- Press and hold down the left mouse button and move the mouse to rotate.
- Press and hold down the central mouse button and move the mouse to translate.
- Use the mouse wheel to zoom in or out.
- Click on any atom on your molecule and the corresponding residue label will be placed beneath the viewer.
| Color | ConSurf Grade |
| No Conservation data available | |
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 | |
| 6 | |
| 7 | |
| 8 | |
| 9 |
Index: link id, click on each number to highlight the corresponding link in the 3D visualization.
Node1 Node2: the two nodes of the corresponding link.
Int. Strength: the interaction strength between the two nodes.
Hub1?, Hub2?: "Yes" if the corresponding node has more than 3 links, otherwise "No".
Community: the id of the community the link belong to, otherwise 0.
ConSurf1, ConSurf2: these columns report the ConSurf conservation grades of the two nodes involved in a link.
| Index | Node1 | Node2 | Int. Strength | Hub1? | Hub2? | Community | ConSurf1 | ConSurf2 |
|---|---|---|---|---|---|---|---|---|
| 1 | R:R:E34 | R:R:V30 | 5.7 | No | No | 0 | 4 | 2 |
| 2 | R:R:E34 | R:R:S31 | 10.06 | No | No | 0 | 4 | 4 |
| 3 | R:R:L32 | R:R:Y205 | 7.03 | No | No | 0 | 4 | 3 |
| 4 | R:R:Q221 | R:R:W33 | 16.43 | No | No | 0 | 4 | 3 |
| 5 | L:L:?1 | R:R:W33 | 13.49 | Yes | No | 0 | 0 | 3 |
| 6 | R:R:P90 | R:R:T35 | 8.74 | No | No | 0 | 4 | 5 |
| 7 | R:R:V36 | R:R:W214 | 9.81 | No | Yes | 0 | 2 | 1 |
| 8 | R:R:L217 | R:R:V36 | 4.47 | No | No | 0 | 4 | 2 |
| 9 | R:R:Q221 | R:R:Q37 | 7.68 | No | No | 0 | 4 | 4 |
| 10 | R:R:K38 | R:R:W87 | 8.12 | No | No | 0 | 4 | 3 |
| 11 | R:R:R43 | R:R:W39 | 5 | Yes | Yes | 1 | 4 | 5 |
| 12 | R:R:E68 | R:R:W39 | 9.81 | No | Yes | 1 | 4 | 5 |
| 13 | R:R:W39 | R:R:Y88 | 15.43 | Yes | Yes | 1 | 5 | 5 |
| 14 | R:R:W214 | R:R:W39 | 11.25 | Yes | Yes | 1 | 1 | 5 |
| 15 | R:R:R40 | R:R:R44 | 12.79 | No | No | 0 | 2 | 3 |
| 16 | R:R:L218 | R:R:R40 | 15.79 | No | No | 0 | 3 | 2 |
| 17 | R:R:E41 | R:R:Q45 | 11.47 | No | No | 0 | 4 | 5 |
| 18 | R:R:F66 | R:R:Y42 | 4.13 | Yes | Yes | 1 | 7 | 6 |
| 19 | R:R:C71 | R:R:Y42 | 8.06 | No | Yes | 1 | 9 | 6 |
| 20 | R:R:P86 | R:R:Y42 | 8.34 | No | Yes | 1 | 8 | 6 |
| 21 | R:R:W87 | R:R:Y42 | 4.82 | No | Yes | 0 | 3 | 6 |
| 22 | R:R:Y42 | R:R:Y88 | 6.95 | Yes | Yes | 1 | 6 | 5 |
| 23 | R:R:F66 | R:R:R43 | 6.41 | Yes | Yes | 1 | 7 | 4 |
| 24 | R:R:E68 | R:R:R43 | 10.47 | No | Yes | 1 | 4 | 4 |
| 25 | R:R:L218 | R:R:R43 | 7.29 | No | Yes | 0 | 3 | 4 |
| 26 | R:R:Q45 | R:R:R48 | 10.51 | No | No | 0 | 5 | 4 |
| 27 | R:R:C46 | R:R:F66 | 5.59 | No | Yes | 1 | 8 | 7 |
| 28 | R:R:C46 | R:R:C71 | 7.28 | No | No | 1 | 8 | 9 |
| 29 | R:R:L50 | R:R:S49 | 4.5 | No | No | 0 | 4 | 2 |
| 30 | R:R:L50 | R:R:R64 | 14.58 | No | Yes | 0 | 4 | 4 |
| 31 | R:R:E52 | R:R:T51 | 5.64 | No | No | 0 | 2 | 4 |
| 32 | R:R:D53 | R:R:R64 | 22.63 | No | Yes | 2 | 3 | 4 |
| 33 | R:R:D53 | R:R:D74 | 3.99 | No | Yes | 2 | 3 | 3 |
| 34 | R:R:F61 | R:R:P56 | 11.56 | No | Yes | 0 | 3 | 1 |
| 35 | R:R:D74 | R:R:P56 | 17.71 | Yes | Yes | 0 | 3 | 1 |
| 36 | R:R:E76 | R:R:P56 | 4.72 | No | Yes | 0 | 1 | 1 |
| 37 | R:R:F61 | R:R:P77 | 10.11 | No | No | 0 | 3 | 4 |
| 38 | R:R:C104 | R:R:C62 | 7.28 | No | No | 0 | 9 | 9 |
| 39 | R:R:P73 | R:R:R64 | 7.21 | No | Yes | 0 | 5 | 4 |
| 40 | R:R:D74 | R:R:R64 | 7.15 | Yes | Yes | 2 | 3 | 4 |
| 41 | R:R:D67 | R:R:T65 | 4.34 | No | No | 0 | 9 | 5 |
| 42 | R:R:E68 | R:R:F66 | 12.83 | No | Yes | 1 | 4 | 7 |
| 43 | R:R:C71 | R:R:F66 | 5.59 | No | Yes | 1 | 9 | 7 |
| 44 | R:R:D67 | R:R:W72 | 5.58 | No | Yes | 1 | 9 | 9 |
| 45 | R:R:D67 | R:R:R121 | 15.48 | No | Yes | 1 | 9 | 6 |
| 46 | R:R:Y69 | R:R:Y88 | 3.97 | Yes | Yes | 1 | 6 | 5 |
| 47 | R:R:R121 | R:R:Y69 | 9.26 | Yes | Yes | 1 | 6 | 6 |
| 48 | R:R:L123 | R:R:Y69 | 12.89 | No | Yes | 0 | 5 | 6 |
| 49 | R:R:H212 | R:R:Y69 | 6.53 | No | Yes | 1 | 3 | 6 |
| 50 | R:R:A70 | R:R:W72 | 3.89 | No | Yes | 0 | 5 | 9 |
| 51 | R:R:R102 | R:R:W72 | 31.99 | No | Yes | 0 | 8 | 9 |
| 52 | R:R:R121 | R:R:W72 | 6 | Yes | Yes | 1 | 6 | 9 |
| 53 | R:R:P73 | R:R:V81 | 5.3 | No | No | 0 | 5 | 6 |
| 54 | R:R:E76 | R:R:P77 | 7.86 | No | No | 0 | 1 | 4 |
| 55 | R:R:E76 | R:R:S79 | 4.31 | No | No | 0 | 1 | 5 |
| 56 | R:R:F103 | R:R:G78 | 6.02 | Yes | No | 0 | 4 | 7 |
| 57 | R:R:F103 | R:R:F80 | 36.44 | Yes | No | 0 | 4 | 4 |
| 58 | R:R:V81 | R:R:V83 | 4.81 | No | No | 0 | 6 | 5 |
| 59 | R:R:H99 | R:R:N82 | 10.2 | No | No | 0 | 4 | 4 |
| 60 | R:R:H99 | R:R:S84 | 4.18 | No | No | 0 | 4 | 4 |
| 61 | R:R:C126 | R:R:C85 | 7.28 | No | No | 0 | 9 | 9 |
| 62 | R:R:P86 | R:R:Y88 | 4.17 | No | Yes | 1 | 8 | 5 |
| 63 | R:R:L89 | R:R:Y88 | 5.86 | No | Yes | 0 | 5 | 5 |
| 64 | R:R:H212 | R:R:Y88 | 6.53 | No | Yes | 1 | 3 | 5 |
| 65 | R:R:W214 | R:R:Y88 | 3.86 | Yes | Yes | 1 | 1 | 5 |
| 66 | R:R:L89 | R:R:W91 | 10.25 | No | No | 0 | 5 | 4 |
| 67 | R:R:S94 | R:R:W91 | 4.94 | No | No | 0 | 4 | 4 |
| 68 | R:R:E128 | R:R:W91 | 20.72 | No | No | 0 | 3 | 4 |
| 69 | R:R:S94 | R:R:V95 | 4.85 | No | Yes | 0 | 4 | 5 |
| 70 | R:R:W120 | R:R:Y101 | 6.75 | Yes | No | 0 | 5 | 5 |
| 71 | R:R:E125 | R:R:Y101 | 8.98 | No | No | 0 | 5 | 5 |
| 72 | R:R:F103 | R:R:W110 | 22.05 | Yes | No | 0 | 4 | 9 |
| 73 | R:R:L109 | R:R:Q112 | 6.65 | No | Yes | 0 | 5 | 3 |
| 74 | R:R:L111 | R:R:W110 | 35.31 | No | No | 3 | 4 | 9 |
| 75 | R:R:W110 | R:R:W120 | 21.56 | No | Yes | 3 | 9 | 5 |
| 76 | R:R:L111 | R:R:W120 | 10.25 | No | Yes | 3 | 4 | 5 |
| 77 | R:R:Q112 | R:R:S116 | 7.22 | Yes | No | 0 | 3 | 5 |
| 78 | R:R:L118 | R:R:Q112 | 3.99 | No | Yes | 0 | 4 | 3 |
| 79 | R:R:P119 | R:R:R121 | 7.21 | No | Yes | 0 | 6 | 6 |
| 80 | R:R:D122 | R:R:W120 | 8.93 | No | Yes | 0 | 7 | 5 |
| 81 | R:R:D122 | R:R:S124 | 8.83 | No | No | 0 | 7 | 5 |
| 82 | R:R:Q140 | R:R:S135 | 11.55 | No | No | 0 | 4 | 1 |
| 83 | R:R:E139 | R:R:S136 | 4.31 | No | No | 0 | 2 | 1 |
| 84 | R:R:E138 | R:R:E139 | 19.03 | No | No | 0 | 2 | 2 |
| 85 | L:L:?1 | R:R:E138 | 7.48 | Yes | No | 0 | 0 | 2 |
| 86 | R:R:L141 | R:R:L144 | 5.54 | No | No | 0 | 5 | 6 |
| 87 | L:L:?1 | R:R:L141 | 9.37 | Yes | No | 0 | 0 | 5 |
| 88 | R:R:F143 | R:R:I147 | 3.77 | No | Yes | 0 | 4 | 6 |
| 89 | R:R:Y145 | R:R:Y148 | 4.96 | No | No | 0 | 5 | 7 |
| 90 | R:R:D198 | R:R:Y145 | 12.64 | No | No | 0 | 7 | 5 |
| 91 | L:L:?1 | R:R:Y145 | 14.55 | Yes | No | 0 | 0 | 5 |
| 92 | R:R:F385 | R:R:I147 | 7.54 | Yes | Yes | 0 | 6 | 6 |
| 93 | R:R:V194 | R:R:Y148 | 5.05 | No | No | 0 | 7 | 7 |
| 94 | R:R:L388 | R:R:Y148 | 9.38 | No | No | 0 | 7 | 7 |
| 95 | R:R:A191 | R:R:Y152 | 5.34 | No | No | 0 | 7 | 7 |
| 96 | R:R:V194 | R:R:Y152 | 10.09 | No | No | 0 | 7 | 7 |
| 97 | R:R:F156 | R:R:I188 | 5.02 | No | No | 0 | 7 | 7 |
| 98 | R:R:F184 | R:R:L159 | 7.31 | No | No | 0 | 8 | 9 |
| 99 | R:R:I188 | R:R:L159 | 4.28 | No | No | 0 | 7 | 9 |
| 100 | R:R:F404 | R:R:I161 | 3.77 | No | No | 0 | 8 | 4 |
| 101 | R:R:F169 | R:R:I165 | 8.79 | No | No | 0 | 7 | 8 |
| 102 | R:R:F413 | R:R:I165 | 7.54 | No | No | 0 | 5 | 8 |
| 103 | R:R:L166 | R:R:L172 | 4.15 | Yes | Yes | 0 | 8 | 9 |
| 104 | R:R:L166 | R:R:N177 | 6.87 | Yes | No | 0 | 8 | 9 |
| 105 | R:R:C403 | R:R:L166 | 4.76 | No | Yes | 0 | 9 | 8 |
| 106 | R:R:L167 | R:R:L181 | 4.15 | No | No | 0 | 3 | 6 |
| 107 | R:R:E412 | R:R:F169 | 8.16 | Yes | No | 0 | 8 | 7 |
| 108 | R:R:H173 | R:R:R170 | 11.28 | No | No | 0 | 7 | 8 |
| 109 | R:R:E412 | R:R:H171 | 13.54 | Yes | No | 0 | 8 | 7 |
| 110 | R:R:L172 | R:R:V409 | 5.96 | Yes | No | 0 | 9 | 9 |
| 111 | R:R:E412 | R:R:L172 | 3.98 | Yes | Yes | 0 | 8 | 9 |
| 112 | R:R:H173 | R:R:Y178 | 13.07 | No | No | 0 | 7 | 6 |
| 113 | R:R:C174 | R:R:R176 | 4.18 | No | Yes | 4 | 8 | 9 |
| 114 | R:R:C174 | R:R:N177 | 4.72 | No | No | 4 | 8 | 9 |
| 115 | R:R:T175 | R:R:Y250 | 6.24 | No | Yes | 0 | 7 | 8 |
| 116 | R:R:F266 | R:R:T175 | 5.19 | Yes | No | 0 | 6 | 7 |
| 117 | R:R:N177 | R:R:R176 | 9.64 | No | Yes | 4 | 9 | 9 |
| 118 | R:R:H180 | R:R:R176 | 4.51 | No | Yes | 0 | 9 | 9 |
| 119 | R:R:R176 | R:R:Y250 | 4.12 | Yes | Yes | 0 | 9 | 8 |
| 120 | R:R:F266 | R:R:Y178 | 4.13 | Yes | No | 0 | 6 | 6 |
| 121 | R:R:E247 | R:R:I179 | 5.47 | Yes | No | 0 | 9 | 8 |
| 122 | R:R:I179 | R:R:Y250 | 4.84 | No | Yes | 0 | 8 | 8 |
| 123 | R:R:F266 | R:R:I179 | 5.02 | Yes | No | 0 | 6 | 8 |
| 124 | R:R:E247 | R:R:H180 | 11.08 | Yes | No | 0 | 9 | 9 |
| 125 | R:R:H180 | R:R:Y402 | 8.71 | No | No | 0 | 9 | 8 |
| 126 | R:R:N182 | R:R:W243 | 18.08 | No | Yes | 5 | 9 | 9 |
| 127 | R:R:N182 | R:R:W274 | 4.52 | No | Yes | 5 | 9 | 9 |
| 128 | R:R:F187 | R:R:L183 | 6.09 | Yes | No | 0 | 8 | 9 |
| 129 | R:R:E247 | R:R:L183 | 6.63 | Yes | No | 0 | 9 | 9 |
| 130 | R:R:F184 | R:R:V398 | 3.93 | No | No | 0 | 8 | 9 |
| 131 | R:R:N240 | R:R:S186 | 5.96 | No | No | 0 | 9 | 9 |
| 132 | R:R:S186 | R:R:W243 | 7.41 | No | Yes | 0 | 9 | 9 |
| 133 | R:R:F187 | R:R:N240 | 16.92 | Yes | No | 0 | 8 | 9 |
| 134 | R:R:F187 | R:R:Q394 | 9.37 | Yes | Yes | 0 | 8 | 9 |
| 135 | R:R:F187 | R:R:G395 | 4.52 | Yes | No | 0 | 8 | 9 |
| 136 | R:R:F187 | R:R:V398 | 3.93 | Yes | No | 0 | 8 | 9 |
| 137 | R:R:R190 | R:R:V237 | 10.46 | Yes | No | 0 | 8 | 6 |
| 138 | R:R:N240 | R:R:R190 | 14.46 | No | Yes | 0 | 9 | 8 |
| 139 | R:R:R190 | R:R:Y241 | 11.32 | Yes | No | 0 | 8 | 8 |
| 140 | R:R:R190 | R:R:T391 | 3.88 | Yes | No | 0 | 8 | 6 |
| 141 | R:R:I196 | R:R:L192 | 7.14 | No | No | 0 | 5 | 3 |
| 142 | R:R:M233 | R:R:S193 | 7.67 | No | No | 0 | 6 | 6 |
| 143 | R:R:C236 | R:R:S193 | 5.16 | No | No | 0 | 5 | 6 |
| 144 | R:R:K197 | R:R:M233 | 5.76 | No | No | 0 | 6 | 6 |
| 145 | L:L:?1 | R:R:K197 | 16.71 | Yes | No | 0 | 0 | 6 |
| 146 | R:R:L201 | R:R:Y205 | 7.03 | No | No | 0 | 7 | 3 |
| 147 | L:L:?1 | R:R:L201 | 10.15 | Yes | No | 0 | 0 | 7 |
| 148 | R:R:L217 | R:R:M204 | 4.24 | No | No | 0 | 4 | 3 |
| 149 | R:R:M204 | R:R:Y220 | 9.58 | No | Yes | 0 | 3 | 3 |
| 150 | R:R:Q210 | R:R:Q211 | 3.84 | No | No | 0 | 4 | 4 |
| 151 | R:R:Q211 | R:R:Q213 | 7.68 | No | No | 0 | 4 | 4 |
| 152 | R:R:H212 | R:R:W214 | 16.93 | No | Yes | 1 | 3 | 1 |
| 153 | R:R:Q221 | R:R:Y220 | 5.64 | No | Yes | 0 | 4 | 3 |
| 154 | R:R:C226 | R:R:Y220 | 9.41 | No | Yes | 0 | 9 | 3 |
| 155 | L:L:?1 | R:R:Y220 | 5.29 | Yes | Yes | 0 | 0 | 3 |
| 156 | R:R:C226 | R:R:C296 | 7.28 | No | No | 0 | 9 | 9 |
| 157 | R:R:D293 | R:R:R227 | 13.1 | No | No | 0 | 7 | 8 |
| 158 | R:R:C296 | R:R:R227 | 4.18 | No | No | 0 | 9 | 8 |
| 159 | R:R:R227 | R:R:W297 | 29.99 | No | Yes | 0 | 8 | 8 |
| 160 | L:L:?1 | R:R:V229 | 4.2 | Yes | No | 0 | 0 | 7 |
| 161 | R:R:F230 | R:R:Q234 | 12.88 | Yes | No | 0 | 5 | 7 |
| 162 | R:R:F230 | R:R:W284 | 6.01 | Yes | Yes | 0 | 5 | 8 |
| 163 | R:R:F230 | R:R:T298 | 7.78 | Yes | No | 0 | 5 | 5 |
| 164 | L:L:?1 | R:R:F230 | 6.87 | Yes | Yes | 0 | 0 | 5 |
| 165 | R:R:L231 | R:R:V281 | 4.47 | No | No | 0 | 5 | 6 |
| 166 | R:R:W274 | R:R:Y235 | 6.75 | Yes | No | 0 | 9 | 9 |
| 167 | R:R:L278 | R:R:Y235 | 7.03 | No | No | 0 | 3 | 9 |
| 168 | R:R:A238 | R:R:F280 | 4.16 | No | Yes | 0 | 6 | 5 |
| 169 | R:R:I313 | R:R:Y241 | 12.09 | No | No | 0 | 7 | 8 |
| 170 | R:R:E364 | R:R:Y241 | 12.35 | Yes | No | 0 | 8 | 8 |
| 171 | R:R:V276 | R:R:Y242 | 5.05 | No | No | 0 | 3 | 6 |
| 172 | R:R:P277 | R:R:Y242 | 9.74 | No | No | 0 | 9 | 6 |
| 173 | R:R:F280 | R:R:Y242 | 10.32 | Yes | No | 0 | 5 | 6 |
| 174 | R:R:V246 | R:R:W243 | 7.36 | No | Yes | 0 | 8 | 9 |
| 175 | R:R:G273 | R:R:W243 | 8.44 | No | Yes | 0 | 9 | 9 |
| 176 | R:R:W243 | R:R:W274 | 24.37 | Yes | Yes | 5 | 9 | 9 |
| 177 | R:R:L244 | R:R:Q394 | 5.32 | No | Yes | 0 | 9 | 9 |
| 178 | R:R:E247 | R:R:L359 | 13.25 | Yes | No | 0 | 9 | 9 |
| 179 | R:R:V249 | R:R:Y269 | 7.57 | No | Yes | 0 | 6 | 7 |
| 180 | R:R:L254 | R:R:Y250 | 11.72 | No | Yes | 0 | 8 | 8 |
| 181 | R:R:Y250 | R:R:Y269 | 5.96 | Yes | Yes | 0 | 8 | 7 |
| 182 | R:R:L251 | R:R:V327 | 4.47 | No | No | 0 | 9 | 9 |
| 183 | R:R:A256 | R:R:Y252 | 8.01 | No | No | 0 | 6 | 8 |
| 184 | R:R:R326 | R:R:Y252 | 5.14 | No | No | 0 | 8 | 8 |
| 185 | R:R:F257 | R:R:T253 | 6.49 | No | No | 0 | 5 | 7 |
| 186 | R:R:T253 | R:R:Y269 | 6.24 | No | Yes | 0 | 7 | 7 |
| 187 | R:R:L255 | R:R:V327 | 4.47 | No | No | 0 | 8 | 9 |
| 188 | R:R:I330 | R:R:L255 | 4.28 | No | No | 0 | 9 | 8 |
| 189 | R:R:Q263 | R:R:R267 | 14.02 | No | No | 0 | 5 | 4 |
| 190 | R:R:I265 | R:R:W264 | 4.7 | No | No | 0 | 5 | 5 |
| 191 | R:R:I265 | R:R:Y269 | 8.46 | No | Yes | 0 | 5 | 7 |
| 192 | R:R:F266 | R:R:V270 | 6.55 | Yes | No | 0 | 6 | 5 |
| 193 | R:R:F280 | R:R:I309 | 3.77 | Yes | No | 0 | 5 | 7 |
| 194 | R:R:F280 | R:R:P312 | 21.67 | Yes | No | 0 | 5 | 8 |
| 195 | R:R:G285 | R:R:W284 | 4.22 | No | Yes | 0 | 5 | 8 |
| 196 | R:R:I308 | R:R:W284 | 8.22 | No | Yes | 0 | 8 | 8 |
| 197 | R:R:I309 | R:R:W284 | 14.09 | No | Yes | 0 | 7 | 8 |
| 198 | R:R:V287 | R:R:Y305 | 6.31 | No | Yes | 0 | 4 | 4 |
| 199 | R:R:E292 | R:R:K288 | 8.1 | Yes | No | 0 | 6 | 8 |
| 200 | R:R:K288 | R:R:W297 | 9.28 | No | Yes | 0 | 8 | 8 |
| 201 | R:R:W297 | R:R:Y289 | 6.75 | Yes | No | 0 | 8 | 3 |
| 202 | R:R:L290 | R:R:Y291 | 4.69 | No | No | 0 | 3 | 3 |
| 203 | R:R:Y291 | R:R:Y305 | 8.94 | No | Yes | 0 | 3 | 4 |
| 204 | R:R:E292 | R:R:S301 | 5.75 | Yes | No | 0 | 6 | 3 |
| 205 | R:R:E292 | R:R:N302 | 3.94 | Yes | No | 6 | 6 | 5 |
| 206 | R:R:E292 | R:R:Y305 | 6.73 | Yes | Yes | 6 | 6 | 4 |
| 207 | R:R:E294 | R:R:G295 | 4.91 | No | No | 0 | 4 | 4 |
| 208 | R:R:E294 | R:R:R299 | 15.12 | No | No | 0 | 4 | 4 |
| 209 | R:R:N300 | R:R:W306 | 6.78 | No | Yes | 0 | 6 | 6 |
| 210 | R:R:N302 | R:R:Y305 | 9.3 | No | Yes | 6 | 5 | 4 |
| 211 | R:R:M303 | R:R:W306 | 5.82 | No | Yes | 0 | 5 | 6 |
| 212 | R:R:L307 | R:R:N304 | 4.12 | No | No | 0 | 7 | 3 |
| 213 | R:R:I309 | R:R:W306 | 4.7 | No | Yes | 0 | 7 | 6 |
| 214 | R:R:R310 | R:R:W306 | 4 | No | Yes | 0 | 6 | 6 |
| 215 | R:R:L307 | R:R:L311 | 9.69 | No | No | 0 | 7 | 4 |
| 216 | R:R:L314 | R:R:R310 | 7.29 | No | No | 0 | 4 | 6 |
| 217 | R:R:F315 | R:R:V319 | 7.87 | No | No | 0 | 4 | 6 |
| 218 | R:R:E364 | R:R:I317 | 6.83 | Yes | No | 0 | 8 | 7 |
| 219 | R:R:L360 | R:R:N320 | 10.98 | Yes | No | 0 | 9 | 9 |
| 220 | R:R:F321 | R:R:V325 | 6.55 | No | No | 0 | 6 | 5 |
| 221 | R:R:F321 | R:R:T362 | 5.19 | No | No | 0 | 6 | 7 |
| 222 | R:R:F321 | R:R:V365 | 9.18 | No | No | 0 | 6 | 6 |
| 223 | R:R:F324 | R:R:I328 | 6.28 | Yes | No | 0 | 9 | 7 |
| 224 | R:R:F324 | R:R:L356 | 7.31 | Yes | No | 0 | 9 | 9 |
| 225 | R:R:F324 | R:R:I357 | 3.77 | Yes | No | 0 | 9 | 8 |
| 226 | R:R:F324 | R:R:T362 | 14.27 | Yes | No | 0 | 9 | 7 |
| 227 | R:R:I328 | R:R:T353 | 4.56 | No | No | 0 | 7 | 9 |
| 228 | R:R:S352 | R:R:V331 | 6.46 | No | No | 0 | 9 | 8 |
| 229 | R:R:L349 | R:R:V332 | 4.47 | No | No | 0 | 9 | 6 |
| 230 | R:R:I345 | R:R:L335 | 5.71 | No | No | 0 | 5 | 8 |
| 231 | R:R:L335 | R:R:R348 | 4.86 | No | No | 0 | 8 | 8 |
| 232 | R:R:D344 | R:R:I345 | 7 | No | No | 0 | 7 | 5 |
| 233 | R:R:K351 | R:R:R348 | 6.19 | No | No | 0 | 8 | 8 |
| 234 | R:R:K351 | R:R:T355 | 4.5 | No | No | 0 | 8 | 8 |
| 235 | R:R:L401 | R:R:T355 | 5.9 | No | No | 0 | 8 | 8 |
| 236 | R:R:L359 | R:R:L360 | 4.15 | No | Yes | 0 | 9 | 9 |
| 237 | R:R:L359 | R:R:Y402 | 19.93 | No | No | 0 | 9 | 8 |
| 238 | R:R:L360 | R:R:T362 | 4.42 | Yes | No | 0 | 9 | 7 |
| 239 | R:R:E364 | R:R:H363 | 11.08 | Yes | No | 0 | 8 | 8 |
| 240 | R:R:F390 | R:R:H363 | 5.66 | No | No | 0 | 6 | 8 |
| 241 | R:R:H363 | R:R:Q394 | 8.65 | No | Yes | 0 | 8 | 9 |
| 242 | R:R:E364 | R:R:F367 | 8.16 | Yes | No | 0 | 8 | 7 |
| 243 | R:R:E387 | R:R:F367 | 16.32 | No | No | 0 | 8 | 7 |
| 244 | R:R:F369 | R:R:M371 | 4.98 | No | No | 0 | 4 | 7 |
| 245 | R:R:H374 | R:R:M371 | 6.57 | Yes | No | 0 | 3 | 7 |
| 246 | R:R:H374 | R:R:L379 | 9 | Yes | No | 0 | 3 | 3 |
| 247 | R:R:H374 | R:R:K383 | 13.1 | Yes | No | 0 | 3 | 7 |
| 248 | R:R:F381 | R:R:T378 | 5.19 | No | No | 0 | 4 | 2 |
| 249 | R:R:F381 | R:R:R380 | 6.41 | No | No | 0 | 4 | 5 |
| 250 | R:R:F385 | R:R:I382 | 6.28 | Yes | No | 0 | 6 | 4 |
| 251 | R:R:E387 | R:R:L384 | 5.3 | No | No | 0 | 8 | 5 |
| 252 | R:R:E387 | R:R:L388 | 6.63 | No | No | 0 | 8 | 7 |
| 253 | R:R:F393 | R:R:S392 | 5.28 | No | No | 0 | 7 | 9 |
| 254 | R:R:I400 | R:R:M397 | 4.37 | Yes | No | 0 | 6 | 5 |
| 255 | R:R:V398 | R:R:Y402 | 3.79 | No | No | 0 | 9 | 8 |
| 256 | R:R:I400 | R:R:L401 | 5.71 | Yes | No | 0 | 6 | 8 |
| 257 | R:R:F404 | R:R:I400 | 3.77 | No | Yes | 0 | 8 | 6 |
| 258 | R:R:C403 | R:R:F404 | 4.19 | No | No | 0 | 9 | 8 |
| 259 | R:R:N406 | R:R:N407 | 4.09 | No | No | 0 | 9 | 6 |
| 260 | R:R:N406 | R:R:V409 | 4.43 | No | No | 0 | 9 | 9 |
| 261 | R:R:E412 | R:R:K415 | 10.8 | Yes | No | 0 | 8 | 7 |
| 262 | R:R:K415 | R:R:R419 | 17.33 | No | No | 0 | 7 | 7 |
| 263 | R:R:W417 | R:R:W420 | 5.62 | No | No | 0 | 5 | 4 |
| 264 | R:R:R421 | R:R:W417 | 8 | No | No | 0 | 4 | 5 |
| 265 | R:R:E418 | R:R:R421 | 9.3 | No | No | 0 | 4 | 4 |
| 266 | L:L:?1 | R:R:P137 | 3.71 | Yes | No | 0 | 0 | 3 |
| 267 | R:R:G151 | R:R:S392 | 3.71 | No | No | 0 | 8 | 9 |
| 268 | R:R:G216 | R:R:S219 | 3.71 | No | No | 0 | 2 | 5 |
| 269 | R:R:F385 | R:R:L144 | 3.65 | Yes | No | 0 | 6 | 6 |
| 270 | R:R:I323 | R:R:Y252 | 3.63 | No | No | 0 | 8 | 8 |
| 271 | R:R:A162 | R:R:C403 | 3.61 | No | No | 0 | 9 | 9 |
| 272 | R:R:P137 | R:R:S31 | 3.56 | No | No | 0 | 3 | 4 |
| 273 | R:R:G248 | R:R:I323 | 3.53 | No | No | 0 | 8 | 8 |
| 274 | R:R:P283 | R:R:V282 | 3.53 | No | No | 0 | 5 | 3 |
| 275 | R:R:E262 | R:R:F260 | 3.5 | No | No | 0 | 6 | 7 |
| 276 | R:R:C46 | R:R:S49 | 3.44 | No | No | 0 | 8 | 2 |
| 277 | R:R:C85 | R:R:V95 | 3.42 | No | Yes | 0 | 9 | 5 |
| 278 | R:R:L231 | R:R:W297 | 3.42 | No | Yes | 0 | 5 | 8 |
| 279 | R:R:L268 | R:R:W264 | 3.42 | No | No | 0 | 2 | 5 |
| 280 | R:R:I357 | R:R:P358 | 3.39 | No | No | 0 | 8 | 9 |
| 281 | R:R:G295 | R:R:Q37 | 3.29 | No | No | 0 | 4 | 4 |
| 282 | R:R:G361 | R:R:Q394 | 3.29 | No | Yes | 0 | 9 | 9 |
| 283 | L:L:?1 | R:R:K202 | 3.18 | Yes | No | 0 | 0 | 4 |
| 284 | R:R:C236 | R:R:L189 | 3.17 | No | No | 0 | 5 | 7 |
| 285 | R:R:A208 | R:R:L217 | 3.15 | No | No | 0 | 4 | 4 |
| 286 | R:R:A316 | R:R:L245 | 3.15 | No | No | 0 | 7 | 8 |
| 287 | R:R:I147 | R:R:S389 | 3.1 | Yes | No | 0 | 6 | 5 |
| 288 | R:R:I328 | R:R:S352 | 3.1 | No | No | 0 | 7 | 9 |
| 289 | R:R:I272 | R:R:V246 | 3.07 | No | No | 0 | 6 | 8 |
| 290 | R:R:K202 | R:R:S206 | 3.06 | No | No | 0 | 4 | 4 |
| 291 | R:R:A209 | R:R:Q210 | 3.03 | No | No | 0 | 1 | 4 |
| 292 | R:R:L181 | R:R:S163 | 3 | No | No | 0 | 6 | 6 |
| 293 | R:R:L123 | R:R:V100 | 2.98 | No | No | 0 | 5 | 7 |
| 294 | R:R:L245 | R:R:V249 | 2.98 | No | No | 0 | 8 | 6 |
| 295 | R:R:L245 | R:R:V319 | 2.98 | No | No | 0 | 8 | 6 |
| 296 | R:R:L278 | R:R:V282 | 2.98 | No | No | 0 | 3 | 3 |
| 297 | R:R:L356 | R:R:V327 | 2.98 | No | No | 0 | 9 | 9 |
| 298 | R:R:I146 | R:R:I147 | 2.94 | No | Yes | 0 | 4 | 6 |
| 299 | R:R:I196 | R:R:M233 | 2.92 | No | No | 0 | 5 | 6 |
| 300 | R:R:D372 | R:R:V370 | 2.92 | No | No | 0 | 5 | 5 |
| 301 | R:R:D198 | R:R:T149 | 2.89 | No | No | 0 | 7 | 7 |
| 302 | R:R:E125 | R:R:V100 | 2.85 | No | No | 0 | 5 | 7 |
| 303 | R:R:I400 | R:R:L396 | 2.85 | Yes | No | 0 | 6 | 7 |
| 304 | R:R:E107 | R:R:T105 | 2.82 | No | No | 0 | 5 | 4 |
| 305 | R:R:E262 | R:R:T175 | 2.82 | No | No | 0 | 6 | 7 |
| 306 | R:R:L154 | R:R:L396 | 2.77 | No | No | 0 | 5 | 7 |
| 307 | R:R:L166 | R:R:L181 | 2.77 | Yes | No | 0 | 8 | 6 |
| 308 | R:R:A375 | R:R:R380 | 2.77 | No | No | 0 | 4 | 5 |
| 309 | R:R:N302 | R:R:N304 | 2.72 | No | No | 0 | 5 | 3 |
| 310 | R:R:K113 | R:R:Q112 | 2.71 | No | Yes | 0 | 4 | 3 |
| 311 | R:R:D74 | R:R:N63 | 2.69 | Yes | No | 0 | 3 | 4 |
| 312 | R:R:E408 | R:R:L172 | 2.65 | No | Yes | 0 | 9 | 9 |
| 313 | R:R:F393 | R:R:S389 | 2.64 | No | No | 0 | 7 | 5 |
| 314 | R:R:F103 | R:R:T105 | 2.59 | Yes | No | 0 | 4 | 4 |
| 315 | R:R:F195 | R:R:T149 | 2.59 | No | No | 0 | 7 | 7 |
| 316 | R:R:A239 | R:R:W274 | 2.59 | No | Yes | 0 | 8 | 9 |
| 317 | R:R:F385 | R:R:T386 | 2.59 | Yes | No | 0 | 6 | 4 |
| 318 | R:R:S155 | R:R:Y152 | 2.54 | No | No | 0 | 9 | 7 |
| 319 | R:R:S223 | R:R:Y220 | 2.54 | No | Yes | 0 | 4 | 3 |
| 320 | R:R:F367 | R:R:I366 | 2.51 | No | No | 0 | 7 | 6 |
| 321 | R:R:S261 | R:R:W264 | 2.47 | No | No | 0 | 7 | 5 |
| 322 | R:R:D222 | R:R:R40 | 2.38 | No | No | 0 | 6 | 2 |
| 323 | R:R:L224 | R:R:Y289 | 2.34 | No | No | 0 | 3 | 3 |
| 324 | R:R:E41 | R:R:W87 | 2.18 | No | No | 0 | 4 | 3 |
| 325 | R:R:G275 | R:R:P277 | 2.03 | No | No | 0 | 6 | 9 |
| 326 | R:R:C104 | R:R:G108 | 1.96 | No | No | 0 | 9 | 9 |
| 327 | R:R:P54 | R:R:P55 | 1.95 | No | No | 0 | 2 | 1 |
| 328 | R:R:P55 | R:R:P56 | 1.95 | No | Yes | 0 | 1 | 1 |
| 329 | R:R:A106 | R:R:P77 | 1.87 | No | No | 0 | 1 | 4 |
| 330 | R:R:G98 | R:R:V95 | 1.84 | No | Yes | 0 | 4 | 5 |
| 331 | R:R:G151 | R:R:V150 | 1.84 | No | No | 0 | 8 | 5 |
| 332 | R:R:A158 | R:R:A399 | 1.79 | No | No | 0 | 7 | 8 |
| 333 | R:R:A162 | R:R:A399 | 1.79 | No | No | 0 | 9 | 8 |
| 334 | L:L:?1 | R:R:A200 | 1.78 | Yes | No | 0 | 0 | 4 |
| 335 | R:R:P90 | R:R:V30 | 1.77 | No | No | 0 | 4 | 2 |
| 336 | R:R:P96 | R:R:V95 | 1.77 | No | Yes | 0 | 3 | 5 |
| 337 | R:R:G318 | R:R:I317 | 1.76 | No | No | 0 | 4 | 7 |
| 338 | R:R:P137 | R:R:T29 | 1.75 | No | No | 0 | 3 | 2 |
| 339 | R:R:A28 | R:R:S135 | 1.71 | No | No | 0 | 2 | 1 |
| 340 | R:R:A158 | R:R:S157 | 1.71 | No | No | 0 | 7 | 3 |
| 341 | R:R:G168 | R:R:L167 | 1.71 | No | No | 0 | 1 | 3 |
| 342 | R:R:G248 | R:R:L360 | 1.71 | No | Yes | 0 | 8 | 9 |
| 343 | R:R:D74 | R:R:G75 | 1.68 | Yes | No | 0 | 3 | 7 |
| 344 | R:R:D215 | R:R:G216 | 1.68 | No | No | 0 | 3 | 2 |
| 345 | R:R:L118 | R:R:P119 | 1.64 | No | No | 0 | 4 | 6 |
| 346 | R:R:S223 | R:R:S225 | 1.63 | No | No | 0 | 4 | 3 |
| 347 | R:R:A368 | R:R:I317 | 1.62 | No | No | 0 | 6 | 7 |
| 348 | R:R:A92 | R:R:L89 | 1.58 | No | No | 0 | 5 | 5 |
| 349 | R:R:A185 | R:R:N182 | 1.56 | No | No | 0 | 3 | 9 |
| 350 | R:R:I161 | R:R:V160 | 1.54 | No | No | 0 | 4 | 3 |
| 351 | R:R:I188 | R:R:V160 | 1.54 | No | No | 0 | 7 | 3 |
| 352 | R:R:A199 | R:R:D198 | 1.54 | No | No | 0 | 3 | 7 |
| 353 | R:R:K334 | R:R:S333 | 1.53 | No | No | 0 | 9 | 7 |
| 354 | R:R:K336 | R:R:V332 | 1.52 | No | No | 0 | 6 | 6 |
| 355 | R:R:L401 | R:R:V405 | 1.49 | No | No | 0 | 8 | 6 |
| 356 | R:R:L349 | R:R:T353 | 1.47 | No | No | 0 | 9 | 9 |
| 357 | R:R:L354 | R:R:T353 | 1.47 | No | No | 0 | 8 | 9 |
| 358 | R:R:I330 | R:R:K334 | 1.45 | No | No | 0 | 9 | 9 |
| 359 | R:R:Q263 | R:R:S261 | 1.44 | No | No | 0 | 5 | 7 |
| 360 | R:R:I146 | R:R:L142 | 1.43 | No | No | 0 | 4 | 3 |
| 361 | R:R:I382 | R:R:L379 | 1.43 | No | No | 0 | 4 | 3 |
| 362 | R:R:Q47 | R:R:T51 | 1.42 | No | No | 0 | 3 | 4 |
| 363 | R:R:C62 | R:R:R102 | 1.39 | No | No | 0 | 9 | 8 |
| 364 | R:R:A153 | R:R:F195 | 1.39 | No | No | 0 | 6 | 7 |
| 365 | R:R:D114 | R:R:K113 | 1.38 | No | No | 0 | 4 | 4 |
| 366 | R:R:L224 | R:R:L228 | 1.38 | No | No | 0 | 3 | 4 |
| 367 | R:R:D114 | R:R:N115 | 1.35 | No | No | 0 | 4 | 4 |
| 368 | R:R:E373 | R:R:M371 | 1.35 | No | No | 0 | 4 | 7 |
| 369 | R:R:L141 | R:R:Q140 | 1.33 | No | No | 0 | 5 | 4 |
| 370 | R:R:N82 | R:R:Y101 | 1.16 | No | No | 0 | 4 | 5 |
| 371 | R:R:M204 | R:R:W203 | 1.16 | No | No | 0 | 3 | 3 |
| 372 | R:R:H374 | R:R:R380 | 1.13 | Yes | No | 0 | 3 | 5 |
| Color | ConSurf Grade |
| No Conservation data available | |
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 | |
| 6 | |
| 7 | |
| 8 | |
| 9 |
Index: hub id, click on each number to highlight the corresponding hub in the 3D visualization.
Hub: the hub being considered.
Avg Int. Strength: the average interaction strength of all the links of the corresponding hub.
Num Of Links: the number of links of the corresponding hub.
Community: the id of the community the link belong to, otherwise 0.
ConSurf: this column reports the ConSurf conservation grades of each hub.
| Index | Hub | Avg Int. Strength | Num Of Links | Community | ConSurf |
|---|---|---|---|---|---|
| 1 | R:R:W39 | 10.3725 | 4 | 1 | 5 |
| 2 | R:R:Y42 | 6.46 | 5 | 1 | 6 |
| 3 | R:R:R43 | 7.2925 | 4 | 1 | 4 |
| 4 | R:R:P56 | 8.985 | 4 | 0 | 1 |
| 5 | R:R:R64 | 12.8925 | 4 | 2 | 4 |
| 6 | R:R:F66 | 6.91 | 5 | 1 | 7 |
| 7 | R:R:Y69 | 8.1625 | 4 | 1 | 6 |
| 8 | R:R:W72 | 11.865 | 4 | 1 | 9 |
| 9 | R:R:D74 | 6.644 | 5 | 2 | 3 |
| 10 | R:R:Y88 | 6.68143 | 7 | 1 | 5 |
| 11 | R:R:V95 | 2.97 | 4 | 0 | 5 |
| 12 | R:R:F103 | 16.775 | 4 | 0 | 4 |
| 13 | R:R:Q112 | 5.1425 | 4 | 0 | 3 |
| 14 | R:R:W120 | 11.8725 | 4 | 3 | 5 |
| 15 | R:R:R121 | 9.4875 | 4 | 1 | 6 |
| 16 | R:R:I147 | 4.3375 | 4 | 0 | 6 |
| 17 | R:R:L166 | 4.6375 | 4 | 0 | 8 |
| 18 | R:R:L172 | 4.185 | 4 | 0 | 9 |
| 19 | R:R:R176 | 5.6125 | 4 | 4 | 9 |
| 20 | R:R:F187 | 8.166 | 5 | 0 | 8 |
| 21 | R:R:R190 | 10.03 | 4 | 0 | 8 |
| 22 | R:R:W214 | 10.4625 | 4 | 1 | 1 |
| 23 | R:R:Y220 | 6.492 | 5 | 0 | 3 |
| 24 | R:R:F230 | 8.385 | 4 | 0 | 5 |
| 25 | R:R:W243 | 13.132 | 5 | 5 | 9 |
| 26 | R:R:E247 | 9.1075 | 4 | 0 | 9 |
| 27 | R:R:Y250 | 6.576 | 5 | 0 | 8 |
| 28 | R:R:F266 | 5.2225 | 4 | 0 | 6 |
| 29 | R:R:Y269 | 7.0575 | 4 | 0 | 7 |
| 30 | R:R:W274 | 9.5575 | 4 | 5 | 9 |
| 31 | R:R:F280 | 9.98 | 4 | 0 | 5 |
| 32 | R:R:W284 | 8.135 | 4 | 0 | 8 |
| 33 | R:R:E292 | 6.13 | 4 | 6 | 6 |
| 34 | R:R:W297 | 12.36 | 4 | 0 | 8 |
| 35 | R:R:Y305 | 7.82 | 4 | 6 | 4 |
| 36 | R:R:W306 | 5.325 | 4 | 0 | 6 |
| 37 | R:R:F324 | 7.9075 | 4 | 0 | 9 |
| 38 | R:R:L360 | 5.315 | 4 | 0 | 9 |
| 39 | R:R:E364 | 9.605 | 4 | 0 | 8 |
| 40 | R:R:H374 | 7.45 | 4 | 0 | 3 |
| 41 | R:R:F385 | 5.015 | 4 | 0 | 6 |
| 42 | R:R:Q394 | 6.6575 | 4 | 0 | 9 |
| 43 | R:R:I400 | 4.175 | 4 | 0 | 6 |
| 44 | R:R:E412 | 9.12 | 4 | 0 | 8 |
| 45 | L:L:?1 | 8.065 | 12 | 0 | 0 |
| Color | ConSurf Grade |
| No Conservation data available | |
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 | |
| 6 | |
| 7 | |
| 8 | |
| 9 |
Index: link id, click on each number to highlight the corresponding link in the 3D visualization.
Node1 Node2: the two nodes of the corresponding link.
Recurrence: the relative Recurrence in the pool of shortest paths.
Int. Strength: the interaction strength between the two nodes.
Hub1?, Hub2?: "Yes" if the corresponding node has more than 3 links, otherwise "No".
Community: the id of the community the link belong to, otherwise 0.
ConSurf1, ConSurf2: these columns report the ConSurf conservation grades of the two nodes involved in a link.
| Index | Node1 | Node2 | Recurrence | Int. Strength | Hub1? | Hub2? | Community | ConSurf1 | ConSurf2 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | L:L:?1 | R:R:Y220 | 99.4663 | 5.29 | Yes | Yes | 0 | 0 | 3 |
| 2 | R:R:M204 | R:R:Y220 | 95.5139 | 9.58 | No | Yes | 0 | 3 | 3 |
| 3 | R:R:L217 | R:R:M204 | 95.116 | 4.24 | No | No | 0 | 4 | 3 |
| 4 | R:R:L217 | R:R:V36 | 94.7053 | 4.47 | No | No | 0 | 4 | 2 |
| 5 | R:R:V36 | R:R:W214 | 94.495 | 9.81 | No | Yes | 0 | 2 | 1 |
| 6 | R:R:W214 | R:R:Y88 | 49.7049 | 3.86 | Yes | Yes | 1 | 1 | 5 |
| 7 | R:R:Y42 | R:R:Y88 | 29.4138 | 6.95 | Yes | Yes | 1 | 6 | 5 |
| 8 | R:R:W214 | R:R:W39 | 26.1454 | 11.25 | Yes | Yes | 1 | 1 | 5 |
| 9 | R:R:R43 | R:R:W39 | 14.2783 | 5 | Yes | Yes | 1 | 4 | 5 |
| 10 | R:R:E68 | R:R:W39 | 11.8962 | 9.81 | No | Yes | 1 | 4 | 5 |
| 11 | R:R:F66 | R:R:R43 | 11.5452 | 6.41 | Yes | Yes | 1 | 7 | 4 |
| 12 | R:R:E68 | R:R:F66 | 11.356 | 12.83 | No | Yes | 1 | 4 | 7 |
| 13 | R:R:F66 | R:R:Y42 | 13.2433 | 4.13 | Yes | Yes | 1 | 7 | 6 |
| 14 | R:R:C71 | R:R:Y42 | 13.1819 | 8.06 | No | Yes | 1 | 9 | 6 |
| 15 | R:R:C46 | R:R:F66 | 34.4174 | 5.59 | No | Yes | 1 | 8 | 7 |
| 16 | R:R:C46 | R:R:C71 | 12.6255 | 7.28 | No | No | 1 | 8 | 9 |
| 17 | R:R:C46 | R:R:S49 | 44.8435 | 3.44 | No | No | 0 | 8 | 2 |
| 18 | R:R:L50 | R:R:S49 | 42.6312 | 4.5 | No | No | 0 | 4 | 2 |
| 19 | R:R:L50 | R:R:R64 | 40.4156 | 14.58 | No | Yes | 0 | 4 | 4 |
| 20 | R:R:D74 | R:R:R64 | 29.2844 | 7.15 | Yes | Yes | 2 | 3 | 4 |
| 21 | R:R:D74 | R:R:P56 | 22.5908 | 17.71 | Yes | Yes | 0 | 3 | 1 |
| 22 | R:R:R121 | R:R:Y69 | 19.8156 | 9.26 | Yes | Yes | 1 | 6 | 6 |
| 23 | R:R:Y69 | R:R:Y88 | 21.8032 | 3.97 | Yes | Yes | 1 | 6 | 5 |
| 24 | R:R:H212 | R:R:Y69 | 17.9785 | 6.53 | No | Yes | 1 | 3 | 6 |
| 25 | R:R:H212 | R:R:W214 | 18.4507 | 16.93 | No | Yes | 1 | 3 | 1 |
| 26 | R:R:L123 | R:R:Y69 | 19.827 | 12.89 | No | Yes | 0 | 5 | 6 |
| 27 | R:R:L123 | R:R:V100 | 18.7062 | 2.98 | No | No | 0 | 5 | 7 |
| 28 | R:R:E125 | R:R:V100 | 17.5613 | 2.85 | No | No | 0 | 5 | 7 |
| 29 | R:R:E125 | R:R:Y101 | 16.413 | 8.98 | No | No | 0 | 5 | 5 |
| 30 | R:R:W120 | R:R:Y101 | 11.7878 | 6.75 | Yes | No | 0 | 5 | 5 |
| 31 | L:L:?1 | R:R:L141 | 20.8701 | 9.37 | Yes | No | 0 | 0 | 5 |
| 32 | R:R:L141 | R:R:L144 | 17.9267 | 5.54 | No | No | 0 | 5 | 6 |
| 33 | R:R:F385 | R:R:L144 | 17.1828 | 3.65 | Yes | No | 0 | 6 | 6 |
| 34 | L:L:?1 | R:R:Y145 | 100 | 14.55 | Yes | No | 0 | 0 | 5 |
| 35 | R:R:Y145 | R:R:Y148 | 99.0976 | 4.96 | No | No | 0 | 5 | 7 |
| 36 | R:R:L388 | R:R:Y148 | 98.2567 | 9.38 | No | No | 0 | 7 | 7 |
| 37 | R:R:E387 | R:R:L388 | 98.0788 | 6.63 | No | No | 0 | 8 | 7 |
| 38 | R:R:E387 | R:R:F367 | 97.7133 | 16.32 | No | No | 0 | 8 | 7 |
| 39 | R:R:E364 | R:R:F367 | 97.3348 | 8.16 | Yes | No | 0 | 8 | 7 |
| 40 | R:R:E364 | R:R:H363 | 86.8553 | 11.08 | Yes | No | 0 | 8 | 8 |
| 41 | R:R:H363 | R:R:Q394 | 86.4058 | 8.65 | No | Yes | 0 | 8 | 9 |
| 42 | R:R:F187 | R:R:Q394 | 85.733 | 9.37 | Yes | Yes | 0 | 8 | 9 |
| 43 | R:R:F187 | R:R:V398 | 42.6263 | 3.93 | Yes | No | 0 | 8 | 9 |
| 44 | R:R:F184 | R:R:V398 | 11.1539 | 3.93 | No | No | 0 | 8 | 9 |
| 45 | R:R:F184 | R:R:L159 | 10.486 | 7.31 | No | No | 0 | 8 | 9 |
| 46 | R:R:F187 | R:R:L183 | 47.6332 | 6.09 | Yes | No | 0 | 8 | 9 |
| 47 | R:R:E247 | R:R:L183 | 47.2273 | 6.63 | Yes | No | 0 | 9 | 9 |
| 48 | R:R:E247 | R:R:H180 | 16.489 | 11.08 | Yes | No | 0 | 9 | 9 |
| 49 | R:R:H180 | R:R:R176 | 31.8881 | 4.51 | No | Yes | 0 | 9 | 9 |
| 50 | R:R:N177 | R:R:R176 | 31.1426 | 9.64 | No | Yes | 4 | 9 | 9 |
| 51 | R:R:L166 | R:R:N177 | 30.004 | 6.87 | Yes | No | 0 | 8 | 9 |
| 52 | R:R:L166 | R:R:L172 | 15.737 | 4.15 | Yes | Yes | 0 | 8 | 9 |
| 53 | R:R:V398 | R:R:Y402 | 32.26 | 3.79 | No | No | 0 | 9 | 8 |
| 54 | R:R:H180 | R:R:Y402 | 16.6928 | 8.71 | No | No | 0 | 9 | 8 |
| 55 | R:R:E247 | R:R:I179 | 17.1375 | 5.47 | Yes | No | 0 | 9 | 8 |
| 56 | R:R:I179 | R:R:Y250 | 12.0385 | 4.84 | No | Yes | 0 | 8 | 8 |
| 57 | R:R:N240 | R:R:S186 | 10.342 | 5.96 | No | No | 0 | 9 | 9 |
| 58 | R:R:C226 | R:R:Y220 | 16.2998 | 9.41 | No | Yes | 0 | 9 | 3 |
| 59 | R:R:C226 | R:R:C296 | 15.5559 | 7.28 | No | No | 0 | 9 | 9 |
| 60 | R:R:C296 | R:R:R227 | 14.8088 | 4.18 | No | No | 0 | 9 | 8 |
| 61 | R:R:R227 | R:R:W297 | 13.3048 | 29.99 | No | Yes | 0 | 8 | 8 |
| 62 | L:L:?1 | R:R:F230 | 14.9317 | 6.87 | Yes | Yes | 0 | 0 | 5 |
| 63 | R:R:F230 | R:R:W284 | 12.6514 | 6.01 | Yes | Yes | 0 | 5 | 8 |
| 64 | R:R:I309 | R:R:W284 | 10.342 | 14.09 | No | Yes | 0 | 7 | 8 |
| 65 | R:R:E247 | R:R:L359 | 20.0178 | 13.25 | Yes | No | 0 | 9 | 9 |
| 66 | R:R:L359 | R:R:Y402 | 18.9504 | 19.93 | No | No | 0 | 9 | 8 |
| 67 | R:R:L359 | R:R:L360 | 37.5645 | 4.15 | No | Yes | 0 | 9 | 9 |
| 68 | R:R:L360 | R:R:T362 | 28.7459 | 4.42 | Yes | No | 0 | 9 | 7 |
| 69 | R:R:F324 | R:R:T362 | 23.6355 | 14.27 | Yes | No | 0 | 9 | 7 |
| 70 | R:R:F324 | R:R:I328 | 10.6331 | 6.28 | Yes | No | 0 | 9 | 7 |
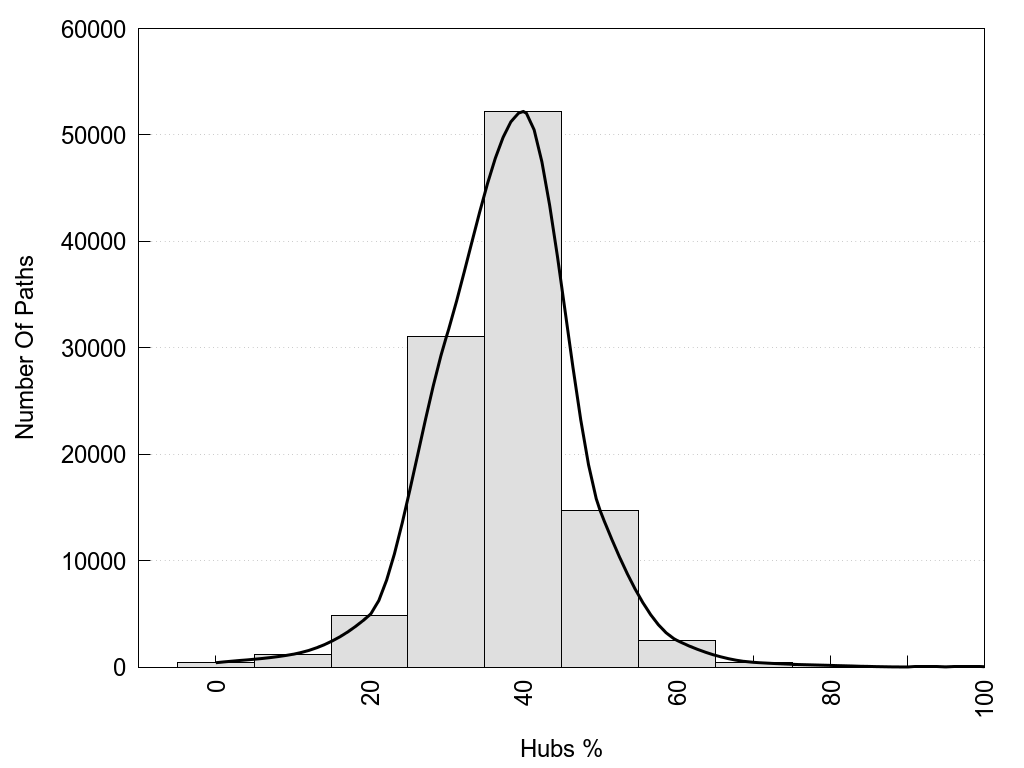
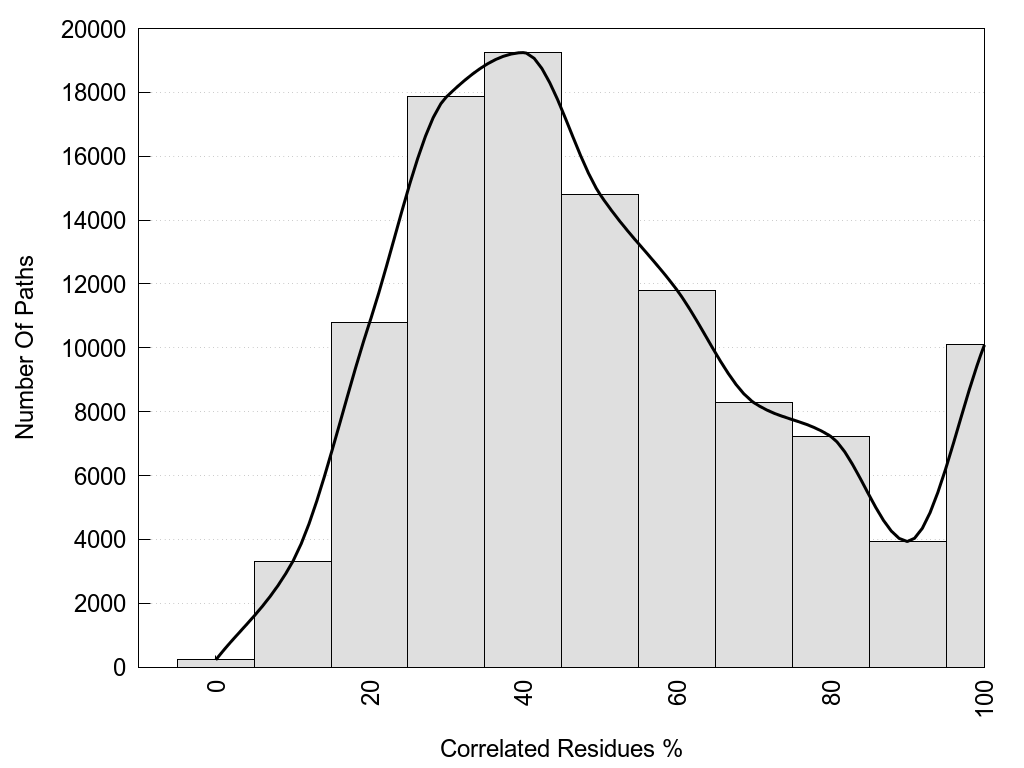
2D representation of the global metapath, ligand(s) interactions and
histograms of path distribution according to several parameters
(click on the image to enlarge it 🔍):

A 2D representation of the global communication in the network.
ConSurf Conservation Grade (See documentation):
n/a 1 2 3 4 5 6 7 8 9
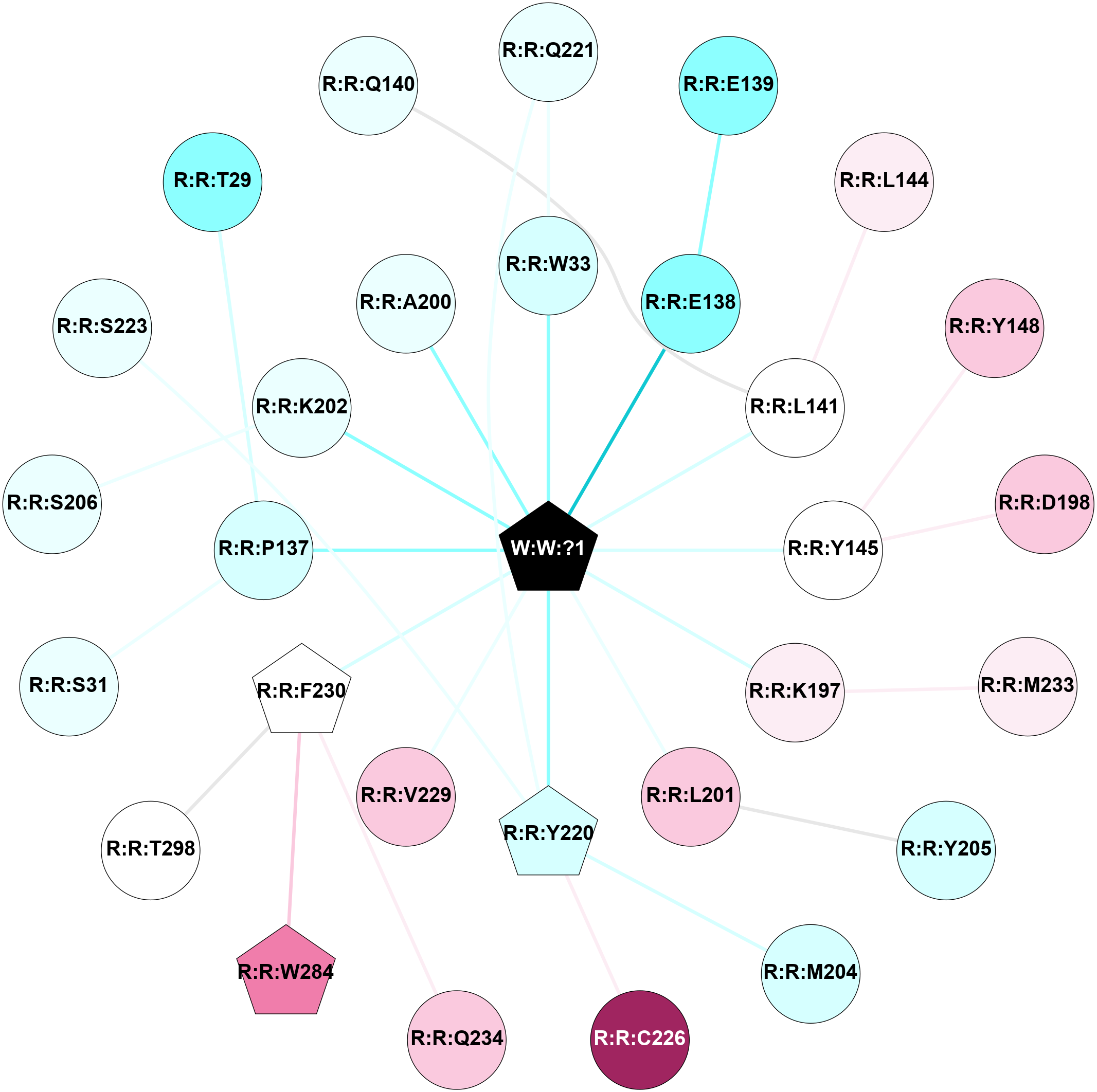
2D representation of the interactions of this orthosteric/allosteric ligand. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Links and nodes colored according to ConSurf Conservation Grade (See documentation): n/a 1 2 3 4 5 6 7 8 9 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Location and physicochemical properties of the interaction partners of this ligand | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Interactions of this ligand | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Similarities between the interactions of this ligand and those of other networks | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|
Details about the values in these tables can be found in the corresponding documentation page .
| |||||||||||||||||||||||||||||||||||
| PDBsum | Open PDBsum Page |
| Chain | R |
| Protein | Receptor |
| UniProt | P43220 |
| Sequence | >6XOX_nogp_Chain_R ATVSLWETV QKWREYRRQ CQRSLTEDP PPFCNRTFD EYACWPDGE PGSFVNVSC PWYLPWASS VPQGHVYRF CTAEGLWLQ KDNSSLPWR DLSECEESS PEEQLLFLY IIYTVGYAL SFSALVIAS AILLGFRHL HCTRNYIHL NLFASFILR ALSVFIKDA ALKWMYSTA AQQHQWDGL LSYQDSLSC RLVFLLMQY CVAANYYWL LVEGVYLYT LLAFSVFSE QWIFRLYVS IGWGVPLLF VVPWGIVKY LYEDEGCWT RNSNMNYWL IIRLPILFA IGVNFLIFV RVICIVVSK LKANLDIKC RLAKSTLTL IPLLGTHEV IFAFVMDEH ARGTLRFIK LFTELSFTS FQGLMVAIL YCFVNNEVQ LEFRKSWER WR Click on each residue to open a popup with some information about it. ConSurf Conservation Grade (See documentation): n/a 1 2 3 4 5 6 7 8 9 |
| This receptor, from the same or other species and bound to the same or other ligands, is also present in the following networks: | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Show | PDB | Class | SubFamily | Type | SubType | Species | Orthosteric Ligand | Other Ligand(s) | Protein Partners | Resolution | Date | DOI |
| 6ORV | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | TT-OAD2 | Gs/β1/γ2 | 3 | 2020-01-08 | doi.org/10.1038/s41586-019-1902-z | |
| 6ORV (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | TT-OAD2 | 3 | 2020-01-08 | doi.org/10.1038/s41586-019-1902-z | ||
| 7E14 | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Orforglipron | PubChem 156022738 | Gs/β1/γ2 | 2.9 | 2021-07-07 | doi.org/10.1038/s41467-021-24058-z | |
| 7E14 (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Orforglipron | PubChem 156022738 | 2.9 | 2021-07-07 | doi.org/10.1038/s41467-021-24058-z | ||
| 7DUQ | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | GLP-1 | PubChem 156022738 | Gs/β1/γ2 | 2.5 | 2021-07-14 | doi.org/10.1038/s41467-021-24058-z | |
| 7DUQ (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | GLP-1 | PubChem 156022738 | 2.5 | 2021-07-14 | doi.org/10.1038/s41467-021-24058-z | ||
| 7DUR | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | PubChem 156022738 | Gs/β1/γ2 | 3.3 | 2021-08-11 | doi.org/10.1038/s41467-021-24058-z | |
| 7DUR (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | PubChem 156022738 | 3.3 | 2021-08-11 | doi.org/10.1038/s41467-021-24058-z | ||
| 7EVM | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | PubChem 156022738 | Gs/β1/γ2 | 2.5 | 2021-08-11 | doi.org/10.1038/s41467-021-24058-z | |
| 7EVM (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | PubChem 156022738 | 2.5 | 2021-08-11 | doi.org/10.1038/s41467-021-24058-z | ||
| 5VEW | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | PF-06372222 | - | 2.7 | 2017-05-24 | doi.org/10.1038/nature22378 | |
| 6LN2 | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | PF-06372222 | - | 3.2 | 2020-03-18 | doi.org/10.1038/s41467-020-14934-5 | |
| 6KJV | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | PF-06372222 | - | 2.8 | 2019-11-13 | doi.org/10.1107/S2052252519013496 | |
| 6KK1 | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | PF-06372222 | - | 2.8 | 2019-11-13 | doi.org/10.1107/S2052252519013496 | |
| 6KK7 | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | PF-06372222 | - | 3.1 | 2019-11-13 | doi.org/10.1107/S2052252519013496 | |
| 5VEX | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | NNC0640 | - | 3 | 2017-05-17 | doi.org/10.1038/nature22378 | |
| 9IVM | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | GLP-1 (Truncated) | LSN3318839 | chim(NtGi1-Gs)/β2/γ2 | 3.22 | 2024-11-13 | doi.org/10.1016/j.apsb.2024.09.002 | |
| 9IVM (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | GLP-1 (Truncated) | LSN3318839 | 3.22 | 2024-11-13 | doi.org/10.1016/j.apsb.2024.09.002 | ||
| 6VCB | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | GLP-1 | LSN3160440 | Gs/β1/γ2 | 3.3 | 2020-07-22 | doi.org/10.1038/s41589-020-0589-7 | |
| 6VCB (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | GLP-1 | LSN3160440 | 3.3 | 2020-07-22 | doi.org/10.1038/s41589-020-0589-7 | ||
| 9IVG | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | GLP-1 (Truncated) | - | chim(NtGi1-Gs)/β1/γ2 | 3 | 2024-11-27 | doi.org/10.1016/j.apsb.2024.09.002 | |
| 9IVG (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | GLP-1 (Truncated) | - | 3 | 2024-11-27 | doi.org/10.1016/j.apsb.2024.09.002 | ||
| 7RTB | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Peptide-19 | - | Gs/β1/γ2 | 2.14 | 2021-10-06 | doi.org/10.1016/j.bbrc.2021.09.016 | |
| 7RTB (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Peptide-19 | - | 2.14 | 2021-10-06 | doi.org/10.1016/j.bbrc.2021.09.016 | ||
| 7KI0 | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Semaglutide | - | Gs/β1/γ2 | 2.5 | 2021-08-04 | doi.org/10.1016/j.celrep.2021.109374 | |
| 7KI0 (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Semaglutide | - | 2.5 | 2021-08-04 | doi.org/10.1016/j.celrep.2021.109374 | ||
| 7KI1 | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Taspoglutide | - | Gs/β1/γ2 | 2.5 | 2021-08-04 | doi.org/10.1016/j.celrep.2021.109374 | |
| 7KI1 (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Taspoglutide | - | 2.5 | 2021-08-04 | doi.org/10.1016/j.celrep.2021.109374 | ||
| 6X18 | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | GLP-1 | - | Gs/β1/γ2 | 2.1 | 2020-09-09 | doi.org/10.1016/j.molcel.2020.09.020 | |
| 6X18 (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | GLP-1 | - | 2.1 | 2020-09-09 | doi.org/10.1016/j.molcel.2020.09.020 | ||
| 6X19 | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | CHU-128 | - | Gs/β1/γ2 | 2.1 | 2020-09-09 | doi.org/10.1016/j.molcel.2020.09.020 | |
| 6X19 (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | CHU-128 | - | 2.1 | 2020-09-09 | doi.org/10.1016/j.molcel.2020.09.020 | ||
| 6X1A | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Danuglipron | - | Gs/β1/γ2 | 2.5 | 2020-09-09 | doi.org/10.1016/j.molcel.2020.09.020 | |
| 6X1A (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Danuglipron | - | 2.5 | 2020-09-09 | doi.org/10.1016/j.molcel.2020.09.020 | ||
| 7LCI | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Danuglipron | - | Gs/β1/γ2 | 2.9 | 2021-01-20 | doi.org/10.1016/j.str.2021.04.008 | |
| 7LCI (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Danuglipron | - | 2.9 | 2021-01-20 | doi.org/10.1016/j.str.2021.04.008 | ||
| 7LCJ | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Danuglipron | - | - | 2.82 | 2021-01-20 | doi.org/10.1016/j.str.2021.04.008 | |
| 7LCK | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Danuglipron | - | - | 3.24 | 2021-01-20 | doi.org/10.1016/j.str.2021.04.008 | |
| 7S15 | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | PubChem 134611223 | - | - | 3.8 | 2022-06-08 | doi.org/10.1021/acs.jmedchem.1c01856 | |
| 9J1P | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | g1:Ox | - | chim(NtGi1-Gs)/β1/γ2 | 2.99 | 2025-02-26 | doi.org/10.1021/jacs.4c12808 | |
| 9J1P (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | g1:Ox | - | 2.99 | 2025-02-26 | doi.org/10.1021/jacs.4c12808 | ||
| 5VAI | B1 | Peptide | Glucagon | GLP-1 | Oryctolagus cuniculus | GLP-1 | - | Gs/β1/γ2 | 4.1 | 2017-05-24 | doi.org/10.1038/nature22394 | |
| 5VAI (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Oryctolagus cuniculus | GLP-1 | - | 4.1 | 2017-05-24 | doi.org/10.1038/nature22394 | ||
| 5NX2 | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | GLP-1 (Truncated) | - | - | 3.7 | 2017-06-14 | doi.org/10.1038/nature22800 | |
| 6B3J | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Exendin-P5 | - | Gs/β1/γ2 | 3.3 | 2018-02-21 | doi.org/10.1038/nature25773 | |
| 6B3J (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Exendin-P5 | - | 3.3 | 2018-02-21 | doi.org/10.1038/nature25773 | ||
| 8WG7 | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | - | Gs/β1/γ2 | 2.54 | 2024-03-06 | doi.org/10.1038/s41421-024-00649-0 | |
| 8WG7 (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | - | 2.54 | 2024-03-06 | doi.org/10.1038/s41421-024-00649-0 | ||
| 7C2E | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | PubChem 149050799 | - | Gs/β1/γ2 | 4.2 | 2020-08-26 | doi.org/10.1038/s41422-020-0384-8 | |
| 7C2E (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | PubChem 149050799 | - | 4.2 | 2020-08-26 | doi.org/10.1038/s41422-020-0384-8 | ||
| 7LLL | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Exendin-4 | - | Gs/β1/γ2 | 3.7 | 2022-01-12 | doi.org/10.1038/s41467-021-27760-0 | |
| 7LLL (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Exendin-4 | - | 3.7 | 2022-01-12 | doi.org/10.1038/s41467-021-27760-0 | ||
| 7LLY | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Oxyntomodulin | - | Gs/β1/γ2 | 3.3 | 2022-01-12 | doi.org/10.1038/s41467-021-27760-0 | |
| 7LLY (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Oxyntomodulin | - | 3.3 | 2022-01-12 | doi.org/10.1038/s41467-021-27760-0 | ||
| 7FIM | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Tirzepatide | - | Gs/β1/γ2 | 3.4 | 2022-03-02 | doi.org/10.1038/s41467-022-28683-0 | |
| 7FIM (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Tirzepatide | - | 3.4 | 2022-03-02 | doi.org/10.1038/s41467-022-28683-0 | ||
| 7VBI | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Non-Acylated Tirzepatide | - | Gs/β1/γ2 | 3 | 2022-03-02 | doi.org/10.1038/s41467-022-28683-0 | |
| 7VBI (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Non-Acylated Tirzepatide | - | 3 | 2022-03-02 | doi.org/10.1038/s41467-022-28683-0 | ||
| 7VBH | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Peptide-20 | - | Gs/β1/γ2 | 3 | 2022-04-06 | doi.org/10.1038/s41467-022-28683-0 | |
| 7VBH (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Peptide-20 | - | 3 | 2022-04-06 | doi.org/10.1038/s41467-022-28683-0 | ||
| 7S1M | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Ex4-D-Ala | - | Gs/β1/γ2 | 2.41 | 2022-01-05 | doi.org/10.1038/s41589-021-00945-w | |
| 7S1M (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Ex4-D-Ala | - | 2.41 | 2022-01-05 | doi.org/10.1038/s41589-021-00945-w | ||
| 7S3I | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | - | Gs/β1/γ2 | 2.51 | 2022-01-05 | doi.org/10.1038/s41589-021-00945-w | |
| 7S3I (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | - | 2.51 | 2022-01-05 | doi.org/10.1038/s41589-021-00945-w | ||
| 6XOX | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Orforglipron | - | chim(NtGi1-Gs)/β1/γ2 | 3.1 | 2020-11-18 | doi.org/10.1073/pnas.2014879117 | |
| 6XOX (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Orforglipron | - | 3.1 | 2020-11-18 | doi.org/10.1073/pnas.2014879117 | ||
| 7RG9 | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | - | chim(NtGi1-Gs)/β1/γ2 | 3.2 | 2022-04-13 | doi.org/10.1073/pnas.2116506119 | |
| 7RG9 (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | - | 3.2 | 2022-04-13 | doi.org/10.1073/pnas.2116506119 | ||
| 7RGP | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Tirzepatide | - | chim(NtGi1-Gs)/β1/γ2 | 2.9 | 2022-04-13 | doi.org/10.1073/pnas.2116506119 | |
| 7RGP (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Tirzepatide | - | 2.9 | 2022-04-13 | doi.org/10.1073/pnas.2116506119 | ||
| 7X8R | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | BOC5 | - | Gs/β1/γ2 | 2.61 | 2022-06-29 | doi.org/10.1073/pnas.2200155119 | |
| 7X8R (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | BOC5 | - | 2.61 | 2022-06-29 | doi.org/10.1073/pnas.2200155119 | ||
| 7X8S | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | WB4-24 | - | Gs/β1/γ2 | 3.09 | 2022-06-29 | doi.org/10.1073/pnas.2200155119 | |
| 7X8S (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | WB4-24 | - | 3.09 | 2022-06-29 | doi.org/10.1073/pnas.2200155119 | ||
| 8JIP | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | MEDI0382 | - | chim(NtGi1-Gs)/β1/γ2 | 2.85 | 2023-09-06 | doi.org/10.1073/pnas.2303696120 | |
| 8JIP (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | MEDI0382 | - | 2.85 | 2023-09-06 | doi.org/10.1073/pnas.2303696120 | ||
| 8JIR | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | SAR425899 | - | chim(NtGi1-Gs)/β1/γ2 | 2.57 | 2023-09-06 | doi.org/10.1073/pnas.2303696120 | |
| 8JIR (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | SAR425899 | - | 2.57 | 2023-09-06 | doi.org/10.1073/pnas.2303696120 | ||
| 8JIS | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Peptide-15 | - | chim(NtGi1-Gs)/β1/γ2 | 2.46 | 2023-11-08 | doi.org/10.1073/pnas.2303696120 | |
| 8JIS (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Peptide-15 | - | 2.46 | 2023-11-08 | doi.org/10.1073/pnas.2303696120 | ||
| 8YW3 | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Retatrutide | - | Gs/β1/γ2 | 2.68 | 2024-09-18 | doi.org/10.1038/s41421-024-00700-0 | |
| 8YW3 (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Retatrutide | - | 2.68 | 2024-09-18 | doi.org/10.1038/s41421-024-00700-0 | ||
| 9EBN | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | - | Gs/β1/γ2 | 3.44 | 2025-03-26 | doi.org/10.1073/pnas.2407574122 | |
| 9EBN (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | - | 3.44 | 2025-03-26 | doi.org/10.1073/pnas.2407574122 | ||
| 9EBO | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | GLP-1 (ACPC18) | - | Gs/β1/γ2 | 3.13 | 2025-03-26 | doi.org/10.1073/pnas.2407574122 | |
| 9EBO (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | GLP-1 (ACPC18) | - | 3.13 | 2025-03-26 | doi.org/10.1073/pnas.2407574122 | ||
| 9EBQ | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | - | Gs/β1/γ2 | 3.16 | 2025-03-26 | doi.org/10.1073/pnas.2407574122 | |
| 9EBQ (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | - | - | 3.16 | 2025-03-26 | doi.org/10.1073/pnas.2407574122 | ||
| 8YWF | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Retatrutide | - | chim(NtGi1-Gs)/β1/γ2 | 2.74 | 2025-04-16 | To be published | |
| 8YWF (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Retatrutide | - | 2.74 | 2025-04-16 | To be published | ||
| 9BYO | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Exendin-3 | - | Gs/β1/γ2 | 2.31 | 2025-06-11 | To be published | |
| 9BYO (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Exendin-3 | - | 2.31 | 2025-06-11 | To be published | ||
| 9C0K | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Exendin-4 | - | Gs/β1/γ2 | 2.72 | 2025-06-11 | To be published | |
| 9C0K (No Gprot) | B1 | Peptide | Glucagon | GLP-1 | Homo sapiens | Exendin-4 | - | 2.72 | 2025-06-11 | To be published | ||
You can download a compressed (zip) file with structure(s), 3D outputs (as PyMol and VMD scripts) and numerical data files (as csv and plain text files).
Download 6XOX_nogp.zipYou can click to copy the link of this page to easily come back here later
or use this QR code to link and share this page.
You can also read or download a guide explaining the meaning of all files and numerical data.