6 | Yes | Yes | 8 | 8 | 1 | 2 | | H:H:?905 | S:S:E82 | 4.76 | 0 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?905 | S:S:N85 | 25.87 | 0 | Yes | No | 0 | 5 | 0 | 1 | | S:S:Y394 | S:S:Y398 | 16.88 | 6 | Yes | Yes | 8 | 8 | 2 | 2 | | R:R:F373 | R:R:P308 | 8.67 | 0 | Yes | No | 5 | 2 | 1 | 2 | | H:H:?905 | R:R:N312 | 28.34 | 0 | Yes | No | 0 | 3 | 0 | 1 | | R:R:F373 | R:R:I376 | 5.02 | 0 | Yes | No | 5 | 3 | 1 | 2 | | H:H:?905 | R:R:F373 | 7.65 | 0 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?905 | R:R:T314 | 2.65 | 0 | Yes | No | 0 | 3 | 0 | 1 | | R:R:F373 | R:R:S372 | 2.64 | 0 | Yes | No | 5 | 4 | 1 | 2 | | R:R:R457 | R:R:T314 | 1.29 | 0 | No | No | 2 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 13.85 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 5.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
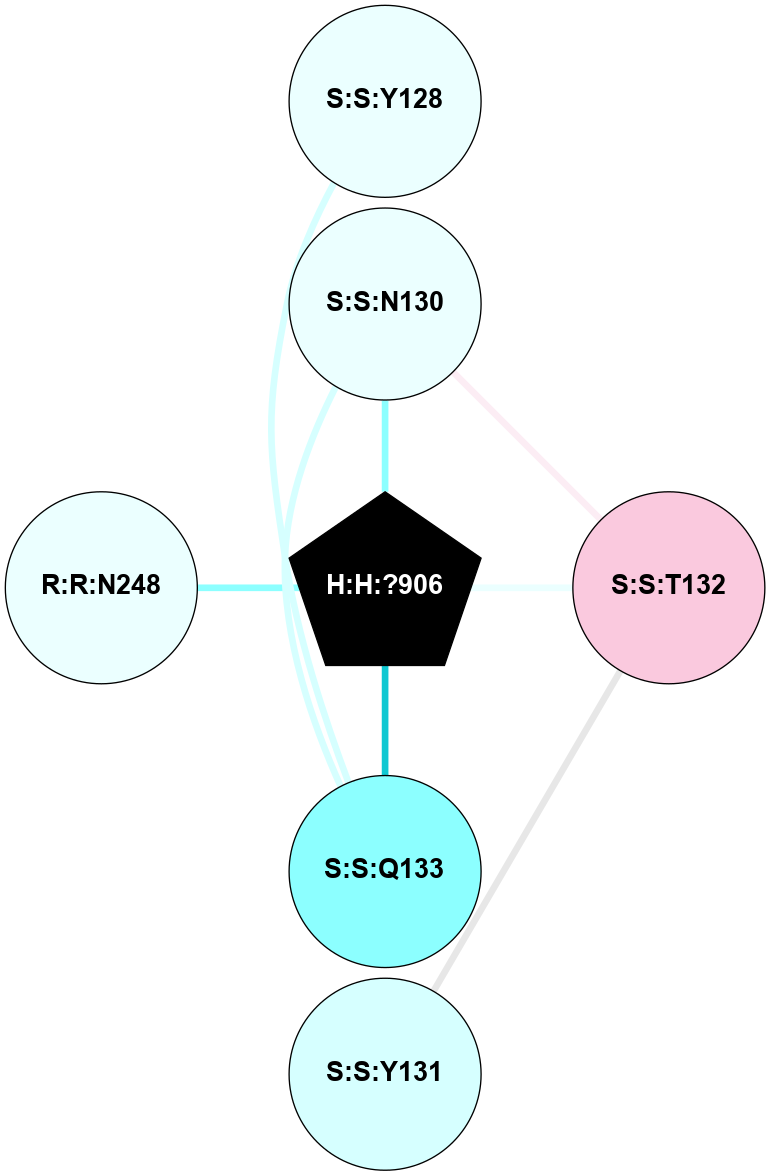
A 2D representation of the interactions of NAG in 9NOS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:Q133 | S:S:Y128 | 9.02 | 21 | No | No | 2 | 4 | 1 | 2 | | S:S:N130 | S:S:T132 | 4.39 | 21 | No | No | 4 | 7 | 1 | 1 | | S:S:N130 | S:S:Q133 | 9.24 | 21 | No | No | 4 | 2 | 1 | 1 | | H:H:?906 | S:S:N130 | 28.34 | 21 | Yes | No | 0 | 4 | 0 | 1 | | S:S:T132 | S:S:Y131 | 8.74 | 21 | No | No | 7 | 3 | 1 | 2 | | H:H:?906 | S:S:T132 | 3.97 | 21 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?906 | S:S:Q133 | 8.36 | 21 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?906 | R:R:N248 | 25.87 | 21 | Yes | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 16.64 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
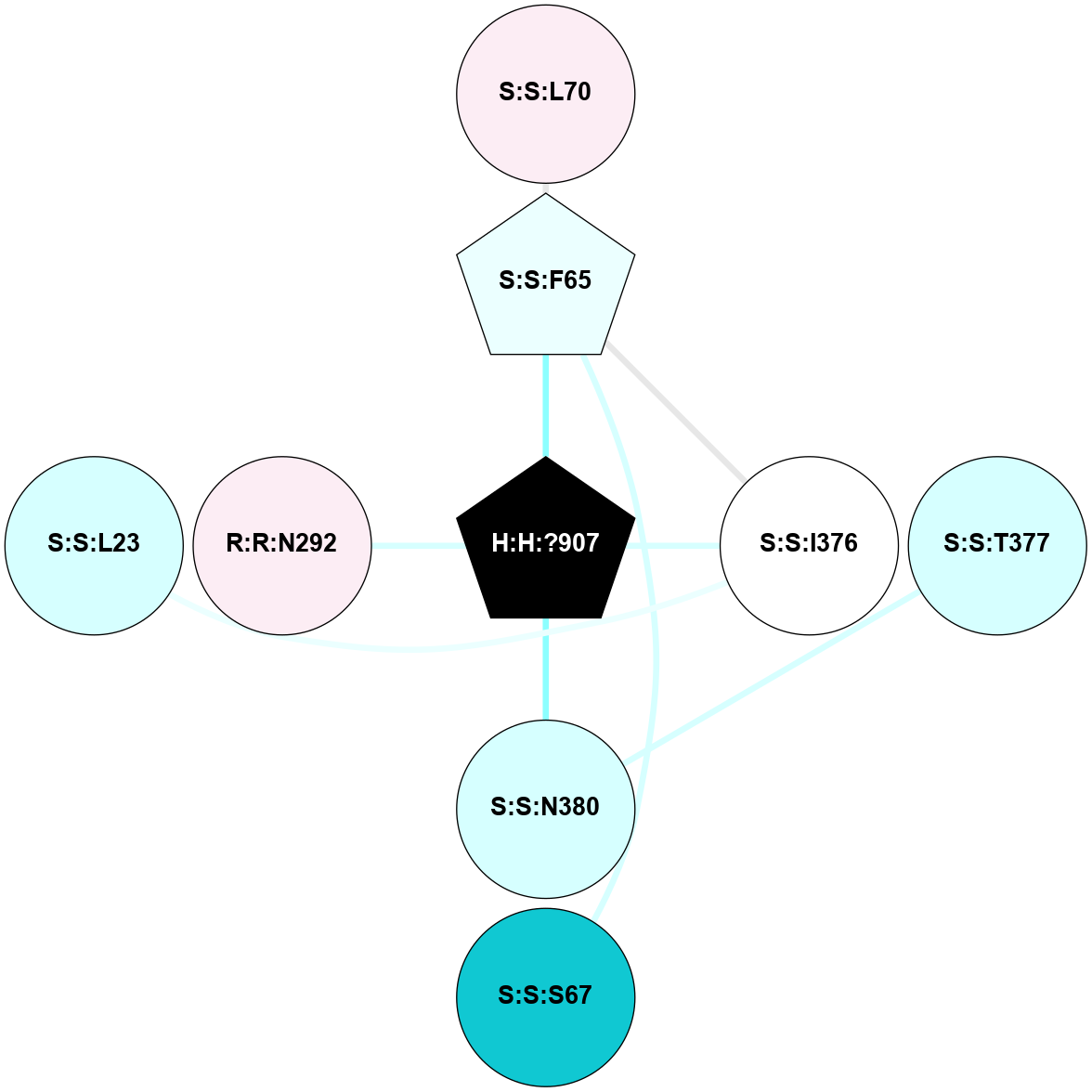
A 2D representation of the interactions of NAG in 9NOS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:F65 | S:S:L70 | 6.09 | 33 | Yes | No | 4 | 6 | 1 | 2 | | S:S:F65 | S:S:I376 | 5.02 | 33 | Yes | No | 4 | 5 | 1 | 1 | | H:H:?907 | S:S:F65 | 4.37 | 33 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?907 | S:S:I376 | 3.84 | 33 | Yes | No | 0 | 5 | 0 | 1 | | S:S:N380 | S:S:T377 | 2.92 | 0 | No | No | 3 | 3 | 1 | 2 | | H:H:?907 | S:S:N380 | 34.5 | 33 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?907 | R:R:N292 | 27.11 | 33 | Yes | No | 0 | 6 | 0 | 1 | | S:S:F65 | S:S:S67 | 2.64 | 33 | Yes | No | 4 | 1 | 1 | 2 | | S:S:I376 | S:S:L23 | 1.43 | 33 | No | No | 5 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 17.45 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9NOS
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?908 | S:S:N432 | 28.34 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?908 | R:R:N368 | 32.03 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:N432 | S:S:R443 | 2.41 | 0 | No | No | 5 | 3 | 1 | 2 | | R:R:L371 | R:R:N368 | 1.37 | 0 | No | No | 1 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 30.19 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9NOT | C | Sensory | Taste1 | TAS1R2; TAS1R3 | Homo Sapiens | - | - | - | 3.8 | 2025-04-09 | doi.org/10.1016/j.cell.2025.04.021 |
|
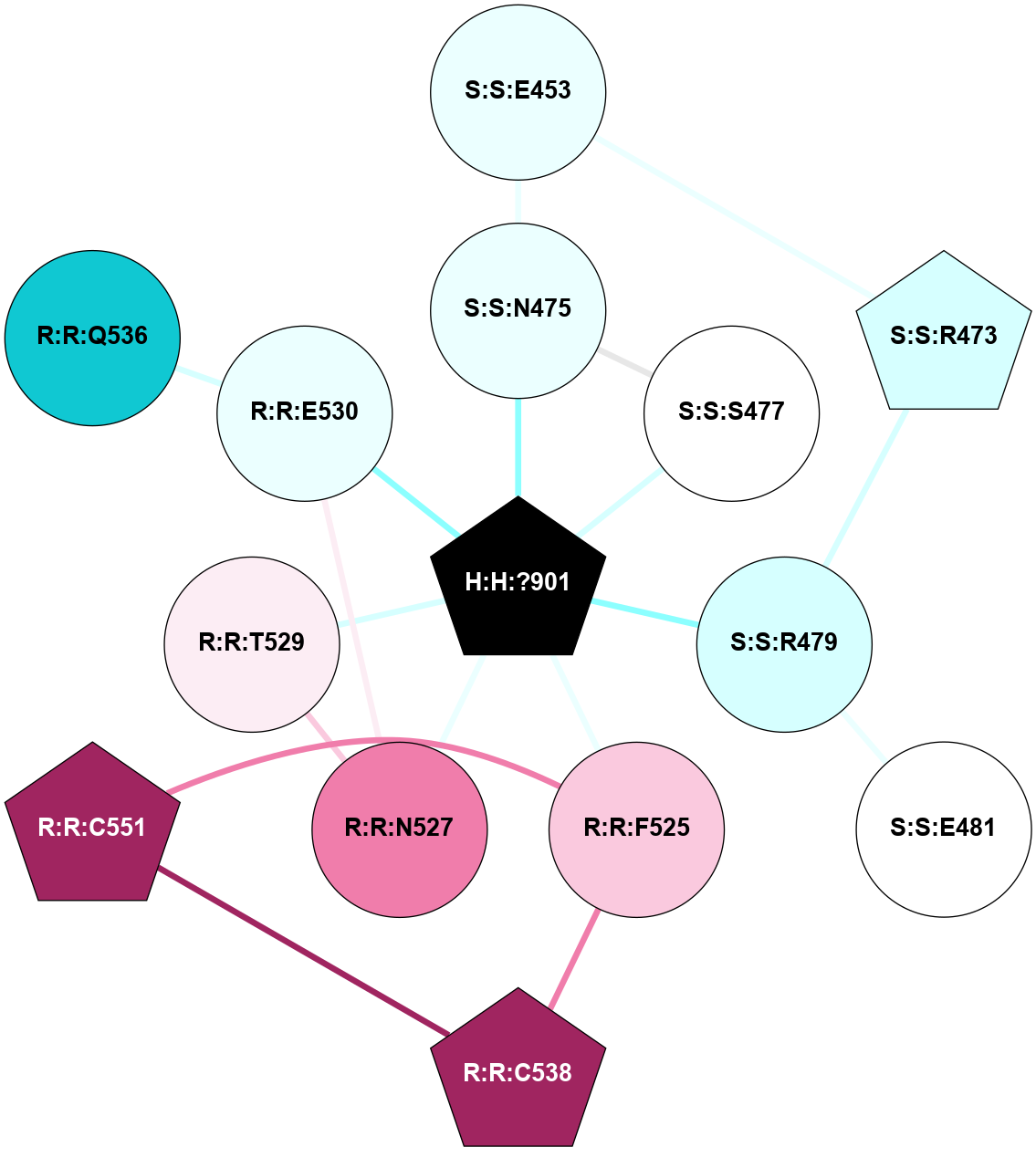
A 2D representation of the interactions of NAG in 9NOT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:E453 | S:S:R473 | 9.3 | 4 | No | Yes | 4 | 3 | 2 | 2 | | S:S:E453 | S:S:N475 | 3.94 | 4 | No | No | 4 | 4 | 2 | 1 | | S:S:R473 | S:S:R479 | 8.53 | 4 | Yes | No | 3 | 3 | 2 | 1 | | S:S:N475 | S:S:S477 | 4.47 | 4 | No | No | 4 | 5 | 1 | 1 | | H:H:?901 | S:S:N475 | 29.57 | 4 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?901 | S:S:S477 | 8.08 | 4 | Yes | No | 0 | 5 | 0 | 1 | | S:S:E481 | S:S:R479 | 9.3 | 0 | No | No | 5 | 3 | 2 | 1 | | H:H:?901 | S:S:R479 | 4.36 | 4 | Yes | No | 0 | 3 | 0 | 1 | | R:R:C538 | R:R:F525 | 5.59 | 4 | Yes | No | 9 | 7 | 2 | 1 | | R:R:C551 | R:R:F525 | 6.98 | 4 | Yes | No | 9 | 7 | 2 | 1 | | H:H:?901 | R:R:F525 | 3.28 | 4 | Yes | No | 0 | 7 | 0 | 1 | | R:R:N527 | R:R:T529 | 7.31 | 4 | No | No | 8 | 6 | 1 | 1 | | R:R:E530 | R:R:N527 | 11.83 | 4 | No | No | 4 | 8 | 1 | 1 | | H:H:?901 | R:R:N527 | 28.34 | 4 | Yes | No | 0 | 8 | 0 | 1 | | H:H:?901 | R:R:T529 | 9.26 | 4 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?901 | R:R:E530 | 17.84 | 4 | Yes | No | 0 | 4 | 0 | 1 | | R:R:C538 | R:R:C551 | 7.28 | 4 | Yes | Yes | 9 | 9 | 2 | 2 | | R:R:E530 | R:R:Q536 | 2.55 | 4 | No | No | 4 | 1 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.39 | | Average Nodes In Shell | 14.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 18.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
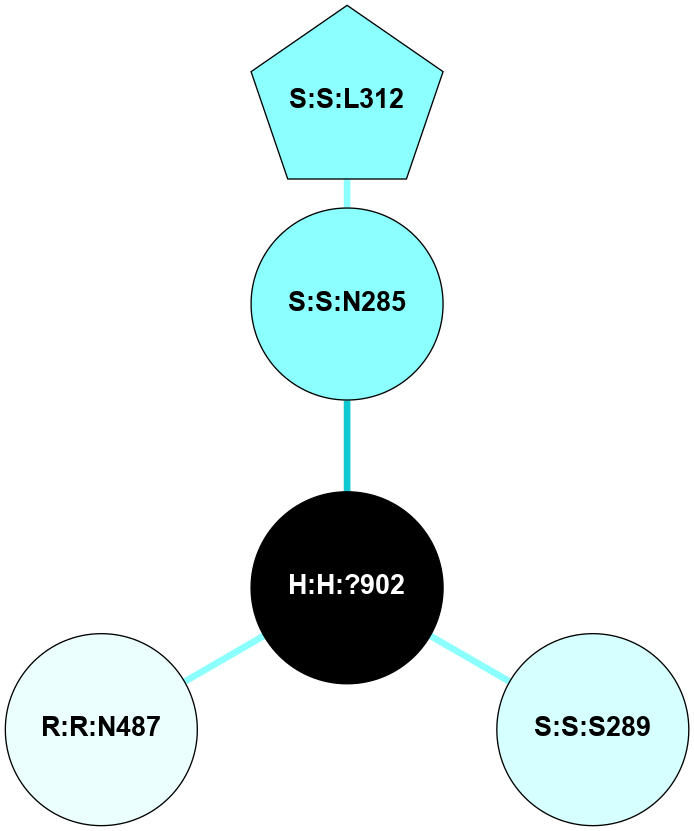
A 2D representation of the interactions of NAG in 9NOT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?902 | S:S:N285 | 27.11 | 0 | No | No | 0 | 2 | 0 | 1 | | H:H:?902 | S:S:S289 | 9.43 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?902 | R:R:N487 | 29.57 | 0 | No | No | 0 | 4 | 0 | 1 | | S:S:L312 | S:S:N285 | 2.75 | 0 | Yes | No | 2 | 2 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 22.04 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 4.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
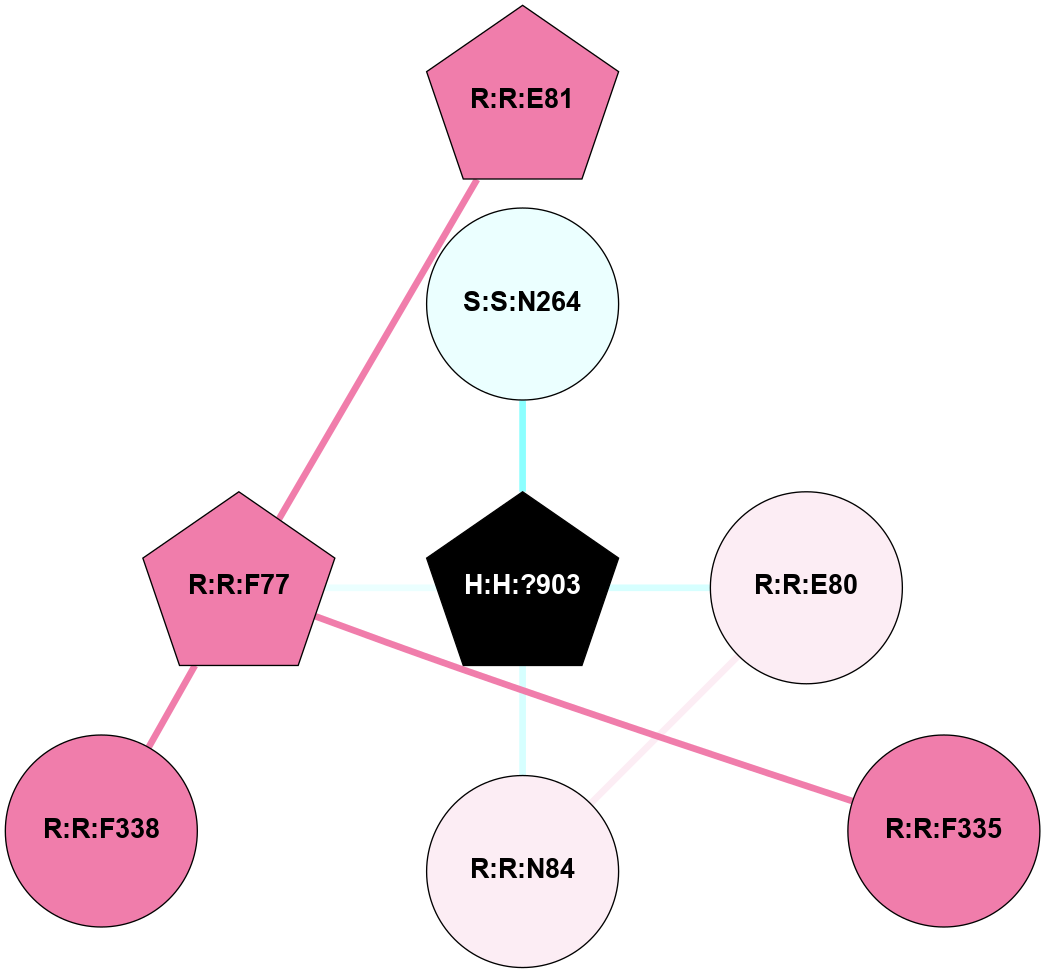
A 2D representation of the interactions of NAG in 9NOT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?903 | S:S:N264 | 24.64 | 27 | Yes | No | 0 | 4 | 0 | 1 | | R:R:E81 | R:R:F77 | 8.16 | 9 | Yes | Yes | 8 | 8 | 2 | 1 | | R:R:F335 | R:R:F77 | 10.72 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:F338 | R:R:F77 | 10.72 | 0 | No | Yes | 8 | 8 | 2 | 1 | | R:R:E80 | R:R:N84 | 11.83 | 27 | No | No | 6 | 6 | 1 | 1 | | H:H:?903 | R:R:E80 | 27.35 | 27 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?903 | R:R:N84 | 27.11 | 27 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?903 | R:R:F77 | 2.19 | 27 | Yes | Yes | 0 | 8 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 20.32 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
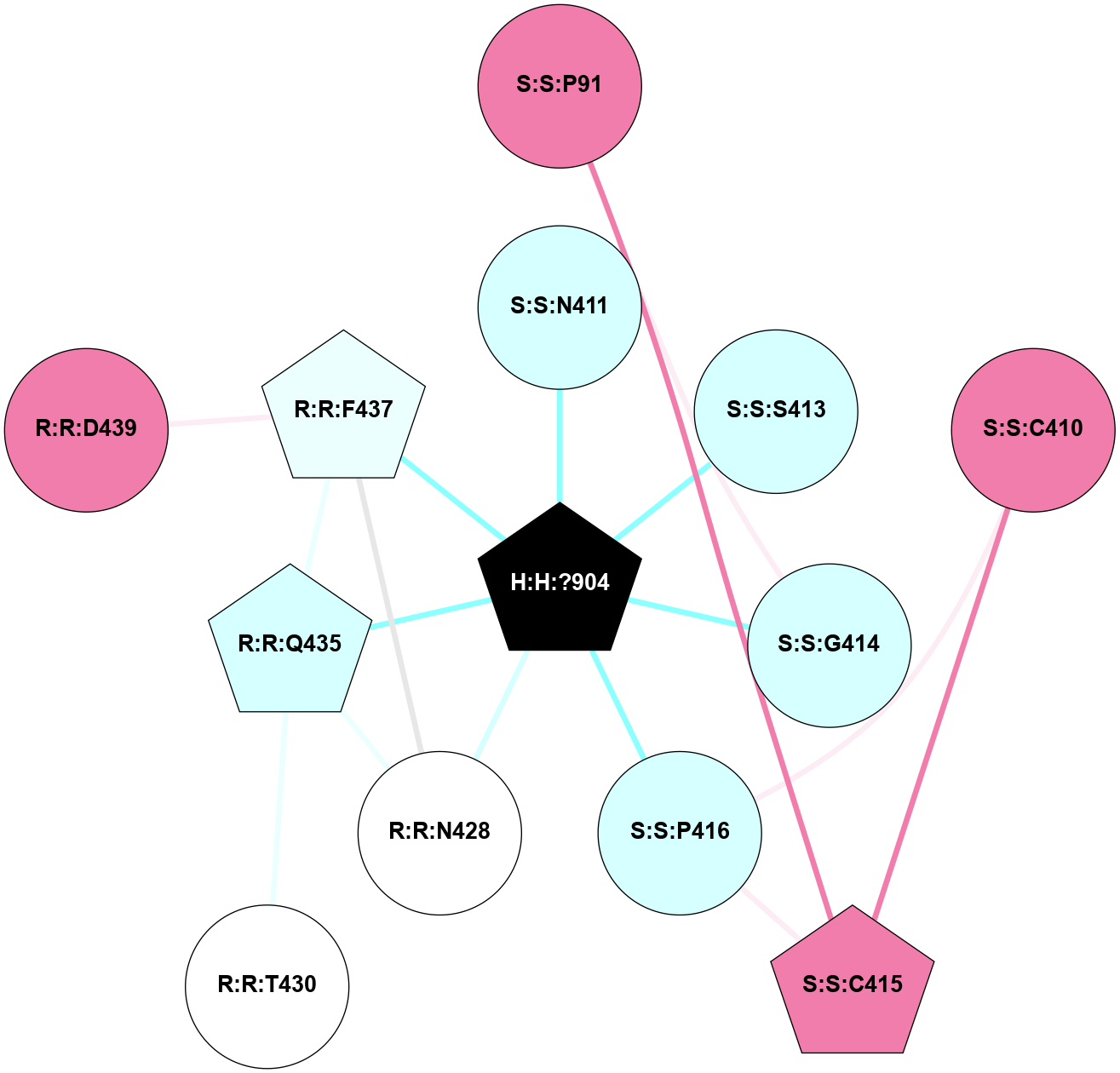
A 2D representation of the interactions of NAG in 9NOT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:G414 | S:S:P91 | 4.06 | 0 | No | No | 3 | 8 | 1 | 2 | | S:S:C415 | S:S:P91 | 3.77 | 1 | Yes | No | 8 | 8 | 2 | 2 | | S:S:C410 | S:S:C415 | 7.28 | 1 | No | Yes | 8 | 8 | 2 | 2 | | S:S:C410 | S:S:P416 | 5.65 | 1 | No | No | 8 | 3 | 2 | 1 | | H:H:?904 | S:S:N411 | 28.34 | 1 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?904 | S:S:S413 | 5.39 | 1 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?904 | S:S:G414 | 3.07 | 1 | Yes | No | 0 | 3 | 0 | 1 | | S:S:C415 | S:S:P416 | 3.77 | 1 | Yes | No | 8 | 3 | 2 | 1 | | H:H:?904 | S:S:P416 | 5.89 | 1 | Yes | No | 0 | 3 | 0 | 1 | | R:R:N428 | R:R:Q435 | 6.6 | 1 | No | Yes | 5 | 3 | 1 | 1 | | R:R:F437 | R:R:N428 | 6.04 | 1 | Yes | No | 4 | 5 | 1 | 1 | | H:H:?904 | R:R:N428 | 25.87 | 1 | Yes | No | 0 | 5 | 0 | 1 | | R:R:Q435 | R:R:T430 | 5.67 | 1 | Yes | No | 3 | 5 | 1 | 2 | | R:R:F437 | R:R:Q435 | 4.68 | 1 | Yes | Yes | 4 | 3 | 1 | 1 | | H:H:?904 | R:R:Q435 | 5.97 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | R:R:D439 | R:R:F437 | 4.78 | 1 | No | Yes | 8 | 4 | 2 | 1 | | H:H:?904 | R:R:F437 | 7.65 | 1 | Yes | Yes | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 11.74 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 17.00 | | Average Links Mediated by Hubs In Shell | 15.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
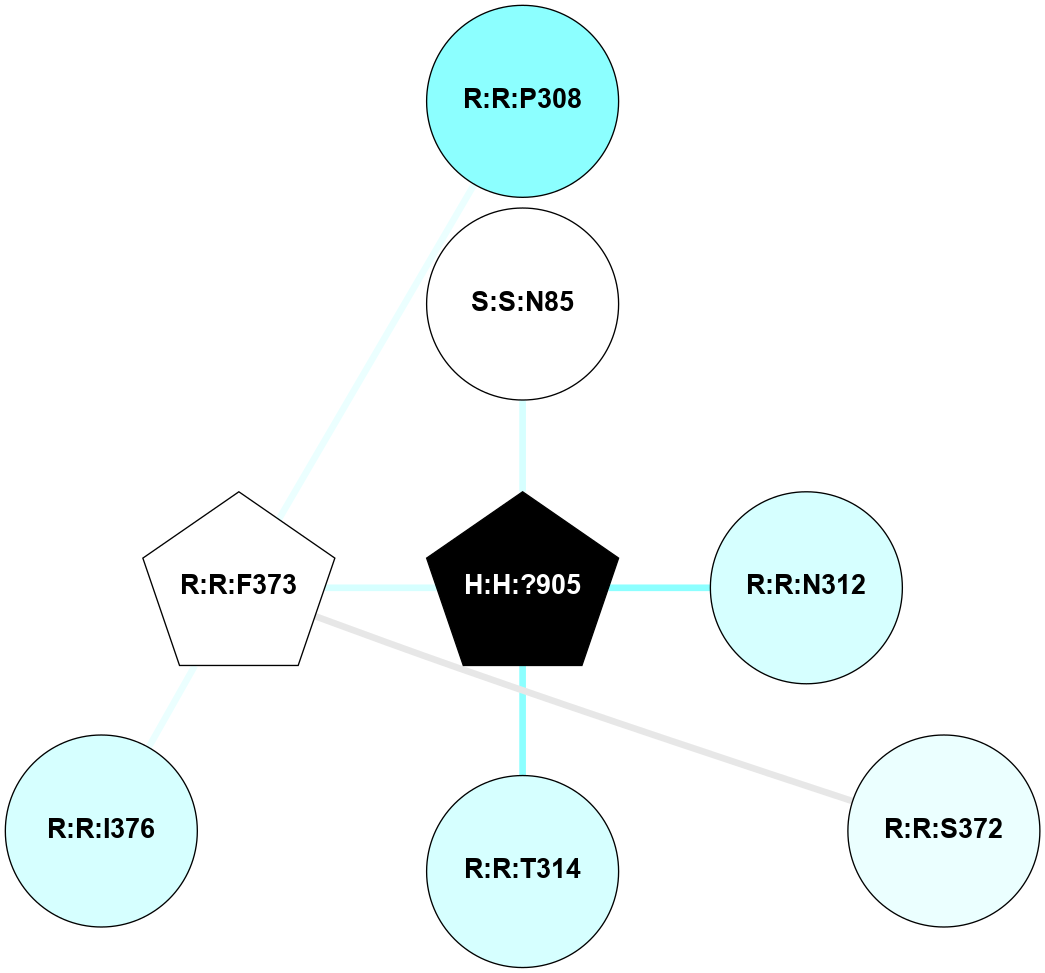
A 2D representation of the interactions of NAG in 9NOT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?905 | S:S:N85 | 27.11 | 0 | Yes | No | 0 | 5 | 0 | 1 | | R:R:F373 | R:R:P308 | 4.33 | 0 | Yes | No | 5 | 2 | 1 | 2 | | H:H:?905 | R:R:N312 | 29.57 | 0 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?905 | R:R:T314 | 9.26 | 0 | Yes | No | 0 | 3 | 0 | 1 | | R:R:F373 | R:R:S372 | 7.93 | 0 | Yes | No | 5 | 4 | 1 | 2 | | R:R:F373 | R:R:I376 | 3.77 | 0 | Yes | No | 5 | 3 | 1 | 2 | | H:H:?905 | R:R:F373 | 6.56 | 0 | Yes | Yes | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 18.12 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
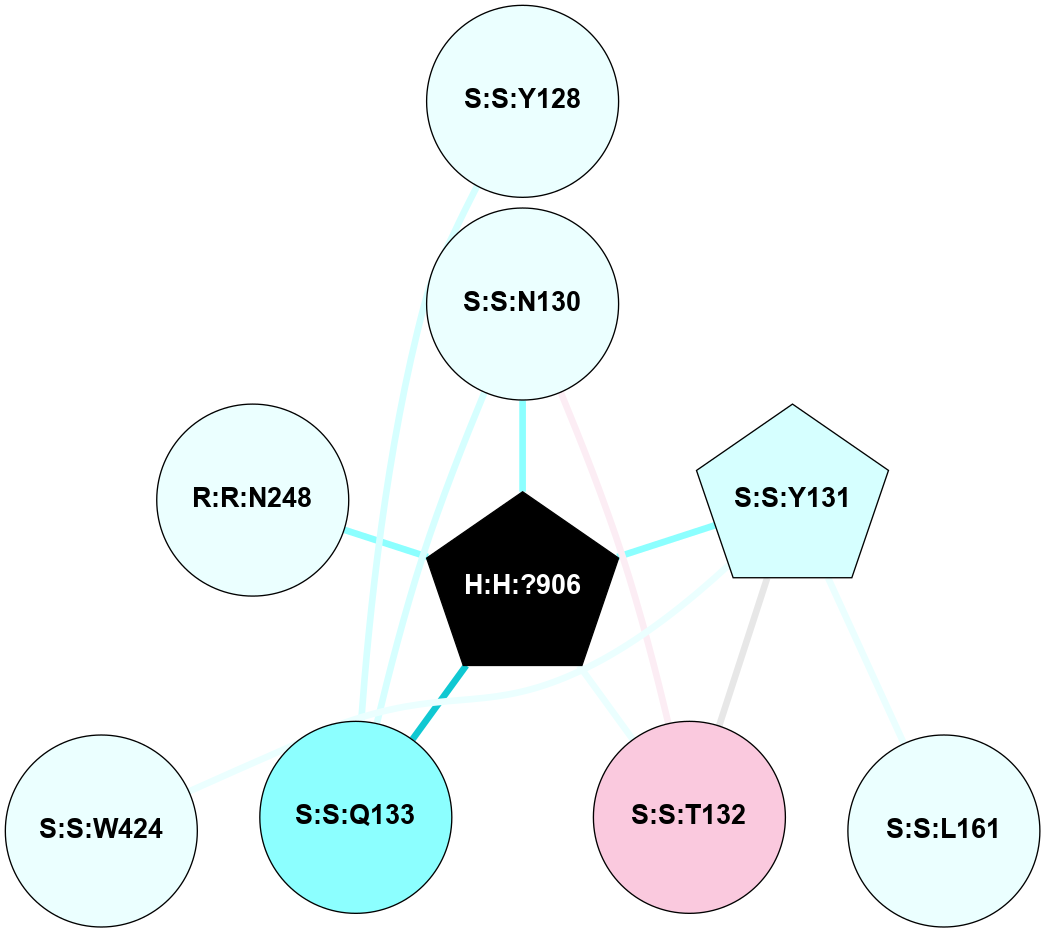
A 2D representation of the interactions of NAG in 9NOT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:Q133 | S:S:Y128 | 9.02 | 1 | No | No | 2 | 4 | 1 | 2 | | S:S:N130 | S:S:T132 | 4.39 | 1 | No | No | 4 | 7 | 1 | 1 | | S:S:N130 | S:S:Q133 | 11.88 | 1 | No | No | 4 | 2 | 1 | 1 | | H:H:?906 | S:S:N130 | 29.57 | 1 | Yes | No | 0 | 4 | 0 | 1 | | S:S:T132 | S:S:Y131 | 11.24 | 1 | No | Yes | 7 | 3 | 1 | 1 | | S:S:L161 | S:S:Y131 | 9.38 | 0 | No | Yes | 4 | 3 | 2 | 1 | | S:S:W424 | S:S:Y131 | 12.54 | 1 | No | Yes | 4 | 3 | 2 | 1 | | H:H:?906 | S:S:Y131 | 3.16 | 1 | Yes | Yes | 0 | 3 | 0 | 1 | | H:H:?906 | S:S:T132 | 3.97 | 1 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?906 | S:S:Q133 | 10.75 | 1 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?906 | R:R:N248 | 27.11 | 1 | Yes | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.91 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
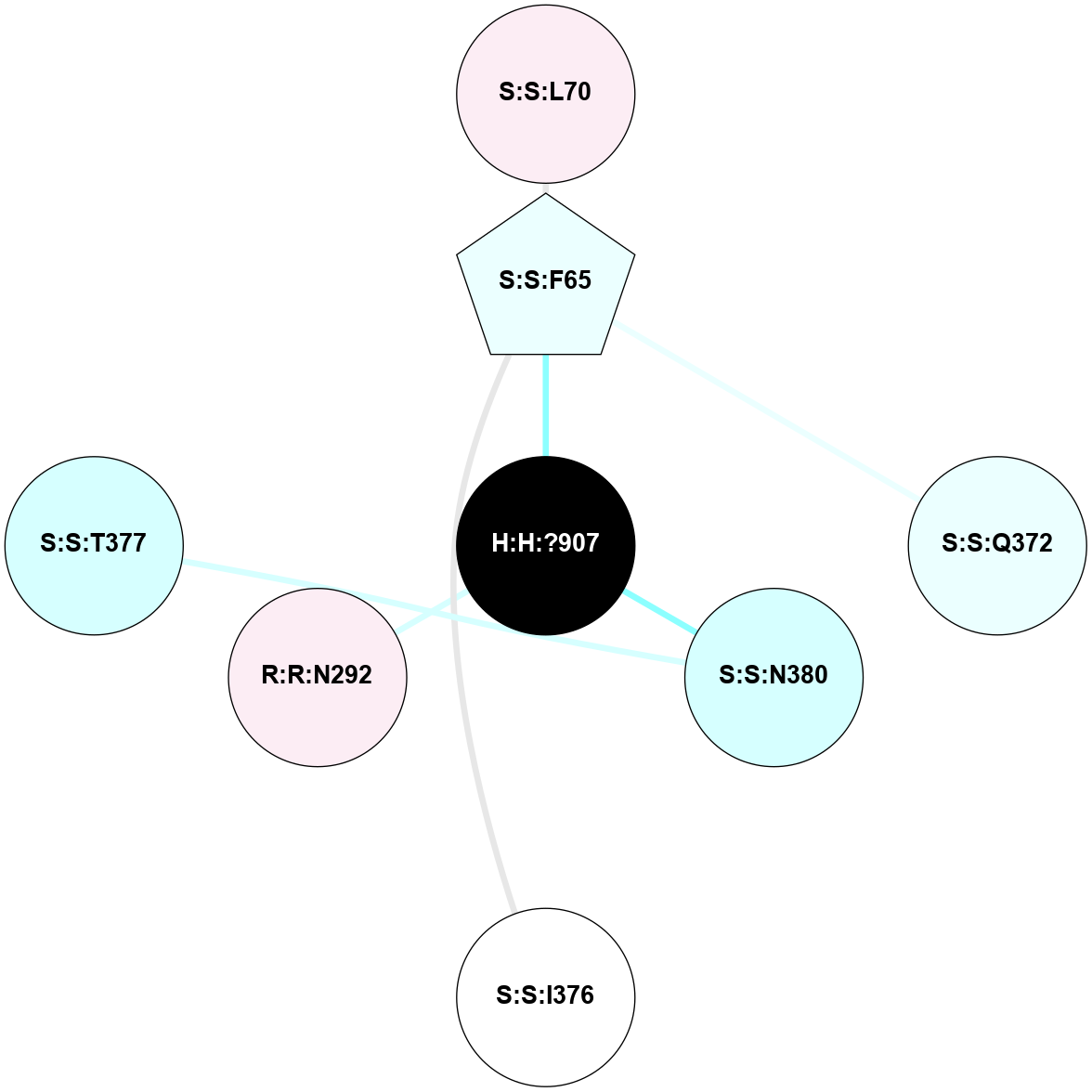
A 2D representation of the interactions of NAG in 9NOT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:F65 | S:S:L70 | 4.87 | 0 | Yes | No | 4 | 6 | 1 | 2 | | S:S:F65 | S:S:Q372 | 3.51 | 0 | Yes | No | 4 | 4 | 1 | 2 | | S:S:F65 | S:S:I376 | 7.54 | 0 | Yes | No | 4 | 5 | 1 | 2 | | H:H:?907 | S:S:F65 | 8.74 | 0 | No | Yes | 0 | 4 | 0 | 1 | | H:H:?907 | S:S:N380 | 33.27 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?907 | R:R:N292 | 28.34 | 0 | No | No | 0 | 6 | 0 | 1 | | S:S:N380 | S:S:T377 | 1.46 | 0 | No | No | 3 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 23.45 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
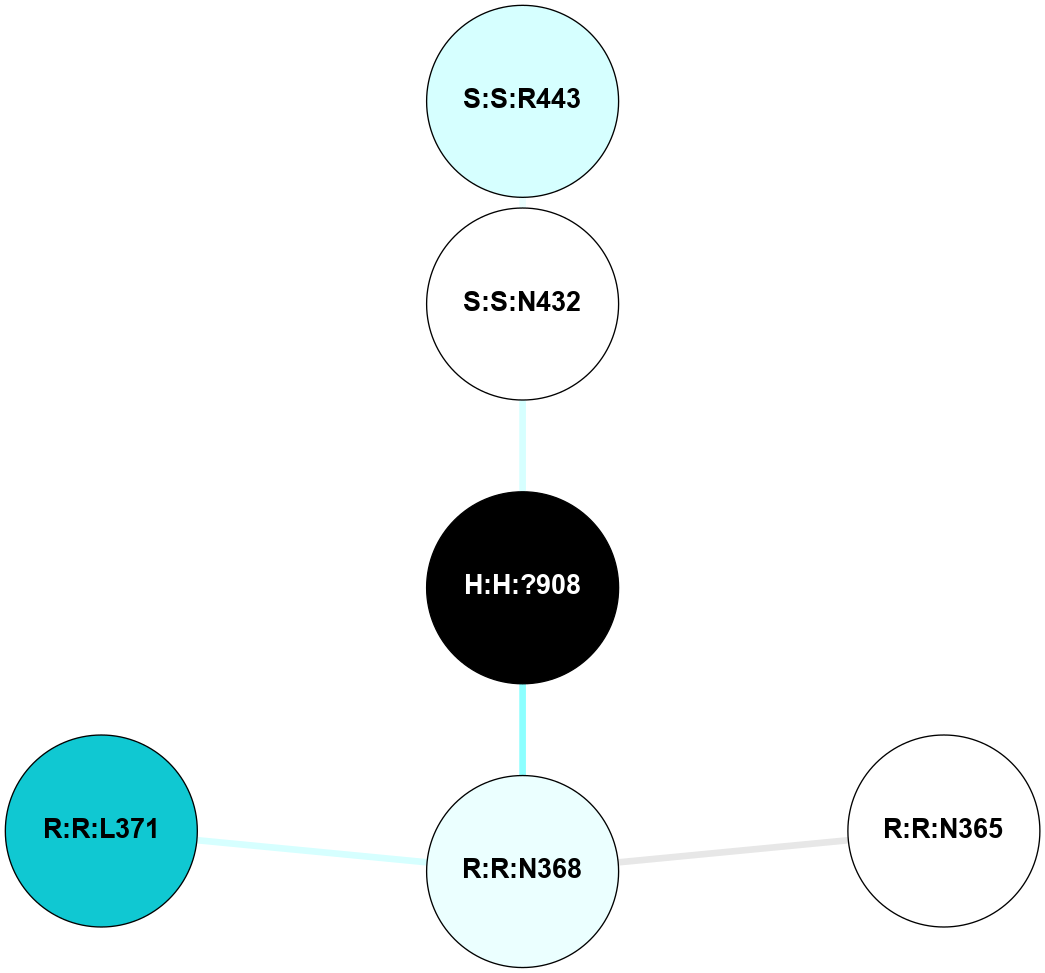
A 2D representation of the interactions of NAG in 9NOT
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:N432 | S:S:R443 | 3.62 | 0 | No | No | 5 | 3 | 1 | 2 | | H:H:?908 | S:S:N432 | 38.2 | 0 | No | No | 0 | 5 | 0 | 1 | | R:R:N365 | R:R:N368 | 5.45 | 0 | No | No | 5 | 4 | 2 | 1 | | H:H:?908 | R:R:N368 | 30.8 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:L371 | R:R:N368 | 1.37 | 0 | No | No | 1 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 34.50 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9NOR | C | Sensory | Taste1 | TAS1R2; TAS1R3 | Homo Sapiens | - | - | - | 3.4 | 2025-04-09 | doi.org/10.1016/j.cell.2025.04.021 |
|
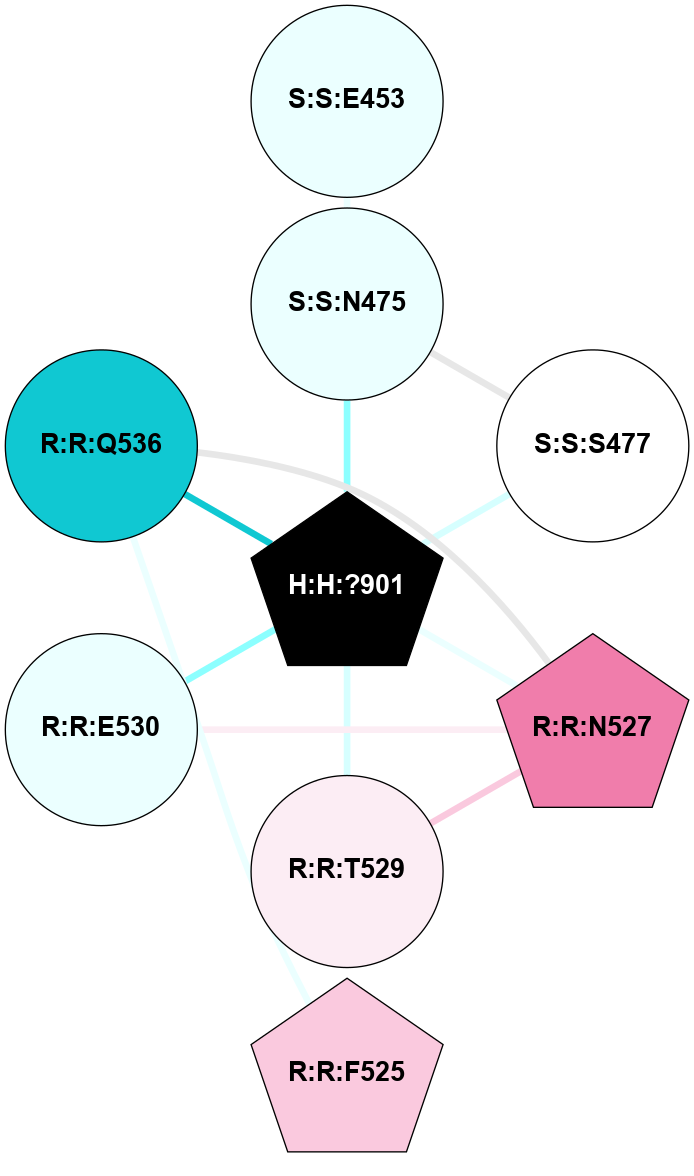
A 2D representation of the interactions of NAG in 9NOR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:E453 | S:S:N475 | 6.57 | 6 | No | No | 4 | 4 | 2 | 1 | | S:S:N475 | S:S:S477 | 2.98 | 6 | No | No | 4 | 5 | 1 | 1 | | H:H:?901 | S:S:N475 | 27.11 | 6 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?901 | S:S:S477 | 5.39 | 6 | Yes | No | 0 | 5 | 0 | 1 | | R:R:F525 | R:R:Q536 | 5.86 | 6 | Yes | No | 7 | 1 | 2 | 1 | | R:R:N527 | R:R:T529 | 7.31 | 6 | Yes | No | 8 | 6 | 1 | 1 | | R:R:E530 | R:R:N527 | 10.52 | 6 | No | Yes | 4 | 8 | 1 | 1 | | R:R:N527 | R:R:Q536 | 9.24 | 6 | Yes | No | 8 | 1 | 1 | 1 | | H:H:?901 | R:R:N527 | 28.34 | 6 | Yes | Yes | 0 | 8 | 0 | 1 | | H:H:?901 | R:R:T529 | 9.26 | 6 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?901 | R:R:E530 | 14.27 | 6 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?901 | R:R:Q536 | 3.58 | 6 | Yes | No | 0 | 1 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.66 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
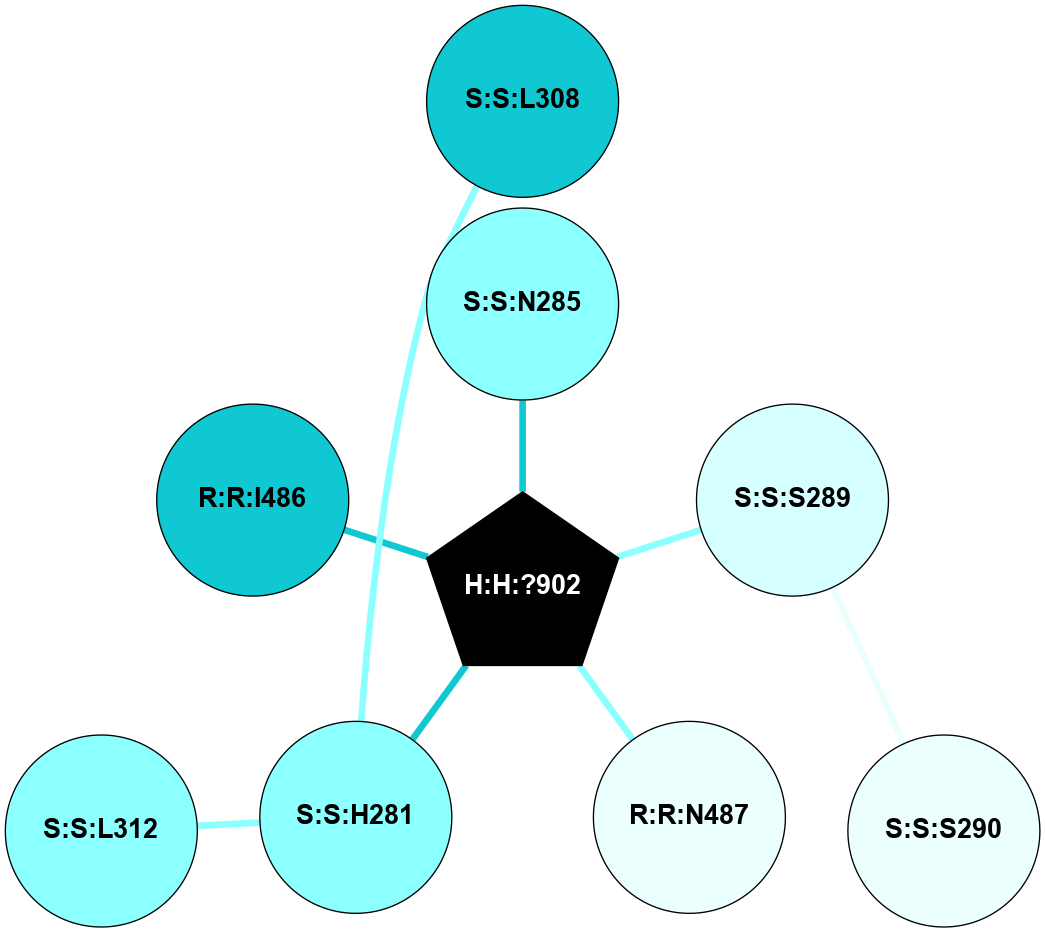
A 2D representation of the interactions of NAG in 9NOR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:H281 | S:S:L308 | 16.71 | 0 | No | No | 2 | 1 | 1 | 2 | | H:H:?902 | S:S:N285 | 29.57 | 0 | Yes | No | 0 | 2 | 0 | 1 | | S:S:S289 | S:S:S290 | 3.26 | 0 | No | No | 3 | 4 | 1 | 2 | | H:H:?902 | S:S:S289 | 6.74 | 0 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?902 | R:R:N487 | 28.34 | 0 | Yes | No | 0 | 4 | 0 | 1 | | S:S:H281 | S:S:L312 | 2.57 | 0 | No | No | 2 | 2 | 1 | 2 | | H:H:?902 | S:S:H281 | 2.31 | 0 | Yes | No | 0 | 2 | 0 | 1 | | H:H:?902 | R:R:I486 | 1.28 | 0 | Yes | No | 0 | 1 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.65 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 8.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
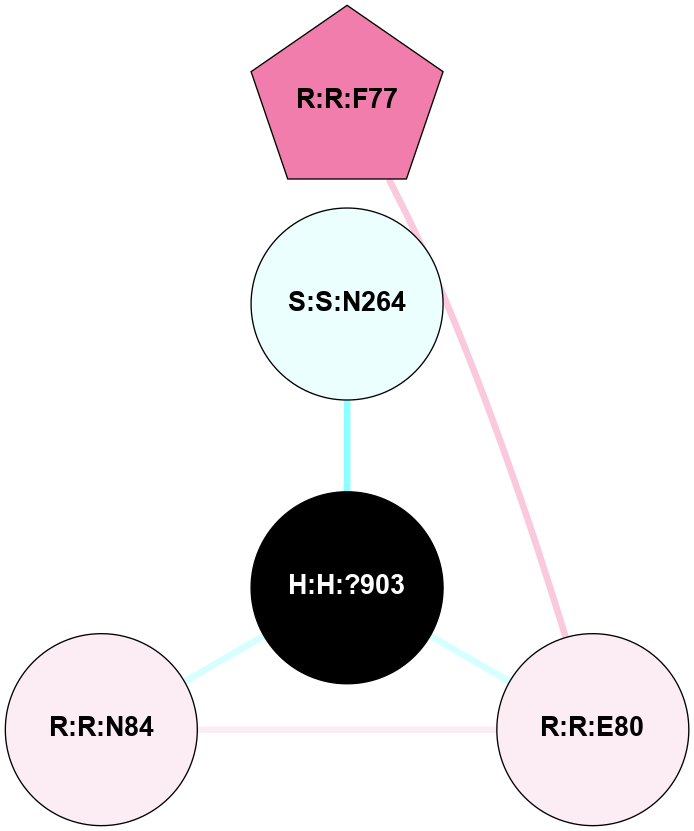
A 2D representation of the interactions of NAG in 9NOR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?903 | S:S:N264 | 27.11 | 38 | No | No | 0 | 4 | 0 | 1 | | R:R:E80 | R:R:N84 | 13.15 | 38 | No | No | 6 | 6 | 1 | 1 | | H:H:?903 | R:R:E80 | 27.35 | 38 | No | No | 0 | 6 | 0 | 1 | | H:H:?903 | R:R:N84 | 25.87 | 38 | No | No | 0 | 6 | 0 | 1 | | R:R:E80 | R:R:F77 | 2.33 | 38 | No | Yes | 6 | 8 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 26.78 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
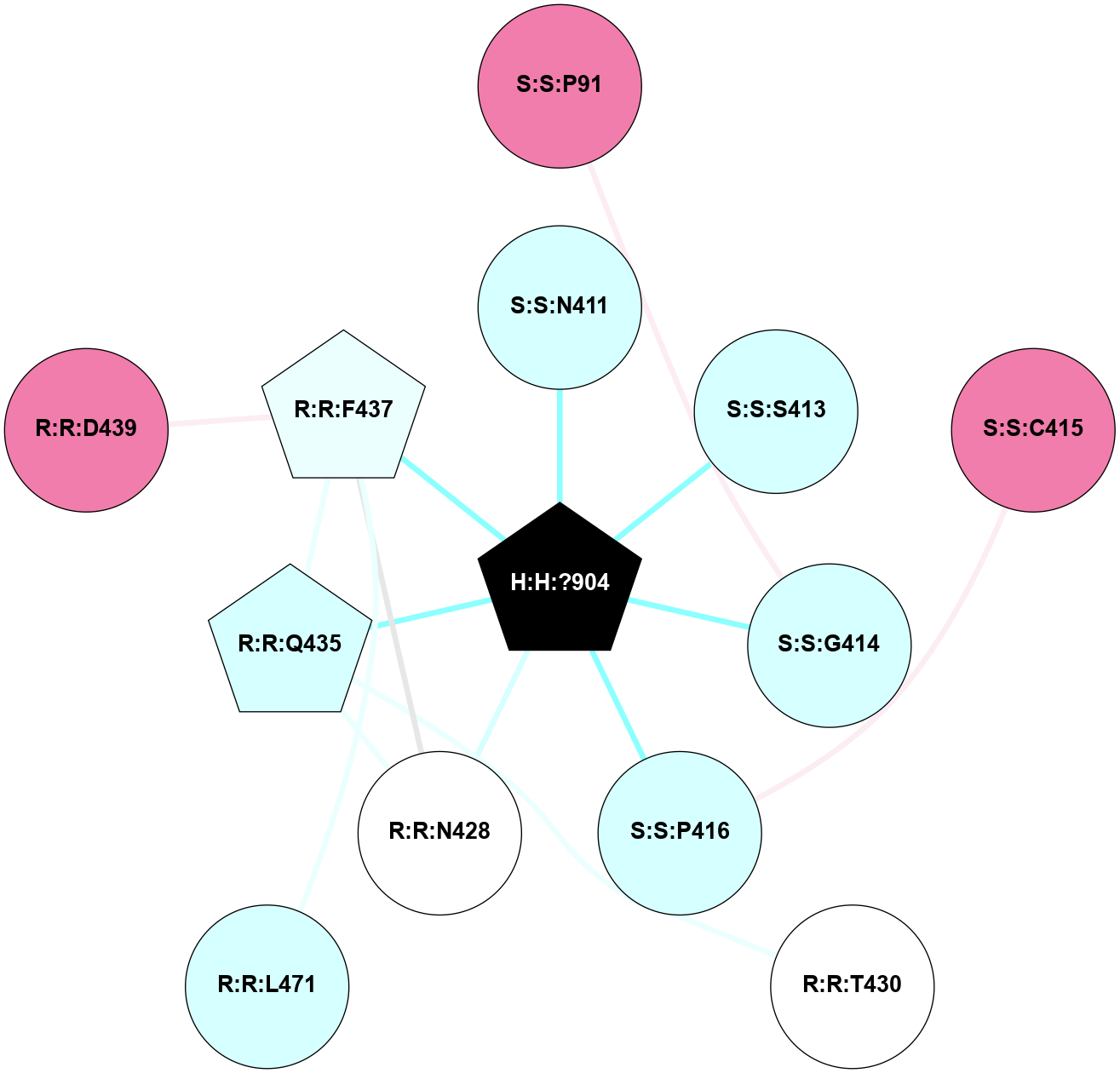
A 2D representation of the interactions of NAG in 9NOR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:G414 | S:S:P91 | 4.06 | 0 | No | No | 3 | 8 | 1 | 2 | | H:H:?904 | S:S:N411 | 28.34 | 25 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?904 | S:S:S413 | 4.04 | 25 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?904 | S:S:G414 | 3.07 | 25 | Yes | No | 0 | 3 | 0 | 1 | | S:S:C415 | S:S:P416 | 3.77 | 0 | No | No | 8 | 3 | 2 | 1 | | H:H:?904 | S:S:P416 | 8.84 | 25 | Yes | No | 0 | 3 | 0 | 1 | | R:R:N428 | R:R:Q435 | 6.6 | 25 | No | Yes | 5 | 3 | 1 | 1 | | R:R:F437 | R:R:N428 | 6.04 | 25 | Yes | No | 4 | 5 | 1 | 1 | | H:H:?904 | R:R:N428 | 25.87 | 25 | Yes | No | 0 | 5 | 0 | 1 | | R:R:F437 | R:R:Q435 | 4.68 | 25 | Yes | Yes | 4 | 3 | 1 | 1 | | H:H:?904 | R:R:Q435 | 5.97 | 25 | Yes | Yes | 0 | 3 | 0 | 1 | | H:H:?904 | R:R:F437 | 4.37 | 25 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:Q435 | R:R:T430 | 2.83 | 25 | Yes | No | 3 | 5 | 1 | 2 | | R:R:F437 | R:R:L471 | 2.44 | 25 | Yes | No | 4 | 3 | 1 | 2 | | R:R:D439 | R:R:F437 | 2.39 | 0 | No | Yes | 8 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 7.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 11.50 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 13.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
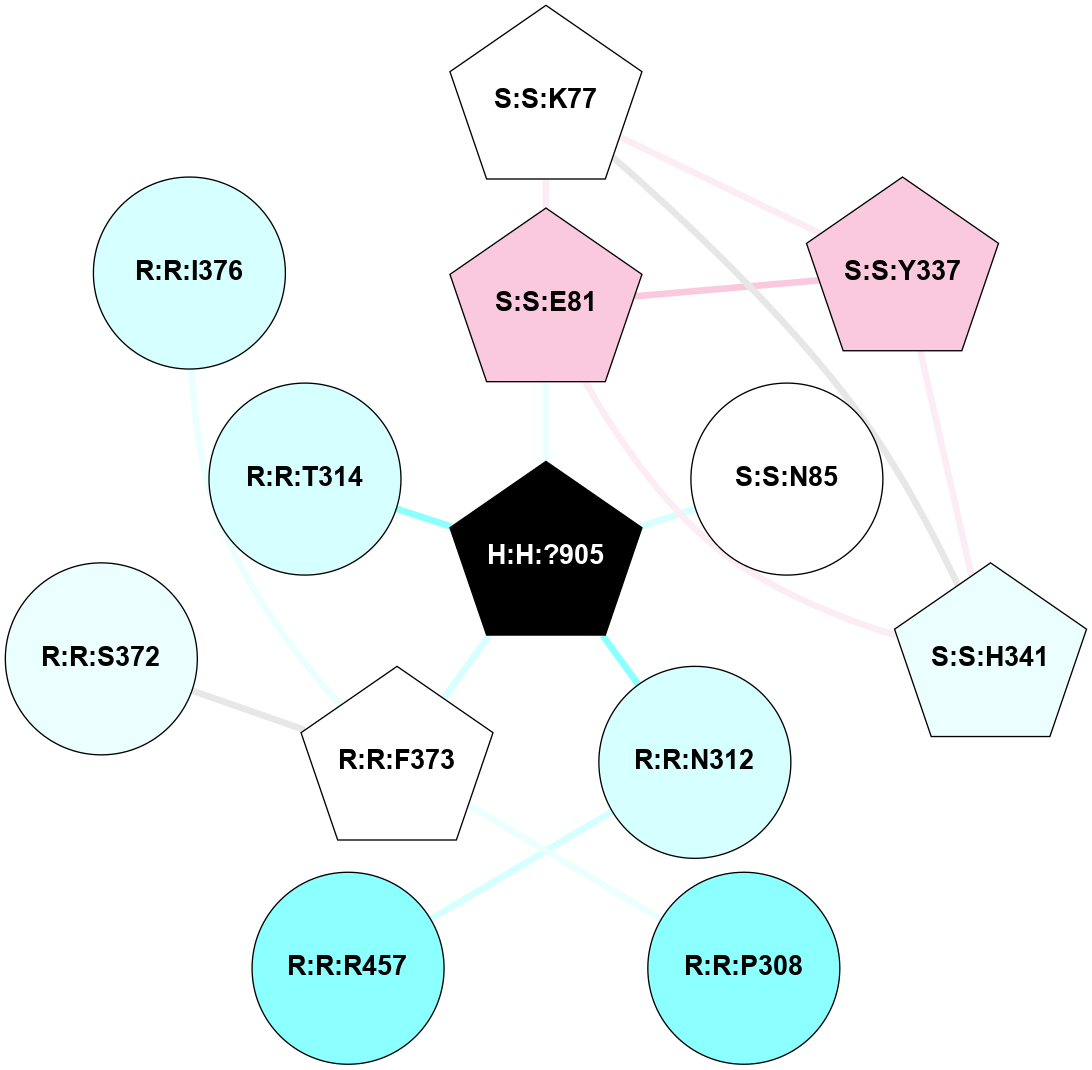
A 2D representation of the interactions of NAG in 9NOR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:E81 | S:S:K77 | 6.75 | 4 | Yes | Yes | 7 | 5 | 1 | 2 | | S:S:K77 | S:S:Y337 | 5.97 | 4 | Yes | Yes | 5 | 7 | 2 | 2 | | S:S:H341 | S:S:K77 | 7.86 | 4 | Yes | Yes | 4 | 5 | 2 | 2 | | S:S:E81 | S:S:Y337 | 4.49 | 4 | Yes | Yes | 7 | 7 | 1 | 2 | | S:S:E81 | S:S:H341 | 6.15 | 4 | Yes | Yes | 7 | 4 | 1 | 2 | | H:H:?905 | S:S:E81 | 3.57 | 0 | Yes | Yes | 0 | 7 | 0 | 1 | | H:H:?905 | S:S:N85 | 25.87 | 0 | Yes | No | 0 | 5 | 0 | 1 | | S:S:H341 | S:S:Y337 | 22.87 | 4 | Yes | Yes | 4 | 7 | 2 | 2 | | R:R:F373 | R:R:P308 | 7.22 | 0 | Yes | No | 5 | 2 | 1 | 2 | | R:R:N312 | R:R:R457 | 10.85 | 0 | No | No | 3 | 2 | 1 | 2 | | H:H:?905 | R:R:N312 | 28.34 | 0 | Yes | No | 0 | 3 | 0 | 1 | | R:R:F373 | R:R:S372 | 3.96 | 0 | Yes | No | 5 | 4 | 1 | 2 | | R:R:F373 | R:R:I376 | 5.02 | 0 | Yes | No | 5 | 3 | 1 | 2 | | H:H:?905 | R:R:F373 | 9.84 | 0 | Yes | Yes | 0 | 5 | 0 | 1 | | H:H:?905 | R:R:T314 | 2.65 | 0 | Yes | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 14.05 | | Average Nodes In Shell | 13.00 | | Average Hubs In Shell | 6.00 | | Average Links In Shell | 15.00 | | Average Links Mediated by Hubs In Shell | 14.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
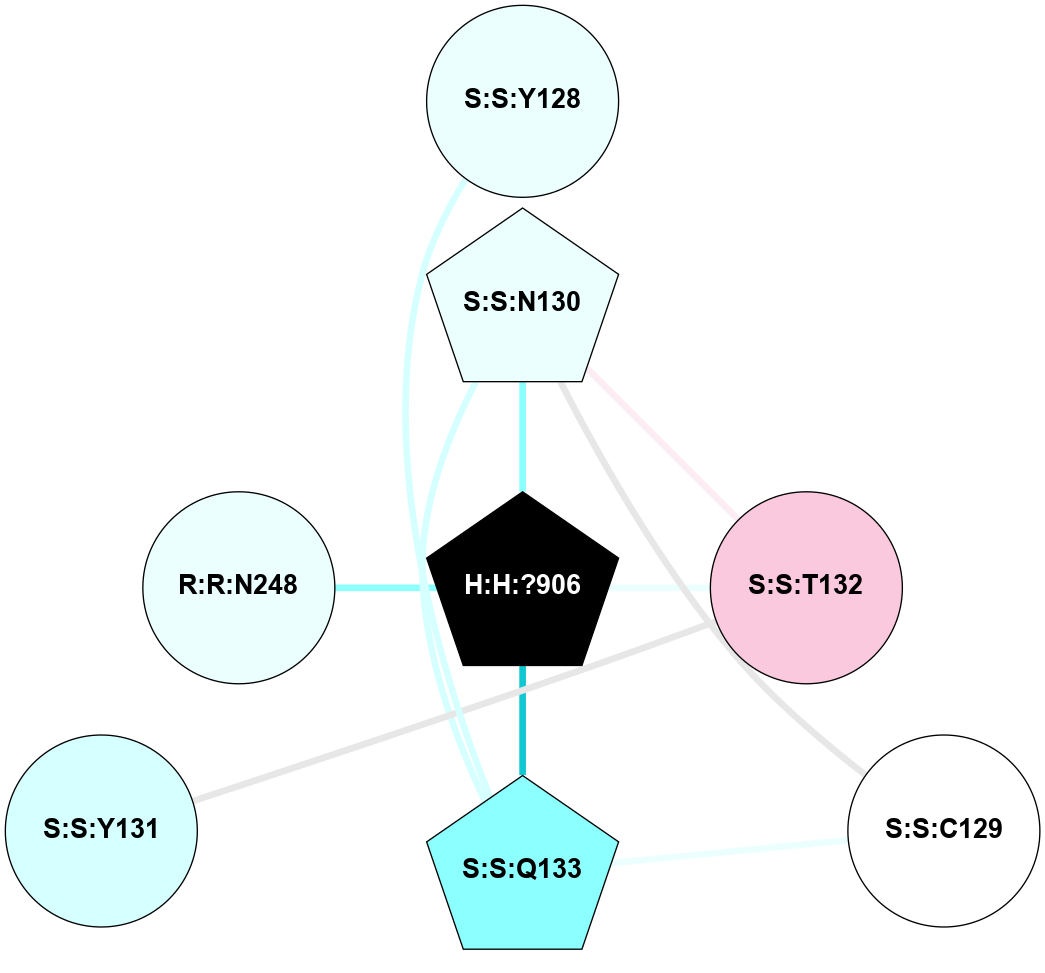
A 2D representation of the interactions of NAG in 9NOR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:Q133 | S:S:Y128 | 7.89 | 19 | Yes | No | 2 | 4 | 1 | 2 | | S:S:C129 | S:S:N130 | 3.15 | 19 | No | Yes | 5 | 4 | 2 | 1 | | S:S:C129 | S:S:Q133 | 4.58 | 19 | No | Yes | 5 | 2 | 2 | 1 | | S:S:N130 | S:S:T132 | 2.92 | 19 | Yes | No | 4 | 7 | 1 | 1 | | S:S:N130 | S:S:Q133 | 10.56 | 19 | Yes | Yes | 4 | 2 | 1 | 1 | | H:H:?906 | S:S:N130 | 28.34 | 19 | Yes | Yes | 0 | 4 | 0 | 1 | | S:S:T132 | S:S:Y131 | 11.24 | 19 | No | No | 7 | 3 | 1 | 2 | | H:H:?906 | S:S:T132 | 3.97 | 19 | Yes | No | 0 | 7 | 0 | 1 | | H:H:?906 | S:S:Q133 | 9.55 | 19 | Yes | Yes | 0 | 2 | 0 | 1 | | H:H:?906 | R:R:N248 | 25.87 | 19 | Yes | No | 0 | 4 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 16.93 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 9.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
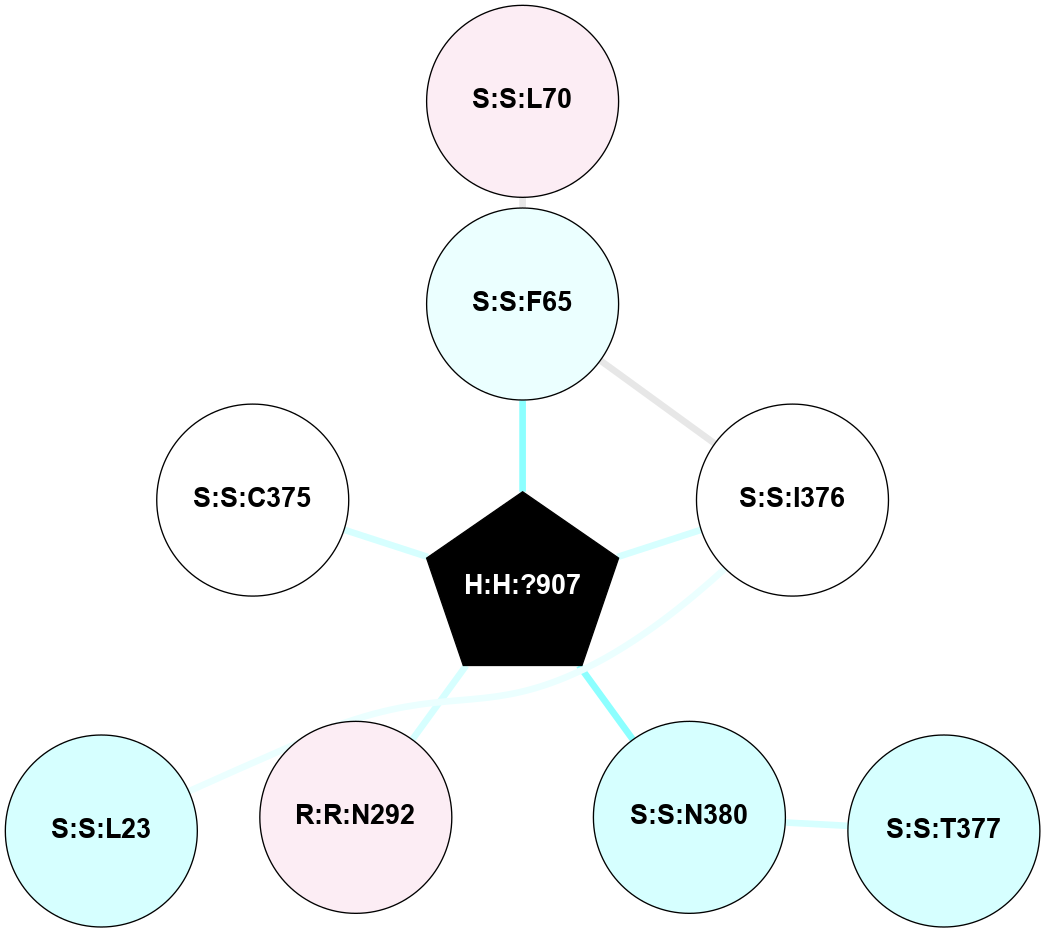
A 2D representation of the interactions of NAG in 9NOR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | S:S:F65 | S:S:L70 | 4.87 | 32 | No | No | 4 | 6 | 1 | 2 | | S:S:F65 | S:S:I376 | 8.79 | 32 | No | No | 4 | 5 | 1 | 1 | | H:H:?907 | S:S:F65 | 3.28 | 32 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?907 | S:S:I376 | 3.84 | 32 | Yes | No | 0 | 5 | 0 | 1 | | S:S:N380 | S:S:T377 | 4.39 | 0 | No | No | 3 | 3 | 1 | 2 | | H:H:?907 | S:S:N380 | 34.5 | 32 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?907 | R:R:N292 | 29.57 | 32 | Yes | No | 0 | 6 | 0 | 1 | | S:S:I376 | S:S:L23 | 1.43 | 32 | No | No | 5 | 3 | 1 | 2 | | H:H:?907 | S:S:C375 | 1.42 | 32 | Yes | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.52 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 9.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
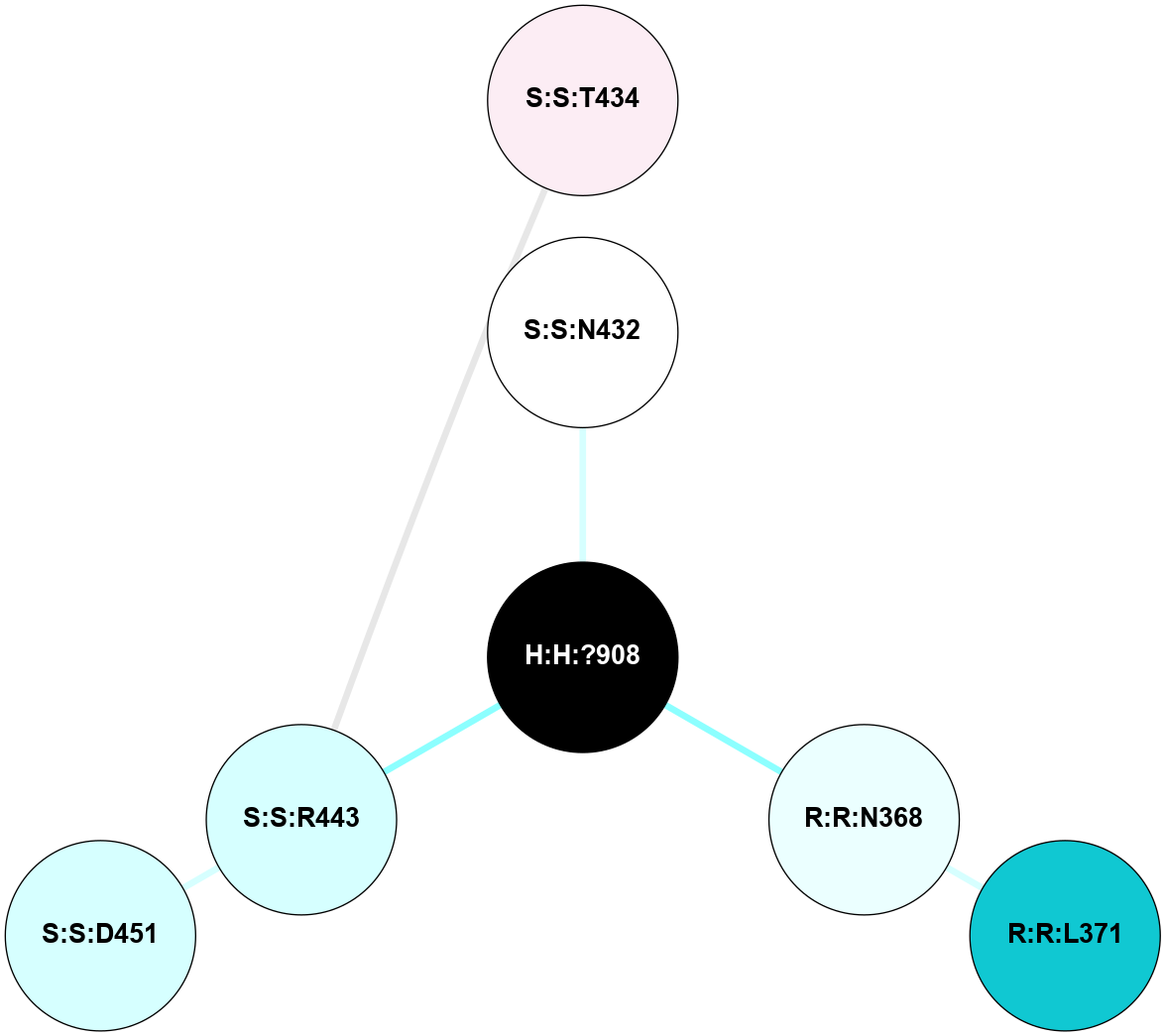
A 2D representation of the interactions of NAG in 9NOR
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?908 | S:S:N432 | 33.27 | 0 | No | No | 0 | 5 | 0 | 1 | | S:S:R443 | S:S:T434 | 5.17 | 0 | No | No | 3 | 6 | 1 | 2 | | H:H:?908 | R:R:N368 | 33.27 | 0 | No | No | 0 | 4 | 0 | 1 | | H:H:?908 | S:S:R443 | 2.18 | 0 | No | No | 0 | 3 | 0 | 1 | | R:R:L371 | R:R:N368 | 1.37 | 0 | No | No | 1 | 4 | 2 | 1 | | S:S:D451 | S:S:R443 | 1.19 | 0 | No | No | 3 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 22.91 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9AU0 | A | Lipid | Prostaglandin | DP1 | Homo Sapiens | BW245C | - | Gs/Beta1/Gamma2 | 2.45 | 2025-08-27 | To be published |
|
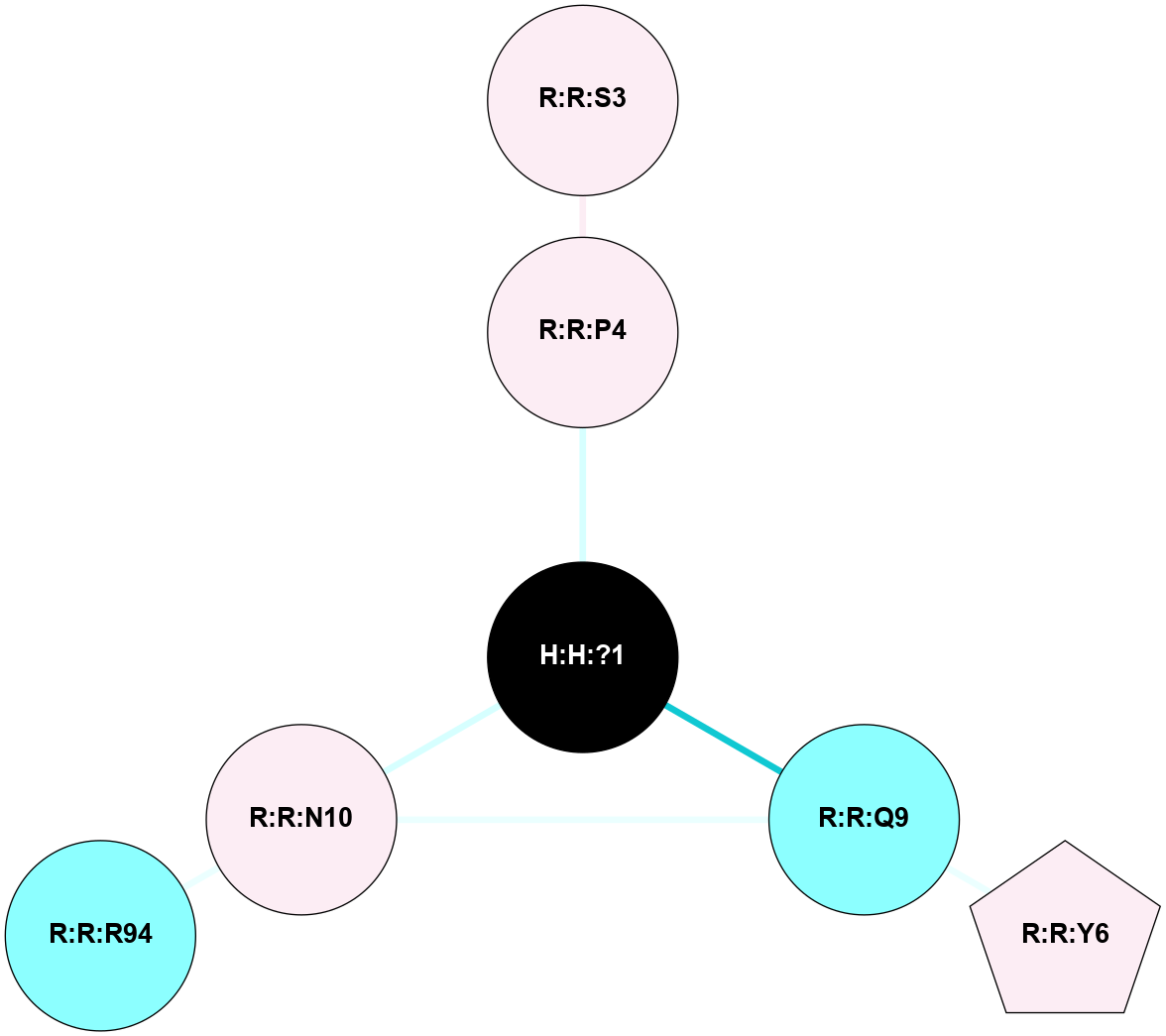
A 2D representation of the interactions of NAG in 9AU0
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:P4 | 10.31 | 1 | No | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:Q9 | 23.89 | 1 | No | No | 0 | 2 | 0 | 1 | | H:H:?1 | R:R:N10 | 27.11 | 1 | No | No | 0 | 6 | 0 | 1 | | R:R:P4 | R:R:S3 | 3.56 | 0 | No | No | 6 | 6 | 1 | 2 | | R:R:Q9 | R:R:Y6 | 3.38 | 1 | No | Yes | 2 | 6 | 1 | 2 | | R:R:N10 | R:R:Q9 | 7.92 | 1 | No | No | 6 | 2 | 1 | 1 | | R:R:N10 | R:R:R94 | 4.82 | 1 | No | No | 6 | 2 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 20.44 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9BP3 | B1 | Peptide | Calcitonin | CT (AMY1) | Homo Sapiens | Cagrilintide | - | Gs/Beta1/Gamma2; RAMP1 | 2.2 | 2025-04-23 | To be published |
|
A 2D representation of the interactions of NAG in 9BP3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?501 | R:R:N125 | 29.57 | 0 | No | No | 0 | 7 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 29.57 | | Average Nodes In Shell | 2.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 1.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
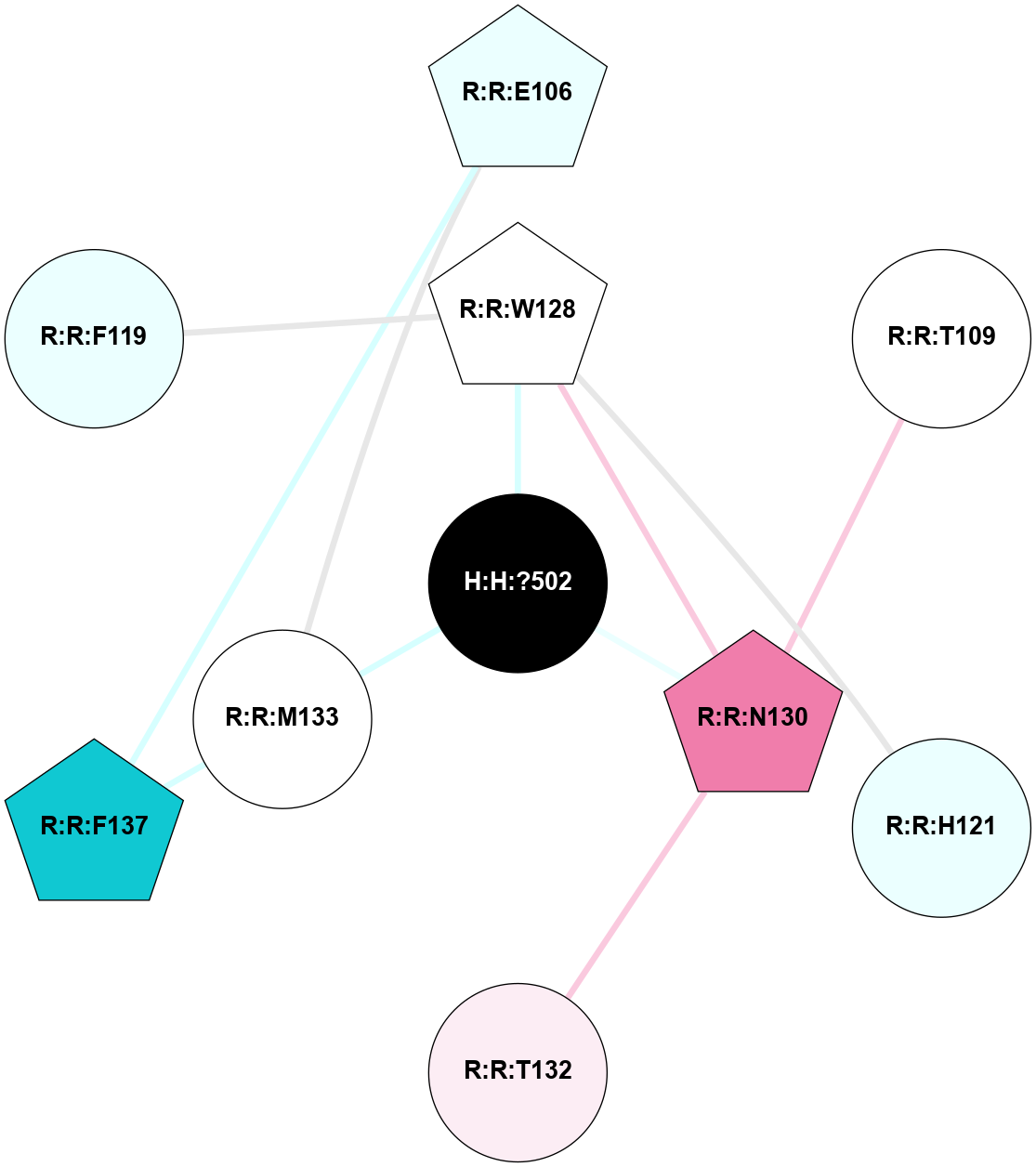
A 2D representation of the interactions of NAG in 9BP3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?502 | R:R:W128 | 6.13 | 12 | No | Yes | 0 | 5 | 0 | 1 | | H:H:?502 | R:R:N130 | 24.64 | 12 | No | Yes | 0 | 8 | 0 | 1 | | H:H:?502 | R:R:M133 | 5.07 | 12 | No | No | 0 | 5 | 0 | 1 | | R:R:E106 | R:R:M133 | 4.06 | 12 | Yes | No | 4 | 5 | 2 | 1 | | R:R:E106 | R:R:F137 | 7 | 12 | Yes | Yes | 4 | 1 | 2 | 2 | | R:R:N130 | R:R:T109 | 4.39 | 12 | Yes | No | 8 | 5 | 1 | 2 | | R:R:H121 | R:R:W128 | 14.81 | 0 | No | Yes | 4 | 5 | 2 | 1 | | R:R:N130 | R:R:W128 | 10.17 | 12 | Yes | Yes | 8 | 5 | 1 | 1 | | R:R:N130 | R:R:T132 | 7.31 | 12 | Yes | No | 8 | 6 | 1 | 2 | | R:R:F137 | R:R:M133 | 3.73 | 12 | Yes | No | 1 | 5 | 2 | 1 | | R:R:F119 | R:R:W128 | 3.01 | 0 | No | Yes | 4 | 5 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 11.95 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 11.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
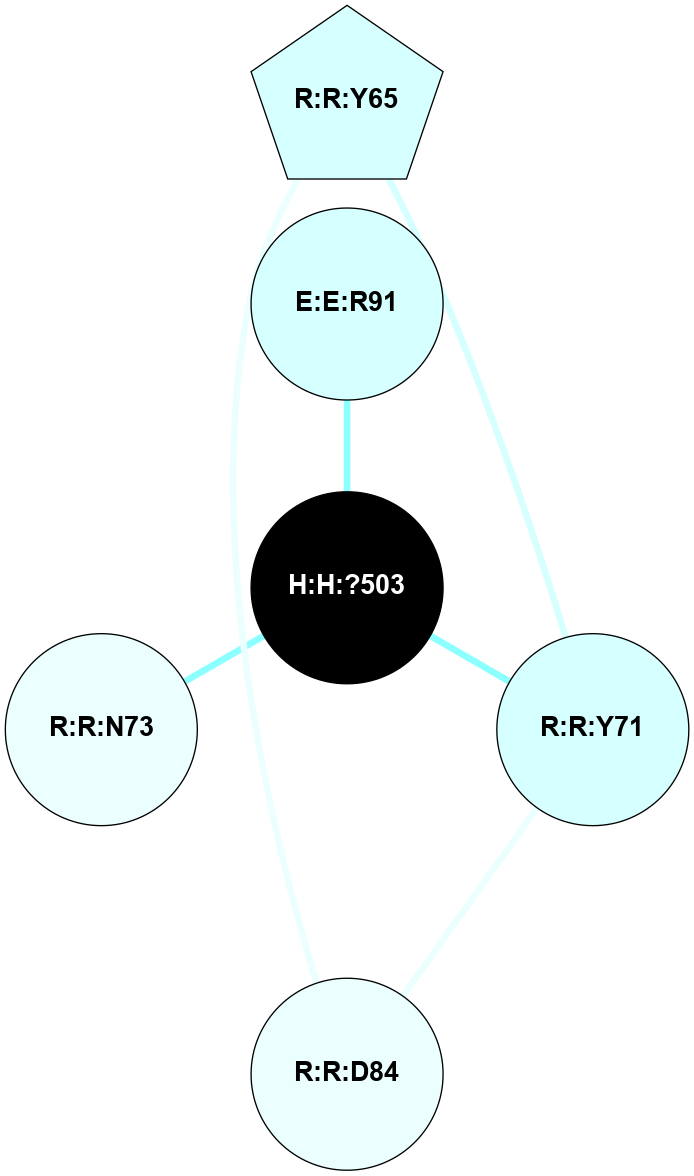
A 2D representation of the interactions of NAG in 9BP3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | E:E:R91 | H:H:?503 | 4.36 | 0 | No | No | 3 | 0 | 1 | 0 | | H:H:?503 | R:R:Y71 | 7.36 | 0 | No | No | 0 | 3 | 0 | 1 | | H:H:?503 | R:R:N73 | 28.34 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:Y65 | R:R:Y71 | 31.77 | 31 | Yes | No | 3 | 3 | 2 | 1 | | R:R:D84 | R:R:Y65 | 5.75 | 31 | No | Yes | 4 | 3 | 2 | 2 | | R:R:D84 | R:R:Y71 | 9.2 | 31 | No | No | 4 | 3 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.35 | | Average Nodes In Shell | 6.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 2.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9C1P | C | Ion | Calcium Sensing | CaS | Homo Sapiens | Ca | Ca; Tryptophan; '6218; PO4 | - | 2.8 | 2024-10-02 | doi.org/10.1126/science.ado1868 |
|

A 2D representation of the interactions of NAG in 9C1P
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N468 | 25.87 | 23 | No | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:Q476 | 8.36 | 23 | No | No | 0 | 3 | 0 | 1 | | H:H:?1 | R:R:T478 | 10.58 | 23 | No | No | 0 | 3 | 0 | 1 | | R:R:N468 | R:R:Q476 | 3.96 | 23 | No | No | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 5.85 | 23 | No | No | 6 | 3 | 1 | 1 | | R:R:Q476 | R:R:T470 | 9.92 | 23 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.94 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
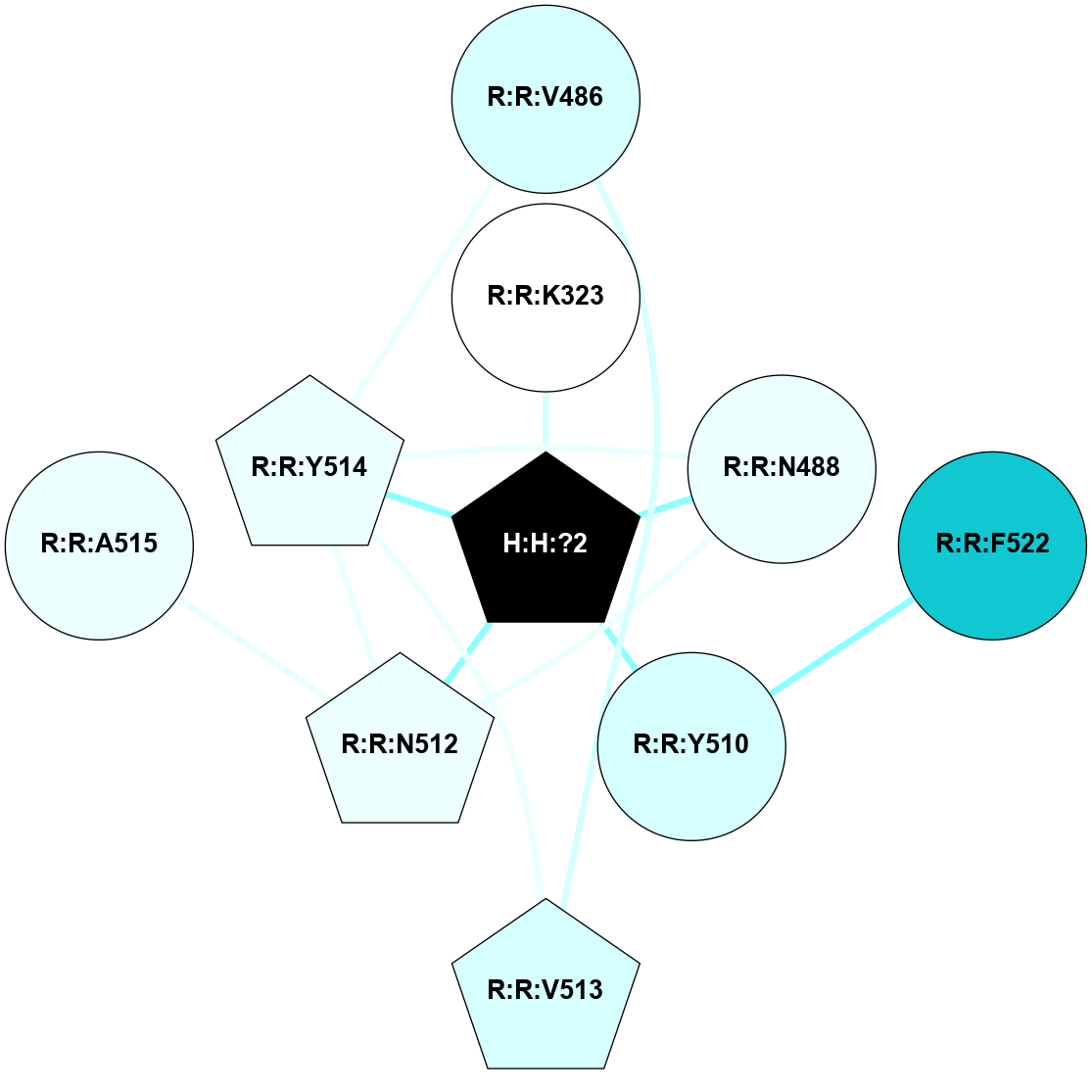
A 2D representation of the interactions of NAG in 9C1P
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?2 | R:R:K323 | 6.33 | 13 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:N488 | 24.64 | 13 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:Y510 | 4.21 | 13 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?2 | R:R:N512 | 13.55 | 13 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?2 | R:R:Y514 | 14.73 | 13 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:V486 | R:R:V513 | 4.81 | 13 | No | Yes | 3 | 3 | 2 | 2 | | R:R:V486 | R:R:Y514 | 3.79 | 13 | No | Yes | 3 | 4 | 2 | 1 | | R:R:N488 | R:R:N512 | 5.45 | 13 | No | Yes | 4 | 4 | 1 | 1 | | R:R:N488 | R:R:Y514 | 3.49 | 13 | No | Yes | 4 | 4 | 1 | 1 | | R:R:F522 | R:R:Y510 | 13.41 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:N512 | R:R:Y514 | 3.49 | 13 | Yes | Yes | 4 | 4 | 1 | 1 | | R:R:V513 | R:R:Y514 | 7.57 | 13 | Yes | Yes | 3 | 4 | 2 | 1 | | R:R:A515 | R:R:N512 | 1.56 | 0 | No | Yes | 4 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 12.69 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 4.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
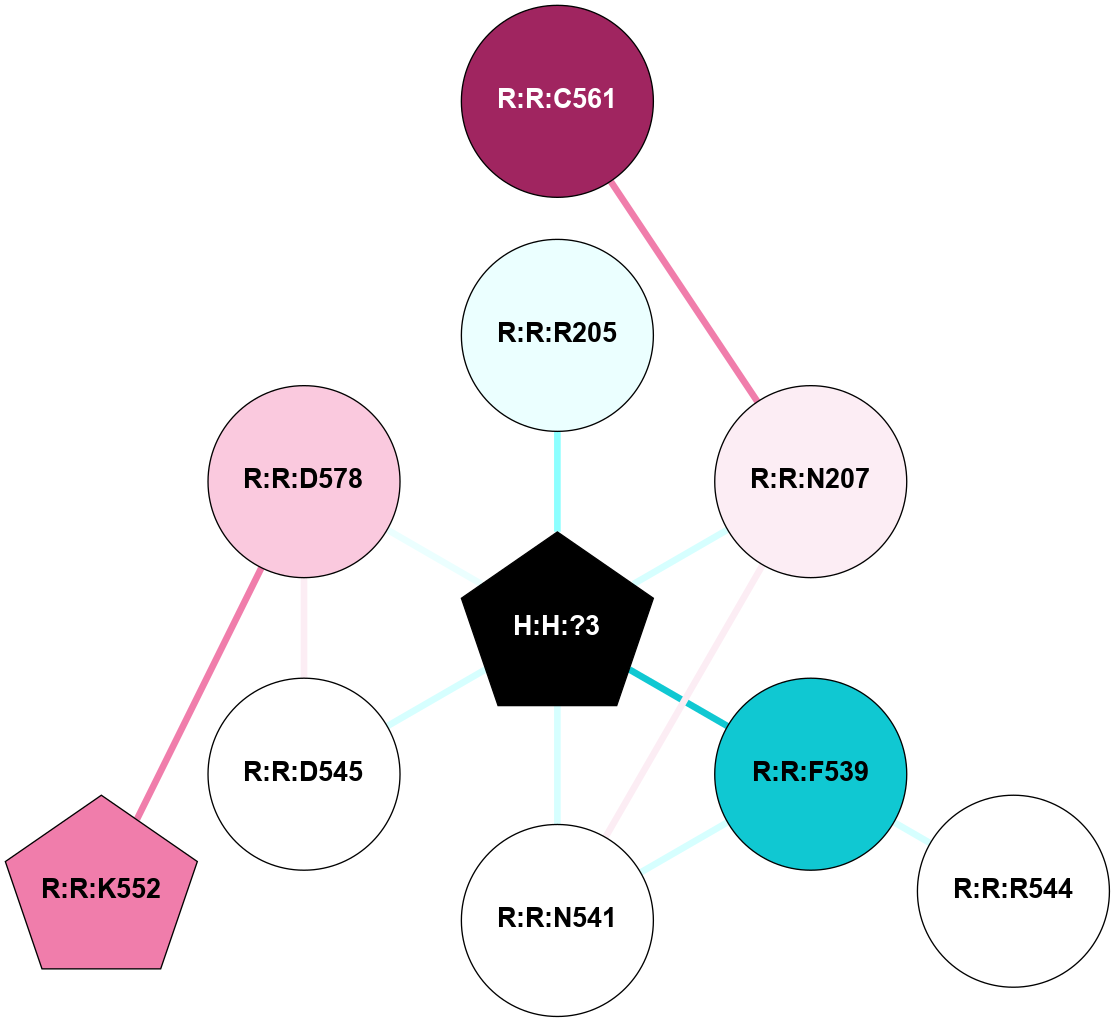
A 2D representation of the interactions of NAG in 9C1P
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | R:R:R205 | 19.62 | 5 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?3 | R:R:N207 | 11.09 | 5 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?3 | R:R:F539 | 4.37 | 5 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?3 | R:R:N541 | 32.03 | 5 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?3 | R:R:D545 | 15.83 | 5 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?3 | R:R:D578 | 6.09 | 5 | Yes | No | 0 | 7 | 0 | 1 | | R:R:N207 | R:R:N541 | 6.81 | 5 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 6.3 | 5 | No | No | 9 | 6 | 2 | 1 | | R:R:F539 | R:R:N541 | 12.08 | 5 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 25.66 | 5 | No | No | 1 | 5 | 1 | 2 | | R:R:D545 | R:R:D578 | 10.65 | 5 | No | No | 5 | 7 | 1 | 1 | | R:R:D578 | R:R:K552 | 9.68 | 5 | No | Yes | 7 | 8 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.84 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 7.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9C1P
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | S:S:N468 | 25.87 | 33 | No | No | 0 | 6 | 0 | 1 | | H:H:?4 | S:S:Q476 | 8.36 | 33 | No | No | 0 | 3 | 0 | 1 | | S:S:N468 | S:S:Q476 | 2.64 | 33 | No | No | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 5.85 | 33 | No | No | 6 | 3 | 1 | 2 | | S:S:Q476 | S:S:T470 | 8.5 | 33 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.12 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9C1P
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | S:S:K323 | 5.06 | 24 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?5 | S:S:N488 | 25.87 | 24 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?5 | S:S:Y510 | 8.42 | 24 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?5 | S:S:N512 | 20.95 | 24 | Yes | Yes | 0 | 4 | 0 | 1 | | H:H:?5 | S:S:Y514 | 14.73 | 24 | Yes | Yes | 0 | 4 | 0 | 1 | | S:S:E475 | S:S:K323 | 2.7 | 0 | No | No | 5 | 5 | 2 | 1 | | S:S:V486 | S:S:Y514 | 3.79 | 0 | No | Yes | 3 | 4 | 2 | 1 | | S:S:N488 | S:S:N512 | 4.09 | 24 | No | Yes | 4 | 4 | 1 | 1 | | S:S:N488 | S:S:Y514 | 4.65 | 24 | No | Yes | 4 | 4 | 1 | 1 | | S:S:F522 | S:S:Y510 | 15.47 | 0 | No | No | 1 | 3 | 2 | 1 | | S:S:N512 | S:S:Y514 | 6.98 | 24 | Yes | Yes | 4 | 4 | 1 | 1 | | S:S:A515 | S:S:N512 | 3.13 | 0 | No | Yes | 4 | 4 | 2 | 1 | | S:S:V513 | S:S:Y514 | 7.57 | 0 | No | Yes | 3 | 4 | 2 | 1 | | S:S:G487 | S:S:Y514 | 1.45 | 0 | No | Yes | 6 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 2.00 | | Average Interaction Strength | 15.01 | | Average Nodes In Shell | 12.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 14.00 | | Average Links Mediated by Hubs In Shell | 12.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
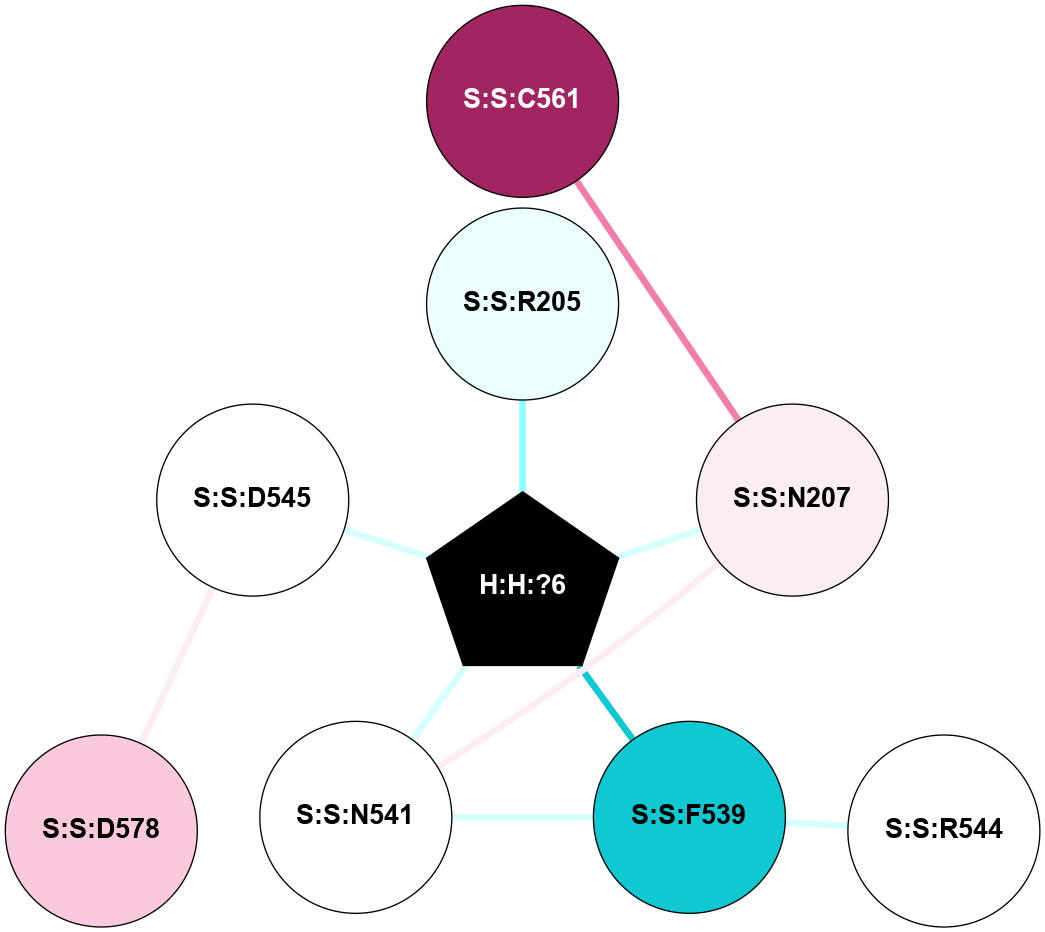
A 2D representation of the interactions of NAG in 9C1P
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | S:S:R205 | 4.36 | 25 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?6 | S:S:N207 | 14.79 | 25 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?6 | S:S:F539 | 5.46 | 25 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?6 | S:S:N541 | 33.27 | 25 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?6 | S:S:D545 | 7.31 | 25 | Yes | No | 0 | 5 | 0 | 1 | | S:S:N207 | S:S:N541 | 6.81 | 25 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 7.87 | 0 | No | No | 9 | 6 | 2 | 1 | | S:S:F539 | S:S:N541 | 13.29 | 25 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 25.66 | 25 | No | No | 1 | 5 | 1 | 2 | | S:S:D545 | S:S:D578 | 6.65 | 0 | No | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 13.04 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9C2F | C | Ion | Calcium Sensing | CaS | Homo Sapiens | Ca | Ca; Ca; Tryptophan; '54149; PO4 | - | 2.8 | 2024-10-02 | doi.org/10.1126/science.ado1868 |
|
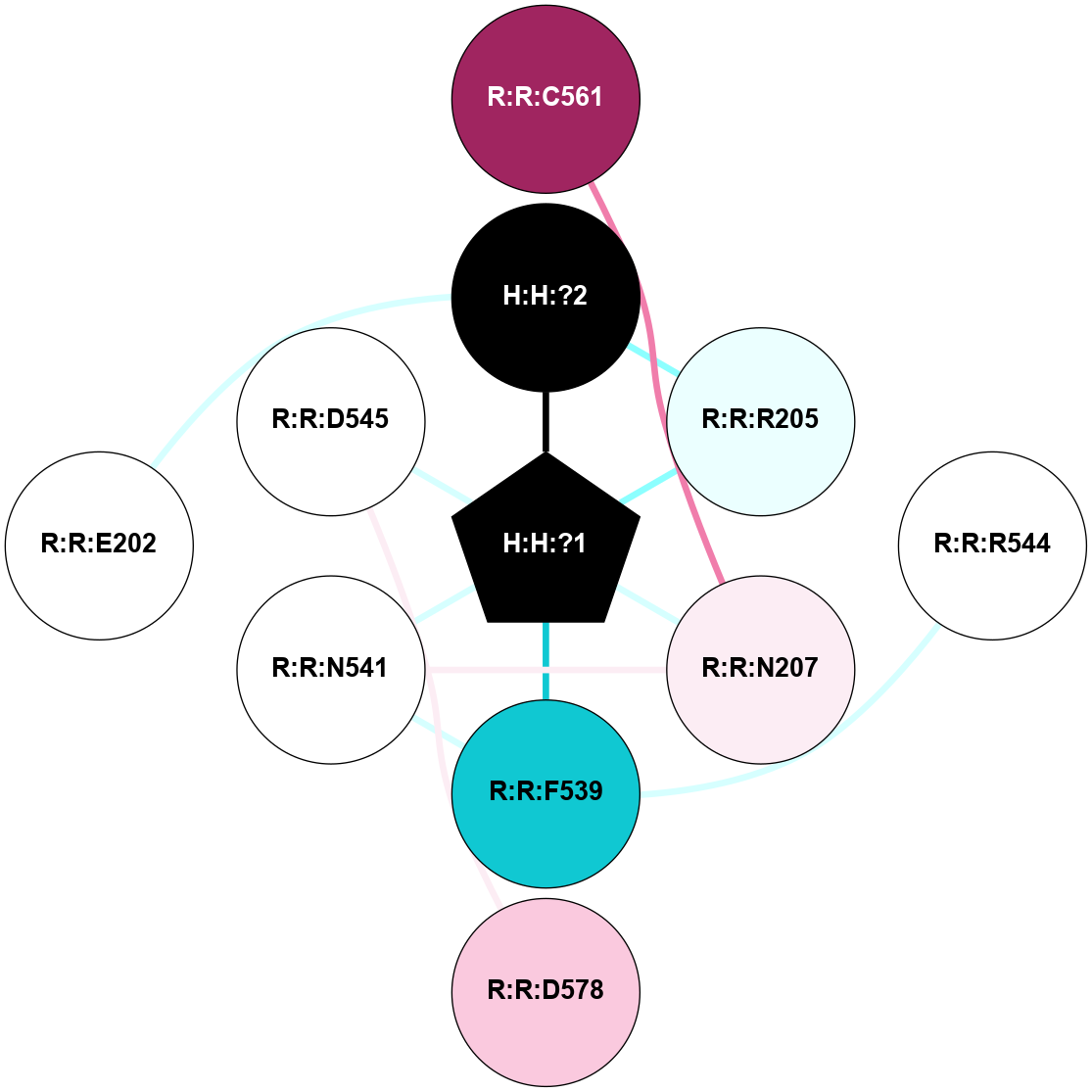
A 2D representation of the interactions of NAG in 9C2F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 33.43 | 15 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:R205 | 5.45 | 15 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?1 | R:R:N207 | 9.86 | 15 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?1 | R:R:F539 | 6.56 | 15 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?1 | R:R:N541 | 29.57 | 15 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:D545 | 4.87 | 15 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?2 | R:R:R205 | 9.81 | 15 | No | No | 0 | 4 | 1 | 1 | | R:R:N207 | R:R:N541 | 14.98 | 15 | No | No | 6 | 5 | 1 | 1 | | R:R:C561 | R:R:N207 | 7.87 | 0 | No | No | 9 | 6 | 2 | 1 | | R:R:F539 | R:R:N541 | 8.46 | 15 | No | No | 1 | 5 | 1 | 1 | | R:R:F539 | R:R:R544 | 26.72 | 15 | No | No | 1 | 5 | 1 | 2 | | R:R:D545 | R:R:D578 | 11.98 | 0 | No | No | 5 | 7 | 1 | 2 | | H:H:?2 | R:R:E202 | 1.19 | 15 | No | No | 0 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 14.96 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 13.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9C2F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | H:H:?2 | 33.43 | 15 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:R205 | 5.45 | 15 | Yes | No | 0 | 4 | 1 | 1 | | H:H:?1 | R:R:N207 | 9.86 | 15 | Yes | No | 0 | 6 | 1 | 2 | | H:H:?1 | R:R:F539 | 6.56 | 15 | Yes | No | 0 | 1 | 1 | 2 | | H:H:?1 | R:R:N541 | 29.57 | 15 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?1 | R:R:D545 | 4.87 | 15 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?2 | R:R:R205 | 9.81 | 15 | No | No | 0 | 4 | 0 | 1 | | R:R:N207 | R:R:N541 | 14.98 | 15 | No | No | 6 | 5 | 2 | 2 | | R:R:F539 | R:R:N541 | 8.46 | 15 | No | No | 1 | 5 | 2 | 2 | | H:H:?2 | R:R:E202 | 1.19 | 15 | No | No | 0 | 5 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 3.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 14.81 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 6.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9C2F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?3 | S:S:N468 | 32.03 | 31 | No | No | 0 | 6 | 0 | 1 | | H:H:?3 | S:S:Q476 | 15.53 | 31 | No | No | 0 | 3 | 0 | 1 | | S:S:N468 | S:S:Q476 | 9.24 | 31 | No | No | 6 | 3 | 1 | 1 | | S:S:N468 | S:S:T478 | 8.77 | 31 | No | No | 6 | 3 | 1 | 2 | | S:S:Q476 | S:S:T470 | 5.67 | 31 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 23.78 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
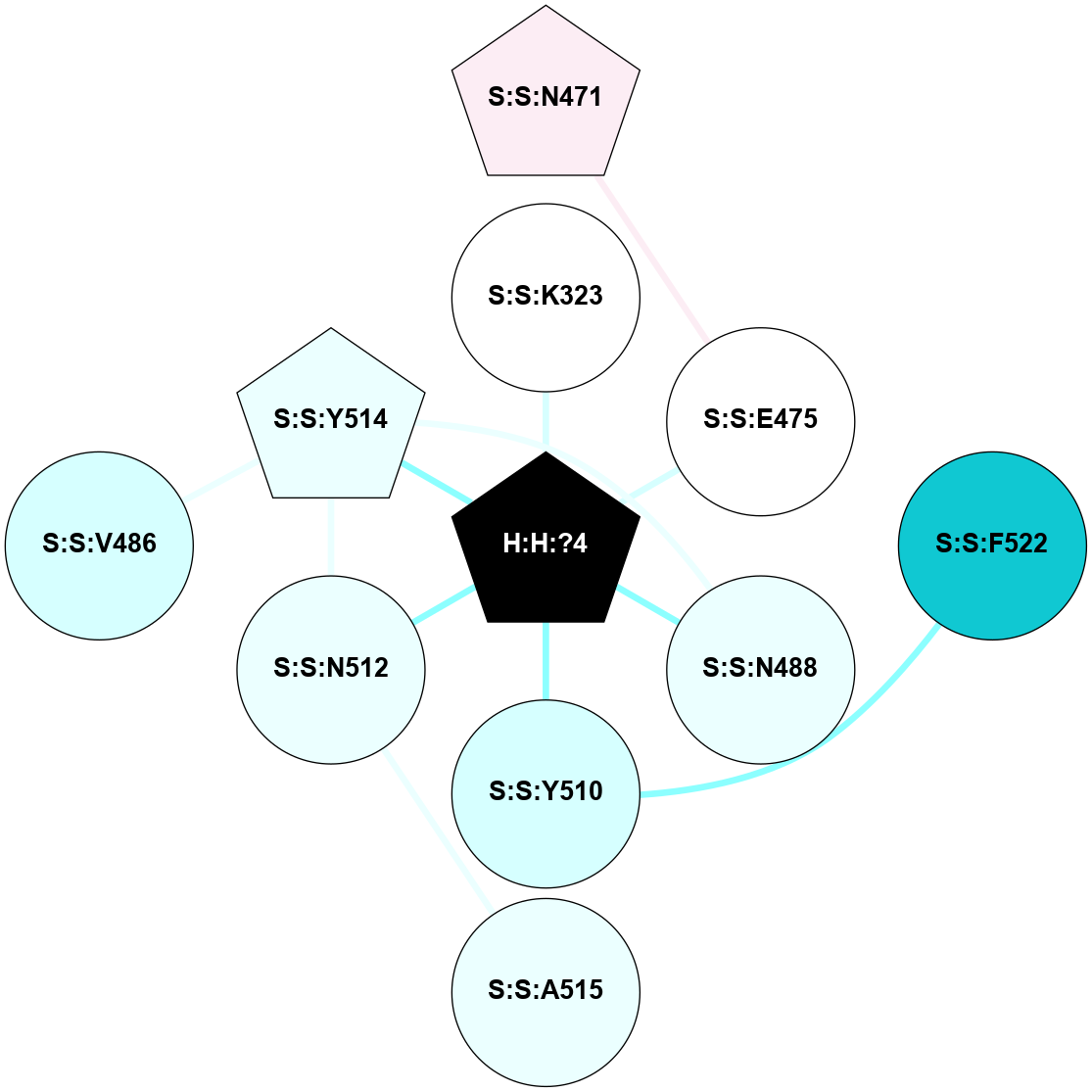
A 2D representation of the interactions of NAG in 9C2F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?4 | S:S:K323 | 5.06 | 25 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?4 | S:S:E475 | 3.57 | 25 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?4 | S:S:N488 | 33.27 | 25 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?4 | S:S:Y510 | 12.62 | 25 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?4 | S:S:N512 | 19.71 | 25 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?4 | S:S:Y514 | 15.78 | 25 | Yes | Yes | 0 | 4 | 0 | 1 | | S:S:E475 | S:S:N471 | 5.26 | 0 | No | Yes | 5 | 6 | 1 | 2 | | S:S:N488 | S:S:Y514 | 5.81 | 25 | No | Yes | 4 | 4 | 1 | 1 | | S:S:F522 | S:S:Y510 | 13.41 | 0 | No | No | 1 | 3 | 2 | 1 | | S:S:N512 | S:S:Y514 | 4.65 | 25 | No | Yes | 4 | 4 | 1 | 1 | | S:S:A515 | S:S:N512 | 4.69 | 0 | No | No | 4 | 4 | 2 | 1 | | S:S:V486 | S:S:Y514 | 2.52 | 0 | No | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 6.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 15.00 | | Average Nodes In Shell | 11.00 | | Average Hubs In Shell | 3.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 10.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
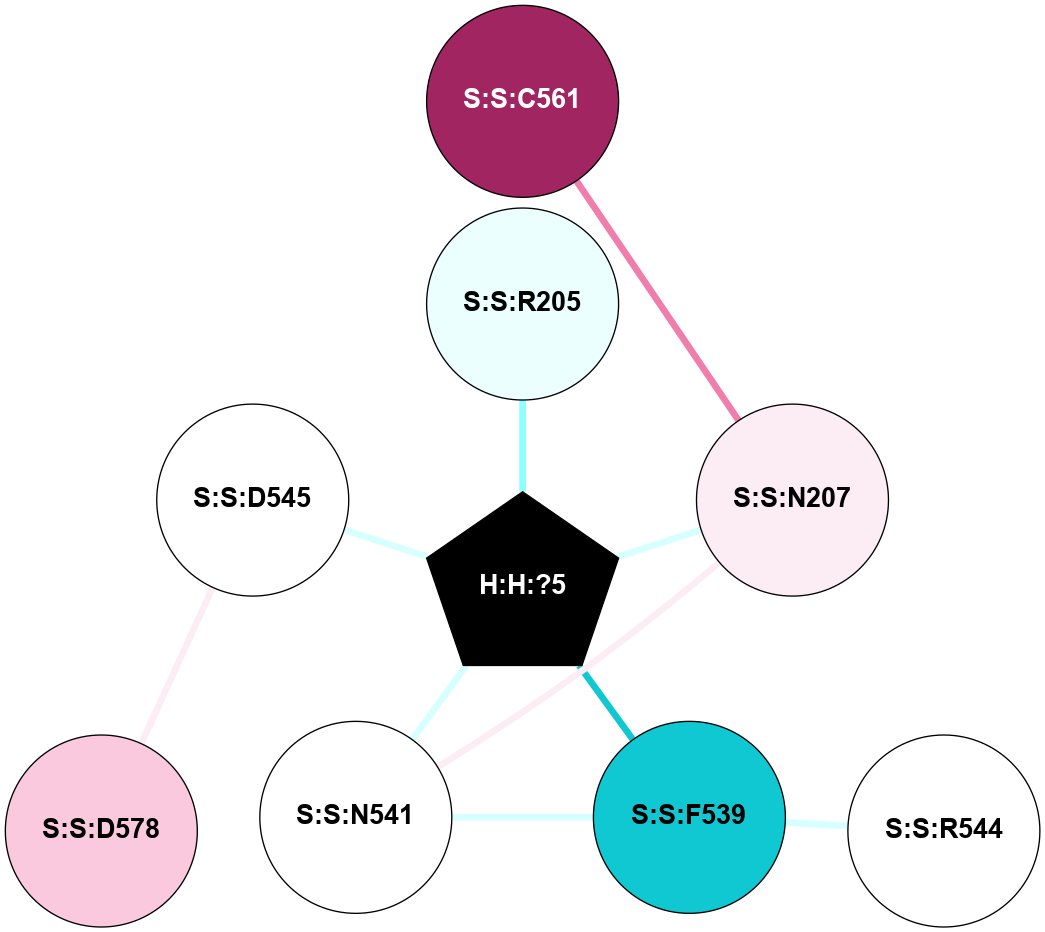
A 2D representation of the interactions of NAG in 9C2F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?5 | S:S:R205 | 3.27 | 26 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?5 | S:S:N207 | 12.32 | 26 | Yes | No | 0 | 6 | 0 | 1 | | H:H:?5 | S:S:F539 | 5.46 | 26 | Yes | No | 0 | 1 | 0 | 1 | | H:H:?5 | S:S:N541 | 27.11 | 26 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?5 | S:S:D545 | 7.31 | 26 | Yes | No | 0 | 5 | 0 | 1 | | S:S:N207 | S:S:N541 | 16.35 | 26 | No | No | 6 | 5 | 1 | 1 | | S:S:C561 | S:S:N207 | 6.3 | 0 | No | No | 9 | 6 | 2 | 1 | | S:S:F539 | S:S:N541 | 8.46 | 26 | No | No | 1 | 5 | 1 | 1 | | S:S:F539 | S:S:R544 | 25.66 | 26 | No | No | 1 | 5 | 1 | 2 | | S:S:D545 | S:S:D578 | 5.32 | 0 | No | No | 5 | 7 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 11.09 | | Average Nodes In Shell | 9.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 10.00 | | Average Links Mediated by Hubs In Shell | 5.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|

A 2D representation of the interactions of NAG in 9C2F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?6 | R:R:N468 | 30.8 | 32 | No | No | 0 | 6 | 0 | 1 | | H:H:?6 | R:R:Q476 | 3.58 | 32 | No | No | 0 | 3 | 0 | 1 | | R:R:N468 | R:R:Q476 | 3.96 | 32 | No | No | 6 | 3 | 1 | 1 | | R:R:N468 | R:R:T478 | 5.85 | 32 | No | No | 6 | 3 | 1 | 2 | | R:R:Q476 | R:R:T470 | 1.42 | 32 | No | No | 3 | 5 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.19 | | Average Nodes In Shell | 5.00 | | Average Hubs In Shell | 0.00 | | Average Links In Shell | 5.00 | | Average Links Mediated by Hubs In Shell | 0.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
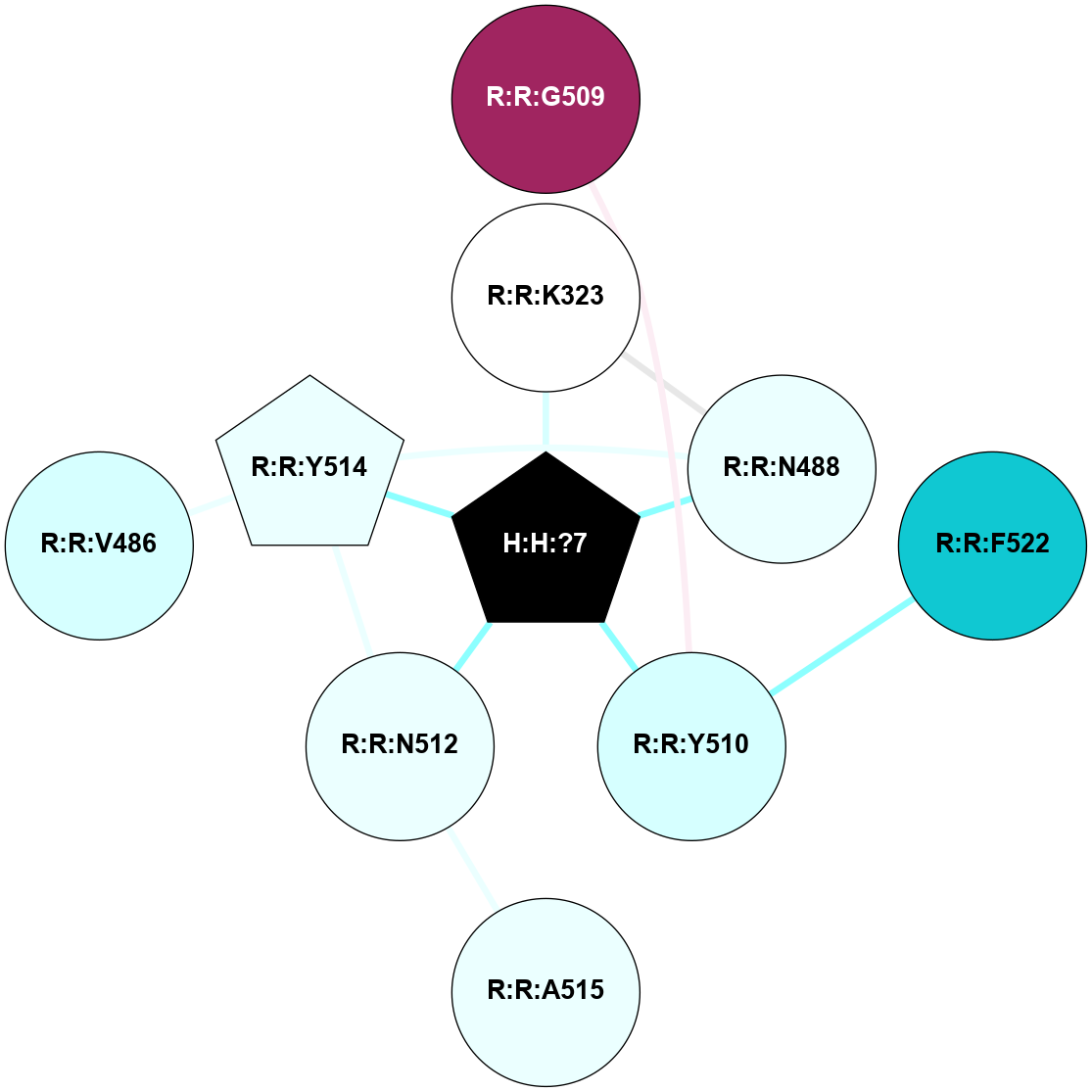
A 2D representation of the interactions of NAG in 9C2F
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?7 | R:R:K323 | 5.06 | 22 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?7 | R:R:N488 | 27.11 | 22 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?7 | R:R:Y510 | 8.42 | 22 | Yes | No | 0 | 3 | 0 | 1 | | H:H:?7 | R:R:N512 | 13.55 | 22 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?7 | R:R:Y514 | 14.73 | 22 | Yes | Yes | 0 | 4 | 0 | 1 | | R:R:K323 | R:R:N488 | 4.2 | 22 | No | No | 5 | 4 | 1 | 1 | | R:R:N488 | R:R:Y514 | 4.65 | 22 | No | Yes | 4 | 4 | 1 | 1 | | R:R:G509 | R:R:Y510 | 2.9 | 0 | No | No | 9 | 3 | 2 | 1 | | R:R:F522 | R:R:Y510 | 13.41 | 0 | No | No | 1 | 3 | 2 | 1 | | R:R:N512 | R:R:Y514 | 3.49 | 22 | No | Yes | 4 | 4 | 1 | 1 | | R:R:A515 | R:R:N512 | 6.25 | 0 | No | No | 4 | 4 | 2 | 1 | | R:R:V486 | R:R:Y514 | 2.52 | 0 | No | Yes | 3 | 4 | 2 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 5.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 13.77 | | Average Nodes In Shell | 10.00 | | Average Hubs In Shell | 2.00 | | Average Links In Shell | 12.00 | | Average Links Mediated by Hubs In Shell | 8.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9GE2 | A | Orphan | Orphan | GPR55 | Homo Sapiens | ML184 | - | chim(NtGi1-G13)/Beta1/Gamma2 | 2.51 | 2025-03-05 | doi.org/10.1038/s41467-025-57204-y |
|

A 2D representation of the interactions of NAG in 9GE2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:H170 | R:R:N16 | 5.1 | 0 | No | No | 5 | 5 | 1 | 2 | | R:R:M167 | R:R:Q87 | 4.08 | 0 | No | No | 3 | 3 | 1 | 2 | | H:H:?2 | R:R:Y166 | 21.04 | 0 | No | No | 0 | 5 | 1 | 2 | | H:H:?1 | R:R:N171 | 25.87 | 0 | Yes | No | 0 | 4 | 0 | 1 | | H:H:?1 | H:H:?2 | 39.01 | 0 | Yes | No | 0 | 0 | 0 | 1 | | H:H:?1 | R:R:H170 | 3.46 | 0 | Yes | No | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:M167 | 2.54 | 0 | Yes | No | 0 | 3 | 0 | 1 | |
| Statistics | Value |
|---|
| Average Number Of Links | 4.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 17.72 | | Average Nodes In Shell | 8.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 7.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
|
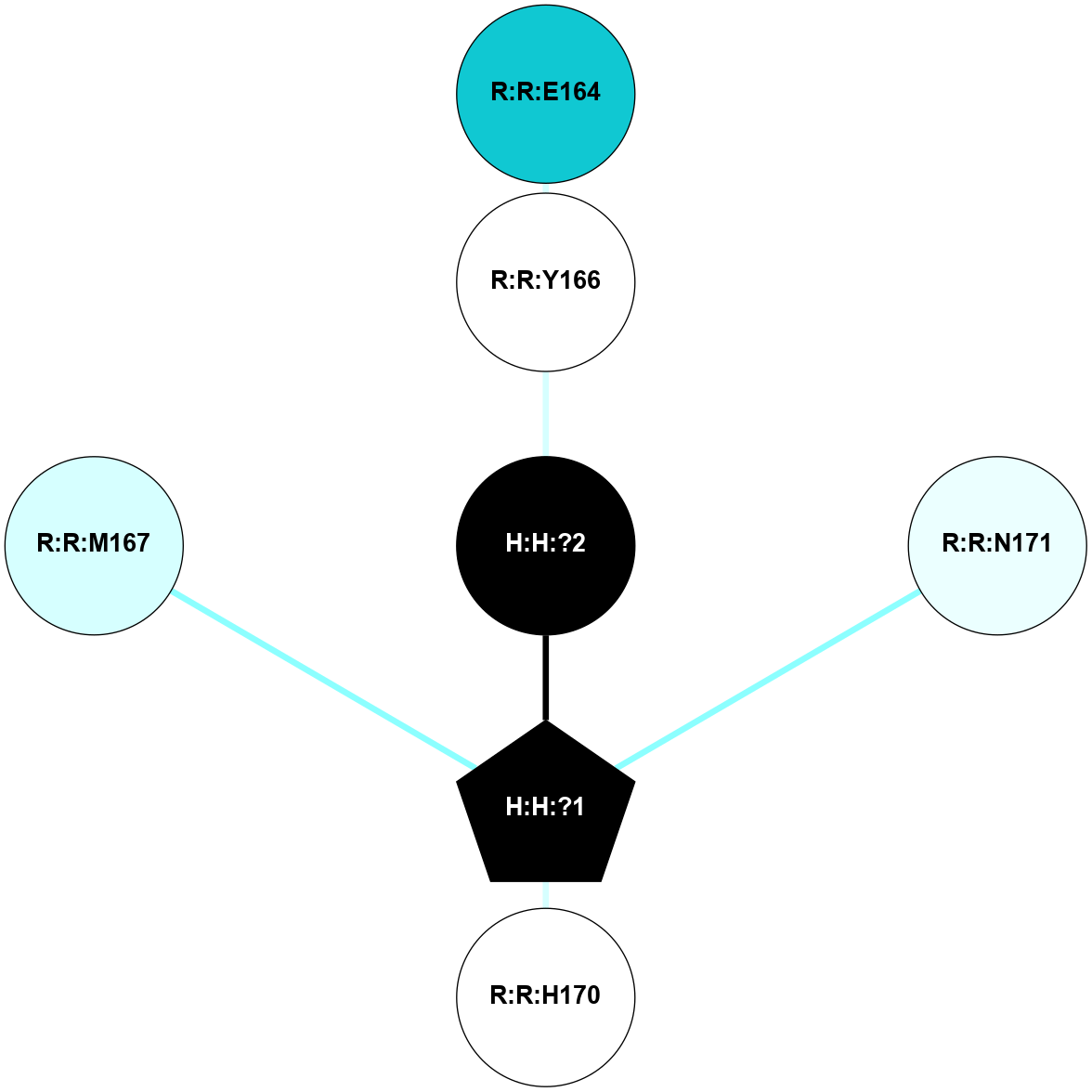
A 2D representation of the interactions of NAG in 9GE2
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | R:R:E164 | R:R:Y166 | 13.47 | 0 | No | No | 1 | 5 | 2 | 1 | | H:H:?2 | R:R:Y166 | 21.04 | 0 | No | No | 0 | 5 | 0 | 1 | | H:H:?1 | R:R:N171 | 25.87 | 0 | Yes | No | 0 | 4 | 1 | 2 | | H:H:?1 | H:H:?2 | 39.01 | 0 | Yes | No | 0 | 0 | 1 | 0 | | H:H:?1 | R:R:H170 | 3.46 | 0 | Yes | No | 0 | 5 | 1 | 2 | | H:H:?1 | R:R:M167 | 2.54 | 0 | Yes | No | 0 | 3 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 2.00 | | Average Number Of Links With An Hub | 1.00 | | Average Interaction Strength | 30.02 | | Average Nodes In Shell | 7.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 6.00 | | Average Links Mediated by Hubs In Shell | 4.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 9GE3 | A | Orphan | Orphan | GPR55 | Homo Sapiens | Lysophosphatidylinositol | - | chim(NtGi1-G13)/Beta1/Gamma2 | 2.87 | 2025-03-05 | doi.org/10.1038/s41467-025-57204-y |
|
A 2D representation of the interactions of NAG in 9GE3
colored according to ConSurf Conservation Grade (See documentation):
(Click to enlarge 🔍) | | Node1 | Node2 | LinkStrength | Comm | IsNode1Hub? | IsNode2Hub? | Node1Cons | Node2Cons | Node1Shell | Node2Shell | | H:H:?1 | R:R:N171 | 27.11 | 0 | No | No | 0 | 4 | 0 | 1 | | R:R:N171 | R:R:R253 | 1.21 | 0 | No | Yes | 4 | 4 | 1 | 2 | |
| Statistics | Value |
|---|
| Average Number Of Links | 1.00 | | Average Number Of Links With An Hub | 0.00 | | Average Interaction Strength | 27.11 | | Average Nodes In Shell | 3.00 | | Average Hubs In Shell | 1.00 | | Average Links In Shell | 2.00 | | Average Links Mediated by Hubs In Shell | 1.00 |
| 
Physico-chemical properties of the nodes interacting with this ligand (click to enlarge 🔍) | 
Location of the nodes interacting with this ligand (click to enlarge 🔍) |
|




















































































































































































































































































































































































































































































































































































































































































































































































































































